 Open Access
Open Access
ARTICLE
Transcriptome Analysis of Inflorescence Development at the Five-Leaf Stage in Castor (Ricinus communis L.)
1 College of Life Science, Baicheng Normal University, Baicheng, 137000, China
2 College of Life Science and Food, Inner Mongolia University for the Nationalities, Tongliao, 028000, China
3 College of Life Science, Northeast Forestry University, Harbin, 150040, China
4 Key Laboratory of Castor Breeding of the State Ethnic Affairs Commission, Inner Mongolia Minzu University, Tongliao, 028043, China
5 Inner Mongolia Industrial Engineering Research Center of Universities for Castor, Inner Mongolia Minzu University, Tongliao, 028043, China
6 Inner Mongolia Key Laboratory of Castor Breeding and Comprehensive Utilization, Inner Mongolia Minzu University, Tongliao, 028000, China
7 Inner Mongolia Engineering Research Center of Industrial Technology Innovation of Castor, Inner Mongolia Minzu University, Tongliao, 028000, China
* Corresponding Authors: Fenglan Huang. Email: ; Fanjuan Meng. Email:
Phyton-International Journal of Experimental Botany 2024, 93(4), 713-723. https://doi.org/10.32604/phyton.2024.047657
Received 13 November 2023; Accepted 12 March 2024; Issue published 29 April 2024
Abstract
The yield of castor is influenced by the type of inflorescence and the proportion of female flowers. However, there are few studies on the genetic mechanism involved in the development and differentiation of castor inflorescences. In this study, we performed transcriptomic analyses of three different phenotypes of inflorescences at the five-leaf stage. In comparison to the MI (complete pistil without willow leaves), 290 and 89 differentially expressed genes (DEGs) were found in the SFI (complete pistil with willow leaves) and the BI (monoecious inflorescence), respectively. Among the DEGs, 104 and 88 were upregulated in the SFI and BI, respectively, compared to the MI. In addition, 186 DEGs and 1 DEG were downregulated in the SFI and BI compared to the MI. Moreover, we conducted GO and KEGG enrichment analyses of the DEGs. In comparison to the MI, the SFI and BI exhibited the enrichment of functional branches in DEGs, specifically in pollen wall assembly, pollen development, and cellular component assembly involved in morphogenesis. In our study, RADL5 showed low expression levels between SFI-vs.-MI types. In addition, we found that the expression of NAC in the SFI differed from that in MI and BI, and some genes related to hormonal signaling changed their expression levels during inflorescence differentiation. These results reveal the genetic mechanism of sex genotypes in castor, which will not only guide researchers in the breeding of castor but also provide a reference for genetic research on other flowering plants.Keywords
Supplementary Material
Supplementary Material FileCastor (Ricinus communis L.) has high economic value and is an important nonfood oil-seed crop used as a biofuel and feedstock [1]. In addition, castor has also been identified as a medicine and fertilizer [2,3]. The global demand for castor has markedly increased, mainly due to the various properties of castor. Furthermore, castor can also grow under various harsh environmental conditions, including those of wastelands, degraded lands, and arid and semiarid lands [4,5]. To date, castor has been widely cultivated in many countries, such as China, India, and Brazil [6]. However, the development of the castor industry has been hindered by its low productivity. This bottleneck of low yield in castor is attributed to the number and ratio of inflorescence differentiation [7,8]. Accordingly, there is a need to develop high-yield castor cultivars by considering different inflorescence types. Unfortunately, studies on the genetic mechanism of inflorescence development and differentiation in castor are scarce.
In general, castor inflorescence type can impact seed purity and yield [9,10]. There are three types of castor inflorescence: MI, SFI, and BI (Fig. 1). MI inflorescences consist solely of pistils. SFI inflorescences feature pistils with willow leaves growing on the inflorescence axis. The BI-type inflorescence is hermaphroditic, with the pistil at the top and the stamen at the bottom of the inflorescence axis. The published literature has indicated that the type of inflorescence affects castor yield [9,11]. However, the formation mechanisms of different inflorescence types remain unclear. Our prior research on flower bud differentiation in three inflorescence types of aLmAB2 castor bean revealed that the 5–6 leaf stage is the transitional period from asexual to sexual reproduction in the inflorescence [12]. Hence, the five-leaf stage is considered an optimal stage for investigating the mechanism of sex differentiation in castor. To dissect the differentiation mechanism of ricin inflorescence, we collected samples at the five-leaf stage for MI, SFI, and BI, and conducted transcriptome experiments to study their molecular mechanisms.
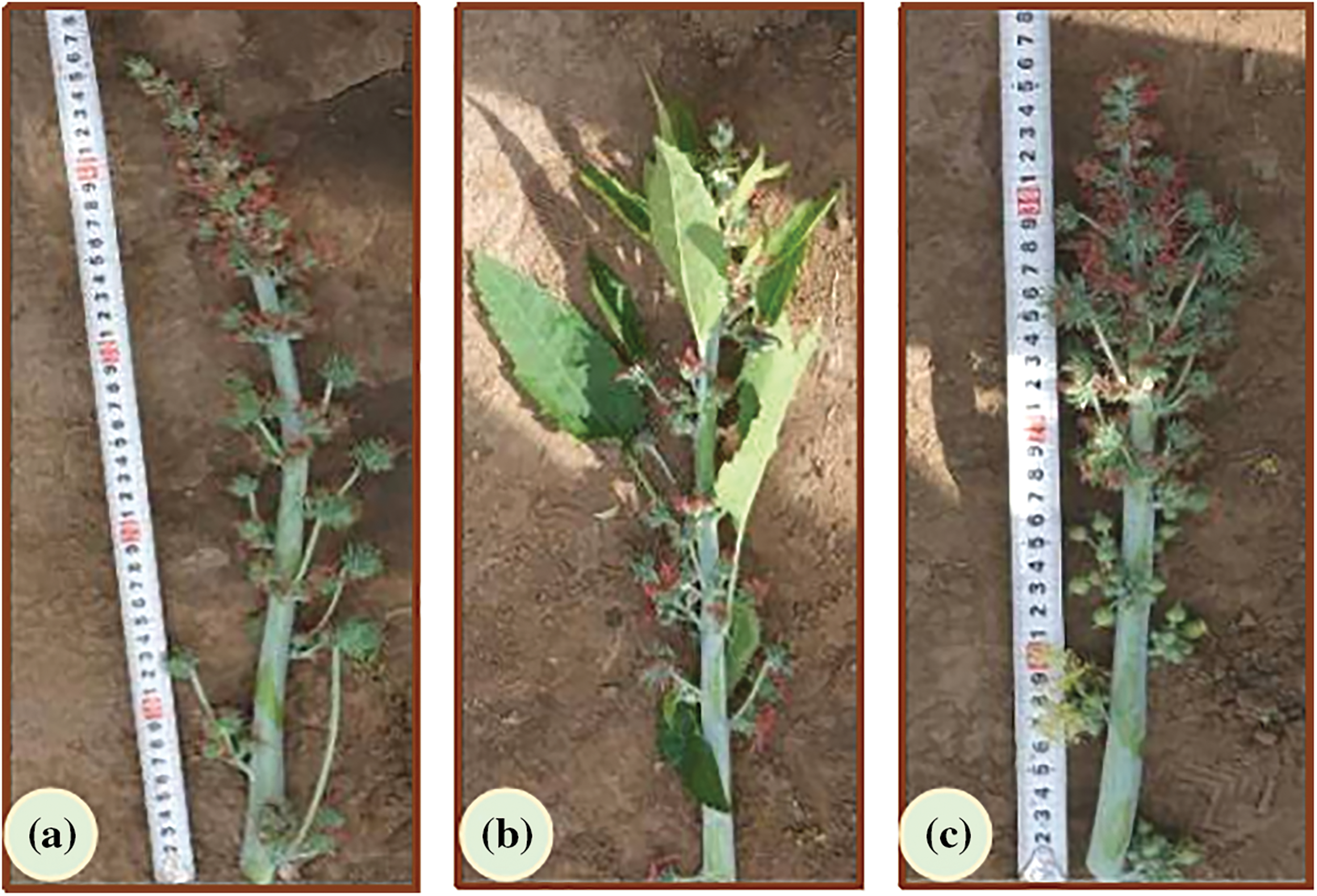
Figure 1: Images of three inflorescence types of Ricinus communis L. (a) Complete pistil without willow leaves (MI). (b) Complete pistil with willow leaves (SFI). (c) Monoecious inflorescence (BI)
The high-throughput RNA-sequencing (RNA-Seq) method based on transcriptome data can detect molecular changes in a wide range of plant species due to its high coverage and low cost [13]. Therefore, transcriptome data have been effectively used in recent years to study plant growth and development. The gene expression profile of RNA sequencing reveals the gene expression patterns of different tissues in castor [14,15]. Currently, RNA-Seq is an effective technique for analyzing the mechanisms of castor development [16]. For example, differentially expressed genes, such as male-abundant or female-specific genes, were identified in castor via RNA-Seq [17]. In addition, transcriptome changes were monitored by RNA-Seq under low temperatures in a cold-tolerant castor variety (Tongbi 5) [18]. However, in castor, the mechanism of inflorescence differentiation has not been extensively investigated. In the present study, we aimed to understand the molecular mechanisms of sex differentiation in castor. At the five-leaf stage of castor, we used RNA-Seq to analyze gene expression in the three different inflorescences. The results reveal the genetic mechanism of the sex genotype of castor, providing not only guidance for breeding castor but also a reference for genetic research on other flowering plants.
aLmAB2 castor (Ricinus communis L.) plants with a row spacing of 80 × 80 cm were grown in the field of the Inner Mongolia Key Laboratory of Castor Breeding. Three castor genotypes were photographed according to sex expression type before sample collection. The three types MI, SFI, and BI, were harvested at the five-leaf stage. Three biological replicates were carried out for each inflorescence type. Nine samples were frozen in liquid nitrogen and stored at −80°C for RNA sequencing.
2.2 RNA Extraction and cDNA Library Preparation
The total RNA of the inflorescences was extracted using a Plant RNA Kit (Beijing Zoman Biotechnology, Co., Ltd., China). To degrade residual genomic DNA, DNase I (Qiagen, Manchester, UK) was added to the RNA. We used a microplate reader to determine the concentration of RNA. cDNA was synthesized from total RNA by using Takara cDNA kit (Code no. 6215A).
2.3 Raw Read Filtering, Assembly, Gene Annotation, and DEG Analysis
RNA-Seq library preparation and sequencing were performed by Baiqu Biotechnology Co., Ltd., Shanghai, China. All libraries were sequenced using the Sanger method on the Illumina 1.9 platform. The raw sequencing data were converted into sequence data through base calling. To ensure data quality, clean reads were filtered to obtain high-quality clean reads for subsequent analysis. The filtering conditions were as follows: (1) removal of reads containing adapters; (2) removal of reads containing >10% N; and (3) removal of low-quality reads (more than 50% of the entire read for bases with a quality value of Q ≤ 20). In addition, we used the short-read matching tool Bowtie to match high-quality clean reads to the ribosome database, removing the reads from the matched ribosomes and using the retained data for transcriptome assembly and analysis. TopHat2 software was used to match reads to the NCBI castor genome (ASM1957865v1), and Cufflinks software was used to assemble the data. Cuffmerge was used to combine and filter the assembly results of multiple samples. |Log2FC (fold change) | ≥ 1 and p value < 0.05 were used as the thresholds to screen DEGs.
2.4 Functional Annotation and Gene Ontology Analysis
Based on the castor genome database, the functional annotations of the differentially expressed genes (with a fold change ≥2) were compared using the BLAST program (http://eggnog5.embl.de/#/app/emapper). The enrichment analysis of GO terms and KEGG annotations was evaluated by Cluster software (v8.0.27).
2.5 Real-Time Quantitative PCR Analysis
qPCR was performed in an ABI 7500 Fast real-time PCR system using SYBR Premix Ex Taq (Takara, Beijing, China). mRNA was extracted from three samples of the different inflorescence types and reverse transcribed to synthesize cDNA. Three biological replicates were performed for each inflorescence type. We used the following conditions for PCR: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 34 s. The primers for the selected qPCR genes are shown in Table S1. 18S was used as an internal reference gene, and the relative quantitative method (2−ΔΔCT) was used to determine gene expression.
3.1 Morphological Observation of Inflorescence Phenotypes
One generation of aLmAB2 castor with three female inflorescence types was used in this study. Among the three castor inflorescences, MI had only female flowers without willow-shaped functional leaves, and SFI developed only female flowers with willow-shaped functional leaves (Figs. 1a and 1b). Compared with the MI and SFI types, BI had male and female flowers simultaneously (Fig. 1c).
3.2 Quality Verification of the Transcriptome Sequencing Data
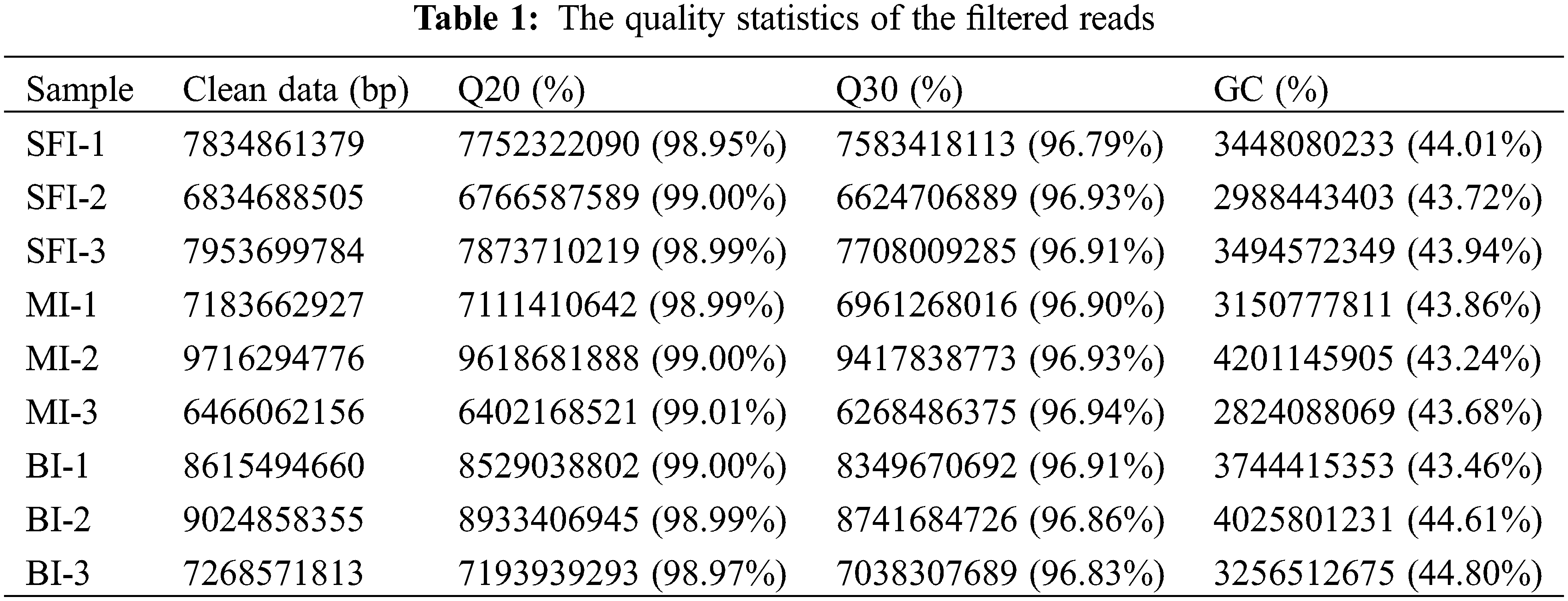
To study the systematic regulatory mechanism of transcriptional responses in the three castor inflorescence types at the five-leaf stage, RNA-Seq experiments were performed in this study. We filtered out the adapters and low-quality reads. Through Illumina high-throughput sequencing, a total of 9 libraries were sequenced. RNA-Seq of 9 castor inflorescence samples yielded 4.4–6.6 million clean reads. The clean reads were filtered to obtain more than 98.58% of the high-quality clean reads (Table 1). In Table 1, 683–971 million clean reads were obtained, and more than 97% of the reads showed high-quality values (≥Q20). The Q30 base distribution percentage ranged from 96.79% to 96.94%. The GC content was above 43%. These data indicated that the quality of the transcriptome sequencing data was relatively high and reliable for further analysis.
3.3 Differential Gene Expression Analysis
According to the transcriptome data obtained for the three inflorescence types, a total of 378 genes were detected according to log2fold change > 1 and FDR < 0.05 (Table S2 and Fig. 2). A total of 290 and 89 differentially expressed genes (DEGs) were found in the SFI and BI, respectively, compared to the MI. Among them, 104 and 88 DEGs were upregulated in the SFI and the BI, respectively, compared to the MI. In addition, 186 DEGs and 1 DEG were downregulated in the SFI and BI groups compared to the MI groups. The number of DEGs identified in the overlapping relationships among the three groups is listed in the Venn diagram of the DEGs (Fig. 2).
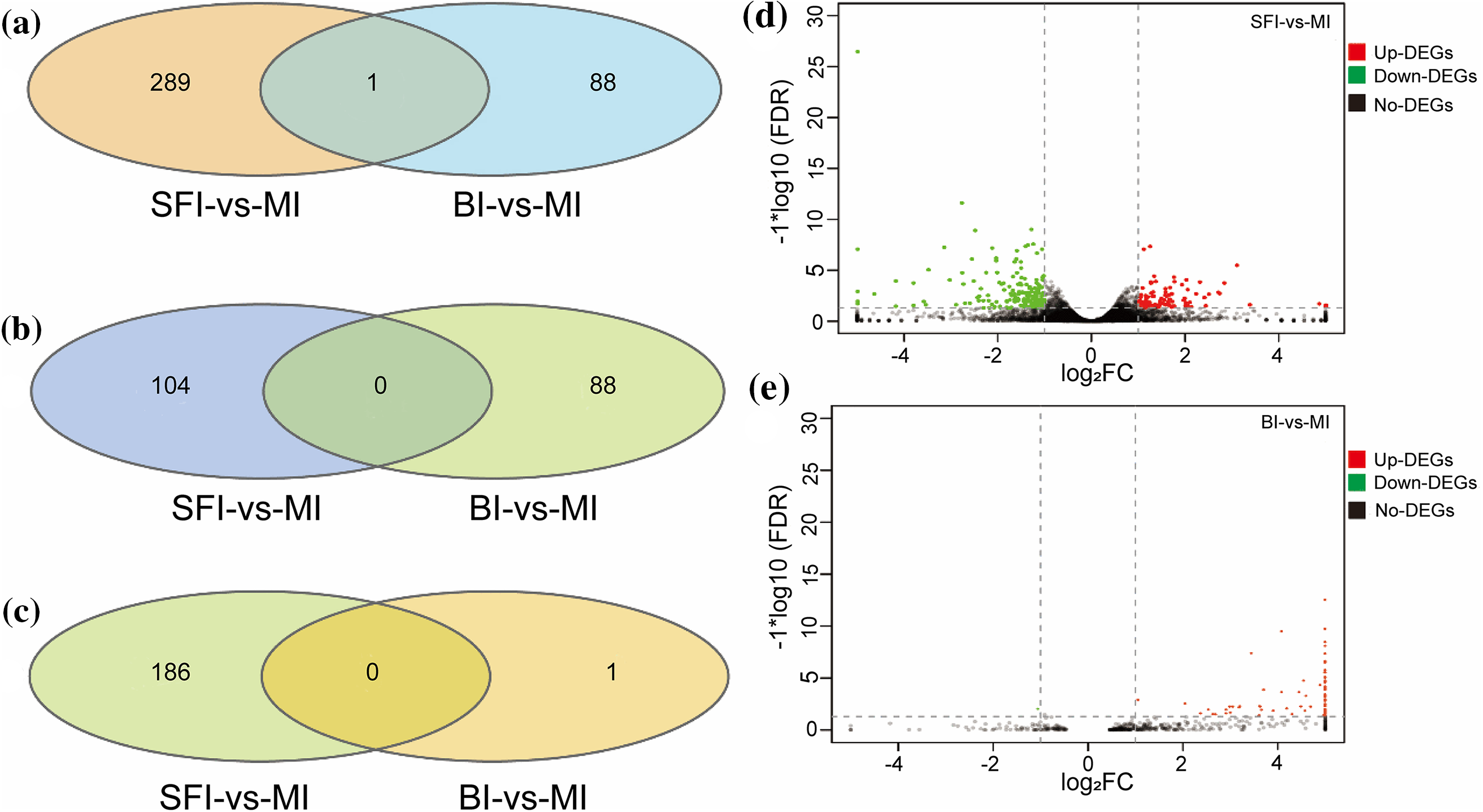
Figure 2: The number of DEGs in the MI, SFI, and BI groups. (a) Venn diagram of all DEGs in the three inflorescence types; (b and c) Venn diagram of upregulated and downregulated DEGs in the three inflorescence types; (d) SFI-vs.-MI volcano map; (e) BI-vs.-MI volcano map
3.4 GO and KEGG Enrichment Analyses of DEGs
To describe gene functions, the DEGs from the three groups (MI, SFI, and BI) were analyzed using GO enrichment (Fig. 3 and Table S3).
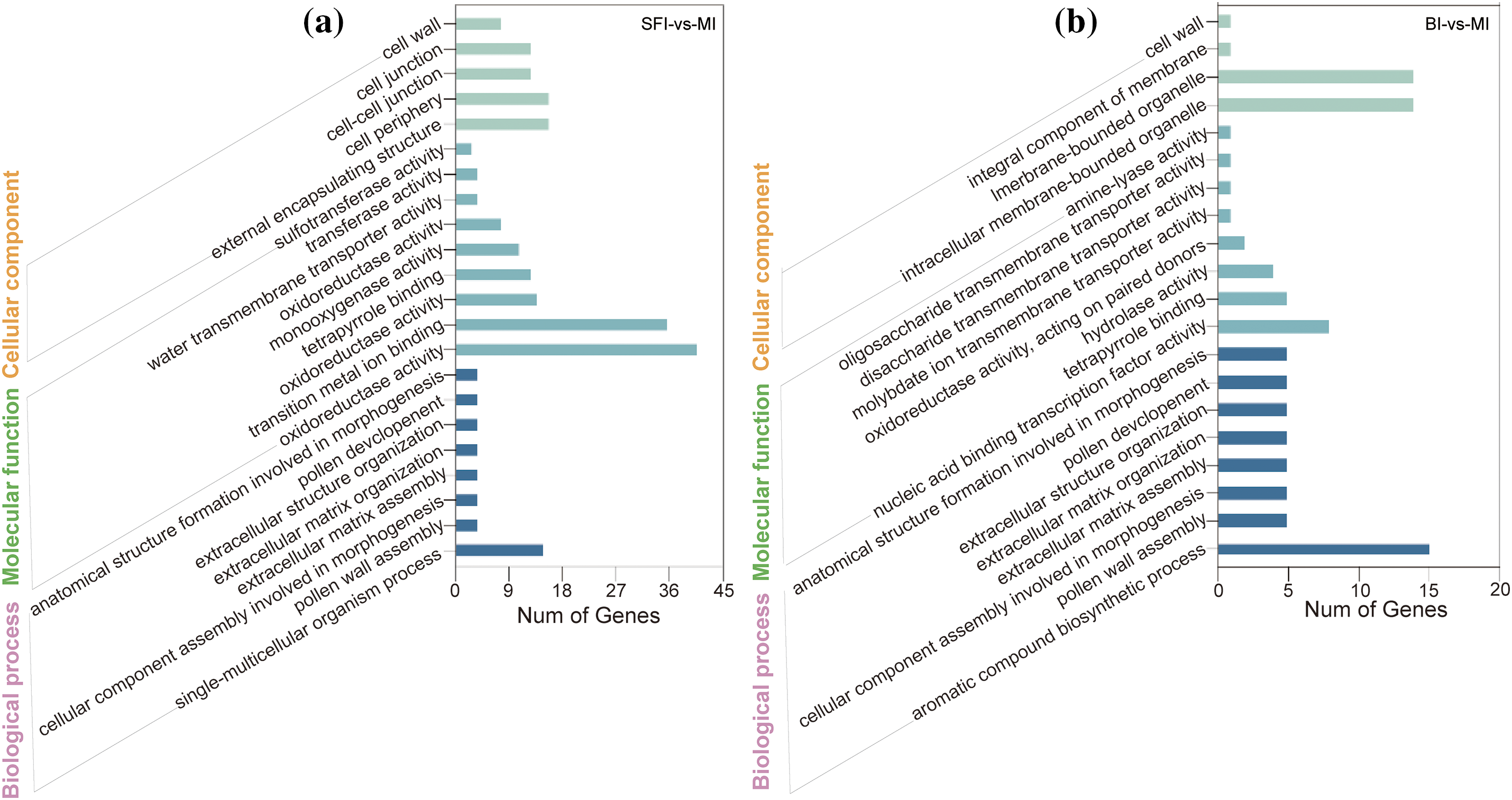
Figure 3: The number of enriched GO terms for the three inflorescence types at the five-leaf stage. (a) GO analysis of DEGs in the SFI-vs.-MI group; (b) GO analysis of DEGs in the BI-vs.-MI group
In the SFI-vs.-MI comparison group, the DEGs enriched in the “single-multicellular organism process” pathway were the most abundant in the term “biological processes”. For molecular functions, transition metal ion binding oxidoreductase activity was most enriched in DEGs. In the cell component category, the DEGs were enriched in five branches, namely the cell wall, cell junction, cell periphery, and external encapsulating structure (Table S3). In the BI-vs.-MI comparison group, the functional pathway most enriched in DEGs was the aromatic compound biosynthetic process biological process category. In the molecular function category, nucleic acid binding transcription factor activity was enriched in DEGs. In addition, the GO classification showed that the DEGs associated with the cellular component category were mainly related to membrane-bound organelles and intracellular membrane-bound organelles (Table S3). Compared to the MI, the SFI and BI had the following functional branches enriched in DEGs: pollen wall assembly, pollen development, and cellular component assembly involved in morphogenesis.
Based on the KEGG pathway data, the biological functions of the genes were determined. As shown in Fig. 4a and Table S4, functions with a Q value ≤ 0.05 were considered significantly enriched. In the SFI-vs.-MI group, some genes were significantly enriched in the biosynthesis of secondary metabolites (ko01110), protein processing in the endoplasmic reticulum (ko04141), and phenylpropanoid biosynthesis (ko00940) (Fig. 4b and Table S4). In the comparison of BI-vs.-MI, the greatest accumulation of DEGs was found in pathways such as metabolic pathways (ko01100) and biosynthesis of secondary metabolites (ko01110).
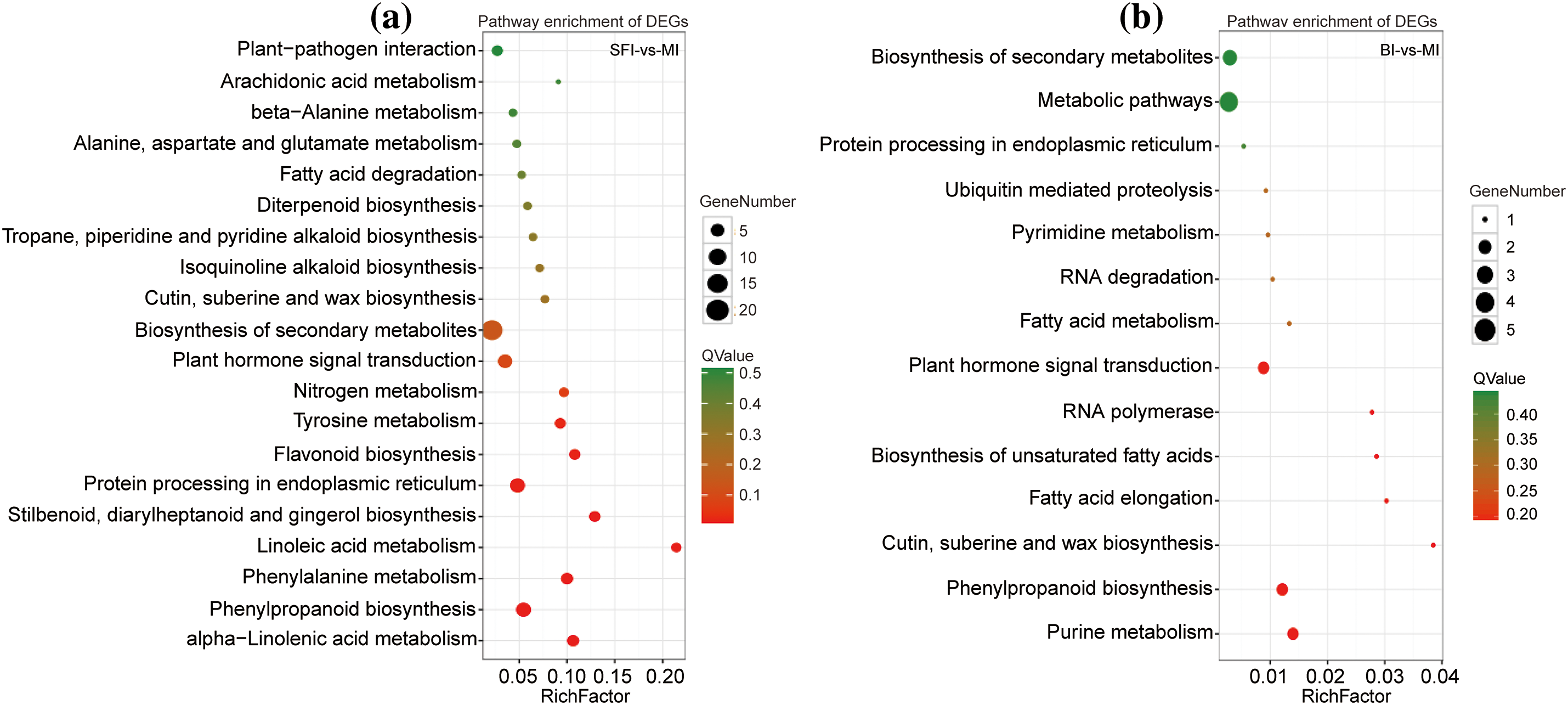
Figure 4: KEGG pathway analysis of DEGs. (a) KEGG enrichment analysis of DEGs in the SFI-vs.-MI group; (b) KEGG enrichment analysis of DEGs in the BI-vs.-MI group
To validate the transcriptomic sequencing results, we selected eight differentially expressed genes for qRT-PCR analysis. Among these eight differentially expressed genes, RADL5 (ncbi_8269992) is a member of the MYB family, and ERF11 (ncbi_8288577), ERF92 (ncbi_8258360), ABAH1 (ncbi_8275341), and G2OX (ncbi_8264113) belonged are hormone response factors. NAC72 (ncbi_8263571), NAC56 (ncbi_8268675), and NAC29 (ncbi_8281834) are members of the NAC family of transcription factors. The eight DEG expression trends were consistent with the transcriptome results, confirming the reliability of our overall transcriptome data, as shown in Fig. 5. RADL5 (protein RADIALIS-like 5), ERF11 (ethylene-responsive transcription factor 1), ERF92 (ethylene-responsive transcription factor 1B), and G2OX (gibberellin 2-beta-dioxygenase) were expressed at high levels in the MI but at low levels in the SFI. In the SFI, ABAH1 (abscisic acid 8-hydroxylase 1) expression was high but was low in the BI. The expression of NAC72, NAC56, and NAC29 decreased in the SFI. These genes may play important roles in the differentiation of castor inflorescences.
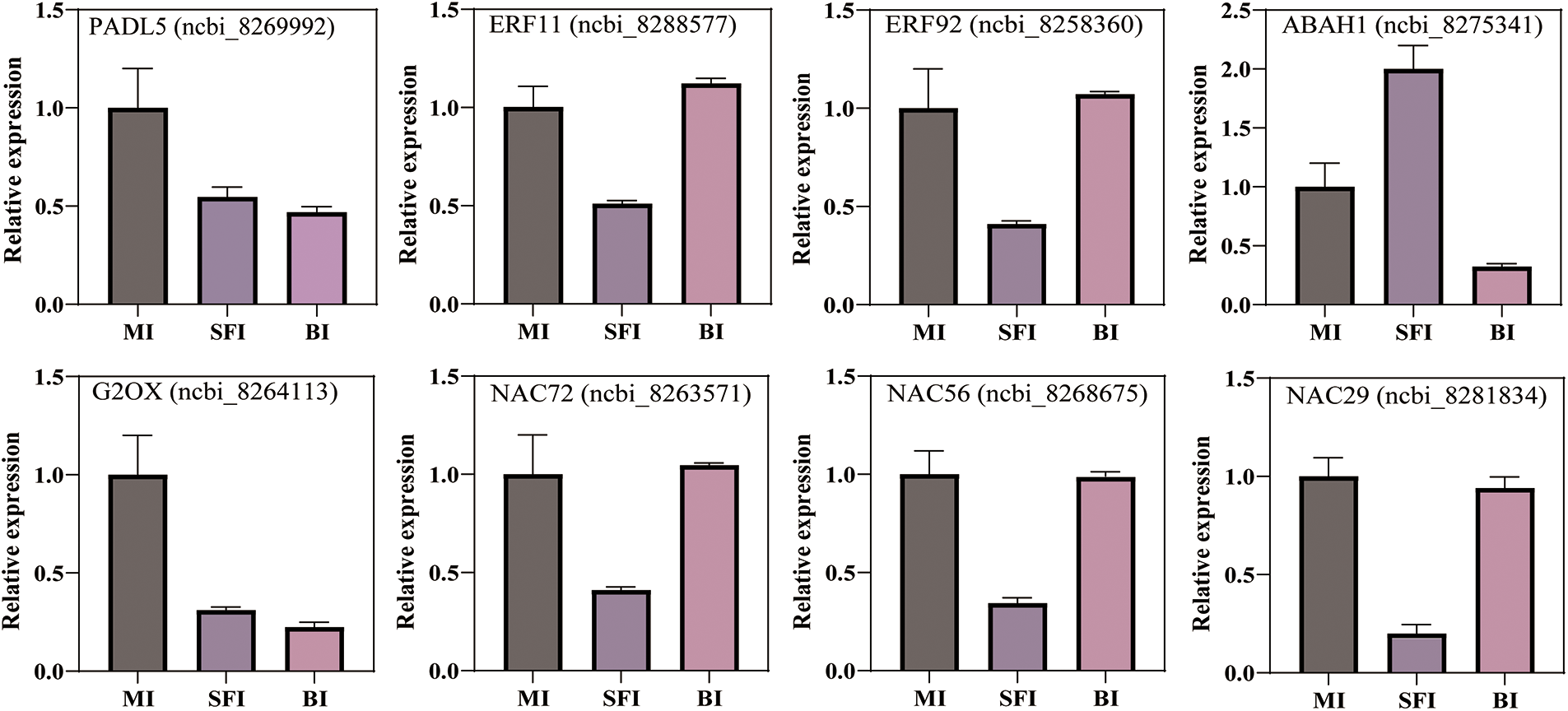
Figure 5: RT-qPCR analysis of DEGs. MI: complete pistil without willow leaves, SFI: complete pistil with willow leaves, BI: monoecious inflorescence. RADL5: protein RADIALIS-like, ERF11: ethylene-responsive transcription factor 1, ERF92: ethylene-responsive transcription factor 1B, G2OX: gibberellin 2-beta-dioxygenase, ABAH1: abscisic acid 8-hydroxylase 1. NAC72, NAC56, and NAC29 are NAC transcription factors
Generally, the transcriptome sequencing method has been widely applied to analyze the key mechanism of growth and development of many plant species [13]. To date, transcriptome information on Ricinus communis is limited [19]. In this study, we compared transcriptome data from three inflorescence types at the five-leaf stage in castor. Here, 683–971 million clean reads were obtained, and more than 97% of the reads showed high-quality values (Table 1). In addition, 21,886 unigenes were successfully and correctly annotated in previous data and in the genome of R. communis (Table S5). Accordingly, these results also suggested that the assembly of the transcriptome was reliable for the castor materials in this study. Therefore, these data can be used to analyze the mechanism of inflorescence development in R. communis. Moreover, the results also provide information for selecting key genes involved in inflorescence sex determination in castor.
Many transcription factors (TFs) in plants play important roles in flower sex differentiation. For example, the MADS-box family of TF genes governs the sex differentiation of male and female flowers in unisexual plants [20], with 176 TFs showing differential expression between male and hermaphroditic flowers [21]. Eight TFs have been found to regulate flavonoid biosynthesis by promoting the expression of multiple structural genes during flower sex differentiation [22]. Analysis of the transcriptome data revealed that PADL5 was expressed at higher levels in MI (Fig. 5 and Table S2). PADL5 is a member of the PAD subfamily of the MYB transcription factor family. RAD plays an important role in the development of flowers. The expression patterns and functions of RAD genes may vary among different plant species. In Arabidopsis, the RAD gene is involved in the formation of dorsal-ventral asymmetry in flowers [23,24]. The RAD gene may interact with the DIV gene and participate in the flower development process in primrose flowers [24]. In addition, we found that different levels of NAC (NAC72, NAC56, NAC29) were expressed in the BI than in the MI (Fig. 5). Therefore, NAC has a regulatory function in flower sex differentiation in castor. This result is consistent with the findings of other reports. For example, NAC transcription factors are associated with pecan flower sex differentiation [25]. NAC plays a crucial role in flower development in Zanthoxylum armatum var. [26]. Moreover, ethylene response factor (ERF) family genes have been identified [27]. In our study, ERF11 and ERF92 exhibited different expression levels between SFI and MI (Fig. 5 and Table S2). ERF11 and ERF92 are classified under the ERF subfamily of ERF transcription factors. Analysis of the transcriptome data and the qRT-PCR data revealed that both ERF11 and ERF92 were expressed at high levels in the BI and at low levels in the SFI. Some research has indicated that some ERF family members can influence sex determination and female flower development by binding to the GCC box to regulate downstream gene expression [28,29]. As one of the largest transcription factor families, ERFs also participate directly or indirectly in multiple processes, such as seed development and flower and fruit organ formation [30]. However, the specific regulation of inflorescence differentiation by the transcription factors MYB, NAC, and ERF in castor needs to be further investigated.
In this study, the expression levels of some genes related to hormone signaling changed during inflorescence differentiation (Table S2). These results may help researchers identify key genes to clarify the mechanisms of inflorescence differentiation in castor. Here, the expression levels of castor inflorescence differentiation genes, such as ABAH1 and G2OX (gibberellin 2-beta-dioxygenase-like), were significantly different expressed at the five-leaf stage, which indicated their key roles in inflorescence differentiation in castor. qRT-PCR analysis revealed that ABAH1 was expressed at the highest level in the SFI, while G2OX was expressed at the highest level in the MI. For example, the expression levels of genes involved in ethylene signal transduction have been shown to positively regulate flower development [31]. Moreover, the, IAA concentration and IAA/ABA ratio play an important roles in flower sexual differentiation in castor beans [19]. Moreover, a high ABA content is beneficial for floral induction and morphological differentiation of flower buds [32]. Notably, ABA levels do not significantly change during flower bud differentiation [33]. A previous study showed that applying the phytohormone gibberellin (GA), a critical hormone for anther development, also effectively promoted lateral flower differentiation [34,35]. In addition, spraying GA3 was found to advance flowering [36]. These findings all indicated that the balance and crosstalk of multiple phytohormones might affect sex differentiation in castor.
In this study, transcriptomic analysis was conducted on three castor inflorescences. A total of 290 and 89 DEGs were found in the SFI and BI, respectively, compared to the MI. Among the DEGs, PADL5YB86 and hormone-related genes (ERF11, ERF92, ABAH1, and G2OX) were significantly differentially expressed in the three inflorescences, which further affected the development of castor inflorescences. However, the specific regulatory mechanism requires further study.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by the following agencies: the Natural Science Foundation of Jilin Province (YDZJ202201ZYTS453), the Scientific Research Project of the Jilin Provincial Department of Education (JJKH20220010KJ), the Program for Innovative Research Team of Baicheng Normal University, the National Natural Science Foundation of China (31860071), the Inner Mongolia Autonomous Region Natural Science Foundation Project (2021MS03008), the Inner Mongolia Autonomous Region Grassland Talent Innovation Team (2022), the 2022 Basic Scientific Research Business Cost Project of Universities Directly under the Autonomous Region (237), the Open Fund Project of Inner Mongolia Castor Industry Collaborative Innovation Center (MDK2021011, MDK2022014, MDK2022008, MDK2021008, MDK2022009, MDK2023003), and Fundamental Research Funds for Universities Directly under the Autonomous Region in 2023 of Inner Mongolia University for Nationalities (225, 227, 243, 244), New Agricultural Science Research and Reform Practice Project of the Ministry of Education (2020114). In 2023, the Science and Technology Department of the Inner Mongolia Autonomous Region approved the construction.
Author Contributions: Yong Zhao, Yaxuan Jiang, and Li Wen performed the experiments. Rui Luo, Guorui Li, and Jianjun Di guided the experimental process. Yong Zhao, Yaxuan Jiang, Mingda Yin, and Zhiyan Wang completed the data analysis. Yong Zhao completed the writing of his paper. Yong Zhao, Fenglan Huang, and Fanjuan Meng designed the experiments and provided funding.
Availability of Data and Materials: All data have been incorporated into the article and its supplementary material.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.047657.
References
1. Zhu GT, Cheng DD, Wang XF, Guo QJ, Zhang Q, Zhang J, et al. Free amino acids, carbon, and nitrogen isotopic compositions responses to cadmium stress in two castor (Ricinus communis L.) species. Plant Physiol Biochem. 2022;184:40–6. [Google Scholar] [PubMed]
2. Elkousy HR, Abd EM, Abu ES. Antiviral activity of castor oil plant (Ricinus communis) leaf extracts. J Ethnopharmacol. 2021;271:113878. [Google Scholar] [PubMed]
3. Sandoval-Salas F, Méndez-Carreto C, Ortega-Avila G, Barrales-Fernández C, Hernández-Ochoa L, Sanchez N. A biorefinery approach to biodiesel production from castor plants. Process. 2022;10(6):151–9. [Google Scholar]
4. Ribeiro PR, Willems LAJ, Mudde E, Fernandez LG, Castro RD, Ligterink W, et al. Metabolite profiling of the oilseed crop Ricinus communis during early seed imbibition reveals a specific metabolic signature in response to temperature. Ind Crop Prod. 2015;67:305–9. [Google Scholar]
5. Zhang HZ, Guo QJ, Yang JY, Shen JX, Chen TB, Zhu GG, et al. Subcellular cadmium distribution and antioxidant enzymatic activities in the leaves of two castor (Ricinus communis L.) cultivars exhibit differences in Cd accumulation. Ecotoxicol Environ Safe. 2015;120:184–92. [Google Scholar]
6. Damodaran T, Hegde DM. Oilseed situation, a statistical compendium. Direct Oilseeds Res. 1999;32:147–53. [Google Scholar]
7. Parveen PA, Ramana J, Prasad RD, Senthilvel S, Ahamed ML, Greeshma K, et al. Waxy bloom on capsules is a major determinant of early infection by gray mold (Amphobotrys ricini (N.F. Buchw.) Hennebert) in castor (Ricinus communis L.). J Phytopathol. 2022;170(5):337–48. [Google Scholar]
8. Gangadhara Rao P, Behera TK, Gaikwad AB, Munshi AD, Jat GS, Boopalakrishnan G. Mapping and QTL analysis of gynoecy and earliness in bitter gourd (Momordica charantia L.) using genotyping-by-sequencing (GBS) technology. Front Plant Sci. 2018;9:1555. [Google Scholar] [PubMed]
9. Tan M, Xue J, Wang L, Huang J, Fu C, Yan X. Transcriptomic analysis for different sex types of Ricinus communis L. during development from apical buds to Inflorescences by digital gene expression profiling. Front Plant Sci. 2016;6:1208. [Google Scholar] [PubMed]
10. Mukhtar A, Masood IA, Sana S, Athar M, Talha J, Shah AN, et al. Sulfur enhancement for the improvement of castor bean growth and yield, and sustainable biodiesel production. Front Plant Sci. 2022;13:905738. [Google Scholar] [PubMed]
11. Wen Y, Luo R, Li L, Liang X, Hu X, Li X, et al. Functional studies of castor (Ricinus communis L.) PLC family genes in Arabidopsis inflorescence development. Phyton-Int J Exp Bot. 2023;92(11):3091–112. doi:10.32604/phyton.2023.030960. [Google Scholar] [CrossRef]
12. Huang FL, Zhu GL, Pan WT, He ZB, Bao CG, Peng M, et al. Comparative study of inflorescence development in three types of castor on microscopy. J Northern Agric. 2014;5(4):10–3. [Google Scholar]
13. Yu Y, Hong Z, Long YP, Yi S, Zhai JX. Plant public RNA-seq database: a comprehensive online database for expression analysis of similar to 45 000 plant public RNA-Seq libraries. Plant Biotechnol J. 2022;20(5):806–8. [Google Scholar] [PubMed]
14. Adrian PB, Kroon J, Swarbreck D, Febrer M, Tony RL, Graham LA, et al. Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One. 2012;7(2):e30100. [Google Scholar]
15. Singh R, Misra AN, Sharma P. Genome-wide transcriptional response of contrasting genotypes of industrial crop castor to As(V) stress: identification of genes and mechanisms associated with As(V) tolerance. Ind Crop Prod. 2022;179:114678. [Google Scholar]
16. Wu ZY, Feng X, Yu L, Ouyang Y, Geng Y. Transcriptome analysis of developing castor bean seeds and identification of ricinoleic acid biosynthesis genes. Biologia Plantarum. 2021;65:273–82. [Google Scholar]
17. Zou Z, Gong J, Huang Q, Mo Y, Yang L, Xie G, et al. Gene structures, evolution, classification and expression profiles of the Aquaporin gene family in castor bean (Ricinus communis L.). PLoS One. 2015;10(10):e0141022. [Google Scholar] [PubMed]
18. Wang X, Wu Y, Sun M, Wei X, Huo H, Yu L, Zhang J. Dynamic transcriptome profiling revealed key genes and pathways associated with cold stress in castor (Ricinus communis L.). Ind Crop Prod. 2022;178(29):114610. [Google Scholar]
19. Lei W, Tan M, Yan MF, Wang LJ, Yan XC. Initial research on inflorescence characteristics and flower bud differentiation of Ricinus communis L. Chin J Oil Crop Sci. 2012;34(5):544–50. [Google Scholar]
20. Fu Q, Niu L, Chen MS, Tao YB, Wang X, He H, et al. De novo transcriptome assembly and comparative analysis between male and benzyladenine-induced female inflorescence buds of Plukenetia volubilis. J Plant Physiol. 2018;221(10):107–18. [Google Scholar] [PubMed]
21. Li W, Zhang L, Ding Z, Wang G, Zhang Y, Gong H, et al. De novo sequencing and comparative transcriptome analysis of the male and hermaphroditic flowers provide insights into the regulation of flower formation in andromonoecious Taihangia rupestris. BMC Plant Biol. 2017;17(1):54. [Google Scholar] [PubMed]
22. Peng J, Li CY, Zhai WH, Dai JY, Zhao YL, et al. Integrative metabolome and transcriptome analysis of flavonoid biosynthesis genes in broussonetia papyrifera leaves from the perspective of sex differentiation. Front Plant Sci. 2022;13:900030. [Google Scholar]
23. Baxter Ca, Costa MR, Coen E. Diversification and co-option of RAD-like genes in the evolution of floral asymmetry. Plant J. 2007;52(1):105–13. [Google Scholar] [PubMed]
24. Zhong JHC, Preston JC, Hileman LC, Kellogg EA. Repeated and diverse losses of corolla bilateral symmetry in the Lamiaceae. Ann Bot. 2017;119(7):1211–23. [Google Scholar] [PubMed]
25. Jia ZM, Wang G, Xuan JS, Zhang J, Zhai M, Jia X, et al. Comparative transcriptome analysis of pecan female and male inflorescences. Russ J Plant Physiol. 2018;65(2):186–96. [Google Scholar]
26. Zhang X, Tang N, Liu XM, Ye JB, Zhang JY, Chen ZX, et al. Comparative transcriptome analysis identified differentially expressed genes between male and female flowers of zanthoxylum armatum var. novemfolius. Agronomy. 2020;10(2):153–9. [Google Scholar]
27. Pan J, Wen H, He HL, Lian HL, Wang G, Pan JS, et al. Genome-wide identification of cucumber ERF gene family and expression analysis in female bud differentiation. Sci Agric Sin. 2020;53(1):133–47. [Google Scholar]
28. Zafar M, Abdul R, Abdul R, Aqsa P, Ghulam M, Mo MJ, et al. Genome wide characterization, identification and expression analysis of ERF gene family in cotton. BMC Plant Biol. 2020;53(1):133–47. [Google Scholar]
29. Pan J, Wen HF, Chen GQ, Lin WH, Du H, Chen Y, et al. A positive feedback loop mediated by CsERF31 initiates female cucumber flower development. Plant Physiol. 2021;21(2):141. [Google Scholar]
30. Qi Z, Jing C, Li L, Zhao MZ, Zhang MP, Wang Y. Research progress on plant AP2/ERF transcription factor family. Biotechnol Bull. 2018;34(8):1–7. [Google Scholar]
31. Shi XF, Wei RF, Huang GY, Zhang Y, Lin L, Han JY, et al. Study on the molecular mechanism of promotion of inflorescence formation of CCC treatment for the second fruiting in grape. J Fruit Sci. 2021;38(2):153–67. [Google Scholar]
32. Wang JX, Luo T, He Z, Shao JZ, Peng JY, Sun JH, et al. Variation of endogenous hormones during flower and leaf buds development in ‘Tianhong 2’ apple. Hortsci. 2020;55(11):1794–8. [Google Scholar]
33. Xu SP, Zhang Y, Yuan XY, Bo C. Explore the key period of floral determination based on the microstructure and photosynthetic characteristics in phalaenopsis. Acta Horticult Sin. 2020;47(7):1359–68. [Google Scholar]
34. Qian Q, Yang YH, Zhang WB, Hu YL, Yuge L, Yu H, et al. A novel Arabidopsis gene RGAT1 is required for GA-mediated tapetum and pollen development. New Phytol. 2021;231(1):137–51. [Google Scholar] [PubMed]
35. Yin YY, Li J, Guo BC, Li TL, Ma GH, Wu KL, et al. Exogenous GA3 promotes flowering in Paphiopedilum callosum (Orchidaceae) through bolting and lateral flower development regulation. Horticult Res. 2022;9:091. [Google Scholar]
36. Zhang YJ, Li A, Lu YF, Sun JX, Liu MX, Zhang JG, et al. Changes in endogenous hormones content and effect of plant growth regulators of phalaenopsis during flowering period. J Trop Subtrop Bot. 2022;30(1):104–10. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools