 Open Access
Open Access
ARTICLE
The Maize WRKY Transcription Factor ZmWRKY25 Respond Drought Stress in Transgenic Tobacco
1 Joint International Research Laboratory of Modern Agricultural Technology, Ministry of Education, Jilin Agricultural University, Changchun, 130118, China
2 College of Agronomy, Jilin Agriculture Science and Technology College, Jilin, 132101, China
3 College of Agronomy, University of Jilin Agricultural, Changchun, 130118, China
4 Agricultural Biotechnology Research Institute, Jilin Academy of Agricultural Sciences (Northeast Innovation Center for Agricultural Science and Technology in China), Changchun, 130033, China
* Corresponding Author: Shuyan Guan. Email:
Phyton-International Journal of Experimental Botany 2024, 93(12), 3617-3635. https://doi.org/10.32604/phyton.2024.052704
Received 11 April 2024; Accepted 29 July 2024; Issue published 31 December 2024
Abstract
The WRKY transcription factors play important roles in various biological processes such as plant development, defense regulation, and stress response. Despite this, there is limited information available on drought-related WRKY genes in maize. Through RNA sequencing (RNA-seq) analysis, a WRKY transcription factor called ZmWRKY25 was identified in this study. ZmWRKY25 belongs to Group II and is localized in the nucleus. Protein interaction analysis revealed that ZmWRKY25 interacts with several proteins involved in the abscisic acid (ABA) signal pathway. Expression of ZmWRKY25 was found to be up-regulated in response to drought, salt, and ABA treatments in tobacco plants. Furthermore, over-expression of ZmWRKY25 in tobacco led to improved tolerance to drought by participating in the ABA signal pathway, regulating the reactive oxygen species (ROS) active oxygen scavenging system, and promoting root growth and development. These findings suggest that ZmWRKY25 plays a role in multiple abiotic stress response pathways and acts as a positive factor under drought stress. Overall, the study highlights the importance of ZmWRKY25 in enhancing plant resilience to drought through its involvement in key physiological pathways.Keywords
Supplementary Material
Supplementary Material FileMaize (Zea mays L.) is a significant food crop and industrial raw material with global germplasm diversity. Maize stands out for its high yield per unit area and straightforward planting and cultivation methods compared to other crops. Maize is a crop that demands a substantial amount of water and is extremely vulnerable to drought. Drought stress in corn is evident through stunted growth, early maturation, and potential sterility, ultimately leading to a notable decrease in yield [1]. Over 50% of China’s territory consists of arid and semi-arid regions [2], with an annual impact of drought on over 62% of crop cultivation [3,4]. Therefore, the identification and utilization of corn drought-resistant genetic resources are of great significance in coping with global climate change and solving the food crisis.
The WRKY transcription factors (TFs) play a significant role in plant biology, being essential for processes such as growth, development, and stress response [5]. In 1994, Ishiguro discovered the first WRKY gene, SPF1, from sweet potato cDNA [6]. Subsequent studies revealed the presence of WRKY family genes in various plant species, with the gene count increasing from lower to higher plants. For instance, Arabidopsis harbors 74 WRKY genes [7], Setaria italica possesses 105 WRKY genes [8], Oryza sativa contains 109 WRKY genes [9], maize encodes 136 WRKY genes [10], while soybean showcases 197 WRKY genes [11]. These TFs feature a distinct WRKYGQK motif at the N-terminal end with slight variations, along with a zinc-finger-like motif at the C-terminal end. These motifs are critical for the binding of WRKY TFs to W-boxes (TTGACT/C; cis-acting elements) [12]. Based on the number of WRKY domains and zinc finger structure characteristics, WRKY proteins can be categorized into three groups. Group I consists of proteins with two WRKY domains and C2H2 zinc finger structures, while Groups II and III have proteins with only one WRKY domain. Group III members typically have C2HC zinc finger structures, while Group II members have C2H2 structures [13]. In plants, the WRKY factor plays a crucial role in regulating the transcriptional mechanism by interacting with other components. This interaction leads to the specific temporal and spatial expression of certain genes, ensuring the correct cellular response to internal and external stressors. This response is essential for plants to maintain basic growth functions across varying levels of water and land environments [14]. The swift activation of the WRKY family plays a crucial role in enhancing plant adaptability and establishing a more efficient signal transduction network, leading to improved plant resistance to stress. Many WRKY family genes with drought-resistance capabilities have been identified in various plant species [15].
In a study focusing on white pears, researchers observed that forty-four PbWRKY genes were upregulated in response to drought treatment [16]. Similarly, in Panicum miliaceum, the expression of ten PmWRKY genes was induced under drought stress [17]. Cai conducted a study where the ZmWRKY58 gene was introduced into rice plants to improve their drought resistance and salt tolerance. Additionally, the heterologous expression of OsWRKY45 from rice led to increased drought resistance in Arabidopsis thaliana, while the heterologous expression of OsWRKY8 in Arabidopsis enhanced osmotic tolerance [18]. The heterologous expression of OsWRKY72 in rice plants was shown to influence root growth and stress tolerance levels [19]. Li successfully transferred the ZmWRKY33 gene into Arabidopsis thaliana, demonstrating its ability to confer drought resistance, salt tolerance, and cold resistance [20]. Liu also introduced the ZmWRKY33 gene into rice plants, confirming its functions in drought resistance, salt tolerance, and low-temperature stress [21]. Moreover, in transgenic tobacco, GhWRKY41/SpWRKY1 was found to enhance drought and salt resistance by regulating reactive oxygen species levels and stomatal conductance [22]. Under normal circumstances, plants will accumulate a large amount of reactive oxygen species after experiencing drought stress. These reactive oxygen species can harm the cell membrane structure and increase the presence of malondialdehyde in the cell membrane components. This damage to the cell membrane can cause cell rupture, leading to a decrease in plant conductivity. If the levels of reactive oxygen species are high, it can result in extensive cell damage, causing yellowing, wilting, and loss of chlorophyll in the leaves. Peroxidase (POD) and SOD exhibit robust activity in drought-tolerant plants, effectively eliminating reactive oxygen species and safeguarding the integrity of the cell membrane. Additionally, the proline content in drought-tolerant plants tends to increase. Therefore, the enzyme activities of POD and SOD, malondialdehyde levels, proline content, chlorophyll levels, and relative conductivity are crucial physiological indicators for assessing the drought resistance of plants. In addition, plants will force their roots to grow in the direction of water sources after suffering drought stress, and well-developed roots are conducive to the plant’s absorption and utilization of water. Therefore, the degree of plant root development is also an important indicator of a plant’s drought tolerance. In this study, the ZmWRKY25 gene was expressed in tobacco and the seed germination rate, POD and SOD enzyme activities, malondialdehyde, proline, chlorophyll content, relative conductivity, root development status, and other indicators were detected under drought stress to verify the effectiveness of ZmWRKY25. Drought resistance function, combined with bioinformatics analysis and related gene expression analysis, preliminarily explored the drought resistance molecular mechanism of ZmWRKY25.
2.1 Transcriptome Sequncing and Bioinformatics Analysis
The leaves of maize drought-tolerant inbred line H8186 and control inbred line B73 were taken at the three-leaf stage and sequenced. The sequencing was entrusted to Mega Biotech. The transcriptome data is not yet public. Data is stored in 10.6084/m9.figshare.12309998.
Ninety Arabidopsis WRKY genes, one hundred twenty-four Maize WRKY genes, and one hundred sixteen Millet WRKY genes were aligned using ClustalW in MEGA 7.0 [23]. A neighbor-joining (NJ) phylogenetic tree was then constructed based on conserved residues with 1000 replicates. In addition, potential WRKY domains including WRKY domain and plant zinc cluster domain were detected using Pfam (https://www.ebi.ac.uk/interpro/) (accessed on 02 April 2024). The ZmWRKY25 protein sequence was fed into the online analysis software String (https://string-db.org/) (accessed on 02 April 2024) for network analysis. In order to further investigate the transcriptional regulation mechanism of ZmWRKY25, the promoter region of ZmWRKY25 was predicted as reported by Zhao et al. [24].
2.2 Test Materials and Reagents
Maize inbred line B73, Tobacco NC89 and nicotiana benthamiana, Escherichia coli strain E. coli DH5α, agrobacterium tumefaciens strain EHA105, Cloning vector pMD18-T, expression vector pCAMBIA3301-Bar, expression vector pCAMBIA1302-GFP, by the Jilin Agricultural University Plant Biotechnology Center Laboratory to provide and save. PCR Kit, Restriction endonucleases (BglII, BstEII, NcoI) were purchased from Thermo Fisher Scientific (Shanghai, China); Construction vector was cloned from Clone Smarter with a seamless cloning kit. The Plant Genome Extraction Kit was purchased from Kang for Century, Primers (Table S1), synthesized by Kumei Corporation.
2.3 Expression Patterns of ZmWRKY25 in Maize
The expression patterns of ZmWRKY25 were analyzed using qRT-PCR with a primer specifically designed for this gene. The study aimed to investigate the expression levels of ZmWRKY25 under various stress conditions, including drought, salt, and ABA. The experimental design and data analysis followed the methodology detailed by Chen et al. [25]. Maize inbred line B73 at the three-leaf stage was utilized to investigate the expression patterns of ZmWRKY25. The seedlings were subjected to various stress treatments—drought treatment using PEG6000 (10%), ABA (120 μM) treatment, and salt stress using NaCl (260 mM). The expression levels of ZmWRKY25 were assessed by collecting leaves at different time points (0, 1, 3, 6, 12 h) during the treatments. Additionally, a tissue-specific expression analysis was performed by sampling roots, stems, and leaves of maize inbred line B73 to examine the expression of ZmWRKY25 in different tissues.
2.4 Cloning of Maize ZmWRKY25 Gene
In this study, following Lagace M’s method of material stress treatment, maize roots were washed and then immersed in a 25% PEG 6000 aqueous solution for 2 h [26]. Total RNA was extracted from young and vigorous maize leaves, followed by reverse transcription into cDNA. Specific primers with homologous arm sequences were designed based on the sequence of the ZmWRKY25 gene in GenBank (accession number NM_001158417) and the expression vector p3301-Bar sequence (Table S1). The cDNA of maize inbred line B73 was used as a template for amplifying the ZmWRKY25 gene coding region sequence. The PCR reaction product was ligated with the cloning vector pMD18-T, then transferred to E. coli DH5α competent cells. These cells were plated on LB-AMP solid medium and cultured overnight, and a single colony was picked. Bacterial plasmids were extracted, and the plasmid was confirmed by PCR before being sent to Changchun Kumei Co. (Changchun, China) for sequencing.
2.5 Construction and Identification of Recombinant Plant Expression Vector
Utilizing the principle of homologous recombination with a seamless kit, the plasmid pMD-18T-ZmWRKY25 served as the template for amplifying the ZmWRKY25 gene using specific primers (Table S1). Subsequently, the expression vector p3301-Bar was subjected to digestion with the restriction endonucleases BglII and BstEII at 37°C for 2 h. The purified gene fragments and the large fragment of the digested product were then recovered. Following the instructions of the seamless cloning connection kit, the recombinant plant expression vector p3301-ZmWRKY25-Bar was constructed. The identity of the plasmid was confirmed through PCR and double digestion before being sent to Changchun Kumei Co., Ltd. (China) for sequencing. In order to determine the subcellular localization of ZmWRKY25, we constructed the pCAMBIA1302-ZmWRKY25-GFP vector. The coding region of ZmWRKY25 was inserted downstream of the 35s promoter in the pCAMBIA1302 vector. The construction process for the pCAMBIA1302-ZmWRKY25-GFP vector followed the same protocol as the pCAMBIA3301 expression vector, involving digestion with BglII and NcoI.
2.6 Genetic Transformation of ZmWRKY25 Gene in Tobacco
The target gene was successfully introduced into tobacco plants using Agrobacterium tumefaciens. For details on the selection and disinfection methods of wild-type tobacco NC89 seeds, refer to the protocol for Agrobacterium tumefaciens-mediated transformation of tobacco leaves and subsequent transformation of tobacco tissue. Additionally, positive plants were selected by culturing transformed seeds on MS medium with 10 mg/mL Bar. Three T3 generation overexpression lines (OE-ZmWRKY25-1, OE-ZmWRKY25-2, OE-ZmWRKY25-3) showing increased expression of ZmWRKY25 were identified through qRT-PCR analysis for further study. For information on tobacco transient transformation, see the work by Huo et al. [27].
2.7 Transgenic Tobacco Phenotype under Stress Treatment
Seed germination rate and root growth experiments were conducted to analyze the impact of drought stress on the phenotype of tobacco plants. Tobacco seeds, including both wild-type (WT) and transgenic varieties, were cultured on MS medium containing either 5% or 10% PEG6000 at 26°C for a duration of one week. To determine root growth, 7-day-old seedlings were moved to MS medium containing 10% PEG6000 for another week, and the tap root length was measured. After 13 days in the same conditions, the survival rates of both transgenic and WT tobacco plants were recorded. The seedling roots after drought stress were washed to remove the medium and put into a root scanner (Epson Perfection V800 Photo) to measure the root phenotype. For the drought treatment, 10-day-old transgenic and WT tobacco seedlings were transferred to soil. After three weeks of incubation at 40% humidity and 26°C (16 h light/8 h dark), watering was stopped for 13 days. The water loss rate and other physiological and biochemical indicators were measured for three periods (before the drought treatment, 13 days after the drought treatment and three days after rewatering). The root morphology of tobacco after 13 days of drought treatment was scanned with a root scanner. All drought stress experiments were repeated 3 times.
2.8 Expression Profile of ABA Signal Pathway Genes
To elucidate the potential molecular mechanism of ZmWRKY25 in the ABA signaling pathway, we utilized qRT-PCR to evaluate the expression levels of NtMYB4a, NtDREB4, NtABF, and NtERF105 genes in transgenic tobacco and wild type plants under normal conditions. The specific primers used for this analysis can be located in Table S1.
2.9 Determination of Physiological and Biochemical Indicators
The peroxidase activity (POD), superoxide dismutase activity (SOD), and malondialdehyde levels (MDA), proline and chlorophyll content (Pro and Chl), as well as relative conductivity (REC) and relative water content (RWC) in the leaves of both transgenic plants and control plants were measured following the protocols outlined by Qin et al. [28] and Zhang [29]. These parameters were assessed at 0, 3, and 13 days post rehydration.
3.1 Transcriptome Sequencing Analysis
In order to identify genes in maize that respond to drought stress, the study halted watering three-leaf seedlings for a period of 7 days and examined the changes in transcription levels through transcriptome sequencing analysis. The results indicated significant alterations in the expression of numerous genes following the drought treatment. These differentially expressed genes were categorized into functional groups using Gene Ontology (GO) and further analyzed to determine the pathways they may be associated with by referencing the KEGG database. As a result, the gene ZmWRKY25 was singled out from the pool of DEGs for additional investigation (Fig. S1). The results of qRT-PCR verification for differential gene expression in the transcriptome are presented in Fig. S2.
3.2 In Bioinformatics Analysis of ZmWRKY 25
Phylogenetic trees show that ZmWRKY25 with AtWRKY11, 17, 15, 39, 74, 21 are phylogenetically close (Fig. 1). A comprehensive analysis of protein sequences was conducted to investigate the correlation between phylogeny and sequence structure among WRKY members. The study revealed a close phylogenetic relationship between ZmWRKY25, AtWRKY11, and AtWRKY17.
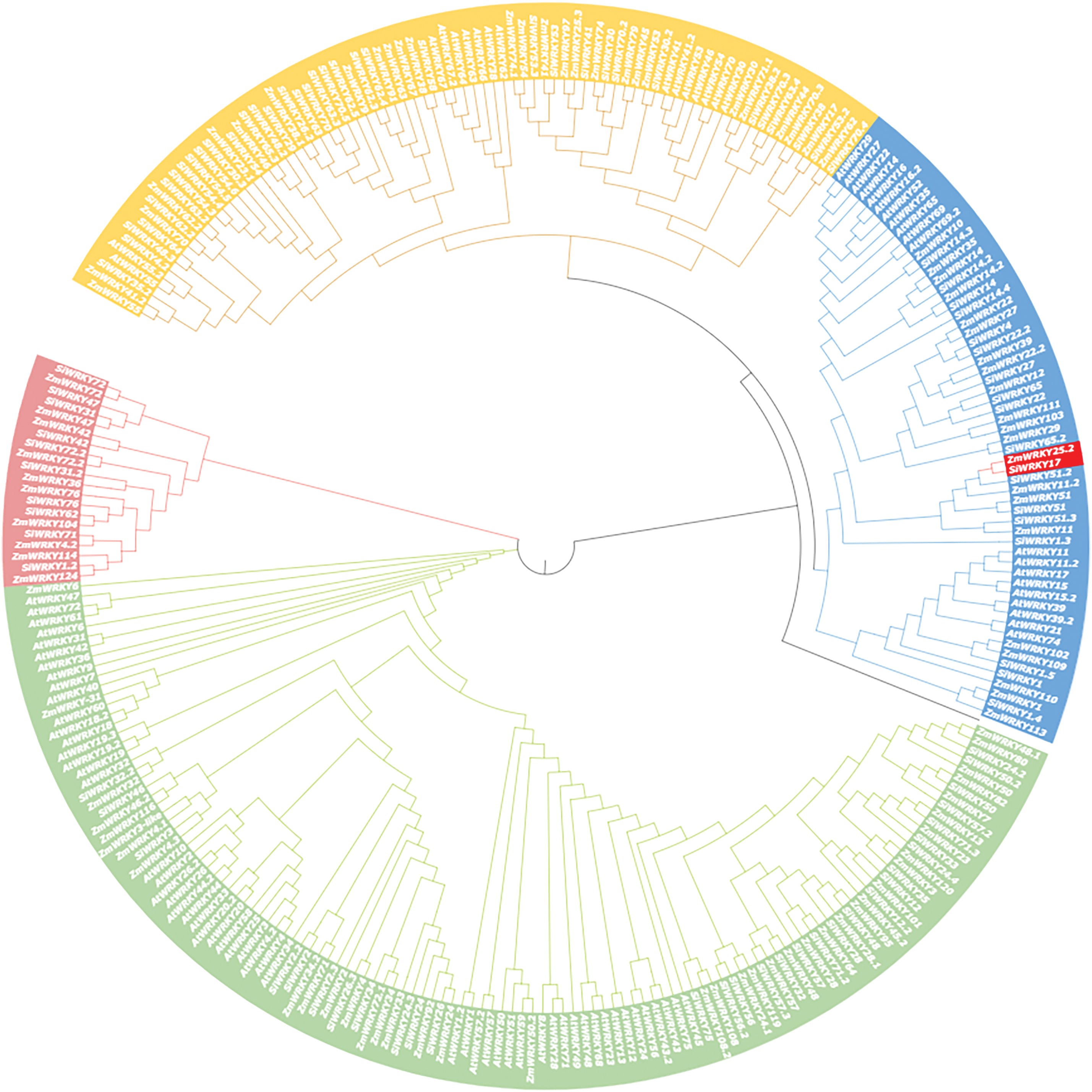
Figure 1: Phylogenetic analysis based on protein sequence. Whereas the red represent ZmWRKY25 and SiWRKY17, other color represent other member of WRKY family
The WRKY domain sequences of WRKY IIb were analyzed by comparing multiple sequences and conducting conserved domain analysis (Fig. 2). Transcription-factor gene families generally contain one or more highly conserved domains that play a role in DNA binding. It is important to note that sequence similarity may decrease beyond these crucial domains (Fig. 2). The monophyletic group formed by AtWRKY11 and AtWRKY17 (Fig. 2) shares a common ancestor with ZmWRKY25, which is supported by the presence of highly conserved motif patterns.
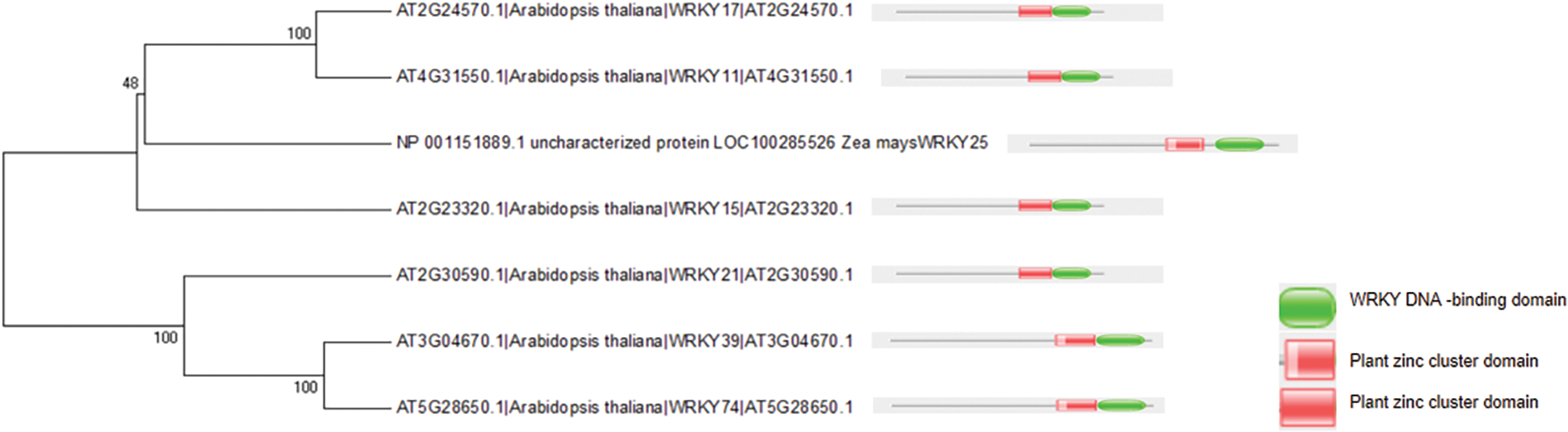
Figure 2: Multiple sequence alignment. Multiple sequence alignment of WRKY domain of ZmWRKY25 and AtWRKY17,11,15,21,39,74
Through protein interaction analysis(Fig. 3, Table S2), ZmWRKY25 protein is mainly associated with EREB61, Ethylene-responsive transcription factor ERF105 interaction. Both EREB61 and ERF105 are involved in the ABA signaling pathway.
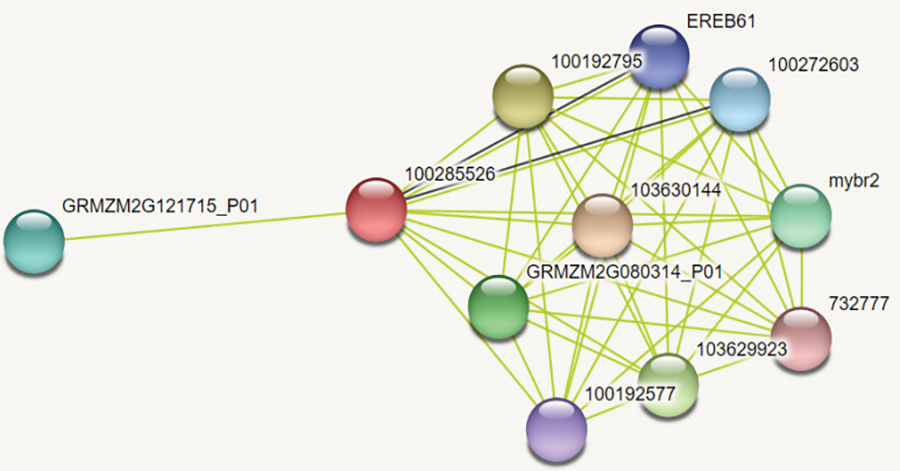
Figure 3: protein interaction analysis. GRMZM2G080314_P01:Protein BREAST CANCER SUSCEPTIBILITY 1-like protein,100285526: ZmWRKY25, 103630144: ATP-dependent6-phosphofructo kinase, 100192795: Transcription initiation factor IIE subunit beta, 103629923: Glycosyltransferase, 100192577: Putative HLH DNA-binding domain superfamily protein isoform 1, GRMZM2G121715_P01: Receptor protein kinase TMK1, 100272603: Ethylene-responsive transcription factor ERF105, mybr2: Putative MYB DNA-binding domain superfamily protein, 732777: Pi starvation-induced transcription factor1
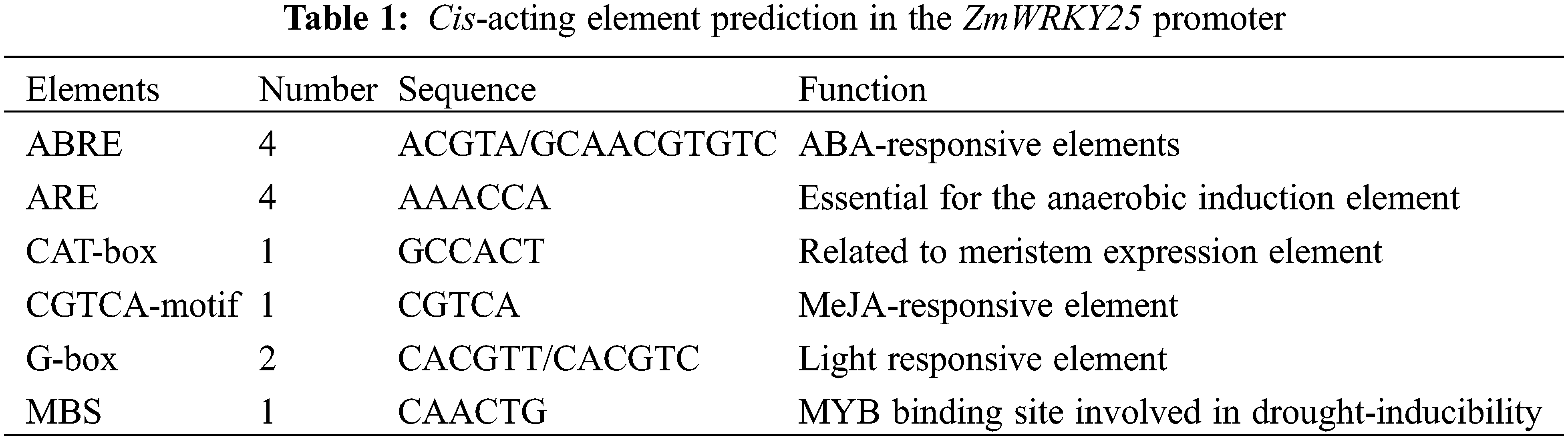
The cis-regulatory element of the ZmWRKY25 gene was identified using the plantCARE online database (available online: http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 02 April 2024). (Table 1). Analysis of Table 1 reveals that the ZmWRKY25 promoter region contains multiple abiotic stress-related elements, such as the MYB binding site involved in drought inducibility (MBS), ABA-responsive elements (ABRE), and the MeJA-responsive element (CGTCA-motif), suggesting that ZmWRKY25 may play a role in drought stress response.
3.3 Verification Results of ZmWRKY25 Cloning and Expression Vector Construction
To confirm the successful construction of the cloning vector and expression vector, we utilized PCR and sequencing techniques for validation. The cloning outcomes of the target gene are depicted in Fig. S2. The RNA extraction, target gene cDNA, and PCR outcomes of the cloning vector exhibit clear and accurate band positions, confirming the successful construction of the cloning vector. The PCR, double restriction enzyme digestion, and sequencing results of the target genes in the pCAMBIA3301 and pCAMBIA1302 plant expression vectors indicated successful vector construction (Figs. S2–S6). Following Agrobacterium-mediated genetic transformation, a total of 6 transgenic lines were obtained in the T3 generation. The qPCR expression detection results of the positive strains are presented in the figure, revealing significantly elevated expression levels of ZmWRKY25 in strains 1, 2, and 3 compared to the control (Figs. S7 and S8).
3.4 ZmWRKY25 Protein Was Localized in the Nucleus
The subcellular localization of the ZmWRKY25 fusion protein was investigated through transient transformation of ZmWRKY25-green fluorescent protein (GFP) recombinant into Nicotiana benthamiana leaves, with the pCambia1302-GFP vector as a control. Fig. 4 clearly shows that the ZmWRKY25-GFP fusion protein is specifically localized in the nucleus, while the pCambia1302-GFP control is distributed throughout the entire cell. These results indicate that the ZmWRKY25-GFP fusion protein has a distinct nuclear localization, suggesting potential interactions with other nuclear proteins or involvement in gene expression regulation within the nucleus. Understanding this subcellular localization is essential for elucidating the function and behavior of ZmWRKY25.
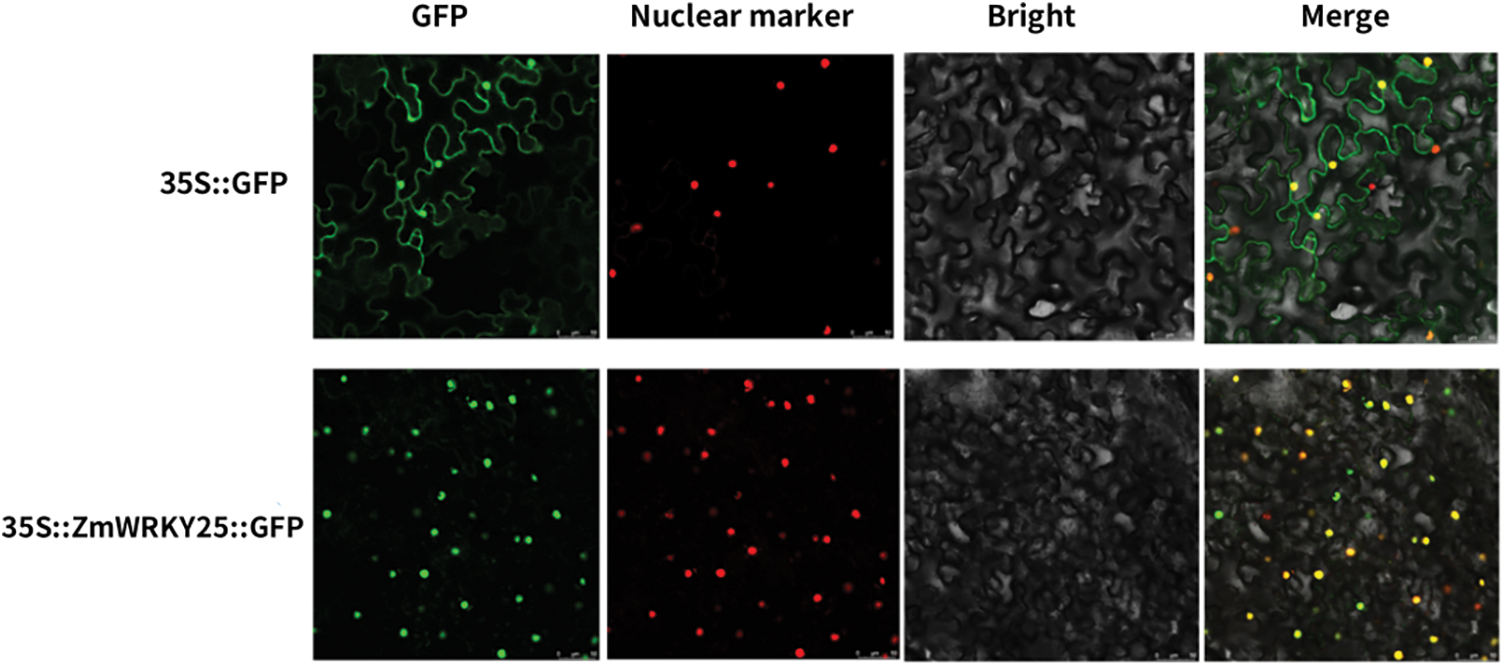
Figure 4: Subcellular localization of ZmWRKY25. ZmWRKY25-green fluorescent protein (GFP) recombinant and pCambia1302GFP control vector were transiently expressed in tobacoo mesophyll cells (Scale bars = 50 μm)
3.5 Expression Patterns of ZmWRKY25 in Maize
The tissue-specific expression of ZmWRKY25 in maize was analyzed using qPCR across roots, stems, and leaves. ZmWRKY25 was mainly expressed in leaves, with lower expression levels observed in roots and stems. Furthermore, the study investigated the effects of drought, salt, and ABA stress on ZmWRKY25 expression patterns. Following ABA treatment, ZmWRKY25 showed its highest expression level after 1 h. Drought stress induced a peak in ZmWRKY25 expression at 3 h, followed by a decrease. Salt stress caused a 2-fold increase in ZmWRKY25 expression at 1 h, reaching its peak at 3 h (Fig. 5A–D).
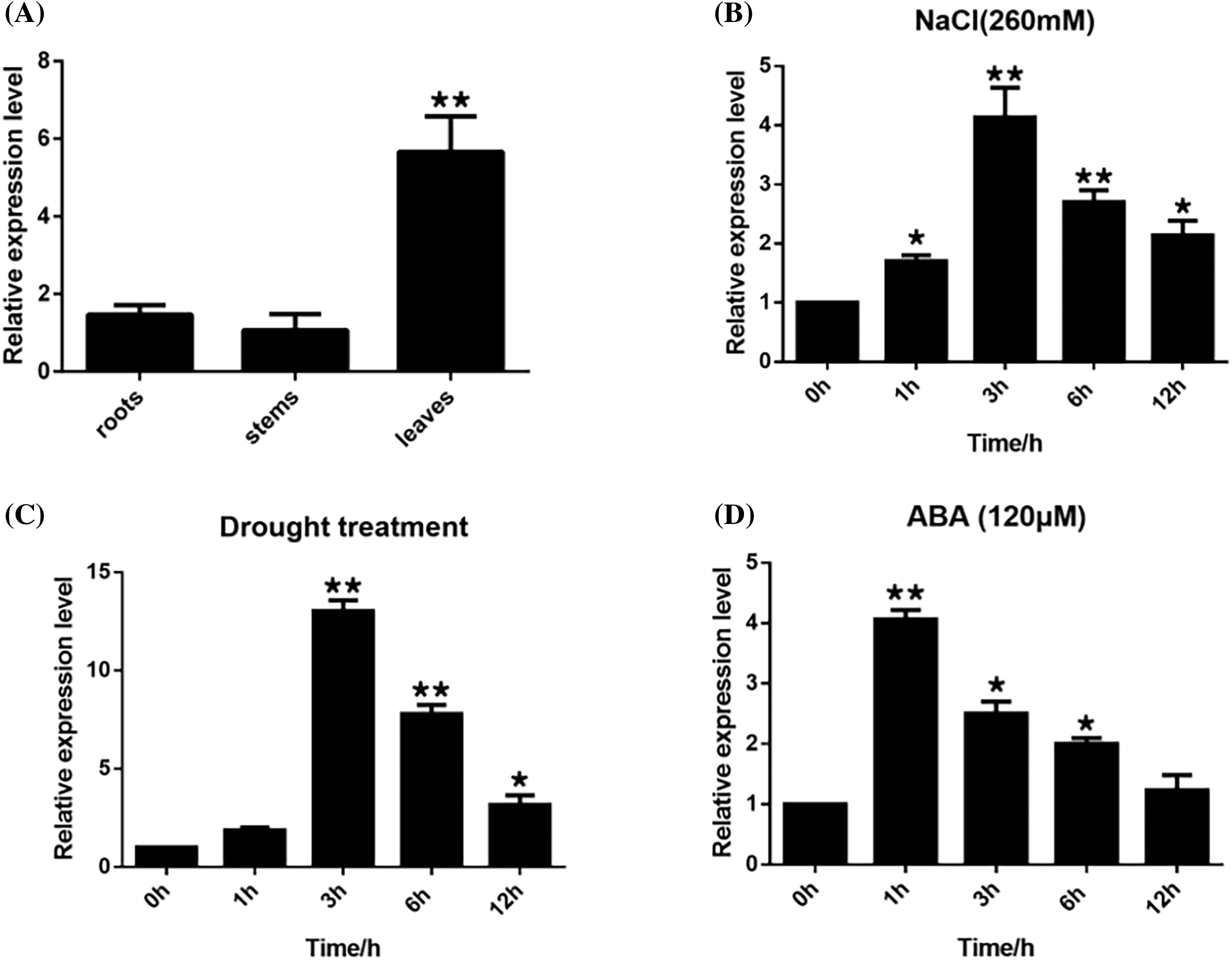
Figure 5: Tissue-specific expression of ZmWRKY25 under drought in maze at roots, steam and leaves (A), Expression patterns of ZmWRKY25 under NaCl (B), Drought (C), exogenous ABA (D). The vertical axes indicate fold changes, while the horizontal axes denote treatment durations. The error bars illustrate standard deviations (SD). The data presented reflect means ± SD from three biological replicates. ABA stands for abscisic acid. Asterisks indicating significant differences. * Significant at 0.05, p < 0.05; ** Significant at 0.01, p < 0.01
3.6 ZmWRKY25 Enhanced Drought Tolerance in Transgenic Plants
WT as a control for genetically modified tobacco, three T3 transgenic tobacco lines that overexpress ZmWRKY25 were selected for analysis. Under normal growth conditions, no significant difference in seed germination rates was observed between wild-type (WT) and transgenic plants. However, when exposed to 5% and 10% PEG6000 stress, the seed germination rate of transgenic plants showed a notable increase compared to WT plants (Fig. 6). There was minimal variation in total root length between the transgenic lines and WT plants. Notably, after a seven-day cultivation period under 10% PEG6000 stress, the taproot lengths of transgenic lines surpassed those of WT plants (Fig. 7). Phenotypic analysis of roots at the seedling stage using a root scanner indicated that the root surface area, total root projected area, root volume, and average root diameter of transgenic plants were significantly larger than those of WT seedlings. Moreover, the total number of root tips and branches in the transgenic lines were notably higher than in WT seedlings (Fig. 8). At the seedling stage, there were no major differences in growth between transgenic plants and WT plants. However, following drought treatment, the transgenic lines showed enhanced resilience compared to WT plants, with a lower water loss rate and a more developed root system (Fig. 9).
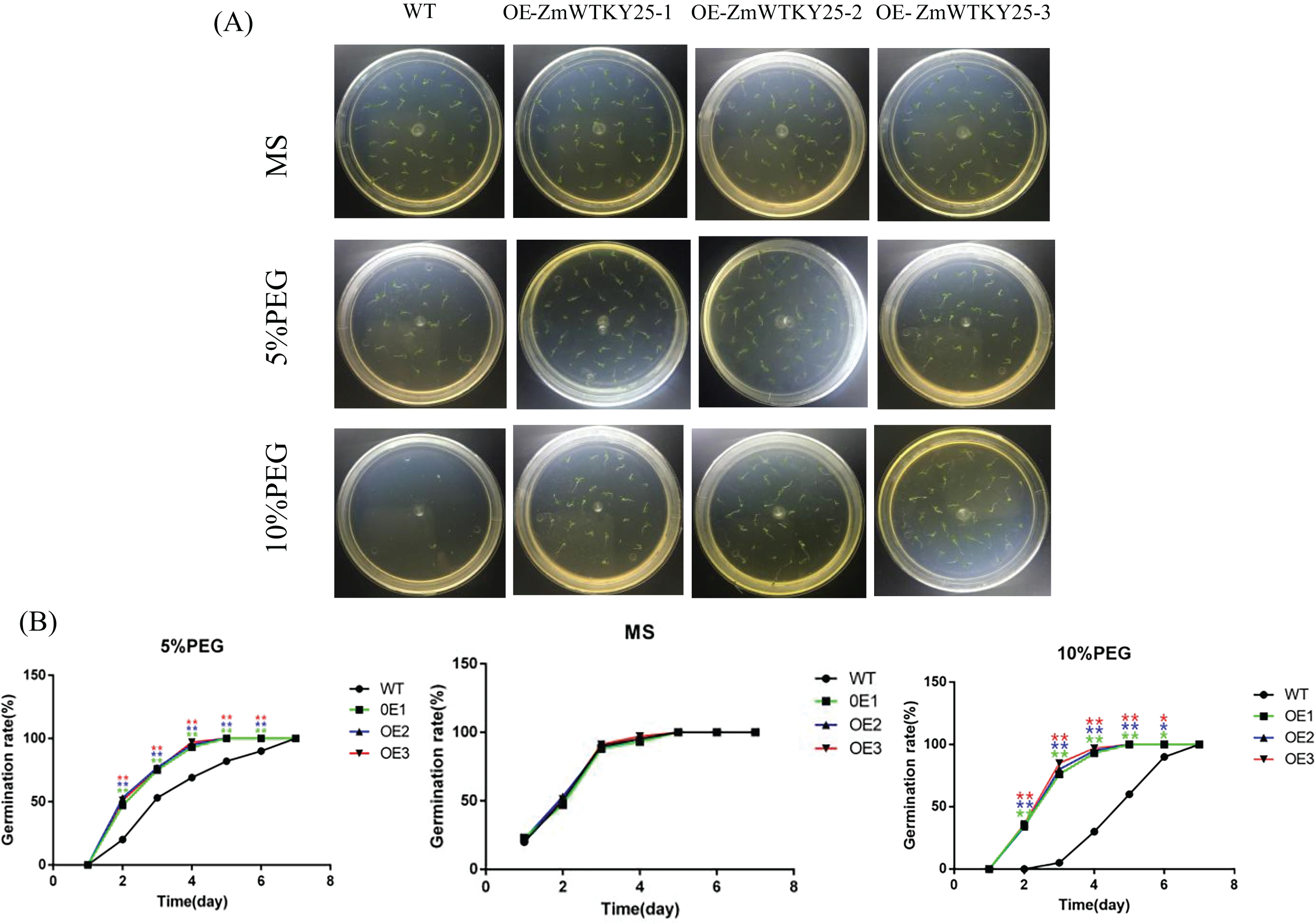
Figure 6: Germ Stress. Seeds from three distinct transgenic lines containing ZmWRKY25 were cultivated on MS medium supplemented with either 5% or 10% PEG6000. (A) The germination rates of seeds grown on standard MS medium compared to those on MS medium with 5% or 10% PEG6000 were observed over a period of 3 days. (B) All data presented represent the means ± standard deviations (SDs) from three independent biological replicates, with asterisks (* or **) indicating significant differences at p < 0.05 or p < 0.01, as determined by Student’s t-test. WT refers to the wild-type
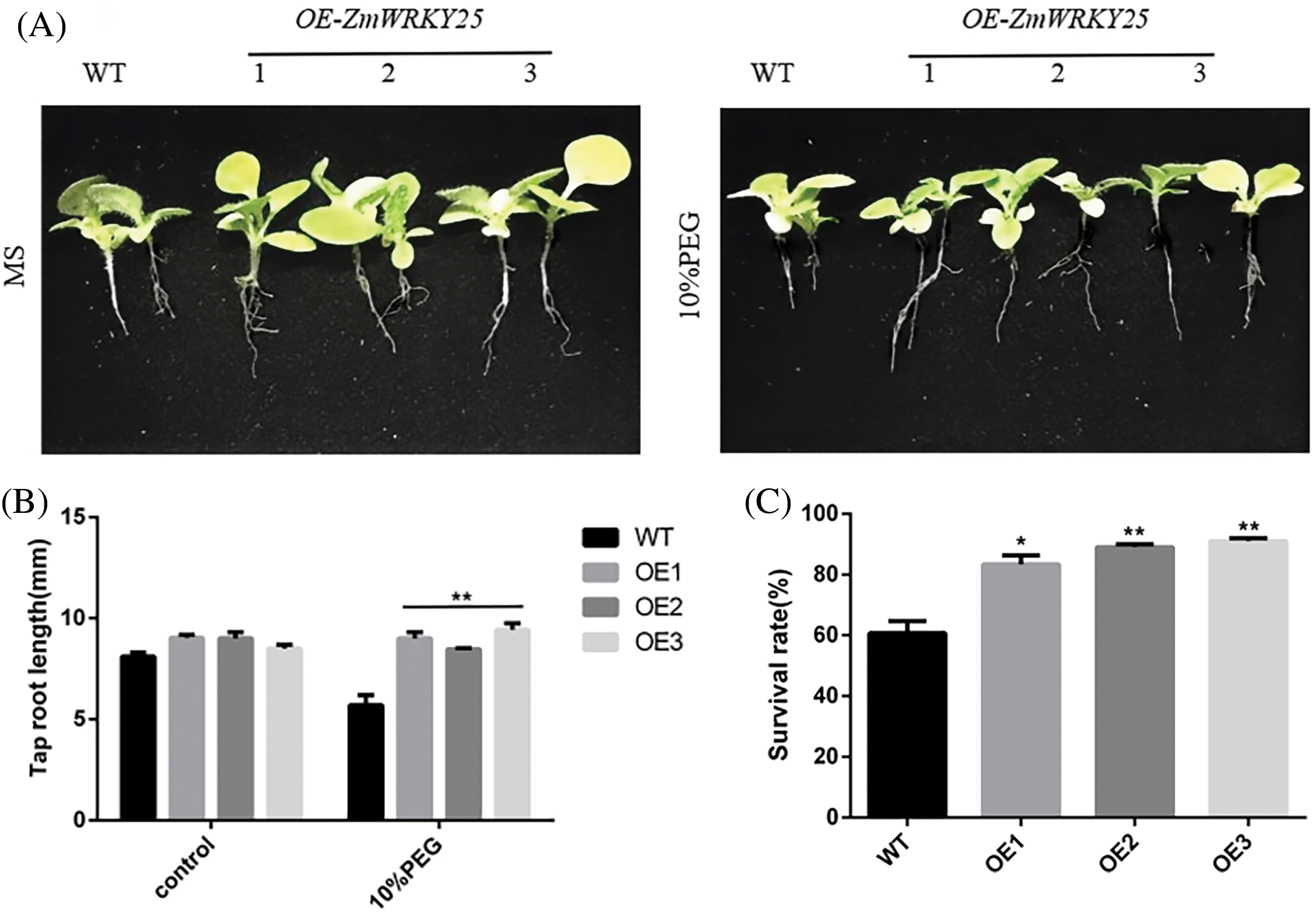
Figure 7: Tap root lengths and survival rates of transgenic Tobacco lines and WT under drought stress. Seven-day-old Tobacco seeding were planted on MS medium with or without 10% PEG6000 for 7 days (A). The tap root length measured (B). The survival rate of WT and transgenic plants were planted MS medium with 10%PEG6000 for 13 days (C). The data are expressed as means ± SD from three separate experiments, with asterisks (* or **) indicating significant differences at p < 0.05 or p < 0.01, respectively, according to the Student’s t-test
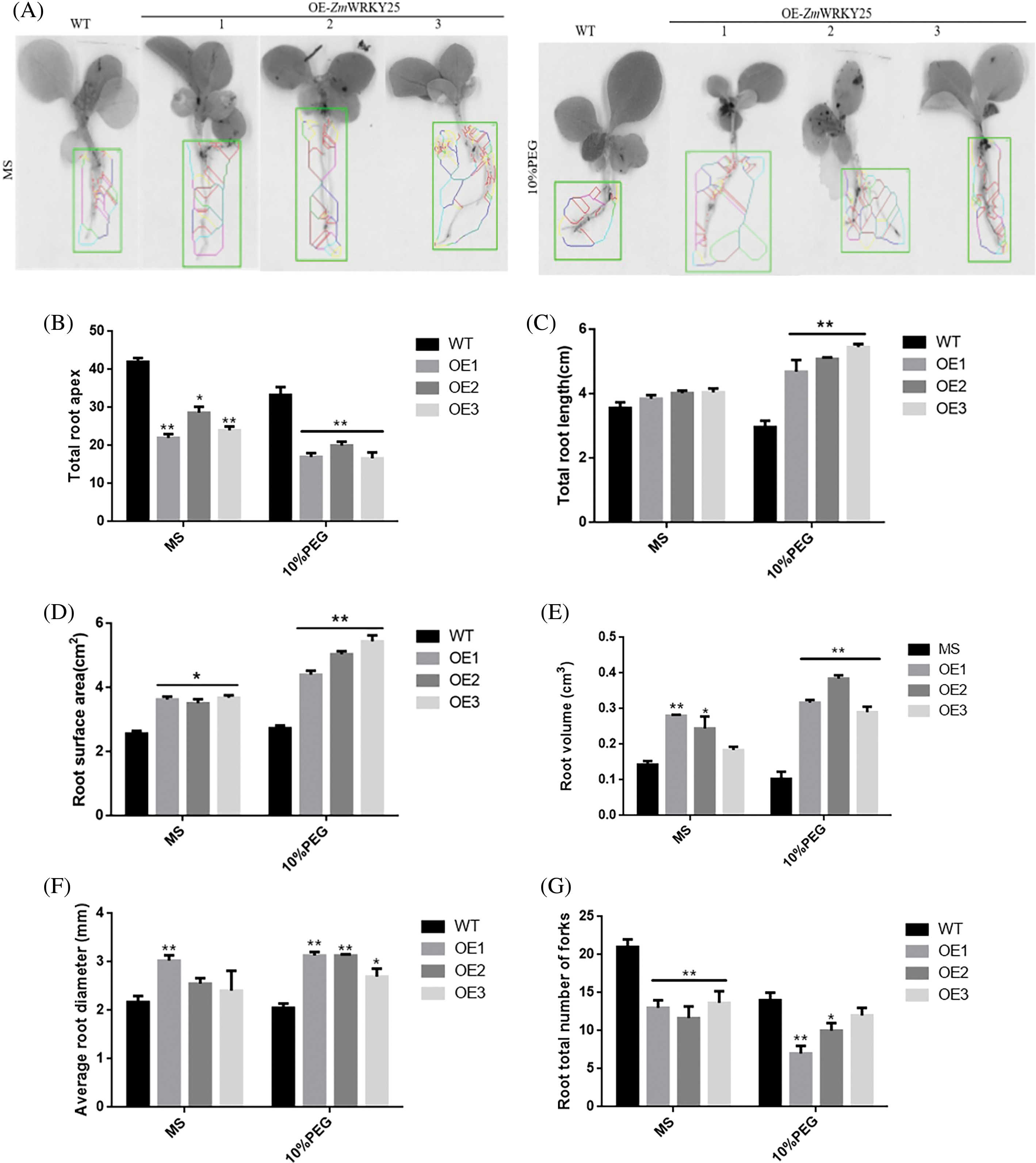
Figure 8: Analysis of root phenotypes in transgenic Tobacco lines and wild type (WT) under conditions of drought stress was conducted. Tobacco seedlings, aged seven days, were cultivated in MS medium supplemented with or without 10% PEG6000 for a duration of seven days (A). Measurements were taken for the total number of root apices (B), overall root length (C), surface area of the root (D), volume of the root (E), average diameter of the root (F), and total fork count of the roots (G). The data presented are means ± SD from three separate experiments, with asterisks (* or **) indicating significant differences at p < 0.05 or p < 0.01, respectively (as determined by Student’s t-test)
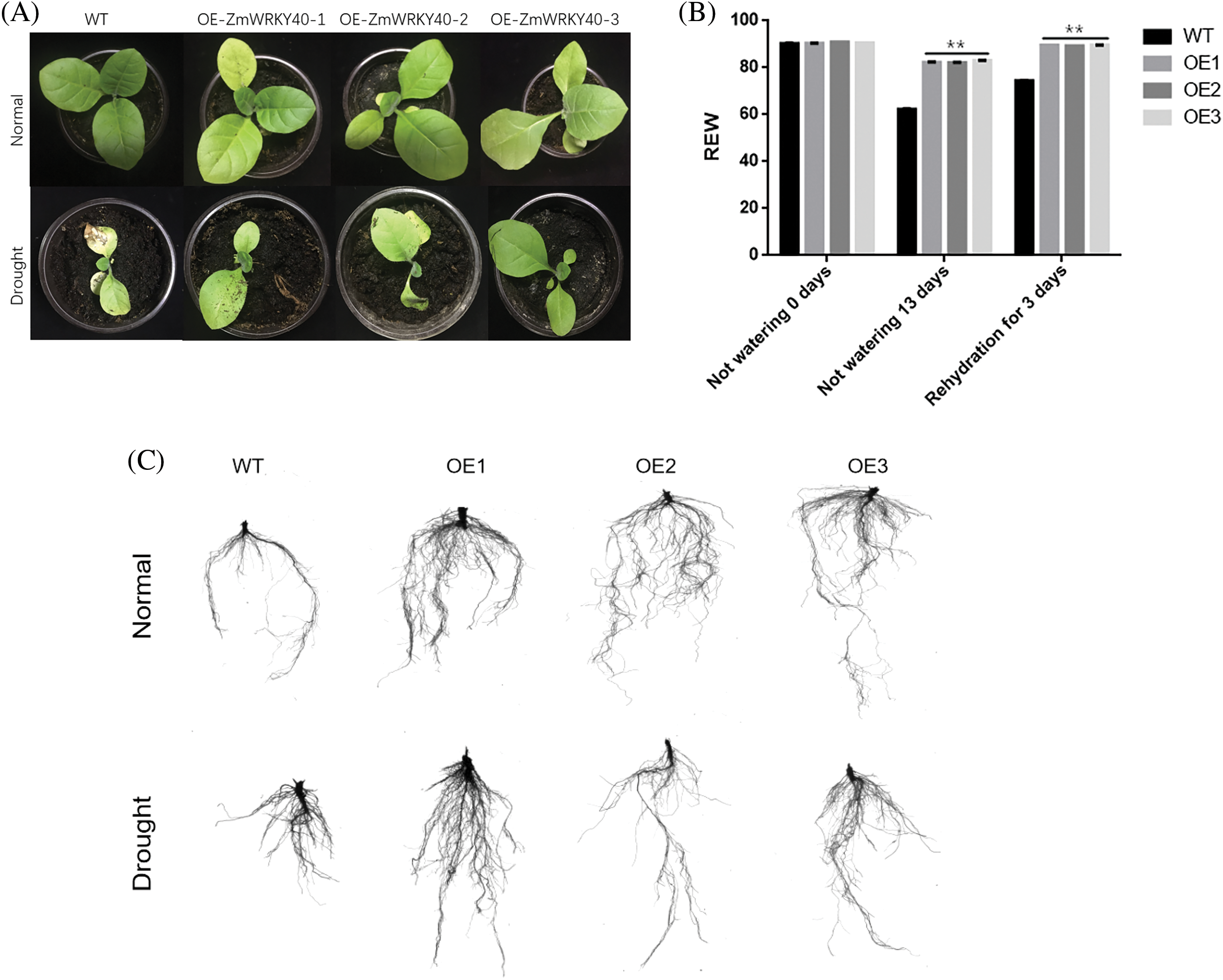
Figure 9: Analysis of phenotype and water loss in transgenic tobacco and wild type (WT) plants subjected to drought treatment. The phenotypic responses of WT and transgenic tobacco were assessed 13 days post-rewatering (A). The rate of water loss in both WT and transgenic tobacco plants during drought conditions was measured (B). Additionally, root phenotype assessments for WT and transgenic tobacco were conducted 13 days after rewatering (C). All data represent means ± SD from three independent experiments, with asterisks (**) indicating significant differences at p < 0.01 (Student’s t-test)
3.7 ZmWRKY25 Chenges the Expression of Stress-Responsive Gene
To investigate the molecular mechanism of ZmWRKY25 stress response, the study examined the gene expression levels of stress response in transgenic and negative controls under normal conditions (see Fig. 10). The results revealed that the expression levels of stress-responsive genes NtMYB4a, NtABF, NtDREB4, and NtERF105 were more than three times higher in transgenic Tobacco compared to the wild-type.
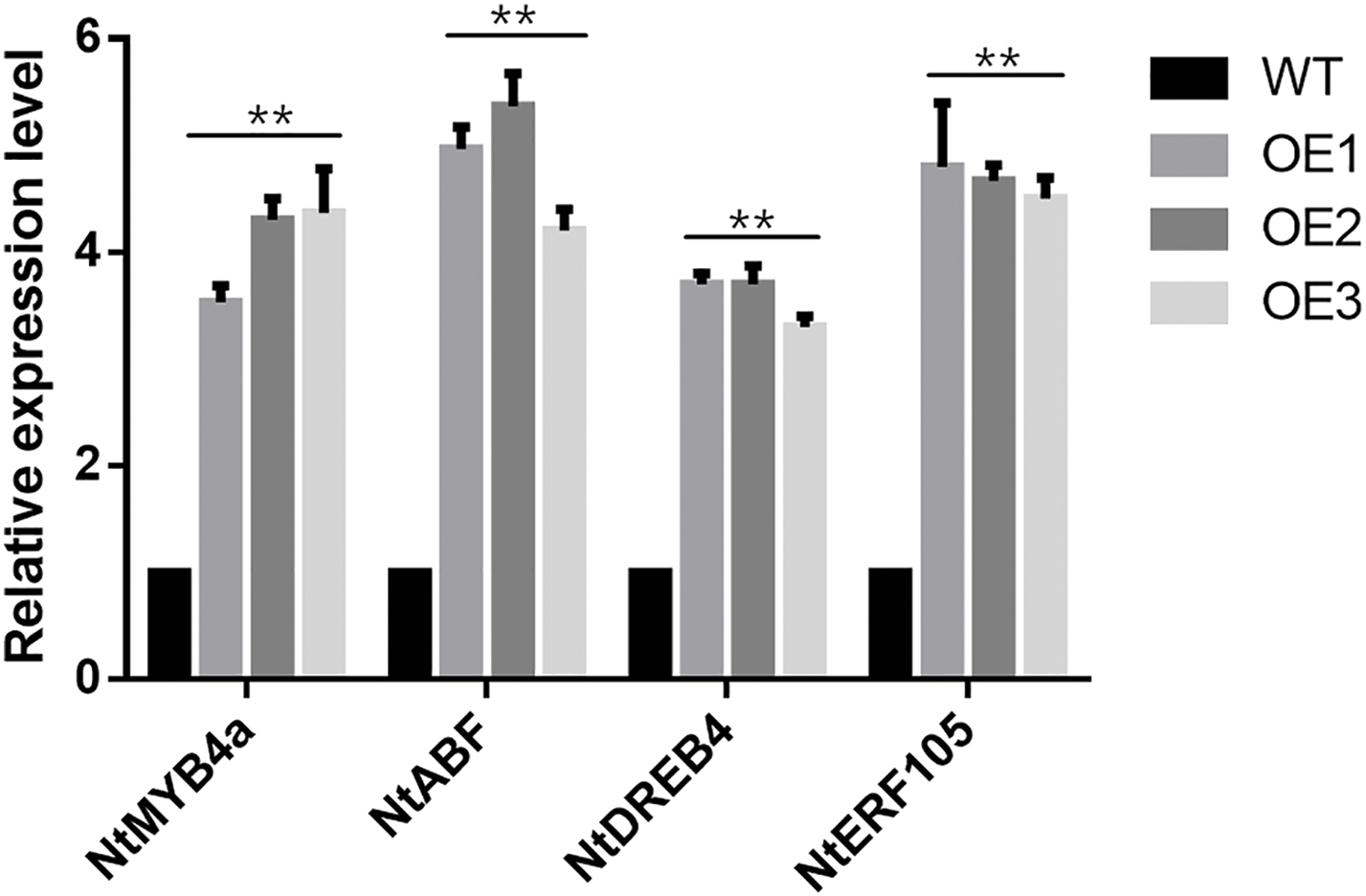
Figure 10: Expression levels of ABA-responsive genes in both wild type and transgenic tobacco under normal conditions. The vertical axis shows fold changes while the horizontal axis indicates gene names. The values presented are means ± SD from three independent replicates, with asterisks (**) denoting significant differences at p < 0.01, according to the Student’s t-test
3.8 ZmWRKY25 Changes Enzyme Activity and the Damage of Active Oxygen to Plant Cells
In order to investigate the role of the ZmWRKY25 gene during drought conditions, a NBT staining experiment was conducted on both WT plants and ZmWRKY25 transgenic plants 13 days after exposure to drought stress. The results indicated a noticeable decrease in reactive oxygen species (ROS) content within the mesophyll cells of ZmWRKY25 transgenic plants compared to WT plants. Additionally, the activities of POD and SOD were assessed under the same conditions. Following 13 days of drought treatment, the POD activity in transgenic plants significantly increased compared to WT plants, while it notably decreased before drought exposure and after three days of rewatering. The changes in SOD enzyme activity closely mirrored those of POD. To further explore the impact of ROS on plant cell damage, the levels of MDA, Pro, chl, and relative conductivity were analyzed under identical conditions. After 13 days of drought treatment and three days of rehydration, the MDA content and relative electrical conductivity in transgenic plants were significantly lower than in WT plants. Conversely, the levels of Pro and chlorophyll in transgenic plants were notably higher than those in WT plants (Fig. 11).
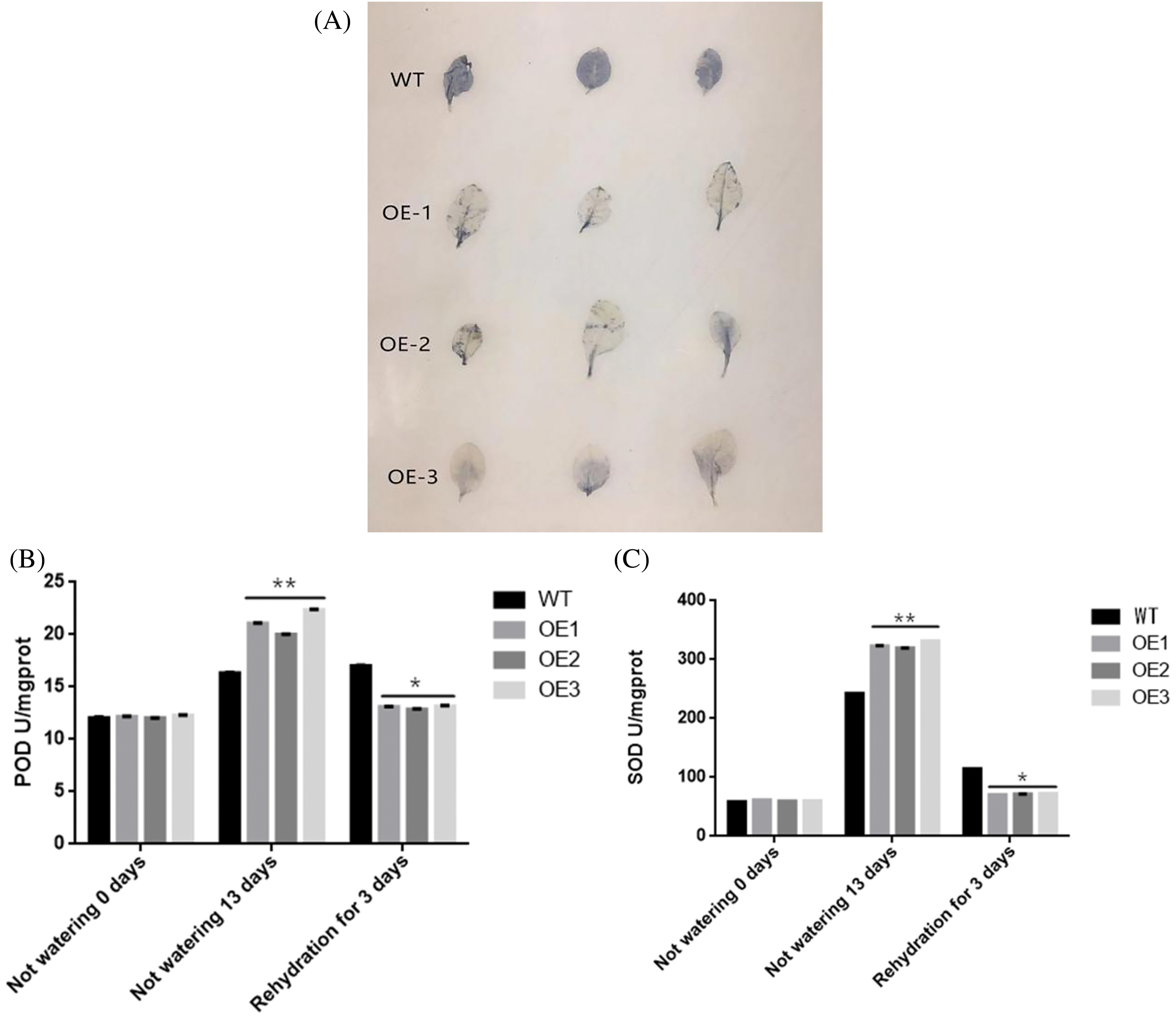
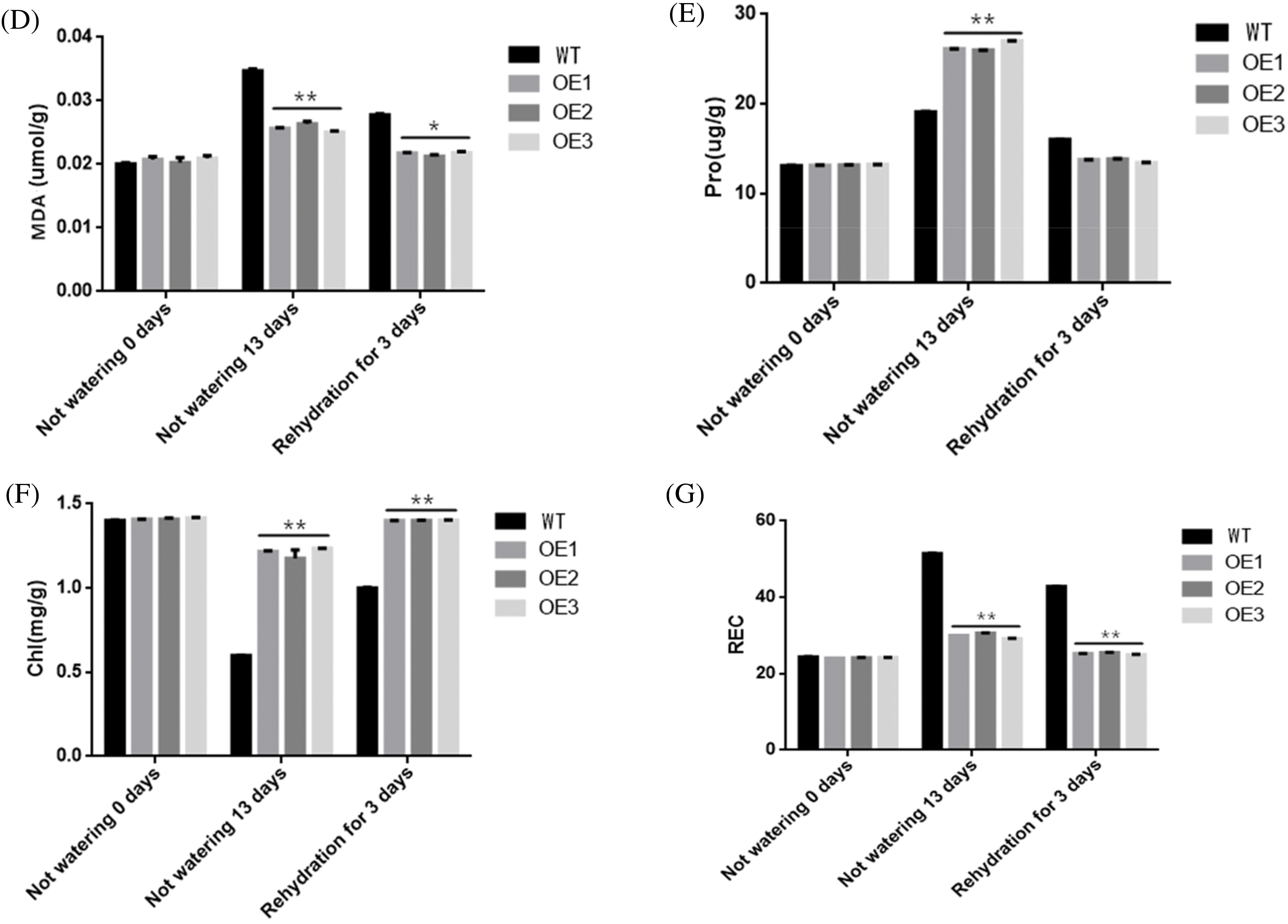
Figure 11: The assessment of physiological and biochemical parameters was conducted under both normal and drought conditions. NBT staining is shown in panel (A). The activities of peroxidase dismutase (POD) (B) and superoxide dismutase (SOD) (C) were evaluated in wild-type (WT) and transgenic lines. Additionally, the relative conductivity (REC) (G), along with the contents of malondialdehyde (MAD) (D), proline (Pro) (E), and chlorophyll (chl) (F) were measured in both WT and transgenic lines. Values represent the means ± SD from three replicates, with asterisks (* or **) indicating significant differences at p < 0.05 or p < 0.01, respectively (Student’s t-test)
The WRKY family of transcription factors was one of the earliest to be identified, with implications in various processes related to responses to abiotic stress [30–32]. In maize, a total of 136 WRKY proteins encoded by 119 genes have been discovered, yet only a limited number of these genes have shown observable functions [33]. To further investigate the functions of the WRKY transcription factor family in maize, we isolated a Group II-d WRKY gene, ZmWRKY25, from maize transcriptome data collected under drought conditions. Our study revealed that ZmWRKY25 is closely related to AtWRKY11 and AtWRKY17 based on phylogenetic tree analyses (Fig. 1). Additionally, an examination of conserved protein domains showed that ZmWRKY25 shares identical conserved domains with AtWRKY11 and AtWRKY17 (Fig. 2). Previous studies have shown that ZmWRKY25 may share functional similarities with AtWRKY11 and AtWRKY17, which are known to respond to salt and osmotic stresses [13,34]. Osmotic stress, including factors like drought stress, can lead to the production of osmotic protectants in the body, resulting in the accumulation of osmotic substances that impact osmotic pressure [35,36]. Interestingly, the promoter region of ZmWRKY25 contains recognition sites for ABRE, MYS, and CGTCA motifs, suggesting its involvement in ABA signal transduction and response to drought. Our study investigated the expression patterns of ZmWRKY25 under salt, drought, and ABA stress conditions (Fig. 5B–D), revealing up-regulation of ZmWRKY25 expression under all stressors, particularly ABA and drought stress. These findings indicate a significant role for ZmWRKY25 in the response to drought stress.
ABA, as a crucial plant hormone, has been identified in studies as having the most significant response to both drought stress and salt stress [37]. In an experiment examining the expression pattern of ZmWRKY25, it was observed that the expression level of ZmWRKY25 peaked within 1 h following ABA stress, whereas it took 3 h for the expression level to reach its peak after PEG6000 simulated drought stress (Fig. 5C–D). This suggests that ABA more directly induces ZmWRKY25, with the response of ZmWRKY25 to drought stress being attributed to the increase in ABA content. Furthermore, gene expression analysis of NtMYB4a, NtABF, NtDREB4, and NtERF105 in the ABA signaling pathway under drought stress indicated that ZmWRKY25 plays a role in drought stress through the ABA signaling pathway (Fig. 10) [38–41].
Overexpression of ZmWRKY25 in tobacco led to an increase in peroxidase (POD) and superoxide dismutase (SOD) activity in the genetically modified tobacco plants (Fig. 11C–B). The transgenic plants exhibited a significant reduction in reactive oxygen species (ROS) levels compared to wild-type plants (Fig. 11A–G). Previous studies have shown that ROS can damage the cell membrane structure, cause membrane breakdown, and elevate malondialdehyde (MDA) content [42,43]. This disruption in cell membrane structure results in the leakage of electrolytes, leading to higher electrical conductivity, decreased chlorophyll levels, and impaired photosynthetic function in mesophyll cells [44,45]. Following drought stress, there was no significant difference in MDA, chlorophyll, and relative electrical conductivity levels in transgenic plants compared to before the stress (Fig. 11D–G). Our study highlights the crucial role of POD and SOD in scavenging ROS, protecting cell membrane integrity, and maintaining optimal photosynthesis in mesophyll cells, enhancing plant resilience to drought stress (Fig. 11A–G). Furthermore, our research revealed that the root system of genetically modified plants exhibited greater strength compared to the wild type (Fig. 9C). Genetically modified plants showed improvements in root characteristics such as increased total root length, expanded root surface area, and a greater abundance of lateral roots, all of which contribute to enhanced water absorption from the soil (Fig. 8B–G). Additionally, the expression of the ERF105 gene in genetically modified plants was significantly upregulated in response to drought stress (Fig. 10). The ERF105 gene plays a key role in regulating processes like cell elongation, division, and root growth and development [16]. It is suggested that ZmWRKY25 may regulate the expression of ERF105 to increase the activity of the root meristem in transgenic plants compared to wild type. Consequently, transgenic plants exhibit faster growth and stronger root systems than wild type, providing them with more water and nutrients to sustain normal plant growth during drought [15]. In conclusion, these findings indicate that ZmWRKY25 could potentially enhance the drought tolerance of genetically modified plants by modulating the removal of reactive oxygen species and promoting root growth and development. Therefore, the function research of ZmWRKY25 in tobacco points out the research direction for maize.
We identified a drought-responsive WRKY gene, from the results of transcriptome sequencing of maize under drought stress, ZmWRKY25. ZmWRKY25 is induced by drought, salt, and ABA treatment, is expressed in the nucleus, and participates in the ABA-dependent pathway. Further research shows that ZmWRKY25 can improve tobacco tolerance to drought. Under drought stress, the overexpression of ZmWRKY25 increases the activity of POD and SOD, reduces the content of active oxygen, and makes the root system stronger and more conducive to water absorption. These results laid the foundation for further research on the function of ZmWRKY25 in maize.
Acknowledgement: This research was supported by Jilin Agricultural University Joint International Research Laboratory of Modern Agricultural Technology.
Funding Statement: Open Project of the International Cooperation Joint Laboratory of the Ministry of Educationon Modern Agricultural Technology (H20230034), Jilin Province Science and Technology Development Plan Project (20230202003NC, 20230508005RC, 20240601060RC).
Author Contributions: Jianbo Fei and Zhibo Liu conceived the study. Jianbo Fei, Zhibo Liu and Piwu Wang designed the experiments and analyzed the data. Jianbo Fei, Jing Qu and Siyan Liu carried out the experimental work and analyzed the data. Yiyong Ma and Shuyan Guan revised the manuscript. Jianbo Fei wrote the manuscript with contributions of Zhibo Liu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.052704.
References
1. Shi J, Habben JE, Archilbald RL, Bruce JD, Mark AC, Robert WW, et al. Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both arabidopsis and maize. Plant Physiol. 2015;169(1):266–82. doi:10.1104/pp.15.00780. [Google Scholar] [PubMed] [CrossRef]
2. Middleton N, Stringer L, Goudle A. The forgotten billion: MDG a chievement in the dry lands. New York: United Nations Development Programme; 2011. [Google Scholar]
3. Zhao J, Li K. Beijing: agricultural drought and agriculture. China Press; 1995. [Google Scholar]
4. Tian T, Wang SH, Yang SP, Yang ZR, Liu SX, Wang YJ, et al. Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat Genet. 2023;55(3):496–506. doi:10.1038/s41588-023-01297-y. [Google Scholar] [PubMed] [CrossRef]
5. Javed T, Gao SJ. WRKY transcription factors in plant defense. Trends Genet. 2023;39(10):787–801. doi:10.1016/j.tig.2023.07.001. [Google Scholar] [PubMed] [CrossRef]
6. Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol Gen Genet. 1994 Sep 28;244(6):563–71. doi:10.1007/BF00282746. [Google Scholar] [PubMed] [CrossRef]
7. Saha B, Nayak J, Srivastava R, Samal S, Kumar D, Chanwala J, et al. Unraveling the involvement of WRKY TFs in regulating plant disease defense signaling. Planta. 2023 Nov 28;259(1):7. [Google Scholar]
8. Hu W, Ren Q, Chen Y, Xu G, Qian Y. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021 Sep 20;21(1):427. doi:10.1186/s12870-021-03206-z. [Google Scholar] [PubMed] [CrossRef]
9. Song H, Cao Y, Zhao L, Zhang J, Li S. Review: WRKY transcription factors: understanding the functional divergence. Plant Sci. 2023;334:111770. doi:10.1016/j.plantsci.2023.111770. [Google Scholar] [PubMed] [CrossRef]
10. Javed T, Zhou JR, Li J, Hu ZT, Wang QN, Gao SJ. Identification and expression profiling of WRKY family genes in sugarcane in response to bacterial pathogen infection and nitrogen implantation dosage. Front Plant Sci. 2022;13:917953. doi:10.3389/fpls.2022.917953. [Google Scholar] [PubMed] [CrossRef]
11. Charvin M, Halter T, Blanc-Mathieu R, Barraud P, Aumont-Nicaise M, Parcy F, et al. Single-cytosine methylation at W-boxes repels binding of WRKY transcription factors through steric hindrance. Plant Physiol. 2023;192(1):77–84. doi:10.1093/plphys/kiad069. [Google Scholar] [PubMed] [CrossRef]
12. Khoso MA, Hussain A, Ritonga FN, Ali Q, Channa MM, Alshegaihi RM, et al. WRKY transcription factors (TFsmolecular switches to regulate drought, temperature, and salinity stresses in plants. Front Plant Sci. 2022;13:1039329. doi:10.3389/fpls.2022.1039329. [Google Scholar] [PubMed] [CrossRef]
13. Desoky ESM, Mansour E, Ali MMA. Exogenously used 24-epibrassinolide promotes drought tolerance in maize hybrids by improving plant and water productivity in an arid environment. Plants. 2021;10(2):354. doi:10.3390/plants10020354. [Google Scholar] [PubMed] [CrossRef]
14. Wang H, Chen W, Xu Z, Chen M, Yu D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023;28(6):630–45. doi:10.1016/j.tplants.2022.12.012. [Google Scholar] [PubMed] [CrossRef]
15. Gupta A, Rico-Medina A, Caño-Delgado AI. The physiology of plant responses to drought. Science. 2020;368(6488):266–9. doi:10.1126/science.aaz7614. [Google Scholar] [PubMed] [CrossRef]
16. Guyomarc’h S, Boutté Y, Laplaze L. AP2/ERF transcription factors orchestrate very long chain fatty acid biosynthesis during Arabidopsis lateral root development. Mol Plant. 2021;14(2):205–7. doi:10.1016/j.molp.2021.01.004. [Google Scholar] [PubMed] [CrossRef]
17. Goyal P, Devi R, Verma B, Hussain S, Arora P, Tabassum R, et al. WRKY transcription factors: evolution, regulation, and functional diversity in plants. Protoplasma. 2023;260(2):331–48. doi:10.1007/s00709-022-01794-7. [Google Scholar] [PubMed] [CrossRef]
18. Cai R. Cloning and anti stress function of maize transcription factor ZmWRKY58 gene (Master Thesis). Anhui Agriculture University: China; 2013. [Google Scholar]
19. Xiong XP, Sun SC, Zhang XY, Li YJ, Liu F, Zhu QH, et al. GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways. Front Plant Sci. 2020 Feb 25;11:69. doi:10.3389/fpls.2020.00069. [Google Scholar] [PubMed] [CrossRef]
20. Li H. Cloning and functional analysis of maize WRKY family transcription factor gene (Master Thesis). Yangzhou University: China; 2011. [Google Scholar]
21. Liu X. Cloning and stress tolerance of maize WRKY transcription factor gene (Master Thesis). Anhui Agriculture University: China; 2011. [Google Scholar]
22. Koentjoro Y, Sukendah, Purwanto E, Purnomo D. The role of silicon on content of proline, protein and abscisic acid on soybean under drought stress. IOP Conf Ser: Earth Environ Sci. 2021;637(1):012086. doi:10.1088/1755-1315/637/1/012086. [Google Scholar] [CrossRef]
23. Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22(7):2444–58. doi:10.1105/tpc.110.075358. [Google Scholar] [PubMed] [CrossRef]
24. Zhao W, Liu YW, Zhou JM, Zhao SP, Zhang XH, Min DH. Genome-wide analysis of the lectin receptor-like kinase family in foxtail millet (Setaria italica L.). Plant Cell Tissue Organ Cult. 2016;127(2):1–12. [Google Scholar]
25. Chen Y, Xu BL, Su N, Ge YQ, Wang SG. Application of real-time fluorescence quantitative PCR in detection of transgenic maize. Acta Agron Sin. 2004;30(6):602–7. [Google Scholar]
26. Lagace M, Matton DP. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta. 2024;219:185–9. [Google Scholar]
27. Huo L, Xiao Y, Wang YC. Optimization of experimental conditions for transient expression mediated by Agrobacterium tumefaciens in tobacco. Mol Plant Breed. 2016;14(1):80–5. [Google Scholar]
28. Qin P, Liu YJ, Yi FH. Effects of drought treatments on the activities of SOD and POD in tobacco leaves. Chin Tobacco Sci. 2005;2:28–30 (In Chinese). [Google Scholar]
29. Zhang F. Genetic transformation and drought resistance detection of ferritin gene from Artemisia annua tobacco (Master Thesis). Lanzhou University: China; 2013. [Google Scholar]
30. Johnson CS. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell Online. 2002;14(6):73–8. [Google Scholar]
31. Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol. 2010;49(6):827–42. [Google Scholar]
32. Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, et al. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genomics. 2013;13(1):75–98. doi:10.1007/s10142-012-0301-4. [Google Scholar] [PubMed] [CrossRef]
33. Wei KF, Chen YF. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013;13(1):148. doi:10.1186/1471-2229-13-148. [Google Scholar] [PubMed] [CrossRef]
34. Journot-Catalino N, Somssich IE, Roby D. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in arabidopsis thaliana. Plant Cell. 2006;18(11):3289–302. doi:10.1105/tpc.106.044149. [Google Scholar] [PubMed] [CrossRef]
35. Ali MA, Azeem F, Nawaz MA. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J Plant Physiol. 2018;226(7):12–21. [Google Scholar] [PubMed]
36. Akos IS, Rafii MY, Ismail MR. Evaluation of inherited resistance genes of bacterial leaf blight, blast and drought tolerance in improved rice lines. Rice Sci. 2021;28(3):10. [Google Scholar]
37. Chaves MM. Understanding plant responses to drought-from gene to the whole plant. Funct Plant Biol. 2003;30:239–64. doi:10.1071/FP02076. [Google Scholar] [PubMed] [CrossRef]
38. Jiang Y, Luo Q, Jiang CY. Functional analysis of tobacco transcription factor NtMYB4a in response to drought, low temperature and methyl jasmonate stress. Crop Res. 2021;35(1):43–9. [Google Scholar]
39. Song LL, Zhang YN, Zhang ZY, Tian JH. Characterization and functional identification of tobacco transcription factor NtDREB4. Mol Plant Breed. 2020;18(15):4918–23 (In Chinese). [Google Scholar]
40. Yang L, Wu YQ, Xie XD. Cloning and bioinformatics analysis of ABF transcription factor gene in tobacco. Tobacco Sci Technol. 2014;6(323):73–92. [Google Scholar]
41. Ren AY, Kong YZ. Identification and expression pattern analysis of ERF transcription factor subfamily members in common tobacco. Chin Tobacco Sci. 2017;38(1):15–22. [Google Scholar]
42. Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87(1):245–313. doi:10.1152/physrev.00044.2005. [Google Scholar] [PubMed] [CrossRef]
43. Bayarmaa J, Purev D. Superoxide dismutase and catalase activities, proline and malondialdehyde contents in extracts of the aerial parts of two species of Pulsatilla native to Mongolia. Mong J Biolog Sci. 2021;19(2):33–8. doi:10.22353/mjbs. [Google Scholar] [CrossRef]
44. Wang N, Tian Y, Chen H. Effects of mesosulfuron-methyl on growth and competitiveness of Aegilops tauschii and Triticum aestivum. Acta Physiol Plant. 2021;43(2):1–10. [Google Scholar]
45. Awaad HA, Abu-Hashim M, Negm AM. Heat stress tolerance, challenges and solutions. In: Awaad, H., Abu-hashim, M., Negm, A., editors, Mitigating environmental stresses for agricultural sustainability in Egypt. Springer, Cham; 2021. doi:10.1007/978-3-030-64323-2_4. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools