 Open Access
Open Access
MINI REVIEW
Interplay between Plants and Microbial Communities: Insights from Holobionts and Environmental Interactions
1 Department of Biology, College of Sciences, Kyung Hee University, Seoul, 02447, Republic of Korea
2 Center for Genome Engineering, Institute for Basic Science, Daejeon, 34126, Republic of Korea
* Corresponding Author: Ho-Seok Lee. Email:
Phyton-International Journal of Experimental Botany 2024, 93(10), 2519-2534. https://doi.org/10.32604/phyton.2024.058012
Received 02 September 2024; Accepted 14 October 2024; Issue published 30 October 2024
Abstract
Plants interact with a complex network of microorganisms, forming a dynamic holobiont that is crucial for their health, growth, and adaptation. This interconnected system is deeply influenced by environmental factors, which modulate the relationships within the plant microbiome. Key environmental drivers such as light, temperature, and moisture can alter the balance of these interactions, impacting plant immunity, resilience, and overall fitness. The traditional disease triangle model, which emphasizes plant-pathogen-environment interactions, is enhanced by incorporating the role of the microbiome, revealing how microbial communities contribute to disease outcomes. This review highlights the importance of shifting focus from studying plants in isolation to embracing an integrated approach that accounts for the intricate interactions between plants, microbes, and their surrounding environments. Comprehending these interactions is pivotal as we explore new approaches, including advanced sequencing technologies and microbiome engineering, to optimize plant-microbe relationships for improved crop resilience. These insights are vital for developing sustainable agricultural practices to address the impacts of climate change and other environmental challenges.Keywords
The concept of the holobiont has profoundly reshaped our understanding of plant biology, emphasizing the intricate and interdependent relationships between plants and their associated microorganisms [1]. A holobiont is defined as a host organism and its microbiota—comprising bacteria, fungi, viruses, and other microorganisms—functioning together as a cohesive ecological unit [2]. This perspective challenges the traditional view of plants as isolated entities, highlighting instead the interconnectedness of life forms within an integrated system. The dynamic nature of the holobiont is shaped by a complex network of interactions driven by both living organisms and environmental conditions. These interactions, which include mutualistic, commensal, and parasitic relationships, play a critical role in the plant’s health, development, and adaptation to various environmental conditions [3]. For instance, under stress conditions like drought, microorganisms that are typically pathogenic may shift to a mutualistic role, helping the plant cope with adverse conditions. This adaptability underscores the resilience of the holobiont and its ability to adjust to changing environments.
A holistic view of the holobiont not only deepens our comprehension of plant biology but also provides a framework for exploring how plants leverage their microbial partners to cope with abiotic stresses and pathogens. Building upon the holobiont concept is the disease triangle model, which has long been fundamental in understanding plant health and disease [4]. Traditionally, this model posits that the occurrence and severity of plant diseases are determined by the interplay between three key factors: the host plant’s vulnerability, the pathogen’s pathogenicity, and the environmental factors. However, recent advances have expanded this model to include the significant influence of the plant-associated microbiome. Pre-existing microbial communities, metabolite signaling, and the recruitment of beneficial microbes can greatly impact the outcomes of pathogen infections, either enhancing the plant’s resistance to disease or exacerbating its vulnerability [5]. This expanded view of the disease triangle provides deeper insights into the intricate relationships among plants, pathogens, and their environment, underscoring the significance of incorporating the microbiome into plant disease management approaches.
This diagram depicts the complex and dynamic interactions within the plant-associated systemic environment, highlighting the interplay between plants and their associated microbiomes. The figure illustrates how various environmental factors, including sunlight, moisture, temperature, and circadian rhythms, influence the relationships between plants and the microorganisms in their surrounding environments. These microorganisms include pathogenic, commensal, and beneficial microbes, which interact with different plant compartments such as roots, leaves, and stems. The figure also emphasizes the role of volatile organic compounds, root exudates, and plant growth-promoting rhizobacteria and fungi in modulating these interactions. Understanding these intricate networks is crucial for advancing our knowledge of plant resilience, stress adaptation, and overall fitness in diverse ecosystems.
These systemic concepts collectively underscore the inseparable relationship between plants and their microbiomes, which is essential for understanding plant responses to environmental stimuli (Fig. 1). Plants, though sessile, are continuously engaged in a dynamic exchange with their environment, mediated by a network of signals and responses. This plant-associated systemic environment is shaped by several biotic and abiotic elements that influence the composition and function of the microbiome, thereby affecting the plant’s overall health and growth. Plants have developed intricate mechanisms to detect and respond to these environmental fluctuations, often through complex signaling networks involving hormones, secondary metabolites, and other molecular signals. The microbiome is essential in regulating these responses, aiding the plant’s adaptation to its surroundings. Recognizing and understanding these interactions are fundamental to enhancing our knowledge of plant biology and ecology, and are vital for developing strategies to improve plant health and productivity, particularly in regard to worldwide challenges such as climate change and intensive agricultural practices [6].
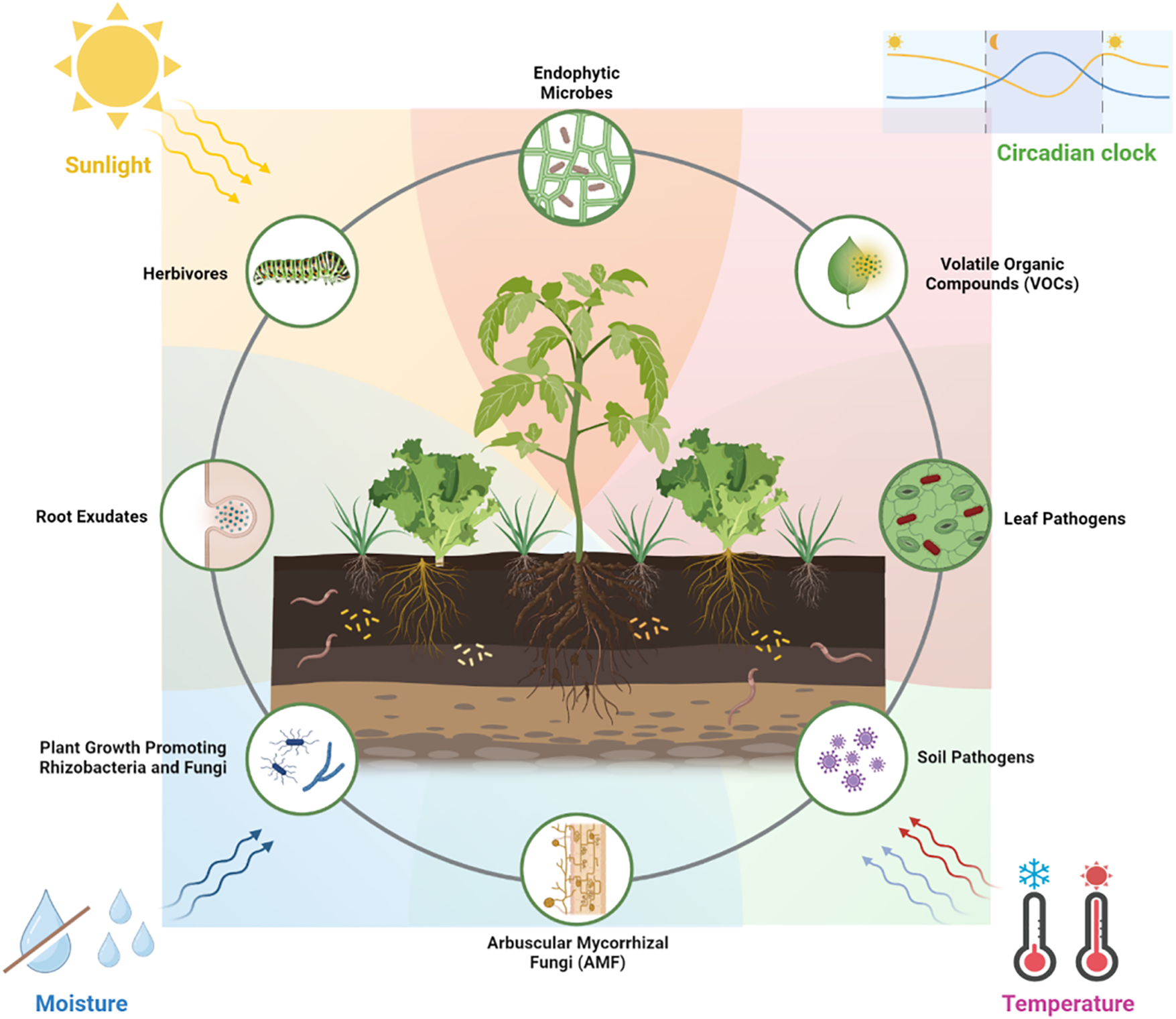
Figure 1: The plant-associated systemic environment
2 Plant Interactions with the Environment
Plants have evolved a range of strategies to survive and thrive in their habitats, including mechanisms for competing for vital resources such as water, nutrients, and light, as well as sophisticated forms of communication with neighboring plants and microorganisms. Such survival strategies are particularly critical in agricultural settings, where they can significantly influence crop productivity and resilience.
A key way in which plants interact with their environment is through the release of root exudates—chemical compounds secreted by roots that can affect the growth and behavior of nearby plants and microorganisms [7]. For instance, root exudates containing phytotoxins can inhibit the growth of competing plants, thereby conferring a competitive advantage to the secreting plant [8,9]. Additionally, root exudates that include growth regulators such as auxins and strigolactones contribute significantly to fostering beneficial microbial communities, which in turn enhance the plant’s capacity for nutrient absorption and cope with environmental stresses [10].
Volatile Organic Compounds (VOCs) are another critical class of signaling molecules that plants utilize to communicate with their environment [11,12]. These compounds play crucial roles in numerous processes, from defending against herbivores to attracting pollinators. Recent studies have shown that VOCs profoundly impact the plant-associated microbiome, affecting the composition and functionality of microbial communities both in the soil and on the plant’s surfaces [8,13]. Root volatiles, in particular, can serve as attractants for rhizosphere bacteria and fungi, guiding them toward nutrient-rich niches for colonization [14].
Plants develop distinct organs above and below the ground, each uniquely adapted to respond to specific environmental factors [15]. Aerial organs, such as leaves and stems, are subjected to variations in light, temperature, and humidity, and serve as the primary interface for defense against herbivores and pathogens. In contrast, subterranean organs, including roots, are primarily involved in the absorption of nutrients and water, and their functions are heavily influenced by soil conditions such as pH level, moisture, and the abundance of microbes [16].
The immune responses of plants to stress are complex and involve multiple, highly coordinated layers of defense. The primary defense mechanism is Pattern-Triggered Immunity (PTI), which is initiated upon the detection of conserved microbial components termed Pathogen-Associated Molecular Patterns (PAMPs) [17–19]. While PTI is often sufficient to thwart infections, some pathogens have evolved sophisticated mechanisms to suppress this response. In these cases, plants rely on a secondary defense mechanism known as Effector-Triggered Immunity (ETI), which is activated by the recognition of specific pathogen effectors. ETI typically induces a more robust response, often culminating in localized cell death—a phenomenon referred to as the Hypersensitive Response (HR)—to contain and limit the spread of the pathogen [20]. Understanding the spatial and temporal dynamics of these immune responses is essential for devising strategies to bolster plant resistance to diseases. For instance, research has shown that circadian rhythms can influence the timing of immune responses, with certain defense genes being more strongly expressed at specific times of the day [21]. Additionally, the effectiveness of these immune responses can vary depending on the developmental stage of the plant; younger plants often exhibit higher susceptibility to infections compared to mature plants [22]. Recognizing these nuances is crucial for optimizing plant defenses against pathogens and improving overall plant health.
3 Dynamics of the Plant-Associated Microbiome
The plant-associated microbiome, comprising the collective genomes and functional repertoires of various microorganisms, serves a fundamental role in determining plant vitality, productivity, and resilience across ecological and agricultural settings. This microbiome comprises diverse microbial communities, including bacteria, fungi, viruses, and archaea, that colonize different plant compartments such as the rhizosphere, root endosphere, phyllosphere, and leaf endosphere. These communities are not merely passive residents; they actively engage in complex interactions with the plant host, influencing and being influenced by the plant’s physiology, immune responses, and environmental conditions [23]. The variability in disease outbreaks, even under seemingly uniform agricultural conditions, can often be traced back to differences in these microbial communities, which are shaped by factors such as pathogen load, plant physiology, and the physicochemical properties of the soil [5].
Historically, plant pathogens have been managed primarily through the application of chemical controls, such as fungicides. However, the rising concerns over environmental sustainability, public health, and the emergence of antibiotic-resistant pathogens have spurred interest in biological control strategies that leverage the plant-associated microbiome. Biological Control Agents (BCAs), for instance, use beneficial bacteria and fungi that can outcompete or inhibit pathogens without damaging the plant itself, offering a more environmentally friendly approach to disease management [24]. However, the efficacy of BCAs is highly dependent on the specific interactions within the plant microbiome and its overall composition, which underscores the need for a deeper understanding of these microbial communities.
A variety of ecological niches provided by plants enable the development and proliferation of microbial communities, enhancing the general fitness and resilience of the plant [23]. Over the past two decades, progress in genomics and bioinformatics has allowed researchers to pinpoint genetic loci associated with crop growth and yield. However, the plant genotype alone does not fully account for trait variations observed in the field. To address this, integrated approaches such as genome-wide association studies, metagenome-wide association studies, and microbiome genome-wide association studies have been developed. These approaches allow for the exploration of complex interactions between plant genotype, phenotype, and associated microbiota, revealing how microbial communities can significantly influence plant traits [25]. Next-generation sequencing technologies, coupled with advanced informatics tools, have further revolutionized our ability to characterize microbiome profiles by analyzing the total gene content involved in various functions crucial for plant health [26].
Research has shown that different plant-associated niches harbor distinct bacterial and fungal communities. For example, the rhizosphere, the confined soil region directly affected by root exudates, is particularly rich in microbial diversity. This diversity is shaped by both abiotic factors, such as soil pH, moisture, and temperature, and biotic factors, encompassing plant root exudates [27] and the plant’s immune responses [28]. For instance, benzoxazinoids released as root exudate alter the rhizosphere microbiota, demonstrating that plants can determine the composition of their associated microbiota [27]. A recent study has revealed that community-level selection of functionally diverse species assemblages during microbiome maturation is linked to the acquisition of resistance traits against pathogen invasion [29]. Re-assembled bacterial consortia, consisting of fungal antagonists and plant growth promoters, exhibited the higher resistance to pathogen invasion and soil-borne diseases [29,30]. The rhizosphere microbiome is vital for nutrient cycling, heavy metal detoxification, plant growth promotion, and disease suppression [31–33]. In contrast, the phyllosphere, which includes the aerial parts of the plant such as leaves and stems, is primarily shaped by environmental factors like sunlight, humidity, and temperature. These compartments also have distinct microbial communities that are specialized to the specific conditions of each niche [28].
The interactions between plants and their microbiomes are highly dynamic, adapting in response to varying environmental conditions [34,35] and the developmental stage of the plant. Microbial relationships within the plant microbiome are not fixed; they can shift between mutualism, commensalism, and parasitism in reaction to shifts in environmental conditions or plant health. For instance, under drought conditions, a pathogen might shift to a mutualistic relationship with its host by aiding the plant in stress adaptation through mechanisms such as hormone-induced signaling pathways and gene expression changes [36]. Furthermore, experimental evolution studies have demonstrated that rhizobia, nitrogen-fixing bacteria that establish symbiotic relationships with legumes, can evolve to become more cooperative with their host plants, particularly when they share an evolutionary history [37]. These findings suggest that plant-microbe interactions are significantly context-dependent, and that the microbiome’s influence on plant fitness and illness is complex and multifaceted.
Despite these advancements, there remain significant gaps in our understanding of the consistency of microbiome functions across different plant species and environments. The majority of existing research has concentrated on the structure of microbial communities—i.e., the diversity and composition of species present—rather than on their functional roles. Understanding how genetic variants in both plants and microbes influence these functions is crucial for developing more effective strategies for manipulating plant-microbe interactions to improve the productivity and health of plants.
4 Environmental Factors Influencing Plant-Microbiome Interactions
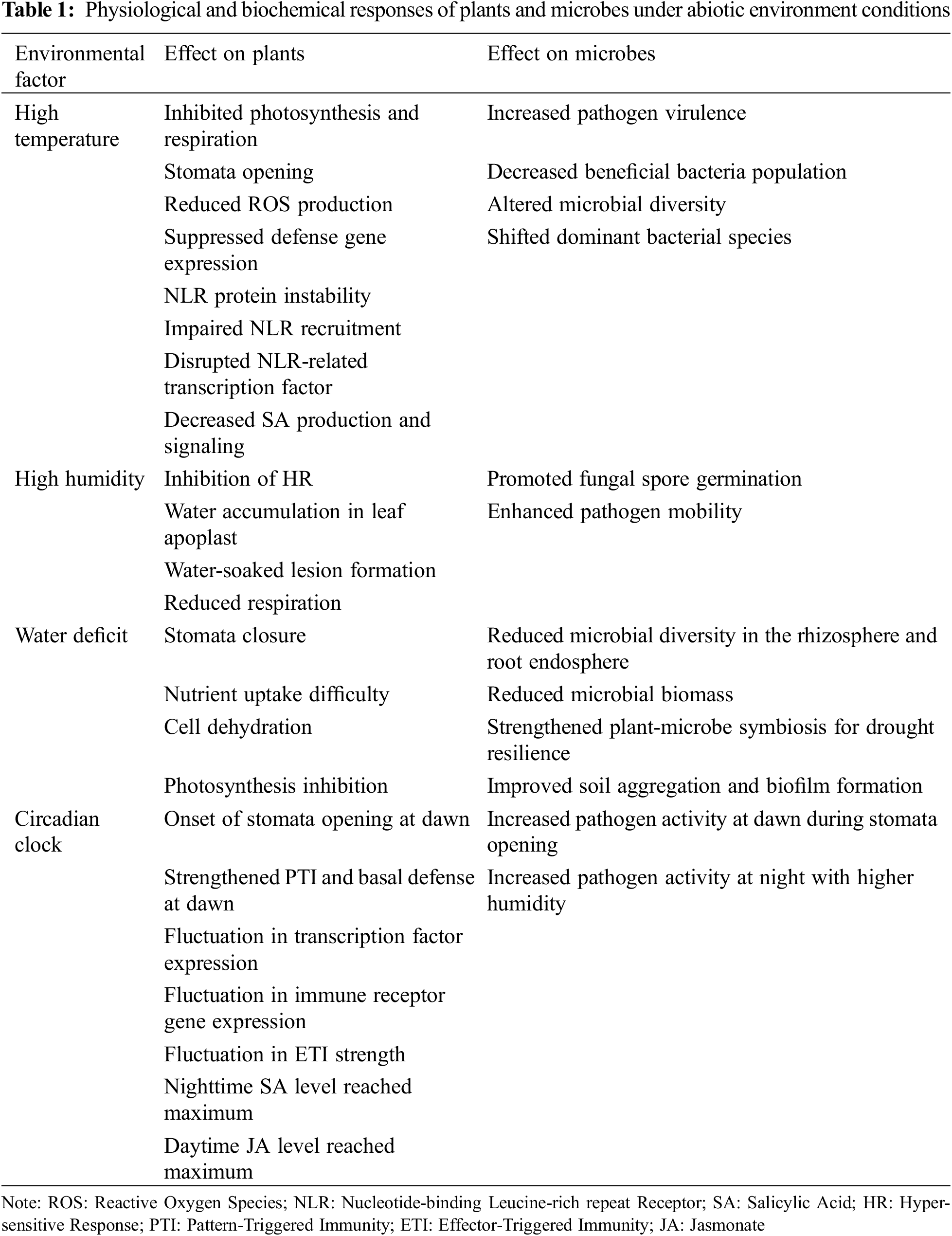
Temperature is among the most critical environmental factors affecting both plant physiology and the dynamics of the plant-associated microbiome. The plant immune system, which is composed of complex signaling networks, transcriptional regulators, and hormonal crosstalk, relies on two primary defense mechanisms: PTI and ETI. PTI is triggered by the identification of PAMPs by pattern-recognition receptors, whereas ETI commences upon the detection of specific pathogen effectors by Nucleotide-binding Leucine-rich repeat Receptors (NLRs) [38]. Both PTI and ETI are essential for the plant’s defense against microbial pathogens, yet they are highly sensitive to temperature fluctuations.
Heat stress, in particular, can severely disrupt plant immune responses, leading to increased susceptibility to pathogens. For instance, high temperatures have been shown to inhibit key PTI responses, such as the production of reactive oxygen species and the transcription of related defense genes [39]. Moreover, heat stress can impair the function of NLR proteins, which are crucial for ETI. Research has demonstrated that the expression and nuclear accumulation of NLR proteins, such as SNC1 and the tobacco N protein, are temperature-sensitive, with elevated temperatures leading to their destabilization and reduced effectiveness in mounting immune responses [40–42]. The impaired recruitment of NLR proteins to their sites of action at elevated temperatures suggests that plants have key components in their immune systems that are inherently temperature-responsive. Additionally, the transcriptional activity of NLR-related transcription factors, such as members of the TEOSINTE BRANCHED1, CYCLOIDEA, and PROLIFERATING CELL FACTOR (TCP) family, can be influenced by temperature, further modulating the plant’s immune responses [43].
The relationship between temperature and plant hormone signaling is also complex. Salicylic Acid (SA) is a key hormone that facilitates the activation of plant defenses, particularly in response to biotrophic pathogens [44]. However, heat stress has been shown to suppress SA biosynthesis and signaling, leading to increased vulnerability to diseases [45,46]. Despite this, some ETI pathways can become temporarily more robust under heat stress, albeit at a potential cost to plant growth and development [47]. Understanding the interplay between temperature, hormone signaling, and plant immunity is crucial for developing strategies to improve plant resilience in the face of global warming and climate change.
Temperature also directly impacts the plant-associated microbiome by influencing microbial survival, activity, and community composition. Pathogens, in particular, may exhibit increased virulence at elevated temperatures, evidenced by the increased susceptibility of barley to powdery mildew and rice to the fungal pathogen Magnaporthe oryzae under heat stress [48,49]. Furthermore, temperature changes can alter the niches available for microbial colonization, affecting the production of molecules related to microbial traits such as virulence and plant growth promotion [46,50]. For example, elevated temperatures have been shown to enhance the expansion and settlement of arbuscular mycorrhizal fungi, possibly attributed to increased carbon allocation to the rhizosphere by the host plant [51]. Conversely, for the phyllosphere microbiome, long-term warming experiments showed a considerable impact on the diversity and phylogenic composition of most abundant species-specific bacteria, although the overall colonization and concentration of leaf-associated bacterial cells remained unchanged [52]. More significantly, warming may increase the possibility of pathogen transmission in grassland ecosystems, as evidenced by the enhancement of potentially pathogenic bacteria (e.g., Enterobacteriaceae, Pseudomonas, and Acinetobacter) and the decrease of beneficial bacteria (e.g., Sphingomonas spp. and Rhizobium spp.) in the phyllosphere [53]. Such shifts in microbiome composition and diversity suggest that plant species may respond differently to temperature changes, ultimately adjusting their fitness. Given that each species maintains unique interactions with its associated microbes, it is likely that even under the same temperature disturbances, plants will adapt in distinct ways that best suit their individual requirements. As part of developing climate-resilient crops, some studies have suggested that inoculation with endophytes can enhance plant thermotolerance, although most of this research has been conducted in controlled laboratory settings, and further field studies are needed to confirm these findings [54].
While much of the present research has focused on the effects of increased temperatures, other temperature-related phenomena, such as chilling-induced immune disruption, remain underexplored. Additionally, the ability of plants to adapt to temperature fluctuations over time suggests that the outcomes of temperature-related studies may vary depending on the specific conditions and temporal scales considered. Therefore, A more thorough comprehension of the effects of temperature on plant-microbiome interactions is crucial for the development of sustainable agriculture in response to climate change.
Water availability is another critical environmental factor that profoundly influences plant physiology and the composition and function of the plant-associated microbiome. The soil-plant-atmosphere continuum regulates water transport from the soil through the plant and into the atmosphere, with key regulatory mechanisms located at the root-soil interface, xylem, and stomata. These mechanisms are crucial for maintaining plant water balance, but they also interact with the plant’s microbiome, which is highly sensitive to changes in soil moisture levels.
Excessive soil moisture can weaken plant immune responses and simultaneously enhance the pathogenicity of microbes, leading to an increased risk of disease. High humidity levels, for example, promote the germination of fungal spores and the motility of bacterial pathogens, facilitating their entry into the plant [55]. These conditions can suppress the HR [56], a key plant defense mechanism against biotrophic pathogens, resulting in the accumulation of water in the leaf apoplast and the formation of water-soaked lesions that create a microenvironment conducive to pathogen proliferation [57]. Such conditions have been shown to favor the success of pathogens like Pseudomonas syringae and Cladosporium fulvum, which thrive under high humidity and weakened plant defenses.
Conversely, drought stress, characterized by low soil moisture levels, triggers a different set of responses in plants and their associated microbiomes. Drought stress reduces microbial diversity in the rhizosphere and root endosphere, affecting the microbial community composition and the competitive interactions among microbes [58,59]. The reduction in microbial biomass and nutrient concentrations in plant tissues further disrupts beneficial plant-microbiome interactions, which are essential for traits such as drought tolerance and disease resistance [60]. However, some plant-microbe relationships may be strengthened under drought conditions, as certain microbial taxa, such as Actinobacteria and Firmicutes, become more abundant and produce metabolites that enhance the host plant’s stress resistance [61].
Moreover, the acclimation of soil microbial communities to dry circumstances can enhance plant fitness by promoting beneficial symbiotic relationships, such as those with mycorrhizal fungi and symbiotic soil bacteria [62]. Various bacterial species, including Bacillus sp., Pseudomonas sp., and Rhizobium sp., aid soil aggregation or biofilm formation around roots to improve plant drought resilience [63–66]. These relationships not only improve plant drought tolerance but also influence plant-soil interactions in ways that may help plants adapt to future climate variability. For example, soils that have experienced long-term drought and high precipitation variability tend to be more resilient to contemporary drought perturbations, with microbial communities quickly regaining activity levels after rewetting [62]. This resilience is likely due to the fast turnover rates of microorganisms, which allow them to respond more quickly to environmental changes than the plants themselves.
Interestingly, drought tolerance traits can be transmitted from maternal plants to their progeny, indicating that both host genetics and the host-associated microbiome play critical roles in determining plant responses to drought. The plant hormone Abscisic Acid (ABA) is central to the plant’s drought response, mediating stomatal closure to decrease transpiration and water loss [67]. However, the interplay between ABA and other hormones, such as SA, which is involved in immune responses, can be antagonistic, complicating the plant’s ability to balance water conservation with pathogen defense. In addition to moisture, other environmental factors such as salinity are connected to these complicated hormonal alterations [68] and each hormone associated with one element can influence responses to other stressors, as they often share common pathways that regulate the plant’s fitness. This complexity underscores the need for further research into how plants integrate moisture-related signals with other environmental cues to optimize their growth and survival.
Finally, the capacity of plant roots to sense and react to water availability, as well as other factors such as nutrient levels and salinity, is essential in shaping plant-microbe interactions. Under drought stress, reduced soil moisture limits nutrient uptake by roots, which in turn affects microbial activity and plant-microbe interactions. In some cases, plants may modify immune reactions to promote the establishment of beneficial microbes that can aid in nutrient acquisition, particularly under conditions of nutrient deficiency [61]. However, the mechanisms underlying these interactions remain poorly explored, and further research is required to elucidate the complex molecular dialogues between plants and their microbiomes in response to moisture stress.
The plant circadian clock, which governs daily physiological processes in response to predictable environmental changes, is a crucial factor influencing plant-microbe interactions. While the circadian clock is primarily regulated by light, it is also responsive to other environmental factors such as temperature and humidity, which can modulate its function [61]. Plants utilize their circadian rhythms to anticipate daily alterations in their environment and adjust their physiological processes accordingly, optimizing their growth, metabolism, and defense mechanisms.
Notably, the circadian clock regulates key aspects of the plant immune system, including PTI and ETI. Research has shown that genes associated with PTI and basal defense are showing increased expression at dawn, a time when many pathogens are most active [69]. This timing aligns with the opening of stomata, which are regulated by circadian rhythms and serve as entry points for pathogens. The transcription factors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) play critical roles in this process by regulating the expression of genes involved in stomatal immunity and enhancing the plant’s ability to defend against pathogens during periods of heightened risk [70].
The circadian system also influences the expression of immune receptor genes, including RECOGNITION OF PERONOSPORA PARASITICA 4 (RPP4), which targets the pathogen Hyaloperonospora arabidopsidis (Hpa). The expression of RPP4 peaks in the morning, coinciding with the peak risk of Hpa infection, demonstrating how the circadian clock optimizes the timing of immune responses to coincide with environmental conditions that favor pathogen activity [71]. Furthermore, the strength of ETI can vary with the circadian cycle, with humidity oscillations at night potentially amplifying immune responses to anticipate increased pathogen pressure during these conditions [72].
Besides its function in regulating immunological responses, the circadian clock also influences the production and signaling of defensive hormones such as SA and Jasmonate (JA). SA levels typically peak at night, while JA levels peak during the day, allowing plants to coordinate their defenses with the daily rhythms of pathogen and herbivore activity [73]. This rhythmic hormone production is crucial for enabling plants to respond effectively to biotic stressors, aligning their defenses with environmental cycles.
The integration of circadian rhythms with environmental factors underscores the complexity of plant-microbe interactions. Although substantial advancements have been achieved in elucidating the role of the circadian clock in regulating plant immunity and microbiome interactions, numerous questions persist. For instance, how do circadian rhythms interact with other environmental stressors, such as drought or temperature extremes, to influence plant-microbe interactions? Addressing these questions will require a multidisciplinary approach that combines molecular biology, ecology, and systems biology to unravel the intricate networks that govern plant health and resilience.
Understanding the connections between plants and their associated microbiomes in relation to environmental factors is necessary for formulating strategies to improve plant health and resilience (Fig. 1 and Table 1) [74–78]. Future research should focus on several key areas.
Most current studies focus on the effects of individual environmental factors. However, in natural environments, plants are often exposed to multiple stressors simultaneously, such as drought combined with high temperatures or nutrient deficiencies. The multifactorial stress combination approach reveals that plants face a more severe immune response and negative microbiome effects under combined stresses. For instance, the interaction of drought and heat has been shown to cause greater reductions in photosynthesis in many experimental crops than either stressor alone [79]. These results are significant in that when two different stresses that induce opposing stomatal responses act together, they reveal new profiles not found with a single stress. Moreover, the case of heat-induced stomatal opening inhibition by viruses [80] suggests to us the need to study the intricate interplay between biotic and abiotic stress responses. Since the combined stress responses remain largely unexplored, future research should explore the combined effects of multiple stressors on plant-microbiome interactions to acquire a more thorough comprehension of how these interactions are modulated in complex environments.
5.2 Advanced Sequencing Technologies
The emergence of high-throughput sequencing technologies has revolutionized our knowledge of the plant-associated microbiome. However, there is still much to learn about the functional roles of different microorganisms and how they interact with each other and with the plant. Future research should leverage advanced sequencing techniques, such as metatranscriptomics and metaproteomics, to study the functional dynamics of the microbiome and how these functions adapt to environmental stressors.
Considering the vital role of the microbiome in plant health, there is increasing interest in the possibility of engineering microbiomes to bolster plant resilience against various stressors. This could involve the manipulation of microbial communities to promote desirable traits, such as increased drought tolerance or disease resistance. Furthermore, genetically engineered bacteria hold significant potential for enhancing crop productivity and resilience. Several examples of genetic improvements include the introduction of antimicrobial enzymes, including chitinase and glucanase, which confer increased resistance to pathogens [81,82]. Singh et al. summarized reports on the use of genetically engineered bacteria for the bioremediation of heavy metals [83]. These approaches provide valuable precedents, allowing for improved design strategies for field application of microbiome engineering. Future research should investigate the practicality and effectiveness of microbiome engineering in different environmental contexts. However, introducing artificial microbiomes risks disrupting existing microbial communities, which can lead to unexpected ecological consequences; hence, caution is necessary when implementing them in actual agricultural settings.
5.4 Natural vs. Controlled Environments
Much of the research on plant-microbiome interactions has been conducted in controlled laboratory settings, where environmental conditions can be tightly regulated. Although these studies offer significant insights, they may not entirely reflect the complex structure of natural ecosystems, where multiple factors interact in unpredictable ways. Future research should prioritize studies conducted in natural settings to better understand how plant-microbiome interactions play out in the real world.
5.5 Integration of Omics Approaches
To gain a deeper understanding of the multifaceted nature of plant-microbiome relationships, future research should integrate multiple omics approaches, including genomics, transcriptomics, proteomics, and metabolomics. By combining these approaches, researchers can gain a more holistic view of the plant-microbiome interactions, from the genetic and molecular levels to the phenotypic and ecological levels.
Many studies on plant-microbiome interactions are short-term, focusing on immediate responses to environmental stressors. However, long-term studies are necessary to comprehend the evolutionary dynamics of these interactions and how they contribute to plant adaptation over time. These investigations could offer critical understanding of the mechanisms that support the resilience of plant-microbiome systems, guiding the development of strategies to boost crop productivity and sustainability amid the challenges posed by climate change.
5.7 Role of Microbial Interactions
While much attention has been given to plant-microbe interactions, the interactions between different microorganisms within the microbiome are also crucial for plant health. Future research should explore the networks of interactions between microorganisms and how these networks influence the overall function of the microbiome and its impact on the plant.
By addressing these areas, future research can deepen our understanding of the complex interplay between plants and their associated microbial communities and how this interplay is shaped by environmental factors. Such understanding is crucial for formulating strategies to strengthen plant health and resilience in a rapidly changing world.
Acknowledgement: The authors would like to express their gratitude to the members of the IMGN lab.
Funding Statement: This research was supported by a grant from the Institute for Basic Science (IBS-R021-D1-2024-a00), the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ho-Seok Lee).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Sejin Choi, Ho-Seok Lee; draft manuscript preparation: Sejin Choi; manuscript review and final approval: Ho-Seok Lee. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Busato S, Gordon M, Chaudhari M, Jensen I, Akyol T, Andersen S, et al. Compositionality, sparsity, spurious heterogeneity, and other data-driven challenges for machine learning algorithms within plant microbiome studies. Curr Opin Plant Biol. 2023;71:102326. doi:10.1016/j.pbi.2022.102326. [Google Scholar] [PubMed] [CrossRef]
2. Hassani MA, Durán P, Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:1–17. doi:10.1186/s40168-018-0445-0. [Google Scholar] [PubMed] [CrossRef]
3. Kavamura VN, Esposito E. Biotechnological strategies applied to the decontamination of soils polluted with heavy metals. Biotechnol Adv. 2010;28(1):61–9. doi:10.1016/j.biotechadv.2009.09.002. [Google Scholar] [PubMed] [CrossRef]
4. Scholthof KB. The disease triangle: pathogens, the environment and society. Nat Rev Microbiol. 2007;5(2):152–6. doi:10.1038/nrmicro1596. [Google Scholar] [PubMed] [CrossRef]
5. Pereira LB, Thomazella DPT, Teixeira P. Plant-microbiome crosstalk and disease development. Curr Opin Plant Biol. 2023;72:102351. doi:10.1016/j.pbi.2023.102351. [Google Scholar] [PubMed] [CrossRef]
6. Rhaman MS, Kibria MG, Hoque A. Climate change and its adverse impacts on plant growth in South Asia: current status and upcoming challenges. Phyton-Int J Exp Bot. 2022;91(4):695–711. doi:10.32604/phyton.2022.018898. [Google Scholar] [CrossRef]
7. Eroglu CG, Bennett AA, Steininger-Mairinger T, Hann S, Puschenreiter M, Wirth J, et al. Neighbour-induced changes in root exudation patterns of buckwheat results in altered root architecture of redroot pigweed. Sci Rep. 2024;14(1):8679. doi:10.1038/s41598-024-58687-3. [Google Scholar] [PubMed] [CrossRef]
8. Brosset A, Blande JD. Volatile-mediated plant-plant interactions: volatile organic compounds as modulators of receiver plant defence, growth, and reproduction. J Exp Bot. 2022;73(2):511–28. doi:10.1093/jxb/erab487. [Google Scholar] [PubMed] [CrossRef]
9. Tang Z, Guo F, Cui L, Li Q, Zhang J, Wang J, et al. Effects of allelochemicals on root growth and pod yield in response to continuous cropping obstacle of peanut. Phyton-Int J Exp Bot. 2023;92(1):17–34. [Google Scholar]
10. Vives-Peris V, de Ollas C, Gomez-Cadenas A, Perez-Clemente RM. Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 2020;39(1):3–17. doi:10.1007/s00299-019-02447-5. [Google Scholar] [PubMed] [CrossRef]
11. Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57:233–66. doi:10.1146/annurev.arplant.57.032905.105159. [Google Scholar] [PubMed] [CrossRef]
12. Dudareva N, Klempien A, Muhlemann JK, Kaplan I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013;198(1):16–32. doi:10.1111/nph.12145. [Google Scholar] [PubMed] [CrossRef]
13. Chai YN, Schachtman DP. Root exudates impact plant performance under abiotic stress. Trends Plant Sci. 2022;27(1):80–91. doi:10.1016/j.tplants.2021.08.003. [Google Scholar] [PubMed] [CrossRef]
14. Schulz-Bohm K, Gerards S, Hundscheid M, Melenhorst J, de Boer W, Garbeva P. Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 2018;12(5):1252–62. doi:10.1038/s41396-017-0035-3. [Google Scholar] [PubMed] [CrossRef]
15. Lee H-S, Choi I, Jeon Y, Ahn H-K, Cho H, Kim J, et al. Chaperone-like protein DAY plays critical roles in photomorphogenesis. Nat Commun. 2021;12(1):4194. doi:10.1038/s41467-021-24446-5. [Google Scholar] [PubMed] [CrossRef]
16. Khalid M, Du B, Tan H, Liu X, Su L, Ali M, et al. Phosphorus elevation erodes ectomycorrhizal community diversity and induces divergence of saprophytic community composition between vegetation types. Sci Total Environ. 2021;793:148502. doi:10.1016/j.scitotenv.2021.148502. [Google Scholar] [PubMed] [CrossRef]
17. Lee DH, Lee HS, Belkhadir Y. Coding of plant immune signals by surface receptors. Curr Opin Plant Biol. 2021;62:102044. doi:10.1016/j.pbi.2021.102044. [Google Scholar] [PubMed] [CrossRef]
18. Colaianni NR, Parys K, Lee H-S, Conway JM, Kim NH, Edelbacher N, et al. A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe. 2021;29(4):635–49. doi:10.1016/j.chom.2021.02.006. [Google Scholar] [PubMed] [CrossRef]
19. Parys K, Colaianni NR, Lee H-S, Hohmann U, Edelbacher N, Trgovcevic A, et al. Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Cell Host Microbe. 2021;29(4):620–34. doi:10.1016/j.chom.2021.02.008. [Google Scholar] [PubMed] [CrossRef]
20. Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–9. doi:10.1038/nature05286. [Google Scholar] [PubMed] [CrossRef]
21. Karapetyan S, Dong X. Redox and the circadian clock in plant immunity: a balancing act. Free Radic Biol Med. 2018;119:56–61. doi:10.1016/j.freeradbiomed.2017.12.024. [Google Scholar] [PubMed] [CrossRef]
22. Gray SB, Brady SM. Plant developmental responses to climate change. Dev Biol. 2016;419(1):64–77. doi:10.1016/j.ydbio.2016.07.023. [Google Scholar] [PubMed] [CrossRef]
23. Trivedi P, Leach JE, Tringe SG, Sa TM, Singh BK. Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18(11):607–21. doi:10.1038/s41579-020-0412-1. [Google Scholar] [PubMed] [CrossRef]
24. Lee J, Kim S, Jung H, Koo B-K, Han JA, Lee H-S. Exploiting bacterial genera as biocontrol agents: mechanisms, interactions and applications in sustainable agriculture. J Plant Biol. 2023;66(6):485–98. doi:10.1007/s12374-023-09404-6. [Google Scholar] [CrossRef]
25. Wang Y, Wang X, Sun S, Jin C, Su J, Wei J, et al. GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype-dependent microbial effects in foxtail millet. Nat Commun. 2022;13(1):5913. doi:10.1038/s41467-022-33238-4. [Google Scholar] [PubMed] [CrossRef]
26. Kang S, Kim KT, Choi J, Kim H, Cheong K, Bandara A, et al. Genomics and informatics, conjoined tools vital for understanding and protecting plant health. Phytopathology. 2022;112(5):981–95. doi:10.1094/PHYTO-10-21-0418-RVW. [Google Scholar] [PubMed] [CrossRef]
27. Hu L, Robert CAM, Cadot S, Zhang X, Ye M, Li B, et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun. 2018;9(1):2738. doi:10.1038/s41467-018-05122-7. [Google Scholar] [PubMed] [CrossRef]
28. Trivedi P, Batista BD, Bazany KE, Singh BK. Plant-microbiome interactions under a changing world: responses, consequences and perspectives. New Phytol. 2022;234(6):1951–9. doi:10.1111/nph.18016. [Google Scholar] [PubMed] [CrossRef]
29. Hu J, Wei Z, Kowalchuk GA, Xu Y, Shen Q, Jousset A. Rhizosphere microbiome functional diversity and pathogen invasion resistance build up during plant development. Environ Microbiol. 2020;22(12):5005–18. doi:10.1111/1462-2920.15097. [Google Scholar] [PubMed] [CrossRef]
30. Lazcano C, Boyd E, Holmes G, Hewavitharana S, Pasulka A, Ivors K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci Rep. 2021;11(1):3188. doi:10.1038/s41598-021-82768-2. [Google Scholar] [PubMed] [CrossRef]
31. Choi S, Jung H, Kim Y, Han JA, Kim EY, Lee H-S. Draft genome sequence of Priestia megaterium strain IMGN3 derived from soil. Microbiol Resour Announc. 2024;13(9):e00458–24. doi:10.1128/mra.00458-24. [Google Scholar] [PubMed] [CrossRef]
32. ur Rahman S, Khalid M, Hui N, Rehman A, Kayani S-I, Fu X, et al. Piriformospora indica alter root-associated microbiome structure to enhance Artemisia annua L. tolerance to arsenic. J Hazard Mater. 2023;457:131752. doi:10.1016/j.jhazmat.2023.131752. [Google Scholar] [PubMed] [CrossRef]
33. Khalid M, Saeed U-R, Hassani D, Hayat K, Pei Z, Nan H. Advances in fungal-assisted phytoremediation of heavy metals: a review. Pedosphere. 2021;31(3):475–95. doi:10.1016/S1002-0160(20)60091-1. [Google Scholar] [CrossRef]
34. Khalid M, Tan H, Ali M, Rehman A, Liu X, Su L, et al. Karst rocky desertification diverged the soil residing and the active ectomycorrhizal fungal communities thereby fostering distinctive extramatrical mycelia. Sci Total Environ. 2022;807:151016. doi:10.1016/j.scitotenv.2021.151016. [Google Scholar] [PubMed] [CrossRef]
35. Khalid M, Liu X, Zheng B, Su L, Kotze DJ, Setälä H, et al. Distinct climatic regions drive antibiotic resistance genes dynamics across public parks and pristine soil ecosystems. J Clean Prod. 2023;409:137275. doi:10.1016/j.jclepro.2023.137275. [Google Scholar] [CrossRef]
36. González R, Butković A, Escaray FJ, Martínez-Latorre J, Melero Í., Pérez-Parets E, et al. Plant virus evolution under strong drought conditions results in a transition from parasitism to mutualism. Proc Nat Acad Sci. 2021;118(6):e2020990118. doi:10.1073/pnas.2020990118. [Google Scholar] [PubMed] [CrossRef]
37. Batstone RT, O’Brien AM, Harrison TL, Frederickson ME. Experimental evolution makes microbes more cooperative with their local host genotype. Science. 2020;370(6515):476–8. doi:10.1126/science.abb7222. [Google Scholar] [PubMed] [CrossRef]
38. Lee HA, Yeom SI. Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief Funct Genom. 2015;14(4):233–42. doi:10.1093/bfgp/elv012. [Google Scholar] [PubMed] [CrossRef]
39. Janda M, Lamparova L, Zubikova A, Burketova L, Martinec J, Krckova Z. Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol Plant Pathol. 2019;20(7):1005–12. doi:10.1111/mpp.2019.20.issue-7. [Google Scholar] [CrossRef]
40. Wang Z, Cui D, Liu J, Zhao J, Liu C, Xin W, et al. Arabidopsis ZED1-related kinases mediate the temperature-sensitive intersection of immune response and growth homeostasis. New Phytol. 2017;215(2):711–24. doi:10.1111/nph.2017.215.issue-2. [Google Scholar] [CrossRef]
41. Mang H-G, Qian W, Zhu Y, Qian J, Kang H-G, Klessig DF, et al. Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell. 2012;24(3):1271–84. doi:10.1105/tpc.112.096198. [Google Scholar] [PubMed] [CrossRef]
42. Zhu Y, Qian W, Hua J. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 2010;6(4):e1000844. doi:10.1371/journal.ppat.1000844. [Google Scholar] [PubMed] [CrossRef]
43. Wang Z, Cui D, Liu C, Zhao J, Liu J, Liu N, et al. TCP transcription factors interact with ZED1-related kinases as components of the temperature-regulated immunity. Plant, Cell Environ. 2019;42(6):2045–56. doi:10.1111/pce.v42.6. [Google Scholar] [CrossRef]
44. Roussin-Leveillee C, Rossi CAM, Castroverde CDM, Moffett P. The plant disease triangle facing climate change: a molecular perspective. Trends Plant Sci. 2024;29(8):895–914. doi:10.1016/j.tplants.2024.03.004. [Google Scholar] [PubMed] [CrossRef]
45. Rossi CAM, Marchetta EJR, Kim JH, Castroverde CDM. Molecular regulation of the salicylic acid hormone pathway in plants under changing environmental conditions. Trends Biochem Sci. 2023;48(8):699–712. doi:10.1016/j.tibs.2023.05.004. [Google Scholar] [PubMed] [CrossRef]
46. Huot B, Castroverde CDM, Velasquez AC, Hubbard E, Pulman JA, Yao J, et al. Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat Commun. 2017;8(1):1808. doi:10.1038/s41467-017-01674-2. [Google Scholar] [PubMed] [CrossRef]
47. Webb KM, Ona I, Bai J, Garrett KA, Mew T, Vera Cruz CM, et al. A benefit of high temperature: increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 2010;185(2):568–76. doi:10.1111/nph.2009.185.issue-2. [Google Scholar] [CrossRef]
48. Mikkelsen BL, Jørgensen RB, Lyngkjær MF. Complex interplay of future climate levels of CO2, ozone and temperature on susceptibility to fungal diseases in barley. Plant Pathol. 2015;64(2):319–27. doi:10.1111/ppa.2015.64.issue-2. [Google Scholar] [CrossRef]
49. Onaga G, Wydra KD, Koopmann B, Séré Y, von Tiedemann A. Elevated temperature increases in planta expression levels of virulence related genes in Magnaporthe oryzae and compromises resistance in Oryza sativa cv. Nipponbare. Funct Plant Biol. 2016;44(3):358–71. [Google Scholar]
50. Hasegawa H, Chatterjee A, Cui Y, Chatterjee A. Elevated temperature enhances virulence of Erwinia carotovora subsp. carotovora strain EC153 to plants and stimulates production of the quorum sensing signal, N-acyl homoserine lactone, and extracellular proteins. Appl Environ Microbiol. 2005;71(8):4655–63. doi:10.1128/AEM.71.8.4655-4663.2005. [Google Scholar] [PubMed] [CrossRef]
51. Compant S, van der Heijden MG, Sessitsch A. Climate change effects on beneficial plant-microorganism interactions. FEMS Microbiol Ecol. 2010;73(2):197–214. doi:10.1111/j.1574-6941.2010.00900.x. [Google Scholar] [PubMed] [CrossRef]
52. Aydogan EL, Budich O, Hardt M, Choi YH, Jansen-Willems AB, Moser G, et al. Global warming shifts the composition of the abundant bacterial phyllosphere microbiota as indicated by a cultivation-dependent and -independent study of the grassland phyllosphere of a long-term warming field experiment. FEMS Microbiol Ecol. 2020;96(8):fiaa087. doi:10.1093/femsec/fiaa087. [Google Scholar] [PubMed] [CrossRef]
53. Zhu YG, Xiong C, Wei Z, Chen QL, Ma B, Zhou SY, et al. Impacts of global change on the phyllosphere microbiome. New Phytol. 2022;234(6):1977–86. doi:10.1111/nph.17928. [Google Scholar] [PubMed] [CrossRef]
54. Shekhawat K, Saad MM, Sheikh A, Mariappan K, Al-Mahmoudi H, Abdulhakim F, et al. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021;22(3):e51049. doi:10.15252/embr.202051049. [Google Scholar] [PubMed] [CrossRef]
55. Dechesne A, Wang G, Gulez G, Or D, Smets BF. Hydration-controlled bacterial motility and dispersal on surfaces. Proc Natl Acad Sci U S A. 2010;107(32):14369–72. doi:10.1073/pnas.1008392107. [Google Scholar] [PubMed] [CrossRef]
56. Wang C, Cai X, Zheng Z. High humidity represses Cf-4/Avr4- and Cf-9/Avr9-dependent hypersensitive cell death and defense gene expression. Planta. 2005;222(6):947–56. doi:10.1007/s00425-005-0036-8. [Google Scholar] [PubMed] [CrossRef]
57. Wright CA, Beattie GA. Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101(9):3269–74. doi:10.1073/pnas.0400461101. [Google Scholar] [PubMed] [CrossRef]
58. Xu L, Naylor D, Dong Z, Simmons T, Pierroz G, Hixson KK, et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc Natl Acad Sci U S A. 2018;115(18):E4284−E93. doi:10.1073/pnas.1717308115. [Google Scholar] [PubMed] [CrossRef]
59. Kaisermann A, de Vries FT, Griffiths RI, Bardgett RD. Legacy effects of drought on plant-soil feedbacks and plant-plant interactions. New Phytol. 2017;215(4):1413–24. doi:10.1111/nph.14661. [Google Scholar] [PubMed] [CrossRef]
60. Pugnaire FI, Morillo JA, Penuelas J, Reich PB, Bardgett RD, Gaxiola A, et al. Climate change effects on plant-soil feedbacks and consequences for biodiversity and functioning of terrestrial ecosystems. Sci Adv. 2019;5(11):eaaz1834. doi:10.1126/sciadv.aaz1834. [Google Scholar] [PubMed] [CrossRef]
61. Cheng YT, Zhang L, He SY. Plant-microbe interactions facing environmental challenge. Cell Host Microbe. 2019;26(2):183–92. doi:10.1016/j.chom.2019.07.009. [Google Scholar] [PubMed] [CrossRef]
62. Rudgers JA, Afkhami ME, Bell-Dereske L, Chung YA, Crawford KM, Kivlin SN, et al. Climate disruption of plant-microbe interactions. Annu Rev of Ecol, Evol, Syst. 2020;51(1):561–86. doi:10.1146/ecolsys.2020.51.issue-1. [Google Scholar] [CrossRef]
63. Sandhya V, Shrivastava M, Ali SZ, Sai Shiva Krishna Prasad V. Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russ Agric Sci. 2017;43(1):22–34. doi:10.3103/S1068367417010165. [Google Scholar] [CrossRef]
64. Khan N, Bano A, Rahman MA, Guo J, Kang Z, Babar MA. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci Rep. 2019;9(1):2097. doi:10.1038/s41598-019-38702-8. [Google Scholar] [PubMed] [CrossRef]
65. Hussain MB, Zahir ZA, Asghar HN, Asgher M. Can catalase and exopolysaccharides producing rhizobia ameliorate drought stress in wheat. Int J Agric Biol. 2014;16(1):3–13. [Google Scholar]
66. Kaci Y, Heyraud A, Barakat M, Heulin T. Isolation and identification of an EPS-producing Rhizobium strain from arid soil (Algeria): characterization of its EPS and the effect of inoculation on wheat rhizosphere soil structure. Res Microbiol. 2005;156(4):522–31. doi:10.1016/j.resmic.2005.01.012. [Google Scholar] [PubMed] [CrossRef]
67. Ng LM, Melcher K, Teh BT, Xu HE. Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol Sin. 2014;35(5):567–84. doi:10.1038/aps.2014.5. [Google Scholar] [PubMed] [CrossRef]
68. Khalid M, Ali M, Hassani D, Rauf A, Jan F, Hui N. Salicylic acid mediated protection of Brassica campestris sp. chinensis from saline stress via SA receptor NPR1 dependent transcriptional regulation and biosynthesis of related biochemicals. Environ Technol Innov. 2021;24:101950. doi:10.1016/j.eti.2021.101950. [Google Scholar] [CrossRef]
69. Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One. 2011;6(10):e26968. doi:10.1371/journal.pone.0026968. [Google Scholar] [PubMed] [CrossRef]
70. Liang T, Yu S, Pan Y, Wang J, Kay SA. The interplay between the circadian clock and abiotic stress responses mediated by ABF3 and CCA1/LHY. Proc Nat Acad Sci. 2024;121(7):e2316825121. doi:10.1073/pnas.2316825121. [Google Scholar] [PubMed] [CrossRef]
71. Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470(7332):110–4. doi:10.1038/nature09766. [Google Scholar] [PubMed] [CrossRef]
72. Mwimba M, Karapetyan S, Liu L, Marques J, McGinnis EM, Buchler NE, et al. Daily humidity oscillation regulates the circadian clock to influence plant physiology. Nat Commun. 2018;9(1):4290. doi:10.1038/s41467-018-06692-2. [Google Scholar] [PubMed] [CrossRef]
73. Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci U S A. 2012;109(12):4674–7. doi:10.1073/pnas.1116368109. [Google Scholar] [PubMed] [CrossRef]
74. Wang X, Li X, Wu Z, Dong S. Transcriptome analysis of soybean in response to different sulfur concentrations. Phyton-Int J Exp Bot. 2022;91(6):1165. doi:10.32604/phyton.2022.019130. [Google Scholar] [CrossRef]
75. Shokri-Gharelo R, Bandehagh A, Hossain MA. Proteomic profiling and protein-protein interaction network reveal the molecular mechanisms of susceptibility to drought stress in canola (Brassica napus L.). Phyton-Int J Exp Bot. 2022;91(7):1403–17. doi:10.32604/phyton.2022.020431. [Google Scholar] [CrossRef]
76. Wang X, Wu Z, Yan C, Ma C, Dong S. Proteomics analysis of soybean seedlings under short-term water deficit. Phyton-Int J Exp Bot. 2022;91(7):1381–401. doi:10.32604/phyton.2022.020251. [Google Scholar] [CrossRef]
77. Wu W-T, Zhang Y-J, Gao Y, Zhang K, Zhu L-Y, Zhang H-X. De novo transcriptome analysis in Leymus mollis to unveil genes involved in salt stress response. Phyton-Int J Exp Bot. 2022;91(8):1629–42. doi:10.32604/phyton.2022.020515. [Google Scholar] [CrossRef]
78. Zhang T, Li P, Wei J. Transcriptome analysis via RNA sequencing reveals the molecular mechanisms underlying the hedera helix response to high temperature. Phyton-Int J Exp Bot. 2022;91(11):2403–17. doi:10.32604/phyton.2022.022421. [Google Scholar] [CrossRef]
79. Rivero RM, Mittler R, Blumwald E, Zandalinas SI. Developing climate-resilient crops: improving plant tolerance to stress combination. Plant J. 2022;109(2):373–89. doi:10.1111/tpj.v109.2. [Google Scholar] [CrossRef]
80. Prasch CM, Sonnewald U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 2013;162(4):1849–66. doi:10.1104/pp.113.221044. [Google Scholar] [PubMed] [CrossRef]
81. Djonović S, Vittone G, Mendoza-Herrera A, Kenerley CM. Enhanced biocontrol activity of Trichoderma virens transformants constitutively coexpressing β-1, 3- and β-1, 6-glucanase genes. Mol Plant Pathol. 2007;8(4):469–80. doi:10.1111/j.1364-3703.2007.00407.x. [Google Scholar] [PubMed] [CrossRef]
82. Downing KJ, Thomson JA. Introduction of the Serratia marcescens chiA gene into an endophytic Pseudomonas fluorescens for the biocontrol of phytopathogenic fungi. Can J Microbiol. 2000;46(4):363–9. doi:10.1139/w99-147. [Google Scholar] [PubMed] [CrossRef]
83. Singh JS, Abhilash P, Singh H, Singh RP, Singh D. Genetically engineered bacteria: an emerging tool for environmental remediation and future research perspectives. Gene. 2011;480(1–2):1–9. doi:10.1016/j.gene.2011.03.001. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools