 Open Access
Open Access
ARTICLE
Genome-Wide Discovery and Expression Profiling of the SWEET Sugar Transporter Gene Family in Woodland Strawberry (Fragaria vesca) under Developmental and Stress Conditions: Structural and Evolutionary Analysis
1 College of Environmental and Biological Engineering, Putian University, Putian, 351100, China
2 College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, 350002, China
3 Fujian Provincial Key Laboratory of Ecology-Toxicological Effects & Control for Emerging Contaminants, Putian University, Putian, 351100, China
4 Key Laboratory of Ecological Environment and Information Atlas (Putian University), Fujian Provincial University, Putian, 351100, China
5 Affiliated Hospital of Putian University, Putian, 351100, China
* Corresponding Authors: Shoukai Lin. Email: ; Fuxiang Qiu. Email:
Phyton-International Journal of Experimental Botany 2024, 93(7), 1485-1502. https://doi.org/10.32604/phyton.2024.050990
Received 24 February 2024; Accepted 27 May 2024; Issue published 30 July 2024
Abstract
The SWEET (sugar will eventually be exported transporter) family proteins are a recently identified class of sugar transporters that are essential for various physiological processes. Although the functions of the SWEET proteins have been identified in a number of species, to date, there have been no reports of the functions of the SWEET genes in woodland strawberries (Fragaria vesca). In this study, we identified 15 genes that were highly homologous to the A. thaliana AtSWEET genes and designated them as FvSWEET1–FvSWEET15. We then conducted a structural and evolutionary analysis of these 15 FvSWEET genes. The phylogenetic analysis enabled us to categorize the predicted 15 SWEET proteins into four distinct groups. We observed slight variations in the exon‒intron structures of these genes, while the motifs and domain structures remained highly conserved. Additionally, the developmental and biological stress expression profiles of the 15 FvSWEET genes were extracted and analyzed. Finally, WGCNA coexpression network analysis was run to search for possible interacting genes of FvSWEET genes. The results showed that the FvSWEET10 genes interacted with 20 other genes, playing roles in response to bacterial and fungal infections. The outcomes of this study provide insights into the further study of FvSWEET genes and may also aid in the functional characterization of the FvSWEET genes in woodland strawberries.Keywords
Woodland strawberry (Fragaria vesca), also known as wild strawberry, is diploid, with two pairs of a base haploid count of seven chromosomes (2n = 14). Woodland strawberries are popular among consumers because of their greater nutritional value than cultivated strawberries [1]. Additionally, because of its small size, short reproductive cycle, simple and efficient genetic transformation system, and abundant genetic resources, the woodland strawberry is considered an important model plant of the Rosaceae family for genomics research [2]. In recent years, researchers have used Pacific Biosciences (PacBio) sequencing technology to sequence the whole genome of woodland strawberries, obtaining a nearly complete genome sequence and updated annotations for this plant [3,4]. The findings of these studies have provided adequate genetic resources for conducting genome-wide analyses of woodland strawberries.
Sugars are the major sources of energy and biological carbon for living organisms [5]. Cellular sugar outflow and absorption across the plasma membrane are the most important processes in growth and development [6]. The SWEET gene family is a recently discovered class of sugar transporters that play crucial roles in plants and mammals [7]. The SWEET proteins are structurally conserved and energy-independent sugar transporter proteins that possess two MtN3/saliva transmembrane domains [8]. The first member of the SWEET family, MtN3, was identified in the interaction between the legume Medicago truncatula and Rhizobium [9]. In subsequent studies, SWEET proteins were identified as sucrose and fructose transporters [7,9].
Physiological, biochemical, and molecular experiments indicate that SWEET protein is involved in many life processes of plants, including growth and senescence [10,11], environmental adaptation [12], seed and pollen development, and tolerance to cold and drought stresses [7,13–16]. For instance, a considerable number of SWEET genes have been identified in Arabidopsis and their functional verification was conducted simultaneously. It has been reported that AT5G40260 (AtSWEET15) is inseparable from the vitality of pollen, and the decrease of its gene expression can lead to the decrease of starch content in pollen grains, thus resulting in male sterility [7]. In view of the fact that AT5G13170 (AtSWEET15) is significantly up-regulated by about 22 times in senescent leaves, it may play a significant role in sugar transport [6]. Overexpression of AT3G16690 (AtSWEET16) has revealed significant changes in plants showing different sugar levels and developmental processes such as germination and growth [17]. The AT4G15920 (AtSWEET17) gene knockout mutant has revealed the phenotypic change of its root system. Furthermore, the expression level of the SWEET gene in plants is also regulated by Botrytis cinerea and some mycorrhizal fungi, resulting in changes in its expression, as shown in other studies [15,18]. Finally, in many plants such as A. thaliana (17) [7], Oryza sativa (21) [12], Vitis vinifera (14) [14], Malus domestica (33) [15] and Solanum lycopersicum (29) [19], SWEET gene has been identified.
In this study, we performed a whole-genome analysis to identify and name the FvSWEET genes in woodland strawberries. In addition, the evolutionary relationships, chromosomal locations, structural features, and interaction genes of the FvSWEET genes were investigated. Then, by using publicly available RNA-seq data from woodland strawberries, the expression patterns of FvSWEET genes during fruit ripening as well as in different tissues in response to different biological stresses were analyzed. Finally, WGCNA coexpression network analysis was performed to search for genes possibly interacting with FvSWEET genes. This research will help us further understand the FvSWEET genes and their roles in fruit ripening and biotic stress responses, laying the groundwork for functional research on FvSWEET genes in woodland strawberries.
2.1 Genome-Wide Identification of FvSWEET Genes
The reference genome (version 4.0) and annotation (version 4.0 a2) of woodland strawberries were obtained from the Genome Database for Rosaceae (GDR) database (https://www.rosaceae.org/, accessed on 18/04/2024). The amino acid sequence files of the A. thaliana SWEET gene family are downloaded from the Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/, accessed on 18/04/2024). Then, using the TBtools [20], the genes of Arabidopsis were mapped to the strawberry genome under the screening condition that the E-value was less than 0.05. Proteins with match ratios of at least 50% were retained, and the redundant proteins were removed. Then, the obtained proteins were verified against the Pfam database (http://pfam.xfam.org/search#tabview=tab1, accessed on 18/04/2024) target proteins, which contain the MtN3_slv domain (PF03083). The FvSWEET family was subsequently confirmed after removing the short sequences and redundant and shorter alternative splicing products. The FvSWEET genes were then renamed according to their gene numbers. Additionally, utilizing ProParam on the ExPASy website (http://web.ExPASy.org/protparam/, accessed on 18/04/2024) analyzes the isoelectric point (pI), amino acid length, and molecular weight of each gene in FvSWEET. PSORT predicted the subcellular localization of these genes (https://psort1.hgc.jp/form.html, accessed on 18/04/2024). All analysis tools were used with default parameters unless otherwise indicated, as also indicated below. The TMHMM website (http://cbs.dtu.dk/services/TMHMM/, accessed on 18/04/2024) was used to predict the number of transmembrane helices (TMHs) in the protein sequences.
A phylogenetic tree of the SWEETs of F. vesca and A. thaliana was constructed using MEGA7.0 software [21], with their amino acid sequences used as inputs. Multiple sequence alignments were performed by ClustalW in MEGA, and then the neighbor-joining (NJ) method with 1000 bootstrap replicates was used to construct the original tree. The output tree was plotted by using the online plotting tool Evolview (https://www.evolgenius.info/evolview/#login, accessed on 18/04/2024).
2.3 Chromosomal Location Analysis
The chromosomal locations of the FvSWEET genes were identified according to the chromosomal information derived from the genome annotation (version 4.0 a2). MG2C (http://mg2c.iask.in/mg2c_v2.0/, accessed on 18/04/2024) was used to locate the positions of the FvSWEET genes on the corresponding chromosomes, thus providing chromosomal location information.
To analyze the replication events of the SWEET genes among species, the genomic data of Arabidopsis (TAIR10) and woodland strawberry (version 4.0 a2) were downloaded from the Arabidopsis (https://www.arabidopsis.org/, accessed on 18/04/2024) and GDR (https://www.arabidopsis.org/) databases, respectively. Finally, collinearity analysis of the SWEET genes among the species was performed by the one-step MCScanX program in TBtools.
2.5 Structural Analysis of FvSWEET Genes and their Encoded Proteins
The MEME tool (http://meme-suite.org/, accessed on 18/04/2024) was utilized to analyze conserved protein motifs in the FvSWEET proteins, with the motif number parameter set to 8. The Gene Structure Display Server (http://gsds.cbi.pku.edu.cn, accessed on 18/04/2024) was used to display the exon-intron structures of the FvSWEET genes according to the genome annotation (version 4.0 a2) of woodland strawberries.
2.6 RNA-seq Data Collection from the NCBI SRA Database
In order to further explore the importance of the FvSWEET gene in F. vesca development and biological stress, we obtained data from the SRA database on the NCBI website (https://www.ncbi.nlm.nih.gov/sra/, accessed on 18/04/2024) download and collect RNA-seq raw data on strawberry growth and related stress, totaling 38. Eighteen of these datasets (SRR5217575–SRR5217592) contained fruit development data for two tissues, namely, the achenes (Ac) and receptacle (Rc); three strawberry strains, namely, Hawaii 4 (HW), yellow wonder (YW), and Reine des Vallées (Rdv); and three stages of development, namely, 10 DPA (days postanthesis), 25 DPA (intermediate), and 35 DPA (ripe) [22]. Other datasets contained biotic stress-related data for F. vesca, including virus (strawberry vein banding virus, SVBV)-infected leaves at 40 dpi (days postinoculation) (SRR1930097 and SRR1930099) [5], fungus (Phytophthora cactorum)-infected roots at 48 hpi (hours postinoculation) (SRR8840916–SRR8840927) [23] and fungus (Botrytis cinerea)-infected white fruits at 14 DPA and red fruits at 21 hpi (SRR3743193–SRR3743198) [24].
2.7 Expression Analysis of FvSWEET Genes
The expression analysis was performed according to Xiong et al.’s method [25]. The SRA Toolkit (v.2.9.6) (https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software, accessed on 18/04/2024) was used to convert the downloaded SRA format data into FASTQ format. Trimmomatic (v.3.0) [26] was used to trim the low-quality reads and Illumina adapter sequences. HISAT2 (v.2.1.0) [27] was utilized to map the clean reads to the F. vesca reference genome (version 4.0). SAMtools (v.1.4) [28] was used to convert and sort the HISAT2 output files. The FPKM values were calculated using Cufflinks (v.2.2.1) [29]. The FPKM values of 15 FvSWEET genes were extracted from FPKM value matrixes and plotted as heatmaps by TBtools [20]. Genes with RPKM values greater than 1 were defined as expressed genes.
Coexpression analysis was conducted with RNA-seq data using the WGCNA R package to construct gene coexpression networks. The log2 (FPKM of each treatment + 1) was used as input, and the soft-thresholding power was set to 6.
GO enrichment analysis was conducted using the online eggNOG mapper (https://www.eggNOG-mapper.embl.de, accessed on 18/04/2024). The default settings of the app were used to annotate the functionality of the strawberry genome. Then, the annotation results were imported into the Omics share cloud platform (https://www.omicshare.com/tools/, accessed on 18/04/2024) to plot the enriched GO terms, and we presented the top 20 enriched terms.
3.1 Identification of FvSWEET Genes
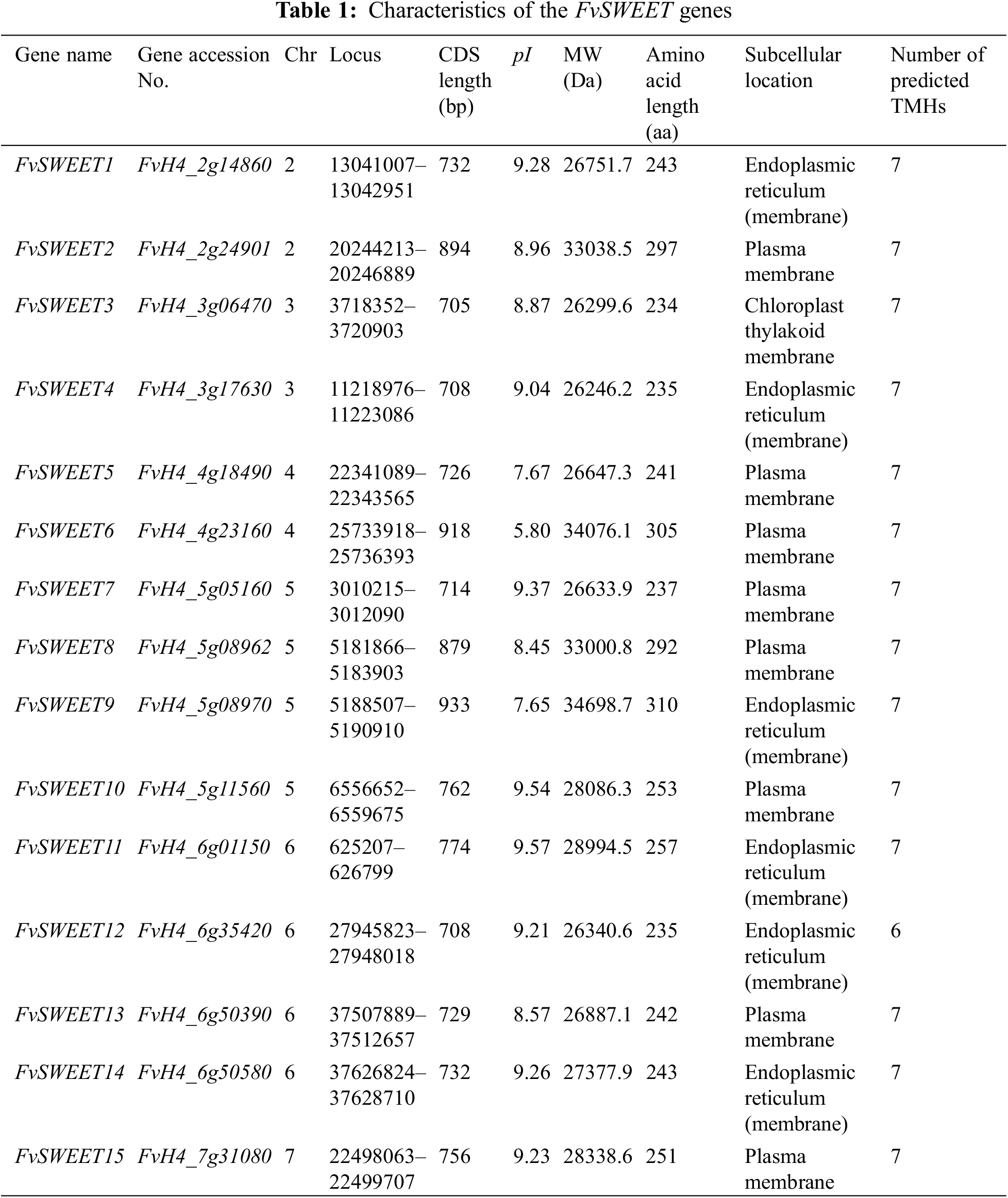
Fifteen typical FvSWEET genes were identified from the F. vesca reference genome (version 4.0) and were renamed FvSWEET1–FvSWEET15. Table 1 summarizes the basic genetic information. The molecular weights (MWs) of the encoded proteins varied from 26246.2 Da (FvSWEET4) to 34698.7 Da (FvSWEET9), the amino acid (aa) lengths ranged from 235 aa (FvSWEET12) to 310 aa (FvSWEET9), and the isoelectric points (pIs) varied from 5.80 (FvSWEET6) to 9.57 (FvSWEET11). Subcellular localization analysis revealed that the FvSWEET proteins were distributed on the plasma membrane (FvSWEET2, FvSWEET5, FvSWEET6, FvSWEET7, FvSWEET8, FvSWEET10, FvSWEET13, and FvSWEET15), endoplasmic reticulum membrane (FvSWEET1, FvSWEET4, FvSWEET9, FvSWEET11, FvSWEET12, and FvSWEET14), and chloroplast thylakoid membrane (FvSWEET3) (Table 1), indicating that they are transmembrane proteins. The prediction results of the transmembrane domains showed that all the SWEET family members had transmembrane domains, with the majority having 7 transmembrane domains, except for FvSWEET12, which contained 6 transmembrane domains.
3.2 Phylogenetic Analysis of FvSWEET and AtSWEET Gene Family Members
To evaluate the evolutionary relationship of SWEET proteins in F. vesca and A. thaliana, a phylogenetic tree was constructed using the neighbor-joining (NJ) approach implemented in MEGA 7.0 software. As shown in Fig. 1, the SWEET gene family is divided into four groups, namely clade I (13), II (8), III (5), and IV (7). Clade I included FvSWEET12, AT5G62850, FvSWEET7, AT3G28007, FvSWEET14, FvSWEET13, FvSWEET10, AT4G10850, AT1G66770, AT5G40260, FvSWEET5, AT4G15920, and AT3G16690. Clade II included FvSWEET15, AT5G53190, FvSWEET1, AT1G21460, FvSWEET3, FvSWEET4, and AT3G14770. Clade III included FvSWEET11, AT2G39060, FvSWEET2, FvSWEET8, and AT5G50790. Clade IV included FvSWEET6, AT5G13170, FvSWEET9, AT5G50800, AT4G25010, AT5G23660, and AT3G48740. There were 7 pairs of SWEET orthologous between F. vesca and A. thaliana, including FvSWEET12 and AT5G62850, FvSWEET15 and AT5G53190, FvSWEET1 and AT1G21460, FvSWEET4 and AT3G14770, FvSWEET11 and AT2G39060, FvSWEET8 and AT5G50790, and FvSWEET6 and AT5G13170. These results suggested that the 7 pairs of SWEET orthologous between F. vesca and A. thaliana may have the same functions.
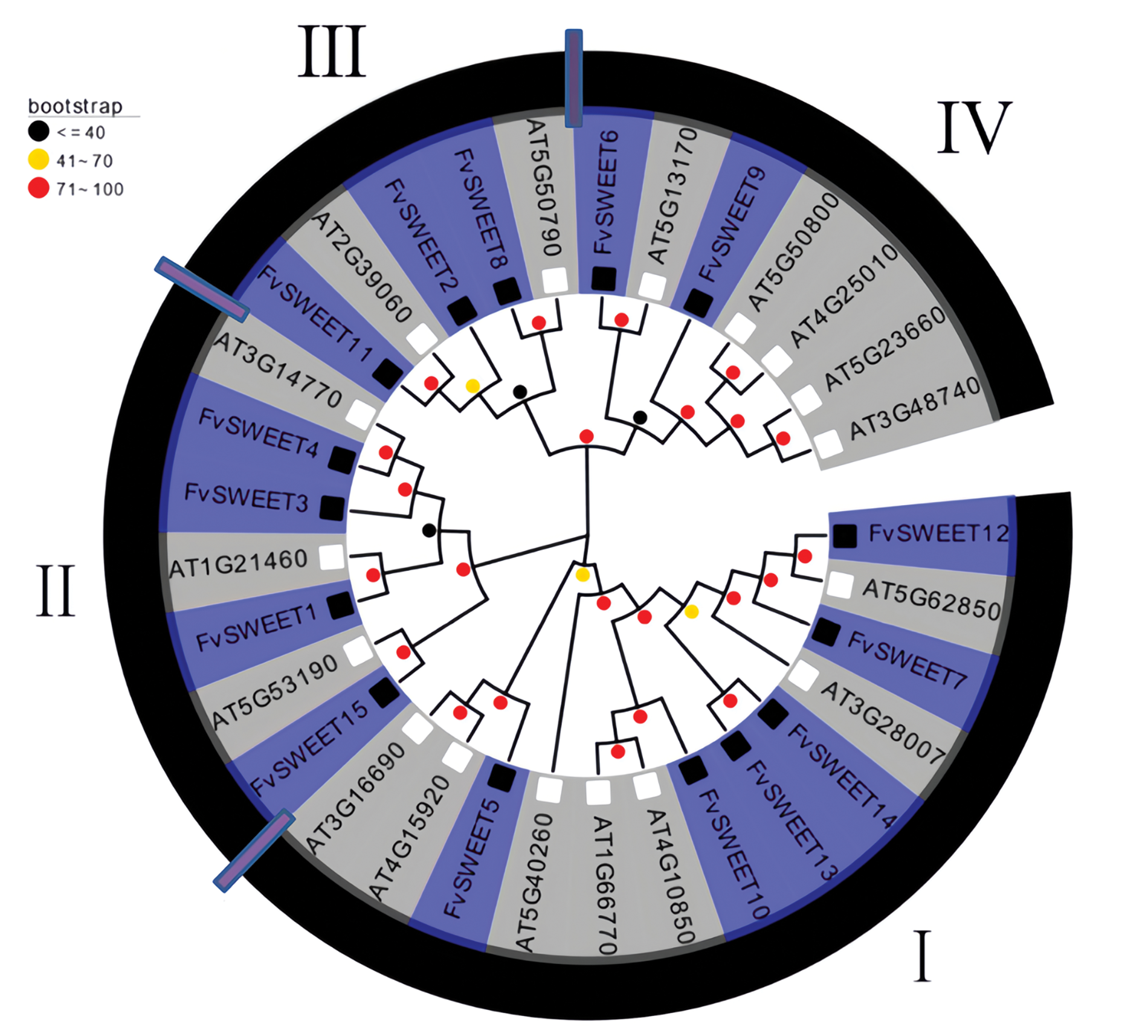
Figure 1: Neighbour-joining phylogenetic analysis (1000 bootstrap replicates) based on SWEET proteins in F. vesca and A. thaliana. Each SWEET protein of woodland strawberry is highlighted in blue, and the SWEET proteins of A. thaliana are highlighted in grey. The tree included four phylogenetic groups, designated I, II, III, and IV. Bootstrap values are shown in the upper left corner. Bootstrap values less than or equal to 40 are represented by black circles, those greater than 41 and less than 70 are represented by yellow circles, and those greater than 71 and less than 100 are represented by red circles. There are 3 black circles and 3 yellow circles, while the rest are red circles
3.3 Chromosomal Localization of FvSWEET Genes
The chromosomal localization of the FvSWEET genes was displayed by MG2C. Fifteen FvSWEET genes are distributed on all chromosomes except chromosome 1. Specifically, FvSWEET1 and FvSWEET2 are located on chromosome 2, FvSWEET3 and FvSWEET4 are located on chromosome 3, FvSWEET5 and FvSWEET6 are located on chromosome 4, four FvSWEET genes (FvSWEET7–FvSWEET10) are located on chromosome 5, four FvSWEET genes (FvSWEET11–FvSWEET14) are located on chromosome 6, and FvSWEET15 is located on chromosome 7. Two pairs of genes (FvSWEET8/FvSWEET9 and FvSWEET13/FvSWEET14) on chromosomes 5 and 6 are tightly linked (Fig. 2).
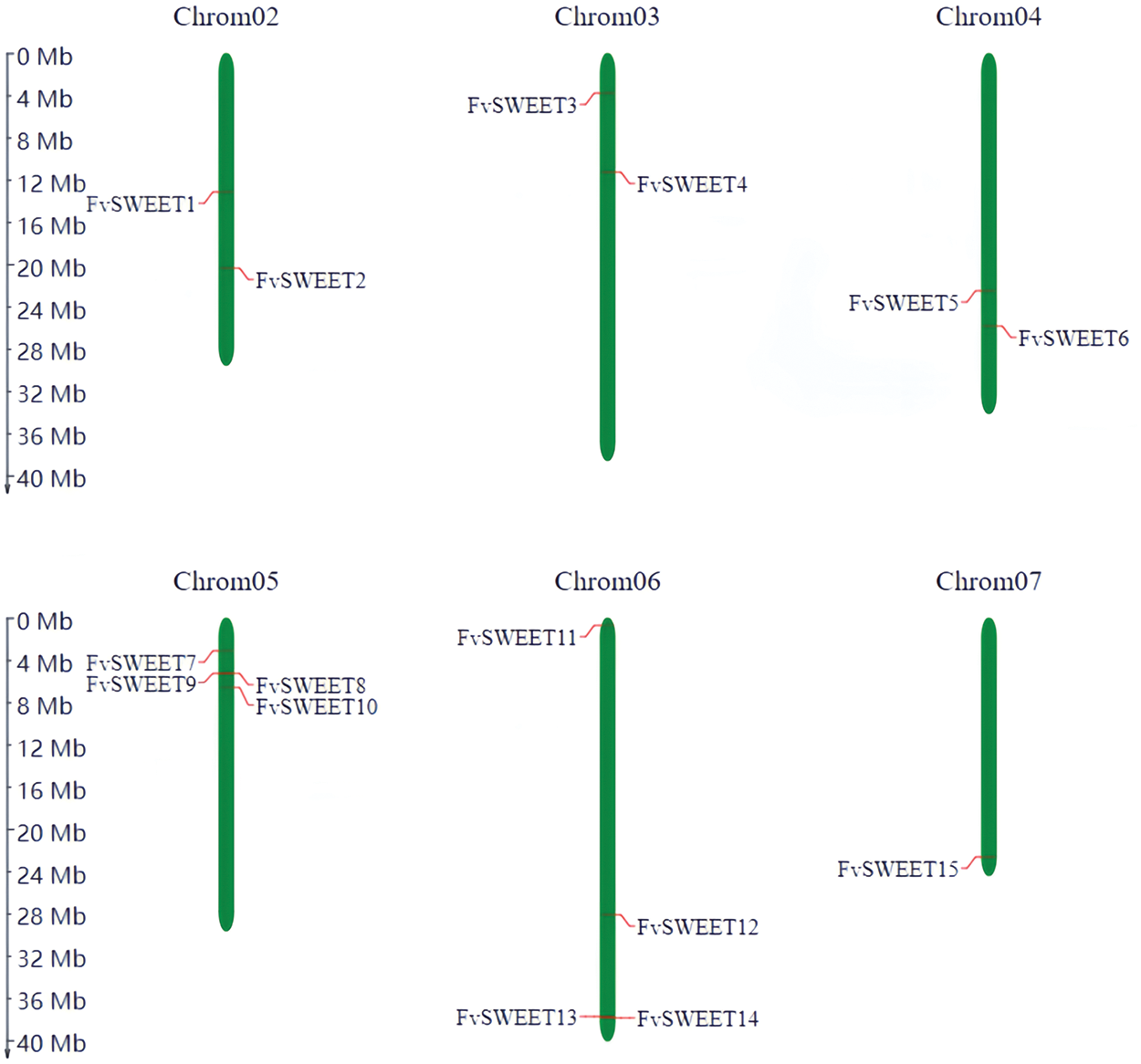
Figure 2: FvSWEET genes are distributed on the six chromosomes of woodland strawberry. The left side of the chromosome is the scale for chromosomal length. Red lines indicate the FvSWEET gene IDs, and green represents the chromosomes
3.4 Analysis of the Collinearity of FvSWEET Genes
According to the MCScanX collinearity analysis, the collinearity groups containing 8 FvSWEET genes and 12 AtSWEET genes were determined and mapped with TBtools software (Fig. 3). A few FvSWEET genes, such as FvSWEET4 and FvSWEET15, have a pair of orthologous genes. There are more than two pairs of other directly homologous FvSWEET genes. These genes may have played a fundamental role in the evolution of the SWEET genes, and they may have originated from a common ancestor and potentially had similar functions.
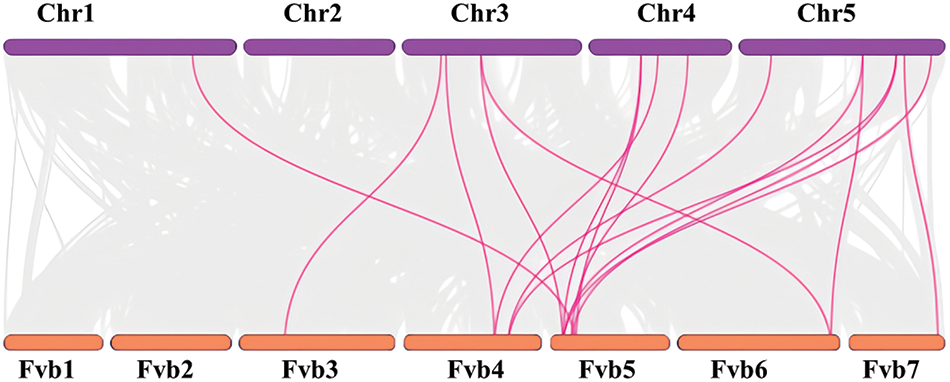
Figure 3: Collinearity analysis of the SWEET family genes in Arabidopsis (chr1–chr5) and woodland strawberry (Fvb1–Fvb7)
3.5 Structural Analysis of FvSWEET Genes and Their Encoded Proteins
The MEME tool was used to identify the conserved motifs of the FvSWEET proteins. Motifs 1–6 were conserved in all the FvSWEET proteins, motif 7 was present in 6 FvSWEET proteins (FvSWEET7, FvSWEET9, and FvSWEET12–FvSWEET15), and motif 8 was present in 5 FvSWEET proteins (FvSWEET5, FvSWEET6, FvSWEET9, FvSWEET13, and FvSWEET14) (Fig. 4a). In addition, the GSDS tool was used to analyze the exon-intron structures of the FvSWEET genes. As shown in Fig. 4b, most of the FvSWEET genes had 5 introns, except for FvSWEET4, FvSWEET10, and FvSWEET13, which had 6, 4, and 4 introns, respectively.
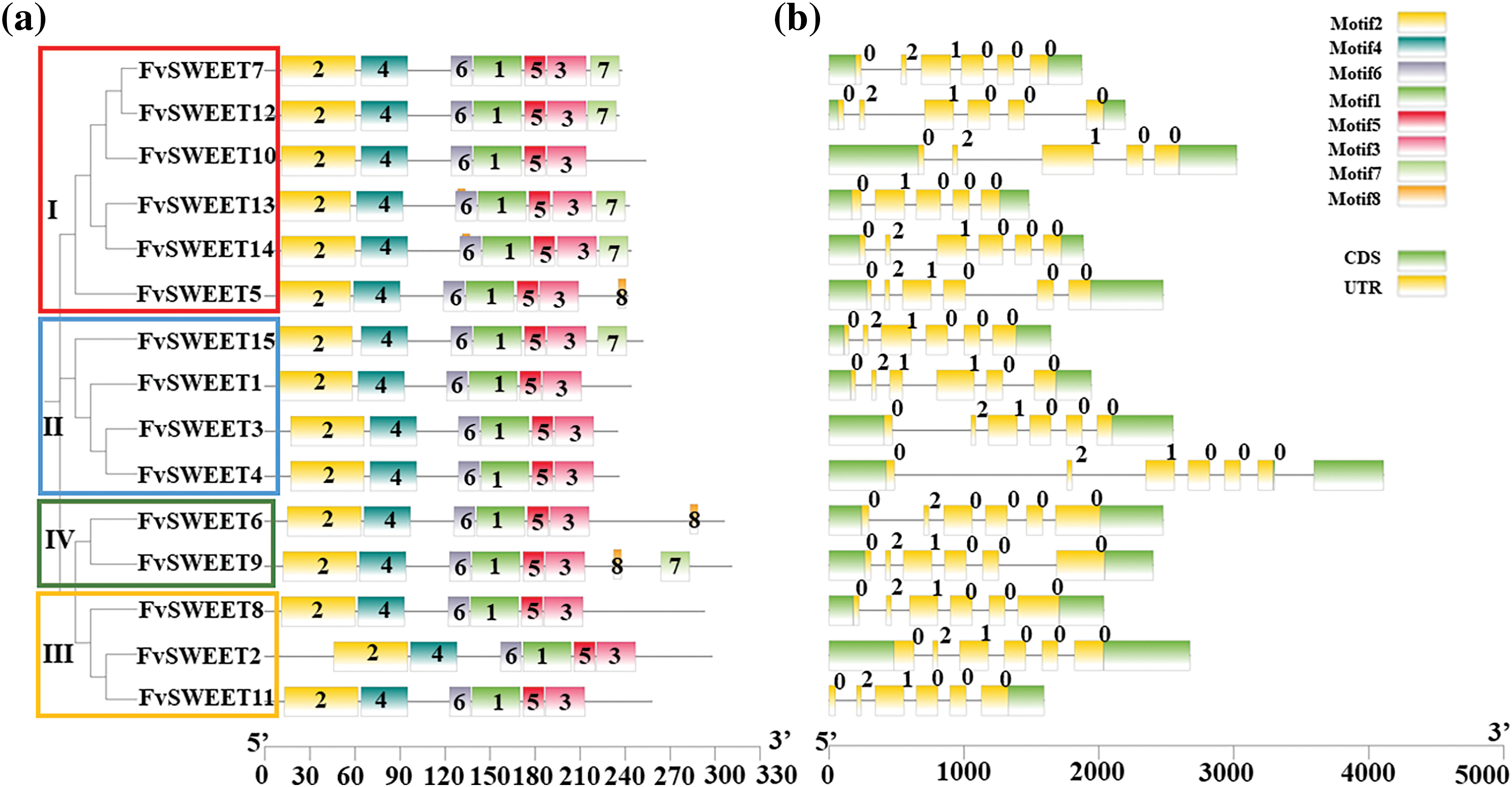
Figure 4: Motif distribution (a) and exon-intron structures (b) of SWEET family members with phylogenetic relationships in woodland strawberries. MEME was used to identify the conserved motifs of the genes. Different motifs are shown in different colors. Exons are shown as yellow boxes, introns are shown as black lines, and UTRs are shown as green boxes
3.6 Analysis of Expression Levels of Woodland Strawberry FvSWEET Genes during Development and Stress Responses
Fig. 5 shows that eight FvSWEET genes, namely, FvSWEET1, FvSWEET2, FvSWEET3, FvSWEET4, FvSWEET6, FvSWEET8, FvSWEET9, and FvSWEET10, were expressed during fruit development in the three strawberry varieties. The 10 DPA (green) and 35 DPA (ripe) samples were clustered together with the achene (AC) and receptacle (RC) tissue samples, which indicated that the expression patterns of the SWEET genes in different woodland strawberry cultivars were specific to young and mature fruits. However, the clustering of the 25 DPA (intermediate) samples was relatively scattered, and the expression patterns of the SWEET genes differed among the varieties, which indicated that the expression timelines of these genes differed among the varieties during ripening and softening. Among these genes, FvSWEET1 was expressed at much greater levels in most samples, especially in mature samples, than were the other genes. The expression of FvSWEET1 was 2.27–9.42 times greater in the 25 DPA samples than in the 10 DPA samples and 26.90–37.42 times greater in the 35 DPA samples than in the 10 DPA samples. Compared with the expression levels in the 10 DPA samples, the expression of FvSWEET1 in the 25 DPA samples increased 2.48–4.50-fold, and that in the 35 DPA samples increased 2.83–4.94-fold. The expression of FvSWEET1 increased continuously in the YW variety but then increased slightly in the HW4 and RDV varieties. These findings indicated that FvSWEET1 is significantly involved in the development and ripening of strawberry plants.
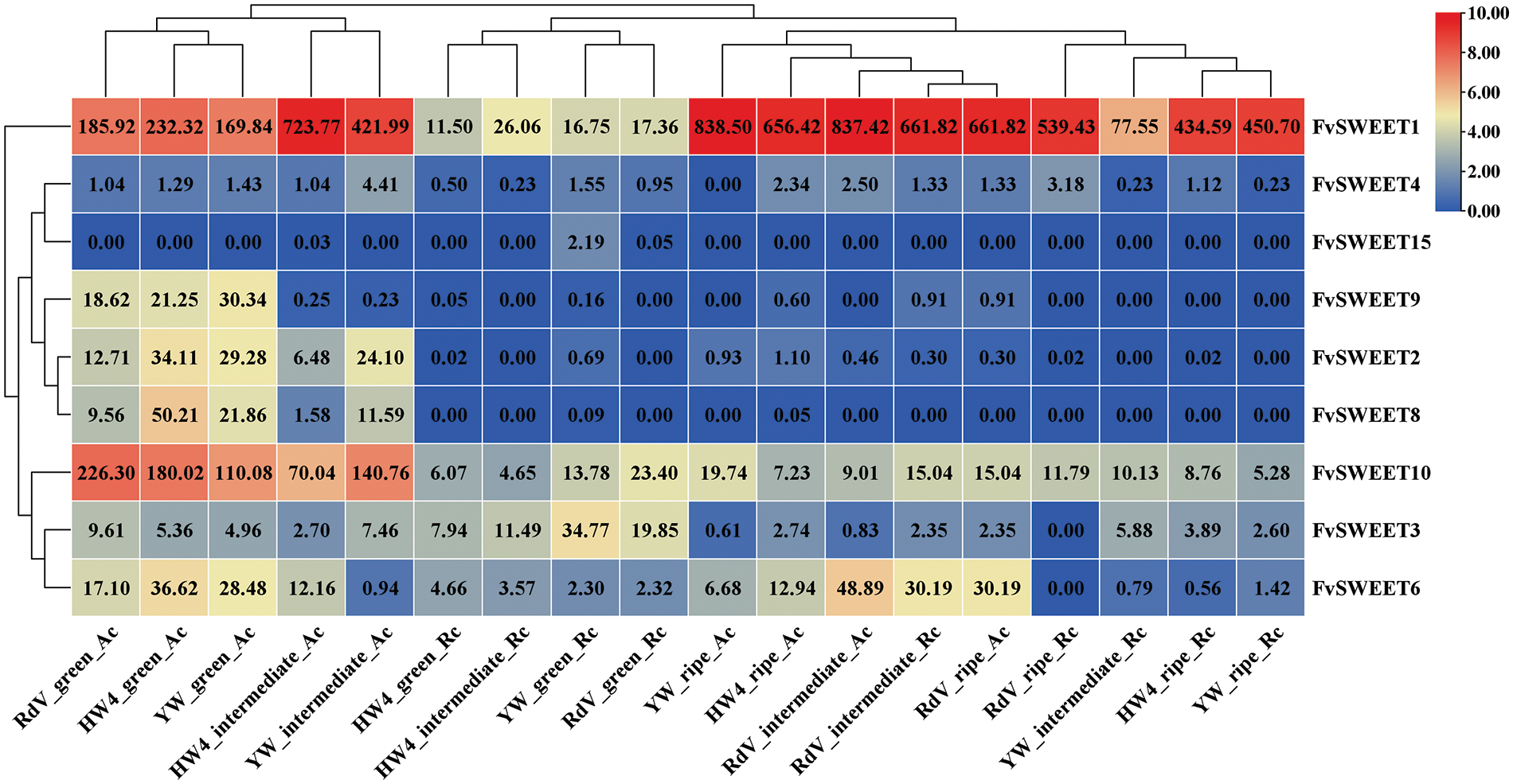
Figure 5: Expression profiles of FvSWEET genes in developing fruits. These fruit development data were obtained from two tissues (achenes (Ac) and receptacles (Rc)), three strawberry strains (Hawaii 4 (HW), yellow wonder (YW) and Reine des values (Rdv)), and three stages of development (10 DPA (days postanthesis), intermediate at 25 DPA, and ripe at 35 DPA)
As shown in Fig. 6, seven FvSWEET genes, namely, FvSWEET1, FvSWEET3, FvSWEET4, FvSWEET5, FvSWEET9, FvSWEET10, and FvSWEET15, were expressed in root tissues. Four FvSWEET genes, namely, FvSWEET1, FvSWEET3, FvSWEET4, and FvSWEET5, were expressed in leaf tissues. Eight FvSWEET genes, namely, FvSWEET1, FvSWEET2, FvSWEET3, FvSWEET4, FvSWEET6, FvSWEET8, FvSWEET9, and FvSWEET10, were expressed in fruit tissues. These results are in agreement with those shown in Fig. 4. These results revealed that the expression profiles of the FvSWEET genes exhibited obvious tissue specificity. Strawberry vein banding virus (SVBV)-infected leaves did not significantly affect the expression of the 4 leaf-specific genes. However, in roots infected with bacteria (Phytophthora cactorum), FvSWEET4 and FvSWEET10 were significantly upregulated, while FvSWEET1, FvSWEET3, FvSWEET5, and FvSWEET15 were significantly downregulated. Similarly, young white fruits infected with the fungus Botrytis cinerea showed significantly upregulated expression of FvSWEET1, FvSWEET6, FvSWEET10, and FvSWEET15 and downregulated expression of FvSWEET2, FvSWEET3, FvSWEET4, and FvSWEET8. In mature red fruit infected with Botrytis cinerea, the expression levels of FvSWEET5 and FvSWEET6 were upregulated, and the expression levels of FvSWEET1, FvSWEET3, and FvSWEET4 were downregulated. FvSWEET genes were responsive to bacterial (Phytophthora cactorum) and fungal (Botrytis cinerea) stress but not to viral (strawberry vein banding virus, SVBV) stress, demonstrating that these SWEET genes were involved in the interactions between woodland strawberry and bacteria and between woodland strawberry and fungi.
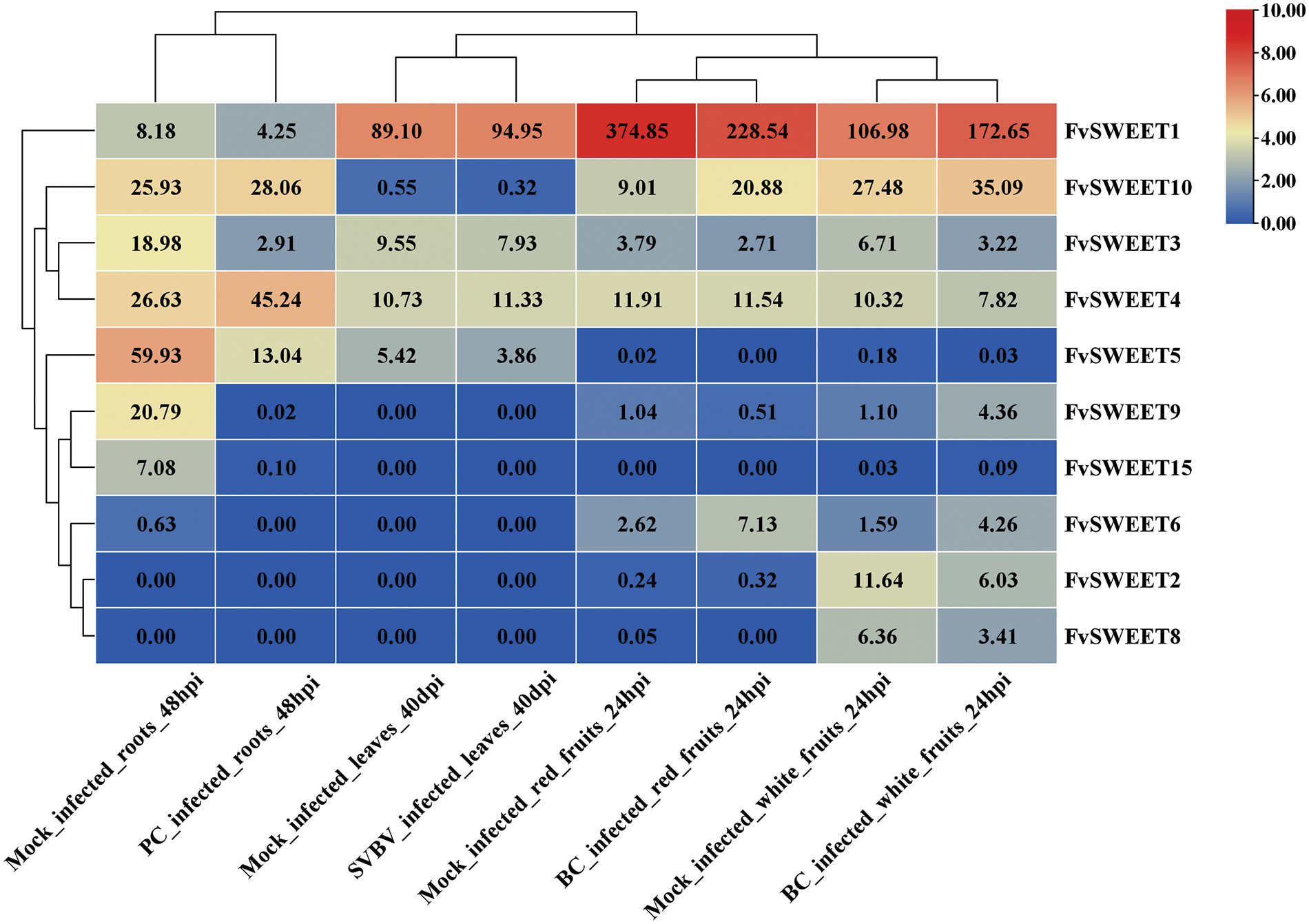
Figure 6: Expression profiles of FvSWEET genes in different tissues under different biotic stresses. These biotic stress data include data from virus (strawberry vein banding virus, SVBV)-infected leaves at 40 dpi (days postinoculation), fungus (Phytophthora cactorum, PC)-infected roots at 48 hpi and fungus (Botrytis cinerea, PC)-infected white fruits at 14 DPA and red fruits at 21 DPA at 24 hpi
3.7 Interaction Analysis of FvSWEET Genes
To study the relationships between genes based on the WGCNA network, all genes were categorized into modules according to their average FPKM value (total FPKM value/sample number) > 10, after which module–trait relationships were obtained. The results showed that when β (soft threshold) = 6 and R2 > 0.8, the average connectivity rate was relatively high, meeting the requirements of network construction (Fig. 7a). Accordingly, a total of 13 modules were obtained. Of the 13 modules, most modules showed negative correlations with the traits, but the maximum positive correlation obtained was 0.91 (Fig. 7c). By setting a correlation threshold of 0.3, possible interactions of the woodland strawberry SWEET genes in the module were identified. As shown in Fig. 7d, twenty genes interacted with FvSWEET10, among which the strongest correlation was observed between FvH4_1g16440 and FvSWEET10, and the weakest correlation was observed between FvH4_4g24340 and FvSWEET10.
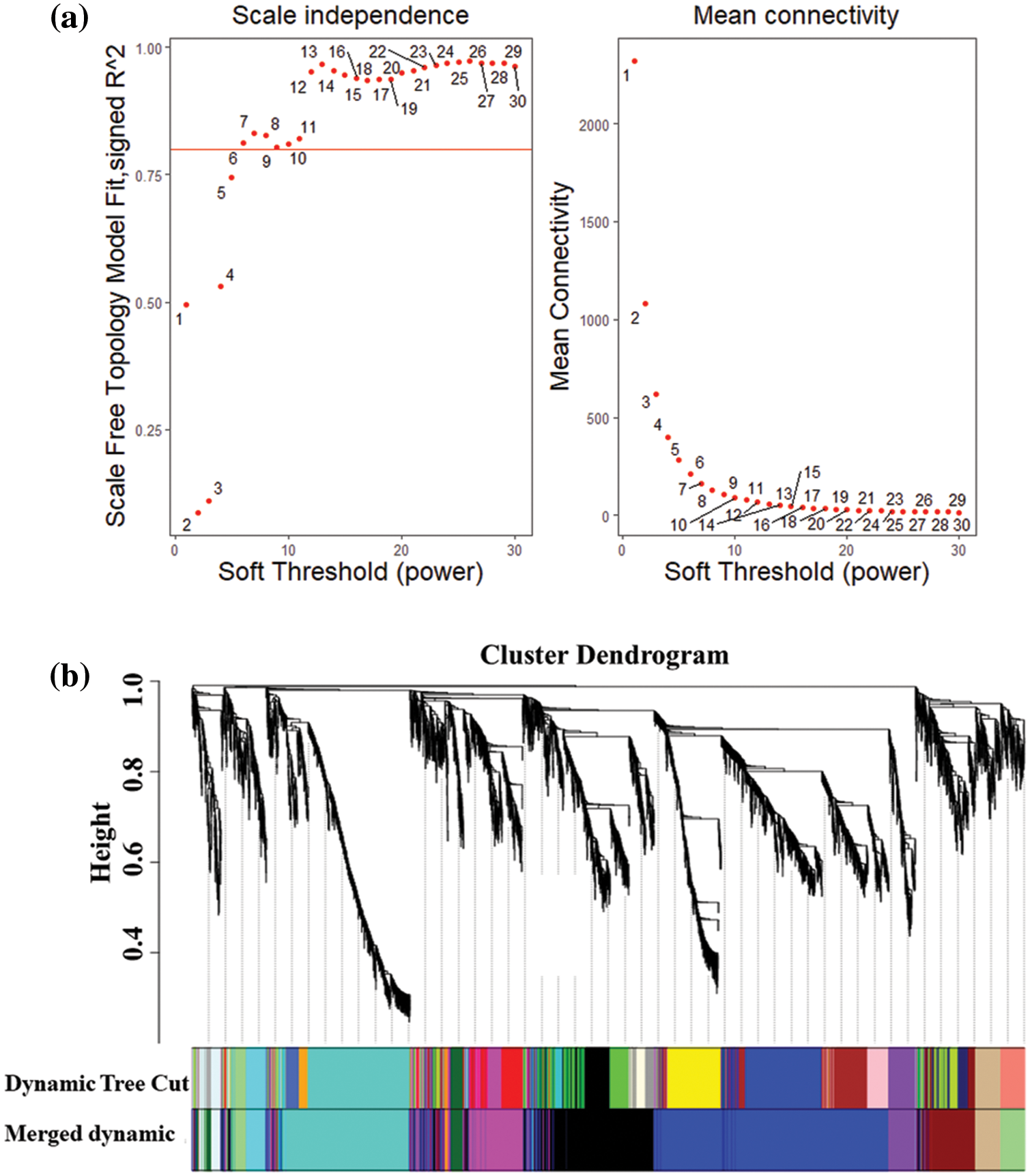
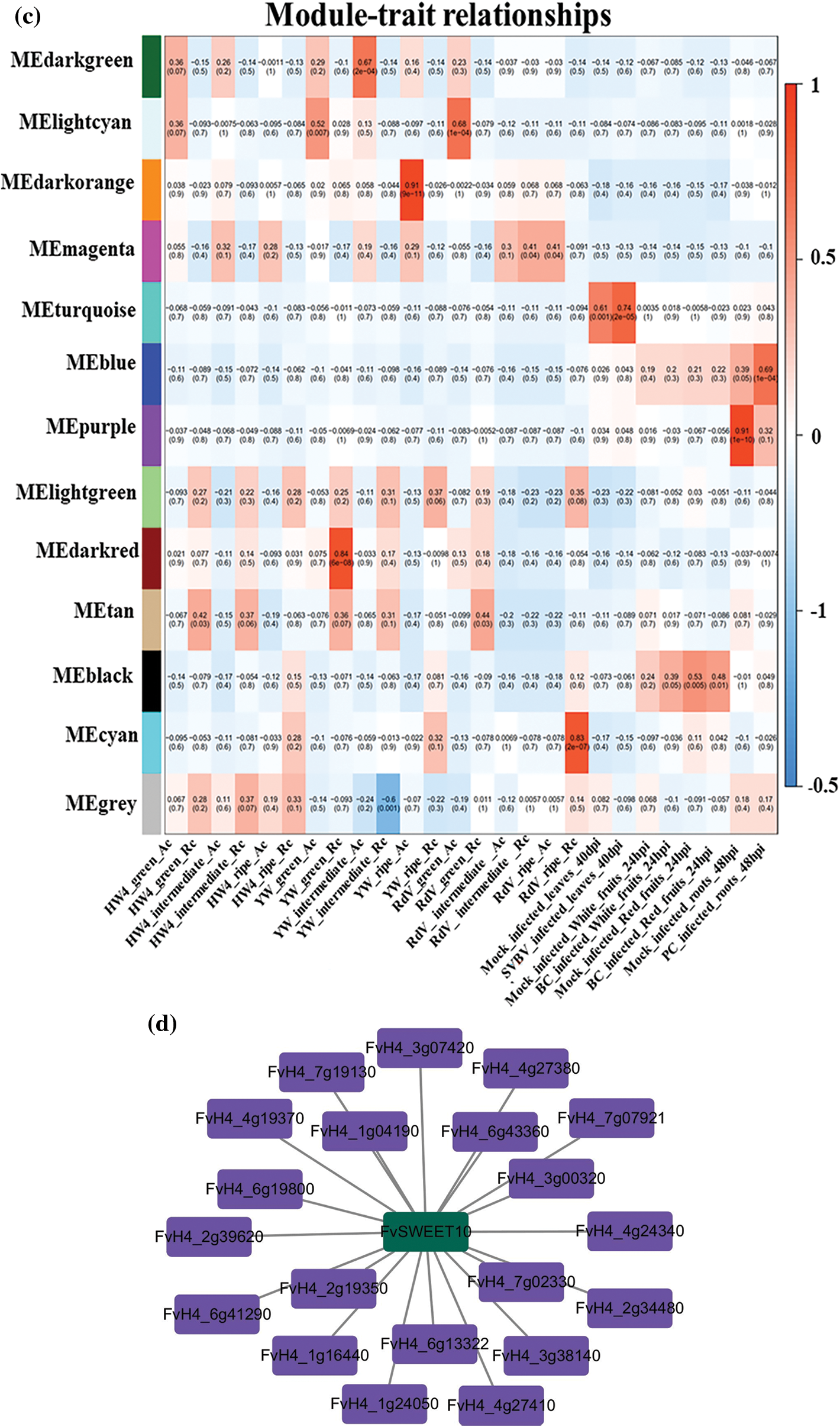
Figure 7: Weighted coexpression gene network analysis. (a) Soft threshold selection. (b) Gene clustering and module cutting. (c) Module trait correlation. At R2 = 0.8 (red line), the soft threshold power was set to 6, and the genes were divided into 26 colored modules. The number represents the correlation and P value between each module and 26 samples. Red represents positive correlations, and blue represents negative correlations. (d) Genes associated with FvSWEET10
3.8 GO Analysis of the WGCNA Genes
To further determine the function of FvSWEET10 and its interacting genes in relation to woodland strawberry disease resistance, GO enrichment analysis was performed. Accordingly, the molecular enriched functions with the lowest p values and the functions involving at least three genes were selected. Overall, three molecular functions were significantly enriched, namely, oxidoreductase activity, glucosyltransferase activity, and UDP-glucosyltransferase activity (Fig. 8). Five genes were related to oxidoreductase activity, namely, the FvH43g38140, FvH4_4g24340, FvH4_6g13322, FvH4_6g41290, and FvH4_7g07921 genes. The genes associated with glucosyltransferase activity included the FvH4_1g24050, FvH4_2g34480, and FvH4_3g07420 genes, and the genes associated with glucosyltransferase activity included the FvH4_1g24050, FvH4_2g34480, and FvH4_3g07420 genes.
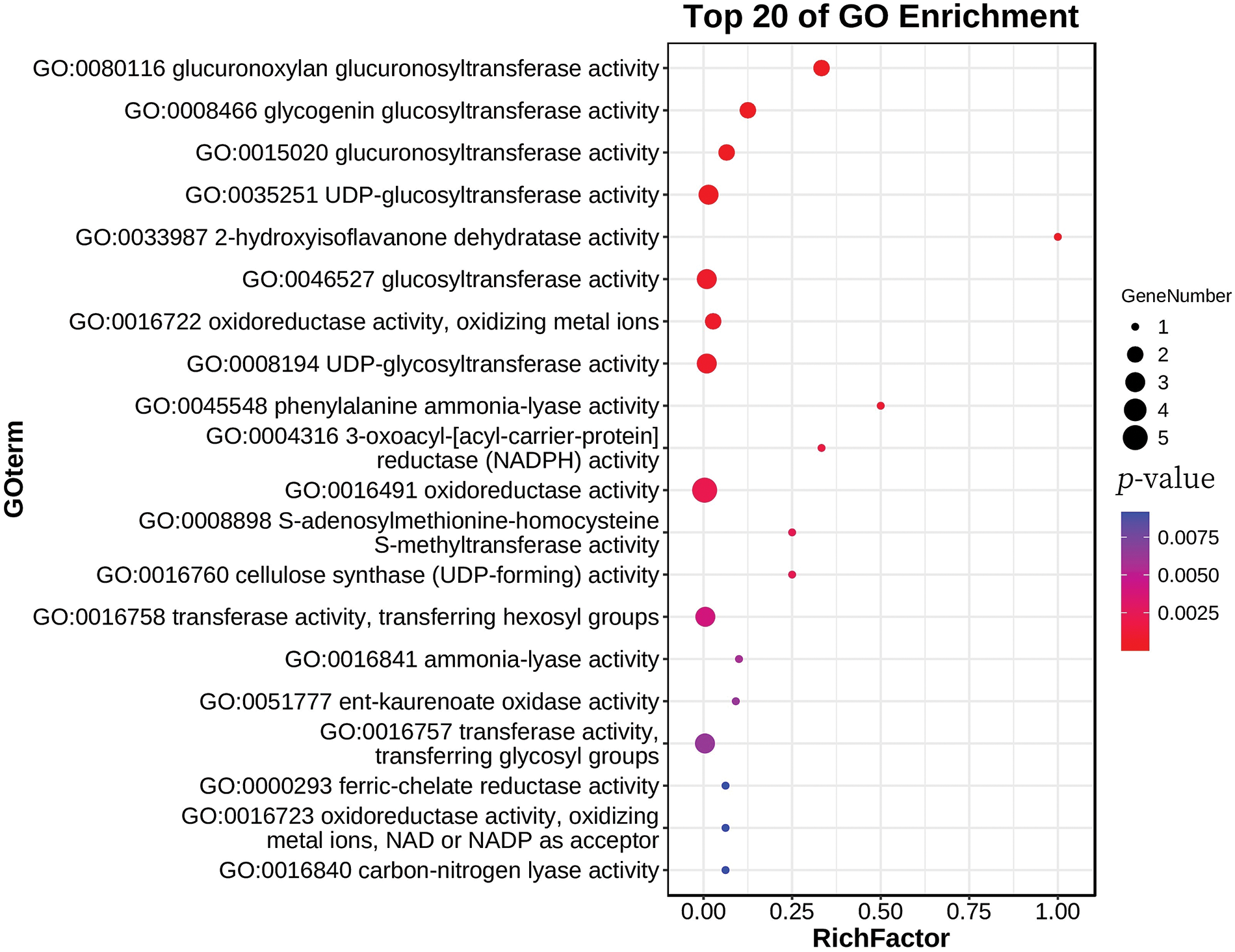
Figure 8: GO enrichment analysis of 21 genes annotated by molecular function
The SWEET gene family is widespread in various organisms and has been implicated in several biological processes, including growth and development, reproduction, host-pathogen interactions, and the abiotic stress response [17]. Its identified number of genes was similar to that of Vitis vinifera (17) and A. thaliana (17), but significantly less than that of Glycine max (52), Solanum tuberosum (35) and Triticum aestivum(108) [7,14,30–32], of which reason may be that species are accompanied by the expansion and contraction of SWEET genes in the process of evolution. Although the number of SWEET gene families was different in different species, it can be found by comparing with the results of phylogenetic tree that SWEET genes can be divided into four clades in Arabidopsis and strawberry; homologous genes are beneficial to the variation of gene families in species, but there may be functional redundancy and diversification among homologous genes. After that, two pairs of closely linked FvSWEETs genes on chromosomes 5 and 6 were identified by chromosomal mapping, indicating possible tandemly repeated genes [30]. The results of eukaryotic SWEET protein containing 7 TMHs reported by previous studies are almost consistent with our findings, except for FvSWEET12, which contains only 6 TMHs [33]. According to previous studies, eukaryotic SWEET genes are produced by copying and fusing archaea genes containing three TMH and bacterial semiSWEET genes, while FvSWEET12 contains only 6 TMH [34], indicating that some structures of genes have undergone other changes in the process of evolution to adapt to environmental changes.
Its inside changed greatly with the development of the fruit; the more mature the fruit, the higher the sugar. In this study, FvSWEET1 was highly expressed in the middle (25 DPA) and late (35 DPA) stages of fruit development in all the strawberry varieties tested. Interestingly, the expression levels of FvSWEET10 in the early stage of fruit development in all strawberry varieties were significantly greater than those of the other FvSWEET genes. In A. thaliana, AtSWEET1 is a plasma membrane hexose transporter, and AtSWEET7 can transport glucose and xylose [35]. As shown in Fig. 1, AtSWEET1 shares strong homology with FvSWEET1, and AtSWEET7 is closely related to FvSWEET10. These findings suggested that the two genes of the strawberry family are important for regulating the growth and development of strawberry plants during different periods.
Most pathogens, pests, weeds, and other major energy substances consist of sugars. In the process of interaction between plants and biological stress, plants often regulate their sugar content by up or downregulating the expression of SWEET genes to respond to various biological stresses. The roles of SWEET genes in response to biological stresses have been studied most thoroughly in rice. The rice OsSWEET11 gene inhibits the growth of blight-causing bacteria by regulating transcription. When a blight-causing bacterium infects rice, the bacterium secretes the Thales transcriptional activator effector through the T3 secretion system, and then the Thales activator combines with the cis-acting element of the upstream promoter of the SWEET gene to regulate the expression of the target SWEET gene, promote the transfer of sugar to the bacterium, and worsen the disease condition [36,37]. As shown in Figs. 6 and 7, FvSWEET1, FvSWEET3, and FvSWEET4 responded to all the types of stress, while FvSWEET10 responded only to bacterial stress (Phytophthora cactorum) and fungal stress (Botrytis cinerea) but not to viral stress (SVBV). However, the viral infection did not induce the expression of FvSWEET10, possibly because the FvSWEET10 gene does not respond to viral stress. It has been reported that bacterial and fungal symbionts and pathogens are capable of inducing the expression of various SWEET genes through the secretion of effector proteins that bind and activate SWEET genes [32].
WGCNA was subsequently performed to identify the genes that interact with FvSWEET10, and 20 genes were identified. These results suggest that the FvSWEET10 gene interacts with other genes to play roles in the interactions between woodland strawberries and bacteria and between woodland strawberries and fungi. The white fruit of strawberry plants is resistant to disease, though the red fruit is susceptible to disease. Grey mould is dormant in white fruit but can parasitize mature red fruit [24]. SWEET gene expression in white fruit infected with fungi (Botrytis cinerea) was mostly downregulated, and that in red fruit infected with fungi (Botrytis cinerea) was mostly upregulated. It is possible that fungi induce SWEET expression in red fruits to obtain energy, which is conducive to infection and results in the susceptibility of red fruits to disease. In contrast, in white fruits, the host actively downregulates SWEET expression, reduces the energy supply, and resists fungal infection, and white fruits exhibit disease resistance. In other studies, transcriptome analysis results showed that the growth and development of plants inoculated with bacteria (Phytophthora cactorum) were affected. The expression levels of most of the SWEET genes in woodland strawberry roots were downregulated, while AtSWEET1 is a plasma membrane hexose transporter that is transported across the plasma membrane [6]. The expression of SWEET might be downregulated in the process of bacterial infection, inhibiting the growth and development of the plant; thus, the host enters the defense mode to resist bacterial infection.
According to the enrichment of GO, there were two genes among the 20 genes that may interact with FvSWEET10 relating to disease resistance, while five were related to oxidordeuctase activity. Oxidordeuctase plays the role of promoting the production of energy and synthesizing various substances needed by plants to cope with environmental stress in the process of growth, making it become significant for directly regulating the production of ROS and oxidative stress, which is particularly evident when plants are infected by pathogens. The role of oxidoreductase in plant immune response has been verified in previous studies; for instance, it has been found that OsTRXm and TRX-like protein OsTDX can be expressed in rice and show the ability to resist harmful pathogen activity. OsTRXm has been identified in other studies as a novel anti-fungal protein that inhibits the growth of a variety of fungi, including the maize pathogen Fusarium moniliforme [38,39]. Similarly, the root-knot nematode Meloidogyne incognita can overcome the resistance of tomato plants by potentially using peroxiredoxin (PRX) in tomatoes to realize infection [40]. Glycosyltransferase has attracted wide attention leveraging own multiple functions, of which functions involving in the changes of plant growth and development and the resistance of biotic and abiotic stresses. Glycosyltransferase plays a key role in plant responses to related stresses through mediating the glycosylation of plant hormones and endogenous secondary metabolites [41]. In addition, the related genes of glycosyltransferase have been identified in different plants. It has been clarified that rice glycosyltransferase gene UGT85E1 is a gene that affects ABA concentration and plant resistance to related stresses, which is involved in the regulation of various hormones in plant stress responses, including auxin [42,43]. According to previous studies, the expression of rice glycosyltransferase OsUGT90A1 gene is induced by low temperature, which is related to the concentration of auxin and able to reduce the damage of cell plasma membrane caused by chilling injury in the case of overexpression of this gene [44]. Glycosyltransferase exerts an indirect effect on stress damage to some extent through glycosylation modification of plant hormones in biological stress. Salicylic acid (SA) is able to mediate the change of plant resistance to biotrophic pathogens, while SA in Arabidopsis thaliana is recognized as substrate by related genes UGT74F1, UGT74F2, UGT75B1 and UGT76B1 [44,45]. The expression of glycosyltransferase gene is affected by the alteration of SA glycosylation, which resulted in different phenotypes of infected plants. The overexpression of UGT74F2 gene in Arabidopsis decreases the level of free SA and changes the resistance to Pseudomonas syringae, but Pseudomonas syringae has been clearly determined as a bio-nutritive pathogen. On the other hand, the knockout of UGT74F2 gene in Arabidopsis increases SA level, which complements the results of overexpression of UGT74F2 gene [46,47]. Based on the above results, it can be concluded that it is possible for FvSWEET10 gene to play multiple roles in plant response to biotic stress and abiotic stress through glycosylation of plant hormones and endogenous secondary metabolites as well as the regulation of ROS/RNS and oxidative stress.
In this study, we identified 15 SWEET genes in strawberries, studied their evolution, chromosomal distribution, and gene structure, and analyzed their contributions to tissue development and resistance to biological stress. Moreover, WGCNA revealed 21 genes in key modules, including FvSWEET10. The findings of this research can serve as a foundation for further understanding the SWEET genes in woodland strawberries.
Acknowledgement: None.
Funding Statement: This research was funded by the Fujian Provincial Science and Technology Project (2021N5014, 2022N5006) and the Key Research Project of the Putian Science and Technology Bureau (2021ZP08, 2021ZP09, 2021ZP10, 2021ZP11, 2023GJGZ001).
Author Contributions: Shoukai Lin and Fuxiang Qiu supervised this study, carried out the bioinformatics analysis, and completed the manuscript. Yifan Xiong performed the RNA-seq analysis of the downloaded data. Shichang Xu, Manegdebwaoaga Arthur Fabrice Kabore and Fan Lin participated in the bioinformatics analysis and revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data presented in this study are available in the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Jin L, Ruan S, Xin Y, Fang X, Li X, Ma H, et al. Comparison on antioxidant capacity in vitro of Fragaria vesca and strawberry cultivars. J Nucl Agric Sci. 2015;29(1):0079–86 (In Chinese). [Google Scholar]
2. Zhou Y, Xiong J, Shu Z, Dong C, Gu T, Sun P, et al. The telomere-to-telomere genome of Fragaria vesca reveals the genomic evolution of Fragaria and the origin of cultivated octoploid strawberry. Hortic Res. 2023;10(4):uhad027. doi:10.1093/hr/uhad027. [Google Scholar] [PubMed] [CrossRef]
3. Edger PP, VanBuren R, Colle M, Poorten TJ, Wai CM, Niederhuth CE, et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience. 2018;7(2):gix124. [Google Scholar] [PubMed]
4. Li Y, Pi M, Gao Q, Liu Z, Kang C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic Res. 2019;6(1):61. doi:10.1038/s41438-019-0142-6. [Google Scholar] [PubMed] [CrossRef]
5. Chen J, Zhang H, Feng M, Zuo D, Hu Y, Jiang T, et al. Transcriptome analysis of woodland strawberry (Fragaria vesca) response to the infection by Strawberry vein banding virus (SVBV). Virol J. 2016;13(1):128. doi:10.1186/s12985-016-0584-5. [Google Scholar] [PubMed] [CrossRef]
6. Chen L, Qu X, Hou B, Sosso D. Sucrose Efflux mediated by SWEET proteins as a key step for phloem transport. 2012;335(6965):207–11. [Google Scholar]
7. Chen L, Hou B, Lalonde S, Takanaga H, Hartung ML. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527–32. doi:10.1038/nature09606. [Google Scholar] [PubMed] [CrossRef]
8. Ji J, Yang L, Fang Z, Zhang Y, Zhuang M, Lv H, et al. Plant SWEET family of sugar transporters: structure, evolution and biological functions. Biomolecules. 2022;12(2):205. doi:10.3390/biom12020205. [Google Scholar] [PubMed] [CrossRef]
9. Gamas P, Niebel FDC, Lescure N, Cullimore JV. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. MPMI. 1996;9(4):223–342. [Google Scholar]
10. Wei X, Liu F, Chen C, Ma F, Li M. The Malus domestica sugar transporter gene family: identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front Plant Sci. 2014;5:569. [Google Scholar] [PubMed]
11. Zhu Y, Tian Y, Han S, Wang J, Liu Y, Yin J. Structure, evolution, and roles of SWEET proteins in growth and stress responses in plants. Int J Biol Macromol. 2024;263:130441. doi:10.1016/j.ijbiomac.2024.130441. [Google Scholar] [PubMed] [CrossRef]
12. Yuan M, Chu Z, Li X, Xu C, Wang S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell. 2010;22(9):3164–76. doi:10.1105/tpc.110.078022. [Google Scholar] [PubMed] [CrossRef]
13. Chen LQ, Lin IW, Qu XQ, Sosso D, McFarlane HE, Londoño A, et al. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell. 2015;27(3):607–19. doi:10.1105/tpc.114.134585. [Google Scholar] [PubMed] [CrossRef]
14. Chong J, Piron MC, Meyer S, Merdinoglu D, Bertsch C, Mestre P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J Exp Bot. 2014;65(22):6589–601. doi:10.1093/jxb/eru375. [Google Scholar] [PubMed] [CrossRef]
15. Liu X, Zhang Y, Yang C, Tian Z, Li J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci Rep. 2016;6(1):24563. doi:10.1038/srep24563. [Google Scholar] [PubMed] [CrossRef]
16. Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet. 2015;47(12):1489–93. doi:10.1038/ng.3422. [Google Scholar] [PubMed] [CrossRef]
17. Yuan M, Wang S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 2013;(3):665–74. [Google Scholar]
18. Perotto S, Rodda M, Benetti A, Sillo F, Ercole E, Rodda M, et al. Gene expression in mycorrhizal orchid protocorms suggests a friendly plant-fungus relationship. Planta. 2014;239(6):1337–49. doi:10.1007/s00425-014-2062-x. [Google Scholar] [PubMed] [CrossRef]
19. Feng L, Frommer WB. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem Sci. 2015;40(8):480–6. doi:10.1016/j.tibs.2015.05.005. [Google Scholar] [PubMed] [CrossRef]
20. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. doi:10.1016/j.molp.2020.06.009. [Google Scholar] [PubMed] [CrossRef]
21. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. doi:10.1093/molbev/msw054. [Google Scholar] [PubMed] [CrossRef]
22. Härtl K, Denton A, Franz-Oberdorf K, Hoffmann T, Spornraft M, Usadel B, et al. Early metabolic and transcriptional variations in fruit of natural white-fruited Fragaria vesca genotypes. Sci Rep. 2017;7(1):45113. doi:10.1038/srep45113. [Google Scholar] [PubMed] [CrossRef]
23. Gijzen M, Toljamo A, Blande D, Kärenlampi S, Kokko H. Reprogramming of strawberry (Fragaria vesca) root transcriptome in response to Phytophthora cactorum. PLoS One. 2016;11(8):e0161078. doi:10.1371/journal.pone.0161078. [Google Scholar] [PubMed] [CrossRef]
24. Haile ZM, Nagpala-De Guzman EG, Moretto M, Sonego P, Engelen K, Zoli L, et al. Transcriptome profiles of strawberry (Fragaria vesca) fruit interacting with botrytis cinerea at different ripening stages. Front Plant Sci. 2019;10:1131. doi:10.3389/fpls.2019.01131. [Google Scholar] [PubMed] [CrossRef]
25. Xiong Y, Lin D, Ma S, Wang C, Lin S. Genome-wide identification of the calcium-dependent protein kinase gene family in Fragaria vesca and expression analysis under different biotic stresses. Eur J Plant Pathol. 2022;164:283–93. doi:10.1007/s10658-022-02560-4. [Google Scholar] [CrossRef]
26. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi:10.1093/bioinformatics/btu170. [Google Scholar] [PubMed] [CrossRef]
27. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. doi:10.1038/nmeth.3317. [Google Scholar] [PubMed] [CrossRef]
28. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi:10.1093/bioinformatics/btp352. [Google Scholar] [PubMed] [CrossRef]
29. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2012;31(1):46–53. [Google Scholar] [PubMed]
30. Gautam T, Saripalli G, Gahlaut V, Kumar A, Sharma PK, Balyan HS, et al. Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.). Mol Biol Rep. 2019;46(2):2327–53. doi:10.1007/s11033-019-04691-0. [Google Scholar] [PubMed] [CrossRef]
31. Manck-Götzenberger J, Requena N. Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato sweet sugar transporter family. Front Plant Sci. 2016;7:191086. [Google Scholar]
32. Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao M, et al. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics. 2015;16(1):520. doi:10.1186/s12864-015-1730-y. [Google Scholar] [PubMed] [CrossRef]
33. Jeena GS, Kumar S, Shukla RK. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol Biol. 2019;100(4–5):351–65. doi:10.1007/s11103-019-00872-4. [Google Scholar] [PubMed] [CrossRef]
34. Tao Y, Mis M, Blazer L, Ustav M, Steinhart Z, Chidiac R, et al. Tailored tetravalent antibodies potently and specifically activate Wnt/Frizzled pathways in cells, organoids and mice. eLife. 2019;8:e46134. doi:10.7554/eLife.46134. [Google Scholar] [PubMed] [CrossRef]
35. Chandran D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life. 2015;67(7):461–71. doi:10.1002/iub.1394. [Google Scholar] [PubMed] [CrossRef]
36. Dai Y, Liu LY, Yang N, Xiang WL, Zhou Y, Huang YC. Research progress of SWEET protein in plants. Mol Plant Breed. 2021;19(4):1128–35 (In Chinese). [Google Scholar]
37. Li MX and Li TT. The effector of bacterial blight, plays a toxic role in rice varieties with OsSWEET11 homologous gene. China Rice Sci. 2020;(4):368–82 (In Chinese). [Google Scholar]
38. Lin B, Zhuo K, Chen S, Hu L, Sun L, Wang X, et al. A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 2015;209:1159–73. [Google Scholar] [PubMed]
39. Park SC, Kim IR, Kim JY, Lee Y, Yoo SH, Jung JH, et al. Functional characterization of a rice thioredoxin protein OsTrxm and its cysteine mutant variant with antifungal activity. Antioxidants. 2019;8(12):12. doi:10.3390/antiox8120598. [Google Scholar] [PubMed] [CrossRef]
40. Li L, Qin Y, Luo E, Qiao Y, Wang D, Tang C, et al. Hypoxia-induced PINK1/Parkin-mediated mitophagy promotes pulmonary vascular remodeling. Biochem Biophys Res Commun. 2021;534:568–75. doi:10.1016/j.bbrc.2020.11.040. [Google Scholar] [PubMed] [CrossRef]
41. Cai M, Liu Z, Zhao Z, Wu H, Xu M, Rao Z. Microbial production of L-methionine and its precursors using systems metabolic engineering. Biotechnol Adv. 2023;69(36):108260. doi:10.1016/j.biotechadv.2023.108260. [Google Scholar] [PubMed] [CrossRef]
42. Liu Q, Dong GR, Ma YQ, Zhao SM, Liu X, Li XK, et al. Rice glycosyltransferase gene UGT85E1 is involved in drought stress tolerance through enhancing abscisic acid response. Front Plant Sci. 2021;12:790195. doi:10.3389/fpls.2021.790195. [Google Scholar] [PubMed] [CrossRef]
43. Meier M, Liu Y, Lay-Pruitt KS, Takahashi H, von Wirén N. Auxin-mediated root branching is determined by the form of available nitrogen. Nat Plants. 2020;6(9):1136–45. doi:10.1038/s41477-020-00756-2. [Google Scholar] [PubMed] [CrossRef]
44. Dean JV, Delaney SP. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol Plant. 2008;132(4):417–25. doi:10.1111/j.1399-3054.2007.01041.x. [Google Scholar] [PubMed] [CrossRef]
45. Chen M, Fang D, Gou H, Wang S, Yue W. Quantitative measurement reveals dynamic volatile changes and potential biochemical mechanisms during green tea spreading treatment. ACS Omega. 2022;7(44):40009–20. doi:10.1021/acsomega.2c04654. [Google Scholar] [PubMed] [CrossRef]
46. Farag MA, Porzel A, Wessjohann LA. Comparative metabolite profiling and fingerprinting of medicinal licorice roots using a multiplex approach of GC-MS, LC-MS and 1D NMR techniques. Phytochemistry. 2012;76:60–72. doi:10.1016/j.phytochem.2011.12.010. [Google Scholar] [PubMed] [CrossRef]
47. Shangguan C, Kuang Y, Gao L, Zhu B, Chen XD, Yu X. Antennae-enriched expression of candidate odorant degrading enzyme genes in the turnip aphid, Lipaphis erysimi. Front Physiol. 2023;14:1228570. doi:10.3389/fphys.2023.1228570. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools