 Open Access
Open Access
ARTICLE
Response of the Ginseng C2H2-Type Zinc Finger Protein Family PgZFPs Gene to Methyl Jasmonate Regulation
1 College of Life Science, Jilin Agricultural University, Changchun, 130118, China
2 Research Center for Ginseng Genetic Resources Development and Utilization, Jilin Province, Jilin Agricultural University, Changchun, 130118, China
3 College of Chinese Medicinal Materials, Jilin Agricultural University, Changchun, 130118, China
* Corresponding Authors: Yi Wang. Email: ; Guang Chen. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Plant Secondary Metabolism and Functional Biology)
Phyton-International Journal of Experimental Botany 2024, 93(11), 3055-3071. https://doi.org/10.32604/phyton.2024.056384
Received 22 July 2024; Accepted 28 October 2024; Issue published 30 November 2024
Abstract
The main active components of ginseng are ginsenosides, which play significant roles in treating cardiovascular diseases, cancer, and providing antioxidant effects. Ginsenosides are primarily synthesized through the mevalonate pathway and the methylerythritol phosphate pathway. Many key enzyme genes involved in this biosynthetic process have been cloned and validated, yet the regulatory functions of transcription factors remain unclear. The C2H2-type zinc finger protein family, one of the largest families of transcription factors, is crucial in plant growth and development, response to biotic and abiotic stresses, and regulation of secondary metabolism. This study, based on the ginseng transcriptome database from Jilin, conducted a correlation analysis between the expression levels of PgZFPs genes in the Jilin ginseng C2H2-type zinc finger protein family and ginsenoside content, a genome-wide association study of PgZFPs, and co-expression analysis of PgZFPs with validated key enzyme genes. Ultimately, five candidate genes involved in ginsenoside biosynthesis were identified. The involvement of PgZFP27 and PgZFP-59-02 genes from the PgZFPs family in the biosynthesis of ginsenosides was validated through in vitro methyl jasmonate (MeJA) induction experiments. This result provides new genetic resources for the biosynthesis of ginsenosides.Keywords
Supplementary Material
Supplementary Material FileGlossary/Nomenclature/Abbreviations
| MeJA | Methyl jasmonate |
| ZFPs | Zinc finger proteins |
| Cys2 | Two cysteine residues |
| His2 | Two histidine residues |
| C2H2 | Cys2/His2 |
| NOT | NOTABILIS |
| SIT | SITIENS |
| FLC | FLACCA |
| MVA | Mevalonate |
| MEP | 2-C-methyl-D-erythritol 4-phosphate |
| GWAS | Genome-wide association studies |
| HPLC | High-performance liquid chromatography |
Zinc finger proteins (ZFPs), as one of the largest families of transcription factors, were first discovered by Miller et al. in the TFIIIA-type transcription factors [1]. Zinc finger proteins are characterized by a CX2–4CX3FX5LX2HX3–5H motif, most of which include a highly conserved extension of six amino acids, QALGGH, within the zinc finger domain [2]. Based on the number and order of cysteine and histidine residues that bind to zinc ions, they can be categorized into C2H2, C2HC, CCCH, C2C2, C2HCC2C2, and C2C2C2C2 types [3]. Among these, C2H2 is the most numerous in eukaryotes [4], containing two cysteine residues (Cys2) and two histidine residues (His2), with zinc ions coordinated by the Cys2/His2 (C2H2) to ensure the stability of the three-dimensional structure [5]. In Arabidopsis thaliana [6], rice (Oryza sativa) [7], poplar (Populus trichocarpa) [8], soybean (Glycine max) [9], tobacco (Nicotiana tabacum), wheat (Triticum aestivum) [10], and ginseng (Panax ginseng) [11], extensive experimental research has found that C2H2-type zinc finger proteins play crucial roles in plant stress resistance, growth regulation, plant hormone signal transduction, and regulation of secondary metabolism. In tomato, the expression of SlZF3 significantly increases after NaCl stress treatment, and concurrently, the ascorbic acid content in tomatoes changes significantly with the high expression of this gene [12]. In Zoysia grass, ZjZFN1 is a key regulatory gene in salt stress tolerance [13]. In poplar, overexpression of PeSTZ1 results in the binding of PeSTZ to the PeSTZ1 promoter, directly regulating the expression of PeAPX2, thereby enhancing the plant’s freeze tolerance [14]. Yin et al. found that overexpression of ZAT18 in Arabidopsis enhances drought tolerance, while mutants of this gene show decreased drought tolerance [15]. In Jilin ginseng, the salt-resistant genes PgZFP31, PgZFP78-01, PgZFP38, and PgZFP39-01 show significant increases in expression under NaCl stress, and concurrently, the lateral root length of ginseng adventitious roots significantly shortens [11]. In pepper, the expression level of the CAZFP1 gene is enhanced during the antagonistic interaction of pepper leaves with the pathogens Anthracnose fungus and Xanthomonas campestris [16]. SlZFP2 inhibits tomato fruit ripening while directly suppressing ABA biosynthesis genes NOTABILIS (NOT), SITIENS (SIT), FLACCA (FLC), and aldehyde oxidase SlAO1, thereby inhibiting ABA biosynthesis during fruit development [17]. In Arabidopsis, ZFP and GIS2 are located downstream of SPINDLY and upstream of GLABROUS1 in the cytokinin signaling pathway, playing a crucial role in the production of trichomes [18]. Increasing evidence suggests that C2H2-ZFPs regulate the expression of stress-responsive genes and serve as an essential component of the abiotic stress response network, exhibiting significant inhibitory activity.
Secondary metabolites are produced in plants in response to external adverse environmental conditions, playing a protective role in harsh environments [19]. Ginsenosides are an important class of secondary metabolites, synthesized through the regulation of both the mevalonate (MVA) pathway and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway [20]. In ginseng, the primary forms of ginsenosides are the tetracyclic triterpenoids and pentacyclic triterpenoids. The tetracyclic triterpenoids mainly include dammarane-type ginsenosides, such as protopanaxadiol ginsenosides (Rb1, Rb2, Rb3, Rc, Rd, Rg3, F2, Rh2, etc.) and protopanaxatriol ginsenosides (Re, Rf, Rg1, Rg2, Rh1, etc.), while the pentacyclic triterpenoids mainly include oleanane-type ginsenosides (R0) [21]. The biosynthesis of ginsenosides is a complex process, regulated by the plant organism as a whole, involving numerous biosynthetic enzymes and regulatory factors [22]. Thus, exploring the regulatory factors involved in the biosynthesis of ginsenosides can help elucidate the accumulation patterns of ginsenosides in ginseng. Current research on the transcriptional regulation of ginsenoside biosynthesis extensively involves transcription factors such as MYB [23–25], bHLH [26], AP2/ERF [27], bZIP [28], HD-Zip [29], WRKY [30], SPL [31], NBS [32], RLK [33], NF-Y [34], GT [35], GRAS [36], NAC [37]. Unfortunately, there are no reported studies on the role of C2H2-type zinc finger proteins in the biosynthesis of ginsenosides.
Inducers refer to factors that can cause rapid growth of plant cells or an increase in secondary metabolites, and are often used to study plant growth and metabolism processes. Inducers can be divided into two types based on their material properties: biotic and abiotic. Biological inducers include fungi, bacteria, etc., while abiotic inducers include various signaling molecules, heavy metal salts, small molecule compounds, etc. Among them, jasmonic acid substances and their derivatives, such as MeJA, have become one of the commonly used inducers in ginseng research due to their significant induction effect and stable action. In ginseng, MeJA is used to study the synthesis of secondary metabolites in various types of culture materials, including hairy roots [38,39], adventitious roots [40,41], and other materials [42]. Previous studies have shown that the ginsengoside content in ginseng culture can be increased by 1.5–28 times under MeJA induction [43]. During this process, most key enzyme genes involved in ginsenoside synthesis are significantly upregulated in response to MeJA induction, such as HMGS, HMGR, PMK, MVD [44], IPP, FPS [45], SS, SE, DDS [46], UGTs [47], CYP450s [48], etc. Therefore, transcriptome analysis and gene expression studies under MeJA induction conditions can be used to identify new functional genes involved in the regulation of ginsenoside synthesis, such as the ginsenoside transport gene PgPDR3 [49], transcription factor PgMYB2 [23], and 21 differentially expressed genes controlling ginsenoside synthesis [50] identified through this method. Li et al. also identified and validated the optimal reference genes for qRT PCR in MeJA induction, providing technical support for the expression analysis of functional genes regulated by MeJA [51]. In summary, MeJA plays an important regulatory role in the synthesis of secondary metabolites in medicinal plants, particularly in the biosynthesis of ginsenosides.
In this study, based on previous research [11], we obtained the Jilin ginseng PgZFPs gene family, consisting of 228 transcripts from 115 genes. Through correlational analysis of PgZFPs gene expression and ginsenoside content, whole-genome association analysis of PgZFPs, and co-expression analysis of PgZFPs with validated key enzyme genes, we identified five candidate genes involved in the biosynthesis of ginsenosides. The involvement of two PgZFPs genes in the biosynthesis of ginsenosides was validated through in vitro induction experiments. Finally, the involvement of 2 PgZFPs genes in the biosynthesis of ginsenosides was verified through MeJA in vitro induction experiments. The results provide new gene resources for the biosynthesis pathway of ginsenosides and also provide basic information for the subsequent analysis of the mechanism of the C2H2 zinc finger protein family PgZFP genes involved in ginsenoside synthesis.
2.1 Acquisition of Plant Material
Four-year-old main roots of ginseng, collected from a ginseng plantation located at 125°25′39″ E, 43°49′9″ N (Ji’an, Tonghua City, China), were sterilized and then induced to form ginseng callus tissues under dark conditions at 22°C–25°C in MS solid medium containing 4 mg/L 2,4-D and 0.2 mg/L 6-BA. Subsequently, well-grown callus tissues were transferred to B5 solid medium containing 5 mg/L IBA and cultured in the dark at 22°C–25°C for 3–4 weeks to obtain ginseng adventitious roots. Vigorously growing clonal lines of ginseng adventitious roots, designated as “seed source clonal lines” were then expanded in liquid B5 medium for subsequent validation of PgZFPs gene functions following MeJA induction treatment. The containers for liquid culture were triangular flasks, and each flask of culture material was one biological repetition. Each flask of liquid culture of ginseng adventitious roots was inoculated with a uniform inoculum of 1 g on the first day, the medium was 250 mL, and 200 μM methyl jasmonate was added into the medium of the ginseng adventitious roots on the 25th day of liquid culture so that the error in fresh weights of the different flasks was generally not more than 1 g on the 25th day. MeJA induction treatment nine-time points were set up, namely 0 h, 6 h, 12 h, 24 h, 36 h, 48 h, 72 h, 96 h and 120 h. Each time point had three biological replicates. 0 h without MeJA was the blank control, and 6–120 h with MeJA was the experimental group.
Three databases were utilized in this study: The first is the Jilin Ginseng Transcriptome Database, which was assembled using the Trinity de novo method from sequencing data obtained via Illumina HiSeqTM2000. This database includes 14 tissue types from four-year-old red fruit phase ginseng and main roots of the DaMaYa variety from Jilin, China, at ages 4 years, 5 years, 8 years, 12 years, and 25 years. The database has been uploaded to NCBI and is publicly available under the SRA accession numbers SRX1445566–SRX1445583 [52]. The second database contains data on the content of individual ginsenoside monomers in the roots of four-year-old ginseng from 42 farmer varieties, along with single nucleotide polymorphisms and insertions/deletions in transcripts assembled from all genes in the Jilin Ginseng Transcriptome Database [53,54]. The third database is the Jilin Ginseng PgZFP Gene Database, composed of 228 transcript sequences and expression levels from 115 PgZFP genes in Jilin Ginseng [11].
2.3 Whole-Genome Identification of PgZFPs Regulating Ginsenoside Biosynthesis
A systematic analysis of the 115 PgZFP genes across 42 varieties was conducted using Databases 1 and 3. The identification of candidate PgZFP genes involved in the biosynthesis of ginsenosides followed three steps. This analysis was based on the premise that gene expression influences traits, and genetic mutations also impact target traits.
Step One: Correlation analysis between PgZFPs gene expression and ginsenoside content. The cloning and functional validation of genes should be closely related to the traits of interest. Thus, correlation analysis between genes and traits should be the initial step in the systematic analysis. Using SPSS 9 (IBM SPSS Statistics 23), Pearson correlation analysis was performed between the expression levels of 115 PgZFPs across 42 farmer varieties and the content of nine individual ginsenosides (including Rg1, Re, Rf, Rb1, Rg2, Rc, Rb2, Rb3, and Rd) as well as the total saponin content in these varieties. Correlation coefficients of PgZFPs under different p-value conditions were calculated, and genes significantly correlated with ginsenoside content at p ≤ 0.05 were identified as Candidate Genes I.
Step Two: Genome-wide association studies (GWAS) of PgZFPs. GWAS were conducted to explore genetic variations affecting quantitative traits by using millions of SNP/InDel molecular genetic markers across the genome. If a gene affects a phenotype, the SNP/InDel mutations in that gene are also likely to influence variations in that trait. SNP/InDels in PgZFP genes were extracted using data from Database 2, and the impact of PgZFPs gene SNP/InDels on ginsenoside content was statistically analyzed. Assuming that the homozygous recessive genotype for an unmutated gene is coded as 0, the heterozygous genotype with mutation as 1, and the dominant homozygous genotype as 2. The 42 farmer varieties are grouped based on their genotypic status. When there are no heterozygotes in the group, the samples are divided into groups 0 and 2, and the impact of SNP/InDels on ginsenoside content is analyzed using an independent samples t-test. When heterozygotes are present, the samples are divided into three groups: 0, 1, and 2, and the effect of SNP/InDels on ginsenoside content is assessed using one-way ANOVA. Genes significantly correlated with ginsenoside content at p ≤ 0.05 are identified as Candidate Genes II. The contribution of gene SNP/InDels to ginsenoside content is calculated using the Formula (1).
where
Step Three: Network of PgZFPs with validated key enzyme genes. Key enzyme genes typically control rate-limiting steps in biosynthetic pathways and can regulate the biosynthesis of metabolic products. In ginseng, ten key enzyme genes (comprising 16 transcripts) currently validated to participate in the synthesis of ginsenosides have been identified [54]. If candidate genes are co-expressed with these key enzyme genes, or if the expression activity of the candidate genes is related to that of the key enzyme genes, it is considered that these candidate genes also contribute to the biosynthesis of ginsenosides. Research by Jiang et al. [50] indicated that the likelihood of forming a co-expression network among transcripts of genes controlling quantitative traits is significantly higher than that for randomly selected unknown genes. Therefore, using SPSS 9 (IBM SPSS Statistics 23), Spearman’s correlation analysis will be conducted between the expression levels of PgZFPs related to ginsenoside content and the expression of validated key enzyme genes involved in ginsenoside synthesis. The expression data for PgZFPs will be extracted from Database 3, and the expression data for key enzyme genes will be drawn from Database 1. Transcript quantification will be carried out using RSEM software [55], and co-expression analysis will be performed using Biolayout Express3D (Version 3.3) [56]. This analysis aims to identify potential interactive relationships between PgZFPs and key enzymes in ginsenoside biosynthesis. PgZFPs that show strong interactive relationships with key enzyme genes will be identified as PgZFP genes involved in the synthesis of ginsenosides.
2.4 Functional Validation of PgZFPs Genes in Ginsenoside Synthesis
Adventitious roots from the same “seed source” [50], which had been grown for 30 days, were inoculated at a standard inoculum size of 1.0 g into 250 mL of liquid B5 medium. The culture conditions were maintained at 22°C–25°C in the dark with a shaking speed of 110 rpm for 25 days. The MeJA induction treatment was set at nine-time points: 0 h, 6 h, 12 h, 24 h, 36 h, 48 h, 72 h, 96 h, and 120 h, with three biological replicates at each time point. On the 25th day, 200 µM final concentration of MeJA was added to the culture bottles at all time points except 0 h. Subsequently, at the nine designated time points, 1 g of adventitious roots was harvested from each replicate, immediately immersed in liquid nitrogen, and stored at −80°C for RNA analysis. The remaining samples were dried for the extraction and analysis of ginsenosides.
The establishment of the RNA library follows the procedures outlined in previous studies [50]. The extraction of ginsenosides is conducted using the Soxhlet extraction method [57], while the detection and analysis of individual ginsenoside monomers are performed using high-performance liquid chromatography (HPLC). The detection and analysis of total ginsenosides are carried out using the vanillin colorimetric method, with all operational steps and conditions adhering to the protocols described in prior research [51]. All experimental samples are set with three biological replicates. The objective is to validate the changes in the average content of ginsenosides in three biological replicates of ginseng adventitious roots after 6 h to 120 h of MeJA induction treatment compared to 0 h. The significance of these changes is analyzed using the independent sample t-test method.
Gene expression of PgZFPs in ginseng adventitious root samples treated with MeJA was detected using the ABI7500 fluorescence quantitative PCR instrument and Kangwei Century’s UltraSYBR Mixture (Low ROX) for qRT-PCR reactions. It was ensured that each treatment level had three biological replicates, each sample was set with three technical replicates, and each gene had a blank control. The relative expression levels of the genes were calculated using the 2−ΔΔCt method. To further verify the genes’ role in controlling the biosynthesis of ginsenosides, the Pearson correlation between transcript levels and ginsenoside content was calculated at each time point. If the correlation between transcript expression and ginsenoside content was significant at a level of p ≤ 0.05 (two-tailed), it was concluded that the transcripts of the gene control the biosynthesis of ginsenosides.
3.1 Genome-Wide Identification of Candidate PgZFPs Genes Controlling Ginsenoside Biosynthesis
Candidate PgZFPs genes controlling ginsenoside biosynthesis were identified through a three-step process across the whole genome. The first step involved the correlation analysis between PgZFPs and individual ginsenosides as well as total saponins. This correlation analysis was the initial step in screening PgZFPs genes related to ginsenoside synthesis. To determine the research method for the correlation analysis, the contents of nine individual ginsenosides and total saponins from 42 farmer varieties were subjected to a normal distribution test. The results indicated that the contents of the nine individual ginsenosides and total saponins in the 42 farmer varieties exhibited the characteristics of a quantitative trait with a continuous normal distribution (Fig. 1). Therefore, in this study, the Pearson correlation analysis method was used to investigate the relationship between the expression of PgZFPs and the content of ginsenosides. Among the 115 PgZFPs genes, 48 were found to correlate with the nine individual ginsenosides and total saponins. Specifically, 17 genes were related to Rg1, 16 to Re, 12 to Rf, 26 to Rb1, 6 to Rg2, 10 to Rc, 11 to Rb2, 9 to Rb3, 21 to Rd, and 26 to total saponins. These 48 genes, corresponding to 79 transcripts, were identified as candidate genes for ginsenoside biosynthesis (Candidate Gene Set I) and were subjected to further analysis.
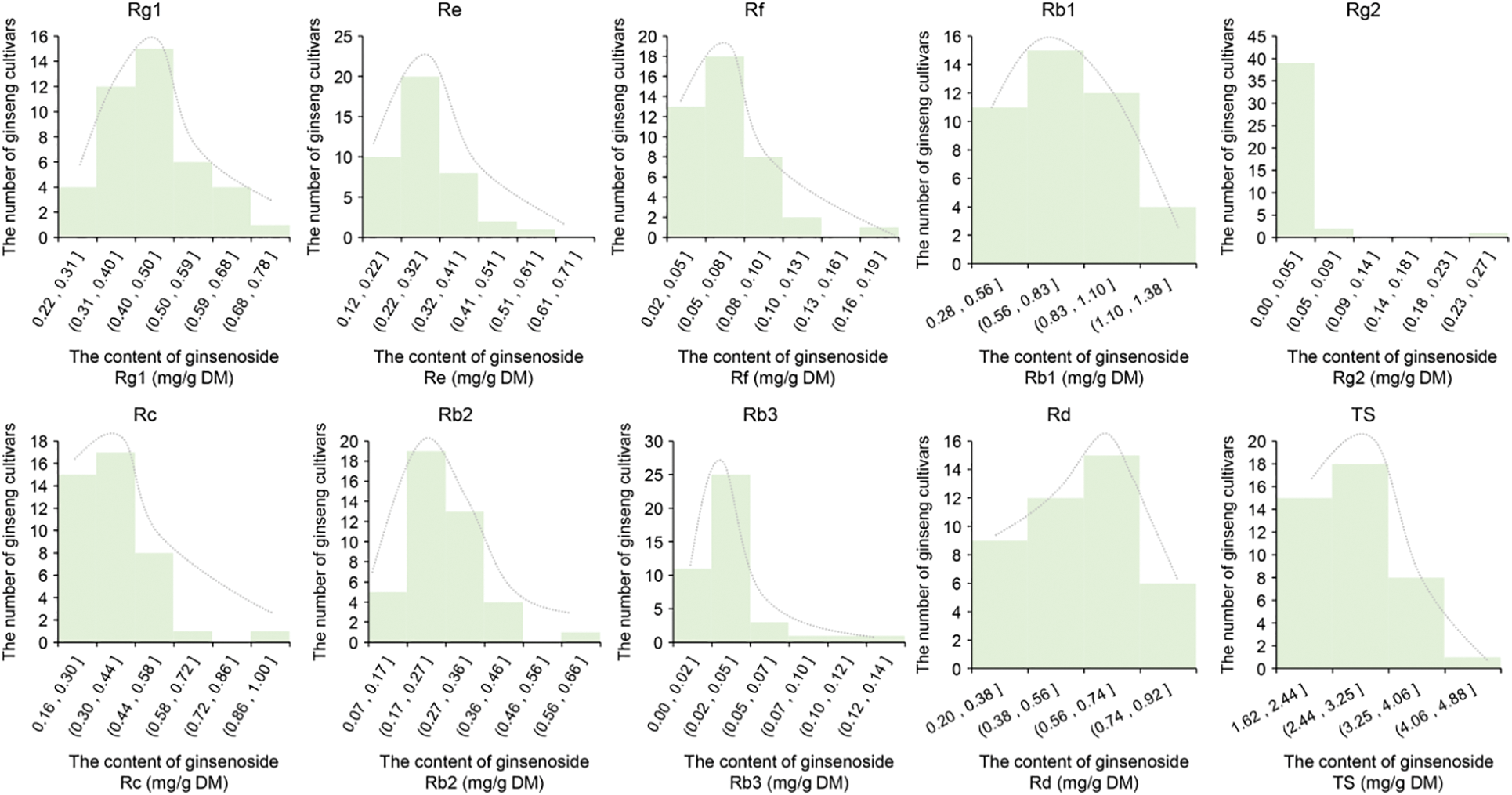
Figure 1: Variation of 9 mono-ginsenoside and total ginsenoside content in four-year-old ginseng plant roots among 42 farm cultivars. DM, dry matter. TS, total saponin
The second step involved statistical analysis of SNP/InDel mutations in the PgZFPs to determine the impact of genetic mutations on ginsenoside content. Among the 48 Candidate Genes Set I, there were a total of 631 SNP/InDel mutations. Of these, 506 SNP/InDels showed no significant effect on the nine individual ginsenosides and total saponins. However, 125 SNP/InDel mutations (110 SNPs and 15 InDels) across 24 genes were significantly correlated with the nine individual ginsenosides and total saponins at a p < 0.05 level (two-tailed), indicating a significant impact on ginsenosides content. These impactful SNP/InDels represented 19.8% of the total mutations, with an average of 5.2 SNP/InDels per gene. The effect of each SNP/InDel on the content of the nine individual ginsenosides and total saponins varied from 2.74% to 530.25% (Fig. 2). Each of the 125 SNP/InDel mutations significantly affected at least one type of individual ginsenoside or total saponins, with some affecting up to five types simultaneously. The 631 SNP/InDel mutations in the 48 genes were identified from 62,034 transcripts in the original transcriptome. With a ginseng genome size of 3.3 Gb, the probability of randomly identifying a SNP/InDel mutation in any of the 48 genes within 1 Mb of the genome affecting any of the nine individual ginsenosides or total saponins was calculated as (48/62,034 × 1/3300) = 2.34 × 10−7. The probability of randomly identifying impactful SNP/InDel mutations in 24 genes affecting the ginsenosides within 1 Mb was (48/62,034 × 1/3300)24 = 4.55 × 10−167, indicating that the findings were highly unlikely to be due to chance. Consequently, the 24 genes with SNP/InDel mutations that significantly impacted the content of the nine individual ginsenosides or total saponins were identified as Candidate Genes Set II for ginsenoside biosynthesis.
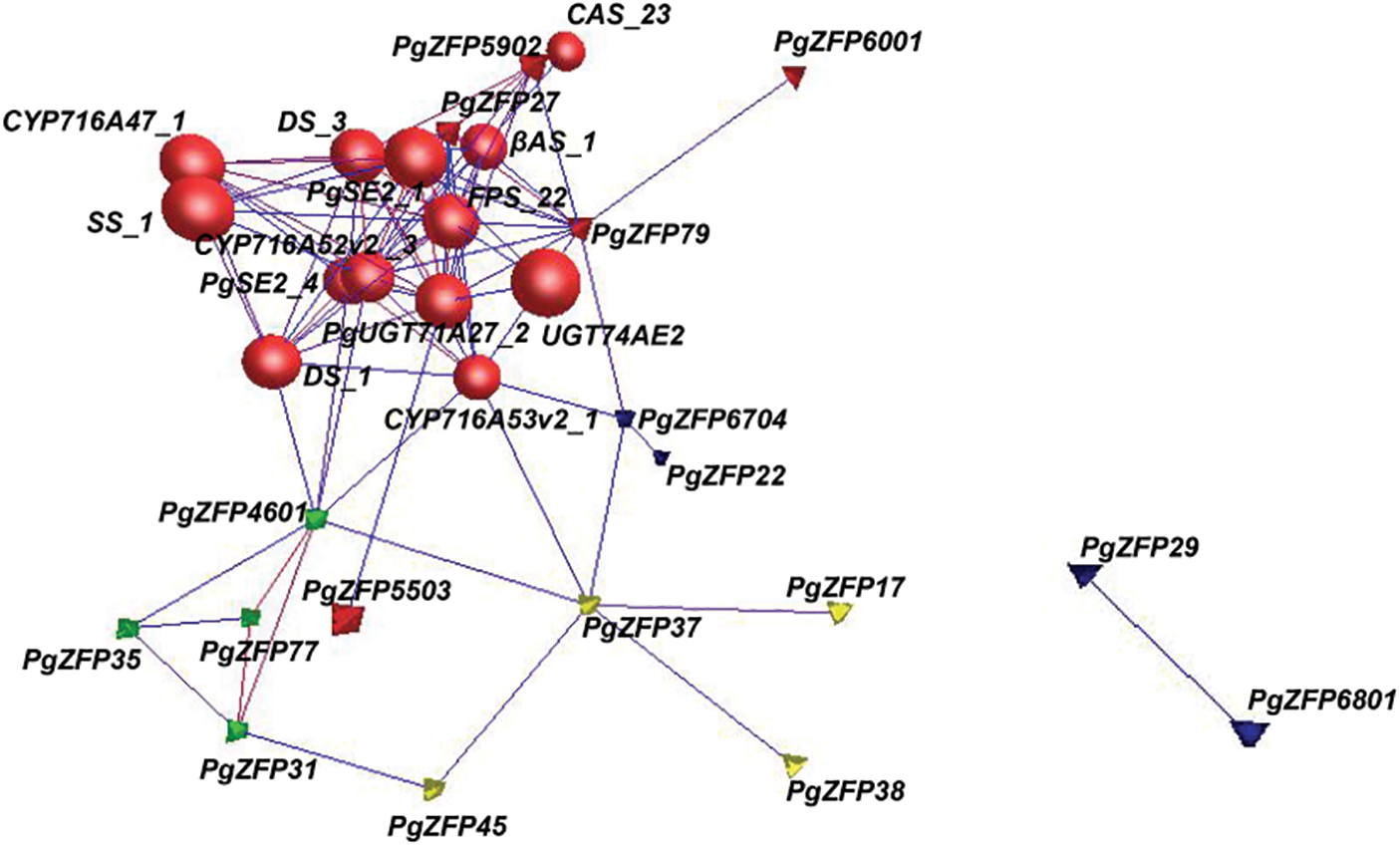
Figure 2: Co-expression analysis of 24 candidate PgZFP genes with 17 validated key enzyme genes. 30 genes form a tight interaction network containing 93 edges with an average connectivity of 6.2 when p ≤ 1.0E-03. Red color indicates cluster1 formed by 18 genes containing 5 PgZFP genes and 13 key enzyme genes, green color indicates cluster 2 containing 4 PgZFP genes, yellow color indicates cluster3 containing 4 PgZFP genes, and blue color indicates no class containing 4 genes. Spheres represent key enzyme genes and tetrahedron represent PgZFP genes
The third step involved a co-expression analysis between the 24 candidate genes and 17 validated key enzyme genes related to ginsenoside biosynthesis. The key enzyme genes reported to be directly associated with ginsenoside synthesis include CAS_23, CAS_22, CAS_21, CAS_11 (AB009029.1) [58], PgUGT71A27_2 (KM491309.1) [59], PgSE2_4, PgSE2_1(FJ393274.2) [60], SS_1 (KP689321.1) [61], β-AS_6 (AB009030.1) [58], β-AS_1(AB265170) [62], CYP716A47_1 (JN604537.1) [63], DS_3 (GU183405.1) [61], DS_1 (AB122080.1) [62], CYP716A52v2 _3 (JX036032.1) [63], CYP716A53v2_1 (JX036031.1) [64], FPS_22(DQ087959.1) [65], UGT74AE2(JX898529.1) [66], etc. These key enzyme genes have been cloned and verified to have a direct relationship with the biosynthesis of ginsenosides. The relationship between Candidate Genes Set II and the key enzymes of ginsenoside synthesis was determined by threshold filtering to establish the extent of correlation. In the interaction network, where the threshold p ≤ 1.0E-03, there were 30 nodes and 93 edges, with an average interaction intensity of 6.2. The interaction network formed by these 30 genes was divided into three network clusters and a no-class group: five PgZFPs Candidate Genes II and 13 key enzyme genes formed a closely interacting network group, cluster 1, while the remaining 12 PgZFPs Candidate Genes II grouped into Cluster 2, Cluster 3, and no class, as shown in Fig. 2. In the interaction network, “connection” refers to the maximum number of connections a gene has with other genes within the network. In Cluster 1, the key enzyme genes have the highest number of connections, reaching up to 14, while the maximum connections for the PgZFPs Candidate Genes II are up to 12. The five PgZFPs candidate genes that share the same cluster with the key enzyme genes are designated as Candidate Genes Set III.
3.2 Analysis of Expression Levels of PgZFPs in Ginseng Adventitious Root Samples before and after MeJA Treatment and Their Correlation with Changes in Ginsenoside Content
MeJA has been widely used in gene function verification studies. In this research, ginseng adventitious roots were treated with MeJA to study the changes in the expression levels of five PgZFPs genes (as shown in Fig. 3). After MeJA treatment, the five PgZFPs genes exhibited varying patterns of expression activity. PgZFP-27 showed varying degrees of upregulation from 6 h to 120 h after treatment, but the changes were not significant compared to the control group, suggesting that this gene consistently responds stably to MeJA regulation. PgZFP-55-03 exhibited a trend of initial upregulation followed by downregulation during the 6 h to 120 h post-treatment period, with significant downregulation at 84 h, compared with the control group, it significantly decreased by 2.1 times. This expression pattern may indicate that the gene has a delayed response to MeJA regulation and may exhibit greater expression activity at specific time points. PgZFP-59-02 showed a highly significant downregulation in expression 6 h after treatment compared to the 0 h control, indicating that MeJA suppressed the expression of this gene. PgZFP-60-01 exhibited highly significant downregulation at 108 h post-treatment. PgZFP-79 showed highly significant downregulation after treatment but demonstrated highly significant upregulation at 120 h. The dramatic changes in PgZFP-59-02 and PgZFP-79 suggest that these genes can rapidly and strongly respond to external stimuli at specific time points or under certain conditions, thus playing a crucial role in key responses. These results indicate that MeJA induction has an effective impact on the expression of these five genes in ginseng adventitious roots.
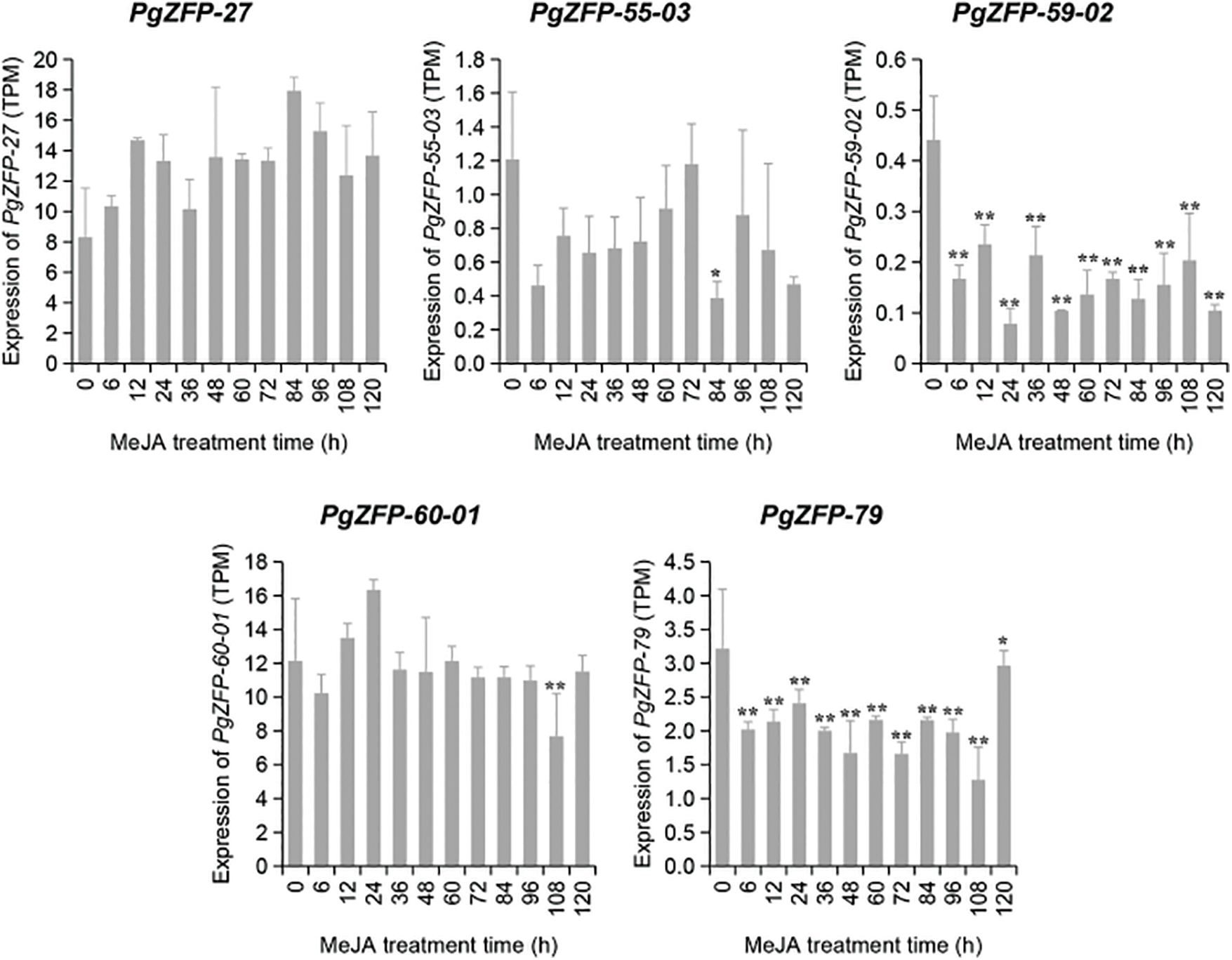
Figure 3: The dynamic expression changes of 5 PgZFPs in ginseng adventitious roots from 0 h to 120 h under the induction of MeJA treatment conditions are shown in the figure. 0 h is the control group without adding MeJA, * represents the significant situation at p < 0.05, and ** represents the extremely significant situation at p < 0.01
After MeJA induction, the content of the nine individual ginsenosides and total saponins in the adventitious roots also varied. The total saponins showed a gradual increase post-MeJA treatment, indicating that MeJA significantly promotes the accumulation of ginsenosides in ginseng adventitious roots. The levels of the individual ginsenosides also showed significant changes; interestingly, the content of certain individual ginsenosides suddenly decreased or increased at specific time points. For example, the content of Rb1 suddenly decreased at 96 h, Rb3 at 60 h, Rf increased suddenly at 72 h, and Rg2 decreased suddenly at 36 h (as detailed in Fig. 4). This suggests that there might be transformations among individual ginsenosides at these time points. The results indicate that the ginsenoside content in ginseng adventitious roots is regulated by MeJA and can respond to MeJA induction.
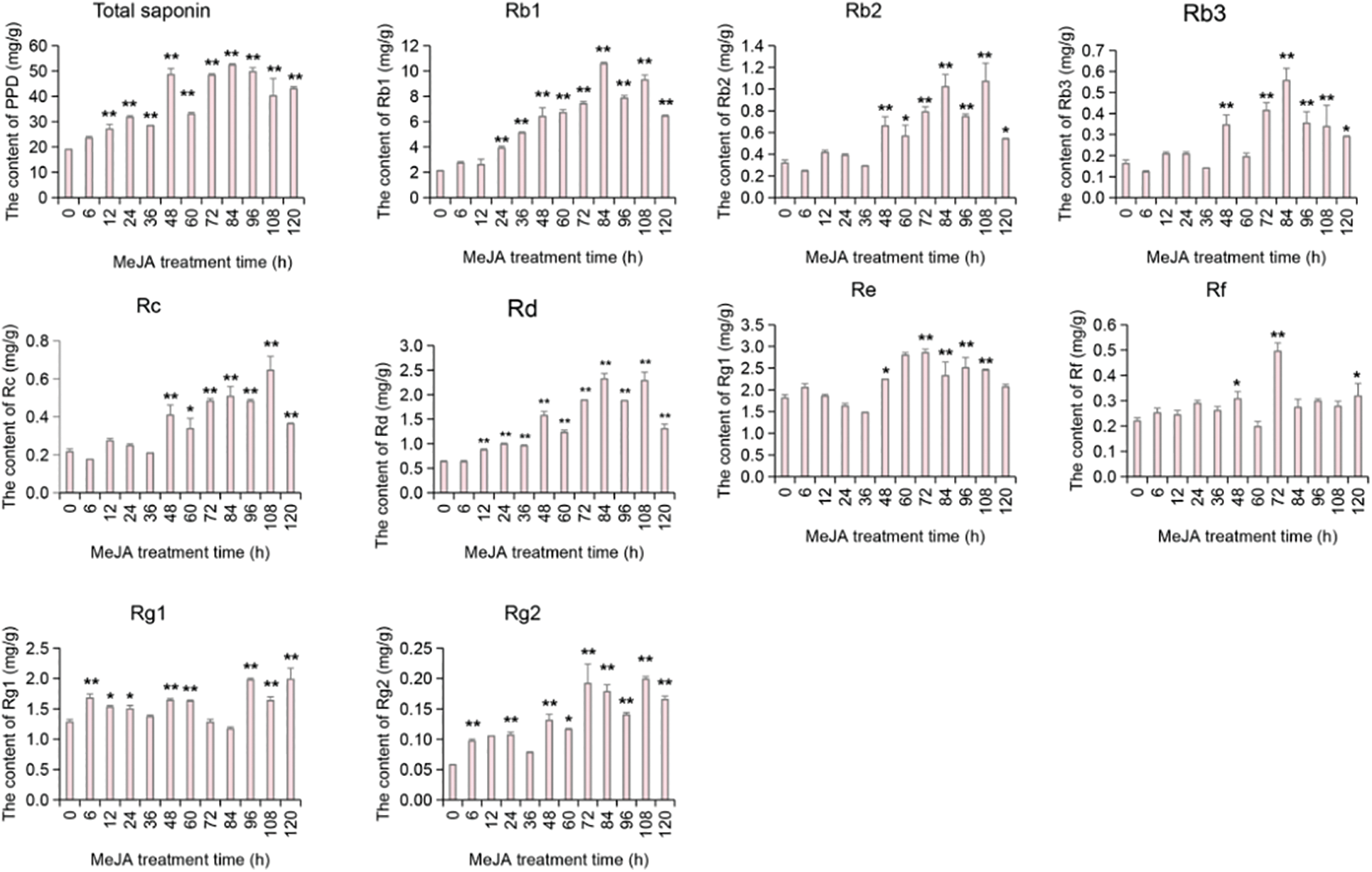
Figure 4: The change trend of ginsenoside content in the root of Panax ginseng after MeJA treatment. The 0 h on the X-axis in the figure represents the average of the ginsenoside content of the sample collected in the negative control group without MeJA treatment. The difference between the average ginsenoside content in the MeJA treatment group and the control group was tested by ANOVA, * represents the significant situation at p < 0.05, and ** represents the extremely significant situation at p < 0.01
Based on the observed changes in gene expression levels and ginsenoside content, we further analyzed the correlation between the expression levels of five PgZFPs and nine mono-ginsenosides, as well as total saponin content (see Table S5 for details). The results showed that PgZFP27 had a significant correlation with ginsenosides Rb1, Rb2, Rd, and Rg2 at the p < 0.05 level, and with Rb3 and total saponins at the p < 0.01 level, with correlation coefficients ranging from 0.618 to 0.759. PgZFP-59-02 showed a significant correlation with total saponins at the p < 0.05 level, with a correlation coefficient of −0.598, as detailed in Fig. 5. The correlation coefficient reflects the degree of association between the gene and the ginsenosides, where the sign of the coefficient indicates positive or negative correlations. The positive correlation between PgZFP27 and ginsenoside suggests that this gene plays an active role in subsequent ginsenoside synthesis, and its function can be validated in gene function verification by overexpressing to increase ginsenoside content. The negative correlation coefficient of PgZFP-59-02 suggests that the function of this gene in ginsenoside synthesis could be validated through methods such as RNAi or gene editing.
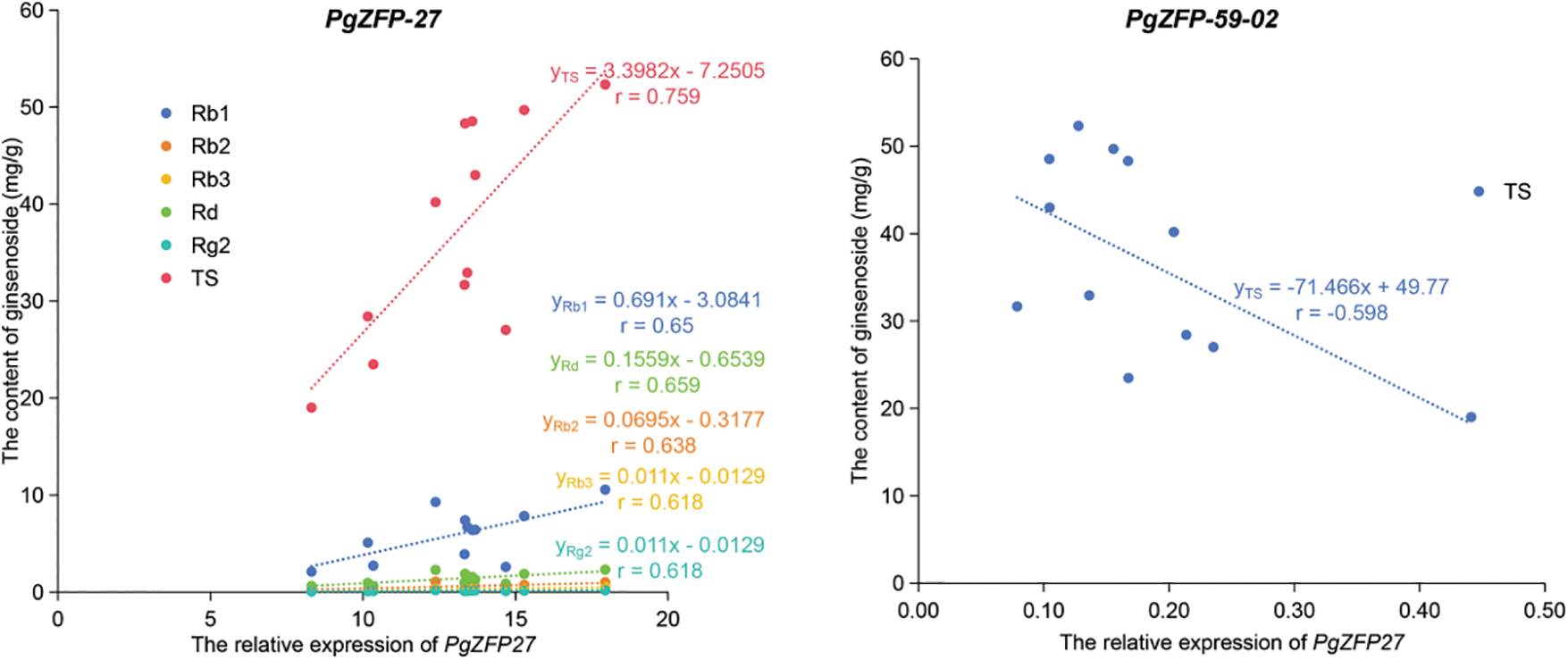
Figure 5: Linear correlation analysis between relative expression of PgZFPs’ and the content of ginsenosid. The figure shows only the linear equation for the significant correlation between relative expression and ginsenoside content, with the names of the individual ginsenoside labeled in the lower corners of the equation, and the correlation between the two indicated by r, which is derived by squaring the linear equation R2
In order to further understand the expression patterns of PgZFP-27 and PgZFP-59-02 with key enzyme genes, Fig. 6 shows a heatmap of the co expression patterns of two PgZFPs and key enzyme genes after MeJA induction treatment. The results showed that although there were differences in expression levels at different induction times, there was a clustering relationship between PgZFP-59-02 and CAS_22, indicating that their expression patterns were more similar. As CAS gene promotes the synthesis of sterols in the ginsenoside synthesis pathway [67], PgZFP-59-02 has a negative correlation with total saponins, that is, this gene plays a negative role in the direction of ginsenoside synthesis. These two results confirm each other. After induction with MeJA, PgZFP-27 was positively correlated with ginsenosides Rb1, Rb2, Rd, Rg2, Rb3 and total saponins, indicating that the expression of PgZFP-27 promoted the synthesis of damaran-type ginsenoside. At the same time, the expression pattern of PgZFP-27 and β-AS_1 was more similar, indicating that PgZFP-27 and β-AS_1 played a similar role in the ginsenoside synthesis pathway, and may play a rate-limiting role in the key branch points of ginsenoside synthesis. Subsequent studies should focus on finding the target genes that this gene regulates downstream monomonin synthesis.
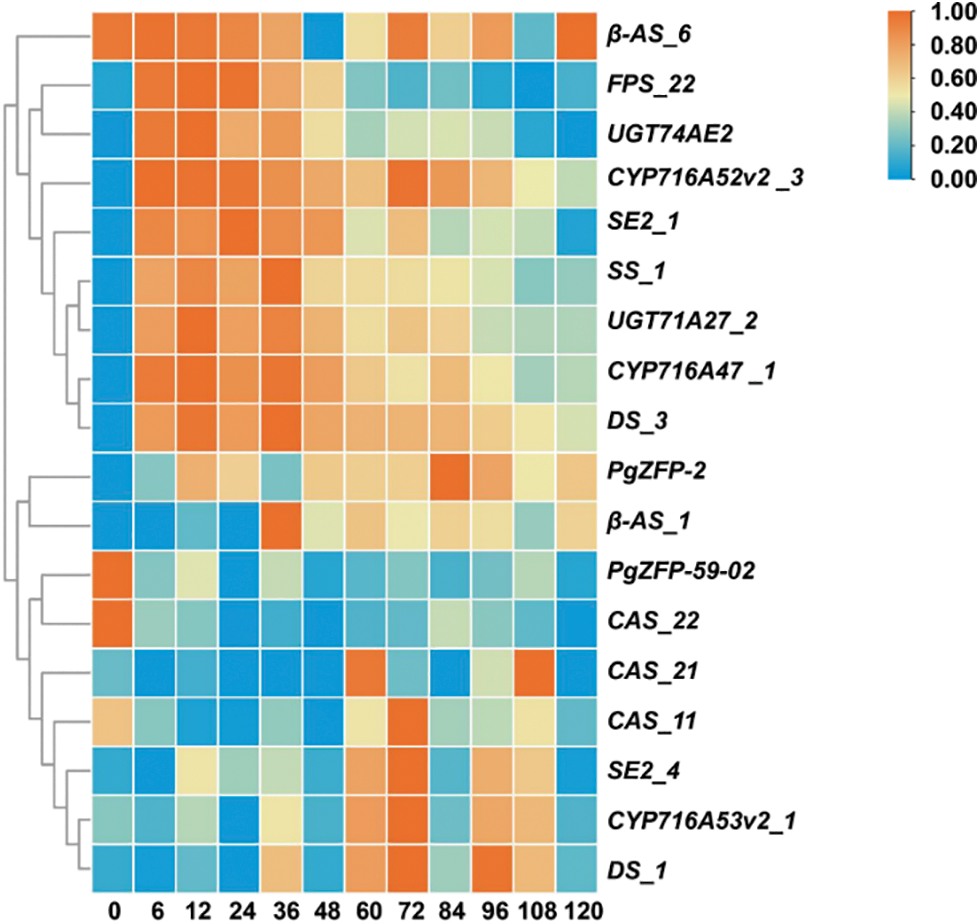
Figure 6: Expression heatmap of the PgZFP genes and key enzyme genes after induction treatment with MeJA
Based on these experimental results, two PgZFP genes involved in ginseng ginsenoside synthesis were identified, namely PgZFP27 and PgZFP-59-02.
The screening and application of important functional genes have always been one of the important directions for biological research and improvement of agronomic traits. However, traditional positional cloning methods are not only costly and time-consuming, but also difficult to apply to the screening of functional genes that affect quantitative traits controlled by multiple genes. This study is based on transcriptome expression databases and SNP mutation databases, combined with corresponding phenotypic traits, and uses statistical methods to screen important functional genes involved in phenotype formation. It is not only fast and efficient but also can calculate the contribution of each gene to trait formation, thereby selecting important functional genes or gene populations. This is of great significance for further research on gene function, a more comprehensive interpretation of the mechanism of important trait formation, and the wide application of functional genes. The function of plant C2H2-type zinc finger proteins has been extensively studied in model plants such as Arabidopsis thaliana, Oryza sativa, and tomato. However, systematic research on C2H2-type zinc finger proteins in ginseng is unprecedented, and studies on their role in the synthesis of plant secondary metabolites are relatively scarce. This study systematically analyzed and identified the C2H2 type zinc finger protein gene family highly correlated with the biosynthesis pathway of ginsenosides. Many biological traits are quantitative traits formed by the participation of multiple genes, and the same gene may also have multiple functions. Therefore, in order to find the main or more influential genes that affect traits, this study used multiple analytical methods for screening. Firstly, based on the correlation analysis between gene expression and ginsenoside content, we aim to identify genes that have a significant impact on phenotype changes. At this point, the correlation between genes and phenotype is p ≤ 0.05. On this basis, correlation analysis was conducted between genes with SNP mutations and phenotypes, where the correlation between genes and phenotypes was p ≤ 1.0E-04. Co-expression analysis was performed between candidate genes and ginsenoside synthesis-related genes, where the correlation between genes and phenotypes was p ≤ 1.0E-07 or higher. The correlation between genes and phenotypes presented in a single analysis or with low p-values may be random and accidental, but there is still a correlation between genes and phenotypes in multiple analysis levels, multiple analysis methods, and extremely strict p-values. It can be basically confirmed that this gene plays a very important role in the formation of traits. However, how this gene affects traits and its specific mechanism of action still needs more experimental verification.
MeJA has been proven to be an effective inducer that can promote the biosynthesis of many high-value medicinal secondary metabolites in various medicinal plants. Ginsenosides, as a plant nutrient, can respond to external stress environments and alleviate the damage caused to plants by the external environment [68]. According to reports, various biotic and abiotic stress conditions can stimulate the accumulation of ginsenosides [42]. Osmotic stress conditions such as salt stress and drought stress can increase the total saponin content of ginseng by 45%–50% [69]. In suspension culture systems, treatment with external hormones such as MeJA can also increase the content of ginsenosides [19]. In this study, the significant changes in the content of ginsenosides after MeJA induction treatment were consistent with previous research results, indicating that ginsenosides are important products for coping with external stress environments and play an important role in regulating plant growth and development. At the same time, the gene function verification method of MeJA induction treatment also verified the rationality and reliability of the gene screening method used in this study.
5 PgZFPs were obtained by a three-step analysis method. Methyl jasmonate was used to treat ginseng adventitious roots, which in turn verified that 2 PgZFPs genes control ginsenoside biosynthesis. The results provide a feasible method for the identification of ginsenoside synthesis genes, as well as new gene resources for ginsenoside biosynthesis.
Acknowledgement: None.
Funding Statement: This work was supported by the Department of Science and Technology of Jilin Province (20240101227JC, 20210402043GH, 20200801063GH, 20190201264JC, 20190103104JH, 20180414077GH, and 20180101027JC), and the Development and Reform Commission of Jilin Province (2016C064 and 2018C047-3).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Guang Chen and Yi Wang; methodology: Yue Jiang and Lingyu Liu; software: Yue Jiang and Lingyu Liu; resources: Kangyu Wang and Yanfang Wang; writing—original draft preparation: Yue Jiang and Lingyu Liu; writing—review and editing: Meiping Zhang; visualization: Mingzhu Zhao, Ping Chen and Jun Lei; project administration: Yi Wang and Guang Chen; funding acquisition: Yi Wang and Guang Chen. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data used for this study have been deposited at Sequence Read Archive (SRA) of National Center for Biotechnology Information (NCBI), BioProject PRJNA302556; and at Gene Expression Omnibus (GEO) of NCBI, SRP066368 and SRR13131364-SRR13131405.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.056384.
References
1. Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–14. doi:10.1002/j.1460-2075.1985.tb03825.x. [Google Scholar] [PubMed] [CrossRef]
2. Zhang H, Sun Z, Feng S, Zhang J, Zhang F, Wang W, et al. The C2H2-type zinc finger protein PhZFP1 regulates cold stress tolerance by modulating galactinol synthesis in Petunia hybrida. J Exp Bot. 2022 Oct 18;73(18):6434–48. doi:10.1093/jxb/erac274. [Google Scholar] [PubMed] [CrossRef]
3. Moore M, Ullman C. Recent developments in the engineering of zinc finger proteins. Brief Funct Genomic Proteomic. 2003;1(4):342–55. doi:10.1093/bfgp/1.4.342. [Google Scholar] [PubMed] [CrossRef]
4. Wang D, Qiu Z, Xu T, Yao S, Chen M, Li Q, et al. Transcriptomic identification of potential C2H2 zinc finger protein transcription factors in Pinus massoniana in response to biotic and abiotic stresses. Int J Mol Sci. 2024;25(15):8361. doi:10.3390/ijms25158361. [Google Scholar] [PubMed] [CrossRef]
5. Takatsuji H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol. 1999;39(6):1073–8. doi:10.1023/a:1006184519697. [Google Scholar] [PubMed] [CrossRef]
6. Traverse KKF, Breselge S, Trautman JG, Dee A, Wang J, Childs KL, et al. Characterization of the ZCTs, a subgroup of Cys2-His2 zinc finger transcription factors regulating alkaloid biosynthesis in Catharanthus roseus. Plant Cell Rep. 2024;43(9):209. doi:10.1007/s00299-024-03295-8. [Google Scholar] [PubMed] [CrossRef]
7. Tong Z, Han X, Duan X, Lin J, Chen J, Xiao J, et al. Genome-wide identification and expression analysis of the Cys2His2 zinc finger protein gene family in Flammulina filiformis. J Fungi. 2024;10(9):644. doi:10.3390/jof10090644. [Google Scholar] [PubMed] [CrossRef]
8. Zhou X, Gao T, Zhang Y, Han M, Shen Y, Su Y, et al. Genome-wide identification, characterization and expression of C2H2 zinc finger gene family in Opisthopappus species under salt stress. BMC Genomics. 2024;25(1):385. doi:10.1186/s12864-024-10273-7. [Google Scholar] [PubMed] [CrossRef]
9. Yuan S, Li X, Li R, Wang L, Zhang C, Chen L, et al. Genome-wide identification and classification of soybean C2H2 zinc finger proteins and their expression analysis in legume-rhizobium symbiosis. Front Microbiol. 2018;9:126. doi:10.3389/fmicb.2018.00126. [Google Scholar] [PubMed] [CrossRef]
10. Liu Y, Khan AR, Gan Y. C2H2 zinc finger proteins response to abiotic stress in plants. Int J Mol Sci. 2022;23(5):2730. doi:10.3390/ijms23052730. [Google Scholar] [PubMed] [CrossRef]
11. Jiang Y, Liu L, Pan Z, Zhao M, Zhu L, Han Y, et al. Genome-wide analysis of the C2H2 zinc finger protein gene family and its response to salt stress in ginseng, Panax ginseng Meyer. Sci Rep. 2022;12(1):10165. doi:10.1038/s41598-022-14357-w. [Google Scholar] [PubMed] [CrossRef]
12. Li Y, Chu Z, Luo J, Zhou Y, Cai Y, Lu Y, et al. The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN5B. Plant Biotechnol J. 2018;16(6):1201–13. doi:10.1111/pbi.12863. [Google Scholar] [PubMed] [CrossRef]
13. Teng K, Tan P, Guo W, Yue Y, Fan X, Wu J. Heterologous expression of a novel Zoysia japonica C2H2 zinc finger gene, ZjZFN1, improved salt tolerance in arabidopsis. Front Plant Sci. 2018;9:1159. doi:10.3389/fpls.2018.01159. [Google Scholar] [PubMed] [CrossRef]
14. He F, Li HG, Wang JJ, Su Y, Wang HL, Feng CH, et al. PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol J. 2019;17(11):2169–83. doi:10.1111/pbi.13130. [Google Scholar] [PubMed] [CrossRef]
15. Yin M, Wang Y, Zhang L, Li J, Quan W, Yang L, et al. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J Exp Bot. 2017;68(11):2991–3005. doi:10.1093/jxb/erx157. [Google Scholar] [PubMed] [CrossRef]
16. Kim SH, Hong JK, Lee SC, Sohn KH, Jung HW, Hwang BK. CAZFP1, Cys2/His2-type zinc-finger transcription factor gene functions as a pathogen-induced early-defense gene in Capsicum annuum. Plant Mol Biol. 2004;55(6):883–904. doi:10.1007/s11103-004-2151-5. [Google Scholar] [PubMed] [CrossRef]
17. Weng L, Zhao F, Li R, Xu C, Chen K, Xiao H. The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol. 2015;167(3):931–49. doi:10.1104/pp.114.255174. [Google Scholar] [PubMed] [CrossRef]
18. Gan Y, Liu C, Yu H, Broun P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development. 2007;134(11):2073–81. doi:10.1242/dev.005017. [Google Scholar] [PubMed] [CrossRef]
19. Kim YJ, Zhang D, Yang DC. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv. 2015;33(6):717–35. doi:10.1016/j.biotechadv.2015.03.001. [Google Scholar] [PubMed] [CrossRef]
20. Xue L, He Z, Bi X, Xu W, Wei T, Wu S, et al. Transcriptomic profiling reveals MEP pathway contributing to ginsenoside biosynthesis in Panax ginseng. BMC Genomics. 2019;20(1):383. doi:10.1186/s12864-019-5718-x. [Google Scholar] [PubMed] [CrossRef]
21. Han JY, Kwon YS, Yang DC, Jung YR, Choi YE. Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 2006;47(12):1653–62. doi:10.1093/pcp/pcl032. [Google Scholar] [PubMed] [CrossRef]
22. Hou M, Wang R, Zhao S, Wang Z. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm Sin B. 2021;11(7):1813–34. doi:10.1016/j.apsb.2020.12.017. [Google Scholar] [PubMed] [CrossRef]
23. Liu T, Luo T, Guo X, Zou X, Zhou D, Afrin S, et al. PgMYB2, a MeJA-responsive transcription factor, positively regulates the dammarenediol synthase gene expression in Panax ginseng. Int J Mol Sci. 2019;20(9):2219. doi:10.3390/ijms20092219. [Google Scholar] [PubMed] [CrossRef]
24. Liu M, Li K, Sheng S, Wang M, Hua P, Wang Y, et al. Transcriptome analysis of MYB transcription factors family and PgMYB genes involved in salt stress resistance in Panax ginseng. BMC Plant Biol. 2022;22(1):479. doi:10.1186/s12870-022-03871-8. [Google Scholar] [PubMed] [CrossRef]
25. Wang X, Niu Y, Zheng Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int J Mol Sci. 2021;22(11):6125. doi:10.3390/ijms22116125. [Google Scholar] [PubMed] [CrossRef]
26. Radani Y, Li R, Korboe HM, Ma H, Yang L. Transcriptional and post-translational regulation of plant bHLH transcription factors during the response to environmental stresses. Plants. 2023;12(11):2113. doi:10.3390/plants12112113. [Google Scholar] [PubMed] [CrossRef]
27. Jiang Y, Zhang Q, Zeng Z, Wang Y, Zhao M, Wang K, et al. The AP2/ERF transcription factor PgERF120 regulates ginsenoside biosynthesis in ginseng. Biomolecules. 2024;14(3):345. doi:10.3390/biom14030345. [Google Scholar] [PubMed] [CrossRef]
28. Fan J, Chen N, Rao W, Ding W, Wang Y, Duan Y, et al. Genome-wide analysis of bZIP transcription factors and their expression patterns in response to methyl jasmonate and low-temperature stresses in Platycodon grandiflorus. PeerJ. 2024;12(4):e17371. doi:10.7717/peerj.17371. [Google Scholar] [PubMed] [CrossRef]
29. Li L, Lv B, Zang K, Jiang Y, Wang C, Wang Y, et al. Genome-wide identification and systematic analysis of the HD-Zip gene family and its roles in response to pH in Panax ginseng Meyer. BMC Plant Biol. 2023;23(1):30. doi:10.1186/s12870-023-04038-9. [Google Scholar] [PubMed] [CrossRef]
30. Li W, Pang S, Lu Z, Jin B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants. 2020;9(11):1515. doi:10.3390/plants9111515. [Google Scholar] [PubMed] [CrossRef]
31. Wang Y, Ruan Q, Zhu X, Wang B, Wei B, Wei X. Identification of Alfalfa SPL gene family and expression analysis under biotic and abiotic stresses. Sci Rep. 2023;13(1):84. doi:10.1038/s41598-022-26911-7. [Google Scholar] [PubMed] [CrossRef]
32. Boopathi V, Subramaniyam S, Mathiyalagan R, Yang DC. Till 2018: a survey of biomolecular sequences in genus Panax. J Ginseng Res. 2020;44(1):33–43. doi:10.1016/j.jgr.2019.06.004. [Google Scholar] [PubMed] [CrossRef]
33. Yan J, Su P, Meng X, Liu P. Phylogeny of the plant receptor-like kinase (RLK) gene family and expression analysis of wheat RLK genes in response to biotic and abiotic stresses. BMC Genomics. 2023;24(1):224. doi:10.1186/s12864-023-09303-7. [Google Scholar] [PubMed] [CrossRef]
34. Liu M, Pan Z, Yu J, Zhu L, Zhao M, Wang Y, et al. Transcriptome-wide characterization, evolutionary analysis, and expression pattern analysis of the NF-Y transcription factor gene family and salt stress response in Panax ginseng. BMC Plant Biol. 2022;22(1):320. doi:10.1186/s12870-022-03687-6. [Google Scholar] [PubMed] [CrossRef]
35. Hu J, Liu T, Huo H, Liu S, Liu M, Liu C, et al. Genome-wide characterization, evolutionary analysis, and expression pattern analysis of the trihelix transcription factor family and gene expression analysis under MeJA treatment in Panax ginseng. BMC Plant Biol. 2023;23(1):376. doi:10.1186/s12870-023-04390-w. [Google Scholar] [PubMed] [CrossRef]
36. Neves C, Ribeiro B, Amaro R, Expósito J, Grimplet J, Fortes AM. Network of GRAS transcription factors in plant development, fruit ripening and stress responses. Hortic Res. 2023;10(12):uhad220. doi:10.1093/hr/uhad220. [Google Scholar] [PubMed] [CrossRef]
37. Chowdhury AT, Hasan MN, Bhuiyan FH, Islam MQ, Nayon MRW, Rahaman MM, et al. Identification, characterization of Apyrase (APY) gene family in rice (Oryza sativa) and analysis of the expression pattern under various stress conditions. PLoS One. 2023;18(5):e0273592. doi:10.1371/journal.pone.0273592. [Google Scholar] [PubMed] [CrossRef]
38. KeeWon Y, WenYuan G, SungHo S, KeeYoeup P. Improvement of ginsenoside production by jasmonic acid and some other elicitors in hairy root culture of ginseng (Panax ginseng C. A. Meyer). Vitro Cell Dev Biol-Plant. 2000;36:424–8. doi:10.1007/s11627-000-0077-4. [Google Scholar] [CrossRef]
39. Palazón J, Cusidó RM, Bonfill M, Mallol A, Moyano E, Morales C, et al. Elicitation of different Panax ginseng transformed root phenotypes for an improved ginsenoside production. Plant Physiol Biochem. 2003;41(11–12):1019–25. doi:10.1016/j.plaphy.2003.09.002. [Google Scholar] [CrossRef]
40. Ali MB, Yu KW, Hahn EJ, Paek KY. Differential responses of anti-oxidants enzymes, lipoxygenase activity, ascorbate content and the production of saponins in tissue cultured root of mountain Panax ginseng C. A. Mayer and Panax quinquefolium L. in bioreactor subjected to methyl jasmonate stress. Plant Sci. 2005;169(1):83–92. doi:10.1016/j.plantsci.2005.02.027. [Google Scholar] [CrossRef]
41. Cao H, Nuruzzaman M, Xiu H, Huang J, Wu K, Chen X, et al. Transcriptome analysis of methyl jasmonate-elicited Panax ginseng adventitious roots to discover putative ginsenoside biosynthesis and transport genes. Int J Mol Sci. 2015;16(2):3035–57. doi:10.3390/ijms16023035. [Google Scholar] [PubMed] [CrossRef]
42. Oh JY, Kim YJ, Jang MG, Joo SC, Kwon WS, Kim SY, et al. Investigation of ginsenosides in different tissues after elicitor treatment in Panax ginseng. J Ginseng Res. 2014;38(4):270–7. doi:10.1016/j.jgr.2014.04.004. [Google Scholar] [PubMed] [CrossRef]
43. Lu M, Wong H, Teng W. Effects of elicitation on the production of saponin in cell culture of Panax ginseng. Plant Cell Rep. 2001;20:674–7. [Google Scholar]
44. Kim YK, Kim YB, Kim JK, Kim SU, Park SU. Molecular cloning and characterization of mevalonic acid (MVA) pathway genes and triterpene accumulation in Panax ginseng. J Korean Soc Appl Biol Chem. 2014;57(3):289–95. doi:10.1007/s13765-014-4008-1. [Google Scholar] [CrossRef]
45. Balusamy SRD, Rahimi S, Sukweenadhi J, Kim YJ, Yang DC. Exogenous methyl jasmonate prevents necrosis caused by mechanical wounding and increases terpenoid biosynthesis in Panax ginseng. Plant Cell Tiss Org. 2015;123:341–8. [Google Scholar]
46. Lee YS, Park HS, Lee DK, Jayakodi M, Kim NH, Koo HJ, et al. Integrated transcriptomic and metabolomic analysis of five Panax ginseng cultivars reveals the dynamics of ginsenoside biosynthesis. Front Plant Sci. 2017;8:1048. doi:10.3389/fpls.2017.01048. [Google Scholar] [PubMed] [CrossRef]
47. Kang KB, Jayakodi M, Lee YS, Nguyen VB, Park HS, Koo HJ, et al. Identification of candidate UDP-glycosyltransferases involved in protopanaxadiol-type ginsenoside biosynthesis in Panax ginseng. Sci Rep. 2018;8(1):11744. doi:10.1038/s41598-018-30262-7. [Google Scholar] [PubMed] [CrossRef]
48. Zeng X, Luo T, Li J, Li G, Zhou D, Liu T, et al. Transcriptomics-based identification and characterization of 11 CYP450 genes of Panax ginseng responsive to MeJA. Acta Biochim Biophys Sin. 2018;50(11):1094–1103. doi:10.1093/abbs/gmy120. [Google Scholar] [PubMed] [CrossRef]
49. Zhang R, Huang J, Zhu J, Xie X, Tang Q, Chen X, et al. Isolation and characterization of a novel PDR-type ABC transporter gene PgPDR3 from Panax ginseng C. A. Meyer induced by methyl jasmonate. Mol Biol Rep. 2013;40(11):6195–204. doi:10.1007/s11033-013-2731-z. [Google Scholar] [PubMed] [CrossRef]
50. Jiang Y, Liu S, Li L, Zang K, Wang Y, Zhao M, et al. Transcriptome and phenotype integrated analysis identifies genes controlling ginsenoside Rb1 biosynthesis and reveals their interactions in the process in Panax ginseng. Int J Mol Sci. 2022;23(22):14016. doi:10.3390/ijms232214016. [Google Scholar] [PubMed] [CrossRef]
51. Li L, Wang K, Zhao M, Li S, Jiang Y, Zhu L, et al. Selection and validation of reference genes desirable for gene expression analysis by qRT-PCR in MeJA-treated ginseng hairy roots. PLoS One. 2019;14(12):e0226168. doi:10.1371/journal.pone.0226168. [Google Scholar] [PubMed] [CrossRef]
52. Wang K, Jiang S, Sun C, Lin Y, Yin R, Wang Y, et al. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C. A. Meyer. Sci Rep. 2015;5(1):18283. doi:10.1038/srep18283. [Google Scholar] [PubMed] [CrossRef]
53. Li L, Wang Y, Zhao M, Wang K, Sun C, Zhu L, et al. Integrative transcriptome analysis identifies new oxidosqualene cyclase genes involved in ginsenoside biosynthesis in Jilin ginseng. Genomics. 2021;113(4):2304–16. doi:10.1016/j.ygeno.2021.05.023. [Google Scholar] [PubMed] [CrossRef]
54. Zhao M, Lin Y, Wang Y, Li X, Han Y, Wang K, et al. Transcriptome analysis identifies strong candidate genes for ginsenoside biosynthesis and reveals its underlying molecular mechanism in Panax ginseng C. A. Meyer. Sci Rep. 2019;9(1):615. doi:10.1038/s41598-018-36349-5. [Google Scholar] [PubMed] [CrossRef]
55. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323. doi:10.1186/1471-2105-12-323. [Google Scholar] [PubMed] [CrossRef]
56. Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout express3D. Nat Protoc. 2009;4(10):1535–50. doi:10.1038/nprot.2009.177. [Google Scholar] [PubMed] [CrossRef]
57. Yue Y, Qiu ZD, Qu XY, Deng AP, Yuan Y, Huang LQ, et al. Discoursing on Soxhlet extraction of ginseng using association analysis and scanning electron microscopy. J Pharm Anal. 2018;8(5):312–7. doi:10.1016/j.jpha.2018.08.003. [Google Scholar] [PubMed] [CrossRef]
58. Kushiro T, Shibuya M, Ebizuka Y. Beta-amyrin synthase—cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur J Biochem. 1998;256(1):238–44. doi:10.1046/j.1432-1327.1998.2560238.x. [Google Scholar] [PubMed] [CrossRef]
59. Khorolragchaa A, Kim YJ, Rahimi S, Sukweenadhi J, Jang MG, Yang DC. Grouping and characterization of putative glycosyltransferase genes from Panax ginseng Meyer. Gene. 2014;536(1):186–92. doi:10.1016/j.gene.2013.07.077. [Google Scholar] [PubMed] [CrossRef]
60. Han JY, In JG, Kwon YS, Choi YE. Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng. Phytochemistry. 2010;71(1):36–46. doi:10.1016/j.phytochem.2009.09.031. [Google Scholar] [PubMed] [CrossRef]
61. Han JY, Kim HJ, Kwon YS, Choi YE. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52(12):2062–73. doi:10.1093/pcp/pcr150. [Google Scholar] [PubMed] [CrossRef]
62. Tansakul P, Shibuya M, Kushiro T, Ebizuka Y. Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 2006;580(22):5143–9. doi:10.1016/j.febslet.2006.08.044. [Google Scholar] [PubMed] [CrossRef]
63. Han JY, Kim MJ, Ban YW, Hwang HS, Choi YE. The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2013;54(12):2034–46. doi:10.1093/pcp/pct141. [Google Scholar] [PubMed] [CrossRef]
64. Han JY, Hwang HS, Choi SW, Kim HJ, Choi YE. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012;53(9):1535–45. doi:10.1093/pcp/pcs106. [Google Scholar] [PubMed] [CrossRef]
65. Kim OT, Bang KH, Jung SJ, Kim YC, Hyun DY, Kim SH, et al. Molecular characterization of ginseng farnesyl diphosphate synthase gene and its up-regulation by methyl jasmonate. Biologia plantarum. 2010;54:47–53. [Google Scholar]
66. Jung SC, Kim W, Park SC, Jeong J, Park MK, Lim S, et al. Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 2014;55(12):2177–88. doi:10.1093/pcp/pcu147. [Google Scholar] [PubMed] [CrossRef]
67. Murthy HN, Georgiev MI, Kim YS, Jeong CS, Kim SJ, Park SY, et al. Ginsenosides: prospective for sustainable biotechnological production. Appl Microbiol Biotechnol. 2014;98(14):6243–54. doi:10.1007/s00253-014-5801-9. [Google Scholar] [PubMed] [CrossRef]
68. Augustin JM, Kuzina V, Andersen SB, Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72(6):435–57. [Google Scholar] [PubMed]
69. Wu JY, Wong K, Ho KP, Zhou LG. Enhancement of saponin production in Panax ginseng cell culture by osmotic stress and nutrient feeding. Enzyme Microb Technol. 2005;36(1):133–8. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools