 Open Access
Open Access
ARTICLE
Exploring the Trait Plasticity of ipa1-2D and qPL6 under Different Nitrogen Treatments and Heading Periods
1 Joint International Research Laboratory of Agriculture and Agri-Product Safety of the Ministry of Education/Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, Yangzhou University, Yangzhou, 225009, China
2 Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops, Jiangsu Key Laboratory of Crop Genetics and Physiology, Yangzhou University, Yangzhou, 225009, China
3 Shanghai Key Laboratory of Bio-Energy Crops, Plant Science Center, School of Life Sciences, Shanghai University, Shanghai, 200444, China
4 Laboratory of Sustainable Agronomy and Crop Protection, Institute of Plantation Studies, Universiti Putra Malaysia, Serdang, Selangor, 43400, Malaysia
* Corresponding Author: Lin Zhang. Email:
# These authors contribute equally to this work
Phyton-International Journal of Experimental Botany 2024, 93(11), 2737-2754. https://doi.org/10.32604/phyton.2024.054649
Received 03 June 2024; Accepted 30 August 2024; Issue published 30 November 2024
Abstract
Panicle size is one of the important factors in shaping yield potential in rice, but it shows plasticity in different environments, which leads to yield fluctuation. Variations in panicle size among varieties are largely determined by quantitative trait loci (QTLs). QTL analysis could elaborate on the environmental impact on trait plasticity using nearly isogenic lines (NILs) of different QTLs. Two QTLs, ipa1-2D and qPL6 are identified to have pleiotropic contributions to panicle size and plant architecture, but their responses to different growth conditions are still unclear. In this study, we developed NILs harboring a single locus or both loci of ipa1-2D and qPL6 and subsequently evaluated these QTL effects under different nitrogen treatments or heading periods. Trait comparison showed that panicle length was highly responsive to the high nitrogen treatment independent of qPL6. At the same time, ipa1-2D reduced the response of plant height, panicle number, and grain yield to the treatment. The background of long heading periods decreased the stem diameter for any genotype combinations but enhanced the performance of ipa1-2D for the panicle primary branch number. Moreover, the middle heading background could better balance the pleiotropic effect of the two QTLs and showed the highest yield potential. In-parallel analysis of the QTL contributions under different nitrogen treatments or heading periods confirmed the significant effect of ipa1-2D in increasing stem diameter, panicle primary branch number, and spikelet number per panicle. We proved that the two individual QTLs had a stable effect in increasing the yield potential but competed to decrease the panicle secondary branch number, panicle number, and yield potential when they were pyramided. This work provides a full view of the plasticity of two QTLs in shaping yield-related traits and lays the foundation for the rational design of rice breeding in the future.Keywords
There is an urgent need to improve grain production, as the present yield potential of major food crops will not satisfy human demands by the 2050s [1]. Rice is one of the major stable foods in the world and it feeds approximately one-half of the world’s population. Rice yield is mainly determined by three factors, namely, panicle number, grain number per panicle, and grain weight [2]. Among these traits, panicle number and grain number per panicle were found to contribute largely to the yield variation of Chinese hybrid rice, and they collectively determine the total grain number per plant. Trait analysis of both indica and japonica varieties bred in China from 1989 to 2018 confirmed that grain number per panicle was the important yield determinant, and the varieties with more panicle grain number but less panicle number still retained the ability to produce high yield [3]. China started a super hybrid breeding program in 1996 to explore its yield potential, and more than 92 super hybrid rice have been bred since then [4]. Yongyou series are well-known super hybrid rice varieties in China, and they could produce a yield of 12 tons per hectare at suitable planting conditions [5]. Large panicles are one of the significant characteristics of super hybrid rice, suggesting breeding varieties with large panicles is the best way to improve yield potential. The panicle structure is highly diversified among different rice varieties and could be characterized by three components, namely, panicle length, and panicle primary and secondary branch number. A range of 50–600 grains can be generated due to different combinations of panicle structure components among varieties [2]. The panicle branch number of super hybrid rice varieties is nearly double that of conventional rice varieties, confirming the important role of optimal panicle structure for high-yield production [6].
Important agronomic traits, including panicle number and panicle grain number, are classified as quantitative traits, and their performances are easily affected by environmental changes such as fertilizer levels. Nitrogen (N) is an important component in compound fertilizers that has a promoting effect both on plant development and physiological processes, as demonstrated in sugar beet [7]. In rice, the panicle length, spikelet number per panicle, and panicle secondary branch number are enhanced by optimized N fertilizer application, and low N treatment would reduce the spikelet number per panicle and grain yield [8,9]. The negative effect of low N application on spikelet number can be alleviated by dense planting, as the increased panicle number contributed to the increase of total spikelet number and balanced the yield components [10]. Grain number per panicle of different varieties had inconsistent responses to N treatment, and the effect of N fertilization strategies on yield improvement was different between cultivars with different panicle weight and panicle number [8,11]. A study showed that the N application rate of super hybrid rice Yongyou 2640 was higher than that of conventional indica and japonica varieties, suggesting that the large panicle variety needed more N to reach its yield potential [12]. Although planting conditions will affect the performance of panicle traits, the intrinsic nature of such change is determined by the genes related to panicle development. Gene expression of MOC1, LAX1, and SP1 was upregulated under optimized N fertilizer, which contributed to the increase of secondary branches and spikelets [8]. Therefore, it is important to identify genes responsible for panicle development to develop breeding programs targeting panicle size.
Quantitative trait loci (QTLs) are major genetic factors determining panicle size variation among varieties. Up to now, several QTLs that are important for panicle structure and grain number have been cloned. Gn1a is the first cloned QTL related to panicle grain number in rice, and it encodes the cytokinin oxidase for cytokinin degradation. Lower expression or mutation of the gene will lead to the accumulation of cytokinin in the panicle primordium, thereby promoting the formation of more secondary branch numbers and grain numbers [13]. DEP1 is another well-known gene for panicle determination, and it encodes a G-protein γ subunit, and mutation of the gene led to dense and erect panicle morphology [14]. Moreover, DEP1 is associated with N response, and japonica varieties with dep1 mutant showed less response to treatments of different N fertilizers compared to the indica variety with DEP1 [15]. dep1 allele has been widely used in the breeding program of japonica in Jiangsu, Liaoning, and Zhejiang provinces in China but rarely used in indica breeding programs due to its negative effect on indica backgrounds. IPA1 determines both plant and panicle architecture, and it encodes SBP-domain transcriptional factor OsSPL14. ipa1-1D and ipa1-2D are two gain-of-function alleles of IPA1, and the former carries an SNP in the coding region of OsSPL14, abolishing the miR156 target sequence and leading to upregulation of the gene; the latter has a tandem repeat upstream of OsSPL14 promoter and specially up-regulates OsSPL14 in the panicle primordium [16,17]. The two gain-of-function alleles of IPA1 greatly improved the stem diameter and panicle size and contributed significantly to the yield improvement in hybrid rice [17]. Genome-wide DNA binding analysis revealed that more than 1000 genes are under the direct control of IPA1, including DEP1, OsTB1, and OsWRKY45, which are associated with panicle length, tiller number formation, and blast disease resistance, and it further demonstrated the important values of superior IPA1 allele in rice breeding [18,19]. SPIKE is a locus identified for its pleiotropic effect, and it is the natural allele of the Nal1 gene, which was first cloned from the narrow leaf mutant [20–22]. The beneficial allele of SPIKE originated from japonica rice and was demonstrated to increase panicle secondary branch number and grain number per panicle. Introgression of SPIKE to indica rice increased yield by 13–36% [21]. In addition, other QTLs with multiple trait contributions were cloned, such as SCM2/qPL6/PBN6 for strong culm, panicle length, and panicle primary branch number as well as SCM3/OsTB1 for panicle number and panicle size [23–26]. Identification of different panicle-related genes provides opportunities to study their genetic effects and breeding values under different planting regimes.
Heading date determines the adaptation of rice plants to local temperature and light conditions, and it is ideal to breed a variety that will fit the local environments. Genes related to the heading date were linked directly to the yield-related traits, especially for panicle structure. Ghd7, which encodes the CCT domain, was associated with simultaneously delayed heading date, improved plant height and stem diameter, as well as increased grain number per panicle [27]. Ghd8 and Ghd7.1 showed a similar phenotypic effect as Ghd7, and Ghd8 encodes the OsHAP3 subunit of the HAP complex while Ghd7.1 encodes the PPR7-like protein [28,29]. On the other hand, Hd1 was associated with photoperiod sensitivity, namely, promoting heading under short days while delaying heading under long days [30]. However, the control of photoperiod sensitivity of Hd1 relied on the function of Ghd7. Hd1 promoted heading under any day length in the background of mutated ghd7, while the mutated hd1 delayed the heading date, increased plant height and spikelet number per panicle, and therefore, was named Ghd6 [31]. Gene knock-out by CRISPR/CAS9 confirmed the effects of the above-mentioned four genes in shaping yield potential, and the combination of different beneficial alleles enhanced the yield-related traits such as panicle branch number [32]. Therefore, prolonging the heading date is a potential way to enhance the panicle size.
Nearly isogenic lines (NILs) are ideal materials to evaluate the genetic effects of QTLs, both for their interactions with the environment and genetic background [33]. Although many panicle-related QTLs have been cloned, their trait contributions are rarely evaluated at different environments or genetic backgrounds, making their value in rice breeding obscure. Our previous studies dissected two important pleiotropic QTLs, namely ipa1-2D and qPL6 that contributed not only to the large panicle formation but also the strong culms of Yongyou series breeding stocks [17,25]. However, it is unclear how the two QTLs shape the traits in response to different N levels and growth durations. In this study, we developed NILs of ipa1-2D and qPL6 in the background of Nipponbare (NIP), a japonica variety with an early heading period. The trait performance of different NILs was compared to determine the responses of these QTLs to N treatment. Another two sets of NILs with the same QTL combinations were selected from plants with middle and late heading periods. The main objective of this study was to explore the trait plasticity of ipa1-2D and qPL6 in response to N treatment and growth duration, which would provide novel insight through environmental adaptation of these two QTLs in achieving the yield potential. Since both QTLs contributed to the formation of large panicles and strong culm, we also studied the additive and competitive effects of the two QTLs under different conditions. Understanding these phenomena would facilitate the efficient application of the two QTLs in breeding programs to produce high-yielding rice in different planting regimes.
2.1 Development of NILs with Different Backgrounds
A breeding stock called YYP1 (indica) showed strong culm and large panicle phenotype, and it contains both ipa1-2D and qPL6 loci [17]. To develop the NILs, YYP1 was crossed with NIP (japonica) to generate F1 plants and then backcrossed with NIP three times to generate BC3F1 plants. Three BC3F2 lines were planted and selfed three times to generate BC3F4 lines. We found that BC3F2-1 derived lines showed an early heading performance similar to NIP, and it contained both ipa1-2D and qPL6 loci. To further purify the genetic background, it was backcrossed with NIP for an additional two times and selfed for two generations to obtain stable lines. During the process, ipa1-2D was selected by a functional marker ipa1-2D.M1 that is dominant for genotyping, and a linkage CAPS marker ipa1-2D.M2 was used to discriminate the homozygous ipa1-2D plants. The PCR product of ipa1-2D.M2 was digested by the Dra I restriction enzyme, and the ipa1-2D sequence was cut into shorter sequences by the enzyme. The linkage InDel marker qPL6.M was used for qPL6 genotyping, and the primer information of different markers are listed in Table 1. Both loci were selected by these markers, and four homozygous lines covering different allele combinations of two loci were obtained, namely, qpl6/IPA1, qPL6/IPA1, qpl6/ipa1-2D and qPL6/ipa1-2D. The other two BC3F4 lines headed later than NIP and one line contains qPL6 while another line contains ipa1-2D. The two late heading lines were crossed to pyramid the two QTLs, and their F1 lines were selfed two times to generate F3 lines with heterozygous ipa1-2D and qPL6. Two types of heading performance were selected from the F3 lines, and designated as middle heading and late heading. The two types of F3 lines were selfed to generate four homozygous lines of different allele combinations as mentioned above. The detailed procedure for NIL development is illustrated in Fig. 1.

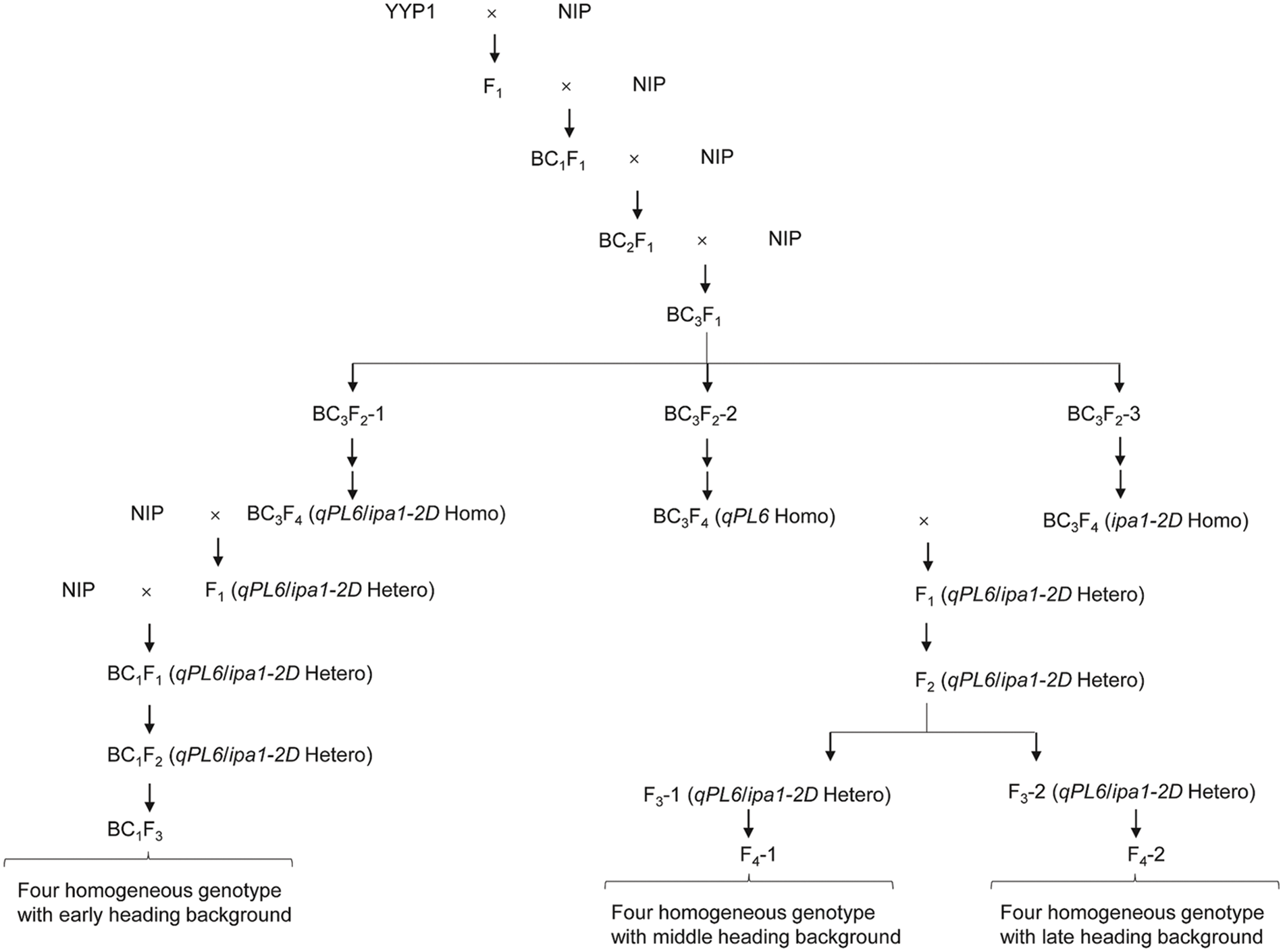
Figure 1: The procedure of NILs development with different heading date backgrounds
2.2 Genome Resequencing of NILs
Three heterozygous lines generating the NILs were subjected to low-coverage genome resequencing to verify the genetic background following a previously published method [34]. In brief, genomic DNA was isolated from the leaf tissues using DNAquick Plant System (DP321, TIANGEN, Beijing, China) following the manufacturer’s procedure and was fragmented and indexed to construct the library for sequencing. The library with indexed DNA was loaded into the Illumina platform for short-read sequencing. After trimming the index, the remaining DNA sequences of the three lines were aligned to the Nipponbare reference genome (IRGSP Releases Build 4.0) and YYP1 pseudogenome to find SNPs. Genotype calling was performed by calculating SNP ratios in the windows of 15 SNPs, and the highest probability of a genotype was obtained. In this way, the whole genome genotype was generated by sliding the window base-by-base along different chromosomes.
All lines were planted in Songjiang experimental station, Shanghai, in large field blocks with six plants in each row, and the distance between rows and between plants was both 15 cm. Early heading NILs were first planted in year 1 to compare the impact of two N treatments. One N treatment consisted of N rate practiced by the local farmers, i.e., 10.7, 6.4, and 4.3 kg urea/667 m2 at seedling, booting, and heading stage, respectively, and it was designated as normal N treatment (NN); another was designated as high N treatment (HN), as we supplemented N at the booting stage (15.5 kg urea/667 m2) and heading stage (6.4 kg urea/667 m2). Twenty kilograms of single superphosphate/667 m2 and 15.9 kg potassium chloride/667 m2 were applied as basal fertilizer before transplanting plants of both treatments. In addition, all three sets of NILs with early, middle, and late heading periods were planted in the second year, and the fertilizer scheme was following the normal practice of local farmers as mentioned above. At the mature stage, the field was equipped with nets to avoid damage to the panicles by birds.
To measure stem diameter and plant height, the main culms were cut from the base portion of the plants. The leaf sheath of the third internode was wiped and the diameter from the major axis and minor axis of the internode was measured by Vernier caliper. The average of two values was used for data analysis. Plant height was measured as the distance from the panicle tip to the base of the main culm. Panicles were cut from the main culms and subjected to panicle trait analysis, while panicle length was measured as the distance from the panicle tip to the peduncle base. Panicle primary and secondary branch numbers were counted manually and all spikelets were detached from panicle rachis and counted by a seed automatic counter (SLY-A, Zhejiang Tuopu, China). Infertile spikelets were counted manually, and the seed setting rate was calculated as follows: (Total spikelet number-Infertile spikelet number)/Total spikelet number. All panicles were collected from a single plant and counted as panicle numbers, and all spikelets were detached from the panicle and weighted as grain yield per plant. After counting all spikelets from a single plant, the average spikelet number per panicle was calculated as the total spikelet number per plant divided by the panicle number. The heading date was recorded manually each day when the first panicle appeared, and the days from planting to the recorded time were calculated as data for analysis. Data of four genotypes under two N treatments or three heading periods were compared simultaneously, and one-way ANOVA analysis was performed followed by the Tukey test at a significant level of 0.05 to obtain statistical significance among lines.
3.1 Genetic Effects of ipa1-2D, qPL6 and Their Combinations under Different Nitrogen Treatments
We obtained a set of NILs with an early heading date similar to NIP, and the overall morphology of the four lines was similar (Fig. 2A). Two QTLs (ipa1-2D, qPL6) were identified to be responsible for panicle size. Apparent differences in panicle morphology among the four lines were observed visually (Fig. 2B). Compared to the plants of control lines (qpl6/IPA1), plants with only qPL6 demonstrated longer panicles, and plants with only ipa1-2D showed highly branched panicles. The lines with both loci showed an accumulated effect of the two loci, namely, more panicle branches and longer panicles. For the comparison of N treatments, eight agronomic traits were assessed, including stem diameter, plant height, panicle length, panicle primary and secondary branch number, spikelet number per main panicle, panicle number, and grain yield per plant (Fig. 2C–J).
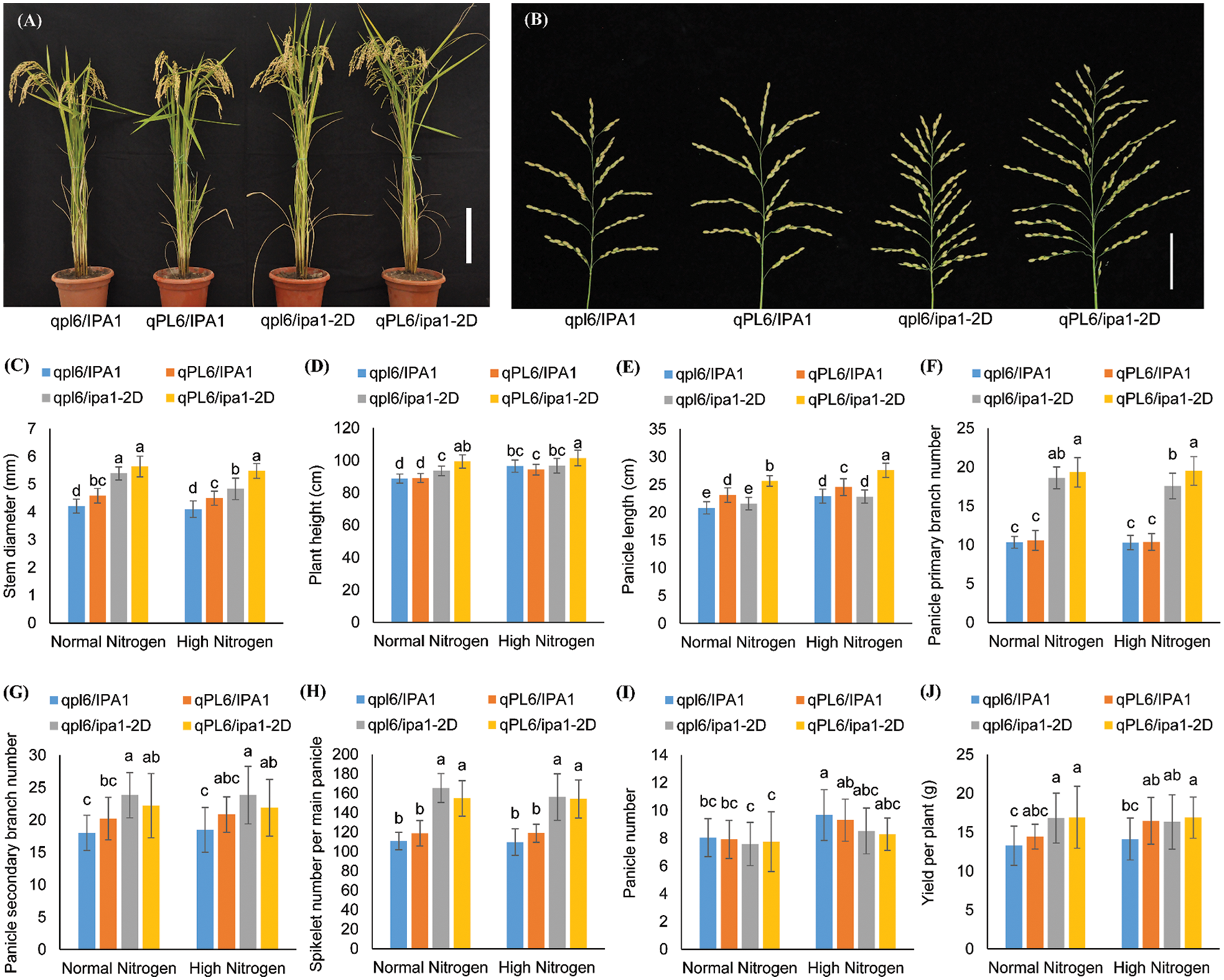
Figure 2: Trait performance of nearly isogenic lines under nitrogen treatments in the early heading background. (A). The overall plant morphology of the NILs under normal nitrogen (NN) conditions. Bar = 20 cm. (B). The overall panicle morphology of the NILs under NN conditions. Bar = 6 cm. C–J: Comparison of eight traits among four genotypes under NN and high nitrogen (HN) conditions, including stem diameter (C), plant height (D), panicle length (E), panicle primary branch number (F), panicle secondary branch number (G), spikelet number per main panicle (H), panicle number (I) and yield per plant (J). Different letters between groups indicate significant differences by Tukey test. Values are means ± SD, and n = 30
The statistical analysis showed that significant differences existed between either genotypes or treatments. Both qPL6 and ipa1-2D plants demonstrated increased stem diameter, but the contribution of ipa1-2D was more than that of qPL6, and the pyramiding of the two loci further improved the trait. Nitrogen treatment had little effect on the performance of stem diameter, and reduction was only found for the genotype qpl6/ipa1-2D under HN conditions (Fig. 2C). Lines with ipa1-2D had a promoting effect on the plant height, especially for NN conditions, while HN conditions would enhance the plant height more for the lines without ipa1-2D (Fig. 2D). Interestingly, panicle length was highly responsive to both genotype changes and N treatments. Plants with qPL6 significantly increased panicle length, but plants with ipa1-2D had little effect on the trait. However, when two loci were combined, the panicle length was further improved compared to the single qPL6 line, suggesting epistatic interaction of two loci for the trait. Moreover, the panicle length of all four genotypes was further enhanced under HN conditions, suggesting the process of panicle elongation was related to N nutrient (Fig. 2E). Plants with ipa1-2D increased both panicle primary and secondary branch number while plants with qPL6 only showed a trend of increasing panicle secondary branch number. It seemed that the pyramiding of qPL6 and ipa1-2D would compete for the secondary branch formation, and a slight reduction was found for the trait in the pyramiding lines under both N levels. Unlike panicle length, panicle branches showed insignificant response to the N treatments (Fig. 2F,G). Consistent with the performance of panicle branches, spikelet number per main panicle was mainly increased in the lines with ipa1-2D and showed no significant difference among the N treatments (Fig. 2H). Panicle number per plant was less affected by the two loci, but could be enhanced by HN conditions, especially for lines without ipa1-2D (Fig. 2I). We finally compared the yield performance of the four lines and found that the QTL-containing lines had higher yield values than the control lines, but only lines with ipa1-2D or both loci under NN conditions and line with both loci under HN conditions showed statistical significance (Fig. 2J).
3.2 Development of NILs with Different Heading Periods
The heading period will affect the overall growth duration of the plants and therefore, the nutrient intake and carbon assimilation accumulation, and it is interesting to determine the QTL effect under different heading periods. We successfully obtained lines with middle and late heading performance, and the four homozygous genotypes were developed from a single plant with heterozygous qPL6 and ipa1-2D (Fig. 1). By low-coverage resequencing, we obtained the genome information of the heterozygous lines with different heading dates, namely, early heading, middle heading and late heading. The result showed that most chromosomes of the three lines were replaced by the recipient parent NIP. However, these three lines still contained other introgressions from the donor parent YYP1 in addition to the heterozygous ipa1-2D and qPL6 region (Fig. 3A–C). In detail, the early heading line has two large YYP1 segments on chromosome 5 and chromosome 10, which were not found in the middle and late heading lines (Fig. 3A–C). The middle heading line has more introgression from YYP1, including segments on chromosomes 1, 4, 6, and 8, and the same segments were also identified in the late heading line (Fig. 3B,C). Moreover, two large segments on chromosomes 2 and 9 were especially harbored by the late heading lines (Fig. 3C). Therefore, the different heading performance might be due to the segment variations identified, and it suggested that high-quality NILs could be generated by selfing the three lines with heterozygous ipa1-2D and qPL6. By genotyping the selfing progeny using respective markers, we finally obtained homozygous lines with all four allele combinations of ipa1-2D and qPL6 in three genotype backgrounds of heading periods (Fig. 3D).

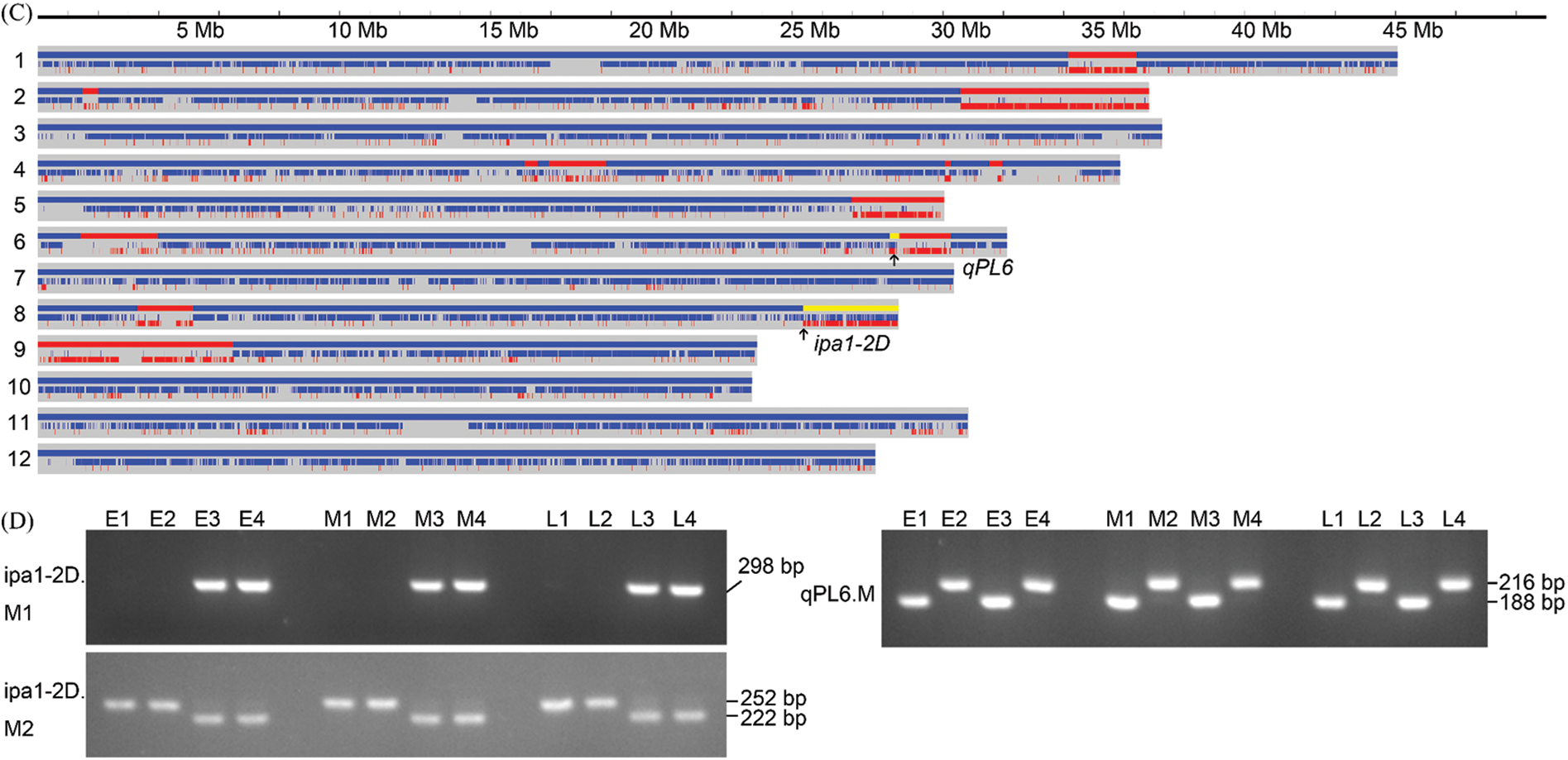
Figure 3: Genetic background of three lines with heterozygous ipa1-2D and qPL6. (A) The genome background of early heading line; (B) The genome background of middle heading line; (C) The genome background of late heading line; The blue and red colors indicates the segments from recipient and donor parents respectively, and yellow color indicates the heterozygous region. Arrows denote the position of qPL6 and ipa1-2D, respectively. (D) The homozygous genotypes of all allele combinations of ipa1-2D and qPL6 in three backgrounds as shown by the PCR-based markers. ipa1-2D.M1 is the dominant functional marker and the appearance of the bands suggest existence of ipa1-2D allele. ipa1-2D.M2 is the CAPS marker tightly linked with ipa1-2D, and qPL6.M is an InDel marker tightly linked with qPL6. The size of markers is indicated as shown
The high-quality NILs were planted for trait comparison, and we found that the plant morphology was similar among the four genotypes but different among three heading lines for each genotype (Fig. 4A–C). The exact heading date was recorded for all the genotypes with different heading periods. The difference in heading date was significantly large among the three heading lines, which were 100 days for early heading lines, 120 days for middle heading lines, and 143 days for late heading lines. A slight difference was found among the four genotypes in different heading lines, while for the early and late heading lines, the pyramiding genotypes headed later than those of the other genotypes. For plants with a middle heading period, the lines with single qPL6 or ipa1-2D headed later than the other two lines (Fig. 4D). Since some genes associated with heading period were reported to affect the plant height and stem diameter, we first compared the variations of the two traits in plants from the three heading lines. The difference in plant height was not as significant as heading date, and only small variation in the trait was observed across all heading lines (Fig. 4E). The potential effect of ipa1-2D in promoting plant height was only identified in the early heading lines, but its contribution to stem diameter could be detected in all the heading lines (Fig. 4E,F). The performance of qPL6 plants in terms of stem diameter was not as significant as the previous season in the early heading lines, but a significant difference was found in the middle heading lines. Interestingly, we found the middle or late heading lines had smaller stem diameters compared with early heading lines, which was stable for all four genotypes (Fig. 4F). These results suggested that the delay in the heading period did not significantly enhance the overall biomass of the plants.
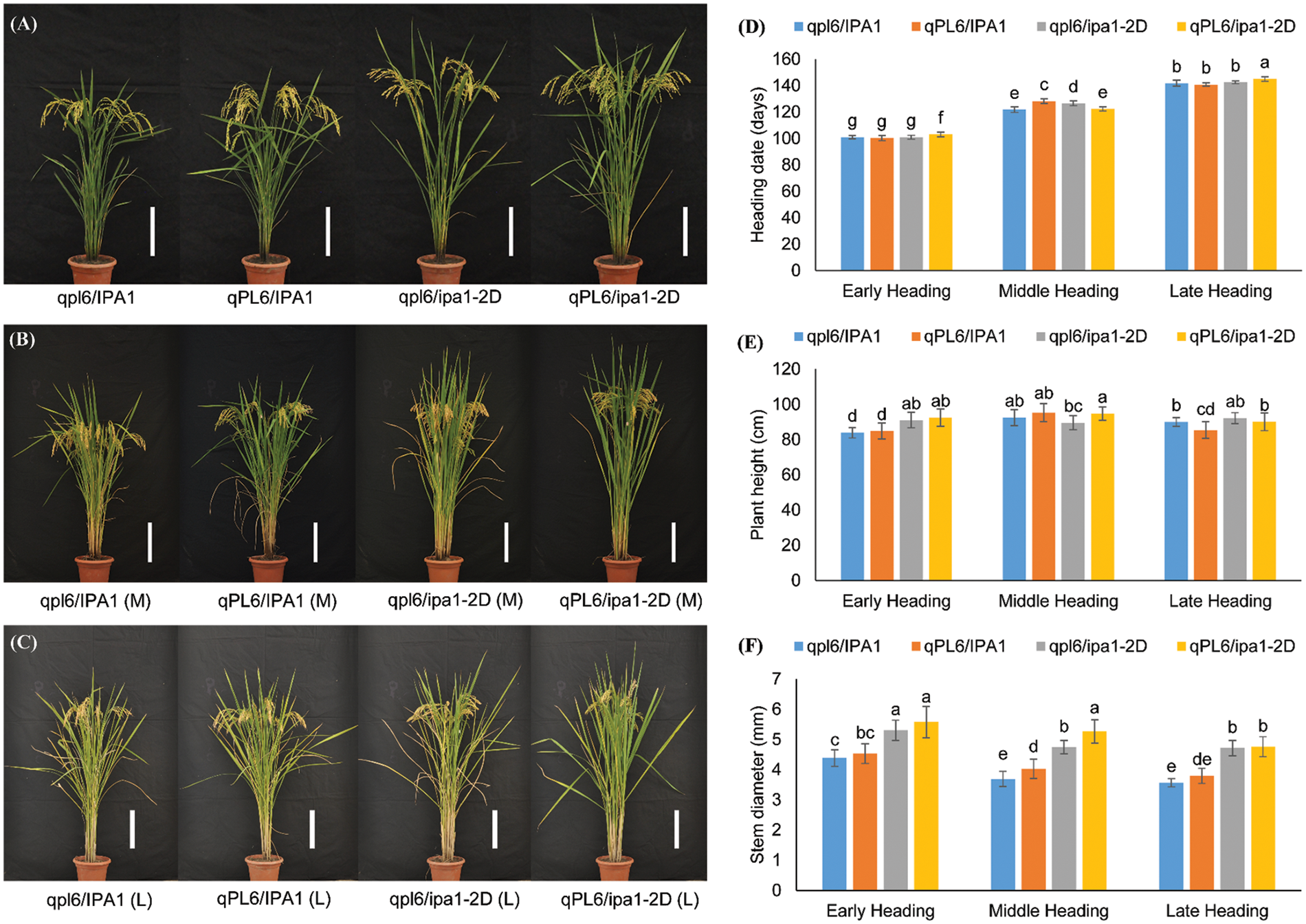
Figure 4: Comparison of plant architecture of NILs with different heading date backgrounds. A–C: The overall plant morphology of different NILs in early (A), middle (B) and late (C) heading backgrounds. Bar = 20 cm. The pictures were taken at the mature stage of the plants, which was 125, 153 and 181 days after planting. (D). Heading date variations among four genotypes of three backgrounds. (E). Plant height variations among four genotypes of three backgrounds. (F). Stem diameter variations among four genotypes of three backgrounds. Different letters between groups indicate significant differences by Tukey-test. Values are means ± SD, and n = 24 for (D) and n = 20 for (E, F)
3.3 Comparison of Panicle and Yield-Related Traits among Three Heading Lines
We then compared the panicle and yield-related traits among three heading lines. The contribution of qPL6 to panicle length was relatively stable among the three heading lines, but the interactive effect of qPL6 with ipa1-2D was not consistent (Fig. 5A). In the early or late heading lines, pyramiding of two loci further improved panicle length, but lines with single qPL6 was not significant in improving this trait. However, single ipa1-2D in the late heading lines showed increased panicle length, which was unprecedented. For the middle heading line, a single qPL6 and the pyramiding of two loci showed similar contributions to the panicle length (Fig. 5A). The contribution of ipa1-2D to panicle primary branch number was consistent in all three heading lines, while the middle and late heading lines enhanced its promoting effect (Fig. 5B). However, the promoting effect of a single ipa1-2D for panicle secondary branch number in the middle or late heading line was not as significant as that in the early heading line, though the values were improved (Fig. 5C). The contribution of a single qPL6 to panicle secondary branch number was demonstrated in the early and middle but not in the late heading lines, and its competitive effect towards ipa1-2D was again found in lines of all three heading periods. It showed that the competition of two loci was weak in the middle heading line, as the value did not decrease compared with lines with a single locus (Fig. 5C). The spikelet number per main panicle showed a consistent change pattern with the panicle branches, and all the lines with single or both loci could increase the traits except for the line with single qPL6 in the late heading background (Fig. 5D). The heading date could affect the performance of spikelet number per main panicle in lines with qPL6, as the trait values were the largest in the middle heading lines compared with the other two heading lines (Fig. 5D).
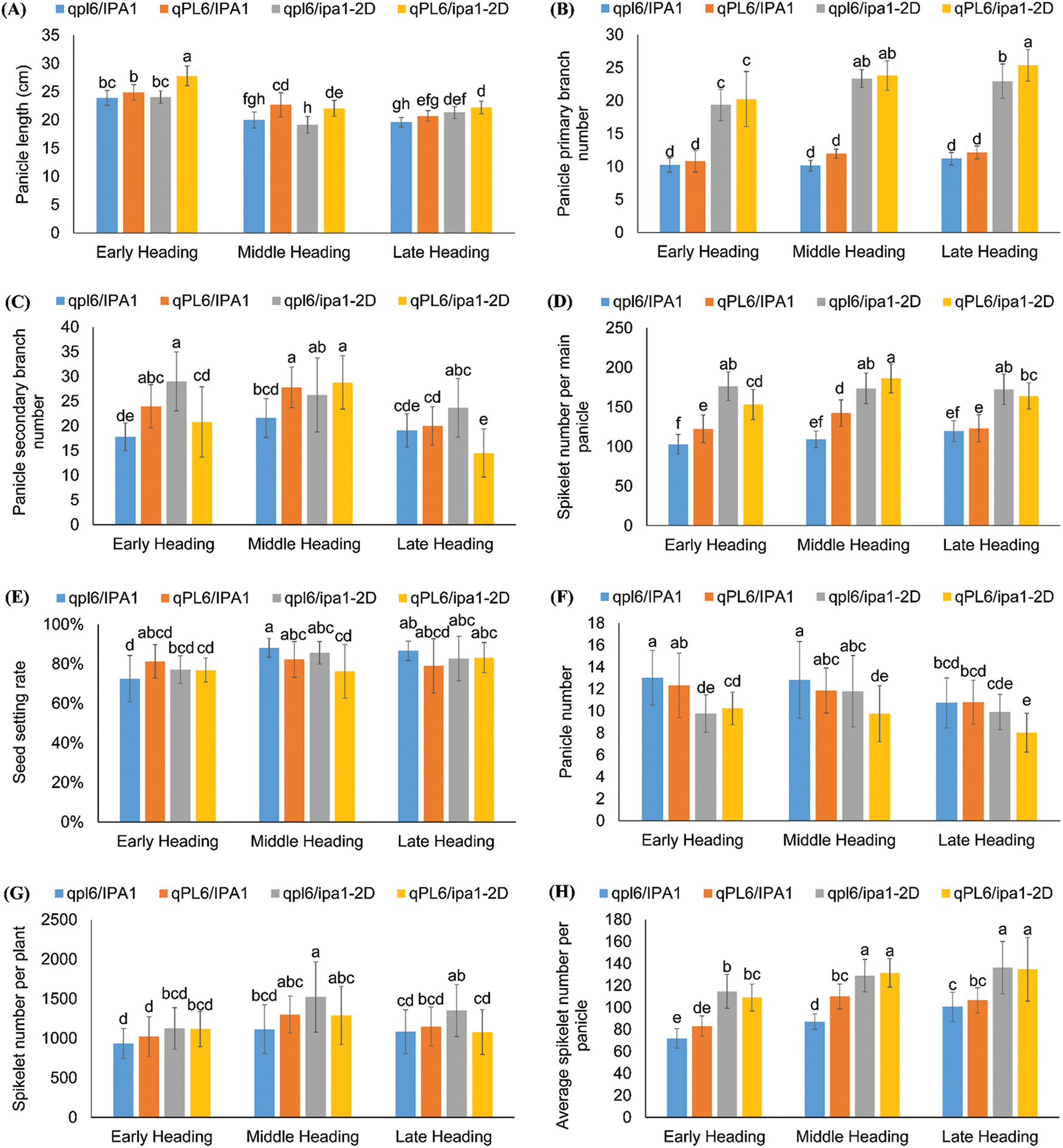
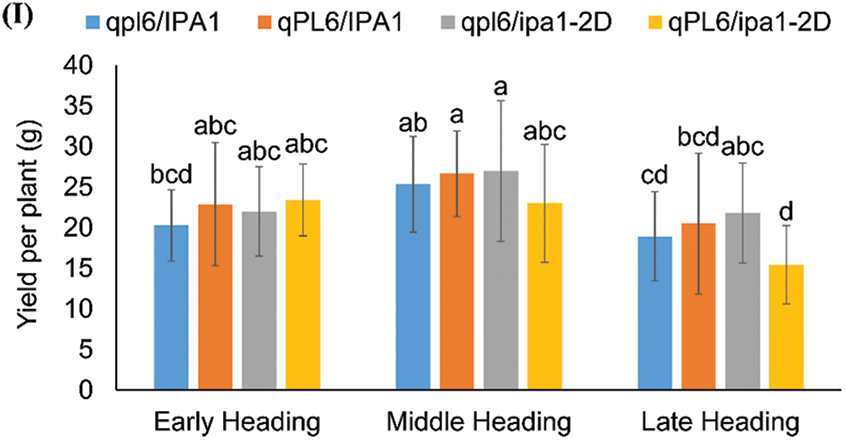
Figure 5: Comparison of panicle and yield-related traits of NILs with different heading date backgrounds. The traits include panicle length (A), panicle primary branch number (B), panicle secondary branch number (C), spikelet number per main panicle (D), seed setting rate (E), panicle number (F), spikelet number per plant (G), average spikelet number per panicle (H) and yield per plant (I). Different letters between groups indicate significant differences by Tukey test. Values are means ± SD, and n = 20 for (A–E) and n = 30 for (F–I)
No significant difference in seed setting rate could be found among the four genotypes in the three heading periods, even though the lines with one or two loci showed a slight increase in the early heading period and a slight decrease in the middle and late heading periods (Fig. 5E). The panicle number was significantly reduced in lines with both ipa1-2D and qPL6 for all three heading periods, indicating that the two loci intensified the depression of tiller formation (Fig. 5F). Due to the balance of panicle number, the increase of spikelet number per plant was not as significant as that of spikelet number per panicle, and lines with single ipa1-2D showed the highest increasing rate in all three heading periods (Fig. 5G). The average spikelet number was calculated from the total spikelet number per plant, and it showed a similar trait change to that of spikelet number per main panicle. However, the mean values could reflect the effect of heading period better, and both middle and late heading periods significantly enhanced the trait performance in comparison to that of early heading period (Fig. 5H). We finally compared the yield performance of different lines and found that the yield traits showed a vast fluctuation among the individuals within lines. All the lines with either qPL6 or ipa1-2D singly increased yield, though the statistical level was insignificant due to the wide trait fluctuation. However, the yields of pyramiding lines with both loci showed decreased yield in the middle and late heading lines. A comparison of three backgrounds suggested that lines with a middle heading period had the best yield performance compared to the other two heading periods (Fig. 5I).
3.4 The in-Parallel Comparison of Trait Contribution Determined by Different Loci
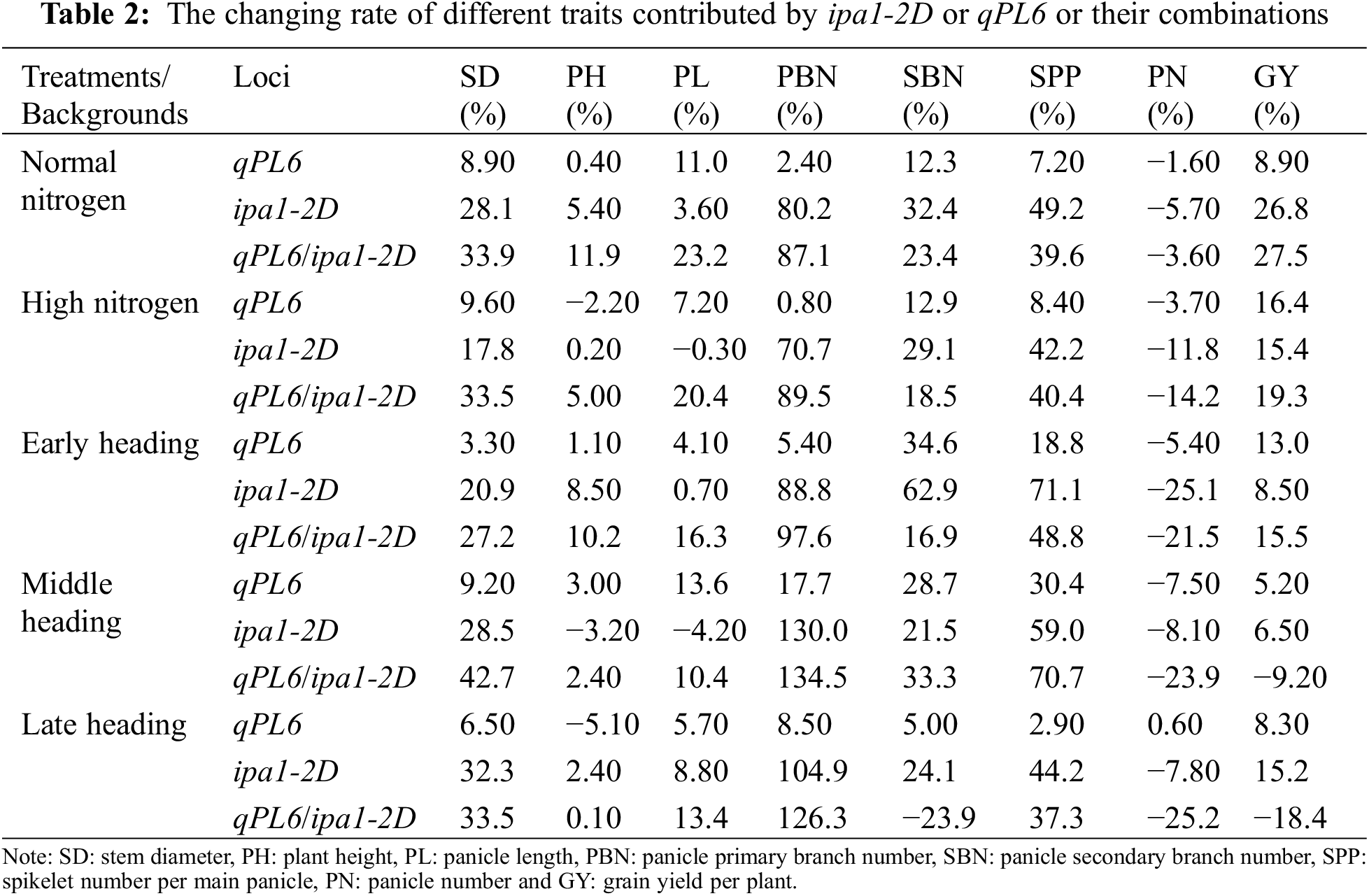
We then compared the changing percentage of different traits contributed by single QTL or their combinations in parallel (Table 2). The stem diameter increased consistently in plants with ipa1-2D, ranging from 20.9% to 32.3%, and its combination with qPL6 further improved the values, ranging from 27.2% to 42.7%. The plant height showed inconsistent change in response to the same genotype under different N treatments and heading periods, suggesting a weak effect of two QTLs for the trait. qPL6 had a stable contribution to generating the phenotype of a long panicle, and the trait was increased by 4.1% to 23.2% when qPL6 was present. Both qPL6 and ipa1-2D increased the panicle branch and spikelet number, especially ipa1-2D. ipa1-2D stably increased the panicle primary branch number by 70.7% to 130%, and qPL6 slightly enhanced the trait performance by 0.8% to 17.7% singly and from 6.9% to 21.4% when combined with ipa1-2D. Lines with single ipa1-2D increased panicle secondary branch number by 21.5% to 62.9%, and lines with single qPL6 increased the trait by 5% to 34.6%. However, the combination of two loci tended to reduce the trait performance by 9% to 48% compared with the line of a single ipa1-2D, except for the case of the middle heading line. Either QTLs increased the spikelet number per main panicle at any conditions, and the increase percentages were 2.9–30.4% for single qPL6, 42.2–71.1% for single ipa1-2D and 37.3–70.7% for their combination, indicating that ipa1-2D had more contribution to the traits. However, both QTLs showed a trend in decreasing panicle number, i.e., 1.6–7.5% for line with single qPL6, 5.7–25.1% for line with single ipa1-2D, and 3.6–25.2% for line of their combination. Despite the negative effect of the two loci regarding panicle number, the grain yield per plant increased under most conditions due to the increase of spikelet number per panicle. The lines with single qPL6 increased yield by 5.2% to 16.4%, and lines with single ipa1-2D increased yield by 6.5% to 26.8%. For the lines with both loci, a yield increase was demonstrated in the early heading lines, ranging from 15.5% to 27.5%, and the yield was reduced by 9.2% and 18.4% in the middle and late heading date lines, respectively. The above analysis presented a full view of the roles of qPL6 and ipa1-2D in shaping different yield-related traits under different N treatments and heading periods.
Though panicle morphology contributes substantially to the yield potential of rice plants, previous research showed that some panicle-related QTLs were only detected once using the same populations in two years [2], suggesting the plasticity nature of the panicle morphology in response to environmental changes. This hypothesis was verified by a study on SPIKE/Nal1 locus using NILs of two genetic backgrounds under shading conditions or different population densities, which showed that panicle secondary branching was a significant source of panicle plasticity affected by both the environment and QTLs [35]. The previous study focused on a single QTL, and it will be beneficial to study the trait plasticity to include more panicle-related QTLs. Two QTLs, ipa1-2D and qPL6, contribute significantly to the panicle size and exert pleiotropic effects on other traits such as stem diameter [17,25]. It is essential to determine the role of these two QTLs in the trait plasticity at different growth conditions.
The panicle morphology-related gene dep1 was reported to participate in N use efficiency, and this gene was confirmed to be upregulated by IPA1/OsSPL14 [15,18]. Moreover, IPA1/OsSPL14 expression was responsive to the variations of N levels [36]. Therefore, we first compared the impact of N treatments on the relevant QTL performance. We found that panicle length was the main contributor to panicle plasticity affected by N treatments, but it had no interaction with qPL6 even though this QTL had a significant impact on the trait, suggesting that other genes might mediate the N response of panicle length. N level could affect the plasticity of panicle branches but was inconsistent among previous reports [37,38]. Luo et al. found that the promoting effect of N on panicle secondary branches and spikelet number per panicle existed only between the lowest N and low N treatments, while the traits could not be further improved by moderate and high N applications [37]. Another study showed that the panicle morphology was enhanced only at the optimal N level while higher N decreased the panicle length and panicle primary and secondary branch number, particularly the panicle secondary branch number. The associate gene, OsAAP15 expression, was induced by N treatments and mediated the panicle response to different N treatments [38]. In our study, we found no change in panicle branch number under the high N treatment, similar to the previous report performed in the paddy field [37]. Although IPA1/OsSPL14 was implicated in N response, we did not find the interactive effect of different IPA1 alleles with N treatment for trait panicle branch number. However, ipa1-2D might change the response of plant height, panicle number, and grain yield to N levels as these traits were less sensitive to N treatments in the presence of this QTL, similar to the performance of dep1 [15], indicating that ipa1-2D locus could improve rice yield potential under limited N input, which needs to be verified further.
The heading period determines the duration of rice growth, and high-yield varieties such as Yongyou series super hybrid rice usually have longer growth periods which guarantees more carbohydrates produced via photosynthesis [39]. However, shortening the maturity duration did not lead to yield penalties in some rice varieties as they contain genes that facilitate N utilization and improve photosynthesis rate [40]. Here, we compared the impact of different growth durations on the performance of ipa1-2D and qPL6 and confirmed the existence of interaction between these QTLs and different growth durations. Though we do not know the underlying genes associated with the variations in the heading period of the three heading lines, we excluded the possibility of some known genes having control over the heading period, including Ghd6, Ghd7, Ghd7.1, and Ghd8, which significantly affect both plant height and panicle spikelet number [27–30]. Therefore, the three sets of NILs were ideal materials to compare only the impact of growth duration on the QTL performance. Compared to Ghd7, which was associated with promoting stem diameter, we found that stem diameter was decreased in the middle or late heading lines, suggesting that a strong culm is not necessarily linked to the late heading performance as assumed before. On the other hand, the panicle length was shortened significantly by the delayed heading, and qPL6 could partially alleviate the impact. The panicle primary branch number was significantly increased by delayed heading only in ipa1-2D lines, suggesting that the extended growth duration might enhance the function of ipa1-2D for primary branch primordium initiation. Nevertheless, the finding showed no consistent changing pattern associated with the panicle secondary branch number among the genotypes. The growth duration of the middle heading lines lasted 153 days, and it fitted well under temperature and light conditions in Shanghai. We found that the well-fitted growth duration could balance the panicle development to generate more spikelets per plant and contribute higher yield potential than plants of the other two heading genotypes. Although the late heading line had more time for vegetative growth, it suffered from colder weather during the grain-filling stage, which could significantly decrease the photosynthesis assimilation and yield potential.
Comparison of ipa1-2D and qPL6 lines in multiple planting conditions and genetic backgrounds provides an opportunity to learn the delicate contribution of two loci in rice breeding. Among the tested traits, we found that the ipa1-2D line has a stable effect on stem diameter, panicle primary branch number, and spikelet number per panicle, which could be used to improve both lodging resistance and yield potential. Strong culm was characterized as a new target for lodging resistance, and only limited loci for the trait were explored due to the weak effect of the loci and their sensitivity to environmental changes. Previous research showed that a NIL with four QTLs could increase stem diameter by 39%, but the plant height also increased significantly, leading to a negative effect on lodging resistance. However, the line with a single locus could only increase the stem diameter by 7% without affecting the plant height [41]. Recently, Gn1a was demonstrated to be an important QTL associated with stem diameter and it could increase the stem diameter by 14.08% singly and 23.31% when it was pyramided with SCM2, another well-known QTL for strong culm [42]. We found that ipa1-2D could increase the stem diameter by 17.8–32.3% singly and 27.2–42.7% when pyramided with qPL6. Therefore, the effect of ipa1-2D was comparable to the combined effect of 2–3 previously reported QTLs without changing the plant height significantly. Moreover, the ipa1-2D line produced nearly double the panicle primary branch number, and the effect was far more than the other QTLs involving panicle primary branch control, such as qPPB3 and PBN6 [24,43], which increased only one or two of the primary branch numbers that could be easily disturbed by environmental changes.
In our study, we demonstrated the role of environmental disturbance in the panicle traits and found that the performance of qPL6 was not stable for all the tested traits. qPL6 for panicle length is allelic to SCM2 for stem diameter and PBN6 for panicle primary branch number, and our previous research confirmed that it also increased the panicle secondary branch number and spikelet number per panicle [23–25]. We found that the panicle length was the most stable trait contributed by qPL6, while the traits stem diameter and panicle branch number were only detected in certain planting conditions or genetic backgrounds. In addition, we found the epistatic interaction between ipa1-2D and qPL6 for panicle secondary branch number, as the trait in pyramiding lines performed worse than that with a single locus. We suggest that the two loci could compete with the branch primordium during inflorescence meristem development and some branch primordium might be degraded during the process. ipa1-2D had a higher possibility for decreasing the panicle number than qPL6, and the trait was further negatively affected when the two loci were combined together. We found that all the lines harboring either ipa1-2D or qPL6 showed a yield increase ranging from 5.2% to 26.8%, confirming their values for yield improvement. However, yield decrease was found in the middle or late heading background when both loci existed. Therefore, it is recommended to utilize either ipa1-2D or qPL6 in the breeding program singly or use both in a compatible genetic background.
5 Conclusion and Future Outlook
The findings in this study confirmed the plasticity of various traits determined by ipa1-2D and qPL6 under different N treatments and growth durations. We found that ipa1-2D has a stable effect on the panicle branch and spikelet number, contributing significantly to the yield improvement. qPL6 could also improve yield performance even though its impact on panicle morphology was easily disturbed. N treatment strongly enhanced the panicle length regardless of the presence of these QTLs, and ipa1-2D seemed to buffer the promoting effect of N on plant height, panicle number, and grain yield. The delayed heading period decreased stem diameter stably but increased the effect of ipa1-2D by increasing the panicle primary branch number. A strong competitive effect was found between the two QTLs in generating tillers and panicle secondary branches, which led to yield loss at longer heading periods. The optimized heading period could better balance the panicle development and contribute more to the yield increase than other backgrounds.
This study lays the foundation to for exploring the breeding value of the two QTLs. As the ipa1-2D line is less sensitive to high N treatment, it is worth testing the value of the ipa1-2D line in retaining yield under limited N input, which might provide the opportunity for developing rice variety that is environmentally friendly by using less chemical fertilizers. Moreover, it is essential to dissect the molecular mechanisms of the two QTLs in their competing roles on panicle secondary branches, and the NILs developed in this study could be used to rapidly investigate the molecular network underlying the competition through transcriptomic analysis in the future.
Acknowledgement: We thank Prof. Huang Xuehui of Shanghai Normal University for support in genome re-sequencing and genotype analysis of the NILs.
Funding Statement: This work was supported by grants from the National Natural Science Foundation of China (32072037), the grant “Postgraduate Research & Practice Innovation Program of Jiangsu Province (Yangzhou University SJCX22_1786)”, the grant from the Ministry of Science and Technology of the People’s Republic of China (DL2022014009L), and the grants from Jiangsu Province Government (BZ2021017 and 20KJA210002).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Lin Zhang, Qiaoquan Liu; data collection: Wenshu Zhuang, Guangyang Jin, Yiting Zou, Zhong Bian, Dong Xie; analysis and interpretation of results: Shuwei Zhang, Hadi Yeilaghi, Liangliang Yu, Xiaolei Fan, Dongsheng Zhao; draft manuscript preparation: Lin Zhang, Muiyun Wong. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used in the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding this study.
References
1. Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS One. 2013;8(6):e66428. doi:10.1371/journal.pone.0066428. [Google Scholar] [PubMed] [CrossRef]
2. Xing Y, Zhang Q. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61(1):61421–442. doi:10.1146/annurev-arplant-042809-112209. [Google Scholar] [PubMed] [CrossRef]
3. Li G, Zhang H, Li J, Zhang Z, Li Z. Genetic control of panicle architecture in rice. The Crop J. 2021;9(3):590–7. doi:10.1016/j.cj.2021.02.004. [Google Scholar] [CrossRef]
4. Cheng S, Zhan X, Cao L. Breeding strategies for increasing yield potential in super hybrid rice. Front Agric Sci Eng. 2015;2(4):277–82. doi:10.15302/J-FASE-2015081. [Google Scholar] [CrossRef]
5. Wei H, Li C, Xing Z, Wang W, Dai Q, Zhou G, et al. Suitable growing zone and yield potential for late-maturity type of Yongyou japonica/indica hybrid rice in the lower reaches of Yangtze River, China. J Integr Agric. 2016;15(1):50–62. doi:10.1016/S2095-3119(15)61082-6. [Google Scholar] [CrossRef]
6. Xu D, Zhu Y, Chen Z, Han C, Hu L, Qiu S, et al. Yield characteristics of japonica/indica hybrids rice in the middle and lower reaches of the Yangtze River in China. J Integr Agric. 2020;19:2394–406. [Google Scholar]
7. Elsayed S, El-Hendawy S, Elsherbiny O, Okasha A, Elmetwalli A, Elwakeel A, et al. Estimating chlorophyll content, production, and quality of sugar beet under various nitrogen levels using machine learning models and novel spectral indices. Agronomy. 2023;13:2743. [Google Scholar]
8. Zhou W, Lv T, Yang Z, Wang T, Fu Y, Chen Y, et al. Morphophysiological mechanism of rice yield increase in response to optimized nitrogen management. Sci Rep. 2017;7(1):17226. [Google Scholar] [PubMed]
9. Gong Y, Lei Y, Zhang X, Yan B, Ju X, Cheng X, et al. Nitrogen rate and plant density interaction enhances grain yield by regulating the grain distribution of secondary branches on the panicle axis and photosynthesis in japonica rice. Photosynthetica. 2022;60(2):179–89. [Google Scholar]
10. Wei H, Meng T, Ge J, Zhang X, Shi T, Ding E, et al. Reduced nitrogen application rate with dense planting improves rice grain yield and nitrogen use efficiency: a case study in east China. The Crop J. 2021;9(4):954–61. [Google Scholar]
11. Zou Y, Zhang Y, Cui J, Gao J, Guo L, Zhang Q. Nitrogen fertilization application strategies improve yield of the rice cultivars with different yield types by regulating phytohormones. Sci Rep. 2023;13(1):21803. doi:10.1038/s41598-023-48491-w. [Google Scholar] [PubMed] [CrossRef]
12. Zhou Q, Yuan R, Zhang W, Gu J, Liu L, Zhang H, et al. Grain yield, nitrogen use efficiency and physiological performance of indica/japonica hybrid rice in response to various nitrogen rates. J Integr Agric. 2023;22(1):63–79. doi:10.1016/j.jia.2022.08.076. [Google Scholar] [CrossRef]
13. Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–5. doi:10.1126/science.1113373. [Google Scholar] [PubMed] [CrossRef]
14. Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41(4):494–7. doi:10.1038/ng.352. [Google Scholar] [PubMed] [CrossRef]
15. Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, et al. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet. 2014;46(6):652–6. doi:10.1038/ng.2958. [Google Scholar] [PubMed] [CrossRef]
16. Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42(6):541–4. doi:10.1038/ng.591. [Google Scholar] [PubMed] [CrossRef]
17. Zhang L, Yu H, Ma B, Liu G, Wang J, Wang J, et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat Commun. 2017;8(1):814789. doi:10.1038/ncomms14789. [Google Scholar] [PubMed] [CrossRef]
18. Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, et al. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell. 2013;25(10):3743–59. doi:10.1105/tpc.113.113639. [Google Scholar] [PubMed] [CrossRef]
19. Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, et al. A single transcription factor promotes both yield and immunity in rice. Science. 2018;361(6406):1026–8. doi:10.1126/science.aat7675. [Google Scholar] [PubMed] [CrossRef]
20. Qi J, Qian Q, Bu Q, Li S, Chen Q, Sun J, et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008;147(4):1947–59. doi:10.1104/pp.108.118778. [Google Scholar] [PubMed] [CrossRef]
21. Fujita D, Trijatmiko KR, Tagle AG, Sapasap MV, Koide Y, Sasaki K, et al. NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc Natl Acad Sci U S A. 2013;110(51):20431–20436. doi:10.1073/pnas.1310790110. [Google Scholar] [PubMed] [CrossRef]
22. Xu X, Wu K, Xu R, Yu J, Wang J, Zhao Y, et al. Pyramiding of the dep1-1 and NAL1NJ6 alleles achieves sustainable improvements in nitrogen-use efficiency and grain yield in japonica rice breeding. J Genet Genomics. 2019;46(6):325–8. doi:10.1016/j.jgg.2019.02.009. [Google Scholar] [PubMed] [CrossRef]
23. Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun. 2010;1132(1):580. doi:10.1038/ncomms1132. [Google Scholar] [PubMed] [CrossRef]
24. Terao T, Nagata K, Morino K, Hirose T. A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor Appl Genet. 2010;120(5):875–93. doi:10.1007/s00122-009-1218-8. [Google Scholar] [PubMed] [CrossRef]
25. Zhang L, Wang J, Wang J, Wang L, Ma B, Zeng L, et al. Quantitative trait locus analysis and fine mapping of the qPL6 locus for panicle length in rice. Theor Appl Genet. 2015;128(6):1151–61. doi:10.1007/s00122-015-2496-y. [Google Scholar] [PubMed] [CrossRef]
26. Yano K, Ookawa T, Aya K, Ochiai Y, Hirasawa T, Ebitani T, et al. Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol Plant. 2014;8(2):303–14. [Google Scholar] [PubMed]
27. Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–7. doi:10.1038/ng.143. [Google Scholar] [PubMed] [CrossRef]
28. Yan W, Liu H, Zhou X, Li Q, Zhang J, Lu L, et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013;23(7):969–71. doi:10.1038/cr.2013.43. [Google Scholar] [PubMed] [CrossRef]
29. Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant. 2011;4(2):319–30. doi:10.1093/mp/ssq070. [Google Scholar] [PubMed] [CrossRef]
30. Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Monna L, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12(12):2473–84. doi:10.1105/tpc.12.12.2473. [Google Scholar] [PubMed] [CrossRef]
31. Zhang Z, Hu W, Shen G, Liu H, Hu Y, Zhou X, et al. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep. 2017;7(1):5388. doi:10.1038/s41598-017-05873-1. [Google Scholar] [PubMed] [CrossRef]
32. Sun K, Zong W, Xiao D, Wu Z, Guo X, Li F, et al. Effects of the core heading date genes Hd1, Ghd7, DTH8, and PRR37 on yield-related traits in rice. Theor Appl Genet. 2023;136(11):227. doi:10.1007/s00122-023-04476-x. [Google Scholar] [PubMed] [CrossRef]
33. Yamamoto T, Yonemaru J, Yano M. Towards the understanding of complex traits in rice: substantially or superficially? DNA Res. 2009;16(3):141–54. doi:10.1093/dnares/dsp006. [Google Scholar] [PubMed] [CrossRef]
34. Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19(6):1068–76. doi:10.1101/gr.089516.108. [Google Scholar] [PubMed] [CrossRef]
35. Adriani DE, Dingkuhn M, Dardou A, Adam H, Luquet D, Lafarge T. Rice panicle plasticity in Near Isogenic Lines carrying a QTL for larger panicle is genotype and environment dependent. Rice. 2016;9(1):28. doi:10.1186/s12284-016-0101-x. [Google Scholar] [PubMed] [CrossRef]
36. Srikanth B, Subhakara Rao I, Surekha K, Subrahmanyam D, Voleti SR, Neeraja CN, et al. Enhanced expression of OsSPL14 gene and its association with yield components in rice (Oryza sativa) under low nitrogen conditions. Gene. 2016;576(1):441–50. [Google Scholar] [PubMed]
37. Luo L, Zhang Y, Xu G. How does nitrogen shape plant architecture? J Exp Bot. 2020;71(15):4415–27. [Google Scholar] [PubMed]
38. Yang X, Yang G, Wei X, Huang W, Fang Z. OsAAP15, an amino acid transporter in response to nitrogen concentration, mediates panicle branching and grain yield in rice. Plant Sci. 2023;330:111640. [Google Scholar] [PubMed]
39. Wei H, Zhang H, Blumwald E, Li H, Cheng J, Dai Q, et al. Different characteristics of high yield formation between inbred japonica super rice and inter-sub-specific hybrid super rice. Field Crops Research. 2016;198:179–87. [Google Scholar]
40. Fang J, Zhang F, Wang H, Wang W, Zhao F, Li Z, et al. Ef-cd locus shortens rice maturity duration without yield penalty. Proc Natl Acad Sci U S A. 2019;116(37):18717–22. [Google Scholar] [PubMed]
41. Kashiwagi T, Togawa E, Hirotsu N, Ishimaru K. Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.). Theor Appl Genet. 2008;117(5):749–57. [Google Scholar] [PubMed]
42. Tu B, Tao Z, Wang S, Zhou L, Zheng L, Zhang C, et al. Loss of Gn1a/OsCKX2 confers heavy-panicle rice with excellent lodging resistance. J Integr Plant Biol. 2022;64(1):23–38. [Google Scholar] [PubMed]
43. Peng Y, Gao Z, Zhang B, Liu C, Xu J, Ruan B, et al. Fine mapping and candidate gene analysis of a major QTL for panicle structure in rice. Plant Cell Rep. 2014;33(11):1843–50. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools