 Open Access
Open Access
ARTICLE
SiRAP2-12, a Positive Regulatory Factor, Effectively Improves the Waterlogging Tolerance of Foxtail Millet (Setaria italica)
1 Institute of Millet Crops, Hebei Academy of Agriculture and Forestry Sciences/Key Laboratory of Genetic Improvement and Utilization for Featured Coarse Cereals (Co-Construction by Ministry and Province), Ministry of Agriculture and Rural Affairs/The Key Research Laboratory of Minor Cereal Crops of Hebei Province, Shijiazhuang, 050035, China
2 School of Life Sciences, Institute of Life Science and Green Development, Hebei University, Baoding, 071002, China
* Corresponding Authors: Shunguo Li. Email: ; Jianfeng Liu. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Abiotic and Biotic Stress Tolerance in Crop)
Phyton-International Journal of Experimental Botany 2024, 93(3), 445-465. https://doi.org/10.32604/phyton.2024.048273
Received 02 December 2023; Accepted 24 January 2024; Issue published 28 March 2024
Abstract
Foxtail millet (Setaria italica) growth was inhibited because of waterlogging stress, which has caused yield reduction. ERF family plays an important role to plant adversity tolerance. In our study, we obtained 19,819 differential expressed genes (DEGs) between the two treatments based on the RNA-seq sequencing of foxtail millet of waterlogging stress. Furthermore, a total of 28 ERF family members were obtained, which have a complete open reading frame. We studied the evolution and function of SiERF family and how they affected the waterlogging tolerance. It was found that SiERF1A/B/C (GenBank ID: OR775217, OR775219, OR775218) and SiRAP2-12 (GenBank ID: OR775216) have similar functions to the known waterlogging tolerance genes of other plants. Among them, the SiRAP2-12 expression was obviously significantly up-regulated in foxtail millet after 5d waterlogging stress. After SiRAP2-12 was silenced, the activity of defense enzymes in millet decreased significantly. In details, superoxide dismutase (SOD), catalase (CAT) and peroxidase (POD), the osmotic regulator proline (Pro), and the activity of the anaerobic respiratory enzyme alcohol dehydrogenase (ADH) content were decreased by 78.61%, 29.52%, 79.95%, 19.41% and 54.77%, respectively. In contrast, the relative electrical conductivity contents (REC), malondialdehyde (MDA), and hydrogen peroxide (H2O2) of the foxtail millet subjected to virus-induced gene silencing clearly increased by 1.03-fold, 36.09%, and 15.21%, respectively. The content of sodium (Na+) in the SiRAP2-12-silenced foxtail millet also increased, but that of potassium (K+) decreased. Interestingly, we found that ethylene content was significantly reduced. Further, the SiAOC1 expression, an essential gene for ethylene synthesis, was inhibited in SiRAP2-12-silenced foxtail millet after waterlogging stress. Taken together, we hypothesized that SiRAP2-12 might be a positive regulator of millet tolerance to waterlogging stress.Keywords
Supplementary Material
Supplementary Material FileFoxtail millet, belongs to an annual gramineous plant and it is widely planted in China [1]. At present, foxtail millet still plays an important role in China [2]. The protein of foxtail millet is more digestible and nutritious than rice (Oryza sativa) and wheat (Triticum aestivum) [3]. The trace elements contents, including iron, zinc, magnesium, and selenium of foxtail millet are higher than rice, wheat, and corn (Zea mays) [4]. The planting period of foxtail millet is generally from April to June, and there is much rain in summer. Some areas often suffer from waterlogging. When foxtail millet grow under waterlogging stress, the metabolism of root cells is disordered, the growth potential energy is decreased, and the yield is also decreased [5,6]. Most of the varieties of foxtail millet grown have a low tolerance to waterlogging, which has caused substantial losses in production [7]. At present, the study on how to improve foxtail millet ability to resist waterlogging stress is still unclear. However, the growth of foxtail millet is seriously affected by waterlogging stress, so there is an urgent need for new methods and ideas on how to improve the tolerance of millet to waterlogging stress.
The waterlogging stress, an abiotic stress, that can seriously damage crop growth and quality, causes hypoxia in the root zone. Long-term hypoxia in plants affects the transmission of electron chains, and the lack of electron acceptors leads to the saturation of redox chains, the accumulation of NAD (P)H, the reduction of ATP synthesis and the decline of plant growth potential energy. The shortage of ATP causes saturation of the electron transfer chain, which results in the formation of excessive reactive oxygen species (ROS) [8]. The imbalance of ROS causes the peroxidation of lipids and disordered plants physiological processes [9]. The internal alteration and external osmotic potential results in the closure of stomata in plant cells, which ultimately impairs root conductivity and decreases the absorption and utilization of nutrients. Plants primarily regenerate energy through anaerobic fermentation under waterlogging stress, which produces lactic acid and causes a decrease in cytoplasmic pH. Eventually,this leads to plant cell apoptosis and cytoplasm acidification [10]. Waterlogging stress also leads to the disintegration of chloroplasts, mitochondria, and nuclei in plants [11].
Transcription factor (TFs) regulates downstream genes expression through sequence-specific binding with cis-acting elements in the target genes promoter, which plays an important role in plants’ response to abiotic stresses and plant growth, including plant resistance to waterlogging stress [12]. The ERF TF is AP2/ERF superfamily. The AP2/ERF family of TFs has been identified in many plants, including 147 family members in Arabidopsis thaliana [13], 167 family members in corn [14], and 164 family members in rice [15]. Studies have shown that all AP2/ERF members contain an AP2 conserved domain, which is consisted of three β-folds and an α-helix containing 60 amino acids [16]. According to the structure or number of domains containing AP2, the AP2/ERF family can be further divide into five subfamilies: AP2, ERF, DREB, RAV and SOLOIST [17]. Among them, the AP2 subfamily plays a role in plant growth, development, and cell differentiation [18]. The DREB and ERF subfamilies are primarily involved in plant defense against abiotic stresses, such as low temperature [19], drought [20], and waterlogging [21]. ERF family is a unique transcription factor family in plants, which participates in ethylene biosynthesis as an ethylene-responsive factor. The ERF regulates the expression of downstream stress-tolerant genes through the cis-acting ethylene response element (ERE), which is called the GCC box [22]. It was found that the JERF1 in tobacco can specifically bind to the DRE cis-acting element in the ABA synthesis gene NtSDR, which led to the increase of ABA content [23]. The overexpression of ERF1 in the ERF subfamily of tomato (Solanum lycopersicum) reduces the damage to its growth that can occur from drought and salt stress [24]. The soybean (Glycine max) GmERF3 responds to drought stress, salt stress, ethylene synthesis, and soybean mosaic virus. The RAV subfamily is involved in the plant responses to abiotic stresses and defends against biotic stresses [25]. Currently, there has been relatively little research on the SOLOIST subfamily, which primarily positively regulates the biosynthesis of salicylic acid [26].
As an important plant hormone, ethylene is closely related to the growth and development of plants and participates in plant growth [27]. ET is involved in ethylene synthesis and is a key signal transduction factor. ET is affected by many signal transduction components, including ERF TF. SAM synthase, ACC synthase (ACS) and ACC oxidase (ACO) play an important role in ethylene synthesis [28,29]. Among them, ACS and ACO are two rate-limiting enzymes that catalyze ethylene synthesis. The ethylene precursor ACC produced in the roots of plants is transported to the stems of plants through the xylem of the roots, and further plays a role. Then it oxidizes to ethylene, thus regulating the waterlogging tolerance of plants [30]. Currently, four members of ethylene response factor (ERF) have been reported in Arabidopsis thaliana, namely AtHER1, AtHER2, AtRAP2.2, and AtRAP2.12 [31–34], which are involved in the regulation of plant hypoxia tolerance. The finding showed that the ethylene receptor ADH1 gene was activated and expressed by the combination of RAP2-12 protein and hypoxia response gene regulatory elements under waterlogging stress [35]. AtRAP2.2 can resist hypoxia stress by regulating genes expression related to hypoxia stress, foe example ADH1 and PDC1, and genes related to ethylene synthesis [36]. In the study of rice, ET induces programmed cell death of epidermal cells in waterlogged areas promoting adventitious root growth [37].
ERF TFs are involved in many abiotic stresses, such as drought, high salt and low temperature. However, the ERF gene effect on the waterlogging stress of millet is still unclear. Therefore, we obtained four genes (SiERF1A-C and SiRAP2-12) related to waterlogging tolerance through gene phylogenetic tree analyses. SiRAP2-12 was a key gene for foxtail millet to resist waterlogging stress. When SiRAP2-12 of foxtail millet was silenced by virus-induced gene sequencing (VIGS), there was decrease significantly in transcription level, defense enzymes, and RWC, however, the REC was increased. Our findings also revealed that the content of ET, a waterlogging-related hormone, and the SiAOC1 expression level, a key gene in ET synthesis, decreased significantly after VIGS silencing SiRAP2-12. Above all, our studies showed that SiRAP2-12 in foxtail millet is a positive regulator in its defense system resistance waterlogging stress.
2.1 Plant Materials and Treatments
Foxtail millet ‘Jigu 168’ was provided by the Hebei Academy of Agriculture and Forestry Sciences, Hebei Province, China (114°29′E, 38°03′N). ‘Jigu 168’ is a new variety with high quality, early maturity, high yield, and no waterlogging tolerance. Seeds with uniform sizes were selected and germinated in a dark incubator at 28°C. All millet plants were planted in a 4-meter-long greenhouse at intervals of 40 cm. The temperature was 20°C during the day and 15°C at night. There were eight rows in each plot, and the plot area was 0.4 × 8 × 4 for a total of 12.8 m2. A total of 40,000 seedlings were planted when they had produced 4–6 leaves. The treatments included watering by supersaturation 5 cm above the ground for 20 d (Waterlogging, LHJ168) and watering normally watering (Control, ZCJ168). The foxtail millet had grown significantly differently, and both samples were used for the transcriptome analysis after 20 d of treatment. Each sample selected 0.1 g of leaves from the same position, and each treatment had three independent biological replicates.
The Total RNA was extracted from the foxtail millet leaves using RNAprep Pure Plant Kit (Tiangen, Beijing, China) and genomic DNA was removed using DNase (Takara), RNA degradation were monitored on 1% agarose gels [38].
2.3 Library Preparation for Transcriptome Sequencing
The total amount of 1 μg RNA was used as the millet detection sample for each millet treatment. Sequencing libraries were generated using the Hieff NGS Ultima Dual-mode mRNA Library Prep Kit for Illumina (Yeasen Biotechnology (Shanghai) Co., Ltd., China) following the manufacturer’s recommendations. The mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. First strand cDNA was synthesized and second strand cDNA synthesis was subsequently performed. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, the NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. The library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA) [39]. Then 3 μL USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The libraries were sequenced on an Illumina NovaSeq platform to generate 150 bp paired-end reads [40].
2.4 Differentially Expressed Genes (DEGs) Analyses
In order to identify DEGs between different samples, each transcript expression level was calculated according to the Reads per Million Transcripts (TPM) method. RSEM is used to quantify gene abundance. In addition, differential expression analysis was performed using DESeq2 or DESeq2. DEGs with |log2FC| ≥ 2 and FDR ≤ 0.05 (DESeq2) or FDR ≤ 0.001 (DESeq2) are considered to be significantly different genes. GO enrichment analysis was realized by the cluster Profiles software based on Wallenius non-central hyper-geometric distribution [41]. KOBAS database and cluster Profilers software were used to test the statistical enrichment of deferentially expressed genes in the KEGG pathway [42].
2.5 Bioinformatics Analysis of the Transcription Factor Family SiERF
The Pfam and SMART analyzed foxtail millet ERF gene family members, which were named based the Setaria viridis ERF gene family (GCA_005286985.1). The molecular weight (MW) and isoelectric point (pI) of the SiERF protein were predicted by the EXPASY Proteomics Server (http://expasy.org/). The Cell-PLOC (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc/) was used to determine the sub-cellular localization of its members. The sequences of the foxtail millet SiERF proteins were aligned using DNAMAN software to analyze their conserved domains. The conserved motif of the ERF protein in foxtail millet was analyzed by MEME (http://meme.nbcz.net/meme/) [43]. We compared multiple sequences of Arabidopsis and Setaria italica SiERF proteins after downloading the sequences of all Arabidopsis ERF proteins. MEGA 6.0. was used to align the sequences. We conducted phylogenetic analysis by maximum likelihood method, and modified the phylogenetic tree with iTOL tool to help classify SiERF family of foxtail millet [38].
The foxtail millet total RNA was extracted according to the manufacturer’s instructions of the RNA Kit (DNase I) (CW2598S, Jingsu Cowin Biotech Co., Ltd., Beijing, China). The RNA was reverse transcribed into cDNA using reverse transcription kit (R2027, US Everbright Inc., Suzhou, China) [44]. The gene expression was determined by real-time fluorescence quantitative (qRT-PCR) experiment with a 2 × AugeGreen qPCR Master Mix kit (S2008L, US Everbright Inc., Suzhou, China). 18sRNA was used as the reference gene. Three-step procedure was used for qRT-PCR and the thermal cycling program was as follows: 95°C for 10 min, followed by 45 cycles of 95°C for 5 s and 56°C for 1 min. The relative expression of the genes were calculated by the 2−ΔΔCT method with three independent biological replicates [45]. All the primer sequences are shown in Table S1.
2.7 VIGS-Mediated Silencing of SiRAP2-12 in Foxtail Millet
The VIGS method used in this study is similar to the previous study, and it is modified accordingly [25]. The SiRAP2-12 fragments that were used to silence TRV: SiRAP2-12 and TRV: SiPDS were cloned from the foxtail millet genome. The TRV vector silencing vector was constructed by General Biomolecule Corporation. The target PCR product was further connected to TRV2 vector. The construction vector was transferred to LB4404 by Agrobacterium tumefaciens transformation. The target PCR product was further ligated into the TRV2: 00 vector. The construct vectors were transferred into LB4404 by electroporation transformation. The infection solution of Agrobacterium tumefaciens pTRV1 and pTRV2, pTRV2-PDS, and pTRV2-SiRAP2-12 were mixed and used to inoculate foxtail millet plants in the 4–6 leaf stage at a ratio of 1:1. In addition, foxtail millet leaves injected with TRV: 00 were used as control materials. The bacteria were inoculated to leaves by rubbing. The conditions required for injection included approximately 25°C and relative humidity levels of approximately 60%–70%. Approximately 6 mL per plant of the infection solution was inoculated onto the leaves by rubbing [46]. After injection, the plants were protected from light for 24 h and then cultured normally. Finally, the transformed leaf samples were collected after 0, 10, and 25 d and stored at −80°C. The foxtail millet that was successfully silenced was treated by waterlogging, and the foxtail millet samples were taken at different times (0, 2, and 5 d) after waterlogging. The sample was quickly frozen in liquid nitrogen and stored in the refrigerator at −80°C as the index sample for subsequent determination.
2.8 Determination of REC, RWC, and MDA Content
Take fresh millet leaves, rinse them with distilled water, and punch holes about 1 cm in the leaves with a puncher. Each treatment takes 0.1 g of leaves as the sample to be tested. Three biological replicates per treatment. The REC was measured by a DDBJ-350 conductivity meter (Shanghai INESA Scientific Instrument Co., Ltd., Shanghai, China) [47]. The leaves RWC was determined using the saturation weighing method described by Wang et al. [48], reference kit for MDA content determination (Nanjing Jiancheng Bioengineering Co., Ltd., China).
The hydrogen peroxide (H2O2) accumulation was determined by staining with diaminobenzidine (DAB) as previously described [49]. The leaves washed with distilled water were immersed in a DAB staining solution and treated for 2 h under the light. The DAB staining solution was extracted completely. Wash the leaves with distilled water for 4–6 times until the dye solution on the leaves is completely washed. Boil the leaves in 95% alcohol for 30 min until the dyeing leaves is observed. Reference Kit for determination of H2O2 content (Nanjing Jiancheng Bioengineering Co., Ltd., China).
2.10 Na+ and K+ Content Measurements
After drying fresh leaf samples, 0.1 g samples from each treatment for digestion, and the digestion solution was used for determining Na+ and K+ ions content. The contents of Na+ and K+ in the foxtail millet tissues were determined using atomic flame photometry (FP6410, Shanghai Yidian, Shanghai, China) as previously described [50].
2.11 Determination of Antioxidant Enzyme Activities
Antioxidant enzymes, including superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), proline (Pro), and alcohol dehydrogenase (ADH), were determined by Ultraviolet spectrophotometer (BioPhotometer, Eppendorf, Germany) according to the instructions of the corresponding kits (Nanjing Jiancheng Bioengineering Co., Ltd., China) [46].
2.12 Extraction and Determination of Hormone
The waterlogging related hormones, such as ET, ABA, IAA, and GA were detected by Zoonbio Biotechnology. The VIGS-treated foxtail millet samples were analyzed with the ZORBAX SB-C18 column (Agilent Technologies) of the high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) method [46].
The error bars represented the SD based on three independent biological replicates. The relative expression of foxtail millet genes and physiological index in the figure were represented the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01. The data were analyzed using OriginPro 2021 (OriginLab, Northampton, MA, USA).
3.1 Quality Assessment of the Transcriptome Data Sequencing
Quality pre-processing analysis was conducted on the raw data using the Baimaike platform. The reference transcriptomes of six samples were sequenced. The GC content ranged from 55.04% to 57.15%, and the Q30 value was >92.45% (Table S2). This indicated that there was high sequencing quality, and the data could be used for subsequent analyses.
In addition, the reads obtained from more than one transcription group were compared, and the ribosomal RNA was removed. The reads after the filtration of ribosomal RNA were compared with the reference genome (setaria_italica _v2.0.genome.fa) of millet. The results of this comparison are shown in Table S3. The rates of alignment of the filtered sequences and foxtail millet reference genomes in this study >86.24%, and there was no significant difference between different samples, which indicated that the sequencing results of each sample were good. Thus, the data obtained in this study could be used for further analyses.
3.2 Analysis of the Differential Genes after Waterlogging Treatments
The data of transcripts processed by different methods were analyzed, and the consistency of biological repeatability among the ZCJ168 and LHJ168 treatments was verified. The results showed that the same treatment was highly repeatable and could be used for subsequent data analysis (Fig. 1A). The LHJ168 treatment showed upregulation of the 918 genes and downregulation of the 712 genes compared with the ZCJ168 treatment (Fig. 1B). Based on the GO function annotation, it was known that the differential genes mainly participate in the Biological Process, such as metabolic process, cellular process and single-organism process. The cellular component was mainly involved in the cell, cell part, and membrane. The molecular function was mainly involved in catalytic activity and binding (Fig. 1C). In addition, 19,819 genes were primarily divided into five pathways, including Cellular Processing, Environmental Information Processing, Genetic Information Processing, Metabolism, and Organismal System. In particular, 45 DEGs were primarily involved in plant hormone signal transfer, 42 DEGs in Carbon Metabolism, and 43 DEGs in Plant-pathogen Interaction (Fig. 1D).
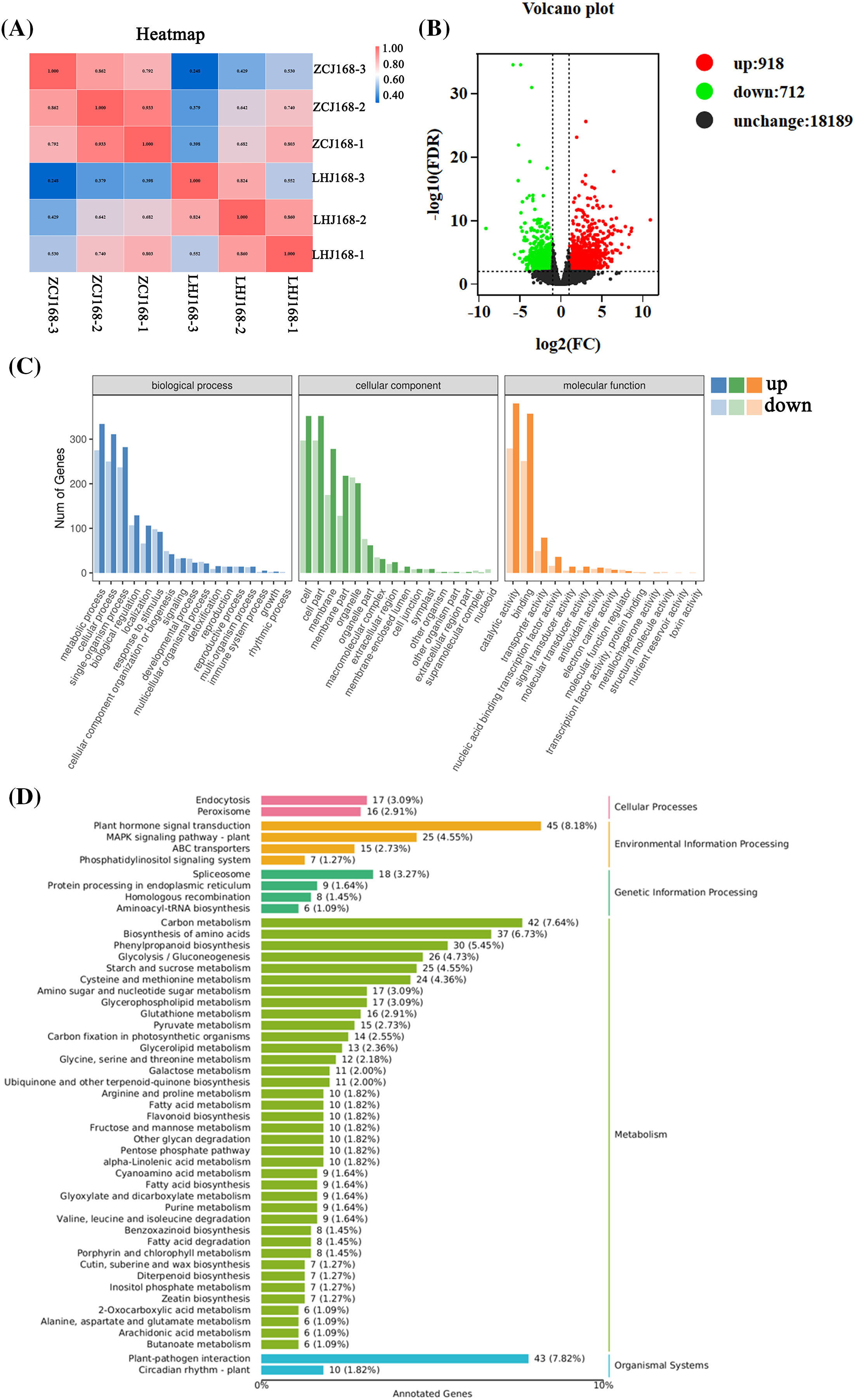
Figure 1: Analysis of differentially expressed genes among different treatments. (A): Correlation heat map analysis between different treatments; (B): Differential gene volcanic map analysis; (C): GO enrichment analysis; (D): KEGG analysis
3.3 The ERF Family in Foxtail Millet Identification
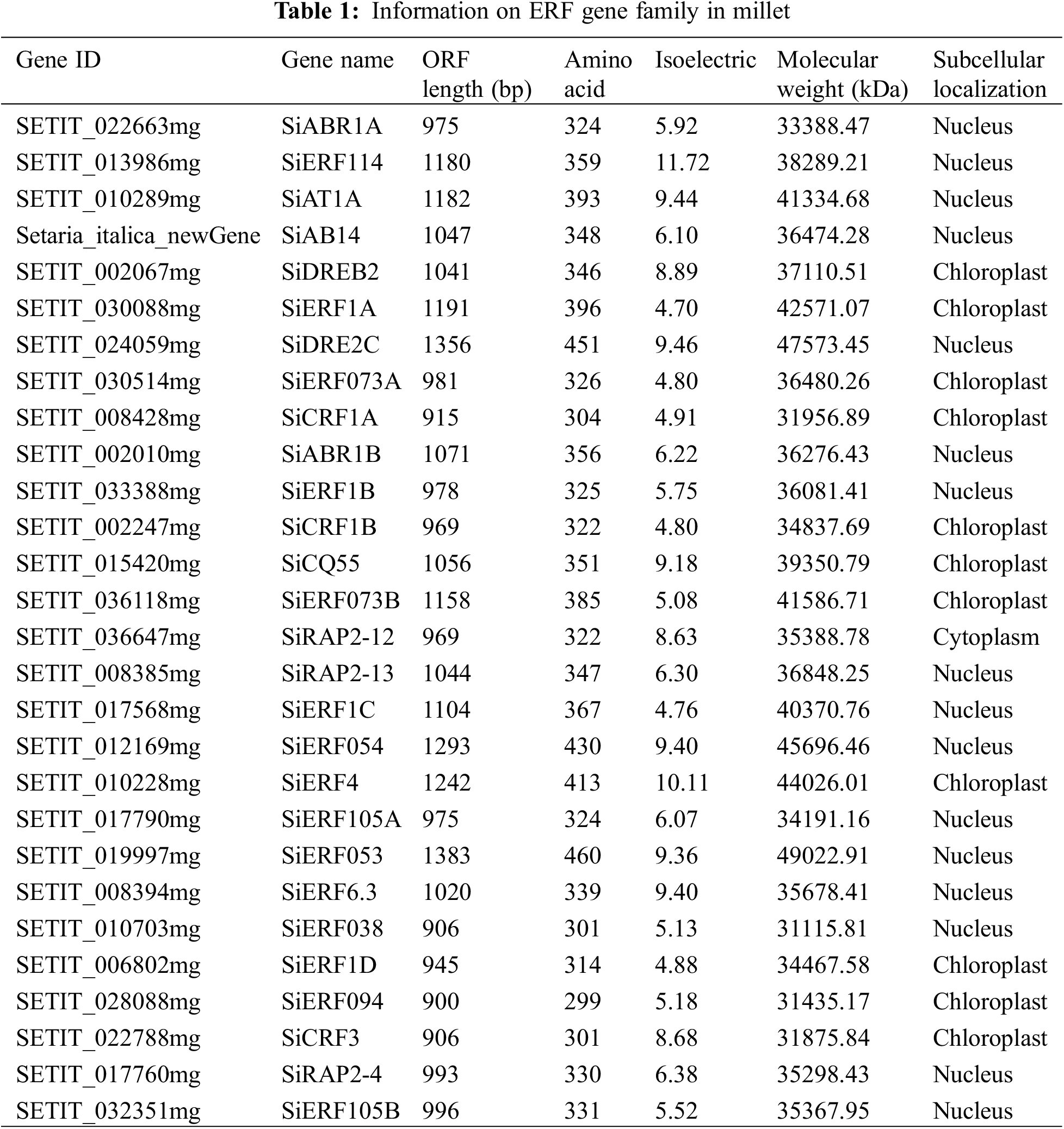
The physicochemical properties of the ERF TFs were analyzed by bioinformatics software, including the length of their amino acids, MW, pI, and cell positioning (Table 1). The amino acid lengths ranged from 299 to 460 aa, with an average length of 352 aa. The shortest protein encoded by SiERF094 was 299~460 aa, and the longest protein encoded by SiERF053 was 455 aa. The pIs were between 4.70 and 11.72, and the MWs were between 31.1 and 49 kDa. Among the 28 ERF proteins screened out, 17 proteins had a pI < 7 and were mostly acidic proteins. The ERF TF was primarily localized to the chloroplast and nucleus (Table 1).
In addition, the conserved motif results showed that there were 11 types of conserved motifs with 11 dissimilar colors. The conserved motifs contained in the family members ranged from two to eight. This indicates that there were functional differentiation among the ERF TFs. SiERF1C contained the most conserved motifs, while SiAB14, SiDREB2, SiERF105B, SiCRF1A, SiERF038, SiERF094, and SiCRF3 contained the least conserved motifs (Fig. 2A). All the members contained moti1 and moti2, and both these motifs together formed the conserved domain of ERF. A multi-sequence alignment showed that all the members of the ERF TFs family had the conserved domain of AP2 (Fig. 2B). They all had three β-folds and a C-terminal α-helix in which the 14th and 19th amino acid residues of the second β-fold determined the types of cis-acting elements, and the C-terminal α-helix consisted of 18 amino acid residues (Fig. 2B).
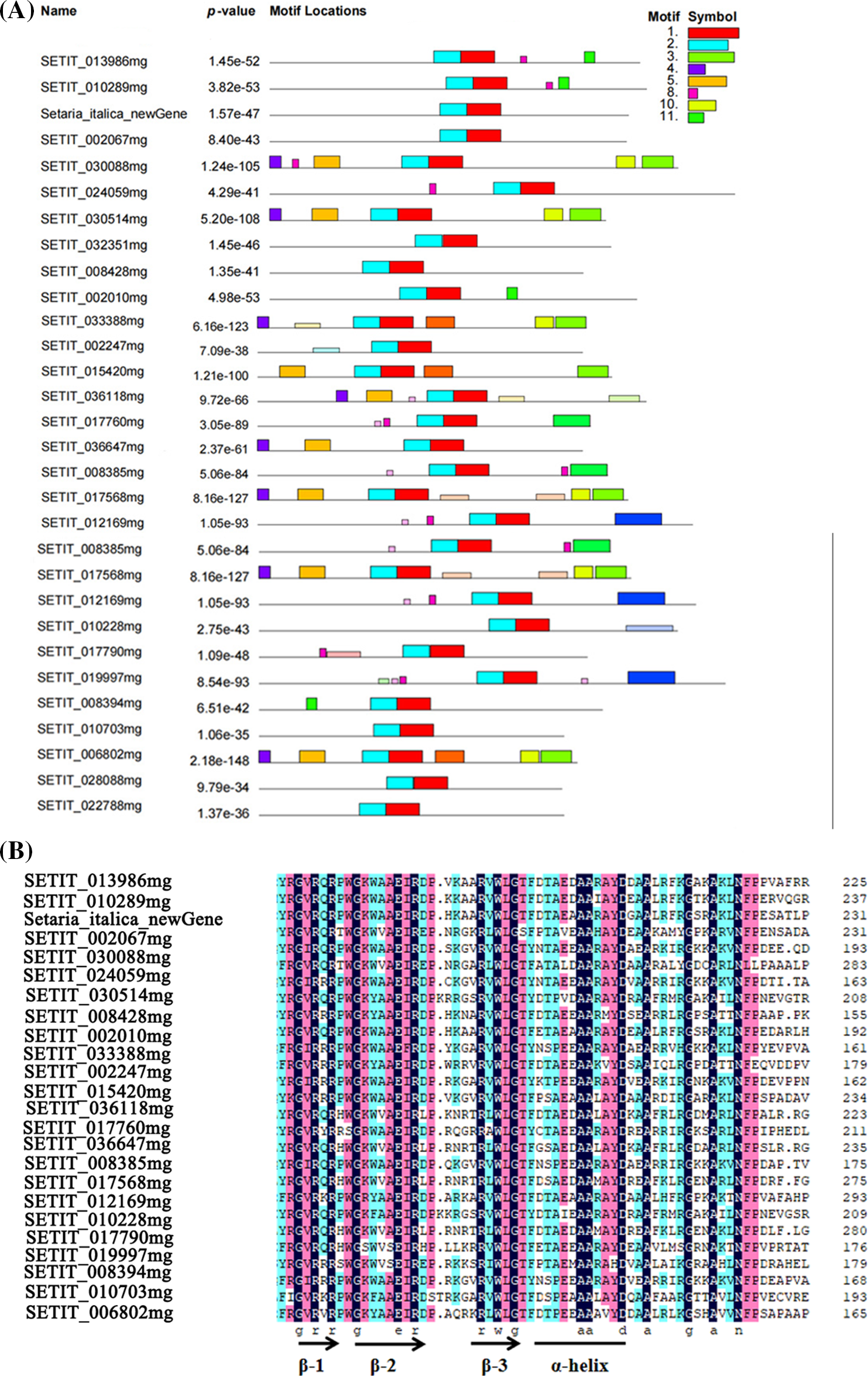
Figure 2: The foxtail millet ERF family conserved domains and motifs analysis. (A) Homology analysis; (B) Motif analysis
3.4 Phylogenetic Analyses of ERF Genes
The phylogenetic tree was furthermore constructed of ERF proteins, including 40 AtERF proteins (from Arabidopsis thaliana) which have been identified and described in detail [13]. This phylogram distinguished 11 groups, namely, groups I to IX. Among them, there was no ERF member distribution in groups II, III, and IV (Fig. 3A).
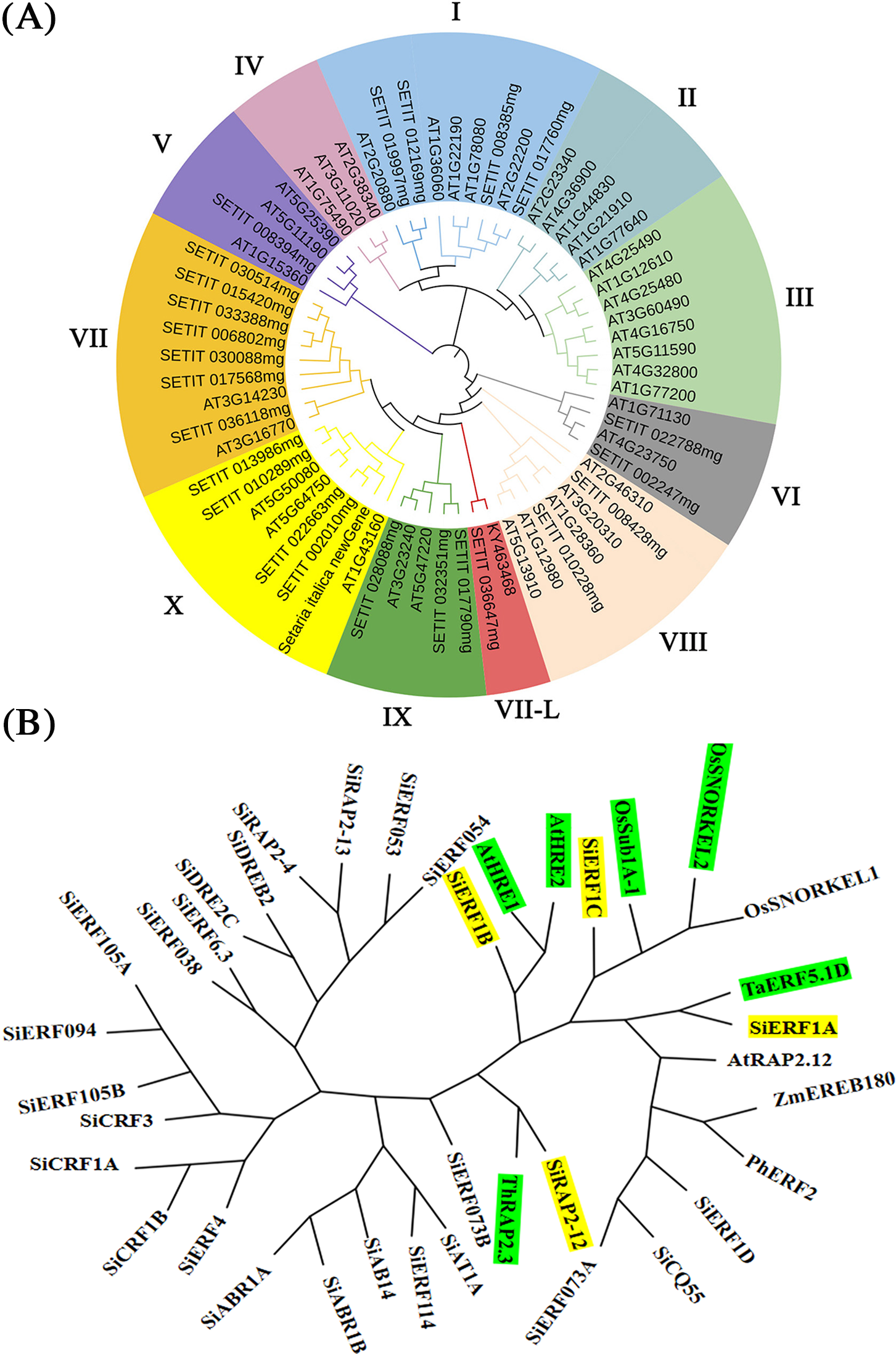
Figure 3: Phylogenetic tree analysis of foxtail millet ERF family. (A): Phylogenetic analysis of foxtail millet ERF family genes. (B): Cluster tree constructed by SiERF of foxtail millet and other plant genes (At: Arabidopsis thaliana; Os: Oryza sativa; Si: Setaria italica; Th: Taxodium hybrid; Ta: Triticum aestivum; Ph: Petunia x hybrida; Zm: Zea mays)
To further screen the key genes involved in waterlogging tolerance, we conducted a clustering analysis on 28 ERF members of foxtail millet and ERF members involved in waterlogging tolerance in other species. SiERF1B was found to be highly homologous to AtHRE1 and AtHRE2. SiERF1C was highly homologous to OsSub1A-1, and OsSNORKE12, SiERF1A, and SiRAP2-12 were highly homologous to TaERF5.1D and ThRAP2.3 (Fig. 3B). Therefore, SiERF1A/B/C and SiRAP2-12 might also be involved in the function of regulating millet to resist waterlogging stress.
3.5 Analysis of the Levels of Expression of the SiERF Genes under Waterlogging Stress
The levels of expression of SiERF1A/C and SiRAP2-12 increased by 35.04%, 13.71%, and 65.72%, respectively, after 20 d of waterlogging stress. Among them, SiRAP2-12 reached the most significant level of expression. In contrast, the expression of SiERF1B decreased by 9.56% compared with the control foxtail millet (Fig. 4). Therefore, SiRAP2-12 was the key gene in the ERF TF of foxtail millet that resisted waterlogging stress.
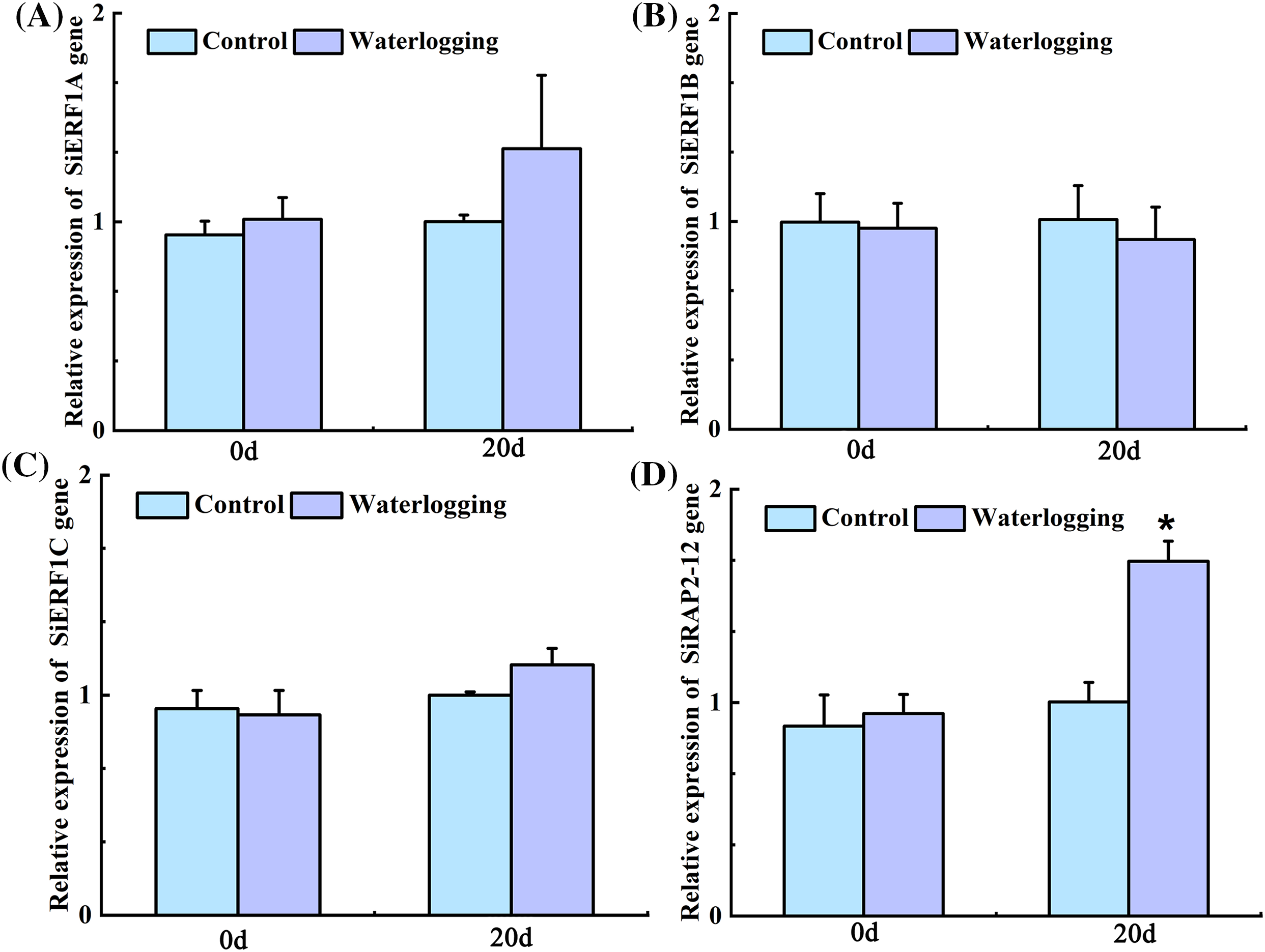
Figure 4: Changes in expression levels of SiERF1A/B/C and SiRAP2-12 genes under waterlogging stress. Values in the figure represent the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01
3.6 Identification of the Silenced SiRAP2-12 in Foxtail Millet
In the blank vector pTRV2, the restriction sites of BamHI and SmaI were located at the 5’ and 3’ ends, respectively. Then restriction sites of SmaI and BamHI were added at the 5’ and 3’ ends to construct the SiRAP2-12 silencing VIGS vector (pTRV2) (Fig. 5C). Further sequencing showed that the silencing gene sequence was successfully linked to TRV2 vector.(Fig. 5A). During the identification of the patterns of enzyme digestion, two enzymes, SmaI and HindIII, were used in the SiRAP2-12 gene silencing vector, and the fragment produced was 2635 bp, which was consistent with the size between SmaI and HindIII in the construction of silencing VIGS vector (Fig. 5B).
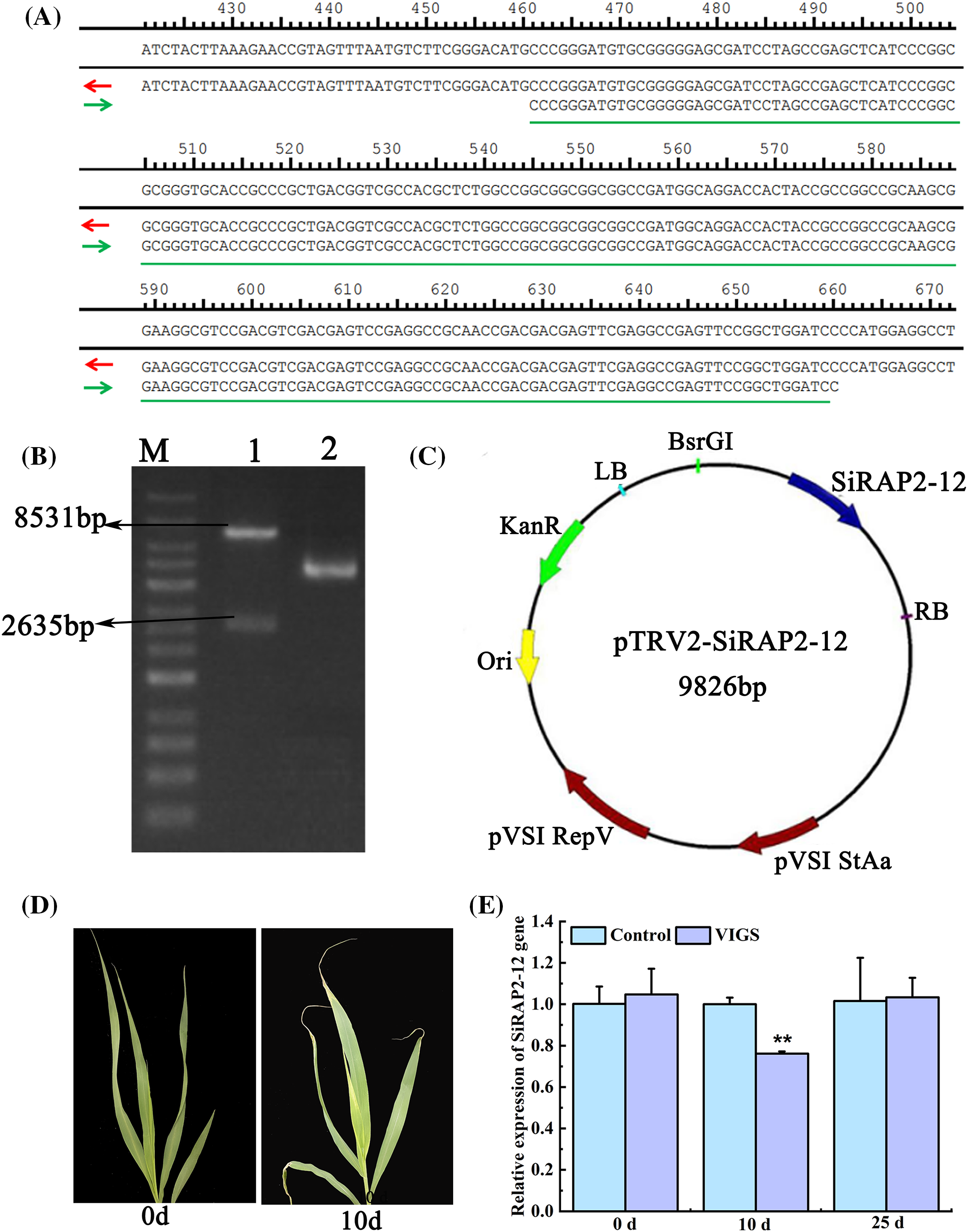
Figure 5: Construction of pTRV2-SiRAP2-12 silencing vector and identification of silencing efficiency. (A): pTRV2-SiRAP2-12 plasmid digested with SmaI-HindIII (M: 1 Kb DNA Marker; 1: SmaI and HindIII digestion products identification; 2: pTRV2 Plasmid); (B): The map of pTRV2-SiRAP2-12 silencing vector; (C): pTRV2-SiRAP2-12 plasmid digested with SmaI-HindIII; (D): Albinism symptom of foxtail millet with SiPDS silenced; (E): Relative expression of SiRAP2-12. Values in the figure represent the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01
We selected SiRAP2-12 for a functional analysis using the VIGS method with phytoene desaturase (SiPDS) which was used as a reporter gene to assess the efficiency of VIGS treatment. The VIGS-treated vector that contained SiRAP2-12 was successfully obtained and identified. The foxtail millet leaves infected with pTRV2-SiPDS showed an albino phenotype after being injected for 10 d. It showed that SiRAP2-12 has been successfully silenced and can be used in subsequent studies. (Fig. 5D). The SiRAP2-12 expression level of the VIGS-treated foxtail millet decreased significantly by 23.88% compared with the control plants after 10 d. The results showed that the SiRAP2-12 gene was successfully silenced (Fig. 5E).
3.7 Silent SiRAP2-12 Enhanced the Sensitivity of Foxtail Millet to Waterlogging
To verify whether SiRAP2-12 was involved in waterlogging stress tolerance of foxtail millet, we identified the waterlogging characteristics of the VIGS-treated foxtail millet to determine the function of SiRAP2-12. The findings showed that the control foxtail millet grew better than the VIGS-treated foxtail millet under waterlogging stress. the leaves were also more intensely yellow after 5 d waterlogging stress (Fig. 6A). We found that the REC of VIGS-treated plants increased significantly by 15.21% compared with that of the control foxtail millet (Fig. 6B), while the height of VIGS-treated millet decreased significantly by 25.12% (Fig. 6C). Above all, the waterlogging tolerance of foxtail millet was significantly reduced, and its growth was more inhibited after silencing SiRAP2-12.
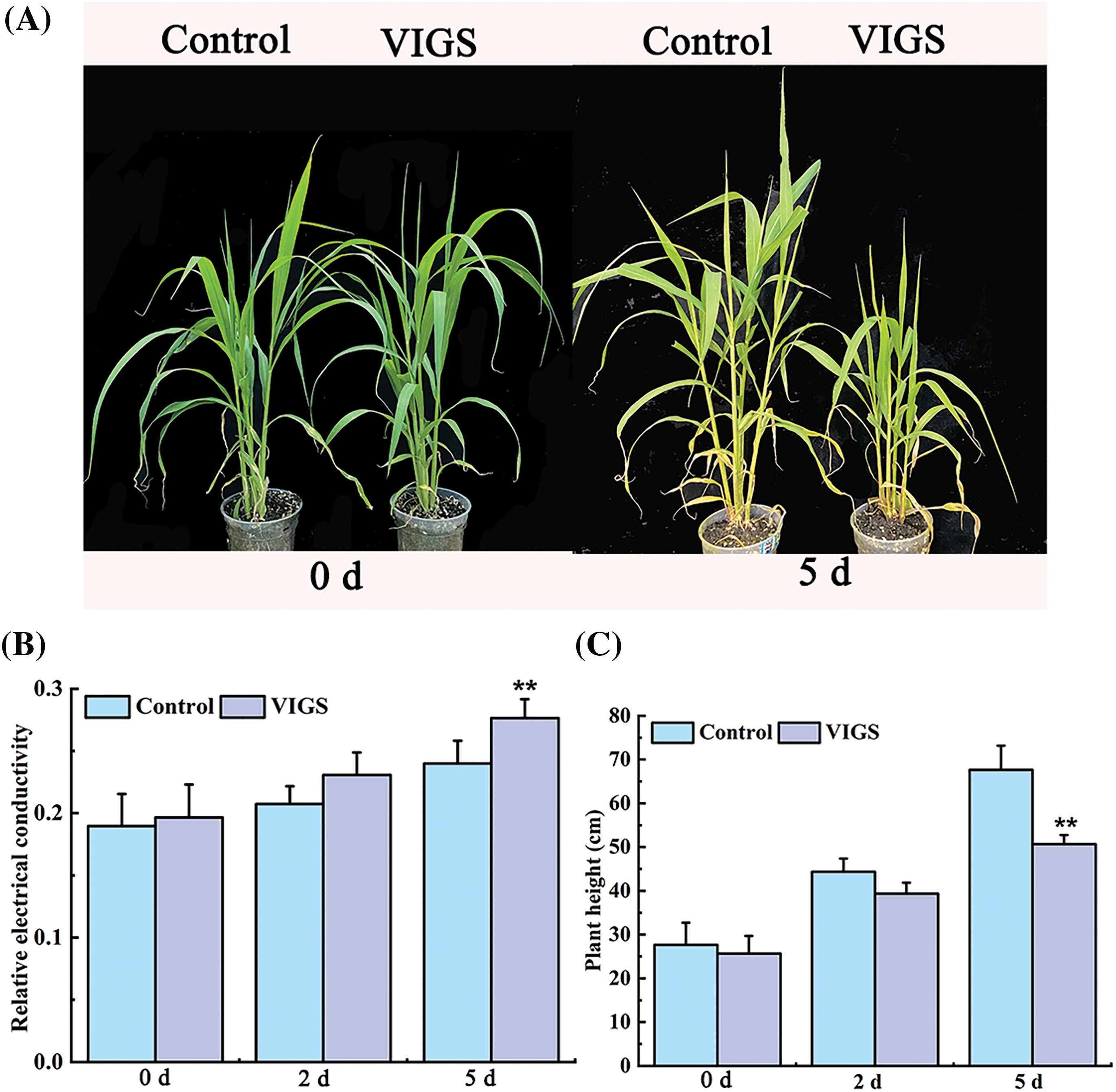
Figure 6: Phenotypic analysis of foxtail millet by silencing SiRAP2-12 gene; (A): Phenotypic changes of millet under VIGS treatment.; (B): Relative conductivity content (REC); (C): Plant height; Values in the figure represent the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01
3.8 Silencing SiRAP2-12 Affected the Antioxidant Enzyme Activity of Foxtail Millet
In order to further explore the effect of silencing SiRAP2-12 on the activities of defense enzymes in millet under saline-alkali stress, we measured the activities of defense enzymes and the content of compounds related to waterlogging metabolism in the silent foxtail millet. We found that the content of MDA in the VIGS-treated foxtail millet increased significantly by 1.03-fold compared with the control plants after 5 d waterlogging stress, while the activities of POD and SOD and the content of Pro decreased significantly by 29.52%, 78.61%, and 19.41%, respectively (Figs. 7B–7E). Alcohol dehydrogenase (ADH) is the key enzyme of anaerobic respiration. The ADH activity of the VIGS-treated millet decreased significantly by 54.77% compared with the control foxtail millet after waterlogging stress (Fig. 7A). This finding showed that the activities of antioxidant enzymes and key enzymes of anaerobic respiration under waterlogging stress will be reduced after SiRAP2-12 has been silenced, and the tolerance of plants to waterlogging stress would finally be reduced.
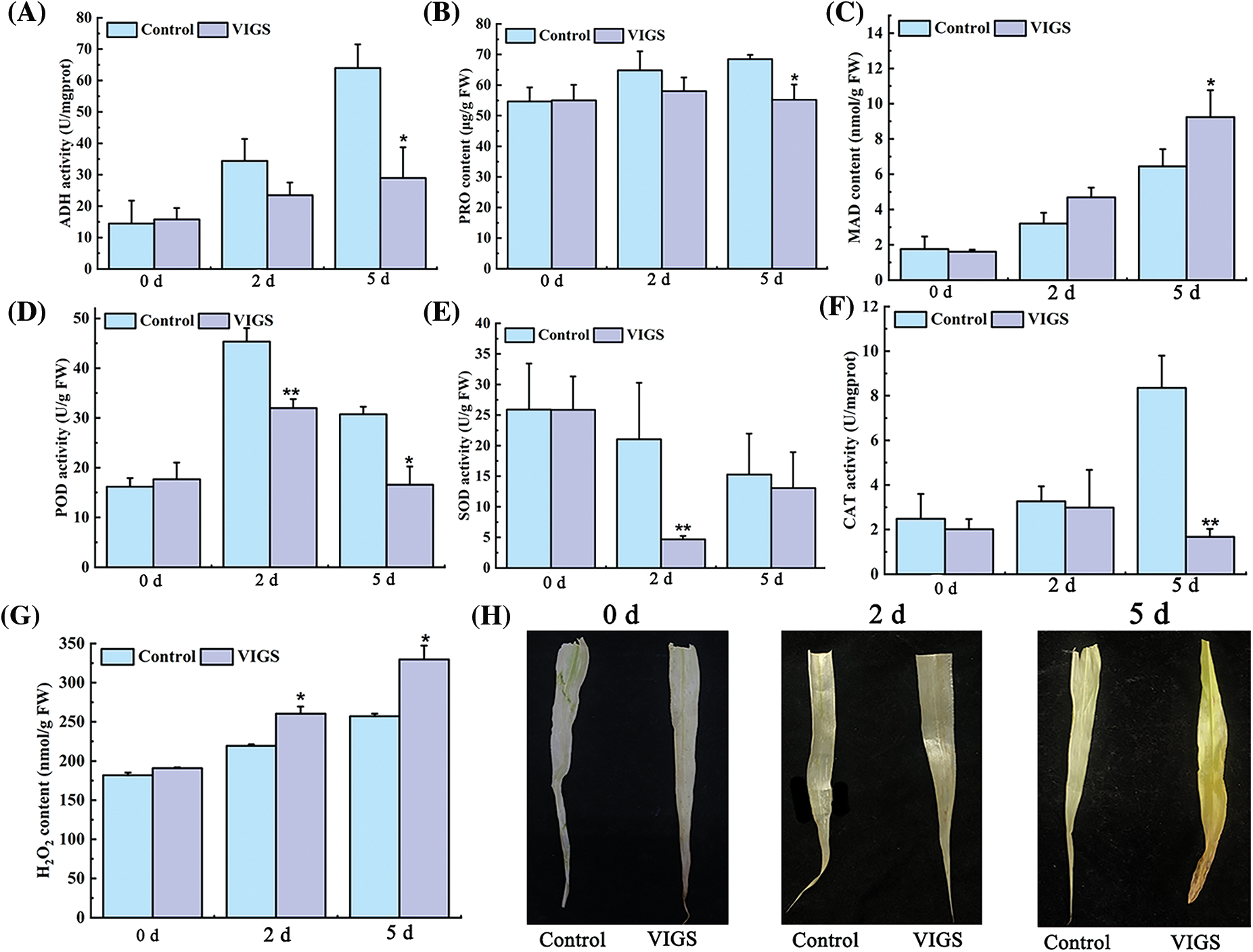
Figure 7: Effect of VIGS-treated on antioxidant enzyme activity of foxtail millet. (A): ADH activity. (B): PRO content. (C): MDA content. (D): POD activity. (E): SOD activity. (F): CAT activity. (G): H2O2 content. (H): DAB staining. Values in the figure represent the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01
CAT could remove the excessive amounts of hydrogen peroxide (H2O2) produced by waterlogging stress in foxtail millet. The findings showed that CAT activity in the VIGS-treated foxtail millet decreased by 79.95%, while the content of H2O2 increased by 36.09% compared with the control foxtail millet (Figs. 7F, 7G). H2O2 reacted with DAB to produce a brown product. In addition, with the extension of time of waterlogging stress, the leaves gradually turned deeper brown. This phenomenon indicated that the H2O2 content of VIGS-treated foxtail millet was higher than that of the control foxtail millet (Fig. 7H).
3.9 Silencing SiRAP2-12 Affects Na+ and K+ Content in Foxtail Millet
The imbalance between the release of Na+ release and the absorption of K+ led to ion poisoning of the plants under waterlogging stress. K+ content of the VIGS-treated foxtail millet leaves decreased significantly by 24.04% compared with the control foxtail millet under waterlogging stress, while Na+ content of the VIGS-treated foxtail millet leaves increased by 1.19-fold compared with the control foxtail millet (Figs. 8A, 8B). The Na+ content of foxtail millet gradually increased, while that of K+ gradually decreased. This phenomenon resulted in a decrease in the K+/Na+ ratio. The K+/Na+ ratio of the VIGS-treated foxtail millet decreased significantly by 65.96% compared with control foxtail millet (Fig. 8C).
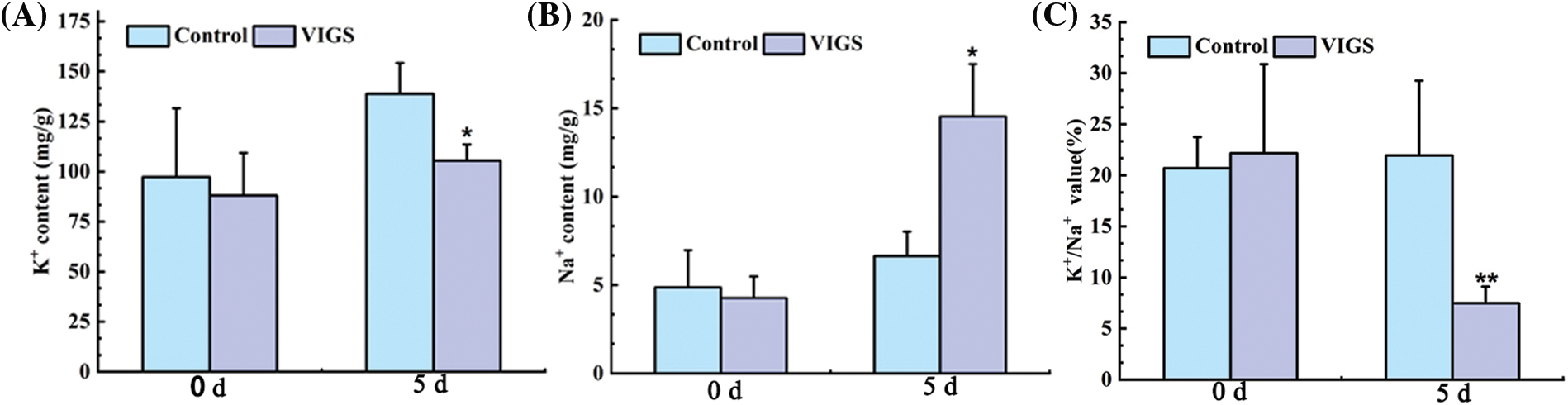
Figure 8: Effect of VIGS-treated on K+ and Na+ content of foxtail millet; (A): K+ content; (B): Na+ content; (C): K+/Na+ value. Values in the figure represent the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01
3.10 Silencing SiRAP2-12 Affected Hormone Content of Foxtail Millet
In order to further verify whether SiRAP2-12 gene can improve the ability of millet to resist waterlogging stress by regulating hormone synthesis, we measured the waterlogging-resistant hormone content of the VIGS-treated foxtail millet. The IAA, GA, ET, and ABA contents of VIGS-treated foxtail millet were lower than those in the control plant (Fig. 9). Especially, the ET content was the most significantly decreased by 38.46% (Fig. 9B). Silencing SiRAP2-12 affected the synthesis of ET, and it was further speculated that this gene was involved in waterlogging tolerance of foxtail millet through regulating ET signaling pathway.
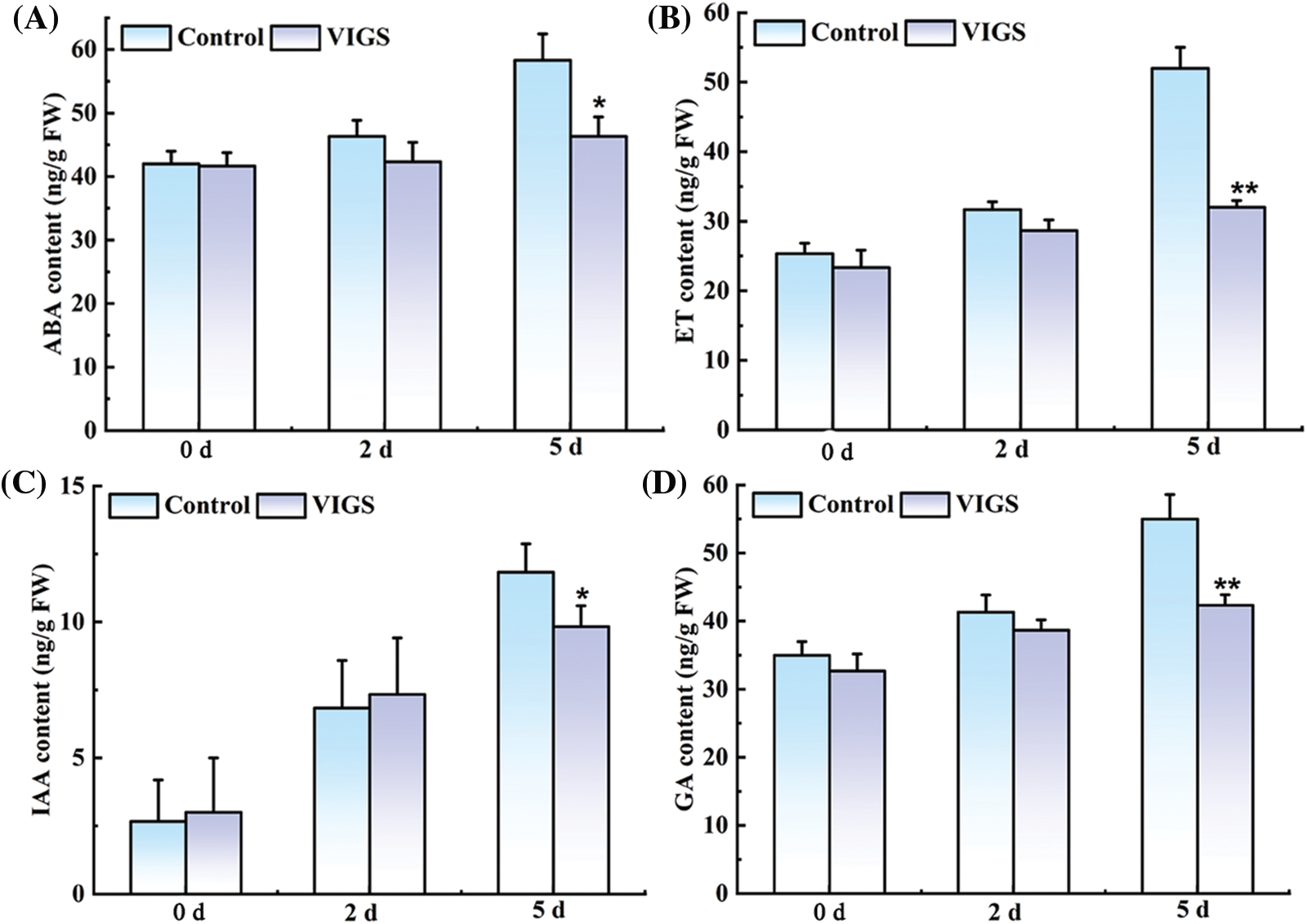
Figure 9: Effect of VIGS-treated on the contents of hormones related to waterlogging tolerance in foxtail millet. (A): ABA content; (B): ET content; (C): IAA content; (D): GA content. Values in the figure represent the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01
3.11 Silencing SiRAP2-12 Regulated Key Genes of the ET Biosynthetic Pathway
Given that SiRAP2-12 influenced ET hormone content in VIGS-treated foxtail millet, the expression of ET biosynthesis-related genes (SiACS1, SiAOC1, SIAOC2, and SiAOC3) was further examined via qRT-PCR (Fig. 10). However, only the SiAOC1 expression was most significantly decreased by 29.91% in VIGS-treated millet than that of control (Fig. 10A). Taken together, we hypothesized that SiRAP2-12 may be involved in regulating the expression of SiAOC1.
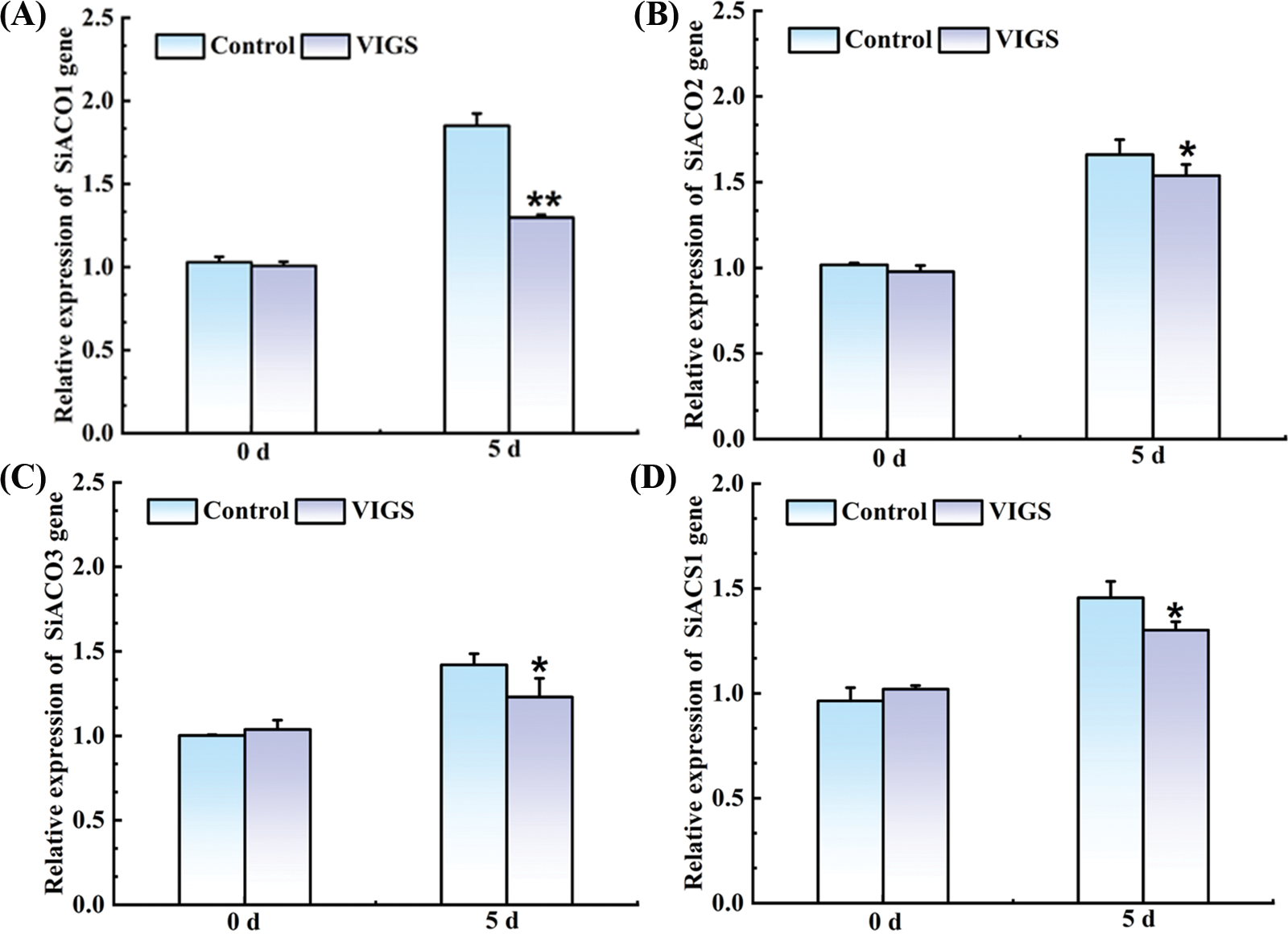
Figure 10: Expression of SiAOC1 (A), SIAOC2 (B), SiAOC3 (C) and SiACS1 (D), associated with the ET biosynthetic pathways. Values in the figure represent the means ± SD of three replicates. Student’s t-test: *p < 0.05 and **p < 0.01
Foxtail millet, as a model plant with a short growth cycle and small genome, has unique advantages for researching the mechanism of stress regulation [51]. However, the growth of foxtail millet and the formation of yield have been seriously affected owing to the influence of extreme weather, such as waterlogging stress. It was known that the ERF TF was involved in regulating plant resistance to abiotic stress. The phylogenetic tree was used to classify members of the foxtail millet ERF TF, and a systematic clustering tree was further constructed [10]. Four differentially expressed genes were isolated from the ERF family under waterlogging stress. Through sequence comparison, it was found that SiRAP2-12, the key gene to resist waterlogging stress in foxtail millet, was similar to ThERF2.3 and ThERF15. The expression of SiRAP2-12, as a positive regulatory factor, was found to be involved in influencing the waterlogging stress of foxtail millet by regulating defense enzymes, ion balance, hormone content, and membrane permeability.
Studies have shown that the ERFVII genes of the AP2/ERF family played a key role in response to flooding stress in Arabidopsis thaliana, rice, corn, and other plants and regulate downstream genes [21,52,53]. Among them, RAP2.2 (At3g14230) of Arabidopsis thaliana ERF subfamily is highly expressed in plants under hypoxia [54]. Up to now, there have been many studies on the regulation of ERF family on abiotic stress in plants, but the effect of waterlogging tolerance in millet remains largely unknown. In our study, the SiRAP2-12 expression increased gradually after waterlogging stress. We further hypothesized that the SiRAP2-12 gene was involved in the process of tolerance to waterlogging in foxtail millet.
When plants suffer from waterlogging stress, the root growth potential energy is blocked and the water absorption capacity is reduced. and it leads to the closure of leaf stomata. This inhibits plant photosynthesis, which leads to the loss of green leaves and a reduction in growth [55]. In this study, the REC of VIGS-treated foxtail millet was significantly higher than control foxtail millet; the leaves were yellow, and the VIGS-treated foxtail millet plants were significantly shorter compared with the control foxtail millet after 5 d of waterlogging. The exchange of CO2 and O2 in plants will be affected after waterlogging stress. The lack of CO2 would result in plants that produce excessive amounts of the strong oxidant superoxide (O2·−), which causes the cell membrane to be destroyed and the osmotic potential to be unbalanced [56]. O2·− and H2O2 are important ROS because of their strong ability to oxidize other compounds, and MDA content can reflect the damage of excessive ROS in plants [57]. This damage manifests as lipid peroxidation. In this study, the VIGS-treated foxtail millet produced more H2O2 and MDA after waterlogging stress. Yordanova et al. [58] showed that plants with higher activities of antioxidant enzymes were more tolerant to waterlogging. This was because POD, CAT, and SOD could effectively remove the H2O2 from plants and reduce the damage of excessive ROS to plants. This is similar to our research. Although the activities of SOD, POD, and CAT in the VIGS-treated millet decreased significantly compared with the control millet, the content of Pro was also significantly lower. Furthermore, waterlogging stress primarily causes root anoxia, and the roots initiate anaerobic respiration to maintain their normal metabolism [59]. In addition, our findings found that the ADH activity of the VIGS-treated foxtail millet was significantly lower than that of the control foxtail millet. ADH, an important anaerobic respiratory enzyme, produces more ethanol and ATP, which is an important regulatory way to improve the tolerance to waterlogging. Therefore, we speculated that it may be due to the regulation of antioxidant enzyme activity by the SiRAP2-12 gene in foxtail millet, which enhanced the ability of foxtail millet to resist waterlogging stress.
Waterlogging stress can damage the root system. Root damage causes an imbalance in the release of Na+ and absorption of K+ by plants, which leads to ion toxicity and disrupts the osmotic balance of the cells [60]. We further analyzed the changes in Na+ and K+ content in foxtail millet under waterlogging stress. This study showed that the content of K+ of the VIGS-treated foxtail millet decreased; the Na+ content increased, and the ratio of K+/Na+ decreased compared with the control foxtail millet after waterlogging stress, which caused the leaves of millet to turn yellow and dry. This treatment finally inhibited the growth of millet. The absorption and secretion of metal ions by roots were inhibited owing to the SiRAP2-12 silencing in the VIGS-treated foxtail millet.
Hormones (ET, IAA, ABA, and GA) are associated with ROS signal interaction, and coordinated regulation of plant stress response to adversity [61]. Exogenous application of GA can reduce MDA content in roots and leaves, thus improving the tolerance to waterlogging stress. ET and IAA regulate CsRBOHB gene expression of cucumber after waterlogging stress, which promotes root generation by inducing ROS signal response [62]. Methylthioadenosine and 1-aminocyclopropane-1-carboxylic acid (ACC) are oxidized to hydrocyanic acid, carbon dioxide (CO2), and ethylene by ACC oxidase (ACO) as precursors of ethylene, and ACO participates in plant resistance to adversity stress [63]. The results showed that ACC oxidase was overexpressed in Arabidopsis thaliana, and the ability to resist waterlogging was significantly improved compared with control plants [64]. This is similar to our results, the ET content decreased significantly in VIGS-treated millet and we also determined the ET hormone-related genes expression (SiAOC1, SiAOC2, SiAOC3, and SiACS1) and the level of SiAOC1 gene expression in silencing plants was most significantly decreased. Moreover, studies have shown that MdERF2 in the apple ERF family can regulate the transcription of ACS, a key gene of the ethylene synthesis pathway, and the cis-acting elements of DRE or GCC-Box on ACO promoter, thus regulating ethylene synthesis.These biological processes may be regulated by ERF gene family [65]. Given this, SiRAP2-12, a member of the ERF subfamily of millet, contains GCC-Box cis-elements, and according to our results, it was found that the expression trends of SiRAP2-12 and SiACO1 genes are consistent. Therefore, we hypothesized that the SiRAP2-12 of the foxtail millet ERF TF might interact with the SiAOC1 gene, a key ethylene synthesis gene, by regulating endogenous hormone content, antioxidant system activity, and ion balance.
In my study, we identified 28 ERF family of the foxtail millet genome and performed a comprehensive bioinformatic analysis for them. It was found that the expression of SiRAP2-12 in foxtail millet was significantly increased after waterlogging stress. The function of SiRAP2-12 was further verified by VIGS silencing, and it was found that SiRAP2-12 could participate in waterlogging resistance of foxtail millet by improving antioxidant enzyme activity, hormone content, and regulating osmotic substance content. In addition, the expression of key genes for ET synthesis also decreased significantly, we speculate that the SiRAP2-12 gene may be involved in the regulation of the SiAOC gene. These results indicated that SiRAP2-12 in the millet ERF TF family might be a positive regulator of millet waterlogging tolerance. This was the first evidence to demonstrate the role of the SiRAP2-12 gene in the foxtail millet ERF TF under waterlogging stress.
Acknowledgement: I am grateful to Ziyi Wang for revising the manuscript.
Funding Statement: This research was supported by the China Agricultural Research System (CARS-06-14.5-A23), HAAFS Basic Science and Technology Contract Project (Grant No. HBNKY-BGZ-02), and Technical System of Foxtail Millet Industry in Hebei Province.
Author Contributions: Xueyan Xia and Xiaohong Fu designed the research and performed the experiments. Xueyan Xia, Xiaohong Fu, Yu Zhao, Jihan Cui, Nuoya Xiao, and Jingxin Wang analyzed the results. Yiwei Lu, Meihong Huang, Cheng Chu, and Mengxin Yang prepared the draft manuscript. Jianfeng Liu and Shunguo Li revised the paper. Xueyan Xia and Xiaohong Fu contributed equally to this research. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.048273.
References
1. Shi ZG, Cheng RH, Liu ZL, Xia XY. Development of foxtail millet cultivar Jigu 22. China: J Hebei Agric Sci. 2007;11(1):95–6. [Google Scholar]
2. Huo DY, Zheng WJ, Li PS, Xu ZS, Zhou YB, Chen M, et al. Identification, classification, and drought response of F-box gene family in foxtail millet. Acta Agronomica Sin. 2004;40(9):1585–94. [Google Scholar]
3. Zhang C, Zhang H, Li JX. Advances of millet research on nutrition and application. China J Chin Cereals Oils Assoc. 2007;22(1):51–5. [Google Scholar]
4. Yan SA, Qian AQ, Song YK, Lin Q, Lin XX. Content of amino acid in cereal protein and its nutritive evaluation. China: Chin Agric Sci Bull. 2009;25(18):113–7. [Google Scholar]
5. Kulkarni SS, Chavan PD. Study of some aspects of anaerobic metabolism in roots of finger millet and rice plants subjected to waterlogging stress. Phyton. 2013;9(2):80–5. [Google Scholar]
6. Liu WH, Lu B, Zhou NJ. Effects of different ecological factors on millet protein study on the influence of fat content. J Shanxi Agric Univ. 1995;(3):244–7 (In Chinese). [Google Scholar]
7. Diao XM. Progresses in stress tolerance and field cultivation studies of orphan cereals in China. Scientia Agricultura Sinica. 2019;52:3943–49 (In Chinese). [Google Scholar]
8. Ashraf M, Rahmatullah, Maqsood MA, Kanwal S, Tahir MA, Ali L. Growth responses of wheat cultivars to rock phosphate in hydroponics. Pedosphere. 2009;19(3):398–402. [Google Scholar]
9. Pucciariello C, Parlanti S, Banti V, Novi G, Perata P. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 2012;159(1):184–96. [Google Scholar] [PubMed]
10. Sweetlove LJ, Dunford R, Ratcliffe RG, Kruger NJ. Lactate metabolism in potato tubers deficient in lactate dehydrogenase activity. Plant Cell Environ. 2000;23(8):873–81. [Google Scholar]
11. Wei HP, Li RQ, Wang JB. Ultrastructural changes in leaf cells of submerged maize. J Integr Plant Biol. 2000;42(8):811–7 (In Chinese). [Google Scholar]
12. Hussain SS, Kayani MA, Amjad M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol Prog. 2011;27(2):297–306. [Google Scholar] [PubMed]
13. Nakano T, Suzuki K, Fujmurn T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140(2):411. [Google Scholar] [PubMed]
14. Li H, Xiao QL, Zhang CX, Du J, Li X, Huang HH, et al. Identification and characterization of transcription factor ZmERFB94 involved in starch synthesis in maize. J Plant Physiol. 2017;216:11–6. [Google Scholar] [PubMed]
15. Wang FJ, Wang CL, Liu PQ, Lei CL, Hao W, Gao Y, et al. Enhanced riceblast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11(4):e0154027. [Google Scholar] [PubMed]
16. Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17(18):5484–96. [Google Scholar] [PubMed]
17. Gou YL, Zhang L, Guo H, Ma H, Bao A. Research progress on the AP2/ERF transcription factor in plants. Pratacultural Science; 2020;37(6):1150–9. [Google Scholar]
18. Jofuku KD, Boer BG, Montagu MV, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6(9):1211–25. [Google Scholar] [PubMed]
19. Zhang ZJ, Huang RF. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol. 2010;73(3):241–9. [Google Scholar] [PubMed]
20. Yu WJ, Wang L, Zhou BR, Wang SJ, Li RH, Jiang TB. Over-expression of poplar transcription factor ERF76 gene confers salt tolerance in transgenic tobacco. J Plant Physiol. 2016;198(2):23–31. [Google Scholar]
21. Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460(7258):1026–30. [Google Scholar] [PubMed]
22. Xu ZS, Xia LQ, Chen M, Chen XG, Zhang RY, Li LC, et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol. 2007;65(6):719–32. [Google Scholar] [PubMed]
23. Wu LJ, Chen XL, Ren HY, Zhang ZJ, Zhang HW, Wang JY, et al. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta. 2007;226(4):815–25. [Google Scholar] [PubMed]
24. Zhang H, Liu WW, Wan L. Functional analyses of ethylene response factor JERF3 with the aim of improving tolerance to drought and osmotic stress in transgenic rice. Transgenic Res. 2010;19(5):809–18. [Google Scholar] [PubMed]
25. Liu JL, Deng ZW, Liang CL, Sun HW, Li DL, Song JK, et al. Genome-wide analysis of RAV transcription factors and functional characterization of anthocyanin-biosynthesis-related RAV genes in pear. Int J Mol Sci. 2010;22(11):5567. [Google Scholar]
26. Wang HB, Gong M, Guo JY, Xin H, Tang LZ, Liu C, et al. Molecular cloning and prokaryotic expression of orphan gene Soloist of AP2/ERF gene family in Jatropha curcas. Sci Silv Sin. 2018;54(9):60–9. [Google Scholar]
27. Bakshi A, Shemansky JM, Chang C, Binder BM. History of research on the plant hormone ethylene. J Plant Growth Regul. 2015;34(4):809–27. [Google Scholar]
28. Pattyn J, Vaughan-Hirsch J, Poel B. The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 2021;229(2):770–82. [Google Scholar] [PubMed]
29. Park C, Han YL, Yoon GM. The regulation of ACC synthase protein turnover: a rapid route for modulating plant development and stress responses. Curr Opin Plant Biol. 2021;63:102046. [Google Scholar] [PubMed]
30. Sasidharan R, Voesenek LA. Ethylene-mediated acclimations to flooding stress. Plant Physiol. 2015;169(1):3–12. [Google Scholar] [PubMed]
31. Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, et al. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010;153(2):757–72. [Google Scholar] [PubMed]
32. Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011;156(1):202–12. [Google Scholar] [PubMed]
33. Gasch P, Fundinger M, Müller JT, Lee T, Bailey-Serres J, Mustroph A. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell. 2016;28(1):160–80. [Google Scholar] [PubMed]
34. Paul MV, Iyer S, Amerhauser C, Lehmann M, van Dongen JT, Geigenberger P. Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 2016;172(1):141–53. [Google Scholar] [PubMed]
35. Licausi F, Kosmacz M, Weits DA. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature. 2011;479(7373):419–22. [Google Scholar] [PubMed]
36. Sun D, Nandety RS, Zhang Y. Apetuniaethylene-re-sponsive element bindingfactor, PhERF2, plays an important role in antiviral RNA silencing. J Exp Bot. 2016;67(11):3353–65. [Google Scholar] [PubMed]
37. Sauter M. Root responses to flooding. Curr Opin Plant Biol. 2013;16(3):282–6. [Google Scholar] [PubMed]
38. Zhao L, Cao DD, Su Z, Fum XH, Li YY, Wei JR, et al. HrTPS12 gene dramatically enhanced insect resistance of sea buckthorn to infection by fruit fly (Rhagoletis batava obseuriosa Kol.). Pest Manag Sci. 2023;79(11):4172–85. doi:10.1002/ps.7614. [Google Scholar] [PubMed] [CrossRef]
39. Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008–17. [Google Scholar] [PubMed]
40. Trapnell C, Williams BA, Pertea G, Mortazavi A. Transcript assembly and quantification by RNA Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2020;28(5):511–15. [Google Scholar]
41. Alexa A, Rahnenfuhrer J. TopGO: enrichment analysis for gene ontology. R package version 2.8. 2010. [Google Scholar]
42. Kanehisa M, Goto S, Kawashima S, Okuno Y. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(suppl_1):D277–80. [Google Scholar] [PubMed]
43. Fu XH, Zhao J, Cao DD, He CX, Wang ZY, Jiang YB, et al. Characteristics and expression of the TCP transcription factors family in Allium senescens reveal its potential roles in drought stress responses. BIOCELL. 2023;47(4):905–17. doi:10.32604/biocell.2023.026930. [Google Scholar] [CrossRef]
44. Zhao L, Wang W, Fu XH, Liu A, Cao JF, Liu JF. Graphene oxide, a novel nanomaterial as soil water retention agent, dramatically enhances drought stress tolerance in soybean plants. Front Plant Sci. 2022;13:810905. [Google Scholar] [PubMed]
45. Fu XH, Bian DH, Gu XY, Cao JF, Liu JF. Combination of graphene oxide and rhizobium improved soybean tolerance in saline-alkali atress. Agron. 2023;13(6):1637. [Google Scholar]
46. Yao Y, Dong LJ, Fu XH, Zhao L, Wei JR, Cao JF, et al. HrTCP20 dramatically enhance drought tolerance of sea buckthorn (Hippophae rhamnoides L). by mediating the JA signaling pathway. Plant Physiol Biochem. 2022;174:51–62. [Google Scholar] [PubMed]
47. Shi H, Ye T, Chen F, Cheng Z, Wang Y, Yang P, et al. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis effect on arginine metabolism and ROS accumulation. J Exp Bot. 2013;64(5):1367–79. [Google Scholar] [PubMed]
48. Wang WW, Lin H, Tang XF, Wei L, Dong XY, Wu GX, et al. Soybean drought resistance related genes expression characteristics under drought. Mol Plant Breed. 2014;12(5):903–8. [Google Scholar]
49. Watanabe N, Lam E. BAX inhibitor-1modulates endoplasmic reticulum stress-mediated programmed cell death in Arabi-dopsis. J Biol Chem. 2008;283(6):3200–10. [Google Scholar] [PubMed]
50. Wang J, Wang HY, Zhou JM, Chen XQ, Du CW. Potassium content in ryegrass extracted by HCl at flame photometry. Chin J Soil Sci. 2013;44(3):624–7. [Google Scholar]
51. Tsai KJ, Lu MYJ, Yang KJ, Li MY, Teng YC, Chen S, et al. Assembling the Setaria italica L. Beauv. genome into nine chromosomes and insights into regions affecting growth and drought tolerance. Sci Rep. 2016;6(1):35076. [Google Scholar] [PubMed]
52. Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, et al. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010;62(2):302–15. [Google Scholar] [PubMed]
53. Yu F, Liang K, Fang T, Zhao HL, Han XS, Cai MJ, et al. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. J Plant Physiol. 2019;17(12):2286–98. [Google Scholar]
54. Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, et al. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 2002;14(10):2481–94. [Google Scholar] [PubMed]
55. Yin DM. Evaluation on waterlogging tolerance and its mechanisms in chrysanthemum morifolium and its related species. China: Nanjing Agricultural University; 2011. [Google Scholar]
56. Ma J, Lv CF, Zhang BB, Wang F, Shen WJ, Chen GX, et al. Comparative analysis of ultrastructure., antioxidant enzyme activities., and photosynthetic performance in rice mutant 812HS prone to photooxidation. Photosynthetica. 2017;55(4):568–78. [Google Scholar]
57. Zhang RD. Physiological mechanism of sorghum response to waterlogging stress and alleviating solutions. Shenyang Agricultural University: China; 2020. [Google Scholar]
58. Yordanova RY, Christov KN, Popova LP. Antioxidative enzymes in barley plants subjected to soil flooding. Environ Exp Bot. 2004;51(2):93–101. [Google Scholar]
59. Pfister SM, Brandle R. Aspects of plant behaviour under anoxia and post-anoxia. Proc Royal Soc B. 1994;102(6):313–24. [Google Scholar]
60. Drew M. Effects of flooding and oxygen deficiency on plant mineral nutrition. Adv Nutr. 1988;3(8):115–59. [Google Scholar]
61. Zhu XC, Zhang HM, Nan WB. Research progress on regulation of ABA in plant root development. J Plant Physiol. 2017;53(7):1123–30. [Google Scholar]
62. Qi XH, Li QQ, Ma XT, Qian CL, Wang HH, Ren NN, et al. Waterlogging-induced adventitious root formation in cucumber is regulated by ethylene and auxin through reactive oxygen species signalling. Plant Cell Environ. 2019;42(5):1458–70. [Google Scholar] [PubMed]
63. Dong JG, Fernandez JC, Yang SF. Purification and characterization of 1-aminocyclopropane-1-carboxylate oxidase from apple fruit. Proc Natl Acad Sci USA. 1992;89(20):9789–93. [Google Scholar] [PubMed]
64. Ramadoss N, Gupta D, Vaidya BN, Joshee N, Basu C. Functional characterization of 1-aminocyclopropane-1-carboxylic acid oxidase gene in Arabidopsis thaliana and its potential in providing flood tolerance. Biochem Biophys Res Commun. 2018;503(1):365–70. [Google Scholar] [PubMed]
65. Yin XR, Allan AC, Xu Q, Burdon J, Dejnoprat S, Chen KS, et al. Differential expression of kiwifruit genes in response to postharvest abiotic stress. Postharvest Biol Technol. 2012;66(2):1–7. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools