 Open Access
Open Access
ARTICLE
Identification and Analysis of the WRKY Transcription Factor Gene Family in Verbena bonariensis
The Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), Guizhou University, Guiyang, 550025, China
* Corresponding Author: Yan Li. Email:
Phyton-International Journal of Experimental Botany 2024, 93(8), 1875-1896. https://doi.org/10.32604/phyton.2024.052190
Received 26 March 2024; Accepted 27 June 2024; Issue published 30 August 2024
Abstract
The WRKY transcription factor gene family is one of the unique gene families in plants. It plays an important role in response to abiotic stresses such as cold and drought, hormone signal transduction, regulation of biosynthesis, leaf senescence seed germination, etc. However, little information is available about WRKY transcription factors in Verbena bonariensis. In this study, 70 VbWRKY genes were identified from the whole genome. The phylogenetic analysis of the WRKY gene family in V. bonariensis and Arabidopsis shows that the WRKY genes in V. bonariensis can be divided into three groups: I, II, and III, which contain 13, 47, and 10 members, respectively. Group II can be further divided into five subclasses: IIa (5), IIb (10), IIc (18), IId (6), and IIe (8). Conservative motif analysis showed that 64 proteins encoded by the VbWRKY gene had conserved motifs 1, 2 and 3, and the same subclass motif elements were approximately the same. The collinearity analysis showed that there were 44 homologous gene pairs among the VbWRKYs, and these homologous gene pairs may have the same function. Promoter sequence analysis showed that the VbWRKY gene has multiple cis-acting elements, including not only cis-acting elements related to low-temperature and light responses, but also cis-acting elements related to hormone regulation, Among them, most VbWRKY genes contain response elements about low-temperature, and 30 VbWRKY genes contain low-temperature response elements (LTR), and 61 VbWRKY genes contain abscisic acid response elements (ABRE), indicating that VbWRKY plays a crucial role in plant growth and abiotic stress. According to the expression of VbWRKY in the cold stress and different tissues transcriptome, 70 VbWRKY genes played their respective roles in various tissues and stages to regulate plant growth, Also, some of them participated in the process of cold stress tolerance, 52 VbWRKYs showed significant differences in expression under cold stress, and 37 VbWRKY genes were up-regulated under cold stress. 9 VbWRKY genes were selected for quantitative real-time PCR (qRT-PCR) analysis under low-temperature stress, and the results showed that all 9 genes were upregulated under low-temperature stress. Ultimately, the present study provides a comprehensive analysis of the predicted V. bonariensis WRKY genes family, which provided a theoretical basis for the study of low-temperature resistance and growth and development of V. bonariensis.Keywords
Supplementary Material
Supplementary Material FileThe WRKY transcription factors are one of the largest families of transcriptional regulators in plants [1], named for the highly conserved WRKY domain. Each WRKY protein contains one or two conserved domains with approximately 60 amino acid residues, including a highly conserved WRKYGQK heptapeptide at the N-terminus and a C2H2 or C2HC zinc finger motif at the C-terminus [2,3]. According to the number of WRKY domains and the type of zinc finger motif, the WRKY family can be divided into three main groups (I, II, and III). Group I contains two WRKY heptapeptide domains and one C2H2 zinc finger motif. Group II contains one WRKY heptapeptide domain and one C2H2 zinc finger motif. Group III contains a WRKY heptapeptide domain and a C2HC-type zinc finger motif. Group II can be divided into five subfamilies (IIa, IIb, IIc, IId, and IIe) based on evolutionary relationships [4,5]. Systematic evolutionary data analysis shows that the WRKY transcription factor family can be more accurately divided into groups I, IIa+IIb, IIc, IId+IIe, and III in higher plants [6].
The first WRKY transcription factor was identified in sweet potatoes in 1994 [7], followed by those identified in potatoes [8], tobacco [9], wheat and barley [10], Arabidopsis [11–13], rice [14], poplar [15], rapeseed [16], cucumber [17], cotton [18], etc. Many studies have shown that WRKY transcription factors have rich biological functions and are closely related to plant growth and development. In plants, WRKY transcription factors mediate defense regulatory functions, mainly in response to various biotic and abiotic stresses [19,20]. At the same time, abiotic stress can induce a large number of WRKY transcription factors to regulate plant tolerance to stress and acquire corresponding resistance [21]. In addition, it is involved in the regulation of plant physiological development, including hormone signal transduction, biosynthesis regulation, leaf senescence, embryo formation, and seed germination [22,23]. In Arabidopsis, AtWRKY33, AtWRKY46, and AtWRKY57 can enhance the tolerance of Arabidopsis to drought and salt stress by regulating the ABA signaling network [24–26]. Chen et al. found that SlWRKY12, SlWRKY13, SlWRKY23, SlWRKY50, and SlWRKY51 were significantly upregulated under cold stress, indicating that they may be involved in the response mechanism of tomato to low-temperature stress [27]. Wang et al. found in their study on Gossypium hirsutum that the GhWRKY22 gene can participate in pollen development through transcriptional regulation [28]. Wheat (Triticum aestivum) WRKY7 is an important regulatory factor for leaf senescence, with its expression continuously increasing during the process of natural leaf senescence [29]. TaWRKY2 and TaWRKY19 enhanced their tolerance to drought, salt, and cold stress in transgenic Arabidopsis. Transgenic wheat overexpressing TaWRKY2 and TaWRKY19 has improved salt tolerance, drought resistance, and frost resistance [30]. Compared with the wild type, overexpression of CsWRKY46 enhances cold resistance in cucumber [26]. OsWRKY71 plays a positive role in cold resistance by regulating downstream target genes in rice [31]. Under cold stress, the induction of VbWRKY32 in V. bonariensis leaves was greater than that in the stems and roots. and overexpression (OE) in V. bonariensis increased cold resistance compared with wild type (WT) [32].
Verbena bonariensis is a perennial herbaceous plant of the Verbena genus in the Verbenaceae family. It is native to Brazil, Argentina, and other regions in South America and is distributed in most parts of East, South, Northwest, and Southwest China. The optimum growing temperature for V. bonariensis is 20°C–30°C. It enjoys light and has strong drought resistance. Seedlings can be obtained by sowing or cuttings. The flowering period is mostly in summer and autumn, and the peak flowering period can reach 3 months. It is an excellent ornamental variety for gardens. The V. bonariensis also has the functions of detoxification, detumescence and spasmolysis, mainly treating symptoms such as dysmenorrhea, vaginal infections, and traumatic swelling and pain. However, in the cultivation of V. bonariensis, the V. bonariensis is not tolerant to the cold and grows slowly when the temperature is below 10°C. The low-temperature environment in winter seriously affects the yield and ornamental value of V. bonariensis [32]. Many studies have suggested that when plants are subjected to cold stress, they will improve their tolerance by regulating the expression of a series of genes. Many transcription factors, including WRKY, ERF, and MYB, are important regulatory factors related to cold stress [33,34]. The transcriptomic data of V. bonariensis under low-temperature stress indicate that WRKY-TFs play a crucial role in helping plants cope with low-temperature stress.
In this study, genome-wide identification of members of WRKY gene family in V. bonariensis was performed using genomic data measured by our research group, and analyzed their phylogeny, classification, chromosome distribution, conserved motifs, gene structure, cis-acting elements. Moreover, we further explored the expression patterns of VbWRKY genes in response to cold stresses. Furthermore, 9 VbWRKYs under cold stress were analyzed by qRT-PCR. The results provide a theoretical foundation for the study of V. bonariensis growth, development, and low-temperature resistance.
2.1 Plant Materials and Stress Treatment
V. bonariensis seedlings were placed in a chamber with a mean temperature of 25.0 ± 1.0°C, relative humidity of 60% ± 10%, and a day/light cycle of 16/8 h. For the cold treatment, V. bonariensis seedlings were placed in low-temperature refrigerator at 4°C and samples were gathered at 0, 3, 6, 9, 12 and 24 h with 0 h as control. The samples were snap frozen in liquid nitrogen and then stored at −80°C freezer to extract total RNA.
2.2 Identification of WRKY Gene Family Members in V. bonariensis
The genome data of V. bonariensis were obtained by our research group (unpublished), and the protein sequences of the Arabidopsis WRKY gene family were downloaded from the Arabidopsis database TAIR (http://www.arabidopsis.org/, accessed on 27 July 2023) [35]. The hidden Markov model (HMM) file of the WRKY domain (PF03106) was downloaded from the Pfam database (http://Pfam.xfam.org/, accessed on 25 July 2023) [36], and used for a search in hmmer (3.0) [37] to obtain the target sequence. At the same time, the Arabidopsis WRKY protein sequence was used as the query sequence, and BLAST program was used to compare the sequences in the V. bonariensis genome database. Merge and remove duplicates to obtain the candidate WRKY transcription factor protein sequence of V. bonariensis. The VbWRKY gene family members were further identified by using the protein conserved domain analysis tools NCBICD-search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 28 July 2023) and Pfam (http://Pfam.xfam.org/, accessed on 28 July 2023) to analyze and screen candidate genes, and ultimately obtain all WRKY transcription factor family members of V. bonariensis.
2.3 Physicochemical Properties of WRKY Gene Family Players in V. bonariensis
The online tool ProtParam (https://web.expasy.org/protparam/, accessed on 27 July 2023) [38] was used to predict the molecular weight and theoretical isoelectric point (pI) of the protein of the WRKY gene family in V. bonariensis. Online tools WoLF PSORT (https://wolfpsort.hgc.jp/, accessed on 19 August 2023) [39] were used to predict the subcellular localization of the VbWRKYs.
2.4 Phylogenetic Analysis and Multiple Sequence Alignment of WRKY Gene Family Members in V. bonariensis
Using the WRKY gene family of Arabidopsis thaliana as the reference sequence, the phylogenetic tree was constructed by using the neighbor-joining (NJ) method of Mega7.0 (set the Bootstrap value to 1000 and other parameters as the default value) [40], cluster analysis of VbWRKY family members was carried out according to the existing grouping of Arabidopsis thaliana, and the evolutionary tree was beautified by using the online website iTOL (http://iTOL.embl.de/, accessed on 21 August 2023) [41].
Software DNAMAN and online software Weblogo3 (http://weblogo.berkeley.edu/logo.cgi, accessed on 19 September 2023) were used to perform multiple sequence alignment of the WRKY domain of V. bonariensis WRKY protein.
2.5 Visualization of Gene Structure and Conserved Motif of WRKY Gene Family Members in V. bonariensis
The MEME (https://meme-suite.org/meme/tools/meme, accessed on 19 September 2023) [42] tool was used to perform conservative motif analysis on the WRKY family protein sequence of V. bonariensis, and the number of motif was set to 10. Based on the V. bonariensis genome GFF3 annotation file, the gene structure of VbWRKY gene was analyzed and visualized by Tb tools software version 2.096 [43].
2.6 Cis-Acting Elements of WRKY Gene Family Members in V. bonariensis
The promoter of the start codon 2000 bp upstream was separated from the genomefile of the V. bonariensis using TBtools software version 2.096 [43], and then the cis-acting elements of the VbWRKYs were found using the online tool PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 5 August 2023) [44] and then visualized using TBtools software version 2.096 [43].
2.7 Collinearity and Chromosome Mapping of WRKY Gene Family Members in V. bonariensis
To identify the pattern of gene duplication, One Step MCScanX from TBtools version 2.096 [43] with default parameters (E-value cut-off < 1 × 10−10 and num of BlastHits with 5) was used to analyze WRKY genes in V. bonariensis. The results were visualized using TBtools version 2.096. To assess the selection pressure of genes encoding WRKY proteins, the ratio of nonsyn-onymous (Ka)/synonymous (Ks) (Ka/Ks is an indicator of selective pressure) was used to evaluate its evolutionary pressure. The values of Ka, Ks, and Ka/Ks were calculated by simple Ka/Ks calculator in TBtools version 2.096 [43].
2.8 Differences in Expression of WRKY Gene Family Members in V. bonariensis
The expression level of V. bonariensis in different tissues and under cold stress was measured by our research group. Take materials from different tissues (flowers, leaves, old stems, and tender stems) of V. bonariensis for transcriptome sequencing. V. bonariensis was treated at low temperature of 4°C, and after 0 and 12 h, take a mixture of uncooled and 4°C cold treated young and old leaves for transcriptome sequencing. The expression levels (FPKM) were calculated with log2 (FPKM+1) [45] and visualized with TBtools software version 2.096 [43]. Analyze the expression changes of WRKY gene in the leaves of V. bonariensis in different tissues and under cold stress.
2.9 qRT‑PCR Analyses of Expression in Response to Cold Stress
Total RNA was extracted using the E.Z.N.A.®Plant RNA Kit (Omega, Xibao Biotech, Shanghai, China) following the manufacture’s instruction. The concentration of the isolated RNA samples were examined in a biophotometer (D30, Eppendorf, Germany). The gene-specific primers used in this study were designed by Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 24 April 2024), and VbActin was applied as reference gene. The first-strand cDNA was synthesized using the StarScript III RT Mix with gDNA Remover StarScript III (GenStar, Beijing, China). 2 × RealStar Fast SYBR qPCR Mix (GenStar, Beijing, China) reagents were used to detect the target sequence. Each PCR mixture (10 µL) contained 1 µL of cDNA, 5 µL of SYBR qPCR Mix, 0.5 µL of each primer (10 µM), and 3 µL of ddH2O. The qRT-PCR were performed using the following program: 95°C for 2 min and 40 cycles of 95°C for 15 s, 57°C for 30 s, and 72°C for 30 s. Each processing is repeated 3 times, relative gene expression was calculated with the 2−△△Ct method, and we used 0 h as an untreated control to calculate the fold change in the expression level of the relevant genes.
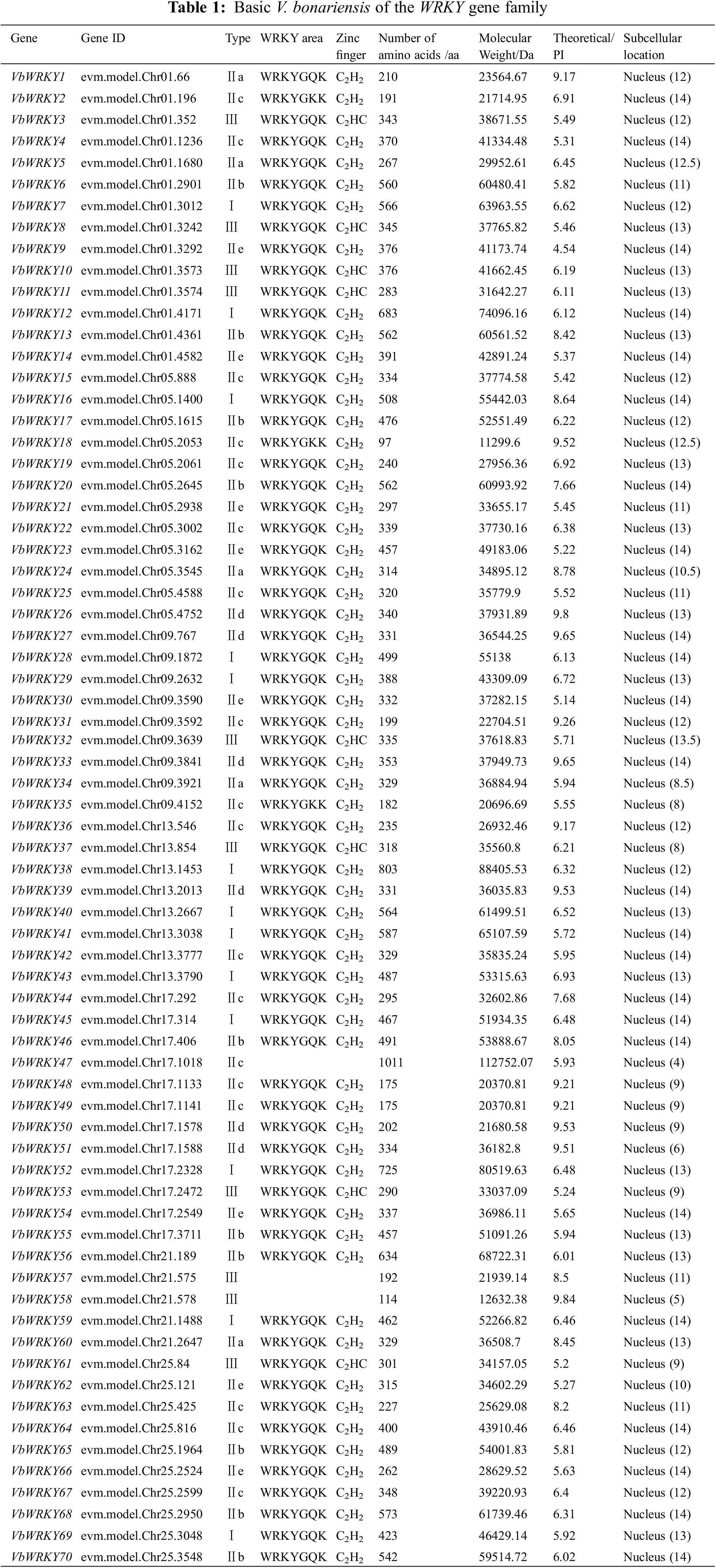
3.1 Identification and Physicochemical Property Analysis of VbWRKYs
Using the Hidden Markov model of the WRKY conserved domain in the Pfam database, HMMER3.0 was used to search for the genome protein sequence of V. bonariensis. Further conserved domain identification by NCBI-CDD and Pfam, finally, 70 members of the VbWRKY gene family were identified. Based on the arrangement order of these genes on chromosomes, they were renamed as VbWRKY1~VbWRKY70 (Table 1). The results of physicochemical property analysis showed that the length of V. bonariensis WRKY protein varied from 98 (VbWRKY18) to 1012 (VbWRKY47) amino acids, and the number of amino acids varied significantly; The relative molecular weight ranges from 11299.6 (VbWRKY18) to 112752.07 (VbWRKY47) Da; The isoelectric point is between 4.54 (VbWRKY9) and 9.84 (VbWRKY58). The predicted subcellular localization results showed that 70 WRKY members of V. bonariensis are located in the nucleus.
3.2 Chromosome Mapping of VbWRKYs
Using TBtools software version 2.096, the VbWRKY gene was mapped to V. bonariensis chromosomes. The results showed that the 70 V. bonariensis WRKY genes were distributed unevenly on all seven chromosomes. Among them, Chr1 chromosome is the most distributed, with 14, Chr2 and Chr5 take second place, each with 12 WRKY genes, Chr7 has 10 WRKY genes, there are 9 WRKY genes distributed on Chr3, 8 WRKY genes distributed on Chr4, and the least distributed on Chr6, with only 5 WRKY genes. According to Holub [46], chromosomal regions containing two or more genes within 200 KB can be defined as gene clusters. In V. bonariensis, a total of 16 VbWRKY genes are clustered into 8 gene clusters, marked in blue in the figure (Fig. 1). The chromosome distribution of gene cluster was irregular, except for Chr7 chromosome, the other 6 chromosomes all had gene cluster. Further analysis of tandem repeats revealed that only one pair of tandem repeats, VbWRKY10 and VbWRKY11, were located on Chr1.
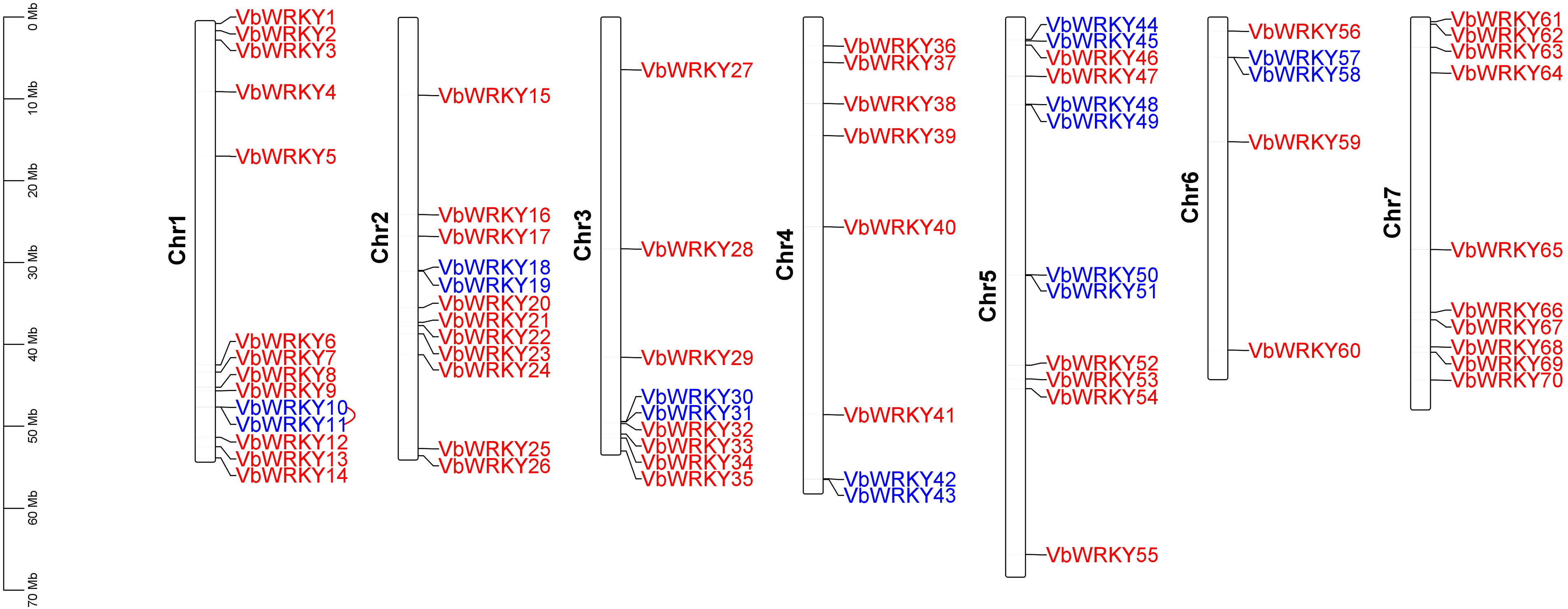
Figure 1: Chromosomal location of WRKY gene family members in V. bonariensis, gene clusters are indicated in blue
3.3 Phylogenetic Analysis and Multiple Sequence Alignment of VbWRKY Gene Family
Perform phylogenetic analysis on the identified 70 WRKY protein sequences of V. bonariensis and 72 known WRKY protein sequences in Arabidopsis. The results showed that the identified 70 V. bonariensis WRKY transcription factors could be classified into three major groups (Fig. 2), which are in line with the definitions of group I, group II, and group III in Arabidopsis by Mangelsen et al. [1]. There are 13 WRKY proteins belonging to group I, 47 WRKY proteins belonging to group II, and 10 WRKY proteins belonging to group III. WRKY members in group II can be further divided into five subclasses: IIa, IIb, IIc, IId, and IIe, with 5, 10, 18, 6, and 8 members. According to evolutionary relationships, the WRKY family II of higher plants can be divided into three subclasses: IIa+IIb, IIc, and IId+IIe, which is consistent with the results presented by the phylogenetic tree.
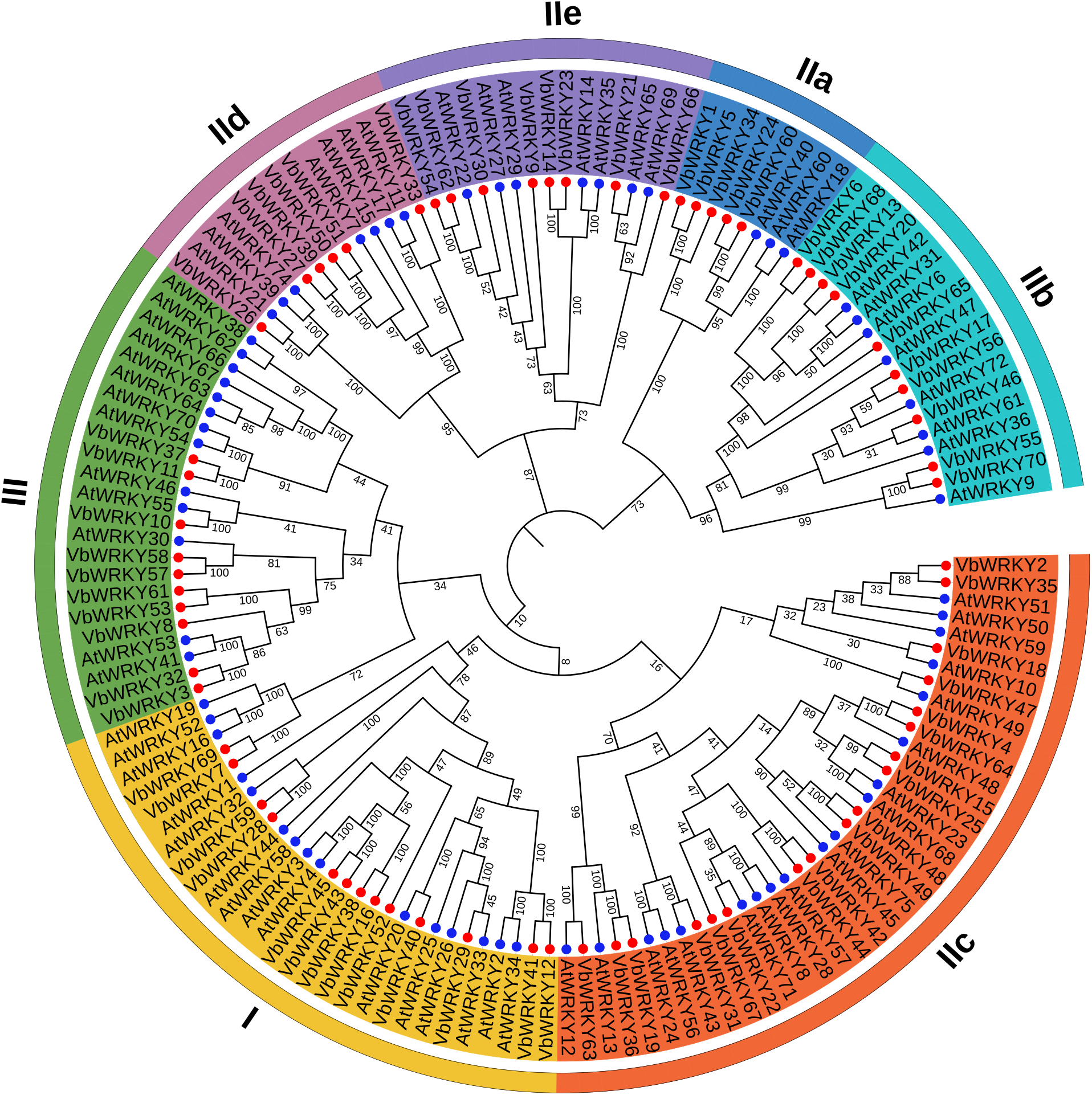
Figure 2: Phylogenetic tree of WRKY gene family members in V. bonariensis and Arabidopsis
The WRKY domain protein sequence of the V. bonariensis WRKY gene was analyzed by DNAMAN sequence analysis software (Fig. 3, Table 1), The results showed that group I contained two WRKY domains, located at the N and C ends of the sequence, including a WRKYGQK sequence and a C2H2-like zinc finger motif, with VbWRKY38 and VbWRKY69 having only one WRKY domain. Group II contained one WRKY domain and a C2H2-like zinc finger motif. Among them, VbWRKY47 of IIc has no conserved WRKY domain and zinc finger motif, VbWRKY18 has a missing zinc finger motif, and WRKYGQK at the conserved sites of VbWRKY2, VbWRKY18, and VbWRKY35 has become WRKYGKK. Group III had a WRKYGQK sequence and the C2HC zinc finger motif. Among them, VbWRKY57 and VbWRKY58 have no conserved WRKY domain and zinc finger motif.
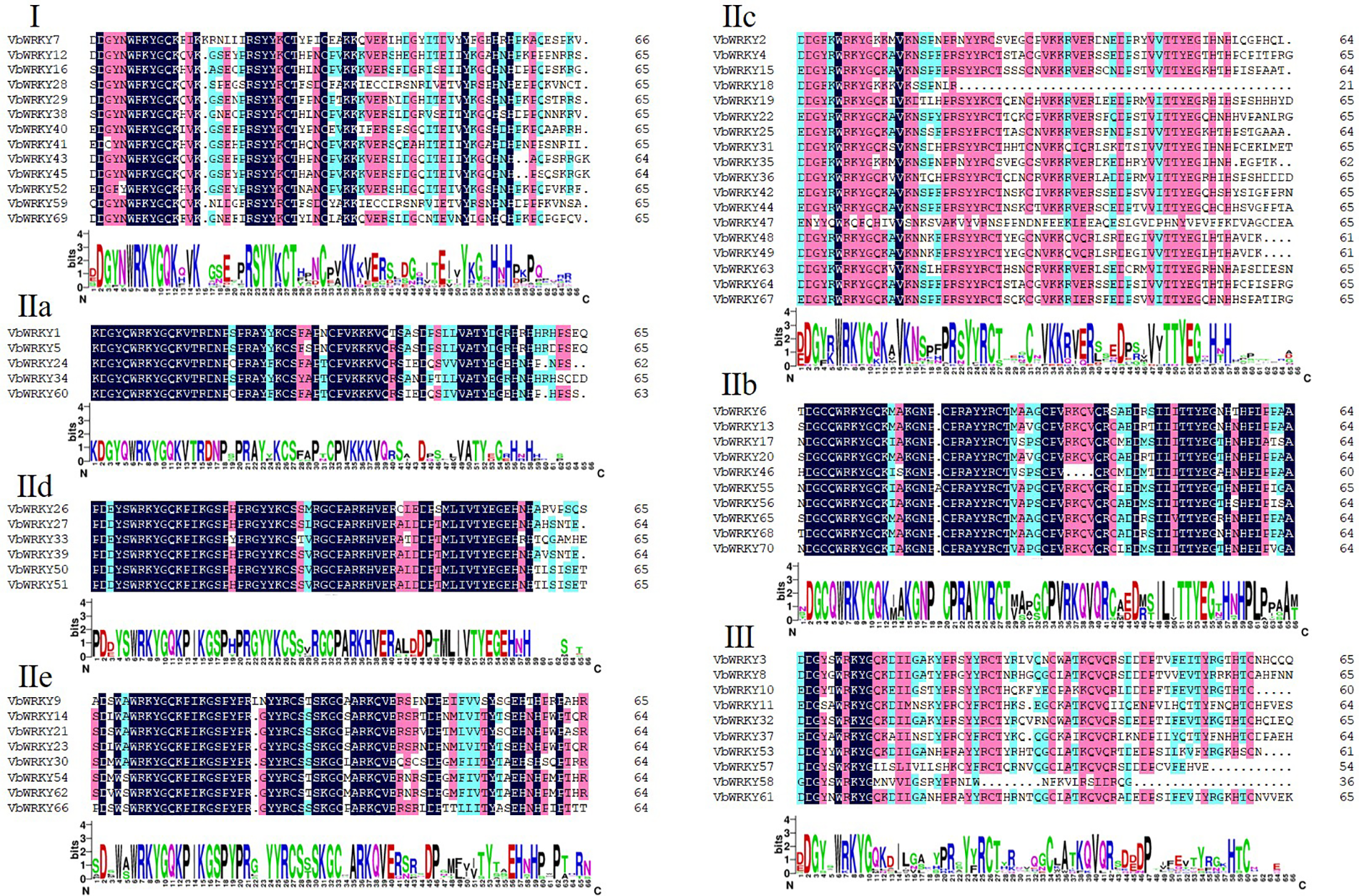
Figure 3: Multiple sequence alignment of VbWRKY family members
3.4 Gene Structure and Conserved Motif Analysis of WRKY Family in V. bonariensis
Using MEME online software to analyze 70 VbWRKY protein sequences, 10 conserved motifs were found (Fig. 4). The frequency of motif occurrence in the WRKY protein determines its importance in the sequence. Among them, motif 1 and motif 8 are conserved sequences of the WRKYGQK heptapeptide segment, motif 2 and motif 5 are zinc finger structural motifs. Motif 1 and motif 2 constitute the C-terminal WRKY box, motif 5 and otimf 8 form the N-terminal WRKY box (Fig. 5). Group I has two WRKY boxes, with motif 1 and motif 2, motif 5 and motif 8, while the other groups only have N-terminal WRKY boxes, which consists of only motif 1 and motif 2, without motif 5 and motif 8. Motif 1, motif 2, and motif 3 are landmark conserved motifs of the VbWRKYs protein in V. bonariensis, with most (64/70) having motif 1, 2, and 3. In general, different subclasses contain different motifs, and the motif elements in the same subclass are roughly the same. Genes with the same motif elements have similar biological functions. The conservative elements in subclasses IIa and IIb, IId and IIe are roughly similar, which also proves that in higher plants, IIa and IIb can be classified into the same subclass, while IId and IIe can be classified into the same subclass. Motif 6 and motif 7 are specifically present in group IIa+b; motif 9 is a unique conservative element in group IIb; motif 10 is a unique conservative element of group I. The number and types of various motifs are relatively fixed, consistent with the phylogenetic tree results (Fig. 2) and identification classification results (Table 1).
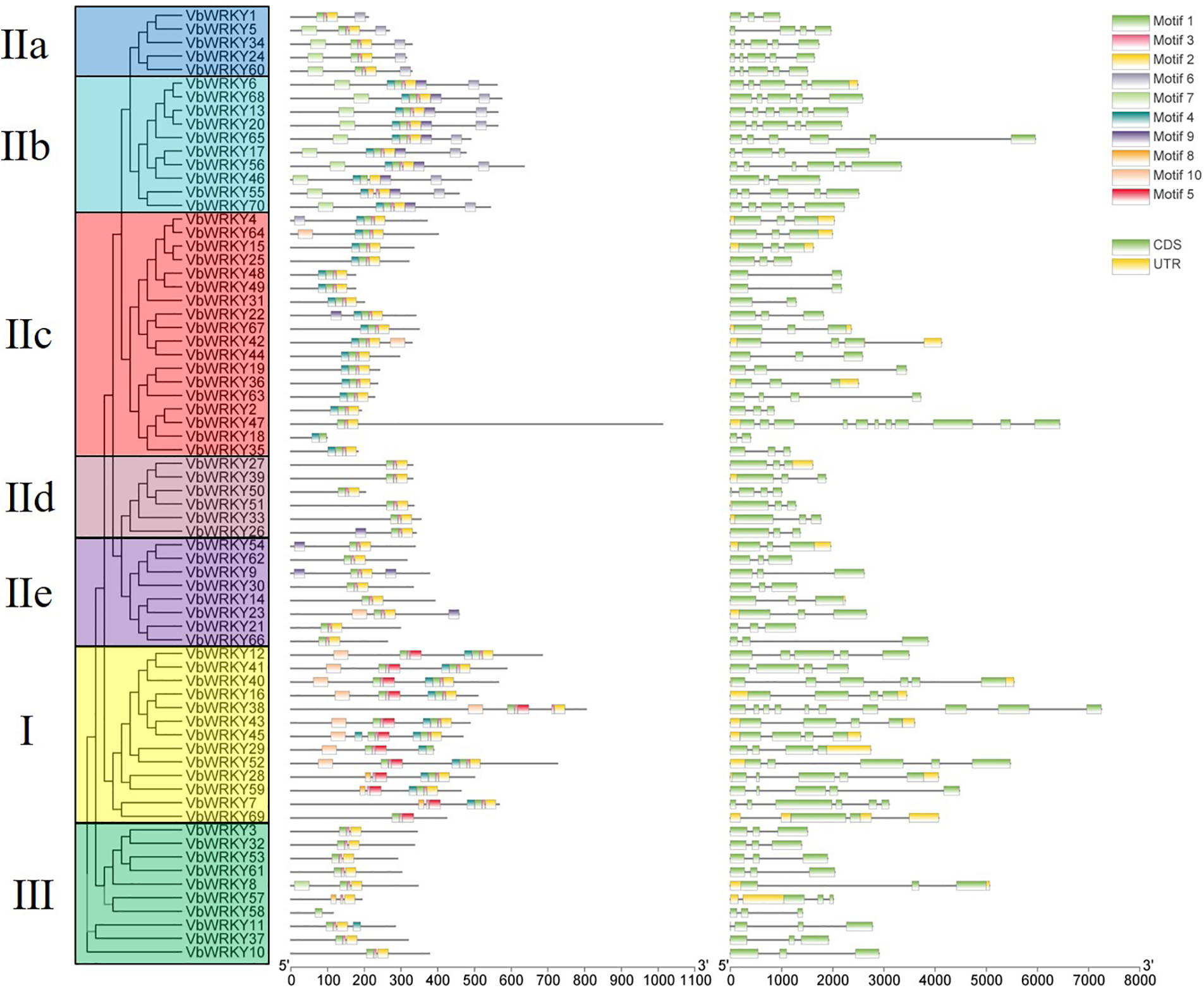
Figure 4: Evolutionary relationship, gene structure, and distribution of conservative motifs of the WRKY gene family in V. bonariensis
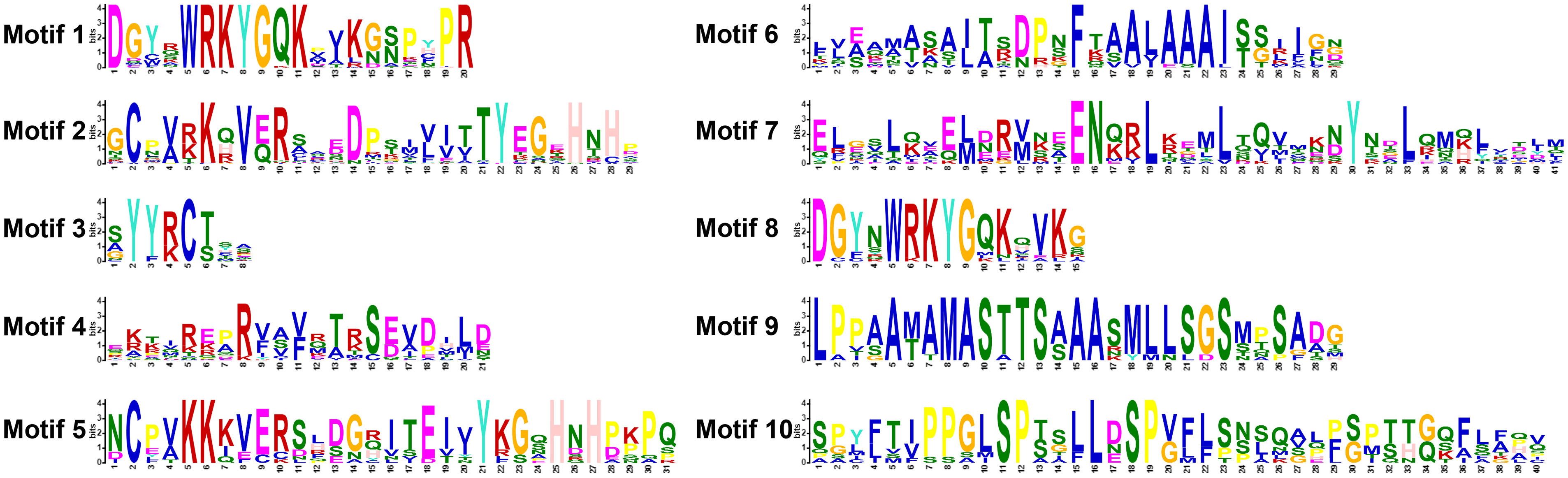
Figure 5: Conservative motif of WRKY gene family members in V. bonariensis
Gene structure analysis showed that most V. bonariensis WRKY genes had 2–6 exons, except that VbWRKY38 and VbWRKY47 had a large number of exons and introns (VbWRKY38 had 10 exons and VbWRKY47 had 11 exons) (Fig. 4). Among them, the number of genes with 3 exons (2 introns) was the most, accounting for 54% (38/70) of all VbWRKY genes, and the number of genes with 2 and 6 exons was the least, with 5 each. In addition, 11 VbWRKY genes contained 4 exons and 9 VbWRKY genes contained 5 exons. All members of group III, IId, and IIe contain 3 exons. group I contains 2–6 exons, and group IIa and IIb contain 3–6 exons, group IIc contains 2–4 exons. The UTR distribution pattern showed that there were 25 VbWRKY genes with UTR region, 7 genes only with 5′ UTR region and 5 genes only with 3′ UTR region, a total of 13 genes contained both 5′ UTR and 3′ UTR regions.
3.5 Cis-Acting Elements Analysis of WRKY Gene Family Members in V. bonariensis
In order to further understand the transcriptional regulation and potential function of VbWRKY, PlantCARE was used to predict the cis-acting elements of the VbWRKY promoter. The results showed that in addition to promoter related elements and WRKY binding site elements, three types of cis-regulatory elements were found to be highly concentrated in the promoter region of VbWRKYs, including light-responsive elements, plant hormone response elements, and environmental stress response related elements (Fig. 6). Among them, The cis-acting elements related to environmental stress response include low-temperature (LTR), drought (MBS), trauma (WUN motif), defense and stress (Tc-rich repeats), and anaerobic induction (ARE) response elements. The number of light-responsive elements is the highest, including Box4, G-box, etc., and 70 VbWRKY genes all contain light-responsive elements. Plant hormone responsive elements include methyl jasmonate responsive elements (CGTCA-Motif and TGACG-Motif), abscisic acid responsive element (ABRE), auxin responsive elements (TGA-element and AuxRR-core), salicylic acid responsive element (MBS), gibberellin responsive element (p-box and TATC-box) and ethylene responsive element (ERE). Specifically, ABRE elements are commonly present in 61 VbWRKY promoters, and 30 VbWRKY genes contain low-temperature responsive elements. These results indicate that most of the cis-acting elements of VbWRKY are related to stress, The analysis of cis-acting elements in the VbWRKY gene family may help to understand the stress response of V. bonariensis, especially under low temperature stress.

Figure 6: Cis-acting elements of the V. bonariensis WRKY gene family
3.6 Collinearity Analysis of WRKY Gene Family Members in V. bonariensis
To explore the conservatism of VbWRKY gene family members during evolution, collinearity analysis of VbWRKY gene family players was carried out. It was found that 44 pairs of highlighted homologous genes existed among the VbWRKY gene family members (Fig. 7). In addition, Ka/Ks values were calculated for all members in each group (Table S1) and found that the vast majority of gene pairs with Ka/Ks values were NaN, this may be due to synonymous mutations in most sites where synonymous mutations can occur [47]. There were no significant differences in Ka/Ks values between the subgroups, all of which were less than 1, indicating strong purification selection of these VbWRKY gene pairs. Among 44 pairs of homologous genes, the Ka/Ks values of VbWRKY3/VbWRKY8, VbWRKY8/VbWRKY32, VbWRKY9/VbWRKY30, VbWRKY10/VbWRKY37, VbWRKY22/VbWRKY67, VbWRKY30/VbWRKY62, VbWRKY8/VbWRKY53, VbWRKY9/VbWRKY54 and VbWRKY7/VbWRKY69, were NaN, the value of Ka/Ks of the other 35 pairs of homologous genes was less than 1, which indicates that the 35 pairs of homologous genes evolved under great purifying selection or negative selection pressure [48].
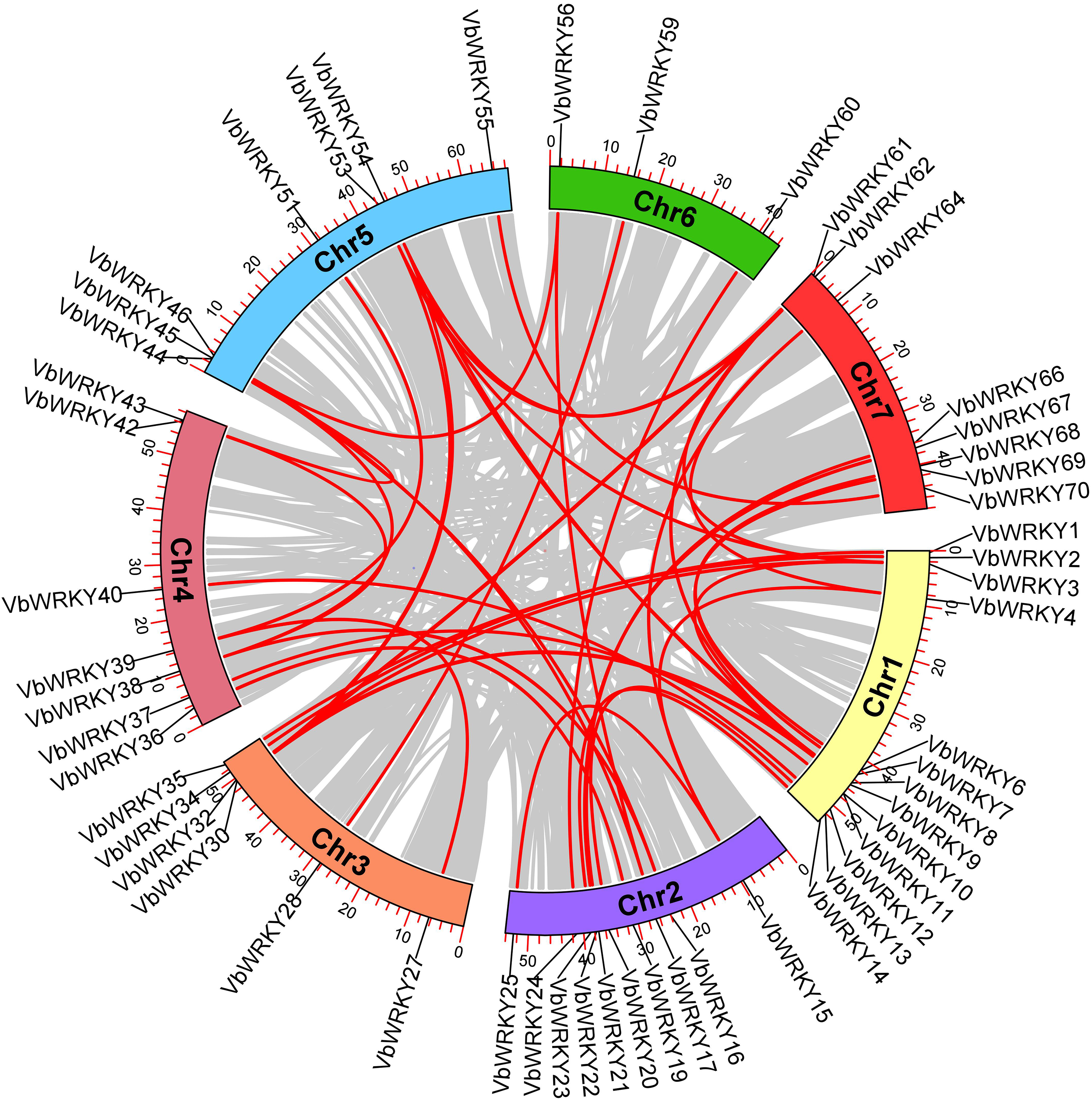
Figure 7: Collinearity relationship of WRKY gene family members in V. bonariensis. The red lines refer to collinear gene pairs
3.7 Differences in Expression of WRKY Gene Family Members in V. bonariensis
To further explore the regulation of VbWRKYs on growth and low temperature stress, transcriptome data (FPKM) of four tissues (flower, leaf, old stem, tender stem) and cold-stressed plants obtained from the V. bonariensis database were calculated and visualized (Figs. 8 and 9). The research found that, except for 7 genes such as VbWRKY6 and VbWRKY17, which are not expressed in different tissues of V. bonariensis, there are differences in the expression levels of the remaining 63 VbWRKY genes in different tissues of V. bonariensis (Fig. 8). VbWRKY33, VbWRKY38, VbWRKY39 and other 17 genes had high expressed in almost all tissues. The expression of 30 genes, including VbWRKY7, VbWRKY23, and VbWRKY70 was very low in almost all tissues. Sixteen genes, including VbWRKY2, VbWRKY24, and VbWRKY52 had highly expressed only in a single tissue. Among them, four genes such as VbWRKY16, VbWRKY25 had highly expressed in flower, and six genes such as VbWRKY13, VbWRK53 had highly expressed in leaf, six genes such as VbWRKY11 and VbWRKY14 had highly expressed in stems.
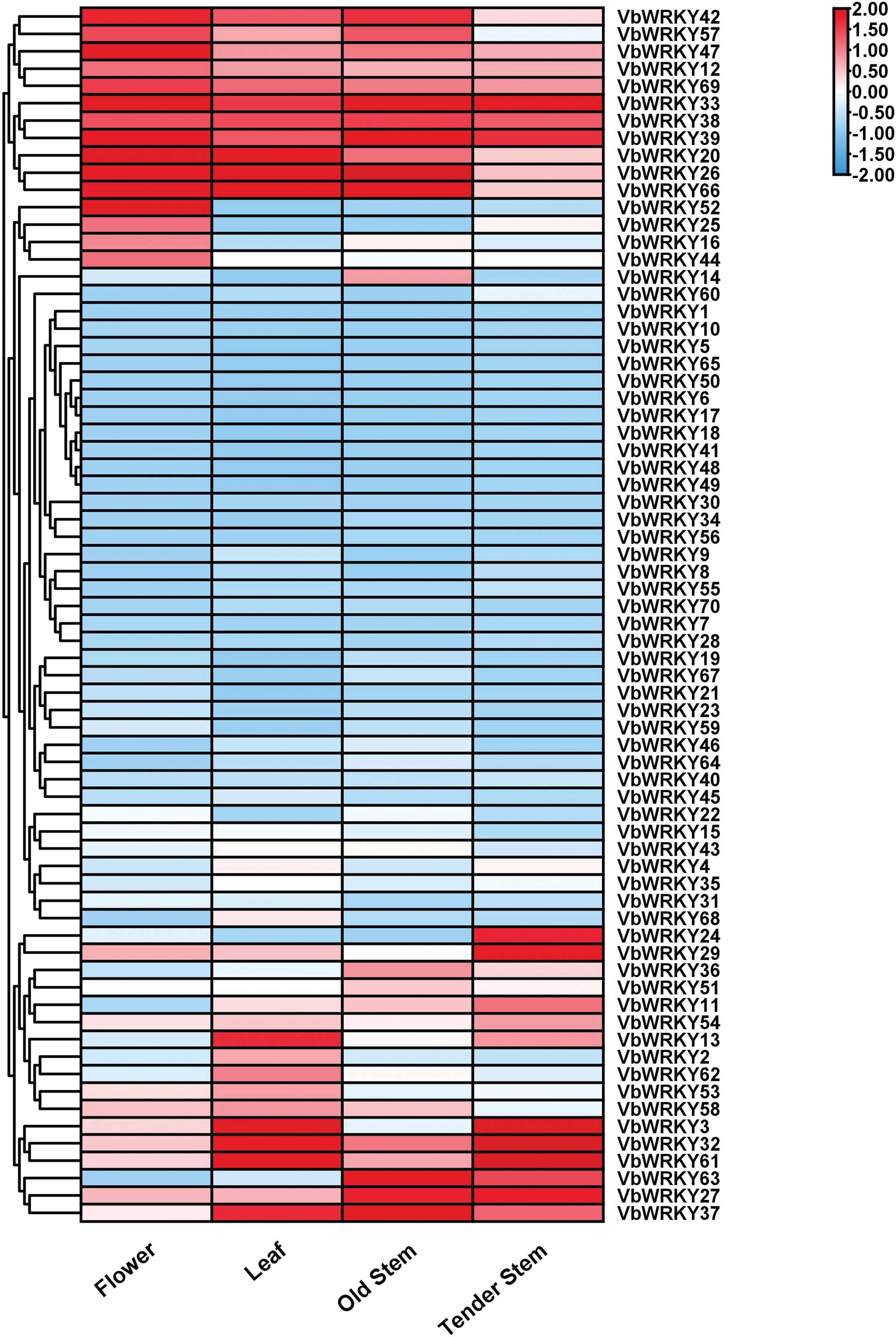
Figure 8: Expression difference of WRKY gene family members in different region of V. bonariensis
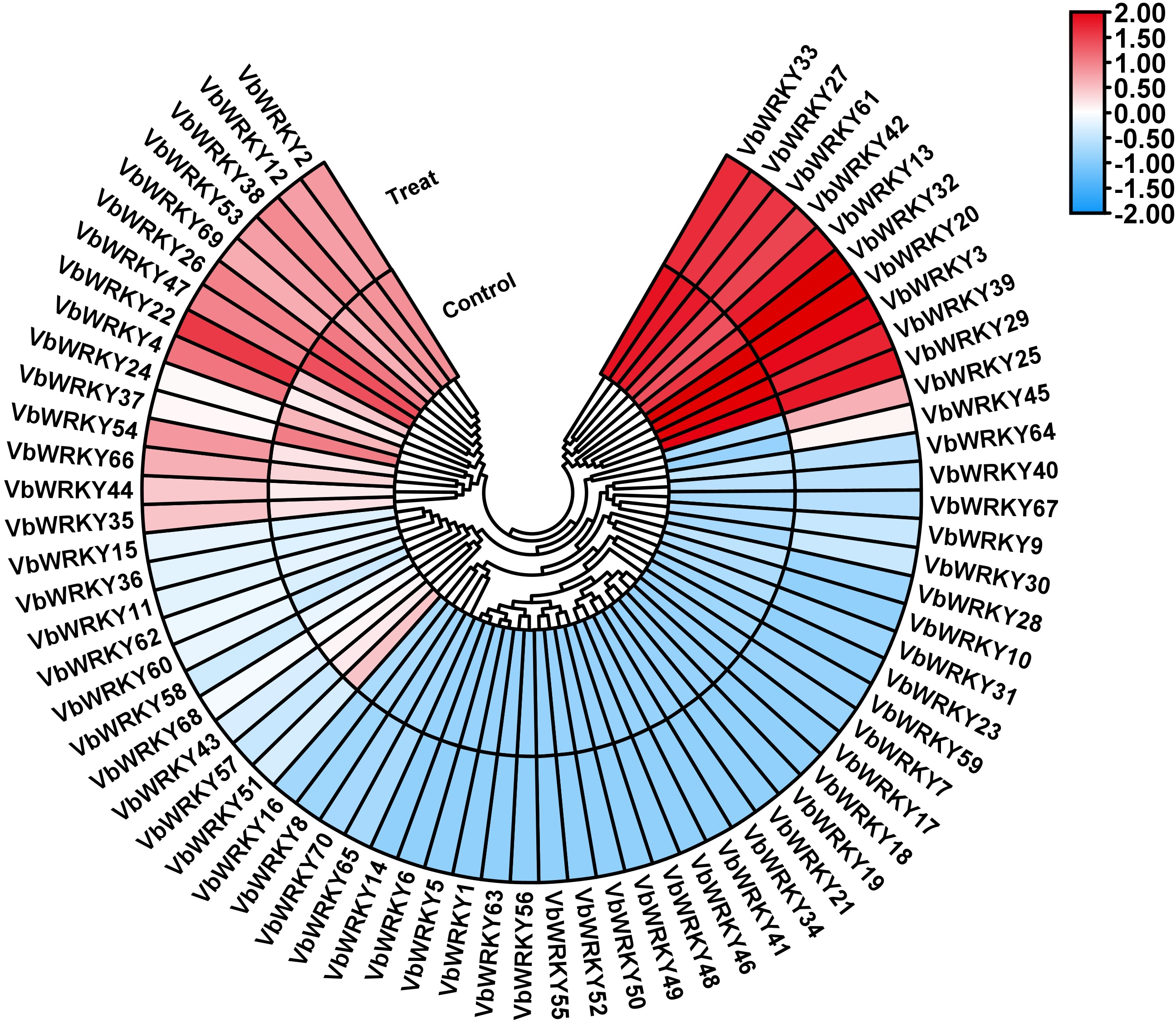
Figure 9: Differences in the expression of WRKY gene family members in V. bonariensis under cold stress. Control, before cold treatment; Treat, after cold treatment
The results showed that 70 VbWRKYs genes were expressed in different degrees under low temperature stress, and they were divided into three types according to their expression patterns under low temperature stress (Fig. 9): the first type includes 37 genes, including VbWRKY2, VbWRKY9, VbWRKY25, VbWRKY40, VbWRKY67, etc., which show increased expression under cold stress, of which 17 are significantly upregulated, with VbWRKY25 and VbWRKY45 being the most significantly upregulated. The second type includes 15 genes, including VbWRKY8, VbWRKY23, VbWRKY31, VbWRKY47, VbWRKY51, etc., which show a decrease in expression under cold stress. The third type includes 18 genes, including VbWRKY1, VbWRKY14, VbWRKY21, VbWRKY48, and VbWRKY63, which exhibit no response under cold stress. 17 genes with upregulated expression under cold stress all possessed cis-acting elements (LTR, W-box and ABRE) related to cold stress, indicating that some VbWRKY members may participate in the process of resistance to cold stress.
3.8 Expression Analysis of VbWRKY Genes under Cold Stress
In order to examine the expression patterns of VbWRKY genes potentially associated with responses to low temperatures, 9 VbWRKY genes from 17 that were significantly upregulated under cold stress were selected and surveyed for their expression levels during different stages of induced low-temperature stress (4°C) (0, 3, 6, 9, 12 and 24 h) (Fig. 10). Under cold treatment, 9 VbWRKYs were induced to present the significant up-regulation at different time points. The highest expression levels in the majority of selected VbWRKY genes (VbWRKY4, VbWRKY9, VbWRKY22, VbWRKY30, VbWRKY45, VbWRKY70) were found after exposure to low temperature for 24 h. The expression of VbWRKY13 was the highest levels at 3 h. The expression of VbWRKY25 was the highest levels at 12 h and the expression of VbWRKY54 was the highest levels at 9 h.
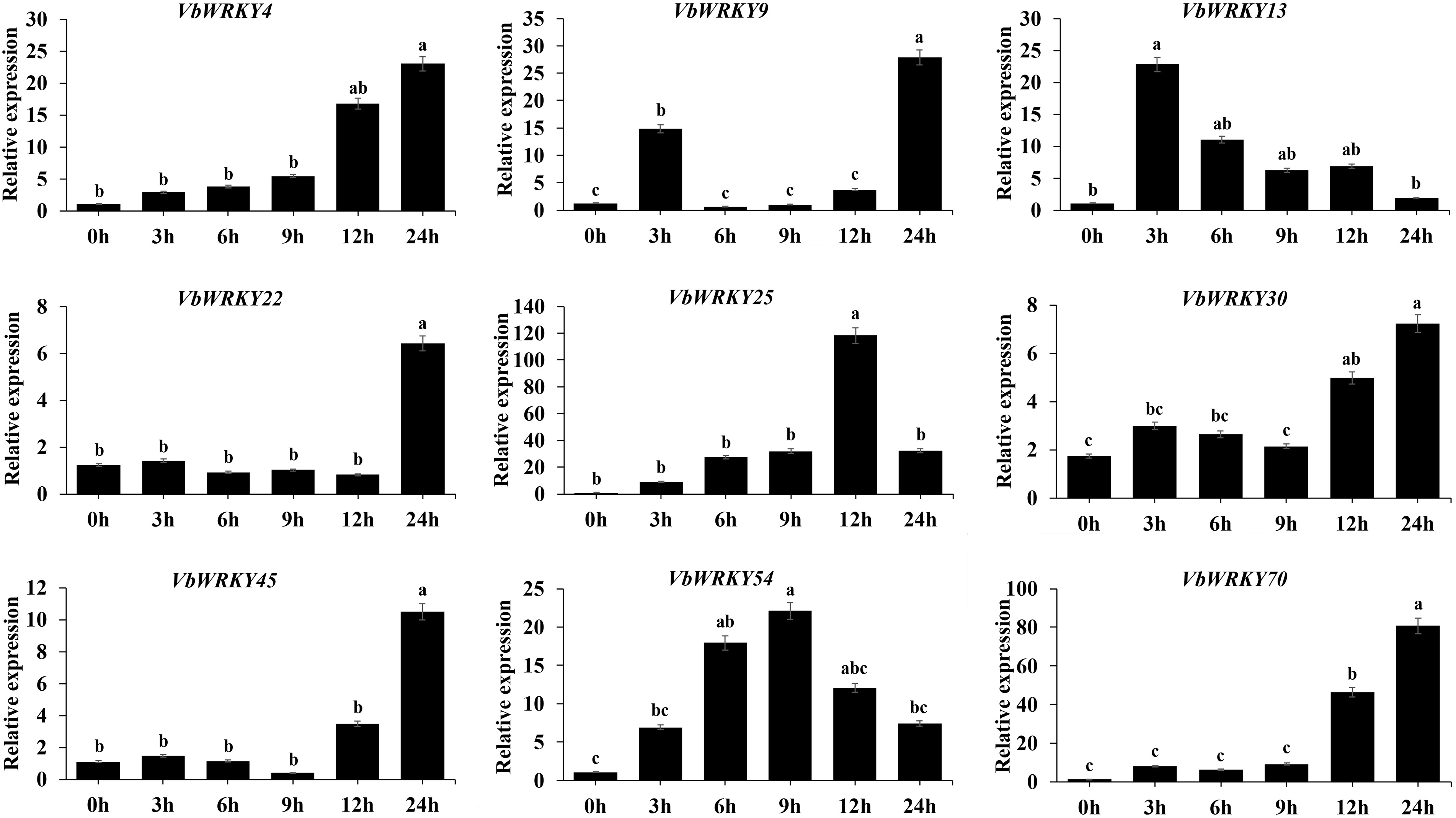
Figure 10: Expression profiles of 9 VbWRKY genes under cold treatment. Gene expression of these VbWRK genes was analyzed by qRT-PCR. Data represents the mean ± SD of three technical repetition. 0 h was used as an untreated control to calculate the fold change in the expression level of the relevant genes (p < 0.05, n = 3)
The WRKY transcription factor family is a class of transcription factors that are specific to plants and regulate gene expression to participate in various signaling pathways in response to biotic and abiotic stresses [49]. With the continuous completion of genome sequencing for different species, the WRKY gene family has been identified in multiple species. Including 72 in Arabidopsis [22], 102 in rice [50], 104 in tomato [51], 139 in Apple [52], 61 in cucumber [53], 89 in Camellia oleifera [54], 59 in grape [55] and 116 in cotton [56]. In this study, 70 VbWRKY members were identified from the genome of V. bonariensis.
In this study, 70 members of the VbWRKY family were divided into three groups (I, II, and III) based on phylogenetic evolution. Group II was further divided into 5 subfamilies. VbWRKY38 and VbWRKY69 in group I lost one WRKY domain during evolution. This phenomenon of WRKY domains loss in group I members is also present in the Arabidopsis genome, for example, AtWRKY10 only contains one WRKY domain [17]. There is a phenomenon of loss and variation of structural domains and zinc finger motifs in groups II and III, such as VbWRKY47 in group IIc and VbWRKY57 and in Group III. Related studies have shown that members of Group II and III evolved from the loss or variation of C-terminus and N-terminus domains and zinc finger motifs in group I during plant evolution [57]. Thus, loss of the VbWRKY47, VbWRKY57, and VbWRKY58 domains may have contributed to the expansion of the V. bonariensis VbWRKY gene family. GmWRKY6 and GmWRKY21, which contain WRKYGKK variants in soybeans, fail to bind W-box normally [58]. The NtWRKY12 of tobacco with the WRKYGKK variant recognizes another binding sequence instead of the normal W-box [59]. In the V. bonariensis, the conserved amino acid motif WRKYGQK in the VbWRKY2, VbWRKY18, and VbWRKY35 encoded proteins of group IIc was altered to WRKYGKK. It is speculated that the WRKY heptapeptide domain variants of V. bonariensis may cause the WRKY protein to lose its ability to recognize and bind to DNA, or to recognize other new motifs and generate new functions. The function of zinc finger motifs is equivalent to chelating agents, and the lack of zinc finger motifs can reduce W-box binding ability or generate new biological functions [60]. The zinc finger motifs have been lost or mutated in VbWRKY18, VbWRKY57, and VbWRKY58 in V. bonariensis, possibly resulting in the loss of their original binding domain function and the emergence of new biological functions.
Gene family members can be distributed in clusters on one chromosome or on different chromosomes. By observing the positions of gene family members on chromosomes, we can judge whether the genes are clustered on the chromosomes. In this study, WRKY members are distributed unevenly on the chromosome, which is consistent with the results of Xu et al. [61] and Mu et al. [62]. At the same time, the analysis of introns, exons and conserved motifs can provide important evidence for further understanding of gene evolution. Gene structure analysis showed that most of the VbWRKY genes (38/70) contained two introns, similar to the reported in cassava (42/85), cucumber (29/61) and maize (78/140) [63,64]. Previous studies have shown that members of the WRKY family have diverse functions in plant growth and various stress responses, but the functions in WRKY genes within the same group or subgroup usually remain similar [65]. The conserved motif distribution pattern is the main basis for the classification of gene family members. Motif analysis found that the VbWRKY genes in the same subgroup had similar conserved motif distribution patterns, while there were differences in the conservative motif distribution patterns of VbWRKY genes in different subgroups. it is inferred that VbWRKY gene has similar or different biological functions due to the difference of conserved motif distribution patterns. Almost all VbWRKYs contain motif 1 and motif 2, with motif 1 being the conserved seven peptide sequence of WRKYGQK and motif 2 being the zinc finger structural motif, which may have been retained as core elements during evolution. The conserved motifs of the IId and IIe subgroups are similar, which may indicate that they established genetic relationships through evolution. This research result is similar to the conservative motif analysis of SsWRKYs genes by Mu et al. [62] and the conservative motif analysis of GhWRKYs genes by Ehsan et al. [66].
The analysis of cis-acting elements shows that the promoter region of the VbWRKY gene in V. bonariensis is rich in various cis-acting elements, such as light responsive elements, hormone responsive elements and stress responsive elements. It is speculated that the VbWRKY gene may be activated and expressed under light signals, hormones, and biotic or abiotic stress, thereby directly or indirectly regulating various biological processes in plants. Gene duplication events played prominent roles in a succession of genomic rearrangements and expansions, and it is also the main motivation of plants evolution [67]. The gene family expansion occurs via three mechanisms: TDs, SDs and transposition events [68]. Gene duplication was found to play a very important role in the expansion of the WRKY gene family. In V. bonariensis, a total of 44 segmental duplication events and a pair of tandem repeat genes are identified in VbWRKYs. Moreover, for all pairs, the Ka/Ks ratios are <1, indicating that the WRKY gene family in V. bonariensis has undergone purifying selection, providing impetus for the evolution of V. bonariensis.
It has been found that the WRKY gene family is constitutively expressed in many plants. In Salix suchowensis, Bi et al. found that some WRKY family genes were expressed in different parts of plants [69]. In tea plant, Pengjie Wang et al. also found that some WRKY genes were expressed in different parts of plants [70]. In this study, we analyzed the expression patterns of 70 VbWRKY genes in four tissues (flower, leaf, old stem and tender stem). The results showed that 17 VbWRKY genes were highly expressed, these 17 genes are presumed to regulate the entire growth and development of V. bonariensis. The expression levels of 16 VbWRKY genes in only one tissue were high, among which four genes, VbWRKY25 and VbWRKY52, were highly expressed in the flowers of V. bonariensis. It is speculated that they are involved in the growth and development process of V. bonariensis flowers; 6 genes, including VbWRKY2 and VbWRKY62, have high expression levels in the leaves, suggesting their involvement in the growth and development process of V. bonariensis leaves.
The WRKY gene family is especially associated with responses to biotic and abiotic stress. Previous studies have shown that WRKY genes function in response to cold stress in many plants. For example, experiments by Qiu et al. [71] showed that 10 OsWRKYs in rice were rapidly induced to express under abiotic stresses such as NaCl, drought, low temperature (4°C) and high temperature (42°C). Holub [46] found that eight TaWRKYs genes in wheat (Triticum aestivum) had rapid responses to low temperature, high salinity, drought and high temperature. Du et al.’s research results showed that the expression levels of most KoWRKY genes, especially KoWRKY16, KoWRKY28, KoWRKY32, KoWRKY43, KoWRKY45, and KoWRKY55, etc., were upregulated after freezing treatment, and their expression increased by more than twice. It was also verified that the expression levels of these 9 genes were upregulated under 4°C low temperature stress [72]. In this study, we investigated the expression patterns of genes in V. bonariensis leaves under low temperature stress, which is a common abiotic stress in the production and cultivation of V. bonariensis, a total of 52 VbWRKY genes were involved in the response to cold stress, 17 (VbWRKY45, VbWRKY25, VbWRKY32, VbWRKY22, VbWRKY4, etc.) of which were significantly up-regulated. All of these 17 genes had cis-acting elements related to cold stress, which further confirmed that these 17 genes were induced by low temperature. 9 VbWRKY genes were surveyed for their response to cold-temperature stress in leaf tissue. The results showed that all had altered expression throughout the experiment, and that 9 VbWRKYs were induced to present the significant up-regulation at different time points. The highest expression levels in the majority of selected VbWRKY genes (VbWRKY4, VbWRKY9, VbWRKY22, VbWRKY30, VbWRKY45, VbWRKY70) were found after exposure to low temperature for 24 h. The expression of VbWRKY13 was the highest levels at 3 h. The expression of VbWRKY25 was the highest levels at 12 h and the expression of VbWRKY54 was the highest levels at 9 h, indicating that these VbWRKY transcription factors may function variously at different periods of the stress response. Although WRKYs have been observed to function in many plants in response to low temperature, the mechanism of how WRKYs respond to cold signals and regulate the expression of downstream genes remains largely unknown. Further research is required to demonstrate the function of these genes in relation to low temperatures and their involvement in cold signal pathways. The function of VbWRKY can be verified through experiments such as cis-acting elements and transgenic experiments to determine its role.
This study utilized the genome data of V. bonariensis previously measured by the research team to identify the members of the WRKY gene family of V. bonariensis, and analyzed their phylogeny, subgroup types, chromosome distribution, conserved motifs, gene structure, cis acting elements, and transcriptome data in different tissues and under low temperature stress, furthermore, 9 VbWRKYs under cold stress were analyzed by qRT-PCR, it provides a basis for further study on the roles of WRKY transcription factors in plant growth and development and in response to low temperature stress, and provides a basis for further study on the functions of V. bonariensis WRKY transcription factor family.
This study identified a total of 70 members of the WRKY gene family in V. bonariensis. By analyzing various bioinformatics information such as the WRKY domain, evolutionary relationships, gene structure, conserved motifs, chromosome distribution, and cis-acting element distribution of the WRKY gene family in V. bonariensis, a comprehensive and systematic identification of the WRKY gene family was conducted. Its structure is highly conservative and participates in many aspects of V. bonariensis growth and development. Analysis of transcriptome data from V. bonariensis under low-temperature stress revealed that some VbWRKY genes can respond to low-temperature stress and exhibit a positive regulatory effect. Expression profiling revealed that most VbWRKY genes were found to have a positive or negative response to cold stresses, and their response changed with the degree of stress. The results of this study provide a theoretical basis for the functional study of the WRKY gene in cold stress and the growth and development of V. bonariensis, and may also contribute to the screening of cold resistance.
Acknowledgement: The authors sincerely thank the anonymous reviewers and editor who made valuable comments on this paper.
Funding Statement: This research was funded by the National Natural Science Foundation of China (32160722) and the Key Research Project of Guizhou Provincial Science and Technology Projects (QKHJC-ZK [2023]ZD-006).
Author Contributions: Study conception and design: Yan Li; data collection: Tao Zhang, Sisi Wang, Xiuliu Yang; analysis and interpretation of results: Dandan Yuan, Ju Cai; draft manuscript preparation: Dandan Yuan. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.052190.
References
1. Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K, Jansson C, et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 2008;9(1):1–17. [Google Scholar]
2. Wang J, Lin Y, Yang J, Zhang Q, Liu M, Hu Y, et al. Solution structure of the DNA binding domain of Arabidopsis transcription factor WRKY11. Biochem Biophys Res Commun. 2023;653:133–9. doi:10.1016/j.bbrc.2023.02.072. [Google Scholar] [PubMed] [CrossRef]
3. Cheng X, Zhao Y, Jiang Q, Yang J, Zhao W, Taylor IA, et al. Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain. Nucleic Acids Res. 2019;47(8):4308–18. doi:10.1093/nar/gkz113. [Google Scholar] [PubMed] [CrossRef]
4. Li H, Xu Y, Xiao Y, Zhu Z, Xie X, Zhao H, et al. Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta. 2010;232:1325–37. doi:10.1007/s00425-010-1258-y. [Google Scholar] [PubMed] [CrossRef]
5. Chanwala J, Satpati S, Dixit A, Parida A, Giri MK, Dey N. Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genom. 2020;21(1):231. doi:10.1186/s12864-020-6622-0. [Google Scholar] [PubMed] [CrossRef]
6. Schluttenhofer C, Yuan L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015;167(2):295–306. doi:10.1104/pp.114.251769. [Google Scholar] [PubMed] [CrossRef]
7. Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet. 1994;244:563–71. doi:10.1007/BF00282746. [Google Scholar] [PubMed] [CrossRef]
8. Dellagi A, Birch PR, Heilbronn J, Avrova AO, Montesano M, Palva ET, et al. A potato gene, erg-1, is rapidly induced by Erwinia carotovora ssp. atroseptica, Phytophthora infestans, ethylene and salicylic acid. J Plant Physiol. 2000;157(2):201–5. doi:10.1016/S0176-1617(00)80191-1. [Google Scholar] [CrossRef]
9. Yoda H, Ogawa M, Yamaguchi Y, Koizumi N, Kusano T, Sano H. Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol Genet Genom. 2002;267:154–61. doi:10.1007/s00438-002-0651-z. [Google Scholar] [PubMed] [CrossRef]
10. Sun C, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15(9):2076–92. doi:10.1105/tpc.014597. [Google Scholar] [PubMed] [CrossRef]
11. Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010;10(1):1–15. [Google Scholar]
12. Hwang SH, Yie SW, Hwang DJ. Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 2011;181(3):316–23. doi:10.1016/j.plantsci.2011.06.007. [Google Scholar] [PubMed] [CrossRef]
13. Zheng Z, Qamar SA, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48(4):592–605. doi:10.1111/tpj.2006.48.issue-4. [Google Scholar] [CrossRef]
14. Jeyasri R, Muthuramalingam P, Satish L, Adarshan S, Lakshmi MA, Pandian SK, et al. The role of OsWRKY genes in rice when faced with single and multiple abiotic stresses. Agronomy. 2021;11(7):1301. doi:10.3390/agronomy11071301. [Google Scholar] [CrossRef]
15. Levee V, Major I, Levasseur C, Tremblay L, MacKay J, Seguin A. Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 2009;184(1):48–70. doi:10.1111/nph.2009.184.issue-1. [Google Scholar] [CrossRef]
16. Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009;9:1–19. [Google Scholar]
17. Ling J, Jiang W, Zhang Y, Yu H, Mao Z, Gu X, et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011;12:1–20. [Google Scholar]
18. Ding M, Chen J, Jiang Y, Lin L, Cao Y, Wang M, et al. Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol Genet Genom. 2015;290:151–71. doi:10.1007/s00438-014-0904-7. [Google Scholar] [PubMed] [CrossRef]
19. Phukan UJ, Jeena GS, Shukla RK. WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci. 2016;7:760. [Google Scholar] [PubMed]
20. Ye J, Wang X, Hu T, Zhang F, Wang B, Li C, et al. An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell. 2017;29(9):2249–68. doi:10.1105/tpc.17.00211. [Google Scholar] [PubMed] [CrossRef]
21. Banerjee A, Roychoudhury A. WRKY proteins: signaling and regulation of expression during abiotic stress responses. Scientif World J. 2015;2015:807560. [Google Scholar]
22. Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–58. doi:10.1016/j.tplants.2010.02.006. [Google Scholar] [PubMed] [CrossRef]
23. Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7(5):491–8. doi:10.1016/j.pbi.2004.07.012. [Google Scholar] [PubMed] [CrossRef]
24. Chu X, Wang C, Chen X, Lu W, Li H, Wang X, et al. The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One. 2015;10(11):e0143022. doi:10.1371/journal.pone.0143022. [Google Scholar] [PubMed] [CrossRef]
25. Duan GF, Li LG, Liu QL. A WRKY transcription factor from Malus domestica negatively regulates dehydration stress in transgenic Arabidopsis. Acta Physiol Plant. 2014;36:541–8. doi:10.1007/s11738-013-1434-3. [Google Scholar] [CrossRef]
26. Zhang Y, Yu H, Yang X, Li Q, Ling J, Wang H, et al. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol Bioch. 2016;108:478–87. doi:10.1016/j.plaphy.2016.08.013. [Google Scholar] [PubMed] [CrossRef]
27. Chen L, Yang Y, Liu C, Zheng Y, Xu M, Wu N, et al. Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem Bioph Res Co. 2015;464(3):962–8. doi:10.1016/j.bbrc.2015.07.085. [Google Scholar] [PubMed] [CrossRef]
28. Wang Y, Li Y, He SP, Gao Y, Wang NN, Lu R, et al. A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol Biochem. 2019;141:231–9. doi:10.1016/j.plaphy.2019.06.005. [Google Scholar] [PubMed] [CrossRef]
29. Zhang H, Zhao M, Song Q, Zhao L, Wang G, Zhou C. Identification and function analyses of senescence-associated WRKYs in wheat. Biochem Bioph Res Co. 2016;474(4):761–7. doi:10.1016/j.bbrc.2016.05.034. [Google Scholar] [PubMed] [CrossRef]
30. Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell. 2012;35(6):1156–70. doi:10.1111/pce.2012.35.issue-6. [Google Scholar] [CrossRef]
31. Kim CY, Vo KTX, Nguyen CD, Jeong DH, Lee SK, Kumar M, et al. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol Rep. 2016;10:13–23. doi:10.1007/s11816-015-0383-2. [Google Scholar] [CrossRef]
32. Wang MQ, Huang QX, Lin P, Zeng QH, Li Y, Liu QL, et al. The overexpression of a transcription factor gene VbWRKY32 enhances the cold tolerance in Verbena bonariensis. Front Plant Sci. 2020;10:1746. doi:10.3389/fpls.2019.01746. [Google Scholar] [PubMed] [CrossRef]
33. Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Experim Botany. 2011;62(14):4731–48. doi:10.1093/jxb/err210. [Google Scholar] [PubMed] [CrossRef]
34. Mitsis T, Efthimiadou A, Bacopoulou F, Vlachakis D, Chrousos GP, Eliopoulos E. Transcription factors and evolution: an integral part of gene expression. World Acad Sci Jo. 2020;2(1):3–8. [Google Scholar]
35. Heidari P, Ahmadizadeh M, Izanlo F, Nussbaumer T. In silico study of the CESA and CSL gene family in Arabidopsis thaliana and Oryza sativa: focus on post-translation modifications. Plant Gene. 2019;19:100189. doi:10.1016/j.plgene.2019.100189. [Google Scholar] [CrossRef]
36. Miller DR, Leek T, Schwartz RM. A hidden Markov model information retrieval system. In: Proceedings of the 22nd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; 1999; Berkley, CA, USA. [Google Scholar]
37. Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(2):W29–37. doi:10.1093/nar/gkr367. [Google Scholar] [PubMed] [CrossRef]
38. Gasteiger E, Hoogland C, Gattiker A, Se D, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. Totowa, NJ, USA: Humana Press; 2005. [Google Scholar]
39. Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24(1):34–5. doi:10.1016/S0968-0004(98)01336-X. [Google Scholar] [PubMed] [CrossRef]
40. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. doi:10.1093/molbev/msw054. [Google Scholar] [PubMed] [CrossRef]
41. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W9. doi:10.1093/nar/gkz239. [Google Scholar] [PubMed] [CrossRef]
42. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(suppl_2):W202–8. doi:10.1093/nar/gkp335. [Google Scholar] [PubMed] [CrossRef]
43. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. doi:10.1016/j.molp.2020.06.009. [Google Scholar] [PubMed] [CrossRef]
44. Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7. doi:10.1093/nar/30.1.325. [Google Scholar] [PubMed] [CrossRef]
45. Zhou T, Chen J, Huang Y, Jin Z, Li J, Li Y, et al. Genome-wide identification and expression analysis of the PIN auxin transporter gene family in Zanthoxylum armatum DC. Agriculture. 2022;12(9):1318. doi:10.3390/agriculture12091318. [Google Scholar] [CrossRef]
46. Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet. 2001;2(7):516–27. doi:10.1038/35080508. [Google Scholar] [PubMed] [CrossRef]
47. Zhang T, Cai J, Wang S, Lv L, Yuan D, Zeng X, et al. Identification and expression analysis of the ethylene response factor gene family in tea plant (Camellia sinensis). Agronomy. 2023;13(7):1900. doi:10.3390/agronomy13071900. [Google Scholar] [CrossRef]
48. Wu H, Ni Z, Yao Y, Guo G, Sun Q. Cloning and expression profiles of 15 genes encoding WRKY transcription factor in wheat (Triticum aestivem L.). Prog Nat Sci. 2008;18(6):697–705. doi:10.1016/j.pnsc.2007.12.006. [Google Scholar] [CrossRef]
49. Huang X, Ding F, Peng HX, Pan JC, He XH, Xu JZ, et al. Research progress on family of plant WRKY transcription factors. Biotechnol Bulletin. 2019;35(12):129. [Google Scholar]
50. Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 2013;18(5):267–76. doi:10.1016/j.tplants.2012.09.001. [Google Scholar] [PubMed] [CrossRef]
51. Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Genet Genom. 2012;287:495–513. doi:10.1007/s00438-012-0696-6. [Google Scholar] [PubMed] [CrossRef]
52. Li MY, Xu ZS, Tian C, Huang Y, Wang F, Xiong AS. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci Rep. 2016;6(1):23101. doi:10.1038/srep23101. [Google Scholar] [PubMed] [CrossRef]
53. Chen C, Chen X, Han J, Lu W, Ren Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020;20:1–19. [Google Scholar]
54. Su W, Zhou Z, Zeng J, Cao R, Zhang Y, Hu D, et al. Genome-wide identification of the WRKY gene family in Camellia oleifera and expression analysis under phosphorus deficiency. Front Plant Sci. 2023;14:1082496. doi:10.3389/fpls.2023.1082496. [Google Scholar] [PubMed] [CrossRef]
55. Guo C, Guo R, Xu X, Gao M, Li X, Song J, et al. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot. 2014;65(6):1513–28. doi:10.1093/jxb/eru007. [Google Scholar] [PubMed] [CrossRef]
56. Dou L, Zhang X, Pang C, Song M, Wei H, Fan S, et al. Genome-wide analysis of the WRKY gene family in cotton. Mol Genet Genom. 2014;289:1103–21. doi:10.1007/s00438-014-0872-y. [Google Scholar] [PubMed] [CrossRef]
57. Chen M, Tan Q, Sun M, Li D, Fu X, Chen X, et al. Genome-wide identification of WRKY family genes in peach and analysis of WRKY expression during bud dormancy. Mol Genet Genom. 2016;291:1319–32. doi:10.1007/s00438-016-1171-6. [Google Scholar] [PubMed] [CrossRef]
58. Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6(5):486–503. doi:10.1111/pbi.2008.6.issue-5. [Google Scholar] [CrossRef]
59. Van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008;146(4):1983–95. doi:10.1104/pp.107.112789. [Google Scholar] [PubMed] [CrossRef]
60. Shen T, Qi H, Luan X, Xu W, Yu F, Zhong Y, et al. The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol J. 2022;20(2):244. doi:10.1111/pbi.13749. [Google Scholar] [PubMed] [CrossRef]
61. Xu Z, Liu Y, Fang H, Wen Y, Wang Y, Zhang J, et al. Genome-wide identification and expression analysis of WRKY gene family in Neolamarckia cadamba. Int J Mol Sci. 2023;24(8):7537. doi:10.3390/ijms24087537. [Google Scholar] [PubMed] [CrossRef]
62. Mu D, Chen W, Shao Y, Wilson IW, Zhao H, Luo Z, et al. Genome-wide identification and expression analysis of WRKY transcription factors in Siraitia siamensis. Plants. 2023;12(2):288. doi:10.3390/plants12020288. [Google Scholar] [PubMed] [CrossRef]
63. Hu W, Ren Q, Chen Y, Xu G, Qian Y. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021;21:1–21. [Google Scholar]
64. Wei Y, Shi H, Xia Z, Tie W, Ding Z, Yan Y, et al. Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front Plant Sci. 2016;7:25. [Google Scholar] [PubMed]
65. Chen LY, Li JJ, Wang B, Du WQ, Gao MX, Liu H, et al. Research progress of WRKY transcription factors in Soybean response to biotic and abiotic stress. J Plant Genet Res. 2022;23(2):10. [Google Scholar]
66. Ehsan A, Naqvi RZ, Azhar M, Awan MJA, Amin I, Mansoor S, et al. Genome-wide analysis of WRKY gene family and negative regulation of GhWRKY25 and GhWRKY33 reveal their role in whitefly and drought stress tolerance in Cotton. Genes. 2023;14(1):171. doi:10.3390/genes14010171. [Google Scholar] [PubMed] [CrossRef]
67. Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–7. doi:10.1126/science.290.5499.2114. [Google Scholar] [PubMed] [CrossRef]
68. Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16(4):510–9. doi:10.1101/gr.4680506. [Google Scholar] [PubMed] [CrossRef]
69. Bi C, Xu Y, Ye Q, Yin T, Ye N. Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. PeerJ. 2016;4:e2437. doi:10.7717/peerj.2437. [Google Scholar] [PubMed] [CrossRef]
70. Wang P, Yue C, Chen D, Zheng Y, Zhang Q, Yang J, et al. Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes & Genom. 2019;41:17–33. doi:10.1007/s13258-018-0734-9. [Google Scholar] [PubMed] [CrossRef]
71. Qiu Y, Jing S, Fu J, Li L, Yu D. Cloning and analysis of expression profile of 13WRKY genes in rice. Chinese Sci Bull. 2004;49:2159–68. [Google Scholar]
72. Du Z, You S, Zhao X, Xiong L, Li J. Genome-wide identification of WRKY genes and their responses to chilling stress in Kandelia obovata. Front Genet. 2022;13:875316. doi:10.3389/fgene.2022.875316. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools