DOI:10.32604/cmc.2022.020523

| Computers, Materials & Continua DOI:10.32604/cmc.2022.020523 |  |
| Article |
Reactions’ Descriptors Selection and Yield Estimation Using Metaheuristic Algorithms and Voting Ensemble
1Department of Computer Science, City University of Hong Kong, Kowloon Tong, Hong Kong SAR
2Information Systems Department, College of Computer Science and Engineering, Taibah University, Tayba, Medina, 42353, Saudi Arabia
3Hong Kong Institute for Data Science, City University of Hong Kong, Kowloon Tong, Hong Kong SAR
*Corresponding Author: Ka-Chun Wong. Email: kc.w@cityu.edu.hk
Received: 28 May 2021; Accepted: 08 July 2021
Abstract: Bioactive compounds in plants, which can be synthesized using N-arylation methods such as the Buchwald-Hartwig reaction, are essential in drug discovery for their pharmacological effects. Important descriptors are necessary for the estimation of yields in these reactions. This study explores ten metaheuristic algorithms for descriptor selection and model a voting ensemble for evaluation. The algorithms were evaluated based on computational time and the number of selected descriptors. Analyses show that robust performance is obtained with more descriptors, compared to cases where fewer descriptors are selected. The essential descriptor was deduced based on the frequency of occurrence within the 50 extracted data subsets, and better performance was achieved with the voting ensemble than other algorithms with RMSE of 6.4270 and R2 of 0.9423. The results and deductions from this study can be readily applied in the decision-making process of chemical synthesis by saving the computational cost associated with initial descriptor selection for yield estimation. The ensemble model has also shown robust performance in its yield estimation ability and efficiency.
Keywords: Buchwald-Hartwig reaction; descriptor selection; machine learning; metaheuristic algorithm; palladium-catalyzed cross-coupling reaction; voting ensemble
Synthesis of chemical reaction has become quite explored in recent research. It is a research area interwoven with biomedical research since cross-coupling chemical reactions are used for natural product synthesis and synthesis of bioactive compounds used in drug discovery and cancer treatment. These interdisciplinary studies in the biological and chemical fields have produced great ideas, innovations, and discoveries [1]. Some chemical compounds possess the ability to inhibit the growth of cancer cells and have anti-tumour capabilities. Harnessing this advantage, Zhou et al. [2] used the Buchwald-Hartwig (B-H) reaction to generate active compounds against breast cancer-causing tumour which is helpful for tumour growth suppression. Buchwald-Hartwig reaction is a significant procedure used for the synthesis of N2-aryl-dG popularly known to cause DNA adduct [3], and phenazine which possess intense antibacterial activity against chlamydia [4]. It is also used to yield poly (arylene–amine) from arylamines polymerisation [5], yield aryl and heteroaryl chlorides in an aqueous condition free from solvent [6], and synthesise iron (II) clathrochelate unit bearing secondary arylamine copolymers due to the strong compounds which it yields. Surveys carried out by Gómez-Bombarelli et al. [7] and Rodrigues et al. [8] shows that chemical space search and optimization, virtual screening, discovery of drug target, the prediction of protein structures, chemical properties, gene-gene interaction, bioactivity of molecules in a compound [9], and toxicity [10], are some of the ways predictive analysis has been applied to the vast data produced in biochemical studies [11].
The search for valuable descriptors necessary for carrying out chemical reactions is related to the idea of feature selection in machine learning. Due to the large number of descriptors often generated for chemical reactions, it is imperative to identify the necessary descriptors which contribute to the estimation of yields and are essential for accurate reactions [12]. Using algorithms for selecting valuable subsets is more superior to using training data that are commercially available and probably contain several unwanted data [13]. Removal and elimination of descriptors that are not relevant to a particular reaction activity can be done before carrying out computational analysis [14–16]. However, exhaustive research and adequate knowledge are needed to know the required descriptors to be selected [17].
Given the rate at which many studies focus on synthesizing bioactive molecules to discover novel pharmaceutical drugs and the yield generated from each reaction, efficiency will be realized if the yield can be estimated within a short computational time. An insight into the parameters which activate Buchwald-Hartwig Pd-catalyzed amination of aryl halides using temperature-scanning reaction protocol gives a rapid description of complicated multistep catalytic reaction [18]. Examples of palladium-catalyzed cross-coupling reaction implemented in some recent studies are shown in Fig. 1.
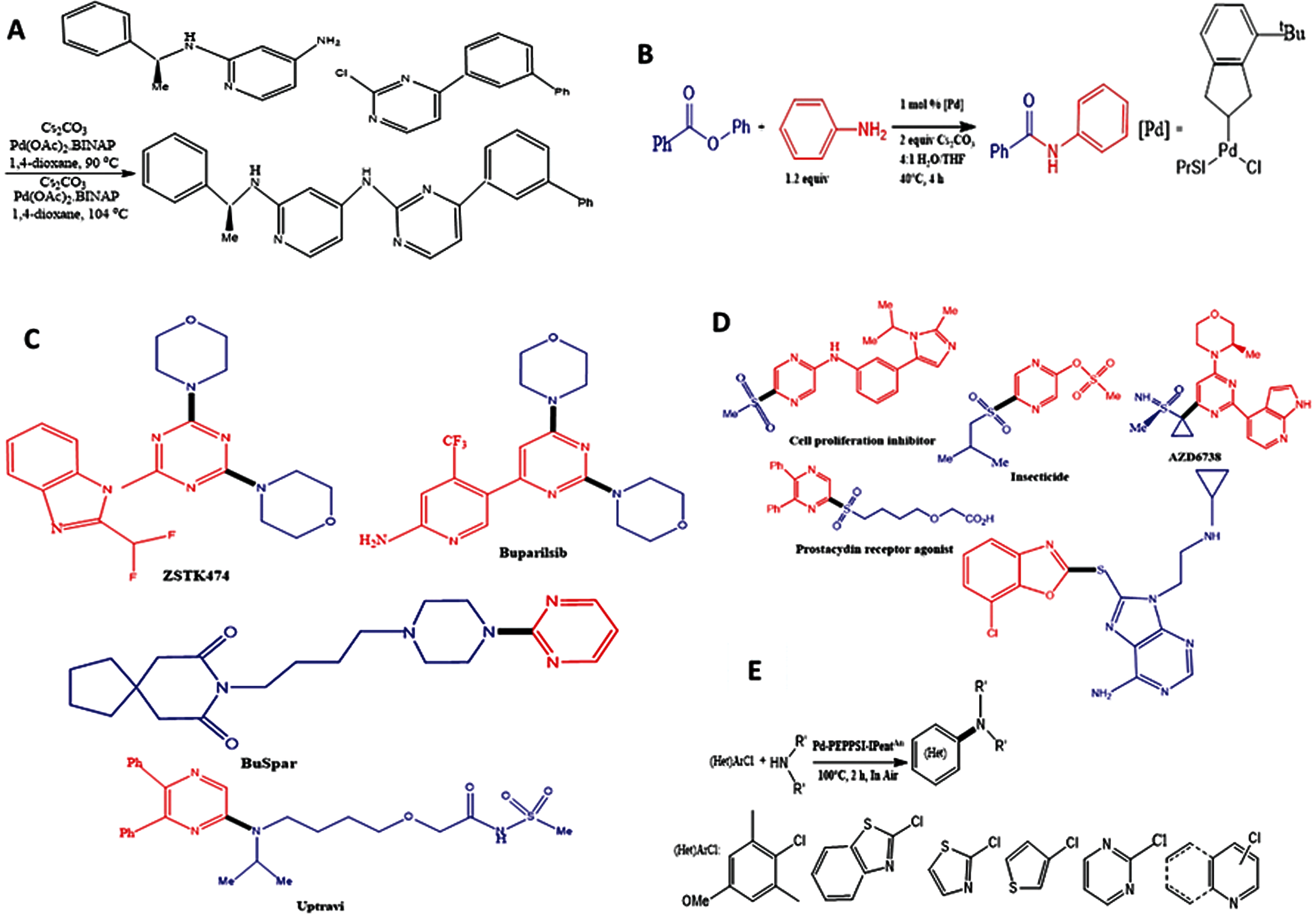
Figure 1: Examples of palladium-catalysed cross-coupling reaction and products. (A) Buchwald-Hartwig amination, where BINAP stands for 2,2′-bis(diphenylphosphino)-1,1′-binaphthyl; (B) Yield generation from Buchwald-Hartwig’s reaction for Phenyl Benzoate under a 40 degrees temperature; (C) Palladium-catalyzed cross-coupling amination of heteroarene in commercially available drugs such as Buparlisib (anticancer drug), ZSTK474 (anticancer drug), Uptravi (hypertension drug), and BuSpar (antidepressants drug); (D) Synthesis of bioactive molecules, pharmaceutical drugs, and cell proliferation inhibitor from thioethers is preferably done using palladium-catalyzed reaction; (E) Palladium-catalyzed amination of (hetero)aryl chloride carried out with moisture, air and mild conditions. The reaction procedure was Pd-PEPPSI (pyridine-enhanced pre-catalyst preparation, stabilization, and initiation)
Nature has inspired the development of several metaheuristic algorithms over the years. These algorithms are modelled after the peculiar characteristics of either animals or plants, and it is an active research area [19]. Due to their ability to efficiently explore ample configuration space, metaheuristic algorithms can achieve near-optimal or optimal solutions [20]. The recent metaheuristic algorithms can be used for optimization with machine learning algorithms [21] and can achieve convergence with few mathematical operations [22] within reasonable computational time [23]. However, quite many metaheuristic algorithms exist, and they cost computational time for descriptor selection even before analysis of reaction for yield estimation.
Given the existence of many metaheuristic algorithms, we would like to explore ten different nature-inspired metaheuristic algorithms for selecting descriptor subsets that are essential in estimating B-H chemical reaction yield and construct a voting ensemble model with these extracted descriptor subsets. The voting ensemble has shown good predictive ability in prediction across several fields [24]. Through this, we can identify the essential descriptors that contribute to the ensemble model's performance in estimating yields. We will further juxtapose the performance of the ensemble model on the extracted descriptor subsets and the full descriptors. The results from our study proffer guidance for subsequent B-H reaction estimations for efficient computational analysis.
2.1 Data Description and Preprocessing
For this study, we have collected cross-coupling Buchwald-Hartwig reaction data from Ahneman et al. [25]. The dataset contains 3724 reactions. For each reaction, it is characterized by 120 descriptors and one resulting yield. The resulting yield is the independent variable for the predictive analysis. These 120 descriptors are the molecular, atomic, and vibrational descriptors that were extracted for constituents of palladium-catalysed Buchwald-Hartwig cross-coupling of aryl halides with 4-methylaniline under the condition of several additives with inhibitory properties. Some of the molecular descriptors used include the surface area, molecular weight, molecular volume, hardness, electronegativity, dipole moment, and ovality. Nuclear Magnetic Resonance (NMR) shift and electrostatic charge are some of the atomic descriptors, while the intensity and frequency are the vibrational descriptors. A Glorius approach was used with Buchwald-Hartwig cross-coupling as the model reaction to determine the structural interaction between heteroaryl halides, aryl and isoxazoles. Together with the yield, a total number of 121 attributes are contained in the entire dataset.
As much as data analysis is concerned, data preprocessing is vital due to the peculiarity of real-world data. Data preprocessing is an essential step before data analysis. The data had some cross-coupling reactions with no yield. Since the number of missing data is small, these reactions with no yields were removed to maintain data consistency with the original dataset. The resulting dataset with which estimation is to be made was left with 3719 reactions. The data was thereafter standardized based on each column. That is, each column will have a mean of 0 and a standard deviation of 1. The standard scaler for a sample x is evaluated based on Eq. (1).
where: x represents a descriptor in an instance,
2.2 Nature-Inspired Metaheuristic Algorithms Used for Selection of Descriptors
Global or near-optimal solutions are attained with metaheuristic algorithms within a reasonable search time and computational cost [26]. In predictive analysis, the optimization of hyperparameters for improved prediction is achieved by deploying metaheuristic algorithms to solve non-convex, complex problems in large space [20]. A recent review reveals that robotics, education, and disease diagnosis are domains where articles on metaheuristics algorithm get frequently published [27]. We therefore examine ten metaheuristic algorithms in the selection of descriptors for this reaction estimation study. They are Ant search algorithm [28], Bat search algorithm [29], Bee search algorithm [30], Cuckoo search algorithm [31], Elephant search algorithm [32], Firefly search algorithm [33], Flower pollination algorithm [34], Genetic algorithm (GA) [35], Rhinoceros search algorithm [36], and Wolf search algorithm [37]. The parameters used for the algorithms are given in Tab. 1. Some of the algorithms have related parameters. Due to the number of parameters associated with each algorithm, we iteratively changed only the population size geometrically and the number of iterations linearly. Other parameters remained constant. Fig. 2 shows the process overview of the study.
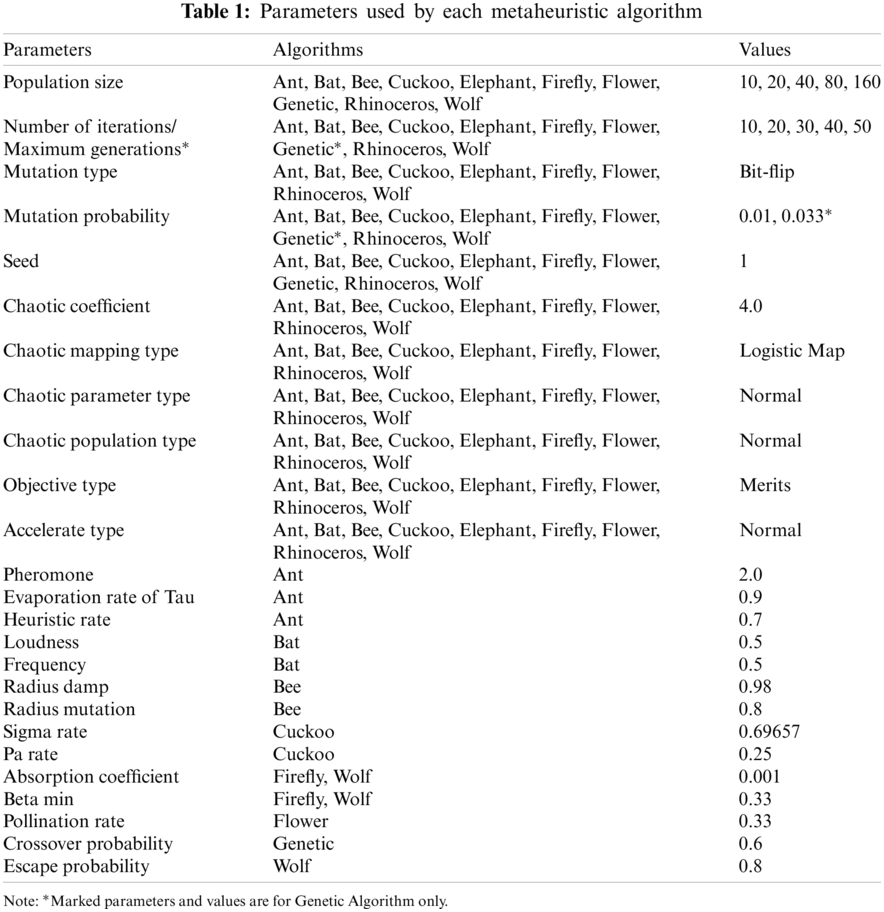
We determine the most important descriptor based on the frequency of appearance in the new data subsets. We evaluated the ensemble model using a 5-fold cross validation which was repeated 5 times and shuffled. Two criteria were used for the evaluation of the models.
Root Mean Square Error (RMSE) which is the square root of the Mean Square Error (MSE) is the standard deviation of the errors from prediction, also known as residuals, that is, the difference between the actual and the observed values, or the distance between the regression line and the data points. The data's concentration around the best fit line or regression line can be determined from the RMSE. It can be determined based on Eq. (4).
where:
R-Squared (R2) is also known as the coefficient of determination. It indicates the explained variability of the data: how the relationship between a factor and another factor can determine its variability. It is a fraction of the total variation in y, which was captured by the model, and also a measure of how close each data point fits the regression line. It analyses how the difference in the second variable can explain the difference in one variable. It is scaled between 0 and 1 and is given by Eq. (5).
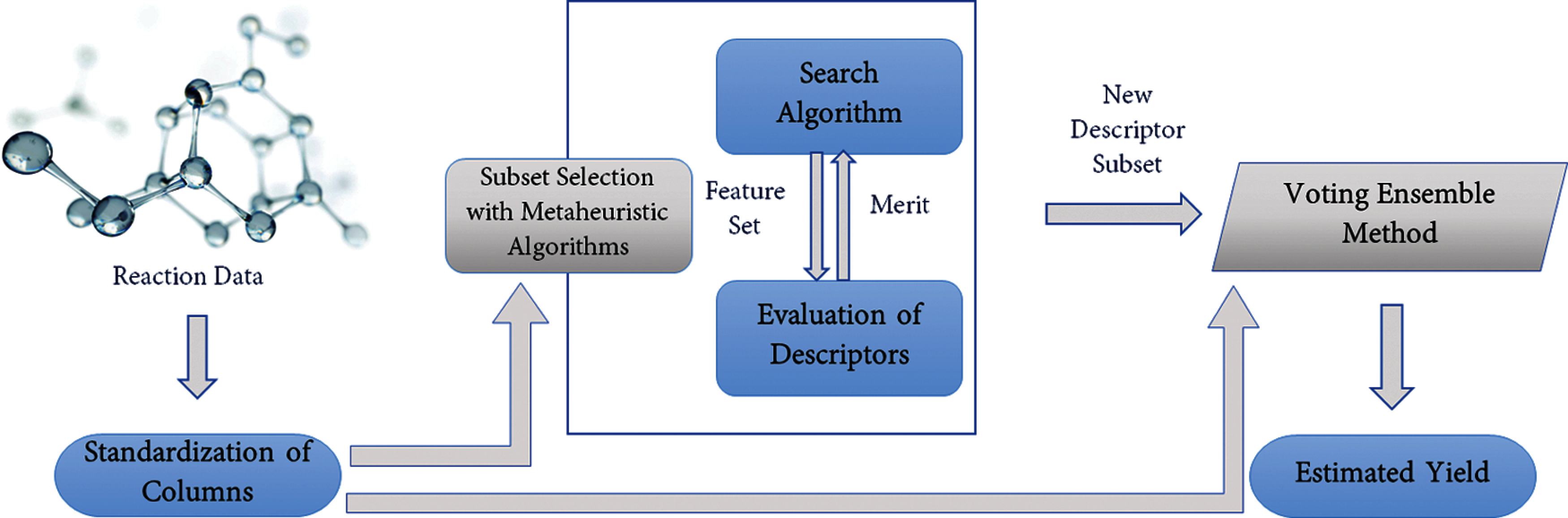
Figure 2: Process overview for descriptor selection with metaheuristic algorithm and yield estimation
We used the Waikato Environment for Knowledge Analysis, Weka 3.9.5 [38] to implement the selection of descriptors based on the algorithms. Weka is a Java-based platform for data analysis and machine learning algorithms. We were able to generate 50 new data with different descriptor subsets. Afterwards, analysis on these new data was implemented using Python 3.6 with Scikit library. We also used MATLAB R2020b for coding and plotting some figures. All analyses in this study were carried out on a Windows 10 64-bit Operating System, x64-based processor computer with 64GB RAM. Processor specification is Intel (R) Core (TM) i7-9700 K CPU @ 3.6 GHz.
3.1 Smaller Iteration and Population Size Generate Large Descriptor Subsets
Using the ten nature-inspired metaheuristic algorithms with the parameters given in Tab. 1, per algorithm, we generated five new data each with different descriptor subsets chosen by the algorithms. We considered the impact of the running time on the number of iterations; hence we increased the number of iterations linearly from 10, 20, 30, 40, to 50, and increase the population size geometrically from 10, 20, 40, 80 to160. We maintained all other parameters for each algorithm but changed the number of iterations and population size five times to create a variety of descriptor subset based on these parameters. A total of 50 different datasets was generated as a result. The resulting number of selected descriptors is given in Tab. 2. The codes, datasets, and equivalent selected descriptors represented by numbers are provided in https://github.com/Olutomilayo/Yield_Estimation.
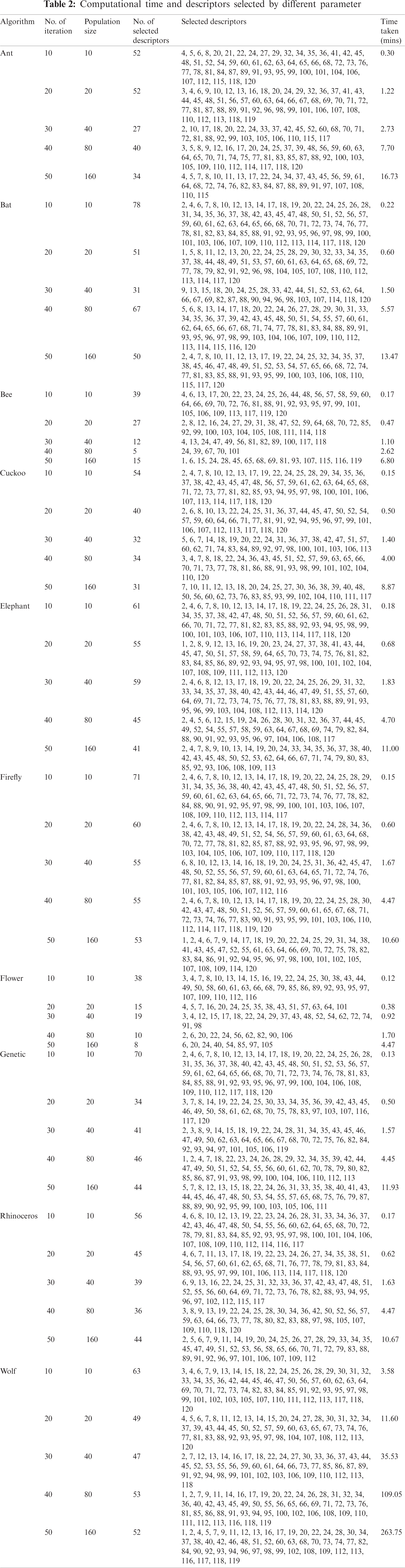
To effectively analyze the time taken by the algorithms, we create a plot of the time taken against the algorithms according to the number of iterations and the population size in Tab. 2. In Fig. 3, we notice that for all the algorithms, a gradual increase in running time occurs according to the increase in population size and number of iterations. However, we discover that the wolf search algorithm takes even longer running time than other algorithms. The time taken for 20 iterations and 20 population size is higher than the time taken for 50 iterations and 160 population size of other algorithms, except ant search algorithm and genetic algorithm. This can be linked to the fact that the search agents in the wolf algorithm require cooperation. Although the search agents search the problem space in random groups, an individual solution is provided to the problems [37], which causes an increased runtime.
Flower pollination algorithm on the other hand has a generally small run time across all examined iterations and population sizes. Fig. 3B shows the consistency in reduced runtime exhibited by this algorithm. Due to the benefit of the insect pollinators travelling long distance, there is an ability to have a larger problem space explored while choosing similar solutions. This improves the rate of convergence [34]. In general, a fair trade-off between the number of iterations, time complexity and performance are encouraged when choosing an algorithm for the selection of descriptors.
Out of a total of 120 descriptors in the entire dataset, we consider the number of descriptors selected by each algorithm based on the different parameters. With this analysis, we can quickly determine the descriptors which are frequently selected by the algorithms. Based on the techniques used by different metaheuristic algorithms, we expect peculiarity in the descriptors selected by each algorithm. We notice a general downward trend in the number of selected descriptors as the number of iterations and population size increases in Fig. 4. Apart from some occasional increase in the number of selected descriptors for some algorithms, we established that for all the algorithms, the number of selected descriptors at ten iterations and ten population size is higher compared to any other number of iterations and population size. The flower pollination algorithm and bee search algorithm having the lowest runtime also has the smallest number of selected descriptors. This will be considered in their yield estimation efficiency.
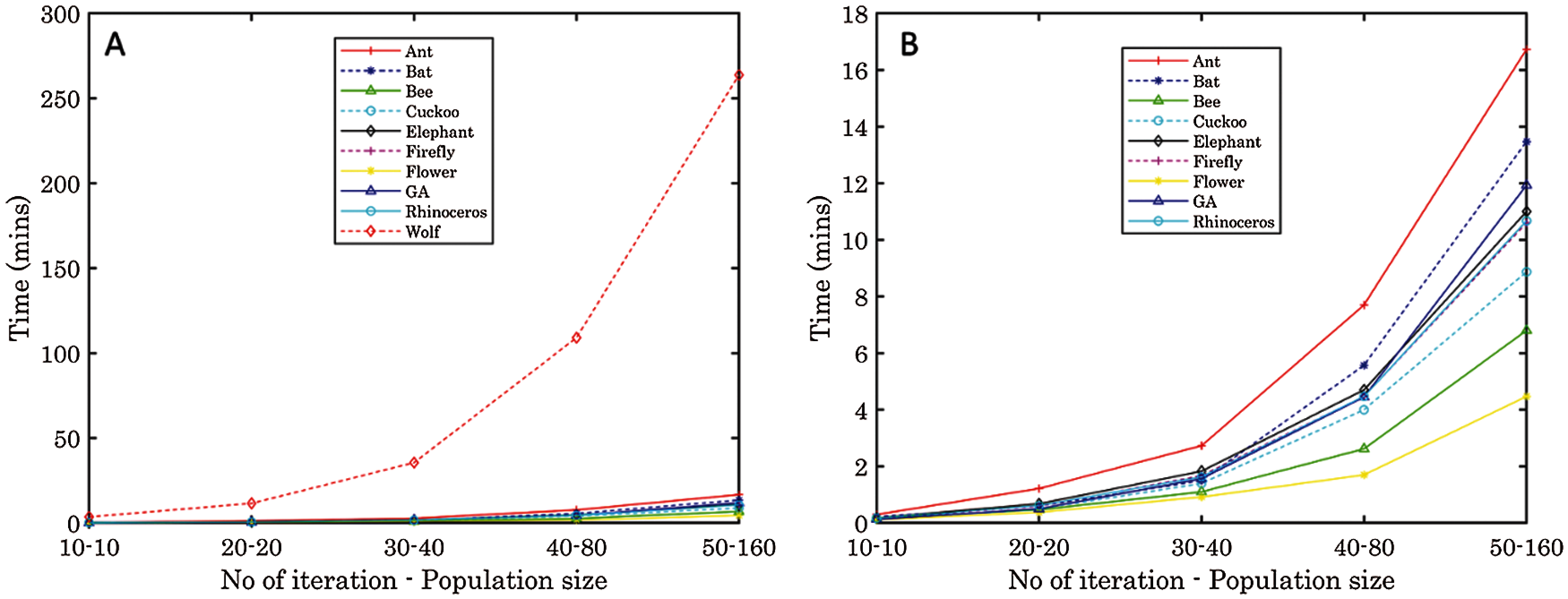
Figure 3: The runtime of each algorithm according to the number of iterations and population size. (A) The high runtime of wolf algorithm is evident, compared to other algorithms. (B) Vivid display of all algorithms except wolf algorithms
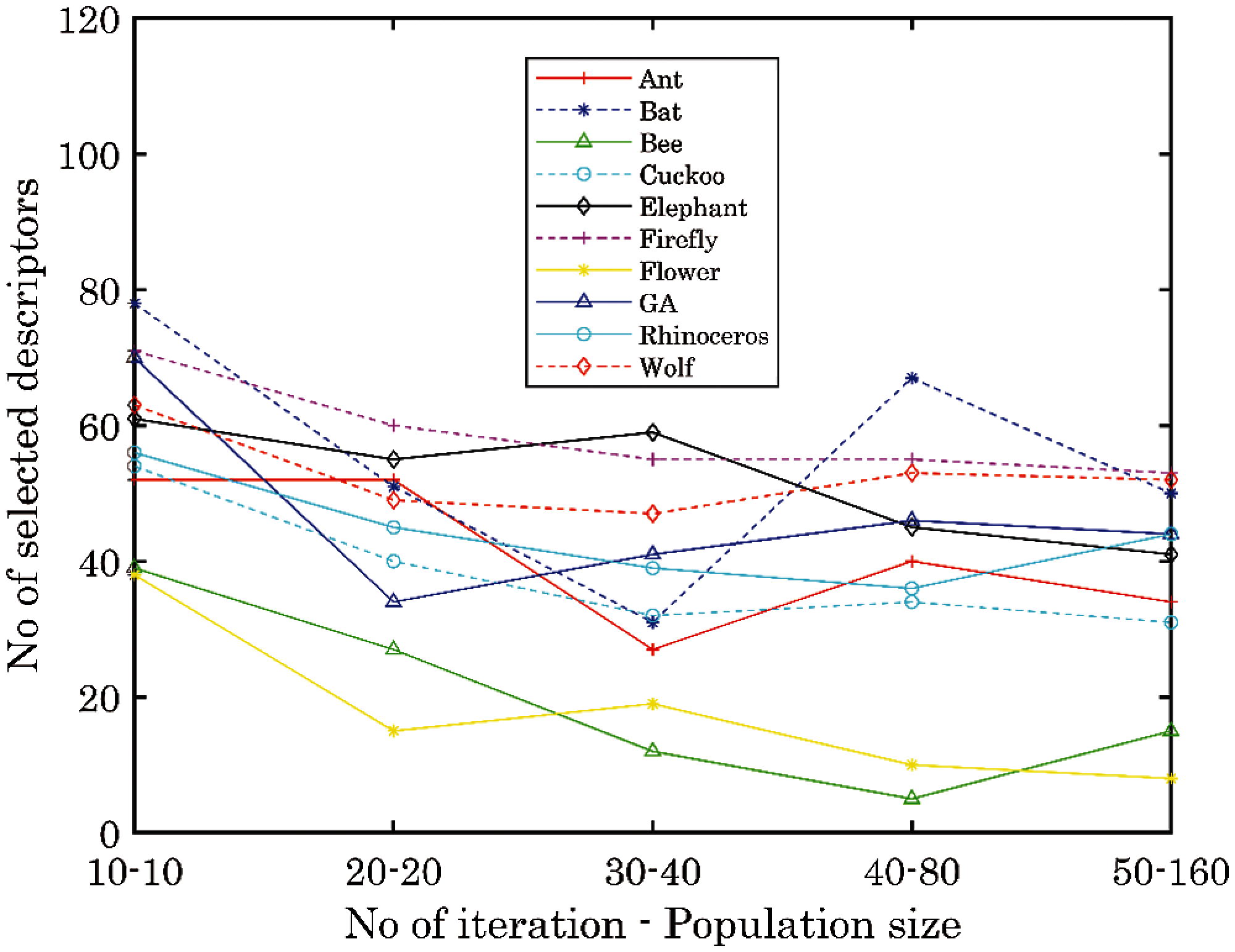
Figure 4: Number of selected descriptors of each algorithm according to parameter change
3.2 Efficacy of Lesser Number of Iterations and Population Size
We investigated the impact of descriptor selection on the estimation of yields. With the 50 new data having different descriptor subsets, we apply the voting ensemble method, which uses the averaging technique for prediction. The voting ensemble was compared with gradient boosting, multilayer perceptron, and random forest, which were also its base regressors. We implemented these using 5-fold cross-validation which we repeated five times. Hence, each model was trained and tested twenty-five times with shuffled data. This repetition enables different sections of the data to be used for training and testing and helps to overcome the introduction of bias in the model. The resulting performance is reported in Tab. 3.
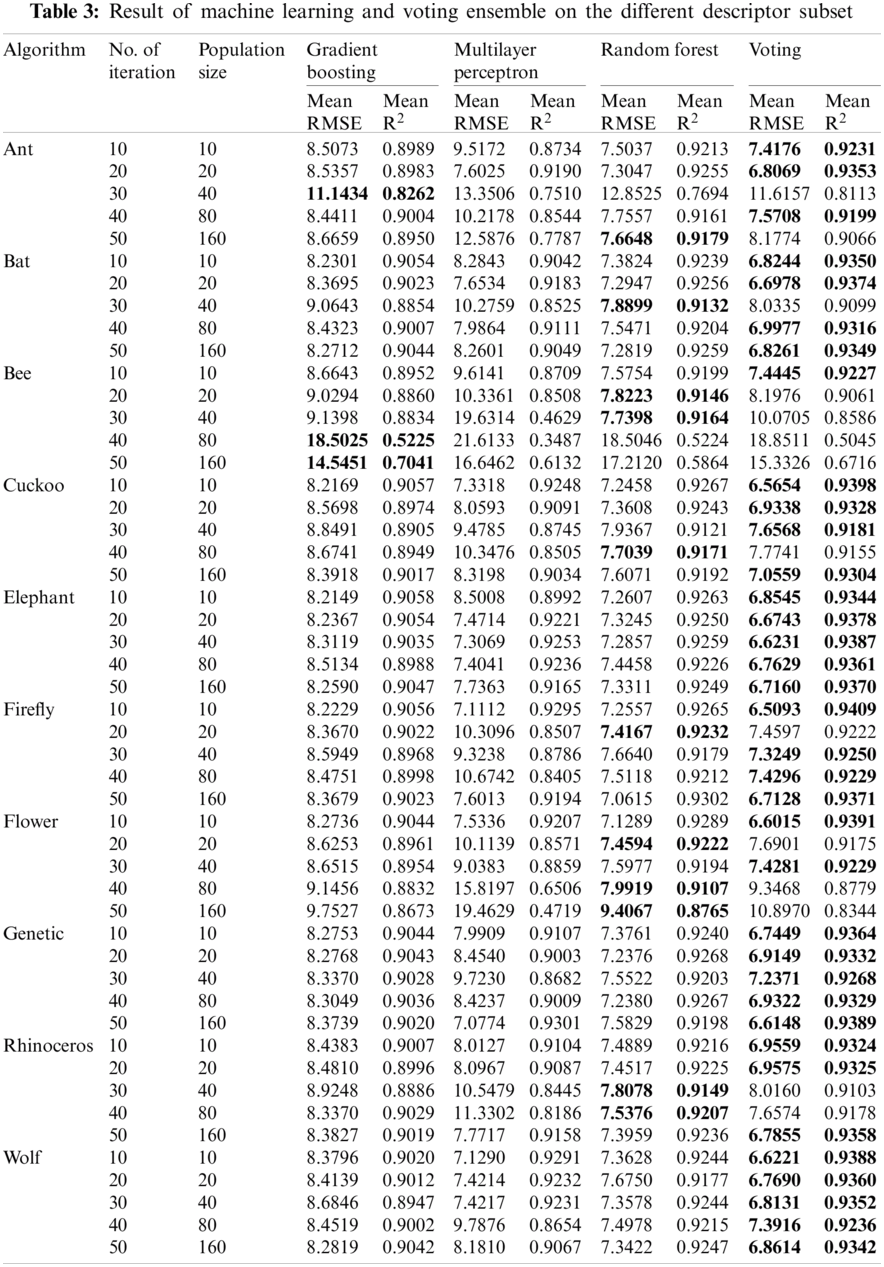
On average, it is revealed that majority of the datasets gotten from metaheuristic algorithms with ten iterations/population size and twenty iterations/population size produced better performance on the machine learning algorithms. In Fig. 4, we have shown that more descriptors were selected with these parameters. These values show the possibility of obtaining optimal solutions even with a lower number of iterations, enabling the option of limiting runtime while achieving optimal performance. We also show the performance of the voting ensemble being suitable for the estimation of yields across the datasets. Comparing the performance with the other base regressors and especially random forest (which is also an ensemble method) used in Science [25], voting ensemble had better performance in estimating the yields. Overly gross error in some of the base regressors affected the performance of voting on some dataset. Therefore, an appropriate combination of regressors should be made when a voting ensemble is used.
3.3 Analysis of Important Descriptors
We ranked the 120 descriptors according to the number of times they were selected in the new data subsets. In other words, according to the newly generated 50 datasets, which comprise of different subsets of descriptors, we rank the descriptors based on the number of times they appear. The name of the descriptors and the corresponding number of appearances is available in a sorted manner in https://github.com/Olutomilayo/Yield_Estimation. The top descriptor appeared in 34 data subsets, while the next one appeared in 25 data subsets. This shows their importance and contribution to yield estimation from reactions. We also used random forest to rank all the descriptors according to their importance. Since random forest splits a tree based on the most important descriptor, it is a good algorithm for ranking descriptors according to their importance and contribution to the models’ performance. The top 20 descriptors are displayed in Fig. 5. From the two analyses, we deduce that aryl halide-based descriptors significantly influence the yield of the reactions. We however note that without the presence of the additional descriptors such as additives, ligands and bases, the model does not produce a good yield. This means that although the aryl halides are essential for the reaction, all other descriptors are also important. Aryl halide, as a class of functional compound, is popularly known and used in medicinal chemistry. It is particularly essential for the arylation and modification of aromatic core and palladium cross-coupling. Cross-coupling of amines with aryl halides or pseudo halides is common [39]. Without the aryl halide, no reaction will be prompted, and the palladium catalyst assists in the instantaneous reaction. Other factors, such as oxidative addition, also influence aryl halides’ reactivity [40]. Base and additives are also crucial in a reaction as well as aryl halide and the catalyst.
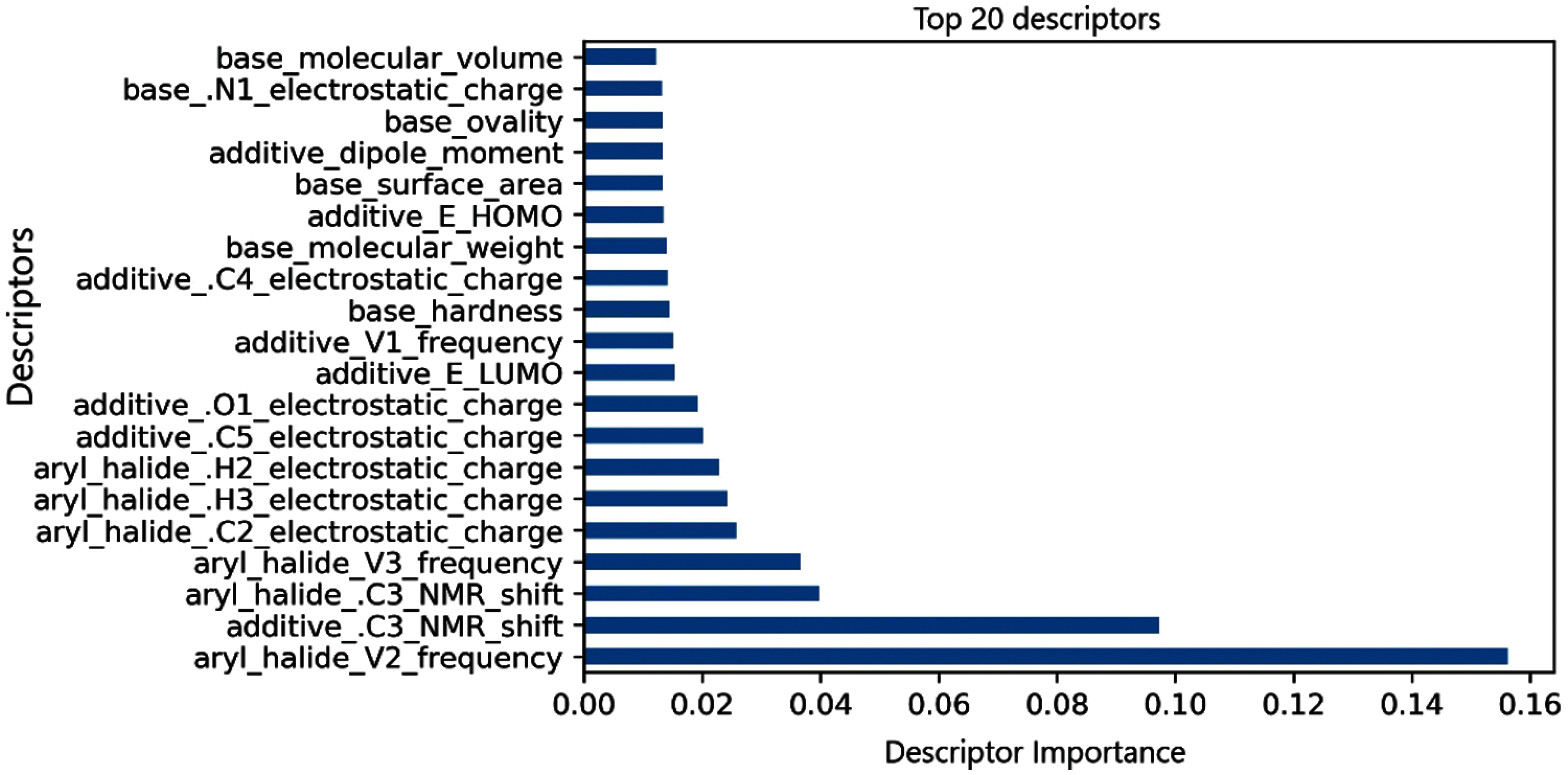
Figure 5: Top 20 descriptors ranked with random forest according to their importance
3.4 More Descriptors are Needed for Yield Estimation
As selecting a specific number of descriptors is quite subjective, we created two other datasets with different descriptor subset for further analysis. From the sorted descriptors, we created a dataset with descriptors having a frequency greater than 11 (that is, out of the 50 combinations of metaheuristic parameters, they were selected at least 12 times). We chose a number slightly greater than half of the original descriptors; hence a new dataset with 70 descriptors was extracted (hereafter referred to as 70_Desc). We also considered the Cuckoo and Firefly algorithm with ten iterations and ten population sizes. Based on the large number of descriptors initially selected by these algorithms and their performances in Tab. 3, we created a dataset with a combination of descriptors from both algorithms (hereafter referred to as CuckFire). Using the same 5-fold cross-validation repeated five times each, we applied the machine learning algorithms and voting ensemble on the entire dataset with the complete descriptors and on two extracted datasets.
Tab. 4 and Fig. 6 shows the performance of the machine learning algorithms and voting ensemble based on the different datasets. We report the consistency of the voting ensemble across the three examined datasets. As published in Science, Random forest recorded a better performance over the other methods with which it was examined [25]. In this study, the voting ensemble has demonstrated even better performance using the same dataset. Considering that RMSE is at its lowest and R2 at its highest when the entire dataset with full descriptors is used, we deduce that the overall performance of yield estimation is better when more descriptors are used. In Skoraczyński, et al. [41], a need for the development of more descriptors was stated. The environment under which a reaction is conducted is based on the catalysts, reagents, solvents used (all of which are the chemical components), and the temperature at which the reaction is carried out [42].
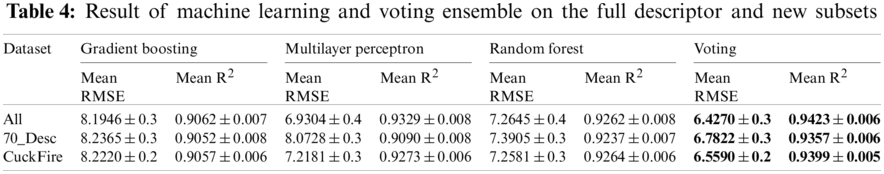
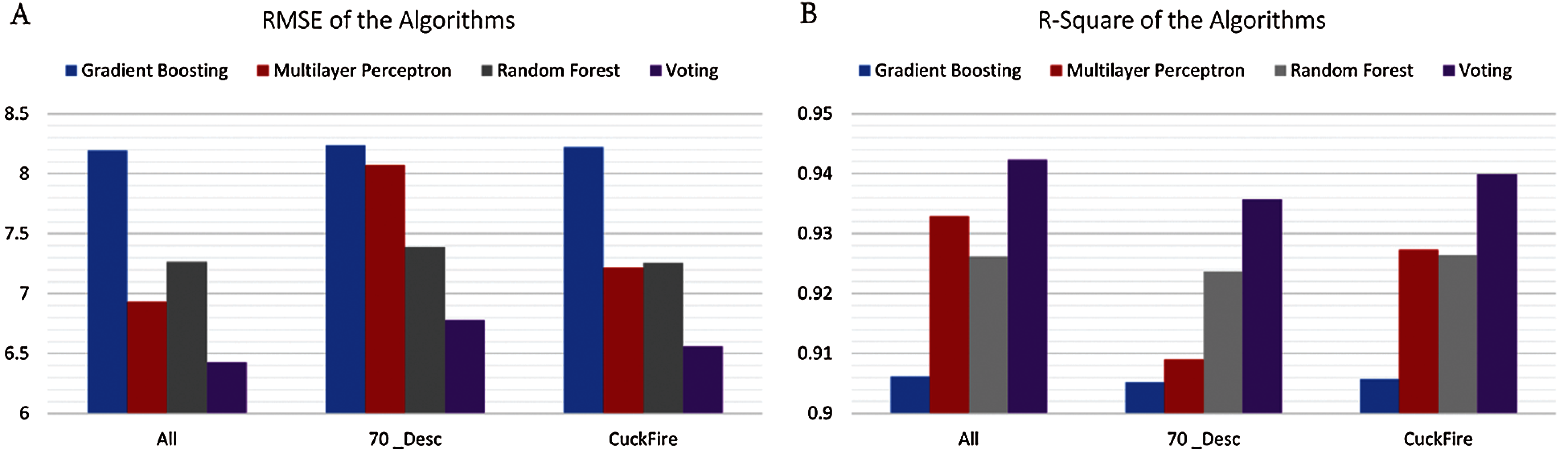
Figure 6: Plot showing the performance of the voting ensemble across three datasets. (A) RMSE of the algorithms. (B) R2 of the algorithms
In this study, we have explored ten nature-inspired metaheuristic algorithms for the selection of descriptors in B-H reaction. We compared these algorithms in terms of time complexity and the number of selected descriptors. We have also identified and enumerated the essential descriptor based on its frequency in the 50 different data subsets. We implemented a voting ensemble and compared it with three other machine learning algorithms.
Based on several analyses which have been conducted in this study, the essential descriptors are identified, and the results establish that more descriptors are essential for estimating the yield of reactions. With the variety of metaheuristic algorithm implemented, the five changes in parameters, the 50 extracted datasets with a variety of descriptors subsets, the dataset extracted with the 70 most selected descriptors, and the combination of descriptors from the Cuckoo and Firefly algorithm, the performance of the machine learning algorithms and voting ensemble were better with the entire dataset having full descriptors. These results show that although some descriptors might be more important for analysis, more descriptors are important and significant for model training in estimating yields from B-H reaction. The voting ensemble method also performed better than other machine learning methods with which it was compared. Although our work considers ten nature-inspired metaheuristic algorithms for selection of descriptor subsets in Buchwald-Hartwig reaction estimation, there exist several other metaheuristic algorithms which can be examined for selection of descriptors. Analysis of different algorithms for other type of chemical reactions can be performed in future work. Voting ensemble can also limited by the base regressors, this necessitates careful selection of base regressors. The results and deductions from this study can be readily applied in chemical synthesis by saving the computational cost associated with initial descriptor selection and making voting ensemble suitable for yield estimation in B-H reactions. We however believe that metaheuristic algorithms may be more suited for high-dimensional datasets and intend to investigate it in further studies.
Funding Statement: The work described in this paper was substantially supported by the grant from the Research Grants Council of the Hong Kong Special Administrative Region [CityU 11200218], one grant from the Health and Medical Research Fund, the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region [07181426], and the funding from Hong Kong Institute for Data Science (HKIDS) at City University of Hong Kong. The work described in this paper was partially supported by two grants from City University of Hong Kong (CityU 11202219, CityU 11203520). This research was substantially sponsored by the research project (Grant No. 32000464) supported by the National Natural Science Foundation of China and was substantially supported by the Shenzhen Research Institute, City University of Hong Kong. The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research with the project number (442/77).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. S. S. Liew, S. Du, J. Ge, S. Pan, S.-Y. Jang et al., “A chemoselective cleavable fluorescence turn-oN linker for proteomic studies,” Chemical Communications, vol. 53, no. 100, pp. 13332–13335, 2017. [Google Scholar]
2. J. Zhou, B. Liao, Y. Deng, X. Guo, J. Zhao et al., “Design and synthesis of imidazo-fused heterocycles derivatives and their anti-tumor activity against breast cancer in mice,” Nan Fang yi ke da xue xue bao= Journal of Southern Medical University, vol. 38, no. 9, pp. 1052–1060, 2018. [Google Scholar]
3. P. P. Ghodke and P. Pradeepkumar, “Synthesis of N2-Aryl-2′-Deoxyguanosine modified phosphoramidites and oligonucleotides,” Current Protocols in Nucleic Acid Chemistry, vol. 78, no. 1, pp. e93, 2019. [Google Scholar]
4. X. Bao, Z. Liu, M. Ni, C. Xia, S. Xu et al., “Synthesis and assessment of 3-substituted phenazines as novel antichlamydial agents,” Medicinal Chemistry (Shariqah (United Arab Emirates)), vol. 16, no. 3, pp. 413–421, 2019. [Google Scholar]
5. R. Grisorio and G. P. Suranna, “Catalyst-transfer polymerization of arylamines by the buchwald–Hartwig cross-coupling,” Polymer Chemistry, vol. 10, no. 15, pp. 1947–1955, 2019. [Google Scholar]
6. Y. Liu, J. Yuan, Z.-F. Wang, S.-H. Zeng, M.-Y. Gao et al., “Application of a 2-aryl indenylphosphine ligand in the buchwald–Hartwig cross-coupling reactions of aryl and heteroaryl chlorides under the solvent-free and aqueous conditions,” Organic & Biomolecular Chemistry, vol. 15, no. 27, pp. 5805–5810, 2017. [Google Scholar]
7. R. Gómez-Bombarelli and A. Aspuru-Guzik, “Machine learning and Big-data in computational chemistry,” Handbook of Materials Modeling: Methods: Theory and Modeling, pp. 1939–1962, 2020. [Google Scholar]
8. J. F. Rodrigues Jr, L. Florea, M. C. de Oliveira, D. Diamond and O. N. Oliveira Jr, “A survey on Big data and machine learning for chemistry,” ArXiv Preprint ArXiv:1904.1 0370, 2019. [Google Scholar]
9. O. O. Petinrin and F. Saeed, “Stacked ensemble for bioactive molecule prediction,” IEEE Access, vol. 7, pp. 153952–153957, 2019. [Google Scholar]
10. A. Mayr, G. Klambauer, T. Unterthiner and S. Hochreiter, “Deeptox: Toxicity prediction using deep learning,” Frontiers in Environmental Science, vol. 3, pp. 80, 2016. [Google Scholar]
11. B. Shine and J. H. Barth, Big data in clinical biochemistry, ed: SAGE publications sage, UK: London, England, pp. 308–309, 2019. [Google Scholar]
12. C. J. Bartel, S. L. Millican, A. M. Deml, J. R. Rumptz, W. Tumas et al., “Physical descriptor for the gibbs energy of inorganic crystalline solids and temperature-dependent materials chemistry,” Nature Communications, vol. 9, no. 1, pp. 1–10, 2018. [Google Scholar]
13. J. J. Henle, A. F. Zahrt, B. T. Rose, W. T. Darrow, Y. Wang et al., “Development of a computer-guided workflow for catalyst optimization. descriptor validation, subset selection, and training Set analysis,” Journal of the American Chemical Society, vol. 142, no. 26, pp. 11578–11592, 2020. [Google Scholar]
14. S. K. Chakravarti and S. R. M. Alla, “Descriptor free QSAR modeling using deep learning with long short-term memory neural networks,” Frontiers in Artificial Intelligence, vol. 2, pp. 17, 2019. [Google Scholar]
15. P. Schwaller, A. C. Vaucher, T. Laino and J.-L. Reymond, “Prediction of chemical reaction yields using deep learning,” Machine Learning: Science and Technology, vol. 2, no. 1, pp. 15016, 2021. [Google Scholar]
16. K. Ogura, T. Sato, H. Yuki and T. Honma, “Support vector machine model for hERG inhibitory activities based on the integrated hERG database using descriptor selection by NSGA-iI,” Scientific Reports, vol. 9, no. 1, pp. 1–12, 2019. [Google Scholar]
17. J. J. Villaverde, C. López-Goti, M. Alcamí, A. M. Lamsabhi, J. L. Alonso-Prados et al., “Quantum chemistry in environmental pesticide risk assessment,” Pest Management Science, vol. 73, no. 11, pp. 2199–2202, 2017. [Google Scholar]
18. O. P. Schmidt and D. G. Blackmond, “Temperature-scanning reaction protocol offers insights into activation parameters in the buchwald–Hartwig Pd-catalyzed amination of aryl halides,” ACS Catalysis, vol. 10, no. 15, pp. 8926–8932, 2020. [Google Scholar]
19. S. Harifi, M. Khalilian, J. Mohammadzadeh and S. Ebrahimnejad, “Emperor penguins colony: A new metaheuristic algorithm for optimization,” Evolutionary Intelligence, vol. 12, no. 2, pp. 211–226, 2019. [Google Scholar]
20. L. Yang and A. Shami, “On hyperparameter optimization of machine learning algorithms: Theory and practice,” Neurocomputing, vol. 415, pp. 295–316, 2020. [Google Scholar]
21. A. Onan, “Biomedical text categorization based on ensemble pruning and optimized topic modelling,” Computational and Mathematical Methods in Medicine, vol. 2018, 2018. [Google Scholar]
22. S. Mirjalili, A. H. Gandomi, S. Z. Mirjalili, S. Saremi, H. Faris et al., “Salp swarm algorithm: A bio-inspired optimizer for engineering design problems,” Advances in Engineering Software, vol. 114, pp. 163–191, 2017. [Google Scholar]
23. W. Dong and M. Zhou, “A supervised learning and control method to improve particle swarm optimization algorithms,” IEEE transactions on systems,” Man, and Cybernetics: Systems, vol. 47, no. 7, pp. 1135–1148, 2016. [Google Scholar]
24. A. Onan, S. Korukoğlu and H. Bulut, “A multiobjective weighted voting ensemble classifier based on differential evolution algorithm for text sentiment classification,” Expert Systems with Applications, vol. 62, pp. 1–16, 2016. [Google Scholar]
25. D. T. Ahneman, J. G. Estrada, S. Lin, S. D. Dreher and A. G. Doyle, “Predicting reaction performance in C–N cross-coupling using machine learning,” Science, vol. 360, no. 6385, pp. 186–190, 2018. [Google Scholar]
26. S. Fong, S. Deb and X.-S. Yang, “How meta-heuristic algorithms contribute to deep learning in the hype of big data analytics,” in Progress in Intelligent Computing Techniques: Theory, Practice, and Applications. Singapore: Springer, pp. 3–25, 2018. [Google Scholar]
27. M. Sharma and P. Kaur, “A comprehensive analysis of nature-inspired meta-heuristic techniques for feature selection problem,” Archives of Computational Methods in Engineering, vol. 28, no. 3, pp. 1–25, 2020. [Google Scholar]
28. M. Dorigo, V. Maniezzo and A. Colorni, “Ant system: Optimization by a colony of cooperating agents,” IEEE transactions on systems, man, and cybernetics,” Part B (Cybernetics), vol. 26, no. 1, pp. 29–41, 1996. [Google Scholar]
29. X.-S. Yang, “A new metaheuristic bat-inspired algorithm,” in Nature Inspired Cooperative Strategies for Optimization (NICSO 2010), Berlin, Hiedelberg: Springer, pp. 65–74, 2010. [Google Scholar]
30. D. T. Pham and M. Castellani, “The bees algorithm: Modelling foraging behaviour to solve continuous optimization problems,” proceedings of the institution of mechanical engineers,” Part C: Journal of Mechanical Engineering Science, vol. 223, no. 12, pp. 2919–2938, 2009. [Google Scholar]
31. X.-S. Yang and S. Deb, “Cuckoo search via lévy flights,” in 2009 world congress on nature & biologically inspired computing (NaBICIeee, pp. 210–214, 2009. [Google Scholar]
32. S. Deb, S. Fong and Z. Tian, “Elephant search algorithm for optimization problems,” in 2015 Tenth Int. Conf. on Digital Information Management (ICDIMJeju Island, South Korea, IEEE, pp. 249–255, 2015. [Google Scholar]
33. X.-S. Yang and X. He, “Firefly algorithm: Recent advances and applications,” International Journal of Swarm Intelligence, vol. 1, no. 1, pp. 36–50, 2013. [Google Scholar]
34. X.-S. Yang, “Flower pollination algorithm for global optimization,” in Int. Conf. on Unconventional Computing and Natural Computation, Berlin, Heidelberg, Springer, pp. 240–249, 2012. [Google Scholar]
35. D. E. Goldberg, “Genetic algorithms in search,” Optimization, and Machine Learning, vol. 3, pp. 95–99, 1989. [Google Scholar]
36. S. Deb, Z. Tian, S. Fong, R. Tang, R. Wong et al., “Solving permutation flow-shop scheduling problem by rhinoceros search algorithm,” Soft Computing, vol. 22, no. 18, pp. 6025–6034, 2018. [Google Scholar]
37. R. Tang, S. Fong, X.-S. Yang and S. Deb, “Wolf search algorithm with ephemeral memory,” in Seventh Int. Conf. on Digital Information Management (ICDIM 2012 Macau, IEEE, pp. 165–172, 2012. [Google Scholar]
38. S. R. Garner, “Weka: The waikato environment for knowledge analysis,” in Proceedings of the New Zealand Computer Science Research Students Conference, vol. 1995, pp. 57–64, 1995. [Google Scholar]
39. M. Fitzner, G. Wuitschik, R. J. Koller, J.-M. Adam, T. Schindler et al., “What can reaction databases teach us about buchwald–Hartwig cross-couplings?,” Chemical Science, vol. 11, no. 48, pp. 13085–13093, 2020. [Google Scholar]
40. A. S. Galushko, D. O. Prima, J. V. Burykina and V. P. Ananikov, “Comparative study of aryl halides in Pd-mediated reactions: Key factors beyond the oxidative addition step,” Inorganic Chemistry Frontiers, vol. 8, no. 3, pp. 620–635, 2020. [Google Scholar]
41. G. Skoraczyński, P. Dittwald, B. Miasojedow, S. Szymkuć, E. Gajewska et al., “Predicting the outcomes of organic reactions via machine learning: Are current descriptors sufficient?,” Scientific Reports, vol. 7, no. 1, pp. 1–9, 2017. [Google Scholar]
42. H. Gao, T. J. Struble, C. W. Coley, Y. Wang, W. H. Green et al., “Using machine learning to predict suitable conditions for organic reactions,” ACS Central Science, vol. 4, no. 11, pp. 1465–1476, 2018. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |