 Open Access
Open Access
ARTICLE
The MtRGF6 Peptide Differentially Regulates Root Development and Symbiotic Nodulation of Medicago truncatula and Lotus japonicus
1 School of Life Sciences, Shanghai Key Laboratory of Bio-Energy Crops, Shanghai University, Shanghai, 200444, China
2 College of Life Science, State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, 230036, China
* Corresponding Authors: Xu Wang. Email: ; Li Luo. Email:
Phyton-International Journal of Experimental Botany 2024, 93(6), 1237-1248. https://doi.org/10.32604/phyton.2024.051517
Received 07 March 2024; Accepted 10 May 2024; Issue published 27 June 2024
Abstract
Rhizobia induces nitrogen-fixing nodules in legumes used in agricultural production, providing a direct source of combined nitrogen to leguminous crops. Small peptides, such as CLAVATA3/EMBRYO SURROUNDING REGION peptides (CLE), are known to regulate the formation and development of nitrogen-fixing nodules in legumes. Root meristem growth factor (RGF) peptides from Medicago truncatula not only regulate root development but also modulate nodulation symbiosis with Sinorhizobium meliloti. However, the impact of RGF peptides from one leguminous species on the others remains unclear. In this study, we investigate the effects of the RGF family peptide MtRGF6p from M. truncatula on nodulation symbiosis and root development in Lotus japonicus. The MtRGF6 gene is predominantly expressed in the root nodules of M. truncatula and shows low identity with RGF homologous genes from L. japonicus. The gene promoter is active in the primordia of root nodules and lateral roots, as well as in young nodules and roots, and the meristem, infection, and nitrogen-fixing regions of the mature nodule. Chemically synthesized MtRGF6p promoted primary root growth in M. truncatula but suppressed the growth of L. japonicus primary roots. The peptide negatively affected the initiation of nodule primordia, the formation of infection threads, and nodulation in both legumes, with a low dosage showing effects on L. japonicus compared to M. truncatula. These results suggest that the MtRGF6 peptide from M. truncatula may serve as an inter-species signal affecting the root organ development of L. japonicus.Keywords
Supplementary Material
Supplementary Material FileThe Root Meristem Growth Factor (RGF), first identified in Arabidopsis thaliana, plays a crucial role in regulating root development [1–4]. RGF is a unique family of small peptides that are typically 13 amino acids in length. These peptides are derived from precursor proteins that contain an N-terminal signal peptide, a variable region in the middle, and a C-terminal active peptide domain with conserved aspartate, tyrosine, and proline residues. The precursor proteins undergo various post-translational modifications such as tyrosine sulfation, proline hydroxylation, and protease cleavages to generate the active peptides [5]. Overexpression of AtRGF genes has been shown to enhance root elongation, while suppressing lateral root formation, resulting in wavy and agravitropic roots [1–4]. The AtRGF peptide binds to a LRR receptor kinase complex, leading to the modulation of downstream gene expression by controlling the gradient formation of PLETHORA (PLT), a key regulator of root development [6–8]. This signaling cascade involves a MAP kinase pathway (DA-MKK4/MKK5-MPK3/MPK6) that connects RGF receptors with PLT1/2 [9,10]. Reactive oxygen species (ROS) have also been implicated as a secondary signal upstream of PLTs [11]. While the signaling pathway of RGF in controlling root development has been extensively studied in A. thaliana, it remains unclear whether RGF peptides from on species can function in others.
In contrast to A. thaliana, leguminous plants such as soybean and alfalfa have the ability to form nitrogen-fixing nodules in symbiosis with rhizobia. The development of these symbiotic root nodules is regulated by nodulation factors (Nod factors) which are lipo-chito-oligosacchrides produced by rhizobia [12]. Nod factors are recognized by LysM receptor kinases such as MtLYK3/MtNFP (Nod Factor Perception) in Medicago truncatula and LjNRF1/LjNRF5 from Lotus japonicus, initiating a signaling cascade known as the Nod factor signaling pathway. This pathway involves key components such as SYMRK (Symbiosis Receptor-like Kinase), CCaMK (Ca2+/Calmodulin dependent Kinase), POLLUX, CASTOR, CYCLOPS, NSP1(Nodulation Signaling Pathway 1), NSP2 and NIN (Nodule Inception), essential for reprogramming of gene expression in response to symbiosis [13]. Furthermore, the autoregulation of nodulation pathway controls the number of nitrogen-fixing root nodules in legumes. This pathway includes CLE (CLAVATA3/EMBRYO SURROUNDINGR REGION) peptides (LjCLE-RS1/RS2 in L. japonicus and MtCLE12/MtCLE13 in M. truncatula) and their receptors (LjHAR1 and MtSUNN) [14–16]. Additionally, the CEP (C-TERMINALLY ENCODED PEPTIDE) family peptide CEP1 with its receptor CRA2 (Compact Root Architecture2) have been implicated in regulating this process in M. truncatula [17–19]. Other peptides such as PSKs (Phytosulfokine) [20–22], RALF1 (Rapid alkalinization factor 1), DVL1 (Devil 1) [23] have also been identified as regulators of symbiotic nodule development and rhizobial infections [24]. Symbiotic functions of RGF homologs were also studied in leguminous plants [25–27].
A genome-wide identification of peptides involved in M. truncatula nodulation revealed that S. meliloti induces the transcription of MtGLV1(Golven 1), MtGLV9, and MtGLV10 in M. truncatula [25]. 15 RGF family genes have been identified, with transcripts of 11 genes detected in different tissues [26]. Our previous study reported that the discovery of a novel RGF peptide, namely MtRGF3p and encoded by MtGLV9, which not only suppressed nodule formation and lateral root emergence but also promoted primary root growth [26]. Overexpression of MtRGF3 in M. truncatula roots consistently led to fewer nodules, while RNAi transgenic roots showed an increased number of nodules [26]. Li et al. found that treatment of M. truncatula seedlings with MtRGF3 peptide resulted in significant repression of key genes in the nodulation factor signaling pathway and activation of defense response-associated genes, as revealed by transcriptome analysis [27]. Additionally, Li et al. observed a notable accumulation of reactive oxygen species (ROS) in M. truncatula roots treated with MtRGF3p [27]. Therefore, MtRGF3p produced by M. truncatula acts as a negative regulator for nodulation symbiosis. The RGF peptide encoded by GLV10 was reported to control organogenesis of nodules and lateral roots in M. truncatula [28]. The question of whether one of RGF peptides from one legume species can function in other legumes remains unanswered. In this study, we investigated the effects of the RGF peptide, MtRGF6p from M. truncatula on the L. japonicus seedlings.
2.1 Preparation of Plant Materials
Medicago truncatula (A17) and Lotus japonicus (MG20) seedlings were cultivated in pots filled with sterilized artificial soil (vermiculite: perlite 3:1) for gene expression analysis of RGFs. A17 and MG20 seedlings were individually grown on Fahraeus medium (FM) and Broughton & Dilworth (BD) agar plates with or without NH4NO3 for peptide activity assays. These seedlings were inoculated with Sinorhizobium meliloti 1021 and Mesorhizobium loti MAFF303099, respectively, in FM and BD liquid medium containing 0.01, 0.1, or 1 μM of the MtRGF6 peptide. The plants were cultivated in a greenhouse environment at 23°C and with 16 h of light per day. To study the expression of MtRGF6 in A17 during the early symbiosis with S. meliloti, roots were collected at various time points post-inoculation, including 1, 3, 5, and 7 days. Additionally, tissues or organs such as roots, leaves, and stems from A17 seedlings grown for 2 weeks in the greenhouse were harvested. Nodules that developed for 3 weeks after rhizobia (2 * 106 cells) inoculation, pods and seeds, as well as young fruits collected at 16 days after pollination (DAP), and fully open flowers were also sampled for analysis.
2.2 RNA Extraction and qRT-PCR
Total RNA was isolated from plant tissues using the RNAprep pure Plant kit (Tiangen Biotech Co., Ltd., Beijing, China, DP437). The synthesis of first-strand cDNA and real-time PCR were carried out with the PrimeScript™ RT reagent kit, which included a gDNA Eraser and a SYBR Green qPCR master mix-SYBR Advantage (Takara, China, RR037Q). For gene expression analysis, qRT-PCR was conducted following the manufacturer’s instructions (Takara, China). House keeping genes such as MtACTIN [26] or LjATPase [20] were utilized, and the primer sequences are provided in Table S1. The expression data of nodulation signaling genes from L. japonicus were subjected to ANOVA analysis.
2.3 Activity Assays of the RGF Peptide
The Scilight-Peptide Company (Beijing, China) synthesized the peptides and dissolved them in sterile water. This peptide solution was then added to FM and BD medium and mixed before being poured into the plates. The assays utilized the following peptides: MtRGF6p, DY(SO3H)AAVKRKP(Hyp)IHN; Random, SHPKY(SO3H)(Hyp)KNSDIRA; MtRG6p unsulfated, DYAAVKRKP(Hyp)IHN; Y(SO3H), sulfated tyrosine; Hyp, hydroxylproline. The peptide of MtRGF3 was described as in the previous study [26]. Seed sterilization and germination of A17 and MG20 were performed following Liu et al. [29] and Wang et al. [20]. Germinated seeds were arranged on solid FM and BD plates with the appropriate peptide (one row per plate, 10 seedlings per row). S. meliloti 1021 and M. loti MAFF303099 were suspended in FM and BD solutions (OD600 = 0.05) and flood-inoculated onto roots. Each peptide (at concentrations of 0.01, 0.1, or 1 μM) was added to FM and BD medium for root phenotype assays of M. truncatula and L. japonicus seedling on plates. Plates with roots were wrapped in black paper bags and placed vertically in a 23°C incubator with 16 h of daylight for either one or three weeks for plant nodulation assays.
For infection thread and nodule primordium assays, seedlings grown on FM and BD solid medium supplemented with appropriate peptides were inoculated with S. meliloli 1021 and M. loti MAFF303099 carrying pXLGD4 (lacZ), respectively, and incubated for 5–7 days as described in previous literature [30]. For the plant nodulation assays, FM and BD medium was supplemented with 0.5 mM of aminoethoxyvinylglycine (AVG, Sigma). To study alternative root growth and development patterns, some seedlings were cultured on FM plates for 1–2 weeks without rhizobia inoculation, but containing peptides and 10 mM of NH4NO3. Data on root length, number of lateral roots, and root nodules were analyzed statistically using student t tests.
The corresponding constructs were introduced into the Agrobacterium rhizogenes strain ARqua1. Hairy root transformation was conducted following the protocols described by Lei et al. [30] and Wang et al. [20], with transgenic roots being identified based on the expression of green fluorescent proteins. The promoter fragment of the MtRGF6 gene was amplified using high fidelity PCR (primer sequences are provided in Table S1), and then inserted the upstream GUS gene in the pBI121 vector, resulting in the creation of pMtRGF6pro:GUS. The number of root nodules were subjected to statistical analysis using student t tests.
The X-gal staining protocol was carried out following the method described by Chen et al. [31] to visualize infection threads and nodule primordia. Subsequently, X-gluc staining was conducted to assess MtRGF6pro:GUS activity in transgenic roots, as outlined in previous studies [30]. The quantification of nodule primordia and infection threads were subjected to statistical analysis using student t tests.
The student t tests were utilized for statistical analysis of data in most experiments, and ANOVA (Analysis of Variance) analysis was conducted for gene expression data.
3.1 The MtRGF6 Gene Is Predominantly Expressed in M. truncatula Root Nodules
Fifteen members of the RGF family (MtRGF1-15) have been identified in M. truncatula [26]. Among them, MtRGF6 showed high expression levels in root nodules at 10, 14, and 28 dpi according to the MtGEA database [26]. The qRT-PCR analysis showed that the MtRGF6 gene exhibited predominant expression in root nodules, while showing lower levels in leaves, roots, seeds, and flowers of M. truncatula (Fig. 1A). The MtRGF6 promoter displayed significant activity in the primordia of lateral roots, young lateral roots, the meristem region of root, nodule primordia, young root nodules, and mature root nodules (Figs. 1B–1G). X-Gluc staining highlighted the highest promoter activity in the enlarged cells with nitrogen fixing zones of mature root nodules (after three weeks inoculation of Sm1021) (Fig. 1H). Based on these expression patterns, it can be inferred that the MtRGF6 gene may be involved in regulating the development of roots and root nodules.
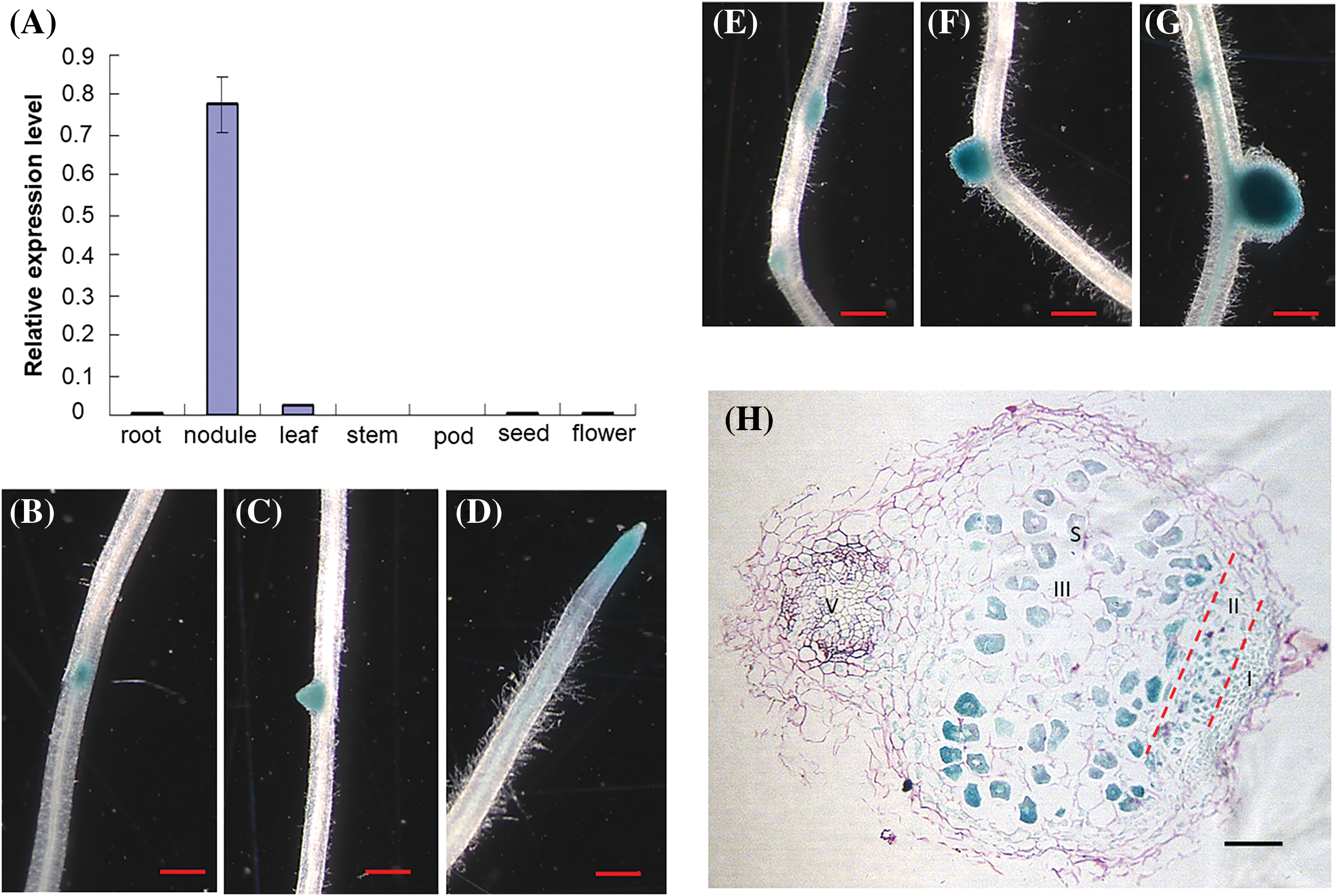
Figure 1: Expression of MtRGF6 in Medicago truncatula. (A) Expression levels of MtRGF6 in various organs of M. truncatula were assessed using quantitative real-time PCR. Three biological replicates were performed, and error bars represent ± SD. (B–F) Promoter activity of MtRGF6 in transgenic M. truncatula roots. The GUS activity was observed in a lateral root primordium (B), a young lateral root, a primary root tip (D), a nodule primordium (E), a young nodule (F), and a mature nodule (G and H). Scale bars range from 1 cm (B–E) to 100 μm (F). I, meristem region; II, infection zone; III, nitrogen fixation zone; S, symbiosome; V, vascular bundle of roots
A total of 7 RGF homologous genes were identified from various databases of L. japonicus, but no high-identity homologs of the MtRGF6 gene and the peptide were found through genomic DNA sequence analysis (Fig. S1). qRT-PCR assays revealed that six of these genes, with the exception of Lj0g3v0191919.1 were transcribed in different tissues. Among them, three genes (Lj6g3v1090130.1, Lj1g3v1222520.2 and Lj1g3v1222540.1) were predominantly expressed in flowers, while three genes (Lj0g3v0278599.2, Lj3g3v1474600.1 and Lj4g3v2775630.1) showed dominant expression in seeds (Fig. S2). Consequently, a gene with similar expression pattern to MtRGF6 was not identified in L. japonicus.
3.2 The MtRGF6 Peptide Has Distinct Effects on Root Development in M. truncatula and L. japonicus
It has been reported that the addition of the MtRGF3 peptide (MtRGF3p) externally promotes the growth of primary roots but suppresses lateral root emergence [3,4,26]. The primary root lengths of M. truncatula were significantly increased by the addition of 0.1 or 1 μM of the chemically synthesized MtRGF6 peptide (MtRGF6p) (Figs. 2A–2C), indicating that an appropriate concentration of MtRGF6p enhances primary root development of M. truncatula. Surprisingly, the inclusion of 0.01 or 0.1 μM of MtRGF6p reduced primary root lengths in L. japonicus seedlings (Figs. 2D–2F), suggesting that exogenous MtRGF6p restricts primary root growth in L. japonicus. Interestingly, the same concentrations of MtRGF3p also suppressed the primary root growth in L. japonicus (Fig. S3). These findings suggest that the RGF peptides produced by M. truncatula may promote primary root growth in their own species but suppress it in L. japonicus.
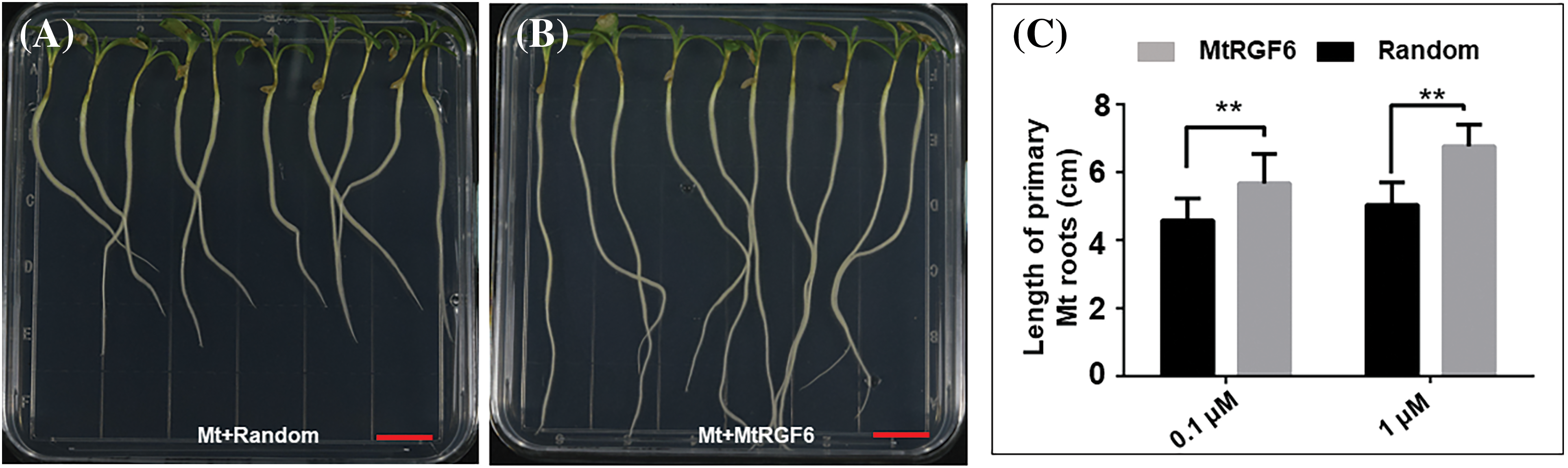
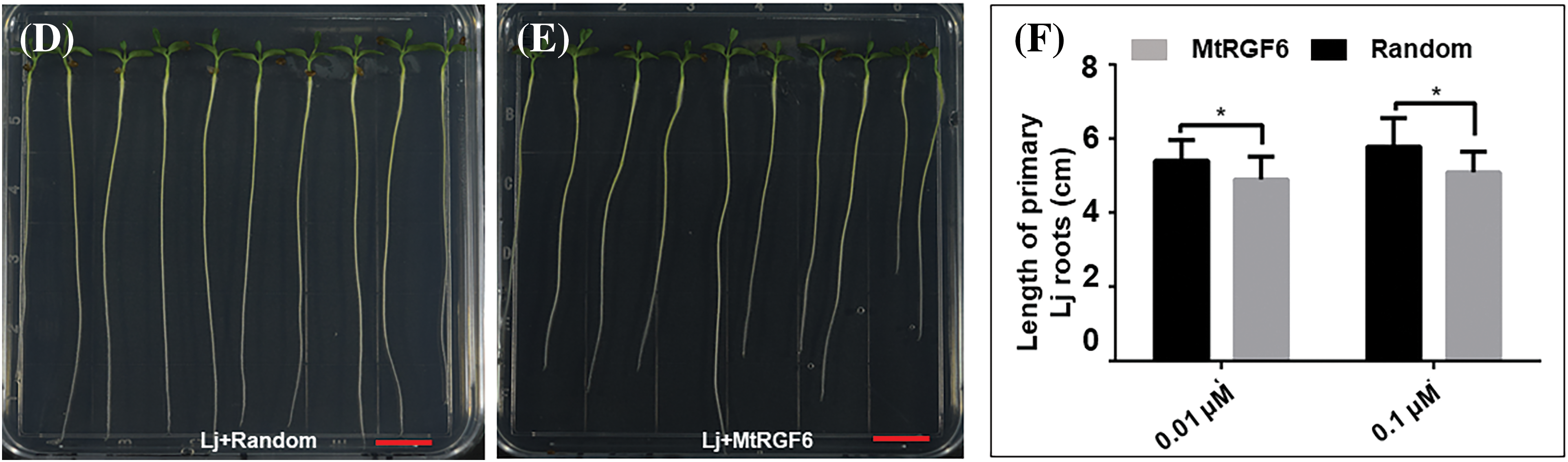
Figure 2: Effects of MtRGF6p on M. truncatula and L. japonicus primary root growth. The effects of 1 μM MtRGF6p and the random peptide on primary root growth of M. truncatula (A17) (A–C) and L. japonicus (MG20) (D–F) seedlings were examined. Scale bars are 1 cm in size. Symbols: Mt–M. truncatula; Lj–L. japonicus; * –Significant differences were observed in t-tests with P < 0.05; ** –P < 0.01; N > 40. Error bars represent ± SE
The counting of lateral root density M. truncatula and L. japonicus after 14 days of treatment with MtRGF6p revealed that concentrations of 0.01 and 0.1μM led to a reduction in the density of lateral roots on the primary roots of both seedlings to a similar extent (Fig. 3). This indicates that MtRGF6p has a comparable suppression effect on the organogenesis of lateral roots in both M. truncatula and L. japonicus.
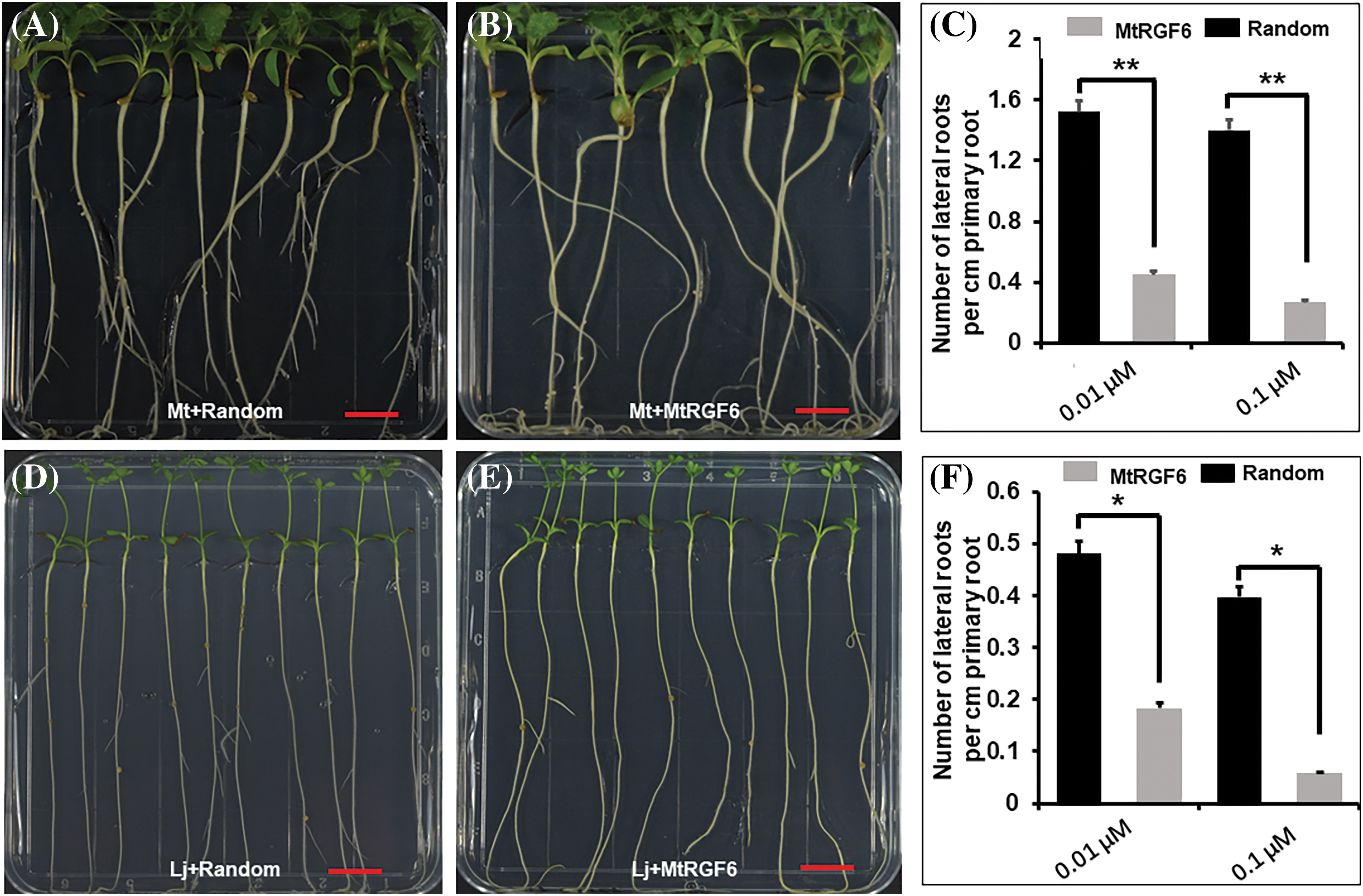
Figure 3: Effects of MtRGF6p on M. truncatula and L. japonicus lateral root formation. The effects of 1 μM of MtRGF6p on lateral root formation of M. truncatula (A–C) and L. japonicus (D–F) seedlings were examined). Symbols: Mt–M. truncatula; Lj–L. japonicus; *Significant differences were observed in t tests with p < 0.05; **p < 0.01; N > 40. Error bars represent ±SE
3.3 MtRGF6p Has Distinct Effects on the Nodulation Symbiosis of M. truncatula and L. japonicus
Given that MtRGR3p suppresses rhizobial infection and symbiotic nodulation in M. truncatula [26], it is reasonable to assume that MtRGF6p may impact the nodulation symbiosis in both M. truncatula and L. japonicus with their respective rhizobia. Nodulation tests on agar plates revealed that treatment with 0.1 and 1 μM of MtRGF6p led to a 30% and 67% reduction in root nodule density in M. truncatula compared to the control peptide (Figs. 4A–4C). Similarly, treatment with 0.1 and 1 μM of MtRGF6p resulted in a 75% and 85% decrease in root nodule density in L. japonicus (Figs. 4D–4F). This suggests that MtRGF6p has a stronger inhibitory effect on nodulation in L. japonicus than in M. truncatula at the same concentrations. Furthermore, the unmodified peptide without the O3H group did not significantly affect nodulation suppression in either legume species (Fig. S4) indicating that the presence of a sulfate group alters peptide activity, with tyrosine being crucial. Interestingly, treatment with 0.01 or 0.1 μM of MtRGF3p did not have a significant impact on nodulation in L. japonicus (Fig. S5), suggesting that the suppression nodulation by MtRGF6p may be specific to L. japonicus.
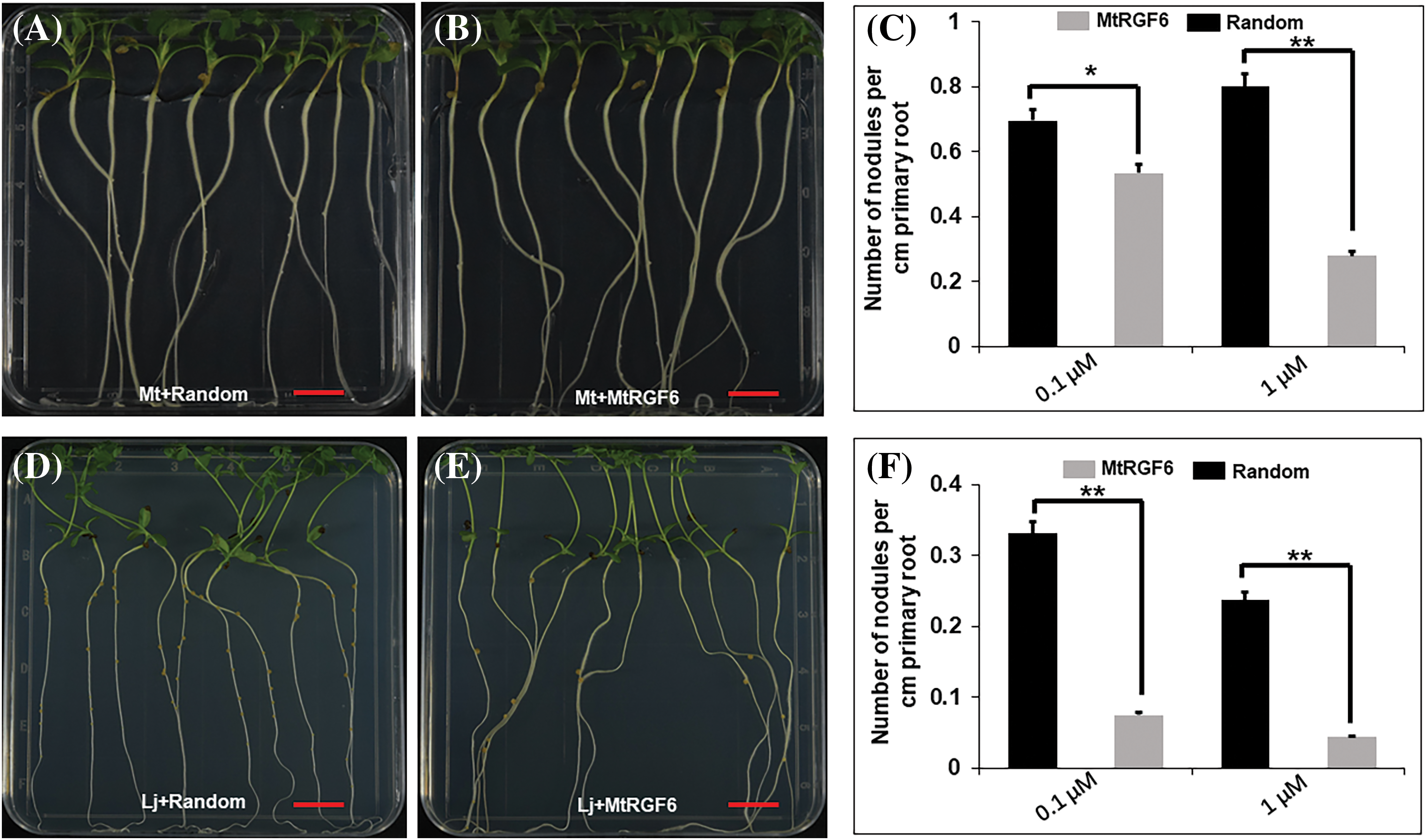
Figure 4: Effects of MtRGF6p on M. truncatula and L. japonicus root nodule formation. The effects of 1 μM MtRGF6p and the random peptide on root nodule formation of M. truncatula (A–C) and L. japonicus (D–F) seedlings were examined. Symbols: Mt–M. truncatula; Lj–L. japonicus; *Scale bars = 1 cm. Significant differences were observed in t tests with p < 0.05; **p < 0.01; N > 40. Error bars represent ±SE
The examination of infection events and nodule primordia revealed that a 70% reduction in infection threads and a 60% decrease in nodule primordia in M. truncatula seedlings after treatment with 1 μM of MtRGF6p (Figs. 5A–5C). Similarly, the addition of 0.1 μM of the peptide resulted in an 80% decrease in infection threads and nodule primordia in L. japonicus seedlings (Figs. 5D–5F). The findings indicate that MtRGF6p modulates early symbiosis events between legumes and rhizobia, with rhizobial infection and nodule initiation in L. japonicus being more sensitivity to MtRGF6p than in M. truncatula.
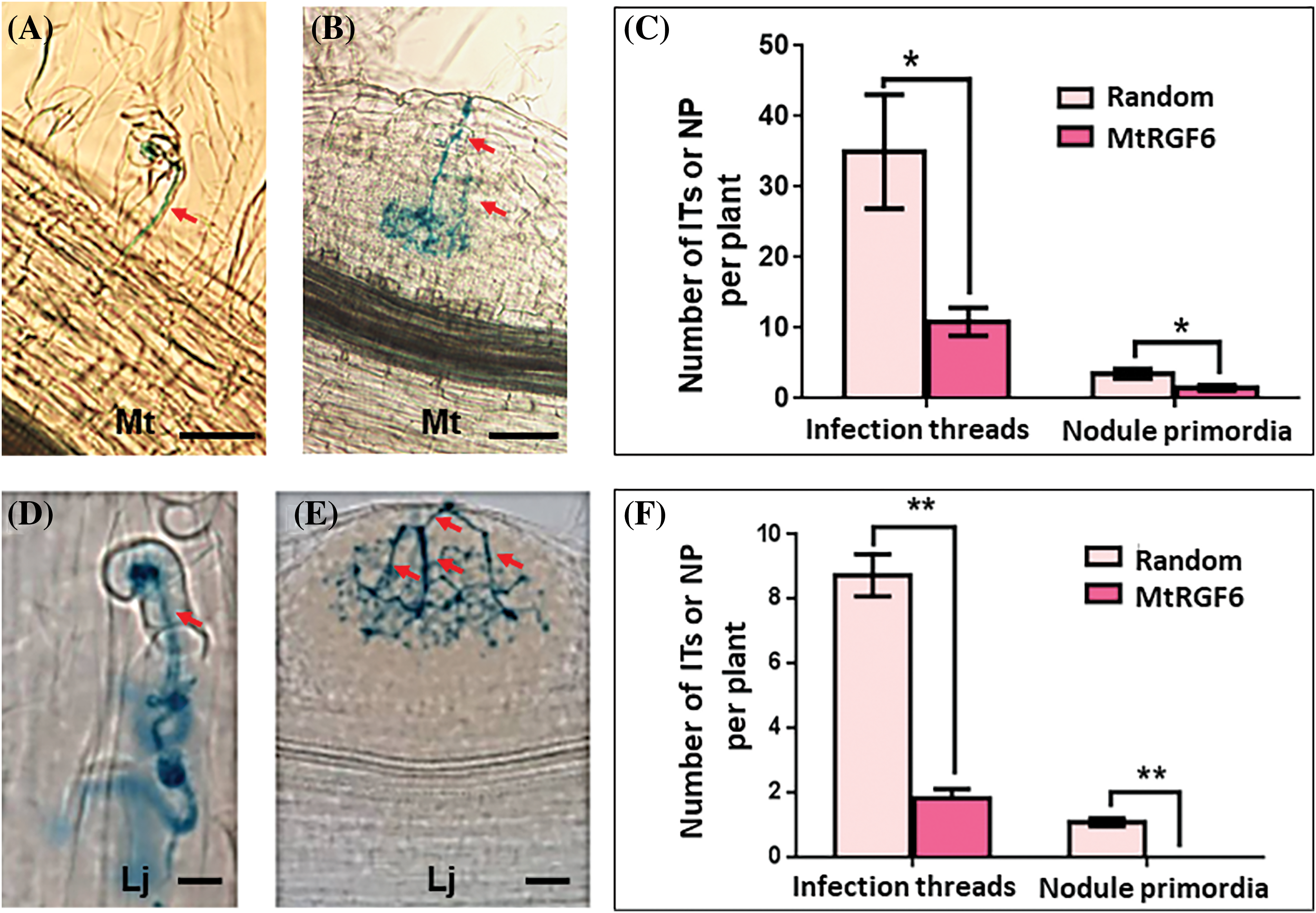
Figure 5: Effects of MtRGF6p on the early symbiosis of M. truncatula and L. japonicus. The effects of 1 μM MtRGF6p on the formation of infection threads and infected nodule primordia of M. truncatula (A–C) and L. japonicus (D–F) seedlings were examined. Symbols: Mt–M. truncatula; Lj–L. japonicus; Red arrows, infection threads; *Significant differences were observed in t tests with p < 0.05; **p < 0.01; N > 40. Error bars represent ±SE
3.4 MtRGF6p Suppressed the Transcriptional Activation of Key Symbiosis-Signaling Genes in L. japonicus Induced by Rhizobia
Similar to MtRGF3 [26], MtRGF6p may suppress the symbiotic nodulation of L. japonicus by downregulating key genes involved in the nodulation factor signaling pathway. qRT-PCR was used to analyze the transcription levels of key nodulation signaling genes (SYMRK, NSP2, NIN, NRF5, NSP1, POLLUX, CCAMK, CASTOR, CYCLOPS and ENOD40-1) in L. japonicus seedlings to validate this possibility, showing that the expression of these genes in L. japonicus was mostly induced by M. loti (Fig. 6) as reported in literatures [32]. However, treatment with 0.1 μM MtRGF6p suppressed the activation of these genes (excluding LjENOD40) after inoculation with M. loti (Fig. 6). This result suggests that MtRGF6p may interfere nodulation signaling to reduce the formation of infection threads and nodule primordia.
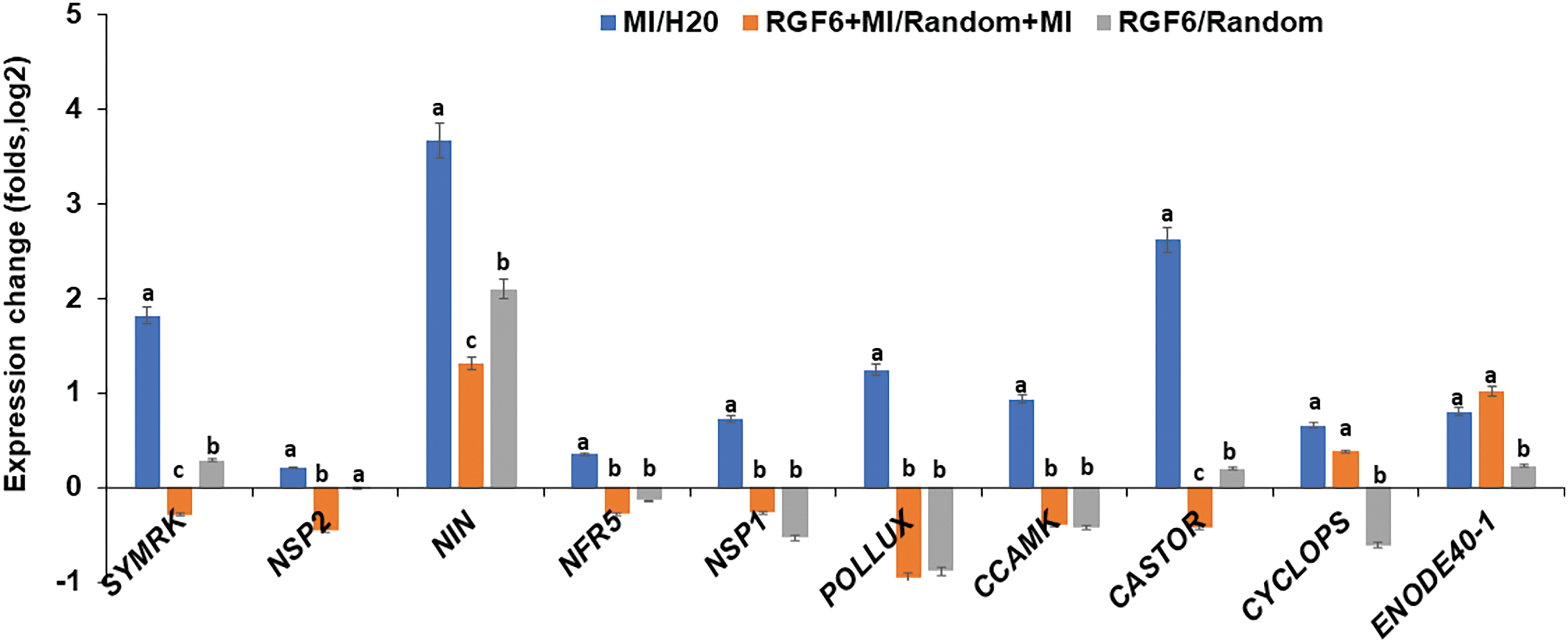
Figure 6: Expression levels of key genes in the nodulation factor signaling pathway in L. japonicus roots were analyzed by qRT-PCR analysis. Ml, M. loti-inoculated samples; H2O, the blank control; Ml/H2O representing the division of the data between the two. Samples treated with both MtRGF6 peptide and M. loti were denoted as RGF6 + Ml, while samples treated with Random peptide and M. loti simultaneously were labeled as Random + Ml. The ratio of RGF6 + Ml to Random+Ml was represented as RGF6 + Ml/Random + Ml, and the ratio of samples treated with MtRGF6 peptide to those treated with Random peptide was denoted as RGF6/Random. Three biological replicates were performed, with one representative shown. Each experiment involved treatment of >30 independent roots. The error bar represents ±SD. The ANOVA program was used for statistical evaluation
While some peptide signals, such as CLE, PSK, and RGF, have been found to regulate the development of root organs in leguminous species like M. truncatula, it was previously unknown if these signals would have the same effect on other legume species. In this study, we demonstrate for the first time that the RGF peptides from M. truncatula impact the development of root organs (primary roots, lateral roots and root nodules) in another leguminous species, L. japonicus. Surprisingly, both MtRGF6p and MtRGF3p suppress the growth of primary roots in L. japonicus, while promoting growth in M. truncatula (Fig. 2 and Fig. S3). This finding suggests that these peptides play a crucial role in inter-species interactions among legumes. MtRGF6p suppresses the emergence of lateral roots of two leguminous model plants (Fig. 3), while also functioning as a negative regulator of nodulation symbiosis on these plants (Figs. 4 and 5). Interestingly, a lower concentration of MtRGF6p was sufficient to suppress nodulation in L. japonicus compared to M. truncatula (Figs. 4 and 5), suggesting that L. japonicus is more sensitive to the peptide. Overall, our results demonstrate that the RGF peptide can modulate the development of root organs in L. japonicus, supporting the idea that it serves as a signal for inter-species interactions among legumes. The expression pattern of MtRGF6 suggests that the encoded peptide may not only regulate the differentiation of meristem cells to affect the development of root organs, but also may be involved in functions related to housing rhizobia in symbiosome. In contrast to MtRGF6, MtRGF3 is predominantly expressed in leaves, with its transcription being induced in roots through rhizobial inoculation or Nod factor treatment [25].
In some plant species, the functions of the RGF peptides are redundant, as the RGF genes form a multi-gene family in A. thaliana, M. truncatula, L. japonucus, and other plants [33]. This redundancy makes it challenging to distinguish the phenotypes of single gene mutants genetically, and overexpression lines only show minor defects in growth and development, such as the nodulation phenotype observed in MtRGF3 [26] and MtRGF6 (Fig. S6). One possible explanation for this is the high background levels of RGF peptides in a plant organ, where multiple RGF genes are redundantly expressed, including constitutive and inducible expression. Consequently, mutations and overexpression of a single RGF gene may not significantly alter the level of RGF peptides in a plant organ. Another explanation could be the restriction of RGF peptide maturation in the overexpression line, as certain processing enzymes may be specifically expressed in certain organs or cells.
The RGF peptides are recognized by the kinase receptors, RGFRs, which form a multi-gene family and interact with MAP kinases and ROS to modulate the activity of the transcriptional factors, PLT1/PLT2, controlling root development in A. thaliana [6–8,34]. Through analysis of genomic DNA, transcriptomic (such as RGFRs, MAP kinases and PLTs) and physiological data (such as ROS), it is suggested that M. truncatula and L. japonicus share these signaling elements with A. thaliana [9–11]. Therefore, the mechanism by which RGF peptides regulate root development, including primary roots and lateral roots in both model legumes (Fig. 2), may be conserved compared to A. thaliana., although further studies are required to confirm this. MtRGF6 and MtRGF3 peptides have opposing effects on primary root development in M. truncatula and L. japonicus, which may be due to differences in their recognition receptors/co-receptors, and downstream elements such as Ca2+, ROS and transcriptional factors. Further research is needed to validate this hypothesis. The process of leguminous root nodule development involves nodule initiation and rhizobium infection [13]. The signaling pathway of RGF peptides regulating nodule development may be derived from roots, as the nodule primordium differentiates from cortical meristem cells, similar to lateral root primordia. The formation of infection threads is regulated by leguminous immune responses, with ROS dynamics modulated by RGF peptides potentially contributing to plant immunity [11,27]. The initiation of nodules and rhizobial infection are influenced by Nod factor signal transduction, which is suppressed by MtRGF6p (Figs. 4–6), as well as MtRGF3p [26]. This conclusion is supported by the expression data of the Nod factor signaling pathway genes (Fig. 6) [27]. Furthermore, the difference in nodulation symbiosis sensitivity to MtRGF6p between M. truncatula and L. japonicus may be linked to the distinct locations of nodule initiation (root endodermis and pericycle for M. truncatula, but root exodermis for L. japonicus). Investigating how the RGF signaling pathway interacts with Nod factor signal transduction is a crucial area for future research.
This study shows that the peptide of MtRGF6, along with MtRGF3p is shown to initially suppress primary root growth in L. japonicus but enhance it in M. truncatula. While the development of lateral roots is uniformly suppressed in both model legumes, the impact on nodulation symbiosis varies, with MtRGF6p selectively dampening the activation of the nodulation signaling pathway. This suggests that RGF peptides could serve as an inter-species signal to modulate plant growth and development.
Acknowledgement: We appreciate the help provided by Dr. Liangliang Yu during the experiments.
Funding Statement: This research was supported by the grant from the Natural Science Foundation of China (No. 31900214 to JY) and the Open Fund of State Key Laboratory of Tea Plant Biology and Utilization (SKLTOF20210113 to LL).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Li Luo and Junhui Yan; data collection: Yawen Wang and Qiong Li; analysis and interpretation of results: Junhui Yan, Yawen Wang and Qiong Li; draft manuscript preparation: Li Luo, Yu Zhou and Xu Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data are contained within the article and supplementary materials. Readers can request relevant materials from the corresponding author, Li Luo.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflict of interest.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.051517.
References
1. Fernandez A, Drozdzecki A, Hoogewijs K, Vassileva V, Madder A, Beeckman T, et al. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. J Exp Bot. 2015;66(17):5245–56. doi:10.1093/jxb/erv329. [Google Scholar] [PubMed] [CrossRef]
2. Matsuzaki Y, Ogawa-Ohnishi R, Mori R, Matsubayashi R. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329(5995):1065–7. doi:10.1126/science.1191132. [Google Scholar] [PubMed] [CrossRef]
3. Meng L, Buchanan BB, Feldman LJ, Luan S. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109(5):1760–5. doi:10.1073/pnas.1119864109. [Google Scholar] [PubMed] [CrossRef]
4. Whitford R, Fernandez A, Tejos R, Perez AC, Kleine-Vehn J, Vanneste S, et al. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell. 2012;22(3):678–85. doi:10.1016/j.devcel.2012.02.002. [Google Scholar] [PubMed] [CrossRef]
5. Matsubayashi Y. Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 2011;52(1):5–13. doi:10.1093/pcp/pcq169. [Google Scholar] [PubMed] [CrossRef]
6. Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, et al. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res. 2016;26:686–98. doi:10.1038/cr.2016.63. [Google Scholar] [PubMed] [CrossRef]
7. Shinohara H, Mori A, Yasue N, Sumida, K, Matsubayashi Y. Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc Natl Acad Sci U S A. 2016;113:3897–902. doi:10.1073/pnas.1522639113. [Google Scholar] [PubMed] [CrossRef]
8. Song W, Liu L, Wang J, Wu Z, Zhang H, Tang J, et al. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 2016;26:674–85. doi:10.1038/cr.2016.62. [Google Scholar] [PubMed] [CrossRef]
9. Lu X, Shi H, Ou Y, Cui Y, Chang J, Peng L, et al. RGF1-RGI1, a peptide-receptor complex, regulates arabidopsis root meristem development via a MAPK signaling cascade. Mol Plant. 2020;13:1594–607. doi:10.1016/j.molp.2020.09.005. [Google Scholar] [PubMed] [CrossRef]
10. Shao Y, Yu X, Xu X, Li Y, Yuan W, Xu Y, et al. The YDA-MKK4/MKK5-MPK3/MPK6 cascade functions downstream of the RGF1-RGI ligand-receptor pair in regulating mitotic activity in root apical meristem. Mol Plant. 2020;13:1608–23. doi:10.1016/j.molp.2020.09.004. [Google Scholar] [PubMed] [CrossRef]
11. Yamada M, Han X, Benfey PN. RGF1 controls root meristem size through ROS signalling. Nature. 2020;577:85–8. doi:10.1038/s41586-019-1819-6. [Google Scholar] [PubMed] [CrossRef]
12. Limpens E, Bisseling T. Signaling in symbiosis. Curr Opin Plant Biol. 2003;6:343–50. doi:10.1016/S1369-5266(03)00068-2. [Google Scholar] [PubMed] [CrossRef]
13. Ghantasala S, Roy Choudhury S. Nod factor perception: an integrative view of molecular communication during legume symbiosis. Plant Mol Biol. 2022;110:485–509. doi:10.1007/s11103-022-01307-3. [Google Scholar] [PubMed] [CrossRef]
14. Mortier V, den Herder G, Whitford R, van de Velde W, Rombauts S, D’Haeseleer K, et al. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153:222–37. doi:10.1104/pp.110.153718. [Google Scholar] [PubMed] [CrossRef]
15. Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun. 2013;4:2191. doi:10.1038/ncomms3191. [Google Scholar] [PubMed] [CrossRef]
16. Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–9. doi:10.1038/nature01231. [Google Scholar] [PubMed] [CrossRef]
17. Mohd-Radzman NA, Laffont C, Ivanovici A, Patel N, Reid D, Stougaard J, et al. Different pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol. 2016;171:2536–48. doi:10.1104/pp.16.00113. [Google Scholar] [PubMed] [CrossRef]
18. Mohd-Radzman NA, Binos S, Truong TT, Imin N, Mariani M, Djordjevic MA. Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. J Exp Bot. 2015;66:5289–300. doi:10.1093/jxb/erv008. [Google Scholar] [PubMed] [CrossRef]
19. Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot. 2013;64:5395–409. doi:10.1093/jxb/ert369. [Google Scholar] [PubMed] [CrossRef]
20. Wang C, Yu H, Zhang Z, Yu L, Xu X, Hong Z, et al. Phytosulfokine is involved in positive regulation of Lotus japonicus nodulation. Mol Plant Microbe Interact. 2015;28:847–55. doi:10.1094/MPMI-02-15-0032-R. [Google Scholar] [PubMed] [CrossRef]
21. Di Q, Li Y, Zhang DP, Wu W, Zhang L, Zhao X, et al. A novel type of phytosulfokine, PSK-epsilon, positively regulates root elongation and formation of lateral roots and root nodules in Medicago truncatula. Plant Signal Behav. 2022;17(1):2134672. doi:10.1080/15592324.2022.2134672. [Google Scholar] [PubMed] [CrossRef]
22. Yu L, Di Q, Zhang D, Liu Y, Li X, Mysore KS, et al. A legume-specific novel type of phytosulfokine, PSK-δ, promotes nodulation by enhancing nodule organogenesis. J Exp Bot. 2022;73:2698–713. doi:10.1093/jxb/erac051. [Google Scholar] [PubMed] [CrossRef]
23. Combier JP, Küster H, Journet EP, Hohnjec N, Niebel A. Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol Plant Microbe Interact. 2008;21:1118–27. doi:10.1094/MPMI-21-8-1118. [Google Scholar] [PubMed] [CrossRef]
24. Kereszt A, Mergaert P, Montiel J, Endre G, Kondorosi E. Impact of plant peptides on symbiotic nodule development and functioning. Front Plant Sci. 2018;9:1026. doi:10.3389/fpls.2018.01026. [Google Scholar] [PubMed] [CrossRef]
25. de Bang TC, Lundquist PK, Dai X, Boschiero C, Zhuang Z, Pant P, et al. Genome-wide identification of medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 2017;175:1669–89. doi:10.1104/pp.17.01096. [Google Scholar] [PubMed] [CrossRef]
26. Li Q, Li M, Zhang D, Yu L, Yan J, Luo L. The peptide-encoding MtRGF3 gene negatively regulates nodulation of Medicago truncatula. Biochem Biophys Res Commun. 2020;523:66–71. doi:10.1016/j.bbrc.2019.12.017. [Google Scholar] [PubMed] [CrossRef]
27. Li Q, Shan D, Zheng W, Wang Y, Lin Z, Jin H, et al. MtRGF3 peptide activates defense responses and represses the expressions of nodulation signaling genes in Medicago truncatula. Acta Biochim Biophys Sin. 2023;55:1319–22. [Google Scholar] [PubMed]
28. Roy S, Torres-Jerez I, Zhang S, Liu W, Schiessl K, Boschiero C, et al. The peptide GOLVEN10 controls nodule and lateral root organogenesis and positioning along the longitudinal root axis. bioRxiv. 2022;5:490929. [Google Scholar]
29. Liu W, Chen AM, Luo L, Sun J, Cao LP, Yu GQ, et al. Characterization and expression analysis of Medicago truncatula ROP GTPase family during the early stage of symbiosis. J Integr Plant Biol. 2010;52:639–52. doi:10.1111/jipb.2010.52.issue-7. [Google Scholar] [CrossRef]
30. Lei MJ, Wang Q, Li X, Chen A, Luo L, Xie Y, et al. The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and nod factor-induced root hair deformation. Plant Cell. 2015;27:806–22. doi:10.1105/tpc.114.135210. [Google Scholar] [PubMed] [CrossRef]
31. Chen AM, Wang YB, Jie S, Yu AY, Luo L, Yu GQ, et al. Identification of a TRAP transporter for malonate transport and its expression regulated by GtrA from Sinorhizobium meliloti. Res Microbiol. 2010;161:556–64. doi:10.1016/j.resmic.2010.05.004. [Google Scholar] [PubMed] [CrossRef]
32. Montiel J, Reid D, Grønbæk TH, Benfeldt CM, James EK, Ott T, et al. Distinct signaling routes mediate intercellular and intracellular rhizobial infection in Lotus japonicus. Plant Physiol. 2021;185:1131–47. doi:10.1093/plphys/kiaa049. [Google Scholar] [PubMed] [CrossRef]
33. Furumizu C, Sawa S. The RGF/GLV/CLEL family of short peptides evolved through lineage-specific losses and diversification and yet conserves its signaling role between vascular plants and bryophytes. Front Plant Sci. 2021;12:703012. doi:10.3389/fpls.2021.703012. [Google Scholar] [PubMed] [CrossRef]
34. Ou Y, Li J. Three divergent approaches identified the same RGF1 receptors in Arabidopsis thaliana. Sci China Life Sci. 2017;60:1040–3. doi:10.1007/s11427-017-9009-x. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools