 Open Access
Open Access
ARTICLE
A Novel Hybrid Model Based on Machine and Deep Learning Techniques for the Classification of Microalgae
1
Department of Computer Engineering, Faculty of Engineering and Architecture, Erzincan Binali Yıldırım University, Erzincan,
24100, Türkiye
2
Department of Biology, Faculty of Arts and Science, Erzincan Binali Yıldırım University, Erzincan, 24100, Türkiye
* Corresponding Author: Özge Zencir Tanır. Email:
Phyton-International Journal of Experimental Botany 2023, 92(9), 2519-2534. https://doi.org/10.32604/phyton.2023.029811
Received 09 March 2023; Accepted 05 May 2023; Issue published 28 July 2023
Abstract
Classification and monitoring of microalgae species in aquatic ecosystems are important for understanding population dynamics. However, manual classification of algae is a time-consuming method and requires a lot of effort with expertise due to the large number of families and genera in its classification. The recognition of microalgae species has become an increasingly important research area in image recognition in recent years. In this study, machine learning and deep learning methods were proposed to classify images of 12 different microalgae species in order to successfully classify algae cells. 8 Different novel models (MobileNetV3Small-Lr, MobileNetV3SmallRf, MobileNetV3Small-Xg, MobileNetV3Large-Lr, MobileNetV3Large-Rf, MobileNetV3Large-Xg, MobileNetV3Small-Improved and MobileNetV3Large-Improved) have been proposed to classify these microalgae species. Among these proposed model structures, the best classification accuracy rate was 92.22% and the loss rate was 0.72, obtained from the MobileNetV3Large-Improved model structure. In addition, as a result of the experimental results obtained, metrics such as the confusion matrix, which can meet the experts in the correct diagnosis of microalgae species, were also evaluated. This research may in the future open a new avenue for the development of a cost-effective, highly sensitive computer-based system for the use of image analysis and deep learning techniques for the identification and classification of different microalgae.Keywords
Microalgae are simple microscopic and photosynthetic organisms that can grow in almost all ecological systems, including fresh and salt waters, and produce a wide variety of bioactive substances, some of which are classes of toxins, ranging from unicellular to multicellular forms [1]. Microalgae, which produce about half of atmospheric oxygen, can be used as a rich food source by humans because they synthesize organic substances (carbohydrate, protein, oil). Microalgae contain Omega-3 and Omega-6 substances that are not produced in the human body and vitamins important for human health. Microalgae monitoring is essential for aquaculture and the aquatic environment to determine the abundance and species of algae populations in the aquatic environment. Because of all these features, it has many uses, and it is gaining new usage areas day by day. They are widely used for a variety of applications such as human and animal nutrition, biofuel production, CO2 capture, cosmetics, pharmaceuticals and nutrient recovery from wastewater [2,3].
The most common method typically used for monitoring and diagnosing microalgae is the use of an optical microscope. It is analyzed by looking at the morphological differences of microalgae species. However, this method is a labor-intensive, time-consuming and knowledge-intensive method that usually takes a few hours to a few days to obtain analysis results from water samples. Since manual identification of microalgae requires expertise and effort, machine learning techniques and especially artificial neural networks (ANN), which is a computer-based automated system with high accuracy, has gained popularity thanks to increasing computational capacity and large volumes of data that can be produced and managed today. Deep learning is widely preferred in microalgae to perform various tasks such as pattern recognition, classification, prediction and feature extraction [4]. These techniques are only capable of inferring models based on data, require less in-depth knowledge of the system, and can adapt appropriately to different conditions. These models can utilize different types of data such as categories, numeric values or images depending on the problem to be solved [5]. A computer-based automatic recognition system for the diagnosis and classification of microalgae will definitely reduce the burden of taxonomists. In addition, it will allow many people to identify and know the types of microalgae without having any knowledge about microalgae [6].
For the classification and recognition of algae species, different machine learning and deep learning studies have been proposed so far. Promdaen et al. [7] revealed 97.22% classification accuracy for 12 microalgae found in water resources of Thailand by an automated recognition system based on feature combination approach with Sequential Minimal Optimization (SMO) technique. Li et al. [8] exhibited 97% classification accuracy on a dataset contain 10,463 algae images by Mueller matrix imaging system based on convolutional neural networks (CNNs). Deglint et al. [9] investigated the effectiveness of automatic classification using a deep residual convolutional neural network and achieve a classification accuracy of 96% in an experiment conducted with six different algae types. Salido et al. [10] used a CNN-based deep learning and achieved a classification accuracy of 99.51% with the AlexNet network and a detection efficiency of 86% with the YOLO network. The macroalgae segmentation study with using three different CNN (MobileNetV2, Resnet18, Xception) were performed with highresolution images [11]. Then, accuracy rates were compared to each other. ResNet18 provided the highest accuracy of 91.9% thanks to the distinguishable textures and colors of macroalgae. The recognition results of five CNN models was proposed, and the accuracy with transfer learning reached 94.0% [12].
In this study, machine learning and deep learning methods were used to classify images of 12 different microalgae species in order to successfully classify algae cells and contribute to the literature in this field. Logistic regression, random forest and XGBoost methods from machine learning methods, MobileNetV3Small and MobileNetV3Large methods from deep learning methods were discussed. As a result of the fact that these methods are both hybrid in themselves and different improvements, 8 novel models have been proposed. These proposed models were trained and tested separately using the the algae cell images data. The contributions of this study can be summarized as follows:
1. For the classification of microalgae, 6 novel hybrid models were applied by performing classification operations with deep learning methods, feature extraction, machine learning methods.
2. By adding auxiliary layers to deep learning methods, 2 novel improved transfer learning models were applied.
3. 13 different models were trained and compared separately using machine learning, deep learning and proposed methods.
4. High classification success with MobileNetV3Large-Improved model.
The remaining arrangements are as follows. Detailed information about the materials and methods used in the classification of algae cells is presented in Section 2. Experimental analyses, results and discussions are given in Section 3. In Section 4, information about the results obtained in the study and future studies are given.
In this section, dataset and preprocessing, system configuration, model structures and training parameters are given in order to get successful results from machine learning and deep learning methods.
2.1 Dataset and Image Pre-Processing
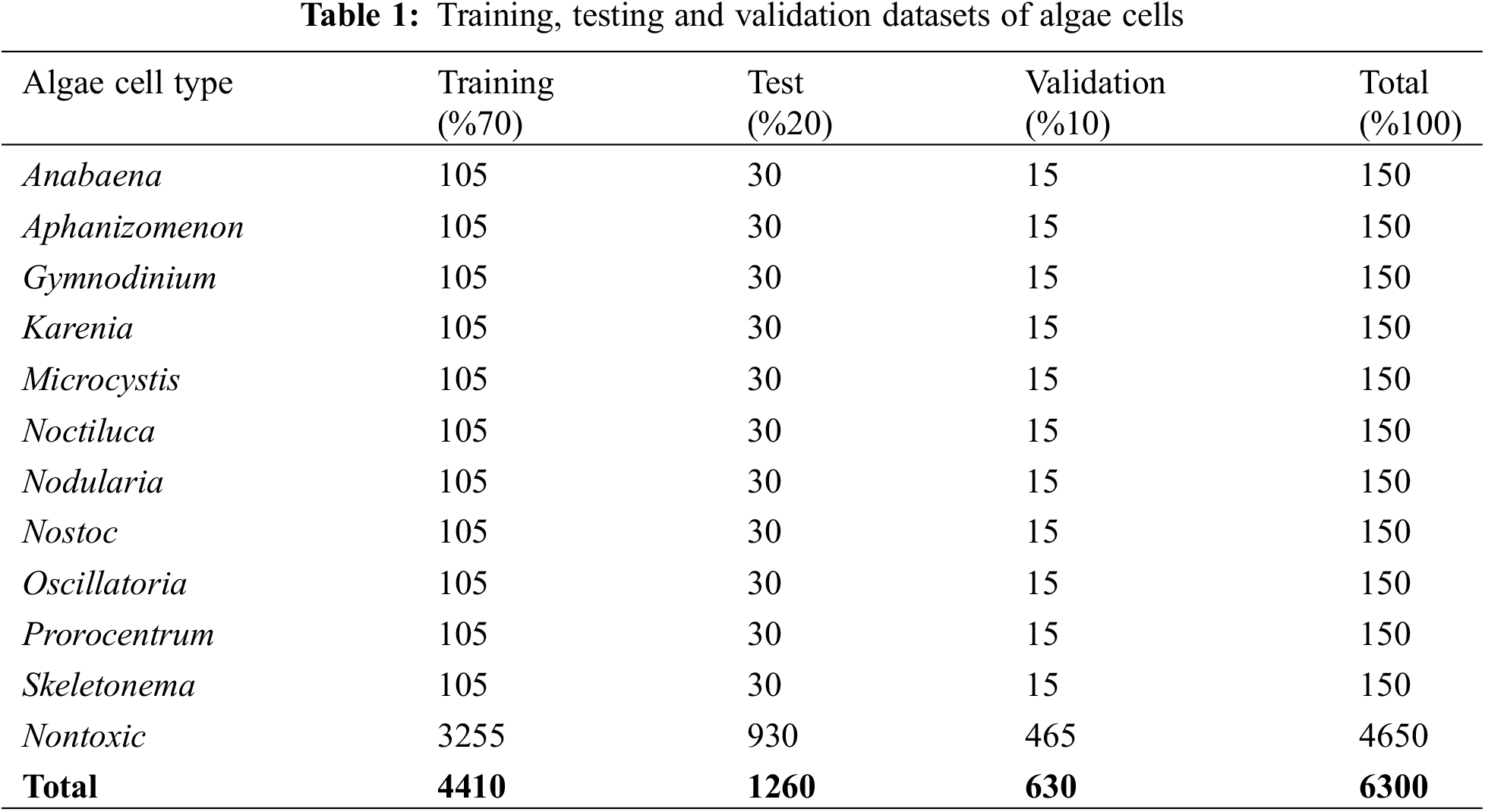
In this study, the 12-class the algae cell images database, which is open to the public, was used for the successful classification of algae cells [13]. The dataset includes Anabaena, Aphanizomenon, Gymnodinium, Karenia, Microcystis, Noctiluca, Nodularia, Nostoc, Oscillatoria, Prorocentrum, Skeletonema, Nontoxic algae cell types. There are 4650 images of algae cells from the nontoxic species and 150 images from each of the other species. There are a total of 6300 images of these algae cell types in different pixel sizes. In order to achieve the desired success from machine learning and deep learning models, all algae cell images in the dataset were converted to 224 × 224 × 3 pixels by preprocessing. Then, in order to extract good features from the dataset in the models discussed in the study, the dataset was divided into 70% train, 20% test and 10% validation dataset. Sample images of each algae cell contained in this dataset are shown in Fig. 1, and the diagram of the data pretreatment and dataset separation stage is shown in Fig. 2.
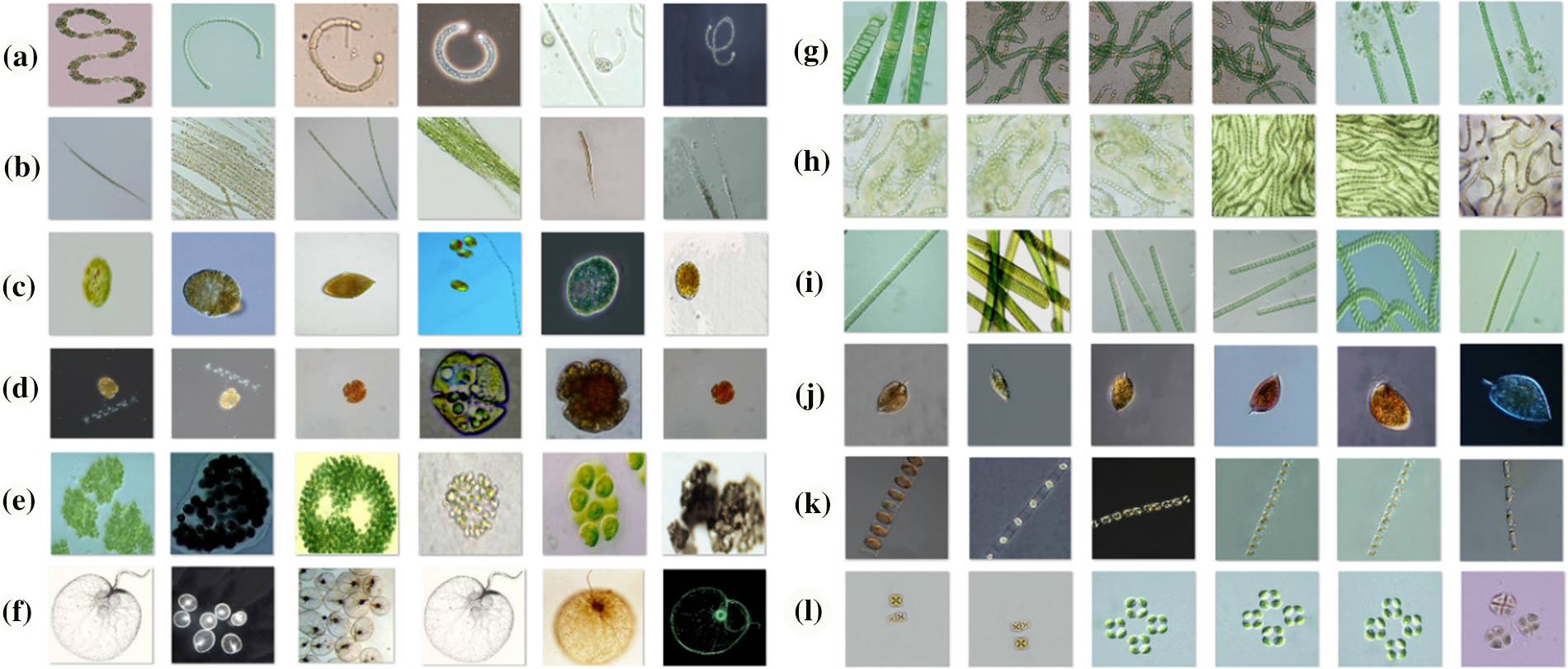
Figure 1: Sample images of each algae cell in the dataset (a) Anabaena, (b) Aphanizomenon, (c) Gymnodinium, (d) Karenia, (e) Microcystis, (f) Noctiluca, (g) Nodularia, (h) Nostoc, (i) Oscillatoria, (j) Prorocentrum, (k) Skeletonema, (l) Nontoxic
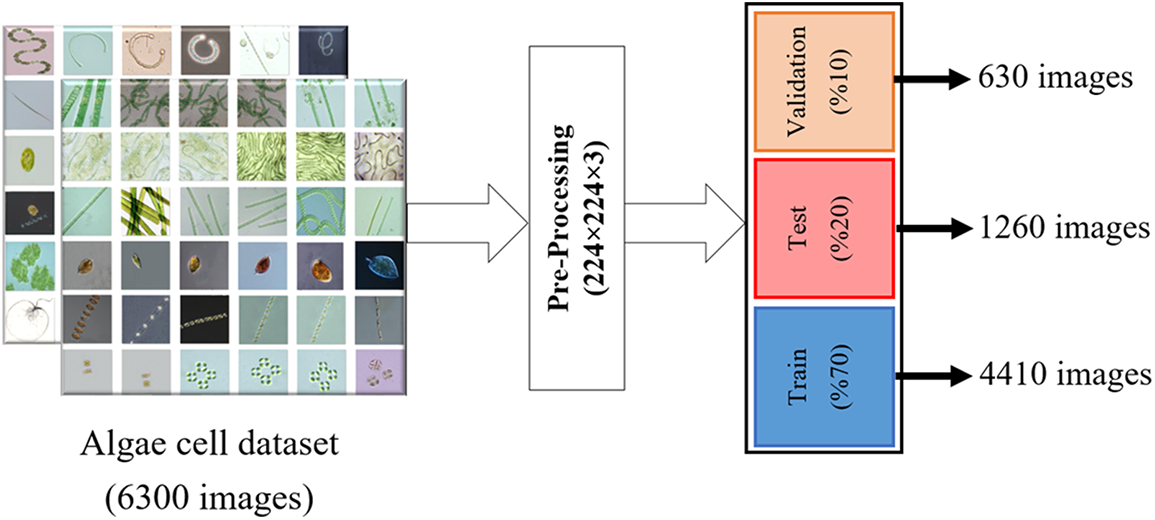
Figure 2: Diagram of the pre-processing of the algae cells in the dataset and the dataset separation stage
The Python programming language was used to analyze the machine learning and deep learning models discussed in the study. In order to compile Python codes and obtain analysis results, Google Colaboratory [14] environment, which is a cloud-based system and has NVIDIA Tesla K80 graphics processor, was used.
2.3 Proposed Hybrid Model Structures
In the study, machine learning and deep learning based model structures were used in order to obtain successful results by making good attribute extraction from algae cell types. Logistic regression from machine learning models [15], random forest [16] and XGBoost [17], and one of the deep learning models is MobileNetV3 [18], two versions of MobileNetV3Small and MobileNetV3Large models are preferred.
8 different novel models have been proposed using these model structures. 6 novel hybrid models have been proposed by using the pre-trained weights of the deep learning methods MobileNetV3Small and MobileNetV3Large methods, feature extraction from machine learning methods, logistic regression, random forest and XGBoost methods and classification processes. In addition, 2 novel improved models named MobileNetV3Small-Improved and MobileNetV3Large-Improved with transfer learning have been proposed by adding auxiliary layers to the MobileNetV3Small and MobileNetV3Large models. The suggested models are: MobileNetV3Small-Lr, MobileNetV3Small-Rf, MobileNetV3Small-Xg, Mobilenetv3Large-Lr, Mobilenetv3Large-Rf, Mobilenetv3Large-Xg, MobileNetV3Small-Improved and Mobilenetv3Large-Improved. The flow diagram of the proposed models for classifying algae cells is shown in Fig. 3.
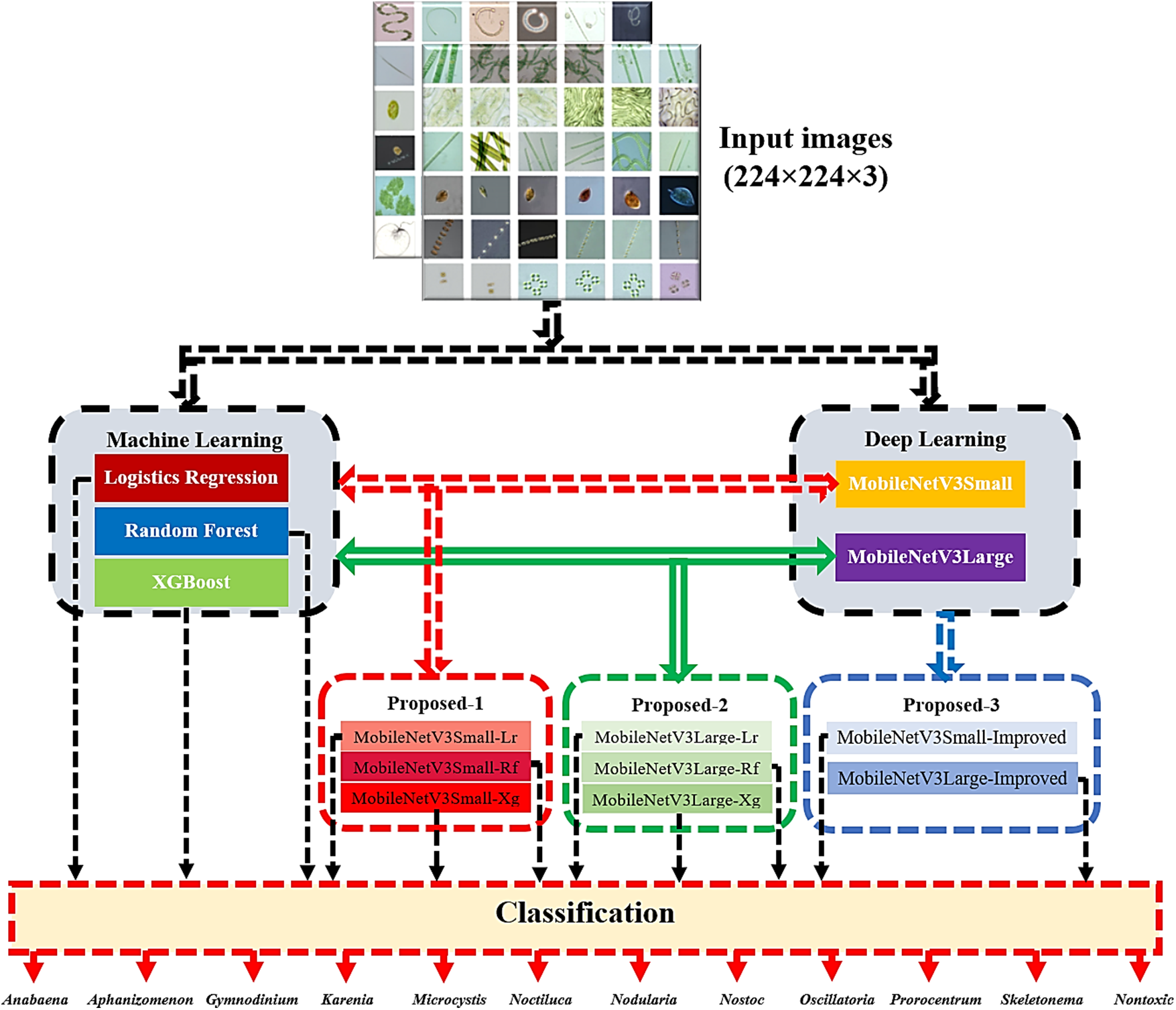
Figure 3: Proposed hybrid model structure
Logistic regression, random forest and XGBoost methods, which are machine learning methods, were first used to classify the algae cell image in the flow diagram given in Fig. 3. Secondly, MobileNetV3Small and MobileNetV3Large methods from deep learning methods were used. A total of 6 hybrid methods, including 3 novel methods by hybridizing with MobileNetV3Small method, logistic regression, random forest and XGBoost methods, and 3 novel methods by hybridizing with MobileNetV3Large method, logistic regression, random forest and XGBoost methods, have been proposed. Then, 2 more improved models were proposed by adding 512 convolution and 25% forget-me-not auxiliary layers to the last layers of the MobileNetV3Small and MobileNetV3Large methods. Using each of these models separately, 12 classed algae cell types were classified.
In order to extract good features from each model structure discussed in the study, each algae cell is divided into training, test and validation datasets (Table 1). The training parameters given in Table 2 were used to compare each model structure using these datasets.

In the study, 8 proposed models were trained and tested together with 3 machine learning and 2 deep learning methods. All the findings obtained as a result of the educational test were analyzed comparatively. Figs. 4, 6–8, 10 illustrate the confusion matrix of Logistic Regression, Random Forest, XGBoost, MobileNetV3Small, MobileNetV3Large, MobileNetV3Small-Lr, MobileNetV3Small-Rf, MobileNetV3Small-Xg, MobileNetV3Large-Lr, MobileNetV3Large-Rf, MobileNetV3Large-Xg, MobileNetV3Small-Improved, and MobileNetV3Large-Improved, respectively. The confusion matrix is a tabular representation and summary of the predicted true and false values of each model in the classification. It provides a better idea of the performance of the models. The confusion matrix is a two-dimensional matrix that shows the actual classes and the predicted classes.
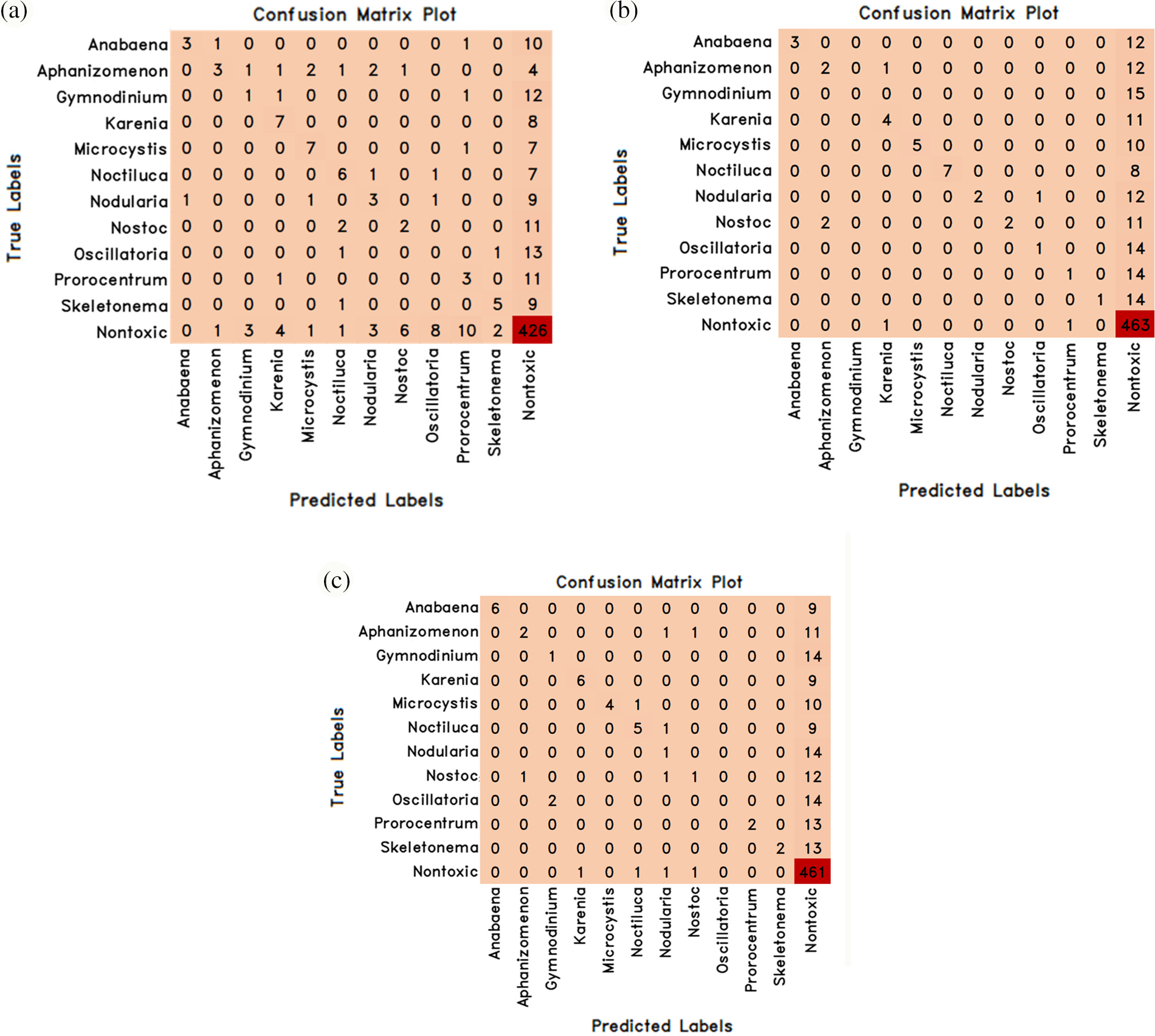
Figure 4: Some the confusion matrix values of machine learning methods in classifying algae cells by normal learning are (a) Logistic Regression (b) Random Forest (c) XGBoost
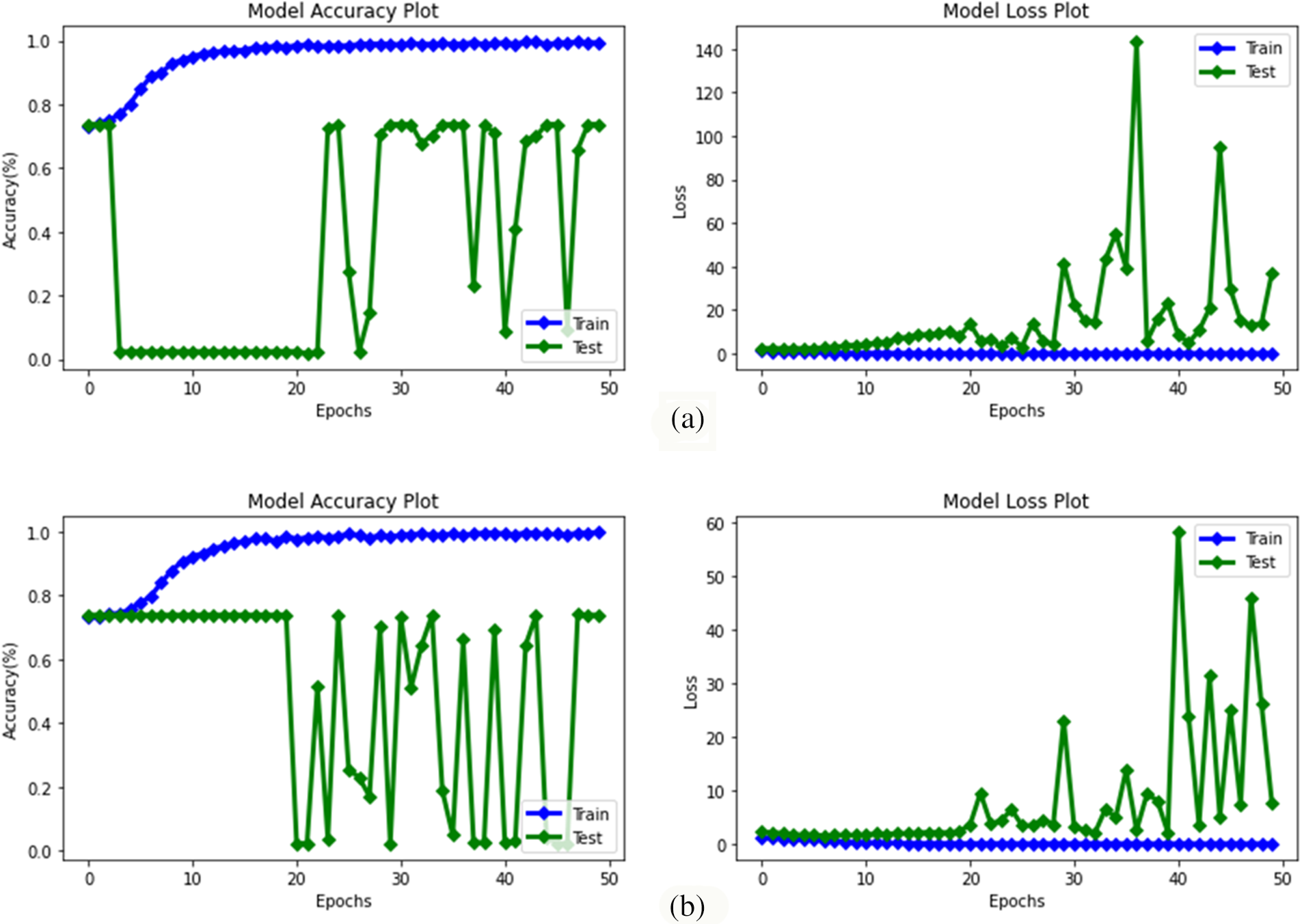
Figure 5: Accuracy and loss graphs of deep learning methods obtained as a result of training test by performing normal learning (a) MobileNetV3Small (b) MobileNetV3Large
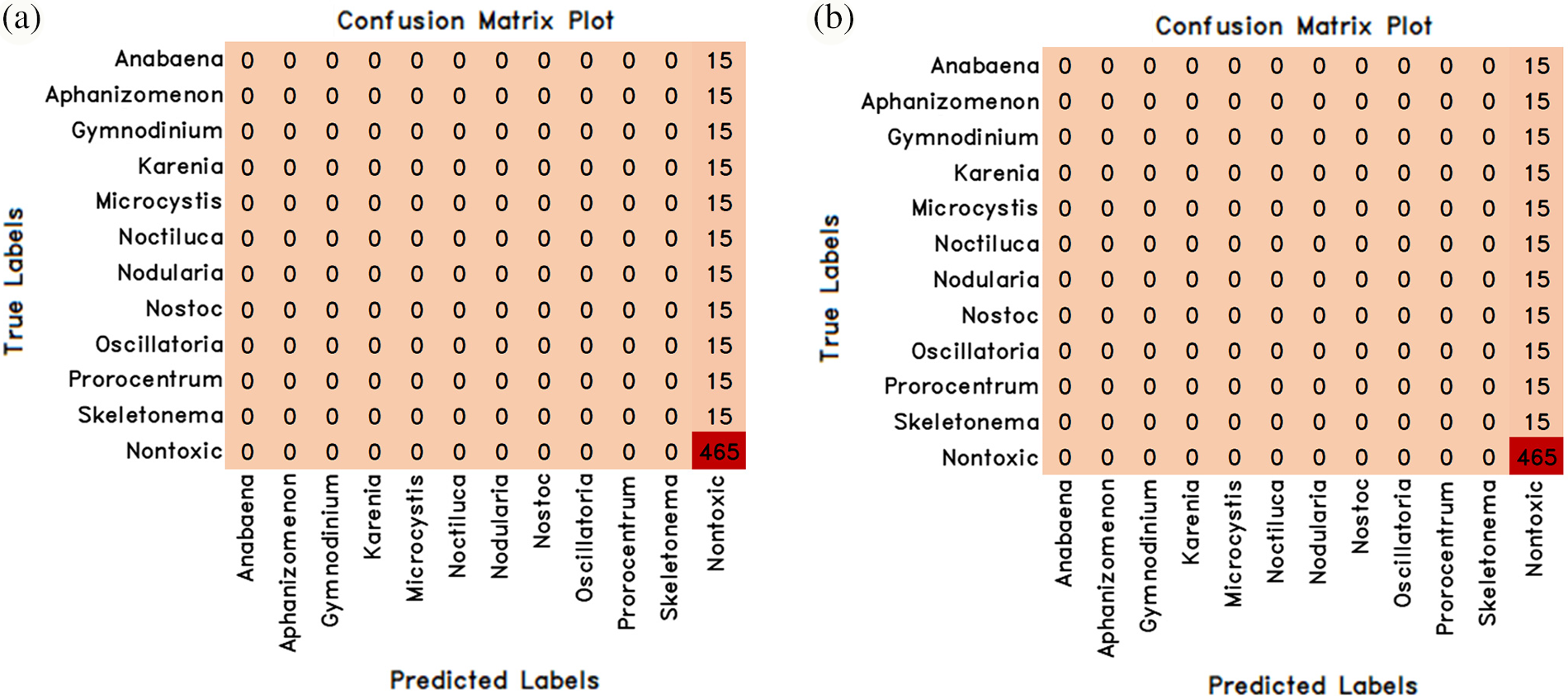
Figure 6: Confusion matrix values of deep learning methods in classifying algae cells by normal learning (a) MobileNetV3Small (b) MobileNetV3Large
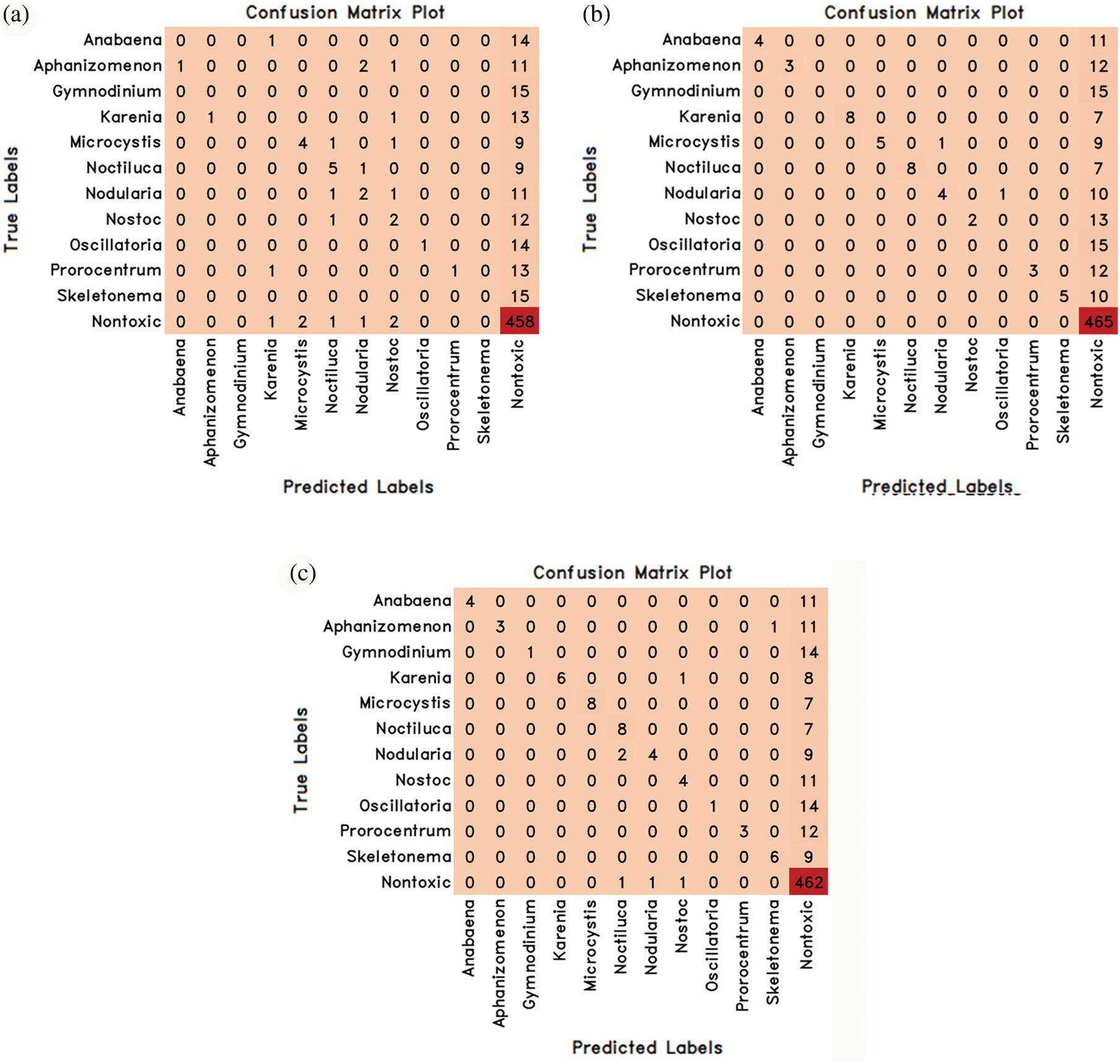
Figure 7: The confusion matrix values of the hybrid models proposed by combining MobileNetV3Small and Machine learning methods are (a) MobileNetV3Small-Lr (b) MobileNetV3Small-Rf (c) MobileNetV3Small-Xg
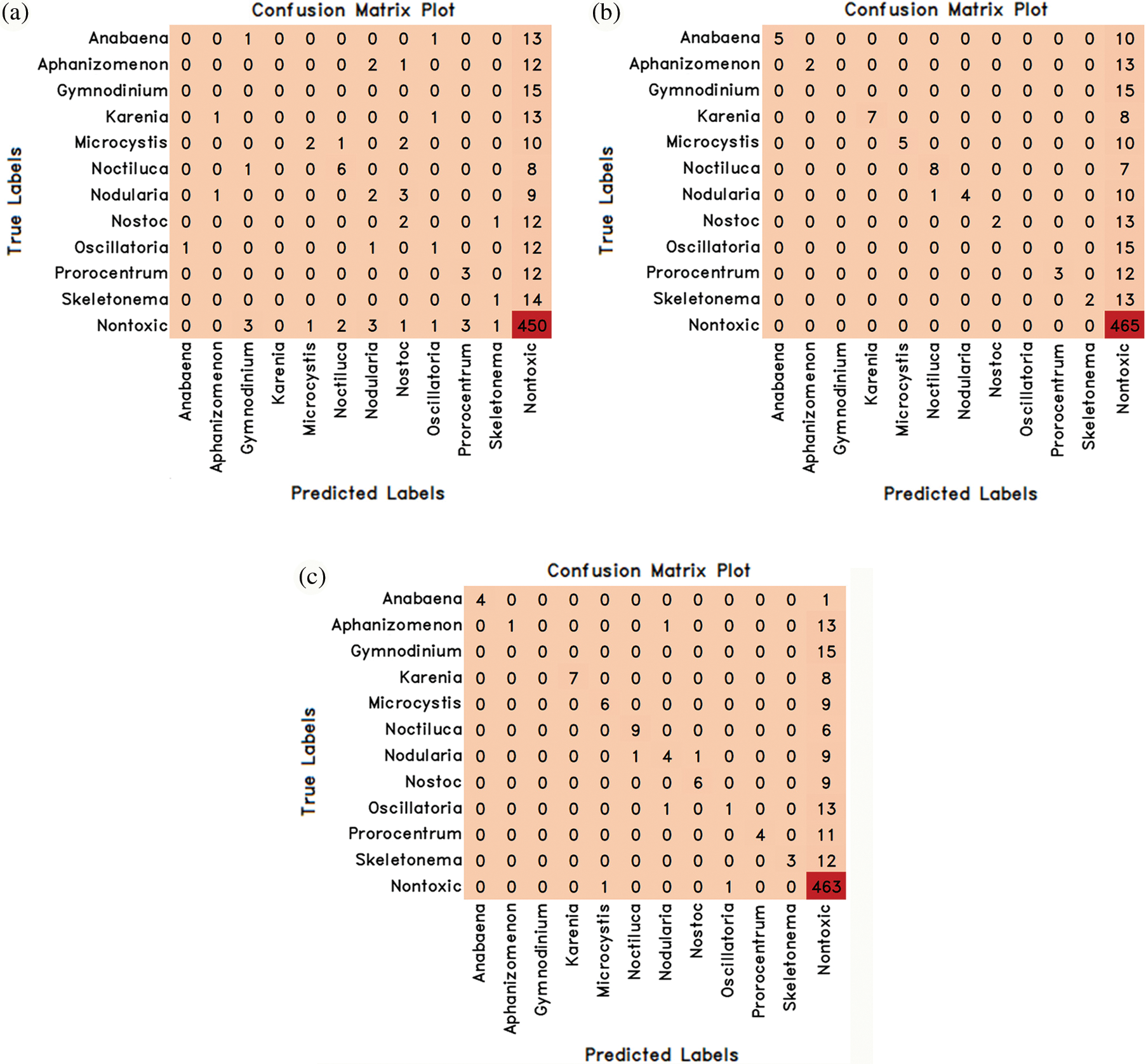
Figure 8: The confusion matrix values of the hybrid models proposed by combining MobileNetV3Large and Machine learning methods are (a) MobileNetV3Large-Lr (b) MobileNetV3Large-Rf (c) MobileNetV3Large-Xg
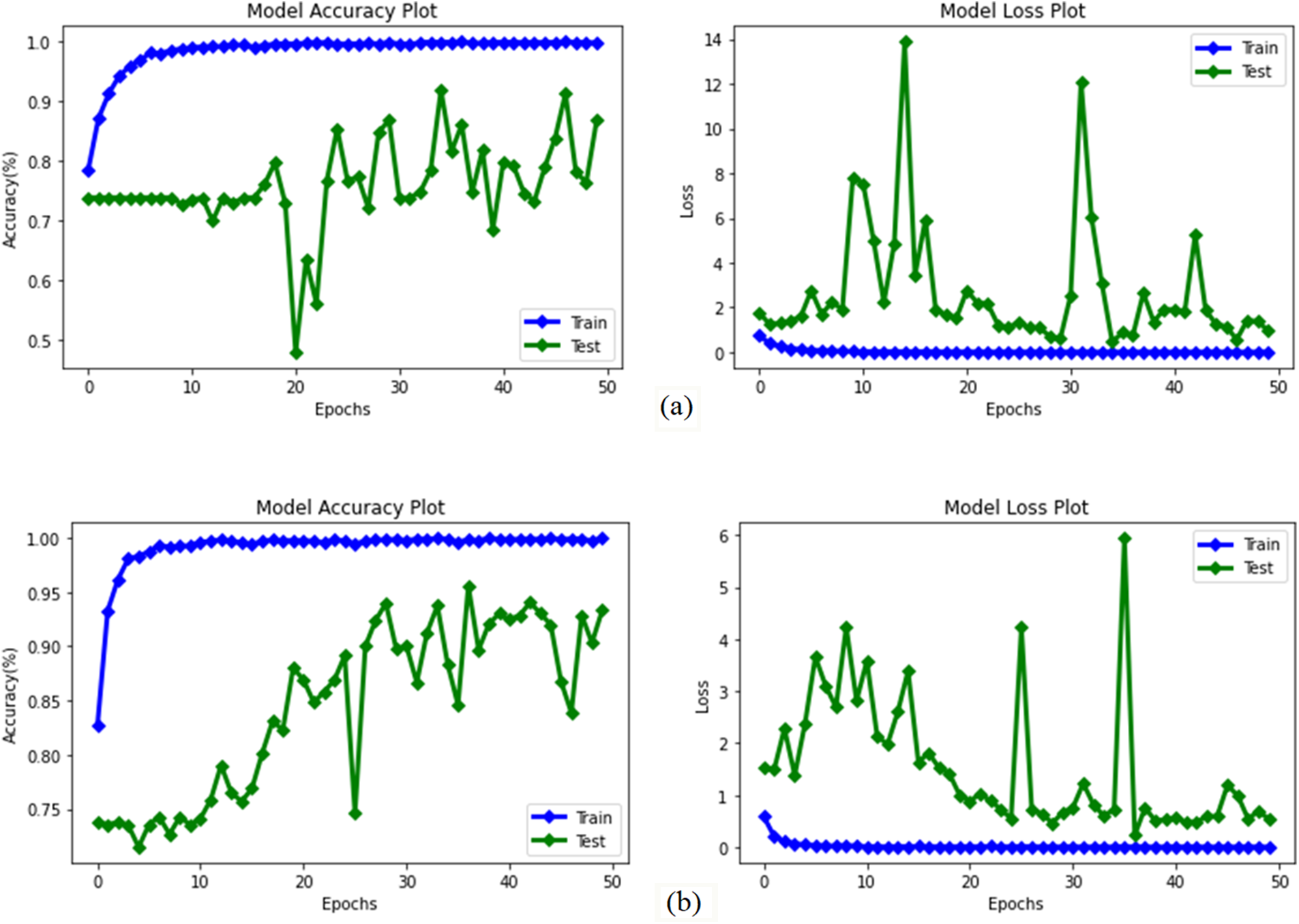
Figure 9: Accuracy and loss graphs obtained as a result of training tests of improved transfer learning models proposed by adding auxiliary layers to the previously trained weights of MobileNetV3Small and MobileNetV3Large deep learning methods (a) MobileNetV3Small-Improved (b) MobileNetV3Large-Improved
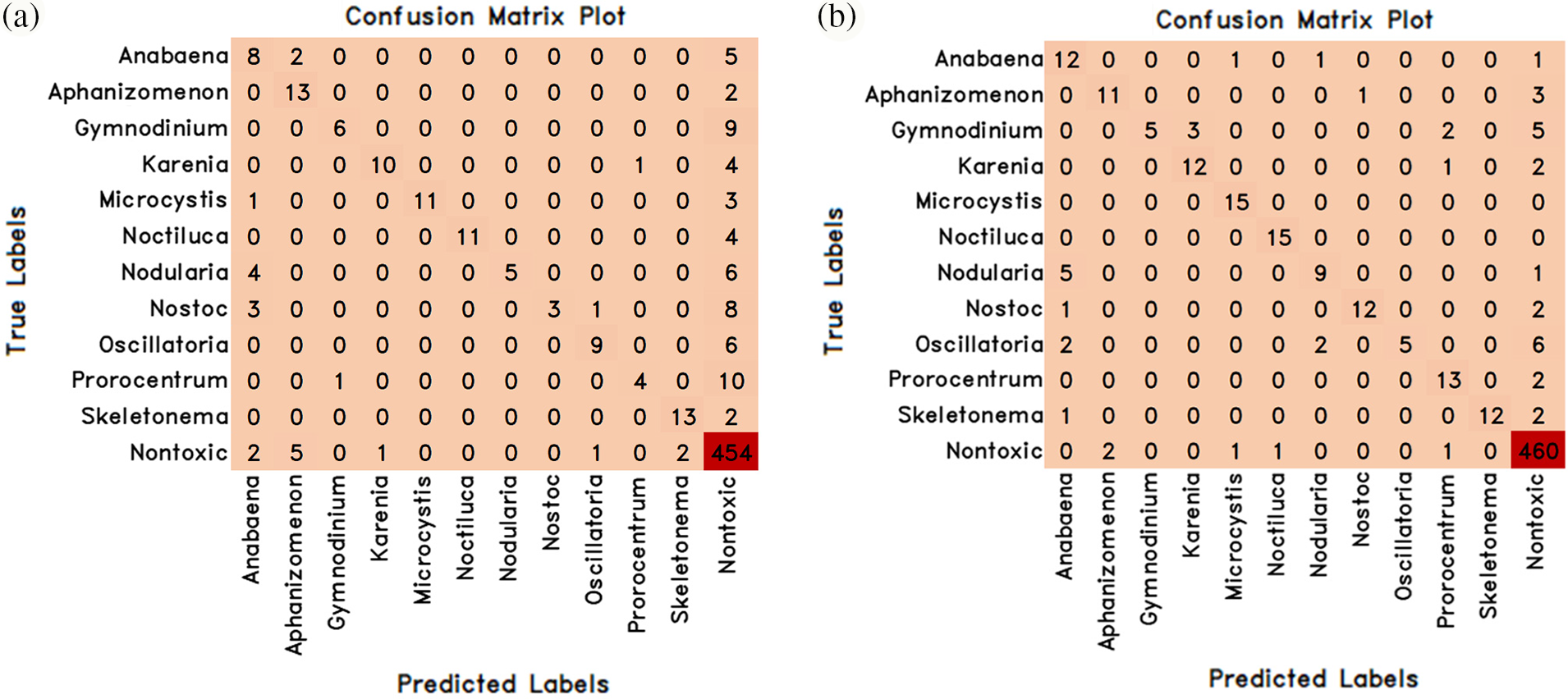
Figure 10: The confusion matrix values of the improved transfer learning models proposed by adding auxiliary layers to the previously trained weights of MobileNetV3Small and MobileNetV3Large deep learning methods are (a) MobileNetV3Small-Improved (b) MobileNetV3Large-Improved
Logistic regression, random forest and XGBoost methods, one of the machine learning methods, were used for normal learning first and the success results in classifying algae cells were obtained and the results obtained are given in Table 3 comparatively. However, the confusion matrix values obtained from these 3 models are given in Fig. 4.
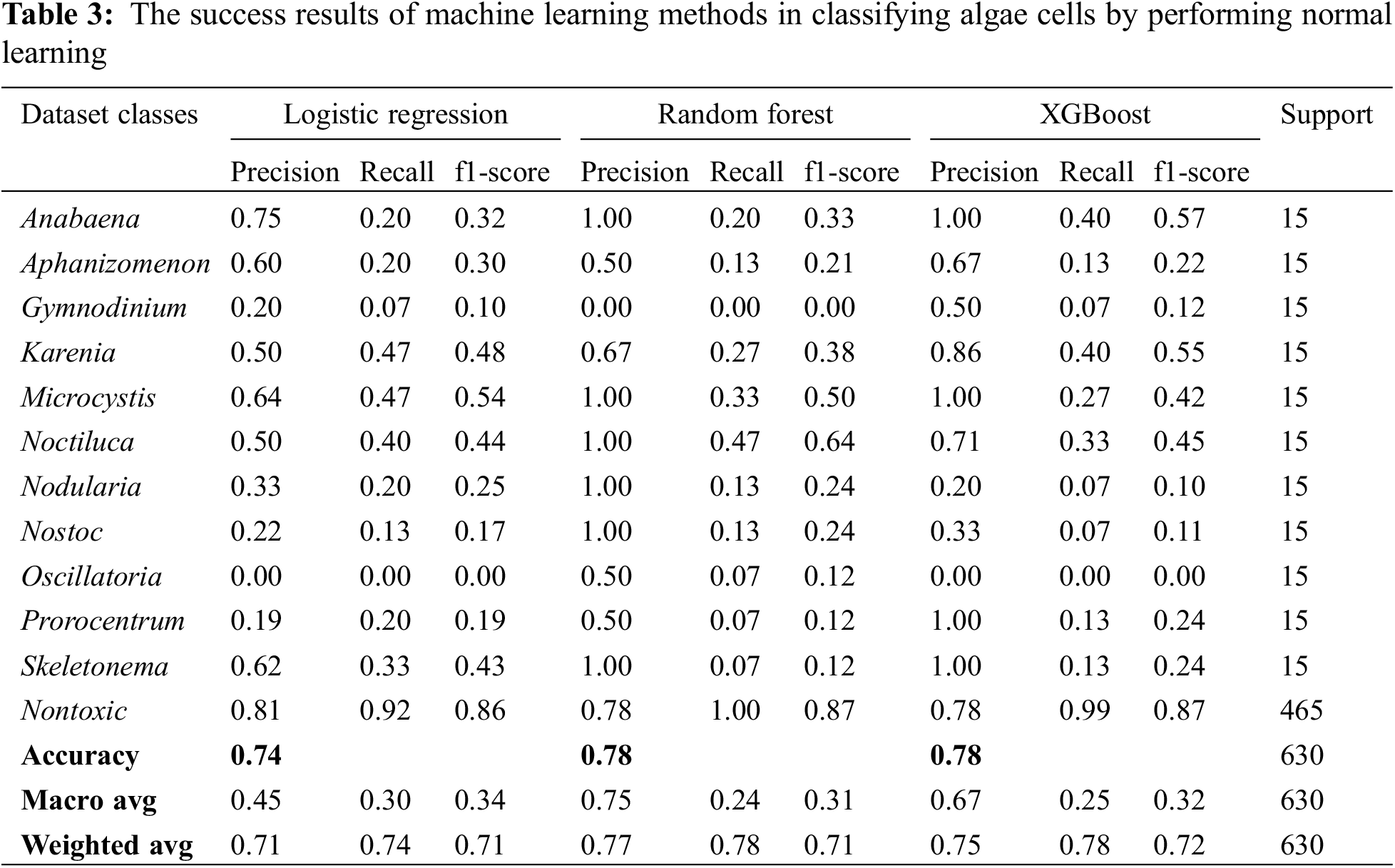
The success rates of logistic regression, random forest and XGBoost methods in classifying algae cells as a result of normal learning; 73.97% success was achieved with the logistic regression method, 77.94% with the random forest method and 77.94% with the XGBoost method (Table 3). In algae cell classification, random forest and XGBoost methods have been found to be better than logistic regression method by providing the same success rate.
The success results of the deep learning methods MobileNetV3Small and MobileNetV3Large in classifying algae cells by using normal learning are given in Table 4 comparatively, and the accuracy and loss graphs obtained from the training test result are given in Fig. 5. However, the confusion matrix values obtained from these 2 models are also given in Fig. 6.
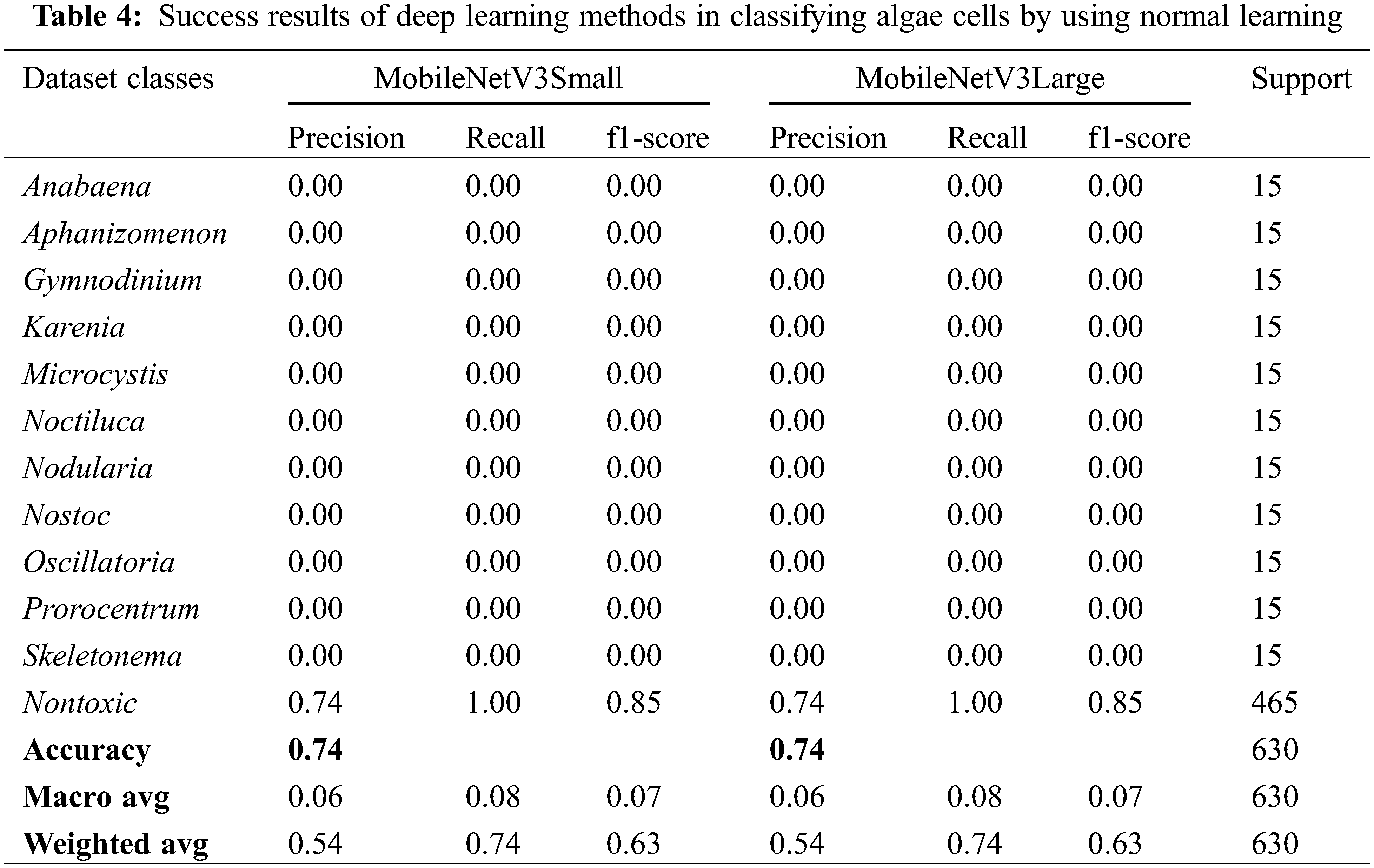
MobileNetV3Small and MobileNetV3Large methods success rates in classifying algae cells without transfer learning were achieved by 73.81% success both by MobileNetV3Small method and MobileNetV3Large method (Table 4). That being said, a loss rate of 37.41% was obtained by the MobileNetV3Small method and 7.98% by the MobileNetV3Large method. However, it was found that MobileNetV3Small and MobileNetV3Large methods did not perform a successful training with normal learning in algae cell classification.
In the MobileNetV3Small deep learning model, attribute extraction was performed only from pre-trained weights without training the entire network, and 3 novel transfer-learning hybrid models named MobileNetV3Small-Lr, MobileNetV3Small-Rf and MobileNetV3Small-Xg were proposed by combining them with machine learning methods. These proposed models have been trained and tested separately. The experimental results obtained as a result of training and testing are given in Table 5 comparatively. The confusion matrix values obtained from these 3 hybrid models are given in Fig. 7.
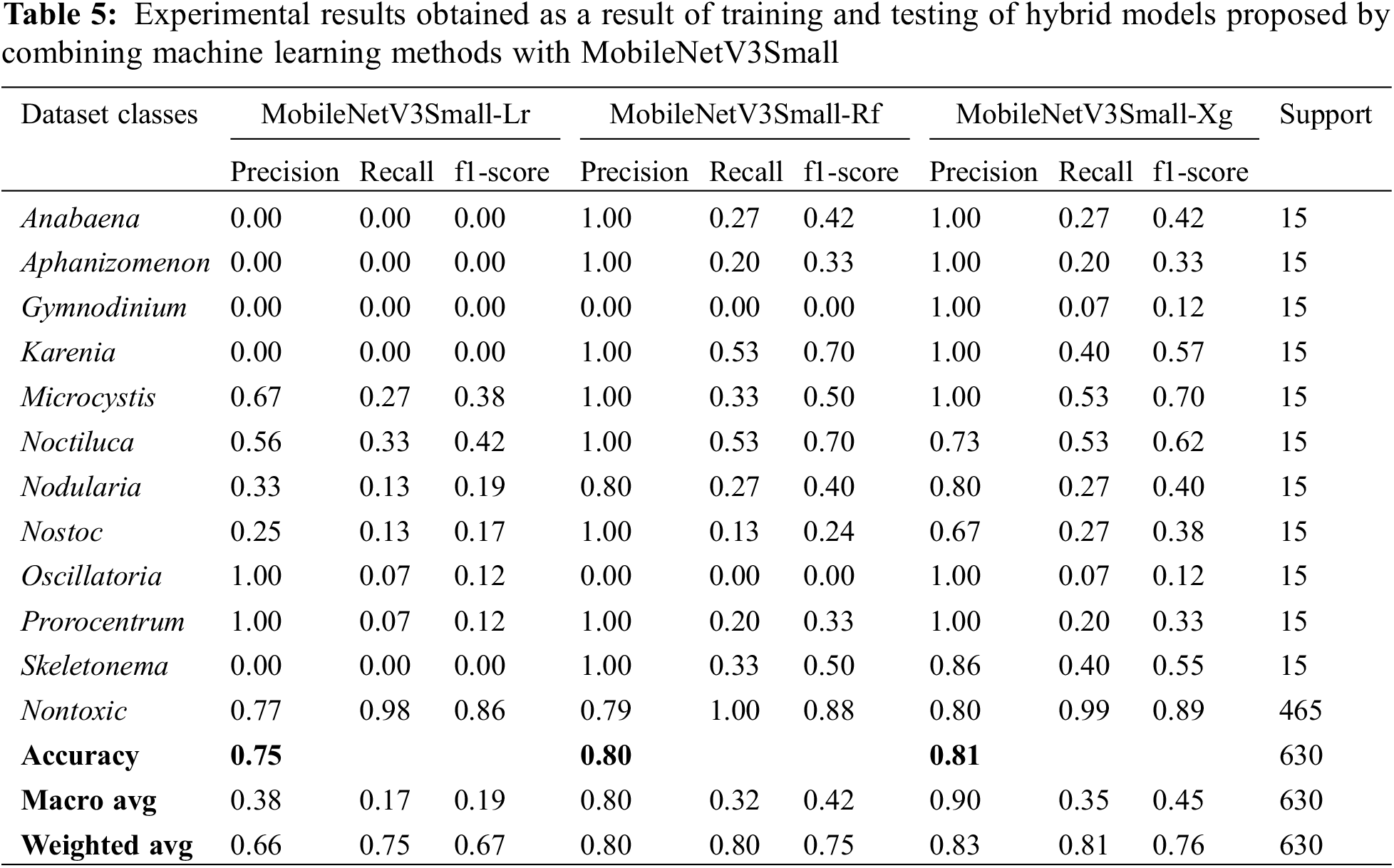
MobileNetV3Small-Lr, MobileNetV3Small-Rf and MobileNetV3Small-Xg hybrid transfer learning methods achieved success rates in classifying algae cells by MobileNetV3Small-Lr method 75.08%, MobileNetV3Small-Rf method 80.48% and MobileNetV3Small-Xg method 80.95% (Table 5). Therefore, it has been seen that the MobileNetV3Small-Xg method is better than the other two hybrid methods in algae cell classification. In Table 4 and Fig. 5, it was seen that learning occurred as a result of combining the MobileNetV3Small model, which gives unsuccessful results with normal learning, with machine learning methods.
Similarly, in the MobileNetV3Large deep learning model, attribute extraction was performed only from pre-trained weights without training the entire network, and 3 novel transfer-learning hybrid models named MobileNetV3Large-Lr, mobilenetv3Large-Rf and mobilenetv3Large-Xg were proposed by combining them with machine learning methods. These proposed models have been trained and tested separately. The experimental results obtained as a result of training and testing are given in Table 6 comparatively. The confusion matrix values obtained from these 3 hybrid models are given in Fig. 8.
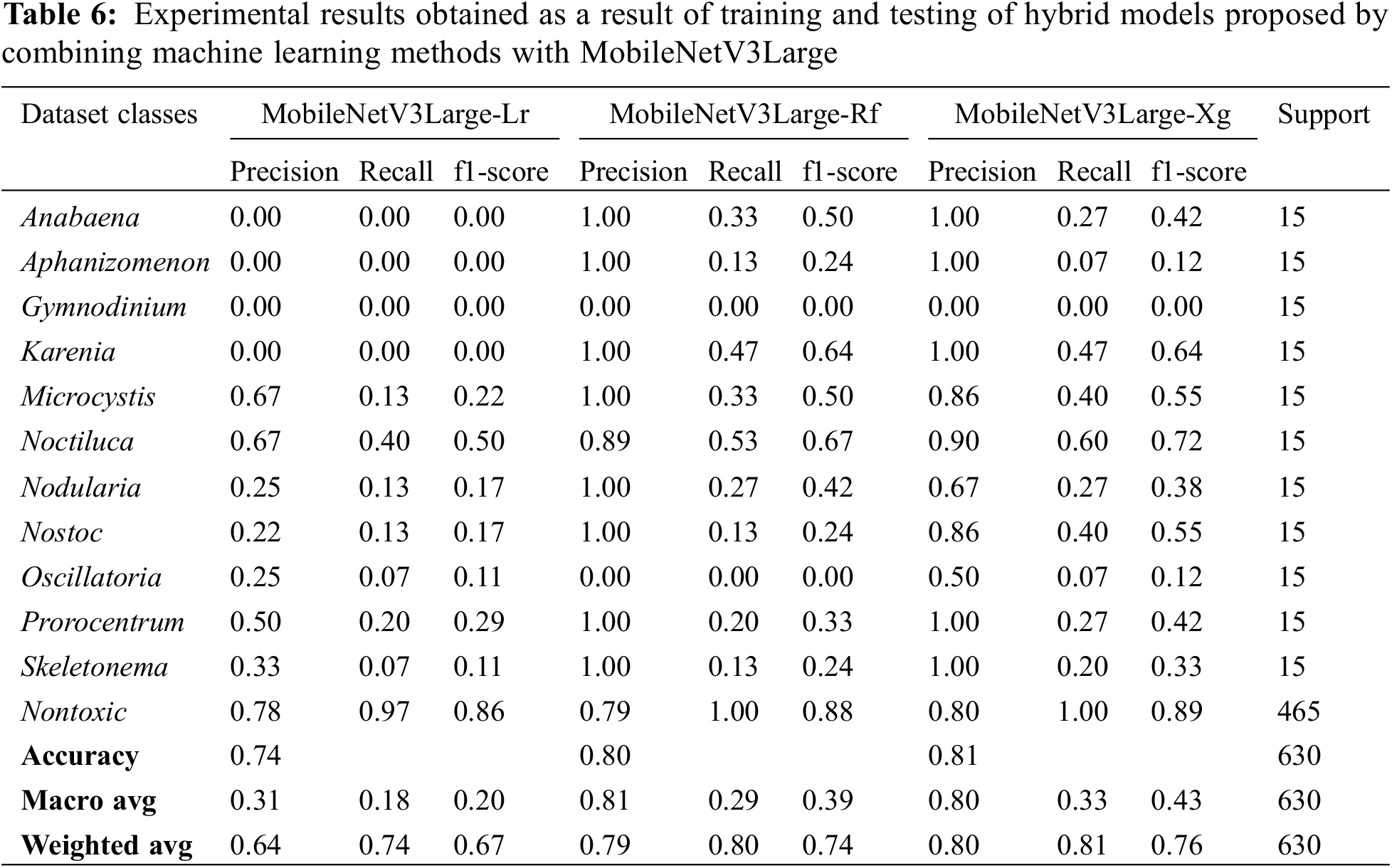
MobileNetV3Large-Lr, mobilenetv3Large-Rf and MobileNetV3Large-Xg hybrid transfer learning methods achieved success rates in classifying algae cells by MobileNetV3Large-Lr method 74.13%, MobileNetV3Large-Rf method 79.89% and MobileNetV3Large-Xg method 80.63% (Table 6). Therefore, it has been seen that the MobileNetV3Large-Xg method is better than the other two hybrid methods in algae cell classification. In Table 4 and Fig. 5, it was seen that learning occurred as a result of combining the MobileNetV3Large model, which gives unsuccessful results with normal learning, with machine learning methods.
In addition, the entire network was trained using pre-trained weights in the MobileNetV3Small and MobileNetV3Large deep learning models, and 2 novel improved transfer learning models called MobileNetV3Small-Improved and MobileNetV3Large-Improved were proposed by adding 1 512 convolution layer and 25% dropout auxiliary layer to the last layers of these models. These proposed models have been trained and tested separately. The experimental results obtained as a result of training and testing are given comparatively in Table 7, and the success and loss graphs are given in Fig. 9. The confusion matrix values obtained from these 2 improved transfer learning models are given in Fig. 10.
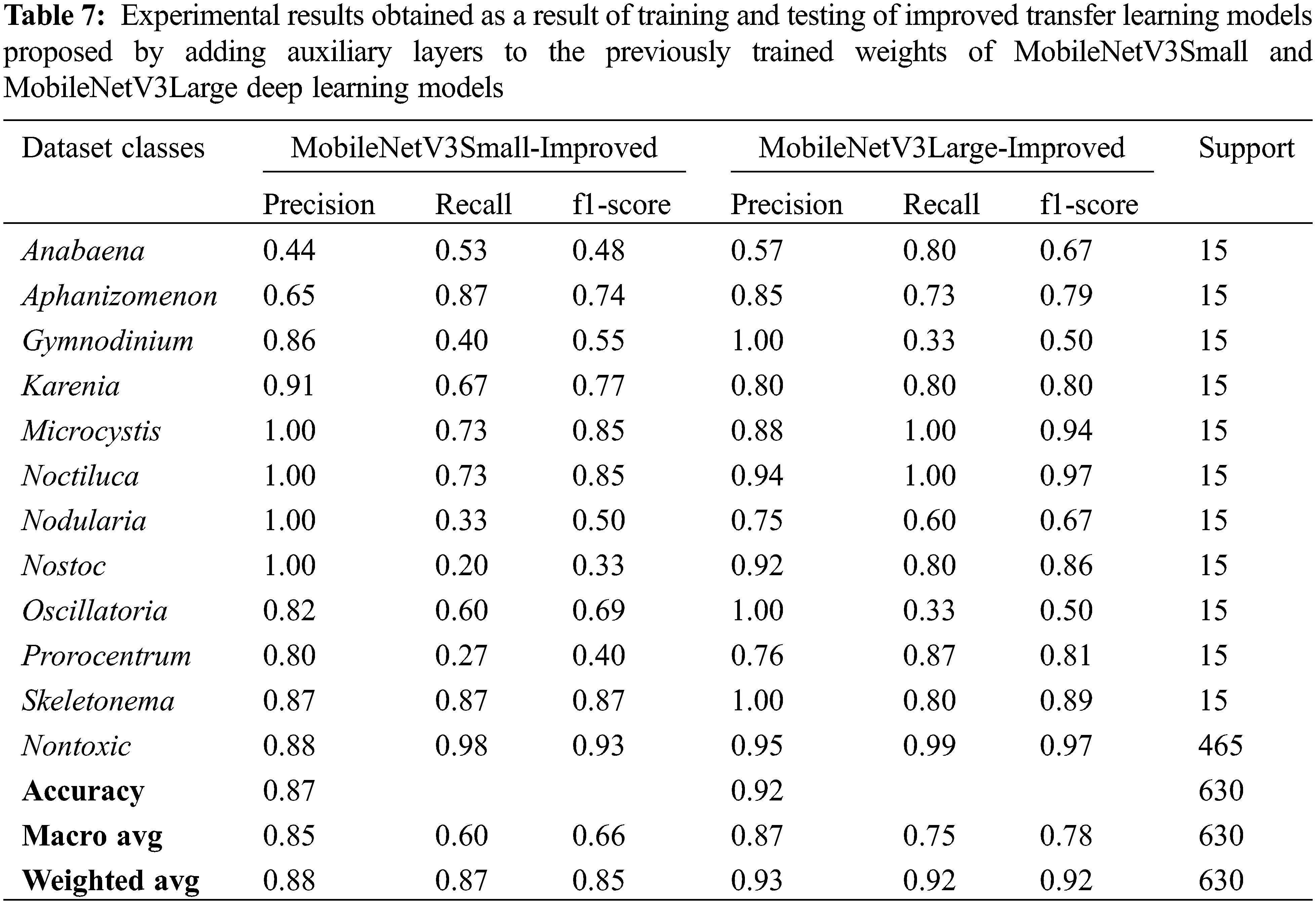
The success rates of MobileNetV3Small-Improved and MobileNetV3Large-Improved methods with improved transfer learning in classifying algae cells were 86.83% with the MobileNetV3Small-Improved method and 92.22% with the MobileNetV3Large-Improved method (Table 7). Therefore, it has been found that the MobileNetV3Large-Improved method is better than the MobileNetV3Small-Improved method in algae cell classification. However, a loss ratio of 1.08 was obtained by the MobileNetV3Small-Improved method and 0.72 by the MobileNetV3Small-Improved method.
Therefore, the machine learning and deep learning methods considered in the study were used both without improvement and by making improvements, experimental results were obtained on algae cells, and the success rates obtained are given comparatively in Table 8.
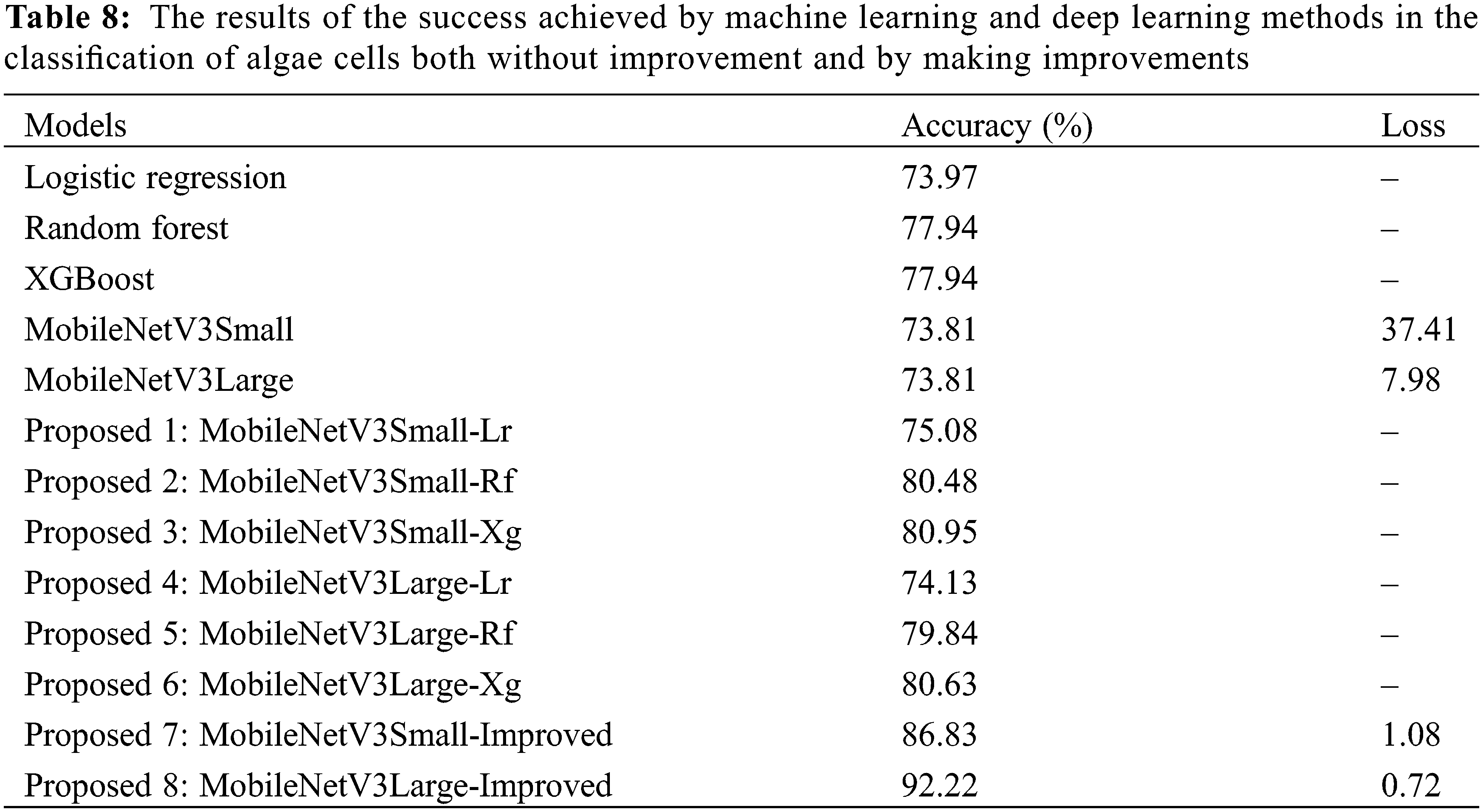
As a result, the classification success rates obtained of hybrid deep learning methods and machine learning methods have shown a significant increase compared to the algae cell classification success rates made using only machine learning methods or only deep learning methods. However, it was found that the MobileNetV3Small-Improved and MobileNetV3Large-Improved models, which were improved by adding an auxiliary layer, also showed better classification success than hybrid models.
According to all the experimental findings obtained, it has been seen that the MobileNetV3Large-Improved model structure is the model that provides the best success rate in classifying algae cells. In this model, it was found that the success rate of classifying algae cells was 92.22% and the loss rate was 0.72. Therefore, it is seen that the success rate is provided well as a result of the 8 different methods proposed using 3 machine learning and 2 deep learning methods based on classifying algae cells, however, the best model is MobileNetV3Large-Improved.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: V.K., İ.A., Ö.Z.T.; data collection: Ö.Z.T., V.K.; analysis and interpretation of results: İ.A., V.K.; draft manuscript preparation: V.K., Ö.Z.T. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Yadav, D., Jalal, A., Garlapati, D., Hossain, K., Goyal, A. et al. (2020). Deep learning-based ResNeXt model in phycological studies for future. Algal Research, 50, 102018. https://doi.org/10.1016/j.algal.2020.102018 [Google Scholar] [CrossRef]
2. Oktor, K. (2018). The role of microalgae in environmental technologies. In: Environmental, science and technology, pp. 330–343. Turkey: Güven Plus Grup AŞ Press. [Google Scholar]
3. Sisman-Aydın, G. (2019). Microalgae technology and its environmental use. Harran University Journal of Engineering, 4, 81–92. [Google Scholar]
4. Aslan, M. F., Unlersen, M. F., Sabanci, K., Durdu, A. (2021). CNN-based transfer learning-bilstm network: A novel approach for COVID-19 infection detection. Applied Soft Computing, 98, 106912. https://doi.org/10.1016/j.asoc.2020.106912 [Google Scholar] [PubMed] [CrossRef]
5. Otálora, P., Guzmán, J. L., Acién, F. G., Berenguel, M., Reul, A. (2021). Microalgae classification based on machine learning techniques. Algal Research, 55, 102256. https://doi.org/10.1016/j.algal.2021.102256 [Google Scholar] [CrossRef]
6. Santhi, N., Pradeepa, C., Subashini, P., Kalaiselvi, S. (2013). Automatic identification of algal community from microscopic images. Bioinformatics and Biology Insights, 7, 327–334. [Google Scholar] [PubMed]
7. Promdaen, S., Wattuya, P., Sanevas, N. (2014). Automated microalgae image classification. Procedia Computer Science, 29, 981–1992. https://doi.org/10.1016/j.procs.2014.05.182 [Google Scholar] [CrossRef]
8. Li, X., Liao, R., Zhou, J., Leung, P. T. Y., Yan, M. et al. (2017). Classification of morphologically similar algae and cyanobacteria using Mueller matrix imaging and convolutional neural networks. Applied Optics, 56(23), 6520–6530. https://doi.org/10.1364/AO.56.006520 [Google Scholar] [PubMed] [CrossRef]
9. Deglint, J. L., Jin, C., Wong, A. (2019). Investigating the automatic classification of algae using the spectral and morphological characteristics via deep residual learning. In: Karray, F., Campilho, A., Yu A. (Eds.). Lecture notes in computer science, vol. 11663, pp. 269–280. [Google Scholar]
10. Salido, J., Sánchez, C., Ruiz-Santaquiteria, J., Cristóbal, G., Blanco, S. et al. (2020). A low-cost automated digital microscopy platform for automatic identification of diatoms. Applied Sciences, 10(17), 6033. https://doi.org/10.3390/app10176033 [Google Scholar] [CrossRef]
11. Balado, J., Olabarria, C., Martínez-Sánchez, J., Rodríguez-Pérez, J. R., Pedro, A. (2020). Semantic segmentation of major macroalgae in coastal environments using high-resolution ground imagery and deep learning. Remote Sensing, 42(5), 1785–1800. https://doi.org/10.1080/01431161.2020.1842543 [Google Scholar] [CrossRef]
12. Yang, M., Wang, W., Gao, Q., Zhao, C., Li, C. et al. (2022). Automatic identification of harmful algae based on multiple convolutional neural networks and transfer learning. Environmental Science and Pollution Research, 30, 15311–15324. https://doi.org/10.1007/s11356-022-23280-6 [Google Scholar] [PubMed] [CrossRef]
13. Kaggle (2023). https://www.kaggle.com/datasets/mengyuy/the-algae-cell-images [Google Scholar]
14. Colab (2023). Google Colaboratory. https://colab.research.google.com [Google Scholar]
15. Hosmer Jr, D. W., Lemeshow, S., Sturdivant, R. X. (2013). Applied logistic regression, vol. 398. USA: John Wiley & Sons. [Google Scholar]
16. Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32. https://doi.org/10.1023/A:1010933404324 [Google Scholar] [CrossRef]
17. Chen, T., He, T., Benesty, M., Khotilovich, V., Tang, Y. et al. (2015). XGBoost: Excessive gradient enhancement. R Package Version 0.4-2, 1(4), 1–4. [Google Scholar]
18. Howard, A., Sandler, M., Chu, G., Chen, L. C., Chen, B. et al. (2019). Searching for MobileNetV3. 2019 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 1314–1324. Seoul, Korea (South). [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools