 Open Access
Open Access
ARTICLE
Biological Analysis and Response to Low Phosphate Stress of Phosphate Transporter Family 1 (PHT1)) Genes in Solanum tuberosum L.
1 Key Laboratory of Crop Germplasm Innovation and Sustainable Utilization, Collage of Agronomy and Biotechnology, Yunnan Agricultural University, Kunming, 650201, China
2 Laboratory of Fruit and Vegetable Preservation and Processing, College of Food Science and Technology, Yunnan Agricultural University, Kunming, 650201, China
* Corresponding Author: Xinyong Zhang. Email:
Phyton-International Journal of Experimental Botany 2023, 92(8), 2325-2348. https://doi.org/10.32604/phyton.2023.027642
Received 07 November 2022; Accepted 07 April 2023; Issue published 25 June 2023
Abstract
Inorganic phosphate (Pi) is one of the main nutrients necessary for plant growth. Phosphate transporters mediate the acquisition, transport and recycling of phosphate, which is essential for plant growth and development. Although PHT1 has been reported in many plants at home and abroad, it is rarely studied in potato. Therefore, it is of great significance to study the PHT1 family members in order to understand the molecular response mechanism of potato in low phosphate state. In this study, a total of 6 potato PHT1 genes were identified and isolated. It was found that after the expression of different members of potato PHT1 gene, there were certain differences in amino acids and proteins, and the transmembrane domains ranged from 6 to 12. The difference in the secondary and tertiary protein structure of potato PHT1 also led to a difference in protein morphology. In addition, the expression of the PHT1 gene in potato increased obviously during 3~9 h of Pi deficiency stress. Overall, the expression levels of different genes in roots, stems and leaves are distinctly different, but the expression levels of the StPHT1;6 and StPHT1;10 genes are very high in roots, stems and leaves, indicating that these two genes may participate in the absorption of Pi in potato and play a role in Pi translocation. These two genes play a major role in the regulation of expression under short-term Pi deficiency stress. Our results provide an important reference for further understanding the evolution and function of potato phosphate transporters, and have important significance for improving the ability of potato to tolerate low Pi.Keywords
Plant growth requires a large amount of P, which is not only an important component of nucleic acids, phospholipids, ATP, ADP and sugars, but also participates in various metabolic and material transportation pathways [1–3]. Phosphorus, as one of the most limiting nutrient elements for plant growth and crop production in the world [4–7], plays an irreplaceable role in plant growth. Although the total amount of P in the soil is very high, most of it exists in the form of an organic state that cannot be absorbed by plants [8], and the mobility of P in the soil is low, the effective utilization rate is poor, and it is easy to be adsorbed and fixed by cations or organic compounds in the soil that convert P into insoluble and organic forms that cannot be absorbed and utilized by plants [9–17]. Phosphate transporters are the main transporters involved in the uptake of Pi (refers to H2PO4− in this paper) from the soil and its reuse and distribution in plants [18,19], they can also enable plants to resist the damage caused by various stresses by regulating the absorption and transportation of Pi under a variety of abiotic stress conditions to maintain the normal growth and development of plants [20].
The phosphate transporter family contains many members. According to the different gene protein structures, subcellular localizations and functions in plants, they can be divided into four subfamilies, named PHT1, PHT2, PHT3, PHT4 [15,21–26]. Different families of phosphate transporter genes play different roles in the process of plant growth and development [1,26,27]. PHT1 is the family with the most research results at present. It plays an important role in the process of Pi absorption, transport, distribution and reuse in plants [9,28–31]. PHT1 subfamily proteins belong to the 9th branch of the major facility superfamily (MFS) in structure. They usually have 12 transmembrane-spanning domains [8,15,16,32,33]. They are located on the plasma membrane and use the H+-gradient at the plasma membrane to provide energy to drive the absorption of Pi by plants [15,16]. In general, the absorption and transport of one Pi by PHT1 family genes requires 2~4 protons to enter the plasma membrane together with the change in membrane potential [16,34].
The first plant phosphate transporter gene (PHT1) was cloned from Arabidopsis [35]. It is a high-affinity phosphate transporter gene located on the cytoplasmic membrane and has a structure and function similar to those of PHO84 cloned from Saccharomyces cerevisiae [8,23,33,36]. With increasing attention given to phosphate transporter genes, many related studies have been conducted, and this type of phosphate transporter gene has been found in rice, maize, tomato, sorghum, soybean, millet, poplar, wheat, pepper, tobacco, potato and other plants [3,19,21,23,26,37–44]. Among them, 13 PHT1 genes were identified in rice (Oryza sativa) and maize (Zea mays) [40,45,46], 8 PHT1 genes in tomato (Solanum lycopersicum) [43], 11 PHT1 genes in sorghum (Sorghum bicolor) [42], 14 PHT1 genes in soybean (Glycine max) [44], 12 PHT1 genes in millet (Setaria italica) [3,47], 14 PHT1 genes in poplar (Populus trichocarpa) [23], 35 PHT1 genes in wheat (Triticum aestivum) [21]and 49 PHT1 genes in oilseed rape (Brassica napus) [48], and 8 PHT1 genes in potato (Solanum tuberosum) [19,49]. Different phosphate transporter genes involve different functions. At present, the molecular mechanism of the PHT1 genes in Arabidopsis and rice is relatively clear. AtPHT1;1, AtPHT1;2 AtPHT1;3 and AtPHT1;4 genes in Arabidopsis, are mainly involved in the uptake of Pi; AtPHT1;5 gene is responsible for the mobilization of Pi from the source to sink organs; AtPHT1;8 and AtPHT1;9 genes are likely to act sequentially in the interior of the plant during the root-to-shoot translocation of Pi [8,22,25,50–53]. In rice, OsPHT1; 4 is mainly expressed in roots and leaves, OsPHT1; 8 is mainly expressed in roots and stems, among which OsPHT1;1, OsPHT1;4 and OsPHT1;8 all plays in rice Pi uptake and translocation [25,26,37]. At the same time, OsPHT1; 8 also plays an important role in redistribution of Pi from source to sink organs [38].
The potato (Solanum tuberosum L.) is an annual herb of Solanaceae. It is the fourth largest food crop in the world after rice, wheat and maize. Because of its high yield, wide distribution and strong adaptability, it has become the main food crop in many countries and regions. Phosphorus plays a key role in potato root development, tuber formation, nutritional quality and resistance to some diseases [54,55]. The lack of available phosphorus in the soil will inhibit the growth and development of potatoes and reduce the yield and quality of potatoes. However, at present, there are few studies on the function of potato PHT1 gene. Therefore, the study of potato PHT1 gene is of great significance to explore the molecular response mechanism of Pi stress and improve the utilization efficiency of Pi in potato. In this study, potato hydroponic seedlings were used as experimental materials to clone potato phosphate transporter PHTI family member StPHT1;1/1;6/1;9/1;10/1;12/1;13 and analyzed the expression pattern of potato PHTI family gene in different tissues and organs under Pi stress. The purpose of this study was to elucidate the function of potato phosphate transporter, explore the molecular response mechanism of potato under Pi stress, screen efficient Pi germplasm resources and provide basis for molecular breeding.
2.1 Test Materials and Treatment Methods
The plant material selected in this study was potato (Solanum tuberosum L.), and the variety is Atlantic (DXY); Prepare 60 potato tissue culture seedlings with neat and consistent growth and put them in an illuminated culture room (temperature: 25°C light/22°C dark, light cycle: 12 h light/12 h dark, light intensity: 240 μmol·m−2·s−1, relative humidity: 75%~80%). Full nutrient solution (1/2 Hoagland nutrient solution) (Pi = 1 mM) and nutrient solution lacking Pi (Pi = 0 mM) were prepared (Table S1), and the tissue culture seedlings were transplanted into full-nutrient solution (Pi = 1 mM), and the nutrient solution was changed every 5 days. After 20 days, half of the materials were transferred to nutrient solution lacking Pi, and Pi treatment was initiated (Pi = 1 mM and Pi = 0 mM). The roots, stems and leaves of 0, 3, 6, 9, 12, 24, 48 and 72 h were frozen in liquid nitrogen and stored at −80°C for later use, with 3 biological replicates per treatment.
2.2 RNA Extraction and Reverse Transcription
After grinding the tissue sample in liquid nitrogen, extract the total RNA according to the Easypure® Plant RNA Kit and store it at −80°C for standby; Refer to the instructions of the reverse transcription kit of Beijing TransGen Biotech Co., Ltd. (China), and use 20 μL Reverse Transcription 2 μg total RNA, and reverse transcribed cDNA were detected and stored at −20°C for standby.
2.3 Gene Cloning and Entry Vector Construction
According to the screening results of the NCBI gene library, and referring to the CDS sequence of the Arabidopsis PHT1 gene, the homologous CDS sequence of the corresponding potato was found from the potato genome database (http://solanaceae.plantbiology.msu.edu/integrated_searches.shtml) using the homologous conservative sequence, and the cloning primer design (Table S2) was performed using Primer Premier 5.0 software. According to the predicted sequence, there are 12 PHT1 members in potato genome, which are named StPHT1;1, StPHT1;2, StPHT1;3, StPHT1;4.5, StPHT1;6, StPHT1;7, StPHT1;8, StPHT1;9, StPHT1;10, StPHT1;11, StPHT1;12 and StPHT1;13, respectively. The 12 PHT1 gene family members are obtained by PCR. The target gene is connected to the entry vector (pGWCm-T) that has been enzyme-digested (enzyme digestion site: BamHI) by in-fusion method, which is named pGWCPHT1;1, pGWCPHT1;6, pGWCPHT1;9, pGWPHT1;10, pGWCPHT1;12 and pGWCPHT1;13, respectively. The PCR reaction procedure is: pre-denatured at 95°C for 2 min; Denatured at 94°C for 30 s; Anneal at 55°C for 30 s, extend at 72°C for 2 min, and cycle for 25 times, extend 72°C for 5 min. The total system of In-fusion connection reaction is: The PCR amplification product (1.5 μL), the enzyme digestion vector pGWC-T (2.5 μL), and 5 × Ligation-Free cioning MasterMix (1 μL) were mixed to undergo ice bath for 1 h, and then transformed into E. coli DH5α. The samples were smears on LB solid screening plates containing 34 μM chloramphenicol, and cultured inverted at 37°C for 12 to 16 h. Single colonies were selected for PCR identification. See the appendix for the corresponding primers (Table S3).
2.4 Bioinformatics Analysis of Target Genes
First, the ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.htlm) of NCBI was used to find the ORF. The GenBank BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and ClustalW software (http://www.ebi.ac.uk/clustalw) were used for sequence homology analysis. In EXPASY, the isoelectronic points and molecular weight of the encoded protein were predicted by using the Protparem program (http://www.expasy.org/tools/pi_tool.html). The hydrophobicity of the encoded protein was predicted and analyzed with the ProtScale program (http://www.expasy.ch/tools/protscale). Using the online transmembrane-spanning domains prediction software TMHMM 2.0 provided by ExPASY (http://www.cbs.dtu.dk/services/TMHMM/), the transmembrane structure of the encoded protein was analyzed. CFSSP online software was used to predict the secondary structure of protein (http://www.biogem.org/tool/chou-fasman/). Phyre 2 protein tertiary structure model construction software was used to predict protein tertiary structure (http://www.sbg.bio.ic.ac.uk/phyre2/phyre2_output/87fecb0d043f5d4d/summary.html).
According to the obtained potato PHT1 phosphate transporter gene (StPHT1;1, StPHT1;6, StPHT1;9, StPHT1;10, StPHT1;12 and StPHT1;13) Specific primers were designed (Table S4), and the expression characteristics of genes in potato roots, stems and leaves at different times were detected by RT-qPCR. The RT-qPCR reaction system was based on the TransScript® All-in-One kit instructions, and the reaction was performed on QuantStudio 7 Flex qPCR instrument. The total reaction system was 20 μL, including 0.5 μL cDNA template, 10 μL EvaGreen 2XqPCR Mastermix, Forwared Primer (10 μM) and Reverse Primer (10 μM) 0.4 μL, respectively. 8.3 μL Nuclease-free H2O. RT-qPCR procedure: pre-denaturation 94°C for 30 s, Denatured 94°C for 5 s, annealing 55°C for 35 s, 45 cycles. All test samples were repeated for 3 times. 2−ΔΔCt was used to calculate the relative gene expression.
RT-qPCR data analysis was performed using Excel 2013, LinRegPCR and SPSS 25 to statistically analyze the results and calculate the average expression amount and standard error of genes. GraphPad Prism 5, Adobe Illustrator CS6 and Origin 2021 software were used for drawing.
3.1 Cloning and Vector Construction of PHT1 Gene
The PHT1 genes were cloned (Fig. S1). The results showed that 10 members of the predicted 12 PHT1 gene family could be cloned by designed primer PCR. They were StPHT1;1, StPHT1;3, StPHT1;4, StPHT1;6, StPHT1;7, StPHT1;8, StPHT1;9, StPHT1;10, StPHT1;12, and StPHT1;13. By comparison with the marker, we can roughly see that the sequence length of these 10 members is approximately 1500 to 2000 bp. The construction of the entry vector for the PHT1 gene (Fig. S2) showed that the base fragment sizes of pGWCPHT1;1, pGWCPHT1;6, pGWCPHT1;9, pGWCPHT1;10, pGWCPHT1;12 and pGWCPHT1;13 were 1901, 1944, 1789, 1787, 1645 and 987 bp, respectively (Table S5).
3.2 Bioinformatics Analysis of the PHT1 Gene
The transmembrane-spanning domains of PHT1 protein were analyzed (Fig. 1). The results showed that the potato PHT1 protein was a transmembrane protein, where StPHT1;1 and StPHT1;6 have 12 transmembrane-spanning domains; StPHT1;9 has 10 transmembrane-spanning domains; StPHT1;10 has 11 transmembrane-spanning domains; and StPHT1;12 and StPHT1;13 have 9 and 6 transmembrane-spanning domains, respectively. The protein secondary structures of PHT1, such as α-helix β-sheet β-turns and random coils, were analyzed (Fig. 2, Table 1). The results showed that the protein secondary structure of potato PHT1 had certain differences among different members. Among the α-helix structures, the StPHT1;1 protein accounted for the greatest proportion, reaching 84.4%; the StPHT1;10 protein accounted for the smallest proportion, reaching 74.3%; the β-sheet in the StPHT1;6 protein accounted for the most, up to 72.3%; the StPHT1;13 protein accounted for the least, reaching 52.4 %; β-turn in the StPHT1;12 protein accounted for the most, reaching 9.7%; and the StPHT1;10 proportion was the smallest, at 8.4%. The protein tertiary structure of PHT1 was analyzed (Fig. 3, Table S6). The results showed that in the predicted protein 3D model, the number of amino acid residues of four PHT1 proteins, StPHT1;1, StPHT1;6, StPHT1;9 and StPHT1;10, is large in potato, and the protein tertiary structure is dense. Because the number of amino acid residues of StPHT1;12 and StPHT1;13 proteins is small, the protein tertiary structure of StPHT1;12 and StPHT1;13 proteins is relatively loose.
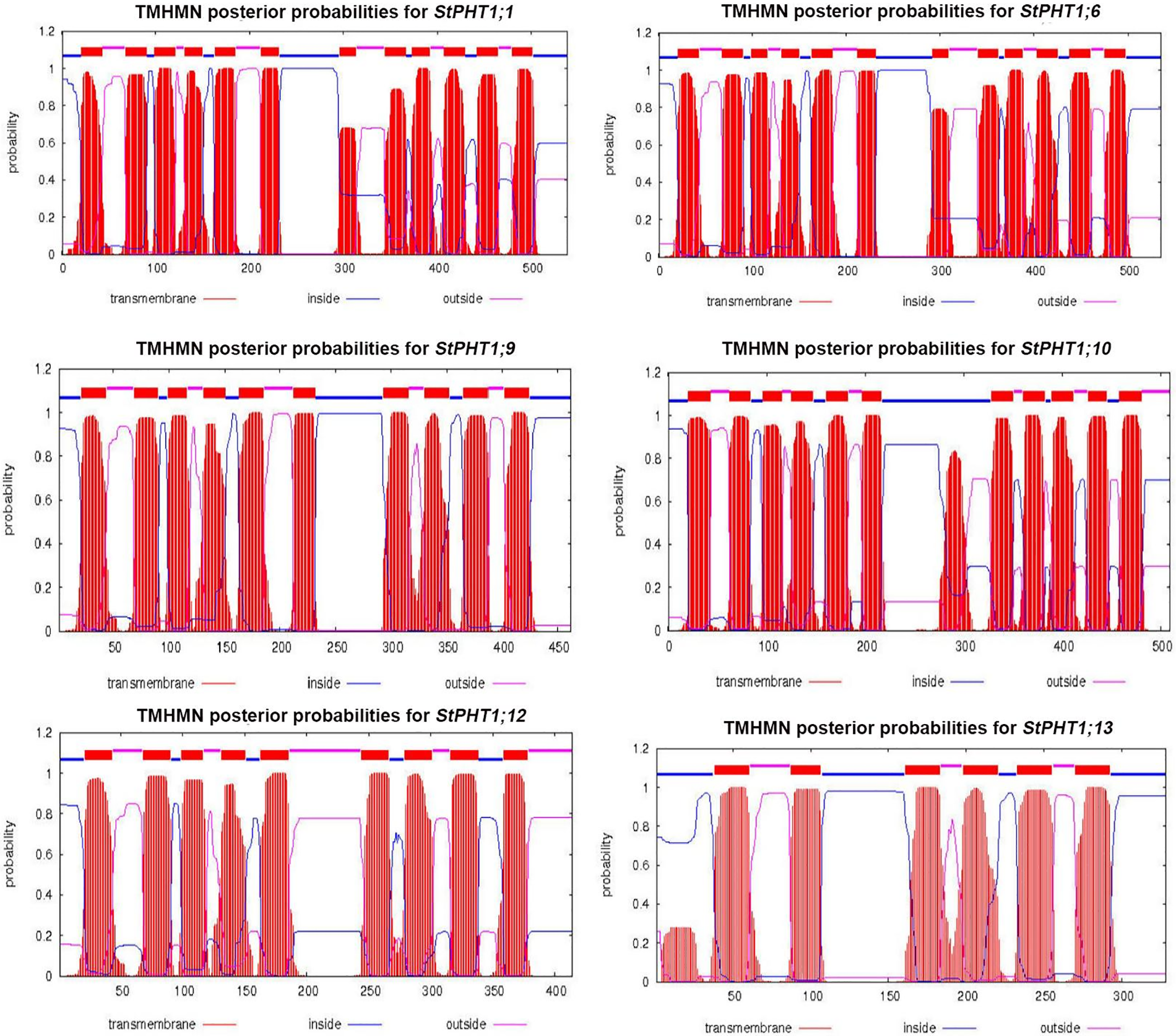
Figure 1: Transmembrane-spanning domain analysis of potato PHT1 protein
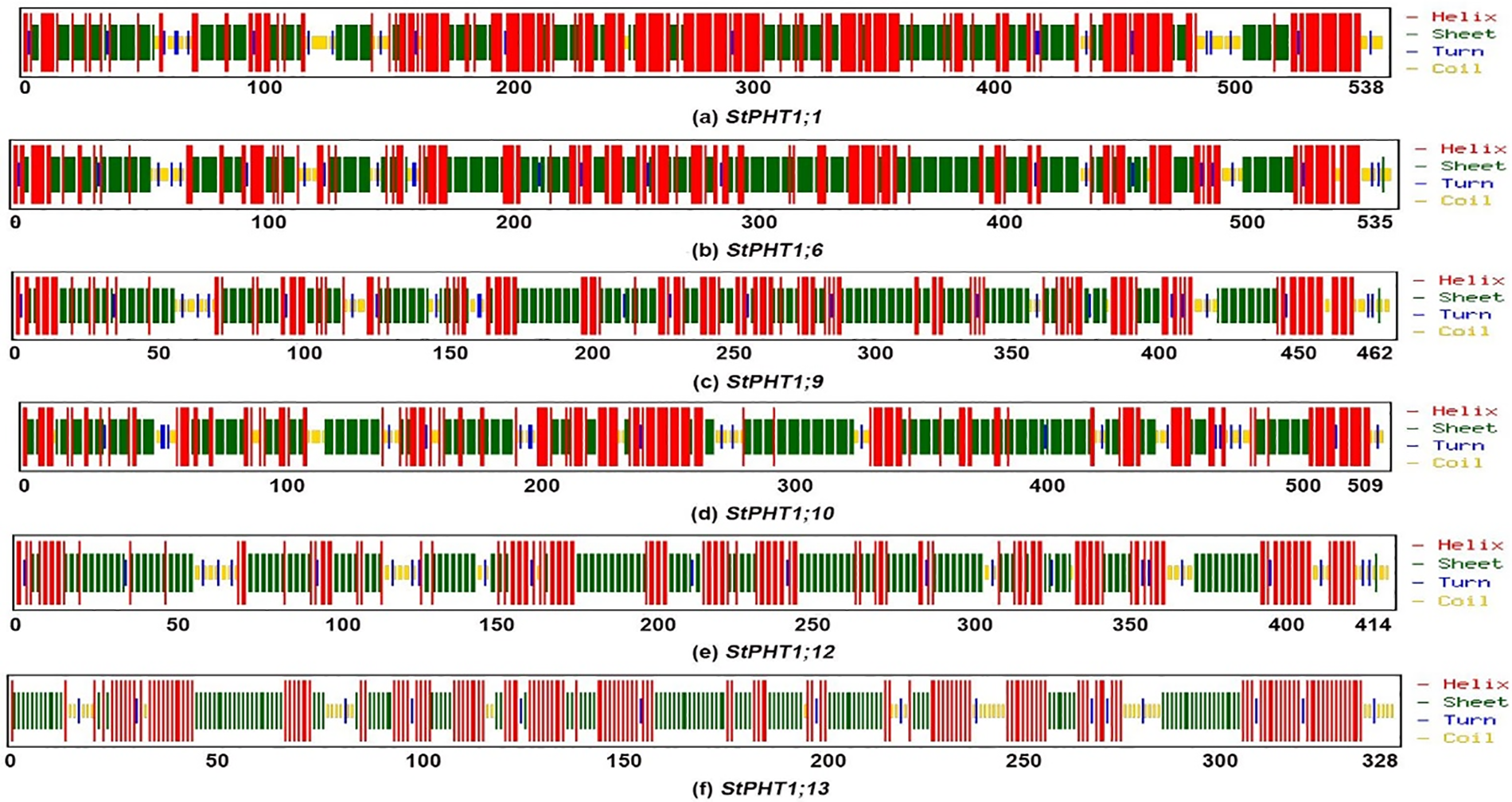
Figure 2: Potato PHT1 protein secondary structure prediction diagram
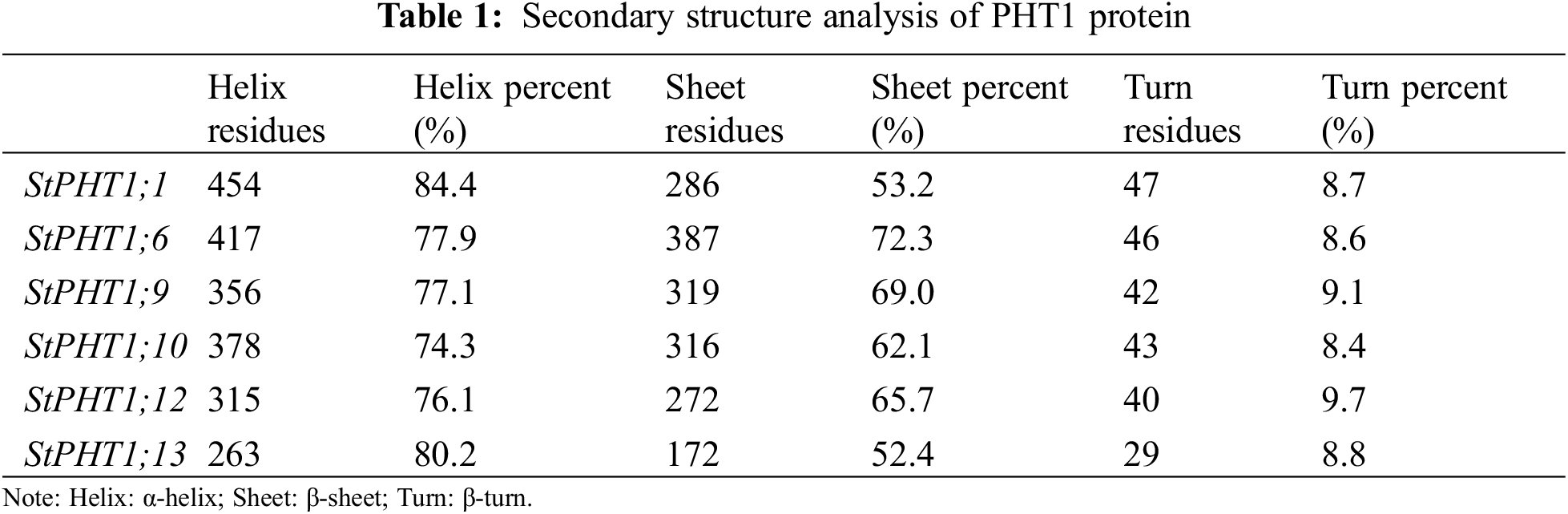
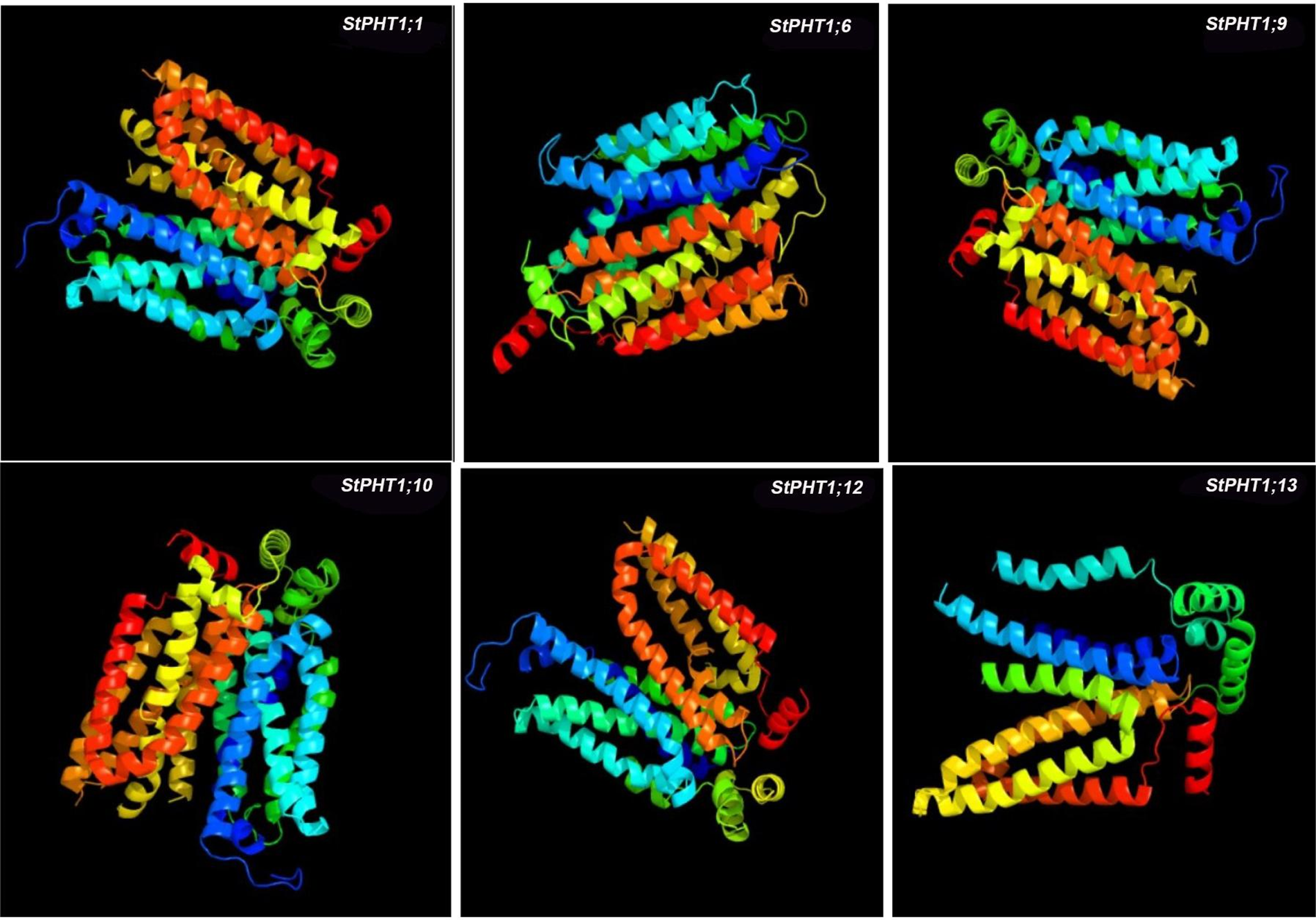
Figure 3: Prediction of the tertiary structure of the PHT1 protein
3.3 Expression Analysis of the PHT1 Gene in Potato under Short-Term Phosphate-Deficiency Stress
The expression of the PHT1 gene in potato under short-term Pi deficiency stress was analyzed (Fig. 4). The results showed that under short-term Pi-deficiency stress, the expression of the StPHT1;1, StPHT1;6, StPHT1;9, StPHT1;10, and StPHT1;12 genes in potato increased at 3~9 h, while the expression of the StPHT1;13 gene increased at 48~72 h. Among them, the expression of the StPHT1;1 gene reached a maximum in the leaves under Pi-deficiency stress for 3 h, which was 71 times more than that of the control group, 28 times and 31 times more than that of the control group in the stems under stress for 6 and 9 h, and 26 times more than that of the control group in the roots under stress for 6 h. The expression of the StPHT1;6 gene reached the maximum value in the stems under Pi-deficiency stress for 9 h, which was 59 times that of the control group; the maximum value in the leaves under Pi-deficiency stress for 3 h, which was 48 times higher than that of the control group; and the maximum value in the roots under Pi-deficiency stress for 72 h, which was 25 times higher than that of the control group. In addition, there was also a high level of expression in stems under stress for 3 and 6 h, which was 40~45 times higher than that of the control group and 18 times higher than that of the control group in roots under stress for 6 and 9 h. The expression of the StPHT1;9 gene in stems and leaves under Pi stress for 3 h reached a maximum, which was 39 and 20 times higher than that of the control group, respectively. The expression in roots under Pi stress for 24 h was the maximum, which was 25 times higher than that of the control group. The expression of the StPHT1;10 gene reached a maximum value of 91 and 50 times higher than that of the control group in the stems and leaves under Pi stress for 6 h and more than 100 times that of the control group in the roots at 72 h. In addition, there was also high expression in the roots at 6 and 9 h, which was approximately 60 times higher than that of the control group. The expression of the StPHT1;12 gene was the largest in stems and leaves under Pi stress for 3 h, approximately 80 times that of the control group, but there was little change in the expression of the StPHT1;12 gene in roots. The expression of the StPHT1;13 gene was higher in the leaves and stems under Pi stress for 3 and 72 h, but its peak appeared in the roots and leaves under stress for 48 h, which was about 60 times higher than that of the control group.
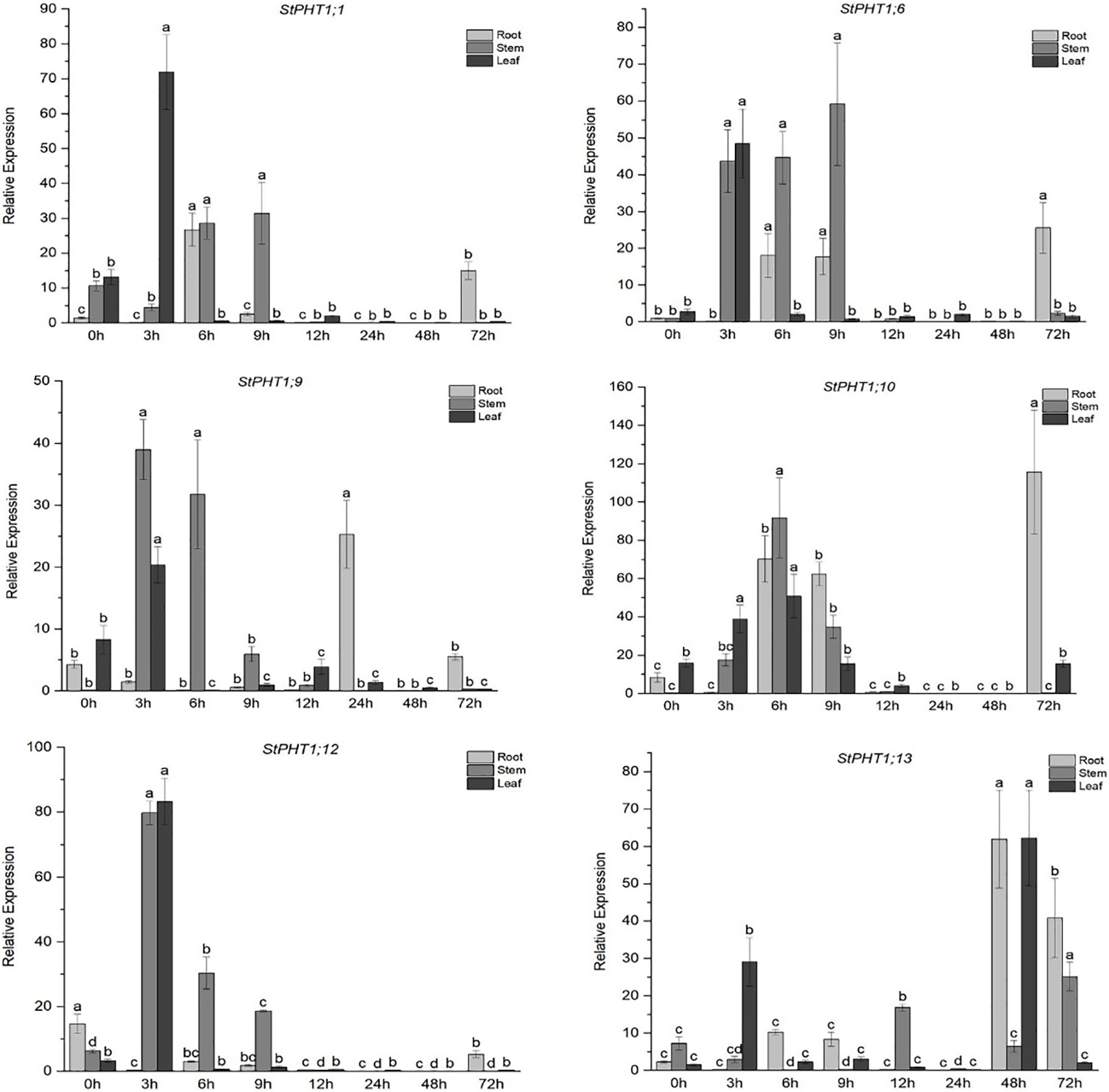
Figure 4: Transient expression of PHT1 under a lack of phosphate
Phosphorus plays an important role in plant growth, and the Pi concentration in soil solution is usually less than 2 μM. However, the concentration of Pi in plant tissue can be greater than 10 mM [10,12,50]. It is precisely because of the gradient difference in the concentration of Pi in soil and plant tissue that the phosphate transporter is indispensable. The PHT1 subfamily is the most widely studied family in the current research on phosphate transporter family. However, the functions of many other members of potato PHT1 family are still unclear. Therefore, the isolation and cloning of potato phosphate transporter genes is of great significance to explore the molecular response mechanism of potato under Pi stress. In this study, we cloned 6 members of potato PHT1 gene family (StPHT1;1, StPHT1;6, StPHT1;9, StPHT1;10, StPHT1;12, StPHT1;13), Bioinformatics analysis results showed that potato PHT1 protein contain 9~11 transmembrane-spanning domains (Fig. 1), which is consistent with the results of previous studies which demonstrated that the 8 members of the potato PHT1 gene contain 9~11 transmembrane-spanning domains [19,49]. As a phosphate transporter, the multi-transmembrane region means the improvement of transport efficiency. At the same time, the number of transmembrane-spanning domains of the potato PHT1 gene is similar to that of maize [56], wheat [57], rice [45,58,59] and indicating that the protein is highly conserved in the number of transmembrane-spanning domains of different plants. In the analysis of protein secondary structure of PHT1 (Fig. 2, Table 1), Our results show that α-helix accounts for most of the amino acid residues of the protein, which is consistent with previous research results [49,60], which is key to the folding of the protein to form the domain. In the analysis of protein tertiary structure of PHT1 (Fig. 3), we can also clearly see that all proteins except StPHT1;13 have a large number of helical structures. The coiled coil of proteins makes the tertiary structure of proteins compact and orderly. while StPHT1;13 protein looks looser than other proteins, which may be due to StPHT1;13 proteins have less secondary structure due to containing fewer amino acid residues, which results in less interaction between various forces. The resulting protein tertiary structure is relatively loose. Overall, the morphology of the PHT1 protein members is relatively similar, which reflects the conservation of the PHT1 protein structure.
Most PHT1 genes are transcribed through Pi starvation induction, and are regulated by Pi-starvation signals, and the expression level will be significantly up-regulated under Pi deficiency conditions [61–67]. In Brassica napus, Pi deficiency can significantly upregulate the expression of approximately 80% of BnaPHT1 gene in roots [48]. In potato, Pi deficiency can also increase the expression of most PHT1 genes in leaves and roots, including StPHT1;4, StPHT1;7 and StPHT1;8 genes were upregulated in potato roots, StPHT1;1, StPHT1;4 and StPHT1;5 genes were upregulated in potato leaves [19]. In Arabidopsis, PHT1;9 gene was highly expressed in Pi deficiency roots [33]. In rice, the transcript level of OsPHT1;4 increased significantly both in shoots and roots with a long time Pi deprivation, the expression of OsPHT1;8 was significantly upregulated only in roots [25,26]. In millet, Pi deficiency will lead to PHT1;2, PHT1;3, PHT1;4 and PHT1;9 genes were overexpressed in roots [47]. In barley, HvPHT1;1 and HvPHT1;2 under the condition of Pi deficiency, expression was induced by up to 5-fold [65]. In this study, we found that the relative expression of potato phosphate transporter gene under short-term Pi stress changed with time, which may be because the concentration of Pi in the culture medium changed with the growth of the plant, thus affecting the gene expression. On the whole, StPHT1;1, StPHT1;6, StPHT1;9, StPHT1;10, StPHT1;12 and StPHT1;13 genes were up-regulated under the condition of short Pi deficiency, indicating that these PHT1 genes were positive regulatory inducible genes in response to low Pi stress, with the characteristics of PHT1 family gene expression, consistent with the results of Arabidopsis [22] and Brassica napus [48]. Secondly, under the condition of low Pi, the expression of StPHT1;1 and StPHT1;12 genes decreased in the underground part, but increased in the aboveground part, and the expression gradually decreased after reaching the maximum at 3 h. It is consistent with previous studies that the expression of OsPT1 and OsPT12 genes in rice PHT1 family members decreased in the underground part and increased in the aboveground part of OsPT1 under low Pi treatment [68].
PHT1 genes, as high-affinity phosphate transporters [21,28,69], are strongly expressed in roots, especially in root epidermal and outer cortical cells, and are related to the uptake of Pi, while the expression in other parts may also be related to the transfer of Pi [16]. In this study, we found that PHT1 gene was detected in roots, stems and leaves from 0 to 72 h, but the time of maximum expression of different genes was different, and the expression of most genes increased to a great extent from 3 to 9 h, it has also been reported in the relevant literature that 6 and 9 h are key points for the expression of some genes in plants. This phenomenon may indicate that the expression of the PHT1 gene was improved in a short time under Pi deficiency. Among them, the up-regulated expression of StPHT1;9 and StPHT1;13 genes in the roots were higher than that in the shoots after low Pi stress, which indicates that StPHT1;9 and StPHT1;13 was mainly expressed in the roots. It is speculated that these genes may participate in the uptake of Pi in potato, which is consistent with the result that ZmPHT1;9 gene was mainly expressed in the root under low Pi stress found in previous studies in maize [56]; After low Pi stress, the up-regulated expression of StPHT1;1 and StPHT1;12 genes in the shoots were higher than that in the roots, indicating that StPHT1;1 and StPHT1;12 genes were mainly expressed in the shoots. It is speculated that these genes may participate in the transport of Pi in potato; While StPHT1;6 and StPHT1;10 genes were expressed in roots, stems and leaves, and the expression level was very high, which indicates that StPHT1;6 and StPHT1;10 genes not only play an important role in the process of Pi absorption from roots, but also may participate in the balance regulation of Pi transport between roots and shoots, and play a major role in the expression regulation under short-term Pi deficiency stress. It is consistent with the results that the expression of OsPT2, OsPT6, OsPT9 and OsPT10 genes in roots and shoots of PHT1 genes induced by Pi deficiency increased exponentially in previous studies in rice [70], and these results also indicate that PHT1 family genes may play a similar role in Pi uptake and transport of different crops. The situation that multiple genes cooperated to improve the expression level can show the interaction and synergistic expression of phosphate transporter genes. It is the synergy of phosphate transporters that can ensure the distribution of Pi in specific tissues, cells and organelles and maintain the growth and development of plants even under stress conditions. Therefore, different phosphate transporters provide an opportunity for genetic engineering to improve Pi utilization to optimize the absorption and distribution of Pi in plants.
In addition, under short-term Pi deficiency stress, the change in gene expression in the stems and leaves of potato plants was the most obvious. This situation is caused by the fact that in the short-term Pi deficiency state, plants must first maintain their own homeostasis and regulate their own Pi transport in the body. Therefore, the gene expression level in the stems and leaves increased sharply. However, with the passage of time, at 72 h of Pi-deficiency stress, the expression level of the gene in the root was greatly improved, which indicates that over time, the plant itself begins to absorb Pi from the external environment to maintain the demand for Pi in the body, so PHT1 gene expression in the plant root increases. However, the expression of the PHT1 gene after more than 72 h and the effect of long-term Pi deficiency on the expression of the PHT1 gene in potato need to be further verified by experiments.
Although people have some knowledge of potato PHT1 gene, it is still in the preliminary stage. Its molecular regulation mechanism and the maintenance of Pi dynamic balance need further research, in order to provide theoretical and technical reference for potato production and research.
Six PHT1 genes were isolated and identified in the potato genome, all of which were induced by low Pi stress. Among them, StPHT1;6, StPHT1;9, StPHT1;10 and StPHT1;13 genes were strongly expressed in the roots under low Pi conditions, StPHT1;6, StPHT1;9, StPHT1;10 and StPHT1;12 genes were strongly expressed in the stems under low Pi conditions, StPHT1;1, StPHT1;6, StPHT1;10, StPHT1;12 and StPHT1;13 genes were strongly expressed in the leaves under low Pi conditions, and StPHT1;6 and StPHT1;10 genes might be potential candidate genes for producing transgenic potatoes with improved Pi efficiency. All these results indicate that PHT1 genes play an important role in the uptake and transport of plant Pi, and phosphate transporters may play a role in complex combinations in the uptake and transport of Pi. The identification of PHT1 gene is helpful to improve the Pi efficiency of potato and obtain Pi efficient germplasm resources.
Acknowledgement: Thanks to Xinyong Zhang for his suggestions on this article.
Funding Statement: This study was supported by the National Natural Science Foundation of China (31460367).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: XZ; data collection: LZ, QT; analysis and interpretation of results: YD, LZ, YB; draft manuscript preparation: YD, YB. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its Supplementary Materials).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Rausch, C., Bucher, M. (2002). Molecular mechanisms of phosphate transport in plants. Planta, 216(1), 23–37. https://doi.org/10.1007/s00425-002-0921-3 [Google Scholar] [PubMed] [CrossRef]
2. Raghothama, K. G. (1999). Phosphate acquisition. Annual Review Plantphysiology and Plant Molecular Biology, 50(1), 665–693. https://doi.org/10.1146/annurev.arplant.50.1.665 [Google Scholar] [PubMed] [CrossRef]
3. Ceasar, S. A., Hodge, A., Baker, A., Baldwin, S. A. (2014). Phosphate concentration and arbuscular mycorrhizal colonisation influence the growth, yield and expression of twelve PHT1 family phosphate transporters in foxtail millet (Setaria italica). PLoS One, 9(9), e108459. https://doi.org/10.1371/journal.pone.0108459 [Google Scholar] [PubMed] [CrossRef]
4. Schachtman, D. P., Reid, R. J., Ayling, S. M. (1998). Phosphorus uptake by plants: From soil to cell. Plant Physiology, 116(2), 447–453. https://doi.org/10.1104/pp.116.2.447 [Google Scholar] [PubMed] [CrossRef]
5. Bulgarelli, R. G., Oliveira, V. H. D., Andrade, S. A. L. (2020). Arbuscular mycorrhizal symbiosis alters the expression of PHT1 phosphate transporters in roots and nodules of P-starved soybean plants. Theoretical and Experimental Plant Physiology, 32(3), 243–253. https://doi.org/10.1007/s40626-020-00185-8 [Google Scholar] [CrossRef]
6. Maharajan, T., Ceasar, S. A., Krishna, T. P. A., Ramakrishnan, M., Duraipandiyan, V. et al. (2018). Utilization of molecular markers for improving the phosphorus efficiency in crop plants. Plant Breeding, 137(1), 10–26. https://doi.org/10.1111/pbr.12537 [Google Scholar] [CrossRef]
7. Sun, Y., Cui, X., Liu, F. (2015). Effect of irrigation regimes and phosphorus rates on water and phosphorus use efficiencies in potato. Scientia Horticulturae, 190, 64–69. https://doi.org/10.1016/j.scienta.2015.04.017 [Google Scholar] [CrossRef]
8. Zhuo, J., Lu, M., Zhang, C., Qu, X., Liu, Y. et al. (2020). Isolation and functional characterisation of the PHT1 gene encoding a high-affinity phosphate transporter in Camellia oleifera. Journal of Horticultural Science & Biotechnology, 95(5), 553–564. https://doi.org/10.1080/14620316.2019.1703562 [Google Scholar] [CrossRef]
9. Nussaume, L., Kanno, S., Javot, H., Marin, E., Pochon, N. et al. (2011). Phosphate import in plants: Focus on the PHT1 transporters. Frontiers in Plant Science, 2(83), 83–95. https://doi.org/10.3389/fpls.2011.00083 [Google Scholar] [PubMed] [CrossRef]
10. Sun, C., Xiao, K., Han, S., Zhao, F., Bao, J. (2011). Research progress on molecular mechanism of phosphorus uptake and transport in plants. Journal of Agricultural Science and Technology, 13(2), 17–24. https://doi.org/10.3969/j.issn.1008-0864.2011.02.03 [Google Scholar] [CrossRef]
11. Grennan, A. K. (2008). Phosphate accumulation in plants: Signaling. Plant Physiology, 148(1), 3–5. https://doi.org/10.1104/pp.104.900269 [Google Scholar] [PubMed] [CrossRef]
12. Bieleski, R. L. (1973). Phosphate pools, phosphate transport, and phosphate availability. Annual Review of Plant Physiology, 24(1), 225–252. https://doi.org/10.1146/annurev.pp.24.060173.001301 [Google Scholar] [CrossRef]
13. Raghothama, K. G. (2000). Phosphate transport and signaling. Current Opinion in Plant Biology, 3(3), 182–187. https://doi.org/10.1016/S1369-5266(00)80063-1 [Google Scholar] [CrossRef]
14. White, P. J., Hammond, J. P. (2008). The ecophysiology of plant-phosphorus interactions. UK: Springer Netherlands. [Google Scholar]
15. Deng, M., Wang, F., Mao, C. (2017). Plant phosphate transporter and its molecular regulation mechanism. Plant Physiology Journal, 53(3), 377–387. https://doi.org/10.13592/j.cnki.ppj.2017.0003 [Google Scholar] [CrossRef]
16. Dong, X., Wang, X., Shi, L., Cai, H. M., Xu, F. S. et al. (2017). Research progress of plant phosphorus transporter Pht1 family. Journal of Plant Nutrition and Fertilizers, 23(3), 799–810. https://doi.org/10.11674/zwyf.16224 [Google Scholar] [CrossRef]
17. Hirsch, J., Marin, E., Floriani, M., Chiarenza, S., Richaud, P. et al. (2006). Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie, 88, 1767–1771. https://doi.org/10.1016/j.biochi.2006.05.007 [Google Scholar] [PubMed] [CrossRef]
18. Roch, G. V., Maharajan, T., Ceasar, S. A., Ignacimuthu, S. (2019). The role of PHT1 family transporters in the acquisition and redistribution of phosphorus in plants. Critical Reviews in Plant Sciences, 38(1/3), 171–198. https://doi.org/10.1080/07352689.2019.1645402 [Google Scholar] [CrossRef]
19. Liu, B., Zhao, S., Wu, X., Wang, X., Nan, Y. et al. (2017). Identification and characterization of phosphate transporter genes in potato. Journal of Biotechnology, 264(25), 17–28. https://doi.org/10.1016/j.jbiotec.2017.10.012 [Google Scholar] [PubMed] [CrossRef]
20. Li, W. (2020). Identification of phosphorus transporter family in potatoes (Solanum tuberosum) and study on PHO1 subfamily to improve heat tolerance of transgenic potatoes (Ph.D. Thesis). Northwest A&F University, Xianyang, China. https://doi.org/10.27409/d.cnki.gxbnu.2020.001459 [Google Scholar] [CrossRef]
21. Zhang, Y., Hu, L., Yu, D., Xu, K., Zhang, J. et al. (2019). Integrative analysis of the wheat PHT1 gene family reveals a novel member involved in arbuscular mycorrhizal phosphate transport and immunity. Cells, 8(5), 490–516. https://doi.org/10.3390/cells8050490 [Google Scholar] [PubMed] [CrossRef]
22. Li, R., Wang, J., Xu, L., Sun, M., Yi, K. et al. (2020). Functional analysis of phosphate transporter OsPHT4 family members in rice. Rice Science, 27(6), 493–503. https://doi.org/10.1016/j.rsci.2020.09.006 [Google Scholar] [CrossRef]
23. Zhang, C., Meng, S., Li, M., Zhao, Z. (2016). Genomic identification and expression analysis of the phophate transporter gene family in poplar. Frontiers in Plant Science, 7, 1398–1412. https://doi.org/10.3389/fpls.2016.01398 [Google Scholar] [PubMed] [CrossRef]
24. Wang, F., Deng, M., Xu, J., Zhu, X., Mao, C. (2018). Molecular mechanisms of phosphate transport and signaling in higher plants. Seminars in Cell & Developmental Biology, 74, 114–122. https://doi.org/10.1016/j.semcdb.2017.06.013 [Google Scholar] [PubMed] [CrossRef]
25. Ye, Y., Yuan, J., Chang, X., Yang, M., Zhang, L. et al. (2015). The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS One, 10(5), e0126186. https://doi.org/10.1371/journal.pone.0126186 [Google Scholar] [PubMed] [CrossRef]
26. Jia, H., Ren, H., Gu, M., Zhao, J., Sun, S. et al. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiology, 156(3), 1164–1175. https://doi.org/10.1104/pp.111.175240 [Google Scholar] [PubMed] [CrossRef]
27. Wang, G., Zhang, C., Battle, S., Lu, H. (2014). The phosphate transporter PHT4;1 is a salicylic acid regulator likely controlled by the circadian clock protein CCA1. Frontiers in Plant Science, 5, 701–711. https://doi.org/10.3389/fpls.2014.00701 [Google Scholar] [PubMed] [CrossRef]
28. López-Arredondo, D. L., Leyva-González, M. A., González-Morales, S. I., López-Bucio, J., Herrera-Estrella, L. (2014). Phosphate nutrition: Improving low-phosphate tolerance in crops. Annual Review of Plant Biology, 65(1), 95–123. https://doi.org/10.1146/annurev-arplant-050213-035949 [Google Scholar] [PubMed] [CrossRef]
29. Ding, G., Chen, S., Shi, L., Cai, H., Ye, X. (2013). Research progress on genetic regulation mechanism of plant tolerance to low phosphorus stress. Journal of Plant Nutrition and Fertilizers, 19(3), 733–744. https://doi.org/10.11674/zwyf.2013.0325 [Google Scholar] [CrossRef]
30. Baker, A., Ceasar, S. A., Palmer, A. J., Paterson, J. B., Qi, W. et al. (2015). Replace, reuse, recycle: Improving the sustainable use of phosphorus by plants. Journal of Experimental Botany, 66(12), 3523–3540. https://doi.org/10.1093/jxb/erv210 [Google Scholar] [PubMed] [CrossRef]
31. Chiou, T. J., Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology, 62(1), 185–206. https://doi.org/10.1146/annurev-arplant-042110-103849 [Google Scholar] [PubMed] [CrossRef]
32. Wang, P., Chen, A., Yu, L., Xu, G. (2006). Research progress of plant phosphorus transporter gene and its expression regulation. Journal of Plant Nutrition and Fertilizers, 12(4), 584–591. [Google Scholar]
33. Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Correia, I. S. et al. (2012). The PHT1;9 and PHT1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytologist, 195(2), 356–371. https://doi.org/10.1111/j.1469-8137.2012.04167.x [Google Scholar] [PubMed] [CrossRef]
34. Preuss, C. P., Huang, C. Y., Gilliham, M., Tyerman, S. D. (2010). Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiology, 152(3), 1431–1441. https://doi.org/10.1104/pp.109.152009 [Google Scholar] [PubMed] [CrossRef]
35. Muchhal, U. S., Pardo, J. M., Raghothama, K. G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 93(19), 10519–10523. https://doi.org/10.1073/pnas.93.19.10519 [Google Scholar] [PubMed] [CrossRef]
36. Bun-Ya, M., Nishimura, M., Harashima, S., Oshima, Y. (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Molecular and Cellular Biology, 11(6), 3229–3238. https://doi.org/10.1128/MCB.11.6.3229 [Google Scholar] [CrossRef]
37. Sun, S., Gu, M., Cao, Y., Huang, X., Zhang, X. et al. (2012). A constitutive expressed phosphate transporter, OsPht1;1 modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiology, 159(4), 1571–1581. https://doi.org/10.1104/pp.112.196345 [Google Scholar] [PubMed] [CrossRef]
38. Li, Y., Zhanga, J., Zhang, X., Fana, H., Gu, M. et al. (2015). Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Science, 230, 23–32. https://doi.org/10.1016/j.plantsci.2014.10.001 [Google Scholar] [PubMed] [CrossRef]
39. Zhang, F., Sun, Y., Pei, W., Jain, A., Sun, R. et al. (2015). Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. The Plant Journal, 82, 556–569. https://doi.org/10.1111/tpj.12804 [Google Scholar] [PubMed] [CrossRef]
40. Liu, F., Xu, Y., Jiang, H., Jiang, C., Du, Y. et al. (2016). Systematic identification, evolution and expression analysis of the zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. International Journal of Molecular Sciences, 17(6), 930–948. https://doi.org/10.3390/ijms17060930 [Google Scholar] [PubMed] [CrossRef]
41. Sisaphaithong, T., Kondo, D., Matsunaga, H., Kobae, Y., Hata, S. (2012). Expression of plant genes for arbuscular mycorrhiza-inducible phosphate transporters and fungal vesicle formation in sorghum, barley, and wheat roots. Bioscience, Biotechnology, and Biochemistry, 76(12), 2364–2367. https://doi.org/10.1271/bbb.120782 [Google Scholar] [PubMed] [CrossRef]
42. Walder, F., Brule, D., Koegel, S., Wiemken, A., Boller, T. et al. (2015). Plant phosphorus acquisition in a common mycorrhizal network: regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. The New Phytologist, 205, 1632–1645. https://doi.org/10.1111/nph.13292 [Google Scholar] [PubMed] [CrossRef]
43. Chen, A., Chen, X., Wang, H., Liao, D., Gu, M. et al. (2014). Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biology, 14, 61–76. https://doi.org/10.1186/1471-2229-14-61 [Google Scholar] [PubMed] [CrossRef]
44. Qin, L., Guo, Y., Chen, L., Liang, R., Gu, M. et al. (2012). Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS One, 7(10), e47726. https://doi.org/10.1371/journal.pone.0047726 [Google Scholar] [PubMed] [CrossRef]
45. Paszkowski, U., Kroken, S., Roux, C., Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences of the United States of America, 99(20), 13324–13329. https://doi.org/10.1073/pnas.202474599 [Google Scholar] [PubMed] [CrossRef]
46. Goff, S. A., Ricke, D., Lan, T., Presting, G., Wang, R. et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296(5565), 92–100. https://doi.org/10.1126/science.1068275 [Google Scholar] [PubMed] [CrossRef]
47. Maharajan, T., Ceasar, S. A., Krishna, T. P. A., Ignacimuthu, S. (2019). Phosphate supply influenced the growth, yield and expression of PHT1 family phosphate transporters in seven millets. Planta, 250(5), 1433–1448. https://doi.org/10.1007/s00425-019-03237-9 [Google Scholar] [PubMed] [CrossRef]
48. Li, Y., Wang, X., Zhang, H., Wang, S., Ye, X. et al. (2019). Molecular identification of the phosphate transporter family 1 (PHT1) genes and their expression profiles in response to phosphorus deprivation and other abiotic stresses in Brassica napus. PLoS One, 14(7), e0220374. https://doi.org/10.1371/journal.pone.0220374 [Google Scholar] [PubMed] [CrossRef]
49. Cao, M. X. (2020). Cloning and functional analysis of StPHT1;7 the potato (Solanum tuberosum L.) phosphate transporter related genes (Master Thesis). Northwest A&F University, Xianyang, China. https://doi.org/10.27409/d.cnki.gxbnu.2020.000751 [Google Scholar] [CrossRef]
50. Lapis-Gaza, H. R., Jost, R., Finnegan, P. M. (2014). Arabidopsis phosphate transportere1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biology, 14, 334–353. https://doi.org/10.1186/s12870-014-0334-z [Google Scholar] [PubMed] [CrossRef]
51. Shin, H., Shin, H. S., Dewbre, G. R., Harrison, M. J. (2004). Phosphate transport in Arabidopsis: PHT1;1 and PHT1;4 play a major role in phosphate acquisition from both low-and high-phosphate environments. The Plant Journal, 39(4), 629–642. https://doi.org/10.1111/j.1365-313X.2004.02161.x [Google Scholar] [PubMed] [CrossRef]
52. Nagarajan, V. K., Jain, A., Poling, M. D., Lewis, A. J., Raghothama, K. G. et al. (2011). Arabidopsis PHT1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiology, 156, 1149–1163. https://doi.org/10.1104/pp.111.174805 [Google Scholar] [PubMed] [CrossRef]
53. Ayadi, A., David, P., Arrighi, J., Chiarenza, S., Thibaud, M. et al. (2015). Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiology, 167(4), 1511–1526. https://doi.org/10.1104/pp.114.252338 [Google Scholar] [PubMed] [CrossRef]
54. Kelling, K. A., Rosen, C. J., Stark, J. C., Essah, S. Y. C. (2014). Proceedings from the 2012 PAA symposium: Potato phosphorus management and utilization for today and tomorrow. American Journal of Potato Research, 91, 119–120. https://doi.org/10.1007/s12230-014-9373-0 [Google Scholar] [CrossRef]
55. Chen, Y., Sun, L., Yuan, L., Nie, X. (2021). Effects of phosphorus fertilizer on physicochemical properties and yield of potato starch. Journal of Plant Nutrition and Fertilizers, 27(9), 1603–1613. https://doi.org/10.11674/zwyf.2021110 [Google Scholar] [CrossRef]
56. Su, S., Wu, F., Liu, D., Wu, L., Gao, S. (2013). Cloning and characterization of a phosphate transporter gene of Pht1 family in maize. Journal of Nuclear Agricultural Sciences, 27(7), 885–894. [Google Scholar]
57. Liu, F., Zhang, L. J., Gu, J. T., Li, X. J., Guo, C. J. et al. (2012). Cloning and molecular characterization analysis of TaPT4, a phosphate transporter gene in wheat (Triticum aestivum L.). Journal of Hebei Agricultural University, 35(3), 1–7. [Google Scholar]
58. Yang, S. Y., Grønlund, M., Jakobsen, I., Grotemeyer, M. S., Rentsch, D. et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. The Plant Cell, 24(10), 4236–4251. https://doi.org/10.1105/tpc.112.104901 [Google Scholar] [PubMed] [CrossRef]
59. Jia, H. F. (2011). Function analysis of a phosphate transporter gene Ospht1;8 in rice (Ph.D. Thesis). Nanjing Agricultural University, Nanjing, China. [Google Scholar]
60. Zhou, J. Q. (2014). Cloning and functional expression of PHT1 gene in Camellia oleifera (Master Thesis). Central South University of Forestry and Technology, Changsha, China. [Google Scholar]
61. Wu, P., Shou, H., Xu, G., Lian, X. (2013). Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Current Opinion in Plant Biology, 16(2), 205–212. https://doi.org/10.1016/j.pbi.2013.03.002 [Google Scholar] [PubMed] [CrossRef]
62. Wang, J., Sun, J., Miao, J., Guo, J., Shi, Z. et al. (2013). A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Annals of Botany, 111(6), 1139–1153. https://doi.org/10.1093/aob/mct080 [Google Scholar] [PubMed] [CrossRef]
63. Mukatira, U. T., Liu, C., Varadarajan, D. K., Raghothama, K. G. (2001). Negative regulation of phosphate starvation-induced genes. Plant Physiology, 127(4), 1854–1862. https://doi.org/10.1104/pp.127.4.1854 [Google Scholar] [CrossRef]
64. Hammond, J. P., Bennett, M. J., Bowen, H. C., Broadley, M. R., Eastwood, D. C. et al. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology, 132(2), 578–596. https://doi.org/10.1104/pp.103.020941 [Google Scholar] [PubMed] [CrossRef]
65. Schünmann, P. H. D., Richardson, A. E., Smith, F. W., Delhaize, E. (2004). Characterization of promoter expression patterns derived from the PHT1 phosphate transporter genes of barley (Hordeum vulgare L.). Journal of Experimental Botany, 55(398), 855–865. https://doi.org/10.1093/jxb/erh103 [Google Scholar] [PubMed] [CrossRef]
66. Schünmann, P. H. D., Richardson, A. E., Vickers, C. E., Delhaize, E. (2004). Promoter analysis of the barley PHT1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiology, 136(4), 4205–4214. https://doi.org/10.1104/pp.104.045823 [Google Scholar] [PubMed] [CrossRef]
67. Yi, K., Wu, Z., Zhou, J., Du, L., Guo, L. et al. (2005). OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiology, 138(4), 2087–2096. https://doi.org/10.1104/pp.105.063115 [Google Scholar] [PubMed] [CrossRef]
68. Wu, N. (2011). Physiological function identification of rice phosphorus transporter OsPT4 (Master Thesis). Nanjing Agricultural University, Nanjing, China. [Google Scholar]
69. Gao, J., Liu, X., Liu, L., Dai, L. (2009). Advance of rice phosphate transporters Pht1. Chinese Agricultural Science Bulletin, 25(15), 31–34. [Google Scholar]
70. Ye, Y. (2018). Temporal and spatial expression analysis of phosphate transporter gene of rice PHT1 family and functional study of OsPT4 in rice (Ph.D. Thesis). Huazhong Agricultural University, Wuhan, China. [Google Scholar]
Supplementary Materials
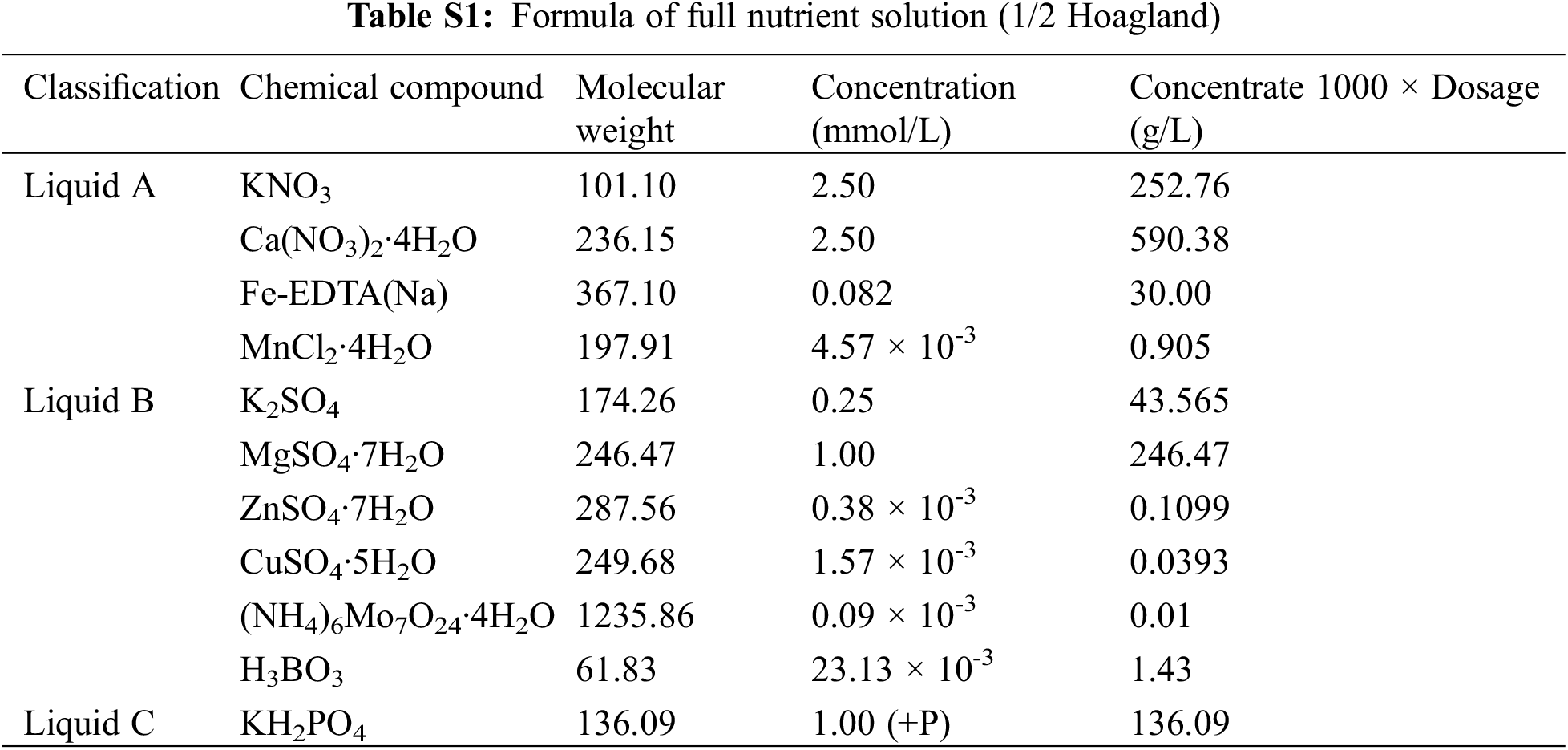

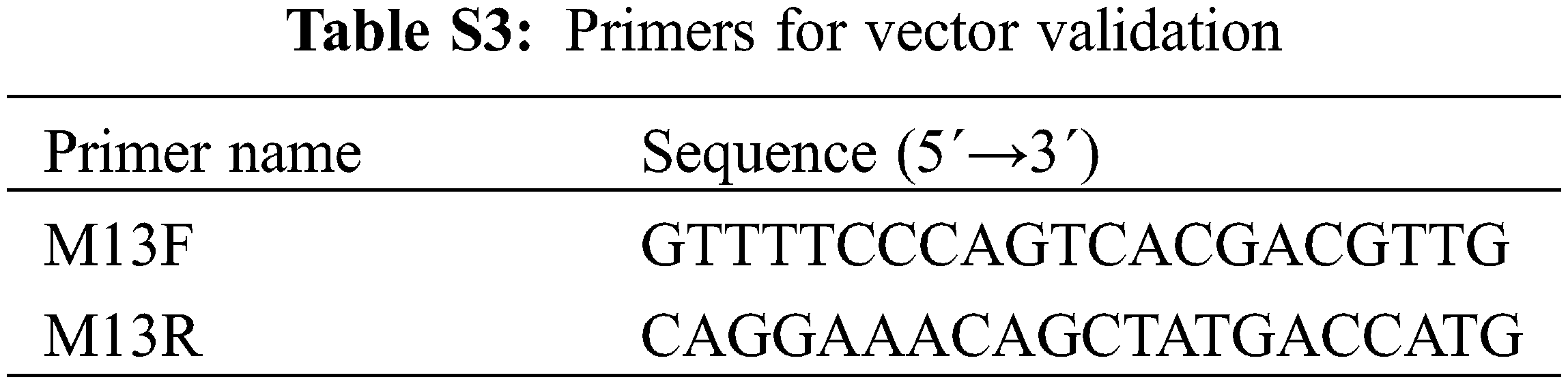
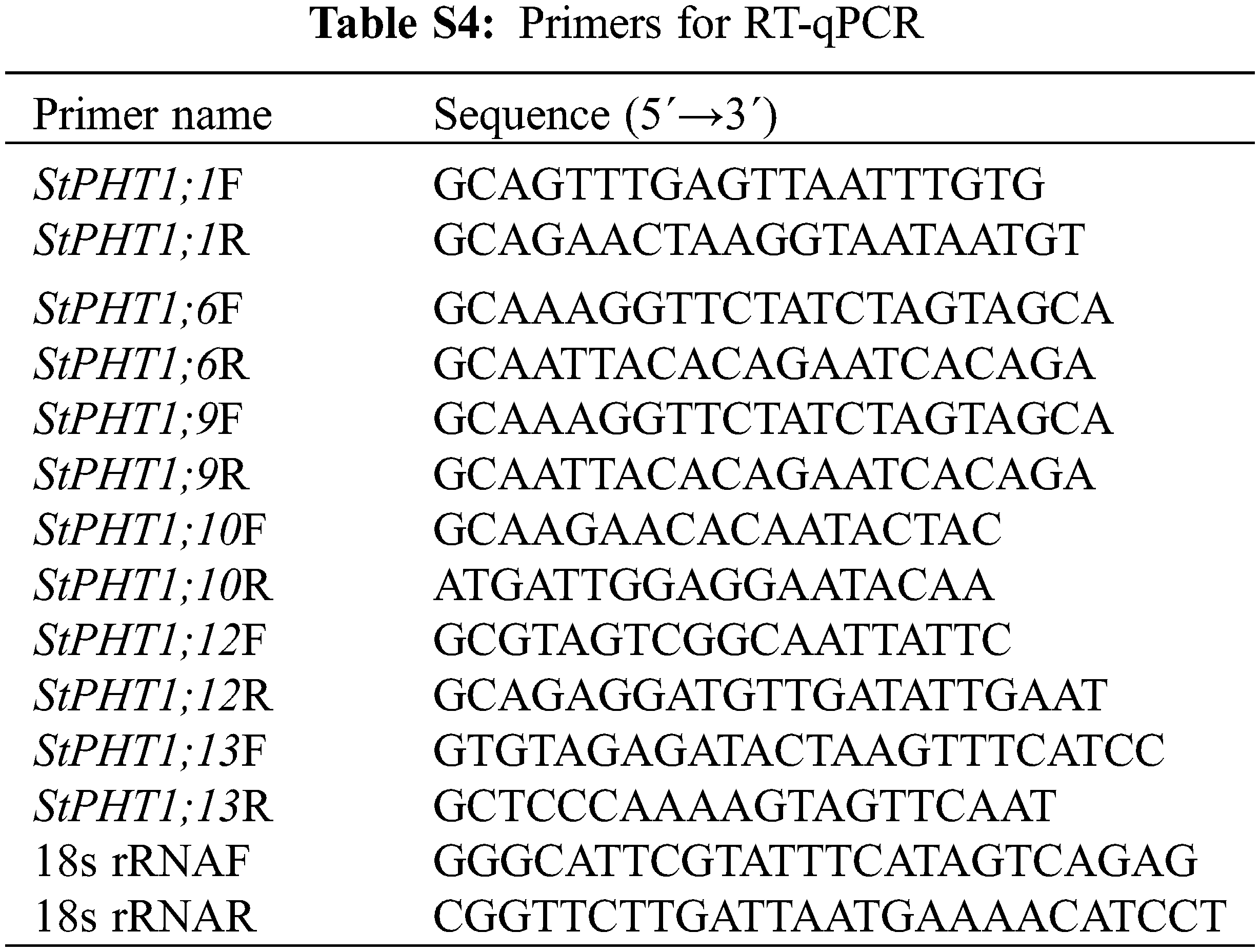
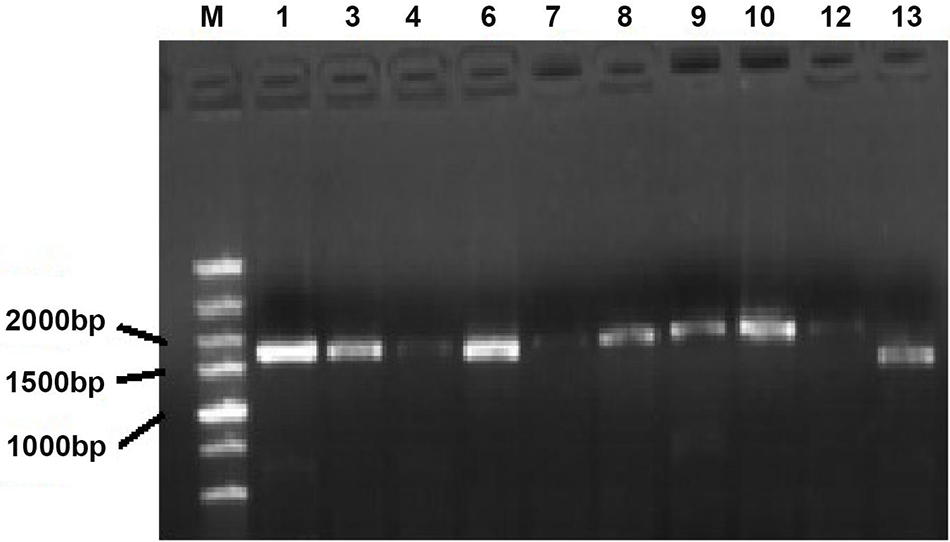
Figure S1: PCR electrophoresis of potato PHT1 gene family members. M: DNA Marker; electrophoresis bands from 1 to 13 followed by StPHT1;1, StPHT1;3, StPHT1;4, StPHT1;6, StPHT1;7, StPHT1;8, StPHT1;9, StPHT1;10, StPHT1;12, StPHT1;13
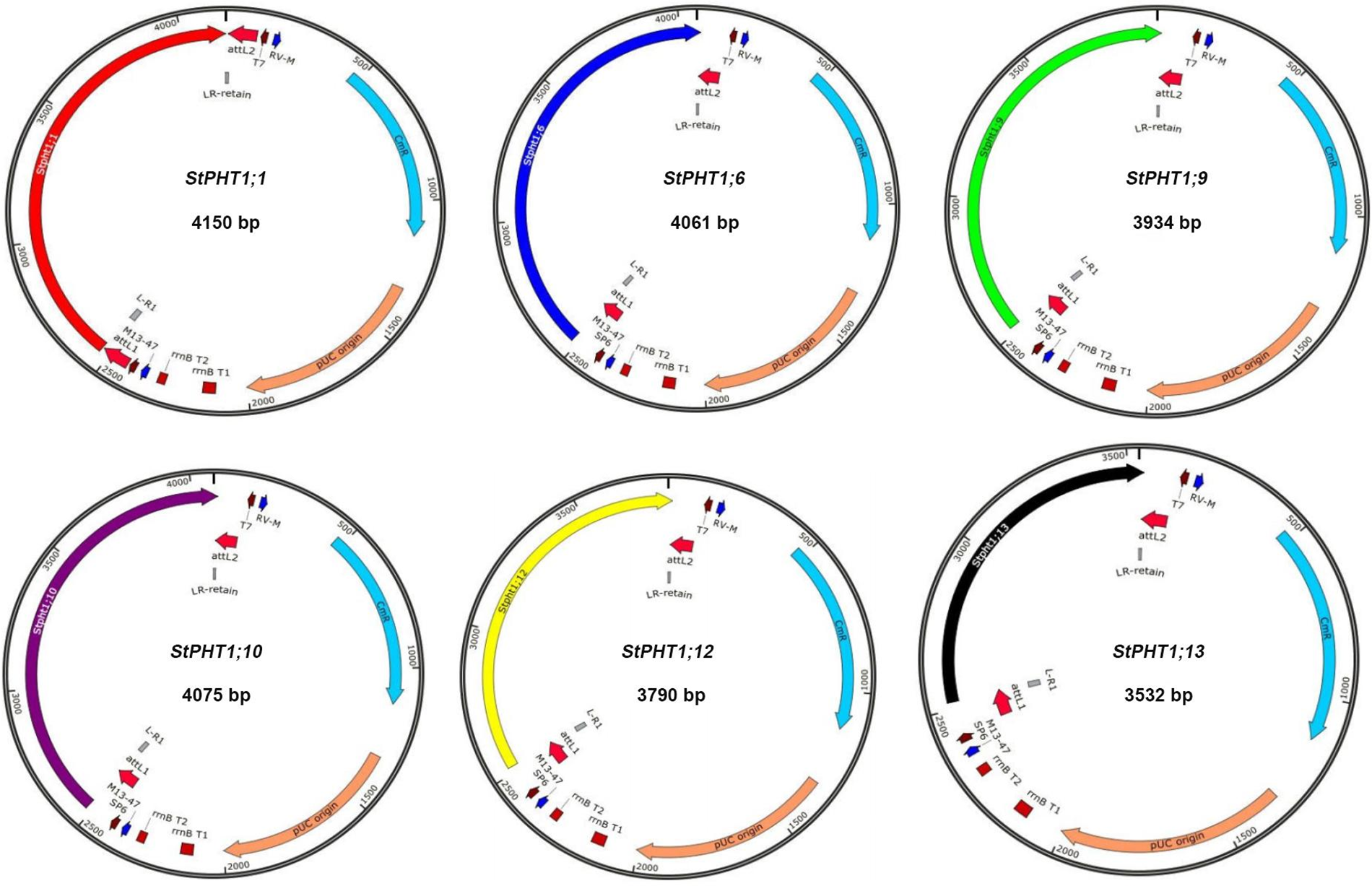
Figure S2: Schematic diagram of the entry vector construction


Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools