 Open Access
Open Access
ARTICLE
Structural Characterization of Chloroplast Genome in Alpinia japonica (Thunb.) Miq., a Medicinal Plant of the Genus Alpinia
Department of Biological Technology, Nanchang Normal University, Nanchang, 330032, China
* Corresponding Author: Quan Kuang. Email:
(This article belongs to the Special Issue: Recent Research Trends in Genetics, Genomics, and Physiology of Crop Plants)
Phyton-International Journal of Experimental Botany 2024, 93(8), 1897-1911. https://doi.org/10.32604/phyton.2024.052395
Received 01 April 2024; Accepted 28 June 2024; Issue published 30 August 2024
Abstract
The analysis of chloroplast gene characteristics in Alpinia japonica (Thunb.) Miq. is of great significance for developing relevant genetic resources. The high-throughput sequencing and bioinformatic research were performed to analyze the chloroplast genome characteristics of A. japonica. The total chloroplast genome length of A. japonica was 161,906 bp, with a typical circular tetrameric structure. And 133 genes were annotated, comprising 87 protein-coding, 38 tRNA, and 8 rRNA genes. Furthermore, 22 genes contained two copies, and 18 genes owned introns. Repeat sequence analysis showed that it contains 321 simple sequence repeats (SSRs) and 37 long segment repeats. Compared with the chloroplast genomes of eight representative plants in the genus Alpinia, the gene structure, type, and quantity were relatively conservative. Rps12 was the highest variation site in the entire chloroplast gene. A phylogenetic tree showed that the genus Alpinia was the most closely related to the genus Amomum. Meanwhile, A. japonica is the most closely related to Alpinia chinensis belonging to the genus Alpinia. Overall, the chloroplast genome of a new species was reported in the genus Alpinia, and a basis was provided for the utilization of Alpinia plants as a medical resource.Keywords
Abbreviations
| LSC | Large single copy |
| SSC | Small single copy |
| IRs, IRa and IRb | Inverted repeats |
| F | Forward repeats |
| R | Reverse repeats |
| C | Complement repeats |
| P | Palindromic repeats |
| Pi | The nucleotide diversity |
| RSCU | The relative synonymous codon usage |
| ML | Maximum likelihood |
| JLB | The IRb-LSC area |
| JSB | The IRb-SSC area |
| JSA | The IRa-SSC area |
| JLA | The IRa-LSC area |
| Met | Methionine |
| Trp | Tryptophan |
| Leu | Leucine |
Alpinia japonica (Thunb.) Miq. is a perennial herbaceous plant of the genus Alpinia in the Zingiberaceae family that has medicinal and spice use value. This plant is an important traditional Chinese medicinal resource, distributed in the southeastern and southwestern regions of China [1]. The core medicinal ingredients in the roots of A. japonica are flavonoids, which dispel dampness and swelling, regulate qi-flowing to relieve pain, and promote blood circulation to remove meridian obstruction. They are often used to treat diseases, for example, dyspepsia, rheumatoid arthritis, traumatic injury and types of tumors [2]. Therefore, it is considered as a medicinal plant with significant potential for drug development.
Chloroplasts are semi-autonomous organelles that convert light energy into chemical energy within green plant cells [3]. This chloroplast genome usually contains a tetrad circular structure, typically consisting of a large single copy (LSC) region, a small single copy (SSC) region, and two inverted repeats (IRs, IRa and IRb) regions. By comparing the chloroplast sequences of plants from the same genus, a phylogenetic tree can be built to comprehensively evaluate the phylogenetic position of the species [4]. And the relatively low rate of nucleic acid substitutions in the chloroplast genome offers a necessary condition for studying the deep-level systematic evolution of plants [5]. Furthermore, an increasing number of chloroplast sequences are being widely utilized in the systematic research of different families, orders, and even entire angiosperms [6–7]. The chloroplast genome sequencing revealed significant sequence and structural differences within and between plant species, which are valuable for understanding the adaptability of economical crops, promoting the breeding of closely related species, and identifying and protecting valuable traits [8]. By utilizing the polymorphisms of the complete chloroplast genome comparison, we have explored complex genetic relationships, which have now been transmitted to families and reached the order level [9]. The complete chloroplast sequence also provides useful information for molecular breeding and the development of DNA barcode labeling, and it has been applied in the protection and utilization of plant germplasm resources [10–11].
As a medicinal resource, A. japonica has been evaluated and utilized in the pharmaceutical, medical, and chemical fields, focusing on its cultivation techniques [12], chemical composition [13], elemental content [14], flavonoids [15], pharmacological effects [16], antibacterial functions and its bactericidal activities of A. japonica extract [17–19]. However, none of the studies have reported genomic evaluation for this species. The chloroplast genome is an important genetic information carrier source that can play a crucial role in analyzing species origins, phylogenetic relationships, genetic diversity, and species identification [20]. Therefore, this is the first to sequence, assemble, and annotate the A. japonica chloroplast genome, further analyze its chloroplast genome characteristics, and build a phylogenetic tree with published chloroplast genomes of the Alpinia and Zingiberaceae plants, thereby elucidating the evolutionary relationship between A. japonica and other Zingiberaceae species, and their position in phylogeny studies. The findings of this work present new ideas for further genetic evolution research on the Alpinia plants, as well as will be a ready reference for the utilization of germplasm resources and chloroplast genetic engineering research on A. japonica.
The individual plants were planted on the Changbei Campus of Nanchang Normal University (Nanchang, China) and were identified by Dr. Tonglin Zhang from the School of Life Sciences as A. japonica (Thunb.) Miq., belonging to the Zingiberaceae family.
2.2 Chloroplast Genome DNA Annotation
The genomic DNA of A. japonica from tender leaves was extracted with the CTAB method. After passing the detection, it was sequenced on the platform of Illumina NovaSeq 6000. Raw sequencing data were obtained through the fastp v0.20.0 software, and valid data were assembled with SPAdes v3.10.1 [21]. During the assembly process, A. pumila (NC_048462.1) was set as a reference. This study validated two types of annotation methods to revise incorrect or redundant annotation results, and establish boundaries for exons. Firstly, to improve the annotation accuracy, Prodigal v2.6.3 software was utilized for protein coding gene annotation [22], Hmmer v3.1b2 was set for rRNA gene prediction (http://www.hmmer.org/, accessed 20 May 2024, up to now), and tRNA gene prediction was performed with Aragorn v1.2.3 [23]. Then, the assembly results were validated in the NCBI database using BLAST v2.6. And the complete genome map was drawn by OGDRAW software [24].
The repetitive sequences were conducted research using two different software programs in A. japonica chloroplast genome. The MicroSAtellite identification tool was utilized to discriminate simple sequence repeats (SSRs), with parameters of repeat type and number set to 1 (10), 2 (5), 3 (4), 4 (3), 5 (3), and 6 (3). And the minimum distance between two SSRs is equal to 100 bp. If the distance was <100 bp, the two SSRs were regarded as a composite microsatellite marker [25]. Meanwhile, the REPuter software was utilized to check long repetitive sequences, comprising of complement repeats (C), forward repeats (F), palindromic repeats (P), and reverse repeats (R). The maximum number of repeating sequences was 100, and the minimum repeating size was 22 bp [26].
2.4 Comparative Genomic Studies on Chloroplasts of the Genus Alpinia
Eight reported chloroplast genomes from the same genus were downloaded from the NCBI website, including A. oxyphylla (NC:035895.1), A. hainanensis (MK262728.1), A. pumila (NC:048462.1), A. officinarum (MT254526.1), A. chinensis (NC:050165.1), A. galanga (MK058682.1), A. nigra (NC_062463.1), and A. kwangsiensis (MZ066612.1). These genomes were compared with the A. japonica chloroplast genome. The chloroplast genome structure of the nine plants were evaluated with the CGView software [27]. Mauve v2.3.1 was utilized to analyze the colinearity relationship of chloroplast sequences [28]. The gene nucleotide diversity (Pi) value was counted using DNAsp v5.0 [29]. The boundary information of IR, SSC, and LSC regions was visualized using SVG packets in Perl (https://perlmaven.com/scalable-vector-graphics-with-perl, accessed 20 May 2024). CodonW1.4.2 (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::CodonW, accessed 20 May 2024) software was utilized to statistically calculate the relative synonymous codon usage (RSCU) of A. japonica chloroplast genome. When RSCU > 1, it shows that the codon is used more frequently; when RSCU = 1, it demonstrates that the codon has no preference; and when RSCU < 1, it reveals that the frequency of codon usage is low.
2.5 Molecular Phylogeny of the Genus Alpinia
Relying on the classification system of the Zingiberaceae family in the Flora of China, this study obtained representative species of the tested genera in the Zingiberaceae family from NCBI (https://www.ncbi.nlm.nih.gov/, accessed 20 May 2024), including two species of genera Boesenbergia, Amomum, Costus, Curcuma, Hedychium, Kaempferia, Roscoea, and Zingiber, and as well as one species each of Cautleya, Globba, Pommereschea, and Rhynchanthus. The sequence alignment was performed using MAFFT [30], according to the 29 chloroplast genome sequences in the Zingiberaceae family including A. japonica. And the arrangement results were optimized using trimAl software [31]. The maximum likelihood (ML) phylogenetic tree was built using IQ tree 1.6.12, with Canna indica (MN832865.1) of the Cannaceae family as the outer group and a bootstrap value of 1000. The built-in Model Finder of IQTREE selects the optimal tree construction module based on the optimized comparison results [32].
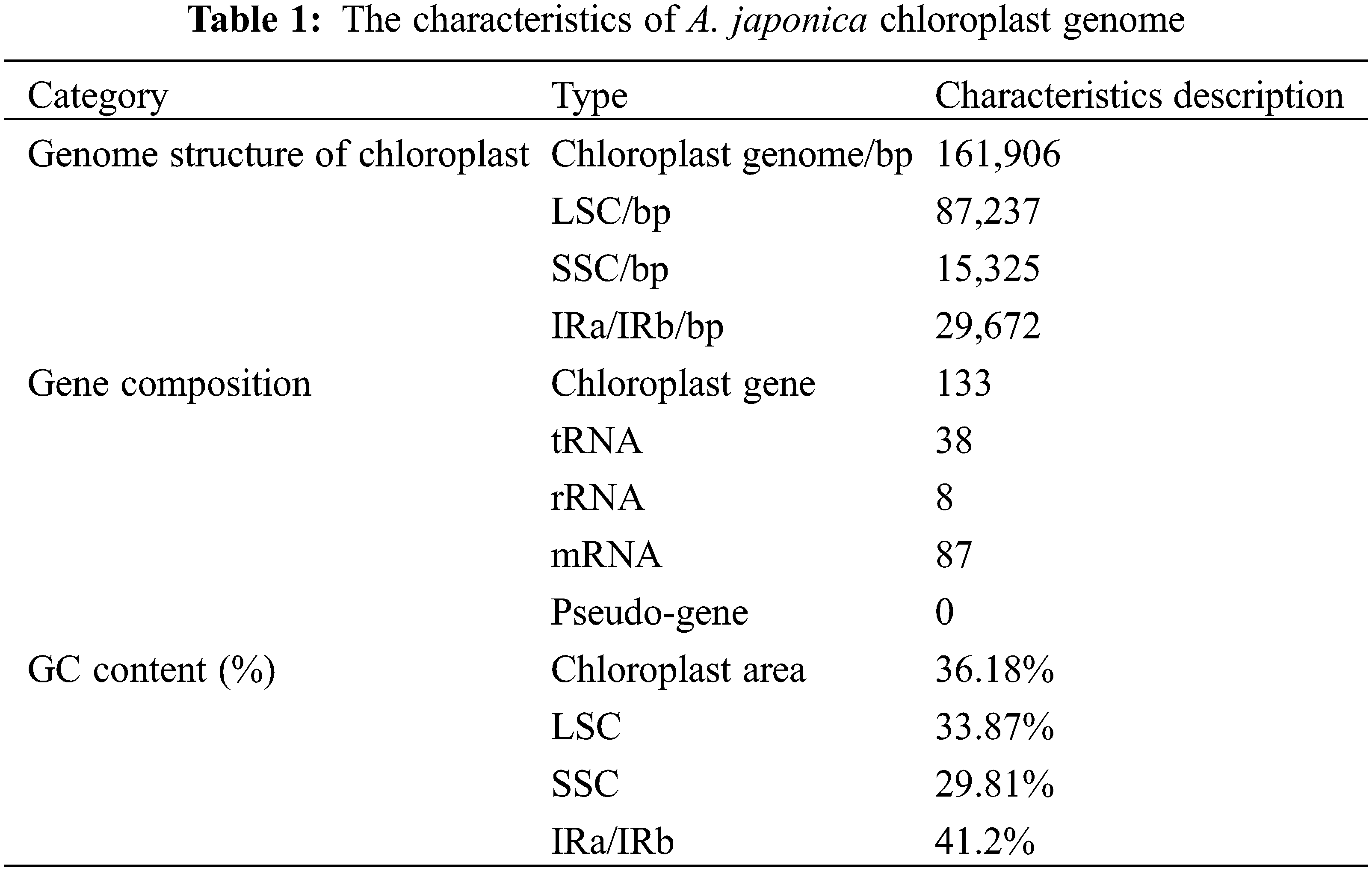
3.1 The Chloroplast Genome of A. japonica
The A. japonica chloroplast genome is structurally similar to that of other plants in the genus Alpinia [33–35]. Its chloroplast genome structure is a covalently closed circular molecule with 161,906 bp length size, and is a typical tetrad structure. The genome includes a LSC region (87,237 bp), a SSC region (15,325 bp), and two IR regions (29,672 bp each). The GC content is 36.18%, with the highest GC content (41.2%) in the IR region, 33.87% in the LSC region and 29.81% in the SSC region (Fig. 1). The complete genome consists of 133 genes, owning 38 tRNA, 8 rRNA, and 87 protein coding genes (Table 1). The rps12 gene has a trans-splicing structure, with its 5′ end located in LSC region and its 3′ end located in IR region (Fig. 1).
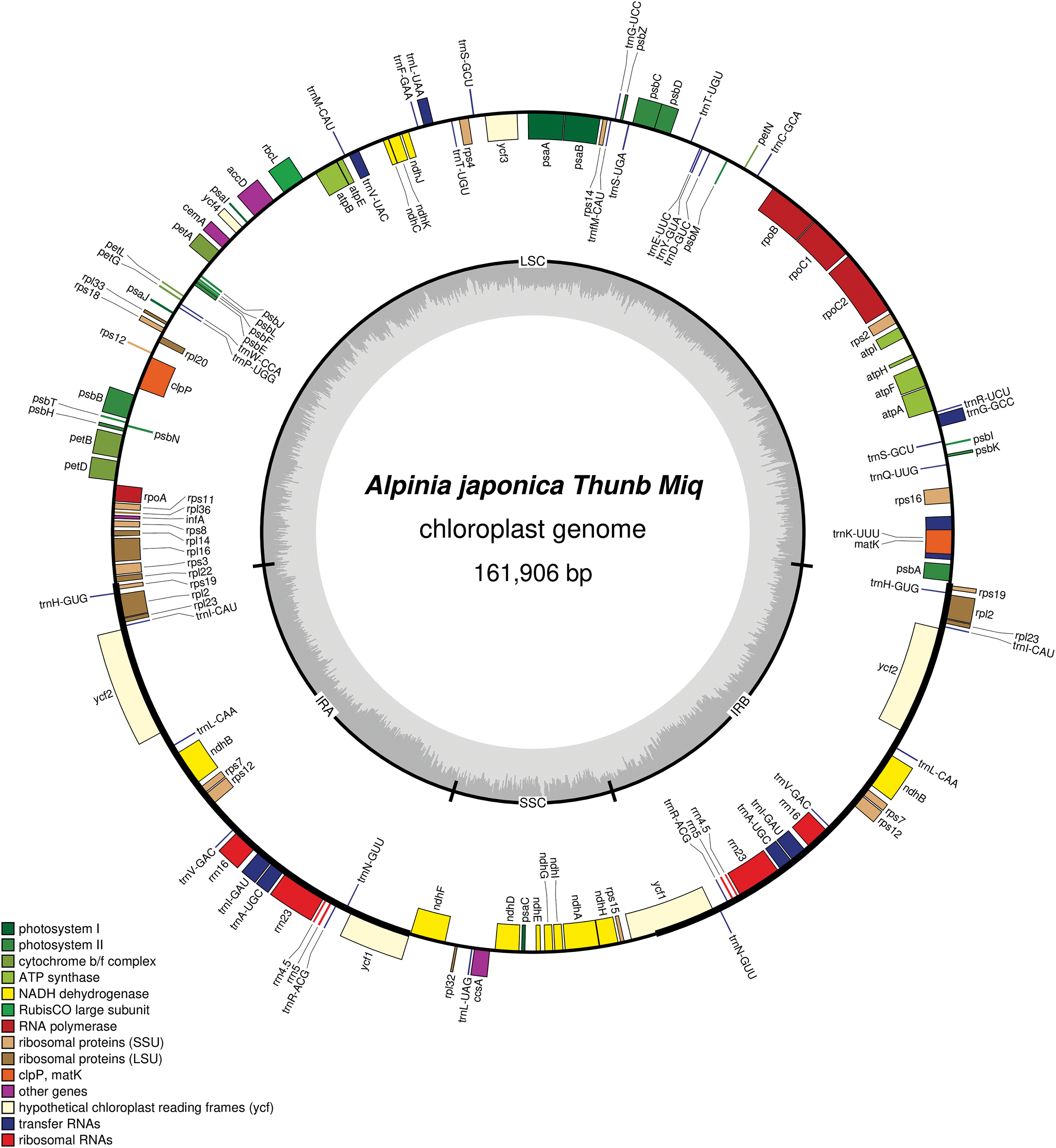
Figure 1: The chloroplast genome map of A. japonica in the genus Alpinia
3.2 Protein Coding Genes of A. japonica Chloroplast Genome
In total, 44 genes were related to photosynthesis, including five photosynthetic system I genes, 15 photosynthetic system II, six cytochrome b/f complex coding, five ATP synthase subunit, 12 NADH dehydrogenase, and one ribulose diphosphate carboxylase large subunit gene; except rRNA and tRNA genes, self-replication related genes also include 15 ribosomal small subunit, 11 ribosomal large subunit, and four DNA-dependent RNA polymerase genes; in addition, there are six other functional and five unknown functional genes (Table 2).
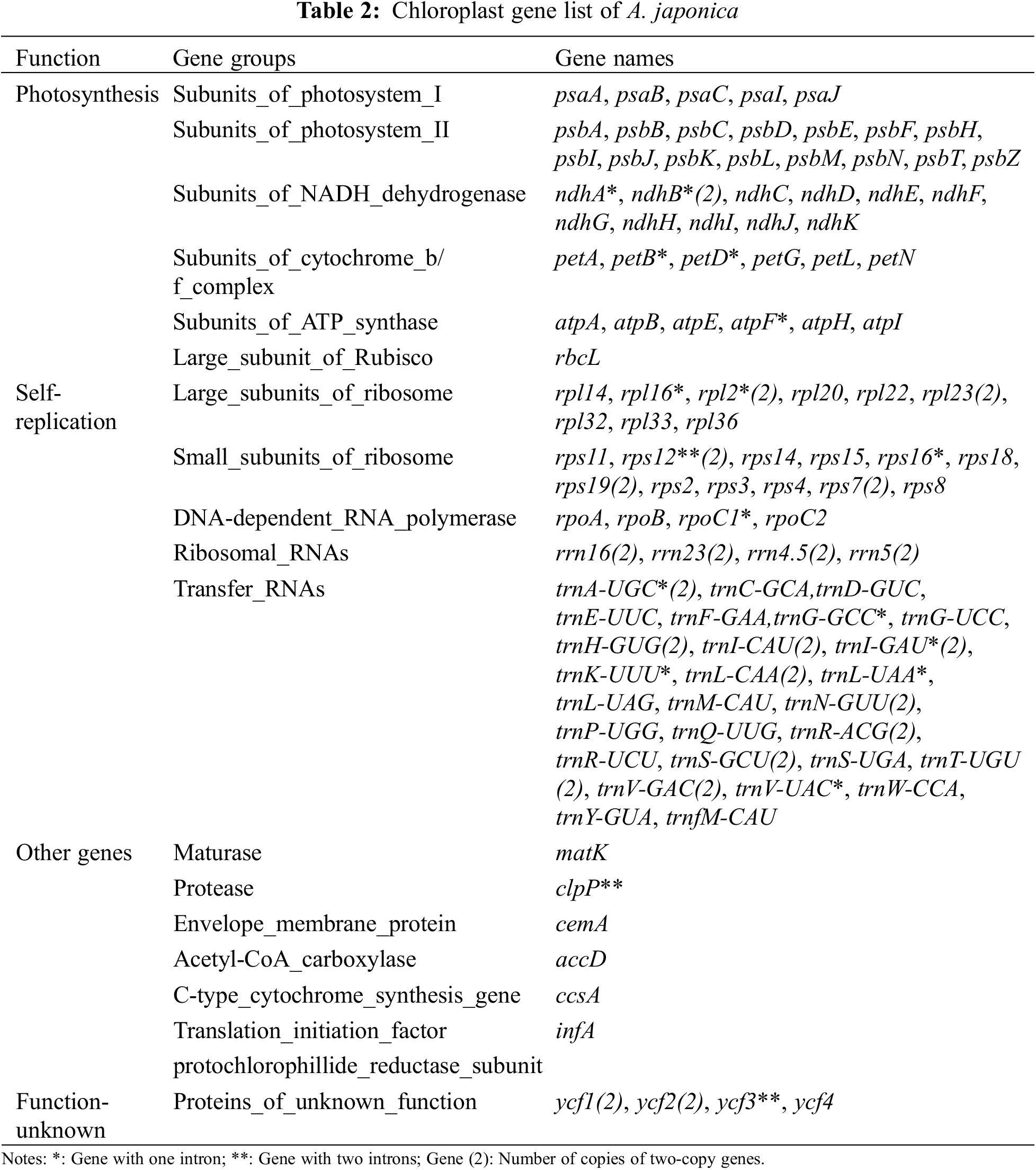
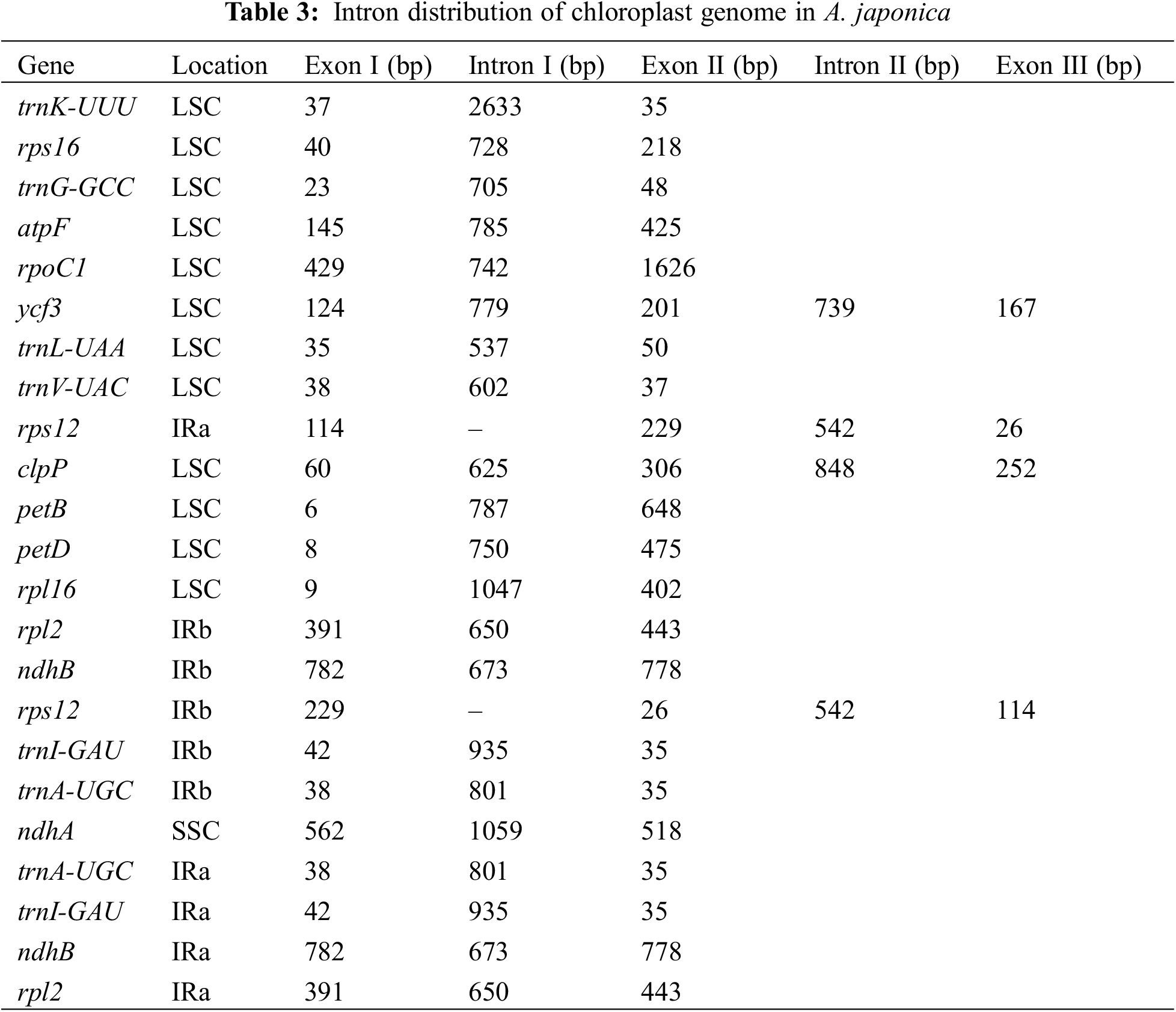
Sequence analysis showed that protein coding genes (ndhB, rpl2, rpl23, rps12, rps19, rps7, rrn16, rrn23, rrn4.5, and rrn5), tRNA genes (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnS-GCU, trnT-UGU, and trnV-GAC), and functionally unknown genes (ycf1 and ycf2) contained two copies. In addition, ndhA, ndhB, petB, petD, atpF, rpl16, rpl2, rps16, rpoC1, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-CAA, trnL-UAA, and trnV-UAC have a single intron, whereas the clpP and ycf3 genes contain two introns (Table 3). And the trnK-UUU gene contains the largest intron (2,633 bp), whereas rps12 had the smallest intron (26 bp). This research result is consistent with the reported chloroplast genomes of the Alpinia plants such as A. oxyphylla [34], A. chinensis [36], A. galanga, and A. kwangsiensis [37].
3.3 Codon Bias in A. japonica Chloroplast Genome
The RSCU values and codon usage preferences were investigated in eight species of the genus Alpinia. A heatmap was built according to the RSCU values of the eight plants (Fig. 2), and the results showed that 64 codons can be separated into two categories. The boundary line is defined by the RSCU value of GUG being zero, with a cluster greater than one on the left and a cluster less than one on the right. Codons that end with both A and T bases have a higher coding rate, except for ATA (isoleucine), CTA (leucine) and TGA (termination codon). And the RSCU of codons ending in A or T is greater than one, while the RSCU of codons ending in C or G is less than one. Although some variations have been observed, most amino acids owned two synonymous codons at least, including arginase, leucine and serine have a total of six codons. The RUSC values of tryptophan (UGG), serine (UCC), and isoleucine (AUA) are all one. According to the RSCU value, A. japonica and A. pumila form a branch, while the other six species (A. kwangsiensis, A. hainanensis, A. officinarum, A. oxyphylla, A. chinensis, A. nigra) converge into another branch.
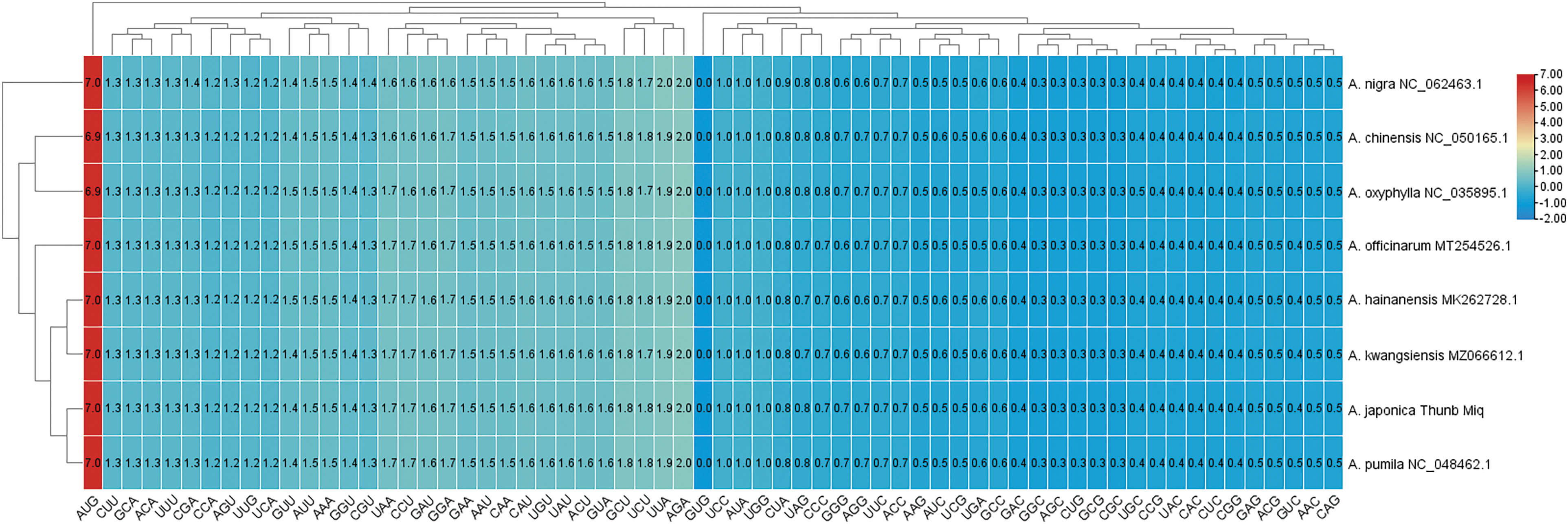
Figure 2: The heatmap of RSCU values in the eight species of the genus Alpinia chloroplast genome
3.4 Differences in Chloroplast Genome Boundaries in the Genus Alpinia
An analysis was conducted on the boundaries of the IRb-LSC (JLB), IRb-SSC (JSB), IRa-SSC (JSA), and IRa-LSC (JLA) areas of the genus Alpinia registered in the NCBI (Fig. 3). It was indicated that the genes at the boundaries were the same, and that the diversity was due to differences in the distribution positions of adjacent genes and the distances between base pairs. The eight chloroplast genomes of the genus Alpinia all contain the rpl22 gene at the JLB area, except for A. officinarum, which spans the LSC and IRb regions, whereas in other species, the length of rpl22 expansion towards the LSC region ranges from 47 to 120 bp. At the JSB area, the ycf1 gene crosses this boundary, and the length of ycf1 extension in the SSC region is between 1–90 bp. At the JSA area, there were varying degrees of rps15 gene expansion, with lengths ranging from 1979 to 2798 bp. At the boundary of the JLA area, the length of rps19 gene extension ranges from 119 to 148 bp. The existing results indicate that there is little difference in the chloroplast genome boundaries.
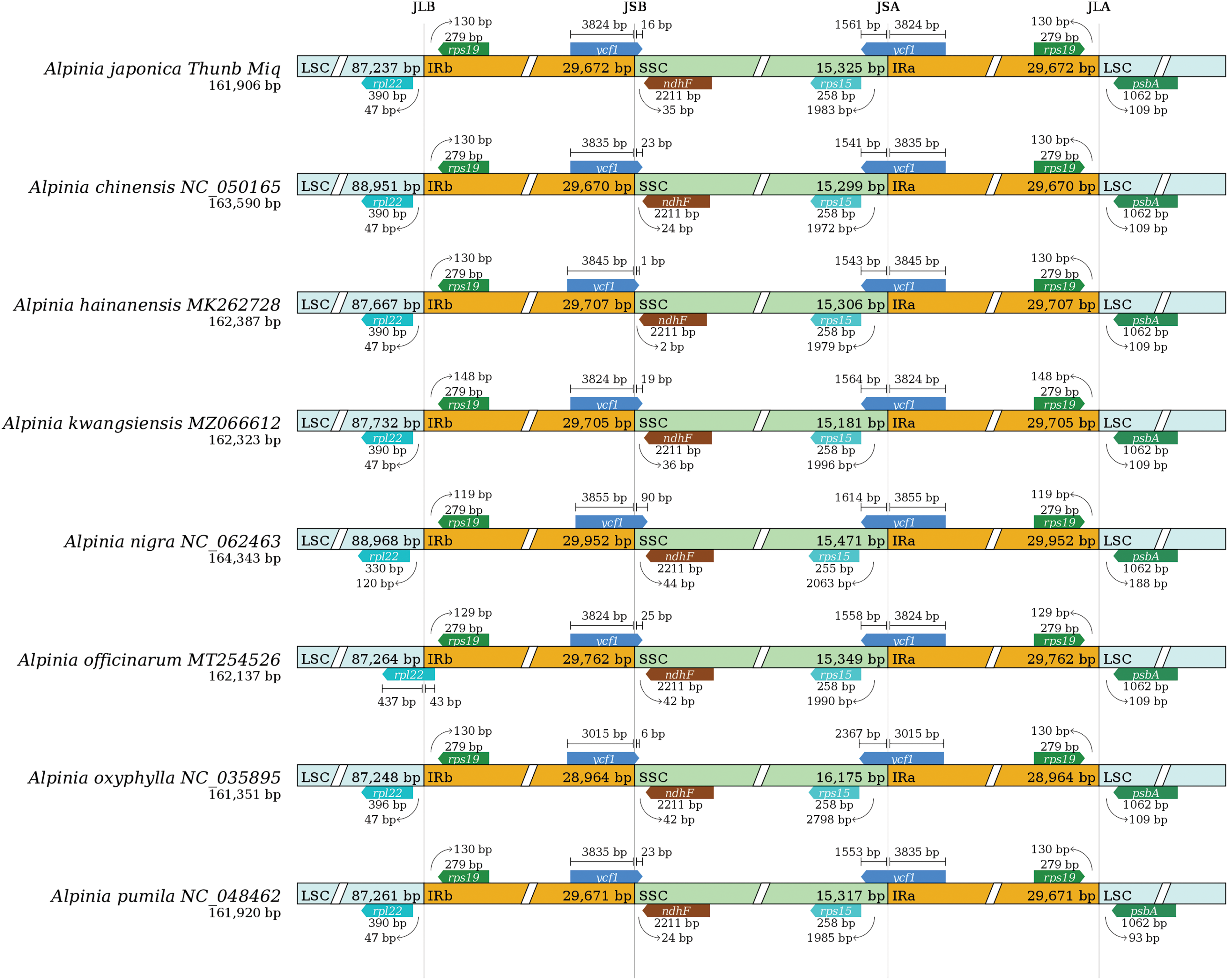
Figure 3: Boundary comparison among LSC, SSC and IR regions of chloroplast genome in the genus Alpinia
3.5 Repeat Sequences Distribution in A. japonica
Through SSR analysis of A. japonica, 321 SSRs were detected, and only four types were detected; among them, single nucleotide repeat was the most numerous type, with a quantity of 188; the number of dinucleotide and trinucleotide repetitions is 31 and 87, followed by a minimum of 15 tetranucleotide repetitions (Fig. 4). This result is similar to the SSR composition of the chloroplasts genome in the genus Alpinia plants [38]. And the A/T repeat was predominant among the single nucleotide repeat types. In the other types of repetitions, bases A and T bases also accounted for the largest proportion. In the SSR distribution area, the number detected by LSC (194) was higher than that detected by SSC (47) and IR regions (80). In the gene regions of SSR distribution, the number distribution of non-coding gene regions (164) and gene-coding regions (157) was relatively uniform.
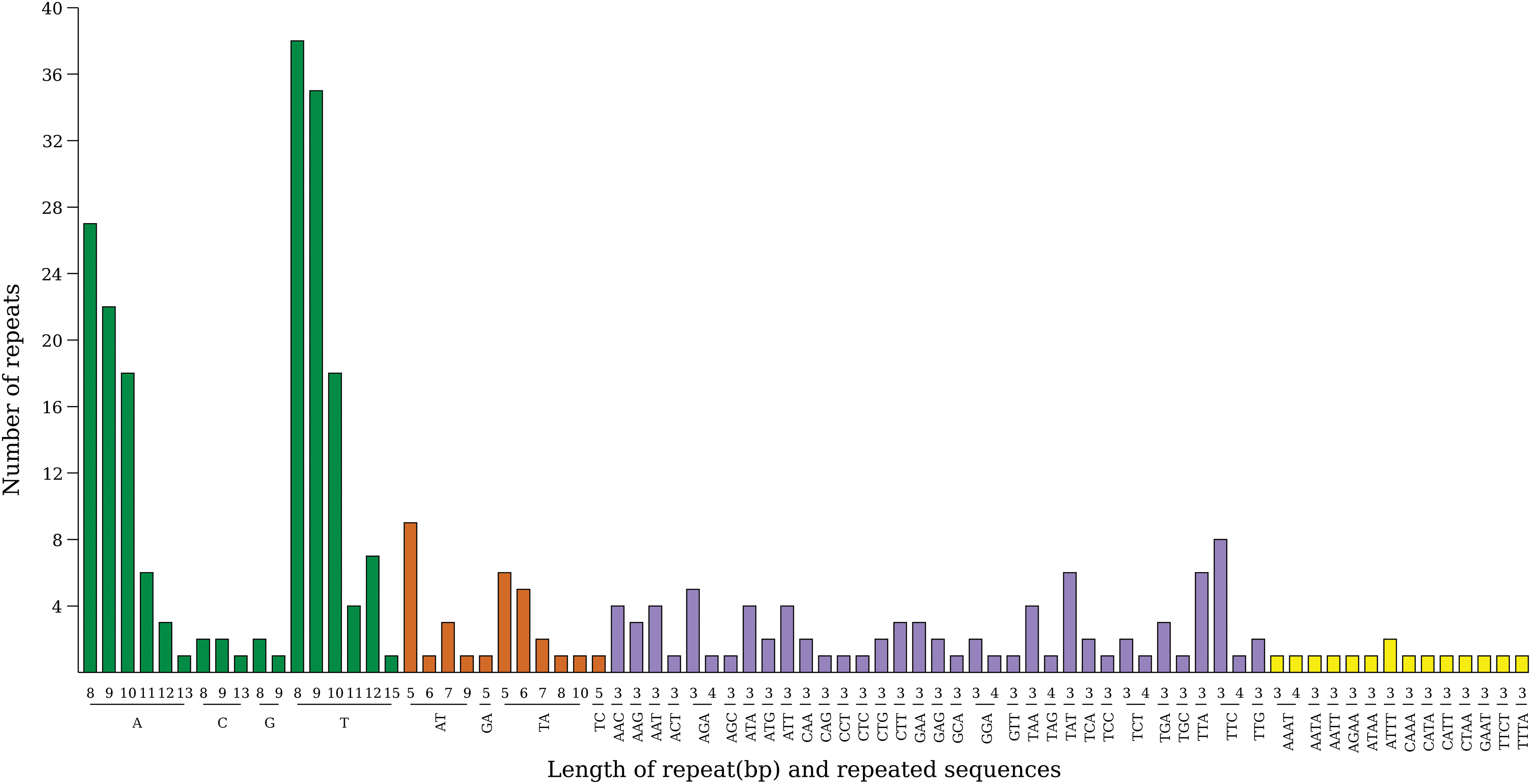
Figure 4: SSR type in the chloroplast genome of A. japonica
The chloroplast genome of A. japonica contains 37 long segment repeat sequences, including 11 F-, 25 P- and one C-type repeat (Fig. 5). In the range of repetitive sequence distribution, the genome contains a 29,672 bp long P-type sequence, whereas other types are distributed in the range of 30–53 bp.
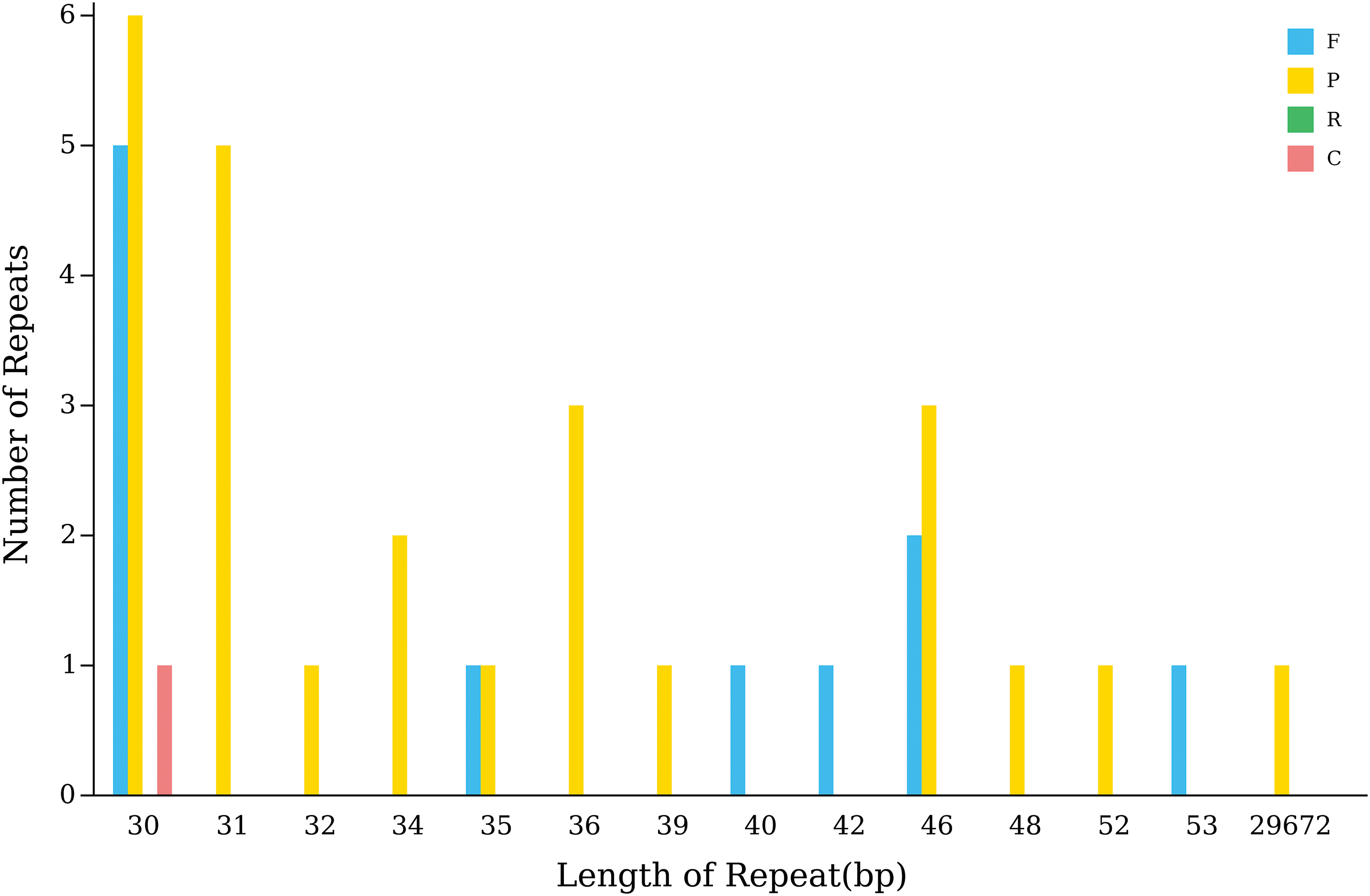
Figure 5: Long fragment repeat distribution in the A. Japonica chloroplast genome. Note: F: forward repeats; P: palindromic repeats; R: reverse repeats and complement repeats
3.6 Gene Differences in A. Japonica Chloroplast Genomes
The degree of nucleotide difference is represented by Pi value: the larger the Pi value, the greater the difference [29]. By examining the variation sites of A. Japonica, it was found that more abundant variation sites in the LSC and SSC regions than in the IR region (Fig. 6). In the chloroplast genome of A. Japonica, rps12 located in the LSC region, owned a Pi value of 0.07747, showing the greatest variation, followed by trnI-GAU, rps18, rpl36, rps3, ndhA, and rps15 with Pi values of 0.01389, 0.01044, 0.01065, 0.01011, 0.01042, and 0.01597, respectively, indicating that these are all in the mutational hot spots.
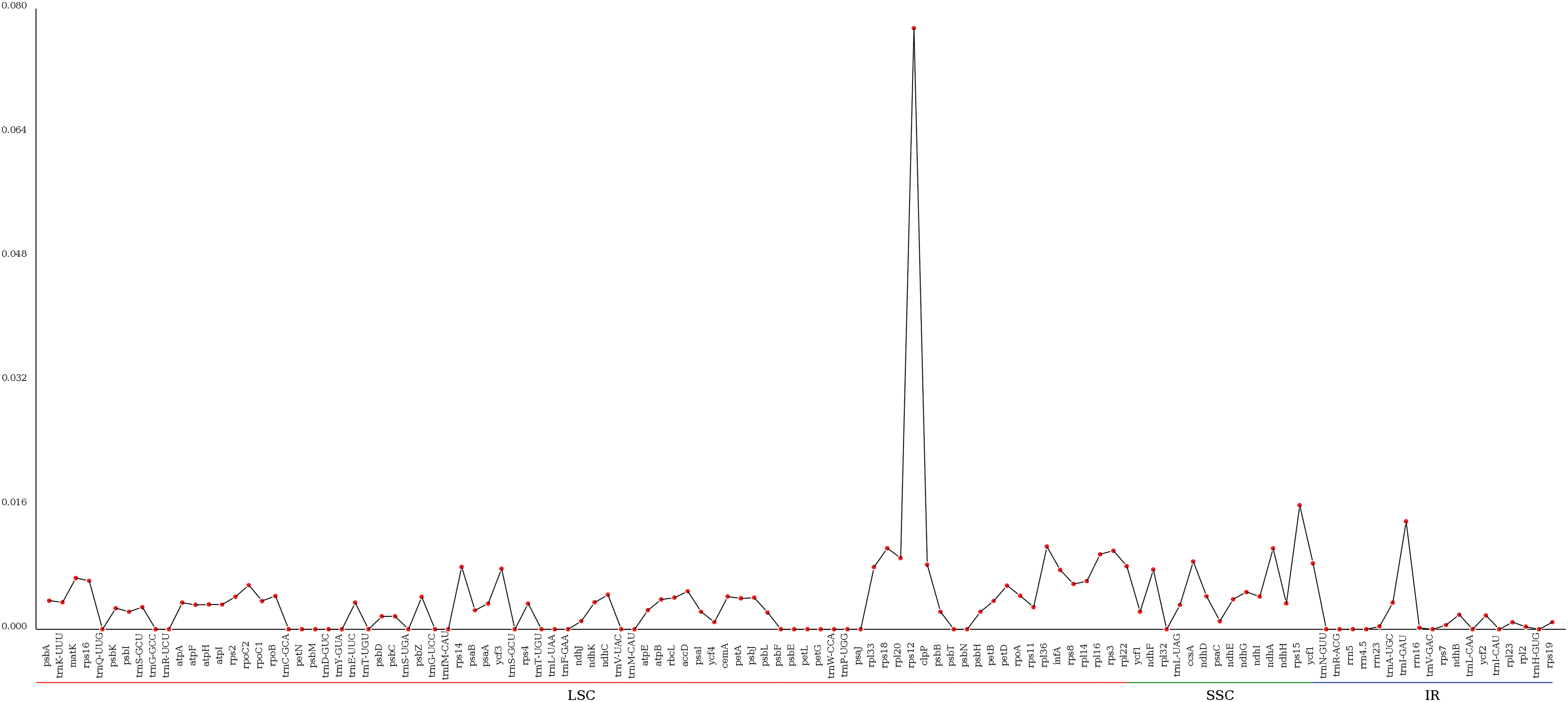
Figure 6: Divergent hot-spot nucleotide sites in Alpinia species’ chloroplast genomes
3.7 Molecular Phylogenetic Trees of Zingiberaceae Plants
Based on the entire chloroplast genome, a phylogenetic tree of 29 species of the Zingiberaceae family was constructed using Canna indica (MN832865.1) as the outer group, including eight reported species from the genus Alpinia and representative species of other genera reported in NCBI (Fig. 7). The main branch step size test value of the constructed ML phylogenetic tree was greater than 90, indicating a high level of credibility for the phylogenetic relationships in this genus. Through phylogenetic tree analysis, it can be concluded that plants of the Zingiberaceae family are fallen into three major categories. The first category is the genus Costus plants; the second type includes plants of the genera Alpinia and Amomum plants; the third category includes the genera Boesenbergia, Cautleya, Curcum, Globba, Hedychium, Kaempferia, Pommereschea, Rhynchanthus, Roscoea, and Zingiber plants. At the inter-genus level, the genus Alpinia had the closest genetic relationship with the genus Amomum. And it was also shown that the genetic relationship between A. japonica and A. chinensis was the closest.
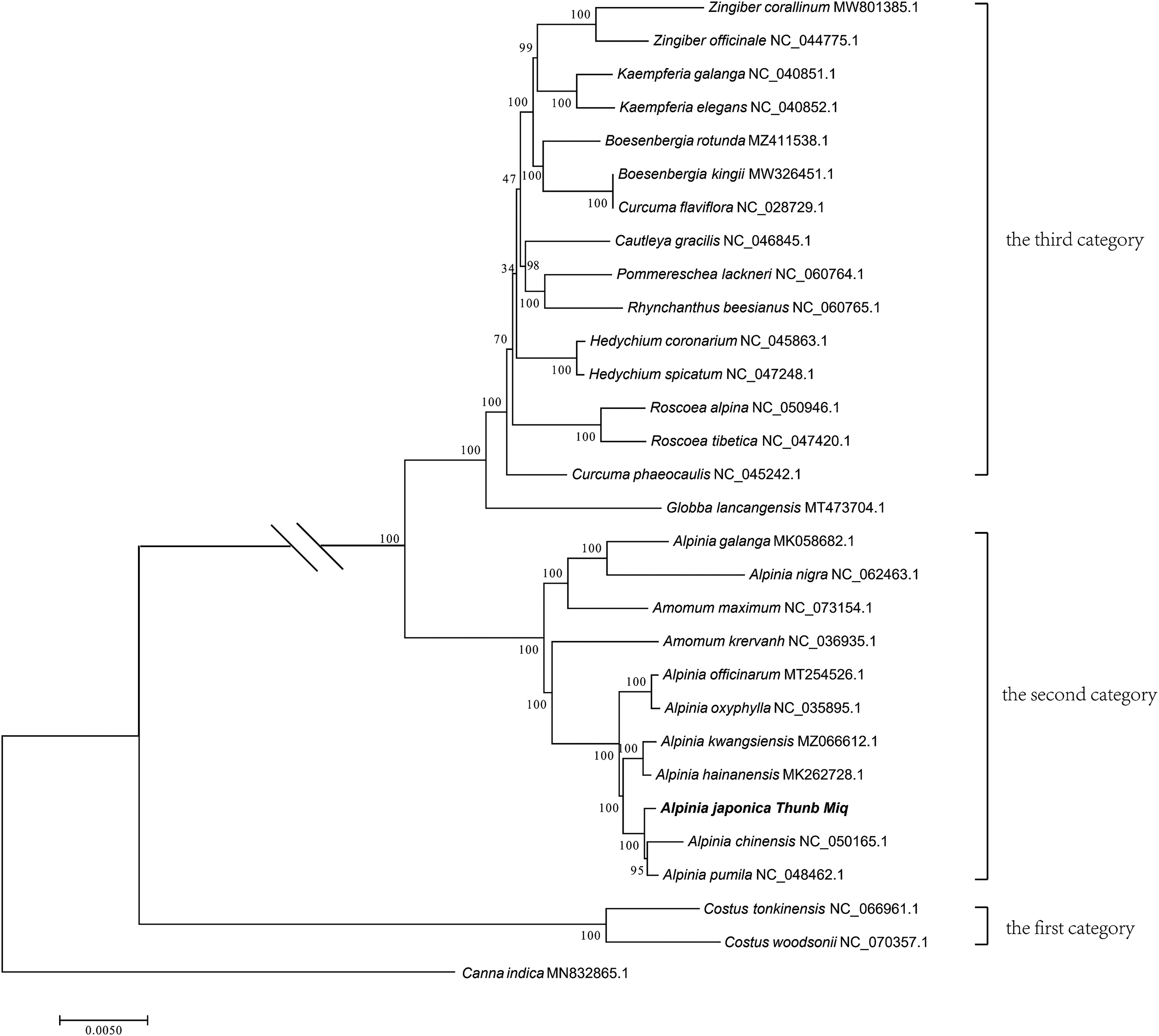
Figure 7: The phylogenetic tree of Zingiberaceae species based on whole chloroplast genome sequences
4.1 Structural Characteristics of Chloroplast Genome in the Genus Alpinia
The chloroplast genome from angiosperms is typically a circular four-part structure, with genome size ranging from 115 to 165 kb [39]. The results showed that the chloroplast genome of A. japonica exhibited a typical loop structure, with the SSC and LSC regions divided by the IR region, a size of 161,906 bp, and 133 genes, which is consistent with the common chloroplast genome characteristics of the genus Alpinia [37]. The reported chloroplast genome of Alpinia oxyphylla owns 132 genes, containing 86 protein coding, eight rRNA, and 38 tRNA genes [34]. Similarly, the chloroplast genome of A. officinarum has 132 genes, and their gene composition and structure are similar with those in A. japonica [35]. Differences in chloroplast genome size were found only in non-coding gene sequences in the genus Alpinia, and the types and quantities of coding genes were consistent, indicating the conservation of genome structure and composition in this genus. Characterization of the structural boundaries within the genome of the genus Alpinia revealed marginal difference in the LSC, SSC, and IR boundaries, which is similar to the chloroplast genomes in this family [7].
In the chloroplast SSR types of the genus Alpinia, A/T types are widely distributed and have high AT content, which further confirms that SSRs in plant chloroplast genomes are often comprised of poly A or poly T repeats [38]. Furthermore, we found that in this species, compared to other plants, this genus of plants contained fewer repetitive types, exhibiting species specificity. Codon usage bias is an important genome evolutionary feature [6], and the RSCU value is often made as an important codon preference indicator [9]. Among the 20 amino acids present in nature, which are encoded by unique codons, all other amino acids correspond to 2–6 synonymous codons except for tryptophan (Trp) and methionine (Met). Owing to frequency differences of synonymous codons used, the frequency of plant codons varies, exhibiting certain preferences. This is the result of multiple factors, among which mutations and natural selection are the two main factors, working together [34–35]. In this study, codon preference analysis indicated that leucine (Leu) had the amino acid with the highest proportion in the A. japonica chloroplast genome, and all 30 codons with RSCU > 1 ended in A/U, which is similar to previously published Alpinia plants [37]. This may be because of the abundance of A/U bases in the chloroplast genome of this plant.
4.2 Polymorphism of Chloroplast Genome in the Genus Alpinia
Compared to nuclear genes, chloroplast genomes contain more diverse variation sites throughout the genome, resulting in higher efficiency in species identification. The chloroplast sequence provides valuable genetic information for identifying species, analyzing species genetic diversity, elucidating phylogenetic relationships, and genetic engineering [9]. Many Alpinia species are edible, and they also have medicinal, horticultural, and aesthetic value. Notably, many plants have been cultivated in many varieties, leading to difficulties in species identification and classification. Zhong et al. [40] has identified the polymorphic site ycf1 gene through chloroplast genome research, but could not distinguish the research groups. This study identified polymorphic loci such as trnI-GAU, rps18, rpl36, rps3, ndhA, and rps15, with rps12 showing the highest variation. The most polymorphic loci identified in A. oxyphylla were rpl16, rpoC1, rps8, and ycf2, whereas in A. oxyphylla, ndhB and ndhC, photosystem II (psbZ), and ATP synthesis (atpE and atpF) were identified, indicating their species specificity. Because of the polymorphic loci already obtained for the species belonging to this genus, the plant groups of this genus can be expanded for locus validation, and these differential sequences can be used to carry out collaborative research.
4.3 Phylogenetic Relationships of A. japonica
As the most widely distributed genus in Zingiberaceae, the genus Alpinia includes up to 230 species. Its famous plant species include A. officinarum, A. oxyphylla, A. katsumadai, A. zerumbet, A. galanga, A. chinensis, A. officinarum and A. japonica, with a distribution ranges spanning both tropical and subtropical regions. Given the frequent occurrence of Chinese medicine adulteration problems among the plants of the genus Alpinia, it is necessary to clarify out its phylogenetic relationships. According to [41], this genus has the following four subgenera: Alpinia, Catimbium, Dielamalpinia, and Probolocalix [41]. Smith [42] and Kress et al. [43] suggested that the genus Alpinia is a complex polyphyletic group that needs revision. Qiao et al. [44] studied the species of the genus Alpinia using ITS and the single chloroplast gene matK, but the classification results did not support Smith’s [42] revision of the classification of this genus, indicating the need to develop new molecular markers for classification research of this species. Zhang et al. [37] conducted a chloroplast genome sequencing analysis for A. galanga and A. kwangsiensis, and also believed that the Alpinia species were not monophyletic genera. Mei et al. [36] and Li et al. [38] conducted a phylogenetic analysis of the entire sequence matrix of Alpinia species and discriminated the phylogenetic relationships of some Alpinia species.
However, the inconsistency in the above classification results may be due to the inconsistent evolutionary rates of different sequences such as ITS, chloroplast genes, and nuclear gene sequences, resulting in different phylogenetic tree relationships [39]. The entire chloroplast genome was analyzed in this research and the genus Alpinia was found to have the closest phylogenetic relationship with Amomum, whereas in the genus Alpinia, A. japonica has the closest phylogenetic relationship with the plant A. chinensis. Therefore, to obtain a more scientific and accurate phylogenetic relationship for this genus, we should continue to expand the high-throughput sequencing of the genome of Alpinia. In the next step of this research, we will use the sequence loci that have obtained high mutation hotspots to conduct out genetic research of this genus resource.
In conclusion, the A. japonica chloroplast genome was assembled and compared for the first time, analyzing not only the genetic characteristics, structural variations, genetic diversity, and phylogenetic relationships, but also the genetic resources of the genus Alpinia. Our findings provide a ready reference for subsequent research on the genetic variation, population evolution, and genetic breeding of this genus.
Acknowledgement: This research was supported by the National Natural Science Foundation of China, the Natural Science Foundation of Jiangxi, the Special Program of Science and Technology Cooperation of Jiangxi Provincial Department of Science and Technology, the Education Reform Program of Jiangxi Provincial Department of Education, Jiangxi Province Key Laboratory of Oil Crops Biology and Education Reform Program of Nanchang Normal University.
Funding Statement: The Natural Science Foundation of China (32060078), the Natural Science Foundation of Jiangxi (20171BAB214024, 20202BABL 203044), the Special Program of Science and Technology Cooperation of Jiangxi Provincial Department of Science and Technology (20212BDH81022), The Education Reform Program of Jiangxi Provincial Department of Education (JXJG-22-23-3, JXJG-23-23-5), the “Biology and Medicine” Discipline Construction Project of Nanchang Normal University (100/20149), Jiangxi Province Key Laboratory of Oil Crops Biology (YLKFKT202203) and Education Reform Program of Nanchang Normal University (NSJG-21-25).
Author Contributions: Wentao Sheng, Xi Lei, Xinjie Chen, and Quan Kuang wrote original draft preparation. Wentao Sheng conducted data collection and analysis. Wentao Sheng and Quan Kuang wrote review, and supervised funding acquisition. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All the data are included in this article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Wu TL. Phylogeography of the Zingiberaceae. J Trop Subtrop Bot. 1994;2(2):1–14 (In Chinese). [Google Scholar]
2. Qin HZ, He RK, Dai QL, Yin Y, Chen JQ. Research progress of flavonoids in Chinese medicine of Alpinia. J Tradit Chin Med. 2022;35(1):145–8 (In Chinese). [Google Scholar]
3. Chen H, Li T, Chen X, Qu T, Zheng X, Luo J, et al. Insights into comparative genomics, structural features, and phylogenetic relationship of species from Eurasian Aster and its related genera (Asteraceae: Astereae) based on complete chloroplast genome. Front plant sci. 2024;15:1367132. doi:10.3389/fpls.2024.1367132. [Google Scholar] [PubMed] [CrossRef]
4. Sloan DB, Triant DA, Forrester NJ, Bergner LM, Wu M, Taylor DR. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae). Mol Phylogen Evol. 2014;72:82–9. doi:10.1016/j.ympev.2013.12.004. [Google Scholar] [PubMed] [CrossRef]
5. Nazareno AG, Carlsen M, Lohmann LG. Complete chloroplast genome of Tanaecium tetragonolobum: the first Bignoniaceae plastome. PLoS One. 2015;10(6):e0129930. doi:10.1371/journal.pone.0129930. [Google Scholar] [PubMed] [CrossRef]
6. Li DM, Li J, Wang DR, Xu YC, Zhu GF. Molecular evolution of chloroplast genomes in subfamily Zingiberoideae (Zingiberaceae). BMC Plant Biol. 2021;21(1):558. doi:10.1186/s12870-021-03315-9. [Google Scholar] [PubMed] [CrossRef]
7. Liang H, Zhang Y, Deng JB, Gao G, Ding CB, Zhang L, et al. The complete chloroplast genome sequences of 14 Curcuma species: insights into genome evolution and phylogenetic relationships within Zingiberales. Front Genet. 2020;11:802. doi:10.3389/fgene.2020.00802. [Google Scholar] [PubMed] [CrossRef]
8. Kathriarachchi H, Hoffmann P, Samuel R, Wurdack KJ, Chase MW. Molecular phylogenetics of Phyllanthaceae inferred from five genes (plastid atpB, matK, 3’ndhF, rbcL, and nuclear PHYC). Mol Phylogen Evol. 2005;36(1):112–34. doi:10.1016/j.ympev.2004.12.002. [Google Scholar] [PubMed] [CrossRef]
9. Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17(1):134. doi:10.1186/s13059-016-1004-2. [Google Scholar] [PubMed] [CrossRef]
10. Liu CK, Deng JJ, Zhou RX, Song BN, Zhou SD, He XJ. Plastid phylogenomics provide evidence to accept two new members of Ligusticopsis (Apiaceae, Angiosperms). Int J Mol Sci. 2023;24:382. [Google Scholar]
11. Abdullah MF, Rahim A, Heidari P, Ahmed I, Poczai P. Comparative plastome analysis of Blumea, with implications for genome evolution and phylogeny of Asteroideae. Ecol. 2021;11(12):7810–26. [Google Scholar]
12. Zhan RT, Liu JM, Chen LK, Han ZZ, Lu YY, Chen WD, et al. Investigation on the production status of Lingnan herbal medicine in Guangdong Province. J Guangzhou Univ Tradit Chin Med. 2020;37(9):1836–43 (In Chinese). [Google Scholar]
13. Ai W, Duan YZ, Li Y, Zhao JS. Trace elements and nutritional components of Alpinia Chinensis from different producing areas of Dehong. J Baoshan Univ. 2018;37(2):7–9 (In Chinese). [Google Scholar]
14. Jiang YF, Luo L, Lin P, Liu LJ, Hang HX. Determination of trace elements in three gingers. J Mianyang Nor Univ. 2012;31(2):46–50 (In Chinese). [Google Scholar]
15. Zhang WJ, Luo JG, Kong LY. The genus Alpinia: a review of its phytochemistry and pharmacology. World J Tradit Chin Med. 2016;2(1):26–41 (In Chinese). doi:10.15806/j.issn.2311-8571.2015.0026. [Google Scholar] [CrossRef]
16. Fan XJ, Chen J, Zhang YJ, Wang SY, Zhong WQ, Yuan HP, et al. Alpinetin promotes hair regeneration via activating hair follicle stem cells. Chin Med. 2022;17:63 (In Chinese). doi:10.1186/s13020-022-00619-2. [Google Scholar] [PubMed] [CrossRef]
17. Wang ZR, Wu MJ, Xu R, Xu TJ, Xiang L, Liang YM, et al. Preliminary evaluation on the safety of aqueous extract of the Alpinia japonica rhizome in Tujia medicine. J Jishou Univ (Nat Sci Ed). 2022;43(5):14–9 (In Chinese). [Google Scholar]
18. Wu JL, Xiong L, Luo L, Gao YZ. Effects of alpinetin on proliferation, invasion and inflammation response of pancreatic cancer cells BXPC-3 by mediating NF-κB signaling pathway. Chin Clin Oncol. 2023;28(5):405 (In Chinese). [Google Scholar]
19. Jiang C, Yang BW, Tian T. Effects of alpinetin on lipopolysaccharids-induced colon NCM460 cell damage and VDR/Nrf2/HO-1 pathway. Drugs & Clinic. 2023;38(6):1289–96 (In Chinese). [Google Scholar]
20. Robles P, Quesada V. Organelle genetics in plants. Int J Mol Sci. 2021;22:2104. doi:10.3390/ijms22042104. [Google Scholar] [PubMed] [CrossRef]
21. Bankevich A, Nurk S, Antipov D, Gurevich AA, Alexander MD, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. doi:10.1089/cmb.2012.0021. [Google Scholar] [PubMed] [CrossRef]
22. Hyatt D, Chen GL, Locasico PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi:10.1186/1471-2105-11-119. [Google Scholar] [PubMed] [CrossRef]
23. Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–6. doi:10.1093/nar/gkh152. [Google Scholar] [PubMed] [CrossRef]
24. Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–5. doi:10.1093/bioinformatics/bth352. [Google Scholar] [PubMed] [CrossRef]
25. Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–5. doi:10.1093/bioinformatics/btx198. [Google Scholar] [PubMed] [CrossRef]
26. Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–42. doi:10.1093/nar/29.22.4633. [Google Scholar] [PubMed] [CrossRef]
27. Stothard P, Grant JR, Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform. 2019;20(4):1576–82. doi:10.1093/bib/bbx081. [Google Scholar] [PubMed] [CrossRef]
28. Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–403. doi:10.1101/gr.2289704. [Google Scholar] [PubMed] [CrossRef]
29. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. doi:10.1093/bioinformatics/btp187. [Google Scholar] [PubMed] [CrossRef]
30. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. doi:10.1093/molbev/mst010. [Google Scholar] [PubMed] [CrossRef]
31. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. doi:10.1093/bioinformatics/btp348. [Google Scholar] [PubMed] [CrossRef]
32. Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74. doi:10.1093/molbev/msu300. [Google Scholar] [PubMed] [CrossRef]
33. Craig FB, Jerrold ID, Leebens-Mack Jim, John GC, Dennis WS. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Ladistics. 2012;29(1):65–87. [Google Scholar]
34. Gao BM, Yuan L, Tang TL, Hou J, Pan K, Wei N. The complete chloroplast genome sequence of Alpinia oxyphylla Miq. and comparison analysis within the Zingiberaceae family. PLoS One. 2019;14(6):e218817. [Google Scholar]
35. Huang QL. Complete sequencing and analysis of chloroplast genome from Alpinia officinarum Hance. Chin J Trop Crop. 2021;42(1):1–6 (In Chinese). [Google Scholar]
36. Mei Y, Cai SK, Xu SQ, Gu Y, Zhou F, Wang JH. The complete chloroplast genome sequence of medicinal plant Alpinia chinensis (Retz.) Rosc. Mitochondrial DNA Part B. 2020;5(3):3328–9. doi:10.1080/23802359.2020.1817805. [Google Scholar] [PubMed] [CrossRef]
37. Zhang Y, Song MF, Li Y, Zhang LX. Complete chloroplast genome analysis of two important medicinal Alpinia species: Alpinia galanga and Alpinia kwangsiensis. Front Plant Sci. 2021;12:705892. doi:10.3389/fpls.2021.705892. [Google Scholar] [PubMed] [CrossRef]
38. Li DM, Zhu GF, Xu YC, Ye YJ, Liu JM. Complete chloroplast genomes of three medicinal Alpinia species: genome organization, comparative analyses and phylogenetic relationships in family Zingiberaceae. Plants. 2020;9:286. doi:10.3390/plants9020286. [Google Scholar] [PubMed] [CrossRef]
39. Daniell H, Jin SX, Zhu XG, Gitzendanner MA, Soltis DE, Soltis PS. Green giant-a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol J. 2021;19:430–47. doi:10.1111/pbi.v19.3. [Google Scholar] [CrossRef]
40. Zhong ZM, Lai XP, Huang S, Zhang GF. Identification and clustering analysis of Zingiberaceae plant based on ycf1 barcode. China J Tradit Chin Med Pharm. 2018;33:4089–92. [Google Scholar]
41. Encyclopedia of Plants. Flora of China. Beijing, China: Science Press Publisher; 1981. vol. 16, no. 2, p. 92 (In Chinese). [Google Scholar]
42. Smith RM. ALPINIA (ZINGIBERACEAEa proposed new infrageneric classification. Redinb J Bot. 1990;47(1):1–75. [Google Scholar]
43. Kress WJ, Liu AZ, Newman MF, Mark N, Li QJ. The molecular phylogeny of Alpinia (Zingiberaceaea complex and polyphyletic genus of gingers. Am J Bot. 2005;92:167–78. doi:10.3732/ajb.92.1.167. [Google Scholar] [PubMed] [CrossRef]
44. Qiao CF, Han QB, Zhao ZL. Sequence analysis based on ITS1 region of nuclear ribosomal DNA of Amomum villosum and ten species of Alpinia. J Food Drug Anal. 2009;17(2):142–5. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools