 Open Access
Open Access
ARTICLE
Identification of the SERK Gene Family in Paulownia fortunei and Its Involvement in the Response to Biotic and Abiotic Stresses
1 Institute of Paulownia, Henan Agricultural University, Zhengzhou, 450002, China
2 College of Forestry, Henan Agricultural University, Zhengzhou, 450002, China
* Corresponding Author: Guoqiang Fan. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(9), 2473-2488. https://doi.org/10.32604/phyton.2023.029469
Received 21 February 2023; Accepted 24 May 2023; Issue published 28 July 2023
Abstract
Somatic embryogenesis receptor-like kinases (SERKs) are receptor-like proteins that contain leucine-rich repeats and are involved in various signaling pathways. This study identified SERK family members in the Paulownia fortunei genome and analyzed their characteristics and expression profiles using bioinformatics methods. We identified 12 SERK genes with relatively conserved gene structures and motifs that were distributed unevenly on eight Paulownia chromosomes. The gene promoters contained various cis-acting elements that regulated the expression of the PfSERK genes in response to hormones and abiotic stresses. Synteny analysis indicated that 10 segmental duplication events had occurred during evolution of the PfSERK family. The expression profile of PfSERKs in various tissues of Paulownia fortunei was detected by quantitative real-time PCR (qRT-PCR). Many PfSERK genes can respond to drought and salt stress. Combined with RNA-seq and protein interaction network, it is speculated that PfSERK3/11 may participate in the occurrence of Paulownia witches’ broom (PaWB) by regulating the plant height of Paulownia.Keywords
Somatic embryogenesis receptor-like kinases (SERKs) belong to a subfamily of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) family. SERK genes generally contain 11 exons that encode seven functional domains in the SERK proteins, namely the SP, ZIP, LRR, SPP, TM, cytoplasmic Ser/Thr kinase domain, and the carboxyl-terminal domain [1,2]. These conserved domains are very important in SERK proteins. For example, ZIP domain is the key domain for the specific binding of SERK [3,4], LRR domain is the key domain for the binding of SERK to plasma membrane [5], and TM domain is the key domain for the separation of intracellular region and extracellular region of SERK proteins [6]. SPP domain is essential in the interaction between protein and cell wall, and is generally considered to be important for the identification of SERKs [3]. SERK gene was first identified in carrots in 1997 [7]. Subsequently, SERK homologues were identified in Arabidopsis, rice, Triticum aestivum, Malus × domestica, and Orchidaceae [3,8–12].
Studies have shown that SERKs not only participates in somatic embryogenesis, but also play important roles in disease resistance, hormone signal transduction, and defense response. For example, of the five SERK proteins in A. thaliana, AtSERK1 is involved in somatic and zygotic embryogenesis [13]; AtSERK2 is involved in spore formation, differentiation and male gametophyte development [14]; AtSERK3–5 are involved in brassinosteroid (BR) signaling [13–16]. Additionally, overexpression of AtSERK3 or AtSERK4 was found to lead to stem elongation [17]. In addition to SERK involvement in the BR pathway, SERK3 also forms a complex with FLS2, another LRR-RLK family protein, that is involved in the innate immune response in plants [18,19]. SERK1, SERK3, and SERK4 participate in regulating the BR signaling pathway, and SERK3 and SERK4 participate in regulating cell death [15,20,21]. Overexpression of OsSERK1 led to plant dwarfing and OsSERK2 was associated with the immune response of rice to bacterial blight and rice blast [9,22–24]. Heterologous expression of wheat TaSERK1–5 in Arabidopsis led to an increased height phenotype [10]. Twelve SERK members were identified in apple, among which MdSERK4, MdSERK6, and MdSERK10 could respond to salt stress [11]. Previous studies that have focused mainly on A. thaliana, Z. mays, O. sativa, and other model plants, and rarely on tree species with great economic value, such as Paulownia [1,3,8,9]. Economically valuable tree species are important contributors to the economic growth of many countries worldwide. Understanding the role of SERK genes involved in the economic tree species will contribute to further research that may lead to breakthroughs in the prevention and control of diseases and pests that help to improve the utilization value of these tree species. Therefore, strengthening the research on growth and development of economic tree species is urgently needed.
With the characteristics of wide adaptation area, fast growth, and good material, Paulownia can play an important role in alleviating wood shortage, ensuring food security and improving the ecological environment [25,26]. However, PaWB disease can causes axillary bud sprouting, shortened internodes, yellowing and smaller leaves, clumping branches, and dwarfing of Paulownia, and leads to the death of young trees and reduction of timber yield [27–29]. In the previous study of our laboratory, it was found that the expression of PfSERK changed in phytoplasma-infected P. fortunei [28,30]. These findings indicated that PfSERKs probably influenced key traits of Paulownia. Understanding the functions of PfSERKs will further enrich and improve the understanding of the response of Paulownia to PaWB infection at the molecular level and may help in breeding programs to improve the economic value of Paulownia. We identified the members of PfSERK gene family at the whole genome level, and analyzed their physiochemical properties and expression patterns to determine the function of SERK in Paulownia.
P. fortunei (PF) and PaWB-infected P. fortunei (PFI) were used as experimental materials, and the seedling age was 30 days. Paulownia infected with witches’ broom disease showed symptoms such as a large number of axillary buds sprouting and clustered branches and leaves. The plantlets were grown based on the method proposed by Fan et al. [31].
2.2 Identification of the P. fortunei PfSERK Gene Family
BlastP search was conducted in the genome database of Paulownia using the known AtSERK1-5 protein sequence, and the candidate protein sequence of PfSERK was screened out (E-value < 1e-5). Further identification of PfSERK gene family members using NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd, accessed on 14 December 2021) and SMART (https://smart.embl.de, accessed on 16 December 2021). ExPASy (https://web.expasy.org/compute_pi/, accessed on 19 January 2022) was used to predicte the physiochemical properties of the PfSERK proteins. The subcellular localization of the PfSERKs was predicted via the Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 17 March 2022).
The genome annotation file and the ID file of all the PfSERKs were submitted to TBtools to obtain a gene structure map. Construction of phylogenetic tree based on SERK proteins from Paulownia and other five species by MEGA7.0.
2.4 Protein Structure Analysis of PfSERKs
The MEME program (https://meme-suite.org/meme/tools/meme, accessed on 22 June 2022) was applied to identify conserved motifs. The motif number was set to 10. The secondary structures of the PfSERKs were predicted using SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html, accessed on 17 February 2023). PfSERK protein spatial models were developed using SWISS-MODEL (https://swissmodel.expasy.org/interactive, accessed on 06 February 2023).
2.5 Collinearity Analysis of the PfSERKs
The Multiple Collinearity Scan toolkit (MCScanX) was applied for collinearity analysis of SERK genes. The Ka/Ks Calculator in TBtools was used for non-synonymous/synonymous (Ka/Ks) analysis of the PfSERKs.
2.6 Identification of Cis-Acting Elements
In the region of 2000 bp upstream of PfSERK promoters, the cis-acting elements were screened by PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 02 July 2022).
2.7 Analysis of the Expression Pattern of PfSERKs
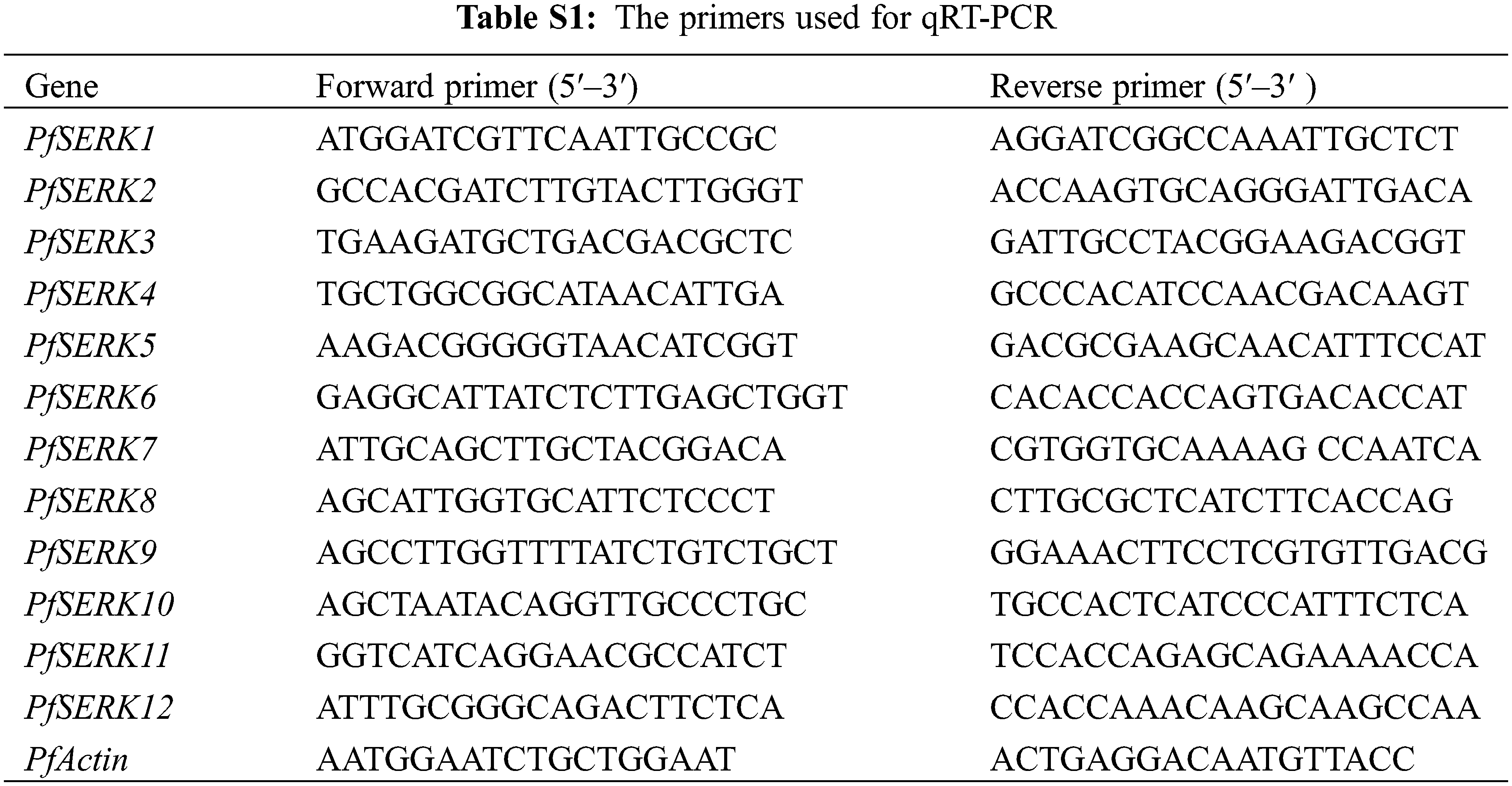
Total RNA was extracted from buds, leaves, nodes, internodes, and roots of PF. cDNA was synthesized using StarScript III All-in-one RT Mix with gDNA Remover (GenStar, Beijing), and the 2 × RealStar Fast SYBR qPCR Mix (Low ROX) (GenStar, Beijing) was used for qRT-PCR. The primers of PfSERK were designed by NCBI Primer-BLAST (Supplementary Table S1). PfActin was applied as the internal reference gene, and there were 3 biological replicates. The 2−ΔΔCt method was applied to determine the expression of PfSERK [32,33].
2.8 Analysis of Expression of PfSERKs under Phytoplasma Infection
The transcriptome data of PF, PFI, and 30 mg L-1 Rifampicin (Rif) treated PFI for 15 days and 30 days were downloaded from NCBI database (accession NO, SRR11787938, SRR11787927, SRR11787916, SRR11787905, SRR11787894, SRR11787883, SRR11787960S, RR11787949, SRR11787971, and SRR11787942–SRR11787944) [27,34]. A heat map of PfSERK expression levels before and after phytoplasma infection was drawn using TBtools. 4 PfSERKs were selected randomly for verification of the transcriptome data by qRT-PCR.
2.9 SERK Protein Interaction Networks
The function of the SERK gene in Arabidopsis has been well studied [13–15,17,18]. We used STRING software (http://string-db.org/, accessed on 03 February 2023) to construct SERK protein interaction network using homologous proteins of Arabidopsis.
2.10 Analysis of Expression of PfSERKs under Salt/Drought Stress
Based on the transcriptome data of healthy seedlings of P. fortunei treated by drought and salt in our laboratory, we downloaded the transcriptome data from NCBI database (accession number: PRJNA221355, PRJNA289582, SRP060682). The heat map of PfSERK expression levels before and after salt/drought stress was drawn using TBtools.
3.1 Identification and Characterization of the PfSERKs
A total of 12 PfSERK protein sequences were obtained by searches of the Paulownia genome using the known AtSERK1–5 protein sequences. The PfSERKs were named PfSERK1–PfSERK12 according to the order of their positions on the chromosomes (Fig. 1). The 12 genes that encode these proteins were distributed on eight Paulownia chromosomes; one was detected on each of chromosomes 2, 4, 7, 11, and 15; two were detected on each of chromosomes 6 and 17; and three were detected on chromosome 13, which had the largest number of PfSERKs among all the chromosomes.
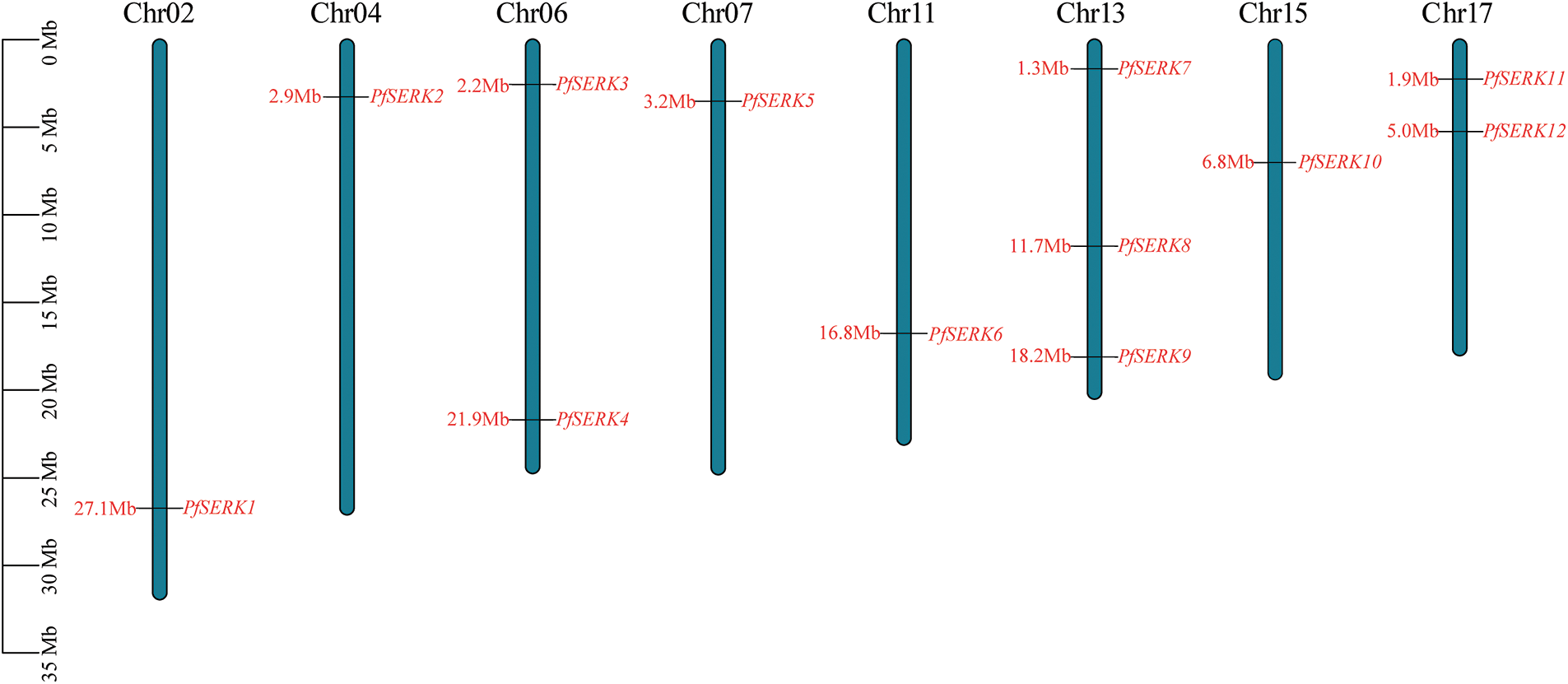
Figure 1: Chromosome distribution of PfSERK gene family members
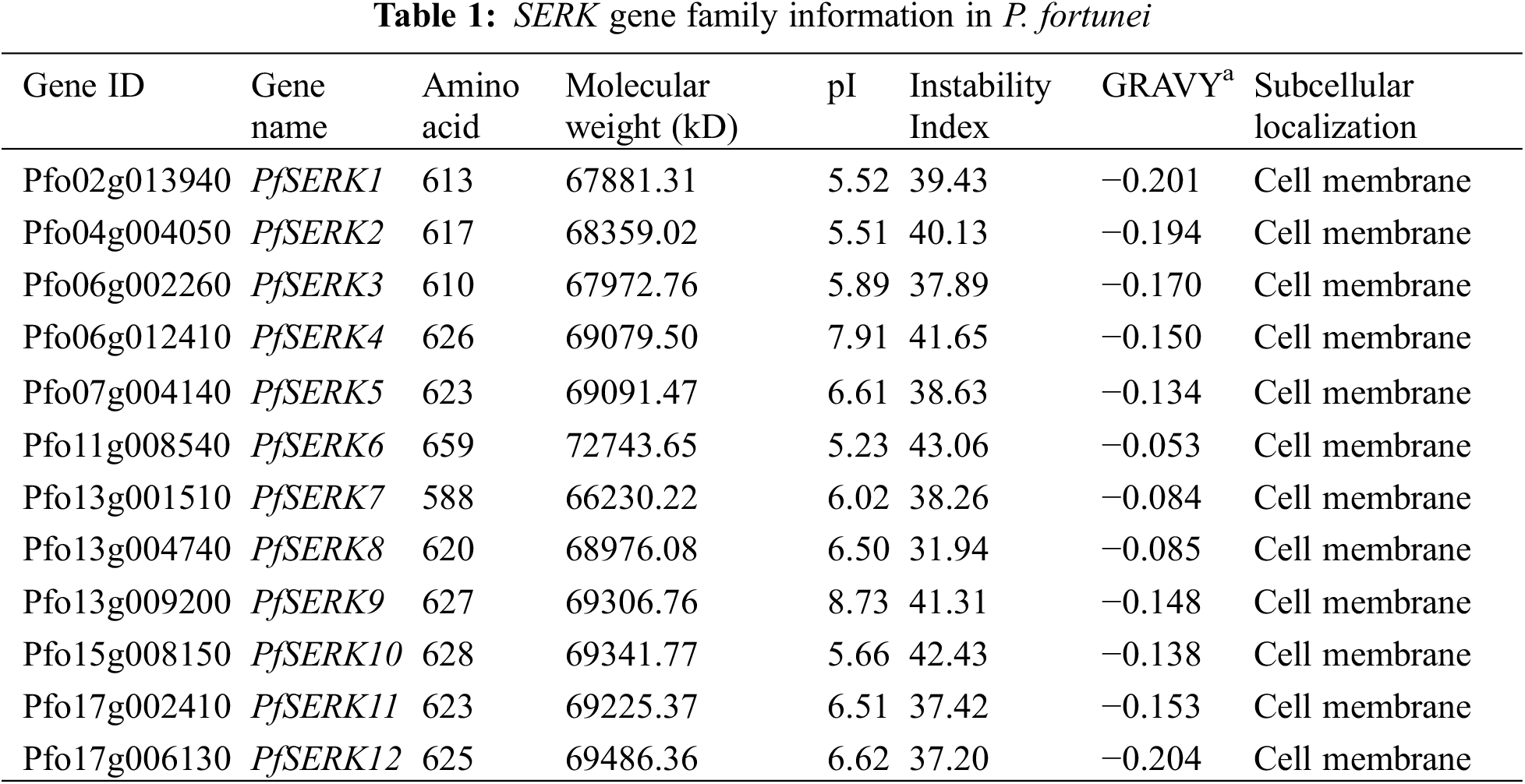
The lengths of the 12 PfSERKs ranged from 588 aa–659 aa; PfSERK6 was the longest and PfSERK7 was the shortest. Their molecular weights ranged from 66230.22 to 72743.65 and their pI ranged from 5.23 to 8.73. Except for PfSERK2, PfSERK4, PfSERK6, PfSERK9, and PfSERK10, the other seven PfSERKs were predicted to be stable (instability index < 40). The predicted results showed that all the 12 PfSERK proteins were hydrophilic proteins, and they were all located on the cell membrane (Table 1).
3.2 Phylogenetic and Gene Structure Analysis
Phylogenetic analysis of the SERK sequences of P. fortunei, A. thaliana, Z. mays, O. sativa, M. truncatula, and Malus × domestica showed that the 12 PfSERKs clustered in three subfamilies (I, II, and III) in the phylogenetic tree (Fig. 2A). Subfamily I had the largest number of proteins, and included all the A. thaliana, Z. mays, O. sativa, and M. truncatula SERKs as well as 4 SERKs from Malus × domestica and 4 SERKs from P. fortunei. Subfamily II contained 7 SERKs from Malus × domestica and 5 SERKs from P. fortunei. Subfamily III contained 4 SERKs, one from Malus × domestica and 3 from P. fortunei. Analysis of the gene structure of the PfSERKs may be useful for studying their evolutionary relationships. We found that except for PfSERK7, which contained 12 exons, the other 11 PfSERKs contained 11 exons, and the exons of the PfSERKs that shares a close evolutionary relationship were similar in length and position (Fig. 2B).
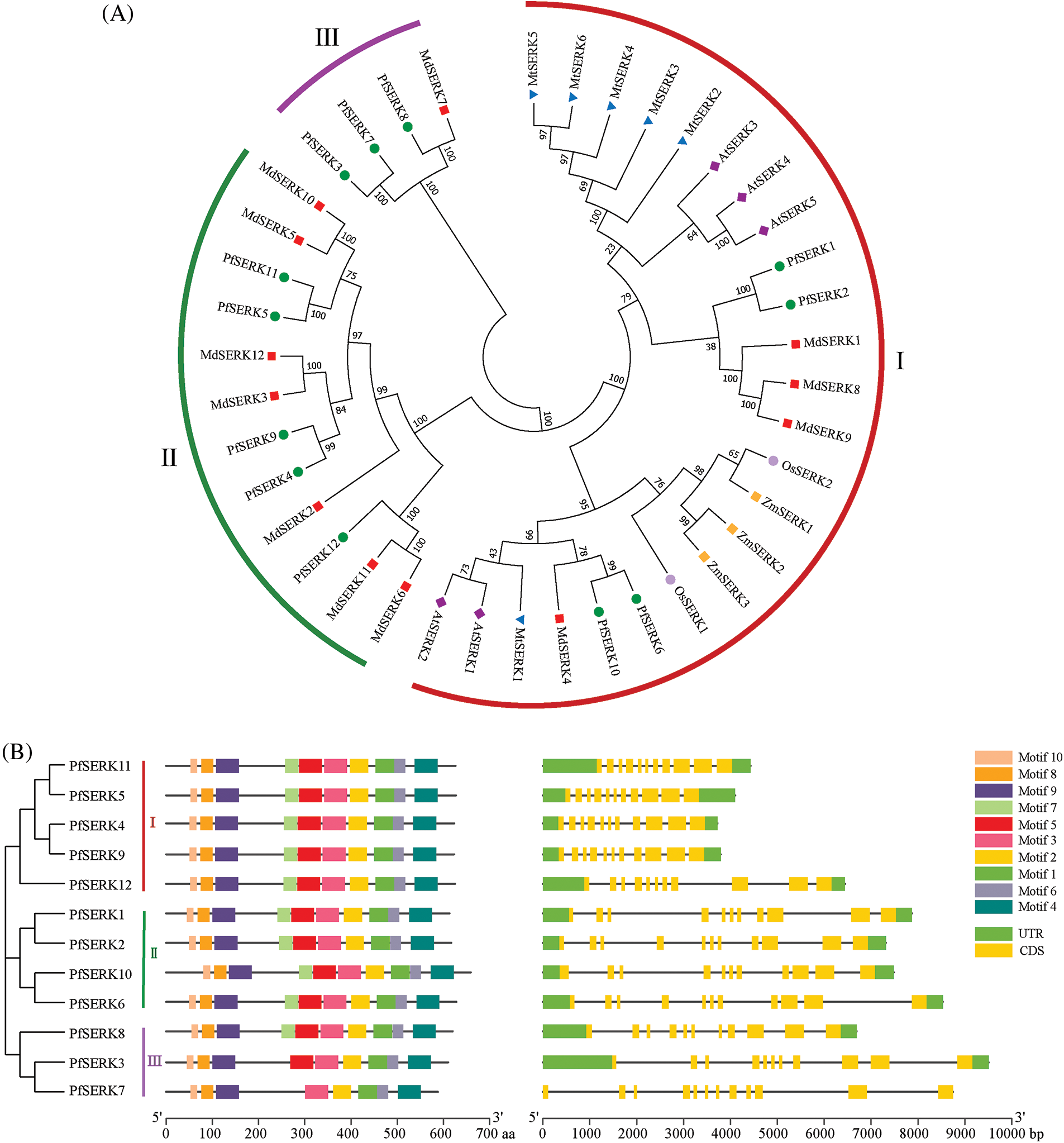
Figure 2: The analysis of phylogenetic, conserved motifs and gene structure. (A) Phylogentic analysis of SERKs from P. fortunie, A. thaliana, Z. mays, O. sativa, M. truncatula, and Malus × domestice. (B) The motif composition of PfSERKs (left) and the exon-intron structure of PfSERKs (right)
3.3 Structure of the PfSERK Proteins
The MEME analysis of the conserved domains of the PfSERKs showed that, except for PfSERK3 and PfSERK7, all the other PfSERKs contained 10 conserved motifs that were similar in length and location (Fig. 2B). The predicted secondary structures of PfSERK protein showed that they had similar secondary structures, mainly α-helix (40.35%–46.43%) and random curl (36.07%–40.89%), followed by extended strand (12.14%–14.56%) and β-turn (3.74%–5.90%) (Fig. 3A). The predicted tertiary structures of PfSERK protein showed that they also have similar three-dimensional spatial structure of protein, and some differences are found in their tertiary structures, such as the length of α-helix, β-turn and random coil (Fig. 3B).
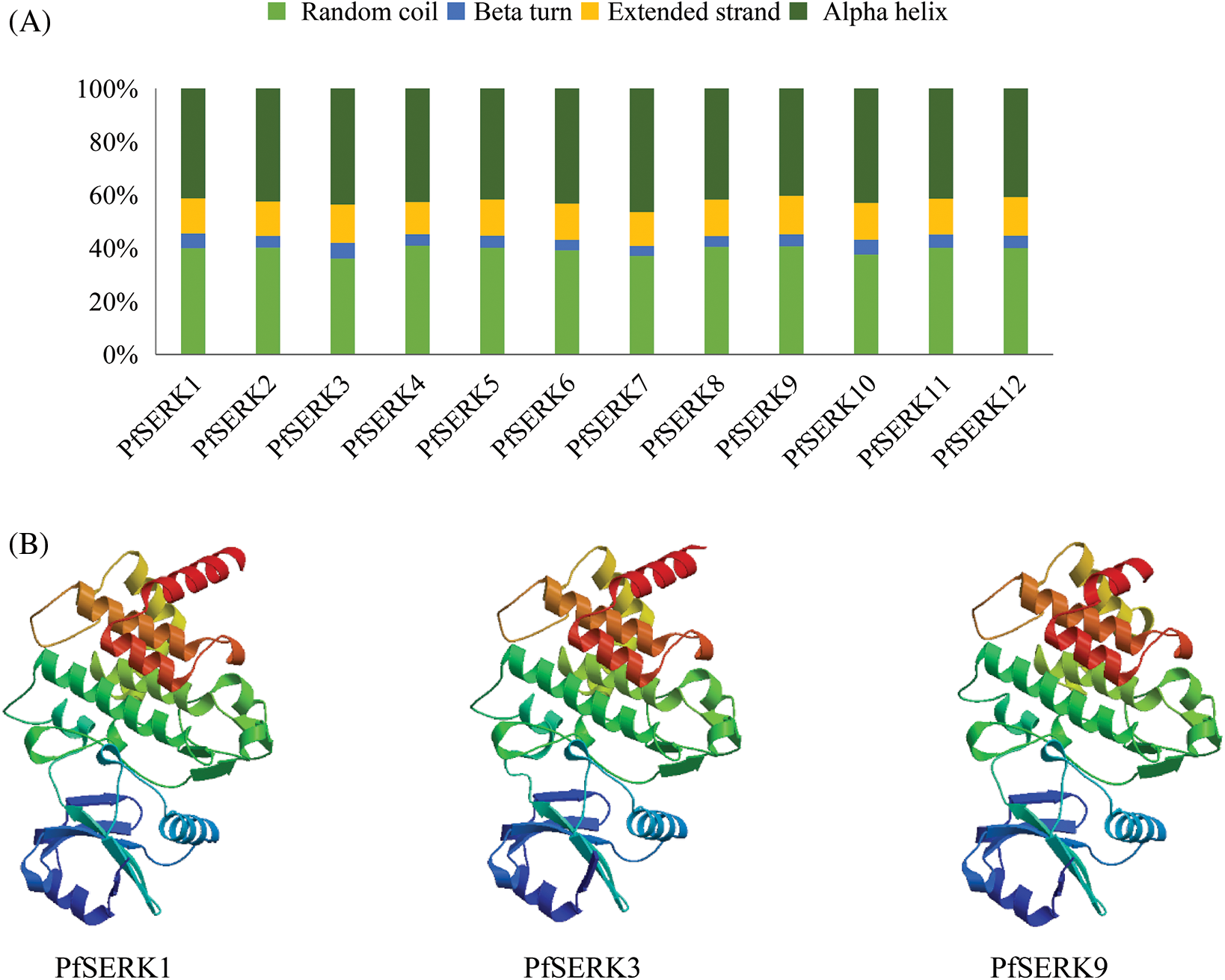
Figure 3: Protein structure of PfSERKs. (A) Secondary structure percentage diagram of PfSERK proteins. (B) Tertiary structure of PfSERK proteins
3.4 Collinearity Analysis of the PfSERK
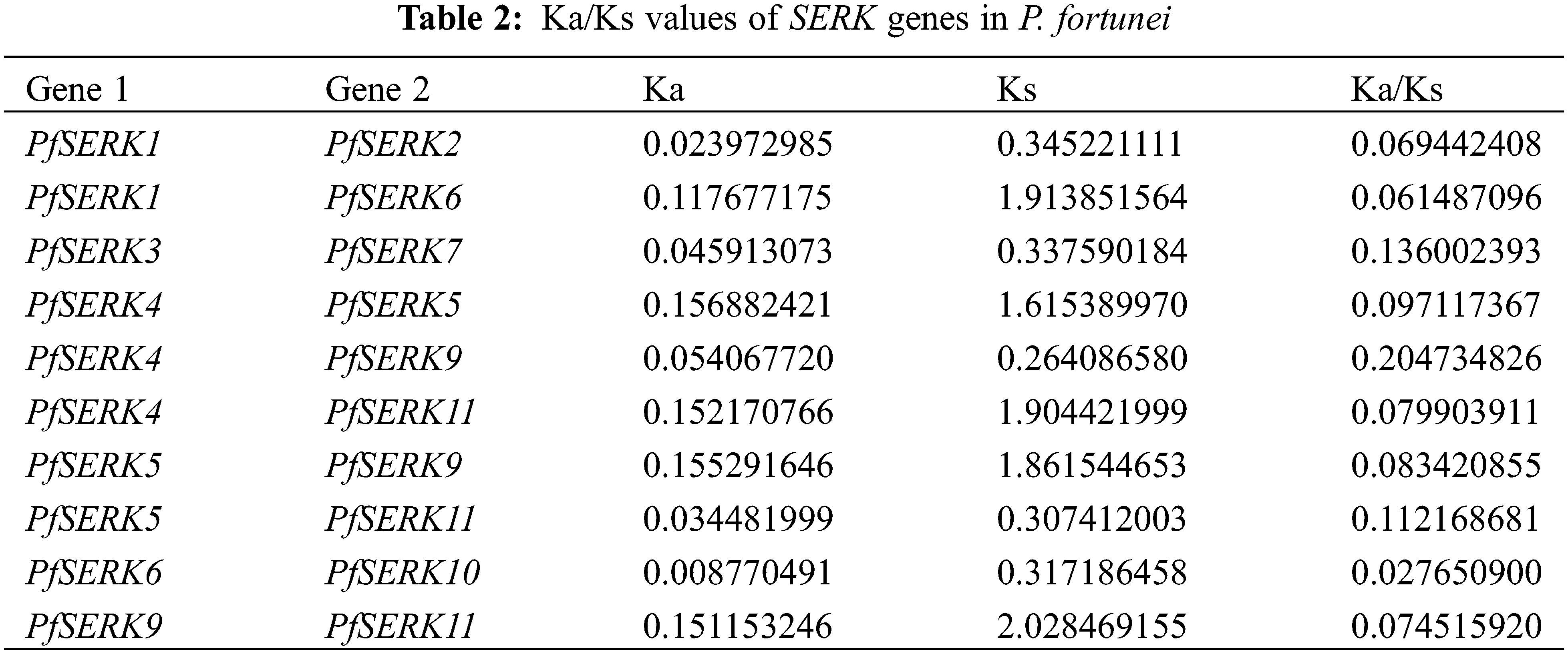
Gene duplication events occur during plant genome evolution. We searched for PfSERK duplicates and detected 10 segmental duplications among the PfSERKs, namely PfSERK1 and PfSERK2/6; PfSERK3 and PfSERK7; PfSERK4 and PfSERK5/9/11; PfSERK5 and PfSERK9/11; PfSERK6 and PfSERK10; and PfSERK9 and PfSERK11 (Fig. 4). This finding indicates that amplification of the PfSERK gene family was largely influenced by segmental duplication. According to the collinear map of AtSERKs and PfSERKs, two homologous genes of SERK, PfSERK6 and AtSERK1/2, were found (Fig. 5). We calculated the Ka/Ks ratio of the selection pressure of PfSERKs (Table 2), the Ka/Ks of all PfSERK family members were much smaller than 1, these resuits indicating that they were subject to purifying selection during the evolutionary process.
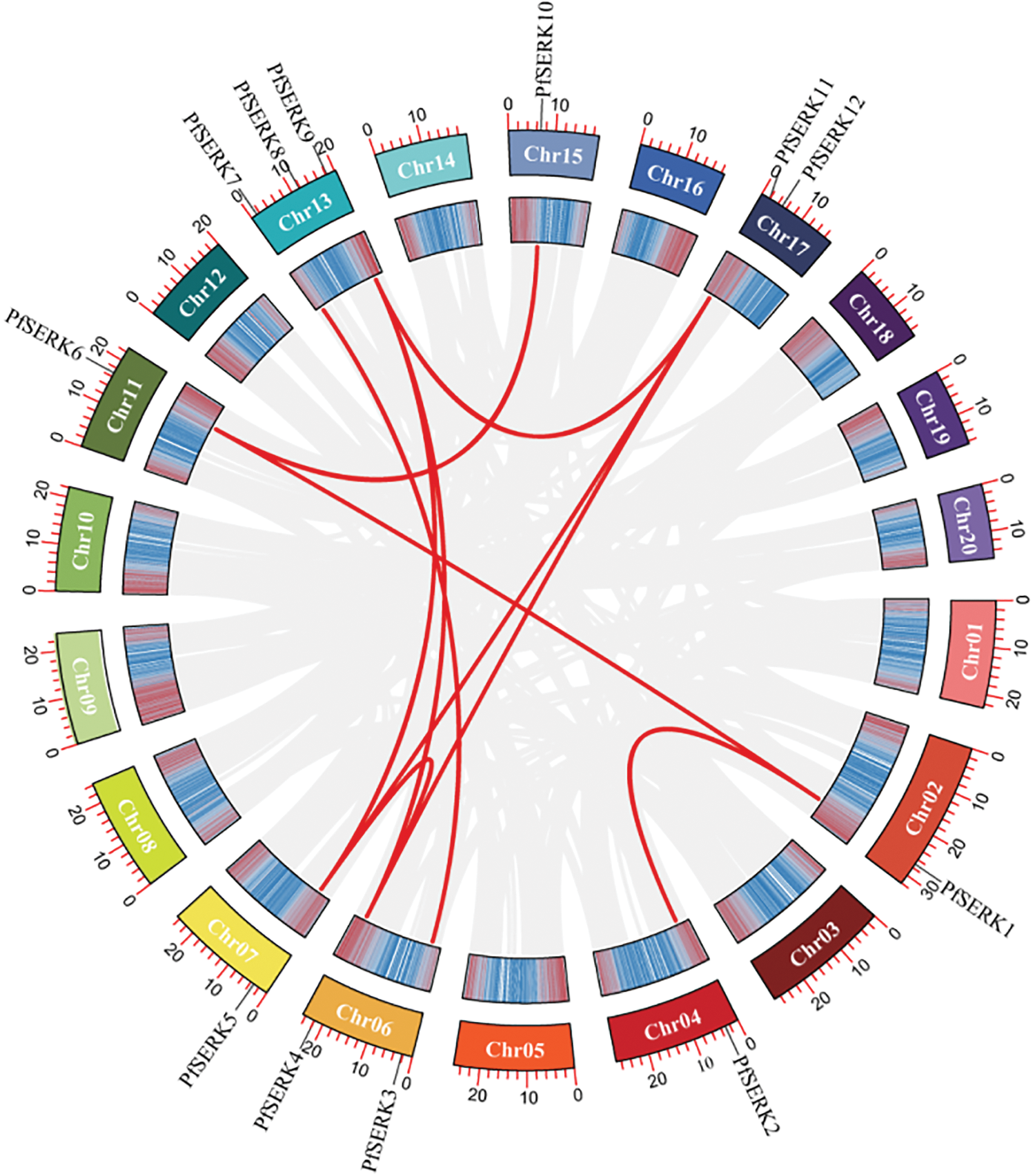
Figure 4: Synteny of PfSERK genes. The red lines refer to collinear gene pairs
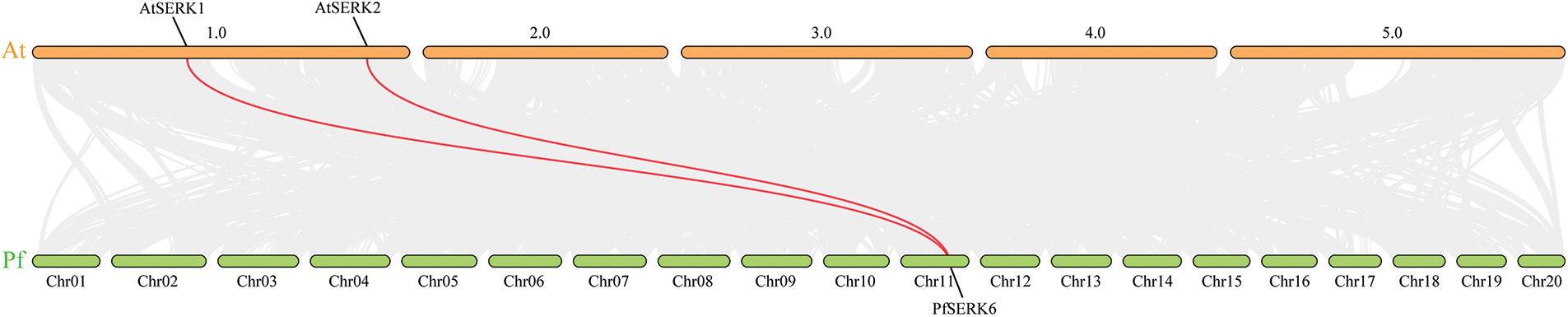
Figure 5: Synteny between PfSERKs and AtSERKs. The red lines refer to collinear gene pairs
3.5 Analysis of Cis-Acting Elements
The prediction of cis-acting elements showed that light response elements were the most common and made up the largest proportion (Fig. 6). Hormone elements such as abscisic acid and auxin were detected, suggesting that the corresponding PfSERKs may affect the growth and development of P. fortunei by participating in different hormonal regulatory pathways. Drought, low temperature, anaerobic, and other abiotic stress-related response elements were also detected, suggesting that the corresponding PfSERKs can respond to stress.
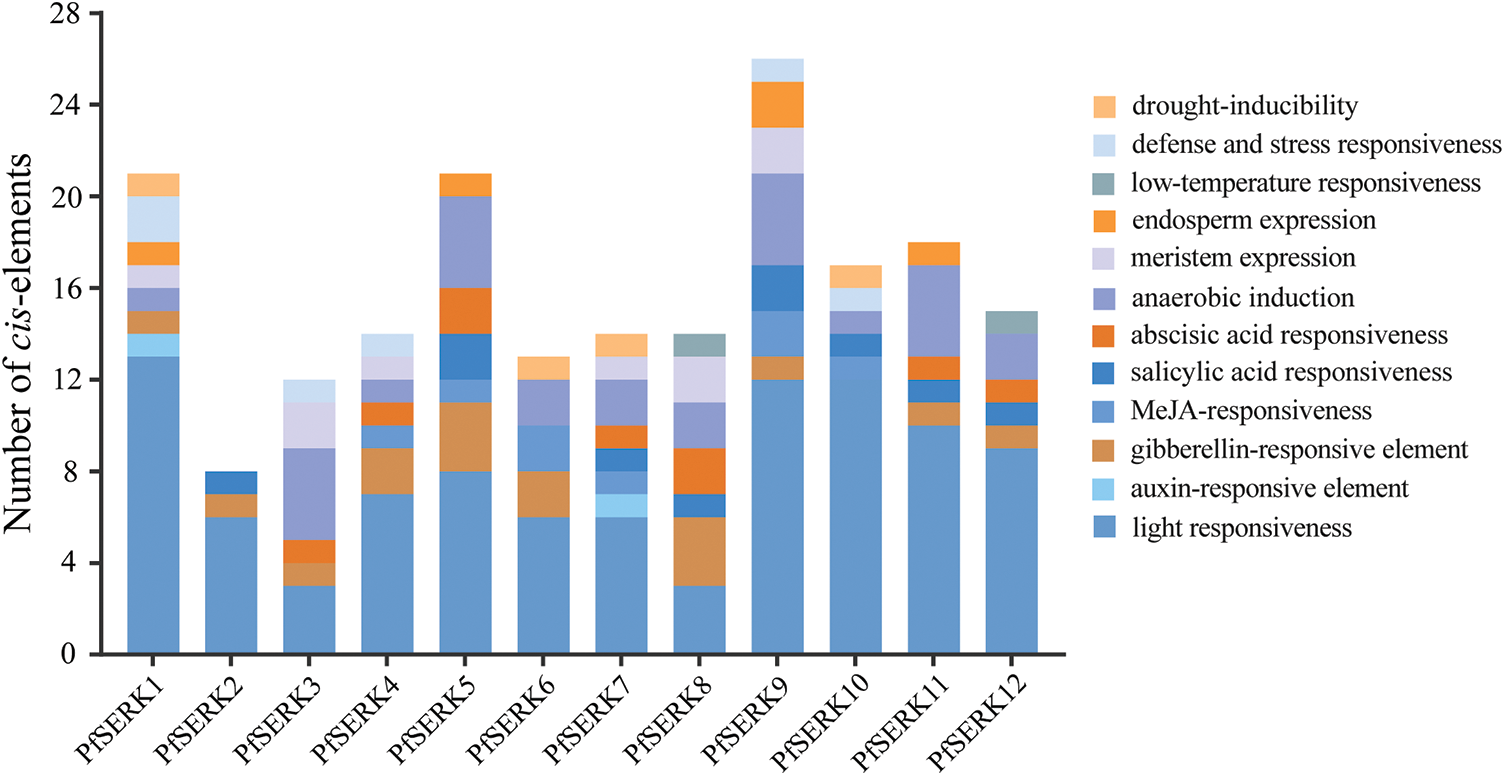
Figure 6: Cis-acting elements of PfSERK genes
3.6 Expression of PfSERKs in Different P. fortunei Tissues
Expression profiles of 12 PfSERKs in different parts of Paulownia showed that the expression levels of PfSERK2/4/7/8/9/11/12 were highest in buds, subsequent in leaves, and lowest in roots; the expression level of PfSERK1 was highest in buds, intermediate in leaves and internodes, and lowest in nodes; PfSERK3 and PfSERK6 were the most expressed in buds; PfSERK5 was expressed more in leaves and roots than in other parts; the expression of PfSERK10 was the highest in nodes (Fig. 7). The above results showed that PfSERK gene was expressed in many tissues of Paulownia, which might affect the growth and development of Paulownia trees.
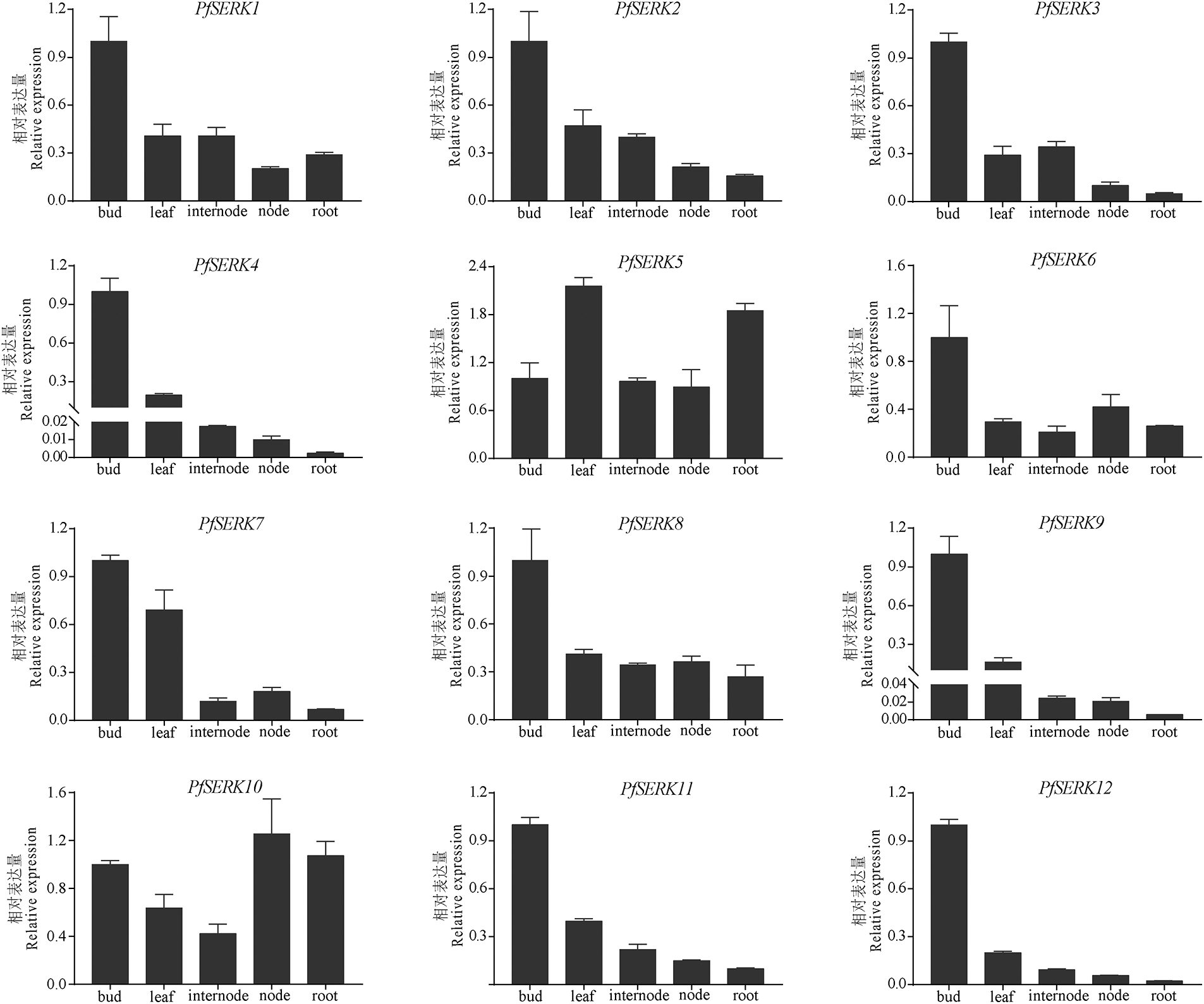
Figure 7: Expression patterns of PfSERKs in P. fortunei. qRT-PCR analysis of SERK gene expression levels in buds, leaves, internodes, nodes, and roots. Bars show mean ± SD (n = 3)
3.7 Analysis of PfSERK Expression under Phytoplasma Infection
To understand the expression pattern of PfSERKs with phytoplasma infected, the transcriptome data of the PfSERKs in PF and PFI was analyzed. All the PfSERKs were expressed in both healthy and PaWB-infected P. fortunei. As shown in Fig. 8A, PfSERK3, PfSERK8, and PfSERK11 were down-regulated and PfSERK4, PfSERK9, and PfSERK12 were up-regulated in the PFI vs. PF comparison, it is indicated that they were associated with the occurrence of PaWB.
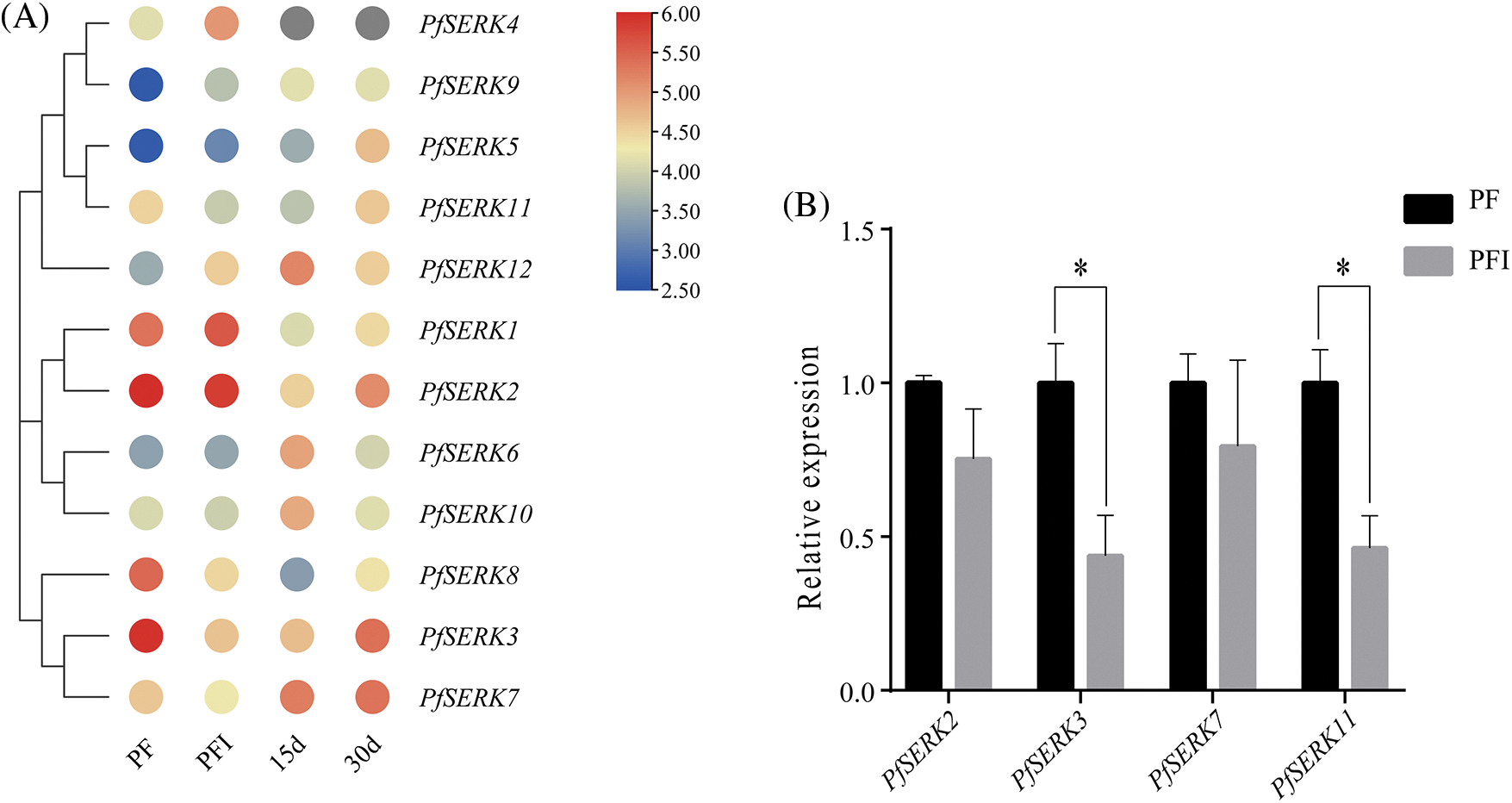
Figure 8: Expression analysis of PfSERK genes under phytoplasma infection. (A) PfSERK genes expression in PF, PFI and PFI treated with Rif. (B) results of qRT-PCR validation. Bars show mean ± SD (n = 3). p < 0.05 was marked with asterisk (*)
Furthermore, after treatment with 30 mg L−1 Rif, the expression pattern of PfSERK3 and PfSERK11 were in accordance with the morphological changes in the simulated system of PaWB (Fig. 8A), indicating that PfSERK3 and PfSERK11 may be related to PaWB. qRT-PCR analysis of 4 PfSERK genes showed that the gene expression levels were consistent with RNA-seq data, indicating that the transcriptome sequencing data was reliable (Fig. 8B).
3.8 Interaction Network of PfSERKs and Other Proteins
In order to further explore the function of PfSERK, an interaction PfSERK–protein network was constructed using A. thaliana orthologs (Fig. 9). The predicted results show that PfSERK1/3/6 may interact with BRI1, and PfSERK4/6/11/12 may interact with BAK1 and SAC52. It has been proved that BAK1, BRI1 and SAC52 can regulate plant height [15,17,35] so it is speculated that PfSERK1/3/4/6/11/12 may be involved in the regulation of plant height. Further combined with transcriptome data analysis, it was found that PfSERK3 and PfSERK11 may associated with the dwarfing of Paulownia witches’ broom disease plant.
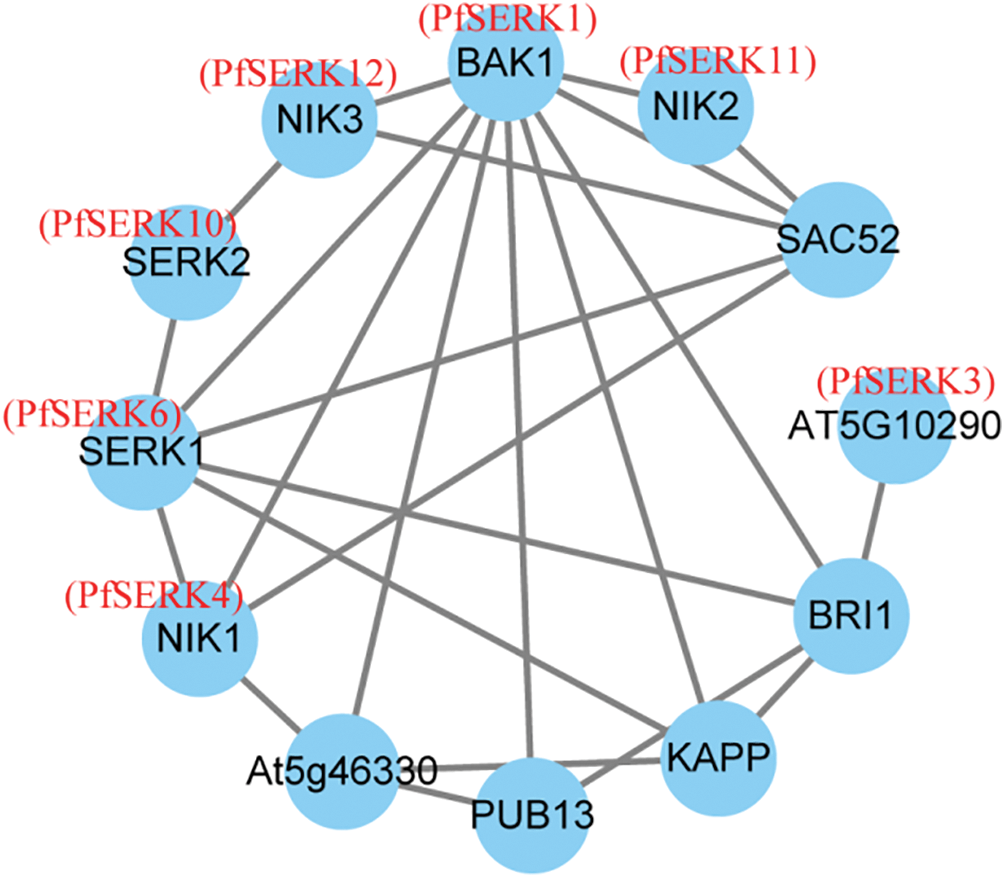
Figure 9: SERK protein interaction network in P. fortunei
3.9 Expression of PfSERKs under Drought and Salt Stress
To understand the expression of the PfSERKs under abiotic stress, we downloaded and analyzed the RNA-seq data of Paulownia under drought and salt stress (Fig. 10). The expression of PfSERK1, PfSERK2, PfSERK4, PfSERK5, PfSERK8, PfSERK9, and PfSERK10 were significantly changed after drought treatment of P. fortunei. In which, the gene expression level of PfSERK1 was up-regulated 2 times after drought stress, and the gene expression levels of PfSERK4, PfSERK5, PfSERK8, PfSERK9, and PfSERK10 were significantly down-regulated 2–3 times, indicating that PfSERK1, PfSERK4, PfSERK5, PfSERK8, PfSERK10, PfSERK9, and PfSERK10 could respond to drought stress of Paulownia. 11 PfSERK genes were expressed after control and salt treatment, among them, the expression level of PfSERK3 changed significantly after salt stress. The gene expression level of PfSERK3 was down-regulated by 2 times after salt stress, indicating that PfSERK3 could respond to salt stress of Paulownia.
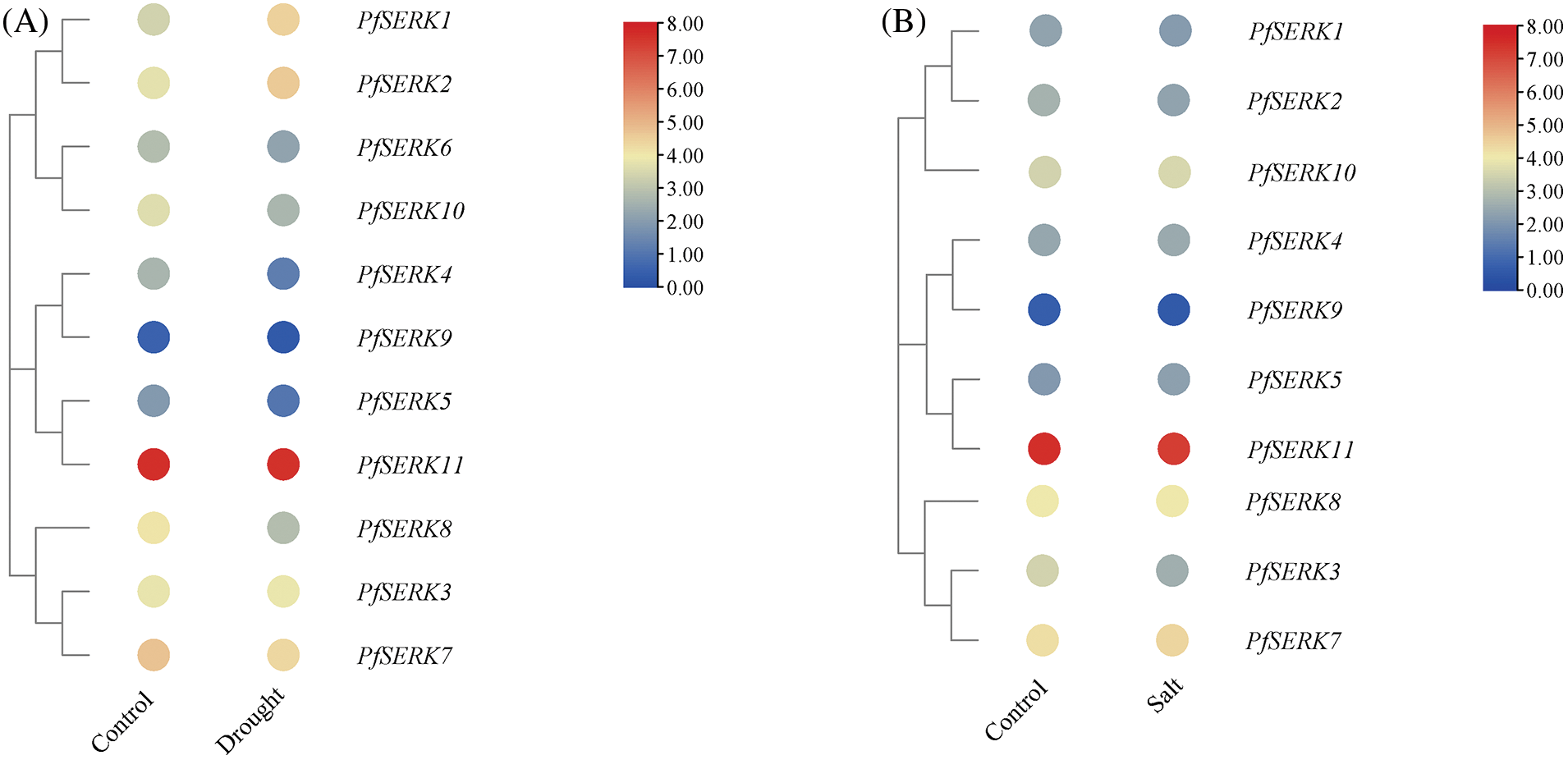
Figure 10: Expression pattern of PfSERKs under drought (A) and salt (B) stress
SERK is a highly conserved class of protein kinases that play regulatory role in the process of stress response [14,36–38]. We identified 12 members of PfSERK family, and made a preliminary prediction of their subcellular localization. It was found that these members all located on the cell membrane. The analysis of physiochemical properties of PfSERK found that most PfSERKs were similar in length and molecular weight to AtSERKs, and more than half of PfSERKs were stable. The numbers of exon in the PfSERKs was consistent with the numbers found in SERKs of other species such as A. thaliana, Malus × domestica, which indicates that SERKs were relatively conserved in the evolutionary process of plant species [3,11,24]. The PfSERKs contained similar domains as SERKs in other species, indicating that they had a close evolutionary relationship. The structure of PfSERK protein is highly conserved in α-helix, β-turns, extended strand, and random coil, which indicates that PfSERKs have similar secondary structures, and the function of protein is determined by its structure, so it is speculated that PfSERKs may have similar functions.
The promoters of several PfSERKs contained hormone-related and stress-related response elements, indicating that these genes are closely associated with the growth of Paulownia and the relevant response to stress. Gene duplication is important for expanding the number of gene family members and increasing their functional diversity [39,40]. We identified 10 gene pairs among the 12 PfSERKs that may have been involved in fragment replication events, and speculated that fragment replication may be very important in PfSERK gene expansion. In our research, two SERK collinear gene pairs were identified from P. fortunei and A. thaliana, namely, PfSERK6 was identified as the homologous gene of AtSERK1 and AtSERK2, so it is speculated that PfSERK6 may have similar functions to AtSERK1 and AtSERK2.
The 12 analyzed PfSERK genes were expressed in buds, leaves, internodes, nodes and roots, and the expression levels were different, indicating that the PfSERK family may be associated with the Paulownia growth and development. It has been reported that SERK can respond to abiotic stress. For example, overexpression of OsSERK2 can obviously increase the grain size of rice and enhance its salt tolerance [41]; MdSERK4, MdSERK6, MdSERK7 can respond to salt stress in apple [11]; HvSERK1 and HvSERK3 have regulatory effect on salt tolerance in barley [42]. Multiple PfSERK genes were able to respond to drought and salt stress, among which PfSERK1/4/5/8/9/10 were sensitive to drought stress and only PfSERK3 was sensitive to salt stress.
The prediction of the PfSERK–protein interaction network showed that PfSERK3 may interact with BRI1, PfSERK11 may interact with SAC52, and BRI1 and SAC52 are both known to be involved in regulating plant height [15,35]. We also found that the expression of PfSERK3 and PfSERK11 genes differed significantly before and after the occurrence of PaWB, and they were expressed in nodes and internodes to varying degrees. Combined with the expression patterns of PfSERK3 and PfSERK11 and the prediction results of protein interaction, we speculated that PfSERK3 and PfSERK11 may be involved in the occurrence of PaWB by regulating the plant height of Paulownia. However, the functions of the PfSERKs need to be confirmed experimentally in future research. The functions of the PfSERKs can be verified by, for example, transgenic experiments to determine their specific roles in the infection process by PaWB.
This is the first time to analyze the SERK gene family of P. fortunei in the whole genome. We conducted detailed bioinformatics analyses of the PfSERKs, including gene chromosome localization, chemical signatures, phylogeny, synteny, gene structure, promoter sequences, and SERK–protein interactions. The expression of PfSERKs in different parts of P. fortunei and in response to biotic and abiotic stresses was also analyzed. These findings are benefit to further understanding the fuction of PfSERKs in Paulownia.
Acknowledgement: We thank Margaret Biswas, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
Funding Statement: This has obtained the support from the Academic Scientist Fund for Zhongyuan Scholars of Henan Province (Grant No. 2018(185)) and Project of Central Plains Science and Technology Innovation Leading Talents of Henan Province (224200510010).
Author Contributions: Study conception and design: Guoqiang Fan; data collection: Tongman Zhao, Yabing Cao, Xiaogai Zhao; analysis and interpretation of results: Tongman Zhao, Yujie Fan; draft manuscript preparation: Tongman Zhao, Yujie Fan. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest for the present study.
References
1. Baudino, S., Hansen, S., Brettschneider, R., Hecht, V. F., Dresselhaus, T. et al. (2001). Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta, 213(1), 1–10. https://doi.org/10.1007/s004250000471 [Google Scholar] [PubMed] [CrossRef]
2. Nolan, K. E., Kurdyukov, S., Rose, R. J. (2011). Characterisation of the legume SERK-NIKgene superfamily including splice variants: Implications for development and defence. BMC Plant Biology, 11(1), 1–16. https://doi.org/10.1186/1471-2229-11-44 [Google Scholar] [PubMed] [CrossRef]
3. Hecht, V., Vielle-Calzada, J. P., Hartog, M. V., Schmidt, E. D., Boutilier, K. et al. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiology, 127(3), 803–816. https://doi.org/10.1104/pp.010324 [Google Scholar] [CrossRef]
4. Landschulz, W. H., Johnson, P. F., McKnight, S. L. (1988). The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science, 240(4860), 1759–1764. https://doi.org/10.1126/science.3289117 [Google Scholar] [PubMed] [CrossRef]
5. Shah, K., GadellaJr, T. W., van Erp, H., Hecht, V., de Vries, S. C. (2001). Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. Journal of Molecular Biology, 309(3), 641–655. https://doi.org/10.1006/jmbi.2001.4706 [Google Scholar] [PubMed] [CrossRef]
6. Shah, K., Russinova, E., Gadella, T. W., Willemse, J., de Vries, S. C. (2002). The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes & Development, 16(13), 1707–1720. https://doi.org/10.1101/gad.220402 [Google Scholar] [PubMed] [CrossRef]
7. Schmidt, E. D., Guzzo, F., Toonen, M. A., de Vries, S. C. (1997). A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development, 124(10), 2049–2062. https://doi.org/10.1242/dev.124.10.2049 [Google Scholar] [PubMed] [CrossRef]
8. Ito, Y., Takaya, K., Kurata, N. (2005). Expression of SERK family receptor-like protein kinase genes in rice. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression, 1730(3), 253–258. https://doi.org/10.1016/j.bbaexp.2005.06.007 [Google Scholar] [PubMed] [CrossRef]
9. Park, H. S., Ryu, H. Y., Kim, B. H., Kim, S. Y., Yoon, I. S. et al. (2011). A subset of OsSERK genes, including OsBAK1, affects normal growth and leaf development of rice. Molecules and Cells, 32(6), 561–569. https://doi.org/10.1007/s10059-011-0178-4 [Google Scholar] [PubMed] [CrossRef]
10. Singh, A., Khurana, P. (2017). Ectopic expression of Triticum aestivum SERK genes (TaSERKs) control plant growth and development in Arabidopsis. Scientific Reports, 7(1), 12368. https://doi.org/10.1038/s41598-017-10038-1 [Google Scholar] [PubMed] [CrossRef]
11. Zheng, L., Ma, J., Mao, J., Fan, S., Zhang, D. et al. (2018). Genome-wide identification of SERK genes in apple and analyses of their role in stress responses and growth. BMC Genomics, 19(1), 1–18. https://doi.org/10.1186/s12864-018-5342-1 [Google Scholar] [PubMed] [CrossRef]
12. Ghai, D., Alok, A., Upadhyay, S. K., Sembi, J. K. (2020). Genome wide characterization of the SERK/SERL gene family in Phalaenopsis equestris, Dendrobium catenatum and Apostasia shenzhenica (Orchidaceae). Computational Biology and Chemistry, 85, 107210. https://doi.org/10.1016/j.compbiolchem.2020.107210 [Google Scholar] [PubMed] [CrossRef]
13. Karlova, R., Boeren, S., Russinova, E., Aker, J., Vervoort, J. et al. (2006). The Arabidopsis somatic embryogenesis receptor-like kinase1 protein complex includes brassinosteroid-insensitive1. The Plant Cell, 18(3), 626–638. https://doi.org/10.1105/tpc.105.039412 [Google Scholar] [PubMed] [CrossRef]
14. Albrecht, C., Russinova, E., Kemmerling, B., Kwaaitaal, M., de Vries, S. C., (2008). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and-independent signaling pathways. Plant Physiology, 148(1), 611–619. https://doi.org/10.1104/pp.108.123216 [Google Scholar] [PubMed] [CrossRef]
15. He, K., Gou, X., Yuan, T., Lin, H., Asami, T. et al. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Current Biology, 17(13), 1109–1115. https://doi.org/10.1016/j.cub.2007.05.036 [Google Scholar] [PubMed] [CrossRef]
16. Jin, Y. L., Tang, R. J., Wang, H. H., Jiang, C. M., Bao, Y. et al. (2017). Overexpression of Populus trichocarpa CYP 85A3 promotes growth and biomass production in transgenic trees. Plant Biotechnology Journal, 15(10), 1309–1321. https://doi.org/10.1111/pbi.12717 [Google Scholar] [PubMed] [CrossRef]
17. Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E. et al. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell, 110(2), 213–222. https://doi.org/10.1016/s0092-8674(02)00812-7 [Google Scholar] [PubMed] [CrossRef]
18. Hosseini, S., Schmidt, E. D., Bakker, F. T. (2020). Leucine-rich repeat receptor-like kinase II phylogenetics reveals five main clades throughout the plant kingdom. The Plant Journal, 103(2), 547–560. https://doi.org/10.1111/tpj.14749 [Google Scholar] [PubMed] [CrossRef]
19. Kemmerling, B., Schwedt, A., Rodriguez, P., Mazzotta, S., Frank, M. et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Current Biology, 17(13), 1116–1122. https://doi.org/10.1016/j.cub.2007.05.046 [Google Scholar] [PubMed] [CrossRef]
20. Gou, X., Yin, H., He, K., Du, J., Yi, J. et al. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genetics, 8(1), e1002452. https://doi.org/10.1371/journal.pgen.1002452 [Google Scholar] [PubMed] [CrossRef]
21. Kauschmann, A., Jessop, A., Koncz, C., Szekeres, M., Willmitzer, L. et al. (1996). Genetic evidence for an essential role of brassinosteroids in plant development. The Plant Journal, 9(5), 701–713. https://doi.org/10.1046/j.1365-313X.1996.9050701.x [Google Scholar] [CrossRef]
22. Chen, X., Zuo, S., Schwessinger, B., Chern, M., Canlas, P. E. et al. (2014). An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Molecular Plant, 7(5), 874–892. https://doi.org/10.1093/mp/ssu003 [Google Scholar] [PubMed] [CrossRef]
23. Hu, H., Xiong, L., Yang, Y. (2005). Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta, 222(1), 107–117. https://doi.org/10.1007/s00425-005-1534-4 [Google Scholar] [PubMed] [CrossRef]
24. Zuo, S., Zhou, X., Chen, M., Zhang, S., Schwessinger, B. et al. (2014). OsSERK1 regulates rice development but not immunity to Xanthomonas oryzae pv. oryzae or Magnaporthe oryzae. Journal of Integrative Plant Biology, 56(12), 1179–1192. https://doi.org/10.1111/jipb.12290 [Google Scholar] [PubMed] [CrossRef]
25. Yadav, N. K., Vaidya, B. N., Henderson, K., Lee, J. F., Stewart, W. M. et al. (2013). A review of Paulownia biotechnology: A short rotation, fast growing multipurpose bioenergy tree. American Journal of Plant Sciences, 4(11), 2070–2082. https://doi.org/10.4236/ajps.2013.411259 [Google Scholar] [CrossRef]
26. Aloi, F., Riolo, M., La Spada, F., Bentivenga, G., Moricca, S. et al. (2021). Phytophthora root and collar rot of Paulownia, a new disease for Europe. Forests, 12(12), 1664. https://doi.org/10.3390/f12121664 [Google Scholar] [CrossRef]
27. Cao, Y., Fan, G., Wang, Z., Gu, Z. (2019). Phytoplasma-induced changes in the acetylome and succinylome of Paulownia tomentosa provide evidence for involvement of acetylated proteins in witches’ broom disease. Molecular & Cellular Proteomics, 18(6), 1210–1226. https://doi.org/10.1074/mcp.RA118.001104 [Google Scholar] [PubMed] [CrossRef]
28. Cao, Y., Sun, G., Zhai, X., Xu, P., Ma, L. et al. (2021). Genomic insights into the fast growth of paulownias and the formation of Paulownia witches’ broom. Molecular Plant, 14(10), 1668–1682. https://doi.org/10.1016/j.molp.2021.06.021 [Google Scholar] [PubMed] [CrossRef]
29. Fan, G, Q., Cao, Y, B., Deng, M, J., Zhai, X, Q., Zhao, Z, L. et al. (2017). Identification and dynamic expression profiling of microRNAs and target genes of Paulownia tomentosa in response to Paulownia witches’ broom disease. Acta Physiologiae Plantarum, 39(1), 28. https://doi.org/10.1007/s11738-016-2326-0 [Google Scholar] [CrossRef]
30. Cao, X., Fan, G., Zhao, Z., Deng, M., Dong, Y. (2014). Morphological changes of Paulownia seedlings infected phytoplasmas reveal the genes associated with witches’ broom through AFLP and MSAP. PLoS One, 9(11), e112533. https://doi.org/10.1371/journal.pone.0112533 [Google Scholar] [PubMed] [CrossRef]
31. Fan, G., Cao, X., Zhao, Z., Deng, M. (2015). Transcriptome analysis of the genes related to the morphological changes of Paulownia tomentosa plantlets infected with phytoplasma. Acta Physiologiae Plantarum, 37(10), 1–12. https://doi.org/10.1007/s11738-015-1948-y [Google Scholar] [CrossRef]
32. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262 [Google Scholar] [PubMed] [CrossRef]
33. Zhao, J., Guo, R., Guo, C., Hou, H., Wang, X. et al. (2016). Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Frontiers in Plant Science, 7, 376. https://doi.org/10.3389/fpls.2016.00376 [Google Scholar] [PubMed] [CrossRef]
34. Wang, Z. (2020). Research of ceRNA related to the occurrence of Paulownia fortunei witches’ broom. Journal of Henan Agricultual University. https://doi.org/10.27117/d.cnki.ghenu.2020.000001 [Google Scholar] [CrossRef]
35. Imai, A., Komura, M., Kawano, E., Kuwashiro, Y., Takahashi, T. (2008). A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. The Plant Journal, 56(6), 881–890. https://doi.org/10.1111/j.1365-313X.2008.03647.x [Google Scholar] [PubMed] [CrossRef]
36. Aan den Toorn, M., Albrecht, C., de Vries, S. (2015). On the origin of SERKs: Bioinformatics analysis of the somatic embryogenesis receptor kinases. Molecular Plant, 8(5), 762–782. https://doi.org/10.1016/j.molp.2015.03.015 [Google Scholar] [PubMed] [CrossRef]
37. Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A. et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. The Plant Cell, 23(6), 2440–2455. https://doi.org/10.1105/tpc.111.084301 [Google Scholar] [PubMed] [CrossRef]
38. Wu, X. M., Kou, S. J., Liu, Y. L., Fang, Y. N., Xu, Q. et al. (2015). Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: Micro RNA- and si RNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnology Journal, 13(3), 383–394. https://doi.org/10.1111/pbi.12317 [Google Scholar] [PubMed] [CrossRef]
39. Albrecht, C., Russinova, E., Hecht, V., Baaijens, E., de Vries, S. (2005). The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. The Plant Cell, 17(12), 3337–3349. https://doi.org/10.1105/tpc.105.036814 [Google Scholar] [PubMed] [CrossRef]
40. Colcombet, J., Boisson-Dernier, A., Ros-Palau, R., Vera, C. E., Schroeder, J. I. (2005). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. The Plant Cell, 17(12), 3350–3361. https://doi.org/10.1105/tpc.105.036731 [Google Scholar] [PubMed] [CrossRef]
41. Dong, N., Yin, W., Liu, D., Zhang, X., Yu, Z. et al. (2020). Regulation of brassinosteroid signaling and salt resistance by SERK2 and potential utilization for crop improvement in rice. Frontiers in Plant Science, 11, 621859. https://doi.org/10.3389/fpls.2020.621859 [Google Scholar] [PubMed] [CrossRef]
42. Li, Y., Liu, C., Guo, G., He, T., Chen, Z. et al. (2017). Expression analysis of three SERK-like genes in barley under abiotic and biotic stresses. Journal of Plant Interactions, 12(1), 279–285. https://doi.org/10.1080/17429145.2017.1339836 [Google Scholar] [CrossRef]
Supplementary Materials
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools