 Open Access
Open Access
ARTICLE
Physiological and Transcriptome Analysis Illuminates the Molecular Mechanisms of the Drought Resistance Improved by Alginate Oligosaccharides in Triticum aestivum L.
1 Institute of Plant Nutrition, Agricultural Resource and Environmental Science, Henan Academy of Agricultural Sciences, Zhengzhou, 450002, China
2 Henan Key Laboratory of Agricultural Eco-Environment, Henan Academy of Agricultural Sciences, Zhengzhou, 450002, China
* Corresponding Author: Yunhong Zhang. Email:
(This article belongs to the Special Issue: New Approaches to Mitigate Abiotic and Biotic Stresses for Improving Crop Productivity and Quality)
Phyton-International Journal of Experimental Botany 2024, 93(2), 185-212. https://doi.org/10.32604/phyton.2023.046811
Received 16 October 2023; Accepted 06 December 2023; Issue published 27 February 2024
Abstract
Alginate oligosaccharides (AOS) enhance drought resistance in wheat (Triticum aestivum L.), but the definite mechanisms remain largely unknown. The physiological and transcriptome responses of wheat seedlings treated with AOS were analyzed under drought stress simulated with polyethylene glycol-6000. The results showed that AOS promoted the growth of wheat seedlings and reduced oxidative damage by improving peroxidase and superoxide dismutase activities under drought stress. A total of 10,064 and 15,208 differentially expressed unigenes (DEGs) obtained from the AOS treatment and control samples at 24 and 72 h after dehydration, respectively, were mainly enriched in the biosynthesis of secondary metabolites (phenylpropanoid biosynthesis, flavonoid biosynthesis), carbohydrate metabolism (starch and sucrose metabolism, carbon fixation in photosynthetic organisms), lipid metabolism (fatty acid elongation, biosynthesis of unsaturated fatty acids, alpha-linolenic acid metabolism, cutin, suberine and wax biosynthesis), and signaling transduction pathways. The up-regulated genes were related to, for example, chlorophyll a-b binding protein, amylosynthease, phosphotransferase, peroxidase, phenylalanine ammonia lyase, flavone synthase, glutathione synthetase. Signaling molecules (including MAPK, plant hormones, H2O2 and calcium) and transcription factors (mainly including NAC, MYB, MYB-related, WRKY, bZIP family members) were involved in the AOS-induced wheat drought resistance. The results obtained in this study help underpin the mechanisms of wheat drought resistance improved by AOS, and provides a theoretical basis for the application of AOS as an environmentally sustainable biological method to improve drought resistance in agriculture.Keywords
Supplementary Material
Supplementary Material FileWheat (Triticum aestivum L.) is one of the most important staple crops worldwide, accounting for 30% of the world’s total harvested cereal area and approximately 26% of total cereal production [1], but its production in many regions is severely impacted by drought [2,3]. For example, a third of the UK wheat crop is grown on drought prone soil, resulting on average in a 10% to 20% loss in total production, valued at £72 million [4]. The North China Plain, which accounts for more than 60% of China’s wheat production, is usually affected by drought, and the intensity, frequency and extent of droughts have increased in recent decades [3,5]. Up to 60% of the wheat cultivation area will be impacted by severe water shortages by the end of the century without the adoption of climate adaptation or drought mitigation measures [6]. Wheat is readily damaged as a consequence of drought, owing to its poor drought resistance [7]. Continuous drought stress triggers various physiological responses, inhibits growth and photosynthesis, and results in increased plant death and reduced yield [8,9]. With global warming and increasingly variable precipitation, drought has become a major factor affecting wheat production [10], so it is important to improve wheat drought tolerance in order to ensure food security and sustainable agricultural development.
In agricultural production, measures that can be taken to improve wheat drought resistance include drought resistant breeding, the application of chemical agents, rational fertilization, and so on [11–13]. Chemical reagents to improve drought resistance are, relatively speaking, convenient to apply. At present, the drought resistance agents used are generally absorbents, exogenous abscisic acid (ABA), nutrient elements, fungicides, or rooting agents, which leave residues in the soil and lead to environmental pollution [14–16]. Hence, it is necessary to find an environmentally sustainable drought resistance agent, such as a biological compound, to replace existing chemical regents.
Alginate oligosaccharides (AOS), which are produced from alginate, an abundant compound, is composed of β-D-mannuronic acid (M), α-L-guluronic acid (G) or both heterozygous segments [15,17–20]. Studies have shown that AOS with a low molecular weight and excellent solubility, promoted root growth, increased crop yield and enhanced the tolerance of crops to abiotic stress [17,18]. It was also shown that AOS enhanced the drought resistance of a wide variety of crops such as wheat, cucumber and Arabidopsis, which was reflected in the improvement of agronomic traits (including shoot and root length, biomass, etc.) and increases in the photosynthetic capacity and yield [15,19,20]. Therefore, AOS may be used as a new environmentally sustainable biological drought resistant agents in agriculture, in order to reduce the need to apply chemical agents, and to reduce the contamination of agricultural products and the environment. However, the application of AOS is currently limited due to a lack of a clear understanding of the mechanisms of action of AOS.
Plants respond and adapt to drought stress by perceiving the stimulus, receiving and transmitting a signal, and then initiating various defense mechanisms [21]. Reactive oxygen species (ROS), such as superoxide anion, hydrogen peroxide, and hydroxyl radical, will accumulate excessively in plants while plants are under drought stress, which causes the disorganization of cellular membranes, accompanied with an increase in malondialdehyde (MDA) content and photosynthetic apparatus [22,23]. However, the toxicity of ROS is balanced by integrated antioxidant systems, including enzymes such as superoxide dismutase (SOD), peroxidase, catalase and non-enzymatic antioxidants such as glutathione, ascorbate, flavonoids, and α-tocopherol [24]. Previous research showed that spraying AOS water solution (1 g L−1, w/v) enhanced the activities of antioxidative enzymes, and reduced the MDA content in wheat leaves under drought stress simulated by adding polyethylene glycol-6000 (PEG-6000) water solution (150 g L−1, w/v) [15]. Additionally, ABA as a major signaling molecule was involved in drought response by inducing antioxidant defenses and regulating stomatal closure [25,26]. Some carbohydrates and secondary metabolism products such flavonoids were also regulated by ABA signals in plant vegetative tissues [27,28]. The drought resistant related genes involved in the ABA signal pathway, such as the late embryogenesis abundant protein 1 gene (LEA1), psbA gene, sucrose non-fermenting 1-related protein kinase 2 gene (SnRK2) and pyrroline-5-carboxylate synthetase gene (P5CS) were up-regulated by AOS in wheat seedlings [15]. Further research showed that AOS induced ABA synthesis mainly by up-regulating the expression of ABA accumulation biosynthesis-related genes, i.e., TaNCED, TaAOX and TaBG [29]. These results preliminarily showed that ABA was involved in wheat drought resistance enhanced by AOS. However, the molecular mechanisms of AOS-induced drought resistance in wheat are still unclear.
To date, studies on the mechanisms of plant tolerance to drought stress have shifted the focus from the physiological level to the molecular levels, especially at the omics levels. RNA sequencing (RNA-Seq) technology, as a tool to analyze gene functions and interactions at the omics level, has seen extensive usage in various fields of genetics and biology, including the analysis of growth characteristics and stress responses mechanism of plants [30,31]. Although the mechanisms of drought-tolerance have been reported recently [15,32], the transcriptional regulatory network of wheat response to drought stress has not been fully understood. PEG is a non-toxic and water soluble polymer, and can reduce the water potential, resulting in water deficiency in plants and causing physiological disorders. Hence, PEG has often been used to mimic drought, which is a common method to study plant responses to water stress [15,33]. On the basis of previous studies, transcriptome sequencing was performed on shoots and roots of wheat seedlings treated with or without AOS under drought stress simulated by PEG-6000, and the Illumina Novaseq 6000 platform was used to sequence the AOS-responsive genes. Gene ontology (GO) annotation and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis were conducted to reveal the biological processes involved in the regulation of wheat drought resistance by AOS, which will enable a better understanding of the underlying mechanisms, and provide a scientific basis for the application of AOS as a new environmentally sustainable biological drought resistance agent in agriculture.
The alginate (sample No. 20100927004; viscosity, 1,010 mPas; molecular formula, (C6H7O6Na)n; molecular weight, 32,000–2,002,000; M/G ratio, 7:3) was obtained from Qingdao Bright Moon Seaweeds Group Co., Ltd. (Qingdao, Shandong, China) and degraded with an alginate lyase (purchased from Shanghai Nagase Trading Co., Ltd., Shanghai, China) from Sphingomonas sp. at 45°C for 10 h. Higher molecular weight components were precipitated by adding 80% (v/v) ethanol (4 volumes) and centrifuging at 8,000 rpm for 15 min. The supernatant was freeze-dried and identified by electrospray ionization mass spectrometry, and mainly composed of dimers, trimers and tetramers (the average molecular weight, 724; viscosity, 180 mPas) (Fig. 1). Other reagents were analytical grade (Sinopharm Chemical Reagent Beijing Co., Ltd., Beijing, China), and all solutions were prepared using deionized water (Resistivity > 16 MΩ cm at 25°C).
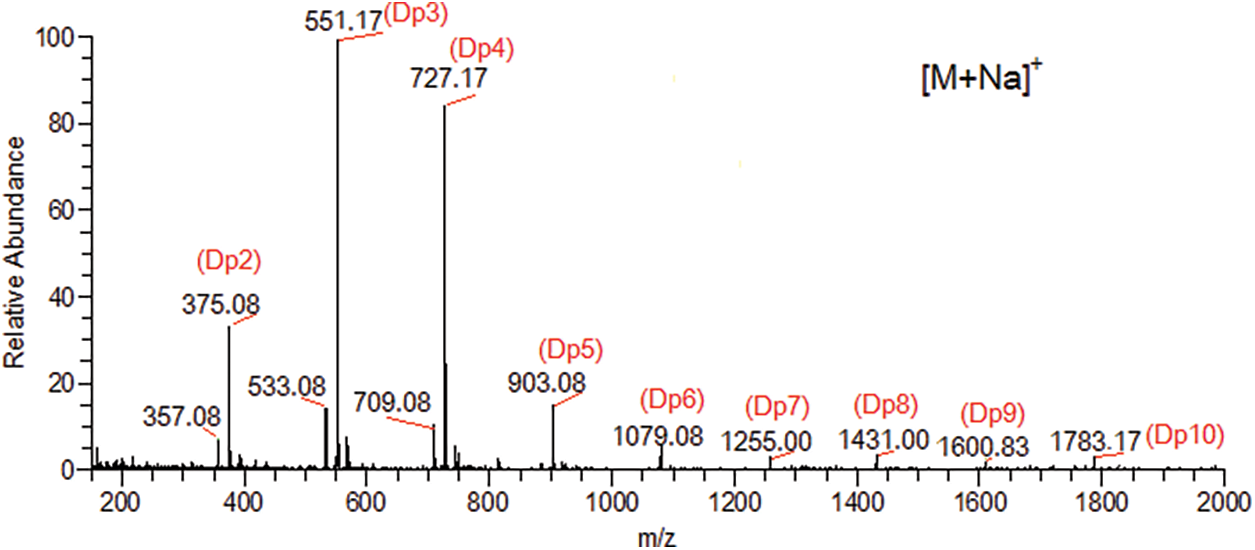
Figure 1: Electrospray ionizations mass spectrum(ESI-MS) of alginate oligosaccharides (AOS). AOS, alginate oligosaccharides; ESI-MS, electrospray ionization mass spectrum; Dp, degree of polymerization
Wheat seeds used in the experiment were of a drought sensitive wheat cultivar ‘Xinong979’, which was bred by Northwest A&F University (Shanxi, China) [15].
2.2 Plant Culture and Experimental Design
Wheat seeds were sterilized with 75% (v/v) ethanol for 5 min and washed extensively with deionized water. To obtain more uniformly germinated seeds, wheat seeds were soaked in deionized water for 8 h at 25°C in the dark, and then transferred to a 20 cm Petri dish containing one sheet of filter paper moistened with deionized water and cultured for 3 days at 25°C/20°C (12 h/12 h day/night cycle), with a relative humidity of 60% and a light intensity of 800 μmol m−2 s−1. Four independent experiments were conducted using 3-day-old wheat seedlings. In experiment 1, similar wheat seedlings were selected and transferred to 150 g L−1 (w/v) PEG water solution (simulating drought stress). A preliminary study showed that spraying 1 g L−1 (w/v) AOS water solution was optimal for enhancing wheat drought tolerance (Data not shown). Deionized water (the control) or AOS solution (the AOS treatment) was sprayed onto the seedlings leaf surfaces after dehydration for 1 h, respectively. Twice a day (morning and evening), 5 mL was sprayed once. The growth parameters and physiological characteristics of wheat seedlings were analyzed at 72 h after the treatment. In experiment 2, the above experiment was repeated, and samples of wheat shoots and roots were harvested for RNA-Sep analysis at 24 and 72 h after the treatment, respectively, and then quick-frozen in liquid nitrogen and stored at −80°C until processing. In experiment 3, the above experiment was repeated again, and samples of wheat shoots were collected at different time points (3, 5, 12, 24, 48, and 72 h) after treatment and analyzed with quantitative real-time RT-PCR (qRT -PCR). In experiment 4, mitogen-activated protein kinase (MAPK) inhibitor PD 98059 was used to study the role of the MAPK signal pathway in AOS-induced drought resistance in wheat. 3-day-old wheat seedlings were first root-dipped in 50 μM (w/v) PD 98059 for 6 h, and then cultured in 150 g L−1 (w/v) PEG water solution and treated with AOS. Samples of wheat shoots and roots were collected at 3, 24, 48, and 72 h and analyzed by qRT-PCR. All the experiments were carried out in three independent repetitions (20 seedlings per biological replicate) in this study.
2.3 Growth Parameters and Physiology Characters
The lengths of shoots and roots of wheat seedlings were estimated using a ruler after harvesting. To determine the relative water content (RWC, %), fresh leaves were weighed immediately to obtain the fresh weight (FW), then these leaves were soaked in distilled water for 2 h and the saturated weight (SW) was determined, and finally these leaves were dried for 24 h at 75°C to determine the dry weight (DW). RWC was calculated as below: RWC(%) = (FW − DW)/(SW − DW) × 100%. Chlorophyll (Chl) was extracted from the fresh leaves using 95% (v/v) ethanol, and absorption of the filtered extract was measured at 665, 649 and 470 nm, respectively. For the enzyme analysis, samples of the leaves (0.5 g FW for each seedling) were homogenized in 50 mM phosphate buffer (pH 7.8) with ice-bath. The homogenate was then centrifuged at 4°C for 10,000 g 10 min, and the supernatant was used for enzyme activity determination. The SOD activity was measured with the nitroblue tetrazolium-reduction method. The POD activity was measured with the guaiacol method. The free proline content was measured with the acid ninhydrin method. The MDA content was measured with the thiobarbituric acid colourimetry method. The above determinations were performed according to the methods of Wang et al. [34]. The data obtained from the experiment were analyzed using SPSS software (version 17.0; IBM, USA). Duncan’s pairwise comparison test was used to determine significant differences at the 5% confidence level (p < 0.05) and the different lowercase letters denote a significant difference.
2.4 Tissue RNA Extraction and Quality Inspection
Total RNA was extracted from three biological replicates of wheat shoots and roots using TRIZOL reagent (Trans GEN, Beijing, China), and its concentration and purity were detected with Nanodrop 2000 (Thermo Fisher Scientific Inc., Waltham, USA), and the RNA integrity value was evaluated using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, USA). The total amount and concentration of RNA were more than 1 μg and 35 ng μL−1, respectively, and the values of OD260/280 and OD260/230 were more than 1.8 and 1.0, respectively, which were suitable for downstream processes.
2.5 Library Construction and Transcriptome Sequencing
To prepare the cDNA library, oligo-(dT) beads were used to collect poly (A) mRNA from total RNA. Fragmentation buffer was added to cut the long mRNA into short fragments of about 300 bp, and then these short mRNA fragments were used as templates to synthesize the first strand of cDNA. The second strand of cDNA was synthesized using DNA polymerase I (New England Biolabs), RNase H (Invitrogen), dNTPs and buffer. After purification with a QiaQuick polymerase chain reaction (PCR) kit (Qiagen, CA, USA) and washing with EB buffer (10 mM Tris-HCl, pH 8.5) for end repairing, poly(A) tails and adaptors were added to the fragments. The fragments of suitable size were selected by agarose gel electrophoresis and enriched by PCR amplication to construct the cDNA library. The constructed libraries were sequenced on Illumina Novaseq 6000 platform. The obtained raw image data were converted into sequence data by base calling to yield the raw reads, which were stored in FASTQ file format. In order to ensure the accuracy of subsequent bioinformatics analysis, low-quality and repetitive sequences were removed to obtain high-quality sequencing data (clean data) for downstream analysis.
2.6 Transcriptome Expression Analysis
The clean data were aligned to the wheat reference genome (http://plants.ensembl.Org/Triticum_aestivum/Info/Index) using TopHat (v2.0.13). Gene expression levels were quantified using RNA-Sep by Expectation-Maximization (http://www.biomedcentral.com/1471-2015/12/323). The fragments per kilo base of transcript per million mapped reads (FPKM) was used to calculate the expression levels [35]. For the samples with biological replicates, differential expression analysis of two groups was performed using DESeq2 R package, and the screening threshold of the differentially expressed unigenes (DEGs) was |log2 fold-change| ≧ 1 and p-value < 0.001. DEGs have been designated as “up-regulated” or “down-regulated” based on whether their expression levels were higher or lower than those of the untreated state.
2.7 GO Classification and KEGG Enrichment
The GO database (http://www.geneontology.org/) was used to understand the functions and mechanisms of differential genes, and the results were divided into three major categories, including biological processes (BP), cellular components (CC) and molecular functions (MF). GO annotation was performed for the DEGs in the two groups of wheat tissues. Taking one of the samples as a control, the results obtained from the other sample were drawn in a GO annotation histogram. KEGG pathway enrichment analyses were performed using the KEGG orthology based annotation system 2.0 (http://bioinfo.org/kobas/genelist/) with Fisher’s exact test. In order to control the false positive rate, the false discovery rate method for multiple testing was used with a threshold of a corrected p-value < 0.05. The KEGG pathways meeting this condition were defined as significantly enriched pathways.
2.8 Real-Time Quantitative PCR Validation of Genes
The expression pattern of some DEGs related to signaling transduction pathway were obtained with qRT-PCR to validate the RNA-Sep data. 1 μg of RNA was reverse transcribed to cDNA using the PrimeScript RT reagent Kit (Takara Bio, Inc., Shiga, Japan). The qRT-PCR was performed using SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology Co., Ltd., Hunan, China) on a real-time fluorescent quantitative PCR instrument (Bio-Rad Laboratories Inc., Hercules, CA, USA). All primers used in the qRT-PCR were designed using NCBI Primer-BLAST, and were listed in Table 1. Relative gene expression levels were calculated by the 2−ΔΔCt method, using wheat GAPDH gene as the internal reference gene. qRT-PCR analysis of each sample was performed in triplicate using three biological replicates per treatment.
3.1 Analysis of the Growth Parameters and Physiological Characters of Wheat Seedlings
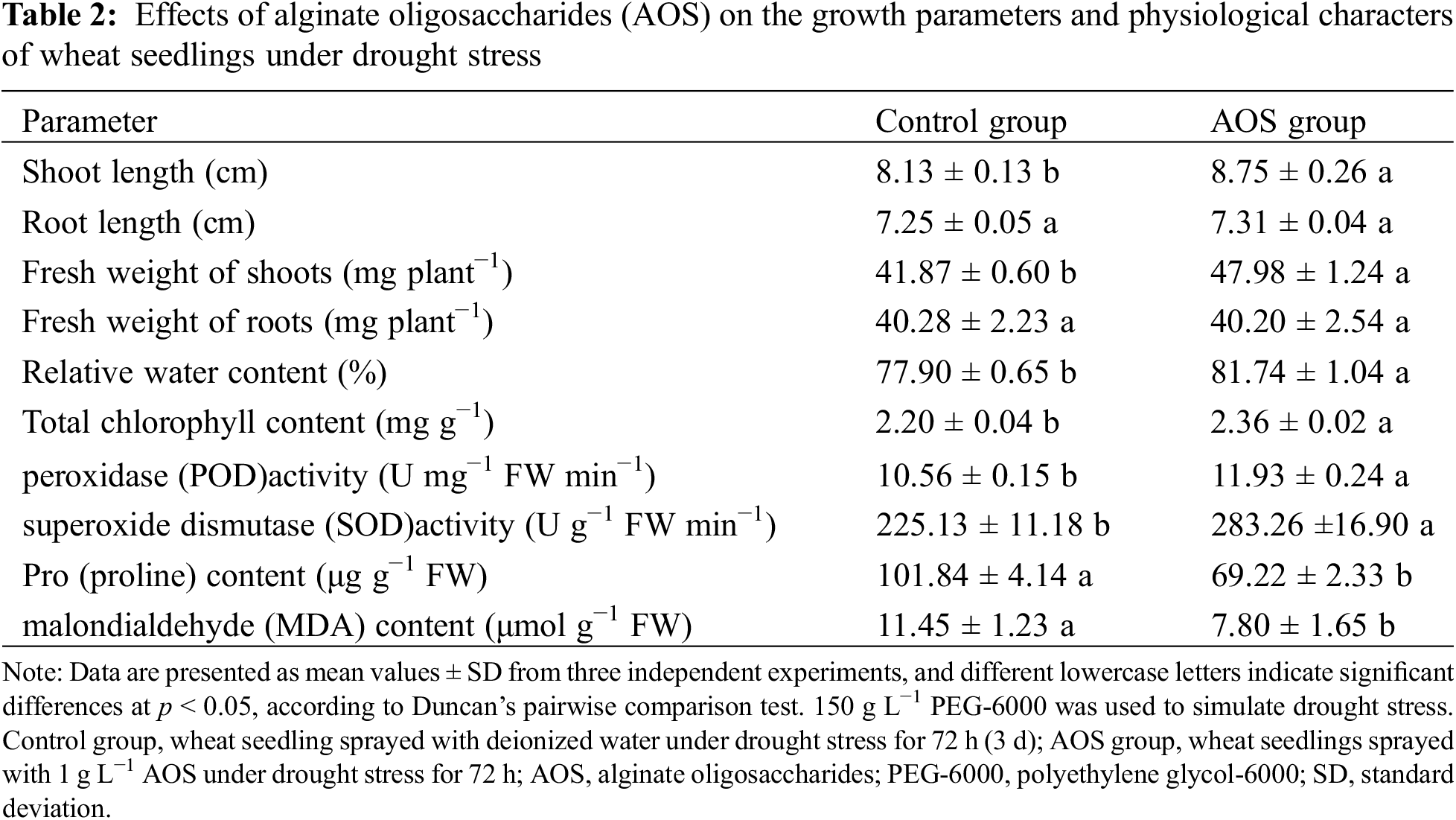
To examine the effects of AOS on the growth of wheat seedlings under drought stress, the growth parameters and physiological characters of wheat seedlings were analyzed (Table 2). Compared with the control, the length and FW of wheat shoots treated with AOS for 72 h were significantly increased by 7.6% and 14.6%, respectively (p < 0.05). RWC and total Chl contents in leaves were increased by 4.9% and 7.3%, and the activities of POD and SOD were also raised by 13.0% and 25.8%, but the contents of MDA and free proline were reduced by 32.0% and 31.9%. These results indicated that AOS reduced oxidative damage caused by drought stress and promoted the growth of wheat seedlings.
3.2 Quality Evaluation of Transcriptome Sequencing Data
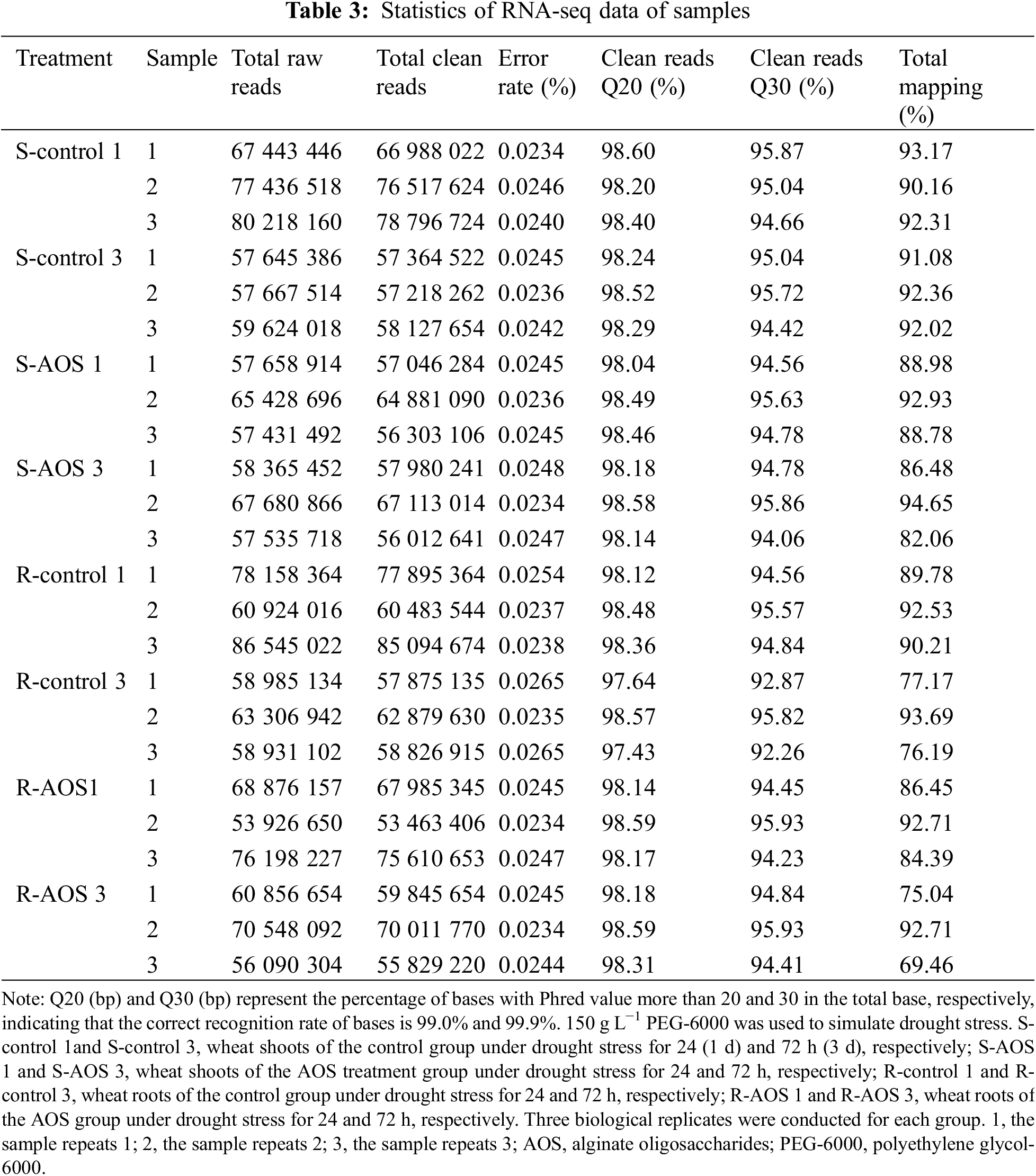
To further understand the molecular mechanisms of AOS-induced drought resistance in wheat, we examined the global gene expression profile of wheat seedlings treated with or without AOS using RNA-Seq analysis. RNA-Seq libraries of three biological replicates for the control and AOS group were prepared and sequenced. Firstly, the quality of the transcriptome sequencing data from all samples were evaluated (Table 3). After filtering out low quality data, the average number of total clean reads obtained from the control group of wheat seedlings were 74, 100, 790 and 57, 570, 146 (in shoots) as well as 74, 491, 194 and 59, 860, 560 (in roots) at 24 and 72 h after dehydration respectively, and those of the AOS treatment group were 59, 410, 160 and 60, 368, 632 (in shoots) as well as 65, 686, 468 and 61, 895, 548 (in roots). The error rates of all samples were less than 0.03%, and the Q20 and Q30 percentages were greater than 97% and 92%, respectively. The match ratio of the effective sequences obtained were more than 75% compared to the wheat reference genome. The results indicated that the quality of the sequencing results met the demands of the subsequent bioinformatics analysis.
3.3 Comparative Analysis of DEGs
In order to analyze the mechanisms of wheat drought resistance enhanced by AOS, DEGs in shoots and roots of wheat seedlings treated with AOS were detected, with reference to the control (Figs. 2A–2D). A total of 5,185 (3,250 up-regulated genes and 1,935 down-regulated genes) and 4,827 DEGs (1,956 up-regulated genes and 2,871 down-regulated genes) in wheat shoots were screened out between the AOS treatment and control samples at 24 and 72 h after dehydration, respectively (Figs. 2A and 2B). However, a total of 4,879 (1,777 up- regulated genes and 3,102 down-regulated genes) and 10,381 DEGs (8,504 up-regulated genes and 1,877 down-regulated genes) in wheat roots were obtained (Figs. 2C and 2D). The results suggested that most DEGs were up-regulated in wheat shoots (at 24 h after dehydration) and roots (at 72 h after dehydration), compared to the control group.
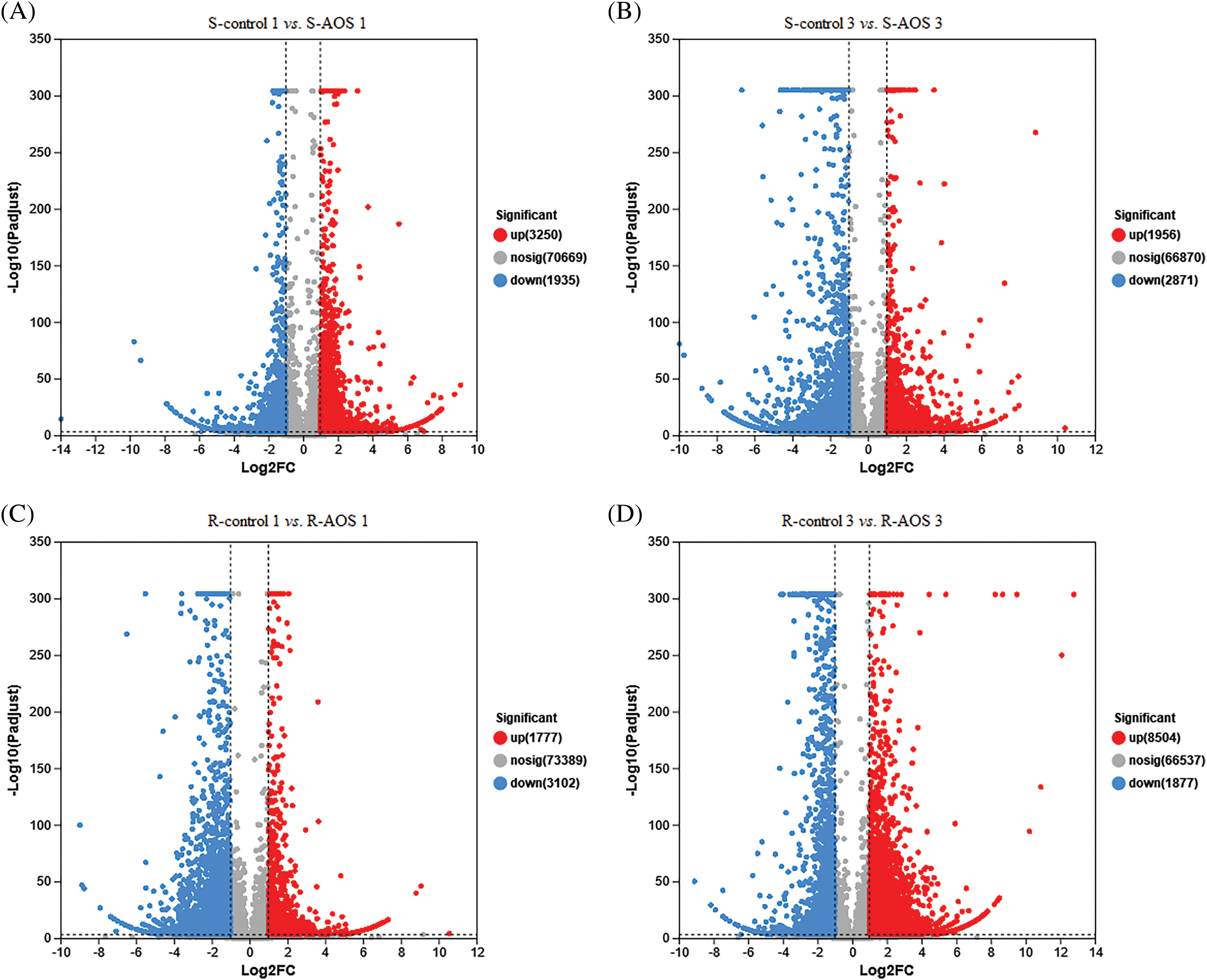
Figure 2: Volcano plot of differentially expressed genes (DEGs) between pairwise comparisons of alginate oligosaccharides (AOS) groups to control groups under drought stress. (A) S-control 1 vs. S-AOS 1; (B) S-control 3 vs. S-AOS 3; (C) R-control 1 vs. R-AOS 1; (D) R-control 3 vs. R-AOS 3. Gray dots indicate no significant difference sense, the red dots indicate significant difference up-regulated genes, the green dots indicate significant difference down-regulated genes. 150 g L−1 PEG-6000 was used to simulate drought stress. S-control 1and S-control 3, wheat shoots of the control group under drought stress for 24 (1 d) and 72 h (3 d), respectively; S-AOS 1 and S-AOS 3, wheat shoots of the AOS treatment group under drought stress for 24 and 72 h, respectively; R-control 1 and R-control 3, wheat roots of the control group under drought stress for 24 and 72 h, respectively; R-AOS 1 and R-AOS 3, wheat roots of the AOS group under drought stress for 24 and 72 h, respectively; AOS, alginate oligosaccharides; DEGs, differentially expressed genes; PEG-6000, polyethylene glycol-6000
3.4 GO Function Annotation Analysis of DEGs
GO enrichment analyses were conducted to describe the biological functions of DEGs between the AOS treatment and control groups (Figs. 3A–3D). These DEGs in wheat shoots were annotated to 20 BP, 14 CC, 13 MF categories and 21 BP, 14 CC and 14 MF categories at 24 and 72 h after dehydration, respectively. However, those in wheat roots were annotated to 19 BP, 14 CC, 14 MF categories and 20 BP, 14 CC, 15 MF categories, respectively. In terms of the BP category, the DEGs were mainly involved in metabolic process, cellular process, biological regulation, response to stimulus, cellular components organization or biogenesis, localization, developmental process, reproduction process, and multi-organism process. For the CC category, the DEGs were mainly associated with cell components, membrane components, organelle, membrane, organelle part, protein-containing complexes, extracellular regions and cell junction. The main terms of MF were binding, catalytic activity, transporter activity, transcription regulator activity, molecular function regulation and antioxidant activity.
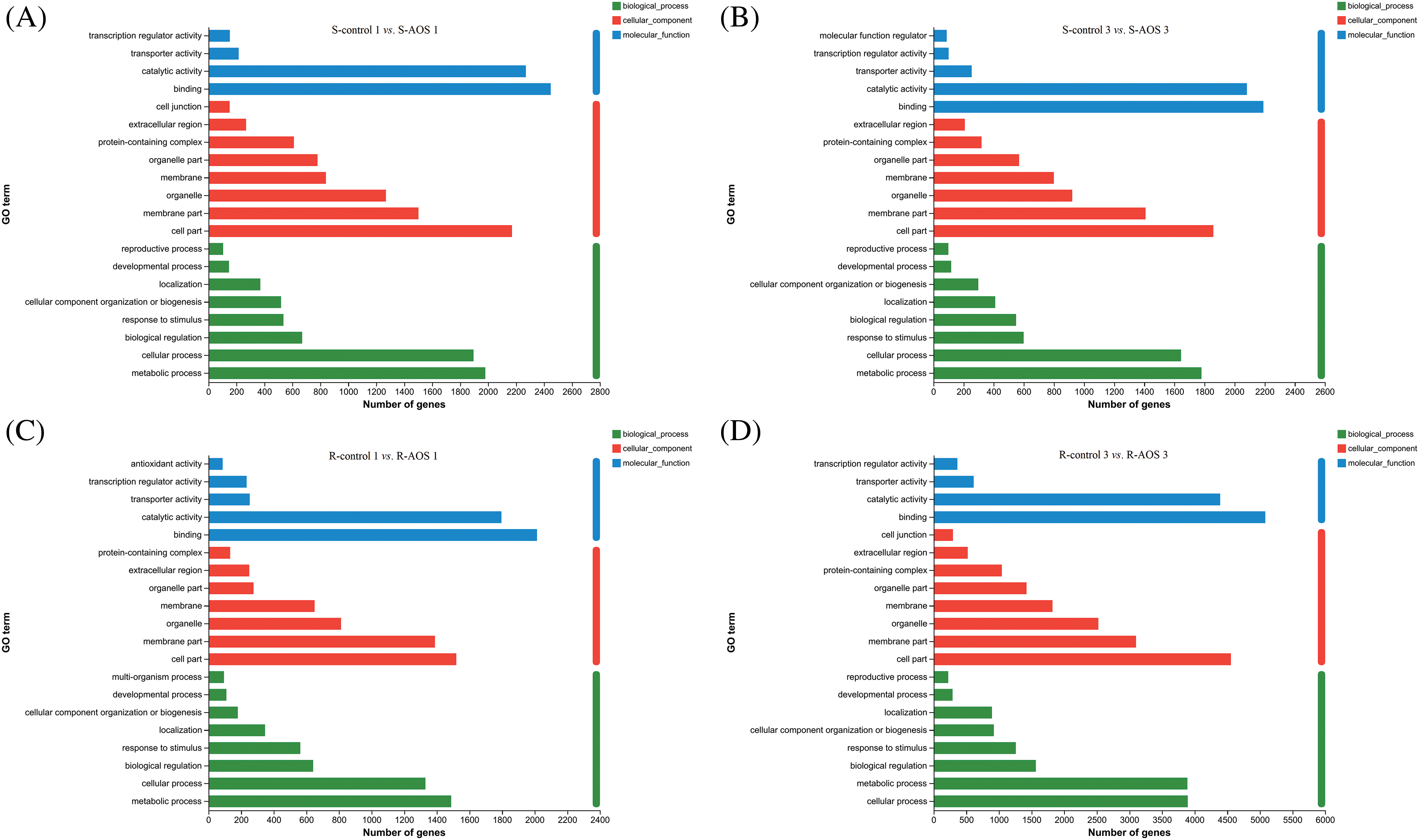
Figure 3: Gene ontology (GO) functional enrichment analysis of the differentially expressed genes (DEGs) between pairwise comparisons of alginate oligosaccharides (AOS) groups to control groups under drought stress. (A) S-control 1 vs. S-AOS 1; (B) S-control 3 vs. S-AOS 3; (C) R-control 1 vs. R-AOS 1;(D) R-control 3 vs. R-AOS 3. These DEGs were mainly involved in biological process, cellular component and molecular function. 150 g L−1 PEG-6000 was used to simulate drought stress. S-control 1and S-control 3, wheat shoots of the control group under drought stress for 24 (1 d) and 72 h (3 d), respectively; S-AOS 1 and S-AOS 3, wheat shoots of the AOS treatment group under drought stress for 24 and 72 h, respectively; R-control 1 and R-control 3, wheat roots of the control group under drought stress for 24 and 72 h, respectively; R-AOS 1 and R-AOS 3, wheat roots of the AOS group under drought stress for 24 and 72 h, respectively; AOS, alginate oligosaccharides; DEGs, differentially expressed genes; GO, gene ontology; PEG-6000, polyethylene glycol-6000
3.5 The KEGG Pathway Annotation and Enrichment Analysis of DEGs
To further identify metabolic or signal transduction pathways in which the DEGs are likely to be involved in wheat drought resistance enhanced by AOS, KEGG pathway annotation and enrichment analysis were carried out (Table S1; Figs. 4A–4D). The results showed that the DEGs were mainly involved in carbohydrate metabolism, biosynthesis of other secondary metabolites, lipid metabolism, amino acid metabolism, energy metabolism and signal transduction (Table S1). The KEGG pathway enrichment analysis indicated that these pathways, including photosynthesis-antenna proteins and cutin, suberine and wax biosynthesis, were highly enriched in wheat shoots (Figs. 4A and 4B). Besides these, the enriched pathways of DEGs in wheat shoots included phenylpropanoid biosynthesis, DNA replication, flavonoid biosynthesis, fatty acid elongation, cyanoamino acid metabolism, biosynthesis of unsaturated fatty acids, flavone and flavonol biosynthesis and so on, at 24 h after dehydration (Fig. 4A). These pathways were enriched, including glycolysis/gluconeogenesis, glutathione metabolism, starch and sucrose metabolism, pyruvate metabolism, glyoxylate and dicarboxylate metabolism, carbon fixation in photosynthetic organisms, peroxisome, glycine, serine and threonine acid metabolism, fatty acid degradation, alpha-linolenic acid metabolism and so on, at 72 h after dehydration (Fig. 4B).
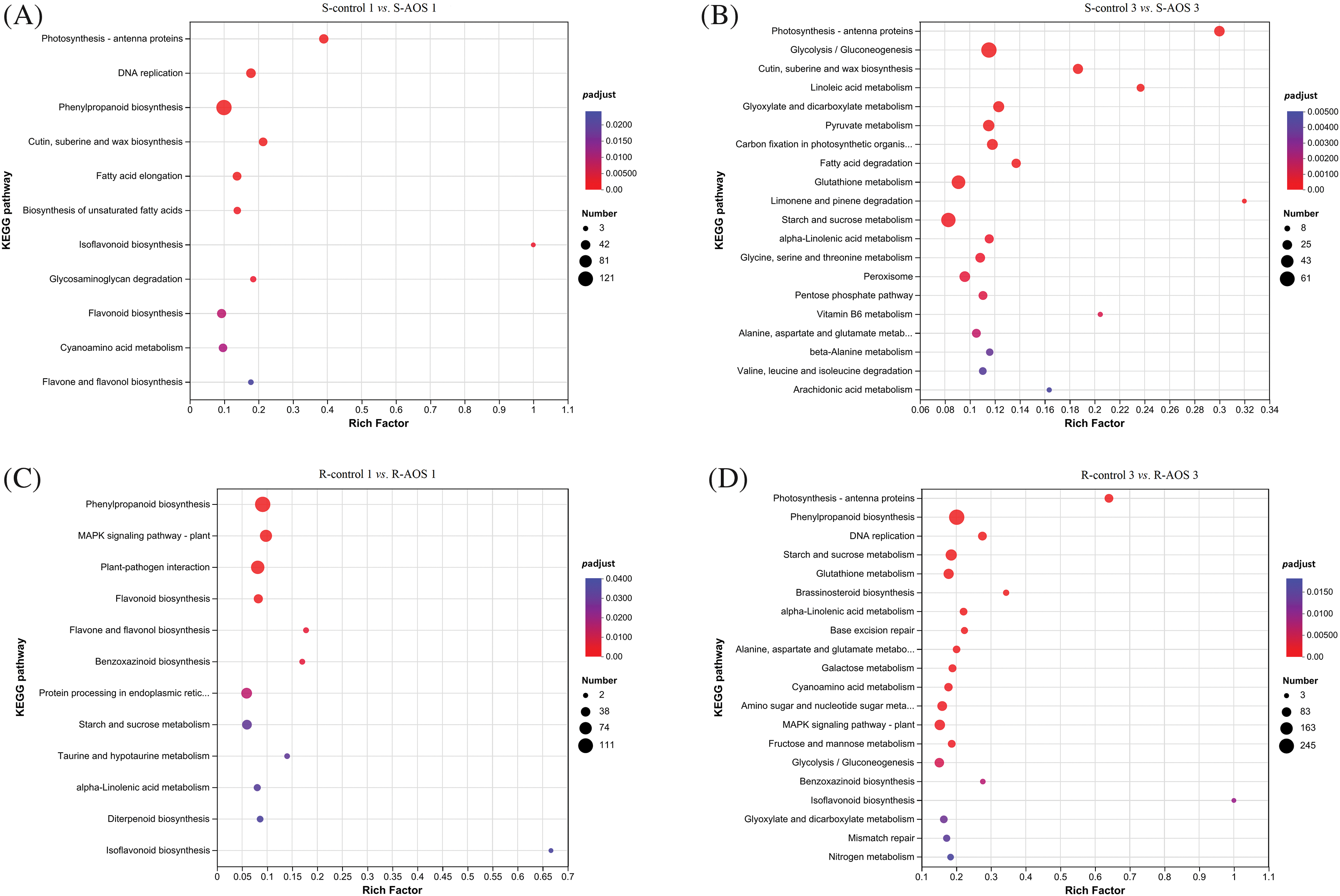
Figure 4: Kyoto encyclopedia of genes and genomes (KEGG) classification enrichment map of the differentially expressed genes (DEGs) between pairwise comparisons of alginate oligosaccharides (AOS) groups to control groups under drought stress. (A) S-control 1 vs. S-AOS 1; (B) S- control 3 vs. S-AOS 3; (C) R-control 1 vs. R-AOS1; (D) R-control 3 vs. R-AOS 3. 150 g L−1 PEG-6000 was used to simulate drought stress. S-control 1 and S-control 3, wheat shoots of the control group under drought stress for 24 (1 d) and 72 h (3 d), respectively; S-AOS 1 and S-AOS 3, wheat shoots of the AOS treatment group under drought stress for 24 and 72 h, respectively; R-control 1 and R-control 3, wheat roots of the control group under drought stress for 24 and 72 h, respectively; R-AOS 1 and R-AOS 3, wheat roots of the AOS group under drought stress for 24 and 72 h, respectively; AOS, alginate oligosaccharides; DEGs, differentially expressed genes; PEG-6000, polyethylene glycol-6000.The choser the p-value is to zero, the closer the color is to red, the more significant the enrichment is
In wheat roots, these pathways, including phenylpropanoid biosynthesis, starch and sucrose metabolism, alpha-linolenic acid metabolism, MAPK signaling pathway-plant, benzoxazinoid biosynthesis and isoflavonoid biosynthesis, were enriched (Figs. 4C and 4D). Besides these, the enriched pathways of DEGs in wheat roots included plant-pathogen interaction, protein processing in endoplasmic reticulum, flavonoid biosynthesis, and so on, at 24 h after dehydration (Fig. 4C). These pathways, including glycolysis/gluconeogenesis, DNA replication, photosynthesis-antenna proteins, cyanoamino acid metabolism, fructose and mannose metabolism, brassinosteroid biosynthesis, and so on, were enriched at 72 h after dehydration (Fig. 4D). The results indicated that carbohydrate metabolism, secondary metabolites synthesis, lipid metabolism, amino acid metabolism, energy metabolism and signal transduction were involved in the AOS-induced drought resistance in wheat.
3.6 Gene Analysis of the Enriched KEGG Pathways
DEGs in the main enriched KEGG pathways involved in carbohydrate metabolism, secondary metabolites synthesis, and lipid metabolism were analyzed (Tables 4 and 5). The results showed that the up-regulated genes in wheat shoots, which were involved in phenylpropanoid biosynthesis (mainly including peroxidase, phenylalanine ammonia lyase, β-glucosidase, 4-coumarate: coenzyme A ligase, phenylalanine deaminase and 4-α-glucanotransferase), photosynthesis-antenna proteins (chlorophyll a-b binding protein), fatty acid elongation (mainly including very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase and 3-ketoacyl-CoA synthase), cutin, suberine and wax biosynthesis (mainly including fatty acyl-CoA reductase, caleosin and cytochrome P450), flavonoid biosynthesis (mainly including glycosyltransferase and flavone synthase), and biosynthesis of unsaturated fatty acids, were more than the down-regulated genes at 24 h after dehydration. Moreover, most DEGs involved in glycolysis /gluconeogenesis and glutathione metabolism in wheat shoots were up-regulated at 72 h after dehydration.


In wheat roots, the up-regulated genes, which were involved in phenylpropanoid biosynthesis, starch and sucrose metabolism (mainly including amylosynthease, phosphotransferase, amylase, trehalose-6-phosphate phosphatase, 1,4-alpha-glucan branching enzyme 3 and endoglucanase), glycolysis/gluconeogenesis, MAPK signaling pathway-plant (mainly including WRKY transcription factor, endochitinase, heavy metal P-type ATPase, mitogen-activated protein kinase, serine/threonine protein kinase, pathogen-associated protein, catalase, calcium binging protein, calmodulin, ribonucleoside diphosphate kinase and ethylene receptor), glutathione metabolism (mainly including TSO2, glutathione synthetase, ribonucleoside diphosphate kinase reductase, hypothetical L-ascorbate peroxidase 6, R-glutamyl cyclotransferase, glucose-6-phosphate dehydrogenase, ascorbate peroxidase, glutathione-S-transferase and hexosyltransferase), amino sugar and nucleotide sugar metabolism, photosynthesis-antenna protein, and alpha-linolenic acid metabolism (mainly including allene oxide synthase, allene oxide cyclase, and alcohol dehydrogenase) were more than the down-regulated genes at 72 h after dehydration. Moreover, most DEGs involved in the phenylpropanoid biosynthesis, flavonoid biosynthesis and alpha-linolenic acid metabolism were up-regulated at 24 h after dehydration.
The results indicated that, most DEGs in the main enriched KEGG pathways, including photosynthesis-antenna protein, starch and sucrose metabolism, phenylpropanoid biosynthesis, flavonoid biosynthesis, fatty acid elongation, cutin, suberine and wax biosynthesis, glutathione metabolism, were up-regulated by AOS, which appeared earlier in wheat shoots than in the roots. These genes were mainly related to chlorophyll a-b binding protein, amylosynthease, phosphotransferase, peroxidase, phenylalanine ammonia lyase, flavone synthase, glutathione synthetase. Signaling molecules, such as MAPK, plant hormones, H2O2 and calcium, were involved in the AOS-induced drought resistance in wheat.
3.7 Differential Expression Analysis of Transcription Factor (TF) Genes
TFs play important roles in response to drought stress, hence the expression of TFs genes were analyzed (Figs. 5A–5D and Table 6). A total of 80 and 68 differentially expressed TF genes in wheat shoots were obtained between the AOS treatment and control samples at 24 and 72 h after dehydration, and the top 10 TF families were FAR1, NAC, MYB, MYB-related, CATA, WRKY, bZIP, GRAS, CAMTA, C3H, and B3, bHLH, MYB, ERF, bZIP, HSF, ARF, WRKY, MYB-related, NAC, respectively (Figs. 5A and 5B). Among which, 49 and 51 TF genes that regulated plant stress response (accounting for 31.25% and 75.0% of all differential TF genes) were obtained, mainly including FAR1, NAC, MYB, MYB-related, WRKY, bZIP, GRAS family members (Table 6). In wheat roots, there were 88 and 78 differentially expressed TF genes at 24 and 72 h after dehydration, and the top 10 families were WRKY, MYB, bHLH, HSF, ERF, MYB-related, B3, bZIP, NAC, GRAS and MYB, bHLH, bZIP, HB-other, MYB-related, WRKY, ERF, C3H, HSF, B3, respectively (Figs. 5C and 5D). Among which, 68 and 51 TF genes that regulated plant stress response (accounting for 77.27% and 65.38% of all differential TF genes) were obtained, mainly including WRKY, MYB, bHLH, MYB-related, NAC, bZIP, B3, ERF, HSF family members (Table 6). These results indicated that many TFs were involved in the regulation of AOS on wheat drought resistance, mainly the NAC, MYB, MYB-related, WRKY, bZIP families.
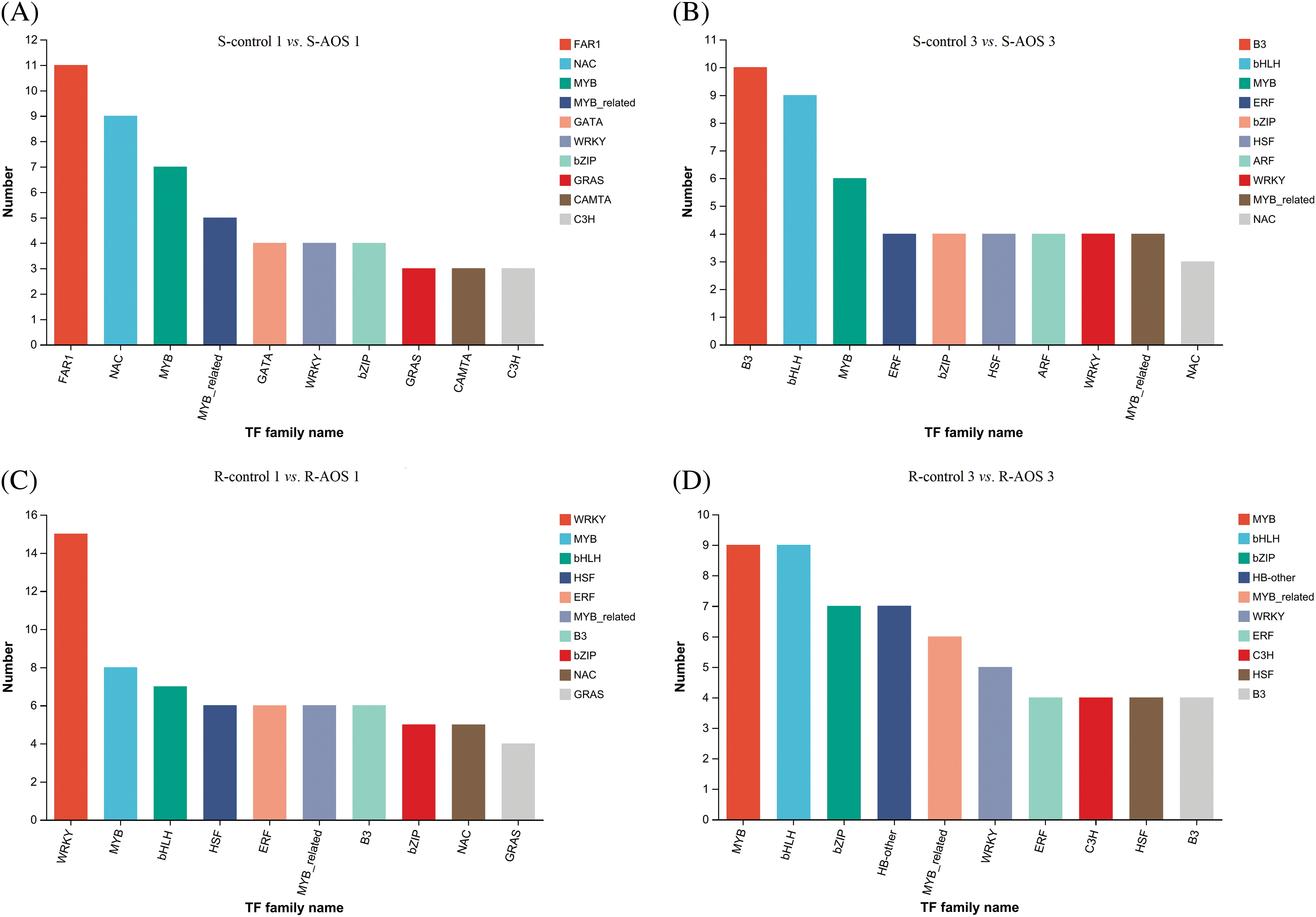
Figure 5: Top 10 transcription factor (TF) families of differentially expressed genes between pairwise comparisons of alginate oligosaccharides (AOS) groups to control groups under drought stress. (A) S-control 1 vs. S-AOS 1; (B) S-control 3 vs. S-AOS 3; (C) R-control 1 vs. R -AOS 1; (D) R-control 3 vs. R-AOS 3. 150 g L−1 PEG-6000 was used to simulate drought stress. S-control 1 and S-control 3, wheat shoots of the control group under drought stress for 24 (1 d) and 72 h (3 d), respectively; S-AOS 1 and S-AOS 3, wheat shoots of the AOS treatment group under drought stress for 24 and 72 h respectively; R-control 1 and R-control 3, wheat roots of the control group under drought stress for 24 and 72 h, respectively; R-AOS 1 and R-AOS 3, wheat roots of the AOS group under drought stress for 24 and 72 h, respectively; AOS, alginate oligosaccharides; PEG-6000, polyethylene glycol-6000. TF: transcription factor
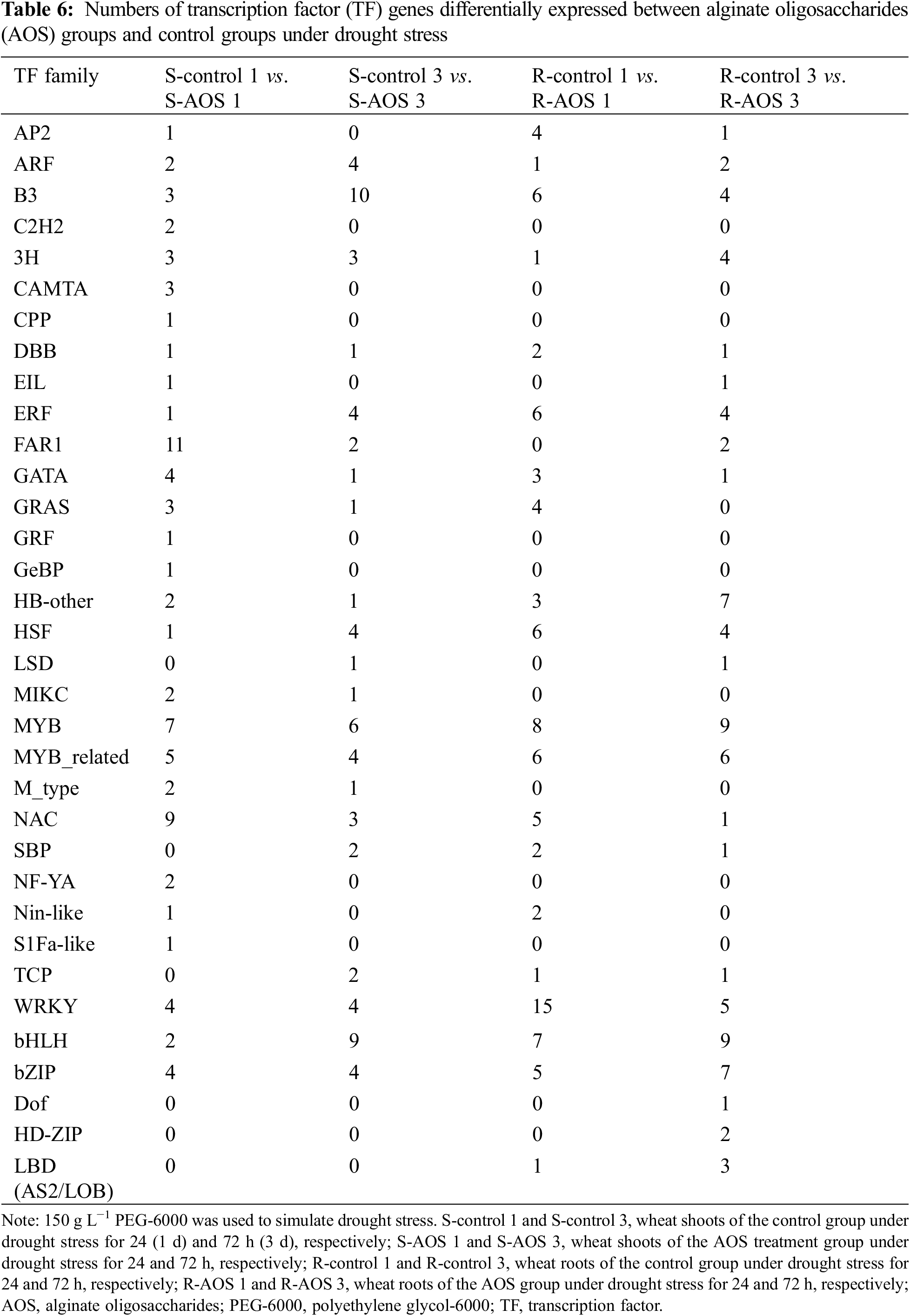
3.8 Verification of Some Key Genes Related to Signaling Transduction
To further clarify the signal transduction mechanism of AOS in regulating wheat drought resistance, the key genes related to signaling transduction were tested by qRT-PCR in view of the key roles of ABA in drought resistance. The results showed that, at 72 h after dehydration, the expression levels of ABA biosynthesis genes TaZEP, and ABA receptor genes TaPYR1, TaPYR2 and TaPYR3 in wheat shoots treated with AOS were significantly higher than those in the control (Fig. 6), indicating that AOS could positively regulate ABA synthesis and signaling in wheat responding to drought stress.
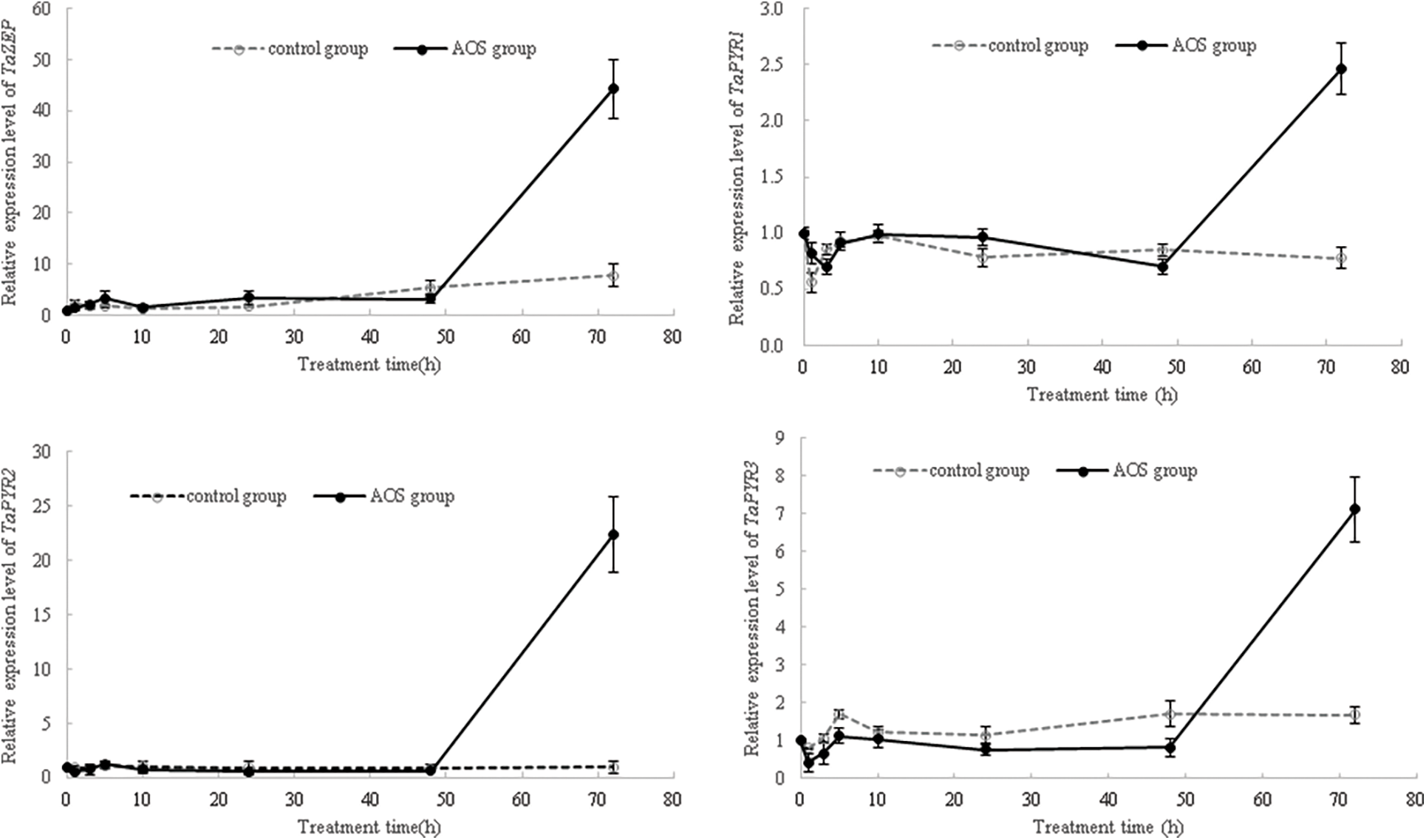
Figure 6: Effects of alginate oligosaccharides (AOS) on the expression of key genes involved in ABA signaling in wheat seedlings under drought stress. 150 g L−1 PEG-6000 was used to simulate drought stress. Control group, wheat seedling sprayed with deionized water under drought stress for 72 h (3 d); AOS group, wheat seedlings sprayed with 1 g L−1 AOS under drought stress for 72 h; AOS, alginate oligosaccharides
Then, after the MAPK signaling pathway was blocked by MAPK inhibitor PD 98059, the expression levels of ABA-responsive genes in wheat seedlings were again analyzed. The results showed that, at 24 and 48 h of treatment, the expression levels of TaPYR1, TaPYR2, TaPYR3, TaERF1 and TaNAC69 in wheat shoots under the PD 98059+PEG+AOS treatment were lower than those of the PD 98059+PEG treatment (Fig. 7), while the opposite trend was observed in wheat roots at 24 h. At 72 h of treatment, the expression levels of TaPYR1, TaPYR2, TaPYR3, and TaNAC69 in wheat shoots under the PD 98059+PEG+AOS treatment were higher than those of the PD 98059+PEG treatment, while the opposite trend was observed in wheat roots. Additionally, the expression levels of TaERF1 and TaNAC69 in wheat root treated with PD 98059+PEG+AOS were also lower than those of the PD 98059 PEG treatment at 48 h of treatment. These results further indicated that MAPK cascades were involved in AOS-induced drought resistance mediated by plant hormones in wheat, showing a time-effect relationship in different wheat tissues.
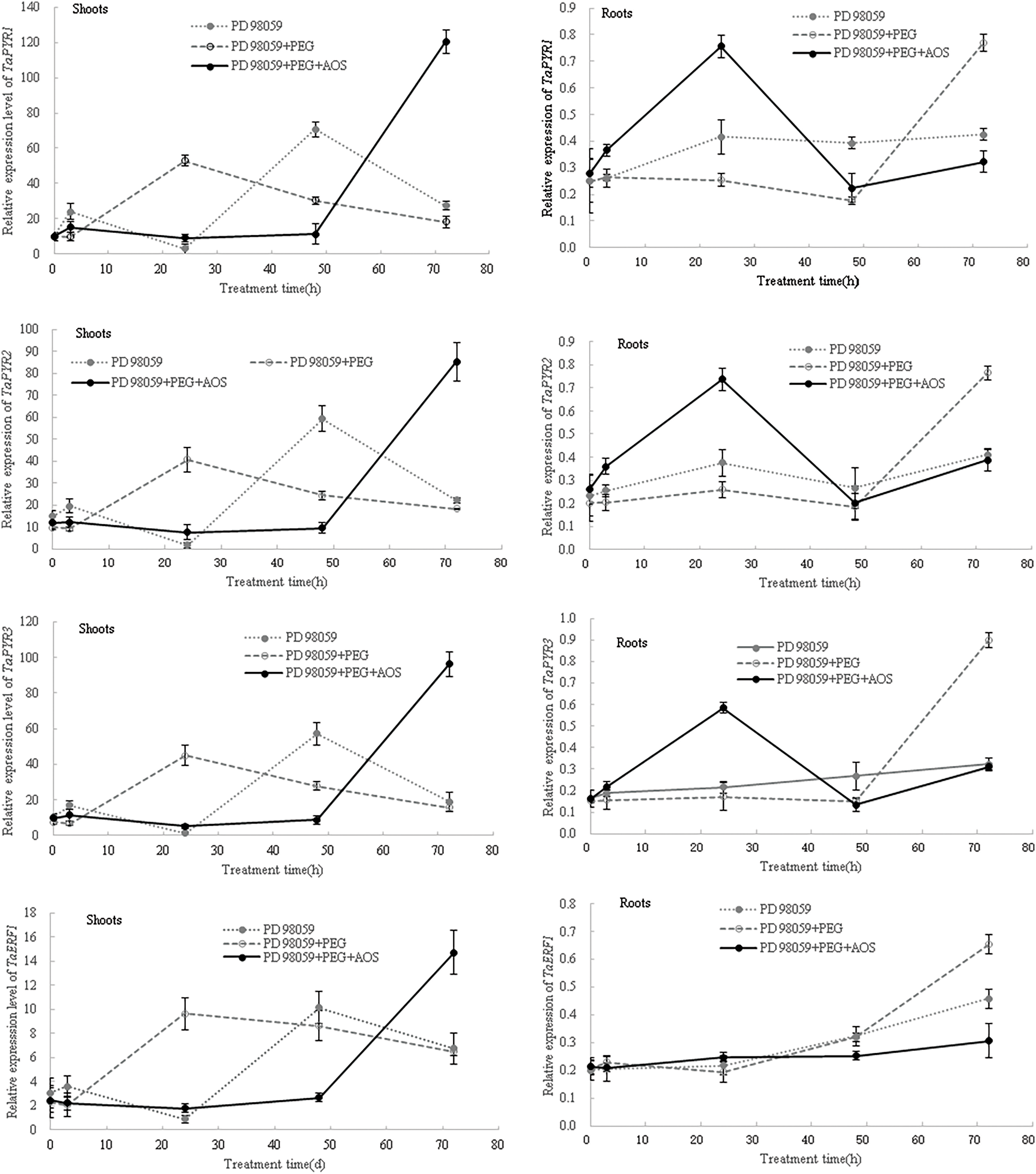
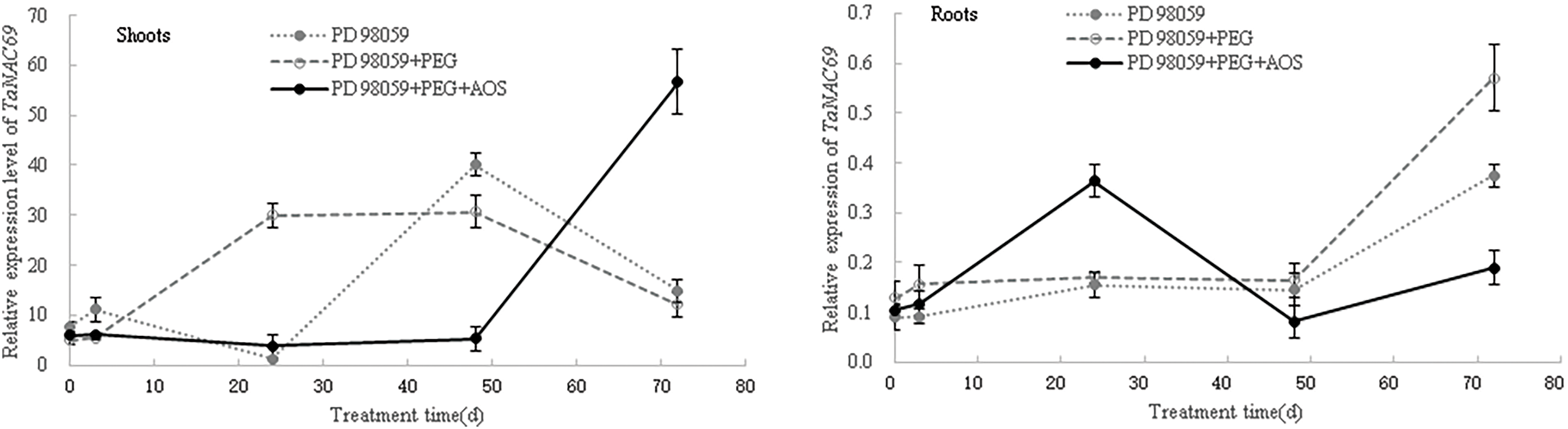
Figure 7: Effects of mitogen-activated protein kinases (MAPK) inhibitor PD 98059 on the expression of ABA-responsive genes induced by alginate oligosaccharides (AOS) in wheat seedlings under drought stress. 150 g L−1 PEG-6000 was used to simulate drought stress. PD 98059 group, wheat seedlings spayed with deionized water for 72 h (3 d) after being root -dipped in 50 μM PD 98059 for 6 h; PD98059+PEG group, wheat seedling sprayed with deionized water under drought stress for 72 h after being root-dipped in 50 μM PD 98059 for 6 h; PD 98059+PEG+AOS group, wheat seedlings sprayed with 1 g L−1 AOS under drought stress for 72 h after being root-dipped in 50 μM PD 98059 for 6 h. AOS, alginate oligosaccharides; MAPK, mitogen-activated protein kinases; PD 98059, a specific inhibitor of the MAPK/extracellular signal-regulated kinase 1/2 (ERK1/2) signaling pathway, without significant inhibitory activity of MAPK itself; PEG-6000, polyethylene glycol-6000.
In this study, the wheat seedlings were treated with 1 g L−1 AOS for 72 h, based on previous studies. The morphology of the wheat seedlings cultured was dramatically changed by AOS under drought stress, which was reflected in the increase of the length and FW of wheat shoots. Total Chl content and the activities of POD and SOD in leaves were also improved by AOS under drought stress, but the MDA content was reduced, compared with the only PEG treatment (Table 2). These results indicated that AOS reduced oxidative damage induced by drought stress, and therefore promoted the growth of wheat seedlings, in agreement with previous studies [15]. Similar results were also obtained for cucumber [18,19].
The fundamental means by which AOS affected wheat morphology and physiology under drought stress were by inducing gene changes at the molecular level. The transcriptome analysis of wheat seedlings reflected on the dynamic changes in AOS-induced genes and proteins directly. Compared with the control, a total of 10,064 (including 5,185 DEGs in shoots and 4,879 DEGs in roots) and 15,208 DEGs (including 4,827 DEGs in shoots and 10,381 DEGs in roots) in AOS-treated wheat seedlings were obtained at 24 and 72 h after dehydration, respectively (Figs. 2A–2D). Go and KEGG pathway enrichment analysis showed that these DEGs, which were annotated to BP (metabolic process, cellular processes, biological regulation and response to stimulus), CC (cell components, membrane components, organelle and membrane) and MF categories (binding, catalytic activity, transporter activity and transcription regulator activity), and were mainly involved in carbohydrate metabolism (starch and sucrose metabolism, carbon fixation in photosynthetic organisms, amino sugar and nucleotide sugar metabolism, glycolysis/gluconeogenesis, pyruvate metabolism, glyoxylate and dicarboxylate metabolism), secondary metabolites synthesis (phenylpropanoid biosynthesis, flavonoid biosynthesis) and lipid metabolism (fatty acid elongation, biosynthesis of unsaturated fatty acids, alpha-linolenic acid metabolism, cutin, suberine and wax biosynthesis), and signaling transduction (Table S1; Figs. 4A–4D). Further analysis showed that most DEGs in the main enriched KEGG pathways were up-regulated by AOS in wheat shoots (at 24 h after dehydration) and roots (at 72 h after dehydration). These key genes were related to chlorophyll a-b binding protein, amylosynthease, phosphotransferase, peroxidase, phenylalanine ammonia lyase, flavone synthase, glutathione synthetase and so on. Signaling molecules, such as MAPK, plant hormones, H2O2 and calcium, were involved in the AOS -induced wheat drought resistance (Tables 4 and 5).
Photosynthesis is the primary metabolic sink for plant growth and the basis of crop yield formation; however, it is a complex metabolic process that includes light absorption, energy conversion, electron transfer, adenosine triphosphate synthesis, and carbon dioxide fixation [36]. In this study, most DEGs related to photosynthesis–antenna proteins (chlorophyll a-b binding protein) in wheat shoots (at 24 h after dehydration) and roots (at 72 h after dehydration) were up-regulated by AOS (Table 5). The key genes involved in starch and sucrose metabolism, including amylosynthease, phosphotransferase and trehalose-6-phosphate phosphatase, were also up-regulated in wheat roots at 72 h (Table 5). These results suggested that AOS enhanced the photosynthetic ability of wheat seedlings and accumulated more photosynthate under drought stress, which was confirmed by the increase in total Chl content and FW of wheat shoots (Table 2). Other findings for flowering Chinese cabbage showed that AOS raised the net photosynthetic rate, carboxylation efficiency of carbon dioxide and light saturation point in functional leaves, but reduced the light compensation point, resulting in increases in sucrose and starch accumulation and yield [37]. Leaf photosynthesis and above-ground growth of maize seedlings were improved by AOS under drought conditions, which was reflected in increases to the net photosynthetic rate, relative Chl content (SPAD value) and dry matter accumulation [38]. Similar results were also obtained for Catharanthus roseus L [39]. Our results therefore agreed with the findings of the previous studies at the transcriptional level.
The biomembrane system is the initial site of injury, particularly in terms of its structure, function, stability, and enzyme activity, resulting in substantial metabolic imbalance, which affects plant growth and development [40]. Phospholipids, as major components of the cell membrane, participate in many cellular biological processes by controlling and regulating cellular function [41]. Glycerolipid metabolism of plants responds to the changing environments, leading to the modification of membrane lipid composition to ensure optimal biochemical and physical properties [42]. In our study, most DEGs enriched in the biosynthesis of unsaturated fatty acids and alpha-linolenic acid metabolism were up-regulated (Fig. 4 and Tables 4, 5), suggesting that AOS altered the membrane lipid composition of wheat seedlings under drought stress. The results of flowering Chinese cabbage showed that AOS raised linoleic acid (C18:2) content while reduced the contents of palmitic acid (C16:1) and stearic acid (C18:0), resulting in an increase in the index of unsaturated fatty acid (IUFA) of thylakoid membrane lipids [43]. Furthermore, IUFA was positively correlated with photosystem II electron transport activity, affecting thylakoid membrane fluidity and membrane-bound enzyme activity [44]. Therefore, the AOS-induced changes in membrane lipid composition were beneficial for improving the photosynthetic capacity of wheat leaves under drought stress (Figs. 4A, 4B and Tables 2, 4). Moreover, cutin, suberine and wax, a family of complex biopolyesters composed of fatty acids and aromatic compounds, limited water loss through non-stomatal transpiration under drought stress, and their biosynthesis began with fatty acid synthesis and elongation [45,46]. Therefore, most DEGs enriched in fatty acid elongation and cutin, suberine and wax biosynthesis were up-regulated in wheat shoots (Fig. 4A and Tables 4, 5), helping to improve RWC and FW of wheat shoots under drought stress (Table 2).
Secondary metabolites of plants perform a vital role in response to drought stress. For example, phenylpropanoid metabolism yields more than 8,000 metabolites (including flavonoids, hydroxycinnamic acid esters, hydroxycinnamic acid amides, and the precursors of lignin and tannins) contributing to plant development and plant–environment interplay [47]. Among them, flavonoids function as antioxidants that reduce the oxidative damage caused by ROS accumulation under abiotic stresses such as drought [48]. In our study, most DEGs involved in phenylpropanoid biosynthesis and flavonoid biosynthesis were up-regulated, mainly including peroxidase, phenylalanine ammonia lyase, β-glucosidase, 4-coumarate: coenzyme A ligase, phenylalanine deaminase, 4-α-glucanotransferase, glycosyltransferase and flavone synthase in wheat shoots at 24 h after dehydration (Tables 4 and 5), which was beneficial for alleviating the oxidative damage caused by drought stress, and improving the growth of wheat seedlings. Additionally, glutathione, a small intracellular thiol molecule considered to be a strong non-enzymatic antioxidant, plays an important role in antioxidant defense in plants [49]. Most DEGs involved in glutathione metabolism were up-regulated, mainly including glutathione synthetase, ascorbate peroxidase and glutathione-S-transferase in wheat roots at 72 h after dehydration (Tables 4 and 5), helping to reduce the oxidative damage caused by drought, which was also confirmed by the decrease of the MDA content in AOS-treated wheat leaves under drought stress (Table 2).
ABA acts as a major signaling molecule in response to drought, by inducing antioxidant defense and regulating stomatal closure, in conjunction with ROS, Ca2+, and sugars [25,26,50,51]. MAPK cascades also played important roles in transducing signals initiated by diverse internal and external cues involving in growth, development, and responses to biotic and abiotic stresses [52]. Our results confirmed that signaling molecules, such as MAPK, plant hormones, H2O2 and calcium, were involved in the AOS-induced drought resistance in wheat (Fig. 4 and Tables 4, 5). AOS induced ABA synthesis and signaling in wheat responding to drought stress, by up-regulating the expression levels of TaZEP, TaPYR1, TaPYR2 and TaPYR3 (Fig. 6). However, after blocking the MAPK signaling pathway, the ABA-responsive genes were inhibited first and then induced in wheat shoots treated with AOS under drought stress, while the opposite trend was observed in wheat roots (Fig. 7), which further indicated that MAPK cascades were involved in AOS-induced drought resistance mediated by ABA in wheat, showing a time-effect relationship in different wheat tissues. These results were basically consistent with the results of transcriptome data; however, the detailed signal transduction mechanisms still need further investigation.
Among all the stress-resistance systems in plants, many TFs related to plant tolerance to stresses, including bZIP, NAC, AP2, WRKY, PHD, DREB, ERF and MYB families, have been identified as playing important roles in abiotic stress by binding specific cis-acting elements to form a complex regulatory network [32,53,54]. In our study, differentially expressed TF genes that regulated plant stress response mainly include the NAC, MYB, MYB-related, WRKY, bZIP, bHLH families (Table 5). MYB and bHLH TFs played a central regulatory role in phenylpropanoid metabolism, especially the regulation of the structural genes of lignin and flavonoid biosynthesis [54,55]. TaNAC5D-2, TaNAC48 and TaNAC69 acted in the ABA-mediated transcriptional regulation in response to drought stress in wheat [32,53]. The expression levels of TaNAC2, TabZIP15 and TaSIM were negatively correlated with MDA content, but positively correlated with RWC under drought stress [56]. TaERF1 induced by exogenous hormones (ABA, ethylene, auxin and salicylic acid) increased multiple stresses tolerance including drought [57]. AOS changed the expression profile of the above TFs genes in our study (Table 5 and Fig. 7), which may regulate the morphology and physiology of wheat seedlings in adapting to drought stress. However, not much is known about the complex molecular network that brings about drought resistance induced by AOS, thus it is necessary to study the interactions of the different signaling pathways induced by AOS, for example using the gene chip and laser scanning confocal microscope technique in a follow-up study.
In summary, physiological and transcriptome analysis preliminarily confirmed that AOS reduced oxidative damage caused by drought stress, and thereby promoted the growth of wheat seedlings. This appeared to be a cumulative outcome of DEGs mainly involved in the biosynthesis of secondary metabolites, carbohydrate metabolism, lipid metabolism, amino acid metabolism, and signaling transduction. Signaling molecules (MAPK, plant hormones, H2O2 and calcium) and transcription factors (NAC, MYB, MYB-related, WRKY, bZIP family members) were involved in the AOS-induced drought resistance in wheat. These results help us to understand the mechanisms of the improved drought resistance by AOS at the transcriptome level in wheat, and provide a theoretical basis for the exploitation and application of AOS as a type of environmentally sustainable biological agent for improving the drought resistance of agricultural crops.
Acknowledgement: We thank Dr. Darrell W.S. Tang (Wageningen University) for their great efforts and time to provide constructive comments and edit the language that greatly improved the quality of this manuscript.
Funding Statement: This research was funded and supported by the National Natural Science Foundation of China (Grant Number 32001443), Zhengzhou Major Science and Technology Innovation Project of Henan Province of China (Grant Number 2020CXZX0085), and Science and Technology Inovation Team of Henan Academy of Agricultural Sciences (Grant Number 2024TD2).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Y.Z. and Y.Y.; data collection and analysis: J.M.; draft manuscript preparation: Y.Z. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The transcriptome raw data that has been used is confidential. The other datasets supporting the conclusions of the article are included within the article.
Ethical Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2023.046811.
References
1. Food and Agriculture Organization of the United Nations. The state of food security and nutrition in the World 2021: transforming food systems, ensuring food security, improving nutrition and ensuring affordable healthy diets for all. Available from: https://www.who.int/publications/m/item/the-state-of-food-security-and-nutrition-in-the-world-2021. [Acessed 2022]. [Google Scholar]
2. Wu BF, Ma ZH, Boken VK, Zeng HW, Shang JL, Igor S, et al. Regional differences in the performance of drought mitigation measures in 12 major wheat-growing regions of the world. Agr Water Manage. 2002;273(1):107888. [Google Scholar]
3. Zhuo W, Fang SB, Wu D, Wang L, Li MQ, Zhang J, et al. Integrating remotely sensed water stress factor with a crop growth model for winter wheat yield estimation in the North China Plain during 2008–2018. Crop J. 2022;10(5):1470–82. [Google Scholar]
4. Ober ES, Clark CJA, Perry A. Traits related to genotypic differences in effective water use and drought tolerance in UK winter wheats. Aspects Appl Biol. 2010;105:13–22. [Google Scholar]
5. Zhang Q, Yu HQ, Sun P, Singh VP, Shi PJ. Multisource data based agricultural drought monitoring and agricultural loss in China. Glob Planet Change. 2019;172:298–306. [Google Scholar]
6. Trnka M, Feng S, Semenov MA, Olesen JE, Kersebaum KC, Rötter RP, et al. Mitigation efforts will not fully alleviate the increase in water scarcity occurrence probability in wheat-producing areas. Sci Adv. 2019;5(9):eaau2406. [Google Scholar] [PubMed]
7. Xu H, Jiang SM, Yuan HW, Liu J, Jin JL. Sensitivity of winter wheat to drought occurring at different growth stages. J Irrig Drain. 2021;40(8):66–72 (In Chinese). [Google Scholar]
8. Javed A, Ahmad N, Ahmed J, Hameed A, Ashraf MA, Zafar SA, et al. Grain yield, chlorophyll and protein contents of elite wheat genotypes under drought stress. J King Saud Univ Sci. 2022;34(7):102279. [Google Scholar]
9. Mu Q, Xu JT, Yu M, Guo ZJ, Dong MQ, Cao YX, et al. Physiological responses of winter wheat (Triticum aestivum L.) during vegetative growth to gradual, persistent and intermittent drought. Agr Water Manage. 2022;274(1):107911. [Google Scholar]
10. Chen QM, Liu YJ, Ge QS, Pan T. Impacts of historic climate variability and land use change on winter wheat climatic productivity in the North China Plain during 1980–2010. Land Use Policy. 2018;76:1–9. [Google Scholar]
11. Boudjabi S, Kribaa M, Chenchouni H. Sewage sludge fertilization alleviates drought stress and improves physiological adaptation and yield performances in Durum wheat (Triticum duruma double-edged sword. J King Saud Univ Sci. 2019;31(3):336–44. [Google Scholar]
12. Mwadzingeni L, Shimelis H, Dube E, Laing MD, Tsilo TJ. Breeding wheat for drought tolerance: progress and technologies. J Integr Agr. 2016;15(5):935–43. [Google Scholar]
13. Pavia I, Roque J, Rocha L, Ferreira H, Castro C, Carvalho A, et al. Zine priming and foliar application enhances photoprotection mechanisms in drought-stressed wheat plants during anthesis. Plant Physiol Bioch. 2019;140:27–42. [Google Scholar]
14. Fernando TN, Ariadurai SA, Disanayaka CK, Kulathunge S, Aruggoda AGB. Development of radiation grafted super absorbent polymers for agricultural applications. Enrgy Proced. 2017;127:163–77. [Google Scholar]
15. Liu H, Zhang YH, Yin H, Wang WX, Zhao XM, Du G. Alginate oligosaccharides enhanced Triticum aestivum L. tolerance to drought stress. Plant Physiol Bioch. 2013;62:33–40. [Google Scholar]
16. Wang YX, Suo B, Zhao PE, Qu XF, Yuan LG, Zhao XJ, et al. Effect of exogenous abscisic acid on psbA expression at grain filling stage in two wheat cultivars under drought stress. Acta Agronomica Sinica. 2011;37(8):1372–7. [Google Scholar]
17. Vasudevan UM, Lee OK, Lee EY. Alginate derived functional oligosaccharides: recent developments, barriers, and future outlooks. Carbohyd Polym. 2021;267:118158. [Google Scholar]
18. Zhang CG, Wang WX, Zhao XM, Wang HY, Yin H. Preparation of alginate oligosaccharides and their biological activities in plants: a review. Carbohyd Res. 2020;494:108056. [Google Scholar] [PubMed]
19. Li JQ, Wang XY, Lin XP, Yan GF, Liu L, Zheng H, et al. Alginate-derived oligosaccharides promote water stress tolerance in cucumber (Cucumis sativus L.). Plant Physiol Bioch. 2018;130:80–8. [Google Scholar]
20. Santaniello A, Scartazza A, Gresta F, Loreta F, Biasone A, Di Tommaso D, et al. Ascophyllum nodosum seaweed extract alleviates drought stress in Arabidopsis by affecting photosynthetic performance and related gene expression. Front Plant Sci. 2017;8:1362. [Google Scholar] [PubMed]
21. Singhal RK, Kumar V, Bose B. Improving the yield and yield attributes in wheat crop using seed priming under drought stress. J Pharmacogn Phytochem. 2019;8(2):214–20. [Google Scholar]
22. Wang XP, Liu HL, Yu FL, Jia Y, Sha HJ, Zhao HW. Differential activity of the antioxidant defense system and alteration in the accumulation of osmolyte and reactive oxygen species under drought stress and recovery in rice (Oryza sativa L.) tillering. Sci Rep. 2019;9(1):8543. [Google Scholar] [PubMed]
23. Saha D, Choyal P, Mishra UN, Dey P, Bose B, Prathibha MD, et al. Drought stress responses and inducing tolerance by seed priming approach in plants. Plant Stress. 2022;4:100066. [Google Scholar]
24. Zhao YJ, Zhang YY, Bai XY, Lin RZ, Shi GQ, Du PP, et al. TaNF-YB11, a gene of NF-Y transcription factor family in Triticum aestivum, confers plant drought tolerance via modulating osmolyte accumulation and active oxygen species homeostasis. J Integr Agr. 2022;11:3114–30. [Google Scholar]
25. Parwez R, Aftab T, Gill SS, Naeem M. Abscisic acid signaling and crosstalk with phytohormones in regulation of environmental stress responses. Environ Exp Bot. 2022;199:104885. [Google Scholar]
26. Wasilewska A, Vlad F, Sirichandraa C, Redko Y, Jammes F, Valon C, et al. An update on abscisic acid signaling in plants and more. Mol Plant. 2008;1(2):198–217. [Google Scholar] [PubMed]
27. Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124(4):509–25. [Google Scholar] [PubMed]
28. Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57(1):781–803. [Google Scholar] [PubMed]
29. Liu H, Feng LQ, Liu XJ. Effects of oligosaccharides on abscisic acid synthesis in wheat. Acta Agriculturae Zhejianggensis. 2015;27(1):12–5 (In Chinese). [Google Scholar]
30. Guo JJ, Zhang RY, Cheng SQ, Fu ZQ, Jia P, Luan H, et al. Physiological and transcriptomic analysis reveal the crucial factors in heat stress response of red raspherry ‘Polka’ seedings. Front Plant Sci. 2023;14:1233448. [Google Scholar] [PubMed]
31. Lamin-Samu AT, Zhuo SB, Ali M, Lu G. Long non-coding RNA transcriptome landscape of anthers at different developmental stages in response to drought stress in tomato. Genomics. 2022;114(4):110383. [Google Scholar] [PubMed]
32. Chen J, Gong Y, Gao Y, Zhou YB, Chen M, Xu Z, et al. TaNAC48 positively regulates drought tolerance and ABA responses in wheat (Triticum aestivum L.). Crop J. 2021;9(4):785–93. [Google Scholar]
33. Búfalo J, Rodrigues TM, de Almeida LF, Tozin LR, Marques MO, Boaro CSF, et al. PEG-induced osmotic stress in Mentha piperita L. structural features and metabolic responses. Plant Physiol Biochem. 2016;105:174–84. [Google Scholar]
34. Wang XK, Huang JL. Principles and techniques of plant physiological biochemical experiment (In Chinese). China: Higher Education Press; 2015. [Google Scholar]
35. Fan GQ, Cao XB, Niu SY, Deng MJ, Zhao ZL, Dong YP. Transcriptome, microRNA, and degradome analyses of the gene expression of Paulownia with phytoplamsa. BMC Genom. 2015;16:896. [Google Scholar]
36. Leister D. Enhancing the light reactions of photosynthesis: strategies, controversies, and perspectives. Mol Plant. 2023;16:4–22. [Google Scholar] [PubMed]
37. Zhang YH, Yin H, Wang WX, Zhao XM, Du YG, Wu L. Enhancement in photosynthesis characteristics and phytohormones of flowering Chinese cabbage (Brassica campestris L.var. utilis Tsen et Lee) by exogenous alginate oligosaccharides. J Food Agric Environ. 2013;11(1):669–75. [Google Scholar]
38. Guo YL, Huang GM, Wei ZX, Feng TY, Zhang K, Zhang M, et al. Exogenous application of coronatine and alginate oligosaccharide to maize seedlings enhanced drought tolerance at seedling and reproductive stages. Agr Water Manage. 2023;279:108185. [Google Scholar]
39. Naeem M, Aftab T, Ansari AA, Idrees M, Ali A, Khan MMA, et al. Radiolytically degraded sodium alginate enhances plant growth, physiological activities and alkaloids production in Catharanthus roseus L. J Radiat Res Appl Sci. 2015;8(4):606–16. [Google Scholar]
40. Zhang H, Dong JL, Zhao XH, Zhang YM, Ren JY, Xing LT, et al. Research progress in membrane lipid metabolism and molecular mechanism in peanut cold tolerance. Front Plant Sci. 2019;10:838. [Google Scholar] [PubMed]
41. Champeyroux C, Stoof C, Rodriguez-Villalon A. Signaling phospholipid in plant development: small couriers determining cell fate. Plant Biol. 2020;57:61–71. [Google Scholar]
42. Szymanski J, Brotman Y, Willmitzer L, Cuadros-Inostroza Á. Linking gene expression and membrane lipid composition of Arabidopsis. Plant Cell. 2014;26(3):915–28. [Google Scholar] [PubMed]
43. Zhang YH, He AL, Sun KG, Hu NH, Wu LS. Effects of alginate-derived oligosaccharides on composition and characteristic of thylakois membrane of Brassica chinensis. Acta Agriculturae Boreali-occidentalis Sinica. 2016;25(1):129–35 (In Chinese). [Google Scholar]
44. Vijayan P, Browse J. Photoinhibition in mutants of Arabidopsis deficient in thylakoid unsaturation. Plant Physiol. 2002;129(2):876–85. [Google Scholar] [PubMed]
45. Kunst L, Samuels AL. Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res. 2003;42(1):51–80. [Google Scholar] [PubMed]
46. Pollard M, Beisson F, Li YH, Ohirogge JB. Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 2013;13(5):236–46. [Google Scholar]
47. Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J Integr Plant Biol. 2021;63(1):180–209. [Google Scholar] [PubMed]
48. Nakabayashi R, Saito K. Integrated metabolomics for abiotic stress responses in plants. Curr Opin Plant Biol. 2015;24:10–6. [Google Scholar] [PubMed]
49. Michael T, Helena S. The glutathione system as a stress marker in plant ecophysiology is a stress-response concept valid. J Exp Bot. 2004;55:1955–62. [Google Scholar]
50. Jia KP, Mi JN, Ali S, Ohyanagi H, Moreno JC, Ablazov A, et al. An alternative, zeaxanthin epoxidase-independent abscisic acid biosynthetic pathway in plants. Mol Plant. 2021;15:151–66. [Google Scholar] [PubMed]
51. Nakashima K, Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013;32:959–70. [Google Scholar] [PubMed]
52. Zhang MM, Su JB, Zhang Y, Xu J, Zhang SQ. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr Opin Plant Biol. 2018;45:1–10. [Google Scholar] [PubMed]
53. Ma JH, Tang XX, Sun B, Wei JT, Ma LY, Yuan M, et al. A NAC transcription factor, TaNAC5D-2, acts as a positive regulator of drought tolerance through regulating water loss in wheat (Triticum aestivum L.). Environ Exp Bot. 2022;196:104805. [Google Scholar]
54. Yang WL, Li N, Fan YX, Dong BY, Song ZH, Cao HY, et al. Transcriptome analysis reveals abscisic acid enhancing drought resistance by regulating genes related to flavonoid metabolism in pigeon pea. Environ Exp Bot. 2021;191:104627. [Google Scholar]
55. Ohtani M, Demura T. The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr Opin Biotech. 2019;56:82–7. [Google Scholar] [PubMed]
56. Shen LJ, Ni ZY, Shi SB, Yu YH. Gene expression and function analysis of drought-related transcription factor in wheat. Mol Plant Breed. 2024;22(2):349–355 (In Chinese). [Google Scholar]
57. Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol. 2007;65(6):719–32. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools