 Open Access
Open Access
ARTICLE
FLOWERING LOCUS T (FT) in Photosensitive Type Chrysanthemum Accelerates Flowering in Arabidopsis
College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, China
* Corresponding Author: Yanhong Guo. Email:
Phyton-International Journal of Experimental Botany 2024, 93(4), 819-830. https://doi.org/10.32604/phyton.2024.049150
Received 29 December 2023; Accepted 21 March 2024; Issue published 29 April 2024
Abstract
FLOWERING LOCUS T (FT) is an important factor for integrating flowering signals through the photoperiod pathway, which significantly promotes flowering. Most chrysanthemum varieties are short-day plants, sensitive to the photoperiod, which limits their ornamental and annual production supply. In this study, we aim to analyze the nutrients and flowering-related genes of chrysanthemums with different photoperiod types and to clone and verify the function of the flowering-related gene CmFT. We found that the formation of floral buds requires the accumulation of starch while consuming soluble sugars and the expression patterns of flowering-related genes GIGANTEA (GI), CONSTANS (CO), and FT in C.morifolium ‘Zilian’ and C. morifolium ‘Zihongtuogui’ had a certain synchronization during floral buds differentiation according to our quantitative validation, and the expression levels of CmGI, CmCO and CmFT in C. morifolium ‘Zihongtuogui’ were higher than those in C. morifolium. ‘Zilian’ in the later stage of differentiation. CmFT was cloned from photosensitive chrysanthemums-C. morifolium ‘Zihongtuogui’ and polypeptide alignment and phylogenetic analysis showed that CmFT was clustered in FT-like subfamily. In further functional verification, we obtained two Arabidopsis transgenic lines. Our results showed that CmFT transgenic ft mutant lines can significantly accelerate flowering in Arabidopsis. Thus, we can initially confirm that CmFT plays an important role in promoting flowering, which may be the key reason for the photosensitivity of C. morifolium ‘Zihongtuogui’. Overall, the results of this study are of great importance in revealing the flowering mechanism of different photoperiod types of chrysanthemums.Keywords
Supplementary Material
Supplementary Material FileNomenclature
| LD | Long day |
| SD | Short day |
| pI | Isoelectric point |
Floral transformation and floral organ formation in higher plants play a central role in species development, reproduction, and evolution. The initiation of flowering in plants is jointly determined by endogenous and environmental signals and also is controlled by intricate regulatory networks [1]. In the model plant Arabidopsis thaliana, at least six major pathways were identified that have been implicated in the control of flowering time: the photoperiod pathway, vernalization pathway, gibberellin-acid pathway, temperature pathway, autonomous pathway, and age pathway [2]. The six pathways finally converge flowering signals to ‘floral integrator genes’, which activate the expression of floral meristem genes and ensure the development of flower organs [3,4].
The flowering of many plants is regulated by photoperiod induction and the photoperiod pathway is one of the most complete and clear approaches. Several genes have been implicated in the photoperiod and occur in a network of the photoperiod pathway, which include GIGANTEA (GI), CONSTANS (CO), PHYTOCHROME A (PHYA), EARLY FLOWERING (ELF3), and FT [5,6]. Of these genes, CO and FT are the two genes that play a central role and participate in multiple biological processes. CO is the first member of the CONSTANS-LIKE (COL) gene found in A. thaliana, which acts as a bridge gene between the circadian clock and the control of flowering [7]. Research showed that the flowering time was related to the natural variation of CTTTACA repeat sequence number of CO promoter in Arabidopsis thaliana. CO encodes a putative transcription factor that is required to promote flowering under LD (Long-day) but not under SD (Short-day) conditions. Several DNA BINDING WITH ONE FIN GER (DOF) transcription factors homologous to CYCLING DOF FACTORs (CDFs) could bind to the CTTT site of CO gene promoter and participate in the inhibition of CO gene transcription in the LD condition [8]. In plant leaves, the photoperiodic pathway mainly transmits signals through GI and CO. As the upstream gene of CO, GI regulates the expression of CO by forming GIFKF1 complex with F-BOX 1 (FKF1) protein, which could reduce the activity of CYCLING DOE FAC-TOR 1 (CDF1), the inhibitor of CO [9,10].
The concept of florigen was first proposed by Chailakhyan in the research on chrysanthemums in 1936. After more than 70 years of continuous exploration and research, FT protein, a factor with the nature of florigen, was finally found in the late flowering mutant of Arabidopsis [11–13]. FT is now a well-known floral integrator gene and acts as the early activation target gene of CO. The function of FT as a floral inducer is conserved in many plant species. In photoperiod-insensitive plants, an orthologue of FT, such as SFT/SP3D could induced in mature leaves and trigger flowering in tomatoes [14]. In addition, as a mobile protein, FT also plays a certain role in plant growth, morphogenesis, and fruit setting [12,14]. Oda isolated three FT-like genes from Chrysanthemum seticuspe and overexpression of CsFTL3 in chrysanthemum could induce flowering under non-inductive conditions, which indicated its potential to induce early flowering [15].
Chrysanthemum morifolium is one of the most important ornamental plants in the world and plays a very important role in the international flower trade. However, chrysanthemum is usually sensitive to the photoperiod, and its flowering period is short and relatively concentrated, which seriously limits its ornamental value and annual production and supply. In this study, C.morifolium ‘Zilian’ (the photoperiod insensitive type) and C. morifolium ‘Zihongtuogui’ (the photosensitive type) were used as test materials to study the effects of nutrients on floral bud differentiation. Specifically, we focused on the molecular mechanism of the photoperiod pathway’s ability to regulate flowering. The results are of great significance in revealing the flowering mechanism of different photoperiod types of chrysanthemums, clarifying the law of the chrysanthemums’ photoperiod and floral bud differentiation, and providing a theoretical basis for precise regulation of florescence and cultivation of new varieties in chrysanthemum production.
2.1 Plant Materials and Growth Conditions
C. morifolium ‘Zilian’ (photoperiod insensitive type) and C.morifolium ‘Zihongtuogui’ (photoperiod sensitive type), crossbred from the same parent, were used as plant materials (Fig. 1). An LED lamp was used to supplement the light for 4 h at night from the time of planting. After rooting, the consistent plants were transferred to pots containing the same substrate. The plants were cultivated in a controlled environment (22 ± 2°C, 16 h of light and 8 h of darkness, 200 μmol·m−2·s−1 light intensity, and 70% relative humidity). Floral buds of two chrysanthemums at 0, 3, 6, 9, 12, and 15 d (Fig. S1) were taken and frozen in liquid nitrogen, then stored in a refrigerator at −80°C for subsequent processing. As samples, the apical buds were taken at about 10 a.m. each time, and each of the three independent plants was mixed into a sample. All plants were grown at the experimental station in Zhongkai University of Agriculture and Engineering, Guangzhou, China.

Figure 1: Characterization of two Chrysanthemum strains. (A) C. morifolium ‘Zilian’ (photoperiod insensitive type). (B) C. morifolium ‘Zihongtuogui’ (photoperiod sensitive type)
2.2 Determination of Soluble Sugar and Starch Contents
We referred to Liu et al. [16] and Debebe et al. [17] to determine the soluble sugar and starch contents of the apical buds with improved anthrone colorimetry. Three repetitions were set for each sample.
2.3 Isolation of CmFT and Sequence Analysis
The sequence information of CmFT was obtained from the C. nankingense genome database (http://210.22.121.250:8880/asteraceae/homePage). The full-length ORF sequence of CmFT (525 bp) was cloned with the primer pair CmFT-F and CmFT-R from the cDNA of C. morifolium ‘Zihongtuogui’. All primers used in this study (Table S1) were designed using the DNAMAN software. Amino acid sequence alignment with functionally reported genes AtFT, AaFT, DzFT, EcFT, GhFT, HaFT, RrFT, and TcFT was performed using DNAMAN software. The construction of a phylogenetic tree was carried out using the MEGA7 software.
2.4 Vector Construction of pNC-Green-SubN-CmFT
The CmFT open reading frame (ORF) was amplified using the primer pair pNC-CmFT-F/R (Table S1). The PCR products and pNC-Green-SubN vector were combined. The generated vector pNC-Green-SubN-CmFT was then transferred into Escherichia coli DH5 α. Kanamycin was used to screen for positive clones. After the sequence was proved correct, we extracted the high-concentration plasmid using E.Z.N.A. Plasmid Mini kit I D6943 (Omega, USA), then transformed the plasmid into Agrobacterium tumefaciens containing pSoup. Finally, we generated constructs, 35S::GFP-CmFT, and stored them in a refrigerator at −80°C.
2.5 Subcellular Localization of CmFT
To determine the localization of CmFT, the constructed vector, pNC-Green-SubN-CmFT was introduced into A. tumefaciens GV3101 via injection and then inoculated into leaves of Nicotiana tabacum. After being cultured for 48–56 h under 16 h light/8 h dark, infiltrated leaf sections were observed under the laser-scanning microscope (TCS SP8, Lecia, Germany) to detect the GFP activity.
2.6 Quantitative Real-Time PCR (qRT-PCR) Analysis
qRT-PCR was used to determine the transcript levels during the different stages of flower bud formation and different tissues at the blooming stage. The cDNA was synthesized from 1 μg of total RNA according to the instructions of the PrimeScriptTM RT reagent Kit with the gDNA Eraser Perfect Real Time (Takara, Dalian, China). The CmEF1α gene was used as the internal reference (Table S1). The SYBR The qRT-PCR reaction system was as follows: 10 μL of SYBR Green Master Mix, 0.4 μl of primer F/R (10 μM), 1 μl of template cDNA, and 8.2 μl of ddH2O. The 2−∆∆Ct method was used to calculate gene relative expression levels and the qRT-PCR procedure was consistent with that used in Zhang et al. [18]. Three biological replicates were used per sample. The sequences of all the gene-specific primers are listed in Table S1.
2.7 Transgenic Arabidopsis Phenotype Observation
A full-length coding sequence of CmFT was cloned into the vector pNC-Cam1304-MCS35S. The floral dip method via A. tumefaciens GV3101-mediated freeze-thaw transformation was used to transform the flora of wild-type Arabidopsis or ft mutants. The presence of transgenes was verified by PCR amplification with targeted primers (Table S1). T2 transgenic Arabidopsis was sown on 1/2 MS solid medium with 50 mg/L of hygromycin, and wild-type Arabidopsis (Col-0) was sown on 1/2 MS without hygromycin as a control. After being placed at 4°C for 2 days, the donor plants were transplanted to the cultivation room under long days (16 h light/8 h dark) at 22 ± 2°C. Transgenic and control plants were grown in a cultivation chamber under 12 h light/12 h dark at 22°C from germination until flowering. To measure flowering time, the number of rosette leaves at bolting was counted.
The contents of soluble sugar and starch and the expression levels of CmFT in different tissues and different stages were determined by statistical variance analysis using the IBM SPSS Statistics 21.0 software. Data were analyzed for significance using Duncan’s method. The differences between mean values were separated at a level of p < 0.05. Origin 17.0 was used for mapping.
3.1 Changes in Soluble Sugar, Starch, and Soluble Protein Contents during Flower Bud Formation
To identify the potential role of soluble sugars, starches, and soluble protein in the process of flower bud morphological differentiation, floral buds of two chrysanthemums at 0, 3, 6, 9, 12, and 15 d (Fig. S1) were taken to determine the soluble sugar, starch, and soluble protein contents. The data showed that the content of soluble sugar in apical buds of C. morifolium ‘Zilian’ rose first and then declined, while that of C. morifolium ‘Zihongtuogui’ also rose first and then declined, which indicates that a large amount of soluble sugar was consumed or transferred in the process of flower bud morphological differentiation (Fig. 2A). The starch content in apical buds of C. morifolium ‘Zilian’ was first decreased and then increased, and the starch contents after 9 d were significantly higher than those before 6 d. The starch content in apical buds of C. morifolium ‘Zihongtuogui’ decreased first, then increased, and finally decreased (Fig. 2B). These results demonstrate that the insensitive type consumes or transfers more soluble sugar and accumulates more starch. The sensitive type also consumes or transfers soluble sugar, while the starch content decreases instead of increasing, which indicates that it needs to consume more starch during flower bud morphological differentiation.
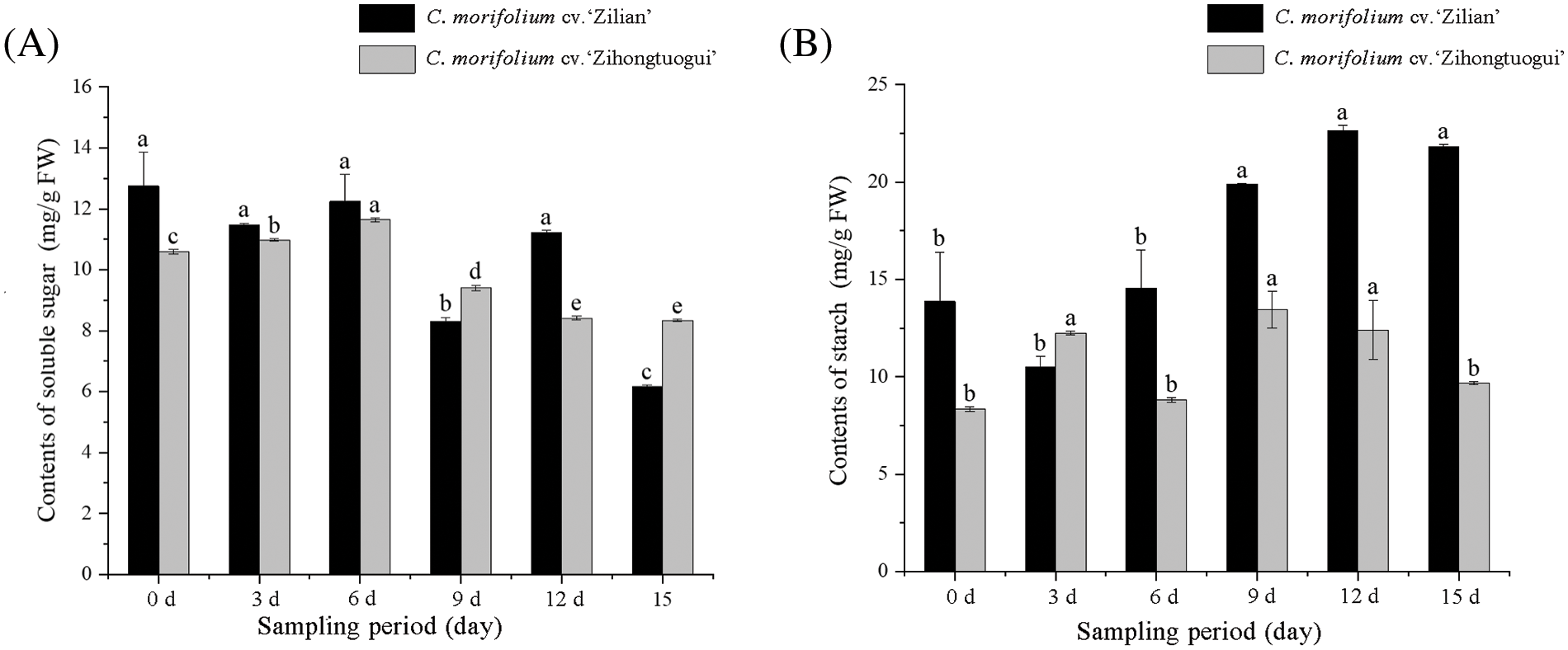
Figure 2: Changes in soluble sugar and starch contents during the flower bud formation in two chrysanthemum varieties. (A) Changes in soluble sugar contents during flower bud formation of C. morifolium cv. ‘Zilian’ and C. morifolium ‘Zihongtuogui’. (B) Changes in starch contents during flower bud formation of C. morifolium ‘Zilian’ and C. morifolium ‘Zihongtuogui’. Different lowercase letters within the same variety indicate significant differences at the 0.05 level during different periods (p < 0.05), the same as below
3.2 Expression Pattern Analysis of CmGI, CmCO and CmFT
Floral buds of two chrysanthemums at 0, 3, 6, 9, 12, and 15 d were used to analyze the gene expression levels of CmGI, CmCO, and CmFT via qRT-PCR. Fig. 3 shows that the expression patterns of CmGI, CmCO, and CmFT are similar, showing a trend of rising first and then falling, in which the expression of CmCO and CmFT reached the highest at 9 d. In general, the expression of CmGI, CmCO, and CmFT in C. morifolium ‘Zihongtuogui’ was higher than that in C. morifolium ‘Zilian’ after 9 d, which indicates that these three genes play an important role in flower bud morphological differentiation in sensitive type.
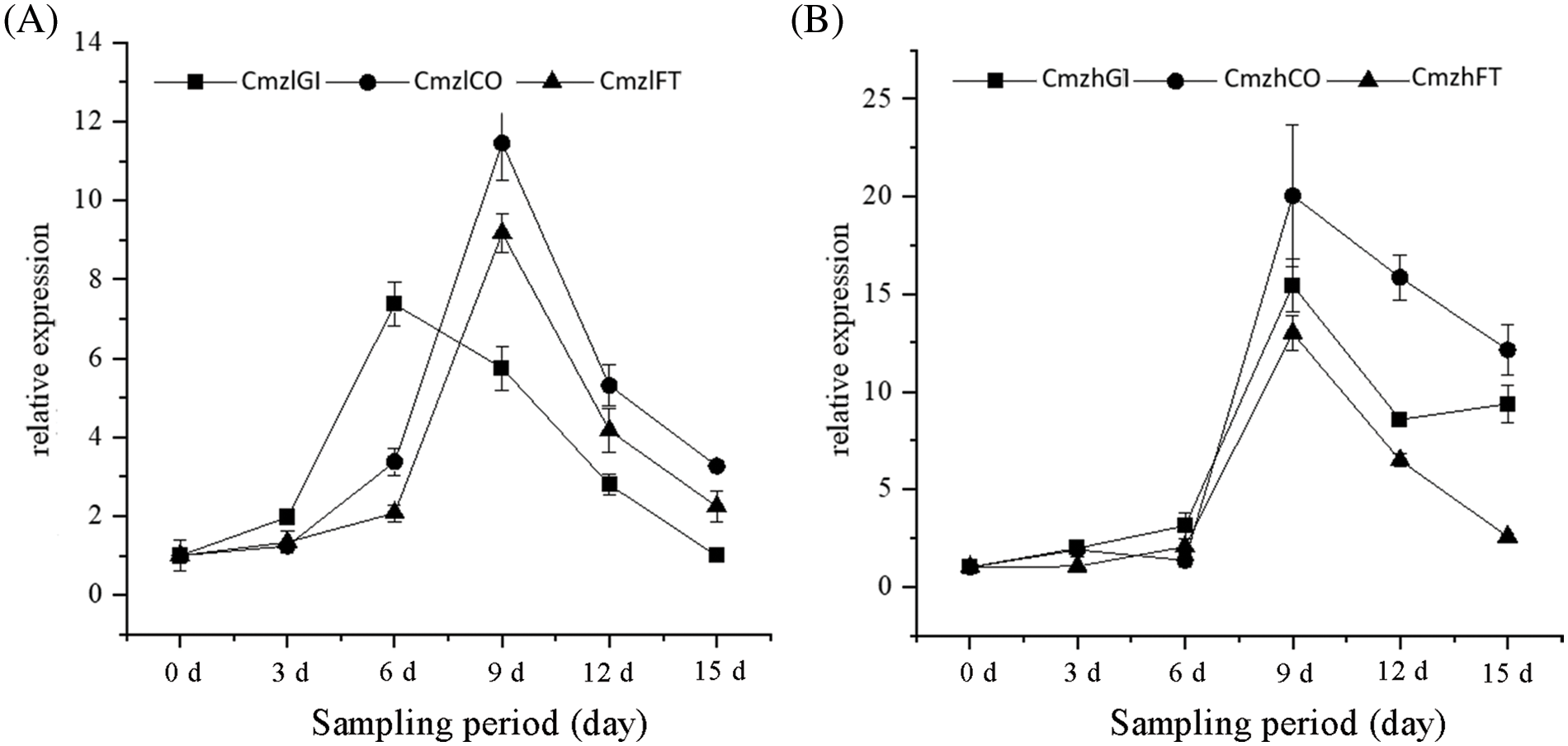
Figure 3: Expression pattern of three flowering-related genes during floral formation in two chrysanthemum varieties. (A) Expression pattern of CmGI, CmCO, and CmFT during floral formation in C. morifolium ‘Zilian’. (B) Expression pattern of CmGI, CmCO, and CmFT during floral formation in C. morifolium ‘Zihongtuogui’. Data are presented as means ± SE (n = 3)
Moreover, samples from the roots, stems, leaves, and flowers at the blooming stage of C.morifolium ‘Zilian’ and C. morifolium ‘Zihongtuogui’ were taken to analyze the gene expression levels of CmGI, CmCO, and CmFT by qRT-PCR. As shown in Fig. 4, CmGI, CmCO, and CmFT were expressed in the roots, stems, leaves, and flowers of two chrysanthemums, and the relative expression levels of different tissues were significantly different. The expression of CmGI was the highest in the roots of both chrysanthemums, while CmCO was the highest in the leaves. Similarly, the expression of CmFT was the highest in leaves, but also in roots. In general, the expression levels of CmFT in roots and leaves of the sensitive type are higher than those of the insensitive type.
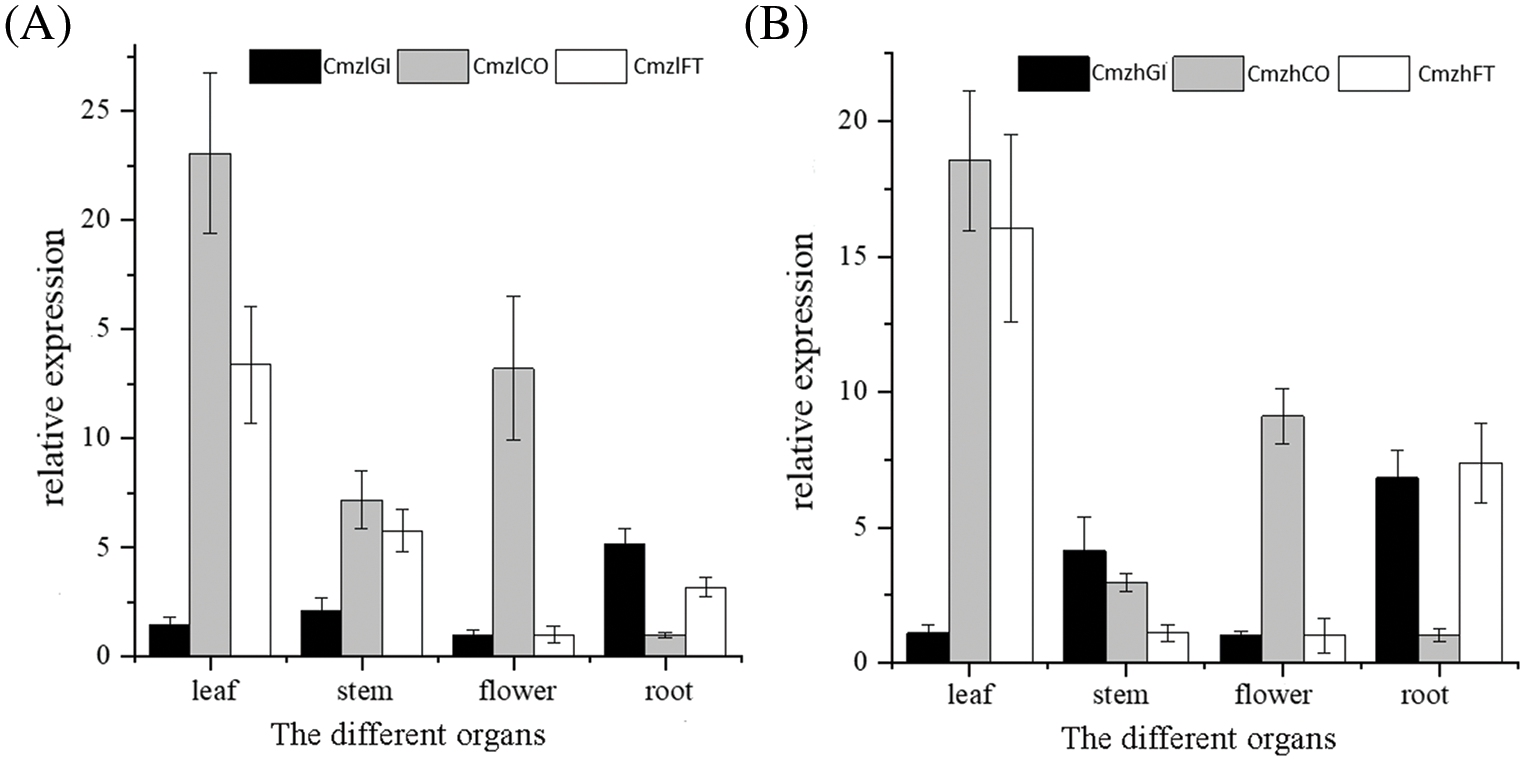
Figure 4: Expression pattern of three flowering-related genes of different tissues at blooming stage in two chrysanthemum varieties. (A) Expression pattern of CmGI, CmCO, and CmFT of different tissues at a blooming stage in C. morifolium ‘Zilian’. (B) Expression pattern of CmGI, CmCO, and CmFT of different tissues at a blooming stage in C. morifolium ‘Zihongtuogui’. Data are presented as means ± SE (n = 3)
3.3 Isolation and Characterization of CmFT
As shown in Fig. 5A, the full-length CmFT sequence was cloned by specific primers (CmFT-F and CmFT-R, Table S1) from C. morifolium ‘Zihongtuogui’ cDNA. Sequence analysis revealed that the open reading frame (ORF) of CmFT was 525 bp, which was predicted to encode 174 amino acid residues with a molecular weight of 19638.25 Da and an isoelectric point (pI) of 6.83. The predicted amino acid sequence of CmFT has a high homology with the protein of other species, and the homology is more than 86%, which is shown in Fig. 5B. A phylogenetic analysis implied that CmFT was most closely related to protein AtFT and AtTSF in A. thaliana (Fig. 5C).
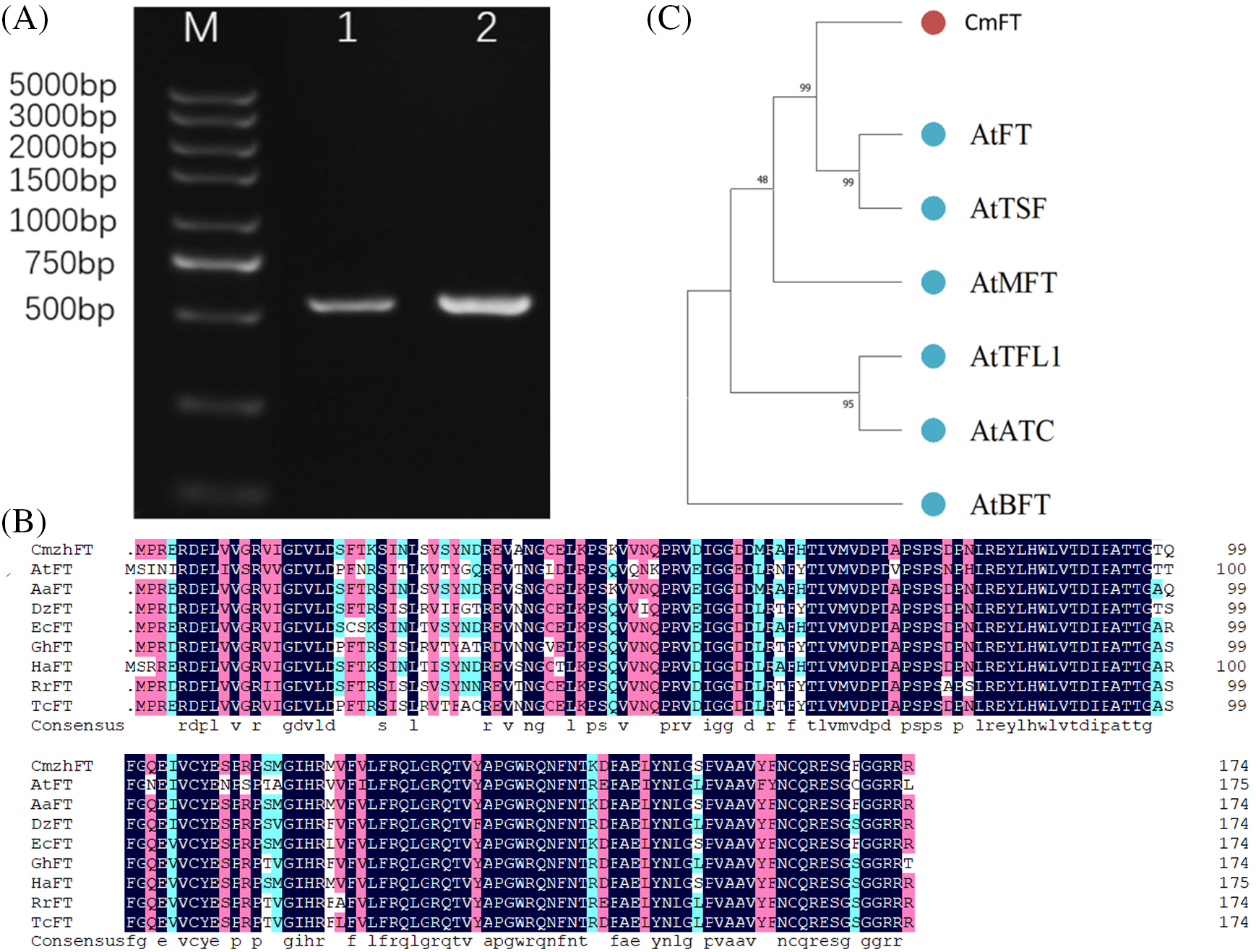
Figure 5: Cloning of the CmFT and sequence analysis of CmFT as well as related FTs. (A) PCR amplification results of CmFT. (B) Polypeptide alignment of CmFT with FTs from A. thaliana (At), Artemisia annua (Aa), Durio zibethinus (Dz), Erigeron canadensis (Ec), Gossypium hirsutum (Gh), Helianthus annuus (Ha), Rhamnella rubrinervis (Rr) and Theobroma cacao (Tc). (C) Phylogenetic tree of CmFT. The amino sequences were subjected to phylogenetic analysis using the neighbor-joining method in MEGA7 software
3.4 Subcellular Localization of CmFT
To determine the localization of the CmFT protein in living cells, we constructed the pNCGreen-CmFT vector, delivered it into Nicotiana tabacum L. inner epidermis cells, and observed GFP signals. The signals were only observed in the nucleus (Fig. 6).
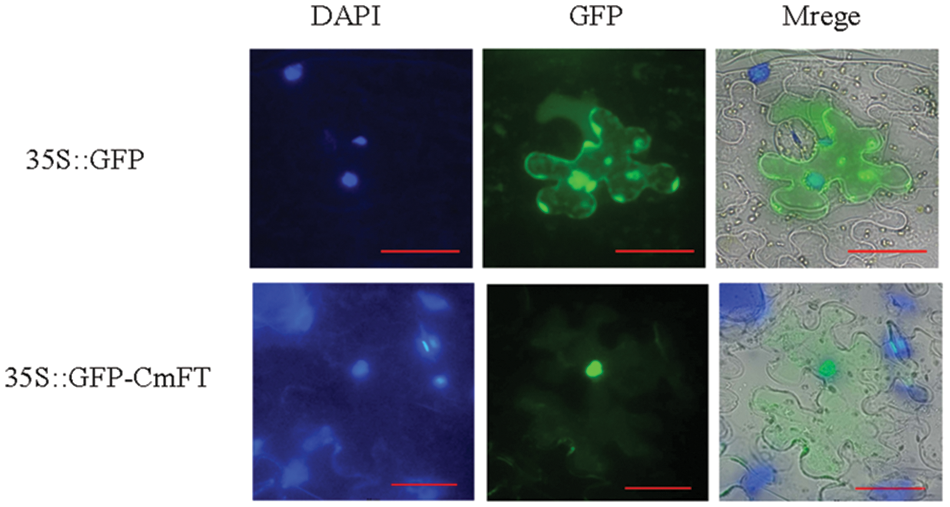
Figure 6: Subcellular localization of CmFT. DAPI: nuclear positioning signal. GFP: green fluorescence signal. Merge Overlay plot. Bar: 50 μm
3.5 Heterologous Expression of CmFT Restored the Delay of Flowering Time in Arabidopsis ft Mutant
To test whether CmFT can change flowering time, an Arabidopsis ft mutant resulting from T-DNA insertion and wild-type Arabidopsis (Col-0) were transformed with CmFT separately. Multiple T0 transgenic lines were obtained from two Arabidopsis. These T0 lines were selfed, and multiple T1 transgenic lines were produced. T2 transgenic Arabidopsis lines were selected randomly to record flowering time and traits (Table 1).
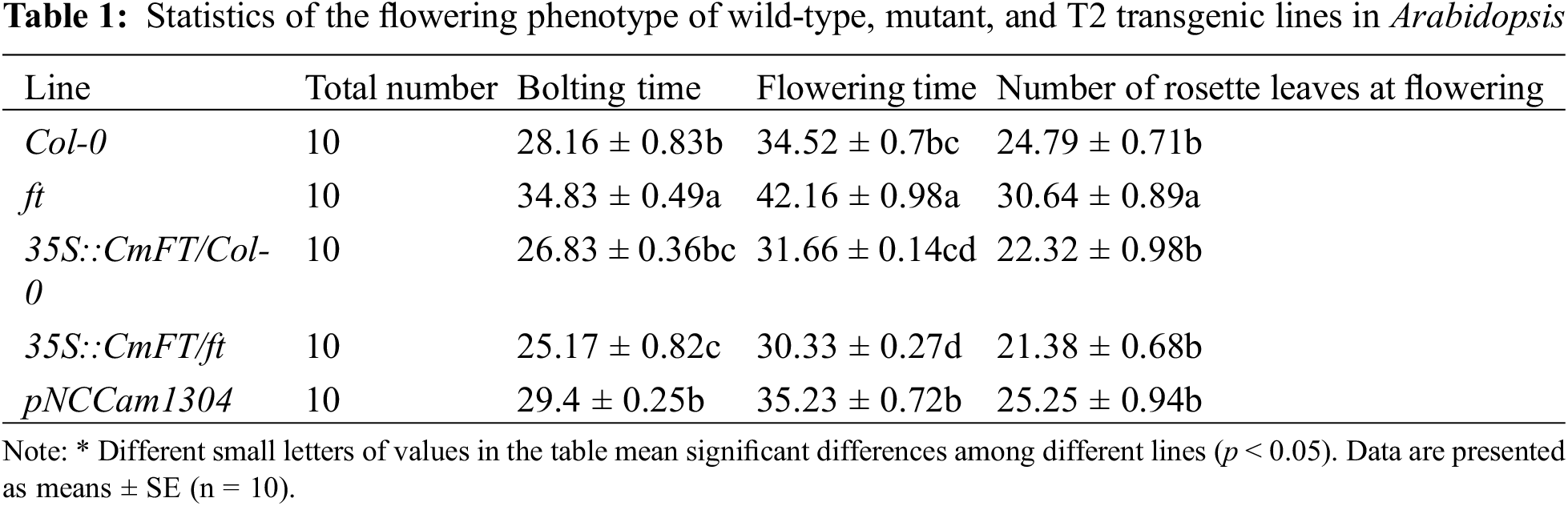
The Arabidopsis ft mutant had a delay in flowering, which was much later than that of wild-type Arabidopsis (34.52 d). CmFT transgenic Arabidopsis lines flowered at 31.66 d, approximately 3 days earlier than wild-type Arabidopsis. Moreover, CmFT transgenic ft mutant lines demonstrated excellent performance and the flowering time was 11.83 d earlier than Arabidopsis ft mutant and 1.33 d earlier than CmFT transgenic Arabidopsis. In addition, while the Arabidopsis ft mutant had only grown pods 41.5 days after sowing, CmFT transgenic ft mutant lines had already grown plump pods (Fig. 7). These results demonstrate that the heterologous expression of CmFT can restore the delay of flowering time in Arabidopsis ft mutant.

Figure 7: Flowering phenotypes of mutant, transgenic lines, and wild-type in Arabidopsis. ft: Arabidopsis ft mutant. 35S::CmFT/ft: CmFT transgenic ft mutant lines. Col: wild-type Arabidopsis (Col-0)
Soluble sugars and starches are important metabolites for plant growth, development, and physiological metabolism, and their accumulation is conducive to flower bud differentiation [19]. It is believed that a high content of soluble sugar, starch, and soluble protein can promote flower bud formation and flowering [20,21]. In this study, a large amount of soluble sugar was consumed to provide energy to synthesize other substances during the initiation of flower bud differentiation of photoperiod insensitive type, C.morifolium ‘Zilian’, which led to the decrease of soluble sugar and starch contents in the apical buds. Carbohydrates may then be transferred from leaves, and the soluble sugar and starch contents of apical buds increase. After that, floral organ formation consumed plenty of soluble sugar, which led to its decrease again. At the beginning of flower bud differentiation, the floral buds of the photosensitive type, C. morifolium ‘Zihongtuogui’, also consumed a lot of nutrients, but the higher starch hydrolysis rate may increase the soluble sugar contents. After that, the content of soluble sugar decreased and the accumulation of starch increased with the differentiation of floral buds. At the later stage of flower bud formation, the starch contents in the apical buds decreased again, so it can be inferred that the photosensitive type also consumes soluble sugar. During the process of flower bud differentiation, more starch needs to be consumed, while the accumulation of starch is beneficial for the differentiation.
The floral bud differentiation of most chrysanthemum varieties is regulated by the photoperiod pathway, among which GI, CO, and FT genes play a crucial role. GI, as an important gene outputting by the biological rhythm clock, positively regulates the expression of CO, which is coding transcription factors acting on FT. The expression products of FT act on other flowering integration factors to promote plant flowering [5,12,22–25]. Previous studies showed that the expression of AgFT1 was the highest in leaves and the lowest in roots [26]; and the expression of ClCOL5 of C. lavandulifolium was the highest in leaves, which was much higher than that in other tissues [27]. In this study, the expression pattern analysis of specific tissues by qRT-PCR showed that the expression of CmGI was the highest in the roots of the two chrysanthemums and the lowest in the flowers, while CmCO and CmFT had the highest expression levels in leaves. Therefore, we speculated that CmFT is activated and expressed in leaves.
Moreover, qRT-PCR analysis of different floral bud formation and differentiation stages in two chrysanthemums showed that CmGI was first expressed, while CmCO and CmFT were delayed, which reached their peak in the floret primordium differentiation stage. It was speculated that CmGI might first receive the biological rhythm clock signal in the leaves, and then start the expression of CmCO and CmFT, thus promoting floral bud differentiation. In the later stage of differentiation, floral bud formation may be mainly controlled by downstream integration factors [12,28,29]. In the photosensitive type, C. morifolium ‘Zihongtuogui’, CmGI, CmCO, and CmFT still maintained a high expression level at the later stage of floral bud differentiation, which may be due to the regulation of plant heat tolerance response or the regulation of the biological clock’s response to sucrose [30,31]. In conclusion, the expression levels of CmGI, CmCO, and CmFT may have a certain degree of synchronization.
The relationship is complicated between plant nutrient contents and flowering-related genes. In plants, the production of carbohydrates through photosynthesis is the key metabolic output of the biological clock, and there is feedback regulation between the circadian rhythm genes and carbohydrate metabolism [31–34]. In addition, studies have found that the biological clock of Arabidopsis was sensitive to nitrogen, and under low nitrogen conditions can up-regulate the expression of flowering-related genes (GI and CO), and increase several key components (CCA1, LHY, and TOC1) of the central oscillator of the biological clock [35,36]. It is believed that the change in nitrate supply can regulate the expression dynamics of FT and SOC1 [37]. In this study, we found that the change in the expression levels of CmGI was similar to that of soluble sugar contents during floral bud formation, suggesting that soluble sugar metabolism might be affected by the regulation of the circadian clock genes. This study only preliminarily connected nutrient-metabolism genes to flowering-related genes, while the potential molecular mechanism of how nutrients participate in regulating flowering time needs to be further analyzed.
In this study, CmFT were cloned from C. morifolium ‘Zihongtuogui’. Polypeptide alignment and phylogenetic analysis showed that CmFT was clustered in FT-the like subfamily and had high homology with FTs of other comparison species, indicating that CmFT was relatively conservative in phylogeny. In addition, the subcellular localization of CmFT in N. tabacum showed that CmFT was nuclear localization. It is speculated that the expression products of CmFT entered the nucleus to regulate the transcription and expression of downstream target genes. Previous studies showed that heterologously expressing ClFT from C. lavandulifolium in Arabidopsis demonstrated the function of flowering in advance [38]. In this study, we transformed an Arabidopsis ft mutant resulting from T-DNA insertion and wild-type Arabidopsis (Col-0) with CmFT separately. The results showed that CmFT transgenic ft mutant lines can significantly advance the flowering time. Therefore, we speculated that CmFT plays an important role in promoting flowering, which, combined with quantitative validation in our study, may be the key reason for the photosensitivity of C. morifolium ‘Zihongtuogui’. In conclusion, this study has laid a foundation for further studying the function of flowering-related genes of chrysanthemum through the photoperiod pathway and provides new clues on promoting flowering of FT in different species.
Insensitive Chrysanthemum consumes more soluble sugar compared with the photosensitive type, while Flowering-related genes CmFT plays an important role in promoting flowering, which may be the key reason for the photosensitivity of C. morifolium ’Zihongtuogui’. In conclusion, this study has laid a foundation for further studying the function of flowering-related genes of chrysanthemum through photoperiod pathway and provides new clues on promoting flowering of FT in different species.
Acknowledgement: We thank Dr. Yan Pu of the Chinese Academy of Tropical Agricultural Sciences for sharing the subcellular localization vector pNC-Green-SubN, and the vector pNC-Cam1304-MCS35S used in the process of functional verification.
Funding Statement: This research was funded by the Major Technological Innovation of Guangdong Province of China, Grant Number 2020B020220009, and the Science and Technology Projects, Grant Number 202201011833.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Fenglan Wang, Zhenyuan Cai; data collection: Qing Wu, Hanhan Xia; analysis and interpretation of results: Zhimei Li, Shilong Zhang, Honghui Luo; draft manuscript preparation: Zhenyuan Cai, Yanhong Guo. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are provided in this published article and its supplementary data files or it will be provided upon a reasonable request.
Ethics Approval: No applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.049150.
References
1. Sachin T, Guiliang T. To bloom or not to bloom: role of MicroRNAs in plant flowering. Mol Plant. 2015;8(3):359–77. [Google Scholar]
2. Martina B, Nadine D, Christian J. Flowering time regulation in crops-What did we learn from Arabidopsis? Curr Opin Biotech. 2015;32:121–9. [Google Scholar]
3. Frédéric B, Guillaume L, Pierre T, Claire P. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016;44(D1):D1167–71. [Google Scholar]
4. Lae-Hyeon C, Jinmi Y, Gynheung A. The control of flowering time by environmental factors. Plant J. 2017;90:708–19. [Google Scholar]
5. Jae SS, Akane K, Takato I. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2017;173(1):5–15. [Google Scholar]
6. Young HS, Jae SS, Hannah AK, Takato I. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–64. [Google Scholar]
7. Paula SL, Kay W, Fran R, Hitoshi O, Federico V, Geogre C. CONSTANS mediates between the circadian clock and the control of-owering in Arabidopsis. Nature. 2001;410:1116. [Google Scholar]
8. Chattha WS, Atif RM, Iqbal M, Shafqat W, Farooq MA, Shakeel A. Cenome-wide identification and evolution of Dof transcription factor family in cultivated and ancestral cotton species. Genomics. 2020;112(6):4155–70. [Google Scholar] [PubMed]
9. Tsuyoshi M, Louisa W, Sumire F, Frédéric C, Karen L, Hitoshi O, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 2005;17(8):2255–70. [Google Scholar]
10. Mariko S, Steve AK. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Science. 2011;108(28):11698–11703. [Google Scholar]
11. Chailakhyan MK, Krikorian AD. Forty years of research on the hormonal basis of plant development—some personal reflections. Bot Rev. 1975;41:1–29. [Google Scholar]
12. Cristina N, José AA, Eduard CO, Carlos AC, Shojiro T, Javier S, et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478(7367):119–32. [Google Scholar]
13. Philip AW, Min CK, Katja EJ, Wolfgang B, Markus S, Jan UL, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5780):1056–9. [Google Scholar]
14. Eliezer L, Tamar E, Alexander R, Akiva S, Alexander G, Ziva A, et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103(16):6398–6403. [Google Scholar]
15. Atsushi O, Takako N, Tuoping L, Takumi K, Yohei H, Katsuhiko S, et al. CsFTL3, a Chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in Chrysanthemums. J Exp Bot. 2012;63(3):1461–77. [Google Scholar]
16. Liu HY, Wang HH, Cui CH, Wang M, Guo JJ, Wen ZP, et al. Improvement of the experiment on the determination of soluble sugar (anthrone method). Lab Sci. 2013;16(2):19–20 (In Chinese). [Google Scholar]
17. Debebe A, Temesgen S, Redi-Abshiro M, Chandravanshi BS, Ele E. Improvement in analytical methods for determination of sugars in fermented alcoholic beverages. Janal Methods Chem. 2018;2018:4010298. [Google Scholar] [PubMed]
18. Zhang TX, Sun M, Guo YH, Shi XJ, Yang YJ, Chen JT, et al. Overexpression of LiDXS and LiDXR from Lily (Lilium ‘Siberia’) enhances the terpenoid content in tobacco flowers. Front Plant Sci. 2018;9. doi:10.3389/fpls.2018.00909. [Google Scholar] [PubMed] [CrossRef]
19. Chen H, He J, He S, Chen Y. Changes in nutritive materials in off-season mango in flower bud differentiation stage. Agricul Biotech. 2016;5(6):13–5. [Google Scholar]
20. Chen NMH, Qiao Z, Xu JP, Wei KH, Li MH. Physiological mechanism and developmental events in differentiating floral buds of Sophora tonkinensis Gagnep. Pharmacogn Mag. 2020;16(67):83–91. [Google Scholar]
21. Zhang SY, Zhang MM, Zhang T, Ye QH, Xie Q, Chen QX. Observation on the process of flower bud morphological differentiation and its physiological and biochemical characteristics in Syzygium cumini (L.). J Botany Northwest China. 2021;41(8):1347–54 (In Chinese). [Google Scholar]
22. Yohei H, Takako N, Atsushi O, Yoshihiro N, Katsuhiko S, Seiichi F, et al. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in Chrysanthemums. Proc Natl Acad Sci. 2013;110(42):17137–42. [Google Scholar]
23. Galopin G, Crespel L, Mauget JC, Morel P. Analysis of the impact of climatic conditions on floral transformation in Hydrangea macrophylla ‘Leuchtfeuer’. HortScience. 2011;46(10):1373–6. [Google Scholar]
24. Andre D, Marcon A, Lee KC, Goretti D, Zhang B, Delhomme N, et al. Flowering Locus T paralogs control the annual growth cycle in Populus trees. Curr Biol. 2022;32(13):2988. [Google Scholar] [PubMed]
25. Yang LW, Fu JX, Qi S, Hong Y, Huang H, Dai SL. Molecular cloning and function analysis of ClCRY1a and ClCRY1b, two genes in Chrysanthemum lavandulifolium that play vital roles in promoting floral transition. Gene. 2017;617:32–43. [Google Scholar] [PubMed]
26. Chen FY, Li Y, Li XY, Li WL, Xu JM, Cao H, et al. Ectopic expression of the Arabidopsis florugen gene FLOWERING LOCUS T in seeds enhances seed dormancy via the GA and DOG1 pathway. Plant J. 2021;107(3):909–24. [Google Scholar] [PubMed]
27. Kwon YJ, Hong MJ, Park M, Jeon D, Lim GH, Chang S, et al. Conserved and unique features of pepper FLOWERING LOCUS T-like genes revealed by comparative analysis among solanaceous crops. Sci Hortic. 2023;326:112710. [Google Scholar]
28. Borovsky Y, Mohan V, Shabtai S, Paran I. CaFT-like is a flowering promoter in pepper and functions as florigen in tomato. Plant Sci. 2020;301:110678. [Google Scholar] [PubMed]
29. Shim JS, Kubota A, Imaizumi T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2017;173(1):5–15. [Google Scholar] [PubMed]
30. Cao SQ, Ye M, Jiang ST. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005;24(11):683–90. [Google Scholar] [PubMed]
31. Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci. 2011;108(12):5104–9. [Google Scholar] [PubMed]
32. Feng JQ, Zhang W, Zhang SB. Floral bud dormancy is responsible for the different flowering tomes of six Pleione species. Glob Ecol Conserv. 2023;46:e02597. [Google Scholar]
33. Michael JH, Olga M, Fiona CR, Katharine EH, Alex AR. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502:689–92. [Google Scholar]
34. Katarzyna G, Ji HA. Isoprenoid-derived metabolites and sugars in the regulation of flowering time: does day length matter? Front Plant Sci. 2021;12:765995. [Google Scholar]
35. Yuan S, Zhang ZW, Zheng C, Zhao ZY, Wang Y, Feng LY, et al. Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc Natl Acad Sci. 2016;113(27):7661–6. [Google Scholar] [PubMed]
36. Sanagi M, Aoyama S, Kubo A, Lu Y, Sato Y, Ito S, et al. Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of flowering BHLH 4 in Arabidopsis. Proc Natl Acad Sci. 2021;118(19):e2022942118. [Google Scholar] [PubMed]
37. Yan FH, Zhang LP, Cheng F, Yu DM, Hu JY. Accession-specific flowering time variation in response to nitrate fluctuation in Arabidopsis thaliana. Plant Divers. 2021;43(1):78–85. [Google Scholar] [PubMed]
38. Fu JX, Wang LL, Wang Y, Yang LW, Yang YT, Dai SL. Photoperiodic control of FT-like gene ClFT initiates flowering in Chrysanthemum lavandulifolium. Plant Physiol Bioch. 2014;74:230–8. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools