 Open Access
Open Access
ARTICLE
Genome-Wide Identification of the MYB Gene Family and Screening of Potential Genes Involved in Fatty Acid Biosynthesis in Walnut
1 College of Horticulture and Gardening, Yangtze University, Jingzhou, 434025, China
2 Lhasa Municipal Forestry and Grassland Bureau, Lhasa, 850000, China
3 Agro-Tech Extension and Service Center, Shannan, 856000, China
* Corresponding Author: Feng Xu. Email:
(This article belongs to the Special Issue: Plant Secondary Metabolism and Functional Biology)
Phyton-International Journal of Experimental Botany 2024, 93(9), 2317-2337. https://doi.org/10.32604/phyton.2024.055350
Received 24 July 2024; Accepted 19 August 2024; Issue published 30 September 2024
Abstract
The multifaceted roles of MYB transcriptional regulators are pivotal in orchestrating the complex processes of secondary metabolism, stress tolerance mechanisms, and life cycle progression and development. This study extensively examined the JrMYB genes using whole genome and transcriptomic data, focusing on identifying putative MYB genes associated with fatty acid metabolism. 126 MYB genes were identified within the walnut genome, characterized by hydrophilic proteins spanning lengths ranging from 78 to 1890 base pairs. Analysis of cis-acting elements within the promoter regions of MYB genes revealed many elements linked to cell development, environmental stress, and phytohormones. Transcriptomic data was utilized to examine the role of JrMYB genes in the biosynthesis of fatty acids in walnuts. The results revealed diverse expression of these genes across various tissue sites, displaying varying levels and distinct expression patterns. Furthermore, by integrating the results of the phylogenetic tree with the correlation of expression levels, a total of 10 genes potentially involved in the regulation of fatty acid synthesis were screened. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was conducted on these 10 genes and further identified 4 candidate genes, and a transcription regulatory network involved in fatty acids metabolism was constructed. This study presents a systematic analysis of JrMYB genes, laying the groundwork for an in-depth exploration of the JrMYB genes family’s function in regulating fatty acid synthesis.Graphic Abstract
Keywords
Supplementary Material
Supplementary Material FileThe walnut (Juglans regia L.), belonging to the Juglandaceae family, represents an economically significant forest species cultivated worldwide. It holds the distinction of being one of the “four nuts” globally due to its abundant and nutritious nuts, as well as its widespread distribution. Renowned for its delectable and aromatic kernel rich in protein, crude fat, 17 distinct amino acids, and Vitamins B1 and B2, walnut is a healthful dietary choice. Its benefits include anti-aging properties, blood lipid regulation, and cardiovascular disease prevention [1]. The fatty acid content of walnuts plays a pivotal role in cellular functions and intracellular metabolic processes in humans [2]. Unsaturated fatty acids, which are necessary for human health [3], are prevalent in walnuts [4], surpassing the levels found in conventional edible oils [5,6]. Consequently, the quality of walnuts is significantly affected by their fatty acid composition.
The MYB gene family is one of the most functionally diverse and abundant transcription factor families in plants. The defining structural property of plant MYB transcription factors is the N-terminal MYB structural domain, which uniquely interacts with the eukaryotic gene promoter regions to regulate gene expression [6]. This domain typically consists of 50–55 bases and adopts a helix-turn-helix (HTH) structure, often featuring 1–4 incomplete repeats in the R structure. MYB transcription factors are categorized into four subclasses, namely, 1R-MYB, 2R-MYB, 3R-MYB, and 4R-MYB, based on the number of structural domains present within the MYB gene [7].
The biological functions of MYB genes have been revealed through studies in Arabidopsis thaliana and a variety of other plant species. Notably, the MYB transcription factor ScMYB52-1 in sugarcane exhibits an inducible response under stressful conditions of exogenous ABA and NaCl [8]. The Arabidopsis transcription factors AtMYB20, AtMYB42, AtMYB43, and AtMYB85 serve as pivotal regulators that control the synthesis of phenylalanine and the subsequent polymerization of lignin during the maturation phase of lignification and cellulose deposition in secondary walls [9]. The PqMYB4 gene plays a regulatory role in anthocyanin synthesis and metabolism in the leaves of Paeonia qiui [10], whereas GlMYB4 and GlMYB88 positively govern flavonoid synthesis in Glycyrrhiza uralensis [11]. Additionally, numerous studies have revealed a strong correlation between MYB family members and the regulation of fatty acid synthesis. For example, AtMYB96 induces the expression of genes associated with fatty acid elongation in seeds and leaves, leading to the accumulation of long-chain fatty acids [12,13]. Similarly, overexpression of AtMYB92 in tobacco leaves promotes the expression of BCCP2 and significantly enhances fatty acid accumulation [14]. Furthermore, MaMYB4 can regulate the production of unsaturated fatty acids in Musa acuminata by controlling the expression of ω-3 fatty acid desaturases (FADs) [15].
Previous studies have made substantial progress in understanding the biological roles of the MYB gene family, particularly with the completion of whole-genome sequencing across various plant species. However, further investigation is needed to fully elucidate how MYB family transcription factors influence fatty acid synthesis in walnuts, as the existing literature on this subject is limited. In this study, we applied bioinformatics techniques to systematically identify members of the JrMYB genes within the genome. Our analysis explored their physicochemical characteristics, evolutionary relationships, conserved motifs, chromosomal locations, and other relevant features. In conjunction with the transcriptomic data and qRT-PCR analysis, our objective was to identify potential JrMYB genes related to fatty acid synthesis. This study provided a theoretical basis for future investigations into the roles of walnut MYB transcription factors in the regulation of fatty acid synthesis.
2.1 Identification and Protein Characterization of JrMYB Genes
The walnut genome data (No. PRJCA002070) were sourced from National Genomics Data Center (https://ngdc.cncb.ac.cn/?lang=zh, accessed on 18 June 2024), whereas the protein sequences of the all AtMYB genes were obtained from the TAIR database (https://www.arabidopsis.org/, accessed on 22 April 2023). To identify potential MYB candidate genes in the walnut genome, a multistep approach was employed. At the outset of investigation, we utilized the HMM file (PF00249), corresponding to the evolutionarily conserved structural motifs of the MYB transcription factor family in the Pfam database, to conduct a comprehensive screen. Subsequently, a local BLAST comparison was performed, utilizing the protein sequence of AtMYB genes as a query sequence to re-identify MYB candidate genes within the walnut genome. Following these screenings, the candidate genes obtained from both steps were further scrutinized using NCBI’s Batch CDD search tool. This manual curation process aimed to eliminate duplicated sequences and prediction errors, resulting in a refined and reliable compilation of the JrMYB genes.
The physicochemical properties of MYB transcription factor proteins, including isoelectric point, stability, and amino acid length, were analyzed. The analysis was conducted using Prot Param, an online tool available through ExPASy (https://web.expasy.org/protparam, accessed on 26 June 2023). Additionally, the subcellular localization and signal peptides of the members of the JrMYB genes were predicted. These predictions were made using Wolf Psort (https://web.expasy.org/protparam, accessed on 26 June 2023) and SignalIP-5.0 (https://web.expasy.org/protparam, accessed on 17 July 2023) online tools.
2.2 Collinearity Analysis and Chromosomal Localization of JrMYB Genes
The genome sequences along withand annotation files for Arabidopsis (Arabidopsis thaliana) and pecan (Carya illinoinensis) were obtained from the TAIR (https://www.arabidopsis.org/, accessed on 22 April 2023) and Ensemble plant databases (http://plants.ensembl.org/index.html, accessed on 22 April 2023). For the purpose of quantifying the collinearity within and between the walnut, Arabidopsis, and pecan genomes, we utilized the bioinformatics tool MCScanX, followed by graphical representation of the findings using TBtools. Using TBtools software, we extracted the genomic positions of JrMYB genes and identified covariance gene pairs spanning the entire walnut genome. These covariance associations were further depicted and visualized using the Advanced Circos software.
2.3 Genetic Structures, Conserved Motifs, and Characteristic Conserved Domains Analysis of JrMYB Genes
We conducted a thorough examination of the conserved protein motifs present across the 126-member JrMYB genes by employing the MEME suite’s online analytical tool (https://meme-suite.org/meme/, accessed on 19 July 2023). Following the motif discovery, TBtools was utilized to map and visualize these motifs, with default parameters configured to recognize a maximum of 10 conserved motifs. For the prediction of conserved structural domains within the JrMYB protein, both the NCBI’s Batch CDD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 24 April 2023) and the InterPro resource (https://www.ebi.ac.uk/interpro/, accessed on 25 April 2023) were engaged. Moreover, the exon-intron structure of the JrMYB genes was subjected to a detailed analysis and graphically displayed using TBtools software.
2.4 Phylogenetic Analysis of JrMYB Genes
The phylogenetic relationships were elucidated by the neighbor-joining method in MEGA-X software, using a bootstrap of 1000 replications, the p-distance model for sequence evolution, and pairwise deletion to accommodate missing data points. The protein sequences of AtMYB genes were extracted from the TAIR database (https://www.arabidopsis.org/, accessed on 27 April 2023), whereas the MYB protein sequences from walnut were subjected to multiple sequence comparisons. The organization and visualization of the phylogenetic trees were facilitated using the EvolView online tool (https://www.evolgenius.info/evolview-v2, accessed on 12 May 2023).
2.5 Cis-Acting Components Analysis of JrMYB Genes
To delineate the cis-acting regulatory elements in the proximal 2000 base pairs of the promoter sequences preceding the JrMYB genes, the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 06 May 2023) served as a predictive platform. Subsequently,the data were subjected to visualization and analysis using the heat map tool available in TBTools.
2.6 Screening of JrMYB Genes Involved in Fatty Acid Synthesis and Their Expression Analysis
Expression data for 126 JrMYB genes in the roots, stems, and leaves of walnut were compiled from previously collected transcriptomic data [16]. Subsequently, data processing and correlation analyses were conducted using Excel 2019. For visualization, a heat map tool within TBtools was used. In addition, JrMYB genes were compared to previously reported genes associated with fatty acid synthesis. To construct the phylogenetic tree, AtMYB92 (AT5G62470) [14], CaMYB340 (XP_016563145.2) [17], JcMYB1(AEI73171.1) [18] were used as reference points.
2.7 Quantitative Real-Time PCR and Determination of Fatty Acid
Total RNA was extracted from the roots, stems, and leaves of the walnut using MiniBEST Plant RNA Extraction Kit (Beijing, China), and cDNA templates were synthesized following the manufacturer’s instructions provided by HiScript cDNA Synthesis Kit (Nanjing, China) for use in subsequent quantitative real-time PCR (qRT-PCR) assays. For normalization purposes in our walnut gene expression assays, the GAPDH gene was selected as a housekeeping gene to provide an internal reference for normalization [19]. The relative quantification of target gene transcripts was performed using the 2−ΔΔCt formula [20], whichaccounts for variations in amplification efficiency, and the data were visualized with Origin software. Each sample was processed with three biological replicates and three technical replicates. Quantitative primers were designed using the Primer Premier 6 software, and all primers used in this study are listed in Supplementary Table S5. The fatty acid content in the roots, stems, and leaves was measured using gas chromatography-mass spectrometry (GC-MS) [21].
2.8 Construction of the Regulatory Network for Enzyme Genes and Transcription Factors Involved in Fatty Acid Biosynthesis
The Plant TF Motifs Shift plugin of TBtools was utilized, with Arabidopsis as a reference, to identify the binding motifs for JrMYB genes. Enzyme genes involved in the fatty acid biosynthesis pathway [16] were selected, and their promoter region sequences were extracted. A gene regulatory network was constructed using Fimo Search and visualized with Cytoscape software.
3.1 Identification of JrMYB Genes and Analysis of Their Physicochemical Properties
Examination of the walnut genome led to the discovery of 126 JrMYB genes, denoted as JrMYB001–JrMYB126 (Table S1). Notably, the proteins encoded by these family members exhibit significant variations in the size and quantity of amino acids. The amino acid sequence lengths ranged from 78 amino acids (JrMYB015) to 1890 amino acids (JrMYB003), with corresponding relative molecular weights ranging from 8.936 to 207.542 kDa.
Regarding the isoelectric point (pI) of the 126 walnut MYB family proteins, it ranged from 4.34 (JrMYB036) to 10.06 (JrMYB079). Interestingly, all MYB proteins exhibited negative Gravy values, which indicated their hydrophilic nature. According to ExPasy’s stability prediction software, 11 JrMYB proteins were found to be stable (with an instability coefficient <40), whereas the remaining 115 proteins were classified as unstable (with an instability coefficient >40). In terms of subcellular localization, JrMYB proteins were primarily located in the nucleus, with a minority of members also being localized in other subcellular regions such as the mitochondria and cytoplasm, as predicted for the subcellular localization of JrMYB genes. Notably, only JrMYB003, JrMYB076, and JrMYB081 were found to possess signal peptides according to signal peptide prediction, whereas the other genes lacked this feature.
3.2 Chromosomal Localization Analysis of JrMYB Genes
The distribution of 126 JrMYB genes across the walnut genome revealed an uneven spread, with genes dispersed across 16 chromosomes. The gene count on the individual chromosomes ranged from 4 to 13. The highest number of genes was observed on chromosomes 1 and 9, each hosting 13 genes, whereas chromosomes 8 and 12 had the lowest gene count, with only four MYB genes each.
Specifically, chromosome 8 housed four genes, namely JrMYB033, JrMYB030, JrMYB118, and JrMYB036, whereas chromosome 12 accommodated another set of four genes, consisting of JrMYB116, JrMYB084, JrMYB043, and JrMYB123. Chromosomes 2, 6, 7, and 10 (Fig. 1) also exhibited a relatively high number of genes distributed across them. Notably, the majority of genes were located toward the proximal or distal ends of the chromosomes, with fewer genes found in the central regions.
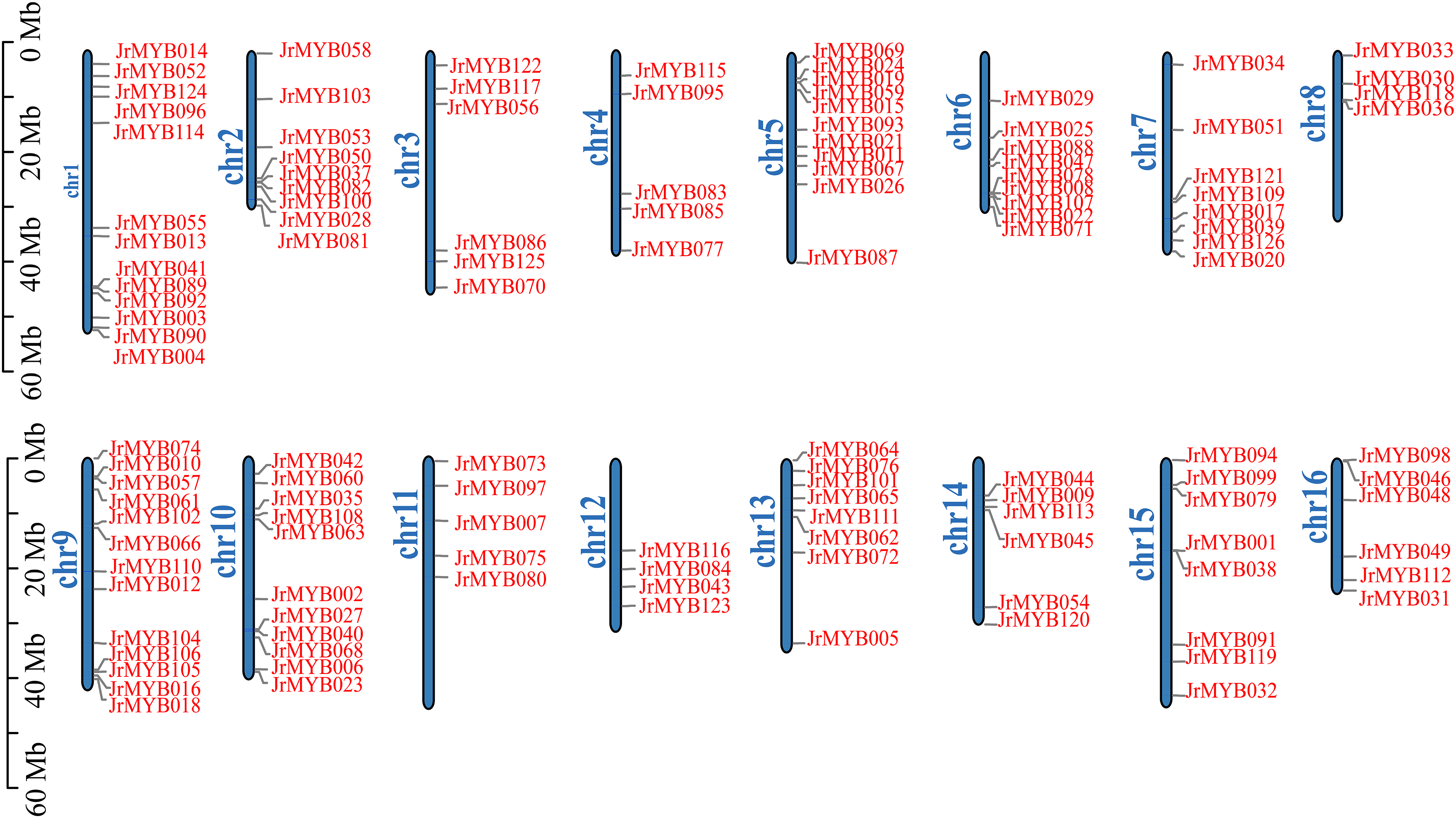
Figure 1: The distribution of JrMYB genes on chromosomes
3.3 Intraspecific and Interspecific Covariance Analysis of JrMYB Genes
Analysis of covariance included 50 gene family members within the JrMYB genes, and their distribution was observed across 11 chromosomes. Notably, chromosome 1 harbored the highest number of genes, totaling 11 (22% of the total), followed by chromosome 10, with seven genes (14%). The remaining chromosomes hosted varying numbers of JrMYB genes, ranging from 2 to 6 (Fig. 2). Concurrently, there exists a pronounced collinearity among segments across various chromosomes, which strongly suggests that a whole-genome duplication event has transpired within the species of walnut [22].
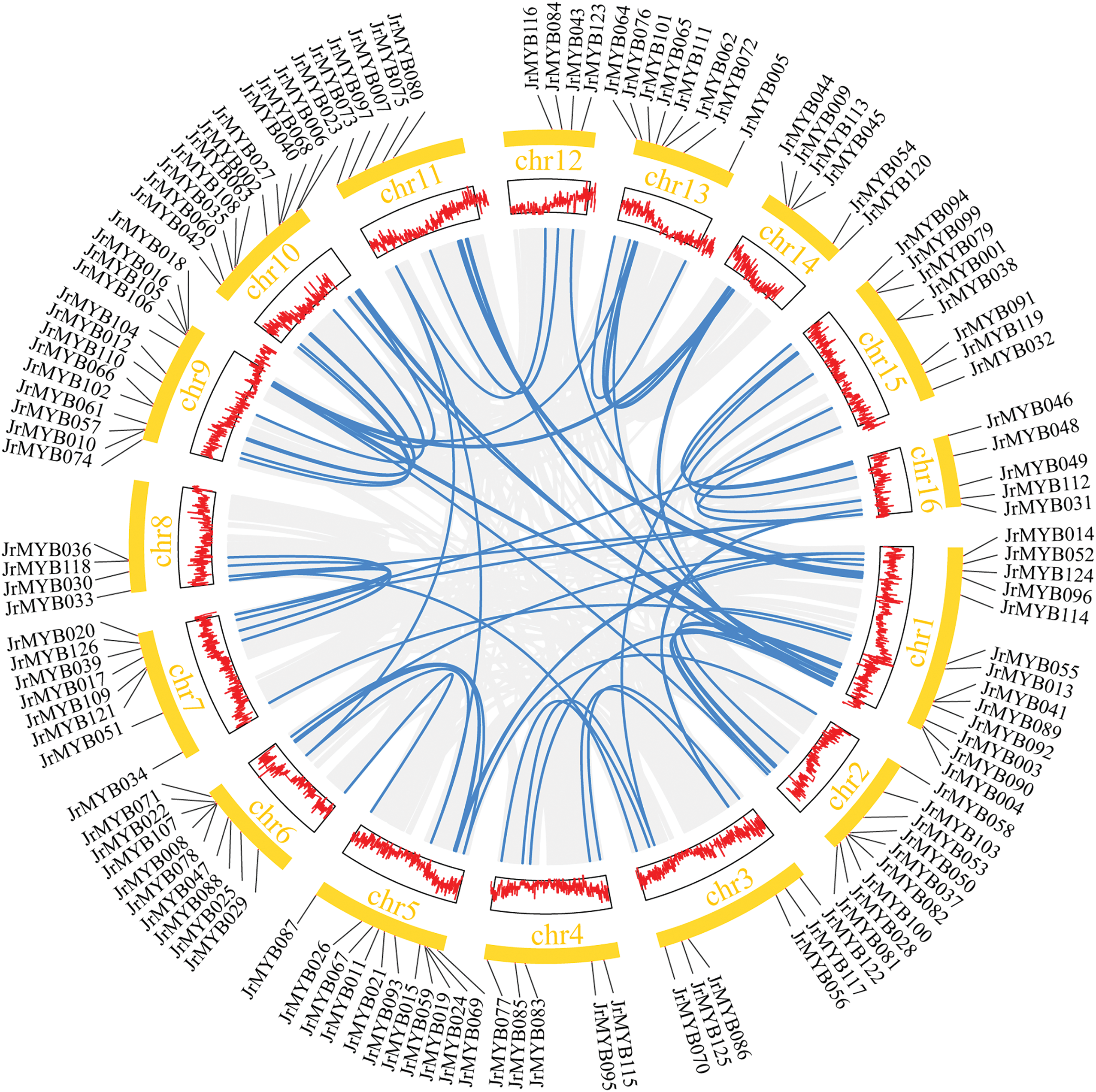
Figure 2: Collinearity analysis of JrMYB genes. The gray lines within the circular representation delineate the regions of collinear blocks, while the blue lines specifically indicate the pairs of collinear JrMYB genes. The gene density is visually depicted in the heatmap enclosed within the centralsquare, with the surrounding orange square illustrating the chromosomal organization
In the covariance analysis, 89 genes exhibited covariance between walnut and both Arabidopsis and pecan (Table S2), while 121 genes displayed covariance solely between walnut and pecan (Table S3). This observation suggests a close evolutionary relationship between the genomes of walnut and pecan (Fig. 3).
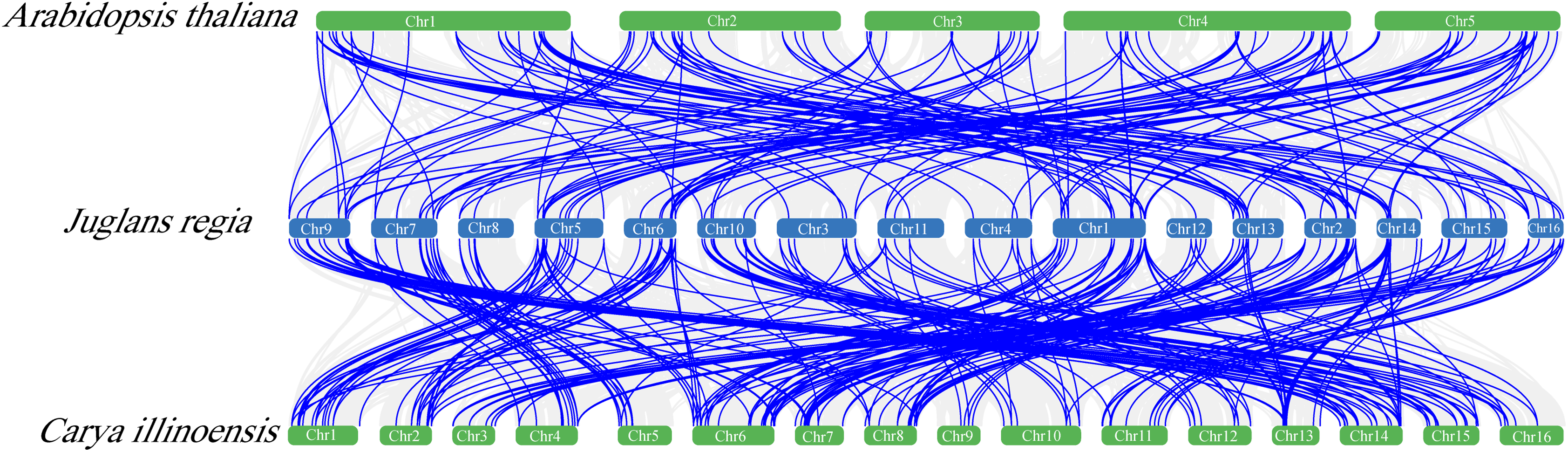
Figure 3: Collinear relationship between A.thaliana, Juglans regia L. and Carya illinoinensis
3.4 Analysis of Gene Evolution and Classification of JrMYB Genes
The JrMYB genes has been categorized into four groups, discerned through structural characterization of distinctive conserved domains within MYB proteins [7] and determination of the number of structural domains in InterPro. These groupings were as follows: 1R-MYB/MYB-related (102 members), R2R3-MYB (19 members), 3R-MYB (4 members), and 4R-MYB (1 member) (Fig. 4 and Table S4).
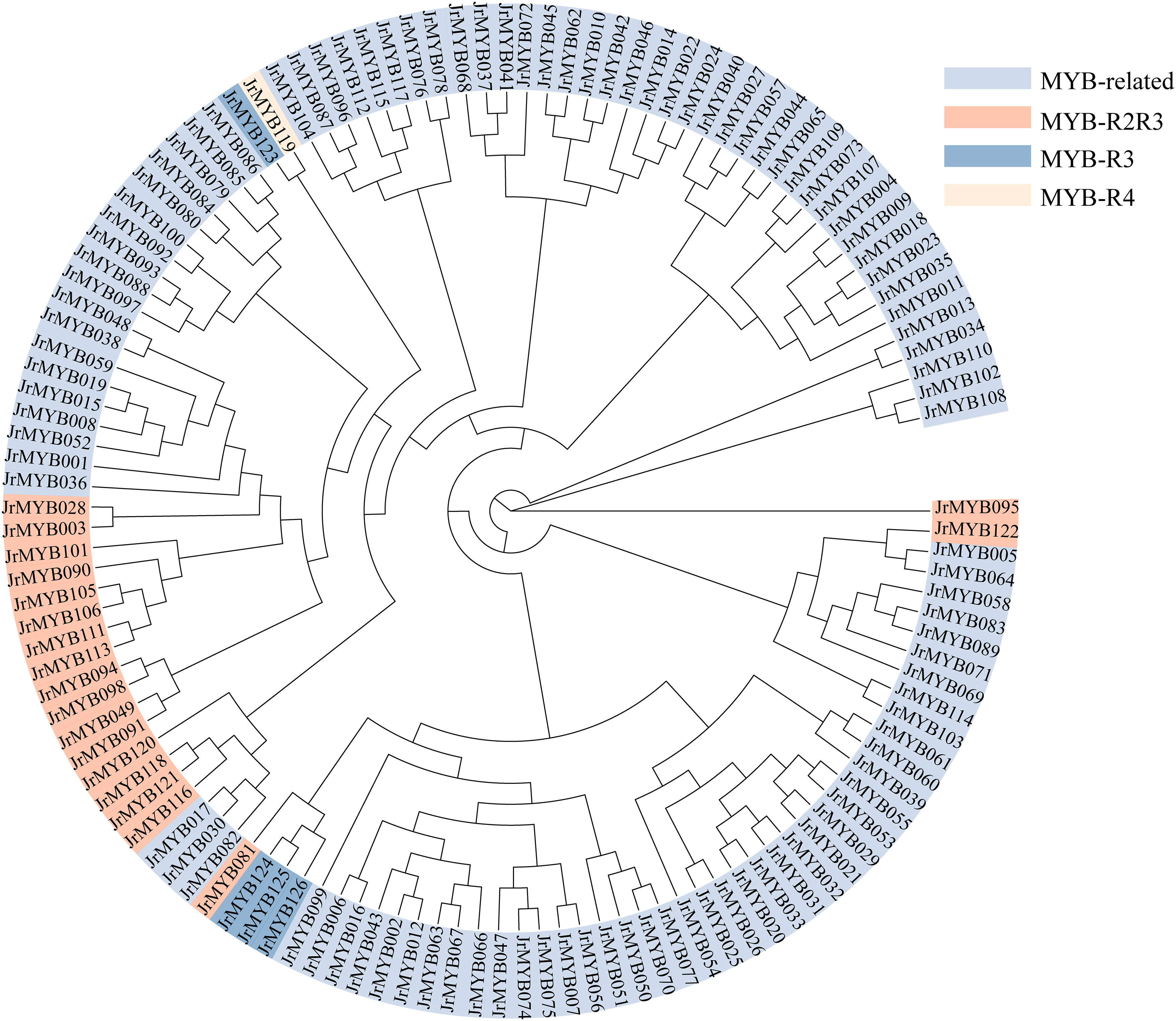
Figure 4: Phylogenetic analysis of JrMYB genes
To investigate the evolutionary relationships among JrMYB genes, phylogenetic trees were constructed using MEGA-X. These trees included 126 JrMYB gene family protein sequences and 126 AtMYB gene family protein sequences of known subgroups (Fig. 5).
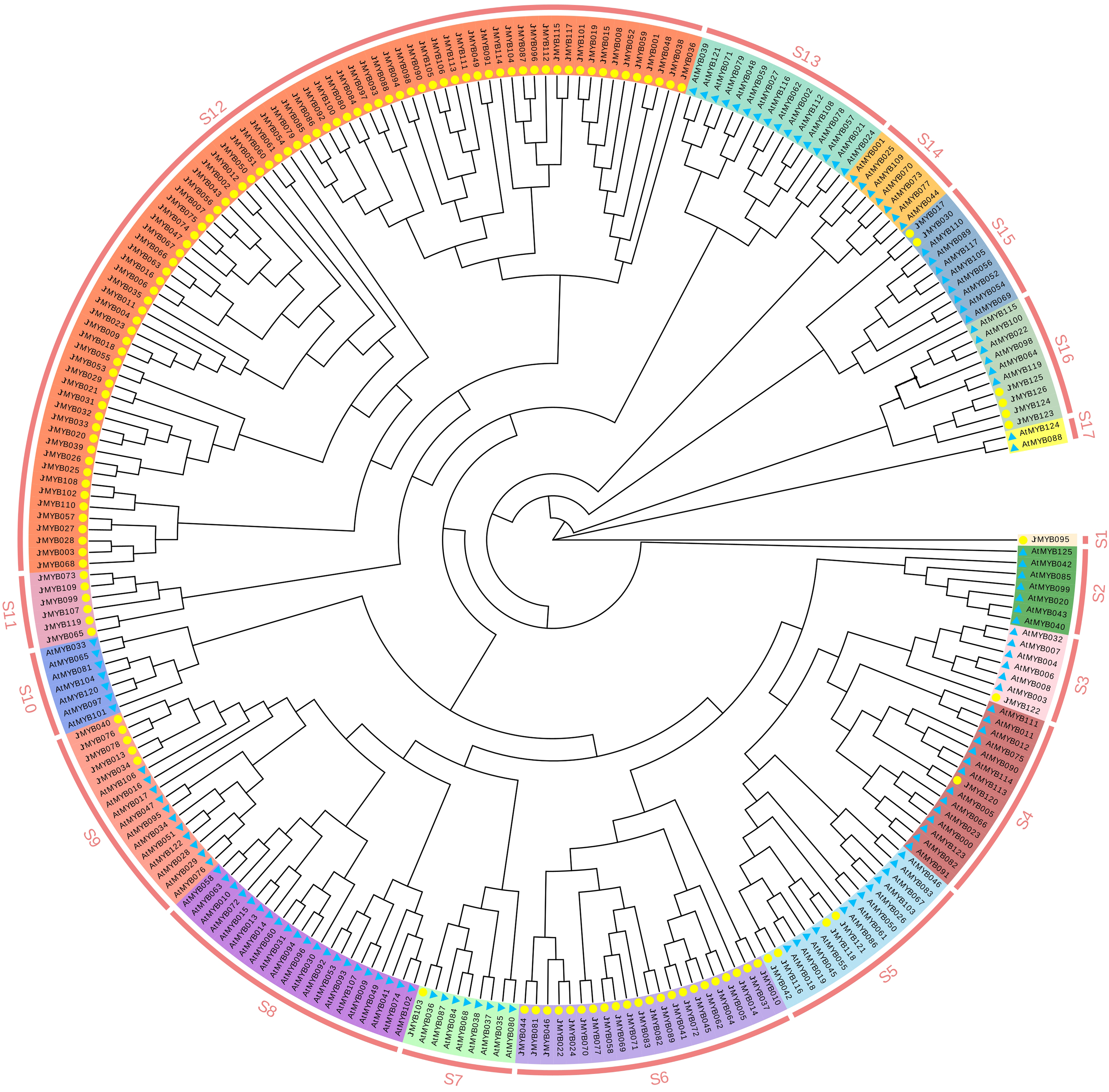
Figure 5: Phylogenetic relationship between Juglans regia L. and A. thaliana MYB families. The yellow circle represents Juglans regia L., blue triangle represents A. thaliana
JrMYB proteins were further divided into 17 subgroups, denoted as S1–S17, following the criteria used for classifying the Arabidopsis MYB gene family [7]. Among these subgroups, seven (S3, S4, S5, S7, S9, S15, and S16) included JrMYB genes that clustered with AtMYB genes. The remaining four subgroups, designated S1, S6, S11, and S12, contained JrMYB genes that were individually clustered. Notably, S1 contained only one gene, JrMYB095.
3.5 Genetic Structures, Conserved Motifs, and Characteristic Conserved Domains Analysis of JrMYB Genes
In this study, MEME software was used to scrutinize the conserved motifs present in the protein sequences encoded by the JrMYB genes, with the aim of gaining deeper insights into the structural characteristics of the 126 JrMYB genes. Ten conserved motifs, designated as motifs 1 through 10, were identified (Fig. 6). Among these, motif 1 was found in all 126 JrMYB genes, indicating that it was the most highly conserved motif within the family. Motifs 1 and 5 were detected in 73 JrMYB genes, highlighting their significance in shaping MYB protein sequences. The graphical representation of the data highlighted that protein subclasses exhibiting comparable affinities for their targets were more conserved at the sequence level and displayed common structural elements. For instance, 17 family members situated in the same branch, characterized by motifs 1, 6, and 7, were likely to have similar structural functions. Likewise, 10 family members possessing motifs 1, 6, 8, 9, and 10 may share a common structural function. In contrast, the number and type of motifs varied among the different evolutionary branches.
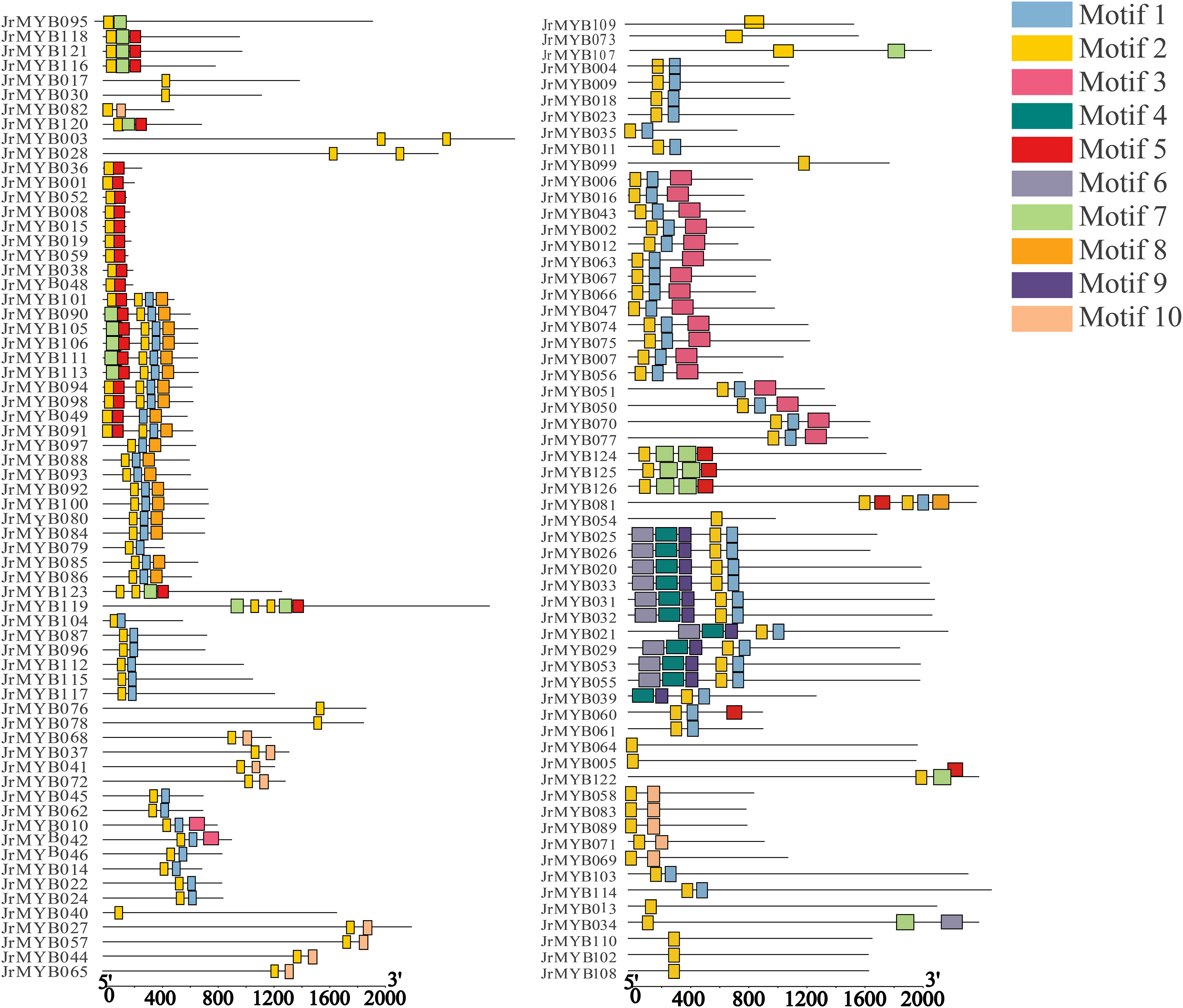
Figure 6: Conserved motifs of 126 JrMYB genes. Different colors signify different motiftypes
Analysis of the walnut MYB protein sequence revealed the presence of ten structural domains, including MYB_DNA-binding, SANT, and MYB_CC-LHEQLE (Fig. 7). A comprehensive examination of 126 amino acid sequences using the CDD-search tool in NCBI and InterPro indicated that various members of the sequence possessed varying numbers and types of structural domains. The SANT and MYB_SHAOKYF domains were among the most frequently observed.
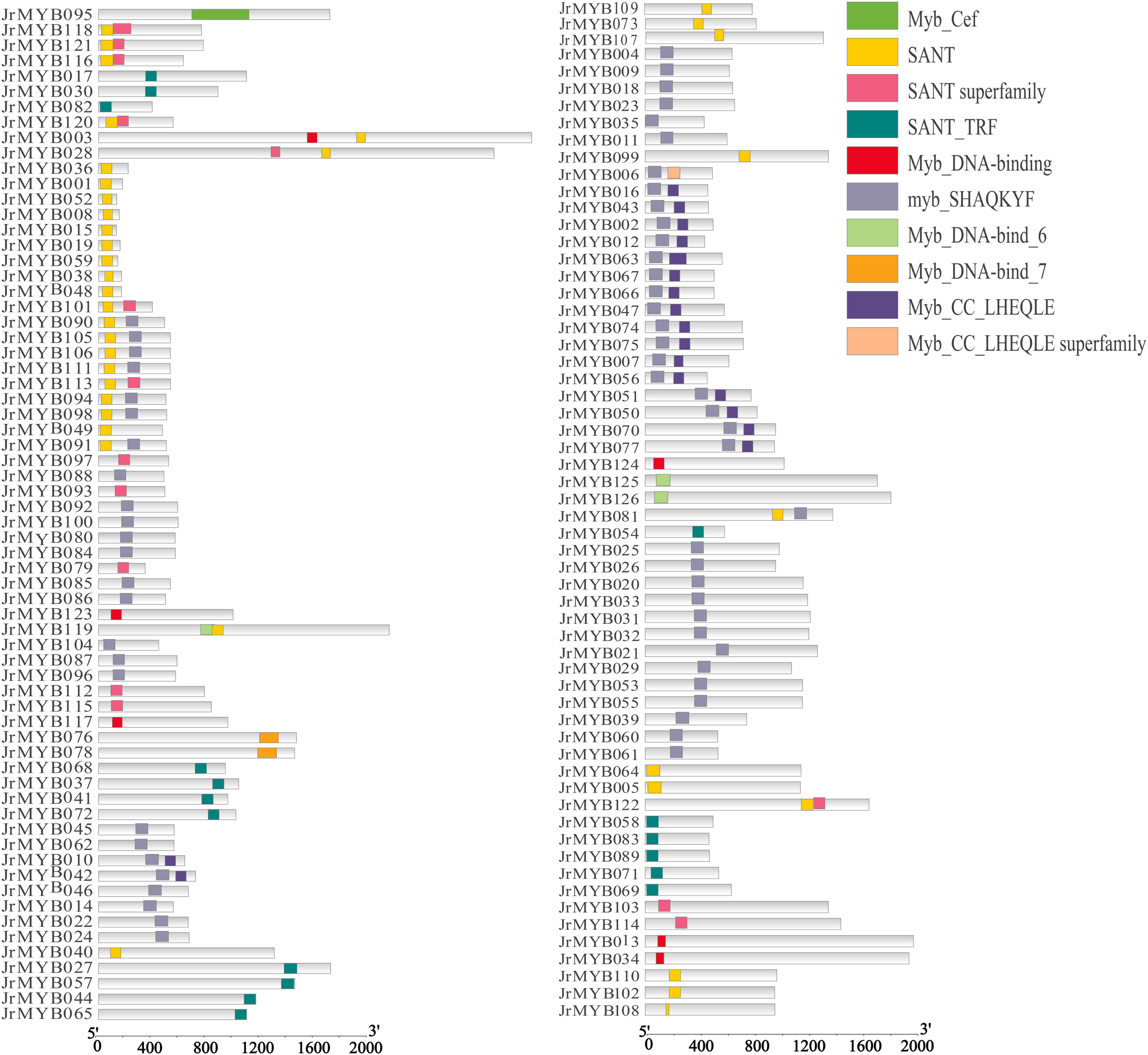
Figure 7: Characteristic conserved domains of 126 JrMYB genes
Furthermore, detailed gene structure analysis was conducted of all JrMYB genes (Fig. 8). The results revealed substantial variations in the number, distribution, and length of coding sequences among members, ranging from 1 to 20 in number and exhibiting diverse lengths. For instance, JrMYB013 and JrMYB034 featured 20 coding sequence regions, whereas JrMYB052 and JrMYB008 contained only one region. This variation underscores distinct evolutionary trajectories within the JrMYB genes, leading to diverse gene structures.
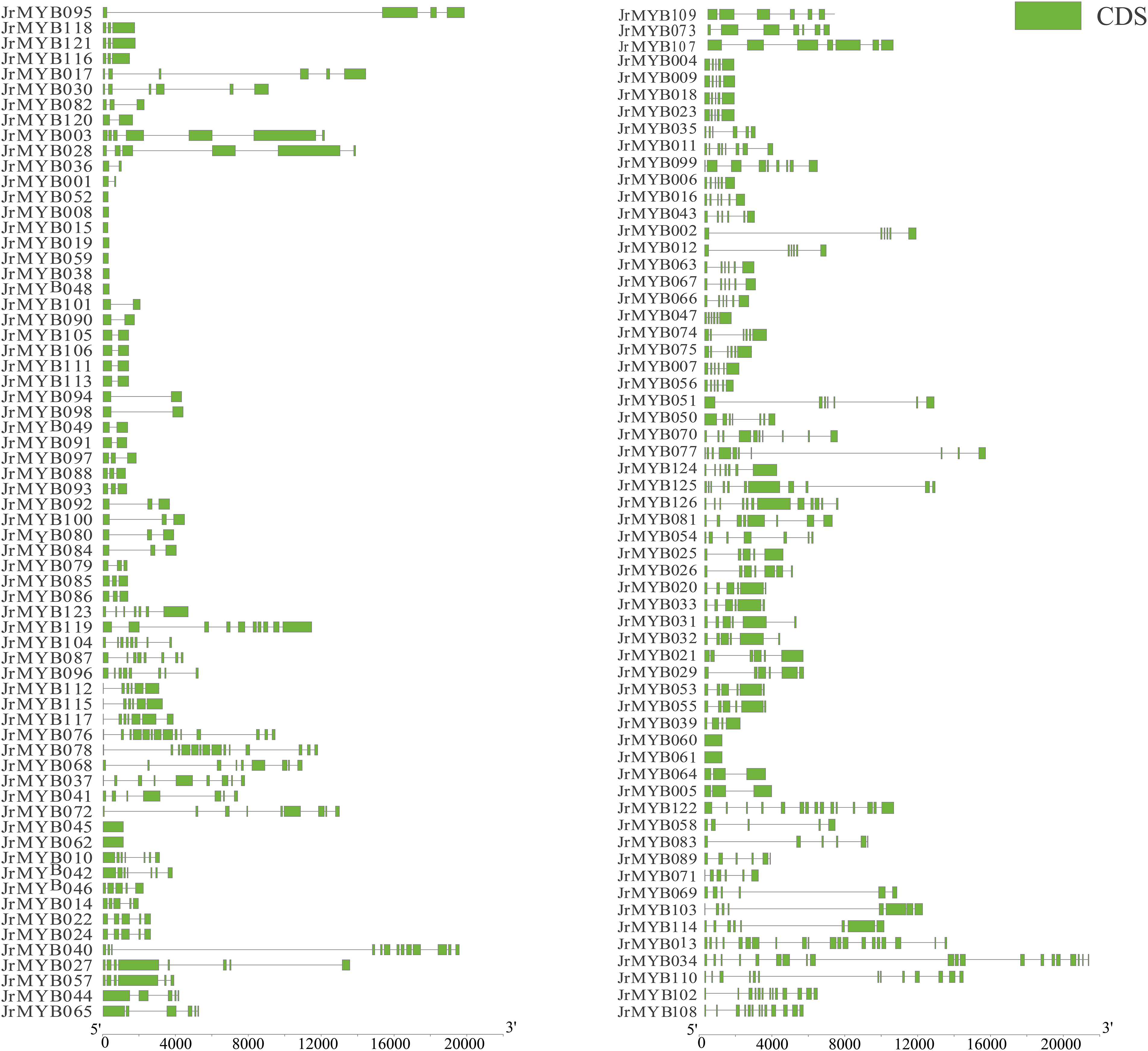
Figure 8: Genetic structures of 126 JrMYB genes
3.6 Cis-Acting Components Analysis of Promoters of JrMYB Genes
The findings revealed that the JrMYB genes predominantly featured four types of cis-acting components associated with cell development, light responsiveness, environmental stress responses, and phytohormone regulation (Fig. 9). Specifically, the four principal cis-acting components linked to cell development included the RY element, MSA-like, CAT-box, and GCN4_motif. These components were distributed across 66 genes, with CAT-box being the most prevalent, present in 56 genes.
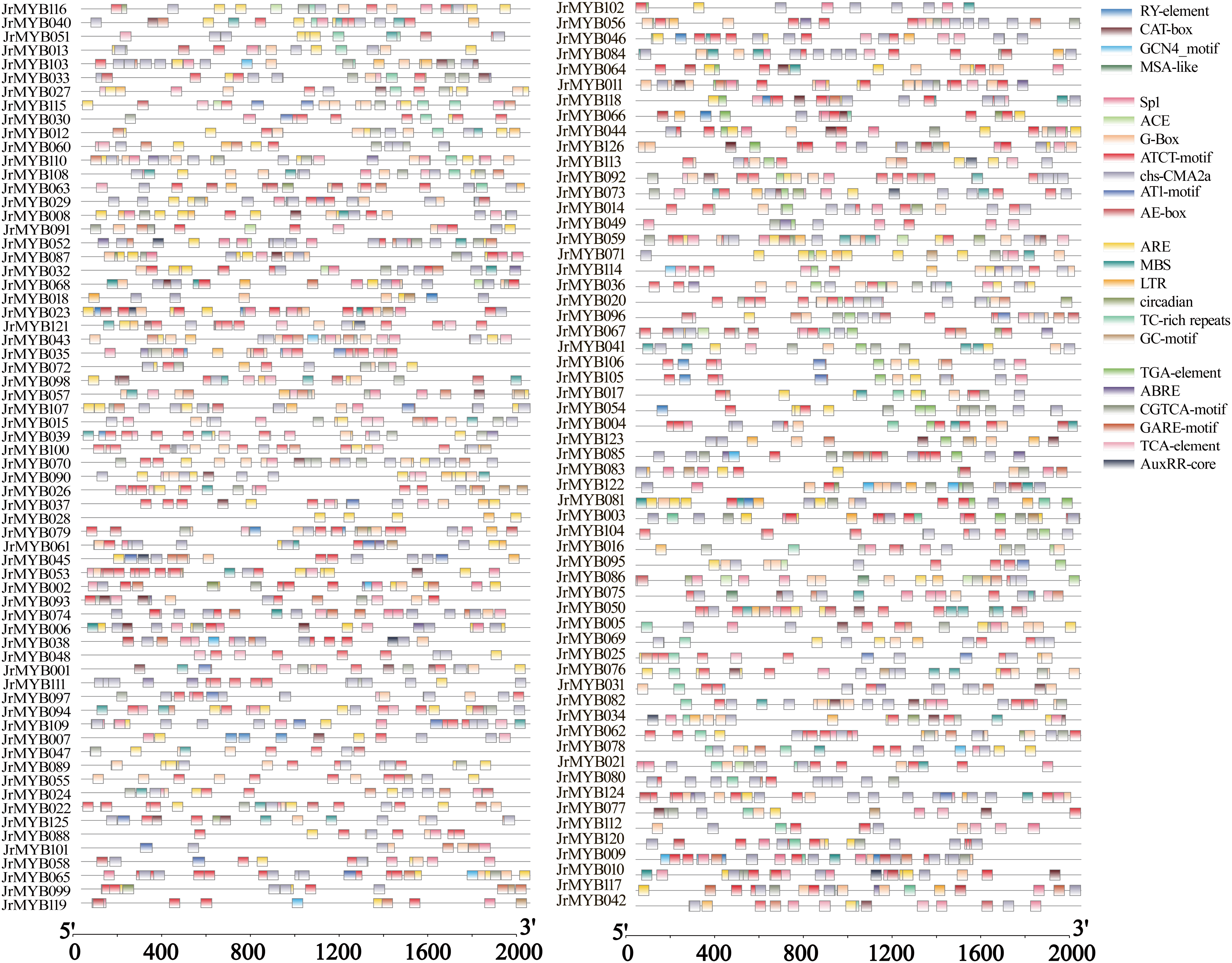
Figure 9: Analysis of cis-acting components of JrMYB genes
For light responsiveness, seven cis-acting components were used, including ACE, AE-box, AT1-motif, ATCT-motif, chs-CMA2a, Sp1, and G-box. Except for JrMYB019, all genes exhibited various light-responsive components. Notably, the analysis identified nine ATCT motifs in three genes (JrMYB023, JrMYB065, and JrMYB100) and JrMYB080 and JrMYB100 contained nine chs-CMA2a elements.
Six components, namely ARE, circadian rhythm, GC-motif, LTR, TC-rich repeats, and MBS, were associated with external defense and stress responses. Stress-related elements were identified in 120 genes, with ARE being the most prevalent and found in 102 genes. In the context of hormone response, six cis-acting components were observed: TCA element, TGA element, GARE motif, CGTCA motif, AuxRR core, and abscisic acid response element (ABRE). These components were primarily linked to responses involving abscisic acid (ABA), growth hormones (indole-3-acetic acid, IAA), methyl jasmonate (MeJA), and gibberellin (GA). Only nine JrMYB genes contained the AuxRR core element related to the growth hormone response, whereas 100 JrMYB genes featured the cis-acting component ABRE associated with the abscisic acid response.
3.7 Screening of JrMYB Genes Involved in Fatty Acid Synthesis
Using GC-MS to analyze the fatty acid content in the roots, stems, and leaves of walnuts, the data showed that the fatty acid content in the walnut roots was the lowest, at 32.43 mg/g, followed by the leaves at 36.39 mg/g, while the stems had the highest fatty acid content, at 61.73 mg/g. And the content of fatty acid Transcriptomic data [16] were used to explore the potential role of JrMYB genes in walnut fatty acid synthesis. The expression patterns of 103 JrMYB genes were analyzed across the major tissues of the roots, stems, and leaves, following the removal of 23 genes with low expression levels. A clustered heat map was employed to visually represent the results, successfully delineating the unique expression patterns observed, as shown in Fig. 10.
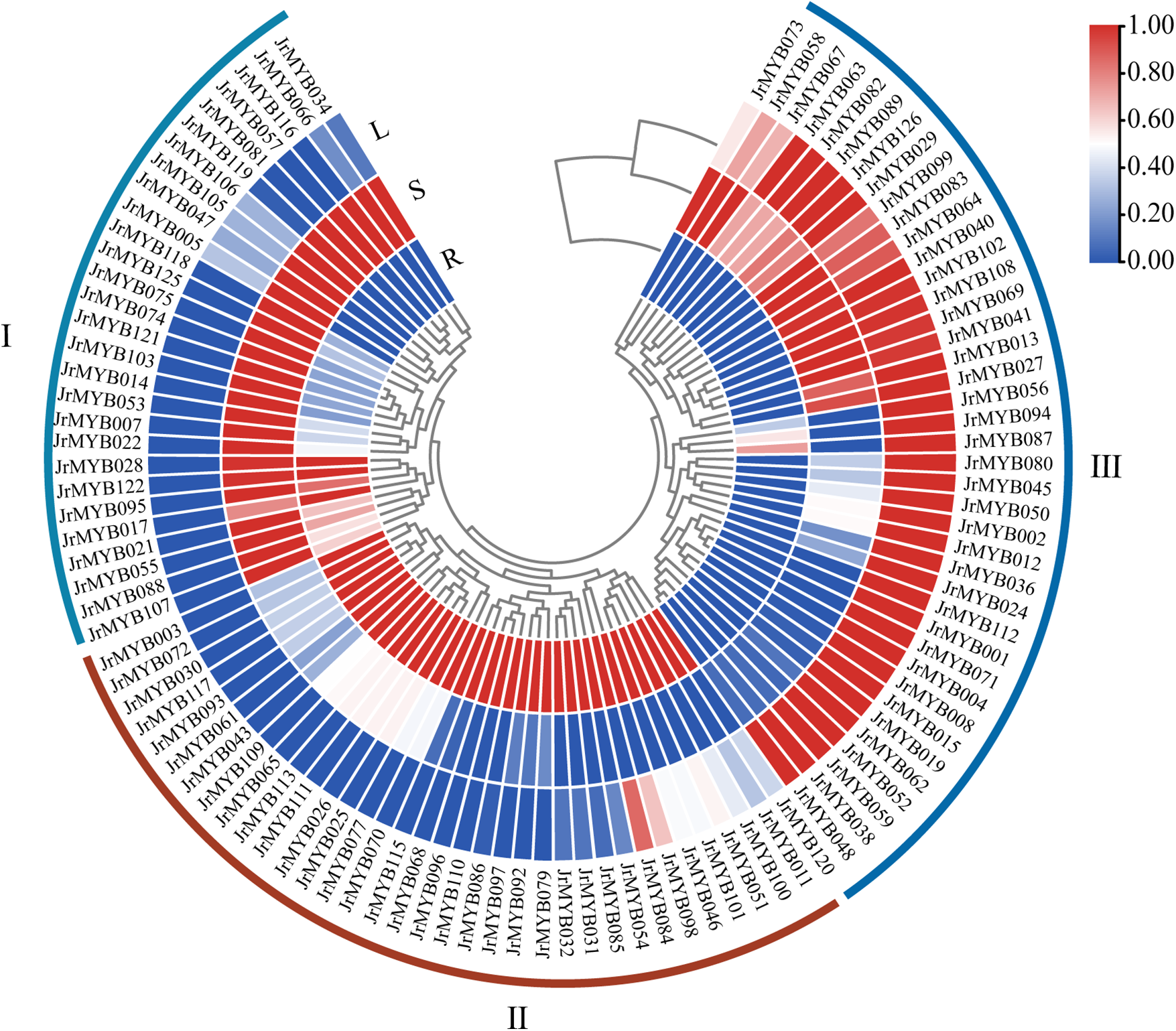
Figure 10: Fatty acid expression profile of JrMYB genes in different tissues. R: root; S: stem; L: leaf. Red areas indicate regions of high gene expression, and blue areas indicate regions of low expression in the corresponding tissues. The color coding for expression levels is shown on the right. The color scale, indicating expression levels, is provided on the the right of the heatmap
The JrMYB genes were categorized into three branches based on their expression patterns: Branch I: Comprising 28 genes that exhibited high expression in stems and low expression in leaves. Branch II: Encompassing 35 genes with high expression primarily in roots and low expression primarily in stems. Branch III: Consisting of 40 genes that were highly expressed mainly in leaves, with exceptions like JrMYB073 and JrMYB058. The other 38 genes displayed little to no expression in the stems, and no gene displayed high expression in all three tissues, reflecting the distinct tissue-specific expression patterns within the JrMYB genes. Heat map analysis further revealed that 16 genes exhibited opposite expression patterns in roots, stems, and leaves, with a “high-middle-low” sequential decrease in expression. Conversely, 26 genes displayed similar expression patterns across these tissues, demonstrating a “low-middle-high” increase in expression. In the root-stem-leaf context, 16 genes exhibited the opposite expression pattern, transitioning from “high-middle-low.” Among these JrMYB genes, 44 genes were found negative correlations with the content of fatty acid, 59 genes showed positive correlations. Especially, 23 JrMYB genes were found highly correlated with fatty acid at the level of |r| ≥ 0.9. For example, JrMYB084 had a correlation of 0.9999, and JrMYB066 had a correlation of 0.9996 (Table S6). Additionally, we further constructed a phylogenetic tree of the JrMYB genes involved in fatty acid metabolism in other plants, showing that 24 JrMYB genes cluster with MYB genes of known functions, suggesting that these JrMYB genes may be involved in fatty acid metabolism. Based on the intersection of the results from both analyses, we have selected 10 candidate JrMYB genes that may be involved in the fatty acid metabolism of walnut (Fig. 11).
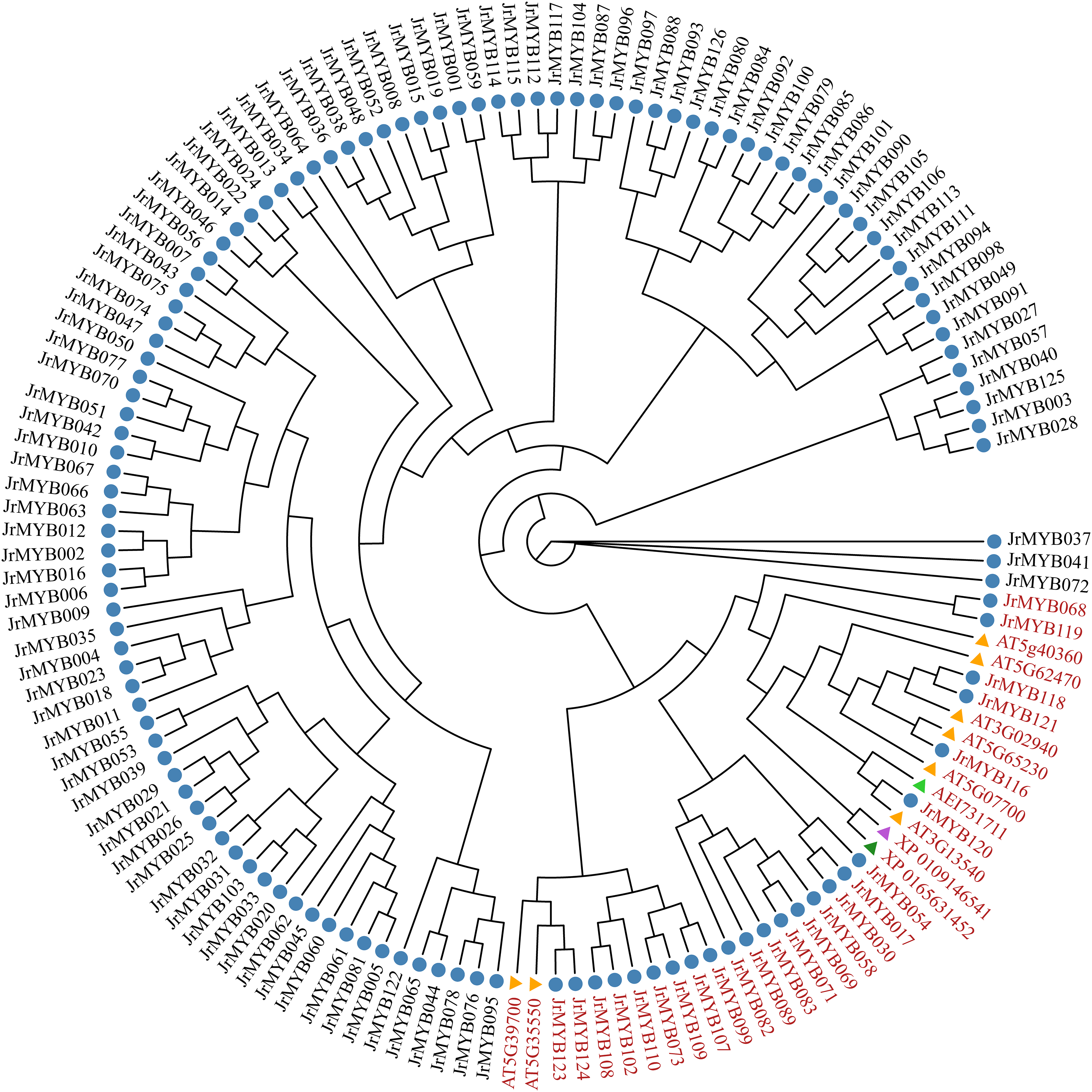
Figure 11: Phylogenetic relationship between JrMYB genes and multispecies fatty acid synthesis related MYB genes. Genes marked in red are related genes that may be involved in fatty acid synthesis. Blue circles represent JrMYB genes, yellow triangles represent AtMYB genes (A. thaliana), green triangles represents JcMYB (Jatropha curcas), purple triangles represents EgMYB (Elaeis guineensis Jacq.), and blue triangles represents CaMYB (Capsicum annuum L.)
3.8 qRT-PCR Verification and Construction of the Transcription Regulatory Network
To further study the expression of walnut JrMYB genes in various tissues, we performed qRT-PCR on ten genes selected from transcriptomic data [16] and phylogenetic analysis, assessing their expression in walnut roots, stems, and leaves. As shown in the Fig. 12A, a correlation analysis between qRT-PCR data and transcriptome FPKM values yielded 3 genes with high correlation coefficients (R2 > 0.7), which are considered key candidate genes for enhancing the fatty acid content in walnuts. To further investigate the regulatory mechanisms of JrMYB genes on fatty acid synthesis in walnuts, we projected the regulatory network Based on the MYB binding motifs (Table S7) found in the promoter regions of enzyme genes, we have identified three potential target genes that may be regulated by JrMYB genes (Fig. 12B,C).
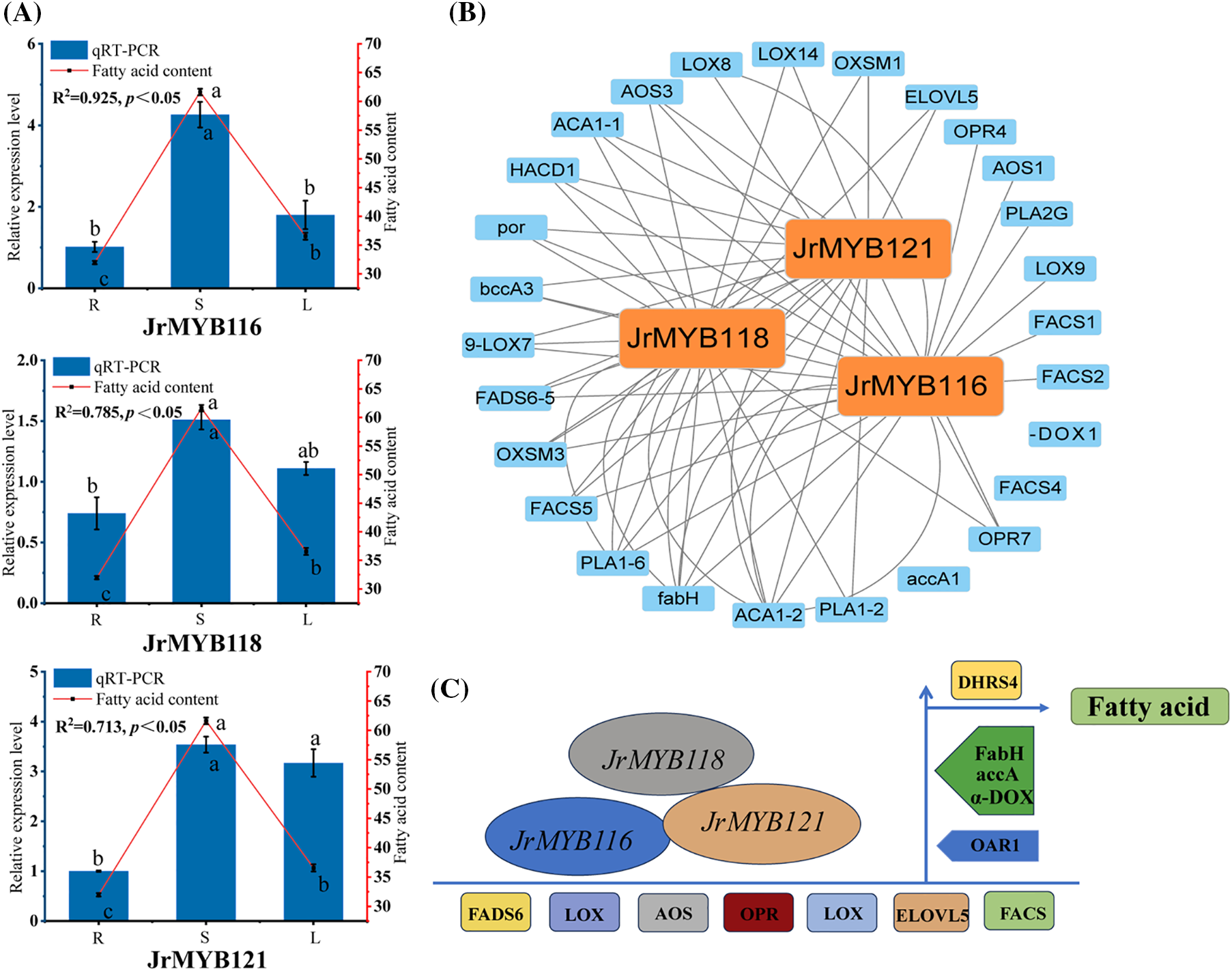
Figure 12: (A). Relative expression level of JrMYB genes in the roots, stems, and leaves of the walnut. (B). The transcription regulatory network of JrMYB genes involving in fatty acid biosynthesis. (C). Model for JrMYB genes and enzyme genes regulating fatty acid biosynthesis
The results indicated that JrMYB120 has a targeting relationship with the promoter regions of 20 enzyme genes, JrMYB121 and JrMYB118 have targeting relationships with the promoter regions of 17 enzyme genes. The targeted enzyme genes are predominantly concentrated in ACA1 (acetyl-CoA acyltransferase 1), FADS6 (omega-6 fatty acid desaturase), and ELOVL5 (elongation of very long chain fatty acids protein 5), among others.
MYB gene family is a pivotal group of transcription factors in plants that plays a significant role in the regulation of plant biology. Although MYB genes have been extensively studied in various species, including Arabidopsis [7], soybean [23], and citrus [24], their investigation in walnut has been relatively limited. In this study, we identified 126 members of the MYB family from the walnut genome. Through bioinformatics, we conducted a comprehensive analysis of the physicochemical properties, chromosomal localization, collinearity relationships, evolutionary relationships, gene structures, and cis-acting elements of these genes. By integrating transcriptomic data with qRT-PCR results, we ultimately selected 3 JrMYB genes that are likely involved in the synthesis of fatty acids in walnuts and predicted the transcription regulatory network involved in fatty acid biosynthesis. These results provide genetic resources for the selection of superior walnut strains with high fatty acid content, identifying candidate genes for the regulatory network of fatty acid production in walnut, and providing a theoretical foundation for further in-depth investigations in this area.
4.1 The Abundant Walnut MYB Genes Exhibit Diverse Functions
The number of JrMYB genes identified in walnut was similar to that in Arabidopsis thaliana (168) and Jatropha curcas (115). However, there were significant differences when compared with the number of MYB family members in peanut (Arachis hypogaea, 31) and torch pine (Pinus taeda, 35), where the number of MYB family members displayed considerable variation. These differences could potentially be attributed to chromosome rearrangements, fusions, and other genomic modifications as well as genome-wide duplication events [25]. Notably, all MYB genes in the walnut family are characterized as hydrophilic proteins. Assessment of instability coefficients indicated that the majority of walnut MYB family members were categorized as unstable proteins, with the exception of 11 stable genes. These findings align with similar observations made for MYB genes in other species such as Lonicera macranthoides [26], and Lysimachia christinae [27], suggesting that the MYB gene family has maintained a relatively conserved nature throughout evolution.
Furthermore, subcellular localization predictions revealed [28] that approximately 80% of MYB transcription factors were likely localized to the nucleus. This prediction aligns with the known functions of transcription factors, which play a pivotal role in controlling the expression of downstream genes [29]. Analysis of the conserved motifs and gene structures of JrMYB genes indicated that members within the same evolutionary tree branch of walnut shared similar conserved structural domains and motif compositions [30]. This observation lent credibility to the phylogenetic tree constructed in this study and, to some extent, laid the foundation for predicting gene function [31]. Additionally, the distribution of the majority of genes at both ends of the 16 chromosomes in walnut aligns with the chromosome distribution patterns observed in MYB family members of other plants such as potato [32], bell cherry [33], and cocoa [34].
4.2 Phylogenetic Relationship Analysis and Functional Exploration of JrMYB Genes
In general, genes that share evolutionary relationships are expected to have functional similarities [35], and phylogenetic analysis serves as a valuable reference for family level investigations. Based on the results of phylogenetic analysis, certain JrMYB genes exhibited clustering patterns with specific AtMYB genes, suggesting potential functional similarities.
JrMYB017 and JrMYB030 were found in the same branch as AtMYB052, a gene known to control the expression of ABA-responsive genes, and consequently, cell wall biosynthesis [36]. This suggests that JrMYB017 and JrMYB030 may play a role in regulating cell wall biosynthesis. JrMYB040, JrMYB076, JrMYB017, JrMYB013, and JrMYB034, which are grouped in the same branch as AtMYB17, might share functions related to early inflorescence development and seed germination [37], similar to AtMYB17. JrMYB120 and AtMYB111, situated in the same branch, may have comparable functions in promoting genes within the phenylpropane pathway, potentially enhancing flavonol production [38]. JrMYB017 and JrMYB030 clustered together and could be associated with fatty acid synthesis, as AtMYB089 is involved in seed oil and major fatty acid biosynthesis [39]. Similarly, AtMYB076 [40], which belongs to the S9 group of the evolutionary tree, was linked to the regulation of fatty acid synthesis, suggesting that JrMYB040, JrMYB076, JrMYB078, JrMYB013, and JrMYB034 may play a role in fatty acid synthesis regulation.
S1, S6, S11, and S12 represent walnut species-specific groupings, which differ from the classification of the Arabidopsis MYB family. These distinctions may be attributed to variations or divergences that arose during evolutionary processes in response to environmental adaptations between walnut and Arabidopsis thaliana [41]. It is hypothesized that the functions of MYB family members within these branches are specific to walnut, a characteristic also observed in cherry MYB genes.
Furthermore, the distribution of multiple JrMYB genes within most subgroup branches suggests a potential functional redundancy within these MYB transcription factors. This redundancy implies that multiple genes in the same branch may have similar or overlapping functions, and silencing or inactivating a single gene may not significantly affect the overall function or expression of this particular group of genes [42].
4.3 Functional Exploration of Promoter Regions of JrMYB Genes
In this study, cis-acting components were analyzed by examining the 2000 bp genomic sequence upstream of the genes and classifying the identified cis-acting components into four main categories. This analysis aimed to gain insights into the functions of JrMYB genes, as the distribution and types of cis-acting components in the promoter region play a crucial role in determining gene expression and function [43]. Six dealt with hormone response, six with stress response, seven with light response, and four with cell development. The findings from this analysis suggest that JrMYB genes may play significant roles in the light and stress response mechanisms in walnut. Notably, almost all genes except JrMYB018 were found to have light response elements in their promoter regions. Furthermore, stress-related components were abundant and present in all 120 genes, indicating their potential involvement in the stress response pathways.
Among the hormone-related cis-acting components, ABRE was identified in 100 genes, highlighting its importance in the regulation of JrMYB genes expression. However, the growth hormone response-related AuxRR-core element was found in only nine genes, suggesting that these specific genes may be associated with the regulation of growth hormone metabolism. We analyzed the cis-acting elements of 10 potential genes and found that both JrMYB054 and JrMYB118 contain the RY-element (CATGCATG). Previous studies [44] have shown that the AtLEC2 can bind to the RY-G-Box and RY elements in the upstream promoter regions of the storage protein gene At2S3 [45], thereby regulating downstream genes and enhancing oil content in seeds. This implies that the RY-element may be a key component in the regulation of fatty acid synthesis.
Additionally, this study revealed the presence of various cis-acting components related to hormone responses, and their distribution varied considerably among the JrMYB genes. This diversity in response elements underscores the complexity and specificity of the regulation of JrMYB genes expression [46].
4.4 Mining and Identification of JrMYB Genes Related to Fatty Acid Synthesis in Walnut
In A.thaliana, Brassica napus, and Elaeis guineensis, the role of MYB in modulating fatty acid production has been extensively documented. However, limited attention has been directed towards its role in walnut. The co-regulation of AtAAD2 and AtAAD3 expression in A. thaliana by AtMYB118, along with its homolog AtMYB115, has been found to significantly enhance monounsaturated fatty acid production in the endosperm [47]. Subsequent research has indicated that EgMYB108 can effectively bind to the promoters of EgLACS and EgKCS, resulting in enhanced fatty acid content [48]. Similarly, overexpression of CaMYB340 in sweet orange resulted in a reduction in fatty acid desaturation [17].
In this study, a comprehensive examination of 103 genes related to fatty acid synthesis was conducted using correlation analysis. Among these genes, 54 JrMYB genes exhibited a positive correlation, whereas 49 JrMYB genes displayed a negative correlation. The gene expression patterns revealed that 126 JrMYB genes lacked a common highly expressed gene in the three tissues studied, indicating tissue-specific expression heterogeneity among the JrMYB genes. To further elucidate the role of JrMYB genes in fatty acid synthesis, phylogenetic analyses were performed using the aforementioned MYB genes (AtMYB92 (AT5G62470) [14], CaMYB340 (XP_016563145.2) [17], JcMYB1 (AEI73171.1) [18], EgMYB108 (XP_ 010914654.1) [49]). This analysis allowed for the identification of 24 genes that are potentially involved in fatty acid synthesis. Among these 24 genes, ten exhibited high correlation coefficients. Notably, JrMYB116, JrMYB118, JrMYB119, and JrMYB121 displayed correlation coefficients exceeding 0.9, suggesting that they were important genes involved in fatty acid synthesis.
Following qRT-PCR verification of 10 candidate genes, we identified 3 key genes. The data show distinct differences in the expression levels of most genes in the roots, stems, and leaves of the walnut, aligning with the transcriptomic data [16] and further indicating that JrMYB genes have tissue specificity [50]. The transcription regulatory network suggests that JrMYB116, JrMYB118 and JrMYB121 can bind to the promoter regions of enzyme genes involved in the fatty acid biosynthetic pathway, thereby influencing gene expression and enhancing the fatty acid content in walnuts. This finding is in line with this finding is in line with the research conducted by Xu et al. [47], who demonstrated that the overexpression of EgMYB108 can elevate lipid levels in fruit. Their study further confirmed that EgMYB108 is capable of binding to the promoters of EgLACS and EgKCS, which in turn enhances the content of fatty acids.
In this study, the walnut genome served as the foundation for the identification of 126 JrMYB genes, which underwent a comprehensive analysis of their physicochemical properties, conserved motifs, gene structure, and gene architecture using bioinformatic methodologies. To establish phylogenetic relationships, Arabidopsis MYB family genes were employed, resulting in the categorization of JrMYB genes into 17 distinct subgroups. The chromosomal distribution of these 126 JrMYB genes across 16 walnut chromosomes was elucidated through chromosomal localization. Results from interspecies covariance analysis revealed a close relationship between walnut and pecan, with a covariance analysis of the 126 JrMYB genes identifying 50 family members displaying covariance within the walnut species. Cis-acting component analysis indicated that the promoters of these 126 JrMYB genes primarily contained four types of cis-acting components, each of which was predicted to play various roles in walnut growth and development.
By employing correlation analysis, gene expression profiling, phylogenetic systematics, and qRT-PCR analysis, we identified 3 JrMYB genes that are likely involved in fatty acid synthesis and constructed a transcription regulatory network. Collectively, the findings of this study provide a solid foundation for the functional identification and mechanistic study of MYB gene regulation in fatty acid biosynthesis in walnuts, as well as for enhancing the content of unsaturated fatty acids in walnuts.
Acknowledgement: Not applicable.
Funding Statement: This research was funded by the Shannan City Science and Technology Plan Project (2022BJKJJHXM-001) and Science and Technology Project of Tibet Autonomous Region (XZ202201YD0005C).
Author Contributions: Conceptualization, Dongxue Su and Feng Xu; methodology, Dongxue Su; validation, Jiarui Zheng, Qijian Wang, Weiwei Zhang and Feng Xu; formal analysis, Shuyuan Zhang; investigation, Luxin Feng; resources, De Qiong and Danzeng Quzhen; data curation, Yuwei Yi; writing—original draft preparation, Dongxue Su; writing—review and editing, Qijian Wang and Jiarui Zheng; project administration, Feng Xu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data are available in this article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.055350.
References
1. Ni ZJ, Zhang YG, Chen SX, Thakur K, Wang S, Zhang JG. Exploration of walnut components and their association with health effects. Crit Rev Food Sci Nutr. 2022;62:5113–29. doi:10.1080/10408398.2021.1881439. [Google Scholar] [PubMed] [CrossRef]
2. Lunn J, Theobald HE. The health effects of dietary unsaturated fatty acids. Nutr Bulletin. 2006;31:178–224. doi:10.1111/j.1467-3010.2006.00571.x. [Google Scholar] [CrossRef]
3. Tutunchi H, Ostadrahimi A, Saghafi-Asl M. The effects of diets enriched in monounsaturated oleic acid on the management and prevention of obesity: a systematic review of human intervention studies. Adv Nutr. 2020;11:864–77. doi:10.1093/advances/nmaa013. [Google Scholar] [PubMed] [CrossRef]
4. Gao P, Liu R, Jin Q, Wang X. Comparative study of chemical compositions and antioxidant capacities of oils obtained from two species of walnut: Juglans regia and Juglans sigillata. Food Chem. 2019;279:279–87. doi:10.1016/j.foodchem.2018.12.016. [Google Scholar] [PubMed] [CrossRef]
5. Schulze MB, Minihane AM, Saleh RNM, Risérus U. Intake and metabolism of omega-3 and omega-6 polyunsaturated fatty acids: nutritional implications for cardiometabolic diseases. The Lancet Diabetes Endocrinol. 2020;8:915–30. doi:10.1016/S2213-8587(20)30148-0. [Google Scholar] [PubMed] [CrossRef]
6. Zhao EP, Cheng YF, Yang X. Research progress of MYB transcription factors in Solanaceae plants. Mol Plant Breed. 2021;19:1522–30. [Google Scholar]
7. Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–81. doi:10.1016/j.tplants.2010.06.005. [Google Scholar] [PubMed] [CrossRef]
8. Lin YX, Yu Q, Feng MC, Lin JY, Zhao MM, Ou QY, et al. Cloning and expression analysis of an MYB transcription factor gene ScMYB52-1 from sugarcane. Chin J Trop Crops. 2022;43:1328–37 (In Chinese). [Google Scholar]
9. Geng P, Zhang S, Liu JY, Zhao CH, Wu J, Cao YP, et al. MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol. 2020;182:1272–83. doi:10.1104/pp.19.01070. [Google Scholar] [PubMed] [CrossRef]
10. Huo D, Liu X, Zhang Y, Duan J, Zhang Y, Luo J. A novel R2R3-MYB transcription factor PqMYB4 inhibited anthocyanin biosynthesis in Paeonia qiui. Int J Mol Sci. 2020;21:5878. doi:10.3390/ijms21165878. [Google Scholar] [PubMed] [CrossRef]
11. Li Y, Chen X, Wang J, Zou G, Wang L, Li X. Two responses to MeJA induction of R2R3-MYB transcription factors regulate flavonoid accumulation in Glycyrrhiza uralensis Fisch. PLoS One. 2020;15:e0236565. doi:10.1371/journal.pone.0236565. [Google Scholar] [PubMed] [CrossRef]
12. Lee HG, Kim H, Suh MC, Kim HU, Seo PJ. The MYB96 transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in Arabidopsis seeds. Plant Cell Physiol. 2018;59:1432–42. doi:10.1093/pcp/pcy073. [Google Scholar] [PubMed] [CrossRef]
13. Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 2011;23:1138–52. doi:10.1105/tpc.111.083485. [Google Scholar] [PubMed] [CrossRef]
14. To A, Joubès J, Thueux J, Kazaz S, Lepiniec L, Baud S. AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J. 2020;103:660–76. doi:10.1111/tpj.14759. [Google Scholar] [PubMed] [CrossRef]
15. Song CB, Yang YY, Yang TW, Ba LJ, Zhang H, Han YC, et al. MaMYB4 recruits histone deacetylase MaHDA2 and modulates the expression of ω-3 fatty acid desaturase genes during cold stress response in banana fruit. Plant Cell Physiol. 2019;60:2410–22. doi:10.1093/pcp/pcz142. [Google Scholar] [PubMed] [CrossRef]
16. Gou YY, Cheng SY, Ye JB, Zhou X, Xu F, Chen ZX, et al. Comparative transcriptome analysis reveals the potential molecular mechanism involved in fatty acids biosynthesis of Juglans regia. Sci Hortic. 2020;269:109388. doi:10.1016/j.scienta.2020.109388. [Google Scholar] [CrossRef]
17. Ge WY, Luo ML, Sun HJ, Wei BD, Zhou X, Zhou Q, et al. The CaMYB340 transcription factor induces chilling injury in post-harvest bell pepper by inhibiting fatty acid desaturation. Plant J. 2022;111:800–18. doi:10.1111/tpj.15854. [Google Scholar] [PubMed] [CrossRef]
18. Khan K, Kumar V, Niranjan A, Shanware A, Sane VA. JcMYB1, a Jatropha R2R3MYB transcription factor gene, modulates lipid biosynthesis in transgenic plants. Plant Cell Physiol. 2019;60:462–75. doi:10.1093/pcp/pcy223. [Google Scholar] [PubMed] [CrossRef]
19. Yang K, Wu J, Wang X, Li H, Jia P, Luan H, et al. Genome-wide characterization of the mitogen-activated protein kinase gene family and their expression patterns in response to drought and colletotrichum gloeosporioides in walnut (Juglans regia). Plants. 2023;12:586. doi:10.3390/plants12030586. [Google Scholar] [PubMed] [CrossRef]
20. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. doi:10.1006/meth.2001.1262. [Google Scholar] [PubMed] [CrossRef]
21. Orsavova J, Misurcova L, Vavra AJ, Vicha R, Mlcek J. Fatty acids composition of vegetable oils and its contribution to dietary energy intake and dependence of cardiovascular mortality on dietary intake of fatty acids. Int J Mol Sci. 2015;16:12871–90. doi:10.3390/ijms160612871. [Google Scholar] [PubMed] [CrossRef]
22. Zhang JP, Zhang WT, Ji FY, Qiu J, Song XB, Bu DH, et al. A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol J. 2020;18:1848–50. doi:10.1111/pbi.13350. [Google Scholar] [PubMed] [CrossRef]
23. Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi:10.1186/1471-2229-12-106. [Google Scholar] [PubMed] [CrossRef]
24. Hou XJ, Li SB, Liu SR, Hu CG, Zhang JZ. Genome-wide classification and evolutionary and expression analyses of citrus MYB transcription factor families in sweet orange. PLoS One. 2014;9:e112375. doi:10.1371/journal.pone.0112375. [Google Scholar] [PubMed] [CrossRef]
25. Zhao P, Li Q, Li J, Wang L, Ren Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol Genet Genom. 2014;289:1183–207. doi:10.1007/s00438-014-0879-4. [Google Scholar] [PubMed] [CrossRef]
26. Zeng J, Long YQ, Fu XS, Wang L, Liu ZX, Zhou RB, et al. Mining and identification of members of MYB transcription factor family in Lonicera macranthoides. China J Chin Mater Med. 2023;48:2103–15 (In Chinese). doi:10.19540/j.cnki.cjcmm.20230115.103. [Google Scholar] [PubMed] [CrossRef]
27. Yang JT, Li H, Zhao H, Long H, Zhang YS, Liu XH. Excavation and identification of members of MYB transcription factor family in Lysimachia christinae. Chin Tradit Herb Drugs. 2022;53:5149–58 (In Chinese). [Google Scholar]
28. Tuğrul M, Paixão T, Barton NH, Tkačik G. Dynamics of transcription factor binding site evolution. PLoS Genet. 2015;11:e1005639. doi:10.1371/journal.pgen.1005639. [Google Scholar] [PubMed] [CrossRef]
29. Li Z, Schulz MH, Look T, Begemann M, Zenke M, Costa IG. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019;20:45. [Google Scholar] [PubMed]
30. Wang J, Liu Y, Tang BQ, Dai XZ, Xie LL, Liu F, et al. Genome-wide identification and capsaicinoid biosynthesis-related expression analysis of the R2R3-MYB gene family in Capsicum annuum L. Front Genet. 2020;11:598183. doi:10.3389/fgene.2020.598183. [Google Scholar] [PubMed] [CrossRef]
31. Ruprecht C, Proost S, Hernandez-Coronado M, Ortiz-Ramirez C, Lang D, Rensing AS, et al. Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 2017;90:447–65. doi:10.1111/tpj.13502. [Google Scholar] [PubMed] [CrossRef]
32. Sun W, Ma Z, Chen H, Liu M. MYB gene family in potato (Solanum tuberosum L.): genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int J Mol Sci. 2019;20:4847. doi:10.3390/ijms20194847. [Google Scholar] [PubMed] [CrossRef]
33. Qu YF, Shen X, Jiang DY, Li YG, Sun ZS, Chen BQ, et al. Genome-wide identification and expression analysis of the MYB transcription factor family in Prunus campanulata. Mol Plant Breed. 2022;1–12. [Google Scholar]
34. Du JH, Zhang QQ, Hou SJ, Chen J, Meng JQ, Wang C, et al. Genome-wide identification and analysis of the R2R3-MYB gene family in theobroma cacao. Genes. 2022;13:1572. doi:10.3390/genes13091572. [Google Scholar] [PubMed] [CrossRef]
35. Ren MH, Zhang YP, Xu T, Zhu HS, Cen HF. Identification and expression analyses of R2R3-MYB subfamily in alfalfa under drought stress. Acta Agrestia Sinica. 2023;31:972–83. doi:10.11733/j.issn.1007-0435.2023.04.005. [Google Scholar] [CrossRef]
36. Park MY, Kang J, Kim SY. Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol Cells. 2011;31:447–54. doi:10.1007/s10059-011-0300-7. [Google Scholar] [PubMed] [CrossRef]
37. Zhang Y, Cao G, Qu LJ, Gu H. Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genet Genom. 2009;36:99–107. doi:10.1016/S1673-8527(08)60096-X. [Google Scholar] [PubMed] [CrossRef]
38. Pandey A, Misra P, Bhambhani S, Bhatia C, Trivedi PK. Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci Rep. 2014;4:5018. doi:10.1038/srep05018. [Google Scholar] [PubMed] [CrossRef]
39. Li D, Jin CY, Duan SW, Zhu YN, Qi SH, Liu KG, et al. MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 2017;173:1211–25. doi:10.1104/pp.16.01634. [Google Scholar] [PubMed] [CrossRef]
40. Duan SW, Jin CY, Li D, Gao CH, Qi SH, Liu K, et al. MYB76 inhibits seed fatty acid accumulation in Arabidopsis. Front Plant Sci. 2017;8:226. doi:10.3389/fpls.2017.00226. [Google Scholar] [PubMed] [CrossRef]
41. Milioni D, Katsareli E, Valassakis C. Molecular and functional characterization of Arabidopsis thaliana VPNB1 gene involved in plant vascular development. Plant Sci. 2018;11:11–9. doi:10.1016/j.plantsci.2018.09.006. [Google Scholar] [PubMed] [CrossRef]
42. Aboussekhra A, Thoma F. Nucleotide excision repair and photolyase preferentially repair the nontranscribed strand of RNA polymerase III-transcribed genes in Saccharomyces cerevisiae. Genes Dev. 1998;12:411–21. doi:10.1101/gad.12.3.411. [Google Scholar] [PubMed] [CrossRef]
43. He Y, Gan S. Identical promoter elements are involved in regulation of the OPR1 gene by senescence and jasmonic acid in Arabidopsis. Plant Mol Biol. 2001;47:595–605. doi:10.1023/a:1012211011538. [Google Scholar] [PubMed] [CrossRef]
44. Kroj T, Savino G, Valon C, Giraudat J, Parcy F. Regulation of storage protein gene expression in Arabidopsis. Development. 2003;130:6065–73. doi:10.1242/dev.00814. [Google Scholar] [PubMed] [CrossRef]
45. Angeles-Núñez JG, Tiessen A. Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J Plant Physiol. 2011;168:1891–900. doi:10.1016/j.jplph.2011.05.003. [Google Scholar] [PubMed] [CrossRef]
46. Millard PS, Kragelund BB, Burow M. R2R3 MYB transcription factors—functions outside the DNA-binding domain. Trends Plant Sci. 2019;24:934–46. doi:10.1016/j.tplants.2019.07.003. [Google Scholar] [PubMed] [CrossRef]
47. Zhang Y, Cao G, Qu LJ, Gu H. Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep. 2009;28:337–46. doi:10.1007/s00299-008-0644-4. [Google Scholar] [PubMed] [CrossRef]
48. Xu X, Li M, Zou JX, Zheng YS, Li DD. EgMYB108 regulates very long-chain fatty acid (VLCFA) anabolism in the mesocarp of oil palm. Plant Cell Rep. 2022;41:1449–60. doi:10.1007/s00299-022-02868-9. [Google Scholar] [PubMed] [CrossRef]
49. Xu X, Zheng YS, Li DD. Identification and expression analysis of MYB transcription factor family in pericarp of Elaeis guineensis Jacq. Mol Plant Breed. 2021;19(19):6277–89. [Google Scholar]
50. Xing HT, Shi JY, Yin SQ, Wu QH, Lv JL, Li HL. The MYB family and their response to abiotic stress in ginger (Zingiber officinale Roscoe). BMC Genom. 2024;25:460. doi:10.1186/s12864-024-10392-1. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools