 Open Access
Open Access
ARTICLE
The Identification of Phenylalanine Ammonia-Lyase (PAL) Genes from Pinus yunnanensis and an Analysis of Enzyme Activity in vitro
1 Key Laboratory of National Forestry and Grassland Administration on Biodiversity Conservation in Southwest China, Southwest Forestry University, Kunming, 650224, China
2 College of Forestry, Southwest Forestry University, Kunming, 650224, China
* Corresponding Author: Junrong Tang. Email:
Phyton-International Journal of Experimental Botany 2024, 93(3), 503-516. https://doi.org/10.32604/phyton.2024.048786
Received 12 November 2023; Accepted 25 January 2024; Issue published 28 March 2024
Abstract
Phenylalanine ammonia lyase (PAL) is the rate-limiting and pivotal enzyme of the general phenylpropanoid pathway, but few reports have been found on PAL genes in Pinus yunnanensis. In the present study, three PAL genes were cloned and identified from P. yunnanensis seedlings for the first time, namely, PyPAL-1, PyPAL-2, and PyPAL-3. Our results indicated that the open-reading frames of PyPAL genes were 2184, 2157, and 2385 bp. Phylogenetic tree analysis revealed that PyPALs have high homology with other known PAL genes in other plants. In vitro enzymatic analysis showed that all three PyPAL recombinant proteins could catalyze the deamination of L-phenylalanine to form trans-cinnamic acid, but only PAL1 and PAL2 can catalyze the conversion of L-tyrosine to ρ-coumaric acid. Three PyPAL genes were expressed in different tissues in 1-year-old P. yunnanensis, and such genes had different expression patterns. This study lays a foundation for further understanding of the biosynthesis of secondary metabolites in P. yunnanensis.Keywords
Supplementary Material
Supplementary Material FileIn the plant kingdom, phenylpropanoids are among the most widely distributed and common secondary metabolites with a wide range of structural classes and biological functions [1]. Phenylpropanoids are also precursors of flavonoids, isoflavones, coumarins, and stilbenes. They are important compounds in cell walls, serving as shields against intense light and UV radiation, acting as natural defense mechanisms against herbivores and pathogens, and functioning as pigments that facilitate interactions between plants and pollinators [2]. For example, lignin, which is an aromatic polymer synthesized from phenylpropanoid precursor, is a major structural component of secondary thickened cell walls in vascular plants [3]. It is essential for mechanical support; long-distance water, mineral, and photosynthetic product transport; and pathogen defense in plants [4].
So far, a series of enzymes involved in phenylpropanoid biosynthesis have been identified, and phenylalanine ammonia-lyase (PAL; E.C 4.3.1.5), first discovered by Koukal and Conn, has been extensively studied [5]. The PAL in dicotyledons converts L-phenylalanine to the corresponding trans-cinnamic acid, whereas the PAL in monocotyledons is active against L-phenylalanine and L-tyrosine [6]. PAL, as the first key enzyme and rate-limiting enzyme in the phenylpropanoid synthesis pathway, is an important regulation point between primary and secondary metabolism [1,7]. In plants, PAL is encoded by a multigene family and the properties, structure, regulation, expression, and catalysis of PALs have been well-studied with a wide range of organisms in recent decades. There are four members in Arabidopsis thaliana [8–10], six in Camellia sinensis [11], one in Hordeum vulgare [5], four in Nicotiana tabacum [12,13], one in Ocimum basilicum [14], nine in Oryza sativa [15], fifteen in Brassica napus [16], seven in Brassica rapa [17], twelve in Juglans Regia [18], five in Populus simonii × P. nigra [19], six in Coffea eugenioides [20], two in P. pinaster [21], one in P. taeda [22], and seven in Carya illinoinensis [23]. In many plants, PAL and its functions are redundant and divergent [14]. For example, two PALs have been identified in Rubus idaeus, and the expression of RIPAL1 was related to the early ripening of fruit, while the expression of RIPAL2 was more closely related to the late stage of flower and fruit development [24]. Compared with angiosperms, the structure and function of PALs in gymnosperms have not been fully studied. The PAL genes of gymnosperms are distributed in three main branches, whereas those of all characterized angiosperms form a branch in the PAL gene tree, suggesting that gymnosperms have more phylogenetic diversity of PAL genes than angiosperms [25]. However, only 17 species constitute gymnosperms because of their large genome sizes (>10 Gb on average), complexity, and low species richness [26]. These limitations have restricted the advances in the study of PAL and other functional genes in gymnosperms. In addition, the lack of model plants and genetic transformation systems limits advances in the study of functional genes in gymnosperms [27]. Therefore, studying the PAL of Pinus yunnanensis, the most important gymnosperm plant in Yunnan Province, China, is challenging but necessary.
P. yunnanensis forests are an important constituent of the forest ecosystem in central Yunnan, China, and P. yunnanensis is commercially (resin and timber), culturally, and economically important [28]. These forests are distributed in southwestern China at altitude of 700–3000 m [29]. At present, the declining genetic quality of P. yunnanensis is a serious problem, and the proportion of short and twisted individuals is gradually increasing, which seriously affects the wood properties of P. yunnanensis [30]. Among them, the appearance of twisted P. yunnanensis may be related to glutathione metabolism, phenylpropanoid biosynthesis, and hormone-related pathways, as revealed by transcriptome and metabolome analyses [31]. In addition, whether the excellent properties of P. yunnanensis, including its fast growth, strong drought resistance, and tolerance of low-fertility soil, are related to the biosynthesis of phenylpropanoid are also essential issues that need to be explored.
Therefore, one-year-old P. yunnanensis seedlings were used as experimental materials, and the PAL candidate genes were screened based on the PALs of A. thaliana [10], N. tabacum [13], Z. mays [32], P. pinaster [21], P. taeda [22], etc., as well as the transcriptome of P. yunnanensis. The PyPAL genes were then cloned, and bioinformatics, tissue expression analysis, in vitro enzymatic analysis, and enzyme kinetics analysis were performed on the selected PyPAL genes. This study will serve as a foundation for further study on the function of PyPAL genes and the biosynthesis of phenylpropanoid in P. yunnanensis.
The different tissues (buds, tender stems, stems, roots, and needles) of one-year-old P. yunnanensis were gathered from the Southwest Forestry University greenhouse (October 17, 2022). The samples were cleaned with running water, blotted dry, flash frozen in liquid nitrogen and stored at −80°C until use.
2.2 Total RNA Extraction and cDNA Synthesis
Total RNA extraction from different tissues was performed using an RNAprep Pure Plant Plus kit (TianGen Bio-chemical Technology Co., Ltd., Beijing, China). The mass of RNA was determined using gel electrophoresis (1% agarose), while the RNA concentration was tested by a NanoDrop 2000 spectrophotometer (Orion Thermo Fisher Scientific, Massachusetts, USA). The RNA samples to be used for the subsequent tests were immediately stored at −80°C. The first-strand cDNA was generated from 2 μg of overall RNA by using a reverse transcription kit (OMEGA Bio-Tek, Georgia, USA), according to the manual.
2.3 Screening and Cloning of PyPAL-1, PyPAL-2, and PyPAL-3 Based on Transcriptome Data
A phenylalanine ammonia-lyase HMMER model has been built based on the PAL amino acid sequences that have been logged into GeneBank for Z. mays (accession number: Q8VXG7.1), A. thaliana (accession number: NP_181241.1, NP_187645.1, NP_190894.1, NP_196043.2, NP_001190223.1), N. tabacum (accession number: NP_001312473.1), P. taeda (P52777.1), and P. pinaster (accession number: AAT66434.1) [33]. Based on the transcriptome data for P. yunnanensis (sequencing and assembly based on the nucleic acid sequence files in the assembly results obtained by Wuhan Metwell Biotechnology Co., Ltd., Hubei, China), the local blast database was constructed using BioEdit (version 7.0.9.0) [34], and the local blast was performed. The PAL amino acid sequences of P. yunnanensis were screened using HMMER3.0 (e-value > 1e-5) software. Combined with the transcriptome data in the Swiss-Prot functional database, sequence integrity, homologous alignment, conserved domain analysis, and expression, the homologous sequences of three PyPAL genes were screened according to the GO annotation procedure finally and named PyPAL-1, PyPAL-2, and PyPAL-3, respectively [35,36]. Based on the cDNA templates for P. yunnanensis, six pairs of primers of PyPAL-1-F/R, PyPAL-2-F/R, and PyPAL-3-F/R were designed using SnapGene software for PCR amplification. (Supplementary Table S1).
PCR amplification was conducted using Phanta Max Super-Fidelity DNA Polymerase (Vazyme Biotech Co., Ltd., Nanjing, China), and the amplified products were analyzed by gel electrophoresis (1% agarose) and subsequently subjected to purification using a DNA gel extraction kit (Vazyme Biotech Co., Ltd.). The amplified products were connected using the Clon Express II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) and introduced into E. coli DH5α competent cells (Solarbio Biotech, Beijing, China). And used restriction enzymes BamH I and Xho I to construct the recombinant vector (pET-30a). Positive clones were checked by PCR with the universal primers pET30a-forward and pET30a-reverse (Supplementary Table S1), and bacterial fluids with expected band sizes were selected and sent to sequencing. The E.Z.N.A Plasmid Miniprep Protocol (Omega Bio-Tek, Georgia, USA) was used to extract plasmid DNA for PyPAL-1, PyPAL-2, and PyPAL-3.
2.4 Bioinformatics Analysis of PyPALs
The nucleotide sequences of PyPAL-1, PyPAL-2, and PyPAL-3 were submitted to NCBI GenBank (OR714894, OR714895, and OR714896). The ORFs (open reading frames) of PyPAL genes were identified using the online ORF-Finder tool at NCBI (https://www.ncbi.nlm.nih.gov/orfnder/, accessed on 14 June 2023). The physicochemical properties and hydrophobicity of the protein were analyzed using ExPASy Protparam (https://web.expasy.org/protparam/, accessed on 14 June 2023) [37] and PredictProtein (https://ppopen.rostlab.org/, accessed on 15 June 2023). Protein secondary structures were predicted through the utilization of SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html, accessed on 15 June 2023), and SWISS-MODEL was used to predict the 3D structure of three PyPAL proteins (http://swissmodel.expasy.org/, accessed on 16 June 2023). Multiple-sequence alignment and phylogenetic tree construction of proteins were conducted using DNAMAN v8 and MEGA v11, respectively [38].
The usage characteristic parameters of the codon were found using CodonW 1.4.2, and these included A3s, U3s, C3s, G3s, relative synonymous codon usage RSCU, effective number of codons, ENC, codon adaptation index CAI, frequency of optimal codons FOP, GC content, and GC content at codon 3 (GC3s).
2.5 Construction of Recombinant Plasmid pET-30a-PyPALs
The recombinant plasmids pET-30a-PyPAL-1, pET-30a-PyPAL-2, and pET-30a-PyPAL-3 were transformed into E. coli DH5α competent cells, then the plasmid was extracted by the E.Z.N.A Plasmid Miniprep Protocol (OMEGA Bio-Tek, Georgia, USA), and the positive transformants were identified by PET30a-F/R amplification and sequencing (Supplementary Table S1). Then, the plasmids were transformed into BL21 (DE3) E. coli cells for expression.
2.6 Expression and Purification of Recombinant PyPALs
Recombinant strains were inoculated in LB-kanamycin medium (50 mg·L−1 kanamycin) and cultured at 37°C with shaking at 180 rpm for about 10 h to produce seed cultures. Under the same culture conditions, 30 mL seed cultures were expanded into 1L LB-kanamycin medium until the OD600 reached between 0.6 and 0.8, and the protein expression levels of PyPAL-1, PyPAL-2, and PyPAL-3 were induced with isopropylthio-β-galactoside (0.5 mM) at 28°C at 180 rpm. The 1L culture was centrifuged at 8000 rpm at 4°C for 15 min, and the supernatant was discarded. The sediment was resuspended in 10 mL buffer (1 mM β-Mercaptoethanol, 10 mM imidazole, 50 mM NaH2PO4, 300 mM NaCl). Then, the samples were broken using an ultrasonic cell crusher for 10 min (ultrasonic on 6 s; ultrasonic off 10 s; total time: 10 min; alarm temperature: 40°C; power ratio: 11%; horn: 6). Collect the proteins in the supernatant and store at −80°C.
The supernatants were loaded separately onto the Ni Sepharose 6FF to purify the PyPAL proteins. All supernatants were mixed with 4 mL Ni Sepharose 6FF washed three times using different concentrations of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM β-mercaptoethanol, 20 mM/80 mM/125 mM imidazole).Then, the target protein was obtained using M-elution buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM β-mercaptoethanol, and 500 mM imidazole). All purification steps were performed at 4°C or on ice. The target protein was separately dialyzed using buffer 1 (20 mM NaH2PO4, 500 mM NaCl, pH 7.4), buffer 2 (16 mM NaH2PO4, 400 mM NaCl, 32 mM imidazole, and 0.8 mM β-mercaptoethanol; pH 7.4), buffer 3 (12 mM NaH2PO4, 300 mM NaCl, 24 mM imidazole, and 0.6 mM β-mercaptoethanol; pH 7.4), buffer 4 (8 mM NaH2PO4, 200 mM NaCl, 16 mM imidazole, 0.4 mM β-mercaptoethanol; pH 7.4), and buffer 5 (ddH2O), and dialysis was performed in each buffer for 4 h.
The sample proteins were used for 12% were used for 12% SDS-PAGE. Protein concentration was determined using a BCA Protein Assay Kit (Sangon Biotech, Shanghai, China).
2.7 Enzyme Activity Assay of PyPAL-1, PyPAL-2, and PyPAL-3
A 1 mL enzymatic reaction system consisting of purified PyPALs (10 ug), 0.1 M Tris-HCl (pH 8.5), and 10 mM L-phenylalanine/10 mM L-tyrosine was incubated for 30 min at 37°C, and then 6 M Tris-HCl (100 mL) was used to terminate the reaction. The enzymatic reaction supernatant solutions, trans-cinnamic acid, and ρ-coumaric acid were filtered through a 0.22 μm Millipore filter before HPLC. Compounds were determined using an Agilent 1260 series liquid chromatography device (Agilent, Beijing, China). The mobile phase consisted of acetonitrile (phase A) and acetic acid-water (0.1%, phase B). The gradient elution was composed of 98% mobile phase A + 2% mobile phase B for 3 min, 95% mobile phase A + 5% mobile phase B for 5 min, and 40% mobile phase A + 60% mobile phase B for 9 min. And using gradient elution at a flow rate of 1 mL−1 min, with the column oven maintained at 30°C. Trans-cinnamic acid and ρ-coumaric acid were determined at 260 and 290 nm, respectively.
A 1 mL enzymatic reaction system consisting of purified PyPALs (10 μg), 100 μL 0.01 mol L−1 L-phenylalanine and 0.1 mol L−1 Tris-HCl with different pH levels (pH 6, 7, 8, 9, and 10) The optimal pH of PyPALs was determined at OD290 by using a microplate reader. At the optimal pH, the recombinant protein of PyPALs was determined at different temperatures (37°C, 47°C, 57°C, 67°C, 77°C, and 87°C). The OD290 of the maximum enzyme activity was defined as 100%. And the relative activity of PyPALs was calculated at different pH levels and temperatures. In 0.1 mol L−1 Tris-HCl buffer (pH 8.5), 10 μg PyPAL recombinant protein was added to L-phenylalanine substrate with concentrations of 0, 0.2, 0.4, 0.6, 0.8, 1, 2, 3, 4, 5, and 6 mM. The activity of PyPAL-1, PyPAL-2, and PyPAL-3 was determined at 37°C for 30 min.
2.8 Expression Pattern Analysis
Total RNA was extracted from different tissues of P. yunnanensis, including buds, tender stems, stems, roots, and needles, using the same method used in Section 2.2. The cDNA was synthesized using the cDNA Synthesis Kit (Yeasen Biotechnology Co., Ltd., Shanghai, China). The 2−ΔΔCt method was used to calculate the relative expression of PyPALs [39].
Significant differences for different samples for each PyPAL were calculated via one-way ANOVA in SPSS 22.0, and the original graphics were generated using GraphPad Prism (version 10.0, GraphPad Software, Inc. USA). The PyPAL’s enzyme kinetic properties were calculated and plot curves with GraphPad Prism “Michaelis–Menten”(version 10.0).
3.1 Molecular Cloning and Physicochemical Properties of the PyPALs
Three PyPAL coding sequences were identified by PCR amplification and sequencing (Fig. 1). The total RNA of P. yunnanensis showed that the bands of 28S and 18S were clear and had no diffuse tails, and the A260/A280 ratios of the total RNA were 1.8~2.0, which indicated that the total RNA was of good quality and was by the requirements for gene cloning (Fig. 1a). The PyPAL-1, PyPAL-2, and PyPAL-3 genes were amplified using reverse-transcription polymerase chain reaction reverse transcription (RT-PCR) with specific primers after the total RNA was reverse transcribed into cDNA. All the primers are listed in Supplementary Table S1. The ORF of the PyPAL-1, PyPAL-2, and PyPAL-3 genes was about 2500 bp (Fig. 1b). The cloning vectors for PyPAL-1, PyPAL-2, and PyPAL-3 were constructed and sequenced. The results showed that the ORFs of PyPAL-1, PyPAL-2, and PyPAL-3 were 2184, 2157, and 2385 bp, respectively (Figs. 1b and 1c).
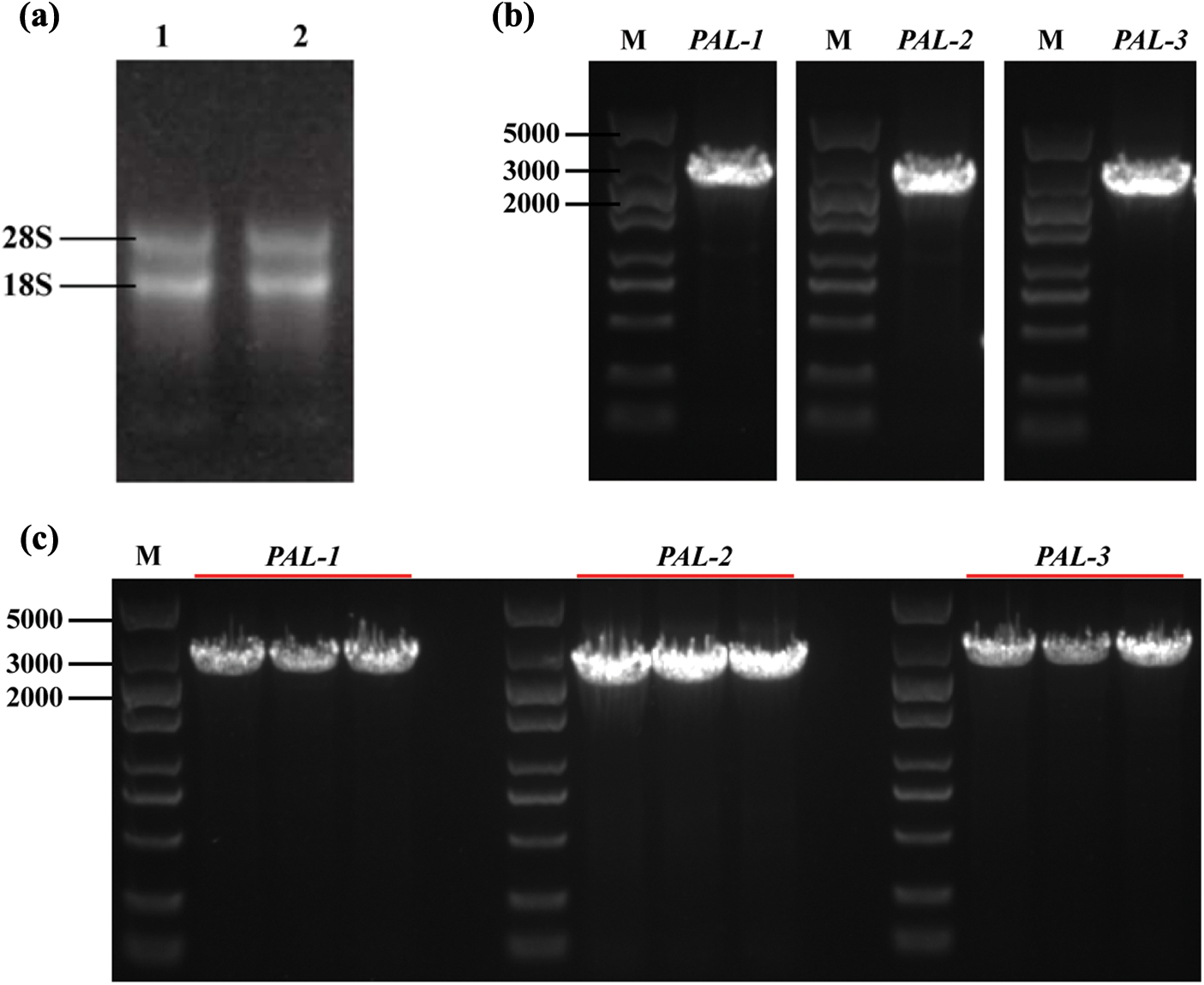
Figure 1: PCR amplification and identification of PyPAL genes from P. yunnanensis. (a) 1, 2, total RNA of P. yunnanensis seedlings; (b) PyPAL-1~PyPAL-3 DNA amplification; (c) identification via bacterial fluid PCR
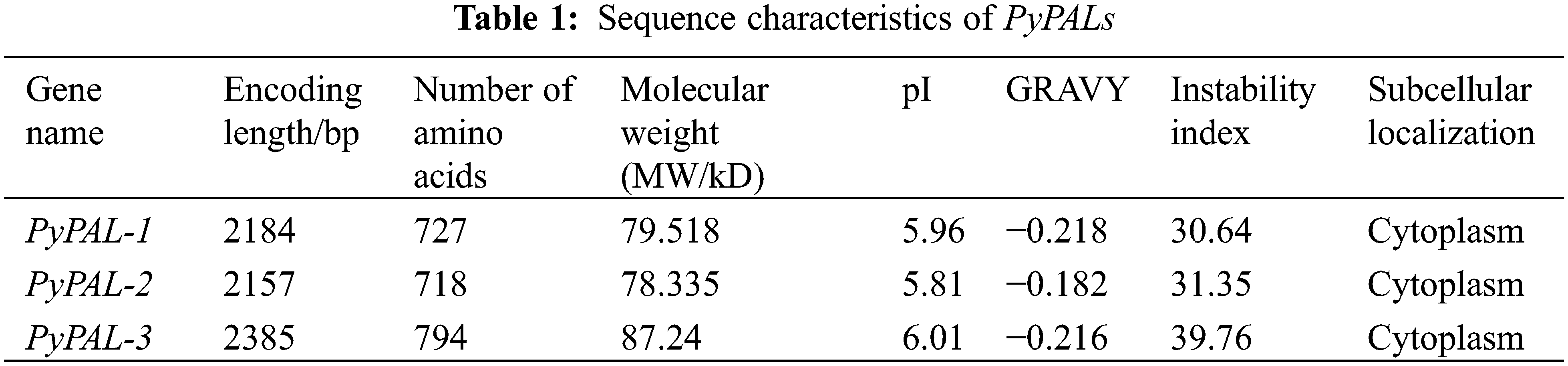
The numbers of amino acid residues of the PyPALs were 727 aa, 718 aa, and 794 aa, and the isoelectric points (PIs) were 5.96, 5.81, and 6.01, respectively; these results suggest that PyPALs may encode acidic proteins (Table 1). The instability indices of all three proteins are less than 40, indicating that they are stable proteins in vitro. The subcellular localization showed that PyPAL proteins are all located in the cytoplasm.
3.2 Characterization of the PyPAL Proteins
The analysis of conserved domains in PyPALs was performed using the conserved domains database (CDD) indicating that both PAL proteins contained active sites of the phenylalanine ammonia-lyase superfamily (GTITASGDLVPLSYIAG, Fig. 2a). The α-helices of PyPAL-1, PyPAL-2, and PyPAL-3 were 54.33%, 56.27%, and 55.29%; the β-rotation was 6.33%, 6.27%, and 5.54%; the extension chain was 9.35%, 9.33%, and 7.43%; and the irregular curl was 29.99%, 28.13%, and 31.74%, respectively (Fig. 2b). The three-dimensional structures of PyPAL-1, PyPAL-2, and PyPAL-3 were predicted using the SWISS-MODEL program. The results showed that the similarity of the PyPAL-1, PyPAL-2, and PyPAL-3 proteins to the template was 65.27%, 67%, and 64.14%, respectively; the GMQE was 0.82, 0.84, and 0.74; and the QMEAND values were 0.79, 0.80, and 0.78, respectively (Fig. 2c). At the same time, the GC contents of the PyPAL genes of P. yunnanensis were 54.53%, 49.47% and 54.97%, respectively, and the average content was 53.06%. The average contents of GC1, GC2, and GC3 were 56.87%, 41.66%, and 60.66%, respectively. The order of size is GC3 > GC1 > GC2. The ENC values ranged from 48.97% to 51.79%. The PyPALs had a weak preference for 57 codons and a low frequency (RSCU < 1), and the PALs of P. yunnanensis had no codon preference (Supplementary Fig. S1, Supplementary Table S2).
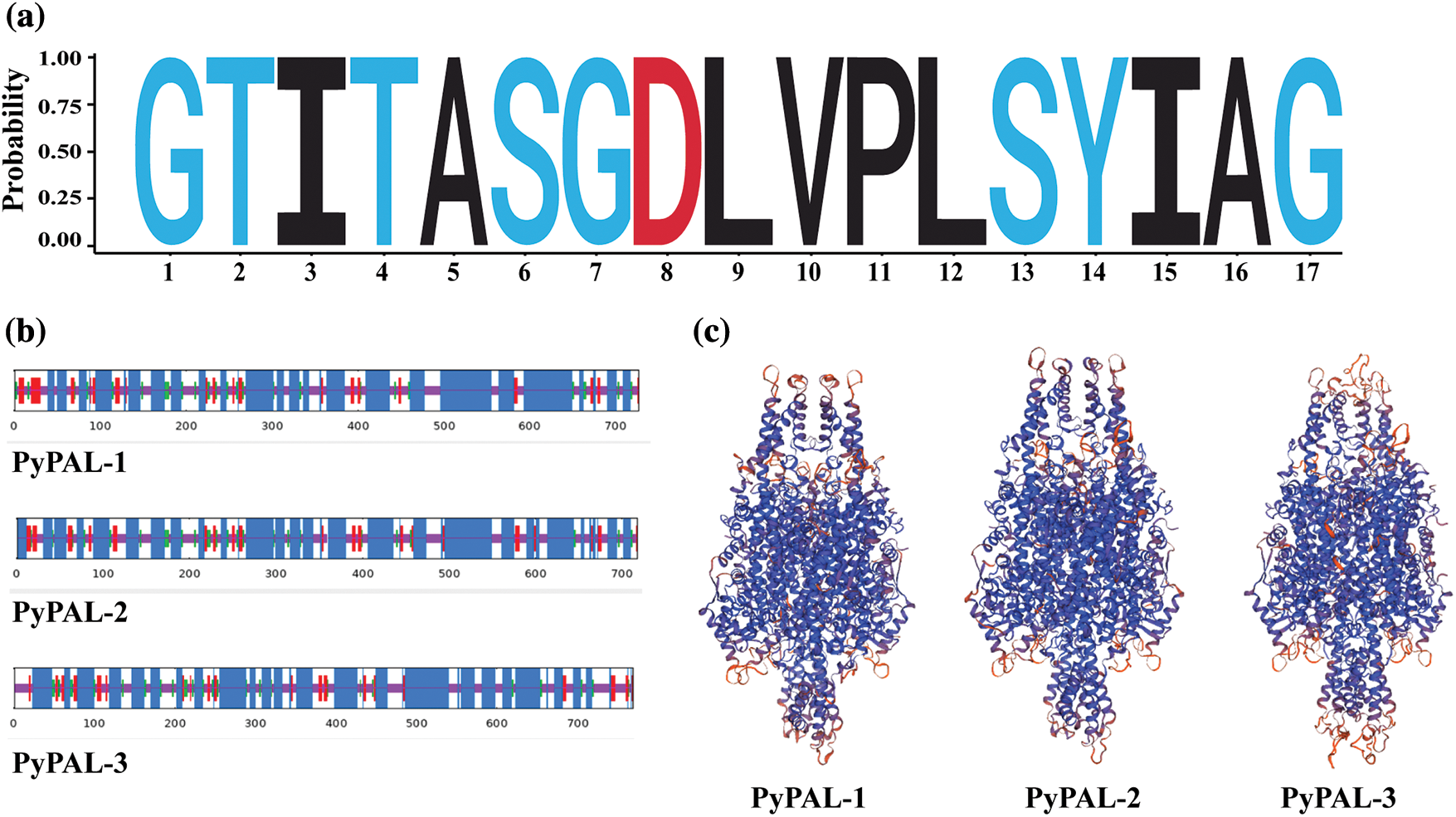
Figure 2: Structural characterization of PyPAL proteins. (a) The sequence logo of active sites of the phenylalanine ammonia-lyase superfamily, (b) secondary structure of PyPAL proteins, and (c) protein tertiary structure models of PyPAL proteins
3.3 Homology and Phylogenetic Relationships of PyPALs
The PyPALs’ amino acid sequences of P. yunnanensis were compared with those of A. thaliana, Z. mays, N. tabacum, and P. taeda using DNAMAN 8 software, with a similarity of 75.79%. The results also showed that PyPAL-1, PyPAL-2, and PyPAL-3 contain the active site of the phenylalanine ammonia-lyase superfamily (Fig. 3, red frame: GTITASGDLVPLSYIAG), which can catalyze the conversion of L-phenylalanine to trans-cinnamic acid (Fig. 3). Based on the comparison results, a phylogenetic tree was constructed using the amino acid sequences of PyPALs and PALs from other plants and MEGA 11.0. All angiosperms and P. banksiana PAL2 and PAL5 clustered in one group. The other gymnosperms were clustered in a clade. PyPAL-2 clustered in a clade with the two P. taeda PALs, while PyPAL-1 and PyPAL-3 were divided into two gymnosperm clades, respectively clustered with P. banksiana PAL4 and PAL3 (Fig. 4). These results suggest that the PyPAL gene shares high homology with other known PAL genes in angiosperms and gymnosperms, which may be associated with some secondary metabolites or functions.
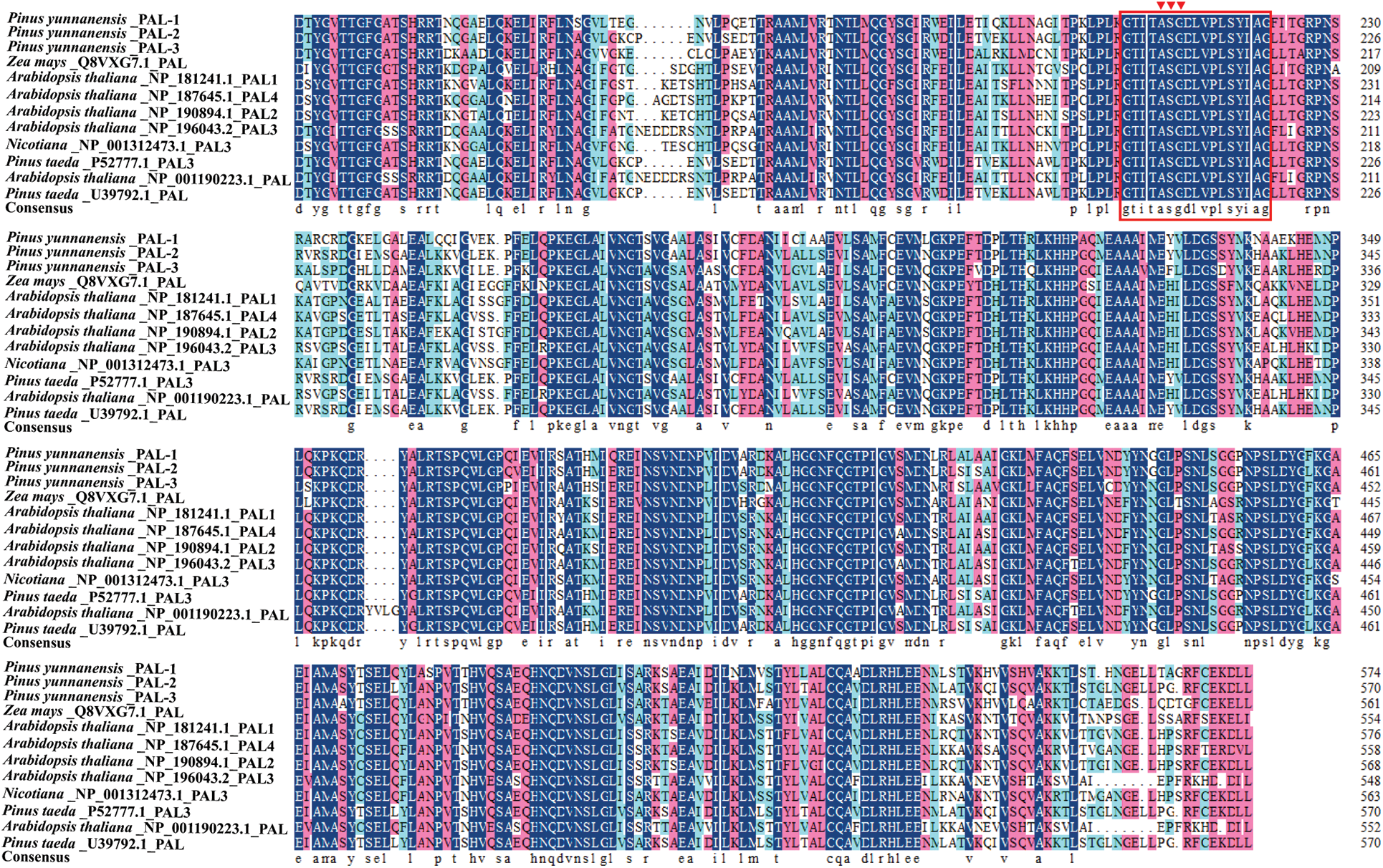
Figure 3: Multiple alignments of deduced amino acid sequences of PALs from P. yunnanensis and other plants. The conserved motifs were labeled in red boxes on the sequence
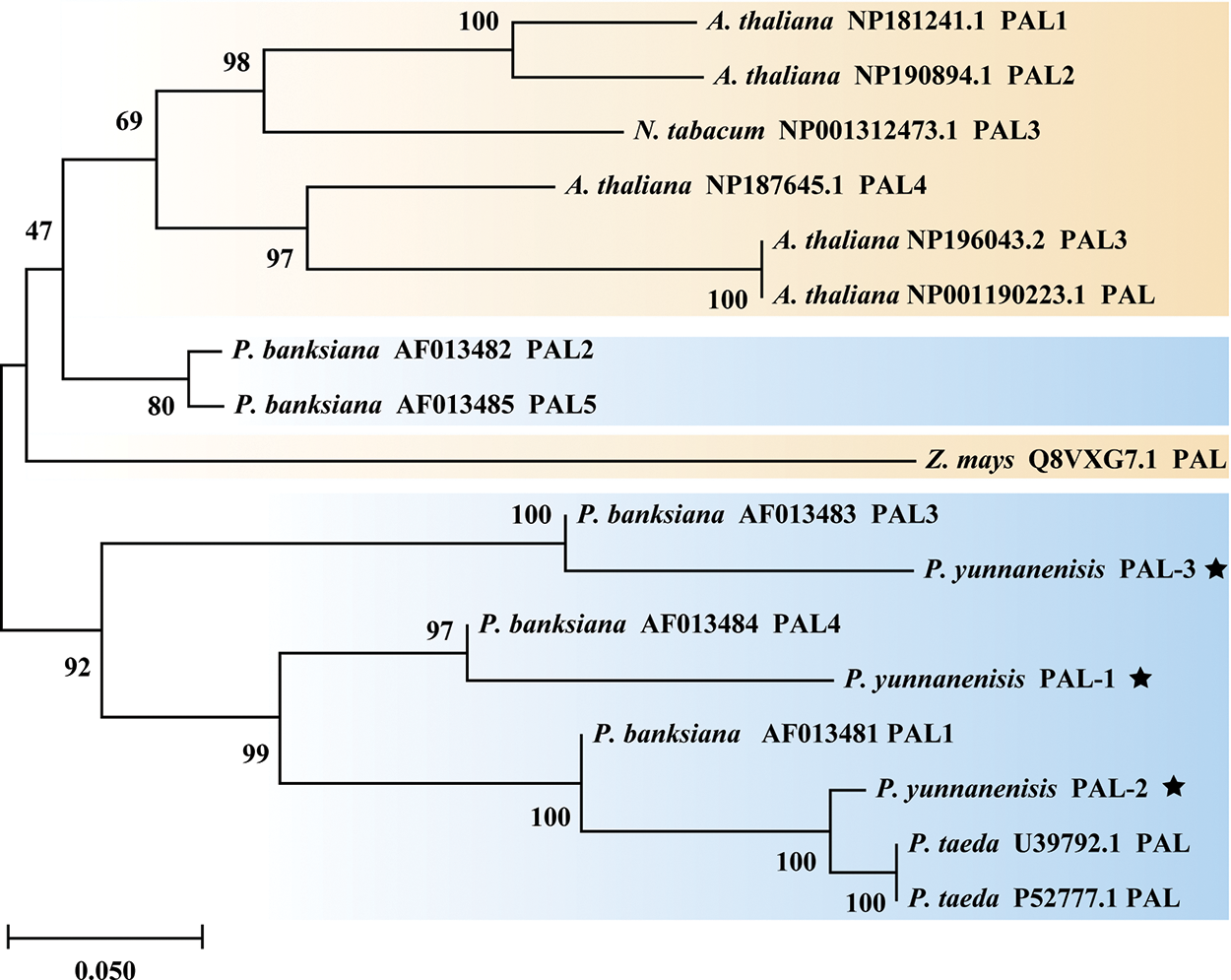
Figure 4: Phylogenetic tree of PALs from P. yunnanensis and other species. The star marker represents the PALs of P. yunnanensis. The PALs of gymnosperms are marked in blue, Orange marks the PALs of angiosperms
3.4 PyPAL Proteins’ Functional Verification and Kinetic Parameters Analysis
To further verify the function of PyPALs, the proteins were first expressed and purified (Supplementary Fig. S2). Then, the purified enzyme’s catalytic functions were determined in vitro via HPLC after protein purification. The results showed that PyPALs catalyzed the generation of trans-cinnamic acid from L-phenylalanine (Fig. 5a). However, PyPALs have a poor ability to catalyze the conversion of L-tyrosine to ρ-coumaric acid (Fig. 5b). PyPAL-1 and PyPAL-2 showed slight catalytic activity, and PyPAL-3 showed no catalytic activity for L-tyrosine.
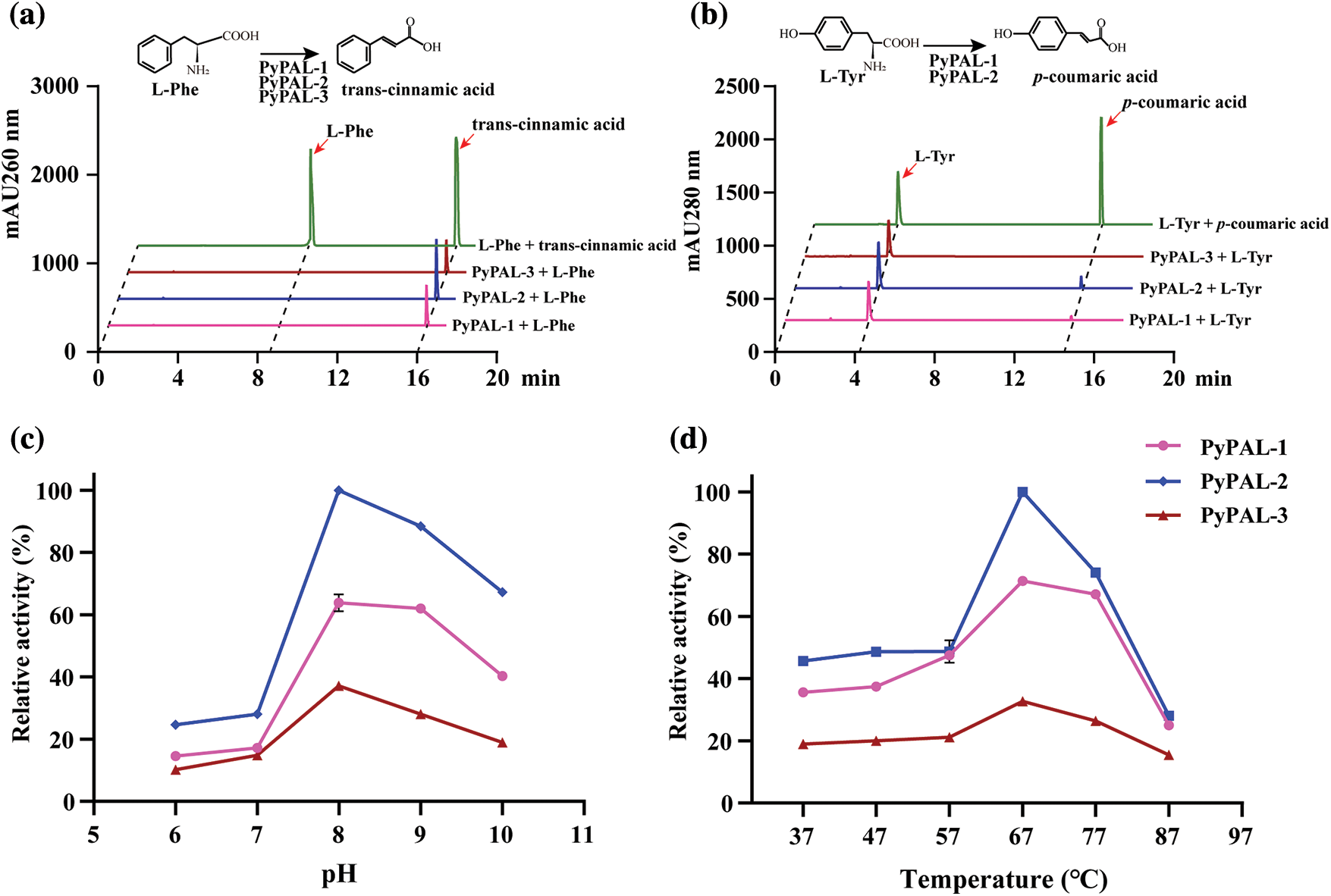
Figure 5: Functional verification and tissue‑specific expression profile of PyPAL proteins. (a) and (b) are the PALs’ ability to catalyze the conversion of L-phenylalanine and L-tyrosine, respectively. (c) and (d) are the L-phenylalanine catalytic activities of PALs at different pH levels and temperatures, respectively
Under the conditions of pH 8.5 and a temperature of 37°C, the kinetic parameters of the L-phenylalanine enzyme with PyPALs were determined, and Miemann’s kinetic curve of PyPALs was fitted. The PyPAL-1, PyPAL-2, and PyPAL-3 Km values were 1.861, 2.364, and 1.049 mmol L−1, respectively, and the Vmax values were 28.80, 22.20, and 8.827 nKat mg−1 (Supplementary Fig. S3). Then, we further explored the optimal conditions for the catalytic conversion of L-phenylalanine by PyPALs, and the results showed that different PyPALs exhibited a single-peak pattern at continuous pH and temperatures. PyPALs exhibited the highest catalytic activity at pH 8 and a reaction temperature of 67°C (Figs. 5c and 5d).
3.5 Tissue-Specific Expression Profile of PyPALs
Finally, the expression profiles of PyPALs in different tissues of one-year-old P. yunnanensis were explored (Fig. 6a). Three PyPALs were all expressed in the buds, tender stems, stems, roots, and needles of P. yunnanensis, but the expression patterns were different. PyPAL-1 expression was the highest in needles and significantly higher than in other tissues, and then in buds, which was also significantly higher than that found in stems and roots. The expression of PyPAL-2 was the highest in the bud, followed by the root, while the expression in the stem was the lowest (p < 0.05). The highest expression of PyPAL-3 was found in the root, followed by the tender stem, stem, and bud, while the lowest expression was found in the needles (Figs. 6b–6d).
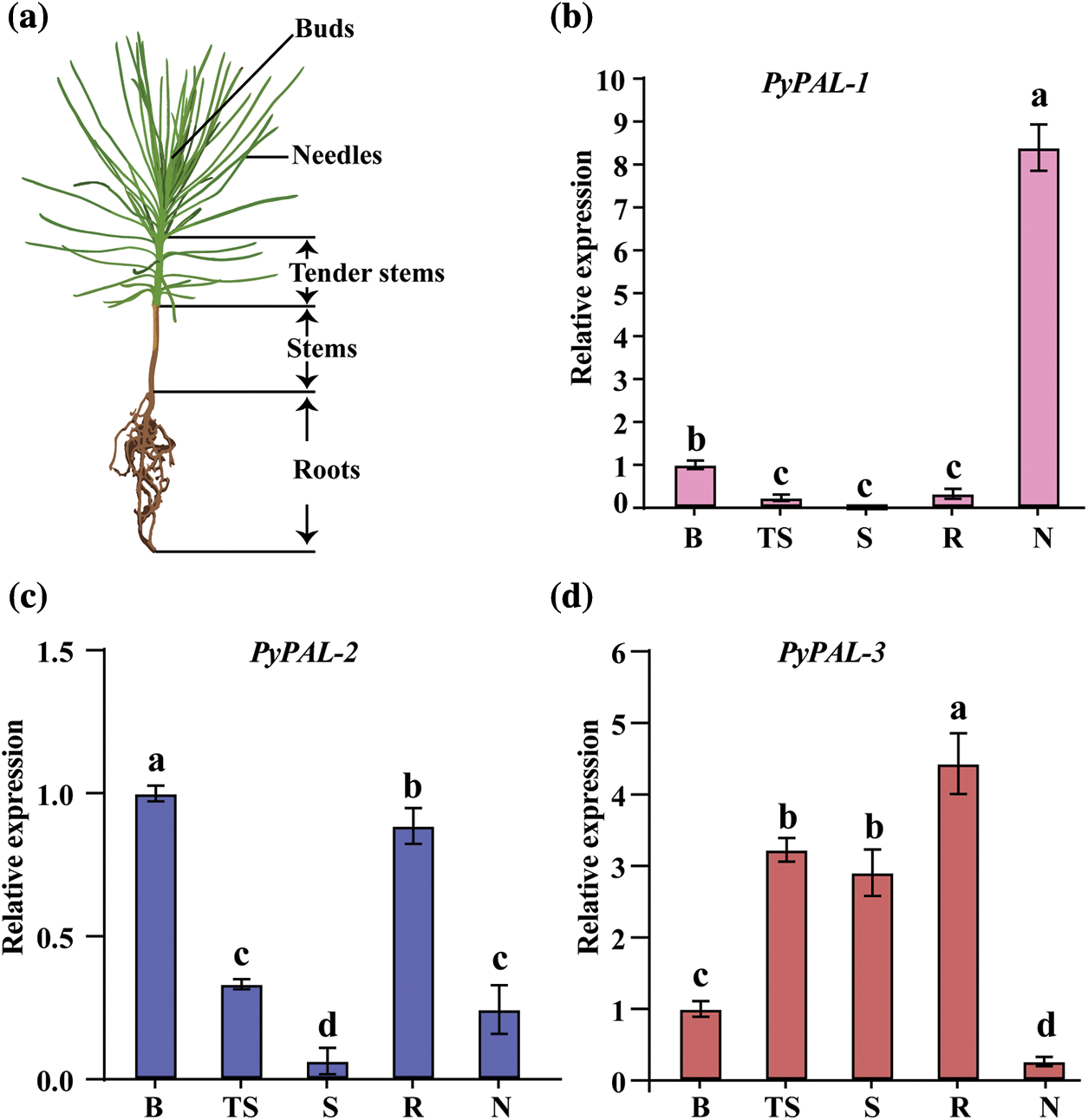
Figure 6: Relative expression of PyPALs in P. yunnanensis. (a) Schematic diagram of sample collection. (b), (c), and (d) are the expression profiles of PALs in different tissues of one-year-old P. yunnanensis, which were normalized by the expression in buds. B: buds; TS: tender stems; S: stems; R: roots; N: needles. Values with different lowercase letters show significant differences among different tissues at the 0.05 probability level
Phenylpropanoids are precursors of flavonoids, isoflavones, coumarins, and stilbenes, which are related to lignin, the main component of woody plants, and are related to plant resistance [2,40]. Therefore, PAL has been thoroughly studied in angiosperms [1,38]. In this study, three PAL genes of P. yunnanensis were identified, and their structures showed active sites of the PAL superfamily. In all plants studied to date, PAL, which is encoded by a multigene family, is a tetrameric enzyme in vivo with around 725 amino acid residues in P. taeda [8,25,41]. Phylogenetically, the PALs of P. yunnanensis and gymnosperms such as P. taeda and P. pinaster were clustered into one clade, whereas the PALs of other angiosperm species such as A. thaliana, Z. mays, and N. tabacum were clustered into another clade. Four PAL genes of P. taeda were expressed in the gymnosperm group [25]. This result is different from the result of He et al., who reported that PALs of Ce. hainanensis clustered with other alkaloid-producing plants belonging to angiosperms [42]. In addition, the three PyPALs were not clustered into one clade, but they were divided into three different clades. Among them, PyPAL-2 clustered into one clade with the PALs of other pines, then PyPAL-1 joined them, and PyPAL-3 was the furthest away from the other two clades. Whether this process is related to the divergent PAL function still needs to be studied in other gymnosperms, along with further study in P. yunnanensis.
Functionally, in vitro enzymatic analysis showed relatively strict substrate specificity, that is, high catalytic activity for L-phenylalanine and low catalytic activity for L-tyrosine [14,43]. Moreover, the highest enzyme activity was observed at pH 8 and 67°C for all the PyPALs (Fig. 5). Although PyPAL-1–3 showed higher L-phenylalanine catalytic activity, PyPAL-1 completely failed to catalyze the conversion of L-tyrosine, whereas PyPAL-2 and PyPAL-3 showed lower catalytic conversion activity for L-tyrosine. These features indicated that PyPALs are more similar to PAL from dicotyledonous and can effectively deaminate L-phenylalanine, while yeast and some monocots can convert L-phenylalanine and L-tyrosine to their corresponding trans-cinnamic acid and ρ-coumaric acid [44]. Meanwhile, PyPAL-2 had the highest Km value, and PyPAL-3 had the lowest Km value. PyPAL-3 also showed differences in substrate affinity. On the contrary, the three PAL genes of P. yunnanensis exhibited different catalytic characteristics in catalytic reactions, indicating that their functions in plants may also have different emphases.
Finally, PyPAL gene expression in different tissues of P. yunnanensis was profiled via quantitative real-time PCR. The results indicated that gene expression and function were different in different tissues. PyPAL-1 was mainly expressed in needles, and PyPAL-2 was mainly expressed in buds and roots, whereas PyPAL-3 was highly expressed in tender stems, stems, and roots. A search of literature shows that the activity of PAL is highly controlled at the transcriptional level by developmental stages; this trend is due to changes in PAL activity related to the different roles play by its different secondary metabolites during biosynthesis, such as plant development and tolerance to abiotic stimulants [14,45,46]. In this study, at least one of the five different tissues showed high expression of PyPALs at the transcriptional level, in which PyPAL-2 and PyPAL-3 were highly expressed in the root compared with other tissues. These results indicate that the function of PAL is redundant and divergent in gymnosperms [14].
PAL is a rate-limiting enzyme encoded by multiple gene families in plants and is involved in the biosynthesis of flavonoids, lignin, and many other compounds. In this study, three PyPALs, namely, PyPAL-1, PyPAL-2, and PyPAL-3 were cloned, confirming that the PAL in P. yunnanensis belongs to a multigene family. In addition, the results of the enzyme activity test in vitro showed that all three PyPALs could catalyze the conversion of L-phenylalanine to the corresponding trans-cinnamic acid, but such genes had no or low catalytic effect on L-tyrosine. The three PyPALs were expressed at the transcriptional level in different P. yunnanensis tissues. Therefore, the study on PyPALs will be helpful to further analyze the key role of PAL in the growth and development of P. yunnanensis, especially in the biosynthesis of lignin.
Acknowledgement: We are very grateful to the editor and reviewers’ comments, which is helpful for improving our manuscript during the manuscript preparation.
Funding Statement: This study received financial support from the Youth Talents Special Project of Yunnan Province, “Xingdian Talents Support Program” (XDYC-QNRC-2022-0203); Southwest Forestry University Scientific Research Start-Up Funds (112116).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Junrong Tang, Lin Chen, and Shi Chen; data collection: Dejin Mu, Heze Wang, and Zhaoliu Hu; analysis and interpretation of results: Nianhui Cai, Sihui Chen, and Dejin Mu; draft manuscript preparation: Dejin Mu, Yulan Xu, and Junrong Tang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.048786.
References
1. Huang JL, Gu M, Lai ZB, Fan BF, Shi K, Zhou YH, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153(4):1526–38. [Google Scholar] [PubMed]
2. Deng YX, Lu SF. Biosynthesis and regulation of phenylpropanoids in plants. Crit Rev Plant Sci. 2017;36(4):257–90. [Google Scholar]
3. Ding LP, Chen YJ, Wang HZ, Wei JH. Efficacy of Cry1Ac protein against gypsy moth and fall webworm in transgenic poplar (Populus davidiana × Populus bolleana) by bioassay. Crit Rev Plant Sci. 2018;98(4):844–50. [Google Scholar]
4. Campbell MM, Sederoff RR. Variation in lignin content and composition (mechanisms of control and implications for the genetic improvement of plants). Plant Physiol. 1996;110(1):3–13. [Google Scholar] [PubMed]
5. Koukol J, Conn EE. The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J Biol Chem. 1961;236:2692–8. [Google Scholar] [PubMed]
6. Zhang X, Liu CJ. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol Plant. 2015;8:17–27. [Google Scholar] [PubMed]
7. Tong ZZ, Xie J, Yin MZ, Wu JX, Zha LP, Chu SS, et al. Cloning, characterization and prokaryotic expression analysis of two phenylalanine ammonia-lyase genes from Peucedanum Praeruptorum Dunn. Braz J Bo. 2022;45(3):897–907. [Google Scholar]
8. Wanner LA, Li GQ, Ware D, Somssich IE, Davis KR. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol Biol. 1995;27(2):327–38. [Google Scholar] [PubMed]
9. Zhang XB, Gou MY, Liu CJ. Arabidopsis kelch repeat F-Box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell. 2013;25(12):4994–5010. [Google Scholar] [PubMed]
10. Cochrane FC, Davin LB, Lewis NG. The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry. 2004;65:1557–64. [Google Scholar] [PubMed]
11. Wu YL, Wang WZ, Li YZ, Dai XL, Ma GL, Xing DW, et al. Six phenylalanine ammonia-lyases from Camellia sinensis: evolution, expression, and kinetics. Plant Physiol Biochem. 2017;118:413–21. [Google Scholar] [PubMed]
12. Reichert AI, He XZ, Dixon RA. Phenylalanine ammonia-lyase (PAL) from Tobacco (Nicotiana tabacumcharacterization of the four Tobacco PAL genes and active heterotetrameric enzymes. Biochem J. 2009;424(2):233–42. [Google Scholar] [PubMed]
13. Tomoko FA, Kung SD, Watson JC. Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana. Plant Mol Biol. 1996;30:711–22. [Google Scholar]
14. Khakdan F, Alizadeh HS, Ranjbar M. Molecular cloning, functional characterization and expression of a drought Inducible phenylalanine ammonia-lyase gene (ObPAL) from Ocimum basilicum L. Plant Physiol Bioch. 2018;130:464–72. [Google Scholar]
15. Hamberger B, Ellis M, Friedmann M, Souza CDA, Barbazuk B, Douglas CJ. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene familiesthis. Can J Bot. 2007;85(12):1182–201. [Google Scholar]
16. Zhang HY, Zhang XH, Zhao HX, Hu J, Wang ZY, Yang GS, et al. Genome-wide identification and expression analysis of phenylalanine ammonia-lyase (PAL) family in rapeseed (Brassica napus L.). BMC Plant Biol. 2023;23(1):481. [Google Scholar] [PubMed]
17. Tian J, Xu R, Chang KZ, Yuan S, Huang CX, Wang JW, et al. Identification of PAL gene in purple cabbage and functional analysis related to anthocyanin synthesis. Horticulturae. 2023;9(4):469. [Google Scholar]
18. Yan F, Li HZ, Zhao P. Genome-wide identification and transcriptional expression of the PAL gene family in common walnut (Juglans regia L.). Genes. 2019;10(1):46. [Google Scholar] [PubMed]
19. Yang CJ, Jin M, Zhang YQ. Bioinformatics and expression analysis of PAL gene family of Poplar plant under different nitrogen treatments. J Anim Plant Sci. 2023;33:1193–1203. [Google Scholar]
20. Huang X, Bai XH, Xie ZH, Fahad S, Gbokie T. De novo transcriptome assembly of Coffea liberica reveals phylogeny and expression atlas of phenylalanine ammonia-lyase genes in Coffea species. Ind Crop Prod. 2023;192:116029. [Google Scholar]
21. Craven-Bartle B, Pascual MB, Canovas FM, Avila C. A Myb transcription factor regulates genes of the phenylalanine pathway in maritime pine. Plant J. 2013;74(5):755–66. [Google Scholar] [PubMed]
22. Osakabe Y, Osakabe K, Chiang VL. Characterization of the tissue-specific expression of phenylalanine ammonia-lyase gene promoter from loblolly pine (Pinus taeda) in Nicotiana tabacum. Plant Cell Rep. 2009;28:1309–17. [Google Scholar] [PubMed]
23. Zhang C, Yao X, Ren H, Wang K, Chang J. Genome-wide identification and characterization of the phenylalanine ammonia-lyase gene family in pecan (Carya illinoinensis). Sci Hortic. 2022;295:110800. [Google Scholar]
24. Kumar A, Ellis BE. The phenylalanine ammonia-lyase gene family in raspberry. structure, expression, and evolution. Plant Physiol. 2001;127(1):230–9. [Google Scholar] [PubMed]
25. Bagal UR, Leebens-Mack JH, Walter Lorenz W, Dean JFD. The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage. BMC Genom. 2012;13(Suppl 3):S1. [Google Scholar]
26. Butland SL, Chow ML, Ellis BE. A diverse family of phenylalanine ammonia-lyase genes expressed in pine trees and cell cultures. Plant Mol Biol. 1998;37:15–24. [Google Scholar] [PubMed]
27. Campbell MM, Ellis BE. Fungal elicitor-mediated responses in pine cell cultures: I. Induction of phenylpropanoid metabolism. Planta. 1992;186:409–17. [Google Scholar] [PubMed]
28. Tang C, Shen LQ, Han PB, Huang DS, Li SF, Li YF, et al. Forest characteristics, population structure and growth trends of Pinus yunnanensis in Tianchi National Nature Reserve of Yunnan, Southwestern China. Vegetation Classification and Survey. 2020;1:7–20. [Google Scholar]
29. Wang BS, Mao JF, Zhao W, Wang XR. Impact of geography and climate on the genetic differentiation of the subtropical Pine Pinus yunnanensis. PLoS One. 2013;8:e67345. [Google Scholar] [PubMed]
30. Li P, Zong D, Gan P, Li HL, Wu ZY, Li FH, et al. Comparison of the diversity and structure of the rhizosphere microbial community between the straight and twisted trunk types of Pinus yunnanensis. Front Microbiol. 2023;14:1066805. [Google Scholar] [PubMed]
31. Gan PH, Li PL, Zhang XL, Li HL, Ma SJ, Zong D, et al. Comparative transcriptomic and metabolomic analyses of differences in trunk spiral grain in Pinus yunnanensis. Int J Mol Sci. 2023;24(19):14658. [Google Scholar] [PubMed]
32. Zang Y, Jiang T, Cong Y, Zheng ZJ, Ouyang J. Molecular characterization of a recombinant Zea mays phenylalanine ammonia-lyase (ZmPAL2) and its application in trans-cinnamic acid production from L-phenylalanine. Appl Biochem Biotech. 2015;176(3):924–37. [Google Scholar]
33. Isaza S, Houtgast E, Gaydadjiev G. Hmmer performance model for multicore architectures. Conf Digit Syst Design. 2011;14:257–61. [Google Scholar]
34. Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acid Symp Ser. 1999;41:95–8. [Google Scholar]
35. Manzoor MA, Li G, Abdullah M, Han W, Wenlong H, Zhang Y, et al. Genome-wide investigation and comparative analysis of MATE gene family in Rosaceae species and their regulatory role in abiotic stress responses in Chinese pear (Pyrus bretschneideri). Physiol Plant. 2021;173(3):1163–78. [Google Scholar] [PubMed]
36. Manzoor MA, Sabir IA, Shah IH, Wang H, Yu Z. Comprehensive comparative analysis of the GATA transcription factors in four rosaceae species and phytohormonal response in Chinese pear (Pyrus bretschneideri) fruit. Int J Mol Sci. 2021;22(22):12492. [Google Scholar] [PubMed]
37. Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the expasy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa: Humana Press; 2005. p. 571–607. [Google Scholar]
38. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. [Google Scholar] [PubMed]
39. Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2−ΔΔCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL transcripts. Diagn Mol Pathol. 2006;15:56–61. [Google Scholar] [PubMed]
40. Morse AM, Peterson DG, Islam-Faridi MN, Smith KE, Magbanua Z, Garcia SA, et al. Evolution of genome size and complexity in Pinus. PLoS One. 2009;4(2):e4332. [Google Scholar]
41. Ronald P, Söderhäll K. Phenylalanine ammonia lyase and peroxidase activity in mycorrhizal and nonmycorrhizal short roots of scots Pine, Pinus sylvestris L. New Phytol. 1985;101:487–94. [Google Scholar] [PubMed]
42. He YD, Zhong XH, Jiang XF, Cong H, Sun H, Qiao F, et al. Characterisation, expression and functional analysis of PAL gene family in Cephalotaxus hainanensis. Plant Physiol Biochem. 2020;156:461–70. [Google Scholar] [PubMed]
43. Whetten RW, Sederoff RR. Phenylalanine ammonia-lyase from loblolly pine: purification of the enzyme and isolation of complementary DNA clones. Plant Physiol. 1992;1992(98):380–6. [Google Scholar]
44. Zhan C, Li YT, Li H, Wang MR, Gong SJ, Ma DF, et al. Phylogenomic analysis of phenylalanine ammonia-lyase (PAL) multigene family and their differential expression analysis in wheat (Triticum Aestivum L.) suggested their roles during different stress responses. Front Plant Sci. 2022;13:982457. [Google Scholar] [PubMed]
45. Rösler J, Krekel F, Amrhein N, Schmid J. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 1997;113:175–9. [Google Scholar]
46. Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MS, Wang L. The phenylpropanoid pathway and plant defence–a genomics perspective. Mol Plant Pathol. 2002;3:371–90. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools