 Open Access
Open Access
REVIEW
Application of Transgenic Technology in Identification for Gene Function on Grasses
Qinghai University, Qinghai Academy of Animal and Veterinary Sciences, Qinghai Provincial Key Laboratory of Adaptive Management on Alpine Grassland, Key Laboratory of Superior Forage Germplasm in the Qinghai-Tibetan Plateau, Xining, 810000, China
* Corresponding Authors: Ying Liu. Email: ; Yushou Ma. Email:
(This article belongs to the Special Issue: Recent Research Trends in Genetics, Genomics, and Physiology of Crop Plants)
Phyton-International Journal of Experimental Botany 2024, 93(8), 1913-1941. https://doi.org/10.32604/phyton.2024.052621
Received 09 April 2024; Accepted 05 July 2024; Issue published 30 August 2024
Abstract
Perennial grasses have developed intricate mechanisms to adapt to diverse environments, enabling their resistance to various biotic and abiotic stressors. These mechanisms arise from strong natural selection that contributes to enhancing the adaptation of forage plants to various stress conditions. Methods such as antisense RNA technology, CRISPR/Cas9 screening, virus-induced gene silencing, and transgenic technology, are commonly utilized for investigating the stress response functionalities of grass genes in both warm-season and cool-season varieties. This review focuses on the functional identification of stress-resistance genes and regulatory elements in grasses. It synthesizes recent studies on mining functional genes, regulatory genes, and protein kinase-like signaling factors involved in stress responses in grasses. Additionally, the review outlines future research directions, providing theoretical support and references for further exploration of (i) molecular mechanisms underlying grass stress responses, (ii) cultivation and domestication of herbage, (iii) development of high-yield varieties resistant to stress, and (iv) mechanisms and breeding strategies for stress resistance in grasses.Keywords
Biologists have long relied on traditional genetic methods, utilizing mutants to study positional cloning, identify plant sequences, and confirm gene function through genetic transformation. However, with rapid advancements in structural genomics and high-throughput analysis, the field of functional genomics has emerged [1]. The aim of this study was to analyze the overall functions of genes, including coding genes, small RNA molecules, non-coding RNA, and metabolites, to explain how they interact and influence biological phenotypes. as well as growth, development, and metabolism. The selection and identification of functional genes (including coding and non-coding genes) form the basis for studying how genes regulate biological processes and variations. However, the application of functional genomics relies on the development and utilization of new techniques and methods.
Typically, two approaches are typically used to isolate the functional genes responsible for specific phenotypes or biological processes. Firstly, the traits are studied, and positive genetic screening methods are used to identify functional genes. Researchers using this approach typically focus on specific phenotypes exhibited by individual organisms or cells and aim to identify genes associated with these phenotypes and reveal their functions; therefore, the method requires the presence of well-defined biological phenotypes. Candidate genes are obtained through molecular biology or genetics methods, followed by evaluation and functional validation.
Alternatively, related traits and gene functions can also be studied using reverse from the perspective of genes (i.e., an identification method based on reverse genetics) [2]. Reverse genetics directly examines the function of a gene or DNA sequence by altering its expression and identifying the resulting mutant phenotype. Numerous methods are used to screen functional genes, such as cDNA library technology [3], RNA interference (RNAi) library technology [4,5], antisense RNA technology, targeted knockout, miRNA-based gene silencing [6], CRISPR/Cas9 screening technology [7], viral-induced gene silencing (VIGS) technology, insertion mutation technology [7], and other model biological screening technologies [8]. Multiple reverse genetics methods have been used to identify functional genes in various grasses.
Recent advancements in techniques have led to an increased recognition of resistance genes in grasses. Several resistance-related functional genes have been studied in grasses, such as Festuca elata [9], Elymus dahuricus [10,11], Agropyron cristatum [12], Agrostis stolonifera [13], Cynodon dactylon [14], Zoysia japonica [15] and Paspalum notatum [16]. These genes can be categorized based on function. The first category comprises functional genes that encode proteins directly protecting grasses under stress, such as COMT [9] and other genes affecting biomacromolecules and membrane structures. The second category includes regulatory genes, such as resistance regulatory genes, protein kinase-like signaling factors under stress [14], and transcription factors (TFs) like bZIP [16], MYB/MYC [17] and WRKY [18,19]. Among these, the TF regulatory family is the most studied [20–26]. The proteins encoded by these genes play a role in signal transduction pathways and regulate stress-related gene expression. Studying the genes with key roles in stress responses, understanding the functions of these resistance-related genes in grasses, and analyzing gene function have significant theoretical importance. This research may also lead to applications in cultivating and domesticating herbage and developing new varieties with high yield and resistance to stress.
2 Screening and Identification of Functional Genes in Grasses
2.1 Screening of Functional Genes Based on Forward Genetics Screening Strategy
Functional genes are typically screened using a forward genetic approach, which focuses on specific phenotypes exhibited by individual organisms or cells to identify genes associated with these phenotypes and elucidate their functions [27]. This strategy requires well-defined biological phenotypes, followed by obtaining candidate genes through molecular biology or genetic methods, and subsequently evaluating and validating their functions. Specifically, the screening of functional genes includes five categories: (i) traditional genetic analysis [28–32] (Table 1). (ii) Gene mutation analysis, based on DNA/RNA sequencing [33–37]. (iii) Whole-genome sequencing technology or genome-wide association studies [38–41]. (v) Expression profile analyses [42–45]. (iv) Bioinformatics technology [46–49]. This approach has been extensively used to explore stress-responsive genes in various species, such as Secale strictum (C. Presl) [50], Agrostis stolonifera [51], Avena sativa L. [52], Paspalum vaginatum, and Paspalum fasciculatum [53,54]. Studies that have used these analytical techniques can serve as valuable references for future investigations of grass gene functions.
2.2 Identification of Functional Genes Based on Reverse Genetics
Recently, the complete genome sequences of model plants (e.g., Arabidopsis thaliana) and other plants have allowed for the establishment of large-scale databases of genome sequence information, and the term “reverse genetics” has been coined, with numerous methods available for screening functional genes using reverse genetics. At present, it mainly includes cDNA library technology [3], RNA interference (RNAi) technology [4], antisense RNA technology, CRISPR/Cas9 screening technology [7], virus-induced gene silencing (VIGS) technology, Agrobacterium-mediated transgenic method [55], and insertion mutation technology [8]. The following is a brief description of the application scope, advantages and disadvantages of three more widely used gene function identification techniques.
2.2.1 CRISPR/Cas Screening and Identification Strategies
Among CRISPR/Cas screening and identification strategies, the CRISPR/Cas9 system stands out as the most efficient method [7,56,57] (Table 2). Numerous studies have shown that transgenic technologies, such as CRISPR/Cas and VIGS techniques, are helpful in identifying plant gene function, among which RNAi is a significant reverse genetics tool for analyzing plant gene function and improving genetic quality [56]. Currently, CRISPR/Cas9 technology finds applications in the treatment of human genetic diseases [58], personalized cell therapy [59], new drug development [60], and improving crop genetics [61]. More than 30 plant species and hundreds of genes have been successfully edited in research aimed at improving crop stress resistance, disease resistance, and insect pests [62,63].
2.2.2 Virus-Induced Gene Silencing Techniques (VIGS)
VIGS represents a novel method for suppressing plant gene expression without requiring a stable transformation regimen or the establishment of large mutant populations (Table 2). It can be readily applied to species that are challenging to transform stably or when limited genetic material from a small research population is available. Moreover, it proves beneficial in the study of non-model plant species [64,65]. Currently, VIGS techniques employing host induction [66,67], virus induction [23,26,27], and spray induction [68,69] have been used to elucidate gene function. Research has showcased the diverse roles of related genes in disease resistance, stress responses, cell signaling and secondary metabolic biosynthesis pathways. Additionally, VIGS has been widely utilized to identify of gene functions in vegetable crops and other species [70,71], leading to the establishment of new correlation models [70,71].
2.2.3 Agrobacterium-Mediated Transgenic Method
The Agrobacterium-mediated transgenic method, a prominent model biological screening strategy, is widely favored due to its convenient, fast, economical, and easy-to-operate characteristics (Table 2). Researchers frequently utilize transgenic approaches to overexpress target genes in plants for functional verification purposes. Using Agrobacterium-mediated transgenic technology as an example, the process of gene function verification involves identifying target functional genes through transcriptome sequencing and bioinformatics analysis. Additionally, it facilitates comprehensive exploration of the gene families of target plants in response to abiotic stress at the genome-wide level. Transgenic experiments validate the responses and functions of each gene under abiotic stress conditions [72], with several grass species serving as subjects for gene studies [73–76].
3 Application of the Latest Methods of Gene Function Identification in Grass Stress
Stress-inducing factors in grasses include drought, injury, reactive oxygen species (ROS), phosphorus deficiency, salinity, heat, cold, flooding, and heavy metals [77]. Grasses primarily adapt to harsh environmental conditions through dynamic adjustments at morphological, physiological, and biochemical levels, particularly during nutritional and reproductive growth stages. Therefore, exploring, studying, and utilizing the stress-resistance mechanisms of grasses using genomics and molecular biology is crucial. For example, many low-temperature stress response genes have been identified in A. thaliana, with most cold-responsive (COR) genes enriched in the starch-sucrose [78]. Studies have demonstrated that proanthocyanidins are important in providing resistance to various stresses (e.g., UV damage, low-temperature damage, drought, flood stress, and bacterial attack).
Numerous studies have highlighted the limitations of single-omics techniques, whereas multi-omics analysis techniques, combining transcriptomics and metabolomics, offer a comprehensive approach to analyzing plant resistance [79]. The multi-technology combination mode of bioinformatics analysis, transcriptomic sequencing analysis, and transgenic technology has been used to identify grass gene function [80]. The combinations of these technologies have been applied to warm-season turf grasses (e.g., C. dactylon [81], Z. japonica [82–88], and P. notatum [89]) and cool-season turfgrasses (e.g., F. elata [35–37], E. dahuricus [9], A. cristatum [13] and A. stolonifera [90]). Cold-season turfgrass stress-response genes have been identified and studied [91] Categorizing these studies reveals that researchers usually choose to analyze the resistance mechanism for diverse gene types (Table 3). These categories are summarized below.
Functional genes of grasses include three categories (Table 3): The first category is growth regulatory genes, functional identification is primarily performed using transgenic technology or a combination of transgenic technology and gene silencing technology [9,16]. The second group is genes for enzymes that scavenge ROS, these exert detoxification and antioxidant effects [89,92]. The third category is active genes that protect biomolecules, such as osmoregulatory protein-related genes, dehydration protein genes, aquaporin genes, plum genes, and late-embryogenetic-rich proteins [56].
Grass regulatory genes encompass resistance regulatory genes [10,11], transcription factors (TFs) [93], and protein kinase signaling factors [14] and other protein functional genes involved in stress responses (Table 3). Transgenic technology is commonly employed for identifying these functional genes, often in combination with gene silencing or knockout techniques for functional validation. The following section provides an overview of the most extensively studied grass regulatory genes in grasses.
3.2.1 Resistance Regulatory Gene
Transgenic technology is the main method for studying resistance-regulatory genes in grasses. Agrobacterium-mediated transgenic technology was used to study the H+- pyrophosphatases (PP) enzyme gene AtAVP1 from E. dahuricus; it was found that H+-PP enhances low-nitrogen stress tolerance in transgenic Arabidopsis and wheat by interacting with a receptor-like protein kinase [10]. This technology was used to transfer genes from wheat and tobacco plants to explore the action mechanism of the type I H+-PP gene EdVP1 from E. dahuricus under low potassium stress. The results showed that overexpression of the V-type H+-PP gene EdVP1 increased the yield and potassium uptake of transgenic wheat [11].
3.2.2 Transcription Factors (TFs)
TFs are central regulators of changes in gene expression [93] and can be categorized based on the characteristics of their structural domains into the following families: WRKY (WRKYGQ), MYB/MYC, NAC (NAM, ATAF, and CUC), Apetala2/Ethylene Responsive Factor (AP2/ERF/EREBP), SQUAMOSA promoter-binding protein-like (SPL), TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP), basic helix-loop-helix (bHLH), basic leucine zipper (bZIP), CBF/dehydration responsive element protein 1 (CBFs/DREB1), HB, Zn-C2H2, MADS, ARF-Aux/IAA, APETALA1/FRUITFULL (AP1/FUL) and others [22]. TFs regulate plant metabolism, development, biosynthesis, other life processes, and response to abiotic stresses; therefore, they represent excellent potential candidate genes that could identify resistant plants [94]. The function of TFs genes is typically determined through transgenic technology. In grasses, the function of TFs genes is usually related to the plant’s response to abiotic stress.
WRKY-Like Transcription Factors
TFs in the WRKY family are plant-specific and play crucial roles in responses to abiotic stresses, such as cold, salt, and drought [95,96]. WRKYs are also instrumental in growth and development processes, including seed germination [97], flowering [98,99], anthocyanin biosynthesis [100], and leaf senescence [101]. Most WRKYs positively influence cold tolerance. For instance, in Bermuda grass, more than 100 WRKYs were identified using full-length transcriptome data [99], and 23 displayed cold-induced expression patterns [102]. Using transgenic technology and VIGS, Bermuda grass CdWRKY2 positively regulated cold tolerance by coordinately activating the CdSPS1-involved sucrose synthesis and CdCBF1-dependent signaling pathways. Similarly, rice WRKYs, such as OsWRKY71 and OsWRKY76, have been reported to enhance cold resistance [19]. Overexpression of VaWRKY12 and VaWRKY33 consistently conferred cold resistance in transgenic Arabidopsis and Vitis vinifera [103,104]. Additionally, transgenic Arabidopsis plants overexpressing TaWRKY19 showed improved freezing tolerance. These gene identification techniques have also been applied to identify WRKY-like TFs in wheat [105,106].
Researchers have also demonstrated that the Bermuda grass CdWRKY50 gene negatively regulates plant responses to salt stress using transgenic technology and VIGS [18]. Similar results have been reported in other plants. For instance, in Arabidopsis, sodium chloride treatment upregulated the transcripts of two closely related WRKYs, and both Atwrky33 and Atwrky25 mutants exhibited salt-sensitive phenotypes, whereas overexpression of AtWRKY25 and AtWRKY33 enhanced salt tolerance [107]. Overexpression GmWRKY54 was reported to improve the salinity and drought resistance of transgenic plants, through the regulation of STZ/Zat10, whereas GmWRKY13 demonstrated different functions [108]. Overexpression FcWRKY40 enhanced salt tolerance in plants by modulating ion balance and proline accumulation [109]. Additionally, researchers utilized transgenic technology to study the regulatory mechanism of AmWRKY-like [94] genes in response to drought stress in Agropyron mongolicum, showing that AmWRKY-like genes were negatively regulated by natural drought-induced expression. Conversely, AtWRKY34, which may be involved in the C-repeat binding factor (CBF) signaling cascade, negatively modulated the cold response of mature pollen [95].
MYB/MYC-Like Transcription Factors
The MYB family is one of the largest TFs families in plants. Studies have shown that TaLHY, a 1R-MYB in Triticum aestivum, plays a defensive role against stripe rust infection [110]. Rice OsMYB48-1, a MYB-related TFs, is involved in drought and salt stress responses by regulating stress-induced abscisic acid (ABA) synthesis [111]. Recent research highlights the key role of MYB/MYC TFs in various stress responses in grasses, such as cold, salt, and drought [112,113]. For example, using transgenic technology, the TFs gene LcMYB4 was identified as a positive regulator of cold and frost tolerance in transgenic A. thaliana, despite its previously unknown function in Sheepgrass (Leymus chinensis (Trin.)) [17]. Another study using Agrobacterium-mediated transgenic method and a homologous sequence cloning demonstrate that the AmrMYB1 and AmrR2R3MYB genes of Agropyron michnoi Roshev positively regulated drought and salt stress [114]. Studies on the MwMYB4 gene of A. mongolicum Keng using transgenic technology showed that MwMYB4 overexpression improved drought and low-temperature tolerances [112]. Additionally, a novel MYC-type bHLH TF ZjICE2 from Z. japonica was studied by transgene transfer into Arabidopsis [15,82]. Its overexpression enhanced cold, drought, and salt stress tolerances, and was closely related to ICE homologs. In wheat, transgenic technology has been used to study the functions of these TFs [115–117].
bZIP-Like Transcription Factors
bZIP TFs regulate gene activities involved in various biological processes related to plant growth and development; the gene ABI5 is a member of this family. The abscisic acid (ABA) signaling pathway is crucial during the plant’s response to salt stress, with ABI5 serving as an important TFs in the ABA signal transduction pathway. Transgenic overexpression studies in tobacco were conducted to evaluate the role of Elymus sibiricus L. abscisic acid-insensitive 5 (EsABI5) in the ABA-dependent regulation of the response of Siberian wildrye to salt stress. These studies demonstrated that EsABI5 is involved in anti-salt responses and acts as a negative regulator during salt stress [118]. Jia et al. [119] identified 32 bZIP TFs using a combination of transgenic and gene knockout technologies. Knockout of the homologous gene of the most downregulated auxin-responsive TabZIP6D_20 in Arabidopsis (AtHY5) negatively affected the auxin signaling pathway. The auxin-responsive TabZIP gene likely has several essential functions in wheat leaf structure in response to auxin. Another study reported that the Arabidopsis TF bZIP29 is involved in leaf and root development by altering cell number in the leaf and root meristem [120].
HEMA-Like Transcription Factors
The HEMA family encodes GluTRs in higher plants. Genetic manipulation of Bermuda grass photosynthetic biosynthesis through Agrobacterium-mediated transformation established a highly efficient transformation method by screening crucial embryogenic calli. The approach also improved infection efficiency and photosynthetic capacity assessment by CdHEMA1 overexpression [80]. The overexpression of CdHEMA1 enhanced the photosynthetic pigment content, OJIP curve, and reaction center density (RC/CSo), as well as absorption parameters (ABS/CSo, ABS/CSM). Consequently, the performance indices PIABS and PICS were improved compared to the wild type.
bHLH-Like Transcription Factors
The bHLH proteins belong to a superfamily of eukaryotic TFs, which are widely distributed in the animal and plant kingdoms, with various functions in plant development and metabolism [121]. A variety of bHLH TFs involved in plant abiotic stress response pathways have been identified [122]. Zuo et al. [15] identification of bHLH genes showed that antisense expression of ZjbHLH076/ZjICE1 influences tolerance to low temperature and salinity in Z. japonica.
AP2/ERF-Like Transcription Factors
The AP2/ERF, a plant-specific TF family, influences various developmental processes and responses to abiotic stresses, such as drought, low temperature, hypoxia, and high salt [123]. Recently, more research has shown that plant hormone-mediate cascade signaling is closely related to stress response, with AP2/ERF TFs playing a key role [124]. The response and adaptation of plants to salt stress require the coordination of many hormones [125], including ABA, which participates in responses to various abiotic stresses. The functions of these TFs in transgenic rice and A. thaliana have been identified. Overexpression of Z. japonica ZjABR1/ERF10 was shown to regulate plant growth and salt tolerance in transgenic O. sativa [86]. Jiang et al. [126] reported that a novel transcriptional regulator, HbERF6, regulates the HbCIPK2-coordinated pathway, enhancing salt tolerance in halophytic Hordeum brevisubulatum.
CBFs/DREB1-Like Transcription Factors
A myriad of TFs involved in cold signaling networks has been identified, among which the CBFs/DREB1 playing a central role in cold resistance by directly activating COR gene expression [127]. In Arabidopsis, three CBFs (CBF1, CBF2, and CBF3) are essential for cold acclimation, enabling plants to acquire freezing tolerance upon exposure to advanced low non-freezing temperatures [128,129]. The CBFs, along with their transcriptional activators, including inducers of CBF expression 1 (ICE1), constitute the ICE1CBF-COR transcriptional cascade, a well-known cold signaling pathway [127]. Additionally, TFs such as MYB15, CAMTA3, PIF3, EIN3, and BZR1 have been identified as upstream regulators of CBFs [128]. However, most research on the regulatory networks of cold and salt stress responses has focused on model plants, indicating a need for studies on the application of these networks in grass.
TCP-Like Transcription Factors
The TCP TFs are named after the first characterized members, such as Teosinte Branched1 (TB1) of maize [24], CYCLOIDEA (CYC) of Antirrhinum [25], and Proliferating Cell Factors one-half (PCF1/2) of rice [26]. TCP proteins (old TFs that appeared approximately 650–800 Mya) have been crucial role in the evolution of plant morphology [130]. Studies have demonstrated the role of TCP proteins in various plant growth and developmental processes [131]. In A. thaliana (At), AtTCP14 and AtTCP15 regulate embryonic growth and influence seed germination [130]. Meanwhile, AtTCP5 inhibits the initiation and growth of leaf serrations [132]. Under normal temperatures, AtTCP17 binds to Cryptochrome 1 (CRY1), repressing the formation of Phytochrome-Interacting Factors 4 (PIF4) AtTCP17 dimer. However, under high temperatures, CRY1 releases AtTCP17, promotes the interaction between the AtTCP17 and PIF4, increases the transcriptional activity of PIF4, and regulates thermomorphogenesis [130,133]. Furthermore, Arabidopsis TCP20 and TCP22 regulate the circadian rhythm by binding to the CIRCADIAN CLOCK ASSOCIATED1 (CCA1) promoter and activating CCA1 expression [134]. In rice, PCF1/2 modulates the expression of PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) and regulates cell cycle processes [26]. In addition, the CYC-like homolog RETARDED PALEA1 (REP1) regulates palea identity and development in rice [135]. Zhan et al. [20] used transgenic technology to express target genes instantaneously in tobacco leaves, the responses of ScTCPs and sctcp-mir319-3p in rye (Secale cereale L.) to low temperature and drought were studied. The results showed that rye ScTCP2, ScTCP5, and ScTCP19 are target genes of sctcp-mir319-3p, which affects the stress resistance or tolerance of rye by regulating the expression of these target genes.
Zn-C2H2 Like Transcription Factors
The C2H2-type zinc-finger protein (ZFP) family is one of the largest TFs families in the plant kingdom, with members engage in plant growth, development, and stress responses. Some DgZFPs were predicted to function in abiotic stress responses or the regulation of flowering time. The tolerance of the C2H2 ZEP gene family DgZFPs of D. glomerata (Dg-ZFP125) to osmotic and salt stress was studied using transgenic technology for transplanting yeast. The results indicated that Dg-ZFP125 enhances the permeability and salt tolerance of D. glomerata [136].
AP1/FUL genes play crucial roles in vernalization induction, promote plant flowering, determining spikelet primordium, and specifying floral organ identity. In monocotyledonous plants, AP1/FUL genes underwent two significant duplications: the first during monocot formation, giving rise to the FUL3/OsMADS18 branch, and the second during gramineous plant formation, resulting in the FUL1/OsMADS14 and FUL2/OsMADS15 branches [137]. Consequently, studies on these gene have proliferated. For instance, CRISPR/Cas9 was employed to knockout BdID1 by targeting different exons of the B. distachyon ID transcription factor (BdID1) genes in dicotyledonous short-stalked grasses. Subsequent genotyping of transgenic progeny confirmed complete loss of BdID1 function, highlighting its unique role in the Poaceae family and its association with regulating flowering time pathways [137–139].
3.2.3 Protein Kinase-Like Signaling Factors under Stress of Adversity
MAPKK Genes Mitogen-Activated Protein Kinases (MAPKs)
During long-term adaptive evolution, plants have developed signaling networks to sense and transmit signals. Among stress-activated molecular pathways, MAPK signaling cascades are important signaling modules with critical roles in plant developmental processes, stress responses, and signal transduction [140]. The MAPK cascade consists of three protein kinases: MAPK, MAPK kinase (MAPKK), and MAPK kinase (MAPKKK) [14]. Wang et al. [14] studied the CdMAPKKKs gene of Bermuda (C. dactylon) grass and showed that CdMAPKKKs are commonly associated with regulating numerous biological processes. This technique illustrated how MAPK cascades modulate cold response by modulating ICE1 protein stability. Other studies based on transgenic technology have shown that the ABA-insensitive five gene of E. sibiricus L. engages in the ABA-dependent salt response [140]. The ABA signal transduction and MAPK cascade pathways are important regulators of plant responses to salt stress. The MAPK cascade directly or indirectly participates in the ABA signal transduction pathway, and the ABA signaling pathway regulates the expression of related genes in the MAPK cascade pathway [141,142]. Studies on the responses of other plants (such as the Vitis vinifera (VvMKK2)) and Gossypium hirsutum (GhMAP3K14-GhMKK11-GhMPK31) to drought and salt stress, targeting MAPK genes through transgenic and VIGS technologies, have also been reported [143].
The CBL-CIPK signaling system, comprising calcineurin B proteins (CBLs) and their interacting protein kinases (CIPKs), forms a crucial regulatory network for abiotic stress responses. The system regulates plant responses to abiotic stress by initiating phosphorylation and interpreting calcium (Ca2+) signals [144]. Transgenic technology was used to conduct experimental studies on the regulation of plant salt tolerance by the CdCBL4-CdCIPk5 complex in C. dactylon and rice. Five CBL genes from C. dactylon were cloned by homologous cloning using the millet genome as a reference sequence. The interaction between CdCIPKS and CdCBL4 was verified using complementary yeast two-hybrid and bimolecular fluorescence assays. The CdCBL4-CdCIPKS complex was found to regulate Na+ absorption, transport, and excretion under salt stress by modulating the expression of Na+ transporters and maintaining intracellular sodium and potassium ion balance, thereby enhancing salt tolerance [145].
The Sucrose Non-Fermenting-1 Related Protein Kinase 2 (SnRK2) Family
Members of the SnRK2 family play crucial roles in abiotic stress responses. Xiang et al. [12] studied the function of the SnRK2 protein kinase gene (AcSnRK2.11) in the protection of A. cristatum against stress using transgenic technology to construct expression vectors of yeast and tobacco. AcSnRK2.11 overexpression increased chlorophyll yield, superoxide dismutase activities, ROS content, peroxidase levels, and soluble carbohydrate levels. Additionally, overexpression also enhanced freezing tolerance in transgenic yeast and tobacco, suggesting that AcSnRK2.11 acts as a regulatory factor involved in cold-response pathways.
Chlorophyll Degradation Enzymes (CCEs)
CCEs interact with photosynthetic organs. During leaf senescence, LHCII interacts with five chlorophyll-degrading enzymes (NYC, NOL, PPH, PAO, and RCCR) [146]. Agrobacterium-mediated transgenic technology was used to transfer Agrobacterium gene into A. thaliana or tobacco. Researchers studied the functional characteristics of the chlorophyll-b reductase gene NYC1 were associated with chlorophyll degradation and photosynthesis in Z. japonica (ZjNYC1) [84]. Similarly, functional characterization of ZjPPH, the pheophytinase gene from Z. japonica, was conducted to understand its influence chlorophyll degradation and photosynthesis [83]. Overexpression of ZjNYC1 and ZjPPH decreased the chlorophyll content and promoted senescence. Likewise, Guan et al. [87] used this method to study the mechanism of Z. japonica Chlorophyll b Reductase Gene NOL (ZjNOL) participation in chlorophyll degradation and photosynthesis. They found that ZjNOL simultaneously led to the excessive accumulation of ROS, antioxidant enzyme activation, oxidative stress generation, and consequently accelerating senescence. Similarly, the ectopically expressed chlorophyll-b reductase gene from Z. japonica changed the leaf color and chlorophyll morphology in A. stolonifera [13]. Functional identification of the H+-PP enzyme gene family in E. dahuricus showed that V-type H+-PP gene EdVP1 overexpression from E. dahuricus increased the yield, potassium uptake, and low-nitrogen stress tolerance of transgenic plants [10,11].
A study on A. thaliana revealed that plants overexpressing NOL exhibited significantly lower chlorophyll content compared to wild-type plants These plants also displayed a reduced photosystem II antenna size, indicating that NOL was engaged in photosystem II antenna size control [147]. Phylogenetic analysis of the rice (Oryza sativa) OsNOL gene revealed that the closest protein to OsNOL was OsNYC1. Rice nol mutants exhibited a phenotype like that of the nyc1 mutant, with significant suppression of chlorophyll b degradation. Additionally, LHCII was selectively preserved throughout senescence, and thylakoid grana were retained even at the late stage of senescence [148]. In perennial ryegrass (Lolium perenne), RNA interference of LpNOL (NOLi) significantly inhibited chlorophyll breakdown during leaf senescence and altered leaf color, suggesting that NOL functions as a chlorophyll b reductase and is associated with the middle color phenotype of this perennial herb [149]. However, many studies have relied on transgenic technology for functional identification, highlighting the need for further research using alternative methods to explore the functional of chlorophyll-degrading enzyme CCE genes.
Hydroxycinnamoyl-CoA Shikimate/Quinate Hydroxycinnamoyl Transferase (HCT)
HCT is an essential enzyme in regulating lignin biosynthesis and composition. Researchers have used transgenic technology to investigate the expression mechanism of the Hydroxycinnamoyl-CoA Shikimate/Quinate Hydroxycinnamoyl Transferase 4 gene from Z. japonica (ZjHCT4) [88]. The expression of ZjHCT4 resulted in A. stolonifera resulted in excessive elongation and changes in lignin composition by altering the phenylpropionic acid metabolic pathway and hormone response.
Other Protein Functional Genes
In grasses, functional protein genes primarily include those of the calmodulin-like (CML) family [150] and Domain of Unknown Function (DUF) 1644 family [85], with transgenic technology often employed to identify these genes. The CML family serves as an important Ca2+ sensor in plants, affecting responses to various stressors. The CML gene PvCMLs in Paspalums vaginatium has been shown to play a role in the response to salt and cold stress. Additionally, DUF proteins have a specific conserved structural domain, the function of which remains unclear [150,151]. Researcher investigated the function of ZmDUF1644, transferred from the halophytic Zoysia matrella to the non-halophytic A. thaliana, under salt stress (150 mM NaCl). The results showed that ZmDUF1644 overexpression enhanced the salt tolerance of A. thaliana [85].
In summary, transgenic technology is frequently used to identify gene function in grasses by transferring the target gene to A. thaliana [152] or Nicotiana tabacum [153,154] for gene function verification. However, a single method often does not suffice for gene function identification in practical applications. The gene function identification techniques described here can be jointly applied or combined with multi-omics analysis techniques (such as transcriptomics and metabolomics) to provide complement insights. This approach aims to rapidly and systematically evaluate gene function and uncover the potential value of functional genes in human health and ecology. Additionally, gene editing using CRISPR/Cas9 technology has been performed in Avena sativa breeding [155]. An efficient screening system for the optimal target sites of CRISPR/Cas9 gene editing in A. sativa was established using the gene gun method, resulting in two mutant plants from Cas9 editing. These results provide a basis for applying gene editing technology in the molecular breeding of grasses. Furthermore, CRISPR/Cas9 technology has been used to identify stress-resistance genes (such as Sh1, a gene associated with grain shedding) in A. mongolicum, and has shown potential in inhibiting the spread of plant DNA viruses [156].
4 Applications in Genes Functional Identification of Other Plant
Currently, gene silencing, transgenic techniques, and gene knockout techniques are the most widely used methods for identifying gene function. These techniques have been increasingly applied to identify functional genes not only in grasses but also in various economic crops such as cotton (G. hirsutum) [157–161], soybean [162], common bean [163], maize [164–166], Arachis hypogaea L. [167], sugarcane [168], potato [169], and alfalfa [170], as well as in vegetables like Chinese cabbage (Brassica rapa L.) [171,172] and chili peppers (Capsicum annuum L.) [173]. They are also used to study flower-forming genes in species such as Paeonia lactiflora [174], Malus halliana [175], Phyllostachys edulis [176], and Dianthus caryophyllus [177], resistance genes associated with fruit ripening in grape (V. vinifera L.) [178] and sweet cherry [179], and disease-resistant genes against pathogens [180–182] and plant antiviral genes [183].
For example, using A. thaliana as a medium, the heterologous overexpression of a gene family in A. thaliana identified the function of PIP5K family genes related to the development of female inflorescences in Ricinus communis Lm [152] and strawberry SVP dormant-related genes [153]. Other studies determined the functional identification of homologous genes and the molecular regulatory network of the floral pathway of the BrcSOC1 gene in Brassica rapa ssp. chinensis in response to vernalization [171]. Using N. tabacum as a carrier, researchers investigated the function of the SWEET gene involved in sugar transport in Hemerocallis fulva in response to low-temperature stress [154]. Additionally, gene function analysis and upstream transcription factor prediction of the key candidate gene HaF4D2 (controlling the fatty acid content of sunflowers) showed that seeds of transgenic strains had higher oil content than those of the wild type [72]. In another study, the adenosine triphosphate (ATP) citrate lyase (ACL) gene was cloned by mining the genes related to flower color formation in P. lactiflora. The function of PlACL2 in flower color formation was determined by overexpression in N. tabacum and transient silencing in P. lactiflora [174]. Simultaneously, CRISPR/Cas9, as a new gene editing system, has successfully edited hundreds of genes in more than 30 plant species and has been widely used in researching crop stress [61], disease, and pest resistance [62,63]. For instance, a CRISPR/Cas9 screening strategy was applied to suppress the gamma response 1 (TaSOG1) gene in wheat, which is involved in the DNA damage response (DDR). The mechanism of action of the TaSOG1 gene in wheat DDR was also investigated [184].
Combining gene function identification techniques is more effective for analyzing plant resistance. For instance, Deng [157] identified the function of Verticillium wilt resistance genes in upland cotton by combining gene silencing technology with transgenic technology and other methods. Furthermore, two resistance response genes from naturally mature tetraploid Poncirus trifoliata, specifically the LEA family gene (PtrLEA7) and the R2R3-type MYB (PtrMYB3), were studied for drought and salt resistance using a tobacco-mediated transgenic technique and VIGS technique, respectively [185]. Studies have shown that when a certain technology is limited in research, other aspects such as innovation, application, and selecting more suitable methods for identifying target gene function need consideration. For example, Zhao [183] first used an easily available vector construction method to study Papaya leaf distortion mosaic virus (PLDMV) and constructed a variety of PLDMV vectors with reporter genes using the E. coli-free method. The method was then used to construct viral infectious clones quickly, efficiently, and stably. Compared with traditional methods of in vitro transcription or yeast homologous recombination, it had a higher success rate, greater efficiency, and improved cost. The resulting plant virus expression vector construction method will significantly advance subsequent scientific research. Moreover, this method does not require large equipment for nucleic acid extraction or isothermal amplification and provides an effective new approach for constructing unstable plant virus infectious clones in E. coli cells. Such methods can be applied to identifying gene functions in grasses.
5 Prospects of Future Research
With the rapid development of high-throughput sequencing technology, an increasing number of whole-genome sequences of plants have been published. However, sequencing efforts for herbage plants remain limited, constraining technical means for gene function research, genetic improvement, and molecular breeding [57]. These factors greatly hinder the development of molecular breeding and functional genomics research in herbage plants. Therefore, it is important to actively explore advanced technologies for identifying gene functions in grasses to facilitate genetic improvement and molecular breeding. Recent studies have highlighted the potential applications of CRISPR/Cas9 technology, transgenic methods, ethyl methanesulfonate (EMS) technology, and other emerging technologies in this regard.
5.1 Genetic Function Identification of Herbage by Ethyl Methanesulfonate (EMS) Technology
EMS-induced mutagenesis is a potent tool for generating genetic resources to uncover novel genes and characterize their functions, thereby elucidating the molecular basis of important agronomic traits [186]. Moreover, EMS mutagenesis has facilitated multi-omics combined analysis and enabled the identification of related genes through reverse genetics. Gene editing and recombination are innovative breeding methods that significantly enhance plant genetics [187], increasingly becoming primary approaches for genetically improving model plants. Furthermore, EMS mutagenesis is applicable across most plant species [186]. It has been successfully employed in crops, vegetables, and fruits to breed varieties with superior traits [188]. Various plant tissues have been used for mutagenesis, mutagens can cause abundant mutations in different plant organs, such as seeds, bulbs, calluses, and pollen [186]. Specific mutagenesis sites in plants can be selected based on experimental requirements. Recent advances have demonstrated the effectiveness of mutagenesis breeding technology in enhancing resistance traits in Arabidopsis, rice, soybean, and other plants [189], underscoring its potential applicability in grasses.
5.2 Transgenic Technology Is Combined with Other Technologies for the Study of Gene Function in Grasses
In corn, rice, wheat, and other gramineous plants, the transformation rate of plant transgenic techniques, such as Agrobacterium-mediated methods, requires improvement [92,190]. Future studies should explore transgenic methods with higher transformation efficiencies, more controllable gene integration, and wider applicability. Combining various genetic transformation methods will enable more in-depth and extensive research on plant genomes and their genetic transformations.
5.3 Virus-Induced Gene Silencing Techniques (VIGS)
VIGS is used to transform plant cells with viral genomes either by direct inoculation or through infection with a modified strain of Agrobacterium rhizogenes [157,191]. This technique is extensively studied in functional genomics due to its advantages, such as simple construction, low cost, and short cycles [192]. VIGS can also be used for comparative genomes studies between distinct species. Currently, VIGS techniques have demonstrated the diversity of related gene functions in studies of disease resistance, stress response, cell signaling, and secondary metabolic biosynthesis pathways. This has led to the construction of new correlative models [65], expanding the genetic toolbox for non-model organisms. Additionally, VIGS facilitates virus-induced overexpression (VOX), virus-induced genome editing (VIGE), and host-induced gene silencing (HIGS), and it has been widely used for identifying gene functions in vegetable crops and other species [70,71]. This technique is valuable for identifying gene functions in grasses.
5.4 Deepen the Wide Application of CRISPR/Cas9 Technology in the Identification of Gene Function in Grasses
Gene editing via the CRISPR/Cas9 system is widely used for targeted genome modifications. As an emerging plant gene editing technology [193,194], it has great research potential in functional genomics. Recently, CRISPR/Cas9 technology has improved forage varieties of grass plants. Researchers have proposed various effective grassland ecological restoration methods, highlighting the role of CRISPR/Cas9 in restoring grassland vegetation. This technology can identify genes related to stress resistance, high yield, and quality of native species according to local conditions Additionally, collecting, developing, and utilizing superior grassland plants and restoring grassland ecological functions can significantly shorten the breeding cycle. Therefore, CRISPR/Cas9 is a promising method for identifying grass gene function in the future.
5.5 Other New Gene Editing Techniques Are Used to Identify the Function of Grasses Genes
In addition to the Cas9 protein family, studies have shown that the Argonaute (Ago) protein family (e.g., TtAgo [160] and PfAgo [195]), displays gene editing capabilities. Some studies have confirmed that NgAgo-gDNA has potential as a new tool for gene knockouts [196], which can reduce mRNA without producing detectable DNA double-strand breaks to cut RNA instead of DNA. Furthermore, the CRISPR/Cas13RNA editor system can be reprogrammed and applied to eukaryotes to combat pathogenic RNA viruses or regulate gene expression, making it useful for identifying grass gene function [197–199]. Recent studies have shown that the CRISPR/Cas12i project [200,201] achieves therapeutic genome editing through efficient immune cell editing. While primarily used in medical science, this technology could be applied to plant genome research. Future studies should explore this potential.
In summary, identifying and understanding the functional genes that confer stress resistance in grasses is crucial for breeding resilient grass varieties. Advances in functional genomics and transgenic technologies are revolutionizing this field, providing powerful tools for gene function identification and validation. Key functional genes in grasses include disease resistance genes (such as NBS-LRR genes and PRR genes), biotic and abiotic stress resistance genes (such as DREB/CBF transcription factors, HSP (Heat Shock Proteins), and WRKY transcription factors), and genes related to growth and yield (such as GA signaling genes and photosynthesis-related genes). Common methods for identifying functional genes include Genome-Wide Association Studies (GWAS), Quantitative Trait Loci (QTL) Mapping, CRISPR/Cas9 Gene Editing, RNA Sequencing (RNA-Seq), proteomics and metabolomics, and transgenic technology. Among these, transgenic technology is the most used method in grass gene function identification and is crucial for validating the function of resistance genes in grasses. It involves introducing, modifying, or silencing genes to assess their role in resistance. Future research in grass gene function identification should leverage advanced genomic tools and transgenic technologies. Methods such as GWAS, QTL mapping, CRISPR/Cas9, and transcriptomics will be crucial in identifying and characterizing resistance genes. Transgenic approaches will play a vital role in validating these genes’ functions and developing grass varieties with enhanced resistance to biotic and abiotic stresses. However, single molecular identification techniques are often limited in studying gene function in grasses. Future advancements in the field will benefit from integrating omics and molecular biology technologies, multiple gene function mining and identification methods, and emerging gene function research tools. This integrated approach will be key to ensuring sustainable agricultural productivity and food security in the face of global challenges. This review of the latest gene function identification methods provides theoretical support for the germplasm development and molecular breeding of resistant grass plants.
Acknowledgement: The authors would like to extend their sincere appreciation to the Chief Scientist Program of Qinghai Province of China, and the authors sincerely thank the anonymous reviewers who made valuable comments on this paper.
Funding Statement: This work was supported by the Chief Scientist Program of Qinghai Province (2024-SF-101).
Author Contributions: Conceptualization, Lijun Zhang and Ying Liu; writing—original draft preparation, Lijun Zhang and Xinyou Wang; writing—review and editing, Lijun Zhang, Xinyou Wang and Ying Liu; supervision, Yanlong Wang, Yushou Ma and Ying Liu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data supporting this research were obtained from relevant literature in public databases such as NCBI and Web of Science, which are open to all researchers. Anyone needing these data can download them independently. Additionally, the data supporting the arguments of this review can be requested from the corresponding authors upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
1. Nagy LG, Zsolt M, Botond H, Balázs B. Novel phylogenetic methods are needed for understanding gene function in the era of mega-scale genome sequencing. Nucleic Acids Res. 2020;5:5. [Google Scholar]
2. Liu X, Chang D, Wang J. Advantages, disadvantages and research progress on strategies for screening and identifying functional genes. Basic Med and Clinic. 2022;42(06):969–74 (In Chinese). [Google Scholar]
3. Wong FS, Karttunen J, Dumont C, Wen L, Visintin I, Pilip IM, et al. Identification of an MHC class I-restricted autoantigen in type 1 diabetes by screening an organ-specific cDNA library. Nat Med. 1999;5(9):1026–31. doi:10.1038/12465. [Google Scholar] [PubMed] [CrossRef]
4. Mamta B, Rajam MV. RNAi technology: a new platform for crop pest control. Physiol Mol Biol Plants. 2017;2(7):587. [Google Scholar]
5. Padilla-Roji I, Ruiz-Jiménez L, Bakhat N, Vielba-Fernández A, Pérez-García A, Ferández-Ortuño D. RNAi technology: a new path for the research and management of obligate biotrophic phytopathogenic fungi. Int J Mol Sci. 2023;24(10):558–73. [Google Scholar]
6. Amritha PP, Shah JM. Can genetic engineering-based methods for gene function identification be eclipsed by genome editing in plants? A comparison of methodologies. Mol Genet Genomics. 2021;296(3):485–500. doi:10.1007/s00438-021-01769-y. [Google Scholar] [PubMed] [CrossRef]
7. Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2013;16(5):299–311. [Google Scholar]
8. Beckmann PJ, Largaespada DA. Transposon insertion mutagenesis in mice for modeling human cancers: critical insights gained and new opportunities. Int J Mol Sci. 2020;21(3):1172. doi:10.3390/ijms21031172. [Google Scholar] [PubMed] [CrossRef]
9. Singh S, Sharma N. Biochemical and in silico molecular study of caffeic acid-O-methyltransferase enzyme associated with lignin deposition in tall fescue. Amino Acids. 2023;24:s00726. [Google Scholar]
10. Zhang HJ, Chen M, Xu CJ, Liu RB, Tang WS, Chen K, et al. H+-pyrophosphatases enhance low nitrogen stress tolerance in transgenic Arabidopsis and wheat by interacting with a receptor-like protein kinase. Front Plant Sci. 2023;14(10):1096091. [Google Scholar] [PubMed]
11. Zhou YB, Li Y, Qi XL, Liu RB, Dong JH, Jing WH, et al. Overexpression of V-type H+ pyrophosphatase gene EdVP1 from Elymus dahuricus increases yield and potassium uptake of transgenic wheat under low potassium conditions. Sci Rep. 2020;10(1):5020. doi:10.1038/s41598-020-62052-5. [Google Scholar] [PubMed] [CrossRef]
12. Xiang DJ, Man LL, Cao S, Liu P, Li ZG, Wang XD. Heterologous expression of an Agropyron cristatum SnRK2 protein kinase gene (AcSnRK2.11) increases freezing tolerance in transgenic yeast and tobacco. 3 Biotech. 2020;10(5):209. doi:10.1007/s13205-020-02203-7. [Google Scholar] [PubMed] [CrossRef]
13. Dong D, Yang ZX, Ma Y, Li SW, Wang MD, Li YRZ, et al. Expression of a Chlorophyll b Reductase gene from Zoysia japonica causes changes in leaf color and Chlorophyll morphology in Agrostis stolonifera. Int J Mol Sci. 2022;23(11):6032. doi:10.3390/ijms23116032. [Google Scholar] [PubMed] [CrossRef]
14. Wang W, Shao A, Amombo E, Fan SG, Xu X, Fu JM. Transcriptome-wide identification of MAPKKK genes in bermudagrass (Cynodon dactylon L.) and their potential roles in low temperature stress responses. PeerJ. 2020;8:e10159. doi:10.7717/peerj.10159. [Google Scholar] [PubMed] [CrossRef]
15. Zuo ZF, Sun HJ, Lee HY, Kang HG. Identification of bHLH genes through genome-wide association study and antisense expression of ZjbHLH076/ZjICE1 influence tolerance to low temperature and salinity in Zoysia japonica. Plant Sci. 2021;313:111088. doi:10.1016/j.plantsci.2021.111088. [Google Scholar] [PubMed] [CrossRef]
16. Pozzi FI, Acuna CA, Quarin CL, Felitti SA. GG13: a candidate gene related to seed development and viability from apomictic Paspalum notatum. Euphytica. 2021;217(7):148. doi:10.1007/s10681-021-02881-1. [Google Scholar] [CrossRef]
17. Li XX, Jia JT, Zhao PC, Guo XF, Chen SY, Qi DM. LcMYB4, an unknown function transcription factor gene from sheepgrass, as a positive regulator of chilling and freezing tolerance in transgenic Arabidopsis. BMC Plant Biol. 2020;20(1):238. doi:10.1186/s12870-020-02427-y. [Google Scholar] [PubMed] [CrossRef]
18. Huang XB, Amee M, Chen L. Bermudagrass CdWRKY50 gene negatively regulates plants’ response to salt stress. Environ Exp Bot. 2021;188(1):128–39. [Google Scholar]
19. Huang XB, Cao LW, Fan JB, Ma GJ, Chen L. CdWRKY2-mediated sucrose biosynthesis and CBF-signalling pathways coordinately contribute to cold tolerance in bermudagrass. Plant Biotechnol J. 2022;20(4):660–75. doi:10.1111/pbi.v20.4. [Google Scholar] [CrossRef]
20. Zhan WM, Cui LH, Guo GH, Zhang YP. Genome-wide identification and functional analysis of the TCP gene family in rye (Secale cereale L.). Gene. 2023;854(2):147104. [Google Scholar] [PubMed]
21. Singh KB, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5(5):430–6. doi:10.1016/S1369-5266(02)00289-3. [Google Scholar] [PubMed] [CrossRef]
22. Lehti-Shiu MD, Panchy N, Wang P, Uygun S, Shiu S-H. Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. BBA-Gene Regul Mech. 2017;18(60):3–20. [Google Scholar]
23. Pilar C, Nick L, John D, Enrico C. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999;18(2):215–22. doi:10.1046/j.1365-313X.1999.00444.x. [Google Scholar] [PubMed] [CrossRef]
24. Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–8. doi:10.1038/386485a0. [Google Scholar] [PubMed] [CrossRef]
25. Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383(6603):794–9. doi:10.1038/383794a0. [Google Scholar] [PubMed] [CrossRef]
26. Ohashi KY. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9(9):1607–19. [Google Scholar] [PubMed]
27. Geng W, Li Z, Hassan MJ, Peng Y. Chitosan regulates metabolic balance, polyamine accumulation, and Na+ transport contributing to salt tolerance in creeping bentgrass. BMC Plant Biol. 2020;20(1):506. doi:10.1186/s12870-020-02720-w. [Google Scholar] [PubMed] [CrossRef]
28. Brown JM, Yu XW, Holloway HMP, DaCosta M, Bernstein RP, Lu J, et al. Differences in proteome response to cold acclimation in Zoysia japonica cultivars with different levels of freeze tolerance. Crop Sci. 2020;60(5):2744–56. doi:10.1002/csc2.v60.5. [Google Scholar] [CrossRef]
29. You J, Ye L, Wang D, Zhang Y, Xiao W, Wei M, et al. Mapping and candidate gene analysis of QTLs for grain shape in a rice chromosome segment substitution line Z485 and breeding of SSSLs. Mol Breeding. 2024;44(6):895–915. [Google Scholar]
30. Chen L, He W, Yu Y, Wang Y, Zhai X, Ling X, et al. Molecular mapping and candidate gene identification of two major quantitative trait loci associated with silique length in oilseed rape (Brassica napus L.). Mol Breeding. 2024;44(4):26. doi:10.1007/s11032-024-01464-x. [Google Scholar] [PubMed] [CrossRef]
31. Zhou FY, Meng ZW, Han YJ, Zhang Y. Studies on the callus induction and Agrobacterium-mediated transformation of Asia minor bluegrass (Polypogon fugax). J Plant Biochem Biot. 2023;3:32. [Google Scholar]
32. Wu X, Li J, Xu H, Dong L. Factors affecting seed germination and seedling emergence of Asia Minor Bluegrass (Polypogon fugax). Weed Sci. 2015;8:221–36. [Google Scholar]
33. Wang YH, Zhuang LL, Zhang XX, Rossi S, Huang BR. Antioxidant regulation of iron as a repressor for salt-induced leaf senescence in perennial grass species. Plant Growth Regul. 2021;94(3):287–301. doi:10.1007/s10725-021-00716-6. [Google Scholar] [CrossRef]
34. Zhou J, Li Y, Wang X, Liu YJ, David-Schwartz R, Weissberg M, et al. Analysis of Elymus nutans seed coat development elucidates the genetic basis of metabolome and transcriptome underlying seed coat permeability characteristics. Front Plant Sci. 2022;13:970957. doi:10.3389/fpls.2022.970957. [Google Scholar] [PubMed] [CrossRef]
35. Zhang JQ, Ma HL, Liu Y. Analysis on characteristics of female gametophyte and functional identification of genes related to inflorescences development of Kentucky bluegrass. Protoplasma. 2022;259(4):1061–79. doi:10.1007/s00709-021-01720-3. [Google Scholar] [PubMed] [CrossRef]
36. Zhang JQ, Ma HL. Identification and expression analysis of the MADS-box genes of Kentucky bluegrass during inflorescence development. Physiol Mol Biol Pla. 2022;28(7):1359–74. doi:10.1007/s12298-022-01216-1. [Google Scholar] [PubMed] [CrossRef]
37. Bushman BS, Robbins MD, Thorsted K, Robins JG, Warnke SE, Martin R, et al. Transcript responses to drought in Kentucky bluegrass (Poa pratensis L.) germplasm varying in their tolerance to drought stress. Environ Exp Bot. 2021;190:104571. doi:10.1016/j.envexpbot.2021.104571. [Google Scholar] [CrossRef]
38. Buniello A, Macarthur J, Cerezo M, Harris L, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics. Nucleic Acid Res. 2019;47(D1):D1005–D12. doi:10.1093/nar/gky1120. [Google Scholar] [PubMed] [CrossRef]
39. Siekmann D, Jansen G, Zaar A, Kilian A, Fromme FJ, Hackauf B. A genome-wide association study pinpoints quantitative trait genes for plant height, heading date, grain quality, and yield in rye (Secale cereale L.). Front Plant Sci. 2021;12:718081. doi:10.3389/fpls.2021.718081. [Google Scholar] [PubMed] [CrossRef]
40. Zhang DJ, Cheng YC, Lu ZY, Wang JG, Ye XS, Zhang XQ, et al. Global insights to drought stress perturbed genes in oat (Avena sativa L.) seedlings using RNA sequencing. Plant Signal Behav. 2021;16(2):1845934. doi:10.1080/15592324.2020.1845934. [Google Scholar] [PubMed] [CrossRef]
41. Xu L, Feng GY, Yang ZF, Xu XH, Huang LK, Yang QC, et al. Genome-wide AP2/ERF gene family analysis reveals the classification, structure, expression profiles and potential function in orchardgrass (Dactylis glomerata). Mol Biol Rep. 2020;47(7):5225–41. doi:10.1007/s11033-020-05598-x. [Google Scholar] [PubMed] [CrossRef]
42. Li Z, Tang MY, Cheng BZ, Han LB. Transcriptional regulation and stress-defensive key genes induced by gamma-aminobutyric acid in association with tolerance to water stress in creeping bentgrass. Plant Signal Behav. 2021;16(3):1858247. doi:10.1080/15592324.2020.1858247. [Google Scholar] [PubMed] [CrossRef]
43. Liang LL, Cao YQ, Wang D, Peng Y, Zhang Y, Li Z. Spermine alleviates heat-induced senescence in creeping bentgrass by regulating water and oxidative balance, photosynthesis, and heat shock proteins. Biol Plantarum. 2021;65:184–92. doi:10.32615/bp.2021.008. [Google Scholar] [CrossRef]
44. Xu XH, Feng GY, Huang LK, Yang ZF, Liu QX, Shuai Y, et al. Genome-wide identification, structural analysis and expression profiles of GRAS gene family in orchardgrass. Mol Biol Rep. 2020;47(3):1845–57. doi:10.1007/s11033-020-05279-9. [Google Scholar] [PubMed] [CrossRef]
45. Li YT, Liu X, Xiao YX, Wen Y, Li KK, Ma ZL, et al. Genome-wide characterization and function analysis uncovered roles of wheat LIMs in responding to adverse stresses and TaLIM8-4D function as a susceptible gene. Plant Genome. 2022;15(3):e20246. doi:10.1002/tpg2.v15.3. [Google Scholar] [CrossRef]
46. Liu X, Wu J, Zhang D, Bing Z, Tian J, Ni M, et al. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Front Genet. 2018;9:265. doi:10.3389/fgene.2018.00265. [Google Scholar] [PubMed] [CrossRef]
47. Xie L, Gong X, Yang K, Huang Y, Zhang S, Shen L, et al. Technology-enabled great leap in deciphering plant genomes. Nature Plants. 2024;10(4):551–66. doi:10.1038/s41477-024-01655-6. [Google Scholar] [PubMed] [CrossRef]
48. Liu YX, Zhang WQ, Huang DX. The new sequencing technology-Ion Torrent PGM and its forensic application. Chinese J of Forensic Med. 2015;29(8):163–76 (In Chinese). [Google Scholar]
49. Mihai P, Steven L, Salzberg. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;58:661–79. [Google Scholar]
50. Szakacs E, Szoke-Pazsi K, Kalapos B, Schneider A, Ivanizs L, Rakszegi M, et al. 1RS arm of secale cereanum ‘Kriszta’ confers resistance to stripe rust, improved yield components and high arabinoxylan content in wheat. Sci Rep. 2020;10(1):1792. doi:10.1038/s41598-020-58419-3. [Google Scholar] [PubMed] [CrossRef]
51. Li Z, Zeng WH, Cheng BZ, Huang T, Peng Y, Zhang XQ. γ-aminobutyric acid enhances heat tolerance associated with the change of proteomic profiling in preeping bentgrass. Molecules. 2020;25(18):4270. doi:10.3390/molecules25184270. [Google Scholar] [PubMed] [CrossRef]
52. Mahmoud M, Zhou Z, Kaur R, Bekele W, Tinker NA, Singh J. Toward the development of Ac/Ds transposon-mediated gene tagging system for functional genomics in oat (Avena sativa L.). Funct Integr Genomic. 2022;22(4):669–81. doi:10.1007/s10142-022-00861-9. [Google Scholar] [PubMed] [CrossRef]
53. Hao JS, Xing JF, Hu X, Wang ZY, Tang MQ, Liao L. Distribution pattern of N6-methyladenine DNA modification in the Seashore Paspalum (Paspalum vaginatum) genome. Front Plant Sci. 2022;13:922152. doi:10.3389/fpls.2022.922152. [Google Scholar] [PubMed] [CrossRef]
54. Salas-Moreno M, Castillejo MA, Lopez-Hidalgo C, Marrugo-Negrete J, Rodriguez-Cavallo E, Mendez-Cuadro D, et al. LC-MS/MS shotgun proteomics reveals biochemical mechanisms of Paspalum fasciculatum tolerance to Pb-stress. Biol Plantarum. 2022;66:188–200. doi:10.32615/bp.2022.016. [Google Scholar] [CrossRef]
55. Weber J, Saa JD, Grove C, Rad R. PiggyBac transposon tools for recessive screening identify B-cell lymphoma drivers in mice. Nat Commun. 2019;10(1):1415. doi:10.1038/s41467-019-09180-3. [Google Scholar] [PubMed] [CrossRef]
56. Fengmin S. Functional study of MwLEA3 gene and cloning of Mongolica Wheatgrass (Ph.D. Thesis). Inner Mongolia Agricultural University: Inner Mongolia, China; 2011 (In Chinese). [Google Scholar]
57. Zhu LZ, Wang F, Wang YL, He J, Li YL, Ying T. Gene editing technology and CRISPR/Cas system in applications in the development of grassland plants. Jiangsu Agr Sci. 2021;49(20):22–30. [Google Scholar]
58. Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi:10.1126/science.1232033. [Google Scholar] [PubMed] [CrossRef]
59. Sürün D, Schw B, Tomasovic J, Ehling A, Stein R, Kurrle S, et al. High efficiency gene correction in hematopoietic cells by donor-template-free CRISPR/Cas9 genome editing. Mol Ther Nucl Acids. 2018;10:1–8. doi:10.1016/j.omtn.2017.11.001. [Google Scholar] [PubMed] [CrossRef]
60. Ramirez JC. Gene editing and CRISPR therapeutics: strategies taught by cell and gene therapy. Prog Mol Biol Transl. 2017;152(3):115–30. [Google Scholar]
61. Jing RC, Liu H. The development of CRISPR/Cas9 system and its application in crop genome editing. Chinese Agr sci. 2016;49(7):11 (In Chinese). [Google Scholar]
62. Yang SH. Use of CRISPR/Cas9 for crop improvement in legume. Prog Mol Biol Transl Sci. 2021;149(27):27–46. [Google Scholar]
63. Holger FP. Transforming plant biology and breeding with CRISPR CRISPR/Cas9, Cas12 and Cas13. FEBS Letters. 2018;592(12):112–25. [Google Scholar]
64. Wei ZX, Sun H, Xiang YT, Jiang HZ, Sha AH, Liu LJ. Application of virus-induced gene silencing in legumes. Chinese J Oil Crop Sci. 2022;44(3):497–502. [Google Scholar]
65. Hao M, Hang Q, Shi G. Application and prospect of VIGS-induced gene silencing in crop gene function research. China Agr Sci and Technolo Herald. 2022;24(1):1–13. [Google Scholar]
66. Xiao Y, Wang J, Li L, Wang Y, Zhang C, Sun G. Advances in the researches and applications of host-induced gene silencing technology. J Plant Pro. 2020;47(1):11–7. [Google Scholar]
67. Huang C, Nie X, Shen C, You C, Li W, Zhao W, et al. Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol J. 2017;15(11):1374–86. doi:10.1111/pbi.2017.15.issue-11. [Google Scholar] [CrossRef]
68. Mo Q, Jiang W, Chen Y, LÜ B. Advances in the development of spray-induced gene silencing for the management of plant pathogenic fungal. Chinese J Bio Control. 2022;38(5):1316–24 (In Chinese). [Google Scholar]
69. Chen X, Shi T, Chen C, Tang T. Advances in the control of plant diseases via spray-induced gene silencing. Plant Protect. 2022;48(5):15–22. [Google Scholar]
70. Zhang Q, Ji Z, Zhu F. Application advance of virus-induced gene silencing technology in dicotyledons. Henan Agr Sci. 2020;49(2):1–8. [Google Scholar]
71. Li J, Yu Z, Feng B. Advances in research and application of virus induced gene silencing in plants. Mol Plant Breeding. 2019;17(5):1537–42 (In Chinese). [Google Scholar]
72. Liu Y. Whole transcriptome analysis of sunflower seeds with high oleic acid at different developmental stages and functional identification of HaFAD2 (Ph.D. Thesis). Northeast Forestry University: Heilongjiang, China; 2021. [Google Scholar]
73. Li MY, Wang JL, Yao T, Wang ZL, Zhang HR, Li CN. Isolation and characterization of cold-adapted PGPB and their effect on plant growth promotion. J Microbiol Biotechn. 2021;31(9):1218–30. doi:10.4014/jmb.2105.05012. [Google Scholar] [PubMed] [CrossRef]
74. Cheng Y, Meng R, Niu S, Peng H, Jing M. Protocol for quantitative evaluation of misfolded protein degradation using Agrobacterium-mediated expression system in Nicotiana benthamiana. STAR Protoc. 2024;5(2):998–1016. [Google Scholar]
75. Gao C, Long D, Lenk I, Nielsen KK. Comparative analysis of transgenic tall fescue (Festuca arundinacea Schreb.) plants obtained by Agrobacterium-mediated transformation and particle bombardment. Plant Cell Rep. 2008;27(10):1601–9. doi:10.1007/s00299-008-0578-x. [Google Scholar] [PubMed] [CrossRef]
76. Dai S, Zheng P, Marmey P, Zhang S, Tian W, Chen S, et al. Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol Breeding. 2001;7(1):25–33. doi:10.1023/A:1009687511633. [Google Scholar] [CrossRef]
77. Chen L, Yu S, Li S, Zhang L, Zou C, Yu D. The role of WRKY transcription factors in plant abiotic stresses. BBA-Gene Regul Mech. 2012;1819(2):120–8. [Google Scholar]
78. Wang K, Bai ZY, Liang QY, Liu QL, Zhang L, Pan YZ, et al. Transcriptome analysis of chrysanthemum (Dendranthema grandiflorum) in response to low temperature stress. BMC Genomics. 2018;19(1):319. doi:10.1186/s12864-018-4706-x. [Google Scholar] [PubMed] [CrossRef]
79. Sun S. Exploration of mechanism underlying the freezing tolerance in Actinidia arguta and functional identification of two low cold response genes (Ph.D. Thesis). Huazhong Agricultural University: Wuhan, China; 2022 (In Chinese). [Google Scholar]
80. Xu X, Liu WW, Liu XY, Cao YP, Li XN, Wang GY, et al. Genetic manipulation of bermudagrass photosynthetic biosynthesis using Agrobacterium-mediated transformation. Physiol Plantarum. 2022;174(3):e13710. doi:10.1111/ppl.v174.3. [Google Scholar] [CrossRef]
81. Gan L, Chen MH, Zhang JX, Fan JB, Yan XB. A novel beta-glucosidase gene for plant type was identified by genome-wide association study and gene co-expression analysis in widespread Bermudagrass. Int J Mol Sci. 2022;23(19):11432. doi:10.3390/ijms231911432. [Google Scholar] [PubMed] [CrossRef]
82. Zuo ZF, Kang HG, Hong QC, Park MY, Sun HJ, Kim J, et al. A novel basic helix-loop-helix transcription factor, ZjICE2 from Zoysia japonica confers abiotic stress tolerance to transgenic plants via activating the DREB/CBF regulon and enhancing ROS scavenging. Plant Mol Biol. 2020;102(4–5):447–62. [Google Scholar] [PubMed]
83. Teng K, Yue YS, Zhang H, Li H, Xu LX, Han C, et al. Functional characterization of the pheophytinase gene, ZjPPH, from Zoysia japonica in regulating Chlorophyll degradation and photosynthesis. Front Plant Sci. 2021;12:786570. doi:10.3389/fpls.2021.786570. [Google Scholar] [PubMed] [CrossRef]
84. Teng K, Tan PH, Guan J, Dong D, Liu LY, Guo YD, et al. Functional characterization of the chlorophyll b reductase gene NYC1 associated with chlorophyll degradation and photosynthesis in Zoysia japonica. Environ Exp Bot. 2021;191(11):104607. [Google Scholar]
85. Li GS, Yin QH, Chen YT, Li XX, Chen XL, Deng H. Overexpression of ZmDUF1644 from Zoysia matrella enhances salt tolerance in Arabidopsis thaliana. Plant Growth Regul. 2023;23:s10725. [Google Scholar]
86. Guo T, Wang SM, Fan B, Zou SH, Chen S, Liu W, et al. Overexpression of the Zoysia japonica ZjABR1/ERF10 regulates plant growth and salt tolerance in transgenic Oryza sativa. Environ Exp Bot. 2023;206:105171. doi:10.1016/j.envexpbot.2022.105171. [Google Scholar] [CrossRef]
87. Guan J, Teng K, Yue YS, Guo YD, Liu LY, Yin SX, et al. Zoysia japonica Chlorophyll b reductase gene NOL participates in Chlorophyll degradation and photosynthesis. Front Plant Sci. 2022;13:906018. doi:10.3389/fpls.2022.906018. [Google Scholar] [PubMed] [CrossRef]
88. Dong D, Yang ZX, Ma Y, Li SW, Wang MD, Li YRZ, et al. Expression of a hydroxycinnamoyl-CoA Shikimate/Quinate Hydroxycinnamoyl transferase 4 gene from Zoysia japonica (ZjHCT4) causes excessive elongation and lignin composition changes in Agrostis stolonifera. Int J Mol Sci. 2022;23(16):9500. doi:10.3390/ijms23169500. [Google Scholar] [PubMed] [CrossRef]
89. Kozak L, Szilagyi Z, Toth L, Pocsi I, Molnar I. Functional characterization of the idtF and idtP genes in the Claviceps paspali indole diterpene biosynthetic gene cluster. Folia Microbiol. 2020;65(3):605–13. doi:10.1007/s12223-020-00777-6. [Google Scholar] [PubMed] [CrossRef]
90. Benkeblia N. Insights on fructans and resistance of plants to drought stress. Front Sustain Food S. 2022;6:827758. doi:10.3389/fsufs.2022.827758. [Google Scholar] [CrossRef]
91. Xia X. Study on resistance mechanism of metamifopin in echinochloa crus-galli (Ph.D. Thesis). Huazhong Agricultural University: Wuhan, China; 2017 (In Chinese). [Google Scholar]
92. Rahman SU, Khan MO, Ullah R, Ahmad F, Raza G. Agrobacterium-mediated transformation for the development of transgenic crops; present and future prospects. Mol Biotechnol. 2023;5(4):118–29. [Google Scholar]
93. Li J, Han G, Sun C, Sui N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal Behav. 2019;14(8):1613131. doi:10.1080/15592324.2019.1613131. [Google Scholar] [PubMed] [CrossRef]
94. Chen Y. Clonging and functional verification of AmWRKY1-like in Agropyron mongolicum Keng (Master’ Degree Thesis). Inner Mongolia Agricultural University: Inner Mongolia, China; 2020 (In Chinese). [Google Scholar]
95. Zou C, Jiang W, Yu D. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot. 2010;61(14):3901–14. doi:10.1093/jxb/erq204. [Google Scholar] [PubMed] [CrossRef]
96. Fan F, Wang Q, Li H, Ding G, Wen X. Transcriptome-wide identification and expression profiles of masson pine WRKY transcription factors in response to low phosphorus stress. Plant Mol Biol Rep. 2021;39(1):101. [Google Scholar]
97. Jiang W, Yu D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009;9(1):96–100. doi:10.1186/1471-2229-9-96. [Google Scholar] [PubMed] [CrossRef]
98. Zhenbing M, Li W, Wang H, Yu D. WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age-mediated flowering. J Integr Plant Biol. 2020;62(11):21–35. [Google Scholar]
99. Zhang LP, Chen L, Yu D. Transcription factor WRKY75 Interacts with DELLA proteins to affect flowering. Plant Physiol. 2018;176(1):790–803. doi:10.1104/pp.17.00657. [Google Scholar] [PubMed] [CrossRef]
100. An J-P, Zhang X-W, You C-X, Bi S-Q, Wang X-F, Hao Y-J. MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol. 2019;224(1):380–95. doi:10.1111/nph.v224.1. [Google Scholar] [CrossRef]
101. Niu F, Cui X, Zhao P, Sun M, Jiang Y. WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 2020;104(1):171–84. doi:10.1111/tpj.v104.1. [Google Scholar] [CrossRef]
102. Chen L, Fan J, Hu L, Hu Z, Xie Y, Zhang Y, et al. A transcriptomic analysis of bermudagrass (Cynodon dactylon) provides novel insights into the basis of low temperature tolerance. BMC Plant Biol. 2015;15(216):2015–20. [Google Scholar]
103. Sun X, Zhang L, Wong D, Wang Y, Xin H. The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 2019;99(5):988–1002. doi:10.1111/tpj.v99.5. [Google Scholar] [CrossRef]
104. Niu CF, Wei W, Zhou QY, Tian AG, Chen SY. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35(6):1156–70. doi:10.1111/pce.2012.35.issue-6. [Google Scholar] [CrossRef]
105. Niu X, Luo TL, Zhao HY, Su YL, Ji WQ, Li HF. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene. 2020;740:144514. doi:10.1016/j.gene.2020.144514. [Google Scholar] [PubMed] [CrossRef]
106. Ding CH. Molecular characterization of WRKY genes TaWRKY72b-2 and TaWRKY72b-3 in wheat and their functions in regulating plant tolerance to low phosphorus stress (Ph.D. Thesis). Agricultural University of Hebei: Hebei, China; 2013 (In Chinese). [Google Scholar]
107. Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233(6):1237–52. doi:10.1007/s00425-011-1375-2. [Google Scholar] [PubMed] [CrossRef]
108. Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6(5):486–503. doi:10.1111/pbi.2008.6.issue-5. [Google Scholar] [CrossRef]
109. Dai W, Min W, Gong X, Liu J. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018;219(3):972–89. doi:10.1111/nph.2018.219.issue-3. [Google Scholar] [CrossRef]
110. Wei Q, Zhang F, Sun F, Luo Q, Wang R, Hu R, et al. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 2017;265:112–23. doi:10.1016/j.plantsci.2017.09.020. [Google Scholar] [PubMed] [CrossRef]
111. Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, et al. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS One. 2014;9(3):e92913. doi:10.1371/journal.pone.0092913. [Google Scholar] [PubMed] [CrossRef]
112. Zhao Y, Han HJ, Zhang R, Tong XM, Gao CP. Functional identification of MwMYB4 gene from Agropyron mongolicum Keng. J Northwestern Botany. 2020;40(4):565–71. [Google Scholar]
113. Bienias A, Goralska M, Masojc P, Milczarski P, Myskow B. The GAMYB gene in rye: sequence, polymorphisms, map location, allele-specific markers, and relationship with alpha-amylase activity. BMC Genomics. 2020;21(1):578. doi:10.1186/s12864-020-06991-3. [Google Scholar] [PubMed] [CrossRef]
114. Han HJ. Cloning and functional preliminary studies of AmrMYBR1 and AmrR2R3MYB genes from Agropyron Michnoi Roshev (Ph.D. Thesis). Inner Mongolia Agricultural University: Inner Mongolia, China; 2022 (In Chinese). [Google Scholar]
115. Kamyab S, Alami-Saeid K, Eslahi M, Moradi M. cDNA-AFLP technique discloses differential gene expression in response to salinity in wheat (Triticum aestivum L.). Genet Resour Crop Ev. 2021;68(6):2299–311. doi:10.1007/s10722-020-01098-w. [Google Scholar] [CrossRef]
116. Bai XX, Zhan GM, Tian SX, Peng H, Cui XY, Islam MA, et al. Transcription factor BZR2 activates chitinase Cht20.2 transcription to confer resistance to wheat stripe rust. Plant Physiol. 2021;187(4):2749–62. doi:10.1093/plphys/kiab383. [Google Scholar] [PubMed] [CrossRef]
117. Hu CH, Li BB, Chen P, Shen HY, Xi WG, Zhang Y, et al. Identification of CDPKs involved in TaNOX7 mediated ROS production in wheat. Front Plant Sci. 2023;13:1108622. doi:10.3389/fpls.2022.1108622. [Google Scholar] [PubMed] [CrossRef]
118. De Y, Shi FL, Gao FQ, Mu HB, Yan WH. Siberian wildrye (Elymus sibiricus L.) abscisic acid-insensitive 5 gene is involved in abscisic acid-dependent salt response. Plants. 2021;10(7):1351. doi:10.3390/plants10071351. [Google Scholar] [PubMed] [CrossRef]
119. Jia ZY, Zhang MJ, Ma C, Wang ZQ, Wang ZH, Fang Y, et al. Identification and functional validation of auxin-responsive tabzip genes from Wheat Leaves in Arabidopsis. Int J Mol Sci. 2023;24(1):756. doi:10.3390/ijms24010756. [Google Scholar] [PubMed] [CrossRef]
120. Leene JV, Blomme J, Kulkarni SR, Cannoot B, Winne ND, Eeckhout D, et al. Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development. J Exp Bot. 2016;67(19):5825–40. doi:10.1093/jxb/erw347. [Google Scholar] [PubMed] [CrossRef]
121. Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr Opin Plant Biol. 2000;3(5):423–34. doi:10.1016/S1369-5266(00)00107-2. [Google Scholar] [PubMed] [CrossRef]
122. Heim MA, Marc J, Martin W, Cathie M, Bernd W, Bailey PC. The basic helix-loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–47. doi:10.1093/molbev/msg088. [Google Scholar] [PubMed] [CrossRef]
123. Chandler JW. Class vIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci. 2018;23(2):151–62. [Google Scholar] [PubMed]
124. Müller M, Munné-bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169(1):32–41. doi:10.1104/pp.15.00677. [Google Scholar] [PubMed] [CrossRef]
125. Zhao C, Zhang H, Song C, Zhu JK, Shabala S. Mechanisms of plant responses and adaptation to soil salinity. Innovation. 2020;1(1):41. [Google Scholar]
126. Jiang Y, Zhang HW, Li Y, Chang CC, Wang YX, Feng H, et al. A novel transcriptional regulator HbERF6 regulates the HbCIPK2-coordinated pathway conferring salt tolerance in halophytic Hordeum brevisubulatum. Front Plant Sci. 2022;13:927253. doi:10.3389/fpls.2022.927253. [Google Scholar] [PubMed] [CrossRef]
127. Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends in Plant Sci. 2007;12(10):444–51. doi:10.1016/j.tplants.2007.07.002. [Google Scholar] [PubMed] [CrossRef]
128. Jiang J, Ma S, Ye N, Ming J, Zhang J. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2016;59(2):86. [Google Scholar]
129. Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu JK. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016;171(4):2744–259. doi:10.1104/pp.16.00533. [Google Scholar] [PubMed] [CrossRef]
130. Resentini F, Felipo-Benavent A, Colombo L, Blázquez MA, Masiero S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol Plant. 2014;8(3):482–5. [Google Scholar] [PubMed]
131. Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, et al. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell. 2010;22(12):3921–34. [Google Scholar] [PubMed]
132. Yu H, Zhang L, Wang W, Tian P, Wang W, Wang K, et al. TCP5 controls leaf margin development by regulating KNOX and BEL-like transcription factors in Arabidopsis. J Exp Bot. 2021;27(5):5. [Google Scholar]
133. Selahattin D. TCP Transcription factors at the interface between environmental challenges and the plant’s growth responses. Front Plant Sci. 2016;7(406):1930. [Google Scholar]
134. Wu JF, Tsai HL, Joanito I, Wu YC, Chang CW, Li YH, et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat Commun. 2016;13(7):13181. [Google Scholar]
135. Yuan Z, Gao S, Xue DW, Luo D, Li LT, Ding SY, et al. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 2009;149(1):235–44. doi:10.1104/pp.108.128231. [Google Scholar] [PubMed] [CrossRef]
136. Shuai Y, Feng GY, Yang ZF, Liu QX, Han JT, Xu XH, et al. Genome-wide identification of C2H2-type zinc finger gene family members and their expression during abiotic stress responses in orchardgrass (Dactylis glomerata). Genome. 2022;65(4):189–203. doi:10.1139/gen-2020-0201. [Google Scholar] [PubMed] [CrossRef]
137. Li Q. Functional analysis of APETALA1/FRUITFULL subfamily and INDETERMINATE1 gene in Brachypodium distachyon L (Ph.D. Thesis). Shandong Agricultural University: Shandong, China; 2015 (In Chinese). [Google Scholar]
138. Zhang W, Rong X, Tian Q, Li T, Wu Z, Liu H, et al. Editing of fallen grain related genes Sh1 of Agropyron mongolicum Keng using CRISPR/Cas9 teachnology. Mol Plant Breeding. 2019;17(15):5(In Chinese). [Google Scholar]
139. Wang Y. Functional analyses of APETALA1/FRUITFULL transcription factor subfamily genes in Branchypodium distachyon L (Ph.D. Thesis). Shandong Agricultural University: Shandong, China; 2017 (In Chinese). [Google Scholar]
140. Zhao C, Wang P, Si T, Hsu CC, Wang L, Zayed O, et al. MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell. 2017;43(5):618–29. doi:10.1016/j.devcel.2017.09.024. [Google Scholar] [PubMed] [CrossRef]
141. Zong XJ, Li DP, Gu LK, Li DQ, Liu LX, Hu XL. Abscisic acid and hydrogen peroxide induce a novel maize group CMAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta. 2009;229(3):485–95. doi:10.1007/s00425-008-0848-4. [Google Scholar] [PubMed] [CrossRef]
142. Xing Y, Jia W, Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J: for Cell and Mole Biology. 2008;54(3):440–51. doi:10.1111/tpj.2008.54.issue-3. [Google Scholar] [CrossRef]
143. Wang G, Liang YH, Zhang JY, Cheng ZM. Cloning, molecular and functional characterization by overexpression in Arabidopsis of MAPKK genes from grapevine (Vitis vinifera). BMC Plant Biol. 2020;20(1):194. doi:10.1186/s12870-020-02378-4. [Google Scholar] [PubMed] [CrossRef]
144. Xie LL, Wang JL, Wu GQ. Regulation mechanism of plant CBL-CIPK signaling system in response to abiotic stress. Chinese Bullet Bot. 2021;56(5):614–26 (In Chinese). [Google Scholar]
145. Huang S. Functional analysis of CdCIPK5 and OsCIPK5 of Cynodon spp. and rice (Ph.D. Thesis). South China Agricultural University: Guangzhou, China; 2019 (In Chinese). [Google Scholar]
146. Sakuraba Y, Kim YS, Yoo SC, Rtensteiner S, Paek NC. 7-Hydroxymethyl chlorophyll a reductase functions in metabolic channeling of chlorophyll breakdown intermediates during leaf senescence. Biochem Bioph Res Co. 2013;430(1):32–7. doi:10.1016/j.bbrc.2012.11.050. [Google Scholar] [PubMed] [CrossRef]
147. Jia T, Ito H, Tanaka A. The chlorophyll b reductase NOL participates in regulating the antenna size of photosystem II in Arabidopsis Thaliana. Procedia Chem. 2015;14(5):422–7. [Google Scholar]
148. Sato Y, Morita R, Katsuma S, Nishimura M, Kusaba M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2010;57(1):120–31. [Google Scholar]
149. Yu G, Xie Z, Zhang J, Lei S, Huang B. NOL ediated functional stay traits in perennial ryegrass (Lolium perenne L.) involving multifaceted molecular factors and metabolic pathways regulating leaf senescence. Plant J. 2021;106(5):1219–32. doi:10.1111/tpj.v106.5. [Google Scholar] [CrossRef]
150. Yang MZ, Chen JJ, Liu TT, Xiang LL, Zhou BF. Genome-wide identification and expression analysis of calmodulin-like gene family in Paspalums vaginatium revealed their role in response to salt and cold stress. Curr Issues Mol Biol. 2023;45(2):1693–711. doi:10.3390/cimb45020109. [Google Scholar] [PubMed] [CrossRef]
151. Smith PJ, O’Neill MA, Backe J, York WS, Pea MJ, Urbanowicz BR. Analytical techniques for determining the role of domain of unknown function 579 proteins in the synthesis of O-methylated plant polysaccharides. SLAS Technol. 2020;25(4):345–55. doi:10.1177/2472630320912692. [Google Scholar] [PubMed] [CrossRef]
152. Liang T. Functional identification of PIP5K family genes related to inflorescence development in female line of Castor Lm (Master Thesis). Inner Mongolia University for Nationalities: Inner Mongolia, China; 2020 (In Chinese). [Google Scholar]
153. Wang Y. Functional identification of SVP-like genes of strawberry (Master’ Degree Thesis). Shenyang Agricultural University: Shenyang, China; 2022 (In Chinese). [Google Scholar]
154. Wang DM. Transcriptome analysis and functional identification of SWEET in daylily under low temperature stress (Master Thesis). Shanghai University of Technology: Shanghai, China; 2022. [Google Scholar]
155. Wu Z. Cloning of CT region of Acetyl-Coenzyme a carboxylase gene from Avena sativa L and establishment of CRISPR/Cas9 technique system (Ph.D. Thesis). Inner Mongolia Agricultural University: Inner Mongolia, China; 2018 (In Chinese). [Google Scholar]
156. Zhang DW, Zhang CF, Dong F, Huang YL, Zhang Y, Zhou H. Application of CRISPR/Cas9 system in breeding of new antiviral plants germplasm. J Genet. 2016;38(9):10. [Google Scholar]
157. Deng Y. Mining and functional identification of Verticillium wilt resistance genes of in upland cotton (Ph.D. Thesis). Xinjiang Agricultural University: Xinjiang, China; 2022 (In Chinese). [Google Scholar]
158. Mustafa R, Iqbal MJ, Hamza M, Rehman AU, Buzdar I, Kamal H, et al. Functional identification of G. hirsutum genes for their role in normal plant development and resistance against Verticillium dahliae using virus-induced gene silencing. Eur J Plant Pathol. 2021;161(4):917–31. doi:10.1007/s10658-021-02376-8. [Google Scholar] [CrossRef]
159. Wang WP, Cui H, Xiao XF, Wu BJ, Sun JL, Zhang YX, et al. Genome-wide identification of cotton (Gossypium spp.) trehalose-6-phosphate phosphatase (TPP) gene family members and the role of GhTPP22 in the response to drought stress. Plants. 2022;11(8):1079. doi:10.3390/plants11081079. [Google Scholar] [PubMed] [CrossRef]
160. Wang XY, Chen BZ, Ma CK, Qiao KK, Li ZS, Wang JS, et al. Systematical characterization of YUCCA gene family in five cotton species, and potential functions of YUCCA22 gene in drought resistance of cotton. Ind Crop Prod. 2021;162:926–37. [Google Scholar]
161. Yu WC, Xue ZH, Zhao XZ, Zhang R, Liu JP, Guo SD. Glyphosate-induced GhAG2 is involved in resistance to salt stress in cotton. Plant Cell Rep. 2022;41(4):1131–45. doi:10.1007/s00299-022-02844-3. [Google Scholar] [PubMed] [CrossRef]
162. Liu SY, Liu JF, Zhang YZ, Jiang YS, Hu SW, Shi AD, et al. Cloning of the Soybean sHSP26 gene and analysis of its drought resistance. Phyton-International J Exp Bot. 2022;91(7):1465–82. [Google Scholar]
163. Wu L, Chang YJ, Wang LF, Wang SM, Wu J. The aquaporin gene PvXIP1;2 conferring drought resistance identified by GWAS at seedling stage in common bean. Theor Appl Genet. 2022;135(2):485–500. doi:10.1007/s00122-021-03978-w. [Google Scholar] [PubMed] [CrossRef]
164. Mei XP, Liu CX, Nan J, Zhao ZK, Bai Y, Dong EF, et al. Overexpression of ZmNF-YC13 confers ER stress tolerance in maize. J Plant Biol. 2021;64(4):337–48. doi:10.1007/s12374-021-09307-4. [Google Scholar] [CrossRef]
165. Hu WJ, Ren QY, Chen YL, Xu GL, Qian YX. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021;21(1):427. doi:10.1186/s12870-021-03206-z. [Google Scholar] [PubMed] [CrossRef]
166. Wang F, Yan HP, Fang P, Ji XZ, Peng YL. Heterologous overexpression of ZmHDZIV13 enhanced drought and salt tolerance in Arabidopsis and tobacco. Agronomy. 2022;12(10):233–6. [Google Scholar]
167. Tu JQ, Feng LL, Hong YB, Liu QY, Huang X, Li Y. Prokaryotic expression of phosphoenolpyruvate carboxylase fragments from peanut and analysis of osmotic stress tolerance of recombinant strains. Plants. 2021;10(2):365. doi:10.3390/plants10020365. [Google Scholar] [PubMed] [CrossRef]
168. Zhu K, Huang C, Phan TT, Yang LT, Zhang BQ, Xing YX, et al. Overexpression of SoACLA-1 gene confers drought tolerance improvement in sugarcane. Plant Mol Biol Reporter. 2021;39(3):489–500. doi:10.1007/s11105-020-01263-6. [Google Scholar] [CrossRef]
169. Yue L, Zhuang Y, Gu YY, Li H, Tu SB, Yang XC, et al. Heterologous expression of Solanum tuberosum NAC1 gene confers enhanced tolerance to salt stress in transgenic Nicotiana benthamiana. J Plant Biol. 2021;64(6):531–42. doi:10.1007/s12374-021-09327-0. [Google Scholar] [CrossRef]
170. Zhou Z. Regulatiory analysis of cytokinin response and function charaterization of IPT genes in Medicago truncatula (Ph.D. Thesis). Beijing Forestry University: Beijing, China; 2020. [Google Scholar]
171. Rui L. Screening of gene in response to vernalization in varieties with different vernalization demands and functional verification of BrcSOC1 in pak choi (Ph.D. Thesis). Shanxi Agricultural University: Shanxi, China; 2021 (In Chinese). [Google Scholar]
172. Wu Y, Zhang LT, Nie LB, Zheng YS, Zhu SD, Hou JF, et al. Genome-wide analysis of the DREB family genes and functional identification of the involvement of BrDREB2B in abiotic stress in wucai (Brassica campestris L.). BMC Genomics. 2022;23(1):598. doi:10.1186/s12864-022-08812-1. [Google Scholar] [PubMed] [CrossRef]
173. Gou BD, Duan PP, Wei M, Zhao SF, Wang YF, Yang N, et al. Silencing CaTPS1 increases the sensitivity to low temperature and salt stresses in pepper. Agronomy. 2023;13(2):319. doi:10.3390/agronomy13020319. [Google Scholar] [CrossRef]
174. Wang X. Transcriptomic analysis and functional identification of PlACL2 gene of the Inner and outer petals in the double-colored Paeonia lactiflora (Ph.D. Thesis). Yangzhou University: Yangzhou, China; 2022. [Google Scholar]
175. Wang SC, Zhang R, Zhang ZX, Zhao T, Zhang D, Sofkova S, et al. Genome-wide analysis of the bZIP gene lineage in apple and functional analysis of MhABF in Malushalliana. Planta. 2021;254(4):78. doi:10.1007/s00425-021-03724-y. [Google Scholar] [PubMed] [CrossRef]
176. Li LC, Yang KB, Wang SN, Lou YF, Zhu CL, Gao ZM. Genome-wide analysis of laccase genes in moso bamboo highlights PeLAC10 involved in lignin biosynthesis and in response to abiotic stresses. Plant Cell Rep. 2020;39(6):751–63. doi:10.1007/s00299-020-02528-w. [Google Scholar] [PubMed] [CrossRef]
177. Sun YY, Hu DD, Xue PC, Wan XL. Identification of the DcHsp20 gene family in carnation (Dianthus caryophyllus) and functional characterization of DcHsp17.8 in heat tolerance. Planta. 2022;256(1):2. doi:10.1007/s00425-022-03915-1. [Google Scholar] [PubMed] [CrossRef]
178. Yu YH, Yang SD, Bian L, Yu KK, Meng XX, Zhang GH, et al. Identification of C3H2C3-type RING E3 ubiquitin ligase in grapevine and characterization of drought resistance function of VyRCHC114. BMC Plant Biol. 2021;21(1):422. doi:10.1186/s12870-021-03162-8. [Google Scholar] [PubMed] [CrossRef]
179. Zhao XH, Qu DH, Wang L, Gao YH, An NN, Wang AP, et al. Genome-wide identification of cysteine-rich receptor-like kinases in sweet cherry reveals that PaCRK1 enhances sweet cherry resistance to salt stress. Plant Cell Rep. 2022;41(10):2037–88. doi:10.1007/s00299-022-02907-5. [Google Scholar] [PubMed] [CrossRef]
180. Chen X. Functional analysis of the RNDXcc gene in Aanthomonas campestris PV. Campestris (Master’ Degree Thesis). Guangxi University: Guangxi, China; 2021. [Google Scholar]
181. Zuo D. Cloning and functional characterization of wheat genes induced by deoxynivalenol mycotoxin (Ph.D. Thesis). Huazhong Agricultural University: Wuhan, China; 2016 (In Chinese). [Google Scholar]
182. Gang L. Expression and functional characterization of wheat antimicrobial peptide genes induced by deoxyniniolenol (Ph.D. Thesis). Huazhong Agricultural University: Wuhan, China; 2019 (In Chinese). [Google Scholar]
183. Zhao GY. Application of papaya malformation Mosaic virus in host gene function identification and cross protection (Ph.D. Thesis). Hainan University: Hainan, China; 2020. [Google Scholar]
184. Du MY. Cloning and functional identification of TaSOG1 gene in Wheat (Ph.D. Thesis). Shandong Agricultural University: Shandong, China; 2022 (In Chinese). [Google Scholar]
185. Wei T. Dissection of mechanisms underlying theenhanced stress tolerance in autotetraploid Poncirus trifoliata and functionalidentification of two stress responsive genes (Ph.D. Thesis). Huazhong Agricultural University: Wuhan, China; 2020 (In Chinese). [Google Scholar]
186. Chen LZ, Duan L, Sun MH, Yang Z, Li HY, Hu KM, et al. Current trends and insights on EMS mutagenesis application to studies on plant abiotic stress tolerance and development. Front Plant Sci. 2023;13(4):1052569. [Google Scholar] [PubMed]
187. Singh R, Kumar G. Analyzing frequency and spectrum of chlorophyll mutation induced through gamma ray and combination treatment (gamma plus EMS) on genetic paradigm of Artemisia annua L. Caryologia. 2022;75(1):15–27. [Google Scholar]
188. Karaman K, Kizil S, Basak M, Uzun B, Yol E. Development of EMS-induced mutagenized groundnut population and discovery of point mutations in the ahFAD2 and ara h 1 genes by TILLING. J Oleo Sci. 2021;70(11):1631–40. doi:10.5650/jos.ess21075. [Google Scholar] [PubMed] [CrossRef]
189. Liu Y, Wang J, Liu B, Xu ZY. Dynamic regulation of DNA methylation and histone modifications in response to abiotic stresses in plants. J Integr Plant Biol. 2022;64(12):2252–74. doi:10.1111/jipb.v64.12. [Google Scholar] [CrossRef]
190. Ye RL, Yang X, Rao YC. Genetic engineering technologies for improving crop yield and quality. Agronomy. 2022;12(4):1–15. [Google Scholar]
191. Kim BM, Inaba JI, Masuta C. Virus induced gene silencing in Antirrhinum majus using the Cucumber mosaic virus vector: functional analysis of the AINTEGUMENTA (Am-ANT) gene of A. majus. Hortic, Env, and Biotechn. 2011;52(2):176–82. doi:10.1007/s13580-011-0172-y. [Google Scholar] [CrossRef]
192. Li J, Luo J, Yang P. Reasearch advances of applying virus-induced gene silencing in vegetable. Chinese Agr Sci. 2021;54(10):2154–66 (In Chinese). [Google Scholar]
193. Wang YH, Tang QL, Pu L, Zhang HW, Li XH. CRISPR-Cas technology opens a new era for the creation of novel maize germplasms. Front Plant Sci. 2022;13(8):992–1014. [Google Scholar]
194. Kumari S, Singh SK, Sharma VK, Kumar R, Mathur M, Upadhyay TK, et al. CRISPR-Cas: a continuously evolving technology. Indian J Agr Sci. 2021;91(9):1274–9. [Google Scholar]
195. Swarts DC, Hegge JW, Ismael H, Masami S, Ellis MA, Justin D, et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015;43(10):5120–9. doi:10.1093/nar/gkv415. [Google Scholar] [PubMed] [CrossRef]
196. Dong ZJ, Chen X, Zhuo R, Li YY, Zhou ZH, Sun Y, et al. Efficient manipulation of gene expression using Natronobacterium gregoryi Argonaute in zebrafish. BMC Biol. 2023;21(1):95. doi:10.1186/s12915-023-01599-x. [Google Scholar] [PubMed] [CrossRef]
197. Kordys M, Sen R, Warkocki Z. Applications of the versatile CRISPR-Cas13 RNA targeting system. Wires RNA. 2022;13(3):e1694. [Google Scholar] [PubMed]
198. Wu SM, Tian PF, Tan TW. CRISPR-Cas13 technology portfolio and alliance with other genetic tools. Biotechnology Adv. 2022;61:108047. doi:10.1016/j.biotechadv.2022.108047. [Google Scholar] [PubMed] [CrossRef]
199. Quansah E, Chen YH, Yang SJ, Wang JY, Sun DH, Zhao YX, et al. CRISPR-Cas13 in malaria parasite: diagnosis and prospective gene function identification. Front Microbiol. 2023;14:1076947. doi:10.3389/fmicb.2023.1076947. [Google Scholar] [PubMed] [CrossRef]
200. Hsieh FK. Engineering of CRISPR Cas12i to enable therapeutic genome editing. Mol Ther. 2022;30(4):268–8. [Google Scholar]
201. Sengupta S, Haswell JR, Keston-Smith E, Wessells Q, Garrity AJ, Yan P, et al. Efficient Immune cell editing with engineered CRISPR-Cas12i for ex vivo cell therapy. Mol Ther. 2022;30(4):450. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

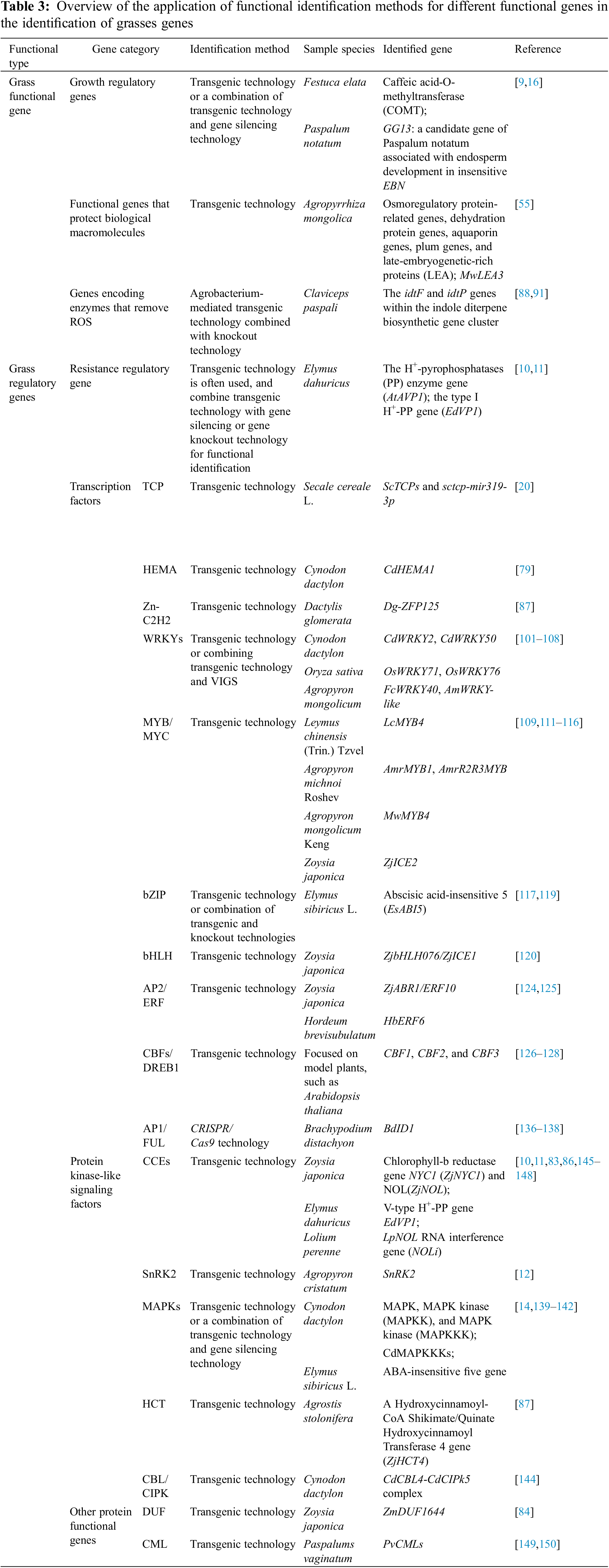
 Downloads
Downloads
 Citation Tools
Citation Tools