 Open Access
Open Access
ARTICLE
Integrated GC-MS and LC-MS-Based Untargeted Metabolomics Reveals Diverse Metabolites in Fermented Pine Needles
1 Hunan Provincial Institute of Product and Goods Quality Inspection, Changsha, 410004, China
2 College of Food Science and Engineering, Central South University of Forestry and Technology, Changsha, 410004, China
* Corresponding Author: Wenhua Zhou. Email:
Phyton-International Journal of Experimental Botany 2024, 93(9), 2367-2382. https://doi.org/10.32604/phyton.2024.047949
Received 23 November 2023; Accepted 20 February 2024; Issue published 30 September 2024
Abstract
The diversity of metabolites produced in fermented pine needles at different stages of fermentation has rarely been investigated. In the present study, untargeted metabolomic analysis using GC-MS and LC-MS was performed to detect metabolites in fermented pine needles at different fermentation stages. A total of 30 samples of pine needles fermented first with yeast (Saccharomyces cerevisiae) and then with a mixed bacterial culture of Lactobacillus fermentum CECT5716 and Bifidobacterium Breve M16V, were used to detect differential metabolites at different stages of fermentation. Pearson’s correlation analysis was used to determine correlations between metabolites and key microbial communities. A total of 708 differential metabolites (430 from LC-MS and 278 from GC-MS analysis) were identified. The PCA and OPLS-DA revealed distinct differences between metabolites at different fermentation stages. Key differential metabolites identified through GC-MS analysis included; Phosphoric acid, D-Fructose, 2-O-alpha-mannosyl-D-glycerate, 1,3-dihydroxyacetone dimer, Galactosylglycerol, 2-Isopropylmalic acid, alpha-D-Galactose, Citrate, 4-Hydroxycinnamic acid, and Shikimate. Similarly, key differential metabolites identified through LC-MS included; 2-Phenlyethanol, Dimethlglycine, 2-Hydroxybenzaldehyde, 3-Aminoisobutanoic acid, p-Cresol, Triethylamine, 2-Ketobutyric acid, Cytosine, Benzaldehyde, and Creatinine. Annotation of differential metabolites to KEGG pathway enrichment analysis revealed the association of these metabolites with phenylpropanoid, flavonoid, and secondary metabolite biosynthesis. Furthermore, the results showed that three bacterial (Firmicutes, Actinobacteria, and Lactobacillus) and three fungal genera (Penicillium, Candida, and Basidiomycota) significantly correlated with differential metabolites showing synergistic effects. Our study reveals a comprehensive comparison of metabolites at different fermentation stages and provides practical insights into the mechanism of metabolite enrichment in fermented pine needles.Keywords
Supplementary Material
Supplementary Material FilePinus massoniana is a species of coniferous evergreen needle-leaved trees (family Pinaceae), widely distributed in East Asia including Japan, Korea, North-Eastern China, and South-East Russia [1]. Various parts of the pine tree, such as pine needles, cones, cortex, and pollen, have been used as folk medicine or food [2,3]. Pine needles contain diverse phytochemical compounds including phenolics, cinnamic acids, terpenoids, carboxylic acids, fatty acids, and flavonoids [4]. Owing to these bioactive compounds, pine needles possess a wide array of biological activities such as antimicrobial, antimutagenic, antioxidant, anti-neurodegenerative, and cardioprotective properties [5–7]. In addition to secondary metabolites, pine needles also contain some significant minerals (calcium 28 mg/100 g), amino acids (glutamic acid, lysine, phenylalanine, and leucine), and vitamins (niacin, beta carotene, riboflavin, and thiamine) [8]. Despite of rich nutrient profile and bioactive compounds, substantially high concentration of condensed tannins and antinutritional factors in pine needles may interfere with nutrient absorption and protein digestibility in the gut [9]. In this regard, fermentation with probiotics is one of the best ways to remove antinutritional factors (e.g., tannins) and improve the flavor, palatability, digestibility, and nutrient availability of pine needles [10,11]. Fermentation can also enhance the shelf life of food while improving biochemical activities like antioxidant and antibacterial properties [11].
Fermented pine needle drink has been utilized as a functional beverage in the countries of Asia-Pacific region particularly Taiwan, Korea, and China owing to its various biological properties. Studies have reported that fermented pine needle extract is rich in phenolics, essential oils, flavonoids, and terpenoids, with potential health-promoting potential [1,10]. Self-fermented pine needle extract has been shown to decrease blood lipid levels and regulate gastrointestinal motility [1]. Additionally, pine needles are used as a health-improving agent in teas, extracts, and some alcoholic beverages [12]. Ethanolic extracts of pine needles have also shown antidepressant activities and hence can be used to amend unidentified clinical symptoms, such as fatigue, anxiety, depression, and sleep disorders [13–15]. Moreover, the potent antioxidant activities of pine needles have been widely reported which are responsible for their excellent free radical scavenging potential and cytotoxicity against cancer cells [8,16]. These antioxidant properties of pine needles are mainly attributed to their rich content of proanthocyanidins, catechins, and essential oils, which are considered to be potent antioxidants and thus may play an important role as anti-aging compounds and in the prohibiting of chronic diseases including cardiovascular disorders, cancer, diabetes, and hypertension [11,17,18]. Therefore, fermented pine needles have been traditionally processed and used to treat a variety of diseases.
It is worth mentioning that pine needles have gained much attention in recent years for their use as an ingredient in herbal teas and various food recipes, owing to their potential health benefits [18]. To better exploit the potential of pine needles as a functional food and its nutraceutical value, it is imperative to comprehensively analyze the diversity of metabolites of fermented pine needles. Various fermentation conditions including probiotic strains, temperature, and duration of incubation substantially affect the quantity and quality of metabolites in pine needles. Hence metabolomic study of fermented pine needles is essentially required to comprehensively analyze metabolites at a given time or under given conditions of fermentation [19]. Spectral analysis of metabolites can be performed by gas chromatography or liquid chromatography combined with mass spectrometry (GC-MS and LC-MS, respectively) [20]. In this regard, nontargeted metabonomic analysis is a desirable approach to get as much information as possible from biological samples like fermented pine needles [21]. Recently, a study has determined the total phenols and flavonoid content of pine needles fermented with Lactobacillus plantarum SK4315, Saccharomyces cerevisiae, and their co-culture through ELISA quantification [19]. But still, no study has performed metabolomic characterization of fermented pine needles to identify individual compounds which is essentially required to understand their potential health benefits and potential applications.
Therefore, in the present study, we evaluated the variation in metabolites at different stages of the fermentation process of pine needles through untargeted GC-MS and LC-MS analysis. Additionally, the correlation of metabolites with the microbiome was also investigated. The present study envisioned to provide novel insights into the mechanism of improvement in the quality of fermented pine needles and their potential utility for developing traditional fermented foods.
2.1 Preparation of Pine Needle Fermentation Extract
Pine needle fermentation extract was obtained as described in our previous study [22]. Briefly, the fresh pine needles were collected from pine trees (Pinus massoniana) at Cha’en Temple, Shaoshan, China. The Pinus massoniana trees in Shaoshan’s Cha’en Temple were harvested to collect fresh pine needles. The needles were ground to a fineness of 0.5 to 1.2 mm and air-dried for three days in dry shade. This was followed by a 1:10 ratio of the needles to water. The mixture was added and evenly dissolved, consisting of 22% brown sugar and 33% rock sugar. Finally, 0.8% active dry yeast was added, and the mixture was left to incubate for 18 days at 22°C. After that, a mixed bacterial solution containing 6% Lactobacillus plantarum and 1.0:1.5 Bifidobacterium Breve M16V was added to inoculate it. Following 7 days at 37°C, the fermentation process was shifted to room temperature (about 25°C) and kept for a total of 180 days. The chemicals used in the creation of the extract were purchased by Tianjin Jinteng Experimental Equipment Co., Ltd. (Tianjin) in China. The chemicals included glucose and bovine serum albumin, both of which had purities of more than 98%. Shanghai Aladdin Biochemical Technology Co., Ltd., Shanghai, China provided the 2-Chloro-L-phenylalanine (internal standard material), d3H2O (Millipore, St Louis, MO, USA), and methanol (purity > 98%, Thermo Fisher, Waltham, MA, USA).
2.2 Sampling of Fermented Pine Needle Extract
Starting from day 0 of fermentation with unfermented pine needles, samples of pine needle fermentation juice were collected regularly on day 1 (GD1), 3 (GD3), 9 (GD9), and 18 (GD18) of yeast fermentation and after mixed bacterial fermentation at day 2nd (HD2) and 7th (HD7) for metabolite detection through GC-MS analysis. The same samples were also used for the detection of metabolites through LC-MS analysis (coded as LD1, LD3, LD9, and LD18 for yeast fermentation and LHD2 and LHD7 for mixed bacterial fermentation). These samples were stored at −80°C until used for further processing. To ensure the reproducibility of the experiment, approximately 5 kg of pine needles were loaded into each fermentation tank. During the fermentation process, a total of 5 parallel groups were set up, and 3 samples were randomly selected for analysis. A total of 5 samples were analyzed for each fermentation stage.
The pine needle fermentation extract was thawed, and then well mixed for one minute before being transferred to a two-milliliter centrifuge tube and concentrated until it was dry. Subsequently, 500 μL of methanol (stored at −20°C) was added to the sample tube, vortexed for 1 min, and centrifuged at 12,000 rpm at 4°C for 10 min. The sample was then concentrated until it was dry after the whole supernatant was transferred to a fresh 2 mL centrifuge tube. The dried material was then dissolved in 150 μL of a 4-ppm solution of 2-chloro-L-phenylalanine made with 80% methanol, and the resulting supernatant was then passed through a 0.22 μM membrane filter. For LC-MS detection, the filtrate was subsequently introduced to the detection bottle. A Vanquish UHPLC System (Thermo Fisher Scientific, Waltham, MA, USA) and an ACQUITY UPLC® HSS T3 (150 mm × 2.1 mm, 1.8 µm) column (Waters, Milford, MA, USA) were used for the LC-MS analysis. At a temperature of 40°C, a 2 μL sample was injected into the column at a flow rate of 0.25 mL/min. The mobile phase in the positive ion mode was made up of 0.1% formic acid in acetonitrile (B2) and 0.1% formic acid in water (A2). The gradient elution procedure was used as reported previously [23]. Using an electrospray ion source (ESI)-equipped Q Exactive Focus equipment (Thermo Fisher Scientific), mass spectrometry was used to identify metabolites. A capillary temperature of 325°C, a sheath gas flow rate of 30 arb, and an auxiliary gas flow rate of 10 arb were the additional parameters. Full-scan MS analysis (m/z 100–1000) yielded a resolution of 70,000 FWHM for all metabolites. The resolving power for dd-MS2 was configured at 17,500 FWHM. A collision energy of 30 eV was used for collision-induced dissociation (CID), and superfluous MS/MS information was eliminated via dynamic exclusion [24].
About 100 μL of the sample and 1 mL of solution (containing acetonitrile, isopropanol, and water at 3:3:3:2 v/v/v) were combined in a 2 mL EP tube (−20°C) to analyze the metabolites in the pine needle extract. After 30 s of vortex, the mixture was exposed to ultrasonication for 5 min. After centrifuging the mixture for two minutes at 12,000 rpm, 500 μL of the supernatant was taken in a 2 mL EP tube. After that, the sample was concentrated under vacuum until it was totally dry, which usually took eight to ten hours. Subsequently, a 20 mg/mL methoxy pyridine solution was added to 80 μL of the dried and concentrated sample, stirred for 30 s, and incubated for 60 min at 60°C. After 30 s of mixing, 100 μL of BSTFA-TMCS (99:1) derivatization reagent was incubated at 70°C for 90 min. After that, the mixture was centrifuged for three min at 14,000 rpm. Approximately 90 to 100 μL of the supernatant was transferred to the detection vial following centrifugation. For testing, the material was kept in a sealed cup, and the Gas Chromatography time-of-flight detection was finished in a day.
To separate the derivatives, gas chromatography was performed on DB-5MS capillary column (Agilent J&W Scientific, Folsom, CA, USA, 30 m × 250 m i.d., 0.25 m film thickness), with a constant-flow (1 mL/min) of helium. Using an automated injector with a split ratio of 1:10, 1 µL of sample was injected. Using an automated injector with a split ratio of 1:10, 1 µL of sample was injected. The transmission line and ion source temperatures were used as reported previously [25].
2.5 Data Processing and Multivariate Analysis
The original raw data was converted to mzXML format using MSConvert in the ProteoWizard program (v3.0.8789) [26]. After that, it was processed using XCMS [27] for features, alignment, and retention time correction. Metabolites were identified based on precise molecular mass and MS/MS data by comparing with HMDB [28], massbank [29], LipidMaps [30], mzcloud [31], and KEGG [32] databases. Robust LOESS signal correction (QC-RLSC) [24] was employed for data normalization to address any systematic bias. To ensure precise metabolite identification, only ion peaks with relative standard deviations in quality control less than 30% were retained after normalization. Metabolome data were analyzed by the ropls R package as reported previously [33]. MetaboAnalyst was used to identify differential metabolites [34]. To improve the overall understanding of the detected metabolites, pathway enrichment, and topological studies were integrated. The KEGG pathway was then mapped to the detected metabolites, which made it easier to comprehend higher-level systemic biological activities. Using the KEGG Mapper program, the metabolites and associated pathways were visualized.
Differential metabolites were defined as those that matched the following criteria: VIP > 1, p-value < 0.05, fold change ≥ 2, or FC ≤ 0.5. When the ratio criteria of x/n > y/N is met, it indicates enrichment in the metabolic pathway enrichment analysis. Additionally, if the p-value was < 0.05 of a metabolic pathway, it was considered enriched statistically.
Pearson correlation was used to elucidate the relationship between metabolites and microbiome as described in our previous study [22]. The heatmap was employed to show significant positive or negative correlations between metabolites and microbiome. Data of major bacterial and fungal genera were employed from our companion study to perform correlation [22]. The correlation between differential metabolites and microbial communities was analyzed by employing the cor function in R version 4.0.3 (R Project for Statistical Computing, Vienna, Austria). The significance of the correlation between differential metabolites was determined by cor. Mtest() function and plotted by corrplot package of R. The p-value of less than 0.05 was considered as the statistical significance.
3.1 Untargeted Metabolomic Analysis
The base peak chromatogram of negative ion mode (ESI−), and positive ion mode (ESI+), in LC/MS metabolite profiles are shown in Fig. S1. The unsupervised principal component analysis (PCA) was employed to evaluate the quality of the GC/MS and LC/MS data. The distribution of metabolic profiles for the test and QC samples in PCA is shown in Fig. 1. Results of PCA indicated overall good reproducibility of results. A clear group separation between GD1, GD3, and other fermentation stages was observed in the PCA score plots (Fig. 1). The PC1 and PC2 explained 46.4% and 11.6% of the total variance in ESI− mode, while 45.2% and 13.3% of the total variance of all samples in ESI+ mode, respectively.
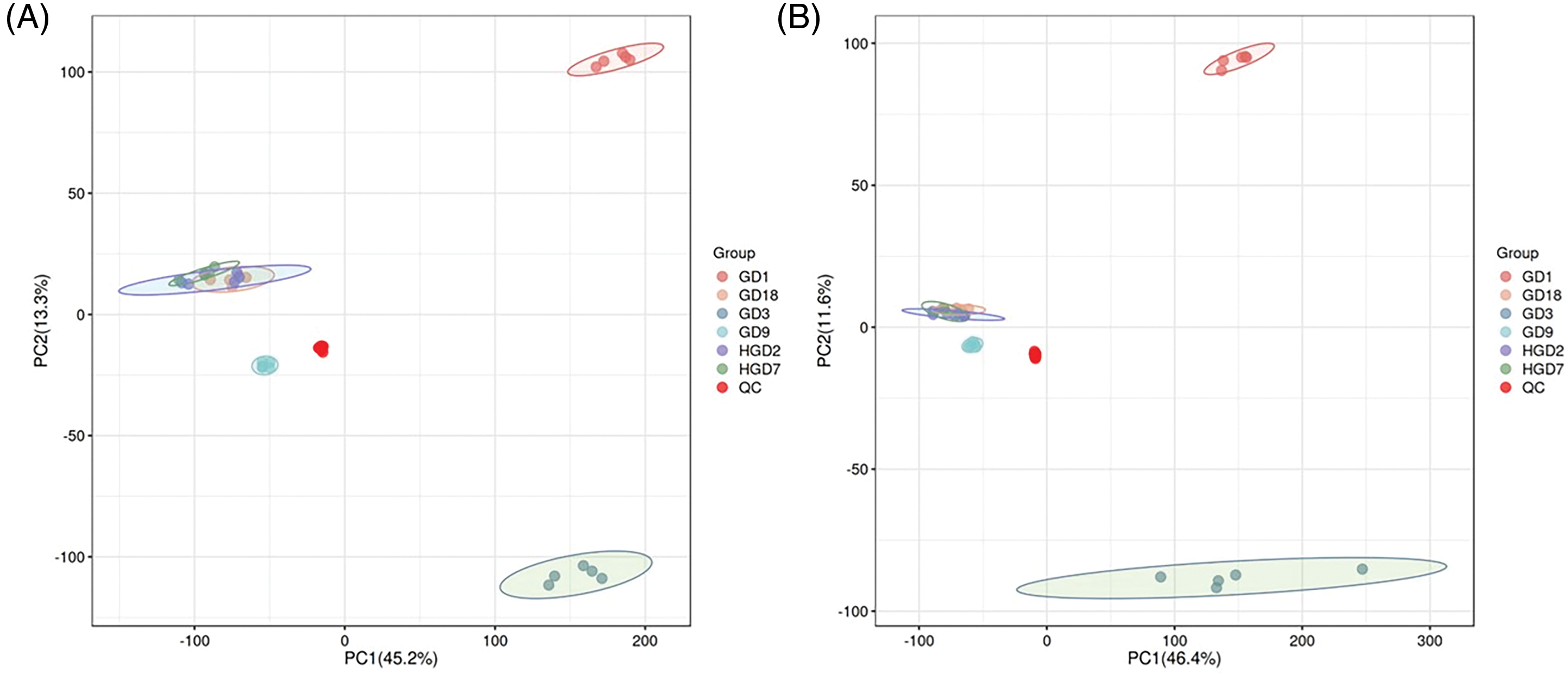
Figure 1: The PCA score scatter plot under positive ion mode (A) and negative ion mode (B)
3.2 Identification of Differential Metabolites
A supervised OPLS-DA was conducted to identify group variables. The OPLS-DA clearly separated GD1, GD3, and other fermentation stages (Fig. 2). We used VIP > 1 and p-value < 0.05 to identify differential metabolites between different fermentation stages. The results revealed a total of 708 differential metabolites identified by LC-MS (430) and GC-MS (278) analyses (Tables 1 and 2). In addition, differential metabolites identified through LC-MS and GC-MS at different stages of fermentation of pine needles are shown in Fig. S2A,B, respectively.
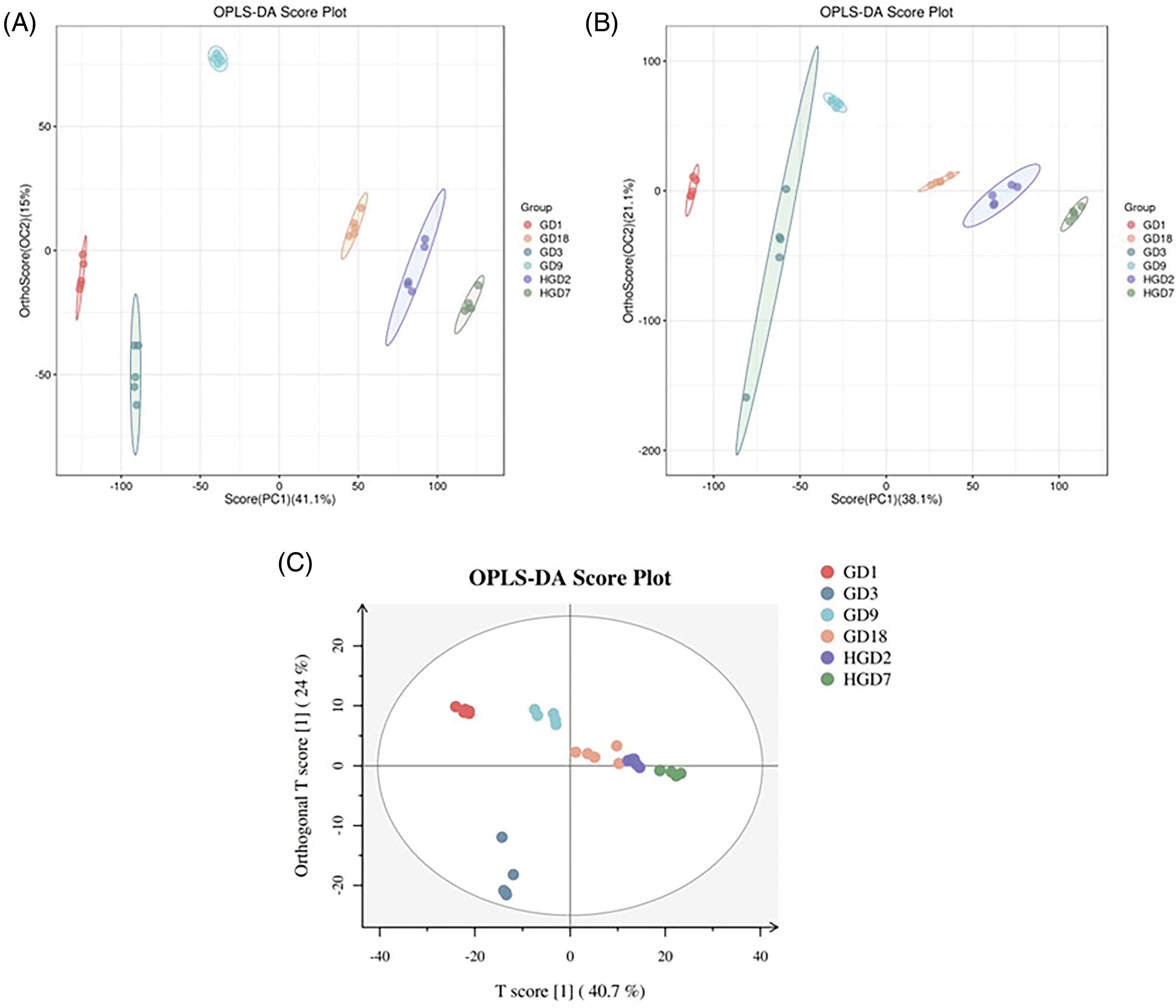
Figure 2: The OPLS-DA score scatter plot under positive ion mode (A) and negative ion mode (B) and OPLS-DA score plot (C)
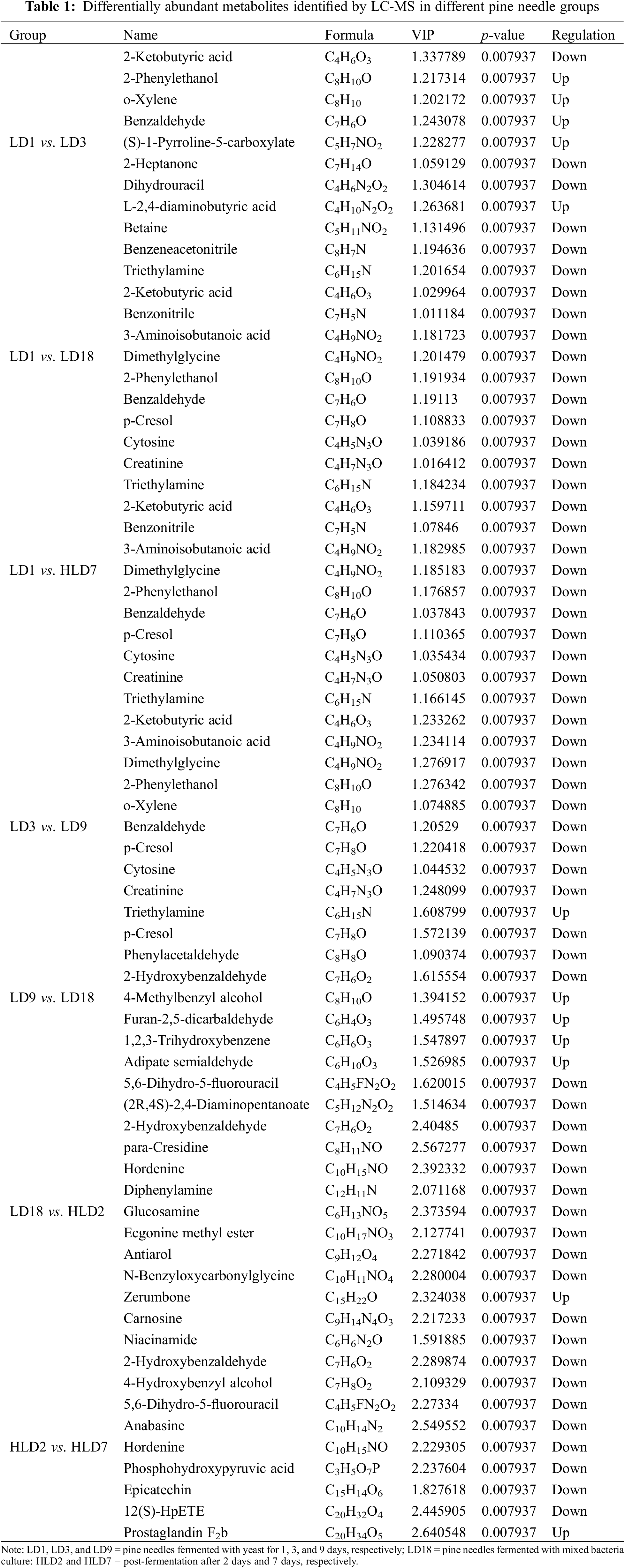
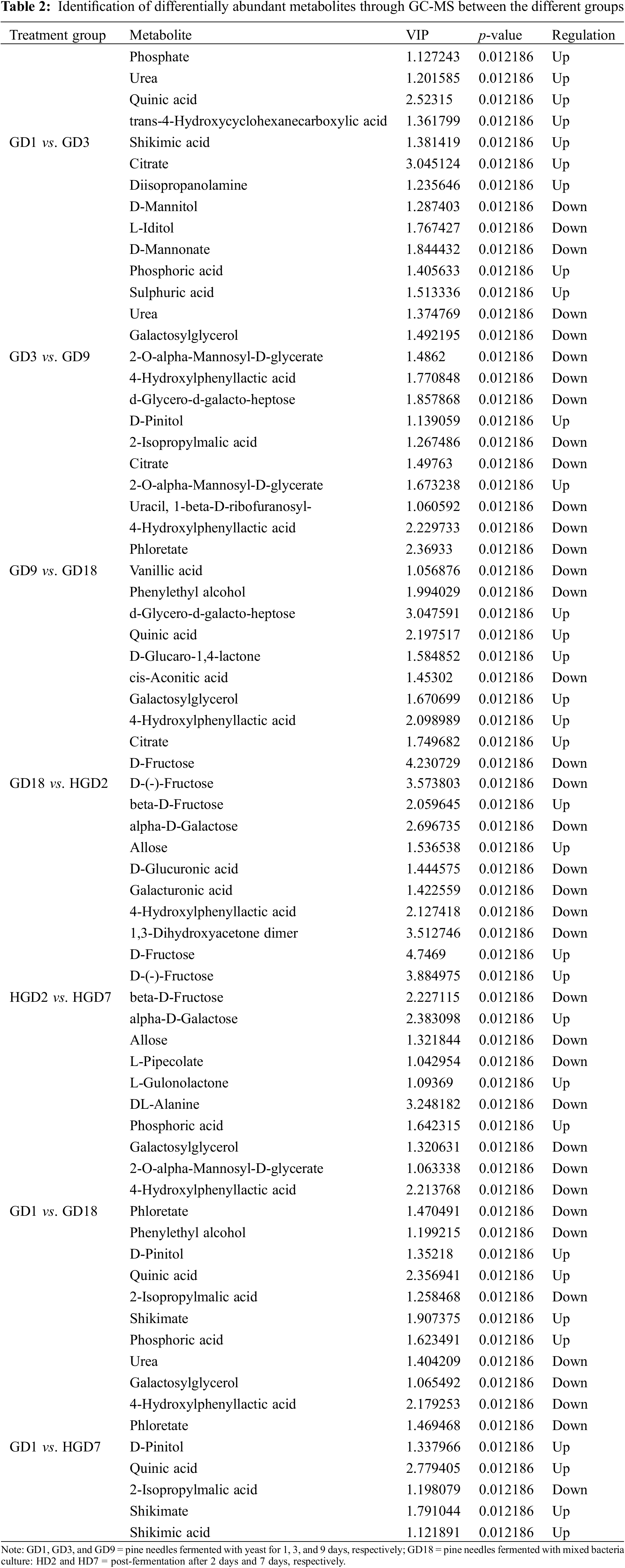
3.3 Metabolic Pathway of Differentially Abundant Metabolites
The MetaboAnalyst 4.0 was used to identify the pathways for differential metabolites among different fermentation stages through KEGG pathway analysis. All the different metabolites were annotated to the biological pathways listed in the KEGG database (Fig. 3). These differentially enriched pathways are grouped into phenylpropanoid biosynthesis, flavonoid biosynthesis, and secondary metabolite biosynthesis. Results revealed seven pathways with impact values greater than 0.1 (threshold value for enrichment and topology analysis) among these differentially enriched pathways.
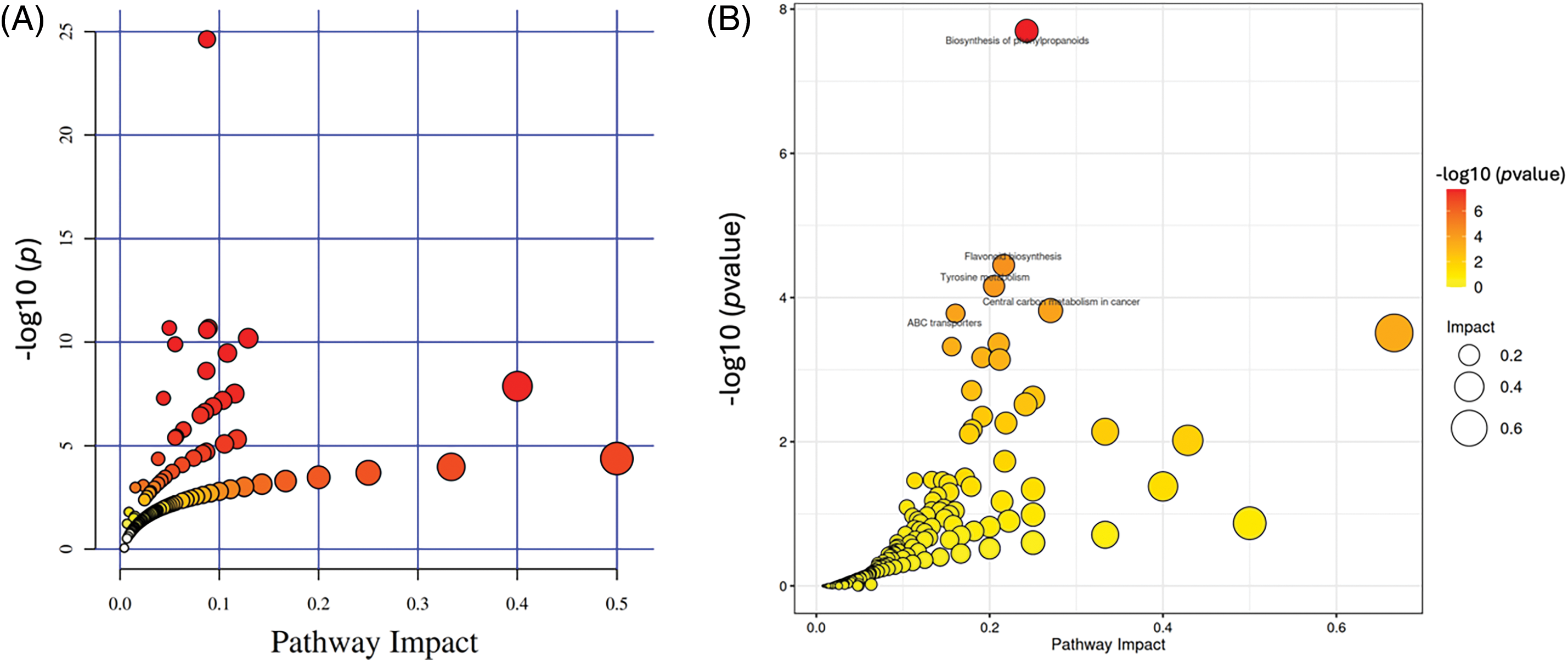
Figure 3: Summary of pathway analysis with MetaboAnalyst. (A) LC-MS; (B) GC-MS
3.4 Association of Metabolites with Microbial Communities in Fermented Pine Needles
Major differential metabolites with high VIP values (top 20) and key abundant bacteria and fungi genera (top 10) observed in fermented pine needles were selected to determine the correlation between microbial communities and metabolites (Fig. 4). Results revealed that Firmicutes, Lactobacillus, and Actinobacteria positively correlated (p < 0.05) with dimethylglycine, 2-ketobutyric acid, cyto-sine, 2-O-alpha-mannosyl-D-glycerate and 1,3-dihydroxyacetone dimer but negatively correlated (p < 0.05) with phosphoric acid. However, Proteobacteria negatively correlated with galactosylglycerol, 1,3-dihydroxyacetone dimer, 2-O-alpha-mannosyl-D-glycerate, dimethylglycine, 2-phenylethanol, creatinine, triethylamine, cytosine, and 2-ketobutyric acid, while positively correlated with phosphoric acid. Lactobacillus was positively associated with dimethylglycine, 2-ketobutyric acid, cytosine, 1,3-dihydroxyacetone dimer, and 2-O-alpha-mannosyl-D-glycerate but negatively correlated with phosphoric acid. Regarding fungal taxa, three fungal genera including Penicillium, Candida and Basidiomycota exhibited a negative correlation (p < 0.05) with the top ten metabolites identified with LC-MS while Schizoasaccharomyces and Ascomycota showed a positive correlation with these metabolites (Fig. 4D). However, Penicillium, Candida, and Basidiomycota exhibited a negative correlation with galactosylglycerol, 1,3-dihydroxyacetone dimer, 2-O-alpha-mannosyl-D-glycerate, dimethylglycine, 2-phenylethanol, creatinine, triethylamine, cytosine, and 2-ketobutyric acid, while positively correlated with phosphoric acid. Both Schizoasaccharomyces and Ascomycota positively correlated with almost all top 10 metabolites (identified through GC-MS) but negatively correlated with D-Fructose and Phosphoric acid.
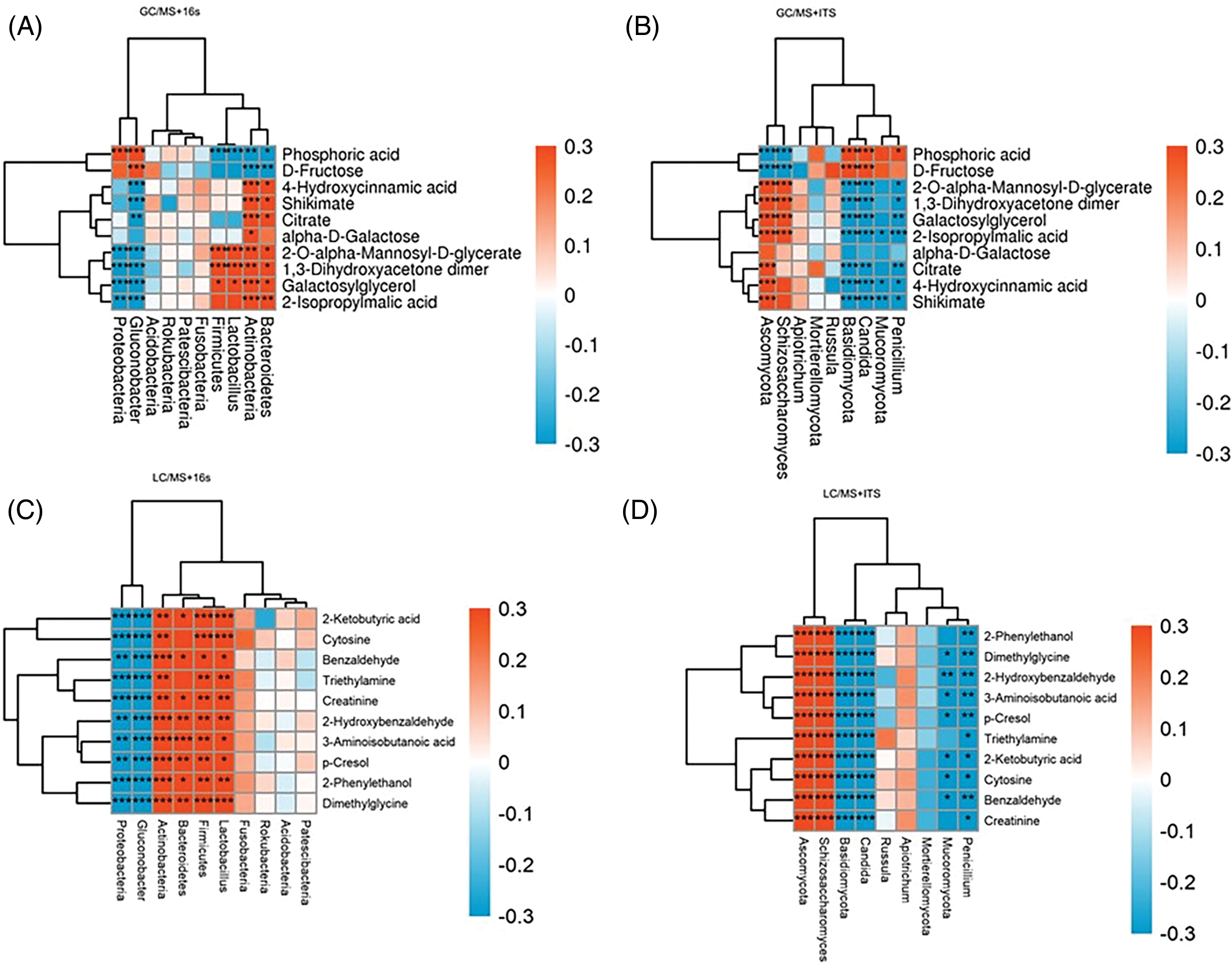
Figure 4: Correlation of bacterial (A and C) and fungi (B and D) with key metabolites of fermented pine needles identified through GC-MS and LC-MS analysis, respectively
The untargeted metabonomic analysis (through LC-MS and GC-MS) of fermented pine needles was conducted to detect a wider range of metabolites because each analytical platform can analyze different types of metabolites. Wider use of GC-MS for metabonomic analysis compared to LC-MS is attributed to available metabolite databases [31]. However, both these analyses are preferred for metabonomic analysis. In the present study, untargeted metabolomic analysis of fermented pine needles successfully detected 207 metabolites by GC-MS, and 136 metabolites by LC-MS analysis.
Differential metabolites observed in the present study are attributed to the fact that fermentation time and inoculant can greatly affect the metabolite contents as more polar phytochemicals are released during the fermentation process [19]. Moreover, different microbial strains produce diverse metabolites during fermentation depending upon the availability of respective substrates. Fermentation of pine needles with Bacillus subtilis exhibited metabolite contents rich in phenolics and flavonoids produced through various enzymatic reactions that triggered the synthesis of new phenolic compounds [10,35]. Similarly, stronger antioxidant activity for P. morrisonicola and its fermented products has been reported compared to its non-fermented counterparts owing to its rich metabolites including phenolic compounds, flavonoids, essential oils, and terpenoids. Even self-fermented pine needles have shown potential bioactivities to address hypercholesterolemia and its associated conditions owing to their rich metabolites [1,36]. A diverse range of metabolites was identified at different fermentation stages of fermented pine needles, with some metabolites having potential health-promoting effects and economic significance. These findings are in agreement with a recent study reporting a substantial increase in total polyphenolic and flavonoid contents after the fermentation of pine needles with Lactobacillus plantarum SK4315 fermentation and Saccharomyces cerevisiae SK3587 leading to higher antioxidant, antimicrobial, and enzymatic activities compared to natural fermented group [19]. It is evident from various studies that metabolite contents and biological activities largely depend on the type of fermentation, fermentation time, and inoculant owing to the enrichment of metabolite contents [11,35].
The present study revealed that differentially enriched metabolites were observed in different fermentation groups. For instance, an aromatic alcohol 2-phenylethanol (2-PE) was found upregulated in yeast fermented pine needles but it decreased with the passage of fermentation time. This compound has a characteristic rose-like odor and is an important raw material in the cosmetics, perfumery, and food industries. Smaller amounts of 2-PE are used in the food industry to enhance the taste of soft drinks, candy, cookies, and other foods [37]. Nevertheless, 2-PE is an important aroma compound found in low concentrations in fermented foods and beverages, such as cheese and wine. It is also reported that 2-PE produced by Pichia anomala can inhibit the growth of Aspergillus flavus [38,39]. In our study, 2-PE contents significantly differed among the different fermentation groups (LD1 vs. LD3, LD1 vs. LD18, LD1 vs. HLD7, LD3 vs. LD9) as its synthesis was downregulated with the passage of fermentation. Proteobacteria, Candida, and Basidiomycota negatively correlated with 2-PE contents, which may be the main reason that influences the fragrance in the different fermentation stages of fermented pine needles. The present study revealed that synthesis of Shikimic acid and Shikimate was upregulated during the fermentation as it positively correlated with Bacteroides and Actinobacteria while negatively correlated with Gluconobacter (Proteobacteria). Shikimic acid is essential to the health of gut bacteria and has been shown to modulate microbiota and NF-κB/MAPK signaling pathway to impart beneficial effects on intestinal integrity and inflammatory conditions [40,41]. Hence, the Shikimic contents of fermented pine needles are crucial for their positive health benefits.
The findings of the present study demonstrated that Lactobacillus, Firmicutes, Bacteroidetes and Actinobacteria positively correlated with key metabolites including 2-Phenlyethanol, Dimethlglycine, 2-Hydroxybenzaldehyde, 3-Aminoisobutanoic acid, p-Cresol, Triethylamine, 2-Ketobutyric acid, Cytosine, Benzaldehyde, and Creatinine. It means that fermentation with Lactobacillus along with other bacteria can increase the contents of major metabolites leading to major bioactivities as reported earlier [1,10]. Moreover, these bacteria revealed a negative association with D-Fructose and Phosphoric acid. Lactobacillus degrades carbohydrates to produce lactic acid which is a precursor for 3-hydroxy-2-butanone and other acids. Additionally, owing to their strong fermentation ability of Lactobacillus, it derives volatile and non-volatile flavor contents with the release and degradation of free amino acids in fermented products. The amino acid transferases produced by Lactobacillus, can convert phenylalanine to phenylacetaldehyde and benzaldehyde; and convert leucine to generate 3-methyl-1-butanol, etc. These metabolites are major flavor compounds produced during the fermentation of pine needles. Moreover, controlled fermentation with Lactobacillus has been shown to increase the antimicrobial activity of Chinese chives, which is attributed to the production of metabolite concentration [11]. Like bacteria strains mentioned above, Candida and Basidiomycota also showed a positive association with key metabolites in fermented pine needles. These fungi catalyze the formation of esters. These substances then interact to produce the distinct fermented flavor of pine needles [42]. In conclusion, diverse metabolites produced in the fermented pine needles is attributed to the harmonious effect of bacteria and fungi.
In the present study, KEGG data illustrated that the ‘biosynthesis of phenylpropanoid, flavonoids, and secondary metabolites’ were the core functional pathways in fermented pine needles. Phenylpropanoid is the largest class of natural products, including flavonoids, anthocyanins, lignans, and tannins. It has a variety of functions, including photosynthesis, nutrient absorption, growth regulation, cell division, maintenance of redox homeostasis, and biological and abiotic stress reactions [43]. Flavonoids is a class of compounds abundantly present in plants kingdom and are responsible to regulate growth and development of plants in addition to other bioactivities with potential industrial implications [44]. Anthocyanins, chalcones aurones, and flavonols are major pigments (which endow plants with various colors) also belong to flavonoids class. Flavonoids possess a variety of uses in daily living, including food and medicine. Proanthocyanidins and anthocyanins, in particular, play an important role as edible pigments and flavor-modifying agents in wine and food [45]. Beyond their culinary applications, plant flavonoids have shown promise in slowing down the aging process in several body systems, including the skin, neurological system, reproductive system, liver, and immunological organs when employed as active ingredients. Furthermore, they are involved in the prevention of illnesses including Alzheimer’s, breast cancer, osteoporosis, and cardiovascular disorders [46]. Our findings indicate that differential metabolites observed in fermented pine needles are attributed to the interaction between the number of microorganisms increased by fermentation and fermenting microorganisms in pine needles at different fermentation stages.
The present study concluded that untargeted metabolomic analysis identified the differential metabolite contents of the fermented pine needles from LC-MS (430 metabolites) and GC-MS (278 metabolites) analyses. Major metabolites identified were associated with pathways including biosynthesis of phenylpropanoids, flavonoids, and secondary metabolites. Furthermore, the results demonstrated that major bacterial (Firmicutes, Proteobacteria, and Lactobacillus) and fungal (Penicillium, Candida, and Basidiomycota) taxa significantly correlated with the differential metabolites which might be attributed to the synergistic effect of both microbial taxa. Our findings provide practical insights into the mechanism of enrichment of metabolites in fermented pine needles and laid down the basis for developing traditional fermented foods with nutraceutical value.
Acknowledgement: Not applicable.
Funding Statement: This work was financially supported by the Hunan Provincial Market Supervision and Administration Bureau Science and Technology Plan Project (Project No. 2020kJJH20), the Program for Food Safety Monitoring and Early Warning Open Project of Hunan Provincial Key Laboratory (2020KFJJ01), and the Scientific Research Project of Hunan Education Department (20C1910).
Author Contributions: Conceptualization, Yinyu Xu, and Wenhua Zhou; methodology, Yinyu Xu, Zheng Li, Qian Wen, Hao He; software, Yinyu Xu, Zheng Li; validation, Yinyu Xu, Qian Wen, and Pengtu Shi; formal analysis, Yinyu Xu, Zheng Li, Qian Wen; investigation, Yinyu Xu, Qian Wen, Hao He, Zheng Li, Pengtu Shi; resources, Wenhua Zhou; data curation, Qian Wen; writing—original draft preparation, Yinyu Xu; writing—review and editing, Wenhua Zhou; visualization, Yinyu Xu; supervision, Wenhua Zhou; project administration, Wenhua Zhou; funding acquisition, Wenhua Zhou. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Datasets used for this manuscript are available within the text and its supplementary files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org (accessed on 14 October 2023)) via the iProX partner repository with the dataset identifier PXD047484.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.047949.
References
1. Park G, Paudyal DIP, Park Y, Lee C, Hwang I, Tripathi GR, et al. Effects of pine needle extracts on plasma cholesterol, fibrinolysis, and gastrointestinal motility. Biotechnol Bioprocess Eng. 2008;13:262–8. doi:10.1007/s12257-008-0039-y. [Google Scholar] [CrossRef]
2. Cheong H, Paudyal DP, Jun JY, Yeum CH, Yoon PJ, Park CG, et al. Effects of pine needle extract on pacemaker currents in interstitial cells of cajal from the murine small intestine. Mol Cells. 2005;20(2):235–40. doi:10.1016/S1016-8478(23)13222-5. [Google Scholar] [CrossRef]
3. Wang W, Zheng B, Wu J, Lv W, Lin P, Gong X. Determination of the dissociation constants of 16 active ingredients in medicinal herbs using a liquid-liquid equilibrium method. Separations. 2021;8:4. [Google Scholar]
4. Kato-Noguchi H. Bioactive compounds involved in the formation of the sparse understory vegetation in pine forests. Curr Org Chem. 2021;25(15):1731–8. doi:10.2174/1385272825666210526092443. [Google Scholar] [CrossRef]
5. Kim Y-S, Shin D-H. Volatile components and antibacterial effects of pine needle (Pinus densiflora S. and Z.) extracts. Food Microbiol. 2005;22:37–45. doi:10.1016/j.fm.2004.05.002. [Google Scholar] [CrossRef]
6. Kwak CS, Moon SC, Lee MS. Antioxidant, antimutagenic, and antitumor effects of pine needles (Pinus densiflora). Nutr Cancer. 2006;56(2):162–71. doi:10.1207/s15327914nc5602_7. [Google Scholar] [PubMed] [CrossRef]
7. Ferreira-Santos P, Zanuso E, Genisheva Z, Rocha CMR, Teixeira JA. Green and sustainable valorization of bioactive phenolic compounds from pinus by-products. Molecules. 2020;25(12):2931. doi:10.3390/molecules25122931. [Google Scholar] [PubMed] [CrossRef]
8. Kang Y-H, Park Y-K, Oh S-R, Moon K-D. Studies on the physiological functionality of pine needle and mugwort extracts. Korean J Food Sci Technol. 1995;27(6):978–84. [Google Scholar]
9. Robbins C, Hanley TA, Hagerman A, Hjeljord O, Baker DL, Schwartz CC, et al. Role of tannins in defending plants against ruminants: reduction in protein availability. Ecology. 1987;68(1):98–107. doi:10.2307/1938809. [Google Scholar] [CrossRef]
10. Chiu H, Wang H, Shen Y, Venkatakrishnan K, Wang C. Anti-inflammatory properties of fermented pine (Pinus morrisonicola Hay.) needle on lipopolysaccharide-induced inflammation in RAW 264.7 macrophage cells. J Food Biochem. 2019;43(11):e12994. [Google Scholar] [PubMed]
11. Lee W, Kothari D, Moon SG, Kim J, Kim KI, Ga GW, et al. Evaluation of non-fermented and fermented Chinese chive juice as an alternative to antibiotic growth promoters of broilers. Animals. 2022;12(20):2742. doi:10.3390/ani12202742. [Google Scholar] [PubMed] [CrossRef]
12. Kim K-Y, Chung H-J. Flavor compounds of pine sprout tea and pine needle tea. J Agric Food Chem. 2000;48(4):1269–72. doi:10.1021/jf9900229. [Google Scholar] [PubMed] [CrossRef]
13. Ichikawa S, Takigawa H, Nara S. Effects of Sho-ju-sen, a herbal medicine, on unidentified clinical syndrome. J Clin Pharmacol New Drugs. 1998;47:879–87. [Google Scholar]
14. Kuribara H, Iwata H, Tomioka H, Takahashi R, Goto K, Murohashi N, et al. The anxiolytic effect of Sho-ju-sen, a Japanese herbal medicine, assessed by an elevated plus-maze test in mice. Phyther Res. 2001;15(2):142–7. doi:10.1002/ptr.v15:2. [Google Scholar] [CrossRef]
15. Bolandghamat S, Moghimi A, Iranshahi M. Effects of ethanolic extract of pine needles (Pinus eldarica Medw.) on reserpine-induced depression-like behavior in male Wistar rats. Pharmacogn Mag. 2011;7(27):248. doi:10.4103/0973-1296.84240. [Google Scholar] [PubMed] [CrossRef]
16. Jeong JB, Seo EW, Jeong HJ. Effect of extracts from pine needle against oxidative DNA damage and apoptosis induced by hydroxyl radical via antioxidant activity. Food Chem Toxicol. 2009;47(8):2135–41. doi:10.1016/j.fct.2009.05.034. [Google Scholar] [PubMed] [CrossRef]
17. Packer L, Rimbach G, Virgili F. Antioxidant activity and biologic properties of a procyanidin-rich extract from pine (Pinus maritima) bark, pycnogenol. Free Radic Biol Med. 1999;27(5–6):704–24. [Google Scholar] [PubMed]
18. Koutsaviti A, Toutoungy S, Saliba R, Loupassaki S, Tzakou O, Roussis V, et al. Antioxidant potential of pine needles: a systematic study on the essential oils and extracts of 46 species of the genus Pinus. Foods. 2021;10(1):142. doi:10.3390/foods10010142. [Google Scholar] [PubMed] [CrossRef]
19. Hwang YA, Lee W, Kim J, Kim S, Choi MG, On JY, et al. In vitro fermentation characteristics of pine needles (Pinus densiflora) as feed additive. Fermentation. 2023;9(5):415. doi:10.3390/fermentation9050415. [Google Scholar] [CrossRef]
20. Hall RD. Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol. 2006;169(3):453–68. doi:10.1111/nph.2006.169.issue-3. [Google Scholar] [CrossRef]
21. De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc. 2007;2(4):778–91. doi:10.1038/nprot.2007.95. [Google Scholar] [PubMed] [CrossRef]
22. Xu Y, Wen Q, He H, Shi H, Chen H, Shi P, et al. Evaluation of bacterial and fungal communities during the fermentation of pine needle. FEMS Microbiol Lett. 2023;370:fnad032. doi:10.1093/femsle/fnad032. [Google Scholar] [PubMed] [CrossRef]
23. Zelena E, Dunn WB, Broadhurst D, Francis-McIntyre S, Carroll KM, Begley P, et al. Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Anal Chem. 2009;81(4):1357–64. doi:10.1021/ac8019366. [Google Scholar] [PubMed] [CrossRef]
24. Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS, et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc. 2013;8(1):17–32. doi:10.1038/nprot.2012.135. [Google Scholar] [PubMed] [CrossRef]
25. Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y, et al. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 2008;53(4):691–704. doi:10.1111/tpj.2008.53.issue-4. [Google Scholar] [CrossRef]
26. Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45(7):703–14. doi:10.1002/jms.v45:7. [Google Scholar] [CrossRef]
27. Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, et al. LMSD: lIPID MAPS structure database. Nucleic Acids Res. 2007;35(suppl_1):D527–32. doi:10.1093/nar/gkl838. [Google Scholar] [PubMed] [CrossRef]
28. Abdelrazig S, Safo L, Rance GA, Fay MW, Theodosiou E, Topham PD, et al. Metabolic characterisation of Magnetospirillum gryphiswaldense MSR-1 using LC-MS-based metabolite profiling. RSC Adv. 2020;10(54):32548–60. doi:10.1039/D0RA05326K. [Google Scholar] [PubMed] [CrossRef]
29. Gagnebin Y, Tonoli D, Lescuyer P, Ponte B, de Seigneux S, Martin PY, et al. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: impact of normalization strategies. Anal Chim Acta. 2017;955:27–35. doi:10.1016/j.aca.2016.12.029. [Google Scholar] [PubMed] [CrossRef]
30. Thévenot EA, Roux A, Xu Y, Ezan E, Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J Proteome Res. 2015;14(8):3322–35. doi:10.1021/acs.jproteome.5b00354. [Google Scholar] [PubMed] [CrossRef]
31. Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743–60. doi:10.1038/nprot.2011.319. [Google Scholar] [PubMed] [CrossRef]
32. Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6(7):1060–83. doi:10.1038/nprot.2011.335. [Google Scholar] [PubMed] [CrossRef]
33. Boulesteix A-L, Strimmer K. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform. 2007;8(1):32–44. [Google Scholar] [PubMed]
34. Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemom. 2002;16:119–28. doi:10.1002/cem.v16:3. [Google Scholar] [CrossRef]
35. Juan M-Y, Chou C-C. Enhancement of antioxidant activity, total phenolic and flavonoid content of black soybeans by solid state fermentation with Bacillus subtilis BCRC 14715. Food Microbiol. 2010;27(5):586–91. doi:10.1016/j.fm.2009.11.002. [Google Scholar] [PubMed] [CrossRef]
36. Chen YH, Hsieh PC, Mau JL, Sheu SC. Antioxidant properties and mutagenicity of Pinus morrisonicola and its vinegar preparation. LWT-Food Sci Technol. 2011;44(6):1477–81. doi:10.1016/j.lwt.2011.01.016. [Google Scholar] [CrossRef]
37. Fabre CE, Blanc PJ, Goma G. 2-Phenylethyl alcohol: an aroma profile. Perfum Flavor. 1998;23:43–5. [Google Scholar]
38. Hua SST, Beck JJ, Sarreal SBL, Gee W. The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 2014;30(2):71–8. doi:10.1007/s12550-014-0189-z. [Google Scholar] [PubMed] [CrossRef]
39. Chang PK, Hua SST, Sarreal SBL, Li RW. suppression of aflatoxin biosynthesis in aspergillus flavus by 2-phenylethanol is associated with stimulated growth and decreased degradation of branched-chain amino acids. Toxins. 2015;7(10):3887–902. doi:10.3390/toxins7103887. [Google Scholar] [PubMed] [CrossRef]
40. Koriem KMM, Abdeen AMA. Shikimic acid recovers diarrhea and its complications in SD rats fed lactose diet to induce diarrhea. Lab Anim Res. 2023;39:28. doi:10.1186/s42826-023-00179-y. [Google Scholar] [PubMed] [CrossRef]
41. Li X, Mo K, Tian G, Zhou J, Gong J, Li L, et al. Shikimic acid regulates the NF-κB/MAPK signaling pathway and gut microbiota to ameliorate DSS-induced ulcerative colitis. J Agric Food Chem. 2023;71(23):8906–14. doi:10.1021/acs.jafc.3c00283. [Google Scholar] [PubMed] [CrossRef]
42. Petrovici AR, Ciolacu DE. Natural flavours obtained by microbiological pathway. Gen Aromas Flavours. 2018;76785:33–52. [Google Scholar]
43. Ramaroson ML, Koutouan C, Helesbeux JJ, Le Clerc V, Hamama L, Geoffriau E, et al. Role of phenylpropanoids and flavonoids in plant resistance to pests and diseases. Molecules. 2022;27(23):8371. doi:10.3390/molecules27238371. [Google Scholar] [PubMed] [CrossRef]
44. Liu W, Feng Y, Yu S, Fan Z, Li X, Li J, et al. The flavonoid biosynthesis network in plants. Int J Mol Sci. 2021;22(23):12824. doi:10.3390/ijms222312824. [Google Scholar] [PubMed] [CrossRef]
45. Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–93. doi:10.1104/pp.126.2.485. [Google Scholar] [PubMed] [CrossRef]
46. Selvakumar P, Badgeley A, Murphy P, Anwar H, Sharma U, Lawrence K, et al. Flavonoids and other polyphenols act as epigenetic modifiers in breast cancer. Nutrients. 2020;12(3):761. doi:10.3390/nu12030761. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools