 Open Access
Open Access
ARTICLE
Construction of a Yeast Hybrid Library and Identification of Proteins Regulating CaABI3/VP1-1 Expression in Capsicum annuum var. conoides
College of Agriculture, Guizhou University, Guiyang, 550025, China
* Corresponding Authors: Guangdong Geng. Email: ; Suqin Zhang. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Abiotic Stress Tolerance in Crop Plants: Physio-biochemical and Molecular Mechanisms)
Phyton-International Journal of Experimental Botany 2024, 93(12), 3273-3291. https://doi.org/10.32604/phyton.2024.058638
Received 17 September 2024; Accepted 11 December 2024; Issue published 31 December 2024
Abstract
Hot pepper (Capsicum annuum var. conoides) is a significant vegetable that is widely cultivated around the world. Currently, global climate change has caused frequent severe weather events, and waterlogging stress harms the pepper industry by affecting the planting period, growth conditions, and disease susceptibility. The gene CaABI3/VP1-1 could improve pepper waterlogging tolerance. In order to explore the upstream regulatory mechanism of CaABI3/VP1-1, a high-quality standardized yeast hybrid library was successfully constructed for yeast one-, two-, and three-hybrid screening using pepper ‘ZHC2’ as the experimental material, with a library recombinant efficiency of up to 100%. The length of inserted fragments varied from 650 to 5000 bp, the library titer was 5.18 × 106 colony-forming units (CFU)·mL−1, and the library capacity was 1.04 × 107 CFU of cDNA inserts. The recombinant bait plasmid was used to successfully identify 78 different proteins through the yeast one-hybrid system, including one transcription factor within the ethylene-responsive factor family and the other within the growth-regulating factor family. The interaction happened between LOC124895848 and CaABI3/VP1-1 promoter by point-to-point yeast one-hybrid experiment. The expression level of the 12 selected protein-coding genes was then evaluated by quantitative real-time polymerase chain reaction. Results indicated the protein coding genes showed different responses to waterlogging stress and that the activity of the CaABI3/VP1-1 promoter could be inhibited or activated by up-regulating or down-regulating gene expression, respectively. The identification of these proteins interacting with the promoter provides a new perspective for understanding the gene regulatory network of hot pepper operating under waterlogging stress and provides theoretical support for further analysis of the complex regulatory relationship between transcription factors and promoters.Keywords
Peppers (Capsicum annuum var. conoides) are the important vegetables in the Solanaceae family. The fruits have a bright color and unique taste, and they are widely used fresh or after processing or extraction. Peppers are used in medicine, as well as food, because they contain a variety of important secondary metabolites. Due to current global climate change, extreme environments occur more frequently, resulting in reduced crop yields and economic losses. During the pepper growth cycle, plants are susceptible to various abiotic stresses, resulting in the stunting of their growth and development. Waterlogging stress is one of the main environmental factors restricting pepper crop growth and development. Excessive soil moisture can lead to growth inhibition, or even death in severe cases, causing huge economic losses, especially for shallow-rooted plants such as peppers. When their organs are surrounded by water (especially still water), it often leads to plant hypoxia, resulting in gas homeostatic imbalance and hypoxia, causing the accumulation of reactive oxygen species, which can harm cell membranes and aggravate the damage caused by the primary stress [1,2]. Therefore, it is particularly important to take various measures to reduce the damage caused by waterlogging stress. Based on our previous study, pepper CaABI3/VP1-1 is an important gene due to its role in waterlogging tolerance [3].
ABI3 (ABSCISIC ACID-INSENSITIVE 3) is a plant-specific transcription factor that contains the B3 (the Basic3 DNA-binding) domain and is involved in the abscisic acid (ABA) signaling pathway. Since the first discovery of the B3 domain in the maize gene VIVIPAROUS1 (VP1), it has been found in 118 Arabidopsis genes and 91 rice genes [4]. ABI3 participates in different ABA signaling cascades, and, in Arabidopsis, appears to interact with pre-existing ABA signaling pathways in differentiating vegetative tissues to enable ABA to transactivate seed-specific promoters [5]. Many transcription factors containing the B3 domain regulate a range of biological processes in plants, influencing nutrition and reproductive development [6,7]. For ABI3/VP1 genes, research has been mainly focused on processes in plant seeds, such as seed development and dormancy [8–10]. ABI3/VP1 genes have various functions, such as regulating the growth of axillary meristems, controlling flowering, and regulating chloroplast differentiation [11]. There have been several reports of stress tolerance associated with ABI3/VP1 genes. Under low oxygen conditions, OsGF14h interacts with two transcription factors, OsHOX3 and OsVP1, of the ABI3/VP1 subfamily, to block the ABA receptor OsPYL5. This reduces ABA sensitivity while activating gibberellin biosynthesis promotes the germination and emergence of weedy rice seeds, and enhances rice adaptability to anaerobic environment [12]. The heterologous transformation of AtABI3 into cotton showed that the transgenic cotton had greater tolerance to drought stress in greenhouse and field conditions, and enhanced photosynthetic efficiency by promoting root growth and leaf area expansion in cotton [13]. The ABI3 gene is a key factor in regulating dehydration stress signaling in Arabidopsis thaliana and reveals the genetic and epigenetic factors required for ABI3 gene expression [14].
The gene transcriptional expression pattern is mediated by cis-regulatory elements (CREs), which include DNA sequences that specifically recognize and bind to transcription factors [15]. As a CRE, the promoter can ensure the normal expression of genes at different times and environments, which helps plants respond in a timely manner to growth, development, and stress, and reduces the damage caused by adverse environments. Promoters can control gene expression by binding to transcription factors; therefore, it is important to determine whether there is interaction between a promoter and transcription factors to understand gene function. The yeast one-hybrid (Y1H) assay is a novel system derived from the yeast two-hybrid system [16]. The Y1H assay is a direct technique to research the interaction between DNA promoters and transcription factors through the screening of yeast cDNA libraries and prey proteins based on the sequence of the bait DNA promoter [17]. This technique is widely used in promoter studies of pepper [18,19], tomato [20], strawberry [21], lily [22], wheat [23], and Arabidopsis [24].
CaABI3/VP1-1, a key stress-response gene in peppers, has been shown to significantly increase the waterlogging tolerance of peppers [3], but the CaABI3/VP1-1 upstream regulatory mechanisms are still unclear. Therefore, a high-capacity yeast hybrid cDNA library was constructed using pepper ‘ZHC2’. Specifically, the CaABI3/VP1-1 promoter was used as the bait DNA, and the proteins interacting with CaABI3/VP1-1 were identified by screening using the Y1H assay. The purpose of this research was to reveal the regulatory switches operating in response to waterlogging stress, investigate the CaABI3/VP1-1 upstream regulatory mechanism, and assess its significance for breeding waterlogging-tolerant pepper cultivars.
The plant material used was the waterlogging-tolerant hot pepper pure line ‘ZHC2’. Full and uniformly sized ‘ZHC2’ seeds were selected and surface sterilized by immersing in 75% (v/v) alcohol for 1 min. The seeds were then rinsed with distilled water and allowed to germinate in an incubator at 25°C. After germination, the seeds were sown in 8 cm diameter pots containing quartz sand of three different degrees of coarseness: the lower (70–140-mesh), middle (4–6-mesh), and upper (20–40 mesh) layers of quartz sand, 1:1:1 (v/v/v), and then put in a culture room at a 25°C/20°C day/night temperature, 270 μmol·m−2·s−1 light intensity, and 10 h/14 h light/dark photoperiod. During the period from cotyledon expansion to the third-leaf expansion, 1/2-strength Hoagland’s solution was utilized for irrigation, and full-strength Hoagland’s solution was utilized for irrigation from the third-leaf stage onwards. At the 6-leaf, 8-leaf, and fruit stages of pepper plant development, the roots, leaves, and fruits were collected, rapidly snap-frozen in liquid nitrogen, and stored at −80°C before cDNA library construction and promoter cloning.
2.2 Total RNA Extraction, mRNA Isolation, and Purification
Equal parts (by weight) of the roots, leaves, and fruits from hot pepper ‘ZHC2’ were evenly mixed, and then total pepper RNA was extracted for subsequent library construction using Cetyltrimethylammonium Bromide (CTAB) method [25]. mRNA isolation and purification were performed according to the procedure described by the manufacturer of the FastTrack MAG mRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). RNA concentration and quality were assessed by electrophoresis on 1% agarose gel and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), respectively.
2.3 Construction of the cDNA Primary Library
The mRNA was reverse-transcribed into double-stranded cDNA and was then ligated to the attB1 recombinant adapter. Then, the double-stranded cDNA was normalized with the Trimer-Direct Trimmer Direct cDNA Normalization Kit (Evrogen, Moscow, Russia). After separation and collection, the normalized cDNA was introduced into the pDONR222 vector using the attB and attP site recombination reaction. The recombinant vector was electrotransformed into competent Escherichia coli strain DH10B cells. After transformation, 2 mL of Super Optimal broth with Catabolite repression medium was added. After shaking and culturing for 1 h at 37°C, the cDNA library solution was obtained. An aliquot (50 μL of 1/1000× diluted suspension was plated on Luria Broth (LB) medium for library capacity identification. The remaining bacterial suspension was mixed with a volume of 60% (v/v) glycerol (2:1 ratio) to obtain the cDNA primary library suspension and stored at −80°C.
2.4 Construction of the Yeast Secondary Library
The library plasmid was obtained from the cDNA primary library, and the attL and attR site recombination reaction system was operated. The library plasmid was recombined with the pGADT7-DEST vector, and the recombinant plasmid was electroporated into competent E. coli DH10B cells. After culture, the yeast secondary library bacterial suspension was obtained, to which was added glycerol to a final concentration of 20% (v/v), followed by storage at −80°C.
2.5 Library Capacity and Insert Fragment Identification
Aliquots (50 μL) of diluted 1/1000× primary library suspension and secondary library suspension were plated on LB medium and then cultured overnight at 37°C. Twenty-four single clones were randomly selected for identification by PCR. The primers are listed in Table 1. The pDONR222 primers were used for primary library identification, and the ADR and T7 primers for secondary library identification. The amplified products were detected by electrophoresis on 1% agarose gels and the library capacity was calculated according to the following equations:
Library, titer (CFU·mL−1) = number of clones on the medium/volume of bacterial solution × n × 103 (n is dilution fold).
library capacity (CFU) = library titer (CFU·mL−1) × total volume of library bacterial solution (mL).
library recombination rate (%) = number of clones with inserted fragments/total number of clones × 100.

2.6 Cloning of CaABI3/VP1-1 Promoter
The 2200-bp sequence before the initiation codon of the CaABI3/VP1-1 gene was retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/ (accessed on 10 December 2024)) as a template for cloning the promoter. Genomic DNA from ‘ZHC2’ leaves was extracted by the CTAB method, and the CaABI3/VP1-1 promoter was cloned with 2 × Phanta Max Master Mix (Vazyme, Nanjing, China) and the primers CaABI3/VP1-1-F and CaABI3/VP1-1-R (Table 1).
2.7 Construction of the Bait Plasmid
The CaABI3/VP1-1 promoter sequence was ligated with the Y1H bait plasmid pHIS2 vector (digested with EcoRI and MluI) by seamless cloning with ClonExpress Ultra One Step Cloning Kit (Vazyme). The bait plasmid was transformed into competent E. coli DH5α cells and screened on LB medium with 50 ug·mL−1 of kanamycin. Twelve E. coli transformants were randomly selected and inoculated into a liquid LB medium. After shaking culture at 37°C and 250 rpm for 16 h (or overnight), PCR detection was performed. When sequencing showed the correct insertion fragment size, subsequent experiments were carried out.
2.8 Auto-Activation Detection of the Bait Plasmid
Different plasmid combinations were transferred into the recipient yeast strain Y187 by the LiAC method [26] (Table 2), which were inoculated on the SD-LWH (10 mM 3-AT (3-amino-1,2,4-triazole), SD-LWH (30 mM 3-AT), and SD-LWH (60 mM 3-AT) defective screening media. 3-AT is a competitive inhibitor and controls the leaky HIS3 reporter gene expression. The bait reporter gene yeast strain was cultured for 4 days at 30°C, after which the minimum 3-AT concentration that could significantly inhibit yeast strain growth was selected for later library screening.
2.9 Y1H Analysis and Screening of Interacting Proteins
The Y187 yeast transformants containing the pHIS2-CaABI3/VP1-1 bait plasmid were used as the recipient bacteria into which the PGADT7-pepper cDNA library plasmid was transferred. An aliquot (20 μL) of the suspension was taken and coated on three SD-LW media (diluted 10×, 100×, and 1000×) to test the library transformation efficiency. The remaining suspension was plated on SD-LWH medium and incubated for 3–4 days at 30°C to observe the transformation results. In order to confirm the positive interaction, the screened positive clones were transferred to a fresh SD-LWH medium for colony PCR detection, followed by plasmid extraction, sequencing, and BLAST comparison. Gene Ontology (GO) and protein-protein interaction (PPI) analyses of the interacting proteins were carried out.
2.10 Point-to-Point Y1H Experiment
The gene LOC124895848 was cloned and ligated with the pGADT7 vector to generate the recombinant plasmid pGADT7-LOC124895848 by seamless cloning with ClonExpress Ultra One Step Cloning Kit (Vazyme). pGADT7-LOC124895848 and pHIS2-CaABI3/VP1-1 were co-transformed into yeast Y187. The transformed yeast was cultured on SD-LWH (with 3-AT) medium. All transformants were incubated for 3–4 days at 30°C. Primers used for Y1H are shown in Table 1.
2.11 Quantitative Real-Time PCR ( qPCR) Analysis of Gene Expression in Response to Waterlogging
‘ZHC2’ seedlings were cultured as described in Section 2.1 above to the 6-leaf stage, at which point seedlings were exposed to waterlogging stress according to Guo et al. [27]. The leaves and roots were harvested at 0 and 24 h after waterlogging stress was applied and after 1 h of recovery from 24 h of waterlogging. Total RNA was isolated from 0.1 g of plant materials with RNAsimple Total RNA Kit (Tiangen, Beijing, China), and cDNA was gotten with HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme), following the manufacturer’s instructions. Twelve protein-coding genes were selected according to high identity (>95%) to the reference gene and functional annotation for the qPCR assay. Specific primers were designed by Primer Premier5 (Premier Biosoft, San Francisco, CA, USA) (Table 1), with GAPDH as the internal reference gene [28]. The qPCR reaction was carried out using the 2 × RealStar Fast SYBR qPCR Mix (Genestar, Beijing, China). The relative expression level for quantification was calculated according to the 2−ΔΔCt formula method [29]. Three biological replicates and three technical replicates for each of the three samples were included in this experiment.
Three biological replicates of each sample were subjected to a one-way analysis of variance (ANOVA), and the significant differences (p < 0.05) were determined by the least significant difference (LSD) test. Data analysis was carried out with SPSS 26.0 software (IBM Inc., Armonk, NY, USA). GraphPad graphical software (Origin 2021, Northampton, MA, USA) was applied for the construction of the figures.
3.1 Cloning of CaABI3/VP1-1 Promoter
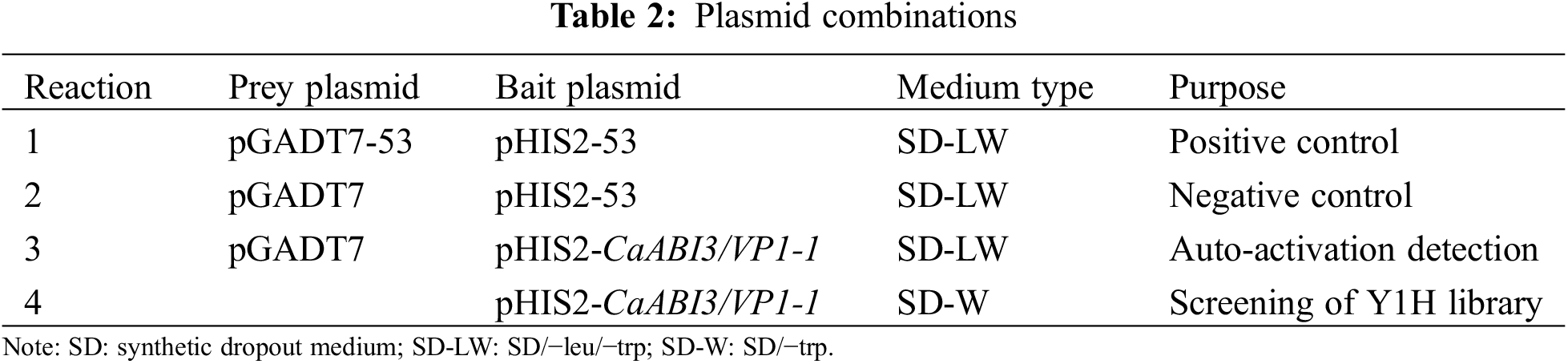
Genomic DNA was extracted from ‘ZHC2’ leaves and agarose gel electrophoresis showed that the extracted DNA bands were clear and bright, reflecting high DNA quality (Fig. 1A). The extracted DNA was utilized as a template for PCR amplification with specific primers. The result of agarose gel electrophoresis and sequencing indicated that a promoter 2143 bp in length had been successfully cloned from the pepper (Fig. 1B). There was a 1-bp difference in length between the CaABI3/VP1-1 promoter in ‘ZHC2’ and the reference sequence of C. annuum ‘Zunla1’ by DNAMAN (Lynnon Biosoft, San Ramon, CA, USA) comparison.
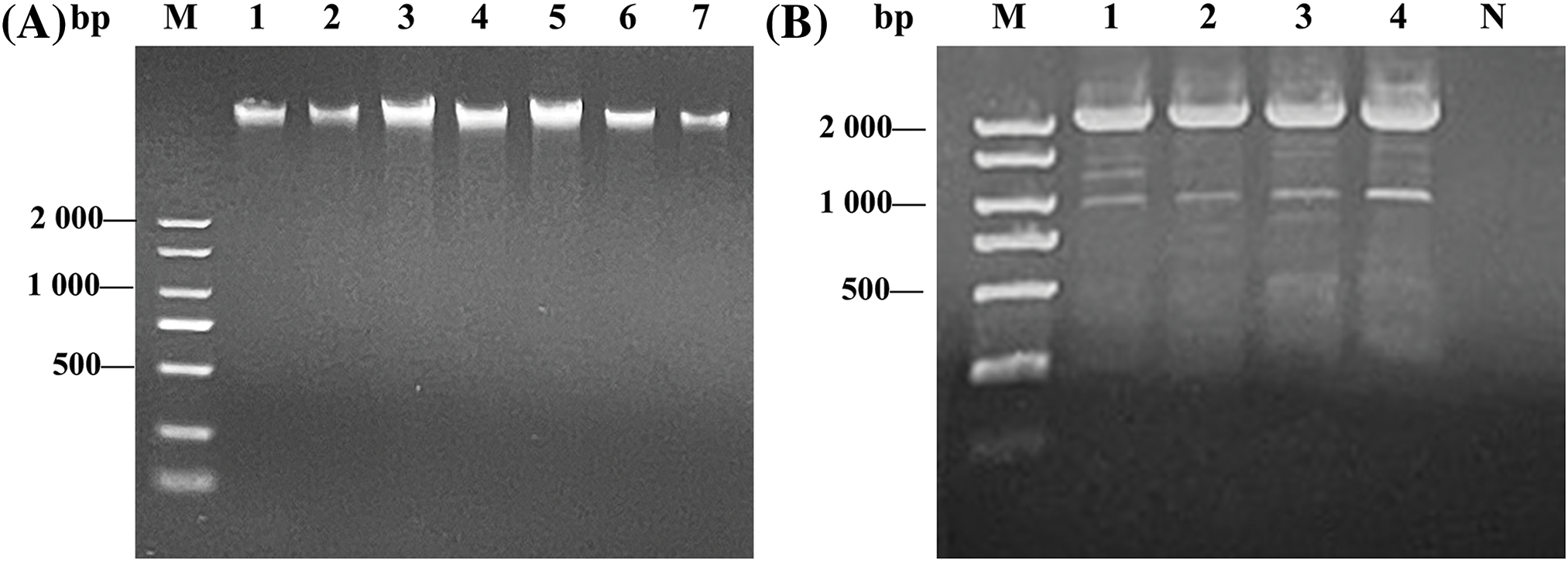
Figure 1: Results of pepper ‘ZHC2’ genomic DNA and promoter cloning. (A) Pepper ‘ZHC2’ genomic DNA. M: DNA marker, 1–7: Genomic DNA. (B) CaABI3/VP1-1 promoter clones, 1–4: CaABI3/VP1-1 promoter, N: Negative control
3.2 Pepper Total RNA Extraction and mRNA Isolation
After total RNA of the pepper was extracted by the CTAB method, agarose gel electrophoresis showed that the total RNA presented clear 18S and 28S rRNA bands, indicating that the extracted RNA had not degraded and showed high integrity (Fig. 2A). The RNA concentration determined spectrophotometrically was 1060.9 ng·μL−1 and the A260/A280 ratio was 2.03, indicating that the RNA was basically free of protein contamination, with high quality and purity. Total RNA was isolated and purified to obtain the mRNA fraction. Agarose gel electrophoresis showed that the mRNA presented a uniform diffuse state without obvious 18 and 28S bands, was mainly distributed above 850 bp in length without impurity or contamination (Fig. 2B), and the quality of isolated mRNA was high enough to meet the requirements of subsequent library construction.
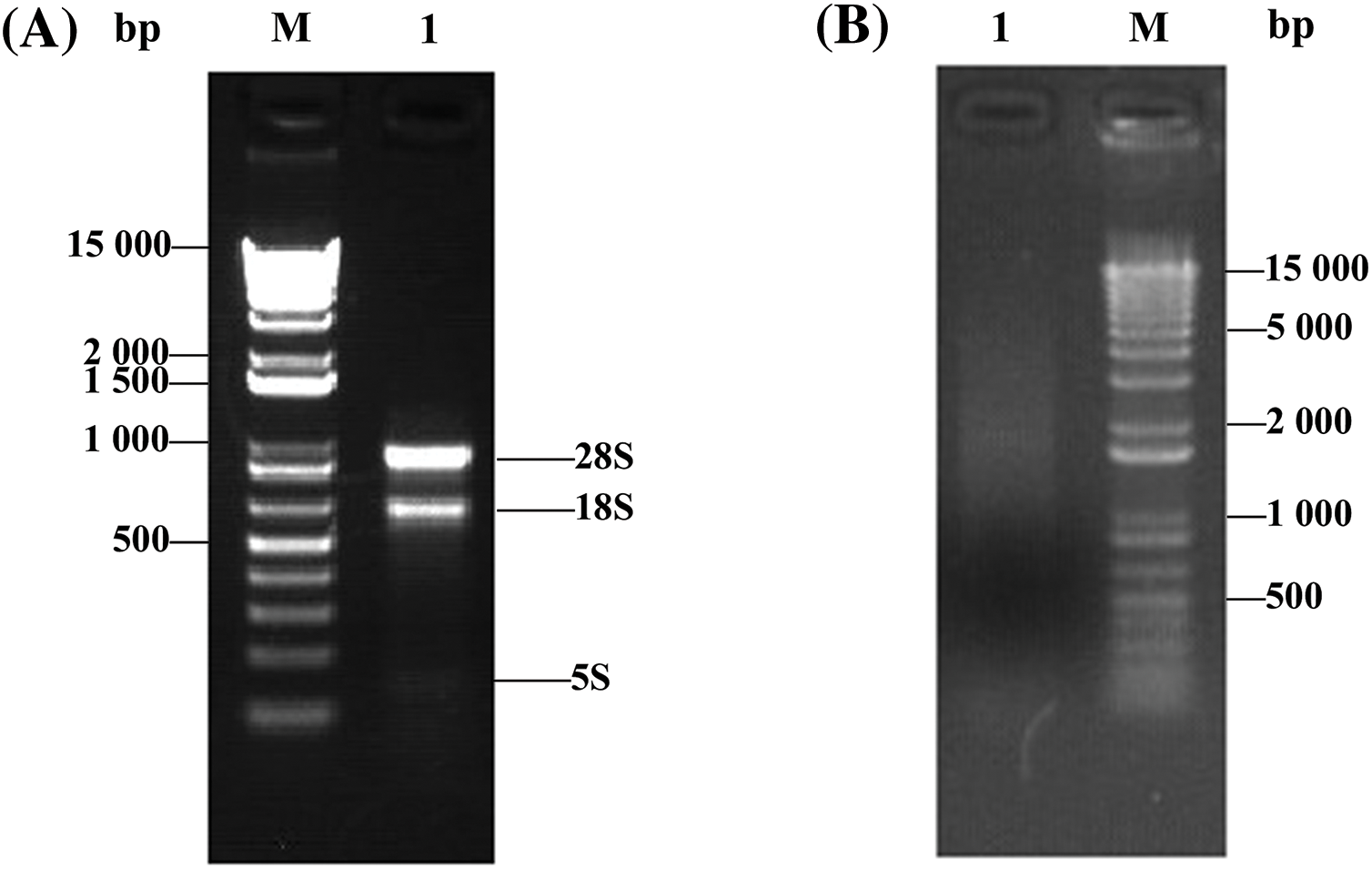
Figure 2: Results of total RNA extraction and mRNA isolation. (A) ‘ZHC2’ total RNA extraction, M: DNA marker, 1: Total RNA. (B) ‘ZHC2’ mRNA isolation, 1: mRNA
3.3 Construction and Quality Assessment of Normalized cDNA Primary Library
The full-length enriched cDNAs obtained were normalized, which reduces the proportion of high-abundance transcripts and balances transcript levels in a cDNA sample. The results of agarose gel electrophoresis (Fig. 3B) showed that the normalized cDNAs presented more uniform and more consistent intensities than the non-normalized cDNAs (Fig. 3A). Therefore, the normalized cDNAs successfully reduced the abundance differences of the different genes in the library. These normalized cDNAs were used to construct a cDNA library and provided a reliable basis for subsequent research.
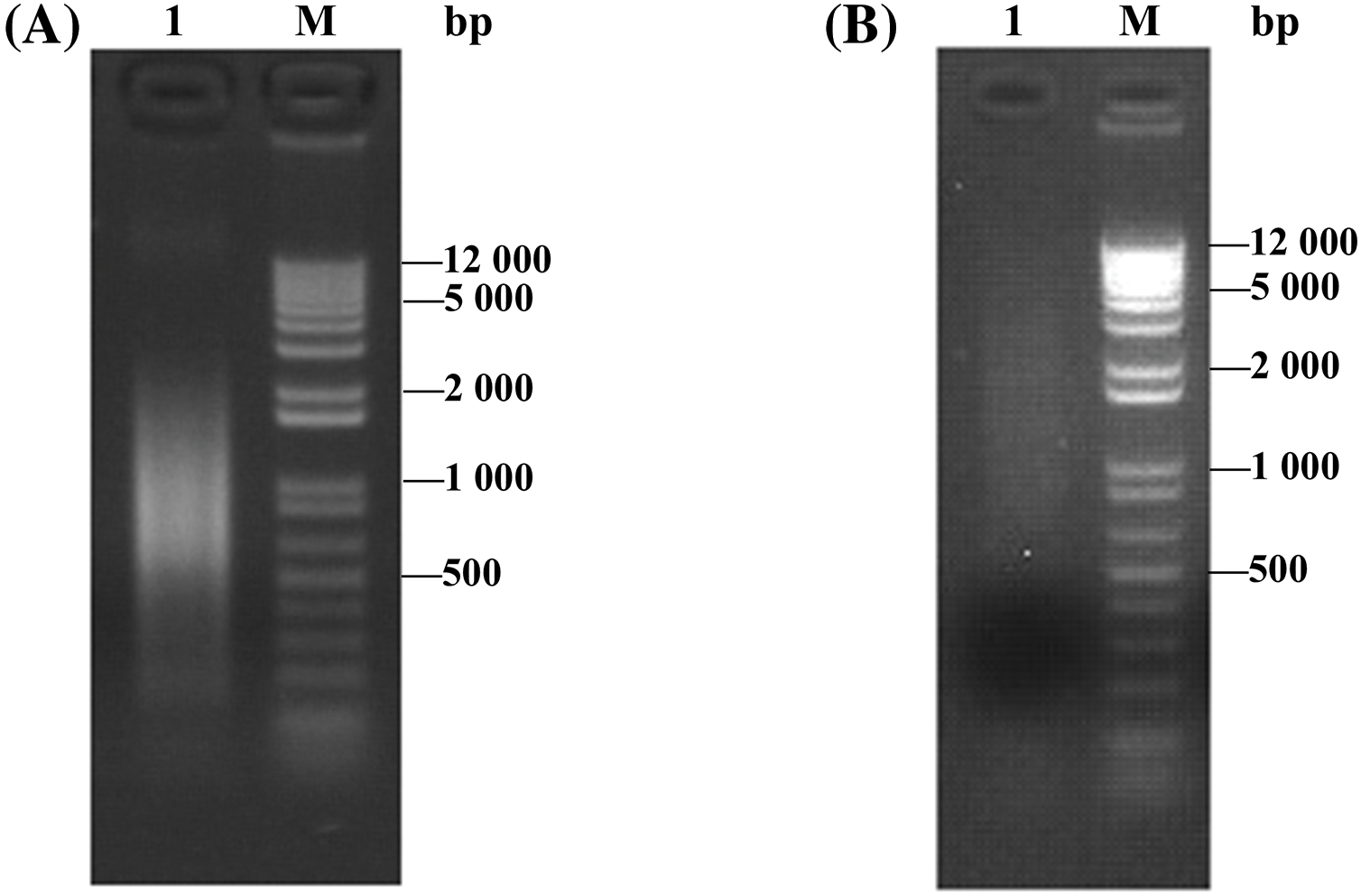
Figure 3: cDNA normalization results. (A) Before normalization, M: DNA marker, 1: cDNA before normalization. (B) After normalization, 1: cDNA after normalization
The normalized cDNAs were recombined with the pDONR222 vector through the attB and attP site reaction and then transformed into E. coli DH10B to gain the primary library bacterial suspension. An aliquot (1 μL) of the original bacterial suspension after transformation was diluted 1000× and a 50 μL aliquot was plated on LB medium. Two hundred and forty-one clones in total were acquired on the medium (Fig. 4A). The calculated library titer was 4.82 × 106 CFU·mL−1 and the library capacity was 9.64 × 106 CFU. Twenty-four single clones were at random selected for colony PCR testing. Agarose gel electrophoresis showed (Fig. 4B) that the size of the amplified fragments varied significantly. The size of the inserted amplified fragments ranged from 300 to 5000 bp, and the average length was greater than 1000 bp, with the library recombination rate being 100%. This shows that the library had considerable richness and the insertion fragments were diverse and homogeneous, covering more genetic information and reaching the standards of a high-quality primary library.
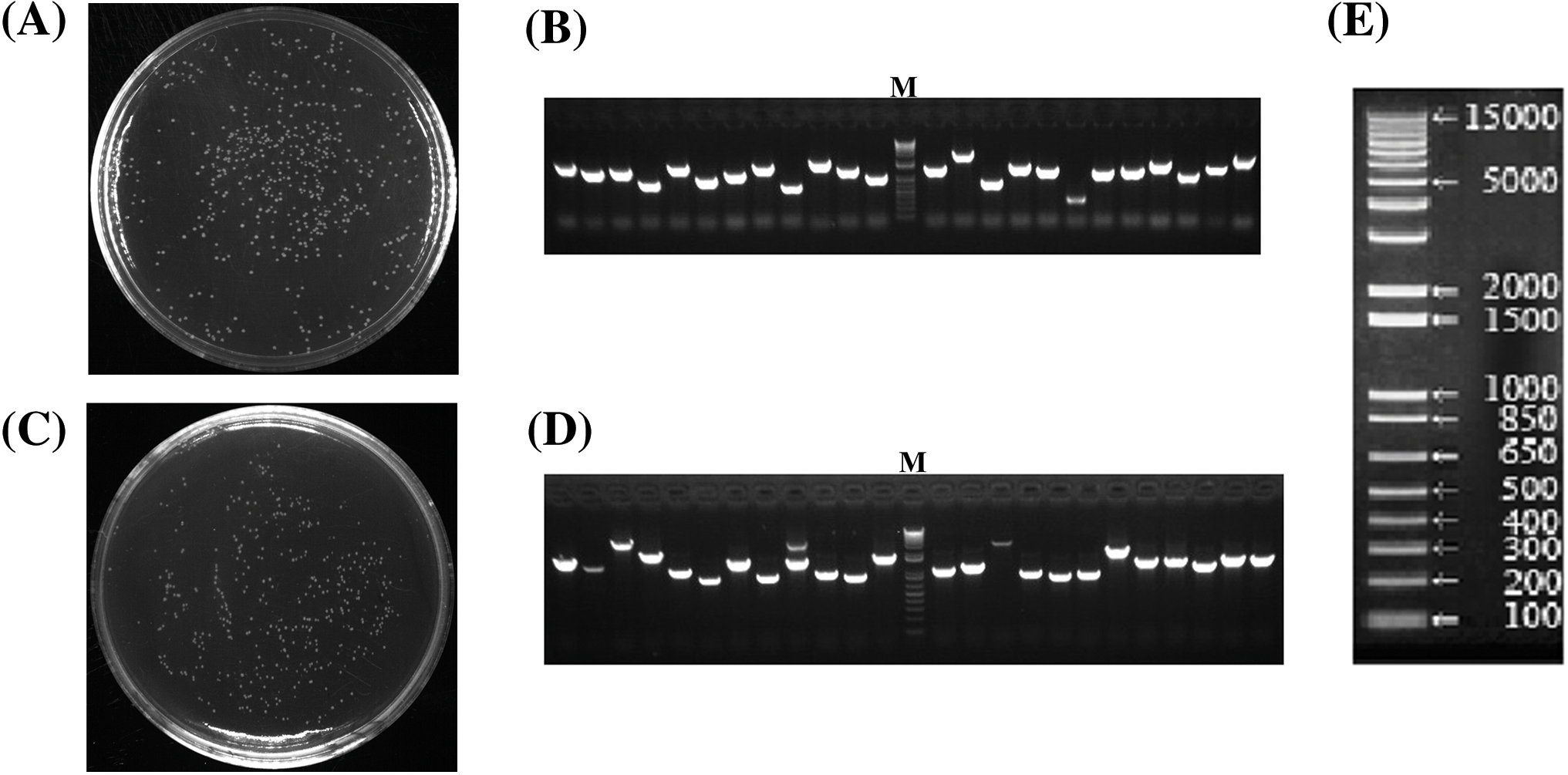
Figure 4: Results of the quality assessment of primary and yeast libraries. (A) Primary library capacity assessment. (B) Primary library insert fragment PCR assessment, M: DNA marker. (C) Yeast library capacity assessment. (D) Yeast library insert fragment PCR identification. (E) Magnified DNA marker
3.4 Construction and Quality Assessment of the Yeast Secondary Library
The plasmid was taken out from the previously obtained primary library suspension; the attL and attR site reaction was then performed with the pGADT7 vector, and the recombinant product was transformed into E. coli DH10B to gain the secondary library suspension. Two hundred and fifty-nine clones in total were collected on the medium (Fig. 4C). The library titer was 5.18 × 106 CFU·mL−1 and the library capacity was 1.04 × 107 CFU. The 24 monoclones for PCR were at random chosen. Agarose gel electrophoresis showed (Fig. 4D,E) that the insert fragment size ranged from 650 to 5000 bp, with the average length being more than 1000 bp. The recombination rate of the secondary library was 100%, which achieves the standard of a high-quality secondary library for yeast one-, two-, and three-hybrid screening.
3.5 The Auto-Activation Detection of the Bait Vector
The promoter segment of CaABI3/VP1-1 was successfully ligated to the pHIS2 vector using the seamless cloning technology, and the sequencing results verified that the pHIS2-CaABI3/VP1-1 bait vector had been successfully constructed. The results of bait vector auto-activation showed (Fig. 5) that the control strains grew normally on SD-LW deficient medium, the positive control grew normally on medium at the different concentrations of 3-AT, and the growth of the pGADT7+pHIS2-CaABI3/VP1-1 transformant on SD-LWH (30 mM 3-AT) defective medium was significantly inhibited; furthermore, the growth status was broadly similar to that of the negative control, indicating that the HIS3 reporter gene was not activated. As a consequence, Y1H library screening was then performed on the 30 and 60 mM 3-AT concentrations in this experiment.
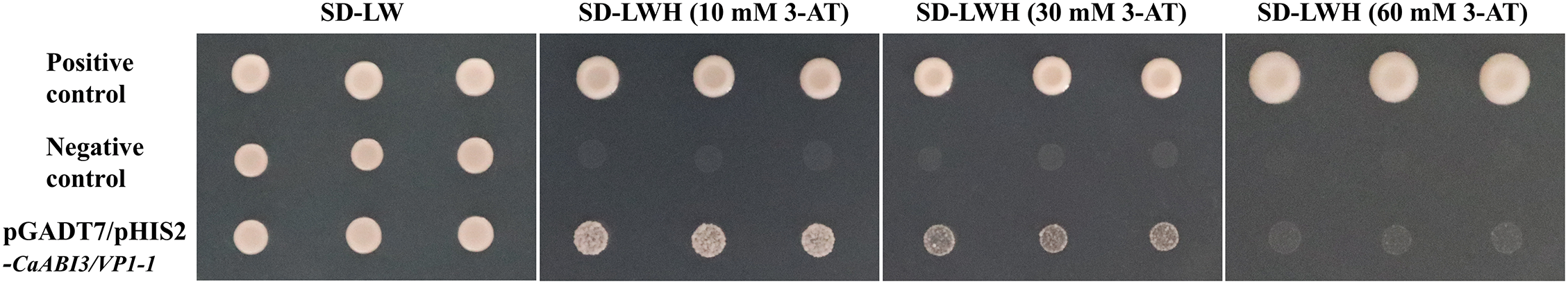
Figure 5: CaABI3/VP1-1 auto-activation assay results at three concentrations of 3-AT. SD: synthetic defined medium; SD-LW: SD/−leu/−trp; SD-LWH: SD/−leu/−trp/−his; 3-AT: 3-amino-1,2,4-triazole
3.6 Y1H Library Screening and Identification of Interacting Proteins
After gradient dilution (1/10×, 1/100×, and 1/1000×) of the pHIS2-CaABI3/VP1-1 yeast cell suspension, 14957, 8352, and 1026 clones were obtained on SD-LW medium, respectively (Fig. 6). The total number of transformed clones was 2.68 × 107, and the transformation efficiency was 1.07 × 107·μg−1, indicating that the efficiency of the library screening was high.
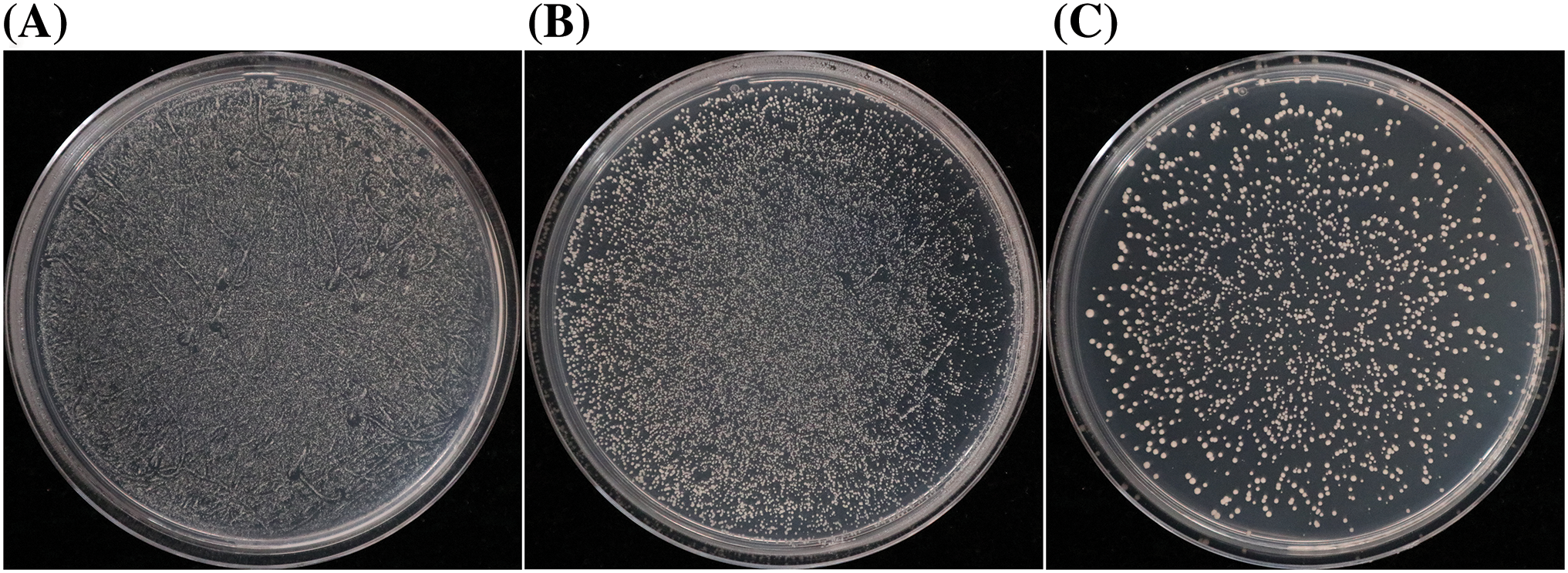
Figure 6: Library transformation efficiency assay. (A) Aliquot (20 μL) diluted with a 10× bacterial suspension. (B) Aliquot (20 μL) diluted with a 100× bacterial suspension. (C) Aliquot (20 μL) diluted with a 1000× bacterial suspension
Overall, 82 initial positive clones were obtained from the SD-LWH (60 mM 3-AT) medium (Fig. 7). To further determine whether there was interaction between them, they were transferred to fresh SD-LWH (60 mM 3-AT) medium for culture and then PCR detection and DNA sequencing were performed on the colonies, and BLAST comparison analysis was carried out against sequences in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 10 December 2024)). The detection results from the HIS3 reporter gene showed that all 82 initial positive clones grew on SD-LWH (60 mM 3-AT) medium, suggesting that the HIS3 reporter gene in all clones was activated and interacted with the CaABI3/VP1-1 promoter. Finally, these proteins were analyzed by BLAST and 78 different proteins were identified, with four duplicate clones. Among these 78 unique clones, there were two transcription factors and two non-coding RNAs, with one transcription factor within the ethylene-responsive factor (ERF) family and the other within the growth-regulatory factor (GRF) family.
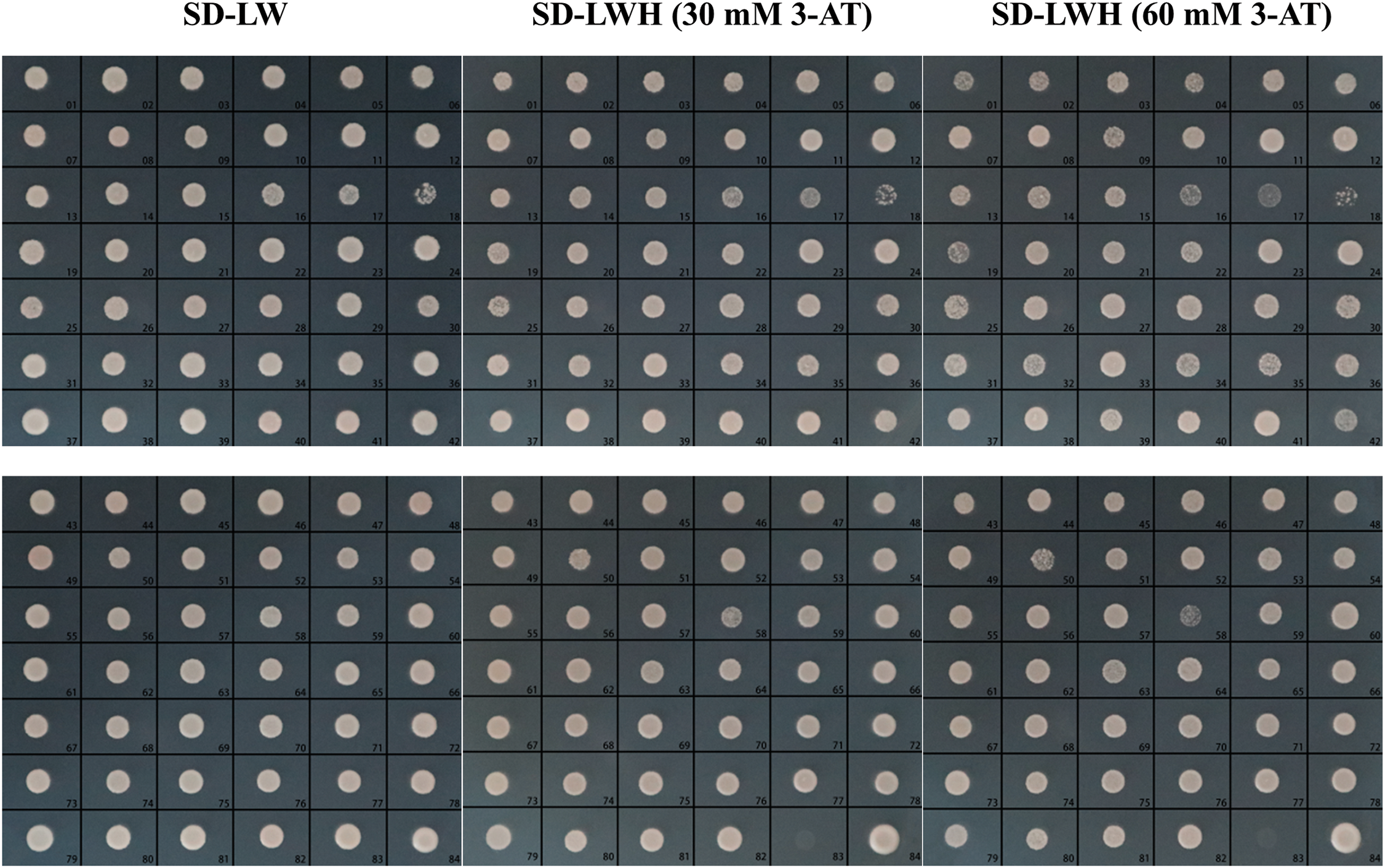
Figure 7: HIS3 reporter gene detection of transformant-positive clones. 1–82: 82 initial positive clones screened; 83: negative control; 84: positive control. SD: synthetic dropout medium; SD-LW: SD/−leu/−trp; SD-LWH: SD/−leu/−trp/−his; 3-AT: 3-amino-1,2,4-triazole
3.7 GO and PPI Analyses of Interacting Proteins
Using the GO enrichment analysis (Fig. 8A), three proteins were discovered to be enriched in each of the terms “cell wall”, “cell wall organization”, “cell wall biogenesis”, “xyloglucan metabolic process”, “xyloglucan: xyloglucosyl transferase activity”, “apoplast”, “plasmodesma”, “peroxisome”, “hydrogen peroxide catabolic process”, and “hydrolase activity, and hydrolyzing O-glycosyl compounds”. In addition, there were two proteins in each of the “response to reactive oxygen species”, “reductive pentose-phosphate cycle”, “ribulose-1,5-bisphosphate carboxylase activity”, “photorespiration”, “4-coumarate-CoA ligase activity”, and “Hsp90 protein binding” terms.
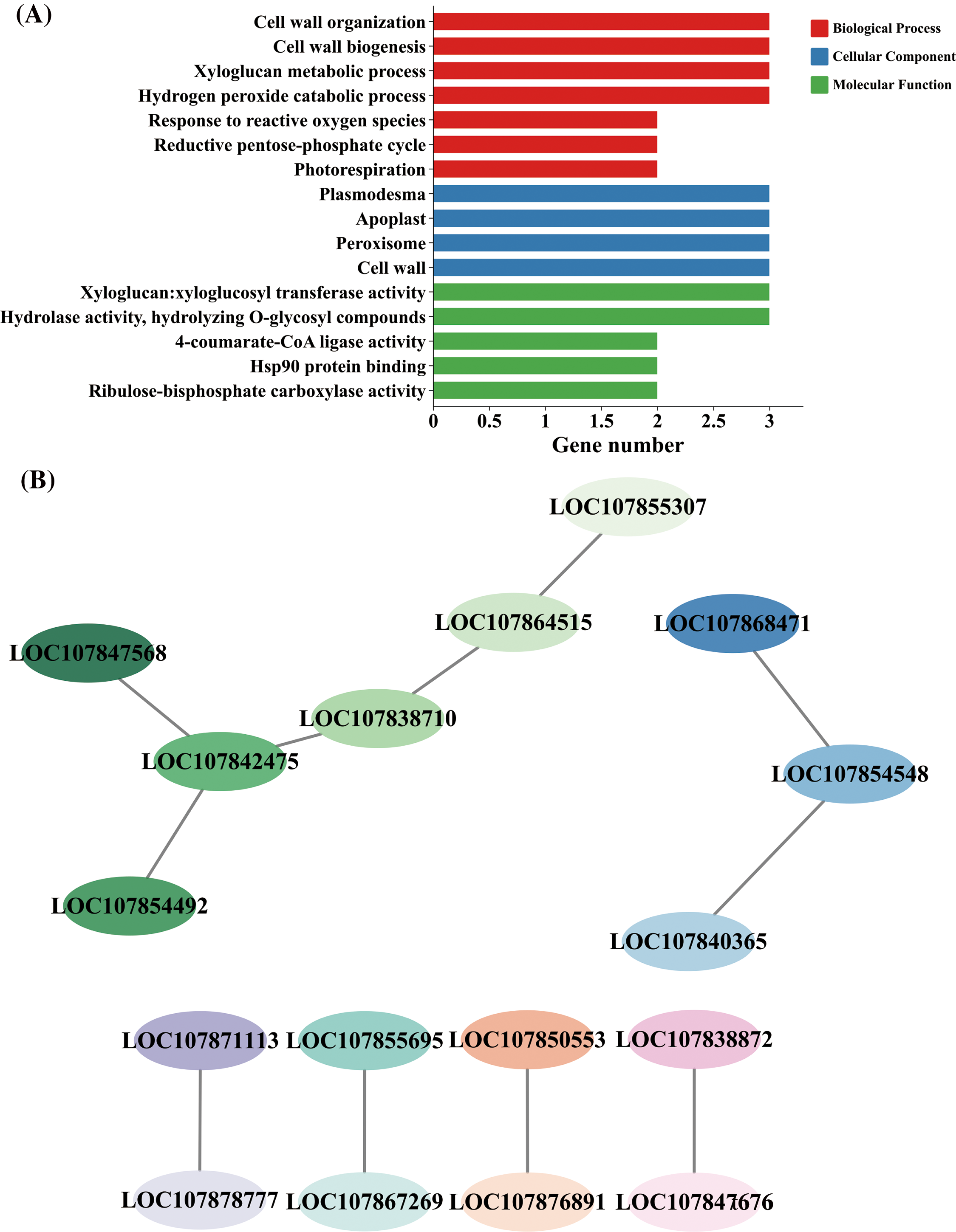
Figure 8: Results of GO (Gene Ontology) and PPI (protein–protein interaction) analysis of interacting proteins. (A) GO enrichment analysis. (B) PPI analysis
PPI network analysis of interacting proteins showed that 17 proteins may interact with each other (Fig. 8B). LOC107855695 and LOC107867269 were related to “Hsp90 protein binding”, while LOC107847568, LOC107864515, and LOC107855307 were annotated to “4-coumarate-CoA ligase activity” or “peroxisome” terms.
3.8 Interaction between LOC124895848 and CaABI3/VP1-1 Promoter
Point-to-point Y1H experiment was used to determine the interaction between the transcription factor LOC124895848 and the CaABI3/VP1-1 promoter. The positive control could grow on the deficient medium with different concentrations of 3-AT, while the negative control could not. The transformed yeast could grow on the medium, indicating that the interaction happened between LOC124895848 and CaABI3/VP1-1 promoter (Fig. 9).
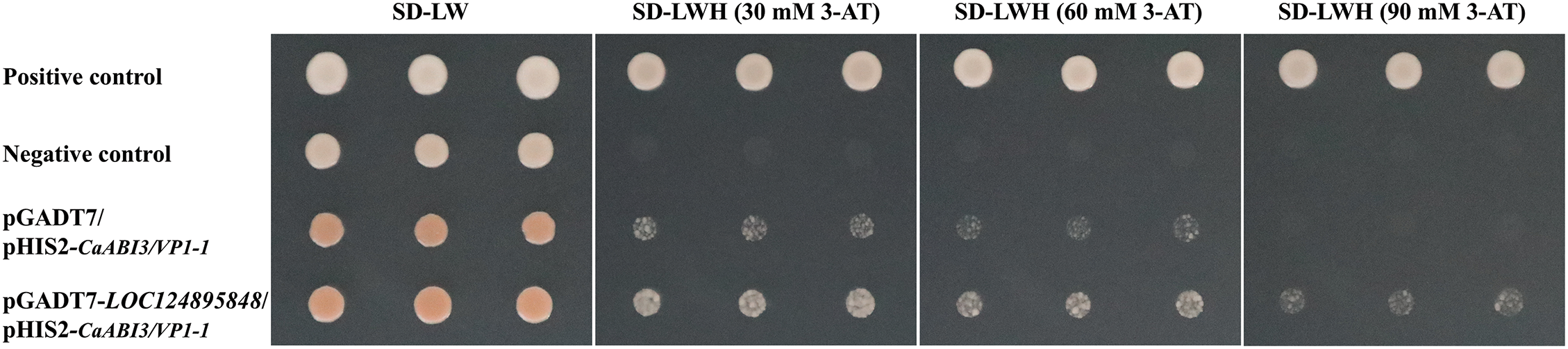
Figure 9: Point-to-point yeast one-hybrid assay of the interaction between LOC124895848 and CaABI3/VP1-1 promoter at three concentrations of 3-AT. SD: synthetic defined medium; SD-LW: SD/−leu/−trp; SD-LWH: SD/−leu/−trp/−his; 3-AT: 3-amino-1,2,4-triazole
To explore whether the screened interacting proteins played a certain role under waterlogging stress, 12 protein-coding genes (including LOC124895848 and LOC107867263, encoding two transcription factors) that might be potentially related to waterlogging stress response, were selected for examination of their expression patterns in different tissues of pepper under waterlogging stress. In leaves, the expression levels of LOC107840027, LOC107850312, LOC107841799, LOC107863737, LOC107866274, CaLTPIII, and LOC124895848 were upregulated after waterlogging and expression continued to increase after recovery (Fig. 10). The expression levels at the three time periods (before waterlogging, after waterlogging, and after recovery) were significantly different from each other, with CaLTPIII having the highest upregulation (fold) level, followed by LOC107840027. The expression levels of LOC107843092, LOC107877537, and LOC107889881 under waterlogging and recovery conditions were significantly higher than that of the control, but there was no marked difference in expression between the waterlogging and recovery stages. The expression levels of LOC107867269 and LOC107867263 after 1 h recovery were markedly higher than that of the controls or in the waterlogging period, but there was no marked difference in expression for either gene under control and waterlogging conditions.
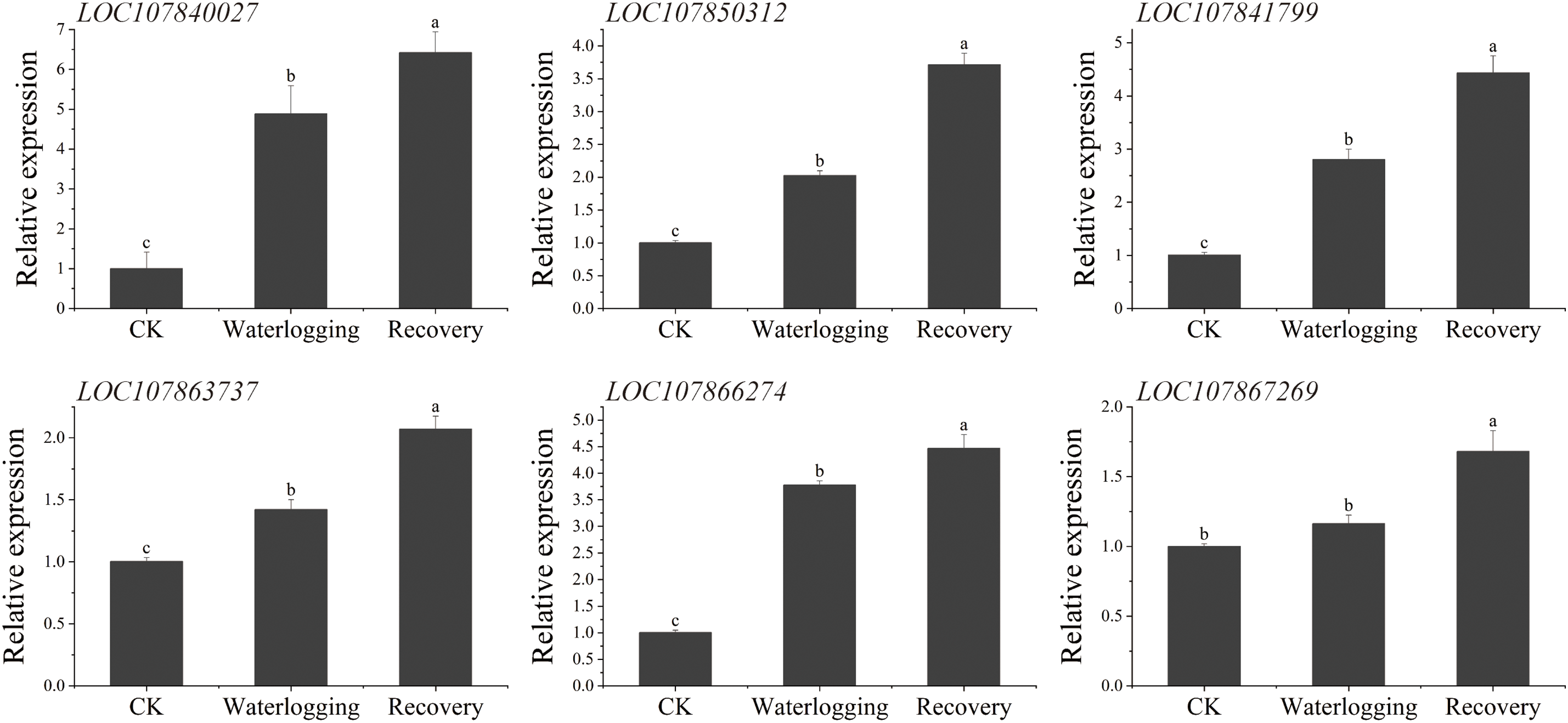
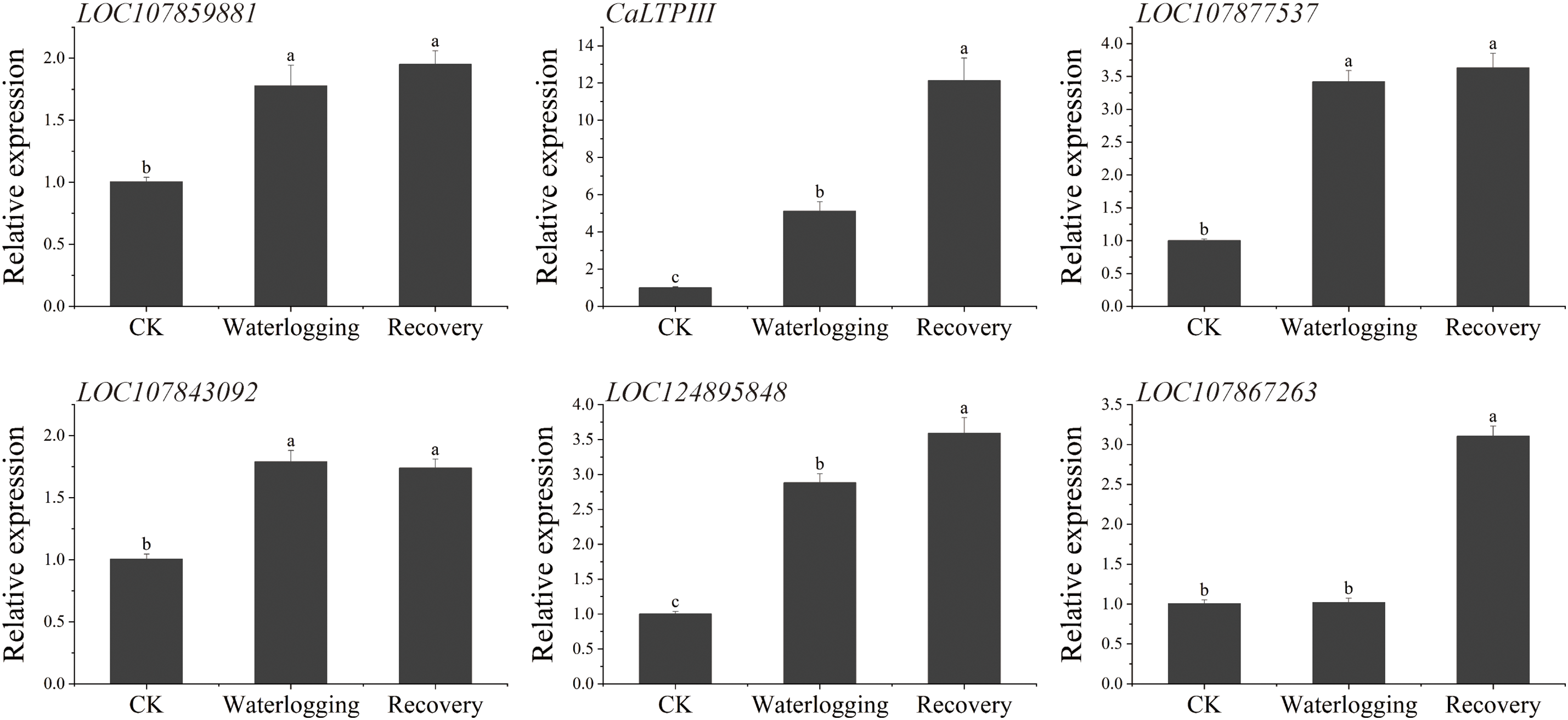
Figure 10: The relative expression levels of CaABI3/VP1-1-interacting protein-coding genes in ‘ZHC2’ leaves at waterlogging stress and recovery stages. CK indicates normal (control) culture. The error bars refer to standard errors. After analysis by one-way analysis of variance (ANOVA) and multiple pairwise comparison by the LSD test, any two bars for the same gene with a shared letter were not markedly different (p > 0.05)
In roots, the expression patterns of LOC107840027, LOC107866274, and LOC107863737 in response to waterlogging were similar to those in leaves, the only difference being that expression of LOC107863737 was not significantly different between waterlogging and recovery conditions (Fig. 11). The expression pattern of LOC107850312 and LOC107877537 in roots showed an opposite trend to that in leaves. The expression level was significantly lower after waterlogging than in the CK treatment and was significantly higher in the recovery phase than under waterlogging stress but was still significantly lower than that of the control. The expression level of LOC107841799 showed no significant change during the three experimental stages. The expression levels of LOC107843092, LOC107889881, and LOC107867263 increased significantly after recovery, compared with the control and waterlogging stress. On the contrary, the expression level of LOC124895848 after recovery was markedly lower than that at the other two stages. CaLTPIII and LOC107867269 showed the same expression pattern, increasing first (from CK to waterlogging) and then decreasing (from waterlogging to recovery), with significant differences in expression among the three stages for each gene.
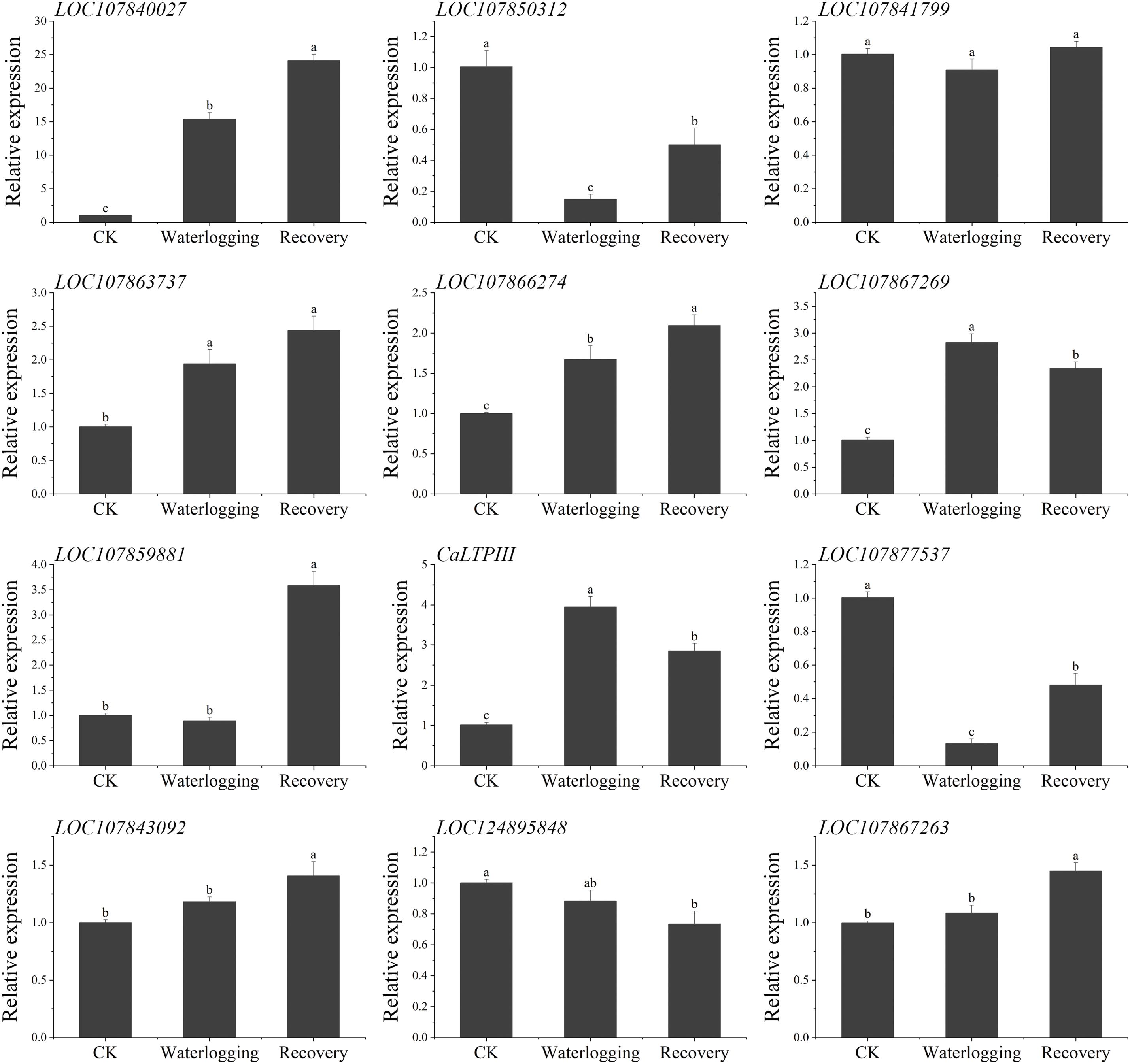
Figure 11: Analysis of the expression levels of CaABI3/VP1-1 interacting protein-coding genes in ‘ZHC2’ roots at waterlogging stress and recovery stages. CK indicates normal (control) culture. The error bars refer to standard errors. After analysis by one-way analysis of variance (ANOVA) and multiple pairwise comparison by the LSD test, any two bars for the same gene with a shared letter were not markedly different (p > 0.05)
Expression levels of several genes, such as LOC107840027 and CaLTPIII, were constantly upregulated under waterlogging stress and recovery conditions, suggesting that such genes might have a critical role in both waterlogging stress and recovery responses. The expression levels of other genes, such as LOC107867263, increased significantly after recovery, indicating that they might act primarily in the recovery period to help plants recover from stress. However, the opposite expression patterns of LOC107850312 and LOC107877537 in leaves and roots suggest that their expression was tissue-specific. Specifically, the response patterns of these genes to waterlogging stress varied between different pepper tissues, suggesting that these genes might have different regulatory roles in the response to waterlogging stress and recovery and that the findings may reflect plant complex regulatory mechanisms in response to environmental stresses.
In recent years, plant science has entered the post-genomic era. With the rapid development of functional genomics, the interaction between proteins and DNA has become a research focus in the field of molecular biology. The Y1H assay technique is one of the most effective strategies by which to study the interaction between proteins and DNA. To use the Y1H technology to screen for potential interacting proteins, the establishment of a high-quality and comprehensive Y1H library is the prerequisite. In addition, the same experiment may produce inconsistent results at different times and under different conditions, leading to unreliable experimental results. Therefore, it is crucial to ensure the quality of the cDNA library in the experiment. The key elements for constructing a high-quality Y1H library include the concentration, integrity, and purity of mRNA, while the evaluation criteria for yeast libraries include transformation efficiency, recombination efficiency, and library capacity, with the latter needing to be ≥1 × 106 CFU [22,30–32]. To avoid constructing a cDNA library with incomplete genetic information due to limited genes and different spatiotemporal expression patterns, this study extracted total RNA from pepper roots, leaves, and fruits to expand the scope of screening for interacting proteins. The A260/A280 ratio of the RNA sample used for the cDNA library construction was 2.03. In agarose gel electrophoresis analysis, the 18 and 28S RNA bands looked clear, indicating that the RNA was of high quality and integrity. Meanwhile, the mRNA also showed clear and distinct state, indicating high quality. The primary library capacity was 9.64 × 106 CFU. The yeast secondary library capacity was 1.04 × 107 CFU, the library transformation efficiency was 1.07 × 107·μg−1, and the recombination rate was 100%. The insertion fragments (650 to 5000 bp) covered more genetic information, which could improve the accuracy of identification of any interacting proteins. These results shown that the yeast library had complete genetic information, high capacity, and high quality, which laid the foundation for the subsequent screening for interacting proteins.
The Y1H assay system was based on the yeast two-hybrid technique. It is mainly used to study the interaction between DNA and proteins and is a powerful tool to reveal the complexity of gene regulation and protein/DNA interactions. Using the Y1H assay system, researchers can screen candidate interacting proteins from cDNA libraries and identify whether specific transcription factors can bind to specific DNA sequences (such as promoter regions). The practical application of this technique is of great value in understanding gene regulatory mechanisms, cell functions, and developmental processes [33–35]. In recent years, the Y1H assay system has made significant progress in plant genetic engineering. Y1H screening, using a library composed of only Arabidopsis transcription factors, further improves the efficiency of identifying specific DNA-sequence-bound transcription factors, marking an important advance in the field [24]. The one-hybrid interaction drives the expression of the HIS3 gene on the pHIS2 vector, allowing the host cells to grow on a histidine-deficient medium. The HIS3 expression level is extremely low if not activated. During library screening, 3-AT is supplemented to the culture medium to control the leaky HIS3 expression. It can be used to observe how much 3-AT the transformant can withstand to distinguish between “strong” and “weak” interaction phenotypes [33]. pGADT7 and the bait vector were co-transformed into Y187, and then the auto-activation activity was detected under different 3-AT concentrations. The goal was to find the lowest 3-AT concentration that could effectively inhibit the expression of HIS3. The Y187 strain lacked the HIS reporter gene in the genome, unlike the AH109 strain, to reduce background expression. This experiment showed that the growth of pGADT7+pHIS2−CaABI3/VP1-1 transformants exhibited marked differences in SD-LWH medium containing different 3-AT concentrations (Fig. 5). Under 30 mM 3-AT conditions, the transformant growth was significantly inhibited, whereas, under 60 mM 3-AT conditions, its growth status was similar to that of the negative control because the HIS3 reporter gene was not activated. The results showed that the bait vector constructed could be used for further library screening, laying the foundation for screening for proteins interacting with the CaABI3/VP1-1 promoter, studying CaABI3/VP1-1 response to waterlogging stress, and constructing a regulatory network of waterlogging tolerance in pepper.
In this study, the pHIS2-CaABI3/VP1-1 bait vector was used to screen for interacting proteins in the pepper cDNA library, and a total of 78 interacting proteins were detected, including two transcription factors from important families, namely the ERF and GRF families, and two non-coding RNAs with potential functions. This finding provided new insights into the CaABI3/VP1-1 role in pepper growth, development, and stress tolerance. Expression analysis of 12 genes showed that they all responded to waterlogging stress to varying degrees. They might activate or inhibit the activity of the CaABI3/VP1-1 promoter by up-regulating or down-regulating gene expression, thereby affecting CaABI3/VP1-1 expression. However, it is not excluded that the gene expression itself can change plant waterlogging tolerance.
However, although some potential regulatory factors were initially screened out through the Y1H assay technique, due to the characteristics of the Y1H system, there is a probability of false positives among the screened interacting proteins. In order to confirm whether they have an interactive relationship in plants, further experiments are needed [36].
The ERFs are important transcription factors that mediate ethylene reactions and belong to a large family of plant-specific transcription factors related to the regulation of abiotic stress responses, which play a key role in activating gene expression in abiotic stress responses by binding to specific CREs in promoters, such as dehydration response elements/C-repeat [37]. ERFs from different plant species have been demonstrated to play a significant role in adaptation to abiotic stresses [38–42]. Ethylene plays a vital role in adaptation to plant hypoxia and in plant metabolism under waterlogging stress. Tomato ERFs can enhance plant adaptability to waterlogging stress [43], while Arabidopsis AtERF71/HRE2 plays a key regulatory role in osmotic and hypoxic stress responses. Loss-of-function mutations of AtERF71/HRE2 show increased sensitivity to osmotic stress, while overexpression improves tolerance to waterlogging, drought, and salt [44]. Fan et al. [45] screened three waterlogging stress-related genes, namely ThERF15, ThERF39, and ThRAP2.3, by transcriptome sequencing, among which ThRAP2.3 was considered to be the key gene in downstream response to ERF and ethylene signal output generated by waterlogging stress. GRF transcription factors are a type of plant-specific transcription factor that contributes to rice leaves and stems [46], and have also been shown to be involved in plant growth, development, flowering, and stress tolerance [47–50]. Currently, most studies focus on the association between the GRFs and stress responses other than waterlogging, and there are few studies on the relationship between the GRFs and waterlogging stress. However, in Arabidopsis, the GRFs were found to be indirectly associated with waterlogging stress [47]. Our results offer new clues for the role of GRFs in plant response to waterlogging stress.
A high-quality pepper yeast hybrid library was successfully constructed, with a library recombination efficiency of 100%, library titer of 5.18 × 106 CFU·mL−1, library capacity of 1.04 × 107 CFU, and diverse insert fragment sizes. The pepper CaABI3/VP1-1 promoter sequence was cloned, and its interacting proteins were identified by screening, using the Y1H assay system. Seventy-eight proteins that interacted with the CaABI3/VP1-1 promoter were successfully identified by screening and included two non-coding RNAs and two transcription factors, belonging to the important ERF and the GRF families. The expression patterns of 12 selected promoter-interacting protein-coding genes were analyzed by qPCR. They were upregulated or downregulated to varying degrees in response to waterlogging stress. The upregulation or downregulation of gene expression might affect the CaABI3/VP1-1 promoter activity. This study revealed how pepper responds and adapts to waterlogging stress through specific molecular mechanisms. It also provided a reference point for further analysis of the complex regulatory relationship between transcription factors and promoters in response to waterlogging stress in peppers. Manipulation of this relationship could help to increase tolerance of this commercially important stress in peppers.
Acknowledgement: We thank Guangdong Geng and Suqin Zhang for their guidance and support and we are grateful to the College of Agriculture at Guizhou University for providing the experiment sites and equipment.
Funding Statement: This research was funded by the National Natural Science Foundation of China (grant no. 32260760), the Science and Technology Program of Guizhou Province (grant no. 20201Z002), and the Platform Construction Project of Engineering Research Center for Protected Vegetable Crops in Higher Learning Institutions of Guizhou Province (Qianjiaoji [2022] No. 040).
Author Contributions: Study conception and design: Huiru Deng, Guangdong Geng; data collection: Huan Tian, Liuyan Yang; analysis and interpretation of results: Siyang Ou, Hui Wang; draft manuscript preparation: Suqin Zhang, Huiru Deng. All authors reviewed the drafts and approved the final version of the manuscript.
Availability of Data and Materials: The authors declare that the data supporting the findings of this study are available within the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Mfarrej MFB, Wang X, Hamzah Saleem M, Hussain I, Rasheed R, Arslan Ashraf M, et al. Hydrogen sulphide and nitric oxide mitigate the negative impacts of waterlogging stress on wheat (Triticum aestivum L.). Plant Biol. 2022;24(4):670–83. doi:10.1111/plb.v24.4. [Google Scholar] [CrossRef]
2. Voesenek LACJ, Bailey-Serres J. Flood adaptive traits and processes: an overview. New Phytol. 2015;206(1):57–73. doi:10.1111/nph.13209. [Google Scholar] [PubMed] [CrossRef]
3. Xiong XW, Tian H, Fan GL, Tian HZ, Wang H, Geng GD, et al. Characterization and transformation of a novel ABI3/VP1-1 gene from hot pepper to enhance waterlogging tolerance. Environ Exp Bot. 2024;220:105708. doi:10.1016/j.envexpbot.2024.105708. [Google Scholar] [CrossRef]
4. Swaminathan K, Peterson K, Jack T. The plant B3 superfamily. Trends Plant Sci. 2008;13(12):647–55. doi:10.1016/j.tplants.2008.09.006. [Google Scholar] [PubMed] [CrossRef]
5. Parcy F, Giraudat J. Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J. 1997;11(4):693–702. doi:10.1046/j.1365-313X.1997.11040693.x. [Google Scholar] [PubMed] [CrossRef]
6. Zhang YF, Clemens A, Maximova SN, Guiltinan MJ. The Theobroma cacao B3 domain transcription factor TcLEC2 plays a duel role in control of embryo development and maturation. BMC Plant Biol. 2014;14:106. doi:10.1186/1471-2229-14-106. [Google Scholar] [PubMed] [CrossRef]
7. Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One. 2009;4(6):5791. doi:10.1371/journal.pone.0005791. [Google Scholar] [PubMed] [CrossRef]
8. Wang J, Deng QW, Li YH, Yu Y, Liu X, Han YF, et al. Transcription factors Rc and OsVP1 coordinately regulate preharvest sprouting tolerance in red pericarp rice. J Agric Food Chem. 2020;68(50):14748–57. doi:10.1021/acs.jafc.0c04748. [Google Scholar] [PubMed] [CrossRef]
9. Guo XL, Hou XM, Fang J, Wei PW, Xu B, Chen ML, et al. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 2013;75(3):403–16. doi:10.1111/tpj.12209. [Google Scholar] [PubMed] [CrossRef]
10. Wang WB, Ao T, Zhang YY, Wu D, Xu W, Han B, et al. Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis). Plant Divers. 2021;44(2):201–12. doi:10.1016/j.pld.2021.06.008. [Google Scholar] [PubMed] [CrossRef]
11. Rohde A, Kurup S, Holdsworth M. ABI3 emerges from the seed. Trends Plant Sci. 2000;5(10):418–9. doi:10.1016/S1360-1385(00)01736-2. [Google Scholar] [PubMed] [CrossRef]
12. Sun J, Zhang GC, Cui ZB, Kong XM, Yu XY, Gui R, et al. Regain flood adaptation in rice through a 14-3-3 protein OsGF14h. Nat Commun. 2022;13(1):5664. doi:10.1038/s41467-022-33320-x. [Google Scholar] [PubMed] [CrossRef]
13. Mittal A, Gampala SS, Ritchie GL, Payton P, Burke JJ, Rock CD. Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol J. 2014;12(5):578–89. doi:10.1111/pbi.12162. [Google Scholar] [PubMed] [CrossRef]
14. Bedi S, Nag Chaudhuri R. Transcription factor ABI3 auto-activates its own expression during dehydration stress response. FEBS Lett. 2018;592(15):2594–611. doi:10.1002/1873-3468.13194. [Google Scholar] [PubMed] [CrossRef]
15. Marand AP, Eveland AL, Kaufmann K, Springer NM. cis-regulatory elements in plant development, adaptation, and evolution. Annu Rev Plant Biol. 2023;74:111–37. doi:10.1146/annurev-arplant-070122-030236. [Google Scholar] [PubMed] [CrossRef]
16. Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262(5141):1870–4. doi:10.1126/science.8266075. [Google Scholar] [PubMed] [CrossRef]
17. Ouwerkerk PB, Meijer AH. Yeast one-hybrid screens for detection of transcription factor DNA interactions. Methods Mol Biol. 2011;678:211–27. doi:10.1007/978-1-60761-682-5. [Google Scholar] [CrossRef]
18. Chen R, Yang C, Gao H, Shi CM, Zhang ZY, Lu GY, et al. Induced mutation in ELONGATED HYPOCOTYL5 abolishes anthocyanin accumulation in the hypocotyl of pepper. Theor Appl Genet. 2022;135(10):3455–68. doi:10.1007/s00122-022-04192-y. [Google Scholar] [PubMed] [CrossRef]
19. Wang YH, Liu SJ, Wang HR, Zhang YX, Li WJ, Liu JK, et al. Identification of the regulatory genes of UV-B-induced anthocyanin biosynthesis in pepper fruit. Int J Mol Sci. 2022;23(4):1960. doi:10.3390/ijms23041960. [Google Scholar] [PubMed] [CrossRef]
20. Spyropoulou EA, Haring MA, Schuurink RC. Expression of Terpenoids 1, a glandular trichome-specific transcription factor from tomato that activates the terpene synthase 5 promoter. Plant Mol Biol. 2014;84(3):345–57. doi:10.1007/s11103-013-0142-0. [Google Scholar] [PubMed] [CrossRef]
21. Valenzuela-Riffo F, Delgado C, Morales-Quintana L, Figueroa CR. The strawberry transcription factor FaWRKY48 transactivates the FaNCED1 promoter as revealed by yeast-one hybrid and Nicotiana benthamiana transfection assays. Sci Hortic. 2024;323:112545. doi:10.1016/j.scienta.2023.112545. [Google Scholar] [CrossRef]
22. Cao YW, Bi MM, Yang PP, Song M, He GR, Wang J, et al. Construction of yeast one-hybrid library and screening of transcription factors regulating LhMYBSPLATTER expression in Asiatic hybrid lilies (Lilium spp.). BMC Plant Biol. 2021;21(1):563. doi:10.1186/s12870-021-03347-1. [Google Scholar] [PubMed] [CrossRef]
23. Liu H, Yang Y, Zhang LS. Identification of upstream transcription factors and an interacting PP2C protein of dehydrin WZY2 gene in wheat. Plant Signal Behav. 2019;14(12):1678370. doi:10.1080/15592324.2019.1678370. [Google Scholar] [PubMed] [CrossRef]
24. Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, et al. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 2010;51(12):2145–51. doi:10.1093/pcp/pcq161. [Google Scholar] [PubMed] [CrossRef]
25. Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–5. doi:10.1093/nar/8.19.4321. [Google Scholar] [PubMed] [CrossRef]
26. Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):31–4. doi:10.1038/nprot.2007.13. [Google Scholar] [PubMed] [CrossRef]
27. Guo H, Tian H, Wang YQ, Xiong XW, Tian HZ, Zhang SQ, et al. Comparative analysis of growth, physiological and transcriptomic response revealed mechanisms of waterlogging tolerance of hot pepper (Capsicum annuum var. conoides). Plant Breed. 2022;141(6):807–19. doi:10.1111/pbr.v141.6. [Google Scholar] [CrossRef]
28. Wan LY, Zhang JF, Zhang HW, Zhang ZJ, Quan RD, Zhou SR, et al. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS One. 2011;6(9):25216. doi:10.1371/journal.pone.0025216. [Google Scholar] [PubMed] [CrossRef]
29. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–8. doi:10.1006/meth.2001.1262. [Google Scholar] [PubMed] [CrossRef]
30. Ohara O, Temple G. Directional cDNA library construction assisted by the in vitro recombination reaction. Nucleic Acids Res. 2001;29(4):E22. doi:10.1093/nar/29.4.e22. [Google Scholar] [PubMed] [CrossRef]
31. Gao XH, Jia RY, Wang MS, Zhu DK, Chen S, Lin M, et al. Construction and identification of a cDNA library for use in the yeast two-hybrid system from duck embryonic fibroblast cells post-infected with duck enteritis virus. Mol Biol Rep. 2014;41(1):467–75. doi:10.1007/s11033-013-2881-z. [Google Scholar] [PubMed] [CrossRef]
32. Xu YQ, Zhou JJ, Liu QQ, Li KP, Zhou Y. Construction and characterization of a high-quality cDNA library of Cymbidium faberi suitable for yeast one- and two-hybrid assays. BMC Biotechnol. 2020;20(1):4. doi:10.1186/s12896-020-0599-2. [Google Scholar] [PubMed] [CrossRef]
33. Reece-Hoyes JS, Marian Walhout AJ. Yeast one-hybrid assays: a historical and technical perspective. Methods. 2012;57(4):441–7. doi:10.1016/j.ymeth.2012.07.027. [Google Scholar] [PubMed] [CrossRef]
34. Ji XY, Wang LQ, Zang DD, Wang YC. Transcription factor-centered yeast one-hybrid assay. Methods Mol Biol. 2018;1794:183–94. doi:10.1007/978-1-4939-7871-7. [Google Scholar] [CrossRef]
35. Kang SE, Breton G, Pruneda-Paz JL. Construction of Arabidopsis transcription factor ORFeome collections and identification of protein-DNA interactions by high-throughput yeast one-hybrid screens. Methods Mol Biol. 2018;1794:151–82. doi:10.1007/978-1-4939-7871-7. [Google Scholar] [CrossRef]
36. Wang JW, Wang JX, Zhu Y, Zhu Y, Liu CZ, Chen YY, et al. Building an improved transcription factor-centered yeast one hybrid system to identify DNA motifs bound by protein comprehensively. BMC Plant Biol. 2023;23(1):236. doi:10.1186/s12870-023-04241-8. [Google Scholar] [PubMed] [CrossRef]
37. Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):86–96. doi:10.1016/j.bbagrm.2011.08.004. [Google Scholar] [PubMed] [CrossRef]
38. Cheng MC, Liao PM, Kuo WW, Lin TP. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162(3):1566–82. doi:10.1104/pp.113.221911. [Google Scholar] [PubMed] [CrossRef]
39. Zhu ZG, Shi JL, Xu WR, Li HE, He MY, Xu Y, et al. Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways. J Plant Physiol. 2013;170(10):923–33. doi:10.1016/j.jplph.2013.01.017. [Google Scholar] [PubMed] [CrossRef]
40. Debbarma J, Sarki YN, Saikia B, Boruah HPD, Singha DL, Chikkaputtaiah C. Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR-Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol Biotechnol. 2019;61(2):153–72. doi:10.1007/s12033-018-0144-x. [Google Scholar] [PubMed] [CrossRef]
41. Wu Y, Li X, Zhang JN, Zhao HQ, Tan SL, Xu WH, et al. ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front Plant Sci. 2022;13:1042084. doi:10.3389/fpls.2022.1042084. [Google Scholar] [PubMed] [CrossRef]
42. Nie SM, Wang D. AP2/ERF transcription factors for tolerance to both biotic and abiotic stress factors in plants. Tropical Plant Biol. 2023;16:105–12. doi:10.1007/s12042-023-09339-9. [Google Scholar] [CrossRef]
43. Klay I, Gouia S, Liu M, Mila I, Khoudi H, Bernadac A, et al. Ethylene response factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 2018;274:137–45. doi:10.1016/j.plantsci.2018.05.023. [Google Scholar] [PubMed] [CrossRef]
44. Park HY, Seok HY, Woo DH, Lee SY, Tarte VN, Lee EH, et al. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem Biophys Res Commun. 2011;414(1):135–41. doi:10.1016/j.bbrc.2011.09.039. [Google Scholar] [PubMed] [CrossRef]
45. Fan WC, Yang Y, Wang ZQ, Yin YL, Yu CG, Shi Q, et al. Molecular cloning and expression analysis of three ThERFs involved in the response to waterlogging stress of Taxodium ‘Zhongshanshan406’, and subcellular localization of the gene products. PeerJ. 2018;6:4434. doi:10.7717/peerj.4434. [Google Scholar] [PubMed] [CrossRef]
46. van der Knaap E, Kim JH, Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000;122(3):695–704. doi:10.1104/pp.122.3.695. [Google Scholar] [PubMed] [CrossRef]
47. Kim JS, Mizoi J, Kidokoro S, Maruyama K, Nakajima J, Nakashima K, et al. Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell. 2012;24(8):3393–405. doi:10.1105/tpc.112.100933. [Google Scholar] [PubMed] [CrossRef]
48. Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, et al. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014;79(3):413–26. doi:10.1111/tpj.2014.79.issue-3. [Google Scholar] [CrossRef]
49. Kim JH, Tsukaya H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot. 2015;66(20):6093–107. doi:10.1093/jxb/erv349. [Google Scholar] [PubMed] [CrossRef]
50. Cheng ZH, Wen SQ, Wu YK, Shang LN, Wu L, Lyu DQ, et al. Comparatively evolution and expression analysis of GRF transcription factor genes in seven plant species. Plants. 2023;12(15):2790. doi:10.3390/plants12152790. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools