 Open Access
Open Access
ARTICLE
Genome-Wide Identification and Expression Pattern Analysis of LONELY GUY Gene Family in Walnut (Juglans regia)
1 State Key Laboratory of North China Crop Improvement and Regulation/College of Forestry, Hebei Agricultural University, Baoding, 071000, China
2 Department of Agriculture, Abdul Wali Khan University, Mardan, 23200, Pakistan
* Corresponding Authors: Guohui Qi. Email: ; Peng Jia. Email:
(This article belongs to the Special Issue: Integrative Approaches to Plant Stress Responses under Changing Climate Conditions)
Phyton-International Journal of Experimental Botany 2024, 93(12), 3331-3346. https://doi.org/10.32604/phyton.2024.059402
Received 07 October 2024; Accepted 21 November 2024; Issue published 31 December 2024
Abstract
LONELY GUY (LOG) is a pivotal cytokinin-activating enzyme that plays an important role in plant growth, development, and stress responses. Walnut (Juglans regia), an important woody oilseed species, has not yet undergone systematic identification of its LOG gene family. In this study, we identified 17 JrLOG genes in the walnut genome, which are unevenly distributed across 11 chromosomes. JrLOG gene expansion was primarily driven by gene duplication, along with purifying selection. Members of the JrLOG family were categorized into five groups, each exhibiting analogous gene structures, featured motifs, and conserved domains. Transcriptome and quantitative real-time PCR analyses revealed that JrLOG gene expression was tissue-specific, developmentally regulated, and responsive to stress situations. Notably, JrLOG3, which localizes to the cell membrane, exhibited high expression levels in leaves and responded to both cold and pathogen infection treatments. This study represents the first comprehensive identification of the LOG gene family in walnut, offering essential data for further functional studies of this gene family.Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| LOG | LONELY GUY |
| CKs | Cytokinin |
| qPCR | Quantitative real-time PCR |
Cytokinins (CKs), one of the six major plant hormones, play a crucial role in regulating various biological processes. Their primary function is to stimulate cell division, which is vital for plant growth and tissue formation [1]. CKs also work in conjunction with auxins to control cell differentiation, tissue formation, and stress response [2]. Additionally, CKs interact either antagonistically or synergistically with other hormones to modulate growth patterns, including branching and the balance between root and shoot growth [3]. CKs also delay leaf senescence by preserving chlorophyll [4]. Under environmental stress, CK levels fluctuate, enabling plants to adapt and enhancing their stress tolerance [5].
CKs are predominantly synthesized in the meristematic tissues of roots, especially at the root apices, and are transported to other parts of the plant via the xylem. The homeostasis of CKs is tightly regulated to ensure normal physiological activities. Several enzymes participate in the synthesis, activation, modification, and degradation of CKs. Among them, LONELY GUY (LOG) plays a pivotal role by directly activating CKs [6] and regulating meristem activity [7]. LOG was first identified in rice mutants with defects in meristem maintenance [8]. In plants, LOG encodes a cytokinin riboside 5′-monophosphate phosphoribohydrolase, which converts inactive cytokinin nucleotides into active free bases [9]. Furthermore, LOG has been demonstrated to exist in all living organisms [10]. In model plants such as Arabidopsis thaliana (Arabidopsis) and Oryza sativa (rice), as well as crops like Triticum aestivum (wheat), LOG genes have been characterized for their roles in regulating plant growth and in responding to environmental stresses [9,11]. Arabidopsis contains nine LOG genes, seven of which have been functionally investigated. Various mutations and overexpression techniques have been employed to elucidate the functions of distinct members of the LOG gene family at various stages of the plant life cycle and in specific tissues. For instance, the log3log4log5 triple mutant demonstrated a reduction in shoot apical meristem size, leading to smaller inflorescences, fewer flowers, and seed pods [12]. Mutations in AtLOG resulted in shorter root systems [13,14]. In contrast, LOG overexpression caused more subtle phenotypes [12]. In poplar, overexpression of LOG1 significantly increased xylem proliferation [15]. The GY3 locus, encoding a rice LOG protein, notably enhanced grain yield [11]. Similarly, overexpression of VlLOG11 from grapevine in tomatoes increased fruit yield and activated CK signaling-related gene expression [16]. In tomatoes, LOG1 stimulated axillary meristem activation, leading to ectopic branching [17]. While knockout of Solanum melongena SmLOG1 inhibited the formation of spines on leaves, stems, and fruit sepals [18].
Beyond its role in development, LOG is also critical in mediating responses to environmental and biotic stresses. Overexpression of Ricinus communis RcLOG5 significantly enhanced drought, salt, and cold tolerance in Arabidopsis [19]. Gossypium hirsutum (cotton) GhLOG3 overexpression enhanced salt tolerance, whereas its downregulation increased sensitivity to salt stress [20]. Under drought and salinity stress, OsLOG-overexpressing rice lines showed reduced H2O2 accumulation and elevated antioxidant enzyme activity [21]. Additionally, OsLOGL1 contributed to yield maintenance under high nighttime temperatures [7], and OsLOG5 regulated rice yield under various environmental and nutrient stress conditions [22].
Walnut (Juglans regia) is a widely cultivated nut tree belonging to the family Juglandaceae, valued for its nutritional benefits and high-quality wood, making it commercially significant. However, walnut growth and development are frequently challenged by abiotic stresses such as extreme temperature fluctuations and biotic stresses like anthracnose disease. While several genes involved in regulating these processes have been well-characterized, research on the role of LOG genes, which are critical for CK activity in plants, remains limited in walnut. Therefore, this study aims to identify the LOG gene family members in walnut on a genome-wide scale and systematically analyze their expression profiles, providing fundamental data for further investigation into their functional roles in walnut.
2.1 Identification of LOG Genes in the Walnut Genome
LOG protein sequences from Arabidopsis were downloaded from the TAIR database and utilized as query sequences to conduct a BLAST search against the protein-coding genes in the walnut genome sequenced by Huang Xuehui’s Lab (http://www.xhhuanglab.cn/ (accessed on 20 November 2024)), with an E-value threshold set at 1E−5. The obtained sequences were further examined using Pfam (http://pfam-legacy.xfam.org/ (accessed on 20 November 2024)) and CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 20 November 2024)) to verify the presence of specific domains. The identified candidate genes were renamed according to their chromosomal locations. The physicochemical properties of these walnut LOG (JrLOG) proteins were examined online using the ExPASy (https://www.expasy.org/ (accessed on 20 November 2024)).
2.2 Phylogenetic Analysis and Chromosomal Localization
The LOG protein sequences from rice, Arabidopsis, and walnut were used to perform a multiple sequence alignment analysis. The resulting alignment was used to construct a phylogenetic tree using MEGA7.0 software, employing the Neighbor-Joining (N-J) statistical method with 1000 bootstrap replicates and the Gamma distribution model. Based on genome annotation data, the chromosomal locations of each JrLOG gene were visualized using TBtools software.
2.3 Gene Structure and Conserved Domain Analysis
The gene structures of JrLOG genes, including coding sequences and introns, were visualized using the GSDS online tool (https://gsds.cgrpoee.top/ (accessed on 20 November 2024)). Conserved motifs in the JrLOG proteins were detected using the MEME suite (https://meme-suite.org/meme/ (accessed on 20 November 2024)) (Table S1).
2.4 Gene Collinearity Analysis
Collinearity relationships among walnut chromosomes, as well as between walnut, Arabidopsis, and rice, were analyzed using the MCScanX program. Gene duplication events for JrLOG in walnut were visualized with Advanced Circos. The nonsynonymous (Ka) and synonymous (Ks) substitution rates, along with their ratio (Ka/Ks), were calculated using the KaKs Calculator software (Table S2).
2.5 Promoter Cis-Regulatory Element Analysis
The 1500 bp upstream sequences from the start codon of each JrLOG gene were extracted from the walnut genome and submitted to the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 20 November 2024)) for the prediction of cis-regulatory elements. The number and distribution of these elements were visualized after statistical analysis.
2.6 Plant Materials and Treatments
Two-year-old, uniformly growing Juglans regia ‘Lvling’ walnut trees were used as experimental materials and randomly divided into two groups. For cold treatments, one group was placed in a growth chamber at 4°C, while the control group was kept at 25°C. Walnut leaves were collected at 0.5, 1, and 2 h post-treatment, immediately frozen in liquid nitrogen and stored at −80°C.
The expression profiles of JrLOG genes in various walnut tissues were based on RNA-Seq data from 19 different tissues [23]. The expression patterns of JrLOG in walnut endopleura during different developmental stages were derived from transcriptomic data covering 35 to 147 days post-flowering [24]. The expression levels of JrLOG in response to Colletotrichum gloeosporioides infection were analyzed using published RNA-Seq data [25]. The response of JrLOG to cold stress was further explored using quantitative real-time PCR (qPCR) following the protocol described by Zheng et al. [26]. The primers used are listed in Table S1.
2.8 Gene Cloning, Sequence Analysis, and Subcellular Localization
The coding sequence of JrLOG3 was cloned from walnut leaf cDNA following the method outlined by Jia et al. [27]. The predicted amino acid sequence was employed for multiple sequence alignment and phylogenetic analysis, as described in Section 2.2, by comparing it with LOG proteins from different species with known functions. The JrLOG3 coding region was inserted into the pCambia2300 vector, creating a JrLOG3-EGFP expression cassette driven by the 35S constitutive promoter. The recombinant vector was then introduced into tobacco (Nicotiana benthamiana) epidermal cells via Agrobacterium-mediated transformation. After 3 days of dark incubation, the transformed tobacco cells were examined for green fluorescent protein (GFP) signals using confocal microscopy, following the previously described procedure [26].
3.1 Identification of Walnut LOG Genes
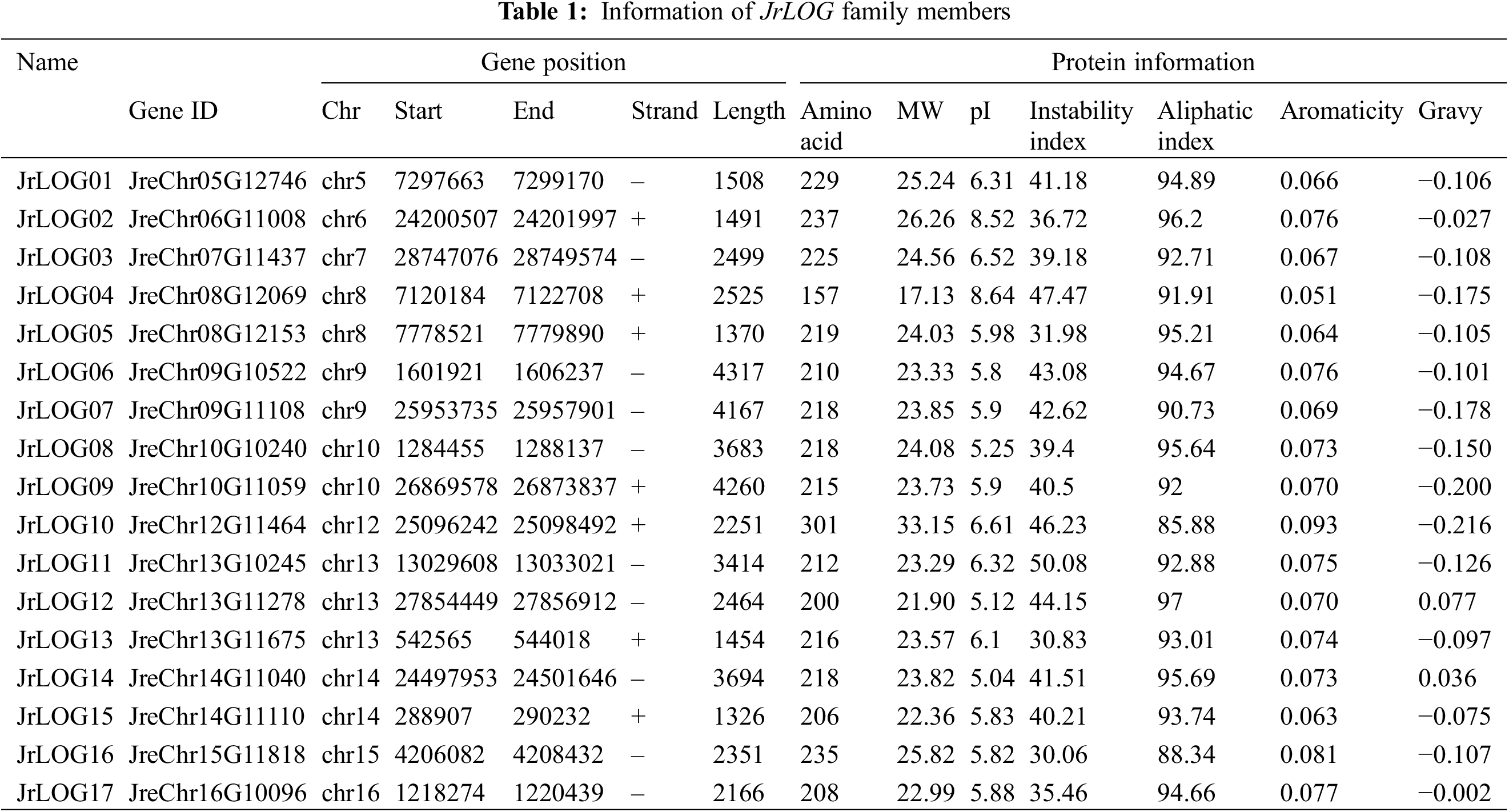
Nine AtLOG genes were identified and documented in the Arabidopsis thaliana genome. BLASTP searches were performed against the walnut genome protein database to discover LOG genes using the nine AtLOG protein sequences. Seventeen potential JrLOG genes were identified by manual inspection and verification using the NCBI conserved domain database (Table 1). These JrLOG genes were designated sequentially based on their chromosomal locations (JrLOG01–JrLOG17). The 17 JrLOG genes were distributed across 11 chromosomes of the walnut genome, with chromosome 13 containing the highest number of LOG genes (3), while chromosomes 8, 9, 10, and 14 each contained two genes. Chromosomes 5, 6, 7, 12, 15, and 16 each housed a single JrLOG gene (Table 1).
The relationships among the LOG genes were elucidated by constructing a neighbor-joining tree based on the LOG protein sequences from Arabidopsis, rice, and walnut. According to the phylogenetic tree (Fig. 1), these proteins were classified into six groups, labeled Group I to Group VI, with all walnut LOG proteins clustering into Groups I to V.
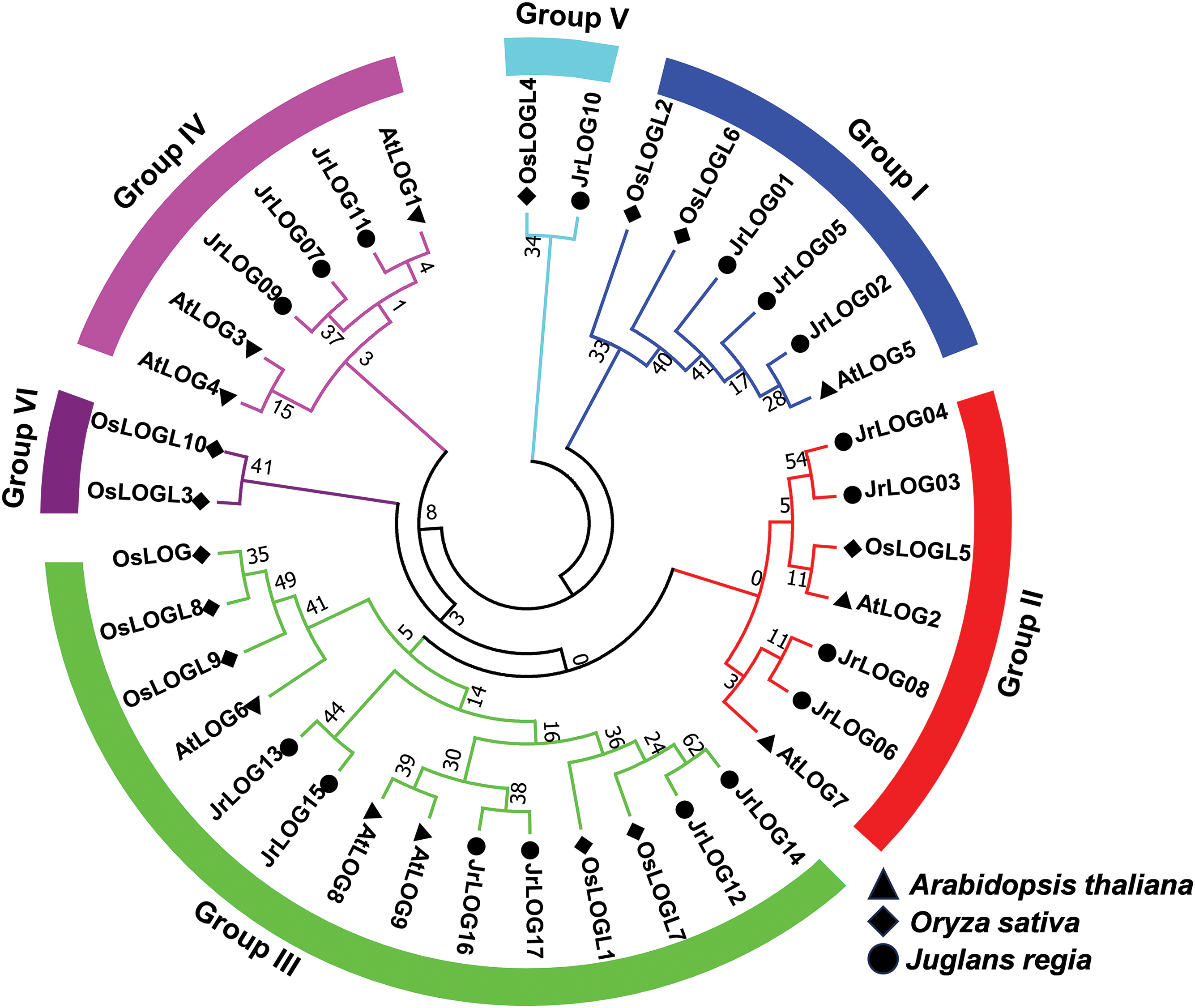
Figure 1: Neighbor-joining tree among LOG genes from walnut, rice, and Arabidopsis
3.2 Features and Structural Analysis of JrLOG Genes
The lengths of JrLOG genes ranged from 1326 to 4317 bp. All members of the JrLOG gene family contained 4 to 6 introns. Specifically, JrLOG10 had 4 introns, JrLOG4 contained 5, while the remaining members had 6 introns. The number and distribution of introns were relatively conserved within each phylogenetic cluster (Fig. 2A,B). For instance, the introns in Group I genes were notably short, while those in Group IV were much longer. This conservation of gene structure within evolutionary clades suggested that these genes have been subjected to similar evolutionary pressures.
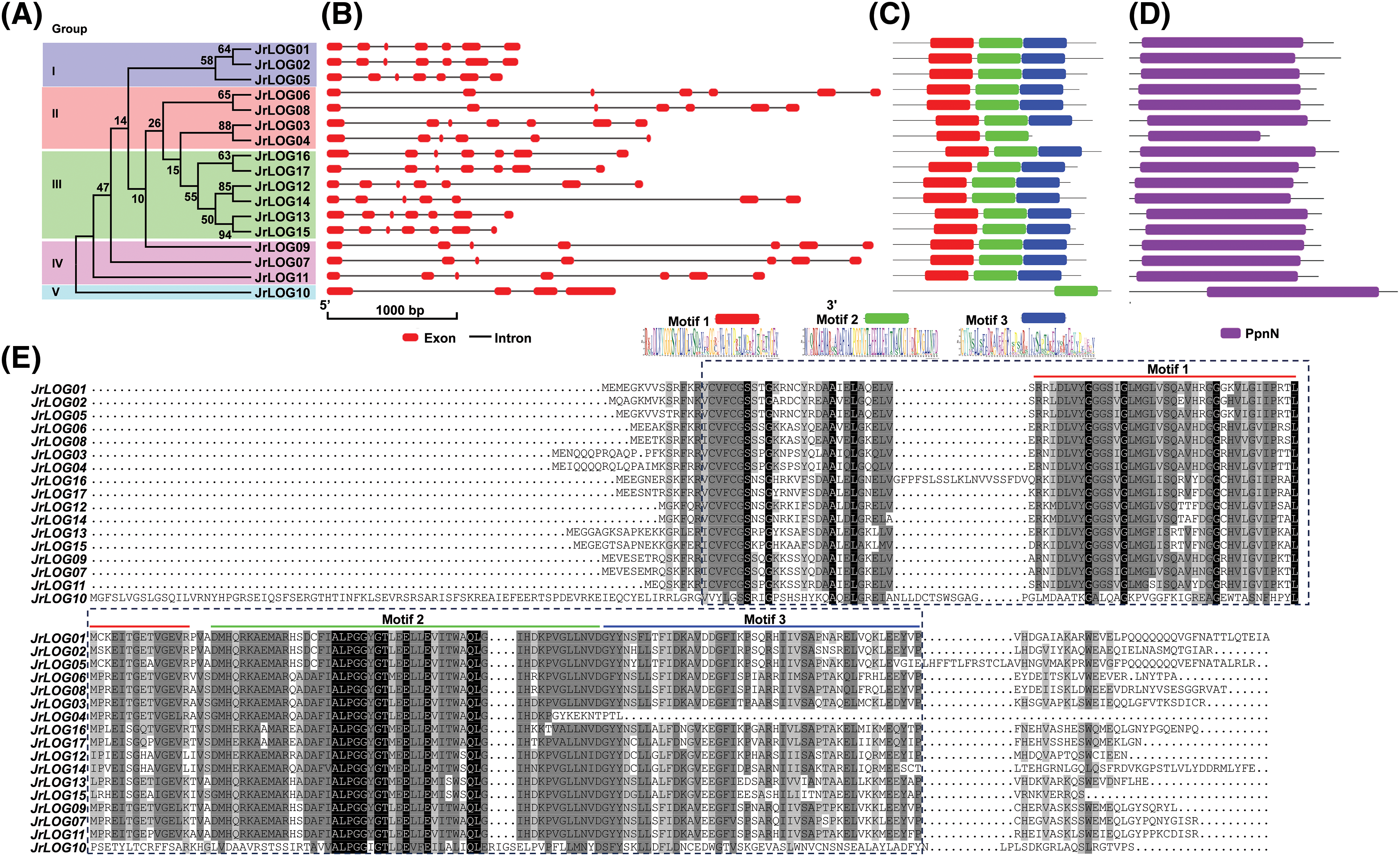
Figure 2: Phylogenetic relationship, gene, and protein features analysis of the JrLOG. (A) Neighbor-joining tree among LOG members from walnut. (B) and (C) show the protein feature motif and conserved domain, respectively. (D) Gene structure. (E) Protein multiple sequence alignment
Physicochemical property analysis of the JrLOG proteins revealed that their amino acid lengths ranged from 157 to 302, with estimated molecular weights between 17.13 and 33.15 kDa. The theoretical isoelectric points (pI) ranged from 5.04 to 8.61, encompassing both acidic and basic values. The instability index of these proteins varied between 30 and 50, with more than half of the members exhibiting an index greater than 40, indicating that JrLOG proteins were generally unstable (Table 1).
MEME analysis indicated that, except JrLOG10 and JrLOG4, all members possessed three characteristic motifs. JrLOG4 contains two specific structural domains, while JrLOG10 had only one (Fig. 2C). Notably, protein domain analysis further revealed that all members contained a conserved PpnN domain, a core component of nucleotide monophosphate nucleosidases (Fig. 2D). Multiple sequence alignment confirmed that the amino acid sequences within these three motifs were highly conserved, except in JrLOG10 and JrLOG4 (Fig. 2E).
Gene collinearity analysis was performed to uncover the conservation and evolutionary patterns of gene family members across different species. Although JrLOG members were distributed across various chromosomes in differing numbers (Fig. 3A), nine pairs of collinear genes were detected among JrLOG members in walnut (Fig. 3B). Except for JrLOG10, all other members exhibited collinear relationships. Notably, these collinear genes were not located on the same chromosome. Additionally, collinearity relationships were observed among the three Group I members, JrLOG01, JrLOG02, and JrLOG05.
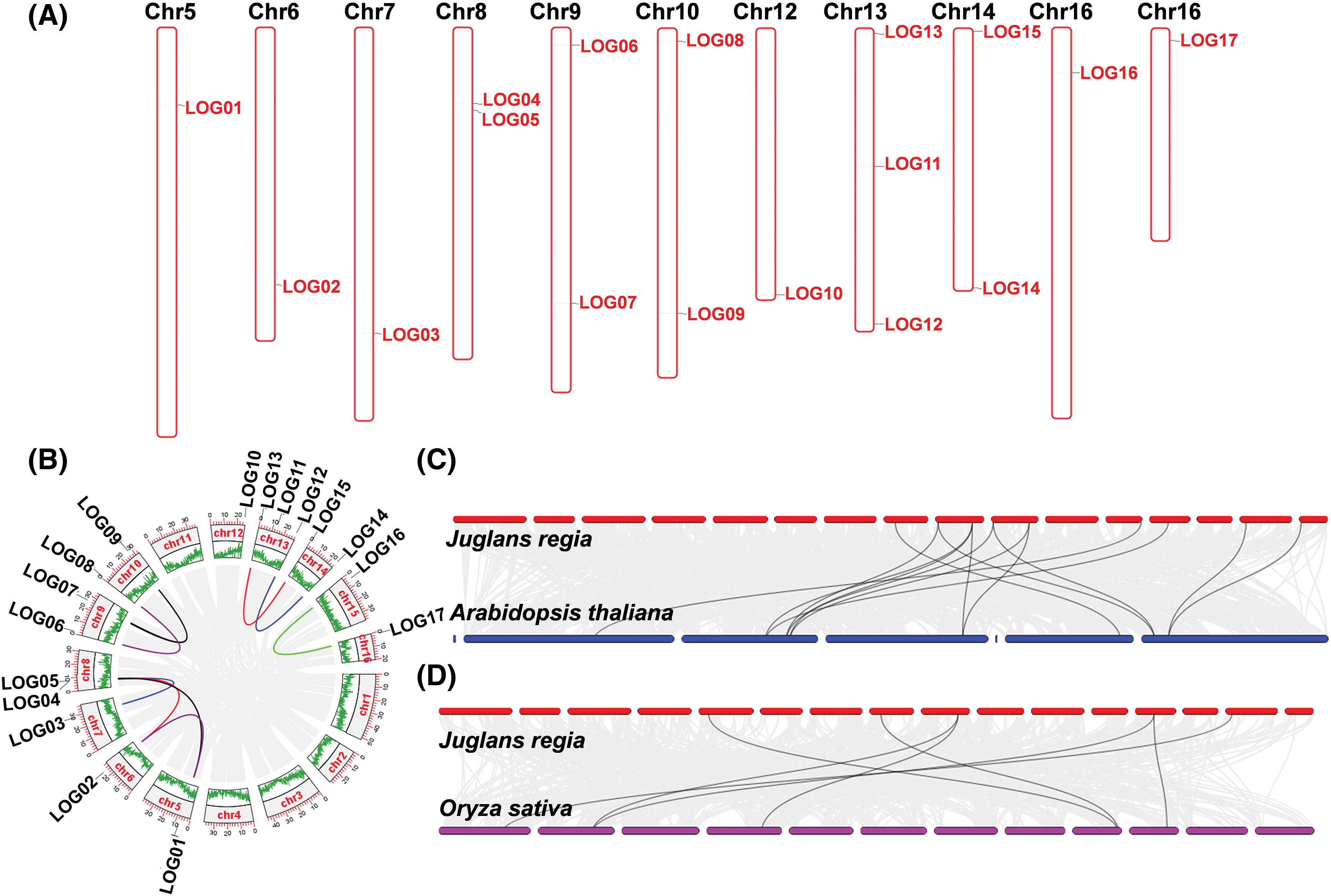
Figure 3: Chromosome localization and collinearity analysis of LOG genes. (A) Diagram showing the physical location of JrLOG genes on walnut chromosomes. (B) Collinearity analysis among JrLOG gene members in walnut. (C) Collinearity analysis of LOG genes between walnut and Arabidopsis. (D) Collinearity analysis of LOG genes between walnut and rice
Thirteen pairs of collinear LOG genes were identified between walnut and Arabidopsis (Fig. 3C), while only seven collinear pairs were detected between walnut and rice (Fig. 3D). This suggests that LOG genes in walnut share a closer evolutionary relationship with those in Arabidopsis than with rice.
3.4 Cis-Regulatory Element Analysis of the Promoter Region
Cis-regulatory elements (CREs) play a critical role in understanding gene regulation, phenotype expression, and plant adaptability. To gain insights into the regulatory mechanisms of JrLOG genes, we analyzed the potential CREs in their promoter regions (Fig. 4).
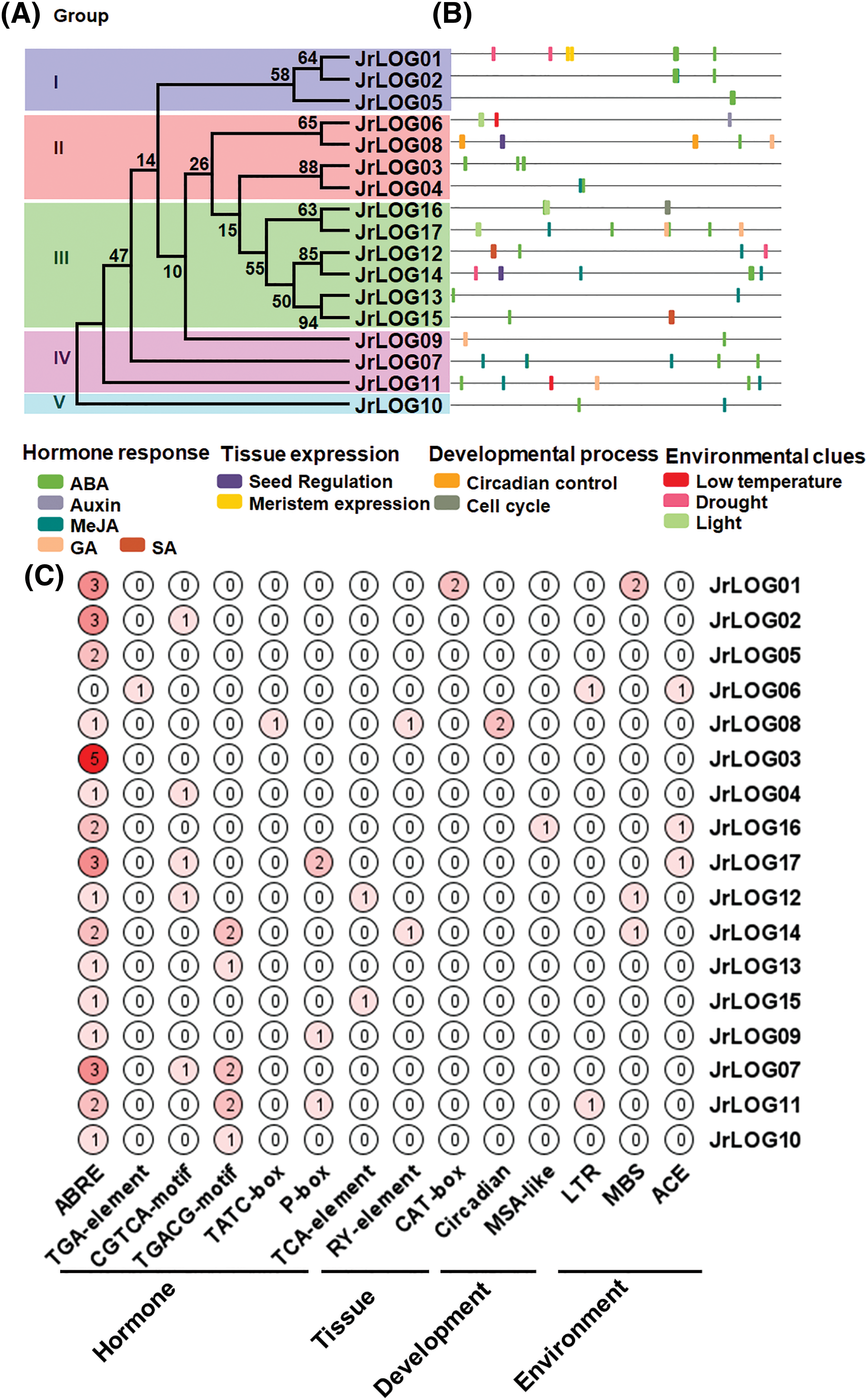
Figure 4: Predicted cis-regulatory elements (CREs) within the JrLOG promoters. (A) and (B) illustrate the clades of LOG family members and the distribution of diverse CREs within their promoters, represented by various colors in the graph. (C) presents a heatmap displaying the number of different CREs elements within different JrLOG promoters. ABRE, ABA-responsive element; TGA-element, auxin-responsive element; CGTCA-motif/TGACG-motif, MeJA-responsiveness; TATC-box/P-box, gibberellin-responsiveness; TCA-element, Salicylic acid responsiveness; RY-element, Seed-specific regulation; CAT-box, Meristem expression; Circadian, Circadian control; MSA-like, Cell cycle; LTR, Low-temperature responsiveness; MBS, Drought-inducibility; ACE, Light responsiveness
Several elements associated with plant hormone responses were identified, including those responsive to ABA (ABRE), auxin (TGA-element), MeJA (TGACG-motif and CGTCA-motif), GA (P-box and TATC-box), and SA (TCA-element), which were present in many members of the gene family. Notably, all members, except JrLOG6, contained ABA-responsive elements, with JrLOG3 exhibiting the highest number of ABRE motifs. Interestingly, auxin-responsive elements were found exclusively in the JrLOG6 promoter. Additionally, tissue-specific expression elements were identified, such as seed-specific expression elements (RY-element) in JrLOG08 and JrLOG14, and meristem-specific elements (CAT-box) in JrLOG01. CREs related to developmental processes were also detected, including two circadian control elements in JrLOG08 and cell cycle-related MSA-like elements in JrLOG16. Moreover, several CREs associated with environmental responses were predicted, such as those related to low temperature (LTR), drought (MBS), and light response (ACE-element). Interestingly, the distribution of CREs was not conserved among members of the same evolutionary clade, suggesting that JrLOG genes may be involved in a diverse array of biological processes.
3.5 Tissue-Specific and Developmental Stage Expression Analysis of JrLOG Genes
Tissue-specific expression analysis (Fig. 5A) revealed that JrLOG3, JrLOG10, JrLOG11, and JrLOG16 were highly expressed in the leaf mature (LM), leaves (LE), and leaf young (LY). JrLOG04 exhibited notably high expression in the pistillate flower (FL), while JrLOG07 showed strong expression exclusively in vegetative buds (VB). JrLOG01 and JrLOG09 were highly expressed in hull dehiscing (HU) and hull peel (HP), respectively, whereas JrLOG13 had high expression in hull immature (HL). Additionally, JrLOG08 and JrLOG17 were strongly expressed in catkins (CK). JrLOG12 was the only gene with high expression in both the callus exterior (CE) and callus interior (CI). JrLOG02 and JrLOG06 exhibited elevated expression levels in somatic embryo (SE), while JrLOG05 and JrLOG14 were specifically highly expressed in roots (RT).
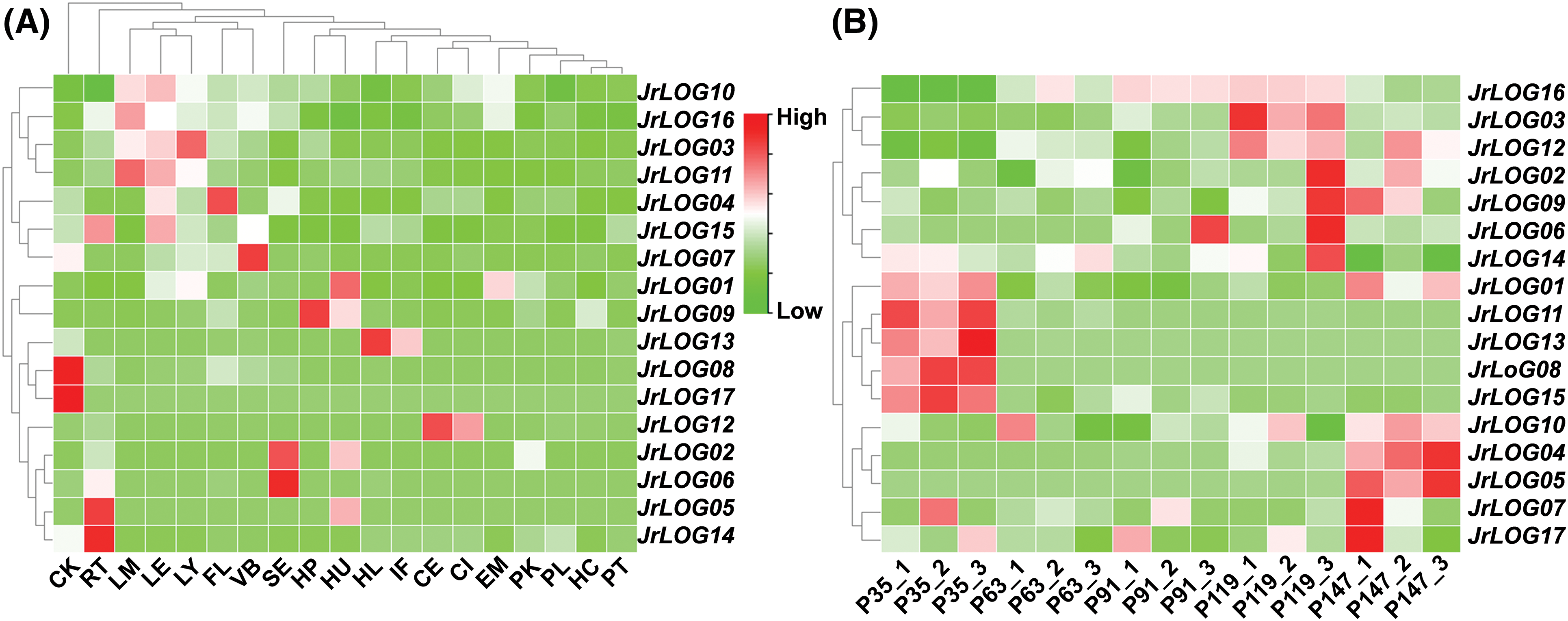
Figure 5: Expression characteristics of JrLOG gene in different tissues and development stage. (A) JrTLP expression profiles in different tissues. CE, callus exterior; CI, callus interior; CK, catkins; EM, embryo; FL, pistil late flower; HC, hull cortex; HL, hull immature; HP, hull peel; HU, hull dehiscing; IF, fruit immature; LE, leaves; LM, leaf mature; LY, leaf young; PK, packing tissue mature; PL, pellicle; PT, packing tissue immature; RT, root; SE, somatic embryo; VB, vegetative bud. (B) JrTLP expression profiles during the developmental stages of walnut endopleura. The heat map was generated based on the transcriptomic data
The walnut endopleura stores nutrients, protects the seed, and influences germination. Therefore, the expression profiles of JrLOG genes during seed endopleura development were also analyzed (Fig. 5B). JrLOG01, JrLOG08, JrLOG11, JrLOG13, and JrLOG15 displayed high expression during the early stages of endopleura development, whereas the remaining members exhibited increased expression closer to fruit maturation. Notably, JrLOG3 was significantly upregulated after the fruit hardening stage.
3.6 JrLOG Genes Expression in Responses to Anthracnose Infection
Upon inoculation with anthracnose, the expression levels of JrLOG genes fluctuated noticeably (Fig. 6). JrLOG05 was upregulated in genotype F423 genotype only 48 h post-infection, whereas, in the F26 genotype, its upregulation was delayed until 72 h and showed only a slight increase. In contrast, JrLOG10 in F26 exhibited two distinct expression peaks at 48 and 120 h post-infection, whereas in F423, it exhibited a consistently low level of expression. Additionally, JrLOG17 in F26 exhibited significant induction at both 24 and 48 h post-infection compared to F423. Notably, despite a general downregulation in transcription, JrLOG3 maintained relatively high expression in F26 at 24 h post-infection compared to its expression in F423. These findings suggest potential differences in the stress response mechanisms between the two genotypes.
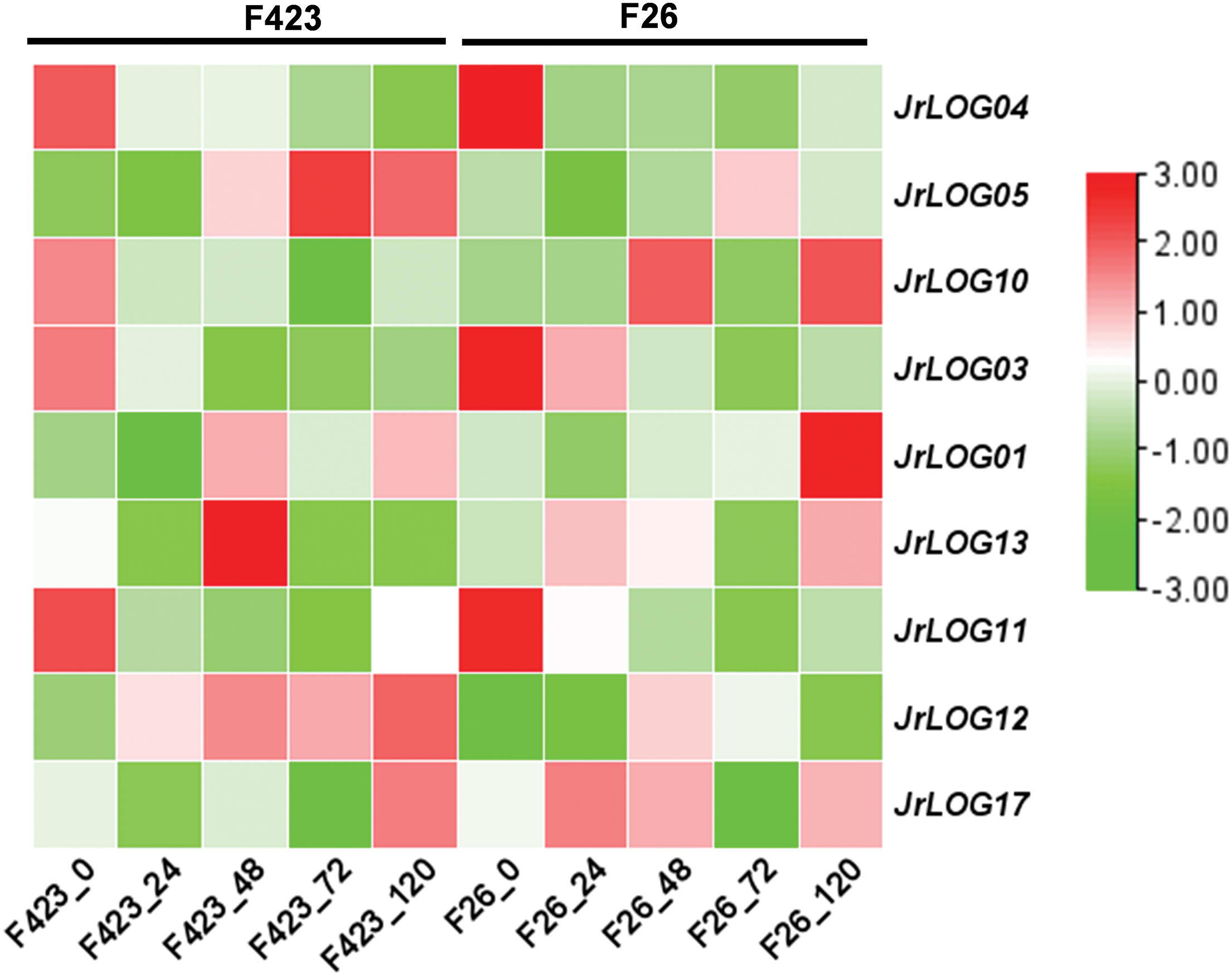
Figure 6: JrLOG gene expression profiles in F26 and the F423 fruits in response to anthracnose infection. The heat map was generated based on the transcriptomic data
3.7 JrLOG Genes Expression in Responses to Cold Stress
Under cold stress, the expression of JrLOG gene family members exhibited substantial variations (Fig. 7). JrLOG5, JrLOG8, JrLOG14, and JrLOG15 appeared to be unresponsive to cold treatment, while JrLOG6, JrLOG12, and JrLOG13 showed a general tendency towards downregulation in their expression levels after cold treatment, despite also experiencing fluctuations. Additionally, some genes displayed fluctuating expression patterns without any specific trend, including JrLOG6, JrLOG9, JrLOG10, JrLOG16, and JrLOG17. No significant differences in transcription levels were detected for JrLOG1, JrLOG2, JrLOG4, and JrLOG7 at 0.5 h and 1 h post-cold treatment; however, significant induction in expression was observed at 2 h. Notably, JrLOG3 and JrLOG11 exhibited strong transcriptional activation as early as 0.5 h post-cold treatment and were induced at all sampling time points.
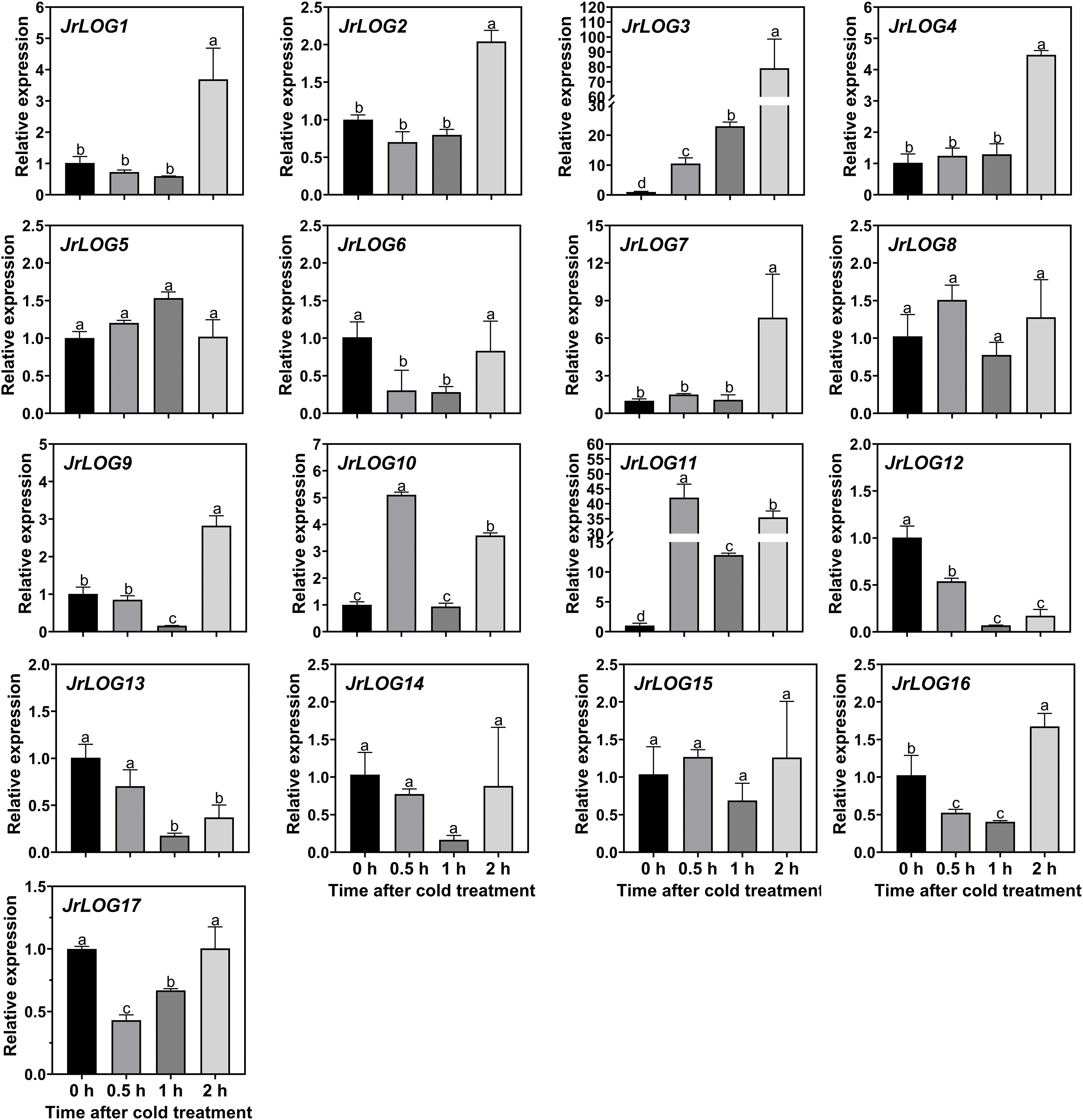
Figure 7: JrLOG gene expression in response to cold stress. Samples were collected at 0, 0.5, 1, and 2 h after cold at 4°C. Each value represents the mean ± SD of three replicates. Different letters mean significant difference at the 0.05 level
3.8 Gene Cloning and Subcellular Localization of JrLOG3
Using leaf tissue as the material, the expected band size for JrLOG3 was successfully amplified through via transcription PCR (Fig. 8A). Sequencing confirmed that the coding region of JrLOG3 is 678 bp in length, encoding a 225-amino-acid protein with a predicted molecular weight of 25.4 kDa. Phylogenetic analysis indicated that JrLOG3 was closely related to LOG proteins from other species, notably GhLOG3 from cotton and SlycLOG1a from tomato (Fig. 8B). Multiple sequence alignment demonstrated that JrLOG3 shares a high degree of conservation with these LOG proteins, including the catalytic sequence ‘PGGxGTxE’, a key feature situated at the core of the protein’s three-dimensional structure. This catalytic motif was entirely conserved across all analyzed proteins (Fig. 8C,D). Subcellular localization analysis revealed that GFP fluorescence driven by JrLOG3 was exclusively expressed in the cell membrane (Fig. 8E).
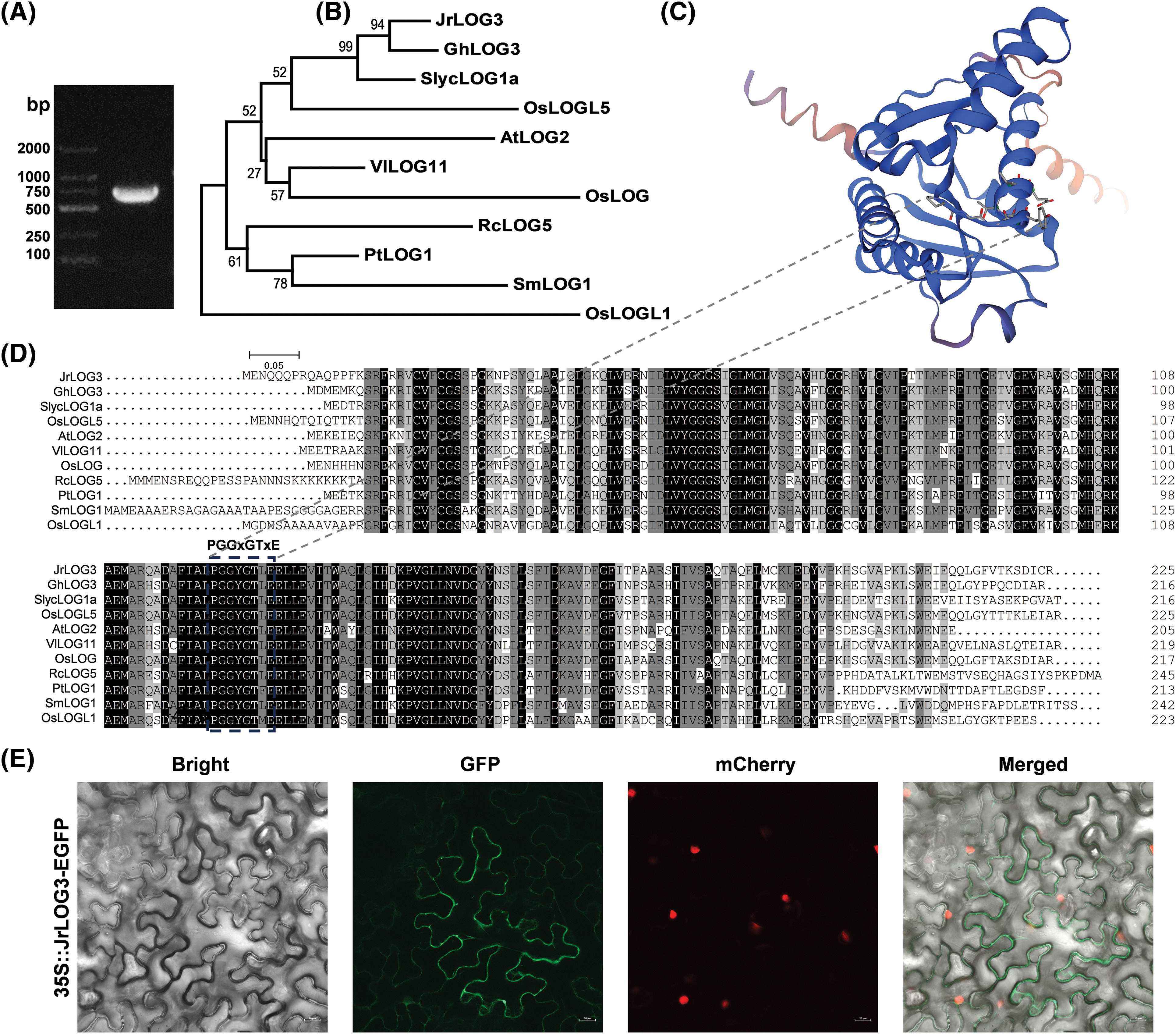
Figure 8: Gene cloning and analysis of JrLOG3. (A) Cloning of the JrLOG1 gene, with agarose gel electrophoresis showing the position of the target band. (B) Phylogenetic analysis of JrLOG3 concerning functionally characterized LOG proteins across diverse species. (C) Three-dimensional structure prediction of JrLOG3 protein. (D) Multiple sequence alignment analysis of JrLOG3 about functionally characterized LOG proteins across diverse species. (E) Subcellular localization analysis of JrLOG3 in tobacco epidermal cells. Scale bar, 20 μm
4.1 Identification of JrLOG Genes
LOG genes have been identified in several plant species; however, the LOG gene family in walnut (Juglans regia), an economically important woody oil tree species, has not been systematically explored. In this study, we identified 17 LOG-encoding genes in the walnut genome through homology-based comparisons. The JrLOG members were categorized into five subgroups using phylogenetic analysis (Fig. 1 and Fig. 2A). It appears that these genes have undergone duplication events (Fig. 3), likely associated with a whole-genome duplication approximately 20 million years ago [28]. Consequently, As a result, JrLOG genes are located on several chromosomes, comprising nine pairs of syntenic genes (Fig. 3B). Similarly, the expansion of the RcLOG gene family in castor bean (Ricinus communis) was driven by gene duplication events [19]. In walnut, this expansion was accompanied by purifying selection, as indicated by the Ka/Ks ratios of all syntenic gene pairs being much less than 1 (Table S2). Closely grouped JrLOG genes display similar gene architectures, amino acid sequences, conserved motifs, and domain characteristics (Fig. 2). Nonetheless, JrLOG10 lacks synteny with other members, suggesting it may have evolved as an independent lineage, leading to the loss of at least three exons and the absence of two characteristic motifs.
Moreover, closely related members, such as JrLOG3 and JrLOG4 (Fig. 2A), although forming a pair of syntenic genes (Fig. 3B), exhibited exon loss during gene duplication (Fig. 2B). This loss resulted in JrLOG4 lacking a C-terminal motif (Fig. 2C) and retaining only a diminished PpnN domain (Fig. 2D). Similarly, some members of the KNOX gene family have evolved without the C-terminal domain, forming distinct clades with functions similar to those of other members [29]. Notably, even closely related JrLOG genes displayed variation in the number and types of CREs in their promoters (Fig. 4), potentially explaining the balance between conservation and variability within the JrLOG family. Thirteen pairs of syntenic LOG genes were identified between walnut and Arabidopsis (Fig. 3C), whereas only seven pairs were found between walnut and rice (Fig. 3D), likely due to the closer evolutionary relationship among dicotyledonous plants compared to that between dicots and monocots.
4.2 Expression Analysis of JrLOG Genes
The LOG gene family plays a critical role in plant growth and development [12]. Thus, we systematically characterized the expression profiles of the JrLOG genes across various tissues and developmental stages. The abundance of JrLOG3, JrLOG10, JrLOG11, and JrLOG16 at various phases of leaf development indicates that these genes are essential for this process. Their expression may enhance endogenous CK levels, which could improve leaf size and regulate plant architecture [30]. Its possible function in floral development and reproduction was indicated by the specific and strong expression of JrLOG04 in pistillate flowers, an organ that is essential for walnut output. These results are in agreement with those from Arabidopsis, whose LOG genes control the development of floral meristems and are expressed in flowers [12]. In rice, LOG1 influences CK levels in young panicles, which modulates grain yield [11].
Seed coats, essential for seed germination, respond positively to exogenous CK treatments [31]. The high expression of JrLOG1, JrLOG9, and JrLOG13 in the hull suggests their possible involvement in hormone regulation related to seed germination [32]. CK application has been shown to enhance shoot regeneration efficiency in peanuts (Arachis hypogaea L.) [33] and to support callus growth by elevating endogenous CK levels [34]. JrLOG12 was specifically expressed in callus tissue, implicating it in tissue regeneration and cellular reprogramming [35]. Additionally, in Medicago truncatula, MtLOG1 and MtLOG2 regulate lateral root formation [36], raising the possibility that the root-specific expression of JrLOG5 and JrLOG14 in walnut may serve a similar function.
CKs also play a critical role in plant immunity against pathogens. For instance, infection by Verticillium longisporum alters CK levels, while in legumes, CK responses vary following bacterial inoculation [37]. In disease-resistant walnut germplasm, JrLOG3 maintained high expression 24 h after pathogen infection, suggesting its potential role in walnut disease resistance (Fig. 6). Under cold stress, JrLOG3 and JrLOG11 were upregulated, with JrLOG03 exhibiting rapid and continuous activation (Fig. 7). Conversely, several JrLOG genes were downregulated and displayed fluctuating expression patterns under cold stress, possibly as a strategy to slow growth in adverse conditions [38]. Given the significant expression of JrLOG3 in leaves and its responsiveness to both biotic and abiotic stresses, we cloned and analyzed this gene further. The cloned JrLOG3 is closely related to GhLOG3 from cotton (Fig. 8B), which induces adversity stress resistance [20]. While transient and stable expression of LOG fusion proteins in rice, Arabidopsis, and Chlamydomonas reinhardtii indicated localization in the cytoplasm and nucleus [9], JrLOG3 exhibited subcellular localization to the cell membrane (Fig. 8E). Similar to other LOG proteins, JrLOG3 contains the conserved “PGGxGTxxE” sequence, which contributes to AMP stability, indicating a shared catalytic mechanism. Consequently, JrLOG3 is a strong candidate for further investigation into its role in cytokinin regulation, organ development (e.g., leaf expansion), and stress tolerance.
This study identified 17 JrLOG genes from the walnut (Juglans regia) genome through bioinformatics analysis and categorized them into five distinct groups. The gene structures, amino acid sequences, unique motifs, and conserved domains demonstrated considerable conservation within each group. Across multiple tissues and developmental stages, as well as in response to various biotic and abiotic stressors, the JrLOG genes displayed distinct yet varied expression patterns. JrLOG3 was localized to the cell membrane, demonstrated elevated expression in leaves, and reacted to both abiotic and biotic stresses. These findings establish a robust basis for the continued investigation of JrLOG genes and underscore potential candidates for genetic modification to improve stress tolerance in walnut.
Acknowledgement: The authors would like to express their gratitude to the members of Chen Peng’s Lab.
Funding Statement: This work was supported by the Special Scientific Research Project for the Introduction of Talents in Hebei Agricultural University (YJ2021026), and the Construction of Innovation Team of Modern Agricultural Industry Technology System in Hebei Province (HBCT2021100211).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Yuan Wang, Guohui Qi, and Peng Jia; data collection: Tianle Zhang, Xinfeng Zeng, Jiale Liu, and Siyu Li; analysis and interpretation of results: Siyu Yang, Shengnan Zhao, Abdullah Shah, and Muhammad Saif Ullah; draft manuscript preparation: Yuan Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Authors confirm that the data supporting the findings of this study are available within article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.059402.
References
1. Li SM, Zheng HX, Zhang XS, Sui N. Cytokinins as central regulators during plant growth and stress response. Plant Cell Rep. 2021;40(2):271–82. doi:10.1007/s00299-020-02612-1. [Google Scholar] [PubMed] [CrossRef]
2. Tiwari M, Kumar R, Subramanian S, Doherty CJ, Jagadish SVK. Auxin-cytokinin interplay shapes root functionality under low-temperature stress. Trends Plant Sci. 2023;28(4):447–59. doi:10.1016/j.tplants.2022.12.004. [Google Scholar] [PubMed] [CrossRef]
3. Puig J, Pauluzzi G, Guiderdoni E, Gantet P. Regulation of shoot and root development through mutual signaling. Mol Plant. 2012;5(5):974–83. doi:10.1093/mp/sss047. [Google Scholar] [PubMed] [CrossRef]
4. Zwack PJ, Rashotte AM. Cytokinin inhibition of leaf senescence. Plant Signal Behav. 2013;8(7):e24737. doi:10.4161/psb.24737. [Google Scholar] [PubMed] [CrossRef]
5. Cortleven A, Leuendorf JE, Frank M, Pezzetta D, Bolt S, Schmülling T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019;42(3):998–1018. doi:10.1111/pce.13494. [Google Scholar] [PubMed] [CrossRef]
6. Nayar S. Exploring the role of a cytokinin-activating enzyme LONELY GUY in unicellular Microalga Chlorella variabilis. Front Plant Sci. 2020;11:611871. doi:10.3389/fpls.2020.611871. [Google Scholar] [PubMed] [CrossRef]
7. Sandhu J, Irvin L, Chandaran AK, Oguro S, Paul P, Dhatt B, et al. Natural variation in LONELY GUY-Like 1 regulates rice grain weight under warmer night conditions. Plant Physiol. 2024;196(1):164–80. doi:10.1093/plphys/kiae313. [Google Scholar] [PubMed] [CrossRef]
8. Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445(7128):652–5. doi:10.1038/nature05504. [Google Scholar] [PubMed] [CrossRef]
9. Chen L, Jameson GB, Guo Y, Song J, Jameson PE. The LONELY GUY gene family: from mosses to wheat, the key to the formation of active cytokinins in plants. Plant Biotechnol J. 2022;20(4):625–45. doi:10.1111/pbi.v20.4. [Google Scholar] [CrossRef]
10. Samanovic MI, Tu S, Novák O, Iyer LM, McAllister FE, Aravind L, et al. Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol Cell. 2015;57(6):984–94. doi:10.1016/j.molcel.2015.01.024. [Google Scholar] [PubMed] [CrossRef]
11. Wu B, Meng J, Liu H, Mao D, Yin H, Zhang Z, et al. Suppressing a phosphohydrolase of cytokinin nucleotide enhances grain yield in rice. Nat Genet. 2023;55(8):1381–9. doi:10.1038/s41588-023-01454-3. [Google Scholar] [PubMed] [CrossRef]
12. Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, et al. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21(10):3152–69. doi:10.1105/tpc.109.068676. [Google Scholar] [PubMed] [CrossRef]
13. Matsumoto-Kitano M, Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Václavíková K, Miyawaki K, et al. Cytokinins are central regulators of cambial activity. Proc Natl Acad Sci U S A. 2008;105(50):20027–31. doi:10.1073/pnas.0805619105. [Google Scholar] [PubMed] [CrossRef]
14. Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, et al. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012;69(2):355–65. doi:10.1111/tpj.2011.69.issue-2. [Google Scholar] [CrossRef]
15. Rauschendorfer J, Yordanov Y, Dobrev P, Vankova R, Sykes R, Külheim C, et al. Overexpression of a developing xylem cDNA library in transgenic poplar generates high mutation rate specific to wood formation. Plant Biotechnol J. 2020;18(6):1434–43. doi:10.1111/pbi.v18.6. [Google Scholar] [CrossRef]
16. Wang LL, Shi Q, Jing P, Wang R, Zhang H, Liu Y, et al. VlMYB4 and VlCDF3 co-targeted the VlLOG11 promoter to regulate fruit setting in grape (Vitis vinifera L). Plant Cell Rep. 2024;43(8):194. doi:10.1007/s00299-024-03279-8. [Google Scholar] [PubMed] [CrossRef]
17. Eviatar-Ribak T, Shalit-Kaneh A, Chappell-Maor L, Amsellem Z, Eshed Y, Lifschitz E. A cytokinin-activating enzyme promotes tuber formation in tomato. Curr Biol. 2013;23(12):1057–64. doi:10.1016/j.cub.2013.04.061. [Google Scholar] [PubMed] [CrossRef]
18. Zhang L, Zhang R, Yan P, Zeng L, Zhao W, Feng H, et al. PE (Prickly Eggplant) encoding a cytokinin-activating enzyme responsible for the formation of prickles in eggplant. Hortic Res. 2024;11(7):uhae134. doi:10.1093/hr/uhae134. [Google Scholar] [PubMed] [CrossRef]
19. Li Y, Zhu G, Sun H, Xiang D, Zhang C, Li Z, et al. Genome-wide analysis of LOG family genes in castor and RcLOG5 enhances drought, salt, and cold stress tolerance in Arabidopsis thaliana. Gene. 2024;913:148398. doi:10.1016/j.gene.2024.148398. [Google Scholar] [PubMed] [CrossRef]
20. Wang R, Liu L, Kong Z, Li S, Lu L, Nosheen K, et al. Identification of GhLOG gene family revealed that GhLOG3 is involved in regulating salinity tolerance in cotton (Gossypium hirsutum L.). Plant Physiol Biochem. 2021;166:328–40. doi:10.1016/j.plaphy.2021.06.011. [Google Scholar] [PubMed] [CrossRef]
21. Rathore RS, Mishra M, Pareek A, Singla-Pareek SL. Concurrent improvement of rice grain yield and abiotic stress tolerance by overexpression of cytokinin activating enzyme LONELY GUY (OsLOG). Plant Physiol Biochem. 2024;211:108635. doi:10.1016/j.plaphy.2024.108635. [Google Scholar] [PubMed] [CrossRef]
22. Wang C, Wang G, Gao Y, Lu G, Habben JE, Mao G, et al. A cytokinin-activation enzyme-like gene improves grain yield under various field conditions in rice. Plant Mol Biol. 2020;102(4–5):373–88. [Google Scholar] [PubMed]
23. Martínez-García PJ, Crepeau MW, Puiu D, Gonzalez-Ibeas D, Whalen J, Stevens KA, et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016;87(5):507–32. doi:10.1111/tpj.2016.87.issue-5. [Google Scholar] [CrossRef]
24. Ji F, Ma Q, Zhang W, Liu J, Feng Y, Zhao P, et al. A genome variation map provides insights into the genetics of walnut adaptation and agronomic traits. Genome Biol. 2021;22(1):300. doi:10.1186/s13059-021-02517-6. [Google Scholar] [PubMed] [CrossRef]
25. Fang H, Liu X, Dong Y, Feng S, Zhou R, Wang C, et al. Transcriptome and proteome analysis of walnut (Juglans regia L.) fruit in response to infection by Colletotrichum gloeosporioides. BMC Plant Biol. 2021;21(1):249. doi:10.1186/s12870-021-03042-1. [Google Scholar] [PubMed] [CrossRef]
26. Zheng G, Zhang T, Liu J, Yan R, Wang W, Wang N, et al. Identification and expression profiles of Tubby-like proteins coding genes in walnut (Juglans regia L.) in response to stress and hormone treatments. Plant Stress. 2024;12:100472. doi:10.1016/j.stress.2024.100472. [Google Scholar] [CrossRef]
27. Jia P, Liu J, Yan R, Yang K, Dong Q, Luan H, et al. Systematical characterization of the AT-Hook gene family in Juglans regia L. and the functional analysis of the JrAHL2 in flower induction and hypocotyl elongation. Int J Mol Sci. 2023;24(8):7244. doi:10.3390/ijms24087244. [Google Scholar] [PubMed] [CrossRef]
28. Zhang J, Zhang W, Ji F, Qiu J, Song X, Bu D, et al. A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol J. 2020;18(9):1848–50. doi:10.1111/pbi.v18.9. [Google Scholar] [CrossRef]
29. Magnani E, Hake S. KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell. 2008;20(4):875–87. doi:10.1105/tpc.108.058495. [Google Scholar] [PubMed] [CrossRef]
30. Lagoutte, ilella F-V. Effect of cell size and cytokinins on growth of petunia plants. Phyton-Int J Exp Bot. 2009;78:31–6. doi:10.32604/phyton.2009.78.031. [Google Scholar] [CrossRef]
31. Boschi CL, Palazuelos M, Gandolfo E. Effect of immersion in solutions with 6-benzylaminopurine on the germination and growth of seeds of Ginkgo biloba L. Phyton-Int J Exp Bot. 2014;83:341–6. doi:10.32604/phyton.2014.83.341. [Google Scholar] [CrossRef]
32. Wang Y, Li L, Ye T, Zhao S, Liu Z, Feng YQ, et al. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 2011;68(2):249–61. doi:10.1111/tpj.2011.68.issue-2. [Google Scholar] [CrossRef]
33. Lamboro A, Han X, Yang S, Li X, Yao D, Song B, et al. Combination of 6-benzylaminopurine and thidiazuron promotes highly efficient shoot regeneration from cotyledonary node of mature peanut (Arachis hypogaea L.) cultivars. Phyton-Int J Exp Bot. 2022;91(12):2619–31. doi:10.32604/phyton.2022.021404. [Google Scholar] [CrossRef]
34. Jia P, Wang Y, Sharif R, Ren X, Qi G. MdIPT1, an adenylate isopentenyltransferase coding gene from Malus domestica, is involved in branching and flowering regulation. Plant Sci. 2023;333:111730. doi:10.1016/j.plantsci.2023.111730. [Google Scholar] [PubMed] [CrossRef]
35. Ikeuchi M, Shibata M, Rymen B, Iwase A, Bågman A-M, Watt L, et al. A gene regulatory network for cellular reprogramming in plant regeneration. Plant Cell Physiol. 2018;59(4):770–82. doi:10.1093/pcp/pcy013. [Google Scholar] [PubMed] [CrossRef]
36. Mortier V, Wasson A, Jaworek P, De Keyser A, Decroos M, Holsters M, et al. Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol. 2014;202(2):582–93. doi:10.1111/nph.2014.202.issue-2. [Google Scholar] [CrossRef]
37. Soyano T, Akamatsu A, Takeda N, Watahiki MK, Goh T, Okuma N, et al. Periodic cytokinin responses in Lotus japonicus rhizobium infection and nodule development. Science. 2024;385(6706):288–94. doi:10.1126/science.adk5589. [Google Scholar] [PubMed] [CrossRef]
38. Hare PD, Cress WA, van Staden J. The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regul. 1997;23(1):79–103. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools