 Open Access
Open Access
REVIEW
SPATULA as a Versatile Tool in Plant: The Progress and Perspectives of SPATULA (SPT) Transcriptional Factor
Shanghai Key Laboratory of Bio-Energy Crops, Research Center for Natural Products, Plant Science Center, School of Life Sciences, Shanghai University, Shanghai, 200444, China
* Corresponding Author: Xiangyang Hu. Email:
Phyton-International Journal of Experimental Botany 2024, 93(3), 517-531. https://doi.org/10.32604/phyton.2024.049277
Received 02 January 2024; Accepted 04 February 2024; Issue published 28 March 2024
Abstract
With the rapid development of modern molecular biology and bioinformatics, many studies have proved that transcription factors play an important role in regulating the growth and development of plants. SPATULA (SPT) belongs to the bHLH transcription family and participates in many processes of regulating plant growth and development. This review systemically summarizes the multiple roles of SPT in plant growth, development, and stress response, including seed germination, flowering, leaf size, carpel development, and root elongation, which is helpful for us to better understand the functions of SPT.Keywords
SPATULA, as an important class of bHLH transcription factors, can combine light signals, auxin signals, and gibberellin (GA) signals to participate in various stress responses and thus regulate many processes of plant growth and development [1–3]. In addition to regulating pistil development, the Arabidopsis SPT gene has roles in organs such as cotyledons, leaves, roots, and fruits, and also plays a role in seed germination [4–9]. Thus, the SPT is involved throughout the plant life cycle. With global warming and the constant destruction of the earth, the outer environment has been altered, which thereby affects the growth and development of plants. Therefore, it is very essential to study the role of transcription factor SPT in plant growth and development.
SPT is a transcription factor encoding a family of bHLH proteins that play an essential role in plant growth and development [10]. Transcription factors, also called trans-acting factors, contain many functional structural domains, among which the DNA binding domain plays a vital role in regulating the transcription activity of genes [11]. The activity and function of transcription factors are also regulated by these functional domains. A transcription factor contains at least one DNA recognition site, also known as a motif, the transcription factor binds to a specific DNA sequence of the target gene through this site, thereby improving or repressing the transcriptional activity of the gene [12,13]. In turn, the expression of a gene is often co-regulated by multiple transcription factors.
The bHLH transcription factor contains a basic region and a helix-loop-helix region. The basic region is rich in basic amino acids and binds to the E-box (CANNTG), and the helix-loop-helix region contains two amphiphilic a-helices and a hydrophobic loop [14–16]. The hydrophobic loop is the key factor in this process that the bHLH transcription factors function by forming homo-or heterodimers with other proteins. The bHLH transcription factor was first identified in the E4 and E12 transcription factors of the mouse, whereas in plants, the study of bHLH transcription factor has been slower, and a total of more than 500 bHLH proteins, which are classified into 26–32 subfamilies, and members of the same family of transcription factors are often functionally redundant [17,18]. In addition, bHLH proteins play essential roles in signal transduction, hormone synthesis, as well as the plant’s resistance to abiotic stresses [19–21].
2 SPT Is Involved in the Regulation of Seed Germination
Seed dormancy and germination are significant stages in the process of plant growth and development, it was shown that SPT is also involved in regulating the germination of newly harvested seeds [1]. In Arabidopsis, the germination of dormant seeds is controlled by light and low temperatures [22]. This signaling pathway is regulated by the gibberellin oxidase GA3ox. GA3ox regulates gibberellin biosynthesis and it plays a vital role in the process of gibberellin biosynthesis [23]. Seed dormancy is the inability of viable seeds to germinate for intrinsic reasons despite suitable ecological conditions [24,25]. GA is necessary for seed germination and its application breaks seed dormancy. Bioactive GA content in seeds is also regulated by brightness and cold [26–28]. SPT, a multifunctional transcription factor, plays a vital role in light-and temperature-controlled seed germination by regulating the transcription of GA oxidase GA3ox and controlling the seed response to cold stratification. In the absence of light and low-temperature stratification, newly harvested wild-type Ler seeds exhibited dormancy, but the mutant spt-10, in contrast to Ler, showed higher germination. These researches suggest that SPT is involved in the regulation of seed germination. By RT-qPCR analysis of the expression level of GA3ox1 and GA3ox2 in the Ler ecotype and spt-10 mutant, it was found that there was a remarkable increase in the expression level of GA3ox1 and GA3ox2 in the spt-10 mutant as compared to the Ler ecotype [1]. These results suggest that SPT negatively regulates seed germination by negatively regulating the expression of GA3ox1 and GA3ox2.
Freshly harvested seeds usually exhibit initial dormancy, which severely affects seed germination, which is co-regulated by a balance of phytohormone ratios and environmental factors, including abscisic acid (ABA) and gibberellin (GA) [29]. ABA transcription level is regulated by the ABA transcription factors ABI3, ABI4, and ABI5 [30–33]. SPT plays a vital role in seed germination, however, it has been proved that SPT exhibits opposite regulatory effects on seed germination in two different ecotypes, Col-0 and Ler. By observing the phenotypes of seed germination of the spt mutant and overexpression materials in the Col-0 and Ler ecotypes, it was found that the SPT overexpression plants promoted seed dormancy in the Col-0 ecotype, but SPT inhibited seed dormancy in the Ler ecotype. The results showed that the mutant spt-2 in the Ler ecotype exhibited less seed dormancy than the wild type under conditions of unstratified low-temperature stratification, but the mutant spt-11 had higher germination rates than the Col-0 ecotype [34]. These results illustrate the importance of different ecotypes for SPT regulation of the seed germination process. This physiological phenomenon can be explained by the regulation of gene expression by SPT. Evaluation of the abundance of transcripts of genes related to ABA and GA biosynthesis during seed germination proved that the expression level of ABA1 was notably increased in spt-2 (Ler), but the ABA1 expression level of the mutant spt-12 was lower than that of Col-0. This also well explains the contrary regulatory effects of SPT on seed germination in Col-0 and Ler. It has been proved that SPT can bind to the G-box (CACGTG) on the promoter sequences of target genes and thus regulate their expression level [35–37]. Further analysis by ChIp and RT-q-PCR to confirm whether SPT binds specifically to the promoters of the target genes proved that only ABI5, REPRESSOR OF GA (RGA), and MOTHER OF FT AND TFL1 (MFT) were enriched in G-Box specific amplicons, on which suggests that SPT could bind to the promoter of MFT, ABI5, and RGA. Moreover, SPT promotes the expression of the ABI5 gene, while the expression of the RGA and MFT gene was suppressed by SPT. This physiological process takes place in two different ecotypes, Col-0 and Ler, resulting in converse seed dormancy phenomena in Col-0 and Ler.
In addition, MFT, as a phosphatidylethanolamine-binding protein, has been shown to play a vital role in multiple growth signaling pathways in plants. In wheat, cold-induced up-regulation of MFT results in strong seed dormancy [38,39]. Similarly, in Arabidopsis, MFT also promotes seed dormancy. It was shown that under far-red light conditions, the expression of SOM was promoted by PIF1, which led to the increase of GA. The ABA level was notably decreased in the mft mutant compared with the wild type under far-red light conditions, suggesting that the inhibitory effect of MFT on seed germination acts downstream of ABA, which also implies that MFT inhibits seed germination under far-red light by positively regulating ABA signaling. The transcription level of MFT was notably increased under far-red light conditions and was lower in the pif1-1 mutant than in the wild type, which also implies that far-red light promotes MFT expression through the PIF1 signaling pathway. However, the expression level of the MFT gene was found to be up-regulated in the spt-2, which indicated that SPT repressed the expression of MFT. Meanwhile, under far-red light conditions, the expression level of SPT was extremely reduced in the Col-0 ecotype, but the expression level of SPT was remarkably increased in the pif1-1 mutant, which implies that far-red light inhibits the expression of SPT through the PIF1 pathway [22].
3 SPT Is Involved in the Regulation of Flowering Time in Arabidopsis thaliana
The transition of plants from nutrient growth to reproductive growth is co-regulated by both external and internal factors, such as light, temperature, and plant hormones [10,40]. CONSTANS (CO) and Flowering Locus C (FLC) were found to be involved in the regulation of flowering time in Arabidopsis [41,42]. CO regulates photoperiodic flowering by binding to the promoter of FLOWERING LOCUS T (FLT) through the TGTG (N2-3) ATG motif. FLC inhibits flowering by binding to the promoter of the SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1) and inhibiting its transcriptional activity [43–45].
In addition, photoperiod is also involved in regulating the flowering process of plants [46]. At least three classes of photoreceptors are known to exist: photoreceptors: sensing red and far-red light, cryptochromes and phototropins as well as proteins such as the ZTL family, which sense blue and near-ultraviolet light, and UVR8, which belongs to the ultraviolet B (UV-B) receptor and senses the UV-B region [47–49]. Among them, phytochromes play a vital role in regulating plant growth [50]. PHYB is one of the phytochromes family and is involved in regulating plant’s response to light [51,52]. SPT, as a multifunctional transcription factor, also plays a vital role in regulating the flowering process in Arabidopsis thaliana [7]. It was shown that the SPT overexpression plant has the phenotype of long hypocotyls, whereas phyb mutants likewise exhibit long hypocotyls, suggesting that SPT also plays a part in the regulation of photomorphogenesis by PHYB [1]. It was shown that PHYB participates in the process of SPT regulating flowering time and PHYB is also involved in the process of SPT response to light [7]. Under long-day conditions, the SPT overexpression plant promoted flowering compared with the Ler ecotype, and conversely, the spt mutant suppressed flowering, and the phyb mutant promoted flowering. However, in the spt/phyb mutant, flowering time was intermediate between the spt mutant and the phyb mutant. This implies that the regulation of the flowering process by SPT is dependent on PHYB. By the RT-qPCR analysis of the transcription level of key flowering-related genes (FLC/SHORT VEGETATIVE PHASE (SVP)/FT/SOC1), among which FT and SOC1 promoted flowering, while FLC and SVP inhibited flowering. The results showed that in the spt-2 mutant, the transcription level of the FLC and SVP were both remarkably higher than those of the Ler ecotype, whereas those of the FT and SOC1 were notably lower than those of the Ler ecotype. Conversely, in the phyb mutant, the transcription level of the FLC and SVP was extremely lower than those of the Ler ecotype, whereas the transcription level of the FT and SOC1 was remarkably higher than those of the Ler ecotype. These results suggest that SPT regulates the flowering process of plants by modulating the expression level of key flowering-related genes, which is dependent on PHYB.
4 SPT Is Involved in the Regulation of Carpel Development
The morphogenesis of plants is regulated by a cascade of different genes. Pistils are determined by the transcription factors AGAMOUS (AG) and SEPALLATA (SEP) [53]. The development of specific tissues at the marginal placentation is regulated by specific genes such as LEUNIG (LUG), AINTEGUMENTA (ANT), NO TRANSMITTING TRACT (NTT), SHI RELATED SEQUENCE 1 (STY1) and SHI RELATED SEQUENCE 2 (STY2) [54–56]. In addition, SPT is also involved in the regulation of carpel development in Arabidopsis, and its expression was observed in both margin and pollen tract tissues throughout Arabidopsis pistil development, implying that SPT promotes the growth of tissues at the margins of the carpels, such as septa, styles, and stigmas [5]. When SPT is mutated, it leads to disruption of the septa and transmission bundles of the style, resulting in impaired development of the carpels [4]. The SPT protein contains two structural domains: an acidic structural domain and an amphipathic helix. SPT requires an acidic structural domain to mediate carpel development [57,58]. To investigate the function of SPT in pistil and fruit development, structural domains were truncated or deleted from transgenes inserted into the spt mutant. The results showed that the fruit pods of the spt-2 mutant were notably shorter and set few seeds compared with Ler, but in the SPT overexpression plant, the fruit pods were almost as same as those of the Ler ecotype. This implies the importance of SPT in pistil development [59]. For example, STY2 is one of the genes promoted by SPT, but when SPT was mutated, STY2 was still expressed in the style, suggesting that it may be regulated in conjunction with other proteins [56].
The function of SPT is also regulated by the level of auxin inside the pistil. It was shown that in Arabidopsis, the formation of stigma, style, ovary, and the short stem from the tip to the base of the pistil is regulated by the concentration of auxin [60]. When the auxin transport at the top of the growing pistil is damaged, the number of styles and ovaries will be reduced. However, in contrast to Col-0, all but the apical portion of the pistil was not affected by the damaged polar transport of auxin in the spt mutant, suggesting that SPT is also involved in the polar transport of auxin from the apical to the basal portion of the pistil.
It was shown that SPT is regulated by the AUXIN RESPONSE TRANSCRIPTION FACTOR 3 (ETT), which senses the concentration of auxin in the pistil, and that the marginals between the style and ovary as well as between the ovary and pistil has been determined by ETT [61]. When ETT is mutated, gynoecium development is also affected; however, in spt/ett double mutant plants, gynoecium development is significantly less affected compared with Col-0, implying that ETT function is SPT-dependent [62].
In addition, the fusion of marginal tissues during gynoecium development is also influenced by gene interactions. It has been shown that SPT is required for carpel development, but the promotion of carpel development by SPT is hampered by CUC1 and CUC2, CUP-SHAPED COTYLEDON genes encoding NAC transcription factors [63,64]. SPT promotes carpel fusion at the pistil tip by negatively regulating CUC1 and CUC2 [65]. By constructing CUC1 and CUC2 promoter-driven GUS vectors and observing their GUS expression positions, it was found that in wild pistils, strong GUS activity was detected on the adaxial side of the medial region, but in spt mutants, strong GUS activity was not detected on the adaxial side, implying that the SPT is involved in the modulation of CUC2 promoter activity, which leads to accumulation of its mRNAs on the abaxial side through the modulation of the transcription of CUC2.
In addition, SPT, CUC1, and CUC2 are involved in ovule formation in the ovary. This also implies a synergistic role of SPT and CUC1/CUC2, and SPT expression was not observed in CUC1/CUC2 mutants, which also implies that SPT plays a role in the downstream of CUC1/CUC2 [4,66]. The effect of SPT on CUC1/CUC2 expression was also ectopic, in the apical region of the pistil, the GUS staining activity of CUC1 and CUC2 was inhibited by SPT, but in the basal region, SPT did not modulate the GUS staining of CUC1/CUC2, instead SPT and CUC1/CUC2 acted synergistically to promote the formation of carpel margin-derived organs.
SPT could also interact with red/far-red light reversible photo-receptors and participate in photo-regulatory processes [10]. It was shown that SPT is also closely related to photo-regulatory processes regulated by PHYB and PIFs. For example, the phenotype of the spt mutant in pistils was inhibited in the spt-11/phyb-9 double mutant [67]. In contrast, the SPT overexpression plant disrupted light signaling in seedlings and promoted hypocotyl elongation, and the phyb mutant also exhibited longer hypocotyls than the Ler ecotype [1]. SPT inhibits seed germination by suppressing the transcription of the gibberellin oxidase GA3ox, which is consistent with PIF1. These growth responses are caused by low red/far-red light ratios, which characterize plant shading [68]. SPT was shown to be a photo-regulatory transcription factor and shares several specific target genes with other photo-regulatory transcription factors such as the PIF transcription factor, which are involved in the regulation of carpel development [69]. For example, SPT plays a vital role in carpel development in Arabidopsis through the activation of genes associated with shade avoidance, such as ATHB2 and ATHB4, which promote cell elongation and are mainly involved in the regulation of shade response and carpel development [4,70,71]. Further studies revealed that the original pistil-deficient phenotypes of the spt-2 mutant and spt-11 mutant were restored under low-red/far-red light conditions compared with the wild type, which also implies that the process of SPT regulation of carpel development was related to the plant shade avoidance response. In addition, there is also a genetic relationship between SPT and PHYB, and by observing the pistil development of the spt-2 and the phyb mutant, it was found that the spt-2 mutant exhibited a significant defect in carpel development, but this defect was significantly ameliorated in the spt-2/phyb mutants. Similarly, by analyzing the pistil development phenotypes of the spt-11 mutant, the spt/pif4-101 mutant, the spt/pif5 mutant, and the spt/pif4-101/pif5 mutant, it was found that under low-red/far-red light conditions, the spt-11 mutant and the spt/pif4-101 mutant, as well as the spt/pif5 mutant, could be recovered to the normal gynoecium developmental phenotype, but the gynoecium developmental phenotype of the spt/pif4-101/pif5 mutant did not, which implies that the PIF4 and PIF5 are also involved in the regulation of SPT on the development of the carpel and that there is functional redundancy between PIF4 and PIF5 [69].
In addition, SPT is also involved in regulating the carpel and fruit development of strawberries [72]. FaSPT is the homologous gene of Arabidopsis SPT in strawberry. After the SPT gene was mutated in strawberries, it was found that the shape and size of the mutant fruit changed. Moreover, strawberry fruit is developed from many separated carpels, which also implies that SPT plays a vital role in the carpel and fruit development of strawberries.
5 SPT Is Involved in the Regulation of Organ Size and Leaf Size in Arabidopsis thaliana
In plants, the leaves are the basic organs of the lateral, leaves possess intrinsic information that determines the ultimate size of the plant, and the cell proliferation is maintained by the chloroplast, which starts at the periphery of the phloem tissue at the stem tip [73,74]. The number and size of leaf pulp cells determine the leaf size. Past studies on leaf size have identified several inhibitors of leaf cell proliferation, e.g., AUXIN RESPONSE TRANSCRIPTION FACTOR 2 (ARF2), which negatively regulates ANT expression [75]. There is also BIG BROTHER (BB), and two ubiquitin receptors, DA1 and DAR [76,77]. Previous studies have also identified several cell proliferation-promoting factors, such as ANGUSTIFOLIA3 (AN3), and GROWTH-REGULATING FACTOR 5 (AtGRF5), that facilitate cell proliferation and enlarge final leaf size [78,79]. In a past study, transcription factor SPT has also been proven to play a vital role in the regulation of leaf size. SPT was shown to inhibit leaf size by limiting the size of meristematic tissues in the leaf primordium [3]. RT-qPCR analysis of SPT transcription revealed that SPT was found to be strongly expressed in young leaf primordia and lightly expressed in mature leaves. This also implies the role of SPT in regulating the process of young leaf primordia.
Observation of the leaf phenotypes of the spt mutant revealed that the spt mutant leaves were significantly larger and had longer petioles, and the total cell number in the petiole was notably higher than that of the Col-0 ecotype, suggesting that SPT regulates Arabidopsis leaf size by inhibiting leaf proliferation. The number of cells has been increased by the loss of SPT function. In conclusion, SPT functions throughout the life cycle of the plant, and the differences between leaf and flower phenotypes of spt mutants are significant.
It has also been proved that SPT is a growth inhibitory factor that inhibits the size of root meristematic tissue by regulating the transport of auxin, thereby inhibiting root size, spt mutants exhibit larger cotyledons and leaves [2,3,80]. In cotyledons, SPT co-represses cell expansion through GA-inhibitory DELLA, and SPT also receives negative regulation from GA, and it has been shown that mutant plants tend to exhibit less accumulation of PIN auxin transporter proteins compared to the Col-0 ecotype [81]. In contrast, in leaves, SPT inhibited cell proliferation by regulating auxin accumulation. SPT expression was also found in the proliferative zone. Moreover, in the spt mutant, there were also significantly more meristematic tissues in the leaf primordia compared with the wild type [8,82]. It also implies that the expansion of meristematic tissues leads to larger leaves in spt mutants. To sum up, SPT negatively regulates cell proliferation in Arabidopsis thaliana.
Higher expression of SPT was found in regions with high auxin concentrations, which also implies that the expression of SPT is correlated with auxin distribution [5,83]. By applying the auxin inhibitor NPA to the spt-11 mutant, it was found that the phenotype of the spt-11 mutant with defective carpel development was restored, which also implies that SPT may also be involved in the regulation of auxin transport [60,67]. In addition, SPT is also involved in regulating the auxin redirection pathway [84].
6 SPT Is Involved in the Regulation of Stomata, Epidermal Hairs, and Anthocyanin Biosynthesis in Arabidopsis
Many new functions of the transcription factor SPT remain to be discovered. Transcriptome analysis of SPT revealed the existence of target genes for stomatal development, epidermal hair formation, and anthocyanin synthesis. SPT was shown to negatively regulate stomatal development and positively regulate epidermal hair formation as well as anthocyanin biosynthesis [9]. Stomatal development requires the combined action of multiple transcription factors, such as the bHLH transcription factor, the transcription factor FAMA, and MUTE, their helix-loop-helix regions can form heterodimers with SCREAM1 (SCRM1) and SCRM2 [85–88]. Three polypeptide receptor kinases (EPF1, FPF2, and EPFL9) are involved in the regulation of stomatal pattern formation in Arabidopsis, where EPF1 is involved in the negative regulation of the MUTE-SCRM module, EPF2 in the negative regulation of the SPCH-SCRM module, and EPFL9 in the positive regulation of the SPCH-SCRM module [89,90].
Plant epidermal hairs are hair-like appendages extending from the epidermal tissue of the above-ground part of the plant. Epidermal hairs increase the thickness of the epidermal layer of the plant, construct a natural physical barrier between the epidermis and the environment, reduce the loss of water and excessive accumulation and consumption of heat in the plant body, and to a certain extent mitigate pests, freezing, ultraviolet rays, and mechanical damage; in addition, the epidermal hairs with glands can secrete alkaloids, aromatic oils, and resins, and other chemical substances to defend against biotic and abiotic stresses and signaling [91,92]. On the other hand, the light-leafed, light-shelled phenotype of crop rice with missing epidermal trichomes (light-leaved rice) facilitates crop harvesting and subsequent processing and thus has been widely utilized in breeding practices. Therefore, the study of trichome formation is of great importance for agricultural breeding.
Trichome formation begins with the internal replication of the protoplast, after which the trichome begins to expand and branch outwards, finally forming a mature trichome [93]. The formation of trichomes of stomata helps the plant to exchange gases with the outside world for better photosynthesis [94]. In addition, SPT plays a vital role in the biosynthesis of anthocyanins in Arabidopsis thaliana. Anthocyanins are glycosylated polyphenolic compounds, also known as anthocyanidin glycosides, which are an important class of plant flavonoid compounds. Anthocyanins are widely found in plant organs such as flowers, fruits, and leaves and are the basis for the colorful appearance of plants [95]. Anthocyanin biosynthesis is regulated by temperature, light, hormones, sugars, and so on [96,97]. Several structural genes are involved in the regulation of anthocyanin biosynthesis, which is categorized as early biosynthetic genes (EBG): CHS, CHI, F3H, and F3H: late biosynthetic genes (LBG): F3, S’H, DFR, ANS, and UFGT, etc. The expression of these structural genes is controlled by environmental factors and transcription factors, which mainly include the three major categories of MYB, bHLH, and WD40 [98,99]. They could regulate structural genes individually or combine to form MBW complexes to co-regulate structural genes.
Transcriptomic analysis of SPT revealed a high enrichment of genes associated with leaf tissue, and leaf structure, implying the role played by SPT in regulating leaf structure. Moreover, genes related to defense cells were also enriched, implying the role of SPT in regulating stomatal development [9]. In addition, the transcription factors GLABRA 2 (GL2) and TESTA 8 (TT8), which are associated with trichome formation, and PAP1, which is involved in anthocyanin production, were also identified. This also implies a role for SPT in regulating trichome formation and anthocyanin synthesis. Observation of the stomatal number of spt mutants revealed that the number of stomata was notably increased in the spt-12 mutant compared with the Col-0 ecotype, whereas the SPT overexpression plant was not remarkably different from the Col-0 ecotype. This suggests that SPT inhibits stomatal production in Arabidopsis thaliana. This process may be regulated by STOMGEN, which is involved in stomatal development and is also the target gene of SPT [100]. In the spt-12 mutant, however, the number of trichomes was notably decreased compared with the Col-0 ecotype, which also implies that SPT is a positive regulator of Arabidopsis trichomes. This may be related to GL2, which has been shown to be involved in the regulation of trichome development [101].
SPT also plays a vital role in sucrose-mediated anthocyanin biosynthesis. Previous studies have revealed the association between SPT and anthocyanin synthesis and genes related to anthocyanin synthesis [102]. By observing anthocyanin synthesis in SPT overexpression plant and spt-2 mutant distributed in 1% sucrose and 5% sucrose, no obvious differences were found between SPT overexpression plants and spt-2 and Ler in 1% sucrose, but in 5% sucrose, it was found that the color intensity of anthocyanin in SPT overexpression plants was remarkably higher than that of Ler, and that of the spt-2 mutant had the notably lower color intensity of anthocyanins than Ler. This process may be regulated by DFR or UF3GT, which are involved in regulating anthocyanin biosynthesis and are also the target genes of SPT [103,104]. This demonstrates that SPT promotes sucrose-mediated anthocyanin biosynthesis.
7 SPT Is Involved in Regulating the Auxin-Cytokinin Signaling Pathway in the Pistil
Auxins and cytokinins are involved in plant growth, and the interaction of auxins and cytokinins plays a vital role in many physiological processes in plants, such as stem cell maintenance, vascularisation, and root development [105–110]. It was shown that in the medial region of the pistil, the auxin biosynthesis gene TAA1 and the auxin efflux transporter protein gene PIN3 are promoted by cytokinin signaling. A recent study has shown that TIR1 and AFB signals of auxin also participate in regulating the growth of root acid growth [111]. At the same time, some studies have proved that auxin regulates its functions in flowers and fruits by integrating the processes of biosynthesis, and signal transduction [112]. Auxin also regulates the formation of plant lateral roots. For example, the formation of plant lateral roots also requires the activation of auxin-dependent transcription factors ARF7, ARF19, and LBD16 [113]. Auxin signaling is also involved in regulating cambium, the stem cell niche that mediates wood formation [114]. In addition, the bHLH family member SPT promotes cytokinin signaling in the medial region of the pistil by regulating the TYPE-B ARABIDOPSIS RESPONSE REGULATOR (ARR) [5,115]. By crossing the auxin response reporter gene line DR5::GFP with SPT overexpressing plants and spt mutant plants, respectively, to observe the expression of DR5 signal, it was shown that in the stigma region of the style of the Ler ecotype, DR5 signal was expressed predominantly in the form of a “ring” around the styles at stage 9, but this ring-shaped pattern of DR5 signal was not observed in the spt mutant. In contrast, a notably enhanced DR5 signal was found in the stigma of pistil styles in SPT overexpression plants compared with Ler, suggesting that SPT is involved in the regulation of Arabidopsis auxin signaling. Auxins and cytokinins act in complex ways [115,116]. By treating the cytokinin reporter gene line TCS::GFP with the auxin IAA, it was found that in the wild type, the application of IAA enhanced TCS signaling. However, TCS signaling was not observed in the spt mutant. It implies that auxin enhances cytokinin expression in an SPT-dependent pathway. Similarly, the application of IAA also enhanced DR5 signaling in plants with the spt mutant, suggesting that exogenous auxin acts independently of SPT in the gynoecium [117].
8 SPT Is Involved in Regulating Plant Response to Abiotic Stress in Cucumber
Abiotic stress is an abiotic environmental condition that is not conducive to the survival, and growth of plants and even leads to injury, destruction, and death [118–120]. In nature, plants often face different kinds of abiotic stresses, such as alkali stress, salt stress, high-temperature stress, and so on [121]. Abiotic stress has seriously endangered plant growth and crop yield, so we must pay attention to the influence of abiotic stress on plant growth [122–124]. High-temperature stress also affects the growth of plants [125,126]. It has been indicated that bHLH transcription factors are involved in regulating the process of plants responding to high-temperature stress [127,128]. Recently, SPT was found to be involved in regulating plant responses to high ambient temperature stress in cucumbers [129]. Through RNA sequencing technology, it has been found that the expression of 75 genes changed after high-temperature treatment. At the same time, after high-temperature treatment, the leaves of the spt mutant became withered and the photosystem and chloroplast activity of the spt mutant were also seriously damaged, which proved that SPT positively regulated the heat resistance of cucumber. In addition, GO enrichment analysis showed that compared with wild-type plants, high temperature induced more photosynthesis and chloroplast-related genes in Csspt mutant plants, which proved that SPT regulated plant heat tolerance by regulating photosystem heat sensitivity.
In conclusion, this paper reviews the multiple roles of the bHLH family transcription factor SPT in the growth and development of Arabidopsis thaliana. SPT is a versatile transcription factor, and the helix-loop-helix region of SPT can form protein complexes with its family members as well as with members of other proteins. This paper mainly reviews the important roles of SPT in regulating flowering, seed germination, carpel development, leaf size, stomatal development, trichome formation, anthocyanin synthesis, and auxin-cytokinin interactions in Arabidopsis thaliana, which is helpful for us to deeply understand the role of transcription factor SPT in plant growth (summarized in Fig. 1). In addition, we also summarized the role of SPT in regulating the response of cucumbers to high-temperature stress, which also suggested the potential application of SPT in regulating the response of plants to abiotic stress.
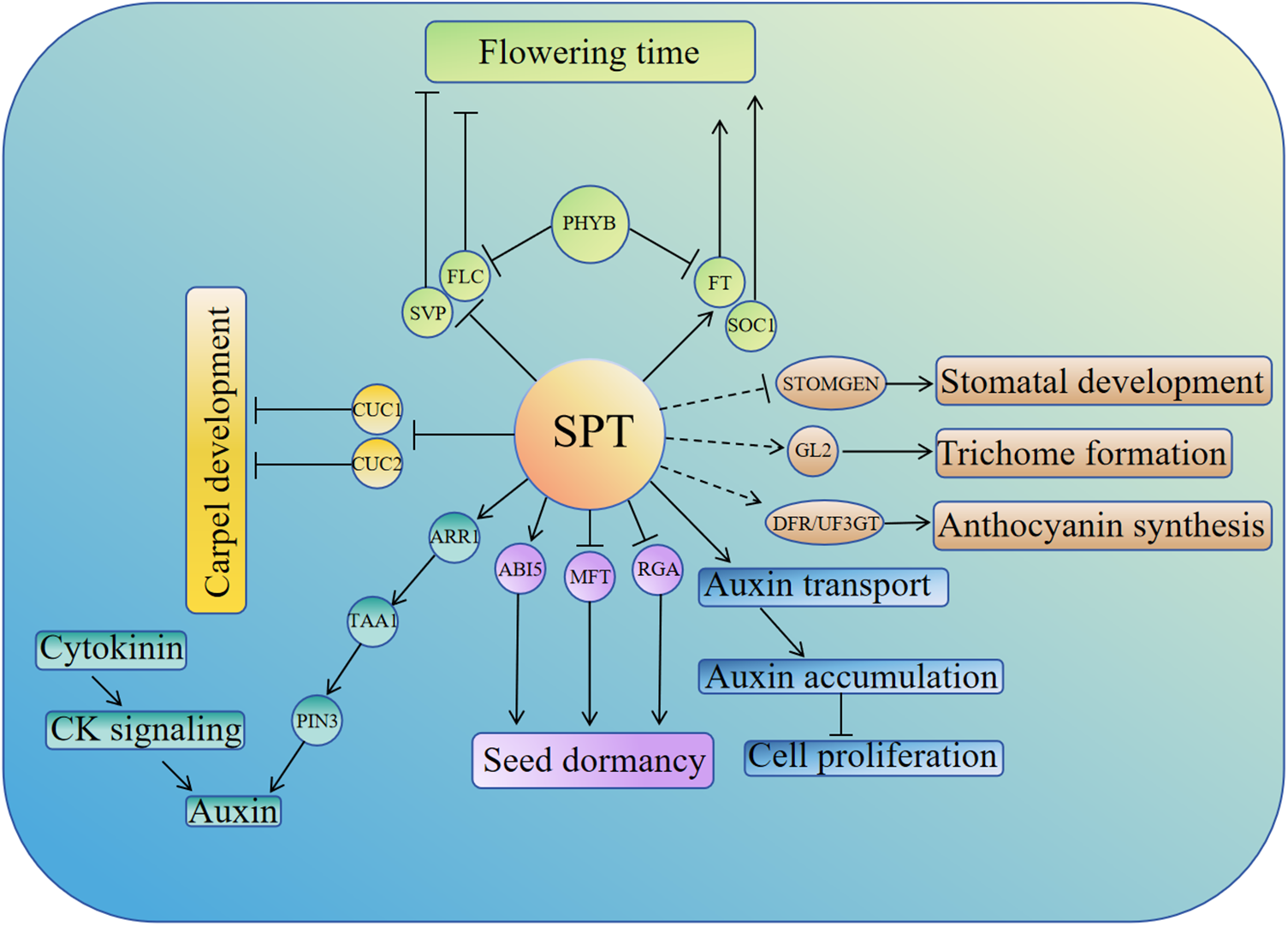
Figure 1: Schematic representation of SPT regulation of the growth and development processes in Arabidopsis, including flowering time, seed germination, leaf size, carpel development, stomatal development, epidermal hair synthesis, anthocyanin biosynthesis, etc. Dashed lines indicate targets that are not direct targets
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm their contribution to the paper as follows: Xiangyang Hu designed the research and edited the manuscript together. Lei Liang obtained the related references and wrote the manuscript together. Both authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol. 2005;15(22):1998–2006. [Google Scholar] [PubMed]
2. Lee KS, Josse EM, Brown A, Gan Y, Halliday KJ, Graham IA, et al. SPATULA links daytime temperature and plant growth rate. Curr Biol: CB. 2010;20(16):1493–7. [Google Scholar]
3. Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H. The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 2010;51(2):252–61. [Google Scholar] [PubMed]
4. Alvarez J, Smyth DR. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Develop. 1999;126(11):2377–86. [Google Scholar]
5. Heisler MG, Atkinson A, Bylstra YH, Walsh R, Smyth DR. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Develop. 2001;128(7):1089–98. [Google Scholar]
6. Girin T, Paicu T, Stephenson P, Fuentes S, Körner E, O’Brien M, et al. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell. 2011;23(10):3641–53. [Google Scholar] [PubMed]
7. Wu M, Upreti S, Yan A, Wakeel A, Wu J, Ge S, et al. SPATULA regulates floral transition and photomorphogenesis in a PHYTOCHROME B-dependent manner in Arabidopsis. Biochem Biophys Res Commun. 2018;503(4):2380–5. [Google Scholar] [PubMed]
8. Makkena S, Lamb RS. The bHLH transcription factor SPATULA is a key regulator of organ size in Arabidopsis thaliana. Plant Signal Behav. 2013;8(5):e24140. [Google Scholar] [PubMed]
9. Bernal-Gallardo JJ, Zuñiga-Mayo VM, Marsch-Martinez N, de Folter S. Novel roles of SPATULA in the control of stomata and trichome number, and anthocyanin biosynthesis. Plants. 2023;12(3):596. [Google Scholar] [PubMed]
10. Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–70. [Google Scholar] [PubMed]
11. Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006;46(4):601–12. [Google Scholar] [PubMed]
12. Kilwinski J, Baack M, Heiland S, Knippers R. Transcription factor Oct1 binds to the AT-rich segment of the simian virus 40 replication origin. J Virol. 1995;69(1):575–8. [Google Scholar] [PubMed]
13. Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Sci. 1997;276(5320):1865–8. [Google Scholar]
14. Hao Y, Zong X, Ren P, Qian Y, Fu A. Basic Helix-Loop-Helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int J Mol Sci. 2021;22(13):7152. [Google Scholar] [PubMed]
15. Xu F, Tang J, Wang S, Cheng X, Wang H, Ou S, et al. Antagonistic control of seed dormancy in rice by two bHLH transcription factors. Nat Genet. 2022;54(12):1972–82. [Google Scholar] [PubMed]
16. Weber D, Wiese C, Gessler M. Hey bHLH transcription factors. Curr Top Dev Biol. 2014;110:285–315. [Google Scholar] [PubMed]
17. Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, et al. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218(2):129–35. [Google Scholar] [PubMed]
18. Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153(3):1398–1412. [Google Scholar] [PubMed]
19. Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell. 2014;26(2):828–41. [Google Scholar] [PubMed]
20. Yamamura C, Mizutani E, Okada K, Nakagawa H, Fukushima S, Tanaka A, et al. Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 2015;84(6):1100–13. [Google Scholar] [PubMed]
21. Raissig MT, Abrash E, Bettadapur A, Vogel JP, Bergmann DC. Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc Natl Acad Sci USA. 2016;113(29):8326–31. [Google Scholar] [PubMed]
22. Vaistij FE, Barros-Galvão T, Cole AF, Gilday AD, He Z, Li Y, et al. MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2018;115(33):8442–7. [Google Scholar] [PubMed]
23. Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10(12):2115–26. [Google Scholar] [PubMed]
24. Munir J, Dorn LA, Donohue K, Schmitt J. The effect of maternal photoperiod on seasonal dormancy in Arabidopsis thaliana (Brassicaceae). Am J Bot. 2001;88(7):1240–9. [Google Scholar] [PubMed]
25. Laserna MP, Sánchez RA, Botto JF. Light-related loci controlling seed germination in Ler x Cvi and Bay-0 x Sha recombinant inbred-line populations of Arabidopsis thaliana. Ann Bot. 2008;102(4):631–42. [Google Scholar] [PubMed]
26. Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet. 1980;58(6):257–63. [Google Scholar] [PubMed]
27. Groot SP, Karssen CM. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta. 1987;171(4):525–31. [Google Scholar] [PubMed]
28. Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004;16(2):367–78. [Google Scholar] [PubMed]
29. Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ. Molecular mechanisms of seed dormancy. Plant, Cell Environ. 2012;35(10):1769–86. [Google Scholar] [PubMed]
30. Clerkx EJ, Vries HB, Ruys GJ, Groot SP, Koornneef M. Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiol. 2003;132(2):1077–84. [Google Scholar] [PubMed]
31. Daszkowska-Golec A, Wojnar W, Rosikiewicz M, Szarejko I, Maluszynski M, Szweykowska-Kulinska Z, et al. Arabidopsis suppressor mutant of abh1 shows a new face of the already known players: ABH1 (CBP80) and ABI4-in response to ABA and abiotic stresses during seed germination. Plant Mol Biol. 2013;81(1–2):189–209. [Google Scholar] [PubMed]
32. Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12(4):599–609. [Google Scholar] [PubMed]
33. Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell. 1998;10(6):1043–54. [Google Scholar] [PubMed]
34. Vaistij FE, Gan Y, Penfield S, Gilday AD, Dave A, He Z, et al. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci USA. 2013;110(26):10866–71. [Google Scholar] [PubMed]
35. Williams ME, Foster R, Chua NH. Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell. 1992;4(4):485–96. [Google Scholar] [PubMed]
36. Loake GJ, Faktor O, Lamb CJ, Dixon RA. Combination of H-box [CCTACC(N)7CT] and G-box (CACGTG) cis elements is necessary for feed-forward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proc Natl Acad Sci USA. 1992;89(19):9230–4. [Google Scholar] [PubMed]
37. Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20(12):506–10. [Google Scholar] [PubMed]
38. Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, et al. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell. 2011;23(9):3215–29. [Google Scholar] [PubMed]
39. Chono M, Matsunaka H, Seki M, Fujita M, Kiribuchi-Otobe C, Oda S, et al. Molecular and genealogical analysis of grain dormancy in Japanese wheat varieties, with specific focus on MOTHER OF FT AND TFL1 on chromosome 3A. Breed Sci. 2015;65(1):103–9. [Google Scholar] [PubMed]
40. Shrestha R, Gómez-Ariza J, Brambilla V, Fornara F. Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann Bot. 2014;114(7):1445–58. [Google Scholar] [PubMed]
41. Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80(6):847–57. [Google Scholar] [PubMed]
42. Rouse DT, Sheldon CC, Bagnall DJ, Peacock WJ, Dennis ES. FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 2002;29(2):183–91. [Google Scholar] [PubMed]
43. Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187(1):57–66. [Google Scholar] [PubMed]
44. Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–84. [Google Scholar] [PubMed]
45. Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot. 2010;61(9):2247–54. [Google Scholar] [PubMed]
46. Johansson M, Staiger D. Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot. 2015;66(3):719–30. [Google Scholar] [PubMed]
47. Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. Embo J. 2002;21(10):2441–50. [Google Scholar] [PubMed]
48. Kang ES, Ju JW, Kim JS, Ahn HK, Lee JK, Kim JH, et al. Multi-wavelength emitting InGan/GaN quantum well grown on V-shaped gan1101 microfacet. J Nanosci Nanotechnol. 2007;7(11):4053–6. [Google Scholar] [PubMed]
49. Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Sci. 1998;282(5393):1488–90. [Google Scholar]
50. Inoue K, Nishihama R, Kohchi T. Evolutionary origin of phytochrome responses and signaling in land plants. Plant Cell Environ. 2017;40(11):2502–8. [Google Scholar] [PubMed]
51. Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25(3):413–27. [Google Scholar] [PubMed]
52. Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, et al. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell. 2002;14(7):1541–55. [Google Scholar] [PubMed]
53. Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99(2):199–209. [Google Scholar] [PubMed]
54. Liu Z, Franks RG, Klink VP. Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell. 2000;12(10):1879–92. [Google Scholar] [PubMed]
55. Crawford BC, Ditta G, Yanofsky MF. The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr Biol. 2007;17(13):1101–8. [Google Scholar] [PubMed]
56. Kuusk S, Sohlberg JJ, Long JA, Fridborg I, Sundberg E. STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Develop. 2002;129(20):4707–17. [Google Scholar]
57. Roeder AH, Ferrándiz C, Yanofsky MF. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol. 2003;13(18):1630–5. [Google Scholar] [PubMed]
58. Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34(5):733–9. [Google Scholar] [PubMed]
59. Groszmann M, Paicu T, Smyth DR. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. Plant J. 2008;55(1):40–52. [Google Scholar] [PubMed]
60. Nemhauser JL, Feldman LJ, Zambryski PC. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Develop. 2000;127(18):3877–88. [Google Scholar]
61. Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Develop. 1997;124(22):4481–91. [Google Scholar]
62. Alvarez J, Smyth DR. Genetic pathways controlling carpel development in Arabidopsis thaliana. J Plant Res. 1998;111(2):295–8. [Google Scholar]
63. Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18(11):2929–45. [Google Scholar] [PubMed]
64. Larue CT, Wen J, Walker JC. A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J. 2009;58(3):450–63. [Google Scholar] [PubMed]
65. Nahar MA, Ishida T, Smyth DR, Tasaka M, Aida M. Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 2012;53(6):1134–43. [Google Scholar] [PubMed]
66. Ishida T, Aida M, Takada S, Tasaka M. Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 2000;41(1):60–7. [Google Scholar] [PubMed]
67. Foreman J, White J, Graham I, Halliday K, Josse EM. Shedding light on flower development: phytochrome B regulates gynoecium formation in association with the transcription factor SPATULA. Plant Signal Behav. 2011;6(4):471–6. [Google Scholar] [PubMed]
68. Martínez-García JF, Gallemí M, Molina-Contreras MJ, Llorente B, Bevilaqua MR, Quail PH. The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome a and B differentiates vegetation proximity and canopy shade. PLoS One. 2014;9(10):e109275. [Google Scholar]
69. Reymond MC, Brunoud G, Chauvet A, Martínez-Garcia JF, Martin-Magniette ML, Monéger F, et al. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell. 2012;24(7):2812–25. [Google Scholar] [PubMed]
70. Sharman AC. Some new terms for duplicated genes. Semin Cell Dev Biol. 1999;10(5):561–3. [Google Scholar] [PubMed]
71. Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martínez-García JF. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009;59(2):266–77. [Google Scholar] [PubMed]
72. Tisza V, Kovács L, Balogh A, Heszky L, Kiss E. Characterization of FaSPT, a SPATULA gene encoding a bHLH transcriptional factor from the non-climacteric strawberry fruit. Plant Physiol Biochem. 2010;48(10–11):822–6. [Google Scholar] [PubMed]
73. Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215(2):407–19. [Google Scholar] [PubMed]
74. Tsukaya H. Mechanism of leaf-shape determination. Annu Rev Plant Biol. 2006;57:477–96. [Google Scholar] [PubMed]
75. Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Develop. 2006;133(2):251–61. [Google Scholar]
76. Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol. 2006;16(3):272–9. [Google Scholar] [PubMed]
77. Li Y, Zheng L, Corke F, Smith C, Bevan MW. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008;22(10):1331–6. [Google Scholar] [PubMed]
78. Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA. 2004;101(36):13374–13379. [Google Scholar] [PubMed]
79. Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43(1):68–78. [Google Scholar] [PubMed]
80. Josse EM, Gan Y, Bou-Torrent J, Stewart KL, Gilday AD, Jeffree CE, et al. A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. Plant Cell. 2011;23(4):1337–51. [Google Scholar] [PubMed]
81. Willige BC, Isono E, Richter R, Zourelidou M, Schwechheimer C. Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell. 2011;23(6):2184–95. [Google Scholar] [PubMed]
82. Ichihashi Y, Kawade K, Usami T, Horiguchi G, Takahashi T, Tsukaya H. Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiol. 2011;157(3):1151–62. [Google Scholar] [PubMed]
83. Groszmann M, Bylstra Y, Lampugnani ER, Smyth DR. Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J Experiment Botany. 2010;61(5):1495–1508. [Google Scholar]
84. Makkena S, Lamb RS. The bHLH transcription factor SPATULA regulates root growth by controlling the size of the root meristem. Bmc Plant Biol. 2013;2(13):1. [Google Scholar]
85. MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nat. 2007;445(7127):537–40. [Google Scholar]
86. de Marcos A, Houbaert A, Triviño M, Delgado D, Martín-Trillo M, Russinova E, et al. A mutation in the bHLH domain of the SPCH transcription factor uncovers a BR-dependent mechanism for stomatal development. Plant Physiol. 2017;174(2):823–42. [Google Scholar] [PubMed]
87. Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18(10):2493–505. [Google Scholar] [PubMed]
88. Pillitteri LJ, Bogenschutz NL, Torii KU. The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant & Cell Physiol. 2008;49(6):934–43. [Google Scholar]
89. Zoulias N, Harrison EL, Casson SA, Gray JE. Molecular control of stomatal development. Biochem J. 2018;475(2):441–54. [Google Scholar] [PubMed]
90. Wang S, Zhou Z, Rahiman R, Lee GSY, Yeo YK, Yang X, et al. Light regulates stomatal development by modulating paracrine signaling from inner tissues. Nat Commun. 2021;12(1):3403. [Google Scholar] [PubMed]
91. Serna L, Martin C. Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 2006;11(6):274–80. [Google Scholar] [PubMed]
92. Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–86. [Google Scholar] [PubMed]
93. Steets JA, Takebayashi N, Byrnes JM, Wolf DE. Heterogeneous selection on trichome production in Alaskan Arabidopsis kamchatica (Brassicaceae). Am J Bot. 2010;97(7):1098–108. [Google Scholar] [PubMed]
94. Fambrini M, Pugliesi C. The Dynamic Genetic-hormonal regulatory network controlling the trichome development in leaves. Plants. 2019;8(8):253. [Google Scholar] [PubMed]
95. Li C, Yu W, Xu J, Lu X, Liu Y. Anthocyanin biosynthesis induced by MYB transcription factors in plants. Int J Mol Sci. 2022;23(19):11701. [Google Scholar] [PubMed]
96. Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139(4):1840–52. [Google Scholar] [PubMed]
97. Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol. 2008;165(8):886–94. [Google Scholar] [PubMed]
98. Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12(12):2383–94. [Google Scholar] [PubMed]
99. Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–27. [Google Scholar] [PubMed]
100. Weng X, Zhu L, Yu S, Liu Y, Ru Y, Zhang Z, et al. Carbon monoxide promotes stomatal initiation by regulating the expression of two EPF genes in Arabidopsis cotyledons. Front Plant Sci. 2022;13:1029703. [Google Scholar] [PubMed]
101. Chen S, Wang S. GLABRA2, a common regulator for epidermal cell fate determination and anthocyanin biosynthesis in Arabidopsis. Int J Mol Sci. 2019;20(20):4997. [Google Scholar] [PubMed]
102. Bulgakov VP, Avramenko TV, Tsitsiashvili GS. Critical analysis of protein signaling networks involved in the regulation of plant secondary metabolism: focus on anthocyanins. Crit Rev Biotechnol. 2017;37(6):685–700. [Google Scholar] [PubMed]
103. Kumar V, Yadav SK. Pyramiding of tea Dihydroflavonol reductase and Anthocyanidin reductase increases flavan-3-ols and improves protective ability under stress conditions in tobacco. 3 Biotech. 2017;7(3):177. [Google Scholar] [PubMed]
104. Li T, Jia KP, Lian HL, Yang X, Li L, Yang HQ. Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem Bioph Res Commun. 2014;454(1):78–83. [Google Scholar] [PubMed]
105. Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, et al. Hormonal control of the shoot stem-cell niche. Nat. 2010;465(7301):1089–92. [Google Scholar]
106. Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nat. 2008;453(7198):1094–7. [Google Scholar]
107. Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, et al. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol. 2011;21(11):917–26. [Google Scholar] [PubMed]
108. de Rybel B, Adibi M, Breda AS, Wendrich JR, Smit ME, Novák O, et al. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Sci. 2014;345(6197):1255215. [Google Scholar]
109. McLaughlin HM, Ang ACH, Østergaard L. Noncanonical auxin signaling. Cold Spring Harb Perspect Biol. 2021;13(5):a039917. [Google Scholar] [PubMed]
110. Ma Y, Wolf S, Lohmann JU. Casting the net-connecting auxin signaling to the plant genome. Cold Spring Harb Perspect Biol. 2021;13(11):a040006. [Google Scholar] [PubMed]
111. Ang ACH, Østergaard L. Save your TIRs-more to auxin than meets the eye. New Phytol. 2023;238(3):971–6. [Google Scholar] [PubMed]
112. Sundberg E, Østergaard L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol. 2009;1(6):a001628. [Google Scholar] [PubMed]
113. Stitz M, Kuster D, Reinert M, Schepetilnikov M, Berthet B, Reyes‐Hernández J, et al. TOR acts as a metabolic gatekeeper for auxin-dependent lateral root initiation in Arabidopsis thaliana. Embo J. 2023;42(10):e111273. [Google Scholar] [PubMed]
114. Brackmann K, Qi J, Gebert M, Jouannet V, Schlamp T, Grünwald K, et al. Spatial specificity of auxin responses coordinates wood formation. Nat Commun. 2018;9(1):875. [Google Scholar] [PubMed]
115. Reyes-Olalde JI, Zúñiga-Mayo VM, Serwatowska J, Chavez Montes RA, Lozano-Sotomayor P, Herrera-Ubaldo H, et al. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 2017;13(4):e1006726. [Google Scholar] [PubMed]
116. Marsch-Martínez N, Reyes-Olalde JI, Ramos-Cruz D, Lozano-Sotomayor P, Zúñiga-Mayo VM, de Folter S. Hormones talking: does hormonal cross-talk shape the Arabidopsis gynoecium? Plant Signal Behav. 2012;7(12):1698–1701. [Google Scholar]
117. Reyes-Olalde JI, Zúñiga-Mayo VM, Marsch-Martínez N, de Folter S. Synergistic relationship between auxin and cytokinin in the ovary and the participation of the transcription factor SPATULA. Plant Signal Behav. 2017;12(10):e1376158. [Google Scholar] [PubMed]
118. Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, et al. Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci. 2020;63(5):635–74. [Google Scholar] [PubMed]
119. Chang YN, Zhu C, Jiang J, Zhang H, Zhu JK, Duan CG. Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol. 2020;62(5):563–80. [Google Scholar] [PubMed]
120. Saijo Y, Loo EP. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020;225(1):87–104. [Google Scholar] [PubMed]
121. Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–24. [Google Scholar] [PubMed]
122. Kopecká R, Kameniarová M, Černý M, Brzobohatý B, Novák J. Abiotic stress in crop production. Int J Mol Sci. 2023;24(7):6603. [Google Scholar]
123. Loudet O, Hasegawa PM. Abiotic stress, stress combinations and crop improvement potential. Plant J. 2017;90(5):837–8. [Google Scholar] [PubMed]
124. Jia Y, Kang L, Wu Y, Zhou C, Li D, Li J, Pan C. Review on pesticide abiotic stress over crop health and intervention by various biostimulants. J Agric Food Chem. 2023;71(37):13595–611. [Google Scholar] [PubMed]
125. Zhao J, Lu Z, Wang L, Jin B. Plant responses to heat stress: physiology, transcription, noncoding RNAs, and epigenetics. Int J Mol Sci. 2020;22(1):117. [Google Scholar] [PubMed]
126. Zhu Z, Dai Vu L, Balasubramanian S. Editorial: plant response to high ambient temperature. Front Plant Sci. 2022;13:971480. [Google Scholar] [PubMed]
127. Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19(5):408–13. [Google Scholar] [PubMed]
128. Wei H, Song Z, Xie Y, Cheng H, Yan H, Sun F, et al. High temperature inhibits vascular development via the PIF4-miR166-HB15 module in Arabidopsis. Curr Biol. 2023;33(15):3203–14.E4. [Google Scholar]
129. Liang Y, Yang C, Ming F, Yu B, Cheng Z, Wang Y, et al. A bHLH transcription factor, CsSPT, regulates high-temperature resistance in cucumber. Horticul Plant J. 2023;10(2):503–514. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools