 Open Access
Open Access
ARTICLE
Genome-Wide Identification and Expression Analysis of GS and GOGAT Gene Family in Pecan (Carya illinoinensis) under Different Nitrogen Forms
1 Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, 210037, China
2 College of Forestry, Nanjing Forestry University, Nanjing, 210037, China
* Corresponding Author: Fangren Peng. Email:
(This article belongs to the Special Issue: Recent Research Trends in Genetics, Genomics, and Physiology of Crop Plants)
Phyton-International Journal of Experimental Botany 2024, 93(9), 2349-2365. https://doi.org/10.32604/phyton.2024.056655
Received 27 July 2024; Accepted 27 August 2024; Issue published 30 September 2024
Abstract
Ammonium nitrogen (NH4+-N) is one of the main forms of nitrogen absorbed and utilized by plants, and mastering the regulatory mechanism of plant ammonium assimilation is a key way to improve the efficiency of plant nitrogen utilization. Glutamine synthetase (GS) and glutamate synthase (GOGAT), two key enzymes for ammonium assimilation, have rarely been studied in pecan. In this study, GS and GOGAT family members of pecan were identified and analyzed using bioinformatics methods. The results indicated that 6 GS and 4 GOGAT genes were identified. The cis-acting elements can be broadly categorized into light-responsive, hormone-responsive, and stress-responsive elements. The findings from the analysis of homologous evolution revealed that neither of the two gene families experienced tandem duplication events. Additionally, different ratios of ammonium to nitrate nitrogen were set to analyze the activities of GS and GOGAT enzymes and expression levels in pecan. The results demonstrate differences in the activities of GS and GOGAT enzymes and the gene expression levels in various tissues of pecan under different nitrogen form ratios. This study established a foundation for further mastering the molecular regulatory mechanism of nitrogen assimilation in pecan, and provided a theoretical basis for enhancing the ability of pecan to absorb and utilize nitrogen.Keywords
Nitrogen (N) is one of the three major elements in fertilizers, and N fertilizer plays a significant role in promoting crop yield. However, recent studies have found that the majority of N fertilizer is applied in the form of urea, which not only leads to significant nitrogen loss but also causes environmental pollution [1]. To reduce the impact of nitrogen fertilizer runoff on both plants and the environment, it is essential to adopt reasonable fertilization practices. Doing so can not only increase plant yields but also decrease environmental pollution. Plants primarily absorb and utilize nitrogen in the forms of Ammonium nitrogen (NH4+) and nitrate nitrogen (NO3−) [2], and they have varying preferences for these two forms of nitrogen. Research has shown that Pseudotsuga menziesii seedlings have a higher potential for absorbing NO3− than NH4+ [3], and pecans also tend to prefer NH4+ [2]. When inorganic N sources enter the higher plant, they must be converted into NH4+ and then transformed into organic N through N assimilation [4]. In this process, glutamine synthetase (GS) and glutamate synthase (GOGAT) play important roles, forming the “GS-GOGAT cycle” [5].
Pecan (Carya illinoensis (Wangenh.) K. Koch), a member of the genus Carya in the family Juglandaceae, is native to Eastern North America. Pecan kernels are rich in protein and have a high oil content, with 90% of it being unsaturated fatty acids [6]. The straight trunk of the pecan tree makes its wood suitable for furniture and landscaping [7]. The pecan contains various phytochemicals [8], such as phenolic compounds. Reports indicate that pecans contain phenolic compounds and other antioxidants, which have a significant effect on degenerative diseases [9]. Due to its nutritional value, economic value, and medicinal value, pecan is widely popular both domestically and internationally [10].
GS is tasked with the conversion of NH4+ absorbed by plants into organic N, playing a pivotal role in the nitrogen assimilation pathway of plants [11]. Studies have also explored the relationship between GS and stresses such as drought [12]. Currently, research on members of the GS family has been conducted in a variety of plants, such as Arabidopsis thaliana [13], wheat (Triticum aestivum L.) [14], rice (Oryza sativa L.) [15], etc. In most plants, GS exists in two forms, including cytoplasmic GS1 and chloroplasts GS2 [16]. The cytosolic GS1 is more abundant in the companion cells of vascular tissues in plant leaves, especially in aging leaves, and it participates in the activation of N during the aging period of plant leaves, which is most significant in small grain crops [16]. GS2 is primarily involved in the assimilation of N during photorespiration and the reduction of nitrate [17]. The GS gene is regulated at various levels, including genetic level and transcription factors [18]. For example, the NIN-Like Protein7 (NLP7) transcription factor can induce the expression of the GS2 gene [19].
GOGAT is the rate-limiting enzyme in the “GS-GOGAT cycle” [20]. GS catalyzes the conversion of NH4+ into glutamate, which is then further converted into glutamate again under the action of GOGAT [21]. In higher plants, GOGAT mainly includes two types: Ferredoxin-Dependent Glutamate Synthase (Fd-GOGAT) and Nicotinamide adenine dinucleotide-Dependent Glutamate Synthase (NADH-GOGAT). Different forms of the GOGAT gene are expressed in different plant tissues, and they also play different roles at various stages of plant growth and development [22]. Fd-GOGAT primarily functions in photosynthetic tissues, where it assimilates NH4+ produced by photorespiration. In contrast, NADH-GOGAT plays an important role in non-photosynthetic ammonia assimilation. There have been few studies on GOGAT in woody plants, but Cao et al. [23] for were the first to use bioinformatics methods study the GOGAT in poplar (Populus trichocarpa Torr. & Gray). They analyzed the response of GOGAT to carbon-nitrogen (C-N) treatment, which provides important clues for exploring the mechanism of regulating C-N balance in poplar.
During the N assimilation in process plants, GS and GOGAT play crucial roles. The gene families of GS and GOGAT have been deeply studied in many plants, but their functional research in pecan is relatively limited. This study identified 6 members of the GS gene family and 4 members of the GOGAT gene family in pecan and analyzed their physicochemical properties, gene structure, and gene duplication. This study also analyzed their expression patterns under different N forms. This research provides a solid theoretical foundation for addressing environmental pollution caused by N and improving the N utilization efficiency of pecans.
2.1 Plant Materials and Experimental Design
The experimental site is located at the Pecan Experimental Base of Nanjing Forestry University (30°15′50″N, 119°09′06″E). The experiment was conducted from 15 May, 2022, to 30 September, 2023. The 14-year-old ‘Pawnee’ pecan varieties in uniform growth conditions were selected as the experiment material. The experiment consisted of six treatments, CK (no nitrogen application), T1 (NH4+:NO3− = 0:100), T2 (NH4+:NO3− = 25:75), T3 (NH4+:NO3− = 50:50), T4 (NH4+:NO3− = 75:25), and T5 (NH4+:NO3− = 100:0). Six trees were selected for each treatment for a total of three biological replications. The fertilizers used in the experiment were NH4HCO3 (17.1% N content) and Ca(NO3)2 (11% N content) at an annual rate of 700 g N per plant. Add dicyandiamide (2% DCD-N) as a nitrification inhibitor at the same time as fertilization [24].
The fertilization experiment was carried out in three stages. The first fertilization conducted in mid-May represents 50% of the total annual fertilizer application. The early June application of fertilizer constituted 30% of the yearly total fertilizer usage. The third fertilization in mid to late June accounted for 20% of the total annual fertilizer application. During the second and third fertilizations, potassium sulfate (with 50% K2O) 150 g/plant and calcium superphosphate (with 12% P2O5) 625 g/plant were applied simultaneously. In the early morning of October, roots, leaves, and kernels were collected from the four cardinal directions of each tree. The collected experimental samples are placed in a laboratory refrigerator set at −80°C.
2.2 Identification of CiGS and CiGOGAT Family Members of Pecan
Firstly, all protein sequences for pecan were downloaded from the Phytozome database (Phytozome (doe.gov), accessed on 20 September 2023). Then, the hidden Markov model files for the structural domains Gln-synt_C (PF00120), Gln-synt_N (PF03951), GATase_2 (PF00310), Glu_synthase (PF01645), Glu_syn_centra (PF04898), GXGXG (PF01493), Pyr_redox_2 (PF07992), and Fer4_20 (PF14691) were downloaded from the Pfam database (Pfam is now hosted by InterPro (xfam.org), accessed on 20 September 2023) [25]. The hmmsearch program from the HMMER3.0 software suite was utilized to search through all protein sequences of pecan (E-Value < 0.001) and obtained candidate members of the gene families [26]. Next, the protein sequences of GS and GOGAT family members in Arabidopsis were downloaded from the TAIR database (TAIR—Home (arabidopsis.org), accessed on 20 September 2023), and then compared with the pecan sequence file (E-Value < 0.001). Afterward, the NCBI-CDD (Home—Conserved Domains—NCBI (nih.gov), accessed on 20 September 2023) was used to validate the protein sequences for their structural domains once again, ultimately determining the members of the gene family. Using the online tool ExPASy (SIB Swiss Institute of Bioinformatics | Expasy), accessed on 20 September 2023 the physicochemical properties of predicted GS and GOGAT proteins were forecast, including amino acid length (AA), molecular weights (MWs), instability index, and theoretical isoelectric points (pIs) [27]. Additionally, predictions regarding the subcellular localization of the GS and GOGAT proteins were made utilizing the Cell-PLoc 2.0 (Plant-mPLoc server (sjtu.edu.cn), accessed on 20 September 2023).
2.3 Phylogenetic Analysis of CiGS and CiGOGAT Genes
Downloaded the GS and GOGAT protein sequences from wheat, maize (Zea mays L.), poplar, walnut (Juglans regia L.), and rice in the Phytozome database (Phytozome v13 (doe.gov), accessed on 20 September 2023). Using Jones-Taylor-Thornton (JTT) model in MEGA-X software and the Neighbor-joining (NJ) method (bootstrap = 1000) to constructing phylogenetic relationships between wheat, maize, poplar, walnut, rice, Arabidopsis and pecan.
2.4 Conserved Domains and Conserved Motifs Analysis
Identification of conserved structural domains of GS and GOGAT in pecan was performed using Pfam. To understand the conserved motifs of GS and GOGAT protein, on online MEME (MEME—Submission form (meme-suite.org), accessed on 20 September 2023) website analysis was conducted, with the number of motifs set to 10. Then, TBtools was used to Visualize the conserved domains and motifs [28].
2.5 Analysis of Promoter Cis-Acting Elements
In order to predict cis-acting elements of pecan GS and GOGAT, the sequence is extracted from the 2.0 kb DNA upstream of the coding sequences (CDS) start site and submitted to be analyzed in the PlantCARE database (PlantCARE, a database of plant promoters and their cis-acting regulatory elements (ugent.be), accessed on 20 September 2023). Visualize the analysis results using TBtools [29].
2.6 Gene Duplication Analysis and Ka/Ks Value Calculation
Performed co-linearity analysis between pecan and walnut, and between pecan and Arabidopsis using the built-in plugin One Step MCScanX in TBtools [28]. To understand the evolutionary status of the GS and GOGAT genes, the ratio of non-synonymous (Ka) to synonymous substitution (Ks) rates was calculated [30].
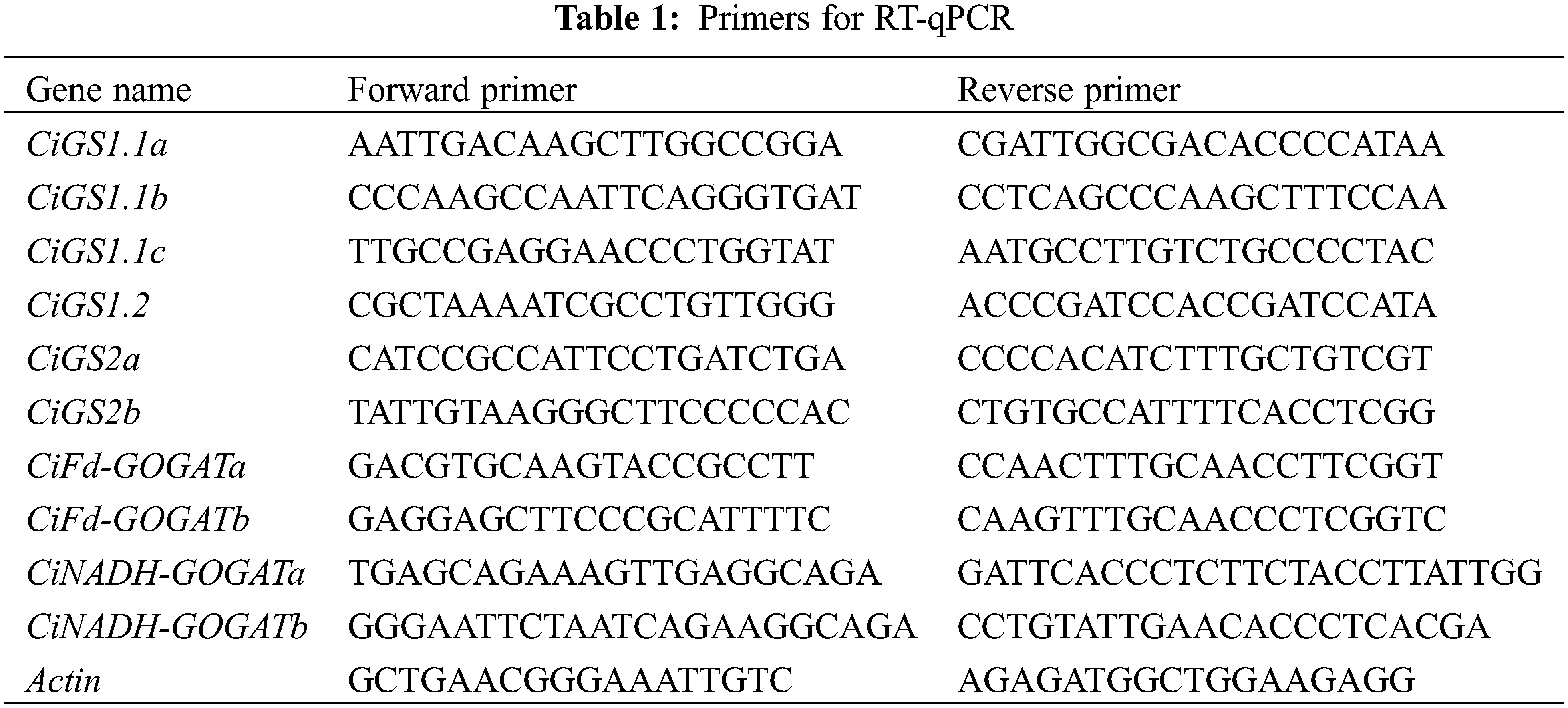
2.7 RNA Extraction and RT-qPCR Analysis
Total RNA was extracted from pecan leaves using a Plant Total RNA Extraction Kit (Vazyme, Nanjing, China), and the cDNA was generated using the reverse transcription PCR kit (TransGen, Beijing, China). The quantitative real-time PCR (RT-qPCR) was performed using Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). The specific sequences of the primers are detailed in the Table 1. The stably expressed pecan actin (CiPaw.03G124400) was used as an internal reference, and the RT-qPCR parameters were as follows: 95°C for 30 s, followed by 40 cycles of 5 s at 95°C and 15 s at 60°C. The relative expression levels of the GS and GOGAT genes in pecan were determined using the 2−∆∆Ct method [31].
2.8 Determination of CiGS and CiGOGAT Enzyme Activities
The enzyme activities of CiGS and CiGOGAT were measured using GS and GOGAT kits (Keming, Suzhou, China).
Performed an analysis on the activities of GS and GOGAT enzyme, as well as the relative expression levels of genes in pecan using one-way ANOVA. Significance was analyzed using the LSD method, with p < 0.05 indicating a significant difference. Performed statistical analysis the SPSS (Chicago, IL, USA, version 23.0) software. Created bar charts using GraphPad Prism 8 software.
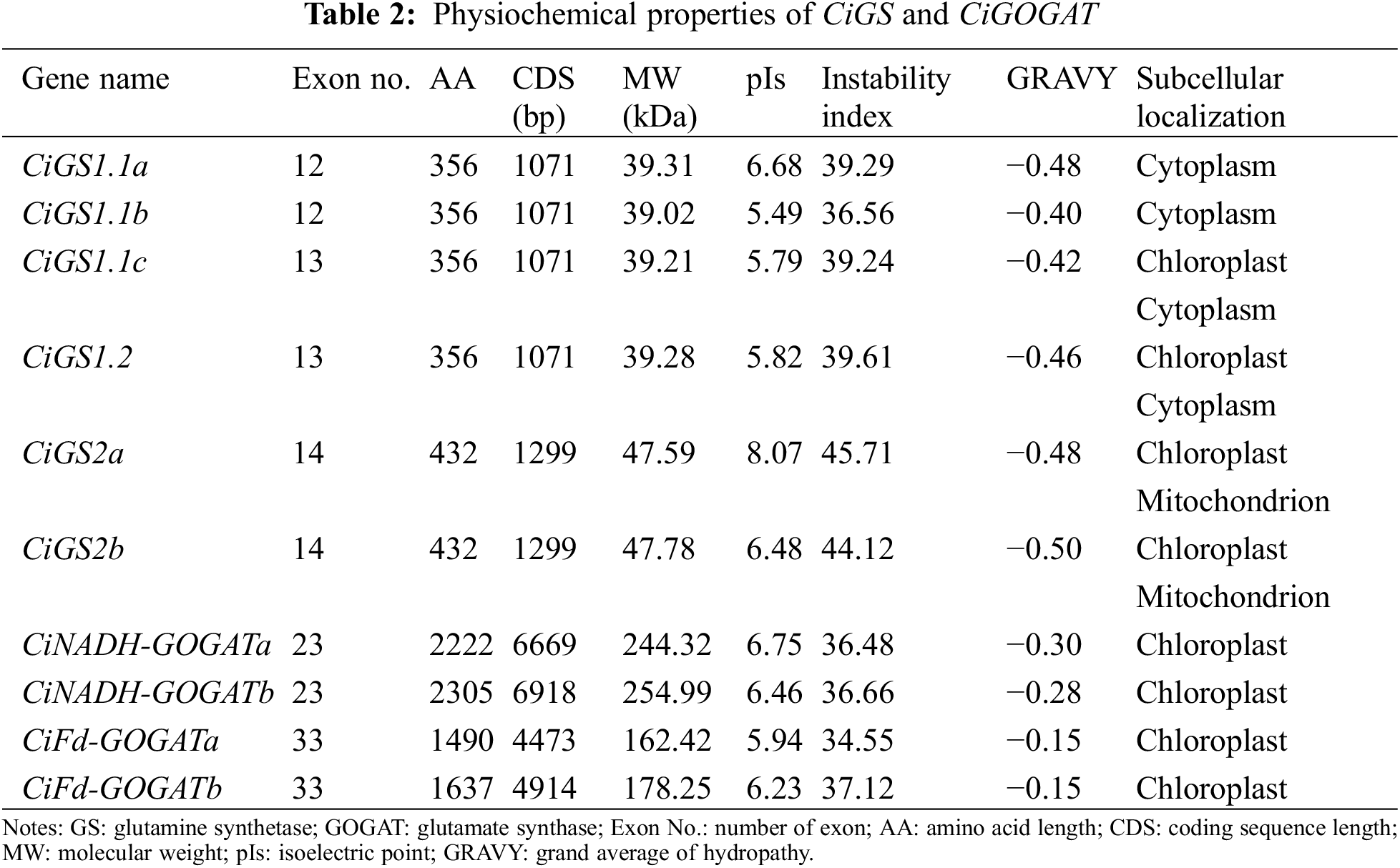
3.1 Identification and Analysis of CiGS and CiGOGAT Family Members
The identification and physicochemical properties analysis of the pecan GS and GOGAT family members (Table 2) indicated that CiGS genes had 12 to 14 exons, while CiGOGAT genes had 23 to 33 exons. The CDS length of CiGS genes varied between 1071 and 1299 bp, while the CDS length of the CiGOGAT genes ranged from 4473 to 6669 bp. The sequence of CiGS1s encompasses 356 amino acids, while that of CiGS2s spans 432 amino acids. The CiNADH-GOGATs feature a range of 2222 to 2305 amino acids, and the CiFd-GOGATs contain between 1490 and 1637 amino acids The pIs of the CiGSs ranged from 5.49 to 8.07, while the pIs of the CiGOGATs ranged from 5.94 to 6.75. Most CiGSs (4/6) and all CiGOGATs (4/4) were stable proteins (instability index <40). The GRAVY value being less than 0 indicates that both GS and GOGAT proteins are hydrophilic. CiGS1.1a and CiGS1.1b are predicted by subcellular localization analysis to be positioned within the cytoplasm, CiGS1.1c and CiGS1.2 were located in chloroplast and cytoplasm, CiGS2a and CiGS2b were located in mitochondrion and the cytoplasm, and all CiGOGATs are located in chloroplast.
3.2 Phylogenetic Analysis of CiGS and CiGOGAT
According to the phylogenetic tree (Figs. 1 and 2), the CiGS and CiGOGAT gene families were classified into three different evolutionary branches (GroupI-a ~GroupI-c). From the GS phylogenetic tree, it can be observed that the CiGS genes can be divided into two subfamilies: GS1 and GS2. The CiGS1 subfamily includes members CiGS1.1a, CiGS1.1b, CiGS1.1c, and CiGS1.2, whereas the CiGS2 subfamily encompasses CiGS2a and CiGS2b. The CiGS1 subfamily is located entirely in GroupI-a, and the CiGS2 subfamily is located entirely in GroupI-c. The phylogenetic tree of GOGAT shows that the GOGAT family can be divided into two subfamilies: NADH-GOGAT and the Fd-GOGAT, with each subfamily containing two GOGAT genes in pecan. CiNADH-GOGAT is located in GroupI-b, whereas CiFd-GOGAT is located in GroupI-c.
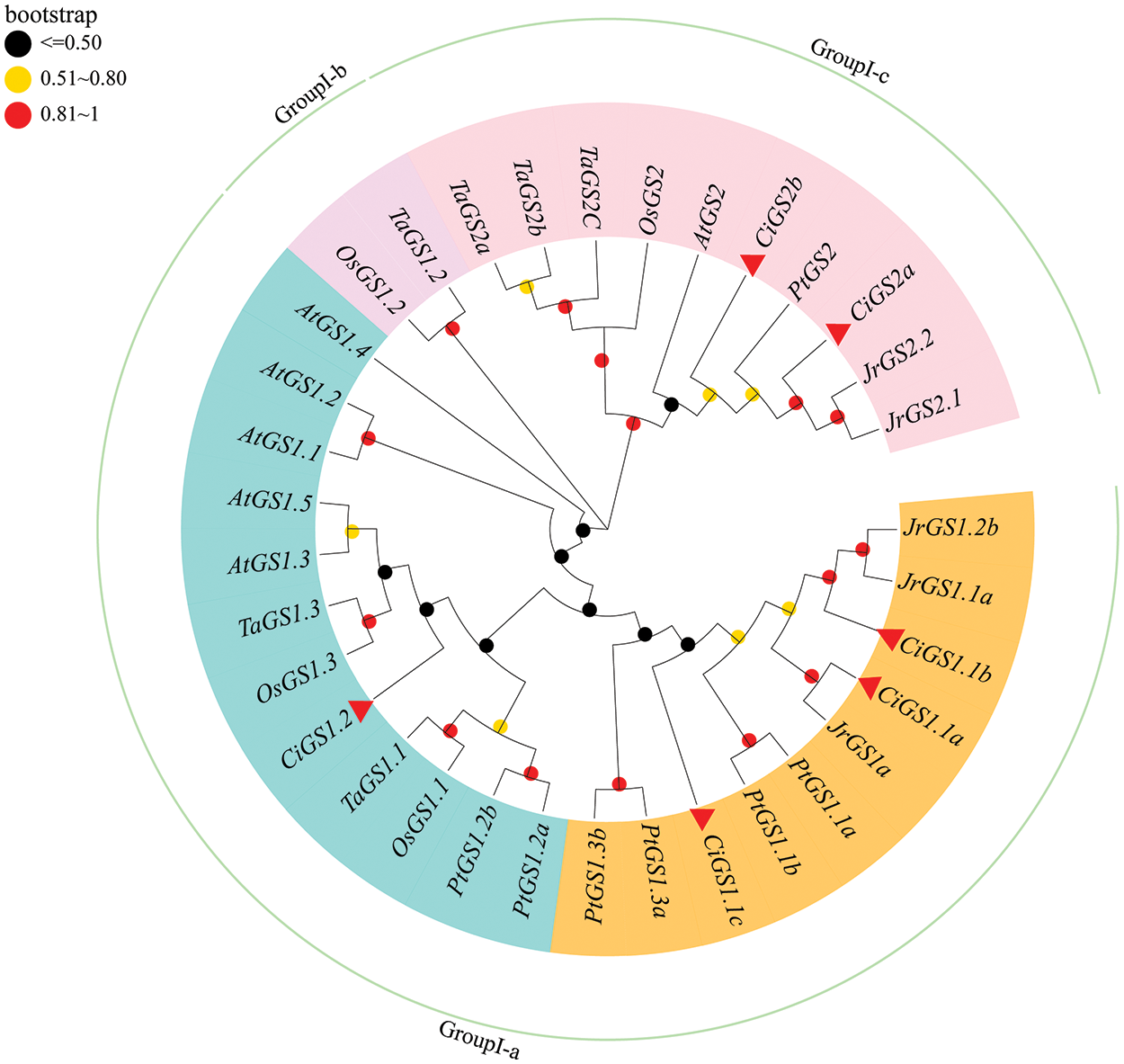
Figure 1: Phylogenetic tree of GS in C. illinoinensis, A. thaliana, P. trichocarpa, T. aestivum, Z. mays, O. sativa, and J. regia. Different annular sector colors represent different subfamilies. The range of Bootstrap values is displayed with circles of different colors. GS: glutamine synthetase
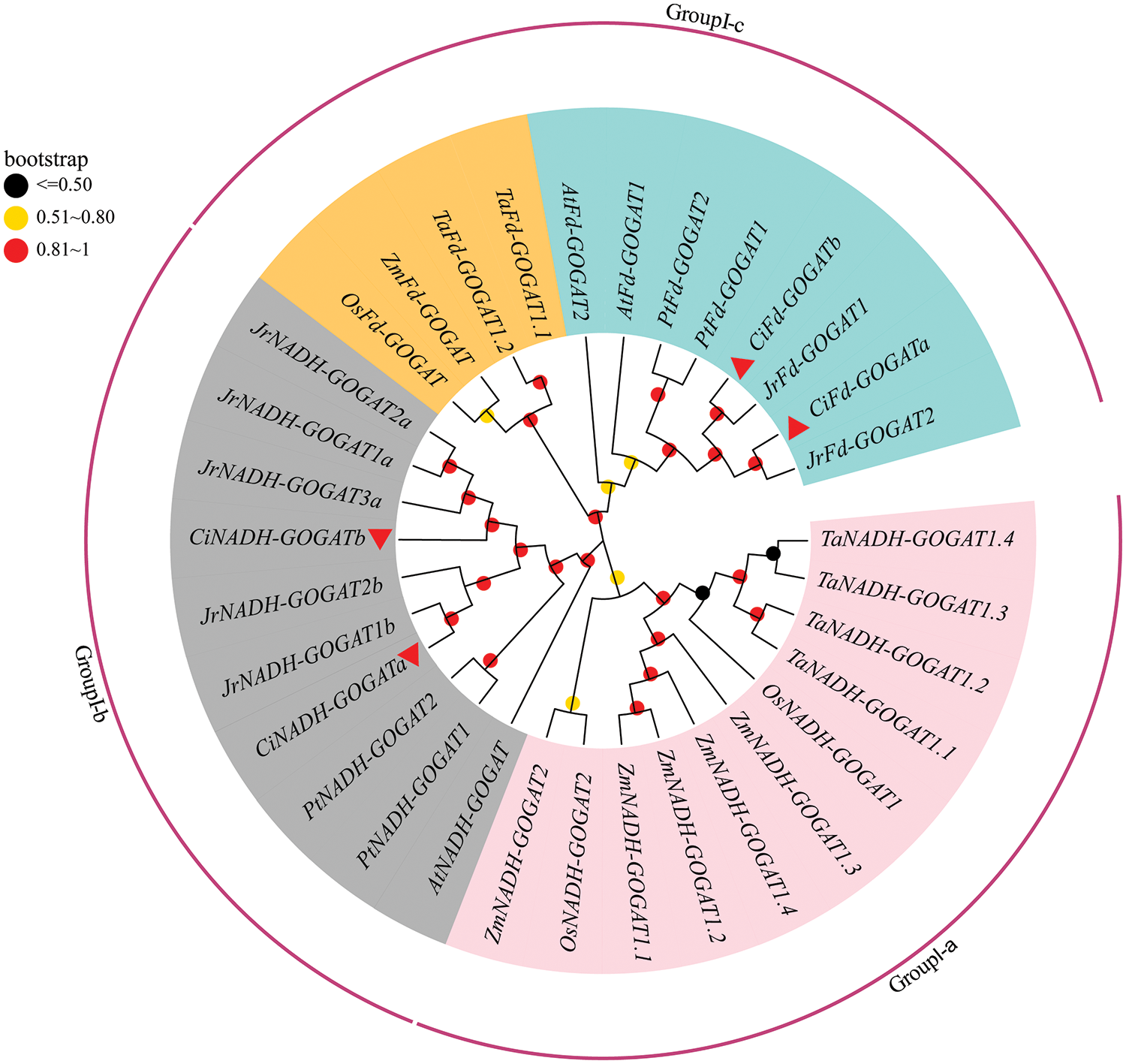
Figure 2: Phylogenetic tree of GOGAT in C. illinoinensis, A. thaliana, P. trichocarpa, T. aestivum, Z. mays, O. sativa, and J. regia. The range of Bootstrap values is displayed with circles of different colors. GOGAT: glutamate synthase
3.3 Conserved Motif and Conserved Domain Analysis of CiGS and CiGOGAT
The analysis of conserved motifs in GS and GOGAT in pecan (Figs. 3 and 4) has found that CiGS and CiGOGAT in pecan contain all the motifs, indicating that the protein functions of CiGS and CiGOGAT may have similarities in some aspects. The analysis of conserved structural domains revealed that the majority of CiGS1s (3/4) possessed the Gln-synt_C domain, and all CiGS2s contained the Gln-synt_N domain. The conserved structural domains of CiFd-GOGAT and CiNADH-GOGAT proteins were analysed and the results showed that CiFd-GOGAT has two less conserved structural domains than CiNADH-GOGAT, specifically Pyr_redox_2 and Fer4_20. This suggests that the protein function of CiNADH-GOGAT may be more complex than that of CiFd-GOGAT.
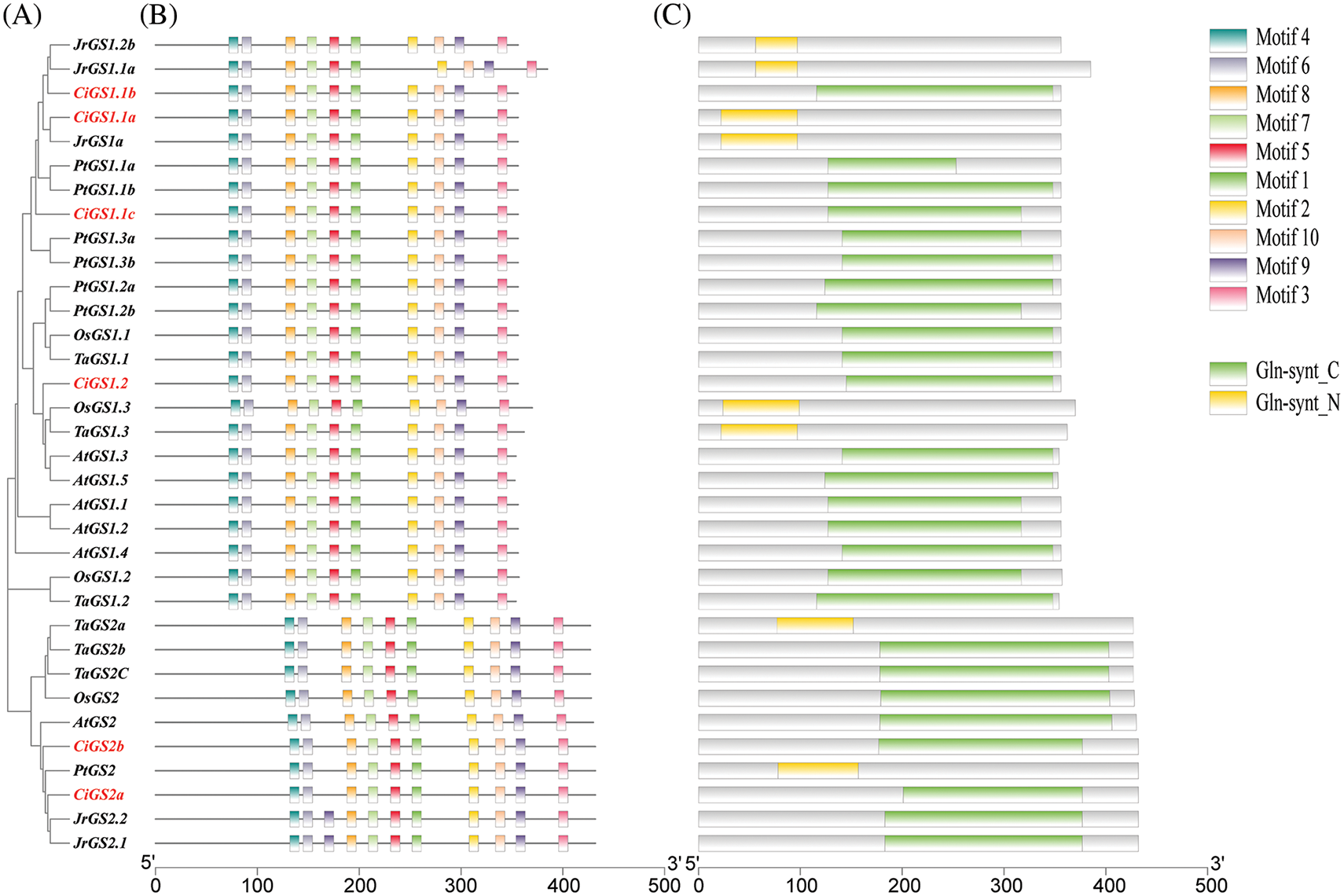
Figure 3: Conserved motif and conserved domain of GS. (A) Evolutionary tree. (B) Conserved motif. (C) Conserved domain. At: Arabidopsis thaliana, Jr: Juglans regia, Ci: Caray illinoensis, Pt: Populus trichocarpa, Os: Oryza sativa, Ta: Triticum aestivum
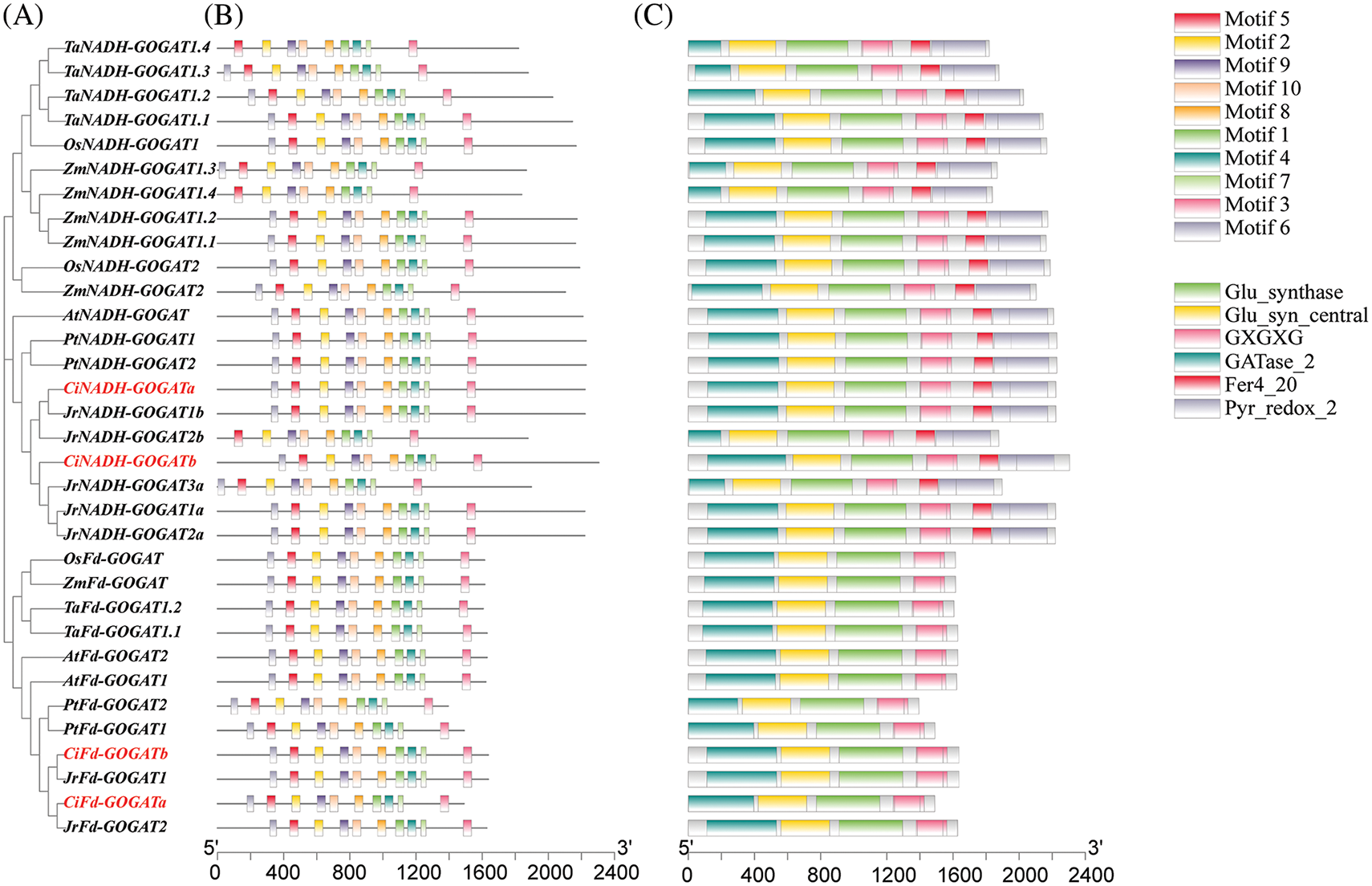
Figure 4: Conserved motif and conserved domain of GOGAT. (A) Evolutionary tree. (B) Conserved motif. (C) Conserved domain. At: Arabidopsis thaliana, Ta: Triticum aestivum, Jr: Juglans regia, Os: Oryza sativa, Ci: Caray illinoensis, Pt: Populus trichocarpa, Zm: Zea mays
3.4 Analysis of Cis-Acting Elements of CiGS and CiGOGAT
Selecting the 2 kb upstream promoter regions of CiGS and CiGOGAT genes in pecan (Fig. 5) for analysis revealed that the cis-acting elements in the promoter regions of CiGS and CiGOGAT genes mainly consist of light-responsive, hormone-responsive, and stress-responsive elements. The abundance of light-responsive elements suggests that CiGS and CiGOGAT genes are regulated by light exposure. The hormone-responsive elements in the promoter regions of CiGS and CiGOGAT genes encompass a range of triggers, including gibberellin and auxin response elements related to plant growth, as well as abscisic acid response elements related to stress response, and salicylic acid and jasmonic acid response elements related to pathogen defense. Additionally, the presence of stress-related functional elements such as low-temperature and defense stress-responsive elements indicates a significant role for CiGS and CiGOGAT genes in the plant’s stress response system. Studying the cis-acting elements for CiGS and CiGOGAT genes can reveal the significant roles these proteins play in various physiological processes of plants.
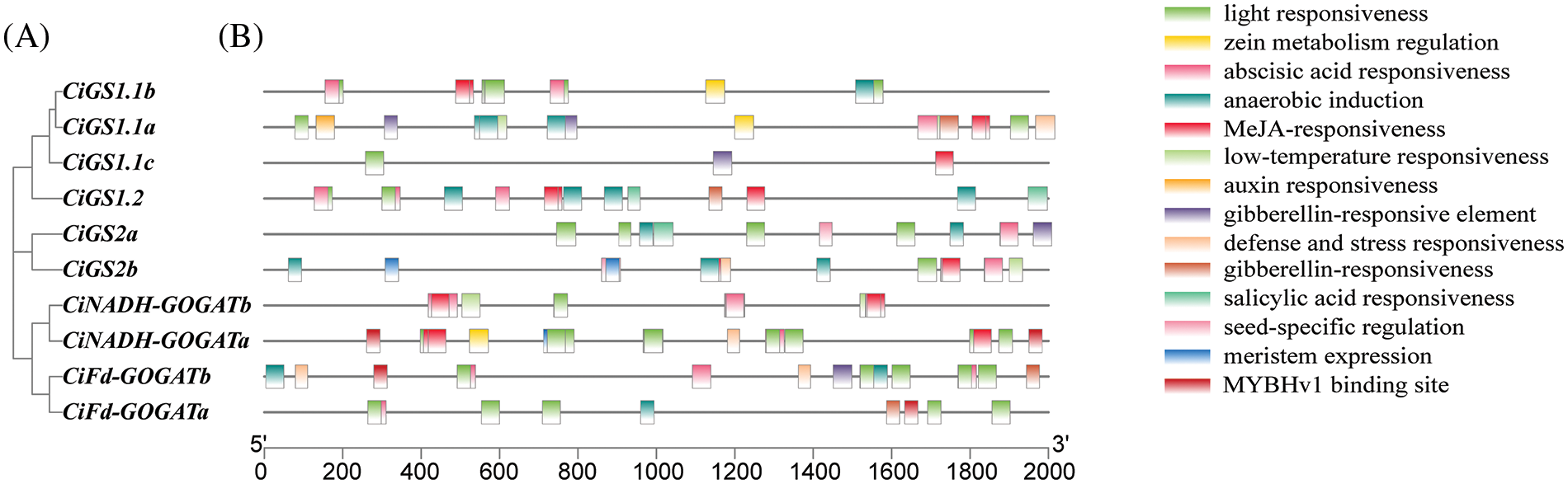
Figure 5: Distribution of cis-acting elements of CiGS and CiGOGAT genes family (A) Evolutionary tree. (B) Cis-acting elements of GS and GOGAT in pecan. Different colors indicate different Cis-regulatory element
3.5 Duplication Events and Syntenic Analysis of CiGS and CiGOGAT
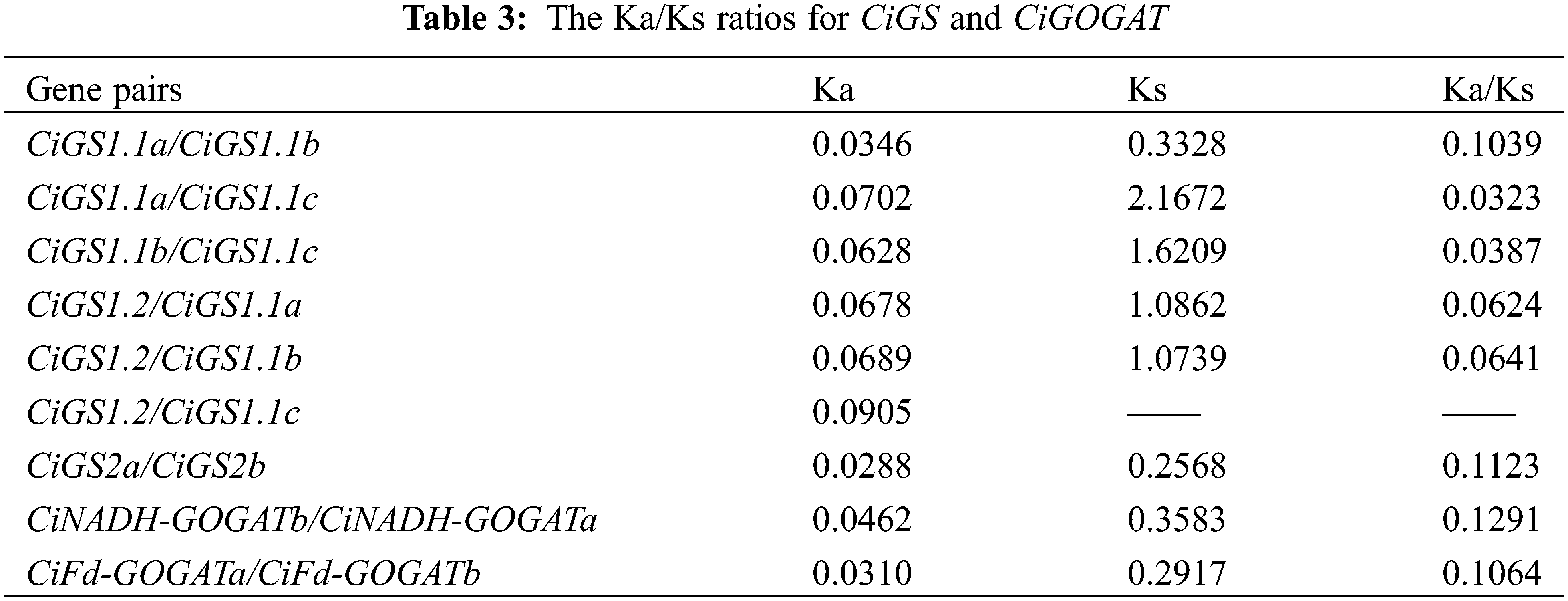
A genome-wide collinearity analysis of pecan, walnut, and Arabidopsis revealed that 6 CiGS genes from pecan are collinear with 14 JrGS and 10 AtGS genes. Additionally, 4 CiGOGAT genes from pecan show collinearity with 7 JrGS and 3 AtGS genes (Fig. 6). This result indicates that pecan and walnut have a relatively close phylogenetic relationship. The Ka/Ks result analysis (Table 3) indicates that the Ka/Ks ratio ranges from 0.03 to 0.13, suggesting that the members of the GS and GOGAT families have been subjected to relatively weak selective pressure, and that GS and GOGAT are under purifying selection.
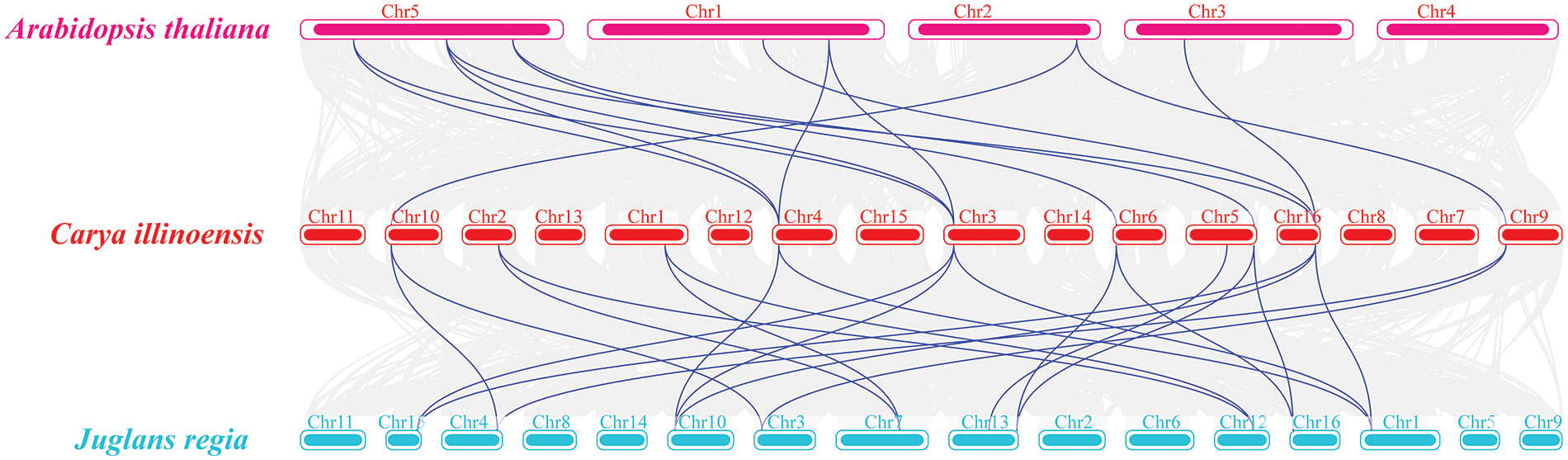
Figure 6: Pecan and walnut, as well as pecan and Arabidopsis, share collinear relationships among GS and GOGAT genes
3.6 Effects of N Forms on CiGS and CiGOGAT Enzyme Activities of Pecan
Analysis of CiGS and CiGOGAT enzyme activities in various tissues show that were under the T4 treatment significantly increased the GS enzyme activities in roots. The NADH-GOGAT enzyme activities in roots significantly increased under T2 treatment, while it significantly decreased under T3, T4, and T5 treatments. The Fd-GOGAT enzyme activities in roots did not differ significantly among treatments. In leaves the GS enzyme activities were significantly increased under T1, T3, and T4 treatments, while T2 and T5 treatments resulted in a significant decrease in GS enzyme activities. The enzyme activities of Fd-GOGAT significantly decreased under T2, T3, and T4 treatments, while the enzyme activities of GOGAT with NADH as the electron donor significantly increased under T2 and T4 treatments. T4 and T5 treatments significantly increased the GS enzyme activities in pecan kernels, while it was significantly decreased under T2 treatment. The Fd-GOGAT enzyme activities were significantly increased under T4 and T5 treatments, and the NADH-GOGAT enzyme activities were significantly decreased under T2 and T4 treatments (Fig. 7).
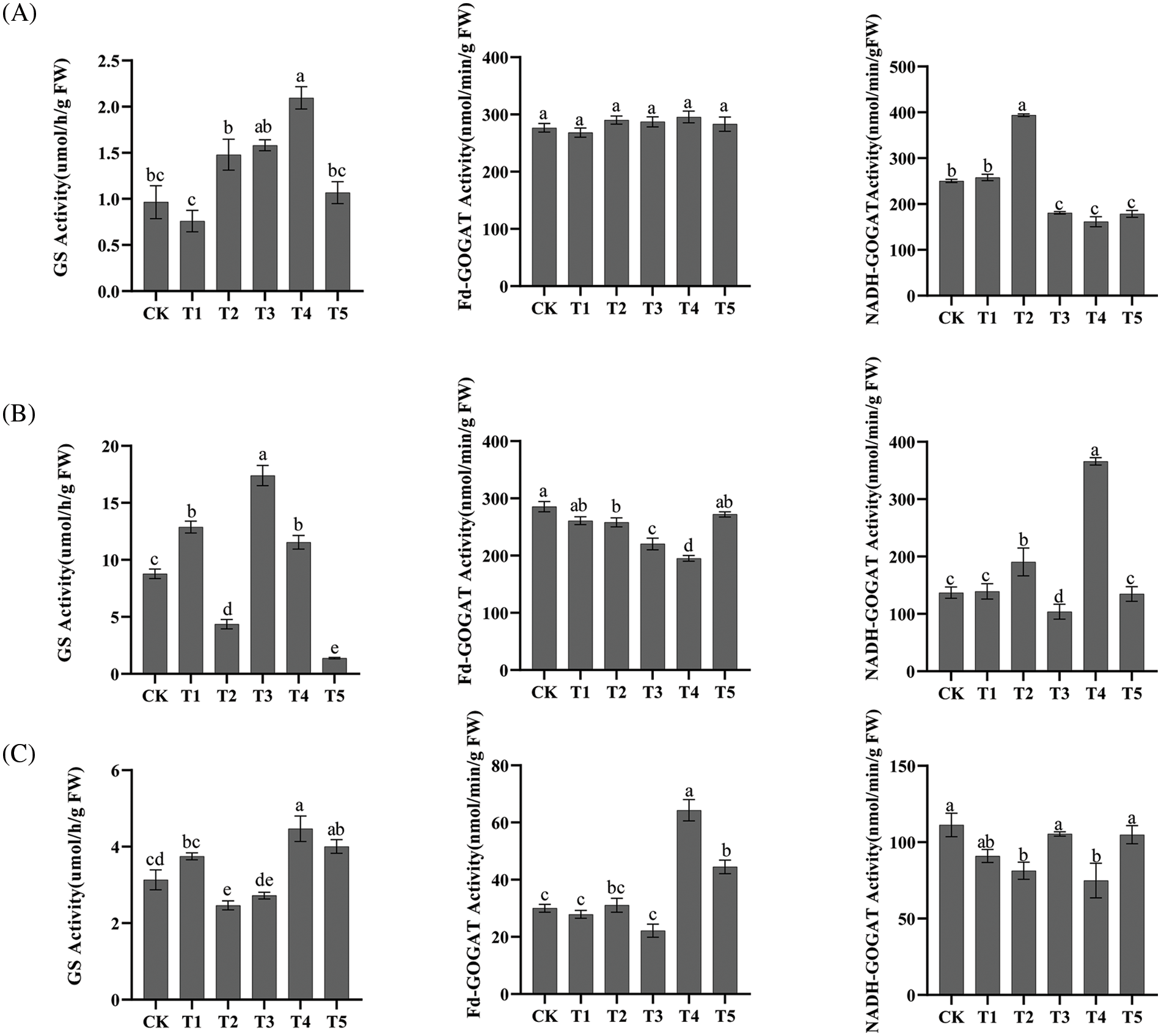
Figure 7: GS and GOGAT enzyme activities under NH4+:NO3− ratios in pecan. (A–C) Bar charts of CiGS and CiGOGAT enzyme activities in roots, leaves and kernels, respectively. Distinct lowercase letters signify statistically significant disparities among the groups (p < 0.05)
3.7 Effect of N Forms on CiGS and CiGOGAT Expression Levels
The expression levels of CiGS and CiGOGAT genes under N form treatment indicates that T1, T2, and T5 treatments significantly upregulated the expression levels of CiGS1.1a and CiNADH-GOGATa genes in roots. The expression levels of CiGS1.1b and CiGS2b genes were significantly upregulated under T2, T3, and T5 treatments, while the expression levels of CiGS1.1c, CiGS1.2, CiGS2a, and CiFd-GOGATb genes in roots were significantly upregulated under T2 and T5 treatments. Except for the T1 treatment, all other N form treatments significantly upregulated the expression levels of the genes CiFd-GOGATa and CiNADH-GOGATb genes in roots. In leaves, the gene CiGS1.1a exhibited significant increase in expression under T1, T3, and T4 treatments. A notable upregulation of CiGS1.1b and CiFd-GOGATb was observed exclusively under T4 treatment in the leaves. The CiGS1.1c gene showed significant upregulation across T2, T4, and T5 treatments. T2 and T4 treatments significantly upregulated the expression levels of CiGS1.2, CiGS2b, and CiFd-GOGATa genes, while the expression levels of CiGS2a gene was significantly downregulated under T3 treatment. CiNADH-GOGATa displayed significant upregulation under T4 and T5 treatments, and CiNADH-GOGATb showed a significant increase in expression under T3 treatment. In the kernels, the expression levels of the CiGS1.1a and CiGS1.1b genes were significantly downregulated in all treatments except for T2, and the expression level of the CiGS1.1c gene was significantly downregulated in all treatments except for T5. In the T4 treatment, the expression level of the CiGS1.2 gene in kernels was significantly upregulated, while T2, T3, and T5 treatments significantly downregulated it. The CiGS2a gene showed a significant upregulated expression under T1 and T2 treatments, but a significant downregulated under T4 treatment. Expression of CiGS2b gene was significantly upregulated under T1 treatment. Both CiFd-GOGATa and CiFd-GOGATb genes were significantly upregulated under T3 treatment. While T1, T2, and T4 treatments all significantly downregulated the expression levels of CiFd-GOGATb and CiNADH-GOGATa genes. The expression levels of CiNADH-GOGATb gene were significantly downregulated under all treatments (Fig. 8).
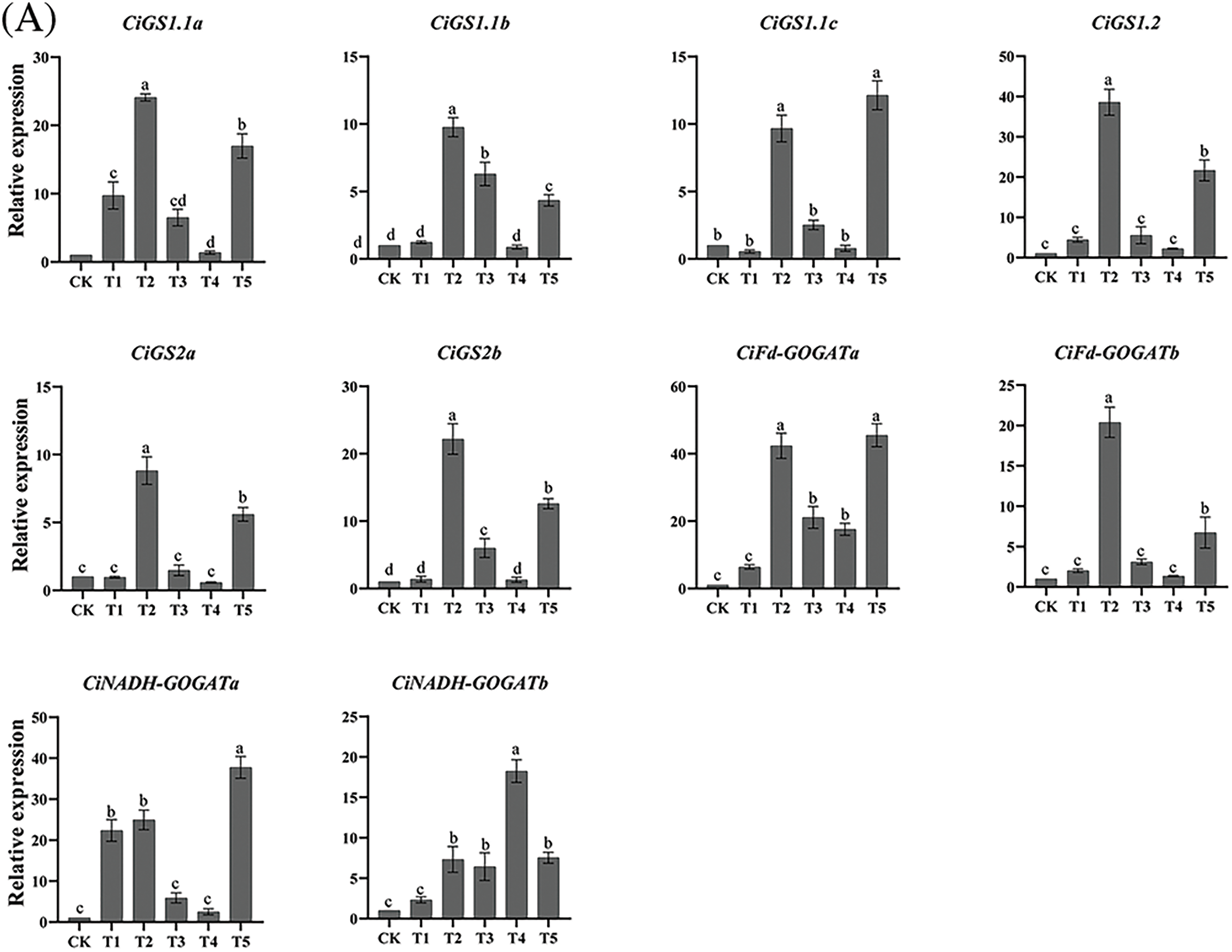
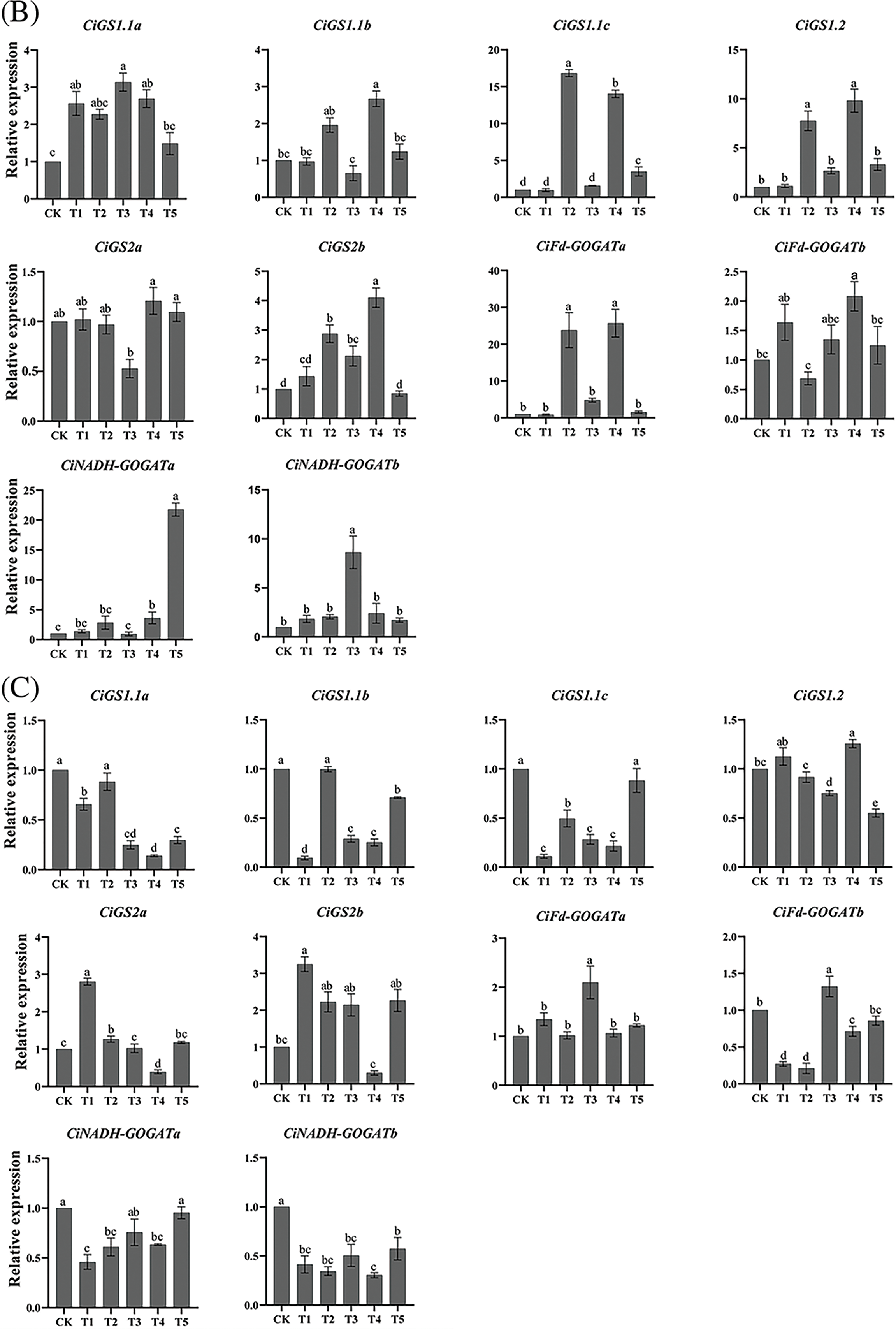
Figure 8: RT–qPCR analysis of pecan CiGS and CiGOGAT genes under varying NH4+:NO3− ratios. (A−C) Bar charts of CiGS and CiGOGAT genes expression in roots, leaves, and kernels, respectively. Distinct lowercase letters signify statistically significant disparities among the groups (p < 0.05)
Nitrogen serves as an essential nutrient for the growth and development of plants. The supply of nitrogen has a direct impact on the plant growth rate and developmental stages. However, the metabolism of nitrogen requires the participation of multiple enzymes, among which the GS/GOGAT cycle is crucial [32]. Cytoplasmic GS1 and chloroplast GS2 have different molecular weights, GS1 genes are 38–40 kDa and are encoded by three to five gene, GS2 genes are 44–45 kDa and its encoding is typically the responsibility of a single gene [13]. A similar identification result was obtained for the GS family members in pecan. Motifs are conserved sequences in protein and are important parts of protein structure. Studying conserved motifs to identify the conserved domains of unknown proteins can further analyze the characteristics and functions of proteins [33]. Some studies have indicated that the function of the conserved motifs of GS have a significant role in regulating the metabolism of NH4+ metabolism [34,35]. CiGS2 and CiNADH-GOGAT contain more structural domains and have a higher number of introns and exons, indicating that these two genes have complex structures [36], which ultimately result in their diverse biological functions. The promoter contains specific sequences that can control gene expression, and the analysis of promoter can help to elucidate the regulatory and responsive mechanisms of gene expression [37]. The results of the cis-acting elements analysis of the GS and GOGAT family members in pecan shows that the promoter sequences of these two gene families contain multiple cis-elements related to hormone and stress responses, indicating that GS and GOGAT in pecan are involved in hormone response and stress tolerance regulation. The same situation is also found in studies on poplar [23] and wheat [38]. The number of genes for GS and GOGAT varies among different species, indicating that they may have experienced different selective pressures [39]. The collinearity results indicate that the compared with Arabidopsis, pecan has a higher degree of gene homology with walnut, and there are more gene duplications, suggesting that the GS and GOGAT family members of pecan and walnut are more closely related, and function similarly in the process of nitrogen assimilation.
Numerous studies on GS and GOGAT isozymes have shown that GS2 and Fd-GOGAT are predominantly present in the chloroplasts, where they primarily assimilate NH4+ produced by photorespiration and ammonia generated from the reduction of nitrite. NADH-GOGAT is primarily expressed in non-photosynthetic tissues, and in non-leguminous plants it may be responsible for the reassimilation of NH4+ from amino acid catabolism [21]. Nitrogen forms have an important influence on nitrogen metabolism in plants, especially the ammonium assimilation process [40]. In the present study, it was discovered that nitrogen form is significantly related to the GS enzyme activities in pecan. Compared to the nitrogen-deficient treatment, GS enzyme activities in roots and kernels were significantly higher under T4 treatment. In the leaves of pecan, it was observed that GS enzyme activities were significantly higher under T3 treatment, but decreased under the all-ammonium treatment, possibly due to the NH4+ toxicity [41]. It was also found that leaves GS enzyme activities of pecans were significantly increased under full nitrate nitrogen treatment, which may be due to the reduction of nitrate absorbed by roots and transported to the leaves to produce ammonium ions [42], which increased the concentration of the GS enzyme substrate, and consequently, the GS enzyme activities. GS enzyme activities of different species respond differently to nitrogen forms. For example, GS enzyme activities of pepper were highest under NH4+:NO3− = 25:75 and NH4+:NO3− = 37.5:62.5 [43]. In studies on the GS enzyme activity in tomato roots, it has been found that the GS enzyme activities were enhanced under the treatment of NH4+:NO3− = 50:50 [35]. It has been shown that during ammonium assimilation by most plants, NADH-GOGAT enzyme in roots and Fd-GOGAT enzyme in leaves catalyze the formation of glutamate using NADH and Fd as electron donors, respectively [44], which in turn affects plant growth and development. The GOGAT enzyme activities in leaves and kernels had opposite trends under T4 treatment, where Fd-GOGAT enzyme activities were the lowest but NADH-GOGAT enzyme activities were highest in leaves, whereas Fd-GOGAT enzyme activities were the highest but NADH-GOGAT enzyme activities were the lowest in kernels, suggesting that the enzyme activities of the two forms of GOGAT may play complementary roles under this treatment [45]. This “GS-GOGAT cycle” is crucial in the process of ammonium assimilation in plants as GS enzyme is able to respond rapidly to nitrogen availability to provide substrate for GOGAT enzyme, and GOGAT enzyme in turn provides glutamate to GS enzyme [46].
The regulation of GS and GOGAT in plant ammonium assimilation is a complex process, and they are differentially expressed at different developmental periods and organs of the plant [47]. Isotope labeling technology can analyze the transport of a certain nutrient element within the plant body. Some studies have shown that application of 15N labelled nitrogen to the roots of peanut was found to be 60-65% taken up by the roots and 30%–35% was found in the seed [48]. In poplar, PtGS1.1 is preferentially expressed in leaves, PtGS1.2 is more abundantly expressed in the root system of young trees, and PtGS1.3 showed the highest expression in stems and petioles [49]. In the analysis of the expression levels of CiGSs and CiGOGATs, it was found that the expression levels in kernels were lower than in roots and leaves, which may be due to the fact that most of the nitrogen was assimilated in roots and leaves, and only a small amount of nitrogen was transferred to the kernels for assimilation, which in turn led to organizational differences in the expression of CiGSs and CiGOGATs. Compared to nitrogen-deficient treatments, the expression levels of GSs, Fd-GOGATs, and NADH-GOGATa in pecan roots significantly increased under T2 and all-ammonium treatments. In contrast, under all-nitrate treatment, except for CiGS1.1a and CiNADH-GOGATa, the expression levels of other GSs and GOGATs did not show significant changes, indicating that the expression of GSs and GOGATs in pecan roots is mainly induced by NH4+. In addition, in pecan leaves, the expression level of CiFd-GOGATa significantly increased under T2 and T4 treatments, while the expression level of CiFd-GOGATb gene only showed a significant increase under T4 treatment. This indicates that in pecan leaves, the ammonium-dominant mixed nitrogen was more conducive to promoting the expression level of CiFd-GOGAT genes. However, Loulakakis et al. [50] found that nitrate had a significant induction effect on the transcription level of Arabidopsis Fd-GOGAT, which might be related to species-specific preferences for inorganic nitrogen. In pecan kernels, CiGS2 genes exhibit higher expression levels compared to CiGS1 genes, indicating that CiGS2 genes are primarily responsible for regulating the process of ammonium assimilation within the kernels. Additionally, it was found that the CiGS2 genes were greatly affected by the all-nitrate treatment, indicating that NO3−may be involved in inducing the expression of the CiGS2 genes.
The “GS-GOGAT cycle” is the primary pathway for NH4+ assimilation, but its regulatory mechanism in pecan remains not fully understood. This study identified 6 GS and 4 GOGAT genes in pecan. Subsequent bioinformatics analysis along with the assessment of enzyme activities and gene expression levels under different nitrogen forms was conducted. Bioinformatics analysis can provide a better understanding of the characteristics, and functions of GS and GOGAT genes and proteins, and their roles in biological evolution. The expression of GS and GOGAT gene families in pecan is significantly affected by the ratio of nitrogen forms, and specific nitrogen source ratios can optimize the expression of these genes and the activity of related enzymes, which may enhance the plant’s ability to absorb and utilize nitrogen. These results provide an important molecular basis for further study of the nitrogen assimilation mechanism in pecan and offer potential strategies for improving the nitrogen utilization efficiency of pecan and other crops by adjusting the ratio of nitrogen fertilizer.
Acknowledgement: None.
Funding Statement: This study was financially supported by Jiangsu Provincial Forestry Bureau, Central Financial Forestry Science and Technology Promotion Demonstration Fund Project (Su [2022] TG04).
Author Contributions: Experimental design, Zhenbing Qiao, Mengyun Chen, and Fangren Peng; sample collection. Zhenbing Qiao, Wenjuan Ma and Mengyun Chen; data analysis and writing—original draft preparation, Zhenbing Qiao, Kaikai Zhu and Juan Zhao; writing—review and editing, Mengyun Chen, Jiaju Zhu and Pengpeng Tan; project administration and funding acquisition, Fangren Peng. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The bioinformatics data involved in this study are provided with specific access links in the Materials and Methods section, the enzyme activity and relative expression data in this study are personal research results that cannot be publicly shared, but data will be provided according to reasonable requirements.
Ethics Approval: Not appliable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Guha T, Gopal G, Mukherjee A, Kundu R. Fe3O4-urea nanocomposites as a novel nitrogen fertilizer for improving nutrient utilization efficiency and reducing environmental pollution. Environ Pollut. 2022;292:118301. doi:10.1016/j.envpol.2021.118301. [Google Scholar] [PubMed] [CrossRef]
2. Chen M, Zhu K, Tan P, Liu J, Xie J, Yao X, et al. Ammonia-nitrate mixture dominated by NH4+–N promoted growth, photosynthesis and nutrient accumulation in pecan (Carya illinoinensis). Forests. 2021;12(12):1808. doi:10.3390/f12121808. [Google Scholar] [CrossRef]
3. Boczulak SA, Hawkins BJ, Roy R. Temperature effects on nitrogen form uptake by seedling roots of three contrasting conifers. Tree Physiol. 2014;34(5):513–23. doi:10.1093/treephys/tpu028. [Google Scholar] [PubMed] [CrossRef]
4. De Carvalho Fernandes G, Turchetto-Zolet AC, Pereira Passaglia LM. Glutamine synthetase evolutionary history revisited: tracing back beyond the last universal common ancestor. Evolution. 2022;76(3):605–22. doi:10.1111/evo.v76.3. [Google Scholar] [CrossRef]
5. Bernard SM, Habash DZ. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009;182(3):608–20. doi:10.1111/nph.2009.182.issue-3. [Google Scholar] [CrossRef]
6. Clermont K, Graham CJ, Lloyd SW, Grimm CC, Randall JJ, Mattison CP. Proteomic analysis of pecan (Carya illinoinensis) nut development. Foods. 2023 Feb 17;12(4):866. doi:10.3390/foods12040866. [Google Scholar] [PubMed] [CrossRef]
7. Liu J, Tang Y, Bao J, Wang H, Peng F, Chen M, et al. Pecan plantation age influences the structures, ecological networks, and functions of soil microbial communities. Land Degrad Dev. 2022;33(16):3294–309. doi:10.1002/ldr.v33.16. [Google Scholar] [CrossRef]
8. Del C Reyes-Vázquez N, de la Rosa LA, Morales-Landa JL, García-Fajardo JA, García-Cruz MA. Phytochemical content and potential health applications of pecan [Carya illinoinensis (Wangenh) K. Koch] Nutshell. Curr Top Med Chem. 2022;22(2):150–67. doi:10.2174/1568026622666220105104355. [Google Scholar] [PubMed] [CrossRef]
9. Juhaimi FA, Özcan MM, Uslu N, Doğu S. Pecan walnut (Carya illinoinensis (Wangenh.) K. Koch) oil quality and phenolic compounds as affected by microwave and conventional roasting. J Food Sci Technol. 2017;54(13):4436–41. doi:10.1007/s13197-017-2921-0. [Google Scholar] [PubMed] [CrossRef]
10. Zhu K, Fan P, Liu H, Tan P, Ma W, Mo Z, et al. Insight into the CBL and CIPK gene families in pecan (Carya illinoinensis): identification, evolution and expression patterns in drought response. BMC Plant Biol. 2022;22(1):221. doi:10.1186/s12870-022-03601-0. [Google Scholar] [PubMed] [CrossRef]
11. Lebedev VG, Korobova AV, Shendel GV, Kudoyarova GR, Shestibratov KA. Effect of glutamine synthetase gene overexpression in birch (Betula pubescens) plants on auxin content and rooting in vitro. Dokl Biochem Biophys. 2018;480(1):143–5. doi:10.1134/S1607672918030043. [Google Scholar] [PubMed] [CrossRef]
12. Pascual L, Solé-Medina A, Faci I, Giraldo P, Ruiz M, Benavente E. Development and marker-trait relationships of functional markers for glutamine synthetase GS1 and GS2 homoeogenes in bread wheat. Mol Breeding. 2023;43(2):8. doi:10.1007/s11032-022-01354-0. [Google Scholar] [PubMed] [CrossRef]
13. Moreira E, Coimbra S, Melo P. Glutamine synthetase: an unlikely case of functional redundancy in Arabidopsis thaliana. Plant Biol J. 2022;24(5):713–20. doi:10.1111/plb.v24.5. [Google Scholar] [CrossRef]
14. Bernard SM, Møller ALB, Dionisio G, Kichey T, Jahn TP, Dubois F, et al. Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol Biol. 2008;67(1–2):89–105. [Google Scholar] [PubMed]
15. Fujita T, Beier MP, Tabuchi-Kobayashi M, Hayatsu Y, Nakamura H, Umetsu-Ohashi T, et al. Cytosolic glutamine synthetase GS1;3 is involved in rice grain ripening and germination. Front Plant Sci. 2022;13:835835. doi:10.3389/fpls.2022.835835. [Google Scholar] [PubMed] [CrossRef]
16. Swarbreck SM, Defoin-Platel M, Hindle M, Saqi M, Habash DZ. New perspectives on glutamine synthetase in grasses. J Exp Bot. 2011;62(4):1511–22. doi:10.1093/jxb/erq356. [Google Scholar] [PubMed] [CrossRef]
17. Nigro D, Blanco A, Anderson OD, Gadaleta A. Characterization of ferredoxin-dependent glutamine-oxoglutarate amidotransferase (Fd-GOGAT) genes and their relationship with grain protein content QTL in wheat. PLoS One. 2014;9(8):e103869. doi:10.1371/journal.pone.0103869. [Google Scholar] [PubMed] [CrossRef]
18. Peterman TK, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light-regulation and differential expression in leaves, roots and seeds. Mol Gen Genet. 1991;230(1–2):145–54. [Google Scholar] [PubMed]
19. Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4(1):1713. doi:10.1038/ncomms2650. [Google Scholar] [PubMed] [CrossRef]
20. Joy KW, Blackwell RD, Lea PJ. Assimilation of nitrogen in mutants lacking enzymes of the glutamate synthase cycle. J Exp Bot. 1992;43(2):139–45. doi:10.1093/jxb/43.2.139. [Google Scholar] [CrossRef]
21. García-Gutiérrez Á, Cánovas FM, Ávila C. Glutamate synthases from conifers: gene structure and phylogenetic studies. BMC Genomics. 2018;19(1):65. doi:10.1186/s12864-018-4454-y. [Google Scholar] [PubMed] [CrossRef]
22. Lu Y, Luo F, Yang M, Li X, Lian X. Suppression of glutamate synthase genes significantly affects carbon and nitrogen metabolism in rice (Oryza sativa L.). Sci China Life Sci. 2011;54(7):651–63. doi:10.1007/s11427-011-4191-9. [Google Scholar] [PubMed] [CrossRef]
23. Cao L, Xu C, Sun Y, Niu C, Leng X, Hao B, et al. Genome-wide identification of glutamate synthase gene family and expression patterns analysis in response to carbon and nitrogen treatment in Populus. Gene. 2023;851:146996. doi:10.1016/j.gene.2022.146996. [Google Scholar] [PubMed] [CrossRef]
24. Serna MD, Legaz F, Primo-Millo E. Improvement of the N fertilizer efficiency with dicyandiamide (DCD) in citrus trees. Fert Res. 1996;43(1–3):137–42. [Google Scholar]
25. El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–32. doi:10.1093/nar/gky995. [Google Scholar] [PubMed] [CrossRef]
26. Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi:10.1093/nar/gkr367. [Google Scholar] [PubMed] [CrossRef]
27. Wang Y, Fan Z, Zhai Y, Huang H, Vainstein A, Ma H. Polygalacturonase gene family analysis identifies FcPG12 as a key player in fig (Ficus carica L.) fruit softening. BMC Plant Biol. 2023;23(1):320. doi:10.1186/s12870-023-04315-7. [Google Scholar] [PubMed] [CrossRef]
28. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an Integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. doi:10.1016/j.molp.2020.06.009. [Google Scholar] [PubMed] [CrossRef]
29. Tao YT, Chen LX, Jin J, Du ZK, Li JM. Genome-wide identification and analysis of bZIP gene family reveal their roles during development and drought stress in Wheel Wingnut (Cyclocarya paliurus). BMC Genomics. 2022;23(1):743. doi:10.1186/s12864-022-08978-8. [Google Scholar] [PubMed] [CrossRef]
30. Chen M, Zhu K, Xie J, Liu J, Tan P, Peng F. Genome-wide identification and expression analysis of AMT and NRT gene family in pecan (Carya illinoinensis) seedlings revealed a preference for NH4+-N. Int J Mol Sci. 2022;23(21):13314. doi:10.3390/ijms232113314. [Google Scholar] [PubMed] [CrossRef]
31. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034.1. doi:10.1186/gb-2002-3-7-research0034. [Google Scholar] [PubMed] [CrossRef]
32. Banerjee S, Subramanian A, Chattopadhyay J, Rup Sarkar R. Exploring the role of GS-GOGAT cycle in microcystin synthesis and regulation–a model based analysis. Mol Biosyst. 2017;13(12):2603–14. doi:10.1039/C7MB00342K. [Google Scholar] [PubMed] [CrossRef]
33. Moons A. Transcriptional profiling of the PDR gene family in rice roots in response to plant growth regulators, redox perturbations and weak organic acid stresses. Planta. 2008;229(1):53–71. doi:10.1007/s00425-008-0810-5. [Google Scholar] [PubMed] [CrossRef]
34. Mondal R, Kumar A, Chattopadhyay SK. Structural property, molecular regulation, and functional diversity of glutamine synthetase in higher plants: a data-mining bioinformatics approach. Plant J. 2021;108(6):1565–84. doi:10.1111/tpj.v108.6. [Google Scholar] [CrossRef]
35. Liu G, Du Q, Li J. Interactive effects of nitrate-ammonium ratios and temperatures on growth, photosynthesis, and nitrogen metabolism of tomato seedlings. Sci Hortic. 2017;214:41–50. doi:10.1016/j.scienta.2016.09.006. [Google Scholar] [CrossRef]
36. Huang DL, Qin CX, Gui YY, Zhao LH, Chen ZL, Wang M, et al. Role of the SPS gene families in the regulation of sucrose accumulation in sugarcane. Sugar Tech. 2017;19(2):117–24. doi:10.1007/s12355-016-0454-x. [Google Scholar] [CrossRef]
37. Zhao Y, Cai M, Zhang X, Li Y, Zhang J, Zhao H, et al. Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS One. 2014;9(4):e94126. doi:10.1371/journal.pone.0094126. [Google Scholar] [PubMed] [CrossRef]
38. Parveen R, Vaish S, Gupta D, Basantani MK. Bioinformatics characterization of patatin-related phospholipase A (pPLA) gene family in agriculturally important crops viz Vigna radiata, Vigna angularis, and Glycine max. Biologia. 2022;77(5):1429–46. doi:10.1007/s11756-022-01026-6. [Google Scholar] [CrossRef]
39. Wang H, He T, Huang C, Wang K, Shi D, Si X, et al. Genome-wide identification of KCS gene family in Carya illinoinensis and their roles under abiotic stress conditions. Sci Hortic. 2023;321:112343. doi:10.1016/j.scienta.2023.112343. [Google Scholar] [CrossRef]
40. Coleto I, Marín-Peña AJ, Urbano-Gámez JA, González-Hernández AI, Shi W, Li G, et al. Interaction of ammonium nutrition with essential mineral cations. J Exp Bot. 2023;74(19):6131–44. doi:10.1093/jxb/erad215. [Google Scholar] [PubMed] [CrossRef]
41. Jia Z, Zhang J, Jiang W, Wei M, Zhao L, Li G. Different nitrogen concentrations affect strawberry seedlings nitrogen form preferences through nitrogen assimilation and metabolic pathways. Sci Hortic. 2024;332:113236. doi:10.1016/j.scienta.2024.113236. [Google Scholar] [CrossRef]
42. Fortunato S, Nigro D, Lasorella C, Marcotuli I, Gadaleta A, De Pinto MC. The role of glutamine synthetase (GS) and glutamate synthase (GOGAT) in the improvement of nitrogen use efficiency in cereals. Biomolecules. 2023;13(12):1771. doi:10.3390/biom13121771. [Google Scholar] [PubMed] [CrossRef]
43. Zhang J, Lv J, Dawuda MM, Xie J, Yu J, Li J, et al. Appropriate ammonium-nitrate ratio improves nutrient accumulation and fruit quality in pepper (Capsicum annuum L.). Agronomy. 2019;9(11):683. doi:10.3390/agronomy9110683. [Google Scholar] [CrossRef]
44. Zhang W, Ni K, Long L, Ruan J. Nitrogen transport and assimilation in tea plant (Camellia sinensis): a review. Front Plant Sci. 2023;14:1249202. doi:10.3389/fpls.2023.1249202. [Google Scholar] [PubMed] [CrossRef]
45. Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, et al. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 2006;140(2):444–56. doi:10.1104/pp.105.071910. [Google Scholar] [PubMed] [CrossRef]
46. Schulz AA, Collett HJ, Reid SJ. Nitrogen and carbon regulation of glutamine synthetase and glutamate synthase in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol Lett. 2001;205(2):361–7. doi:10.1111/fml.2001.205.issue-2. [Google Scholar] [CrossRef]
47. Stanford AC, Larsen K, Barker DG, Cullimore JV. Differential expression within the glutamine synthetase gene family of the model legume Medicago truncatula. Plant Physiol. 1993;103(1):73–81. doi:10.1104/pp.103.1.73. [Google Scholar] [PubMed] [CrossRef]
48. Inanaga S, Utunomiya M, Horiguchi T, Nishihara T. Behaviour of fertilizer-N absorbed through root and fruit in peanut. Plant Soil. 1990;122(1):85–9. doi:10.1007/BF02851913. [Google Scholar] [CrossRef]
49. Castro-Rodríguez V, García-Gutiérrez A, Canales J, Avila C, Kirby EG, Cánovas FM. The glutamine synthetase gene family in Populus. BMC Plant Biol. 2011;11(1):119. doi:10.1186/1471-2229-11-119. [Google Scholar] [PubMed] [CrossRef]
50. Loulakakis K, Primikirios N, Nikolantonakis M, Roubelakis-Angelakis K. Immunocharacterization of Vitis vinifera L. ferredoxin-dependent glutamate synthase, and its spatial and temporal changes during leaf development. Planta. 2002;215(4):630–8. doi:10.1007/s00425-002-0785-6. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools