 Open Access
Open Access
ARTICLE
Landscape of Sequence Variations in Homologous Copies of FAD2 and FAD3 in Rapeseed (Brassica napus L.) Germplasm with High/Low Linolenic Acid Trait
College of Agronomy, Northwest A&F University, Yangling, 712100, China
* Corresponding Author: Chengyu Yu. Email:
# The two authors contributed equally
§ Present address: Jiangsu Cowin Biotech Co., Ltd., Zhenjiang, 212000, China
Phyton-International Journal of Experimental Botany 2024, 93(3), 627-640. https://doi.org/10.32604/phyton.2024.050321
Received 02 February 2024; Accepted 26 February 2024; Issue published 28 March 2024
Abstract
Genetic manipulation (either restraint or enhancement) of the biosynthesis pathway of α-linolenic acid (ALA) in seed oil is an important goal in Brassica napus breeding. B. napus is a tetraploid plant whose genome often harbors four and six homologous copies, respectively, of the two fatty acid desaturases FAD2 and FAD3, which control the last two steps of ALA biosynthesis during seed oil accumulation. In this study, we compared their promoters, coding sequences, and expression levels in three high-ALA inbred lines 2006L, R8Q10, and YH25005, a low-ALA line A28, a low-ALA/high-oleic-acid accession SW, and the wildtype ZS11. The expression levels of most FAD2 and FAD3 homologs in the three high-ALA accessions were higher than those in ZS11 and much higher than those in A28 and SW. The three high-ALA accessions shared similar sequences with the promoters and CDSs of BnFAD3.C4 and BnFAD3.A3. In A28 and SW, substitution of three amino acid residues in BnFAD2.A5 and BnFAD2.C5, an absence of BnFAD2.C1 locus, and a 549 bp long deletion on the BnFAD3.A3 promoter were detected. The profile of BnFAD2 mutation in the two low-ALA accessions A28 and SW is different from that reported in previous studies. The mutations in BnFAD3 in the high-ALA accessions are reported for the first time. In identifying the sites of these mutations, we provide detailed information to aid the design of molecular markers for accelerated breeding schemes.Keywords
Supplementary Material
Supplementary Material FileOil rapeseed (Brassica napus. L. AACC, 2n = 38) is the second leading edible oil crop in the world. The nutritional value of rapeseed to human beings has been greatly improved since the creation of canola oil, which contains a low level of erucic acid and has the great advantage of very high content of three major unsaturated fatty acids with reasonable ratio, i.e., approximately 65% oleic acid (OA, C18:1, ω-9), 20% linoleic acid (LA, C18:2, ω-6), and 9% α-linolenic acid (ALA, C18:3, ω-3) [1]. The production of rapeseed which is either high or low in ALA is now an important priority for breeders, based on the two major requirements of oxidative stability and high ω-3 nutrient content [1,2]. Because of the oxidative instability of ALA during long-term storage and high-temperature cooking, breeders have developed dozens of high-OA/low-ALA (HOLL) varieties [3–8]. However, defects such as poor resistance, late ripening, and low seed vigor have been reported in some low-ALA germplasms [9]. To better manipulate OA and ALA biosynthesis, therefore, it is necessary to find new genetic loci with fewer adverse effects on the biological and agronomic traits [10]. All previous studies on the genetic control of rapeseed ALA content have been based on plant accessions with low and medium levels of ALA content, but not high-ALA genotypes [4,5]. Nowadays, high-ALA trat is also desired by consumers because ALA is an essential nutrient that cannot be synthesized by the human body and must be obtained from foods [11]. ALA has been recommended by the Food and Agriculture Organization as a vegetable ω-3 fatty acid for dietary supplementation, to increase the ratio of ALA: LA (ω-3:ω-6) in food [11]. However, the availability of the main vegetable ω-3 sources, i.e., oilseed flax and Perilla, is restrained by stringent cultivation conditions. In light of this, an economical and effective alternative method may be to supplement ALA directly into people’s diets through high-ALA canola oil [1–2]. It is possible to obtain high-ALA B. napus germplasm because high-ALA germplasm exists in some other Brassica species [12]. In recent years, several new germplasms of B. napus with ALA content levels of 15% to 21%, for example, two yellow-seeded B. napus inbred lines YH25005 and R8Q10, have been created through various breeding methods [1,2,13,14].
For better-quality canola breeding, therefore, the genetic mechanisms underpinning the formation of either high- or low-ALA resources in B. napus must be comprehensively understood, and more knowledge is required about the genetic network that regulates ALA synthesis. ALA biosynthesis in Arabidopsis seed is carried out by two types of fatty acid desaturase (FAD). The ω-6 type FAD2 and FAD6 catalyze the first desaturation step of OA to LA; the ω-3 type FAD3, FAD7, and FAD8 catalyze the further desaturation of LA to ALA. The FAD2 and FAD3 proteins work in the endoplasmic reticulum and determine the content of the three major unsaturated fatty acids in seed oil; their isoenzymes FAD6, FAD7, and FAD8 are located on the chloroplast membrane and exert less influence on seed oil deposition. Dysfunction of FAD2 and FAD3 is the main method used to increase OA content or reduce ALA content [3–5]; hence, previous studies have mainly focused on the relationship between the traits of low ALA or high OA and coding sequence (CDS) variations in FAD2 or FAD3 [3–8]. However, the promoter information of BnFAD2/BnFAD3 and the regulation of their transcription levels have been less studied. Higher ALA content and lower OA content can be achieved both by overexpression and by increasing the copy numbers of Arabidopsis FAD3 [15]. The upregulation of FAD3 expression in flax seeds has been associated with high ALA levels [16]. In one recent study, researchers found that more ALA was accumulated in a transgenic B. napus plant with overexpressed BnFAD3; however, the seeds also shriveled, with decreased oil content and lower levels of seed germination [17].
Although FAD2 and FAD3 are the key factors in controlling OA reduction and ALA biosynthesis, the inheritance of OA and ALA content in the allotetraploid species rapeseed is more complex than the model plant Arabidopsis because most B. napus germplasm maintain four or six homologous copies of FAD2 and FAD3 [5] from its progenitors. The duplicated genes, such as BnFAD2.A5 and BnFAD2.C5, often have redundant functions and show additive effects on the accumulation of OA and ALA [18]. Therefore, OA or ALA content in rapeseed is controlled by multiple copies of BnFAD2 and BnFAD3 and function variation in different copies may affect the phenotype of OA or ALA content. For instance, in the progenies of the two crosses ‘YH25005 × ZH9’ and ‘R8Q10 × D636’ (lines ZH9 and D636 are wildtypes containing about 9% ALA), the inheritances of OA and ALA content are controlled by a lot of minor-effect genes, following a genetic model of ‘additive-dominant-epistatic polygene’ [14]. However, the results of our subsequent study [1] showed that, if ZH9 and D636 were replaced by a HOLL cultivar SW, the inheritances of OA and ALA changed to another genetic model of ‘two pairs of additive major genes + additive-dominant polygenes’. It indicates that the HOLL parent SW might contain two loss-of-function mutations in BnFAD2 and/or BnFAD3 and the mutations had a strong effect of ‘two pairs of additive major genes’ to reduce ALA and increase OA content [1].
In the present study, we focused on the mutation and expression regulation of BnFAD2 and BnFAD3, which are the most critical factors in controlling ALA content. The expression levels of multiple copies of these genes were detected by real-time quantitative PCR (qPCR). We cloned the coding sequences (CDSs) and promoters of all copies of BnFAD2 and BnFAD3 in six B. napus accessions with high-, medium- or low-ALA traits to find important mutations including SNPs (single-nucleotide polymorphisms) and indels (insertions/deletions) in the CDS region, affecting protein functions, and also in promoter cis-regulatory elements (CREs), affecting changes in expression levels.
Six plant accessions (Table 1) were included in the study: three high-ALA inbred lines (R8Q10, 2006L, and YH25005); an elite cultivar ZS11 whose genome had been sequenced (representing a wildtype genotype); a low-ALA line A28; and a special-line SW with high-OA and low-ALA traits. The plants were grown in the autumn of the year 2021 in the experimental field of Northwest A&F University. In the next spring, several batches of flowers opening on the same day were selected in the six accessions and bagged to obtain their self-pollinated seeds. The content of fatty acid components in mature seeds of the six plant accessions was determined using a GC-2010 Plus gas chromatographer (Shimadzu, Kyoto, Japan) as it had been described [1].

2.2.1 Gene Cloning and Sequencing
Different reference genes for BnFAD2 and BnFAD3 were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov) and the B. napus pan-genome database (BnPIR) (http://cbi.hzau.edu.cn/bnapus). For example, the ZS11 genome possesses four copies of BnFAD2 (BNaA01g00369500zs, BNaA05g0427800zs, BNaC01g00461200zs and BNaC05g0480500zs) and six copies of BnFAD3 (BNaA03g0143900zs, BNaA04g0019190zs, BNaA05g0136600zs, BNaC03g0167300zs, BNaC0480200zs, and BNaC0496200zs). These were henceforth referred by chromosome, i.e., FAD2.A1, FAD2.C1, FAD2.A5, FAD2.C5, FAD3.A3, FAD3.A4, FAD3.A5, FAD3.C3, FAD3.C4, and FAD3.C4- (antisense strand of C4). The PCR primers (Table 2) were designed for full-length or overlapping partial fragments of gDNA, CDSs, and promoter sequences of different gene copies using the online Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).

DNA was extracted from seedlings of the six genotypes using a standard protocol using hexadecyl trimethylammonium bromide lysis buffer. RNAs in the seed samples at 20, 27, and 34 DAP (days after pollination) were extracted by using E.Z.N.A.® Plant RNA Kit (Omega Bio-tek Inc., GA, USA) and were transcribed into cDNA for cloning of CDS region. PCR amplification was performed on a thermal cycler using Phanta® Max Super Fidelity DNA polymerase (Vazyme Biotech, Nanjing, China). The PCR products were separated via agarose gel electrophoresis, and the target fragments were extracted using the D2500 Gel Extraction Kit (Omega Bio-tek Inc, GA, USA). The fragments were inserted into pEASY ® Blunt Zero Cloning Vector (TransGen Biotech., Beijing, China). The positive clones were identified by PCR using specific primers, and the molecular weights of PCR products were confirmed again using agarose gel electrophoresis. The DNA sequencing was conducted by SANGON Biotech (Shanghai, China) and conformed with the results of two or three clones per gene copy. The CDS region of each gene was identified after sequence alignment using MEGA 7.0 (https://www.megasoftware.net/) and Vector NTI 11.0 (Invitrogen, CA, USA). The 1.5–2 kb sequence before the first open reading frame (ORF) of a gene was defined as the candidate promoter region. Each promoter was named by adding ‘P-’ before the corresponding gene. The prediction of promoter structures and cis-regulatory elements (CREs) was conducted on the PLANTCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
2.2.2 Detection of Gene Expression Levels by qPCR
Immature seeds from the six plant accessions were quickly collected at 20, 27, and 34 DAP, immersed in liquid nitrogen, and then stored at −80°C. Each plant accession had three biological replications. Total RNAs were extracted using RNA Isolation Kit (Omega Bio-tek Inc., GA, USA) and then transcribed into cDNA. The primers designed for the various homologous copies of FAD2 and FAD3 (Table 3) were used to detect their expression levels by qPCR. BnFAD2.A1, encoding a truncated protein without function [5,18], was not detected here in light of that very low abundance in the transcriptome of developing seeds [18]. The PCR reaction system consisted of NovoStart SYBR qPCR Supermix plus (Novoprotein, Shanghai, China) with SYBR Green I, Rox II, primers, cDNA template, and RNase-free water. The Rapeseed beta-actin7 gene was used as the internal reference. The PCR reactions were completed on a QuantStudio 7 Flex PCR thermal cycler (ABI, CA, USA).

3.1 Expression Levels of BnFAD2 and BnFAD3 Copies during Seed Development
qPCR was carried out to detect the expression levels of individual copies of BnFAD2 and BnFAD3 in immature seeds (20, 27 and 34 DAP) of 2006L, R8Q10, YH25005, ZS11, A28 and SW (Fig. 1). The expression levels of FAD2 copies, especially BnFAD2.C1, in the two low-ALA accessions SW and A28 were significantly lower than in the others. The expression of individual BnFAD3 copies in the three high-ALA accessions was higher than in the low-ALA seeds of A28 and SW. The expression of BnFAD3.A4 and BnFAD3.A5 in the three high-ALA accessions was significantly higher than in ZS11.
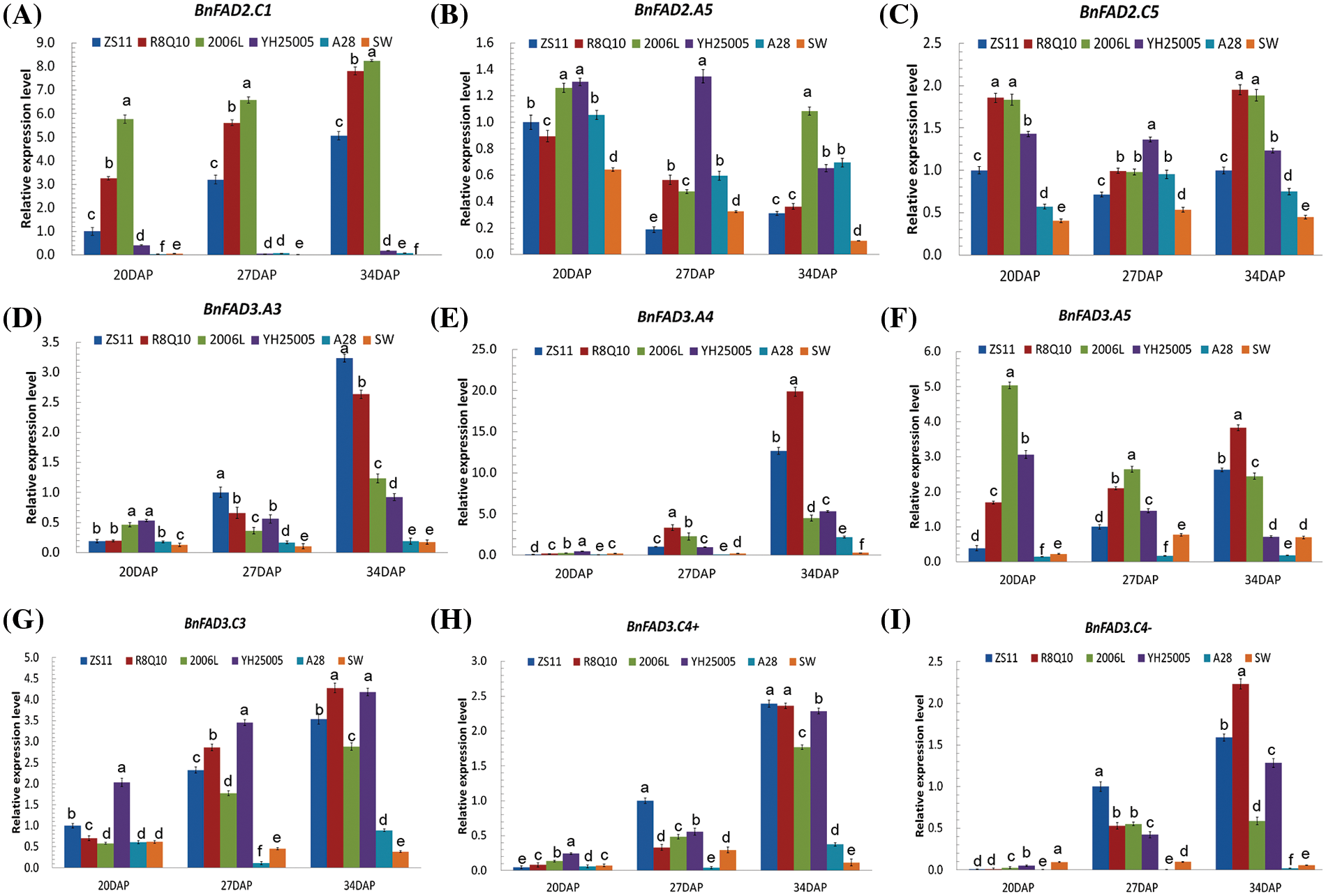
Figure 1: Expression level of BnFAD2 and BnFAD3 copies in the six genotypes with different ALA contents. The letters a, b, c, etc., indicate significant differences at a 95% confidence level
3.2 CDS Region Variations in BnFAD2 Copies
Due to the close similarity of the four BnFAD2 copies, they were amplified by PCR using two forward primers (HY267 and HY267-2) and one reverse primer (HY268) [5], and then identified after monoclonal sequencing. The BnFAD2.A1 (BNaA01g00369500zs in ZS11 genome deposited in BnPIR website http://cbi.hzau.edu.cn/bnapus/) is a short pseudogene encoding a truncated protein without enzyme function [5,6]. An identical 1155 bp long CDS region of BnFAD2.C1 (BNaC01g00461200zs) was found in ZS11, 2006L, and R8Q10, but was not detected in SW, A28, and YH25005. This copy is also absent in the Darmor genome in the BnPIR database, in line with a previous report of BnaC.FAD2.b absence in cv. Cabriolet [7], indicating the possible deletion of this copy in many germplasm during the evolutionary process. Both obtained sequences of BnFAD2.A5 and BnFAD2.C5 were 1155 bp long and encoded 385 amino acids, in line with the results of previous studies [5–7]. BnFAD2.A5 (NCBI accession No. OR676828) of SW had a G > A SNP at nucleotide 316 of the CDS region, resulting in E (Glu) > K (Lys) substitution in the 106th residue (abbreviated as E106K) of the amino acid (Table 4, Fig. S1) compared to ZS11. This was an amino acid mutation from acidic to alkane residue which may have caused the defect in this protein function. BnFAD2.C5 (OR676829) in both SW and A28 had a G > A SNP at 908 bp of the CDS (Fig. S2), resulting in a mutation from G (Gly) to E (Glu) in the 303rd residue (Fig. S3). Finally, BnFAD2.C5 (OR676830) in A28 had a C > T mutation at 1103 bp of the CDS, resulting in a mutation P (Pro) > L (Leu) in the 368th residue (Fig. S3).

3.3 Sequence Comparison of BnFAD3 Copies
There are six copies of the BnFAD3 gene in the ZS11 reference genome, and these exhibited 99%, 97.6%, and 85.7% nucleotide identity in a pairwise comparison of the homologous copies of chromosome A5 vs. C4, A4 vs. C4-, and A3 vs. C3, respectively. Thus, three pairs of specific primers with main differences at the 3-prime end were designed for the three groups of BnFAD3 copies, and the PCR products using each pair of primers obtained two different nucleotide sequences. The six accessions had the same CDSs as BnFAD3.A3 (Bna03g0143900ZS) and BnFAD3.A5 (Bna05g0136600ZS) in the BnPIR database, but different CDSs of the other copies.
The CDSs of BnFAD3.A4 in the six accessions were 1134 bp long and encoded 378 amino acids. The CDS sequences of BnFAD3.A4 in R8Q10, 2006L, and SW were identical to those of BnFAD3.A4 in ZS11 (BNaA04g0019190zs). The BnFAD3.A4 CDS in YH25005 had a T > C SNP at 309 bp, compared with that of ZS11; however, this was a synonymous mutation. BnFAD3.A4 in A28 (OR676831) had three SNPs at 309, 620, and 870 bp of the CDS region, compared with that of ZS11 (Fig. S4). The shift of C > T at the 620th base led to the substitution of Ser (S) with Leu (L) in the 207th residue (Fig. S5), but the other two mutations were synonymous.
The CDS sequences of BnFAD3.C3 in YH25005 and SW were as same as in ZS11 (BnaC03g0167300zs), with 1152 nucleotides coding for 384 amino acids. However, 2006L, R8Q10, and A28 (OR676832) had another CDS haplotype (Table 4, Fig. S6) of BnFAD3.C3, the same as that of cv. Tapidor (BnaC03t0127700ta), Westar (BnaC03g0107200we), and No. 2127 (BnaC03t0094100no) in the BnPIR database. There were six SNPs in the BnFAD3.C3 CDS of 2006L, R8Q10, and A28 when compared with the CDS of ZS11; three of these SNPs were synonymous mutations, and the remainder caused amino acid substitution (Fig. S7). Notwithstanding this result, it may be that these SNPs do not affect ALA synthesis, because both cv. Tapidor and Westar have a common phenotype of ALA content.
The CDS of the BnFAD3.C4 gene is also 1152 bp long and encodes 384 amino acids. Compared to ZS11, the BnFAD3.C4 CDS of R8Q10 (OR676834), 2006 L and YH2500 had one T > G SNP at 16 bp, resulting in the mutation of the sixth residue from Y (Tyr) to D (Asp) (Table 4, Fig. S8). BnFAD3.C4 in A28 (OR676833) had one C > T SNP at 200 bp of the CDS region, resulting in Ala (A) to Val (V) mutation at the 67th residue (Fig. S8).
The CDS of the BnFAD3.C4- gene of YH25005 is the same as that of ZS11, with 1143 bp encoding 381 amino acids (Fig. S9). In comparison, the BnFAD3.C4- CDS of R8Q10 (OR676837) has one G > T SNP at 26 bp, resulting in a mutation of the ninth amino acid from Ser (S) to Ile (I) (Fig. S10). There were two G > T SNPs at 152 and 157 bp in BnFAD3.C4- CDS of 2006L (OR676838), resulting in the mutation of the 51st residue from Ser (S) to Ile (I), and of the 53rd residue from Val (V) to Phe (F). The BnFAD3.C4- CDS of SW (OR676836) had an SNP of G > C at 16 bp, resulting in the mutation of the sixth amino acid from D (aspartic acid) to Y (tyrosine) (Table 4).
3.4 Variations in BnFAD2 Promoter Sequences
The promoter sequences of BnFAD2.A5 (OR676843) and BnFAD2.C5 (OR676846) obtained from the six accessions were 100% identical, except a depletion of three bp in P-BnFAD2.A5 of A28 (OR676845) and insertion of seven bp in P-BnFAD2.A5 of Yh25005 (OR676847). P-BnFAD2.A1 of SW (OR676839) and A28 (OR676840) showed five SNPs and a deletion of four nucleotides (TAAT) compared to ZS11 and P-BnFAD2.A1 of 2006L and R8Q10 (OR676841) possessed three SNPs. However, these SNPs are not located on any known CREs and all known CREs in the six accessions were arranged in the same pattern in the promoter. The promoter of BnFAD2.C1 was successfully amplified by designing a specific nucleotide at the 3’ end of the primer. After separation via agarose gel electrophoresis, a band with identical molecular weight of ca. 2 kb—as well as an identical sequence (OR676844)—was produced in R8Q10, 2006L, and ZS11; however, no band was produced in YH25005, A28, and SW (Fig. 2). The divergence of two alleles of BnFAD2.C1 promoters was consistent with their CDS sequence being absent, as mentioned above, indicating that this copy of R8Q10, 2006L, and ZS11 possibly has an ancestry different from that of YH25005, A28, and SW.
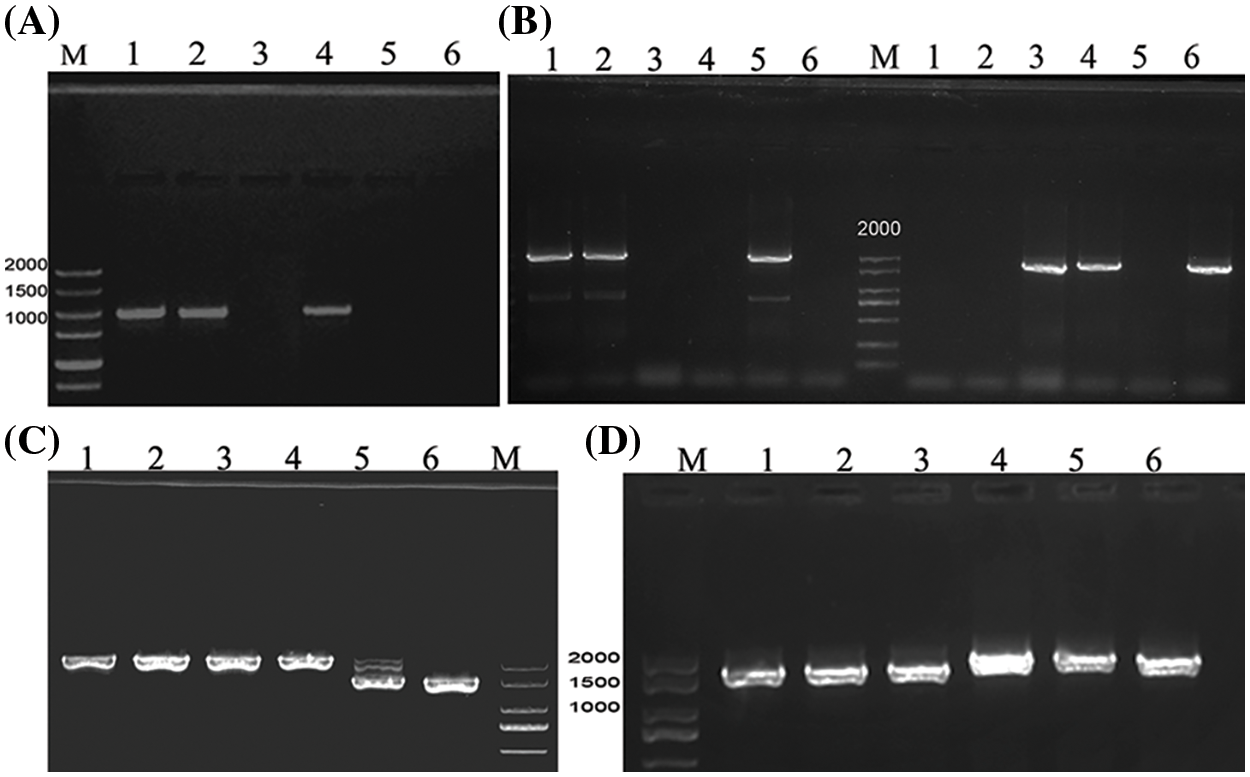
Figure 2: Electrophoretic profile of PCR products for the promoter P-BnFAD2.C1 (A), P-BnFAD3.C3 (B) using primer pairs PC3-DF/PC3-R (left panel) and PC3-ZF/PC3-R (right panel), P-BnFAD3.A3 (C), and P-BnFAD3.C4- (D), respectively. M: DNA Marker; Lane 1: R8Q10; Lane 2: 2006L; Lane 3: YH25005; Lane 4: ZS11; Lane 5: A28; Lane 6: SW
3.5 Variations in BnFAD3 Promoter Sequences
There are great differences in the transcription start sites in the promoter sequences of the six BnFAD3 copies of the ZS11 reference genome, so we designed six pairs of specific PCR primers to amplify them. The similarity in the promoter sequences of BnFAD3.A4, BnFAD3.A5, and BnFAD3.C4- (OR676854-OR676866) within the six accessions was very high, i.e., about 99%, with a few SNPs located not on any known CREs.
According to two different promoter sequences of the BnFAD3.C3 gene found in the public database, two pairs of specific primers were designed, i.e., PC3-DF/PC3-R and PC3-ZF/PC3-R; agarose gel for electrophoresis resulted in amplification bands for R8Q10, 2006L, and A28 in the case of PC3-DF/PC3-R, while bands for YH25005, ZS11, and SW (OR676868) were obtained in the case of PC3-ZF/PC3-R (Fig. 2). The promoter sequences of the three accessions R8Q10 (OR676867), 2006L, and A28 (Fig. S11) were the same as those for the corresponding fragment of P-BnFAD3.C3 of cv. Tapidor, Westar, and No. 2127 in the BnPIR database. Comparing the two types of promoter sequences, except the 322 bp before the transcription initiation site, the remaining promoter sequences were found to be very different. In the prediction of CREs, there was also a large gap between the two alleles (Fig. 3).
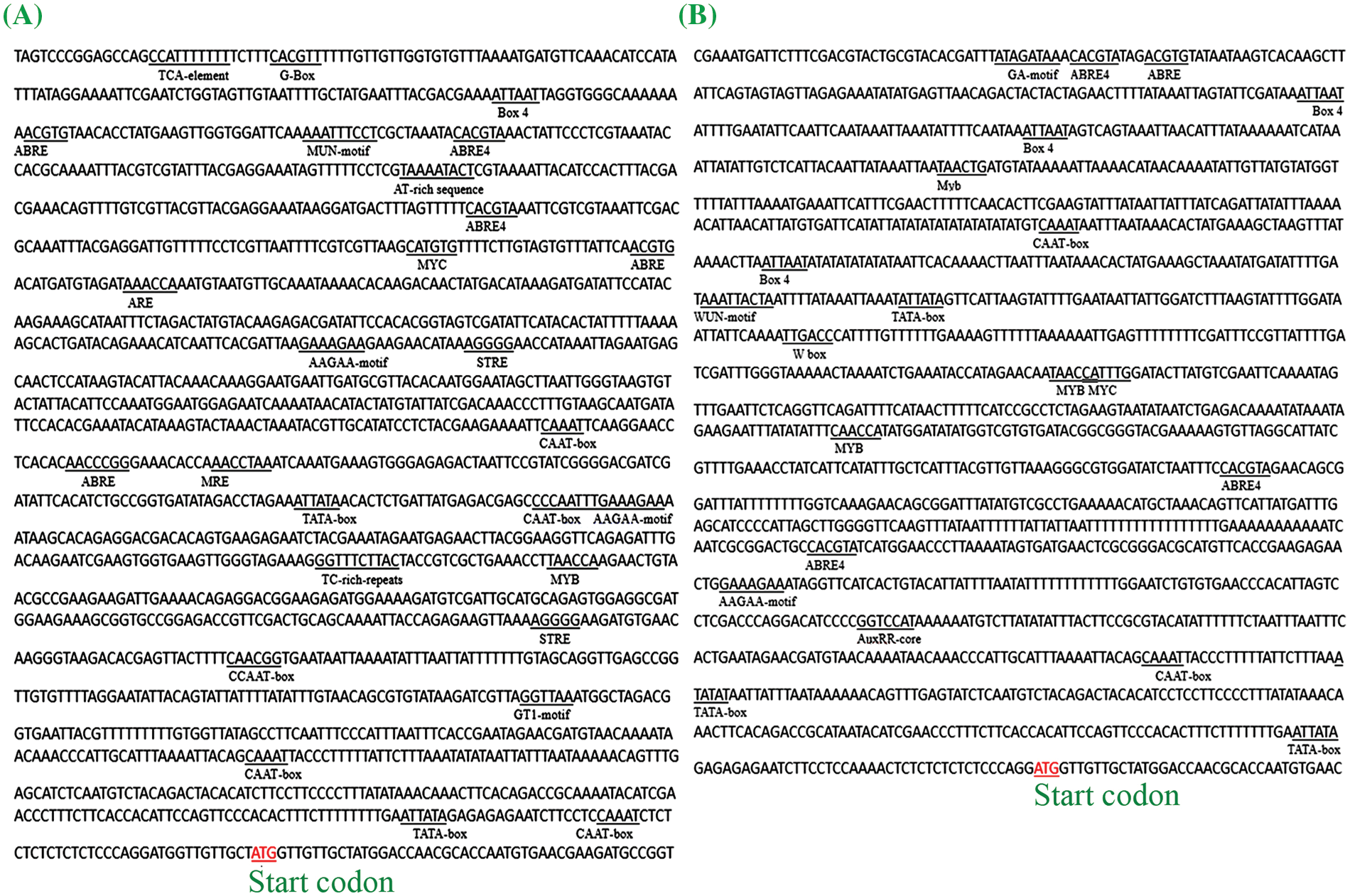
Figure 3: The predicted CREs on the promoter of BnFAD3.C3 of R8Q10 (A) and ZS11 (B)
The PCR products for P-BnFAD3.A3 of A28 and SW had a smaller size of 1470 bp on gel electrophoresis (Fig. 2) than the band of 2016 bp in ZS11, YH25005 (OR676851), 2006L, and R8Q10 (OR676848). P-BnFAD3.A3 of A28 (OR676849) and SW (OR676850) had a 549 bp fragment deletion and an (AT) insertion (Fig. S12). The depletion included two key CREs, namely, TATA-box (TATAAAT) and A-box (CCGTCC) of polymerase II transcription, and several important CREs such as the MYB binding site MBSI (AAAAAACCGATATGTA), the abscisic acid response element ABRE4 (CACGTA), the auxin response element TGA element (AACGAC), the WRKY homologous action element W-box (TTGACC), and the stress response element STRE (AGGGG) (Fig. 4).

Figure 4: The predicted CREs on the promoter of BnFAD3.A3 of ZS11. The red region was deleted in SW and A28
The PCR products for P-FAD3.C4 of six accessions showed two bands of slightly different size, separated by agrarose gel electrophoresis (Fig. 2). The shorter band produced by R8Q10 (OR676852), YH25005 and 2006L was 1750 bp long, identical to cv. Darmor and Tapidor in the BnPIR database; however, A28 and SW (OR676853) shared with ZS11 a longer sequence of 2018 bp (Fig. S13). The divergent patterns of the two alleles, with a low similarity of 83.0%, are consistent with the CDS differences mentioned above, further indicating that the BnFAD3.C4 gene of these six genotypes has two different ancestries. Compared with ZS11, the promoter sequences of the three high-ALA accessions lacked the module ACE and the MYB-binding site MRE involved in light response, as well as the MYB-binding site MBSI involved in flavonoid biosynthesis regulation (Fig. 5).

Figure 5: The predicted CREs on the promoter of BnFAD3.C4 of R8Q10 (A) and ZS11 (B)
4.1 Both BnFAD2 and BnFAD3 are Hotspots of Genetic Modification in the Production of High-OA or Low-ALA Phenotypes
The in-tandem steps of OA and ALA biosynthesis are contradictory when total levels of unsaturated fatty acid are fixed. Therefore, when producing a high-OA and/or a low-ALA phenotype, either FAD2 or FAD3 will be the hotspot of genetic modification [3,5,8,18–20]. The results of the present study show that, compared to ZS11, the expression of most BnFAD3 copies was upregulated in the three high-ALA accessions, but downregulated dramatically in the low-ALA line A28 and the HOLL line SW, at either one or two of the three stages of seed development. Generally, the different expression levels of these genes under the same background may be caused by differences in their promoter sequences. For example, the obvious differences between high and low-ALA plant groups in terms of indels and SNPs in the BnFAD3.C4 promoter may explain their differences in expression levels. Compared with ZS11, the promoter of BnFAD3.A3 in the low-ALA SW and A28 has a 549 bp deletion and a 2 bp (AT) insertion. The causal relationship between the promoter and the level of gene expression level may be further analyzed by investigating the activity of a mutated promoter tagged by a reported gene such as GUS or GFP. The activity of some upstream transcription factors such as FUS3 or bZIP67 should also be studied because these can regulate the expression of FAD3 or even FAD2 [21–24]. In addition to considerable changes in gene expression levels, some non-synonymic amino acid residue mutations may also affect the catalytic function of enzymes. Protein and enzyme functions can be validated by expressing the mutated CDS of BnFAD2 and BnFAD3 in yeast and then measuring the catalytic speed of extracted proteins on their substrate OA or LA. In addition, the association between these SNP or indel mutations and phenotype can be validated in various segregation populations and thus there need for many allele-specific populations to keep unanimous genetic background (at least the same background of non-target BnFAD2 and BnFAD3 copies) in each population.
4.2 Relationship between Variations in BnFAD2/BnFAD3 and the Low-ALA Phenotype
Loss-of-function mutation in FAD2 leads to an increase in OA but a reduction in ALA. In a previous study, it was found that the high-OA mutant M604-855 harbors SNPs at nucleotides 270, 1044, and 1062 in a BnFAD2 copy, leading to the production of a stop codon [3]. In another high-ALA rapeseed accession FC81, it was found a C > T substitution at nucleotide 421 of BnFAD2.A5, leading to His141Tyr mutation in the amino acid residue, as well as a G > A substitution at nucleotide 1073 of BnFAD2a.C5, leading to Arg358Lys mutation [25]. In the HOLL cv. SW-hickory, Yang, et al. [5] found that a 4 bp insertion at the 567 bp of BnaA.FAD2 (chromosome A5) resulted in a frameshift that led to a misreading. They also found a C > T substitution in the seventh exon, resulting in a missense mutation of BnaA.FAD3.b, as well as a G > A transition in the 5’ splice donor site of the sixth intron, resulting in abnormal splicing in BnaC.FAD3.b. Two identical C >T and G > A SNPs were also validated in another low-ALA germplasm [19].
The mutation sites of BnFAD2 and BnFAD3 found in the two low-ALA accessions A28 and SW (Table 4 and Figs. 3–5) are different from those described in previous studies [3–5]. In the present study, the HOLL accession SW had four obvious mutation sites including the absence of BnFAD2.A1, Glu106Lys mutation of FAD2.A5 protein, Gly303Glu mutation of BnFAD2.C5, and Asp6Tyr mutation of BnFAD3.C4, leading to stringent limitations in LA and ALA synthesis and very high OA accumulation. Compared to SW, the low-ALA accession A28 also had an absence of BnFAD2.A1 and a mutation of Gly303Glu in BnFAD2.C5 but a normal copy of BnFAD2.A5. The normal BnFAD2.A5 can synthesize LA, resulting in a limited accumulation of OA (67.10%) (Table 1).
Compared with 2006L and R8Q10, SW, YH25005, and A28 all lacked a copy of BnFAD2.C1, which has a normal CDS encoding 384 amino acids like the A5 and C5 copies, and thus may have a certain enzymic function. The results of qPCR detection of BnFAD2.C1 showed that it is expressed in 2006L, R8Q10, and ZS11 but not in SW, YH25005, and A28. In addition, ALA content in YH25005 is slightly lower—by two to four percentage points—compared with R8Q10 and 2006L. Meanwhile, the content of OA plus LA in YH25005 is slightly higher than R8Q10 and 2006L. We speculated, therefore, that BnFAD2.C1 also plays a minor role in ALA biosynthesis.
4.3 Upstream Transcription Factors May Regulate the Expression of Various BnFAD3 Copies
The variation in BnFAD2 and BnFAD3 promoters and the corresponding transcriptional regulation may be an important reason for the elevated expression levels of BnFAD2 and BnFAD3 in the three high-ALA accessions. However, the SNP and indel variations in the promoters of the three BnFAD3 copies (chromosomes A3, C3, and C4) in the three high-ALA accessions (Fig. 5) cannot explain the upregulation of the other copies on chromosomes A4, A5 and C4-. Therefore, there may be an activation of upstream transcription regulatory factors (for example transcription factor), which may play important roles both in the upregulation of the former three copies (chromosomes A3, C3, and C4) and the latter three copies chromosomes (A4, A5 and C4-). Previous studies have found that some Transparent Testa mutations in Arabidopsis have significant effects on key transcription factors related to embryonic development and that such transcription factors as FUS3, LEC1, ABI3, and bZIP67 can regulate the expression of FAD3 [21–24]. Our high-ALA plants, R8Q10, 2006L, and YH25005, all have yellow-seed phenotypes with lowered expression of many Transparent Testa homologs and upregulation of the genes encoding transcription factors including FUS3, LEC1, ABI3, and bZIP67 in the three accessions [26]. We found that, in thousands of rapeseed samples, the yellow-seeded population often had an ALA content around two percentage points higher than that of their black- or brown-seeded siblings (unpublished). Thus, yellow-seededness may also be an important indirect factor in the elevation of ALA content. However, this does not mean that yellow seedness is necessary for the elevation of ALA content because some other high-ALA accessions have brown-colored seeds [2,13].
In the present study, detailed sequence mutation profiles for the CDSs and promoter regions of BnFAD2 and BnFAD3 in six genotypes with high- or low-ALA phenotypes were produced. Most of these have not previously been reported in the literature. The profile of BnFAD2 mutation in the two low-ALA accessions A28 and SW is different from that reported in previous studies. The CDSs of BnFAD2.C5 and BnFAD2.A5 in SW and A28 are quite different from those in the wild type and may lead to significant defects in their catalytic function. Some mutations in BnFAD3 in the high-ALA accessions are reported for the first time here. The SNPs or indels in the BnFAD3 promoters of high-, medium-, and low-ALA accessions may affect their binding situation concerning corresponding transcription factors and transcriptional levels. The influence on the enzymic activity by residue substitution of several BnFAD3 CDSs of 2006L, R8Q10, and YH25005 needs a further study. The mutation sites may be considered useful information for the design of molecular markers for our next breeding program.
Acknowledgement: None.
Funding Statement: The study was financially supported by Projects from Shaanxi Province (2021LLRH-07-03-01 and 2023-ZDLNY-07) and Yangling Seed Industry Innovation (YLzy-yc2021-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions: Study conception and design: C.Y.Y.; data collection: H.X.W., X.H.Z., X.Y.C. and K.L.; analysis and interpretation of results: H.X.W., X.H.Z., X.Y.C., K.L., J.G.D., A.X.X., Z.H. and C.Y.Y.; draft manuscript preparation: C.Y.Y., H.X.W., and X.H.Z. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are contained in the paper and the additional information. The CDS and promoter sequences of BnFAD2 and BnFAD3 copies have been deposited in the NCBI database and the accession numbers are provided in the paper.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Ethics Approval: Not applicable.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.050321.
References
1. Zhang XH, Lian JL, Dai CY, Wang XL, Zhang MZ, Su X, et al. Genetic segregation analysis of unsaturated fatty acids content in the filial generations of high-linolenic-acid rapeseed (Brassica napus). Oil Crop Sci. 2021;6:169–74. [Google Scholar]
2. Li DR, Zhang YW, Zhang WX. A major breakthrough in the breeding of Brassica napus with high linolenic acid content, which linolenic acid content of germplasm resources exceeding 21% and of hybrids about 15%. J Plant Sci. 2019;3(2):165–6. [Google Scholar]
3. Guan CY, Liu CL, Chen SY, Pen Q, Li X, Guan M. High oleic acid content materials of rapeseed (Brassica napus) produced by radiation breeding. Acta Agron Sin. 2006;32(11):1625–9. [Google Scholar]
4. Hu XY, Sullivan-Gilbert M, Gupta M, Thompson SA. G-to-A mutation at the 5′ splice site of fad3c caused impaired splicing in a low linolenic mutant of canola (Brassica napus L.). Plant Biotech. 2007;24:397–400. [Google Scholar]
5. Yang Q, Fan C, Guo Z, Qin J, Wu J, Li Q, et al. Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic and low linolenic acid contents. Theor Appl Genet. 2012;125:715–29. [Google Scholar] [PubMed]
6. Lee K, In Sohn S, Jung JH, Kim SH, Roh KH, Kim J, et al. Functional analysis and tissue-differential expression of four FAD2 genes in amphidiploid Brassica napus derived from Brassica rapa and Brassica oleracea. Gene. 2013;531:253–62. [Google Scholar] [PubMed]
7. Wells R, Trick M, Soumpourou E, Clissold L, Morgan C, Werner P, Gibbard C, Clarke M, Jennaway R, Bancroft I. The control of seed oil polyunsaturate content in the polyploid crop species Brassica napus. Mol Breed. 2014;33:349–62. doi:10.1007/s11032-013-9954-5. [Google Scholar] [PubMed] [CrossRef]
8. Singer SD, Weselake RJ, Rahman H. Development and characterization of low α-linolenic acid Brassica oleracea lines bearing a novel mutation in a ‘class a’ FATTY ACID DESATURASE 3 gene. BMC Genet. 2014;15:94. [Google Scholar] [PubMed]
9. Bai S, Engelen S, Denolf P, Wallis JG, Lynch K, Bengtsson JD, et al. Identification, characterization and field testing of Brassica napus mutants producing high-oleic oils. Plant J. 2009;98(1):33–41. [Google Scholar]
10. Zhao Q, Wu J, Cai G, Yang QY, Shahid M, Fan CC, et al. A novel quantitative trait locus on chromosome A9 controlling oleic acid content in Brassica napus. Plant Biotech J. 2019;7(12):2313–24. [Google Scholar]
11. Fats and Fatty Acids in Human Nutrition. Report of an expert consultation. FAO Food Nutr Pap. 2010;91:1–166. [Google Scholar]
12. Sharafi Y, Majidi MM, Goli SAH, Rashidi F. Oil content and fatty acids composition in Brassica species. Inter J Food Propert. 2015;18(10):2145–54. [Google Scholar]
13. Chen ZZ, Zheng HG. High linolenic acid producing brassica plants; 2016. Patent Application Number: US20160010096A1. [Google Scholar]
14. Lian JL, Zhang XH, Dai CY, Wang XL, Gong Q, Yu CY. Genetic analysis of three unsaturated fatty acids in high linolenic acid rapeseed. J Jiangsu Agric Sci. 2020;48(21):84–92 (In Chinese). doi:10.15889/j.issn.1002-1302.2020.21.01. [Google Scholar] [CrossRef]
15. O’Neill CM, Baker D, Bennett G, Clarke J, Bancroft I. Two high linolenic mutants of Arabidopsis thaliana contain megabase-scale genome duplications encompassing the FAD3 locus. Plant J. 2011;68(5):912–8. [Google Scholar] [PubMed]
16. Banik M, Duguid S, Cloutier S. Transcript profiling and gene characterization of three fatty acid desaturase genes in high, moderated, and low linolenic acid genotypes of flax (Linum usitatissimum L.) and their role in linolenic acid accumulation. Genome. 2011;54(6):471–83. [Google Scholar] [PubMed]
17. Yin Y, Guo Z, Chen K, Tian T, Tan J, Chen X, et al. Ultra-high α-linolenic acid accumulating developmental defective embryo was rescued by Lysophosphatidic Acid Acyltransferase 2. Plant J. 2020;103(6):2151–67. [Google Scholar] [PubMed]
18. Huang H, Cui T, Zhang L, Yang Q, Yang Y, Xie K, et al. Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor Appl Genet. 2020;33(8):2401–11. [Google Scholar]
19. Bocianowski J, Mikołajczyk K, Bartkowiak-Broda I. Determination of fatty acid composition in seed oil of rapeseed (Brassica napus L.) by mutated alleles of the FAD3 desaturase genes. J Appl Genet. 2012;53:27–30. [Google Scholar] [PubMed]
20. Tian E, Zeng F, MacKay K, Roslinsky V, Cheng B. Detection and molecular characterization of two FAD3 genes controlling linolenic acid content and development of allele-specific markers in yellow mustard (Sinapis alba). PLoS One. 2014;9(5):e97430. [Google Scholar] [PubMed]
21. Deng W, Chen G, Peng F, Truksa M, Snyder CL, Weselake RJ. Transparent testa16 plays multiple roles in plant development and is involved in lipid synthesis and embryo development in canola. Plant Physiol. 2012;160:978–89. [Google Scholar] [PubMed]
22. Chen MX, Xuan LJ, Wang Z, Zhou LH, Li ZL, Du X, et al. TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol. 2014;165:905–16. [Google Scholar] [PubMed]
23. Zhai Y, Yu K, Cai S, Hu L, Amoo O, Xu L, et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotech J. 2020;18(5):1153–68. [Google Scholar]
24. Xie T, Chen X, Guo T, Rong H, Chen Z, Sun Q, et al. Targeted knockout of BnTT2 homologs for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J Agric Food Chem. 2020;68:5676–90. [Google Scholar] [PubMed]
25. Fu Y, Zhang YF, Zhang DQ, Yu HS, Lin BG. A high oleic acid allelic mutation of rapeseed BnFAD2 gene and development and application of the primers for SNP marker; 2019. China Invention Patent: ZL2019112120007. [Google Scholar]
26. Chen XY, Wu HX, Zhang XH, Guo RH, Li Ket al. Comparative transcriptomics uncovers upstream factors regulating BnFAD3 expression and affecting linolenic acid biosynthesis in yellow-seeded rapeseed (Brassica napus L.). Plants. 2024;13(6):760. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools