 Open Access
Open Access
ARTICLE
Transcriptomic Analysis Reveals the Formation Mechanism of Anthocyanins Light-Independent Synthesis in Chrysanthemum
1 Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Key Laboratory of Ornamental Plants Germplasm Innovation & Molecular Breeding, National Engineering Research Center for Floriculture, Beijing Laboratory of Urban and Rural Ecological Environment, Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants of Education Ministry, School of Landscape Architecture, Beijing Forestry University, Beijing, 100083, China
2 Urban Construction School, Beijing City University, Beijing, 100083, China
* Corresponding Authors: Xia Li. Email: ; He Huang. Email:
(This article belongs to the Special Issue: Plant Secondary Metabolism and Functional Biology)
Phyton-International Journal of Experimental Botany 2024, 93(7), 1599-1621. https://doi.org/10.32604/phyton.2024.051386
Received 04 March 2024; Accepted 29 May 2024; Issue published 30 July 2024
Abstract
Chrysanthemum × morifolium is a horticultural crop which plays a vital role in the flower industry with significant economic value and has a cultivation history of over three thousand years in China. The accumulation of anthocyanins is always affected by light. Here, we revealed that anthocyanin accumulation is highly dependent on light in ‘2021135’ genotype chrysanthemum, while it is light-independent in ‘2001402’ genotype chrysanthemum. However, no literature has been reported regarding the non-photosensitive chrysanthemum in anthocyanins light-independent synthesis pathways. Through the phenotype analysis of 44 F generations, we found that light-independence is a dominant trait which can be stable inherited by progeny. The transcriptome of the ray florets of ‘2021135’ and ‘2001402’ under light and bagging treatment were sequenced and analyzed. Based on weighted gene co-expression network analysis (WGCNA), K-means analysis, and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) analysis, 16 genes were highly correlated with the anthocyanin content. The anthocyanin content of ray florets treated with different light-quality conditions indicated that blue light significantly affected anthocyanin accumulations. Through Yeast one-hybrid analysis, CmBIC1.1 and CmBIC1.2 can directly regulate the anthocyanin structural gene CmCHS2. In our study, we revealed the important characteristics of light-independent anthocyanin synthesis in chrysanthemums and screened regulatory factors in light-dependent and light-independent anthocyanin synthesis pathways. The results laid the groundwork for subsequent analysis of the molecular mechanism involved in the light-independent synthesis of anthocyanins in chrysanthemums.Keywords
Supplementary Material
Supplementary Material FileBright-colored petals are typically the most appealing aspects of ornamental plants. The commercial value of many ornamental plants is affected by flower color. The stability of color is is a primary breeding objective for many horticulturists, aiming to elevate the ornamental appeal of plants. Anthocyanins are biosynthesized through the flavonoid pathway, which is catalyzed stepwise by structural genes coding for enzymes has been well studied. The regulation of the anthocyanin pigment pathway is governed by a group of transcription factors, including R2R3-MYB, basic helix-loop-helix (bHLH), and WD repeat proteins (WDR). These factors have been demonstrated to form a MYB-bHLH-WDR (MBW) complex that enhances the expression of genes involved in anthocyanin biosynthesis [1–3].
Light signaling plays an essential role in the control of anthocyanin synthesis. The influence of light on anthocyanin accumulation has been widely studied in cherries, tomatoes, strawberries, grapes, and other crops, in which anthocyanins decrease under low light or shade conditions and increase under strong light conditions [4–8]. The range of wavelengths of sunlight from 280 to 800 nm impacts plant biological processes, encompassing ultraviolet (UV) light (UV-A and UV-B), visible light, and far-red light (710–810 nm). The visible light spectrum ranges from 400–710 nm and is subdivided into blue (400–495 nm), green (495–570 nm), yellow (570–590 nm), and red (590–710 nm) wavelengths. The exposure to blue and ultraviolet (UV) light exhibits a stimulatory effect on the enhancement of anthocyanin biosynthesis, whereas far-red light demonstrates a suppressive influence [9–13].
Photoreceptors and light signal transduction elements play a crucial role in the light-dependent anthocyanin accumulation of plants. Light activates various photoreceptors in plants, including phytochrome (PHYA-PHYE), cryptochromes (CRYs), phototropins (PHOTs) and UV resistance locus 8 (UVR8) [14]. Phytochromes (PHYA-PHYE) are red and far-red light receptors that can regulate the growth and development of plants, such as phytochromes regulate nitrogen-, phosphate- and cold-induced anthocyanin accumulation in shoots of Arabidopsis [15,16]. CRYs, PHOTs, and three zeitlupe proteins are characterized as UV-A/blue photoreceptors [17]. The induction of anthocyanin synthesis in eggplant and tomato fruit epidermis has been reported through the activation of anthocyanin biosynthesis enzymes by CRYs [18,19]. PHOTs play a crucial role in the synthesis of secondary metabolites mediated by blue light, including the regulation of pigment synthesis within plant cells. It was demonstrated that FaPHOT2 functioned in detecting blue light and facilitating anthocyanin production in strawberry fruits [20]. The blue-light photoreceptor Flavin-binding, Kelch Repeat, F-box1 (FKF1) can interact with Constitutive Photomorphogenic 1 (COP1) and reduce COP1 activity in a day-length-dependent manner, thereby promoting photomorphogenesis and anthocyanin accumulation in Arabidopsis [21]. UVR8 mediates UV-B-induced changes in gene expression, and photomorphogenic responses include induction of flavonoid biosynthesis and accumulation of flavonoid compounds [17,22]. In Arabidopsis uvr8 mutants, the expression of Chalcone synthase (CHS) and the content of anthocyanin accumulation is significantly stimulated [23]. It has also been reported in plants such as grapes and apples that UVR8 serves as a significant photoreceptor to promote anthocyanin accumulation [22,24,25].
Downstream of these photoreceptors, COP1 emerges as a pivotal suppressor, instrumental in catalyzing ubiquitination and subsequent degradation of anthocyanin biosynthesis’s positive regulators, including elongated hypocotyl 5 (HY5), COP1-interacting protein (CIP7) [3,21,25]. A quantitative evaluation of anthocyanins in the fruit peel showed that the hy5 mutant fruit exhibited reduced levels of anthocyanins [26]. Research indicated that CIP7 plays an essential role in regulating fruit color development; SmCIP7 nhances the expression of structural genes to increase anthocyanin accumulation in eggplant [27]. Phytochrome rapidly regulated 1 (PAR1) was first recognized as an early downregulated gene in the phyA-mediated far-red and phyB-mediated red light signaling pathways, which was observed to be inhibited by photoreceptors [28]. These findings suggest that PAR1 is a photomorphogenesis-promoting factor functioning across various photoreceptor signaling pathways [29]. There are two negative regulators to CRYs called Blue light inhibitors of cryptochromes (BICs, BIC1, and BIC2) in the blue light signaling pathway. Studies showed that BICs (blue-light inhibitor of cryptochromes) interact with CRY2 to suppress the photomorphogenesis [30,31]. The involvement of two WD40-repeat proteins, Repressor of UV-B photomorphogenesis 1 (RUP1) and RUP2, is essential in the UV-B light signaling pathway. These proteins play a vital role in providing negative feedback regulation and deactivating UVR8 by aiding in its redimerization [32].
Light-dependent anthocyanin accumulation is influential in the pigmentation of plants. Light-independent pigmentation has also been reported, such as the ‘Mantianhong’ pear and ‘Ruby’ mango, which can still accumulate a significant amount of anthocyanins even when bagged [33,34]. The accumulation of light-independent anthocyanins has also been found in blood oranges and carrots, in which mutations of the R2R3-MYB promoter were discovered, resulting in constitutive expression of MYB and potentially leading the fruits to exhibited elevated levels of anthocyanin pigmentation when cultivated in darkness [35,36].
Chrysanthemum × morifolium, native to China, is an ornamental plant with a wide range of attractive traits and high economic value. However, weak light stress during greenhouse cultivation can decrease its ornamental quality and landscape effect, leading to higher production costs and insufficient energy conservation in today’s horticultural production. Multiple MYB transcription factors that regulate anthocyanin biosynthesis have been identified in chrysanthemums. Transcription factors such as CmMYB6 and CmMYB9a can enhance anthocyanin accumulation in chrysanthemum flowers, while CmMYB4, CmMYB#7, and CmMYB012 suppress anthocyanin accumulation [13,37–39]. Similar to other crops, light exposure also affects the coloration of chrysanthemum. Under UV-B treatment, the content of anthocyanins in chrysanthemums significantly increases [40]. The mechanism of fine-tuning the dynamic balance of anthocyanin pigments in chrysanthemum ray florets can be influenced by different R/FR through the regulation of CmbHLH16 [41].
Additionally, during the coloration process of light-induced non-photosensitive chrysanthemum, expression of structural genes and biosynthesis of anthocyanin pigments are regulated by light-responsive transcription factors such as CmMYB4, CmMYB5-1, CmMYB6, CmbHLH24, and CmMYB7-1 [42–44]. CmHY5 in chrysanthemum has also been reported to regulate the expression of CmFNSII-1 directly, thereby regulating the biosynthesis of flavonoids. However, there are currently no reports on the light-independent synthesis of anthocyanins in chrysanthemum, which indicates that the molecular mechanism of the light-independent synthesis of anthocyanins in chrysanthemum is still not well understood.
In this study, we obtained the non-photosensitive chrysanthemum ‘2001402’. Through transcriptome analysis, Y1H and Y2H assays, light signal transduction elements CmBIC1.1 and CmBIC1.2 may act as the key factors in light-independent anthocyanins synthesis in chrysanthemum. The results laid the groundwork for the subsequent analysis of the molecular mechanism underlying anthocyanin biosynthesis in non-photosensitive chrysanthemum.
2.1 Plant Materials and Treatment
We conducted an experiment at the Greenhouse of Beijing Forestry University to investigate the light regulatory characteristics of anthocyanin synthesis in chrysanthemum. A total of 103 chrysanthemum genotypes were selected for this study. Three materials were selected as biological replicates. We covered the 10 μm-thick tin foil around the capitulum to block the light during the bud stage cultivation (Fig. 1a).

Figure 1: The shading treatment method and the grading standard: (a) Chrysanthemum treated with bagging; (b) Developmental stages of ray florets development. Scale bar = 1 cm; (c) Part of light quality treatments with 120 LEDs
Five stages were divided according to the following definitions [45]. S1, 0–0.5 cm in ray florets length; S2, 0.5–0.8 cm in ray florets length; S3, 0.8–1.2 cm in ray florets length; S4, 1.2–1.8 cm in ray florets length; and S5, 1.8–2.5 mm in ray florets length (Fig. 1b).
The capitulums of different cultivars were bagged before the S1 stage until the S5 stage. The non-photosensitive chrysanthemum ‘2001402’ and photosensitive chrysanthemum ‘2021135’ are the samples for the RNA sequencing and gene expression analyses. For these two genotypes, the five distinct phases of ray florets development under light and bagged conditions were cryogenically preserved in liquid nitrogen and kept at −80°C for later analysis of pigment levels and RNA extraction. Besides, a consistent light intensity (120 LEDs) treatment was applied to ‘2021135’ before the S1 stage until the S3 stage. Eight light quality treatments were conducted in 2 years, using far red (peak wavelength = 730 nm), red (peak wavelength = 660 nm), blue (peak wavelength = 460 nm), UV-A (peak wavelength = 340 nm), UV-B light (peak wavelength = 313 nm) in the first years with white light was used as the control while blue light (80 LEDs) supplemented with UV-A (40 LEDs) or UV-B (40 LEDs) were using in the second year. White and blue light was used as the control (Fig. 1c). All the light treatments were arranged over an area of 150 cm × 60 cm.
2.2 Hybridization Experiment between Non-Photosensitive and Photosensitive Chrysanthemum
The non-photosensitive chrysanthemum ‘2001402’, as well as the photosensitive chrysanthemum B200 obtained through years of field experiments and pigment analysis, were used as parents to create the reciprocal hybrids. Bagging was applied following the method as stated before. After bagging the capitulum until the S5 stage, the bags were opened to assess the accumulation of anthocyanins in the ray florets. The number of chrysanthemums with different degrees of photosensitivity was recorded.
2.3 Anthocyanin Extraction and Measurement
The anthocyanin content in the ray florets of chrysanthemum was quantified using UV spectrophotometry methods utilized in prior research. A total of 0.1 g of tissue was finely ground to powder using liquid nitrogen, combined with 5 mL of anthocyanin extracts [methanol:water:formic acid:trifluoroacetic acid (70:27:2:1, v/v/v/v)], and incubated at 4°C for 24 h in the absence of light. The supernatant was then taken for measurement. Using a UV-visible spectrophotometer (TU-1901; Beijing Puxi Co., Ltd., Beijing, China), the absorbance was measured at 520 nm with anthocyanin extracts as the control. The content of anthocyanins was calculated based on the extinction coefficient (98.2) of total anthocyanins. The average value was calculated based on three replicates of each sample. The formula for the calculation of anthocyanin content is as follows: MF = (A × V)/(98.2 × M), where MF stands for total anthocyanin content (mg/g), A represents the absorbance value at the maximum wavelength, V is the final volume (mL), and M is the weight of the sample (g).
2.4 RNA Sequencing, Functional Annotation, and Data Processing
Ray florets at S2–S4 stages of 2001402 and 2021135 were selected for RNA-Seq analysis (Each stage of sample sequencing is repeated three times). In brief, total RNA was extracted from ray florets of S2–S4 stages and cDNA libraries were synthesized by Biomarker Technologies Corporation (Beijing, China). The data processing is same as a previous study by Lu et al. [46].
2.5 Identification and Analysis of DEGs
The transcript abundances of all unigenes were calculated using the FPKM method with RSEM [47]. Analysis of all DEGs using Weighted Gene Co-Expression Network Analysis (WGCNA) and K-means clustering analysis. We used the EBSeq technique for identifying differentially expressed genes in an RNA-Seq experiment comparing two biological conditions [48]. In addition, we can correct the p-values using the Benjamini-Hochberg method and use this indicator to filter differentially expressed genes between the two libraries. Strict criteria for gene selection include a False Discovery Rate (FDR) less than 0.05 and a fold change (FC) of at least 1.5. Further screening of Unigenes involved in phototransduction and anthocyanin biosynthesis pathways is carried out based on NR and Swissprot annotations. To ensure the comprehensiveness and accuracy of differential gene screening, homologous genes are selected for transcription factors reported to design phototransduction pathways.
2.6 RT-PCR (Semi-Quantitative Reverse Transcriptase-Polymerase Chain Reaction) and RT-qPCR (Real-Time Quantitative Polymerase Chain Reaction) Validation of Differentially Expressed Genes
RT-PCR was conducted to analyze the expression patterns of selected genes sourced from the RNA-Seq database. Total RNAs from ray florets at S1–S5 stages of both non-photosensitive chrysanthemum ‘2001402’ and photosensitive chrysanthemum ‘2021135’ were isolated using a Plant RNA Rapid Extraction Kit (HUAYUEYANG Biotechnology, Beijing, China). These RNAs were then utilized to produce cDNA for RT-PCR through a transcription kit. The semi-quantitative RT-PCR protocol adhered to the methodology outlined in a previous study by Lu et al. [46], utilizing 26S as a reference gene. The primer sequences are documented in Table S1.
To validate the accuracy of the RT-PCR findings, RT-qPCR reactions were carried out using a CFX Connect Realtime System (Bio-Rad, Hercules, CA, USA) along with SYBR Premix Ex Taq (TaKaRa, Kyoto, Japan), following the protocol detailed by Huang et al. [49]. The primer sequences are provided in Table S1. Each data point from RT-qPCR was obtained from three biological and three technical replicates. The relative gene expression levels were standardized by comparing them to the expression of PP2A in Chrysanthemum × morifolium [50], and the analysis was conducted using the 2−ΔΔCT method [51].
2.7 Gene Isolation and Yeast Two-Hybrid Assay
According to the information from the transcriptome analysis and gene expression analysis, we cloned two light receptors of the blue light signaling pathway and two light signal transduction components from the non-photosensitive chrysanthemum ‘2001402’ and the photosensitive chrysanthemum ‘2021135’, as well as four anthocyanin synthesis-related transcription factors. The forward and reverse (F/R) primers for these genes (primer sequences are shown in the Table S1) were designed and used to clone the open reading frame (ORF) of these genes from the light-independent and photosensitive chrysanthemum into the pCE3 Blunt Vector (Vazyme, Nanjing, China), and sequenced (Sangon Biotech, Shanghai, China). The amino acid sequences of the genes of other species were obtained from the National Center of Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/, accessed on 20 April 2024) database. The phylogenetic tree for the proteins was constructed using MEGA V11.0.
The genes encoding components involved in blue light signaling and transcription factors related to anthocyanin biosynthesis were integrated into pGADT7 and pGBKT7 vectors to create recombinant vectors. These recombinant vectors were co-transformed into competent yeast cells (strain Y2H Gold). After verifying no self-activation activity, the recombinant vectors were spread on SD/−Trp−Leu medium. Subsequently, a singular colony was selected and diluted by factors of 1x, 10x, 102x, and 103x-fold to inoculate the SD/−Trp/−Leu/−His/−Ade screening medium. The complete list of primers utilized in this research is detailed in Table S1.
2.8 Isolation of Promoter Sequences Using the Chrysanthemum Genome Database and Yeast One-Hybrid Assay
The promoter sequences of CmCHS2 and CmBIC1 were cloned from genomic DNA of ‘2001402’ and ‘2021135’ ray florets using the Chrysanthemum Genome Database (http://210.22.121.250:8880/asteraceae/homePage, accessed on 20 April 2024). The prediction and analysis of cis-elements in promoters were performed using PlanCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html, accessed on 20 April 2024). The light-responsive elements (LRE) in the promoter were amplified and inserted into the pAbAi vector.
The promoter of the critical genes CmCHS2 and CmBIC1 in this study were cloned, and yeast one-hybrid experiments were performed with other key light signal transduction factors, CmBIC1, CmHY5, and CmWRKY40. The yeast one-hybrid assay was performed as described previously [52]. pAbAi-pCmCHS2 was converted into Y1H Gold, and the minimal inhibitory concentration of aureobasidin A (AbA) for the bait strain was examined on SD/-Ura medium. Subsequently, the hybrid vectors and AD empty vector (control) were introduced into yeast cells harboring the recombinant pAbAi plasmid. The modified yeast cells were diluted sequentially by ten-fold and spotted on SD/-Leu medium containing the least AbA concentration. The yeast cells were then cultivated in an incubator at 30°C for 2–3 days.
3.1 Classification and Screening of Different Light-Dependent Types of Chrysanthemum
After the materials were bagged for 30 days, compared with different treatments, the petal anthocyanin of 103 chrysanthemums had different accumulations. Most chrysanthemums showed obvious fading; 51 (49.5%) chrysanthemum genotypes completely faded to white or yellow, belonging to photosensitive chrysanthemum (Fig. 2a); 40 (38.8%) chrysanthemum genotypes showed partial fading, belonging to light-semi-dependent chrysanthemum (Fig. 2b). There were 12 (11.7%) genotypes continued to exhibit a significant level of anthocyanin synthesis even after bagging were classified as non-photosensitive chrysanthemum (Fig. 2c). Among the 12 non-photosensitive chrysanthemum genotypes, the pink genotype ‘2001402’ with a darker color and a larger number of ray florets, is more suitable for this study than the orange and red strains due to the absence of interference from carotenoids. Therefore, ‘2001402’ was selected as the non-photosensitive chrysanthemum for pigment content determination, while the completely faded photosensitive chrysanthemum ‘2021135’ was selected as the control. A three-year sequential shade experiment on both chrysanthemum genotypes was carried out, and the phenotypes of the ray florets following shading displayed repeatability, suggesting the two kinds’ consistent photosensitivity features.
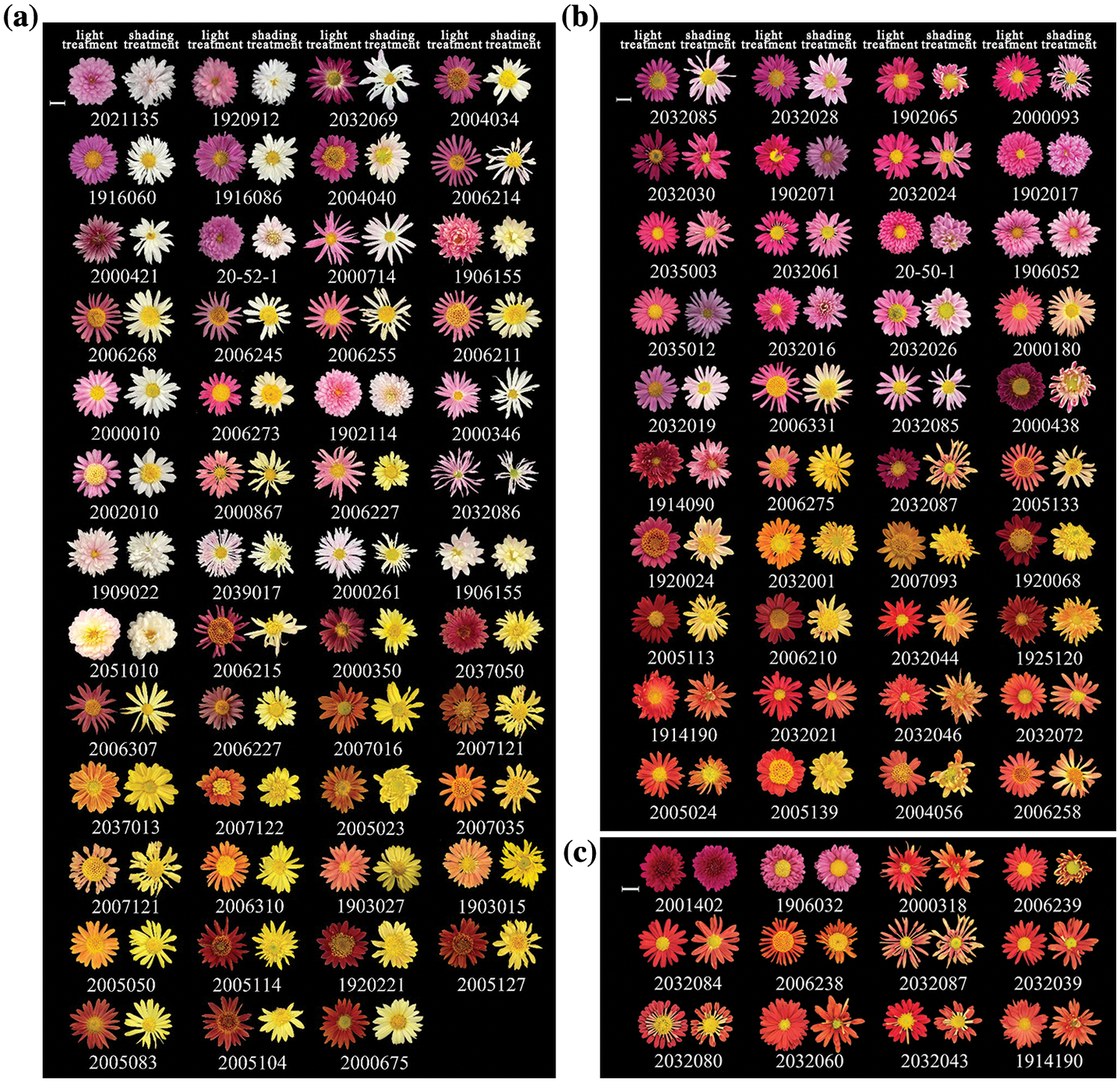
Figure 2: Phenotypic changes of 103 chrysanthemum genotypes before and after shading: (a) Phenotypic changes of chrysanthemum completely light-dependent anthocyanin accumulation; (b) Phenotypic changes of chrysanthemum light-semi-dependent anthocyanin accumulation; (c) Phenotype of non-photosensitive chrysanthemum. Note: Scale bar = 1 cm
3.2 Light Treatment and Determination of Anthocyanin Content
Through the determination of pigment content (Fig. 3a), the photosensitive chrysanthemum ‘2021135’ had almost no presence of anthocyanin in the ray florets. In contrast, the non-photosensitive chrysanthemum ‘2001402’ showed the synthesis of anthocyanin both before and after shading. The variance analysis of anthocyanin content revealed that darkness exerted a subtle influence on the biosynthesis of anthocyanins in non-photosensitive chrysanthemums; however, the ray florets of ‘2001402’ were evenly colored and remained at a relatively high level (Fig. 3b).
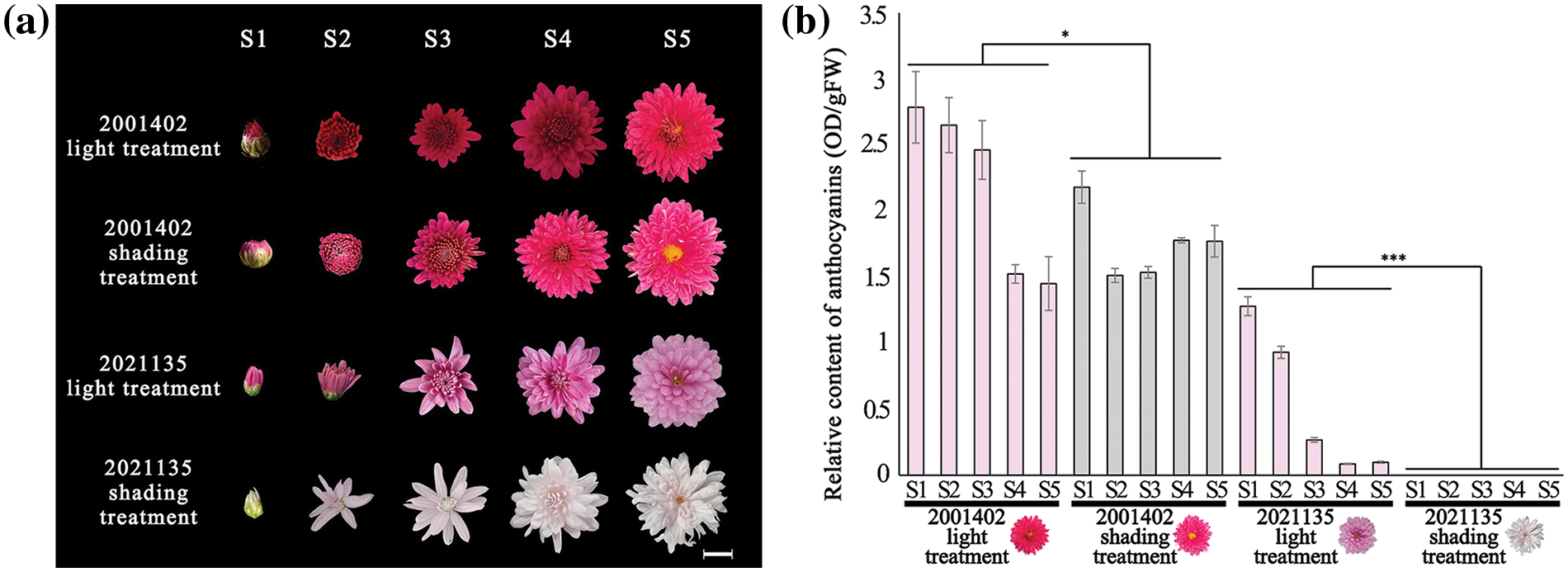
Figure 3: (a) Flower phenotypes at various developmental stages after light and shading treatments; (b) Anthocyanin content at various developmental stages after light and shading treatments (*p < 0.05; ***p < 0.001). Note: Scale bar = 1 cm, the histograms are the mean of n = 3, the +/− vertical bars on the upper part of the histograms mean standard deviation
According to the measurement of anthocyanin content, it was found that shading significantly affected the synthesis of anthocyanins in ‘2021135’ chrysanthemums (Table S2). Therefore, materials ‘2001402’ and 2021135 were selected for further experiments.
Through the light quality treatment on photosensitive chrysanthemum ‘2021135’ (Fig. 4a), the results of the treatment in the first year at equal intensity showed that the single light quality treatment caused a decrease in anthocyanin content of the chrysanthemum ray florets to varying degrees, with the most significant accumulation of anthocyanin under blue light irradiation. Therefore, we believe that the most crucial light quality condition affecting chrysanthemum anthocyanin biosynthesis is blue light (Fig. 4b, Table S3). Thus, in the second year, we chose blue light supplemented with UV light. The results showed that a proper amount of UV-A and UV-B can enhance the promoting effect of blue light on chrysanthemum anthocyanin to a certain extent (Fig. 4c, Table S4). In general, significantly higher anthocyanin in ray florets treated with blue light supplemented with UV-A and UV-B light were observed than that of the control (blue light only).
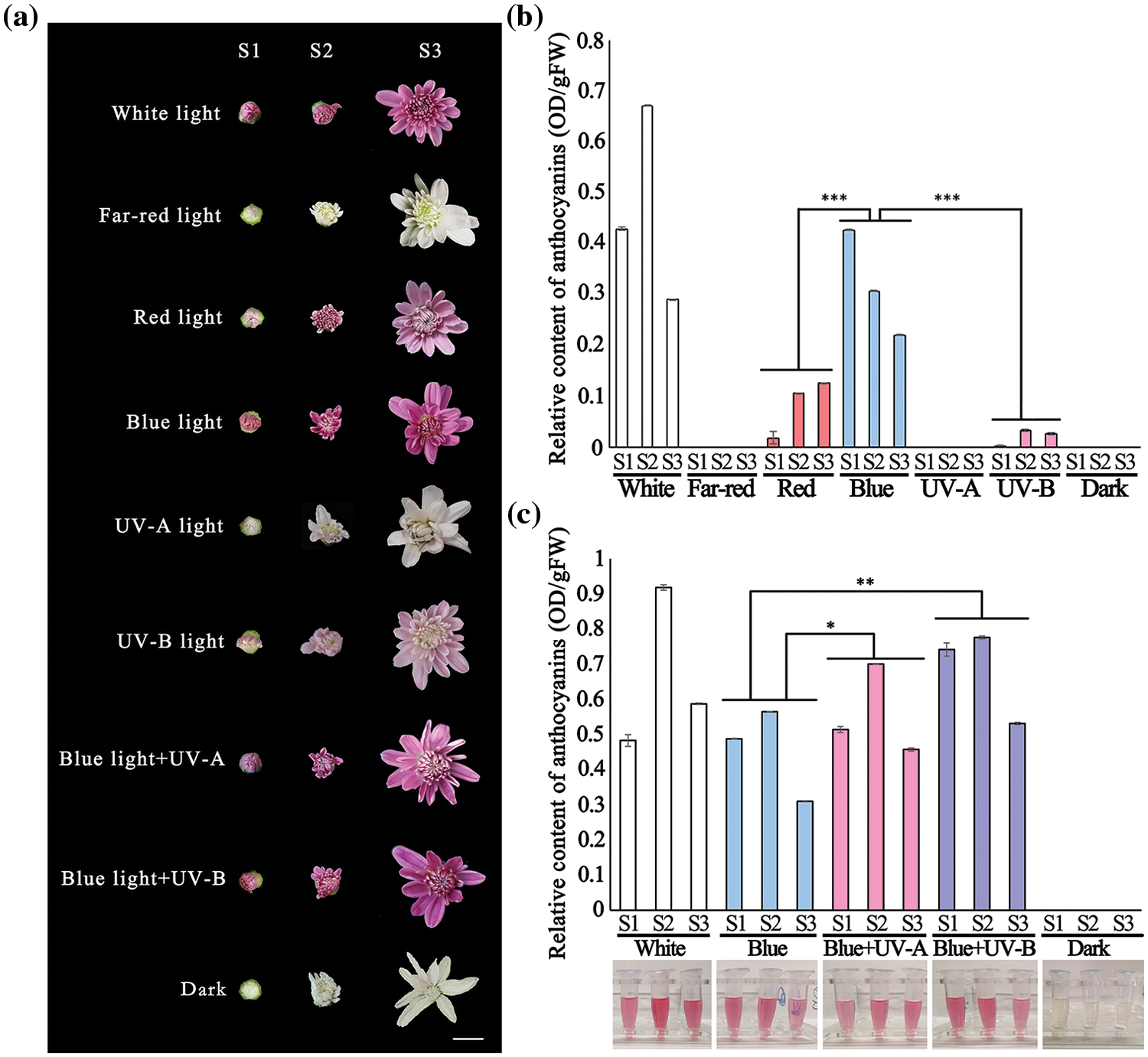
Figure 4: (a) Flower phenotypes at S1–S3 stages under different light quality treatments; (b) Anthocyanin content at various developmental stages under light quality treatment in the first year; (c) Anthocyanin content at various developmental stages under light quality treatment in the second year. (*p < 0.05; **p < 0.01; ***p < 0.001). Note: Scale bar = 1 cm, the histograms are the mean of n = 3, the +/− vertical bars on the upper part of the histograms mean standard deviation
3.3 Genetic Analysis of Light-Independent Properties
The results of the shading test indicated significant differences in anthocyanin synthesis between the non-photosensitive chrysanthemum ‘2001402’ and the photosensitive chrysanthemum. The fluorescence of 44 F1 generation of ‘2001402’ and photosensitive chrysanthemum ‘B200’ was in late October 2019, and all of it was capable of light-independent anthocyanin synthesis (Fig. 5). Based on the results, we can infer that the light-independent characteristics of chrysanthemums are a stable inherited dominant trait.
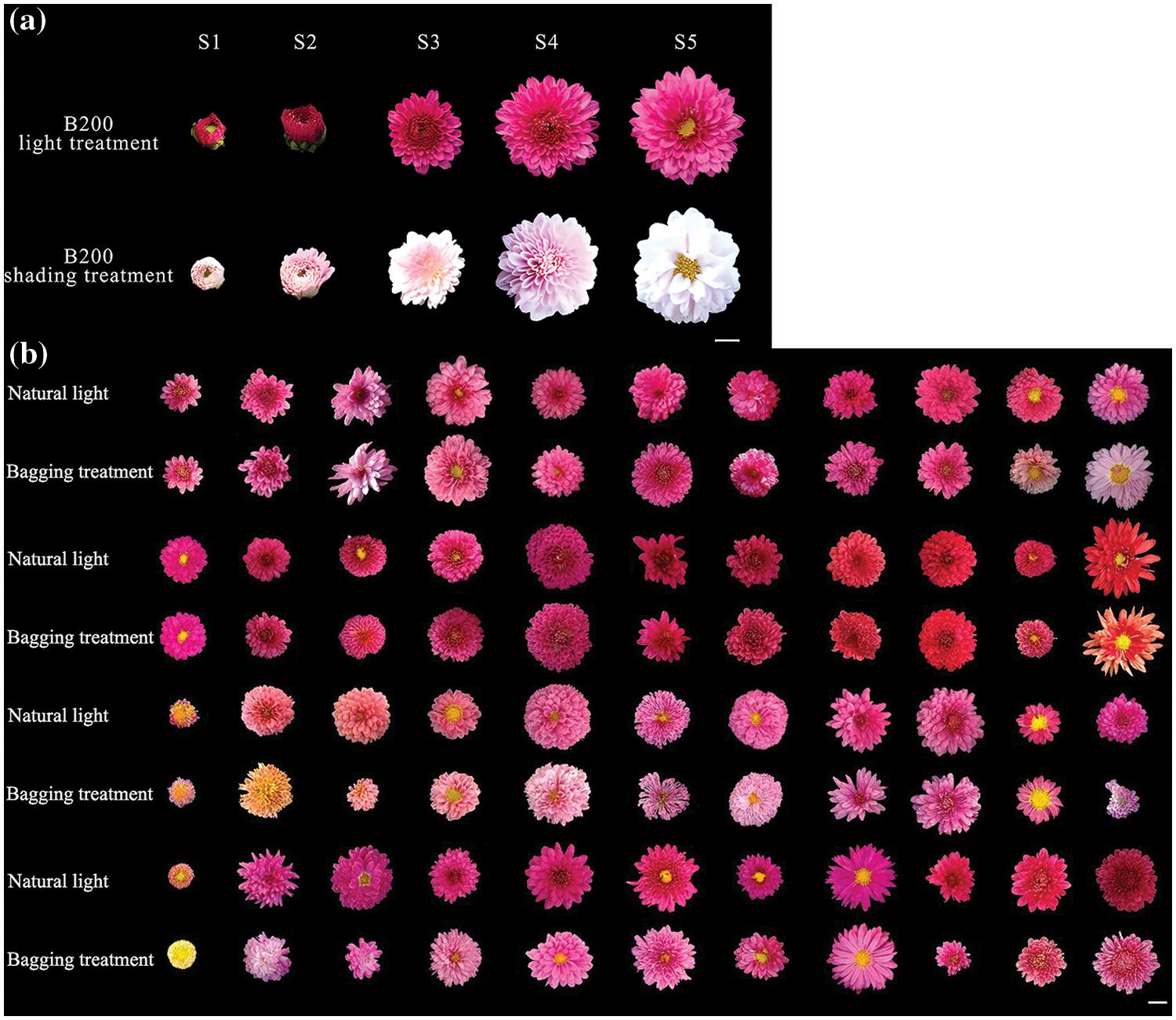
Figure 5: (a) Flower phenotypes at various developmental stages after light and shading treatments. (b) Phenotypic changes of the hybrid offspring between ‘2001402’ and ‘B200’ chrysanthemums before and after shading. Note: Scale bar = 1 cm
3.4 De novo Assembly, Analyses of RNA-Seq Data, and Sequences Functional Annotation
RNA-Seq of the ray floret in the S2 stage–S4 stage of two types of chrysanthemums was carried out with 36 samples were sequenced and filtered. A total of 223.01 Gb Clean Data was generated with a Q30 (percentage of sequences with sequencing error rates lower than 0.1%) of 92.89%–94.4%. The statistics of 36 samples sequencing data evaluations are shown in Table S5 and Fig. S1.
100,132 high-quality unigenes were obtained after filtration, and 26,576 unigenes were above 1 kb in length. Transcript abundance (estimated from fragments per Kilobase of exon model per Million mapped fragments, FPKM) was highly correlated between replicate samples were highly correlated with each other. Unigenes with an average FPKM value less than 0.3 across all samples are considered non-expressed genes [53]. Subsequent filtering will be applied to the remaining high-quality Unigenes.
A total of 44,862 unigenes were annotated based on BLASTx (E-value < 1 × 10−5) and HMMER (E-value < 1 × 10−10) searches against public databases, including COG, GO, KEGG, KOG, Pfam, Swiss-Port, eggNOG, and Nr (Fig. S2). 21,608 (48.17%) were annotated in KOG, 25,630 (57.13%) were annotated in Pfam, 24,275 (54.11%) were annotated in Swissprot, 30,726 (68.49%) were annotated in eggNOG, and 42,221 (94.11%) were annotated in Nr database.
3.5 Screening for DEGs Regulating Anthocyanins Light-Independent Synthesis through WGCNA
In this study, we used three developmental stages (Fig. 3) of ray florets of ‘2001402’ and ‘2021135’ as materials to build transcriptome libraries and explored the potential transcription factors that regulated anthocyanin accumulation in chrysanthemum using WGCNA combined with the analysis of dynamic changes of anthocyanin total contents in twelve different developmental stages in different light condition.
The expression patterns of DEGs obtained from transcriptome sequencing were analyzed by WGCNA. The genes with similar expression patterns were grouped into modules, and a total of 14 modules were identified (Figs. 6a and S3). Because the pattern of gene expression was often associated with phenotypic changes, such as pigment accumulation in higher plants [46], we analyzed the correlation between the expression patterns of each module and the changes of the anthocyanin total contents. The results showed that the Pierson correlation coefficient of the module ‘darkgreen’ with the anthocyanin total contents was the highest, which was 0.88 (Fig. 6b), indicating that the module ‘darkturquoise’ was closely related to carotenoid accumulation in chrysanthemum. Furthermore, the Pierson correlation coefficient of the modules ‘paleturquoise’, ‘saddlebrown’, ‘orange’, and ‘lightsteelblue1’ are also highly correlated.
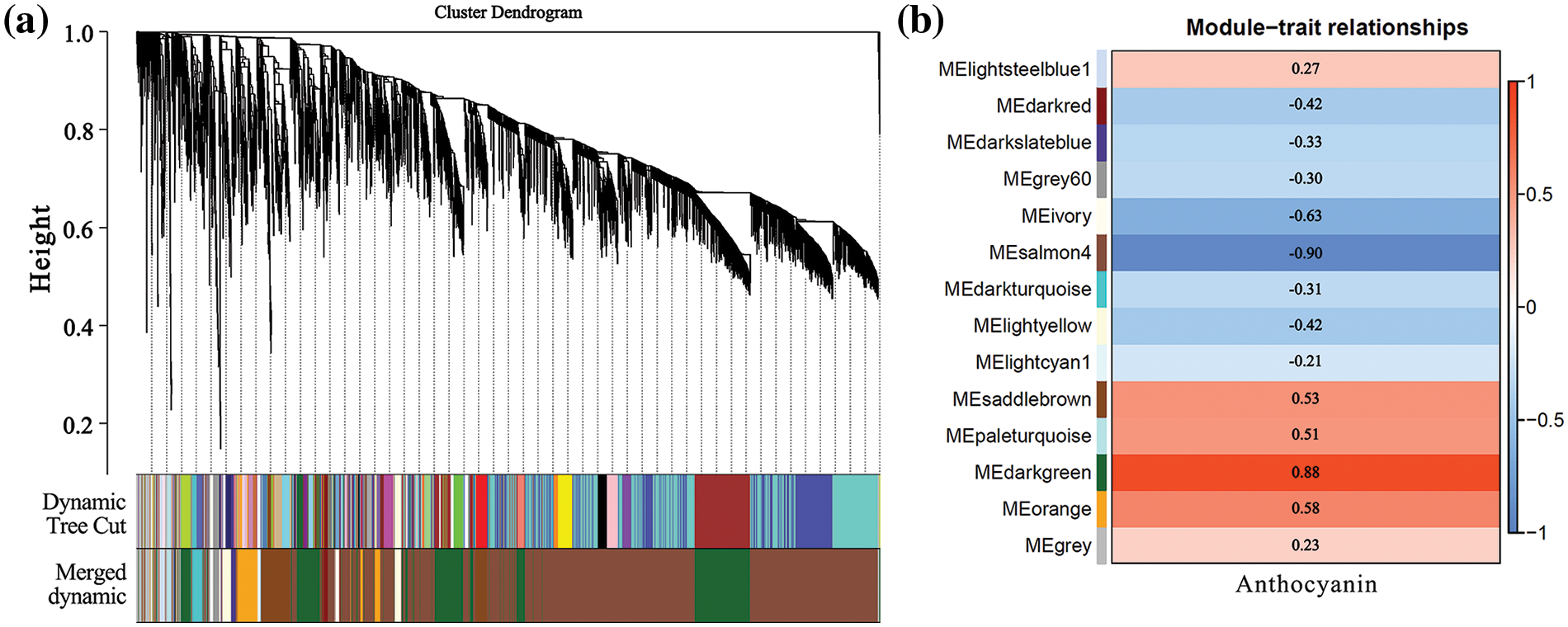
Figure 6: Network analysis dendrogram showing modules identified by weighted gene co-expression network analysis (WGCNA). (a) Dendrogram plot with color annotation; (b) Module-anthocyanin weight correlations and corresponding p-values. The left panel shows the 14 modules. The color scale on the right shows module-trait correlation from −1 (blue) to 1 (red). The internal reading means the pierson correlation coefficient of the module with the anthocyanin
Functional and pathway enrichment analyses were performed for the genes in ‘darkgreen’, ‘paleturquoise’, ‘saddlebrown’, ‘orange’, and ‘lightsteelblue1’ modules, in which the genes related to anthocyanin biosynthesis in chrysanthemum. Deep analysis revealed that the ‘darkturquoise’ module contained a total of 1,381 genes, out of which 896 are annotated with functions, including PHYB-1 (c106435.graph_c0), PAR1 (c95398.graph_c0) involved in light signaling reception, as well as MYB3 (c108549.graph_c1), MYB13 (c76800.graph_c0), and bHLH2 (c119981.graph_c1) associated with anthocyanin synthesis. It also includes the anthocyanin synthesis structural genes CHS1 (c114859.graph_c0), CHI-1 (c108890.graph_c0), and ANS (c113803.graph_c0). The ‘paleturquoise’ module contains 52 genes, out of which 33 are functionally annotated, but no genes directly related to light signaling or anthocyanin synthesis pathways were found in this module. The module ‘saddlebrown’ contains 495 genes, among which 350 genes have functional annotations, but there was one gene, UVR8-1 (c116302.graph_c0), involved in the light signaling reception. The module ‘orange’ consists of 329 genes, in which 250 genes have functional annotations, including BIC1 (c104215.graph_c0) involved in the light signaling pathway, and the structural genes CHS2 (c88987.graph_c0) and CHI-2 (c115206.graph_c1) for anthocyanin synthesis. In the ‘lightsteelblue1’ module, there are 91 genes, out of which 73 have functional annotations, including CIP7 (c125178.graph_c0) involved in the light signaling pathway, and WRKY40 (c111000.graph_c0) and MYB6 (c93023.graph_c0) involved in anthocyanin synthesis pathway. Additionally, genes such as CHS3 (c111705.graph_c0), F3H (c121497.graph_c0), F3’H-1 (c102320.graph_c0), DFR (c122995.graph_c0), and 3MT (c118954.graph_c0) related to anthocyanin synthesis were found in this module. In conclusion, a total of 20 genes related to chrysanthemum anthocyanin biosynthesis and light signaling pathway were selected through WGCN screening (Table S6).
3.6 Screening for Transcription Factors Regulating Anthocyanins Light-Independent Synthesis through Cluster Analysis
The overall expression pattern of DEGs was shown on the clustering map with K-means cluster analysis; 5530 DEGs were classified into 18 clusters (Fig. S4), of which 7 clusters matched the expression trends we were expecting (Fig. 7).
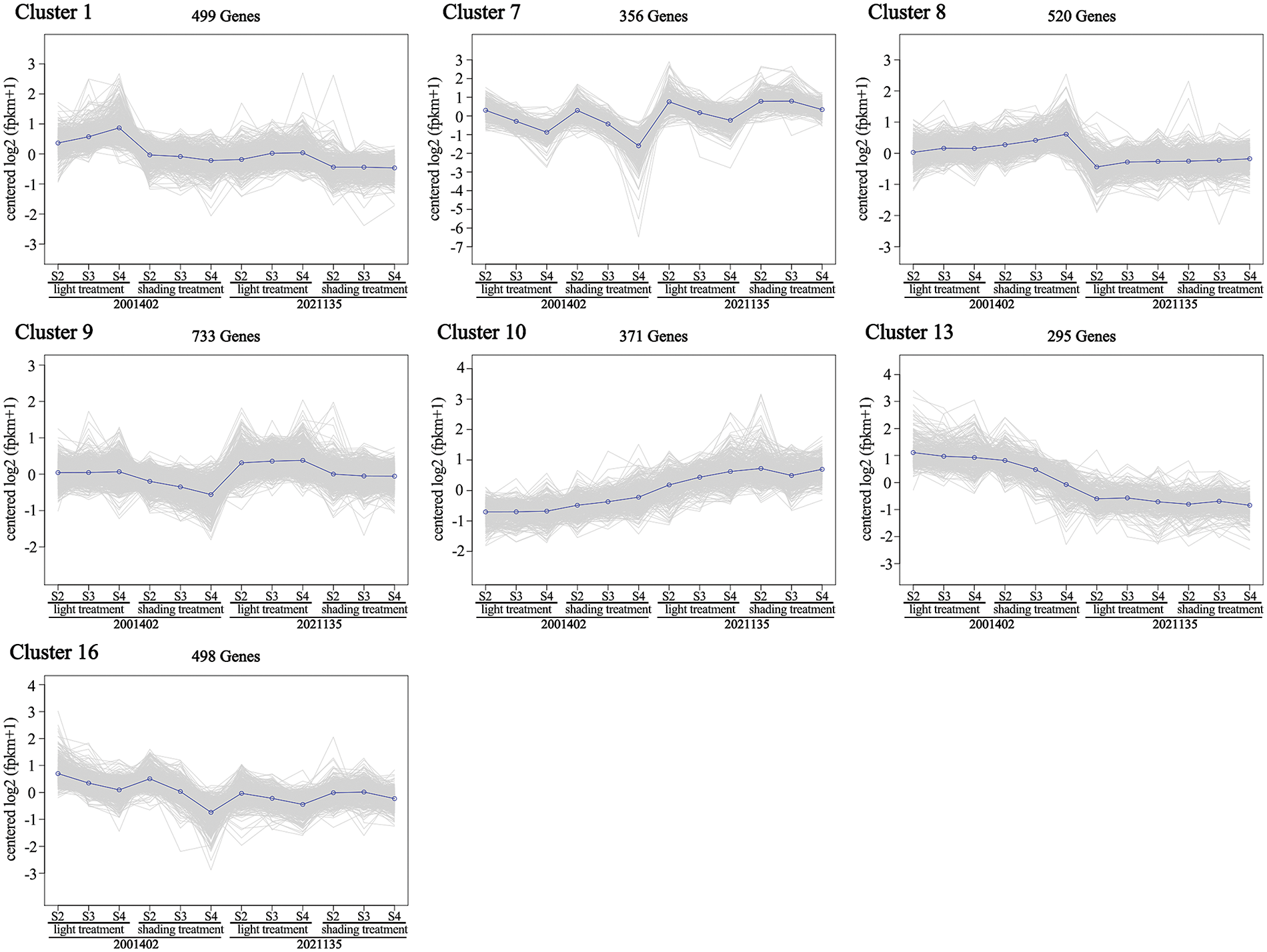
Figure 7: Analysis of DEGs in 7 clusters matched the expected expression trends
The expression trends of Cluster 1 (499 DEGs) decreased much more obviously in ‘2001402’ than in ‘2021135’ after shading. In Cluster 7 (356 DEGs), the genes were down-regulated in the ‘2001402’ after bagging treatment, while they were up-regulated after shading in ‘2021135’. In Cluster 8 (520 DEGs), the genes showed an upward expression trend after shading in ‘2001402’, while there was no significant change in the ‘2021135’. In two types of chrysanthemums under light treatment, the genes of Cluster 9 (733 DEGs) did not show any significant changes in expression across three developmental stages. However, in ‘2001402’ under bagging treatment, the gene expression levels showed a clear decreasing trend with the progression of development, while the gene expression in ‘2021135’ remained unchanged. In Cluster 10 (371 DEGs), the genes showed a consistent low expression under light in ‘2001402’. However, after bagging treatment, the expression was up-regulated. In contrast, the genes in the ‘2021135’ showed an up-regulation in expression with light exposure and no significant changes in expression under bagging treatment. Cluster 13 (295 DEGs) and Cluster 16 (498 DEGs) were similar, showing lower expression under bagging treatment than light treatment in ‘2001402’, while showed no significant changes in expression under both treatments in ‘2021135’.
In the K-means clustering analysis, a total of 18 DEGs related to the light signaling pathways and anthocyanin synthesis pathway were finally detected, including PHYA (c124786.graph_c0), PHYB-1 (c106435.graph_c0), PHYB-2 (c122807.graph_c0), CRY1b-1 (c107160.graph_c0), FKF1 (c119423.graph_c0), UVR8-1 (c116302.graph_c0), UVR8-2 (c121876.graph_c0), UVR8-3 (c122272.graph_c0), HY5 (c120662.graph_c0), SPA1 (c122020.graph_c0), RUP2 (c120802.graph_c0), WRKY40 (c111000.graph_c0), MYB3 (c108549.graph_c1), MYB13 (c76800.graph_c0), CHS1 (c114859.graph_c0), CHI-2 (c115206.graph_c1), F3’H-2 (c113373.graph_c0), and ANS (c113803.graph_c0). The results are shown in Table S6. Thus, we have obtained a total of 30 key genes with differential expression through transcriptome analysis.
3.7 Expression Pattern of Flower Development-Related Genes in Light Influencing Anthocyanin Biosynthesis
To avoid missing genes in the transcriptome analysis clustering algorithm, we further filtered genes annotated to the light signal pathway and anthocyanin synthesis pathway in the transcriptome, resulting in 16 genes related to the light signal pathway or anthocyanin synthesis.
According to the annotations of NR and Swissprot, we identified two unigenes annotated as red light receptors PHYs, respectively PHYC (c111957.graph_c0) and PHYE (c118794.graph_c0). Four unigenes annotated as blue light receptors CRYs, including CRY1 (c107160.graph_c1), CRY1a (c124901.graph_c0), CRY1b-2 (c107160.graph_c3), and CRY2 (c118543.graph_c0). We also found five unigenes annotated as UV-B receptor UVR8, namely UVR8-4 (c120777.graph_c0), UVR8-5 (c123052.graph_c0), UVR8-6 (c112558.graph_c0), UVR8-7 (c115587.graph_c0), and UVR8-8 (c113730.graph_c0). There is one unigene annotated as the inhibitor of photomorphogenesis COP1 (c106643.graph_c0). Additionally, we identified three PIF family transcription factors, PIF1 (c117264.graph_c0), PIF3 (c121983.graph_c0), and PIF7 (c104014.graph_c0), as well as one WRKY transcription factor, WRKY65 (c98625.graph_c0). Combining the three gene selection methods, we obtained a total of 46 differentially expressed genes (results are shown in Table S6).
RT-PCR analysis obtained 23 genes that appeared to be differentially expressed in ray florets of non-photosensitive chrysanthemum and photosensitive chrysanthemum (gels are presented in Fig. S5). These genes were selected for further validation by RT-qPCR in non-photosensitive chrysanthemums (20001402) and photosensitive chrysanthemums (2021135, Fig. 8). From the RT-qPCR results, seven genes were found not to be differentially expressed in two genotypes (Fig. S6a). Finally, we obtained 16 genes that appeared to be differentially expressed in light-independent and light-dependent floret (Fig. S6b).
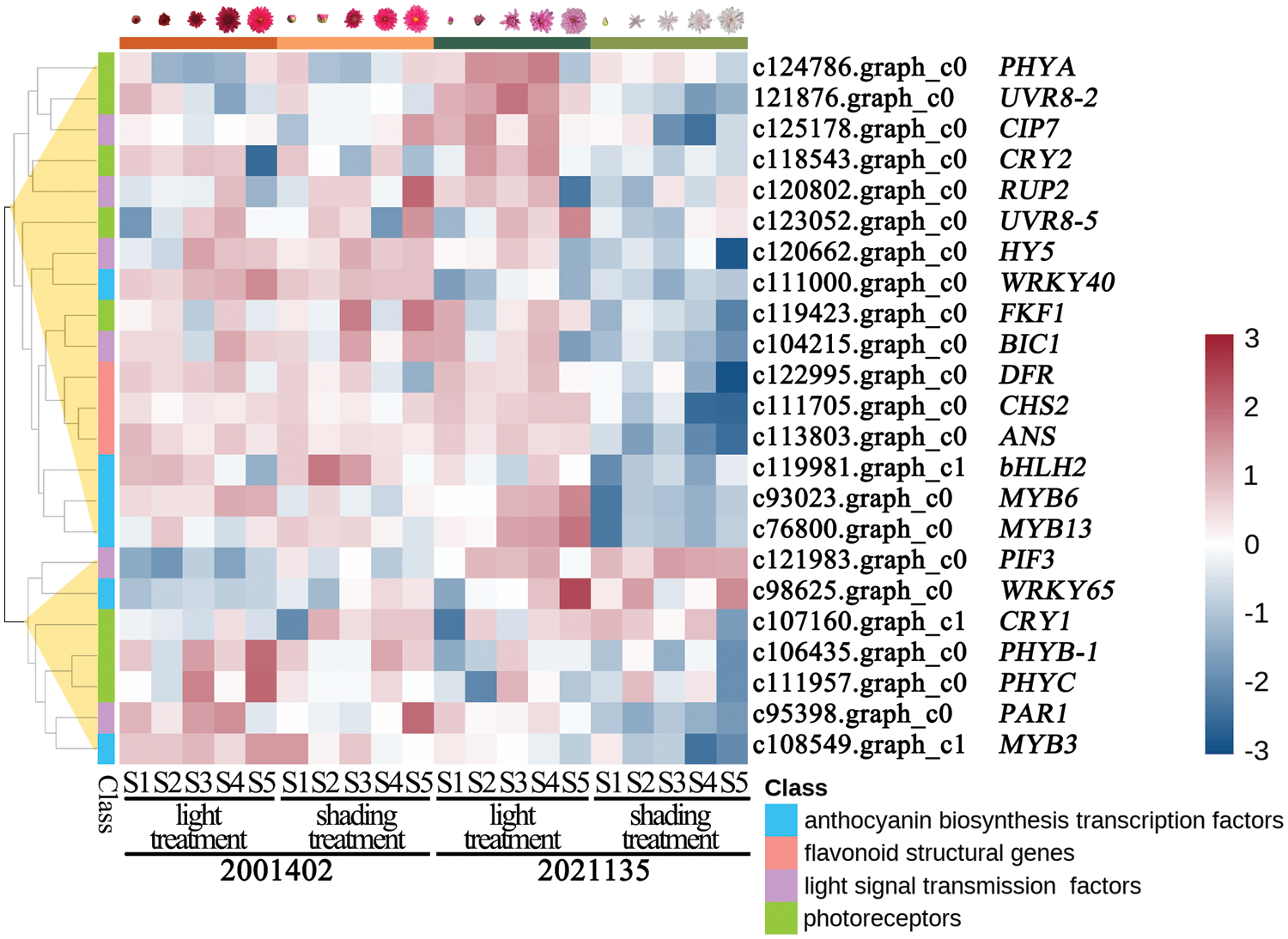
Figure 8: Expression patterns of 23 genes in non-photosensitive chrysanthemum (2001402) and photosensitive chrysanthemum (2021135) under light and dark conditions, respectively. The color scale represents the Log2. Genes that are up-regulated appear in red, and those that are down-regulated appear in blue
To determine the blue light signaling pathway and anthocyanin synthesis transcription factors’ function of anthocyanins light-independent synthesis, we isolated full-length cDNA sequences of CmCRY2, CmFKF1, CmBIC1, CmHY5, CmWRKY40, CmMYB6, CmMYB13 and CmbHLH2 in the ‘2001402’ and ‘2021135’ ray florets. Except for CmBIC1, no differences were observed in these genes amino acid sequences of the ‘2001402’ and ‘2021135’ ray florets (Figs. S7–S13). In 2001402, we identified two CmBIC1 sequences (Fig. S10b). The longer one was the same as it in 2021135, so we named the longer one CmBIC1.1, while the shorter one was CmBIC1.2.
3.8 Experimental Verification of the Regulatory Relationship between Anthocyanins Light-Independent Synthesis-Related Genes and Target Gene CmCHS2
Investigating the interaction mechanisms of transcription factors, CmCRY2 and CmFKF1 were found to interact with each other. Additionally, it was discovered that CmbHLH2 can interact with CmMYB6 and CmMYB13, forming the MBW complex (Fig. 9a). We have isolated and characterized the promoters of anthocyanin biosynthesis structural genes CmCHS in chrysanthemum. There were two light responsive elements, Box 4 and G-box. Additionally, A variety of cis-regulatory elements have been identified in the promoter sequences of CmCHS2, including those responsive to abscisic acid responsive (ABRE), methyl jasmonate responsive (CGTCA-motif, TGACG-motif), salicylic acid response element (CCATCTTTTT), auxin response element (TGA-box), and low-temperature response element (LTR). This suggests that the activity of the CmCHS2 promoter may be influenced by light, abscisic acid, methyl jasmonate, salicylic acid, auxin and low-temperature. To investigate the regulatory role of CmBIC1 in the anthocyanin biosynthetic pathway, yeast hybrid experiments were conducted with CmBIC1.1 and CmBIC1.2 and the promoter of CmCHS2. The yeast one-hybrid assay revealed that two CmBICs directly participant in anthocyanin synthesis by binding to the promoter of CmCHS2 (Fig. 9b).
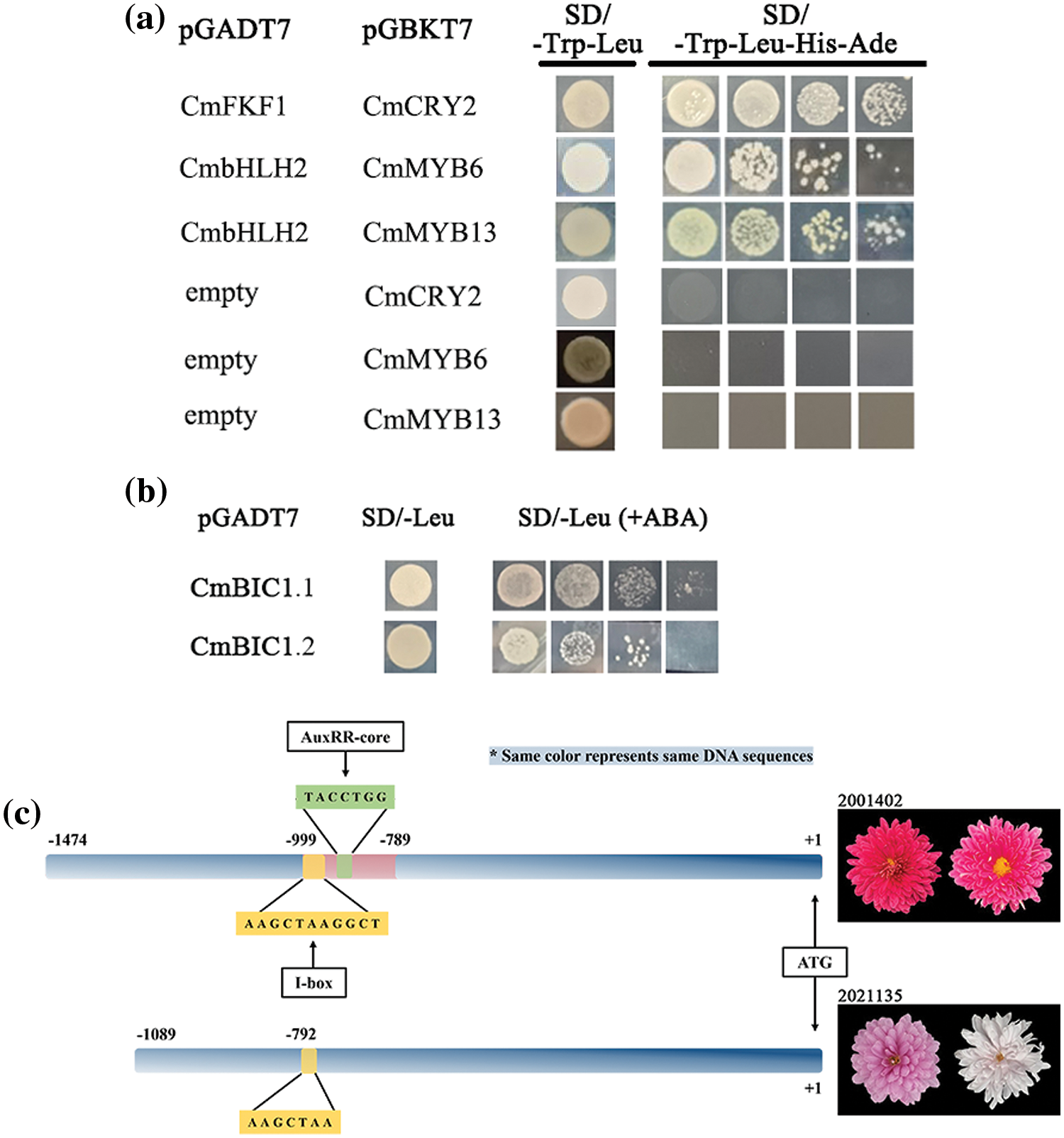
Figure 9: Yeast hybrid assays. (a) Y2H assays revealed interactions between CmFKF1 and CmCRY2, CmMYB6 or CmMYB13 with bHLH2; (b) Y1H assays revealed the binding of CmBIC1.1 and CmBIC1.2 with the anthocyanin biosynthesis gene CmCHS2 promoter; (c) Schematic of the differences between the CmBIC1 promoters in 2001402 and 2021135 showing positions of the ATG translation start site, 210 bp segment, the I-box, and the AuxRR-core in 2021135. To the right of the schematics are representative phenotypes
We further isolated the promoter of CmBIC1 in two non-photosensitive chrysanthemums ‘2001402’ and ‘GB200’, as well as in five photosensitive chrysanthemums ‘2021135’, ‘2004034’, ‘2032086’, ‘1907086’ and ‘1902077’. By conducting a multiple sequence alignment of the CmBIC1 promoters in these seven genotypes, we discovered a 210 bp segment exclusively present in non-photosensitive chrysanthemums (Figs. 9c and S14). Due to the insertion of this segment, we identified an additional light-responsive element (I-box) and an auxin-responsive element (AuxRR-core). So we indicated that CmBIC1 in non-photosensitive chrysanthemum might participate in the regulation of auxin.
4.1 The Impact of Light on the Coloration of Ray Florets Varies among Different Chrysanthemum Genotypes
Anthocyanins, belonging to the flavonoid family, are the main pigment in many flowers. Light plays a vital role in regulating anthocyanin biosynthesis [53]. In this study, shading treatment was applied to the capitulum of 103 chrysanthemum genotypes, and we found that most of the flowers under dark would fade considerably (Fig. 2a,b). As a result, 12 chrysanthemums were identified as light-independent anthocyanin accumulation, 40 as light semi-dependent anthocyanin accumulation, and 51 as light-dependent anthocyanin accumulation. This feature of photosensitive chrysanthemum that anthocyanins biosynthesis occurs in light-grown plants and does not occur in dark-grown plants was consistent with Arabidopsis and apple [54]. Similarly, The pigmentation of the ray florets of the chrysanthemum genotype ‘Purple Reagan’ is influenced by light [42].
Higher plants detect distinct ranges of light wavelengths, including ultraviolet-B to far-red, through various photoreceptors. The exposure of plants to various qualities of light can trigger distinct developmental reactions, such as the production of anthocyanin. In tomatoes, exposure to blue light and the overexpression expression of CRY1a have been found to be associated with increased accumulation of anthocyanin [19], while the total anthocyanin content in strawberry after treatment with the blue films was observed to be significantly lower than that of other treatment [55]. Here, we report that the photosensitive chrysanthemum ‘2021135’ is significantly responsive to blue light, and blue light supplemented with UV light results in higher anthocyanin accumulation in the ray florets. Moreover, when the chrysanthemums were exposed to both blue and UV-A/B light simultaneously, the accumulation of anthocyanin significantly increased compared to the anthocyanin accumulation when exposed to either blue or UV-A/B light alone. This suggests a synergistic effect between blue and UV-B light exposure. Similar synergistic effects have been observed in turnip seedlings [56].
The coloration of light-independent is of significant research importance for all plant species, such as the discovery of the sunlight-independent synthesis of anthocyanins in grapes ‘Jingyan’ [57]; turnip “Tsuda” (Brassica rapa) accumulates anthocyanin in a light-dependent manner [58]; anthocyanin accumulation in the dark-red ‘Hongdeng’ fruits is minimally influenced by light conditions [4]. Through the pigment content determination, the anthocyanin biosynthesis of the ray floret of ‘2001402’ remained at a relatively high level. This further demonstrates the important effect of the production of research on non-photosensitive chrysanthemums.
All tomato plants displayed an anthocyaninless phenotype under low-light conditions, regardless of their genetic background [5]. In contrast, We performed reciprocal crosses using the non-photosensitive chrysanthemum ‘2001402’ and the photosensitive chrysanthemum ‘B200’ as parents. After bagging, all 44 hybrids were able to synthesize anthocyanin, confirming the dominant nature of the light-independent trait in chrysanthemums. As the ability of the hybrids to accumulate anthocyanin in dark conditions varied, we speculate that the light-independent trait in chrysanthemums is determined by more than one dominant gene. This is different from the genetic analysis, which showed that the non-photosensitivity phenotype in eggplant is controlled by a singular dominant nuclear gene [59]. This result indicates that the light-independent synthesis of anthocyanin in chrysanthemums is complex.
4.2 Regulation of Anthocyanin Light-Independent Biosynthesis in Chrysanthemum by Light Signal Transduction Factors
By the RNA-Seq analysis, RT-PCR, and RT-qPCR analysis, we analyzed the regulation of light-independent for chrysanthemum. Our study discovered three types of light receptors: the blue light receptors CRY2 and FKF1, the red and far-red light receptors phytochromes phyA, and two UV-B light receptors UVR8.
Cryptochromes are the most widespread photoreceptors in plants that control growth and development. The CRY apoprotein contains two domains: the N-terminal PHR domain and the C-terminal CCE domain [60]. CmCRY2 in this research showcases highly conserved PHR and CCE domains at both termini, indicating similar functions in regulating plant growth. The CCE domain, as a cryptochrome effector domain, interacts with COP1 via its WD40 domain. In dark-grown Arabidopsis plants, COP1 plays a pivotal role in light signal transduction by engaging with light receptors and target proteins, and anthocyanin biosynthesis failed to occur [61]. Aside from the WD40 domain, COP1 features an RING-finger domain accountable for degrading various photomorphogenesis-promoting transcription factors [62]. COP1 exhibits E3 ubiquitin ligase activity and the ability to interact with light receptors upstream.
HY5, a bZIP protein, functions as a controller of photomorphogenic growth and acts as a light-controlled developmental molecular toggle; this conclusion has also been verified in eggplant [18,63]. In this study, the expression level of CmHY5 shows a similar trend to the relative content of anthocyanins and the expression levels of anthocyanin biosynthetic structural genes CmCHS2, CmDFR, and CmANS (Figs. 3b and 7b), indicating that CmHY5 involved in non-photosensitive chrysanthemums.
In Arabidopsis thaliana, AtFKF1 can interact with and negatively regulate COP1. This is similar to the expression tendencies of CmFKF1 in our study, in which the high expression of CmFKF1 reduces the activity of the COP1 complex, releasing a large amount of CmHY5 and further promoting anthocyanin synthesis. During the process of FKF1 expression change, only the protein-level function of the COP1 complex is affected, while the transcription level may remain unchanged. We speculate that this explains the role of COP1 as a pivotal mediator in the light signal transduction pathway, yet the expression tendencies in different photosensitivity types of chrysanthemums are irregular. For COP1 in dark conditions, it first interacts with one or more specific promoter binding proteins, and then binds to the target promoter to regulate transcriptional activity [25]. Except for HY5, COP1 actively polyubiquitinates many regulators, such as HFR1, PAPs, and CIPs, leading to suppressed light signal transmission and photomorphogenesis in the absence of light [64]. The CmCIP7 discovered in this study is a positive regulator of photomorphogenesis and anthocyanin formation, whose expression trend was consistent with the changes in anthocyanin content. In eggplant, SmCIP7-RNAi fruits exhibited a noticeable reduction in the presence of anthocyanins [27]. Based on the above analysis, we speculated that the irregular expression pattern of CmCOP1 may be due to the regulation of multiple regulatory factors and their complex mutual regulatory interactions.
In the intricate orchestration of the UVR8-mediated UV-B response, numerous molecular components play pivotal roles. Upon UV-B irradiation, UVR8 homodimers monomerize instantaneously to active monomers and activate the UV-B signaling pathway, thereby regulating the transcriptional expression of a series of downstream target genes. In the intricate orchestration of the UVR8-mediated UV-B response, numerous molecular components play pivotal roles. Upon exposure to UV-B irradiation, UVR8 homodimers promptly transform into active monomers, thereby initiating the UV-B signaling cascade. This activation modulates the transcriptional expression of a diverse array of downstream target genes. Notably, the RUP1 and RUP2 proteins, both belonging to the WD40-repeat protein family, they achieve this by suppressing the expression of HY5 which governs the expression of genes involved in anthocyanin biosynthesis [65]. The RLL4 gene in red lettuce leaves encodes a WD40 transcription factor, homologous to the RUP genes in Arabidopsis. However, a mis-sense mutation within the rll4 gene diminishes its suppressive function, ultimately resulting in a significant accumulation of anthocyanins [66]. Here, the expression pattern of CmRUP2 correlates with changes in anthocyanin accumulation, suggesting that its structural domain might also have a mutation, which led to the loss of its inhibitory effect on light signaling transduction and anthocyanin biosynthesis. RT-qPCR validation showed that UVR8 has different expression patterns among the two types of chrysanthemums (Fig. 7b). Two CmUVR8s with expression patterns consistent with anthocyanins were identified in this study. Similarly, SmUVR8 is a key photoreceptor for its anthocyanin synthesis in non-photosensitive eggplants [67]. Extant research has demonstrated that UVR8 possesses the capacity to stimulate anthocyanin synthesis in radish sprouts and Arabidopsis, and this synthesis process is contingent upon the presence on light [68,69].
Despite being the primary receptor for UV-B, estimates suggest that the photon absorption of UVR8 in sunlight extends into the blue spectral region. Consequently, it has been observed that UVR8 exerts an influence on blue light-induced gene expression [70]. In our study, blue and UV-A co-irradiation can significantly promote anthocyanin accumulation in chrysanthemums. So we predict that CmUVR8s also play an essential role in anthocyanins light-independent synthesis in chrysanthemum.
4.3 Blue Light Signal Transduction of Anthocyanins Light-Independent Synthesis in Chrysanthemum
We subjected the photosensitive chrysanthemum ‘2021135’ to different light qualities. As a result, the accumulation of anthocyanins in the ray florets of the chrysanthemum was highest under blue light treatment (λ = 460 nm). Furthermore, we observed differences in the sequence of both the ORF and promoter of CmBIC1, indicating that the blue light-mediated BIC1 in chrysanthemum has significant implications for non-light-dependent processes. Therefore, we further investigated the light signal transduction pathway mechanism involving the blue light receptors CmFKF1, CmCRY2, and the blue light signaling factor CmBIC1. Yeast two-hybrid experiments indicated that besides the interaction between CmCRY2 and CmFKF1, both CmBIC1 variants were unable to interact with other transcription factors under light and dark conditions (Fig. S15). However, in Arabidopsis, both BIC1 variants could interact with the CID (CRY-Interacting Domain) and PHR domains of CRY2 under light and dark conditions [30].
We cloned the promoter of CmBIC1, and through multiple sequence alignment of CmBIC1 promoters from 7 genotypes, we identified a 210 bp segment that is unique to non-photoperiodic chrysanthemums. This insertion led to the presence of an additional light-responsive element I-box and an auxin-responsive element AuxRR-core in the CmBIC1 promoter of non-photoperiodic chrysanthemums. I-box is speculated to be the binding site for HY5 [71]. In Gynandropsis gynandra, in vivo binding of light-responsive elements such as G-box and I-box motifs were associated with the rapid increase in transcripts of C4 genes, and the analysis confirmed that regions containing G-boxes and I-boxes were necessary for high expression [72]. Many studies suggest a significant role for auxin in light-mediated plant growth, for instance Arabidopsis CRY1 physically interacts with AUX/IAAs in a blue light-dependent way to hinder auxin signaling and hypocotyl elongation [6]. We hypothesize that similar mechanisms exist in non-photosensitive chrysanthemums. Given that the putative HY5 binding site I-box and auxin-responsive element was discovered in the promoters, we propose that the regulation of CmBIC1 in non-photosensitive chrysanthemum by HY5 and auxin, which allows high expression of CmBIC1s under darkness, thereby promoting the transcription of downstream anthocyanin structural genes.
Through yeast one-hybrid experiments, it has been demonstrated that CmBIC1 can bind to the downstream CHS promoter. In eggplant and tomato, the anthocyanin biosynthesis activator CRY promotes the accumulation of anthocyanins [18,19]. Both CmCRY2 and CmBIC1 in chrysanthemum show consistent changes in anthocyanin content, and CmBIC1 can bind to the downstream CHS promoter, similar to the functional study of BIC in eggplant [31]. Therefore, we have reason to speculate that CmBIC1 binds to CmCHS2 promoter and activates its expression, leading to the accumulation of anthocyanins.
4.4 Key Genes Were Involved in Anthocyanin Synthesis
Through transcriptome comparison and expression pattern validation, we found that CmCHS2, CmDFR, and CmANS were down-regulated after shading in photosensitive chrysanthemums, but in non-photosensitive chrysanthemums, there is no significant decrease in expression, and they still had a high expression level in dark conditions.
On the one hand, structural genes of anthocyanin biosynthesis are simultaneously regulated by the activated transcription factors. We also screened for 4 TFs with the same expression pattern as the structural genes, which contained 2 MYBs, 1 bHLH, and 1 WRKY (Fig. 7b). On the other hand, upon receiving a light signal, photoreceptors will proceed to adjust the activity of transcription factors (TFs), such as MYBs, bHLHs, and WD40. This process regulates the expression of structural genes, thereby exerting a profound impact on anthocyanin biosynthesis subsequently. At present, numerous R2R3-MYB transcription factors have been identified as key regulators involved in modulating anthocyanin synthesis, and some of them have been found to respond to light [9]. Sunlight exposure enhanced, while bagging suppressed the expression of LcMYB1 and accumulation of anthocyanin in the pericarp [73]. All SG6 R2R3-MYB genes control early anthocyanin biosynthesis in vegetative tissues [74,75]. After silencing ScMYB6 in Senecio cruentus, the leaves failed to accumulate anthocyanins, and the expression of structural genes decreased significantly [52]. Exposure to high light intensities triggers the expression of MYB12/PFG1 and MYB111/PFG3, members of the R2R3-MYB family derived from subgroup 7 (SG7) [76].
Based on differentially expressed genes between two types of chrysanthemums, we screened out CmMYB6 and CmMYB13 from the S6 and S7 subfamily, respectively, and CmbHLH2 from bHLH gene family, whose expression patterns correspond to changes in anthocyanin content. The CmMYB6 gene has been previously reported in chrysanthemum that over-expressed CmMYB6 could not separately induce anthocyanin in ‘Jimba’ petals because its transactivation was bHLH-dependent. Until now, only CmbHLH2 has been identified as a co-regulator with CmMYB6 in chrysanthemum [38]. In this study, we also found CmbHLH2 could interact with CmMYB6, which proved that MYB transcription factor and MBW complex play a key role in chrysanthemum color (Fig. 8a). In addition to CmMYB6, we observed that CmMYB13 can interact with CmbHLH2 to form a complex. Within the same subfamily as CmMYB13, CmMYB7-1 emerges as a potential participant in the negative regulation of anthocyanin biosynthesis [42]. However, CmMYB13 presented an expression trend consistent with the changes in anthocyanin content in our study (Fig. 7b). However, the expression trend of CmMYB13 was consistent with the change of anthocyanins in this study (Fig. 7b), so we believe that CmMYB13 may have unknown functions in the light-independent accumulation of anthocyanins in chrysanthemum. Subfamilies 2, 5 and 24 have been identified as regulators that play crucial roles in modulating flavonoid or anthocyanin metabolism [42]. In our study, the CmbHLH2 was from subfamily 5.
WRKY transcription factors are one of the most prominent transcription factor families in plants, and they are an integral part of the signaling network regulating many plant processes, including anthocyanin metabolism. For example, in eggplant, SmWRKY44 has been found to significantly increase anthocyanin accumulation, while in apple, MdWRKY40 can decrease the inhibition of anthocyanin synthesis by MdWRKY111, and simultaneously enhance the promotion of downstream structural genes by MdMYB1, thereby increasing anthocyanin accumulation. Similarly, in Arabidopsis, ectopic expression of BnWRKY41-1 can directly promote anthocyanin biosynthesis [31,5,77]. In this study, a CmWRKY40 was screened, and its expression pattern was consistent with changes in anthocyanin content. However, phylogenetic analysis indicated that CmWRKY40 is homologous to AtWRKY40, which is an important component of WRKY-mediated ABA signal transduction. Exogenous abscisic acid (ABA) treatment reduced its transcription and translation [78]. ABA also participates in controlling anthocyanin biosynthesis [79]. Therefore, it is speculated that chrysanthemum CmWRKY40 positively regulates anthocyanin accumulation through a similar pathway.
This study applied comparative transcriptome and yeast two/one-hybrid verification to elucidate the underlying molecular mechanism for anthocyanins light-independent synthesis in chrysanthemum. The comprehensive analyses have elucidated that the structural genes and transcription factors associated with photosensitivity in chrysanthemums undergo significant downregulation following exposure to shading conditions. In contrast, the expression levels in non-photosensitive chrysanthemums under dark conditions did not show significant down-regulation, and some genes even showed up-regulation. We believe that PHYA, CRY2, FKF1, UVR8-1, UVR8-2, HY5, CIP7, BIC1 (BIC1.1 and BIC1.2), RUP2, WRKY40, bHLH2, MYB6, and MYB13 may be involved in the regulation of light-independent anthocyanin biosynthesis in chrysanthemums. Through yeast one-hybrid verification, we found that a key light signal transduction factor, CmBIC1 can directly promote the expression of the anthocyanin structural gene CmCHS2. At the same time, the blue light receptor CmCRY2 can interact with CmFKF1, and CmbHLH2 can interact with CmMYB6 and CmMYB13. Our findings provide crucial insights that enhance the understanding of the formation mechanism of anthocyanins light-independent synthesis in chrysanthemum and provide a new strategy for energy-saving and environmental protection in horticultural production.
Acknowledgement: We thank the Beijing Botanical Garden for providing the greenhouse for the cultivation of different chrysanthemum cultivars.
Funding Statement: This project is supported by General Project of 2021 Science and Technology Plan of Beijing Municipal Education Comission (KM202111418001) and Beijing Natural Science Foundation, China (Grant No. 6212022).
Author Contributions: Xia Li, He Huang, Fangye Liu and Jiaping Qu conceived and designed this study. Fangye Liu, Jiaping Qu, Yajun Li, Jingwen Wu and Xueqi Gu performed the experiments. Fangye Liu and Jiaping Qu carried out the data analysis. Fangye Liu, Jiaping Qu, Jiawei Fan and Yumeng Cui wrote this manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request. All data generated in this study are included in this manuscript and its supplementary information.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.051386.
References
1. Gonzalez A, Zhao MZ, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–27. [Google Scholar] [PubMed]
2. Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20(3):176–85. [Google Scholar] [PubMed]
3. Li J, Ren L, Gao Z, Jiang MM, Liu Y, Zhou L, et al. Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Cell Environ. 2017;40(12):3069–87. [Google Scholar] [PubMed]
4. Guo X, Wang YT, Zhai ZF, Huang TJ, Zhao D, Peng X, et al. Transcriptomic analysis of light-dependent anthocyanin accumulation in bicolored cherry fruits. Plant Physiol Biochem. 2018;130:663–77. [Google Scholar] [PubMed]
5. Zhang YJ, Li Y, Li WP, Hu ZL, Yu XH, Tu Y, et al. Metabolic and molecular analysis of nonuniform anthocyanin pigmentation in tomato fruit under high light. Hortic Res. 2019;6:56. [Google Scholar] [PubMed]
6. Xu PB, Zawora C, Li Y, Wu J, Liu LC, Liu ZC, et al. Transcriptome sequencing reveals role of light in promoting anthocyanin accumulation of strawberry fruit. Plant Growth Regul. 2018;86(1):121–32. [Google Scholar]
7. Ma AN, Wang D, Lu HL, Wang HC, Qin YH, Hu GB, et al. LcCOP1 and LcHY5 control the suppression and induction of anthocyanin accumulation in bagging and debagging litchi fruit pericarp. Sci Hortic. 2021;287:110281. [Google Scholar]
8. Sun L, Li SC, Tang XP, Fan XC, Zhang Y, Jiang JF, et al. Transcriptome analysis reveal the putative genes involved in light-induced anthocyanin accumulation in grape ‘Red Globe’ (V. vinifera L.). Gene. 2020;728:144284. [Google Scholar] [PubMed]
9. Zoratti L, Karppinen K, Escobar AL, Häggman H, Jaakola L. Light-controlled flavonoid biosynthesis in fruits. Front Plant Sci. 2014;5:534. [Google Scholar] [PubMed]
10. Henry-Kirk RA, Plunkett B, Hall M, McGhie T, Allan AC, Wargent JJ, et al. Solar UV light regulates flavonoid metabolism in apple (Malus × domestica). Plant Cell Environ. 2018;41(3):675–88. [Google Scholar] [PubMed]
11. Castagna A, Dall’Asta C, Chiavaro E, Galaverna G, Ranieri A. Effect of post-harvest UV-B irradiation on polyphenol profile and antioxidant activity in flesh and peel of tomato fruits. Food Bioprocess Technol. 2014;7(8):2241–50. [Google Scholar]
12. Zhao Y, Dong WQ, Wang K, Zhang B, Allan AC, Wang KL, et al. Differential sensitivity of fruit pigmentation to ultraviolet light between two peach cultivars. Front Plant Sci. 2017;8:01552. [Google Scholar]
13. Zhou LJ, Wang YX, Wang YG, Song AP, Jiang JF, Chen SM, et al. Transcription factor CmbHLH16 regulates petal anthocyanin homeostasis under different lights in chrysanthemum. Plant Physiol. 2022;190(2):1134–52. [Google Scholar] [PubMed]
14. Kong SG, Okajima K. Diverse photoreceptors and light responses in plants. J Plant Res. 2016;129(2):111–4. [Google Scholar] [PubMed]
15. Oh S, Warnasooriya SN, Montgomery BL. Mesophyll-localized phytochromes gate stress-and light-inducible anthocyanin accumulation in Arabidopsis thaliana. Plant Signal Behav. 2014;9(3):e28013. [Google Scholar] [PubMed]
16. Legris M, Ince YÇ, Fankhauser C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun. 2019;10:5219. [Google Scholar] [PubMed]
17. Rai N, Morales LO, Aphalo PJ. Perception of solar UV radiation by plants: photoreceptors and mechanisms. Plant Physiol. 2021;186(3):1382–96. [Google Scholar] [PubMed]
18. Jiang MM, Ren L, Lian HL, Liu Y, Chen HY. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016;249:46–58. [Google Scholar] [PubMed]
19. Liu CC, Chi C, Jin LJ, Zhu J, Yu JQ, Zhou YH. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 2018;41(8):1762–75. [Google Scholar] [PubMed]
20. Kadomura-Ishikawa Y, Miyawaki K, Noji S, Takahashi A. Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria × ananassa fruits. J Plant Res. 2013;126(6):847–57. [Google Scholar] [PubMed]
21. Ponnu J. Molecular mechanisms suppressing COP1/SPA E3 ubiquitin ligase activity in blue light. Physiol Plant. 2020;169(3):418–29. [Google Scholar] [PubMed]
22. Hu JF, Fang HC, Wang J, Yue XX, Su MY, Mao ZL, et al. Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. 2020;292:110377. [Google Scholar] [PubMed]
23. Moriconi V, Binkert M, Costigliolo C, Sellaro R, Ulm R, Casal JJ. Perception of sunflecks by the UV-B photoreceptor UV resistance locus8. Plant Physiol. 2018;177(1):75–81. [Google Scholar] [PubMed]
24. Ma CQ, Liang BW, Chang B, Yan JY, Liu L, Wang Y, et al. Transcriptome profiling of anthocyanin biosynthesis in the peel of ‘Granny Smith’ apples (Malus domestica) after bag removal. BMC Genomics. 2019;20:353. [Google Scholar] [PubMed]
25. Yamamoto YY, Matsui M, Ang LH, Deng XW. Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell. 1998;10(7):1083–94. [Google Scholar] [PubMed]
26. Qiu ZK, Wang HJ, Li DJ, Yu BW, Hui QL, Yan SS, et al. Identification of candidate HY5-dependent and -independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol. 2019;60(3):643–56. [Google Scholar] [PubMed]
27. Li Y, Xing MH, Yang Q, Wang Y, Jiang J, Zhao YK, et al. SmCIP7, a COP1 interactive protein, positively regulates anthocyanin accumulation and fruit size in eggplant. Int J Biol Macromol. 2023;234:123729. [Google Scholar] [PubMed]
28. Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, et al. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 2004;38(5):725–39. [Google Scholar] [PubMed]
29. Zhou P, Song MF, Yang QH, Su L, Hou P, Guo L, et al. Both phytochrome rapidly regulated1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 2014;164(2):841–52. [Google Scholar] [PubMed]
30. Wang Q, Zuo ZC, Wang X, Gu LF, Yoshizumi T, Yang ZH, et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science. 2016;354(6310):343–7. [Google Scholar] [PubMed]
31. He YJ, Li DL, Li SH, Liu Y, Chen HY. SmBICs inhibit anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Cell Physiol. 2021;62(6):1001–11. [Google Scholar] [PubMed]
32. Podolec R, Demarsy E, Ulm R. Perception and signaling of Ultraviolet-B radiation in plants. Annu Rev Plant Biol. 2021;72:793–822. [Google Scholar] [PubMed]
33. Shi B, Wu HX, Zheng B, Qian MJ, Gao AP, Zhou KB. Analysis of light-independent anthocyanin accumulation in Mango (Mangifera indica L.). Horticulturae. 2021;7(11):423. [Google Scholar]
34. Qian MJ, Zhang D, Yue XY, Wang SK, Li XG, Teng YW. Analysis of different pigmentation patterns in ‘Mantianhong’ (Pyrus pyrifolia Nakai) and ‘Cascade’ (Pyrus communis L.) under bagging treatment and postharvest UV-B/visible irradiation conditions. Sci Hortic. 2013;151:75–82. [Google Scholar]
35. Butelli E, Licciardello C, Zhang Y, Liu JJ, Mackay S, Bailey P, et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24(3):1242–55. [Google Scholar] [PubMed]
36. Xu ZS, Yang QQ, Feng K, Yu X, Xiong AS. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol J. 2020;18(7):1585–97. [Google Scholar] [PubMed]
37. Xiang LL, Liu XF, Li H, Yin XR, Grierson D, Li F, et al. CmMYB#7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J Exp Bot. 2019;70(12):3111–23. [Google Scholar] [PubMed]
38. Xiang LL, Liu XF, Shi YN, Li YJ, Li WD, Li F, et al. Comparative transcriptome analysis revealed two alternative splicing bHLHs account for flower color alteration in chrysanthemum. Int J Mol Sci. 2021;22(23):12769. [Google Scholar] [PubMed]
39. Wang YG, Zhou LJ, Wang YX, Geng ZQ, Liu SH, Chen CW, et al. CmMYB9a activates floral coloration by positively regulating anthocyanin biosynthesis in chrysanthemum. Plant Mol Biol. 2022;108(1–2):51–63. [Google Scholar] [PubMed]
40. Yao XQ, Chu JZ, He XL, Si C. The effects of UV-B radiation intensity on biochemical parameters and active ingredients in flowers of Qi chrysanthemum and Huai chrysanthemum. Photochem Photobiol. 2014;90(6):1308–13. [Google Scholar] [PubMed]
41. Zhou LJ, Geng ZQ, Wang YX, Wang YG, Liu SH, Chen CW, et al. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic Res. 2021;8(1):248. [Google Scholar] [PubMed]
42. Hong Y, Tang XJ, Huang H, Zhang Y, Dai SL. Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum. BMC Genomics. 2015;16:202. [Google Scholar] [PubMed]
43. Hong Y, Yang LW, Li ML, Dai SL. Comparative analyses of light-induced anthocyanin accumulation and gene expression between the ray florets and leaves in chrysanthemum. Plant Physiol Biochem. 2016;103:120–32. [Google Scholar] [PubMed]
44. Hong Y, Li ML, Dai SL. Ectopic expression of multiple chrysanthemum (Chrysanthemum × morifolium) R2R3-MYB transcription factor genes regulates anthocyanin accumulation in tobacco. Genes. 2019;10(10):777. [Google Scholar] [PubMed]
45. Huang H, Hu K, Han KT, Xiang QY, Dai SL. Flower colour modification of Chrysanthemum by suppression of F3′H and overexpression of the exogenous Senecio cruentus F3′5′H gene. PLoS One. 2013;8(11):e74395. [Google Scholar]
46. Lu CF, Pu Y, Liu YT, Li YJ, Qu JP, Huang H, et al. Comparative transcriptomics and weighted gene co-expression correlation network analysis (WGCNA) reveal potential regulation mechanism of carotenoid accumulation in Chrysanthemum × morifolium. Plant Physiol Biochem. 2019;142:415–28. [Google Scholar] [PubMed]
47. Li B, Dewer CN. RSEM: accurate transcript quantification from RNA seq data with or without a reference genome. BMC Bioinform. 2011;12:323. [Google Scholar]
48. Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–43. [Google Scholar] [PubMed]
49. Huang H, Wang Y, Wang SL, Wu X, Yang K, Niu YJ. Transcriptome-wide survey and expression analysis of stress-responsive NAC genes in Chrysanthemum lavandulifolium. Plant Sci. 2012;193–194:18–27. [Google Scholar] [PubMed]
50. Hong Y, Dai SL. Selection of reference genes for real-time quantitative polymerase chain reaction analysis of light-dependent anthocyanin biosynthesis in chrysanthemum. J Am Soc Hortic Sci. 2015;140(1):68–77. [Google Scholar]
51. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Methods. 2013;25(4):402–8. [Google Scholar]
52. Cui YM, Fan JW, Lu CF, Ren JS, Qi FT, Huang H, et al. ScGST3 and multiple R2R3–MYB transcription factors function in anthocyanin accumulation in Senecio cruentus. Plant Sci. 2021;313:111094. [Google Scholar] [PubMed]
53. Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z. Genome-scale transcriptomic insights into early-stage fruit development in woodland Strawberry Fragaria vesca. Plant Cell. 2013;25(6):1960–78. [Google Scholar] [PubMed]
54. Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, et al. MdCOP1 Ubiquitin E3 Ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012;160(2):1011–22. [Google Scholar] [PubMed]
55. Miao LX, Zhang YC, Yang XF, Xiao JP, Zhang HQ, Zhang ZF, et al. Colored light-quality selective plastic films affect anthocyanin content, enzyme activities, and the expression of flavonoid genes in strawberry (Fragaria × ananassa) fruit. Food Chem. 2016;207:93–100. [Google Scholar] [PubMed]
56. Wang Y, Zhou B, Sun M, Li YH, Kawabata S. UV-A light induces anthocyanin biosynthesis in a manner distinct from synergistic blue plus UV-B light and UV-A/blue light responses in different parts of the hypocotyls in turnip seedlings. Plant Cell Physiol. 2012;53(8):1470–80. [Google Scholar] [PubMed]
57. Zheng Y, Li JH, Xin HP, Wang N, Guan L, Wu BH, et al. Anthocyanin profile and gene expression in berry skin of two red Vitis vinifera grape cultivars that are sunlight dependent versus sunlight independent. Aust J Grape Wine Res. 2013;19(2):238–48. [Google Scholar]
58. Yang JF, Chen YZ, Kawabata S, Li YH, Wang Y. Identification of light-independent anthocyanin biosynthesis mutants induced by ethyl methane sulfonate in turnip “Tsuda” (Brassica rapa). Int J Mol Sci. 2017;18(7):1288. [Google Scholar] [PubMed]
59. He YJ, Li SH, Dong YX, Zhang XT, Li DL, Liu Y, et al. Fine mapping and characterization of the dominant gene SmFTSH10 conferring non-photosensitivity in eggplant (Solanum melongena L.). Theor Appl Genet. 2022;135(7):2187–96. [Google Scholar] [PubMed]
60. Liu HT, Liu B, Zhao CX, Pepper M, Lin CT. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16:684–91. [Google Scholar] [PubMed]
61. Hoecker U, Quail PH. The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem. 2001;276(41):38173–8. doi:10.1074/jbc.M103140200. [Google Scholar] [PubMed] [CrossRef]
62. Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17(10):584–93. doi:10.1016/j.tplants.2012.05.004. [Google Scholar] [PubMed] [CrossRef]
63. Xu DQ, Jiang Y, Li JG, Lin F, Holm M, Deng XW. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci U S A. 2016;113(27):7655–60. doi:10.1073/pnas.1607687113. [Google Scholar] [PubMed] [CrossRef]
64. Han X, Huang X, Deng XW. The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun. 2020;1(3):100044. doi:10.1016/j.xplc.2020.100044. [Google Scholar] [PubMed] [CrossRef]
65. Ren H, Han JP, Yang PY, Mao WW, Liu X, Qiu LL, et al. Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc Natl Acad Sci U S A. 2019;116(10):4722–31. doi:10.1073/pnas.1816268116. [Google Scholar] [PubMed] [CrossRef]
66. Su WQ, Tao R, Liu WY, Yu CC, Yue Z, He SP, et al. Characterization of four polymorphic genes controlling red leaf colour in lettuce that have undergone disruptive selection since domestication. Plant Biotechnol J. 2020;18(2):479–90. [Google Scholar] [PubMed]
67. He YJ, Chen H, Zhou L, Liu Y, Chen HY. Comparative transcription analysis of photosensitive and non-photosensitive eggplants to identify genes involved in dark regulated anthocyanin synthesis. BMC Genomics. 2019;20(1):678. [Google Scholar] [PubMed]
68. Mao K, Wang L, Li YY, Wu RL. Molecular cloning and functional analysis of UV Resistance Locus 8 (PeUVR8) from Populus euphratica. PLoS One. 2015;10(7):e0132390. [Google Scholar] [PubMed]
69. Wu Q, Su NN, Zhang XY, Liu YY, Cui J, Liang YC. Hydrogen peroxide, nitric oxide and UV RESISTANCE LOCUS8 interact to mediate UV-B-induced anthocyanin biosynthesis in radish sprouts. Sci Rep. 2016;6:29164. [Google Scholar] [PubMed]
70. Rai N, O’Hara A, Farkas D, Safronov O, Ratanasopa K, Wang F, et al. The photoreceptor UVR8 mediates the perception of both UV-B and UV-A wavelengths up to 350 nm of sunlight with responsivity moderated by cryptochromes. Plant Cell Environ. 2020;43(6):1513–27. [Google Scholar] [PubMed]
71. Wang X, Wang Q, Han YJ, Liu Q, Gu LF, Yang ZH, et al. A CRY-BIC negative-feedback circuitry regulating blue light sensitivity of Arabidopsis. Plant J. 2017;92(3):426–36. [Google Scholar] [PubMed]
72. Singh P, Tevenson SR, Dickinson PJ, Reyna-Llorens I, Tripathi A, Reeves G, et al. C4 gene induction during de-etiolation evolved through changes in cis to allow integration with ancestral C3 gene regulatory networks. Sci Adv. 2003;29(13):eade9756. [Google Scholar]
73. Lai B, Li XJ, Hu B, Qin YH, Huang XM, Wang HC, et al. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PLoS One. 2014;9(1):e86293. [Google Scholar] [PubMed]
74. Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–81. [Google Scholar] [PubMed]
75. Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013;587(10):1543–7. [Google Scholar] [PubMed]
76. Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 2015;169(3):1862–80. [Google Scholar] [PubMed]
77. Duan SW, Wang JJ, Gao CH, Jin CY, Li D, Peng DS, et al. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018;268:47–53. [Google Scholar] [PubMed]
78. Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, et al. The Mg-chelatase h subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22(6):1909–35. [Google Scholar] [PubMed]
79. Lacampagne S, Gagné S, Gény L. Involvement of abscisic acid in controlling the proanthocyanidin biosynthesis pathway in grape skin: new elements regarding the regulation of tannin composition and leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) activities and expression. J Plant Growth Regul. 2010;29(1):81–90. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools