 Open Access
Open Access
ARTICLE
Overexpression of the LcPIN2 and LtPIN2 Gene in Arabidopsis thaliana Promotes Root Elongation
Key Laboratory of Forest Genetics & Biotechnology of Ministry of Education, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, 210037, China
* Corresponding Authors: Jinhui Chen. Email: ; Zhaodong Hao. Email:
Phyton-International Journal of Experimental Botany 2023, 92(8), 2383-2397. https://doi.org/10.32604/phyton.2023.029845
Received 10 March 2023; Accepted 26 April 2023; Issue published 25 June 2023
Abstract
The auxin polar transporter, PIN-FORMED 2 (PIN2) plays an important role in root development. However, it remains unclear whether PIN2 genes form two Liriodendron species, L. chinense (LcPIN2) and L. tulipifera (LtPIN2), are both involved in root development and whether and to what extent these two genes diverge in function. Here, we cloned and overexpressed LcPIN2 and LtPIN2 in Arabidopsis thaliana wild-type (WT) and Atpin2 mutant. Phylogenetic and sequence analysis showed a small degree of differentiation between these two Liriodendron PIN2 genes. Tissue-specific gene expression analysis indicated that both Liriodendron PIN2 genes were highly expressed in roots, implying a potential role in root development. Finally, heterologous overexpression of LcPIN2 and LtPIN2 in Arabidopsis both significantly increased the root length compared to wild-type and empty vector. Furthermore, the root length defect in Atpin2 was complemented both by LcPIN2 and LtPIN2. However, heterologous overexpression of LcPIN2 and LtPIN2 cannot rescue the defect in root gravitropism of Atpin2 mutants. Taken together, our findings unravel PIN2 genes from the magnoliids plant Liriodendron were functionally conserved with AtPIN2 in the dicotyledonous plant Arabidopsis in regard to the regulation of root length, but not root gravitropism. This study also provides a potential target for genetic improvement of the root system in these valuable forest trees Liriodendron.Keywords
Auxin is a class of endogenous hormones that play important roles in almost every aspect of plant growth and development [1–4]. Auxin controls many aspects of plant growth and development [5–8]. Membrane transport processes [9], such as secretion and transmembrane transport [10], influence plant growth and development by establishing concentration gradients of auxin, such as apical dominance [11] and root elongation [12–14]. The auxin activity in various biological processes is mainly determined by its biosynthesis, transport, and signaling [15–18]. Among them, directional auxin transport mediated by auxin influx and efflux carriers helps to establish an auxin concentration gradient, regulating embryo development, root patterning, organ formation, etc. [19–21]. For auxin import, AUXIN1/LIKE-AUX1 (AUX1/LAX) proteins are the major auxin influx carriers, whereas PIN-FORMED (PIN), PIN-LIKES (PILS) and ATP-BINDING CASSETTE subfamily B (ABCB) proteins are major auxin efflux carriers that function in auxin export [22,23]. A total of eight PIN proteins were identified in Arabidopsis thaliana, five of which AtPIN1-4 and 7 are localized at the plasma membrane and act as auxin efflux carriers whereas the other three AtPIN5, 6, and 8 are localized in the endoplasmic reticulum (ER) and facilitate intracellular auxin movement between the cytosol and ER [24,25].
PIN-mediated auxin polar transport plays a crucial role in establishing a directional auxin flow in the root [13,22,26]. Specifically, auxin is first transported to the root tip by PIN1, 3, and 7 in the stele and then laterally transported by PIN3 and 7 to the epidermis where PIN2 guides auxin flow upwards to the root elongation zone [19,27–29]. Thus, PIN2 is involved in the proper root development via establishing a directional auxin flow in the root. As a result, mutations in PIN2 led to gravitropic root growth by affecting the redistribution of auxin from the stele toward the root elongation zone in A. thaliana [28]. And in rice, the pin2-1 and pin2-2 exhibited curly root phenotypes and altered lateral root formation patterns due to the defect in proper auxin distribution. Endogenous overexpression of PIN2 contributed to aluminum resistance both in A. thaliana and Oryza sativa [30]. In addition, overexpression of OsPIN2 in O. sativa caused less sensitivity in root response to phosphate deficiency and N-1-naphthylphthalamic acid treatment [31].
Liriodendron, an ancient relict genus, is comprised of two woody plants, i.e., L. tulipifera and L. chinense [32]. L. chinense is naturally distributed in the east of Asia while L. tulipifera grows in the east of North America, comprising a pair of vicarious species with a well-known classical intercontinental disjunct distribution [33,34]. Although this species pair has been diverged since 10–16 million years ago [35], they are similar in morphological and phenological characteristics in general and interspecific hybrids with obvious heterosis were created by artificial pollination approaches. Genome-wide of the PIN gene family has been identified using the genome of L. chinense and LcPIN2 was highly expressed in roots [36–38]. However, the potential role of LcPIN2 in roots and whether PIN2 genes form L. chinense and L. tulipifera diverge in function is still largely unclear. Here, we explored the above issues by cloning the full-length sequence of PIN2 genes from L. chinense (LcPIN2) and L. tulipifera (LtPIN2) and overexpressing them in A. thaliana.
2.1 Gene Identification in Liriodendron Species
The sequence of LcPIN2 was obtained from the previous report [37]. The Arabidopsis PIN2 protein sequence was downloaded from The Arabidopsis Information Resource (TAIR) [39]. Then, AtPIN2 was used as a query to search the protein database of L. tulipifera (unpublished data) using the NCBI Blastp program [40]. The candidate PIN gene from L. tulipifera was further submitted to the TAIR database for searching potential homologous genes, resulting in AtPIN2. Thus, LcPIN2 and LtPIN2 sequences were used as references to design primers for gene cloning.
2.2 Phylogenetic Analysis and Gene Sequence Analysis
The PIN2 protein sequences of ten species (Solanum tuberosum, Salix purpurea, Theobroma cacao, Vitis vinifera, Anacardium occidentale, Corymbia citriodora, Cucumis sativus, Citrus sinensis, Glycine soja, Carya illinoinensis Pawnee) were downloaded from Phytozome (https://phytozome-next.jgi.doe.gov/) [41]. The PIN2 protein sequence was analyzed using Blastp in the National Center for Biotechnology Information (NCBI). The PIN2 protein structure was analyzed using DNAMAN (v6.0.3.40). We use TMHMM to predict the transmembrane domains of PIN2 proteins. The phylogenetic tree of PIN2 was reconstructed using the Neighbor-Joining algorithm with default parameters and a bootstrap value of 1000, using MEGA 7.0 software [42].
2.3 RNA Extraction and cDNA Synthesis
The total RNA was isolated from L. chinense and L. tulipifera tissue by using the FastPure Plant Total RNA Isolation Kit of Vazyme following the instructions of the manufacturer (Nanjing, China). RNA was quantified by measuring absorbance at 260 nm, and its integrity was checked through denaturing agarose gel electrophoresis. The cDNA (complementary DNA) was synthesized from 500 ng of the total RNA by using the Evo M-MLV RT Premix (Accurate Biology, Changsha, China) for RT-qPCR according to the manufacturer’s protocol.
2.4 Quantitative Real-Time PCR Analysis
The plant materials used in the quantitative analysis of PIN2 expression across different tissues were collected from the Baima Forest Farm of Nanjing Forestry University (119.18E, 31.61N). Different tissues were obtained from adult plants and immediately frozen in liquid nitrogen and then stored in a refrigerator at −80°C (We took the tissue from 15-year-old trees). The expression of the PIN2 gene in specific tissues, i.e., root, stem, leaf, petal, pistil, and stamen, of L. chinense and L. tulipifera were quantified by RT-qPCR analysis. Specific primers were designed according to the sequences of LcPIN2 and LtPIN2 genes, and quantitative experiments were performed with three biological replicates and three technical replicates. The qRT-PCR data were analyzed using the 2−ΔΔCt method (Livak method).
2.5 Plant Materials and Growth Conditions
A. thaliana Columbia ecotype (Col-0) wild-type (WT) plants were provided by Prof. Thomas Laux (Signalling Research Centres BIOSS and CIBSS, Faculty of Biology, University of Freiburg, Germany). The Atpin2 mutant was derived from Arashare Corporation (https://www.arashare.cn/index/Product/index.html) and the company storage number is AS016. Seeds were sown on 1/2 MS medium after disinfection and then stored at 4°C for 3 days for vernalization. Next, they were transferred into a homoeothermic incubator (22°C) with a 16/8 h (light/dark) photoperiod. When seedlings reached the four leaves stage, they were transferred to the mixed matrix with peat soil, vermiculite, and perlite ratio of 5:1:1 in a greenhouse.
2.6 Heterologous Overexpression in A. thaliana
pBI121 was used to construct overexpression vectors of Liriodendron PINs, i.e., 35S: LcPIN2 and 35S: LtPIN2, which were confirmed to be correct after Sanger sequencing. The constructed overexpression vectors were respectively introduced into Agrobacterium GV3101, and the positive strains were screened by PCR through the medium containing kanamycin resistance. After that, the agrobacterium solution was inoculated in an LB medium. After centrifugation, the strains were collected, and the optical density value reached 0.6 by shaking the bacterial solution with 5% sucrose solution. Using Arabidopsis plants in good condition, we cut off the pod, leaving the bud unopened. After that, the bacterial solution was used to infect the Arabidopsis inflorescence, soak for 45 s, hide from light for 24 h, then grow normally, then collect mature seeds. The positive plants were screened by 1/2MS medium containing Kan,PCR, and RT-qPCR.
2.7 Phenotypic Identification of Transgenic A. thaliana
The collected T1 generation seeds of 35S: LcPIN2 and 35S: LtPIN2 were sterilized and placed in a 1/2MS medium containing Kana for screening. After a week, the positive plants were transferred to the nutrient soil and cultured in a light incubator. A week later, leaf DNA was extracted for PCR verification, and RNA was extracted for qRT-PCR to determine the expression level of the PIN2 gene. After normal culture, T2 generation seeds were obtained, T2 generation seeds were cultured to T3 generation, and then T3 generation seeds were used to observe the phenotype.
3.1 Gene Cloning, and Sequence Analysis of Liriodendron PIN2 Genes
LtPIN2 gene was identified using LcPIN2 and AtPIN2 as queries to search the protein database of L. tulipifera (unpublished data) by the NCBI blast program. Two pairs of primers were designed to clone the predicted gene sequences. The length and sequences of LcPIN2 and LtPIN2 genes were further determined by Sanger sequencing. Results showed that the length of LcPIN2 and LtPIN2 was both 1,848 bp in length, encoding 616 amino acids. The molecular weight of LcPIN2 and LtPIN2 was predicted to be 66,771.05 Da and 66,771.18 Da, respectively, and the isoelectric point was 9.49 and 9.55, Grand average of hydropathicity (GRAVY) was 0.063 and 0.062, respectively (Table 1). The prediction results of subcellular protein showed that both Liriodendron PIN2 belonged to the integral membrane protein, which is consistent with the auxin output function of PIN proteins.
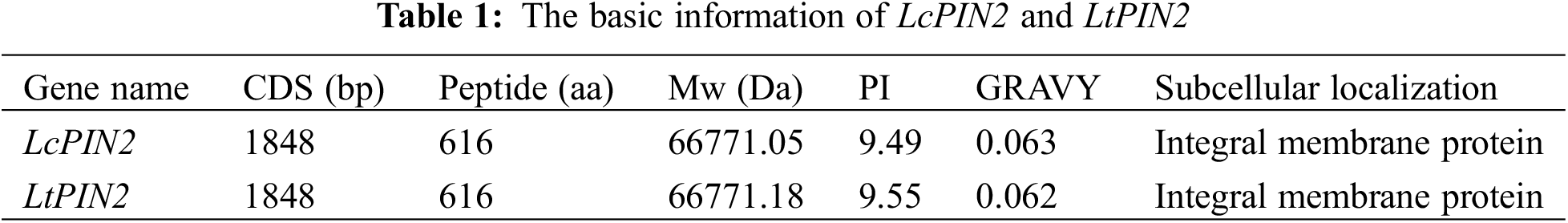
3.2 Phylogenetic Analysis of Liriodendron PIN2 Genes
We used DNAMAN to perform multiple sequence alignment of two Liriodendron PIN2 together with PIN2 proteins from Arabidopsis and the other ten species. Extremely conserved regions were found at both ends of the protein sequences, referring to the transmembrane domain (Fig. 1). Further analysis confirmed the transmembrane domain of LcPIN2 and LtPIN2 (Figs. 2B and 2C), both with five transmembrane structures at the N-terminal and four transmembrane regions at the C-terminal. Phylogenetic analysis showed that Liriodendron PIN2 proteins were closely clustered together (Fig. 2A). Furthermore, motif analysis indicated that two Liriodendron PIN2 proteins shared almost the same motif arrangement except the missing motif 5 located at the C-terminal compared with PIN2 proteins from plant species (Fig. 2D).
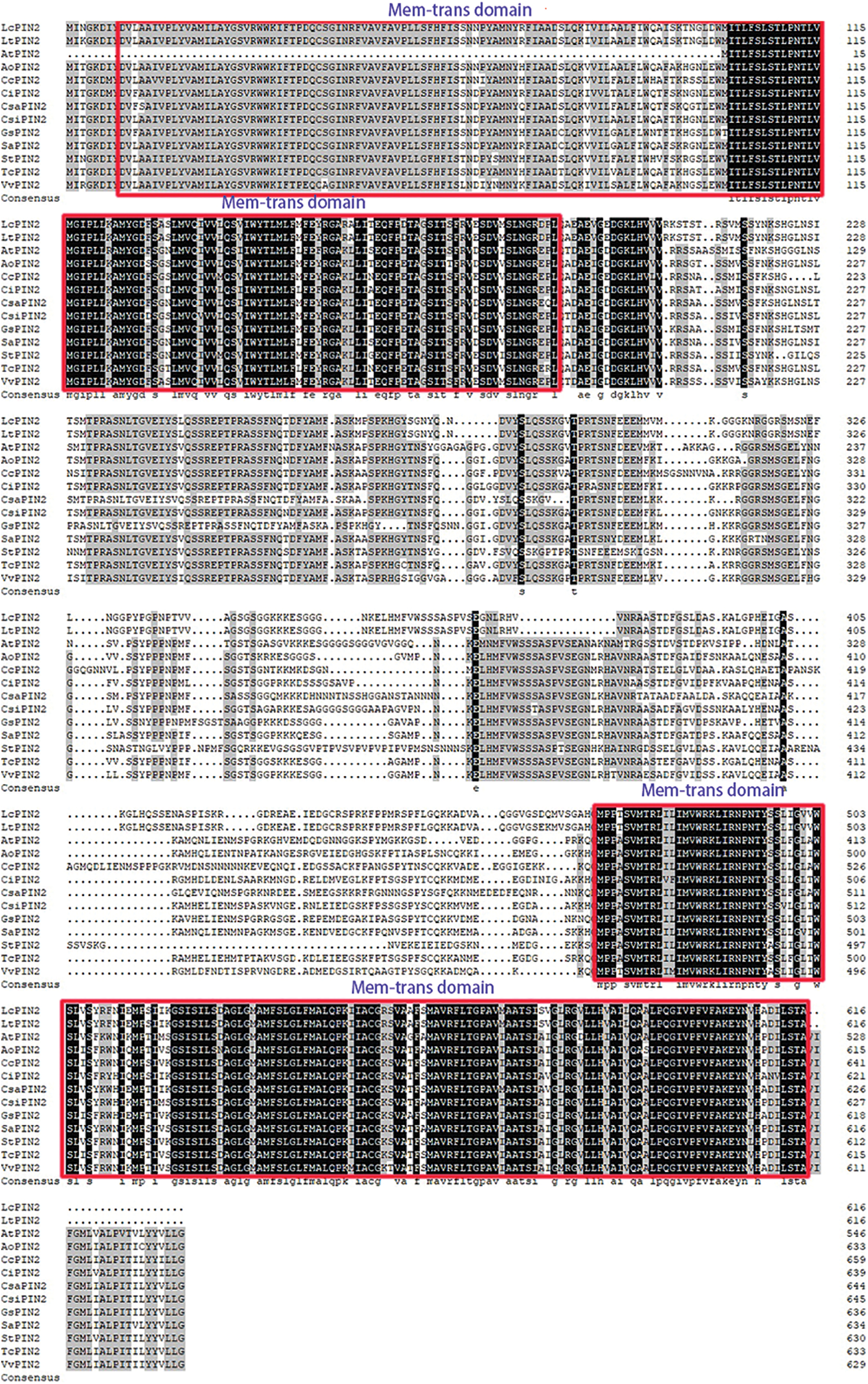
Figure 1: LcPIN2 and LtPIN2 genes amino acid sequence multiple alignments. The red box shows the transmembrane domain. Anacardium occidentale (Ao), Arabidopsis thaliana (At), Corymbia citriodora (Cc), Carya illinoinensis (Ci), Cucsa sativus (Csa), Citrus sinensis (Csi), Glycine soja (Gs), Salix purpurea (Sp), Solanum tuberosum (St), Theobroma cacao (Tc), Vitis vinifera (Vv)
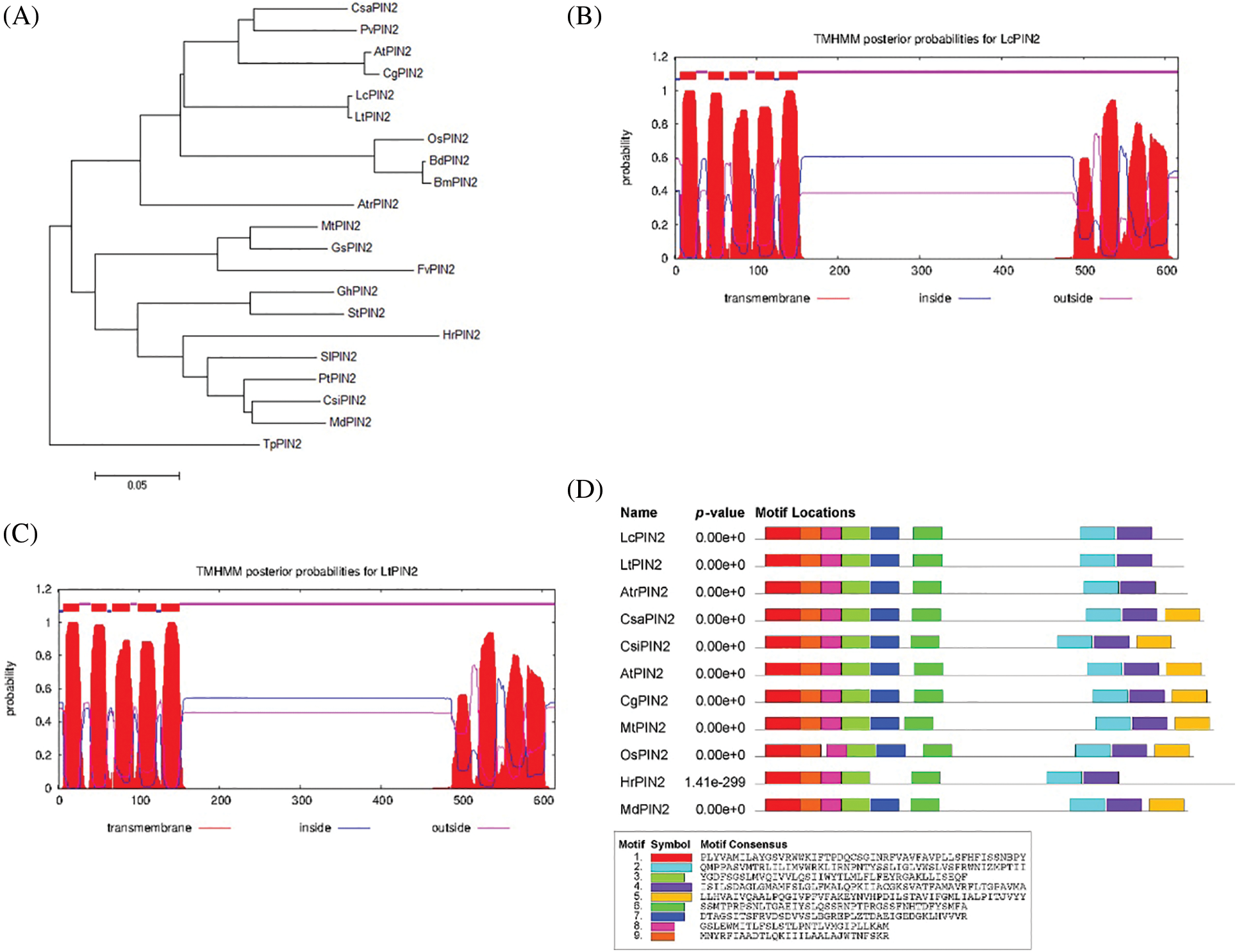
Figure 2: Evolution and structural analysis of LcPIN2 and LtPIN2. (A) Phylogenetic tree of PIN2 from Cucumis sativus (Csa), Phaseolus vulgaris (Pv), Arabidopsis thaliana (At), Capsella grandiflora (Cg), Oryza sativa (Os), Brachypodium distachyon (Bd), Brachypodium mexicanum (Bm), Liriodendron chinense (Lc), Liriodendron tulipifera (Lt), Amborella trichopoda (Atr), Thuja plicata (Tp), Medicago truncatula (Mt), Glycine soja (Gs), Citrus sinensis (Csi), Fragaria vesca (Fv), Gossypium hirsutum (Gh), Hordeum vulgare (Hv), Populus trichocarpa (Pt), Solanum lycopersicum (Sl), Malus domestica (Md), Solanum tuberosum (St), Triticum aestivum (Ta), Zea mays (Zm). (B) Prediction of the transmembrane domain of the LcPIN2 gene. (C) Prediction of the transmembrane domain of the LtPIN2 gene. (D) Motif analysis of LcPIN2 and LtPIN2
3.3 Liriodendron PIN2 Genes Were Both Highly Expressed in Roots
We quantified the tissue expression profiling of LcPIN2 and LtPIN2 across different tissues, i.e., root, stem, leaf, petal, pistil, and stamen, in L. chinense and L. tulipifera, respectively (Fig. 3). The results showed that both LcPIN2 and LtPIN2 were highly expressed in roots compared with other tissues. And, in L. tulipifera, the expression level of LtPIN2 was also relatively high in the pistil with no similar results in L. chinense. These results implied that Liriodendron PIN2 genes might both function in the root development, reflecting the functional conservation. However, LcPIN2 and LtPIN2 have diverged at least in tissue-specific expression patterns.
Figure 3: Expression patterns of LcPIN2 and LtPIN2 across different tissues. (A) Schematic diagram of Liriodendron sampling site. The root, stem, and flower are shown in enlarged pictures. The purple arrow represents the leaf, the cyan arrow represents the pistil, the red arrow represents the stamen and the yellow arrow represents the petal. (B) Gene expression patterns of LcPIN2 and LtPIN2 in different tissues of L. chinensis and L. tulipifera, respectively
3.4 Overexpressing Liriodendron PIN2 Genes Promotes Root Elongation in A. thaliana
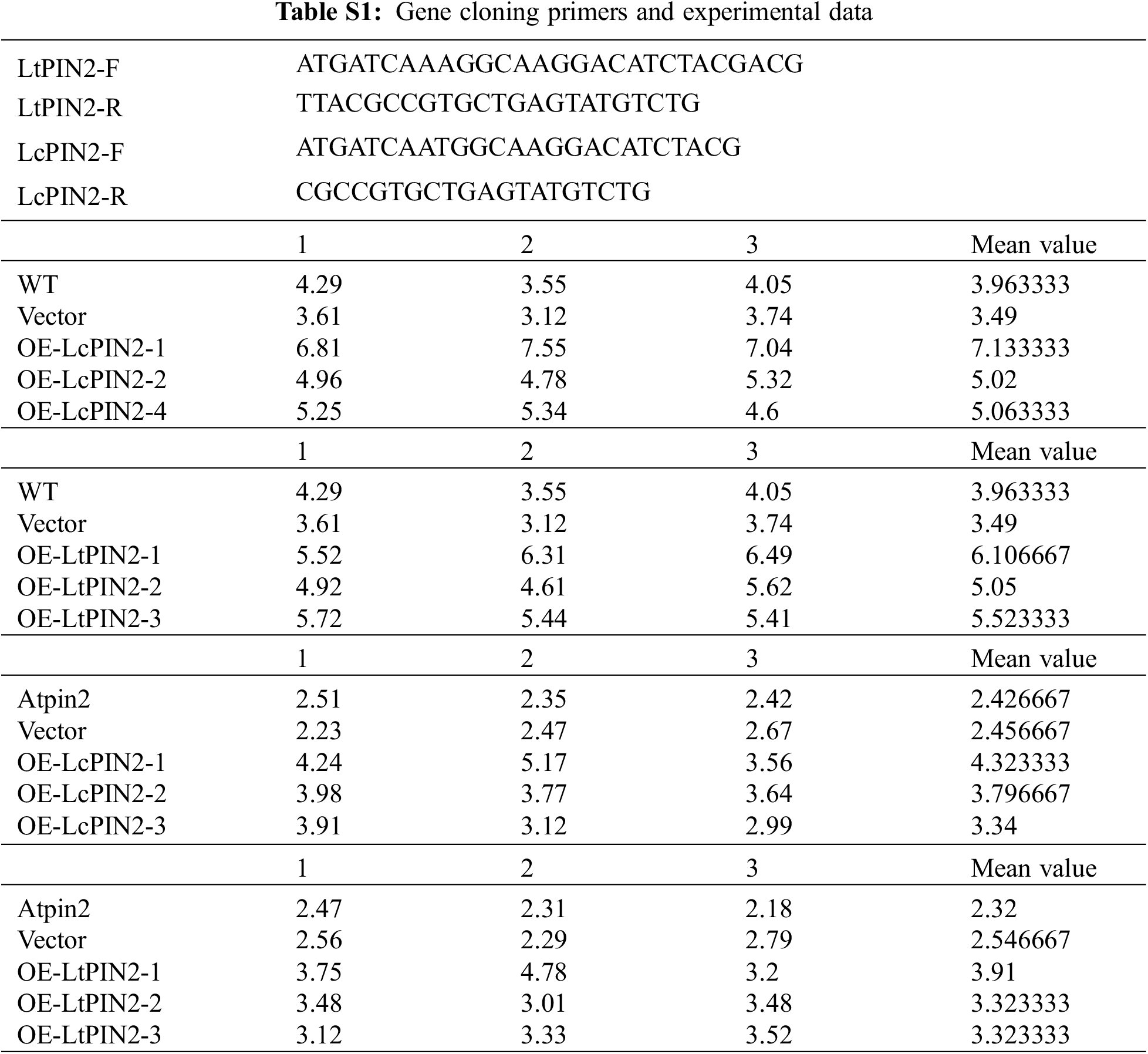
To explore the potential role of Liriodendron PIN2 genes in root development, we overexpressed LcPIN2 and LtPIN2 in A. thaliana. We used kanamycin to screen positive transgenic lines. The transgenic lines were further confirmed by qRT-PCR (Fig. S1), indicating an obvious heterogeneous overexpression of LcPIN2 or LtPIN2 compared to WT and empty vector (Fig. 4B). We did not find significant changes in root density of lateral roots of A. thaliana in overexpressed strains (Fig. S2).Overexpression of LcPIN2 and LtPIN2 significantly increased the root length compare to WT and empty vector whereas no significant difference was found between WT and empty vector (Fig. 4). Since we performed these two experiments independently, we removed potential batch effects and then compared the effects of two Liriodendron PIN2 genes in promoting root elongation. Results showed that no significant difference existed between the root length of p35S: LcPIN2 and p35S: LtPIN2. To investigate whether LcPIN2 and LtPIN2 promote the function of wild-type A. thaliana differently, we calculated and compared the root length of wild-type A. thaliana overexpressing LcPIN2 and LtPIN2 (Fig. 4D). The results showed that strain 1 overexpressing LcPIN2 was significantly different from other strains, but there was no significant difference between strains 2 and 4 and lines overexpressing LtPIN2. So we determined that LcPIN2 and LtPIN2 may be consistent in their ability to promote wild-type A. thaliana. In summary, we concluded that there was no significant difference between LcPIN2 and LtPIN2 in promoting the root length of wild-type A. thaliana.
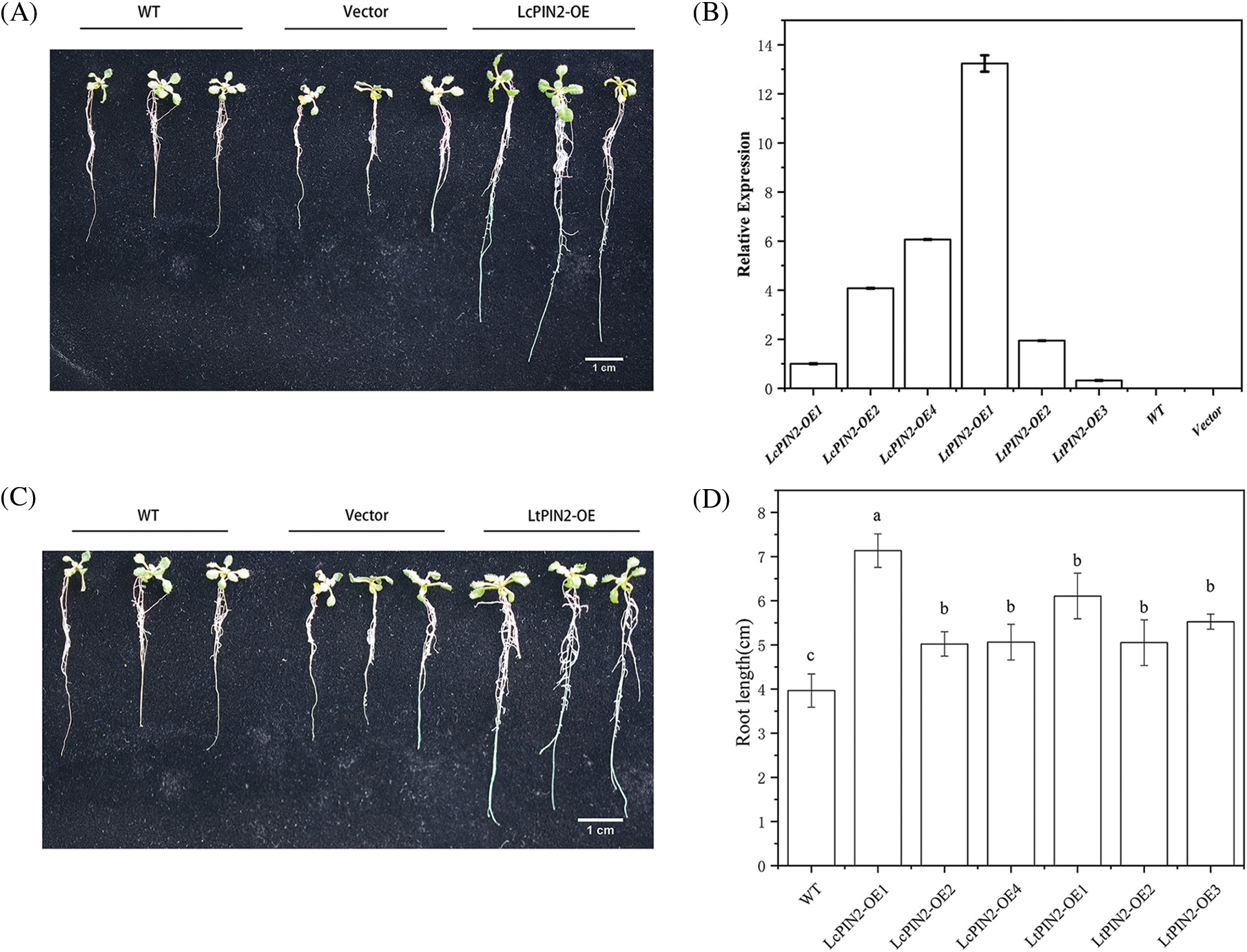
Figure 4: Overexpression of PIN2 promotes root growth in A. thaliana plants. (A) Representative photographs of A. thaliana wild type (WT), LcPIN2-1, LcPIN2-2, and LcPIN2-4 plants, grown for 12 d on plates. (B) Expression of PIN2 gene in transgenic wild A. thaliana. The PIN2 gene expression of LcPIN2-OE1 was 1. (C) Representative photographs of Arabidopsis wild-type (WT), LtPIN2-1, LtPIN2-2, and LtPIN2-3 plants, grown for 12 d on plates. (D) Comparison of root length of wild-type A. thaliana overexpressing LcPIN2 and LtPIN2 genes. The three plants on the left are wild-type Arabidopsis, the three plants in the middle are no-load controls, and the three plants on the right are 35S: PIN2 transgenic A. thaliana. Bar = 1 cm. All experiments were normalized using A. thaliana Actin as an internal reference, and three independent experiments were performed. The error bars in the charts indicate the standard deviation from the mean of each triplicate treatment
3.5 Overexpressing Liriodendron PIN2 Genes Rescue Defects in Root Length, But Not Root Gravitropism, of Atpin2 Mutants
The phenotype of the Atpin2 mutant is a loss of gravitropism and a decrease in the amount of root elongation [28,43]. To determine whether the Liriodendron PIN2 gene has the function of restoring the gravitropism of the Atpin2 mutant, we overexpressed the LcPIN2 and LtPIN2 genes in the A. thaliana mutant. After qRT-PCR identification (Figs. 5B and S3) and T3 generations are produced by self-crossing, it was found that the overexpressed Atpin2 mutant under vertical culture conditions, the roots of Atpin2 mutant did not return to gravity, and the roots remained curled without a definite downward growth direction (Figs. 5A and 5C). We found no significant change in lateral root density in overexpressed AtPIN2 (Fig. S4). Therefore, we believe that LcPIN2 and LtPIN2 do not restore gravitropism in Arabidopsis Atpin2 mutants. The root length of the Atpin2 mutant was measured 18 days after the mutant grew (Fig. 5D, Table S1). It was found that the root of the Atpin2 mutant overexpressing LcPIN2 and LtPIN2 genes was longer than that of the WT, and the change was statistically significant after statistical analysis. This result is similar to that of the wild-type A. thaliana with overexpression of the Liriodendron PIN2 gene. To investigate whether LcPIN2 and LtPIN2 promote the function of the Atpin2 mutant differently, we calculated and compared the root length of the Atpin2 mutant overexpressing LcPIN2 and LtPIN2 (Fig. 5D). LcPIN2 and LtPIN2 genes were overexpressed in the Atpin2 mutant, and statistical analysis showed that there was no significant difference between overexpressed LcPIN2 and LtPIN2 in A. thaliana. In summary, we concluded that there was no significant difference between LcPIN2 and LtPIN2 in promoting the root length of the Atpin2 mutant.
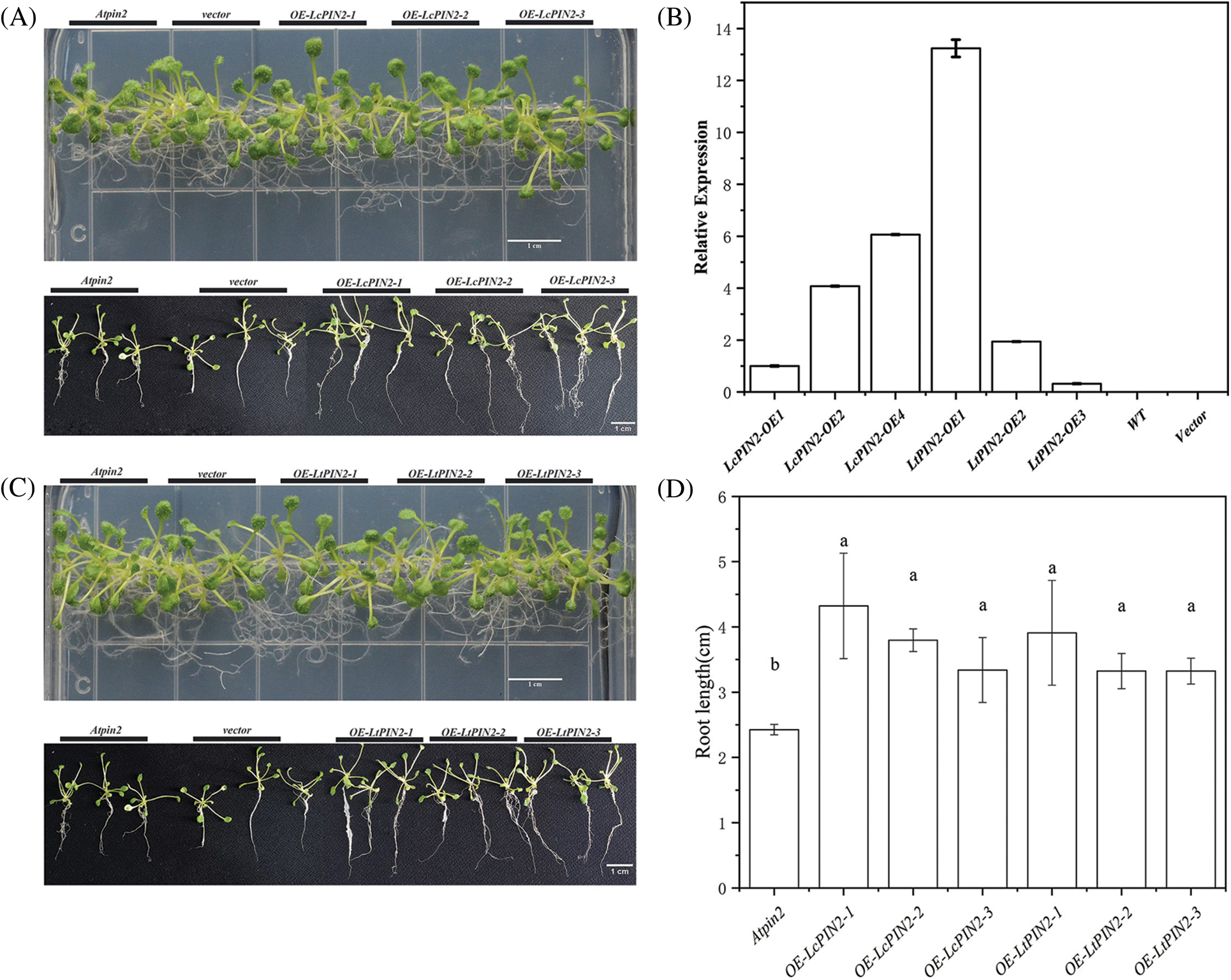
Figure 5: Overexpression of PIN2 promotes root growth in A. thaliana plants. (A) Representative photographs of Atpin2 mutant, different strains that overexpress LcPIN2 (OE-LcPIN2-1, OE-LcPIN2-2, OE-LcPIN2-3), grown for 18 d on plates. (B) Expression of PIN2 gene in transgenic Atpin2 mutant. The PIN2 gene expression of OE-LcPIN2-1 was 1. (C) Representative photographs of Atpin2 mutant, different strains that overexpress LtPIN2 (OE-LtPIN2-1, OE-LtPIN2-2, OE-LtPIN2-3), grown for 18 d on plates. (D) Comparison of root length of Atpin2 mutant overexpressing LcPIN2 and LtPIN2 genes. The three plants on the left are Atpin2 mutants, the three plants in the Second group on the left are no-load controls, and the three plants on the right are 35S: PIN2 transgenic A. thaliana. Bar = 1 cm. All experiments were normalized using A. thaliana Actin as an internal reference, and three independent experiments were performed. The error bars in the charts indicate the standard deviation from the mean of each triplicate treatment
PIN proteins, as the major auxin efflux carriers, function in the auxin polar transport, thus playing an important role in the various biological process [22]. In plant roots, the combined action of several PINs constructs a bidirectional auxin flow in the root which affects the root development [22]. Here, we cloned two PIN2 genes from two intercontinental vicariate species in L. chinense and L. tulipifera. Phylogenetic and sequence analysis indicated that these two genes showed a small degree of differentiation at least in the levels of protein sequence, motif, and domain (Figs. 1 and 2). Furthermore, the relatively high expression in roots suggested a conserved function of these two Liridoendron PIN genes (Fig. 3). However, the tissue expression analysis also indicated a potential functional divergence in expression intensity in roots and expression in pistil between LcPIN2 and LtPIN2 (Fig. 3). Further heterologous overexpression in Arabidopsis confirmed the conserved fucntion of these two Liridoendron PIN2 genes in promoting root elongation (Figs. 4 and 5). This is opposite to the phenotype of overexpression of OsPIN2 in rice which led to an obvious inhibition in root length [31,44]. This might be the reflection of the evolutionary divergence since Lirirodenron belongs to the magnoliids which have diverged from other flowering plants during early angiosperm evolution [36]. Previous studies have shown that the PIN2 gene in A. thaliana can compensate for the geotropism of Atpin2 deletion [45]. To further study the function of Liriodendron PIN2, we overexpressed LcPIN2 and LtPIN2 in the Atpin2 mutant. The results showed that LcPIN2 and LtPIN2 could not compensate for the loss of geotropism in the mutant, but they could promote the root growth of the Atpin2 in terms of root length, indicating that Liriodendron PIN2 could compensate for the reduced root length of the Atpin2 mutant. Therefore, the PIN2 gene can partially compensate for the missing phenotype of the Atpin2 mutant. We compared the effects of LcPIN2 and LtPIN2 on root elongation of WT and Atpin2 mutants, and found that LcPIN2 and LtPIN2 had similar effects on root elongation, indicating that LcPIN2 and LtPIN2 are highly conserved and have similar functions.
In this study, the LtPIN2 gene was first reported, and the similarities and differences between two Liridoendron PIN genes, LcPIN2 and LtPIN2, were illustrated. Heterologous overexpression in Arabidopsis suggested that these two Liriodendron PIN2 genes both functioned in promoting root elongation with no obvious functional divergence. Overexpression of LcPIN2 and LtPIN2 in the Atpin2 mutant indicated that the PIN2 gene of Liriodendron could partially compensate for the Atpin2 mutant phenotype. Thus, our study provides the first insights into how LcPIN2 and LtPIN2 genes may be used in the improvement of the root system in Liriodendron.
Funding Statement: This research was supported by the Youth Foundation of the Natural Science Foundation of Jiangsu Province (BK20210614), the Nature Science Foundation of China (32071784), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX21_0921).
Author Contributions: Z. D. H. and J. H. C. were the lead investigators of this research program. Z. D. H. and Z. J. C. designed the experiments and coordinated the project. Z. J. C. performed all the experiment works. Z. D. H. and Z. J. C. wrote and edited most of the manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Pacurar, D. I., Perrone, I., Bellini, C. (2014). Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiologia Plantarum, 151(1), 83–96. https://doi.org/10.1111/ppl.12171 [Google Scholar] [PubMed] [CrossRef]
2. Zhang, Y., Ma, Y., Zhao, D., Tang, Z., Zhang, T. et al. (2022). Genetic regulation of lateral root development. Plant Signaling & Behavior, 2081397. https://doi.org/10.1080/15592324.2022.2081397 [Google Scholar] [PubMed] [CrossRef]
3. Gu, H., Ding, W., Shi, T., Ouyang, Q., Yang, X. et al. (2022). Integrated transcriptome and endogenous hormone analysis provides new insights into callus proliferation in Osmanthus fragrans. Scientific Reports, 12(1), 7609. https://doi.org/10.1038/s41598-022-11801-9 [Google Scholar] [PubMed] [CrossRef]
4. Adamowski, M., Friml, J. (2015). PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell, 27(1), 20–32. https://doi.org/10.1105/tpc.114.134874 [Google Scholar] [PubMed] [CrossRef]
5. Ludwig-Muller, J. (2015). Bacteria and fungi controlling plant growth by manipulating auxin: Balance between development and defense. Journal of Plant Physiology, 172, 4–12. https://doi.org/10.1016/j.jplph.2014.01.002 [Google Scholar] [PubMed] [CrossRef]
6. Li, T., Dai, Z., Zeng, B., Li, J., Ouyang, J. et al. (2022). Autocatalytic biosynthesis of abscisic acid and its synergistic action with auxin to regulate strawberry fruit ripening. Horticulture Research, 9, uhab076. https://doi.org/10.1093/hr/uhab076 [Google Scholar] [PubMed] [CrossRef]
7. Weijers, D. (2016). Phyllotaxis: A Matthew effect in auxin action. Current Biology, 26(23), R1233–R1235. https://doi.org/10.1016/j.cub.2016.10.019 [Google Scholar] [PubMed] [CrossRef]
8. Pattison, R. J., Csukasi, F., Catala, C. (2014). Mechanisms regulating auxin action during fruit development. Physiol Plant, 151(1), 62–72. https://doi.org/10.1111/ppl.12142 [Google Scholar] [PubMed] [CrossRef]
9. Xie, P., Cath, T. Y., Ladner, D. A. (2021). Mass transport in osmotically driven membrane processes. Membranes, 11(1). https://doi.org/10.3390/membranes11010029 [Google Scholar] [PubMed] [CrossRef]
10. Yin, S., Duan, M., Fang, B., Zhao, G., Leng, X. et al. (2022). Zinc homeostasis and regulation: Zinc transmembrane transport through transporters. Critical Reviews in Food Science and Nutrition, 1–11. https://doi.org/10.1080/10408398.2022.2048292 [Google Scholar] [PubMed] [CrossRef]
11. Tanaka, M., Takei, K., Kojima, M., Sakakibara, H., Mori, H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant Journal, 45(6), 1028–1036. https://doi.org/10.1111/j.1365-313X.2006.02656.x [Google Scholar] [PubMed] [CrossRef]
12. Hussain, S., Wang, W., Ahmed, S., Wang, X. T., Adnan et al. (2021). PIP2, an auxin induced plant peptide hormone regulates root and hypocotyl elongation in Arabidopsis. Frontiers in Plant Science, 12. https://doi.org/10.3389/fpls.2021.646736 [Google Scholar] [PubMed] [CrossRef]
13. Inahashi, H., Shelley, I. J., Yamauchi, T., Nishiuchi, S., Takahashi-Nosaka, M. et al. (2018). OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiologia Plantarum, 164(2), 216–225. https://doi.org/10.1111/ppl.12707 [Google Scholar] [PubMed] [CrossRef]
14. Qi, Y. H., Wang, S. K., Shen, C. J., Zhang, S. N., Chen, Y. et al. (2012). OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytologist, 193(1), 109–120. https://doi.org/10.1111/j.1469-8137.2011.03910.x [Google Scholar] [PubMed] [CrossRef]
15. Teale, W. D., Paponov, I. A., Palme, K. (2006). Auxin in action: Signalling, transport and the control of plant growth and development. Nature Reviews Molecular Cell Biology, 7(11), 847–859. https://doi.org/10.1038/nrm2020 [Google Scholar] [PubMed] [CrossRef]
16. Tivendale, N. D., Belt, K., Berkowitz, O., Whelan, J., Millar, A. H. et al. (2021). Knockdown of succinate dehydrogenase assembly factor 2 induces reactive oxygen species-mediated auxin hypersensitivity causing pH-dependent root elongation. Plant and Cell Physiology, 62(7), 1185–1198. https://doi.org/10.1093/pcp/pcab061 [Google Scholar] [PubMed] [CrossRef]
17. Street, I. H., Mathews, D. E., Yamburkenko, M. V., Sorooshzadeh, A., John, R. T. et al. (2016). Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development, 143(21), 3982–3993. https://doi.org/10.1242/dev.132035 [Google Scholar] [PubMed] [CrossRef]
18. Song, P., Xu, H., Zhang, J. X., Chen, H. T., Li, L. et al. (2021). Functional analysis of indole 3-hexanoic acid as a novel auxin from Arabidopsis thaliana. Planta, 254(4). https://doi.org/10.1007/s00425-021-03719-9 [Google Scholar] [PubMed] [CrossRef]
19. Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T. et al. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell, 108(5), 661–673. https://doi.org/10.1016/S0092-8674(02)00656-6 [Google Scholar] [PubMed] [CrossRef]
20. Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D. et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell, 115(5), 591–602. https://doi.org/10.1016/S0092-8674(03)00924-3 [Google Scholar] [PubMed] [CrossRef]
21. Liu, Y., Mu, C., Du, D., Yang, Y., Li, L. et al. (2022). Alkaline stress reduces root waving by regulating PIN7 vacuolar transport. Frontiers in Plant Science, 13, 1049144. https://doi.org/10.3389/fpls.2022.1049144 [Google Scholar] [PubMed] [CrossRef]
22. Petrasek, J., Friml, J. (2009). Auxin transport routes in plant development. Development, 136(16), 2675–2688. https://doi.org/10.1242/dev.030353 [Google Scholar] [PubMed] [CrossRef]
23. Geisler, M. M. (2021). A retro-perspective on auxin transport. Frontiers in Plant Science, 12, 756968. https://doi.org/10.3389/fpls.2021.756968 [Google Scholar] [PubMed] [CrossRef]
24. Vieten, A., Sauer, M., Brewer, P. B., Friml, J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends in Plant Science, 12(4), 160–168. https://doi.org/10.1016/j.tplants.2007.03.006 [Google Scholar] [PubMed] [CrossRef]
25. Naramoto, S. (2017). Polar transport in plants mediated by membrane transporters: Focus on mechanisms of polar auxin transport. Current Opinion in Plant Biology, 40, 8–14. https://doi.org/10.1016/j.pbi.2017.06.012 [Google Scholar] [PubMed] [CrossRef]
26. Zhang, M., Wang, C. P., Lin, Q. F., Liu, A. H., Wang, T. et al. (2015). A tetratricopeptide repeat domain-containing protein SSR1 located in mitochondria is involved in root development and auxin polar transport in Arabidopsis. Plant Journal, 83(4), 582–599. https://doi.org/10.1111/tpj.12911 [Google Scholar] [PubMed] [CrossRef]
27. Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I. et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature, 433(7021), 39–44. https://doi.org/10.1038/nature03184 [Google Scholar] [PubMed] [CrossRef]
28. Muller, A., Guan, C., Galweiler, L., Tanzler, P., Huijser, P. et al. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO Journal, 17(23), 6903–6911. https://doi.org/10.1093/emboj/17.23.6903 [Google Scholar] [PubMed] [CrossRef]
29. Friml, J., Benkova, E., Mayer, U., Palme, K., Muster, G. (2003). Automated whole mount localisation techniques for plant seedlings. Plant Journal, 34(1), 115–124. https://doi.org/10.1046/j.1365-313X.2003.01705.x [Google Scholar] [PubMed] [CrossRef]
30. Wu, D., Shen, H., Yokawa, K., Baluska, F. (2014). Alleviation of aluminium-induced cell rigidity by overexpression of OsPIN2 in rice roots. Journal of Experimental Botany, 65(18), 5305–5315. https://doi.org/10.1093/jxb/eru292 [Google Scholar] [PubMed] [CrossRef]
31. Sun, H., Guo, X., Xu, F., Wu, D., Zhang, X. et al. (2019). Overexpression of OsPIN2 regulates root growth and formation in response to phosphate deficiency in rice. International Journal of Molecular Science, 20(20). https://doi.org/10.3390/ijms20205144 [Google Scholar] [PubMed] [CrossRef]
32. Long, X. F., Weng, Y. H., Liu, S. Q., Hao, Z. D., Sheng, Y. et al. (2019). Genetic diversity and differentiation of relict plant liriodendron populations based on 29 novel EST-SSR markers. Forests, 10(4). https://doi.org/10.3390/f10040334 [Google Scholar] [CrossRef]
33. Hao, R. (1995). Geographical distribution of Liriodendron chinense in China and its significance. Journal of Plant Resources and Environment (China), 4(1), 1–6. [Google Scholar]
34. Parks, C. R., Miller, N. G., Wendel, J. F., McDougal, K. M. (1983). Genetic divergence within the genus Liriodendron (Magnoliaceae). Annals of the Missouri Botanical Garden, 70(4), 658–666. [Google Scholar]
35. Parks, C. R., Wendel, J. F. (1990). Molecular divergence between Asian and North American species of Liriodendron (Magnoliaceae) with implications for interpretation of fossil floras. American Journal of Botany, 77(10), 1243–1256. [Google Scholar]
36. Chen, J., Hao, Z., Guang, X., Zhao, C., Wang, P. et al. (2019). Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nature Plants, 5(1), 18–25. https://doi.org/10.1038/s41477-018-0323-6 [Google Scholar] [PubMed] [CrossRef]
37. Hu, L., Wang, P., Long, X., Wu, W., Zhang, J. et al. (2021). The PIN gene family in relic plant L. chinense: Genome-wide identification and gene expression profiling in different organizations and abiotic stress responses. Plant Physiology and Biochemistry, 162(2), 634–646. https://doi.org/10.1016/j.plaphy.2021.03.030 [Google Scholar] [PubMed] [CrossRef]
38. Li, R., Pan, Y., Hu, L. F., Yang, D. J., Yuan, M. J. et al. (2022). PIN3 from liriodendron may function in inflorescence development and root elongation. Forests, 13(4), 568. https://doi.org/10.3390/f13040568 [Google Scholar] [CrossRef]
39. Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C. et al. (2012). The Arabidopsis Information Resource (TAIRImproved gene annotation and new tools. Nucleic Acids Research, 40, 1202–1210. https://doi.org/10.1093/nar/gkr1090 [Google Scholar] [PubMed] [CrossRef]
40. Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2 [Google Scholar] [PubMed] [CrossRef]
41. Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D. et al. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research, 40, 1178–1186. https://doi.org/10.1093/nar/gkr944 [Google Scholar] [PubMed] [CrossRef]
42. Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. https://doi.org/10.1093/molbev/msw054 [Google Scholar] [PubMed] [CrossRef]
43. Tan, C., Wang, H., Zhang, Y., Qi, B., Xu, G. et al. (2011). A proteomic approach to analyzing responses of Arabidopsis thaliana root cells to different gravitational conditions using an agravitropic mutant, pin2 and its wild type. Proteome Science, 9, 72. https://doi.org/10.1186/1477-5956-9-72 [Google Scholar] [PubMed] [CrossRef]
44. Chen, Y., Fan, X., Song, W., Zhang, Y., Xu, G. (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnology Journal, 10(2), 139–149. https://doi.org/10.1111/j.1467-7652.2011.00637.x [Google Scholar] [PubMed] [CrossRef]
45. Abas, L., Benjamins, R., Malenica, N., Paciorek, T., Wisniewska, J. et al. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nature Cell Biology, 8(3), 249–256. https://doi.org/10.1038/ncb1369 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
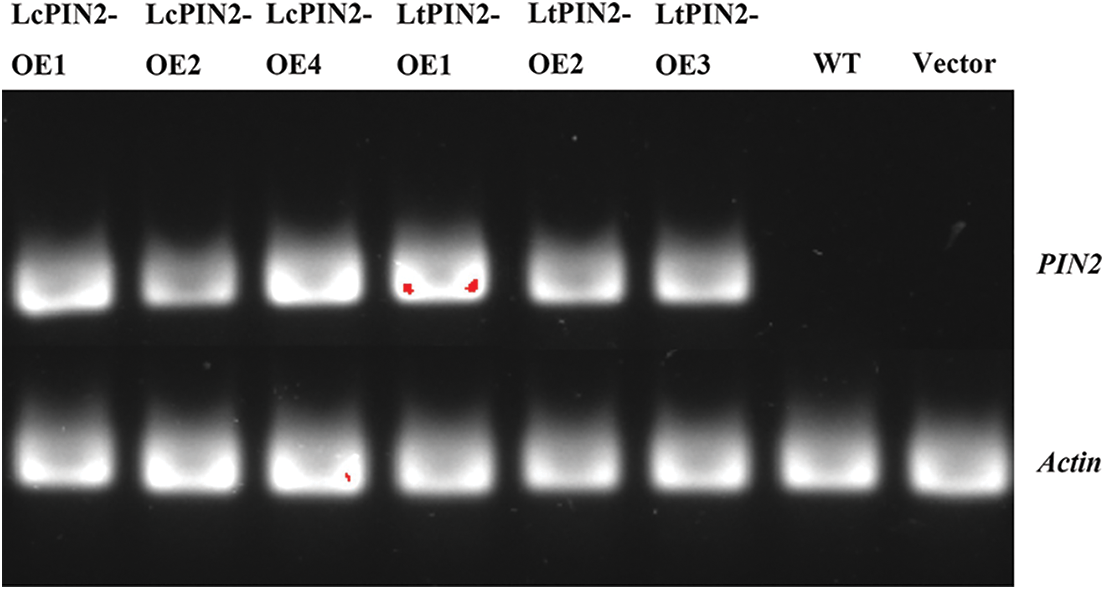
Figure S1: Semi-quantitative map of overexpression of LcPIN2 and LtPIN2 in A. thaliana
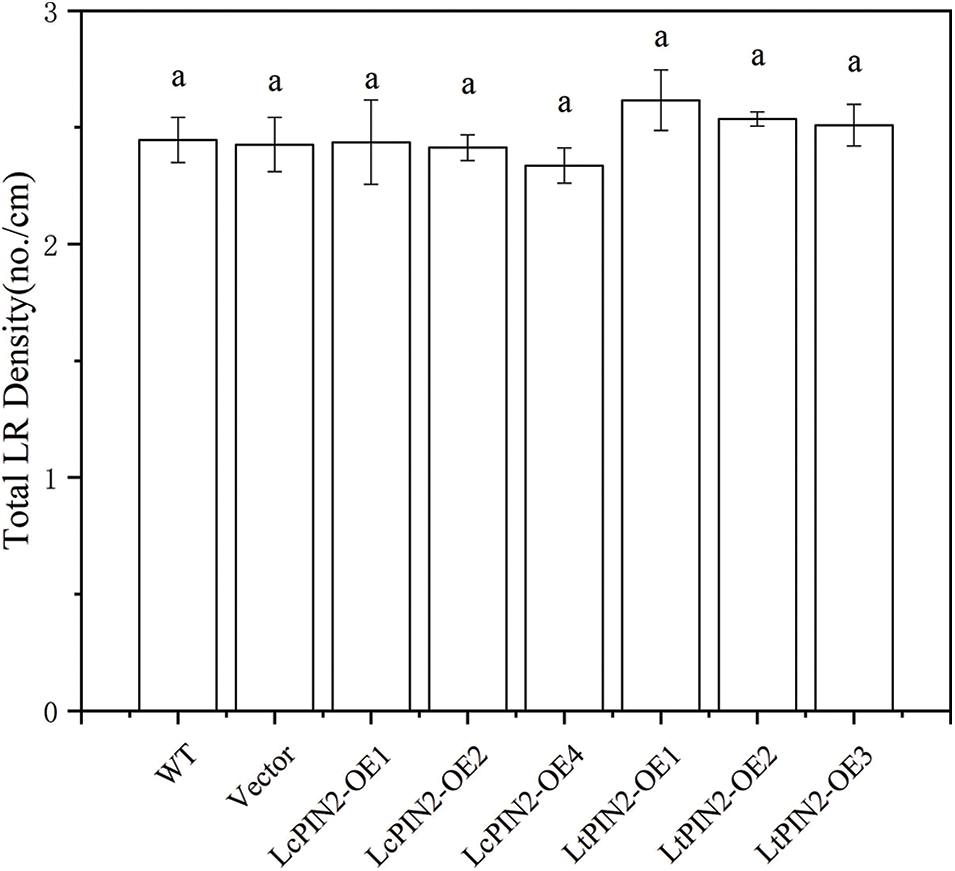
Figure S2: Lateral root density of overexpressed LcPIN2 and LtPIN2 in A. thaliana
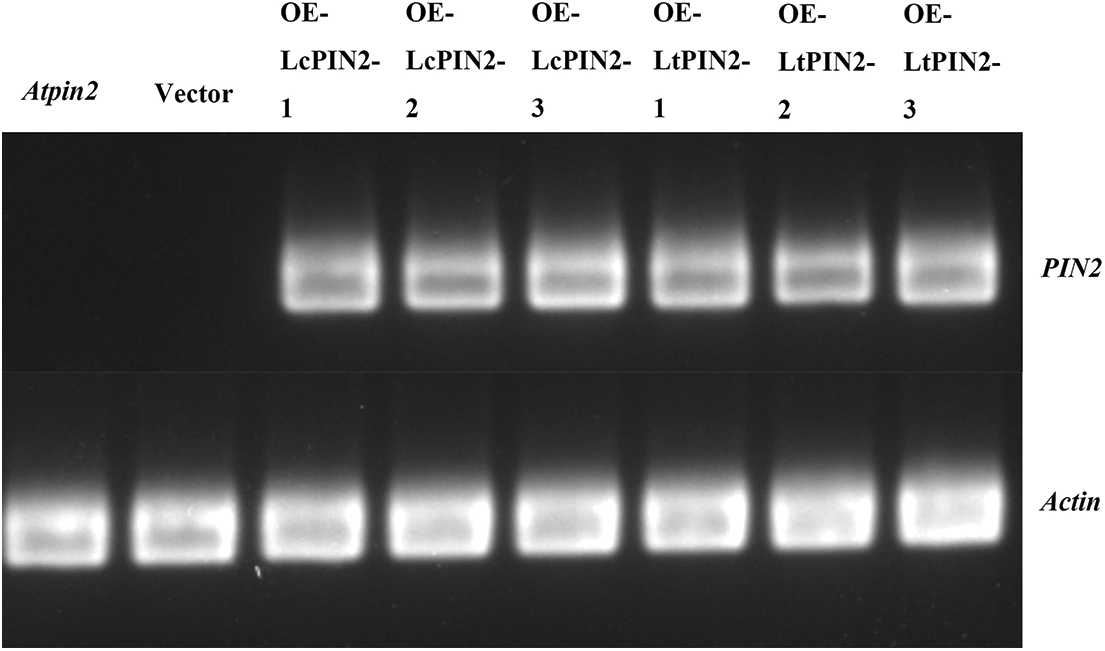
Figure S3: Semi-quantitative map of overexpression of LcPIN2 and LtPIN2 in AtPIN2
Figure S4: Lateral root density of overexpressed LcPIN2 and LtPIN2 in AtPIN2
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools