 Open Access
Open Access
ARTICLE
Identification and Expression Analysis of Abscisic Acid Signal Transduction Genes in Hemp Seeds
1 School of Chemistry, Chemical Engineering and Life Sciences, Wuhan University of Technology, Wuhan, 430070, China
2 Yunnan Hemp Industrial Investment Co., Ltd., Kunming, 650000, China
* Corresponding Authors: Xia Liu. Email: ; Linlin Dong. Email:
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(7), 2087-2103. https://doi.org/10.32604/phyton.2023.029041
Received 28 January 2023; Accepted 28 March 2023; Issue published 29 May 2023
Abstract
Abscisic acid (ABA) is involved in regulating diverse biological processes, but its signal transduction genes and roles in hemp seed germination are not well known. Here, the ABA signaling pathway members, PYL, PP2C and SnRK2 gene families, were identified from the hemp reference genome, including 7 CsPYL (pyrab-actin resistance1-like, ABA receptor), 8 CsPP2CA (group A protein phosphatase 2c), and 7 CsSnRK2 (sucrose nonfermenting1-related protein kinase 2). The content of ABA in hemp seeds in germination stage is lower than that in non-germination stage. Exogenous ABA (1 or 10 μM) treatment had a significant regulatory effect on the selected PYL, PP2C, SnRK2 gene families. CsAHG3 and CsHAI1 were most significantly affected by exogenous ABA treatment. Yeast two-hybrid experiments were performed to reveal that CsPYL5, CsSnRK2.2, and CsSnRK2.3 could interact with CsPP2CA7 and demonstrate that this interaction was ABA-independent. Our results indicated that CsPYL5, CsSnRK2.2, CsSnRK2.3 and CsPP2CA7 might involve in the ABA signaling transduction pathway of hemp seeds during the hemp seed germination stages. This study suggested that novel genetic views can be brought into investigation of ABA signaling pathway in hemp seeds and lay the foundation for further exploration of the mechanism of hemp seed germination.Keywords
Hemp (Cannabis sativa L.) has been cultured since thousands of years ago and is distributed all over the world [1,2]. It is a multipurpose crop which can be used for food, feed, cosmetic and medicine [3,4]. Based on the concentration of THC (delta-9-tetrahydrocannabinol), drug type (THC > 0.3%) and non-drug type (THC < 0.3%) are characterized [5]. More attention was paid to cannabis products over time. Hemp seeds are a source of medicine and food. They can generate oils rich in polyunsaturated fatty acids including linoleic and α-linolenic acids and a small amount of other fatty acids that are not commonly found in vegetable oils, such as γ-linolenic acid and stearidonic acid, which are believed to have a positive effect on human health [6]. Hemp seeds are also known to be an excellent source of digestible protein, which can be absorbed due to the large proportion of storage protein in seed [3]. Efficacy and wide application are the main reasons for their popularity. As is well known, seed is an important organ in higher plants. And it is an important organ for plant survival and species dispersion [7]. Seed germination is a fundamental biological process affecting crop production [8]. The success of seedling cultivation depends largely on successful germination [9]. The research on the germination mechanism of Arabidopsis, rice, and maize seeds is very comprehensive [10]. Abscisic acid (ABA) is a main regulator of seed germination. Many studies have demonstrated that ABA is tightly associated with seed germination, and that the mutation or overexpression of genes related to ABA biosynthesis and degradation often result in germination-associated phenotypes [11–13]. The levels of endogenous ABA are tightly associated with seed germination. After seed imbibition, the levels of endogenous ABA rapidly decrease to a low level so that the subsequent germination processes can occur [14]. In addition to endogenous ABA levels, ABA signaling is another inhibiting factor for seed germination [14]. In plants, phytohormone ABA-dependent signaling pathway plays a central role in regulating seed dormancy and germination, seedling development and plant response to abiotic stress [15,16]. It has at least three crucial components, namely, PYL (ABA receptors), PP2CA (negative regulator) and SnRK2 (positive regulator) [17–19]. In the presence of ABA, PYL can interact with PP2CA and inhibit PP2CA from dephosphorylating SnRK2. The activated SnRK2s are then phosphorylating target substrates and activate multiple ABA-responsive element-binding factors (AREBs/ABFs) [15,18,20,21]. In Arabidopsis, 14 PYR/PYL/RCARs, 9 PP2CAs, and 10 SnRK2s have been identified [17,22,23]. Other studies had identified these genes in many plants including alfalfa (Medicago sativa L.), banana (Musa nana Lour.) and Soybean (Glycine max (L.) Merr) [24–26]. Numerous studies support the function of PYL, PP2CA, SnRK2 during germination process of seeds. In Arabidopsis, two AtPYLs, AtPYL11 and AtPYL12 positively modulate seed germination [27]. Ectopic expression of FsPP2C1 in Arabidopsis indicated that FsPP2C1 could negatively regulate ABA signaling [28]. In Arabidopsis, seed germination was significantly affected by SnRK2.2 and SnRK2.3 [22]. And, the expression of one member of CsPP2C gene CsPP2C-1 in embryos were verified. The results indicated that in vitro cultivation of embryos leaded to the increase of expression of CsPP2C-1 and the activation of ABA signaling pathways [29]. However, the systematical investigation about PYL, PP2CA, SnRK2 gene family in hemp has not yet been reported. Previous studies mainly focused on the response to stresses during the hemp seed germination, especially under saline and alkaline stress [9]. The identification and functional analysis of PYL, PP2CA, SnRK2 genes offer information of the mechanism of ABA signal pathway regulating seed germination, which can help us increase seed germination rate by regulating phytohormones.
In this study, members of genes families PYLs, PP2CAs and SnRK2s were identified from the hemp genome. Molecular biological approaches were used to reveal their evolutionary structures and investigate their expression patterns during the process of hemp seed germination. Germination assays on hemp seeds were performed to determine the ABA content during hemp seed germination. The interaction among hemp core ABA signaling components was validated by yeast two-hybrid experiments. This investigation provides useful information of the components of ABA signaling related to seed germination and provide foundation for the further research of hemp seeds.
2.1 Identification of Genes Involved in ABA Signal Transduction during the Development Stages of Hemp Seed Germination
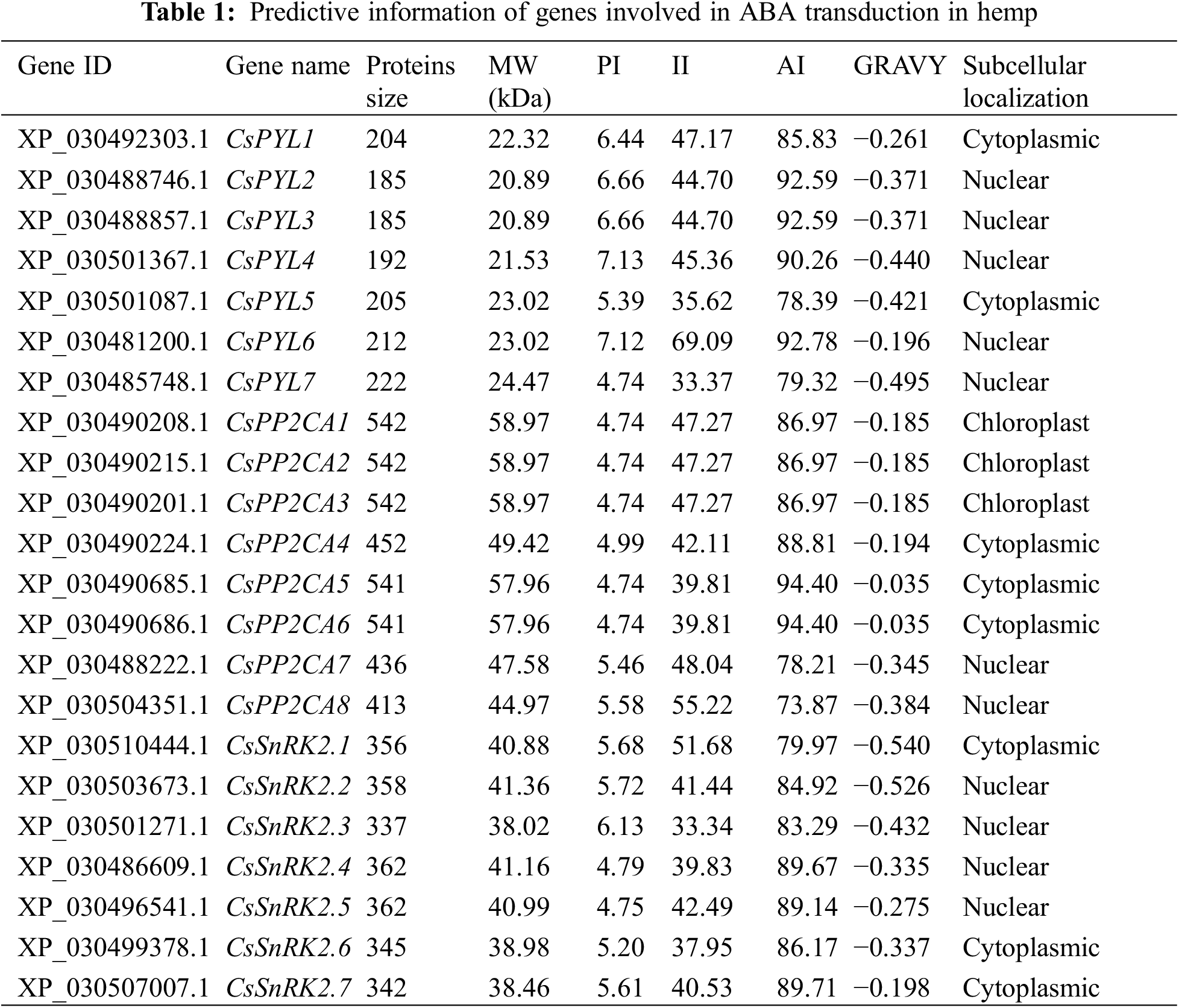
To identify the PYL, PP2CA and SnRK2 gene families in hemp, multiple database searches were performed. The Cannabis sativa (GCF_900626175.2_cs10_genomic) genome sequence was downloaded from the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/). Arabidopsis PYL, PP2C and SnRK2 protein sequences were downloaded from TAIR (http://www.arabidopsis.org/). Glycine max (L.) Merr. and Oryza sativa L. PYL, PP2C and SnRK2 protein sequences were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). They were used as the queries to perform a BLASTP search against the local protein database of Cannabis sativa. The E-value threshold was set at of 1e−5. Meanwhile, the PYL (PF10604), PP2C (PF00481) and SnRK2 (PF00069) typical domains were downloaded from the PFAM database (http://pfam.xfam.org/). HMMER 3.0 was used to search for the proteins with those domains in the local C. sativa protein database with an E-value of 1e−5. Protein sequences identified by two methods are combined and de-duplicated to obtain candidate hemp PYL, PP2CA and SnRK2 proteins. The candidates were finally submitted to InterPro (http://www.ebi.ac.uk/interpro/) and NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd/) to verify the typical conserved domain. The molecular weight (Mw), isoelectric point (pI), instability index (II), aliphatic index (AI), and grand average of hydropathicity (GRAVY) of these identified proteins were investigated with ExPASy (http://web.expasy.org/protparam/) online software. Subcellular localization was predicted with CELLO v2.5 software (http://cello.life.nctu.edu.tw/).
2.2 Chromosome Location of CsPYL, CsPP2C and CsSnRK2 Gene Families
Using the annotation file (gff3 file) of the PYL, PP2CA and SnRK2 genes in the C. sativa genome database, the distribution of C. sativa PYL, PP2CA and SnRK2 genes on the 10 chromosomes of C. sativa were analyzed. The chromosome physical location of the PYL, PP2CA and SnRK2 genes was displayed by MapChart v2.32 (https://www.wur.nl/en/show/Mapchart.htm).
2.3 Multiple Sequence Alignment and Phylogenetic Analysis
The identified PYL, PP2CA, SnRK2 proteins of hemp and PYL, PP2CA, SnRK2 proteins of Arabidopsis were used for multiple sequence alignment generated by MUSCLE 3.6 and BioEdit software. The neighbor-joining (NJ) tree was constructed for PYL, PP2C, SnRK2 proteins of hemp, Arabidopsis, Glycine max (L.) Merr. and Oryza sativa L. by MGEA 5.2 software with bootstrap value of 1000 [30].
Hemp seeds of cultivars Yunnan Eight (Y8) were provided by Yunnan Hemp Industrial Investment Co., Ltd. (China). During the seed harvest period, mature seeds were collected during the harvest period in 2019. After collection, all seeds were cleaned and air-dried under shade at 23°C–25°C for 2 weeks and then stored at 4°C for about 1 month until use in the experiment.
2.5 Germination Experiment on Hemp Seed
Hemp seeds were incubated in Petri dish containing moistened paper towels. All the seeds were incubated at 25°C and in continuous darkness. Seeds were sterilized with 75% ethanol solution for 1 min and cleaned with distilled water. Before being used in experiments, the seeds must be air dried. 25 treated seeds per group were evenly placed in Petri dish. It contained two layers of sterile filter paper soaked in equal volume of distilled water (the control) or distilled water with ABA (treatments). The solutions of ABA concentrations were 1 or 10 µM. The sterile filter paper was replaced every day. The developmental morphology of seeds was observed every 12 h from the beginning of the experiment. Four seed germination stages were defined and used as the sampling points for further analysis. The four seed germination stages were as follows: S0, quiescent hemp seeds stored for experiment; S1, the emergence of the radicles up to 1 mm; S2, the length of the radicles reached 1.5–3 cm; and S3, the emergence of two well-developed cotyledons (Fig. 1A). Seeds up to seed germination stages S1–S3 were collected for further analysis. Three replicates of 25 seeds were performed for each treatment.
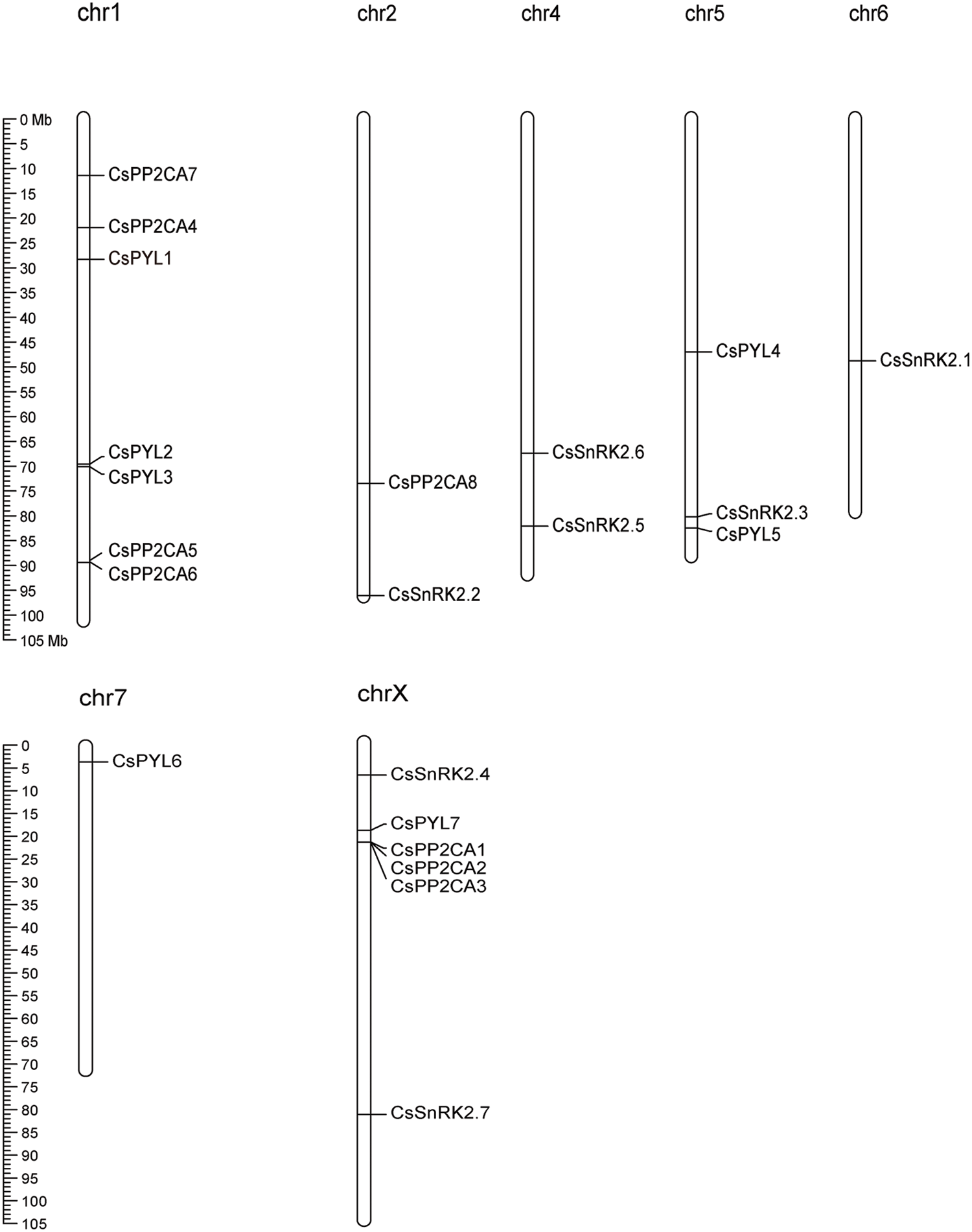
Figure 1: Distribution of the identified 7 CsPYLs, 8 CsPP2CAs and 7 CsSnRK2s genes across the hemp genome. All hemp chromosomes are drawn to scale based on their actual physical lengths
2.6 The Measurement of ABA Content in Seeds
Hemp seeds incubated in distilled water at three seed germination stages S1, S2 and S3 were collected for the determination of ABA content during the germination process of seeds. Additionally, quiescent hemp seeds acted at stage S0 were collected for further analysis of the ABA content as the control. The seed coats of hemp seed samples at stage S0–S3 were removed. To extract ABA, 10 g of seed samples were ground into a powder in liquid nitrogen, following which 0.05 g was accurately weighed and 5 mL 90% methanol was added; the samples were mixed by vortexing [31]. The resultant mixture was incubated overnight at 4°C, extracted by ultrasonication for 30 min, and centrifuged at 12000 rpm for 10 min. The supernatant was filtered through a 0.22 μm membrane and the filtrate was used to determine ABA content. 1 μL of the filtrate was used to determine the ABA content on Agilent 1290 liquid chromatograph and Agilent 6410 triple quadrupole mass spectrometer [32]. The conditions of HPLC-MS/MS system (Agilent, USA) were as follows, A C18 column (3.0 mm × 100 mm, 1.8 μm) was used for chromatographic separation at 30°C. Mobile phases A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile) were used for separation under the following gradient elution program: 0–14 min, 70%–80% B; 14–17 min, 80%–100% B; and 17–20 min, 100% B. The flow rate was 0.25 mL·min−1. The scanning mode was positive ionization mode, the capillary voltage was 4000 V(+), the desolvation temperature was 350°C, the desolvation gas flow was 10 l/min, and the nebulizer pressure was 35 psi. The detection method was multiple reaction monitoring (MRM). Dose-dependent calibration curves of ABA standards (Sigma-Aldrich, St. Louis, MO, USA; A1049) and internal standard were used to determine the concentrations of the components. Repeat the measurement three times for each sample.
2.7 Expression Analysis of PYL, PP2C, SnRK2 Genes under Exogenous ABA Treatment
Hemp seeds incubated in distilled water and distilled water with ABA at three seed germination stages S1, S2 and S3 were collected for the gene expression analysis. Some representative genes from the three families were selected to investigate the influence of ABA on seed germination and the expression levels of PYL, PP2C, SnRK2 genes under exogenous ABA treatment by qPCR analysis. Total RNA was extracted from samples (100 mg) frozen in liquid nitrogen using the Quick RNA Isolation Kit (Waryoung, China) following the manufacturer’s instructions. RNA was reverse transcribed using the FastQuant RT Kit (Tiangen, China) according to the manufacturer’s instructions. Primers used are listed in Table S1. Three biological replicates per treatment, each with three technical replicates were tested. For relative quantification of gene expression, CsEF1α was used as an internal control [33]. Quantification of gene expression was carried out using the 2−ΔΔCT method [34] and reported as relative expression levels, compared with control conditions as internal calibrator.
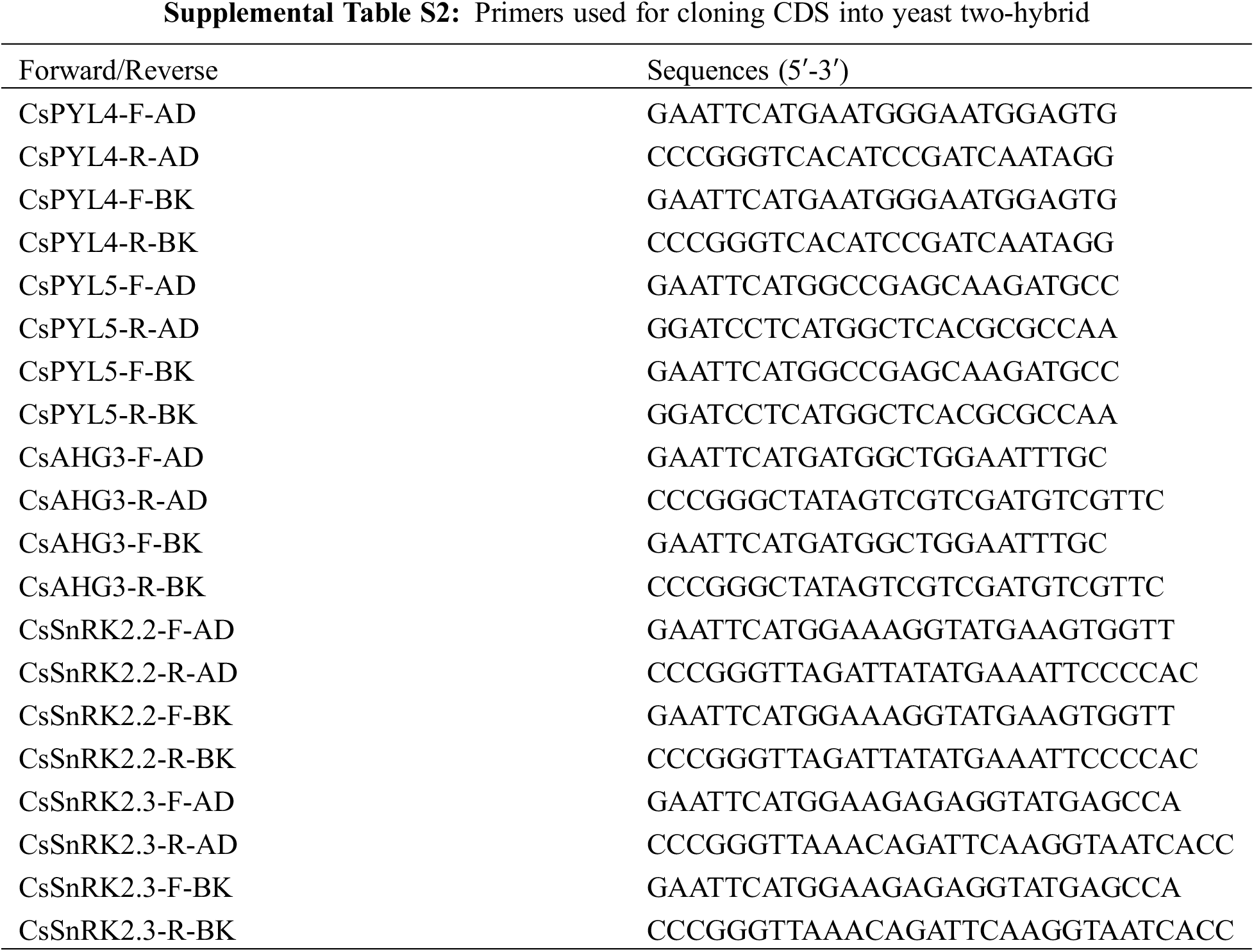
For yeast two-hybrid experiments, cDNA was used to PCR amplify the full-length coding sequence of CsPYLs, CsPP2CAs and CsSnRK2s. And cDNA is obtained by reverse transcription of RNA extracted from seeds. Primers used for PCR amplification of the mentioned genes are listed in Table S2. The vectors pGADT7 and pGBKT7 were transformed into yeast strain AH109 (Clontech, USA) using the Lithium acetate/Polyethylene glycol method [35]. CsPYL4 and CsPYL5 were cloned into vectors pGADT7 and pGBKT7 between EcoRI and SmaI, EcoRI and BamHI restriction sites respectively. CsAHG3 was cloned into vectors pGADT7 and pGBKT7 between EcoRI and SmaI restriction sites. CsSnRK2.2 and CsSnRK2.3 were cloned into vectors pGADT7 and pGBKT7 EcoRI and SmaI, EcoRI and BamHI restriction sites, respectively. Transformed colonies containing pGADT7 and pGBKT7 vectors were selected on synthetic medium lacking tryptophan and leucine (-W/-L). Active transformed colonies were picked up and diluted with distilled water to form bacterial fluid. Bacterial fluid was transferred to the synthetic medium lacking tryptophan, leucine, histidine and adenine (-W/-L/-H/-A). ABA was added to the synthetic medium (-W/-L/-H/-A) to obtain synthetic medium (-W/-L/-H/-A, +ABA). If the proteins encoded by two genes can interact with each other, some colonies will appear on the synthetic medium. Only when the vectors linked by the two genes are exchanged and the same positive results are still displayed in the synthetic medium, these two genes are supposed to interact with each other. Positive and negative controls were also performed as indicated in the figure legend.
SPSS 22.0 statistical software was used to conduct an analysis of variance (ANOVA). A one-way and two-way ANOVA using development stages of hemp seeds and ABA concentrations as the factors was employed to test the differences among gene expression levels under different treatments. Tukey tests were performed to estimate the significant difference between the treatment means (p < 0.05).
3.1 Identification of Genes Involved in ABA Signal Transduction
A total of 22 putative ABA signal transduction core components, namely, 7 ABA receptor genes (CsPYLs), 8 ABA signal transduction negative regulatory genes (CsPP2CAs), and 7 ABA signal transduction positive regulatory genes (CsSnRKs), were identified from the genome of C. sativa (Table 1). Chromosome localization found that the identified 7 CsPYLs, 8 CsPP2CAs and 7 CsSnRK2s were unevenly distributed in 7 out of 10 hemp chromosomes (Fig. 1). The gene number is based on the position on the chromosome. Based on the protein sequence of genes, several physical properties of these genes were predicted. The results were shown in Fig. 1. The prediction of subcellular localization indicated that members of the CsPYL and CsSnRK2 gene families could only localize to the cytoplasm or nucleus. Members of the CsPP2CA gene family may also be located in chloroplasts.
3.2 Phylogenetic Analysis and Multiple Alignment of ABA Signal Transduction Genes
To investigate the phylogenetic relationship among PYL-PP2CA-SnRK2 family members from C. sativa, an unrooted phylogenetic tree was constructed with MAGA 5.2 according to multiple alignments of the predicted protein sequences of 7 CsPYLs, 8 CsPP2CAs and 7 CsSnRK2s from C. sativa, 14 AtPYLs, 9 AtPP2CAs, and 10 AtSnRK2s from Arabidopsis, 21 GmPYLs, 35 GmPP2C, 21 GmSnRK2s from Glycine max (L.) Merr., 13 OsPYL, 11OsPP2C, 10 OsSnRK2 form Oryza sativa L. (Fig. 2). Based on the classification of subfamilies in Arabidopsis, 7 CsPYLs were classified into three subfamilies (Fig. 2A). Subfamily I contained CsPYL5 and CsPYL7. Subfamily II contained CsPYL1 and CsPYL6. Subfamily III contains CsPYL2, CsPYL3, and CsPYL4. PYL, PP2CA, SnRK2 proteins of the same subfamily may have similar biological functions. To predict the functions of PYL, PP2CA, SnRK2 genes, multiple deduced amino acid sequence alignments were conducted to verify the functional residues of the PYL, PP2CA, SnRK2. The results showed that the functional residues of CsPYLs and AtPYLs were very conserved (Fig. 3A). Members of CsPYLs might have the same function with AtPYLs that were in the same clade with them. CsPYL5 clustered in the classical subfamily I, and was in a clade with AtPYL1 and AtPYL14. CsPYL4 clustered in the classical subfamily III and was in a clade with AtPYL8. Thus, CsPYL5 might have the same function with AtPYL1 and AtPYL14, and CsPYL4 might have the same function with AtPYL8. Eight genes encoding PP2CA proteins were classified into two subfamilies (Fig. 2B). The first subfamily included CsPP2CA7 and CsPP2CA8. They were clustered with Arabidopsis AIP1, HAI3, HAI1, AHG3, and AHG1. Another subfamily contained CsPP2CA1, CsPP2CA2, CsPP2CA3, CsPP2CA4, CsPP2CA5 and CsPP2CA6. And they were grouped with Arabidopsis ABI1, ABI2, HAB2, and HAB1. We focused our studies on two CsPP2C representatives, CsPP2CA7 and CsPP2CA8. Fig. 3B showed that CsPP2CA7 and CsPP2CA8 had the necessary functional residues for Mn/Mg++ interaction and the interaction with PYLs. And the functional residues of CsPP2CA7, CsPP2CA8 and Arabidopsis PP2CAs were well conserved. Similar to AtAHG3, CsPP2CA7 lacked the glutamic acid that other clade A PP2Cs had to interact with PYL genes (E122 in AtHAI1, E88 in AtHAI3) and replaced it with aspartic acid (D138 in CsPP2CA7 and D115 in AtAHG3). Thus, CsPP2CA7 was named as CsAHG3. Three amino acids in other clade A PP2Cs had to interact with PYL genes were not conserved in CsPP2CA8 and same as in AtHAI1. Thus, CsPP2CA8 was named as CsHAI1. Seven genes encoding CsSnRK2s were clustered into three well-supported sub-families (Fig. 2C). CsSnRK2.1 and CsSnRK2.2 belonged to subfamily I; CsSnRK2.4 and CsSnRK2.5 belonged to subfamily II; CsSnRK2.3, CsSnRK2.6, and CsSnRK2.7 belonged to subfamily III. Multiple sequence alignment of hemp and Arabidopsis SnRK2 proteins was performed. We identified one conserved kinase region, Ser/Thr kinase activity domain and two functional subdomains, Domain I and Domain II within SnRK2 (Fig. 3C). Meanwhile, one conserved kinase region, Ser/Thr kinase activity domain and two functional subdomains, Domain I and Domain II within SnRK2 were identified. Domain I is important in response to abiotic stress, and is conserved in all SnRK2s [36]. Kinase activity in domain II determined whether SnRK2 protein can be activated by ABA [37]. SnRK2 proteins could interact with PP2CA and response to ABA. This function is mainly due to the Domain II in the C-terminal region of the SnRK2 proteins [38]. Seven hemp SnRK2s (CsSnRK2.3, CsSnRK2.4, CsSnRK2.5, CsSnRK2.6, and CsSnRK2.7) belonged to subgroups II and III; CsSnRK2.1 and CsSnRK2.2 clustered with subgroup I (Figs. 2C and 3C). Based on the kinase activity brought by Domain II (Fig. 3C), CsSnRK2.3, CsSnRK2.4, CsSnRK2.5, CsSnRK2.6, and CsSnRK2.7 could be activated by ABA with multiple strengths. CsSnRK2.1 and CsSnRK2.2 could not be activated by ABA.
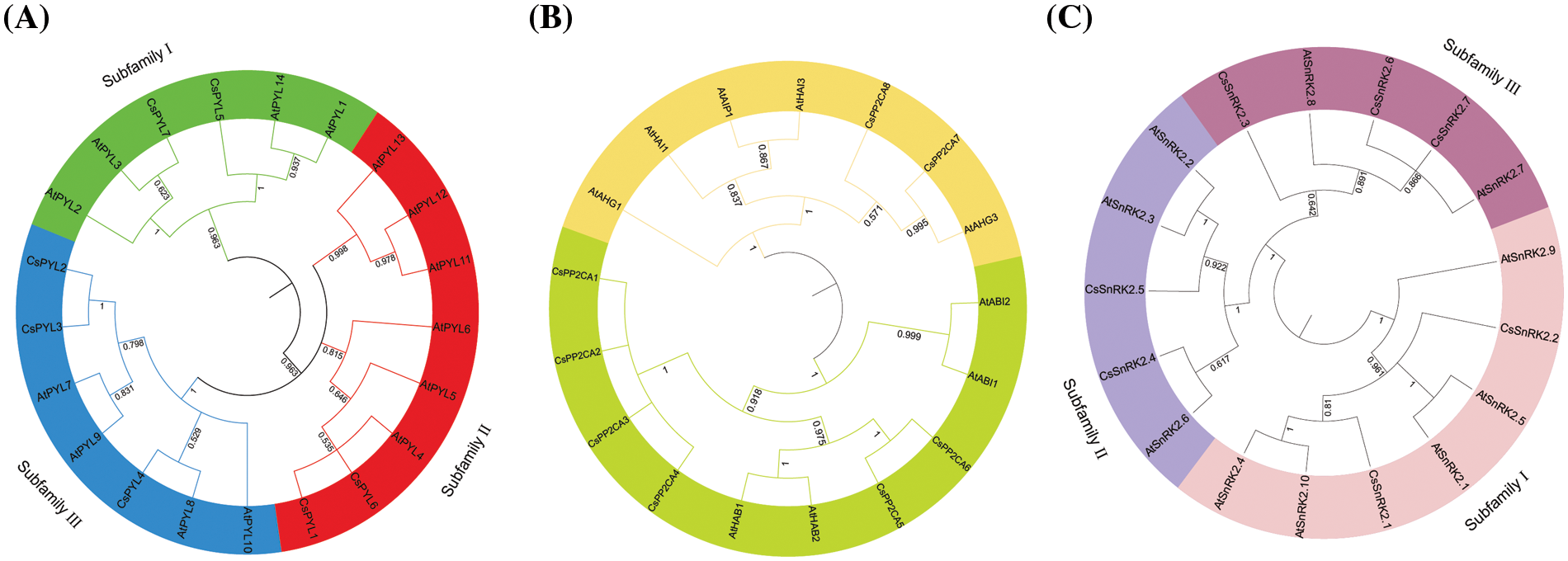
Figure 2: Phylogenetic analysis of the PYLs (A), PP2CAs (B) and SnRK2s (C) from A. thaliana, C. sativa., Glycine max (L.) Merr. and Oryza sativa L. The accession numbers of all C. sativa. PYL-PP2CA-SnRK2 genes used in the present study are listed in Table 1
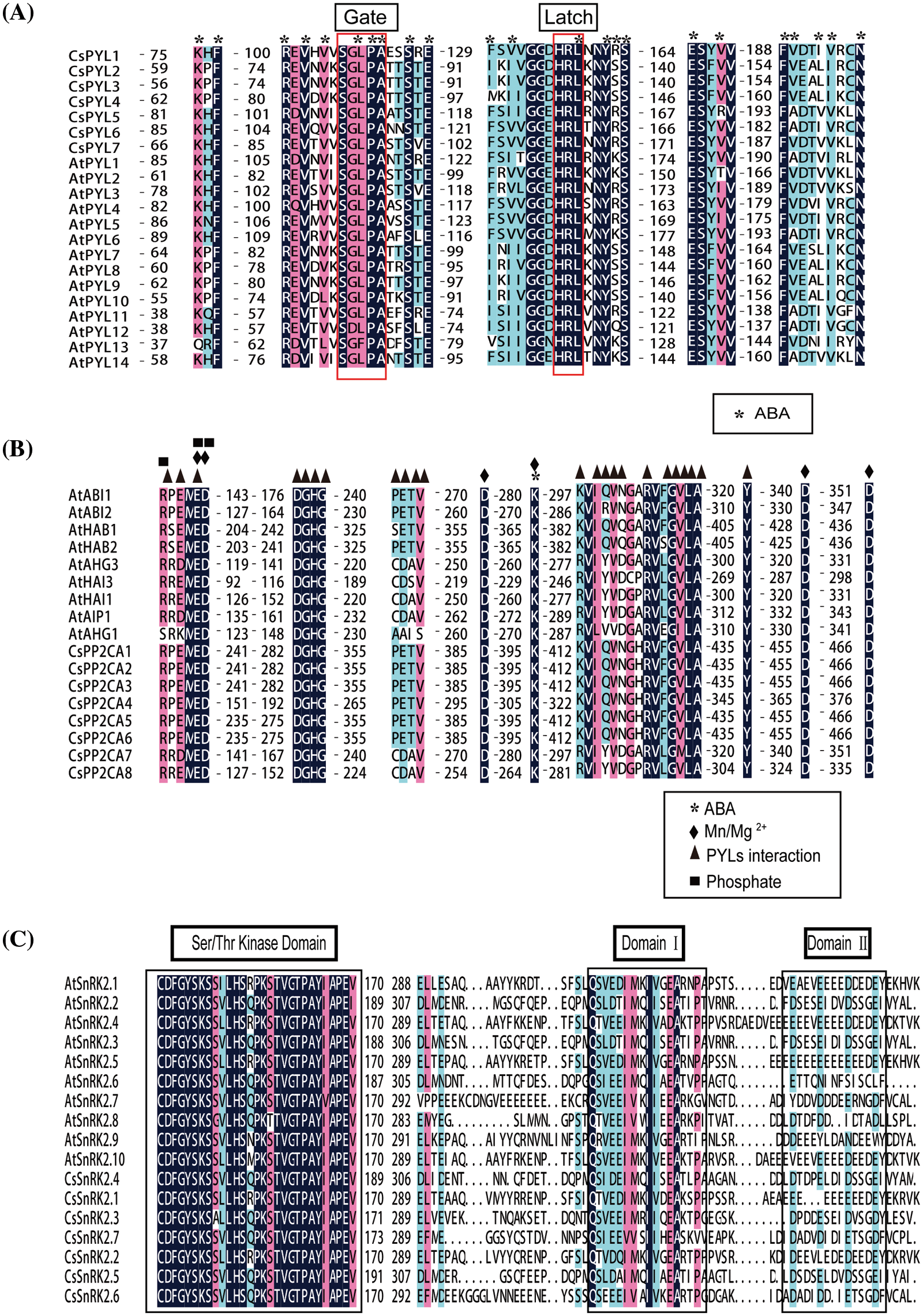
Figure 3: Sequence alignment and structural analysis of PYLs, PP2Cs and SnRK2s in A. thaliana and hemp. (A) Alignment of 14 A. thaliana PYLs and 7 hemp PYLs with characteristic Gate and Latch domains labeled. (B) Alignment of 9 A. thaliana PP2CAs and 8 hemp PP2CAs. Residues interacting with ABA, PYLs, and Mn/Mg2+ irons are marked with asterisks, black triangles, and black diamonds, respectively. Phosphatase sites are marked with black squares. (C) Alignment of 10 A. thaliana SnRK2s and 7 hemp SnRK2s. Functional residues are marked with corresponding symbols in the figure
3.3 ABA Content during the Development Stages of Hemp Seed Germination
ABA plays a pivotal role in various aspects of plant growth and development [32]. The ABA content of well-developed and undamaged seeds during the germination process of hemp seeds was investigated. The three developmental morphologies of hemp seeds are shown in Fig. 4A, and the time when the seeds reached three developmental stages was recorded. These stages were as follows: emergence of the radicles up to 1 mm after 24 h of germination experiment (Fig. 4A); when the length of the radicles reached 1.5–3.0 cm after 48 h of germination experiment and most of the seeds developed two complete cotyledons after 72 h of germination experiment. The ABA content in seeds at three development stages was measured. The ABA content in dormant seeds was measured and used as control. The content of ABA in dormant seeds was 4–10 times higher than the content of ABA in the three other stages of seed germination. As the seeds started to absorb water and then germinate, the content of ABA reached the minimum after 24 h. The content of ABA gradually increased from 24 to 72 h (Fig. 4B).
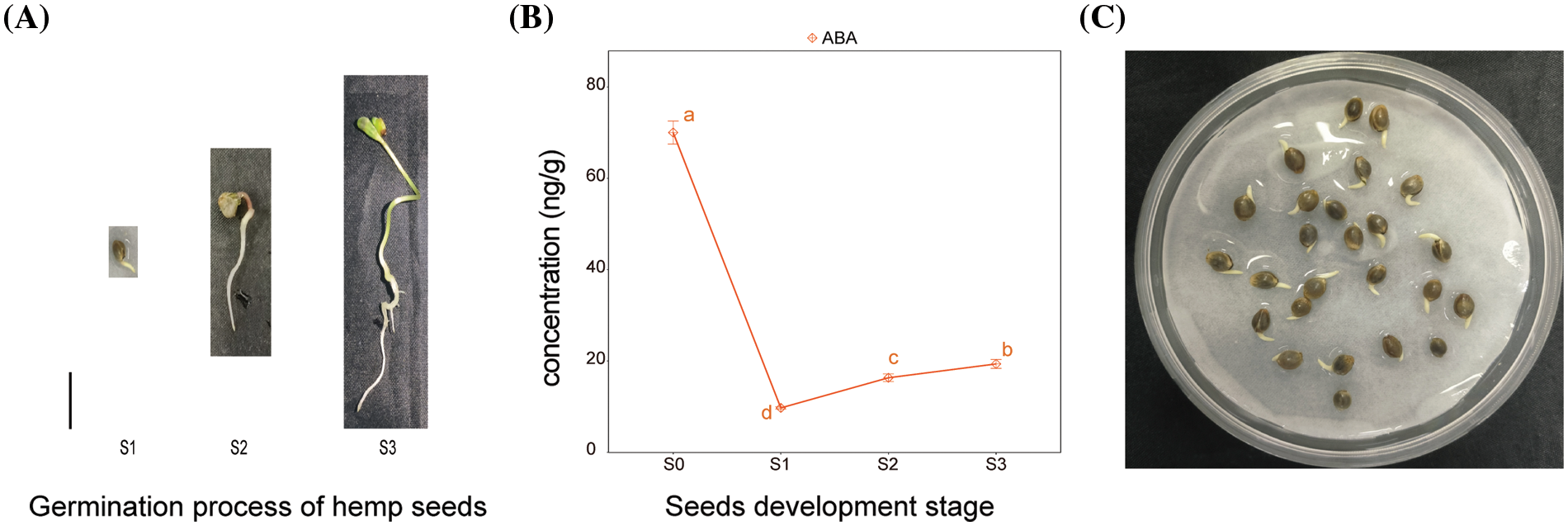
Figure 4: (A)Three development stages of hemp seeds incubated in water irrespective of the presence of ABA. S1: The emergence of the radicles up to 1 mm. S2: The length of the radicles reached 1.5–3 cm. S3: The emergence of two well-developed cotyledons. (B) ABA content in dormant seeds (S0) and germinated seeds at the three development stages. Values are means ± SD (n = 3). (C) Hemp seeds incubated in water with 10 µM ABA for 24 h
3.4 Expression of PYL, PP2C, and SnRK2 Regulated by ABA
To verify the expression pattern of CsPYLs, CsPP2Cs and CsSnRK2s in hemp seeds under exogenous ABA treatment, we analyzed the germination and growth of hemp seeds treated by ABA in the germination experiment. On the basis of the results of phylogenetic analysis and multiple alignment, CsPYL4 and CsPYL5 may have the same function as AtPYL1 and AtPYL8 during the germination of seeds. CsAHG3 and CsHAI1 may have the same function as AtHAI1 and AtAHG3 in the ABA response. Thus, CsPYL4, CsPYL5, CsAHG3 and CsHAI1 were selected for gene expression analyses. Three SnRK2s were selected, one for each subfamily (CsSnRK2.2, CsSnRK2.3 and CsSnRK2.5). The expression of selected genes in hemp seeds incubated in water with ABA (0, 1, and 10 µM) for 0, 24, 48, and 72 h was investigated to determine the potential functions of PYL, PP2CA, and SnRK2 genes during the germination of hemp seeds. The expression levels of 7 genes under different treatments are shown in Fig. 5. High concentrations of exogenous ABA treatment changed the expression levels of CsPYL5. Low concentrations of exogenous ABA treatment influenced the expression levels of CsPYL4 (Figs. 5A and 5B). High concentrations of exogenous ABA treatment had a significant effect on the increase in expression of CsAHG3 and CsHAI1 (Figs. 5C and 5D). High concentrations of exogenous ABA treatment had a significant effect on the increase in CsSnRK2.2 expression from stage S2 to stage S3 (Fig. 5E). Exogenous ABA treatment had a significant effect on the increase in CsSnRK2.3 expression. The expression of CsSnRK2.3 in seeds under 10 µM ABA treatment was significantly higher than that in seeds incubated in water or water with 1 µM ABA at stages S1, S2, and S3 (Fig. 5F). The expression of CsSnRK2.5 in seeds incubated in water was significantly higher than that in seeds under 1 or 10 µM ABA treatment at stage S1. The expression of CsSnRK2.5 in seeds under 10 µM ABA treatment was significantly higher than that in seeds incubated in water at stage S3 (Fig. 5G). This result revealed that the expression levels of selected genes at the same seed germination stages were significantly affected by exogenous ABA. The effect of exogenous ABA on the expression of genes was both positive and negative and was related to the concentration of exogenous ABA.
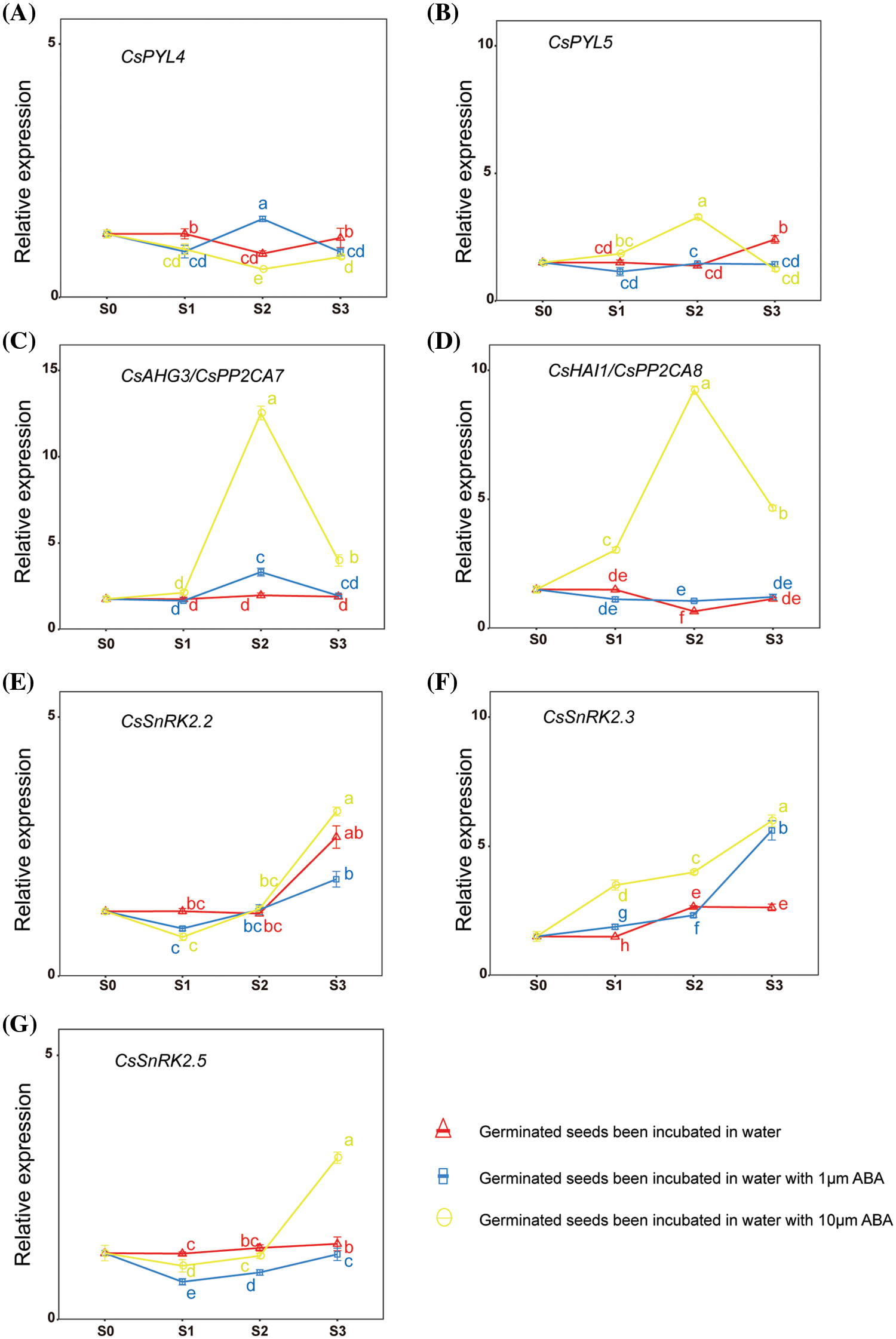
Figure 5: Expression of CsPYL, CsPP2CA, and CsSnRK2 genes at different stages of hemp seed germination under different treatments. Values are means ± SD of three biological replicates (n = 3). Means within a column with different letters are significantly different (p < 0.05)
3.5 Interaction Among PYLs, PP2Cs, and SnRK2s
The key function of ABA receptors upon ABA binding is their ability to interact with and inhibit PP2CAs, releasing SnRK2s from inhibition. Therefore, we tested whether selected CsPYLs, CsPP2CAs, and CsSnRK2s, which showed seed development-related expression patterns, were capable of interacting with one another through yeast two-hybrid assay. The full-length coding sequences of CsPYL4, CsPYL5, CsAHG3, CsSnRK2.2 and CsSnRK2.3 were amplified by PCR and cloned into pGBKT7 and pGADT7. They were used to test multiple combinations. Yeast two hybrid experiment among CsPYL4, CsPYL5, CsAHG3, CsSnRK2.2 and CsSnRK2.3 showed ABA-independent interactions (Fig. 6). CsPYL5 could interact with CsAHG3 irrespective of the presence of ABA, whereas CsPYL4 did not interact with CsAHG3 (Figs. 6A and 6B). CsAHG3 could interact with CsSnRK2.2 and CsSnRK2.3 regardless of the presence of ABA, too (Figs. 6A and 6B).
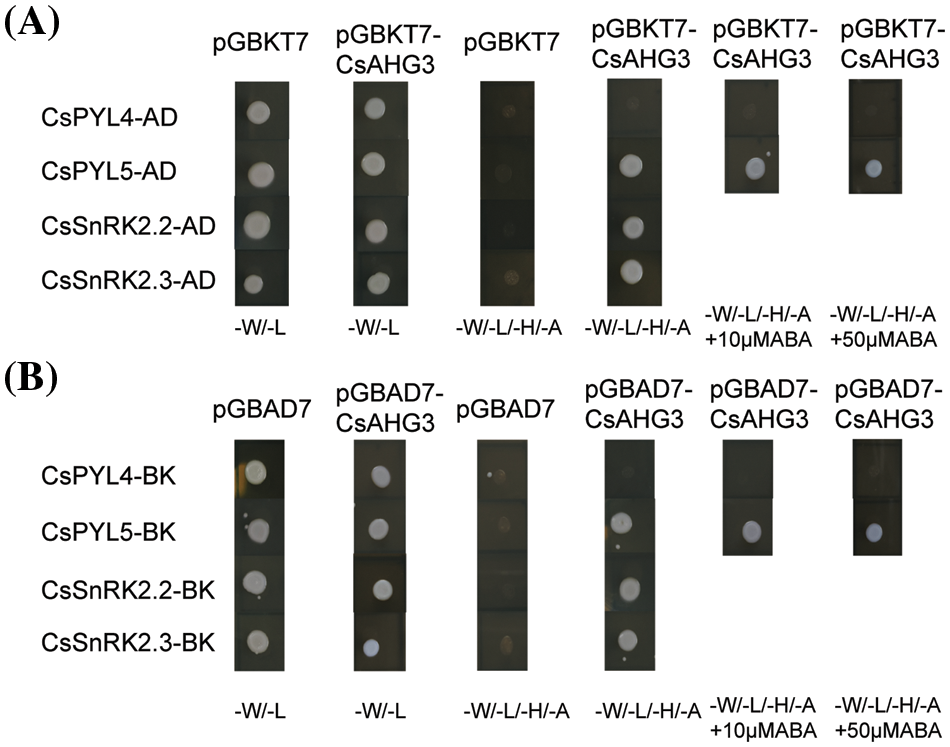
Figure 6: Interaction test of PP2Cs with PYLs and SnRK2s in the yeast two-hybrid assay. (A) The pGADT7 vector was used to express the CsPP2CA gene CsAHG3 and the pGBKT7 vector was used to express the CsSnRK2s and CsPYLs including CsPYL4, CsPYL5, CsSnRK2.2, and CsSnRK2.3. (B) The pGBKT7 vector was used to express the CsPP2CA gene CsAHG3 and the pGADT7 vector was used to express the CsSnRK2s and CsPYLs including CsPYL4, CsPYL5, CsSnRK2.2, and CsSnRK2.3. Co-transformations of the hemp PYL, SnRK2, PP2CA genes with appropriate complementary empty vectors are shown as negative controls. Yeast cells grown on synthetic media lacking tryptophan and leucine (-W/-L) and on synthetic, selective media lacking tryptophan, leucine, histidine and adenine (-W/-L/-H/-A) or, where indicated, with 10 or 50 µM ABA (-W/-L/-H/-A+ABA) are shown. Pictures were taken after 3 days of incubation at 25°C
In this study, the ABA content in quiescent hemp seeds and seeds in three germination stages was measured. The ABA content in quiescent seeds was significantly higher than that in the three stages of seed germination. These results were consistent with the view that ABA is the primary effector of the regulation of seed germination, and the amount of ABA quickly reduces during seed germination [39]. Seed germination is adversely affected by ABA [40]. Given the germination process of hemp seeds depend on ABA signaling, the crucial components of the ABA pathway in hemp must be investigated. Here, 7 PYLs, 8 PP2CAs, and 7 SnRK2s were identified from the C. sativa genome. Based on the evolutionary relationship, they were divided into 3, 2, and 3 subfamilies, respectively (Fig. 2). This result was consistent with previous evolutionary classifications of PYL, PP2CA, or SnRK2 in Arabidopsis [41]. CsPYLs, CsPP2CAs, and CsSnRK2s included well-conserved functional domains for each gene family (Fig. 3). CsPYL5 was in a clade with AtPYL1 and AtPYL14, and CsPYL4 was in a clade with AtPYL8. Thus, CsPYL5 might have the same function as AtPYL1 and AtPYL14 and CsPYL4 might have the same function as AtPYL8.Arabidopsis PYL1, PYL2, PYL4, PYL8, and PYL14 play an important role in the regulation of the germination process of seeds [18,42,43]. AtHAI1 and AtAHG3 played a major role among AtPP2CAs in the ABA response in seeds [44,45]. CsAHG3 and CsHAI1 were similar to AtHAI1 and AtAHG3, respectively (Fig. 3B). Based on the kinase activity brought by Domain II (Fig. 3C), CsSnRK2.3, CsSnRK2.4, CsSnRK2.5, CsSnRK2.6, and CsSnRK2.7 could be activated by ABA with multiple strengths. CsSnRK2.1 and CsSnRK2.2 could not be activated by ABA.
ABA treatments resulted in changes in the expression of genes involved in ABA signaling during hemp seed germination stages. Quantification of gene expression of selected genes under different treatments showed differences among family members (Fig. 5). The sensitivity of CsPYL4 to exogenous ABA was different in three seed germination stages. It indicated that CsPYL4 may play diverse roles in the regulation of hemp seed germination. It is similar to the results of another study of PYL genes in pear [16]. CsAHG3, CsHAI1 and CsPYL5 were sensitive to high concentrations of ABA, and they had the same expression trend (Figs. 5A–5D). This result is similar to that of similar experiments in other species, such as pepper and maize [46,47]. Yeast two-hybrid assay showed that CsPYL5 interacted with CsAHG3 irrespective of the presence of ABA (Figs. 6A and 5B). A series of studies reported the negative regulatory function of PP2CAs [20,48]. Thus, CsAHG3 and CsPYL5 may involve in the regulation of the normal development of seeds under high concentrations of ABA treatment. CsSnRK2.3 was sensitive to exogenous ABA, and the sensitivity of CsSnRK2.3 to exogenous ABA was positively related to its concentration. The previous study revealed the sensitivity of SnRK2s to exogenous ABA in many crops [47]. CsAHG3 could interact with CsSnRK2.3 irrespective of the presence of ABA (Figs. 5A and 5B). The previous study reported that PP2CAs can directly interact with groups II and III of SnRK2s in some plants [49]. Although CsSnRK2.3 had a different expression pattern from CsAHG3 and CsPYL5, it might form a regulatory network with CsAHG3 and CsPYL5 to play a role in the ABA signaling pathway during germination of hemp seeds. This is a common way of action of the ABA signaling pathway obtained in previous studies [20,41]. Our predictions based on evolutionary relationships, presence of these conserved residues, quantification of gene expression under different treatments and interaction between CsPYL5, CsSnRK2.2, and CsSnRK2.3 with CsAHG3/CsPP2CA7 observed in yeast suggested that CsPYL5 and CsAHG3/CsPP2CA7 might be functional ABA receptors.
In conclusion, the ABA content was lower in hemp seeds during the germinating stages compared with that during the non-germinating stage. Seven CsPYL, 8 CsPP2CA, and 7 CsSnRK2 genes were identified based on the genome of C. sativa and confirmed to be homologous to Arabidopsis PYL, PP2CA, and SnRK2 genes, respectively. The expression patterns and levels of CsPYL, CsPP2CA, and CsSnRK2 genes varied during the germination stages of seeds, depending on the concentration of exogenous ABA. CsPYL4, CsPYL5, and CsSnRK2.2 were insensitive to the exogenous ABA. Yeast two-hybrid assays demonstrated that CsPYL5, CsSnRK2.2, and CsSnRK2.3 could interact with CsAHG3 irrespective of the presence of ABA. CsPYL5, CsAHG3, and CsSnRK2.2 may play a role in the ABA signaling transduction pathway of hemp seeds during the germination stages. These results should help improve our understanding of molecular biological information involved in the ABA transduction pathway during the germination process of hemp seeds. This work can lay a foundation for the further study about germination of hemp seeds.
Funding Statement: This study was funded by the Fundamental Research Funds for the Central Public Welfare Research Institutes (ZZ13-YQ049), the Scientific Research Project of Hainan Academician Innovation Platform (SQ2021PTZ0052), and the National Key R&D Program of China from the Ministry of Science and Technology of China (No. 2019YFC1711100).
Author Contributions: The authors confirm contribution to the paper as follows: Cong Hou and Kang Ning provided experimental design and supervision. Cong Hou and Yufei Cheng performed the experiments and manuscript preparation. Haibin Yu and Xiuye Wei prepared the plant materials. Huatao Yu, Xia Liu and Linlin Dong reviewed and modified this manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest: Haibin Yu is employed by Yunnan Hemp Industrial Investment CO. LTD. And equipment, drugs, or supplies were provided by Yunnan Hemp Industrial Investment CO. LTD. Other authors have no conficts of interest to declare.
References
1. Andre, C. M., Hausman, J. F., Guerriero, G. (2016). Cannabis sativa: The plant of the thousand and one molecules. Frontiers in Plant Science, 7, 19. [Google Scholar] [PubMed]
2. Wei, H., Chen, S., Niyitanga, S., Liu, T., Qi, J. et al. (2022). Genome-wide identification and expression analysis response to GA3 stresses of WRKY gene family in seed hemp (Cannabis sativa L.). Gene, 822, 146290. [Google Scholar] [PubMed]
3. Farinon, B., Molinari, R., Costantini, L., Merendino, N. (2020). The seed of industrial hemp (Cannabis sativa L.Nutritional quality and potential functionality for human health and nutrition. Nutrients, 12(7), 1935. [Google Scholar] [PubMed]
4. Schluttenhofer, C., Yuan, L. (2017). Challenges towards revitalizing hemp: A multifaceted crop. Trends in Plant Science, 22(11), 917–929. [Google Scholar] [PubMed]
5. Kovalchuk, I., Pellino, M., Rigault, P., van Velzen, R., Ebersbach, J. et al. (2020). The genomics of cannabis and its close relatives. Annual Review of Plant Biology, 71, 713–739. [Google Scholar] [PubMed]
6. Simopoulos, A. P. (2008). The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Experimental Biology and Medicine, 233(6), 674–688. [Google Scholar] [PubMed]
7. Rodriguez-Gacio Mdel, C., Matilla-Vazquez, M. A., Matilla, A. J. (2009). Seed dormancy and ABA signaling: The breakthrough goes on. Plant Signaling & Behavior, 4(11), 1035–1049. [Google Scholar]
8. Penas, E., Martinez-Villaluenga, C. (2020). Advances in production, properties and applications of sprouted seeds. Foods, 9(6), 790. [Google Scholar] [PubMed]
9. Hu, H., Liu, H., Liu, F. (2018). Seed germination of hemp (Cannabis sativa L.) cultivars responds differently to the stress of salt type and concentration. Industrial Crops and Products, 123, 254–261. [Google Scholar]
10. Han, C., Yang, P. (2015). Studies on the molecular mechanisms of seed germination. Proteomics, 15(10), 1671–1679. [Google Scholar] [PubMed]
11. Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F. et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant Journal, 70(3), 501–512. https://doi.org/10.1111/j.1365-313X.2011.04887.x [Google Scholar] [PubMed] [CrossRef]
12. Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S. et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. European Molecular Biology Organization, 23(7), 1647–1656. https://doi.org/10.1038/sj.emboj.7600121 [Google Scholar] [PubMed] [CrossRef]
13. Lindgren, L. O., Stalberg, K. G., Hoglund, A. S. (2003). Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiology, 132(2), 779–785. https://doi.org/10.1104/pp.102.017053 [Google Scholar] [PubMed] [CrossRef]
14. Vishal, B., Kumar, P. P. (2018). Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Frontiers in Plant Science, 9, 838. https://doi.org/10.3389/fpls.2018.00838 [Google Scholar] [PubMed] [CrossRef]
15. Leng, P., Yuan, B., Guo, Y. (2014). The role of abscisic acid in fruit ripening and responses to abiotic stress. Journal of Experimental Botany, 65(16), 4577–4588. https://doi.org/10.1093/jxb/eru204 [Google Scholar] [PubMed] [CrossRef]
16. Wang, G., Qi, K., Gao, X., Guo, L., Cao, P. et al. (2022). Genome-wide identification and comparative analysis of the PYL gene family in eight Rosaceae species and expression analysis of seeds germination in pear. BMC Genomics, 23(1), 233. https://doi.org/10.1186/s12864-022-08456-1 [Google Scholar] [PubMed] [CrossRef]
17. Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E. et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell, 24(6), 2483–2496. https://doi.org/10.1105/tpc.112.098574 [Google Scholar] [PubMed] [CrossRef]
18. Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H. et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324(5930), 1068–1071. https://doi.org/10.1126/science.1173041 [Google Scholar] [PubMed] [CrossRef]
19. Yoshida, T., Nishimura, N., Kitahata, N., Kuromori, T., Ito, T. et al. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiology, 140(1), 115–126. https://doi.org/10.1104/pp.105.070128 [Google Scholar] [PubMed] [CrossRef]
20. Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M. et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant & Cell Physiology, 51(11), 1821–1839. https://doi.org/10.1093/pcp/pcq156 [Google Scholar] [PubMed] [CrossRef]
21. Chen, H. H., Qu, L., Xu, Z. H., Zhu, J. K., Xue, H. W. (2018). EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Molecular Plant, 11(5), 706–719. [Google Scholar] [PubMed]
22. Long, T., Xu, B., Hu, Y., Wang, Y., Mao, C. et al. (2021). Genome-wide identification of ZmSnRK2 genes and functional analysis of ZmSnRK2.10 in ABA signaling pathway in maize (Zea mays L). BMC Plant Biology, 21(1), 309. [Google Scholar] [PubMed]
23. Nie, L., Xu, Z., Wu, L., Chen, X., Cui, Y. et al. (2022). Genome-wide identification of protein phosphatase 2C family members in Glycyrrhiza uralensis Fisch. and their response to abscisic acid and polyethylene glycol stress. Journal of Taibah University for Science, 15(1), 1260–1268. [Google Scholar]
24. Nian, L., Zhang, X., Yi, X., Liu, X., Ain, N. U. et al. (2021). Genome-wide identification of ABA receptor PYL/RCAR gene family and their response to cold stress in Medicago sativa L. Physiology and Molecular Biology of Plants, 27(9), 1979–1995. [Google Scholar] [PubMed]
25. Hu, W., Yan, Y., Shi, H., Liu, J., Miao, H. et al. (2017). The core regulatory network of the abscisic acid pathway in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. BMC Plant Biology, 17(1), 145. [Google Scholar] [PubMed]
26. Zhang, Z., Ali, S., Zhang, T., Wang, W., Xie, L. (2020). Identification, evolutionary and expression analysis of PYL-PP2C-SnRK2s gene families in soybean. Plants, 9(10), 1356. https://doi.org/10.3390/plants9101356 [Google Scholar] [PubMed] [CrossRef]
27. Zhao, H., Nie, K., Zhou, H., Yan, X., Zhan, Q. et al. (2020). ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytologist, 228(2), 596–608. https://doi.org/10.1111/nph.16713 [Google Scholar] [PubMed] [CrossRef]
28. Gonzalez-Garcia, M. P., Rodriguez, D., Nicolas, C., Rodriguez, P. L., Nicolas, G. et al. (2003). Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiology, 133(1), 135–144. https://doi.org/10.1104/pp.103.025569 [Google Scholar] [PubMed] [CrossRef]
29. Král, D., Šenkyřík, J. B., Ondřej, V. (2022). Expression of genes involved in ABA and auxin metabolism and LEA gene during embryogenesis in hemp. Plants, 11(21), 2995. https://doi.org/10.3390/plants11212995 [Google Scholar] [PubMed] [CrossRef]
30. Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Molecular Biology and Evolution, 30(5), 1229–1235. https://doi.org/10.1093/molbev/mst012 [Google Scholar] [PubMed] [CrossRef]
31. Zhu, P., Cai, Y., Yu, J., Zhao, A., Liang, Y. et al. (2017). Characterization and expression of abscisic acid signal transduction genes during mulberry fruit ripening. Acta Physiologiae Plantarum, 39(7), 149. https://doi.org/10.1007/s11738-017-2442-5 [Google Scholar] [CrossRef]
32. Zeng, W., Tan, B., Deng, L., Wang, Y., Muzammal Aslam, M. et al. (2020). Identification and expression analysis of abscisic acid signal transduction genes during peach fruit ripening. Scientia Horticulturae, 270, 109402. https://doi.org/10.1016/j.scienta.2020.109402 [Google Scholar] [CrossRef]
33. Guo, R., Guo, H., Zhang, Q., Guo, M., Xu, Y. et al. (2018). Evaluation of reference genes for RT-qPCR analysis in wild and cultivated Cannabis. Bioscience, Biotechnology, and Biochemistry, 82(11), 1902–1910. https://doi.org/10.1080/09168451.2018.1506253 [Google Scholar] [PubMed] [CrossRef]
34. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262 [Google Scholar] [PubMed] [CrossRef]
35. Chang, B., Stephen, J. E. (1997). Gene identification using the yeast two-hybrid system. Methods in Enzymology, 283, 141–156. https://doi.org/10.1016/S0076-6879(97)83013-3 [Google Scholar] [PubMed] [CrossRef]
36. Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F. et al. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. Journal of Biological Chemistry, 281(8), 5310–5318. [Google Scholar] [PubMed]
37. Umezawa, T., Sugiyama, N., Mizoguchi, M., Myougae, F., Hayashi, S., Yamaguchi-Shinozakic, K. et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciencesof the United States of America, 106, 17588–17593. [Google Scholar] [PubMed]
38. Wu, Z., Cheng, J., Hu, F., Qin, C., Xu, X. et al. (2020). The SnRK2 family in pepper (Capsicum annuum L.Genome-wide identification and expression analyses during fruit development and under abiotic stress. Genes Genomics, 42(10), 1117–1130. [Google Scholar] [PubMed]
39. Chen, H., Ruan, J., Chu, P., Fu, W., Liang, Z. et al. (2020). AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant Journal, 101(2), 310–323. [Google Scholar]
40. Yan, A., Chen, Z. (2017). The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Reports, 36(5), 689–703. [Google Scholar] [PubMed]
41. Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid: Emergence of a core signaling network. Annual Review of Plant Biology, 61(1), 651–679. https://doi.org/10.1146/annurev-arplant-042809-112122 [Google Scholar] [PubMed] [CrossRef]
42. Li, X., Li, G., Li, Y., Kong, X., Zhang, L. et al. (2018). ABA receptor subfamily III enhances abscisic acid sensitivity and improves the drought tolerance of Arabidopsis. International Journal of Molecular Sciences, 19(7), 1938. https://doi.org/10.3390/ijms19071938 [Google Scholar] [PubMed] [CrossRef]
43. Saavedra, X., Modrego, A., Rodriguez, D., Gonzalez-Garcia, M. P., Sanz, L. et al. (2010). The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiology, 152(1), 133–150. https://doi.org/10.1104/pp.109.146381 [Google Scholar] [PubMed] [CrossRef]
44. Nee, G., Kramer, K., Nakabayashi, K., Yuan, B., Xiang, Y. et al. (2017). DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nature Communications, 8(1), 72. https://doi.org/10.1038/s41467-017-00113-6 [Google Scholar] [PubMed] [CrossRef]
45. Bhaskara, G. B., Nguyen, T. T., Verslues, P. E. (2012). Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiology, 160(1), 379–395. https://doi.org/10.1104/pp.112.202408 [Google Scholar] [PubMed] [CrossRef]
46. Ruggiero, A., Landi, S., Punzo, P., Possenti, M., van Oosten, M. J. et al. (2019). Salinity and ABA seed responses in pepper: Expression and interaction of ABA core signaling components. Frontiers in Plant Science, 10, 304. https://doi.org/10.3389/fpls.2019.00304 [Google Scholar] [PubMed] [CrossRef]
47. Wang, Y. G., Fu, F. L., Yu, H. Q., Hu, T., Zhang, Y. Y. et al. (2018). Interaction network of core ABA signaling components in maize. Plant Molecular Biology, 96(3), 245–263. https://doi.org/10.1007/s11103-017-0692-7 [Google Scholar] [PubMed] [CrossRef]
48. Nishimura, N., Sarkeshik, A., Nito, K., Park, S. Y., Wang, A. et al. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant Journal, 61(2), 290–299. https://doi.org/10.1111/j.1365-313X.2009.04054.x [Google Scholar] [PubMed] [CrossRef]
49. Liu, S., Lu, C., Jiang, G., Zhou, R., Chang, Y. et al. (2022). Comprehensive functional analysis of the PYL-PP2C-SnRK2s family in Bletilla striata reveals that BsPP2C22 and BsPP2C38 interact with BsPYLs and BsSnRK2s in response to multiple abiotic stresses. Front Plant Sciencce, 13, 963069. https://doi.org/10.3389/fpls.2022.963069 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools