 Open Access
Open Access
ARTICLE
Genome-Wide Identification of ABCC Gene Subfamily Members and Functional Analysis of CsABCC11 in Camellia sinensis
1 The Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), College of Life Science, Guizhou University, Guiyang, 550025, China
2 College of Tea Science, Institute of Plant Health & Medicine, Guizhou University, Guiyang, 550025, China
3 Wengfu Group, Guiyang, 550025, China
4 Huaneng Clean Energy Research Institute, Guiyang, 550000, China
* Corresponding Authors: Hu Tang. Email: ; Litang Lu. Email:
(This article belongs to the Special Issue: Plant Secondary Metabolism and Functional Biology)
Phyton-International Journal of Experimental Botany 2024, 93(8), 2019-2036. https://doi.org/10.32604/phyton.2024.052938
Received 19 April 2024; Accepted 15 July 2024; Issue published 30 August 2024
Abstract
The ATP-binding cassette (ABC) transporter is a gene superfamily in plants. ATP-binding cassette subfamily C (ABCC) protein is a multidrug resistance-associated (MRP) transporter. They play various roles in plant growth, development, and secondary metabolite transport. However, there are few studies on ABCC transporters in tea plants. In this study, genome-wide association study (GWAS) analysis of epigallocatechin gallate (EGCG) content in 108 strains of Kingbird revealed that CsABCCs may be involved in EGCG transport. We identified 25 CsABCC genes at the genomic level of the tea plant, their phylogenetic tree, gene structure, targeted miRNA and other bioinformatics were analyzed. The expression patterns of CsABCCs in eight different tissues and abiotic stress indicate that they have potential roles in regulating the growth, development, and defense of tea plants. The correlation analysis revealed that the expression of the CsABCC11 gene was closely related to the EGCG content in tea buds of 108 strains of the Kingbird, and the subcellular localization experiments in tobacco showed that CsABCC11 protein was localized on the plasma membrane. The virus-induced gene silencing (VIGS) strategy in tea plants further verified that CsABCC11 was involved in EGCG accumulation. Our study laid a foundation for studying the biological function of CsABCC and provided a new candidate molecular marker gene for further EGCG-related variety breeding, which will be of great interest to breeders.Keywords
Supplementary Material
Supplementary Material FileThe tea plant (Camellia sinensis (L.) O. Kuntze) is a significant cash crop cultivated worldwide due to its economic importance. The plant ATP-binding cassette (ABC) protein is an important membrane protein [1]. Plant ABC transporters have multiple functions, such as participating in transporting plant secondary metabolites. It is involved in heavy metal detoxification, stress [2–4], accumulation and metabolism [5], mediating antibiotic resistance and plant hormone transport such as cytokinin and abscisic acid. Most plant ABC transporters exist in the cell membrane and participate in organ growth, plant nutrition, plant development, and other processes [6]. It also plays an important role in plant gametogenesis, seed development, germination, organ formation, and secondary growth. It can be seen that the ABC protein can act as an input, an output, and a channel [7]. In animals, ABC transporters have been shown to have the functions of ion channels and channel regulators [8,9]. ABC transporter gene family has been characterized in many species, such as pigeon pea (Cajanus cajan (L.) Druce) [10], corn (Zea mays L.) [2], flax (Linum usitatissimum L.) [11], pineapple (Ananas comosus L.) [12,13], wheat (Triticum aestivum L.), soybean (Glycine max (L.) Merr.). In the plant genome, ABC transporters are divided into eight different subfamilies (ABCA-ABCG and ABCI). ABCC subfamily proteins are involved in the detoxification of metal ions and the transport of plant secondary metabolites. Studies have indicated that the maize ABCC subfamily proteins play a role in vesicle transport and seed phytic acid anthocyanin content [14]. However, plant ABC transporters have been well studied in other species, but it remain to be studied in tea plants.
The quality of tea depends on the secondary metabolites in tea. Catechins are a class of phenolic active substances extracted from tea plants [15]. Catechins include eight monomers, catechins, epicatechins, epigallocatechin gallate, and eight other monomers. EGCG is the most effective active ingredient in tea polyphenols. It is the monomer with the highest proportion of catechins, accounting for about 60%. EGCG is the main component that determines the color, aroma, and taste of tea, and its content is the highest in green tea. EGCG has many functions in medicine and health care, such as antioxidant [16], anti-free radical DNA damage [17], anti-inflammatory and anti-tumor effects [18,19]. It has made good progress in anti-cervical cancer research [20]. Some studies have shown that EGCG is used as a special functional preservative and skin care agent on daily chemical products to scavenge free radicals, resist the effects of pollution, sun exposure, and smoking, and prevent skin aging and wrinkles. In recent years, research has found that EGCG has a potential protective effect on COVID-19, so the author concluded that drinking green tea has a potential preventive effect on COVID-19 [21]. The anti-tumor effect of EGCG has been recognized by relevant researchers in various countries and has been listed as a potential new anti-cancer drug in many countries such as China and the United States. A large number of clinical medical experiments have proved that EGCG is no less than paclitaxel in the treatment of various cancers, and its toxicity is much lower than paclitaxel. In particular, there are many reports on the application of skin cancer [22], which has now entered the clinical stage. It can be said that EGCG is an effective and non-toxic ideal drug for the treatment of cancer, which has great development value [23,24].
The role of epigallocatechin gallate (EGCG) in a variety of abiotic stresses has become a topic of concern in recent literature. Studies have shown that exogenous application of EGCG can alleviate a variety of abiotic stresses, such as high temperature, salt, and drought, thereby enhancing stress tolerance [25]. The study also reported that EGCG levels increased during drought stress, suggesting that this compound may play a role in mitigating the effects of water deficit on plants [26]. In addition, catechin synthesis (EGCG is its derivative) has been found to be sensitive to environmental disturbances such as drought and salinity, which highlights the importance of these compounds in plant stress responses [27]. In addition, the signaling pathways of plants in response to cold, salt, and drought stress have also been extensively studied, providing clues for revealing the molecular mechanism of stress tolerance. EGCG may play a role in regulating these pathways, potentially helping plants treated with this compound to enhance stress tolerance [28], and the compound EGCG found in green tea may have a protective effect on plants against drought and salt stress. More research is needed to elucidate how EGCG enhances stress resistance and to explore its potential applications in agriculture and crop improvement.
In our previous research, the EGCG content of the 108 strains of Kingbird was analyzed, and it was found that there were differences in EGCG content among different strains, and then GWAS was mapped to a gene CsABCC11. In this study, we performed a genome-wide identification of CsABCCs in tea plants, and the chromosome mapping, phylogeny classification, conserved motifs, promoter components, miRNA, and expression pattern were analyzed. Subcellular localization experiments confirmed the subcellular localization of CsABCC11 protein in tobacco. Subsequently, correlation analysis revealed that the expression of the CsABCC11 gene was closely related to the EGCG content of tea plant buds, and the VIGS strategy carried out in tea plants further verified that CsABCC11 was an indispensable part in the formation of EGCG. Therefore, it is highly significant to investigate the ABCC transporter in tea plants to enhance the EGCG content, providing relevant insights for further research.
The tea cultivar ‘FudingDabai’ was planted in the experimental field of Guizhou Academy of Agricultural Sciences (Guiyang, Guizhou, China). The buds of 108 strains (‘Kingbird’(Niaowangzhong)) were taken from Guiding Yunwu (Guiding, Guizhou, China). Tobacco for subcellular localization was planted in our laboratory.
2.2 Identification of CsABCC Subfamily Genes in Tea Plant
Firstly, the Arabidopsis AtABCC gene is derived from the Arabidopsis genome database (Table S1). Taking the Arabidopsis thaliana gene sequence as a reference, the tea tree genome annotation obtained in the tea tree genome database was used. The BLASTP alignment was performed using the ABCC gene-specific conserved domain model [29], and the threshold was set to (E-value < 10−5) to obtain the hypothetical CsABCCs. The protein characteristics of the CsABCCs were analyzed by the NCBI-CDD database to ensure that the CsABCCs only had its domainI (Accession: cl33195) [30]. The obtained sequence was sorted and the repetitive items were removed, and finally, the CsABCC gene family members of the tea plant were obtained. The amino acid length of ABCC protein in tea plants was analyzed according to the Perl script of the downloaded tea plant genome database. The relative molecular mass, isoelectric point and subcellular localization were identified by Expasy and Plant-mPloc online [31].
2.3 Phylogenetic Analysis of ABCCs in Different Plant Species
MEGA X was used to analyze the evolutionary relationship of proteins from tea plants and Arabidopsis thaliana based on the adjacency method. The Bootstrap test is repeated 1000 times [32]. Use the FigTree software (version 1.4.2) to display the phylogenetic tree. The protein sequence of Arabidopsis thaliana was obtained from Arabidopsis thaliana Information Resource (TAIR).
2.4 Analysis of Chromosome Position, Gene Structure and Conserved Motif of CsABCC Genes in Tea Plant
The chromosome location information of the CsABCCs (including chromosome distribution, length, start, and end position) was downloaded from TPIA and visualized by TBtools. TBtools software was used to visualize the exon-intron structure of the CsABCCs based on genome annotation. The conserved protein motifs of CsABCC protein were analyzed by the MEME program.
2.5 Analysis of Cis-Regulatory Elements of CsABCC Genes Promoter
The promoter sequences of all CsABCC genes were obtained from TPIA and analyzed using PlantCARE and visualized by TBtools.
2.6 Prediction of Putative miRNA Targeting CsABCC Genes
Based on the miRNA of Arabidopsis thaliana, the hypothetical miRNA targeting CsABCC was predicted by online software (https://www.zhaolab.org/psRNATarget/analysis, accessed on 21 June 2024), and the miRNA-target gene network was constructed by online software (https://www.bioinformatics.com.cn, accessed on 18 June 2024).
2.7 Expression Pattern Analysis of CsABCC Genes
The CsABCCsTranscriptome data of eight tissues (top bud, young leaf, mature leaf, old leaf, stem, root, flower and fruit) and salt, drought and cold stress of tea variety “Shuchazao” were downloaded from open database (TPIA) and heat maps were created using TBtools.
The primers of CsABCC11 gene were designed by IDT. The cetyltrimethylammonium bromide (CTAB) method was used to extract the total RNA from the bud heads of 7 groups of 108 strains of VIGS and Kingbird, the retrotranscription system is as follows: 50 ng–5 µg Total RNA, 1 µL Oligo (dT) 18 (0.5 μg/µL), 3.6–4 µL 5 × TRUE Reaction Mix, RNase free H2O to final volume 20 µL. The cDNA was then used as a template for qRT-PCR analysis. Each reaction system contained 10 μL SYBR Green qPCR Mix, 1 μL primers, 3 μL cDNA template, and 5 μL H2O. The reaction process is as follows: 1 cycle at 95°C for 30 s; 95°C 10 s, 60°C 30 s, 40 cycles; after 40 cycles of 72°C 60 s, The relative expression level was calculated with CsGAPDH gene as reference.
The PCAMBIA1300-35S-CsABCC11-GFP fusion expression vector was constructed (Fig. S1). he constructed plant fusion expression vector PCAMBIA1300-35S-CsABCC11-GFP and the empty vector PCAMBIA1300-35S-GFP were transformed into GV3101 competent cells, and co-expressed and injected into the lower epidermis of 4–6-week-old N. benthamiana [33]. After 2 days of low light culture, the distribution of green fluorescence signal was observed under laser confocal microscope [34].
2.10 Virus-Mediated VIGS and HPLC Determination of EGCG Content
The pTRV2-CsABCC11 vector was constructed, and pTRV1, pTRV2 and pTRV2-CsABCC11 were introduced into competent cells GV3101. Agrobacterium strains were inoculated into solid YEP medium (yeast powder 10 g/L, peptone 10 g/L, NaCl 5g/L, Rif ampicin 50 mg/L, Kanamycin 100 mg/L), and the single colony was cultured in the corresponding liquid YEP medium. The bacterial liquid was collected and resuspended. The pTRV1 and pTRV2, pTRV2-CsABCC11 bacterial liquid were mixed at 1:1, and then the tea tree cuttings were vacuum infiltrated [35]. Stored in dark for 3 days, cultured at 25 degrees Celsius, the light duration and dark duration were 16 and 8 h, respectively. The content of EGCG in VIGS buds in silenced cuttings was determined by high performance liquid chromatography (HPLC). The sample preparation method is as follows. Weigh 0.2 g tea powder (accurate to 0.0001 g) through 40 mesh sieve, place it in a 15 mL centrifuge tube, add 10 mL of 70% methanol. The extraction was carried out in 70°C water bath for 10 min (shake once every 2 min), centrifuge it in a high-speed centrifuge (low temperature 4°C, 6000 r/min) for 5 min, and transfer the supernatant to a 25 mL measuring bottle. The residue was repeatedly extracted with 5 mL 70% methanol, and the supernatant was combined. The supernatant was diluted with stable liquid to 25 mL, shaken, and filtered through 0.22 μm microporous membrane. The detection method of EGCG content was based on the national standard GB/T8313.
2.11 Data Analysis and Mapping
All results were expressed as mean ± standard deviation of at least 3 biological replicates. SPSS and Excel software were used for data analysis, drawing with Graphpad Prism and TBtools.
3.1 Identification and Phylogenetic Tree Analysis of CsABCC Subfamily Members in Tea Plant
A total of 25 CsABCC genes were identified, all containing a common domain (MRP). All CsABCC protein sequences are provided in Fig. S1. According to its position on the chromosome, it was named CsABCC1 to CsABCC25. The basic information of CsABCC genes was analyzed (Table 1). The protein range was 778 to 1556 bp, the molecular weight was 86.15 to 173.39 kDa, and the isoelectric point was 5.45~8.61. The 25 CsABCC proteins are located in the plasma membrane, which shows that the ABCC transporter is located on the plasma membrane and plays a role in transport.
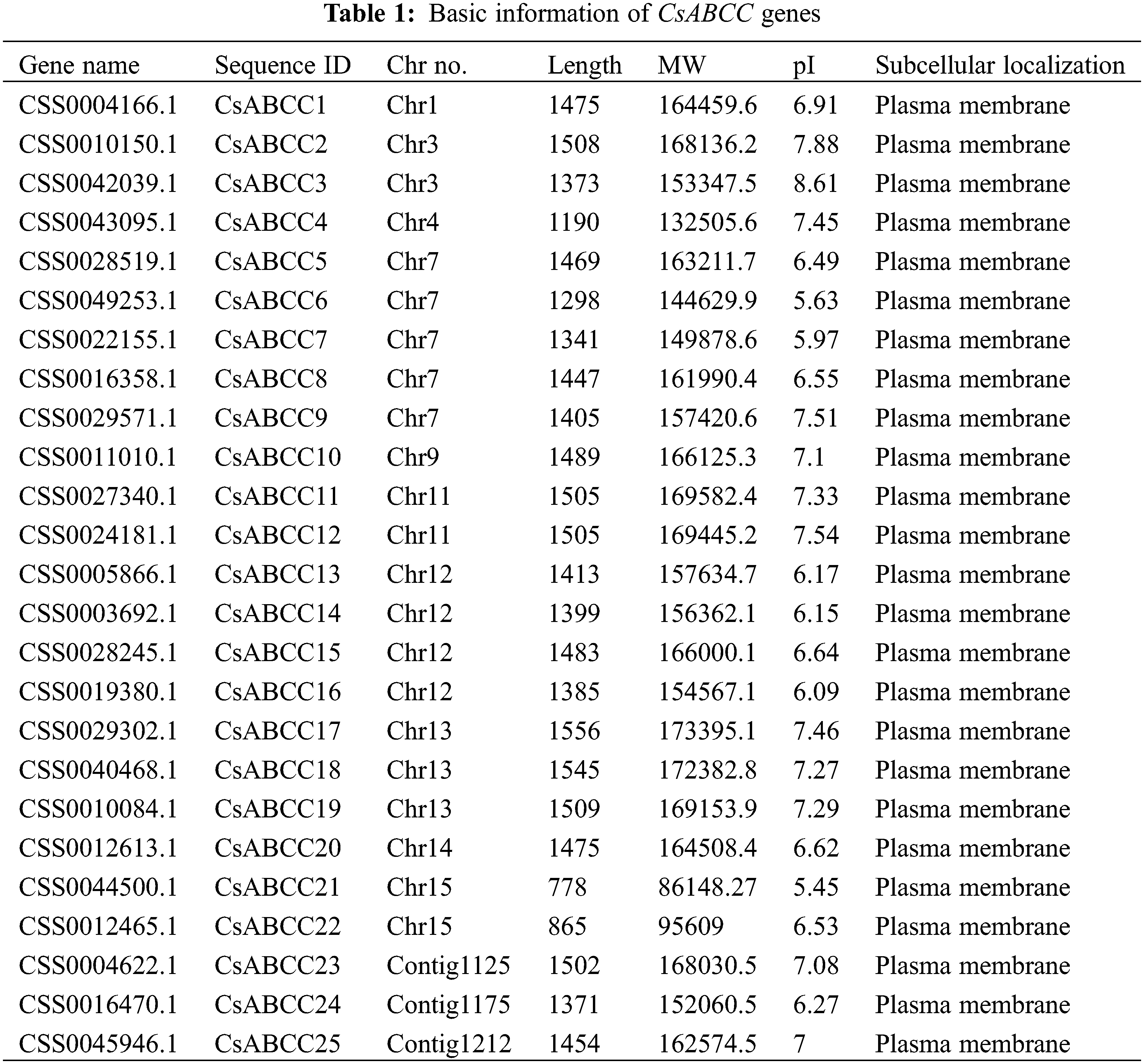
In order to explore the evolutionary relationship between the ABCC gene of tea plant and other plant homologues, the phylogenetic tree of tea plant and Arabidopsis thaliana was constructed by neighbor-joining method. According to the conserved motifs between them, all ABCC proteins were divided into four groups. As shown in Fig. 1, there were 13 members in the first and second groups, and 7 members in the third and fourth groups, respectively.
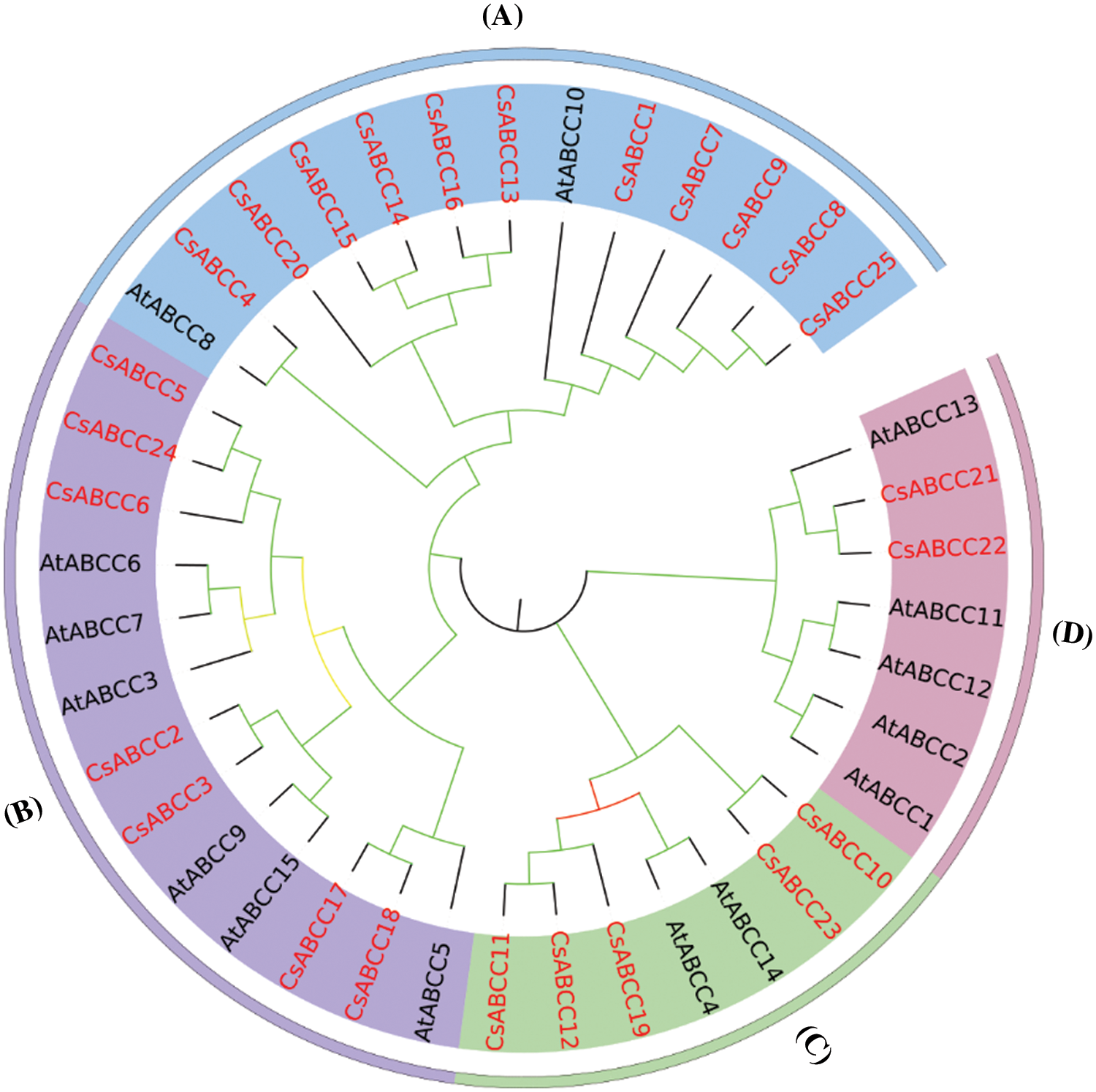
Figure 1: The phylogenetic relationship and classification of Camellia sinensis, Arabidopsis thaliana, and ABCC proteins. All ABCC proteins were divided into three groups. Gene ID is represented by different colors, red represents the tea plant, and black represents Arabidopsis thaliana
3.2 Analysis of Conserved Motifs of Genes
To obtain the conserved domain of CsABCC proteins, we used the MEME program to analyze it. A total of 10 conserved motifs are predicted, containing about 34–50 conserved amino acids (Fig. 2), and CsABCC showed highly similar motif composition in the same subfamily. It shows that these motifs are conserved during the evolution of tea plants. Majority proteins have the same conserved motifs, but the number of motifs is different. All CsABCC genes have more than 9 conserved motifs, up to 15. There are motif 1, motif 2, motif 3, motif 4, motif 5 and motif 6, motif 9 and motif 10 in each CsABCC sequence. There is no motif 7 and motif 8 in CsABCC2 and CsABCC19. CsABCC1, CsABCC3, CsABCC5, CsABCC7, CsABCC16, CsABCC17, CsABCC20, CsABCC24 and CsABCC25 have the same motifs, including 12 conserved motifs, which also indicates that the same CsABCC gene subfamily members have similar functions.
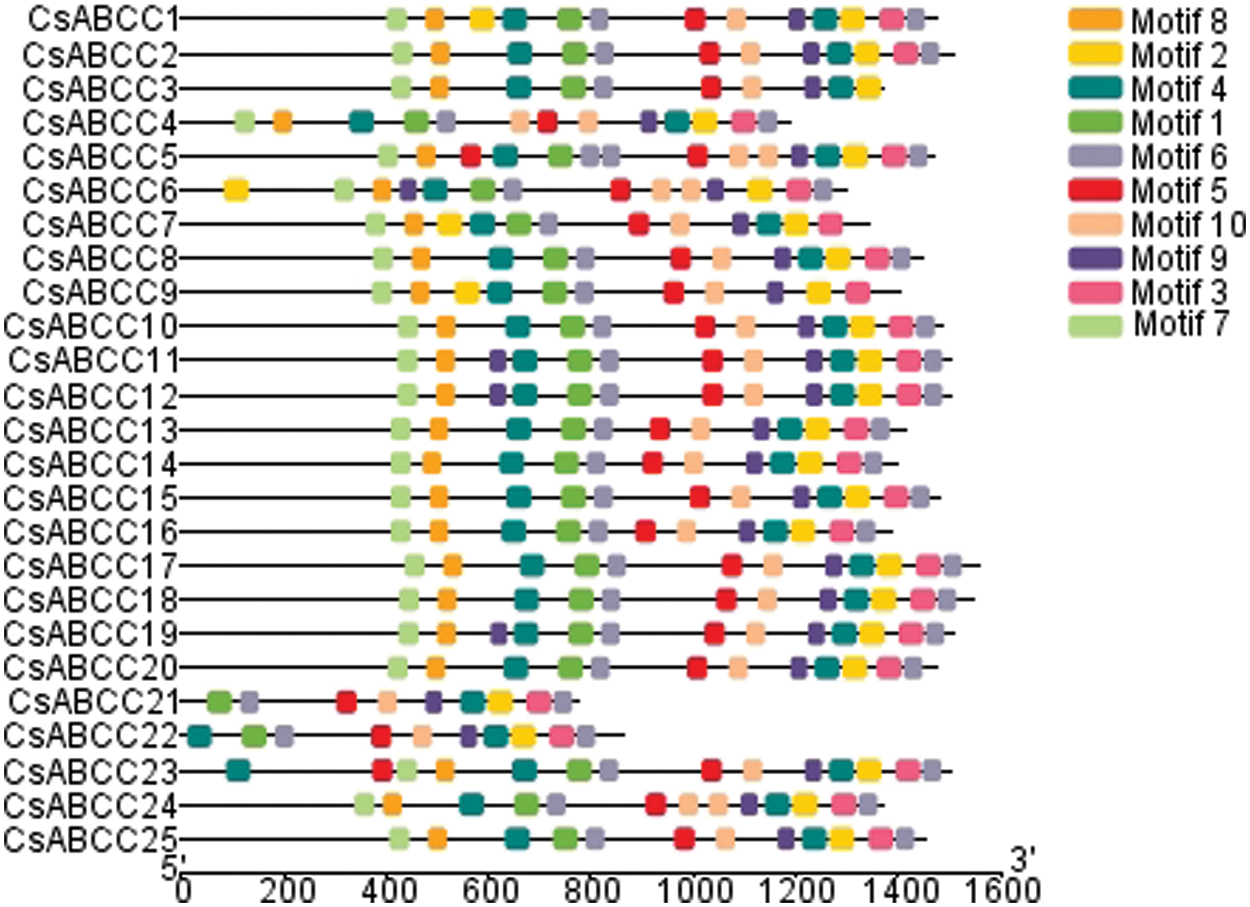
Figure 2: Conserved motifs of CsABCCs in tea plant. The left side represents the position of the motif on the gene, the right side represents the motif name, and different color blocks represent different motifs
3.3 Gene Structure and Chromosome Location of CsABCC
In order to further study the effect of gene evolution on the CsABCC genes, the chromosome localization of the CsABCC gene in tea plants was analyzed by TBtools software. As shown in Fig. 3, the results show that 22 of the 25 members of CsABCC are unevenly distributed on 10 chromosomes (Fig. 3), and 3 CsABCC genes were located on the unanchored scaffold. The largest number of CsABCC genes is on chromosome 7, containing 5 genes, which is the largest of all chromosomes. Chromosomes 3, 11 and 15 contained the same number of CsABCC genes, namely 2, and chromosomes 1, 4, 9 and 14 also contained the same number of CsABCC gene, namely 1.
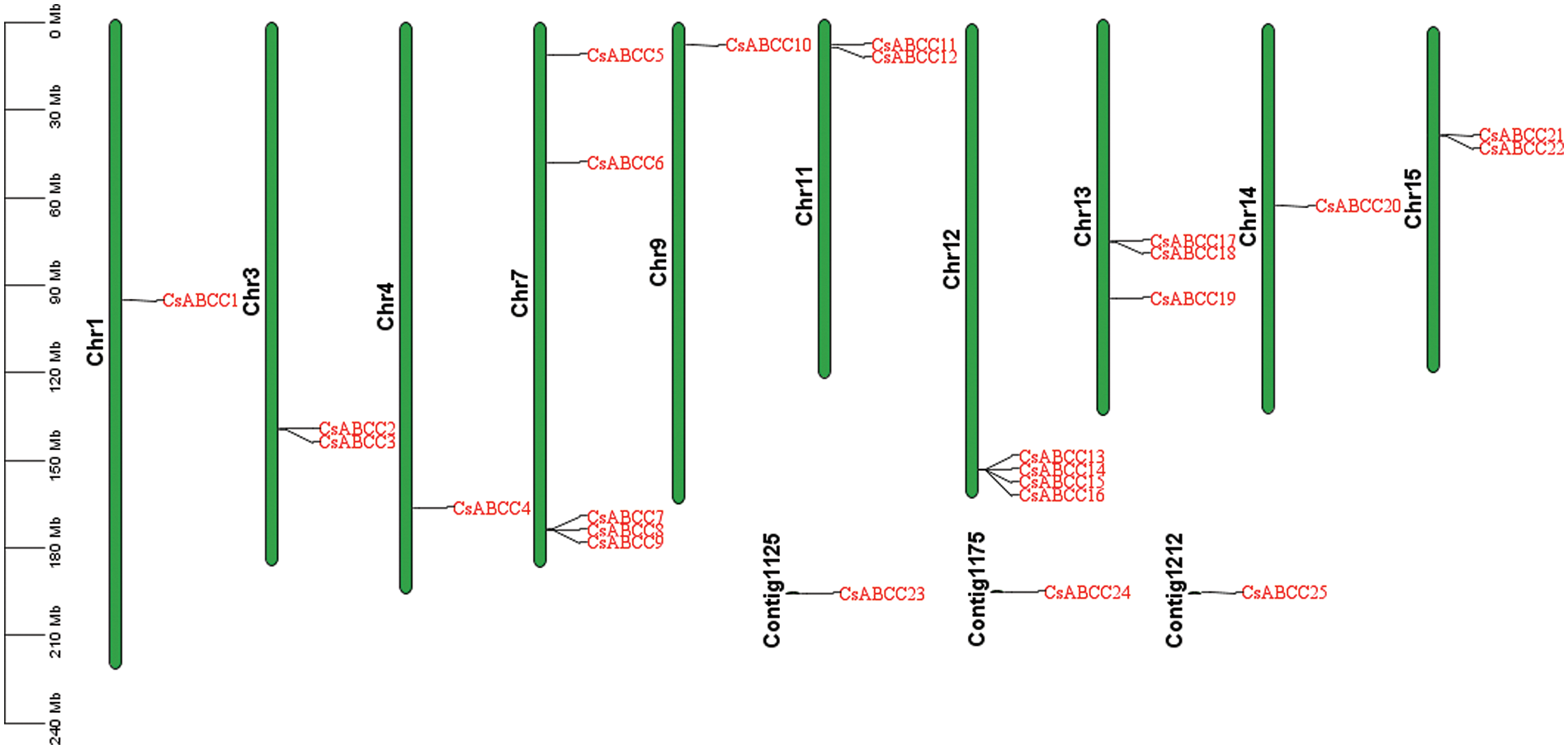
Figure 3: Chromosome distribution of CsABCC genes. The numbering of the CsABCC genes is shown on the right side of the chromosome. A ruler describes the relative length of a chromosome, denoted by megabases (Mb)
We analyzed the distribution of exons and introns in the CsABCC genes sequence. The exons of CsABCC ranged from 8 to 18 (Fig. 4). Among them, CsABCC22 and CsABCC21 have the most exons, with 18 and 17 exons, respectively, while CsABCC3 contains only 8 exons, which is the least. There were 17 CsABCC genes without UTR. To better understand the evolutionary relationship of CsABCC, we constructed a phylogenetic tree of ABCC proteins, and divided all ABCC proteins into four groups. As shown in Fig. 4, there are 9 pairs of homologous pairs in CsABCCs, and all 9 pairs of Bootstrap values are 100, indicating that these 9 pairs of CsABCCs are closely related.
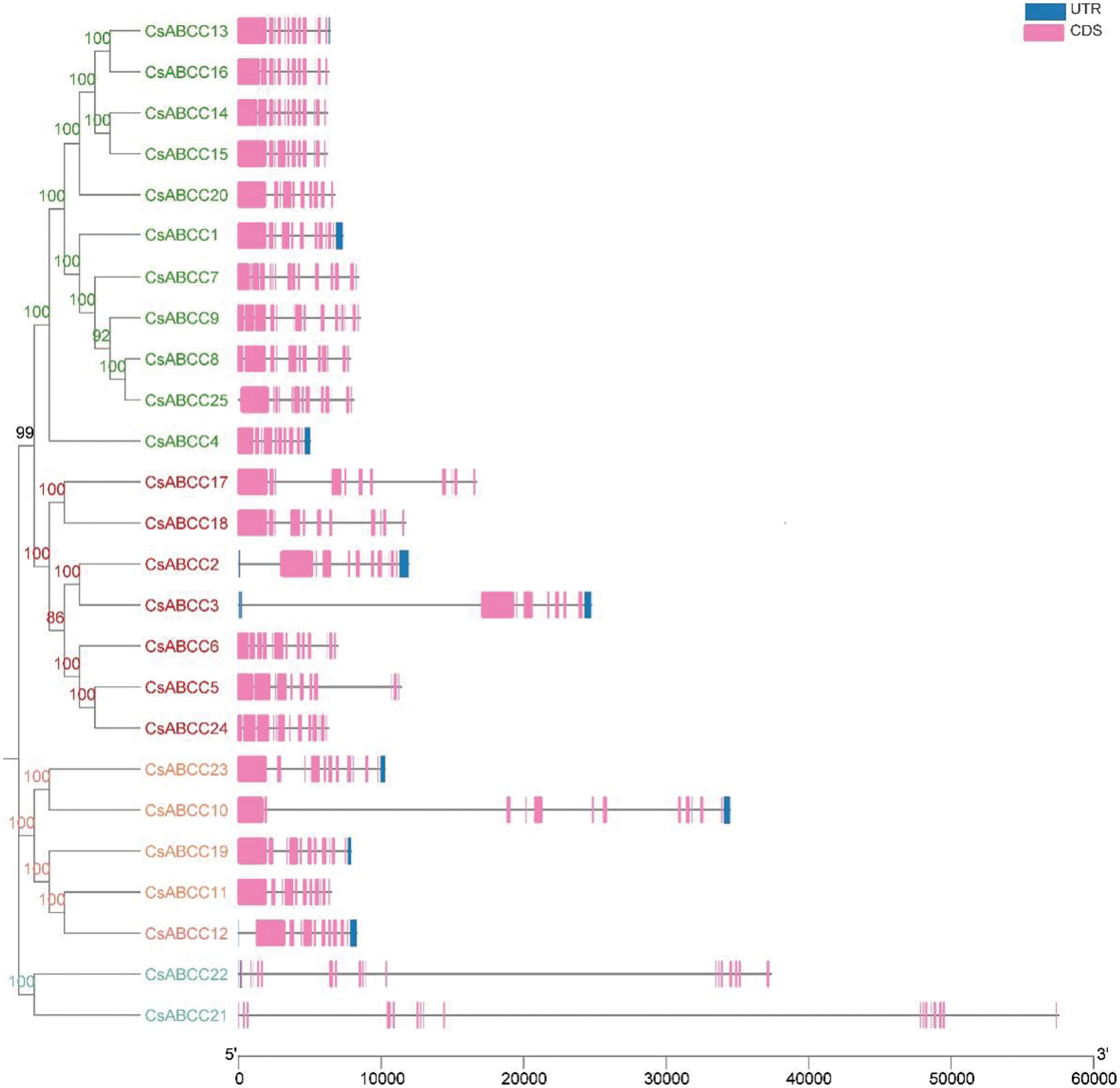
Figure 4: CsABCC phylogenetic tree of tea plant (left) gene structure (right), purple box, blue box and black line represent exons, UTR and introns, respectively
3.4 Analysis of Cis-Acting Elements of CsABCC Genes Promoter
In this study, 2000 bp upstream of the transcription start site ATG in the genome of 25 CsABCC genes was extracted and submitted to the online database plantCARE. The analysis results showed that the promoter region contained a large number of promoter cis-acting elements as shown in Fig. 5. Light response, plant hormone response, plant growth and development, adversity stress and so on. These results indicate that CsABCC genes plays a role in different tissues and stress environments.
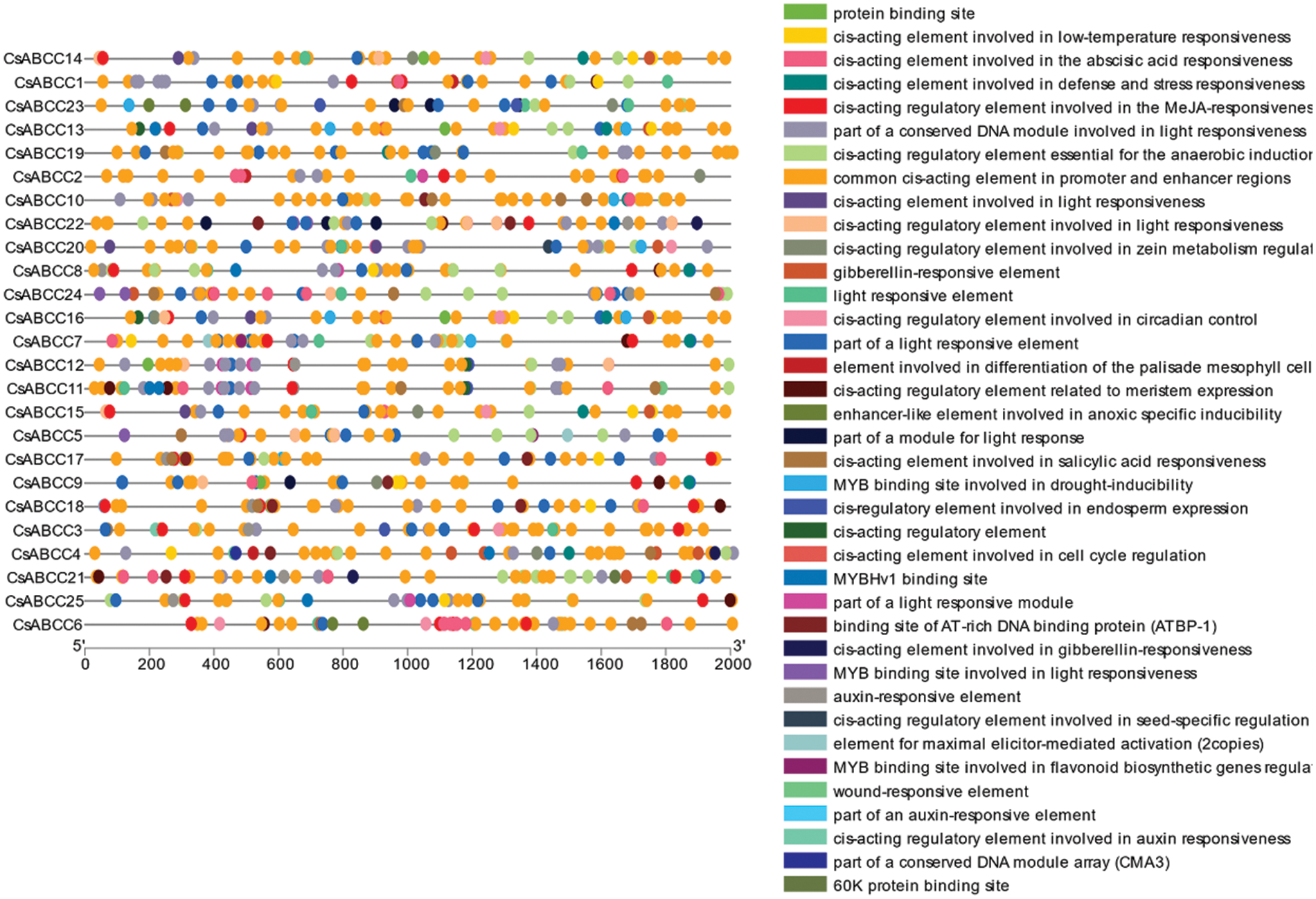
Figure 5: Analysis of CsABCC genes promoter region. The left circle represents the position of the cis-acting element in the promoter region, the right box represents the name of the cis-acting element
3.5 Prediction of Putative miRNA Targeting CsABCC Gene
As a class of non-coding small RNA (sRNA), microRNA (miRNA) is regulated at the post-transcriptional level by binding to target genes. Based on the miRNA of Arabidopsis thaliana, the miRNA targeting CsABCC genes was predicted (Table S2). The network diagram was drawn by online software (Fig. 6). Among them, the miRNA targeting CsABCC7 gene was the least, with 12 miRNAs, and the miRNA targeting CsABC19 gene was the most, with 49 miRNAs. There were 44 genes targeting CsABCC11. A large number of miRNAs binding to CsABCCs may regulate the accumulation of secondary metabolites in tea plants.
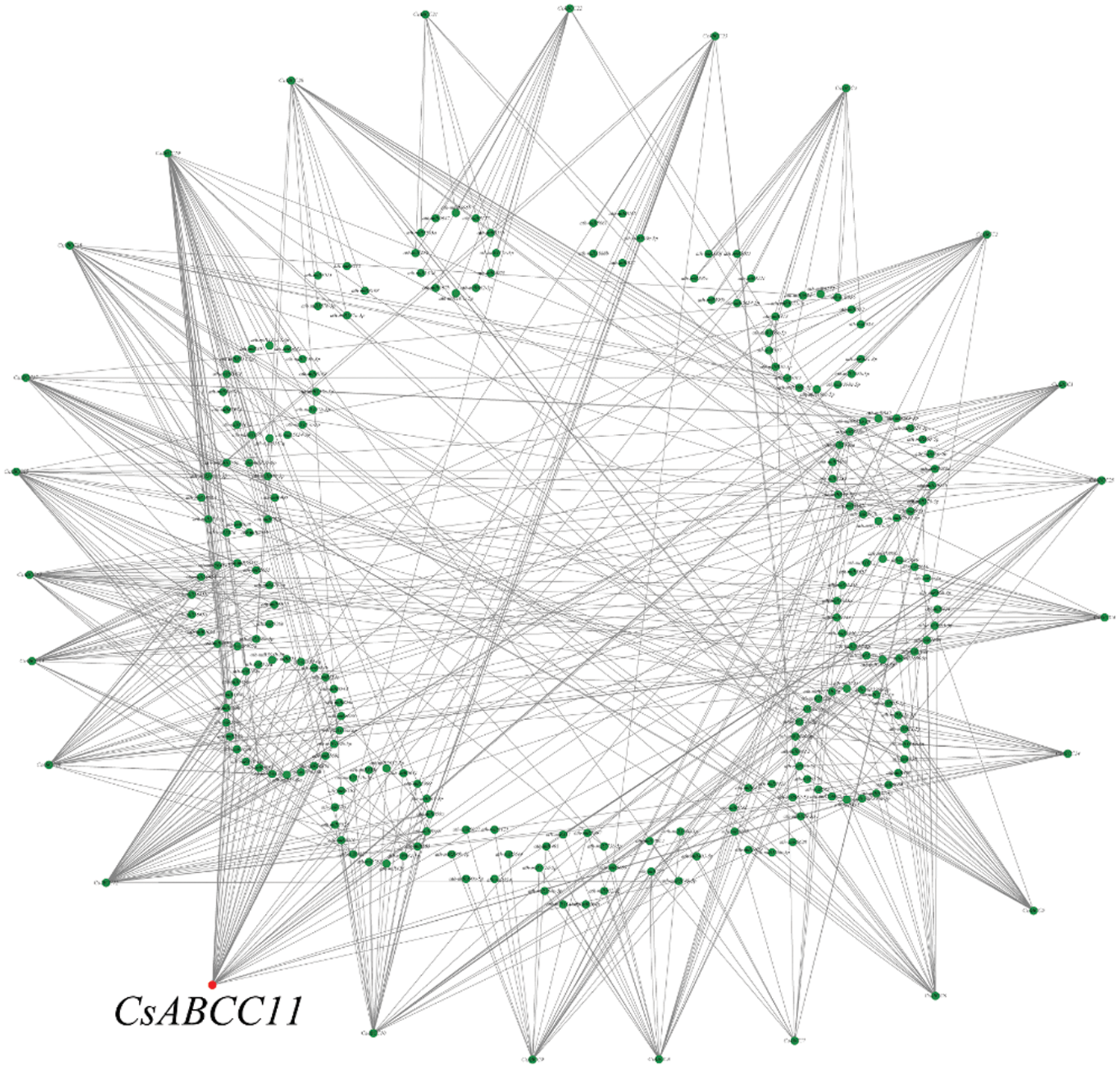
Figure 6: miRNA-CsABCC network diagram, the inner circle represents the name of miRNA, and the outer circle represents the name of CsABCCs
3.6 Analysis of CsABCC Genes Expression in Different Tissues and under Different Stress Conditions
The differential expression of 25 CsABCC genes in eight different tissues of tea plant and their potential role in stress response were studied. The CsABCCs expression data of 8 different tissues of tea plant and cold acclimation, drought stress and salt stress were obtained. The 25 CsABCC genes showed different expression patterns in eight different tissues. Most CsABCC genes were lowly expressed in apical buds. Only CsABCC7 gene was significantly expressed in flowers. CsABCC5 and CsABCC24 genes were significantly expressed in fruits. The expression of CsABCC3 gene was higher in young leaves. Most CsABCC genes were highly expressed in mature and old leaves. CsABCC13 gene was significantly expressed in roots, while CsABCC9 and CsABCC23 genes were highly expressed. CsABCC24 gene was significantly expressed in stems (Fig. 7A). Among them, CsABCC11 was expressed in different degrees in terminal buds, young leaves, old leaves, roots, flowers and fruits, indicating that the expression of CsABCC11 may play a significant role in the transport of substances in tea plants. The changes in expression patterns indicate that different CsABCCs play a key role in the growth and development of tea plants.
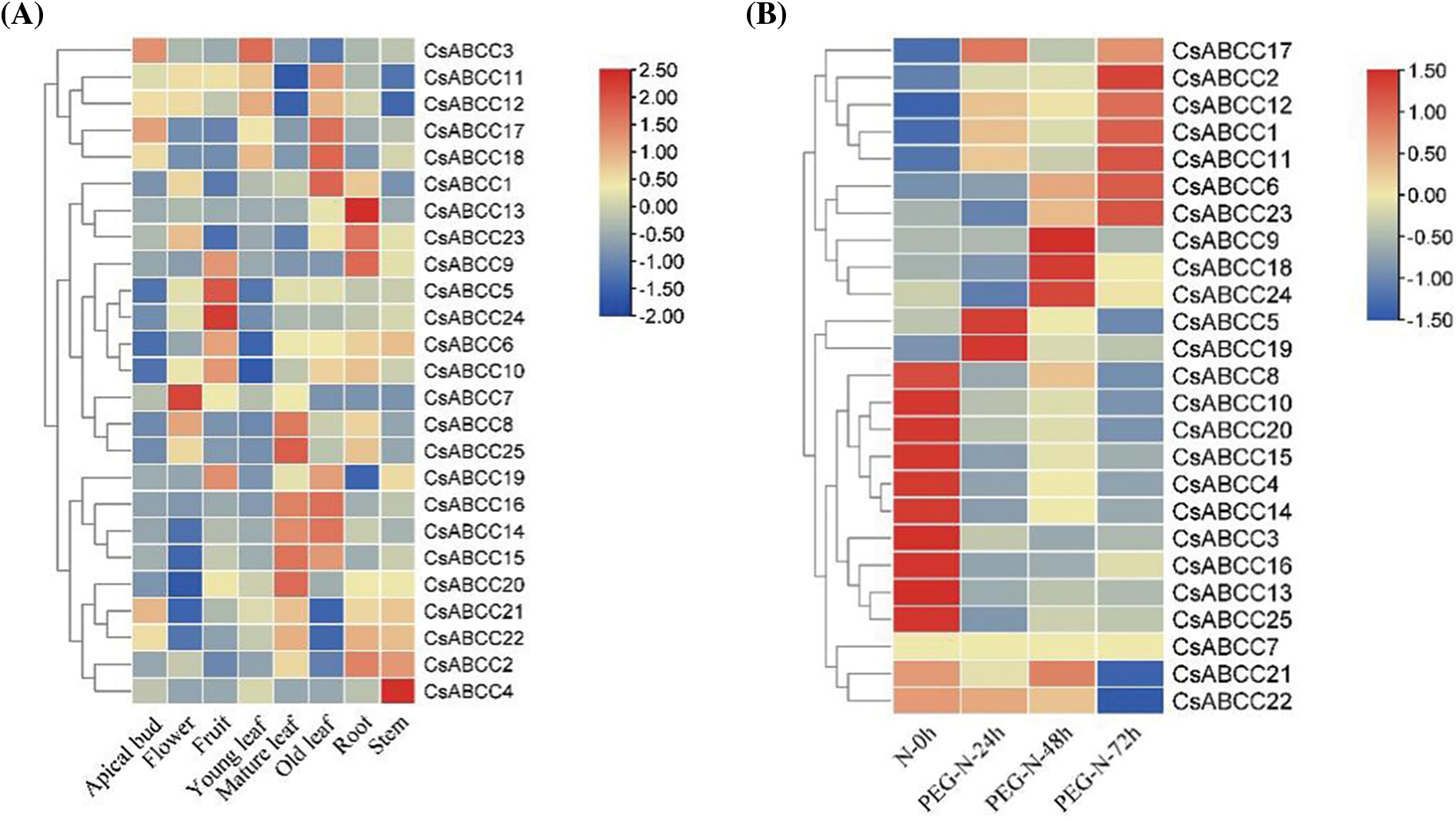
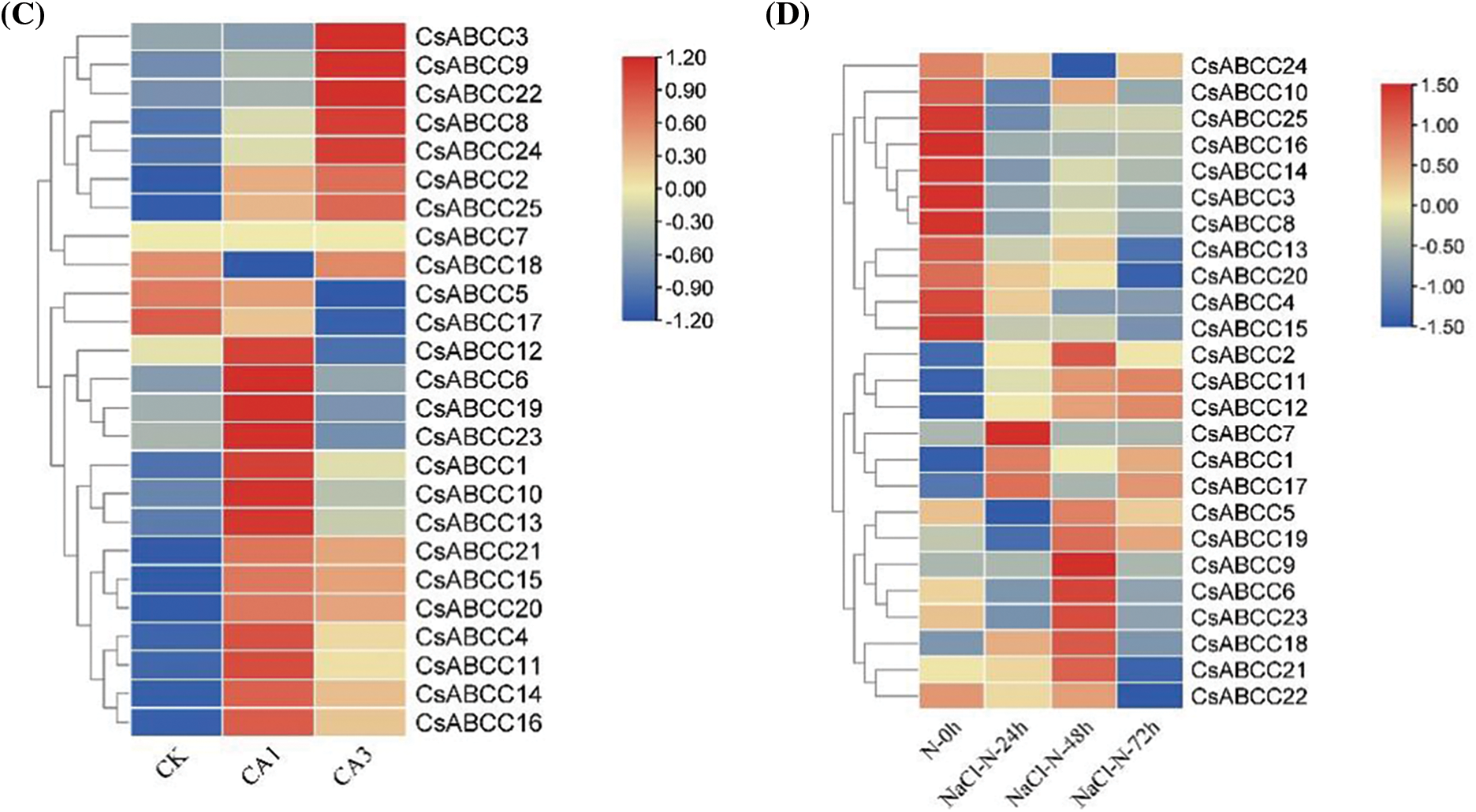
Figure 7: Expression profiles of CsABCC genes in 8 tissues and under different stress conditions. (A) Heat map of CsABCC expression in different tissues of tea plant. (B) Expression heat map of CsABCC under drought treatment. (C) Expression heat map of CsABCC under cold stress. (D) Expression heat map of CsABCC under salt stress
Under drought stress, the expression of three CsABCC genes (5,17,19) was significantly up-regulated at 24 h, the expression of four CsABCC genes (9,18,21,24) was significantly up-regulated at 48 h, and the expression of seven CsABCC genes (1,2,6,11,12,17,23) was up-regulated at 72 h, and the expression of CsABCC6 and CsABCC23 was continuously up-regulated (Fig. 7B). Under cold conditions, most CsABCC genes were significantly up-regulated in CA1 and CA3 states (Fig. 7C). Under salt treatment, the expression of three CsABCC genes (1,7,17) was significantly up-regulated at 24 h, the expression of 10 CsABCC genes (2,5,6,9,11,12,18,19,21,23) was up-regulated at 48 h, and the expression of three CsABCC genes (11,12,17) was up-regulated at 72 h (Fig. 7D). These results indicate that CsABCC gene plays an important role in different stress environments.
3.7 The Expression Level of CsABCC11 Was Closely Related to the Content of EGCG in Tea Buds
To explore the relationship between CsABCC11 and EGCG content in buds of Kingbird, according to the EGCG content from low to high, the EGCG content of 108 strains of Kingbird was divided into 7 groups for qRT-PCR expression analysis. The primer sequence of CsABCC11 gene is shown in Table S3. It can be seen from Fig. 8 that with the increase of EGCG content, the expression of the CsABCC11 gene in tea plants increased gradually, indicating that EGCG content was positively correlated with CsABCC11 expression, which also indicated that CsABCC11 transporter promoted the synthesis of EGCG in the buds of Kingbird.
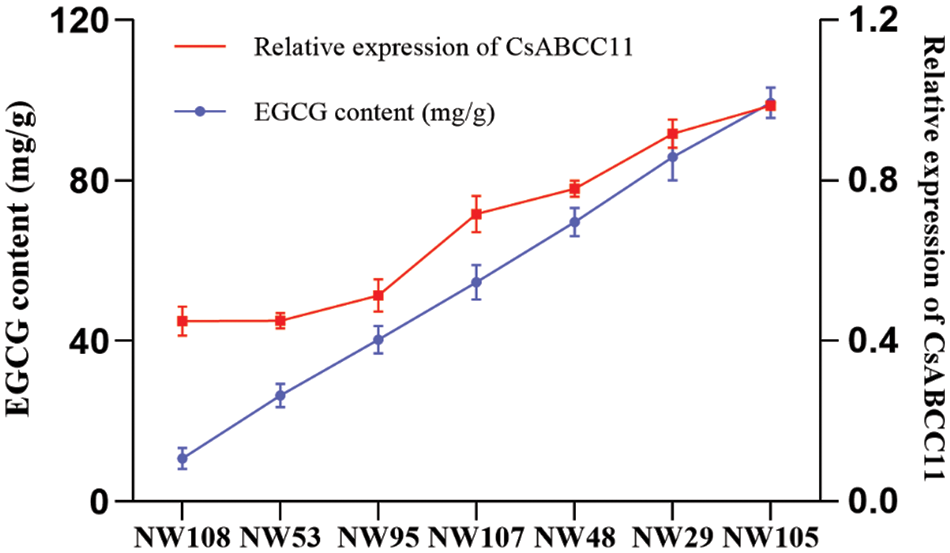
Figure 8: Correlation analysis of EGCG in 108 strains of CsABCC11 Kingbird. The blue line represents the EGCG content, the red line represents the relative expression of CsABCC11, and NW represents the number of Kingbird. SPSS was used for correlation analysis, r = 0.9767, p = 0.0002 < 0.05
3.8 Subcellular Localization Results
ABC transporters are usually membrane localization proteins [36]. In order to find the subcellular location of CsABCC11 protein, we fused the GFP empty protein with the CsABCC11 protein transiently expressed in tobacco leaves. As shown in the Fig. 9, the empty vector PCAMBIA1300-35S-GFP can detect green fluorescence signals in tobacco epidermal cell membrane and nucleus, and the plant fusion expression vector PCAMBIA1300-CsABCC11-35S-GFP can detect green fluorescence signals in tobacco epidermal plasma membrane, which proves that CsABCC11 protein is located in the plasma membrane.
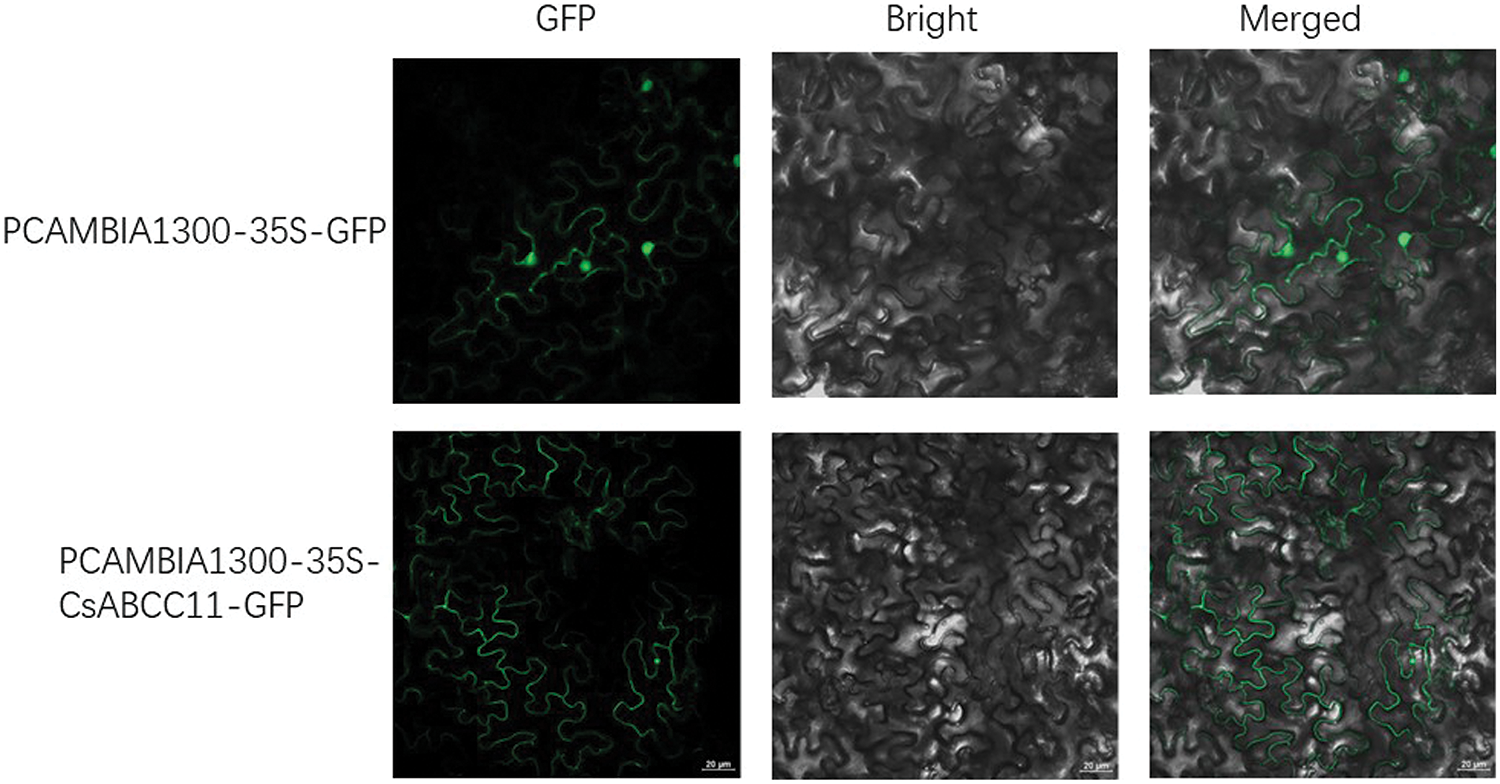
Figure 9: Subcellular localization of CsABCC11 protein. Confocal microscopy was used to obtain the CsABCC11-GFP protein image in N. benthamiana mesophyll cells, and the empty vector GFP was used as a control. Bright represents the bright field picture, and Merged represents the superimposed field picture
3.9 The EGCG Content of Fudingdabai Tea Buds Decreased after CsABCC11 Silencing
The latest established VIGS technology [35] was used to detect the change of EGCG content in tea plant cuttings, and the relative expression level of CsABCC11 was verified by qRT-PCR (Fig. 10A). The relative expression of CsABCC11 was significantly decreased after silencing. The results showed that CsABCC11 was silenced in tea cuttings, and VIGS results showed that CsABCC11 gene silencing inhibited the accumulation of EGCG in tea buds. Compared with WT and cuttings infected with pTRV1 + pTRV2, the EGCG content of cuttings infected with pTRV2-CsABCC11 was significantly reduced, and pTRV2-CsABCC11 was about 7.0 times lower than that of WT and no-load (Fig. 10B). Silencing CsABCC11 significantly inhibited the accumulation of EGCG. These results indicate that CsABCC11 is required for EGCG intracellular transport in tea plants.
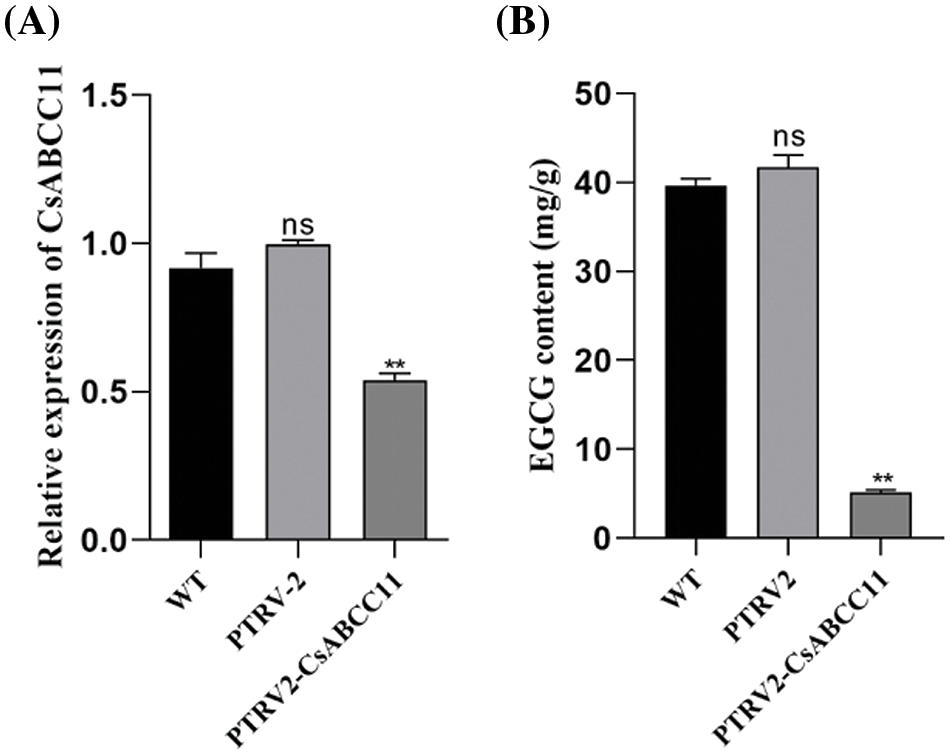
Figure 10: Using the VIGS system to silence CsABCC11 in tea leaves. A shows the expression of CsABCC11 after VIGS. B shows the content of EGCG, TRV, tobacco brittle fracture virus after VIGS; WT, wild type. pTRV2, pTRV1 + pTRV2 agrobacterium-infected tea cuttings; pTRV2-CsABCC11, pTRV1 + pTRV2-CsABCC11 agrobacterium-infected tea cuttings. “**” indicates that there are significant differences between different samples. “ns” indicates that there is no significant difference
There are many reports on the study of ABCC gene in other species, but there are few reports on tea plants. EGCG is the main component that determines the color, aroma and taste of tea. EGCG is the highest catechin content in green tea, which can reach 6%–10% of the dry weight of tea. We used bioinformatics analysis, correlation analysis, subcellular localization, qRT-PCR and VIGS-mediated gene silencing strategy to clarify that CsABCC11 is involved in the transport of EGCG in tea plant buds.
4.1 Bioinformatics Analysis of CsABCC Genes Subfamily and Its Relationship with Environmental Response
We identified 25 CsABCC genes at the genomic level of tea plant (Table 1). According to the conserved domain of ABCC transporter protein, it contains a common domain (MRP), which is divided into four main phylogenetic groups (Fig. 4). It can be seen that the homology between each group is very high, which also indicates that the functions of the same subfamily may be similar. Multi-species phylogenetic tree analysis shows that the ABCC gene family is evolving in multiple directions (Fig. 1). CsABCC protein showed highly identical motif composition in the same subfamily (Fig. 2). It means that the evolution of these motifs is conservative.
The diversity of gene structure can provide important information about the evolutionary history of gene families and provide additional help for phylogenetic classification. The number of introns of CsABCC genes is between 7 and 17 (Fig. 4), which is much more than that of other species such as Arabidopsis [37]. This phenomenon indicates that CsABCC family genes exhibit high genetic structure diversity. The members of the same subgroup showed closely related intron-exon distribution patterns; this also proves the functional similarity between members of the same evolutionary group.
ABCC gene is involved in a variety of plant abiotic stress (such as salt, metal stress) has been widely studied [38]. In Arabidopsis thalian, Sedum alfredii, etc., ABCC genes have been identified to respond to various abiotic stresses. We analyzed the cis-elements of CsABCC genes, the promoter region contained a large number of promoter cis-acting elements such as Light response, plant hormone response, plant growth and development, adversity stress and so on. As a class of non-coding small RNA (sRNA), microRNA (miRNA) is regulated at the post-transcriptional level by binding to target genes.The miRNAs have been shown to be involved in the regulation of plant secondary metabolites [39]. The miRNA-CsABCC network diagram (Fig. 6) showed that a large number of miRNA binding to CsABCCs may regulate the connotation of tea plant substances. The expression pattern study found that CsABCC had important contributions to the growth and development of tea plants and different stress environments (Fig. 7). These results suggest that CsABCC genes may have a response in the above pathways, and those that also typically induce the synthesis of EGCG for the growth or resistance to stress of the tea plant itself.
4.2 CsABCC11 Were Related to the Accumulation of EGCG in Tea Plant Buds
The tea plant is the most EGCG-containing plant in nature. EGCG has a very special stereochemical structure and strong antioxidant activity [40]. EGCG has a broad-spectrum anti-viral effect. It can not only inhibit coronavirus, but also has obvious inhibitory effects on influenza virus, adenovirus, HIV (AIDS virus), HPV (human papillomavirus), EB virus and other viruses [41,42]. It is expected to become a potential broad-spectrum anti-viral drug. Through correlation analysis, we found that the EGCG content of 108 strains of the Kingbird was positively correlated with the expression of CsABCC11 (Fig. 8). This shows that the content of EGCG is highly correlated with the expression of CsABCC11.
In plants, ABCC transporters are involved in the transport of secondary metabolites. Many studies have shown that MRP is located in the plasma membrane and participates in the accumulation of secondary metabolites, suggesting that MRP is involved in the vacuolar transport of anthocyanins. The bz2 mutant showed that MRP (ABCC) was involved in the vacuolar transport of phenolic glycosides [43]. Such as Arabidopsis. and carnation. Gao et al. provided further strong evidence for the involvement of MRP protein in anthocyanin accumulation through reverse genetics research. The ZmMRP3 is localized to the vacuolar membrane [13]. It is necessary for the accumulation of anthocyanin in maize. Interestingly, the above plant ABCC proteins were localized on the tonoplast, while the subcellular localization of CsABCC11 protein in our study was predicted to be in the plasma membrane, and the subcellular localization of CsABCC11 protein was confirmed by tobacco subcellular (Fig. 9) localization experiments.
To verify that CsABCC11 gene is involved in the accumulation of EGCG content in tea plant buds, we used the newly developed virus-mediated gene silencing (VIGS) technology to silence CsABCC11. The findings indicated a significant reduction in EGCG content in silent branches compared to non-silent branches. qRT-PCR further showed that CsABCC11 was silenced. These findings confirm the involvement of CsABCC11 in the accumulation of EGCG in tea buds. Virus-induced gene silencing led to a significant decrease in EGCG accumulation (Fig. 10). CsABCC11 appears to play a pivotal role in the accumulation of EGCG in tea plants. This may be due to ABCC transporters are known to facilitate the transport of multiple substrates across biofilms, which is a very important ability for the biosynthesis of EGCG in tea plant. Our study laid a foundation for studying the biological function of CsABCC and provided a new candidate molecular marker gene for further EGCG-related variety breeding, which will be of great interest to breeders.
In the early stage of this study, a gene CsABCC11 was located by GWAS of EGCG content in 108 strains of bird king species, and we identified 25 CsABCC genes at the genomic level of tea plant. Bioinformatics analysis and expression pattern analysis were performed. Correlation analysis showed that the expression of CsABCC11 was positively correlated with EGCG content. Subcellular localization experiments confirmed that the CsABCC11 protein was localized on the plasma membrane. Silencing CsABCC11 in tea plants by VIGS strategy reduced the content of EGCG in tea buds. In summary, this study found that CsABCC11 was closely related to EGCG content in tea plant buds, which provided a solid groundwork for further research into the function of CsABCC and the biosynthesis and transportation mechanism of EGCG.
Acknowledgement: The authors would like to extend their sincere appreciation to the researchers.
Funding Statement: This work was supported by the Guizhou University Talent Introduction Program ([2021]05); Guizhou University Cultivation Program ([2020]48); Institute of Technology of YF ([2022] 017); Guizhou Province High-Level Innovative Talents “Hundred” Level Talent Project (Qiankehe Platform Talent) GCC[2023]014; Supported by the earmarked fund for GZMARS-Tea and Research on the Planting Technology of China HUANENG Photovoltaic Tea Garden (Project No. HNKJ2022-H135).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Litang Lu, Mingyuan Luo; data collection: Mingyuan Luo; analysis and interpretation of results: Mingyuan Luo, Hu Tang, Xinzhuan Yao; Zifan Yang, Huagen Hao, Yue Wan; draft manuscript preparation: Mingyuan Luo, Zhouzhuoer Chen, Shiyu Tian, Fei Liu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All the data and materials supporting the findings of this study are included in this article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.052938.
References
1. Geisler M, Hegedűs T. A twist in the ABC: regulation of ABC transporter trafficking and transport by FK506-binding proteins. FEBS Lett. 2020;594(23):3986–4000. doi:10.1002/feb2.v594.23. [Google Scholar] [CrossRef]
2. Guo Z, Yuan X, Li L, Zeng M, Yang J, Tang H, et al. Genome-wide analysis of the ATP-binding cassette (ABC) transporter family in Zea mays L. and its response to heavy metal stresses. Int J Mol Sci. 2022;23(4):2109. doi:10.3390/ijms23042109. [Google Scholar] [PubMed] [CrossRef]
3. Lee J, Yang J, Zhitnitsky D, Lewinson O, Rees D. Structural and functional characterization of a heavy metal detoxifying ABC transporter (997.2). FASEB J. 2014;28(S1):997–2. [Google Scholar]
4. He G, Tian W, Qin L, Meng L, Wu D, Huang Y, et al. Identification of novel heavy metal detoxification proteins in Solanum tuberosum: insights to improve food security protection from metal ion stress. Sci Total Environ. 2021;779:146197. doi:10.1016/j.scitotenv.2021.146197. [Google Scholar] [PubMed] [CrossRef]
5. Ren H, Li X, Guo L, Wang L, Hao X, Zeng J. Integrative transcriptome and proteome analysis reveals the absorption and metabolism of selenium in tea plants [Camellia sinensis (L.) O. Kuntze]. Front Plant Sci. 2022;13:848349. doi:10.3389/fpls.2022.848349. [Google Scholar] [PubMed] [CrossRef]
6. Wang H, Liu Y, Peng Z, Li J, Huang W, Liu Y, et al. Ectopic expression of poplar ABC transporter PtoABCG36 confers Cd tolerance in Arabidopsis thaliana. Int J Mol Sci. 2019;20(13):3293. doi:10.3390/ijms20133293. [Google Scholar] [PubMed] [CrossRef]
7. Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8(1):67–113. doi:10.1146/cellbio.1992.8.issue-1. [Google Scholar] [CrossRef]
8. Ben Saad A, Bruneau A, Mareux E, Lapalus M, Delaunay J-L, Gonzales E, et al. Molecular regulation of canalicular ABC transporters. Int J Mol Sci. 2021;22(4):2113. doi:10.3390/ijms22042113. [Google Scholar] [PubMed] [CrossRef]
9. Bieczynski F, Painefilú JC, Venturino A, Luquet CM. Expression and function of ABC proteins in fish intestine. Front Physiol. 2021;12:791834. doi:10.3389/fphys.2021.791834. [Google Scholar] [PubMed] [CrossRef]
10. Mall MS, Shah S, Singh S, Singh N, Singh N, Vaish S, et al. Genome-wide identification and characterization of ABC transporter superfamily in the legume Cajanus cajan. J Appl Genet. 2023;64(4):615–44. doi:10.1007/s13353-023-00774-8. [Google Scholar] [PubMed] [CrossRef]
11. Khan N, You FM, Datla R, Ravichandran S, Jia B, Cloutier S. Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genomics. 2020;21(1):722. doi:10.1186/s12864-020-07121-9. [Google Scholar] [PubMed] [CrossRef]
12. Chen P, Li Y, Zhao L, Hou Z, Yan M, Hu B, et al. Genome-wide identification and expression profiling of ATP-binding cassette (ABC) transporter gene family in pineapple (Ananas comosus (L.) Merr.) Reveal the role of AcABCG38 in pollen development. Front Plant Sci. 2017;8:2150. doi:10.3389/fpls.2017.02150. [Google Scholar] [PubMed] [CrossRef]
13. Gao X, Zou R, Sun H, Liu J, Duan W, Hu Y, et al. Genome-wide identification of wheat ABC1K gene family and functional dissection of TaABC1K3 and TaABC1K6 involved in drought tolerance. Front Plant Sci. 2022;13:991171. doi:10.3389/fpls.2022.991171. [Google Scholar] [PubMed] [CrossRef]
14. Goodman CD, Casati P, Walbot V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. The Plant Cell. 2004;16(7):1812–26. doi:10.1105/tpc.022574. [Google Scholar] [PubMed] [CrossRef]
15. Sun L, Wang Y, Ding Z, Liu F. The dynamic changes of catechins and related genes in tea (Camellia sinensis) flowers. Acta Physiol Plant. 2019;41(2):30. doi:10.1007/s11738-019-2822-0. [Google Scholar] [CrossRef]
16. Liu B, Yan W. Lipophilization of EGCG and effects on antioxidant activities. Food Chem. 2019;272:663–9. doi:10.1016/j.foodchem.2018.08.086. [Google Scholar] [PubMed] [CrossRef]
17. Yan X, Zhang X, McClements DJ, Zou L, Liu X, Liu F. Co-encapsulation of epigallocatechin gallate (EGCG) and curcumin by two proteins-based nanoparticles: role of EGCG. J Agric Food Chem. 2019;67(48):13228–36. doi:10.1021/acs.jafc.9b04415. [Google Scholar] [PubMed] [CrossRef]
18. Wei Y, Chen P, Ling T, Wang Y, Dong R, Zhang C, et al. Certain (−)-epigallocatechin-3-gallate (EGCG) auto-oxidation products (EAOPs) retain the cytotoxic activities of EGCG. Food Chem. 2016;204:218–26. doi:10.1016/j.foodchem.2016.02.134. [Google Scholar] [PubMed] [CrossRef]
19. Romano A, Martel F. The role of EGCG in breast cancer prevention and therapy. Mini-Rev Med Chem. 2021;21(7):883–98. doi:10.2174/1389557520999201211194445. [Google Scholar] [PubMed] [CrossRef]
20. Wang Y-Q, Lu J-L, Liang Y-R, Li Q-S. Suppressive effects of EGCG on cervical cancer. Molecules. 2018;23(9):2334. doi:10.3390/molecules23092334. [Google Scholar] [PubMed] [CrossRef]
21. Zhang Z, Zhang X, Bi K, He Y, Yan W, Yang CS, et al. Potential protective mechanisms of green tea polyphenol EGCG against COVID-19. Trends Food Sci Technol. 2021;114:11–24. doi:10.1016/j.tifs.2021.05.023. [Google Scholar] [PubMed] [CrossRef]
22. Marwah M, Perrie Y, Badhan RKS, Lowry D. Intracellular uptake of EGCG-loaded deformable controlled release liposomes for skin cancer. J Liposome Res. 2020;30(2):136–49. doi:10.1080/08982104.2019.1604746. [Google Scholar] [PubMed] [CrossRef]
23. Karimi-Shahri M, Alalikhan A, Hashemian P, Hashemzadeh A, Javid H. The applications of epigallocatechin gallate (EGCG)-nanogold conjugate in cancer therapy. Nanotechnology. 2023;34(21):212001. doi:10.1088/1361-6528/acaca3. [Google Scholar] [PubMed] [CrossRef]
24. Chu C, Deng J, Man Y, Qu Y. Green tea extracts epigallocatechin-3-gallate for different treatments. Biomed Res Int. 2017;2017:5615647. [Google Scholar] [PubMed]
25. Li X, Li Y, Ahammed GJ, Zhang X-N, Ying L, Zhang L, et al. RBOH1-dependent apoplastic H2O2 mediates epigallocatechin-3-gallate-induced abiotic stress tolerance in Solanum lycopersicum L. Environ Exp Bot. 2019;161:357–66. doi:10.1016/j.envexpbot.2018.11.013. [Google Scholar] [CrossRef]
26. Koech RK, Malebe PM, Nyarukowa C, Mose R, Kamunya SM, Joubert F, et al. Functional annotation of putative QTL associated with black tea quality and drought tolerance traits. Sci Rep. 2019;9(1):1465. doi:10.1038/s41598-018-37688-z. [Google Scholar] [PubMed] [CrossRef]
27. Ahammed GJ, Wu Y, Wang Y, Guo T, Shamsy R, Li X. Epigallocatechin-3-Gallate (EGCGa unique secondary metabolite with diverse roles in plant-environment interaction. Environ Exp Bot. 2023;209:105299. doi:10.1016/j.envexpbot.2023.105299. [Google Scholar] [CrossRef]
28. Gai Z, Wang Y, Ding Y, Qian W, Qiu C, Xie H, et al. Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Sci Rep. 2020;10(1):12275. doi:10.1038/s41598-020-69080-1. [Google Scholar] [PubMed] [CrossRef]
29. Zhou H, Zhou W, Yao X, Zhao Q, Lu L. Genome-wide investigation and functional analysis reveal that csgebp4 is required for tea plant trichome formation. Int J Mol Sci. 2023;24(6):5207. doi:10.3390/ijms24065207. [Google Scholar] [PubMed] [CrossRef]
30. Zhou T, Chen J, Huang Y, Jin Z, Li J, Li Y, et al. Genome-wide identification and expression analysis of the PIN auxin transporter gene family in Zanthoxylum armatum DC. Agriculture. 2022;12(9):1318. doi:10.3390/agriculture12091318. [Google Scholar] [CrossRef]
31. Zhang T, Cai J, Wang S, Lv L, Yuan D, Zeng X, et al. Identification and expression analysis of the ethylene response factor gene family in tea plant (Camellia sinensis). Agronomy. 2023;13(7):1900. doi:10.3390/agronomy13071900. [Google Scholar] [CrossRef]
32. Mo L, Yao X, Tang H, Li Y, Jiao Y, He Y, et al. Genome-wide investigation and functional analysis reveal that CsKCS3 and CsKCS18 are required for tea cuticle wax formation. Foods. 2023;12(10):2011. doi:10.3390/foods12102011. [Google Scholar] [PubMed] [CrossRef]
33. Han Z, Zhang C, Zhang H, Duan Y, Zou Z, Zhou L, et al. CsMYB transcription factors participate in jasmonic acid signal transduction in response to cold stress in tea plant (Camellia sinensis). Plants. 2022;11(21):2869. doi:10.3390/plants11212869. [Google Scholar] [PubMed] [CrossRef]
34. Yao X, Chen H, Ai A, Wang F, Lian S, Tang H, et al. The transcription factor CsS40 negatively regulates TCS1 expression and caffeine biosynthesis in connection to leaf senescence in Camellia sinensis. Hortic Res. 2023;10(9):uhad162. doi:10.1093/hr/uhad162. [Google Scholar] [PubMed] [CrossRef]
35. Li G, Li Y, Yao X, Lu L. Establishment of a virus-induced gene-silencing (VIGS) system in tea plant and its use in the functional analysis of CsTCS1. Int J Mol Sci. 2023;24(1):392. [Google Scholar]
36. Kashiwayama Y, Seki M, Yasui A, Murasaki Y, Morita M, Yamashita Y, et al. 70-kDa peroxisomal membrane protein related protein (P70R/ABCD4) localizes to endoplasmic reticulum not peroxisomes, and NH2-terminal hydrophobic property determines the subcellular localization of ABC subfamily D proteins. Exp Cell Res. 2009;315(2):190–205. doi:10.1016/j.yexcr.2008.10.031. [Google Scholar] [PubMed] [CrossRef]
37. Kolukisaoglu ÜH, Bovet L, Klein M, Eggmann T, Geisler M, Wanke D, et al. Family business: the multidrug-resistance related protein (MRP) ABC transporter genes in Arabidopsis thaliana. Planta. 2002;216(1):107–19. doi:10.1007/s00425-002-0890-6. [Google Scholar] [PubMed] [CrossRef]
38. Chu HD, Le QN, Nguyen HQ, Le DT. Genome-wide analysis of genes encoding methionine-rich proteins in Arabidopsis and soybean suggesting their roles in the adaptation of plants to abiotic stress. Int J Genom. 2016;2016:5427062. [Google Scholar]
39. Li H, Lin Q, Yan M, Wang M, Wang P, Zhao H, et al. Relationship between secondary metabolism and miRNA for important flavor compounds in different tissues of tea plant (Camellia sinensis) as revealed by genome-wide miRNA analysis. J Agric Food Chem. 2021;69(6):2001–12. doi:10.1021/acs.jafc.0c07440. [Google Scholar] [PubMed] [CrossRef]
40. Ahmed NA, Radwan NM, Aboul Ezz HS, Salama NA. The antioxidant effect of green tea mega EGCG against electromagnetic radiation-induced oxidative stress in the hippocampus and striatum of rats. Electromagn Biol Med. 2017;36(1):63–73. [Google Scholar] [PubMed]
41. Bachar SC, Mazumder K, Bachar R, Aktar A, Al Mahtab M. A review of medicinal plants with antiviral activity available in bangladesh and mechanistic insight into their bioactive metabolites on SARS-CoV-2, HIV and HBV. Front Pharmacol. 2021;12:PF7. doi:10.3389/fphar.2021.732891. [Google Scholar] [PubMed] [CrossRef]
42. da Silva-Júnior FE, Silva RL. Multi-target approaches of epigallocatechin-3-O-gallate (EGCG) and its derivatives against influenza viruses. Curr Top Med Chem. 2022;22(18):1485–500. doi:10.2174/1568026622666220127112056. [Google Scholar] [PubMed] [CrossRef]
43. Marrs KA, Alfenito MR, Lloyd AM, Walbot V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature. 1995;375(6530):397–400. doi:10.1038/375397a0. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools