 Open Access
Open Access
ARTICLE
Genome-Wide Analysis of the F3′5′H Gene Family in Blueberry (Vaccinium corymbosum L.) Provides Insights into the Regulation of Anthocyanin Biosynthesis
1 College of Life Sciences, Huizhou University, Huizhou, 516007, China
2 College of Forestry, Guizhou University, Guiyang, 550025, China
3 School of Architecture and Civil Engineering, Huizhou University, Huizhou, 516007, China
4 Guizhou Institute of Subtropical Crop., Guizhou Academy of Agricultural Sciences, Xingyi, 562400, China
* Corresponding Author: Delu Wang. Email:
Phyton-International Journal of Experimental Botany 2023, 92(9), 2683-2697. https://doi.org/10.32604/phyton.2023.029820
Received 10 March 2023; Accepted 08 June 2023; Issue published 28 July 2023
Abstract
The F3′5′H gene family plays an important role in regulating anthocyanin biosynthesis, abiotic stress, and hormone signaling. In this study, 14 F3′5′H genes were identified from the blueberry genome. The chromosomal distribution, physicochemical properties, F3′5′H domain, conserved motifs, cis-acting elements, and intron/exon compositions were analyzed. The functional prediction analysis of these VcF3′5′Hs indicated that their biological functions included light response and other secondary metabolites. The results of qRT-PCR showed that VcF3′5′Hs (especially VcF3′5′H4) were highly expressed at the ripening stage. Subcellular localization revealed that VcF3′5′H4 may be located in the endoplasmic reticulum. Co-expression analysis showed that the VcF3′5′H gene family was related to anthocyanin. This research provides an overview of the blueberry F3′5′H family and helps verify the role of these genes in regulating anthocyanin biosynthesis.Keywords
Supplementary Material
Supplementary Material FileThe blueberry bush is a commercially significant shrub in North America and a popular fruit tree. Blueberry refers to the blue fruit produced by member species within the genus Vaccinium (Ericaceae). In China, 347,200 tons of blueberries were harvested from 66,400 hm2 of cultivated area [1]. Guizhou Province has a blueberry planting area of 15,000 hm2, with an annual output of 85,000 tons [1]. A growing number of studies have demonstrated the positive effects of fruit metabolites (e.g., anthocyanins) on human health, which drives the market expansion of this so-called “superfood” [2,3]. Anthocyanins are used to improve visual acuity and immunity, and have certain anti-inflammatory, anti-ultraviolet radiation, anti-pathogen infection, anti-aging, and anti-cancer effects [4–7]. On this basis, the economic value of blueberries can be improved by increasing their anthocyanin contents.
The blueberry anthocyanin synthesis pathway has been studied [8]. The purple pigment in blueberries is anthocyanin, mainly consisting of delphinidin-3-O-arabinoside [9,10]. Our preliminary research discovered that delphinidin accumulation is associated with the VcF3′5′Hs gene, which encodes the flavonoid 3′5′-hydroxylase (F3′5′H) enzyme [11]. In the anthocyanin metabolic pathway, F3′5′H, belonging to the cytochrome P450 (CYP) enzyme family, is the main enzyme in the synthesis of blue delphinium and crystalizes the hydroxylation of the 3′ and 5′ positions of the B ring of dihydroflavonols [12]. F3′5′H is also known as the blue gene, and its gram long offers the possibility of generating blue pigment [13]. In this sense, F3′5′H is thought to be the major enzyme gene that regulates blueberry anthocyanin production.
F3′5′H was first cloned from petunia and was found to be involved in pigment synthesis [14,15]. Its homologs were subsequently cloned from various plants, including gentian [16], Arabidopsis [17], Solanum tuberosum [18], tomato [19], Ginkgo [20], and Aconitum [21]. F3′5′H genes are linked to anthocyanin synthesis. The overexpression of the F3′5′H gene can promote the accumulation of poplar anthocyanins and enhance the hydroxylation of flavonoids [22]. The hydroxylation pattern of flavonoids is largely determined by F3′5′H genes. For example, cloning the F3′5′H genes can change the color of roses, carnations, and other plants without F3′5′H activity from lavender to blue [23–25]. Some F3′5′H genes have been used in breeding new varieties of blue ornamental plants [16,26,27]. In sum, the cloning and characterization of the F3′5′H gene are of special significance.
In the past few decades, many studies on anthocyanin biosynthesis have given an understanding of the F3′5′H pathway. Anthocyanins are natural water-soluble secondary metabolites belonging to the flavonoid metabolism branch of the phenylpropanoid pathway [28]. Many genes involved in this pathway, such as phenylalanine ammonia-lyase, 4-coumaroyl: CoA ligase, and chalcone synthase (CHS), have been identified. However, few studies have been conducted to analyze the F3′5′H gene family associated with blueberry anthocyanin biosynthesis. The biosynthetic pathway is relatively conserved among different species, and the genes regulating anthocyanin biosynthesis differ in organs and tissues. The MYB gene appears to play a key role in regulating anthocyanin biosynthesis. In blueberries, VcMYBPA1.1 can activate VcCHS, VcF3′5′H, VcDFR, VcANS, and VcGST promoters and facilitate the anthocyanin synthesis [29]. Silencing VmMYBPA1.1 in bilberry results in the decreased expression of VmCHS, VmF3′5′H, VmANS, VmLAR1a, and anthocyanin loss [30]. The R2R3-MYB transcription factor SmMYB35 can bind to promoters, such as SmCHS, SmF3H, SmDFR, and SmANS, to enhance their activity to promote anthocyanin synthesis in eggplant [31]. Studies have shown that the F3′5′H gene is regulated by MYB or bHLH transcription factors, thereby promoting anthocyanin synthesis. The study of the blueberry F3′5′H gene family is significant for the cultivation of blueberry varieties with high anthocyanin contents.
Blueberry (Vaccinium spp.) is one of the most important economic berries in the world. The finding of the whole blueberry genome sequence enables the performance of whole-genome F3′5′H gene analysis [32]. F3′5′H family research showed that F3′5′H members are related to anthocyanin biosynthesis, abiotic stress, and hormone signaling [10,33,34]. Anthocyanin accumulation at different developmental stages of blueberries resulted in color-phenotype differentiation. However, few studies have explored the genetic basis of the peel and pulp color. The genetic basis of anthocyanin regulation in blueberries can be further understood by combining phenotypic, genomic, and transcriptome analyses. In this study, a genome-wide analysis of F3′5′H in blueberries was performed, and 14 F3′5′H genes were identified. A comprehensive investigation of the gene structure, cis-acting element analysis, and conserved domains provided evidence for the functional annotation. Furthermore, subcellular localization and gene expression analyses indicate that the VcF3′5′H4 gene is crucial in regulating blueberry anthocyanin synthesis. This study lays a foundation for exploring the role of the F3′5′H gene in anthocyanin biosynthesis in blueberries and promotes further functional characterization of these regulators.
Five-year-old rabbiteye blueberries ‘fenlan’ (Vaccinium ashei: Reade) were grown in Guiyang, Guizhou, China, with a planting distance of 30 cm × 30 cm. The growing region was located at 106°27′−106°52′ east longitude and 26°11′−26°34′ north latitude. Three plants of uniform growth were selected for each treatment as biological replicates. Fruit samples were collected at 80 d (S1), 95 d (S2), and 110 d (S3) after the flowering (Fig. 1). All samples were immediately frozen in liquid nitrogen and stored at −80°C until use.
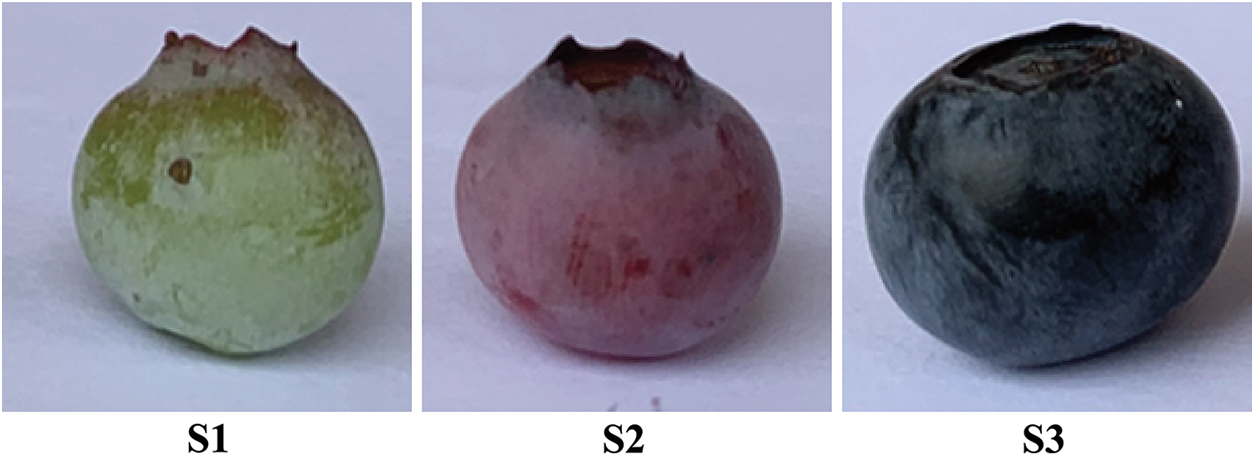
Figure 1: Fruit sampling period
2.2 Identification of VcF3′5′H Members and Phylogenetic Analysis
The blueberry genome file was aligned with the F3′5′H sequence mentioned by Liu et al. [35]. The alignment method was BLAST with an E-value of 1.0 × 10−5, and 17 VcF3′5′H were identified by default parameter settings. P450 database download site was as follows: https://drnelson.uthsc.edu/. All protein sequences were aligned by the ClustalW program in MEGA 7, and then NJ phylogenetic tree was constructed based on the Poisson model with a bootstrap value of 1,000.
2.3 Motif Identification, Gene Structures, and Protein Motif Compositions of VcF3′5′H Members
The gene structures of 14 blueberry F3′5′H genes were extracted from the gff3 structural annotation file by Perl script. Online tools were employed for analysis. Specifically, GSDS 2.0 (http://gsds.cbi.pku.edu.cn/) was used for domain visualization [36], and meme (http://memesuite.org/) [37] was adopted for comparing the VcF3′5′H conserved motif with 10 conserved motif parameters and other default parameters; TBtools (https://github.com/CJ-Chen/TBtools) was used for motif visualization analysis [38], and molecular weight, stability coefficient, and hydrophobicity were analyzed using ExPASy [39].
2.4 Sequence Alignments, Chromosome Mapping, Profiles of VcF3′5′H Conserved Amino Acids, and Collinear Analysis of Anthocyanin
SOPMA was used to predict the secondary structure of VcF3′5′H family proteins. DNAMAN was employed for the protein alignment of the identified 14 VcF3′5′H genes. The structure files of F3′5′H genes in blueberries were extracted by TBtools [39] to determine their starting and ending positions in the chromosomes. Co-expression network analysis was performed on the VcF3′5′H gene and anthocyanins. The analysis tool was Cytoscape 3.7.2, and R ≥ 0.9 was considered significantly correlated [40]. Anthocyanins were detected using LC-MS/MS. All data are shown in Supplementary Table S1.
2.5 Cis-Acting Elements Located in VcF3′5′H Gene Promoters
A 2000 bp upstream sequence of the gene was extracted by Bedtools [41], and was analyzed by the online tool PlantCARE [42], and the online tool GSDS 2.0 was used for the visual analysis of functional elements.
A total of 14 VcF3′5′Hs samples at three periods (S1, S2, and S3) were selected for qRT-PCR analysis. The primers for the genes were designed using Primer Premier 5.0. Total RNA was isolated from green, pink, and purple fruits by CTAB, and was checked on 1.2% agarose gel under UV light without smearing before the concentration detection using spectrophotometry. One microgram of total RNA was used to synthesize cDNA using a PrimeScript™ RT reagent kit with gDNA Eraser (TaKaRa, Japan) following the manufacturer’s instructions. The primers were designed for PCR amplification (Supplementary Table S2). QRT-PCR was performed in a real-time PCR system (QuantStudio 3D digital PCR system, ABI) using SYBR Real Master Mix (TransGen, Beijing, China) under the following PCR thermal cycling conditions: pre-denaturation at 95°C for 30 s, and 40 cycles at 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s, respectively. GAPDH (AY123769) bilberry genes were used as the housekeeping genes. Tree biological replicates were performed for each gene, and the standard curve method was applied in statistical analysis. The relative mRNA expression data were analyzed using 2–ΔΔCt [43]. Excel v. 2010 (Microsoft Corp., Redmond, WA, USA) and SPSS v. 25 (IBM Corp., USA) program was used to analyze the data using one-way analysis of variance (ANOVA). Duncan’s new multiple range test was used to separate means where ANOVA indicated significant differences at p < 0.05.
2.7 VcF3′5′H 4 Gene Cloning and Subcellular Localization Analysis
The CDS of the VcF3′5′H4 gene was obtained by searching the reference genome (http://gigadb.org/dataset/100537) according to the gene number, and primers were designed for PCR amplification (Supplementary Table S3). PCR products were cloned into a pBWA(V)HS-ccdb-GLosgfp and sequenced (Sangon Biotech, Shanghai, China).
The VcF3′5′H4 CDS with Bsal sites but without termination codons were amplified from the pBWA(V)HS-ccdb-GLosgfp plasmid template with Taq DNA polymerase (TaKaRa Biotechnology Co., Ltd., Japan). It was then inserted immediately upstream of the green fluorescent protein (GFP) coding sequence, and framed in the pBWA(V)HS-35s-GLosgfp vector, which had been digested with Bsal to generate pBWA(V)HS-VcF3′5′H4-GLosgfp vector with the cauliflower mosaic virus 35S promoter. The construct was subsequently introduced into Agrobacterium tumefaciens strain GV3101. To investigate the subcellular localization of VcF3′5′H4 protein, 35S: VcF3′5′H4-GFP and negative control 35S: GFP plasmids were transformed into N. benthamiana as described previously (Johansen and Carrington, 2001) and cultured in a growth chamber for 48 h (25°C, 16 h light/8 h dark). Fluorescence visualization of GFP was performed using a Zeiss confocal microscope (LSM510; Carl Zeiss, Thornwood, NY, USA) with an excitation wavelength of 480 ± 20 nm and an emission wavelength of 510 ± 20 nm.
3.1 Identification, Conserved Domain, and Gene Structure Analysis of the VcF3′5′H Gene Family
A total of 17 blueberry VcF3′5′H genes were identified using the BLAST alignment with reference to the F3′5′H gene sequence provided by Liu et al. [35] (Supplementary Figure S1). Only 14 F3′5′H sequences and a CYP75 subtribe were discovered to be together when we examined 17 samples with P450 subfamily sequences for an evolutionary study (Supplementary Figure S1). KEGG annotation was used to further verify that the 14 genes were blueberry F3′5′H genes. All of them were mapped to pseudochromosomes (VaccDscaff) and renamed from VcF3′5′H1 to VcF3′5′H14 according to the order of locations on the pseudochromosomes. Gene characteristics were analyzed (Table 1). The result showed that the shortest proteins were VcF3′5′H11, VcF3′5′H13, and VcF3′5′H14 (508 amino acids), and the longest one was VcF3′5′H1 (601 amino acids). According to the analysis, the molecular weights of the 14 VcF3′5′H proteins ranged from 56.7 to 66.4 kDa, and the isoelectric points ranged from 7.14 to 9.19. The hydrophilicity coefficient was between −0.207 and 0.053. Except for VcF3′5′H1, VcF3′5′H6, and VcF3′5′H11, which were hydrophobins, other family members were hydrophilic proteins. Secondary structure prediction revealed that the secondary structures of the VcF3′5′H gene family were dominated by the α-helix, ranging from 41.73% to 46.09%, followed by the random coil (30.27%–33.07%); the extended strand accounted for 14.17%–17.58%, and the β-turn was relatively less (7.32%–10.83%). As shown in Fig. 2C, the numbers of exons in VcF3′5′H genes were between 2 and 4, with an average of 2.4. Among all VcF3′5′H genes, 11 contained two exons and accounted for approximately 65% of VcF3′5′H gene family members, whereas only 3% of VcF3′5′H genes had more than three exons (Fig. 2C). The differences in protein sequences among the blueberry VcF3′5′H were analyzed using Multiple Expectation Maximization for Motif Elicitation online tool.
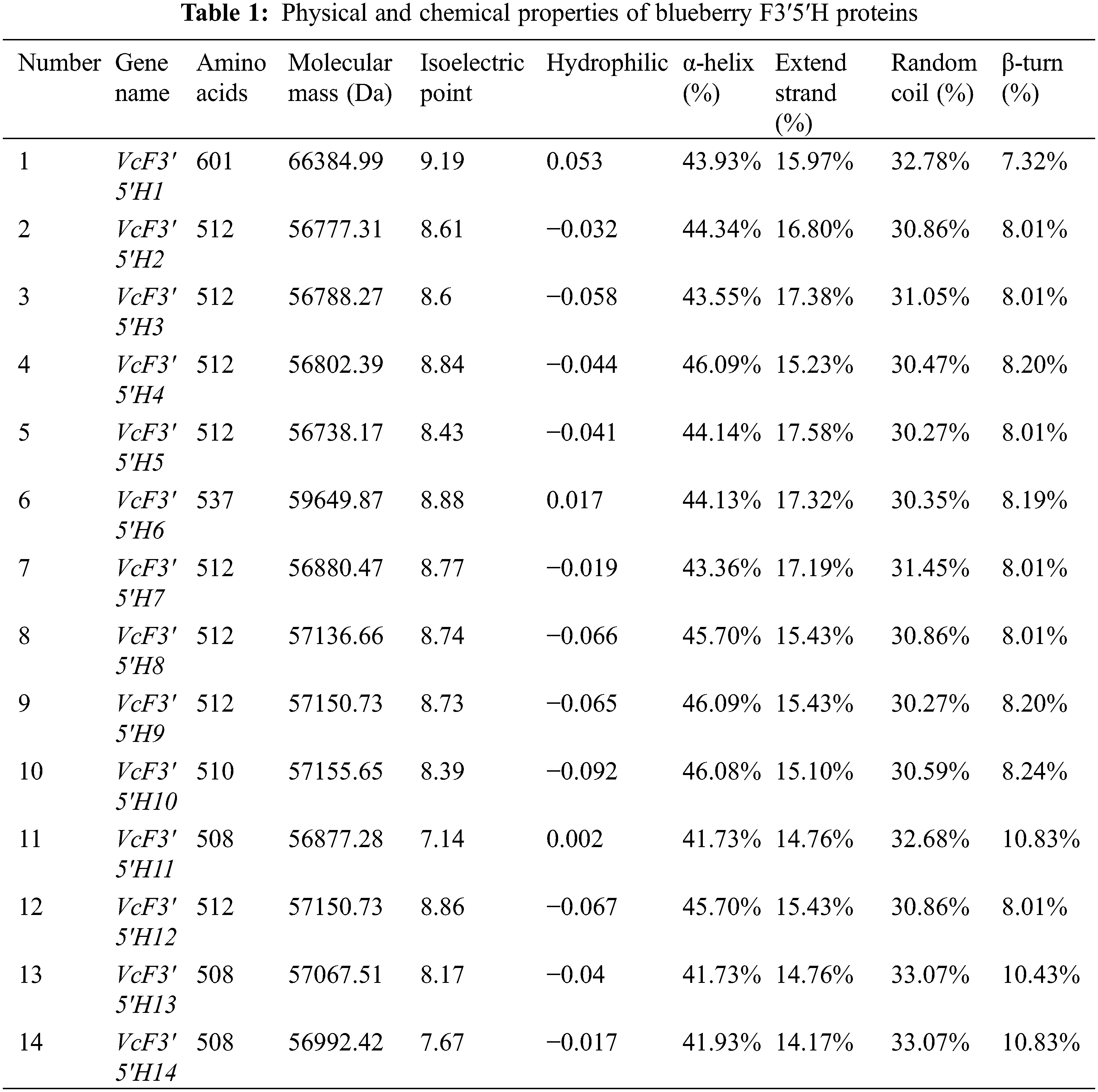
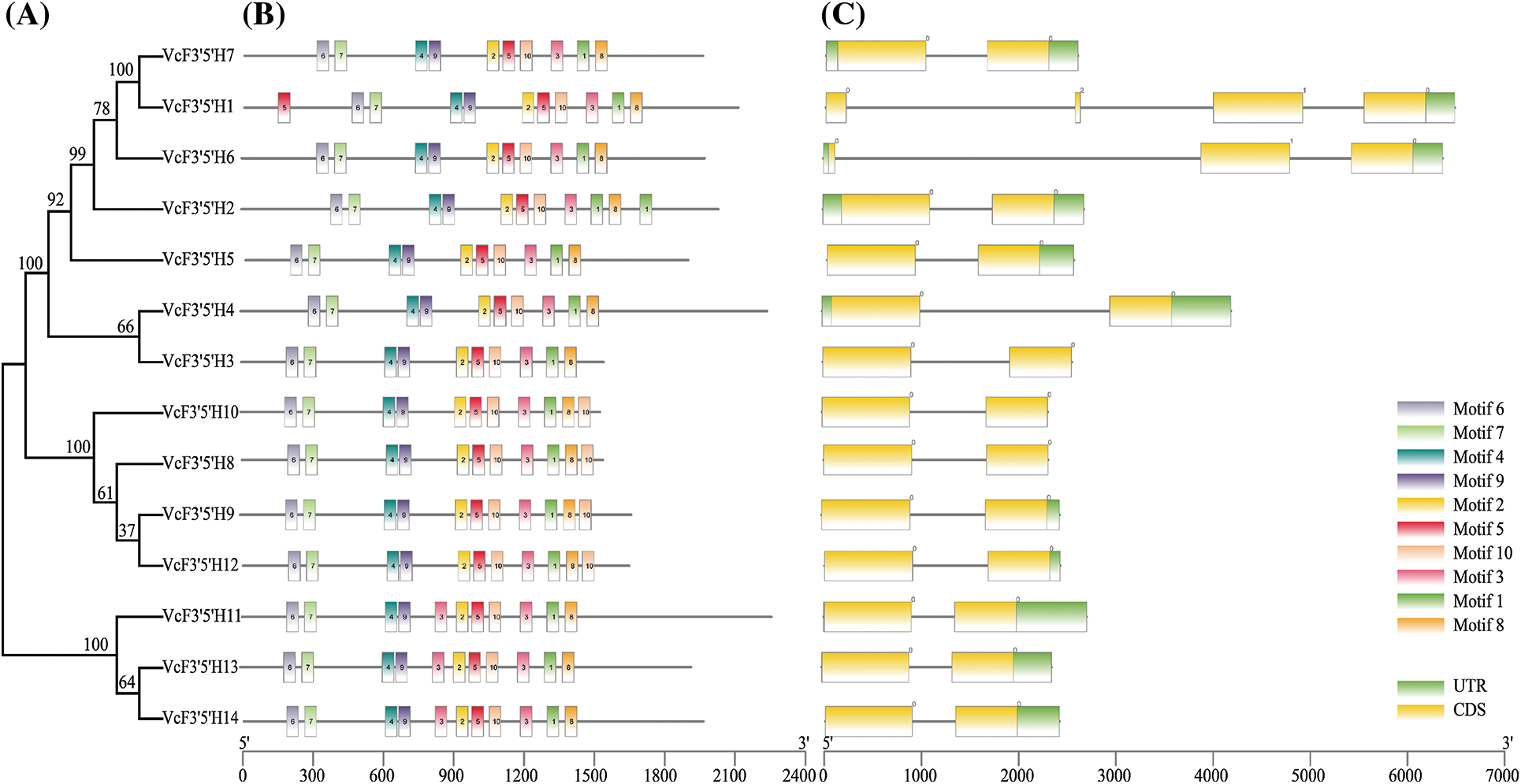
Figure 2: Phylogenetic tree (A), conserved motifs (B), and gene structure analysis (C) of the blueberry F3′5′H gene family. A neighbor-joining phylogenetic tree was constructed by aligning the full-length amino acid sequences of the 14 F3′5′H genes in blueberries
3.2 Sequence Alignments and Chromosomal Distribution of the VcF3′5′H Genes
As shown in Fig. 3A, the proline-rich region is regarded as a “hinge” required for the optimal orientation of the P450 enzyme. The under-labeled asterisk motif, which formed a binding pocket for oxygen molecules needed for catalytic activities, was present in the blueberry F3′5′H genes. The absolutely conserved EXXR motif used to stabilize the core structure, and the most characteristic P450 consensus sequence FxGxRxCxG (heme-binding domain), which was responsible for carbon monoxide-binding ability, were also found in the blueberry F3′5′H genes. The LPPGP domain is involved in a variety of biological processes, including protein transport, receptor signal transduction, mRNA splicing, apoptosis regulation, and stress responses to biotic and abiotic stresses. These conserved domains may be associated with the specific functions of the members of this family. The 14 F3′5′H genes were mapped to nine scaffolds of the Vaccinium corymbosum genome sequence assembly based on their localization information (Fig. 3B). These genes were relatively uniformly distributed. For example, except for scaffold5, scaffold8, and scaffold9, which had two or three genes, there is only one scaffold.
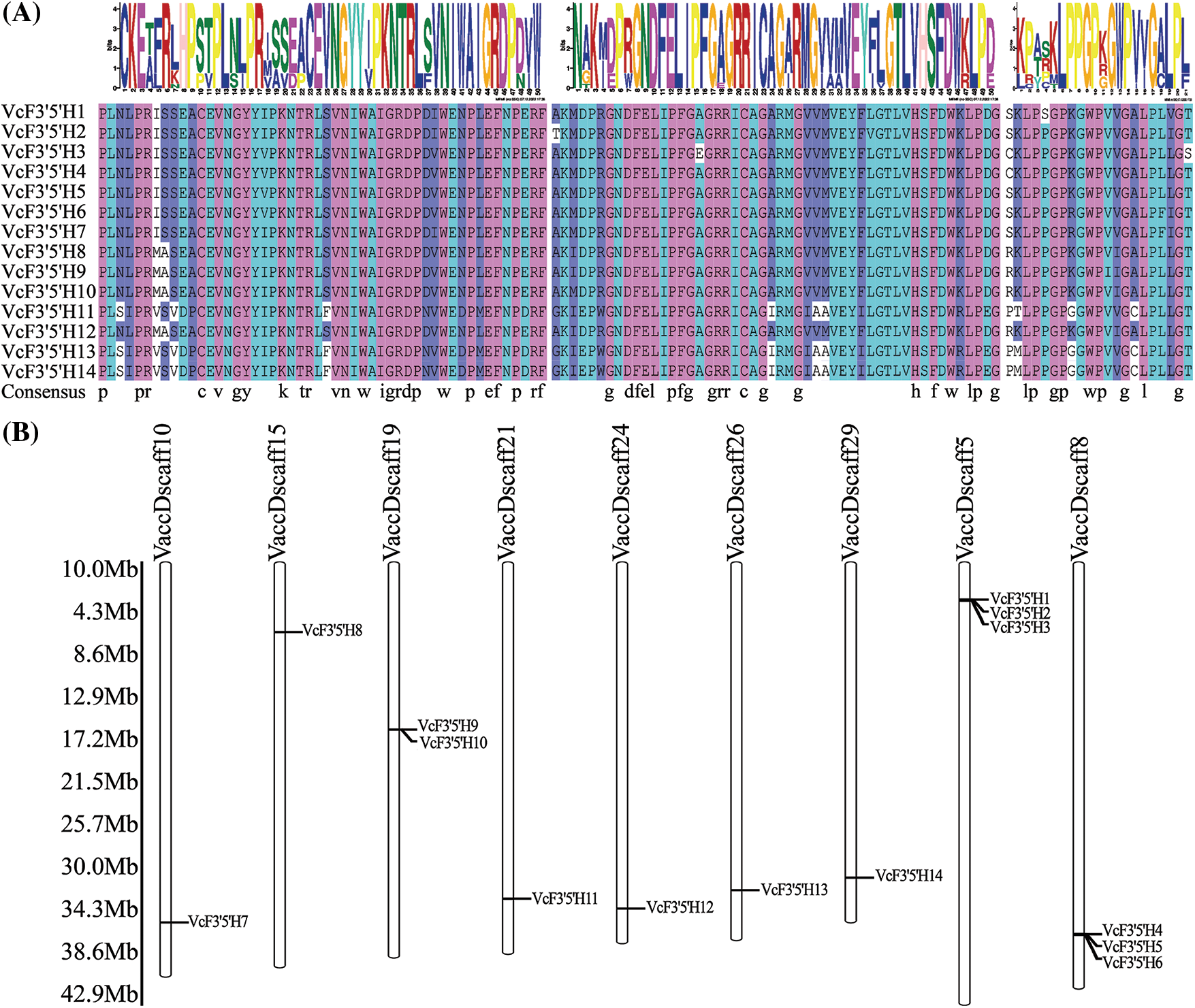
Figure 3: Sequence alignment analysis of VcF3′5′H family members (A); chromosomal map of the position of VcF3′5′H related genes on chromosomes (B)
Note: Figure A shows the EXXR, FxGxRxCxG, and LPPGP domains from left to right.
3.3 Analysis of VcF3′5′H cis-Elements
Using the Bedtools software, the 2000 bp upstream of the blueberry VcF3′5′H genes was extracted, predicted, and analyzed (Fig. 4). The results showed that VcF3′5′H contained numerous basic elements, such as CAAT-box and TATA-box, and various cis-acting elements, including ABRE for abscisic acid responses, ARE for anaerobic responses, CGTCA-motif involved in methyl jasmonate, ACE, G-box and AE-box involved in the light-responsive element, O2-site involved in the zein metabolic regulatory element, TC-rich repeats involved in the defense and stress response, MBS involved in Drought-induced MYB binding site, as well as AC-I, AP-I, and other motifs. In summary, many cis-acting elements upstream of VcF3′5′H were found to be involved in different regulatory processes, indicating that the VcF3′5′H gene family has multiple functions.
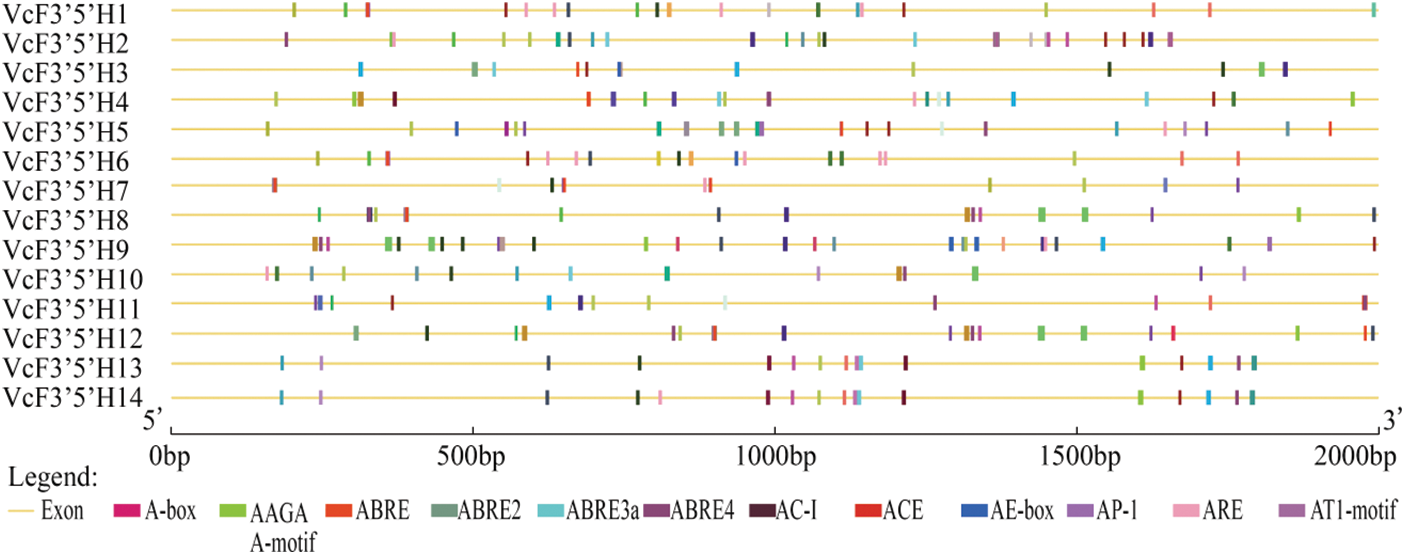
Figure 4: Analysis of the cis-acting elements of VcF3′5′H genes
Note: This figure only shows elements with large numbers.
3.4 Identification of VcF3′5′H Genes Regulating Anthocyanins in Blueberries
As shown in Fig. 5, the co-expression network analysis of F3′5′H and anthocyanins revealed that VcF3′5′H3 and VcF3′5′H4 were both connected to anthocyanins (R > 0.9). In particular, VcF3′5′H4 was related to the synthesis of six different types of anthocyanins, with Petunidin-3-O-arabinoside and Delphinidin-3-O-arabinoside being the most prominent. According to the data, VcF3′5′H4 was essential for the synthesis of anthocyanins in blueberries.
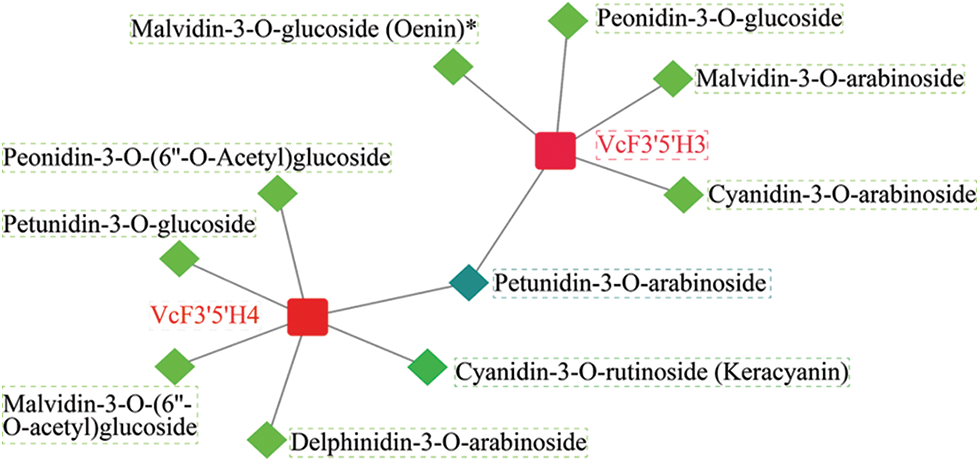
Figure 5: Analysis of the anthocyanin content in blueberries and co-expression analysis of F3′5′H. The correlation coefficient of the F3′5′H gene and anthocyanin content is 0.90
The F3′5′H gene family is related to fruit coloration. To explore the relationship between the F3′5′H gene family and the fruit coloration of blueberries, we detected the expression levels of the F3′5′H gene family in three stages using qRT-PCR. The results showed that except VcF3′5′H8, VcF3′5′H10, VcF3′5′H11, VcF3′5′H12, and VcF3′5′H13, the genes were highly expressed at ripening stage (Fig. 6). In particular, VcF3′5′H4 might play a key role in the formation of fruit color. These results indicate that the F3′5′H gene family is indeed involved in the color formation of purple fruit.
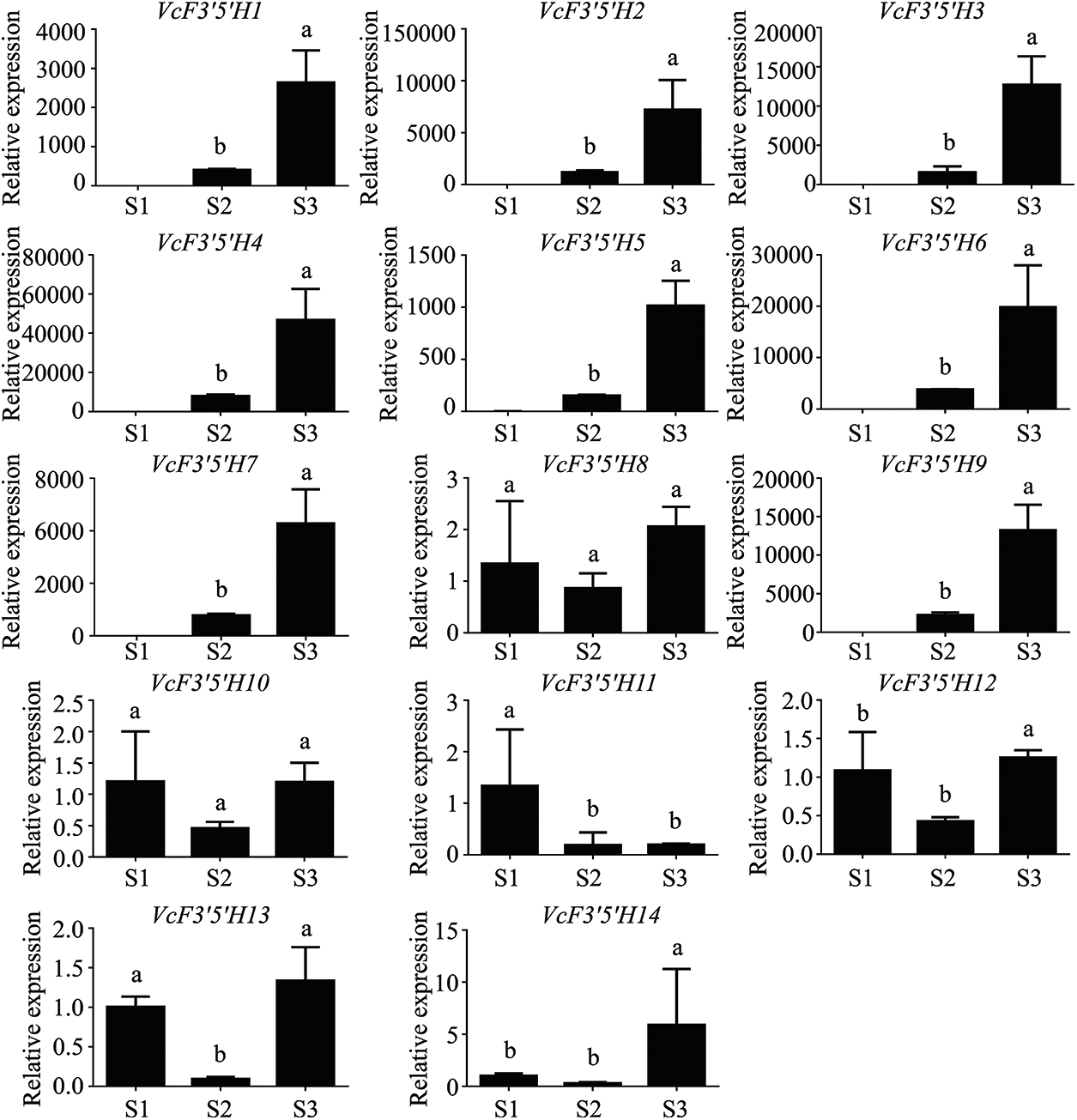
Figure 6: Expression of F3′5′H genes at different developmental stages
3.6 Cloning and Subcellular Localization of the VcF3′5′H4 Gene
The plant expression vector of VcF3′5′H4, pBWA(V)HS-VcF3′5′H4-GFP, was constructed and injected in N. benthamiana leaves. The fluorescence expression was observed under a laser confocal microscope (Fig. 7). Combining the observation results with a bioinformatics predictor (Cell-PLoc 2.0), we speculated that VcF3′5′H4 was located in the endoplasmic reticulum.
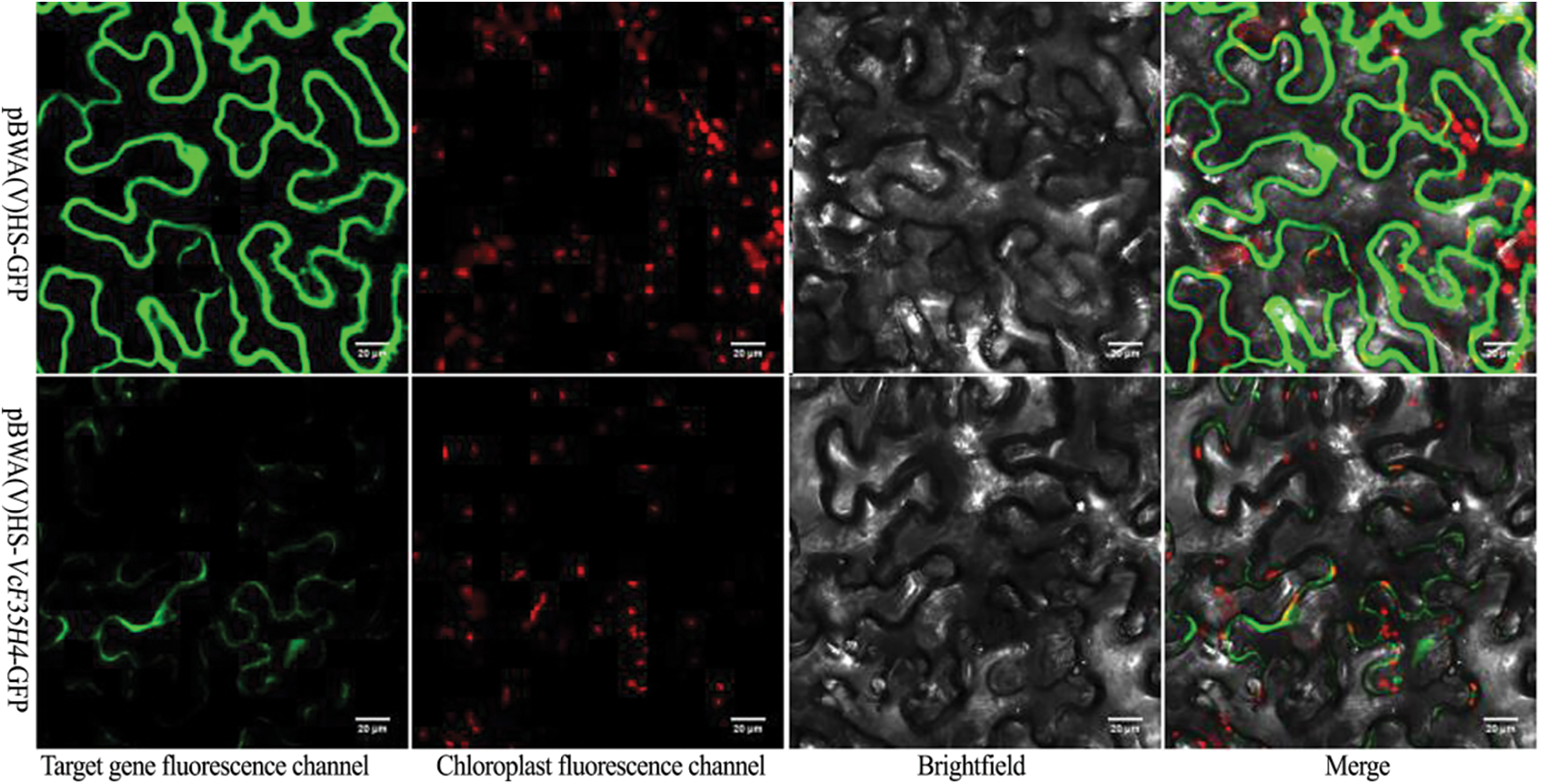
Figure 7: Map of VcF3′5′H4 protein subcellular locations
Note: pBWA(V)HS-VcF3′5′H4-GFP fusion protein and pBWA(v)HS-GFP after the injection of N. benthamiana for 48 h were observed under a laser confocal microscope, and pBWA(v)HS-GFP was used as a control.
4.1 Analysis of the Basic Characteristics of Blueberry F3′5′H
Cytochrome P450 protein family contains 10 subfamilies [44], and F3′5′H belongs to the CYP75A subfamily [33,45]. The members of the other nine families are mainly involved in the metabolism of sterols, fatty acids and hormones, while the members of the CYP75A subfamily are involved in the synthesis of secondary compounds, such as phenylpropanes, flavonoids, isoflavones, and alkaloids. For example, the overexpression of F3′5′H genes can promote the accumulation of poplar anthocyanins and enhance the hydroxylation of flavonoids [22]. The hydroxylation pattern of flavonoids is largely determined by F3′5′H genes. The cloning and characterization of F3′5′H genes are of great significance. For example, cloning F3′5′H genes can change the color of plants, such as roses and carnations that do not naturally have F3′5′H activities, from lavender to blue [23–25].
Some studies focused on F3′5′H genes in plants [20,21,46]. In these studies, blueberry genes were identified with bioinformatics analyses. It was found that these proteins were basically hydrophilic proteins with 508–601 amino acids. The members were relatively conserved and had flavonoid hydroxyl enzyme-specific conserved domains, such as proline-rich “hinge” regions, EXXR motif heme-binding domains, and substrate recognition sites [30,47,48]. In particular, the LPPGP domain was involved in various biological processes, including protein trafficking, receptor signal transduction, mRNA splicing, apoptosis regulation, and stress responses to biotic and abiotic stresses [49]. This result is consistent with that of previous studies on tobacco, petunia, moss, and tea trees [33,35,50]. For example, a study showed that most genes from the same gene family have similar numbers of exons and introns but very different lengths of introns [51]; our experiment showed that the genes in the VcF3′5′H gene family mostly have two exons, with some genes having three or four. In addition, some research has demonstrated that F3′5′H proteins are mainly located in the endoplasmic reticulum membrane and are involved in the metabolic pathways of fatty acids, sterols, hormones, and secondary metabolites [33]; the results of our study are consistent with those of previous research, and the conclusion can be drawn that the structural genes of the anthocyanin synthesis pathway generally function on the endoplasmic reticulum [52]. It has also been reported that the amino acid sequences of F3′5′H are diverse, but their folding and structures remain highly conserved throughout evolution. The results of our study are in agreement with those of previous studies [53,54].
4.2 Functional Analysis of Blueberry F3′5′H
Numerous studies have shown that F3′5′H genes are involved in the synthesis of anthocyanins [22,55–57]. The synthesis pathway of anthocyanins needs to be induced by light, and light-inducible genes often contain cis-acting elements, such as GT-1 motif and G-box. Some researchers believed that when the light time prolonged, the expression levels of PAP1 (MYB) and TT8 (bHLH) in Arabidopsis thaliana seedlings were increased, improving the expression levels of key synthesis enzyme genes for anthocyanins, such as CHS, F3H, and DFR. The probable reason is that these candidate genes have three cis-acting elements, including MRE (light-responsive MYB binding site), ACE, and G-box, which can regulate gene expression levels [58–60]. This also indicates that light-responsive elements, such as GT-1 motif, G-box, and ACE, can regulate the synthesis of anthocyanins through light intensity. Wang et al. [61] found that the CsF3′5′H gene promoter of the tea plants contained multiple light-responsive elements by cloning it. The results in our study are consistent with the findings in the previous research, indicating that this promoter may synthesize anthocyanins mediated by light.
Sun et al. [57] confirmed that the heterologous expression of PCFH belonging to the CYP75B subfamily could produce a kind of delphinidin derivative (3′,5′-hydroxylated anthocyanidins) in vivo and its encoded protein had the 3′,5′-hydroxylation function. Takatori et al. [62] found that F3′5′H genes were involved in the synthesis of delphinium in Eustoma grandiflorum (Raf.) Shinn, and of other types of anthocyanins, such as cyanidin-type anthocyanin and pelargonidin-type anthocyanin that produce various flower colors. In our study, the co-expression network analysis of F3′5′H and anthocyanins revealed that VcF3′5′H3 and VcF3′5′H4 were both connected to anthocyanins (R > 0.9). In particular, VcF3′5′H4 was related to the synthesis of six different types of anthocyanins, with Petunidin-3-O-arabinoside and Delphinidin-3-O-arabinoside being the most prominent. In line with the results in previous studies, our findings indicate that the VcF3′5′H4 genes mainly induce the synthesis of delphinium in blueberries.
In this research, we identified 14 VcF3′5′H genes. The VcF3′5′H gene and anthocyanin content co-expression confirmed that the VcF3′5′H family was closely related to the synthesis of anthocyanins. In particular, VcF3′5′H4 played a key role. Based on the prediction using Cell-PLoc 2.0, we hypothesized that VcF3′5′H4 was localized in the endoplasmic reticulum. This study lays a theoretical foundation for further research on the function of the F3′5′H gene family in blueberries.
Funding Statement: This research was funded by the National Natural Science Foundation of China (Grant No. 31760205), Guangdong Basic and Applied Basic Research Foundation of Guangdong Province-Yuehui Joint Foundation (Grant No. 2022A1515111095), and the Professor and Doctoral Research Project (Grant No. 2022JB021) in Huizhou University, the Project of the Department of Education of Guangdong Province (Grant No. 2018KTSCX214).
Author Contributions: XG and DW designed the project. JH and XG prepared the samples. XG and SY analyzed the data. XG wrote the manuscript. All authors contributed to the article and approved the submitted version.
Availability of Data and Materials: The following information was supplied regarding data availability: Raw data are available in the supplemental files.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material iavailable online at https://doi.org/10.32604/phyton.2023.029820.
References
1. Li, Y. D., Pei, J. B., Chen, L. (2021). Annual report on China’s blueberry industry 2020. Journal of Jilin Agricultural University, 43(1), 1–8. [Google Scholar]
2. Shi, M., Loftus, H., Mcainch, A. J., Su, X. Q. (2017). Blueberry as a source of bioactive compounds for the treatment of obesity, type 2 diabetes and chronic inflammation. Journal of Functional Foods, 30, 16–29. https://doi.org/10.1016/j.jff.2016.12.036 [Google Scholar] [CrossRef]
3. Govers, C., Berkel, K. M., Sluis, A. A., Mes, J. J. (2018). Review of the health effects of berries and their phytochemicals on the digestive and immune systems. Nutrition Reviews, 76, 29–46. https://doi.org/10.1093/nutrit/nux039 [Google Scholar] [PubMed] [CrossRef]
4. Tapas, A. R., Sakarkar, D. M., Kakde, R. B. (2008). Flavonoids as nutraceuticals: A review. Tropical Journal of Pharmaceutical Research, 7, 1089–1099. https://doi.org/10.4314/tjpr.v7i3.14693 [Google Scholar] [CrossRef]
5. Martin, C., Butelli, E., Petroni, M., Tonelli, C. (2011). How can research on plants contribute to promoting human health? The Plant Cell, 23, 1685–1699. https://doi.org/10.1105/tpc.111.083279 [Google Scholar] [PubMed] [CrossRef]
6. Zhao, Y., Dong, W. Q., Wang, K., Zhang, B., Allan, A. C. et al. (2017). Differential sensitivity of fruit pigmentation to ultraviolet light between two peach cultivars. Frontiers in Plant Science, 8, 1552. https://doi.org/10.3389/fpls.2017.01552 [Google Scholar] [PubMed] [CrossRef]
7. He, Y. J., Chen, H., Zhou, L., Liu, Y., Chen, H. Y. (2019). Comparative transcription analysis of photosensitive and non-photosensitive eggplants to identify genes involved in dark regulated anthocyanin synthesis. BMC Genomics, 20, 678. https://doi.org/10.1186/s12864-019-6023-4 [Google Scholar] [PubMed] [CrossRef]
8. Yang, L., Wang, Y. H., Tan, H., Li, D. N. et al. (2018). Comparative transcriptome analysis of genes involved in anthocyanin synthesis in blueberry. Plant Physiology and Biochemistry, 127, 561–572. https://doi.org/10.1016/j.plaphy.2018.04.034 [Google Scholar] [PubMed] [CrossRef]
9. Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends in Plant Science, 18, 477–483. [Google Scholar] [PubMed]
10. Li, T., Yamane, H., Tao, R. (2021). Preharvest long-term exposure to UV-B radiation promotes fruit ripening and modifies stage-specific anthocyanin metabolism in highbush blueberry. Horticulture Research, 8, 67. https://doi.org/10.1038/s41438-021-00503-4 [Google Scholar] [PubMed] [CrossRef]
11. Guo, X. L., Shakeel, M., Wang, D. L. (2022). Metabolome and transcriptome profling unveil the mechanisms of light-induced anthocyanin synthesis in rabbiteye blueberry (vaccinium ashei: Reade). BMC Plant Biology, 22, 223. [Google Scholar] [PubMed]
12. Hirsch, C. D. (2015). A systems biology approach to understanding anthocyanin biosynthesis and regulation in potato. Plant and Animal Genome XX III Conference, Plant and Animal Genome, 0843. California, USA. [Google Scholar]
13. Tanaka, Y. (2006). Flower colour and cytochromes P450. Phytochemistry Reviews, 5(2–3), 283–291. https://doi.org/10.1007/s11101-006-9003-7 [Google Scholar] [CrossRef]
14. Brugliera, F., Barri-Rewell, G., Holton, T. A. (1999). Isolation and characterization of a flavonoid 30-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant Journal, 19(4), 441–451. https://doi.org/10.1046/j.1365-313x.1999.00539.x [Google Scholar] [PubMed] [CrossRef]
15. Holton, T. A., Brugliera, F., Lester, D. R., Tanaka, Y., Hyland, C. D. et al. (1993). Cloning and expression of cytochrome P450 genes controlling flower colour. Nature, 366(6452), 276–279. https://doi.org/10.1038/366276a0 [Google Scholar] [PubMed] [CrossRef]
16. Tanaka, Y., Yonekura, K., Fukuchi-Mizutani, M., Fukui, Y., Fujiwara, H. et al. (1996). Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentiana triflora. Plant and Cell Physiology, 37(5), 711–716. https://doi.org/10.1093/oxfordjournals.pcp.a029004 [Google Scholar] [PubMed] [CrossRef]
17. Schoenbohm, C., Martens, S., Eder, C., Forkmann, G., Weisshaar, B. (2000). Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biological Chemistry, 381(8), 749–753. https://doi.org/10.1515/bc.2000.095 [Google Scholar] [PubMed] [CrossRef]
18. Jung, C. S., Griffiths, H. M., de Jong, D. M., Cheng, S. P., Bodis, M. et al. (2005). The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theoretical and Applied Genetics, 110(2), 269–275. https://doi.org/10.1007/s00122-004-1829-z [Google Scholar] [PubMed] [CrossRef]
19. Olsen, K. M., Hehn, A., Jugdé, H., Slimestad, R., Larbat, R. et al. (2010). Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biology, 10, 21. https://doi.org/10.1186/1471-2229-10-21 [Google Scholar] [PubMed] [CrossRef]
20. Wu, Y. Q., Wang, T. L., Xin, Y., Wang, G. B., Xu, L. A. (2020). Overexpression of GbF3′5′H1 provides a potential to improve the content of epicatechin and gallocatechin. Molecules, 25, 4836. https://doi.org/10.3390/molecules25204836 [Google Scholar] [PubMed] [CrossRef]
21. Nguyen, T. N. L., Hoang, T. H. H., Nguyen, H. Q., Tran, T. H., Vu, T. T. T. et al. (2021). Agrobacterium tumefaciens-mediated genetic transformation and overexpression of the flavonoid 3′5′-hydroxylase gene increases the flavonoid content of the transgenic Aconitum carmichaelii Debx. plant. In Vitro Cellular & Developmental Biology-Plant, 58, 93–102. https://doi.org/10.1007/s11627-021-10190-4 [Google Scholar] [CrossRef]
22. Ma, D. W., Tang, H., Reichelt, M., Piirtola, E. M., Salminen, J. P. et al. (2021). Poplar MYB117 promotes anthocyanin synthesis and enhances flavonoid B-ring hydroxylation by up-regulating the flavonoid 3′,5′-hydroxylase gene. Journal of Experimental Botany, 72(10), 3864–3880. https://doi.org/10.1093/jxb/erab116 [Google Scholar] [PubMed] [CrossRef]
23. Tanaka, Y., Tsuda, S., Kusumi, T. (1998). Metabolic engineering to modify flower color. Plant Cell Physiology, 39, 1119–1126. https://doi.org/10.1093/oxfordjournals.pcp.a029312 [Google Scholar] [CrossRef]
24. Forkmann, G., Martens, S. (2001). Metabolic engineering and applications of flavonoids. Current Opinion in Biotechnology, 12, 155–160. https://doi.org/10.1016/S0958-1669(00)00192-0 [Google Scholar] [PubMed] [CrossRef]
25. Fukui, Y., Tanaka, Y., Kusumi, T., Iwashita, T., Nomoto, K. (2003). A rationale for the shift in colour towards blue in transgenic carnation flowers expressing the flavonoid 3′,5′-hydroxylasegene. Phytochemistry, 63(1), 15–23. [Google Scholar] [PubMed]
26. Tanaka, Y., Brugliera, F. (2013). Flower colour and cytochromes P450. Philosophical Transactions of the Royal Society B: Biological Sciences, 368(1612), 20120432. https://doi.org/10.1098/rstb.2012.0432 [Google Scholar] [PubMed] [CrossRef]
27. Noda, N., Yoshioka, S., Kishimoto, S., Nakayama, S., Douzono, M. et al. (2017). Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Science Advances, 3(7), e1602785. https://doi.org/10.1126/sciadv.1602785 [Google Scholar] [PubMed] [CrossRef]
28. Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology, 126, 485–493. https://doi.org/10.1104/pp.126.2.485 [Google Scholar] [PubMed] [CrossRef]
29. Lafferty, D. J., Espley, R. V., Deng, C. H., Günther, C. S., Plunkett, B. et al. (2022). Hierarchical regulation of MYBPA1 by anthocyanin and proanthocyanidin-related MYB proteins is conserved in Vaccinium species. Journal of Experimental Botany, 73(5), 1344–1356. https://doi.org/10.1093/jxb/erab460 [Google Scholar] [PubMed] [CrossRef]
30. Karppinen, K., Lafferty, D. J., Albert, N. W., Mikkola, N., McGhie, T. et al. (2021). MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytologist, 232, 1350–1367. https://doi.org/10.1111/nph.17669 [Google Scholar] [PubMed] [CrossRef]
31. Li, Z., Li, S. H., Ge, H. Y., Shi, S. L., Li, D. L. et al. (2022). A light-responsive transcription factor SmMYB35 enhances anthocyanin biosynthesis in eggplant (Solanum melongena L.). Planta, 255, 12. https://doi.org/10.1007/s00425-021-03698-x [Google Scholar] [PubMed] [CrossRef]
32. Colle, M., Leisner, C. P., Wai, C. M., Ou, S. J., Bird, K. A. et al. (2019). Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. GigaScience, 8(3), giz012. https://doi.org/10.1093/gigascience/giz012 [Google Scholar] [PubMed] [CrossRef]
33. Guo, L. N., Gao, L. P., Ma, X. B., Guo, F., Ruan, H. X. (2019). Functional analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylases from tea plant (Camellia sinensisinvolved in the B-ring hydroxylation of flavonoids. Gene, 717, 144046. https://doi.org/10.1016/j.gene.2019.144046 [Google Scholar] [PubMed] [CrossRef]
34. Kukk, K. (2023). Identification, characterisation and recombinant expression of flavonoid 3′,5′-hydroxylases and cytochrome P450 reductases from Vaccinium species. bioRxiv-Synthetic Biology, 12(7), 1086. https://doi.org/10.1101/2023.01.09.523147 [Google Scholar] [CrossRef]
35. Liu, S. G., Ju, J. F., Xia, G. G. (2014). Identification of the flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase genes from Antarctic moss and their regulation during abiotic stress. Gene, 543, 145–152. https://doi.org/10.1016/j.gene.2014.03.026 [Google Scholar] [PubMed] [CrossRef]
36. Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J. C. et al. (2014). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics, 31(8), 1296–1297. https://doi.org/10.1093/bioinformatics/btu817 [Google Scholar] [PubMed] [CrossRef]
37. Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E. et al. (2009). MEME Suite: Tools for motif discovery and searching. Nucleic Acids Research, 37, W202–W208. https://doi.org/10.1093/nar/gkp335 [Google Scholar] [PubMed] [CrossRef]
38. Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 [Google Scholar] [PubMed] [CrossRef]
39. Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G. et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research, 40(W1), W597–W603. https://doi.org/10.1093/nar/gks400 [Google Scholar] [PubMed] [CrossRef]
40. Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., Demchak, B. (2009). Cytoscape automation: Empowering workflow-based network analysis. Genome Biology, 20(1), 1–15. https://doi.org/10.1186/s13059-019-1758-4 [Google Scholar] [PubMed] [CrossRef]
41. Quinlan, A. R., Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–842. https://doi.org/10.1093/bioinformatics/btq033 [Google Scholar] [PubMed] [CrossRef]
42. Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y. et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30(1), 325–327. https://doi.org/10.1093/nar/30.1.325 [Google Scholar] [PubMed] [CrossRef]
43. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262 [Google Scholar] [PubMed] [CrossRef]
44. Nelson, D. R., Schuler, M. A., Paquette, S. M., Werck-Reichhart, D., Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiology, 135, 756–772. https://doi.org/10.1104/pp.104.039826 [Google Scholar] [PubMed] [CrossRef]
45. Toguri, T., Umemoto, N., Kobayashi, O. (1993). Activation of anthocyanin synthesis genes by white light in eggplant hypocotyl tissues, and identification of an inducible P-450 cDNA. Plant Molecular Biology, 23(5), 933–946. https://doi.org/10.1007/BF00021810 [Google Scholar] [PubMed] [CrossRef]
46. Seitz, C., Eder, C., Deiml, B., Kellner, S., Martens, S. et al. (2006). Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Molecular Biology, 61, 365–381. https://doi.org/10.1007/s11103-006-0012-0 [Google Scholar] [PubMed] [CrossRef]
47. Falginella, L., Castellarin, S. D., Testolin, R., Gambetta, G. A., Morgante, M. (2010). Expansion and subfunctionalisation of flavonoid 3′,5′-hydroxylases in the grapevine lineage. BMC Genomics, 11(1), 562. https://doi.org/10.1186/1471-2164-11-562 [Google Scholar] [PubMed] [CrossRef]
48. Huang, W., Sun, W., Wang, Y. (2012). Isolation and molecular characterisation of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes from a traditional Chinese medicinal plant, Epimedium sagittatu. Gene, 497, 125–130. https://doi.org/10.1016/j.gene.2011.11.029 [Google Scholar] [PubMed] [CrossRef]
49. Li, X. Y., Hou, Y. M., Xie, X., Li, H. X., Li, X. D. (2020). Blueberry MIR156a/SPL12 module coordinates the accumulation of chlorophylls and anthocyanins during fruit ripening. Journal of Experimental Botany, 71(19), 5976–5989. https://doi.org/10.1093/jxb/eraa327 [Google Scholar] [PubMed] [CrossRef]
50. Shimada, Y., Nakanoshimada, R., Ohbayashi, M., Okinaka, Y., Kiyokawa, S. (1999). Expression of chimeric P450 genes encoding flavonoid-3′,5′-hydroxylase in transgenic tobacco and petunia plants. FEBS Letters, 461(3), 241–245. [Google Scholar] [PubMed]
51. Wang, W., Zhou, Y., Wu, Y., Dai, X. L., Liu, Y. J. (2018). Insight into Catechins metabolic pathways of Camellia sinensis based on genome and transcriptome analysis. Journal of Agricultural and Food Chemistry, 66(16), 4281–4293. https://doi.org/10.1021/acs.jafc.8b00946 [Google Scholar] [PubMed] [CrossRef]
52. Zhao, J., Dixon, R. A. (2010). The ‘ins’ and ‘outs’ of flavonoid transport. Trends in Plant Science, 15(2), 72–80. https://doi.org/10.1016/j.tplants.2009.11.006 [Google Scholar] [PubMed] [CrossRef]
53. Hasemann, C., Kurumbail, R., Boddupalli, S., Peterson, J. A., Deisenhofer, J. (1995). Structure and function of cytochromes P450: A comparatives analysis of three crystal structures. Structure, 3, 41–62. https://doi.org/10.1016/S0969-2126(01)00134-4 [Google Scholar] [PubMed] [CrossRef]
54. Rupasinghe, S., Baudry, J., Schuler, M. A. (2003). Common active site architecture and binding strategy of four phenylpropanoid P450s from Arabidopsis thaliana as revealed by molecular modelling. Protein Engineering, 16(10), 721–731. https://doi.org/10.1093/protein/gzg094 [Google Scholar] [PubMed] [CrossRef]
55. Shimada, Y., Ohbayashi, M., Nakano-Shimada, R., Okinaka, Y., Kiyokawa, S. (2001). Genetic engineering of theanthocyanin biosynthetic pathway with flavonoid-3′,5′-hydroxylase: Specific switching of the pathway in petunia. Plant Cell Reports, 20, 456–462. https://doi.org/10.1007/s002990100319 [Google Scholar] [CrossRef]
56. Okinaka, Y., Shimada, Y., Nakano-Shimada, R., Ohbayashi, M., Kiyokawa, S. (2003). Selective accumulation ofdelphinidin derivatives in tobacco using a putative F3′5′H-hydroxylase cDNA from Campanula medium. Bioscience, Biotechnology and Biochemistry, 67(1), 161–165. https://doi.org/10.1271/bbb.67.161 [Google Scholar] [PubMed] [CrossRef]
57. Sun, J., Liu, Y., Zhao, Y., Ou, Y., Pang, S. H. (2011). Optimization of analytical condition of determining anthocyanins content in blueberry wine by pH-differential method. China Brew, 30(11), 171–174. [Google Scholar]
58. Shin, J., Park, E., Choi, G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. The Plant Journal, 49, 981–994. https://doi.org/10.1111/j.1365-313x.2006.03021.x [Google Scholar] [PubMed] [CrossRef]
59. Cominelli, E., Gusmaroli, G., Allegra, D., Galbiati, M., Wade, H. K. (2008). Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. Journal Plant Physiology, 165(8), 886–894. https://doi.org/10.1016/j.jplph.2007.06.010 [Google Scholar] [PubMed] [CrossRef]
60. Sato, M., Kawabe, T., Hosokawa, M., Tatsuzawa, F., Doi, M. (2011). Tissue culture-induced flower-color changes in Saintpaulia caused by excision of the transposon inserted in the flavonoid 3′,5′H ydroxylase (F3′5′H) promoter. Plant Cell Reports, 30(5), 929–939. https://doi.org/10.1007/s00299-011-1016-z [Google Scholar] [PubMed] [CrossRef]
61. Wang, Y. S., Xu, Y. J., Gao, L. P., Yu, O., Wang, X. Z. (2014). Functional analysis of flavonoid 3′,5′-hydroxylase from tea plant (Camellia sinensisCritical role in the accumulation of catechins. BMC Plant Biology, 14, 347. https://doi.org/10.1186/s12870-014-0347-7 [Google Scholar] [PubMed] [CrossRef]
62. Takatori, Y., Shimizu, K., Yauwapaksopon, D., Nakamura, Y., Oshima, H. et al. (2022). Flavonoid 3′,5′-Hydroxylase (F3′5′H) gene polymorphisms Co-segregate with variation in anthocyanin composition in the flower petals of lisianthus [Eustoma grandiflorum (Raf.) Shinn.]. Horticulture Journal, 91(2), 229–239. https://doi.org/10.2503/hortj.utd-320 [Google Scholar] [CrossRef]
Supplementary Materials
Table S1: 14 VcF3′5′H genes co-expressed with anthocyanin
Table S2: Quantitative primer sequences
Table S3: VcF3′5′H4 primer sequence
Figure S1: 17 VcF3′5′H genes and P450 subfamily sequences for Phylogenetic tree
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools