 Open Access
Open Access
ARTICLE
Protein Disulfide Isomerase and Its Potential Function on Endoplasmic Reticulum Quality Control in Diatom Phaeodactylum tricornutum
1 Zhangzhou Institute of Technology, Zhangzhou, China
2 Guangdong Provincial Key Laboratory of Marine Biotechnology, Guangdong Provincial Key Laboratory of Marine Disaster Prediction and Prevention, College of Sciences, Shantou University, Shantou, China
* Corresponding Author: Xiaojuan Liu. Email:
# These authors contributed equally to this work
Phyton-International Journal of Experimental Botany 2024, 93(1), 137-150. https://doi.org/10.32604/phyton.2023.044996
Received 14 August 2023; Accepted 28 November 2023; Issue published 26 January 2024
Abstract
PDI is a molecular chaperone and plays an important role in Endoplasmic Reticulum quality control (ERQC). PDI participates in the refolding of the misfolded/unfolded proteins to maintain cellular homeostasis under different stresses. However, bioinformatic characteristics and potential functions of PDIs in diatom Phaeodactylum tricornutum (Pt) are still unknown so far. Hence, the genome-wide characteristics of PtPDI proteins in P. tricornutum were first studied via bioinformatic and transcriptomic methods. 42 PtPDI genes were identified from the genome of P. tricornutum. The motif, protein structure, classification, number of introns, phylogenetic relationship, and the expression level of 42 PtPDI genes under the tunicamycin stress were analyzed. A pair of tandem duplicated genes (PtPDI15 and PtPDI18) was observed in P. tricornutum. The 42 PtPDIs with different gene characteristics were divided into three independent clades, indicating different evolutional relationships and functions of these PtPDIs. The 14 up-regulated PtPDI genes under the tunicamycin treatment might have a positive effect on the ER quality control of the unfolded/misfolded proteins, while the 7 down-regulated PtPDIs might negatively affect the ERQC. The characteristics of all 42 PtPDIs and their proposed working model here provide a comprehensive understanding of the PtPDIs gene family. The differential expression of 21 PtPDIs will be useful for further functional study in the ERQC.Keywords
Supplementary Material
Supplementary Material FileMolecular chaperones are the guardians of protein homeostasis [1]. They widely exist in eubacteria archaebacteria and eukaryotic cells [2]. Molecular chaperones could interact with another protein to form its functional active conformation [3]. Previous studies have already shown that molecular chaperones are essential for cellular protein folding, refolding, and the ubiquitin-proteasome system, playing an important role in removing the misfolded and aggregated proteins timely under stresses [4]. Additionally, molecular chaperones could promote cell survival in a non-physiological state and stressful conditions by preventing aggregation and promoting refolding of proteins. It was reported that these functions are ubiquitous and conserved in all organisms [1].
Protein disulfide isomerases (PDIs), a kind of 55 kDa proteins, are important molecular chaperones in the Endoplasmic Reticulum quality control (ERQC). ERQC is known as a calreticulin/calnexin cycle and plays a key role in glycoprotein folding, secretion, and maturity [5,6]. The ERQC recognizes the misfolded/unfolded proteins on the way to their secretory pathway [7]. In the ERQC, together with the ER lectins calnexin (CNX) and/or calreticulin (CRT), molecular chaperones (e.g., PDIs) combine to the N-linked mono-glucosylated glycan of glycoproteins and aid the folding of glycoproteins [5]. The structure of PDIs was widely studied in plants. The PDIs are composed of five domains, including a, b, b’, a’, and c domains. There are two active sites in the a and a’ domains, both have two cysteine residues [8]. The a-type domain carrying the “Cys-Gly-His-Cys” motif contains catalytic active sites and plays a role in the reduction-oxidation cycle. The b-type domain is enriched in hydrophobic residues, involving substrate binding. It was reported that PDIs are a kind of thiol oxidoreductase chaperones located in the ER [9]. It was shown that PDIs are divided into three classes in microalgae. The first class contains a central Trx module with a canonical YAPWCGHC active site sequence. The second class harbors an N-terminal Trx domain with a variable active site sequence. The third class has two domains, a N-terminal Trx domain and a long α-helix domain at the C-terminus [10]. For example, Rb60 in Chlamydomonas reinhardtii, the third class of PDI protein, contains two thioredoxin-like domains with a putative catalytic site of “-Cys-Gly-His-Cys-” [11]. Although characteristics of PDIs were compared between plants and chromalveolates [12], however, the structure of PDIs and their characteristics in microalgae, including the conserved motifs and domains, classification, the number of introns-exons, and phylogenetic relationship are still unknown so far.
Additionally, PDIs display different functions in plants. The classical function of PDI proteins is responsible for the introduction or isomerization of disulfide bonds in nascent ER proteins, aiding the refolding of misfolded/unfolded proteins, and finally retaining the protein homeostasis [8,11,13]. For example, AtPDIs located in chloroplasts of leaves are associated with starch granule biosynthesis in Arabidopsis thaliana [14]. In Soybean DG330, the upregulated expression of the GmPDIL7 gene helps the formation of disulfide bonds in nascent proteins under ER stress [15]. SlPDI (Protein disulfide isomerases of Solanum Lycopersicum) promotes the tomato resistance to TYLCV virus by reducing hydrogen peroxide (H2O2) levels, increasing the activity of superoxide dismutase (SOD) and peroxidase (POD), and enhancing antioxidant activity. It was indicated that SlPDI plays an essential role in enhancing the protein folding function of ER and promoting the synthesis and conformation of antioxidant-related proteins [16]. In addition to plants, the function of PDIs in microalgae was also studied. In C. reinhardtii, Rb60 regulates the translation of proteins and can maintain photosynthesis. CrPDI2 is a redox-active protein in C. reinhardtii. It was found that CrPDI2 participates in the regulation of the circadian signaling pathway [17]. So far, a lot of PDI studies have been focused on Chlorophyta, such as C. reinhardtii. However, the expression characteristics and function of the PDI gene family in diatom remain unknown.
Phaeodactylum tricornutum, a diatom, is one of the most important primary producers in the ocean [18]. In this study, P. tricornutum was used to study the conserved motifs, gene structures and the distribution of genes on the chromosomes, evolutionary relationship, and putative function of PtPDIs via bioinformatic analysis and molecular biotechnology. Tunicamycin is an effective inhibitor of protein N-glycosylation to disrupt the N-glycosylation modification of proteins in organisms [19]. N-glycosylation is an important co- and post-translational event in the modification of proteins in eukaryotes. N-glycosylation plays a vital role in the function of protein. In P. tricornutum, the pathway of protein N-glycosylation mainly takes place in the ER and Golgi apparatus [14]. The stress of tunicamycin will disrupt protein homeostasis. Therefore, it is valuable to study the expression characteristics of PDIs and propose their potential functions under tunicamycin stress. The results obtained in this study will provide an important foundation for clarifying the mechanism of PDIs proteins in protein homeostasis [20].
2.1 Microalgae and Growth Conditions
P. tricornutum (Pt1, obtained from Oil Crops Research Institute of Chinese Academy of Agricultural Sciences, China) was cultured in f/2 medium and placed in a 16 h light and 8 h dark culture environment with a speed of 100 rpm and a light intensity of 30–40 μmol photons·m−2·s−1 at 25 ± 1°C.
2.2 Bioinformatic Analysis of PtPDI Gene Family
2.2.1 Mining and Identification of PtPDI Gene Family in P. tricornutum
cDNA, PEP (phospholipid exchange proteins), CDS (Coding sequence), and GFF (Generic Feature Format) files were downloaded from the Ensembl database (http://www.protists.ensembl.org). The hidden Markov model (HMM) file of the PtPDI gene family (Pfam number: PF0085) was downloaded from the Pfam database (http://pfam.xfam.org/). Based on these data, PDI candidates were screened via the domain search (e-value < 3.8e-06) from the P. tricornutum genome. The PDI candidates were manually validated on the NCBI, CDD (https://www.ncbi.nlm.nih.gov/CDD/), and SMART (http://www.smart.embl.de) websites to ensure that each gene has the thioredoxin domain.
2.2.2 Phylogenetic Tree and Conserved Motif Domain, Gene Structure Analysis of PtPDIs
The multi-sequence alignment of PtPDIs was performed by mega 7.0 software (http://www.megasoftware.net). The multi-sequence alignment results were subsequently analyzed by the phylogenetic tree with 1000 bootstraps, and the phylogenetic tree was constructed to obtain the correlation among all PtPDIs. The evolutionary tree was finally optimized by Evolview (https://www.evolgenius.info/evolview-v2/#login). The motif structures of the PtPDI gene family were analyzed by multiple Em for motif elicitation website (MEME, http://meme-suite.org). The parameters were set to any number of repetitions. The optimal motif length was 6~10 amino acids. The time was 18,000 s. The maximum size was 6,000,000. The number of motifs was 10 and other parameters were the default values. The GFF file of P. tricornutum was used to screen the location information of CDS and UTR (untranslated region) of PtPDI genes on chromosomes. Finally, the software TBtools was used to visually integrate and improve the evolutionary tree, domain information, CDS, and UTR of PtPDI genes. The software map chart was used (http://www.mapchart.net) to draw the position of PtPDI genes on the chromosome.
2.2.3 Bioinformatic Prediction of PtPDIs
Signal peptide 3.0 (https://services.healthtech.dtu.dk/service.php?SignalP-3.0) and signal peptide 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) were applied to predict the signal peptide of PtPDIs. The transmembrane domain was predicted by TMHMM (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0). The N-glycosylation site of PtPDI proteins was predicted by netnglyc-1.0 (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0) and the subcellular localization of PtPDI proteins was determined by cell PLOC 2.0 website (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) and WoLF PSORT (https://wolfpsort.hgc.jp/), and further validated by Diatom specific HECTAR v1.3 (https://webtools.sb-roscoff.fr/root?tool_id=abims_hectar).
2.2.4 Tunicamycin Treatment and the Analysis of PtPDIs Expression in P. tricornutum
The 0.3 μmol L−1 tunicamycin was used to treat the logarithmic growth stage of P. tricornutum. Transcriptomic analysis was carried out after 24 h of culture. The expression level of genes under the tunicamycin stress was analyzed and compared with that in the wild type. Genes’ expression was processed by the Z-score method. The transcriptomic data was visualized by Heatmapper (http://www.heatmapper.ca/). Sequence reads are available on the NCBI sequence read archives [GSE209809]. The data was published in our recent paper. Genes with |fold change| ≥ 1.5 and FDR < 0.01 (adjusted p-value, determined by the Benjamini and Hochberg multiple-testing correction implemented in the ‘p. adjust’ method of R) were defined as differentially expressed genes. The value of FPKM was the average of the three biological replicates. Sequence reads are available on the NCBI sequence read archives [GSE209809]. The more detailed methods were reported in our recent paper [12].
3.1 Identification and Classification of PtPDIs in P. tricornutum
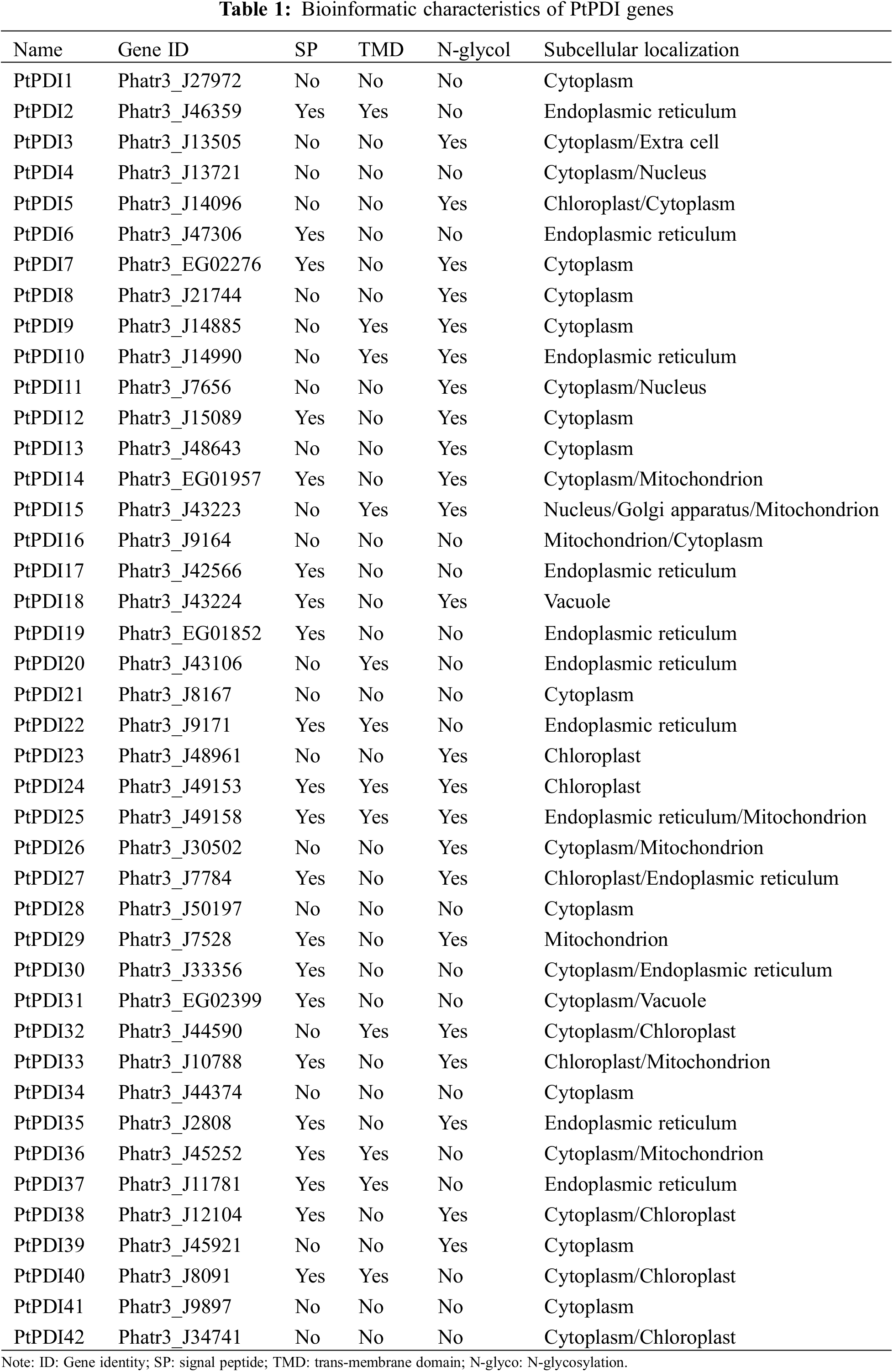
Through the HMM search, 45 putative PtPDIs corresponding to the Pfam-thioredoxin family were obtained from the whole genome of P. tricornutum. After the manual confirmation by smart, NCBI CDD, and Pfam, 42 genes containing the thioredoxin domain were identified as PtPDIs. The 42 PtPDIs were named PtPDI1~PtPDI42, respectively (Table 1). The gene ID, signal peptide, transmembrane domain, N-glycosylation, and subcellular localization of these PtPDIs are shown in Table 1. Among them, 21 PtPDI proteins were predicted to contain signal peptides, while the other 21 PtPDI proteins did not have signal peptides. According to TMD prediction, the 12 of 42 PtPDI proteins had transmembrane domains. It was predicted that 23 of 42 PtPDI proteins contained N-glycosylation sites. PtPDI proteins were mainly predicted to target to endoplasmic reticulum, cytoplasm, chloroplast, and extracellular matrix. Some of them might be located in more than one subcellular organelle. For example, PtPDI25-27 might be located in the endoplasmic reticulum, mitochondrion, and cytoplasm.
3.2 Phylogenetic Analysis of PtPDI Genes
To investigate the evolutionary relationship among PtPDIs, the protein sequence of PtPDIs was used to map the phylogenetic tree through MEGA 7.0. Each PtPDI protein contained one or more thioredoxin domains. It was found that 42 PtPDI genes were divided into three branches according to the proteins’ homology. In the pink branch, most PtPDI proteins had a thioredoxin domain. The homology of PtPDI17 and PtPDI19 was highest (94%). The homology of PtPDI34 and PtPDI42 and PtPDI36 was 95%. In the green branch, most PtPDI proteins had more than one thioredoxin domain. PtPDI39, PtPDI11, PtPDI13, and PtPDI29 had the highest homology, at about 80%. In the yellow branch, there were 3 PtPDI proteins with one thioredoxin domain and 3 PtPDI proteins with multiple thioredoxin domains (Fig. 1).
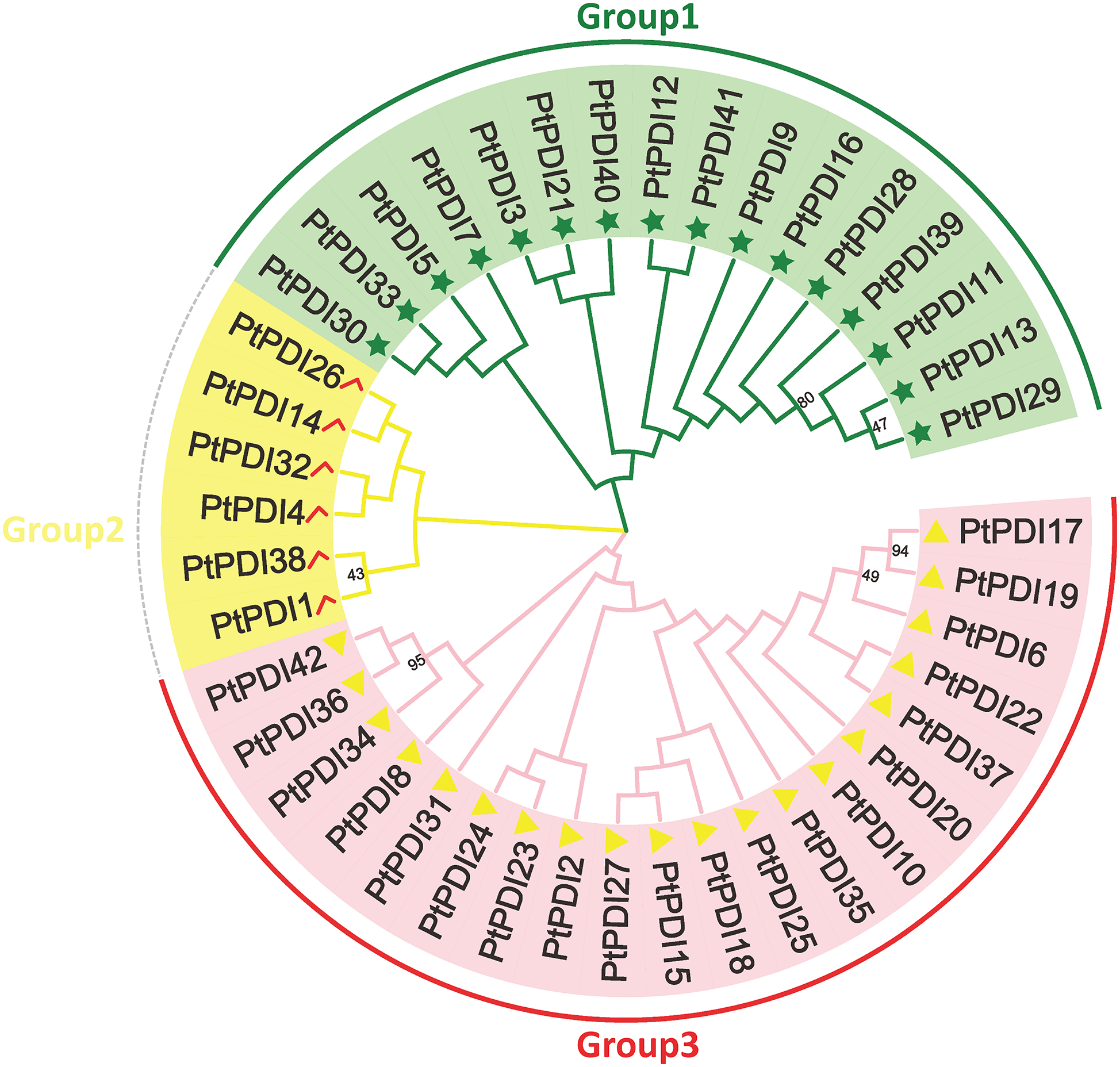
Figure 1: The phylogenetic tree of PtPDIs. Multi-sequence alignment was performed by Clustal W. Phylogenetic tree was constructed by Mega 7.0 software. And the multi-sequence alignment was analyzed by 1000 bootstrap
3.3 Characteristics of Conserved Domains in P. tricornutum
The analysis of gene structure and location, function, and evolution were shown. The whole genome of P. tricornutum was compared with the cDNA of PtPDI proteins to select the domain information and the coding sequences (CDS) (Fig. 2). It was observed that most PtPDI genes of P. tricornutum had multiple domains (Fig. 2). Except PtPDI1 without motif1, PtPDI28 and PtPDI32 without motif 2, all the other PtPDI genes contained motifs 1 and 2. Additionally, some PtPDI genes harbored some specific motifs, such as PtPDI20 and PtPDI35 with motifs 5, 6, and 9. 42 PtPDI genes had CDS structure (Fig. 2). Most of the genes had more than one exon. PtPDI35, PtPDI18, PtPDI6 and PtPDI13 genes contained three exons. The PtPDI20 gene harbored four exons. Based on the conserved domain, the 42 PtPDIs were classified into PDI-F (69%, 29/42), PDI-D (9.5%, 4/42), PDI-M (4.7%, 2/42), PDI-L (4.7%, 2/42), PDI-A (4.7%, 2/42), PDI-B (2.3%, 1/42) and PDI-C (2.3%, 1/42).
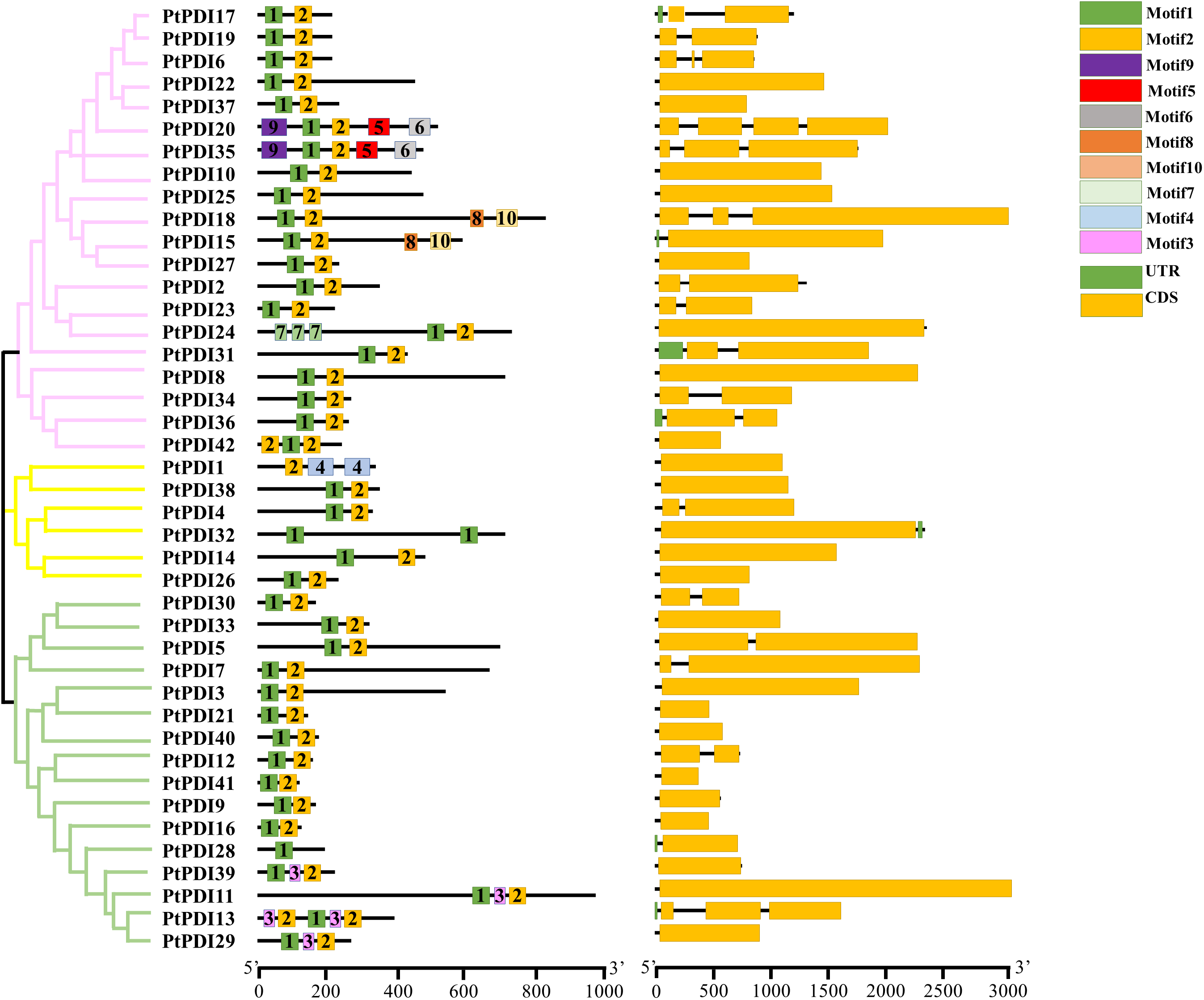
Figure 2: Phylogenetic tree, motif, and CDS sequence analysis of 42 PtPDIs. A Phylogenetic tree was optimized via mega 7.0 software with 1000 bootstrap. B, the motif structure of PtPDIs was shown by the MEME website. Each motif was represented by a color. The number in the box is the serial number of the motif type. The length of PtPDI genes was observed through the scale line at the bottom. C, CDS was represented in the yellow box, and UTR was represented in the green box
3.4 The Analysis of PtPDI Genes on the Chromosome
It was shown that the 42 PtPDI genes were distributed on 22 of 33 chromosomes of P. tricornutum (Fig. 3). There were 8 PtPDI genes located on chromosome 1. Moreover, the localization distance of PtPDI 15 and 18 genes was close on the chromosome. On chromosomes 4, 6, and 18, there were three PtPDI genes. In chromosome 1, there was a pair of tandem duplicate genes (PtPDI15 and PtPDI18 genes). On chromosome 6, there was a pair of PtPDI genes, named PtPDI42 and PtPDI37. There were 2 PtPDI genes on chromosomes 2, 3, 10, 13, 14, 17, 18 and 21. The location of the two PtPDI genes on chromosomes 14, 17, and 21 was adjacent. There was only one PtPDI gene on the other chromosomes.
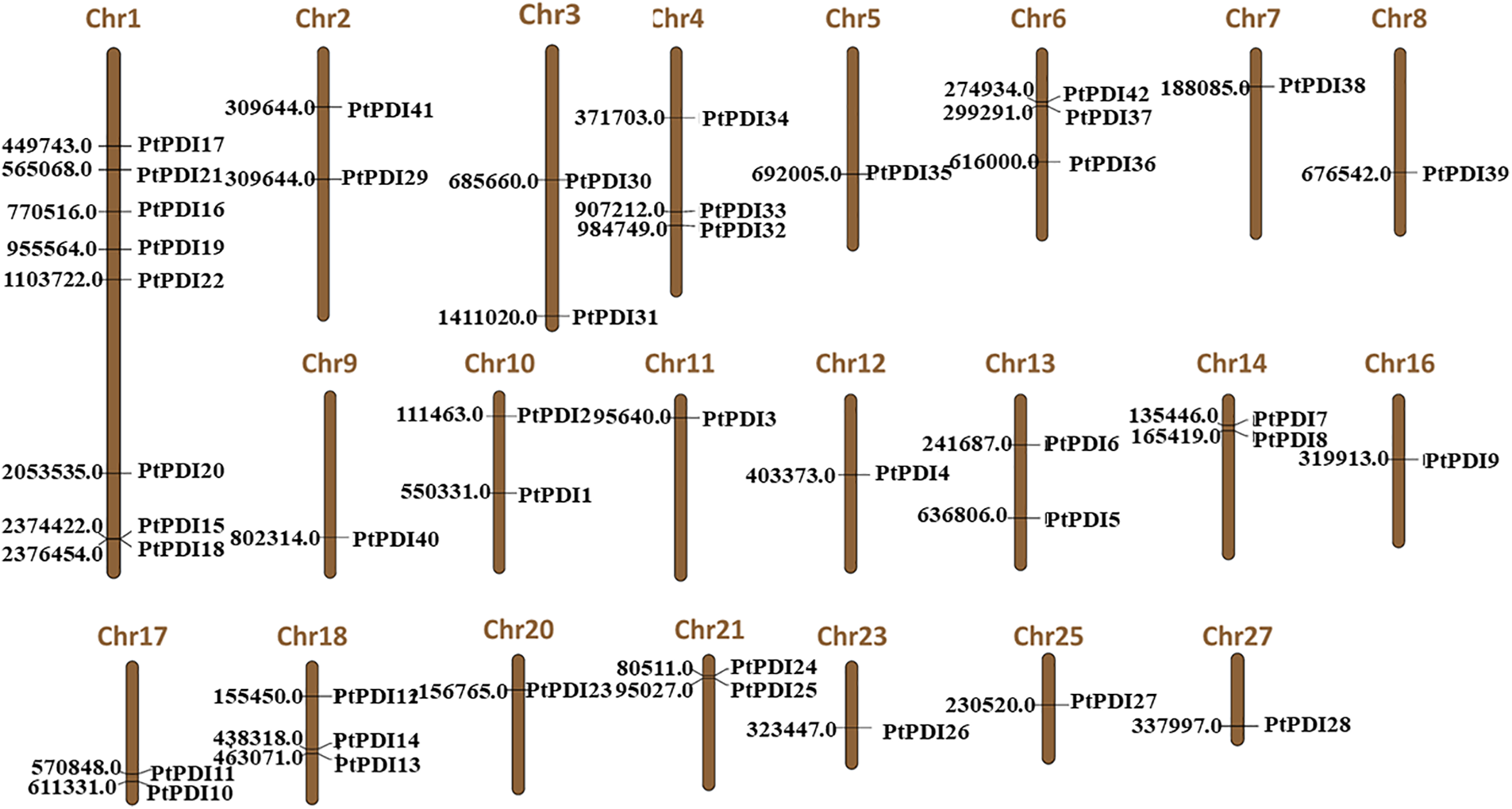
Figure 3: Localization of PtPDI genes on chromosomes. The number of each chromosome was shown on the top of the chromosome. The number on the left was the position of PtPDI genes on the chromosome
3.5 Expression Characteristics of PtPDI Genes under Tunicamycin Stress
To further analyze the expression of PtPDI genes, the transcription level of PtPDI genes was studied under the 0.3 μmol L−1 tunicamycin treatment. The expressional heatmap is shown in Fig. 4. Compared to the expression of PtPDIs in the wild type, 14 PtPDI genes were up-regulated. They were PtPDI1, PtPDI4-5, PtPDI7, PtPDI11-12, PtPDI15, PtPDI17, PtPDI22, PtPDI29, PtPDI35-37 and PtPDI42 genes. The fold change of upregulated PtPDI11 and PtPDI42 genes was more than 1.5. Conversely, 7 genes were significantly down-regulated after the treatment of 0.3 μmol L−1 tunicamycin. They were PtPDI6, PtPDI13, PtPDI16, PtPDI31, PtPDI33, PtPDI40 and PtPDI41 genes.
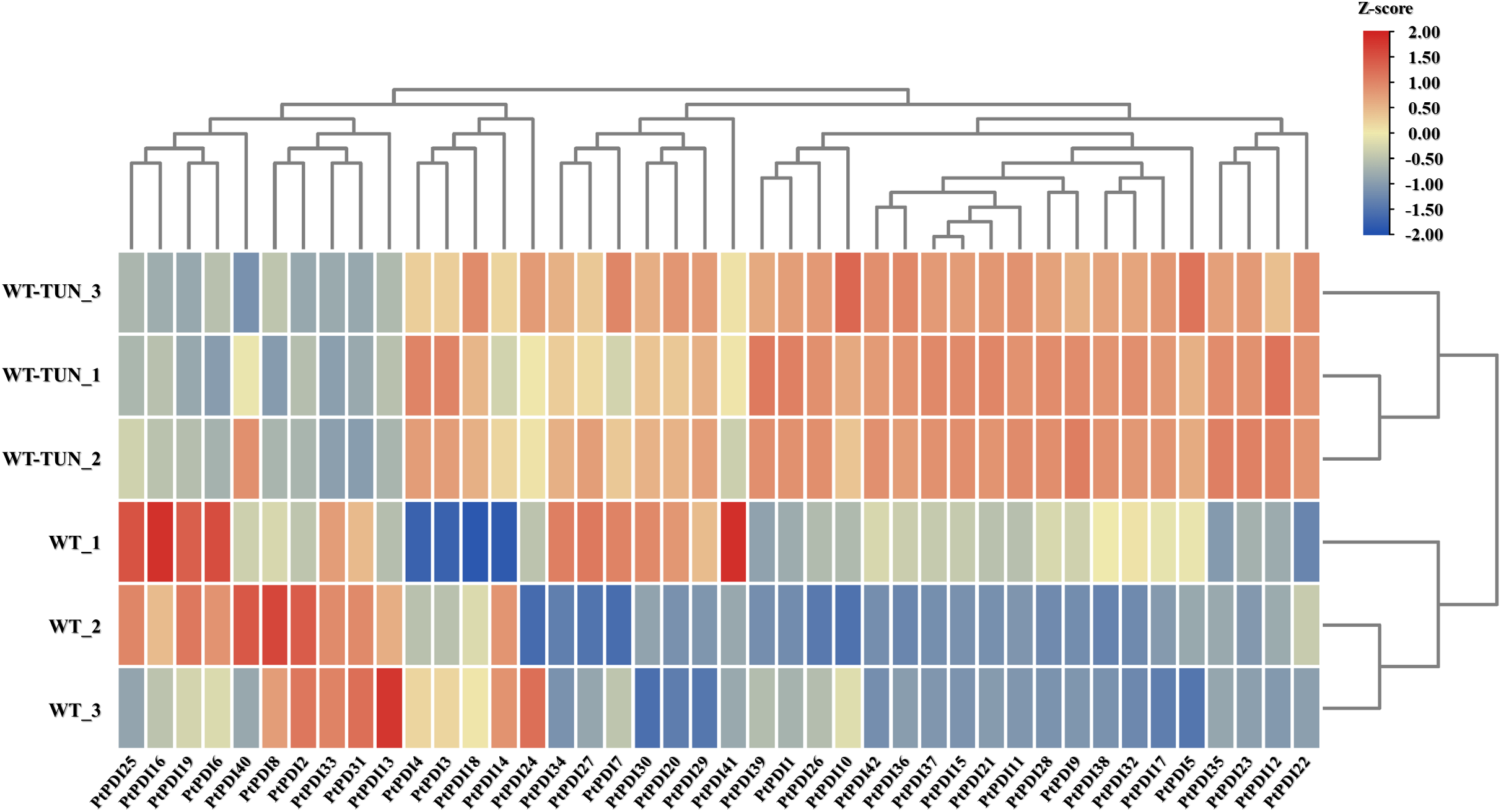
Figure 4: Transcription level of PtPDI genes under the treatment of 0.3 μmol L−1 tunicamycin in P. tricornutum. WT represents wild-type, and WT-TUN represents 0.3 μmol L−1 tunicamycin treatment. The expression of genes was processed by the Z-score method. Genes with |fold change| ≥ 1.5 and FDR < 0.01 (adjusted p-value, determined by the Benjamini and Hochberg multiple-testing correction implemented in the ‘p. adjust’ method of R) were defined as differentially expressed genes
In this study, 42 PtPDI candidates were identified from P. tricornutum. However, only 8 CrPDIs were identified from the genome of C. reinhardtii, and 10 OsPDIs were observed in Ostreococcus lucimarinus [21]. The different number of PDIs might be dependent on the algal species. Many more PtPDI in P. tricornutum were identified here, indicating that Diatom might need more PDIs to regulate the complex ERQC pathway. In high plants, PDI proteins were classified into nine classes, including PDI-A, PDI-S, PDI-E, PDI-B, PDI-C, PDI-M, PDI-D, and PDI-F. The difference between these 9 PDI classes was the number and the position of the Trx domain and the presence of additional domains [14]. Previous reports showed that PDI-A in Arabidopsis thaliana could bind an iron-sulfur (Fe−S) cluster. It was certificated that this binding scavenged the free iron to prevent the production of reactive oxygen [22]. PDI-M from plant Populus trichocarpa x Populus deltoides had the ability of oxidation-reduction regulation to activate the NADP-MDH [23]. This indicated that different kinds of PDIs played diverse roles. It was reported that PDI-F was found in microalgae, such as C. reinhardtii, Thalassiosira pseudonana, and Emiliania huxleyi [14,21,24]. Previous papers showed that three PDI-F proteins were found in diatom T. pseudonana [14,21]. Differently, 27 PtPDI-F subfamily was found in P. tricornutum in this study. These results enriched the PDI-F subfamily, and it was indicated that the number of PDIs was species-specific. At the same time, it was found that PDIs were divided into different groups. For example, in Brassica rapa ssp. Pekinensis, 32 PDIs were clustered into PDI-A (15.6%, 5/32), PDI-L (34.3%, 11/32), PDI-C (18.7%, 6/32), PDI-S (6.25%, 2/32), PDI-M (6.25%, 2/32), PDI-D (6.25%, 2/32), and PDI-E (12.5%, 4/32) [25]. In Arabidopsis thaliana, 21 PDIs were grouped into PDI-A (19%, 4/21), PDI-L (33.3%, 7/21), PDI-C (14.28%, 3/21), PDI-S (4.7%, 1/21), PDI-M (9.5%, 2/21), PDI-D (9.5%, 2/21) and PDI-E (9.5%, 2/21) [25]. 17 PDIs from Medicago truncatula belonged to PDI-L (29.4%, 5/17), PDI-S (23.5%, 4/17), PDI-C (17.6%, 3/17), PDI-B (5.8%, 1/17), PDI-A (5.8%, 1/17), PDI-E (5.8%, 3/17) [25,26]. 8 PDIs identified from C. reinhardtii were clustered into PDI-E (12.5%, 1/8), PDI-C (12.5%, 1/8), PDI-L (12.5%, 1/8), PDI-S (12.5%, 1/8), PDI-M (12.5%, 1/8), PDI-D (12.5%, 1/8) and PDI-F (25%, 2/8) [21]. In a previous paper, 7 PtPDIs were identified by blast search in the genome, including 1 PtPDI-B, 2 PtPDI-C, 1 PtPDI-E, and 4 PtPDI-F [14]. Many more PtPDIs (4 PtPDI-B, 4 PtPDI-C, 2 PtPDI-E, 27 PtPDI-F, 5 PtPDI-L) were identified via the Pfam number in this study, indicating a further method was important for the genome wild research. It was also proposed that more PtPDIs in P. tricornutum were needed to adapt to complex environments during secondary endosymbiosis.
Except PtPDI1, all the other PtPDIs contained motif 1 and motif 2 in P. tricornutum in this study, indicating that these two motifs were critical for the function of PtPDIs. Similarly, motif 1 and motif 2 were also highly conserved in rice [27]. Additionally, it was reported that motif 1 and motif 2 were responsible for the resistance of plant Panax ginseng to environmental stresses, such as drought [27,28]. Therefore, it was speculated that these two motifs in P. tricornutum might have a similar function under environmental pressures. In this study, 42 PtPDIs were located in different subcellular organelles, including cytoplasm, endoplasmic reticulum, Golgi apparatus, chloroplast, and nucleus. Consistent with the previous reports [14,29], ER-localized PtPDIs also contained ER retention signal, for example, PtPDI17 contained KEEL at the C-terminus, and PtPDI19 had EDEL motif. The subcellular localization of PtPDIs in C. reinhardtii was similar to that in P. tricornutum [30]. This suggested that PtPDIs were widely distributed into different subcellular organelles and played different functions in these diverse structures. However, all these putative localizations needed experiments to certify, such as via the observation of PtPDIs-eGFP fluorescence in confocal laser scanning microscopy. It was also found that PtPDIs played a vital role in forming disulfide bonds and modifying misfolded proteins in ER [31]. Moreover, it has been shown that PDIs could regulate thiol-disulfide switches and help repair the cytoskeleton in cell surface and cytosol [32]. Hence, ER-localized PtPDIs might also participate in the formation of disulfide bonds in proteins and regulate the correct folding of misfolded/unfolded proteins to maintain protein homeostasis in the cell. However, the exact function of these PtPDIs needs more experiments to clarify, such as by the overexpression, knock-out, and/or knockdown of genes in P. tricornutum.
A previous study reported that gene-duplicated events played an important role in the rapid expansion and evolution of gene families [33]. Gene-duplicated events usually happen among most plants and algae [34]. A pair of tandem duplicated genes (PtPDI15 and PtPDI18 genes) was observed in the genome of P. tricornutum. At the same time, it was observed that the gene structure, motifs, and homology of these two genes were highly conserved, indicating that these two PtPDIs might have similar functions in P. tricornutum. Compared to the reported homologous gene PDI-L5-1 (At1g04980) with two CXXC motifs in Arabidopsis thaliana [14,35], both PtPDI15 and PtPDI18 contained three CXXC motifs, indicating the structure specificity among different organisms. WP1 in yeast Rhodotorula gramini, a homologous gene of PtPDI15 and PtPDI18, could increase resistance and tolerance against a wide range of biotic and abiotic stresses, such as oxidative stress and heavy metals [34]. The homologous genes of PtPDI15 and PtPDI18 in the fungus Wallemia played an important role in osmoregulation under the hyperosmotic environment [36]. These studies suggested that PtPDI15 and PtPDI18 genes might also be important for the responses of P. tricornutum under the biotic and abiotic stresses, such as tunicamycin stress in this study.
It was known that PDIs play a crucial role in aiding the correct folding and assembly of nascent proteins in ERQC to maintain protein and cellular homeostasis. ERQC is an important pathway in ER (Fig. 5) [7]. Two ER-located PDI proteins were reported to be related to ER stress in Soybeans [15,37]. In this paper, PtPDI6, 17, 19, 20, 22, 35, and 37 with ER retention motif were predicted to be located in ER, therefore, it was reasonable to propose that these 7 PtPDIs might be involved in the ERQC pathway to resist ER stress and maintain cellular protein and cellular homeostasis. Moreover, it was found that the expression of these PtPDI genes was different under the stress of tunicamycin. Tunicamycin is an effective protein inhibitor, affecting the N-glycosylation modification of proteins and causing the accumulation of unfolded and/or misfolded proteins in the ER lumen [20,38]. PtPDI genes were differentially expressed to respond to the tunicamycin stress. Similar results were observed in tomato and Chlamydomonas [39]. The differential expression of PDI genes was dependent on the activation or repression of transcription factors. It was reported that bZIPs played a key role in the regulation of expression of PDI genes [29], therefore, it was speculated that the differentially expressed PtPDIs were regulated by different bZIP transcription factors. In this paper, it was suggested that the up-regulated PtPDIs (PtPDI1, PtPDI4-5, PtPDI7, PtPDI11-12, PtPDI15, PtPDI17, PtPDI22, PtPDI29, PtPDI35-37, and PtPDI42) might be involved in maintaining cellular protein homeostasis as positive regulatory factors. This suggestion was verified by that the homologous genes of PtPDI15 played important roles in cellular homeostasis, especially under oxidative, heavy metal, and hyperosmotic stresses [40]. The homologous gene Rb60/PDI1A of PtPDI22 in C. reinhardtii was also strongly upregulated in response to tunicamycin treatment, and it was also found that both Rb60/PDI1A and PtPDI22 were putatively targeted to ER [39,41]. The second homologous gene PDI6 in C. reinhardtii of PtPDI1 was also dramatically upregulated in tunicamycin-treated cells, indicating that this gene was involved in the cellular response to this stress [39]. The homologous relationship is shown in Supplementary Fig. 1. However, the downregulated PtPDIs (PtPDI6, PtPDI13, PtPDI16, PtPDI31, PtPDI33, PtPDI40 and PtPDI41) might act as negative regulatory factors. However, the negative regulatory mechanism of PtPDIs in P. tricornutum was still unknown. The homologous genes AtPDI2/5 of PtPDI6/16 showed the same differentially expressed profiles, indicating that these two PtPDIs might have synergistically functioned as AtPDI2/5 on the formation of a disulfide bond [42]. The differentially expressed PtPDIs could also be explained by that tunicamycin induced the unfolded protein response (UPR), and then UPR affected the transcription of PtPDIs [29]. In our recent paper, it was concluded that the differentially expressed PDI genes in C. reinhardtii might also play an important role in the response to Pb stress [21]. Anyway, it was proposed that these PtPDI genes might act as a catalyst of disulfide bond formation, helping stabilize protein folding’s tertiary and quaternary structures and maintaining cellular homeostasis during various stresses.
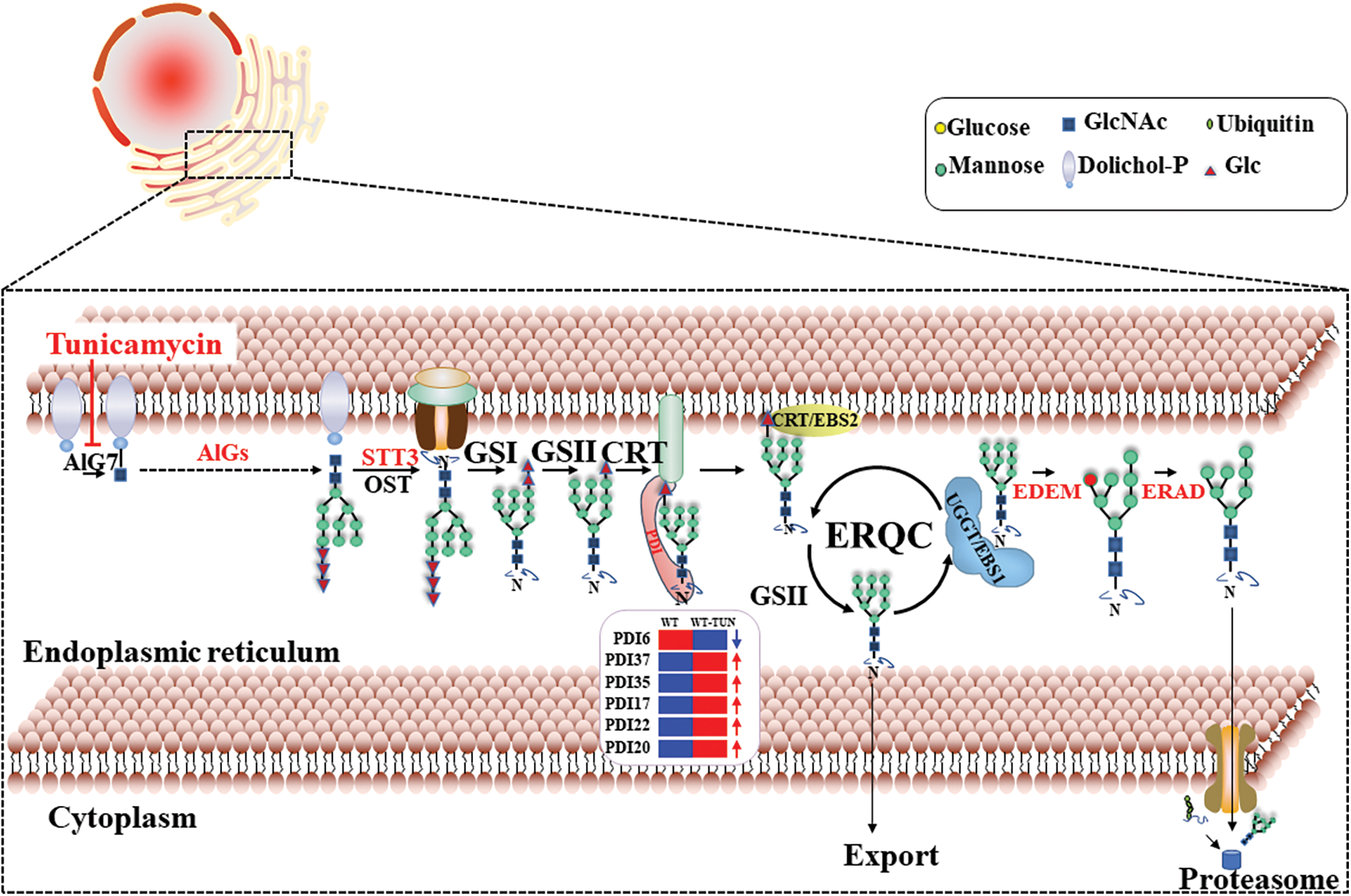
Figure 5: The working model of PtPDIs during the ERQC pathway in P. tricornutum. PtPDI6, 17, 19, 20, 22, 35, and 37 genes were predicted to be located in ER. The blue arrow means the downregulation and the red arrows mean the upregulation of PtPDI genes under the tunicamycin treatment
Acknowledgement: Thanks to the funding of Educational and Scientific Research Projects for Young and Middle-Aged Teachers in Fujian Province, Natural Science Foundation of Guangdong Province, the Program for University Innovation Team of Guangdong Province, National Natural Science Foundation of China, and the Innovation and Entrepreneurship Project of Shantou.
Funding Statement: Thanks to the funding of Educational and Scientific Research Projects for Young and Middle-Aged Teachers in Fujian Province (Grant Number: 2022JAT220693), Natural Science Foundation of Guangdong Province (Grant Number: 2022A1515012141), the Program for University Innovation Team of Guangdong Province (Grant Number: 2022KCXTD008), National Natural Science Foundation of China (92158201 and 42376001) and the Innovation and Entrepreneurship Project of Shantou (201112176541391).
Author Contributions: X. Liu designed the experiments and supervised the project. Y. Lin, H. Du, and Z. Ye performed the experiments and analysed the data. Y. Lin, H. Du, S. Wang, Z. Wang and X. Liu wrote and revised the manuscript. All authors discussed the results and implications and commented on the manuscript at all stages.
Availability of Data and Materials: All data generated or analysed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declares that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2023.044996.
References
1. Edkins AL, Boshoff A. General structural and functional features of molecular chaperones. Advan Experi Med Biol. 2021;1340:11–73. [Google Scholar]
2. Lee S, Tsai FT. Molecular chaperones in protein quality control. J Biochem Molecul Biol. 2005;38:259–65. [Google Scholar]
3. Winter J, Jakob U. Beyond transcription--new mechanisms for the regulation of molecular chaperones. Critical Reviews Biochem Molecul Biol. 2004;39:297–317. [Google Scholar]
4. Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. [Google Scholar] [PubMed]
5. Lia A, Gallo A, Marti L, Roversi P, Santino A. EFR-mediated innate immune response in Arabidopsis thaliana is a useful tool for identification of novel ERQC modulators. Genes. 2018;10:9–10. [Google Scholar]
6. Marti L, Lia A, Reca IB, Roversi P, Santino A et al. Planta preliminary screening of ER glycoprotein folding quality control (ERQC) modulators. IJMS. 2018;19:9–7. [Google Scholar]
7. Tax G, Lia A, Santino A, Roversi P. Modulation of ERQC and ERAD: A broad-spectrum spanner in the works of cancer cells? J Oncol. 2019;2019:8384913. [Google Scholar] [PubMed]
8. Hashimoto S, Okada K, Imaoka S. Interaction between bisphenol derivatives and protein disulphide isomerase (PDI) and inhibition of PDI functions: Requirement of chemical structure for binding to PDI. J Biochem. 2008;144:335–42. [Google Scholar] [PubMed]
9. Oliveira PVS, Garcia-Rosa S, Sachetto ATA, Moretti AIS, Debbas V et al. Protein disulfide isomerase plasma levels in healthy humans reveal proteomic signatures involved in contrasting endothelial phenotypes. Redox Biol. 2019;22:101142. [Google Scholar] [PubMed]
10. Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: Substrate interactions and functional properties. Embo Reports. 2005;6:28–32. [Google Scholar] [PubMed]
11. Laurindo FR, Pescatore LA, Fernandes Dde C. Protein disulfide isomerase in redox cell signaling and homeostasis. Free Radi Biol Med. 2012;52:1954–69. [Google Scholar] [PubMed]
12. Chen J, Du H, Liu Z, Li T, Du H et al. Endoplasmic reticulum-quality control pathway and endoplasmic reticulum-associated degradation mechanism regulate the N-glycoproteins and N-glycan structures in the diatom Phaeodactylum tricornutum. Microb Cell Fact. 2022;21:219. [Google Scholar] [PubMed]
13. Firrincieli A, Otillar R, Salamov A, Schmutz J, Khan Z et al. Genome sequence of the plant growth promoting endophytic yeast Rhodotorula graminis WP1. Fronti Microbiol. 2015;6:978. [Google Scholar] [PubMed]
14. Selles B, Jacquot JP, Rouhier N. Comparative genomic study of protein disulfide isomerases from photosynthetic organisms. Genom. 2011;97:37–50. [Google Scholar]
15. Okuda A, Matsusaki M, Masuda T, Urade R. Identification and characterization of GmPDIL7, a soybean ER membrane-bound protein disulfide isomerase family protein. Febs J. 2017;284:414–28. [Google Scholar] [PubMed]
16. Wai AH, Waseem M, Khan A, Nath UK, Lee DJ et al. Genome-wide identification and expression profiling of the PDI gene family reveals their probable involvement in abiotic stress tolerance in tomato (Solanum lycopersicum L.). Genes. 2020;12:6–7. [Google Scholar]
17. Araie H, Shiraiwa Y. Selenium utilization strategy by microalgae. Molecules. 2009;14:4880–91. [Google Scholar] [PubMed]
18. Hempel F, Lau J, Klingl A, Maier UG. Algae as protein factories: Expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS One. 2011;6:e28424. [Google Scholar] [PubMed]
19. Faheem M, Arshad M, Xiao JIN, Wang H, Li Y et al. Cloning and functional assessment of the protein disulfide isomerase (PDI-V) gene and its promoter sequences from Haynaldia villosa. Turkish J Bot. 2018;42:9–10. [Google Scholar]
20. Xiao H, Smeekens JM, Wu R. Quantification of tunicamycin-induced protein expression and N-glycosylation changes in yeast. Analyst. 2016;141:3737–45. [Google Scholar] [PubMed]
21. Du H, Zheng C, Aslam M, Xie X, Wang W et al. Endoplasmic reticulum-mediated protein quality control and endoplasmic reticulum-associated degradation pathway explain the reduction of N-glycoprotein level under the lead stress. Front Plant Sci. 2020;11:598552. [Google Scholar] [PubMed]
22. Remelli W, Santabarbara S, Carbonera D, Bonomi F, Ceriotti A et al. Iron binding properties of recombinant class a protein disulfide isomerase from Arabidopsis thaliana. Biochem. 2017;56:2116–25. [Google Scholar]
23. Selles B, Zannini F, Couturier J, Jacquot JP, Rouhier N. Atypical protein disulfide isomerases (PDIComparison of the molecular and catalytic properties of poplar PDI-A and PDI-M with PDI-L1A. PLoS One. 2017;12:e0174753. [Google Scholar] [PubMed]
24. Obata T, Shiraiwa Y. A novel eukaryotic selenoprotein in the haptophyte alga Emiliania huxleyi. J Biol Chem. 2005;280:18462–8. [Google Scholar] [PubMed]
25. Kayum MA, Park JI, Nath UK, Saha G, Biswas MK et al. Genome-wide characterization and expression profiling of PDI family gene reveals function as abiotic and biotic stress tolerance in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2017;18:885. [Google Scholar]
26. Meng Z, Zhao Y, Liu L, Du X. Genome-wide characterization of the PDI gene family in Medicago truncatula and their roles in response to endoplasmic reticulum stress. Genome. 2021;64:599–614. [Google Scholar] [PubMed]
27. Zhang Q, Shen L, Ren D, Hu J, Zhu L et al. Characterization of the CRM gene family and elucidating the function of OsCFM2 in rice. Biomole. 2020;10:6–7. [Google Scholar]
28. Bae KS, Rahimi S, Kim YJ, Devi BSR, Khorolragchaa A et al. Molecular characterization of lipoxygenase genes and their expression analysis against biotic and abiotic stresses in Panax ginseng. Eur J Plant Pathol. 2016;145:331–43. [Google Scholar]
29. Gupta D, Tuteja N. Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal Behav. 2011;6:232–6. [Google Scholar] [PubMed]
30. Reyes S, Moreno A. The endoplasmic reticulum role in the plant response to abiotic stress. Front Plant Sci. 2021;12:1–7. [Google Scholar]
31. Määttänen P, Gehring K, Bergeron JJ, Thomas DY. Protein quality control in the ER: The recognition of misfolded proteins. Seminars Cell Developl Biol. 2010;21:500–11. [Google Scholar]
32. Soares Moretti AI, Martins Laurindo FR. Protein disulfide isomerases: Redox connections in and out of the endoplasmic reticulum. Arch Biochem Biophys. 2017;617:106–19. [Google Scholar] [PubMed]
33. Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Ann Rev Plant Biol. 2009;60:433–53. [Google Scholar] [PubMed]
34. Qiao X, Li Q, Yin H, Qi K, Li L et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 201920:38. [Google Scholar] [PubMed]
35. Zhou Y, Fan F, Han Y, Lu D. Arabidopsis PDI11 interacts with lectin molecular chaperons calreticulin 1 and 2 through its D domain. Biochem Biophys Research Comm. 2022;588:55–60. [Google Scholar]
36. Padamsee M, Kumar TK, Riley R, Binder M, Boyd A et al. The genome of the xerotolerant mold Wallemia sebi reveals adaptations to osmotic stress and suggests cryptic sexual reproduction. Fungal Genet Biol. 2012;49:217–26. [Google Scholar] [PubMed]
37. Okuda A, Matsusaki M, Masuda T, Morishima K, Sato N et al. A novel soybean protein disulphide isomerase family protein possesses dithiol oxidation activity: Identification and characterization of GmPDIL6. The J Biochem. 2020;168:393–405. [Google Scholar] [PubMed]
38. McCormick LM, Urade R, Arakaki Y, Schwartz AL, Bu G. Independent and cooperative roles of N-glycans and molecular chaperones in the folding and disulfide bond formation of the low-density lipoprotein (LDL) receptor-related protein. Biochem. 2005;44:5794–803. [Google Scholar]
39. Pérez-Martín M, Pérez-Pérez ME, Lemaire SD, Crespo JL. Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol. 2014;166:997–1008. [Google Scholar]
40. Zhao C, Shono M, Sun A, Yi S, Li M et al. Constitutive expression of an endoplasmic reticulum small heat shock protein alleviates endoplasmic reticulum stress in transgenic tomato. J Plant Physiol. 2007;164:835–41. [Google Scholar] [PubMed]
41. Levitan A, Trebitsh T, Kiss V, Pereg Y, Dangoor I et al. Dual targeting of the protein disulfide isomerase RB60 to the chloroplast and the endoplasmic reticulum. PNAS. 2005;102:6225–30. [Google Scholar] [PubMed]
42. Fan F, Zhang Q, Zhang Y, Huang G, Liang X et al. Two protein disulfide isomerase subgroups work synergistically in catalyzing oxidative protein folding. Plant Physiol. 2022;188:241–54. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools