 Open Access
Open Access
ARTICLE
4-Hydroxy-2-Oxoglutaric Acid, A Key Metabolite Involved in Trypsin-Regulation of Arginine Metabolism in Hylocereus undatus during Storage
1 College of Food and Bioengineering, Henan University of Science and Technology, Luoyang, 471023, China
2 Henan Engineering Research Center of Food Microbiology, Luoyang, 471023, China
3 National Demonstration Center for Experimental Food Processing and Safety Education, Luoyang, 471000, China
* Corresponding Author: Xin Li. Email:
(This article belongs to the Special Issue: Multi-omics Approach to Understand Plant Stress Tolerance)
Phyton-International Journal of Experimental Botany 2024, 93(5), 885-900. https://doi.org/10.32604/phyton.2024.050450
Received 06 February 2024; Accepted 28 March 2024; Issue published 28 May 2024
Abstract
Trypsin, a novel superoxide scavenger, significantly enhances the storage quality of Hylocereus undatus (H. undatus). To elucidate the preservation mechanism of trypsin on H. undatus, a widely targeted metabolomic analysis, and transcriptomics analysis were conducted. Firstly, a total of 453 metabolites were identified, with organic acids and their derivatives constituting the largest proportion (25%). Amino acids and their metabolites, prominent among organic acids, were further analyzed. Among them, 73 metabolites were associated with amino acids, and 37 exhibited significant differences. The most enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was arginine biosynthesis (map00220), with polyamine metabolites showing the most pronounced differences, particularly spermine (FC = 1.7594). Compared with the control group, 4-hydroxy-2-oxoglutaric acid was significantly upregulated (FC = 2.117) in the process of spermine biosynthesis. Furthermore, the results of Gene Ontology (GO) and KEGG enrichment analysis of the H. undatus transcriptome profile revealed that trypsin treatment led to 187 differentially expressed genes associated with arginine. Both GO and KEGG analyses exhibited significant enrichment in the spermine biosynthetic process (GO:0006597) (map:00220) within the arginine biosynthesis pathway. Moreover, most enzymes and metabolites within the spermine biosynthesis pathway in H. undatus were upregulated. The results of the PPI network highlighted that ADC, SPDS, and SAMDC, among others, were pivotal proteins involved in trypsin-regulated arginine metabolism and spermine synthesis. This study revealed that trypsin could significantly delay postharvest senescence of H. undatus at room temperature. This effect might be attributed to trypsin triggering the synthesis of 4-hydroxy-2-oxoglutaric acid in the fruit peel, thereby promoting the biosynthesis of spermine and other polyamines.Keywords
Supplementary Material
Supplementary Material FileHylocereus undatus (H. undatus), belonging to the Cactaceae family, contained plant-based albumin and anthocyanins, along with abundant vitamins and water-soluble dietary fiber—components rarely found in most plants. It boasted effects such as preventing arteriosclerosis, detoxifying and protecting the stomach, as well as skin whitening and weight loss [1]. These qualities have earned it significant popularity among consumers. However, after being harvested, H. undatus often succumbed to a range of diseases, leading to fruit damage and a loss of commercial value [2]. Therefore, research aimed at enhancing postharvest fruit quality serves as a reference for future studies on the storage of fruits and vegetables.
Most researchers considered reactive oxygen species (ROS) to be the primary cause of inducing fruit decay and deterioration [3]. Under normal circumstances, the production and elimination of ROS within plant cells remained balanced, preventing damage to the fruit. However, as the storage period lengthens, the content of oxygen free radicals increases, disrupting the equilibrium of ROS within cells. This led to lipid peroxidation of membranes, damage to biomolecules such as proteins and nucleic acids, and affected the normal physiological metabolism of the fruit. Ultimately, this resulted in the decay and deterioration of the fruit [4,5]. Plants had complex antioxidant mechanisms to eliminate excessive ROS, and the strength of their antioxidant capacity primarily depended on the content of various small molecules and the activity of antioxidant enzymes [6]. Research indicated that polyamines (PAs), such as putrescine, spermidine, and spermine, can function as antioxidants within plants under certain stressful conditions [7].
The antioxidant effects of PAs were attributed to their inherent polycationic nature, which enabled them to participate in regulating cellular ion balance and interacting with polyanionic molecules. This interaction prevented the degradation of large molecules and protected cells from oxidative damage. They collectively mitigated plant cell aging caused by superoxide radicals. Additionally, Liu et al. [8] demonstrated that PAs enhance plant drought tolerance by modulating K+ channels to regulate stomatal opening; Cuevas et al. [9] used Arabidopsis mutants to show that endogenous PAs levels can enhance a plant cell’s response to low-temperature conditions; Roy et al. [10] found that rice transformed with the oat arginine decarboxylase (ADC) gene led to PAs accumulation in line with increased ADC activity, subsequently enhancing plant salt tolerance. However, as of now, the influence of PAs compounds present in H. undatus peel on its quality during storage remains to be elucidated.
Amino acids or certain PAs synthesized from amino acid precursors could enhance a plant’s adaptability to stress conditions. Amino acids are important physiological substances within plants, playing a vital regulatory role in growth processes. When faced with stress and adversity, plants control the absorption, synthesis, and degradation of amino acids to mitigate the harm caused by these conditions. In recent years, research on how amino acids enhance plant stress resistance has garnered widespread attention. Proline and amino butyric acid, in particular, have been extensively studied, and some progress has been made in understanding their mechanisms [11]. While arginine was typically considered a crucial nitrogen storage nutrient for reuse, recent studies have indicated that arginine’s enhancement of the abiotic stress tolerance of plants may be achieved through the production of PAs and nitric oxide (NO) through metabolism [7]. Furthermore, nitrogen nutrition could be supplied by other amino acids, but these other amino acids couldn’t generate PAs and NO [11]. Nitric oxide synthase (NOS), ADC, and arginase were key enzymes in L-arginine catabolism. The relative strengths of the activities of these three enzymes could determine the metabolic direction of arginine. Arginine is transformed through metabolism to form important signaling molecules, potentially holding more significance for plant growth, development, and tolerance formation compared to being just a regular nitrogen nutrient [12,13]. From this perspective, it became evident that both arginine and its metabolic products play a crucial physiological role throughout the entire lifecycle of plants.
In recent years, preservation methods could be categorized into two main groups: physical [14,15] and chemical methods [16]. However, as awareness of food safety increased, postharvest storage and environmentally friendly preservation of fruits and vegetables became especially important, leading to the gradual development of biological preservation techniques. Our laboratory discovered that trypsin has a positive effect on superoxide scavenging [17,18]. Trypsin, an enzyme belonging to the serine protease family, has been reported to effectively scavenge superoxide anions and provide excellent protection to membrane lipids, thereby preventing cell damage [19–21]. Additionally, trypsin could enhance the biocontrol function of pseudohyphal yeast, inhibit mold growth on the surface of fruits and vegetables, and extend their shelf life. However, whether amino acid metabolism is involved in the process of trypsin-mediated preservation of fruits and vegetables has not been addressed in any reports.
This study integrated metabolic and transcriptomic data from H. undatus peel samples treated with trypsin and control groups, across different storage times. Within the metabolic dataset, amino acid-related differentially expressed metabolites were screened. Variable Importance in Projection (VIP) value analysis using various algorithms was employed to showcase the expression patterns of metabolites across different samples, as well as the VIP values in multivariate statistical analysis and the p-values in univariate statistics. This analysis was conducted to uncover significant metabolic pathways and their crucial differential metabolites, and functional analysis of these metabolites was performed. In combination with transcriptomic data, core regulatory genes were identified, and interaction networks among proteins were studied. At the RNA level, the regulatory mechanism of trypsin-induced amino acid metabolism was investigated. This research aimed to further elucidate the preservation mechanism of H. undatus postharvest by trypsin, providing additional theoretical support to the field of biopreservation techniques.
2.1 H. undatus Treatment Methods
Thirty fruits (approximately 15 cm in diameter) of H. undatus were divided into two groups: the trypsin group and the control group. The pericarp of each fruit was evenly sprayed with either a trypsin solution (2.41 × 10−6 mol/L) or distilled water. Each group, including the experimental and control groups, consisted of three biological replicates. The samples were placed at room temperature (25°C) and observed, photographed, and recorded, the water loss rate was measured, and color difference was assessed every 24 h. After one week of storage, H. undatus pericarp samples were immediately frozen in liquid nitrogen and stored at −80°C for subsequent analysis.
The weight loss rate and hardness of samples were detected and recorded during 10 days of storage at 25°C. The color of H. undatus peels was determined at room temperature by colorimeter (X-Rite Color i5, Germany). The value of ΔΕ was calculated as the following formula [20,21]:
Sample collection, sequencing, compound identification for omics analysis, selection of differential compounds and differential genes, and the analysis methods for data such as Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were conducted following the descriptions outlined in our previously published papers and those of other researchers [20,21].
2.4 Protein-Protein Interaction (PPI) Network Analysis
Using a network modeling approach, a protein-protein interaction network was constructed to elucidate the relationships between proteins. Starting from a network perspective, topology attributes of the network were analyzed to extract significant protein interaction relationships from complex biological data. The protein interaction network construction was carried out using the Majorbio Cloud platform, and the resulting network was visualized using Cytoscape [22,23]. The MCODE and CytoHubba plugins in Cytoscape were employed for cluster analysis and the identification of key nodes.
2.5 Quantitative Real-Time PCR Analysis
Reverse transcription-qPCR (RT-qPCR) was used to check the result of RNA-seq as described by Yang et al. [24]. Gene β-actin was used as an internal control to normalize gene expression in H. undatus. The relative copy numbers of the genes were calculated by the 2−∆∆Ct method [24,25].
To assess differences between groups, Partial Least Squares Discriminant Analysis (PLS-DA) was performed, and metabolites were selected based on p(corr) combinations. VIP values were calculated using OPLS-DA analysis. All experiments were conducted in triplicate. Statistical analysis was conducted using the SPSS statistical software package (version 11.0.1). Paired sample t-tests were used to determine the significance of differences between samples at specific time points. Significance levels were evaluated as p < 0.05 for significant differences and p < 0.01 for highly significant differences.
3.1 Influence of Trypsin on the Storage Quality of H. undatus
During the initial stages of storage, all groups of H. undatus exhibited good quality with vibrant color (Fig. 1A). After 6 days of storage, the control group’s fruit scales were completely dried out, appearing dull in color, and the fruit bodies were visibly infected by pathogens, showing extensive diseased spots (Fig. 1A). In contrast, the trypsin-treated group displayed partially dried scales, particularly noticeable at the top; however, the fruit skin maintained its vibrant color without evident signs of fungal growth (Fig. 1A).
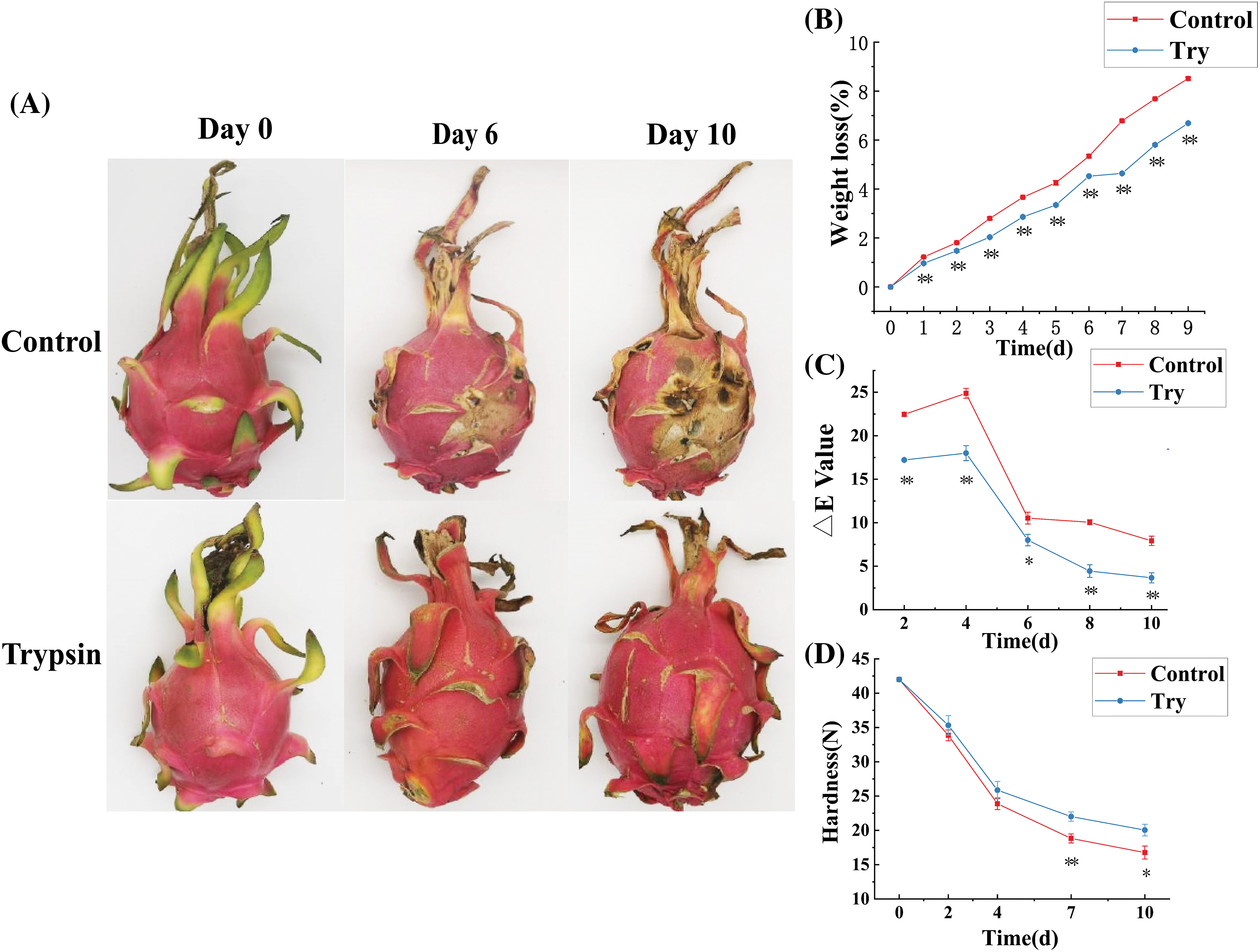
Figure 1: Effect of trypsin treatment on the storage quality of H. undatus. (A) The appearance changes of H. undatus stored at 25°C. (B) Weight loss of H. undatus fruit stored with or without trypsin for 9 d. (C) Changes of the E value of H. undatus fruit stored with or without trypsin at 25°C for 10 days. (D) The hardness of H. undatus fruit stored with or without trypsin for 10 d. Bars represent standard error. Respectively, * represents p < 0.05, and ** represents p < 0.01
As the storage time extended, the weight loss rate for all groups exhibited a significant upward trend (Fig. 1B). The average fresh weight reduction of the control group’s fruit was relatively high, decreasing by 0.95% per day, while the trypsin-treated group showed weight reduction rates of 0.74% per day (Fig. 1B). The difference between the trypsin-treated and control groups was notably distinct. Throughout the storage process, the rate of change of the E value increased during the initial stages of storage and began to decline after the 4th day. Comparative color analysis indicated that the E value of trypsin-treated fruit changed at a slower rate, particularly in the first 4 days of storage, highlighting the favorable effect of trypsin treatment on maintaining the color stability of H. undatus under storage conditions (Fig. 1C). Trypsin treatment suppressed the loss of hardness during the entire storage (Fig. 1D).
The study results suggested that trypsin can effectively delay fruit dehydration, maintain higher hardness, significantly improve fruit quality, and extend shelf life.
3.2 Widely Targeted Metabolomics Analysis
In this study, metabolomics analysis was conducted to investigate amino acid-related metabolic pathways and identify key metabolites, to determine the regulatory effect of trypsin on their synthesis pathways.
3.2.1 Overview of Metabolomics Data
Based on the number of metabolites, a Human Metabolome Database (HMDB) classification was performed (Fig. S1A), where different colors in the pie chart represented various categories of metabolites, and the area depicted the relative proportion of metabolites within each category. Among these, organic acids and their derivatives accounted for 25% of the total, followed by heterocyclic compounds at 17.50%, lipids and lipid-like molecules at 15.83%, aromatic compounds at 10.00%, nucleosides, nucleotides, and their analogs at 10.00%, organic oxygen compounds at 9.17%, phenylpropanes and polyketides at 5.83%, organic nitrogen compounds at 4.17%, alkaloids and their derivatives at 1.67%, and organic sulfur compounds at 0.83%. Organic acids and their derivatives were the largest category, with amino acids and their metabolites being the primary components. Consequently, amino acid-related metabolites within the category of organic acids were selected for further analysis. A total of 453 differential metabolites were identified, with 219 significantly different metabolites (p-Value < 0.05). The volcano plot illustrated these differences (Fig. S1B), where red circles indicated upregulated metabolites (113), blue circles represented downregulated metabolites (106), and gray circles indicated non-significant differences. To analyze differences between groups, the variability in metabolite composition and abundance among samples was quantitatively analyzed using correlation data between samples. In Fig. S1C, each cell represented the correlation between two samples, with high correlation values close to one indicating high similarity in metabolite composition and abundance between samples. The PLS-DA score plot visually demonstrates the classification effect of the model (Fig. S1D), where the contribution rate of PC1 (trypsin-treated group) was 70.20% and that of PC2 (control group) was 19.70%. The distinct separation of metabolite expression levels and group relationships indicated a significant classification effect.
3.2.2 Analysis of Amino Acid-Related Differential Metabolites
From the category of organic acids and their derivatives, a total of 73 amino acid-related metabolites were screened (Fig. 2A), with 56 metabolites upregulated and 17 metabolites downregulated. Among these, 37 metabolites showed significant differences. As shown in Fig. 2B, there was a positive correlation between L-(−)-tyrosine, spermine, histidine, and glutamine. Butyric acid, methylmalonic acid, citric acid, isopropylidene malic acid, and succinic acid showed a positive correlation, while they displayed a negative correlation with other amino acids.
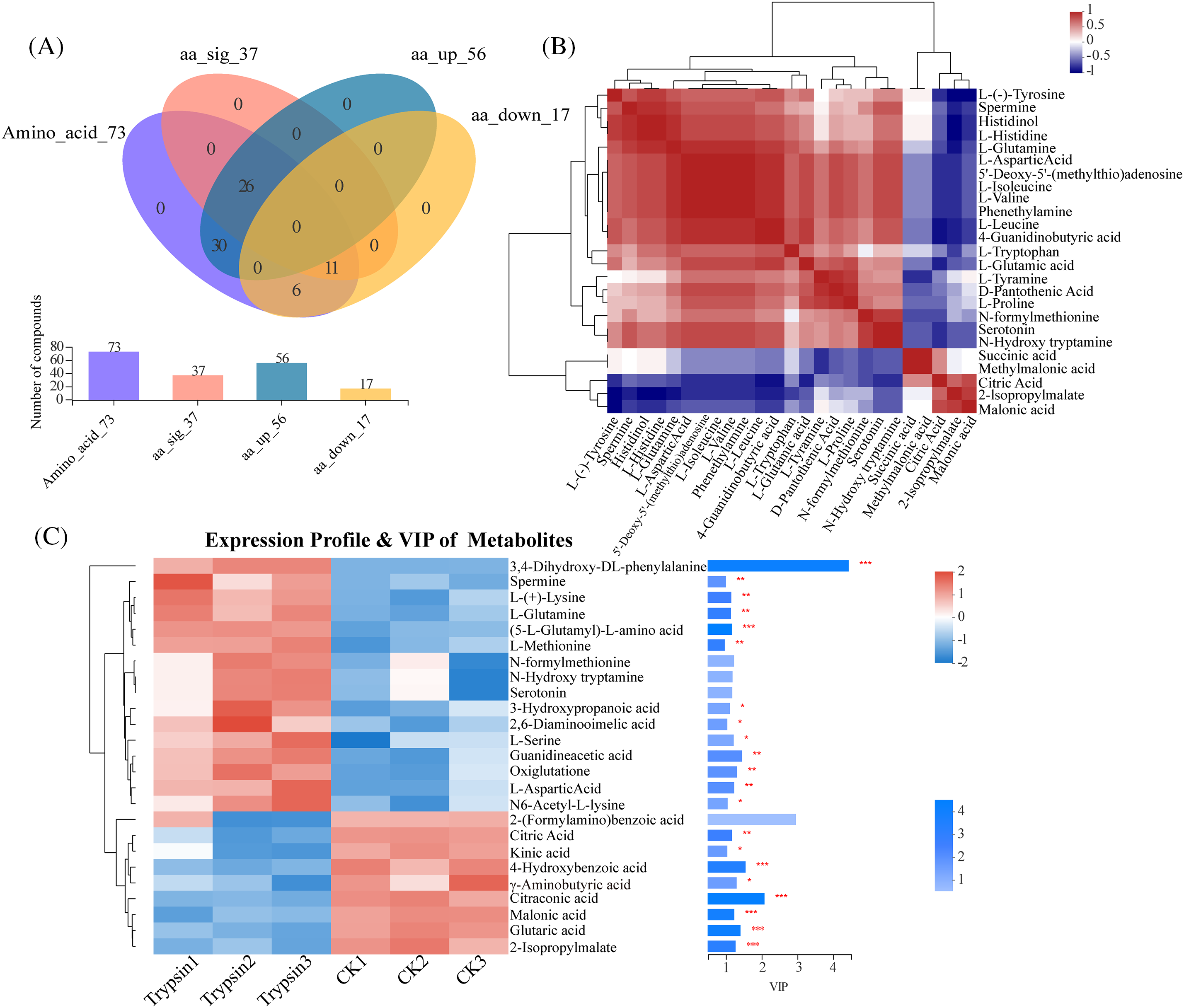
Figure 2: Metabonomic analyses of metabolites regulated by trypsin. (A) Metabolic set analysis venn diagram. (B) Metabolite correlation analysis diagram. (C) 25 significantly different metabolites (VIP ≥ 0.9) expression profile and VIP value. Respectively. * represents p < 0.05, and ** represents p < 0.01, and *** represents p < 0.001. Within the heatmap, red represents a positive correlation, and blue represents a negative correlation
Based on the threshold criteria of |log2(fold change)| ≥ 1 and VIP ≥ 0.9, the top 25 differential metabolites were identified (Fig. 2C). Among these 25 differentially expressed metabolites, 16 amino acids were upregulated in the trypsin-treated group compared to the control group. These included 3,4-dihydroxy-DL-phenylalanine, spermine, L-(+)-lysine, glutamine, (5-L-glutaminyl)-L-amino acid, L-asparagine, N-formylmethionine, N-hydroxytryptamine, serotonin, 3-hydroxypropionic acid, 2,6-diaminopropionic acid, L-serine, guanidine acetic acid, pentanedial, aspartic acid, and N6-acetyl-L-lysine. On the other hand, 9 amino acids were downregulated, including 2-(methylamino) benzoic acid, citric acid, isoleucine, 4-hydroxybenzoic acid, γ-aminobutyric acid, citric acid, succinic acid, glutaric acid, and isopropylidene malic acid.
3.2.3 Amino Acid-Related KEGG Enrichment Analysis
Through KEGG enrichment analysis of all amino acid-related metabolic sets (Table 1; Fig S2), the most enriched metabolic pathway was arginine biosynthesis (map00220) with 10 metabolites, followed by alanine, aspartate, and glutamate metabolism (map00250) with 10 metabolites, valine, leucine, and isoleucine degradation (map00410) with 9 metabolites, aminoacyl-tRNA biosynthesis (map00970) with 14 metabolites, and arginine and proline metabolism (map00330) with 17 metabolites (Fig. S2). Among these, the arginine and proline metabolism pathways involved the highest number of metabolites.
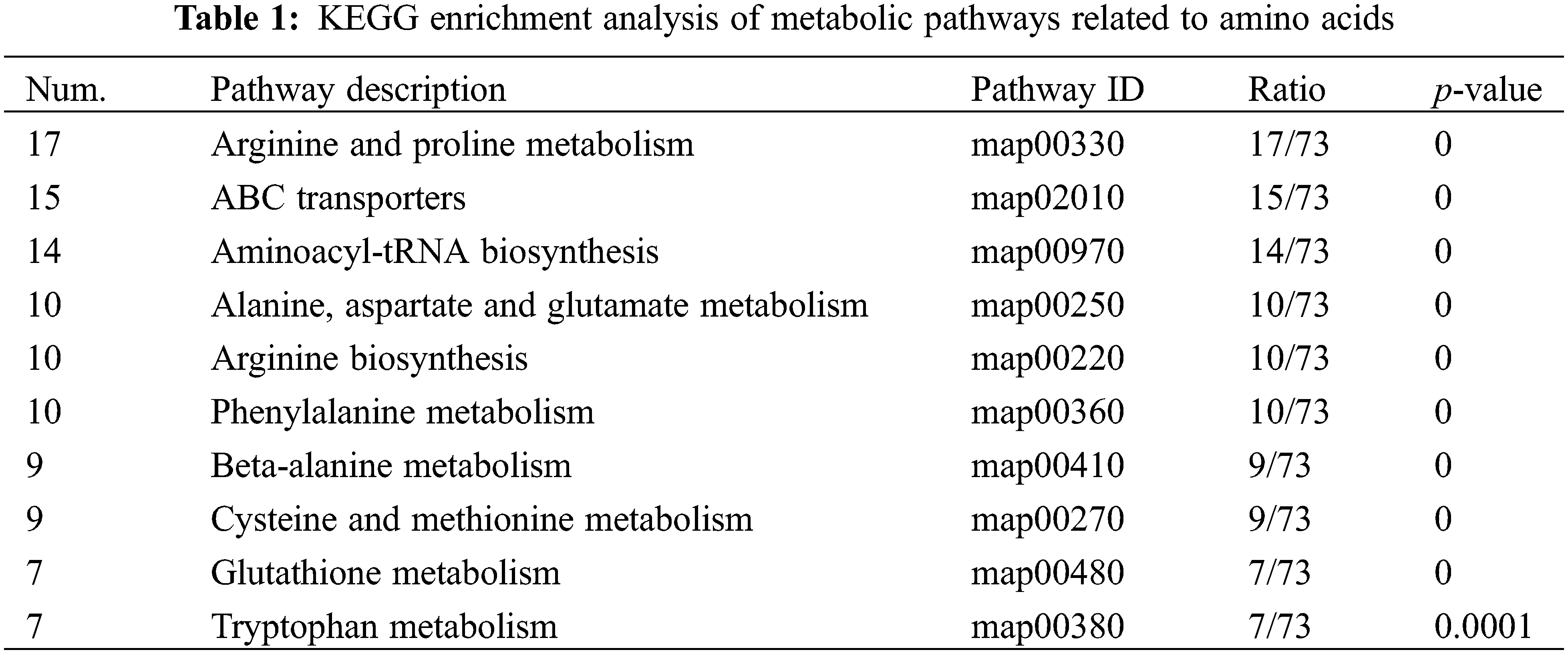
The results indicated that not only the arginine metabolism pathway was significant, but its biosynthesis pathway was also highly significant. We speculated that arginine might be a key amino acid regulated by trypsin. Arginine metabolism leads to the production of PAs and NO. Generally, the accumulation of PAs decreases as fruits mature and age [26]. However, we found that compared to untreated H. undatus fruits, trypsin-treated H. undatus showed significant upregulation of spermine (VIP = 0.9872) during storage (Fig. 2C). This suggested that spermine could play an important role in the preservation effects of trypsin on H. undatus, as it is involved in regulating various physiological processes.
3.3.1 Sequencing Quality Analysis
Through the analysis of amino acid-related metabolites in the metabolomics data, we observed significant changes in the arginine and proline metabolism pathways. However, the differences in metabolites related to the proline pathway were not significant. In contrast, the arginine metabolism pathway, including its biosynthesis, showed significant changes. This led us to speculate that arginine might be a key amino acid regulated by trypsin. To further investigate how trypsin regulates the biosynthesis and metabolism of arginine to achieve its preservation mechanism, we conducted an in-depth analysis of the transcriptome data.
In this study, we constructed two groups, an experimental group and a control group, each with three biological replicates, totaling six transcriptome libraries for transcriptional analysis. After sequencing, the Q20 (probability of base sequencing accuracy over 99.9%) accounted for over 98% of the entire read length, and Q30 (probability of base sequencing accuracy over 99.99%) accounted for over 94% of the entire read length. The GC content was 45%, and the sequencing error rate was below 0.1%. The sequencing results exhibited high quality, meeting the requirements for subsequent analyses.
3.3.2 GO Enrichment Analysis of Arginine-Related Genes
We conducted a GO annotation analysis on the set of arginine-related genes (ARGs) (Fig. 3), which included a total of 187 differentially expressed genes (DEGs). The DEGs were annotated using GO terms from categories including biological processes, cellular components, and molecular functions. More than 100 DEGs were associated with biological biosynthesis processes (GO:0009058) and metabolic processes (GO:0008152). The most significantly enriched GO term was the spermine biosynthetic process (GO:0006597), followed by spermine metabolic process (GO:0008215), S-adenosylmethionine decarboxylase activity (GO:0004014), spermine metabolic process (GO:0008216), spermine biosynthetic process (GO:0008295), and arginine biosynthetic process (GO:0006526), among others.
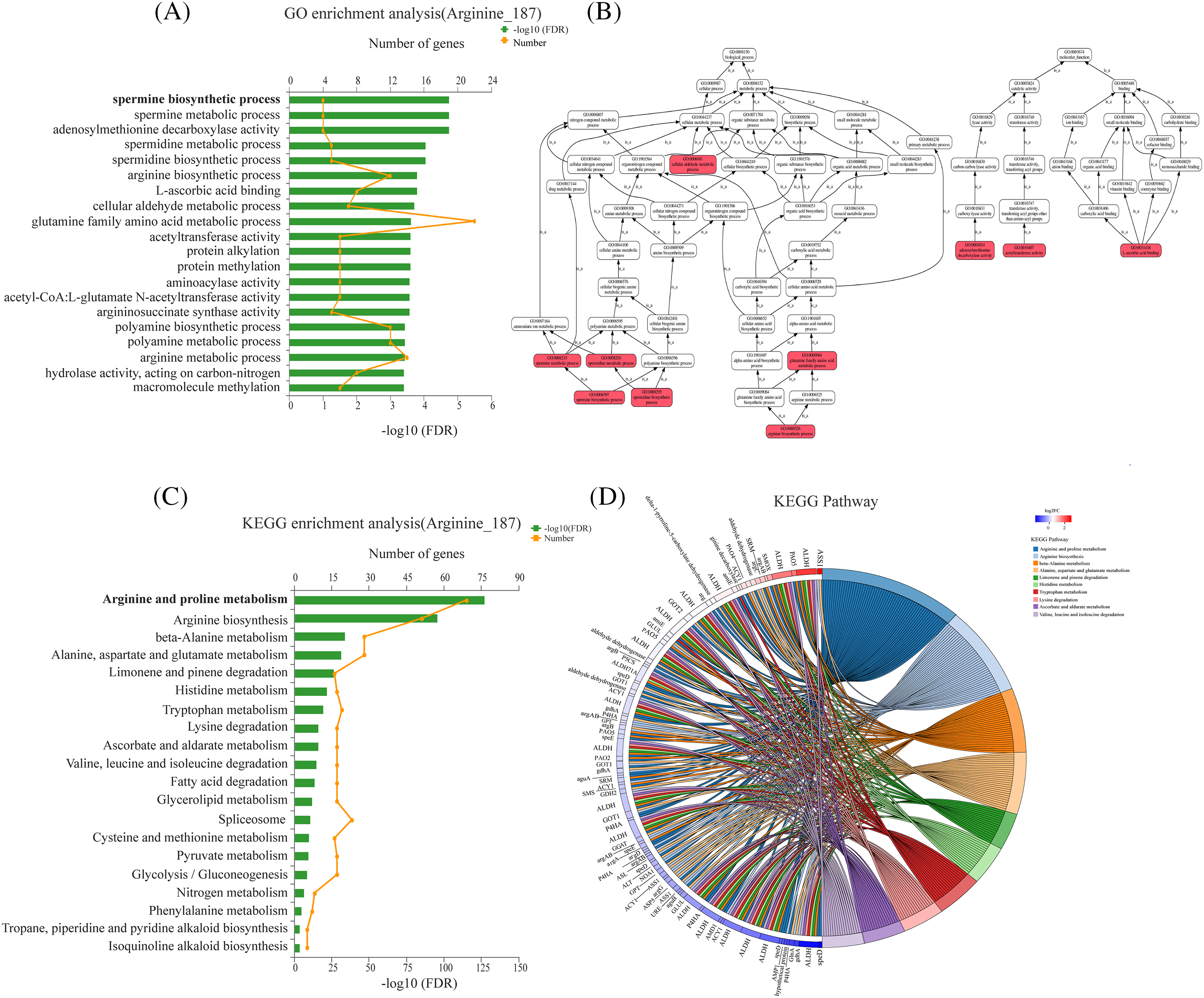
Figure 3: GO and KEGG enrichment analysis of ARGs. (A) GO enrichment analysis diagram of ARGs. (B) Relationships between GO terms in a directed acyclic graph (DAG). (C) KEGG enrichment analysis diagram of ARGs. (D) Chordal graph of KEGG enrichment of ARGs
The GO terms of the ARGs regulated by trypsin were organized into a Directed Acyclic Graph (DAG) that depicted the relationships between GO terms (Fig. 3B). The relationships between terms are described as “is_a” and “part_of.” For instance, the GO biological process “arginine biosynthetic process (GO:0006526)” is a parent term of “arginine metabolic process (GO:0006525),” which, in turn, is a parent term of “ spermine metabolic process (GO:0008215)” and “spermine metabolic process (GO:0008216).” Similarly, the MF term “S-adenosylmethionine decarboxylase activity (GO:0004014)” is a child term of “carboxy-lyase activity (GO:0016831).” The root term for BP ontology is traced back to the ancestral term GO:0008150.
3.3.3 KEGG Enrichment Analysis of Arginine-Related Genes
By conducting a combined KEGG enrichment analysis on both transcriptomic and metabolomic data, we identified the top 10 enriched KEGG pathways related to arginine (p < 0.01) (Fig. 3C, Table 2). The most enriched pathway in the transcriptomic data was “Arginine and proline metabolism (map00330)” with 69 genes enriched, followed by “Arginine biosynthesis (map00220)” with 51 genes enriched, which was consistent with the metabolomic results.
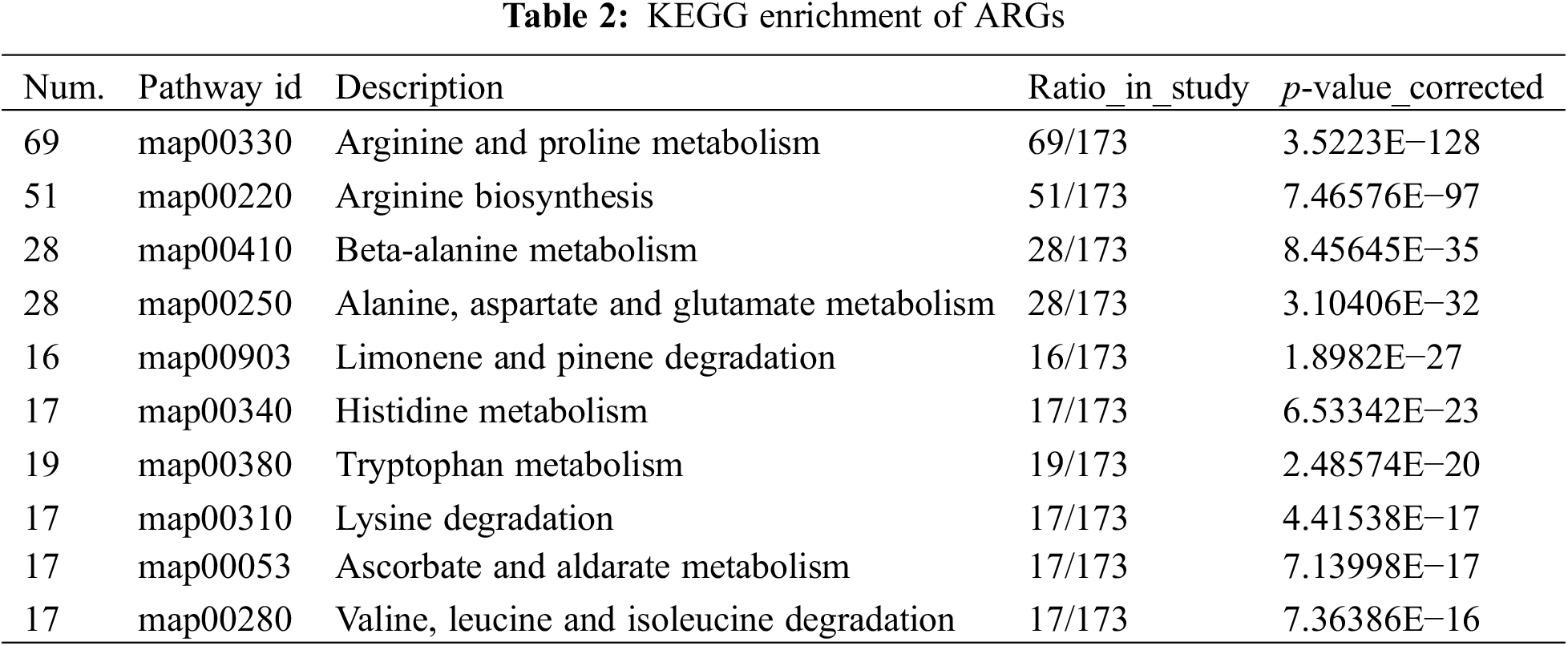
Furthermore, the expression changes of genes in the transcriptomic data within the enriched KEGG pathways were depicted in the chord diagram of the KEGG enrichment analysis (Fig. 3D). In the arginine and proline metabolism pathway (map00330), genes such as argininosuccinate synthase (ASS), spermidine synthase (SPDS), and S-adenosylmethionine decarboxylase precursor (SAMDC), which were involved in the biosynthesis of spermine, were significantly regulated either up or down, suggesting their pivotal roles as central nodes and complex regulatory mechanisms through interactions in signaling pathways. Other enriched pathways included “Arginine biosynthesis (map00220),” “Beta-alanine metabolism (map00410),” “Alanine, aspartate, and glutamate metabolism (map00250),” “Limonene and pinene degradation (map00903),” “Histidine metabolism (map00340),” “Tryptophan metabolism (map00380),” “Lysine degradation (map00310),” “Ascorbate and aldarate metabolism (map00053),” and “Valine, leucine, and isoleucine degradation (map00280).”
3.4 Integrated Metabolomic-Transcriptomic Analysis
Throughout plant evolution, various strategies have evolved to cope with stress, and among them, amino acids play a crucial role as soluble substances in maintaining cellular osmotic pressure, stabilizing cell membranes, and balancing water levels. Protease enzymes, such as trypsin, can impact plant secondary metabolites, especially amino acids, PAs, and other compounds found in H. undatus. Our understanding of the effects of trypsin on amino acids in H. undatus is still limited. Integrative multi-omics analysis can help uncover potential molecular mechanisms.
Our study revealed that trypsin promotes the production of metabolites related to arginine metabolism in H. undatus. Fig. 4 depicts the biosynthesis and metabolism of arginine, demonstrating that trypsin enhances both the biosynthesis and metabolism of arginine. Metabolites associated with arginine metabolism were significantly elevated, including arginine, spermine, agmatine, and putrescine, with spermine showing the most pronounced upregulation. Trypsin increased its abundance by a fold change (FC) of 1.7594. During the biosynthesis of spermine, the upregulation of 4-hydroxy-2-oxoglutaric acid exhibits the most significant change (FC = 2.117). Further VIP value analysis was performed on the 15 metabolites involved in spermine biosynthesis (Fig. S3), and 4-hydroxy-2-oxoglutaric acid had the highest VIP value (VIP = 1.1788), with p < 0.01, indicating that 4-hydroxy-2-oxoglutaric acid plays a crucial role as a key metabolite in the regulation of spermine metabolism by pancreatic protease. PAs are important regulators that play a crucial role in plant growth, morphogenesis, delaying senescence, and environmental adaptation. Under trypsin treatment, the PAs content in H. undatus pericarp was higher compared to the control group.
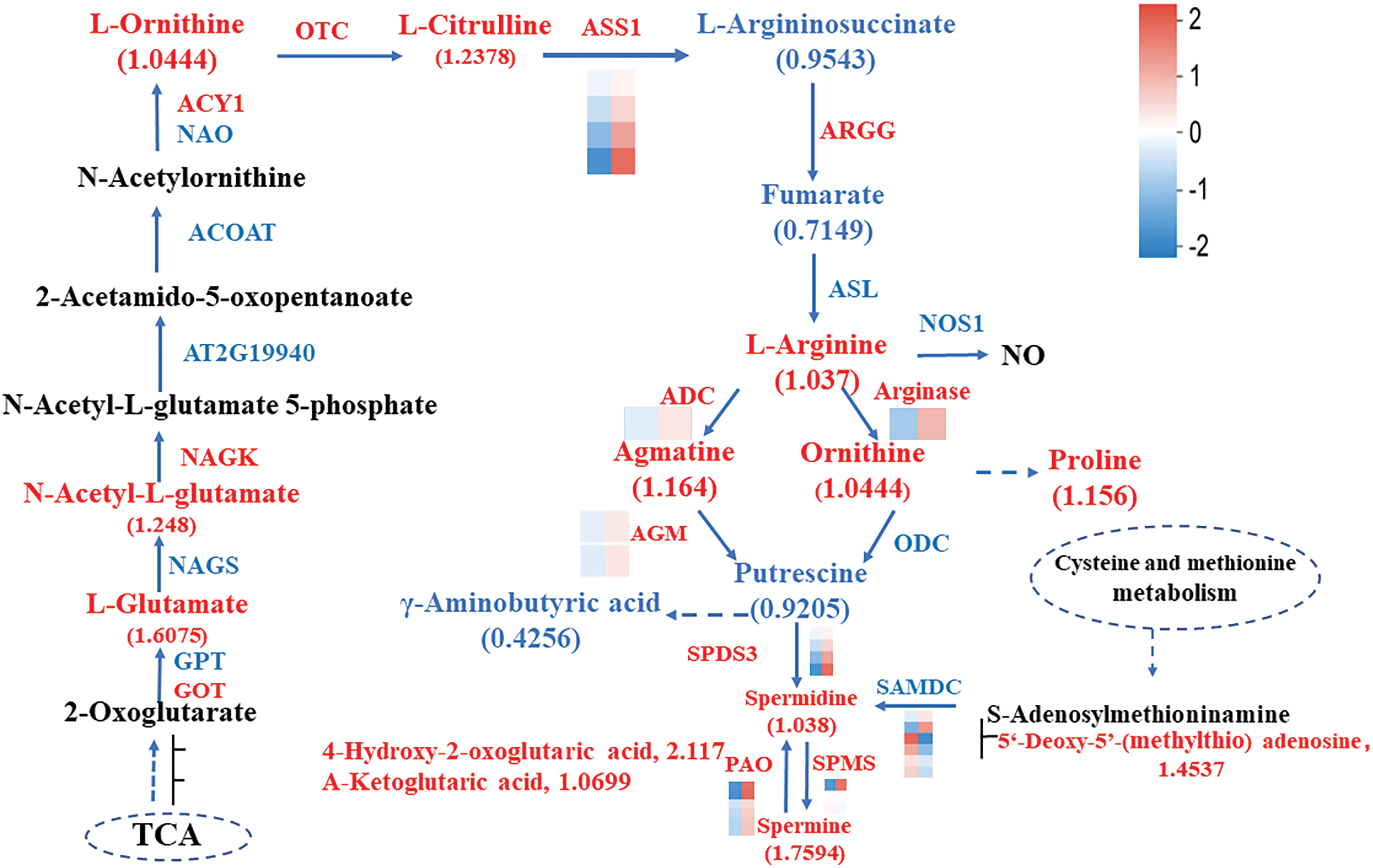
Figure 4: Schematic representation of the arginine biosynthesis pathway as affected by trypsin in H. undatus. Note: red represents up-regulation and blue represents down-regulation. Each column of the color legend in the figure represents a sample, from left to right are the control group and the treatment group. For the specific change trend of the expression amount, see the number label under the color bar at the top right
These findings suggested that trypsin promotes arginine metabolism in H. undatus pericarp, leading to an increase in PAs content, with spermine being notably affected. The biosynthesis and metabolism of PAs involve complex processes. In the arginine metabolism pathway (Fig. 4), three key enzymes were involved: ADC, NOS, and arginase. In the transcriptomic data, we observed significant upregulation of ADC and arginase compared to the control group, which determined the direction of arginine metabolism. Therefore, it was speculated that during H. undatus storage, trypsin regulation mainly promotes arginine metabolism, subsequently stimulating the synthesis of related proteins, thereby achieving a preservation effect.
3.5 Protein-Protein Interaction (PPI) Network Analysis
3.5.1 PPI Network Analysis of Proteins Related to Arginine
Next, protein-protein interaction (PPI) network analysis was employed to investigate the mechanisms of interaction among proteins encoded by ARGs. The analysis aimed to identify key proteins of arginine-polyamine metabolism involved in the regulation of trypsin.
A PPI subnetwork was constructed, consisting of the arginine-related proteins (ARPs) encoded by 187 ARGs. This subnetwork comprised 92 nodes and 799 edges, highlighting 49 upregulated nodes and 43 downregulated nodes, represented by triangles and rounded rectangles, respectively (Fig. 5A).
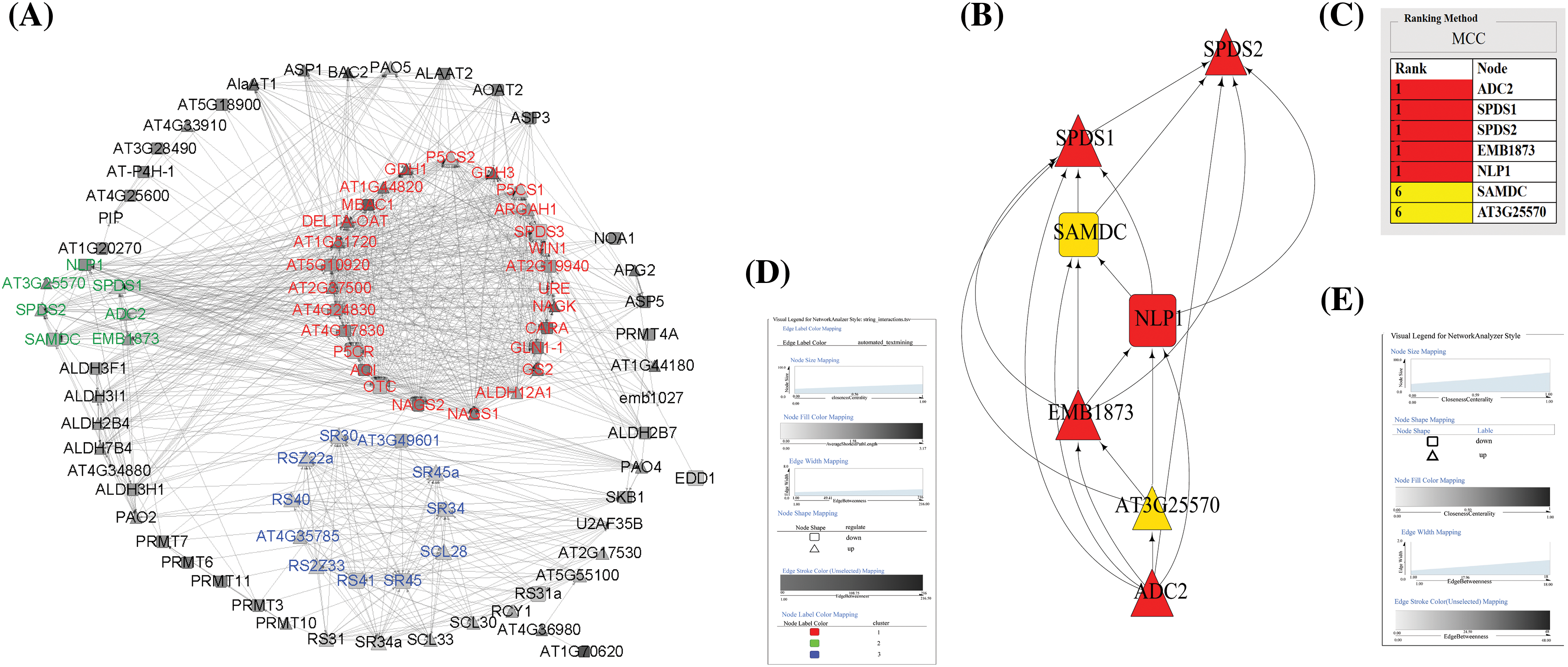
Figure 5: Expression correlation network analysis. (A) PPI networks of trypsin-induced ARPs constructed by Cytoscape. (B) PPI network of differentially expressed genes in cluster 3. (C) Cluster ranking information. (D and E) Legends for the figure. Note: red indicates cluster 1, blue indicates cluster 2, and green indicates cluster 3
3.5.2 Cluster Analysis in the PPI Network of ARGs
Cluster analysis was performed using the Cytoscape plugin “MCODE,” resulting in the identification of 8 clusters within the ARGs’ PPI network. These clusters contained 27, 11, 7, 6, 6, 5, 3, and 3 nodes, respectively. For analysis, we focused on the nodes from the top three protein clusters, which were marked with different colors (Fig. 5A).
Cluster 1 primarily consisted of enzymes related to arginine and proline biosynthesis, such as arginine succinyltransferase (AT4G), ornithine carbamoyltransferase (OTC), pyrroline-5-carboxylate reductase (P5CR), and others. Cluster 2 involved serine/arginine-rich proteins (SR proteins) enriched in serine and arginine residues, which are important splice factors in plants’ response to abiotic stress. Notable proteins in this cluster included serine/arginine-rich splicing factor (SR45) and serine/arginine-rich SC35-like splicing factor (SCL28). Cluster 3 was associated with enzymes involved in the generation of PAs during arginine metabolism, including SPDS1, ADC, guanidinobutyrase (EMB1873), and others.
The Cytoscape plugin “MCODE” aided in illustrating the affiliation of ARGs and DEGs within the PPI network. It was evident that many key enzymes related to arginine biosynthesis and metabolisms, such as arginine succinate synthase (ASS), arginine succinate lyase (AT5G10920), ADC, SPDS, and SAMDC were clustered in protein clusters 1 and 3.
3.5.3 Core Protein Interaction Network Analysis of ARGs
To delve into the potential regulatory mechanisms of trypsin, it was crucial to identify the central proteins within the ARPs network. Studying the interactions among ARPs can provide insights into their roles in trypsin-induced arginine metabolism.
To explore the hub of ARPs metabolism, the nodes from cluster 3 within the DEG overall network were ranked using the cytoHubba method “Maximum Clique Centrality (MCC).” The results from cluster 3 indicated a two-tier structure (Fig. 5B): ADC2, SPDS1, SPDS2, guanidinobutyrase (EMB1873), and N-acetylpolyamine oxidase (NLP) comprised the first tier, while SAMDC and AT3G25570 constituted the second tier. This ordering, based on the cytoHubba plugin, further underscored the central roles of ADC, SPDS, and SAMDC within the ARPs metabolism network.
In plants, the initial reaction of PAs biosynthesis involves arginine and ornithine undergoing decarboxylation by ADC and ornithine decarboxylase (ODC) to form putrescine. Subsequently, putrescine is converted to spermidine by SPDS3, and spermine is synthesized by adding an aminopropyl group derived from S-adenosylmethionine decarboxylation. From the sorting results in Fig. 5, we can observe that ADC and SPDS were the core genes of cluster 3 and are both significantly upregulated.
Based on this, we can speculate that trypsin regulates arginine metabolism and polyamine biosynthesis primarily through core proteins like ADC and SPDS. Many other proteins were controlled by these core proteins, which amplified trypsin’s preservative effect on fruit skin, thus extending the storage period of the fruit.
3.6 Accuracy of the RNA-Seq Data Verification by RT-qPCR
The accuracy of transcriptome data of four key genes was checked by RT-qPCR (Fig. 6A). The square value of R (R2 = 0.9389) was obtained between RT-qPCR and transcriptome spectrum (Fig. 6B). Table S1 showed the RT-qPCR experimental information of four key genes.
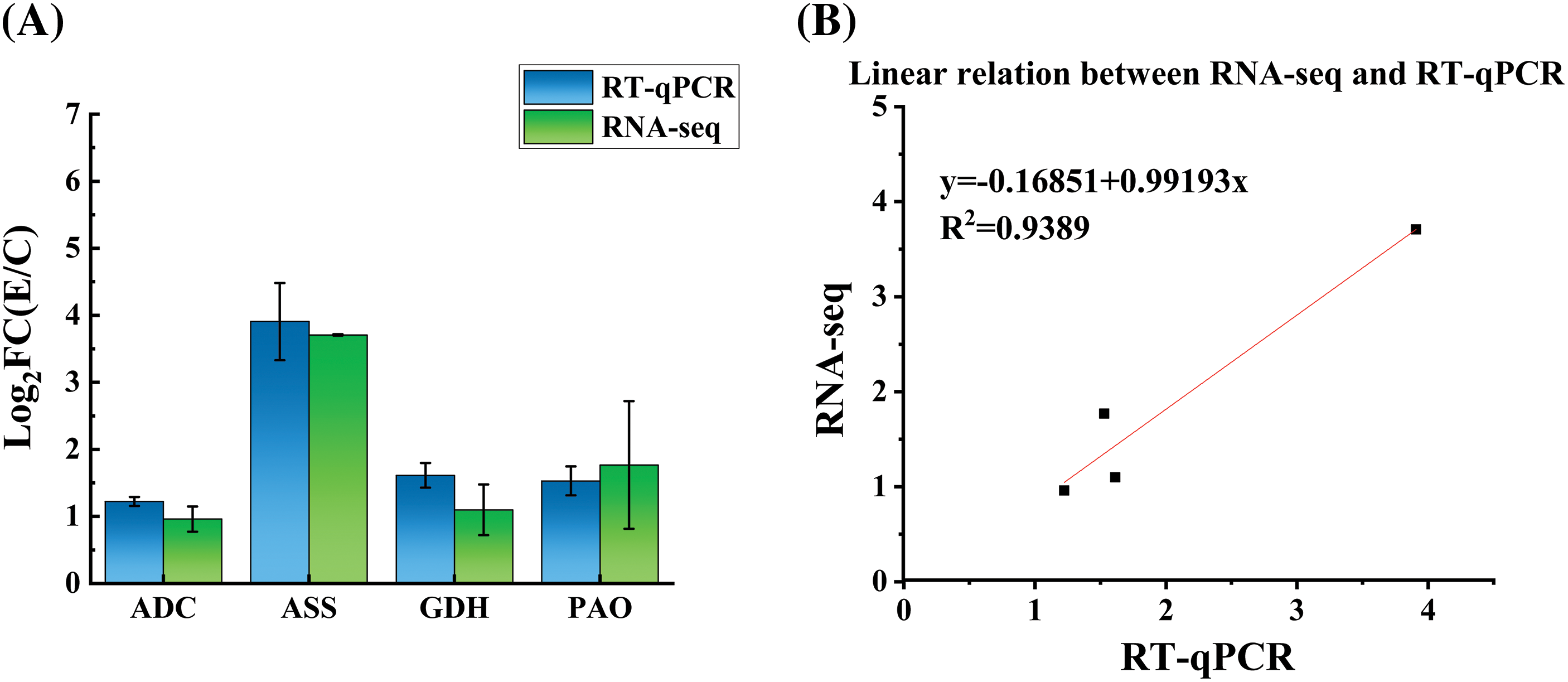
Figure 6: Expression of four gens treated with trypsin in transcriptome data and RT-qPCR. (A) Expression patterns of the selected 4 genes, as determined by transcriptomic and RT-qPCR. (B) Linear relationship of the expression ratio of selected genes measured by transcriptomic and RT-qPCR
In this study, we investigated the effect of trypsin on the storage quality of H. undatus and elucidated its regulatory mechanisms through widely targeted metabolomics and transcriptomics analysis. We found that trypsin treatment can significantly reduce the dehydration rate of H. undatus, and maintain the color of the fruit skin, thereby improving the storage quality of the fruit. By integrating metabolomics and transcriptomics data, we revealed the potential mechanisms underlying the enhancement of H. undatus preservation through trypsin-induced arginine metabolism and PAs biosynthesis.
Our study indicated that the fruit preservation effect of trypsin may be mainly attributed to the promotion of arginine metabolism and the increase of PAs biosynthesis. The most important plant PAs are putrescine, spermidine, and spermine [7]. Compared with putrescine and spermidine, spermine has more amino groups and higher physiological activity in plants. Based on the results of widely targeted metabolomics studies, we reached a similar conclusion: during trypsin-regulated arginine metabolism, putrescine was downregulated, while spermidine and spermine showed an upregulation trend, with spermine showing a significant increase (FC = 1.7594). The increased activity of spermine inhibits the respiration of the fruit, exerting an antioxidant effect [7], thereby further extending the storage time of H. undatus. 4-hydroxy-2-oxoglutaric acid showed the most significant upregulation (FC = 2.117; VIP = 1.1788) in spermine biosynthesis, indicating that 4-hydroxy-2-oxoglutaric acid is the most critical metabolite in trypsin-regulated arginine metabolism.
Through transcriptomic analysis, we have extensively studied the regulatory mechanisms of trypsin treatment on the arginine metabolism pathway. Transcriptomic data revealed that under trypsin treatment, the key gene ADC in the arginine metabolism pathway was upregulated. ADC catalyzes the cleavage of the carboxyl group in arginine molecules, resulting in the production of spermine and carbon dioxide. This further confirmed the important role of arginine metabolism in H. undatus preservation. We speculated that trypsin may promote the synthesis of 4-hydroxy-2-oxoglutaric acid by regulating arginine metabolism, thereby facilitating the production of spermine and various other polyamines. Which in turn participates in the regulation of protein synthesis and degradation, maintaining cellular metabolic balance, enhancing plant antioxidant activity, and mitigating damage to further delay fruit senescence.
In the biosynthesis pathway of spermine, 4-hydroxy-2-oxoglutaric acid is one of the precursors which is eventually transformed into spermine via a series of enzyme-catalyzed reactions. Therefore, the accumulation level of 4-hydroxy-2-oxoglutaric acid can indirectly reflect the activity and efficiency of spermine biosynthesis. Researchers found that under drought stress conditions, 4-hydroxy-2-oxoglutaric acid accumulated significantly in tobacco roots by analyzing the metabolite composition [27]. This result indicated that the metabolic state of plants changes under drought stress, and the accumulation of 4-hydroxy-2-oxoglutaric acid may be an adaptive strategy to cope with the metabolic and physiological changes caused by drought stress. It has also been suggested that 4-hydroxy-2-oxoglutaric acid may serve as a useful marker for drought stress in Solanaceae plants [28]. However, the role of 4-hydroxy-2-oxoglutaric acid in post-harvest fruit senescence is still poorly understood and further research is needed to elucidate its mode of action and interaction with other metabolic pathways during fruit senescence. Our study provides new insights into the role of trypsin-regulated spermine metabolism during the storage of H. undatus and contributes to a better understanding of the multifunctionality of 4-hydroxy-2-oxoglutaric acid in different biological processes.
In summary, our study revealed the potential mechanism by which trypsin promotes the preservation of H. undatus. We speculate that 4-hydroxy-2-oxoglutaric acid regulates the biosynthesis of polyamines, including spermine, enhancing the plant’s adaptability to abiotic stress. Spermine plays a role in abiotic stress through various mechanisms, such as regulating stomatal closure, alleviating oxidative damage, and maintaining cell osmotic balance [29,30]. This provides a new perspective and theoretical support for research in the field of fruit preservation. However, further research is needed to elucidate the molecular details of trypsin regulation and its application prospects under different plant and environmental conditions.
Among the 453 compounds analyzed, a total of 75 differential metabolites related to amino acids were identified. The arginine metabolism pathway emerged as the primary route through which trypsin regulates amino acids, contributing to the quality improvement of H. undatus during storage. Transcriptional analysis identified 186 ARGs. The most prominently regulated amino acid-related metabolite was 4-hydroxy-2-oxoglutaric acid. The “arginine biosynthesis and metabolism pathway” appeared to be the central route through which trypsin regulates amino acids, as demonstrated by the results from the PPI network protein clusters, including ADC, SPDS, and SAMDC, which served as pivotal proteins in trypsin-regulated arginine metabolism. This observation aligned well with the findings from KEGG enrichment chord diagrams.
As a novel bio-preservative, trypsin promotes arginine metabolism, and increases the content of 4-hydroxy-2-oxoglutaric acid, thereby regulating the biosynthesis of polyamines including spermine, and extending the shelf life of fruits. The results of this study provided valuable theoretical support for the post-harvest preservation of H. undatus.
Acknowledgement: We are grateful for the free online platform of Majorbio I-Sanger Cloud Platform (www.majorbio.com).
Funding Statement: This work was supported by the National Key Research and Development Program of China (2017YFC1600802).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Bairu Li, Xin Li; data collection: Jingyu Jia; analysis and interpretation of results: Hemin Wang, Jiaju Sun, Enyan Chen; draft manuscript preparation: Bairu Li, Xin Li. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data and materials used in this research were publicly available. Raw sequence data from this study have been submitted to the NCBI sequence read archive under the BioProject Accession (PRJNA 509494).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.050450.
References
1. Arivalagan M, Karunakaran G, Roy TK, Dinsha M, Sindhu BC, Shilpashree VM, et al. Biochemical and nutritional characterization of dragon fruit (Hylocereus species). Food Chem. 2021;353:129426. doi:10.1016/j.foodchem.2021.129426 [Google Scholar] [PubMed] [CrossRef]
2. Guo LW, Wu YX, Mao ZC, Ho HH, He YQ. Storage rot of dragon fruit caused by Gilbertella persicaria. Plant Dis. 2012;96(12):1826. doi:10.1094/PDIS-07-12-0635-PDN [Google Scholar] [PubMed] [CrossRef]
3. Tian S, Qin G, Li B. Reactive oxygen species involved in regulating fruit senescence and fungal pathogenicity. Plant Mol Biol. 2013;82(6):593–602. doi:10.1007/s11103-013-0035-2 [Google Scholar] [PubMed] [CrossRef]
4. Chomkitichai W, Chumyam A, Rachtanapun P, Uthaibutra J, Saengnil K. Reduction of reactive oxygen species production and membrane damage during storage of ‘Daw’ longan fruit by chlorine dioxide. Scientia Hortic. 2014;170:143–9. doi:10.1016/j.scienta.2014.02.036. [Google Scholar] [CrossRef]
5. Sun J, Lin H, Zhang S, Lin Y, Wang H, Lin M, et al. The roles of ROS production-scavenging system in Lasiodiplodia theobromae (Pat.) Griff. & Maubl.-induced pericarp browning and disease development of harvested longan fruit. Food Chem. 2018;247:16–22. doi:10.1016/j.foodchem.2017.12.017 [Google Scholar] [PubMed] [CrossRef]
6. Belozerskaia TA, Gessler NN. Reactive oxygen species and the strategy of the antioxidant defense in fungi: a review. Prikl Biokhim Mikrobiol. 2007;43(5):565–75. doi:10.1134/S0003683807050031. [Google Scholar] [CrossRef]
7. Alcázar R, Bueno M, Tiburcio AF. Polyamines: small amines with large effects on plant abiotic stress tolerance. Cells. 2020;9(11):2373. doi:10.3390/cells9112373 [Google Scholar] [PubMed] [CrossRef]
8. Liu K, Fu H, Bei Q, Luan S. Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol. 2000;124(3):1315–26. doi:10.1104/pp.124.3.1315 [Google Scholar] [PubMed] [CrossRef]
9. Cuevas JC, López-Cobollo R, Alcázar R, Zarza X, Koncz C, Altabella T, et al. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 2008;148(2):1094–105. doi:10.1104/pp.108.122945 [Google Scholar] [PubMed] [CrossRef]
10. Roy M, Wu R. Arginine decarboxylase transgene expression and analysis of environmental stress tolerance in transgenic rice. Plant Sci. 2001;160(5):869–75. doi:10.1016/s0168-9452(01)00337-5 [Google Scholar] [PubMed] [CrossRef]
11. Cai J, Aharoni A. Amino acids and their derivatives mediating defense priming and growth tradeoff. Curr Opin Plant Biol. 2022;69:102288. doi:10.1016/j.pbi.2022.102288 [Google Scholar] [PubMed] [CrossRef]
12. Gong X, Zhang J, Hu J, Wang W, Wu H, Zhang Q, et al. FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 2015;38(11):2248–62. doi:10.1111/pce.12539 [Google Scholar] [PubMed] [CrossRef]
13. Shi HT, Chan ZL. In vivo role of Arabidopsis arginase in arginine metabolism and abiotic stress response. Plant Signal Behav. 2013;8(5):e24138. doi:10.4161/psb.24138 [Google Scholar] [PubMed] [CrossRef]
14. Song L, Benjiao L, Binrong M, Yong J, Qiang W, Zheming Zeditors. Research progress on physical storage methods of Toona Sinensis. In: E3S Web of Conferences, 2021; Guangzhou, China. doi:10.1051/e3sconf/202123302050. [Google Scholar] [CrossRef]
15. Vanden Abeele C, Raes K, Sampers I. Effect of mild heat treatment on browning-related parameters in fresh-cut Iceberg lettuce. J Food Biochem. 2019;43(7):e12906. doi:10.1111/jfbc.12906 [Google Scholar] [PubMed] [CrossRef]
16. Wei H, Seidi F, Zhang T, Jin Y, Xiao H. Ethylene scavengers for the preservation of fruits and vegetables: a review. Food Chem. 2021;337:127750. doi:10.1016/j.foodchem.2020.127750 [Google Scholar] [PubMed] [CrossRef]
17. Li X, Tang Z, Pang X, Zhao C, Li X, Liu Y. Trypsin slows the aging of mice due to its novel superoxide scavenging activity. Appl Biochem Biotechnol. 2017;181(4):1549–60. doi:10.1007/s12010-016-2301-7 [Google Scholar] [PubMed] [CrossRef]
18. Li X, Zhong Y, Zhao C. Trypsin binding with copper ions scavenges superoxide: molecular dynamics-based mechanism investigation. Int J Environ Res Public Health. 2018;15(1):139. doi:10.3390/ijerph15010139 [Google Scholar] [PubMed] [CrossRef]
19. Li Q, Wei Q, Yuan E, Yang J, Ning Z. Interaction between four flavonoids and trypsin: effect on the characteristics of trypsin and antioxidant activity of flavonoids. Int J Food Sci Tech. 2014;49(4):1063–9. doi:10.1111/ijfs.12401. [Google Scholar] [CrossRef]
20. Zhang Y, Li B, Zhang M, Jia J, Sun S, Chen X, et al. Transcriptome analyses and virus-induced gene silencing identify HuWRKY40 acting as a hub transcription factor in the preservation of Hylocereus undatus by trypsin. J Food Biochem. 2022;46(12):e14437. doi:10.1111/jfbc.14437 [Google Scholar] [PubMed] [CrossRef]
21. Li X, Liu X, Yin Y, Yu H, Zhang M, Jing H, et al. Transcriptomic analysis reveals key genes related to antioxidant mechanisms of Hylocereus undatus quality improvement by trypsin during storage. Food Funct. 2019;10(12):8116–28. doi:10.1039/c9fo00809h [Google Scholar] [PubMed] [CrossRef]
22. Wahab Khattak F, Salamah Alhwaiti Y, Ali A, Faisal M, Siddiqi MH. Protein-protein interaction analysis through network topology (oral cancer). J Healthc Eng. 2021;2021:6623904. doi:10.1155/2021/6623904 [Google Scholar] [PubMed] [CrossRef]
23. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11 [Google Scholar] [PubMed] [CrossRef]
24. Yang A, Yu L, Chen Z, Zhang S, Shi J, Zhao X, et al. Label-free quantitative proteomic analysis of chitosan oligosaccharide-treated rice infected with southern rice Black-streaked dwarf virus. Viruses. 2017;9(5):115. doi:10.3390/v9050115 [Google Scholar] [PubMed] [CrossRef]
25. Li X, Zhang Y, Zhao S, Li B, Cai L, Pang X. Omics analyses indicate the routes of lignin related metabolites regulated by trypsin during storage of pitaya (Hylocereus undatus). Genomics. 2021;113(6):3681–95. doi:10.1016/j.ygeno.2021.08.005 [Google Scholar] [PubMed] [CrossRef]
26. Brauc S, de Vooght E, Claeys M, Geuns JM, Höfte M, Angenon G. Overexpression of arginase in Arabidopsis thaliana influences defence responses against Botrytis cinerea. Plant Biol. 2012;14(s1):39–45. doi:10.1111/j.1438-8677.2011.00520.x [Google Scholar] [PubMed] [CrossRef]
27. Rabara RC, Tripathi P, Reese RN, Rushton DL, Alexander D, Timko MP, et al. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genom. 2015;16(1):484. doi:10.1186/s12864-015-1575-4 [Google Scholar] [PubMed] [CrossRef]
28. Rabara RC, Tripathi P, Rushton PJ. Comparative metabolome profile between tobacco and soybean grown under water-stressed conditions. Biomed Res Int. 2017; 2017:3065251. doi:10.1155/2017/3065251 [Google Scholar] [PubMed] [CrossRef]
29. Misra BB, Acharya BR, Granot D, Assmann SM, Chen S. The guard cell metabolome: functions in stomatal movement and global food security. Front Plant Sci. 2015;6:334. doi:10.3389/fpls.2015.00334 [Google Scholar] [PubMed] [CrossRef]
30. Agurla S, Gayatri G, Raghavendra AS. Polyamines increase nitric oxide and reactive oxygen species in guard cells of Arabidopsis thaliana during stomatal closure. Protoplasma. 2018;255(1):153–62. doi:10.1007/s00709-017-1139-3 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools