 Open Access
Open Access
ARTICLE
Analysis and Verification of the Conserved MYB Binding Element in the DFR Promoter in Compositae
1 State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University, Xining, 810016, China
2 School of Ecol-Environmental Engineering, Qinghai University, Xining, 810016, China
3 Key Laboratory of Landscape Plants of Qinghai Province, Qinghai University, Xining, 810016, China
* Corresponding Author: Tao He. Email:
(This article belongs to the Special Issue: Plant Secondary Metabolism and Functional Biology)
Phyton-International Journal of Experimental Botany 2024, 93(2), 343-353. https://doi.org/10.32604/phyton.2024.047429
Received 05 November 2023; Accepted 10 January 2024; Issue published 27 February 2024
Abstract
Anthocyanins, ubiquitous in the Compositae family, are regulated by MYB (v-myb avian myeloblastosis viral oncogene homolog), playing an important role in anthocyanin synthesis. In this study, we analyzed the regulation pathway in which the MYB protein of subgroup 6 promotes dihydroflavonol reductase (DFR) expression in Compositae, and validated this law in Saussurea medusa through yeast one-hybrid experiments. Our results showed that MYB and DFR underwent purification selection, DFR promoter analysis revealed the presence of MYB binding site (GAGTTGAATGG) and bHLH binding site (CANNTG) at the sense strand of 84–116 nucleotide residues from the start codon. These two motifs were separated by 9–10 nucleotide residues, as existed in the DFR promoters of many Compositae plants. Furthermore, the yeast one-hybrid experiment demonstrated that SmMYB1 can activate the promoter of SmDFR. Our results provide a reference for further functional study of DFR in Compositae.Keywords
Supplementary Material
Supplementary Material FileThe MYB gene family is large, functionally diverse, and represented in all eukaryotes. In plants, many MYB genes, acting in complex with basic helix–loophelix (bHLH) and WD40 partners, are key factors in regulatory networks controlling development, metabolism, and responses to biotic and abiotic stresses [1]. Dihydroflavonol 4-reductase (DFR) catalyzes the NADPH-dependent reduction of dihydroflavonols into leucoanthocyanidins, acting as the key enzyme committed to anthocyanin and proanthocyanidin biosynthesis [2]. The overexpression of DFR genes of different species in tobacco can induce the accumulation of pigments in the flower [3], and gene silencing of IbDFR in sweet potato (Ipomoea batatas) can induce the decrease of anthocyanin content [4]. Anthocyanin biosynthesis is mainly regulated at the transcriptional level [5]. MYB protein plays an important role in the regulation of anthocyanin synthesis. Different structures in the DFR promoter of Malus crabapple have influences on MYB binding, which Affects the accumulation of anthocyanins in crabapple cultivars [6].
The Asteraceae family (Compositae) is a widespread family of flowering plants [7]. It is the largest family of angiosperms due to the developed sexual reproduction. Several Compositae plants have ornamental, medicinal, and edible values, while Mikania micrantha is a destructive invasive plant in China [8]. Through studying anthocyanins components in Asteraceae, Saigo et al. [9] found that delphinidin is the most abundant in Asterales, followed by cyanidin, pelargonin, and malvain; Sareedenchai et al. [10] found 9 types of anthocyanins, including 6 pelargonins, 2 cyanidins, and 1 malvain, in the tribe Cichorieae of Asteraceae [11]; The flower color of dahlia is mainly determined by xanthein, anthocyanins, and other flavonoids [12]; anthocyanins act as nutrients and improve the quality of Asteraceae vegetables Lactuca sativa and Gynura bicolor DC [13–15].
Comparative transcriptome analysis reveals mechanisms of adaptation to extreme environments among Three Species of Compositae [16]. The regulation pathway through which the MYB protein of subgroup 6 promotes DFR expression has been verified in an increasing number of Asteraceae plants, suggesting that such a pathway is highly conserved in Compositae (Table 1). At the same time, the publication of many genomic data has made evolutionary analysis of this pathway possible This study used Asteraceae plants as the research object, conducted selection pressure analysis of MYB and DFR protein, analyzed the conserved elements of DFR promoter, and verified it by yeast one-hybrid experiment in Saussurea medusa. The findings can provide a theoretical basis for the evolution and transcriptional control of anthocyanin metabolism in Compositae. Anthocyanin is an active substance with high ornamental and nutritional value; this element may be used as a molecular marker related to anthocyanins in Compositae plants.
2.1 Conservation Analysis of DFR and MYB Protein
The Compositae plants were summarized based on published literature (Table 1). The composition of anthocyanins in Asteraceae plants was searched in the KNApSAcK database (http://kanaya.naist.jp/KNApSAcK/). The Compositae DFR protein sequences (Table S1) were retrieved from the NCBI and the OrthDB website (http://www.orthodb.org/). BLASTp search was performed against Compositae plants with published genome sequences. DNAMAN was used to perform multiple sequence alignments.
The MYB of subgroup 6 in Gynura bicolor DC, Gerbera hybrida, Chrysanthemum morifolium, and Centaurea cyanus has been experimentally demonstrated to positively regulate DFR expression [11,14,17,18]. DNAMAN was used to perform multiple sequence alignments of these MYB proteins.
The amino acid sequence was submitted to ColabFold v1.5.5 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) using the default settings. The resulting models were evaluated using the pLDDT score.
2.2 Conservation Analysis of DFR Gene Promoter
The Compositae plants were summarized based on published literature (Table 1). The composition of anthocyanins in Asteraceae plants was searched in the KNApSAcK database The 1000 bp DFR promoter sequences were retrieved with the published genome (Table S1). The DFR promoters (Table S1) were analyzed on the MEME website (https://meme-suite.org/meme/doc/overview.html) to find the conserved motifs. The conserved motifs were annotated by the PLACE website (https://www.dna.affrc.go.jp/PLACE/?action=newplace). The existence of the conserved motifs in the Asteraceae DFR promoter was further analyzed.
2.3 Cloning of Compositae DFR Genome Sequence and Yeast One-Hybrid Experiment
The DFR promoter sequence of Saussurea medusa was amplified by the method of hi-TAIL PCR [31,32]. Primers used in the experiment are listed in Table 2. Three nested downstream-specific primers were designed according to the SmDFR gDNA sequence (GenBank: EF672726.1). The pGADT7 vector was double-digested with EcoRI and BamHI, and the pHIS2 vector was double-digested with EcoRI and MluI, both incubated at 30°C for 2 h followed by 65°C for 10 min. The enzyme-cut mixture was separated by electrophoresis. The pGADT7-SmMYB1 yeast expression vector and the pSmDFR-HIS2 yeast reporter vector were constructed by in-fusion cloning. Yeast-competent cells were prepared by the LiAc method, and the pGADT7-SmMYB1 and pHIS2-pSmDFR vectors were co-transformed into yeast strains and cloned into SD/-Trp-Leu-His triple deficiency medium with a 3-AT concentration gradient.

2.4 Evolutionary Analysis of MYB and DFR Genes
Multiple sequence alignments were performed through the MUSCLE (CONDON) option of MEGA7. The neighbor-joining method was used to construct a phylogenetic tree with the alignment sequences. The selection pressure analysis was performed using EasyCodeML [33]. The sequence alignments and tree files were imported into EasyCodeML to estimate the selection pressures based on the ratio of non-synonymous to synonymous substitution rates (Omega = dN/dS).
3.1 DFR and MYB Protein Conservation Analysis
According to the OrthDB website, both Helianthus annuus and Lactuca sativa contain one DFR gene (Table S1). By searching for BLASTp in the published genomes of Compositae plants, we found that Cynara cardunculus contains one DFR gene, while Artemisia annua contains two DFR genes (Table S1). Compositae DFR protein multiple sequence alignment reveals that the sequences at positions 9 to 332 are highly conserved (Fig. 1). DFR is divided into Asn/Asp type (cannot convert dihydrokaempferol to leucopelarginin), and non-Asn/Asp type according to the type of amino acid at position 134. All of the isolated DFR genes in Asteraceae are Asn-type DFR. The modeling results show that only the DFR protein is highly consistent (Fig. 2).
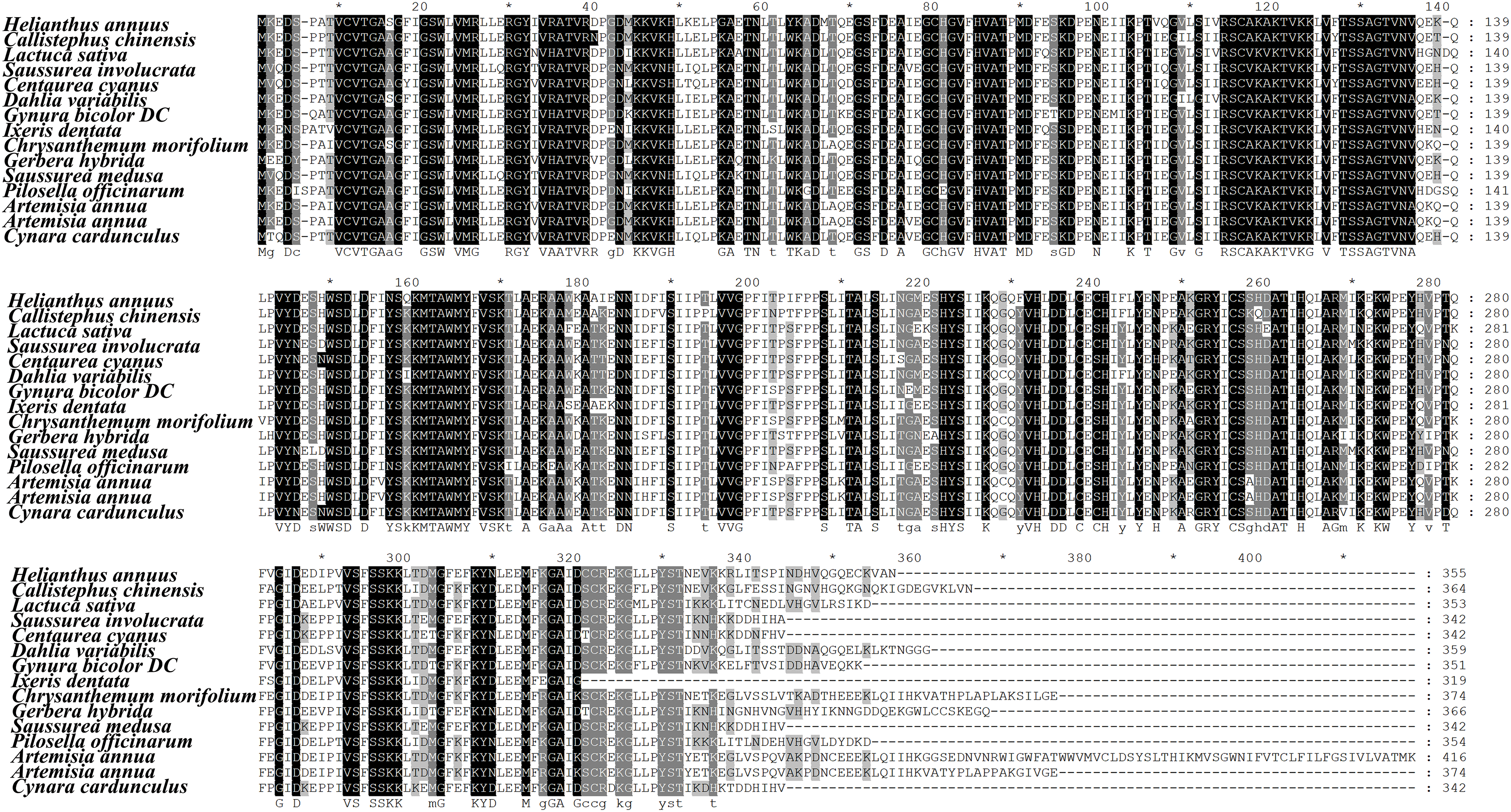
Figure 1: Multiple sequence alignment of DFR protein
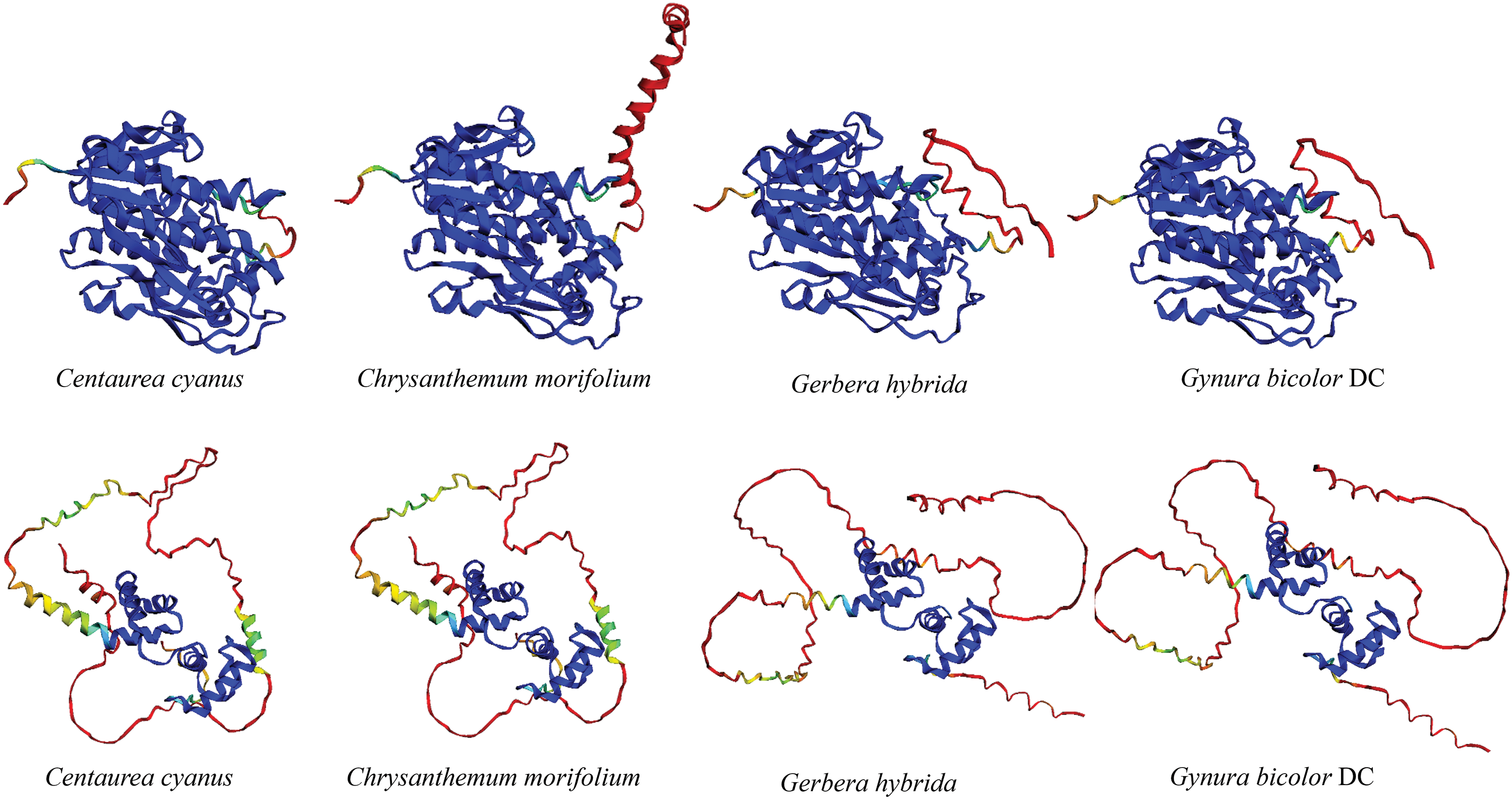
Figure 2: Protein structure of DFR and MYB
Note: The uplink is DFR protein, and the downlink is MYB protein. Dark blue means very high (greater than 80), green means moderate (70), yellow means low (60), and red means very low (less than 50).
MYB protein contains the conserved R2R3-MYB domain, which contains two motifs, [DE]Lx(2)[RK]x(3)Lx(6)Lx(3)R and ANDV (Fig. 3). The modeling results show that only the R2R3-MYB domain is structurally similar (Fig. 2).

Figure 3: Multiple sequence alignment of MYB protein
3.2 Conservation Analysis of DFR Gene Promoter
Three ubiquitous motifs were found in the promoters of Centaurea cyanus, Chrysanthemum morifolium, and Gynura bicolor (Fig. 4A), including GMYB10 binding site (GAGTTGAATGG), bHLH binding site (CACGTG), and ELM2 (AACGG) binding site. The MYB binding site and the bHLH binding site are separated by 9–10 nucleotide residues and relatively conserved in the sense strand 84–111 bp from the start codon (Fig. 4A).
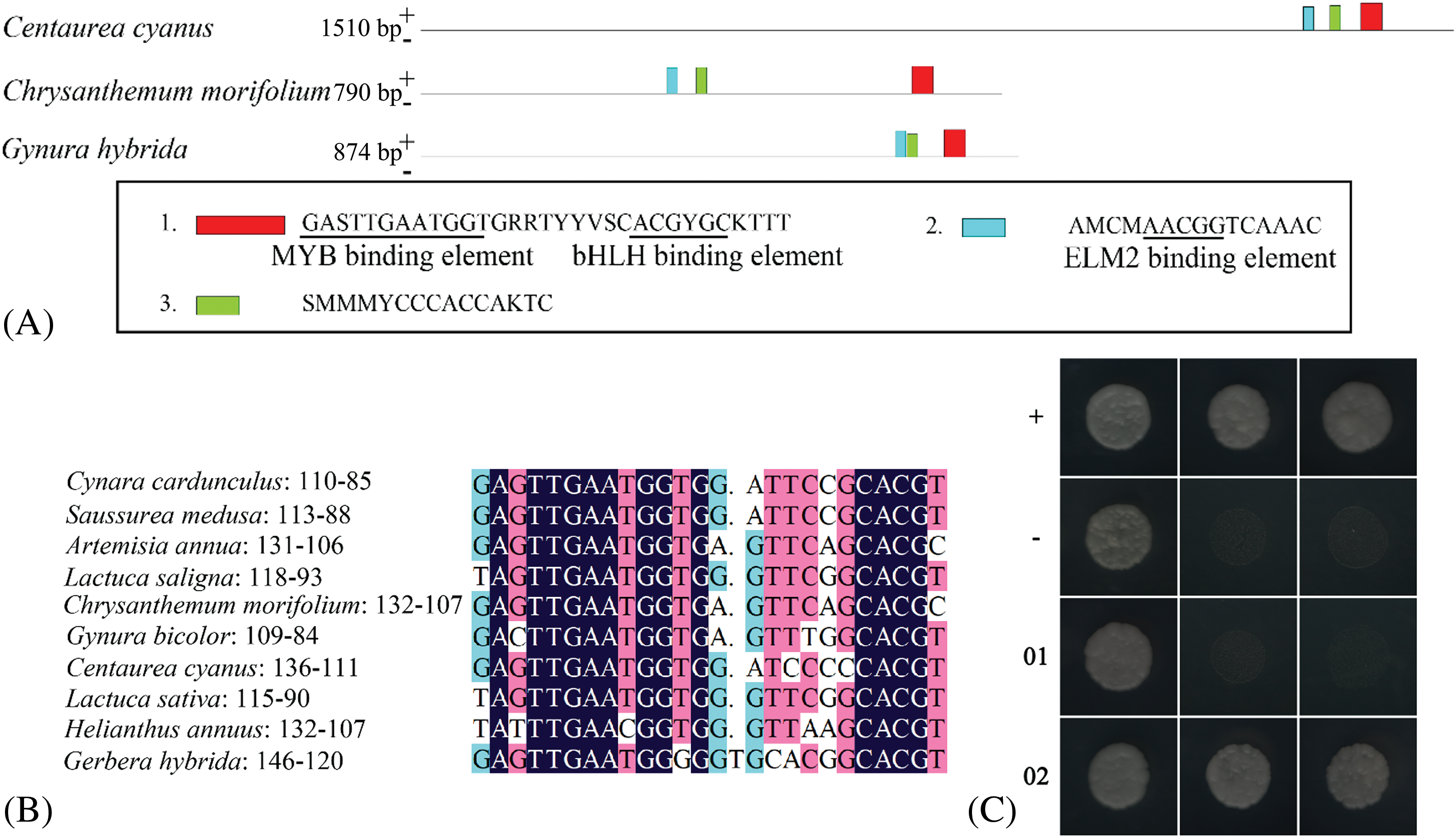
Figure 4: Conservation analysis of DFR gene promoter
Note: (A) Conserved motif of DFR gene promoter in Compositae. (B) Alignment of conserved elements in compositae, the number after the species is the number of bases from the start codon of the elements. (C) Yeast one-hybrid, left column: SD/-Leu/-Trp, middle column: SD/-His/-Leu/-Trp+2.5mM 3-AT, right column: SD/-His/-Leu/-Trp+5mM 3-AT, +: positive control, -: negative control, 01: pSmDFR-HIS2/pGADT7, 02: pGADT7-SmMYB1+pSmDFR-HIS2.
It was found that the DFR promoter of Cynara cardunculus conformed to the above-mentioned law. Lactuca sativa, Lactuca saligna, and Artemisia annua also conformed to such rule except for one base substitution (Fig. 4B). These findings indicate that the binding elements of MYB and bHLH are ubiquitous in the DFR promoters, and their positions are located at 87–119 bp from the start codon (Fig. 4B).
3.3 Cloning of Saussurea Medusa DFR Genome Sequence and Yeast One-Hybrid Experiment
According to the article by Elomaa et al. [11], particle bombardment analysis indicated that the 276-bp fragment of GDFR2 confers similar levels of reporter gene activity as the full-length promoter. Also, mutations at the site resembling an MYB-binding site (GAGTTGAATGGGG) reduced reporter gene activities to approximately 20% to 40% of the full-length promoter in Gerbera hybrida [34].
The 389 bp SmDFR promoter sequence (GenBank: MT634229) was amplified, and conserved MYB and bHLH binding elements (GAGTTGAATGGGG) were found at 88–113 bp from the start codon (Fig. 4C). All the yeast strains co-transfected with pHIS2 and pGADT7 vectors could grow on SD/-Leu/-Trp double deficiency medium, indicating successful transformation of the two vectors. The grown clones were picked and transferred to SD/-His/-Leu/-Trp triple deficiency medium with 2.5 mM 3-AT. We found that the experimental group grew better than the self-activation group (pSmDFR-HIS2+pGADT7). The clones were then picked and transferred to SD/-His/-Leu/-Trp triple deficiency medium with 5 mM 3-AT, where the growth of the self-activating group (pSmDFR-HIS2+pGADT7) was found inhibited. Meanwhile, the experimental group (pGADT7-SmMYB1+pSmDFR-HIS2) could grow normally (Fig. 4C). These results indicate that the SmMYB1 protein potentially binds with the SmDFR promoter.
3.4 Evolutionary Analysis of MYB and DFR Genes
The MYB of subgroup 6 in Gynura bicolor DC, Gerbera hybrida, Chrysanthemum morifolium, Centaurea cyanus, and Saussurea medusa has been experimentally demonstrated to positively regulate DFR expression [11,14,17,35]. These MYB sequences were collected and carried out the selection pressure analysis by EasyCodeML, and the likelihood ratio test showed no significant difference between the one-ratio vs. free-ratio model of the branch model This indicates the existence of the same selection pressure among different branches. The selection pressure was 0.40885 under the one-ratio model, indicating that the gene was mainly selected by purification. The likelihood ratio test of these DFR sequences showed no significant difference between the one-ratio vs. free-ratio model of the branch model. Once again, this indicates the existence of the same selection pressure among different branches. The selection pressure was 0.24999 under the one-ratio model, indicating that the gene was mainly selected by purification.
No DFR and MYB gene was found in the published Asteraceae genomes of Mikania micrantha (Genbank: GCA_009363875.1), Erigeron canadensis (Genbank: GCA_010389155.1), Stevia rebaudiana (Genbank: GCA_009936405.2), Chrysanthemum seticuspe (Genbank: GCA_019973895.1), and Silphium perfoliatum (Genbank: GCA_900538075.1). Meanwhile, no anthocyanins were found in the above species by searching the KNApSAcK database.
DFR genes are closely related to anthocyanin synthesis in most of the Compositae plants (Table 1). The DFR of Saussurea medusa, Gerbera hybrida, and Callistephus chinensis can catalyze the divergent conversion of dihydroflavonols to proanthocyanidins [19,24,25]. Meanwhile, all the Compositae plants discussed above-contained cyanidin, indicating a flow of cyanidin towards the branch in the late stage of anthocyanin synthesis, and all of the isolated DFR genes in Asteraceae were Asn-type DFR. Such a branching pathway has been previously reported in Lactuca sativa [26], in which DFR is required to catalyze dihydroquercetin to leucocyanidin simultaneously.
The MYB of the sixth subfamily can interact with the bHLH partner to induce DFR expression in Gynura bicolor DC, Gerbera hybrida, Chrysanthemum morifolium, and Centaurea cyanus [11,14,17,35]. The selection pressure analysis showed that the MYB and DFR genes are mainly subjected to purification selection. The DFR promoter analysis revealed the existence of conserved MYB binding sites (GAGTTGAATG) and bHLH binding sites (CANNTG). Zhu et al. [36] have previously identified ANCNNCC for MYB recognizing elements and CACN(A/C/T)(G/T) for bHLH recognizing elements within anthocyanin gene promoters in at least 35 species, including gymnosperms and angiosperms. Although the MYB binding element (ANCNNCC) is not identical to the antisense strand sequence CCATTCAACTC of the MYB binding element (GAGTTGAATGG) of the Compositae family in this study, they are both rich in A and C bases. The DFR promoter of Saussurea medusa was found to conform to this law, and the yeast one-hybrid experiment demonstrated that SmMYB1 can activate the SmDFR promoter. This regulatory pathway, in which MYB protein can activate the promoter of DFR, is predicted to be ubiquitous in the Compositae plants.
Wheeler et al. [37] have found that the transcription factors exhibit faster rates of molecular evolution (dN/dS) than their targets, with the highly specialized MYB genes evolving the fastest in the flavonoid pigment pathway of Petunieae. In Compositae, MYB is lowly conserved with a selection pressure of 0.40885 while DFR is highly conserved with a selection pressure of 0.25000; this is consistent with the evolution model in Petuniaeae. This may be because transcription factors only function in the DNA-binding domain and the transcriptional activation domain. Enzymes, on the other hand, require the entire protein to catalyze the reaction.
However, no DFR gene was found in the genomic sequences of Mikania micrantha, Erigeron canadensis, Stevia rebaudiana, Chrysanthemum seticuspe, Silybum marianum, Carthamus tinctorius, and Silphium perfoliatum. OsDFR is defective in rice varieties that do not produce anthocyanins [38]. These results indicate that this pathway is not necessary in all species. The above-mentioned findings reveal that the expression and regulation of DFR still follow certain rules in some species, though there still is a great variation of the DFR gene in Compositae.
Through evolutionary analysis of several Compositae plants in which MYB has been confirmed to regulate DFR, the MYB gene was found under purification selection, and conserved MYB and bHLH binding elements were identified in the DFR promoter (such elements have been verified in Gerbera hybrida). This indicates the important role of such elements in the anthocyanin pathway. This element may be used as a molecular marker related to anthocyanins in Asteraceae plants. The MYB gene was found to be expanded in some Compositae plants with published genomes, and the conserved MYB and bHLH binding elements were also identified in their DFR promoters. MYB was speculated to regulate DFR in these Compositae plants, which was further verified in Saussurea medusa by a yeast one-hybrid experiment. However, these genes are lost in some species, suggesting a different evolutionary direction.
Acknowledgement: We thank Prof. Dai Silan (Beijing Forestry University, Beijing, China) for providing sequence information on Centaurea cyanus.
Funding Statement: This work was financially supported by the National Natural Science Foundation of China (31960222, 31360095).
Author Contributions: Jialei Guo: Conceptualization; Methodology; Investigation; Writing-original draft. Fengzhen Li, Guomin Shi, and Weimin Zhao: Investigation; Data curation. Tao He: Supervision; Review and Editing.
Availability of Data and Materials: The data supporting the findings of this study are available within the supplementary materials.
Ethics Approval: None.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.047429.
References
1. Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–81. doi:10.1016/j.tplants.2010.06.005. [Google Scholar] [PubMed] [CrossRef]
2. Martens S, Knott J, Seitz C, Janvari L, Yu S, Forkmann G. Impact of biochemical pre-studies on specific metabolic engineering strategies of flavonoid biosynthesis in plant tissues. Biochem Eng J. 2003;14(3):227–35. doi:10.1016/S1369-703X(02)00224-3. [Google Scholar] [CrossRef]
3. Luo P, Ning G, Wang Z, Shen Y, Jin H, Li P, et al. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front Plant Sci. 2015;6:1257. doi:10.3389/fpls.2015.01257. [Google Scholar] [PubMed] [CrossRef]
4. Wang H, Fan W, Li H, Yang J, Huang J, Zhang P. Functional characterization of Dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS One. 2013;8(11):e78484. doi:10.1371/journal.pone.0078484. [Google Scholar] [PubMed] [CrossRef]
5. Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20(3):176–85. doi:10.1016/j.tplants.2014.12.001. [Google Scholar] [PubMed] [CrossRef]
6. Tian J, Chen M, Zhang J, Li K, Song T, Zhang X. Characteristics of dihydroflavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hort Res. 2017;4:17070. doi:10.1038/hortres.2017.70. [Google Scholar] [PubMed] [CrossRef]
7. Rodriguez-Flores CI, Ornelas JF, Wethington S, Arizmendi MDC. Are hummingbirds generalists or specialists? Using network analysis to explore the mechanisms influencing their interaction with nectar resources. PLoS One. 2019;14(2):e0211855. doi:10.1371/journal.pone.0211855. [Google Scholar] [PubMed] [CrossRef]
8. Zeng H, Liu X, Zhang L, Li Y, Zhu M, Chen D. Educational approaches help bridge perception gaps of invasive alien species (Mikania micrantha) between managers and non-managers. Environ Manage. 2021;68(3):340–52. doi:10.1007/s00267-021-01505-7. [Google Scholar] [PubMed] [CrossRef]
9. Saigo T, Wang T, Watanabe M, Tohge T. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr Opin Plant Biol. 2020;55:93–9. doi:10.1016/j.pbi.2020.04.001. [Google Scholar] [PubMed] [CrossRef]
10. Sareedenchai V, Zidorn C. Flavonoids as chemosystematic markers in the tribe cichorieae of the asteraceae. Biochem Syst Ecol. 2010;38(5):935–57. doi:10.1016/j.bse.2009.09.006. [Google Scholar] [CrossRef]
11. Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RA, Teeri TH. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol. 2003;133(4):1831–42. doi:10.1104/pp.103.026039. [Google Scholar] [PubMed] [CrossRef]
12. Walliser B, Lucaciu CR, Molitor C, Marinovic S, Nitarska DA, Aktas D, et al. Dahlia variabilis cultivar ‘Seattle’ as a model plant for anthochlor biosynthesis. Plant Physiol Biochem. 2021;159:193–201. doi:10.1016/j.plaphy.2020.12.016. [Google Scholar] [PubMed] [CrossRef]
13. Šuštar-Vozlič J, Ugrinović K, Maras M, Křístková E, Lebeda A, Meglič V. Morphological and genetic diversity of Slovene lettuce landrace ‘Ljubljanska ledenka’ (Lactuca sativa L.). Genet Resour Crop EV. 2020;68(1):185–203. doi:10.1007/s10722-020-00978-5. [Google Scholar] [CrossRef]
14. Shimizu Y, Maeda K, Kato M, Shimomura K. Co-expression of GbMYB1 and GbMYC1 induces anthocyanin accumulation in roots of cultured Gynura bicolor DC. plantlet on methyl jasmonate treatment. Plant Physiol Bioch. 2011;49(2):159–67. doi:10.1016/j.plaphy.2010.11.006. [Google Scholar] [PubMed] [CrossRef]
15. Su W, Tao R, Liu W, Yu C, Yue Z, He S, et al. Characterization of four polymorphic genes controlling red leaf colour in lettuce that have undergone disruptive selection since domestication. Plant Biotechnol J. 2020;18(2):479–90. doi:10.1111/pbi.13213. [Google Scholar] [PubMed] [CrossRef]
16. Zhao W, Li F, Shi G, Guo J, He G. Comparative transcriptome analysis reveals different mechanisms of adaptation to environment among three species of saussurea DC. Phyton-Int J Exp Bot. 2022;91(7):1517–28. doi:10.32604/phyton.2022.019630. [Google Scholar] [CrossRef]
17. Lockowandt L, Pinela J, Roriz CL, Pereira C, Abreu RMV, et al. Chemical features and bioactivities of cornflower (Centaurea cyanus L.) capitula: the blue flowers and the unexplored non-edible part. Ind Crop Prod. 2019;128:496–503. doi:10.1016/j.indcrop.2018.11.059. [Google Scholar] [CrossRef]
18. Deng C, Wang J, Lu C, Li Y, Kong D, Hong Y, et al. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower. Plant Physiol Biochem. 2020;151:271–83. doi:10.1016/j.plaphy.2020.03.024. [Google Scholar] [PubMed] [CrossRef]
19. Bashandy H, Pietiainen M, Carvalho E, Lim KJ, Elomaa P, Martens S, et al. Anthocyanin biosynthesis in gerbera cultivar ‘Estelle’ and its acyanic sport ‘Ivory’. Planta. 2015;242(3):601–11. doi:10.1007/s00425-015-2349-6. [Google Scholar] [PubMed] [CrossRef]
20. Han K, Zhao L, Tang X, Hu K, Dai S. The relationship between the expression of key genes in anthocyanin biosynthesis and the color of chrysanthemum. Acta Hortic Sinica. 2012;39:516–24. [Google Scholar]
21. Lim S, Park B, Kim D, Park S, Yang J, Jung J, et al. Cloning and functional characterization of dihydroflavonol 4-reductase gene involved in anthocyanin biosynthesis of chrysanthemum. Int J Mol Sci. 2020;21(21):7960. doi:10.3390/ijms21217960. [Google Scholar] [PubMed] [CrossRef]
22. Xiang L, Liu X, Li X, Yin X, Grierson, Li F, D, et al. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS One. 2015;10(11):e0143892. doi:10.1371/journal.pone.0143892. [Google Scholar] [PubMed] [CrossRef]
23. Cheng L, Xu Y, Grotewold E, Jin Z, Wu F, Fu C, et al. Characterization of anthocyanidin synthase (ANS) gene and anthocyanidin in rare medicinal plant-Saussurea medusa. Plant Cell, Tissue Organ Cult. 2007;89(1):63–73. doi:10.1007/s11240-007-9211-x. [Google Scholar] [CrossRef]
24. Li H, Qiu J, Chen F, Lv X, Fu C, Zhao D, et al. Molecular characterization and expression analysis of dihydroflavonol 4-reductase (DFR) gene in Saussurea medusa. Mol Biol Rep. 2012;39(3):2991–9. doi:10.1007/s11033-011-1061-2. [Google Scholar] [PubMed] [CrossRef]
25. Zhang Y, Xu S, Cheng Y, Ya H, Han J. Transcriptome analysis and anthocyanin-related genes in red leaf lettuce. Genet Mol Res. 2016;15(1):gmr.15017023. doi:10.4238/gmr.15017023. [Google Scholar] [PubMed] [CrossRef]
26. Zhang Y, Xu S, Cheng Y, Peng Z, Han J. Transcriptome profiling of anthocyanin-related genes reveals effects of light intensity on anthocyanin biosynthesis in red leaf lettuce. PeerJ. 2018;6:e4607. doi:10.7717/peerj.4607. [Google Scholar] [PubMed] [CrossRef]
27. Zhang J, Pan D, Zhou Y, Wang Z, Sui F. Cloning and expression of genes involved in anthocyanins synthesis in ornamental sunflower. Acta Hortic Sinica. 2009;36:73–80. [Google Scholar]
28. Ohno S, Hosokawa M, Hoshino A, Kitamura Y, Morita Y, Park KI, et al. A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis). J Exp Bot. 2011;62(14):5105–16. doi:10.1093/jxb/err216. [Google Scholar] [PubMed] [CrossRef]
29. Chen S, Li C, Zhu X, Deng Y, Sun W, Wang L, et al. The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol Plantarum. 2012;56(3):458–64. doi:10.1007/s10535-012-0069-3. [Google Scholar] [CrossRef]
30. Witte S, Moco S, Vervoort J, Matern U, Martens S. Recombinant expression and functional characterisation of regiospecific flavonoid glucosyltransferases from Hieracium pilosella L. Planta. 2009;229(5):1135–46. doi:10.1007/s00425-009-0902-x. [Google Scholar] [PubMed] [CrossRef]
31. Liu Y, Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTech. 2007;43(5):649–50. doi:10.2144/000112601. [Google Scholar] [PubMed] [CrossRef]
32. Tan J, Gong Q, Yu S, Hou Y, Zeng D, Zhu Q, et al. A modified high-efficiency thermal asymmetric interlaced PCR method for amplifying long unknown flanking sequences. J Genet Genomics. 2019;46(7):363–66. doi:10.1016/j.jgg.2019.05.002. [Google Scholar] [PubMed] [CrossRef]
33. Gao F, Chen C, Arab DA, Du Z, He Y, Ho S. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol. 2019;9(7):3891–98. doi:10.1002/ece3.5015. [Google Scholar] [PubMed] [CrossRef]
34. Elomaa P, Mehto M, Kotilainen M, Helariutta Y, Nevalainen L, Teeri T. A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae). Plant J. 1998;16(1):93–9. doi:10.1046/j.1365-313x.1998.00273.x. [Google Scholar] [PubMed] [CrossRef]
35. Liu X, Xiang L, Yin X, Grierson D, Li F, Chen K. The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chrysanthemum flowers. Sci Hortic. 2015;194:278–85. doi:10.1016/j.scienta.2015.08.018. [Google Scholar] [CrossRef]
36. Zhu Z, Wang H, Wang Y, Guan S, Wang F, Tang J, et al. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J Exp Bot. 2015;66(13):3775–89. doi:10.1093/jxb/erv173. [Google Scholar] [PubMed] [CrossRef]
37. Wheeler LC, Walker JF, Ng J, Deanna R, Dunbar-Wallis A, Backes A, et al. Transcription factors evolve faster than their structural gene targets in the flavonoid pigment pathway. Mol Biol Evol. 2022;39(3):msac044. doi:10.1093/molbev/msac044. [Google Scholar] [PubMed] [CrossRef]
38. Zhu Q, Yu S, Zeng D, Liu H, Wang H, Yang Z, et al. Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol plant. 2017;10(7):918–29. doi:10.1016/j.molp.2017.05.008. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF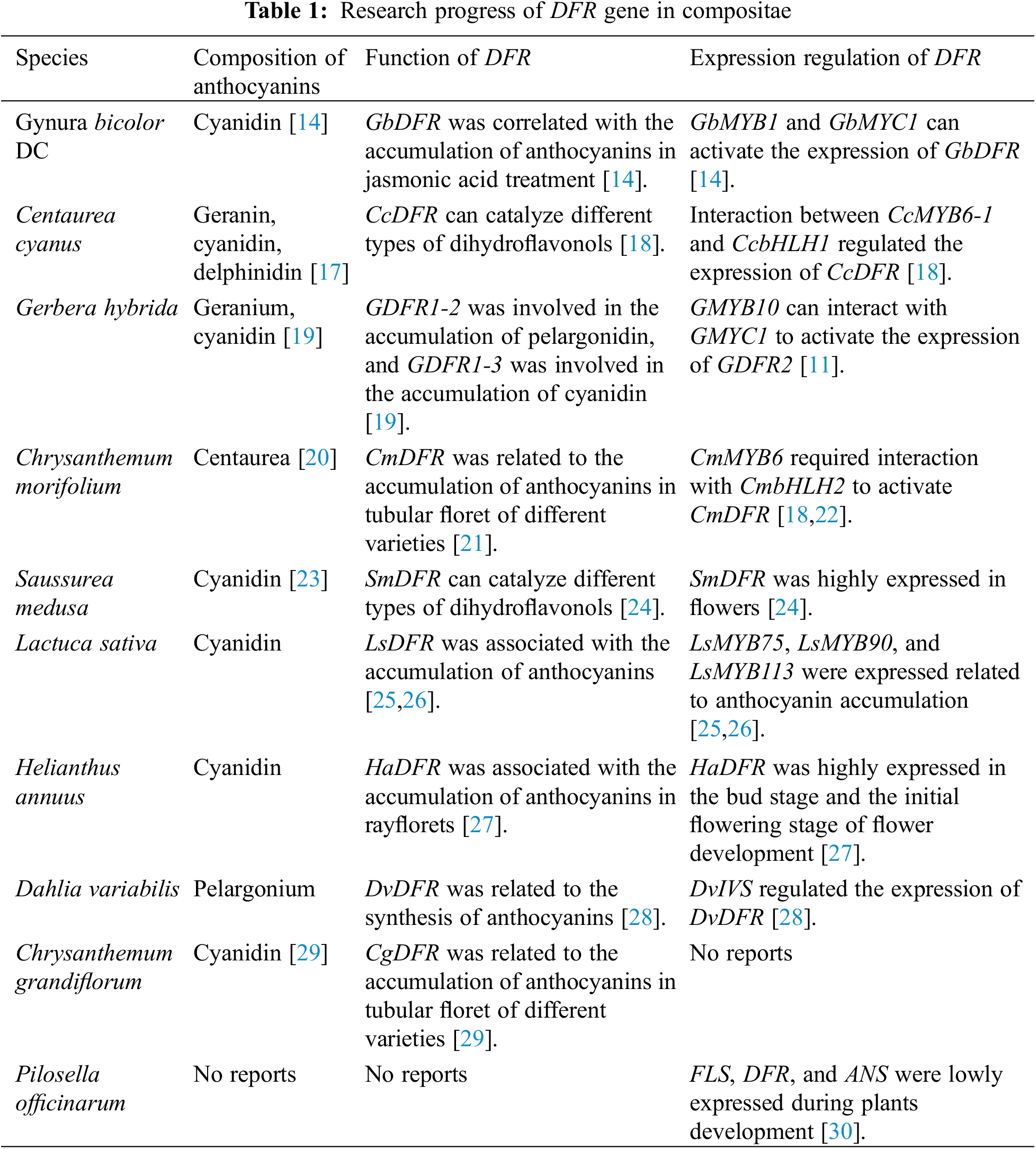
 Downloads
Downloads
 Citation Tools
Citation Tools