 Open Access
Open Access
ARTICLE
Genetic Diversity and Combining Ability of Developed Maize Lines to Realize Heterotic and High Yielding Hybrids for Arid Conditions
1 Department of Agronomy, Faculty of Agriculture, Kafrelsheikh University, Kafr El-Sheikh, 33516, Egypt
2 Department of Biology, College of Science, Princess Nourah bint Abdulrahman University, Riyadh, 11671, Saudi Arabia
3 Department of Biology, Faculty of Science, Umm Al-Qura University, Makkah, 24372, Saudi Arabia
4 Date Palm Research Center of Excellence, King Faisal University, Al-Ahsa, 31982, Saudi Arabia
5 Central Laboratory for Date Palm Research and Development, Agriculture Research Center, Giza, 12511, Egypt
6 Biology Department, College of Science, King Khalid University, Abha, 61321, Saudi Arabia
7 Department of Biotechnology, College of Sciences, Taif University, Taif, 21944, Saudi Arabia
8 Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
9 Immunology Unit, King Fahd Medical Research Center, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
10 Department of Plant Production (Genetic Branch), Faculty of Environmental Agricultural Sciences, Arish University, El-Arish, 45511, Egypt
11 Genetics Department, Faculty of Agriculture, Zagazig University, Zagazig, 44511, Egypt
12 Department of Biology, College of Science, Buraydah, 51452, Saudi Arabia
13 National Authority for Remote Sensing and Space Sciences (NARSS), Al-Nozha Al-Gedida, Cairo, P.O. Box 1564, Egypt
14 Department of Crop Science, Faculty of Agriculture, Zagazig University, Zagazig, 44519, Egypt
* Corresponding Authors: Imen Ben Abdelmalek. Email: ; Elsayed Mansour. Email:
(This article belongs to the Special Issue: Integrative Approaches to Plant Stress Responses under Changing Climate Conditions)
Phyton-International Journal of Experimental Botany 2024, 93(12), 3465-3485. https://doi.org/10.32604/phyton.2024.058628
Received 17 September 2024; Accepted 11 November 2024; Issue published 31 December 2024
Abstract
Developing high-yield maize hybrids is critical for sustaining maize production, especially in the face of rapid climate changes and the growing global population. Exploring the genetic diversity and combining ability in parental inbreds is needed for developing such high-yielding hybrids. Consequently, this study aimed at evaluating parental genetic diversity employing simple sequence repeats (SSR) markers, estimating effects of general (GCA) and specific (SCA) combining abilities for grain yield and yield contributing characters, identifying high yielding hybrids, and evaluating the association of SCA effects and performance of hybrids with genetic distance. Half-diallel mating scheme was utilized to develop 21 F1 hybrids from seven diverse maize inbred lines. The F1 hybrids along with check hybrid (SC-10), were investigated in a field trial over two growing seasons under arid conditions. The assessed F1 hybrids displayed significant genetic variations across all recorded traits. The inbreds P1 and P3 were detected as effective combiners to develop early maturing hybrids. Additionally, P3 and P4 were recognized as better combiners for improving grain yield and yield attributed characters. The hybrids P1 × P5 and P4 × P7 displayed significant SCA effects coupled with favorable agronomic performance. These hybrids are recommended for further evaluation and release as variety for arid environments to increase total maize production and contribute to food security. The alleles per locus differed between 2 and 5, with average of 3.5 alleles/locus. The polymorphic information content (PIC) altered between 0.21 to 0.74, with a mean of 0.56. Unweighted neighbor-joining tree grouped the inbred lines into three clusters, providing a valuable tool to decrease the crosses needed to be assessed in the trial field. Parental genetic distance varied from 0.63 to 0.90, averaging 0.79. The relationship between genetic diversity assessed through SSR markers and SCA effects was insignificant for all considered traits. Otherwise, SCA demonstrated a significant correlation with hybrid performance, suggesting that SCA serves as a reliable predictor for hybrid performance. The assessed maize inbred lines and developed hybrids revealed substantial genetic variability, offering valuable resources for enhancing maize productivity under arid conditions. The identified promising inbred lines (P1, P3, and P4) might be regarded as effective combiners for developing early-maturing genotypes and excellent combiners for enhancing yield attributes. Notably, the developed hybrids P1 × P5 and P4 × P7 possessed significant SCA alongside superior yield traits. SCA demonstrated a significant correlation with hybrid performance, suggesting its potential as a reliable predictor for the performance of developed hybrids.Keywords
Maize is among the most vital cereal crops in total production and cultivated area [1]. Its grains contain about 70% starch, 8.5% dietary fiber, 10% protein, 4.8% fat, and 3.0% sugar without antinutrients [2]. Maize is globally cultivated for various purposes involving edible oil, animal feed, fuel, and human food [3,4]. The global maize cultivation covers approximately 206 million hectares, yielding around 1.21 billion tons of grain [5]. In Egypt, maize production is lacking in order to meet the growing population; accordingly, around nine million tons of maize are imported annually [5]. The disparity between production and consumption is widening due to current and projected climate fluctuations. Subsequently, breeding high-yielding hybrids has become essential for maintaining maize production, principally with climate change and population growth.
Identifying suitable parents and selecting desirable traits is critical for implementing appropriate breeding strategies [6–9]. Exploring the general (GCA) and specific (SCA) combining abilities and understanding inheritance patterns of important agronomic characters is vital. Diallel mating design provides valuable insights into the effects of both GCA and SCA and genetic mechanisms governing the inheritance of targeted attributes during early generations [10–13]. In addition, this analysis is useful for distinguishing between non-additive and additive effects of the analyzed traits. SCA is typically associated with dominance effects, while GCA is linked to additive effects [14–16]. Additive and non additive gene actions are demonstrated to be vital in inheritance of yield traits of maize [17–19]. However, numerous previously published reports exposed the dominance of non additive gene effects governing yield traits in maize hybrids [20–22]. Due to the contrasting results depicted in previous studies and utilizing different inbred lines for genetic analyses, it is necessary to further study inheritance mode of maize grain yield by exploiting diverse inbred lines with varying genetic backgrounds.
Understanding molecular genetic distance (GD) is crucial for maize breeders, as it helps accelerate the discovery of vigorous hybrids without assessing all potential parent combinations [23–25]. In recent years, many DNA markers have successfully uncovered maize inbred lines genetic distance (GD). Among the most valuable molecular markers are simple sequence repeats [26,27]. The informative, multiallelic, codominant, and reproducible markers are vital characteristics of SSRs [28,29]. The association between SSR based genetic distance of parent and both combining ability and hybrid (F1) performance especially for grain yield has contrasting results ranging from not significant [30,31] to significantly vital [32–34]. However, using molecular markers to determine the association between combining ability and hybrid performance in maize is still inconclusive in Egypt [35]. Therefore, this study was performed to (i) explore GCA for evaluated inbred and specific combining ability of their hybrids, (ii) depict inheritance nature of studied agronomic characters, (iii) identify high-yielding hybrids, and (iv) explore the genetic diversity of parental lines and association of genetic diversity with SCA and performance of hybrids.
2.1 Experimental Layout and Plant Materials
Seven genetically distinct maize inbred lines (Zea mays L.) were utilized in the current study, obtained from CIMMYT and the Agricultural Research Center Egypt. The source and pedigree information for these lines are detailed in Table A1. The parental inbreds were crossed applying half-diallel mating scheme to develop twenty one F1 single crosses during summer 2021. The generated F1 hybrids, along with commercial hybrid SC-10, were assessed in a two-year field trial conducted during the summer seasons of 2022 and 2023 at a private farm in El-Mahmoudia, Egypt (31°15′N, 30°29′E). The experimental site features an hot and arid climate (Fig. 1). In Egypt, where no summer rainfall, maize cultivation depends on irrigation. The soil characteristics were investigated and presented in Table A2 for both seasons. Randomized Complete Block Design was performed in three replications at each season. The plots contained 5 m long three rows with 0.65 m wide. Potassium, phosphorus, and nitrogen fertilization were added at 116 kg/K2O, 76 kg P2O5/ha, and 290 N/ha.
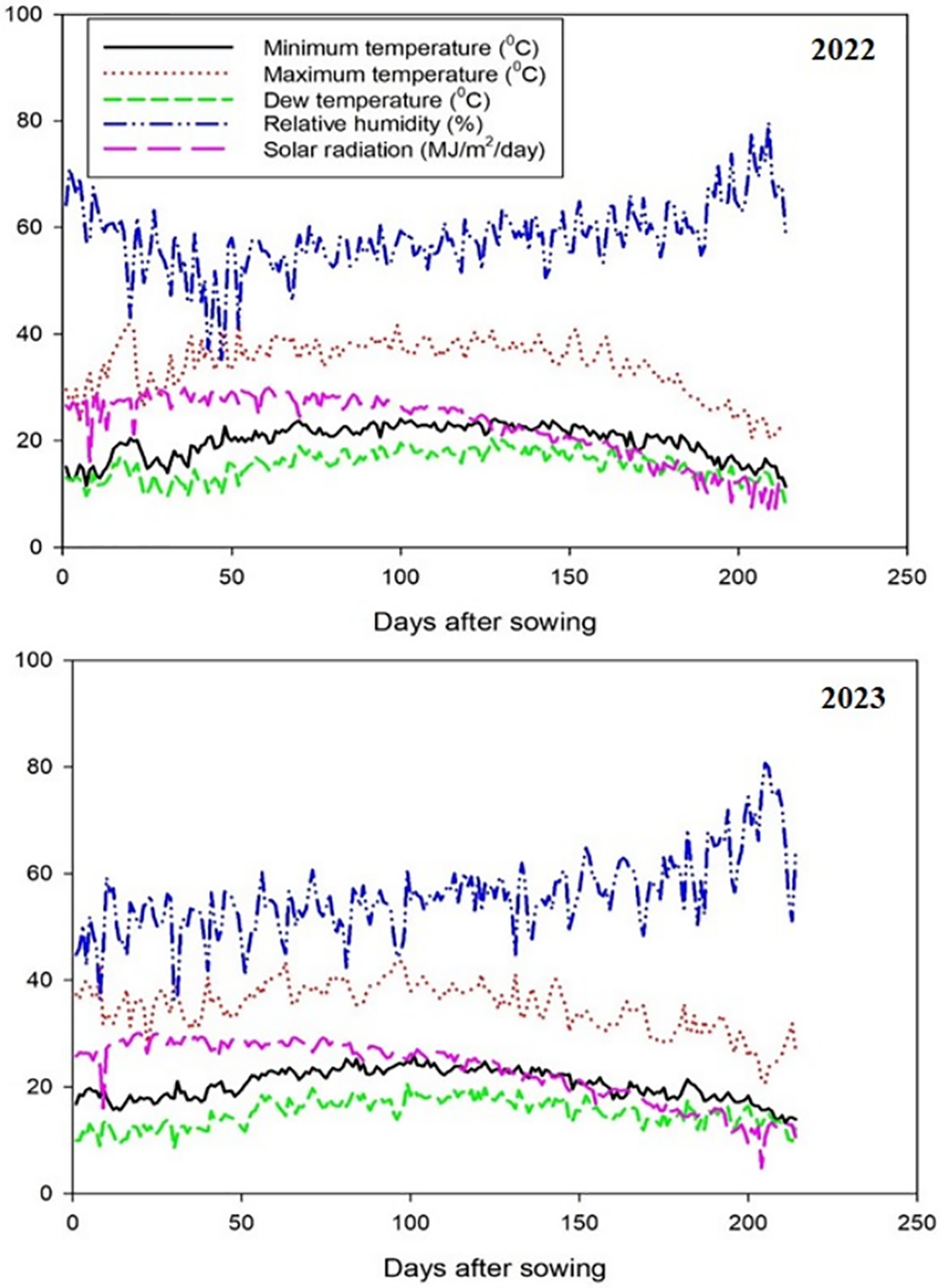
Figure 1: Meteorological characterization for the 2022 and 2023 growing seasons
The height of plants (cm) was recorded from soil level to top of first branch of tassel. Days to silking were documented when 50% of plants started in silk appearing. Ten ears were randomly collected at harvest from each experimental unit to estimate no. of rows/ear, ear length, weight of thousand kernels and no. of kernels/row. The plots were manually harvested, and grain yield/plant was estimated by adjusting the weight of the shelled grain at 15.5% moisture content.
Variance analysis for obtained data was conducted using R software 4.1.1. The least significant difference (LSD) test (p < 0.05) was applied to explore significance of differences among averages of assessed hybrids. Combining abilities were estimated according to Griffing’s method 4 model 1 [36]. Standard heterosis was determined by comparing the mean of F1 hybrids over the standard check (SC) hybrid using the following formulae: Heterosis over the standard check = [(F1 − SC)/SC] × 100. Hierarchical cluster analysis was conducted to categorize assessed hybrids based on yield traits. In addition, principal components and heatmap analyses were performed to study interrelationships among evaluated characters.
DNA was isolated from 200 mg of assessed lines leaves employing the CTAB protocol [37]. Nano Drop spectrophotometer was used to assess DNA consistency and quantity. Fourteen SSR primer pairs were used, with sequences detailed in Table A3, obtained from Maize-GDB database. PCR was conducted in 10 μL reaction volume comprising 1 μL of genomic DNA template (20 ng/μL), 1 unit of Taq DNA polymerase, 2 mM MgCl2, 0.2 mM dNTPs, and 0.5 μM each of forward and reverse primers. The PCR protocol included an initial denaturation at 94°C for 2 min, followed by cycles of 94°C for 30 s, 55°C for 30 s for annealing, and 72°C for 30 s for extension, ending with a final extension at 72°C for 3 min. Amplified products were subjected to analysis on 1.5% agarose gel. Bands were recorded per SSR marker, creating a binary data matrix where band presence was marked as 1 and absence as 0. According to Jaccard [38], genetic distances of the parental lines were determined using PAST software.
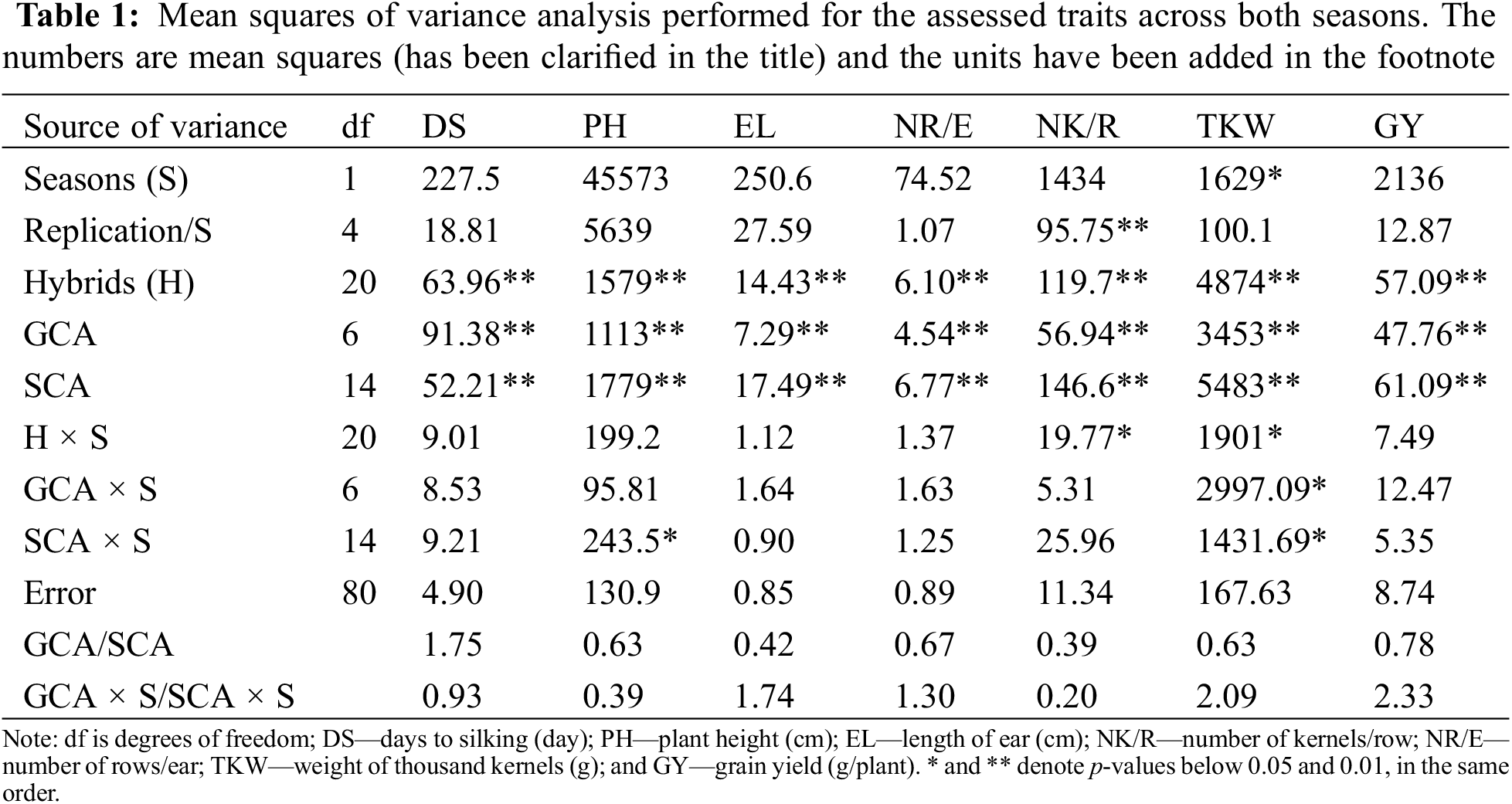
Analysis of variance exhibited an insignificant effect due to growing season (S) for most traits. In addition, the interaction of season with tested hybrids (H × S), with GCA, and SCA were not significant for nearly all the recorded characters. Otherwise, the assessed hybrids displayed highly significant variation for all evaluated characters (Table 1). Moreover, the hybrid effect, specifically the separation into SCA and GCA components, revealed that SCA and GCA showed significant or highly significant impacts on all traits. The ratio of GCA/SCA was lower than one for all characters except days to silking. Besides, the magnitude of GCA × S interaction was less than SCA × S for all evaluated characters except 1000 kernel weight, ear length, number of rows/ear, and grain yield (Table 1).
3.2 Performance and Classification of Hybrids
The hybrids differed significantly for all the traits (Figs. 2 and 3). Days to silking fluctuated from 58.7 to 69.2 days with an average of 63.8 days. The earliest hybrid was P1 × P7, while the latest hybrid was P4 × P6. Plant height averaged 231.2 cm and varied from 195.8 to 261.7 cm (Fig. 2A). The hybrid P1 × P5 obtained the uppermost values of plant height, whereas hybrid P1 × P7 displayed the lowest values (Fig. 2B). Ear length fluctuated from 16.6 to 22.3, averaging 19.5 cm. The hybrid P4 × P6 produced the most extended ear, but the hybrid P1 × P7 had the shortest ear (Fig. 2C). Similarly, number of rows/ear fluctuated from 12.7 to 16.3, averaging 14.6. The cross P3 × P5 resulted in the highest number of rows, whereas P3 × P6 registered the minimum number (Fig. 3A). The average number of kernels/row was 36.9, varying from 29.0 to 44.0. P4 × P7 exhibited the uppermost values; meanwhile, the lowest values were observed in P1 × P7 (Fig. 3B). In the same context, the mean of 1000-kernel weight diverged from 251.7 to 361.0 g, averaging 322.7. The hybrid P4 × P7 recorded the heaviest TKW, whereas P4 × P6 had the lightest kernel weight (Fig. 3C). Grain yield per plant ranged from 117.7 to 129.5, averaging 124.7 g. The hybrids P4 × P7, P1 × P5, and P3 × P5 produced the uppermost grain yield, but the hybrid P1 × P7 displayed the lowermost grain yield (Fig. 3D).
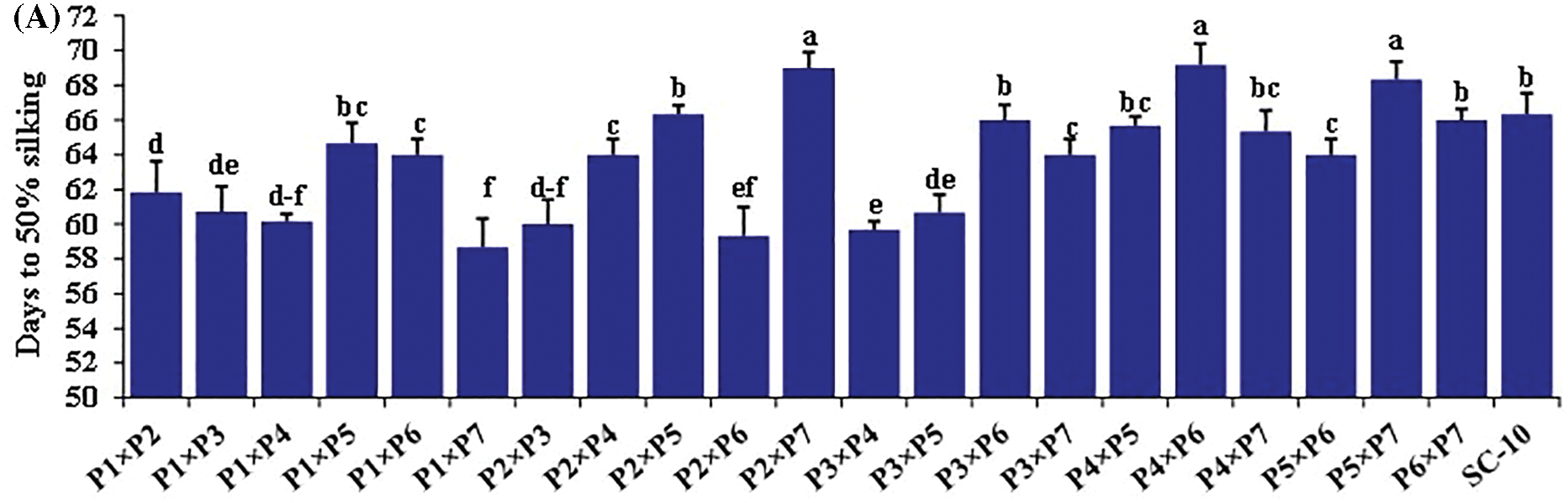
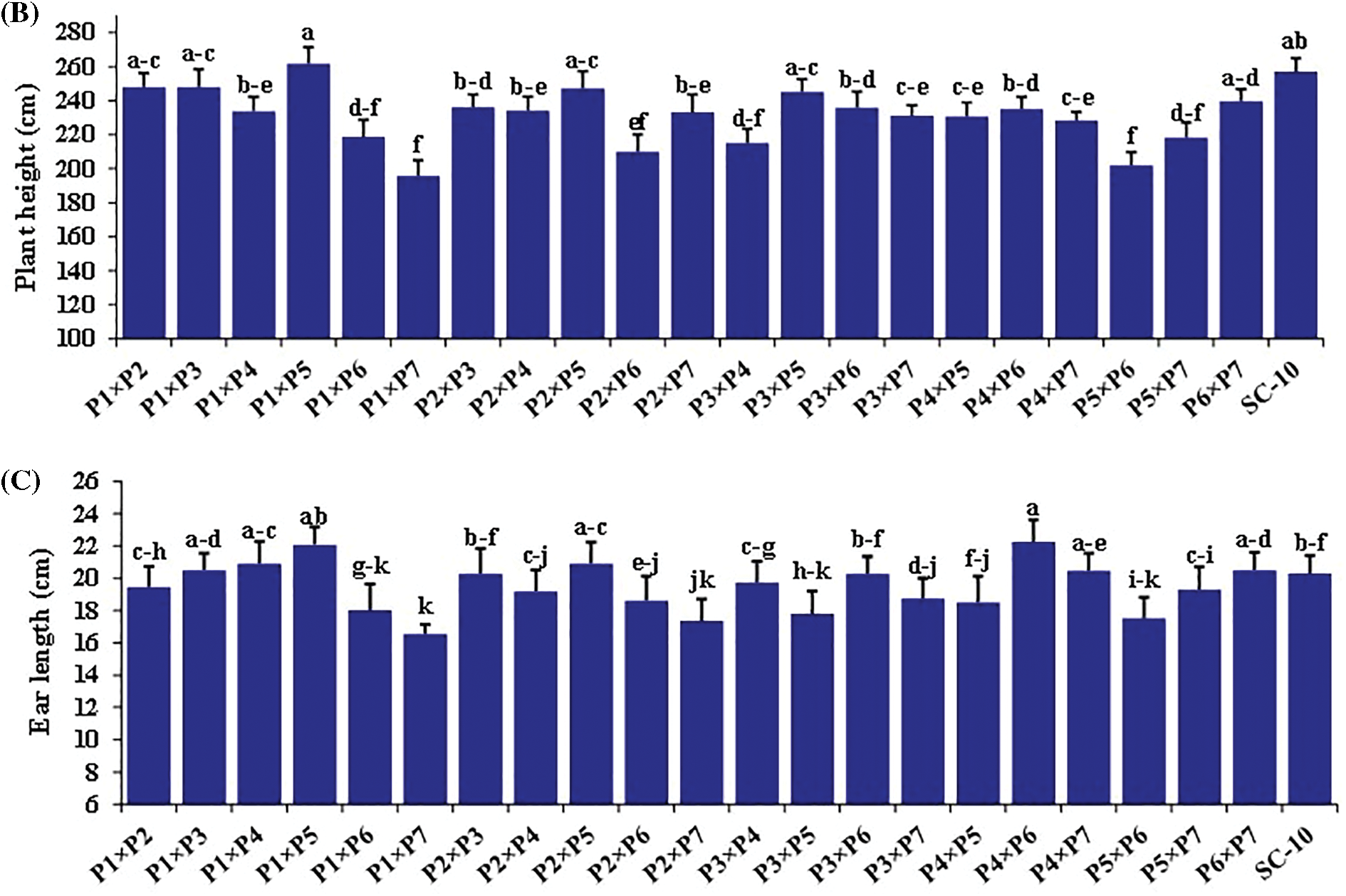
Figure 2: Performance evaluation of twenty one F1 hybrids and check hybrid for (A) days to silking, (B) plant height and (C) ear length. Bars above columns indicate SE. Columns with different letters represent significant differences as determined by LSD with a p-value of less than 0.05
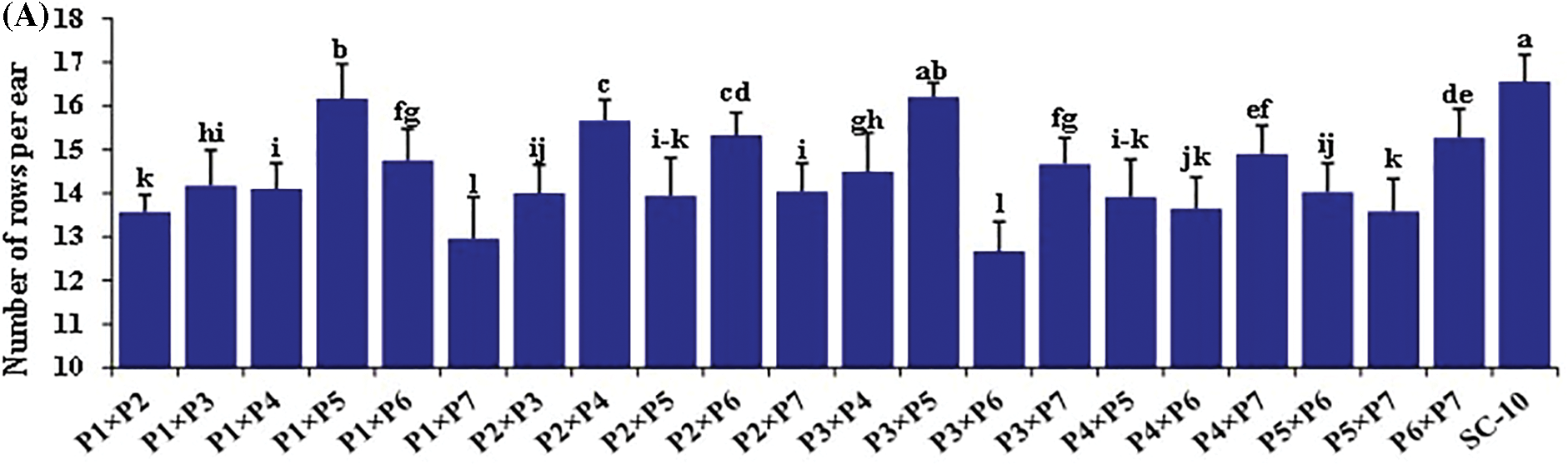
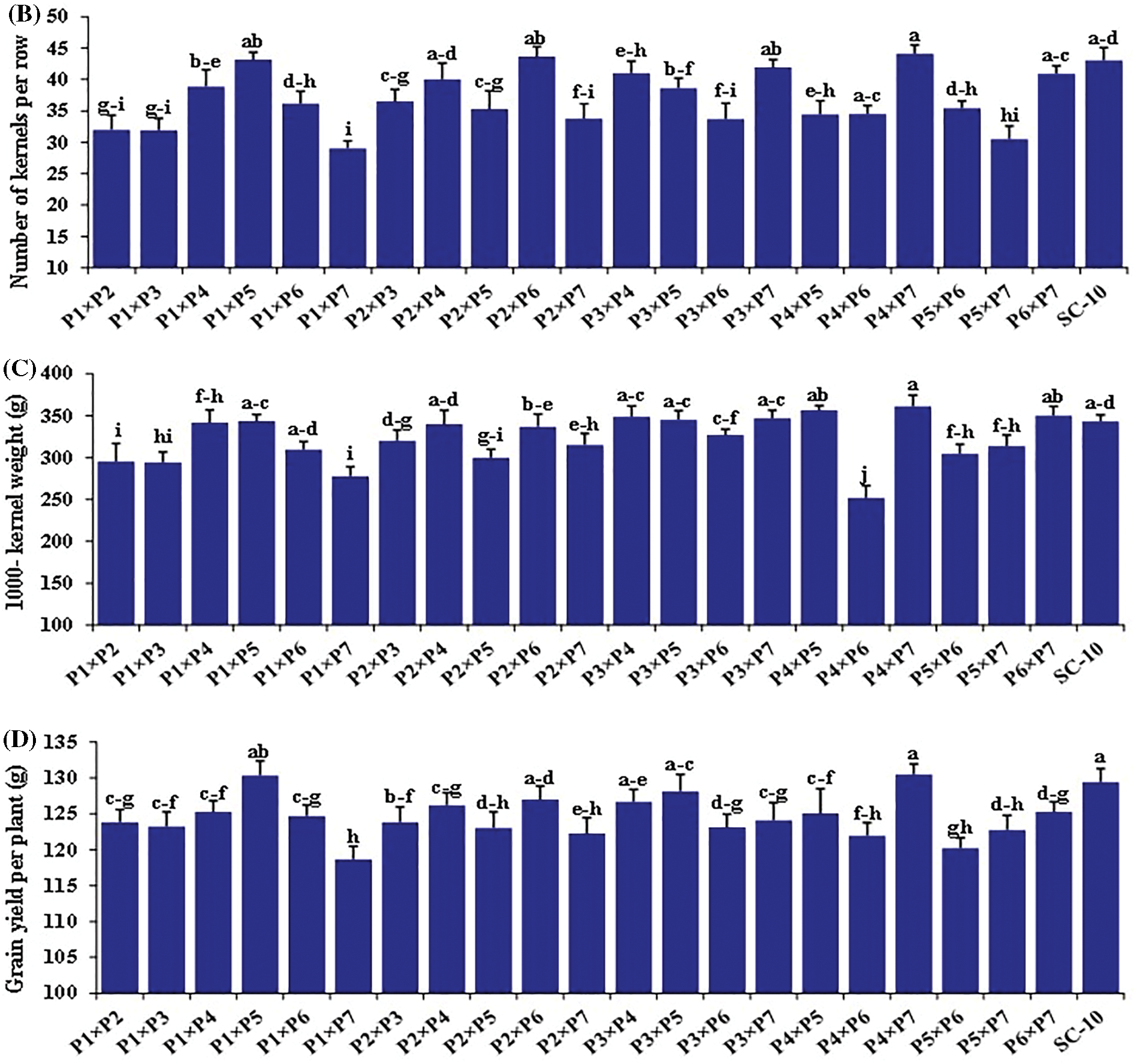
Figure 3: Performance evaluation of twenty one F1 hybrids and the check hybrid for (A) Number of rows/ear, (B) Number of kernels/row, (C) 1000 kernel weight, and (D) grain yield per plant. Bars above columns indicate SE. Columns with different letters imply significant differences as determined by LSD with a p-value of less than 0.05
The F1 hybrids, along with the check SC-10, were classified based on yield traits. These hybrids were grouped into five distinct clusters as illustrated in Fig. 4. Group-A contained three hybrids (P4 × P7, P1 × P5, and SC-10), which exhibited superior yield and contributing traits. Group-B comprised 4 hybrids (P3 × P5, P3 × P4, P2 × P6, and P2 × P4) depicted by high-yielding traits. Groups-C and D comprised seven and six hybrids, respectively, showing intermediate-high and intermediate-low yield traits. Finally, Group-E comprised the two hybrids with the lowermost agronomic performance (P5 × P6 and P1 × P7).
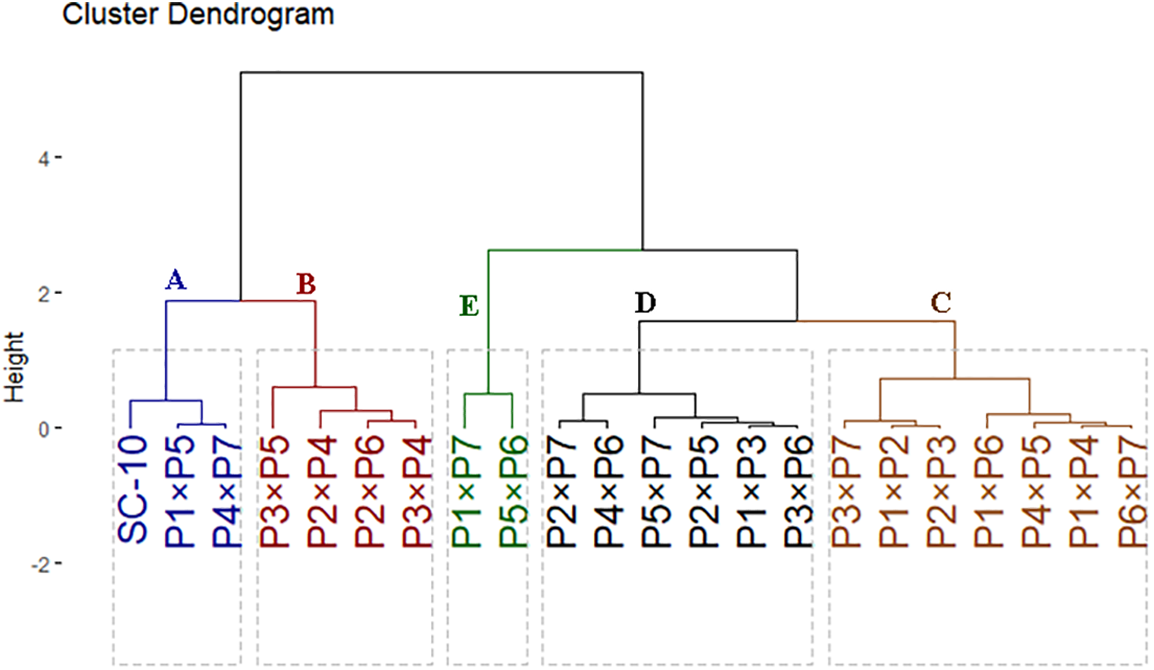
Figure 4: Dendrogram illustrating the phenotypic distances among the 21 F1 hybrids and check SC-10, based on the studied yield characters
Standard heterosis over the commercial check SC10 has been given in Fig. 5. Positive heterotic effects are required for evaluated characters excepting days to silking and plant height for which the negative heterotic effects are favorable. Earliness is a crucial factor that considerably affects maize productivity in arid environments. The cross combinations P1 × P4, P1 × P3, P1 × P7, P1 × P2, P2 × P6, P2 × P3, P3 × P4, and P3 × P5 recorded negative heterosis for days to silking. The negative and considerable heterosis for days to silking is required due to its association with earliness. Fifteen hybrids showed negative heterotic effects for plant height towards shortness. The most pronounced negative heterotic effects were detected in P1 × P6, P3 × P5, P1 × P7, P2 × P6, P2 × P7, P5 × P6, P3 × P7, and P4 × P5. In contrast, the uppermost positive substantial heterosis for ear length was illustrated by P1 × P5 and P4 × P6. The best heterotic combinations for number of rows/ear were P2 × P4, P1 × P5, and P3 × P5. Likewise, two hybrids P2 × P6 and P4 × P7 manifested positive heterosis for no. of kernels/row. The crosses with the best heterotic effects for 1000-kernel weight were P4 × P7 and P4 × P5. The cross combinations P1 × P5 and P4 × P7 disclosed the superior positive heterosis for grain yield. Owing to their benefits for one or more associated features, these heterotic combinations appear superior in grain yield.
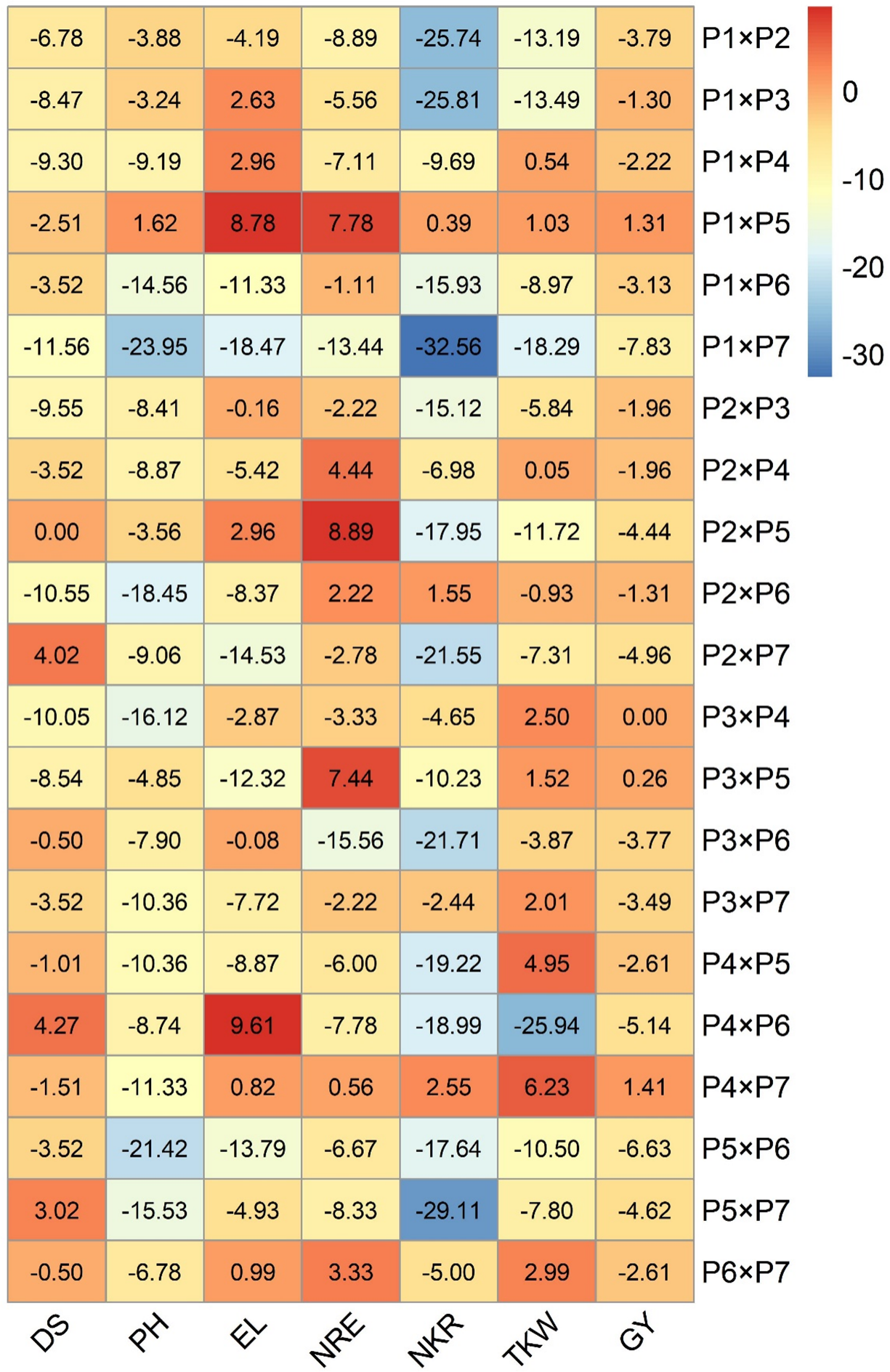
Figure 5: Heatmap depicting heterosis over the standard check (SH %)
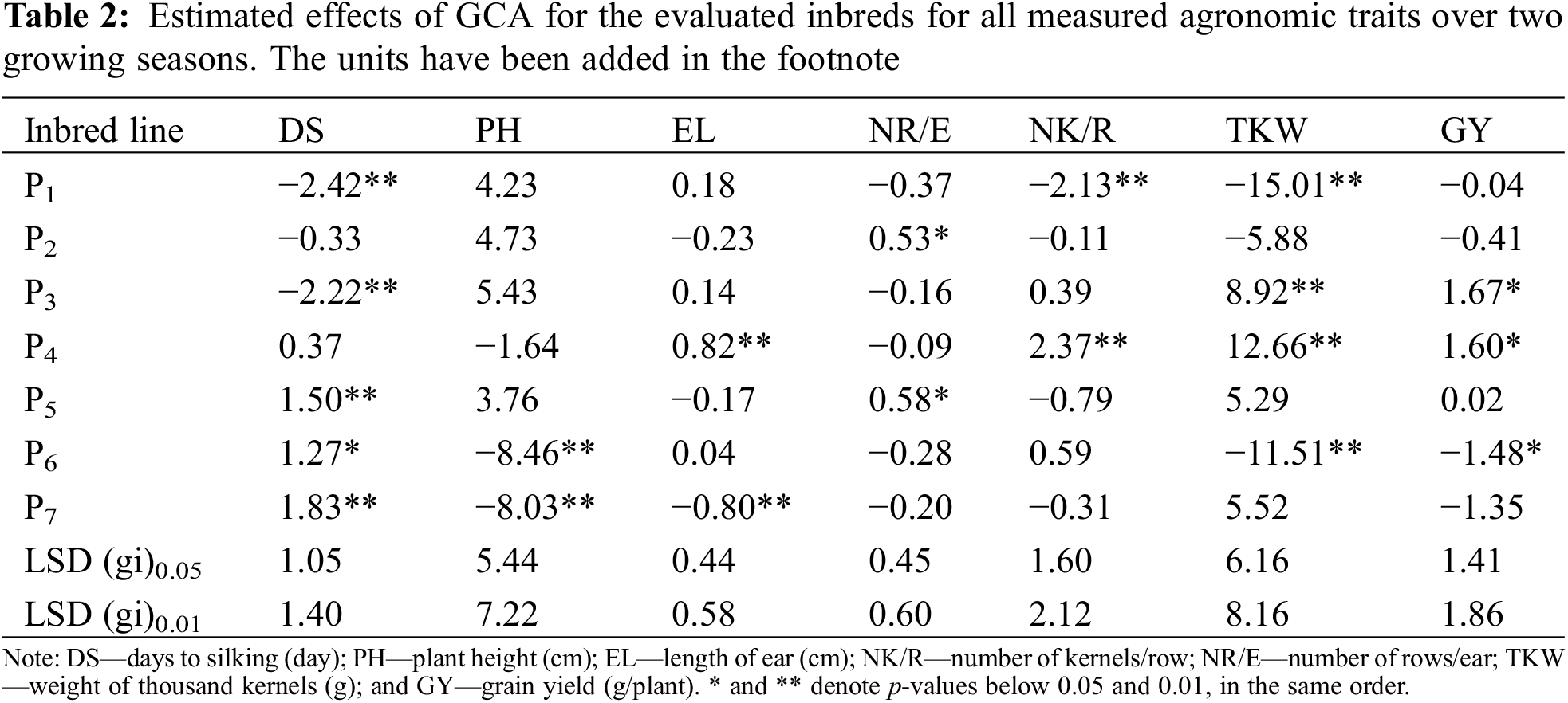
3.4 General Combining Ability (GCA)
All recorded agronomic characters except days to silking and plant height were desired to have highly substantial positive GCA effects. Table 2 explained the GCA effects of assessed inbreds. The lines P1 and P3 exhibited the superior negative GCA estimates for days to silking, while P6 and P7 displayed substantially negative GCA estimates for plant height. Otherwise, specific lines reflected positive GCA effects for diverse traits. Specifically, P4 mirrored a notably superior positive GCA effect for ear length, number of kernels/row. In addition, inbred P2 and P5 had positive GCA effect for no. of rows per ear and both P3 and P4 manifested significant GCA estimates for grain yield and 1000-kernel weight.
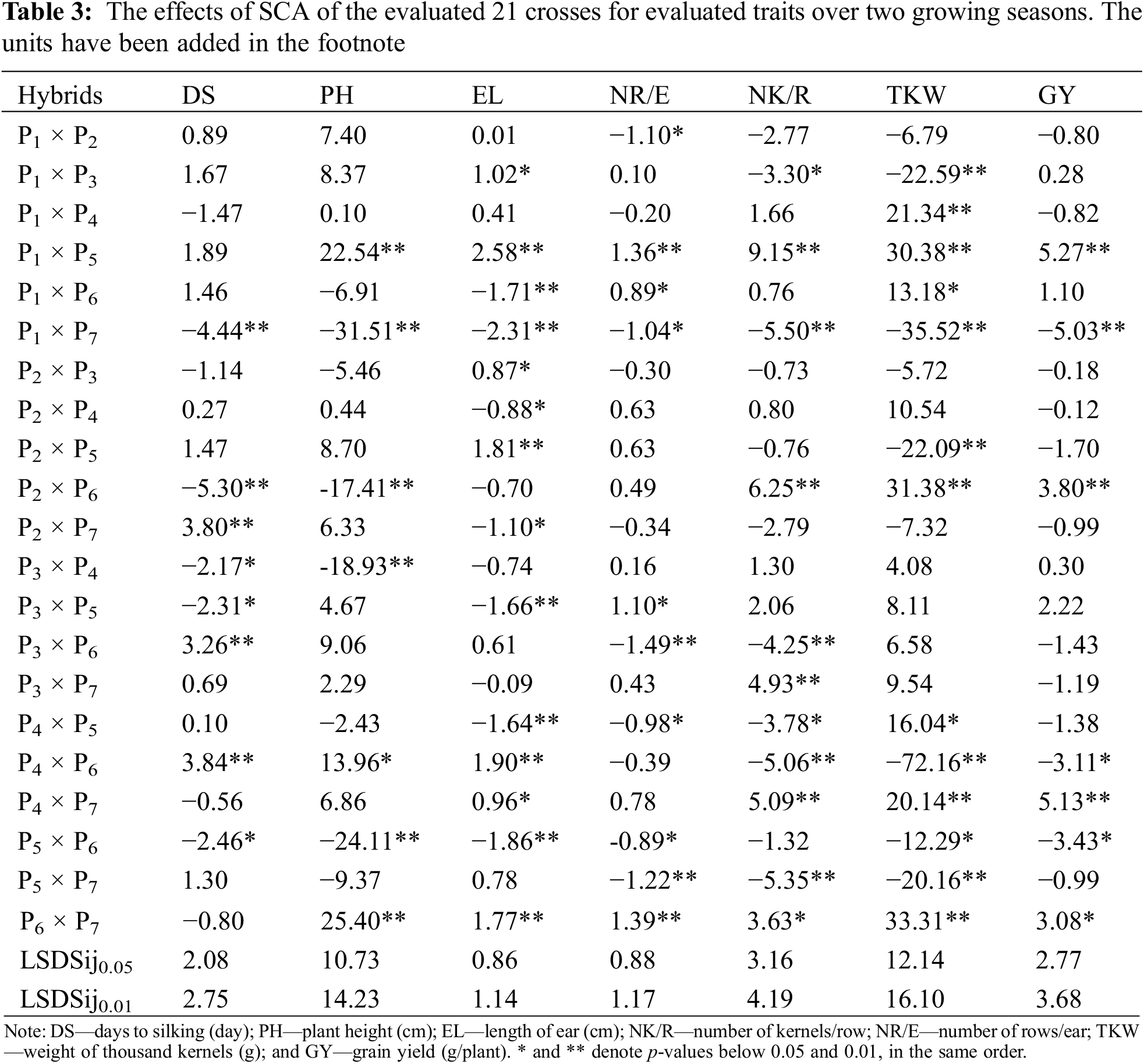
3.5 Specific Combining Ability
The effects of SCA showed that the hybrids P3 × P4, P5 × P6, P3 × P5, P2 × P6, and P1 × P7 obtained desirable substantial negative effects for days to silking (Table 3). Likewise, favorable substantial negative SCA estimates for plant height were assigned for P3 × P4, P1 × P7, P2 × P6, and P5 × P6. Otherwise, the highest positive SCA effects for ear length were displayed by P4 × P6, P1 × P3, P2 × P5, P1 × P5, P4 × P7 and P6 × P7. Moreover, hybrids P3 × P5, P1 × P5 and P6 × P7 recorded substantially positive SCA estimates for no. of rows/ear. Meanwhile, hybrids P2 × P6, P3 × P7, P1 × P5, and P4 × P7 displayed most substantial and positive SCA estimates for no. of kernels/row. The advantageous SCA effect for 1000-kernel weight was noticed in P1 × P5, P2 × P6, P1 × P4, P1 × P6, P4 × P7, P4 × P5, and P6 × P7. The hybrids P1 × P5, P2 × P6, P4 × P7, and P6 × P7 displayed the topmost substantial positive SCA for grain yield. None of evaluated hybrids recorded desirable SCA estimates for all characters. However, certain hybrids demonstrated desirable effects for grain yield and beneficial SCA effects for its related traits. Notably, hybrids P1 × P5 and P4 × P7 exhibited favorable SCA effects for no. of grain/row, ear length, no. of rows/ear, grain yield and 1000 kernel weight.
3.6 Interrelation among Hybrids and Characters
The association between developed hybrids and evaluated characters was analyzed employing principal component analysis. The first two PCAs displayed a considerable percentage of the total variance, with PC-1 explaining 52.27% and PC-2 accounting for 20.35%. Therefore, these PCs were utilized to create the biplot as shown in Fig. 6. Notably, PC-1 exhibited greater variation and was closely associated with the evaluated hybrids. This component effectively separated the hybrids into those on the negative and positive sides of PC-1. The evaluated traits were aligned with hybrids on positive side of PC-1, indicating that these hybrids, particularly P4 × P7, P1 × P5, and SC-10, demonstrated superior agronomic performance. Conversely, the hybrids on negative side of PC-1, such as P5 × P6 and P1 × P7, exhibited lower agronomic performance. Vectors proximity suggested a positive robust association among studied characters. The grain yield was associated positively with no. of kernels/row, 1000 grain weight, no. of rows/ear, and ear length. Furthermore, heatmap based on the evaluated agronomic traits categorized hybrids into distinctive groups (Fig. 7). Hybrids P1 × P5, P4 × P7, and SC-10 exhibited highest values across all characters (indicated in blue), whereas hybrids P5 × P6 and P1 × P7 had lowest performance (marked in red).
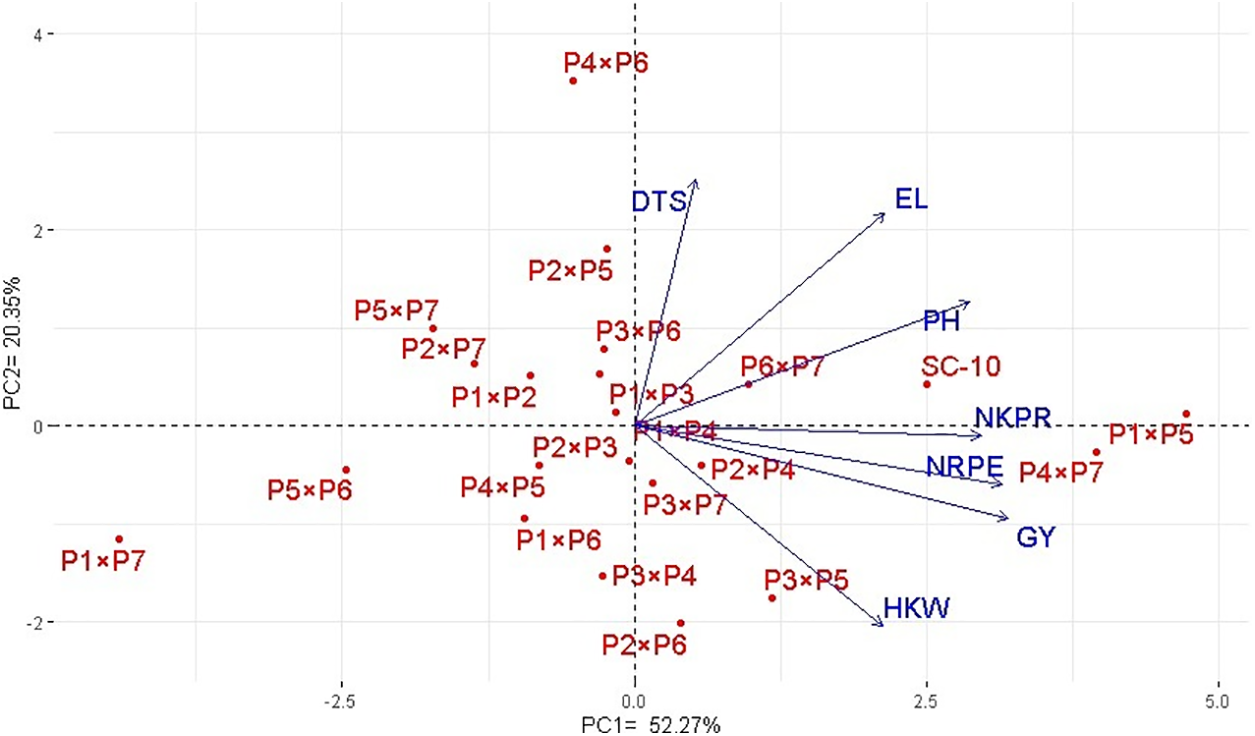
Figure 6: PC biplot for studied characters and the assessed maize hybrids
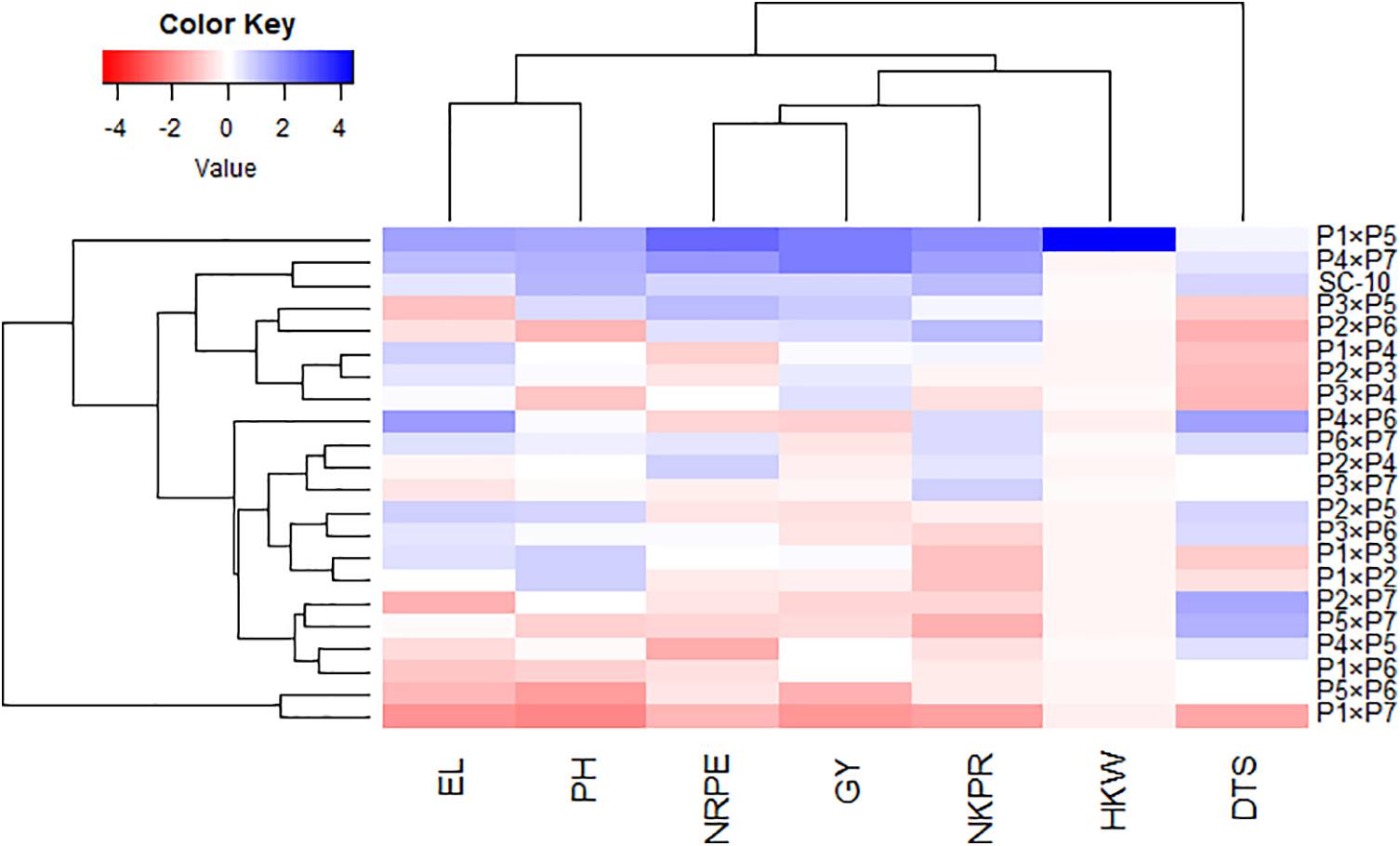
Figure 7: Heatmap categorized the assessed maize hybrids into distinct clusters based on the studied agronomic traits. In this representation, red denotes high values, while blue implies low values for the corresponding attributes
3.7 Genetic Diversity among Parental Lines
Genetic diversity was studied employing fourteen SSR markers to evaluate genetic variability among seven parental lines. Ten markers were found to be polymorphic and were used to further investigate the genetic diversity (Fig. A1). Number of alleles/locus varied, ranging from 2 (observed in markers phi-9610 and phi-453121) to 5 (observed in marker umc-1033), averaging 3.5 alleles per locus (as shown in Table 4). The primary allele frequency averaged 0.45, ranging from 0.29 to 0.73. Gene variability varied between 0.24 (phi-96100) and 0.78 (umc-1033), with overall average of 0.62. Additionally, polymorphic information content averaged 0.56, with values ranging from 0.21 (phi-96100) to 0.74 (umc-1033).
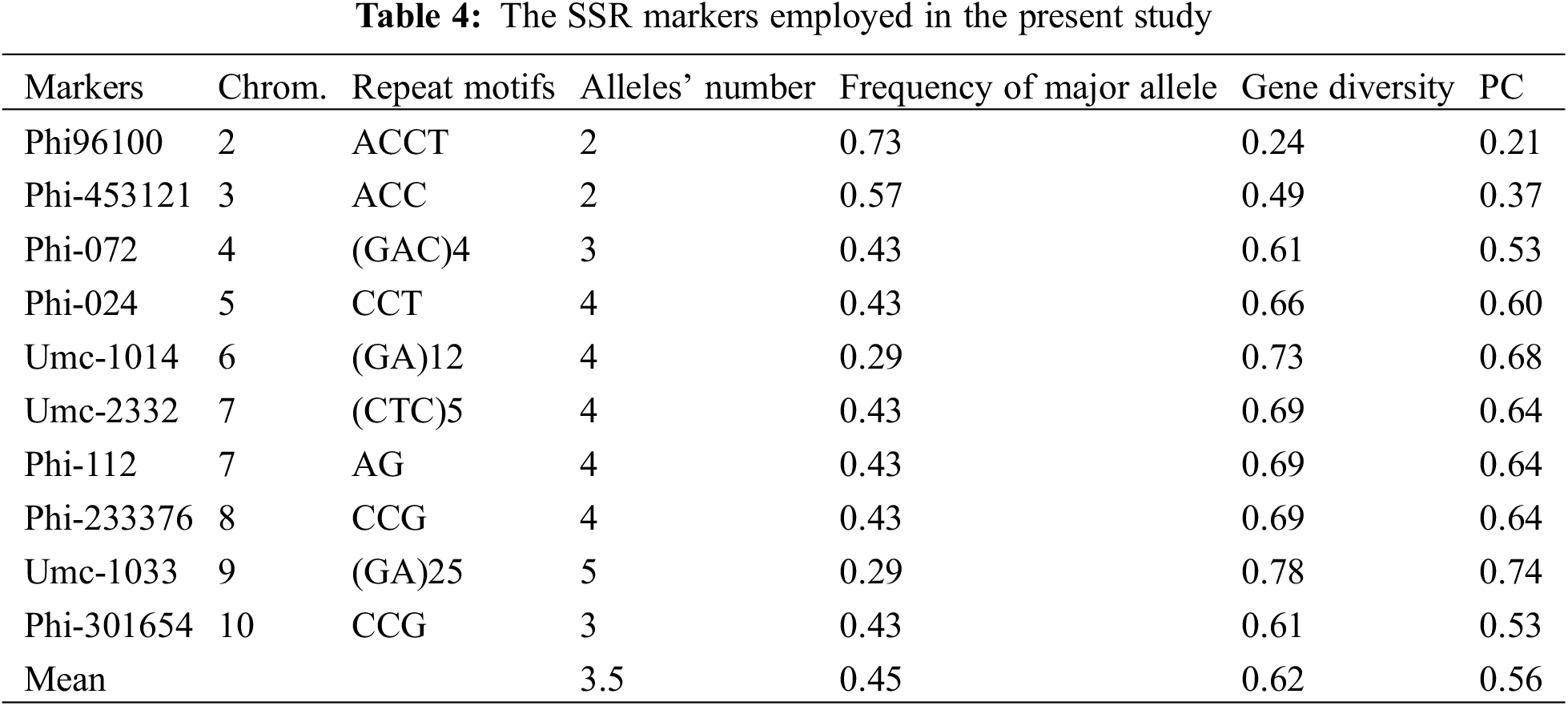
Genetic distance measured utilized SSR markers differed between 0.63 and 0.90, averaging 0.79 (Table 5). Lowest genetic distance was found between two lines P1 and P2 (0.63), while greatest distance was observed between P6 and P4 and also between P4 and P6 (0.90). The neighbor-joining tree constructed using a genetic distance matrix, divided the parental lines to three main groups with internal subgroups that displayed variable levels of diversity (Fig. 8). Group-A contained P6 and P7, Group-B included P3 and P5, and Group-C comprised P1, P2, and P4.
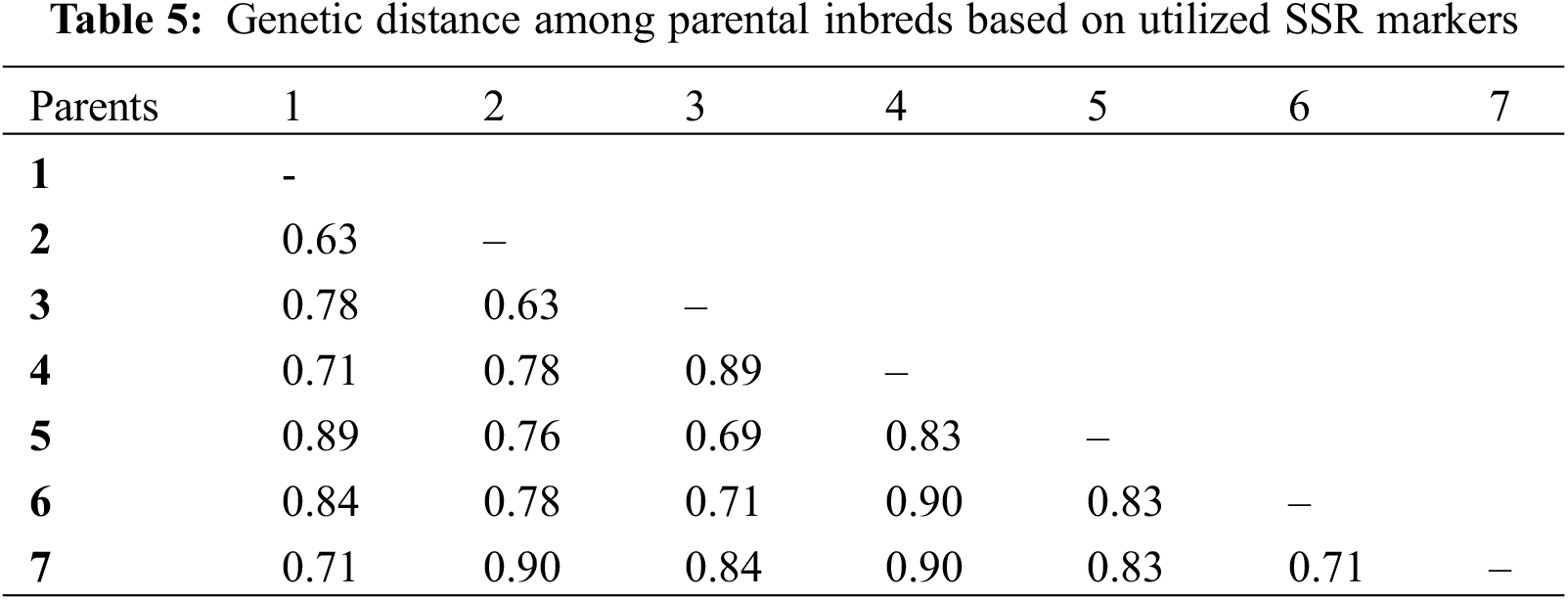
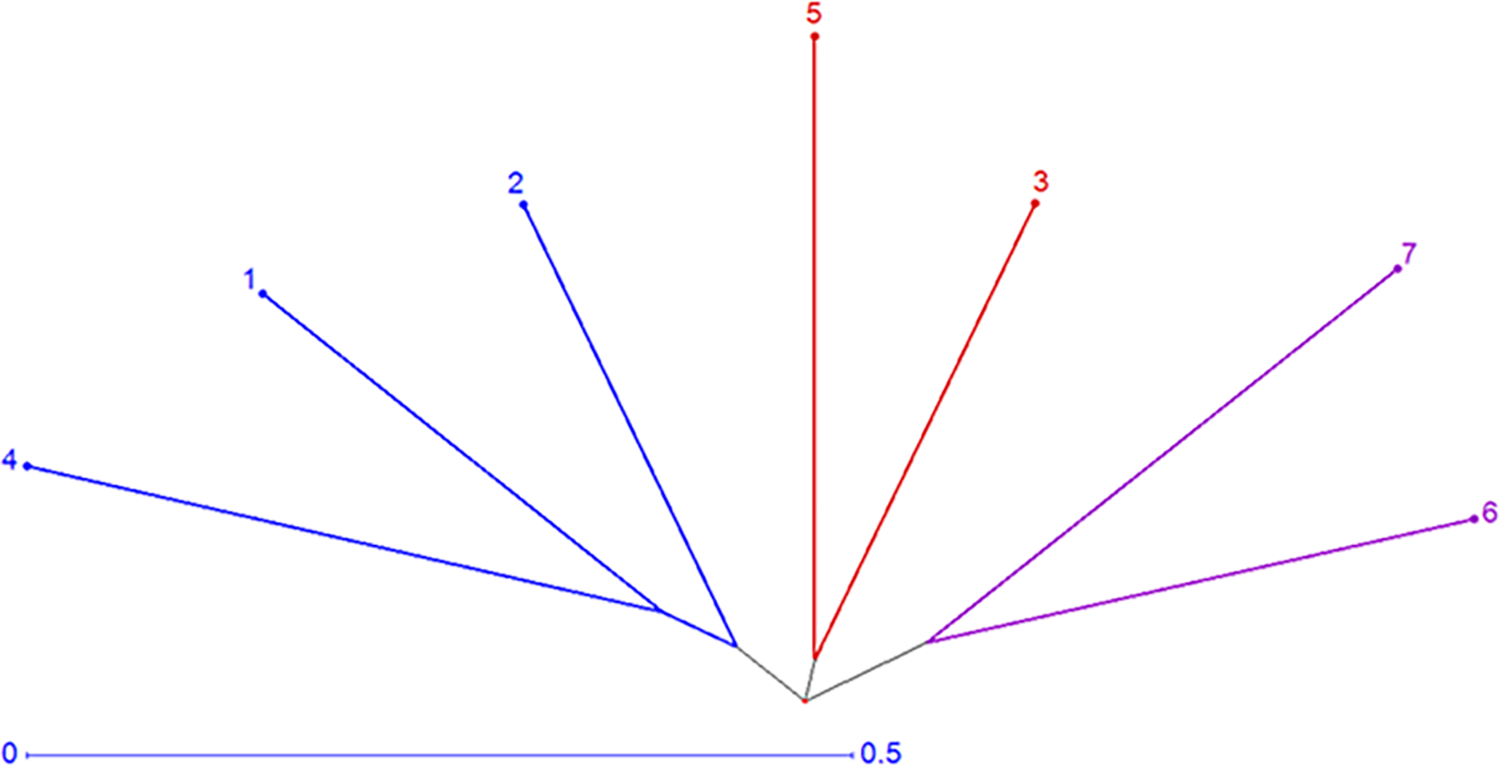
Figure 8: Neighbor-joining tree of the estimated lines based on applied SSR markers
3.8 Relationship between Genetic Diversity of Inbreds, SCA, and Hybrid Performance
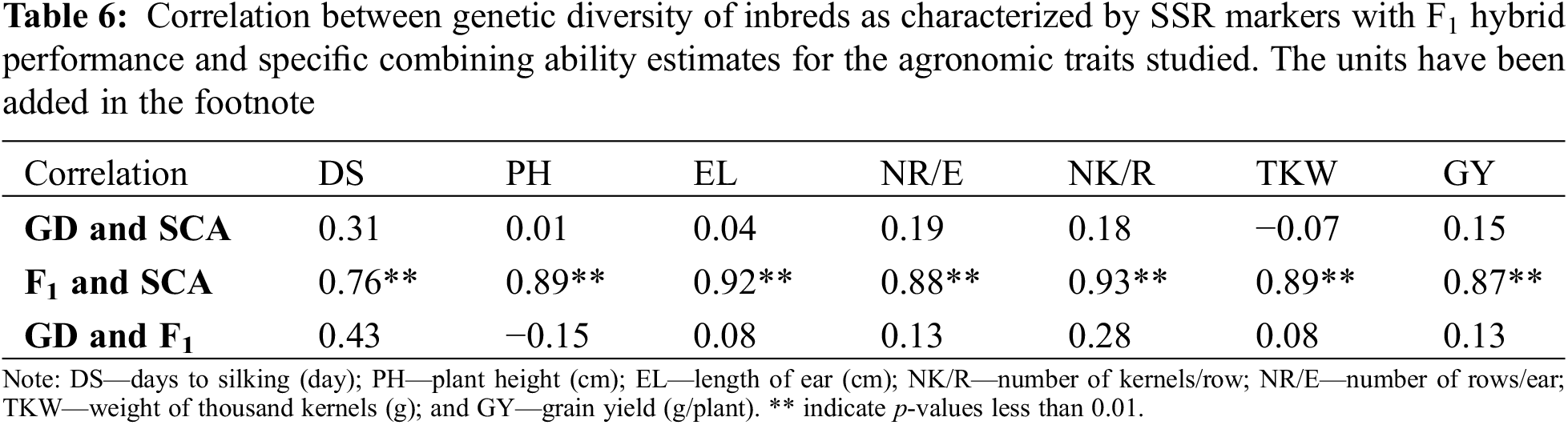
The relationship between genetic diversity based on SSR markers and SCA effects was insignificant for studied agronomic traits (Table 6). Similarly, genetic diversity of parental lines marked a nonsignificant association with F1 hybrid performance. Conversely, F1 hybrid performance strongly correlated with SCA effects for all evaluated traits.
Maize production faces considerable challenges in arid environments attributed to diverse environmental stresses. The main problems encountered in arid environments include high temperatures, high evaporation rates, and poor soil fertility. Additionally, the unpredictable climatic conditions in arid regions pose further challenges to sustainable agriculture [39–41]. These harsh conditions often result in reduced crop yields and limit the cultivation of traditional maize hybrids. The aim of current study wat to explore combining ability and genetic diversity of newly developed maize lines specifically adapted for arid environments. Using developed high-yielding maize hybrids adapted to arid climates could enhance maize production and enhance food security in arid environments.
The development of high-yielding maize hybrids largely depends on selecting suitable parental lines. Consequently, recognizing diverse parental inbred with advantageous alleles is essential to confirm their effective transfer to the offspring [35]. The favorable GCA estimates detected in inbred lines P1 and P3 for days to silking suggest their potential for enhancing earliness. Parental lines P6 and P7 were considered valuable combiners for reducing plant height as a critical factor in improving lodging tolerance. Additionally, P3 and P4 exhibited desirable GCA estimates for grain yield, 1000-kernel weight, ear length, no. of rows/ear, and no. kernels/row. This indicates that these inbreds possess valuable alleles that can be passed on to their offspring to produce high-yielding maize hybrids [42,43]. The derived hybrids displayed significant variations in days to silking, plant height, ear length, weight of thousand kernel, no. of rows/ear, no. of kernels/row, and grain yield. The detected significant genetic variation supports the selection of desirable hybrids. The crosses P1 × P5 and P4 × P7 reflected outstanding agronomic performance with grain yield greater than the check hybrid. This finding implies that these hybrids have the potential to compete with check hybrids and may serve as favorable hybrids for valuable utilization after additional evaluation. Additionally, they can be utilized in generating new and superior lines [44].
Similarly, previously published reports confirmed considerable genetic variability for different characters in developed maize hybrids such as Babic et al. [45], Oluwaseun et al. [46], Amegbor et al. [47], Ajala et al. [48]. The identification of desirable hybrids is accomplished through the use of SCA estimates. In this study, a significant number of evaluated hybrids demonstrated the favorable effects of SCA for at least one trait. The cross combinations P2 × P6, P1 × P5, and P4 × P7 were determined as the greatest particular combiner for breeding high yielding maize hybrids compared to the others. The great SCA effect appeared as a result of nonadditive gene effects. Two hybrids of the assessed crosses, P1 × P5 and P4 × P7, marked favorable SCA coupled with superior grain yield. Consequently, these maize hybrids could be exploited in breeding to enhance maize productivity under arid conditions.
Understanding inheritance pattern of agronomic characters is fundamental for breeding programs [49,50]. According to considerable GCA and SCA effects, additive and nonadditive gene actions were shown to be proportionately relevant in inheritance of assessed agronomic character in the current work. However, ratio of GCA to SCA was less than one for nearly all measured characters except for days to silking. This suggests that non additive gene action substantially regulates inheritance of evaluated traits [51–53]. Therefore, crossing method is greatly efficient in elevating these traits and employing heterosis effects. These findings are in consonance with Makumbi et al. [54], Derera et al. [55], which also depicted significance of nonadditive gene action in inheritance of agronomic characters in maize. Otherwise, Annor et al. [56] and Oyetunde et al. [57] elucidated that additive gene action played decisive role in inheritance of yield characters in maize. The nonsignificant GCA × S interaction and also SCA × S interaction indicated a lack of environmental influence on determining gene expression of the studied traits [35].
Molecular markers are an effective tool for exploring genetic variability among maize lines [2,58,59]. These markers can assist maize breeders in recognizing parental lines with desirable characters for generating high yielding hybrids. The results presented genetic variabilitylevels among parental lines employing SSR markers. The genetic diversity analysis utilizing SSR markers indicated an average of 3.5 alleles/locus, fluctuating from 2 to 5 alleles. This coincides with previous results of Wegary et al. [60], and Kamara et al. [35]. Variations in allele numbers across studies may be influenced by factors such as the specific SSR markers employed and the population size. The PIC values of SSR markers were employed to evaluate their ability to discriminate between parental genotypes. The markers umc-1014 and umc-1033 exhibited high PIC values of 0.68 and 0.74, respectively, indicating their effectiveness in differentiating parental inbreds based on their genetic associations. The SSR markers used in the analysis divided the parental lines into three different clusters. Crossing distinct lines from distinct groups can produce improved high yielding hybrids. The information obtained from cluster analysis might assist in decreasing crosses needed to be assessed in field trials. The lack of association between genetic diversity through SSR markers with SCA effects and hybrid performance for evaluated traits might be ascribed to small number of markers employed. This is proven by the results reported by Dhliwayo et al. [61].
Contrarily, Phumichai et al. [34] and Reif et al. [62] elucidated a substantial relationship between genetic diversity and performance of maize hybrid. In the study of Phumichai et al. [34] seven parental maize genotypes were evaluated employing a set of sixty-four SSR loci. Also, Reif et al. [62] assessed 336 maize genotypes (48 individuals from seven populations) using eighty-five SSR markers. The increasing number of markers used in these studies enhanced the resolution of genetic diversity assessments. The higher number of markers amplified more loci and provided a broader view of genetic markers across the genomes of the parental lines and more genetic differences. The present findings indicated that the SCA effects are substantially associated with performance of maize hybrids. These results concur with the previous results of Mageto et al. [21], Kamara et al. [35].
The current investigation exhibited substantial genetic variability among the assessed maize hybrids for all evaluated agronomic characters. Nonadditive effects were more prevalent for yield characters. The lines P1 and P3 were determined as effective combiners for developing early maturing genotypes. Besides, P3 and P4 were determined as superior combiners to improve grain yield and most of its contributing attributes. The hybrids P1 × P5 and P4 × P7 possessed favorable SCA along with desirable superior grain yield. These promising hybrids would be additionally assessed for potential commercial exploitation. In the current investigation, SSR-based genetic diversity did not significantly predict SCA effects or performance of hybrids. Notwithstanding, SCA could be substantially exploited to expect performance of developed hybrids.
Acknowledgement: The authors are grateful to the Faculty of Agriculture, Kafrelsheikh University, Egypt, for the support provided to conduct this work. The authors also would like to express their gratitude to Research Supporting Project Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement:: This word was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2024R318), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through Large Research Project under grant number RGP2/342/45. Also, this work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (KFU241870).
Author Contributions:: The authors confirm contribution to the paper as follows: study conception and design: Mohamed M. Kamara, Fatmah A. Safhi, Nora M. Al Aboud, Maha Aljabri, Samah A. Alharbi, Hesham S. Ghazzawy, Mohammed O. Alshaharni, Eman Fayad, Wessam F. Felemban, Diaa Abd El-Moneim, Abdallah A. Hassanin, Imen Ben Abdelmalek, Abdelraouf M. Ali, Elsayed Mansour; data collection: Mohamed M. Kamara, Fatmah A. Safhi, Nora M. Al Aboud, Maha Aljabri, Samah A. Alharbi, Hesham S. Ghazzawy, Mohammed O. Alshaharni, Eman Fayad, Wessam F. Felemban, Diaa Abd El-Moneim, Abdallah A. Hassanin, Imen Ben Abdelmalek, Abdelraouf M. Ali, Elsayed Mansour; analysis and interpretation of results: Nora M. Al Aboud, Maha Aljabri, Samah A. Alharbi, Hesham S. Ghazzawy, Mohammed O. Alshaharni, Eman Fayad, Wessam F. Felemban, Diaa Abd El-Moneim; draft manuscript preparation: Mohamed M. Kamara, Fatmah A. Safhi, Abdallah A. Hassanin, Imen Ben Abdelmalek, Abdelraouf M. Ali, Elsayed Mansour. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Wang H, Ren H, Zhang L, Zhao Y, Liu Y, He Q, et al. A sustainable approach to narrowing the summer maize yield gap experienced by smallholders in the North China Plain. Agric Syst. 2023;204:103541. doi:10.1016/j.agsy.2022.103541. [Google Scholar] [CrossRef]
2. Mathiang EA, Sa KJ, Park H, Kim YJ, Lee JK. Genetic diversity and population structure of normal maize germplasm collected in south sudan revealed by SSR markers. Plants. 2022;11:2787. doi:10.3390/plants11202787. [Google Scholar] [PubMed] [CrossRef]
3. Mukri G, Patil MS, Motagi BN, Bhat JS, Singh C, Jeevan Kumar S, et al. Genetic variability, combining ability and molecular diversity-based parental line selection for heterosis breeding in field corn (Zea mays L.). Mol Biol Rep. 2022;49:4517–24. doi:10.1007/s11033-022-07295-3. [Google Scholar] [PubMed] [CrossRef]
4. Erenstein O, Jaleta M, Sonder K, Mottaleb K, Prasanna BM. Global maize production, consumption and trade: trends and R&D implications. Food Secur. 2022;14:1295–319. doi:10.1007/s12571-022-01288-7. [Google Scholar] [CrossRef]
5. FAOSTAT. Food and agriculture organization of the united nations. statistical database. Available from: http://www.fao.org/faostat/en/#data. [Accessed 2024]. [Google Scholar]
6. Gracia MP, Mansour E, Casas AM, Lasa JM, Medina B, Molina-Cano JL, et al. Progress in the Spanish national barley breeding program. Span J Agric Res. 2012;10:741. [Google Scholar]
7. Alemu A, Åstrand J, Montesinos-Lopez OA, Sanchez JI, Fernandez-Gonzalez J, Tadesse W, et al. Genomic selection in plant breeding: key factors shaping two decades of progress. Mol Plant. 2024;17:552–78. doi:10.1016/j.molp.2024.03.007. [Google Scholar] [PubMed] [CrossRef]
8. Sun L, Lai M, Ghouri F, Nawaz MA, Ali F, Baloch FS, et al. Modern plant breeding techniques in crop improvement and genetic diversity: from molecular markers and gene editing to artificial intelligence–critical review. Plants. 2024;13:2676. doi:10.3390/plants13192676. [Google Scholar] [PubMed] [CrossRef]
9. Ponce-Molina LJ, María Casas A, Pilar Gracia M, Silvar C, Mansour E, Thomas WB, et al. Quantitative trait loci and candidate loci for heading date in a large population of a wide barley cross. Crop Sci. 2012;52:2469–80. doi:10.2135/cropsci2012.01.0029. [Google Scholar] [CrossRef]
10. Riyanto A, Hidayat P, Suprayogi Y, Haryanto TAD. Diallel analysis of length and shape of rice using hayman and griffing method. Open Agric. 2023;8:20220169. doi:10.1515/opag-2022-0169. [Google Scholar] [CrossRef]
11. Abdel-Moneam M, Sultan M, Khalil A, El-Awady HE. Estimation of heterosis, combining ability, and gene action using diallel analysis for some inbred lines of yellow maize. J Plant Pro. 2024;15:111–7. [Google Scholar]
12. Gaballah MM, Attia KA, Ghoneim AM, Khan N, El-Ezz AF, Yang B, et al. Assessment of genetic parameters and gene action associated with heterosis for enhancing yield characters in novel hybrid rice parental lines. Plants. 2022;11:266. doi:10.3390/plants11030266. [Google Scholar] [PubMed] [CrossRef]
13. Verma V, Biswal M, Yadav NK, Gathiy GS. Exploration of diallel method for assessing heterosis and combining ability in maize (Zea mays L.). Int J Plant Soi Sci. 2023;35:218–34. doi:10.9734/ijpss/2023/v35i244313. [Google Scholar] [CrossRef]
14. Heiba H, Mahgoub E, Mahmoud A, Ibrahİm M, Mansour E. Combining ability and gene action controlling chocolate spot resistance and yield traits in faba bean (Vicia faba L.). J Agric Sci. 2023;29:77–88. [Google Scholar]
15. ElShamey EAZ, Sakran RM, ElSayed MAA, Aloufi S, Alharthi B, Alqurashi M, et al. Heterosis and combining ability for floral and yield characters in rice using cytoplasmic male sterility system. Saudi J Biol Sci. 2022;29:3727–38. doi:10.1016/j.sjbs.2022.03.010. [Google Scholar] [PubMed] [CrossRef]
16. Hallauer AR, Carena MJ, Miranda Filho JB. Quantitative genetics in maize breeding. 3rd edNew York, London: Springer; 2010. [Google Scholar]
17. Al-Naggar A, El-Ganayni A, El-Lakany M, El-Sherbeiny H, Soliman M. Mode of inheritance of maize resistance to the pink stem borer, Sesamia cretica Led. under artificial infestation. Egypt J Plant Breed. 2000;4:13–35. [Google Scholar]
18. Akinwale RO, Eze CE, Traore D, Menkir A. Detection of non-additive gene action within elite maize populations evaluated in contrasting environments under rainforest ecology in Nigeria. Crop Breed Genet Genomic. 2021;3(1):e210003. [Google Scholar]
19. Ali S, Khan NU, Gul R, Naz I, Goher R, Ali N, et al. Genetic analysis for earliness and yield traits in maize. Pak J Bot. 2018;50:1395–405. [Google Scholar]
20. Elmyhun M, Liyew C, Shita A, Andualem M. Combining ability performance and heterotic grouping of maize (Zea mays) inbred lines in testcross formation in Western Amhara, North West Ethiopia. Cogent Food Agric. 2020;6:1727625. doi:10.1080/23311932.2020.1727625. [Google Scholar] [CrossRef]
21. Mageto EK, Makumbi D, Njoroge K, Nyankanga R. Genetic analysis of early-maturing maize (Zea Mays L.) inbred lines under stress and nonstress conditions. J Crop Improv. 2017;31:560–88. doi:10.1080/15427528.2017.1315625. [Google Scholar] [CrossRef]
22. Badu-Apraku B, Oyekunle M, Akinwale RO, Aderounmu M. Combining ability and genetic diversity of extra-early white maize inbreds under stress and nonstress environments. Crop Sci. 2013;53:9–26. doi:10.2135/cropsci2012.06.0381. [Google Scholar] [CrossRef]
23. Chakradhar T, Hindu V, Reddy PS. Genomic-based-breeding tools for tropical maize improvement. Genetica. 2017;145:525–39. doi:10.1007/s10709-017-9981-y. [Google Scholar] [PubMed] [CrossRef]
24. Gedil M, Menkir A. An integrated molecular and conventional breeding scheme for enhancing genetic gain in maize in Africa. Front Plant Sci. 2019;10:1430. doi:10.3389/fpls.2019.01430. [Google Scholar] [PubMed] [CrossRef]
25. Singh K, Kumar S, Kumar SR, Singh M, Gupta K. Plant genetic resources management and pre-breeding in genomics era. Indian J Genet Plant Breed. 2019;79:117–30. [Google Scholar]
26. Al-Samarai FR, Al-Kazaz AA. Molecular markers: an introduction and applications. Eur J Mol Biotechnol. 2015;9:118–30. doi:10.13187/issn.2310-6255. [Google Scholar] [CrossRef]
27. Grover A, Sharma P. Development and use of molecular markers: past and present. Crit Rev Biotechnol. 2016;36(2):290–302. doi:10.3109/07388551.2014.959891. [Google Scholar] [PubMed] [CrossRef]
28. Ahmad A, Wang J-D, Pan Y-B, Sharif R, Gao S-J. Development and use of simple sequence repeats (SSRs) markers for sugarcane breeding and genetic studies. Agronomy. 2018;8:260. doi:10.3390/agronomy8110260. [Google Scholar] [CrossRef]
29. Eid M. Validation of SSR molecular markers linked to drought tolerant in some wheat cultivars. J Plant Breed Genet. 2018;6:95–109. doi:10.33687/pbg.006.03.2739. [Google Scholar] [CrossRef]
30. Dermail A, Suriharn B, Chankaew S, Sanitchon J, Lertrat K. Hybrid prediction based on SSR-genetic distance, heterosis and combining ability on agronomic traits and yields in sweet and waxy corn. Sci Hortic. 2020;259:108817. doi:10.1016/j.scienta.2019.108817. [Google Scholar] [CrossRef]
31. Akaogu I, Badu-Apraku B, Adetimirin V, VROH-BI I, Oyekunle M, Akinwale R. Genetic diversity assessment of extra-early maturing yellow maize inbreds and hybrid performance in Striga-infested and Striga-free environments. J Agric Sci. 2013;151(4):519–37. doi:10.1017/S0021859612000652. [Google Scholar] [CrossRef]
32. Gichuru L, Derera J, Tongoona P, Murenga M. Combining ability and molecular genetic distances in tropical maize lines for grain yield and agronomic traits. S Afr J Plant Soil. 2017;34:175–83. doi:10.1080/02571862.2016.1213323. [Google Scholar] [CrossRef]
33. Betran F, Beck D, Bänziger M, Edmeades G. Genetic analysis of inbred and hybrid grain yield under stress and nonstress environments in tropical maize. Crop Sci. 2003;43:807–17. doi:10.2135/cropsci2003.8070. [Google Scholar] [CrossRef]
34. Phumichai C, Doungchan W, Puddhanon P, Jampatong S, Grudloyma P, Kirdsri C, et al. SSR-based and grain yield-based diversity of hybrid maize in Thailand. Field Crops Res. 2008;108:157–62. doi:10.1016/j.fcr.2008.04.009. [Google Scholar] [CrossRef]
35. Kamara M, Ghazy NA, Mansour E, Elsharkawy MM, Kheir AS, Ibrahim KM. Molecular genetic diversity and line × tester analysis for resistance to late wilt disease and grain yield in maize. Agronomy. 2021;11:898. doi:10.3390/agronomy11050898. [Google Scholar] [CrossRef]
36. Griffing B. Concept of general and specific combining ability in relation to diallel crossing systems. Aust J Biol Sci. 1956;9:463–93. doi:10.1071/BI9560463. [Google Scholar] [CrossRef]
37. Doyle J. A rapid total DNA preparation procedure for fresh plant tissue. Focus. 1990;12:13–5. [Google Scholar]
38. Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat. 1908;44:223–70. [Google Scholar]
39. Naorem A, Jayaraman S, Dang YP, Dalal RC, Sinha NK, Rao CS, et al. Soil constraints in an arid environment—challenges, prospects, and implications. Agronomy. 2023;13:220. doi:10.3390/agronomy13010220. [Google Scholar] [CrossRef]
40. Ahmed Z, Gui D, Murtaza G, Liu YF, Ali S. An overview of smart irrigation management for improving water productivity under climate change in drylands. Agronomy. 2023;13:2113. doi:10.3390/agronomy13082113. [Google Scholar] [CrossRef]
41. Ahmad U, Hussain M, Ahmad W, Javed J, Arshad Z, Akram Z. Impact of global climate change on maize (Zea maysphysiological responses and modern breeding techniques. Trends Biotech Plant Sci. 2024;2:62–77. doi:10.62460/TBPS. [Google Scholar] [CrossRef]
42. Olayiwola MO, Ajala SO, Ariyo OJ, Ojo DK, Gedil M. Heterotic grouping of tropical maize inbred lines and their hybrid performance under stem borer infestation and low soil nitrogen condition in West and Central Africa. Euphytica. 2021;217:14. doi:10.1007/s10681-020-02739-y. [Google Scholar] [CrossRef]
43. Udo E, Abe A, Meseka S, Mengesha W, Menkir A. Genetic analysis of zinc, iron and provitamin a content in tropical maize (Zea mays L.). Agronomy. 2023;13:266. doi:10.3390/agronomy13010266. [Google Scholar] [CrossRef]
44. Makinde SA, Badu-Apraku B, Ariyo OJ, Porbeni JB. Combining ability of extra-early maturing pro-vitamin A maize (Zea mays L.) inbred lines and performance of derived hybrids under Striga hermonthica infestation and low soil nitrogen. PLoS One. 2023;18:e0280814. doi:10.1371/journal.pone.0280814. [Google Scholar] [PubMed] [CrossRef]
45. Babic V, Stanisavljevic D, Zoric M, Mikic S, Mitrovic B, Andjelkovic V, et al. Identification of new sources for earliness and low grain moisture at harvest through maize landraces’ test-cross performance. Agronomy. 2022;12:1939. doi:10.3390/agronomy12081939. [Google Scholar] [CrossRef]
46. Oluwaseun O, Badu-Apraku B, Adebayo M, Abubakar AM. Combining ability and performance of extra-early maturing provitamin A maize inbreds and derived hybrids in multiple environments. Plants. 2022;11:964. doi:10.3390/plants11070964. [Google Scholar] [PubMed] [CrossRef]
47. Amegbor IK, Badu-Apraku B, Adu GB, Adjebeng-Danquah J, Toyinbo J. Combining ability of extra-early maize inbreds derived from a cross between maize and Zea diploperennis and hybrid performance under contrasting environments. Agronomy. 2020;10:1069. doi:10.3390/agronomy10081069. [Google Scholar] [CrossRef]
48. Ajala SO, Olayiwola MO, Job AO, Olaniyan AB, Gedil M. Assessment of heterotic patterns of tropical low nitrogen-tolerant maize (Zea mays L.) inbred lines using testcross performance, morphological traits and SNP markers. Plant Breed. 2020;139:1113–24. doi:10.1111/pbr.v139.6. [Google Scholar] [CrossRef]
49. Omar M, Rabie HA, Mowafi SA, Othman HT, El-Moneim DA, Alharbi K, et al. Multivariate analysis of agronomic traits in newly developed maize hybrids grown under different agro-environments. Plants. 2022;11:1187. doi:10.3390/plants11091187. [Google Scholar] [PubMed] [CrossRef]
50. Sánchez-Pérez R, Ortega E, Duval H, Martínez-Gómez P, Dicenta F. Inheritance and relationships of important agronomic traits in almond. Euphytica. 2007;155:381–91. doi:10.1007/s10681-006-9339-5. [Google Scholar] [CrossRef]
51. Feyzian E, Dehghani H, Rezai A, Jalali Javaran M. Diallel cross analysis for maturity and yield-related traits in melon (Cucumis melo L.). Euphytica. 2009;168:215–23. doi:10.1007/s10681-009-9904-9. [Google Scholar] [CrossRef]
52. Abdel-Moneam M, Sultan M, Sadek S, Shalof M. Combining abilities for yield and yield components in diallel crosses of six new yellow maize inbred lines. Int J Plant Breed Genet. 2015;9:86–94. doi:10.3923/ijpbg.2015.86.94. [Google Scholar] [CrossRef]
53. Sedhom SA, El-Badawy MEM, Hosary AAE, Abd El-Latif MS, Rady AM, Moustafa MM, et al. Molecular markers and GGE biplot analysis for selecting higher-yield and drought-tolerant maize hybrids. Agron J. 2021;113:3871–85. doi:10.1002/agj2.v113.5. [Google Scholar] [CrossRef]
54. Makumbi D, Betrán JF, Bänziger M, Ribaut J-M. Combining ability, heterosis and genetic diversity in tropical maize (Zea mays L.) under stress and non-stress conditions. Euphytica. 2011;180:143–62. doi:10.1007/s10681-010-0334-5. [Google Scholar] [CrossRef]
55. Derera J, Tongoona P, Vivek BS, Laing MD. Gene action controlling grain yield and secondary traits in southern African maize hybrids under drought and non-drought environments. Euphytica. 2008;162:411–22. doi:10.1007/s10681-007-9582-4. [Google Scholar] [CrossRef]
56. Annor B, Badu-Apraku B, Nyadanu D, Akromah R, Fakorede MA. Testcross performance and combining ability of early maturing maize inbreds under multiple-stress environments. Sci Rep. 2019;9:13809. doi:10.1038/s41598-019-50345-3. [Google Scholar] [PubMed] [CrossRef]
57. Oyetunde OA, Badu-Apraku B, Ariyo OJ, Alake CO. Efficiencies of heterotic grouping methods for classifying early maturing maize inbred lines. Agronomy. 2020;10:1198. doi:10.3390/agronomy10081198. [Google Scholar] [CrossRef]
58. Zhao M, Shu G, Hu Y, Cao G, Wang Y. Pattern and variation in simple sequence repeat (SSR) at different genomic regions and its implications to maize evolution and breeding. BMC Genom. 2023;24:1–13. doi:10.1186/s12864-023-09156-0. [Google Scholar] [PubMed] [CrossRef]
59. Miyassa MA, Lupini A, Mauceri A, Morsli A, Khelifi L, Sunseri F. Genetic variation and structure of maize populations from saoura and gourara oasis in Algerian Sahara. BMC Genom. 2018;19:51. [Google Scholar]
60. Wegary D, Vivek B, Labuschagne M. Association of parental genetic distance with heterosis and specific combining ability in quality protein maize. Euphytica. 2013;191:205–16. doi:10.1007/s10681-012-0757-2. [Google Scholar] [CrossRef]
61. Dhliwayo T, Pixley K, Menkir A, Warburton M. Combining ability, genetic distances, and heterosis among elite CIMMYT and IITA tropical maize inbred lines. Crop Sci. 2009;49:1201–10. doi:10.2135/cropsci2008.06.0354. [Google Scholar] [CrossRef]
62. Reif J, Melchinger A, Xia X, Warburton M, Hoisington D, Vasal S, et al. Genetic distance based on simple sequence repeats and heterosis in tropical maize populations. Crop Sci. 2003;43:1275–82. doi:10.2135/cropsci2003.1275. [Google Scholar] [CrossRef]
Appendix A

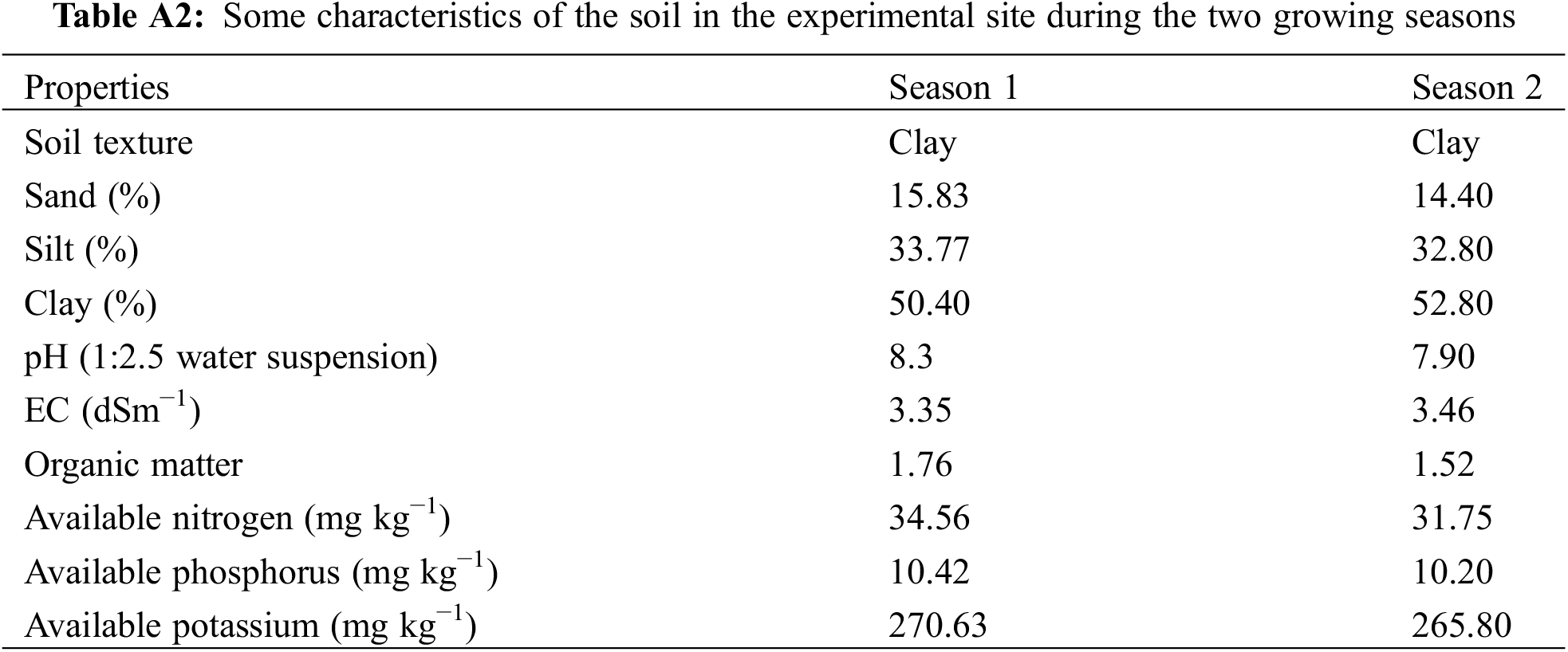

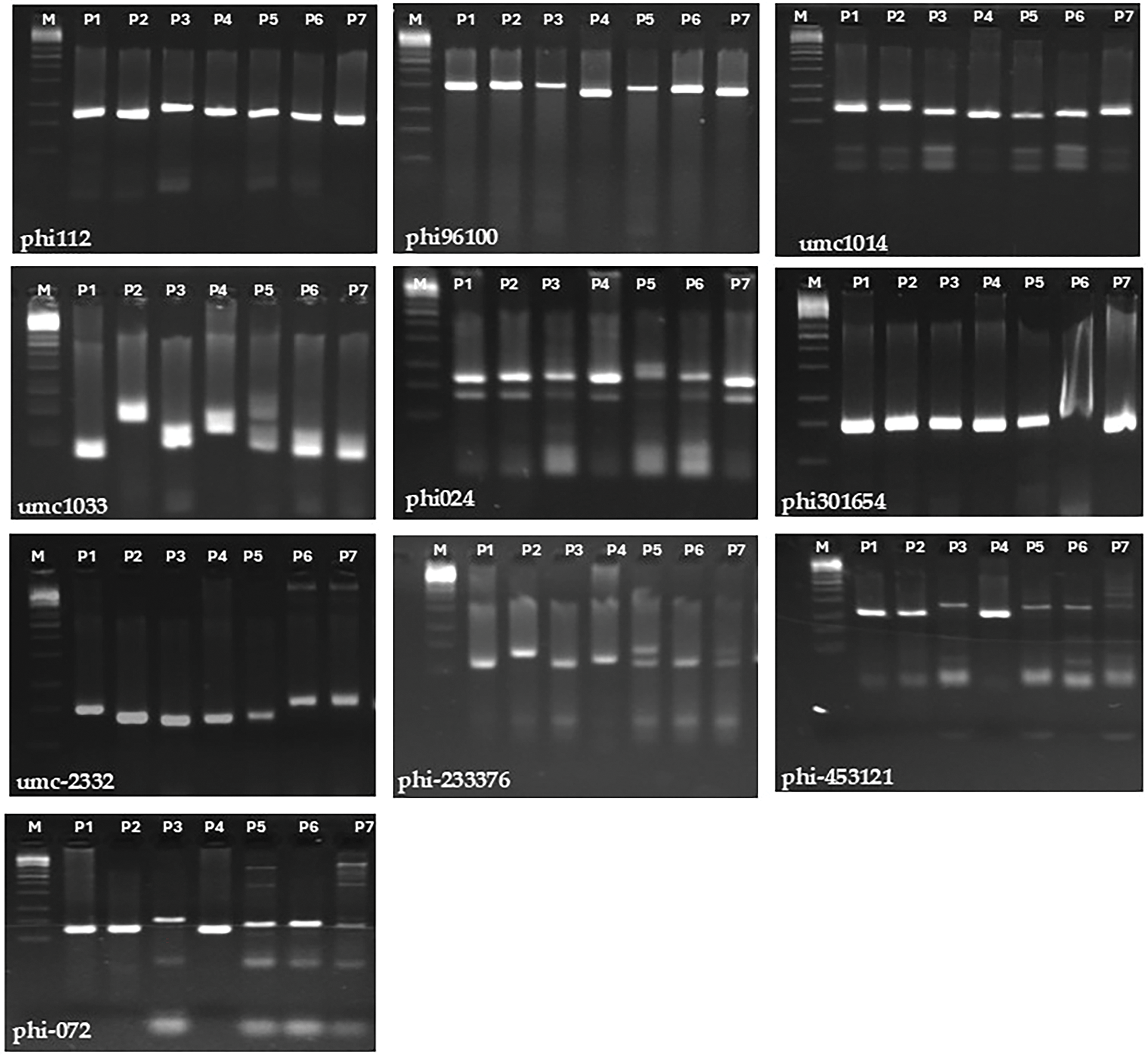
Figure A1: SSR-PCR amplification patterns of seven maize inbred lines using ten SSR primers
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools