 Open Access
Open Access
ARTICLE
Systematic Identification of Acer rubrum bZIP Transcription Factors and Their Potential Role in Anthocyanin Accumulation under Low Temperature with Light
1 School of Forestry & Landscape Architecture, Anhui Agricultural University, Hefei, 230036, China
2 Institute of Agricultural Engineering, Anhui Academy of Agricultural Sciences, Hefei, 230031, China
* Corresponding Authors: Jie Ren. Email: ; Hua Liu. Email:
# Authors contribute equally to this work
(This article belongs to the Special Issue: Recent Research Trends in Genetics, Genomics, and Physiology of Crop Plants)
Phyton-International Journal of Experimental Botany 2024, 93(11), 3109-3130. https://doi.org/10.32604/phyton.2024.056548
Received 25 July 2024; Accepted 30 October 2024; Issue published 30 November 2024
Abstract
Acer rubrum is an important garden color-leafed plant. Its leaves will turn red in autumn, which is of great ornamental value. The leaf color change in Acer rubrum is closely associated with anthocyanins accumulation. In anthocyanin synthesis and accumulation, various transcription factor families play significant regulatory roles, including the basic (region) leucine zipper (bZIP). However, there is no report on the systematic identification and functional analysis of the bZIPs in Acer rubrum. In this study, 137 bZIPs distributed on 29 chromosomes of Acer rubrum were identified and renamed according to their locations on the chromosomes. According to the constructed bZIP phylogenetic tree of Arabidopsis thaliana and Acer rubrum, bZIPs were divided into 13 groups. Two pairs of bZIP genes were involved in tandem duplication, and 106 segmental duplication gene pairs were found. Cis-acting elements in the promoter region of these bZIP genes were analyzed. The results of promoter analysis showed that many elements were closely related to light conditions, hormone responses, and abiotic stress factors. Among them, the cis-acting elements related to light response were most abundant and prominent. The results of anthocyanin determination showed that anthocyanin contents in the leaves of Acer rubrum increased significantly under low temperature with light. In addition, gene expression analysis showed that compared to other ArbZIPs, ArbZIP137, ArbZIP136, ArbZIP114, ArbZIP130, and ArbZIP14 showed a more pronounced increase in gene expression both under low- temperature conditions and under light conditions. From the correlation analysis, there was a high correlation between ArbZIPs and several anthocyanin-regulated transcription factors, including ArMYBs, ArbHLH and ArWD40s. Conclusively, the bZIP genes in Acer rubrum were identified and analyzed, providing a foundational basis for future studies on their function and significantly enhancing our understanding of the color mechanism of Acer rubrum.Keywords
Supplementary Material
Supplementary Material FileThe genus Acer (Aceraceae) species are notable among the colorful forest trees for their brilliant colors and high ornamental properties. It comprises approximately 200 species and is widely distributed across Asia, Europe, and North America [1]. Acer rubrum (red maple), belonging to the genus of Acer, Aceraceae, Sapindales, is one of the most important red-leaved trees. Native to North America, the red maple is mainly found in the Northern United States (e.g, Minnesota and Wisconsin), a few Southern States (e.g., coastal Florida and Texas), and some areas of Canada. Due to its upright trunk shape and attractive leaf color, A. rubrum is increasingly significant in urban greening initiatives and garden design. Whether planted alone or in groups, it can create a stunning scene for the garden, having high ornamental value. In addition to its outstanding applications in landscaping, Acer rubrum is used in furniture production [2] and brings great economic value. Furthermore, the flowers, leaves, and roots of A. rubrum can be used as medicine, indicating their therapeutic potential.
The reddening of the green leaves of the red maple is caused by anthocyanins [3]. Anthocyanin is a common natural pigment in plants. Most significant color-presenting substances in fruits, vegetables, flowers and forest trees are related to them. Anthocyanins are natural coloring agents and antioxidants with high medicinal and economic exploitation value. Anthocyanins, owing to their anti-apoptotic and anti-inflammatory properties, are widely used to prevent and treat various diseases [4]. Therefore, anthocyanins hold substantial research value.
Temperature and light affect anthocyanin accumulation in many plants [5] and cause the leaves to take on different shades of color. Reddening of autumn leaves as temperatures drop is an outward sign of anthocyanin accumulation [6]. The formation of red color in the rind of Pyrus pyrifolia Nakai is influenced by light and temperature [7]. Low temperatures promote the accumulation of anthocyanin, while high temperatures lead to a decrease in its concentration [8]. Moreover, light is also one of the leading environmental factors that impact anthocyanin accumulation, which directly or indirectly activates the expression of relevant genes in the anthocyanin biosynthesis pathway, promoting the synthesis of relevant enzymes and leading to anthocyanin synthesis and accumulation [9]. According to previous research, plants with high anthocyanin usually exhibit greater tolerance to abiotic stresses than green plants [10]. In many plant species, foliar anthocyanins usually accumulate transiently under various abiotic stresses, including bright light, salinity, drought, cold, nutrient deficiency and heavy metal stress, turning the leaves red or purple [11]. Anthocyanins can bring beautiful colors to plants and provide multi-functional, multi-faceted and effective protection to plants under adverse conditions.
Transcription factors (TFs) are important in the plant anthocyanins regulatory network. Many studies have proposed that TFs, like MYB, bHLH, and WD40, are key regulators of anthocyanin biosynthesis. MYBs are categorized into four subfamilies, among which R2R3-MYB is considered the most closely related to anthocyanin biosynthesis [12]. This conclusion has been verified in Arabidopsis previously [13]. In apples, MdMYB10 encodes a regulator of anthocyanin biosynthesis [14]. In lily and poplar, related MYB genes have also been found to be involved in anthocyanin synthesis [15,16]. MdMYB114 can act as an activator of anthocyanin biosynthesis and transport [17]. BHLHs such as CsbHLH89 and VabHLH137 enhance anthocyanin accumulation by binding to a specific promoter region [18,19]. WD40 TFs have been relatively less studied in this field, but OsTTG1 has been identified as an important regulator of anthocyanin synthesis [20]. By forming a conserved MBW (MYB-bHLH-WD40) ternary complex, the proteins of MYB, bHLH, and WD40 can also control the anthocyanin synthesis in different plant species [21]. For instance, TT2 (MYB), TT8 (bHLH), and TTG1 (WD40) can form a stable ternary complex that regulates the expression of the BANYULS (BAN) gene expression, thus affecting anthocyanin synthesis [22]. This phenomenon has been demonstrated in kiwifruit, blueberry, strawberry and persimmon [23–26].
In addition to MYB, bHLH and WD40 TFs, the role of the basic (region) leucine zippers (bZIPs) in anthocyanin accumulation cannot be ignored. The bZIP gene family is a prominent and highly prevalent group of transcription factors in higher plants [27]. It consists of two distinct functional domains: the leucine zipper region and the basic region [28]. The bZIP is distinguished by a highly conserved basic DNA-binding domain. Its structure contains 16–20 basic amino acid residues (e.g., lysine, arginine), alongside an adjacent leucine zipper dimerization motif (e.g., methionine, isoleucine) [29], which are connected by a hinge connector [30]. At the C-terminal end of the basic region, there is a conserved N-x7-R/K structure, which is necessary for sequence-specific DNA binding. At the N-terminal region, the leucine zipper consists of multiple heptapeptide repeats or hydrophobic amino acid residues [31], influencing bZIP protein dimerization before DNA binding [32].
So far, bZIP genes have been discovered in the genomes of several plants. For example, there are 78 bZIPs in Arabidopsis thaliana [33], 101 in Populus tremula (http://planttfdb.cbi.pku.edu.cn) (accessed on 29 October 2024), 103 in Olive [34], 56 in Betula Platyphylla [35], 89 in rice [36] and so on. These TFs play crucial roles in plant growth and development, influencing processes such as germination, seed maturation, and flower development [37]. In addition, they help plants cope with abiotic stresses such as heat, drought, and salinity. For instance, certain bZIPs in Arabidopsis are pivotal in ABA signaling, which affects seed development and stress tolerance [38]. Overexpression of specific bZIP genes has been shown to enhance salt tolerance in Arabidopsis (e.g., AtbZIP11 and AtbZIP53) [39] and wheat (e.g., TabZIP15) [40], while also improving frost tolerance in other species (e.g., TabZIP96 in Arabidopsis) [41]. Moreover, the combined action of DgbZIP3 and DgbZIP2 has been found to enhance low-temperature tolerance in chrysanthemum [42].
In addition to these functions, bZIPs are involved in anthocyanin synthesis. It has been found that bZIPs (especially HY5 and HYH) play a role by regulating the biosynthesis of flavonoid secondary metabolites anthocyanins in response to low-temperature stress in Arabidopsis [43]. Overexpression of SlAREB1, a bZIP gene, significantly increases the expression of key genes related to anthocyanin synthesis. This results in a substantial enhancement in anthocyanin accumulation, particularly under low temperature [44]. HY5 in Arabidopsis modulates anthocyanin biosynthesis by stimulating transcriptional activation of the Arabidopsis MYB75/PAP1. Specifically, HY5 regulates PAP1 expression by directly binding to the promoter region’s light-responsive cis-acting elements G-box and ACE-box [45]. Moreover, bZIP can be combined with MYB to promote anthocyanin synthesis and increase its expression. For instance, in Malus domestica, MdbZIP44 augments MdMYB1 binding to downstream gene promoters under ABA influence, thereby stimulating anthocyanin accumulation [46]. Similarly, MdHY5 regulates the expression of MdMYB10 and other anthocyanin-related genes, thereby promoting anthocyanin accumulation [47]. Thus, bZIPs have a very important role in anthocyanin accumulation, whether directly or indirectly affected.
Although bZIPs have a significant impact on plant growth and development, stress response, and anthocyanin accumulation, there is still a lack of information on bZIPs in landscape color-leafed plants, especially in Acer rubrum. Therefore, in our study, the bZIP gene family members of Acer rubrum (ArbZIP) were systematically identified by bioinformatics methods, and the physicochemical properties, conserved domains, motifs, gene structure, evolutionary relationship, and cis-acting elements of the ArbZIP family were analyzed in depth. We preliminarily explored the ArbZIP family’s response mechanism to low-temperature stress with light in Acer rubrum and further speculated its potential role in anthocyanin accumulation. These findings provide a valuable reference for understanding the regulatory mechanism of bZIPs in Acer rubrum, offering new strategies for genetic improvement and stress resistance breeding of ornamental plants with colored leaves.
The red maple variety ‘Yanhong’, characterized by its reddish leaf color, was utilized in this study. Its seeds, obtained from the Anhui Academy of Agricultural Sciences, were stored at 4°C in a refrigerator. The seeds were placed in sand and maintained at the same temperature for one month. After germination, the seedlings were nurtured on peat moss and maintained in a plant growth chamber, with 22°C during the day and 16°C at night. These seedlings were exposed to light for 12 h and darkness for 12 h each day. Healthy seedlings with uniform growth were selected for the low-temperature stress experiment. Selected seedlings were exposed to 22°C with 24 h of light as a control group. At the same time, some seedlings were placed in complete darkness at 4°C for 24 h and another group of seedlings was placed in complete light for 24 h. These two conditions served as two experimental groups. Three biological samples were collected from each group and immediately immersed in liquid nitrogen. These samples were then stored at −80°C for future analysis and utilization.
2.2 Identification of BZIPs in Acer rubrum
The Arabidopsis thaliana bZIP (AtbZIP) sequences used in this study were downloaded from TAIR (https://www.arabidopsis.org) (accessed on 29 October 2024). To compare and identify bZIP genes of Acer rubrum (ArbZIP), AtbZIP protein sequences were used as queries to search the Acer rubrum genome databases (Accession number: PRJNA741546; https://www.ncbi.nlm.nih.gov/bioproject/?term=prjna741546) (accessed on 29 October 2024) via BLASTP. The protein sequences of red maple bZIP TFs were extracted by TBtools software [48]. Candidate sequences were further validated by screening for conserved domains using the CD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (accessed on 29 October 2024). Subsequently, redundant sequences and incomplete domains were removed to compile the Acer rubrum bZIP family members [49]. The ArbZIP protein sequence file was uploaded to the Protein Parameter Calc program in TBtools to calculate its molecular weight, amino acid number, theoretical isoelectric point and grand average of hydropathicity.
2.3 Phylogenetic Analysis and Cis-Acting Element Predictions
Multiple sequence alignments of ArbZIP proteins and AtbZIP proteins were conducted using MEGA 11 [50]. We used the Neighbor-Joining (NJ) method to construct a phylogenetic tree, and the bootstrap value was set to 1000. The p-distance method is used to calculate the evolutionary distance. The obtained evolutionary tree data were imported into the iTOL website for landscaping. An NJ phylogenetic tree of ArbZIP proteins was constructed using the same methods described above. The 2000 bp sequence upstream of the ArbZIPs served as the promoter region for predicting cis-elements, utilizing the PlantCare website (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 29 October 2024). Based on the functions of these elements, we removed unnamed elements and those with negligible practical significance, then filtered and organized the results. We organized and analyzed the processed data again using R software to categorize the remaining 20 kinds of cis-acting elements according to their functions. Finally, a visual heat map was created using HeatMap of TBtools.
2.4 ArbZIP Gene Structure, Domain and Motif
The GFF file of A. rubrum was parsed to visualize the gene structure of ArbZIP using TBtools. Conserved domains of the ArbZIP protein were searched using the CD-Search tool on the NCBI website. Additionally, conserved motifs within ArbZIP protein were predicted via the MEME program integrated into TBtools software [51], with the default settings used except for the number of motifs set to 20.
2.5 Chromosomal Location, Gene Duplication, and Synteny Analysis
The chromosomal location data for the ArbZIP gene from Acer rubrum genome annotation files included details such as chromosome number, gene orientation, length, start position, and end position. Each ArbZIP gene was named according to its specific physical location. To analyze gene duplication events, we used One Step MCScanX-Super Fast in TBtools with default parameters [52]. Using both the MG2C website (http://mg2c.iask.in/mg2c_v2.1/) (accessed on 29 October 2024) and TBtools, we mapped the chromosomal locations of ArbZIP genes onto the Acer rubrum linkage map using the General Feature Format file [48]. The genomic sequences and GFF files from Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, and Vitis vinifera were acquired from NCBI (https://www.ncbi.nlm.nih.gov/) (accessed on 29 October 2024). TBtools was utilized to examine the syntenic relationships between these sequences and the ArbZIP genes.
2.6 Determination of Anthocyanin Content
Add 5 mL of 1 mol/L HCl-ethanol to 0.1 g of sample. Three replicates were taken for each treatment and 9 samples were hydrolyzed in a 55°C water bath for 2 h. The supernatant was taken as the sample extract. The anthocyanin content was determined by spectrophotometry, measuring the optical density of the extract at 530 nm, 620 nm, and 650 nm, using 1 mol/L HCl-ethanol solution as the reference.
The corrected absorbance was calculated as:
ODλ: the corrected absorbance; ε: 4.62 × 104 (the molar extinction coefficient of the major anthocyanin); V: total volume of the extract (mL); m: quality of the sample (g); M: 287.24 (the molecular weight of the major anthocyanin-cyanidin [3]).
2.7 Expression Analysis of Red Maple BZIPs and qRT-PCR Analysis
The transcriptome gene expression data of Acer rubrum under low temperature stress with light and without light were downloaded in NCBI, and the BioProject accession number was PRJNA1094633. The FPKM values for bZIP were obtained from RNA-seq data of Acer rubrum leaves under varying conditions, including light (all-black environment and light environment under 4°C) and temperatures (4°C and 22°C). After processing the data using R software, ArbZIP gene expression was analyzed and visualized using heatmap in TBtools software, and all FPKM values were processed using row-scale transformation.
The reliability of RNA-seq data was confirmed through Quantitative Real-Time PCR (qRT-PCR). RNA was extracted from red maple leaves using the method followed by Shah et al. [53]. The corresponding cDNA was synthesized using the EasyScript First-Strand cDNA Synthesis kit [54]. The qRT-PCR primers for ArbZIPs were designed with the primer design software Primer 6.0 [55]. All qRT-PCR primers utilized in this research are listed in Table S1. QRT-PCR reactions were conducted using the SYBR Premix Ex Taq (Tli RNaseH Plus; RR420A, TaKaRa, Dalian, China) on a Lightcycle K Real-Time PCR Detection System (BIOER, Hangzhou, China). The qRT-PCR program included an initial denaturation step at 95°C for 30 s, followed by 40 cycles consisting of 95°C for 5 s, 60°C for 20 s, and 72°C for 20 s [4]. ACTIN was employed as the internal control gene. Gene expression levels were assessed using the 2−ΔΔC method developed by Livak et al. [56].
Using R software, code was written to calculate the transcriptome correlations of ArbZIPs and other anthocyanin-related genes based on the Pearson correlation coefficient. Genes with correlations greater than 0.9 were selected and their co-expression network maps were visualized using Cytoscape (v3.10.1).
3.1 Identification and Analysis of ArbZIPs
In Acer rubrum, we identified 137 bZIP genes (Table S2) from the genome, which were named ArbZIP1-ArbZIP137 based on their chromosome locations. According to the results given by TBtools (Table S3), the sequence lengths of all ArbZIP proteins varied from 142 (ArbZIP88) to 700 (ArbZIP38) amino acids, with an average length of 317. The molecular weights of these proteins ranged from 16.1835 kDa (ArbZIP88) to 76.33152 kDa (ArbZIP36). The theoretical isoelectric points varied between 4.32 (ArbZIP16) and 9.96 (ArbZIP104). The overall mean of hydropathicity of all ArbZIP proteins was below zero, suggesting their hydrophobic nature.
3.2 Phylogenetic Analysis and Classification of the ArbZIP Genes
To examine the evolutionary relationships of Acer rubrum bZIP TFs, we compared and analyzed their protein sequences with those of AtbZIPs using the Muscles program within MEGA software. Subsequently, an NJ phylogenetic tree was constructed (Fig. 1). Within the Arabidopsis bZIP gene family, AtbZIP73 (AT2G13115) was identified as a pseudogene [33], while the bZIP domain of AT4G06598 (AtbZIP78) was deemed incomplete. Therefore, they were excluded from the phylogenetic tree, leaving 77 AtbZIPs (AtbZIP1-AtbZIP79) and 137 ArbZIPs for tree construction. As depicted in Fig. 1, the entire evolutionary tree was constructed through deep cluster analysis, using the evolutionary relationships and naming conventions of AtbZIP genes as a Reference [57]. The bZIPs of the two species were categorized into 13 groups, designated as groups A-K, M, and S. The largest group, S, contained 17 AtbZIPs and 28 ArbZIPs. The smallest was group J, with only 1 AtbZIP and 1 ArbZIP. Eleven groups remain, including Group A (26 ArbZIPs), Group B (3 ArbZIPs), Group C (11 ArbZIPs), Group D (27 ArbZIPs), Group E (8 ArbZIPs), Group F (5 ArbZIPs), Group G (5 ArbZIPs), Group H (4 ArbZIPs), Group I (15 ArbZIPs), Group K (2 ArbZIPs), Group M (2 ArbZIPs). Overall, the number of ArbZIPs was greater than that of AtbZIPs in most groups (A, C-F, H, I, K, M, and S), while in Groups B, G, and J, the counts were equivalent between red maple and Arabidopsis.
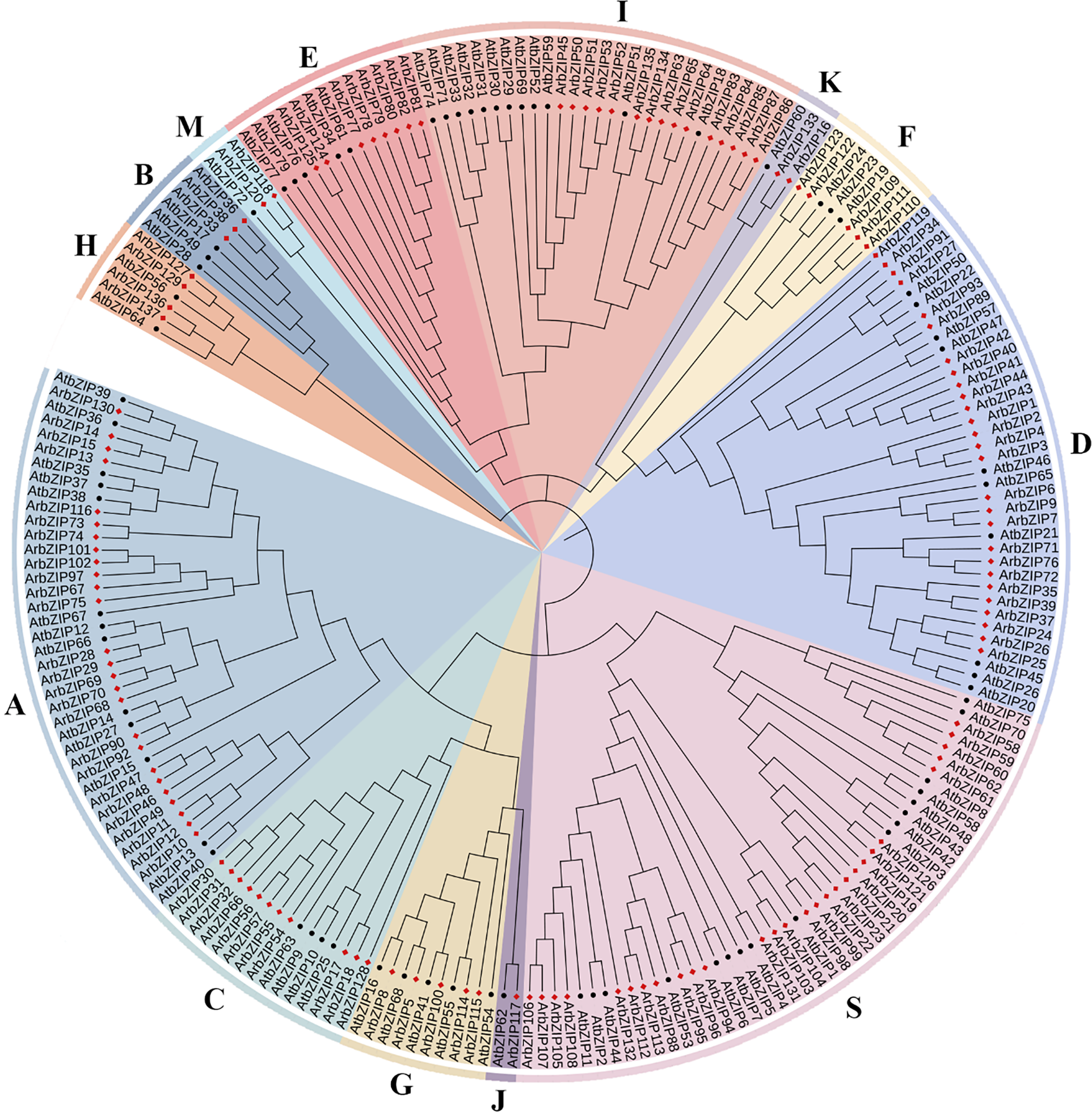
Figure 1: The phylogenetic tree of AtbZIP (Arabidopsis) and ArbZIP (Acer rubrum) proteins
3.3 Analysis of ArbZIP Motif, Domain and Gene Structure
To further investigate the phylogenetic relationships of Acer rubrum bZIP genes, we conducted the motifs, conserved domains, and exon-intron structures of the red maple bZIP proteins. After using the MEME program in TBtools, 20 conserved motifs were identified in 137 Acer rubrum bZIP proteins. The findings were largely in line with the phylogenetic relationships, as individuals with similar conserved motifs were classified together in the relevant phylogenetic groupings, suggesting potential functional similarities [49]. As shown in Fig. 2A, Motif 1 is present in every Acer rubrum bZIP. Most ArbZIP proteins contain multiple motifs, but ArbZIP119 and ArbZIP115 only contain Motif 1. The majority of ArbZIPs contain Motif 8, except for all members of Group D. The most motifs were identified in group D, with most members containing 8 motifs, while members of Groups H and K had the fewest motifs, with the remaining four members, except for ArbZIP136, having only 2 motifs. Furthermore, the conserved domain bZIP, exclusive to the bZIP gene family, is present in all ArbZIP proteins. This conserved domain comprises the BRLZ domain, which is characteristic of the bZIP family’s conserved domains [58], supporting the reliability of the gene identifications (Fig. 2B).

Figure 2: Phylogenetic relationships, motifs, conserved domains, and gene structure of ArbZIP TFs. (A): Motif composition of ArbZIP proteins. Motif composition of ArbZIP proteins was identified using the MEME tool, different motifs represented by modules in various colors (B): Conserved domains of ArbZIP proteins. Conserved structural domains were obtained using CD-research and plotted in TBtools after organizing the data. (C): ArbZIP genes structures. CDS denoted coding sequence and lines denoted introns. The image was drawn using Biosequence Structure Illustrator of TBtools
Since the intron/exon composition and the types and numbers of introns were significant markers in the evolution of some gene families, we investigated the gene structure of ArbZIPs to further understand their characteristics of the bZIP gene structure in Acer rubrum. Visual analysis of the coding regions (CDS) and introns of ArbZIP genes (Fig. 2C) revealed diverse conserved domains, gene structures, and varying numbers of introns/exons among different groups. The intron count ranged from 0 to 11, consistent with expectations. Most Acer rubrum bZIP genes (78.1%) contain at least one intron. Except for one intron in ArbZIP104, no introns existed in the S1 and S2 groups. ArbZIP5, ArbZIP8, ArbZIP114, ArbZIP72, ArbZIP76, and ArbZIP24 have the highest number of introns, with 11 each. The number of introns is also consistent within the same subfamily. For example, B group members have three introns except for ArbZIP52, which has four; the C group has four to seven introns, and the B group has one intron each. Based on the comprehensive results of phylogenetic relationships, gene sequences, gene structures, and conserved domains, it is evident that ArbZIP genes are highly conserved during the long process of evolution.
3.4 Analysis of Chromosomal Positions, Duplications, and Synteny with Other Plants
The hexaploid red maple used in this study had 39 chromosomes. Analysis of the chromosomal locations showed that the 137 ArbZIPs were unevenly distributed among 29 chromosomes (Fig. 3). Chromosomes 14 and 21 each contained only one ArbZIP gene, whereas chromosomes 9, 16, and 24 had the highest number, each with 9 ArbZIPs. Gene duplication, including both tandem and segmental duplication, is crucial for the expansion of gene families [59]. Using One Step MCScanX-Super Fast of TBtools, we analyzed duplication events of the ArbZIP genes in red maple. Two pairs of bZIP genes were tandem duplications (Table S4), and in Fig. 3, each pair of tandemly duplicated ArbZIP genes is labeled in red and connected with a blue line. The study identified 106 segmental duplication gene pairs (Fig. 4, Table S5), suggesting segmental duplications were much higher than tandem duplications.
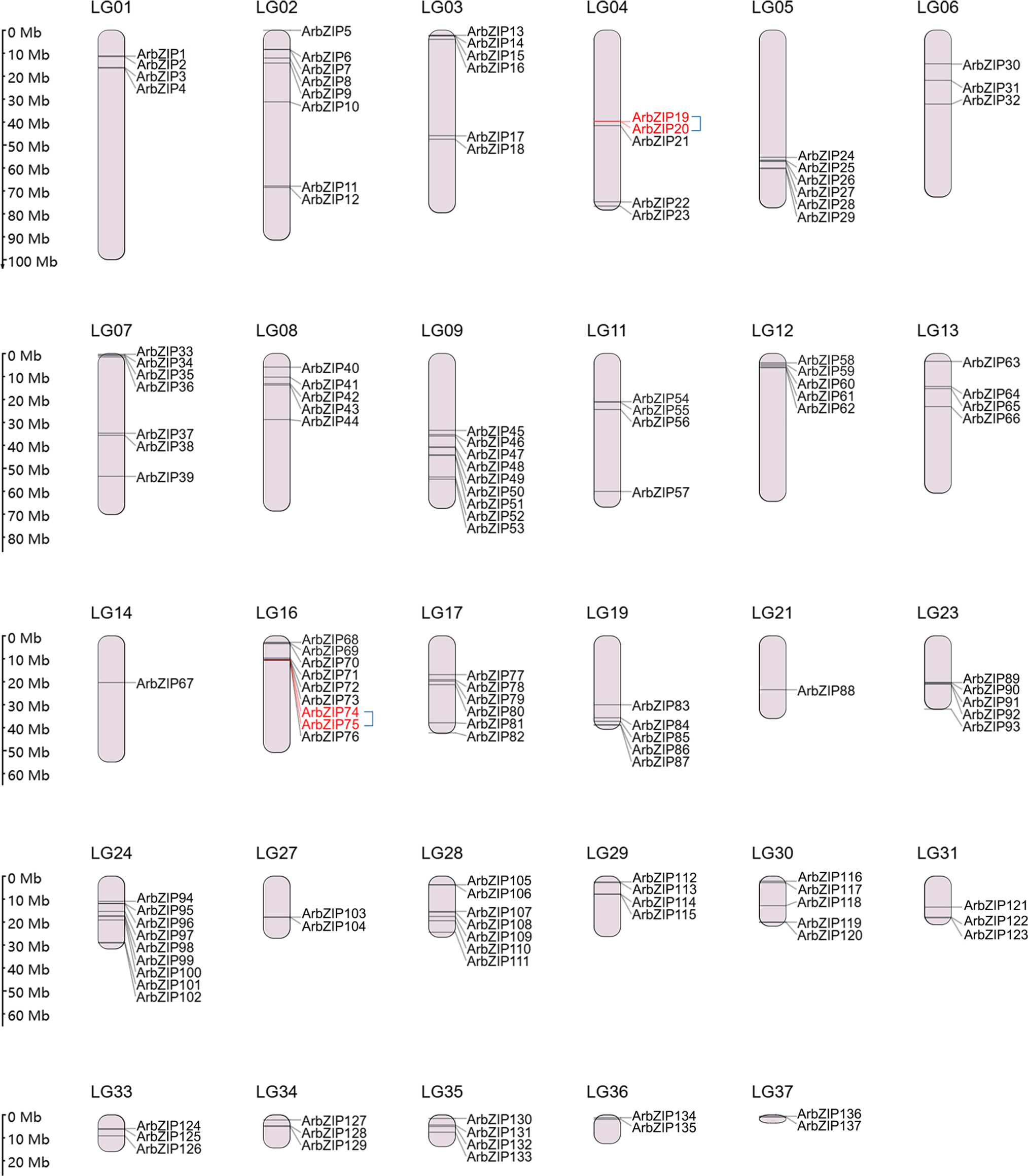
Figure 3: Chromosomal locations of ArbZIP TFs. The distribution of 137 ArbZIP genes was examined across 29 chromosomes using TBtools, based on the General Feature Format (GFF) files. Duplication events of ArbZIP genes were analyzed with One Step MCScanX-Super Fast of TBtools. A total of 2 pairs of bZIP genes (highlighted in red) resulted from tandem duplication, with each pair of tandem duplicated ArbZIP genes connected by a blue line. The lengths of chromosomes and the positions of genes can be deduced from the scale provided on the left side
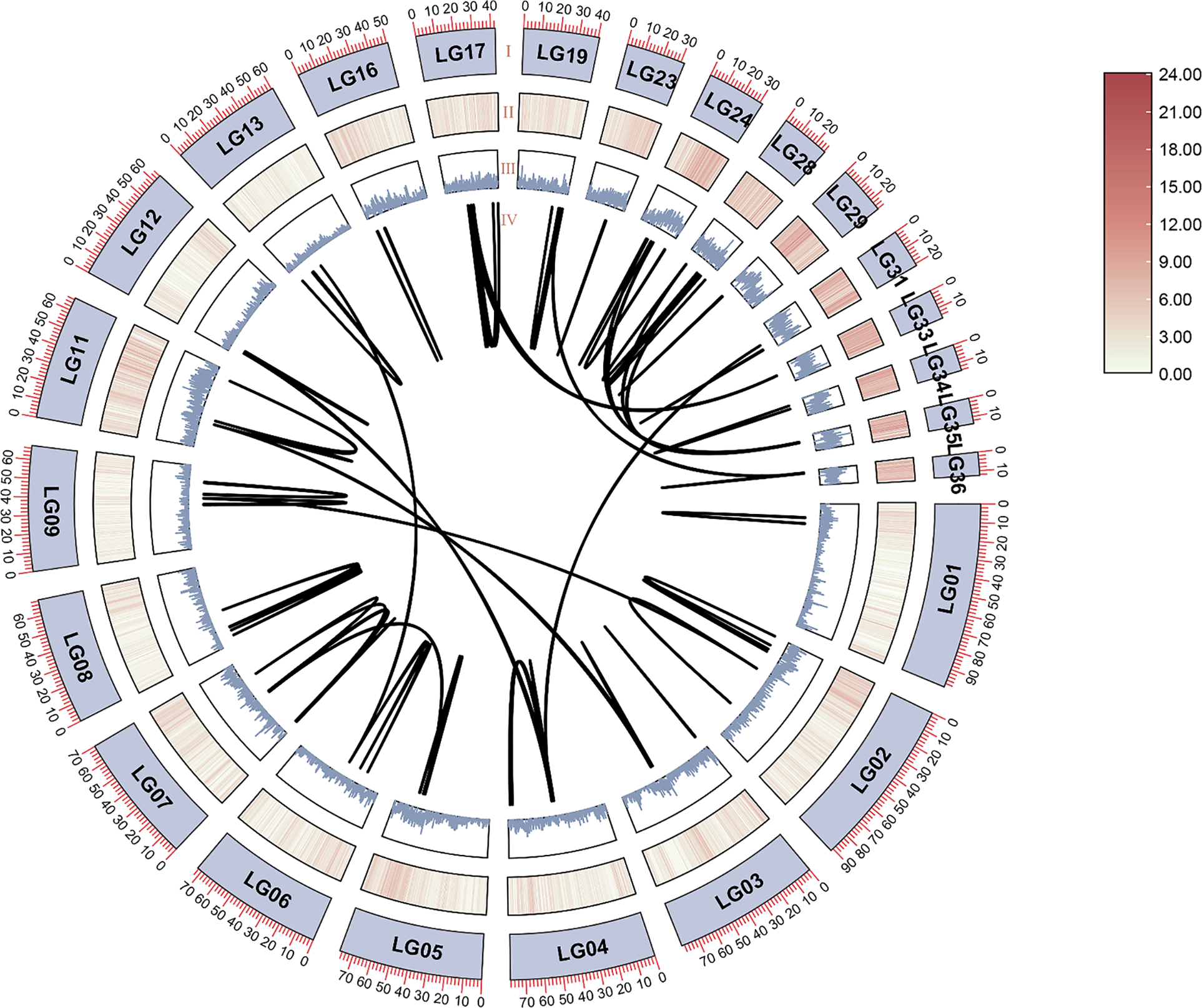
Figure 4: Inter-chromosomal relationships between ArbZIP genes via TBtools. I, Chromosomes (24 with segmentally duplicated) name. II, The heatmap of gene density. III, The line graph of gene density. IV, Segmental duplications in red maple bZIP genes. Using One Step MCScanX-Super Fast of TBtools, 106 segmental duplication gene pairs were found
Collinearity refers to the preserved positional relationships of genes on a chromosome, encompassing the conserved gene type and relative order among different species that have diverged from a common ancestral type. To investigate the relationship between the ArbZIP genes and bZIPs of other plants, we performed whole genome synteny analysis (Fig. 5). Using the One Step MCScanX -Super Fast program in TBtools, bZIP genes in Arabidopsis thaliana, Oryza sativa, Populus trichocarpa and Vitis vinifera were chosen for comparison with red maple, respectively. The findings showed that Arabidopsis thaliana had 80 orthologous gene pairs with red maple, while Populus trichocarpa had 192 orthologous gene pairs, Oryza sativa had 59 orthologous gene pairs, and Vitis vinifera had 135 orthologous gene pairs. Co-linear conservation of bZIP genes can be observed between these plants and Acer rubrum.
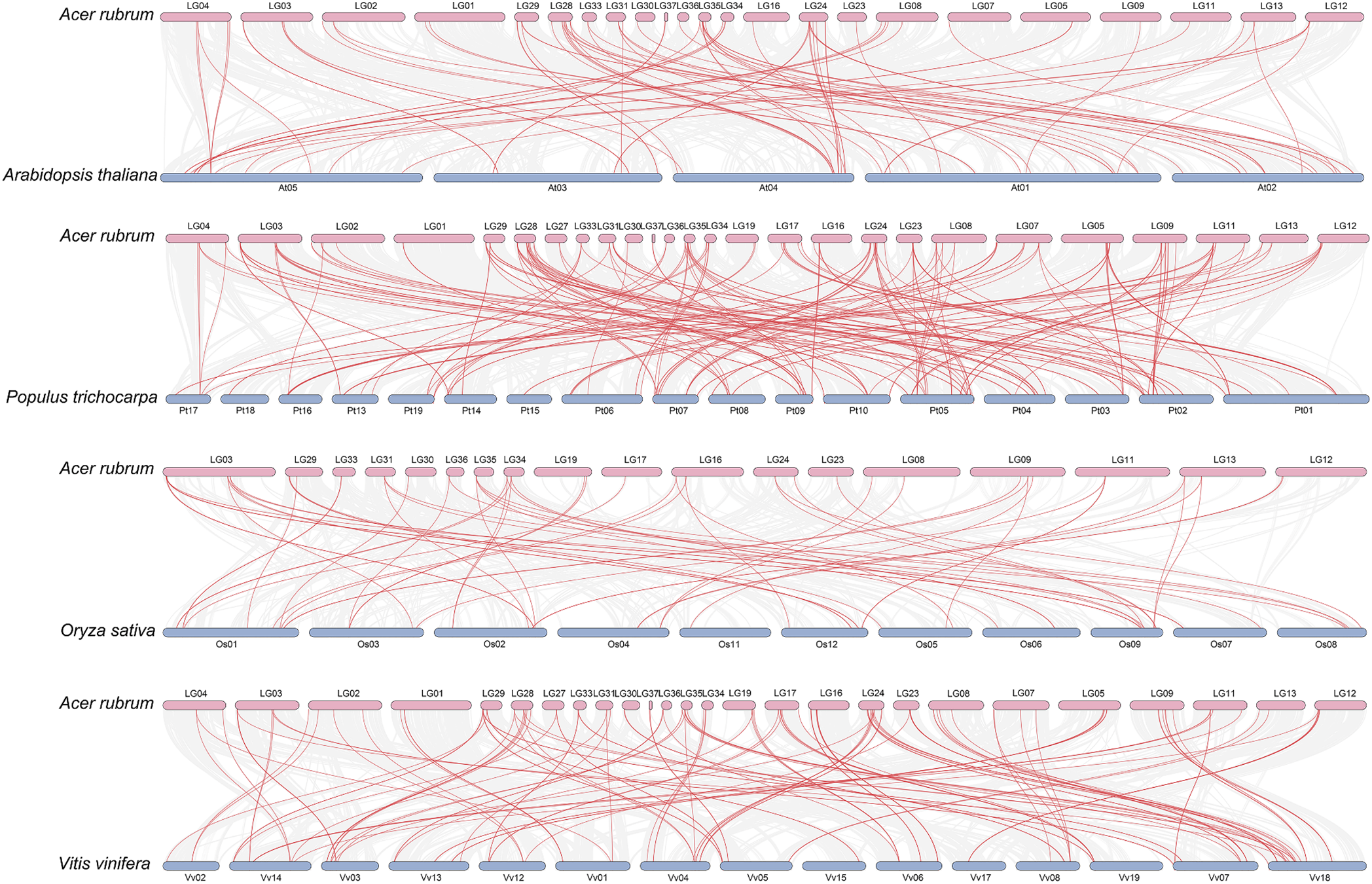
Figure 5: Synteny analyses between the ArbZIPs and four representative plant species. The four plants are Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Vitis vinifera, and their genome sequences and GFF files were acquired from NCBI. Comparisons were made using the One Step MCScanX-Super Fast program from TBtools and visualizations were made using the Dual Systeny Plot program
3.5 Analysis of Cis-Acting Element for ArbZIPs
Cis-acting elements are binding sites for transcriptional regulators, influencing upregulation and downregulation of gene expression [49]. We obtained the cis-acting elements from the PlantCare website to elucidate their potential roles and mechanisms (Fig. 6, Table S6). The results of data processing indicated that the 20 kinds of cis-acting elements of ArbZIP genes can be categorized into five groups: those involved in the regulation of plant circadian control; those involved in phytohormone responsiveness (e.g., CGTCA-motif and TGACG-motif); those related to plant growth and development; those implicated in light responsiveness; and those associated with abiotic stress response (e.g., LTR and TC-rich repeats). The identified 137 ArbZIPs each harbored multiple cis-acting elements, implying their potential association with abiotic stresses. The largest subset of cis-acting elements was related to light responsiveness. Box 4 elements had the highest number, with a total of 419, whereas there were 55 ArbZIPs with one or more LTR responsiveness elements. Interestingly, the MYB gene had a binding site with the bZIP gene, indicating its role in triggering drought, responding to light, and facilitating the biosynthesis of flavonoids in Acer rubrum (Table S7). These findings suggest that the ArbZIP genes might participate in multiple plant biological processes through the formation of gene regulatory networks.
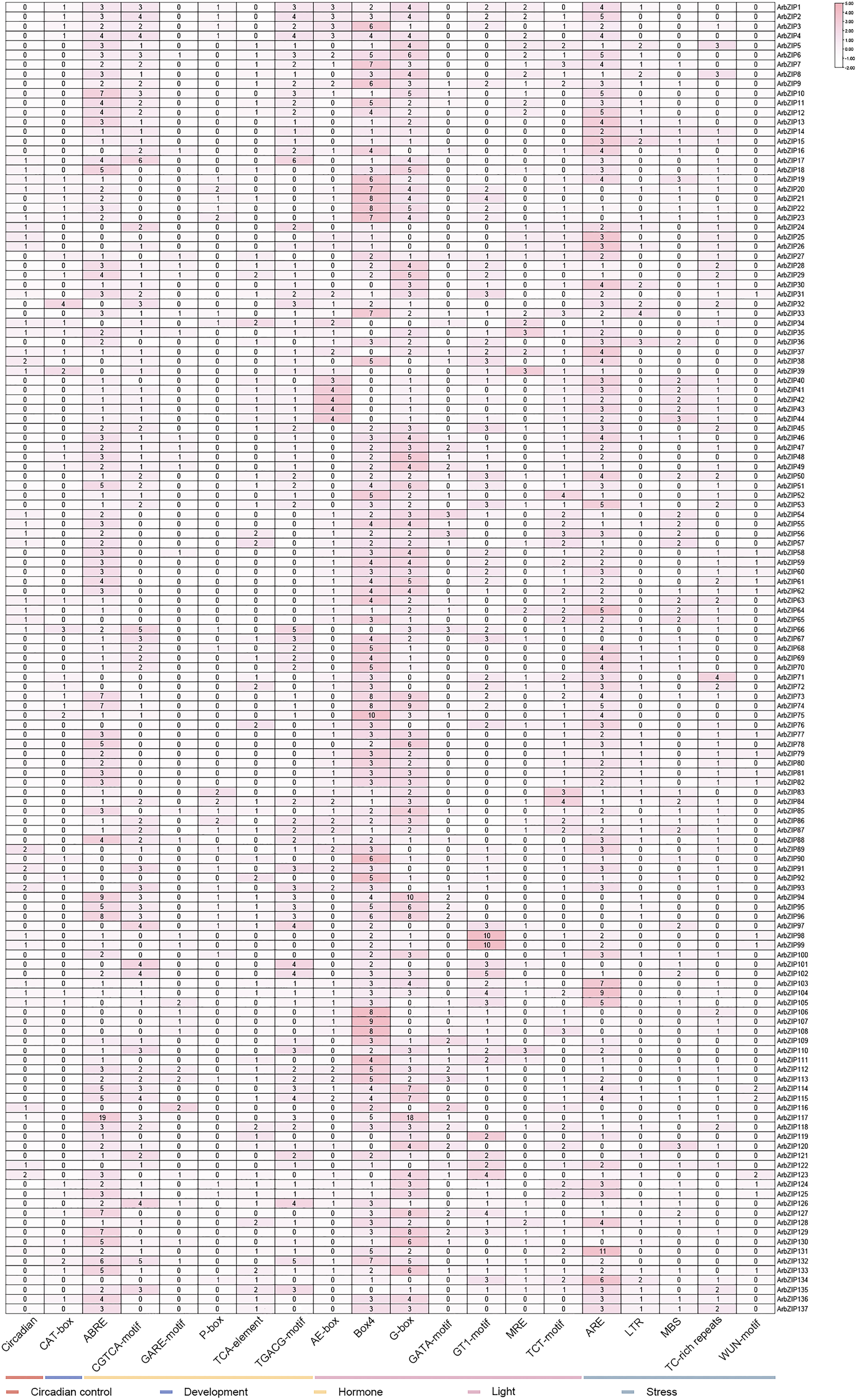
Figure 6: Analysis of cis-acting elements in ArbZIP promoters. The color and number of the square represent the number of cis-acting elements in the corresponding ArbZIP. Data were obtained from PlantCare and then collated. Images were plotted using HeatMap of TBtools
3.6 Analysis of Anthocyanin Content
The leaf color of Acer rubrum under low temperature with light changed significantly (Fig. 7A), and the leaf color only became red under low temperature with light. The results of anthocyanin content determination of leaves were consistent with this performance (Fig. 7B). The highest anthocyanin content of 247.45 μg/g was found in the leaves under low temperature with light, which was 12.26 times higher than the anthocyanin content of leaves in the control group (20.19 μg/g). In contrast, the anthocyanin content of leaves under the low temperature without light was only 3.07 μg/g, which was even lower than that of the control leaves.

Figure 7: Effect of low temperature with light and without light on red maple. (A): Changes in leaf color. (B): Anthocyanin content. The X-axis represents the treatment methods. Control is the control group, AR4C_D represents low temperature without light, and AR4C represents low temperature with light. The Y-axis represents the anthocyanin content. Each bar was the mean ± standard error (SE) from three replicates. Identical letters on error bars indicated no significant difference (p > 0.05), while different letters represent significant differences (p < 0.05)
3.7 Expression Pattern of ArbZIP under Low Temperature with Light and without Light
Gene expression patterns play a crucial part in predicting gene function. We analyzed the expression levels of 137 ArbZIPs using FPKM transcriptome data. Compared with the Control group, the expression of 40 ArbZIP genes increased under low temperature with light (AR4C), suggesting that they might respond to low temperature stress. The expression of 48 ArbZIP genes increased under light conditions (AR4C) compared with dark conditions (AR4C_D), indicating that they might respond to light, and the 25 ArbZIPs with the most pronounced contrasts were selected for mapping (Fig. 8A, Table S8). Based on the comparative results of TF expression, we selected 20 TFs each that were most affected by light and low temperature for comparison. We found that ArbZIP137, ArbZIP136, ArbZIP114, ArbZIP130, and ArbZIP14 were both greatly affected under light and low temperature conditions.
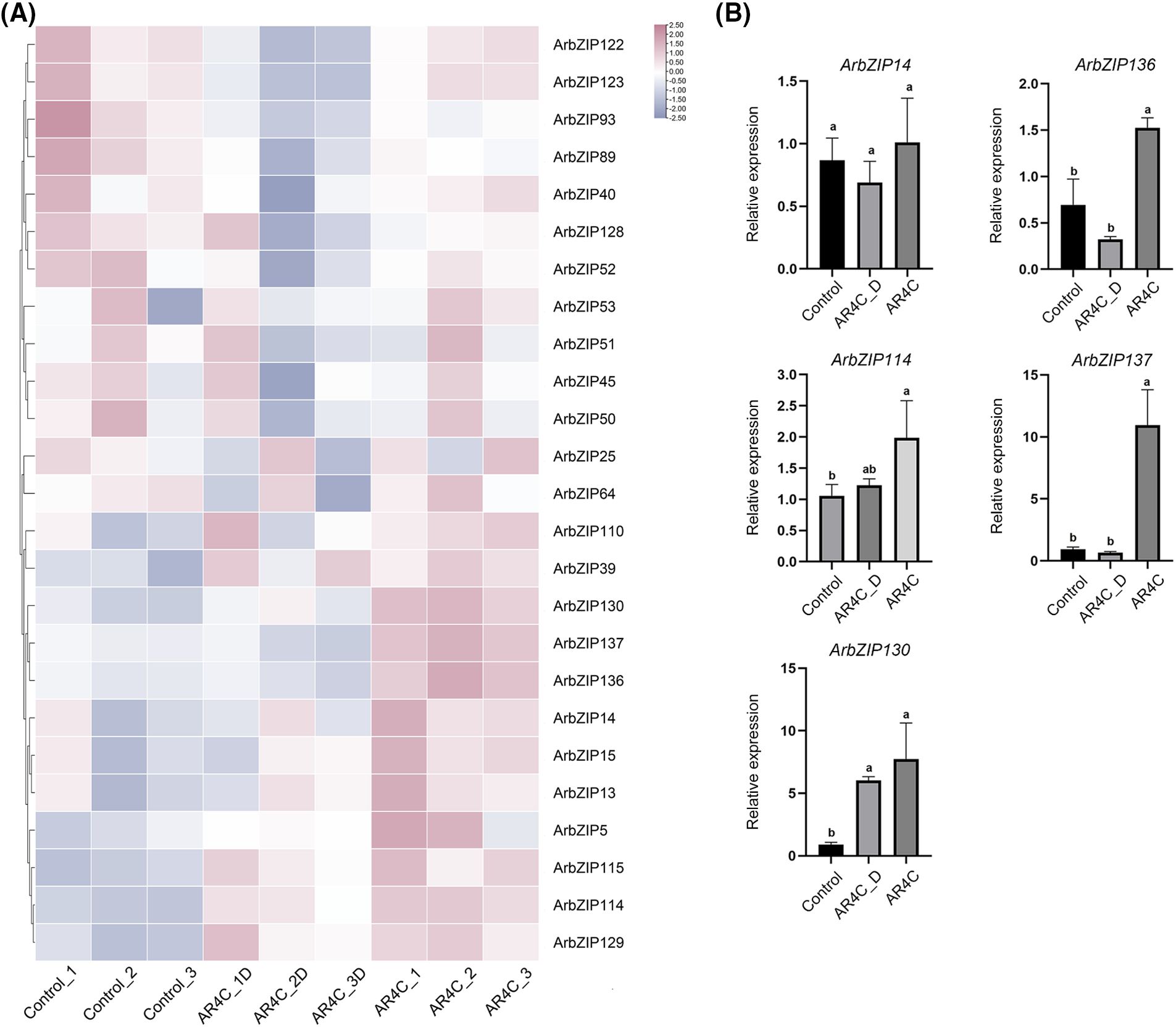
Figure 8: (A): Analysis of the expression pattern of ArbZIP genes under low temperature with light and without light (the 25 ArbZIPs with the most pronounced contrast in expression comparing AR4C vs. AR4C_D group). Control: 22°C with light; AR4C_D: 4°C without light; AR4C: 4°C with light. (B): QRT-PCR analysis of bZIPs in the leaves of Acer rubrum under low temperature with light and without light treatments. The X-axis represents the treatment methods. Control is the control group, AR4C_D represents low temperature without light, and AR4C represents low temperature with light. The Y-axis represents the relative expression level of the genes. In all bar graphs, the data presented show the mean ± standard error from three biological replicates. A one-way ANOVA was used for statistical analysis, followed by Tukey’s HSD test for comparisons. Different letters indicate significance at p < 0.05
The 5 ArbZIPs that are influenced by low temperature with light were validated using qRT-PCR. The results showed that, compared to the Control group, the relative expression levels of ArbZIP137, ArbZIP136, ArbZIP114, and ArbZIP130 in the leaves were significantly elevated under low temperature with light (Fig. 8B). Among these ArbZIPs, ArbZIP137 and ArbZIP136 exhibited significant differences when compared to both the Control group and the low temperature without light group. These results support the accuracy of the RNA-Seq data.
BZIP genes typically form regulatory networks with various TFs to assist plants in coping with abiotic stresses. To investigate the transcriptional regulatory network of ArbZIPs associated with anthocyanins, we analyzed the TFs in the transcriptome of Acer rubrum leaves under low temperature stress with light and without light. The anthocyanin-related transcription factor families MYB, bHLH, and WD40 [60] that have been reported were selected. 393 ArMYB genes were derived from research by Chen et al. [4]. Additionally, 256 ArbHLH genes and 656 ArWD40 genes were identified from the Acer rubrum database, and all genes were named based on their physical chromosome location. We analyzed the expression of TFs belonging to three families under low temperature stress with and without light. Five ArbZIP TFs significantly affected by temperature with light and without light were selected: ArbZIP137, ArbZIP136, ArbZIP114, ArbZIP130 and ArbZIP14. The expression levels of MYB, bHLH, and WD40 family and 5 selected ArbZIPs in the leaves of Acer rubrum under light and without light under low temperature stress were analyzed by R, and the co-expressed TFs of the ArbZIP gene were explored (Table S9). Next, Cytoscape (v3.10.1) was used for visualization (Fig. 9). The 87 genes from 3 families were co-expressed with the 5 ArbZIP genes (correlation > 0.9). The WD40 family had the most significant number of co-expressed TFs (43), followed by MYB (35) and the bHLH family (9).
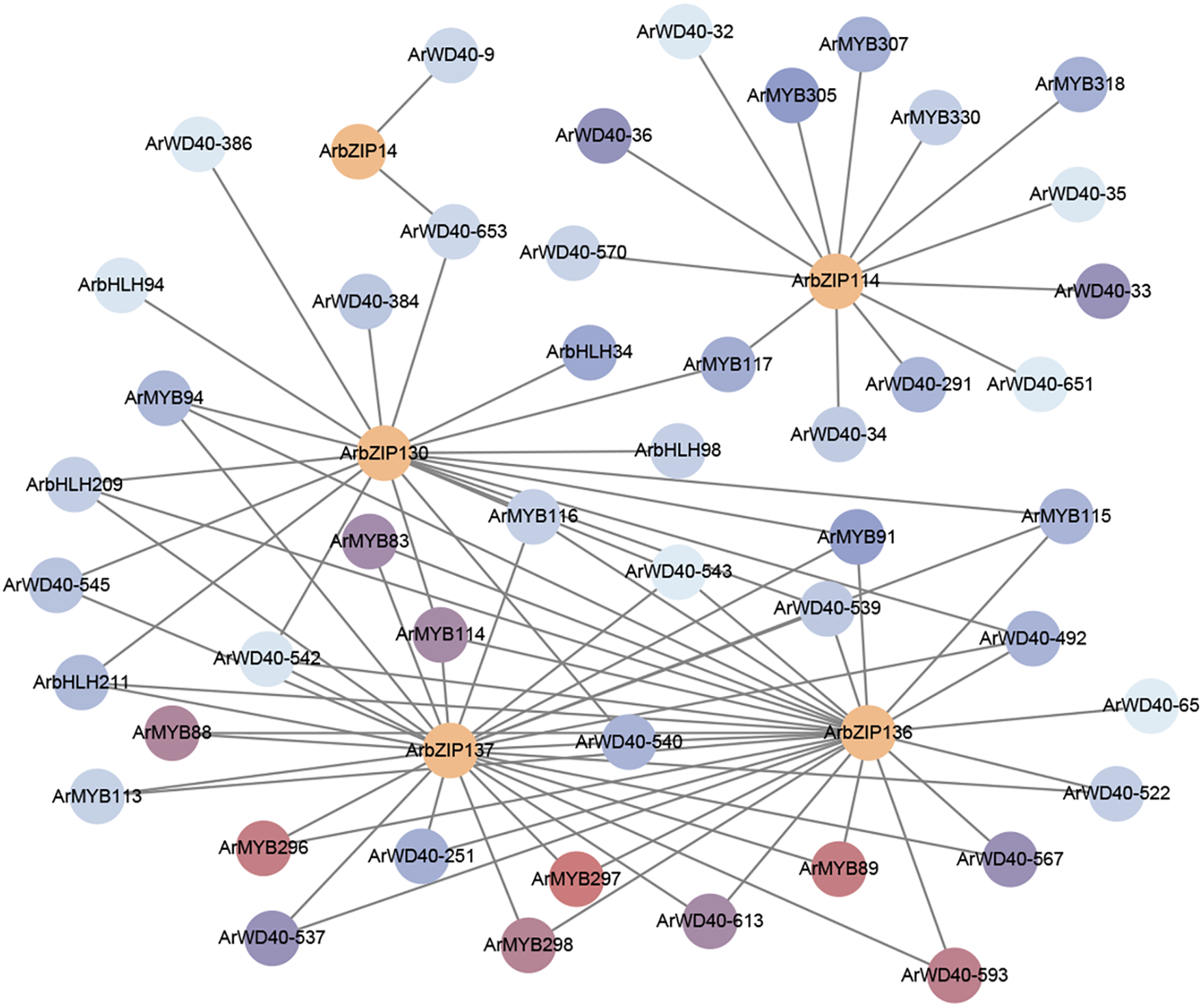
Figure 9: ArbZIP gene co-expression network was created using the transcriptome data of Acer rubrum leaves under low temperature with light and without light conditions. Red represents genes with high correlation, and blue represents genes with low correlation, and the deeper the color, the higher the correlation. Different colors represent different types of genes: orange represents ArbZIP gene, and red and blue represent other TFs
Acer rubrum is among the tree species with red autumn leaves widely used in landscapes. Leaf color change correlates with anthocyanin accumulation, a group of secondary metabolites prevalent in plants belonging to flavonoids. Anthocyanins play an important role in determining plant flower and leaf color expression. The anthocyanin accumulation can make the leaf color more beautiful and help it deal with various environmental stresses. The regulation of TFs (TFs) is critical for the synthesis of plant secondary metabolites. According to the existing studies, bZIP TFs play an important role in the accumulation of anthocyanins. Therefore, it is crucial to study the systematic identification of bZIPs in red maple and its potential role in anthocyanin accumulation.
As one of the largest and most diverse gene families, bZIP genes significantly affect abiotic stress response, plant growth and development, and anthocyanin accumulation. Research on this gene family has predominantly focused on model plants like Arabidopsis thaliana and specific crops, such as AtbZIP18 in Arabidopsis thaliana [61], GsbZIP2 in soybeans [62], TabZIP15 in wheat [40], and MdbZIP44 in apples [46]. In recent years, with improving people’s quality of life, the pursuit of beautiful ecological environments has become increasingly intense. The continuous development of urban parks and enhancing urban quality have also led to an increasing demand for the quantity and quality of ornamental trees. Acer rubrum, as an essential ornamental tree, has beautiful foliage, wide applications, and highly ornamental, economic, and medicinal values. Although the bZIPs has been widely reported in agriculture, there is relatively little research on ornamental trees, especially in Acer species. Therefore, this study, 137 ArbZIP genes were identified using bioinformatics methods after predicting the structural domains and removing incomplete structural domains and redundant sequences using the CD-Search tool. They all have the conserved structural domain bZIP (including the BRLZ structural domain), unique to the bZIP gene family, and are unevenly distributed in 29 of the 39 chromosomes.
Phylogenetic analysis predicts gene functions identifying homologous genes across diverse species. Therefore, we constructed a phylogenetic tree by utilizing the sequences of Acer rubrum bZIP proteins and Arabidopsis thaliana bZIP proteins. The 137 ArbZIP proteins and 77 AtbZIP proteins were classified into 13 subfamilies (A–K, M, and S) based on previous research about Arabidopsis thaliana [33]. In Arabidopsis, conserved motifs specific to the AtbZIPs subfamily critically impact their functional differentiation. For instance, the H group members, such as AtbZIP56/HY5 and AtbZIP64 which have a CKII phosphorylation site and a WD40-interacting structural domain [63], directly bind to promoters of light-induced genes. This regulatory action impacts the elongation of plant cells, the synthesis of chloroplast, hormone production, and anthocyanin biosynthesis [64]. We hypothesized that ArbZIP127, ArbZIP129 (homologous to AtbZIP56), ArbZIP136 and ArbZIP137 (homologous to AtbZIP64), might have similar functions. The study also revealed that the number of bZIP proteins in red maple is more significant than in Arabidopsis thaliana in multiple subfamilies (A, C–F, H, I, K, M, S), indicating that it might possess more complex and diverse functions in responding to environmental changes, and regulating growth and development. This difference might be attributed to the varying ecological niches, growth habits, and life cycles of the two plants. Furthermore, while the bZIP proteins of the two plants are evolutionarily conserved, the differences in the number of members in specific groups reflect their different histories of gene family expansion and divergence, providing insights into the functions of bZIP family members in plant adaptation to the environment and responses to biotic stresses.
Gene structure and conserved motifs are crucial for studying gene evolution and duplication. Acer rubrum bZIP proteins commonly contain motif 1, which might serve as a core functional element as a transcription factor. The different distributions of motif 8 among subgroups reveal potential functional differences. Similar to other plants [65], we found that members of different groups in Acer rubrum exhibit diverse gene structures, conserved domains, and intron/exon numbers, with intron numbers ranging from 0 to 11.
When studying cis-acting elements, we identified 20 kinds of cis-acting elements within the 2000 bp region upstream of the ArbZIPs. These cis-acting elements belonged to five major classes of regulatory functions, including circadian control, growth and development, hormone response, light response, and stress response, which might be related to the regulation of ArbZIP gene expression, a result similar to that of the studies previous studies [57,66]. In particular, the largest number of cis-acting elements are associated with the light response. The light-responsive element Box 4 contains the highest number of elements, with 419, while the G-box is second, with 382 elements. In tomatoes, SlHY5 directly recognizes and binds to the G-box and ACGT elements in the promoter of anthocyanin biosynthetic genes [67]. Therefore, the bZIPs might also regulate anthocyanin biosynthesis in red maple through the light-responsive elements. According to the results given by Plantcare, there are binding sites between the transcription factor MYB and the bZIP gene in red maple, which is similar to blackberry [68]. These binding sites play a role in processes such as light response, drought induction, and flavonoid biosynthesis, suggesting that the ArbZIP gene might form a complex gene regulatory network through interactions with other TFs, such as the MYB gene, and thus be involved in a variety of biological processes in the plant, such as responding to light signals and contributing to anthocyanin accumulation.
Both light and temperature are crucial environmental factors impacting plant growth and development, especially for anthocyanin accumulation. Consequently, we further examined the expression of the red maple bZIPs under low temperature with light and without light. Our study revealed that the synergistic effect of light and temperature significantly affected anthocyanin accumulation in Acer rubrum leaves, which exhibited significant color changes under different experimental conditions (Fig. 7A). Notably, the leaves turned red only under low temperature (4°C) with light, and the detection of anthocyanin content also proved that anthocyanins were significantly accumulated under low temperature with light (Fig. 7B). When studying gene expression patterns, we found that the expression of 40 ArbZIPs increased under low temperature condition, suggesting that they might play key roles in coping with low temperature stress. In apple pericarp, low temperature stimulated genes related to anthocyanin biosynthesis, leading to increased anthocyanin accumulation [69]. The effect of temperature on the fruit color of red Chinese sand pear was similar to that of apple. Low temperature could effectively induce the red coloration of ‘Mantianhong’ and ‘Meirensu’ pears [7]. Therefore, it could be seen that low temperature is more favorable for the synthesis of anthocyanin glycosides in the fruit. Under light conditions, the expression of 48 ArbZIPs increased, including ArbZIP136 and ArbZIP137. ArbZIP137 and ArbZIP136 belong to the Group H of the phylogenetic tree and are highly homologous to AtbZIP64, and the latter is known to belong to Group H of the phylogenetic tree with AtbZIP56 (AtHY5) belongs to Group H, which is involved in the regulation of photomorphogenesis and has a structural domain interacting with WD40. Up to now, considerable research has been done on HY5. In Arabidopsis, HY5 and HYH serve as downstream phytochrome receptors of the light signaling pathway, promoting the accumulation of pigment in a light-dependent manner [70]. Silencing of the SlHY5 gene down-regulates the anthocyanins accumulation in tomato [67]. The WD40 protein also modulates anthocyanin by binding to an action element in the promoter region of the structural gene biosynthesis. Therefore, we supposed that some ArbZIPs might have similar functions in regulating anthocyanin accumulation. Among the expression level data of 137 ArbZIP genes under low temperature stress with light (Table S8), we selected 5 transcription factors, ArbZIP137, ArbZIP130, ArbZIP114, ArbZIP136, and ArbZIP14, which were strongly affected by both low temperature and light. QRT-PCR results confirmed that, under the low temperature conditions with light, the expression levels of ArbZIP137, ArbZIP130, ArbZIP114, ArbZIP136 and ArbZIP14 were significantly increased. These findings suggested that these genes might coordinate the anthocyanin biosynthesis pathway in response to the combined effects of low temperature with light.
Based on the findings of correlation analysis, the selected ArbZIPs have high correlation with some anthocyanin-related TFs. The high correlation between ArbZIP137 and ArMYB89 and between ArbZIP136 and ArMYB89 is particularly noteworthy. Chen et al. have thoroughly demonstrated the role of ArMYB89 in anthocyanin synthesis [4]. Therefore, it can be speculated that ArbZIP136 and ArbZIP137 might also have a similar effect. The 42 MYB TFs were highly correlated with the 5 ArbZIP genes (Table S9), further confirming the close relationship between ArbZIP genes and MYB family members and their potential impact on the anthocyanin synthesis pathway. In addition to the MYB family, we also found a significant correlation between the ArbZIP gene and WD40 family members (such as ArbZIP137 and ArWD40-593). As another critical regulator in the anthocyanin synthesis pathway, the interaction between the WD40 family and the ArbZIP gene might further enrich the complexity of the plant transcriptional regulatory network. A strong correlation (0.98) was observed between ArbHLH211 and ArbZIP136, indicating a relationship between ArbHLH and ArbZIP genes. Combining the results of gene expression analysis and anthocyanin content experiments, we could further speculate that ArbZIP136 and ArbZIP137 might have a particular effect on the accumulation and inheritance of anthocyanin in Acer rubrum under low temperature with light conditions. Conclusively, the ArbZIP gene might be involved in the response of plants to environmental conditions through a complex regulatory network formed by members of multiple transcription factor families like MYB, bHLH, and WD40, affecting anthocyanin synthesis and accumulation, especially ArbZIP136 and ArbZIP137. These findings triggered our interest in exploring the mechanism of ArbZIP136 and ArbZIP137 in anthocyanin accumulation. In the future, we will further study these two TFs.
In summary, in this study, the red maple bZIP TFs were identified at the genome-wide level. Bioinformatics methods identified 137 ArbZIP genes, their bZIP structural domains were determined, and phylogenetic trees of red maple and Arabidopsis thaliana were constructed. The gene structure, chromosomal localization, replication events, cis-acting elements, anthocyanin contents and expression patterns of the red maple bZIP TFs were further analyzed. After treatment, we found that the plant leaf color changed to red and anthocyanin content increased significantly only under low temperature with light. The gene expression analysis showed that some bZIPs in red maple might respond to low temperature with light, which is confirmed by the qRT-PCR results. Combining with the analysis of anthocyanin content, gene expression analysis, correlation analysis and the analysis of existing studies, ArbZIP TFs might promote anthocyanin accumulation under low temperature with light conditions, especially ArbZIP136 and ArbZIP137. In addition, some bZIP TFs might also affect the synthesis and accumulation of anthocyanins through complex regulatory networks. However, the specific function of ArbZIPs in red maple still needs to be experimentally proved in future work. In the future, we might focus on ArbZIP136 and ArbZIP137 to carry out the following work.
Acknowledgement: None.
Funding Statement: This study was funded by the National Natural Science Foundation of China (Project No. 32271914) and the Natural Science Foundation of Anhui Province (2108085MC110).
Author Contributions: Study conception and design: Hua Liu, Jie Ren; data collection: Zhu Chen, Khan Arif Kamal; analysis and interpretation of results: Yue Zhao, Shah Faheem Afzal, Yuzhi Fei, Xin Meng; draft manuscript preparation: Yue Zhao, Shah Faheem Afzal. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All the data supporting the findings of this study are included in this article (and its supplementary information files).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.056548.
References
1. Gelderen DV, Jong PC, Oterdoom H. Maples of the World; 2005. doi:10.5860/choice.32-2727. [Google Scholar] [CrossRef]
2. Lu X, Chen Z, Liao B, Han G, Shi D, Li Q, et al. The chromosome-scale genome provides insights into pigmentation in Acer rubrum. Plant Physiol Biochem. 2022;186:322–33. doi:10.1016/j.plaphy.2022.07.007 [Google Scholar] [PubMed] [CrossRef]
3. Chen Z, Lu X, Xuan Y, Tang F, Wang J, Shi D, et al. Transcriptome analysis based on a combination of sequencing platforms provides insights into leaf pigmentation in Acer rubrum. BMC Plant Biol. 2019;19(1):240. doi:10.1186/s12870-019-1850-7 [Google Scholar] [PubMed] [CrossRef]
4. Chen Z, Lu X, Li Q, Li T, Zhu L, Ma Q, et al. Systematic analysis of MYB gene family in Acer rubrum and functional characterization of ArMYB89 in regulating anthocyanin biosynthesis. J Exp Bot. 2021;72(18):6319–35. doi:10.1093/jxb/erab213 [Google Scholar] [PubMed] [CrossRef]
5. Plunkett B. Environmentally regulated control of anthocyanin biosynthesis in apple. Auckland: The University of Auckland; 2012. Available from: http://hdl.handle.net/2292/12291. [Accessed 2024]. [Google Scholar]
6. Li Z, Ahammed GJ. Hormonal regulation of anthocyanin biosynthesis for improved stress tolerance in plants. Plant Physiol Biochem. 2023;201:107835. doi:10.1016/j.plaphy.2023.107835 [Google Scholar] [PubMed] [CrossRef]
7. Sun Y, Qian M, Wu R, Niu Q, Teng Y, Zhang D. Postharvest pigmentation in red Chinese sand pears (Pyrus pyrifolia Nakai) in response to optimum light and temperature. Postharvest Biol Technol. 2014;91:64–71. doi:10.1016/j.postharvbio.2013.12.015. [Google Scholar] [CrossRef]
8. Shaked-Sachray L, Weiss D, Reuveni M, Nissim-Levi A, Oren-Shamir M. Increased anthocyanin accumulation in aster flowers at elevated temperatures due to magnesium treatment. Physiol Plant. 2002;114(4):559–65. doi:10.1034/j.1399-3054.2002.1140408.x [Google Scholar] [PubMed] [CrossRef]
9. Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol. 2005;57(2):155–71. doi:10.1007/s11103-004-6910-0 [Google Scholar] [PubMed] [CrossRef]
10. Sudheeran PK, Feygenberg O, Maurer D, Alkan N. Improved cold tolerance of mango fruit with enhanced anthocyanin and flavonoid contents. Molecules. 2018;23(7):1832. doi:10.3390/molecules23071832 [Google Scholar] [PubMed] [CrossRef]
11. Jezek M, Allan AC, Jones JJ, Geilfus CM. Why do plants blush when they are hungry? New Phytol. 2023;239(2):494–505. doi:10.1111/nph.v239.2. [Google Scholar] [CrossRef]
12. Chen X, Wang P, Gu M, Lin X, Hou B, Zheng Y, et al. R2R3-MYB transcription factor family in tea plant (Camellia sinensisgenome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics. 2021;113(3):1565–78. doi:10.1016/j.ygeno.2021.03.033 [Google Scholar] [PubMed] [CrossRef]
13. Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13(9):2099–114. doi:10.1105/TPC.010098 [Google Scholar] [PubMed] [CrossRef]
14. Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49(3):414–27. doi:10.1111/tpj.2007.49.issue-3. [Google Scholar] [CrossRef]
15. Yin X, Zhang Y, Zhang L, Wang B, Zhao Y, Irfan M, et al. Regulation of MYB transcription factors of anthocyanin synthesis in lily flowers. Front Plant Sci. 2021;12:761668. doi:10.3389/fpls.2021.761668 [Google Scholar] [PubMed] [CrossRef]
16. Wang H, Wang X, Song W, Bao Y, Jin Y, Jiang C, et al. PdMYB118, isolated from a red leaf mutant of Populus deltoids, is a new transcription factor regulating anthocyanin biosynthesis in poplar. Plant Cell Rep. 2019;38(8):927–36. doi:10.1007/s00299-019-02413-1 [Google Scholar] [PubMed] [CrossRef]
17. Jiang S, Sun Q, Zhang T, Liu W, Wang N, Chen X. MdMYB114 regulates anthocyanin biosynthesis and functions downstream of MdbZIP4-like in apple fruit. J Plant Physiol. 2021;257:153353. doi:10.1016/j.jplph.2020.153353 [Google Scholar] [PubMed] [CrossRef]
18. Zhang K, Lin C, Chen B, Lin Y, Su H, Du Y, et al. A light responsive transcription factor CsbHLH89 positively regulates anthocyanidin synthesis in tea (Camellia sinensis). Sci Hortic. 2024;327:112784. doi:10.1016/j.scienta.2023.112784. [Google Scholar] [CrossRef]
19. Yu D, Wei W, Fan Z, Chen J, You Y, Huang W, et al. VabHLH137 promotes proanthocyanidin and anthocyanin biosynthesis and enhances resistance to Colletotrichum gloeosporioides in grapevine. Hortic Res. 2023;10(2):uhac261. doi:10.1093/hr/uhac261 [Google Scholar] [PubMed] [CrossRef]
20. Yang X, Wang J, Xia X, Zhang Z, He J, Nong B, et al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021;107(1):198–214. doi:10.1111/tpj.v107.1. [Google Scholar] [CrossRef]
21. Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–27. doi:10.1111/tpj.2008.53.issue-5. [Google Scholar] [CrossRef]
22. Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39(3):366–80. doi:10.1111/tpj.2004.39.issue-3. [Google Scholar] [CrossRef]
23. Liu Y, Ma K, Qi Y, Lv G, Ren X, Liu Z, et al. Transcriptional Regulation of Anthocyanin Synthesis by MYB-bHLH-WDR Complexes in Kiwifruit (Actinidia chinensis). J Agric Food Chem. 2021;69(12):3677–91. doi:10.1021/acs.jafc.0c07037 [Google Scholar] [PubMed] [CrossRef]
24. Zhao M, Li J, Zhu L, Chang P, Li L, Zhang L. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes. 2019;10(7):496. doi:10.3390/genes10070496 [Google Scholar] [PubMed] [CrossRef]
25. Yue M, Jiang L, Zhang N, Zhang L, Liu Y, Lin Y, et al. Regulation of flavonoids in strawberry fruits by FaMYB5/FaMYB10 dominated MYB-bHLH-WD40 ternary complexes. Front Plant Sci. 2023;14:1145670. doi:10.3389/fpls.2023.1145670 [Google Scholar] [PubMed] [CrossRef]
26. Gil-Muñoz F, Sánchez-Navarro JA, Besada C, Salvador A, Badenes ML, Naval MDM, et al. MBW complexes impinge on anthocyanidin reductase gene regulation for proanthocyanidin biosynthesis in persimmon fruit. Sci Rep. 2020;10(1):3543. doi:10.1038/s41598-020-60635-w [Google Scholar] [PubMed] [CrossRef]
27. Wang Y, Zhang Y, Zhou R, Dossa K, Yu J, Li D, et al. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS One. 2018;13(7):e0200850. doi:10.1371/journal.pone.0200850 [Google Scholar] [PubMed] [CrossRef]
28. Li H, Chen J, Zhao Q, Han Y, Li L, Sun C, et al. Basic leucine zipper (bZIP) transcription factor genes and their responses to drought stress in ginseng, Panax ginseng C.A Meyer. BMC Genom. 2021;22(1):316. doi:10.1186/s12864-021-07624-z [Google Scholar] [PubMed] [CrossRef]
29. Zhao K, Chen S, Yao W, Cheng Z, Zhou B, Jiang T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021;21(1):122. doi:10.1186/s12870-021-02879-w [Google Scholar] [PubMed] [CrossRef]
30. Zhang Y, Zhang G, Xia N, Wang X-J, Huang L-L, Kang Z-S. Cloning and characterization of a bZIP transcription factor gene in wheat and its expression in response to stripe rust pathogen infection and abiotic stresses. Physiol Mol Plant Pathol. 2008;73(4):88–94. [Google Scholar]
31. Zhang Y, Xu Z-C, Ji A-J, Song J-Y. Regulation of secondary metabolite biosynthesis by bZIP transcription factors in plants. Plant Sci J. 2017;35(1):128–37. [Google Scholar]
32. Liu S-X, Qin B, Fang Q-X, Zhang W-J, Zhang Z-Y, Liu Y-C, et al. Genome-wide identification, phylogeny and expression analysis of the bZIP gene family in Alfalfa (Medicago sativa). Biotechnol Biotechnol Equip. 2021;35(1):905–16. doi:10.1080/13102818.2021.1938674. [Google Scholar] [CrossRef]
33. Dröge-Laser W, Snoek BL, Snel B, Weiste C. The Arabidopsis bZIP transcription factor family–an update. Current Opin Plant Biol. 2018;45:36–49. doi:10.1016/j.pbi.2018.05.001 [Google Scholar] [PubMed] [CrossRef]
34. Rong S, Wu Z, Cheng Z, Zhang S, Liu H, Huang Q. Genome-wide identification, evolutionary patterns, and expression analysis of bZIP gene family in olive (Olea europaea L.). Genes. 2020;11(5):510. doi:10.3390/genes11050510 [Google Scholar] [PubMed] [CrossRef]
35. Dong W, Xie Q, Liu Z, Han Y, Wang X, Xu R, et al. Genome-wide identification and expression profiling of the bZIP gene family in Betula platyphylla and the functional characterization of BpChr04G00610 under low-temperature stress. Plant Physiol Biochem. 2023;198:107676. doi:10.1016/j.plaphy.2023.107676 [Google Scholar] [PubMed] [CrossRef]
36. Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant physiol. 2008;146(2):333–50 [Google Scholar] [PubMed]
37. Li H, Li L, Shangguan G, Jia C, Deng S, Noman M, et al. Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctorius L. Sci Rep. 2020;10(1):15521. doi:10.1038/s41598-020-72390-z [Google Scholar] [PubMed] [CrossRef]
38. Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7(3):106–11. doi:10.1016/S1360-1385(01)02223-3 [Google Scholar] [PubMed] [CrossRef]
39. Lee SS, Yang SH, Berberich T, Miyazaki A, Kusano T. Characterization of AtbZIP2, AtbZIP11 and AtbZIP53 from the group S basic region-leucine zipper family in Arabidopsis thaliana. Plant Biotechnol. 2006;23(3):249–58. doi:10.5511/plantbiotechnology.23.249. [Google Scholar] [CrossRef]
40. Bi C, Yu Y, Dong C, Yang Y, Zhai Y, Du F, et al. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol J. 2021;19(2):209–11. doi:10.1111/pbi.v19.2. [Google Scholar] [CrossRef]
41. Liang Y, Xia J, Jiang Y, Bao Y, Chen H, Wang D, et al. Genome-wide identification and analysis of bZIP gene family and resistance of TaABI5 (TabZIP96) under freezing stress in wheat (Triticum aestivum). Int J Mol Sci. 2022;23(4):2351. doi:10.3390/ijms23042351 [Google Scholar] [PubMed] [CrossRef]
42. Bai H, Liao X, Li X, Wang B, Luo Y, Yang X, et al. DgbZIP3 interacts with DgbZIP2 to increase the expression of DgPOD for cold stress tolerance in chrysanthemum. Hortic Res. 2022;9:uhac105. doi:10.1093/hr/uhac105 [Google Scholar] [PubMed] [CrossRef]
43. Zhang Y, Zheng S, Liu Z, Wang L, Bi Y. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol. 2011;168(4):367–74. doi:10.1016/j.jplph.2010.07.025 [Google Scholar] [PubMed] [CrossRef]
44. Xu Z, Wang J, Ma Y, Wang F, Wang J, Zhang Y, et al. The bZIP transcription factor SlAREB1 regulates anthocyanin biosynthesis in response to low temperature in tomato. Plant J. 2023;115(1):205–19. doi:10.1111/tpj.v115.1. [Google Scholar] [CrossRef]
45. Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013;587(10):1543–7. doi:10.1016/j.febslet.2013.03.037 [Google Scholar] [PubMed] [CrossRef]
46. An JP, Yao JF, Xu RR, You CX, Wang XF, Hao YJ. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018;41(11):2678–92. doi:10.1111/pce.v41.11. [Google Scholar] [CrossRef]
47. An JP, Qu FJ, Yao JF, Wang XN, You CX, Wang XF, et al. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic Res. 2017;4:17023. doi:10.1038/hortres.2017.23 [Google Scholar] [PubMed] [CrossRef]
48. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. doi:10.1016/j.molp.2020.06.009 [Google Scholar] [PubMed] [CrossRef]
49. Wang Z, Zhang Z, Wang P, Qin C, He L, Kong L, et al. Genome-wide identification of the NAC transcription factors family and regulation of metabolites under salt stress in Isatis indigotica. Int J Biol Macromol. 2023;240:124436. doi:10.1016/j.ijbiomac.2023.124436 [Google Scholar] [PubMed] [CrossRef]
50. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–7. doi:10.1093/molbev/msab120 [Google Scholar] [PubMed] [CrossRef]
51. Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43(W1):W39–49. doi:10.1093/nar/gkv416 [Google Scholar] [PubMed] [CrossRef]
52. He P, Zhang J, Lv Z, Cui P, Xu X, George MS, et al. Genome-wide identification and expression analysis of the polygalacturonase gene family in sweetpotato. BMC Plant Biol. 2023;23(1):300. doi:10.1186/s12870-023-04272-1 [Google Scholar] [PubMed] [CrossRef]
53. Shah FA, Wei X, Wang Q, Liu W, Wang D, Yao Y, et al. Karrikin improves osmotic and salt stress tolerance via the regulation of the redox homeostasis in the oil plant Sapium sebiferum. Front Plant Sci. 2020;11:216. doi:10.3389/fpls.2020.00216 [Google Scholar] [PubMed] [CrossRef]
54. Shah FA, Chen Z, Ni F, Kamal KA, Zhang J, Chen J, et al. ArNAC148 induces Acer rubrum leaf senescence by activating the transcription of the ABA receptor gene ArPYR13. Int J Biol Macromol. 2024;279:134950. doi:10.1016/j.ijbiomac.2024.134950 [Google Scholar] [PubMed] [CrossRef]
55. Lalitha S. Primer premier 5. Biotech Softw Internet Rep. 2000;1(6):270–2. doi:10.1089/152791600459894. [Google Scholar] [CrossRef]
56. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–8. doi:10.1006/meth.2001.1262 [Google Scholar] [PubMed] [CrossRef]
57. Zhao P, Ye M, Wang R, Wang D, Chen Q. Systematic identification and functional analysis of potato (Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance. Int J Biol Macromol. 2020;161:155–67. doi:10.1016/j.ijbiomac.2020.06.032 [Google Scholar] [PubMed] [CrossRef]
58. Wang Y, Zhang X, Zhao Y, Yang J, He Y, Li G, et al. Transcription factor PyHY5 binds to the promoters of PyWD40 and PyMYB10 and regulates its expression in red pear ‘Yunhongli No. 1’. Plant Physiol Biochem. 2020;154:665–74. doi:10.1016/j.plaphy.2020.07.008 [Google Scholar] [PubMed] [CrossRef]
59. Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi:10.1186/1471-2229-4-10 [Google Scholar] [PubMed] [CrossRef]
60. Wu Y, Han T, Lyu L, Li W, Wu W. Research progress in understanding the biosynthesis and regulation of plant anthocyanins. Sci Hortic. 2023;321:112374. doi:10.1016/j.scienta.2023.112374. [Google Scholar] [CrossRef]
61. Gibalová A, Steinbachová L, Hafidh S, Bláhová V, Gadiou Z, Michailidis C, et al. Characterization of pollen-expressed bZIP protein interactions and the role of ATbZIP18 in the male gametophyte. Plant Reprod. 2017;30(1):1–17. doi:10.1007/s00497-016-0295-5 [Google Scholar] [PubMed] [CrossRef]
62. Yang Y, Yu TF, Ma J, Chen J, Zhou YB, Chen M, et al. The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int J Mol Sci. 2020;21(2):670. doi:10.3390/ijms21020670 [Google Scholar] [PubMed] [CrossRef]
63. Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001;20(1):118–27. doi:10.1093/emboj/20.1.118 [Google Scholar] [PubMed] [CrossRef]
64. Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10(5):673–83. doi:10.1105/tpc.10.5.673 [Google Scholar] [PubMed] [CrossRef]
65. Fan L, Xu L, Wang Y, Tang M, Liu L. Genome- and transcriptome-wide characterization of bZIP gene family identifies potential members involved in abiotic stress response and anthocyanin biosynthesis in radish (Raphanus sativus L.). Int J Mol Sci. 2019;20(24):6334. doi:10.3390/ijms20246334 [Google Scholar] [PubMed] [CrossRef]
66. Rahman S, Rehman A, Waqas M, Mubarik MS, Alwutayd K, AbdElgawad H, et al. Genome-wide exploration of bZIP transcription factors and their contribution to alkali stress response in Helianthus annuus. Plant Stress. 2023;10:100204. doi:10.1016/j.stress.2023.100204. [Google Scholar] [CrossRef]
67. Liu CC, Chi C, Jin LJ, Zhu J, Yu JQ, Zhou YH. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 2018;41(8):1762–75. doi:10.1111/pce.v41.8. [Google Scholar] [CrossRef]
68. Wu Y, Huang X, Zhang C, Yang H, Lyu L, Li W, et al. Genome-wide characterization and expression of the bZIP family in black raspberry. J Plant Growth Regul. 2024;43(1):259–71. doi:10.1007/s00344-023-11082-0. [Google Scholar] [CrossRef]
69. Ubi BE, Honda C, Bessho H, Kondo S, Wada M, Kobayashi S, et al. Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci. 2006;170(3):571–8. doi:10.1016/j.plantsci.2005.10.009. [Google Scholar] [CrossRef]
70. Gangappa SN, Botto JF. The multifaceted roles of HY5 in plant growth and development. Mol Plant. 2016;9(10):1353–65. doi:10.1016/j.molp.2016.07.002 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools