 Open Access
Open Access
ARTICLE
In silico Prediction and Analysis of Potential Off-Targets and Off-Target Mutation Detection in StERF3-Gene Edited Potato Plants
1
Centre of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture, Faisalabad, 38040, Pakistan
2
Department of Plant Pathology, University of Agriculture, Faisalabad, 38040, Pakistan
* Corresponding Author: Siddra Ijaz. Email:
(This article belongs to the Special Issue: Agricultural Intensification, Climate Change, and Food Security)
Phyton-International Journal of Experimental Botany 2023, 92(8), 2451-2460. https://doi.org/10.32604/phyton.2023.030501
Received 10 April 2023; Accepted 11 May 2023; Issue published 25 June 2023
Abstract
The imperative aspect of the CRISPR/Cas9 system is a short stretch of 20 nucleotides of gRNA that control the overall specificity. Due to the small size, the chance of its multiple occurrences in the genome increases; however, a few mismatches are tolerated by the Cas9 endonuclease activity. An accurate and careful in silico-based off-target prediction while target selection is preferred to address the issue. These predictions are based on a comprehensive set of selectable parameters. Therefore, we investigated the possible off-target prediction and their screening in StERF3 gene-edited potato plants while developing StERF3-loss-of-function mutants using CRISPR/Cas9 approach. The 201 off-targets for the selected targets of the StERF3 gene were predicted, and 79 were filtered as potential off-targets. Of these 79, twenty-five off-targets showed scores with defined cut-off values <0.5 and were analyzed in Sterf3-edited potato plants compared to wild-type plants. No off-targeting was found to have occurred in edited plants.Keywords
Supplementary Material
Supplementary Material FileGenome editing techniques have grown with the use of engineered biomolecules for directing the modification in the genome. These techniques benefited from the natural DNA repair mechanism after inducing a site-directed break in the double-stranded DNA. Repairing involves a non-templated non-homologous end-joining mechanism or, in the case of break-site homologous templates, a homology-directed repair mechanism [1]. These techniques became increasingly popular, displacing transgenics because of their less toxic effect as well as do not pass through tight regulatory policies. Particularly Zinc-finger nucleases (ZFN), TALE nucleases (TALEN), and CRISPR/Cas systems are used to edit an organism’s genome. ZFN and TALEN work as dimers and direct the DNA double-stranded break with the help of the fused non-specific endonuclease domain, FokI [2,3]. There are few reports of successful applications of the ZFN and TALEN in mouse, mammalian, and plant cells [4–6]. However, the limitations of their use include high off-target edits, no ability to target methylated DNA (control several processes, i.e., gene expression, DNA repair, plant growth and development, stress response), and expensive and tedious designing [7–10]. However, the rapid and widespread adoption of genome editing as a preferred technique to target highly specific changes in genomes is due to the development of the robust CRISPR/Cas9 system [11]. This technique is simple, highly specific, less expensive, and does not need structural validation [12].
CRISPR/Cas9 is a prokaryotes, derived system that produces a double-stranded break at the targeted site of DNA, adjacent to the protospacer adjacent motif (PAM) site (5′-NGG-3′ sequence recognized by Cas9 endonuclease of Streptococcus pyogenes). This event is guided by ~20 nucleotide long single guide RNA (sgRNA), and Cas-9 produces the break after protospacer adjacent motif (PAM) recognition. Apart from the on-target site (selected site for targeting), this 20-nucleotide stretch of sgRNA may be found multiple times anywhere in the genome, defined as off-target [13]. However, genotoxicity and the possibility of off-targeting with CRISPR/Cas9 have been documented [14,15]. Off-target editing may have negative consequences like genotoxicity, cytotoxicity, or possible chromosomal rearrangements [16]. Off-target edits can be detected and evaluated to limit undesirable downstream phenotypic consequences. Genome editing for therapeutic purposes in humans strongly focuses on the potential side effects of off-targeting events in the genome. Conversely, crop-specific editing concerns are not directly relevant to mammalian systems [17]. These problems must be considered as they raise safety and regulatory concerns.
Studies based on genome-editing events in crops possibly encounter the most deleterious effect, like off-targeting. The off-targets have been predicted through in silico predictors, CHOPCHOP, CRISPOR, Cas-OFFinder, CCTop, Crisflash, etc. [18]. These predictors identify sgRNA and its efficiency for the targeted site and predict possible off-targets. However, different parameters and algorithms of predictors cause inconsistencies in their results. Overall, off-target predictions, criteria are the presence of a defined PAM site (NGG) and the number and position of mismatches. The activity of Cas9 endonuclease strongly correlates with PAM identification and stable gRNA-DNA heteroduplex. The gRNA complements the target site and controls the system’s specificity. However, sequences with few mismatches are acceptable by sgRNA/Cas9 and result in off-targeting. The double-strand break may be prevented if the overall sequence of sgRNA has >4 bp [8].
Fewer mismatches and their position upstream to PAM associated with unintended CRISPR/Cas9 editing at untargeted genome loci. Cutting efficiency of the CRISPR/Cas9 system depends on several points, including GC contents of the target site, the presence of the PAM site, and mismatched positions in off-target [19]. The off-target sequences with mismatches ≤4 are more likely to be cut during this event [20]. Moreover, the seed region (8–12 bp upstream from the PAM) with ≤2 mismatches could be at high risk of cleavage [21]. Therefore, a detailed analysis predicting possible off-target and on-target edits should be investigated before CRISPR/Cas-9 targeting. The cleavage based on homology other than the target site results in off-target editing, and the related repercussion is challenging for the researchers [18].
Several in silico tools are available to investigate off-target sites. However, the difference among their results has been observed because each follows different criteria and algorithms [22]. The present study aimed to predict possible off-target sites of selected targets of the StERF3 gene in Solanum tuberosum. Our previous study successfully produced StERF3 full allelic edited potato knockouts (PTP2 and PTP3) [23]. The potential off-target sites of selected targets were identified using the three most commonly used online predictors and were screened in PTP2 and PTP3 plants.
2.1 In Silico Analysis of Off-Targets
The target regions of the Sterf3 gene for CRISPR/Cas9 editing were selected using CHOPCHOP and CRISPOR web tools. The location of selected targets was inside the Sterf3 gene. For off-target prediction, off-target site predictors, CHOPCHOP [24], CRISPOR [25], and CC-TOP [26] were employed. Full description is available at https://link.springer.com/article/10.1007/s11033-022-07958-1.
2.1.2 Putative Off-Target Selection
Off-targets were filtered for further analysis following the criterion of conservation of ≤5 nucleotides upstream to Protospacer adjacent motif (PAM), allowing ≤2 nucleotides as maximum core region (seed region) mismatches [13].
The off-target scoring was based on the CCTop scoring algorithm and computed using the formula given by Stemmer et al. [26].
*pos = position of each mismatch from the 5′ end.
2.1.4 Off-Target Sequence Analysis for Mismatch Distribution
Off-target sequences were analyzed for mismatch distribution magnitude through heat map analysis using TBtools-II (Toolbox for Biologists) v1.112.
2.2 Off-Target Analysis in the Sterf3-Edited Potato Plants
2.2.1 DNA Isolation and PCR Analysis
The detected putative off-target sites were probed in Sterf3-edited potato plants to identify off-target events. DNA of fully edited plants (PTP2 &PTP3) and a wild-type plant was isolated using GeneJET Plant Genomic DNA Purification Kit (Thermo Scientific, USA). The PCR amplification was achieved on a thermal cycler, peqSTAR, and products were resolved on 1% agarose gel through gel electrophoresis. The off-target primers for PCR analysis and off-target event identification are in Table S1.
2.2.2 DNA Elution, Cloning, and Sequencing
For each off-target, the DNA band of the required amplicon size was excised from the gel and eluted using Gel Purification Kit (FavorPrep, Taiwan). The eluted product for each off-target was cloned into a cloning vector, pTZ57R/T, using the InsTAclone PCR Cloning Kit (Thermo Scientific, USA) and direct sequenced to Eurofins Genomics DNA Sequencing Services, USA.
The generated sequences were trimmed through BioEdit ver. 7.2.6.1. The alignment of trimmed high-quality sequences of wild type, PTP2, and PTP3 was made using Multiple Alignment using Fast Fourier Transform (MAFFT), a multiple sequence alignment tool.
Off-targeting is the major concern associated with genome editing executed through the CRISPR-Cas system. Therefore scientists have turned their faces to find the solution regarding off-target prediction while selecting the target sites. In silico web-tools are being practiced to address the issue of off-targeting. This study found two hundred and one (201) off-target sequences (Table S2) with canonical PAM for target-1 sgRNA and target-2 sgRNA using the three predictors against the reference Solanum tuberosum genome. CHOPCHOP and CCTop recognized fewer off-target sites for target-1 (2 and 20, respectively) and target-2 (17 and 20, respectively). CRISPOR brought the highest number of predicted off-target sites for target-1 and target-2 (48 and 94, respectively). After filtering for sequences with ≤5 mismatches, 79 sequences (Table 1) were obtained as potential off-targets for both target sites (Fig. 1). The potential off-targets with >4 mismatches prevent double-stranded break induction [13].
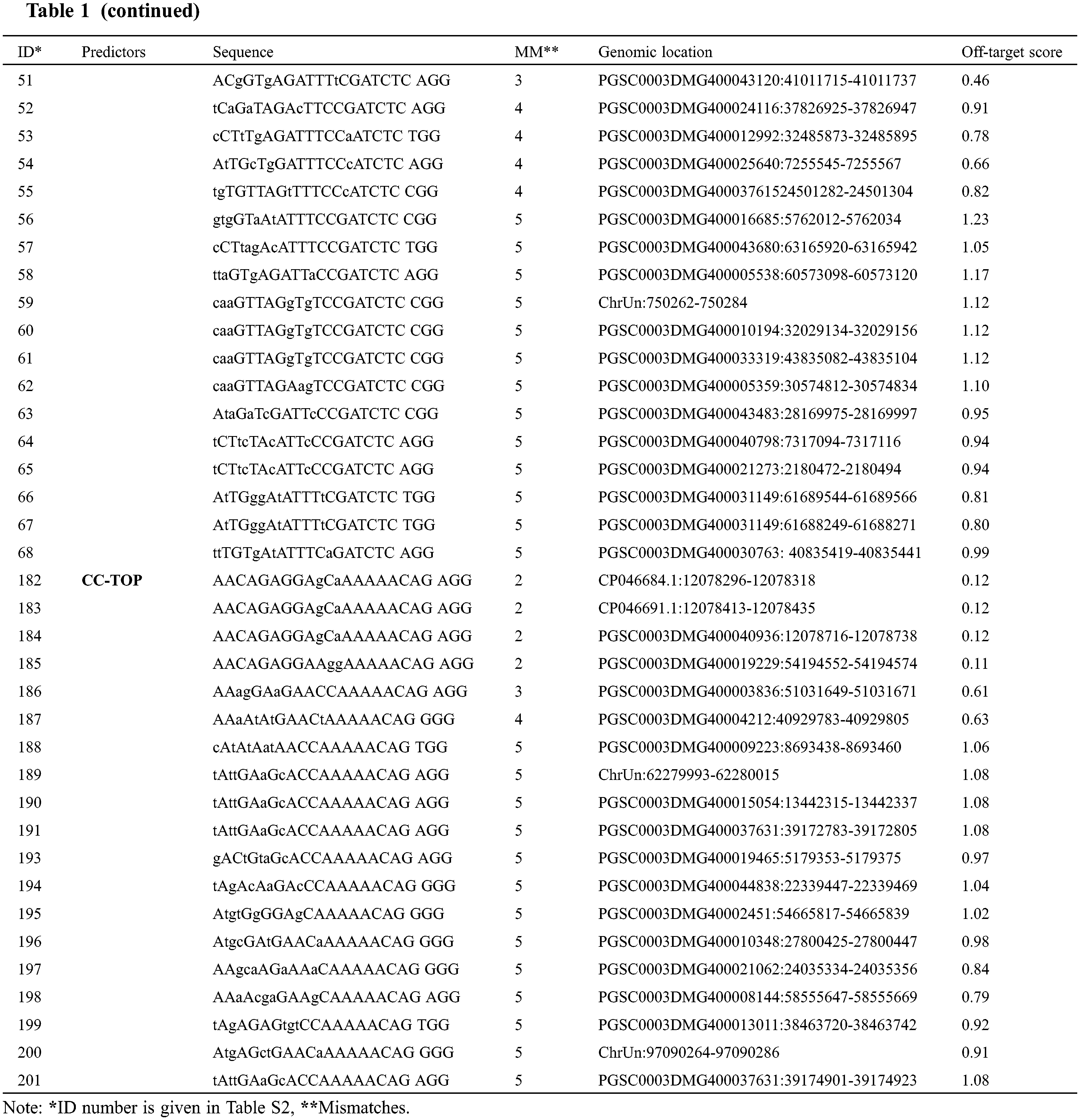
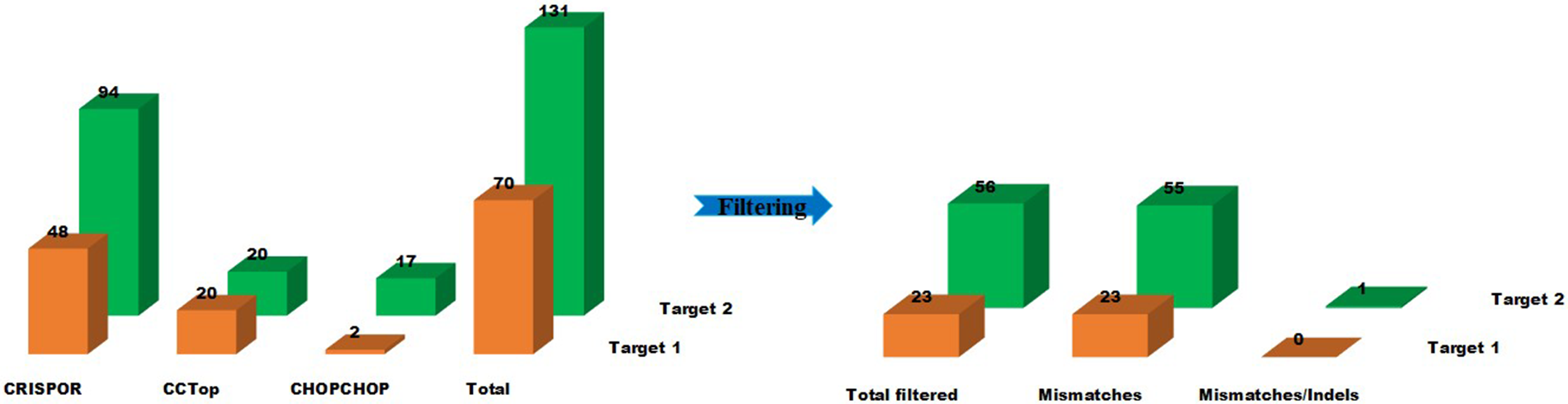
Figure 1: Off-target prediction to target-1 and target-2 of the Sterf3 gene by employing off-target site predictors, CHOPCHOP, CRISPOR, and CC-TOP and filter criteria. Graphically illustrate the off-target predictions of both targets before and after filtering steps. The illustration shows the number of sequences returned for each of the three predictors and the number of sequences after the filtering step
The seed region determines the overall target specificity. The closer the mismatches to PAM, the more chances to abolish double-strand break [13]. Therefore, we selected the off-targets with ≤2 bp mismatches in the seed region because Cas9 can tolerate mismatches up to 5 and does more off-target edits [13,27,28]. Conserving a few base pairs adjacent to PAM is also important [21,29]. Hence, we selected off-target sequences following 5 bp conservation. Similarly, Carneiro et al. [22] also used the criteria of 5 bp conservation upstream to PAM.
In off-target sequences with mismatches only, PAM is located at positions 21–23, while in off-target sequences with mismatches and indels, PAM location was found at positions 20 to 22. The non-random distribution was observed, as most of these mismatches were concentrated at positions 1–12 in off-target sequences (Fig. 2). Fifteen identical off-target sequences were observed more than once in different genomic positions, which could be responsible for this distribution.
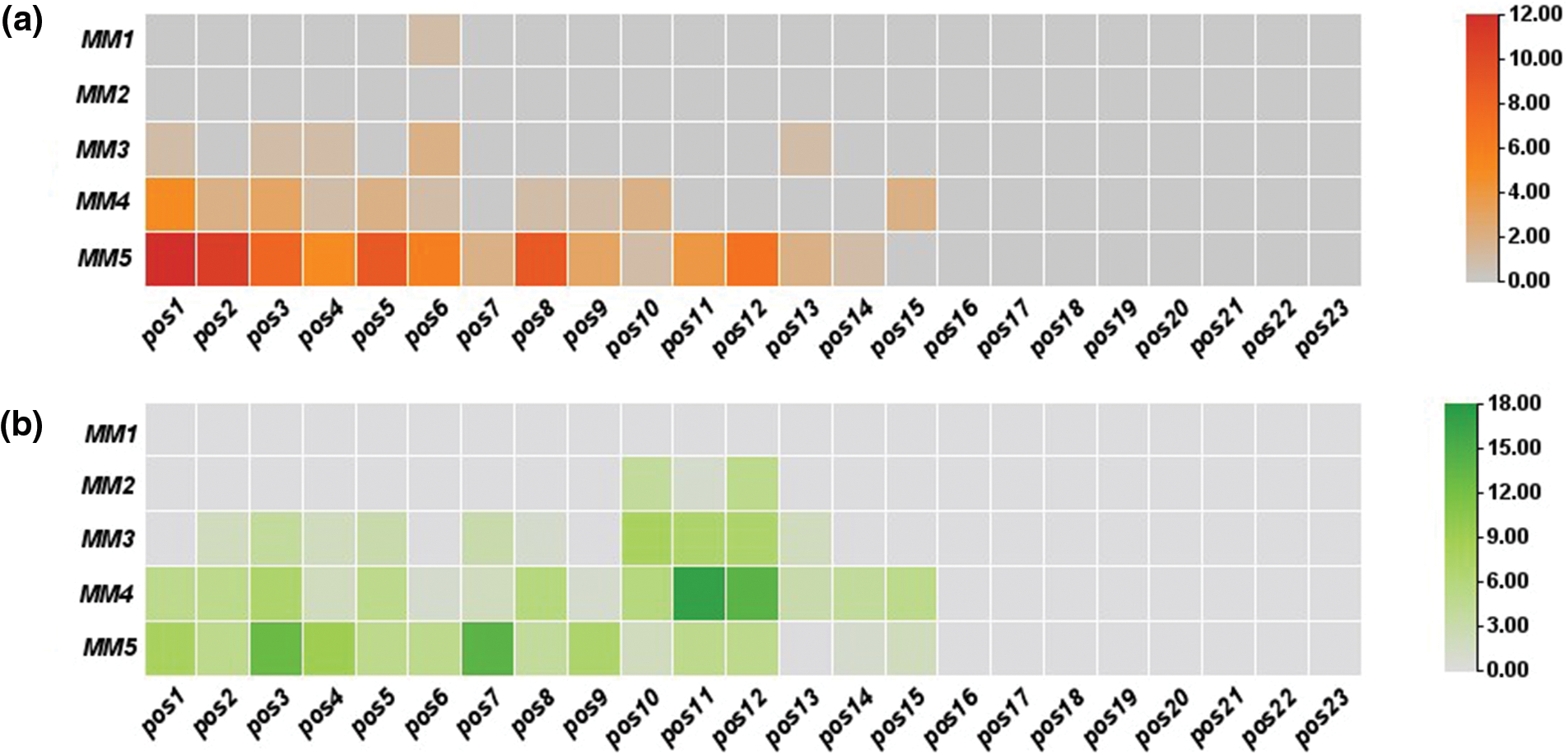
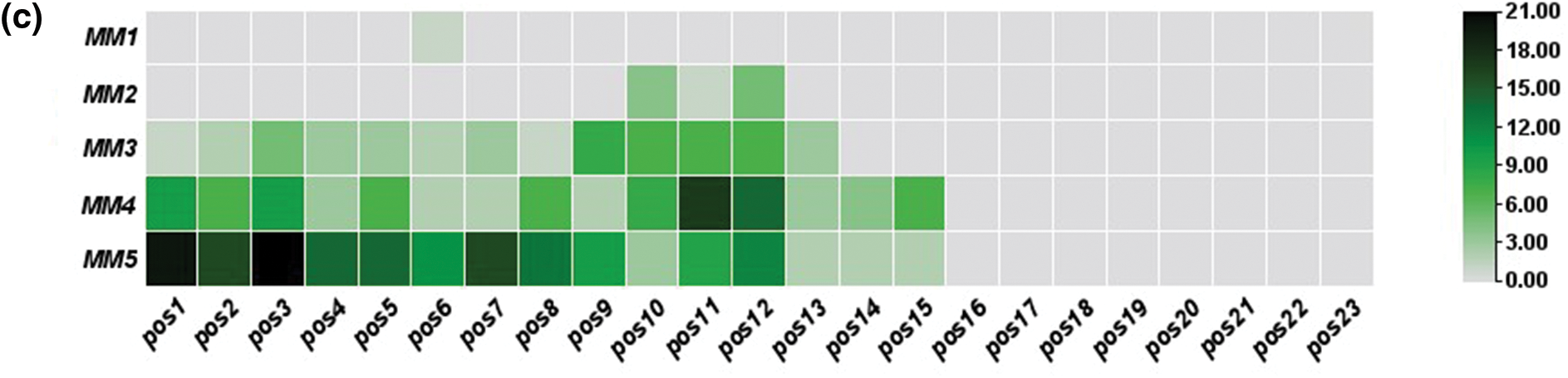
Figure 2: Distribution magnitude and frequency of mismatches in predicted off-targets by heat map analysis; (a) distribution magnitude of mismatches in off-targets of target-1; (b) distribution magnitude of mismatches in off-targets of target-2; and (c) distribution magnitude of mismatches in off-targets of both targets
The cleavage probability for putative off-targets was scored based on the CCTop score algorithm. The higher the CCTop score, the lower the chance of off-targeting [30]. This scoring method is based on the position of mismatched nucleotides from the PAM [26]. Here in this study, we defined 0.5 as a cut-off value, and the off-targets with a score of less than 0.5 were selected for identifying off-target events in fully edited plants of Solanum tuberosum. Of these 79 off-targets, twenty-five off-targets with a score <0.5 were selected for off-targeting analysis of Sterf3-edited potato plants.
Doench et al.’s [30] work for minimizing off-target effects of CRISPR-Cas9 documented that criteria for cut-off value should be based on experimental observation. Carneiro et al. [22] followed Doench et al. [30] and set the cut-off value greater than 0.2. So, for more stringency, we set the cut-off value <0.5 and screened the material for more off-targets. As the cut-off value is high, more off-targets would be selected for analysis to minimize the off-target effects of CRISPR-Cas9 in the edited plants.
3.1 Detection of Off-Target Mutations in Edited Potato Plants
The putative off-target sites of selected targeted sites of the StERF3 gene were probed for off-target events in Sterf3-edited potato plants. DNA fragments with putative off-target sites amplified through PCR amplification (Fig. 3) were cloned in a TA cloning vector for sequencing. The primers to amplify off-target sites were selected following the criteria, 600 bp-sanger reads. Sequencing data of PCR amplicons were aligned, which showed no mutation at the putative off-target sites of StERF3 knockout mutants (Supplementary File).
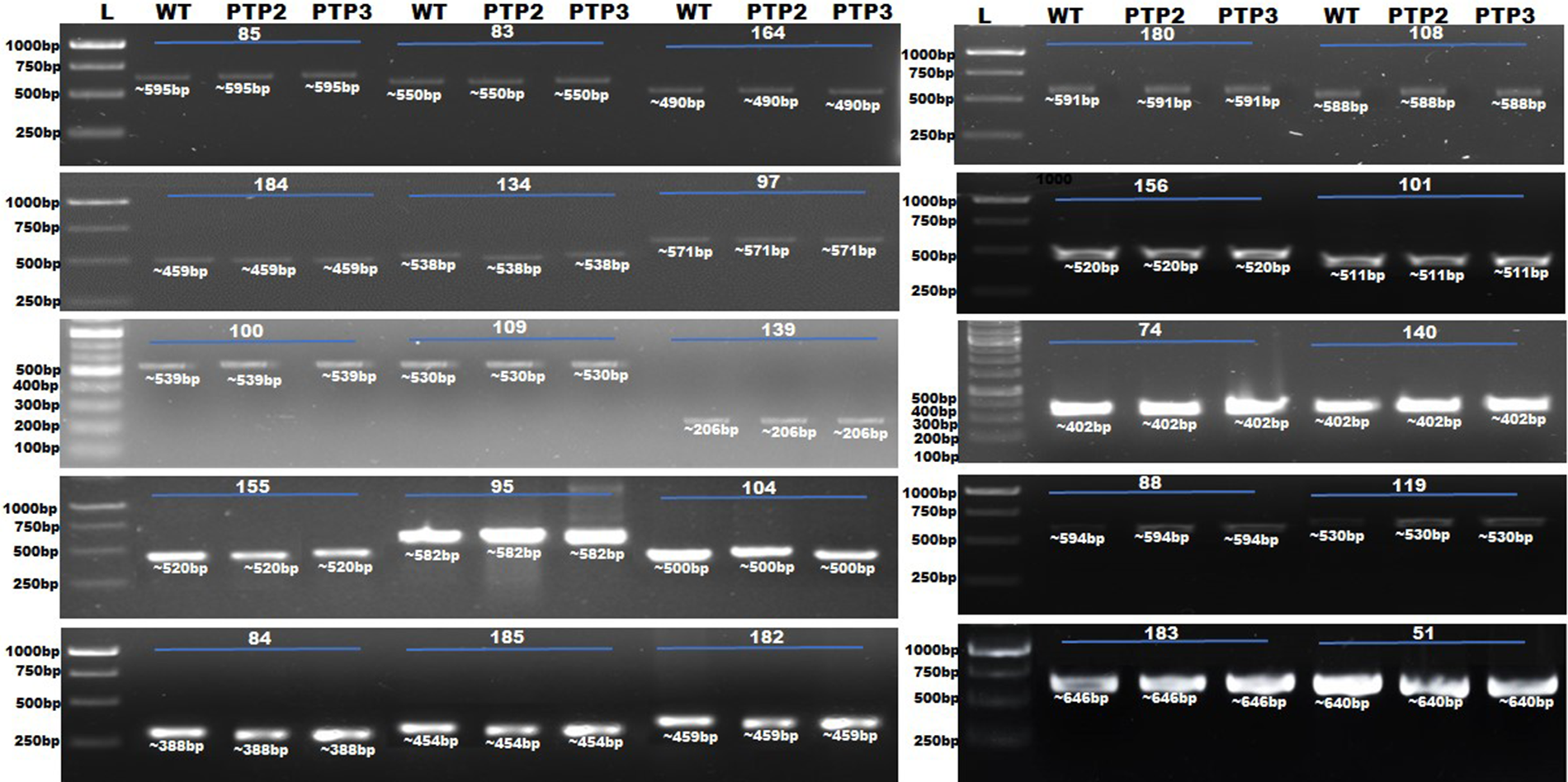
Figure 3: PCR amplification of off-targets in Sterf3-edited potato plants (PTP2 and PTP3) compared to wild-type plants (WT). *L = 1Kb DNA ladder 51 = PGSC0003DMG400043120:41011715-41011737, 74 = CP055244.1:13377863-13377885, 83 = CP023767.1:9582767-9582789, 84 = CP055240.1:38608045-38608067, 85 = CP046684.1: 16274350-16274372, 184 = PGSC0003DMG400040936:12078716-12078738, 88 = CM020976.1 47.24 Mbp_CM020976.1_47243233, 95 = CM020969.1 57.56 Mbp_CM020969.1_57556750, 97 = CM020967.1 54.09 Mbp_CM020967.1_54094651, 100 = CM020976.1 41.23 Mbp_CM020976.1_41234525, 101 = CM020976.1 34.81 Mbp_CM020976.1_34814664, 104 = CM020974.1 10.79 Mbp_CM020974.1_10791170, 109 = CM020969.1 42.66 Mbp_CM020969.1_42660027, 108 = CM020969.1 41.63 Mbp_CM020969.1_41630271, 119 = CM020974.1 46.26 Mbp_CM020974.1_46256016, 134 = CM020973.1 1.32 Mbp_CM020973.1_1324106, 139 = CM020970.1 11.78 Mbp_CM020970.1_11775544, 140 = CM020972.1 602.80 Kbp_CM020972.1_602798, 155 = CM020965.1 84.70 Mbp_CM020965.1_84700489, 156 = CM020965.1 36.17 Mbp_CM020965.1_36174514, 164 = CM020967.1 53.84 Mbp_CM020967.1_53841206, 180 = CM020969.121.34 Mbp_CM020969.1_21341797, 182 = CP046684.1:12078296-12078318, 183 = CP046691.1:12078413-12078435, 185 = PGSC0003DMG400019229:54194552-54194574
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm their contribution to the paper as follows: Study conception and design: Siddra Ijaz and Imran Ul Haq; Data collection: Hafiza Arooj Razzaq, Siddra Ijaz, and Imran Ul Haq; Analysis and interpretation of results: Siddra Ijaz, Imran Ul Haq, and Hafiza Arooj Razzaq; Draft manuscript preparation: Hafiza Arooj Razzaq, Siddra Ijaz, Imran Ul Haq, and Faisal Saeed Awan. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Curtin, S. J., Voytas, D. F., Stupar, R. M. (2012). Genome engineering of crops with designer nucleases. Plant Genome, 5(2), 42–50. https://doi.org/10.3835/plantgenome2012.06.0008 [Google Scholar] [CrossRef]
2. Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., Gregory, P. D. (2010). Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics, 11(9), 636–646. https://doi.org/10.1038/nrg2842 [Google Scholar] [PubMed] [CrossRef]
3. Bortesi, L., Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology Advances, 33(1), 41–52. https://doi.org/10.1016/j.biotechadv.2014.12.006 [Google Scholar] [PubMed] [CrossRef]
4. Dambournet, D., Hong, S. H., Grassart, A., Drubin, D. G. (2014). Tagging endogenous loci for live-cell fluorescence imaging and molecule counting using ZFNs, TALENs, and Cas9. In: Methods in enzymology, vol. 546, pp. 139–160. USA: Academic Press. https://doi.org/10.1016/B978-0-12-801185-0.00007-6 [Google Scholar] [CrossRef]
5. Liang, Z., Zhang, K., Chen, K., Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. Journal of Genetics and Genomics, 41(2), 63–68. https://doi.org/10.1016/j.jgg.2013.12.001 [Google Scholar] [PubMed] [CrossRef]
6. Sato, T., Sakuma, T., Yokonishi, T., Katagiri, K., Kamimura, S. et al. (2015). Genome editing in mouse spermatogonial stem cell lines using TALEN and double-nicking CRISPR/Cas9. Stem Cell Reports, 5(1), 75–82. https://doi.org/10.1016/j.stemcr.2015.05.011 [Google Scholar] [PubMed] [CrossRef]
7. Engler, C., Kandzia, R., Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One, 3(11), e3647. https://doi.org/10.1371/journal.pone.0003647 [Google Scholar] [PubMed] [CrossRef]
8. Pattanayak, V., Lin, S., Guilinger, J. P., Ma, E., Doudna, J. A. et al. (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature Biotechnology, 31(9), 839–843. https://doi.org/10.1038/nbt.2673 [Google Scholar] [PubMed] [CrossRef]
9. Lowder, L. G., Zhang, D., Baltes, N. J., Paul, J. W., Tang, X. et al. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiology, 169(2), 971–985. https://doi.org/10.1104/pp.15.00636 [Google Scholar] [PubMed] [CrossRef]
10. Malzahn, A., Lowder, L., Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell & Bioscience, 7(1), 1–18. https://doi.org/10.1186/s13578-017-0148-4 [Google Scholar] [PubMed] [CrossRef]
11. Gaj, T., Gersbach, C. A., Barbas, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology, 31(7), 397–405. https://doi.org/10.1016/j.tibtech.2013.04.004 [Google Scholar] [PubMed] [CrossRef]
12. Doudna, J. A., Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096. https://doi.org/10.1126/science.1258096 [Google Scholar] [CrossRef]
13. Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S. et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology, 31(9), 827–832. https://doi.org/10.1038/nbt.2647 [Google Scholar] [PubMed] [CrossRef]
14. Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F. et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnology Journal, 12(6), 797–807. https://doi.org/10.1111/pbi.12200 [Google Scholar] [PubMed] [CrossRef]
15. Peterson, B. A., Haak, D. C., Nishimura, M. T., Teixeira, P. J., James, S. R. (2016). Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One, 11(9), e0162169. https://doi.org/10.1371/journal.pone.0162169 [Google Scholar] [PubMed] [CrossRef]
16. Corrigan-Curay, J., O’Reilly, M., Kohn, D. B., Cannon, P. M., Bao, G. et al. (2015). Genome editing technologies: Defining a path to clinic. Molecular Therapy, 23(5), 796–806. https://doi.org/10.1038/mt.2015.54 [Google Scholar] [PubMed] [CrossRef]
17. Wolt, J. D. (2017). Safety, security, and policy considerations for plant genome editing. In: Progress in molecular biology and translational science, vol. 149, pp. 215–241. Netherlands: Elsevier Inc. https://doi.org/10.1016/bs.pmbts.2017.03.005 [Google Scholar] [CrossRef]
18. Bao, X. R., Pan, Y., Lee, C. M., Davis, T. H., Bao, G. (2021). Tools for experimental and computational analyses of off-target editing by programmable nucleases. Nature Protocols, 16(1), 10–26. https://doi.org/10.1038/s41596-020-00431-y [Google Scholar] [PubMed] [CrossRef]
19. Fu, Y., Foden, J. A., Khayter, C., Maeder, M. L., Reyon, D. et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology, 31(9), 822–826. https://doi.org/10.1038/nbt.2623 [Google Scholar] [PubMed] [CrossRef]
20. Cho, S. W., Kim, S., Kim, Y., Kweon, J., Kim, H. S. et al. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research, 24(1), 132–141. https://doi.org/10.1101/gr.162339.113L [Google Scholar] [CrossRef]
21. Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R. et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121), 819–823. https://doi.org/10.1126/science.1231143 [Google Scholar] [PubMed] [CrossRef]
22. Carneiro, P., de Freitas, M. V., Matte, U. (2022). In silico analysis of potential off-target sites to gene editing for Mucopolysaccharidosis type I using the CRISPR/Cas9 system: Implications for population-specific treatments. PLoS One, 17(1), e0262299. https://doi.org/10.1371/journal.pone.0262299 [Google Scholar] [PubMed] [CrossRef]
23. Razzaq, H. A., Ijaz, S., Haq, I. U., Khan, I. A. (2022). Functional inhibition of the StERF3 gene by dual targeting through CRISPR/Cas9 enhances resistance to the late blight disease in Solanum Tuberosum L. Molecular Biology Reports, 49(12), 11675–11684. https://doi.org/10.1007/s11033-022-07958-1 [Google Scholar] [PubMed] [CrossRef]
24. Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B., Valen, E. (2016). CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Research, 44(W1), W272–W276. https://doi.org/10.1093/nar/gkw398 [Google Scholar] [PubMed] [CrossRef]
25. Concordet J.P., Haeussler M. (2018). CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research, 46(W1), W242–W245. https://doi.org/10.1093/nar/gky354 [Google Scholar] [PubMed] [CrossRef]
26. Stemmer, M., Thumberger, T., del Sol Keyer, M., Wittbrodt, J., Mateo, J. L. (2017). Correction: CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One, 12(4), e0176619. https://doi.org/10.1371/journal.pone.0124633 [Google Scholar] [PubMed] [CrossRef]
27. Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature, 507(7490), 62–67. https://doi.org/10.1038/nature13011 [Google Scholar] [PubMed] [CrossRef]
28. Young, J., Zastrow-Hayes, G., Deschamps, S., Svitashev, S., Zaremba, M. et al. (2019). CRISPR-Cas9 editing in maize: Systematic evaluation of off-target activity and its relevance in crop improvement. Scientific Reports, 9(1), 6729. [Google Scholar] [PubMed]
29. Wu, X., Scott, D. A., Kriz, A. J., Chiu, A. C., Hsu, P. D. et al. (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nature Biotechnology, 32(7), 670–676. https://doi.org/10.1038/nbt.2889 [Google Scholar] [PubMed] [CrossRef]
30. Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W. et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature Biotechnology, 34(2), 184–191. https://doi.org/10.1038/nbt.3437 [Google Scholar] [PubMed] [CrossRef]
Appendix
Table S1: List of primers use to amplify twenty-five putative off-targets of two targets (target-1 & target-2)
Table S2: Off-target sites prediction of two targets with three online predictors
Supplementary File: Alignment
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools