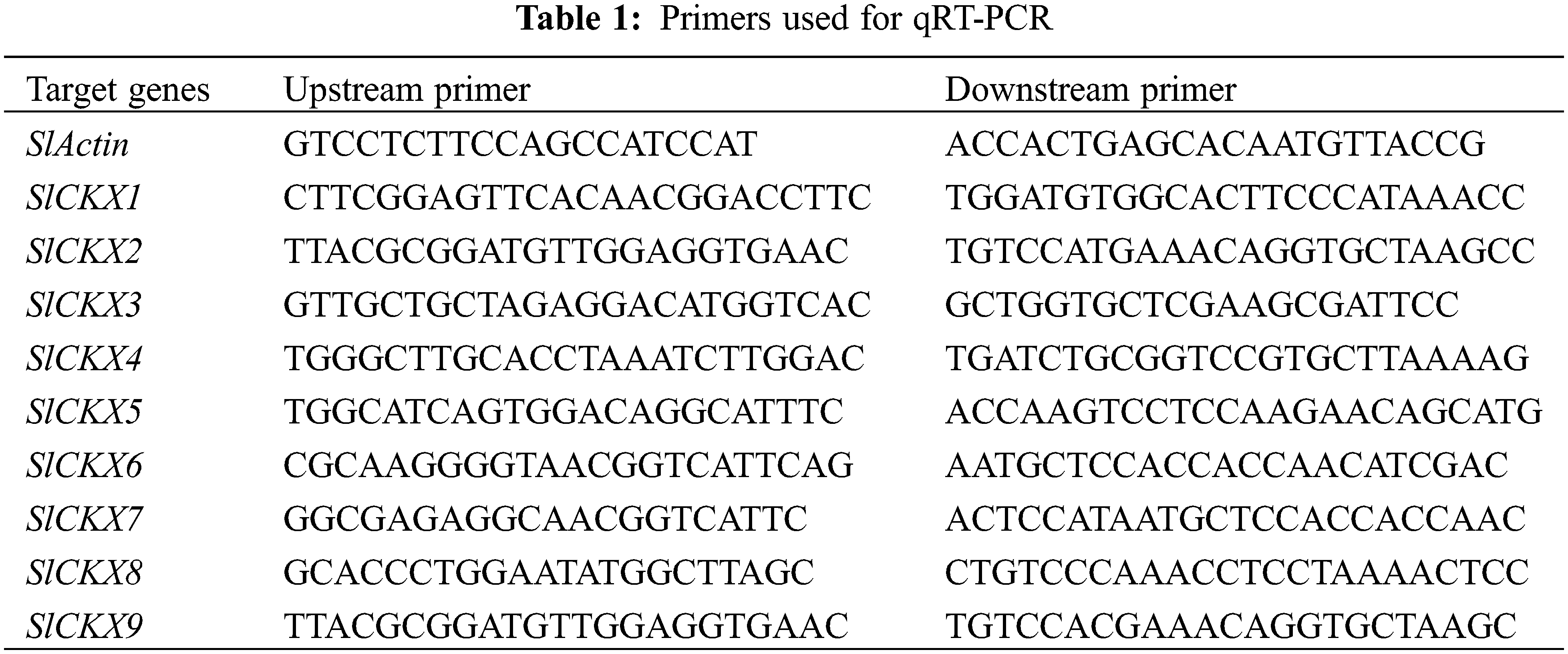
 Open Access
Open Access
ARTICLE
Genome-Wide Identification of Tomato (Solanum lycopersicum L.) CKX Gene Family and Expression Analysis in the Callus Tissue under Zeatin Treatment
Subtropical Agriculture Research Institute, Fujian Academy of Agricultural Sciences, Zhangzhou, 363005, China
* Corresponding Author: Zhou Li. Email:
(This article belongs to the Special Issue: Recent Research Trends in Genetics, Genomics, and Physiology of Crop Plants)
Phyton-International Journal of Experimental Botany 2024, 93(6), 1143-1158. https://doi.org/10.32604/phyton.2024.051207
Received 29 February 2024; Accepted 18 April 2024; Issue published 27 June 2024
Abstract
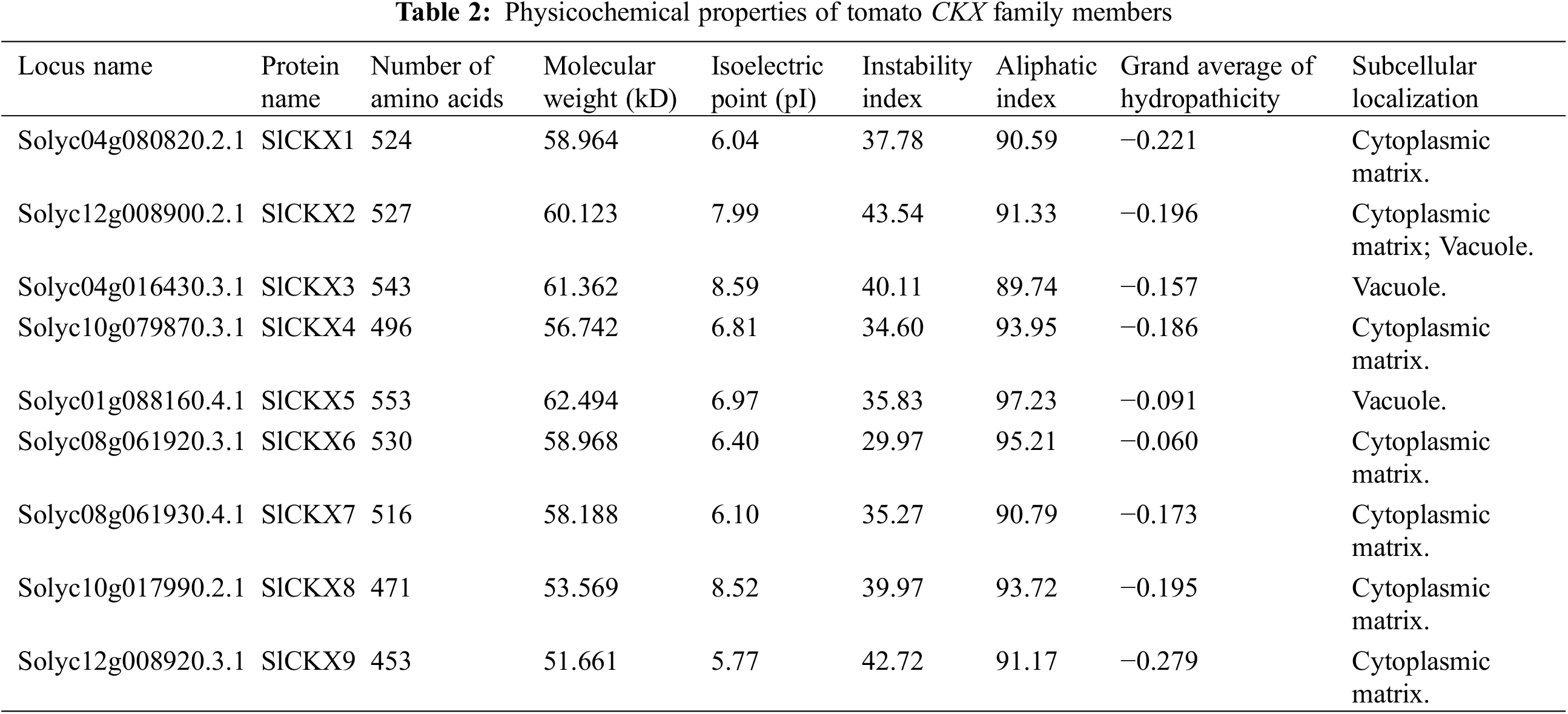
The cytokinin oxidase/dehydrogenase (CKX) enzyme is essential for controlling the fluctuating levels of endogenous cytokinin (CK) and has a significant impact on different aspects of plant growth and development. Nonetheless, there is limited knowledge about CKX genes in tomato (Solanum lycopersicum L.). Here we performed genome-wide identification and analysis of nine SlCKX family members in tomatoes using bioinformatics tools. The results revealed that nine SlCKX genes were unevenly distributed on five chromosomes (Chr.1, Chr.4, Chr.8, Chr.10, and Chr.12). The amino acid length, isoelectric points, and molecular weight of the nine SlCKX proteins ranged from 453 to 553, 5.77 to 8.59, and 51.661 to 62.494 kD, respectively. Subcellular localization analysis indicated that SlCKX2 proteins were located in both the vacuole and cytoplasmic matrix; SlCKX3 and SlCKX5 proteins were located in the vacuole; and SlCKX1, 4, 6, 7, 8, and 9 proteins were located in the cytoplasmic matrix. Furthermore, we observed differences in the gene structures and phylogenetic relationships of SlCKX proteins among different members. SlCKX1-9 were positioned on two out of the three branches of the CKX phylogenetic tree in the multispecies phylogenetic tree construction, revealing their strong conservation within phylogenetic subgroups. Unique patterns of expression of CKX genes were noticed in callus cultures exposed to varying concentrations of exogenous ZT, suggesting their roles in specific developmental and physiological functions in the regeneration system. These results may facilitate subsequent functional analysis of SlCKX genes and provide valuable insights for establishing an efficient regeneration system for tomatoes.Keywords
Supplementary Material
Supplementary Material FileCytokinins (CKs), a type of phytohormone, play a pivotal role in the regulation and modulation of plant growth and development [1]. They induce cytoplasmic division and have diverse applications in plant tissue culture because of their ability to stimulate plant regeneration [2]. The biosynthesis and degradation of CKs control their internal levels in plant cells [3]. Trans-zeatin (ZT), a plant growth factor extracted from maize whiskers, exists in two forms: trans and cis. ZT regulates growth and exhibits anti-aging effects. Two biosynthetic pathways for CKs in plants are the transfer RNA (tRNA) pathway and the AMP (ATP/ADP) pathway. In the tRNA pathway, an isopentenyl adenosine residue can be modified by tRNA isopentenyl transferase (tRNA IPT, EC 2.5.1.8), leading to the subsequent conversion of cis-ZT to active trans-ZT catalyzed by cis-trans isomerase [4–6]. The degradation of CKs is primarily catalyzed by CK oxidase/dehydrogenase (CKX), which is the only known enzyme associated with the irreversible degradation of CKs and is present in all plant tissues [7]. The CKX enzyme is vital for controlling processes that depend on CK [8,9]. The CKX gene families have been discovered in millet [10], maize [11], soybean [12], oilseed rape [13], and wheat [14], with the reported numbers of gene family members being 11, 13, 17, 23 and 11, respectively.
Although CKX isoforms catalyze similar reactions, they exhibit distinct patterns of subcellular localization and tissue specificity in different plant species. In Arabidopsis, AtCKX1 and AtCKX3 are localized in the vacuole and AtCKX2 is localized in the endoplasmic reticulum [15]. Likewise, in maize, ZmCKX1 is found in the apoplast, while ZmCKX10 is situated in the cytosol [16]. Expression analyses in Arabidopsis revealed that AtCKX1 and AtCKX2 are predominantly expressed in the shoot apex; AtCKX4 in stomatal precursor cells, trichomes, and root cap; and AtCKX5 in the root meristem and stamen. In addition, AtCKX6 expression was associated with the gynoecium at various development [4,17]. Likewise, in maize, the genes ZmCKX2, ZmCKX3, ZmCKX4, and ZmCKX5 exhibited high levels of expression in mature tassels, aged leaves, young tassels, and developing ears, respectively [18].
The impact of external phytohormones on the expression levels of CKX in various plant tissues remains poorly understood. For instance, 6-benzylaminopurine (6-BA) could counteract poor growth and decrease chlorophyll content in wheat under low temperatures by modulating endogenous hormone levels [19], In addition, 6-BA could reduce CKX activity, improve antioxidant enzyme activity, promote grain filling, and increase wheat yield [20]. Treating trifoliate orange plants with exogenous 6-BA resulted in increased expression of PsCKX1, PsCKX2, and PsCKX5 in both the leaves and roots. Furthermore, this treatment slightly increased the expression of PsCKX7 in the leaves but not in the roots [21]. Under abscisic acid (ABA) stress, the expression of CKX was increased in Medicago sativa [22], and the heightened expression of MdCKX5.2 caused an increased sensitivity to external ZT which affected the primary root elongation [23]. These findings suggest that different CKXs typically exhibit varying subcellular localization, tissue specificity, and spatial and temporal expression patterns, and the physiological functions of most species remain unexplored.
Tomato (Solanum lycopersicum L.) is a vital horticultural crop in China [24], It has various varieties with broad adaptability to diverse environments, and its production and consumption continue to rise with the discovery of its high nutritional values and medicinal applications [25]. However, the tomato industry faces challenges such as a long growth cycle and susceptibility to pests and diseases during the seedling stage [26]. Cellular engineering offers a promising solution to enhance tomato production efficiency by reducing the effects of season, pest, and disease, and reducing cultivation time. High-quality callus tissue is crucial for cell culture in plant asexual propagation [27]. Thus, investigating tomato CKX family genes and their expression during callus development is crucial for developing efficient tomato in vitro regeneration systems.
This study aimed to identify nine CKX genes in tomato plants, examine their structural features, and evaluate the expression of CKX in callus tissues under ZT treatments through qRT-PCR. The findings of this research establish a foundation for further investigations into the biological roles of CKX genes in tomatoes and offer insights for improving callus induction techniques.
2.1 Identification of CKX Family in Tomato
To identify the CKX gene family members in tomatoes, we conducted a comparison of gene sequences with the tomato genome database found on the Sol Genomics Network website (https://solgenomics.net/), which is derived from the Ensembl Plants database (https://plants.ensembl.org/index.html). This analysis allowed us to establish the total count of tomato CKX gene family members, which were then assigned the names SlCKX1-9.
2.2 Analysis of Physicochemical Properties of SlCKXs Gene
The Expasy website (https://web.expasy.org/protparam/) was utilized to analyze the physicochemical characteristics of members of the SlCKXs family. This analysis encompassed a range of properties such as molecular weights, theoretical isoelectric points, protein instability indexes, and fat indexes (which represent the relative values of aliphatic amino acid content) [28]. Subcellular localizations were predicted using Cell-Ploc 2.0 (www.csbio.sjtu.edu.cn).
2.3 Analysis of SlCKXs Protein Structure
We used the Expasy website (https://swissmodel.expasy.org/interactive) to analyze the secondary structure of the SlCKX proteins and predict their tertiary structures. Additionally, we constructed protein tertiary structure models.
2.4 Analysis of Chromosome Localization, Gene Structure, Conserved Motifs, and Conserved Domains
The MapChart tools (v 2.3.2) were used to visually show the localization of SlCKX genes on the chromosome [29]. By utilizing the MEME online software (https://meme-suite.org/meme/tools/meme), conserved motifs (motif) in the SlCKX protein sequences were identified and then visualized with TBtools.
2.5 Comparison of SlCKX Gene Family Members with CKX Gene Members of Other Species and Collinearity Analysis
We used MEGA 7.0 (www.megasoftware.net) to determine the evolutionary relationships of the SlCKX tree [30], and the results were displayed using evolutionary trees from the online website iTOL (https://itol.embl.de/itol.cgi) [31].
We utilized the MapChart tool (v. 2.3.2) [29] for mapping the SlCKX genes on chromosomes. Furthermore, we employed MCScanX software [32] to study the evolutionary relationship of homologous CKX genes among tomato and Arabidopsis, eggplant, and cabbage. Subsequently, KaKs_Calculator 2.0 was applied to determine the nonsynonymous (Ka) and synonymous (Ks) substitutions for each iteration of the SlCKX gene [33]. The divergence time was calculated using the formula T = Ks/2R, with R representing 1.5 × 108 synonymous substitutions per year.
2.6 Analysis of Cis-Acting Elements
To investigate the cis-acting elements found in the promoter regions of SlCKX family members, 2000-base pair DNA sequences upstream of the start codon of SlCKX family members were obtained through TBtools software. Subsequently, PlantCARE, an online tool available at http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ [34], was employed for analyzing cis-elements.
To establish a sterile seedling culture, seeds of ‘Micro Tom’ were immersed in sterilized water at 55°C for 30 min, and the water temperature was lowered to 30°C for germination by immersion. Under aseptic conditions, the seeds were soaked in 75% alcohol for 30 s, rinsed three times with sterile water, dried on filter paper, soaked in 4%–5% NaClO for 10 min, rinsed four to five times with sterile water, and blotted on filter paper to dry the seed surface. The sterile seeds were placed on MS solid medium (0.7% agar) amended with 3% sucrose and allowed to germinate and grow for 10 days in a constant incubator (25°C, 4,000 lux light intensity, and 12/12 h day/night rhythm).
Young leaves of 10-day-old tomato seedlings were cut into 0.5 cm × 0.5 cm squares on an ultra-clean bench, then inoculated onto the MS co-culture medium (composition: MS nutrition powder 4.42 g·L−1, IAA 1.0 mg·L−1, agar 7 g·L−1, sucrose 30 g·L−1; pH 5.8) supplemented with five different concentrations of ZT (0.1, 0.5, 1.0, 1.5 and 2.0 mg·L−1). 20 explants were cultured in one vial, each replicated three times in a completely randomized design. Whole explants were kept at 25°C, 12 h photoperiod, and the light intensity was adjusted to 3,000 lux. According to the induction of the callus, the growth of the callus was counted regularly.
Callus explants with different ZT concentrations were selected to extract RNA from tomato calli by following the method of the Vazyme Plant Total RNA Extraction Kit (RC401), and the reverse transcription kit (KR118) of the Tiangen Company was then used to synthesize cDNA using RNA as a template. Genstar Biological 2 × RealStar Green Power Mixture (A311-01) was used for the quantitative procedures, and the website Sangon.com was employed to design specific primers for qRT-PCR analysis, with the gene sequence provided in Supplementary S1. A LightCycler 96 fluorescence quantitative PCR system from Roche was utilized for the PCR, with 20 μL of SlActin serving as the reference gene. The gene-specific primers can be found in Table 1. The PCR protocol comprised denaturation at 95°C for 15 s, annealing/extension at 60°C for 15 s, repeated for 40 cycles, followed by the generation of a melting curve. The relative gene expressions were calculated using the 2−∆∆Ct method [35]. Each treatment was conducted with three biological replicates.
Statistical analysis was performed on the collected data using a one-way analysis of variance (ANOVA), followed by Tukey’s test with SPSS (IBMSPSS Statistical Version 24). Statistical significance was determined by a p-value of <0.05. The graphs were created using GraphPad Prism 8 (San Diego, CA, USA).
3.1 Discovery of the CKX Gene Family in Tomato across the Entire Genome
To identify the CKX family proteins in tomatoes, we conducted a query of the tomato genome (v4.0) (https://solgenomics.net/ftp/tomato_genome/assembly/build_4.00/). This search yielded nine SlCKX sequences: SlCKX1 (Solyc04g080820.2.1), SlCKX2 (Solyc12g008900.2.1), SlCKX3 (Solyc04g016430.3.1), SlCKX4 (Solyc10g079870.3.1), SlCKX5 (Solyc01g088160.4.1), SlCKX6 (Solyc08g061920.3.1), SlCKX7 (Solyc08g061930.4.1), SlCKX8 (Solyc10g017990.2.1), and SlCKX9 (Solyc12g008920.3.1). All these sequences contain the typical flavin adenine dinucleotide (FAD) binding and CK-binding domains, which are unique features of CKX family members. Table 2 presents basic information on SlCKXs including loci, chromosome positioning, and open reading frame (ORF) length, the nine members of the SlCKXs gene family exhibit various physicochemical properties, The analysis revealed that these proteins consist of amino acid sequences ranging from 453 to 553 residues, with corresponding relative molecular weights falling within the range of 51.661 to 62.494 kD, and isoelectric points varying from 5.77 to 8.59. Among them, SlCKX1, 4, 5, 6, 7, and 9 are acidic proteins, while SlCKX2, 3, and 8 are basic proteins. The stability coefficient of SlCKX1, 4, 5, 6, 7, and 8 is less than 40, indicating that they are all stable proteins. However, SlCKX2, 3, and 9 have coefficients greater than 40, suggesting that they are unstable proteins.
3.2 Analysis of the Protein Structure of CKX in Tomatoes
To explore the structural characteristics of SlCKX proteins, we used the online tool SOPMA for secondary structure prediction. As shown in Table 3, we determined that all SlCKX proteins β-sheets, extended chains, and random coils. The proportion of α-helices, β-sheets, extended chains, and random coils ranged from 32.26% to 36.98%, from 5.47% to 6.98%, from 17.54% to 20.87%, and from 38.11% to 42.94%, respectively. These structural elements were found to differ among the nine SlCKX proteins. Following the assessment of the secondary structure, we analyzed the tertiary structure (Fig. 1). Protein modeling indicated that the global model quality estimation (GMQE) scores for the SlCKX1-9 protein sequences were all close to 1, indicating a high degree of consistency within the protein family and greater reliability of the related sequences.
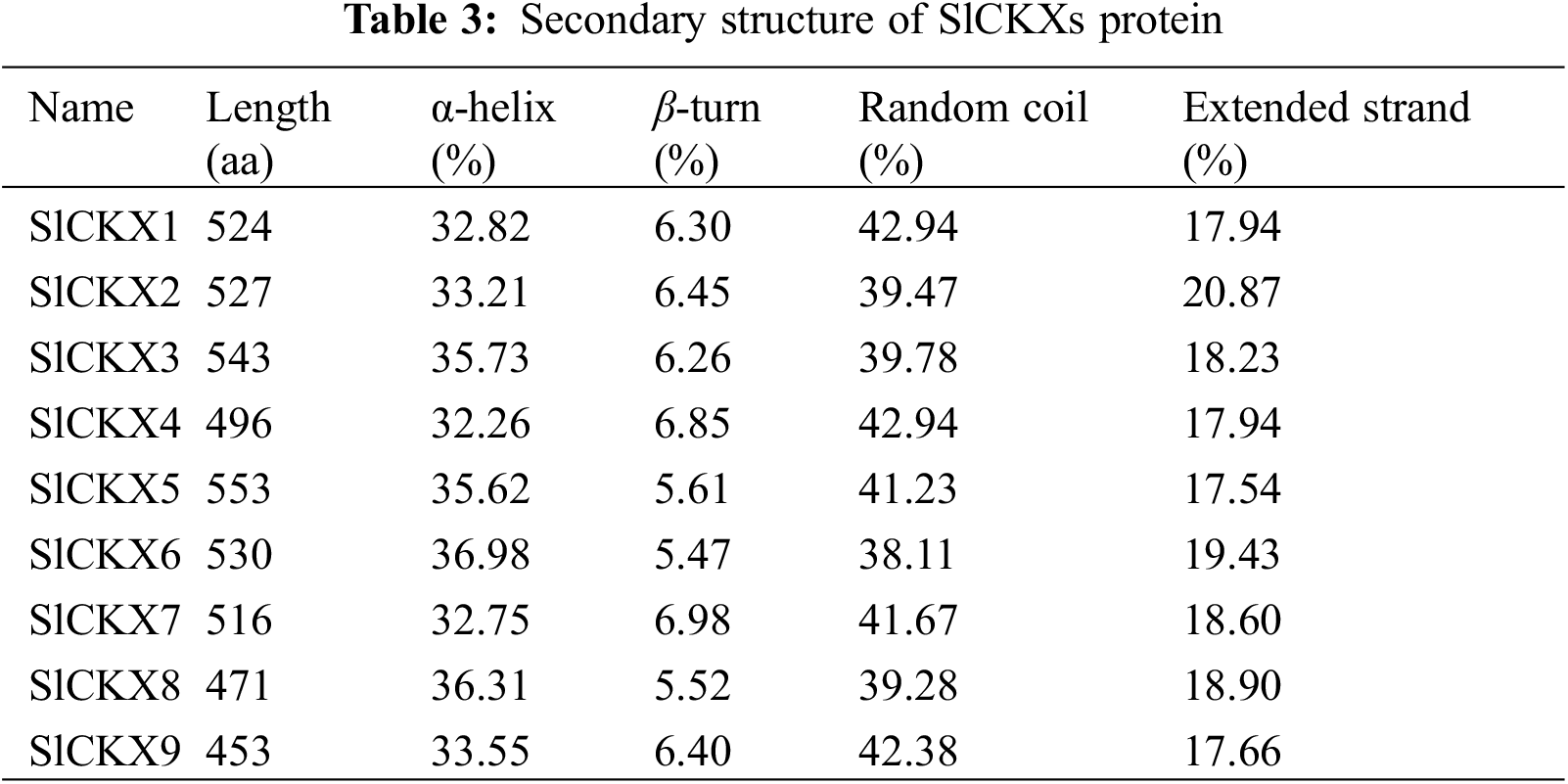
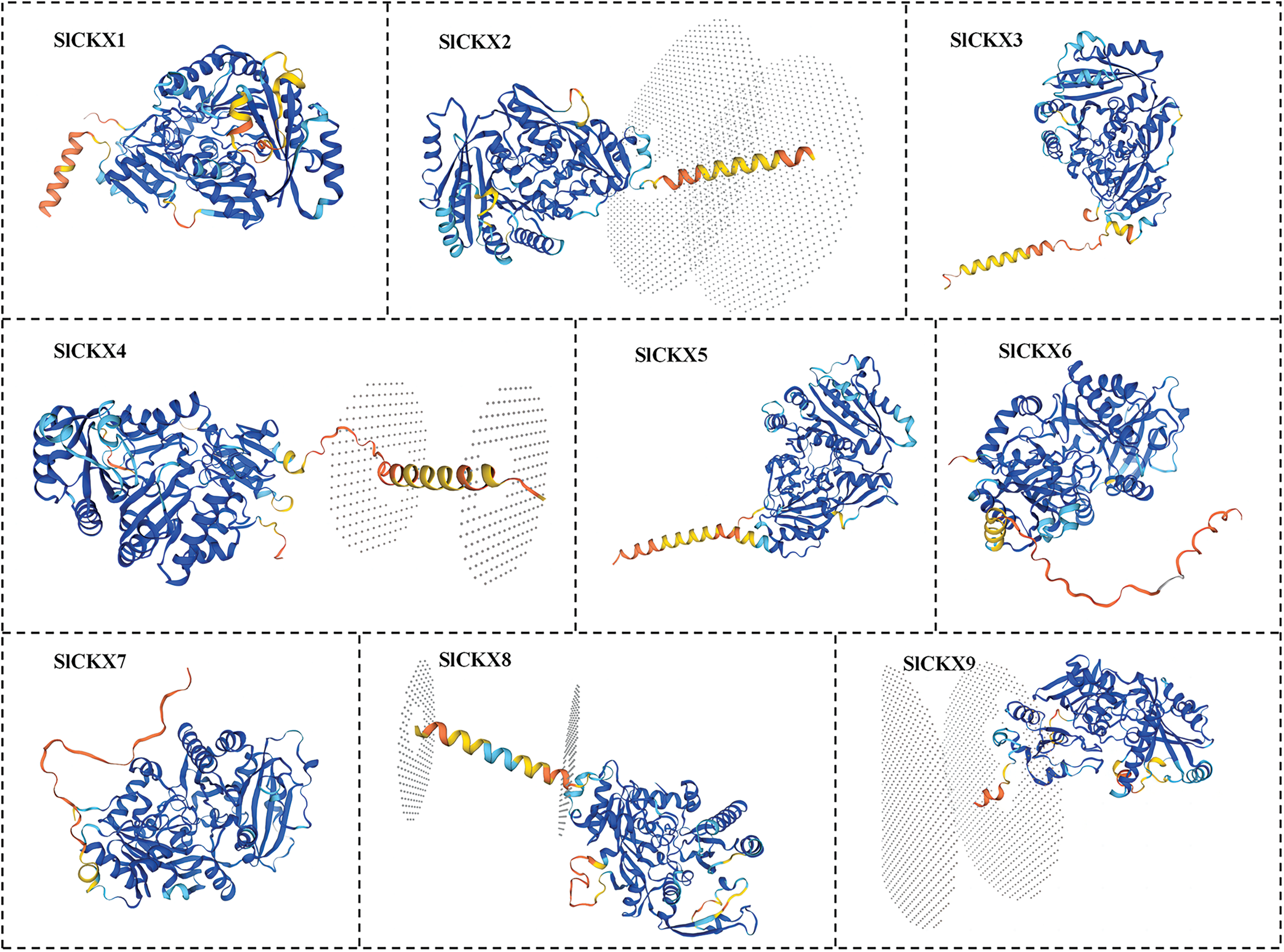
Figure 1: Tertiary structure of SlCKXs protein
3.3 Chromosome Distribution, Gene Structure, Conserved Motifs, and Conserved Domains Analysis
Based on tomato genome annotation, we identified nine members that belonged to the SlCKXs gene family and were distributed across five tomato chromosomes (Fig. 2). Eight of these genes were evenly located on four chromosomes, with SlCKX5 being the only gene located on chromosome 1. Furthermore, SlCKX1 and SlCKX3 were located on chromosome 4, SlCKX6 and SlCKX7 on chromosome 8, and SlCKX4 and SlCKX8 on chromosome 10. Additionally, SlCKX2 and SlCKX9 were found on chromosome 12. Notably, the proximity of SlCKX6 and SlCKX7 on chromosome 8, as well as SlCKX2 and SlCKX9 on chromosome 12, suggests that these genes may be orthologs resulting from segmental duplication events.
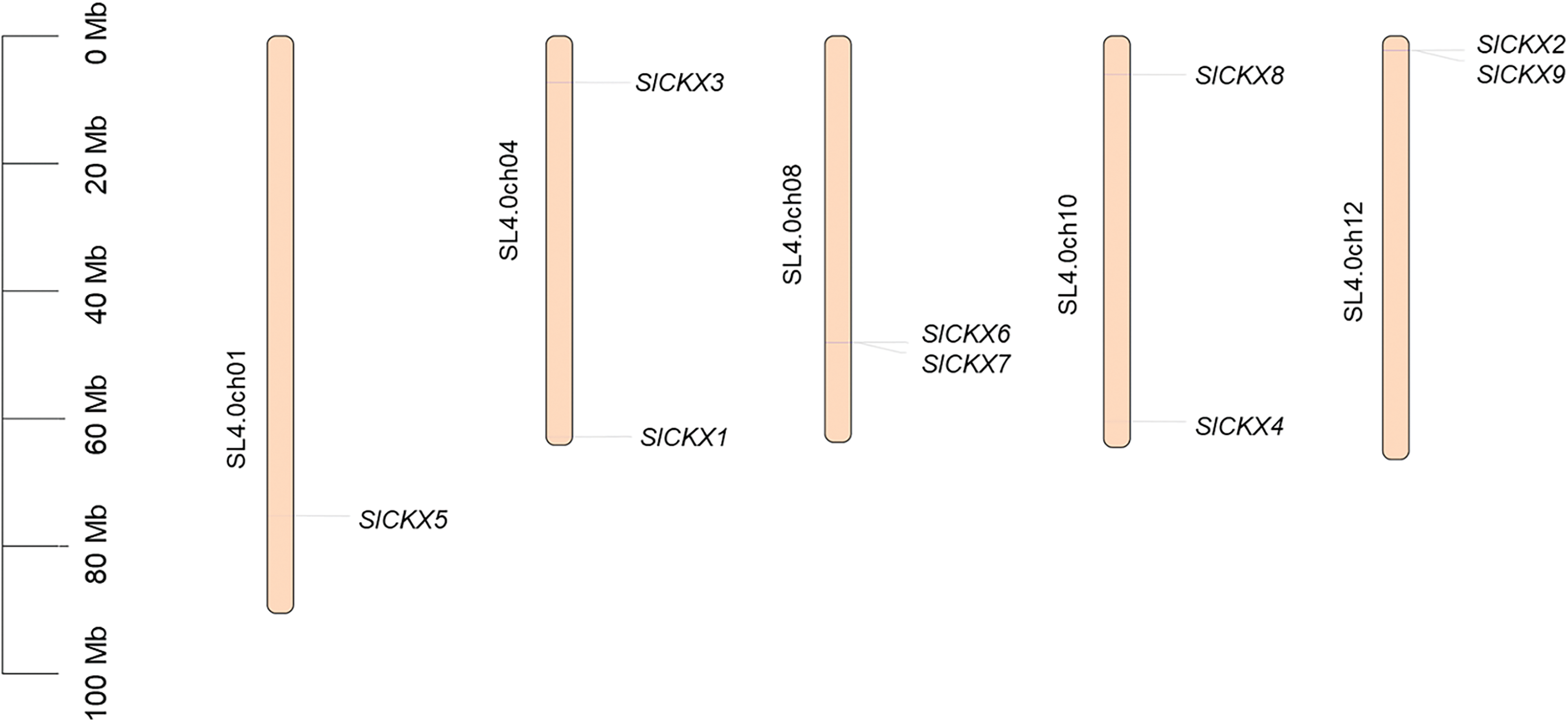
Figure 2: Localization of CKX in chromosomes of tomato
We identified 15 conserved motifs of SlCKX proteins by using MEME software. As depicted in Fig. 3, we observed some similarity in the number and arrangement of conserved motifs among the nine protein sequences of SlCKX. Each protein within the same subfamily exhibited a unique motif pattern, where SlCKX8 lacked motif9, SlCKX9 lacked motif4 and motif7, and SlCKX4 lacked motif2. All nine members of the SlCKX family possessed six highly conserved motifs, including motif1, motif3, motif5, motif6, motif8, and motif10. Furthermore, motif10 was located closer to the 5′-end, whereas motif5 and motif7 were located closer to the 3′-end, suggesting the high conservation of these six motifs during evolution.
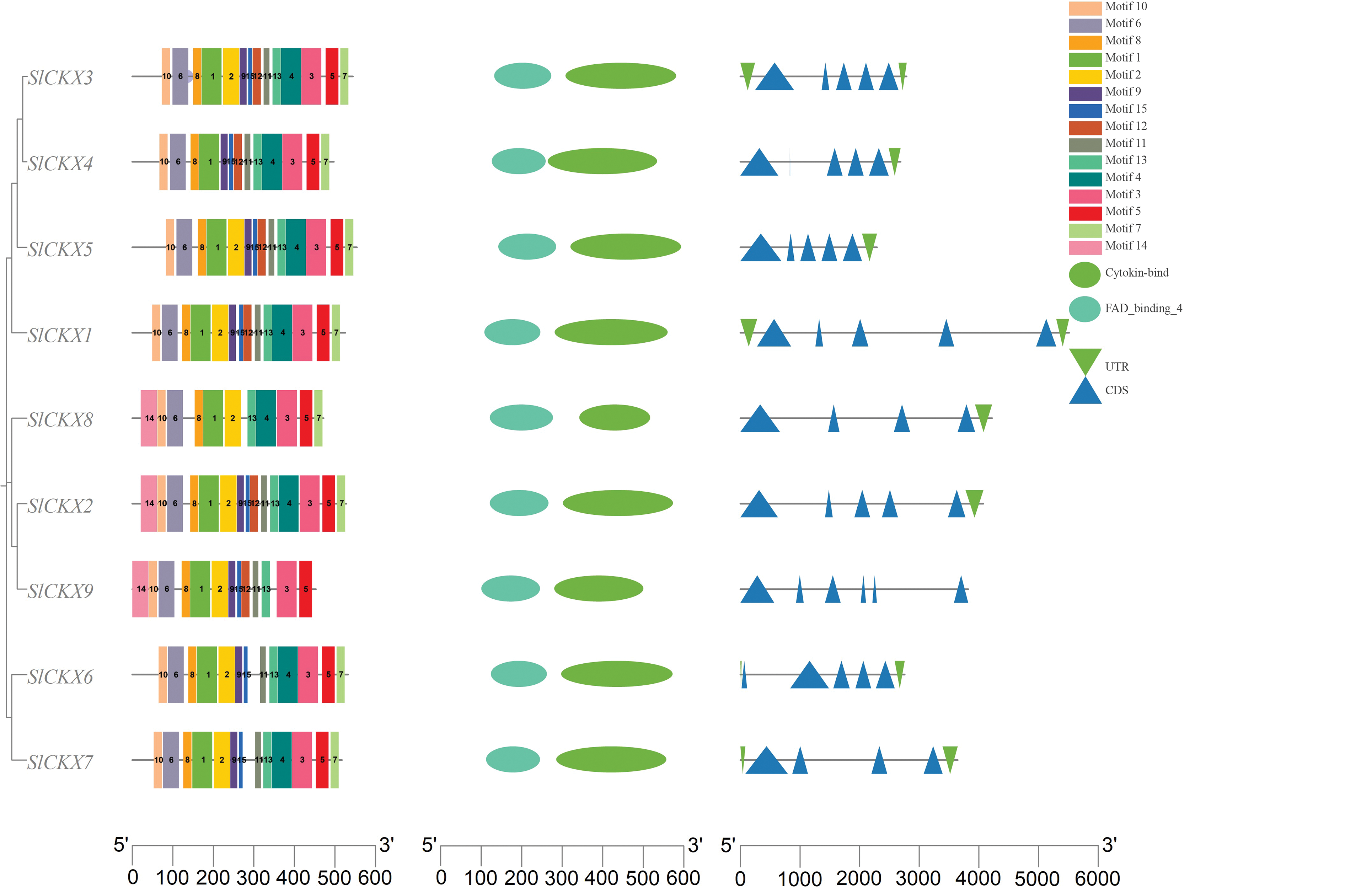
Figure 3: Conserved motif, conserved domain, and gene structure of SlCKXs gene family
All the nine members of the SlCKXs family possess conserved domains for FAD-binding and CK-binding 2, respectively. The FAD-binding domains are located at the 5′-end, whereas the CK-binding domains are situated at the 3′-end. Members within the SlCKXs family exhibit variations in untranslated regions and coding sequences (CDS), with a number of these CDS ranging between 4 and 6. This diversity may arise from evolutionary changes in CKX genes.
3.4 Phylogeny and Synteny Analyses of the CKX Gene Family
To better understand the phylogenetic relationships of CKX genes in tomato, Arabidopsis, eggplant, and nonbulb cabbage, we constructed a phylogenetic tree. This tree was based on conserved domains of the CKX genes, utilizing the neighbor-joining (NJ) method and multiple sequence alignments. Analysis of the results revealed that the 34 CKX genes can be classified into four distinct subgroups: I, II, III, and IV (Fig. 4). Among these groups, groups I and II were the largest, each comprising 12 members. Subgroup I consisted of three tomatoes, two Arabidopsis, four nonbulbous cabbage, and three eggplant species, whereas subgroup II consisted of three tomatoes, three Arabidopsis, five nonbulbous cabbage, and one eggplant species. The phylogeny diagram indicated a high homology between tomato and eggplant likely due to their shared membership in the Solanaceae family. Overall, members in the same subfamily are likely to share similar functions, allowing for further exploration into the potential biological roles of the SlCKX family.
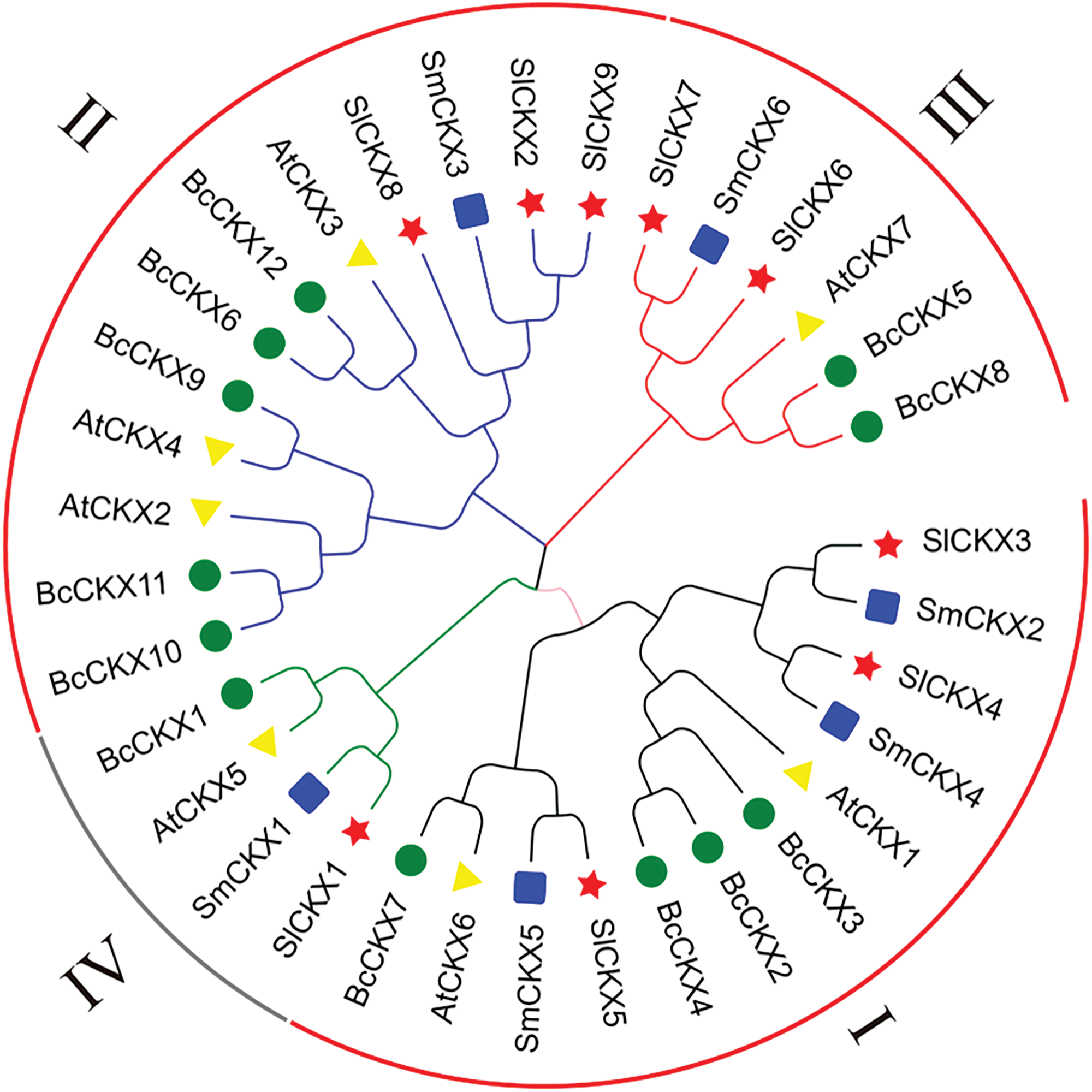
Figure 4: Rootless phylogenetic trees from the CKX family of Arabidopsis thaliana (triangle), Solanum melongena (square), Tomato (pentagram), and Brassica (round) with different colored lines in the outer ring indicating different subgroup
To delve deeper into the evolutionary relationships of the tomato CKX family, a comparative collinear analysis was conducted between the CKX family members of Arabidopsis and tomato. This analysis identified strong collinear relationships between the nine SlCKX family genes and other species and identified the presence of a strong collinear region between tomato and Arabidopsis (Fig. 5). Moreover, The collinearity analysis of AtCKXs and SlCKX revealed that three tomato chromosomes were collinear with five Arabidopsis chromosomes. Moreover, SL4.0ch12 corresponded to three Arabidopsis chromosomes, demonstrating high homology and providing valuable insights into the genetic relationships and gene functions of these species.
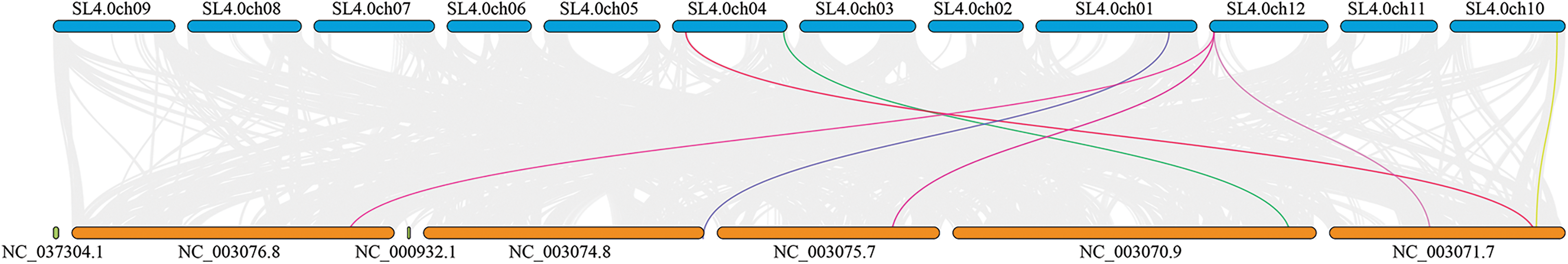
Figure 5: Between-species collinear relationship between tomato and Arabidopsis. Tomato and Arabidopsis chromosomes are marked with different colors, the tomato chromosome in blue and the Arabidopsis chromosome in orange. Chromosome numbers are marked above the chromosome. Different colored lines connect collinear relationships between members of the CKX gene family of different species
3.5 Analysis of Promoter Cis-Acting Elements of the SlCKX Gene Family
We analyzed cis-acting elements located in the promoter region 2000 bp upstream of the SlCKX gene. We found that the SlCKX gene family primarily comprises four categories of cis-acting elements (Fig. 6A): light-response elements, hormone-response elements, stress-response elements, and growth-response elements. The details are provided in the Supplementary S2. The number of hormone-response elements in the SlCKX gene family was five (Fig. 6B), distributed across all eight members except for SlCKX6. The most widely distributed element was the ABA-response element, with the highest number found in SlCKX1 and SlCKX9. Auxin-response elements were found in SlCKX1, 2, and 8; methyl jasmonate-response elements were detected in SlCKX1, 3, 8 and 9; ABA-response elements in SlCKX1, 2, 3, 4, 5, 7, 8, and 9; salicylic acid-response elements in SlCKX2, 5, 7, and 9; and gibberellin-response elements in SlCKX5, 8, and 9, these cis-elements indicated the functional diversity of SlCKX genes, particularly in hormone response, suggesting the involvement of SlCKX in plant growth regulation.
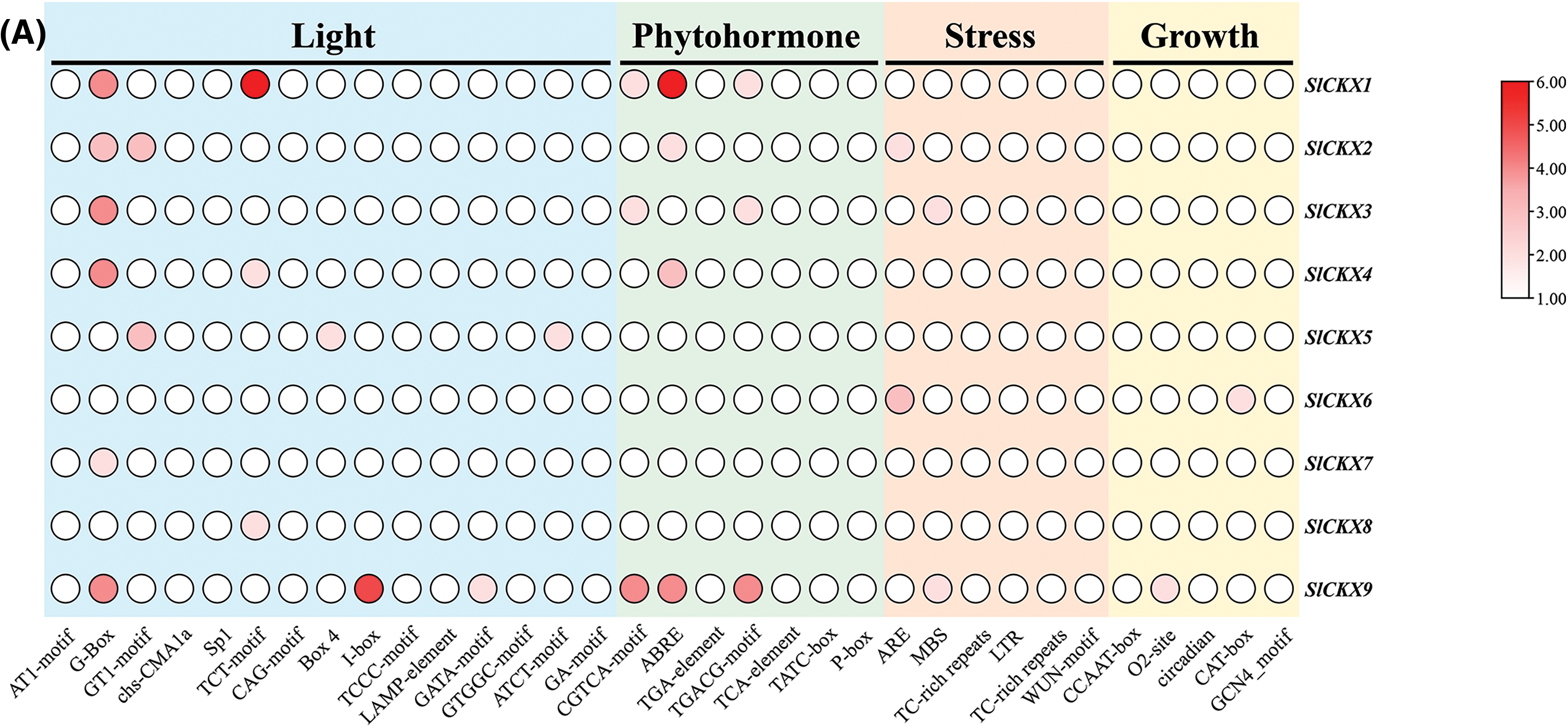
Figure 6: Distribution of cis-acting elements of the SlCKX gene family (Fig. 6A) and hor-mone response elements (Fig. 6B)
3.6 Validation of Expression of Tomato CKX Gene Family in Callus
To further examine the expression patterns of the SlCKX gene family in tomato (Fig. 7), We isolated RNA from the calli exposed to varying concentrations of trans-ZT and subsequently reverse-transcribed into cDNA for quantitative real-time PCR. The findings revealed that the transcriptional activity of SlCKX5 and SlCKX9 genes initially increased and then decreased, whereas the transcriptional activity of SlCKX2, 3, 4, 6, 7 and 8 genes initially increased and then decreased at a concentration of 2.0 mg·L−1. The most pronounced difference was observed in the medium supplemented with 1.0 mg·L−1 IAA and 1.0 mg·L−1 ZT, suggesting that a hormonal concentration of 1.0 mg·L−1 is optimal for the growth of tomato calli.
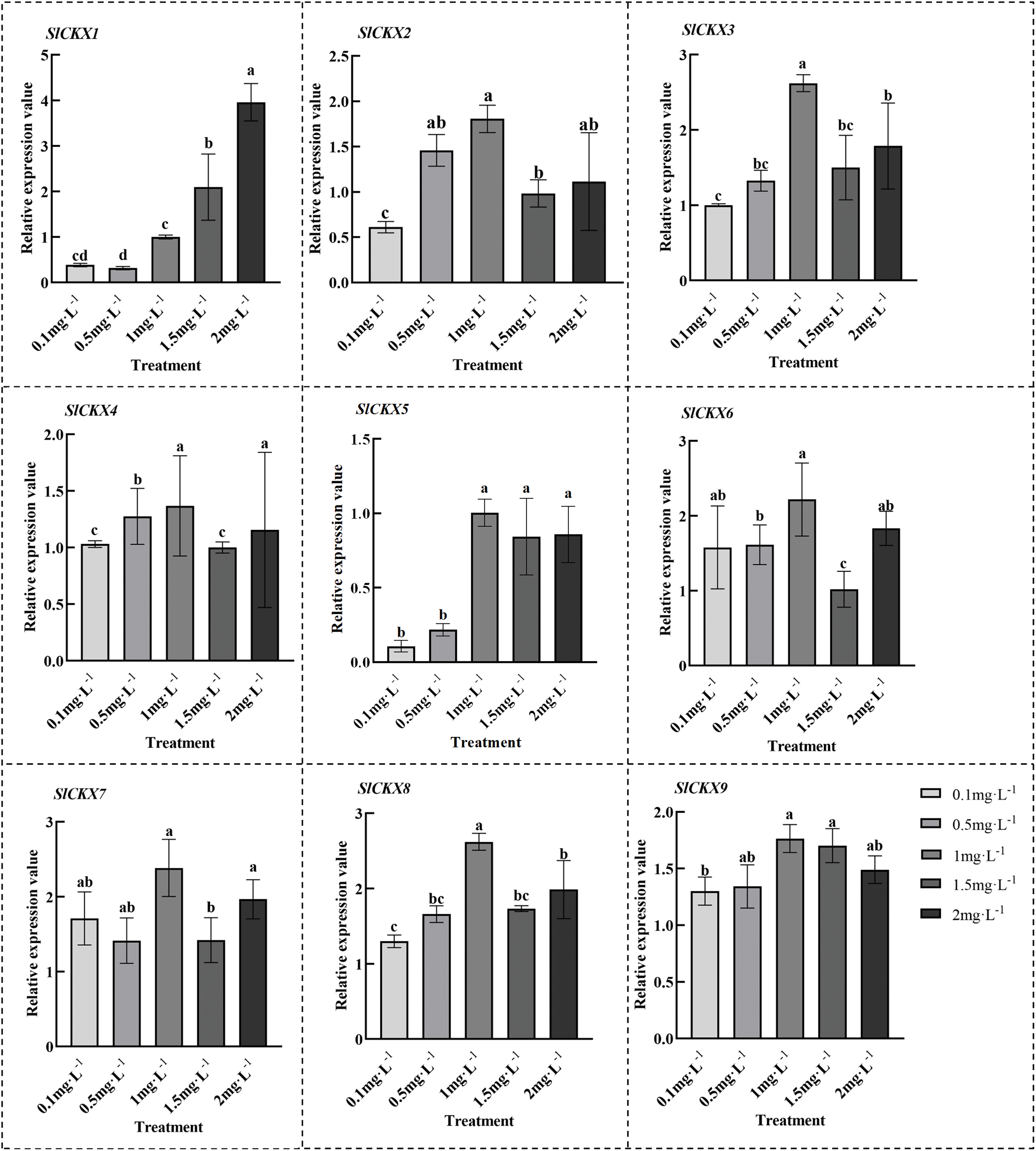
Figure 7: Levels of gene expression for the SlCKX family members in callus were examined under varying zeatin concentrations. The results are displayed as the average ±SD, with a sample size of n = 3. Distinct lowercase letters above the bars indicate statistically significant differences (p < 0.05)
Endogenous hormones play a crucial role in regulating plant growth and development [36], affecting various processes such as seed germination [37], cell differentiation [38], root elongation [39], and fruit ripening and abscission [40]. In plant tissue culture, exogenous hormone application is a standard technique to promote explant regeneration. These exogenous hormones can affect the synthesis and distribution of endogenous hormones, thereby influencing plant growth and development. CK dehydrogenase, encoded by the CKX gene, plays a crucial role in controlling CK homeostasis by degrading CKs. The degradation process is crucial for controlling plant growth, development, stress tolerance, and productivity [41]. They are ensuring an appropriate conducive to maintaining cell division capacity in plant cells.
The SlCKX gene family in tomatoes was found to be larger than in Arabidopsis but notably smaller than in cabbage. This finding indicates that the events of gene expansion and reduction have occurred in the CKX family during plant evolution. The finding is consistent with those reported by Guo et al. [42]. The lengths of the CKX proteins in tomatoes vary from 453 to 553 amino acids, corresponding to molecular weights ranging from 51.66 to 52.49 kDa. The theoretical isoelectric points of these proteins range from 5.77 to 8.59, suggesting their hydrophilic nature. Furthermore, each of these comprises a corresponding CKX gene in eggplant, indicating their common ancestry within the Solanaceae family. Moreover, we observed that motifs1, 3, 5, 6, 8, and 10 are conserved among all the nine SlCKX proteins. This finding indicates that these motifs are essential for the biological functions of CKX proteins in tomatoes.
All nine CKX genes in tomato exhibit conserved FAD-binding and cytokinin-binding 2 domains situated at the 5′ and 3′ termini, respectively. This organization aligns with findings in wheat as reported by Feng et al. [43]. The functional diversity of SlCKX genes is highlighted by the presence of different cis-elements, especially in hormone-response pathways, suggesting their involvement in growth regulation in plants. Further investigation of the expression of tomato genes revealed the effect of CK on callus growth and development. The expression of SlCKX1 increased with trans-ZT concentration. However, the transcriptional activity of SlCKX5 and SlCKX9 genes increased initially and then decreased, whereas the transcriptional activity of SlCKX2, 3, 4, 6, 7, 8 genes initially increased and then decreased at a concentration of 2.0 mg·L−1. The most pronounced difference was observed in the medium supplemented with 1.0 mg·L−1 IAA and 1.0 mg·L−1 ZT, implying that a hormonal concentration of 1.0 mg·L−1 is optimal for the growth of tomato calli.
In this study, we identified nine SlCKX genes in the tomato genome. We constructed a phylogenetic tree of SlCKX proteins from tomato and other species and examined their evolutionary relationships. We investigated the gene structure and conserved motifs of all nine SlCKX proteins, as well as the stress response of functional cis-elements in the promoter region. Furthermore, we analyzed the expression of the SlCKXs gene under different hormonal concentrations with the most significant expression observed in the medium supplemented with 1.0 mg·L−1 IAA and 1.0 mg·L−1 ZT, indicating that 1.0 mg·L−1 is the optimal hormone concentration for tomato callus growth.
Acknowledgement: Not applicable.
Funding Statement: This work was funded by the Special Project for Science and Technology Innovation Platform of Fujian Academy of Agricultural Sciences, China (CXPT2023003), the Freely Explore Scientific and Technology Innovation Program of Fujian Academy of Agricultural Sciences (ZYTS202207), and the Program for Innovative Research Team of Fujian Academy of Agricultural Sciences, China (CXTD2021006-3).
Author Contributions: Z. L. was responsible for designing the experiments, analyzing the results, performing the experiments, and wrote the manuscript. D. L., Y. J. and S. W. performed the experiments. S. Z. and Y. Y. performed the experiments and took part in data analysis. B. L. collected the plant samples. J. H. conceptualized and designed the research. Z. L. interpreted the results and revised the paper.
Availability of Data and Materials: All the data supporting the findings of this study are included in this article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declared that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.051207.
References
1. Mishra DC, Arora D, Budhlakoti N, Solanke AU, Mithra S, Kumar A, et al. Identification of potential cytokinin responsive key genes in rice treated with trans-zeatin through systems biology approach. Front Genet. 2021;12:780599. [Google Scholar] [PubMed]
2. Grzegorczyk-Karolak I, Hnatuszko-Konka K, Krzeminska M, Olszewska MA, Owczarek A. Cytokinin-based tissue cultures for stable medicinal plant production: regeneration and phytochemical profiling of Salvia bulleyana shoots. Biomolecules. 2021;11(10):1513. doi:10.3390/biom11101513. [Google Scholar] [PubMed] [CrossRef]
3. Uniyal S, Bhandari M, Singh P, Singh RK, Tiwari SP. Cytokinin biosynthesis in cyanobacteria: insights for crop improvement. Front Genet. 2022;13:933226. doi:10.3389/fgene.2022.933226. [Google Scholar] [PubMed] [CrossRef]
4. Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyl transferases. Plant Cell Physiol. 2001;42:677–85. doi:10.1093/pcp/pce112. [Google Scholar] [PubMed] [CrossRef]
5. Mok DWS, Mok Machteld C. Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:89–118. doi:10.1146/arplant.2001.52.issue-1. [Google Scholar] [CrossRef]
6. Hluska T, Sebela M, Lenobel R, Frebort I, Galuszka P. Purification of maize nucleotide pyrophosphatase/phosphodiesterase casts doubt on the existence of Zeatin cis-trans isomerase in plants. Front Plant Sci. 2017;8:1473. doi:10.3389/fpls.2017.01473. [Google Scholar] [PubMed] [CrossRef]
7. Gouda G, Gupta MK, Donde R, Kumar J, Vadde R, Mohapatra T, et al. Computational approach towards understanding structural and functional role of cytokinin oxidase/dehydrogenase 2 (CKX2) in enhancing grain yield in rice plant. J Biomol Struct Dyn. 2020;38(4):1158–67. doi:10.1080/07391102.2019.1597771. [Google Scholar] [PubMed] [CrossRef]
8. Gu R, Fu J, Guo S, Duan F, Wang Z, Mi G, et al. Comparative expression and phylogenetic analysis of maize cytokinin dehydrogenase/oxidase (CKX) gene family. J Plant Growth Regul. 2010;29(4):428–40. doi:10.1007/s00344-010-9155-y. [Google Scholar] [CrossRef]
9. Galuszka P, Popelková H, Werner T, Frébortová J, Pospíšilová H, Mik V, et al. Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J Plant Growth Regul. 2007;26(3):255–67. doi:10.1007/s00344-007-9008-5. [Google Scholar] [CrossRef]
10. Wang Y, Liu H, Xin Q. Genome-wide analysis and identification of cytokinin oxidase/dehydrogenase (CKX) gene family in foxtail millet (Setaria italica). Crop J. 2014;2(4):244–54. doi:10.1016/j.cj.2014.05.001. [Google Scholar] [CrossRef]
11. Zalabak D, Galuszka P, Mrizova K, Podlesakova K, Gu R, Frebortova J. Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol Biochem. 2014;74:283–93. doi:10.1016/j.plaphy.2013.11.020. [Google Scholar] [PubMed] [CrossRef]
12. Le DT, Nishiyama R, Watanabe Y, Vankova R, Tanaka M, Seki M, et al. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS One. 2012;7(8):e42411. doi:10.1371/journal.pone.0042411. [Google Scholar] [PubMed] [CrossRef]
13. Liu P, Zhang C, Ma JQ, Zhang LY, Yang B, Tang XY, et al. Genome-wide identification and expression profiling of cytokinin oxidase/dehydrogenase (CKX) genes reveal likely roles in pod development and stress responses in oilseed rape (Brassica napus L.). Genes. 2018;9(3):168. doi:10.3390/genes9030168. [Google Scholar] [PubMed] [CrossRef]
14. Jain P, Singh A, Iquebal MA, Jaiswal S, Kumar S, Kumar D, et al. Genome-wide analysis and evolutionary perspective of the cytokinin dehydrogenase gene family in wheat (Triticum aestivum L.). Front Genet. 2022;13:931659. doi:10.3389/fgene.2022.931659. [Google Scholar] [PubMed] [CrossRef]
15. Werner T, Motyka V, Laucou V, Smets R, van Onckelen H, Schmulling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15(11):2532–50. doi:10.1105/tpc.014928. [Google Scholar] [PubMed] [CrossRef]
16. Smehilova M, Galuszka P, Bilyeu KD, Jaworek P, Kowalska M, Sebela M, et al. Subcellular localization and biochemical comparison of cytosolic and secreted cytokinin dehydrogenase enzymes from maize. J Exp Bot. 2009;60(9):2701–12. doi:10.1093/jxb/erp126. [Google Scholar] [PubMed] [CrossRef]
17. Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. New insights into the biology of cytokinin degradation. Plant Biol. 2006;8(3):371–81. doi:10.1055/s-2006-923928. [Google Scholar] [PubMed] [CrossRef]
18. Massonneau A, Houba-Herin N, Pethe C, Madzak C, Falque M, Mercy M, et al. Maize cytokinin oxidase genes: differential expression and cloning of two new cDNAs. J Exp Bot. 2004;55(408):2549–57. doi:10.1093/jxb/erh274. [Google Scholar] [PubMed] [CrossRef]
19. Panda BB, Sekhar S, Dash SK, Behera L, Shaw BP. Biochemical and molecular characterisation of exogenous cytokinin application on grain filling in rice. BMC Plant Biol. 2018;18(1):89. doi:10.1186/s12870-018-1279-4. [Google Scholar] [PubMed] [CrossRef]
20. Zhang W, Xia L, Peng F, Song C, Manzoor MA, Cai Y, et al. Transcriptomics and metabolomics changes triggered by exogenous 6-benzylaminopurine in relieving epicotyl dormancy of Polygonatum cyrtonema Hua seeds. Front Plant Sci. 2022;13:961899. doi:10.3389/fpls.2022.961899. [Google Scholar] [PubMed] [CrossRef]
21. Ma YY, Zheng L, Xie R, He SL, Deng L. Genome-wide identification and analysis of CKX genes in Poncirus trifoliata. J Hortic Sci Biotech. 2016;91(6):592–602. doi:10.1080/14620316.2016.1194171. [Google Scholar] [CrossRef]
22. Li S, An Y, Hailati S, Zhang J, Cao Y, Liu Y, et al. Overexpression of the cytokinin oxidase/dehydrogenase (CKX) from Medicago sativa enhanced salt stress tolerance of Arabidopsis. J Plant Biol. 2019;62(5):374–86. doi:10.1007/s12374-019-0141-z. [Google Scholar] [CrossRef]
23. Liu Y, Wang X, Wang X, Gao W, You C. Identification and functional characterization of apple MdCKX5.2 in root development and abiotic stress tolerance. Horticulturae. 2022;8(1):62. doi:10.3390/horticulturae8010062. [Google Scholar] [CrossRef]
24. Qi S, Zhang S, Islam MM, El-Sappah AH, Zhang F, Liang Y. Natural resources resistance to tomato spotted wilt virus (TSWV) in tomato (Solanum lycopersicum). Int J Mol Sci. 2021;22:10978. doi:10.3390/ijms222010978. [Google Scholar] [PubMed] [CrossRef]
25. Gerszberg A, Hnatuszko-Konka K, Kowalczyk T, Kononowicz AK. Tomato (Solanum lycopersicum L.) in the service of biotechnology. Plant Cell Tiss Org (PCTOC). 2014;120(3):881–902. [Google Scholar]
26. Gatahi DM. Challenges and opportunities in tomato production chain and sustainable standards. Int J Hortic Sci Technol. 2020;7(3):235–62. [Google Scholar]
27. Hnatuszko-Konka K, Gerszberg A, Weremczuk-Jezyna I, Grzegorczyk-Karolak I. Cytokinin signaling and de novo shoot organogenesis. Genes. 2021;12(2):265. doi:10.3390/genes12020265. [Google Scholar] [PubMed] [CrossRef]
28. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi:10.1093/bioinformatics/btm404. [Google Scholar] [PubMed] [CrossRef]
29. Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Heredity. 2002;93:77–8. doi:10.1093/jhered/93.1.77. [Google Scholar] [PubMed] [CrossRef]
30. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. doi:10.1093/molbev/msw054. [Google Scholar] [PubMed] [CrossRef]
31. Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–9. doi:10.1093/nar/gkz239. [Google Scholar] [PubMed] [CrossRef]
32. Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi:10.1093/nar/gkr1293. [Google Scholar] [PubMed] [CrossRef]
33. Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinf. 2010;8(1):77–80. doi:10.1016/S1672-0229(10)60008-3. [Google Scholar] [PubMed] [CrossRef]
34. Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Peer YVD, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7. doi:10.1093/nar/30.1.325. [Google Scholar] [PubMed] [CrossRef]
35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods. 2001;25(4):402–8. doi:10.1006/meth.2001.1262. [Google Scholar] [PubMed] [CrossRef]
36. Shimotohno A, Aki SS, Takahashi N, Umeda M. Regulation of the plant cell cycle in response to hormones and the environment. Annu Rev Plant Biol. 2021;72:273–96. doi:10.1146/arplant.2021.72.issue-1. [Google Scholar] [CrossRef]
37. Ozden E, Light ME, Demir I. Alternating temperatures increase germination and emergence in relation to endogenous hormones and enzyme activities in aubergine seeds. S Afr J Bot. 2021;139:130–9. doi:10.1016/j.sajb.2021.02.015. [Google Scholar] [CrossRef]
38. Long TA, Benfey PN. Transcription factors and hormones: new insights into plant cell differentiation. Curr Opin Cell Biol. 2006;18(6):710–4. doi:10.1016/j.ceb.2006.09.004. [Google Scholar] [PubMed] [CrossRef]
39. Chen H, Lei Y, Sun J, Ma M, Deng P, Quan JE, et al. Effects of different growth hormones on rooting and endogenous hormone content of two Morus alba L. Cuttings Hortic. 2023;9(5):552. doi:10.3390/horticulturae9050552. [Google Scholar] [CrossRef]
40. Arnao M, Hernández-Ruiz J. Melatonin in flowering, fruit set and fruit ripening. Plant Reprod. 2020;33(2):77–87. doi:10.1007/s00497-020-00388-8. [Google Scholar] [PubMed] [CrossRef]
41. Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, et al. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell. 2015;27(12):3354–66. doi:10.1105/tpc.15.00452. [Google Scholar] [PubMed] [CrossRef]
42. Guo B, Yang H, Dai L, Zhao X, Wang LF. Genome-wide identification and response stress expression analysis of the BES1 family in rubber tree (Hevea brasiliensis Muell. Arg.). PeerJ. 2022;10:e13189. doi:10.7717/peerj.13189. [Google Scholar] [PubMed] [CrossRef]
43. Feng X, Yu Q, Zeng J, He X, Liu W. Genome-wide identification and characterization of GATA family genes in wheat. BMC Plant Biol. 2022;22(1):372. doi:10.1186/s12870-022-03733-3. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools