 Open Access
Open Access
ARTICLE
Transcriptome Profiling of the Salt-Stress Response in Paper Mulberry
1 College of Forestry, Northwest A&F University, Yangling, 712100, China
2 Guangxi Dongmen Farm, Dongmen Forest Farm, Chongzuo, 5321808, China
3 College of Forestry, Henan University of Science and Technology, Luoyang, 471023, China
4 Key Laboratory of Tree Breeding and Cultivation of National Forestry and Grassland Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, 100091, China
* Corresponding Authors: Jianwei Ni. Email: ; Dongmei Wang. Email:
(This article belongs to the Special Issue: Development of New Sensing Technology in Sustainable Farming and Smart Environmental Monitoring)
Phyton-International Journal of Experimental Botany 2023, 92(9), 2591-2610. https://doi.org/10.32604/phyton.2023.028338
Received 14 December 2022; Accepted 06 May 2023; Issue published 28 July 2023
Abstract
Paper mulberry is a high-quality woody feed resource plant with high crude protein content. It is widely distributed in China and has excellent characteristics of salt and alkali tolerance. Paper mulberry has ecological and economic importance. Salt stress has become a critical factor with the increasing degree of soil salinity that restricts plant growth. In the saline-alkali environments, transcriptome expression is altered leading to phenotypic defects in most plants. However, the regulatory mechanism related to paper mulberry’s salt-stress (SS) response is not clearly understood. In the present study de novo transcriptomic assembly was performed, and gene expression levels were measured between different SS and natural conditions (25°C) as a control for paper mulberry plants. According to the results of our study, under NaCl stress conditions, the differential gene expression was observed in the leaves of paper mulberry compared with the control. A total of 2126 differentially expressed unigenes were observed. Among these unigenes the expression of 812 DEGs was up-regulated and the expression of 1,314 DEGs was down-regulated. Additionally, The GO and KEGG analyses regarding differentially expressed unigenes (DEUs) revealed that the observed critical transcriptomic alterations under salt stress (SS) conditions were associated with primary and secondary metabolism, photosynthesis, and plant hormone signaling pathways. Further investigations such as gene function studies regarding the unigenes depicting altered expression under salt stress conditions in paper mulberry will help understand the mechanism of salt tolerance, and this information can be utilized in paper mulberry breeding and improvement programs.Keywords
Supplementary Material
Supplementary Material FilePaper mulberry belongs to the family Moraceae. It is native to Asia and its natural distribution occurs across the eastern Asian and Pacific areas [1,2]. Paper mulberry has the adaptation ability under unfavorable environmental conditions and this is utilized in ecological restoration [2,3]. For instance, the trees survive within the abandoned factories, facilitating mining recovery and salinity control [3–6]. The bark of the paper mulberry is used in papermaking; leaves can be utilized as fodder for livestock, and its fruits and roots are used in traditional Chinese medicine (TCM). Moreover, the entire plant can be used to prepare bioethanol that has high economic value [7–10]. In China, the cultivation of hybrid paper mulberry is getting popularity. Paper mulberry hybrid Broussonetia kazinoki × B. papyifera is the dominant tree species, and this is widely planted and utilized in several regions in China [1,11].
There are large areas of salinized land all over the world; therefore, the identification of the genes associated with salt tolerance is of great importance for environmental protection and economic development. Salt stress (SS) suppresses the protein activities and metabolism in plants leading to retarded plant growth and development [12]. Thus, it is important to understand the mechanism underlying salt tolerance in plants for sustainable agriculture [13]. In plants, SS can induce oxidative stress (OS), osmotic stress, and ion toxicity, leading to cell dehydration, reduced turgor pressure, impaired photosynthesis, and disrupted biochemical processes [14–16]. To survive under SS conditions, plant cells perceive SS and initiate various changes that include (1) changes in regulatory and signaling pathways [17–19] by changing the hormonal levels [20,21]; (2) activating certain transcription factors (TFs) associated with SS response [22]; and (3) regulating the expression of genes that encodes the proteins associated with cell protection, osmoregulation, or acclimation [23–26]. However, different plant species exhibit different adaptive mechanisms to withstand salt stress. Therefore, comprehensive investigations of the transcriptome of major halophytes are necessary for a better understanding of the mechanism involved in salt tolerance. The sequencing studies to understand plant responses to salt stress are mainly focused on model plants and crops, however, very less information is available for tree species of economic importance such as paper mulberry. Considering this, we have planned this study to understand the mechanism of salt tolerance in paper mulberry and the genes involved in salt tolerance.
Although several studies have investigated the salt tolerance mechanism in plants, however the functional gene studies related to salt stress need to be focused particularly on the tree species. In the present study to elucidate the salt stress response mechanism of paper mulberry at the transcriptomic level, salt stress treatment was applied and gene expression profiling and transcriptome sequencing was performed using Illumina sequencing. The candidate genes related to plant development, hormones, and signaling, primary and secondary metabolism were identified. As a result, a comprehensive understanding of the genetic changes within Paper mulberry leaf tissues under salt stress conditions, and a deeper understanding of the molecular mechanisms governing the transcriptome response might help to discover the potential genes and improve salt tolerance.
2.1 Plant Growth Conditions and Treatments
A hybrid cultivar of Broussonetia kazinoki × B. papyifera [1,11] was obtained from the Institute of Botany, Chinese Academy of Sciences (116° 10′-116° 20′ E, 39° 55′-49° 5′ N) and used in the study. The seedlings with uniform growth were cultivated using the cuttings propagation technique which resulted in the plantlets with identical tissue-culture batches. The plants were cultivated for 1 month in a greenhouse (day natural light: about 5500 lx; day/night temperature: 25°C/20°C; relative humidity: 75%). Thereafter, stems with original length and diameter of 25–35 cm and 2–4 mm, respectively, were trimmed separately into 10 cm segments to simulate the field harvesting and trigger re-sprouting.
After growing for 2-week, healthy plants were divided into six groups and exposed to different salt concentration gradients. A half-strength modified Hoagland’s solution was used as a normal nutrient solution, NaCl was added to prepare 0, 50, 100, 200, 300, and 400 mmol/L solution that was coded as CK, Y50, Y100, Y200, Y300, and Y400, respectively. Based on the growth performance and previous experience, the CK, Y50, and Y300 were selected for further studies. The control, and moderate and high salt treatment groups were designated as samples CK, B1 and B2, respectively. For every group, there were three replicates (i.e., 1, 2 and 3) and 20 plants were used per replicate. The nutrient solution with the corresponding NaCl concentration was irrigated for 12 days to maintain the water content of the matrix at approximately 60% to 90%. Significant phenotypic differences were observed after 12 days of salt stress, which was followed by transcriptome and physiology analysis. In this study, perlite was used as pot media to grow the paper mulberry plants. All the other management practices were kept similar except NaCl application.
2.2 RNA Extraction, RNA-Seq Library Construction, and Illumina Sequencing
The leaf samples were collected from the salt-treated and control groups, and the plant RNA Kit (Aidlab China) was employed to extract the total RNA following the specific protocols. The quality of RNA samples was determined using Agilent 2100 Bioanalyzer. For quantification of extracted RNA, NanoDrop Technologies ND-2000 was used followed by the elimination of genomic DNA (gDNA) with Takara DNase I. Each experiment consisted of three biological replicates.
The RNA-Seq library was constructed using an Illumina TruSeq™ sample preparation kit (Illumina, San Diego, CA, USA), following the manufacturer’s instructions. To isolate polyA mRNA from total RNA, oligo (dT) was used, followed by fragmentation. Furthermore, a synthesis kit (Invitrogen, CA, USA) was utilized to synthesize the double-stranded cDNA, and the above-described fragments were utilized as templates. Then, agarose gel electrophoresis (AGE) was performed to select the target fragments, followed by amplification with NEB Phusion DNA polymerase. Finally, to process the eventual library, the Illumina HiSeq platform was employed. Each experiment consisted of three biological replicates.
2.3 The de novo Assembly and Sequence Annotation
The low-quality and adaptor reads were removed from the raw transcriptome data to obtain clean reads. Following preprocessing, the obtained clean reads based on SS-treatments (BP1, BP2) and control (CK) samples were subject to de novo assembly to acquire the unigenes with Trinity [27,28]. The BLASTX software was used to search all the unigenes for NCBI NR, SwissProt, COG, and KEGG public databases with an E-value of less than 1.0 × 10−5. The BLAST2GO (https://www.blast2go.com/) program [29], and KEGG database (http://www.genome.jp/kegg/) [5] provided the Gene Ontology (GO) terms and KEGG pathways enriched by those assembled unigenes.
2.4 The Analysis and Functional Enrichment of DEGs
To measure the transcript expression levels, fragments per kilobase of exon per million mapped fragments (FPKM) values were calculated. Additionally, RSEM (http://deweylab.biostat.wisc.edu/rsem/) was applied in calculating gene expression [30]. Further, the R package EdgeR (Empirical Analysis of Digital Gene Expression) software was used for differential analysis of transcript expression between SS-treatments and CK samples [31]. A significant difference was screened upon the thresholds of |log2FC (CT/CK)| ≥ 1 and false discovery rate (FDR) < 0.05. Then GO analysis on differentially expressed genes (DEGs) was conducted using KOBAS (http://kobas.cbi.pku.edu.cn/expression.php) and Goatools (https://github.com/tanghaibao/Goatools) [32]. Furthermore, the same method was used to analyze KEGG pathways enriched by those identified DEGs. The DEGs were considered to be significantly enriched when a Bonferroni-corrected p-value ≤ 0.05 was obtained.
The expression level of DEGs was measured by RT-qPCR to validate the transcriptomic data. The reaction was performed using specific primers designed by the Platinum SYBR Green qPCR Supermix-UDG (Invitrogen) and Light Cycler 480 (Roche Diagnostics, Germany) using the specific protocols. Three technical replicates were set for each sample, while the amplification specificity was verified based on the dissociation curves. The 2−ΔΔCT method [33] was used for calculating the relative expression of the genes, and EF1 was used as the reference gene [25,19] (Supplementary Table S3).
3.1 Response of Paper Mulberry to Salt Stress
Paper mulberry plants showed visible morphological changes in their growth and development. The paper mulberry plants where NaCl was applied had resulted in drying and drooping of leaves. Additionally, the entire plant showed apparent phenotypic damage as the duration of SS treatment was increased. In response to 400 mmol/L NaCl stress, plants exhibited severe leaf wilting, along with drying of entire leaves or leaf margins (Fig. 1F). Furthermore, complete leaf withering was observed on the 12-day of SS treatment at 400 mmol/L NaCl stress (Fig. 1F).
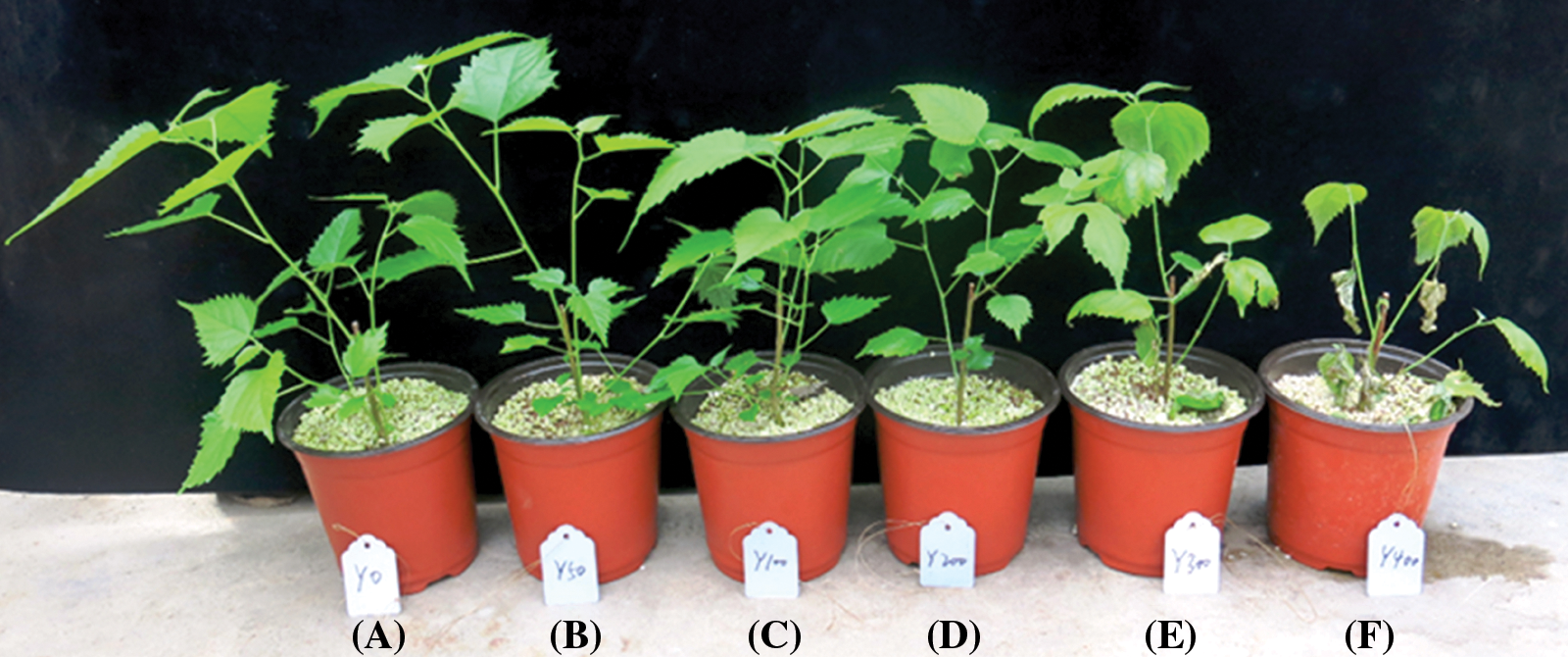
Figure 1: The response of paper mulberry plants grown under different levels of NaCl stress. (A) 0 mmol/L NaCl; (B) 50 mmol/L NaCl; (C) 100 mmol/L NaCl; (D) 200 mmol/L NaCl; (E) 300 mmol/L NaCl; (F) 400 mmol/L NaCl. Pictures were taken after 12 days of SS treatment
3.2 mRNA Sequencing and de novo Transcriptome Assembly
In this study, total leaf RNA was isolated from SS-treated (NaCl) and CK (control) paper mulberry plants. The Illumina platform was employed to perform the RNA-sequencing (RNA-seq) of three replicates in SS-treated and CK groups that were named B1–1, 2, 3, B2–1, 2, 3, and CK-1, 2, 3, respectively. For the control CK samples, 7.76, 8.64, and 7.93 Gb clean data was obtained (Table 1), whereas, from SS-treated samples, 8.31, 7.84, 8.20, and 7.10, 8.77, and 8.49 Gb clean data was obtained (Table 1). Among those nine samples, GC concentration was around 47.51%. Similar data were obtained from the biological replicates (Table 1). Further, the clean transcriptomic data were assembled with the Trinity program for these nine samples [28]. A total of 45,550 transcripts were obtained from the initial assembly, and the N50 was 2,243 bp. The mean length of these transcripts was 1,200 bp, along with the longest one being 17,035 bp (Table 2 and Supplementary Table S1). Meanwhile, a total of 35,068 unigenes (N50 = 2261) were obtained collectively, with the length ranging from 201 to 17,035 bp and the average size being 1,055 bp (Table 2).
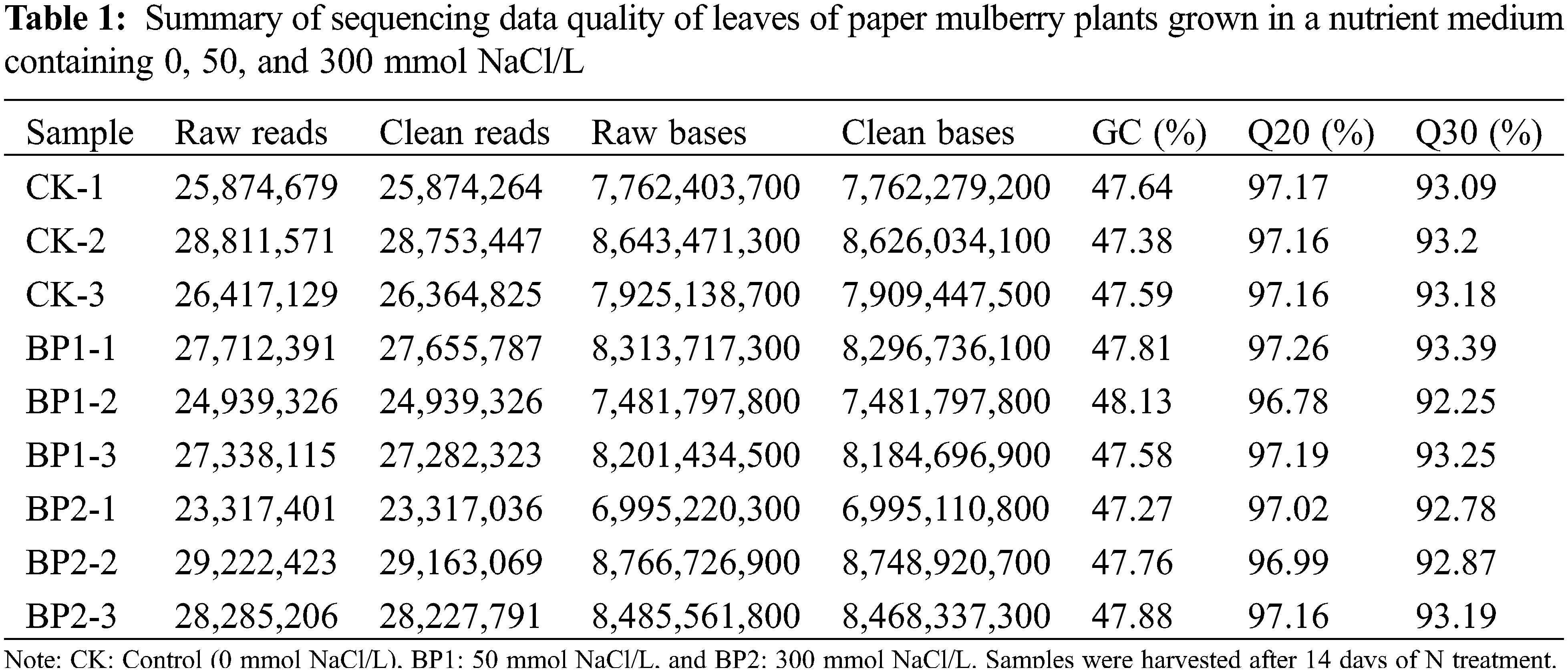
As illustrated in Fig. 2a, many unigenes (35,068; 93.25%) had a length of <3000 bp. In general, the number of unigene reduced with an increase in gene length. Among the obtained unigenes, most have a length of 1,000–2,000 bp (17,035) which suggested a high-quality assembly. All the raw sequence reads were imported into the NCBI short read archive (SRA) (BioProject accession number, PRJNA516316).
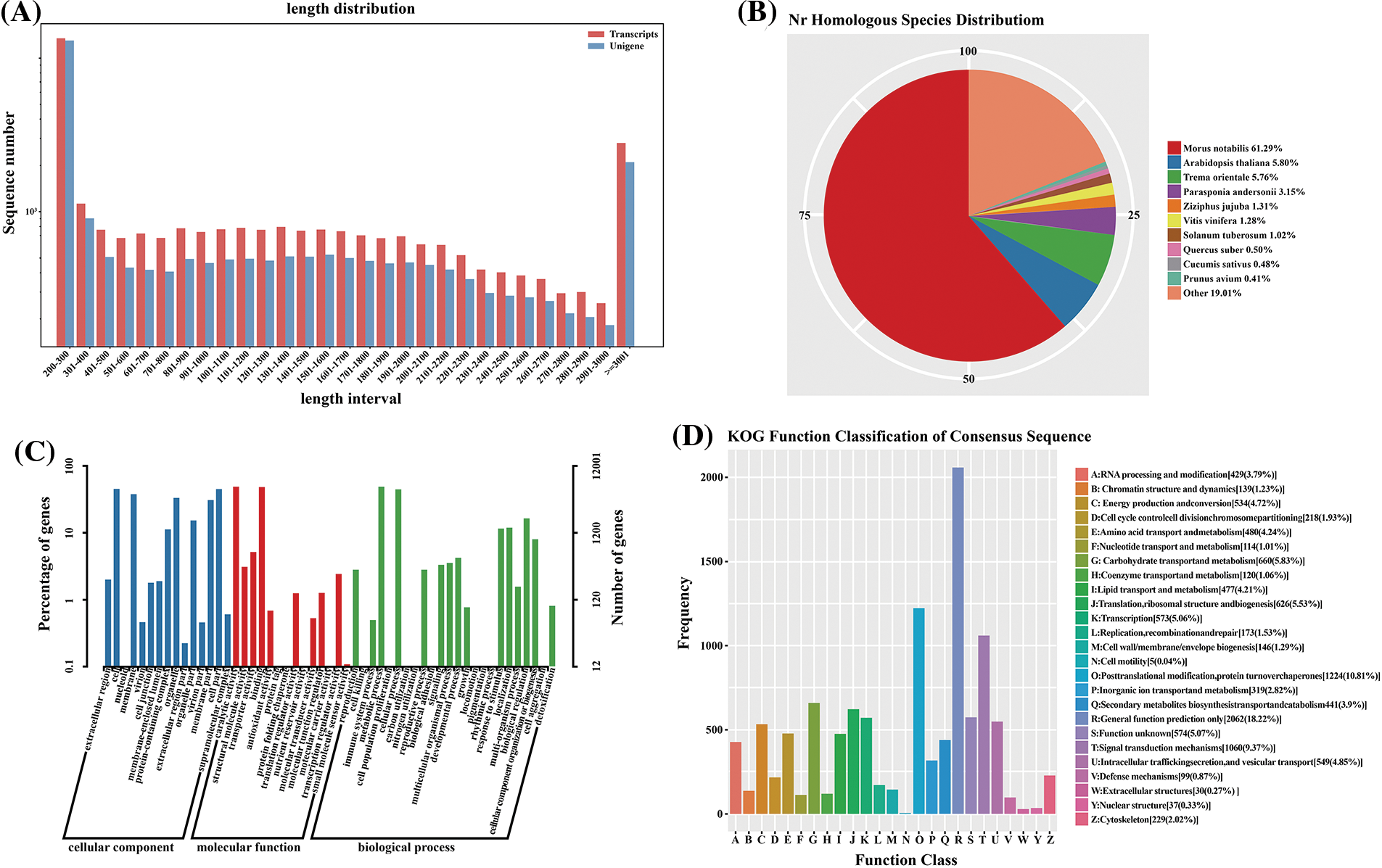
Figure 2: Annotation and classification of the paper mulberry transcriptome unigenes grown in a nutrient medium containing 0, 50, and 300 mmol NaCl/L
3.3 Annotation and Classification of Paper Mulberry Transcriptome Unigenes
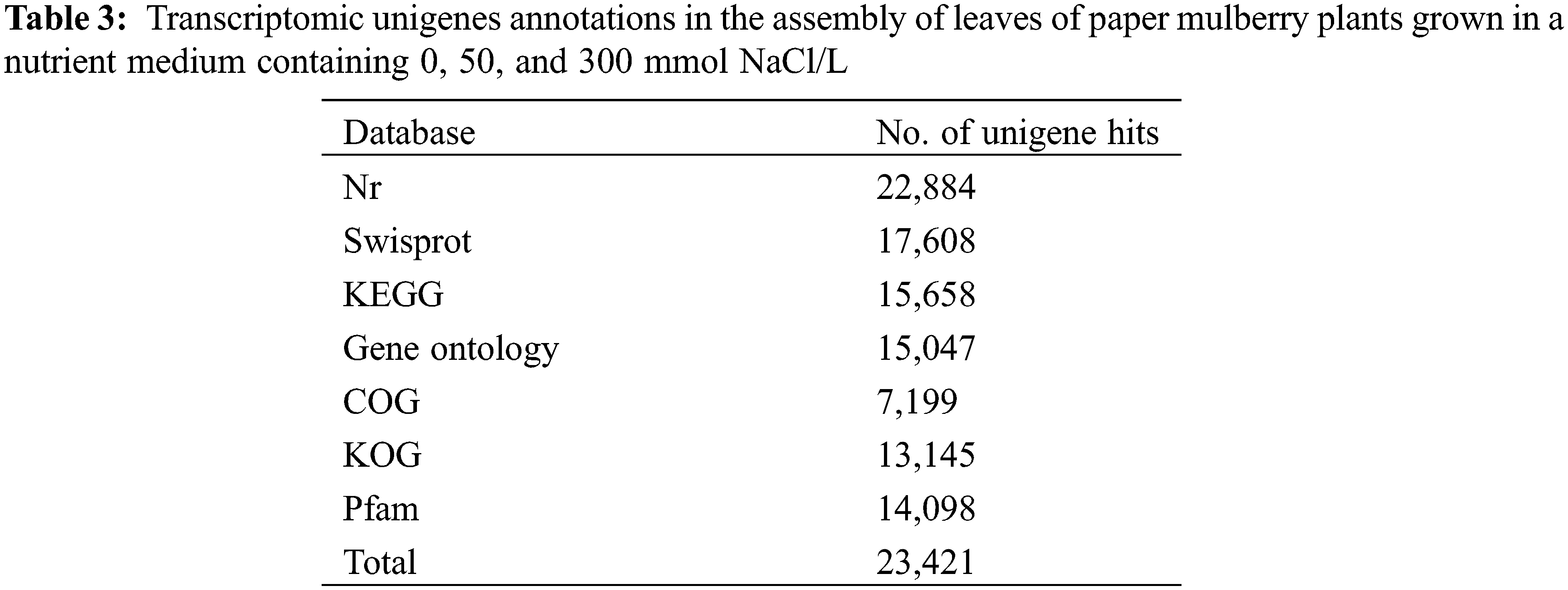
To identify the functions of unigene in paper mulberry, the multiple complimentary annotation approaches were used, and the assembled unigenes were searched in SwissProt, NCBI non-redundant (Nr), GO, KEGG, KOG, COG, and Pfam databases using BLASTX algorithm, and the E-value was set at <1.0 × 10−5. Hence, the known proteins were identified in one or more databases corresponding to 23,421 unigenes (Fig. 2a, Table 3, Supplementary Table S5, and Supplementary Fig. S1). Among these unigenes, 15,047 showed apparent hits (E-value <1e−5) within the GO database, whereas 22,884 were within the Nr database. Thus, the known proteins were identified for 17,608, 7,199, and 15,658 unigenes in SwissProt, COG, and KEGG databases, respectively, whereas 11,647 unigenes remained unmatched with sequences from 7 databases.
A BLASTX similarity search analysis was performed based on the Nr database, and the E-value was set to <1e−5. The greatest annotation proportion (n = 22,884 unigenes) was observed in the Nr database compared with the remaining six databases. Altogether, these unigene sequences showed the closest BLASTx matching against gene sequences to Morus notabilis (57.39%), Trema orientale (5.71%), Arabidopsis thaliana (5.41%), and Parasponia andersonii (3.17%) (Fig. 2b). The paper’s mulberry unigenes were similar to Morus notabilis genes, which indicated that Morus notabilis genomes and transcriptomes may be used as a reference in further research.
The GO analysis was conducted to annotate the unigene functions related to paper mulberry. A total of 15,047 unigenes belonging to 55 functional subgroups were classified collectively (Fig. 2c) and were divided later into 3 GO categories using BLAST2GO [34]. These unigenes were related to biological processes (BPs), molecular functions (MFs), and cellular components (CCs), which were useful for gene annotation. For BP terms, these genes were mainly enriched into the metabolic process (7.490%), cellular process (6.686%) as well as multi-organism-process (2.340%). Furthermore, those genes were mostly enriched into catalytic activity (7.446%), binding (7.234%) together, and transporter activity (8.090%), related to MF terms. The genes associated with CC terms were mainly ‘cell’ (6.656%), ‘cell part’ (6.620%), and ‘organelle’ (2.192%).
In addition, the COG database was searched to predict and classify the unigene functions related to paper mulberry. Under the assumption that every protein from the COG database exhibited independent evolution from a certain ancestral protein, 7,199 unigenes were divided into 25 COG classification subgroups (Fig. 2d). Among these subgroups, post-translational modification, protein turnover, and chaperones (6.480%) occupied the greatest percentage, translation, ribosomal structure, and biogenesis were 5.990%, signal transduction mechanisms were 1.790%, and general function prediction was only 1.108%. The remaining unigenes could be classified as function unknown (2.430%) category. Nonetheless, there were few clusters in nuclear structure (n = 2 unigenes) and cell motility (7 unigenes) categories, separately. Meanwhile, there was no unigene classified under the extracellular structures category.
Further, the KEGG protein database was used to categorize paper mulberry unigene functions with an emphasis on biochemical pathways. For identifying the aforementioned pathways related to paper mulberry, KEGG analysis was conducted for the identified unigenes. As a result, the identified 15,658 unigenes were enriched into 143 KEGG pathways (Supplementary Fig. S1 and Tables 3 and 4 and Supplementary Table S6). Among them, carbon metabolism, plant-pathogen interaction, starch and sucrose metabolism, ribosome and amino acid biosynthesis pathways were most significantly enriched. Thus, these findings indicate that SS tolerance of paper mulberry mainly depends on the metabolism of large amounts of metabolites and energy-related processes.
3.4 Differentially Expressed Genes (DEGs) in Salt-Treated Paper Mulberry Plants
The DEGs between SS-treated and CK samples were examined with de novo assembled transcriptome considered as reference. To comprehensively explore the levels of transcripts associated with SS tolerance of paper mulberry, DEGs were detected between SS-treated and CK paper mulberry samples. A total of 2,126 DEGs (selected upon |log2FC (CT/CK)| ≥ 1 and FDR < 0.05 thresholds) were detected between SS-treated and CK groups using edgeR software. Among these DEGs, 812 genes were up-regulated and 1,314 genes were down-regulated (Fig. 3) in the leaves of paper mulberry exposed to NaCl stress conditions (Supplementary Figs. S2 and S3).
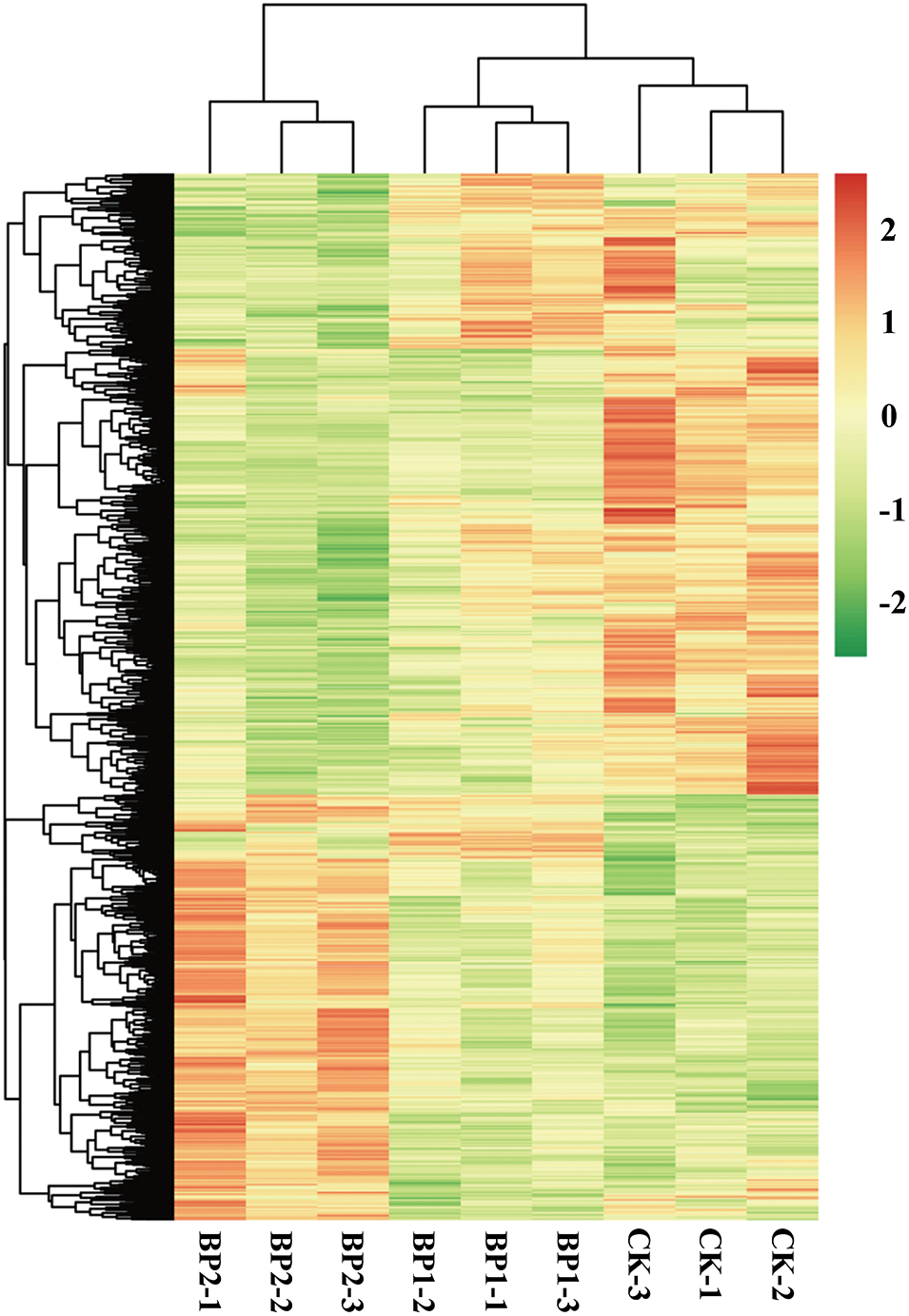
Figure 3: Hierarchical cluster analysis map presenting differential gene expression of the paper mulberry grown in a nutrient medium containing 0, 50, and 300 mmol NaCl/L. The control and salt-treated paper mulberry samples are represented as CK-1, 2, 3, B1–1, 2, 3, and B2–1, 2, 3. The leaf samples of paper mulberry were harvested after 12 days of NaCl treatment
3.5 The KEGG Pathway and GO Enrichment Analysis of DEGs
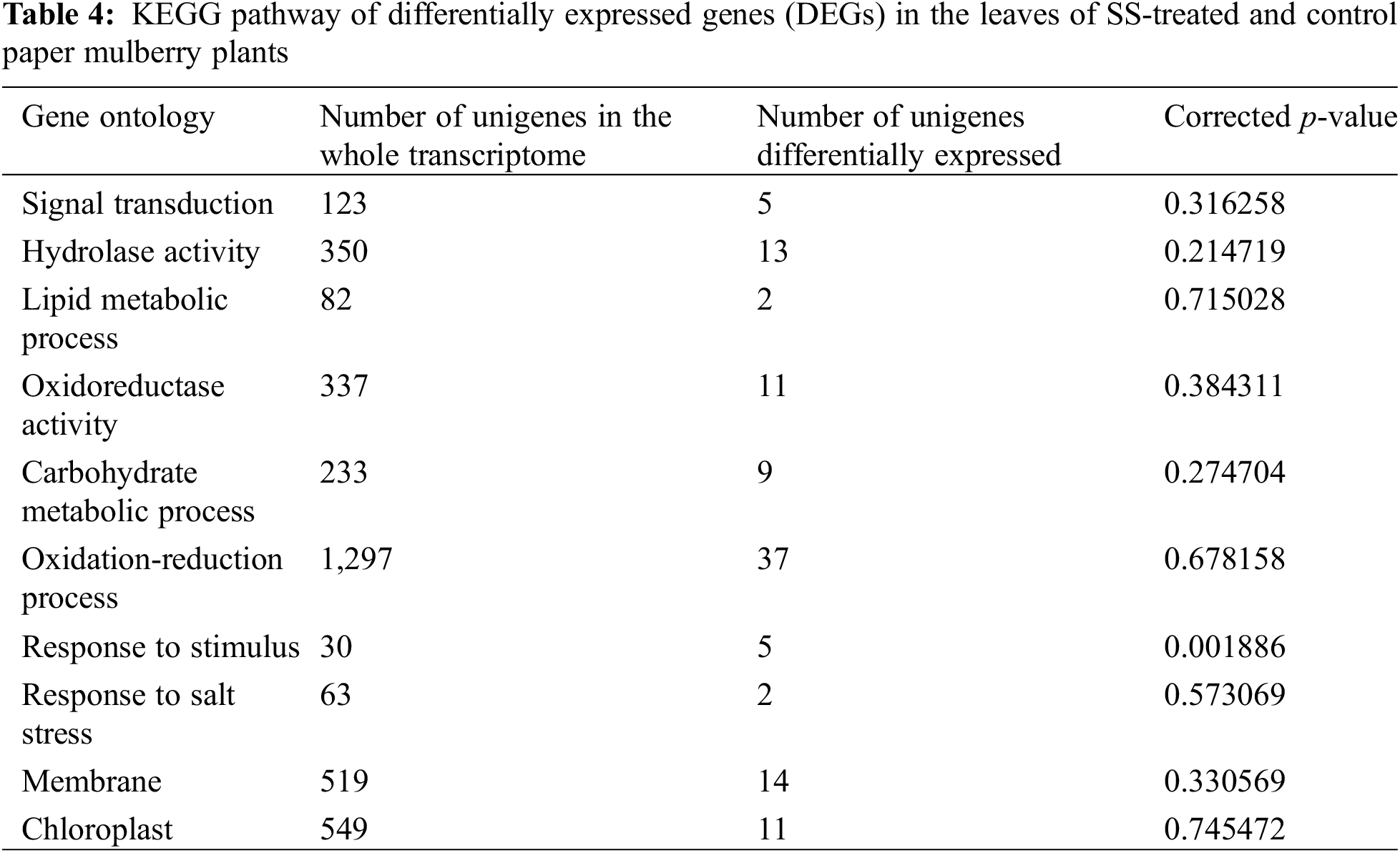
The KEGG database was used to identify the significantly changed pathways (p ≤ 0.05) in the leaves of paper mulberry exposed to SS-treatment. As a result, 15 significant KEGG pathways were discovered (Table 4), including plant-pathogen interaction, plant hormone signal transduction, fatty acid metabolism, starch and sucrose metabolism, and MAPK pathway, which displayed the highest significance.
Most DEGs were enriched into the plant hormone signal transduction pathway (ko04075), which suggests that plant hormones have critical functions in resistance against SS in Paper mulberry. In addition, the plant-pathogen interaction pathway (ko04626) suggests the vulnerability of SS-challenged plants to pathogens. However, based on the MAPK pathway (ko04013), endogenous gene levels within SS-challenged plants are modulated via diverse signaling molecules to adapt to the SS environment. The fatty acid metabolism (ko01212), starch, and sucrose metabolism (ko00500) pathways were significantly enriched, since SS exposure results in altered plant metabolism for improving salt tolerance [35].
To explore the functions of DEGs under SS conditions, GO analysis was performed using Goatools (p ≤ 0.05 by Fisher exact test). The results showed that DEGs were mostly associated with signal transduction, lipid metabolic process, hydrolase activity, carbohydrate metabolic process, oxidoreductase activity, response to stimulus, oxidation-reduction process, chloroplast, single-organism metabolic process, and membranes (Table 5). These findings suggest that DGEs associated with these pathways may also be important for the paper mulberry plants to adapt under NaCl stress conditions.
3.6 Identification of Transcription Factors (TFs), Plant Hormone and Signal Transduction Related Unigenes Involved in Response to Salt Stress
Transcription factors have a major role in the regulation stress-responsive genes. According to a report, TFs have a critical impact on salt stress signaling and trigger diverse downstream transcripts [12]. Based on the Arabidopsis database-predicted TFs belonging to 81 families, several genes encoding TFs belonging to 15 families were detected and were differentially expressed in response to NaCl stress in the leaves of paper mulberry (Supplementary Table S2–S4). The major transcription factor families expressed in this study include bHLH, MYB, NAC, bZIP, and WRKY.
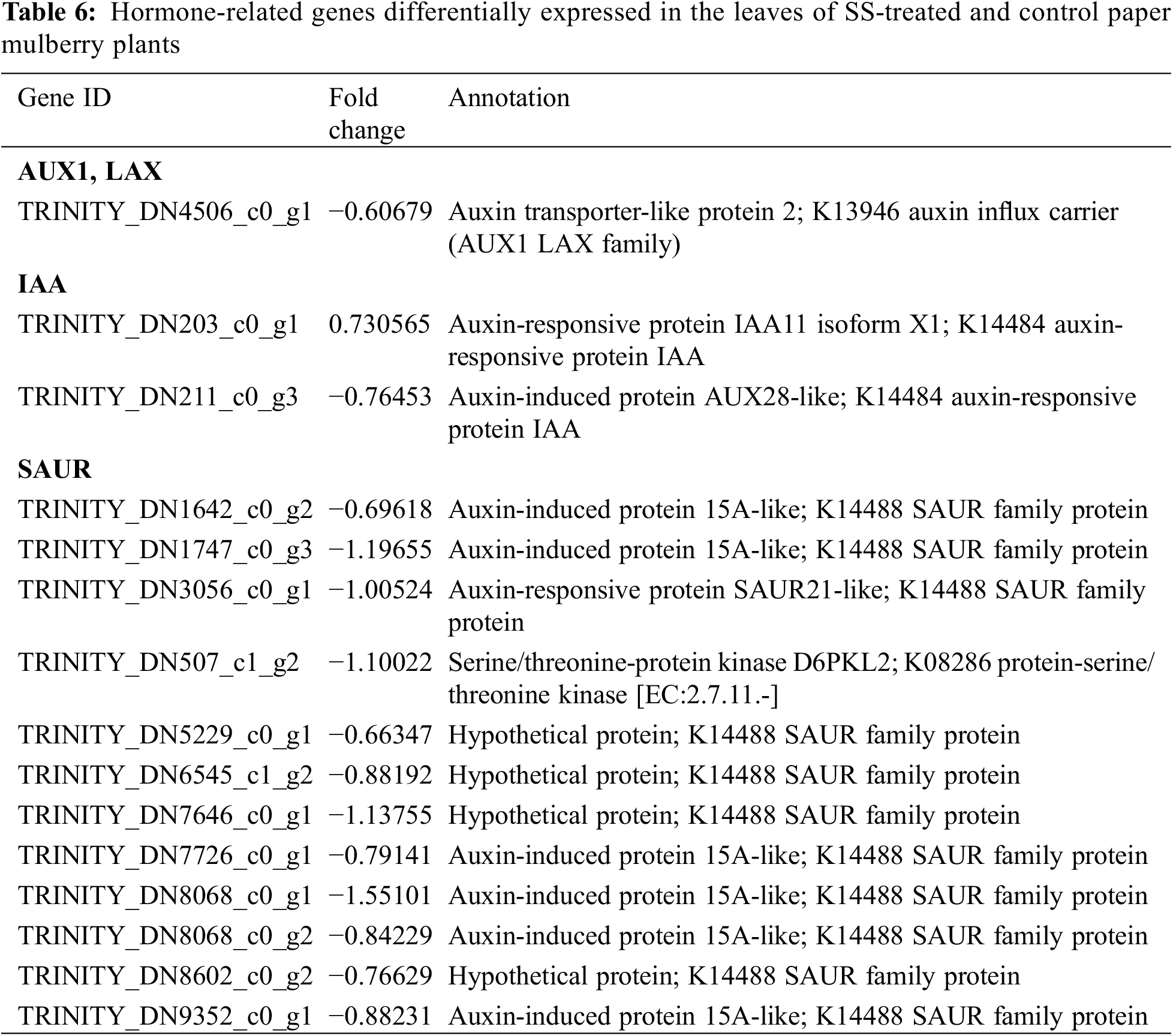
In plants, auxin (indole-3-acetic acid) plays a critical role in growth and development processes such as cell division, leaf expansion, lateral root initiation, and response to stress [36]. According to the results of this study 16 genes related to the auxin pathway were detected and differentially expressed (Table 6); among these one gene encoded the auxin transporter-like protein (AUX), 2 were auxin-responsive GH3-like protein (GH3) genes, 4 were auxin-responsive factors (ARF), and 6 were auxin-induced protein IAA6 (AUX/IAA) genes. These genes were up-regulated under salt stress conditions in the leaves of paper mulberry plants. Moreover, 3 unigenes were also discovered that encoded SAUR; among them, 2 were up-regulated, whereas 1 was down-regulated. These observations suggest that the resistance of paper mulberry to salt stress may be associated with the auxin signaling pathways.
Kinases were previously reported to have critical functions in signaling that is achieved by protein phosphorylation to relay signals [37,1]. In SS-treated paper mulberry plants, 117 genes encode protein kinases that showed diverse expression patterns (Table 4). Additionally, based on the results of KEGG analysis it was observed that SS-treatment induced MAPK pathway (ko04013). Therefore, the abundant MAPK signaling-related genes that were identified from the paper mulberry transcriptome are potential candidate genes and seem to be involved in salt stress adaptation.
3.7 Expression of Genes Involved in Metabolism and Biosynthesis
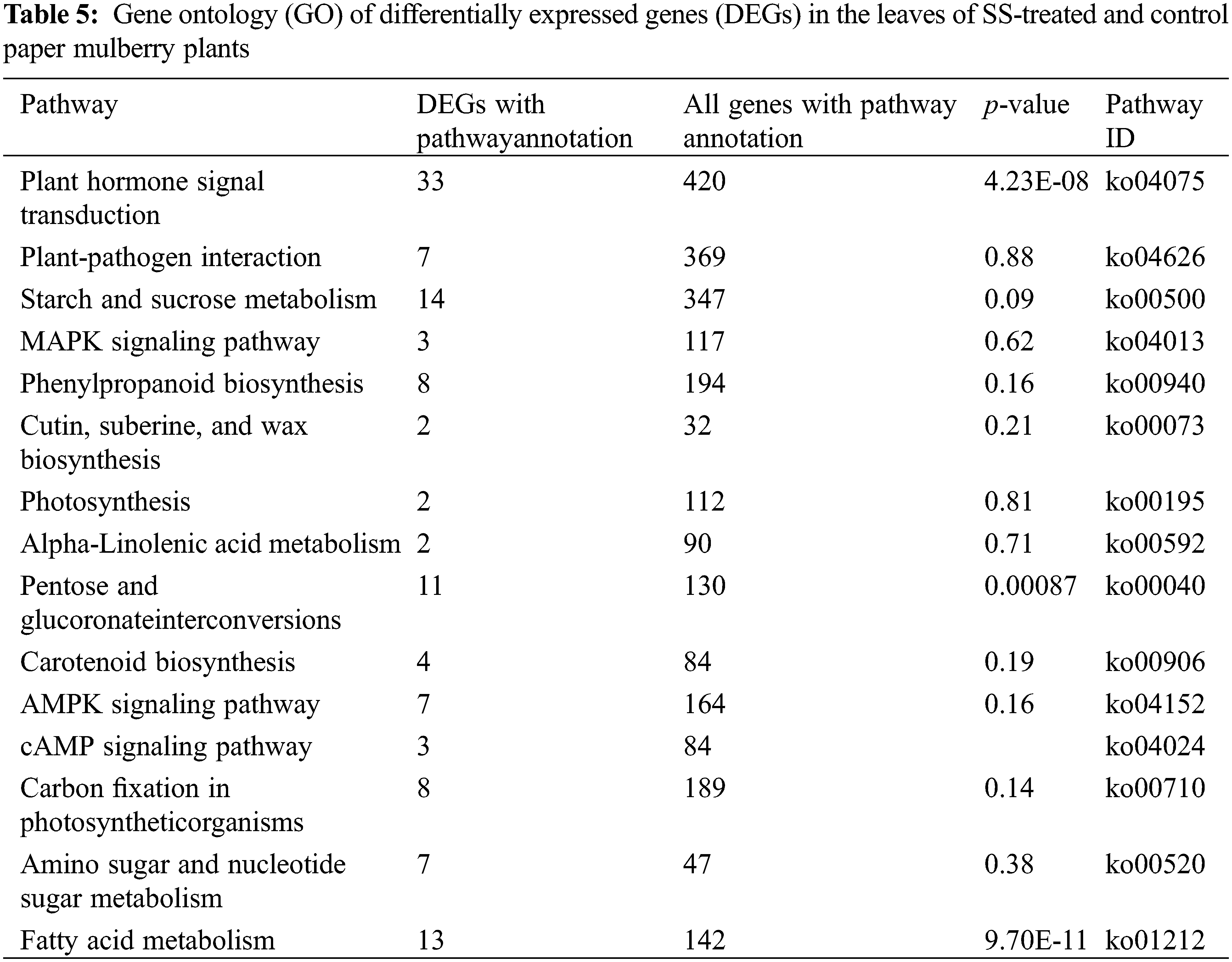
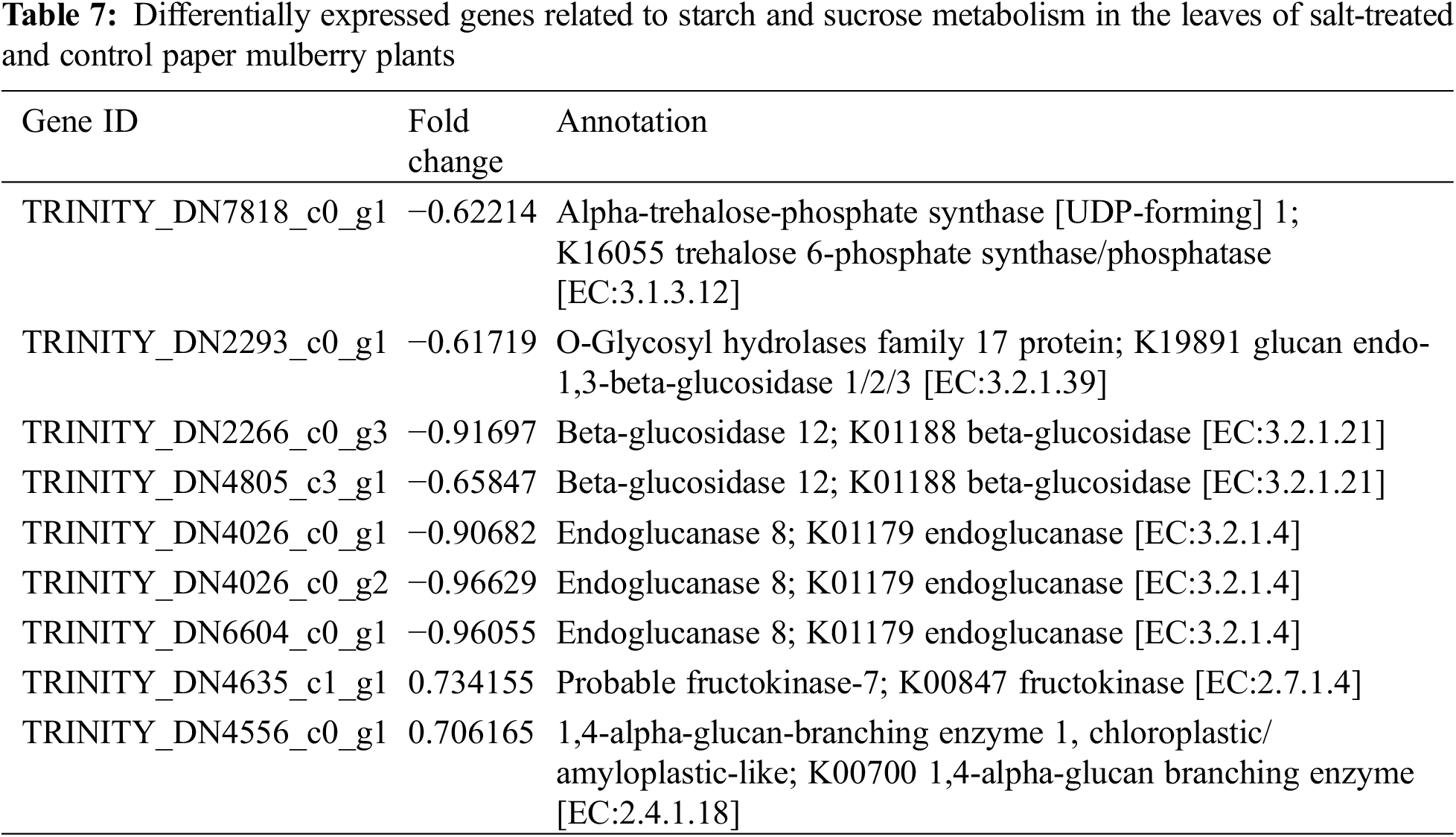
The metabolic processes in plants are changed under salt-stress conditions. According to the result of KEGG analysis, most of the DEGs were related to biosynthesis and metabolism. In this study, the changes in the secondary metabolites were observed when paper mulberry plants were exposed to salt stress; these changes include alteration of flavonoid biosynthesis (ko00941) and phenylpropanoid biosynthesis (ko00940) pathways. Overall, 2 pathways were mainly enriched by nine DEGs (Table 7) under salt stress conditions. Specifically, 2 DEGs that encoded flavonol synthase were enriched in the flavonoid biosynthesis pathway. The expression of one of these was up-regulated and one was down-regulated. Additionally, several genes enriched in the phenylpropanoid biosynthesis pathway showed down-regulation of expression. However, the mechanism of paper mulberry resistance to salt stress had a complicated process and involves several metabolic pathways. Therefore, it is difficult to reach conclusions only based on transcriptome data analysis because this clarifies part of the entire process. Thus, further precise molecular biology, proteomics, and genomic studies are required to clearly explain and verify the relevant results.
3.8 Validation of RNA-Seq-Based DEGs in Salt-Treated Paper Mulberry Plants by qRT-PCR
To validate the reproducibility and accuracy of RNA-Seq data, transcript levels were analyzed for 15 selected DEGs by qRT-PCR. According to the RNA-Seq data, out of these 15 unigenes, the expression of 14 unigenes was up-regulated and one unigenes was down-regulated. The results showed that salt stress-induced changes in the expression of tested genes in the leaf of paper mulberry were similar for RNA-Seq and qRT-PCR data. Thus it was concluded that the qRT-PCR analysis was consistent with RNA-Seq analysis and this confirmed the reliability of RNA-Seq data (Fig. 4, Supplementary Fig. S4).
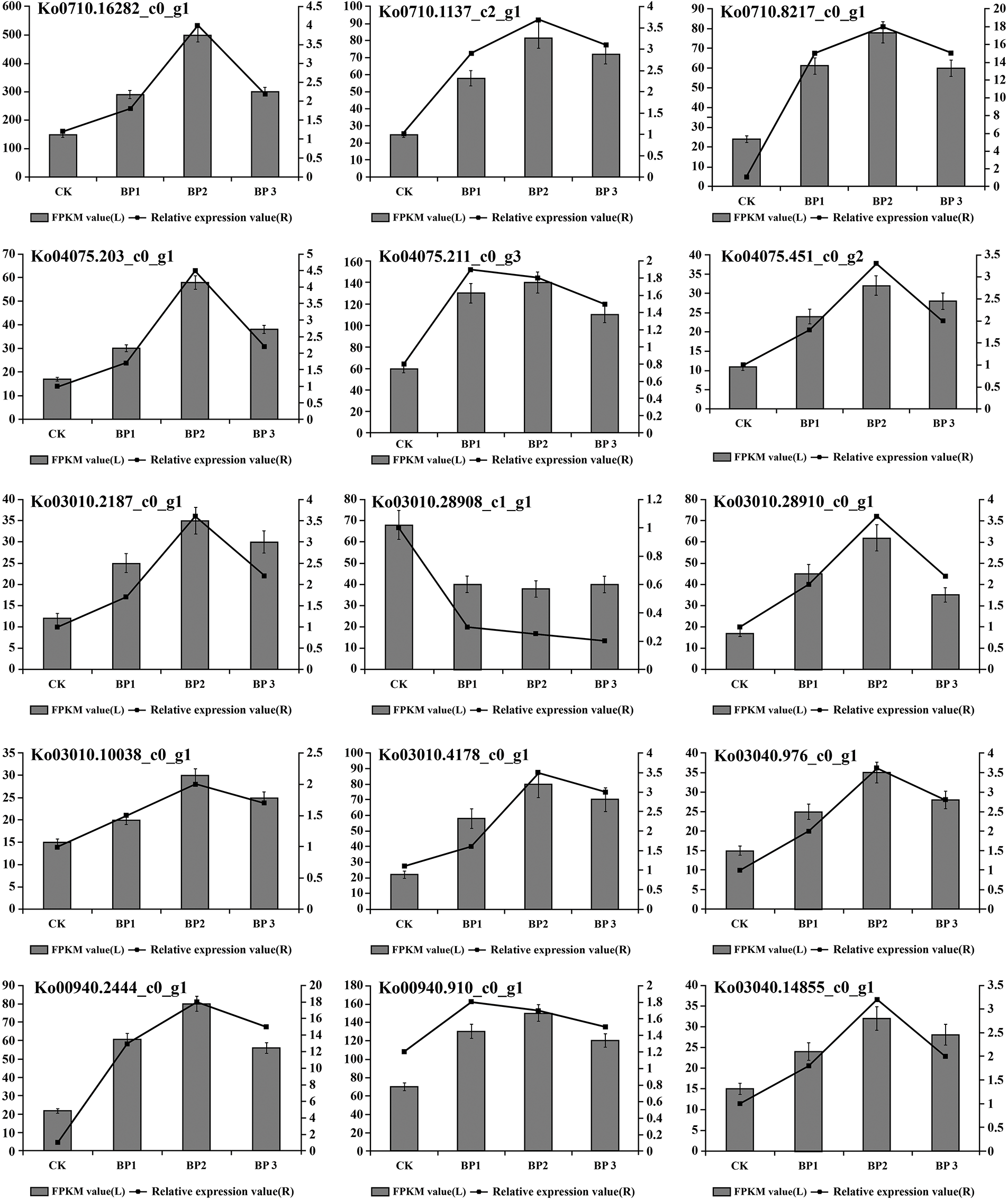
Figure 4: The gene expression changes in the leaves of paper mulberry exposed to different levels of NaCl stress. The samples were harvested after 12 days of NaCl treatment. The numbers on the left side of Y axes of each part of figure represent fragments per kilobase of transcript per million mapped reads (FPKM) values and the numbers on the right side of Y axes of each part of figure represent relative gene expression values
Salinity is an important concern across the world that is affecting plant growth and development from crops and tree species. In this study, transcriptome sequencing was performed on paper mulberry plants that were exposed to different concentrations of NaCl. The sequencing provided more than 73.04 Gb of high-quality clean data and 35,068 unigenes with N50 of 2,261 bp, and an average length of 1,055 bp. The obtained results indicated the high-quality assembly and might contain numerous full-length cDNAs. There were 66.79% homologous genes in the assembled unigenes from one or more publicly accessible databases, with homolog being observed in 65.26% of sequences, as detected by the great probability score from any one of these 4 species namely Morus notabilis (57.39%), Trema orientale (5.71%), Arabidopsis thaliana (5.41%) and Parasponia andersonii (3.17%). Under salt stress conditions, plants improve their salt tolerance by regulating (inducing or inhibiting) gene expression [38]. In this study, 2,126 DEGs were identified in the transcriptome sequencing of paper mulberry seedlings treated with different salt concentrations.
Phytohormones have critical functions in plants and they help improve stress tolerance by regulating different signaling pathways under abiotic stress conditions [39]. For instance, upon environmental stresses, endogenous IAA content is quickly increased in plants, thereby modifying the gene expression levels and activating certain important pathways [40]. In the present study, under salt stress conditions, the gene expression related to several plant hormones involved in signal transduction pathways was changed; among those 16 genes associated with auxin pathway showed significant alterations (Table 6).
According to a report, salt stress mainly targets intracellular auxin transport; however, it does not block each auxin transport pathway [33]. SIZ1, an important factor regulating the salt signal transduction pathway, is responsible for stabilizing ICE1 under salt stress by suppressing the poly-ubiquitination [34]. However, it negatively affects the phosphate starving-mediated root structural remodeling by regulating the auxin biosynthesis and transport [34]. Therefore, further studies are required to determine the role and mechanism of auxin-mediated signaling in response to salt stress in paper mulberry.
In plants, transcription factors (TFs) are involved in abiotic stress responses through different transcription mechanisms [41]. TFs of bHLH, bZIP, GRAS, HSF, WRKY, MYB, NAC, and MYC families modulate the expression of a different gene by coordination of signal transduction or interaction with cis-acting elements in stress-induced gene promoters [37,42]. In our study, salt stress treatment increased the expression of several predicted TFs (Supplementary Table S2–S4). According to a report, AtbZIP62 was involved in the negative regulation of the transcriptional events of SOS pathway genes (AtbZIP18 and AtbZIP69) and modulated the antioxidants responses in Arabidopsis [43]. Although it has been confirmed that these TF family members have significant effects on the salt tolerance of plants [1], however, they synergistic or antagonistic effect on salt tolerance mechanisms of paper mulberry remains unclear and this requires further investigations. Interestingly the transcription factors have got a great significance, and the breeder is targeting transcription factors involved in salt stress signaling pathways to improve the salt stress tolerance in plants [44–46].
Apart from the activation of primary mechanisms in plants that includes lipid metabolism, amino acid biosynthesis, and glucose accumulation; plants also employ secondary mechanisms. For instance, the biosynthesis of secondary metabolites in response to salt stress. In this study, under salt stress conditions, the flavonoid and phenylpropanoid biosynthetic pathways were involved in adaptation. This is evident from the identification of nine genes that were associated with flavonoid synthesis (Table 7). Previously a report suggested the involvement of flavonoid and phenylpropanoid pathways in salt stress tolerance [12] (Supplementary Fig. S5).
The present work provides a comprehensive analysis of the responses of paper mulberry under different salt concentration stress at the transcriptomic level. 812 DEGs were up-regulated and 1,314 DEGs were down-regulated in response to NaCl stress. The GO and KEGG analysis revealed that signaling pathways affected protein kinases and initiated the biosynthesis of different proteins in the downstream TFs family. TFs activation promoted the expression of downstream salt stress-responsive genes, and transcription of these genes regulated the secondary metabolites such as phenylpropanoid biosynthesis and flavonoid biosynthesis. Additionally, several genes were identified encoding proteins with unclear activities that seem specific to paper mulberry, and those genes have a critical role in salt stress signaling and adaptation mechanism.
Acknowledgement: We thank Bioeditas Technology Corporation-Shaanxi for providing valuable technical assistance.
Funding Statement: This work was supported by the National Natural Science Foundation of China (31600552).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Jianwei Ni; data collection: Jie Zhang; analysis and interpretation of results: Jie Zhang, Yingwei Zhao, Hongying Li; draft manuscript preparation: Jie Zhang, Dongmei Wang; editing: Hongying Li, Jianwei Ni. All authors have reviewed the results and approved the final version of the manuscript for submission.
Ethics Approval: Written informed consent was obtained from the individual for the publication of any potentially identifiable images or data included in this article. The studies involving human participants were reviewed and approved by the Northwest Agriculture and Forestry University’s Institutional Research Board. The participants provided their writteninformed consent to participate in this study.
Conflicts of Interest: The authors declare that they have no conflict of interest to report regarding the present study.
References
1. Peng, X., Teng, L., Wang, X., Wang, Y., Sheng, S. (2014). De novo assembly of expressed transcripts and global transcriptomic analysis from seedlings of the paper mulberry (Broussonetia kazinoki × Broussonetia papyifera). PLoS One, 9(9), e97487. [Google Scholar]
2. Saito, K., Linquist, B., Keobualapha, B., Shiraiwa, T., Horie, T. (2009). Broussonetia papyrifera (paper mulberryIts growth, yield, and potential as a fallow crop in slash-and-burn upland rice system of northern Laos. Agroforest Systems, 76, 525–532. [Google Scholar]
3. Bosu, P. P., Apetorgbor, M. M., Nkrumah, E. E., Bandoh, K. P. (2013). The impact of Broussonetia papyrifera (L.) Vent. On community characteristics in the forest and forest-savannah transition ecosystems of Ghana. African Journal of Ecology, 51(4), 528–535. [Google Scholar]
4. Li, B., Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [Google Scholar] [PubMed]
5. Kanehisa, M., Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28(1), 27–30. [Google Scholar] [PubMed]
6. Nagpal, U. M. K., Bankar, A. V., Pawar, N. J., Kapadnis, B. P., Zinjarde, S. S. (2011). Equilibrium and kinetic studies on biosorption of heavy metals by leaf powder of paper mulberry (Broussonetia papyrifera). Water, Air, & Soil Pollution, 215, 177–188. [Google Scholar]
7. Mei, R., Wang, Y., Du, G., Liu, G., Zhang, L. et al. (2009). Antioxidant lignans from the fruits of Broussonetia papyrifera. Journal of Natural Products, 72(4), 621–625. [Google Scholar] [PubMed]
8. Peng, X., Liu, H., Chen, P., Tang, F., Hu, Y. et al. (2019). A chromosome-scale genome assembly of paper mulberry (Broussonetia papyrifera) provides new insights into its forage and papermaking usage. Molecular Plant, 12(5), 661–677. [Google Scholar] [PubMed]
9. Yao, L., Yang, H., Yoo, C. G., Meng, X., Li, M. et al. (2017). Adsorption of cellobiohydrolases I onto lignin fractions from dilute acid pretreated Broussonetia papyrifera. Bioresource Technology, 244, 957–962. [Google Scholar] [PubMed]
10. Zheng, Z., Cheng, K., Chao, J., Wu, J., Wang, M. (2008). Tyrosinase inhibitors from paper mulberry (Broussonetia papyrifera). Food Chemistry, 106(2), 529–535. [Google Scholar]
11. Xu, Z., Yang, G., Dong, M., Wu, L., Zhang, W. et al. (2018). The complete chloroplast genome of an economic and ecological plant, paper mulberry (Broussonetia kazinoki × Broussonetia papyifera). Mitochondrial DNA Part B, 3(1), 28–29. [Google Scholar]
12. Carillo, P., Annunziata, M. G., Pontecorvo, G., Fuggi, A., Woodrow, P. (2011). Salinity stress and salt tolerance. In: Shanker, A. K., Venkateswarlu, B. (Eds.Abiotic stress in plants-mechanisms and adaptations, pp. 21–38. UK: InTech. [Google Scholar]
13. Wu, H. J., Zhang, Z., Wang, J. Y., Oh, D. H., Dassanayake, M. et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proceedings of the National Academy of Sciences, 109(30), 12219–12224. [Google Scholar]
14. Blumwald, E. (2000). Sodium transport and salt tolerance in plants. Current Opinion in Cell Biology, 12, 431–434. [Google Scholar] [PubMed]
15. Møller, I. S., Tester, M. (2007). Salinity tolerance of Arabidopsis: A good model for cereals? Trends Plant Science, 12(4), 534–540. [Google Scholar]
16. Waditee, R., Hibino, T., Nakamura, T., Incharoensakdi, A., Takabe, T. (2002). Overexpression of a Na+/H+ antiporter confers salt tolerance on a freshwater cyanobacterium, making it capable of growth in seawater. Proceedings of the National Academy of Sciences of the United States of America, 99(6), 4109–4114. [Google Scholar] [PubMed]
17. Bohnert, H. J., Nelson, D. E., Jensen, R. G. (1995). Adaptations to environmental stresses. Plant Cell, 7(7), 1099–1111. [Google Scholar] [PubMed]
18. Langridge, P., Paltridge, N., Fincher, G. (2006). Functional genomics of abiotic stress tolerance in cereals. Briefings in Functional Genomics & Proteomics, 4(4), 343–354. [Google Scholar]
19. Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annual Review of Plant Biology, 59, 651–681. [Google Scholar] [PubMed]
20. Ma, Q., Dai, X., Xu, Y., Guo, J., Liu, Y. et al. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiology, 150(1), 244–256. [Google Scholar] [PubMed]
21. Xu, Z. S., Xia, L. Q., Chen, M., Cheng, X. G., Zhang, R. Y. et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Molecular Biology, 65, 719–732. [Google Scholar] [PubMed]
22. Urano, K., Kurihara, Y., Seki, M., Shinozaki, K. (2010). Omics’ analyses of regulatory networks in plant abiotic stress responses. Current Opinion in Plant Biology, 13, 132–138. [Google Scholar] [PubMed]
23. Close, T. J. (1996). Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiologia Plant, 97, 795–803. [Google Scholar]
24. Meiri, D., Breiman, A. (2009). Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant Journal, 59, 387–399. [Google Scholar]
25. Munns, R. (2005). Genes and salt tolerance: Bringing them together. New Phytologist, 167, 645–663. [Google Scholar] [PubMed]
26. Tyerman, S. D., Niemietz, C. M., Bramley, H. (2002). Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell and Environment, 25, 173–194. [Google Scholar]
27. Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A. et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29, 644–652. [Google Scholar] [PubMed]
28. Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D. et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nature Protocols, 8, 1494–1512. [Google Scholar] [PubMed]
29. Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M. et al. (2005). Blast2GO: A universal tool for annotation, visualization, and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [Google Scholar] [PubMed]
30. Li, M., Li, Y., Li, H., Wu, G. (2011). Overexpression of AtNHX5 improves tolerance to both salt and drought stress in Broussonetia papyrifera (L.) Vent. Tree Physiology, 31, 349–357. [Google Scholar] [PubMed]
31. Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [Google Scholar] [PubMed]
32. Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J. et al. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research, 39, 316–322. [Google Scholar]
33. Shibasaki, K., Uemura, M., Tsurumi, S., Rahman, A. (2009). Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. The Plant Cell, 21(12), 3823–3838. [Google Scholar] [PubMed]
34. Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V. et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. The Plant Cell, 19(4), 1403–1414. [Google Scholar] [PubMed]
35. Valluru, R., Vanden, E. W. (2008). Plant fructans in stress environments: Emerging concepts and future prospects. Journal of Experimental Botany, 59(11), 2905–2916. [Google Scholar] [PubMed]
36. Gallavotti, A. (2013). The role of auxin in shaping shoot architecture. Journal of Experimental Botany, 64(9), 2593–2608. [Google Scholar] [PubMed]
37. Liu, J. H., Peng, T., Dai, W. (2014). Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Molecular Biology Reporter, 32(2), 303–317. [Google Scholar]
38. Kusunoki, K., Nakano, Y., Tanaka, K., Sakata, Y., Koyama, H. et al. (2017). Transcriptomic variation among six Arabidopsis thaliana accessions identified several novel genes controlling aluminum tolerance. Plant, Cell & Environment, 40(2), 249–263. [Google Scholar]
39. Tsukagoshi, H. (2016). Control of root growth and development by reactive oxygen species. Current Opinion in Plant Biology, 29, 57–63. [Google Scholar] [PubMed]
40. Hamel, L. P., Nicole, M. C., Sritubtim, S., Morency, M. J., Ellis, M. et al. (2006). Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends in Plant Science, 11, 192–198. [Google Scholar] [PubMed]
41. Lata, C., Yadav, A., Prasad, M. (2011). Role of plant transcription factors in abiotic stress tolerance. In: Shanker, A., Venkateshwarulu, B. (Eds.Abiotic stress response in plants-physiological, biochemical and genetic perspectives, pp. 269–296. UK: InTech. [Google Scholar]
42. Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 1819, 97–103. [Google Scholar] [PubMed]
43. Rolly, N. K., Imran, Q. M., Lee, I. J., Yun, B. W. (2020). Salinity stress-mediated suppression of expression of salt overly sensitive signaling pathway genes suggests negative regulation by AtbZIP62 transcription factor in Arabidopsis thaliana. International Journal of Molecular Sciences, 21(5), 1726. [Google Scholar] [PubMed]
44. Fernando, V. C. D. (2020). Major transcription factor families involved in salinity stress tolerance in plants. In: Wani, S. H. (Ed.Transcription factors for abiotic stress tolerance in plants, pp. 99–109. USA: Academic Press. [Google Scholar]
45. Nawaz, M. A., Chen, C., Shireen, F., Zheng, Z., Sohail, H. et al. (2018). Genome-wide expression profiling of leaves and roots of watermelon in response to low nitrogen. BMC Genomics, 19(1), 456. [Google Scholar] [PubMed]
46. Zhang, X., Long, Y., Chen, X., Zhang, B., Xin, Y. et al. (2021). A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biology, 21(1), 546. [Google Scholar] [PubMed]
Supplementary Materials
Table S1: Details of the nutrient solution used for growing paper mulberry in this study
Table S2: List of parameters and the software used for transcriptome data processing in this study
Table S3: List of genes chosen for validation and the primers used for qPCR
Table S4: Summary of sequences analysis of leaves of paper mulberry plants grown in a nutrient medium containing 0 mmol NaCl/L, 50 mmol NaCl/L, and 300 mmol NaCl/L
Table S5: Summary of unigene annotations of leaves of paper mulberry plants grown in a nutrient medium containing 0 mmol NaCl/L, 50 mmol NaCl/L, and 300 mmol NaCl/L
Table S6: Summary of unigene annotated in KEGG leaves of paper mulberry plants grown in a nutrient medium containing 0 mmol NaCl/L, 50 mmol NaCl/L, and 300 mmol NaCl/L
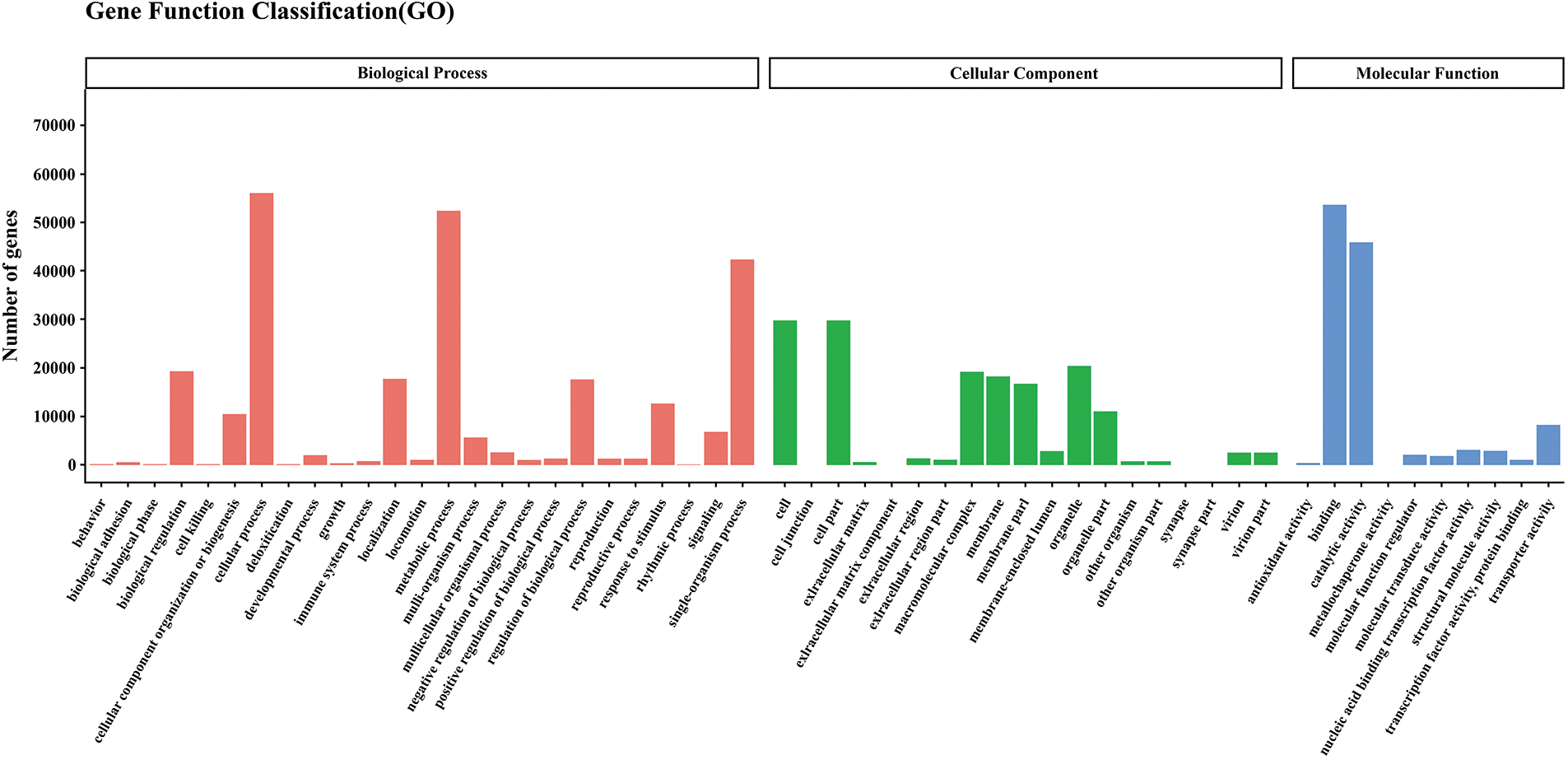
Figure S1: Gene ontology classification of assembled unigenes leaves of paper mulberry plants grown in a nutrient medium containing 0, 50, and 300 mmol NaCl/L. Total 94,099 unigenes were categorized into three main categories: biological process, cellular component, and molecular function
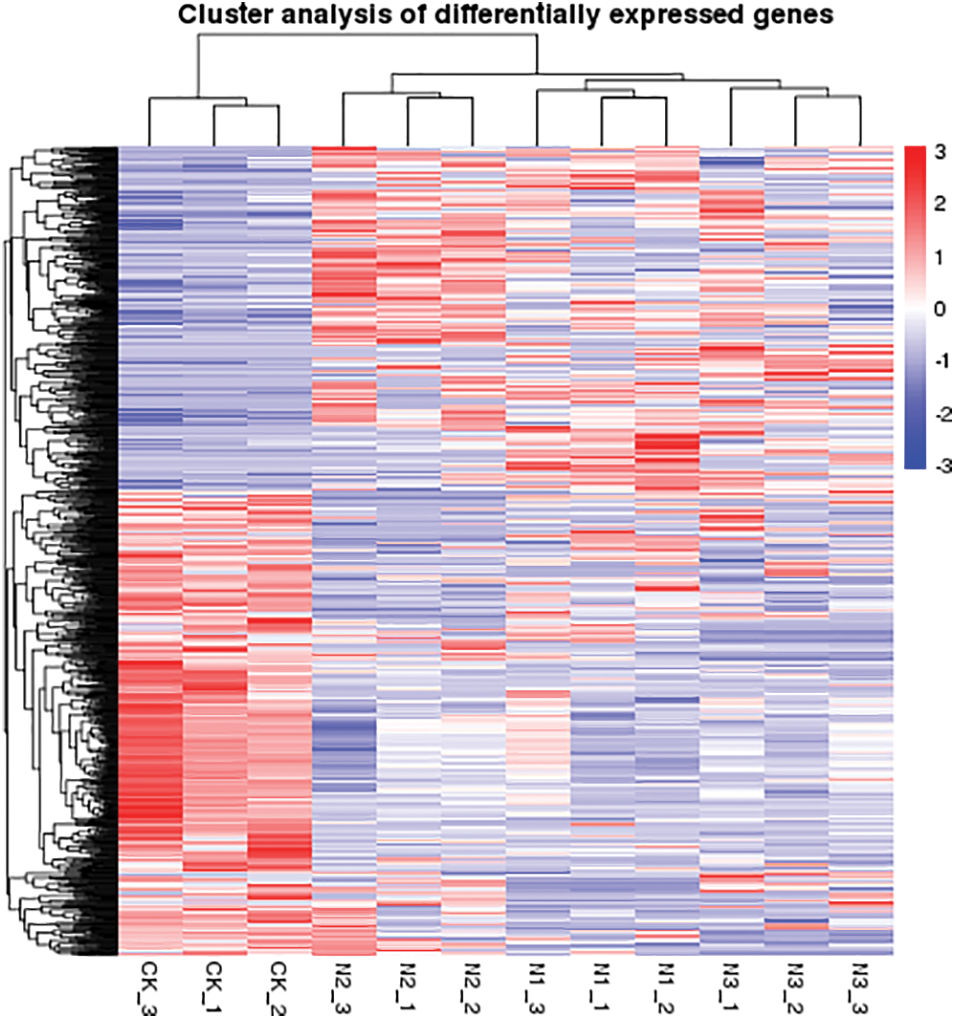
Figure S2: Heatmap of differentially expressed unigenes identified from the leaves of paper mulberry plants grown in a nutrient medium containing 0, 50, and 300 mmol NaCl/L. There were three treatments in this experiment and each group contain three replicates. Expression values were presented as fragments per kilobase of transcript per million mapped reads (FPKM) normalized log2 transformed counts. Red and blue colors indicate up- and down- regulated transcripts, respectively
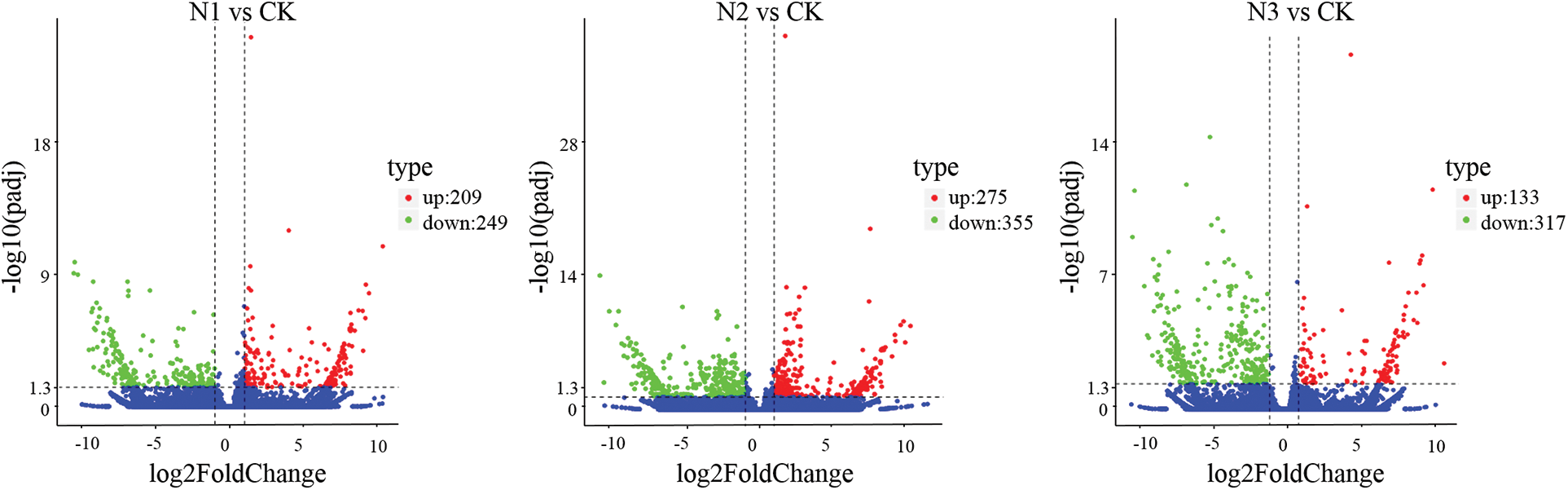
Figure S3: Volcano plot displaying differential expressed genes between N1 vs Ck, N2 vs CK and N3 vs CK sample. The y-axis corresponds to the mean expression value of log 10 (p value), and the x-axis displays the log2 fold change value. The red dots represent the up-regulated transcripts, the green dots represent the down-regulated transcripts
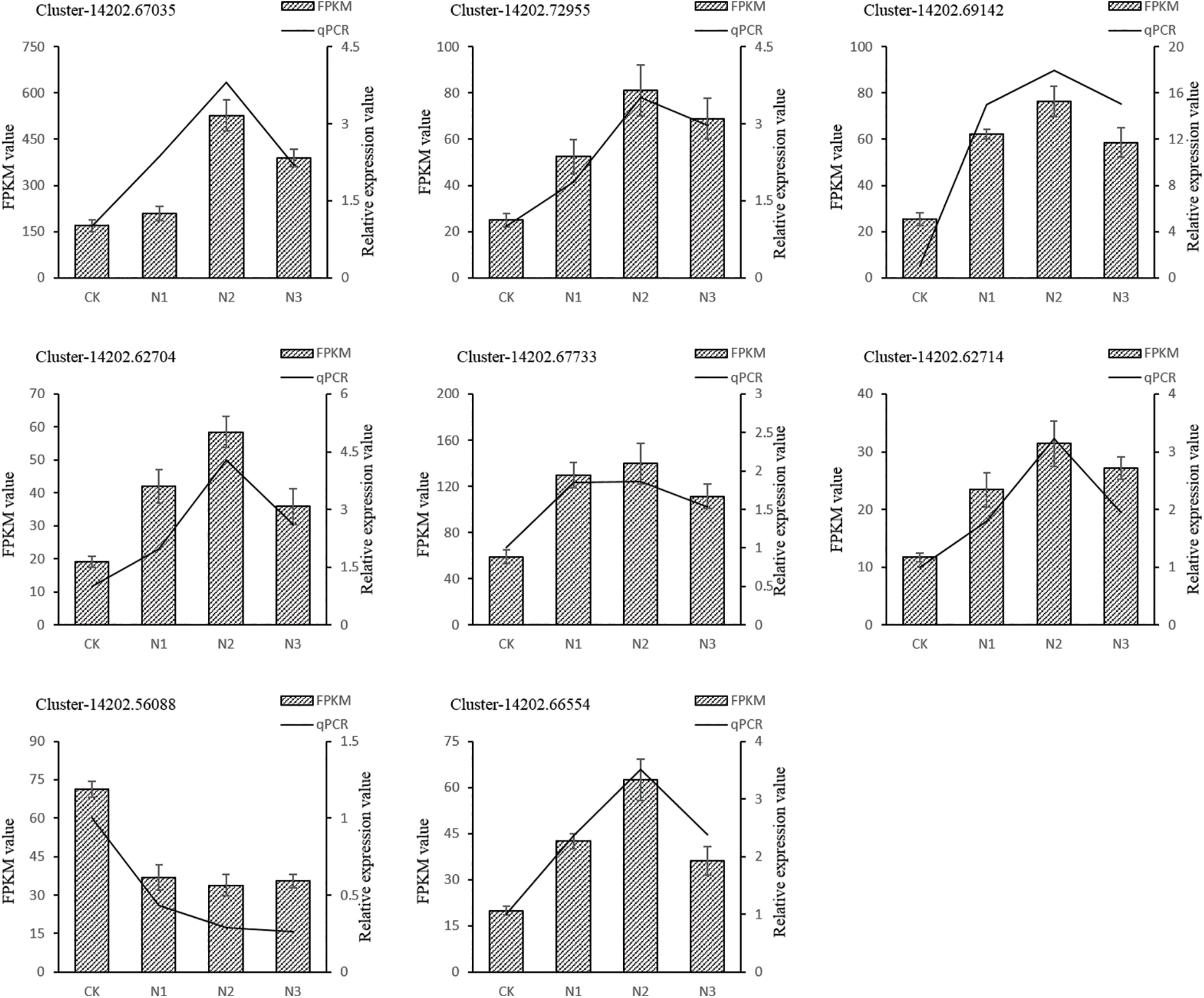
Figure S4: Quantitative realtime PCR validations of transcriptome result. Columns represent the means value of FPKM (Y axis at left). Lines represent the qPCR relative expression value (Y axis at right). Each experiment was performed in triplicate
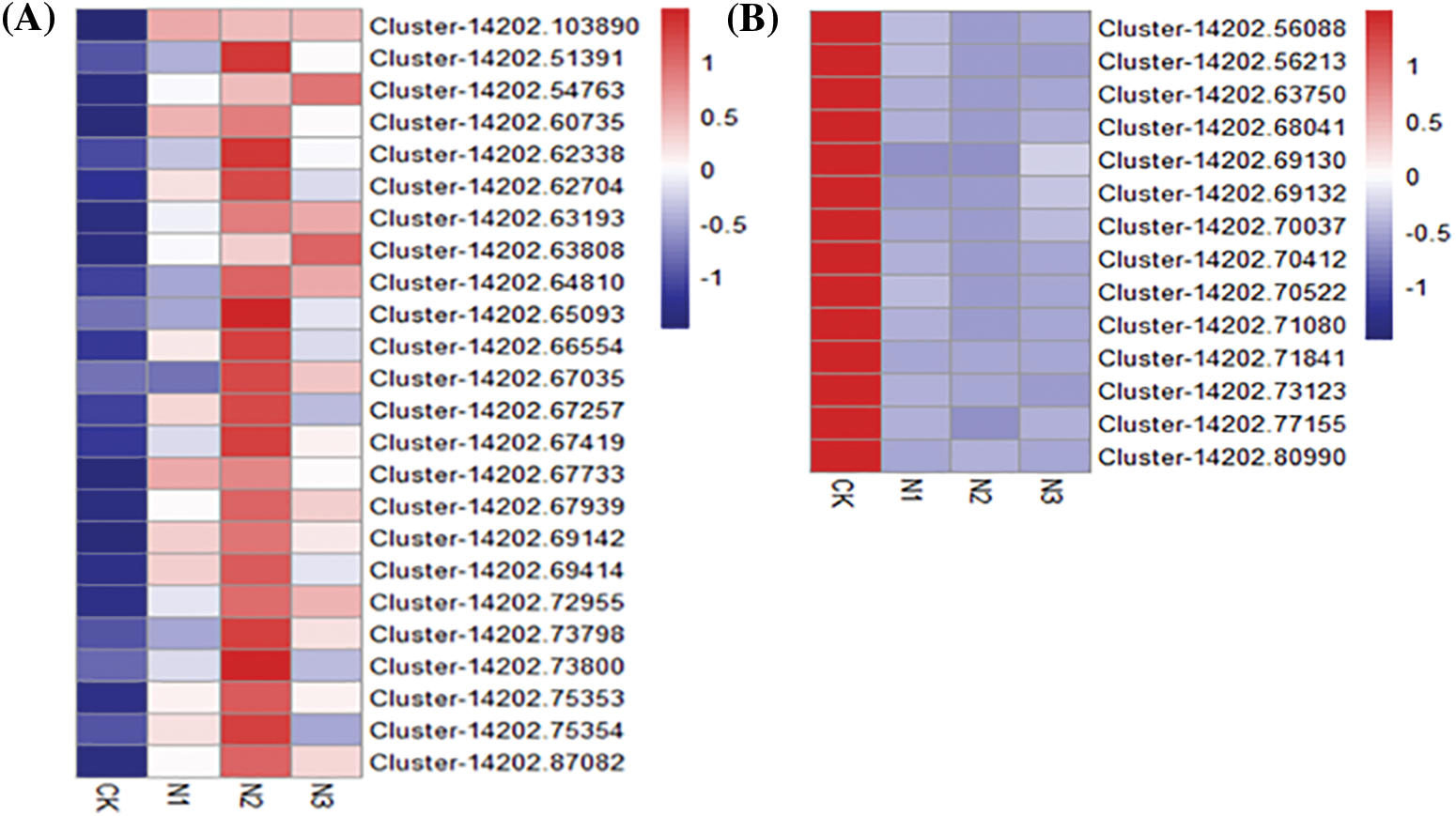
Figure S5: Heatmap of differential expressed genes indentified from the Hub Gene in different modules. A: nitrogen metabolism module (midnightblue module), the Hub Gene is Cluster-14202.68601; B: phosphorus metabolism module (brown module), the Hub Gene is Cluster-14202.71840
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools