 Open Access
Open Access
ARTICLE
Analysis of Intraspecies Diversity of Rice Reveals Variation Patterns of Oryza sativa Basic Leucine Zippers
1 Anhui Provincial Key Laboratory of Molecular Enzymology and Mechanism of Major Diseases, Key Laboratory for Conservation and Use of Important Biological Resources of Anhui Province, College of Life Sciences, Anhui Normal University, Wuhu, 241000, China
2 Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing, 210000, China
* Corresponding Author: Yang Sun. Email:
Phyton-International Journal of Experimental Botany 2024, 93(5), 859-873. https://doi.org/10.32604/phyton.2024.048151
Received 29 November 2023; Accepted 19 March 2024; Issue published 28 May 2024
Abstract
The basic leucine zipper (bZIP) is an important class of transcription factors in plants, playing a critical role in plant growth and development and responses to biotic and abiotic stress. Due to gene presence/absence variations, it is limited to identify bZIP genes based on the reference genome. Therefore, we performed the bZIP gene family analysis in the rice pan-genome. By employing a rice pan-genome, ninety-four OsbZIPs (72 core genes and 22 variable genes) were identified and divided into 11 groups in a phylogenetic tree. Based upon Ka/Ks values in 33 accessions, OsbZIPs were subjected to different selection pressures during domestication. The analysis of the effects of structural variations (SVs) on gene expression, gene structure, and conserved domains showed that SVs could significantly alter the expression levels of certain OsbZIPs, leading to gene truncation and the emergence of numerous atypical genes. Thirty-four differentially expressed OsbZIPs were identified by analyzing RNA-seq data of the Xanthomonas oryzae pv. oryzae (Xoo) infection susceptible (IR24) and resistant (IRBB67) lines under high temperature, and by counting the number of differentially expressed OsbZIPs in different subgroups. These OsbZIPs were found to respond to Xoo infection at an early stage and may not be involved in the mechanism of Xa4 and Xa7 resistance to Xoo. The multiple variation patterns of OsbZIP genes provide new insights into the OsbZIP genes in rice. These results provide new resources and offer new directions for functional studies of OsbZIPs.Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| TFs | Transcription factors |
| JA | Jasmonic acid |
| SA | Salicylic acid |
| ABA | Abscisic acid |
| PAVs | Present/absent variations |
| HMM | Hidden markov model |
| JTT | Jones–Taylor–Thornton |
| BB | Rice bacterial blight |
| bZIP | Basic leucine zipper |
| SNPs | Single nucleotide polymorphisms |
| SVs | Structural variations |
| CNVs | Copy number variants |
| ML | Maximum likelihood |
Plants are regulated by many transcription factors (TFs) during growth and development. Transcription factors are a class of proteins that regulate gene expression by attaching to particular cis-acting promoter elements, thus activating or inhibiting the transcription rate of their target genes [1]. The bZIP transcription factors are ubiquitous in eukaryotic genomes, constituting one of the largest and most conserved gene families discovered so far. The conserved 60–80 amino acid domain of bZIPs is mainly composed of a basic region and a leucine zipper region, in which the basic region is located at the C-terminal and contains a conserved motif N-x7-R/K-x9 consisting of 16–20 amino acid residues. It is mainly involved in nuclear localization and specific binding to DNA [2,3].
The bZIPs are one of the most widely studied transcription factor gene families. A large number of studies have shown that bZIP transcription factors play an important role in plant growth and development. For instance, AtbZIP29 in Arabidopsis thaliana affects root growth and development by regulating the expression of genes related to cell wall formation in root tip meristem cells [4]. Overexpression of Arabidopsis ABF4 in potatoes can significantly increase tuber yield and inhibit tuber germination, thereby improving tuber processing and storage quality [5]. These bZIP transcription factors are involved in plant responses to biotic and abiotic stress. In soybeans, the expression of GmbZIP19 was induced by S. sclerotiorum [6]. Rice OsbZIP1 may boosts resistance to Magnaporthe grisea through jasmonic acid (JA), salicylic acid (SA) and abscisic acid (ABA) signaling pathways [7]. Overexpression of TabZIP15 in wheat significantly improved salt stress tolerance [8]. In tomatoes, the bZIP transcription factor SIAREB is involved in response to drought and salt stress [9]. In Arabidopsis responding to salt stress, AtbZIP17 can activate the expression of downstream genes directly or indirectly, thereby playing an important role in improving salt tolerance [10,11]. Basic leucine zipper transcription factor BnaABF2 in Brassica napus regulated the salt tolerance of plants through the ABA pathway [12].
Comparative genomics studies in plants typically focus on single nucleotide polymorphisms (SNPs) among individuals from different populations [13,14]. However, due to the substantial amount of SVs, including presence/absence variants (PAVs) and copy number variants (CNVs) [15], a solitary reference genome fails to encompass the genomic diversity evident in nature. To capture the entire genomic sequences within a species, including the complete gene set, the concept of pan-genome has been introduced [16]. The tomato pan-genome captured 4,873 novel genes absent in the reference genome and identified a flavor-related allelic gene [17]. Tang et al. constructed the pan-genome of potatoes, revealing a 5.8 Mb inversion associated with carotenoid content in tubers [18]. Qin et al. built a pan-genome based on 33 rice high-quality assemblies, which provided abundant SV and PAV information and provided a foundation for the rice gene family research [19].
Sixty-eight bZIPs were identified in Arabidopsis thaliana [20]. Wang et al. identified 50 and 45 bZIPs, in the Arachis duranensis and Arachis ipaensis genomes of peanut [21]. One hundred and sixty bZIPs were identified in the soybean genome [15,22]. Seventy-six bZIPs were identified in the tomato [23]. Rice is not only a food crop widely cultivated all over the world but also a model species widely used in plant research. Eighty-nine bZIPs have been identified from a single rice reference genome [24], it is limited to identifying and analyzing a gene family based on a specific reference genome. In this study, ninety-four OsbZIPs were identified based on the gene-based rice pan-genome, of which 72 were core genes (present in 33 accessions) and 22 were variable genes (not present in all accessions). By calculating the Ka/Ks values of bZIPs among 33 accessions, OsbZIPs were found subjected to different selection pressures. Most OsbZIPs were subjected to purifying selection. A few of OsbZIPs may be rapidly evolving, such as OsbZIP76. Some OsbZIPs may act as housekeeping genes (e.g., OsbZIP1, OsbZIP37, and OsbZIP85). Gene expression, structures, and conserved domains of some OsbZIPs were altered by SVs. Thirty-four differentially expressed OsbZIPs were identified by analysis of the RNA-seq data of Xoo infection susceptible (IR46) and resistance (IRBB67) lines under high and low temperatures. These results provide new resources for the study of bZIP transcription factors in rice and provide new ideas and directions for the identification and analysis of gene families.
2.1 Identification of Rice bZIP Transcription Factors
The protein sequences of 33 rice accessions were downloaded from the Rice Resource Center (https://ricerc.sicau.edu.cn/), and the Hidden Markov Model (HMM) of the bZIP domain (PF00170) was downloaded from the Pfam database (http://pfam.xfam.org/). Using a threshold E-value of < 1e-5, HMMER (v3.3.2) was used to search the bZIP domain among protein sequence files of 33 rice accessions to extract candidate OsbZIPs. The protein sequences of candidate OsbZIPs were uploaded to SMART (http://smart.embl-heidelberg.de) to confirm the bZIP domain. Finally, according to the gene colinear relationships between 33 rice accessions in the study of Qin et al. [19], the genes that are colinear genes of the confirmed OsbZIPs were also regarded as bZIP gene family members.
2.2 Phylogenetic Analysis of the Rice bZIP Members
The Arabidopsis bZIP protein sequences were obtained from the TAIR database (https://www.arabidopsis.org/). Multiple sequence alignments were conducted using MAFFT v7.490 with the default parameters. Subsequently, a phylogenetic tree was generated using FastTree version 2.1.11 [25] with the Jones–Taylor–Thornton (JTT) model and the maximum likelihood (ML) method and visualized through iTOL (https://itol.embl.de/upload.cgi).
The classification information of core and variable OsbZIPs was extracted from the study of Qin et al. [19] and a heatmap of variable genes constructed using R v4.0.3 ComplexHeatmap and RColorBrewer packages.
2.3 Calculation of Ka/Ks Values of OsbZIPs in 33 Rice Assemblies
The Ka/Ks values of the colinear genes of each OsbZIP in 33 rice assemblies were calculated by KaKs Calculator Toolbox v2.0. The result was visualized using R v4.0.3 with ggridges and ggplot2 packages. The numbers and proportions of OsbZIP pairs with Ka/Ks >1 were counted. A heatmap was generated using R v4.0.3 ComplexHeatmap and RColorBrewer packages.
2.4 Analysis of SVs Overlapping with OsbZIPs
SVs that overlap with OsbZIPs were searched at the Rice Resource Center (https://ricerc.sicau.edu.cn/), and the expression values of genes with SVs and genes without SVs were derived from the study of Qin et al. [19]. The expression levels of OsbZIPs with and without SVs were plotted using GraphPad Prism v8.0.2.
2.5 Analysis of the Gene Structures of SV-Affected OsbZIPs
The gff (general feature format) files were obtained from the Rice Resource Center (https://ricerc.sicau.edu.cn/). The protein sequences significantly affected by SVs were uploaded to MEME Suite v5.4.1 (https://meme-suite.org/meme/tools/meme), with the number of motifs set to 10. The resulting outcomes, along with the gff file of these proteins, were used as input files in TBtools (v1.098696) to obtain the gene structure.
The protein sequences of OsbZIPs in reference (90 OsbZIPs from NIP and 4 from CG14, which were lost in NIP) and OsbZIPs from FS32, which have the most OsbZIPs overlapped with SVs, were submitted to MEME Suite v5.4.1 (https://meme-suite.org/meme/tools/meme) with the number of motifs setting to 10 to obtain the sequence weblogos. The typical genes (containing the complete conserved domain) and atypical genes (not containing the complete conserved domain) in 33 accessions were plotted by ComplexHeatmap and RColorBrewer packages in R v4.03.
Raw RNA-seq data were downloaded from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79011). The fastq-dump of SRA Toolkit v2.10.0 was used to convert the SRA format files to FASTQ format. Fastp v0.20.1 was used to remove the adapter sequences and low-quality sequences. Clean reads were mapped to the reference genome (NIP) with HiSAT2 version 2.1.0 [26]. Fragments per kilobase of transcript per million mapped reads (FPKM) values of genes were calculated as the expression level. Differentially expressed genes were identified using DESeq2 version 1.32.0 [27] with a threshold |log2FoldChange| > 1 and FDR ≤ 0.05. A heatmap was generated with the transformed log2 (FPKM + 1) values using R (version 4.0.3) with the ComplexHeatmap package (version 2.6.2) [28].
3.1 Identification and Phylogenetic Analysis of bZIP in Pan-Genome
Ninety-four OsbZIPs were identified by HMM search and SMART domains analysis (Table S1). These genes were named OsbZIP1-OsbZIP94 according to the order of their location. Among the 94 OsbZIPs, seventy-two OsbZIPs were core genes and twenty-two OsbZIPs were variable genes. Since the reference genome (NIP) does not contain all OsbZIPs, the phylogenetic tree was constructed with OsbZIPs from NIP (90 OsbZIPs) and CG14 (4 OsbZIPs which were lost in the reference genome) as well as AtbZIPs from Arabidopsis (Fig. 1A). According to the division of the AtbZIP phylogenetic tree in the previous study [20], Oryza sativa basic leucine zippers were divided into 11 groups. All the OsbZIPs in the group B, D, E, G, H, and I are core genes, suggesting that these groups were stable during domestication and improvement. Among the other five groups, there are the most variable genes in group S, indicating that the OsbZIPs in this group are more changeable in different accessions, which may be related to the unique traits of some accessions.
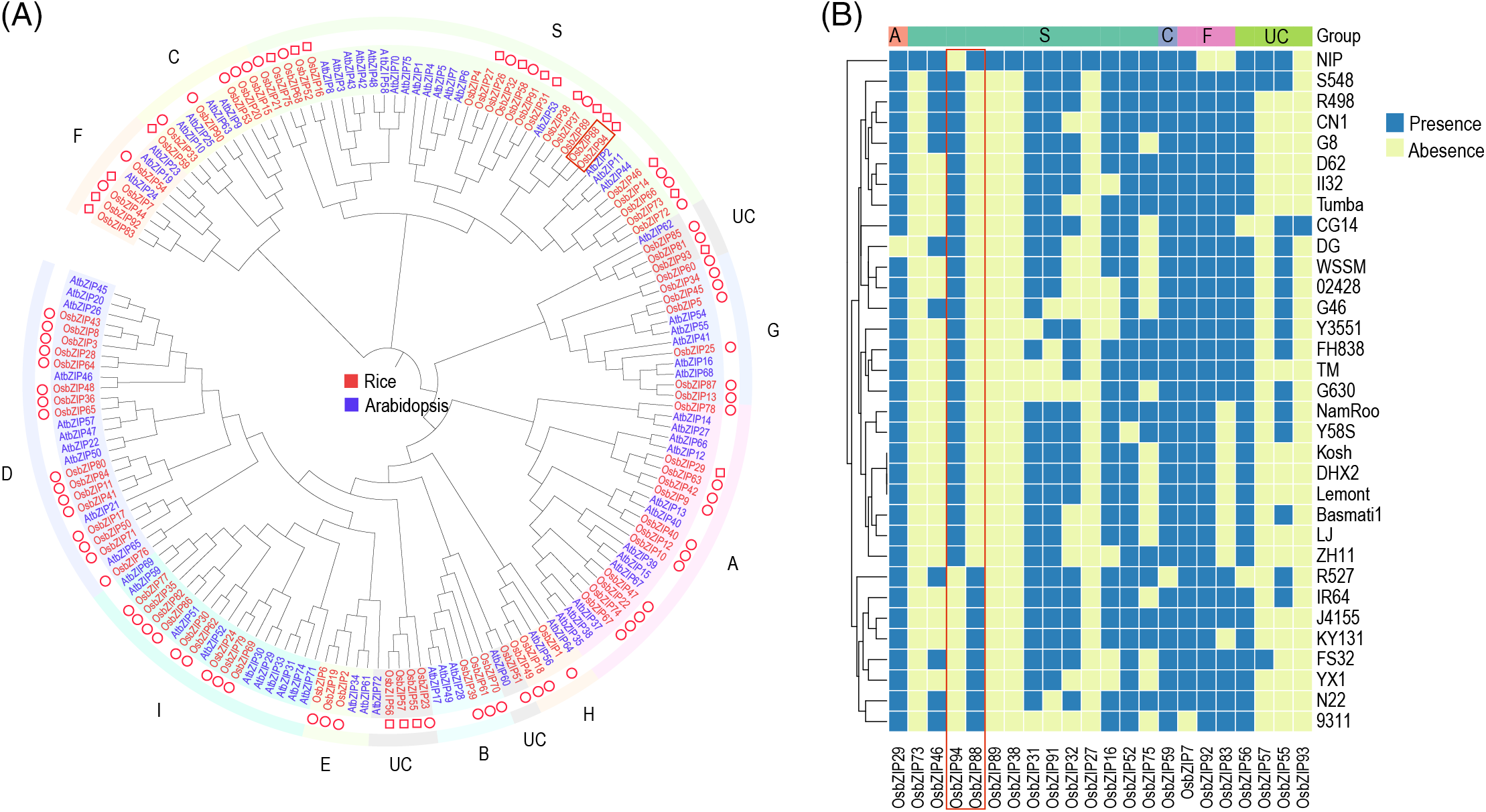
Figure 1: Phylogenetic analysis of bZIP. (A) The phylogenetic tree of the bZIP gene family in rice and Arabidopsis. The square and circle in front of OsbZIP represent variable genes and core genes, respectively. The characters around the tree are the subgroups of bZIPs. (B) The heatmap of 22 variable OsbZIPs present and absent in 33 accessions. The genes in the red box demonstrate the possible mutual exclusivity of PAVs
In the PAV analysis (Fig. 1B), groups A and C contain only one variable gene, the least number in those groups containing variable genes. These two genes are lost in only one accession (OsbZIP29 in group A only lost in DG, and OsbZIP59 in group C only lost in R527), indicating that among all the groups containing variable genes, groups A and C were the most stable groups during domestication and improvement of rice. Several OsbZIPs (OsbZIP73, OsbZIP89, OsbZIP38, and OsbZIP93) exist only in one accession, suggesting that these genes may be related to the specific traits of the accessions. Among the 33 accessions, the absence frequency of OsbZIPs in NIP was the lowest (only 4 OsbZIPs). Thirteen genes were lost in G630 and 9311, and the gene absence frequency was the highest in all varieties, which may lead to the differentiation of OsbZIP-related traits between NIP, G630 and 9311.
3.2 Evolutionary Analysis of bZIPs in 33 Rice Accessions
To explore the selection pressure of OsbZIPs in 33 rice accessions, the Ka/Ks values for the homologous sequence of each OsbZIP in 33 accessions were calculated (Fig. 2). The Ka/Ks values of OsbZIP76 in 02428 and the other 9 accessions are greater than 700; thus, OsbZIP76 was not shown in the figure here, indicating that OsbZIP76 was rapidly evolving due to selection. The Ka/Ks values of most OsbZIPs are between 0 and 1, and the Ka/Ks values of OsbZIP1, OsbZIP37, and OsbZIP85 are far less than 1, indicating that these three genes are stable during evolution and may play a fundamental role in plant growth and development as housekeeping genes; thus, eliminating harmful allelic mutations and under negative purifying selection during evolution. The peaks of Ka/Ks values for OsbZIP10, OsbZIP34, OsbZIP59, OsbZIP62, and OsbZIP84 were located to the right of 1, suggesting that these genes were under positive selection during the domestication of the 33 rice accessions. Out of 94 OsbZIPs, 66 genes do not exhibit Ka/Ks values greater than 1, suggesting that the majority of OsbZIPs underwent purifying selection. The proportion of 28 OsbZIPs with Ka/Ks values greater than 1 in all gene pairs was shown (Fig. 2B). Seven genes (OsbZIP10, OsbZIP14, OsbZIP34, OsbZIP59, OsbZIP62, OsbZIP74, and OsbZIP91) contributes significantly to the proportion of Ka/Ks values greater than 1, suggesting that these genes experienced higher selection pressure during the differentiation process among the 33 accessions and may be undergoing rapid divergence.
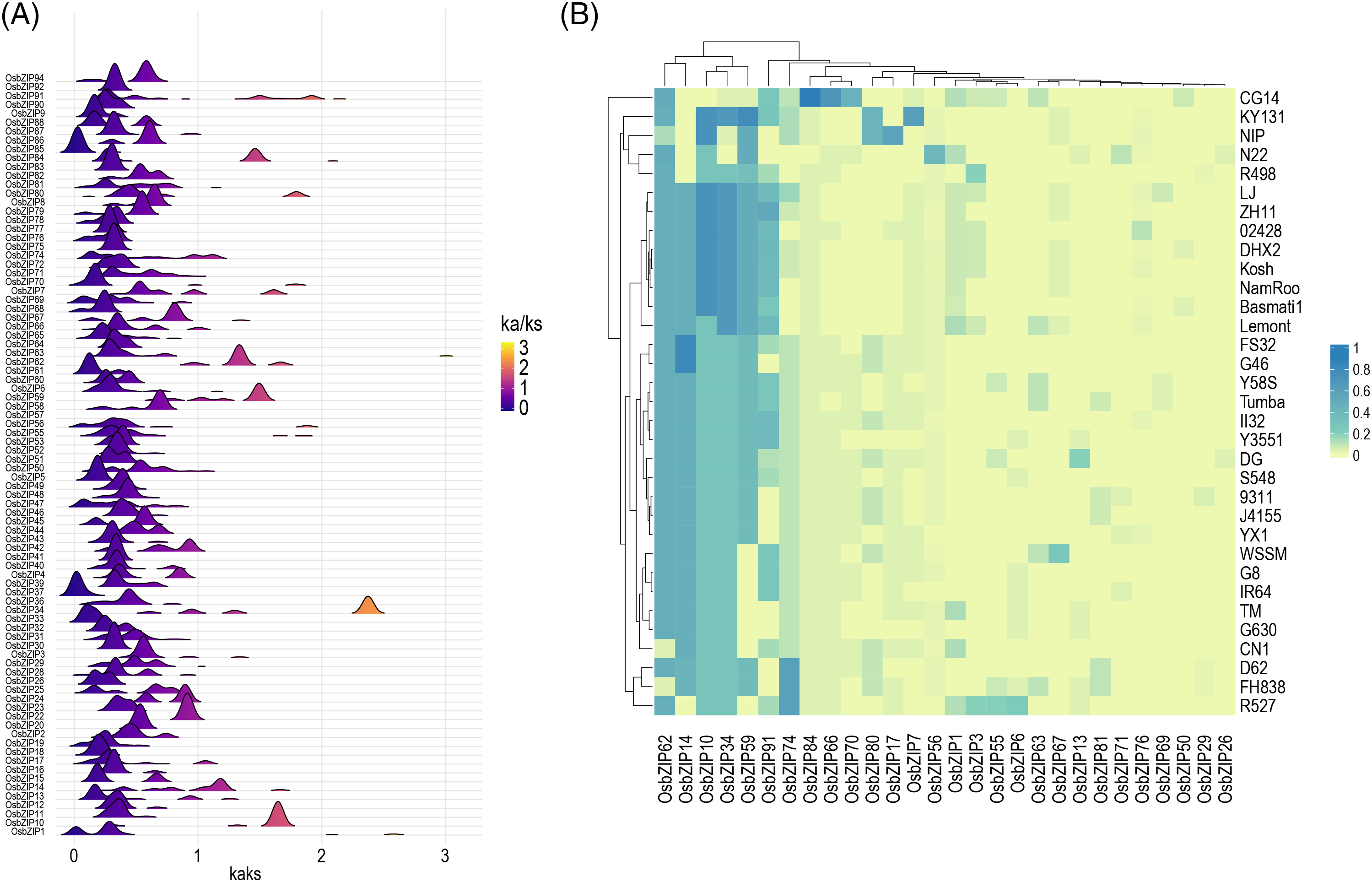
Figure 2: Ka/Ks values of bZIPs in 33 rice accessions. (A) Distribution of Ka/Ks values of bZIPs in 33 accessions. (B) Heatmap of the proportion of bZIPs with Ka/Ks values greater than 1 in all gene pairs
3.3 Analysis of OsbZIPs Expression Level under the Influence of SVs
Structural Variation is the main source of genetic variation and is closely related to various plant phenotypes [29,30]. Qin et al. [19] identified abundant SVs by aligning 32 high-quality rice genomes with the reference genome (NIP). One hundred and seventy SVs overlap with the 2 kbp downstream, 2 kbp upstream, coding region, and intron regions of the 43 OsbZIPs (Table S2). Aligned with the reference genome, these SVs are classified into deletion, insertion, inversion, and translocation. SVs influence the expression of neighboring genes by either modifying gene sequences or impacting regulatory sequences [31,32]. The effect of SVs on the expression of OsbZIPs was shown based on the RNA-seq data from roots and stems of 33 accessions provided in the study of Qin et al. [19] (Fig. 3, Table S3). Insertion or deletion SVs were located in the intro, 2 kbp upstream, 2 kbp downstream, and coding region of OsbZIP17, OsbZIP18, OsbZIP29, OsbZIP36, OsbZIP57, OsbZIP58, and OsbZIP77 (Fig. 3A), leading to notable alterations in the expression of these genes either in the root or shoot (Fig. 3C). Furthermore, the ratio of SVs that significantly modified the expression levels of OsbZIPs to all SVs overlapping with OsbZIPs was calculated. The results revealed that 4% of SVs overlapping with OsbZIPs significantly altered the expression levels of OsbZIPs (Fig. 3B). These results suggested that the occurrence of some SVs can significantly alter gene expression levels.
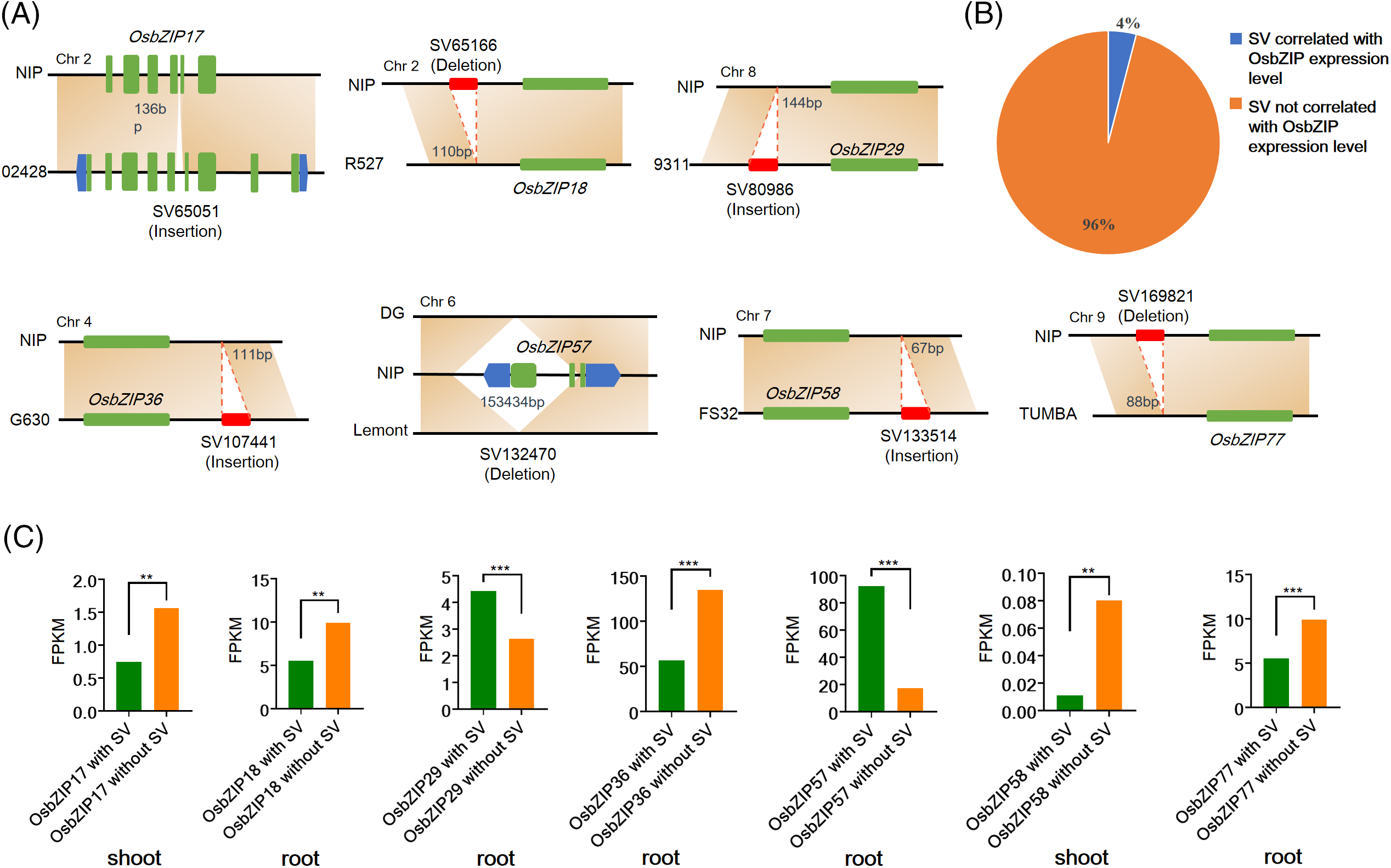
Figure 3: SVs altered the expression level of OsbZIPs. (A) The deletion or insertion of SVs in the OsbZIPs. (B) The proportion of the number of SVs that significantly altered the OsbZIPs expression level to the number of all SVs that overlapped with OsbZIPs. (C) The expression level of OsbZIPs which expression level are significantly altered by SVs (***p < 0.001, **p < 0.01, Student’s t-test)
3.4 Changes in the Gene Structure of the OsbZIP Gene Family under the Influence of SVs
To further investigate the effect of SVs on the gene structure of OsbZIPs, TBtools was used to analyze the gene structure of OsbZIPs whose expression levels were significantly altered by SVs (Fig. 4 and Figure S1). The gene structures of SV-influenced OsbZIPs were different in different varieties. Two genes (OsbZIP29 in 9311 and OsbZIP36 in Tumba) do not contain any motif, and thus these two genes cannot be identified in the reference genome-based gene family identification and would be neglected in functional studies. This problem may be avoided by identifying gene families with the gene-based pan-genome and making the gene family members becoming more comprehensive by colinear relation among 33 rice accessions (Table S4). It could provide powerful information for assigning functions to OsbZIPs. Moreover, due to the occurrence of SVs, some genes, such as OsbZIP29 in D62 and R498, were undergoing gene truncations. This may lead to interesting changes in gene functions, such as pseudogenization or the emergence of new functions [33], which deserve further study.
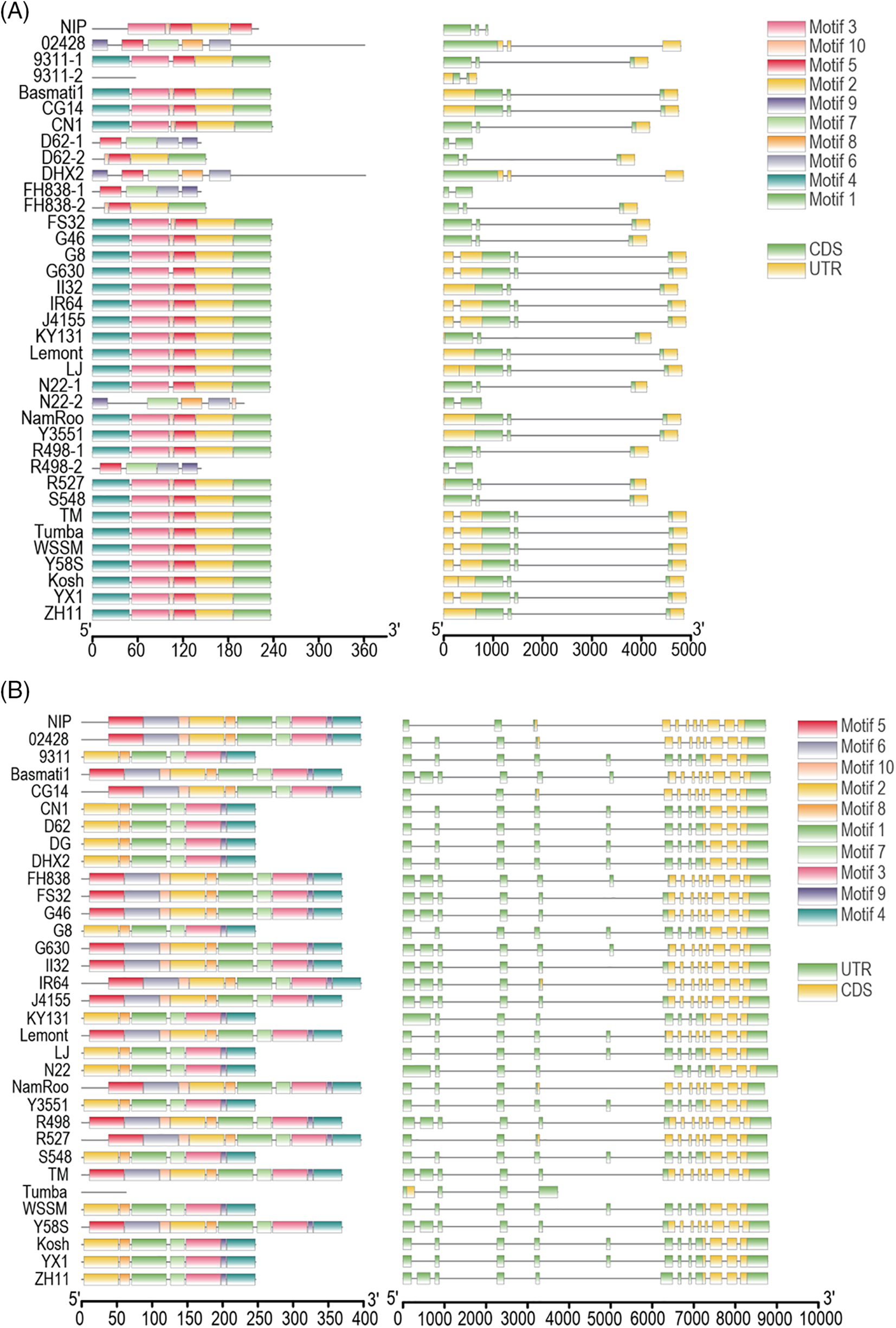
Figure 4: The gene structures of OsbZIP29 (A) and OsbZIP36 (B) in 33 rice accessions
3.5 Motifs of OsbZIPs Were Affected by SVs
To investigate how SVs impact the conserved domains of OsbZIPs across different varieties, the motifs of OsbZIPs in FS32, which has the highest number of SV overlapped OsbZIPs, were displayed (Fig. 5A). Four of the 10 motifs of OsbZIPs in FS32 did not correspond with the reference. The amino acids in each set of mutually corresponding motifs, as well as the consistency of each mutually corresponding amino acid are not the same. These results indicate that the motifs of OsbZIPs were strongly influenced by SVs. As gene family identification relies on conserved domain analysis, alterations in these domains often lead to the neglect of these genes. Hence, we counted typical genes (containing conserved domains) and atypical genes (lacking conserved domains) in each accession (Fig. 5B). Six OsbZIPs (OsbZIP25, OsbZIP31, OsbZIP48, OsbZIP81, OsbZIP88, and OsbZIP90) had both typical and atypical genes in most varieties; while OsbZIP52 was present as atypical genes in 32 varieties except N22 which had both typical and atypical genes. These results suggest that SVs affect the conserved domain of OsbZIPs and lead to the emergence of a large number of atypical genes.
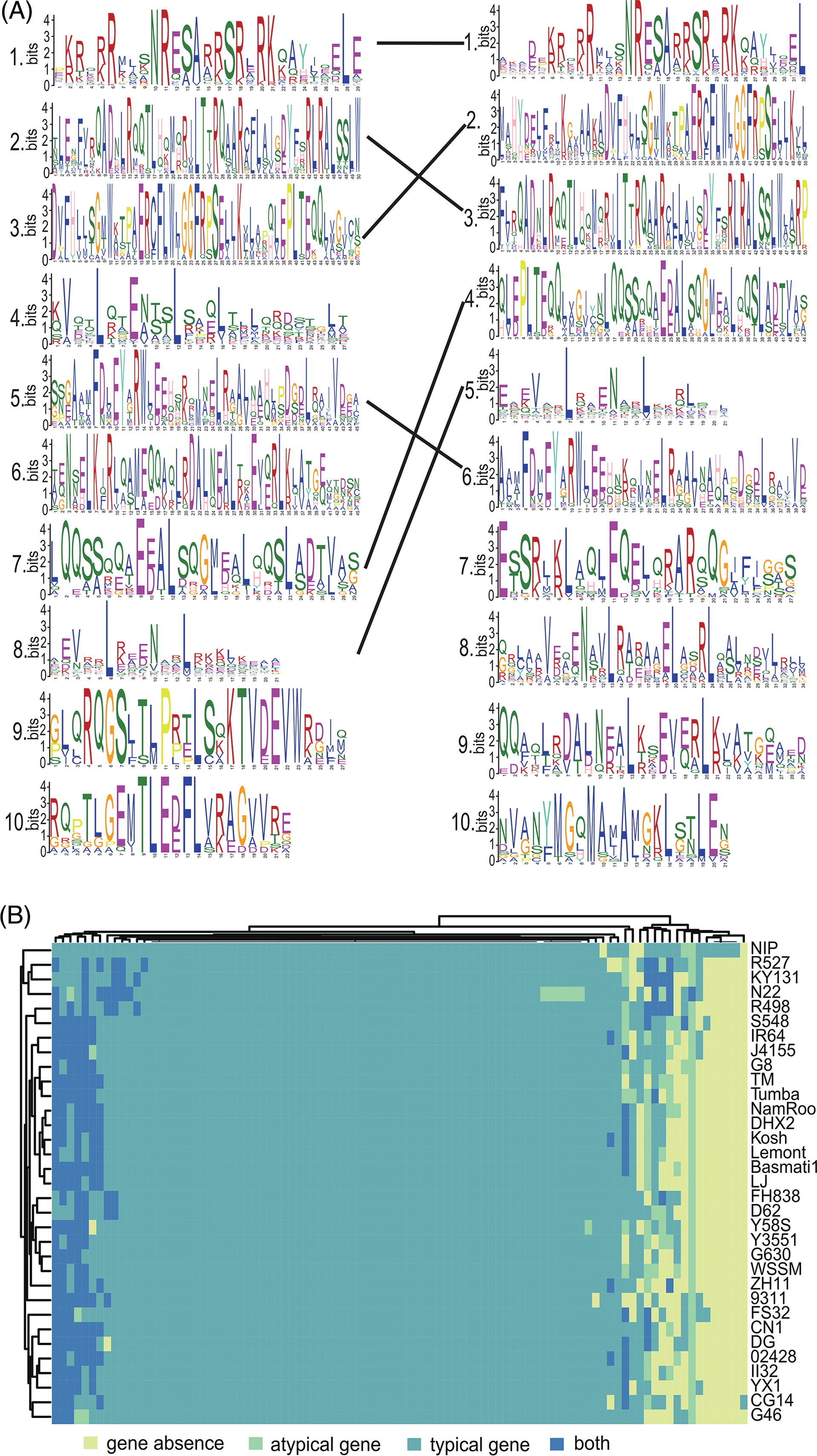
Figure 5: Structural variations affect the conserved domain of OsbZIPs. (A) The weblogos of OsbZIPs in FS32 are shown on the left, and the weblogos of the reference (90 OsbZIPs from NIP and 4 OsbZIPs from CG14) are shown on the right. The weblogos connected by a line represent they are corresponding and the weblogos are arranged in ascending order of E-value. (B) The heatmap showed the OsbZIPs are typical genes or atypical genes or lost in 33 accessions. The term “both” indicates the presence of both typical and atypical genes in each accession
3.6 The Expression of OsbZIPs that Respond to the Combined Stress of Bacterial Blight and High-Temperature
Rice bacterial blight (BB) is caused by Xoo infection, which leads to a reduction in rice yield [34]. Increased temperature due to global warming suppresses host plant defense against the pathogen. Dossa et al. [35] infected a susceptible line (IR24) and a resistant line (IRBB67) with Xoo under high temperature and performed RNA-seq at 3, 72 and 120 hpi. To explore the response of OsbZIPs to BB, the gene expression levels of IR24 and IRB667 at different times of mock and Xoo inoculation were compared and there were 34 OsbZIPs genes were differentially expressed (Fig. 6, Table S5). Compared to the genes with differential expression in Xoo-infected and mock, there were more genes (44) with differential expression at different time points post-infection than Xoo-infected or mock (25), indicating a more pronounced response of OsbZIP to Xoo infection at different infection times (Figs. 6B–6F). In the comparison between Xoo-infected and mock groups across various temperatures and samples at 3 hpi (Figs. 6C–6F), a higher number of differentially expressed OsbZIPs were observed, suggesting a critical response period for certain OsbZIPs to Xoo at 3 hpi. Moreover, Only OsbZIP75 showed differential expression among different varieties, indicating that OsbZIPs’ response to Xoo was minimally influenced by sample variations. Only 3 OsbZIPs exhibited differential expression at 120 hpi in IR24 under high vs. low temperature conditions, suggesting a limited impact of temperature variations on OsbZIPs’ response to Xoo infection in IR24.
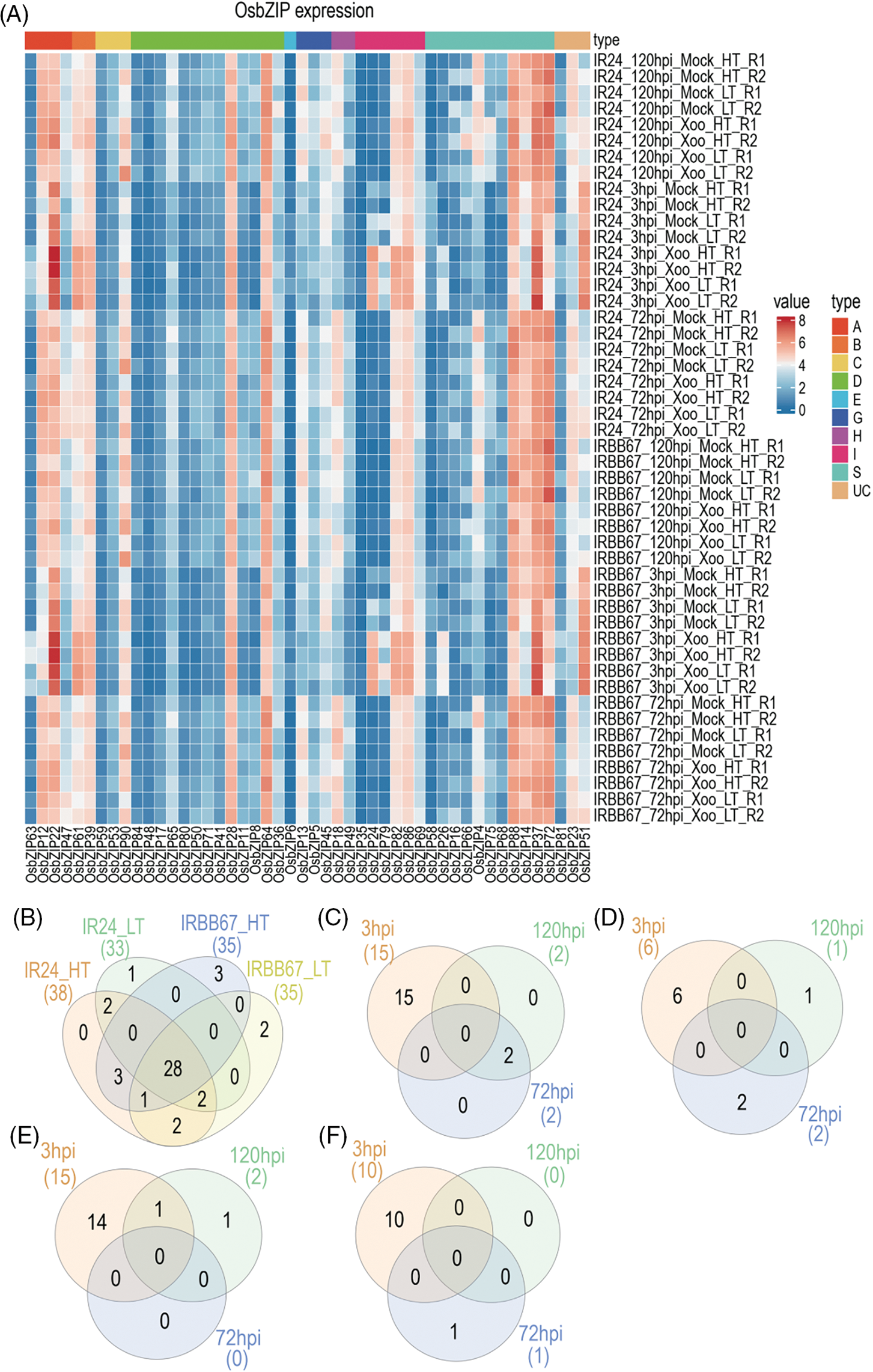
Figure 6: Transcriptome analysis of OsbZIPs responses to Xoo under high temperature. (A) Heatmap of the differentially expressed OsbZIPs. (B–F) respectively present Venn diagrams of: Differential expression of OsbZIPs at different hours points post-infection; IR24 under high temperature after the inoculation of Xoo; IR24 under low temperature after the inoculation of Xoo; IRBB67 under high temperature after the inoculation of Xoo; IRBB67 under low temperature after the inoculation of Xoo
Nijhawan et al. [24] identified 89 bZIP gene family members in rice based on the TIGR (release 5) reference genome. Our study identified 94 OsbZIPs based on the gene-based pan-genome. The higher quality rice genome used as a reference in this study ensured that the identification of the OsbZIP gene family was more accurate compared with the earlier study [24]. Furthermore, four of the newly discovered OsbZIPs were lost from the reference genome. According to the PAV information of the gene, this study found that OsbZIP94 and OsbZIP88 are a pair of absent mutually exclusive genes. Figs. 1A and 1B show that the two genes have a very close evolutionary relationship, so their functions may be very close. They were not absent in more than one variety, which indicates that this pair of genes may complement each other in function. Xu et al. created single, double, three, four, and five mutants of the ALOG gene, which produced a phenotypic continuum with continuous gradient changes in flowering time from normal to extremely early flowering and inflorescence structure from multi-flower to single flower, and finally found five homologous genes in the functional division and cooperation [36]. The functional division and cooperation between OsbZIP94 and OsbZIP88 found in this study are worthy of further study.
A large number of atypical OsbZIP genes were found in the comprehensive study of the colinear relationships of 33 rice genomes [19]. Gene family identification is based on searching for conserved domains in protein-coding genomic sequences. However, atypical genes lacking complete conserved domains cannot be identified based on a single reference genome, hindering subsequent functional studies. For example, AtVRLK1 is a member of the RLK gene family involved in secondary cell wall thickening. Single and triple mutants of AtVRLK1 had a weak phenotype and a strong phenotype only in dominant-negative mutants [37], suggesting that the functional genes may be other RLK gene family members, which could not be identified due to their lack of RLK conserved domains. In chrysanthemums, CmMYB012 suppresses the biosynthesis of flavonoids and anthocyanins in response to high temperatures. This gene was discovered by yeast one-hybrid, not by gene family identification through conserved domain searching, as it does not contain SG7 or SG7–2 conserved sequences [38]. Thus, the large number of atypical OsbZIPs identified in this paper is important for subsequent functional studies.
Gene truncation may lead to phenotype changes; for example, truncation of RBM12 is associated with a psychiatric phenotype [39]. Individuals with microcephaly and mental retardation exhibited gene truncation of ASPM [40]. Gene-truncation-related research to date has focused mainly on medicine, and there is a lack of research on gene truncation in plants. In this study, OsbZIPs were identified based on the gene-based pan-genome. Using SV information contained in the pan-genome, gene truncation was discovered by analyzing the gene structures of OsbZIPs overlapped with SVs. Gene truncation occurred in OsbZIP29 in 9311 and OsbZIP36 in Tumba, and these two genes do not contain any motif (Fig. 4). Since these genes undergo gene truncation leading to changes in conserved domains, their function may dramatically change. However, whether these genes give rise to new functions in rice due to gene truncation requires further functional studies.
Previous studies have shown that the bZIP gene family is involved in a large number of biotic and abiotic stress responses. In grapes, expression of VvbZIP23 was strongly induced by drought, high temperature, low salt, ethylene, JA, and SA [41]. The expression of PPI1, which is homologous with bZIP in pepper, was induced during pathogen infection [42]. To explore the role of OsbZIPs in response to Xoo at high temperatures, the RNA-seq data (accession No. GSE79011) of two rice varieties (a susceptible line IR24 and a resistant line IRBB67) under the different temperatures of mock and Xoo infection at 3, 72 hpi, as well as 120 hpi was analyzed. A large number of differentially expressed OsbZIPs appeared in 3 hpi vs. mock, 3 hpi vs. 72 hpi, and 3 hpi vs. 120 hpi. However, there were few OsbZIPs differentially expressed in 120 hpi vs. 72 hpi and 120 hpi vs. mock (Figure S2). These results indicate that OsbZIPs mainly respond to Xoo infection at an early stage. Studies have shown that high temperatures may alter the expression of resistance genes [43]. High temperature reduced the resistance of IRBB NILs with Xa4 to Xoo, but the resistance of IRBB7 with Xa7, to Xoo was increased [44]. IRBB67, which carries two R genes (Xa4 and Xa7), has broader resistance [45]. In this study, only IR24 showed a few differently expressed OsbZIPs when comparing high temperature with low temperature (Fig. 6C), indicating that the high temperature had a greater effect on OsbZIPs in response to Xoo infection in the susceptible line (IR24). However, the high temperature did not alter the expression of OsbZIPs in IRBB67, suggesting that the mechanism of the OsbZIPs resisting Xoo may not be consistent with that of the R genes (Xa4/Xa7). In conclusion, this study identified abundant atypical members of the rice bZIP gene family within the pan-genome, offering resources for the functional investigation of rice bZIP genes.
Acknowledgement: This work was supported by the National Natural Science Foundation of China, the Major Science and Technology Projects in Anhui Province, Earmarked Fund for China Agriculture Research System, the Special Funds for Supporting Innovation and Entrepreneurship for Returned Oversea-Students in Anhui Province, and Anhui Provincial Natural Science Foundation.
Funding Statement: The National Natural Science Foundation of China (31871964, 31801738, and 32100352), the Major Science and Technology Projects in Anhui Province (202003a06020009), Earmarked Fund for China Agriculture Research System (CARS-01-40), the Special Funds for Supporting Innovation and Entrepreneurship for Returned Oversea-Students in Anhui Province (2020LCX035), and Anhui Provincial Natural Science Foundation (Youth Project, 2008085QC148).
Author Contributions: YS, and XW conceived and designed the experiments. YS, JW, QNW, WQX, and XW contributed to paper writing. YS, WQX, YCH, JW, and QNW conducted the experiment. YS, YCH, XW, and BJ contributed to the data analysis.
Availability of Data and Materials: The datasets used and analyzed during the current study are available in the manuscript and its additional files.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.048151.
References
1. Riechmann AJL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among Eukaryotes. Science. 2000;290:2105–10 [Google Scholar] [PubMed]
2. Landschulz WH, Johnson PF, Mcknight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–64 [Google Scholar] [PubMed]
3. Lee SC, Choi HW, Hwang IS, Choi DS, Hwang BK. Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta. 2006;224:1209–25 [Google Scholar] [PubMed]
4. van Leene J, Blomme J, Kulkarni SR, Cannoot B, de Winne N, Eeckhout D, et al. Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development. J Exp Bot. 2016;67:5825–40 [Google Scholar] [PubMed]
5. Muñiz García MN, Cortelezzi JI, Fumagalli M, Capiati DA. Expression of the Arabidopsis ABF4 gene in potato increases tuber yield, improves tuber quality and enhances salt and drought tolerance. Plant Mol Biol. 2018;98:137–52. [Google Scholar]
6. He Q, Cai H, Bai M, Zhang M, Chen F, Huang Y, et al. A soybean bzip transcription factor gmbzip19 confers multiple biotic and abiotic stress responses in plant. Int J Mol Sci. 2020;21(13):1–19. doi:10.3390/ijms21134701 [Google Scholar] [PubMed] [CrossRef]
7. Meng XB, Zhao WS, Lin RM, Wang M, Peng YL. Identification of a novel rice bZIP-type transcription factor gene, OsbZIP1, involved in response to infection of Magnaporthe grisea. Plant Mol Biol Report. 2005;23(3):301–2. doi:10.1007/BF02772762. [Google Scholar] [CrossRef]
8. Bi C, Yu Y, Dong C, Yang Y, Zhai Y, Du F, et al. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol J. 2021;19(2):209–11. doi:10.1111/pbi.13453 [Google Scholar] [PubMed] [CrossRef]
9. Hsieh AT, Li C, Su R, Cheng C, Tsai C, Chan M, et al. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta. 2010;231(6):1459–73. doi:10.1007/s00425-010-1147-4 [Google Scholar] [PubMed] [CrossRef]
10. Tajima H, Iwata Y, Iwano M, Takayama S, Koizumi N. Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem Biophys Res Commun. 2008;374(2):242–7. doi:10.1016/j.bbrc.2008.07.021 [Google Scholar] [PubMed] [CrossRef]
11. Zou M, Guan Y, Ren H, Zhang F, Chen F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol. 2008;66(6):675–83. doi:10.1007/s11103-008-9298-4 [Google Scholar] [PubMed] [CrossRef]
12. Zhao BY, Hu YF, juan Li J, Yao X, de Liu K. BnaABF2, a bZIP transcription factor from rapeseed (Brassica napus L.enhances drought and salt tolerance in transgenic Arabidopsis. Bot Stud. 2016;57(1):12. doi:10.1186/s40529-016-0127-9 [Google Scholar] [PubMed] [CrossRef]
13. Lai K, Lorenc MT, Lee HC, Berkman PJ, Bayer PE, Visendi P, et al. Identification and characterization of more than 4 million intervarietal SNPs across the group 7 chromosomes of bread wheat. Plant Biotechnol J. 2015;13(1):97–104. doi:10.1111/pbi.12240 [Google Scholar] [PubMed] [CrossRef]
14. Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, et al. A first-generation haplotype map of Maize. Science. 2009;326(5956):1115–7. doi:10.1126/science.1177837 [Google Scholar] [PubMed] [CrossRef]
15. Saxena RK, Edwards D, Varshney RK. Structural variations in plant genomes. Brief Funct Genomics. 2014;13(4):296–307. doi:10.1093/bfgp/elu016 [Google Scholar] [PubMed] [CrossRef]
16. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial pan-genome. Proc Natl Acad Sci. 2005;102:13950–5 [Google Scholar] [PubMed]
17. Gao L, Gonda I, Sun H, Ma Q, Bao K, Tieman DM, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet. 2019;51:1044–51 [Google Scholar] [PubMed]
18. Tang D, Jia Y, Zhang J, Li H, Cheng L, Wang P, et al. Genome evolution and diversity of wild and cultivated potatoes. Nature. 2022;606:535–41 [Google Scholar] [PubMed]
19. Qin P, Lu H, Du H, Wang H, Chen W, Chen Z, et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell. 2021;184:3542–3558.e16 [Google Scholar] [PubMed]
20. Dröge-Laser W, Snoek BL, Snel B, Weiste C. The Arabidopsis bZIP transcription factor family—an update. Curr Opin Plant Biol. 2018;45:36–49. [Google Scholar]
21. Wang Z, Yan L, Wan L, Huai D, Kang Y, Shi L, et al. Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genomics. 2019;20:1–14. [Google Scholar]
22. Zhang M, Liu Y, Shi H, Guo M, Chai M, He Q, et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genomics. 2018;19:1–14. [Google Scholar]
23. Pan Y, Hu X, Li C, Xu X, Su C, Li J, et al. SlbZIP38, a tomato bZIP family gene downregulated by abscisic acid, is a negative regulator of drought and salt stress tolerance. Genes. 2017;8:402 [Google Scholar] [PubMed]
24. Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–50 [Google Scholar] [PubMed]
25. Price MN, Dehal PS, Arkin AP. Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50 [Google Scholar] [PubMed]
26. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60 [Google Scholar] [PubMed]
27. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550 [Google Scholar] [PubMed]
28. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9 [Google Scholar] [PubMed]
29. Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14:125–38 [Google Scholar] [PubMed]
30. Lye ZN, Purugganan MD. Copy number variation in domestication. Trends Plant Sci. 2019;24:352–65 [Google Scholar] [PubMed]
31. Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, et al. Major impacts of widespread structural variation on gene expression and crop improvement in Tomato. Cell. 2020;182(1):145–161.e23. doi:10.1016/j.cell.2020.05.021 [Google Scholar] [PubMed] [CrossRef]
32. Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, et al. Pan-genome of wild and cultivated Soybeans. Cell. 2020;182(1):162–176.e13. doi:10.1016/j.cell.2020.05.023 [Google Scholar] [PubMed] [CrossRef]
33. Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183(3):557–64. doi:10.1111/j.1469-8137.2009.02923.x [Google Scholar] [PubMed] [CrossRef]
34. Mew TW. Focus on bacterial blight of rice. Plant Dis. 1993;77(1):5–12. doi:10.1094/PD-77-0005. [Google Scholar] [CrossRef]
35. Dossa GS, Quibod I, Atienza-Grande G, Oliva R, Maiss E, Vera Cruz C, et al. Rice pyramided line IRBB67 (Xa4/Xa7) homeostasis under combined stress of high temperature and bacterial blight. Sci Rep. 2020;10(1):1–15. doi:10.1038/s41598-020-57499-5 [Google Scholar] [PubMed] [CrossRef]
36. Huang X, Xiao N, Zou Y, Xie Y, Tang L, Zhang Y, et al. Heterotypic transcriptional condensates formed by prion-like paralogous proteins canalize flowering transition in tomato. Genome Biol. 2022;23(1):1–21. doi:10.1186/s13059-022-02646-6 [Google Scholar] [PubMed] [CrossRef]
37. Huang C, Zhang R, Gui J, Zhong Y, Li L. The receptor-like kinase AtVRLK1 regulates secondary cell wall thickening. Plant Physiol. 2018;177(2):671–83. doi:10.1104/pp.17.01279 [Google Scholar] [PubMed] [CrossRef]
38. Zhou LJ, Geng Z, Wang Y, Wang Y, Liu S, Chen C, et al. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic Res. 2021;8(1):248. doi:10.1038/s41438-021-00675-z [Google Scholar] [PubMed] [CrossRef]
39. Steinberg S, Gudmundsdottir S, Sveinbjornsson G, Suvisaari J, Paunio T, Torniainen-Holm M, et al. Truncating mutations in RBM12 are associated with psychosis. Nat Genet. 2017;49(8):1251–4. doi:10.1038/ng.3894 [Google Scholar] [PubMed] [CrossRef]
40. Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, et al. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet. 2003;73(5):1170–7. doi:10.1086/379085 [Google Scholar] [PubMed] [CrossRef]
41. Tak H, Mhatre M. Cloning and molecular characterization of a putative bZIP transcription factor VvbZIP23 from Vitis vinifera. Protoplasma. 2013;250:333–45 [Google Scholar] [PubMed]
42. Lee SJ, Lee MY, Yi SY, Oh SK, Choi SH, Her NH, et al. PPI1: a novel pathogen-induced basic region-leucine zipper (bZIP) transcription factor from pepper. Mol Plant-Microbe Interact. 2002;15:540–8 [Google Scholar] [PubMed]
43. Dossa GS, Torres R, Henry A, Oliva R, Maiss E, Cruz CV, et al. Rice response to simultaneous bacterial blight and drought stress during compatible and incompatible interactions. Eur J Plant Pathol. 2017;147:115–27. [Google Scholar]
44. Webb KM, Oña I, Bai J, Garrett KA, Mew T, Vera Cruz CM, et al. A benefit of high temperature: increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 2010;185:568–76 [Google Scholar] [PubMed]
45. Suh JP, Jeung JU, Noh TH, Cho YC, Park SH, Park HS, et al. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice. 2013;6:1–11. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools