 Open Access
Open Access
ARTICLE
Genome-Wide Identification and Expression Analysis of the GSK3 Gene Family in Sunflower under Various Abiotic Stresses
Department of Ornamental Horticulture, Anhui Agricultural University, Hefei, 230061, China
* Corresponding Author: Xinyi Deng. Email:
# Xianwen Ji and Ziying Jiang contributed equally to this work
(This article belongs to the Special Issue: Abiotic Stress Impacts on Plant Physiology and Their Alleviation)
Phyton-International Journal of Experimental Botany 2024, 93(8), 1839-1850. https://doi.org/10.32604/phyton.2024.052809
Received 16 April 2024; Accepted 24 June 2024; Issue published 30 August 2024
Abstract
Genes in the glycogen synthase kinase 3 (GSK3) family are essential in regulating plant response to stressful conditions. This study employed bioinformatics to uncover the GSK3 gene family from the sunflower genome database. The expressions of GSK3 genes in different tissues and stress treatments, such as salt, drought, and cold, were assessed using transcriptome sequencing and quantitative real-time PCR (qRT-PCR). The study results revealed that the 12 GSK3 genes of sunflower, belonging to four classes (Classes I–IV), contained the GSK3 kinase domain and 11–13 exons. The majority of GSK3 genes were highly expressed in the leaf axil and flower, while their expression levels were relatively lower in the leaf. As a result of salt stress, six of the GSK3 genes (HaSK11, HaSK22, HaSK23, HaSK32, HaSK33, and HaSK41) displayed a notable increase in expression, while HaSK14 and HaSK21 experienced a significant decrease. With regard to drought stress, five of the GSK3 genes (HaSK11, HaSK13, HaSK21, HaSK22, and HaSK33) experienced a remarkable rise in expression. When exposed to cold stress, seven of the GSK3 genes (HaSK11, HaSK12, HaSK13, HaSK32, HaSK33, HaSK41, and HaSK42) showed a substantial increase, whereas HaSK21 and HaSK23 had a sharp decline. This research is of great importance in understanding the abiotic resistance mechanism of sunflowers and developing new varieties with improved stress resistance.Keywords
Supplementary Material
Supplementary Material FileGlycogen synthase kinase 3 (GSK3) is a conserved serine/threonine protein kinase, and is also referred to as SHAGGY-like protein kinase. GSK3 was initially found in rabbit skeletal muscle, and existed in two isoforms (GSK3α and GSK3β) in mammals [1]. Subsequently, GSK3 proteins are identified in various plants, including 10 in Arabidopsis, nine in rice, and nine in potatoes [2,3]. These discoveries suggest a greater diversity of the GSK3 gene family in plants than the corresponding genes in animals.
GSK3 protein contributes to the formation of flowers, stomata, seeds, plant height, flowering time and root systems in plants. BRASSINOSTEROID INSENSITIVE2 (BIN2), a GSK3-like kinase, has been identified to phosphorylate the transcription factors BRI1-EMS-SUPPRESSOR 1 (BES1) and Brassinazole resistant 1 (BZR1), thus inhibiting the signal transduction pathway of brassinolide (BR) and regulating plant growth [4,5]. Research has revealed that BIN2 and BIL1/2 can mediate root growth and seed germination by antagonizing BR and abscisic acid (ABA) pathways [6–8]. Claisse et al. observed that the overexpression of AtSK32-R178A resulted in shorter pedicels and smaller petals than the wild-type controls [9]. GSK3 phosphorylated and regulated Rht-B1b to reduce plant height in wheat [10]. GSK3 could interact with VRN1 and regulate its accumulation to mediate flowering in wheat [11]. GSK3-like kinase ZmSK2, a maize homolog of BIN2 in Arabidopsis, was proven to be involved in embryonic development [12].
GSK3 has additionally been linked to responses to biotic and abiotic stressors [3,13]. Research has demonstrated that AtSK13, AtSK31, and AtSK42 are all activated in response to salt stress [14]. The overexpression of AtSK22 has been associated with improved salt tolerance in plants [15]. Evidence from the research of Dal Santo et al. [16] and Stampfl et al. [17] suggested that increased expression of AtSK11 augmented salt tolerance in plants and decreased their sensitivity to Pseudomonas syringae. Moreover, Yang et al. [18] found that increased expression of OsGSK3 in rice enhanced ABA responsiveness as well as tolerance to salt, cold, drought, and mechanical damage. AtBIN2 has been demonstrated to interact with RD26 and augment the drought tolerance of plants [19]. AtBIN2 can also phosphorylate and impede the activity of SOS2, thereby inhibiting the salt stress tolerance of Arabidopsis [20]. Under cold stress, BIN2 can phosphorylate and bolster the activity of BZR1, CESTA, and ICE1, thereby improving the cold tolerance of seedlings [21–23] and raising the level of C-REPEAT BINDING FACTOR (CBF). Researchers have discovered that the interaction between BIN2 and JAZ proteins lessens plant resistance to Verticillium wilt, as reported by Song et al. [24]. GmSK2-8, which can be strongly induced under high-salt conditions, interacted with GmNSP1a and GmNSP1b to mediate salt-inhibited legume-rhizobiumsymbiosis [25].
Helianthus annuus L. is an essential oil crop and ornamental plant. However, its production is significantly influenced by numerous abiotic and biotic stresses. Although the GSK3 gene family is implicated in many stress response processes of plants, there is limited information about the GSK3 gene family in sunflowers. The present investigation aimed to identify the GSK3 genes of sunflowers and analyze their protein sequence, gene structure, cis-acting elements, and expression traits across diverse tissues and treatments. This research serves as a fundamental basis for further exploring the role of GSK3 genes in stress in plants.
In this study, H. annuus var. ‘huoli’ was used. The sunflower plants were grown in nutrient-rich soil in square pots and put in a tissue culture room under the following growth conditions: 5000 lx, 16/8 h (light/dark), 23 ± 2°C.
2.2 Identification of GSK3 Genes in Sunflower
The protein sequences in the sunflower genome database (https://www.heliagene.org/HanXRQr2.0-SUNRISE/, accessed on 20 May 2024) were screened using the sunflower GSKs as queries. After removing the repetitive sequences and those without the GSK3 domain, the GSK3 genes of the sunflower were determined. To validate sunflower GSKs, the Hidden Markov Model (HMM) was employed. To get the molecular weights, theoretical isoelectric points, and number of amino acids in each sunflower GSK sequence, the sequences were uploaded to ExPASy (http://web.expasy.org/protparam/, accessed on 20 May 2024) for analysis. The sequences of Arabidopsis GSKs were obtained from NCBI.
2.3 Phylogenetic, Gene Structure, and Motif Distribution
The MEME Suite 5.0.5 was utilized to examine the conservative motifs of sunflower GSK3 genes. To validate the GSK3 gene structure, GSDS 2.0 software (http://gsds.cbi.pku.edu.cn/, accessed on 20 May 2024) was employed. To construct a maximum likelihood phylogenetic tree of the GSK3 protein sequence of sunflower and Arabidopsis, Clustal X 1.8 and MEGA 11.0 were employed and bootstrap repeat analysis was conducted 1000 times [26].
2.4 Prediction of the Cis-Regulatory Elements in GSK3 Genes
The upstream flanking sequence (2000 bp) from the transcription start site of each putative sunflower GSK3 gene was downloaded. Utilizing PlantCARE online software, the conserved cis-elements in the promoter regions of sunflower GSK3 genes were studied.
2.5 Expression Profile Analysis of GSK3 Genes
RNA was extracted from the buds, roots, leaves, stems, and flowers of H. annuus var. ‘huoli’. RNA extraction, RNA sequencing, and data analysis were all conducted according to the method described by Dong et al. [27].
H. annuus var. ‘huoli’ seeds were grown in pots with mixed soil (1:1, vermiculite/humus) for a duration of 30 days. Then the seedlings were irrigated with either 300 mM NaCl or 20% PEG 6000. The control group was irrigated with water. Six hours later, the sunflower leaves were harvested for gene expression analysis [28]. The 30-day old seedlings were placed in an incubator at 5°C for cold treatment, while the control group was subjected to 25°C. Six hours later, the leaves were gathered.
Total RNA extraction was performed with TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China) following the manufacturer’s protocol. Utilizing MonScript™ RTIII all-in-one mix with dsDNase (Monad, Wuhan, China), the first cDNA strand was generated from 1 µg of total RNA. Using the SYBR Green PCR Master Mix (Toyobo, Shanghai, China), qRT-PCR assay was conducted on an ABI 7500 Fast real-time PCR system. The reaction procedure was as foIlows: denature at 95°C for 1 min, foIlowed by 45 cycles of 95°C for 20 s, 50°C–60°C for 20 s, 72°C for 30 s.
To obtain accurate results, three biological and technical replicates were employed for every treatment. For each biological replicate, five sunflower plants were utilized. Elongation factor 1 alpha (EF1α) was employed as the reference gene (Table S1). Relative gene expression were calculated based on the 2−ΔΔCt method [29].
Data was reported as mean value ± standard deviation (SD). Student’s t-tests were conducted for statistical analyses.
3.1 Identification and Classification of GSK3 Gene Family in Sunflower
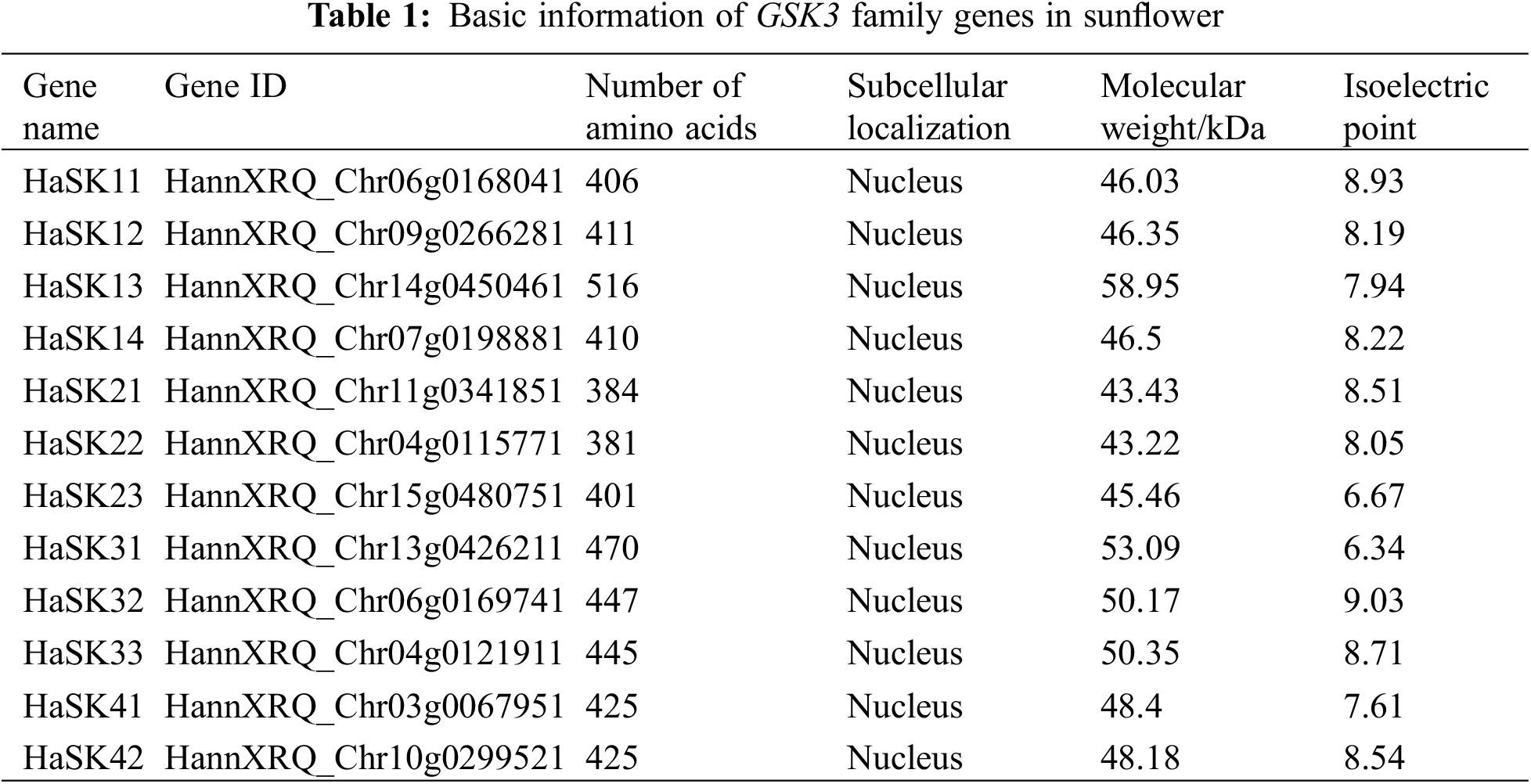
We analyzed the sunflower genomic database and identified 12 GSK3 genes. The proteins encoded by the GSK3 genes displayed a range of lengths from 381 amino acids (HaSK22) to 516 amino acids (HaSK13). Molecular weights ranged from 43.2 kDa (HaSK22) to 59.0 kDa (HaSK13). Isoelectric points from 6.34 (HaSK31) to 9.03 (HaSK32). According to predictions regarding subcellular localization, all GSK3 proteins were present in the nucleus (Table 1).
Sequencing anazlysis showed that the protein sequences of GSK3 members were highly conserved across the kinase domain, while the N- and C-terminal regions are variable. The multiple conserved subdomains of GSK3 proteins included APE, GxGxxG, xAxK, RD, SYICSR, CDFGSAK and the TREE motif (Fig. 1). Among them, the presence of SYICSR and CDFGSAK motifs can distinguish HaSK proteins from other serine/threonine protein kinases, while the TREE motif is a unique feature of plant GSK proteins.

Figure 1: Sequence alignment of sunflower GSK3 proteins. Note: The subdomains of kinase domain were marked with blue frames
3.2 Phylogenetic Analysis and Grouping of GSK3 Proteins
A phylogenetic tree with 10 Arabidopsis GSK3 proteins, nine rice GSK3, and 12 sunflower GSK3 proteins was created to examine (Fig. 2). Using the Arabidopsis and rice GSK3 protein groups as a reference, 12 GSK3 proteins have been divided into four subgroups (I–IV). Group I had four members, including HaSK11, HaSK12, HaSK13, HaSK14. Group II had HaSK21, HaSK22, HaSK23. Group III had HaSK31, HaSK32, and HaSK33. Group IV comprised two members, including HaSK41 and HaSK42.
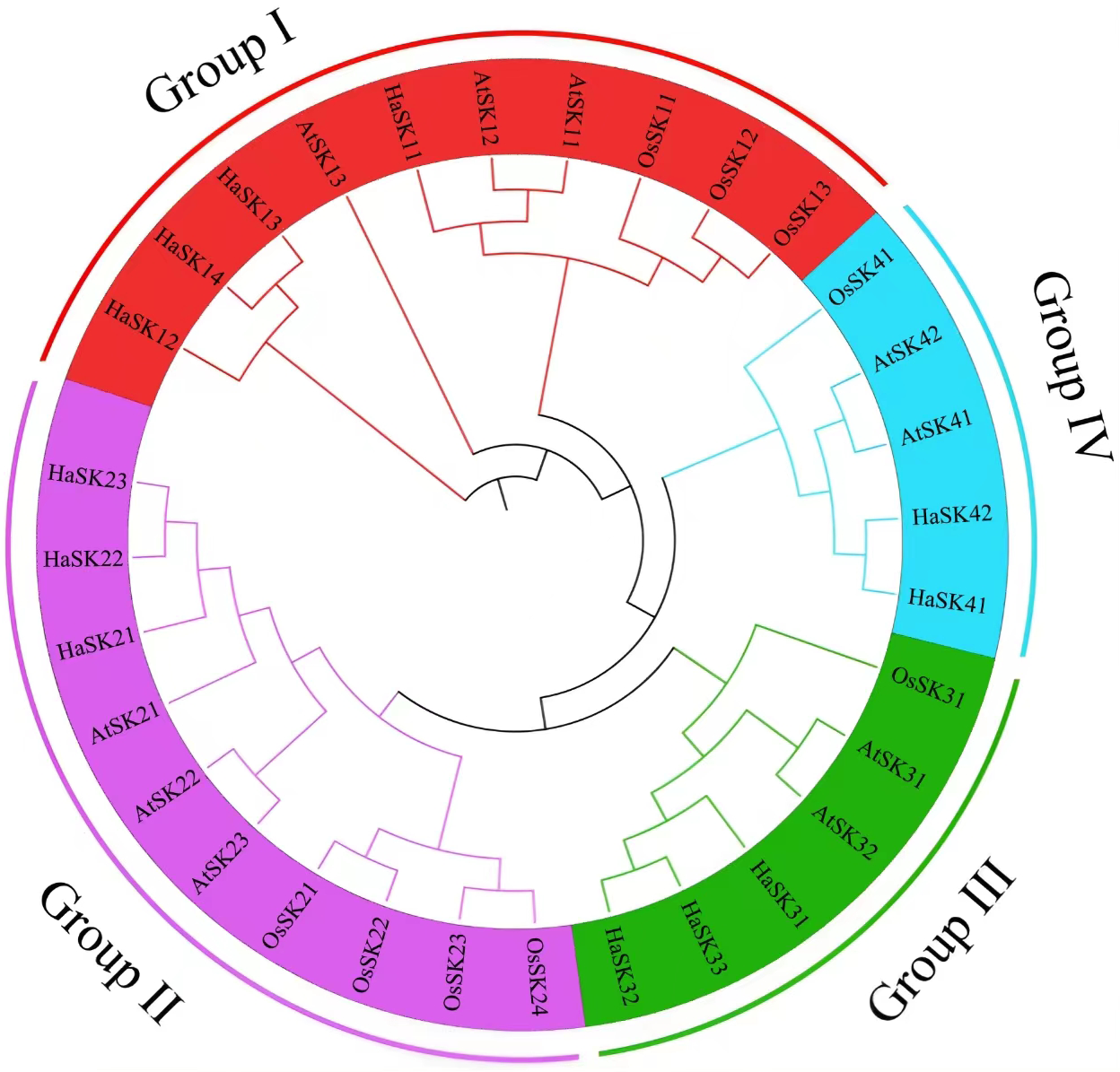
Figure 2: Phylogenetic tree of GSK3s from sunflower (Ha), rice (Os) and Arabidopsis (At). The four groups could be identified using different colors
3.3 Analysis of GSK3 Gene Structure in Sunflower
Analysis of the structure of sunflower GSK3 genes revealed that all HaSK genes contained 11–13 exons, especially all genes in Group IV had 12 exons. Additionally, MEME software identified 13 protein motifs in sunflower GSK3. The majority of GSK3 proteins belonging to the same group shared a motif, indicating that their functions might be comparable. The HaSK proteins all contained motif 1–7 and motif 9, which together make up the GSK3 kinase domain. All proteins of Groups I, II, and IV contained motif 8, whereas members of Groups I (apart from HaSK11), III, and IV primarily contained motif 10. Notably, motif 13 was detected in both Group IV and Group I members (apart from HaSK11 and HaSK13) (Fig. 3).
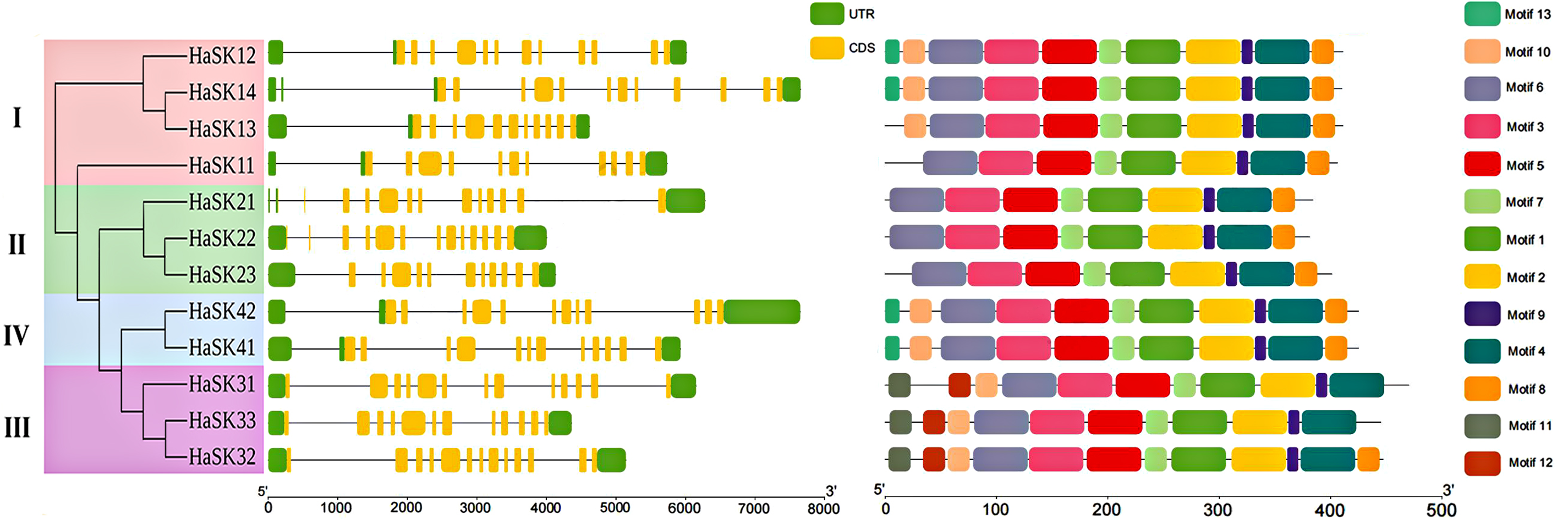
Figure 3: Gene structure, motifs, and phylogenetic relationships of sunflower GSK3s. Exons are represented as yellow boxes, introns are represented as black lines, and the green boxes represent the UTR of sunflower GSK3 genes. The thirteen putative motifs are presented by boxes of different colors
3.4 Analysis of Cis-Acting Elements of GSK3 Gene Promoters in Sunflower
Potential roles for HaSK genes in regulating stress-coping response were identified by analyzing the 2000 bp sequence upstream of the initial codon. Growth and development, stress, hormones, and light response are the four categories into which the cis-regulatory elements were separated. Elements associated with growth and development included MSA-like and CAT-box. Stress-related regulation has been linked to drought-responsive elements (MBS) and low-temperature-responsive elements (LTR, WRE3). The hormone-related elements were jasmonic acid-responsive element (TGACG motif, CGTCA motif), gibberellic acid (GA)-responsive element (GARE motif, P-box), auxin-responsive element (TGA-element), salicylic acid-responsive element (TCA-element), and ABA-responsive element (ABRE). The elements that were responsive to light were Sp1, GT1-motif, MRE, CCAAT-box, and G-box (Fig. 4).
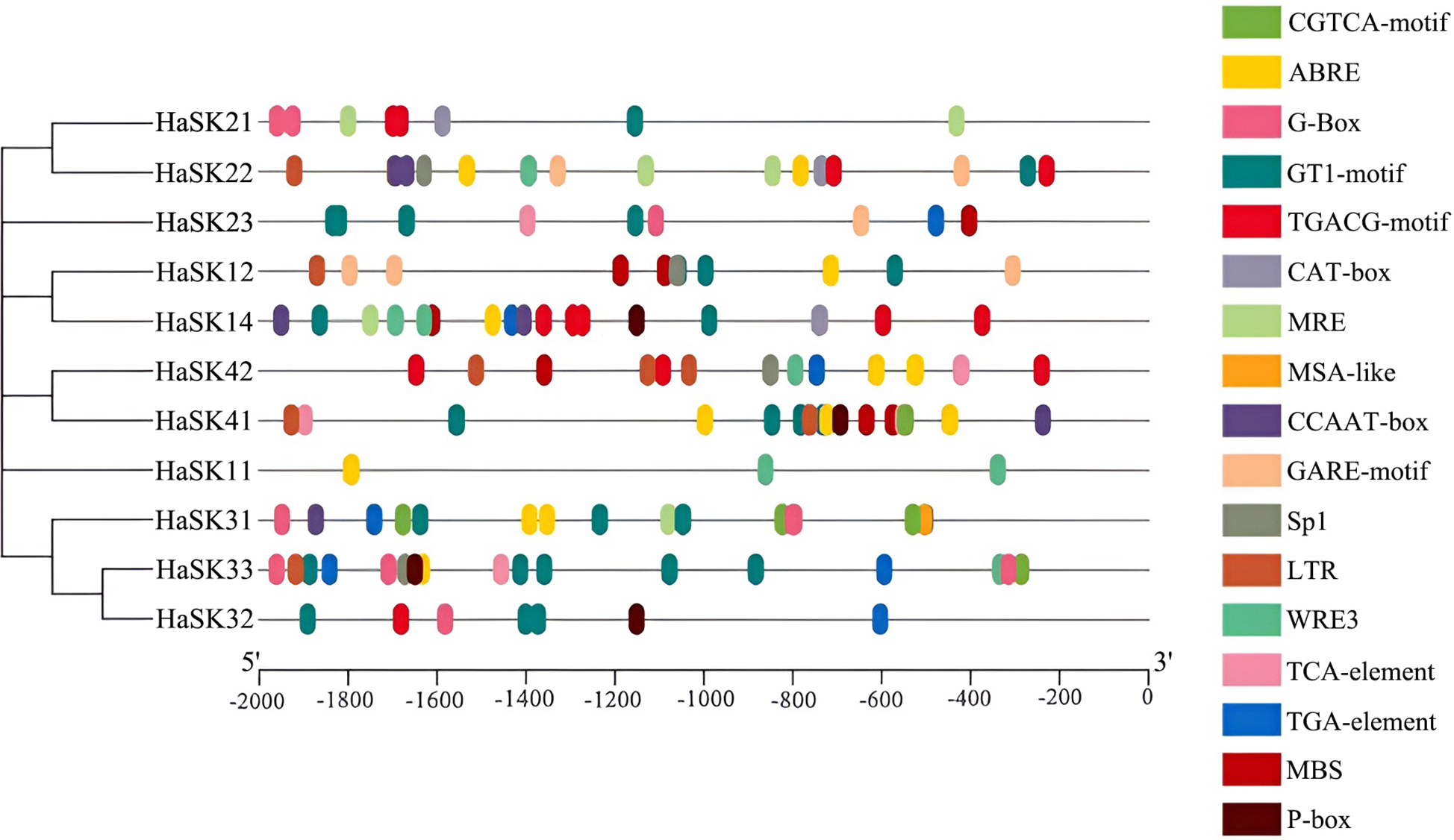
Figure 4: Types of cis-acting elements in promoter regions of sunflower GSK3 genes. The bottom scale can be used to determine the upstream length to the translation start site
3.5 Expression Profiles of GSK3 Genes in Different Tissues of Sunflower
We conducted transcriptome sequencing to investigate the expression levels of the GSK3 genes in sunflowers. Our findings revealed that the expressions of the GSK3 genes were the greatest in flowers (HaSK22, HaSK23, HaSK41, and HaSK42) and leaf axils (HaSK14, HaSK32, HaSK33). In contrast, their expression levels in leaves were comparatively lower (Fig. 5). This suggests that GSK3 genes of sunflowers might be implicated in flower and axillary bud development.
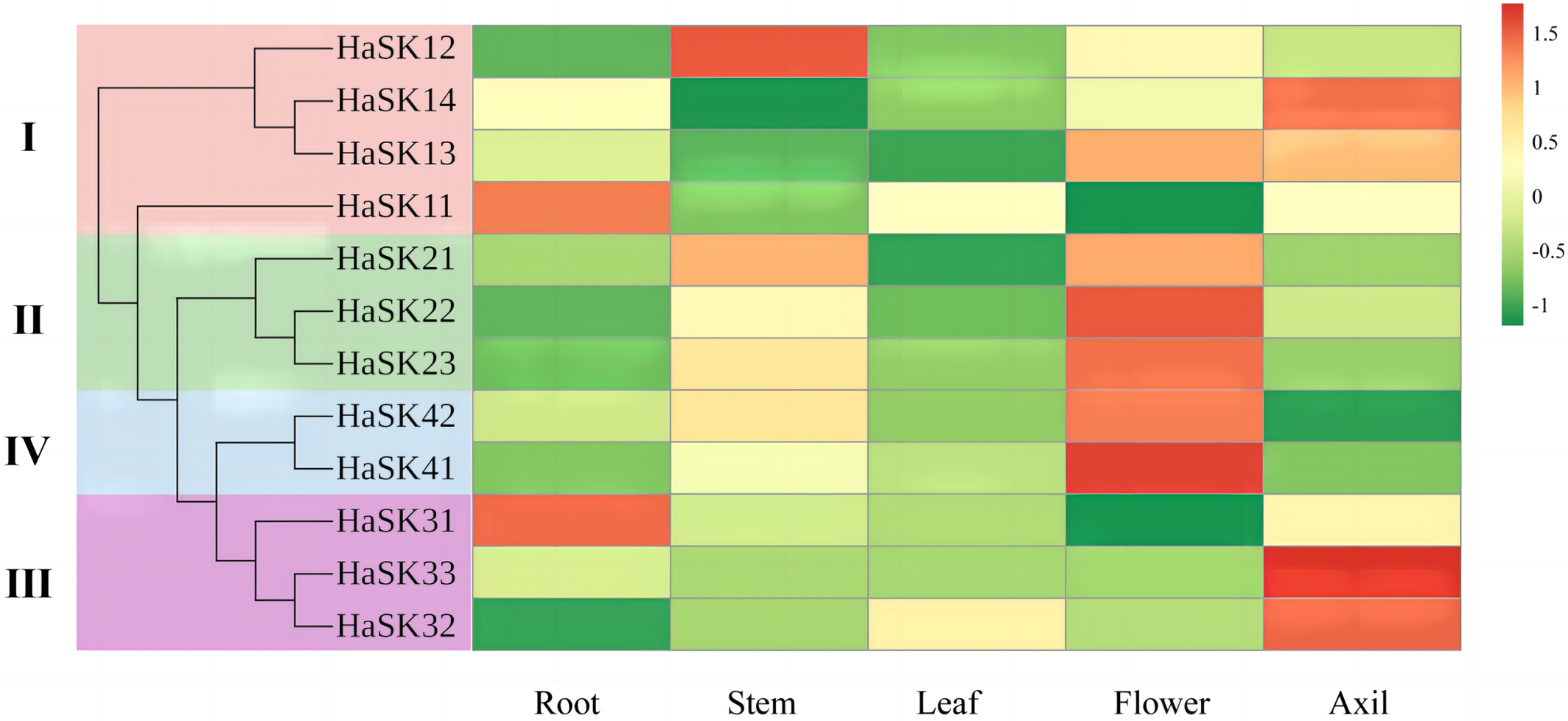
Figure 5: Analysis of GSK3 gene expression in five different sunflower tissues. The FPKM values of the HaSK genes were converted using log2
3.6 Expression Profile of GSK3 Genes in Sunflower under Salt Stress
The expression levels of HaSK11, HaSK22, HaSK23, HaSK32, HaSK33 and HaSK41 were significantly impacted after 6 h of NaCl treatment. The levels of HaSK22 and HaSK23 increased by more than ten times. HaSK14 and HaSK21, on the other hand, showed a marked decline in expression levels (Fig. 6). These results imply that the sunflower’s adaptation to salt stress may be mediated by these genes.
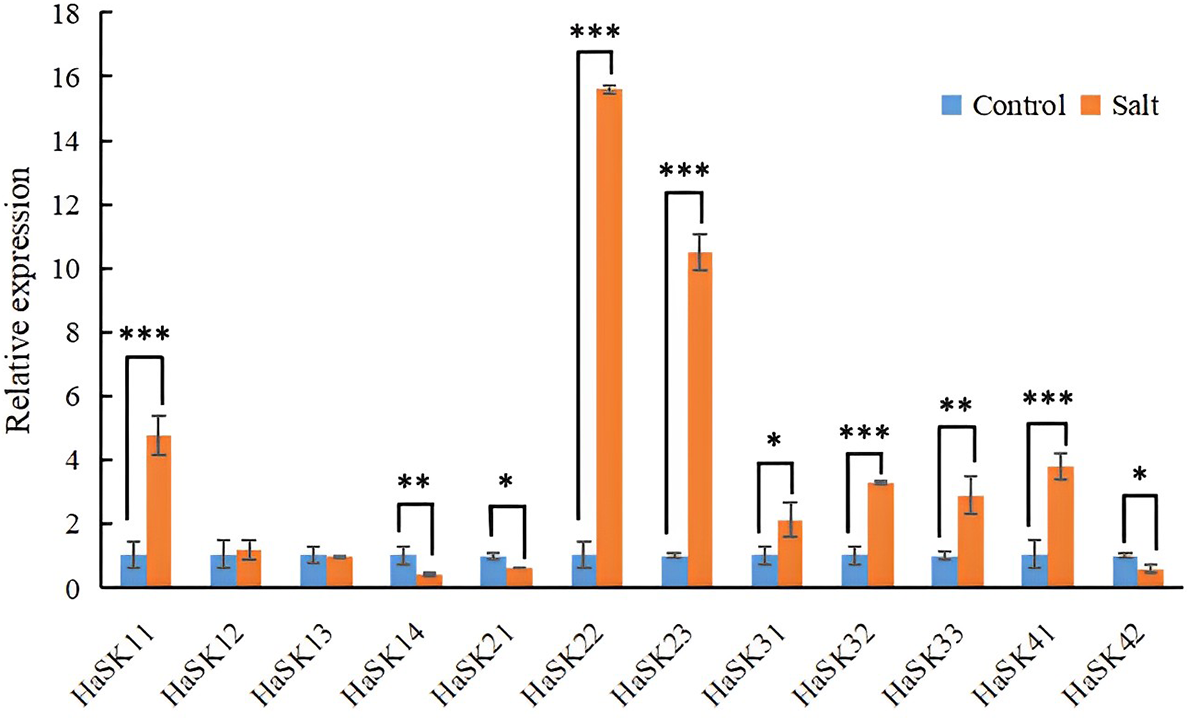
Figure 6: Expressions profile of GSK3 genes in response to NaCl treatment. The results were represented by mean ± SD. ***p < 0.001, **p < 0.01, and *p < 0.05 by t-test
3.7 Expression Profile of GSK3 Genes to Drought Stress in Sunflower
To determine the reaction of HaSK genes to drought stress in sunflowers, 20% PEG 6000 solution was applied. The expression levels of HaSK genes were determined after 6 h. It was observed that the expression levels of HaSK11, HaSK13, HaSK21, HaSK22, and HaSK33 were significantly elevated (Fig. 7). These findings indicate that these genes might be involved the sunflower’s ability to withstand drought conditions.
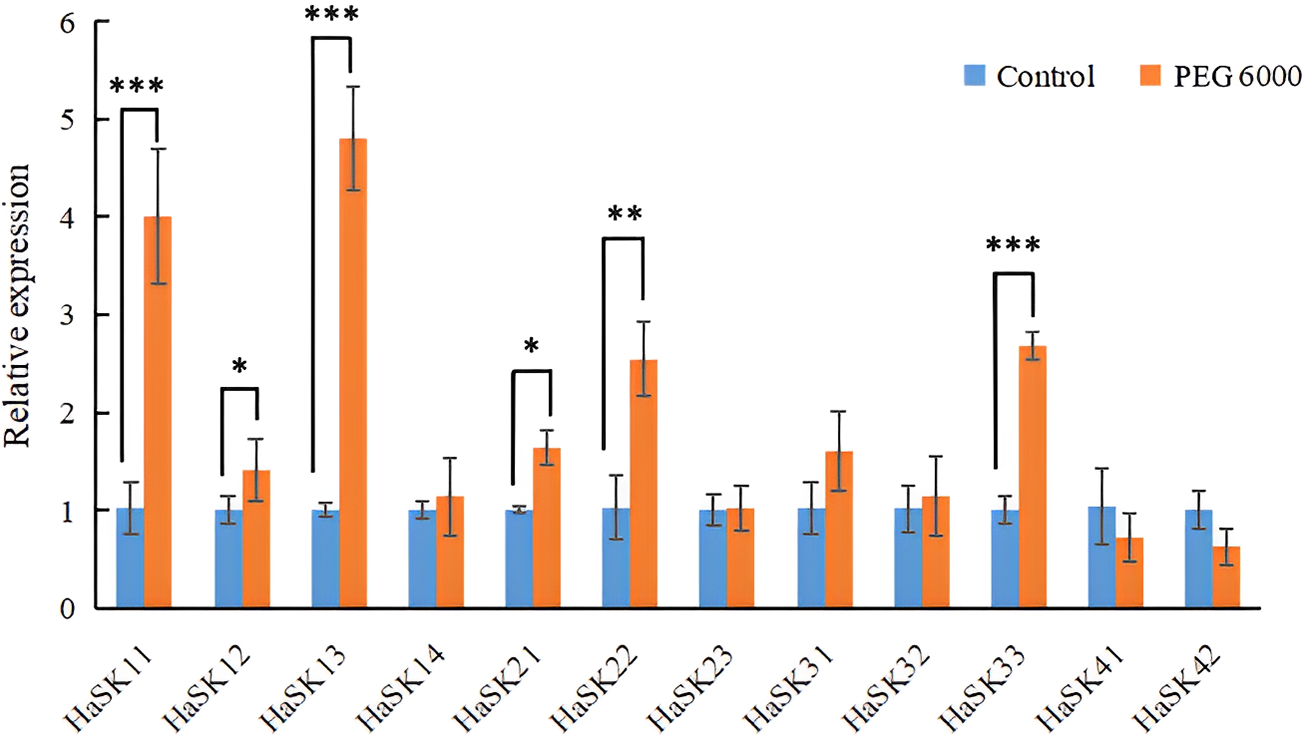
Figure 7: The profiles of GSK3 gene expression in response to drought stress treatment. The results were represented by mean ± SD. ***p < 0.001, **p < 0.01, and *p < 0.05 by t-test
3.8 Expression Profile of GSK3 Genes in Sunflower to Cold Stress
The expression levels of HaSK genes were assessed 6 h after the plants were exposed to a temperature of 5°C to evaluate the effects of cold stress on sunflowers. The results revealed that the expressions of HaSK11, HaSK12, HaSK13, HaSK32, HaSK33, HaSK41, and HaSK42 were significantly increased. Converserly, the levels of HaSK21 and HaSK23 were significantly decreased. These results suggest that these genes might be implicated in the sunflower’s reaction to cold stress (Fig. 8).
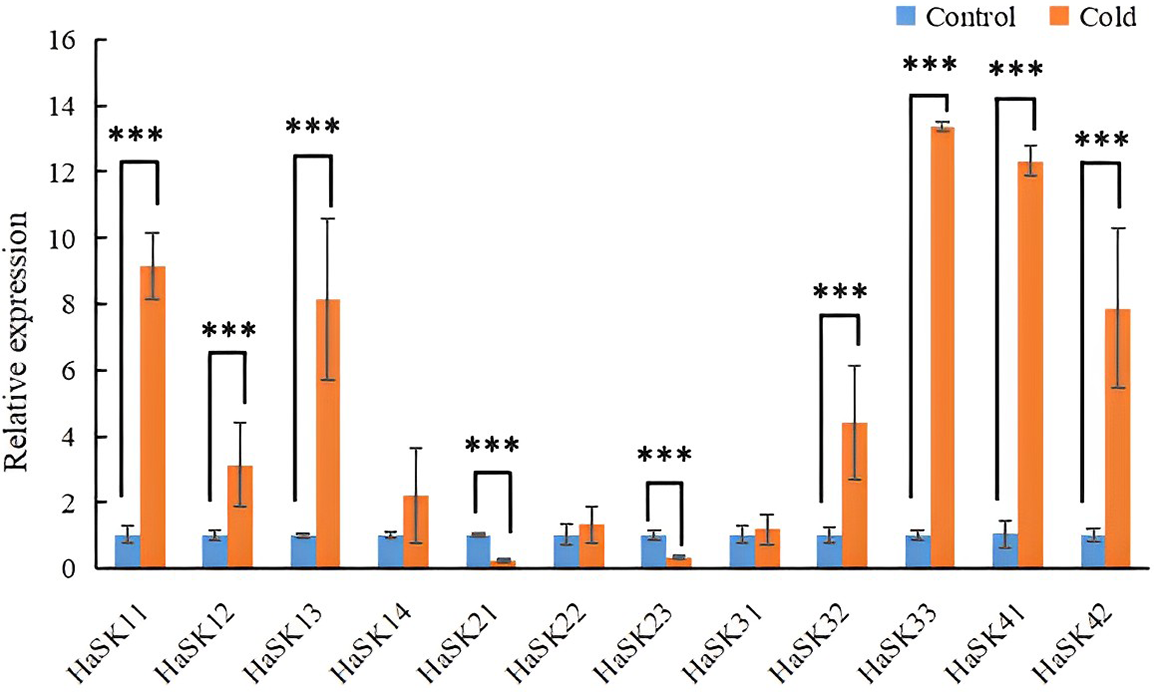
Figure 8: GSK3 gene expression profile in reaction to cold stress. The results were represented by mean ± SD. ***p < 0.001 by t-test
This study identified 12 GSK3 genes in sunflowers, more than those found in rice [30], Arabidopsis [31] and potatoes [3]. To further investigate the GSK3 proteins, phylogenetic analysis, exon/intron structure and conservative motif analysis were conducted. Results revealed that GSK3 proteins were divided into four groups, with a similar gene structure within each group (Fig. 3). Additionally, a low degree of similarity between the N- and C-terminal amino acid sequences was found through comparison, indicating a functional divergence among GSK3 members. Additionally, the homology of the kinase domain was found to be high, which was confirmed by the results of the conservative motif analysis (Fig. 1). This kinase domain consists of multiple conserved subdomains, as previously reported by Bittner et al. [32]. These subdomains are essential for regulating plant growth and maintaining kinase activity. Research by Kim et al. [31] found that a mutation of the lysine (K) to arginine (R) within the xAxK motif decreased kinase activity.
Tissue-specific expression profile analysis showed that Groups II and IV genes were expressed similarly, with the highest expression in flowers and the lowest expression in leaves, roots, and leaf axils (Fig. 5). These results imply that these genes regulate the development of these tissues in a similar manner. However, even though HaSK11, HaSK12, HaSK13, and HaSK14 were classified as subgroup I, their expression patterns varied. For instance, HaSK11, HaSK12, and HaSK14 demonstrated the highest expressions in stems, roots, and leaf axils, respectively (Fig. 5), indicating that there may be functional differences between the genes in the same branch. Together, GSK3 proteins have a multifunctional capacity in plants.
According to an analysis of cis-regulatory elements in the gene, several elements linked to growth responses were found in the sunflower GSK3 gene promoter, stress, hormones, and light, which is similar to findings reported in cotton and potato [3,13]. This implies that the function of this gene family is conserved across different species. ABRE, TGACG motif, and CGTCA motif, associated with ABA and jasmonic acid response, were present in most GSK3 genes. GSK3 genes were found to be associated with various elements, such as GA response, auxin response, salicylic acid response, stress, and growth and development, indicating their diverse roles.
Studies in various species have demonstrated that GSK3 genes, such as AtSK11, AtSK22, ASKα, TaSK5, and GmBIN2, are implicated in multiple of biological stress responses [13,15,16,33,34]. The analysis of the expression level of sunflower GSK3 genes under various abiotic stressors found that HaSK11 and HaSK33 responded to all three stresses. Most GSK3 genes only responded to two of them, such as HaSK13, HaSK22, HaSK32, and HaSK41. Furthermore, it was found that under abiotic stress, most of the genes’ expression levels dramatically increased, and some genes decreased significantly, such as HaSK14, HaSK21, and HaSK23. Notably, the expression level of HaSK22, closely related to GmBIN2 (Fig. S1), was significantly increased in the presence of drought and salt stress.
Transgenic Arabidopsis and peas that overexpress GmBIN2 have improved resistance to salt and drought stress [13]. This finding suggests that HaSK22, known to respond to salt and drought stress, may regulate sunflowers’ salt and drought resistance. Phylogenetic tree analysis revealed that OsGSK3 and GmGSK were closely related to HaSK11 (Fig. S1). Research has indicated that OsGSK3 and GmGSK expression levels can be elevated by salt, drought, and cold stress [18,35]. HaSK11 expression was detected in sunflowers, indicating that HaSK11 is critical in regulating the abiotic stress of sunflowers, and its function is relatively consistent across species. Furthermore, phylogenetic tree analysis showed that StSK21 was closely related to HaSK21 (Fig. S1). The overexpression of StSK21 in Arabidopsis caused a decrease in seed germination rate and increased salt sensitivity of transgenic plants [3]. On the other hand, HaSK21 expression was significantly reduced under salt stress, which is in line with the findings in potatoes. These findings indicate that HaSK21 may negatively regulate salt stress tolerance in sunflowers.
Acknowledgement: Not applicable.
Funding Statement: The research was financed by the Anhui Provincial Central Leading Local Science and Technology Development Special Fund Project (202007d06020021), and Project of Suzhou Science and Technology Bureau (2021143).
Author Contributions: All authors made contributions to this research. The experiments were completed by Xianwen Ji and Ziying Jiang. Jichao Wang was responsible for data collection and analysis. Xinyi Deng and Lili Dong wrote the manuscript and provided experimental guidance. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All of the data developed or examined during this investigation are available in this published article and its supplementary materials, or they can be provided at reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they do not have any conflicts of interest to disclose in relation to this study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.052809.
References
1. Song Y, Wang Y, Zhang J, Ren M. Regulatory network of GSK3-like kinases and their role in plant stress response. Front Plant Sci. 2023;14:1123436. [Google Scholar] [PubMed]
2. Mao J, Li W, Liu J, Li J. Versatile physiological functions of plant GSK3-like kinases. Genes. 2021;12(5):697. [Google Scholar] [PubMed]
3. Huang S, Liu Y, Deng R, Lei T, Tian A, Ren H, et al. Genome-wide identification and expression analysis of the GSK gene family in Solanum tuberosum L. under abiotic stress and phytohormone treatments and functional characterization of StSK21 involvement in salt stress. Gene. 2021;766:145156. [Google Scholar] [PubMed]
4. He J, Gendron JM, Yang Y, Li J, Wang Z. The GSK3-like kinase BIN2 phosphorylates and desta-bilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99(15):10185–90. [Google Scholar] [PubMed]
5. Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130(3):1221–9. [Google Scholar] [PubMed]
6. Hu Y, Yu D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell. 2014;26(11):4394–408. [Google Scholar] [PubMed]
7. Wang H, Tang J, Liu J, Hu J, Liu J, Chen Y, et al. Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol Plant. 2018;11(2):315–25. [Google Scholar] [PubMed]
8. Wang H, Wang X. GSK3-like kinases are a class of positive components in the core ABA signaling pathway. Mol Plant. 2018;11(6):761–3. [Google Scholar] [PubMed]
9. Claisse G, Charrier B, Kreis M. The Arabidopsis thaliana GSK3/Shaggy like kinase AtSK3-2 modulates floral cell expansion. Plant Mol Biol. 2007;64:113–24. [Google Scholar] [PubMed]
10. Dong H, Li D, Yang R, Zhang L, Zhang Y, Liu X, et al. GSK3 phosphorylates and regulates the Green Revolution protein Rht-B1b to reduce plant height in wheat. Plant Cell. 2023;35(6):1970–83. [Google Scholar] [PubMed]
11. Cui G, Li D, Zhang L, Xia C, Kong X, Liu X. GSK3 regulates VRN1 to control flowering time in wheat. J Integr Plant Biol. 2023;65(7):1605–8. [Google Scholar] [PubMed]
12. Wang Y, Xu J, Yu J, Zhu D, Zhao Q. Maize GSK3-like kinase ZmSK2 is involved in embryonic development. Plant Sci. 2022;1(318):111221. [Google Scholar]
13. Wang L, Chen Q, Xin D, Qi Z, Zhang C, Li S, et al. Overexpression of GmBIN2, a soybean glycogen synthase kinase 3 gene, enhances tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots. J Integr Agric. 2018;17(9):1959–71. [Google Scholar]
14. Charrier B, Champion A, Henry Y, Kreis M. Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 2002;130(2):577–90. [Google Scholar] [PubMed]
15. Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I. Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J. 2001;27(4):305–14. [Google Scholar] [PubMed]
16. Dal Santo S, Stampfl H, Krasensky J, Kempa S, Gibon Y, Petutschnig E, et al. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell. 2012;24(8):3380–92. [Google Scholar] [PubMed]
17. Stampfl H, Fritz M, Dal Santo S, Jonak C. The GSK3/Shaggy-like kinase ASKα contributes to pattern-triggered immunity. Plant Physiol. 2016;171(2):1366–77. [Google Scholar] [PubMed]
18. Yang XG, Liang WH, Li F, Ma WS. OsGSK3 is a novel GSK/Shaggy-like gene from Oryza sativa L., involved in abiotic stress signaling. Pak J Bot. 2012;44(5):1491–6. [Google Scholar]
19. Jiang H, Tang B, Xie Z, Nolan T, Ye H, Song G, et al. GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 2019;100(5):923–37. [Google Scholar] [PubMed]
20. Li J, Zhou H, Zhang Y, Li Z, Yang Y, Guo Y. The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev Cell. 2020;55(3):367–80. [Google Scholar] [PubMed]
21. Eremina M, Unterholzner SJ, Rathnayake AI, Castellanos M, Khan M, Kugler KG, et al. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc Natl Acad Sci U S A. 2016;113(40):E5982–91. [Google Scholar] [PubMed]
22. Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant. 2017;10(4):545–59. [Google Scholar] [PubMed]
23. Ye K, Li H, Ding Y, Shi Y, Song C, Gong Z, et al. BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell. 2019;31(11):2682–96. [Google Scholar] [PubMed]
24. Song Y, Zhai Y, Li L, Yang Z, Ge X, Yang Z, et al. BIN2 negatively regulates plant defence against Verticillium dahliae in Arabidopsis and cotton. Plant Biotechnol J. 2021;19(10):2097–112. [Google Scholar] [PubMed]
25. He C, Gao H, Wang H, Guo Y, He M, Peng Y, et al. GSK3-mediated stress signaling inhibits legume-rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol plant. 2021;14(3):488–502. [Google Scholar] [PubMed]
26. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–7. [Google Scholar] [PubMed]
27. Dong L, Wu Y, Zhang J, Deng X, Wang T. Transcriptome analysis revealed hormone pathways and bZIP genes responsive to decapitation in sunflower. Genes. 2022;13(10):1737. [Google Scholar] [PubMed]
28. Ning K, Chen S, Huang H, Jiang J, Yuan H, Li H. Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell. 2017;130:469–81. [Google Scholar]
29. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–8. [Google Scholar] [PubMed]
30. Youn JH, Kim TW. Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol Plant. 2015;8(4):552–65. [Google Scholar] [PubMed]
31. Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11(10):1254–60. [Google Scholar] [PubMed]
32. Bittner T, Campagne S, Neuhaus G, Rensing SA, Fischer-Iglesias C. Identification and character-ization of two wheat glycogen synthase kinase 3/Shaggy-like kinases. BMC Plant Biol. 2013;13:1–15. [Google Scholar]
33. Kempa S, Rozhon W, Šamaj J, Erban A, Baluška F, Becker T, et al. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohy-drate metabolism. Plant J. 2007;49(6):1076–90. [Google Scholar] [PubMed]
34. Christov NK, Christova PK, Kato H, Liu Y, Sasaki K, Imai R. TaSK5, an abiotic stress-inducible GSK3/shaggy-like kinase from wheat, confers salt and drought tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2014;84:251–60. [Google Scholar] [PubMed]
35. Zhang C, Zhao H, Liu Y, Li Q, Liu X, Tan H, et al. Isolation and characterization of a novel glycogen synthase kinase-3 gene, GmGSK, in Glycine max L. that enhances abiotic stress tolerance in Saccharomyces cerevisiae. Biotechnolo Lett. 2010;32(6):861–6. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools