 Open Access
Open Access
ARTICLE
Effects of Heterologously Overexpressing PIP5K-Family Genes in Arabidopsis on Inflorescence Development
1 College of Life Science and Food Engineering, Inner Mongolia Minzu University, Tongliao, 028000, China
2 Key Laboratory of Castor Breeding of the State Ethnic Affairs Commission, Inner Mongolia Minzu University, Tongliao, 028000, China
3 Inner Mongolia Industrial Engineering Research Center of Universities for Castor, Inner Mongolia Minzu University, Tongliao, 028043, China
4 Inner Mongolia Key Laboratory of Castor Breeding and Comprehensive Utilization, Inner Mongolia Minzu University, Tongliao, 028000, China
5 Inner Mongolia Engineering Research Center of Industrial Technology Innovation of Castor, Inner Mongolia Minzu University, Tongliao, 028000, China
* Corresponding Author: Fenglan Huang. Email:
# Mingda Yin and Rui Luo contributed equally to this work and shared co-first authorship
(This article belongs to the Special Issue: Advances in Molecular Genetics and Physiology towards a Better Understanding of Agricultural Crop Plants)
Phyton-International Journal of Experimental Botany 2024, 93(1), 97-117. https://doi.org/10.32604/phyton.2023.031228
Received 23 May 2023; Accepted 21 November 2023; Issue published 26 January 2024
Abstract
Castor is one of the top 10 oil crops in the world and has extremely valuable uses. Castor inflorescences directly affect yield, so the study of inflorescence development is very important in increasing castor yield. Our previous studies have shown that the PIP5K gene family (PIP5Ks) is associated with inflorescence development. In this study, to determine the function of each PIP5K gene in castor, a female Lm-type castor line, aLmAB2, was used to determine the relative expression levels of the PIP5Ks in castor inflorescences. Six PIP5K genes were heterologously overexpressed in Arabidopsis thaliana, the relative expression of each gene and the effect on plants was determined in A. thaliana, and the relationships among the PIP5Ks in castor were inferred. The expression levels of the PIP5Ks in the female Lm-type castor line aLmAB2 were analyzed. The relative expression levels of the PIP5K9 and PIP5K11 genes were high (p < 0.05) in isofemale inflorescences, and those of PIP5K1, PIP5K2, PIP5K6, and PIP5K8 were high (p < 0.05) in female inflorescences but low (p < 0.05) in bisexual inflorescences. The PIP5Ks were heterologously overexpressed in A. thaliana, and T3-generation plants with stable genetic resistance, i.e., AT-PIP5K+ plants (AT-PIP5K1+, AT-PIP5K2+, AT-PIP5K6+, AT-PIP5K8+, AT-PIP5K9+, and AT-PIP5K11+ plants), were obtained. Biological tests of the AT-PIP5K+ plants showed that the growth of the main stem was significantly delayed in AT-PIP5K+ plants compared with Columbia wild-type (WT) A. thaliana plants; the PIP5K1 and PIP5K2 genes promoted lateral stem growth and flower and silique development; and the PIP5K6, PIP5K8, PIP5K9 and PIP5K11 genes inhibited lateral stem growth and flower and silique development. The correlations among PIP5Ks in castor suggest that there may be a synergistic relationship among PIP5K1, PIP5K2, and PIP5K6 in castor inflorescences, and PIP5K8, PIP5K9, and PIP5K11 are complementary to the other three genes.Keywords
In plants, the phosphatidylinositol (PI) signal transduction pathway is involved in regulating seed germination, growth and development, reproduction, and senescence. Phosphatidylinositol phosphate 5-kinase (PIP5K) is a key enzyme in the PI signal transduction pathway that can catalyze the phosphorylation of PI-4-phosphate (PI4P) and synthesize PI 4,5-biphosphate (PI(4,5)P2) [1]. PI(4,5)P2 is a key component in the generation of second messengers via a variety of pathways (including phosphoinositide phospholipase C, phosphoinositide 3-kinase and phospholipase D (PLD)) [2]. PI(4,5)P2 is involved in regulating the actin cytoskeleton and vesicle transport [3].
Castor is an important oil crop, and the traits of its inflorescences directly affect the entire flowering to fruiting process and determine yield [4–6]. The inflorescence development of castor is extremely complex, and there are two main situations: one is the majority of castor female lines in the world, in which the plants appear monoecious (also called female, with all female flowers on each inflorescence axis of the plant) and hermaphroditic (also called monoecious, with male flowers on the base and female flowers on the top of each inflorescence axis of the plant) inflorescences of two types [7]. The other is the castor Lm-type female line (marker female line) with independent intellectual property rights, in which three types of inflorescences, monoecious, marker female (plants bearing all female flowers on each inflorescence axis, together with willow-like functional leaves) and bisexual, occur in the plants [8]. The contents of endogenous indole-3yl-acetic acid (IAA) and abscisic acid (ABA) in the apical buds also change significantly.
Our previous studies have shown that PIP5K family genes (PIP5Ks) are associated with the development of castor inflorescences. Therefore, this study used the castor PIP5Ks as study subjects and analyzed their expression levels at different developmental stages in different inflorescence types in the female castor line aLmAB2, studied the functions of the PIP5Ks through their heterologous overexpression in Arabidopsis thaliana, and analyzed the correlations among PIP5Ks in castor. This study lays the foundation to study the role of PIP5Ks in inflorescence development in the female line aLmAB2 and is of great significance in the breeding of high-yield castor varieties.
The female Lm-type castor line (aLmAB2), Columbia wild-type (WT) A. thaliana, and the plant overexpression vector pCAMBIA1390 were provided by the Key Laboratory of Castor Breeding of the State Ethnic Affairs Commission of the Inner Mongolia University for the Nationalities. The full-length coding sequence (CDS) of each PIP5K gene (PIP5K1, PIP5K2, PIP5K6, PIP5K8, PIP5K9, and PIP5K11) was synthesized by Beijing Qingke Xinye Biotechnology Co., Ltd. (China); polymerase chain reaction (PCR) product purification kits, plant genomic DNA extraction kits, plant tissue total RNA extraction kits, Escherichia coli DH5α, and competent Agrobacterium (GV3101) cells were purchased from Beijing Zoman Biotechnology Co., Ltd. (China); and complementary DNA (cDNA) synthesis kits, microcolumn concentrated gel DNA extraction kits, fluorescent SYBR dye, KpnI restriction endonuclease, and BamHI restriction endonuclease were purchased from Beijing Baori Medical Biotechnology Beijing Co., Ltd. (China). The subcellular localization expression vector pCAMBIA1301-PIP5Ks was provided by our laboratory.
2.2.1 Analysis of the Expression Levels of PIP5Ks in aLmAB2
A total RNA extraction kit was used to extract total RNA from isofemale, female, and bisexual aLmAB2 inflorescences at the four-leaf, five-leaf, main stem spike flowering and secondary branch flowering stages (a total of 12 samples) to synthesize cDNA via reverse transcription using a cDNA synthesis kit. Real-time quantitative PCR (RT-qPCR) was performed using the cDNA as the template and SYBR Green as the fluorescent dye. The primer sequences are shown in the article by one of the authors of this paper, Liang [9]. The 12 samples were subjected to RT-qPCR using a 20 μL reaction system (2 μL cDNA, 10 μL SYBR Premix Ex Taq, 0.4 μL ROX Reference Dye II, 0.4 μL primer S, 0.4 μL primer X, and 6.8 μL ddH2O). The expression levels of the six genes were analyzed using 18S as an internal reference. The experiment was repeated three times. After data processing, the expression levels of the target genes in different samples were analyzed.
2.2.2 Heterologous Overexpression of the PIP5Ks in A. thaliana
Construction of the Heterologous Overexpression Vector pCAMBIA1390-PIP5Ks
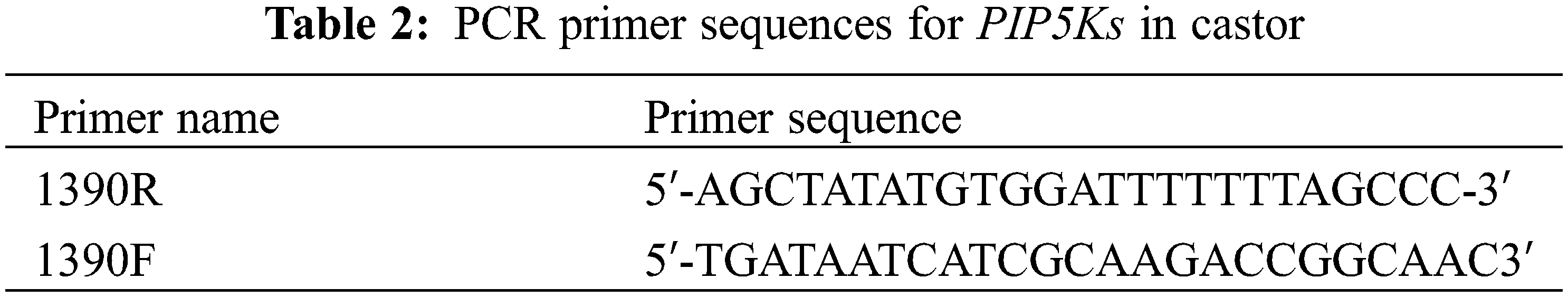
Using SnapGene software (2.3.2), two pairs of primers (PKsR1 and PKsF1; PKsR2 and PKsF2) were designed for the two ends of PIP5K1, PIP5K2, PIP5K6, PIP5K8, and PIP5K9, and the primer sequences are shown in Table 1 (of the six PIP5K genes, the PIP5K11 gene has KpnI and BamHI restriction sites at both ends, so there was no need to manually add KpnI and BamHI restriction sites when designing the primers). The extracted subcellular localization expression vector pCAMBIA1301-PIP5Ks was used as the template, and the designed primers were used for PCR (PCR system: 2 μL of recombinant plasmid, 1 μL of PKsR F, 1 μL of PKsF R, 10 μL of PrimeSTAR buffer, 4 μL of dNTP mixture, 0.5 μL PrimeSTAR HS DNA polymerase, and 31.5 μL of ddH2O). The PCR products were subjected to agarose gel electrophoresis and then purified and extracted using a gel extraction kit. After extraction, digestion was performed at a ratio of 1:1 to obtain two sticky ends, KpnI and BamHI. The overexpression vector pCAMBIA1390 was extracted using a reagent kit, double digestion was performed with KpnI and BamHI, and the double digestion products were purified and extracted (double digestion system: 40 μL plasmid, 8 μL KpnI, 8 μL BamHI, 5 μL 10× K. buffer, and 39 μL ddH2O). The six genes were ligated into the overexpression vector pCAMBIA1390 overnight at 16°C using T4 DNA ligase (ligation system: 5 μL target gene, 2 μL vector, 1 μL 10× T4 DNA ligase buffer, 1 μL T4 DNA ligase, and 1 μL ddH2O), and the ligation product was transformed into E. coli DH5α. PCR of the transformed products was performed on the bacteria in liquid medium. The PCR primer sequences are shown in Table 2. After validation, the obtained recombinant vector pCAMBIA1390-PIP5Ks was transformed into competent Agrobacterium GV3101 cells.

Genetic Transformation of WT Columbia A. thaliana with Heterologous Overexpression Vectors
Using the method of Jiang et al. [10], the WT A. thaliana was genetically transformed, and the obtained T1 generation A. thaliana seedlings were transplanted until they could be harvested. By the T3 generation, six types of resistant Columbia WT A. thaliana plants heterologously overexpressing PIP5Ks (AT-PIP5K+ plants) were obtained.
Molecular Characterization of A. thaliana Plants with Heterologous Overexpression
Genomic DNA from AT-PIP5K+ and WT A. thaliana leaves was extracted using a genomic DNA extraction kit.
PCR detection: Genomic DNA from T3-generation AT-PIP5K+ and WT A. thaliana leaves were used as templates, the PCR product from the recombinant heterologous overexpression pCAMBIA1390-PIP5K vectors were used as positive controls, and the PCR product from genomic DNA from WT A. thaliana leaves was used as a negative control. Each sample was subjected to PCR detection using a 50 μL reaction system (2 μL of recombinant plasmid, 1 μL of PKsR F, 1 μL of PKsF R, 10 μL of PrimeSTAR buffer, 4 μL of dNTP mixture, 0.5 μL of PrimeSTAR HS DNA polymerase, and 31.5 μL of ddH2O). The reaction conditions were as follows: predenaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min 30 s; extension at 72°C for 5 min; and storage at 4°C. The PCR products were detected and observed via agarose gel electrophoresis.
RT-qPCR detection: Total RNA was extracted from inflorescences from T3-generation AT-PIP5K+ and Columbia WT A. thaliana plants. The RNA was diluted to approximately 500 ng/μL, the integrity of the obtained RNA was detected via agarose gel electrophoresis, and RNA images were obtained using a gel imaging system. Total RNA from AT-PIP5K+ and WT A. thaliana inflorescences was reverse transcribed into cDNA according to the cDNA synthesis kit instructions. The PIP5K messenger RNA (mRNA) sequences in the National Center for Biotechnology Information (NCBI) database (GenBank accession number: PIP5K 1 (LOC8274135), PIP5K 2 (LOC8287614), PIP5K 6 (LOC8270872), PIP5K 8 (LOC8285184), PIP5K 9 (LOC8283966), PIP5K 11 (LOC8286714)) were searched, and RT-qPCR primers were designed. The primer sequences for each gene, shown in Table 3, were determined based on preliminary experiments using the same design method. SPSS 19.0 software was used for statistical analysis of the data, and GraphPad was used for data processing and plotting. Independent samples t tests were used to compare the averages for significance analyses, and the obtained results are listed in a histogram of relative expression levels.

Biological Characterization of A. thaliana Plants with Overexpression
WT and AT-PIP5K+ plants that grew to the four-leaf stage were transplanted and grown under the same conditions. For each type of heterologous overexpression and control plant, 30 pots were grown, with one plant per pot, for a total of 30 plants. Representative AT-PIP5K+ and WT plants were selected for image collection. The phenotypic traits of each type of A. thaliana plant were statistically analyzed, and the averages are listed in a table for analysis.
Seeds from WT and T3-generation AT-PIP5K+ plants with stable inheritance were suspended in liquid Murashige and Skoog (MS) medium and inoculated into germination medium. The seeds were cultured under 16 h of light and 8 h of dark per day at 22°C. After 25 d, robust four-leaf A. thaliana seedlings were picked for transplantation.
The T3-generation AT-PIP5K+ plants with stable inheritance were biologically characterized, including studying their development, height, and growth status (the number of florets, the bolting time of the main stem, the number of lateral stems, and the numbers of basal leaves and cauline leaves).
Correlation Analysis of PIP5Ks in Castor
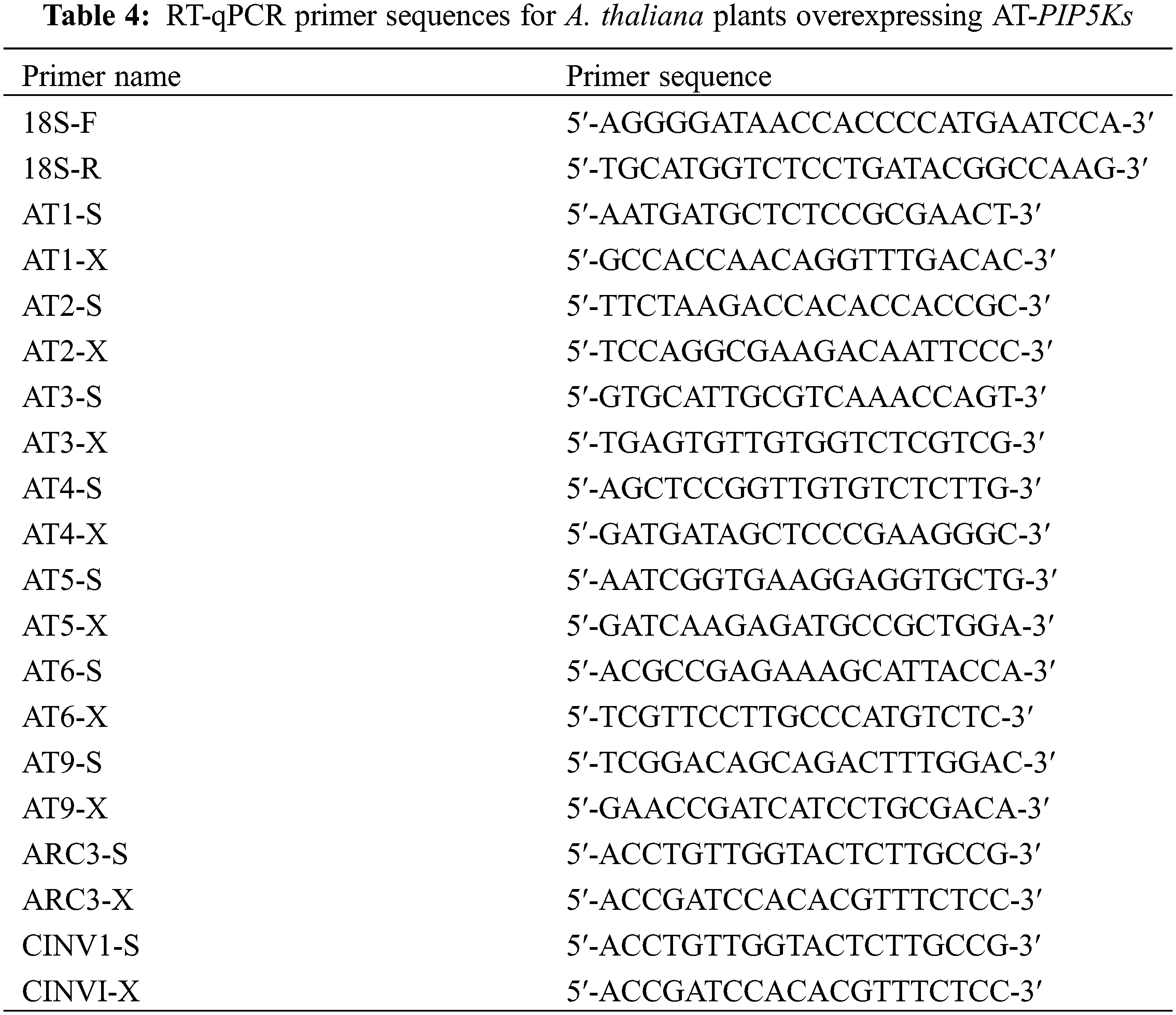
RT-qPCR was performed on heterologously overexpressing T3-generation resistant AT-PIP5K+ A. thaliana plants (six types). The mRNA sequences of the A. thaliana PIP5Ks were queried separately in the NCBI database to determine the primer sequences for each gene, as shown in Table 4. The same design method and experimental procedure were used. RT-qPCR was used to determine the changes in the expression levels of the other five PIP5K genes in A. thaliana plants caused by the overexpression of one PIP5K gene. Based on sequence homology analysis, the castor PIP5K1 gene corresponds to the A. thaliana PIP5K1 gene, the castor PIP5K2 gene corresponds to the A. thaliana PIP5K2 and PIP5K3 genes, the castor PIP5K6 gene corresponds to the A. thaliana PIP5K4 and PIP5K6 genes, the castor PIP5K8 gene corresponds to the A. thaliana PIP5K5 gene, the castor PIP5K9 gene corresponds to the A. thaliana accumulation and replication of chloroplasts 3 (ARC3) and cytosolic invertase 1 (CINV1) genes, and the castor PIP5K11 gene corresponds to the A. thaliana PIP5K 9 gene.
SPSS 19.0 software was used for the statistical analysis of the data, and GraphPad was used for data processing and plotting. Independent samples t tests of the means were used for significance analyses, and the obtained results are listed in a histogram of relative expression levels.
3.1 The Expression Levels of PIP5Ks in Castor Inflorescences
3.1.1 Differential Expression Levels of PIP5Ks in Plants with Different Inflorescence Types at Different Flowering Stages
The PIP5K RT-qPCR results has been shown in Fig. 1 and include several observations.
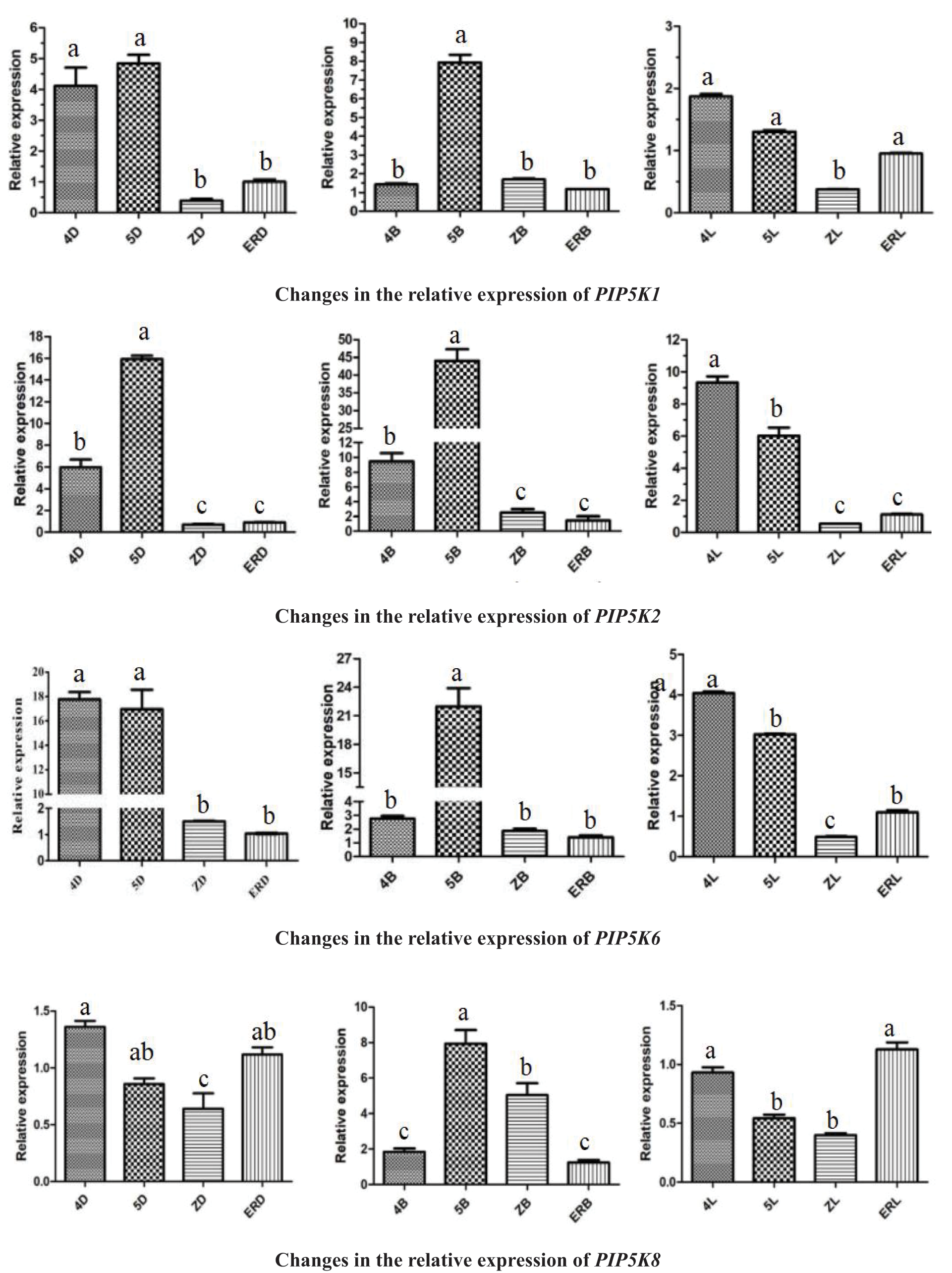
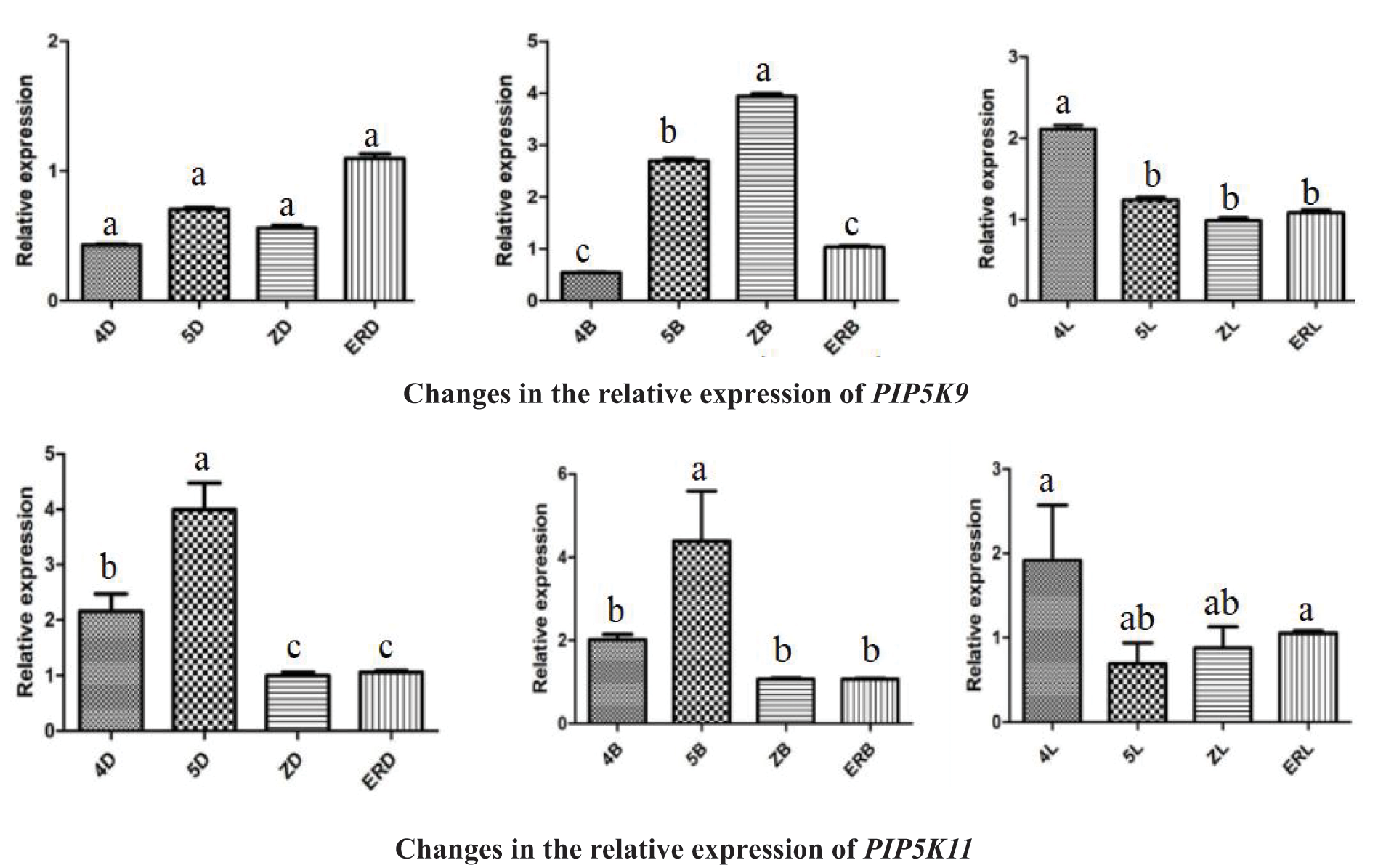
Figure 1: Differential expression levels of PIP5Ks at different flowering stages for the three inflorescence types
Note: 4D: isofemale four-leaf stage; 5D: isofemale five-leaf stage; ZD: isofemale main stem spike flowering stage; ERD: isofemale secondary branch flowering stage; 4B: female four-leaf stage; 5B: female five-leaf stage; ZB: female main stem spike flowering stage; ERB: female secondary branch flowering stage; 4L: bisexual four-leaf stage; 5L: bisexual five-leaf stage; ZL: bisexual main stem spike flowering stage; ERL: bisexual secondary branch flowering period. Different letters indicated different differences, and the significance level was p < 0.05.
The relative expression level of PIP5K1 was high (p < 0.05) at the five-leaf stage and low (p < 0.05) at the main stem spike flowering stage in all three inflorescence types, namely, isofemale, female and bisexual inflorescences. At the five-leaf stage, it was the highest (p < 0.05) in the female inflorescence, and at the main stem spike flowering stage, it was lowest (p < 0.05) in the bisexual inflorescence. The highest relative expression level was approximately 16 times the lowest relative expression level.
The relative expression level of PIP5K2 was high (p < 0.05) at the five-leaf stage and low (p < 0.05) at the main stem spike flowering stage in all three inflorescence types. At the five-leaf stage, it was highest (p < 0.05) in the female inflorescence, and at the main stem spike flowering stage, it was lowest (p < 0.05) in the bisexual inflorescence. The highest relative expression level was approximately 80 times the lowest relative expression level.
The relative expression level of PIP5K6 was high (p < 0.05) at the four-leaf and five-leaf stages and low (p < 0.05) at the main stem spike flowering and secondary branch flowering stages in all three inflorescence types. At the five-leaf stage, it was highest (p < 0.05) in the female inflorescence, and at the main stem spike flowering stage, it was lowest (p < 0.05) in the bisexual inflorescence. The highest relative expression level was approximately 46 times the lowest relative expression level.
The relative expression level of PIP5K8 was high (p < 0.05) at the five-leaf and main stem spike flowering stages in the female inflorescence and at the four-leaf and secondary branch flowering stages in the isofemale and bisexual inflorescences, and it was low (p < 0.05) at the four-leaf and secondary branch flowering stages in the female inflorescence and at the five-leaf and main stem spike flowering stages in the isofemale and bisexual inflorescences. It was the highest (p < 0.05) at the five-leaf stage in the female inflorescence, and it was the lowest (p < 0.05) at the main stem spike flowering stage in the bisexual inflorescence. The highest relative expression level was approximately 16 times the lowest relative expression level.
The relative expression level of PIP5K9 was high (p < 0.05) at the four-leaf, five-leaf, main stem spike flowering and secondary branch flowering stages in the isofemale and bisexual inflorescences and was low (p < 0.05) at the five-leaf and main stem spike flowering stages in all three inflorescence types. It was the highest (p < 0.05) at the main stem spike flowering stage in the female inflorescence and was the lowest (p < 0.05) at the four-leaf stage in the isofemale inflorescence. The highest relative expression level was approximately eight times the lowest relative expression level.
The relative expression level of PIP5K11 was high (p < 0.05) at the five-leaf stage and low (p < 0.05) at the main stem spike flowering and secondary branch flowering stages in all three inflorescence types. It was the highest (p < 0.05) at the five-leaf stage in the female inflorescence and was the lowest (p < 0.05) at the five-leaf stage in the bisexual inflorescence. The highest relative expression level was approximately eight times the lowest relative expression level.
The analysis of the above results showed that in aLmAB2 plants with different inflorescence types, the expression levels of the six PIP5K genes were significantly upregulated and downregulated at different developmental stages. The relative expression levels of PIP5K1, PIP5K2, PIP5K6, and PIP5K8 were the highest (p < 0.05) at the five-leaf stage in the female inflorescence, and those of PIP5K1, PIP5K2, and PIP5K6 were the lowest (p < 0.05) at the main stem spike flowering stage in the bisexual inflorescence.
3.1.2 Differential Expression Levels of PIP5Ks in the Same Inflorescence Types at the Same Flowering Stage in Castor
The RT-qPCR results of the relative expression levels of PIP5Ks in the same inflorescence type at the same flowering stage of this part of castor were in article [9] it and yields several observations.
At the four-leaf stage, the relative expression levels of PIP5K1 and PIP5K2 were high (p < 0.05) for all three inflorescence types, the relative expression levels of PIP5K8 and PIP5K9 were medium (p < 0.05), and the relative expression levels of PIP5K6 and PIP5K11 were low (p < 0.05). The relative expression of PIP5K1 was highest (p < 0.05) in the isofemale inflorescence, and the relative expression of PIP5K11 was lowest (p < 0.05) in the bisexual inflorescence.
At the five-leaf stage, the relative expression levels of PIP5K1 and PIP5K2 were high (p < 0.05) for all three inflorescence types, the relative expression levels of PIP5K8 and PIP5K9 were medium (p < 0.05), and the relative expression levels of PIP5K6 and PIP5K11 were low (p < 0.05). The relative expression of PIP5K2 was highest (p < 0.05) in the bisexual inflorescence, and the relative expression of PIP5K11 was lowest (p < 0.05) in the isofemale inflorescence.
At the main stem spike flowering stage, the relative expression levels of PIP5K1, PIP5K8, and PIP5K9 were high (p < 0.05) for all three inflorescence types, the relative expression of PIP5K2 was medium (p < 0.05), and the relative expression levels of PIP5K6 and PIP5K11 were low (p < 0.05). The relative expression of PIP5K9 was highest (p < 0.05) in the female inflorescence, and the relative expression of PIP5K11 was lowest (p < 0.05) in the female inflorescence.
At the secondary branch flowering stage, the relative expression levels of PIP5K1, PIP5K8, and PIP5K9 were high (p < 0.05) for all three inflorescence types, the relative expression of PIP5K2 was medium (p < 0.05), and the relative expression levels of PIP5K6 and PIP5K11 were low (p < 0.05). The relative expression of PIP5K9 was highest (p < 0.05) in the isofemale inflorescence, and the relative expression of PIP5K11 was lowest (p < 0.05) in the female inflorescence.
In summary, for all three inflorescence types, the expression levels of PIP5K1 and PIP5K2 were relatively high (p < 0.05) at the four-leaf and five-leaf stages; the expression levels of PIP5K1, PIP5K8, and PIP5K9 were relatively high (p < 0.05) at the main stem spike flowering and secondary branch flowering stages; and the expression of PIP5K11 was the lowest (p < 0.05) at all four stages.
3.2 Results of Genetically Transforming A. thaliana Plants Using Heterologous Overexpression Vectors
3.2.1 Obtaining Resistant Heterologous Overexpression A. thaliana Plants
Fig. 2 showed the process of resistance screening to obtain T3-generation AT-PIP5K+ plants. Vigorous WT A. thaliana plants at the four-leaf stage in medium without MS showed well-developed root systems, and there were no withering phenomena, such as yellowing or transparency. A. thaliana plants grown to the two-leaf stage in MS medium supplemented with hygromycin started to yellow and ultimately withered. The transformed A. thaliana plants were screened for resistance. When screening the T1 generation, only 1–2 vigorously growing plants per petri dish reached the four-leaf stage, and most of the plants screened in the T2 generation reached the four-leaf stage. During the screening of the T3 generation, vigorously growing plants at the four-leaf stage were obtained. Resistant overexpressing A. thaliana plants were shown in Fig. 2. The T3-generation AT-PIP5K+ seeds were germinated to obtain A. thaliana plants that were vigorously growing at the four-leaf stage. After transplanting into small pots and growing for 10 d without removing the main stem, the WT plants were approximately 10 cm tall, the resistant AT-PIP5K1+ and AT-PIP5K2+ plants had a main stem of approximately 3.5 cm, and the remaining four AT-PIP5K+ plants had no main stem. The typical growth statuses of the AT-PIP5K+ and WT plants were shown in Fig. 3.
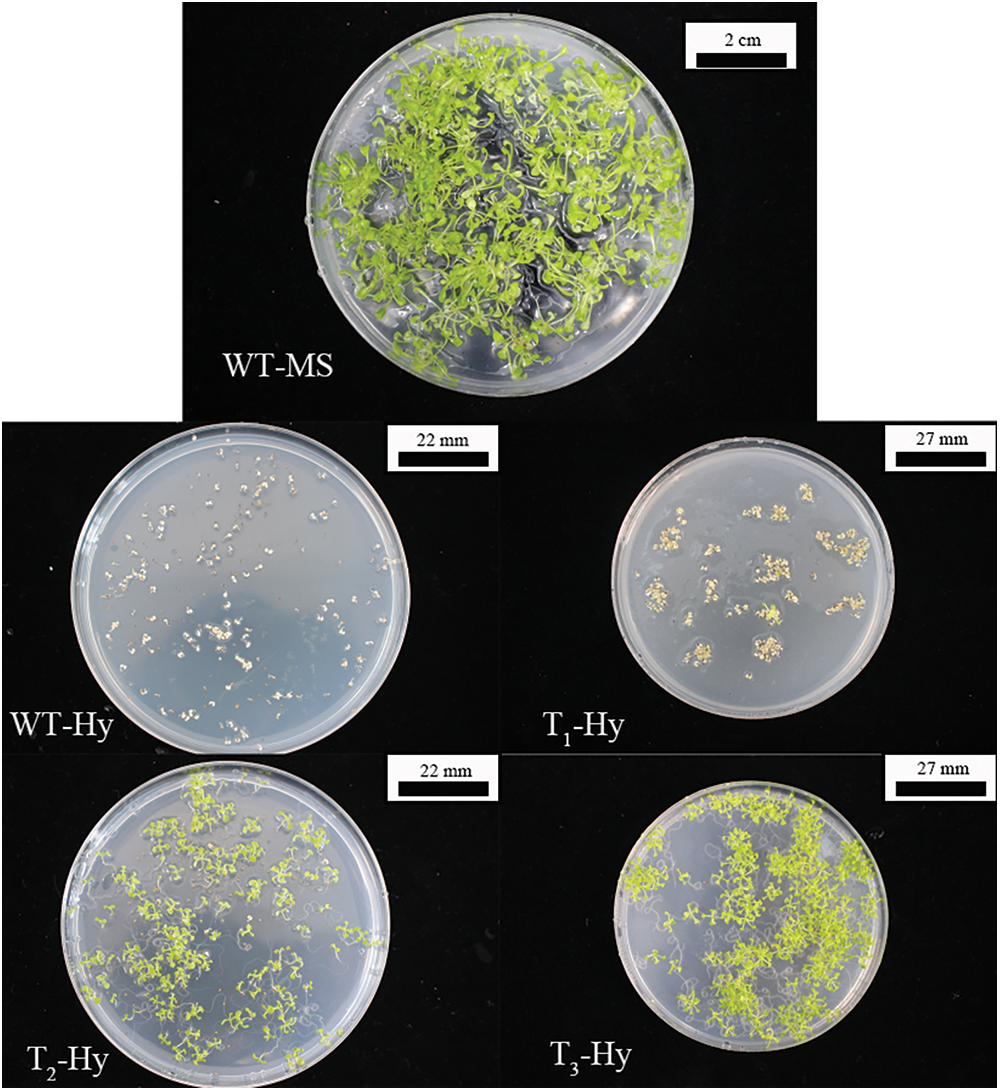
Figure 2: Screening process to identify resistant overexpressing A. thaliana
Note: WT-MS: Columbia WT plants germinated on medium without antibiotics; WT-Hy: Columbia WT plants germinated on medium containing hygromycin; T1-Hy: T1-generation plants germinated on medium containing hygromycin; T2-Hy: T2-generation plants germinated on hygromycin-containing medium; T3-Hy: T3-generation plants germinated on hygromycin-containing medium.

Figure 3: Reproduction and growth processes of transgenic overexpressing A. thaliana plants
Note: 3–1: seed stage; 3–2: reaching the four-leaf stage; 3–3: 10 d after transplanting; 3–4: 20 d after transplanting; WT: Columbia WT A. thaliana plants.
3.2.2 Molecular Characterization of Resistant Heterologous Overexpression A. thaliana Plants
PCR results: The PCR products were subjected to agarose gel electrophoresis. The PCR results from resistant heterologous overexpression A. thaliana plants were shown in Fig. 4. The sizes of the obtained bands were consistent with those of the castor PIP5Ks.7
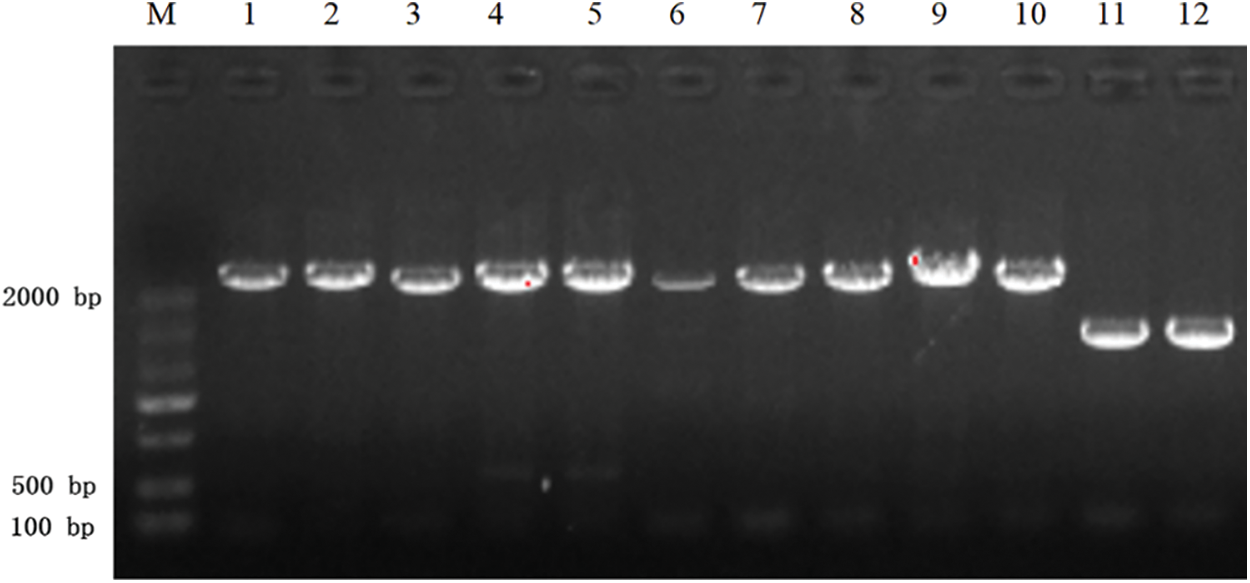
Figure 4: PCR results of transgenic overexpressing A. thaliana plants
Note: M: DL 2000 marker; 1, 2: AT-PIP5K1+ PCR; 3, 4: AT-PIP5K2+ PCR; 5, 6: AT-PIP5K6+ PCR; 7, 8: AT-PIP5K8+ genomic PCR; 9, 10: AT-PIP5K9+ PCR; 11, 12: AT-PIP5K11+ PCR.
RT-qPCR results: RT-qPCR was performed on AT-PIP5K+ plants, and the results were shown in Fig. 5. In AT-PIP5K+ plants, the relative expression levels of the castor PIP5Ks were all upregulated (p < 0.05), indicating successful heterologous overexpression.
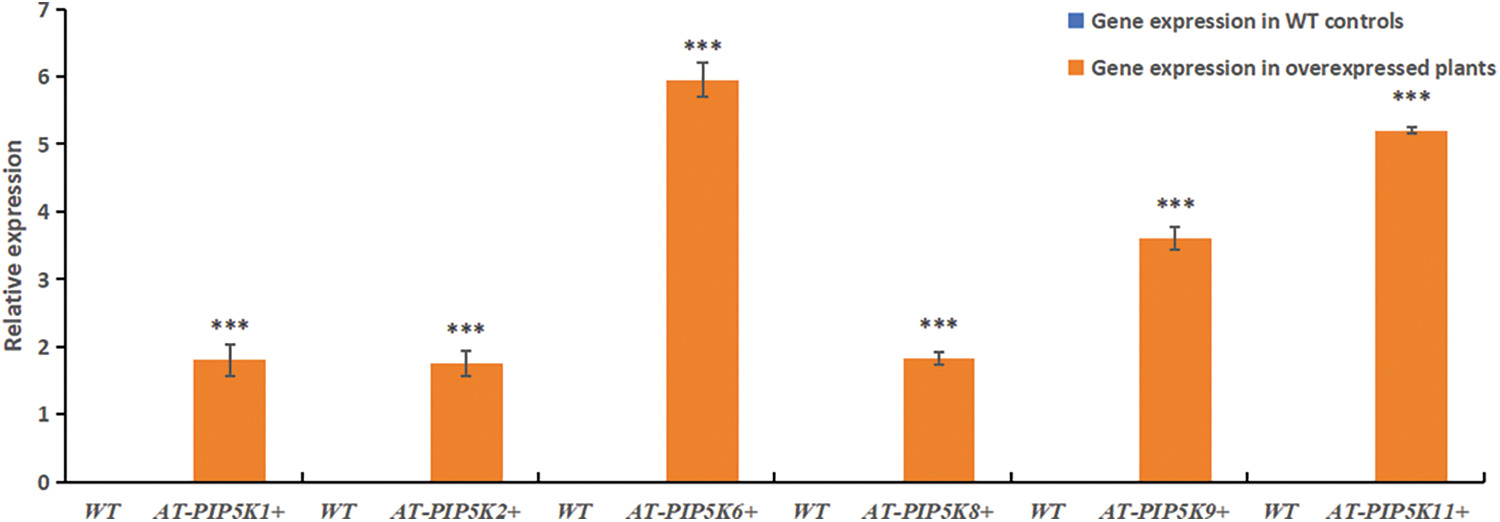
Figure 5: RT-qPCR results of resistant A. thaliana plants with heterologous overexpression
Note: *Significant difference, p < 0.05, **Significant extremely difference, p < 0.01, ***Significant extremely difference, p < 0.001.
3.2.3 Biological Characterization of Resistant Heterologous Overexpression A. thaliana Plants
Phenotypic analysis of A. thaliana overexpression plants at 10 d: WT and AT-PIP5K+ plants at the four-leaf stage were cultured under the same conditions for 10 d (Fig. 3), and the average phenotypic traits were shown in Table 5. A comparison between the WT and overexpressing A. thaliana plants in Fig. 6 and Table 4 showed that when the main stem length of the WT plants was approximately 15 cm, the main stem lengths of the AT-PIP5K1+ and AT-PIP5K2+ plants were 5 and 3 cm, respectively, and the main stems of the AT-PIP5K6+, AT-PIP5K8+, AT-PIP5K9+, and AT-PIP5K11+ plants all tended to sprout but did not bolt, indicating that in the early development of A. thaliana seedlings, the bolting time of the main stem was delayed in the AT-PIP5K+ plants, and significantly so in the AT-PIP5K6+, AT-PIP5K8+, AT-PIP5K9+, and AT-PIP5K11+ plants. The average number of basal leaves was low (p < 0.05) in the WT and AT-PIP5K9+ plants and was high (p < 0.05) in the remaining plants. Cauline leaves grew in the WT, AT-PIP5K1+, and AT-PIP5K2+ plants, and in the WT plants, the main stem was relatively long (p < 0.05), but there were few (p < 0.05) cauline leaves. In contrast, AT-PIP5K6+, AT-PIP5K8+, AT-PIP5K9+, and AT-PIP5K11+ plants did not bolt, and no cauline leaves were observed. On average, four florets grew on the main stem of WT plants, and one floret grew on the AT-PIP5K1+ plants. AT-PIP5K2+ plants grew a main stem but with no florets, and AT-PIP5K6+, AT-PIP5K8+, AT-PIP5K9+, AT-PIP5K11+ plants showed no bolting or florets. Therefore, the overexpression of the PIP5Ks significantly inhibited the early stages of main stem growth in A. thaliana plants.
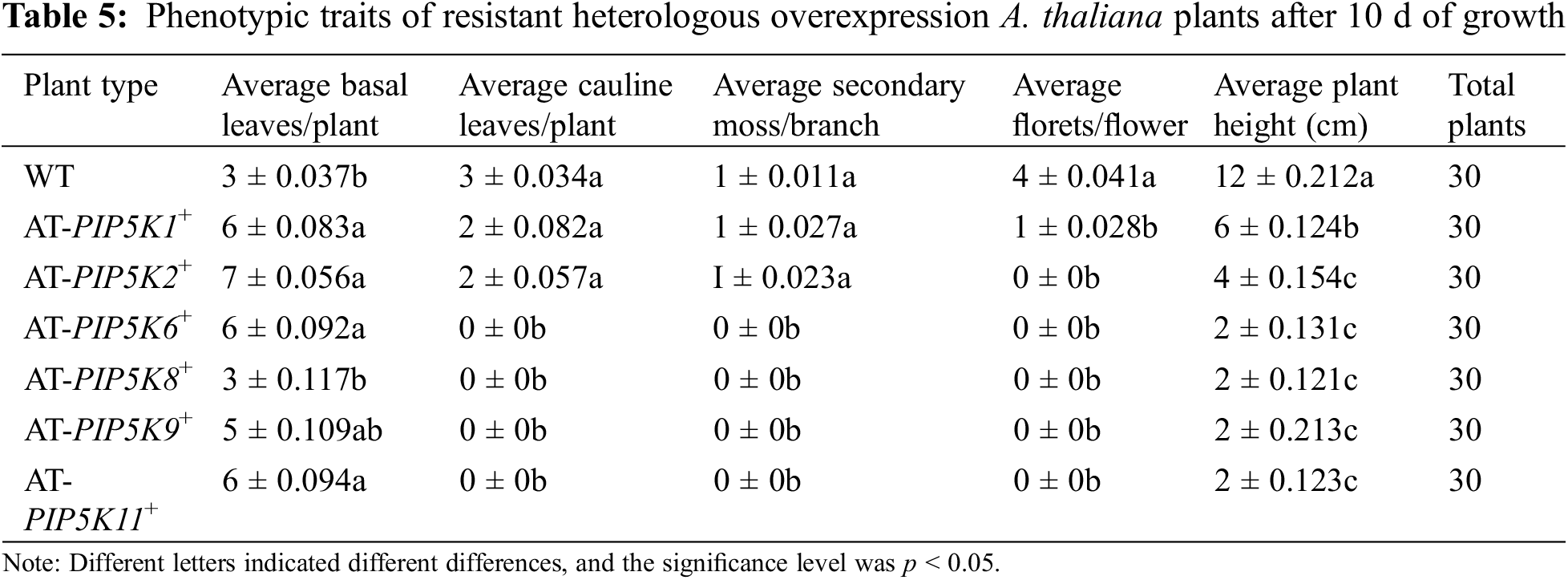

Figure 6: Phenotypes of resistant heterologous overexpression A. thaliana plants after 25 d of growth
Phenotypic analysis of overexpressing A. thaliana plants at 25 d: The phenotypic status of the AT-PIP5K+ plants grown for 25 d was shown in Fig. 6, and the statistical analyses of the plant traits were shown in Table 6. As shown in Fig. 6 and Table 6, comparison of the length of lateral stems of the AT-PIP5Ks+ plants with that of WT plants at 25 d showed the following: 1) the lateral stems of the AT-PIP5K6+, AT-PIP5K8+, and AT-PIP5K9+ plants were the same length as those of the WT plants; 2) the lateral stems of the AT-PIP5K1+ and AT-PIP5K2+ plants were significantly longer (p < 0.05) than those of the WT plants; and 3) the growth of the lateral stems of the AT-PIP5K11+ plants was significantly inhibited, thus reducing (p < 0.05) length. On average, there were few (p < 0.05) basal leaves in WT, AT-PIP5K1+, AT-PIP5K8+, AT-PIP5K9+, and AT-PIP5K11+ plants, while there were many (p < 0.05) in AT-PIP5K2+ and AT-PIP5K6+ plants. On average, there were many (p < 0.05) lateral stems in AT-PIP5K1+ and AT-PIP5K2+ plants; a medium (p < 0.05) amount in AT-PIP5K6+ and AT-PIP5K11+ plants; and few (p < 0.05) in WT, AT-PIP5K8+, and AT-PIP5K9+ plants. The average number of florets in was small p < 0.05) in WT, AT-PIP5K6+, AT-PIP5K8+, AT-PIP5K9+, and AT-PIP5K11+ plants and was large (p < 0.05) in AT-PIP5K1+ and AT-PIP5K2+ plants.
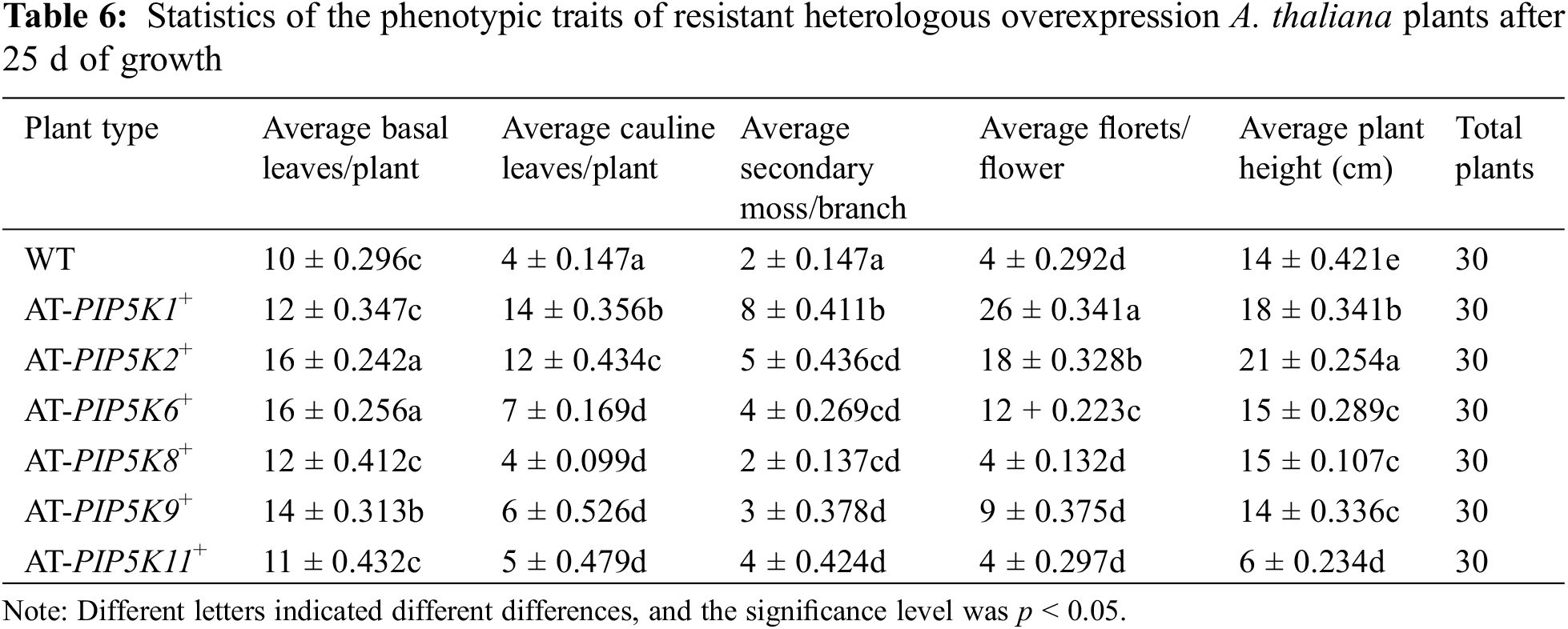
Therefore, in A. thaliana plants, overexpressing the PIP5K1, PIP5K2, and PIP5K6 genes could significantly promote lateral stem growth and flower and silique development, while overexpressing PIP5K8, PIP5K9, and PIP5K11 could significantly inhibit lateral stem growth and flower and silique development.
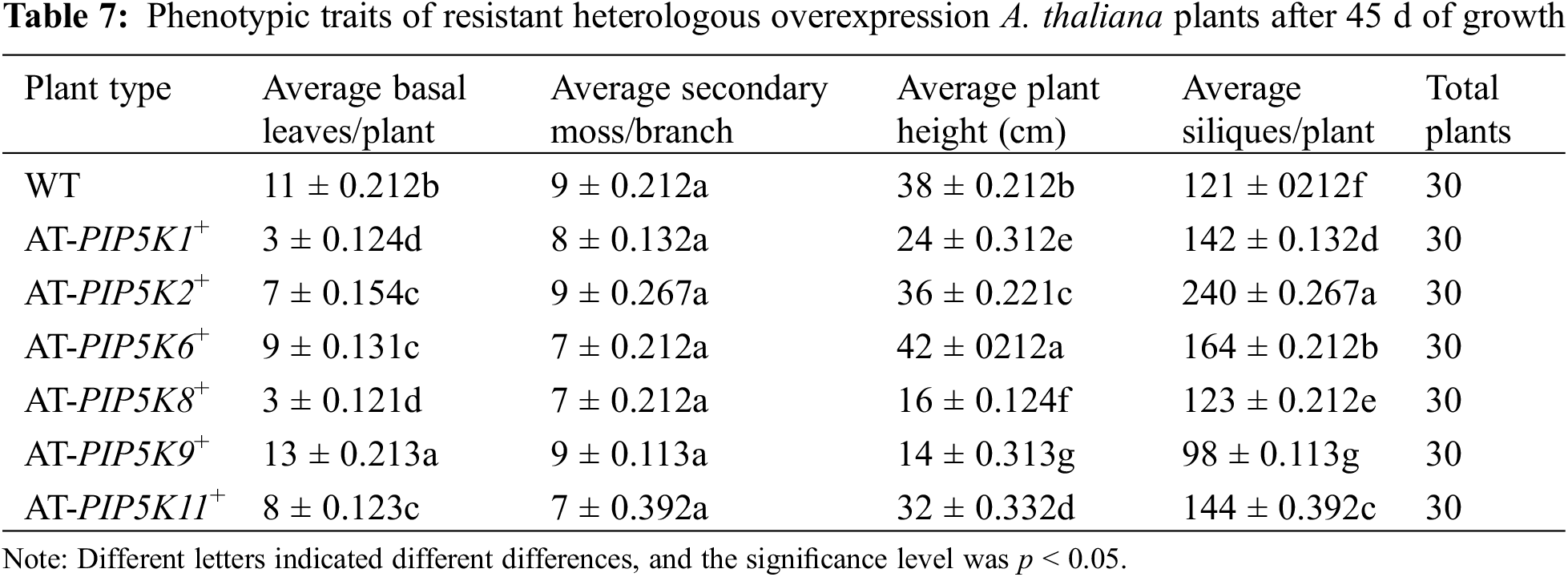
Phenotypic analysis of overexpression A. thaliana plants after 45 d: As shown in Fig. 7 and Table 7, WT, AT-PIP5K2+ and AT-PIP5K6+ plants had similar heights; however, AT-PIP5K2+ was slightly shorter (p < 0.05) (growth was slightly inhibited), and AT-PIP5K6+ was slightly taller (p < 0.05) (growth was slightly promoted) than WT; and the heights of AT-PIP5K1+, AT-PIP5K8+, AT-PIP5K9+, and AT-PIP5K11+ were significantly reduced (p < 0.05), indicating severe growth inhibition. The AT-PIP5K1+, AT-PIP5K2+, AT-PIP5K6+, AT-PIP5K8+, and AT-PIP5K11+ plants showed reduced (p < 0.05) numbers of basal leaves to varying degrees, while in AT-PIP5K9 plants, there were slightly more (p < 0.05) basal leaves than in WT. The AT-PIP5K1+, AT-PIP5K6+, AT-PIP5K8+, and AT-PIP5K11+ plants showed an increase (p < 0.05) in the final number of siliques, and AT-PIP5K2+ plants had the most (p < 0.05) siliques, indicating significant promotion; AT-PIP5K9+ plants had the fewest (p < 0.05) siliques, indicating a significant inhibition of overall plant growth. The AT-PIP5K1+ and AT-PIP5K11+ plants matured significantly earlier (p < 0.05) than the WT plants. Therefore, in A. thaliana, the overexpression of PIP5K1 and PIP5K11 could inhibit maturation and lateral stem growth but promote silique development; the overexpression of PIP5K8 and PIP5K9 could significantly inhibit growth and development; the overexpression of PIP5K2 could significantly promote maturation and silique development but inhibit growth to a certain extent; and the overexpression of PIP5K6 could significantly promote growth and development.

Figure 7: Phenotypes of resistant heterologous overexpression A. thaliana plants after 45 d of growth
3.2.4 Correlation Analysis of Castor PIP5Ks
An RT-qPCR analysis was performed on the 11 A. thaliana PIP5K genes in the A. thaliana AT-PIP5K+ plants, as shown in Fig. 8, and yielded several results.
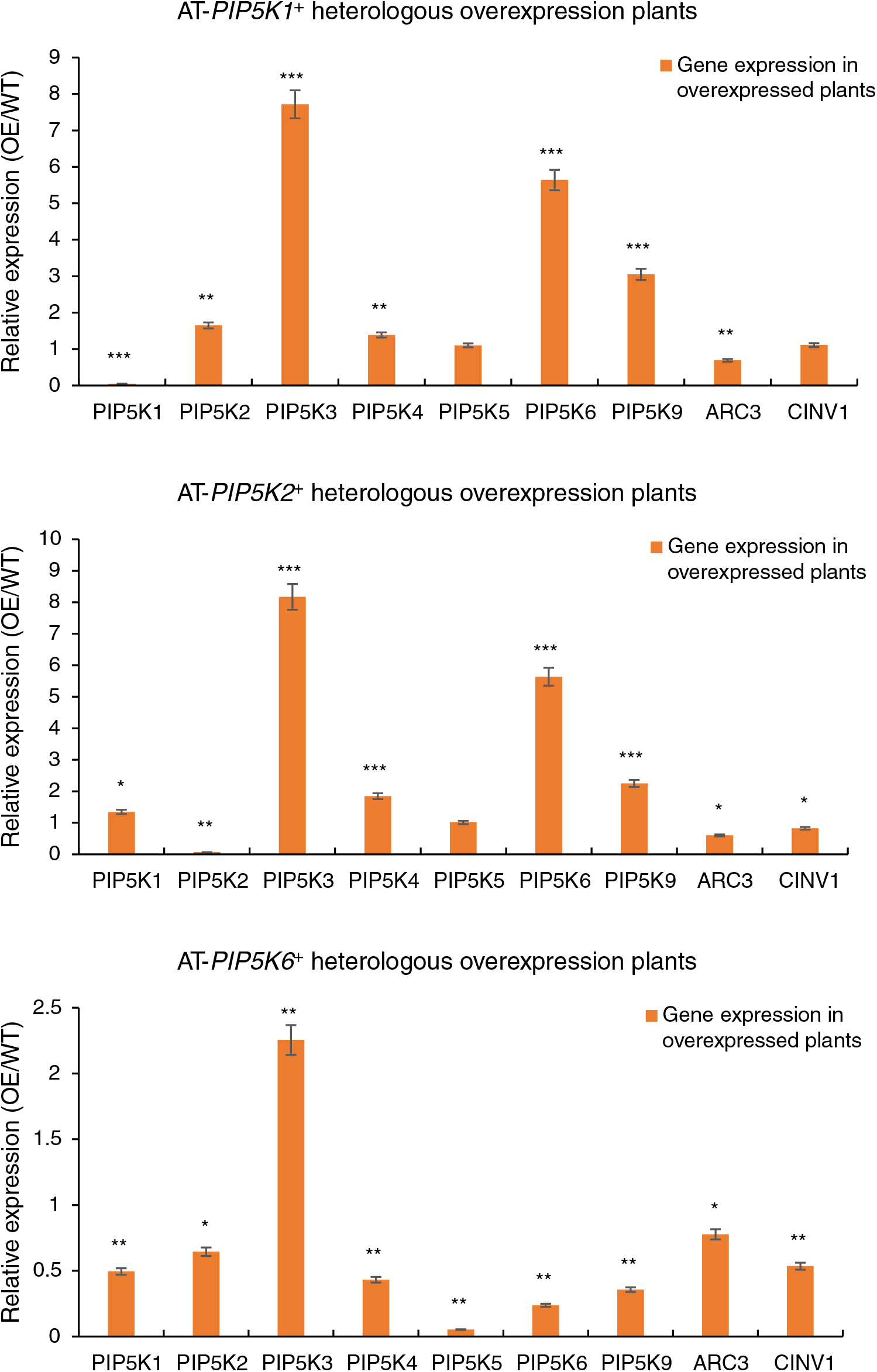
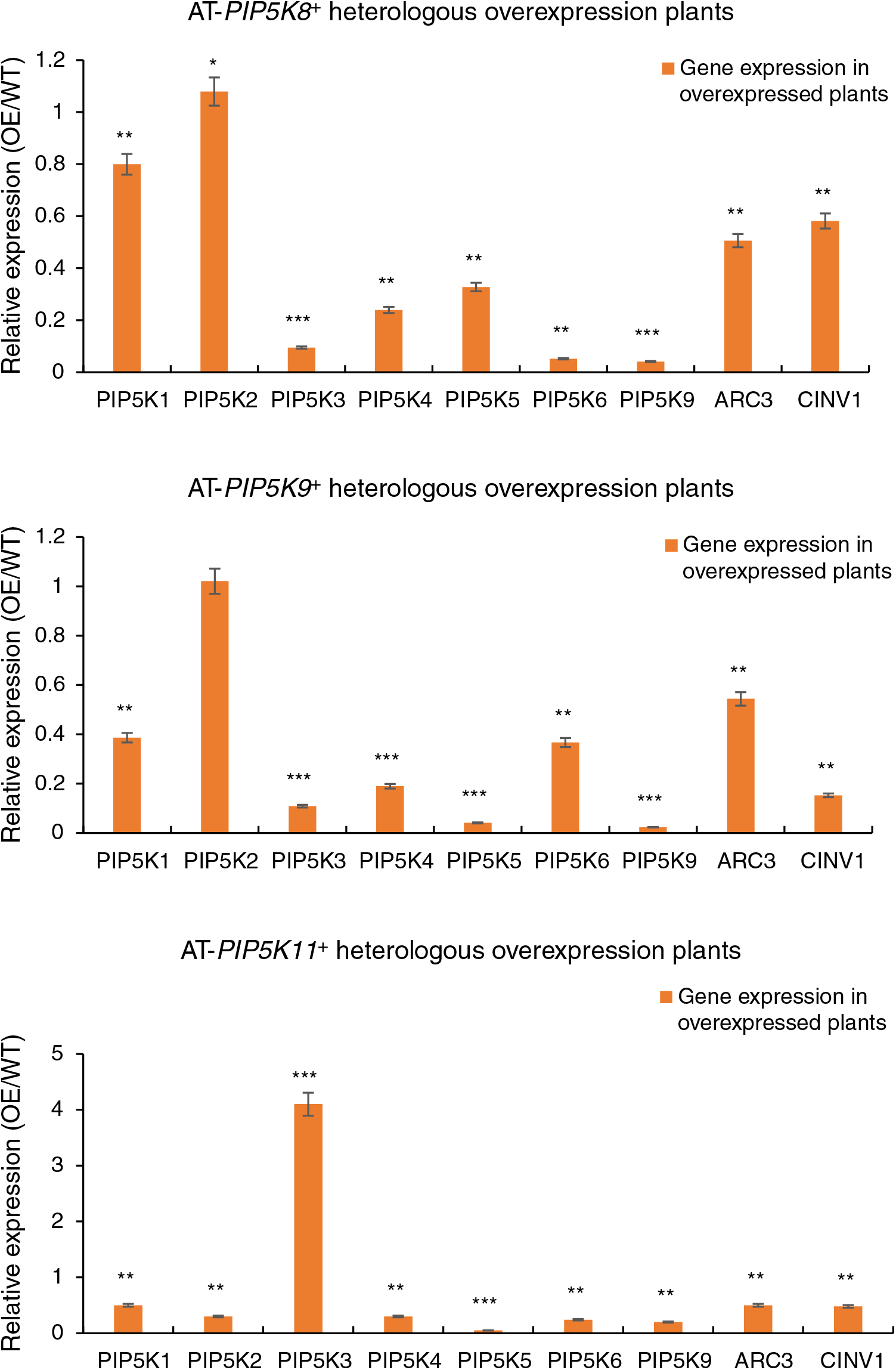
Figure 8: (1) RT-qPCR results from A. thaliana plants heterologously overexpressing castor PIP5Ks; (2) RT-qPCR results of A. thaliana plants heterologously overexpressing castor PIP5Ks
Note: *Significant difference, p < 0.05, **Significant extremely difference, p < 0.01, ***Significant extremely difference, p < 0.001.
When the castor PIP5K1 gene was heterologously overexpressed in A. thaliana, the expression of the homologous A. thaliana PIP5K1 gene decreased (p < 0.05), and the expression levels of the A. thaliana PIP5K3, PIP5K6 and PIP5K9 genes increased (p < 0.05). Therefore, these results combined with the phenotypic results indicated that the PIP5K1 gene may have a synergistic relationship with the PIP5K2 and PIP5K6 genes in castor inflorescences.
When the castor PIP5K2 gene was heterologously overexpressed in A. thaliana, the expression of the homologous A. thaliana PIP5K2 gene decreased (p < 0.05), and the expression levels of the A. thaliana PIP5K3, PIP5K4, PIP5K6, and PIP5K9 genes increased (p < 0.05). Therefore, these results combined with the phenotypic results indicated that the PIP5K2 gene may have a synergistic relationship with the PIP5K6 gene in castor inflorescences.
When the castor PIP5K6 gene was heterologously overexpressed in A. thaliana, the expression levels of the homologous A. thaliana PIP5K4 and PIP5K6 genes decreased (p < 0.05), and the expression of the A. thaliana PIP5K3 gene increased (p < 0.05). Therefore, these results combined with the phenotypic results indicated that the PIP5K6 gene may have a synergistic relationship with the PIP5K2 gene in castor inflorescences.
When the castor PIP5K8 gene was heterologously overexpressed in A. thaliana, the expression of the homologous A. thaliana PIP5K5 gene decreased (p < 0.05), and the expression levels of the A. thaliana PIP5K1, PIP5K3, PIP5K4, PIP5K6, PIP5K9, ARC3, and CINV1 genes also decreased (p < 0.05). Therefore, these results combined with the phenotypic results indicated that the PIP5K8 gene may have a complementary relationship with the PIP5K1, PIP5K2, PIP5K6, and PIP5K9 genes in castor inflorescences.
When the castor PIP5K9 gene was heterologously overexpressed in A. thaliana, the expression levels of the homologous A. thaliana ARC3 and CINV1 genes decreased (p < 0.05), and the expression levels of the A. thaliana PIP5K1, PIP5K3, PIP5K4, PIP5K5, PIP5K6, and PIP5K9 genes also decreased (p < 0.05). Therefore, these results combined with the phenotypic results indicated that the PIP5K9 gene may have a complementary relationship with the PIP5K1, PIP5K2, and PIP5K6 genes in castor inflorescences.
When the castor PIP5K11 gene was heterologously overexpressed in A. thaliana, the expression of the homologous A. thaliana PIP5K5 gene decreased (p < 0.05), and the expression levels of PIP5K1, PIP5K2, PIP5K4, PIP5K6, PIP5K9, ARC3, and CINV1 were also downregulated (p < 0.05). Therefore, these results combined with the phenotypic results indicated that the PIP5K11 gene may have a complementary relationship with the PIP5K1, PIP5K2, PIP5K6, PIP5K8, and PIP5K9 genes in castor inflorescences.
Based on the above results, there may be a synergistic relationship among PIP5K1, PIP5K2, and PIP5K6 in castor inflorescences, and the PIP5K8, PIP5K9, and PIP5K11 genes are complementary to the other three genes.
In the past few years, the biological functions of some PIP5K family members in A. thaliana have been reported, including the involvement of AT-PIP5K1 and AT-PIP5K2 in pollen development [11–13]. A variety of PIP5K isoenzymes had multiple functions in plants. Studies have shown that the lateral roots of AT-PIP5K2 mutants are reduced, and the gravitational pull on the root system is affected [14]. Other studies have shown that double AT-PIP5K1 and AT-PIP5K2 mutations can cause dwarfism and male sterility [15]. In this study, the relative expression levels of PIP5Ks in the female Lm-type castor line aLmAB2 showed that the expression levels of PIP5K1, PIP5K2, PIP5K6, PIP5K8, PIP5K9, and PIP5K11 in different inflorescence types had certain patterns, such as upregulation and downregulation, at different developmental stages of the inflorescences, and the same patterns were maintained in the expression levels of the PIP5Ks in isofemale and bisexual inflorescences at the four-leaf, five-leaf, main stem spike flowering and secondary branch flowering stages. The castor PIP5Ks maintained certain patterns at different stages of inflorescence development or participated in the regulation of castor inflorescence development.
Studies have shown that the PIP5K3 gene is associated with the formation of root hairs in A. thaliana, can be specifically expressed in A. thaliana roots, and can regulate root hair elongation [16,17]. PIP5K4 not only regulated stomatal opening [18] but also, together with PIP5K5, participated in pollen germination and pollen tube growth [19,20]. The PIP5K9 gene negatively regulated sugar-mediated root cell elongation by interacting with CINV1 [21], and RNA interference of the PIP5K6 gene inhibited the growth and development of pollen tubes [22]. Studies on PIP5K7-9 have shown that these genes had little effect on plant growth and development under favorable growth conditions, but they were involved in the adaptation of root growth to osmotic stress [23,24]. In this study, when castor PIP5Ks were heterologously overexpressed in A. thaliana, the relative expression levels of the corresponding A. thaliana PIP5Ks were significantly downregulated (p < 0.05) due to inhibition. Taking the AT-PIP5K1+ plants as an example, when the castor PIP5K1 gene was heterologously overexpressed, the expression of the A. thaliana PIP5K1 gene was significantly downregulated (p < 0.05), and the relative expression levels of the A. thaliana PIP5K3 and PIP5K6 genes were significantly upregulated (p < 0.05). These results, combined with the analysis of the phenotypic changes in plants with heterologous overexpression, showed that the PIP5Ks jointly regulate A. thaliana inflorescence development, and the relative expression of the A. thaliana PIP5K3 gene was upregulated (p < 0.05) when the castor PIP5Ks were all heterologously overexpressed. The PIP5K2 gene corresponded to the A. thaliana PIP5K2 gene. Therefore, there may be a synergistic relationship among the PIP5K1, PIP5K2, and PIP5K6 genes in castor inflorescences, and PIP5K8, PIP5K9, and PIP5K11 are complementary to the other three genes.
An analysis of the expression levels of the PIP5Ks in the female Lm-type castor line aLmAB2 showed that except for PIP5K9 and PIP5K11, which had high (p < 0.05) relative expression levels in isofemale inflorescences, the other four genes all had the highest (p < 0.05) relative expression levels in female inflorescences. Except for PIP5K9, which had a low (p < 0.05) relative expression level in isofemale inflorescences, and PIP5K11, which had a low (p < 0.05) relative expression level in bisexual inflorescences, the other four genes all showed their lowest (p < 0.05) relative expression levels in bisexual inflorescences.
PIP5Ks were heterologously overexpressed in A. thaliana, and PCR was performed on the AT-PIP5K plants, i.e., the six types of T3-generation A. thaliana plants with stable genetic resistance and heterologous overexpression of the PIP5Ks. The RT-qPCR results confirmed that the relative expression levels of the six genes in the AT-PIP5K+ plants were significantly upregulated (p < 0.05) compared to those in Columbia WT plants. The biological characterization of AT-PIP5K+ plants showed that main stem growth was significantly delayed in A. thaliana plants overexpressing the six genes compared with the control; in the early development of the lateral stems, the overexpression of PIP5K1 and PIP5K2 promoted lateral stem growth and flower and silique development, while the overexpression of the other four genes inhibited lateral stem growth and flower and silique development in A. thaliana. In the late growth stage of A. thaliana, the overexpression of the PIP5K1 and PIP5K11 genes significantly promoted maturation; the overexpression of the PIP5K1, PIP5K2, PIP5K8, PIP5K9 and PIP5K11 genes inhibited lateral stem growth and flower and silique development; and the overexpression of PIP5K6 slightly promoted lateral stem growth and flower and silique development. A correlation analysis of the PIP5Ks indicated that there may be a synergistic relationship among the PIP5K1, PIP5K2, and PIP5K6 genes in castor inflorescences, and the PIP5K8, PIP5K9, and PIP5K11 genes are complementary to the other three genes.
Acknowledgement: Not applicable.
Funding Statement: Supported by National Natural Science Foundation of China (31860071); Ministry of Education New Agricultural Research and Reform Practice Program (2020114); Surface Program of Inner Mongolia Natural Science Foundation (2021MS03008); Inner Mongolia Autonomous Region Grassland Talent Innovation Team-Rolling Support Program for Castor Molecular Breeding Research Innovation Talent Teams (2022); 2023 Inner Mongolia Autonomous Region Science and Technology Department Establishes the Project of Key Laboratory Construction of Castor Breeding and Comprehensive Utilization in Inner Mongolia Autonomous Region; Inner Mongolia University for Nationalities 2022 Basic Research Operating Expenses of Colleges and Universities directly under the Autonomous Region Project (237); Open Fund Project of Castor Industry Collaborative Innovation Center of Inner Mongolia Autonomous Region (MDK2021011, MDK2022014).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Rui Luo, Fenglan Huang; data collection: T. Liang, X. Hu, W. Ren, C. Gao; analysis and interpretation of results: Q. Wen, X. Liang, Y. Wen; draft manuscript preparation: M. Yin, Z. Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: These data were derived from the following resources available in the China Dissertations Database: doi: 10.27228/d.cnki.gnmmu.2020.000325. Plant material used in this experiment was provided with “Key Laboratory of Castor Breeding of the State Ethnic Affairs Commission”.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Pertile P, Liscovitch M, Chalifa V, Cantley LC. Phosphatidylinositol 4,5-bisphosphate synthesis is required for activation of phospholipase D in U937 cells (*). J Biol Chem. 1995;270(10):5130–5. [Google Scholar] [PubMed]
2. Yoneda A, Kanemaru K, Matsubara A, Takai E, Shimozawa M, Satow R, et al. Phosphatidylinositol 4, 5-bisphosphate is localized in the plasma membrane outer leaflet and regulates cell adhesion and motility. Biochem Biophys Res Commun. 2020;527(4):1050–6. [Google Scholar] [PubMed]
3. Fíla J, Klodová B, Potěšil D, Juříček M, Šesták P, Zdráhal Z, et al. The beta subunit of nascent polypeptide associated complex plays a role in flowers and Siliques development of Arabidopsis thaliana. Int J Mol Sci. 2020;21(6):2065. [Google Scholar]
4. Ababakeli S, Wei G, Lei J, Zhu YX. Cloning and transactivation identification of Atlg30210, a new member of TCP family high expressed in Flower in arabidopsis. Molecular Plant Breeding = Fen zi zhi wu yu Zhong. 2005;3(1):26–30 (In Chinese). [Google Scholar]
5. Brown AP, Kroon JT, Swarbreck D, Febrer M, Larson TR, Graham IA, et al. Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One. 2012;7(2):e30100. [Google Scholar] [PubMed]
6. Lin JT, Arcinas A, Harden LA. Identification of acylglycerols containing dihydroxy fatty acids in castor oil by mass spectrometry. Lipids. 2009;44(4):359–65. [Google Scholar] [PubMed]
7. Tan M, Xue J, Wang L, Huang J, Fu C, Yan X, et al. Transcriptomic analysis for different sex types of Ricinus communis L. during development from apical buds to inflorescences by digital gene expression profiling. Front Plant Sci. 2016;6:1208. [Google Scholar] [PubMed]
8. Wang L, Tan ML, Yan MF, Wang LJ, Yan XC. Initial research on inflorescence characteristics and flower bud differentiation of Ricinus communis L. Chin J Oil Crop Sci. 2012;34(5):544–550 (In Chinese). [Google Scholar]
9. Liang TN. Bioinformatics analysis and fluorescence quantitative PCR results of Ricinus communis PIP5Ks. Chin Tradit Herb Drugs. 2018;5892–900 (In Chinese). [Google Scholar]
10. Ling J, Yang X, Shan H, Wenjun T, Xiaohong L, et al. Cloning of the soybean GmSDG25 gene and genetic transformation in Arabidopsis thaliana. Mol Plant Breed. 2021;1–16. [Google Scholar]
11. Gerth K, Lin F, Daamen F, Menzel W, Heinrich F, Heilmann M, et al. Arabidopsis phosphatidylinositol 4-phosphate 5-kinase 2 contains a functional nuclear localization sequence and interacts with alpha-importins. Plant J. 2017;92(5):862–78. [Google Scholar] [PubMed]
12. Mikami K, Katagiri T, Iuchi S, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998;15(4):563–8. [Google Scholar] [PubMed]
13. Ugalde JM, Rodriguez-Furlán C, de Rycke R, Norambuena L, Friml J, León G, et al. Phosphatidylinositol 4-phosphate 5-kinases 1 and 2 are involved in the regulation of vacuole morphology during Arabidopsis thaliana pollen development. Plant Sci. 2016;250:10–9. [Google Scholar] [PubMed]
14. Mei Y, Jia WJ, Chu YJ, Xue HW. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res. 2012;22(3):581–97. [Google Scholar] [PubMed]
15. Marhava P, Fandino ACA, Koh SW, Jelínková A, Kolb M, Janacek DP, et al. Plasma membrane domain patterning and self-reinforcing polarity in Arabidopsis. Dev Cell. 2020;52(2):223–35. [Google Scholar] [PubMed]
16. Hirano T, Konno H, Takeda S, Dolan L, Kato M, Aoyama T, et al. PtdIns(3,5)P2 mediates root hair shank hardening in Arabidopsis. Nat Plants. 2018;4(11):888–97. [Google Scholar] [PubMed]
17. Stenzel I, Ischebeck T, Konig S, Hołubowska A, Sporysz M, Hause B, et al. The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. The Plant Cell. 2008;20(1):124–41. [Google Scholar] [PubMed]
18. Lee Y, Kim YW, Jeon BW, Park KY, Suh SJ, Seo J, et al. Phosphatidylinositol 4,5-bisphosphate is important for stomatal opening. Plant J. 2007;52(5):803–1. [Google Scholar] [PubMed]
19. Ischebeck T, Stenzel I, Heilmann I. Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. The Plant Cell. 2008;20(12):3312–30. [Google Scholar] [PubMed]
20. Ischebeck T, Stenzel I, Hempel F, Jin X, Mosblech A, Heilmann I. Phosphatidylinositol-4, 5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J. 2011;65(3):453–68. [Google Scholar] [PubMed]
21. Lou Y, Gou JY, Xue HW. PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell. 2007;19(1):163–81. [Google Scholar] [PubMed]
22. Zhao Y, Yan A, Feijó JA, Furutani M, Takenawa T, Hwang I, et al. Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. The Plant Cell. 2010;22(12):4031–44. [Google Scholar] [PubMed]
23. Kuroda R, Kato M, Tsuge T, Aoyama T. Arabidopsis phosphatidylinositol 4-phosphate 5-kinase genes PIP5K7, PIP5K8, and PIP5K9 are redundantly involved in root growth adaptation to osmotic stress. Plant J. 2021;106(4):913–27. [Google Scholar] [PubMed]
24. Zarza X, van Wijk R, Shabala L, Hunkeler A, Lefebvre M, Rodriguez-Villalón A, et al. Lipid kinases PIP5K7 and PIP5K9 are required for polyamine-triggered K+ efflux in Arabidopsis roots. Plant J. 2020;104(2):416–32. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools