 Open Access
Open Access
ARTICLE
Differential Expression of Genes Related to Fruit Development and Capsaicinoids Synthesis in Habanero Pepper Plants Grown in Contrasting Soil Types
1 Unidad de Bioquímica y Biología Molecular de Plantas, Centro de Investigación Científica de Yucatán, Calle 43 # 130, Chuburná de Hidalgo, Mérida, Yucatán, 97205, México
2 Instituto Politécnico Nacional-Centro de Biotecnología Genómica, Blvd Del Maestro esq. Elias Piña, Reynosa, Tamaulipas, 88710, México
* Corresponding Authors: José Narváez-Zapata. Email: ; Manuel Martínez-Estévez. Email:
# Both contribute equally
Phyton-International Journal of Experimental Botany 2024, 93(2), 151-183. https://doi.org/10.32604/phyton.2023.046943
Received 20 October 2023; Accepted 02 December 2023; Issue published 27 February 2024
Abstract
Habanero pepper (Capsicum chinense Jacq.) is a crop of economic relevance in the Peninsula of Yucatan. Its fruits have a high level of capsaicinoids compared to peppers grown in other regions of the world, which gives them industrial importance. Soil is an important factor that affects pepper development, nutritional quality, and capsaicinoid content. However, the effect of soil type on fruit development and capsaicinoid metabolism has been little understood. This work aimed to compare the effect of soils with contrasting characteristics, black soil (BS) and red soil (RS), on the expression of genes related to the development of fruits, and capsaicinoid synthesis using a transcriptomic analysis of the habanero pepper fruits. Plants growing in RS had bigger fruits and higher expression of genes related to floral development, fruit abscission, and softening which suggests that RS stimulates fruit development from early stages until maturation stages. Fruits from plants growing in BS had enrichment in metabolic pathways related to growth, sugars, and photosynthesis. Besides, these fruits had higher capsaicinoid accumulation at 25 days post-anthesis, and higher expression of genes related to the branched-chain amino acids metabolism (ketol-acid reductisomerase KARI), pentose phosphate pathway and production of NADPH (glucose-6-phosphate-1-dehydrogenase G6PDH), and proteasome and vesicular traffic in cells (26S proteasome regulatory subunit T4 RPT4), which suggest that BS is better in the early stimulation of pathways related to the nutritional quality and capsaicinoid metabolism in the fruits.Keywords
Supplementary Material
Supplementary Material FileNomenclature
| AT3 | Acyltransferase protein |
| BP | Biological process |
| BS | Black soil |
| CC | Cellular component |
| CDS | Coding sequence |
| CS | Capsaicin synthase |
| DAA | Days post-anthesis |
| DGE | Differential gene expression |
| DPT | Days post-transplant |
| DW | Dry weight of fruit |
| FDR | False discovery rate |
| GSEA | Gene set expression analysis |
| LHC | Light-harvesting complex |
| LFC | log2FoldChange |
| MF | Molecular function |
| ORF | Open reading frame |
| RS | Red soil |
| TLC | Thin layer chromatography |
| TPM | Transcripts per million |
The genus Capsicum is a group of plants belonging to the Solanaceae family together with relevant crop species like tomato and tobacco [1]. Capsicum species, also named peppers, produce fruits that are characterized by the pungency sensation that they produce when consumed. They are consumed in fresh or as condiments. Also, peppers have a wide range of industrial and pharmaceutical applications due to the variety of metabolites they produce [2,3]. They are used in the elaboration of a wide variety of products like drugs, cosmetics, pigments, colorants, paints, oleoresins, and antimicrobials [2]. Habanero pepper (C. chinense Jacq.) is a crop with economic and cultural relevance in the Peninsula of Yucatan in the southeast of Mexico [4]. The environmental conditions of this region allowed the development of varieties with particular organoleptic traits, which gave them the denomination of origin “Habanero pepper of the Peninsula of Yucatan” in the year 2010 [5]. The growth environment significantly influences the development, yield, and metabolic composition of habanero pepper fruits, ultimately determining their quality [6–9].
Soils with distinct physical and microbial properties exert an impact on fruit production, as well as the nutritional and metabolic composition of the fruits [6]. In Yucatan, there are different types of soils, like the black soils (BS, Boxlu’um) and the red soils (RS, Haylu’um), that have different physicochemical properties like the texture, organic matter, nitrogen, phosphorus, carbonates, and calcium contents, which tend to be higher in the BS [10,11]. Also, they have different bacterial and fungal compositions [11]. Habanero pepper plants grown in RS and BS have different contents of metabolites, like polyphenols, carotenes, and A, C, and E vitamins in stems, leaves, and peduncles [12]. When the fruit development and production were compared in habanero pepper plants growing in these soils that were irrigated with high conductivity water, it was found that the plants growing in RS were more efficient in the use of some soil nutrients, like nitrogen, phosphorus, and potassium, and produced more and bigger fruits. On the other hand, the plants growing in BS presented a higher retention of sodium and other nutrients, affecting the efficiency in the use of them [8].
Capsaicinoids, a group of metabolites found in pepper fruits, are responsible for providing a distinct pungent sensation upon consumption. These metabolites find applications in the production of condiments, repellents, and a wide range of industrial, medicinal, and pharmaceutical products, owing to their notable bioactive properties [13]. They are composed of a vanillylamine moiety and an acyl moiety composed of 9 to 11 carbons bonded to the vanillylamine by an amide group [14]. The hydrocarbon structure of the acyl moiety gives the identity of the capsaicinoid, with the capsaicin and the dihydrocapsaicin composing about 90% of the total capsaicinoid content [3]. The metabolic pathway of capsaicinoid biosynthesis is composed of the conjunction of the phenylpropanoid pathway that gives the vanillylamine, and the metabolic pathway of the branched-chain aminoacids and fatty acids, that give the acyl group [15], and then, both are condensed in a reaction catalyzed by the capsaicin synthase (CS) acyltransferase protein (AT3) [16]. The capsaicinoids are synthesized and accumulated in epidermic cells vacuoles of the interlocular septum of the placental tissue of the fruits [17]. Also, there is evidence of synthesis and accumulation of capsaicinoids in the pericarp of the fruits, mainly in the hottest varieties of C. chinense like ‘Trinidad Moruga Scorpion Yellow’ [18].
The type of soil affects the capsaicinoid contents in C. chinense fruits, and this is related to soil properties, like nitrogen, phosphorus, potassium contents, and microbial activity [6]. The capsaicinoid content in the fruits was higher if the plants grew in RS instead of BS of Yucatán, comparing the same development stage of the fruits [19]. It is known that factors like light, temperature, mechanical wounding, drought, salinity, nitrogen, and phytohormone content affect the capsaicinoid content in the fruits, and the expression of genes related to the capsaicinoid biosynthesis [20–23]. The study of the genes related to capsaicinoid metabolism is relevant due it gives information for the design of crop or breeding strategies that allow the control of the capsaicinoid content in the fruits by the modulation of the genes that control the differences in the content of capsaicinoids in contrasting environmental conditions. Genes like AT3 (PUN1), the putative aminotransferase (pAMT), and transcriptional factor genes like MYB, are key points that control the content of capsaicinoids [24–26].
Although soil characteristics play a crucial role in fruit development and capsaicinoid content, there is limited research on the influence of soil type on gene expression related to capsaicinoid metabolism. Existing studies often focus on individual genes, such as PUN1, but comprehensive investigations are scarce [27]. In light of the significant impact of soil type on fruit development and metabolism, this study conducted transcriptomic analysis to identify genes involved in or regulating the observed differences in fruit development and capsaicinoid production in habanero pepper cultivated in two contrasting soils of Yucatán, namely RS and BS.
Habanero pepper seedlings (C. chinense Jacq. ‘Jaguar’) with ≈45 d of germination were used. They were transplanted into polyethylene bags of 12 kg of capacity containing BS or RS. There were 15 seedlings for each type of soil. The seedlings were cultivated in greenhouse conditions for 150 days post-transplant (DPT). Information on the average conditions of temperature, relative humidity, and intense light inside the greenhouse during the 5 months of the experiment is in Table S1. The values are averages of the house means of one of the measured parameters. Plants were distributed in blocks of three plants by type of soil, alternating blocks of soil type. The characteristics of the soils used for the experiments are described extensively in Oney-Montalvo et al. [28].
Plants were irrigated with 0.5–1.5 L of water. They were fertilized using Triple-18 fertilizer (0.7713 g/plant) two days per week. Also, micronutrients were applied to leaves, one day per week, using Bayfolan Forte. In the floral period, a growth regulator containing phytohormones (Biozyme) was applied every 15 d.
2.2 Analysis of Flowers and Fruit Production
Flowers were labeled when 50% of the plants had started flowering period. Flowers were labeled on anthesis day during 60 d and the flowers and fruit numbers were daily registered. The total fruit number per plant and the total number of fruits of 45 days post-anthesis (DAA) per plant were determined. Also, the fruits of 45 DAA were divided into four categories according to their length (1–1.99 cm, 2–2.99 cm, 3–3.99 cm, and ≥4 cm), to analyze in detail the effect of the soil type in the growth of the fruits, and the total number of fruits per plant, total fresh weight of fruits per plant and average weigh of fruit per plant were determined in each category. ANOVA was carried out using Tukey analysis (p ≤ 0.05) with n = 8 plants for the analysis of number of flowers and total fruits, n = 13 plants for the flower abortion, and n = 7 plants for the 45 DAA fruit analysis.
2.3 Capsaicinoid Determination
Fruits of 25, 45, and 60 DAA were frozen in liquid nitrogen and stored at −80°C. These times were selected due to what has been observed that corresponds to different development stages of the fruits and have different behavior in the changes of the capsaicinoid content in habanero pepper [9]. We wanted to compare the content of capsaicinoids between soils and to analyze if there are significant differences that could be explored in the transcriptomic analysis. Two pools of five fruits randomly selected for each maturation stage and soil type were ground with a blender in liquid nitrogen, and a lyophilized sample (0.15 g) was used to extract the capsaicinoids in 20 mL anhydride ethanol according to Córdova-Alvarado et al. [29], resuspending the extracts in ethanol grade HPLC. The capsaicinoids were determined by thin-layer chromatography (TLC) according to Córdova-Alvarado et al. [29] by loading 5 μL of resuspended extract as a band of 5.0 mm length with a Linomat 5 equip (CAMAG) from a distance of 10 mm above the bottom edge of the plates, and developing the chromatography without solvent saturation until reaching 7.0 cm from the bottom of the plates. Then the plates were set in a CAMAG TLC Scanner 4 equipment and the capsaicinoids were quantified by densitometry at 282 nm. For the extracts of fruits of 25 and 45 DAA, the determination was made in ten samples (n = 10), and for the extracts of fruits of 60 DAA, the determination was made in twelve samples (n = 12). In parallel, standards of capsaicin were used in a range of 0.25 to 3.0 µg to construct a calibration curve. For the statistical analysis, two-way ANOVA was carried out using Tukey analysis (p ≤ 0.05) with the factors: soil type (BS and RS) and development stage of the fruit (25, 45, and 60 DAA) with n = 10 samples in fruits of 25 and 45 DAA, and n = 12 samples in fruits of 60 DAA.
2.4 RNA Extraction, Library Construction, and Sequencing
We wanted to evaluate the effect of the soil type in the expression of genes of the capsaicinoid synthesis pathway, we selected the fruits of 25 DAA (intermediate development stage) because it has been reported that the fruits in intermediate development stages are very active in the capsaicinoid synthesis, and the genes of the corresponded pathway have high levels of expression [15,22]. Two pools of five fruits of 25 DAA of each type of soil were used to extract the RNA to obtain two biological replicates per soil type (BS1 and BS2 for black soil and RS1 and RS2 for red soil. The fruits, previously grounded, were prepared to RNA-Seq using the Illumina sequencing platform with the services of Macrogen (South Korea).
Total RNA was extracted (0.1 g) using the kit RNeasy Plant Minikit (Qiagen, USA) following the manufacturer’s instructions. The RNA extracted was treated with the kit Turbo DNase (Invitrogen, USA) following the manufacturer’s instructions. RNA integrity was verified by electrophoresis in agarose gel (1.5%) in TAE 1X. RNA was quantified in a NanoDrop 2000 spectrophotometer (ThermoScientific, USA). The RNA was dried with the kit RNAstable/RNAstable LD (Biomatrica, USA) following the manufacturer protocol stored in bags with silica gel (Biomatrica, USA), and then sent to Macrogen for sequencing. The RNA integrity number (RIN) was acceptable for all the samples (8.7 for BS1 and BS2 and 8.2 for RS1 and RS2).
The four RNA samples were prepared for RNA-Seq using the Illumina TruSeq Stranded mRNA LT Sample Prep Kit for library construction of each biological replicate (BS1, BS2, RS1, and RS2), obtaining a total of four libraries. Briefly, the mRNA was purified from the total RNA using poly-T oligo-attached magnetic beads from each biological replicate. The mRNA was randomly fragmented, and then the cleaved RNA fragments were reverse transcribed to obtain the first cDNA strands using random hexamer primers. Then, the second cDNA strands were synthesized using RNase H and DNA Polymerase I. Adapters were ligated onto both ends of the cDNA fragments preparing them for hybridization onto the flow cell. Later, the fragments were amplified by PCR, and those with insert sizes between 200–400 bp were sequenced with the Illumina platform from both ends to obtain 101 nt paired ends reads. The raw reads were deposited in the NCBI database (BioProject under the accession number PRJNA983842).
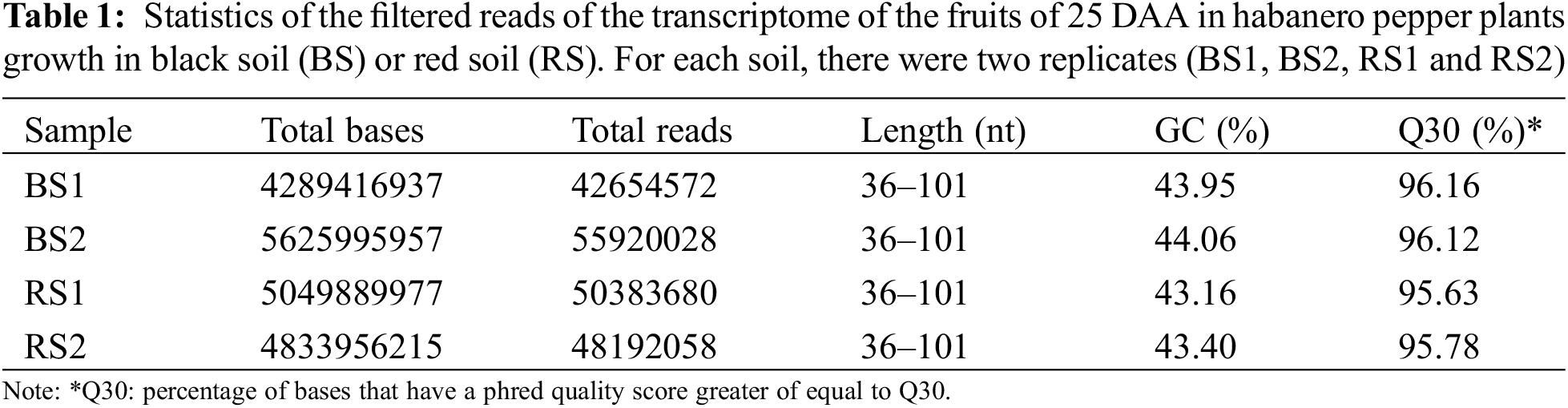
2.5 Reads Quality Filtering, de novo Assembling and Remapping
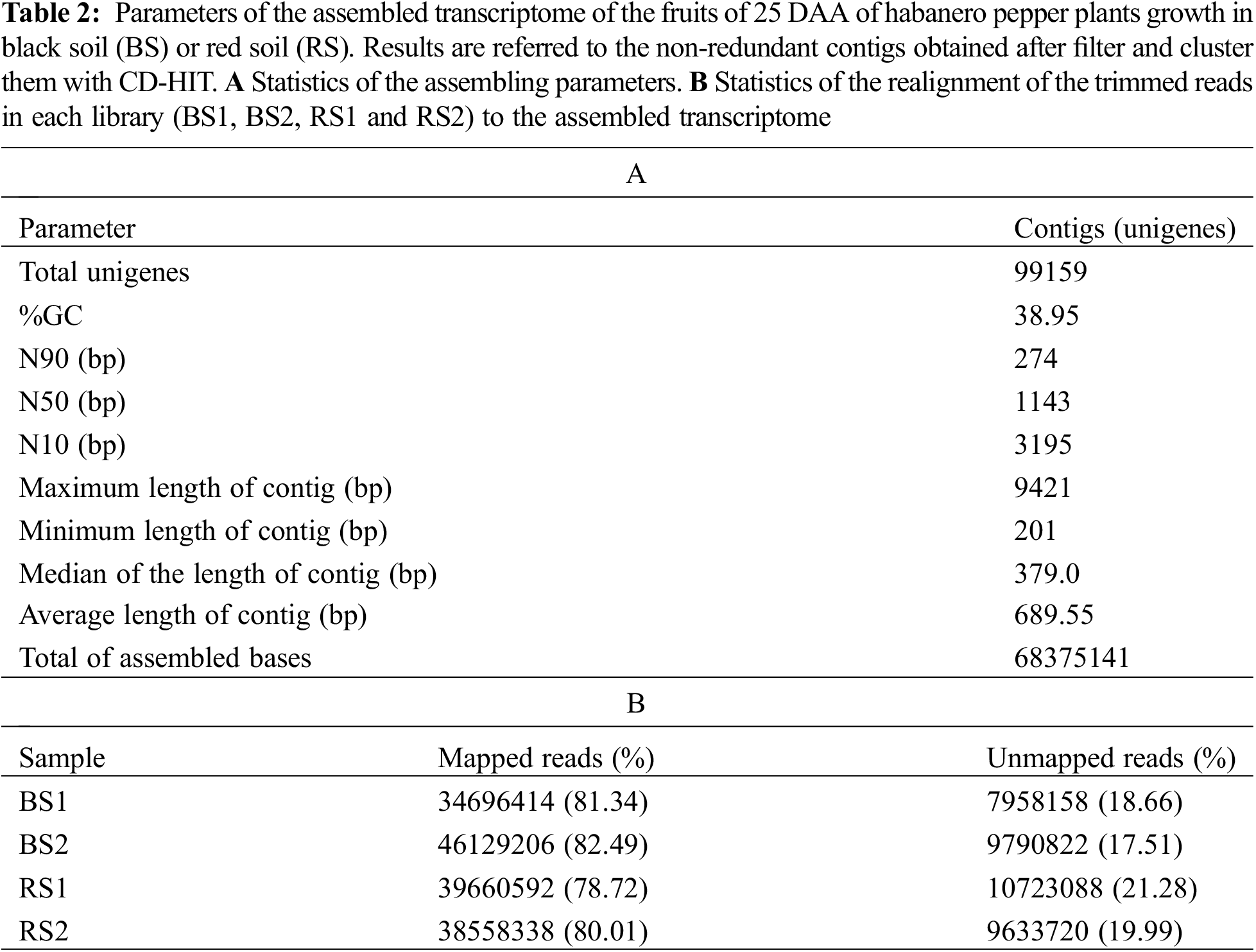
The raw reads were filtered using the software Trimmomatic v.0.38 [30] to eliminate adapter sequences and bases with a quality score of Q < 3 from the ends of the fragments. Also, we used a sliding window of four bases and a mean quality of Q15 to trim bases of low quality from the reads. Afterward, reads with lengths shorter than 36 bp were dropped. With these parameters, trimmed reads of high quality were obtained in the four libraries (Table 1). Statistics of the quality of the raw and trimmed data were obtained with the software FastQC v.0.11.7 [31]. Assembly of the trimmed data was performed using the software Trinity (version trinityrnaseq_r20140717) with default parameters [32] obtaining an acceptable assembled transcriptome statistic (Table 2A). Remapping of the trimmed reads to the assembled contigs was performed using the program Bowtie v.1.1.2 [33]. Finally, the contigs were filtered and clustered into non-redundant representative transcripts (unigenes) using the software CD-HIT v.4.6 [34].
2.6 Open Reading Frames Prediction, Functional Annotation, and Abundance Estimation
The prediction of the open reading frames (ORFs) of at least 100 aa in length was made using the program TransDecoder v.3.0.1 [35] to identify candidate coding regions in the unigenes library.
To estimate the abundance of the unigenes in each sample (BS1, BS2, RS1, and RS2), trimmed reads of each sample were aligned to the assembled unigenes library with Bowtie v.1.1.2, and then, the abundance of the unigenes was calculated in each sample using the software RSEM v.1.3.1 [36], expressing the result in read counts, fragments per kilobase of transcript per million mapped reads (FPKM) and transcripts per million (TPM).
The functional annotation of the unigenes library was performed against the public databases of Gene Ontology (GO, version v20180319), Kyoto Encyclopedia of Genes and Genomes (KEGG KO_EUK, version 20220103), and against the nucleotide (NT, version 20180116) and non-redundant protein (NR, version 20180503) databases of the NCBI. For functional annotation of unigenes on the NT database of the NCBI, we carried out BLASTN (BLAST v.2.9.0) with an e-value cut-off of 1.0e-5. For functional annotation on the NR database of the NCBI, we carried out BLASTX with the software DIAMOND v.0.9.21 [37] with an e-value cut-off of 1.0e-5. For functional annotation on the GO database, we used BLASTX with DIAMOND v.0.9.21 and an e-value cut-off of 1.0e-5. The GO terms of the main categories: biological process (BP), cellular component (CC), and molecular function (MF) were listed. For functional annotation on the KEGG database, we used BLASTX with DIAMOND v.0.9.21 and an e-value cut-off of 1.0e-5. A bi-directional best-hit method was performed to search against the KEGG database to obtain the KO (reference pathway) number of the KEGG annotation.
2.7 Global Differential Expression and Enrichment Metabolic Pathways Analysis
To compare the effect of the soil type in the expression of the genes of the habanero pepper transcriptome, a differential gene expression (DGE) analysis was performed using the program DESeq2 [38] in R v.4.2.0. The comparison was established as RS vs. BS, using two biological replicates for each soil type (RS1 and RS2 vs. BS1 and BS2), using as input data the read counts previously determined in the section of abundance estimation of the unigenes to obtain the log2FoldChange (LFC), p-value and adjusted p-value. A cut-off of LFC ≥ 1.5 was established for the highest differentially expressed genes in RS, and a cut-off of LFC ≤ −1.5 was established for the highest differentially expressed genes in BS. A p-value < 0.00835 was established as a statistical significance value. Additionally, a Benjamini-Hochberg (BH) test was performed to compute the false discovery rate (FDR), and an adjusted p-value < 0.1 was established as a confidence limit for statistical significance [39]. A volcano diagram was used to represent the expression of the genes with LFC values in the x-axe and the −log10 p-value in the y-axe using the ggplot2 package [40] of R. Afterwards, a gene set expression analysis (GSEA) was performed in the transcriptome to obtain enrichment metabolic pathways which were related with the development, quality and the capsaicinoid content in the fruits by the effect of the soil type. First, the read counts of all the unigenes annotated against the same KEGG term were summed for each sample (RS1, RS2, BS1, and BS2), and the sums were used as input data to perform a DGE analysis with DESeq2 comparing the conditions RS vs. BS using the two biological replicates (RS1 and RS2 vs. BS1 and BS2). Then, the LFC values were used to perform the GSEA with the software ClusterProfiler v.4.4.4 [41] with the R packages ggplot2, enrich plot [42] and pathview [43]. GSEA was run with the following parameters: 1000 permutations, a minimum and maximum gene set the size of 3 and 800, respectively, a p-value cut-off of 0.5, without adjusted p-value method, and searching against the KEGG pathway database of C. annuum to obtain a dot-plot of enriched terms.
2.8 Individual Differential Expression Analysis of the Genes of the Enriched Metabolic Pathways and the Biosynthetic Capsaicinoid Pathway
To compare the difference in the individual expression of the genes of the enriched metabolic pathways and the genes of the capsaicinoid biosynthetic pathway by the effect of the soil type, representative genes of the different pathways were selected for DGE analysis between the soil types. Two representative regulated genes of each enriched pathway were selected in the C. annuum KEGG database after the GSEA analysis. The coding sequences (CDS) of the genes were used as query sequences to search homologs against the unigenes library of the transcriptome by local BLAST analysis using the program Bioedit and selecting the transcripts sequences with an e-value cut-off of 0.0. As representative sequences in the capsaicinoid pathway, genes of key steps of the pathway were selected according to the literature to analyze if the soil type affected its expression [15,24,26,44–47]. For AT3-D1 and AT3-D2 [44] the CDS of the respective sequences (PHU26684.1 and PHU26685.1) in the genome of C. chinense were selected. For pAMT, the CDS of the mRNA of the gene in C. chinense (AF085149.1) was selected [45]. For the ketoacyl-ACP synthase (KAS) and acyl-ACP thioesterase (FAT) genes, the mRNA of C. chinense (AF085148.1 and AF318288.1, respectively) were selected [46]. For the ketoacyl-ACP reductase (KR) gene, the mRNA of C. annuum (EU616561.1) [47] was selected. Also, a MYB transcription factor gene that regulates the expression of genes of the biosynthetic capsaicinoid pathway was selected [46], using the CDS of the mRNA of C. annuum and C. chinense MYB homologues (MK074771.1 and MF062086.1) [25,46]. The selected sequences were used as a query in a local BLAST analysis against the unigenes library using Bioedit to search homolog transcripts with an e-value cut-off of 0.0. A DGE analysis was performed with the software DESeq2 in R v.4.2.0 comparing the conditions RS vs. BS for each gene and using the read counts obtained in the abundance estimation section as input data. Two biological replicates were used for each soil type (RS1 and RS2 vs. BS1 and BS2). A BH test was performed to calculate the FDR and an adjusted p-value < 0.05 or adjusted p-value < 0.1 were considered limit values for statistical significance. A heat map was constructed (in TPM units) to represent the expression of the transcripts of the genes between BS and RS, using the gplots package [48] and RColorBrewer [49] in R v.4.2.0. The TPM values were calculated by the software RSEM v.1.3.1 with the read count values [36].
2.9 Expression Analysis of AT3 Copies during the Fruit Development of Habanero Pepper Plants in Black Soil and Red Soil
The expression of AT3-D1 and AT3-D2 genes was determined in habanero pepper fruits at 25, 45, and 60 DAA from plants growing in BS and RS. First, total RNA isolation from fruits (set of five fruits per sample), previously grounded with a blender in liquid nitrogen, was performed by using the Plant/Fungi Total RNA Purification kit (NORGEN BIOTEK CORP, Canada) according to the manufacturer’s instructions. Then, ≈3 μg of total RNA was treated with DNase using the Turbo DNA-free kit (Invitrogen, USA). cDNA synthesis was carried out in ≈1.5 μg of RNA using the SuperScript III Reverse Transcriptase (Invitrogen, USA) following the manufacturer’s instructions.
PCR was made using specific primers for AT3-D1 and AT3-D2 designed by aligning the CDS of AT3-D1 (PHU26684.1) and AT3-D2 (PHU26685.1). Specific primers designed were F-AT3D1 5′-ATCTCTTGCAGAGAGCATAGTTTTG-3′ and R-AT3D1 5′-TGGAACTAAAGTTGTTGTCGTATG-3′ for AT3-D1. For AT3-D2 the specific primers were F-AT3D2 5′-GCTCATGCAGAGGGCATAGTTTTT-3′ and R-AT3D2 5′-AGGAATTAGAGTCGTTGTCATCAT-3′. β-tubulin gene of habanero pepper (PHU20966.1) was used as a constitutive control using the specific primers F-Tub 5′-GGAGGGTGAGTGAGCAGTTC-3′ and R-Tub 5′-CTGCTGTCGCATCCTGGTAT-3′. The PCR reaction was made in 12 μL reactions mix containing 0.2 μL of Taq polymerase (Invitrogen, USA), 1 μL (500 ng) of cDNA, and 0.36 μL (9 pmol) of forward and reverse primers. Gene amplifications were done by an initial denaturalization at 95°C at 30 s; 35 cycles of 30 s at 95°C, 30 s at 53.4°C–55°C and 15 s at 72°C; and a final extension of 72°C at 5 min. Aligning temperature was 53.4°C for AT3-D1, 55°C for AT3-D2, and 54.2°C for the β-tubulin control. Finally, the relative expression of each gene was determined by the comparative method between the target gene and the β-tubulin gene as a reference. Gel-analyzer Software (http://www.gelanalyzer.com/) was used for quantification.
3.1 Flower and Fruit Production and Characterization in Habanero Pepper Plant Growth in Two Soil Types
Habanero pepper plants (C. chinense ‘Jaguar’) were cultivated in two soil types: BS and RS, in a cycle of 150 DPT. In a cycle of flowering of 60 d, the number of flowers and fruit production were measured, comparing both soil types (Fig. 1). There was no difference in the flower production, nor the fruit production between soils. The flower production was 305 and 284 flowers in BS and RS, respectively, and the total fruit production was 40 and 35 fruits per plant, respectively. The flower abortion was 85% for the plants in both soil types, so the fruit production was low. This is expected in habanero pepper cultivated in Yucatan, where the environmental temperature is nearly of 40°C in summer, even when C. chinense is tolerant to water deficit [50]. The total fruit production of 45 DAA was not higher than 35 fruits per plant in none of the soils. If the total production is considered, there was no difference in the production parameters between soils (Fig. 1A).
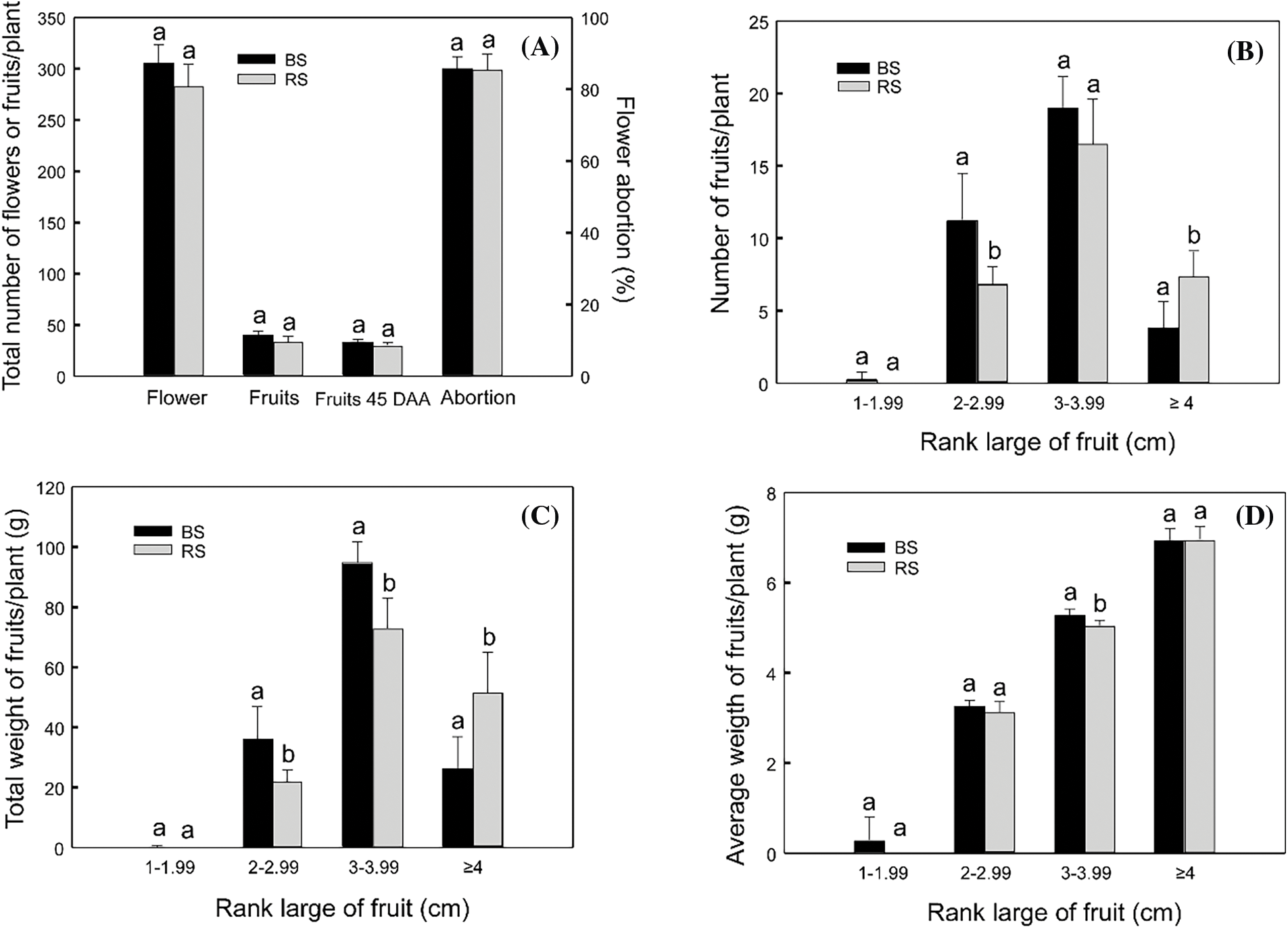
Figure 1: Production parameters of flowers and fruits in habanero pepper plants grown in black soil (BS) or red soil (RS). (A) Total production of flowers, fruits and fruits of 45 DAA in the plants comparing both soil types. (B) Total production of fruits of 45 DAA in ranks of size, comparing both soils. (C) Total weight of fruits of 45 DAA in ranks of size, comparing both soils. (D) Average weight of fruits of 45 DAA in ranks of size, comparing both soils. Different letters indicate significant differences among soils for the same parameter according to Tukey’s test (p ≤ 0.05). Data indicates the average of “n” number of plants (n = 8 for the parameter total flowers and fruits; n = 13 for the parameter of abortion; n = 7 for 45 DAA fruits comparation)
Also, to compare the fruit development, the fruits of 45 DAA, which are starting the maturation stage, were divided into categories according to the fruit large. The production of fruits, total weight, and average weight of the fruits per plant were compared in each rank between soils. The highest number of fruits in this stage is 3–3.99 cm in size, with 19 and 16 fruits per plant for BS and RS, respectively, without differences between soils. The total number of the biggest fruits (≥4 cm in large) was lower (3 and 7 for BS and RS, respectively), however, in RS there was a greater number of biggest fruits than in BS. Also, there was a greater number of fruits between 2–2.99 cm in BS than in RS. There was a similar number of smaller fruits (<2 cm) between soils (Fig. 1B).
When the total weight of the fruits was analyzed (Fig. 1C), it was observed that the fruits between 3–3.99 cm were the heaviest, with 94 g and 73 g of total weight in BS and RS, respectively, and with the fruits in BS with a greater total weight than the fruits of RS. The fruits in the range of 2–2.99 cm also have a greater total weight in BS than in RS, with 36 and 22 g of total weight, respectively. On the contrary, the bigger fruits (≥4 cm) have a greater total weight per plant in RS (51 g) than in BS (26 g). The total weight of the smaller fruits was similar between soils. The tendencies observed in the total weight were like those observed in the total number of fruits (Fig. 1B), which is an expected behavior.
Finally, the average weight of the fruits per plant was analyzed (Fig. 1D). The tendency was an increment in the average weight with the increment in the size of the fruits. The weight was similar in all the ranks, except in the fruits of 3–3.99 cm, in which there was a greater average weight in the fruits of BS (5.30 g) in comparison to RS (5.05 g).
Having all the results into account, for the fruits of 45 DAA, RS is better to harvest a greater number of fruits with bigger size (≥4 cm), and in BS there is a tendency to obtain a major number of smaller fruits (2–2.99 cm). However, both soils have a greater number of fruits of an intermediate size (3–3.99 cm). Also, having into account that the flower abortion, the total flower and fruit production, and the average weights were similar, we conclude that any of both soils is good for cultivating the habanero pepper and harvesting the fruits, even though there is a tendency to obtain bigger fruits in RS. However, this is a little different if we consider that this is part of the fruit development and that the total production is more relevant.
3.2 Capsaicinoid Content in Fruits of Habanero Pepper Plants Growth in Two Soil Types
One of the most important parameters to evaluate the quality of the habanero pepper is the determination of the capsaicinoid content. These compounds add value to the fruits, especially the capsaicin and the dihydrocapsaicin, which represent 90% of the capsaicinoids and give the spicy sensation to the peppers, which is an attribute highly appreciated by consumers and’s a fundamental parameter in the pepper quality [4].
The content of capsaicinoids in the fruits of habanero pepper plants growth in BS and RS were compared between soils and in different development stages (25, 45, and 60 DAA) (Fig. 2). In general, the capsaicinoid content was between 7 to 12 mg of capsaicin/g dry weight of fruit (DW). The analysis shows the evolution in the capsaicinoid content between the two soils during the fruit development and determines if there was a difference in the evolution of the capsaicinoid content and in which soil there was higher capsaicinoid content due to the soil used since in the anterior section we observed that the general production parameters were similar between BS and RS. The difference in the content of capsaicinoid in the peppers by the effect of the type of soil was dependent on the development stage analyzed. At 25 DAA there was a difference in the capsaicinoid content, where BS has a higher content (10.72 mg/g DW), concerning RS (8.92 mg/g DW). At 45 DAA the capsaicinoid content reaches the maximum in both soil types. It was similar between soils, but there was a higher and significant increment in the capsaicinoids in RS (11.48 mg/g) concerning 25 DAA. On the contrary, in BS there was no difference in the capsaicinoid content (11.30 mg/g DW) concerning 25 DAA. At 60 DAA there was a higher capsaicinoid content in RS (9.87 mg/g DW) concerning BS (7.72 mg/g DW), but in both soils, the capsaicinoid content decreased due to the capsaicinoid catabolism in advanced development stages.
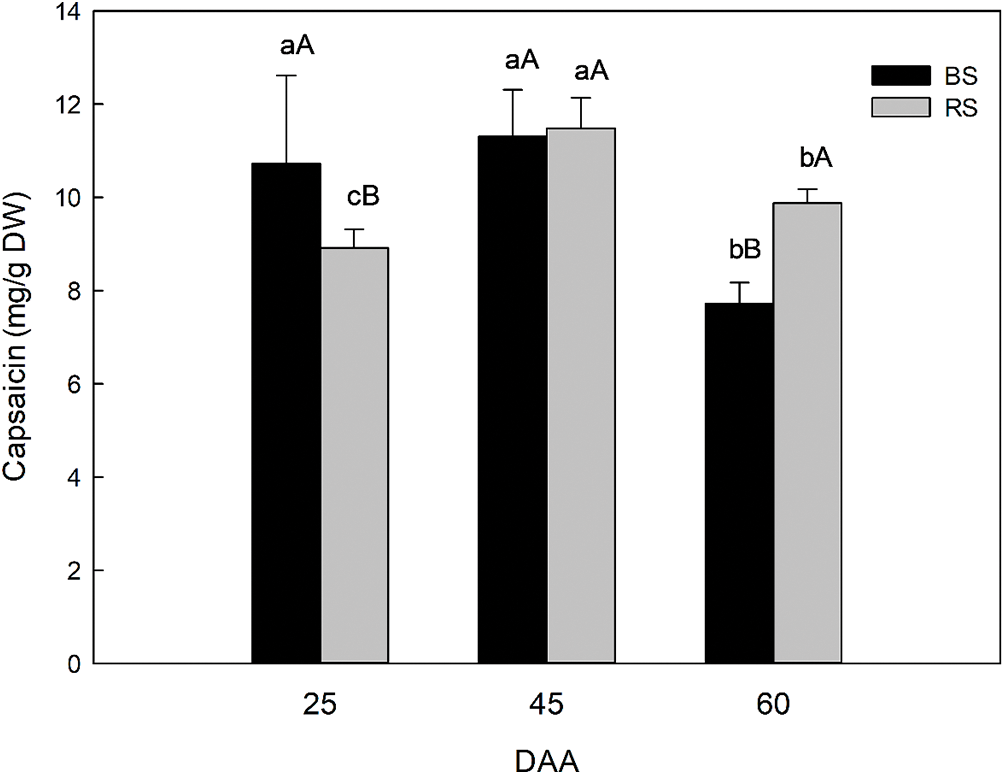
Figure 2: Capsaicinoid content in complete fruits of habanero pepper growth in black soil (BS) or red soil (RS) in different development stages: 25, 45 and 60 days post-anthesis (DAA). The results are expressed as mg of capsaicin/g DW (dry weight of fruit). Different small letters indicate significant difference between development stages in the same soil and different capital letters indicate significant difference between soils in the same development stage, according to Tukey’s test (p ≤ 0.05). Data represents the average of “n” number of samples from capsaicinoid extracts of fruits (n = 10 samples for 25 and 45 DAA and n = 12 samples for 60 DAA)
When we analyzed and compared the evolution in the content of capsaicinoids between soils, the results suggested a difference in the metabolism process during fruit development. In BS, at 25 DAA a maximum was reached without a significant difference to the content at 45 DAA. On the contrary, in RS there was an increment between 25 and 45 DAA, suggesting that the synthesis of capsaicinoids continued predominant in RS until reached 45 DAA. Also, we noted an apparent maximum limit of capsaicinoids at 45 DAA in both soils, and this maximum was reached first in BS at 25 DAA, suggesting that the synthesis was more active in the early development stages in BS. Finally, at 60 DAA, although in both soils there was a decrement in capsaicinoids due to the catabolism predominance, this was smaller in RS concerning 45 DAA, which corresponds to 85.97% concerning the maximum capsaicinoid content at 45 DAA. In comparison in BS at 60 DAA there was a higher decrement respect to the content found at 45 DAA, which corresponds to 68.31% (Fig. 2).
We conclude that if we collect fruits at 45 DAA, the type of soil is not important if we want the capsaicinoid content for industrial applications, and the fruits of 45 DAA can be harvested without waiting for advanced maturation. On the contrary, if we want the complete mature fresh fruit of 60 DAA for consumption or use of the capsaicinoid content in industrial applications, then is better to harvest the fruits in RS, that have a higher capsaicinoid content in complete mature stages.
3.3 General Transcriptomic Analysis in Fruits of Habanero Pepper Plants Growth in Two Soil Types
According to the aim of this study, we wanted to compare the expression of genes of the capsaicinoid biosynthesis pathway between soils. For this purpose, we selected the fruits of 25 DAA for transcriptomic analysis due to it was the difference in the capsaicinoid content between soils in which BS reached a maximum content while in RS the accumulation continued beyond 25 DAA (Fig. 2), suggesting a difference in the level of synthesis metabolism that could be analyzed at transcriptomic level. Also, in intermediate development stages, it is known that the capsaicinoid synthesis is very active in the pepper fruits and the gene expression of the biosynthetic pathway is at high levels [51], so the fruits of 25 DAA were appropriate to compare levels of expression of these genes in the different soils.
For each soil type (RS and BS), two cDNA libraries were generated (biological replicates) having in total four cDNA libraries in paired ends (RS1, RS2, BS1, and BS2). After filtering the raw reads, there were 42.6 a 55.9 million total reads per library. Between 95%–96% of the reads had a base quality of Q30 or higher, and the average quality of the reads was Q36 (Table 1), indicating high quality in the sequences in the four libraries. The reads have 36–101 nt in length, with most of the reads having 101 nt. The size of the libraries was between 4289 to 5625 Mb. The GC composition was similar between samples, which was expected due to the fruits being of the same development stage and pepper variety (Table 1).
After de novo assembling and filtering redundant sequences, a transcriptome of 99159 unigenes was obtained with an average length of 689.55 bp, a composition of 38.95% GC, and 68.37 Mb. The minimum contig length was 201 bp and the maximum was 9421 bp. The N50 was 1143 bp (Table 2A). Similar de novo assembling values have been obtained in transcriptomes of flowers, immature fruits, intermediate mature fruits, and mature fruits in spicy varieties of C. chinense and C. frutescens [52,53]. In the realignment of the reads to the assembled transcriptome, 78%–82% of the reads were mapped in the library of unigenes (Table 2B), which indicates that a lot of the information of the reads is conserved in the assembled transcriptome [54], and allows to use it to search genes of interest, estimate the abundance of the genes and to analyze its levels of expression in the four libraries [32,51,52].
The annotation of the four libraries against the GO database, in the main categories BP, CC, and MF, was made to group the genes in the most abundant GO subcategories (Fig. 3). A total of 30993 unigenes of the transcriptome were annotated against the GO database. The four libraries had a similar proportion of the dominant subcategories. In the BP category, the most abundant subcategories were “cellular process” (15910 unigenes), “metabolic process” (13790), “biological regulation” (7048 unigenes) and “response to stimulus” (5345 unigenes). In CC the most abundant subcategories were “cell part” (24747 unigenes), “organelle” (15865 unigenes), “membrane” (6602 unigenes) and “organelle part” (6304 unigenes). In MF, “catalytic activity” and “binding” (14398 and 12176 unigenes) were the subcategories with more proportion of genes, followed by “transcription regulator activity” (1892 unigenes) and “transporter activity” (1878 unigenes) (Fig. 3).
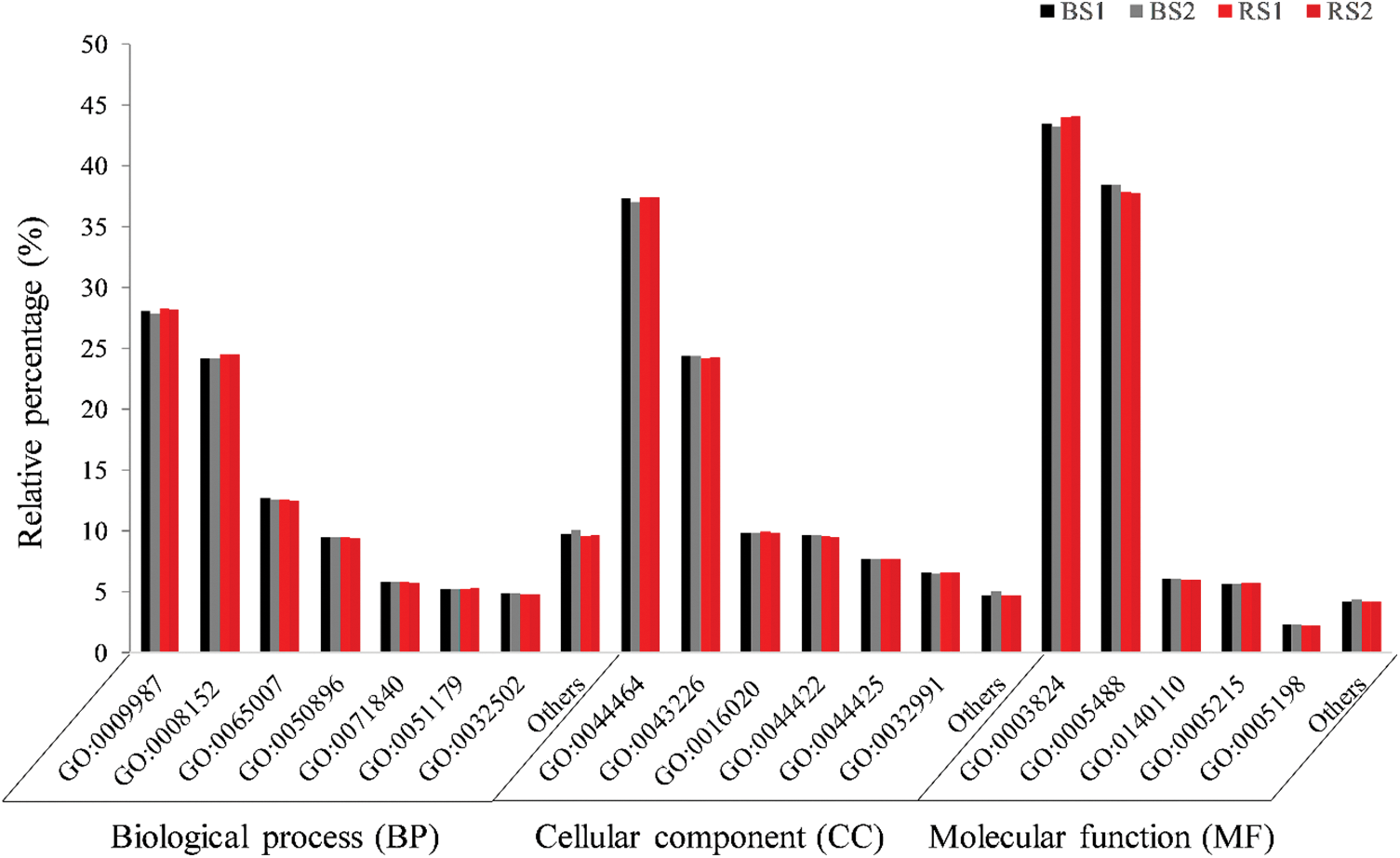
Figure 3: Analysis of the main functional categories GO of unigenes of the four libraries of the transcriptome in the fruits of habanero pepper growth in black soil (BS) or red soil (RS). The relative percentage indicates the number of unigenes in the same main functional category (BP, CC, MF) in each replicate (BS1, BS2, RS1 or RS2). In “others” the subcategories with less than 3% (BP or CC) or less than 1% (MF) of unigenes. GO: 0009987: cellular process, GO: 0008152: metabolic process, GO: 0065007: biological regulation, GO: 0050896: response to stimulus, GO: 0071840: cellular component organization or biogenesis, GO: 0051179: localization, GO: 0032502: developmental process, GO: 0044464: cell part, GO: 0043226: organelle, GO: 0016020: membrane, GO: 0044422: organelle part, GO: 0044425: membrane part, GO: 0032991: protein-containing complex, GO: 0003824: catalytic activity, GO: 0005488: binding, GO: 0140110: transcription regulator activity, GO: 0005215: transporter activity, GO: 0005198: structural molecule activity
3.4 Global Expression Analysis and Analysis of Enriched Metabolic Pathways in Fruits of Habanero Pepper Plants Growth in Two Soil Types
To compare the effect of the soil type in the transcriptome of habanero pepper fruits, a DGE analysis was performed comparing the conditions RS vs. BS. We used the trimmed reads libraries having two biological replicates for soil type (RS1 and RS2 vs. BS1 and BS2) and the assembled unigenes library estimated the abundance of the transcripts in each sample (as reads counts), and then the DGE was performed in the software DESeq2. Of the 99159 unigenes, 6292 had 0 counts in the four libraries and were discarded for further analysis. The other 92867 unigenes were analyzed with a volcano diagram (Fig. 4A), where the comparison of the expression was in function of the LFC value, which shows how much a gene is expressed in a soil concerning the other, and the p-value, which express the statistical significance of the comparison [55]. Due to the comparison performed as RS against BS, the positive values of LFC represent the genes more expressed in fruits of RS, and the negative values represent the genes more expressed in fruits of BS. A total of 533 unigenes had a differential expression between soils, 83 unigenes had higher expression in BS (LFC ≤ −1.5), and 450 unigenes had higher expression in RS (LFC ≥ 1.5) (Fig. 4A). Between the genes with higher expression in RS, there were F-box proteins, extensins, proline-rich proteoglycan, polyubiquitin-like protein, BURP domain-like protein, leucine-rich repeat receptor-like protein kinase, protein mago nashi homolog, calmodulin-lysine N-methyltransferase and 3-ketoacyl-CoA-synthase (Fig. 4A and Table S2a).
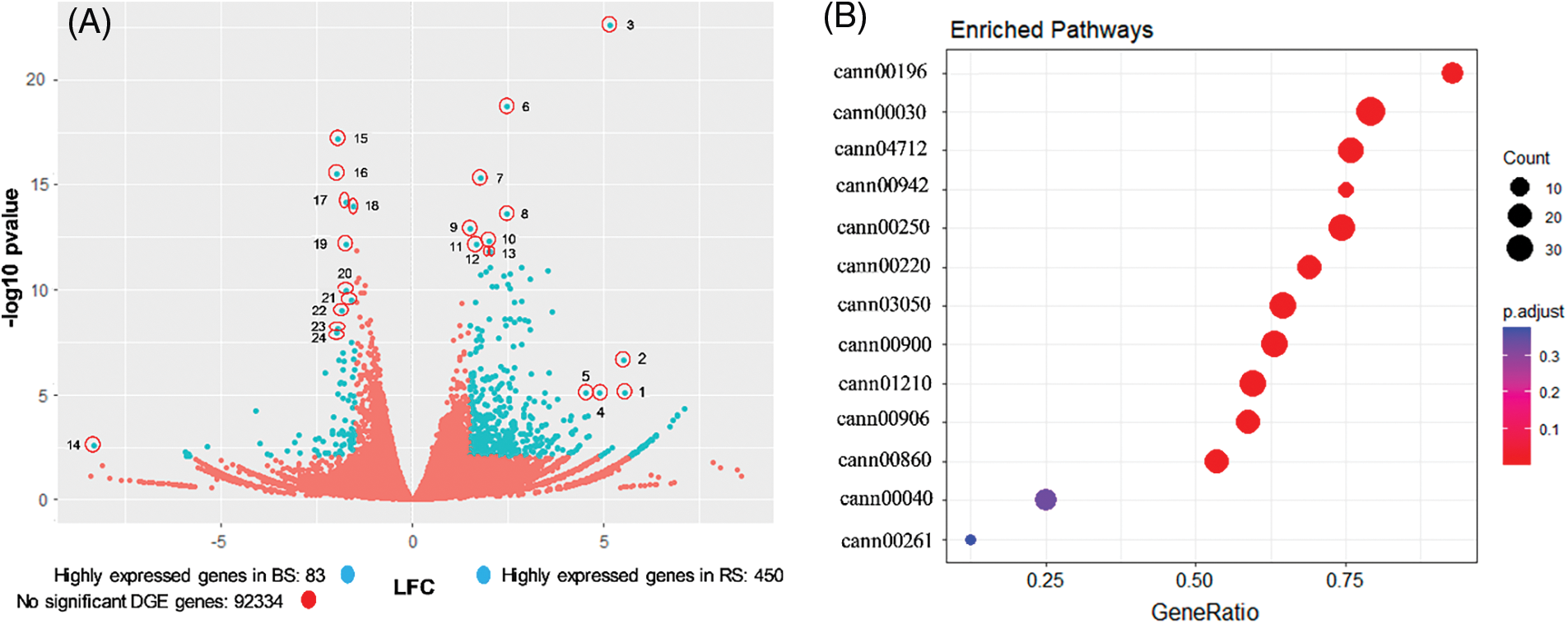
Figure 4: Global analysis of differential gene expression (DGE) analysis and enriched metabolic pathways in the transcriptome of fruits of 25 DAA of habanero pepper growth in black soil (BS) or red soil (RS). (A) Volcano plot that represents the global DGE analysis comparing RS against BS. The magnitude of the comparison log2FoldChange (LFC) and the statistical confidence of the comparison (p-value) are related in the volcano plot. In total, 92867 unigenes were represented (blue and red circles). The blue circles represent those unigenes with higher expression in RS (LFC ≥ 1.5), or BS (LFC ≤ −1.5) with a p-value < 0.00835. The red circle represents those genes without differential expression, according to the selected criteria. The outliers genes selected are signaled into a red circle. The key used (number) correspond to the gene list in the Table S2. (B) Dot-plot that represents the enriched metabolic pathways obtained by a Gene set expression analysis (GSEA) comparing RS against BS. The circle size represents the number of genes associated with the metabolic pathway in a soil type (counts) and the color represents the statistical confidence (p.adjust). The x-axis represents the proportion of genes annotated in that pathway in a soil against the total of genes annotated in the pathway (geneRatio). The y-axis shows the metabolic pathways by its KEGG code. cann00196: photosynthesis-antenna proteins, cann00030: pentose phosphate pathway, cann04712: circadian rhythm-plant, cann00942: anthocyanin biosynthesis, cann00250: alanine, aspartate and glutamate metabolism, cann00220: arginine biosynthesis, cann03050: proteasome, cann00900: terpenoid backbone biosynthesis, cann01210: 2-oxocarboxylix acid metabolism, cann00906: carotenoid biosynthesis, cann00860: porphyrin metabolism, cann00040: pentose and glucuronate interconversions, cann00261: monobactam biosynthesis. The DGE analysis was performed with DESeq2 and the GSEA with ClusterProfiler v4.4.4. Two biological replicates for each soil type (BS1, BS2, RS1 and RS2) were used to perform both analyses
Between the genes with higher expression in BS, there were orthologues of the RuBisCO, chlorophyll-binding proteins, BREVIS RADIX-like, hypothetical proteins of the mitochondrion, albumin-like protein, and RING-H2 finger-like protein (Fig. 4A and Table S2b). We noted a particularly high expression of genes related to photosynthesis and antenna proteins in BS, that was not found in RS, and we concluded that BS is better for activating the expression of the photosynthetic system, which could be favorable for the quality of the fruits.
Posteriorly, the genes were analyzed in sets in function of the metabolic pathways to find metabolic pathways that were influenced by the type of soil. This gene-set analysis has the advantage of analyzing the behavior of the genes of large databases grouping them in categories or sets that allow to analyze tendencies within these categories and that may have a biological significance [56]. In this case, we were interested in finding in which soil there were more represented (enriched), metabolic pathways related to the fruit development, quality, and its capsaicinoid content, which would suggest which soil can be favorable for those metabolic pathways in a transcriptomic level.
The enrichment analysis of metabolic pathways was made taking into account the annotation of the unigenes by its KEGG terms. Of the 99159 unigenes, 42936 were annotated within the KEGG database. A GSEA analysis was performed with a previous DGE analysis, in which all the reads of the contigs annotated within the same KEGG term were summed in each library (RS1, RS2, BS1, and BS2) and comparing the condition RS vs. BS, which allowed us to find metabolic pathways overrepresented in a soil type by the LFC values [41,57]. A total of 11 enriched metabolic pathways were found, with a high statistical significance (p-value) (Fig. 4B). The enriched pathways were photosynthesis-antenna proteins; pentose phosphate pathway; circadian rhythm; anthocyanin biosynthesis; alanine, aspartate, and glutamate metabolism; arginine biosynthesis; proteasome; terpenoid backbone biosynthesis; 2-oxocarboxylic acid metabolism; carotenoid biosynthesis and porphyrin metabolism. The metabolic pathway with the higher number of genes was the pentose phosphate pathway, and the anthocyanin biosynthesis had the lesser number of genes. The pathway with the higher proportion of genes concerning the total number of genes annotated in the pathway (GeneRatio) was the group of the photosynthesis-antenna proteins, and the pathway with minor GeneRatio was the porphyrin metabolism. Other pathways, the pentose and glucuronate interconversions, and monobactam biosynthesis, had low statistical significance and were discarded for further analysis (Fig. 4B).
In the case of RS, the enrichment was found only in the anthocyanin biosynthesis pathway. In BS, most of the pathways were overrepresented. These pathways were related to the photosynthesis (antenna proteins, porphyrin metabolism), sugar metabolism and reduction equivalents (pentose phosphate pathway), biosynthesis and metabolism of secondary metabolites, nitrogen and amino acids (anthocyanins, carotenoids, terpenoids backbone, arginine, alanine, aspartate, glutamate, 2-oxocarboxylic acid) and cellular function related with the protein degradation (proteasome) and circadian rhythm (Fig. 4B). Concerning the capsaicinoid metabolism, the 2-oxocarboxylic acid metabolism is related to the capsaicinoid pathway via the synthesis of the amino acids precursors valine and leucine [15]. However, we did not find an enriched pathway directly related to the capsaicinoids, like the phenylpropanoids pathway or the synthesis of branched-chain fatty acids. This suggests that these pathways were not overrepresented by a particular soil.
The results obtained from the general DGE analysis and the enriched pathways analysis led us to conclude that BS is particularly favorable for a high expression of the photosynthesis genes and the antenna proteins. Also, an important number of metabolic pathways related to primary and secondary metabolism are overrepresented in BS, which is favorable for the quality and the nutrient content of the fruits of habanero pepper plants growth in this soil, at least, in the transcriptomic level at 25 DAA. Taking into account that the global production of fruits was similar in both soils (Fig. 1A), BS would be better for the habanero pepper fruits to stimulate its photosynthetic system and metabolism (porphyrin, sugars, amino acids, terpenoids, carotenoids) which could lead to better nutritional content.
3.5 Individual Expression Analysis of Genes in Fruits of Habanero Pepper Growth in Two Soil Types
According to the aim of the study, we wanted to compare the expression of genes related to fruit development and the capsaicinoid synthesis between the two soil types. The enriched metabolic pathway analysis shows us that in BS various metabolic pathways related to fruit development were overrepresented, but this only allowed us to analyze the effect of the soil in the pathways as a whole, but not the individual expression of the genes of these pathways. This is important, particularly for regulated or key genes. Also, we did not find an overrepresentation of a pathway related to capsaicinoid metabolism. Therefore, we proceeded to perform a DGE analysis of highly regulated genes of the capsaicinoid metabolism and of genes of the enriched pathways comparing the condition RS vs. BS in a similar way as we did for the global expression analysis.
3.5.1 Individual Expression Analysis of Genes of the Metabolic Enriched Pathways and the Capsaicinoid Biosynthetic Pathway in Fruits of Habanero Pepper Plant Growth in Two Soil Types
Regulated representative genes of the capsaicinoid synthesis pathway and at least two regulated genes of each enriched metabolic pathway of the GSEA were selected (Fig. 4B). The search of the genes was realized using reference genes as a query to search homologs by local blast against the unigenes library of the transcriptome of the habanero pepper fruits (Table 3 and Table S3).

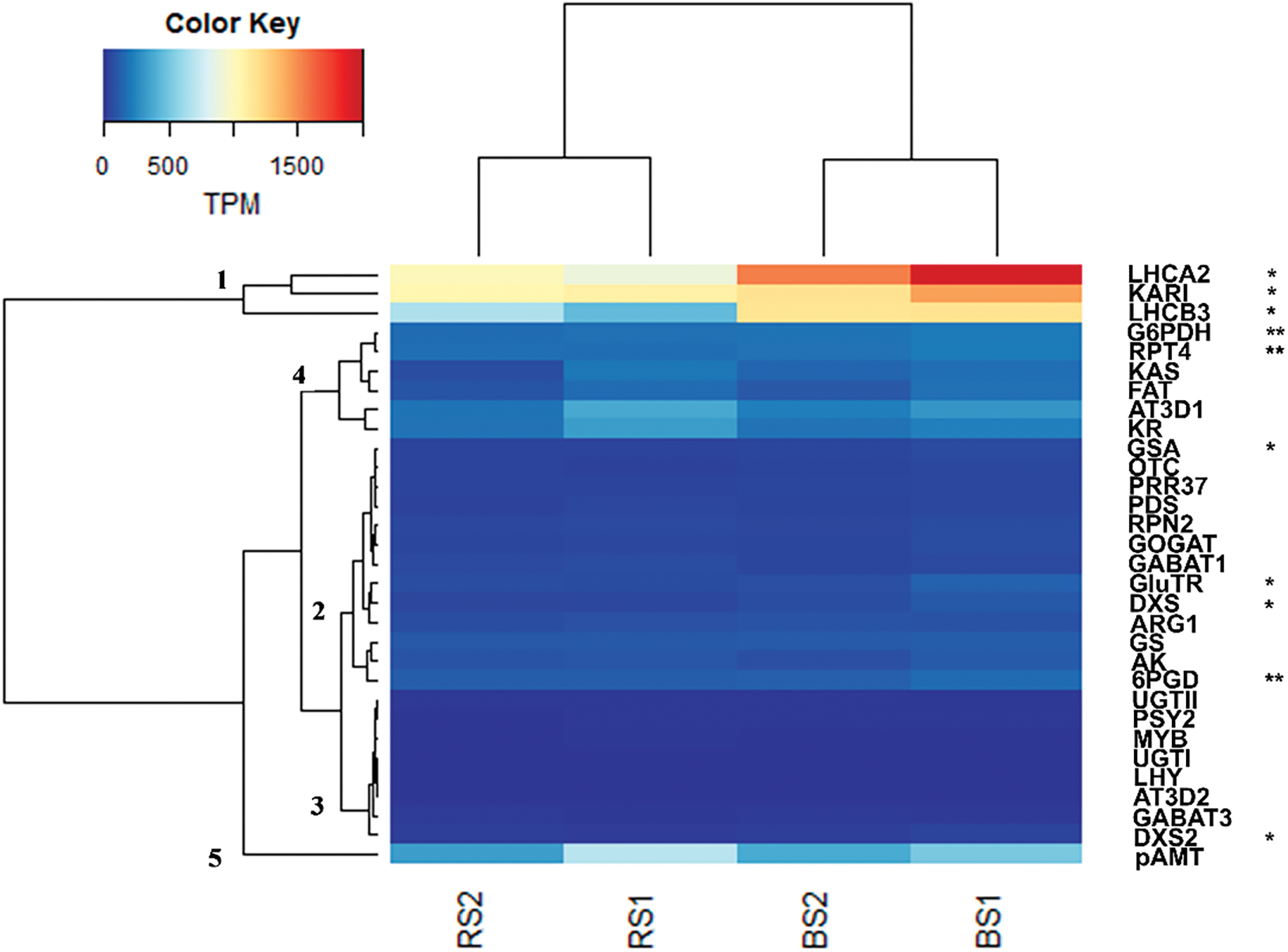
Figure 5: Analysis of the expression of selected genes of the enriched metabolic pathways (Fig. 4B) and of the capsaicinoid biosynthesis metabolism in fruits of 25 DAA of habanero pepper growth in black soil (BS) or red soil (RS). The analysis compares RS against BS. The expression is shown in Transcripts Per Million (TPM). The name of the gene and the number of the clade correspond to the list of the Table 3. Data represents the results of four samples (RS1, RS2, BS1 and BS2) and the statistical analysis was made with DESeq2 having into account the replicates (RS1 and RS2 vs. BS1 and BS2). *indicate genes differentially expressed between soils (adjusted p-value < 0.05), **indicate genes differentially expressed between soils (adjusted p-value < 0.1) according to the DESeq2 analysis
In Fig. 5, the results of the DGE analysis of thirty-one genes are shown for the four samples (BS1, BS2, RS1, RS2) in TPM units. By the expression levels and patterns, the genes were grouped into five principal clades (Fig. 5 and Table 3). Clade 1 included three highly expressed genes, with 450-2000 TPM (Table S3 and Fig. 5). Two of them correspond to genes of the photosynthesis-antenna proteins complex (LHCA2 and LHCB3) (Table 3). The first was a homolog of protein 2 of the light-harvesting complex I (LCHI) and the second corresponded to a homolog of protein 3 of the light-harvesting complex II (LHCII). Also, these genes were more expressed in BS (adjusted p-value < 0.05) (Fig. 5 and Table S3), which shows us that this soil is favorable for a higher expression of genes related to the photosynthesis and of antenna proteins, which was also suggested by the global DGE analysis (Fig. 4A and Table S2b) and analysis of enriched metabolic pathways (Fig. 4B). Also, in this clade a gene of the 2-oxocarboxylix acid metabolism, the ketol-acid reductisomerase (KARI) was more expressed in BS (adjusted p-value < 0.05) (Fig. 5 and Table S3).
Clade 2 included thirteen genes with different levels of expression (38–166 TPM) but with a lower level of expression than clade 1 (Fig. 5 and Table S3). In one subclade we had the genes GSA (glutamate-1-semialdehyde 2,1-aminomutase) of the porphyrin metabolism, OTC (ornithine transcarbamylase) of the metabolism of the arginine, PRR37 (pseudo-response regulator 7) of the circadian rhythm, and PDS (15-cis-phytoene desaturase) of the carotenoid biosynthesis (Table 3). Only GSA was more expressed in BS (adjusted p-value < 0.05) (Fig. 5 and Table S3). Other subclade was included by RPN2 (subunit N2 of the proteasome machinery), GOGAT (glutamate synthase), and GABAT1 (gamma aminobutyrate transaminase 1) (Fig. 5 and Table 3). There was no difference in the expression of those genes between soils. Also, except for PRR37 and PDS, the genes of these two subclades are related or participate in the amino acids metabolism (glutamate) and the nitrogen metabolism. This may suggest a relation in the regulation of its expression. A third subclade included GluTR (glutamyl-tRNA reductase) of the porphyrin metabolism, DXS (1-deoxy-D-xylulose-5-phosphate synthase) of the terpenoid backbone synthesis, and ARG1 (arginase 1) of the arginine metabolism (Table 3 and Fig. 5). GluTR and DXS were more expressed in BS, but there was no difference in the expression of ARG1 by the type of soil (Fig. 5 and Table S3). A fourth subclade included the GS (glutamine synthetase), AK (aspartokinase), and 6PGD (6-phosphogluconate dehydrogenase) of the pentose phosphate pathway (Fig. 5 and Table 3). These genes had a relatively higher expression in clade 2, especially 6PGD (124–166 TPM) (Table S3). Only 6PGD was more expressed in BS (adjusted p-value < 0.1) (Fig. 5). Also, the higher expression of the genes in this subclade may reflect its importance in the metabolic pathways (metabolism of glutamate, aspartate and alanine, 2-oxocarboxylix acid metabolism, and pentose phosphate pathway).
The genes GluTR and GSA, both of the porphyrin metabolism, were more expressed in BS (Fig. 5). This result, together with the results of the individual expression of photosynthesis-antenna proteins genes, the global DGE analysis (RuBisCO and chlorophyll-binding proteins) (Fig. 4A and Table S2b) and the enriched metabolic pathway results (Fig. 4B), suggests that BS is better for stimulating the expression of genes of photosynthetic-related processes in habanero pepper. DXS and 6PDG were also more expressed in BS, suggesting the importance of the selection of soil type for the pathway of the synthesis of terpenoid backbone and the pentose phosphate pathway (Figs. 4B and 5). Although the expression of other genes of clade 2 was not affected by the type of soil, many of these genes related to nitrogen, amino acids, and protein metabolism were grouped in clade 2 (OTC, ARG1, GABAT1, GS, GOGAT, AK, RPN2) suggesting a coregulation of its expression. Also, the corresponded pathways (arginine biosynthesis, aspartate, glutamate, and alanine metabolism, 2-oxocarboxylic acid metabolism and proteasome) were enriched in BS (Fig. 4B). These results suggests that BS could be better in habanero pepper fruits to stimulate at a transcriptome level the metabolism of amino acids and nitrogen.
Clade 3 had eight genes of low expression (0.5–43 TPM) (Table S3) of different metabolic pathways (Fig. 5 and Table 3). The genes were UGT1 and UGTII (UDP-glycosyltransferase), related to anthocyanin biosynthesis, PSY2 (phytoene synthase 2) related to the carotenoid biosynthesis, MYB (transcription factor MYB), LHY (LATE ELONGATED HYPOCOTYL), AT3-D2 (acyltransferase homolog related to AT3-D1), GABAT3 (Gamma aminobutyrate transaminase 3) related to the glutamate, aspartate, and alanine metabolism, and DXS2 (1-deoxy-D-xylulose-5-phosphate synthase 2) of the terpenoid backbone biosynthesis. In this clade, only DXS2 has a higher expression in BS (adjusted p-value < 0.05) (Fig. 5 and Table S3). The other genes were not differentially expressed, although the pathways were enriched when the soils were compared, suggesting that the pathways as a whole are influenced by the soil type (Fig. 4B).
Most of the genes of clade 3 belonged to pathways that also have genes in clades 2 and 4. This was observed in PSY2/PDS, LHY/PRR37, GABAT1/GABAT3, DXS/DXS2 and AT3-D1/AT3-D2 (Fig. 5 and Table 3). These pairs of genes have different levels of expression (Table S3). This may indicate a different regulation of these genes in the same pathway. Also, having into account that the genes of the terpenoid backbone biosynthesis pathway, DXS2, and DXS (in the clade 2), were highly expressed in BS, and the terpenoid backbone biosynthesis pathway was enriched in BS (Fig. 4B) suggests that this soil could be selected if we are interested in stimulate the metabolism of terpenoids for a better fruit quality.
In clades 4 and 5 there were six genes and one gene, respectively (Fig. 5). The levels of expression of these genes were, in general, higher than those of clades 2 and 3, but lower than those of clade 1 (77–696 TPM) (Table S3). Most of the genes belonged to the capsaicinoid biosynthesis pathway (KAS, FAT, KR, AT3-D1 and pAMT) were in this clade. Also, there was a gene related to the proteasome subunit T4 (RPT4) and the glucose-6-phosphate-1-dehydrogenase (G6PDH) of the pentose phosphate pathway (Fig. 5 and Table 3). These last two genes had a higher expression in BS in comparison to RS (adjusted p-value < 0.1) (Fig. 5 and Table S3). The FAT and KAS, both related to the synthesis of fatty acids and the acyl moiety of the capsaicinoids, are grouped in one subclade. The KR and AT3-D1 genes were grouped in another subclade and had the higher expression in clade 4 (192–379 TPM) (Fig. 5 and Table S3).
The higher expression of AT3-D1 was expected in fruits of 25 DAA since is one of the principal genes that determine the pungency in pepper fruits and usually had higher expression in the intermediate development stages of fruits. The gene pAMT had the higher expression between the genes of the capsaicinoid biosynthesis pathway (338–696 TPM) (Fig. 5 and Table S3). This gene, together with AT3-D1, are key genes in the capsaicinoid synthesis pathway and its higher expression is expected in pungent fruits in medium development stages. The position of pAMT in a different clade suggests a different level of regulation from the other genes of the pathway (Fig. 5). None of these genes or the other genes directed related to the capsaicinoid synthesis were differentially expressed in fruits of 25 DAA. This result was in accord with the fact that we did not find an enriched metabolic pathway directly related to the capsaicinoid synthesis (Fig. 4B). We proposed that the differences in the phenotype observed in the level of capsaicinoids (Fig. 2) must be related to processes or pathways different to the capsaicinoid biosynthesis. We observed that the genes RPT4 and G6PDH were grouped with the genes of the capsaicinoid synthesis and both were more expressed in BS (Fig. 5). We suggest that RPT4 participates in the differences in the capsaicinoid regulation between soils via the vacuole formation where the capsaicinoids accumulate in the cells [58]. In the case of G6PDH, we suggest that may participate via the production of NADPH equivalents that may protect the capsaicinoids against catabolic activity of peroxidases [59]. Another gene that could be related to the high capsaicinoid content in BS was the gene KARI, of the 2-oxocarboxylic acid metabolism (clade 1), which was more expressed in BS. It may have contributed to the difference observed in the capsaicinoid content between soils at 25 DAA via the synthesis of valine and leucine precursors [15].
We concluded that the higher accumulation of capsaicinoid in BS not only depends on the anabolic processes. Another mechanism like the accumulation of them in vacuoles (RPT4), the processes that reduce the degradation of the capsaicinoids (G6PDH) and pathways indirectly related to the capsaicinoid biosynthesis, like the 2-oxocarboxilic acid metabolism, are also important.
3.5.2 Expression Analysis of AT3 Copies during the Fruit Development of Habanero Pepper Plant Growth in Two Soil Types
Since the AT3 catalyzes the last key step of the capsaicinoid synthesis pathway [16], we analyzed its expression in more detail during fruit development. We found different AT3 genes, namely AT3-D1 and AT3-D2 that were in different clades, with different levels of expression between them. In the comparison of the CDS and the aminoacidic traduced sequence of AT3-D1 and AT3-D2, we found 89.63% and 86.57% of identity, respectively (results not shown). Also, in a phylogeny analysis, AT3-D1 was grouped with other AT3-D1 orthologues, and AT3-D2 was grouped with AT3-D2 orthologues (results not show), which suggests that they belong to distinct copies of the gene, which was previously reported by Qin et al. [44]. We compared the expression of the AT3-D1 and AT3-D2 genes during the fruit development to deduce the possible role of each copy in the accumulation of capsaicinoids (Fig. 6). RT-PCR expression of the AT3-D1 and AT3-D2 genes regarding constitutive β-tubulin gene and relative expression plot calculated using GelAnalyzer program. In AT3-D1, we found a progressive diminution of the expression during the fruit development, with a higher expression at 25 DAA in both soil types, when capsaicinoid accumulation almost reached a maximum (Fig. 2). At 45 DAA, in BS a higher expression was found respect to RS, but the expression was lower in both soils concerning 25 DAA. This was in agreement with the change in the level of capsaicinoid between 25 and 45 DAA, in which, although at 45 DAA the level of capsaicinoids reached a maximum, the change was minor concerning 25 DAA. Also, the higher level of expression in BS may indicate that this soil stimulates a bit more the expression of AT3-D1 at 45 DAA, and this may be related to the maintenance of a high level of capsaicinoids observed in BS between 25 and 45 DAA (Fig. 2). At 60 DAA the lower expression of AT3-D1 was found in both soil types, which was in agree with the trend of the decrease in the levels of capsaicinoids in this stage (Fig. 2). AT3-D2 expression tendency was different, with the maintenance of a high expression during the fruit development (Fig. 6). This was most notable in the comparison of the expression at 45 and 60 DAA respect to AT3-D1. In these times, the expression of AT3-D2 was higher in both soils. This indicates a different regulation between both AT3 copies during the fruit development and AT3-D1 seems to have a change in expression more in agreement with the changes in capsaicinoid levels.
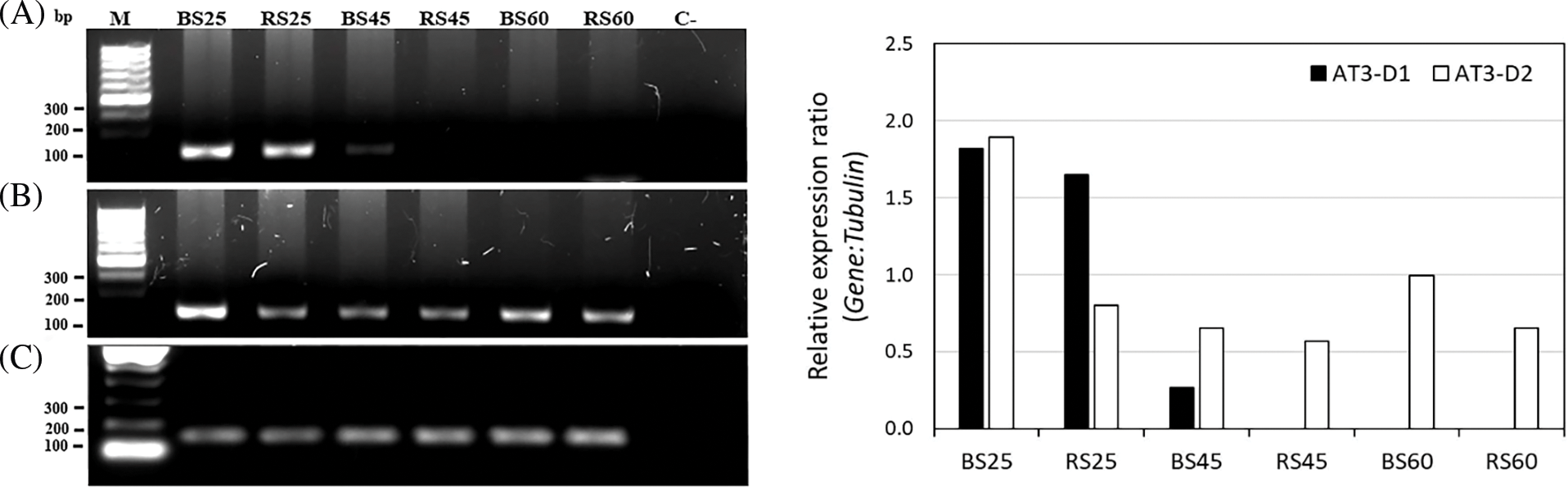
Figure 6: RT-PCR expression of the AT3-D1 and AT3-D2 genes regarding to constitutive β-tubulin gene. Left: Agarose gel (1.5%) showed three replicates by gene. − symbol indicates the negative control. Right: relative expression plot calculated using GelAnalyzer program. M: DNA ladder, C-: negative control
In synthesis, the results of the expression of the genes showed us that in BS there was higher expression of key genes related to important metabolic pathways like photosynthesis, the synthesis of porphyrins and chlorophyll, the pentose phosphate pathway, the 2-oxocarboxylic acid, the metabolism of terpenoid and the proteasome. Also, although other metabolic pathways did not have differences in the expression of regulated genes by the soil type, they were enriched in BS. These results suggest that this soil type is better for stimulating, at the transcriptomic level, important metabolic pathways related to the fruit quality and could lead to obtain better nutritional content by metabolites like sugars, terpenoids or carotenoids in the fruits. It is important to note that these results are based on the transcriptome at 25 DAA in certain cultivation conditions. The understanding of the expression of the genes related to the development and quality of the habanero pepper fruit by the soil type in this intermediate stage is important to propose cultivation strategies to improve the fruit quality, whether one prefers to select BS or RS. Also, the results suggested that the accumulation of capsaicinoids could also be regulated at the transcriptomic level via the expression of genes related to processes like the accumulation of them in vacuoles or the reduction of the catabolism, and not only by the biosynthetic pathway.
4.1 Flower and Fruit Production and Characterization in Habanero Pepper Plant Growth in Two Soil Types
In this work, we reported the effect of two contrasting soil types (BS and RS) of Yucatan on the production of fruits and the capsaicinoid content in habanero pepper. The results of production showed a floral abortion relatively high (85%) in both soils. The total production was, on average, 35 to 40 total fruits and 31 to 33 fruits of 45 DAA, which are fruits that are nearly in the complete maturity stage. Ramírez-Meraz et al. [60] reported that, in general in open sky and protected agriculture systems, that the habanero pepper ‘Jaguar’ had fruits of 6.5 to 10 g and a size of 3.8 to 5.5 cm in large. In the present work, the bigger fruits (≥4 cm) had similar values (6.95 to 6.96 g of average weight).
When the fruits were compared in ranks of size, in RS there were more bigger fruits (≥4 cm), and in BS there were more fruits with a minor size (2–2.99 cm). This result was similar to the obtained by Medina-Lara et al. [8] in habanero pepper ‘Jaguar’ in RS and BS irrigated with water with high conductivity, and obtained a more contrasting result in the differences in the size of the fruits by soil type, with a tendency in BS to obtain smallest fruits (<1 cm). In the present work, we used water of low conductivity. Compared with Medina-Lara et al. [8], we also obtained in RS bigger fruits, but in BS there is still an important number of developed fruits (3–3.99 cm of large), and the global difference in production was considered small between soils.
BS tends to have a higher content of nutrients than RS [10,11]. But it is important to consider the mobility of the nutrients in the soil matrix to the solution of the soil, and the roots of the plants [61]. The more efficient use of the nutrients of the soil has been observed in RS in comparison to the BS despite the lower nutritional content of the former [8], but these observations have been made in habanero pepper irrigated with water with high conductivity, in contrast to the water used in the present work, so the efficiency in the use of the nutrients in habanero pepper in water of low conductivity should be also studied.
The differences in soil texture (composition of sand, silt, and clay) and content of organic matter are also important for the mobility and efficient use of nutrients [8,62]. These differences in composition influence the structure, porosity, and apparent density of the soil, which affects the water filtration and the retention of the components of the soil matrix [63]. RS tends to have higher apparent density and lower porosity [62]. BS tends to have higher organic matter, which should give them a higher proportion of largest pores that filter the soil solution more than in RS, and this affects the diffusion of the nutrients to the plant roots, but also the organic matter helps to the soil structure through stabilization of aggregation, given cohesiveness to the soil particles. On the other hand, RS has more particles of smaller size with a higher superficial area that could retain the soil components, affecting mobility [63]. Also, the chemical form of the nutrients in the soil must be considered. In BS, there has been reported higher content of nitrate than in RS [10], which is a chemical form of the nitrogen that the habanero pepper can assimilate. Other nutrients, like phosphorus, are in low solubility and availability for the plant [64] if we attend to the pH of BS and RS, which are neutral or have little alkalinity [8,11]. Also, the differences in the microbial populations between RS and BS must be considered, which influenced the chemical form of the nutrients and the organic matter of the soils [11]. For all these reasons, the chemical composition of the nutrients and the organic matter must be investigated in more depth to have a better understanding of the availability of the components of the soil for the habanero pepper.
4.2 Capsaicinoid Content in Fruits of Habanero Pepper Plants Growth in Two Soil Types
The capsaicinoids are considered the most important metabolites in the habanero pepper quality for the spicy sensation that they give to consumers and the bioactive properties that are of industrial interest [13]. For this, is important to determine the effect of the soil type over the capsaicinoid content in the pepper fruits to know which soil is more convenient if we want to produce fruits with higher capsaicinoid content. The results showed an increment in the capsaicinoid content between 25 and 45 DAA in RS, and later a decrease at 60 DAA. In BS the maximum capsaicinoid content was reached at 25 DAA and only decreased at 60 DAA. The higher capsaicinoid content was between 11.30 to 11.48 mg/g DW or 169500 to 172200 Scoville Heat Units (SHU) in BS and RS, respectively. This pungency was superior to those showed in other domestic species of Capsicum (C. annuum, C. frutescens, C. pubescens, and C. baccatum) [46], but lower than other varieties of habanero pepper of Yucatan [65]. Also, the capsaicinoid content was similar to the obtained by Medina-Lara et al. [7] for habanero pepper in RS, in which, under urea fertilization, there was not possible to increment the capsaicinoid content after reaching a limit, without difference with the treatment without fertilization [7]. It has been reported that is possible to obtain higher capsaicinoid content under organic fertilization in soils with contrast characteristics, as has been reported by Das et al. [6] in C. chinense ‘Borbhut’ in two different soil types in India, with different characteristics in texture, nutrients and organic matter, similar to the contrast between BS and RS. Comparing the results of the present work, the two soils are good to obtain a similar capsaicinoid content at 45 DAA, which are fruits reaching the maturity stage, without the necessity to apply specific nitrogen fertilization, but considering the results at 60 DAA is more convenient to harvest the fruits in RS to obtain mature fruits with higher capsaicinoid content.
Concerning the evolution in the capsaicinoid content during fruit development, the results suggest an effect in the synthesis, accumulation, and degradation of the capsaicinoids, with different effects in each development stage. Considering that the capsaicinoid content in a determinate point is the reflection of its synthesis and degradation, then, between 25 and 45 DAA in RS predominate the synthesis, increasing the capsaicinoid content. In BS, neither the synthesis, nor the degradation seems to predominate, and the synthesis should occur in early stages, so between 25 and 45 DAA the global capsaicinoid metabolism did not change the capsaicinoid content. At 60 DAA, in both soils, the catabolism of capsaicinoids seems to predominate, so the content of these metabolites decreased, and the effect was higher in BS. This indicates that the soil type affects the metabolism of capsaicinoids. The expression of genes like PUN1 is sensitive to the type of soil [27]. One possibility is that the nitrogen content, its availability [8], or the chemical form in which it is presented in the soils affects the gene expression of the capsaicinoid metabolism of its enzymes [23]. It is known that the nitrate content in the fruits and the placenta of the habanero pepper affects capsaicinoid accumulation [66]. The differences in the organic matter and the humic and fulvic acids between soils also is related to the capsaicinoid content [6], and the differences in organic matter between RS and BS probably are another factor that affects the changes in the capsaicinoid accumulation [8]. According to the results of the present work, in BS the capsaicinoid synthesis probably has been stimulated more strongly in the early stages, reaching a maximum at 25 DAA, before the maximum in RS. So, we selected this development stage for the transcriptomic analysis and to find if the gene expression of the capsaicinoid metabolism was related to the differences in the capsaicinoid accumulation.
4.3 Global Expression Analysis and Analysis of Enriched Metabolic Pathways in Fruits of Habanero Pepper Plants Growth in Two Soil Types
We analyzed the fruits of 25 DAA by transcriptomic to compare the expression of genes related to capsaicinoid accumulation. These intermediate development stages are known to be active in the upregulation of the genes related to capsaicinoid biosynthesis [51,56].
The genes in each library (BS1, BS2, RS1, and RS2) were annotated against the GO database. This allowed us to group them into the three main functional categories: BP, CC, and MF [67]. The proportion of the genes in the subcategories in each one of the three main terms was similar. This suggests a similar functionality in the habanero pepper fruits of 25 DAA, independent of the type of soil, in the main GO categories.
When the global expression of the genes was analyzed and compared between soils, in BS many of the higher expressed genes (LFC ≤ −1.5) were related to the photosynthesis. There were orthologues of chlorophyll binding proteins of the LHC, and RUBisCO. It is known that the expression of genes related to photosynthesis is controlled by the development stage of the fruits, decreasing its expression in fruits in ripening stages with the change in coloration [68,69]. In this work, the fruits of 25 DAA did not show the difference in the coloration between soils, but at 45 DAA, when the fruits were in ripening or nearly in the complete mature stage, in BS there was a higher proportion of fruits with a phenotype of change in coloration and less proportion of green fruits in comparison to RS (results not show). It is possible that in BS the photosynthetic system was more stimulated in fruits of 25 DAA than in RS, and then in advanced development stages the maturation is favored for obtaining fruits with higher sugar and secondary metabolites content [70,71]. Other orthologues with higher expression in BS were RING-H2 finger protein, which is related to the growth, development, and response to abiotic stress [72] and with seed development [73]; BREVIS RADIX, related to the development of the fruits and the number of locules in C. annuum [74]; albumin-like, that has been found in seeds of C. annuum, serves as storage and had antimicrobial properties [75].
In RS, between the higher expressed genes (LFC ≥ 1.5) there was a 3-ketoacyl-CoA synthase, related to the fatty-acid synthesis. Another gene was an orthologue of the calmodulin-lysine N-methyltransferase, where the calmodulin is related to the calcium signaling between the cell wall and the cytoplasm, and in the growth of the pollen tube [76,77]. Other genes with higher expression in RS were orthologues of the mago nashi protein, related to the fertility [78], proline-rich proteoglycan; extensins; F-box proteins; polyubiquitin; and BURP domain protein, which participate in different processes like the response to environmental stimuli, floral development and in the fruit and its cellular wall development, including in the stages of maturation, softening and abscission [79–82].
The main difference in the higher expressed genes between BS and RS was that, although in both soils there were genes related to the general fruit development, RS was more specific in the genes related to growth and development, including the flowering and maturation of the fruits, the metabolism of the cell wall and the softening and the abscission of the fruits, which could indicate that RS is favorable in the growth of the fruits and in the softening of the fruits. This could have influenced that there were bigger fruits of 45 DAA (≥4 cm) in RS than in BS (Fig. 1B). In BS, there was specifically higher expression of genes related to the photosynthesis and the chlorophyll, like RUBisCO and chlorophyll-binding proteins. We suggest that this is related to the higher organic matter and higher content of nutrients in BS which can stimulate the metabolism of habanero pepper fruit and the biosynthetic processes for primary and secondary metabolites [6] more than RS, a soil with less organic matter and nutrients [6,11].
The enrichment analysis of metabolic pathways showed that most of the overrepresented pathways were in BS. Between there, there were essential metabolic functions like photosynthesis, where we found photosynthesis-antenna proteins and the porphyrin metabolism; the pentose phosphate pathway (that generates NADPH and pentoses for the biosynthetic processes); the proteasome; the circadian rhythm; and the nitrogen and amino acids metabolism. The inclusion of overrepresented photosynthetic-related pathways together with the presence of higher expressed genes related to photosynthesis and also the other overrepresented pathways related to primary metabolic processes, suggest that BS is better for the metabolic and physiologic development of the fruits. Also, other overrepresented metabolic pathways were related to the synthesis of metabolites of interest (terpenoids, carotenoids, and amino acids). If we take into account that these metabolites have properties that determine the fruit quality and are beneficial for consumer health [3], the results suggest that BS stimulates the expression of genes of the metabolism of the fruits better than RS, which could lead to obtain better quality in the fruits, enhancing biosynthetic processes and more content in nutrients like sugars, amino acids, terpenoids, carotenoids, and capsaicinoids, which also has been observed in other works that use soils rich in carbon and organic matter, and nutrients like nitrogen and phosphorus [6]. Related to the capsaicinoid biosynthetic pathway, some enriched pathways were indirectly related to the capsaicinoids, like the glutamate metabolism [23], the 2-oxocaborxilic acid metabolism, which is related to the synthesis of the branched-chain amino acids leucine and valine, precursors of the capsaicinoids [83], or the synthesis of anthocyanins, that are part of the phenylpropanoids pathway [84]. In general, these pathways seem to be more related to the fruit development than to the relative accumulation of capsaicinoids at 25 DAA.
4.4 Individual Expression Analysis of Genes in Fruits of Habanero Pepper Growth in Two Soil Types
In line with the study objective, our aim was to compare the precise gene expression patterns associated with development and capsaicinoid synthesis. To achieve this, we carefully selected a comprehensive set of thirty-one genes from enriched pathways and biosynthetic pathways of capsaicinoid production. Subsequently, we conducted a differential gene expression (DGE) analysis for these genes. Additionally, we examined the expression of two copies of AT3, a pivotal gene in the capsaicinoid biosynthesis pathway, across various developmental stages to compare its behavior in different soil conditions.
4.4.1 Individual Expression Analysis of Genes of the Metabolic Enriched Pathways and the Capsaicinoid Biosynthetic Pathway of Habanero Pepper Plant Growth in Two Soil Types
The expression levels of chlorophyll a/b binding proteins (LHCA2 and LHCB3) belonging to the LHCI and II complexes were considerably high in BS. These proteins play a crucial role in light energy absorption within the light-harvesting complex (LHC), facilitating its transfer to photosynthetic reaction centers. Additionally, they contribute to photoprotection [85], the plant’s response to environmental stimuli and stress [86,87]. Regulation of both photosynthesis-related genes [68] and enzymes [88], as well as those involved in chlorophyll and carotenoid metabolism, plays a crucial role in fruit ripening and coloration changes. Another gene that has high expression is an orthologue of KARI, a key enzyme in the synthesis of branched-chain amino acids [89]. Its high expression in BS in the same clade that the photosynthetic-related genes suggest a related regulation, where photosynthesis could be used in the synthesis of valine and leucine in the chloroplasts [90]. Other genes higher expressed in BS were orthologues of GSA and GluTR, key enzymes in the porphyrins, heme, and chlorophyll synthesis; and orthologues of G6PDH and 6PGD, of the pentose phosphate pathway which give NADPH equivalents [91]. Together, these results suggest that BS stimulates the metabolism of the habanero pepper fruit, enhancing processes like the photosynthesis, synthesis of porphyrins and chlorophylls and equivalents of NADPH (reduction power), which could lead to the synthesis of metabolites of interest like sugars, amino acids, terpenoids, carotenoids and capsaicinoids, which has been observed in others soils rich in nutrients and organic matter [6].
The orthologues of DXS and DXS2 (terpenoid biosynthesis) were more expressed in BS, although in different levels, which suggest a different regulation. This finding aligns with previous studies indicating that while both DXS homologues are involved in carotenoid metabolism, DXS1 also plays a role in plant development [92,93], whereas DXS2 is primarily associated with secondary metabolism, including carotenoid production [93]. The DXS enzyme is involved in the beginning of the terpenoid synthesis pathway, which precedes the carotenoid pathway. The difference in expression levels observed in both DXS orthologues across soils suggests distinct roles in carotenoid synthesis. Soils abundant in organic matter and nutrient content, such as nitrogen, have been shown to stimulate the production of carotenoids like β-carotene [6]. At 25 DAA, no significant differences were observed in the expression of PDS and PSY2 genes involved in the carotenoid synthesis. However, the enrichment of this pathway in BS suggests that habanero pepper fruits grown in BS may have higher carotenoid content compared to fruits grown in RS. This could be observed in advanced development stages of the fruits, like in ripening and in the change of coloration, together with a higher expression of the correspondent genes like PDS and PSY2 [56,88]. An analysis of the content of metabolites like sugar, amino acids, terpenoids and carotenoids in the fruits of BS and RS should be addressed to find out if BS have higher content of these metabolites.
All of the genes of the capsaicinoid synthesis pathway had similar expression between two types of soils. Most of them were grouped in the clade 4 (AT3-D1, KAS, KR and FAT), which suggest a coregulation of its expression. The orthologue of pAMT was grouped in the clade 5, suggesting a different regulation among the genes of the pathway. pAMT and AT3-D1 were the first and second most expressed genes of the pathway in both soils. These genes are important key steps in the capsaicinoid pathway which involved in the two final steps of the capsaicinoid synthesis [15] and its expression is generally high in the intermediate development stages of the Capsicum fruits [26,94] which concords with the results obtained in the present work. The regulation of the orthologues of KR, KAS, and FAT is also important. They participate in the synthesis of the different acyl moieties of the capsaicinoids [15,47,83]. Evidence suggests that regulating the relative proportions of various acyl-ACP precursor groups plays a significant role in determining the concentrations of different capsaicinoids formed [83]. Additionally, KR has been identified as an important factor in determining capsaicinoid content and has been reported to be associated with acyl moieties in C. annuum [47].
The separation between pAMT and other genes of the pathway in different clades could be explained in part by the fact that KR, KAS, and FAT are in the same branch of the capsaicinoid pathway, synthetizing the acyl moieties, [15,47], and pAMT is in the other branch of the capsaicinoid pathway, the phenylpropanoid pathway [15], synthetizing the vanillylamine group [95], so they are part of different pathways that only converge in the final condensation of the acyl group and the vanillylamine group by the AT3 [16]. Although a different regulation of pAMT from most of the other genes of the pathway can be expected, it is still part of the capsaicinoid metabolism, and there is evidence of a relation in the regulation between pAMT and AT3 which enzyme catalyzes the consecutive step in the pathway [96].
A gene of a transcription factor MYB was also analyzed. It has a lower expression level than the genes of the capsaicinoid pathway. MYB had a similar level of expression between soils. There is evidence that MYB transcription factors control the expression of different genes of the capsaicinoid pathway, like KAS, FAT, pAMT, and AT3, and also regulate the level of capsaicinoids in spicy fruits [97]. Genes like pAMT, KAS, and AT3 have binding sites for MYB transcription factors in their regulatory region [22,98], and control the expression in response to environmental factors [20], so the control of the expression of these transcription factors by the effect of different soils could be an interesting step to control the capsaicinoid levels. Previous studies have reported lower expression levels of MYB genes compared to other genes in the capsaicinoid pathway, and it has been observed that their expression can be relatively higher during early developmental stages [46].
Although the genes related to the capsaicinoid pathway had a similar expression between soils, in the clade 4, at 25 DAA, in the fruits of BS there was a higher content of capsaicinoids than those of RS (Fig. 2). It was hypothesized that alternative genes, distinct from those involved in the capsaicinoid pathway, might account for the observed differences in the phenotype. The orthologues of G6PDH and RPT4 were in the same clade of the genes of the capsaicinoid pathway, and they were most expressed in BS, and we supposed that these genes could be related to the higher capsaicinoid content in BS. There has been found a relation between the G6PDH activity, the NADPH, and the antioxidant activity in fruits of pepper that could protect the capsaicinoids from catabolism by peroxidases [59]. This suggests that, at 25 DAA, the higher capsaicinoid content in BS could be partially related to a lower degradation of them [9] than in RS. Also, is important to note that the activity of the KR that participates in the synthesis of the acyl moiety of the capsaicinoids requires NADPH [47], which also could be related to the role of G6PDH, which gives NADPH equivalents, favoring the higher content of capsaicinoids in BS. The orthologue of RPT4 of the proteasome had also higher expression in BS. Its relationship with the accumulation of the capsaicinoids could be related to the changes in the cellular walls and the membranes during the vesicular transport and the vacuolar storage in the cells where the capsaicinoids are synthesized and accumulated [17,58]. Also, there has been a co-segregation between the Pun1 locus (which encodes the AT3) and the vacuoles in which the capsaicinoids are accumulated [99]. Another gene that could contribute to the higher accumulation of capsaicinoids in BS could be KARI, by its relationship with the synthesis of valine and leucine which are precursors of the capsaicinoids. This suggests that at 25 DAA the synthesis of valine and leucine could be more stimulating in BS than in RS, which can be produced in the placental tissues of the habanero pepper fruits [100].
Based on these findings, we reached the conclusion that it is vital to investigate not only the genes within the metabolic pathway but also those involved in pathways and cellular processes associated with capsaicinoid accumulation. These include genes responsible for the transport and storage of capsaicinoids, their catabolism, mechanisms that could potentially mitigate their effects, and indirect pathways such as 2-oxocarboxylic acid metabolism.
4.4.2 Expression Analysis of AT3 Copies during the Fruit Development of Habanero Pepper Plants Growth in Two Soil Types
With respect to AT3, since this gene is in a key regulatory step in the capsaicinoid synthesis pathway [16,99,101] and we found two copies (AT3-D1 and AT3-D2) with different expression level at 25 DAA, we decided to compare in more detail the expression of these two copies during the fruit development. The description of the expression of AT3 has been studied generally ignoring the different copies of the gene in Capsicum [44,102]. In a comparative study of AT3-D1 and AT3-D2 expression among various spicy and non-spicy Capsicum varieties, it was observed that AT3-D1 exhibited either undetectable or minimal expression levels in non-spicy varieties, while AT3-D2 showed higher expression in the same varieties. In spicy fruits, the expression differences between the copies varied depending on the developmental stage, with both copies exhibiting higher expression during active stages of capsaicinoid synthesis [44]. Similarly, in our work, we found that at 25 DAA both AT3 copies presented higher expressions in both soil types, which was in agreement with this stage and was most active in capsaicinoid accumulation in the habanero pepper fruits in both, BS and RS. The main difference was found at 45 and 60 DAA, in which AT3-D1 expression decreased progressively at 45 and 60 DAA, which was in agree in which in these stages that were less active in capsaicinoid synthesis, especially at 60 DAA, in which the capsaicinoids levels decreased in soils and the level of expression of AT3-D1 was very low in both soils. The pattern of AT3 expression closely aligning with capsaicinoid accumulation, characterized by an initial increase during early and medium developmental stages followed by a gradual decrease, has been extensively documented by multiple authors [46,51,96,101,103]. This trend is consistent with the observed behavior of the AT3-D1 gene. On the contrary, AT3-D2 presented higher expression at 45 and 60 DAA in both soils. This indicates that both genes have a different regulation during the fruit development. We suggest that the regulation of AT3-D1 is more closely related to the higher active accumulation of capsaicinoids in habanero pepper fruits, observed mainly until the medium development stages. The high expression of AT3-D2 at 25 DAA may also indicate that it can have a complementary role in CS activity together with AT3-D1. Also, its higher expression in mature fruits, especially at 60 DAA, may indicate that AT3-D2 is related to the CS activity observed in habanero pepper in these advanced stages [9]. Another possibility is that each AT3 copy has a role in different fruit tissues since there has been observed that highly spicy pepper varieties have an accumulation of capsaicinoid and expression of genes of the capsaicinoid biosynthetic pathway in placenta and pericarp [46,103]. A deep study in the possible role of the different AT3 copies in different tissues should be addressed.
The analysis of the transcriptome and individual gene expression revealed that BS exerted a greater stimulatory effect on the expression of genes of metabolic processes in habanero pepper fruits compared to RS. This was evident in the enrichment of several metabolic pathways, including photosynthesis-antenna proteins, porphyrin synthesis, the pentose phosphate pathway, amino acid metabolism, the proteasome, 2-oxocarboxylic acid metabolism, terpenoid backbone synthesis, and carotenoid synthesis. Also, there was a higher expression of genes that relate to photosynthesis, antenna proteins, porphyrin synthesis, the pentose phosphate pathway, 2-oxocarboxylic acid, the proteasome and terpenoid synthesis compared to RS. All indications suggest that BS is more efficient in stimulating, at transcriptomic level, photosynthesis and biosynthetic processes for the production of primary and secondary metabolites, rather than promoting fruit growth. This could lead to the obtention of fruits of better quality in the content of metabolites like sugars, terpenoids, carotenoids and aminoacids, resulting in fruits with better nutritional value. The physicochemical properties of the BS, such as the texture composition, contain the higher content of organic matter and the higher content of nutrients (N and P) [10,11,62], making this soil better for stimulating metabolism than RS. Other soils rich in organic matter and nutrients has been demonstrated to be excellent for the obtention of fruits with a high content of metabolites like sugars, carotenoids and capsaicinoids than other least fertile soils [6]. It is important to note that the transcriptome was performed in fruits of 25 DAA. At advanced development stages the tendency could change. This was the case for the capsaicinoid content in the fruits of habanero pepper plants growth in BS, which was higher than those from RS at 25 DAA than but decreased at 60 DAA. In this case, the information of the transcriptome helps to explain that BS stimulate a higher expression of genes related to the capsaicinoid accumulation in the pepper better than RS, leading to obtain an early (25 DAA) higher capsaicinoid content compared to RS. The determination of metabolites of interest in habanero peppers in mature fruits could help to prove if BS is really better for obtain fruits with good quality or only is better for an early stimulation of the metabolism.
It is essential to emphasize that the findings of this study were derived from specific cultivation conditions. Modifying these conditions, such as altering the type of fertilization [6,7], adjusting the irrigation regimen [9], or managing water salinity [8], can impact soil properties by influencing nutrient content, chemical forms of nutrients, and the movement of components within the soil matrix and toward plant roots. Therefore, it is necessary to explore optimal cultivation conditions for habanero pepper cultivation in both BS and RS, determining which is more suitable. This research area is currently being investigated in Yucatan soils and will provide valuable insights to guide producers in utilizing the available soils effectively.
Habanero pepper plants growth in both BS and RS exhibited similar total fruit production, but RS showed a higher proportion of larger fruits. At 25 DAA, the expression of genes related with the growth and development of the fruits was higher in RS. In BS, there was enrichment of metabolic pathways linked to photosynthesis, the pentose phosphate pathway, the proteasome, amino acid metabolism, and secondary metabolites, including capsaicinoids. Moreover, genes related to these pathways displayed higher expression levels. This suggests that, at 25 DAA, there is a stimulating effect in BS on fruit development and metabolism, particularly in relation to photosynthesis and chlorophyll, which enhances fruit quality and nutritional content. Additionally, BS exhibited higher capsaicinoid content at 25 DAA, potentially attributed to the expression of genes associated with the synthesis of valine and leucine (KARI), the vesicular transport and storage of the capsaicinoids (RPT4) and the antioxidant activity and NADPH production in cells (G6PDH). Genes related to the capsaicinoid synthesis pathway were highly expressed in pepper fruits, although in a similar level of expression between soils. The expression patterns of AT3-D1 and AT3-D2 genes during fruit development differed, indicating potential variations in their regulation and roles in CS activity.
Acknowledgement: CONAHCyT Scholarship Burgos-Valencia Eduardo No. 711358, GarcÃŁa-Laynes Federico No. 752767.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: All authors contributed to the study’s conception and design. Material preparation, data collection and analysis were performed by E. Burgos-Valencia, F. García-Laynes, F. Medina-Lara, I. Echevarría-Machado, M. Monforte-González, M. Martínez-Estévez and J. Narváez-Zapata. The first draft of the manuscript was written by E. Burgos-Valencia and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are provided in this published article and its supplementary data files or it will be provided upon a reasonable request. The raw reads used for the transcriptomic analysis are deposited in the NCBI database with the accession number PRJNA983842.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2023.046943.
References
1. Bosland PW, Votava EJ. Taxonomy, pod types and genetic resources. In: Atherton J, editor. Peppers: vegetable and spices capsicum. 2nd ed. USA: CABI; 2012. p. 13–23. [Google Scholar]
2. Akhtar A, Asghar W, Khalid N. Phytochemical constituents and biological properties of domesticated Capsicum species: a review. Bioact Compd Health Dis. 2021;4(9):201–25. [Google Scholar]
3. Antonio AS, Wiedemann LSM, Veiga Junior VF. The genus Capsicum: a phytochemical review of bioactive secondary metabolites. RSC Adv. 2018;8(45):25767–84. [Google Scholar] [PubMed]
4. Ruiz-Lau N, Medina-Lara F, Martínez-Estévez M. El chile habanero: su origen y usos. Ciencia-Acad Mexicana de Ciencias. 2011;62(3):70–8 (In Español). [Google Scholar]
5. González-Estrada T, Zúñiga-Aguilar JJ, Vázquez-Flota F. Mejoramiento genético del chile habanero de la península de yucatán. México: Conacyt, Cicy; 2018 (In Español). [Google Scholar]
6. Das S, Teja KC, Duary B, Agrawal PK, Bhattacharya SS. Impact of nutrient management, soil type and location on the accumulation of capsaicin in Capsicum chinense (Jacq.one of the hottest chili in the world. Sci Horti. 2016;213:354–66. [Google Scholar]
7. Medina-Lara F, Echevarría-Machado I, Pacheco-Arjona R, Ruiz-Lau N, Guzmán-Antonio A, Martinez-Estevez M. Influence of nitrogen and potassium fertilization on fruiting and capsaicin content in habanero pepper (Capsicum chinense Jacq.). Hortic Sci. 2008;43(5):1549–54. [Google Scholar]
8. Medina-Lara F, Souza-Perera R, Martínez-Estévez M, Ramírez-Sucre MO, Rodríguez-Buenfil IM, Echevarría-Machado I. Red and brown soils increase the development and content of nutrients in habanero pepper subjected to irrigation water with high electrical conductivity. Hortic Sci. 2019;54(11):2039–49. [Google Scholar]
9. Zamudio-Moreno E, Echevarría-Machado I, Medina-Lara MDF, Calva-Calva G, Miranda-Ham MDL, Martínez-Estévez M. Role of peroxidases in capsaicinoids degradation in habanero pepper (Capsicum chinense Jacq.) plants grown under water deficit conditions. Aust J Crop Sci. 2014;8(3):448–54. [Google Scholar]
10. Bautista-Zúñiga F, Jiménez-Osornio J, Navarro-Alberto J, Manu A, Lozano R. Microrelieve y color del suelo como propiedades de diagnóstico en Leptosoles cársticos. Terra Latina. 2003;21(1):1–11 (In Español). [Google Scholar]
11. Estrada-Medina H, Canto-Canché BB, Santos-Briones C, O’Connor-Sánchez A. Yucatán in black and red: linking edaphic analysis and pyrosequencing-based assessment of bacterial and fungal community structures in the two main kinds of soil of Yucatán State. Microbiol Res. 2016;188–189:23–33. [Google Scholar] [PubMed]
12. Chel-Guerrero LD, Oney-Montalvo JE, Rodríguez-Buenfil IM. Phytochemical characterization of by-products of habanero pepper grown in two different types of soils from Yucatán, Mexico. Plants. 2021;10(4):779. [Google Scholar] [PubMed]
13. Wang F, Xue Y, Fu L, Wang Y, He M, Zhao L, et al. Extraction, purification, bioactivity and pharmacological effects of capsaicin: a review. Crit Rev Food Sci Nutr. 2022;62(19):5322–48. [Google Scholar] [PubMed]
14. Werner J. Capsaicinoids–properties and mechanisms of pro-health action. In: Analytical methods in the determination of bioactive compounds and elements in food. 2021. p. 193–225. [Google Scholar]
15. Arce-Rodríguez ML, Ochoa-Alejo N. Biochemistry and molecular biology of capsaicinoid biosynthesis: recent advances and perspectives. Plant Cell Rep. 2019;38(9):1017–30. [Google Scholar]
16. Ogawa K, Murota K, Shimura H, Furuya M, Togawa Y, Matsumura T, et al. Evidence of capsaicin synthase activity of the Pun1-encoded protein and its role as a determinant of capsaicinoid accumulation in pepper. BMC Plant Biol. 2015;15(1):1–10. [Google Scholar]
17. Tyler BJ, Peterson RE, Lee TG, Draude F, Pelster A, Arlinghaus HF. ToF-SIMS imaging of capsaicinoids in Scotch Bonnet peppers (Capsicum chinense). Biointerphases. 2016;11(2):2A327. [Google Scholar]
18. Tanaka Y, Watachi M, Nemoto W, Goto T, Yoshida Y, Yasuba K, et al. Capsaicinoid biosynthesis in the pericarp of chili pepper fruits is associated with a placental septum-like transcriptome profile and tissue structure. Plant Cell Rep. 2021;40(10):1859–74. [Google Scholar] [PubMed]
19. Rodríguez-Buenfil I, López-Domínguez C, Morozova K, Schampicchio M, Ramírez-Sucre M. Capsaicinoids and vitamin C in habanero chili (Capsicum chinense) cultivated in different types of soils from Yucatan, Mexico. In: 2nd EuroSciCon Conference on Food Technology, 2018; Rome, Italy. p. 36. [Google Scholar]
20. Arce-Rodríguez ML, Ochoa-Alejo N. An R2R3-MYB transcription factor regulates capsaicinoid biosynthesis. Plant Physiol. 2017;174(3):1359–70. [Google Scholar]
21. Kirke J, Kaplan N, Velez S, Jin XL, Vichyavichien P, Zhang XH. Tissue-preferential activity and induction of the pepper capsaicin synthase PUN1 promoter by wounding, heat and metabolic pathway precursor in tobacco and tomato plants. Mol Biotech. 2018;60(3):194–202. [Google Scholar]
22. Sun B, Zhu Z, Chen C, Chen G, Cao B, Chen C, et al. Jasmonate-inducible R2R3-MYB transcription factor regulates capsaicinoid biosynthesis and stamen development in Capsicum. J Agri Food Chem. 2019;67(39):10891–903. [Google Scholar] [PubMed]
23. Zhang J, Lv J, Xie J, Gan Y, Coulter JA, Jihua Y, et al. Nitrogen source affects the composition of metabolites in pepper (Capsicum annuum L.) and regulates the synthesis of capsaicinoids through the GOGAT-GS pathway. Foods. 2020;9(2):150. [Google Scholar] [PubMed]
24. Egan AN, Moore S, Stellari GM, Kang BC, Jahn MM. Tandem gene duplication and recombination at the AT3 locus in the Solanaceae, a gene essential for capsaicinoid biosynthesis in Capsicum. PLoS One. 2019;14(1):1–24. [Google Scholar]
25. Han K, Jang S, Lee JH, Lee DG, Kwon JK, Kang BC. A MYB transcription factor is a candidate to control pungency in Capsicum annuum. Theor Appl Gene. 2019;132(4):1235–46. [Google Scholar] [PubMed]
26. Tanaka Y, Asano T, Kanemitsu Y, Goto T, Yoshida Y, Yasuba K, et al. Positional differences of intronic transposons in pAMT affect the pungency level in chili pepper through altered splicing efficiency. Plant J. 2019;100(4):693–705. [Google Scholar] [PubMed]
27. Das S, Sarkar S, Das M, Banik P, Bhattacharya SS. Influence of soil quality factors on capsaicin biosynthesis, pungency, yield, and produce quality of chili: an insight on Csy1, Pun1, and Pun12 signaling responses. Plant Physiol Bioch. 2021;166:427–36. [Google Scholar]
28. Oney-Montalvo J, Uc-Varguez A, Ramírez-Rivera E, Ramírez-Sucre, Rodríguez-Buenfil I. Influence of soil composition on the profile and content of polyphenols in habanero peppers (Capsicum chinense Jacq.). Agron. 2020;10(9):1234. [Google Scholar]
29. Córdova-Alvarado AA, Monforte-González M, Rozete-Navarro M, Estrada-Alfaro NA, Vázquez-Flota F. A validated method for the thin-layer chromatography in situ densitometric quantitation of capsaicinoids in Habanero pepper (Capsicum chinense Jacq.). JPC-J Planar Chromat. 2022;35:473–9. [Google Scholar]
30. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinform. 2014;30(15):2114–20. [Google Scholar]
31. Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics; 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/(accessed on 10/01/2022). [Google Scholar]
32. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. [Google Scholar] [PubMed]
33. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. [Google Scholar] [PubMed]
34. Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinform. 2012;28(23):3150–2. [Google Scholar]
35. Haas BJ. TransDecoder (Find coding regions within transcripts). https://github.com/TransDecoder/transdecoder.github.io/blob/master/index.asciidoc(accessed on 10/01/2022). [Google Scholar]
36. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. [Google Scholar]
37. Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using diamond. Nat Method. 2021;18:366–8. [Google Scholar]
38. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [Google Scholar] [PubMed]
39. Su W, Wei Y, Huang B, Ji J. Identification of hub genes and immune infiltration in psoriasis by bioinformatics method. Front Gene. 2021;12:606065. [Google Scholar]
40. Wickham H. Toolbox. In Wickham H (Ed.ggplot2: elegant graphics for data analysis. New York: Springer; 2016. [Google Scholar]
41. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov. 2021;2(3):100141. [Google Scholar]
42. Yu G. enrichplot: visualization of functional enrichment result. R package version 1.16.2. 2022. https://yulab-smu.top/biomedical-knowledge-mining-book/(accessed on 15/07/2022). [Google Scholar]
43. Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinform. 2013;29(14):1830–1. [Google Scholar]
44. Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. PNAS. 2014;111(14):5135–40. [Google Scholar] [PubMed]
45. Tanaka Y, Fukuta S, Koeda S, Goto T, Yoshida Y, Yasuba KI. Identification of a novel mutant pAMT allele responsible for low-pungency and capsinoid production in chili pepper: accession ‘no. 4034’ (Capsicum chinense). Hort J. 2018;87(2):222–8. [Google Scholar]
46. Sun B, Chen C, Song J, Zheng P, Wang J, Wei J, et al. The Capsicum MYB31 regulates capsaicinoid biosynthesis in the pepper pericarp. Plant Physiol Bioch. 2022;176:21–30. [Google Scholar]
47. Koeda S, Sato K, Saito H, Nagano AJ, Yasugi M, Kudoh H, et al. Mutation in the putative ketoacyl-ACP reductase CaKR1 induces loss of pungency in Capsicum. Theor Appl Genet. 2019;132(1):65–80. [Google Scholar] [PubMed]
48. Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, et al. gplots: various R programming tools for plotting data. R package version. 2009;2(4):1. [Google Scholar]
49. Neuwirth E. RColorBrewer: colorbrewer palette. The comprehensive R archive network. 2022. https://cran.r-project.org/package=RColorBrewer(accessed on 21/10/2022). [Google Scholar]
50. Nieto-Garibay A, Barraza A, Caamal-Chan G, Murillo-Amador B, Troyo-Diéguez E, Burgoa-Cruz CA, et al. Habanero pepper (Capsicum chinense) adaptation to water-deficit stress in a protected agricultural system. Funct Plant Biol. 2022;49(3):295–306. [Google Scholar] [PubMed]
51. Zhang ZX, Zhao SN, Liu GF, Huang ZM, Cao ZM, Cheng SH, et al. Discovery of putative capsaicin biosynthetic genes by RNA-Seq and digital gene expression analysis of pepper. Sci Rep. 2016;6:34121. [Google Scholar] [PubMed]
52. Chhapekar SS, Brahma V, Rawoof A, Kumar N, Gaur R, Jaiswal V, et al. Transcriptome profiling, simple sequence repeat markers development and genetic diversity analysis of potential industrial crops Capsicum chinense and C. frutescens of Northeast India. Ind Crop Prod. 2020;154:112687. [Google Scholar]
53. Liu S, Li W, Wu Y, Chen C, Lei J. De Novo transcriptome assembly in chili pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids. PLoS One. 2013;8(1):e48156. [Google Scholar] [PubMed]
54. Raghavan V, Kraft L, Mesny F, Rigerte L. A simple guide to de novo transcriptome assembly and annotation. Brief Bioinform. 2022;23(2):1–30. [Google Scholar]
55. McDermaid A, Monier B, Zhao J, Liu B, Ma Q. Interpretation of differential gene expression results of RNA-seq data: review and integration. Brief Bioinform. 2019;20(6):2044–54. [Google Scholar] [PubMed]
56. Martínez-López LA, Ochoa-Alejo N, Martínez O. Dynamics of the chili pepper transcriptome during fruit development. BMC Genomics. 2014;15:143. [Google Scholar]
57. Nguyen TM, Shafi A, Nguyen T, Draghici S. Identifying significantly impacted pathways: a comprehensive review and assessment. Genome Biol. 2019;20:203. [Google Scholar] [PubMed]
58. Zamski E, Shoham O, Palevitch D, Levy A. Ultrastructure of capsaiclnold-secretlng cells 1n pungent and nonpungent red pepper (Capslcum annuum L.) cultlvars. Bot Gaz. 1987;148(1):1–6. [Google Scholar]
59. Palma JM, Terán F, Contreras-Ruiz A, Rodríguez-Ruiz M, Corpas FJ. Antioxidant profile of pepper (Capsicum annuum L.) fruits containing diverse levels of capsaicinoids. Antioxid. 2020;9(9):878. [Google Scholar]
60. Ramírez-Meraz M, Arcos-Cavazos G, Méndez-Aguilar R. Jaguar: cultivar de chile habanero para México. Rev Mex de Cienc Agric. 2018;9(2):487–92 (In Español). [Google Scholar]
61. Borges-Gómez L, Escamilla-Bencomo A, Soria-Fregoso M, Casanova-Villareal V. Potasio en suelos de Yucatán. Terra Latinoamericana. 2005;23(4):437–45 (In Español). [Google Scholar]
62. Bautista-Zúñiga F, Díaz-Garrido S, Castillo-González M, Zinck JA. Spatial heterogeneity of the soil cover in the Yucatán Karst: comparison of Mayan, WRB, and numerical classifications. Eurasian Soil Sci. 2005;38(Suppl. 1):S81–8. [Google Scholar]
63. Schaetzl RJ, Anderson S. Basics concepts: soil morphology. In: Soils: Genesis and geomorphology. USA: Cambridge University Press; 2005. p. 9–31. [Google Scholar]
64. Borges-Gómez L, Moo-Kauil C, Ruíz-Novelo J. Suelos destinados a la producción de chile habanero en Yucatán: características físicas y químicas predominantes. Agrocienc. 2014;48(4):347–59. [Google Scholar]
65. Canto-Flick A, Balam-Uc E, Bello-Bello J, Lecona-Guzmán C, Solís-Marroquín D, Avilés-Viñas S, et al. Capsaicinoids content in habanero pepper (Capsicum chinense Jacq.hottest known cultivars. HortScience. 2008;43(5):1344–9. [Google Scholar]
66. Monforte-González M, Guzmán-Antonio A, Uuh-Chim F, Vázquez-Flota F. Capsaicin accumulation is related to nitrate content in placentas of habanero peppers (Capsicum chinense Jacq.). J Sci Food Agric. 2010;90(5):764–8. [Google Scholar]
67. Gene Ontology Consortium. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(suppl_1):D258–61. [Google Scholar]
68. Borovsky Y, Monsonego N, Mohan V, Shabtai S, Kamara I, Faignboim A, et al. The zinc-finger transcription factor CcLOL1 controls chloroplast development and immature pepper fruit color in Capsicum chinense and its function is conserved in tomato. Plant J. 2019;99(1):41–55. [Google Scholar] [PubMed]
69. Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, et al. Uniform ripening encodes a golden 2-like transcription factor regulating tomato fruit chloroplast development. Sci. 2012;336(6089):1711–5. [Google Scholar]
70. Lupi ACD, Lira BS, Gramegna G, Trench B, Alves FRR, Demarco D, et al. Solanum lycopersicum GOLDEN 2-LIKE 2 transcription factor affects fruit quality in a light-and auxin-dependent manner. PLoS One. 2019;14(2):e0212224. [Google Scholar] [PubMed]
71. Nguyen CV, Vrebalov JT, Gapper NE, Zheng Y, Zhong S, Fei Z, et al. Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell. 2014;26(2):585–601. [Google Scholar] [PubMed]
72. Han G, Qiao Z, Li Y, Yang Z, Wang C, Zhang Y, et al. RING Zinc finger proteins in plant abiotic stress tolerance. Front Plant Sci. 2022;13:1–16. [Google Scholar]
73. Xu R, Li QQ. A RING-H2 zinc-finger protein gene RIE1 is essential for seed development in Arabidopsis. Plant Mol Bio. 2003;53:37–50. [Google Scholar] [PubMed]
74. Ma X, Qiao YM, Li Y, Yu YN, Gong ZH. Identification of fruit traits related QTLs and a candidate gene, CaBRX, controlling locule number in pepper (Capsicum annuum L.). Horticulturae. 2022;8(2):146. [Google Scholar]
75. Ribeiro SFF, Taveira GB, Carvalho AO, Dias GB, Cunha MD, Santa-Catarina C, et al. Antifungal and other biological activities of two 2S albumin-homologous proteins against pathogenic fungi. Protein J. 2012;31(1):59–67. [Google Scholar] [PubMed]
76. Gong X, Xu Y, Li H, Chen X, Song Z. Antioxidant activation, cell wall reinforcement, and reactive oxygen species regulation promote resistance to waterlogging stress in hot pepper (Capsicum annuum L.). BMC Plant Bio. 2022;22:425. [Google Scholar] [PubMed]
77. Hepler PK, Winship LJ. Calcium at the cell wall-cytoplast interface. J Integr Plant Biol. 2010;52(2):147–60. [Google Scholar] [PubMed]
78. Ihsan H, Khan MR, Ajmal W, Ali GM. WsMAGO2, a duplicated MAGO NASHI protein with fertility attributes interacts with MPF2-like MADS-box proteins. Planta. 2015;241(5):1173–87. [Google Scholar] [PubMed]
79. Leszczuk A, Kalaitzis P, Blazakis KN, Zdunek A. The role of arabinogalactan proteins (AGPs) in fruit ripening—a review. Hort Res. 2020;7:176. [Google Scholar]
80. Park J, Cui Y, Kang BH. AtPGL3 is an Arabidopsis BURP domain protein that is localized to the cell wall and promotes cell enlargement. Front Plant Sci. 2015;6:412. [Google Scholar] [PubMed]
81. Sommer-Knudsen J, Clarke AE, Bacic A. A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J. 1996;9(1):71–83. [Google Scholar] [PubMed]
82. Zhang X, Gonzalez-Carranza ZH, Zhang S, Miao Y, Liu CJ, Roberts JA. F-box proteins in plants. Annu Rev Plant Biol. 2019;2(1):307–27. [Google Scholar]
83. Thiele R, Mueller-Seitz E, Petz M. Chili pepper fruits: presumed precursors of fatty acids characteristic for capsaicinoids. J Agri Food Chem. 2008;56(11):4219–24. [Google Scholar] [PubMed]
84. Liu Y, Tikunov Y, Schouten RE, Marcelis LFM, Visser RGF, Bovy A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: a review. Front Chem. 2018;6:52. [Google Scholar] [PubMed]
85. Lokstein H, Renger G, Götze JP. Photosynthetic light-harvesting (antenna) complexes-structures and functions. Mol. 2021;26(11):3378. [Google Scholar]
86. Silva J, Kim YJ, Sukweenadhi J, Rahimi S, Kwon WS, Yang DC. Molecular characterization of 5-chlorophyll a/b-binding protein genes from Panax ginseng Meyer and their expression analysis during abiotic stresses. Photosynthetica. 2016;54(3):446–58. [Google Scholar]
87. Zhao S, Gao H, Luo J, Wang H, Dong Q, Wang Y, et al. Genome-wide analysis of the light-harvesting chlorophyll a/b-binding gene family in apple (Malus domestica) and functional characterization of MdLhcb4.3, which confers tolerance to drought and osmotic stress. Plant Physiol Bioch. 2020;154:517–29. [Google Scholar]
88. Rödiger A, Agne B, Dobritzsch D, Helm S, Müller F, Pötzsch N, et al. Chromoplast differentiation in bell pepper (Capsicum annuum) fruits. Plant J. 2021;105(5):1431–42. [Google Scholar]
89. Leung EWW, Guddat LW. Conformational changes in a plant ketol-acid reductoisomerase upon Mg2+ and NADPH binding as revealed by two crystal structures. J Mol Biol. 2009;389(1):167–82. [Google Scholar] [PubMed]
90. Chen Y, Zhou B, Li J, Tang H, Tang J, Yang Z. Formation and change of chloroplast-located plant metabolites in response to light conditions. Int J Mol Sci. 2018;19(3):654. [Google Scholar] [PubMed]
91. Nelson DL, Cox MM. Lehninger. In: Principios de bioquímica. 6th edEspaña: Omega; 2014 (In Español). [Google Scholar]
92. García-Alcázar M, Giménez E, Pineda B, Capel C, García-Sogo B, Sánchez S, et al. Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci Rep. 2017;7:45333. [Google Scholar]
93. Zhang S, Ding G, He W, Liu K, Luo Y, Tang J, et al. Functional characterization of the 1-Deoxy-D-Xylulose 5-phosphate synthase genes in morus notabilis. Front Plant Sci. 2020;11:1142. [Google Scholar] [PubMed]
94. Stellari GM, Mazourek M, Jahn MM. Contrasting modes for loss of pungency between cultivated and wild species of Capsicum. Heredity. 2010;104(5):460–71. [Google Scholar] [PubMed]
95. Weber N, Ismail A, Gorwa-Grauslund M, Carlquist M. Biocatalytic potential of vanillin aminotransferase from Capsicum chinense. BMC Biotech. 2014;14:25. [Google Scholar]
96. Arce-Rodríguez ML, Ochoa-Alejo N. Silencing AT3 gene reduces the expression of pAmt, BCAT, Kas, and Acl genes involved in capsaicinoid biosynthesis in chili pepper fruits. Biol Plant. 2015;59(3):477–84. [Google Scholar]
97. Zhu Z, Sun B, Cai W, Zhou X, Mao Y, Chen C, et al. Natural variations in the MYB transcription factor MYB31 determine the evolution of extremely pungent peppers. New Phytol. 2019;223(2):922–38. [Google Scholar] [PubMed]
98. Sun B, Zhou X, Chen C, Chen C, Chen K, Chen M, et al. Coexpression network analysis reveals an MYB transcriptional activator involved in capsaicinoid biosynthesis in hot peppers. Hort Res. 2020;7:162. [Google Scholar]
99. Stewart CJr, Mazourek M, Stellari GM, O’Connell M, Jahn M. Genetic control of pungency in C. chinense via the Pun1 locus. J Exp Bot. 2007;58(5):979–91. [Google Scholar] [PubMed]
100. Baas-Espinola FM, Castro-Concha LA, Vázquez-Flota FA, Miranda-Ham ML. Capsaicin synthesis requires in situ phenylalanine and valine formation in in vitro maintained placentas from Capsicum chinense. Mol. 2016;21(6):799. [Google Scholar]
101. StewartJr C, Kang BC, Liu K, Mazourek M, Moore SL, Yoo EY, et al. The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J. 2005;42(5):675–88. [Google Scholar] [PubMed]
102. Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Gene. 2014;46(3):270–8. [Google Scholar]
103. Tanaka Y, Nakashima F, Kirii E, Goto T, Yoshida Y, Yasuba KI. Difference in capsaicinoid biosynthesis gene expression in the pericarp reveals elevation of capsaicinoid contents in chili peppers (Capsicum chinense). Plant Cell Rep. 2017;36(2):267–79. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools