 Open Access
Open Access
ARTICLE
Genome-Wide Analysis of the KANADI Gene Family and Its Expression Patterns under Different Nitrogen Concentrations Treatments in Populus trichocarpa
1 State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University), School of Forestry, Northeast Forestry University, Harbin, 150040, China
2 Key Laboratory of Saline-Alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin, 150040, China
3 College of Life Science, Northeast Forestry University, Harbin, 150040, China
4 College of Forestry, Guizhou University, Guiyang, 550025, China
* Corresponding Authors: Chunpu Qu. Email: ; Guanjun Liu. Email:
# The first two authors have contributed equally to this work
Phyton-International Journal of Experimental Botany 2023, 92(7), 2001-2015. https://doi.org/10.32604/phyton.2023.029350
Received 14 February 2023; Accepted 22 March 2023; Issue published 29 May 2023
Abstract
KANADI (KAN) is a plant-specific gene that controlled the polarity development of lateral organs. It mainly acted on the abaxial characteristics of plants to make the lateral organs asymmetrical. However, it had been less identified in woody plants. In this study, the members of the KAN gene family in Populus trichocarpa were identified and analyzed using the bioinformatics method. The results showed that a total of 8 KAN family members were screened out, and each member contained the unique GARP domain and conserved region of the family proteins. Phylogenetic analysis and their gene structures revealed that all KAN genes from P. trichocarpa, Arabidopsis thaliana, and Nicotiana benthamiana could be divided into four subgroups, while the eight genes in P. trichocarpa were classified into three subgroups, respectively. The analysis of tissue-specific expression indicated that PtKAN1 was highly expressed in young leaves, PtKAN6 was highly expressed in young leaves and mature leaves, PtKAN2, PtKAN5, and PtKAN7 were highly expressed in nodes and internodes, PtKAN8 was highly expressed in roots, and PtKAN3 and PtKAN4 showed low expression levels in all tissues. Among them, PtKAN2 and PtKAN6, and PtKAN4 and PtKAN5 might have functional redundancy. Under high nitrogen concentrations, PtKAN2 and PtKAN8 were highly expressed in mature stems and leaves, respectively, while PtKAN4, PtKAN5, and PtKAN7 were highly expressed in roots. This study laid a theoretical foundation for further study of the KAN gene-mediated nitrogen effect on root development.Keywords
KANADI (KAN) genes, a subset of the GARP (Golden2, ARR-B, Psr1) family of transcription factors, are key regulators of abaxial identity in the polarity and development of lateral organs [1]. The lateral organs of higher plants included the collateral branches, leaves, inflorescences, flowers, and other organs [2], which were developed from the apical meristem [3]. Almost all the lateral organs of higher plants show polarity development in the adaxial-abaxial fates [4], which eventually leads to the phenotype differences between the upper (adaxial) and the lower (abaxial) characteristics in the leaves and other organs [5]. The KAN gene was first identified in Arabidopsis thaliana in 2001 [1]. Later, members of the KAN family were found in rice [6] and maize [7]. According to the current reports, most of the related studies of this gene family were concentrated in herbaceous plants, and it found in gymnosperms such as ferns [8], few studies were conducted in other woody plants.
KAN was a transcription factor belonging to the GARP family and contained a DNA-binding domain of MYB class. In Arabidopsis, the KAN family included four members (AtKAN1-4) [1], and its nucleic acid sequence structure included 6 exons. In rice, the KAN family contained at least six members, among which, the SLL1 (SHALLOT-LIKE1) gene was the closest to AtKAN1, which was a transcription factor encoding 377 amino acids [6]. In maize, the MWP1 (MILKWEED POD1) gene had high homology with SLL1. MWP1 also consisted of six exons, which encoded a predicted protein of 477 amino acids with an estimated molecular weight of 48.36 kD. The GARP domains of MWP1 and SLL1 were identical to those of AtKAN1 and AtKAN2 proteins, differing only by one amino acid residue [9]. All KAN genes contained a highly conserved GARP domain, which played an important role in chloroplast development, cytokinin signal transduction, phosphorus metabolism, organ polarity regulation, and other biological processes [10].
In plants, KAN genes had controlled embryogenesis, lateral root growth, leaf polarity, and integument formation, and were expressed in the phloem and distal developmental regions of lateral organs during early development [11,12]. In A. thaliana, Kerstetter et al. [1] found that the KAN gene was localized in the nucleus. RNA in situ hybridization showed that the KAN transcription factor was expressed in the distal plane of early globular embryos [13], which confirmed that this gene was involved in regulating the development of the abaxial characteristics of plants. Previous studies showed that AtKAN1 expression was observed at the periphery of lateral root apices by the KAN::GUS fusion system, AtKAN2 expression in lateral roots was consistent with AtKAN1 expression, and AtKAN4 expression was detected at the periphery of root cap and developing lateral root primordia [14]. These results suggested that KAN genes played an important role in the establishment of polarity in the distal surface of leaves and the development of lateral roots. At the same time, overexpression of the KAN gene could lead to defects in shoot apical meristem and vascular development in cotyledons in Arabidopsis. While the transgenic plants expressing KAN ectopic in the paraxial plane had narrow and unable to expand leaves, and the paraxial plane of cotyledons produced structural characteristics of the distal plane [15]. The gain-of-function KAN alleles result in a loss of cambium activity, while the cambium activity in the hypocotyls of seedlings of KAN loss-of-function mutants had increased [16]. In Arabidopsis, the four KAN genes displayed a complex pattern of genetic redundancy. KAN1, KAN2, and KAN3 promote the establishment of leaf polarity, and KAN1 and KAN2 also could promote the establishment of floral organ polarity, including the outer integument. At the same time, single deficiency mutants have no or no obvious defective phenotype, none of the kan1, kan2, or kan3 single mutants exhibited a dramatic loss of polarity, while all lateral organs had gross morphological defects in kan1 kan2 plants, and in kan1 kan2 kan3 leaves, the mature blade expanded in various planes giving rise to long narrow leaves with a fan-like blade at their distal end [15,17], which indicated that there were extensive redundancy relationships among Arabidopsis KANADI genes. In rice, SHALLOT-LIKE1 (SLL1, closest to AtKAN1 among the Arabidopsis KAN members) was crucial in polarity formation and help to direct the development of the leaf abaxial cell layer. SLL1 deficiency suppresses the specification of sclerenchymatous cells in the abaxial mesophyll, while SLL1 overexpression resulted in dwarf plants with twisted and abnormal inner rolled leaves, and the abaxial features of leaves following SLL1 overexpression had been enhanced [6]. The maize MILKWEED POD1 (MWP1), the closest KAN protein to SLL1, was involved in the abaxial-adaxial patterning of leaves, both sll1 and mwp1 show adaxialized sectors of cells in the sheath [9]. To sum up, KAN played an important role in the regulation of lateral organ polarity formation in plants. Previous studies had shown that this gene was regulated by nitrogen, but its mechanism in mediating nitrogen regulation of root development was still unclear.
Populus trichocarpa was a model species of woody plants, whose genome sequence had been widely used in genetic studies since it was sequenced and published in 2006 [18]. In this study, eight putative KAN genes of P. trichocarpa were analyzed by bioinformatics analysis methods. The chromosome distribution, phylogeny, gene structure, and motif composition of family genes were analyzed in detail. And qRT-PCR was used to analyze the tissue-specific expression of the KAN genes in P. trichocarpa under different exogenous nitrogen treatments. This study laid a theoretical foundation for speculating the role of this gene in the regulation of root development mediated by nitrogen.
2.1 Identification and Analysis of KAN Family Genes in P. trichocarpa
The protein sequences of P. trichocarpa were downloaded from phytozome v13.0 genome database (https://phytozome.jgi.doe.gov/pz/portal.html) to identify the KAN protein. The unique Myb DNA-binding (PF00249) domain of the Arabidopsis KAN family was downloaded from the Pfam 35.0 database (https://pfam.xfam.org/) [19]. The Hmmsearch command in HMMER (V3.1) software was used to search the poplar protein database with Myb DNA-binding [20]. The sequences had been integrated and identified, and candidate proteins in the P. trichocarpa protein database had been selected. In addition, the Arabidopsis AtKAN1 sequence was used as a probe to search the P. trichocarpa protein database online using BlastP. Protein sequences obtained using these two methods were further screened. After screening and identification, eight PtKAN amino acid sequences were obtained. The online ExPASy program (https://www.expasy.org) [21] was used to analyze the physical and chemical properties of these amino acid sequences, including predicted molecular weight, isoelectric point, number of amino acids, aliphatic index, and grand average of hydropathicity (GRAVY). The subcellular localizations of PtKANs proteins were predicted by Wolfpsort (https://www.genscript.com/psort/wolf_psort.html) [22].
2.2 Gene Structure and Conserved Motifs Analysis
The organization of the exon-intron of the PtKAN gene family was predicted using Gene Structure Display Server (GSDS2.0, http://gsds.cbi.pku.edu.cn) [23]. The conserved motifs in PtKAN proteins were identified according to Multi Em for Motif Elicitation (MEME V5.0.5; http://meme-suite.org/tools/meme) [24]. Then Toolbox for Biologists (TBtools v1.09876) [25] was used for integrated analysis.
2.3 Multiple Sequence Alignment and Phylogenetic Analysis
The alignment of PtKAN amino acid sequences and conserved GARP domains was performed using the Clustal X program [26]. The KAN family protein sequences of Nicotiana benthamiana were downloaded from Solanaceae Genomics Network (http://solgenomics.net). Phylogenetic trees (No. of bootstrap replications = 1000) were constructed using MEGA V7.0.14 software according to Neighbor-Joining (NJ) method [27].
2.4 Chromosomal Mapping and Synteny Analysis
The chromosome locations of PtKAN gene family members were acquired and analyzed using Phytozome v13.0 and PopGenIE v2.0 (http://popgenie.org/chromosome-diagram) [28]. MG2C tools (MapGene2Chrom web, v2; http://mg2c.iask.in/mg2c_v2.0) was used to construct the chromosome distribution map of the PtKAN gene. P. trichocarpa and other species of chromosome gff3 files were obtained from Ensembl the Plants (https://plants.ensembl.org) [29]. Then, using the multicollinear scanning toolkit (MCScanX; https://github.com/wyp1125/MCScanX) [30] to analyze the collinear relationship and gene replication events among PtKAN orthologous genes and other selected species orthologous genes.
2.5 Tissue-Specific Expression Patterns of Genes
The tissue-specific expression data of the PtKAN gene in mature leaves, young leaves, roots, nodes, and internodes were obtained from PopGenIE V2.0 (http://popgenie.org) and used to generate visual images.
2.6 Plant Materials, Growing Conditions, and Nitrogen Treatments
The experimental material P. trichocarpa was obtained from the State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University (Harbin). The homogeneous seedlings (about 15 cm) after rooting hydroponically were planted in vermiculite in the greenhouse under long-day conditions (16 h light/8 h dark) at 25°C ± 1°C. They were irrigated with a modified 1/2 Hoagland nutrient solution (ammonium nitrate concentration 1 mm) [31]. The nutrient solution had changed every 7 days. After cultivating 28 days, the seedlings were treated with nutrient solution supplemented with different concentrations of nitrogen (0.1, 1, 5, and 10 mM NH4NO3) for 28 days. The control group was cultured with 1 mM NH4NO3. The nutrient solution was updated every 7 days. The roots, leaves, upper stems (stems grown after treatment), and lower stems (stems grown before treatment) were sampled according to Sun et al. [32], frozen immediately with liquid nitrogen, and then stored at −80°C for future analysis. Each treatment had three biological replicates. Each sample also had three technical replicates.
2.7 RNA Extraction and qRT-PCR Analysis
Total RNA of different tissues was extracted using the Hexadecyl Trimethyl Ammonium Bromide (CTAB) method [33]. With the action of DNase I (Fermentas, Waltham, MA, USA), genomic DNA contamination was removed and then using PrimeScript™ RT reagent Kit with gDNA Eraser to reverse transcribed 1 μg of total RNA into cDNA (Takara Bio, Dalian, China). The qRT-PCR was performed using Power Green qPCR Mix reagent (Dongsheng Biotech Co., Ltd., Beijing, China) according to Zuo et al. [34]. UBQ7 gene was used as the reference gene [35], and the relative expression was calculated by the 2−ΔΔCT method [36]. The statistically significant differences (p < 0.05) among expression levels of PtKANs in different samples were tested by the Duncan test. TBtools V1.09876 [24] and GraphPad Prism 8.0.1 were used to generate gene expression maps.
3.1 Identification and Sequence Analysis of PtKAN Gene
In this study, a total of eight PtKAN genes were obtained and named PtKAN1-8 based on their positions on the chromosome, respectively. Their physical and chemical properties including amino acids, molecular weight, isoelectric points, GRAVY, aliphatic Index, chromosome location, and cellular localization were listed in Table 1. The length of encoded protein of PtKANs ranged from 341 (PtKAN2) to 485 (PtKAN4) amino acids. The maximum and minimum molecular weights were 53.87 kDa (PtKAN4) and 37.34 kDa (PtKAN2), respectively. The predicted pI ranged from 6.76 (PtKNA6) to 9.23 (PtKAN1), and the aliphatic index ranged from 56.89 (PtKAN4) to 75.03 (PtKAN6). They all were identified as hydrophilic proteins according to GRAVY scores. What’s more, it was predicted that they were located in the cytoplasm and nucleus. The multiple sequence alignments of the eight PtKAN protein sequences showed that they all had typical GARP domains and conserved structures (Fig. 1).
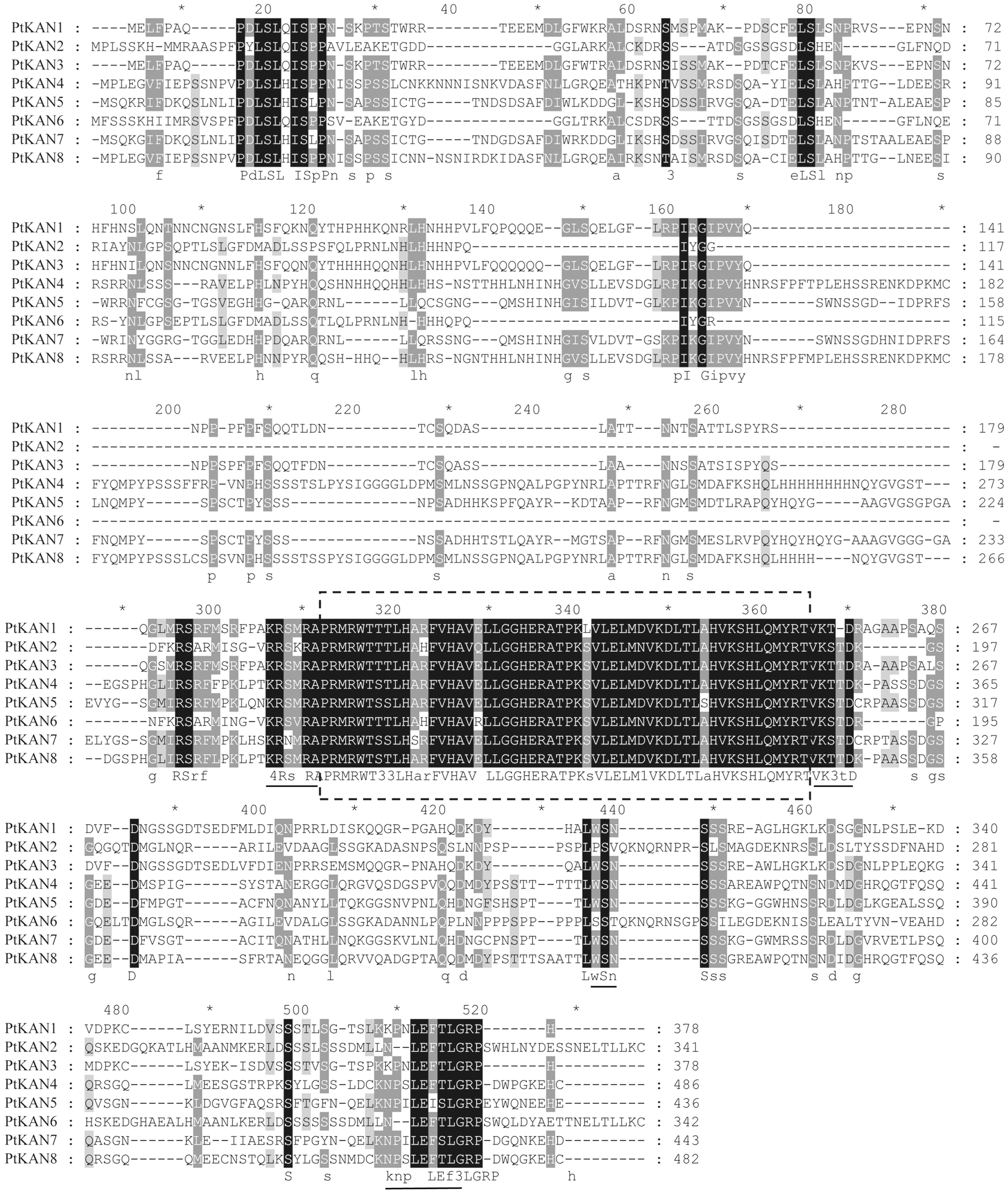
Figure 1: Multiple alignments of 8 putative PtKAN amino acid sequences. The GARP domain was in the box and the conserved region unique to the PtKAN gene family was underlined [37], where * represented the same amino acid
3.2 Gene Structure and Phylogenetic Analysis of PtKAN Gene
According to the full-length sequence of eight PtKAN proteins, the phylogenetic tree was constructed and shown in Fig. 2. Phylogenetic tree analysis indicated that the PtKAN family was clustered into three subfamilies. The first group contained PtKAN4, PtKAN5, PtKAN7, and PtKAN8, and they had 10 motifs. The second group included PtKAN1 and PtKAN3, while PtKAN3 contained motifs and PtKAN1 did not. PtKAN2 and PtKAN6 were the third groups, they both had 6 motifs. Meanwhile, the results of gene structure showed that all the family members had 6 exons, but the length of introns varied significantly.
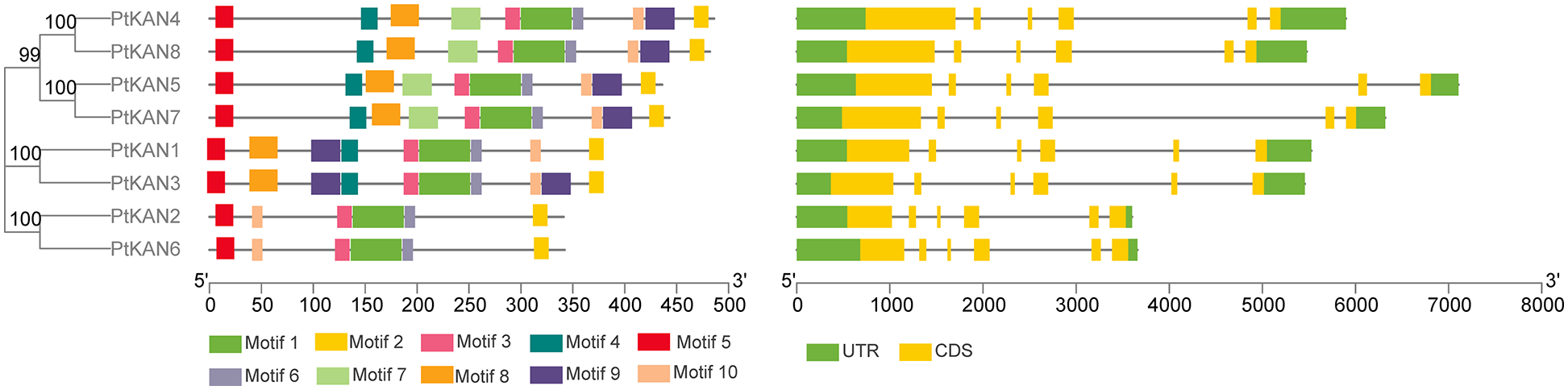
Figure 2: Phylogenetic tree and gene structural analysis of eight PtKAN genes. Phylogenetic trees (left) were constructed based on full-length protein sequences of PtKAN. The motifs (middle) were predicted using the MEME tool. The right was the structure of the corresponding PtKAN gene. Protein coding sequences (CDS) were shown in yellow. The green color indicated upstream/downstream sequences
In order to understand the evolutionary relationship among the KAN gene families of P. trichocarpa, A. thaliana, and Nicotiana benthamiana, MEGA7 software was used to compare their KAN amino acid sequences and constructed phylogenetic trees (Fig. 3). Phylogenetic analysis revealed that all KAN genes could be divided into four subgroups, while the eight genes in P. trichocarpa were classified into three subgroups, respectively. PtKAN4, PtKAN5, PtKAN7, and PtKAN8 of P. trichocarpa were highly homologous to AtKAN1. PtKAN1 and PtKAN3 had similar homology to NbKAN2, while PtKAN2 and PtKAN6 had high homology to AtKAN3, AtKAN4, and NbKAN4.
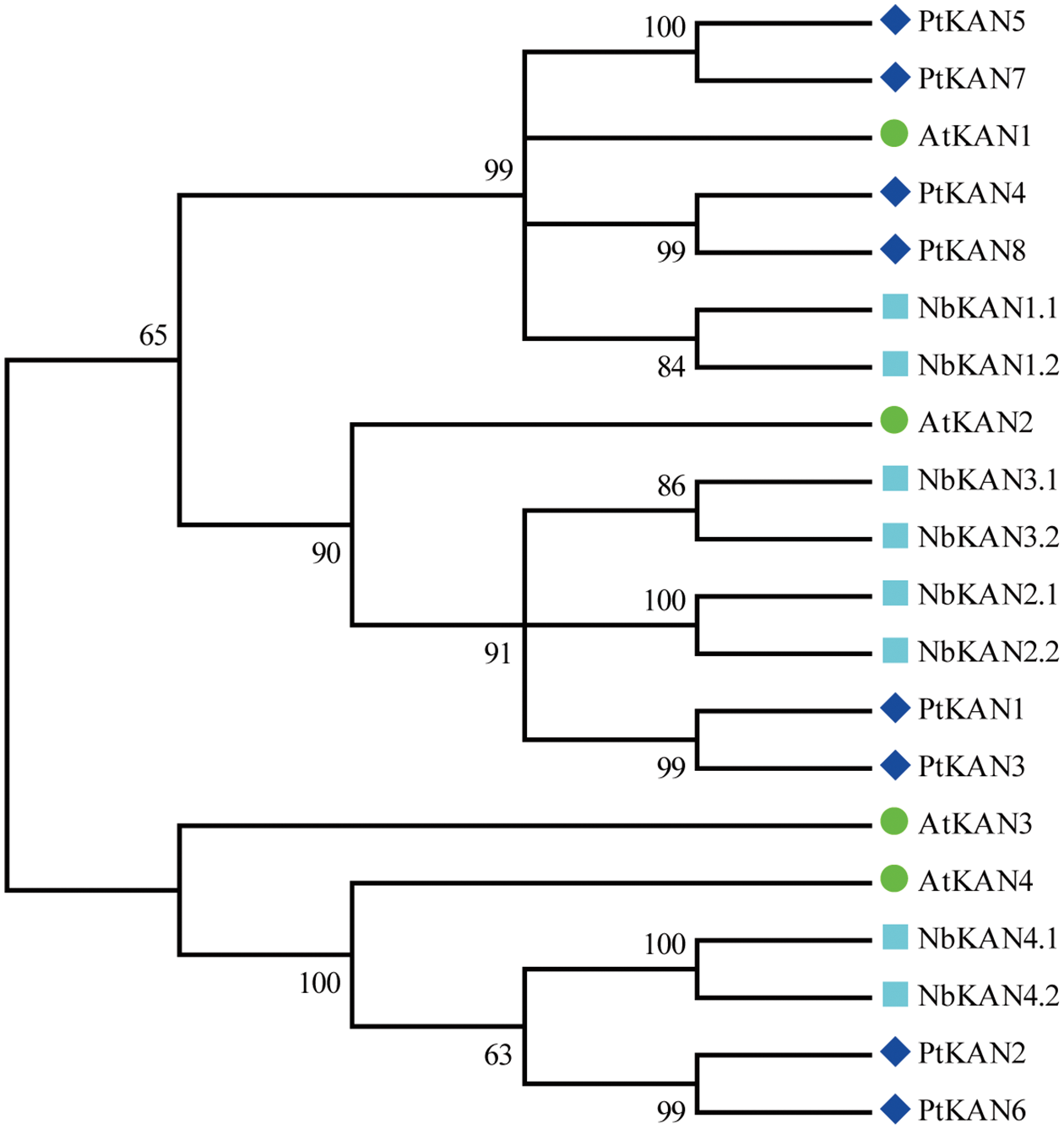
Figure 3: Phylogenetic trees of KAN gene families of P. trichocarpa (Pt), Nicotiana benthamiana (Nb), and Arabidopsis thaliana (At)
3.3 Chromosomal Distribution and Synteny Analysis of PtKAN Gene
The chromosome distributions of these eight PtKAN genes were shown in Fig. 4. The results showed that the eight PtKAN genes were distributed on eight chromosomes, respectively. The MCScanX was used to analyze the segmental duplication events of the PtKAN genes (Fig. 5). Four pairs of segmental duplication events were found in this study, among which there were gene duplication events between PtKAN1 (Chr01) and PtKAN3 (Chr03), and between PtKAN2 (Chr02) and PtKAN6 (Chr14), and between PtKAN4 (Chr04) and PtKAN8 (Chr17), and between PtKAN5 (Chr12) and PtKAN7 (Chr15).
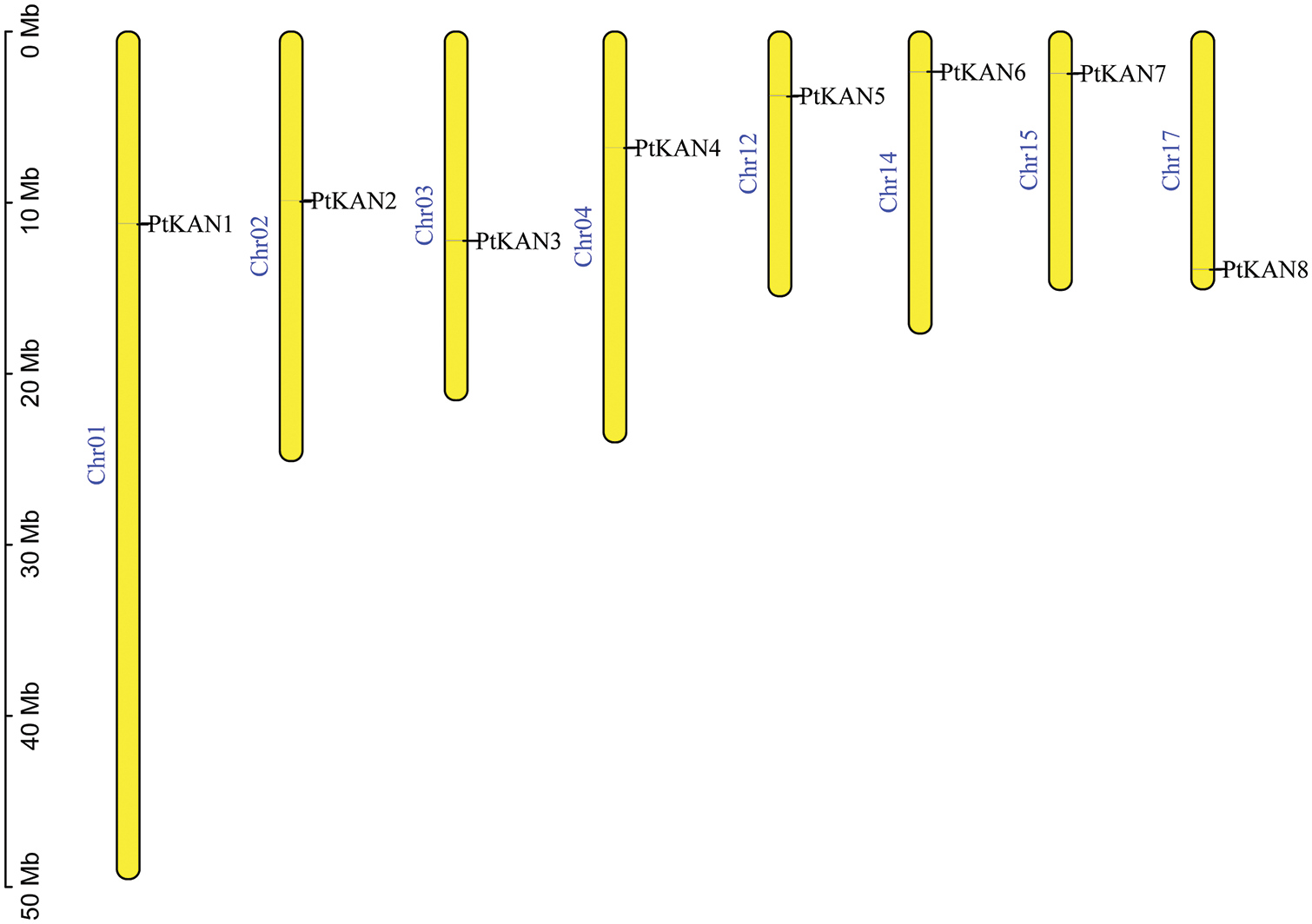
Figure 4: Chromosomal distribution of KAN gene family members in P. trichocarpa
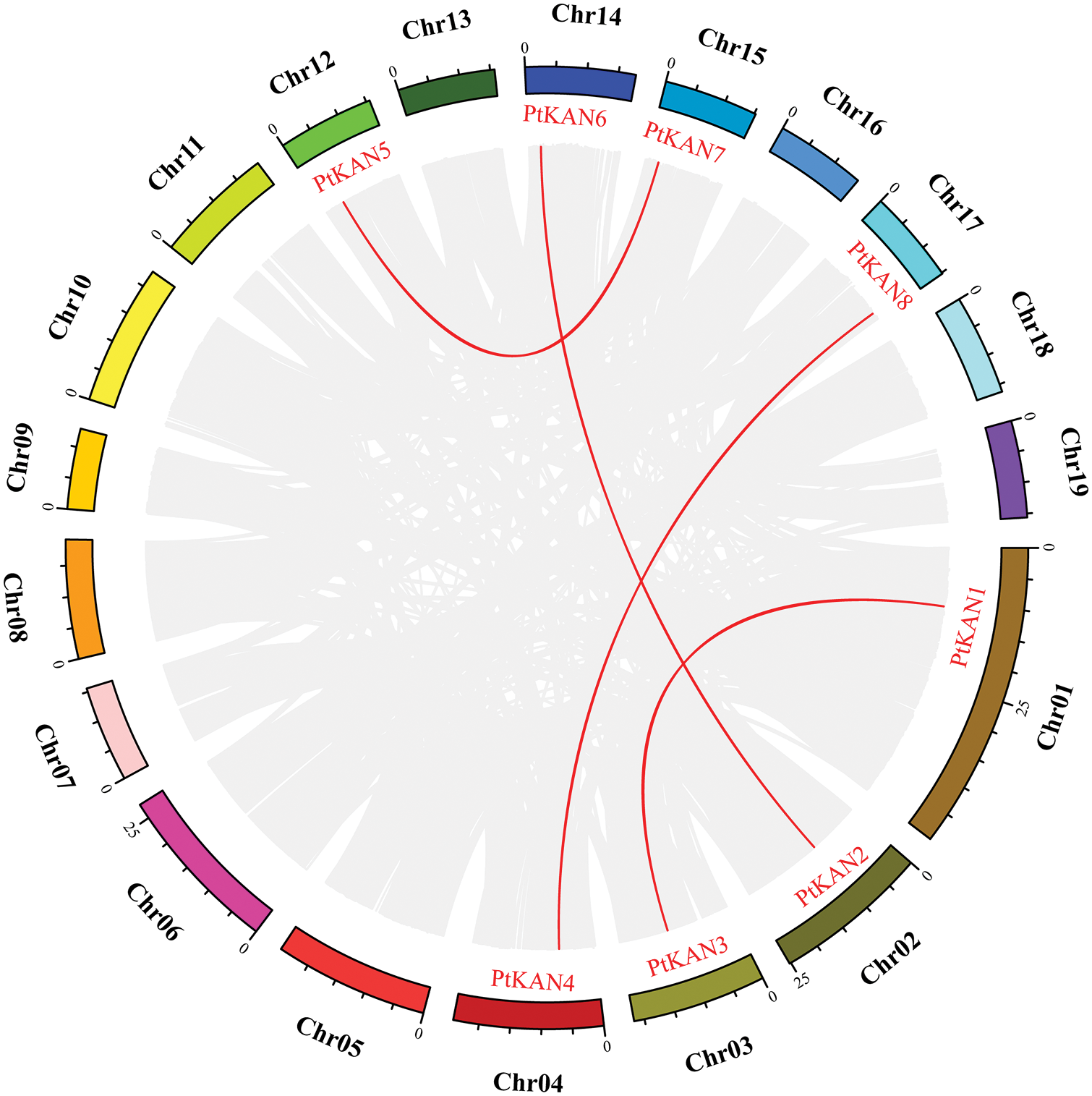
Figure 5: Schematic representations of segmental duplications of PtKAN gene. Gray lines indicate all syntenic blocks between each chromosome in the poplar genome; The red line showed the duplicated PtKAN gene pair. Gene names were shown in red. Chromosome numbers were shown in the middle of each chromosome. The scale of markers on each chromosome indicated chromosome length (Mb)
To further understand the mechanism of gene replication among PtKANs, four comparative syntenic maps related to four representative species, including monocotyledon (Oryza sativa, Fig. 6A) and dicotyledons (A. thaliana, Glycine max, and Medicago truncatula, Fig. 6B) were constructed. The results showed that there were four KAN syntenic gene pairs between poplar and rice. Compared with O. sativa, A. thaliana, and M. truncatula, PtKANs had the most syntenic gene pairs with G. max, with higher homology and closer kinship.
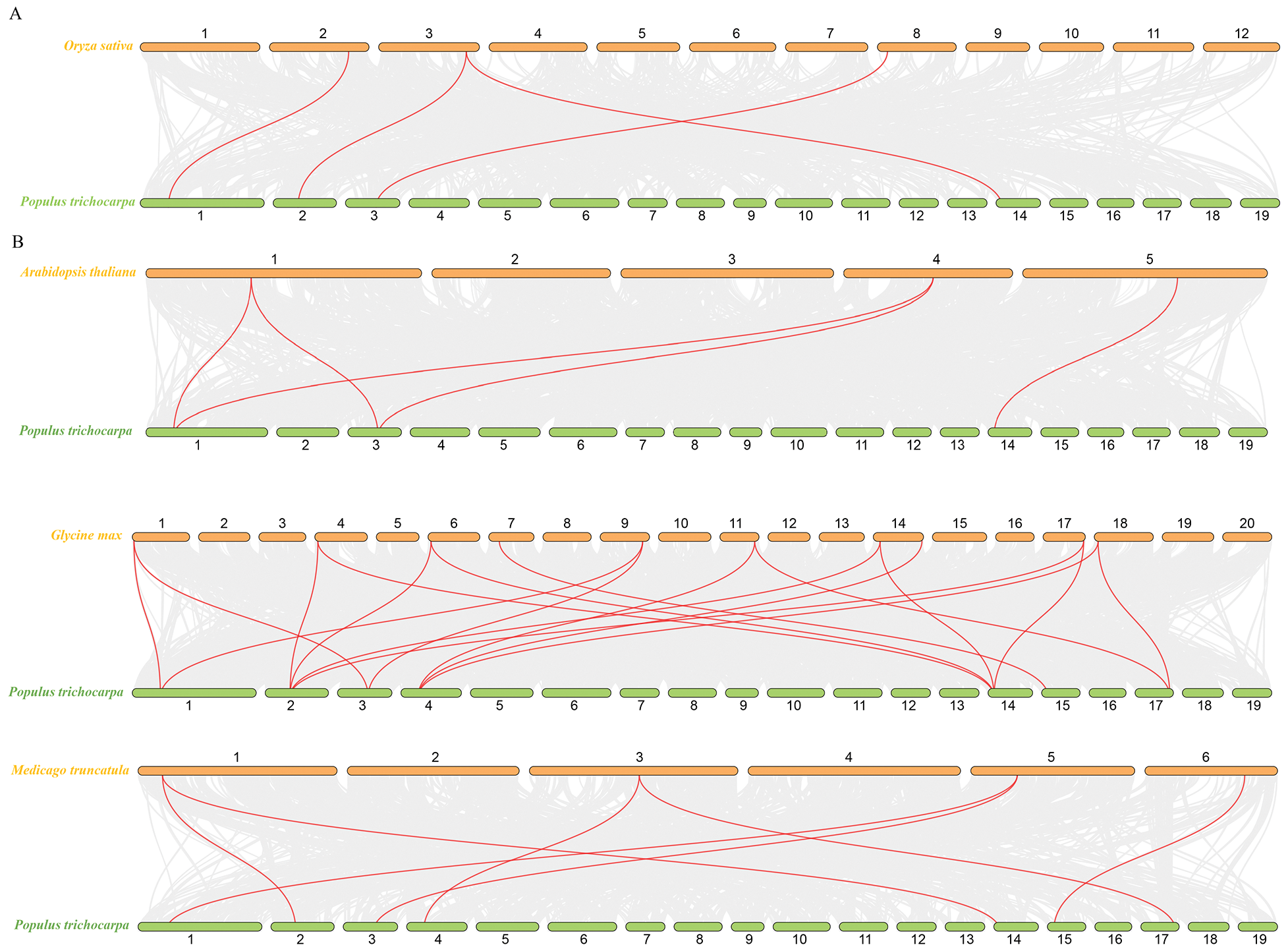
Figure 6: Synteny analysis of KAN genes between poplar and other plants. (A) was monocotyledon. (B) were dicotyledons. The red line showed the KAN syntenic gene pairs. Orange and green bars represented chromosomes, and whose numbers were at the top or bottom
3.4 Tissue-Specific Expression Pattern of PtKAN Genes
The tissue-specific expression data of PtKAN genes in mature leaves, young leaves, roots, nodes, and internodes were obtained from PopGenIE (https://PlantGenIE.org) (Fig. 7). PtKAN1 was highly expressed in young leaves. PtKAN2, PtKAN 5, and PtKAN 7 were highly expressed in nodes and internodes. PtKAN6 was highly expressed in young leaves and mature leaves. PtKAN8 was highly expressed in roots. However, PtKAN3 and PtKAN4 showed low expression levels in all tissues.
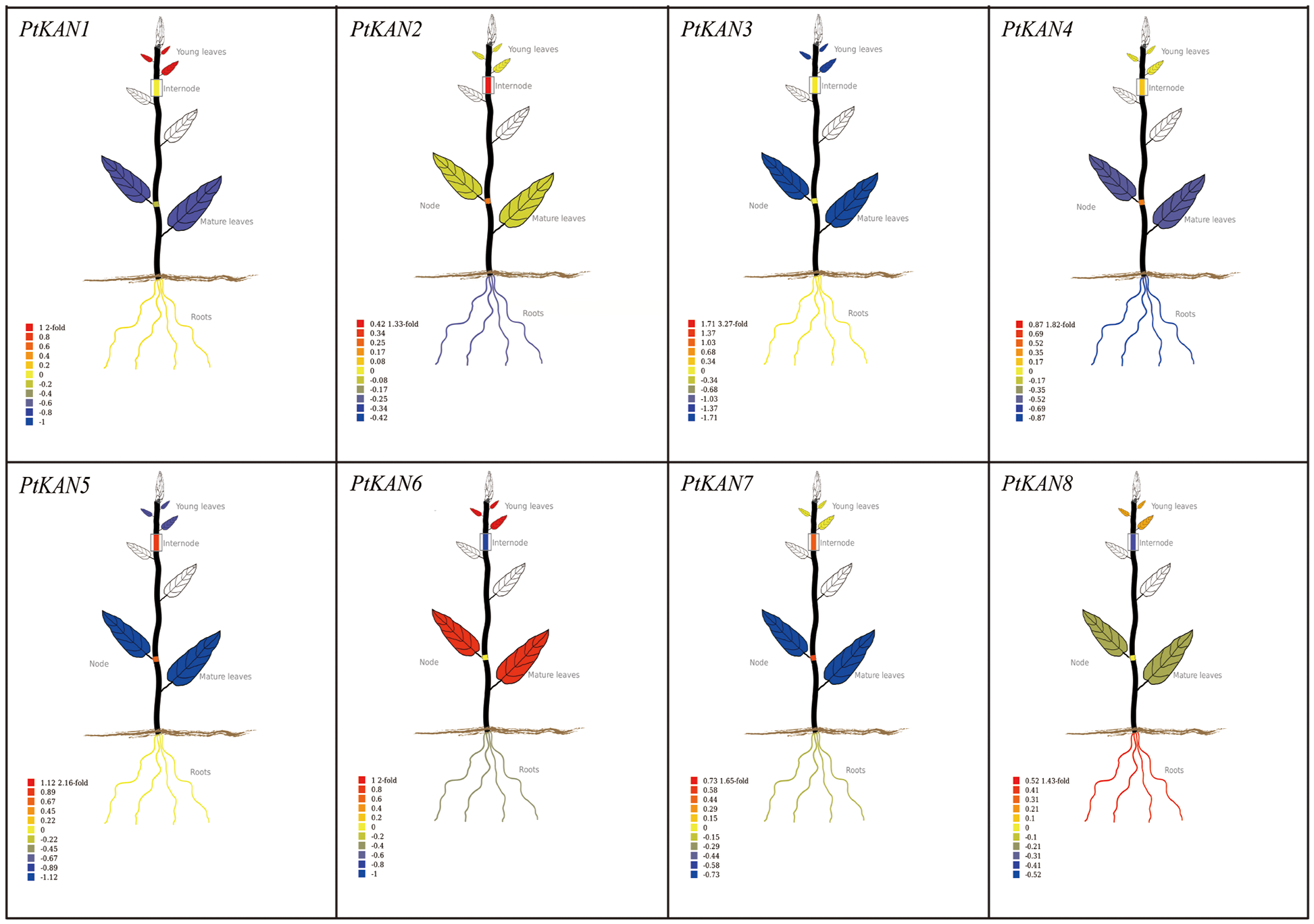
Figure 7: Tissue-specific expression pattern of PtKAN genes. Visual images of PtKAN gene in mature leaves, young leaves, roots, nodes, and internodes were generated using tissue-specific expression data from https://PlantGenIE.org
3.5 Expression Pattern of PtKAN Genes under Different Nitrogen Treatment
The seedlings were treated with different concentrations (0.1, 1, 5, and 10 mM NH4NO3) of nitrogen, and the expression patterns of 8 PtKANs in 4 tissues (upper stems, lower stems, leaves and roots) were determined (Fig. 8). 1 mM NH4NO3 was regarded as control group. In upper stems, the expression levels of PtKAN2 and PtKAN5 were significantly increased under low nitrogen treatment. The expressions of PtKAN2 in upper stems were significantly upregulated under medium and high nitrate treatments, while the transcription levels of other genes were down-regulated. In lower stems, the expression of PtKAN2 was significantly upregulated under low nitrogen treatment, while PtKAN8 was significantly downregulated. PtKAN2, PtKAN4, PtKAN6, and PtKAN8 were significantly downregulated under medium nitrogen treatment. However, PtKAN2 was upregulated under high nitrogen treatment. In leaves, the transcription levels of PtKAN1 and PtKAN2 were significantly downregulated under low nitrogen treatment. Almost all PtKANs were significantly downregulated under medium and high nitrogen treatment. In roots, PtKAN2 was significantly downregulated and PtKAN7 was significantly upregulated under low nitrogen treatment. The relative expression levels of PtKAN2, PtKAN4, PtKAN5, and PtKAN7 were significantly upregulated under medium nitrogen, and the expression levels of PtKAN7 and PtKAN8 were upregulated under high nitrogen treatment.
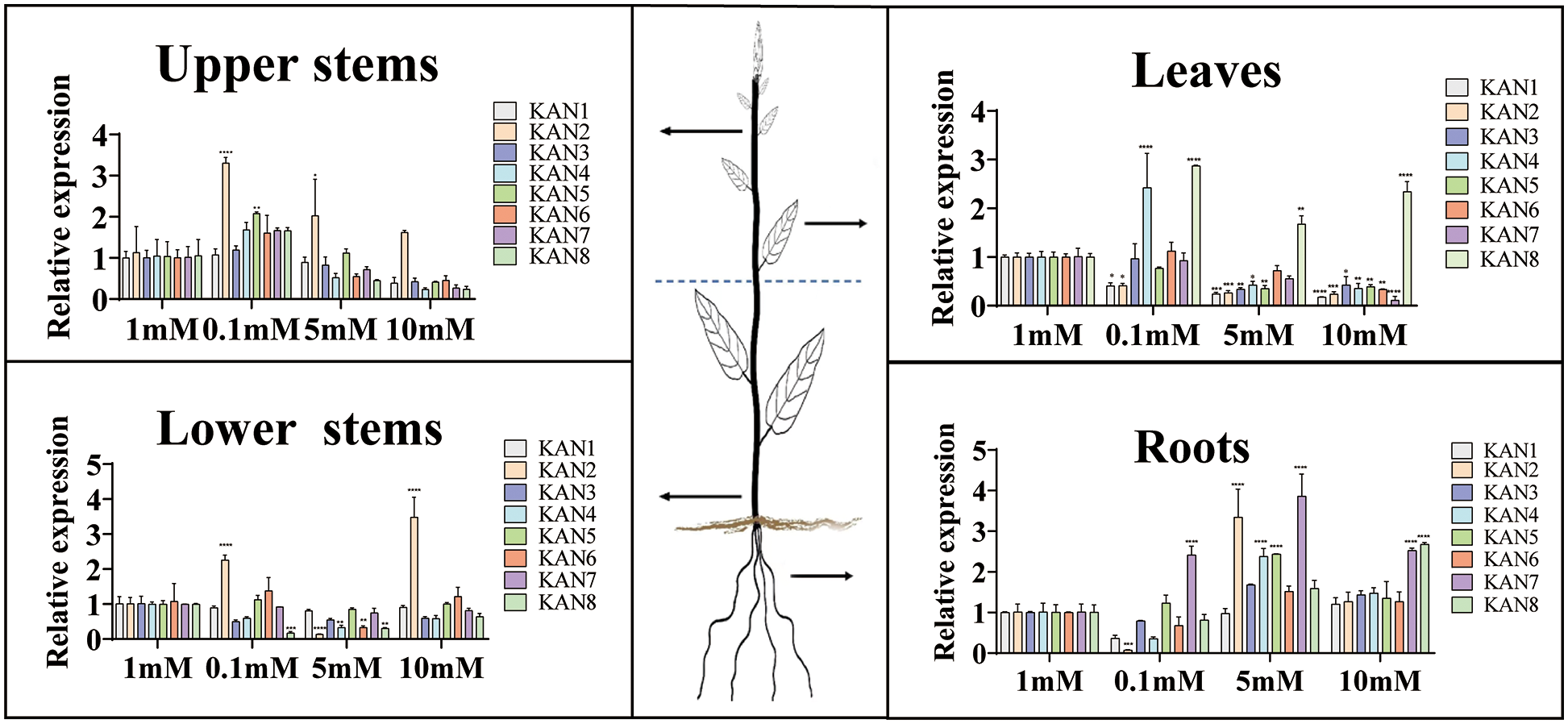
Figure 8: Expression patterns of PtKAN genes in different tissues (upper stems, lower stems, leaves, and roots) under different concentrations NH4NO3 treatments. Control: 1 mM. Low nitrogen: 0.1 mM. Medium nitrogen: 5 mM. High nitrogen: 10 mM. The relative expression of PtKANs was calculated using 2−ΔΔCT. The significance test indicated the statistical analysis of relative expression under different treatments and the gene expression under 1 mM nitrogen treatment. “*”, “**” and “***” indicated significant differences at “p < 0.05”, “p < 0.01” and “p < 0.001” levels, respectively
KAN family genes played an important role in controlling the polar development of lateral organs in plants. In recent years, it had been confirmed that this gene family acted on the formation of leaf and flower polarities in Arabidopsis [38]. In previous studies, our team found that this gene was regulated by nitrogen and might play an important role in the nitrogen-induced root formation of woody plants (data not shown). However, the composition and functions of KAN family members in woody plants were less reported, and the gene family analysis was a necessary condition for further functional verification. Therefore, in this study, eight members of the KAN gene family in P. trichocarpa were identified and analyzed using bioinformatics methods. Meanwhile, the expression of KAN family genes under nitrogen treatment was further studied. It could provide new insight into the response of the KAN gene to nitrogen and also lay an important foundation for exploring the function of KAN family members in the later stage.
In this study, the subcellular localization, protein conserved motifs analysis, and phylogenetic tree analysis of 8 PtKAN genes were conducted. The results showed that both PtKAN2 and PtKAN6 were located in the nucleus, and both of them had 6 identical motifs. Phylogenetic tree analysis showed that they were on the same branch, indicating that they had a high homology relationship. Therefore, it was speculated that there might be functional redundancy between them. At the same time, PtKAN4 and PtKAN5 also had similar characteristics, indicating that there might also be functional redundancy between them. At the same time, the phylogenetic analysis showed that PtKAN2 and PtKAN6 had similar evolutionary relationships with AtKAN4, while PtKAN4 and PtKAN5 had high homology with AtKAN1. Previous studies had shown that Arabidopsis KAN1-4 were functionally redundant genes [1], so it was speculated that there was a similar situation of functional redundancy in PtKAN genes. However, whether this “redundancy” was real or not will be our future research direction.
In this study, a total of 8 PtKAN genes were identified, and the chromosomal position of the genes was analyzed, and it was found that each chromosome contained one gene. MCScanX was used to analyze the segmental repeat events of the PtKAN genes, and a total of 4 pairs of duplicate genes were found (Fig. 5), namely PtKAN1/3, PtKAN2/6, PtKAN4/8, PtKAN5/7, and the PtKANs repeat genes might have been formed during the whole genome doubling of poplars. The occurrence of multiple repeat events indicates that the PtKAN gene family continues to expand during the evolution of the poplar genome. Through collinear analysis, multiple PtKAN genes were homologous to AtKANs and OsKAN were found (Fig. 6), which might be from the same ancestor as the KAN genes of Arabidopsis thaliana and rice, etc., and might have similar functions.
Previous studies had shown that the KAN gene family functions in the apical meristem and that the expression pattern of each gene varies in organs such as leaves, flowers, cotyledons, and embryos [15]. For example, AtKAN1 could promote the development of abaxial leaf polarity [39]. AtKAN1, AtKAN2, and AtKAN3 could promote the establishment of leaf polarity, while AtKAN1 and AtKAN2 also contributed to floral organ polarity [40]. AtKAN4 was expressed in the root, but it had redundant activity with other KAN genes [14]. In this study, we found that KAN genes were expressed in various organs of P. trichocarpa, but their expression levels were different. For example, PtKAN8 was highly expressed in roots, indicating that the KAN gene might play an important regulatory function in the root development of P. trichocarpa.
Nitrogen was an essential mineral nutrient element for tree growth and development, and it played an important role in improving yield [41]. Plant roots required N supply for growth and development. However, in terms of plasticity, lateral roots were more sensitive to N supply at different concentrations. Previous studies had shown that the KAN gene family was expressed in lateral root and acted in lateral root formation [14]. In this study, the expression characteristics of the PtKAN gene in response to nitrogen treatment with different concentrations were investigated. The results showed that the expression of PtKAN2 in stems was significantly upregulated, while it was downregulated in leaves and roots under low nitrogen treatment. The expression of PtKAN7 in roots was significantly upregulated. It was shown that PtKAN2 and PtKAN7 played important roles in stems and roots in the nitrogen starvation state, respectively. Other PtKAN genes were downregulated under low nitrogen treatment, while the PtKAN genes in the roots were upregulated under middle and high nitrogen treatments, which might be the PtKAN gene in roots was upregulated to adapt to high nitrogen environment. With higher nitrogen concentrations, the expression of most KAN genes was inhibited. However, the expression of PtKAN8 was elevated under the conditions of nitrogen deficiency and high nitrogen concentration, indicating that PtKAN8 was sensitive to nitrogen, and it might mainly play a role in lateral organs such as leaves and lateral roots. Therefore, it was possible that PtKAN family played an important role in mediating nitrogen response and root morphological differences. However, the PtKAN gene was down-regulated in stems and leaves to adapt to high nitrogen supply, which might be due to different regulatory mechanisms depending on different tissues in plants. The molecular mechanism by which the PtKAN family affected root development under nitrogen treatment was still unclear. However, further studies needed to be carried out to confirm that.
In this study, the bioinformatics method was used to speculate KAN family members in P. trichocarpa, and their expression patterns under different nitrogen treatments had been analyzed. The results showed that a total of 8 KAN family members were screened out in P. trichocarpa, and each member contained the unique GARP domain and conserved region. Among them, PtKAN2 and PtKAN6, and PtKAN4 and PtKAN5 might have functional redundancy, which needed to be further verified. This study also identified and analyzed the gene expression patterns under nitrogen treatment, aiming to provide an important theoretical basis for the future study of this gene in nitrogen-mediated root development.
Funding Statement: This research was funded by the Natural Science Foundation of Heilongjiang Province, China (ZD2020C004), the Fundamental Research Funds for the Central Universities (2572019CT02).
Author Contributions: Niu MH, Qu CP, and Liu GJ conceived and designed the study. Niu MH performed most of the experiments. Zhang HJ and Hu ZB conducted the sampling. Niu MH, Hu ZB, and Li XY performed bioinformatics analysis. Niu MH and Zhang H processed and analyzed the data, and Niu MH and Zhang H wrote the manuscript. Qu CP, Liu GJ and Xu ZR provided helpful comments in the draft and reviewed and edited the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Kerstetter, R. A., Bollman, K., Taylor, R. A., Bomblies, K., Poethig, R. S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature, 411(6838), 706–709. https://doi.org/10.1038/35079629 [Google Scholar] [PubMed] [CrossRef]
2. Bowman, J. L., Eshed, Y. (2000). Formation and maintenance of the shoot apical meristem. Trends in Plant Science, 5(3), 110–115. https://doi.org/10.1016/S1360-1385(00)01569-7 [Google Scholar] [PubMed] [CrossRef]
3. Kieffer, M., Davies, B. (2001). Developmental programmes in floral organ formation. Seminars in Cell & Developmental Biology, 12(5), 373–380. https://doi.org/10.1006/scdb.2001.0266 [Google Scholar] [PubMed] [CrossRef]
4. Chien, J. C., Sussex, I. M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana. Plant Physiology, 111(4), 1321–1328. https://doi.org/10.1104/pp.111.4.1321 [Google Scholar] [PubMed] [CrossRef]
5. Wu, G., Lin, W. C., Huang, T. B., Poethig, R. S., Spronger, P. S. et al. (2008). KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proceedings of the National Academy of Sciences of the United States of America, 105(42), 16392–16397. https://doi.org/10.1073/pnas.0803997105 [Google Scholar] [PubMed] [CrossRef]
6. Zhang, G. H., Xu, Q., Zhu, X. D., Qian, Q., Xue, H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell, 21(3), 719–735. https://doi.org/10.1105/tpc.108.061457 [Google Scholar] [PubMed] [CrossRef]
7. Henderson, D. C., Zhang, X. L., Brooks, L., Scanlon, M. J. (2006). RAGGED SEEDLING2 is required for expression of KANADI2 and REVOLUTA homologues in the maize shoot apex. Genesis, 44(8), 372–382. https://doi.org/10.1002/dvg.20223 [Google Scholar] [PubMed] [CrossRef]
8. Zumajo-Cardona, C., Ambrose, B. A. (2020). Phylogenetic analyses of key developmental genes provide insight into the complex evolution of seeds. Molecular Phylogenetics and Evolution, 147, 106778. https://doi.org/10.1016/j.ympev.2020.106778 [Google Scholar] [PubMed] [CrossRef]
9. Candela, H., Johnston, R., Gerhold, A., Foster, T., Hake, S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell, 20(8), 2073–2087. https://doi.org/10.1105/tpc.108.059709 [Google Scholar] [PubMed] [CrossRef]
10. Ahmad, R., Liu, Y. T., Wang, T. J., Meng, Q. X., Yin, H. et al. (2019). GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiology, 179(4), 1844–1860. https://doi.org/10.1104/pp.18.01466 [Google Scholar] [PubMed] [CrossRef]
11. Kelley, D. R., Arreola, A., Gallagher, T. L., Gasser, C. S. (2012). ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development, 139(6), 1105–1109. https://doi.org/10.1242/dev.067918 [Google Scholar] [PubMed] [CrossRef]
12. McAbee, J. M., Hill, T. A., Skinner, D. J., Izhaki, A., Hauser, B. A. et al. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant Journal, 46(3), 522–531. https://doi.org/10.1111/j.1365-313X.2006.02717.x [Google Scholar] [PubMed] [CrossRef]
13. Axtell, M. J., Bartel, D. P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell, 17(6), 1658–1673. https://doi.org/10.1105/tpc.105.032185 [Google Scholar] [PubMed] [CrossRef]
14. Hawker, N. P., Bowman, J. L. (2004). Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiology, 135(4), 2261–2270. https://doi.org/10.1104/pp.104.040196 [Google Scholar] [PubMed] [CrossRef]
15. Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development, 131(12), 2997–3006. https://doi.org/10.1242/dev.01186 [Google Scholar] [PubMed] [CrossRef]
16. Ilegems, M., Douet, V., Meylan-Bettex, M., Uyttewaal, M., Brand, L. et al. (2010). Interplay of auxin, KANADI and class III HD-ZIP transcription factors in vascular tissue formation. Development, 137(6), 975–984. https://doi.org/10.1242/dev.047662 [Google Scholar] [PubMed] [CrossRef]
17. Eshed, Y., Baum, S. F., Perea, J. V., Bowman, J. L. (2001). Establishment of polarity in lateral organs of plants. Current Biology, 11(16), 1251–1260. https://doi.org/10.1016/S0960-9822(01)00392-X [Google Scholar] [PubMed] [CrossRef]
18. Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I. et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science, 313(5793), 1596–1604. https://doi.org/10.1126/science.1128691 [Google Scholar] [PubMed] [CrossRef]
19. Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J. et al. (2008). The Pfam protein families database. Nucleic Acids Research, 36, 281–288. https://doi.org/10.1093/nar/gkm960 [Google Scholar] [PubMed] [CrossRef]
20. Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R. et al. (2018). HMMER web server: 2018 update. Nucleic Acids Research, 46(W1), W200–W204. https://doi.org/10.1093/nar/gky448 [Google Scholar] [PubMed] [CrossRef]
21. Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D. et al. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Research, 31(13), 3784–3788. https://doi.org/10.1093/nar/gkg563 [Google Scholar] [PubMed] [CrossRef]
22. Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H. et al. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Research, 35, 585–587. https://doi.org/10.1093/nar/gkm259 [Google Scholar] [PubMed] [CrossRef]
23. Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J. C. et al. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics, 31(8), 1296–1297. https://doi.org/10.1093/bioinformatics/btu817 [Google Scholar] [PubMed] [CrossRef]
24. Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E. et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Research, 37, 202–208. https://doi.org/10.1093/nar/gkp335 [Google Scholar] [PubMed] [CrossRef]
25. Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 [Google Scholar] [PubMed] [CrossRef]
26. Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24), 4876–4882. https://doi.org/10.1093/nar/25.24.4876 [Google Scholar] [PubMed] [CrossRef]
27. Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. https://doi.org/10.1093/molbev/msw054 [Google Scholar] [PubMed] [CrossRef]
28. Sjodin, A., Street, N. R., Sandberg, G., Gustafsson, P., Jansson, S. (2009). The Populus genome integrative explorer (PopGenIEA new resource for exploring the Populus genome. New Phytologist, 182(4), 1013–1025. https://doi.org/10.1111/j.1469-8137.2009.02807.x [Google Scholar] [PubMed] [CrossRef]
29. Bolser, D., Staines, D. M., Pritchard, E., Kersey, P. (2016). Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods in Molecular Biology, 1374, 115–140. https://doi.org/10.1007/978-1-4939-3167-5_6 [Google Scholar] [PubMed] [CrossRef]
30. Tang, H. B., Bowers, J. E., Wang, X. Y., Ming, R., Alam, M. et al. (2008). Synteny and collinearity in plant genomes. Science, 320(5875), 486–488. https://doi.org/10.1126/science.1153917 [Google Scholar] [PubMed] [CrossRef]
31. Cavagnaro, T. R., Smith, F. A., Lorimer, M. F., Haskard, K. A., Ayling, S. M. et al. (2001). Quantitative development of Paris-type arbuscular mycorrhizas formed between Asphodelus fistulosus and Glomus coronatum. New Phytologist, 149, 105–113. https://doi.org/10.1046/j.1469-8137.2001.00001.x [Google Scholar] [PubMed] [CrossRef]
32. Sun, X., Cao, L. N., Zhang, S., Yu, J. J., Xu, X. Y. et al. (2020). Genome-wide analysis of the RGP gene family in Populus trichocarpa and their expression under nitrogen treatment. Gene Expression Patterns, 38, 119142. https://doi.org/10.1016/j.gep.2020.119142 [Google Scholar] [PubMed] [CrossRef]
33. Gambino, G., Perrone, I., Gribaudo, I. (2008). A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochemical Analysis, 19(6), 520–525. https://doi.org/10.1002/pca.1078 [Google Scholar] [PubMed] [CrossRef]
34. Zuo, Z., Sun, X., Cao, L. N., Zhang, S., Yu, J. J. et al. (2021). Genome-wide identification of FRK genes in Populus trichocarpa and their expression under different nitrogen treatments. Physiology and Molecular Biology of Plants, 27(9), 1919–1931. https://doi.org/10.1007/s12298-021-01055-6 [Google Scholar] [PubMed] [CrossRef]
35. Pettengill, E. A., Parmentier-Line, C., Coleman, G. D. (2012). Evaluation of qPCR reference genes in two genotypes of Populus for use in photoperiod and low-temperature studies. BMC Research Notes, 5, 66. https://doi.org/10.1186/1756-0500-5-366 [Google Scholar] [PubMed] [CrossRef]
36. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262 [Google Scholar] [PubMed] [CrossRef]
37. Wang, J. H., Ding, Q., Zeng, J., He, X. Q., Li, W. et al. (2013). Cloning and expression analysis of KANADI gene in Nicotiana benthamiana. Journal of Agricultural Biotechnology, 21(11), 1328–1336. https://doi.org/10.3969/j.issn.1674-7968.2013.11.009 [Google Scholar] [CrossRef]
38. Byrne, M. E. (2006). Shoot meristem function and leaf polarity: The role of class III HD-ZIP genes. PLoS Genetics, 2(6), 89. https://doi.org/10.1371/journal.pgen.0020089 [Google Scholar] [PubMed] [CrossRef]
39. Reinhart, B. J., Liu, T., Newell, N. R., Magnani, E., Huang, T. B. et al. (2013). Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: Ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell, 25(9), 3228–3249. https://doi.org/10.1105/tpc.113.111518 [Google Scholar] [PubMed] [CrossRef]
40. Izhaki, A., Bowman, J. L. (2007). KANADI and Class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell, 19(2), 495–508. https://doi.org/10.1105/tpc.106.047472 [Google Scholar] [PubMed] [CrossRef]
41. Vatter, T., Neuhauser, B., Stetter, M., Ludewig, U. (2015). Regulation of length and density of Arabidopsis root hairs by ammonium and nitrate. Journal of Plant Research, 128(5), 839–848. https://doi.org/10.1007/s10265-015-0733-8 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools