 Open Access
Open Access
ARTICLE
The Expression Characteristics and Potential Functions of Heat Shock Factors in Diatom Phaeodactylum tricornutum
1 Zhangzhou Institute of Technology, Zhangzhou, 363000, China
2 Guangdong Provincial Key Laboratory of Marine Biotechnology and Guangdong Provincial Key Laboratory of Marine Disaster Prediction and Prevention, College of Sciences, Shantou University, Shantou, 515063, China
* Corresponding Author: Xiaojuan Liu. Email:
# These authors contributed equally to this work
Phyton-International Journal of Experimental Botany 2024, 93(10), 2583-2596. https://doi.org/10.32604/phyton.2024.055616
Received 02 July 2024; Accepted 03 September 2024; Issue published 30 October 2024
Abstract
Heat shock transcription factor (HSF) are essential regulators of heat shock protein (HSP) gene expression in plants and algae, contributing to their resilience against biotic and abiotic stresses. However, the localization, structure, phylogenetic relationship, and characteristics of PtHSF genes in microalgae, especially in diatom Phaeodactylum tricornutum, remain largely unexplored. This study presents a comprehensive analysis of the PtHSF gene family in P. tricornutum. A genome-wide analysis identified 68 PtHSF genes, which were classified into two distinct subfamilies: traditional and untraditional. Motif and structure analyses revealed evidence of multiple duplication events within the PtHSF gene family. Expression profiling revealed diurnal patterns, with 34 genes being downregulated during the light period and upregulated during the dark period, while 19 genes exhibited the opposite pattern. These findings suggest that PtHSF genes may have specialized functions during the diurnal cycle and play a crucial role in maintaining cellular homeostasis in response to various stresses. Notably, PtHSF16, 30, and 43 genes exhibited higher expression levels, suggesting their potential importance. This study provides a valuable foundation for future investigations into the specific functions of HSFs under different stress conditions and their regulatory mechanisms in P. tricornutum and other microalgae.Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| HSF | Heat shock transcription factor |
| TFs | Transcription factors |
| HSP | Heat shock protein |
| DBD | DNA-binding domain |
| DREB2A | Dehydration-responsive element binding protein 2A |
| PLAAOx | Periplasmic L-amino Acid Oxidase |
| CSAP | Coccolith Scale Associated Protein |
| HMM | Hidden Markov Model |
| TMD | Transmembrane Domains |
| MEME | Multiple Em for Motif Elicitation |
| NJ | Neighbor-Joining |
| LTR | Long Terminal Repeat |
| IGL | Intron Gain or Loss |
| HSE | Heat Shock Element |
| ABRE | ABA response element |
Transcription factors (TFs) bind with DNA-regulatory sequences enhance or silence the transcriptional level of genes [1]. TFs typically bind to the promoter region of target genes, influencing gene transcription and protein expression, and ultimately change the phenotype of organisms. A typical TF contains four unconnected domains, i.e., the DNA-binding domain (DBD), nuclear localization signal, oligomerization site, and transcription-activation domain [2]. Heat shock transcription factors (PtHSFs) are crucial regulators of cellular homeostasis in Phaeodactylum tricornutum, particularly under different abiotic and biotic stresses [3–5]. HSFs can form trimers in response to stress, activating the expression of molecular chaperones and stress-related proteins [6]. Based on evolutionary relationships, the HSF family was usually divided into two subfamilies, i.e., traditional and untraditional [7]. The traditional subfamily is smaller than untraditional subfamily. Additionally, HSFs within the traditional subfamily exhibit a closer evolutionary relationship among different species, including plants, fungi, and animals [7].
HSF TFs play critical roles in plant stress responses. In Arabidopsis thaliana, HsfA2 is known to protect organelles from oxidative damage and cell death [8]. Additionally, HSF TFs are involved in responses to heat shock, high light, anoxia, salt/osmotic stress, and oxidative stress [8–10]. HsfA3 is a key component of drought stress signaling. Its expression can be induced by drought and heat shock through the action of the dehydration-responsive element binding protein 2A (DREB2A) TF in plants, such as in A. thaliana [11]. Differentially, AtHSFA9 is expected to affect the expression of heat shock protein (HSP) during seed development through the regulation of transcription factor abscisic acid-insensitive 3 (ABI3) [12]. Additionally, HSFs can repress the expression of the heat shock gene by controlling the defensin gene [11]. Recent studies have focused on the the function, structure, and characteristics of HSF TFs in microalgae. For example, PtHSF genes were upregulated during the early stage of phosphate deficiency [13] and simultaneously regulated the expression of 69 genes, including those encoding periplasmic L-amino acid oxidase (PLAAOx) and coccolith scale associated protein (CSAP) encoding genes [14]. HSFs are the key genes for heat stress tolerance in the red alga Neopyropia yezoensis [15]. Recent research has also explored the functions of several PtHSFs in Diatom, especially in P. tricornutum. For example, PtHSF3 has been implicated in fucoxanthin synthesis by modulating the expression of the fucoxanthin synthesis genes [16]. Additionally, PtHSF (Phatr3_J49594) is important for the adaptation of P. tricornutum to laboratory culture temperatures [17]. However, a comprehensive analysis of the HSF gene family in microalgae at the genomic level has been lacking. The evolutionary relationships, gene structures, and chromosomal localization of HSFs in microalgae remain largely unknown. Light is an important element for the growth of diatom, affecting cellular metabolisms and homeostasis, such as lipid, protein, and carbohydrate metabolisms [18,19]. However, the overall responses of HSF TFs to the diurnal cycle in microalgae and their role in maintaining cellular homeostasis have not been extensively investigated.
In this study, P. tricornutum (Pt), a marine photosynthetic model diatom with the whole genome sequenced was used as an example to characterize PtHSF TFs [20]. Phylogenetic analysis, gene structure, chromosomal localization, and duplication events were thoroughly investigated. Additionally, the study compared the relative expression of PtHSF genes during the light and dark periods in P. tricornutum to understand the overall responses and characteristics of PtHSF TFs. Phylogenetic analysis, gene structure, chromosomal localization, and duplication events were thoroughly investigated. Additionally, the study compared the relative expression of PtHSF genes during the light and dark periods in P. tricornutum to understand the overall responses and characteristics of PtHSF TFs. Key PtHSFs were identified and characterized at the genome-wide level. The findings from this study will serve as a valuable foundation for future research on the functions of PtHSFs under various stress conditions and their role in cellular homeostasis.
2.1 The Identification and Fundamental Bioinformatic Analysis of PtHSF Genes
To identify PtHSF proteins within the P. tricornutum genome, we first obtained mRNA, DNA, and protein sequences from the Ensembl database (http://plants.ensembl.org/index.html) (accessed on 02 September 2024). Subsequently, the PtHSF domain Hidden Markov Model (HMM) (PF004477) was downloaded from the Pfam database (http://pfam.xfam.org/) (accessed on 02 September 2024). This HMM model, with a e-value < 1.0, was employed to screen the genomic data for potential PtHSF proteins. Pfam database (http://pfam.xfam.org/) (accessed on 02 September 2024) [21]. To ensure the accuracy of our predictions, the putative PtHSF proteins identified through the HMM search were further validated using following independent platforms, i.e., SMART (http://smart.embl.de/) (accessed on 02 September 2024) and Pfam (http://pfam.xfam.org/) (accessed on 02 September 2024) and NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd/) (accessed on 02 September 2024) [21]. Only sequences confirmed to contain the PtHSF domain by both SMART, Pfam and NCBI CDD database were classified as bona fide members of the PtHSF family. This rigorous approach minimized the possibility of false positives and ensured the reliability of our PtHSF protein identification. And NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd/). (accessed on 02 September 2024).
At the same time, signal peptide 3.0 (http://www.cbs.dtu.dk/services/SignalP-3.0/) (accessed on 02 September 2024) [22] and its improved signal peptide 5.0 (http://www.cbs.dtu.dk/services/SignalP/) (accessed on 02 September 2024) were used to identify the signal peptide of PtHSF proteins. While transmembrane domains (TMDs) of PtHSFs were analyzed by TOPCON (http://topcons.cbr.su.se/) (accessed on 02 September 2024), SOSUI (https://harrier.nagahama-i-bio.ac.jp/sosui/mobile/) (accessed on 02 September 2024), ΔG (https://dgpred.cbr.su.se/index.php?p=fullscan) (accessed on 02 September 2024) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (accessed on 02 September 2024) websites. The N-glycosylation modification of PtHSFs were further explored by NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/) (accessed on 02 September 2024) [21].
2.2 The Analysis of Phylogenetic Tree, Architecture of Conserved Motifs and Gene Structure
The full-length protein sequence of PtHSFs and their corresponding PtHSF domain sequences were aligned using ClustalW software with default parameters, e-value < 1e−5 and maximun target sequences were 5. The aligned sequences were used to construct a phylogenetic tree using software MEGA 7.0. The Neighbor-Joining method was chosen for tree construction, and 1000 bootstrap replicates were performed to assess the reliability of the branching patterns observed within the tree [23]. The resulting phylogenetic tree was further optimized using Evolview software to ensure its accuracy.
The Online program MEME (Multiple Em for Motif Elicitation) (http://meme-suite.org/) (accessed on 02 September 2024) was used to analyze the architecture of the conserved motifs within PtHSF with following parameters: any number of repetitions, an optimum motif length between 10 and 200 amino acids, a maximum analysis time of 18,000 s, a maximum input size of 6,000,000 sequences, and a search for 10 motifs with a minimum length of 6 amino acids and a maximum length of 50 amino acids.
The more detailed parameters were described in our previous study [21]. Additionally, the online program Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) (accessed on 02 September 2024) was utilized to predict the intron and exon structures of PtHSFs gene. Subsequently, the identified conserved motifs, predicted gene structures, and the constructed phylogenetic tree were assembled and visualized using TBtools software to create a comprehensive overview of the PtHSF family in P. tricornutum. The location of PtHSF genes on chromosomes was further analyzed. The maps of PtHSFs on chromosomes were depicted by MapChart software. MCScanX was used to clarify the duplication events of PtHSF genes [24].
2.3 The Analysis of PtHSF Genes Expression
P. tricornutum cells were cultured in axenic f/2 medium under a light/dark cycle of 12 h each, with a light intensity of 80 μmol photons m−2 s−1, at 20°C for 6 days to reach the logarithmic growth phase. Cells were harvested at 0, 4, 8, 12, 16, 20, and 24 h of the light/dark cycle for transcriptome analysis. Detailed data from this analysis can be downloaded from the NCBI Sequence Read Archive under accession number GSE210141. To identify PtHSF genes with changes in expression throughout the diurnal cycle, RNA-seq data was analyzed. Genes with a fold change ≥1.5 and a False Discovery Rate (FDR) < 0.01 were classified as differentially expressed. The reported FPKM values represent the average of three biological replicates [25]. A heatmap depicting the expression of PtHSFs across the diurnal cycle was generated using the online tool Heatmapper (http://www.heatmapper.ca/) (accessed on 02 September 2024) [26].
A total of 68 PtHSF genes from the P. tricornutum genome were identified by HMM search and were named PtHSF1 to PtHSF68, as shown in Table 1. Detailed information regarding the Gene ID, transmembrane domain, signal peptide, and modification of proteins is provided in supplementary material Table S1. Our results of protein modification analysis indicate that most PtHSFs are likely modified by pho sphorylation, N-glycosylation and/or ubiquitination; however, none of PtHSF proteins were predicted to contain signal peptide and transmembrane domain.
The phylogenetic tree depicted in Fig. 1 illustrates the evolutionary relationships among the 68 identified PtHSF proteins in P. tricornutum. These proteins were classified into four distinct groups based on their clustering on the tree. Group 1 includes PtHSF5 and PtHSF22, Group 2 includes PtHSF59, PtHSF60, and PtHSF66, Group 3 includes PtHSF13, PtHSF14, PtHSF29, and PtHSF38, Group 4 comprises all the remaining PtHSFs. The shapes in front of the PtHSF labels indicate the species: stars represent A. thaliana HSFs, while triangles represent P. tricornutum HSFs. The color backgrounds of the PtHSF labels correspond to their respective group: blue for Group 1, green for Group 2, pink for Group 3, and purple for Group 4. Seven PtHSFs in P. tricornutum were clustered within the traditional subfamily, which also includes HSFs from A. thaliana. The remaining 61 PtHSFs were classified into the untraditional subfamily based on their phylogenetic distance from A. thaliana HSFs.
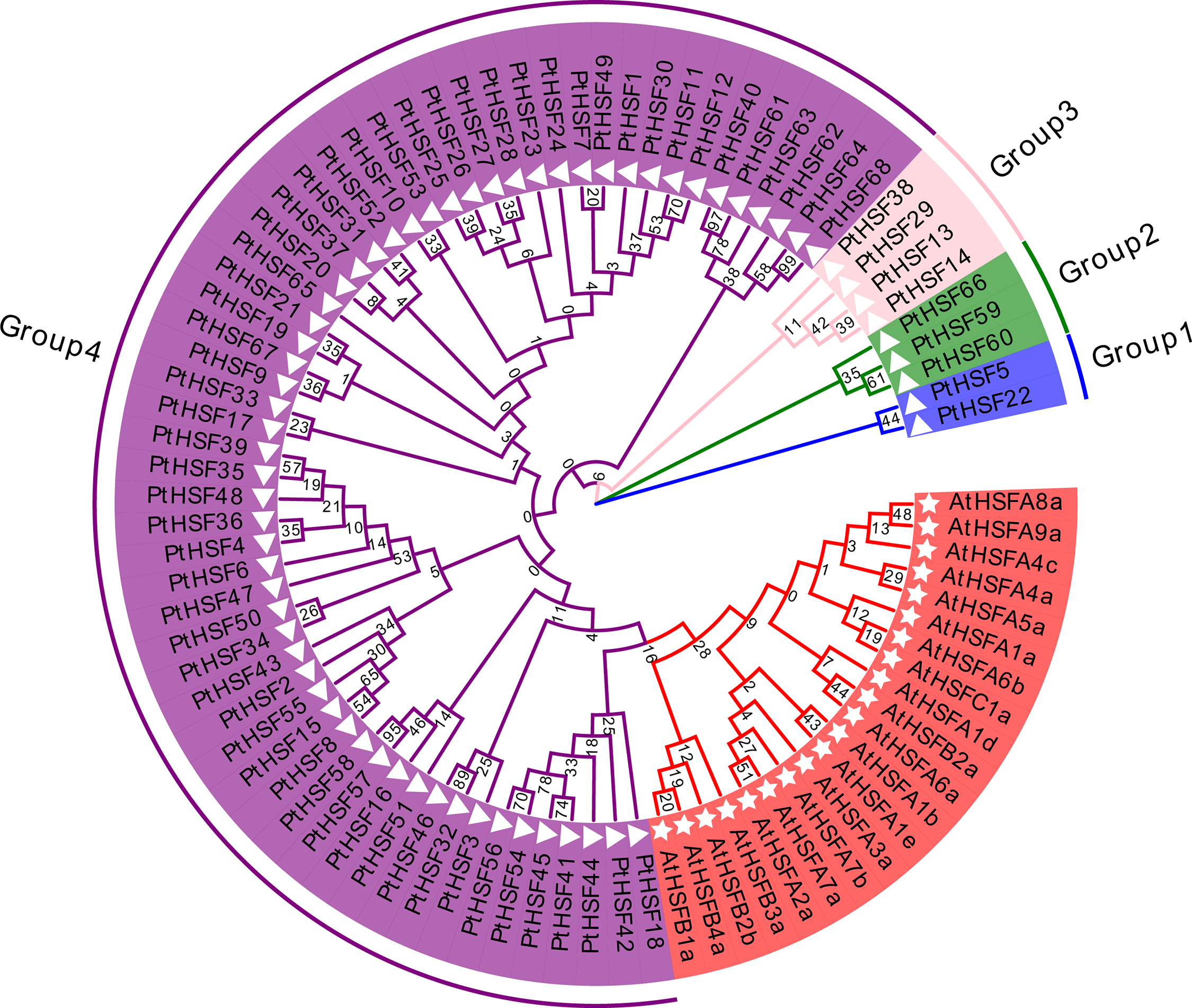
Figure 1: Evolutionary relationship of PtHSF proteins
A Neighbor-Joining (NJ) phylogenetic tree was constructed by using MEGA 7.0 software (Fig. 1). It was shown that all 68 putative PtHSF proteins were divided into 4 groups according to their phylogenetic relationship. PtHSFs on the same branch of phylogenetic tree were classified into one group. Thus, 2 PtHSFs (PtHSF5 and PtHSF22) belonged to Group 1. 3 PtHSFs (PtHSF59, PtHSF60 and PtHSF66) belonged to Group 2. 4 PtHSFs (PtHSF13, PtHSF14, PtHSF29 and PtHSF38) belonged to Group 3. All the rest PtHSFs belonged to Group 4. Among all 68 putative PtHSF proteins, 7 PtHSFs (PtHSF18, PtHSF41-42, PtHSF44-45, PtHSF54 and PtHSF56) in P. tricornutum belonged to the traditional PtHSFs subfamily, while the remaining 61 PtHSFs were grouped into the untraditional PtHSFs subfamily according to the distance from AtHSFs in the phylogenetic tree.
The phylogenetic tree of 68 PtHSF proteins was shown in part A while the conserved motifs of PtHSF proteins were shown in part B. The numbers 1–10 in different color boxes mean 10 different motifs. Additionally, the intron-exon structure of PtHSF genes were displayed in Part C. Black line, green box and yellow box were introns, UTR sequence and exons, respectively.
The phylogenetic tree of 68 PtHSF members was constructed and shown in Fig. 2A. The conserved architecture of motifs was indicated in Fig. 2B. It was found that most of PtHSF proteins contained motif 2 and 3. Besides, some PtHSF proteins harbored specific motifs, such as the motif 8 from PtHSF42, the motif 10 from PtHSF18, and the motifs 6, 8 and 10 from PtHSF41, 44–45, 54 and 56. Anyway, the PtHSF proteins in the same clade of the phylogenetic tree were highly similar in motif architecture. The gene structures of PtHSFs were shown in Fig. 2C. All PtHSFs genes contained 1 to 4 exons. Most of them encoded one, two or three exons, while PtHSF32 and PtHSF18 encoded four exons.
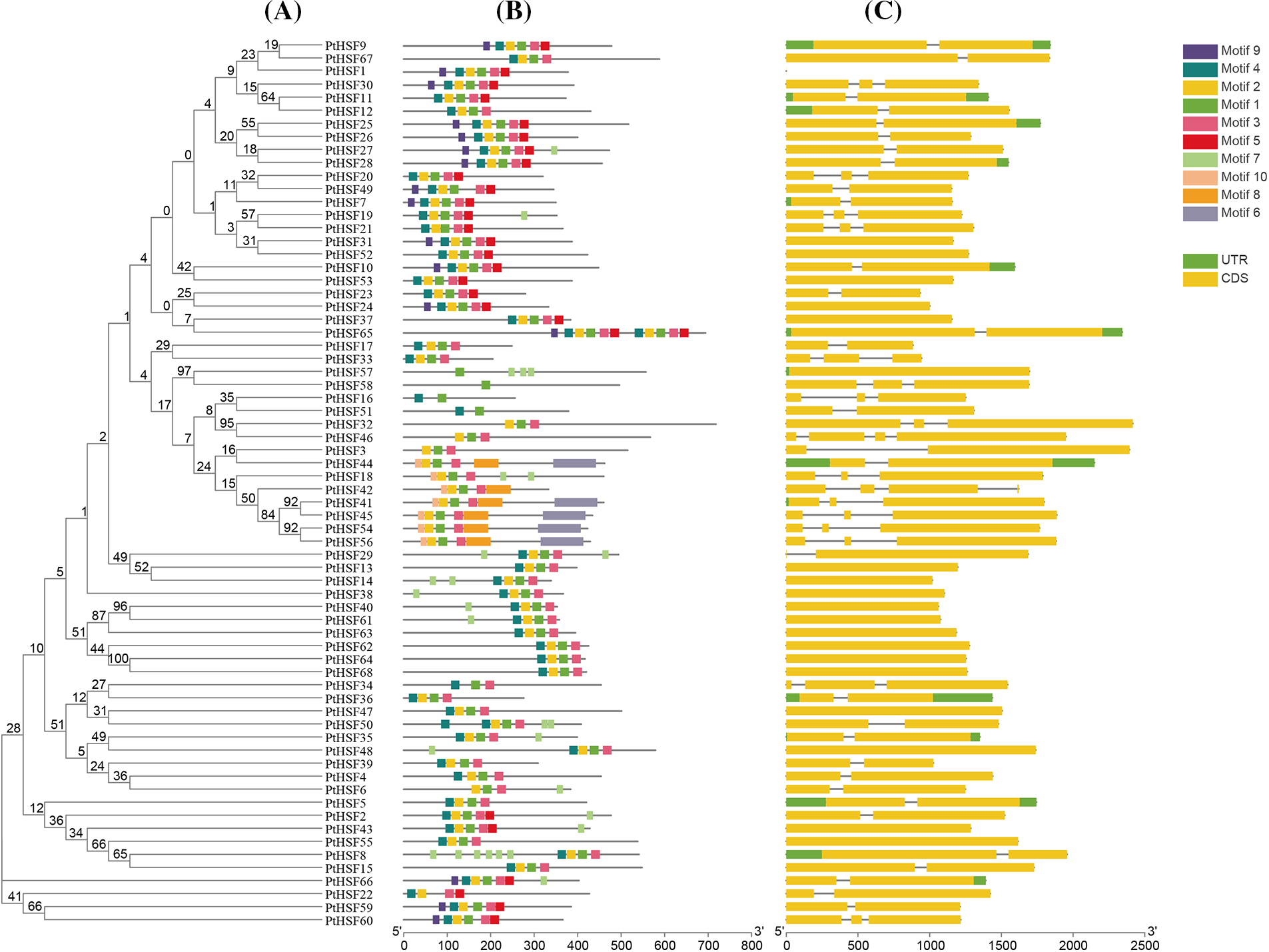
Figure 2: Phylogenetic, motif and gene structure analysis of PtHSFs
It depicts the chromosomal distribution of the 68 PtHSF genes in P. tricornutum. The numbers above the chromosomes indicate the chromosome number, while the numbers on the left side of each chromosome represent the base pair (bp) location of the corresponding PtHSF gene on that chromosome.
The analysis reveals that the 68 PtHSF genes are unevenly distributed across the 21 chromosomes of P. tricornutum, with some chromosomes containing a higher density of PtHSF genes than others. Among 68 PtHSFs, 67 PtHSF genes were disproportionally distributed to 21 chromosomes (out of a total of 33), while PtHSF1 was not associated with any chromosome (Fig. 3). Only one PtHSF gene was found on chromosomes 13, 16, 24, 25 and 30. Two PtHSF genes were located to chromosomes 8, 12, 15, 27 and 32. Three PtHSF genes were observed on chromosomes 2, 5, 6 and 17. Four PtHSF genes were found on chromosomes 4, 18 and 21. In addition, some chromosomes contained more than 4 PtHSF genes. For example, 5 PtHSF genes were found on chromosomes 1. Moreover, 255 tandem-duplicated gene pairs were discovered in the P. tricornutum genome in total (Table S2). Among these 9 pairs were identified as PtHSF gene pairs, including PtHSF68 and PtHSF40, PtHSF41 and PtHSF45, PtHSF56 and PtHSF54, PtHSF20 and PtHSF19, PtHSF38 and PtHSF37, PtHSF63 and PtHSF62, PtHSF60 and PtHSF59, PtHSF28 and PtHSF25, PtHSF24 and PtHSF31. Finally, 140 segmental-duplicated gene pairs were identified in the whole genome according to the bioinformatic analysis (Table S3), but none of these were PtHSF genes.
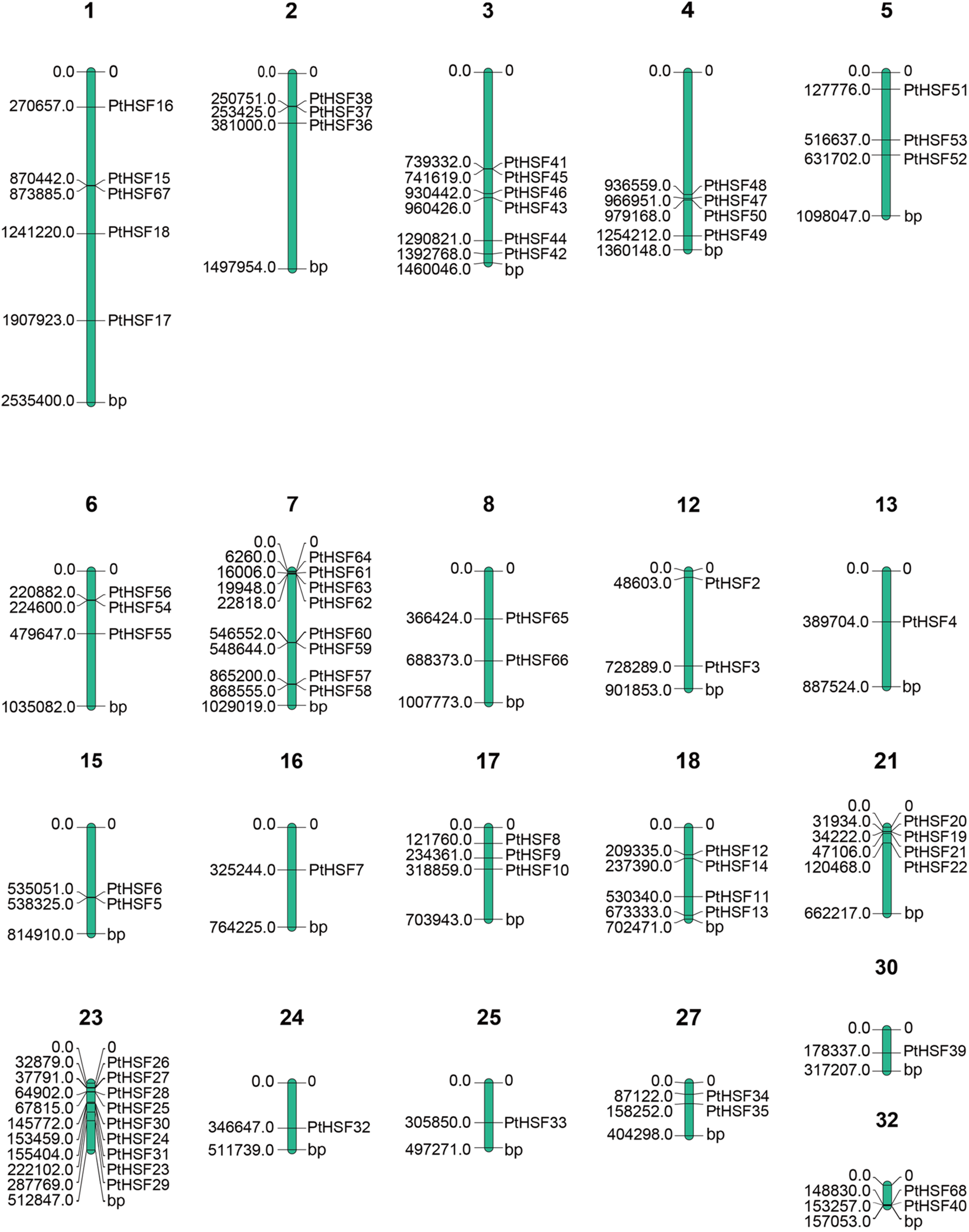
Figure 3: The distribution of PtHSF genes on chromosomes
The expression of PtHSF1 was not detected. However, a heatmap of the remaining 67 PtHSFs was generated by heatmapper software (Fig. 4). The expression level of all 67 PtHSF genes in P. tricornutum were compared (Table S4). It was shown that 35 PtHSF genes (PtHSF4, 8, 11, 15, 17, 19, 27, 28, 29, 33, 34, 36, 39, 40, 41, 42, 43, 46, 47, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, and 68) were down-regulated during the light period and up-regulated during the dark period. In contrast, 19 PtHSF genes (PtHSF2, 6, 7, 10, 12, 13, 16, 21, 22, 24, 25, 30, 31, 32, 37, 38, 44, 49, and 67) were up-regulated in the light and down-regulated in the dark period. The remaining 13 PtHSF genes did not show any specific fluctuation under the same conditions.
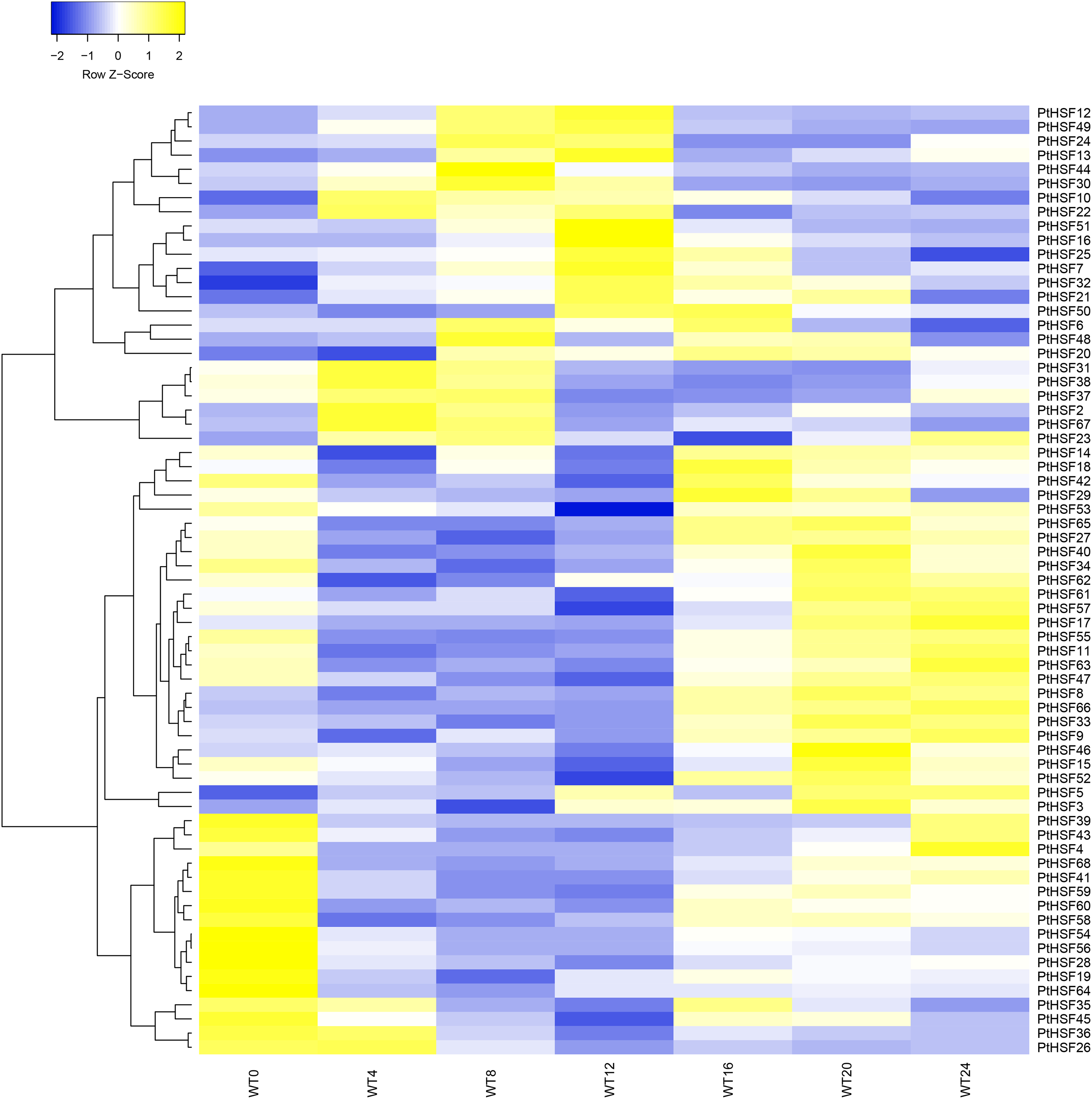
Figure 4: The differentially expression of PtHSF genes during one diurnal cycle. WT represents the wild type of P. tricornutum. The number following WT indicates the time (in hours) throughout the day: a 12-h light period (0–12 h) and a 12-h dark period (13–24 h). The expression patterns of all 68 PtHSFs were classified into three distinct groups: rising during the day and descending at night (Group 1), descending during the day and rising at night (Group 2), and no specific fluctuation (Group 3)
WT represents the wild type of P. tricornutum. The number following WT indicates the time (in hours). WT4 and WT8 occurred during the light period, while WT16 and WT20 occurred during the dark period. Among all 68 PtHSF genes, the expression levels of PtHSF16, 30, and 43 were significantly higher than those of the other PtHSF genes at specific time points.
The range of variation in PtHSF expression is shown in Fig. 5. Except for PtHSF16, 30, and 43, all other PtHSF genes were expressed at low levels. The expression levels of PtHSF16, 30, and 43 changed drastically and differently during the diurnal cycle. PtHSF16 and 30 were upregulated during the light period and subsequently downregulated during the dark period. Notably, the highest expression of PtHSF16 was observed at the 12th h, while PtHSF30 peaked at the 8th hour of the light period. In contrast, PtHSF43 was gradually downregulated during the light period and upregulated during the dark period.
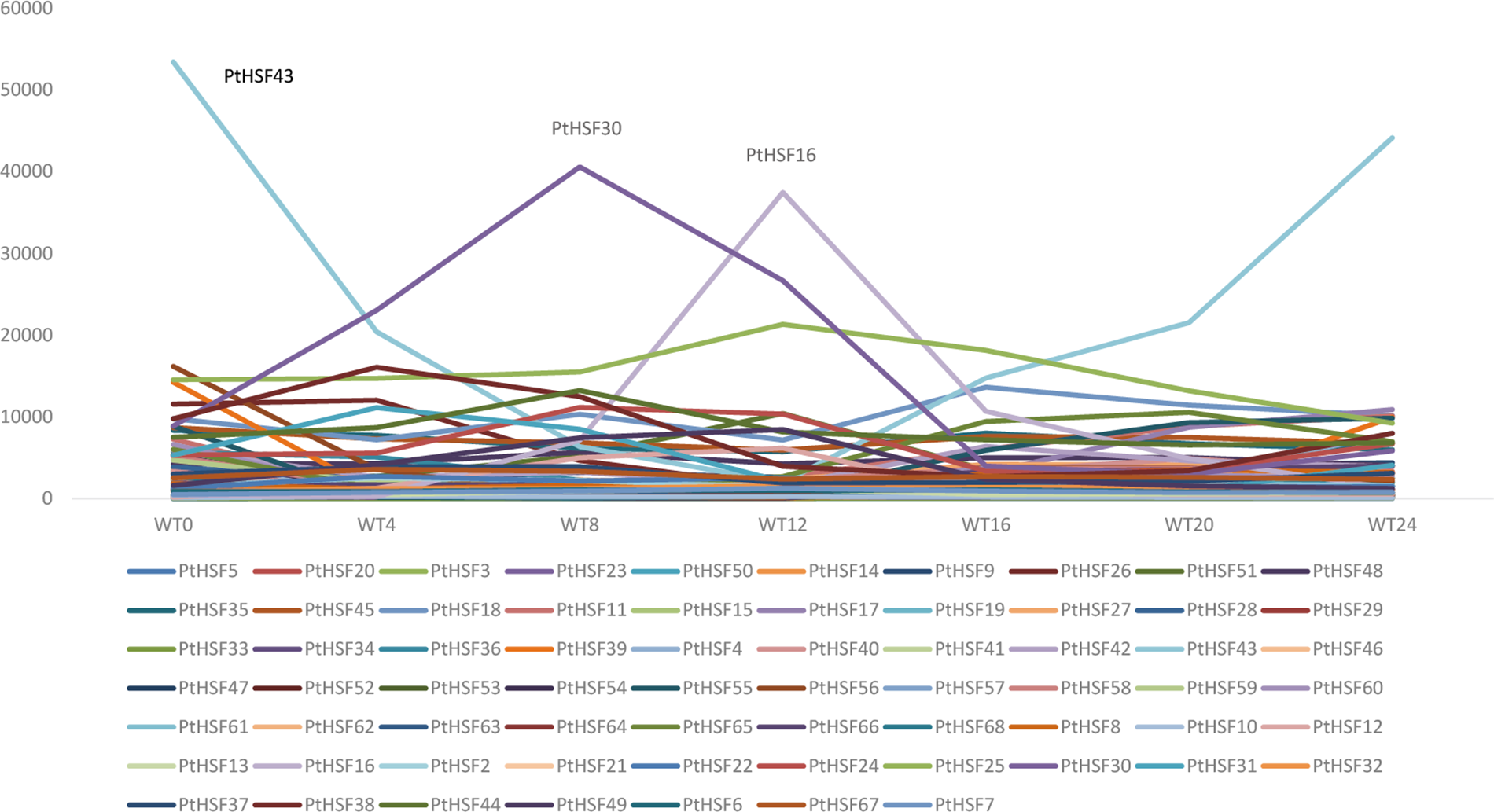
Figure 5: The expression level of PtHSF genes in diurnal cycle
PtHSFs are abundant in the genome of diatom P. tricornutum [7]. In this study, 68 PtHSFs were identified from P. tricornutum. Compared to the reported HSF genes in plants, such as 25 HSF genes in Oryza sativa L. [27] and 21 HSF members in Arabidopsis thaliana [4], the number of PtHSFs in P. tricornutum was significantly higher. Similar to MYB genes in higher plants and diatom [21], it is speculated that numerous duplication events occurred in PtHSF genes during the evolution to adapt to the marine environment. As a result, PtHSFs expanded into a larger gene family. Compared to the previously reported 70 PtHSF TFs [7], 68 PtHSF genes were identified from the genome of P. tricornutum in this study, with 10 members not matching those in the previous research (Table S5). This discrepancy may be due to differences in bioinformatic analysis methods. In the previous research, 7 members PtHSF1.1a, PtHSF1.1b, PtHSF1.1c, PtHSF1.1d, PtHSF1.2a, PtHSF1.2b, PtHSF1.2c) were classified into the traditional HSF subfamily, while the remaining 63 members belonged to the non-traditional HSF subfamily [7]. In this study, 7 PtHSFs (PtHSF18, PtHSF41-42, PtHSF44-45, PtHSF54, and PtHSF56) were found to be more closely related to traditional AtHSFs, indicating that these 7 PtHSFs belong to the traditional PtHSF subfamily. PtHSF18 (also named PtHSF3) has been shown to play a role in fucoxanthin synthesis [16], suggesting that traditional PtHSFs might be involved in fucoxanthin metabolism. However, PtHSF30, from the non-traditional group, plays an important role in the adaptation of algae to laboratory culture [17], leading to the reasonable speculation that PtHSFs in the non-traditional group might be related to growth regulation. These studies suggest that PtHSFs in different subfamilies may have diverse functions. It is noteworthy that PtHSF1, which contains the PtHSF domain, was not found on traditional chromosomes and did not exhibit detectable transcription levels. It is speculated that PtHSF1 may not be located on genomic chromosomes but rather on plastid or mitochondrial chromosomes, or possibly both.
All PtHSFs containing different PtHSF domains were grouped into 4 categories in the phylogenetic tree. Although their evolutionary relationship remains unknown, some speculations have been proposed. For example, has been suggested that Long Terminal Repeat (LTR)-retrotransposon-mediated recombination may have played a crucial role in the amplification of PtHSFs in P. tricoruntum [7]. It is therefore proposed that repeated transposition events led to the evolution and expansion of PtHSFs, ultimately resulting in four distinct phylogenetic groups.
Gene structure analysis identified 10 conserved motifs within PtHSFs. Since motifs 1, 2, 3, and 4 were discovered within the PtHSF domain, it is speculated that these four motifs were necessary for the PtHSF function. More details are provided in Table S6. Motifs 6, 9, and 10 were only found in the traditional subfamily, indicating that these motifs are more conserved. They were the sign of a traditional subfamily to separate from others. Additionally, it was found that 48 (71.6%) PtHSF genes possess 1–3 introns, while the remaining 19 (28.4%) lack introns. Intron gain or loss (IGL) has been primarily analyzed in eukaryotes due to the absence of intron in prokaryotic genes [28]. However, the mechanism of IGL in PtHSFs remains unknown.
The differentially expressed patterns of 68 PtHSF TFs indicated that different PtHSF genes may have different functions under the diurnal cycle. The 68 PtHSFs with different expression patterns were divided into three groups: rise by day and descend by night (Group 1), descend by day and rise by night (Group 2), and no specific fluctuation (Group 3). It is proposed that Group 1 might be related to synthesis metabolism, while Group 2 might be involved in degradation metabolism in response to different stresses and maintaining cellular homeostasis. The PtHSF16, 30, and 43 genes exhibited high expression levels and different expression patterns, suggesting that they may have diverse regulatory mechanisms during the light and dark periods.
In P. tricornutum, carbohydrates, such as laminarin and lipid, were accumulated in the light for energy storage and are consumed in the dark for growth, development, and reproduction. The up-regulated and then down-regulated PtHSF16 and PtHSF30 genes might play an essential role in the accumulation of these carbohydrates during the light period. The down-regulated and subsequently up-regulated PtHSF43 might be crucial for carbohydrate degradation during the dark period. However, the exact mechanism of PtHSFs on the regulation of cellular homeostasis and stress response remains unknown and requires further experiments to verify. For example, it would be valuable to verify the function and regulatory networks of PtHSFs through the isolation and analysis of the PtHSF overexpression and knock-out mutants via CRISPR/CAS9 technology [29] and high-throughput experiments.
Interestingly, PtHSF32 was identified as having a unique AP2 region in this study. Several studies have shown that AP2 is important in hormones and stress stimuli at specific growth stages [30–32]. Research showed that the important cis-elements on the promoter and/or the multiple responsive elements might be the key regulator of the AP2 expression [33]. AtHSFA1 plays a vital role in the stress response of plants by binding with the Heat Shock Element (HSE) and ABA Response Element (ABRE) [34,35]. Since PtHSF32 is highly homologous with AtHSFA1, it is speculated that PtHSF32 might have a similar function to AtHSFA1 in participating in stress response. Additionally, it was found that PtHSF18, 41, and 56 contained a coiled-coil region. This unique structure has extreme thermostability [36]. Thus, it is suggested that PtHSF18, 41, and 56 may act as a messenger to regulate stress reactions in diatoms, such as protecting the cell against heat stress. Given the same fluctuation characteristics of PtHSFs expression pattern and the metabolisms, PtHSF TFs might play an essential role in regulating the cellular homeostasis of P. tricornutum under the diurnal cycle and different stresses. The diversity of PtHSF TF gene expressions suggested that different PtHSF TF might have diverse functions. Thus, clarifying the functions of PtHSF TFs would be of great significance for cellular homeostasis in microalgae, such as in P. tricornutum.
Acknowledgement: Thanks to Muhammad Aslam for English language editing of this manuscript.
Funding Statement: Thanks to the fundings of educational and scientific research projects for young and middle-aged teachers in Fujian Province (Grant number: 2022JAT220693) and Natural Science Foundation of Guangdong Province (Grant numbers: 2024A1515013029 and 2022A1515012141).
Author Contributions: Xiaojuan Liu designed the experiments and supervised the project. Yanhuan Lin, Jiaxin Feng, Hao Fang, Wei Huang, Kanglie Guo and Xiyan Liu performed the experiments and analyzed the data. Yanhuan Lin, Shuqi Wang and Xiaojuan Liu wrote and revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.055616.
References
1. Wang Y, Xu L, Chen YL, Shen H, Gong YQ, Limera C, et al. Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (Pb) stress with next generation sequencing. PLoS One. 2013;8(6):e66539. doi:10.1371/journal.pone.0066539. [Google Scholar] [CrossRef]
2. Wang Z, Tang J, Hu R, Wu P, Hou XL, Song XM, et al. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics. 2015;16(1):17. doi:10.1186/s12864-015-1216-y. [Google Scholar] [CrossRef]
3. Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16(9):2452–62. doi:10.1093/emboj/16.9.2452. [Google Scholar] [CrossRef]
4. Nover L, Bharti K, Döring P, Mishra SK, Ganguli A. Scharf K-D. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6(3):177–89. doi:10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2. [Google Scholar] [PubMed] [CrossRef]
5. Barna J, Csermely P, Vellai T. Roles of heat shock factor 1 beyond the heat shock response. Cell Mol Life Sci. 2018;75(16):2897–916. doi:10.1007/s00018-018-2836-6. [Google Scholar] [CrossRef]
6. Ohama N, Kusakabe K, Mizoi J, Zhao HM, Kidokoro S, Koizumi S, et al. The transcriptional cascade in the heat stress response of arabidopsis is strictly regulated at the level of transcription factor expression. Plant Cell. 2016;28(1):181–201. doi:10.1105/tpc.15.00435. [Google Scholar] [CrossRef]
7. Rayko E, Maumus F, Maheswari U, Jabbari K, Bowler C. Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 2010;188(1):52–66. doi:10.1111/nph.2010.188.issue-1. [Google Scholar] [CrossRef]
8. Zhang LR, Li YS, Xing D, Gao CJ. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J Exp Bot. 2009;60(7):2073–91. doi:10.1093/jxb/erp078. [Google Scholar] [CrossRef]
9. Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, et al. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta. 2008;227(5):957–67. doi:10.1007/s00425-007-0670-4. [Google Scholar] [CrossRef]
10. Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot. 2007;58(12):3373–83. doi:10.1093/jxb/erm184. [Google Scholar] [CrossRef]
11. Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta. 2012;1819(2):104–19. doi:10.1016/j.bbagrm.2011.10.002. [Google Scholar] [CrossRef]
12. Kotak S, Vierling E, Baumlein H, Von Koskull-Doring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell. 2007;19(1):182–95. doi:10.1105/tpc.106.048165. [Google Scholar] [CrossRef]
13. Cruz de Carvalho MH, Sun HX, Bowler C, Chua NH. Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 2016;210(2):497–510. doi:10.1111/nph.2016.210.issue-2. [Google Scholar] [CrossRef]
14. Scarsini M, Thiriet-Rupert S, Veidl B, Mondeguer F, Hu H, Marchand J, et al. The transition toward nitrogen deprivation in diatoms requires chloroplast stand-by and deep metabolic reshuffling. Front Plant Sci. 2021;12:760516. [Google Scholar] [PubMed]
15. Kobayashi Y, Harada N, Nishimura Y, Saito T, Nakamura M, Fujiwara T, et al. Algae sense exact temperatures: small heat shock proteins are expressed at the survival threshold temperature in Cyanidioschyzon merolae and Chlamydomonas reinhardtii. Genome Biol Evol. 2014;6(10):2731–40. doi:10.1093/gbe/evu216. [Google Scholar] [PubMed] [CrossRef]
16. Zhao H, Liu Y, Zhu Z, Feng Q, Ye Y, Zhang J, et al. Mediator subunit MED8 interacts with heat shock transcription factor HSF3 to promote fucoxanthin synthesis in the diatom Phaeodactylum tricornutum. New Phytol. 2024;241(4):1574–91. doi:10.1111/nph.v241.4. [Google Scholar] [CrossRef]
17. Chaumier T, Yang F, Manirakiza E, Ait-Mohamed O, Wu Y, Chandola U, et al. Genome-wide assessment of genetic diversity and transcript variations in 17 accessions of the model diatom Phaeodactylum tricornutum. ISME Commun. 2024;4(1):ycad008. doi:10.1093/ismeco/ycad008. [Google Scholar] [CrossRef]
18. Becker S, Tebben J, Coffinet S, Wiltshire K, Iversen MH, Harder T, et al. Laminarin is a major molecule in the marine carbon cycle. P Natl Acad Sci U S A. 2020;117(12):6599–607. doi:10.1073/pnas.1917001117. [Google Scholar] [CrossRef]
19. Chen J, Yang J, Du H, Aslam M, Wang W, Chen W, et al. Laminarin, a major polysaccharide in stramenopiles. Mar Drugs. 2021;19(10):576. doi:10.3390/md19100576. [Google Scholar] [CrossRef]
20. Kumar Sharma A, Muhlroth A, Jouhet J, Marechal E, Alipanah L, Kissen R, et al. The Myb-like transcription factor phosphorus starvation response (PtPSR) controls conditional P acquisition and remodelling in marine microalgae. New Phytol. 2020;225(6):2380–95. doi:10.1111/nph.v225.6. [Google Scholar] [CrossRef]
21. Wang WN, Fang H, Aslam M, Du H, Chen JC, Luo HD, et al. MYB gene family in the diatom Phaeodactylum tricornutum revealing their potential functions in the adaption to nitrogen deficiency and diurnal cycle. J Phycol. 2022;58(1):121–32. doi:10.1111/jpy.v58.1. [Google Scholar] [CrossRef]
22. Li L, Miao WG, Liu WB, Zhang SJ. The signal peptide-like segment of hpaXm is required for its association to the cell wall in transgenic tobacco plants. PLoS One. 2017;12(1):e0170931. doi:10.1371/journal.pone.0170931. [Google Scholar] [CrossRef]
23. Hao ZM, Liu Y, Liu J, Zhou C, Shen S, Dong J, et al. First report of Rhizoctonia solani AG-4 HG-III causing sheath blight on foxtail millet in China. Plant Dis. 2022;107(7):2223. [Google Scholar]
24. Yu Z, Zhang W, Fu H, Zou X, Zhao M, Liang S, et al. Genomic analysis of Poxviridae and exploring qualified gene sequences for phylogenetics. Comput Struct Biotechnol J. 2021;19:5479–86. doi:10.1016/j.csbj.2021.09.031. [Google Scholar] [CrossRef]
25. Chen J, Du H, Liu Z, Li T, Du H, Wang W, et al. Endoplasmic reticulum-quality control pathway and endoplasmic reticulum-associated degradation mechanism regulate the N-glycoproteins and N-glycan structures in the diatom Phaeodactylum tricornutum. Microb Cell Fact. 2022;21(1):219. doi:10.1186/s12934-022-01941-y. [Google Scholar] [CrossRef]
26. Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–53. doi:10.1093/nar/gkw419. [Google Scholar] [CrossRef]
27. Zhang Y, Wang C, Wang CY, Yun L, Song LH, Idrees M, et al. OsHsfB4b confers enhanced drought tolerance in transgenic arabidopsis and rice. Int J Mol Sci. 2022;23(18):10830. doi:10.3390/ijms231810830. [Google Scholar] [CrossRef]
28. Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, et al. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the arabidopsis circadian clock. Plant Cell. 2016;28(3):696–711. doi:10.1105/tpc.15.00737. [Google Scholar] [CrossRef]
29. Xie X, Yang J, Du H, Chen J, Sanganyado E, Gong Y, et al. Golgi fucosyltransferase 1 reveals its important role in α-1,4-fucose modification of N-glycan in CRISPR/Cas9 diatom Phaeodactylum tricornutum. Microb Cell Fact. 2023;22(1):6. doi:10.1186/s12934-022-02000-2. [Google Scholar] [CrossRef]
30. Feng JX, Liu D, Pan Y, Gong W, Ma LG, Luo JC, et al. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol Biol. 2005;59(6):853–68. doi:10.1007/s11103-005-1511-0. [Google Scholar] [CrossRef]
31. Li H, Wang Y, Wu M, Li LH, Li C, Han ZP, et al. Genome-wide identification of AP2/ERF transcription factors in cauliflower and expression profiling of the ERF family under salt and drought stresses. Front Plant Sci. 2017;8:946. doi:10.3389/fpls.2017.00946. [Google Scholar] [CrossRef]
32. Owji H, Hajiebrahimi A, Seradj H, Hemmati S. Identification and functional prediction of stress responsive AP2/ERF transcription factors in by genome-wide analysis. Comput Biol Chem. 2017;71:32–56. doi:10.1016/j.compbiolchem.2017.09.004. [Google Scholar] [CrossRef]
33. Xie ZL, Nolan TM, Jiang H, Yin YH. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci. 2019;10:228. doi:10.3389/fpls.2019.00228. [Google Scholar] [CrossRef]
34. Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in arabidopsis. Plant Cell Physiol. 2011;52(12):2136–46. doi:10.1093/pcp/pcr143. [Google Scholar] [PubMed] [CrossRef]
35. Liu HC, Liao HT, Charng YY. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011;34(5):738–51. doi:10.1111/pce.2011.34.issue-5. [Google Scholar] [CrossRef]
36. Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11(2):82–8. doi:10.1016/S0962-8924(00)01898-5. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools