 Open Access
Open Access
ARTICLE
Identification of Mulberry Bacterial Blight Caused by Klebsiella oxytoca in Bazhong, Sichuan, China
1
School of Life Sciences, Anhui Agricultural University, Hefei, 230036, China
2
Institute of Sericulture and Tea, Zhejiang Academy of Agricultural Sciences, Hangzhou, 310021, China
3
School of Life Sciences, Anqing Normal University, Anqing, 246133, China
* Corresponding Authors: Junshan Gao. Email: ; Zilong Xu. Email:
Phyton-International Journal of Experimental Botany 2024, 93(8), 1995-2008. https://doi.org/10.32604/phyton.2024.052353
Received 30 March 2024; Accepted 15 July 2024; Issue published 30 August 2024
Abstract
To provide a scientific basis for controlling mulberry bacterial blight in Bazhong, Sichuan, China (BSC), this study aimed to isolate and purify pathogenic bacteria from diseased branches of mulberry trees in the region and to clarify their taxonomic status using morphological observation, physiological and biochemical detection, molecular-level identification, and the construction of a phylogenetic tree. A total of 218 bacterial strains were isolated from samples of diseased mulberry branches. Of these, 7 strains were identified as pathogenic bacteria based on pathogenicity tests conducted in accordance with Koch’s postulates. Preliminary findings from the analysis of the 16S rRNA sequence indicated that the 7 pathogenic bacteria are members of Klebsiella spp. Morphological observation revealed that the pathogenic bacteria were oval-shaped and had capsules but no spores. They could secrete pectinase, cellulase, and protease and were able to utilize D-glucose, D-mannose, D-maltose, and D-Cellobiose. The 7 strains of pathogenic bacteria exhibited the highest homology with Klebsiella oxytoca. This study identifies Klebsiella oxytoca as the causative agent of mulberry bacterial blight in BSC, laying the foundation for the prevention and control of this pathogen and further investigation into its pathogenic mechanism.Keywords
Supplementary Material
Supplementary Material FileMulberry (Morus alba L.) is a perennial woody economic plant belonging to the family Moraceae [1]. It has diverse commercial uses, including traditional sericulture as well as food, medicine, and ecological benefits [2–4]. Mulberry is extensively utilized in the pharmaceutical, food, and building processing industries [5]. In China, mulberry has been cultivated for sericulture for 5000 years and is widely distributed across various geographical regions [6]. Sichuan, located in the southwest of China, is one of the main mulberry planting areas and boasts abundant mulberry germplasm resources [7]. Recently, mulberry bacterial blight has spread rapidly in the mulberry-growing region of Bazhong, Sichuan, China (BSC). The leaves become rotten and blackened, and the top of the mulberry bud wilts, resembling a “rotten head”. Moreover, the upper leaves and stem portions of the mulberry trees wilt and turn black. According to the symptoms of the disease, the disease is initially predicated as black bacterial blight. Bacterial blight is one of the most complex and devastating plant diseases. It commonly infects a wide range of plant communities, leading to significant yield losses in a variety of economically important crops [8,9]. Therefore, it is essential to identify the pathogen, explore biological characteristics, and implement control measures.
Emerging information reveals that the mulberry tree faces multiple threats, ranging from plant pathogens to pests throughout its life cycle [10], with mulberry bacterial disease being one of the most serious. The pathogen Pseudomonas syringae pv. Mori was first identified in France [11]. In 2008, mulberry bacterial blight caused by Enterobacter cloacae was reported in Hangzhou mulberry orchards in Zhejiang, China [12]. From the main silkworm areas in Guangdong and Guangxi provinces of China, 47 samples of mulberry wilt strains were collected, and it was deduced that the 10 selected strains belonged to three genera of Enterobacter, Klebsiella, and Pantoea of the family Enterobacteriaceae [13]. Similarly, Meng et al. collected mulberry tree samples with typical wilt symptoms and soil samples from nine counties and cities in Guangxi, China. They obtained 105 pathogenic strains through isolation and purification, analyzed 48 representative pathogenic strains, and eventually identified them as Enterobacter cloacae (16 strains), E. asburise (13 strains), E. mori (7 strains), Klebsiella oxytoca (3 strains), as well as unidentified species of Enterobacter spp. (3 strains), Klebsiella spp. (6 strains) [14]. Recently, Luo et al. isolated Klebsiella michiganensis from diseased mulberry in Guangxi, China [15].
The above research progress not only highlights the complexity and diversity of the pathogenic bacteria causing mulberry wilt disease but also indicates that these pathogenic bacteria can vary by region. Therefore, we specifically collected diseased branches in BSC for the identification and characteristic analysis of pathogenic bacteria, which is helpful for farmers to accurately diagnose the disease and take measures. The pathogenic bacteria causing mulberry bacterial blight in BSC were identified using physiological and biochemical assays, 16S rRNA molecular-level identification, and phylogenetic tree analysis. This provides a theoretical foundation for the prevention and control of pathogenic bacteria.
The plant materials used in this experiment consisted of 20 diseased branches of “Qiang Sang No. 1”, collected from Enyang District, Bazhong City, Sichuan Province, China (106.4864°E, 31.8168°N). These branches were stored in ice boxes and transported to the laboratory for the isolation of pathogenic bacteria.
2.2 Isolation and Purification of Pathogenic Bacteria
The stem segments of the diseased mulberry branches were first cleaned with sterile water and then placed on a super-clean worktable to dry. Subsequently, the diseased branches were cut into small pieces, each approximately 1 cm in size, and the outer epidermis of the stems was peeled off. The yellow or brown parts of the xylem were ground in a sterilized mortar, adding 1 mL of sterile water for each grinding. The supernatant obtained after grinding was streaked onto an LB (Luria-Bertani) solid medium and incubated in the dark at 30°C for 12 h. A representative single strain from the dominant flora was selected and inoculated into LB liquid medium. The single strain was then incubated overnight at 30°C in a shaker. The strains were stored in a refrigerator at −80°C.
2.3 Morphological Observation of Pathogenic Bacteria
The pathogenic bacteria were stained using a four-step Gram staining procedure [16], which was recorded and photographed. The bacterial suspension of the tested strain was dropped onto a copper carrier mesh, and excess sample was absorbed using filter paper, resulting in the formation of a water film layer on the copper carrier mesh. While the water film was still wet, a drop of 2% phosphotungstic acid staining solution was added to mesh. Any excess staining solution was then absorbed using filter paper, and the mesh was allowed to air-dry naturally at room temperature. Subsequently, the dried mesh was transferred to a HitachiH7650 transmission electron microscope for examination. The morphology of the pathogenic bacteria was photographed and recorded at an accelerating voltage of 80 kV.
2.4 Pathogenicity of Pathogenic Bacteria
To test for pathogenicity, a bacterial suspension (OD600 = 0.5) was prepared. The pinprick method was used for artificial inoculation [17]. Mulberry leaves and stem segments of “Qiang Sang No. 1” plants were punctured with a sterilized inoculation needle, and bacterial suspension was introduced dropwise to assess pathogenicity. Three different parts of the mulberry plant were used for the pathogenicity test.
Firstly, a bacterial suspension was inoculated onto freshly detached mulberry leaves. A sterile needle was used to puncture the back of the leaves, and round paper pieces with a 6 mm diameter were used to seal the punctures. Subsequently, 20 μL of bacterial suspension was added onto the round paper pieces. Similarly, the control group received an equivalent amount of sterile water treatment. The leaves were placed in an incubator at 30°C and 80% humidity, and disease incidence was observed and recorded daily. The experiment was repeated three times.
Secondly, the young hydroponic branches were inoculated with the bacterial suspension. The specific procedure involved puncturing the stem segments, injecting 20 μL of bacterial suspension, and wrapping with cotton, while the control group received sterile water treatment. The branches were also placed in an incubator at 30°C and 80% humidity, and disease incidence was observed and recorded daily.
Thirdly, apical branches of potted mulberry plants and disease-free mulberry trees in the field were inoculated with the bacterial suspension. Three healthy and unaffected mulberry plants were chosen, and two tender apical branches from each tree were injected with 20 μL bacterial suspension and sterile water, respectively. To minimize environmental effects, cotton was used to wrap and seal the inoculation sites. The experiment was also repeated three times and disease incidence was continuously observed and recorded after a week.
If symptoms of disease occur, diseased leaves and branches will be collected, and the pathogenic bacteria will be isolated again to confirm whether the pathogenic bacteria after inoculation are the same as those initially isolated.
2.5 Extracellular Enzyme Assays of Pathogenic Bacteria
The cellulase assay was carried out following the method of Barnard et al. [18]. A 10 μL of the bacteria solution to be tested (OD600 = 1.0) was added to the cellulase assay plate (Table S1). Once the plate was fully dried, it was transferred to an incubator set at 30°C for inverted culture for 12 h. Subsequently, 0.1% Congo red solution was added to the culture dish and allowed to stain for 20 min. The Congo red solution was then decanted, and 1M NaCl solution was added for decolorization for 15 min. Finally, 1M HCl was used for coloring for 5 min. Strains producing cellulase exhibited transparent hydrolytic circles around their colonies against a red background. The diameters of these transparent circles were measured to compare differences in cellulase activity among strains. The experiment was repeated three times.
The pectinase assay was conducted using the method of Barnard et al. [18]. A 10 μL of the bacteria solution to be tested (OD600 = 1.0) was added to the pectinase assay plate (Table S2). Once the plate was fully dried, it was transferred to an incubator set at 30°C for inverted culture for 12 h. The culture dish was then treated with a 7.5% copper acetate solution for staining for 1 h. Afterward, the dye solution was discarded, and the plate was washed four times with sterile water. Pectinase-producing strains formed transparent circles around the growing colonies on the blue and green background of the testing medium. The diameter of the transparent circle around each colony was observed and measured. The experiment was repeated three times.
The protease assay was performed following the method of Barnard et al. [18]. A 10 μL of the bacteria solution to be tested (OD600 = 1.0) was added to the protease testing plate (Table S3). After the plate was fully dried, it was placed in a 30°C incubator for inverted culture for 12 h. The presence and diameter of hydrolytic transparent circles around the colonies were compared. The experiment was repeated three times.
2.6 Identification of Physiological and Biochemical Characteristics of Pathogenic Bacteria
The GN card, designed for identifying Meriaeg negative bacteria (GN card) was selected [19]. After 24 h of incubation, the samples were prepared into 0.5–0.63 McF solution. The cards were then positioned sequentially on the card carrier, and the infusion tube was inserted into the bacterial solution tube. Identification was conducted using the Vitek 2 system (bioMérieux) [20].
2.7 Amplification and Comparison of 16S rRNA Sequences of Pathogenic Bacteria
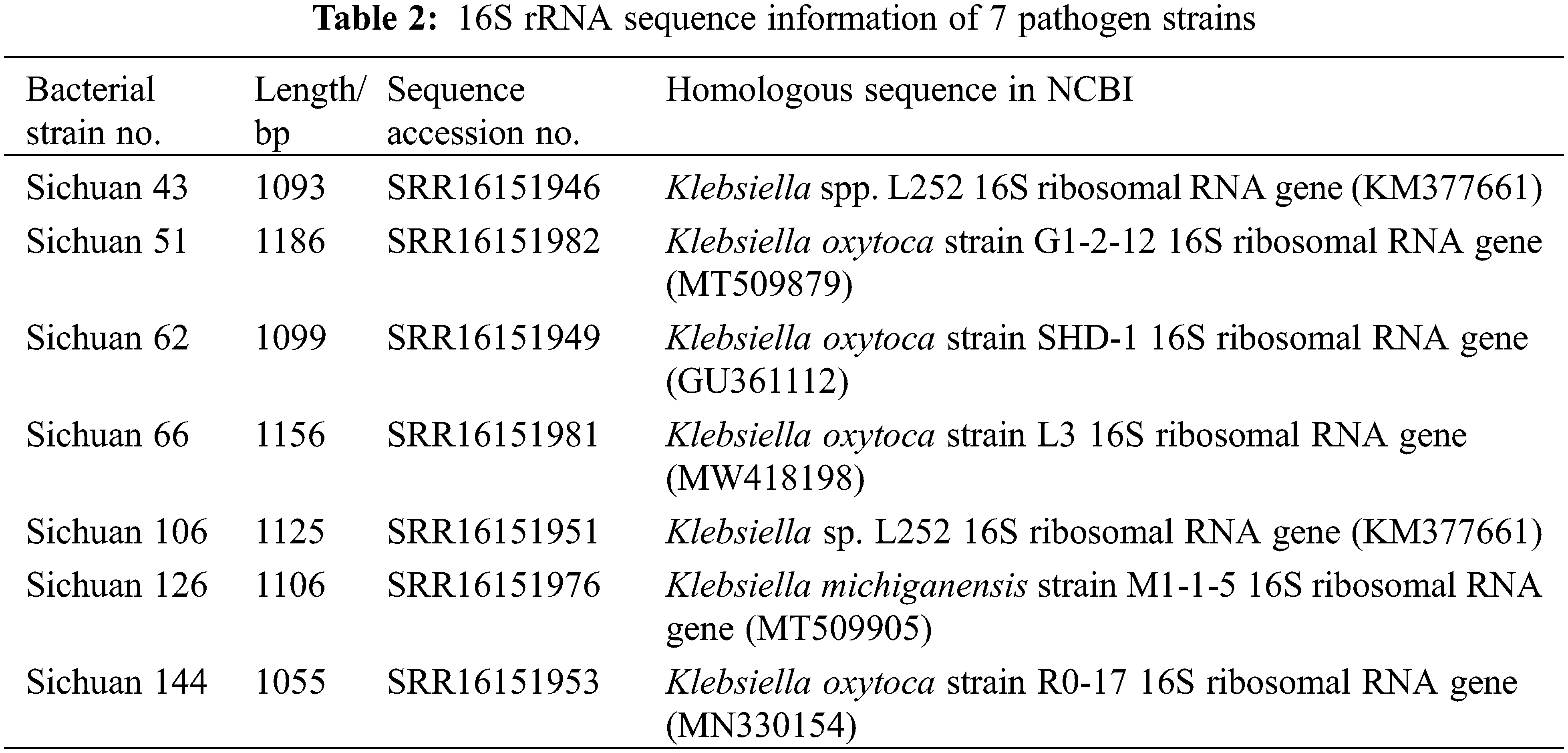
16S rRNA sequences of representative strains were amplified using PCR with universal primers 27F and 1492R, using the bacteria solution as a template [21]. The PCR reaction system consisted of 50 μL, including 25 μL 2 × Heff@ PCR Master Mix (cat#10102ES03), 1.5 μL of each upstream and downstream primers (10 μmol·L−1), 2 μL of template, and 20 μL of ddH2O. The PCR products were electrophoresed on 1.2% agarose gel, then excised and sequenced. Primer synthesis and sequencing were provided by Hangzhou Youkang Biotechnology Company, Hangzhou, China. To preliminarily estimate the species classification of the pathogenic bacteria, the 16S rRNA sequencing data of representative strains were assessed for homology using BLAST software on the NCBI website. The 16S rRNA sequences of the representative strains were selected and uploaded to the Sequence Read Archive (SRA) (Accession Numbers: SRR16151946, SRR16151982, SRR16151949, SRR16151981, SRR16151951, SRR16151976, SRR16151953).
2.8 Phylogenetic Tree Construction of 16S rRNA Sequences of Pathogenic Bacteria
The homologous sequences of 16S rRNA of each strain were downloaded from the NCBI website, and multiple alignment of these sequences was conducted using ClustalX [22]. The phylogenetic tree was constructed using the NJ (neighbor-joining) method with MEGA 5.1 and examined 1000 times using the bootstrap method. The outgroup consisted of 16S rRNA sequences from Bacillus subtilis.
3.1 Symptoms of Bacterial Blight of Mulberry Trees
In the early stages of the disease, the tender leaves of the mulberry tree exhibited yellow spots on the upper surface, accompanied by a few localized wounds. As the disease progressed into the middle and late phases, the upper leaves and stem portions of the mulberry trees wilted and turned black (Fig. 1A,B). The epidermal portion at the branch tip showed black streaks, and the second through fourth leaves from the top tip exhibited localized trauma, spreading downward from the immature apical section. The subcutaneous tissue appeared deteriorated, with no pus overflowing. Upon peeling the epidermis, the internal color was dark yellow, and the subcutaneous tissue could be seen decaying with no pus overflowing (Fig. 1C).

Figure 1: (A and B) Symptoms of mulberry bacterial blight. (C) Stem segments of mulberry bacterial blight
3.2 Isolation and Morphological Observation
Twenty samples of mulberry disease were collected from the main infected area in BSC, and the pathogenic bacteria were isolated. A total of 218 bacterial strains were isolated from the disease branch samples, and 7 pathogenic strains were obtained through culture screening. These strains were then incubated at 30°C for 12 h using LB solid medium. The morphology of the 7 strains on the LB solid medium was similar; the colonies were creamy white, round and bulgy, with a smooth and flat surface, displaying distinct individual morphology. (Fig. 2A). However, the growth rates of the colonies varied slightly among the strains. The morphology of the bacteria was observed using electron microscopy (Fig. 2B), revealing oval-shaped bacteria with capsules but no spores (Fig. 2C). They were stained with Gram stain, and the red staining result indicated that the pathogenic bacteria were gram-negative (Fig. 2D).
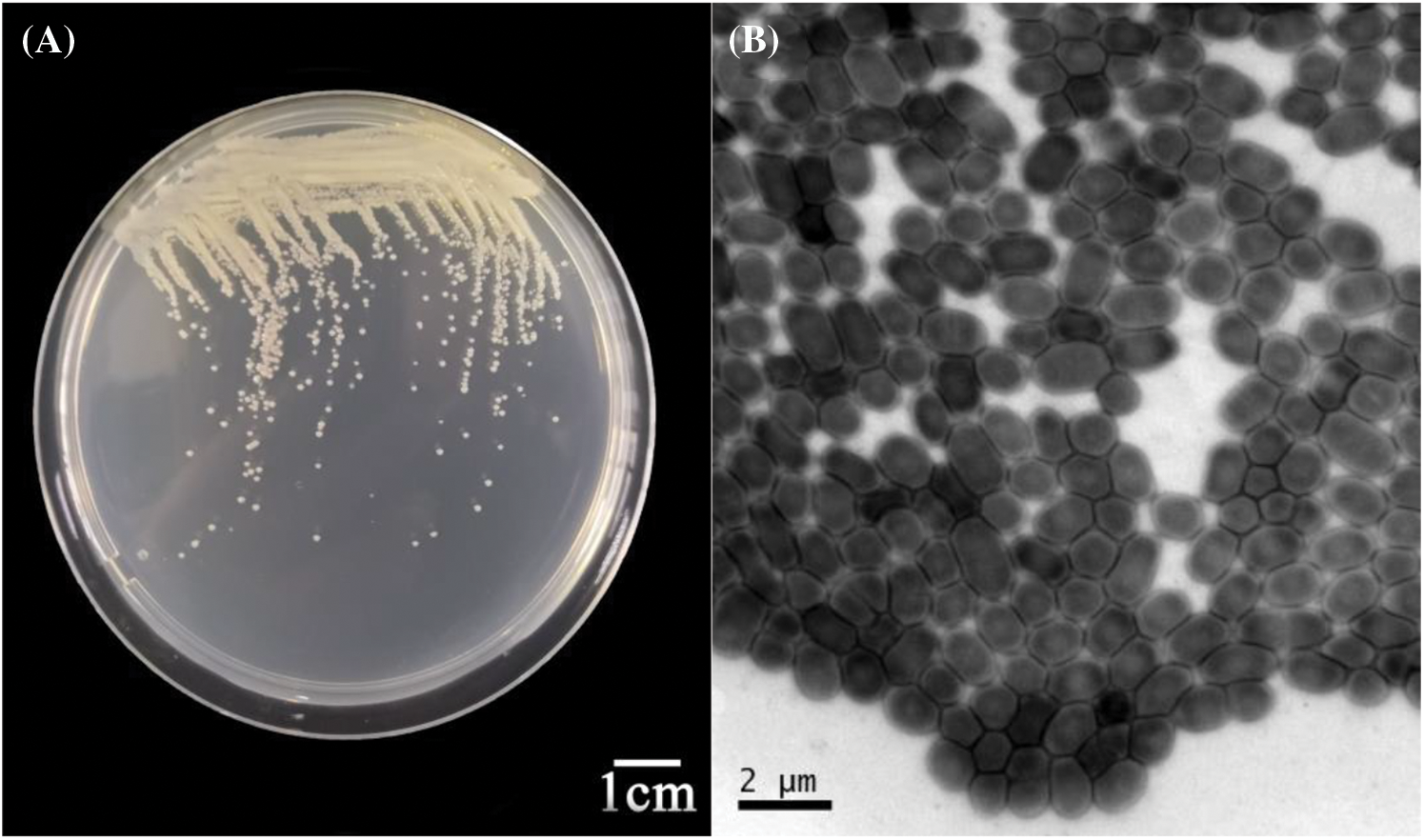
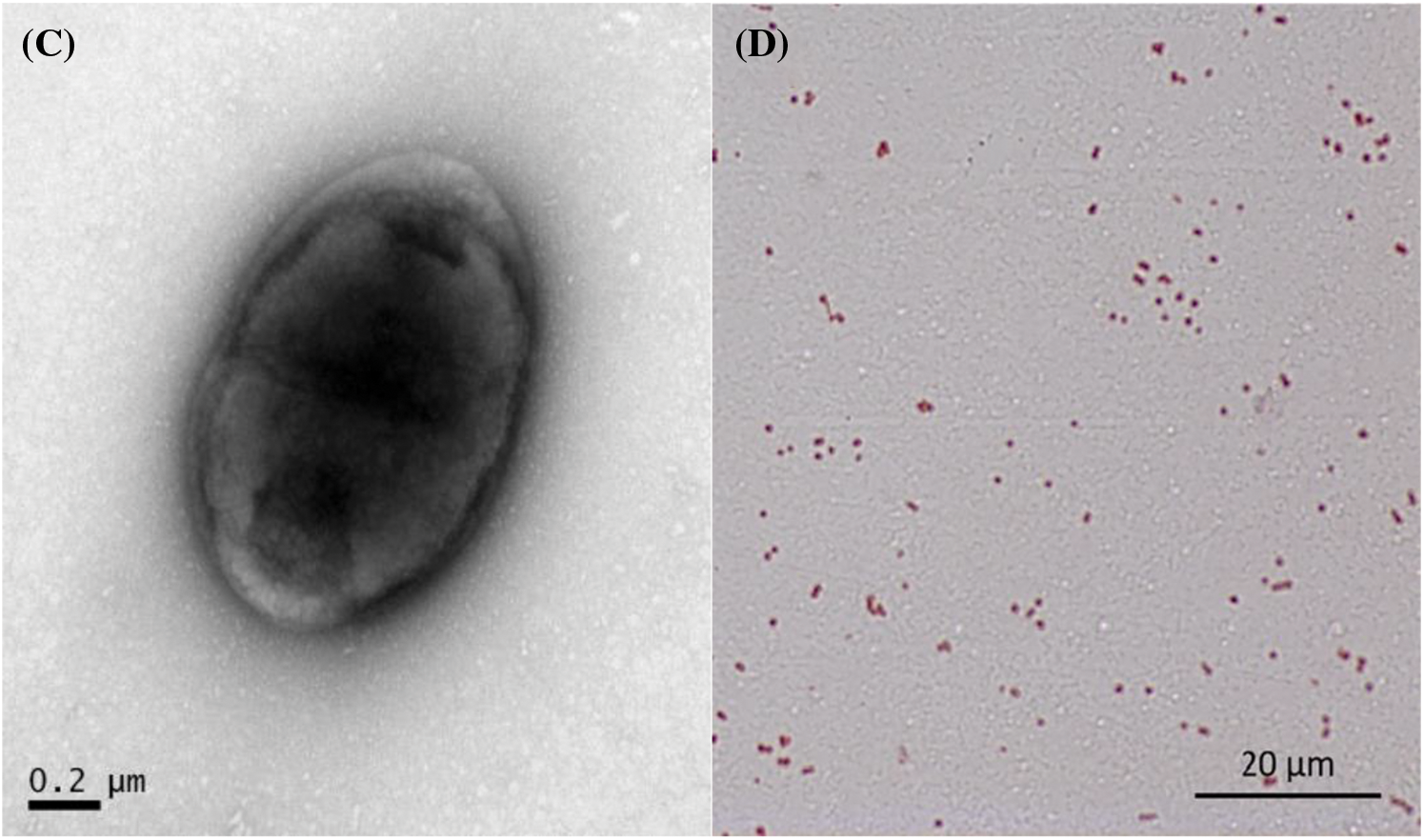
Figure 2: (A) Morphology of the pathogenic bacteria on LB solid medium. (B and C) Morphology of pathogenic organisms observed under an electron microscope. (D) Gram stain image of the pathogenic bacteria
3.3 Pathogenicity of Pathogenic Bacteria
In this experiment, senven strains (Sichuan 43, 51, 62, 66, 106, 126, 144) were selected for pathogenicity determination. The pathogenic bacteria were inoculated onto tender leaves in vitro, hydroponically grown young branches, apical branches of potted mulberry plants and mulberry trees naturally in the field. After inoculation for 48 h, the leaves exhibited evident rotten spots (Fig. 3A), whereas the control group showed no change (Fig. 3B). It was found that the disease spot size caused by Sichuan 106 and Sichuan 144 was consistently larger than those caused by Sichuan 43 and 66. Rotten spots were also observed on the stem segments after 48 h of hydroponic branch inoculation. Approximately a week later, the branches displayed severe rot and blackening, with the leaves drying out, curling up, and turning black (Fig. 3C), while the control group showed no signs of disease (Fig. 3D). Black rot spots appeared on the apical stem segments of potted mulberry plants and naturally growing disease-free apical branches after inoculation for 48 h, and the young apical parts wilted, blackened and rotted after about a week (Fig. 3E,G), exhibiting symptoms similar to those of diseased mulberry trees in BSC. The control group showed no change (Fig. 3F,H). The pathogen was once again isolated, and the morphological traits of the acquired strains were consistent with those of the initially inoculated pathogenic bacteria. According to Koch’s postulates, it was confirmed that the isolated strains were the causal agents of bacterial wilt affecting mulberry trees.
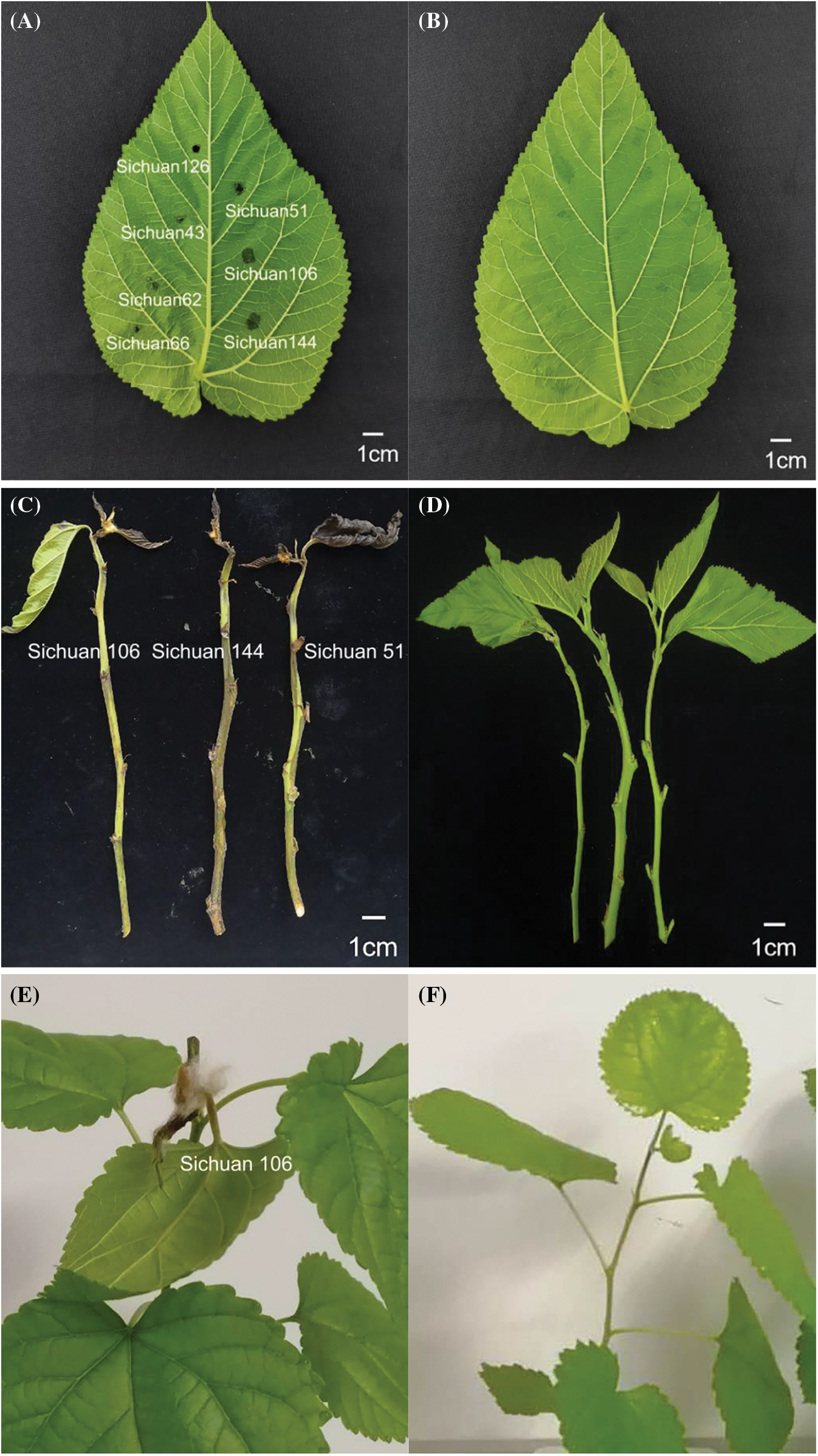

Figure 3: (A) Mulberry leaves were inoculated with bacterial solution.(B) Mulberry leaves were inoculated with sterile water as control. (C) Young branches of the mulberry were inoculated with bacterial suspension. (D) Young mulberry branches were inoculated with sterile water. (E) Potted mulberry plants were inoculated with bacterial suspension. (F) Potted mulberry plants were inoculated with sterile water as control. (G) The pathogen were being reinoculated into the top part of the mulberry tree in the field. (H) Sterile water was injected into the top part of the mulberry tree in the field as a control
3.4 Analysis of Extracellular Enzyme Assays
The extracellular enzyme assays were conducted on seven strains (Sichuan 43, 51, 62, 66, 106, 126, 144), and partial results are shown in Fig. 4A–C. It was found that the average hydrolytic circle diameter of the pathogenic bacteria on the protease test medium was 12.43 ± 3.06 mm. In contrast, the hydrolytic circle diameter of pathogenic bacteria on the pectinase and cellulase test medium was smaller, with an average hydrolytic circle diameter of 2.45 ± 1.01 mm and 2.59 ± 0.40 mm, respectively (Fig. 5). This indicates that the pathogenic bacteria have the capability to secrete pectinase, cellulase and protease, with varying secretion abilities among strains. Detailed results can be found in Table S4. It was found that Sichuan 106 and Sichuan 144 had the stronger secretion capabilities of three extracellular enzymes compared to Sichuan 43 and 66.
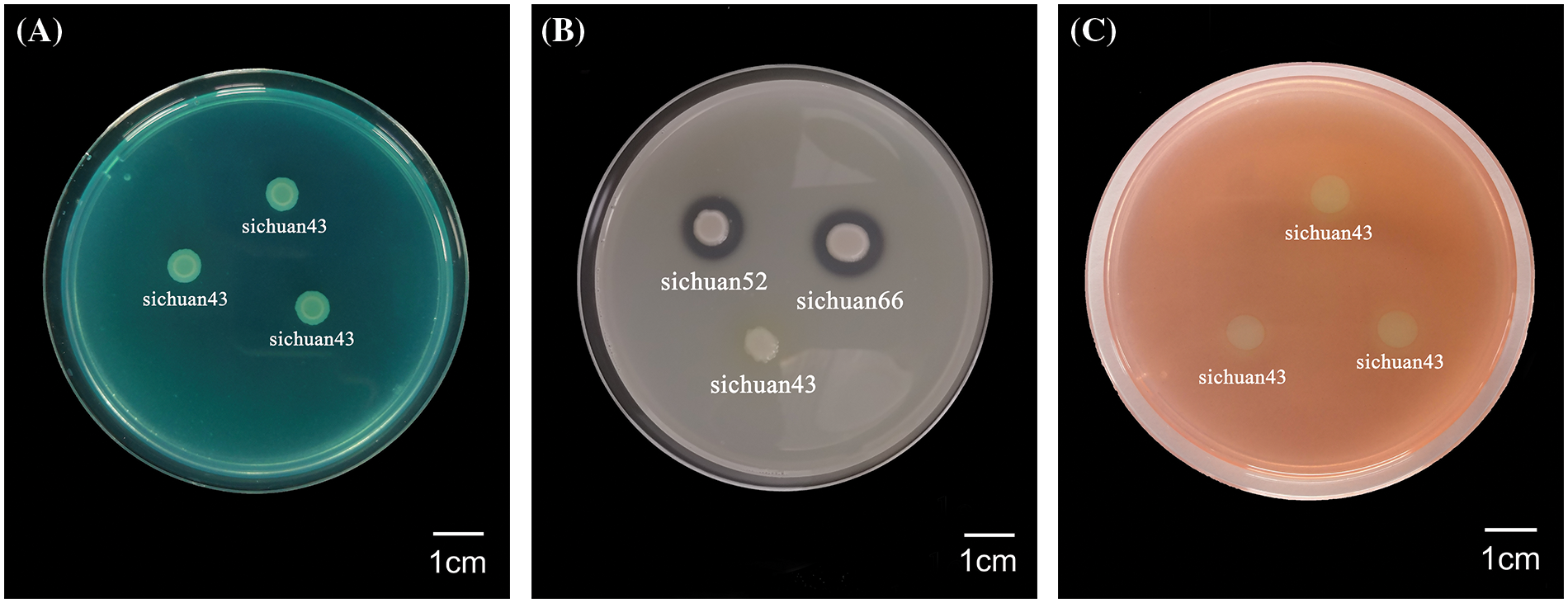
Figure 4: (A) Pectinase assay for strain Sichuan 43. (B) Protease assay results for strains Sichuan 43, 52, 66. (C) Cellulase assay result for strain Sichuan 43
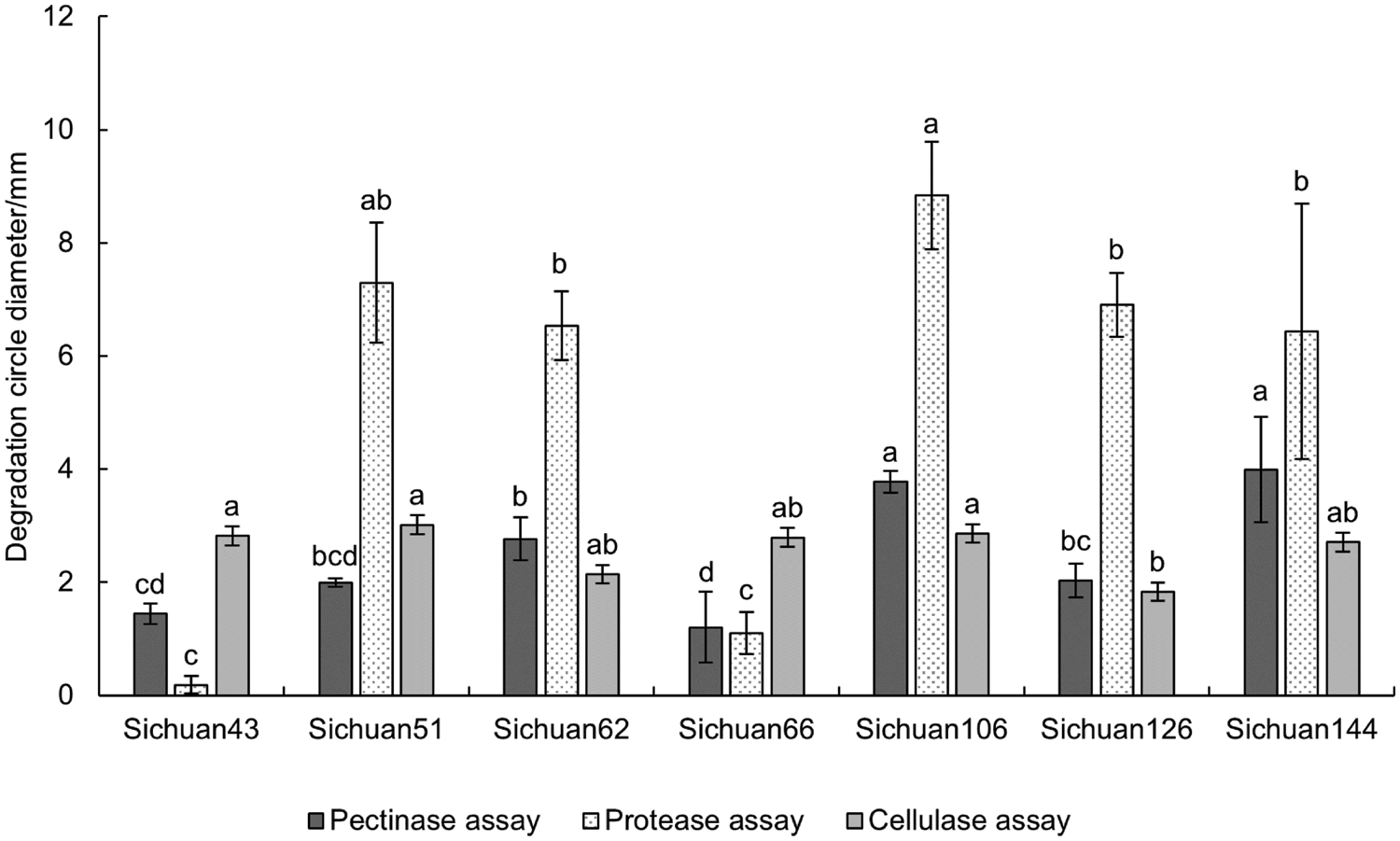
Figure 5: Results of extracellular enzyme assays. The same lowercase letter indicates significant differences (p < 0.05; one-way ANOVA)
3.5 Physiological and Biochemical Characteristics Analysis
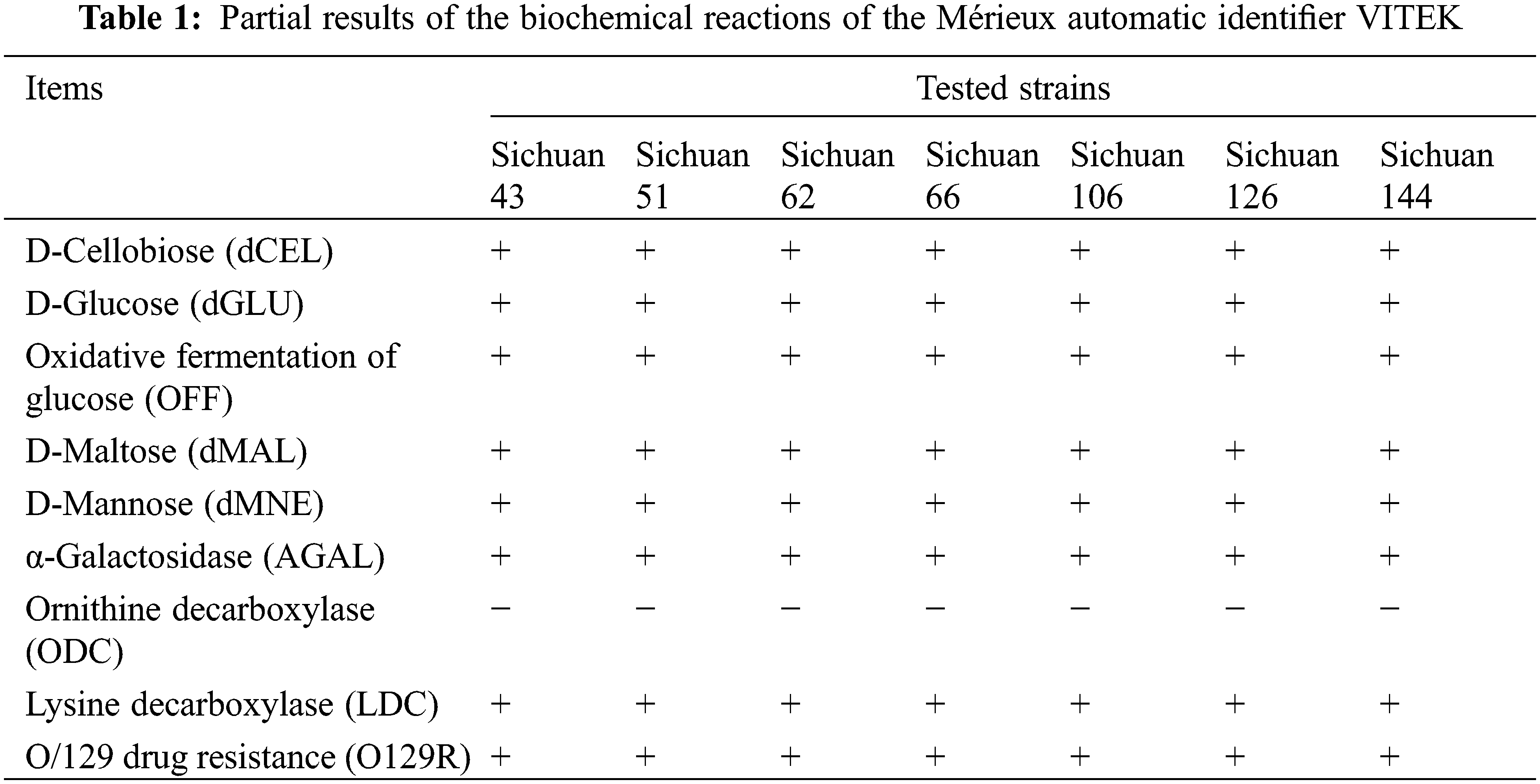
Biochemical testing was conducted on seven representative strains using the Mérieux VITEK 2 system (Table 1). They all exhibited utilization of D-cellobiose, D-glucose, D-maltose, and D-mannose, along with α-galactosidase activity. Additionally, they tested positive for lysine decarboxylation but negative for the ornithine decarboxylation reaction. Furthermore, they showed resistance to O/129. Detailed results can be found in Table S5. The results of biochemical analysis revealed that the seven bacteria had a 96% probability of being classified Klebsiella spp.
3.6 Molecular Identification of Pathogenic Bacteria
The 16S rRNA sequences of seven representative strains (Sichuan 43, 51, 62, 66, 106, 126, 144) were selected for PCR amplification, and the bands obtained by agarose gel electrophoresis were all approximately 1100 bp (Fig. 6). The sequences of the 16S rRNA, obtained through sequencing for these seven strains, were submitted to NCBI (Table 2). Alignment on the NCBI website revealed that they had the highest similarity to Klebsiella spp. The BLAST analysis further confirmed that the 16S rRNA sequences of these seven strains showed the highest homology with those of Klebsiella spp.
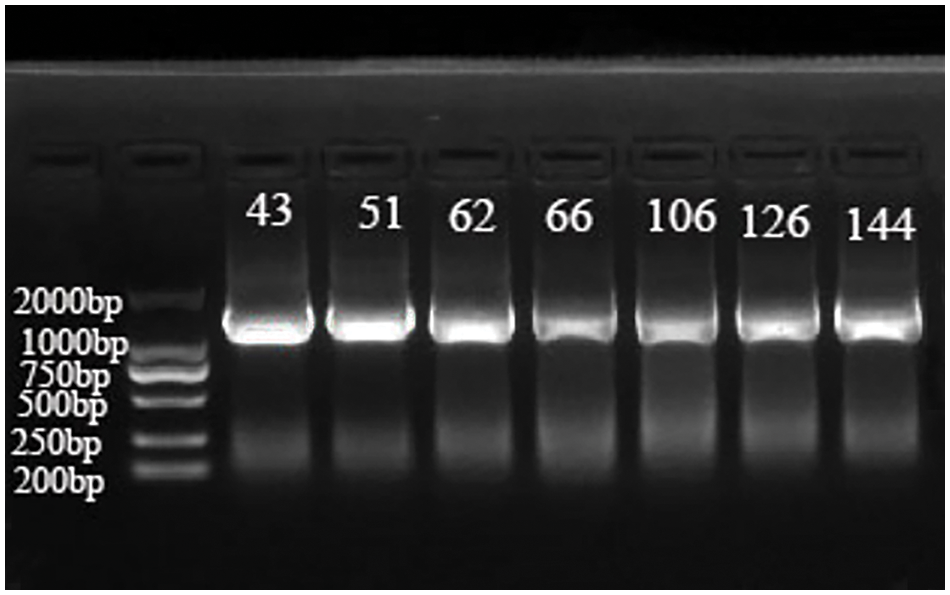
Figure 6: Results of 16S rRNA sequence PCR amplification of 7 pathogen strains
The 16S rRNA sequences reported for Klebsiella spp., including Klebsiella oxytoca, Klebsiella terrigena, Klebsiella michiganensis, Klebsiella pneumoniae, Klebsiella pasteurii, Klebsiella grimontii and Klebsiella kielensis, were downloaded from NCBI. Bacillus subtilis was used as an outgroup for comparison. The NJ (neighbor-joining) method of MEGA 5.1 software was utilized to construct a phylogenetic tree. According to the phylogenetic tree (Fig. 7), Sichuan 43, 51, 62, 66, 106, 126 and 144 clustered with Klebsiella oxytoca. Consequently, all 7 strains were identified as Klebsiella oxytoca.
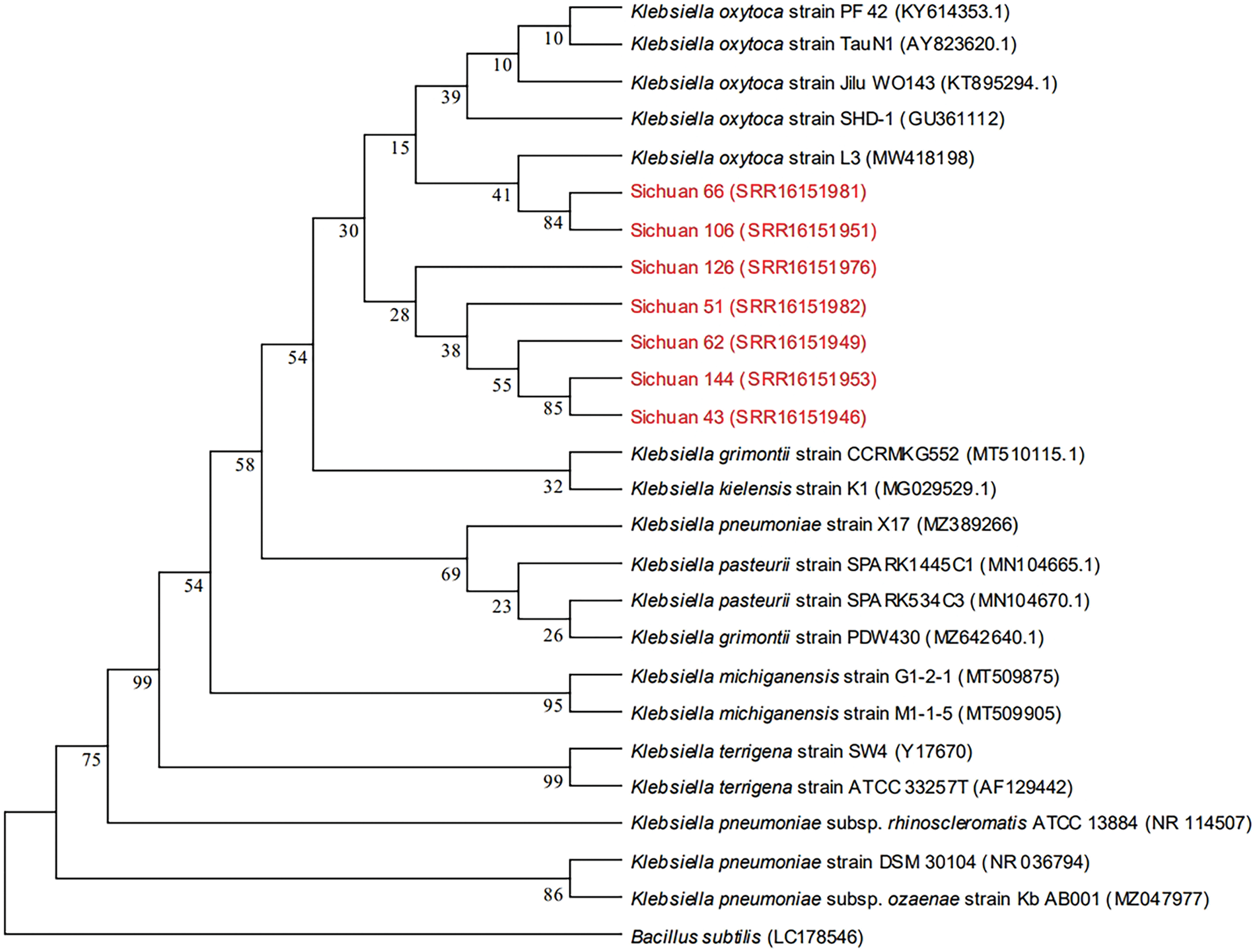
Figure 7: A phylogenetic tree of 7 pathogen strains
In this study, the principal pathogenic bacteria were isolated from diseased branches of mulberry in BSC, and their pathogenicity was verified in accordance with Koch’s postulates. The primary pathogenic bacteria in this region were identified as Klebsiella spp. through physiological and biochemical tests, 16S rRNA molecular identification, and phylogenetic analysis of representative strains.
Klebsiella spp. bacteria inhabit various environments, including insects (in symbiosis), plants, soil and surface water. They can also parasitize the upper respiratory and gastrointestinal tracts of humans and animals [23]. The Klebsiella genus encompasses a variety of species, such as Klebsiella oxytoca, Klebsiella pneumoniae, Klebsiella ornithinolytica, Klebsiella pneumonia, Klebsiella planticola, Klebsiella ozaenae, Klebsiella rhinoscleromatis, and Klebsiella variicola [24]. Huang et al. [23] discovered that Klebsiella pneumoniae can directly infect maize, banana, pomegranate, and sorghum, crossing biological boundaries and causing diseases like maize top rot, banana leaf spot, pomegranate wilt, and sorghum leaf spot. Klebsiella pneumoniae is a significant foodborne pathogen found in the intestinal flora. Therefore, it is crucial to investigate the source of mulberry bacterial blight infestation in BSC. This investigation should explore whether animal excreta used as organic fertilizer in mulberry cultivation areas could be a source and whether Klebsiella spp., known to be pathogenic to mulberry, is closely related to Klebsiella pneumoniae. A comprehensive analysis of the factors contributing to disease occurrence is also necessary. Additionally, factors such as the density of mulberry planting in the area, the duration of continuous mulberry cultivation, and the introduction of disease-carrying seedlings from other regions should be considered.
Plant cell walls are primarily composed of pectin, cellulose and proteins, serving as crucial defenses against the invasion of foreign pathogens [25,26]. The pathogens causing bacterial blight can produce a range of virulence factors, such as pectinases, cellulases, and proteases, to degrade the plant cell walls and facilitate the invasion of the host by the pathogen [27]. In this study, we discovered that Klebsiella spp. could secrete pectinase, cellulase and protease, as characterized enzymatically, with variations in secretion abilities among strains. It is speculated that the pathogen may degrade plant cell walls by secreting pectinase and cellulase, while protease may work in conjunction with these enzymes to infect mulberry cell tissues. Enterobacter cloacae, a noval species of Enterobacter spp., was identified as the pathogenic bacterium causing mulberry bacterial blight through various identification techniques [28]. The pathogenic bacterium induced plant wilt by extensively colonizing the vascular tissues of plant rhizomes, obstructing the ducts, and resulting in leaf water deficiency, thereby exhibiting wilt symptoms. It is speculated that this effect may be due to the secretion of secondary metabolites toxic to plants, leading to decay and death by the pathogenic bacteria [29]. Therefore, it is essential to conduct further investigations into the pathogenic and molecular mechanisms of mulberry bacterial wilt pathogens, considering that the infection pathways of various plant pathogens may differ.
In the initial stage of this study, other pathogenic bacteria such as Serratia spp., Enterbacter spp., and Raoultella spp. were detected. It indicated that the pathogenic bacteria causing mulberry bacterial wilt were diverse and complex, and the disease may be the result of multiple bacterial infections. This is consistent with the findings of Dai et al. [13]. Therefore, identifying the primary pathogenic bacteria of mulberry bacterial blight in different regions is of great significance. Additionally, Meng et al. [14] proposed that the severity of mulberry wilt symptoms varied from location to location, suggesting either a diversity of pathogenic bacteria or pathogenic differentiation within the same bacterial species. Furthermore, their pathogenicity tests confirmed considerable variance in pathogenicity among strains within the same species, demonstrating the genetic diversity of pathogenic bacteria causing mulberry bacterial blight in Guangxi. From our extracellular enzyme detection results, it was found that Sichuan 106 and Sichuan 144 had the stronger secretion capabilities of three extracellular enzymes compared to Sichuan 43 and 66. After inoculation on detached leaves, the disease spot size caused by Sichuan 106 and Sichuan 144 was consistently larger than those caused by Sichuan 43 and 66. Based on the results of the inoculation experiments, we have categorized them into high pathogenicity strains: Sichuan 106, and Sichuan 144; moderate pathogenicity strains: Sichuan 51, Sichuan 126, and Sichuan 62; and weak pathogenicity strains: Sichuan 43 and Sichuan 66. In the extracellular enzyme secretion capability test results, the secretion capability of pectinase from strong to weak is ranked as Sichuan 144 > 106 > 62 > 126 > 51 > 43 > 66; the secretion capability of protease from strong to weak is ranked as Sichuan 106 > 51 > 126 > 62 > 144 > 66 > 43; and the secretion capability of cellulase from strong to weak is ranked as Sichuan 51 > 106 > 43 > 66 > 144 > 62 > 126. We found that the high pathogenicity strains have the strongest secretion capability of pectinase, the moderate pathogenicity strains have a more prominent secretion ability of protease, and the weak pathogenicity strains have the weakest secretion capabilities of both pectinase and protease. Therefore, this study also revealed that the detected pathogenic bacteria in the region were diverse and exhibited varying levels of pathogenicity. It also confirmed that the virulence of a bacterial strain was related to its secretion capacity of cell wall-degrading enzymes; the stronger the virulence, the greater the secretion capacity of these enzymes. Thus, the development of rapid detection techniques for a variety of diseases is necessary at this stage. Strengthening quarantine measures and pathogen detection when introducing new mulberry varieties is essential to prevent the spread of highly pathogenic bacteria that could cause widespread diseases in mulberry-growing areas.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02072-6) and the Natural Science Foundation of Anhui Provincial Education Department (KJ2019A0574).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Zilong Xu, Yuan Huang, Junshan Gao; data collection: Peigang Liu, Yan Zhu, Tianbao Lin, Zhiqiang Lv; analysis and interpretation of results: Jia Wei, Yijun Li, Mei Zong, Yun Zhou; draft manuscript preparation: Yuan Huang, Zilong Xu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The original contributions presented in the study are included the article and in Supplemental Materials. Further inquiries should be directed to the corresponding authors.
Ethics Approval: This article does not contain any studies with animals or humans performed by any of the authors.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.052353.
References
1. Sulochana P. Medicinal values of mulberry–an overview. J Pharm Res. 2012;5(7):3588–96. [Google Scholar]
2. Ghosh A, Gangopadhyay D, Chowdhury T. Economical and environmental importance of mulberry: a review. Int J Plant Environ. 2017;3(2):067–72. [Google Scholar]
3. Zhang J, Zhao Y, Li H, Ni J, Wang D. Transcriptome profiling of the salt-stress response in paper mulberry. Phyton-Int J Exp Bot. 2023;92(9):2591–610. doi:10.32604/phyton.2023.028338. [Google Scholar] [CrossRef]
4. Rahman M, Islam S. Studies on food, health and environmental perspectives in mulberry (Morus spp.)—a review. J Bio-Sci. 2021;29:163–79. [Google Scholar]
5. Wu M, Guo P, Shi Y, Zheng D, Zhang Q, Jin X, et al. Genome-wide analysis of microRNAs induced and regulated by colchicine in mulberry. BIOCELL. 2022;46(8):1935–45. doi:10.32604/biocell.2022.019223. [Google Scholar] [CrossRef]
6. Chen T, Shuang FF, Fu QY, Ju YX, Zong CM, Zhao WG, et al. Evaluation of the chemical composition and antioxidant activity of mulberry (Morus alba L.) fruits from different varieties in China. Molecules. 2022;27(9):2688. doi:10.3390/molecules27092688. [Google Scholar] [PubMed] [CrossRef]
7. Yang WY, Wan DG. Resources of Chinese herbs Sang (Morus sp.) in Sichuan and strategies for development and utilization. China J Chin Mater Med. 2008;33(24):2871–5 (In Chinese). [Google Scholar]
8. Dong Z, Guo Y, Yu C, Zhu Z, Mo R, Wen D, et al. The dynamics in rhizosphere microbial communities under bacterial wilt resistance by mulberry genotypes. Arch Microbiol. 2020;203(3):1107–21. [Google Scholar] [PubMed]
9. Wu G, Rui L, Lu X, Han L, Lv G, Fu X, et al. Characterization and pathogenicity of pseudopestalotiopsis vietnamensis causing gray blight of wuyi rock tea (Camellia sinensis) in China and specific mechanisms of disease infection. Phyton-Int J Exp Bot. 2023;92(1):131–47. doi:10.32604/phyton.2022.021919. [Google Scholar] [CrossRef]
10. Xie S, Vallet M, Sun C, Kunert M, David A, Zhang X, et al. Biocontrol potential of a novel endophytic bacterium from mulberry (Morus) tree. Front Bioeng Biotechnol. 2020;7:488. doi:10.3389/fbioe.2019.00488. [Google Scholar] [PubMed] [CrossRef]
11. Sato M, Takahashi K. Ecological studies on the bacterial blight of mulberry: 1. The overwintering of the pathogen, Pseudomonas mori (BOYER et LAMBERT) STEVENS. J Seric Sci Jpn. 1972;41(4):285–93. [Google Scholar]
12. Wang G, Praphat K, Xie G, Zhu B, Li B, Liu B, et al. Bacterial wilt of mulberry (Morus alba) caused by Enterobacter cloacae in China. Plant Dis. 2008;92(3):483. doi:10.1094/PDIS-92-3-0483B. [Google Scholar] [PubMed] [CrossRef]
13. Dai FW, Luo GQ, Wang ZJ, Li L, Tang CM, Kuang ZS, et al. Isolation and molecular identification of the pathogen causing mulberry bacterial blight in southern sericultural areas of China. Science of Sericulture. 2012;38(6):981–7 (In Chinese). [Google Scholar]
14. Meng JR, Ouyan QF, Cen JL, Zhu LL, Li JQ, Zhu FR, et al. Pathogenicity and genetic diversity of the pathogens causing mulberry bacterial wilt in Gunagxi province. Genom Appl Biol. 2014;33(5):998–1006 (In Chinese). [Google Scholar]
15. Luo L, Huang Y, Liu J. Genome sequence resources of Klebsiella michiganensis AKKL-001, which causes bacterial blight of mulberry. Mol Plant-Microbe Interact. 2022;35(4):349–51. doi:10.1094/MPMI-09-21-0222-A. [Google Scholar] [PubMed] [CrossRef]
16. Froböse NJ, Bjedov S, Schuler F, Kahl BC, Kampmeier S, Schaumburg F. Gram staining: a comparison of two automated systems and manual staining. J Clin Microbiol. 2020;58(12):e01914–20. [Google Scholar]
17. Liu L, Liu X, Liu L, Zhu T, Ye R, Chen H, et al. Clarification of the infection pattern of Xanthomonas citri subsp. citri on citrus fruit by artificial inoculation. Plant Methods. 2024;20(1):65. doi:10.1186/s13007-024-01190-7. [Google Scholar] [PubMed] [CrossRef]
18. Barnard AML, Simpson NJL, Lilley KS. Mutations in rpsL that confer streptomycin resistance show pleiotropic effects on virulence and the production of a carbapenem antibiotic in Erwinia carotovora. Microbiology. 2010;156(4):1030–9. doi:10.1099/mic.0.034595-0. [Google Scholar] [PubMed] [CrossRef]
19. Angaali N, Vemu L, Padmasri C, Mamidi N, Teja VD. Direct identification and susceptibility testing of Gram-negative bacilli from turbid urine samples using VITEK2. J Lab Phys. 2018;10(3):299–303. [Google Scholar]
20. Valenza G, Ruoff C, Vogel U, Frosch M, Abele-Horn M. Microbiological evaluation of the new VITEK 2 Neisseria-Haemophilus identification card. J Clin Microbiol. 2007;45(11):3493. doi:10.1128/JCM.00953-07. [Google Scholar] [PubMed] [CrossRef]
21. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74(8):2461–70. doi:10.1128/AEM.02272-07. [Google Scholar] [PubMed] [CrossRef]
22. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–500. doi:10.1093/nar/gkg500. [Google Scholar] [PubMed] [CrossRef]
23. Huang M, He PB, He PF, Wu YX, Li J, He YQ. Research advances in effects of Klebsiella pneumoniae on plants. Zhi Wu Bing Li Xue Bao. 2022;52(4):511–21 (In Chinese). [Google Scholar]
24. Li MJ, Peng S, Xu SZ, Yu DH, Zhao MF, Wen GS. Application of Klebsiella spp. in agriculture and environmental management. Curr Biotechnol. 2014;4(6):415–20 (In Chinese). [Google Scholar]
25. Perez S, Rodriguez-Carvajal MA, Doco T. A complex plant cell wall polysaccharide: rhamnogalacturonan II. A structure in quest of a function. Biochimie. 2003;1(2):109–21. [Google Scholar]
26. Wan J, He M, Hou Q, Zou L, Yang Y, Wei Y, et al. Cell wall associated immunity in plants. Stress Biol. 2021;1(1):3. doi:10.1007/s44154-021-00003-4. [Google Scholar] [PubMed] [CrossRef]
27. Xiao WC, Fan HY, Bai TT, Wang W, Li HP. Prediction and analysis of the signal peptide and secreted proteins in soft rot bacteria XJ8-3-3 genome ORFs of banana. Int J Fruit Sci. 2014;31(6):1057–64 (In Chinese). [Google Scholar]
28. Zhu B, Wang G, Xie G, Zhou Q, Zhao M, Praphat K, et al. Enterobacter spp.: a new evidence causing bacterial wilt on mulberry. Sci China Life Sci. 2010;53:292–300. doi:10.1007/s11427-010-0048-x. [Google Scholar] [PubMed] [CrossRef]
29. Zhu B. Nominating, molecular detection about bacterial wilt in mulberry and horizontal gene transfer analysis in Enterobacteriaceae. Zhejiang University: China; 2010. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools