 Open Access
Open Access
ARTICLE
HuLBD1 Promotes Flavonoid Biosynthesis Involved in Senescence of Hylocereus undatus by Negatively Regulating HuCHS
1 College of Food and Bioengineering, Henan University of Science and Technology, Luoyang, 471023, China
2 Henan Engineering Research Center of Food Microbiology, Luoyang, 471023, China
3 The National and Local Joint Engineering Laboratory of High Efficiency and Superior-Quality Cultivation and Fruit Deep Processing Technology of Characteristic Fruit Trees in South Xinjiang, Tarim University, Alar, 843300, China
* Corresponding Author: Xin Li. Email:
(This article belongs to the Special Issue: Influence of Biotic and Abiotic Stresses Signals on Plants and their Performance at Different Environments)
Phyton-International Journal of Experimental Botany 2025, 94(3), 825-842. https://doi.org/10.32604/phyton.2025.062015
Received 08 December 2024; Accepted 19 February 2025; Issue published 31 March 2025
Abstract
In plants, transcription factor (TF) family LATERAL ORGAN BOUNDARIES DOMAIN (LBD) has been identified to be involved in the ripening and senescence processes of fruits. However, the function of LBD in Hylocereus undatus (Haw.) Britton & Rose (H. undatus) has not been reported yet. Through transcriptomic analysis, virus-induced gene silencing (VIGS) technology, and RT-qPCR validation, we investigated the role of the LBD TFs in the senescence of H. undatus. Transcriptomic analysis revealed that HuLBD1 is a key transcription factor of the LBD family regulating H. undatus senescence. After silencing HuLBD1, 5075 differentially expressed genes (DEGs) were identified. GSEA results showed that these DEGs were mainly enriched in the phenylpropanoids and flavonoid biosynthesis pathway. The leading subset of genes in these two pathways included 16 DEGs, with four upregulated genes, HuCHS1-2, HuFLS1, HuCYP75B2, and HuCHS5-2 being central in the Protein-Protein Interaction (PPI) network. H. undatus phenotypic experiments confirmed that silencing HuLBD1 significantly increased the flavonoid content in the fruit peel, and RT-qPCR validation showed that the gene expression trends were consistent with the RNA-seq data. The study indicated that HuLBD1 likely delays the fruit senescence process by negatively regulating the expression of four key genes, including HuCHS1-2, thus promoting the synthesis of phenylpropanoids such as flavonoids. In conclusion, our findings suggest that the HuLBD1 TF plays an important role in the senescence of H. undatus fruit, providing a theoretical basis for postharvest senescence control.Keywords
Supplementary Material
Supplementary Material FileThe senescence and decay of fruits is a complex physiological and biochemical process in the life activities of fruits, including changes in quality attributes such as color, flavor, and texture, which are closely related to storage time, environment, and other factors [1,2]. Senescence can also be completed through the accumulation of secondary metabolites and the degradation and disintegration of the cell wall, involving common signaling pathways and regulatory factors, including various transcription factors [3,4].
LATERAL ORGAN BOUNDARIES DOMAIN (LBD) transcription factors (TFs), also known as AS2/LOB, are a class of plant-specific transcription factors characterized by a highly conserved (Lateral Organ Boundaries) domain [5,6]. Studies have indicated that LBD TFs participate in a range of biological activities, which encompass plant growth and development, senescence, hormone regulation, stress response, and metabolism [7,8]. With the advancement of molecular biology, LBD TFs have been identified in multiple species, such as Arabidopsis and tomato [9–11]. Numerous studies have reported the involvement of LBDs in plant growth and development [12–14]. Recent findings have also revealed that LBD genes are crucial for plant secondary metabolism. In apple, MdLBD13 inhibits anthocyanin synthesis and nitrogen utilization through the flavonoid pathway [15]. AtLBD37/38/39 are strongly induced by nitrate and negatively regulate anthocyanin synthesis [16]. Overexpression of OsLBD37 in rice can lead to nitrogen metabolism disruption, premature senescence, and the formation of low cotyledon leaves [17]. These findings suggest that LBD transcription factor family members may be related to the maturation and senescence of fruits and vegetables. However, the specific regulatory mechanisms remain unclear.
Hylocereus undatus (Haw.) Britton & Rose (H. undatus) belongs to the Cactaceae family and the Hylocereus genus. As a tropical fruit known for its many health advantages, it is abundant in carbohydrates, plant proteins, vitamins, and water-soluble dietary fibers [18], and has recently garnered considerable consumer attention. The peel of H. undatus is rich in flavonoid compounds, which establish a strong basis for its anti-senescence characteristics. Consequently, H. undatus is regarded as an outstanding candidate for investigating the mechanisms behind fruit senescence. However, there has been no report on whether LBD TFs are involved in the regulation of senescence in H. undatus.
In this study, the key HuLBD1 TF was first identified through RNA-Seq analysis. Next, the expression of the HuLBD1 gene in H. undatus was silenced using Virus-Induced Gene Silencing (VIGS) technology. Finally, the expression pattern of HuLBD1 during postharvest senescence of H. undatus was preliminarily confirmed through transcriptomic analysis of the HuLBD1-silenced samples. Additionally, downstream target genes regulated by HuLBD1 were screened. The findings of this study provide important theoretical insights for a deeper understanding of the physiological regulatory mechanisms of postharvest senescence in H. undatus.
Fruits were harvested from H. undatus (Vietnam No. 1) 35 days post artificial pollination (DAAP) at Miaoshui Base, located on Luoge Road in Ruyang County, Henan Province, China. These collected fruits served as samples. A total of eighty H. undatus fruits exhibiting consistent shape, size, and color were chosen for the study. The fruits were kept in a controlled environment at 25°C with a relative humidity of 85%. Throughout the storage duration, the exocarp’s flavonoid levels were assessed every three days. Trypsin (bovine source, 5 × 105 units/g) was purchased from Amersco, USA.
2.2 H. undatus Fruit Treatment Methods and Transcriptomic Analyses
Trypsin was used as a preservative, and the fruits were divided into a control group and a trypsin group. Five H. undatus fruits are selected in each group. The treatment conditions for trypsin were the same as those previously reported, with a final concentration of 2.41 × 10−6 mol/L [19]. The control group was soaked in distilled water following the same procedure. Samples were taken for transcriptome sequencing after one week of H. undatus fruits storage. The sequencing process is carried out according to our previous research. Differential expression analysis of the transcriptome was performed using the DESeq2 software (|log2FC| = 1.05, p-value <= 0.05) [20].
2.3 Sequence Analysis of HuLBD1
The molecular weight and theoretical isoelectric point of the protein were predicted using the ExPASy online website (https://web.expasy.org/protparam/) (accessed on 18 February 2025) [21]. Similar sequences were searched in the NCBI database. Multiple amino acid sequences were aligned using DNAMAN software, and a phylogenetic tree was constructed using MEGA software [22].
The total RNA of H. undatus peel was extracted using the RNAprep Pure Micro Kit (DP432, TransGen Biotech Co., Ltd., Beijing, China), and cDNA first strand synthesis was performed using the TransScript® II One-Step gDNA Removal and cDNA Synthesis SuperMix (AH311-02, TransGen Biotech Co., Ltd., Beijing, China). Amplification was conducted using specific primers for HuLBD1. The primers used for VIGS were 5′CCGGAATTCATGGTATATGAAGCGAATGCTC-3′(sense) and 5′CGCGGATCCTCAAGTCCAAAGAGGCTCCC-3′(antisense), which were synthesized by Genscript Biotech Corporation. The PCR product was subjected to agarose gel electrophoresis, and the target band was recovered and ligated with the pEASY®-Blunt Cloning vector (CB101-01, TransGen Biotech Co., Ltd., China). The ligation product was then transformed into Escherichia coli TOP10 competent cells by heat shock. Single colonies that were confirmed as positive by PCR were sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequencing. After confirming the correctness of the sequencing, the product was digested with EcoR I and BamH I restriction enzymes and ligated with T4 DNA Ligase (FL101-01, TransGen Biotech Co., Ltd., China) to the pTRV2 vector [23], constructing the pTRV2-HuLBD1 recombinant plasmid. After PCR verification, the recombinant plasmid was transformed into Agrobacterium GV3101 to induce gene silencing in H. undatus.
2.5 Preparation and Infection of Bacterial Solution
In accordance with the approach described by Zhang et al. [24], a solution known as the infection buffer was created, consisting of 10 mM MES, 10 mM MgCl2, and 200 μM AS. For the injection treatment, the peel of H. undatus was immersed in a bacterial suspension with an OD600 of 0.6 using a 1 mL syringe [3,25]. The fruits infected with a 1:1 Agrobacterium infection solution carrying pTRV1 and pTRV2-HuLBD1 were used as the experimental group (LBD-i), while the fruits infected with a 1:1 Agrobacterium infection solution carrying pTRV1 and empty pTRV2 were used as the control group (CK). Thirty-five H. undatus fruits are selected in each group. Both groups of H. undatus were placed in a 25°C incubator. The color, shape, and dehydration-induced wrinkles or lesions of the fruit were observed every 24 h, photographed, and recorded. The water loss rate was measured [23]. After one week of storage, the treated H. undatus peel was cut with a scalpel, rapidly frozen in liquid nitrogen, and then preserved at −80°C for subsequent transcriptomic sample preparation.
2.6 Detection of Flavonoid Levels, Reducing Sugar Content, Soluble Protein Content, and pH
The total flavonoid content was assessed using the aluminum chloride colorimetric method [26]. The soluble protein and soluble sugar content were determined using methods previously studied [27].
2.7 Gene Set Enrichment Analysis and KEGG Functional Enrichment Analysis
GSEA was used as a statistical method to estimate the enrichment between a query gene set and reference gene features [28]. KEGG enrichment analysis was performed according to the method of Candar-Cakir et al. [29].
2.8 Protein-Protein Interaction (PPI) Network Analyses
The regulatory network between proteins was constructed using Cytoscape software. The CytoHubba plugin was used to score and rank the nodes, while MCODE was used to calculate and collect clusters in the protein-protein interaction network [19].
2.9 Gene Expression Analysis by RT-qPCR
Reverse RT-qPCR was conducted according to the method of Li et al. with β-Actin used as an internal reference for gene expression normalization [30]. The relative copy number of genes was calculated using the method [6].
Statistical analysis was performed using the SPSS (version 12.0.1) statistical software package. The Wilcoxon test, a non-parametric test, was utilized to analyze differences between the entire sample groups. OriginPro 2024 (version 10.1.0.178) analyzed data and drew graphs [31].
3.1 Identification of HuLBD1 Transcription Factors
After trypsin treatment, the transcriptome data showed enrichment for a total of 70 LBD-related genes, including 31 upregulated genes and 39 downregulated genes(Table S1). Among these, TRINITY_DN52324_c0_g1 exhibited the largest change in expression among the downregulated genes. The protein sequence was obtained from NCBI (https://www.ncbi.nlm.nih.gov/) (accessed on 18 February 2025) and analyzed using ExPASy. Its open reading frame (ORF) is 546 bp in length, encoding a polypeptide of 182 amino acids. The calculated molecular weight was 20.06 kDa, with a theoretical isoelectric point of 5.16. Using the Blast online alignment tool in NCBI (Fig. 1A), six proteins with high homology to TRINITY_DN52324_c0_g1 were retrieved, including those from Arabidopsis (NP_172268.1), Oryza sativa (XP_015638880.1), Solanum lycopersicum (XP_004251265.1), Nicotiana tabacum (XP_016487519.1), Amaranthus tricolor (XP_057521184.1), and Chenopodium quinoa (XP_021715798.1). A multiple sequence alignment was performed, and a phylogenetic tree was constructed using MEGA 11.0 software (Fig. 1B). The findings revealed that this protein shares the highest homology with LBD1 from Arabidopsis thaliana. It possesses a relatively conserved Lateral Organ Boundary (LOB) domain at the N-terminus, containing a zinc finger motif responsible for DNA binding (CX2CX6CX3C), a GAS-block motif (Gly-Ala-Ser), and a leucine zipper motif (LX6LX3LX6L) responsible for protein dimerization, which are characteristic features of LBD TFs. Therefore, it was named HuLBD1.
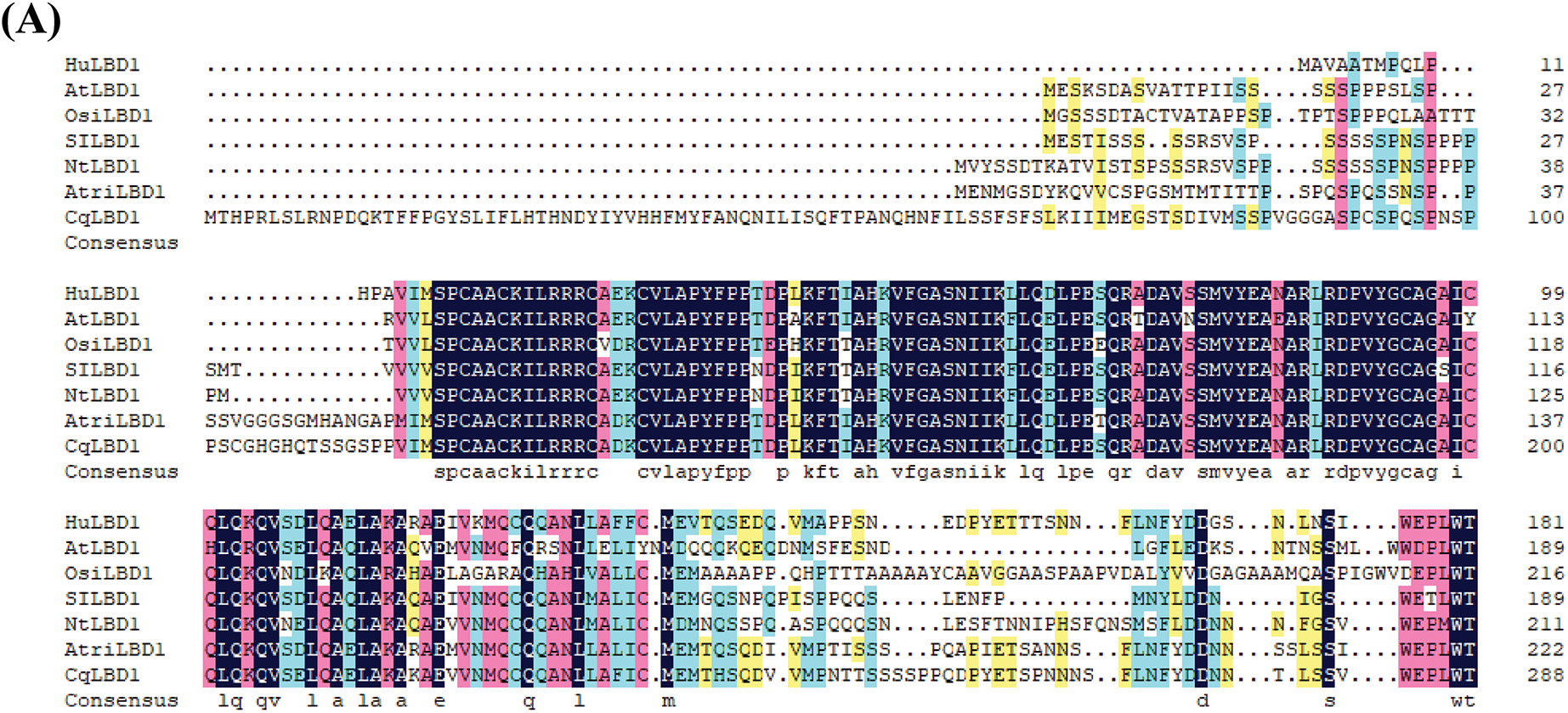
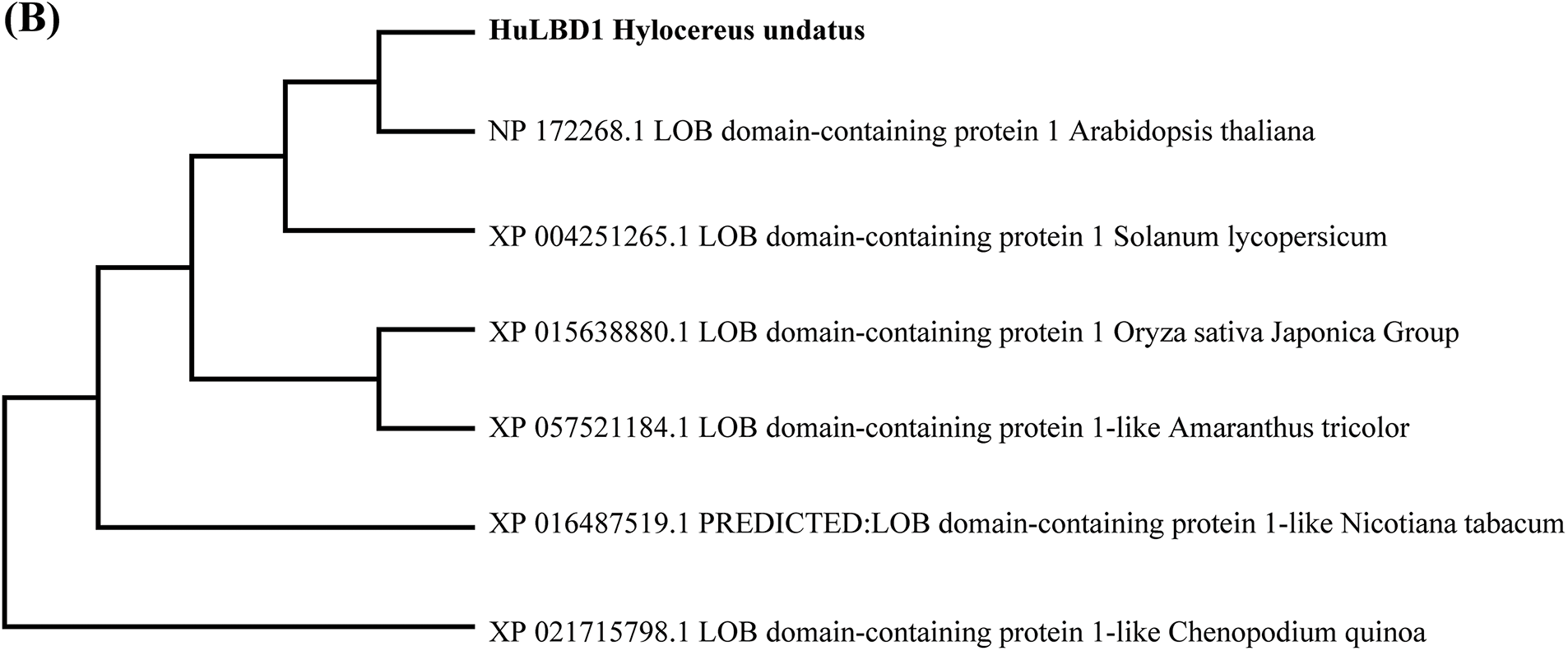
Figure 1: Bioinformatics analysis of HuLBD1. (A), Alignment of HuLBD1 with sequences of other plant LBDs. Identical and similar amino acids are represented in blue and red, respectively. (B), Evolutionary tree of LBD transcription factor systems
3.2 VIGS Technology Verifies the Functionality of HuLBD1
To confirm the function of this gene in preserving fruits, the VIGS technique was utilized to inhibit the expression of HuLBD1 mRNA in H. undatus. Employing cDNA extracted from newly harvested H. undatus as a template, a 312 bp segment of the HuLBD1 gene was successfully obtained through PCR using specific primers. The target fragment was then inserted into the pTRV2 vector (Fig. 2A). The results obtained through RT-qPCR indicated that, compared to the CK group, the expression level of the HuLBD1 gene in the LBD-i group decreased. This finding demonstrated that the silencing of the HuLBD1 gene in H. undatus was effective (Fig. 2B).
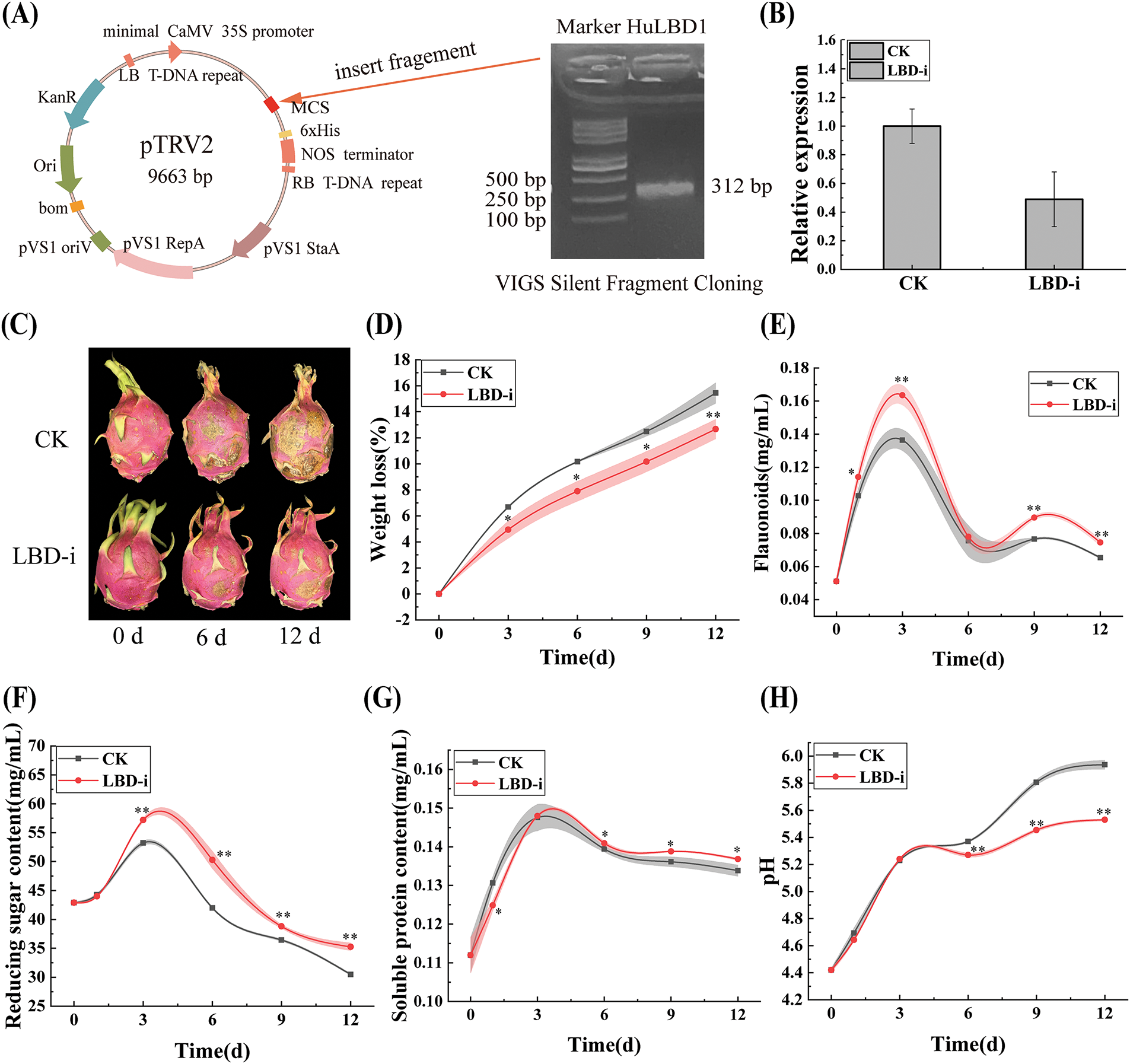
Figure 2: Construction schematic of the VIGS vector and physicochemical index detection of H. undatus. Data are presented as mean values ± SD. The style of connecting is spline. The area under curve is filled. A paired two-tailed t-test was used for all statistical analyses. No adjustments were made for multiple comparisons. * represents p < 0.05, and ** represents p < 0.01. (A), Schematic diagram of the construction of the recombinant expression vector PTRV2-HuLBD1. (B), HuLBD1 gene expression verification. (C), Phenotypic illustration of H. undatus. (D), Weight loss rate of H. undatus. (E), Flavonoid content of H. undatus peel. (F), Content of reducing sugar in H. undatus juice. (G), Content of soluble protein in H. undatus juice. (H), PH changes in H. undatus juice
By observing the phenotypic changes of the fruit, in the early stages of storage, both the CK group and LBD-i group showed high-quality fruit, with bright color (Fig. 2C). After 6 days of storage, the scales of the CK group began to shrivel, and the color became dull, while the LBD-i group maintained a relatively bright color with no obvious fungal spots on the surface. After 12 days, the scales of the CK group were completely dry, with numerous disease spots, while the LBD-i group maintained a favorable fruit color, with only a few disease spots (Fig. 2C).
With an increase in storage duration, an upward trend was observed in the weight loss rate of the H. undatus. Notably, the weight loss rate in the LBD-i group remained consistently lower compared to the CK group. After 12 days of storage (p < 0.01), the weight loss rate of the LBD-i group (12.67%) was markedly lower than that of the CK group (15.44%), indicating that VIGS treatment slowed down the water loss rate of the H. undatus (Fig. 2D).
The amount of flavonoid in the H. undatus peel exhibited a pattern of initially rising and then falling as the storage duration extended (Fig. 2E). On the third day of storage, both groups exhibited a “peak,” but the LBD-i group (0.163 mg/mL) was significantly higher than the CK group (0.136 mg/mL) (p < 0.01). On the ninth and twelfth day of storage, the LBD-i group (0.089 mg/mL) also had a higher flavonoid content than the CK group (0.076 mg/mL) (p < 0.01). Throughout the storage period, the trends of both groups were almost identical, but the LBD-i group consistently had higher levels, indicating that the silencing of HuLBD1 effectively enhanced the flavonoid content in the H. undatus peel.
To clarify the changes in fruit quality during storage, we measured the content of reducing sugars, soluble proteins, and pH in the fruit juice. During the early stage of storage, no significant differences were observed between the two groups for these indicators (p > 0.05). After 3 days of storage, the reducing sugar content and soluble protein content in the juice of both groups showed a decreasing trend (Fig. 2F,G). On the 12th day, the reducing sugar content in the LBD-i group (35.27 mg/mL) was significantly higher than that in the CK group (30.48 mg/mL) (p < 0.01). Similarly, the soluble protein content in the LBD-i group (0.14 mg/mL) was higher than that in the CK group (0.13 mg/mL) (p < 0.05). The pH of the fruit juice in both groups showed an increasing trend, with the CK group showing a significant acceleration in pH increase after day 3, while the LBD-i group remained relatively stable (Fig. 2H).
3.3 Transcriptomic Analysis of H. undatus Following HuLBD1 Silencing Was Performed
HuLBD1, as a plant-specific TF, identification of downstream target genes is particularly crucial for studying the function of LBD genes. Therefore, comparing the transcriptomic data of the CK group with the LBD-i group, and analyzing differentially expressed genes, was essential. The results of sample correlation analysis indicated a strong correlation between biological replicates (Fig. 3A). From the analysis of expression level differences, 5075 significantly differentially expressed genes were identified, comprising 2757 upregulated genes and 2318 downregulated genes (Fig. 3B).
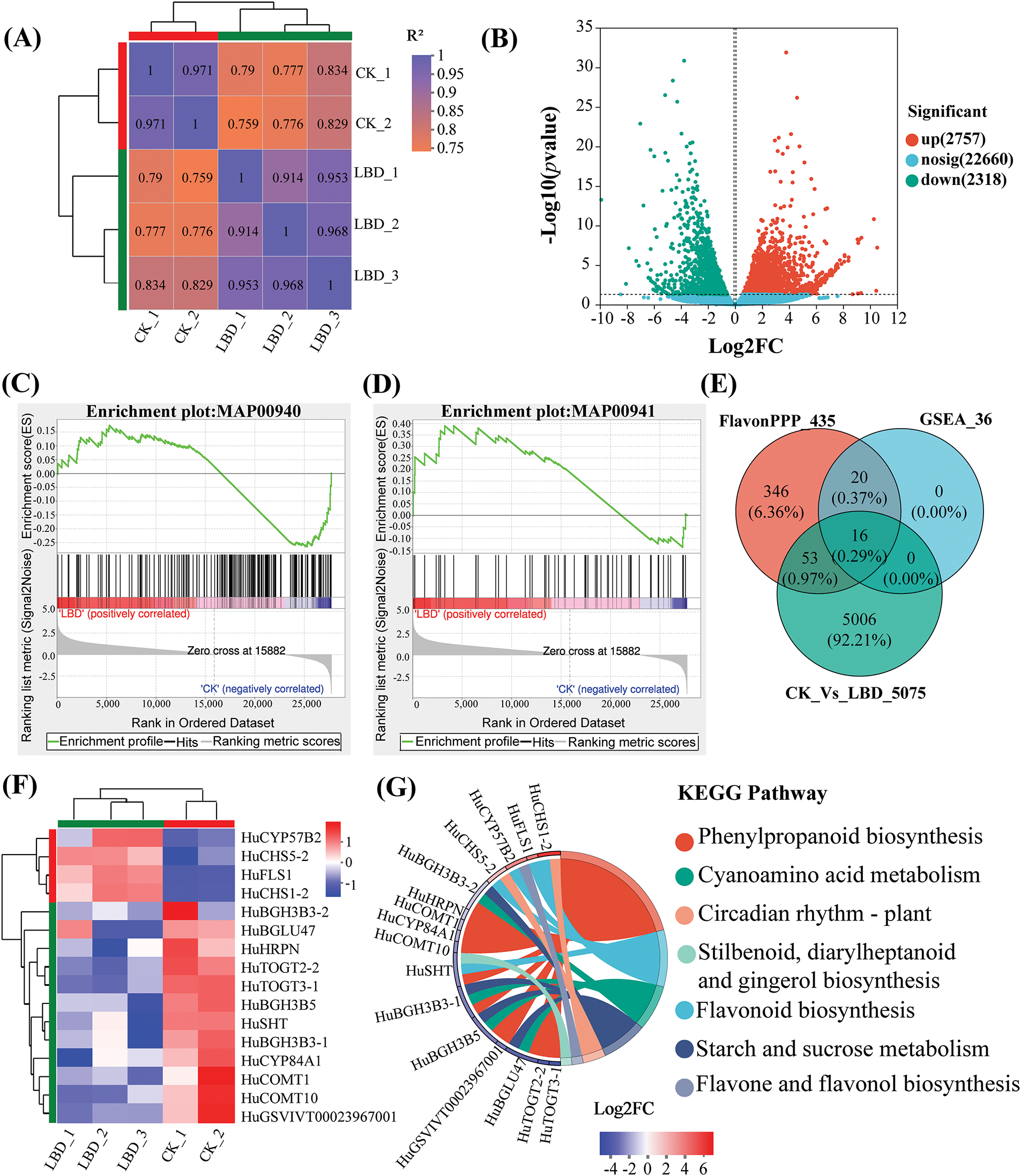
Figure 3: Analysis of differentially expressed genes between LBD-i and CK groups. (A), Correlation analysis between samples. (B), Volcano plot showing differential expression levels. (C), GSEA results for MAP00940. (D), GSEA results for MAP00941. (E), Venn diagram showing differentially expressed genes. (F), Heatmap depicting gene expression patterns. Genes with high and low expression levels are represented in red and blue, respectively. (G), Enrichment string diagram for differentially expressed genes in KEGG pathways
The differentially expressed genes that were identified had been subjected to GSEA to annotate them to five metabolic pathways. As shown in Table S2, these pathways included Phenylpropanoid biosynthesis (MAP00940), Flavonoid biosynthesis (MAP00941), Stilbenoid, diarylheptanoid, and gingerol biosynthesis (MAP00945), Isoquinoline alkaloid biosynthesis (MAP00950), and Tropane, piperidine, and pyridine alkaloid biosynthesis (MAP00960). The biosynthesis pathways of both Phenylpropanoid and Flavonoid originate from phenylalanine and involve a sequence of enzyme-catalyzed reactions, indicating a close relationship between them. MAP00940 and MAP00941 pathways had the most enriched genes. Among them, 26 genes had been clustered in the leading edge of the MAP00940 pathway, while 10 genes had been clustered in the leading edge of the MAP00941 pathway (Fig. 3C,D). As the key genes had already been identified in the GSEA results, they played significant roles in Phenylpropanoid and Flavonoid biosynthesis pathways.
In our transcriptomic data, we identified 435 genes related to flavonoid and other phenylpropanoid synthesis, 5075 differentially expressed genes after HuLBD1 silencing, and 36 leading subsets obtained from GSEA analysis. Functional annotation analysis was performed on the three gene sets mentioned above (Fig. 3E), and a total of 16 genes were annotated to be present in all three gene sets. Hierarchical clustering analysis was conducted on these 16 differentially expressed genes (Fig. 3F), revealing that 4 genes showed significantly increased expression while 12 genes exhibited decreased expression, indicating significant differences between the two groups. Enrichment string analysis indicated that the four upregulated genes (Fig. 3G), HuCHS1-2, HuFLS1, HuCYP75B2 and HuCHS5-2, mainly participated in phenylpropanoid and flavonoid biosynthesis. These results suggest that HuLBD1 may have promoted the accumulation of flavonoid compounds and influenced the senescence process of H. undatus by regulating the expression of genes related to phenylpropanoid and flavonoid biosynthesis, such as HuCHS1-2, HuFLS1, HuCYP75B2, and HuCHS5-2.
3.4 Analysis of PPI Network and Co-Expression Network of Differentially Expressed Genes
To further validate the core genes, 69 genes related to flavonoid and other phenylpropanoid synthesis, screened from transcriptomic data, were used to construct a PPI network (Fig. 4) and performed co-expression network analysis (Fig. 5) using Cytoscape. The PPI network consisted of 36 nodes and 100 edges, with 31 upregulated nodes and 38 downregulated nodes represented by triangles and squares, respectively. Based on the CytoHubba sorting method “Maximal Clique Centrality (MCC),” the Top10 nodes in the overall protein network were ranked (Fig. 4B). The results showed that HuCHS5-2, HuFLS1, and HuCYP75B2 ranked in the top ten, ranking first, third, and ninth, respectively (Fig. 4B), indicating their core roles in the PPI network. MCODE was used to divide the expression-related network into two clusters (Fig. 5) and analyzed the correlation between genes. Cluster 1 comprised genes related to phenylpropanoid synthesis (Fig. 5A), such as phenylalanine ammonia-lyase (PAL), which catalyzed the non-oxidative deamination of phenylalanine to trans-cinnamic acid, a key enzyme in the phenylpropanoid metabolism pathway. β-glucosidase (BGLU) catalyzed the conversion of cinnamic acid to coumarins, while catechol-O-methyltransferase (COMT) catalyzed the conversion of caffeic acid to ferulic acid. Cluster 2 comprised genes related to flavonoid synthesis (Fig. 5B), such as chalcone synthase (CHS), which catalyzed the formation of naringenin chalcone from 4-coumaroyl-CoA and malonyl-CoA, initiating the biosynthesis of flavonoids. Flavonol synthase (FLS), a key enzyme downstream in the flavonoid biosynthesis pathway, promoting the accumulation of flavonol. The clustering results indicated that there was an expression correlation between HuCHS1-2 and HuFLS1, jointly promoting the biosynthesis of phenylpropanoids and flavonoids. The results of PPI network and co-expression network analysis indicated that HuLBD1 primarily regulated the expression of genes related to phenylpropanoid and flavonoid biosynthesis pathways, such as HuCHS1-2, HuFLS1, HuCYP75B2, and HuCHS5-2.
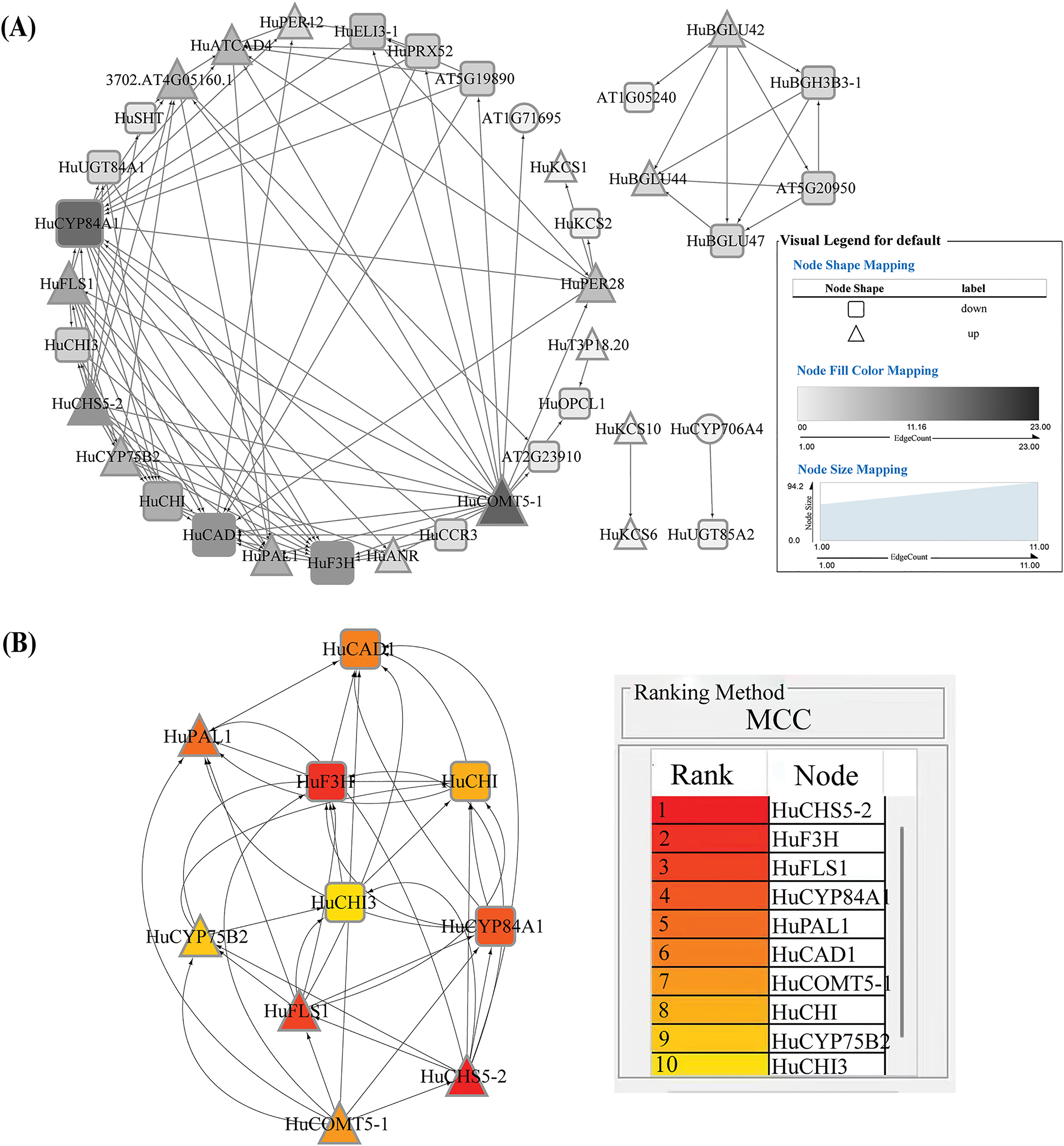
Figure 4: Network of 69 differentially expressed genes related to flavonoid and other phenylpropanoid synthesis. (A), PPI network of differentially expressed genes. (B), Top 10 protein network interaction diagram
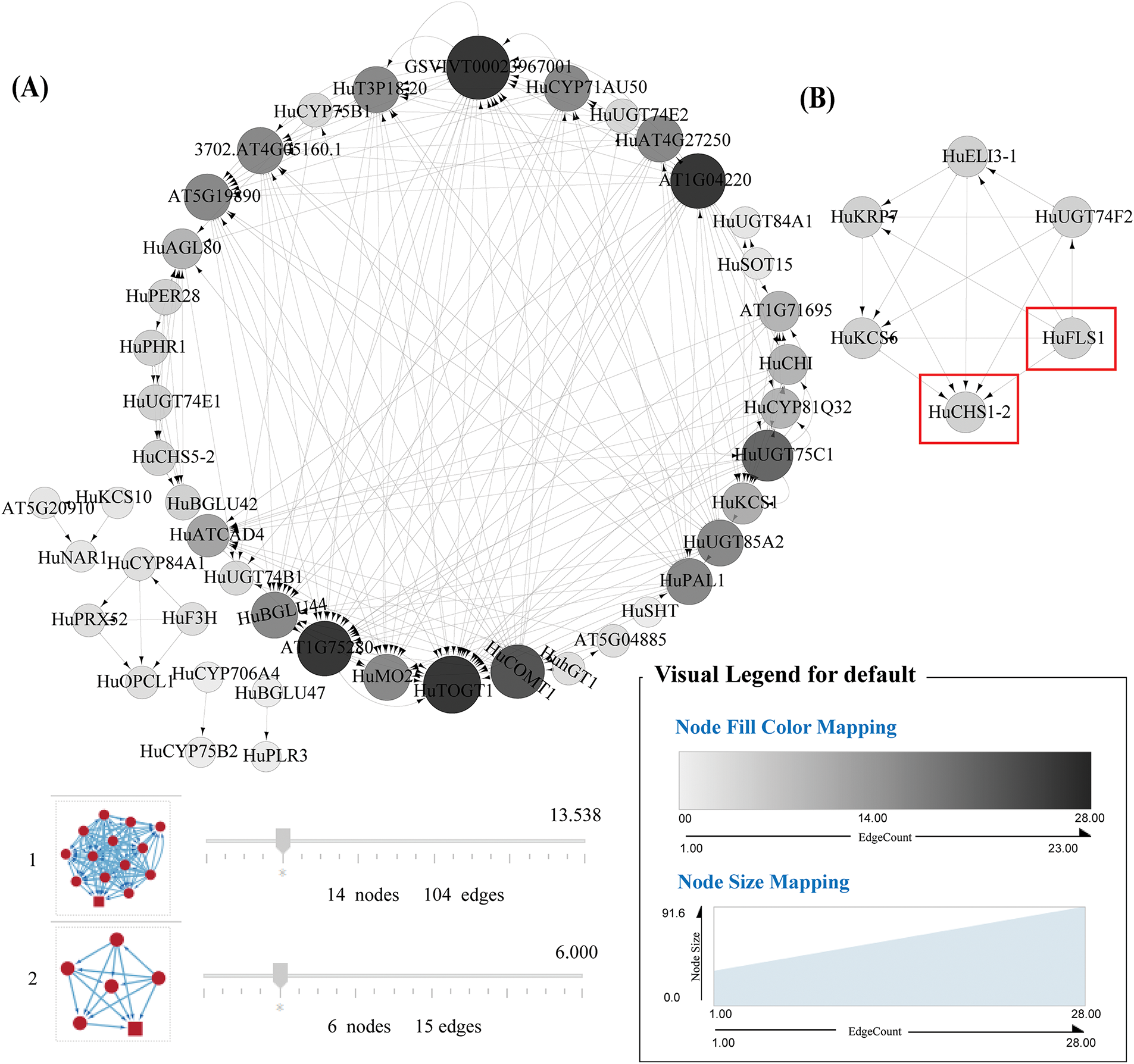
Figure 5: Co expression network diagram of 69 genes related to the synthesis of flavonoids and other phenylpropanoids in differentially expressed genes. Note: The red box in the figure highlights the correlation between HuCHS1-2 and HuFLS1 genes. (A), Cluster 1 comprised genes related to phenylpropanoid synthesis; (B), Cluster 2 comprised genes related to flavonoid synthesis
3.5 Accuracy of the RNA-Seq Data Verification by RT-qPCR
RT-qPCR was performed to detect the expression levels of four upregulated genes, HuCHS1-2, HuCHS5-2, HuFLS1, and HuCYP75B2 (Table 1), using RNA-seq data (Fig. 6). The R-squared value (R2 = 0.9541) between the RT-qPCR results and RNA-seq data indicates a high level of reliability of the transcriptomic data.
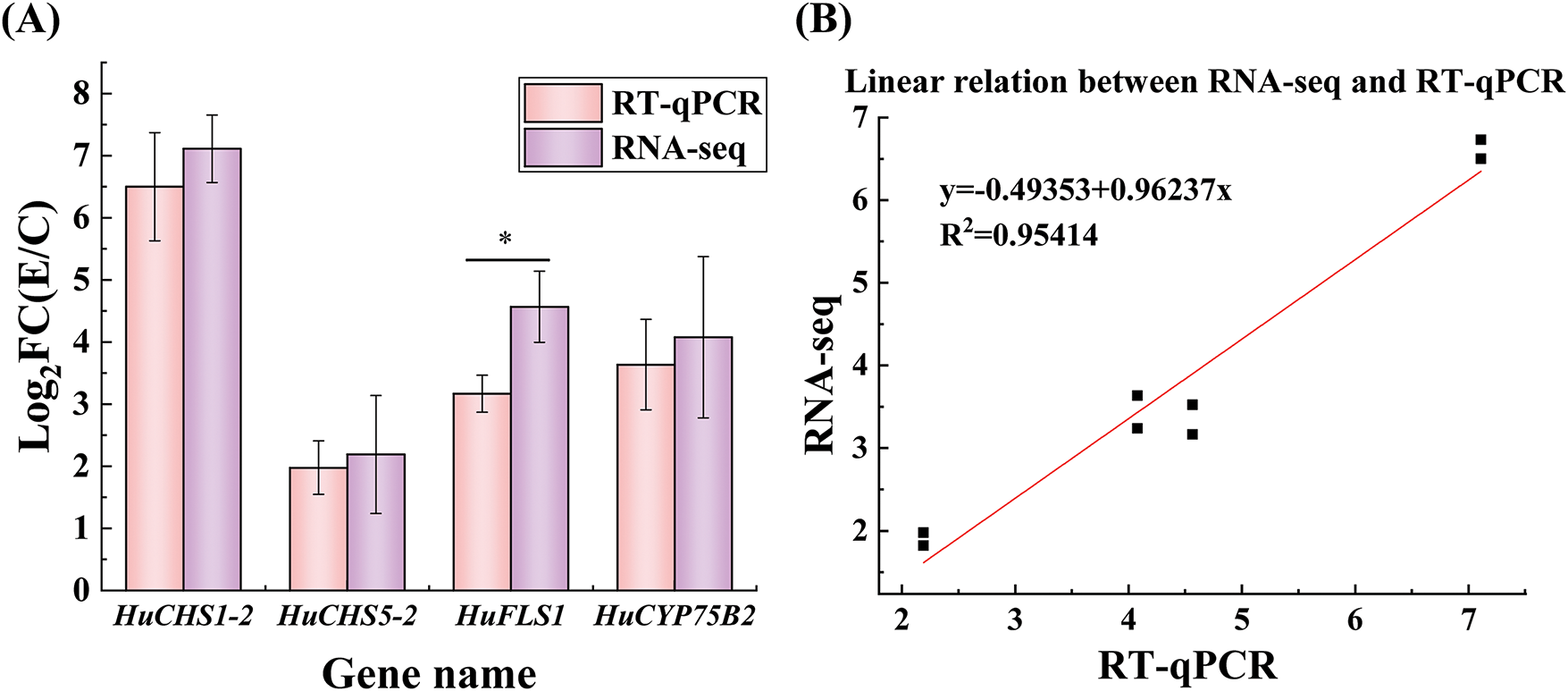
Figure 6: RNA-seq analysis and RT-qPCR validation of four target genes. (A), Expression levels of the selected four genes determined by RNA-seq and RT-qPCR. (B), Linear relationship between the expression ratios of selected genes detected by RNA-seq and RT-qPCR
4.1 Identification of HuLBD1 Transcription Factors
The LBD transcription factor family members are crucial in the genetic evolution of various species, including lateral root development, secondary metabolism, stress response, fruit ripening, and senescence [32]. H. undatus has a high water and sugar content, with tender tissue that is prone to dehydration, wilting, rotting, and rapid senescence, resulting in a short shelf life [33]. However, little is known about the functions of LBDs in H. undatus. In this study, 70 LBD-related genes were identified in H. undatus, of which 17 belong to the LBD transcription factor family. Through analysis of the expression changes of LBD-related genes, the key gene HuLBD1 was screened, and the HuLBD1 TF in H. undatus was identified through bioinformatics analysis.
4.2 The Effect of HuLBD1 Gene Silencing on the Storage Quality of H. undatus
During long-term storage of fruits, water loss is a key factor affecting quality. Research indicates that maintaining ideal water levels in fruits may decrease cellular metabolism and prolong senescence while in storage [34]. In tomato fruits, inhibiting the expression of the SlLOB1 transcription factor significantly reduces water loss during storage and delays fruit ripening [35]. The results of the weight loss measurement in H. undatus in this study were similar to those in tomato fruits. As the storage time increased, the weight loss rate of H. undatus continuously increased, but after silencing the HuLBD1 gene, the weight loss rate increased more slowly. This suggests that HuLBD1 may play an important role in water retention and delaying senescence in H. undatus fruits.
In addition, sugars and organic acids are also key factors in determining the flavor and health value of fruits [36]. Post-harvest, H. undatus undergoes intense metabolic activity, leading to the continuous consumption and breakdown of nutrients such as proteins, sugars, and acids in the flesh. In the current study, it was observed that the content of reducing sugars and soluble proteins initially increased and then decreased, particularly showing a decline on the third day of storage. This could be due to the gradual hydrolysis of starch into soluble sugars during the early stages of storage, leading to an increase in reducing sugar content. Similarly, the soluble protein content also increased due to metabolic activity during the early stages of storage. However, with prolonged storage, fruit respiration and metabolism intensified, resulting in the extensive consumption of these substances, which led to a decline in their levels. A similar trend was also observed in pears, where reducing sugar content increased during the early stages of storage, but started to decrease after four days [37]. After silencing the HuLBD1 gene, the rate of decrease in reducing sugar and soluble protein content was delayed, suggesting that inhibiting the expression of HuLBD1 can slow down the decline in fruit quality. The pH level gradually increased with the extension of storage time. Compared to the control group, the fruit with silenced HuLBD1 showed a smaller change in pH level. A similar finding was reported by Rashid et al. [38], where apples showed smaller pH level changes after preservation treatment. This indicates that the silencing of HuLBD1 affects the physiological metabolic responses in the fruit.
It is noteworthy that flavonoids play an important role in postharvest fruit senescence processes, such as in stress resistance and influencing the quality, flavor, and nutritional value of fruits [39]. It has been reported that in species such as citrus [40,41], flavonoid-related metabolites in the fruit peel change during the ripening process and play a key role in fruit maturation and senescence. This study’s findings distinctly illustrate a bimodal trend in the accumulation of flavonoids as senescence progresses, with the highest flavonoid levels observed on the third day of storage. Compared to the control group, the flavonoid content in the peel of HuLBD1-silenced fruits was significantly increased and maintained at a higher level. This suggests that HuLBD1 plays an important role in the flavonoid biosynthesis process in H. undatus. HuLBD1 may regulate the expression of key genes in the flavonoid biosynthesis pathway, affecting the synthesis and accumulation of flavonoids, thereby delaying the senescence process of the fruit. Therefore, further research is needed to analyze the regulatory mechanism of HuLBD1 in fruit senescence.
4.3 Transcriptomic Analysis of Potential Genes Related to Flavonoid Synthesis Regulated by HuLBD1
Previous research has shown that trypsin can upregulate the expression of CsMYC2 in cucumber (Cucumis sativus), thereby promoting flavonoid biosynthesis to achieve preservation effects [42]. Qi et al. [43] investigated the ripening and softening processes of sweet cherry fruit (Prunus avium L.) and revealed that the expression of PaPG1, which is positively regulated by PaMADS7, facilitates the ripening and softening of sweet cherry fruit. The silencing of PaMADS7 notably hinders the ripening of sweet cherry fruit. As for the LBD transcription factor, the tomato fruit (Solanum lycopersicum) SlLOB1 is upregulated during ripening and activates cell wall-related genes. Inhibiting SlLOB1 in fruit hinders softening, while overexpression of SlLOB1 promotes premature induction of cell wall genes and fruit softening [35]. In other species such as Arabidopsis, Banana, and Citrus, members of the LBD family have been reported to activate downstream cell wall gene expression to participate in the regulation of fruit senescence [44–46]. Based on these existing reports, we speculate that HuLBD1 may participate in the senescence process of H. undatus by regulating the expression of certain genes.
However, unlike the previous understanding of LBD regulation in the maturation and senescence processes of fruit and vegetable cell walls, our results indicate that HuLBD1 acts through negative regulation of the expression of genes involved in flavonoid biosynthesis, promoting the accumulation of flavonoid compounds in H. undatus peel, enhancing fruit resistance, and delaying fruit senescence. In Camellia sinensis, CsLBD can directly bind to cis-elements in the promoters of CsC4H, CsDFR, and CsUGT84A, positively regulating flavonoid synthesis, thus affecting the growth, development, and resistance to abiotic stress in Camellia sinensis [47]. PtrLBD41 promotes the expression of chalcone synthase (CHS) under salt stress conditions, affecting the biosynthesis of flavonoids and enhancing tolerance to salt stress [48]. In this research, inhibiting LBD expression could promote the expression of genes such as HuCHS1-2 (Log2FC = 7.1111), HuCHS5-2 (Log2FC = 2.1918), HuFLS1 (Log2FC = 4.5660), HuCYP75B2 (Log2FC = 4.0781), thereby increasing the flavonoid content in the fruit peel, enhancing the fruit’s disease resistance and stress tolerance, and delaying fruit senescence.
It is worth noting that among the aforementioned genes, CHS showed the most significant expression changes. CHS is the first enzyme in the plant flavonoid biosynthesis pathway and is also one of the key enzymes in plant secondary metabolism, holding significant physiological significance for plants [49]. High expression of GmCHS8 in soybeans can promote the production of isoflavones [50]. Overexpression of the CitCHS gene in citrus leads to a significant increase in flavonoid content [51]. This is consistent with our results. Combining the expression pattern of the HuCHS gene in fruits with the transcriptome data, we observed induced expression of HuCHS in HuLBD1-silenced plants. We consider HuCHS and other genes as a downstream target gene of LBD, capable of promoting flavonoid compound synthesis. By measuring the flavonoid content in the fruit peel, we have demonstrated that the increased expression of chalcone synthase in H. undatus significantly enhances flavonoid content.
4.4 The Mechanism of HuLBD in Fruit Senescence of H. undatus
Previous studies have shown that trypsin can eliminate superoxide anions (O2−) and protect cells, exhibiting anti-senescence effects [52]. H2O2 is a stable element of ROS when compared to its precursor. H2O2 can be transported into the cytoplasm by particular members of the aquaporin (AQP) super family [53] and the accumulation of H2O2 can cause oxidative damage to intracellular macromolecules. Treatment with trypsin significantly reduced the excessive accumulation of endogenous ROS, such as O2− and H2O2, in H. undatus, hindered cell damage, and improved the storage quality of H. undatus [3,19,20]. In this study, the expression of HuLBD1 was inhibited by trypsin. Transcriptome data showed that HuLBD1 promotes the accumulation of flavonoid compounds by negatively regulating the expression of genes such as HuCHS, thereby delaying the postharvest senescence in H. undatus (Fig. 7).
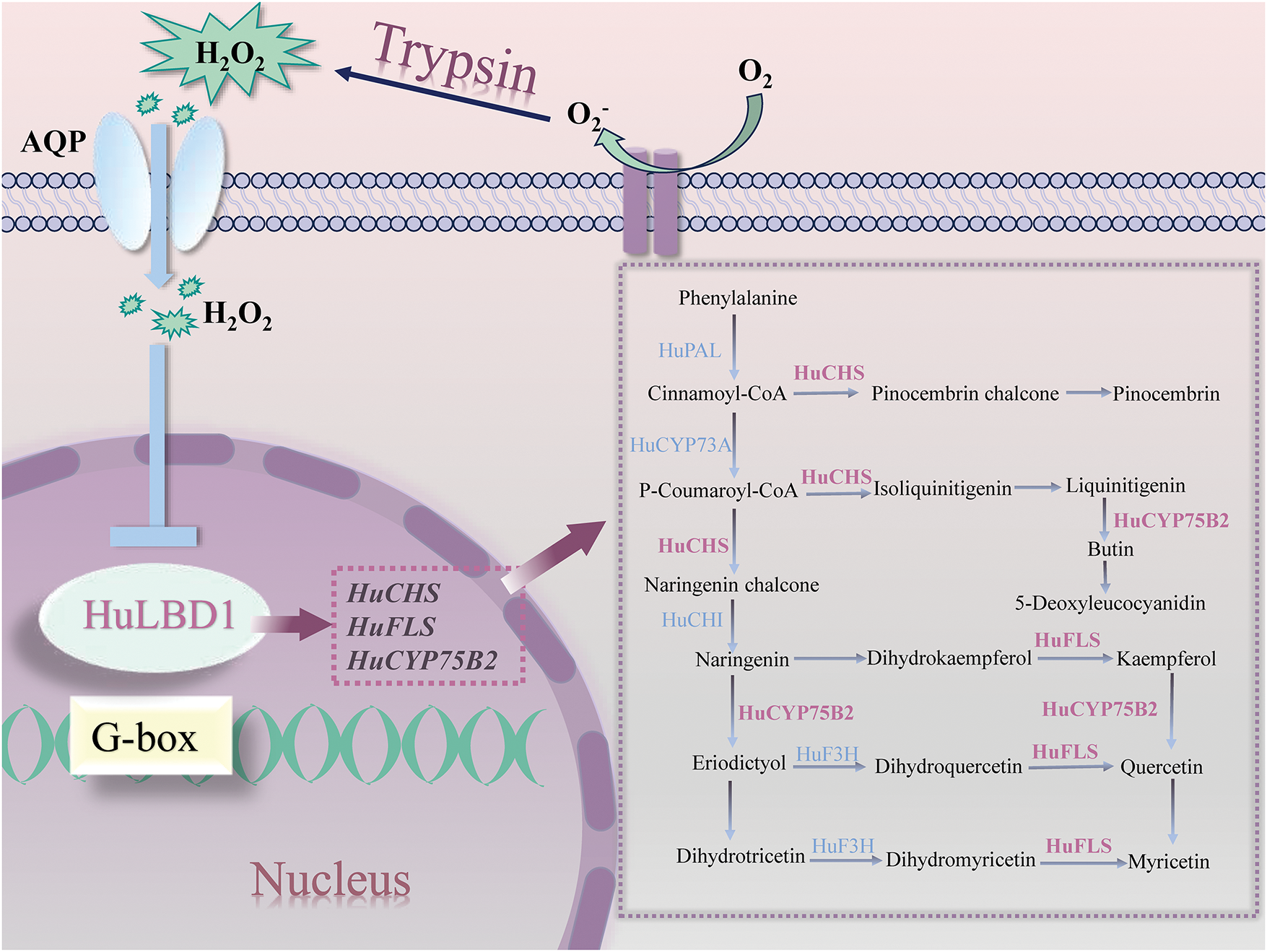
Figure 7: Regulatory model of flavonoid biosynthesis mediated by HuLBD1. AQP denotes aquaporin proteins. HuCHS: chalcone synthase; HuFLS: flavonol synthase; HuCYP75B2: flavonoid 3′-hydroxylase. The purple dashed box depicts the process of flavonoid compound biosynthesis. Purple genes represent genes upregulated by LBD influence
In this study, trypsin treatment of H. undatus fruit resulted in changes in the expression of LBD genes, with the most significant variation observed in the expression of HuLBD1. VIGS results demonstrated significant phenotypic differences between H. undatus with silenced HuLBD1 and the control group, along with an increase in the content of flavonoid compounds. LBD, as a unique transcription factor family in plants, holds significant importance in the metabolism, maturation, and senescence of fruits and vegetables. HuLBD1, acting as a positive transcriptional regulator of fruit maturation and senescence, regulates fruit maturation and senescence through downstream target gene regulation networks. These findings lay the groundwork for subsequent research in H. undatus, such as gene resource exploration and gene function verification, and provide new insights for extending the shelf life of H. undatus and delaying fruit senescence.
Acknowledgement: We are grateful for the free online platform of Majorbio I-Sanger Cloud Platform (www.majorbio.com) (accessed on 18 February 2025).
Funding Statement: This work was supported by the National Key Research and Development Program of China (2017YFC1600802), Henan Provincial Science and Technology Research Project, China (No. 232102110134), and the National and Local Joint Engineering Laboratory of High Efficiency and Superior-Quality Cultivation and Fruit Deep Processing Technology of Characteristic Fruit Trees in South Xinjiang of China (No. FE202303).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Xinxin Chen, Xin Li; data collection: Fuxin Li; analysis and interpretation of results: Jingyu Jia, Yajing Tian; draft manuscript preparation: Xinxin Chen, Xin Li. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data and materials used in this research were publicly available. Raw sequence data from this study have been submitted to the NCBI sequence read archive under the BioProject accession (PRJNA 509494) (PRJNA 886472).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2025.062015.
References
1. Gu C, Xu HY, Zhou YH, Yao JL, Xie ZH, Chen YY, et al. Multiomics analyses unveil the involvement of microRNAs in pear fruit senescence under high- or low-temperature conditions. Hortic Res. 2020;7(1):196. doi:10.1038/s41438-020-00420-y. [Google Scholar] [PubMed] [CrossRef]
2. Zhu Y, Du M, Jiang X, Huang M, Zhao J. Nitric oxide acts as an inhibitor of postharvest senescence in horticultural products. Int J Mol Sci. 2022;23(19):11512. doi:10.3390/ijms231911512. [Google Scholar] [PubMed] [CrossRef]
3. Li X, Li B, Gu S, Pang X, Mason P, Yuan J, et al. Single-cell and spatial RNA sequencing reveal the spatiotemporal trajectories of fruit senescence. Nat Commun. 2024;15(1):3108. doi:10.1038/s41467-024-47329-x. [Google Scholar] [PubMed] [CrossRef]
4. Guo S, Ji Y, Zheng Y, Watkins CB, Ma L, Wang Q, et al. Transcriptomic, metabolomic, and ATAC-seq analysis reveal the regulatory mechanism of senescence of post-harvest tomato fruit. Front Plant Sci. 2023;14:1142913. doi:10.3389/fpls.2023.1142913. [Google Scholar] [PubMed] [CrossRef]
5. Mangeon A, Lin WC, Springer PS. Functional divergence in the Arabidopsis LOB-domain gene family. Plant Signal Behav. 2012;7(12):1544–7. doi:10.4161/psb.22320. [Google Scholar] [PubMed] [CrossRef]
6. Xu M, Liu CL, Luo J, Qi Z, Yan Z, Fu Y, et al. Transcriptomic de novo analysis of pitaya (Hylocereus polyrhizus) canker disease caused by Neoscytalidium dimidiatum. BMC Genomics. 2019;20(1):10. doi:10.1186/s12864-018-5343-0. [Google Scholar] [PubMed] [CrossRef]
7. Derelli Tufekci E. Genome-wide identification and analysis of Lateral Organ Boundaries Domain (LBD) transcription factor gene family in melon (Cucumis melo L.). PeerJ. 2023;11:e16020. doi:10.7717/peerj.16020. [Google Scholar] [PubMed] [CrossRef]
8. Rong M, Gao SX, Wen D, Xu YH, Wei JH. The LOB domain protein, a novel transcription factor with multiple functions: a review. Plant Physiol Biochem. 2024;214:108922. doi:10.1016/j.plaphy.2024.108922. [Google Scholar] [PubMed] [CrossRef]
9. Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009;58(3):525–37. doi:10.1111/j.1365-313X.2009.03797.x. [Google Scholar] [PubMed] [CrossRef]
10. Zhao D, Chen P, Chen Z, Zhang L, Wang Y, Xu L. Genome-wide analysis of the LBD family in rice: gene functions, structure and evolution. Comput Biol Med. 2023;153:106452. doi:10.1016/j.compbiomed.2022.106452. [Google Scholar] [PubMed] [CrossRef]
11. Dong L, Manghwar H. Genome-wide expression analysis of LBD genes in tomato (Solanum lycopersicum L.) under different light conditions. Plant Signal Behav. 2023;18(1):2290414. doi:10.1080/15592324.2023.2290414. [Google Scholar] [PubMed] [CrossRef]
12. Wang Z, Zhang R, Cheng Y, Lei P, Song W, Zheng W, et al. Genome-wide identification, evolution, and expression analysis of LBD transcription factor family in bread wheat (Triticum aestivum L.). Front Plant Sci. 2021;12:721253. doi:10.3389/fpls.2021.721253. [Google Scholar] [PubMed] [CrossRef]
13. Wu M, He W, Wang L, Zhang X, Wang K, Xiang Y. PheLBD29, an LBD transcription factor from Moso bamboo, causes leaf curvature and enhances tolerance to drought stress in transgenic Arabidopsis. J Plant Physiol. 2023;280:153865. doi:10.1016/j.jplph.2022.153865. [Google Scholar] [PubMed] [CrossRef]
14. Li X, Zhang Y, Wu Y, Li B, Sun J, Gu S, et al. Lipid metabolism regulated by superoxide scavenger trypsin in Hylocereus undatus through multi-omics analyses. J Food Biochem. 2022;46(7):e14144. doi:10.1111/jfbc.14144. [Google Scholar] [PubMed] [CrossRef]
15. Li HH, Liu X, An JP, Hao YJ, Wang XF, You CX. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue and Organ Culture. 2017;130(1):47–59. [Google Scholar]
16. Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21(11):3567–84. doi:10.1105/tpc.109.067041. [Google Scholar] [PubMed] [CrossRef]
17. Albinsky D, Kusano M, Higuchi M, Hayashi N, Kobayashi M, Fukushima A, et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol Plant. 2010;3(1):125–42. doi:10.1093/mp/ssp069. [Google Scholar] [PubMed] [CrossRef]
18. Fan QJ, Yan FX, Qiao G, Zhang BX, Wen XP. Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene. 2014;533(1):322–31. doi:10.1016/j.gene.2013.08.098. [Google Scholar] [PubMed] [CrossRef]
19. Pang X, Li X, Liu X, Cai L, Li B, Li X. Transcriptomic analysis reveals Cu/Zn SODs acting as Hub genes of SODs in Hylocereus undatus induced by trypsin during storage. Antioxidants. 2020;9(2):162. doi:10.3390/antiox9020162. [Google Scholar] [PubMed] [CrossRef]
20. Li X, Liu X, Yin Y, Yu H, Zhang M, Jing H, et al. Transcriptomic analysis reveals key genes related to antioxidant mechanisms of Hylocereus undatus quality improvement by trypsin during storage. Food Funct. 2019;10(12):8116–28. doi:10.1039/c9fo00809h. [Google Scholar] [PubMed] [CrossRef]
21. Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: sIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(W1):W597–603. doi:10.1093/nar/gks400. [Google Scholar] [PubMed] [CrossRef]
22. Qian Y, Tong J, Liu N, Wang B, Ji Y, Wu Z. Effect of light on ascorbic acid biosynthesis and bioinformatics analysis of related genes in Chinese chives. PLoS One. 2024;19(8):e0307527. doi:10.1371/journal.pone.0307527. [Google Scholar] [PubMed] [CrossRef]
23. Zhang Y, Li B, Zhang M, Jia J, Sun S, Chen X, et al. Transcriptome analyses and virus-induced gene silencing identify HuWRKY40 acting as a hub transcription factor in the preservation of Hylocereus undatus by trypsin. J Food Biochem. 2022;46(12):e14437. doi:10.1111/jfbc.14437. [Google Scholar] [PubMed] [CrossRef]
24. Zhang Y, Niu N, Li S, Liu Y, Xue C, Wang H, et al. Virus-induced gene silencing (VIGS) in Chinese Jujube. Plants. 2023;12(11):2115. doi:10.3390/plants12112115. [Google Scholar] [PubMed] [CrossRef]
25. Zhang J, Wu Z, Hu F, Liu L, Huang X, Zhao J, et al. Aberrant seed development in Litchi Chinensis is associated with the impaired expression of cell wall invertase genes. Hortic Res. 2018;5:39. doi:10.1038/s41438-018-0042-1. [Google Scholar] [PubMed] [CrossRef]
26. Ghasemzadeh A, Jaafar HZ, Rahmat A. Variation of the phytochemical constituents and antioxidant activities of Zingiber officinale var. rubrum theilade associated with different drying methods and polyphenol oxidase activity. Molecules. 2016;21(6):780. doi:10.3390/molecules21060780. [Google Scholar] [PubMed] [CrossRef]
27. Li S, Zhang Z, Zhang J, Zheng X, Zhang H, Zhang H, et al. Using mathematical models to study the influences of different ratios of chemical nitrogen, phosphorus, and potassium on the content of soluble protein, vitamin C, and soluble sugar in Melon. Int J Environ Res Public Health. 2022;20(1):283. doi:10.3390/ijerph20010283. [Google Scholar] [PubMed] [CrossRef]
28. Park A, Harris LK. Gene expression meta-analysis reveals interferon-induced genes associated with SARS infection in Lungs. Front Immunol. 2021;12:694355. doi:10.3389/fimmu.2021.694355. [Google Scholar] [PubMed] [CrossRef]
29. Candar-Cakir B, Arican E, Zhang B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J. 2016;14(8):1727–46. doi:10.1111/pbi.12533. [Google Scholar] [PubMed] [CrossRef]
30. Nie Q, Gao GL, Fan QJ, Qiao G, Wen XP, Liu T, et al. Isolation and characterization of a catalase gene HuCAT3 from pitaya (Hylocereus undatus) and its expression under abiotic stress. Gene. 2015;563(1):63–71. doi:10.1016/j.gene.2015.03.007. [Google Scholar] [PubMed] [CrossRef]
31. Liu J, Gao Y, Gong F, Hou F, Zhang Z, Cheng X, et al. The transcriptome and metabolome reveal stress responses in sulfur-fumigated cucumber (Cucumis sativus L.). Front Plant Sci. 2021;12:778956. doi:10.3389/fpls.2021.778956. [Google Scholar] [PubMed] [CrossRef]
32. Grimplet J, Pimentel D, Agudelo-Romero P, Martinez-Zapater JM, Fortes AM. The LATERAL ORGAN BOUNDARIES Domain gene family in grapevine: genome-wide characterization and expression analyses during developmental processes and stress responses. Sci Rep. 2017;7(1):15968. doi:10.1038/s41598-017-16240-5. [Google Scholar] [PubMed] [CrossRef]
33. Li C, Wang S, Wang J, Wu Z, Xu Y, Wu Z. Ozone treatment promotes physicochemical properties and antioxidant capacity of fresh-cut red pitaya based on phenolic metabolism. Front Nutr. 2022;9:1016607. doi:10.3389/fnut.2022.1016607. [Google Scholar] [PubMed] [CrossRef]
34. Jadhav PB. Extending the storage and Post-Storage life of dragon fruit using a cold room (Ecofrost). Int J Agric Environ Biotechnol. 2018;11(3):573–7. doi:10.30954/0974-1712.06.2018.19. [Google Scholar] [CrossRef]
35. Shi Y, Vrebalov J, Zheng H, Xu Y, Yin X, Liu W, et al. A tomato LATERAL ORGAN BOUNDARIES transcription factor, SlLOB1, predominantly regulates cell wall and softening components of ripening. Proc Natl Acad Sci U S A. 2021;118(33):e2102486118. doi:10.1073/pnas.2102486118. [Google Scholar] [PubMed] [CrossRef]
36. Neves LC, Tosin JM, Benedette RM, Cisneros-Zevallos L. Post-harvest nutraceutical behaviour during ripening and senescence of 8 highly perishable fruit species from the Northern Brazilian Amazon region. Food Chem. 2015;174(4):188–96. doi:10.1016/j.foodchem.2014.10.111. [Google Scholar] [PubMed] [CrossRef]
37. Dave RK, Ramana Rao TV, Nandane AS. Improvement of post-harvest quality of pear fruit with optimized composite edible coating formulations. J Food Sci Technol. 2017;54(12):3917–27. doi:10.1007/s13197-017-2850-y. [Google Scholar] [PubMed] [CrossRef]
38. Rashid F, Ahmed Z, Ferheen I, Mehmood T, Liaqat S, Ghoneim MM, et al. Effect of fenugreek and flaxseed polysaccharide-based edible coatings on the quality attributes and shelf life of apple fruit during storage. Food Sci Nutr. 2024;12(3):2093–103. doi:10.1002/fsn3.3909. [Google Scholar] [PubMed] [CrossRef]
39. Wang N, Qu C, Jiang S, Chen Z, Xu H, Fang H, et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018;96(1):39–55. doi:10.1111/tpj.14013. [Google Scholar] [PubMed] [CrossRef]
40. Yu EA, Kim GS, Lee JE, Park S, Yi S, Lee SJ, et al. Flavonoid profiles of immature and mature fruit tissues of Citrus grandis Osbeck (Dangyuja) and overall contribution to the antioxidant effect. Biomed Chromatogr. 2015;29(7):1131. doi:10.1002/bmc.3496. [Google Scholar] [PubMed] [CrossRef]
41. Liang D, Shen Y, Ni Z, Wang Q, Lei Z, Xu N, et al. Exogenous melatonin application delays senescence of Kiwifruit leaves by regulating the antioxidant capacity and biosynthesis of flavonoids. Front Plant Sci. 2018;9:426. doi:10.3389/fpls.2018.00426. [Google Scholar] [PubMed] [CrossRef]
42. Wang J, Tian P, Sun J, Li B, Jia J, Yuan J, et al. CsMYC2 is involved in the regulation of phenylpropanoid biosynthesis induced by trypsin in cucumber (Cucumis sativus) during storage. Plant Physiol Biochem. 2023;196:65–74. doi:10.1016/j.plaphy.2023.01.041. [Google Scholar] [PubMed] [CrossRef]
43. Qi X, Liu C, Song L, Li M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 2020;301:110634. doi:10.1016/j.plantsci.2020.110634. [Google Scholar] [PubMed] [CrossRef]
44. Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013;73(2):212–24. doi:10.1111/tpj.12013. [Google Scholar] [PubMed] [CrossRef]
45. Ba LJ, Shan W, Kuang JF, Feng BH, Xiao YY, Lu WJ, et al. The Banana MaLBD (LATERAL ORGAN BOUNDARIES DOMAIN) transcription factors regulate EXPANSIN expression and are involved in fruit ripening. Plant Mol Biol Rep. 2014;32(6):1103–13. doi:10.1007/s11105-014-0720-6. [Google Scholar] [CrossRef]
46. Hu Y, Zhang J, Jia H, Sosso D, Li T, Frommer WB, et al. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci U S A. 2014;111(4):E521–9. doi:10.1073/pnas.1313271111. [Google Scholar] [PubMed] [CrossRef]
47. Zhang X, He Y, He W, Su H, Wang Y, Hong G, et al. Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci Rep. 2019;9(1):15651. doi:10.1038/s41598-019-52027-6. [Google Scholar] [PubMed] [CrossRef]
48. Wang J, Liu Y, Song X. Overexpression analysis of PtrLBD41 suggests its involvement in salt tolerance and flavonoid pathway in Populus trichocarpa. Int J Mol Sci. 2024;25(22):12349. doi:10.3390/ijms252212349. [Google Scholar] [PubMed] [CrossRef]
49. Yuan Y, Zhang J, Liu X, Meng M, Wang J, Lin J. Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics. 2020;112(2):1781–94. doi:10.1016/j.ygeno.2019.10.010. [Google Scholar] [PubMed] [CrossRef]
50. Yi J, Derynck MR, Li X, Telmer P, Marsolais F, Dhaubhadel S. A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 2010;62(6):1019–34. doi:10.1111/j.1365-313X.2010.04214.x. [Google Scholar] [PubMed] [CrossRef]
51. Wang Z, Yu Q, Shen W, El Mohtar CA, Zhao X, Gmitter FG Jr. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 2018;18(1):189. doi:10.1186/s12870-018-1418-y. [Google Scholar] [PubMed] [CrossRef]
52. Li X, Tang Z, Pang X, Zhao C, Li X, Liu Y. Trypsin slows the aging of mice due to its novel superoxide scavenging activity. Appl Biochem Biotechnol. 2017;181(4):1549–60. doi:10.1007/s12010-016-2301-7. [Google Scholar] [PubMed] [CrossRef]
53. Sies H, Berndt C, Jones DP. Oxidative stress. Annu Rev Biochem. 2017;86:715–48. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools