 Open Access
Open Access
ARTICLE
Cloning and Functional Validation of Mung Bean VrPR Gene
1
College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang, 524088, China
2
Innovation Research Center for Saline-Alkali Tolerant Rice, Shenzhen Research Institute of Guangdong Ocean University,
Shenzhen, 518120, China
* Corresponding Authors: Naijie Feng. Email: ; Dianfeng Zheng. Email:
Phyton-International Journal of Experimental Botany 2023, 92(8), 2369-2382. https://doi.org/10.32604/phyton.2023.027457
Received 04 November 2022; Accepted 23 April 2023; Issue published 25 June 2023
Abstract
For the purpose of functional validation, the mung bean (Vigna radiata) VrPR gene was cloned and overexpressed in Arabidopsis thaliana. The findings revealed that the ORF of VrPR contained 1200 bp, in which 399 amino acids were encoded. Bioinformatics analysis showed that the VrPR protein belonged to the NADB Rossmann superfamily, which was one of the non-transmembrane hydrophilic proteins. VrPR was assumed to have 44 amino acid phosphorylation sites and be contained in chloroplasts. The VrPR secondary structure comprised of random coil, α helix, β angle, and extended chain, all of which were quite compatible with the anticipated tertiary structure. Moreover, analysis of the phylogenetic tree indicated that the soybean PR (Glyma.12G222200) and VrPR were closely related. Furthermore, chlorophyll content in leaves is markedly increased in Arabidopsis when VrPR is overexpressed. Our findings will serve as a reference for more functional studies on the PR genes in mung bean.Keywords
Mung bean (Vigna radiata) is an important grain legume that is largely grown in South and East Asia, as a summer species with a short growth period (55–70 days) [1]. Mung bean contains cheap source of carbohydrates (60%–75%), high-quality proteins (20%–27%), folate, and iron [2]. Till now, many people still suffer from hunger and malnutrition, with about 800 million people suffering from hunger in 2021, and the number having risen to 150 million since the outbreak of COVID-19 [3]. Thus, improving the growth and production of mung bean will help solve the food security problem.
In order to enhance the yield of mung beans, a number of studies on the response of associated genes to the regulation of plant growth or the tolerance of environmental stresses have been examined, such as the validation of genes related to salt resistance [4,5], herbicide resistance [6], flowering [7–9], drought resistance [10], cold tolerance in mung bean [11]. Moreover, further researches regarding functional genes mining in mung bean [12,13], genome-wide association analysis [14,15], bioinformatics analysis of key gene families [2,16–18], proteins expression response to diseases [19], somatic cell induction culture [20], seedling germination toxicity [21], and transcriptome analysis in mung bean [22–30] have also been conducted. Furthermore, the whole genome of mung bean has been sequenced in 2014, which has the length of 543 MB [31]. But studies related to gene functions in mung bean are largely unknown.
In this study, we examined the prochlorophylliate reductase gene VrPR, which was associated with the pathway of porphyrin and chlorophyll metabolism production based on the results of the transcriptome sequencing on mung bean leaves [27]. The first enzyme in the de novo synthesis pathway of NAD is L-aspartate oxidase, which has been reported to be encoded by the NADB gene, which belongs to the VrPR family [32–38]. All living organisms require NAD, a vital cofactor and essential molecule [39,40]. And NADB is mandatory for normal NAD biosynthesis [41–43]. The current study used bioinformatics analysis to clone and characterize the VrPR gene. Additionally, VrPR overexpression in Arabidopsis was used to study its functional validation.
In the present study, the VrPR gene was properly cloned using the mung bean genotype Lvfeng no. 2. The overexpressed VrPR gene was functionally analyzed using the Arabidopsis thaliana Columbia-0 (Col) strain.
2.2.1 Total RNA Extraction and PCR Amplification of VrPR
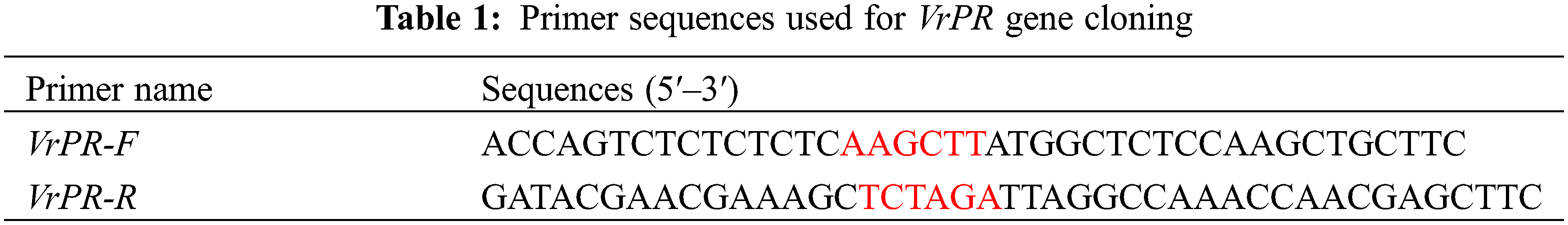
For the purpose of extracting total RNA, mung bean leaves were collected and rapidly frozen in liquid nitrogen. The TaKaRa RNA extraction kit was used to extract the total RNA following the protocol. The complementary DNA (cDNA) was consolidated through the TaKaRa reverse transcription kit. We designed the primers for the gene of VrPR, in which we labeled VrPR-F and VrPR-R using HindIII and XbaI enzymatic sites (Table 1). After that, the cDNA of the VrPR gene was cloned by PCR amplification with the primers. The PCR reactions were performed under the following conditions: 94°C for 4 min, followed by 40 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min.
2.2.2 Construction of VrPR Gene Overexpression Vector
The PCR-amplified VrPR fragment was isolated from an agarose gel and combined with an enzyme-extracted PHB empty vector (Fig. 1). They were then ligated using Tiangen Biologicals’ EasyGeno DNA recombinant technology, and transformed into E. coli DH5α. The positive colonies were chosen for sequencing after 16 h of culture at 37°C. And the accurate plasmids were isolated for further analysis.
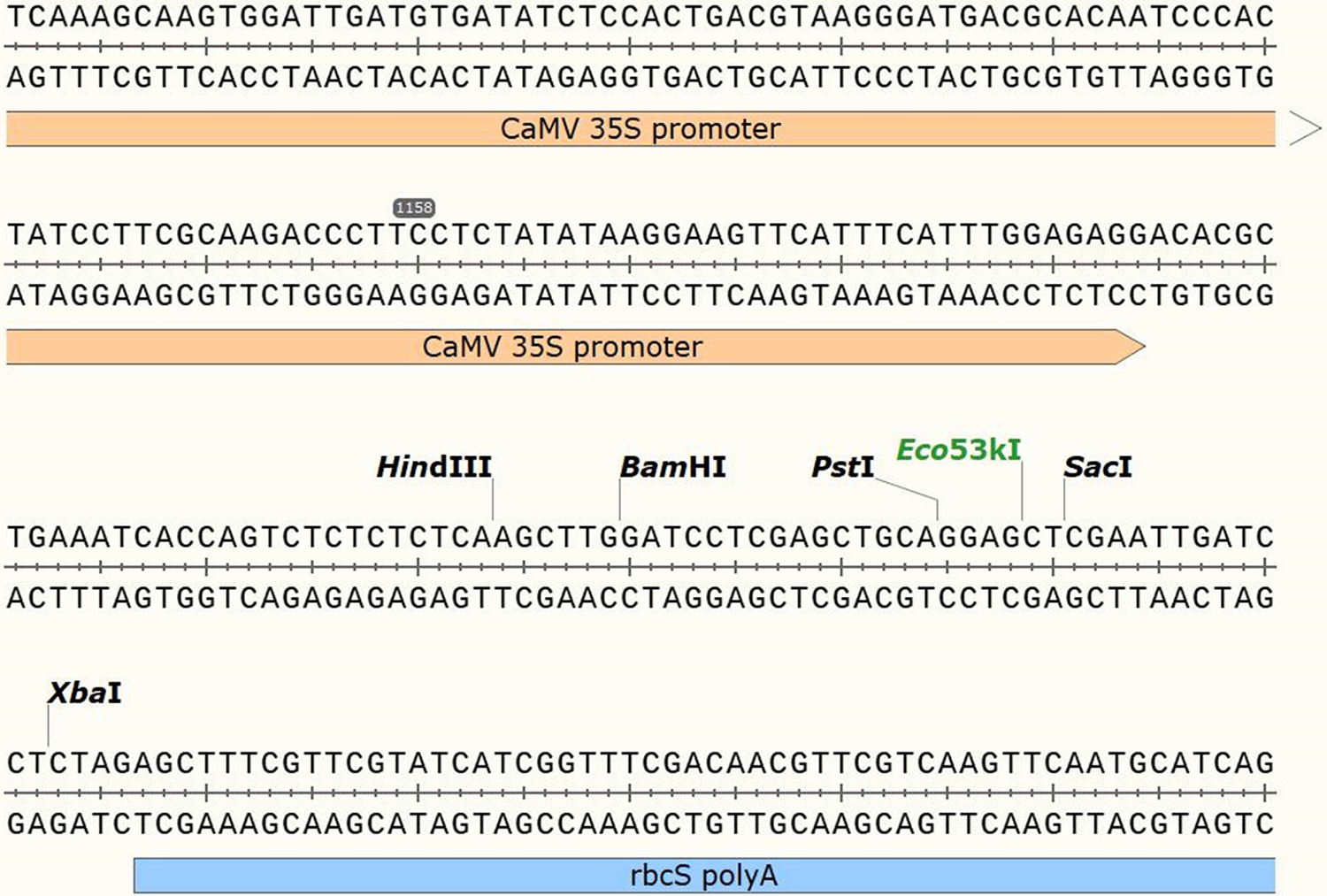
Figure 1: Figure of the structural information of the PHB over-expression vector
2.2.3 Genetic Transformation and Identification of VrPR Gene
In the present study, we transformed the validated VrPR plasmid with a biynary expression vector. We mixed the plasmid with 700 mL of antibiotic-free YEP liquid medium and Agrobacterium tumefacien GV3101 receptor state and incubated it at 28°C for 2–3 h in a shaker at 200 rpm. The bacterial solution was centrifuged at 6000 rpm and resuspended in about 100 μL of supernatant on kanamycin-containing YEP plates. Then we incubated the plates upside down at 28°C in an incubator for 2–3 days. We selected one colony at random for colony PCR identification and used Agrobacterium tumefaciens, which carried the PHB-PR overexpression vector, to infest inflorescences of wild-type Arabidopsis. After two infestations, we collected the seeds at maturity and screened them using an Arabidopsis plant known as the T1 plant that was resistant to thaumatin in YEP solid medium. We identified the positive plants by extracting DNA from the leaves of resistant Arabidopsis with a DNA extraction kit from Tiangen Biologicals and testing through PCR.
2.2.4 Functional Validation of Transgenic Arabidopsis thaliana
We identified the Arabidopsis homolog genes of VrPR through Phytozome (https://phytozome-next.jgi.doe.gov/) and checked the similarity ratio, i.e., 35.80% through DNAMAN software (Fig. 2), which indicated that the Arabidopsis homolog genes had limited influence on the VrPR gene.
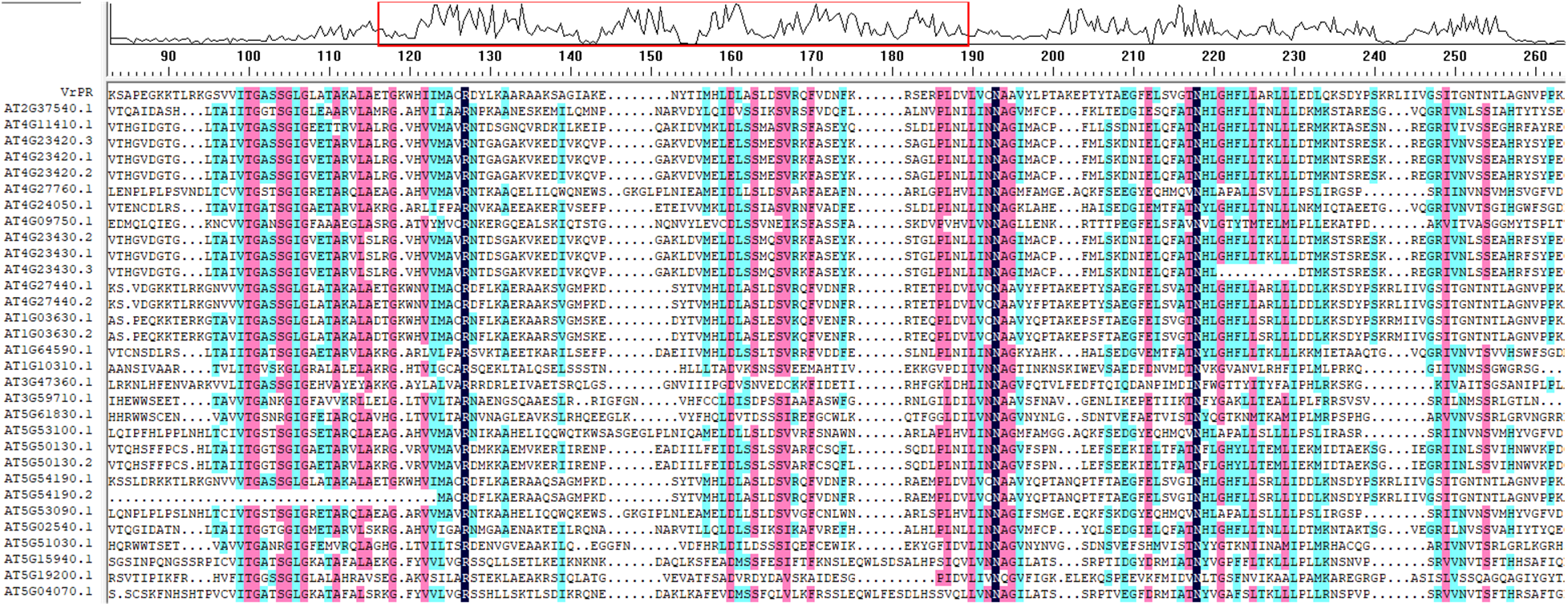
Figure 2: Figure of the sequence similarity alignment between VrPR gene and Arabidopsis homolog gene
Furthermore, we measured and analyzed phenotypic and physiological indexes, such as lateral root number, root length, root fresh weight, and chlorophyll.
2.2.5 Bioinformatics Analysis of VrPR Gene
The physicochemical properties of proteins and other traits were identified by the Protparam tool in ExPASy; The conserved domain search tool in the NCBI database is used to find conserved structural domains of gene-encoded proteins; The ProtScale tool in the ExPASy online server is used to predict protein affinity and hydrophobicity; SignalP 5.0 Server is used to predict proteins’ transmembrane region; SignalP 5.0 Server is used to predict proteins signal peptide; Softberry Server is used to predict proteins’ subcellular region; Softberry Server is used to predict proteins’ subcellular region; TMHMM Server 2.0 is used to predict the transmembrane region of proteins; SignalP 5.0 Server is used to predict the signal peptides of proteins; ProtComp 9.0 tool in Softberry Server is used to predict the subcellular localization of proteins; The SOPMA program is used to predict the secondary structure of proteins; The PHYRE2 program is used to predict the protein’s tertiary structure; Mega 6.0 software is used to build phylogenetic trees.
We processed and analyzed the experiment data using three different software packages: EXCLE, IBM SPSS Statistics 25, and Origin 2021 Pro. The statistical method is one-way ANOVA, and we express the data in mean and standard deviation (n = 3). The significant differences are significant for p-value < 0.05, very significant for p-value < 0.01, highly significant for p-value < 0.001.
3.1 Cloning to Obtain the ORF of VrPR Gene
The information of the VrPR gene was retrieved from NCBI with the annotated function asprochlorophyllate reductase (LOC 106764794). By using the CDS sequence of this gene, full-length amplification primers were designed and amplified the gene fragment of about 1200 bp using cDNA from mung bean (Fig. 3). The specific fragment of this gene was ligated to a PHB overexpression vector and verified by colony PCR, which confirmed the 1200 bp gene length (Fig. 4). The results of the bacteriophage sequencing were consistent with the transcriptome sequencing, which indicated the successful cloning of the VrPR gene.

Figure 3: Figure of VrPR gene ORF sequence and its encoding proteins
Note: * The terminator.
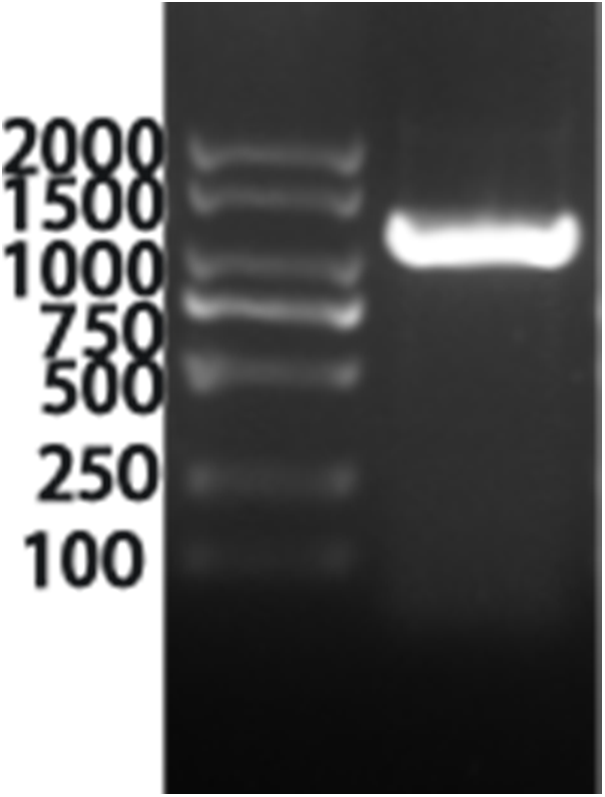
Figure 4: Figure of the positive figure of PCR cloning, marker is 2000 marker (from top to bottom is 2000, 1500, 1000, 750, 500, 250, 100)
3.2 Bioinformatics Analysis of VrPR Protein
3.2.1 Prediction of Conserved Structural Domains of VrPR Protein
Our results showed that the conserved structural domain of VrPR protein was located at the central, non-C-terminal position of the polypeptide chain, which belongs to the NADB Rossmann superfamily of proteins (Fig. 5a). The NCBI blast results confirmed this protein as a protochlorophyllide lipid reductase.
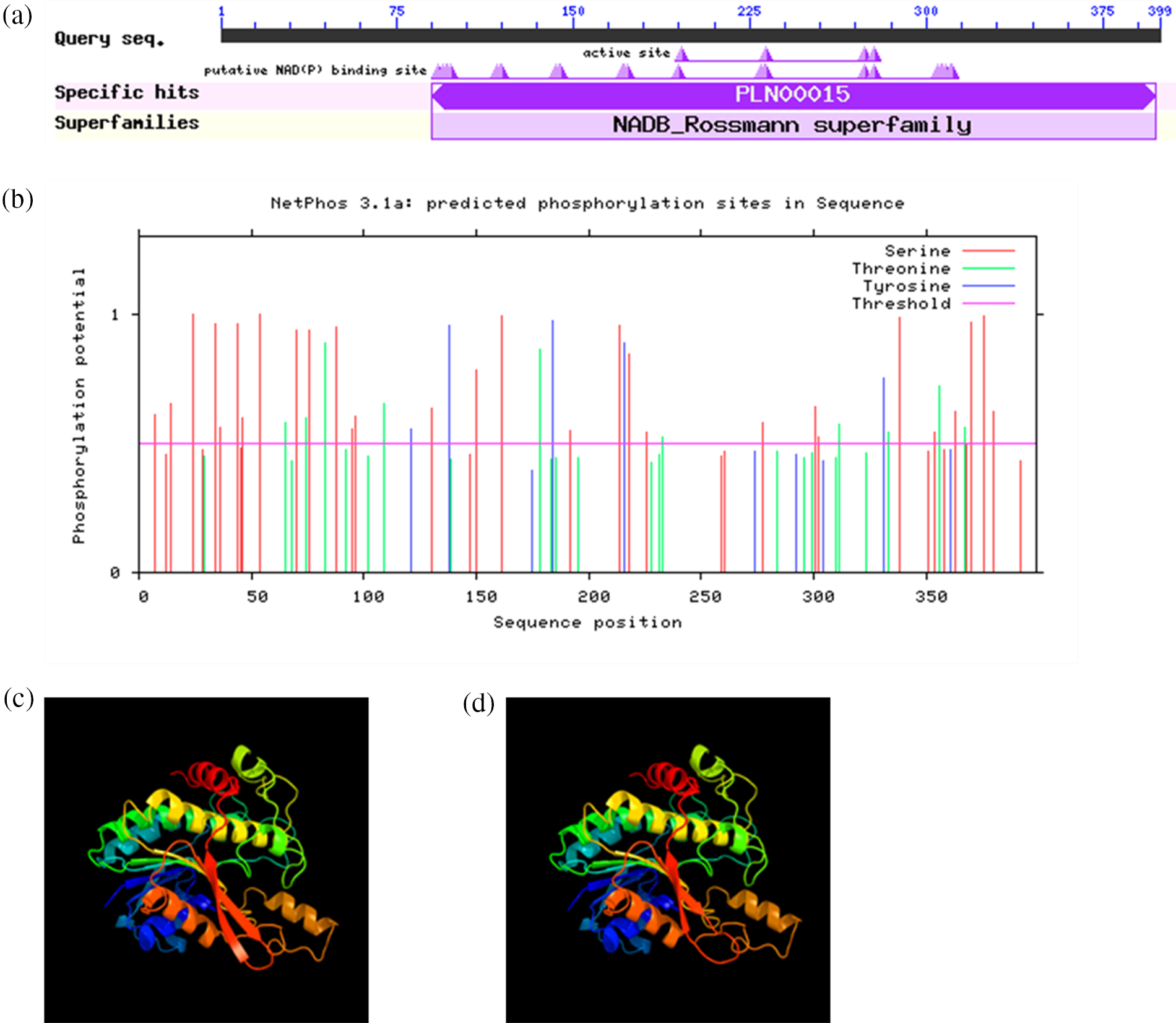
Figure 5: (a) is conservative domain retrieval of VrPR proteins amino acid sequence in NCBI; (b) is the prediction of VrPR protein phosphorylation site; (c) is tertiary protein structure of VrPR gene; (d) is tertiary protein structure of GmPR gene
3.2.2 Prediction of Physicochemical Properties of VrPR-Encoded Protein
Physicochemical analyses of this protein indicated its molecular weight 43101.29 Da with molecular formula C1915H3068N530O577S11 and the total number of atoms is 6101. This protein contained 399 amino acids, among which 40 amino acid residues (Asp+Glu) have a negative charge and 50 amino acid residues (Arg+Glu) have a positive charge. In addition, it had a 9.23 theoretical isoelectric point (PI) value and a 37.25 instability index, which suggested its stable and hydrophilic nature.
3.2.3 Prediction of VrPR Protein Signal Peptide, Transmembrane Structure and Subcellular Localization
The VrPR was found to be a non-secretory and non-transmembrane protein based on signal peptide prediction and transmembrane structure, respectively. Moreover, subcellular localization results showed its location in chloroplasts.
3.2.4 VrPR Protein Phosphorylation Site Prediction
The mung bean VrPR protein had 44 amino acid phosphorylation sites (threshold > 0.5). Among them are 10 phosphorylation sites for threonine, 29 for serine, and 5 for tyrosine, indicating phosphorylation-related activity (Fig. 5b). The phosphorylation of serine sites is predominant in the mung bean VrPR protein.
3.2.5 VrPR Protein Secondary Structure and Tertiary Structure Prediction
The secondary structure of VrPR protein mainly consisted of α-helices (41.60%), irregular coils (35.84%), extended chains (15.04%), and β-turns (7.52%). On the other hand, tertiary structure revealed that VrPR proteins are mainly composed of irregular coils and α-helix. Besides, we performed the soybean gene (GmPR) tertiary structure prediction and showed the structural similarity between VrPR and GmPR (Figs. 5c and 5d). Therefore, the VrPR protein might have the same functions as GmPR.
3.2.6 VrPR Protein Evolutionary Tree Construction and Multiple Sequence Alignment
The amino acid sequences of VrPR were aligned using Phytozome, and phylogenetic analysis of four species sequences showed high concordance. Based on the obtained results, the VrPR was found to be closely related to soybean and Arabidopsis proteins and less related to pineapple and rice proteins (Fig. 6). The DNAMAN software results showed that 15 NADB proteins had high homology, which indicated mung bean VrPR proteins as a typical member of the NADB transcription factor family.
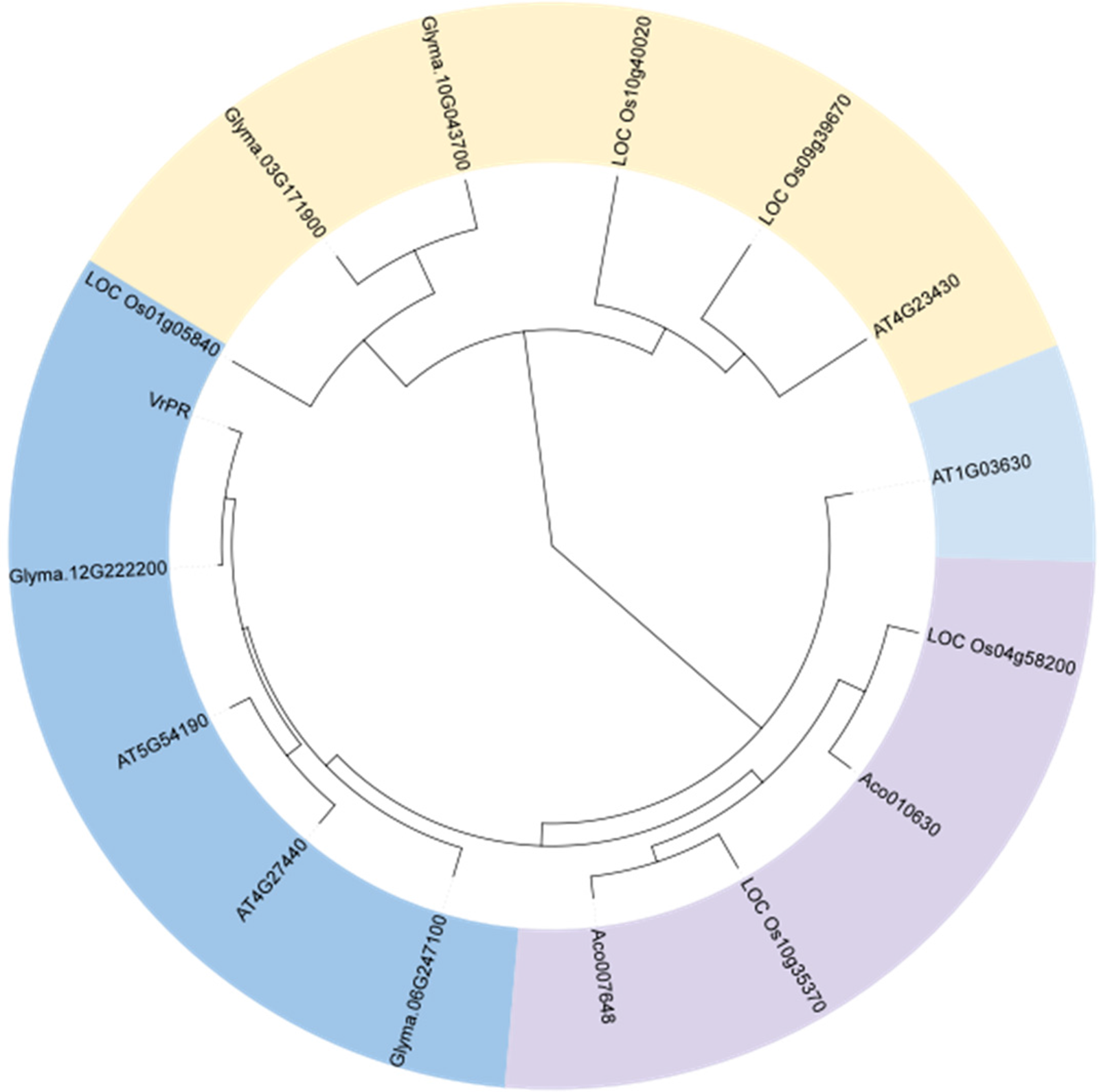
Figure 6: Figure of amino acid sequence alignment of homologous
3.3 Construction of VrPR Gene Overexpression Vector
Cloning of the VrPR gene was achieved by overexpressing the PHB vector by adding HindIII and XbaI enzyme cleavage sites. Later, the targeted gene was ligated to the PHB plasmid with the recombinant enzyme Exnase II. Two pairs of primers, i.e., PRF and PRR, were used to obtain 1200 bp of fragment length through PCR amplification. Thus, an overexpression vector with the targeted gene was constructed.
3.4 Genetic Transformation and Identification of VrPR Gene
The recombinant plasmid PHB-PR was extracted and identified by PCR. Then we purified the plasmid and transferred it to Agrobacterium tumefaciens GV3101 receptor cells and selected the positive colonies. The agrobacterium containing the PHB-PR plasmid was identified by PCR with two pairs of primers, PRR and PRF, HPT-F and HPT-R, and then used to infest Arabidopsis’s inflorescence (T0). The infested seeds were collected for resistance screening to obtain Arabidopsis resistant plants (T1). All 16 randomly selected Arabidopsis plants contained the target gene, indicating that the VrPR gene had been transferred successfully into Arabidopsis (Fig. 7).
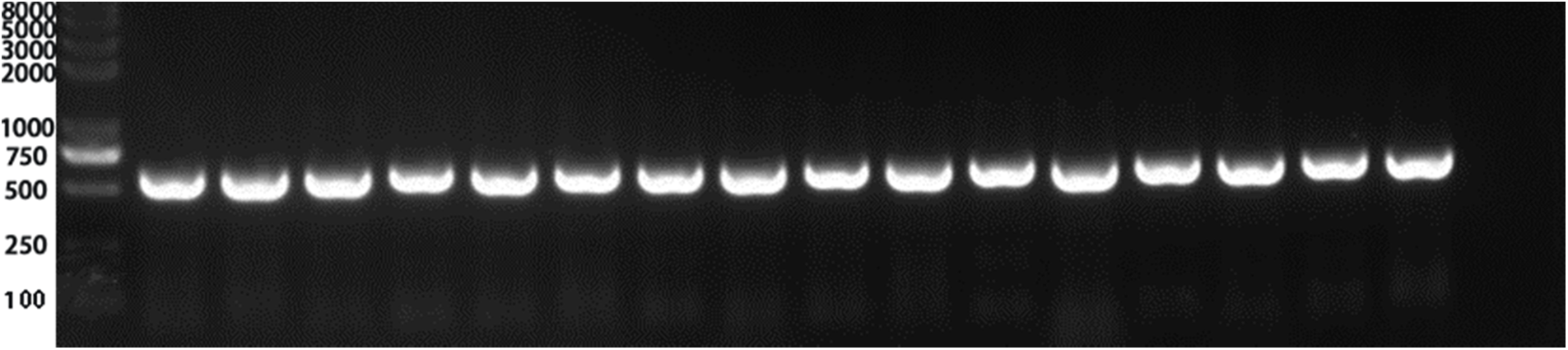
Figure 7: Figure of positive PCR result. Lines 1–15 PCR results (PCR detection of hygromycin resistance gene HPT, 598 bp) showed that 15 lines were positive (Marker: 8000, 5000, 3000, 2000, 1000, 750 (brightest), 500, 250, 100 bp) Note: Only the first 15 lines were detected. The 15 lines was a positive control. The last lane was a negative control.
3.5 VrPR Transgenic Arabidopsis Root System Phenotype
In Fig. 8, we can see that the VrPR transgenic Arabidopsis showed a similar phenotype to the wild type after transformation. On the other hand, no significant differences were found among leaves, root length, and lateral root number, which indicated that genetic transformation did not cause significant phenotypic differences in Arabidopsis.
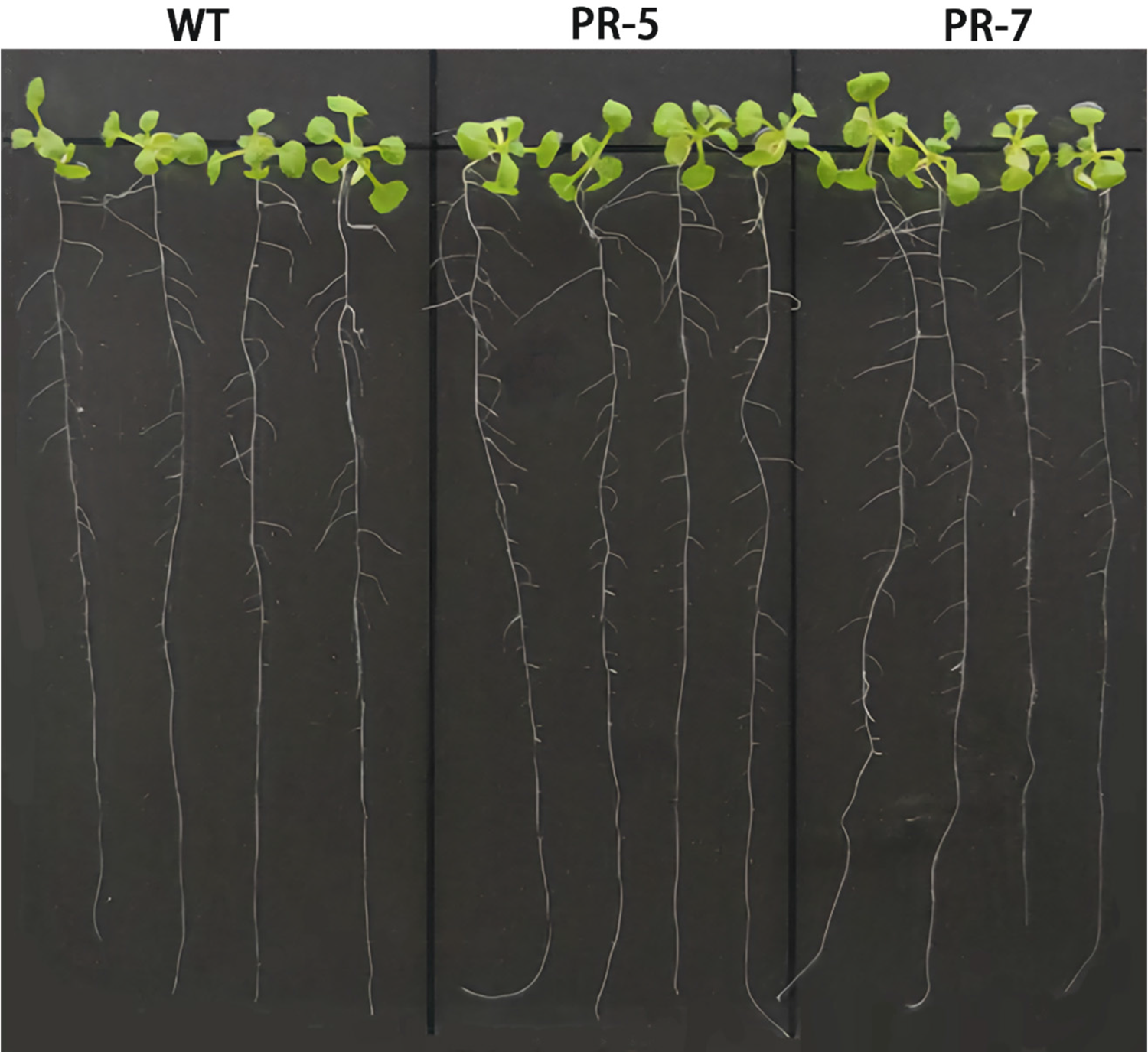
Figure 8: Figure of phenotype picture of Arabidopsis thaliana
3.6 Root Fresh Weight and Chlorophyll Index of VrPR Transgenic Arabidopsis
Fig. 9a showed non-significant differences in fresh root weight between transgenic and wild-type plants. These results suggested that genetic transformation had no significant effects on the phenotype of Arabidopsis. Fig. 9b showed extremely significant differences in chlorophyll content between transgenic and wild-type plants. These results suggested that genetic transformation had significant effects on the chlorophyll content of Arabidopsis.
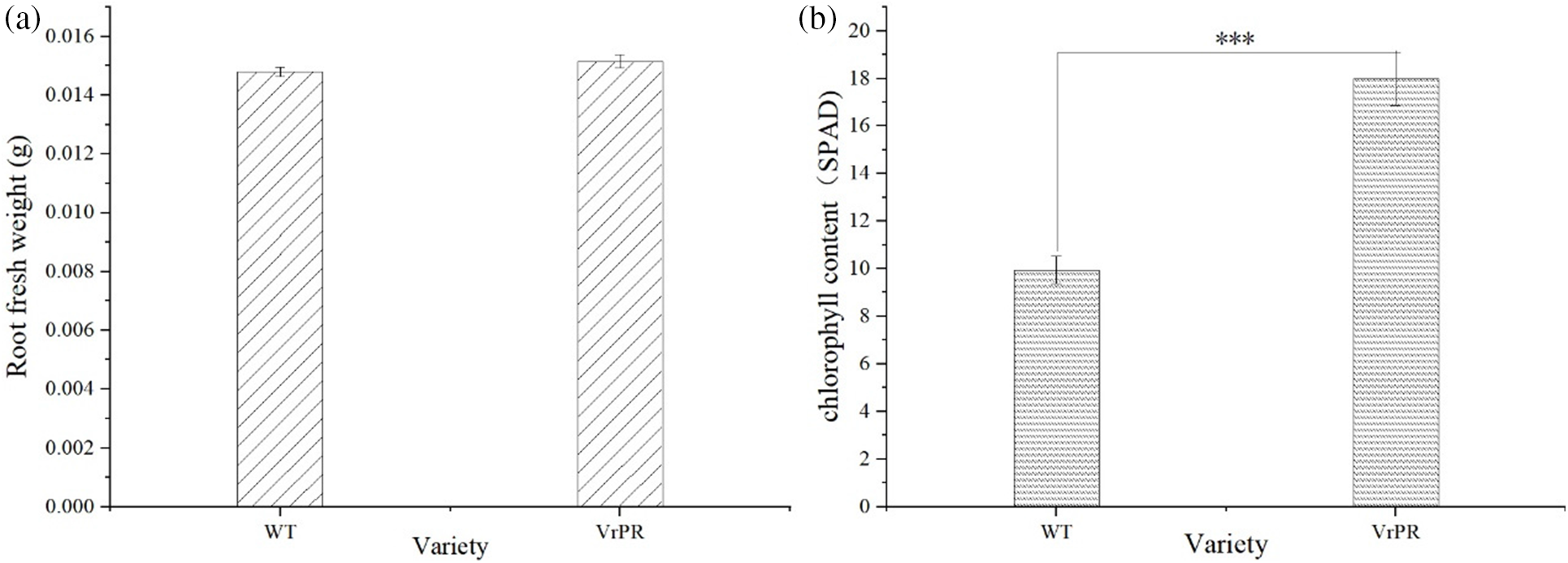
Figure 9: Figure of root fresh weight index of different varieties (a) and figure of chlorophyll index of different varieties (b). The one-way ANOVA was performed between the group and *** indicates that highly significantly; WT indicates wild type of arabidopsis; VrPR indicates transgenic arabidopsis of VrPR
In this experiment, we selected the gene from the mining of transcriptome analysis and attained a 1200 bp ORF fragment by gene cloning. We analyzed the protein sequence of the gene in bioinformatics and found it is a protochlorophyllide, which belongs to the NADB Rossmann superfamily. And it is predicted to be a hydrophilic protein that is located in chloroplasts in subcellular localization. Serine site Phosphorylation was predominant, and the tertiary protein structure was similar to that of the homologous gene in soybean. The evolutionary tree indicated a closer evolutionary relationship with soybean. By constructing the overexpression vectors and genetically transforming Arabidopsis, we obtained T1 generation seeds and T3 generation pure seeds. We then made a further validation and found that the transformation did not significantly affect the root phenotype. Moreover, it was significantly increased in the transgenic Arabidopsis, and we proposed that overexpression of the VrPR gene led to increased chlorophyll in transgenic Arabidopsis.
The VrPR gene is blasted as protochlorophyllide with its enzyme activity and gene expression. This enzyme is a key enzyme for chlorophyll synthesis in cyanobacteria, algae, and multicellular plants and is essential for the growth and development of green plants, which can directly affect the greening ability of yellowing seedlings. VrPR belongs to the NADB Rossmann superfamily and has binding sites for conserved NADB sequences. NADB can promote the synthesis of NAD coenzymes, indirectly provide energy for chlorophyll synthesis, and improve the viability of Arabidopsis. This may be the way that VrPR is involved in increasing the production of chlorophyll.
All of the following studies provided theoretical and data support for the resistance and high yield of mung bean from different directions. For example, research of mining genes related to salt tolerance in mung bean [44], research of genome-wide analysis of different gene families related different stresses in mung bean [2,16,17,45–48], research of validation of mung bean related genes in Arabidopsis [7,49], research of transcriptome analysis of mung bean related different stress [50], and research of reciprocal between different proteins in mung bean [51]. In this experiment, we selected the gene by analyzing the transcriptome [27]. With the bioinformatics analysis of the gene and the functional verification by transgenic Arabidopsis, we found that transgenic Arabidopsis had a significant increase in chlorophyll, and it may be the reason that the overexpression vector promoted Arabidopsis to overexpress the VrPR gene and carry out the secretion of related proteins, thus promoting the increase of chlorophyll. However, we can still do gene bioinformatics more comprehensively and deeply. For example, we can still analyze the promoter cis-element and conserved motifs of the VrPR gene, etc. The functional validation of Arabidopsis is still in the preliminary stage. For further research, we can focus on experiments such as the interactions of related proteins, the oxidative stress response, and photosynthesis in Arabidopsis.
In this experiment, we selected the VrPR gene from the analysis of the transcriptome in the preliminary stage of this experiment and performed a functional validation. Including bioinformatics analysis, overexpression vector construction, genetic transformation of Arabidopsis, and functional validation. The results showed that there is a significant increase in chlorophyll in transgenic Arabidopsis, which may be due to the overexpression of the VrPR gene, which is a protochlorophyllide that promotes the secretion of chlorophyll in transgenic Arabidopsis. We proposed that overexpression of the VrPR gene led to increased chlorophyll in transgenic Arabidopsis, and this can lay the foundation for further experimental research.
Funding Statement: This research was funded by National Natural Science Foundation of China, Grant Number (31871576); National Keypoint Research and Invention Program of the Thirteenth, Grant Number (2019YFD1002205) and The APC was funded by National Keypoint Research and Invention Program of the Thirteenth.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Naijie Feng and Dianfeng Zheng; Data curation, Xiaokui Huang and Yingbin Xue; Formal analysis, Xiaokui Huang and Yingbin Xue; Funding acquisition, Naijie Feng and Dianfeng Zheng; Investigation, Yingbin Xue; Methodology, Yingbin Xue and Dianfeng Zheng; Project administration, Naijie Feng and Dianfeng Zheng; Resources, Hanqiao Hu, Naijie Feng and Dianfeng Zheng; Software, Hanqiao Hu; Supervision, Xiaokui Huang and Yingbin Xue; Validation, Naijie Feng and Dianfeng Zheng; Writing—original draft, Xiaokui Huang; Writing—review & editing, Aaqil Khan and Yingbin Xue. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Noble, T. J., Tao, Y., Mace, E. S., Williams, B., Jordan, D. R. et al. (2017). Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Frontiers in Plant Science, 8, 2102. https://doi.org/10.3389/fpls.2017.02102 [Google Scholar] [PubMed] [CrossRef]
2. Chen, H., Hu, L., Wang, L., Wang, S., Cheng, X. (2022). Genome-wide identification and expression profiles of AP2/ERF transcription factor family in mung bean (Vigna radiata L.). Journal of Applied Genetics, 63(2), 223–236. https://doi.org/10.1007/s13353-021-00675-8 [Google Scholar] [PubMed] [CrossRef]
3. The State of Food Security and Nutrition in the World (2022). Food and Agriculture Organization of the United Nations. [Google Scholar]
4. Mishra, S., Alavilli, H., Lee, B. H., Panda, S. K., Sahoo, L.W. (2014). Cloning and functional characterization of a vacuolar Na+/H+ antiporter gene from mungbean (VrNHX1) and its ectopic expression enhanced salt tolerance in Arabidopsis thaliana. PLoS One, 9(10), e106678. https://doi.org/10.1371/journal.pone.0106678 [Google Scholar] [PubMed] [CrossRef]
5. Sahoo, D. P., Kumar, S., Mishra, S., Kobayashi, Y., Panda, S. K. et al. (2016). Enhanced salinity tolerance in transgenic mungbean overexpressing Arabidopsis antiporter (NHX1) gene. Molecular Breeding, 36(10), 144. https://doi.org/10.1007/s11032-016-0564-x [Google Scholar] [CrossRef]
6. Kumar, S., Kalita, A., Srivastava, R., Sahoo, L. (2017). Co-expression of Arabidopsis NHX1 and bar improves the tolerance to salinity, oxidative stress, and herbicide in transgenic mungbean. Frontiers in Plant Science, 8, 1896. https://doi.org/10.3389/fpls.2017.01896 [Google Scholar] [PubMed] [CrossRef]
7. Zhang, Q. Q., Dong, J., Wang, Z. Q., Wang, Y., Cai, C. M. et al. (2022). Mungbean VrCOL1 regulates flowering time under short-day conditions in Arabidopsis. Plant Cell Tissue and Organ Culture, 148(3), 599–608. https://doi.org/10.1007/s11240-021-02217-w [Google Scholar] [CrossRef]
8. Babar, M., Ijaz, S., Khan, M. S., Ul Haq, I. (2021). Computational genomics based probing of resistance gene analogs (RGAs) in mungbean under cercospora leaf spot disease challenge. Pakistan Journal of Agricultural Sciences, 58(5), 1523–1536. [Google Scholar]
9. Shi, R., Xu, W., Liu, T., Cai, C., Li, S. (2021). VrLELP controls flowering time under short-day conditions in Arabidopsis. Journal of Plant Research, 134(1), 141–149. https://doi.org/10.1007/s10265-020-01235-7 [Google Scholar] [PubMed] [CrossRef]
10. Chen, H., Liu, L., Wang, L., Wang, S., Cheng, X. (2016). VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. Jouranl of Plant Research, 129(2), 263–273. https://doi.org/10.1007/s10265-015-0773-0 [Google Scholar] [PubMed] [CrossRef]
11. Chen, L. R., Ko, C. Y., Folk, W. R., Lin, T. Y. (2017). Chilling susceptibility in mungbean varieties is associated with their differentially expressed genes. Botanical Studies, 58(1), 7. https://doi.org/10.1186/s40529-017-0161-2 [Google Scholar] [PubMed] [CrossRef]
12. Hwang, W. J., Ha, J. M., Lee, T., Jeong, H., Kim, M. Y. et al. (2017). A candidate flowering gene in mungbean is homologous to a soybean Phytochrome A gene. Euphytica, 213(4), 1300. https://doi.org/10.1007/s10681-017-1866-8 [Google Scholar] [CrossRef]
13. Chotechung, S., Somta, P., Chen, J., Yimram, T., Chen, X. et al. (2016). A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theoretical and Applied Genetics, 129(9), 1673–1683. https://doi.org/10.1007/s00122-016-2731-1 [Google Scholar] [PubMed] [CrossRef]
14. Reddy, V. R. P., Das, S., Dikshit, H. K., Mishra, G. P., Aski, M. et al. (2020). Genome-wide association analysis for phosphorus use efficiency traits in mungbean (Vigna radiata L. Wilczek) using genotyping by sequencing approach. Frontiers in Plant Science, 11, 537766. [Google Scholar] [PubMed]
15. Likhith, R. K., Alagarasan, G., Muthurajan, R., Parasuraman, B., Subramanian, R. (2021). Genome wide identification of mungbean (Vigna radiata [L.] R. Wilczek) Late Embryogenesis Abundant (LEA) protein gene family. Israel Journal of Plant Sciences, 69(1–2), 79–86. https://doi.org/10.1163/22238980-bja10049 [Google Scholar] [CrossRef]
16. Xu, W. Y., Liu, T., Zhang, H. Y., Zhu, H. (2021). Mungbean DIRIGENT gene subfamilies and their expression profiles under salt and drought stresses. Frontiers in Genetics, 12, 658148. https://doi.org/10.3389/fgene.2021.658148 [Google Scholar] [PubMed] [CrossRef]
17. Yin, L., Zhang, M., Wu, R., Chen, X., Liu, F. et al. (2021). Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response. BMC Plant Biology, 21(1), 408. https://doi.org/10.1186/s12870-021-03184-2 [Google Scholar] [PubMed] [CrossRef]
18. Parveen, R., Vaish, S., Gupta, D., Basantani, M. K. (2022). Bioinformatics characterization of patatin-related phospholipase A (pPLA) gene family in agriculturally important crops viz Vigna radiata, Vigna angularis, and Glycine max. Biologia, 77(5), 1429–1446. https://doi.org/10.1007/s11756-022-01026-6 [Google Scholar] [CrossRef]
19. Cayalvizhi, B., Nagarajan, P., Raveeendran, M., Rabindran, R., Selvam, N. J. et al. (2015). Unraveling the responses of mungbean (Vigna radiata) to mungbean yellow mosaic virus through 2D-protein expression. Physiological and Molecular Plant Pathology, 90(4), 65–77. https://doi.org/10.1016/j.pmpp.2015.03.001 [Google Scholar] [CrossRef]
20. Sindhujaa, V., Gnanaraj, M., Viji, M., Karuppanapandian, T., Manoharan, K. (2018). Induction of high frequency somatic embryogenesis and analysis of developmental stagewise expression of SERK1 gene during somatic embryogenesis in cultures of Vigna radiata (L.) R.Wilczek. Indian Journal of Experimental Biology, 56(3), 180–193. [Google Scholar]
21. Nair, P. M. G., Chung, I. M. (2015). Physiological and molecular level studies on the toxicity of silver nanoparticles in germinating seedlings of mung bean (Vigna radiata L.). Acta Physiologiae Plantarum, 37(1), 43. https://doi.org/10.1007/s11738-014-1719-1 [Google Scholar] [CrossRef]
22. Liu, M. S., Kuo, T. C., Ko, C. Y., Wu, D. C., Li, K. Y. et al. (2016). Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean (Vigna radiata [L.] R. Wilczek). BMC Plant Biology, 16(1), 46. https://doi.org/10.1186/s12870-016-0736-1 [Google Scholar] [PubMed] [CrossRef]
23. Kumar, S., Ayachit, G., Sahoo, L. (2020). Screening of mungbean for drought tolerance and transcriptome profiling between drought-tolerant and susceptible genotype in response to drought stress. Plant Physiology and Biochemistry, 157(12), 229–238. https://doi.org/10.1016/j.plaphy.2020.10.021 [Google Scholar] [PubMed] [CrossRef]
24. Li, S. W., Leng, Y., Shi, R. F. (2020). Transcriptome characterization of gene profiling during early stage of nitric oxide-induced adventitious rooting in mung bean seedlings. Journal of Plant Growth Regulation, 39(1), 430–455. https://doi.org/10.1007/s00344-019-09993-y [Google Scholar] [CrossRef]
25. Li, S. W., Shi, R. F., Leng, Y., Zhou, Y. (2016). Transcriptomic analysis reveals the gene expression profile that specifically responds to IBA during adventitious rooting in mung bean seedlings. BMC Genomics, 17(1), 43. https://doi.org/10.1186/s12864-016-2372-4 [Google Scholar] [PubMed] [CrossRef]
26. Ha, J., Shim, S., Lee, T., Lee, E., Yang, X. et al. (2020). Transcriptomic and biochemical analyses of the accumulation of sucrose in mungbean (Vigna radiata (L.) Wilczek) leaves after pod removal. Theoretical and Applied Genetics, 133(8), 2355–2362. https://doi.org/10.1007/s00122-020-03603-2 [Google Scholar] [PubMed] [CrossRef]
27. Hu, H., Feng, N., Shen, X., Zhao, L., Zheng, D. (2022). Transcriptomic analysis of Vigna radiata in response to chilling stress and uniconazole application. BMC Genomics, 23(1), 205. https://doi.org/10.1186/s12864-022-08443-6 [Google Scholar] [PubMed] [CrossRef]
28. Li, S. W., Shi, R. F., Leng, Y. (2015). De novo characterization of the mung bean transcriptome and transcriptomic analysis of adventitious rooting in seedlings using RNA-seq. PLoS One, 10(7), e0132969. https://doi.org/10.1371/journal.pone.0132969 [Google Scholar] [PubMed] [CrossRef]
29. Tian, X., Li, S., Liu, Y., Liu, X. (2016). Transcriptomic profiling reveals metabolic and regulatory pathways in the desiccation tolerance of mungbean (Vigna radiata [L.] R. Wilczek). Frontiers in Plant Science, 7, 1921. [Google Scholar] [PubMed]
30. Li, S. W., Leng, Y., Shi, R. F. (2017). Transcriptomic profiling provides molecular insights into hydrogen peroxide-induced adventitious rooting in mung bean seedlings. BMC Genomics, 18(1), 188. https://doi.org/10.1186/s12864-017-3576-y [Google Scholar] [PubMed] [CrossRef]
31. Kang, Y. J., Satyawan, D., Shim, S., Lee, T., Lee, J. et al. (2015). Draft genome sequence of adzuki bean, Vigna angularis. Scientific Reports, 5(1), 8069. https://doi.org/10.1038/srep08069 [Google Scholar] [PubMed] [CrossRef]
32. Li, Y., Ishida, M., Ashida, H., Ishikawa, T., Shibata, H. et al. (2014). A non-NadB type L-aspartate dehydrogenase from Ralstonia eutropha strain JMP134: Molecular characterization and physiological functions. Bioscience, Biotechnology, and Biochemistry, 75(8), 1524–1532. https://doi.org/10.1271/bbb.110216 [Google Scholar] [PubMed] [CrossRef]
33. Saunders, A. H., Griffiths, A. E., Lee, K. H., Cicchillo, R. M., Tu, L. et al. (2008). Characterization of quinolinate synthases from Escherichia coli, Mycobacterium tuberculosis, and Pyrococcus horikoshii indicates that [4Fe-4S] clusters are common cofactors throughout this class of enzymes. Biochemistry, 47(41), 10999–11012. https://doi.org/10.1021/bi801268f [Google Scholar] [PubMed] [CrossRef]
34. Chow, C., Hegde, S., Blanchard, J. S. (2017). Mechanistic characterization of Escherichia coli l-aspartate oxidase from kinetic isotope effects. Biochemistry, 56(31), 4044–4052. https://doi.org/10.1021/acs.biochem.7b00307 [Google Scholar] [PubMed] [CrossRef]
35. Bitew, M. A., Wawegama, N. K., Newton, H. J., Sansom, F. M. (2019). Meso-tartrate inhibits intracellular replication of Coxiella burnetii, the causative agent of the zoonotic disease Q fever. Pathogens and Disease, 77(8), e00175. https://doi.org/10.1093/femspd/ftz066 [Google Scholar] [PubMed] [CrossRef]
36. Tedeschi, G., Nonnis, S., Strumbo, B., Cruciani, G., Carosati, E. et al. (2010). On the catalytic role of the active site residue E121 of E. coli L-aspartate oxidase. Biochimie, 92(10), 1335–1342. https://doi.org/10.1016/j.biochi.2010.06.015 [Google Scholar] [PubMed] [CrossRef]
37. Seifert, J., Kunz, N., Flachmann, R., Laufer, A., Jany, K. D. et al. (1990). Expression of the E. coli nadB gene and characterization of the gene product L-aspartate oxidase. Biological Chemistry Hoppe-Seyler, 371(3), 239–248. https://doi.org/10.1515/bchm3.1990.371.1.239 [Google Scholar] [CrossRef]
38. Teramoto, H., Suda, M., Inui, M., Yukawa, H. (2010). Regulation of the expression of genes involved in NAD de novo biosynthesis in Corynebacterium glutamicum. Applied and Environmental Microbiology, 76(16), 5488–5495. https://doi.org/10.1128/AEM.00906-10 [Google Scholar] [PubMed] [CrossRef]
39. Marinoni, I., Nonnis, S., Monteferrante, C., Heathcote, P., Hartig, E. et al. (2008). Characterization of L-aspartate oxidase and quinolinate synthase from Bacillus subtilis. FEBS Journal, 275(20), 5090–5107. https://doi.org/10.1111/j.1742-4658.2008.06641.x [Google Scholar] [PubMed] [CrossRef]
40. di Martino, M. L., Fioravanti, R., Barbabella, G., Prosseda, G., Colonna, B. et al. (2013). Molecular evolution of the nicotinic acid requirement within the Shigella/EIEC pathotype. International Journal of Medical Microbiology, 303(8), 651–661. https://doi.org/10.1016/j.ijmm.2013.09.007 [Google Scholar] [PubMed] [CrossRef]
41. Bitew, M. A., Khoo, C. A., Neha, N., de Souza, D. P., Tull, D. et al. (2018). De novo NAD synthesis is required for intracellular replication of Coxiella burnetii, the causative agent of the neglected zoonotic disease Q fever. Journal of Biological Chemistry, 293(48), 18636–18645. https://doi.org/10.1074/jbc.RA118.005190 [Google Scholar] [PubMed] [CrossRef]
42. DeVries, C. A., Hassett, D. J., Flynn, J. L., Ohman, D. E. (1995). Genetic linkage in Pseudomonas aeruginosa of algT and nadB: Mutation in nadB does not affect NAD biosynthesis or alginate production. Gene, 156(1), 63–67. https://doi.org/10.1016/0378-1119(95)00028-5 [Google Scholar] [PubMed] [CrossRef]
43. Bouvet, O., Bourdelier, E., Glodt, J., Clermont, O., Denamur, E. (2017). Diversity of the auxotrophic requirements in natural isolates of Escherichia coli. Microbiology, 163(6), 891–899. https://doi.org/10.1099/mic.0.000482 [Google Scholar] [PubMed] [CrossRef]
44. Liu, J., Xue, C., Lin, Y., Yan, Q., Chen, J. et al. (2022). Genetic analysis and identification of VrFRO8, a salt tolerance-related gene in mungbean. Gene, 836(3), 146658. https://doi.org/10.1016/j.gene.2022.146658 [Google Scholar] [PubMed] [CrossRef]
45. Tariq, R., Hussain, A., Tariq, A., Khalid, M. H. B., Khan, I. et al. (2022). Genome-wide analyses of the mung bean NAC gene family reveals orthologs, co-expression networking and expression profiling under abiotic and biotic stresses. BMC Plant Biology, 22(1), 343. https://doi.org/10.1186/s12870-022-03716-4 [Google Scholar] [PubMed] [CrossRef]
46. Liu, C., Zhang, Q., Dong, J., Cai, C., Zhu, H. et al. (2022). Genome-wide identification and characterization of mungbean CIRCADIAN CLOCK ASSOCIATED 1 like genes reveals an important role of VrCCA1L26 in flowering time regulation. BMC Genomics, 23(1), 374. https://doi.org/10.1186/s12864-022-08620-7 [Google Scholar] [PubMed] [CrossRef]
47. Azeem, F., Ijaz, U., Ali, M. A., Hussain, S., Zubair, M. et al. (2022). Genome-wide identification and expression profiling of potassium transport-related genes in Vigna radiata under abiotic stresses. Plants, 11(1), 2. https://doi.org/10.3390/plants11010002 [Google Scholar] [PubMed] [CrossRef]
48. Zhang, W., Ye, S., Du, Y., Zhao, Q., Du, J. et al. (2022). Identification and expression analysis of bZIP members under abiotic stress in mung bean (Vigna radiata). Life, 12(7), 938. https://doi.org/10.3390/life12070938 [Google Scholar] [PubMed] [CrossRef]
49. Lin, Y., Laosatit, K., Chen, J., Yuan, X., Wu, R. et al. (2020). Mapping and functional characterization of stigma exposed 1, a DUF1005 gene controlling petal and stigma cells in mungbean (Vigna radiata). Frontiers in Plant Science, 11, 575922. https://doi.org/10.3389/fpls.2020.575922 [Google Scholar] [PubMed] [CrossRef]
50. Pan, L., Yu, X., Shao, J., Liu, Z., Gao, T. et al. (2019). Transcriptomic profiling and analysis of differentially expressed genes in asparagus bean (Vigna unguiculata ssp. sesquipedalis) under salt stress. PLoS One, 14(7), e0219799. https://doi.org/10.1371/journal.pone.0219799 [Google Scholar] [PubMed] [CrossRef]
51. Zhang, Q., Yan, Q., Yuan, X., Lin, Y., Chen, J. et al. (2021). Two polygalacturonase-inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp.). Journal of Plant Physiology, 258–259(1), 153376. https://doi.org/10.1016/j.jplph.2021.153376 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools