 Open Access
Open Access
REVIEW
The Role of Phytohormones in Alleviating Salt Stress in Rice
1 Jia Sixie Agricultural College, Weifang Institute of Science and Technology, Weifang, 262700, China
2 Shenzhen Hospital, Southern Medical University, Shenzhen, 518100, China
3 Chemometrics Group, Faculty of Science, University of Copenhagen, Frederiksberg, 1958, Denmark
4 Circular Economy/Sustainable Solutions, LAB University of Applied Sciences, Lahti, 15101, Finland
* Corresponding Author: Huiwen Yu. Email:
(This article belongs to the Special Issue: Abiotic Stress Tolerance in Crop Plants: Physio-biochemical and Molecular Mechanisms)
Phyton-International Journal of Experimental Botany 2024, 93(12), 3131-3149. https://doi.org/10.32604/phyton.2024.058622
Received 16 September 2024; Accepted 07 November 2024; Issue published 31 December 2024
Abstract
Rice is a crucial food crop globally. Soil salt stress has adverse effects on the physiology and biochemistry of rice, leading to ionic toxicity and disrupted metabolism. Research aimed at improving salt tolerance and understanding its underlying mechanisms in rice is becoming increasingly important. Phytohormones are crucial in managing rice’s reaction to salt stress by controlling its physiological and biochemical functions. Some phytohormones can improve salt tolerance in rice by affecting gene programming, protein expression, and salt stress signaling, thereby helping rice adapt to salt-stressed environments. This review highlights recent advancements in understanding how various phytohormones—such as abscisic acid (ABA), auxin (IAA), cytokinins (CKs), jasmonates (JA), gibberellins (GAs), melatonin (MT), salicylic acid (SA), ethylene (ETHY) and brassinosteroids (BRs)—help mitigate the detrimental effects of salt stress in rice. Additionally, we explore the current challenges and future research directions for utilizing exogenous phytohormone regulators to boost rice’s resistance to salt stress.Keywords
Rice, known scientifically as Oryza sativa L., is a crucial staple food and cereal crop with global significance. In 2021, global rice production reached 787 million tons [1], making it the second most cultivated cereal crop globally. Given the growing global population, it is projected that the rice supply will need to double by 2050 to satisfy the rising demand for food [2]. However, various factors impact rice growth, including environmental stress, management strategies, and nutrient deficiencies. Among these, abiotic stress, particularly salt stress, is a major influence affecting rice development and yields worldwide [3].
The salinization of soil poses a major environmental challenge to agriculture, putting enormous pressure on agricultural production and hindering the sustainable development of agriculture [4]. On the one hand, soil salinization poses a serious threat to agroecosystem health [5] and is expected to be exacerbated by climate change, such as prolonged droughts and sea level rise [6]. On the other hand, salt stress significantly threatens crop production in irrigated lands, with an estimated one-third of the global irrigated croplands facing this challenge [7]. It adversely affects plant growth and crop yields. The soil affected by salt can lead to nutrient deficiencies, stunted plant growth, and decreased overall productivity. For rice production, it is reported that salt stress results in a 50% reduction in global rice production [8], which has an impact on the global food supply.
In rice, salt stress induces physicochemical changes in grain composition and quality [9] and affects physiological and biochemical properties such as enzyme activity, chlorophyll fluorescence parameters, and photosynthesis speed [10–12]. Salt stress severely impacts rice growth and yield worldwide [13]. Specifically, it causes physiological and biochemical damage, ion toxicity, disruption of normal metabolism, and inhibition of growth, ultimately affecting overall yield [14–16]. To enhance rice tolerance to salt stress, exogenous regulatory substances can be applied in appropriate amounts, helping to alleviate salt stress injury and improve tolerance [17,18]. Phytohormones, which serve as exogenous regulatory substances, are small-molecule chemical compounds vital for the development and growth of crops [19]. Widely used phytohormones are essential for adjusting crop-environment interactions [20], coordinating salt stress responses, and improving plant growth [21,22].
Phytohormones are essential for rice growth by controlling the plant’s reaction to salt stress. The typical mechanisms of plant hormones alleviating rice salt stress are illustrated in Fig. 1. As shown in Fig. 1, phytohormones adapt rice to salt-stress environments by regulating physiological and biochemical processes [23–25]. Also, phytohormones boost rice’s ability to tolerate salt by influencing gene expression [21,26,27], the synthesis of proteins [28], and salt stress signaling pathways [25,29]. Therefore, a systematic summary of the functions of various phytohormones in rice salt stress tolerance is essential for understanding the underlying mechanisms and optimizing the use of phytohormones for sustainable rice production.
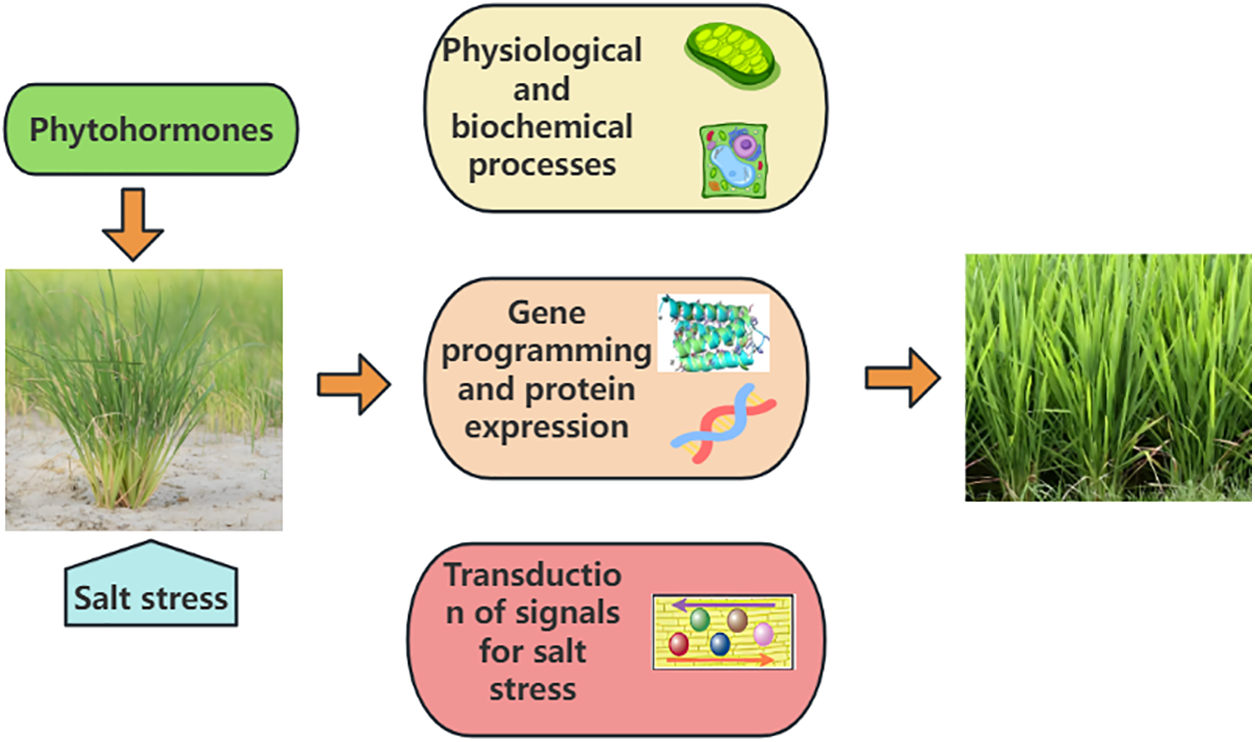
Figure 1: Illustration of typical mechanisms of phytohormones in alleviating salt stress in rice
This review highlights and examines the crucial roles of various phytohormones, such as abscisic acid (ABA), auxin (IAA), cytokinins (CKs), jasmonates (JA), gibberellins (GAs), melatonin (MT), salicylic acid (SA), ethylene (ETHY), and brassinosteroids (BRs), in alleviating the negative impacts of salt stress on rice. We provide a comprehensive overview of the inter-regulatory relationships between these phytohormones and salt stress, aiming to offer valuable insights into the use of exogenous phytohormone regulators to improve salt tolerance of rice and to elucidate the core mechanisms.
2 Phytohormones and Salt Stresses
ABA is a phytohormone that plays a crucial role in helping plants adapt to environmental stresses like drought and salinity [30]. Numerous studies have shown that applying exogenous ABA enhances the salt tolerance of rice, including increasing the survival and growth of rice in saline conditions [31], enhancing rice’s ability to withstand salinity, and boosting the yield of saline-alkali rice [32]. Some studies investigate the physiological mechanisms of how ABA alleviates salt stress in rice. For example, Gurmani et al. [33] showed that ABA promotes rice growth under salt stress by inhibiting the accumulation of sodium (Na+) in rice shoots and reducing the sodium (Na+)/potassium (K+) ratio in rice genotypes. This may be because exogenous ABA improves the ion homeostasis of rice under salt stress by optimizing the ion transport-related signaling pathways [34]. Likewise, Chen et al. [35] demonstrated that applying ABA reduces the harmful impact of salt on rice by improving photosynthesis and modulating the antioxidant defense system, thereby enhancing ion balance and improving salt tolerance. Additionally, endogenous biosynthesis of ABA enhances the ability of rice seedlings to withstand salt stress by facilitating stomatal closure, ion exchange, and bettering water relations [36]. Furthermore, ABA-pretreated rice seedlings exhibited upregulation of 40 protein spots at the molecular level under salt stress conditions, which may be beneficial for enhancing salt tolerance. This may be because of the specific upregulation of many enzymes associated with energy metabolism, primary metabolism, and defense in rice seedlings pretreated with ABA. Moreover, ABA de novo biosynthesis enhances rice’s resilience to salt stress by inducing strong OsLea3 expression in rice roots [37].
Additionally, research has shown that the interactions between rice genes and ABA influence rice’s response to salt stress. For example, the transcription factor OsNAC45 of NAC (NAM, ATAF1/2, CUC2) enhances rice salt tolerance by positively regulating the ABA signaling pathway [38]. Wang et al. [39] illustrated that miR528-AO, one of the target genes of miR528 in rice, can regulate the genetic expression associated with ABA metabolism and biosynthesis (including NCED2, NCED3, NCED5, ZEP, ABA8ox1, ABA8ox2, and ABA8ox3) under saline conditions. This regulation boosts ABA content, increases proline levels, accelerates reactive oxygen species (ROS) removal, and ultimately enhances rice’s salt tolerance. Zhao et al. [40] demonstrated that OsCSLD4 is involved in rice’s response to salt stress via the ABA-mediated osmotic regulation pathway. By overexpressing OsCSLD4, the expression of ABA synthesis genes in rice can be enhanced, ABA content can be increased, and rice’s salt tolerance can be improved. Zhang et al. [41] indicated that OsIAA20, an Aux/IAA protein upregulated under abiotic stress conditions, is essential for rice’s tolerance to salt stress by regulating an ABA-dependent mechanism. Furthermore, to explore the molecular pathways through which ABA enhances rice salt tolerance, Li et al. [42] and Yoshida et al. [43] demonstrated that ABA binding to PYR/PYL/RCAR receptors on the cell membrane inhibits protein phosphatase 2C (PP2C) activity. This inhibition activates SnRK2 kinase and it further phosphorylates the transcription factor ABF/AREB. The phosphorylated transcription factors enter the cell nucleus and initiate the expression of salt stress resistance genes, ultimately boosting rice salt tolerance. Table 1 summarizes the representative studies on ABA and related genes in alleviating salt stress in rice.
The plant hormone auxin, also known as indole-3-acetic acid (IAA), which influences essential aspects of plant cell division, elongation, and differentiation, is strongly correlated with the final shape and function of cells and tissues in plants [44]. In rice, IAA regulates and coordinates growth under stress conditions. IAA’s response to salt stress mediates the regulatory network that links IAA signaling with salt stress [45]. IAA significantly contributes to lateral root development [46]. Meng et al. [47] indicated that IAA accumulation and dependent signaling can be influenced through biosynthesis and polar IAA transport, thereby affecting lateral root formation in rice. IAA is crucial for the growth of rice when exposed to salt stress. On one side, reduced growth and development of rice under salt stress may result from the inhibition of IAA accumulation and response. For example, the reduction in IAA response due to salt stress is linked to inhibited root growth [48]. On the flip side, boosting the IAA content in the root system may enhance the root characteristics of rice under salt stress, thereby promoting growth [49]. The authors further demonstrated that elevated IAA content and a higher buffering capacity of IAA homeostatic processes may be critical for rice to tolerate salinity. Hence, the application of IAA may be a valid method to support the cultivation of rice under salt stress.
Many studies have been dedicated to revealing the intricate mechanisms through which IAA alleviates salt stress in rice. Applying IAA externally under salt stress conditions has been shown to enhance 1000-grain weight, grain filling rate, and yield by boosting the levels of starch, fructose, glucose, and sucrose in rice grains [50]. This may be attributed to the fact that exogenous IAA can mitigate the halophilic response of plant roots to salt [51]. Furthermore, IAA alleviates the harmful effects of salt stress on plants by boosting the antioxidant system and modulating the function of enzymatic antioxidants. Studies have shown that IAA can significantly increase the activity of α-tocopherol and the concentration of hydrogen peroxide (H2O2) in rice plants under salt stress [52]. This reason may lie in that IAA improves rice salt tolerance by optimizing antioxidant enzyme signaling pathways. In addition, some studies have shown that genes related to IAA production can be modulated to strengthen rice’s ability to withstand salinity (Table 1). For instance, the overexpression of RCc3 increases the accumulation of IAA in roots by affecting local IAA biosynthesis and polar transport, thereby improving rice root structure, promoting plant growth, and enhancing tolerance to salt stress [53]. Chai et al. [54] showed that OsLAX and OsABCB auxin transporter genes are crucial in mediating crosstalk between salt stress and IAA signaling pathways, enabling rice to adapt to salt stress by regulating auxin distribution. Furthermore, some strains enhance the expression of the IAA efflux carrier gene OsPIN1 under salt stress, increasing the IAA concentration in rice and mitigating the negative impacts of salt stress, thus enhancing the growth characteristics of rice [55].
CKs are important hormones that primarily influence plant growth and development by controlling cellular division and morphogenesis [56,57]. CKs also affect plant defenses against abiotic stress like salt stress [58]. The relationship between plant resilience to salt stress and CKs has been investigated in various studies. CK biosynthesis and signaling mutants impact tolerance to salt stress, while salt stress affects the transcription of genes involved in CK signaling or homeostasis in rice [59]. Some research has shown that the upregulation of CKs can alleviate the harmful effects of salinity on rice. For instance, the application of exogenous CKs has been reported to enhance rice’s salinity tolerance [50]. Joshi et al. [60] demonstrated that the inflorescence meristem-specific rice CKs oxidase OsCKX2 can regulate flower primordial activity under salt stress conditions by controlling CKs levels, thereby regulating rice grain yield. OsCKX2 knockout rice has higher relative water content and yield by accumulating CKs under salt stress.
However, it has also been shown that lowering CKs levels can favor salt tolerance in rice in some cases. For example, Li et al. [61] showed that the overexpression of AtUGT76C2, a CK glycosyltransferase, in rice reduces endogenous CK levels and promotes root growth, greatly enhancing rice adaptability to salt stress. Wang et al. [62] demonstrated that CKs-induced OsRR9 and OsRR10 genes function as negative regulators in response to salinity in rice. Additionally, the distribution of CKs is crucial in assessing the response of rice to stress. Yin et al. [63] emphasized that the overexpression of ARGONAUTE2 (AGO2) in rice increases CK content in roots and decreases it in shoots, enhancing salt tolerance. This occurs because AGO2 activates the expression of BG3, a potential CK transporter, altering CK distribution and ultimately regulating grain size and salt tolerance in rice. Hence, optimizing the distribution of CKs is a proven technique to enhance both rice yield and salt resistance. Therefore, the role of CKs in salt tolerance in rice varies depending on the CKs species, as well as the related cells and tissue distribution.
GAs, a group of tetracyclic diterpene carboxylic acids, have a major impact on plant development, especially in seed dormancy and the processes of germination, leaf expansion, stem growth, and the formation of flowers and fruits [64]. Research has shown that GAs have a role in reducing salt stress in rice. The application of exogenous GA3 under salt stress conditions can elevate the germination rate, seed vigor, photosynthetic pigment, and protein content of rice, promote the growth of rice seedlings in salty environments, elevate rice yield, and strengthen salt tolerance [65,66].
Furthermore, a wealth of research has uncovered how GA3 helps rice cope with salt stress physiologically. For instance, exogenous GA3 application may counteract the detrimental effects of salt stress by affecting the lipid content and fatty acid unsaturation in rice chloroplast membranes during salt treatment [67], as well as by increasing the permeability of membranes and the nutrient content in rice leaves [65]. GA3 pretreatment helps improve ion balance, delay the loss of pigment accumulation, and enhance the antioxidant system of rice under salt stress, thereby improving the growth of rice seedlings under such conditions [68]. Salt stress reduces alpha-amylase activity by downregulating the expression of the alpha-amylase gene, inhibiting rice seed germination. The application of exogenous bioactive GA can enhance alpha-amylase activity to rescue rice seed germination under salt stress [69]. Moreover, Wen et al. [70] demonstrated that GA can mitigate the growth suppression of rice due to salt stress by regulating certain salt-regulating proteins. Furthermore, GA hormones can promote rice growth in conditions of salt stress by promoting cell multiplication and expansion [24], thereby alleviating the adverse consequences of salt stress on rice and improving its growth and development [71].
In addition, the synergistic effect of GA and certain genes can enhance salt stress resistance in rice (Table 1). For example, the positive effect of OsHAK9 on seed germination in saline conditions is partially based on elevated GA concentration. Disruption of OsHAK9 significantly reduces gibberellin 4 (GA4) levels, and exogenous GA3 partially rescues the oshak9a mutant’s germination-deficient phenotype [72]. Furthermore, overexpressing OsGA2ox5, a gibberellin-metabolizing enzyme, boosts rice’s salt tolerance by regulating GA biosynthesis and signaling [73].
Phytohormone SA belongs to a class of phenolic compounds that function as endogenous growth regulators in plant seed germination, flowering, photosynthesis, the absorption and transport of ions, yield, and heat tolerance [74]. Additionally, SA is essential for plant defense mechanisms against both biotic and abiotic stresses, including salinity [75].
Under salt stress conditions, exogenous SA can promote the accumulation of nitrogen and dry matter in rice stems and sheaths in the short term and in the aboveground organs of rice in the long term [76]. Mostofa et al. [77] demonstrated that exogenous SA could alleviate oxidative damage in rice induced by salt stress by adjusting the antioxidant protection and glyoxalase functions. Furthermore, exogenous SA can increase the growth, germination rate, yield, and nutritional value of rice under saline conditions by inhibiting the accumulation of Na+ and Cl− ions and the increase in the activity of some enzymes (e.g., Superoxide Dismutase (SOD), Catalase (CAD) and Peroxidase (POD)) induced by salt stress [78]. Two later studies yielded similar results. Sheteiwy et al. [79] confirmed that SA attenuates the ionic imbalance caused by salt stress by reducing the accumulation of Na+ and boosting the content of K+ and Ca+ in rice cells, promoting the germination of rice seeds, seedling growth, and nutrient absorption in plants, and enhances photosynthetic pigments, photosynthetic gas exchange, and chlorophyll fluorescence. Liu et al. [80] also concluded that SA alleviates ion toxicity by reducing Na+ content, thereby promoting the maintenance of reactive oxygen species and hormone homeostasis, facilitating starch hydrolysis, generating sufficient energy for seed germination, and ultimately improving the germination of rice seeds under saline conditions. From these studies, it can be inferred that SA may regulate ion homeostasis in rice under salt stress by optimizing ion transport signaling pathways, thereby enhancing rice salt tolerance.
Additionally, SA also induces the expression of several genes (Table 1), such as CAT, SOD, Autophagy Related Genes (ATG), Glutathione Peroxidase (GPX), Mitogen-Activated Protein Kinase 1 (MAPK1), transcription factor WRKY53 and Bax Inhibiotr-1 (BI-1), in rice seedlings. The SA pathway is essential for controlling autophagy and programmed cell death mechanisms, which are essential for salt tolerance in rice [81]. Studies suggest that SA can improve Rice’s resilience to salt stress by upregulating the SA biosynthetic pathway gene (such as OsPAL, OsICS, and OsCM) expression levels under saline conditions [78].
BRs, isoprenoid-derived molecules that serve as unique polyhydroxy steroid hormones in plants, play a vital role in controlling plant metabolism, growth, and size. They are capable of sending endogenous signals that instruct plant growth and expansion [82]. BRs have proven effective in enhancing crop yields in saline environments [83]. The activated BR signaling pathway can reduce the negative impacts of salt stress on rice, and exogenous BRs have been shown to alleviate the inhibitory impact of salt stress on rice seedling development and the germination of seeds [84]. Anuradha et al. [85] showed that applying exogenous BRs can alleviate the harmful effects of salt stress on the germination of rice seeds, root elongation, and other growth processes by restoring pigment content and increasing nitrate reductase function.
Foliar spraying of rice seedlings with 24-epibrassinolide (EBL) can reduce oxidative stress by decreasing protein oxidation and lipid peroxidation and maintaining chlorophyll concentration, thereby restoring the development of rice seedlings under saline conditions [86]. Additionally, 28-homobrassinolide (HBL) and EBL can enhance rice’s ability to withstand salt stress by enhancing growth parameters, protein and proline content, increasing the activity of various antioxidant enzymes, and decreasing malondialdehyde (MDA) content [87,88]. Similar studies have shown that EBL can significantly boost seed germination, and improve seedling morphological traits, and rice yield in conditions of salt stress by increasing rice antioxidant enzymes and K+ maintenance in leaves and decreasing Na+ content in rice roots and MAD content [89]. Therefore, BRs may enhance rice salt tolerance by impacting antioxidant enzyme signaling pathways and ion transport-related signaling pathways. Moreover, Sharma et al. [88] found that EBL can alleviate salt stress in rice by up-regulating BR (OsBRI1) genes. Similarly, Li et al. [90] demonstrated that BR enhances rice salt tolerance by inducing the dephosphorylation of the core transcription factor OsBZR1 under salt stress (Table 1).
JAs, derivatives of fatty acids, are endogenous signaling molecules involved in various plant developmental processes. These include jasmonic acid (JA), methyl jasmonate (MeJA), jasmonic acid isoleucine conjugates (JA-Ile), and other key compounds [91]. Exogenous JA application [92] and the control of JA metabolism [93] have been shown to enhance rice tolerance under saline conditions. Exogenous JA effectively restored rice photosynthetic rate, seedling growth, leaf water potential, and dry matter in conditions of salt stress [92]. Foliar application of methyl jasmonate (MeJA) significantly improved leaf fluorescence, leaf gas exchange traits, and plant agronomic parameters in rice under salt stress [94]. MeJA can mitigate the negative effects of salt stress on rice by increasing plant growth, chlorophyll, and K+ content [95].
Furthermore, exogenous JA application may affect rice salt stress by altering other endogenous hormones (e.g., ABA and GAs) in rice plants. For example, JA can increase the ABA concentration in rice under salt stress and counteract the decrease in gibberellins caused by salt stress, thus enabling better adaptation to salt-stressed environments [96]. In addition, the interaction of rice genes with JA affects the response of rice to salt stress (Table 1). Ye et al. [97] showed that the overexpression of the JIOsPR10 gene, induced by JA, enhances rice’s ability to withstand salt stress. Wu et al. [98] demonstrated that forms a transcriptional regulatory complex with OsNINJA and OsbHLH to control the expression of JA-responsive genes associated with salt tolerance in rice under saline conditions, thereby modulating the tolerance of rice to salt stress. Moreover, JA acts as a signaling molecule for inducing the rice PR10 gene. RSOsPR10, a novel PR10 protein, is induced by salt stress in rice roots through the activation of the JA signaling pathway, thereby enhancing rice’s defense against saline conditions [99].
However, excessive activation of JA signaling can also have adverse effects. Some studies have shown that inhibiting JA signaling is important for enhancing salt tolerance in rice [100]. For instance, OsJAZ8 and its dominant-negative variant OsJAZ8ΔC, when overexpressed in the specific rice tissues, enhance salt tolerance in rice by suppressing JA signaling and other JA-dependent downstream genes under salt stress [101]. RICE SALT SENSITIVE3 (RSS3), a nuclear-localized protein that interacts with the transcription factors OsbHLH089 and OsbHLH094 to form a ternary complex that controls the salt-mediated elongation of rice root cells and promotes cell elongation by inhibiting the root JA response, thereby ensuring the development of rice roots when exposed to salt stress [102,103]. The conflicting results in the literature may be due to differences in the temporal and intensity patterns of JA accumulation and signaling, leading to varied responses to salt stress. Effective fine-tuning of JA signaling determines whether cells will adapt or undergo programmed cell death [104].
ETHY, a gaseous hormone with a simple C2H4 structure, regulates plant growth and development and plays a crucial role in various stress responses. The involvement of ETHY in plant responses to salt stress has been extensively studied. It has been reported that ETHY signaling regulates salinity responses by modulating ROS production and scavenging mechanisms [105]. Appropriate levels of ETHY can enhance plant salt tolerance by improving nutrient uptake, maintaining Na+/K+ homeostasis, regulating ROS, and increasing nitrate and sulfate assimilation under saline conditions [106].
ETHY plays a significant role in mitigating the inhibitory effects of salt stress on rice root growth. Research indicates that applying exogenous Ethrel (an ETHY-releasing agent) can significantly promote the growth of rice seedling root systems under salt stress. The improvement in these growth parameters helps rice better absorb water and nutrients in conditions of salt stress, thereby maintaining normal growth and development. ETHY also mitigates the adverse effects of salt stress by influencing the physiological characteristics of rice. Under salt stress, ETHY increases the activities of antioxidant enzymes in the rice root system, enhances the content of osmotic regulators, and decreases the content of MDA [107]. The improvement in these physiological properties helps rice mitigate oxidative damage and osmotic stress induced by salt stress, thus maintaining the integrity and stability of cell membranes.
In addition, several studies have demonstrated that increasing ETHY levels in rice can enhance its salt tolerance (Table 1). For example, the overexpression of OsARD1 increases the rate of endogenous ETHY release in rice, reducing its sensitivity to salinity [108]. Similarly, Glutamicibacter sp., a novel halotolerant plant growth-promoting rhizosphere bacterial strain, improves rice’s resistance to salt stress by regulating ETHY production and reactive oxygen species accumulation, preserving ion homeostasis, enhancing photosynthetic capacity, and upregulating salt stress-responsive genes [109]. Moreover, rice SALT TOLERANCE1 (SIT1), a lectin receptor-like kinase, maintains the homeostasis of ETHY by enhancing MAPK3/6 protein kinase activity, thereby improving salt tolerance in rice [110].
However, some studies have shown that reducing ETHY levels can enhance salt tolerance in rice [111]. For example, 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing Streptomyces sp. GMKU 336 reduces ETHY through the action of ACCD while helping plants scavenge ROS and balance ion content and osmotic pressure, thereby increasing salinity tolerance in rice [112]. Similarly, Sarkar et al. [113] found that Enterobacter sp. strain P23, a potent salt-tolerant ACC deaminase-containing plant growth-promoting rhizosphere bacterium, effectively protected the growth of rice seedlings under salt stress by reducing stress ETHY and reactive oxygen species, and by enhancing seed germination rate and seed vigor index. The differences in the role of ETHY biosynthesis in rice response to salt may be because only a single gene in the ETHY biosynthesis pathway is usually evaluated rather than the entire genes, and the optimal ETHY level required for normal rice growth may also be different in different stages of rice [114]. The appropriate amount of ETHY and signal intensity in rice response to salt stress may vary at different growth stages. Therefore, precise regulation of ETHY production and signal transduction may be crucial for improving rice salt tolerance. Further studies on the role of ETHY in rice resistance to salt stress are needed to clarify the differences in ETHY synthesis in alleviating salt stress.
MT, a lipophilic and hydrophilic biological indoleamine, plays a multifunctional role in plants. It regulates seed germination, promotes root growth, delays leaf senescence, and aids in plant growth and development under salt stress [115,116]. Exogenous application of MT is an advantageous strategy to improve the salt tolerance of rice plants. MT can increase both the dry and fresh weight of rice under salt stress, significantly reduce the accumulation of reactive oxygen species, and enhance salt tolerance [117].
Recent studies have investigated the physiological mechanisms by which MT enhances salinity tolerance in rice. Under salt stress, MT increases the relative water content, sucrose, and starch content by boosting the net photosynthetic rate and enhancing light energy absorption and transmission [118]. The application of MT increases rice yield by promoting the growth of vigorous green leaves with photosynthetic properties, enhancing the photosynthetic capacity and physiological activity of functional leaves, and protecting leaf cell membranes by preventing reactive oxygen species bursts and enhancing antioxidant activity [119]. Furthermore, MT improves rice plant tolerance to salt stress by (i) Activating antioxidants, regulating phytohormones, and promoting seed germination [120]. (ii) Improving nutrient accumulation and transport and regulating the ionic balance of K+/Na+ and Ca+/Na+ [121]. (iii) Decreasing chlorophyll degradation, inhibiting senescence-related genes, counteracting the accumulation of intracellular H2O2, and delaying leaf senescence and cellular death [122]. (iiii) Improving seed germination quality and root vitality, enhancing CAT and SOD activities in roots and leaves, and reducing Na+ and Cl− contents in roots and leaves [123].
In addition, MT can enhance rice salt tolerance by influencing specific genes (Table 1). For example, MT can increase salt tolerance by regulating the expression of OsHAKs to maintain Na+/K+ balance [119]. It also regulates K+ balance by up-regulating the potassium transporter gene OsHAK5 at the root tip, ensuring better potassium retention and thus improving salt tolerance [124]. Xie et al. [125] showed that MT notably increased the expression levels of a distinct group of transcription factor genes, including 11 AP2/EREBP, 14 HB, and 3 WRKY genes, in rice seedlings under salt stress, activated multiple antioxidant pathways, and improved the abundance of metabolites such as gallic acid, butyraldehyde, AFMK, and isoquercitrin, which resulted in the improvement of salt tolerance in rice. Furthermore, The Salt Overly Sensitive (SOS) signaling pathway plays a crucial role in maintaining cellular ion homeostasis during salt stress. Exogenous MT influences the SOS pathway by regulating the expression of SOS genes, thereby alleviating salt stress in rice [126].
3 Plant Hormone Crosstalk Regulates Rice Tolerance to Salt Stress
The interplay and crosstalk between various plant hormones are important for rice’s tolerance to salt stress. This crosstalk helps rice adapt to such stress by regulating its morphological traits and mediating changes in hormone levels within roots, stems, and leaves [127]. ABA-mediated IAA transport regulates the formation of rice lateral root primordia (LRP), facilitating rice adaptation to mild salt stress conditions [128]. The synergistic effects of ABA and ET can mediate rice adaptation to salt stress, this may be because ABA and ET interact to control different mechanisms of rice root growth [129]. However, ABA and JA have antagonistic effects on salt stress-induced transcripts in rice roots [130]. Moreover, the JA/ET signaling pathway in rice roots is involved in the induction of RSOsPR10 (a root-specific gene) under salt stress, promoting root growth and root mass, thereby improving salt tolerance, while exogenous SA has an antagonistic inhibitory effect on it [131]. However, Liu et al. [69] showed that exogenous SA can improve the germination of rice seeds under salt stress by positively regulating the homeostasis of GA and ABA in rice under salt stress. In addition, exogenous MT boosts salt tolerance in rice by activating the IAA/ABA/BR signaling pathways, as well as the AP2/EREBP–HB–WRKY transcriptional cascade [125]. IAA and CK synergistically enhance carbohydrate accumulation in rice grains under salt stress. Exogenous IAA application increases IAA levels in the grains, subsequently raising CK levels and stimulating cell division, elongation, and enlargement, ultimately boosting grain weight, fruit set, and rice yield [50]. Hence the crosstalk effects of plant hormones are important for understanding and regulating rice response mechanism to salt stress.
Soil salinization poses a significant threat to rice. This study reviewed the role of various phytohormones, including ABA, IAA, CKs, GAs, SA, BRs, JA, MT, and ETHY, in rice’s response to salt stress, both directly and indirectly. In many cases, exogenous applications of these phytohormones mitigate the negative effects of salt on rice. From the perspective of agricultural practice strategies, the type and concentration of hormones associated with the rice’s growth stage and specific needs should always be taken into account, and this is because different hormones have unique mechanisms and effects. The timing and method of hormone application greatly influence its effectiveness, so it is important to determine the optimal application schedule considering the rice growth cycle and weather conditions. Particular attention is that the overuse of plant hormones can negatively impact rice, so the application dosage should be strictly controlled. Many studies on the role of phytohormones in rice response to salt stress have emerged over the years, however, the precise mechanisms by which phytohormones mitigate salt stress in rice remain incompletely understood in the current literature. The pathways, signaling processes and interactions with other physiological processes have not been systematically understood yet. Additionally, research on phytohormones primarily relies on traditional biochemical techniques, which have limitations in revealing the specific mechanisms. Future progress in further understanding the complex role of phytohormones in mitigating salt stress will depend on how to overcome these limitations. Future research could focus on the following directions:
• Mitigation effects of different types of phytohormones on salt stress in rice can vary, and the same hormone may have different effects at different concentrations. Therefore, it is interesting to screen for the most suitable combination of hormone types and concentrations for specific types of plants.
• Various ecological factors, such as soil type, climatic conditions, and irrigation methods, influence the response of rice to salt stress and the effectiveness of phytohormones. Therefore, the effects of these ecological factors are interesting to be examined in the future.
• Exploring the combination of different types of phytohormones and their combined regulatory effects on rice growth will provide valuable guidance for enhancing rice’s resilience to future stress conditions.
• Identifying novel plant hormones or signaling molecules is of importance for regulating rice salt tolerance. Developing hormone analogs based on the structure and function of known hormones, and enhancing their activity and specificity to rice salt tolerance are important for future search.
• Investigations of new technologies and methods are beneficial for advancing plant phytohormone research. It is interesting to explore how to use high-throughput sequencing and gene editing technologies to more efficiently pinpoint and isolate genes and transcription factors associated with salt tolerance. Moreover, integrating smart agricultural technology, precision irrigation, and nutrient management with plant hormone regulation to enhance resource utilization and rice productivity under salt stress is also a promising direction. For further explorations.
Acknowledgement: None.
Funding Statement: The first author was supported by the Basic Research Fund of Weifang Institute of Science and Technology under Grant No. KJRC2023047.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization and methodology: Lili Guo, Huiwen Yu; formal analysis and investigation: Lili Guo, Huiwen Yu, Chenxi Zhang, Mourad Kharbach; resources: Lili Guo, Huiwen Yu; writing—original draft preparation: Lili Guo; writing—review and editing: Lili Guo, Huiwen Yu, Chenxi Zhang, Mourad Kharbach; project administration and funding acquisition: Lili Guo. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The materials used to support the findings of this study can be made available by the authors upon request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Wijayanti EB, Setyoko BH. Dataset analysis and feature characteristics to predict rice production based on eXtreme gradient boosting. J Comput Theories Appl. 2024;1(3):299–310. doi:10.33633/jcta.v2i1. [Google Scholar] [CrossRef]
2. Mitra D, de los Santos-Villalobos S, Cota FIP, Montelongo AMG, Blanco EL, Lira VL, et al. Rice (Oryza sativa L.) plant protection by using dual biological control and plant growth-promoting agents—current scenarios and future prospects: a review. Pedosphere. 2023;33(2):268–86. doi:10.1016/j.pedsph.2022.06.034. [Google Scholar] [CrossRef]
3. Hussain S, Zhang J-H, Zhong C, Zhu L-F, Cao X-C, Yu S-M, et al. Effects of salt stress on rice growth, development characteristics, and the regulating ways: a review. J Integr Agr. 2017;16(11):2357–74. doi:10.1016/S2095-3119(16)61608-8. [Google Scholar] [CrossRef]
4. Mishra AK, Das R, George Kerry R, Biswal B, Sinha T, Sharma S, et al. Promising management strategies to improve crop sustainability and to amend soil salinity. Front Environ Sci. 2023;10:962581. doi:10.3389/fenvs.2022.962581. [Google Scholar] [CrossRef]
5. Singh A. Soil salinity: a global threat to sustainable development. Soil Use Manage. 2022;38(1):39–67. doi:10.1111/sum.v38.1. [Google Scholar] [CrossRef]
6. Negacz K, Malek Ž, de Vos A, Vellinga P. Saline soils worldwide: identifying the most promising areas for saline agriculture. J Arid Eeviron. 2022;203:104775. doi:10.1016/j.jaridenv.2022.104775. [Google Scholar] [CrossRef]
7. Mohanavelu A, Naganna SR, Al-Ansari N. Irrigation induced salinity and sodicity hazards on soil and groundwater: an overview of its causes, impacts and mitigation strategies. Agriculture. 2021;11(10):983. doi:10.3390/agriculture11100983. [Google Scholar] [CrossRef]
8. Ma NL, Che Lah WA, Abd. Kadir N, Mustaqim M, Rahmat Z, Ahmad A, et al. Susceptibility and tolerance of rice crop to salt threat: physiological and metabolic inspections. PLoS One. 2018;13(2):e0192732. doi:10.1371/journal.pone.0192732. [Google Scholar] [PubMed] [CrossRef]
9. Razzaq A, Ali A, Safdar LB, Zafar MM, Rui Y, Shakeel A, et al. Salt stress induces physiochemical alterations in rice grain composition and quality. J Food Sci. 2020;85(1):14–20. doi:10.1111/jfds.v85.1. [Google Scholar] [CrossRef]
10. Lekklar C, Suriya-arunroj D, Pongpanich M, Comai L, Kositsup B, Chadchawan S, et al. Comparative genomic analysis of rice with contrasting photosynthesis and grain production under salt stress. Genes. 2019;10(8):562. doi:10.3390/genes10080562. [Google Scholar] [PubMed] [CrossRef]
11. Rossatto T, do Amaral MN, Benitez LC, Vighi IL, Braga EJB, de Magalhães Júnior AM, et al. Gene expression and activity of antioxidant enzymes in rice plants, cv. BRS AG, under saline stress. Physiol Mol Biol Pla. 2017;23:865–75. doi:10.1007/s12298-017-0467-2. [Google Scholar] [PubMed] [CrossRef]
12. Tsai Y-C, Chen K-C, Cheng T-S, Lee C, Lin S-H, Tung C-W. Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 2019;19:1–17. [Google Scholar]
13. Qin H, Li Y, Huang R. Advances and challenges in the breeding of salt-tolerant rice. Int J Mol Sci. 2020;21(21):8385. doi:10.3390/ijms21218385. [Google Scholar] [PubMed] [CrossRef]
14. Arif Y, Singh P, Siddiqui H, Bajguz A, Hayat S. Salinity induced physiological and biochemical changes in plants: an omic approach towards salt stress tolerance. Plant Physiol Bioch. 2020;156:64–77. doi:10.1016/j.plaphy.2020.08.042. [Google Scholar] [PubMed] [CrossRef]
15. Sarma B, Kashtoh H, Lama Tamang T, Bhattacharyya PN, Mohanta YK, Baek K-H. Abiotic stress in rice: visiting the physiological response and its tolerance mechanisms. Plants. 2023;12(23):3948. doi:10.3390/plants12233948. [Google Scholar] [PubMed] [CrossRef]
16. Mushtaq Z, Faizan S, Gulzar B. Salt stress, its impacts on plants and the strategies plants are employing against it: a review. J Appl Biol Biotechnol. 2020;8:81–91. doi:10.7324/JABB.2020.80315. [Google Scholar] [CrossRef]
17. Abdelhamid MT, Sekara A, Pessarakli M, Alarcón J, Brestic M, El-Ramady H, et al. New approaches for improving salt stress tolerance in rice. Rice research for quality improvement: genomics and genetic engineering. Breed Tech Abiotic Stress Toleran. 2020;1:247–68. [Google Scholar]
18. Feng D, Gao Q, Liu J, Tang J, Hua Z, Sun X. Categories of exogenous substances and their effect on alleviation of plant salt stress. Eur J Ageon. 2023;142:126656. doi:10.1016/j.eja.2022.126656. [Google Scholar] [CrossRef]
19. Pal P, Ansari SA, Jalil SU, Ansari MI. Regulatory role of phytohormones in plant growth and development. In: Plant hormones in crop improvement. Cambridge, MA, USA: Elsevier; 2023. p. 1–13. [Google Scholar]
20. Jha Y. Plant microbiomes with phytohormones: attribute for plant growth and adaptation under the stress conditions. In: Advances in plant microbiome and sustainable agriculture. Singapore: Springer; 2020. p. 85–103. [Google Scholar]
21. Waadt R, Seller CA, Hsu P-K, Takahashi Y, Munemasa S, Schroeder JI. Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Bio. 2022;23(10):680–94. doi:10.1038/s41580-022-00479-6. [Google Scholar] [PubMed] [CrossRef]
22. Zheng Y, Wang X, Cui X, Wang K, Wang Y, He Y. Phytohormones regulate the abiotic stress: an overview of physiological, biochemical, and molecular responses in horticultural crops. Front. Plant Sci. 2023;13:1095363. doi:10.3389/fpls.2022.1095363. [Google Scholar] [PubMed] [CrossRef]
23. Fatma M, Khan MIR, Masood A, Khan NA. Coordinate changes in assimilatory sulfate reduction are correlated to salt tolerance: involvement of phytohormones. Annu Rev Res Biol. 2013;3(3):267–95. [Google Scholar]
24. Khan MIR, Kumari S, Nazir F, Khanna RR, Gupta R, Chhillar H. Defensive role of plant hormones in advancing abiotic stress-resistant rice plants. Rice Sci. 2023;30(1):15–35. doi:10.1016/j.rsci.2022.08.002. [Google Scholar] [CrossRef]
25. Yu Z, Duan X, Luo L, Dai S, Ding Z, Xia G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020;25(11):1117–30. doi:10.1016/j.tplants.2020.06.008. [Google Scholar] [PubMed] [CrossRef]
26. Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43(1):265–85. doi:10.1146/genet.2009.43.issue-1. [Google Scholar] [CrossRef]
27. Hussain S, Hafeez MB, Azam R, Mehmood K, Aziz M, Ercisli S, et al. Deciphering the role of phytohormones and osmolytes in plant tolerance against salt stress: implications, possible cross-talk, and prospects. J Plant Growth Regul. 2024;43(1):38–59. doi:10.1007/s00344-023-11070-4. [Google Scholar] [CrossRef]
28. Anfang M, Shani E. Transport mechanisms of plant hormones. Curr Opin Plant Biol. 2021;63:102055. doi:10.1016/j.pbi.2021.102055. [Google Scholar] [PubMed] [CrossRef]
29. Harrison MA. Cross-talk between phytohormone signaling pathways under both optimal and stressful environmental conditions. In: Phytohormones and abiotic stress tolerance in plants. Berlin, Heidelberg: Springer; 2012. p. 49–76. [Google Scholar]
30. Jones AM. A new look at stress: abscisic acid patterns and dynamics at high-resolution. New Phytol. 2016;210(1):38–44. doi:10.1111/nph.2016.210.issue-1. [Google Scholar] [CrossRef]
31. Sripinyowanich S, Klomsakul P, Boonburapong B, Bangyeekhun T, Asami T, Gu H, et al. Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ Exp Bot. 2013;86:94–105. doi:10.1016/j.envexpbot.2010.01.009. [Google Scholar] [CrossRef]
32. Wei L-X, Lv B-S, Li X-W, Wang M-M, Ma H-Y, Yang H-Y, et al. Priming of rice (Oryza sativa L.) seedlings with abscisic acid enhances seedling survival, plant growth, and grain yield in saline-alkaline paddy fields. Field Crops Res. 2017;203:86–93. doi:10.1016/j.fcr.2016.12.024. [Google Scholar] [CrossRef]
33. Gurmani AR, Bano A, Ullah N, Khan H, Jahangir M, Flowers TJ. Exogenous abscisic acid (ABA) and silicon (Si) promote salinity tolerance by reducing sodium (Na+) transport and bypass flow in rice (Oryza sativa indica). Aust J Crop Sci. 2013;7(9):1219–26. [Google Scholar]
34. Jiang S, Lan Z, Zhang Y, Kang X, Zhao L, Wu X, et al. Mechanisms by which exogenous substances enhance plant salt tolerance through the modulation of ion membrane transport and reactive oxygen species metabolism. Antioxidants. 2024;13(9):1050. doi:10.3390/antiox13091050. [Google Scholar] [PubMed] [CrossRef]
35. Chen G, Zheng D, Feng N, Zhou H, Mu D, Zhao L, et al. Physiological mechanisms of ABA-induced salinity tolerance in leaves and roots of rice. Sci Rep. 2022;12(1):8228. doi:10.1038/s41598-022-11408-0. [Google Scholar] [PubMed] [CrossRef]
36. Shahzad R, Khan AL, Bilal S, Waqas M, Kang S-M, Lee I-J. Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environ Exp Bot. 2017;136:68–77. doi:10.1016/j.envexpbot.2017.01.010. [Google Scholar] [CrossRef]
37. Moons A, De Keyser A, Van Montagu M. A group 3 LEA cDNA of rice, responsive to abscisic acid, but not to jasmonic acid, shows variety-specific differences in salt stress response. Gene. 1997;191(2):197–204. doi:10.1016/S0378-1119(97)00059-0. [Google Scholar] [PubMed] [CrossRef]
38. Zhang X, Long Y, Huang J, Xia J. OsNAC45 is involved in ABA response and salt tolerance in rice. Rice. 2020;13:1–13. [Google Scholar]
39. Wang M, Guo W, Li J, Pan X, Pan L, Zhao J, et al. The miR528-AO module confers enhanced salt tolerance in rice by modulating the ascorbic acid and abscisic acid metabolism and ROS scavenging. J Agric Food Chem. 2021;69(31):8634–48. doi:10.1021/acs.jafc.1c01096. [Google Scholar] [PubMed] [CrossRef]
40. Zhao H, Li Z, Wang Y, Wang J, Xiao M, Liu H, et al. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol J. 2022;20(3):468–84. doi:10.1111/pbi.v20.3. [Google Scholar] [CrossRef]
41. Zhang A, Yang X, Lu J, Song F, Sun J, Wang C, et al. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021;308:110903. doi:10.1016/j.plantsci.2021.110903. [Google Scholar] [PubMed] [CrossRef]
42. Li Q, Zhu P, Yu X, Xu J, Liu G. Physiological and molecular mechanisms of rice tolerance to salt and drought stress: advances and future directions. Int J Mol Sci. 2024;25(17):9404. doi:10.3390/ijms25179404. [Google Scholar] [PubMed] [CrossRef]
43. Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–9. doi:10.1016/j.pbi.2014.07.009. [Google Scholar] [PubMed] [CrossRef]
44. Guo Y, Jiang Q, Hu Z, Sun X, Fan S, Zhang H. Function of the auxin-responsive gene TaSAUR75 under salt and drought stress. Crop J. 2018;6(2):181–90. doi:10.1016/j.cj.2017.08.005. [Google Scholar] [CrossRef]
45. Naser V, Shani E. Auxin response under osmotic stress. Plant Mol Biol. 2016;91(6):661–72. doi:10.1007/s11103-016-0476-5. [Google Scholar] [PubMed] [CrossRef]
46. Yamauchi T, Tanaka A, Inahashi H, Nishizawa NK, Tsutsumi N, Inukai Y, et al. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc Natl Acad Sci U S A. 2019;116(41):20770–5. doi:10.1073/pnas.1907181116. [Google Scholar] [PubMed] [CrossRef]
47. Meng F, Xiang D, Zhu J, Li Y, Mao C. Molecular mechanisms of root development in rice. Rice. 2019;12(1):1. doi:10.1186/s12284-018-0262-x. [Google Scholar] [PubMed] [CrossRef]
48. Liu W, Li R-J, Han T-T, Cai W, Fu Z-W, Lu Y-T. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015;168(1):343–56. doi:10.1104/pp.15.00030. [Google Scholar] [PubMed] [CrossRef]
49. Saini S, Kaur N, Marothia D, Singh B, Singh V, Gantet P, et al. Morphological analysis, protein profiling and expression analysis of auxin homeostasis genes of roots of two contrasting cultivars of rice provide inputs on mechanisms involved in rice adaptation towards salinity stress. Plants. 2021;10(8):1544. doi:10.3390/plants10081544. [Google Scholar] [PubMed] [CrossRef]
50. Ghorbani Javid M, Sorooshzadeh A, Modarres Sanavy SAM, Allahdadi I, Moradi F. Effects of the exogenous application of auxin and cytokinin on carbohydrate accumulation in grains of rice under salt stress. Plant Growth Regul. 2011;65(2):305–13. doi:10.1007/s10725-011-9602-1. [Google Scholar] [CrossRef]
51. Galvan-Ampudia CS, Julkowska MM, Darwish E, Gandullo J, Korver RA, Brunoud G, et al. Halotropism is a response of plant roots to avoid a saline environment. Curr Biol. 2013;23(20):2044–50. doi:10.1016/j.cub.2013.08.042. [Google Scholar] [PubMed] [CrossRef]
52. Saedipour S. Role of exogenous application of auxin on antioxidant enzyme activities in rice under salt stress. J Plant Physiol. 2016;6(2):49–61. [Google Scholar]
53. Li X, Chen R, Chu Y, Huang J, Jin L, Wang G, et al. Overexpression of RCc3 improves root system architecture and enhances salt tolerance in rice. Plant Physiol Bioch. 2018;130:566–76. doi:10.1016/j.plaphy.2018.08.008. [Google Scholar] [PubMed] [CrossRef]
54. Chai C, Subudhi PK. Comprehensive analysis and expression profiling of the OsLAX and OsABCB auxin transporter gene families in rice (Oryza sativa) under phytohormone stimuli and abiotic stresses. Front Plant Sci. 2016;7:593. [Google Scholar] [PubMed]
55. Khan MA, Asaf S, Khan AL, Adhikari A, Jan R, Ali S, et al. Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol. 2020;22(5):850–62. doi:10.1111/plb.v22.5. [Google Scholar] [CrossRef]
56. Kiba T, Takei K, Kojima M, Sakakibara H. Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev Cell. 2013;27(4):452–61. doi:10.1016/j.devcel.2013.10.004. [Google Scholar] [PubMed] [CrossRef]
57. Papon N, Courdavault V. ARResting cytokinin signaling for salt-stress tolerance. Plant Sci. 2022;314:111116. doi:10.1016/j.plantsci.2021.111116. [Google Scholar] [PubMed] [CrossRef]
58. Liu Y, Zhang M, Meng Z, Wang B, Chen M. Research progress on the roles of cytokinin in plant response to stress. Int J Mol Sci. 2020;21(18):6574. doi:10.3390/ijms21186574. [Google Scholar] [PubMed] [CrossRef]
59. Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23(6):2169–83. doi:10.1105/tpc.111.087395. [Google Scholar] [PubMed] [CrossRef]
60. Joshi R, Sahoo KK, Tripathi AK, Kumar R, Gupta BK, Pareek A, et al. Knockdown of an inflorescence meristem-specific cytokinin oxidase-OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell Environ. 2018;41(5):936–46. doi:10.1111/pce.12947. [Google Scholar] [PubMed] [CrossRef]
61. Li Y, Liu F, Li P, Wang T, Zheng C, Hou B. An arabidopsis cytokinin-modifying glycosyltransferase UGT76C2 improves drought and salt tolerance in rice. Front Plant Sci. 2020;11:560696. doi:10.3389/fpls.2020.560696. [Google Scholar] [PubMed] [CrossRef]
62. Wang WC, Lin TC, Kieber J, Tsai YC. Response regulators 9 and 10 negatively regulate salinity tolerance in rice. Plant Cell Physiol. 2019;60(11):2549–63. doi:10.1093/pcp/pcz149. [Google Scholar] [PubMed] [CrossRef]
63. Yin W, Xiao Y, Niu M, Meng W, Li L, Zhang X, et al. ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell. 2020;32(7):2292–306. doi:10.1105/tpc.19.00542. [Google Scholar] [PubMed] [CrossRef]
64. Daviere JM, Achard P. Gibberellin signaling in plants. Development. 2013;140(6):1147–51. doi:10.1242/dev.087650. [Google Scholar] [PubMed] [CrossRef]
65. Krishna K, Mahadevaswamy M. The effect of exogenous application of gibberellic acid on two salt stressed paddy cultivars during seed germination. Int J Res Appl Sci Eng Technol. 2019;7:327–32. doi:10.22214/ijraset. [Google Scholar] [CrossRef]
66. Misratia K, Islam MR, Ismail M, Oad F, Hanafi M, Puteh A. Interactive effects of gibberellic acid (GA3) and salt stress on growth, biochemical parameters and ion accumulation of two rice (Oryza sativa L.) varieties differing in salt tolerance. J Food Agric Environ. 2015;13:66–70. [Google Scholar]
67. Liu X, Wang X, Yin L, Deng X, Wang S. Exogenous application of gibberellic acid participates in up-regulation of lipid biosynthesis under salt stress in rice. Theor Exp Plant Physiol. 2018;30(4):335–45. doi:10.1007/s40626-018-0129-y. [Google Scholar] [CrossRef]
68. Chunthaburee S, Sanitchon J, Pattanagul W, Theerakulpisut P. Alleviation of salt stress in seedlings of black glutinous rice by seed priming with spermidine and gibberellic acid. Not Bot Horti Agrobo. 2014;42(2):405–13. doi:10.15835/nbha4229688. [Google Scholar] [CrossRef]
69. Liu Z, Ma C, Hou L, Wu X, Wang D, Zhang L, et al. Exogenous SA affects rice seed germination under salt stress by regulating Na+/K+ balance and endogenous GAs and ABA homeostasis. Int J Mol Sci. 2022;23(6):3293. doi:10.3390/ijms23063293. [Google Scholar] [PubMed] [CrossRef]
70. Wen F-P, Zhang Z-H, Bai T, Xu Q, Pan Y-H. Proteomics reveals the effects of gibberellic acid (GA3) on salt-stressed rice (Oryza sativa L.) shoots. Plant Sci. 2010;178(2):170–5. doi:10.1016/j.plantsci.2009.11.006. [Google Scholar] [CrossRef]
71. Colebrook EH, Thomas SG, Phillips AL, Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol. 2014;217(1):67–75. doi:10.1242/jeb.089938. [Google Scholar] [PubMed] [CrossRef]
72. Zeng P, Xie T, Shen J, Liang T, Yin L, Liu K, et al. Potassium transporter OsHAK9 regulates seed germination under salt stress by preventing gibberellin degradation through mediating OsGA2ox7 in rice. J Integr Plant Biol. 2024;66(4):731–48. doi:10.1111/jipb.v66.4. [Google Scholar] [CrossRef]
73. Shan C, Mei Z, Duan J, Chen H, Feng H, Cai W. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One. 2014;9(1):e87110. doi:10.1371/journal.pone.0087110. [Google Scholar] [PubMed] [CrossRef]
74. Hernández JA, Diaz-Vivancos P, Barba-Espín G, Clemente-Moreno MJ. On the role of salicylic acid in plant responses to environmental stresses. In: Salicylic acid: a multifaceted hormone. Singapore: Springer; 2017. p. 17–34. [Google Scholar]
75. Bastam N, Baninasab B, Ghobadi C. Improving salt tolerance by exogenous application of salicylic acid in seedlings of pistachio. Plant Growth Regul. 2012;69(3):275–84. [Google Scholar]
76. Sha H-J, Liu H-L, Hu B-W, Gu J-J, Hu W-C, Jia Y, et al. Effect of salicylic acid on the dry matter and nitrogen accumulation, partitioning and translocation in two contrasting rice genotypes under salt stress. Pak J Bot. 2019;51(5):1541–50. [Google Scholar]
77. Mostofa MG, Fujita M, Tran L-SP. Nitric oxide mediates hydrogen peroxide- and salicylic acid-induced salt tolerance in rice (Oryza sativa L.) seedlings. Plant Growth Regul. 2015;77(3):265–77. doi:10.1007/s10725-015-0061-y. [Google Scholar] [CrossRef]
78. Jini D, Joseph B. Physiological mechanism of salicylic acid for alleviation of salt stress in rice. Rice Sci. 2017;24(2):97–108. doi:10.1016/j.rsci.2016.07.007. [Google Scholar] [CrossRef]
79. Sheteiwy MS, An J, Yin M, Jia X, Guan Y, He F, et al. Cold plasma treatment and exogenous salicylic acid priming enhances salinity tolerance of Oryza sativa seedlings. Protoplasma. 2019;256(1):79–99. doi:10.1007/s00709-018-1279-0. [Google Scholar] [PubMed] [CrossRef]
80. Liu C, Mao B, Yuan D, Chu C, Duan M. Salt tolerance in rice: physiological responses and molecular mechanisms. Crop J. 2022;10(1):13–25. doi:10.1016/j.cj.2021.02.010. [Google Scholar] [CrossRef]
81. Khan MS, Akther T, Mubarak Ali D, Hemalatha S. An investigation on the role of salicylic acid alleviate the saline stress in rice crop (Oryza sativa (L)). Biocatal Agr Biotech. 2019;18:101027. doi:10.1016/j.bcab.2019.101027. [Google Scholar] [CrossRef]
82. Khan MTA, Yusuf M, Qazi F, Ahmad A. Brassinosteroids signalling: intervention with phytohormones and their relationship in plant adaptation to abiotic stresses. Singapore: Springer; 2022. [Google Scholar]
83. Amir R, Munir F, Khan M, Iqbal T. Use of plant hormones for the improvement of plant growth and production under salt stress. In: Salt stress, microbes, and plant interactions: causes and solution. Singapore: Springer; 2019. p. 59–90. [Google Scholar]
84. Ryu H, Cho Y-G. Plant hormones in salt stress tolerance. J Plant Biol. 2015;58(3):147–55. doi:10.1007/s12374-015-0103-z. [Google Scholar] [CrossRef]
85. Anuradha S, Seeta Ram Rao S. Effect of brassinosteroids on salinity stress induced inhibition of seed germination and seedling growth of rice (Oryza sativa L.). Plant Growth Regul. 2001;33:151–3. doi:10.1023/A:1017590108484. [Google Scholar] [CrossRef]
86. Reyes Guerrero Y, Martínez González L, Dell`Amico J, Núñez M, Pieters AJ. Reversion of deleterious effects of salt stress by activation of ROS detoxifying enzymes via foliar application of 24-epibrassinolide in rice seedlings. Theor Exp Plant Physiol. 2014;27(1):31–40. [Google Scholar]
87. Sharma I, Bhardwaj R, Pati P. Exogenous application of 28-homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa Basmati-1. J Plant Growth Regul. 2015;34(3):509–18. doi:10.1007/s00344-015-9486-9. [Google Scholar] [CrossRef]
88. Sharma I, Ching E, Saini S, Bhardwaj R, Pati PK. Exogenous application of brassinosteroid offers tolerance to salinity by altering stress responses in rice variety Pusa Basmati-1. Plant Physiol Bioch. 2013;69:17–26. doi:10.1016/j.plaphy.2013.04.013. [Google Scholar] [PubMed] [CrossRef]
89. Chen Y, Ge J, Liu Y, Li R, Zhang R, Li K, et al. 24-epibrassnolide alleviates the adverse effect of salinity on rice grain yield through enhanced antioxidant enzyme and improved K+/Na+ homeostasis. Agronomy. 2022;12(10):2499. doi:10.3390/agronomy12102499. [Google Scholar] [CrossRef]
90. Li L, Yin W, Niu M, Meng W, Zhang X, Tong H. Functional analysis of brassinosteroids in salt stress responses in rice. Chin Bull Bot. 2019;54(2):185. [Google Scholar]
91. Ruan J, Zhou Y, Zhou M, Yan J, Khurshid M, Weng W, et al. Jasmonic acid signaling pathway in plants. Int J Mol Sci. 2019;20(10):2479. doi:10.3390/ijms20102479. [Google Scholar] [PubMed] [CrossRef]
92. Kang DJ, Seo YJ, Lee JD, Ishii R, Kim K, Shin D, et al. Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J Agron Crop Sci. 2005;191(4):273–82. doi:10.1111/jac.2005.191.issue-4. [Google Scholar] [CrossRef]
93. Kurotani K, Hayashi K, Hatanaka S, Toda Y, Ogawa D, Ichikawa H, et al. Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol. 2015;56(4):779–89. doi:10.1093/pcp/pcv006. [Google Scholar] [PubMed] [CrossRef]
94. Hussain S, Zhang R, Liu S, Li R, Wang Y, Chen Y, et al. Methyl jasmonate alleviates the deleterious effects of salinity stress by augmenting antioxidant enzyme activity and ion homeostasis in rice (Oryza sativa L.). Agronomy. 2022;12(10):2343. doi:10.3390/agronomy12102343. [Google Scholar] [CrossRef]
95. Mahmud S, Sharmin S, Lal Das Ch B, Anowar Hos M. Effect of salinity and alleviating role of methyl jasmonate in some rice varieties. Asian J Plant Sci. 2017;16(2):87–93. doi:10.3923/ajps.2017.87.93. [Google Scholar] [CrossRef]
96. Wang J, Song L, Gong X, Xu J, Li M. Functions of jasmonic acid in plant regulation and response to abiotic stress. Int J Mol Sci. 2020;21(4):1446. doi:10.3390/ijms21041446. [Google Scholar] [PubMed] [CrossRef]
97. Ye H, Du H, Tang N, Li X, Xiong L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol. 2009;71(3):291–305. doi:10.1007/s11103-009-9524-8. [Google Scholar] [PubMed] [CrossRef]
98. Wu H, Ye H, Yao R, Zhang T, Xiong L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015;232:1–12. doi:10.1016/j.plantsci.2014.12.010. [Google Scholar] [PubMed] [CrossRef]
99. Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 2004;45(5):550–9. doi:10.1093/pcp/pch063. [Google Scholar] [PubMed] [CrossRef]
100. Trang Nguyen H, Thi Mai To H, Lebrun M, Bellafiore S, Champion A. Jasmonates-the master regulator of rice development, adaptation and defense. Plants. 2019;8(9):339. doi:10.3390/plants8090339. [Google Scholar] [PubMed] [CrossRef]
101. Peethambaran PK, Glenz R, Honinger S, Shahinul Islam SM, Hummel S, Harter K, et al. Salt-inducible expression of OsJAZ8 improves resilience against salt-stress. BMC Plant Biol. 2018;18(1):311. doi:10.1186/s12870-018-1521-0. [Google Scholar] [PubMed] [CrossRef]
102. Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015;20(4):219–29. doi:10.1016/j.tplants.2015.02.001. [Google Scholar] [PubMed] [CrossRef]
103. Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y, et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell. 2013;25(5):1709–25. doi:10.1105/tpc.113.112052. [Google Scholar] [PubMed] [CrossRef]
104. Ismail A, Seo M, Takebayashi Y, Kamiya Y, Eiche E, Nick P. Salt adaptation requires efficient fine-tuning of jasmonate signalling. Protoplasma. 2014;251(4):881–98. doi:10.1007/s00709-013-0591-y. [Google Scholar] [PubMed] [CrossRef]
105. Zhang M, Smith JA, Harberd NP, Jiang C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol Biol. 2016;91(6):651–9. doi:10.1007/s11103-016-0488-1. [Google Scholar] [PubMed] [CrossRef]
106. Riyazuddin R, Verma R, Singh K, Nisha N, Keisham M, Bhati KK, et al. Ethylene: a master regulator of salinity stress tolerance in plants. Biomolecules. 2020;10(6):959. doi:10.3390/biom10060959. [Google Scholar] [PubMed] [CrossRef]
107. Dewei M, Naijie F, Dianfeng Z, Hang Z, Minglong Y, Guanjie C. Ethylene alleviates salt stress damage to the root system of rice seedlings. Molec Plant Breed. 2024;22(7):2271–80. [Google Scholar]
108. Liang S, Xiong W, Yin C, Xie X, Jin YJ, Zhang S, et al. Overexpression of OsARD1 improves submergence, drought, and salt tolerances of seedling through the enhancement of ethylene synthesis in rice. Front Plant Sci. 2019;10:1088. doi:10.3389/fpls.2019.01088. [Google Scholar] [PubMed] [CrossRef]
109. Ji J, Yuan D, Jin C, Wang G, Li X, Guan C. Enhancement of growth and salt tolerance of rice seedlings (Oryza sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity. Acta Physiol Plant. 2020;42(4):1–17. [Google Scholar]
110. Li CH, Wang G, Zhao JL, Zhang LQ, Ai LF, Han YF, et al. The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell. 2014;26(6):2538–53. doi:10.1105/tpc.114.125187. [Google Scholar] [PubMed] [CrossRef]
111. Nadeem SM, Zahir ZA, Naveed M, Ashraf M. Microbial ACC-deaminase: prospects and applications for inducing salt tolerance in plants. Crit Rev Plant Sci. 2010;29(6):360–93. doi:10.1080/07352689.2010.524518. [Google Scholar] [CrossRef]
112. Jaemsaeng R, Jantasuriyarat C, Thamchaipenet A. Molecular interaction of 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing endophytic Streptomyces sp. GMKU 336 towards salt-stress resistance of Oryza sativa L. cv. KDML105. Sci Rep. 2018;8(1):1950. doi:10.1038/s41598-018-19799-9. [Google Scholar] [PubMed] [CrossRef]
113. Sarkar A, Ghosh PK, Pramanik K, Mitra S, Soren T, Pandey S, et al. A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res Microbiol. 2018;169(1):20–32. doi:10.1016/j.resmic.2017.08.005. [Google Scholar] [PubMed] [CrossRef]
114. Tao JJ, Chen HW, Ma B, Zhang WK, Chen SY, Zhang JS. The role of ethylene in plants under salinity stress. Front Plant Sci. 2015;6:1059. [Google Scholar] [PubMed]
115. Arnao MB, Hernandez-Ruiz J. Functions of melatonin in plants: a review. J Pineal Res. 2015;59(2):133–50. doi:10.1111/jpi.2015.59.issue-2. [Google Scholar] [CrossRef]
116. Arnao MB, Hernandez-Ruiz J. Melatonin: a new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019;24(1):38–48. doi:10.1016/j.tplants.2018.10.010. [Google Scholar] [PubMed] [CrossRef]
117. Yan F, Zhang J, Li W, Ding Y, Zhong Q, Xu X, et al. Exogenous melatonin alleviates salt stress by improving leaf photosynthesis in rice seedlings. Plant Physiol Biochem. 2021;163:367–75. doi:10.1016/j.plaphy.2021.03.058. [Google Scholar] [PubMed] [CrossRef]
118. Yan F, Wei H, Ding Y, Li W, Liu Z, Chen L, et al. Melatonin regulates antioxidant strategy in response to continuous salt stress in rice seedlings. Plant Physiol Biochem. 2021;165:239–50. doi:10.1016/j.plaphy.2021.05.003. [Google Scholar] [PubMed] [CrossRef]
119. Chen Y, Li R, Ge J, Liu J, Wang W, Xu M, et al. Exogenous melatonin confers enhanced salinity tolerance in rice by blocking the ROS burst and improving Na+/K+ homeostasis. Environ Exp Bot. 2021;189:104530. doi:10.1016/j.envexpbot.2021.104530. [Google Scholar] [CrossRef]
120. Huangfu L, Zhang Z, Zhou Y, Zhang E, Chen R, Fang H, et al. Integrated physiological, metabolomic and transcriptomic analyses provide insights into the roles of exogenous melatonin in promoting rice seed germination under salt stress. Plant Growth Regul. 2021;95(1):19–31. doi:10.1007/s10725-021-00721-9. [Google Scholar] [CrossRef]
121. Wei L, Zhao H, Wang B, Wu X, Lan R, Huang X, et al. Exogenous melatonin improves the growth of rice seedlings by regulating redox balance and ion homeostasis under salt stress. J Plant Growth Regul. 2021;41(6):2108–21. [Google Scholar]
122. Liang C, Zheng G, Li W, Wang Y, Hu B, Wang H, et al. Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J Pineal Res. 2015;59(1):91–101. doi:10.1111/jpi.2015.59.issue-1. [Google Scholar] [CrossRef]
123. Li X, Yu B, Cui Y, Yin Y. Melatonin application confers enhanced salt tolerance by regulating Na+ and Cl− accumulation in rice. Plant Growth Regul. 2017;83:441–54. doi:10.1007/s10725-017-0310-3. [Google Scholar] [CrossRef]
124. Liu J, Shabala S, Zhang J, Ma G, Chen D, Shabala L, et al. Melatonin improves rice salinity stress tolerance by NADPH oxidase-dependent control of the plasma membrane K+ transporters and K+ homeostasis. Plant Cell Environ. 2020;43(11):2591–605. doi:10.1111/pce.v43.11. [Google Scholar] [CrossRef]
125. Xie Z, Wang J, Wang W, Wang Y, Xu J, Li Z, et al. Integrated analysis of the transcriptome and metabolome revealed the molecular mechanisms underlying the enhanced salt tolerance of rice due to the application of exogenous melatonin. Front Plant Sci. 2021;11:618680. doi:10.3389/fpls.2020.618680. [Google Scholar] [PubMed] [CrossRef]
126. Khan Z, Jan R, Asif S, Farooq M, Jang Y-H, Kim E-G, et al. Exogenous melatonin induces salt and drought stress tolerance in rice by promoting plant growth and defense system. Sci Rep. 2024;14(1):1214. doi:10.1038/s41598-024-51369-0. [Google Scholar] [PubMed] [CrossRef]
127. Formentin E, Barizza E, Stevanato P, Falda M, Massa F, Tarkowskà D, et al. Fast regulation of hormone metabolism contributes to salt tolerance in rice (Oryza sativa spp. Japonica, L.) by inducing specific morpho-physiological responses. Plants. 2018;7(3):75. doi:10.3390/plants7030075. [Google Scholar] [PubMed] [CrossRef]
128. Choudhary P, Pramitha L, Rana S, Verma S, Aggarwal PR, Muthamilarasan M. Hormonal crosstalk in regulating salinity stress tolerance in graminaceous crops. Physiol Plantarum. 2021;173(4):1587–96. doi:10.1111/ppl.v173.4. [Google Scholar] [CrossRef]
129. Aizaz M, Lubna, Jan R, Asaf S, Bilal S, Kim K-M, et al. Regulatory dynamics of plant hormones and transcription factors under salt stress. Biology. 2024;13(9):673. doi:10.3390/biology13090673. [Google Scholar] [PubMed] [CrossRef]
130. Moons A, Prinsen E, Bauw G, Van Montagu M. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. The Plant Cell. 1997;9(12):2243–59. [Google Scholar] [PubMed]
131. Takeuchi K, Gyohda A, Tominaga M, Kawakatsu M, Hatakeyama A, Ishii N, et al. RSOsPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol. 2011;52(9):1686–96. doi:10.1093/pcp/pcr105. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools