 Open Access
Open Access
Analysis of Subcellular Localization and Pathogenicity of Plum Bark Necrosis Stem-Pitting Associated Virus Protein P6
1 College of Life Sciences, Zaozhuang University, Zaozhuang Cherry Virus Disease Rapid Diagnosis and Green Control Technology Innovation Center, Zaozhuang, 277160, China
2 College of Plant Protection, Shandong Agricultural University, Shandong Agricultural Microbiology Key Laboratory, Tai’an, 271018, China
3 Yicheng Agricultural and Rural Comprehensive Service Center, Zaozhuang, 277300, China
* Corresponding Authors: Xuefeng Yuan. Email: ; Deya Wang. Email:
# These authors have equal contribution on this manuscript
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(7), 2079-2085. https://doi.org/10.32604/phyton.2023.028237
Received 06 December 2022; Accepted 16 March 2023; Issue published 29 May 2023
Abstract
Infection of plum bark necrosis stem pitting associated virus (PBNSPaV) has been reported in many Prunus species in several countries, causing significant economic losses. The very small proteins encoded by plant viruses are often overlooked due to their short sequences and uncertain significance. However, numerous studies have indicated that they might play important roles in the pathogenesis of virus infection. The role of small hydrophobic protein P6, encoded by the open reading frame 2 of PBNSPaV, has not been well explored. In this study, we amplified the P6 fragment from a PBNSPaV isolate by RT-PCR using specific primers and found that it is 174 bp long and encodes a protein of approximately 6.3 kD with a transmembrane domain. Subcellular localization analysis of P6 proteins in tobacco leaves showed that P6 localizes to the cytomembrane and nuclear membrane. To further clarify the pathogenicity of P6 proteins, we constructed a PVX-P6 expression vector by inserting the p6 fragment into a potato virus X (PVX)-based vector and transformed it into Agrobacterium tumefaciens GV3101. Infiltration of Nicotiana benthamiana (N. benthamiana) with the PVX vector-transformed A. tumefaciens led to slight mosaic symptoms at 14 days of post-inoculation. Meanwhile, infiltration with the PVX-P6 vector-transformed A. tumefaciens resulted in no significant symptoms. These results demonstrated that heterologous expression of P6 in N. benthamiana could not enhance the pathogenicity of PVX. Our study indicates that P6 may not be a potential pathogenic factor associate with the causing of symptoms, and the mode of action of PBNSPaV-P6 protein remains to be further studied.Keywords
The stem-pitting disease was first discovered in the 1960s in sweet cherry trees in North America. It has also been found in many Prunus species in many countries, including China [1–4]. The disease is mainly caused by plum bark necrosis stem pitting-associated virus (PBNSPaV), which was first reported to infect Japanese Plum cv. ‘Black beaut’ in 1986 [5,6]. Once infected, the trunk will secretes black gelatinous substances, the bark tissues become severely necrotic, and stem pox symptoms will appear in the xylems, resulting in weak tree vigor, flat branches, and other symptoms that seriously endangers plant growth [5]. China is one of the world’s largest Prunus planting and producing countries [7,8]. PBNSPaV infection was first reported in China in 2011, causing significant economic losses [2,9,10]. However, studies on PBNSPaV are still lacking.
PBNSPaV is a non-enveloped, linear, single-stranded RNA virus belonging to the genus Ampelovirus of the family Closteroviridae [5]. The complete PBNSPaV genome comprises seven major open reading frames (ORFs) with a total length of approximately 14 kb. Of the seven ORFs, ORF1a encodes a polyprotein, ORF3 encodes the heat shock protein (HSP) 70, ORF4 encodes a protein of about 61.6 kD, ORF5 and ORF6 encode coat proteins, and ORF2 encodes a 6.3-kD protein located between RdRp and HSP70 with unknown functions [5,6]. Although the encoded proteins have been reported to be primarily involved in viral replication, motility, and mediator transmission [5,11], the functions of some PBNSPaV-encoded proteins, including P6, are yet to be determined. Especially, very small ORFs/proteins are often overlooked due to their short sequences and uncertain significance. Therefore, it is imperative to identify the coding regions and understand their functions to decipher how these small ORFs/proteins contribute to the infection cycle. To analyze the functions of P6 protein, the subcellular localization and pathogenic characteristics of P6 were studied by confocal microscopy and phenotypic observation, respectively. Our results laid a theoretical foundation for further analysis of P6’s functions and mechanism in the interaction between PBNSPaV and its hosts.
2.1 Plant Materials and Bacterial Strain
Sweet cherry leaves infected by PBNSPaV were collected from Shandong Province in 2021. Fluorescent expression vector 35S:GFP, Agrobacterium GV3101, Nicotiana benthamiana (N. benthamiana) seeds, and other plant materials were stored at our laboratory. The PVX vector (pGR107) was a gift from Professor Xiaofei Cheng at Northeast Agricultural University [12].
2.2 P6 Gene Cloning and Sequence Analysis
Total RNAs were extracted from 100 milligrams of cherry leaves using TRIzol reagent following the manufacturer’s instructions and stored at −80°C. Primers used to amplify the P6 gene were designed based on the PBNSPaV sequence (GenBank: MZ221026) to amplify the P6 gene. cDNAs were synthesized by first incubating primers, dNTPs, and total RNAs at 80°C for 5 min, then with PrimeScript reverse transcriptase in 1 × buffer (Takara Inc., Japan) at 42°C for 1.5 h, and subjected to PCR for P6 gene amplification. The PCR product was cloned into the pMD18-T vector and transformed into DH5α competent cells. Positive clones were sequenced at Boshang Biotech Company (Jinan, China) for validation. The sequencing results were aligned to the PBNSPaV genome sequences on the NCBI database. Multiple alignments and homology analyses were performed using ClustalX and BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/), and the transmembrane prediction was performed using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/).
2.3 Subcellular Localization of P6
A P6-GFP plasmid was constructed by cloning the P6 gene into the 35S:GFP vector via BamHI and sequencing validated by Boshang Biotech Company (Jinan, China). The P6-GFP and GFP plasmids were transformed into Agrobacterium tumefaciens GV3101 using the CaCl2-mediated freeze-thaw method, as described previously. Single A. tumefaciens colonies were cultured in LB medium with 50 µg/mL kanamycin and 100 µg/mL rifampin at 28°C until OD600 reached 1.0~2.0. After centrifugation, the pellets were resuspended in the infiltration solution containing 10 mM MgCl2, 10 mM 2-(N-morpholino) and 0.15 mM acetosyringone to OD600 of 1.2 and resuscitated at 28°C for 2 h. N. benthamiana at 5–7 leaf age was selected and treated on the back of leaves with resuscitated A. tumefaciens for 40–48 h using a 1 mL needless syringe. The infiltrated leaves were prepared as slides and observed under a laser confocal microscope (Zeiss, Germany) to check P6-GFP subcellular localization. The treatment was repeated by at least three times.
2.4 Pathogenicity Analysis of P6
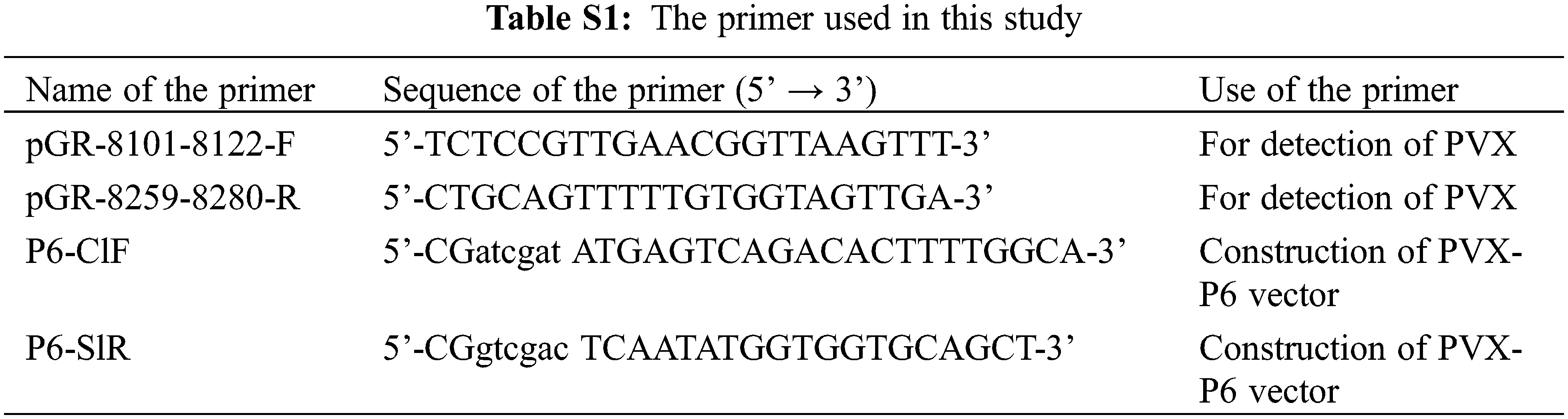
The P6 gene fragment was cloned via ligation using T4 ligase into the PVX expression vector using T4 ligase after the vector was digested with Sal I and Cla I. The resulting PVX heterologous expression vector and recombinant plasmid PVX-P6 and PVX heterologous expression vector were transformed into Agrobacterium GV3101 by the freeze-thaw method and cultured on plates containing 50 mg/L kanamycin and 25 mg/L rifampicin at 28°C. Single colonies were selected for colony PCR identification. After identification, they were propagated overnight in liquid medium containing kanamycin and rifampicin until the OD600 reached 0.6–0.8. The cultures were then used to infiltrated into 5–7-leaf age N. benthamiana. After 7 and 14 days of infiltration, samples were collected, and their symptoms were recorded. Total RNAs were extracted from the upper new leaves (non-invasive leaves) inoculated with PVX and PVX-P6 which were fully ground after liquid nitrogen freezing and used to extract total RNAs. P6 and PVX expression were detected by RT-PCR using primer sets P6-ClF and P6-SlR and primer set PVX-F and PVX-R, respectively.
3.1 Analysis of the Properties of P6 Protein
The obtained P6 gene is 174 bp long, encoding a protein of about 6.3 kD. BLAST analysis with 24 reference isolates in GenBank showed that the P6 protein is highly homologous to other reference proteins with only seven mutations (Fig. 1A). This indicates that P6 genes of PBNSPaV isolates are highly conservative in phylogeny. The P6 protein is a stable hydrophobic protein consisting of 57 amino acid residues and lacks a signal peptide. It has an isoelectric point (pI) of 7.81, and transmembrane prediction indicated that the P6 is a transmembrane protein with transmembrane region composed of amino acid residues located at position 15 to 37 (Fig. 1B). Structural analysis of the Swiss model showed that the P6 protein mainly consists of α-helix (Fig. 1C).
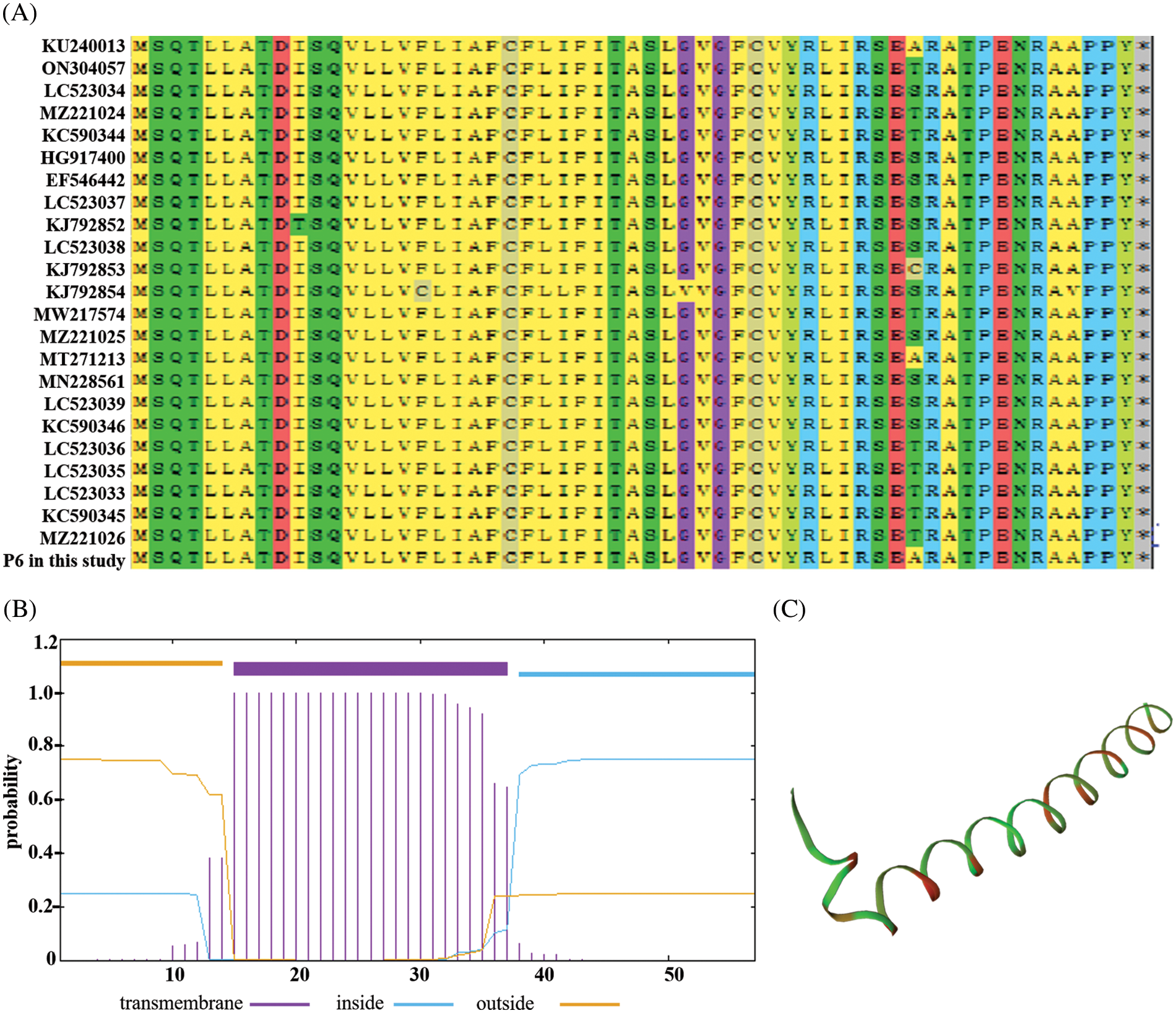
Figure 1: Analysis of the properties of P6 protein. (A) Amino acid sequence homology analysis of P6; (B) Prediction of the transmembrane region in the P6 protein; (C) Spatial structure analysis of the P6 protein
3.2 p6 May Not Be a Potential Pathogenic Factor Associate with the Causing of Symptom
To determine P6 protein’s localization in plant cells, the P6 sequence was fused in-frame to the 5’ end of the GFP gene behind a 35S promoter to create p35S-P6:GFP vector. The expression vectors of GFP, P6-GFP, and CBL1-RFP (a membrane localization protein) were transformed into Agrobacterium GV3101 and used to infiltrate N. benthamiana. Confocal microscopic observation at 48 h of post-infiltration revealed green fluorescence signals in the cytomembrane and nuclear membrane of the epidermal cells of N. benthamiana infiltrated with P6-GFP and overlapped green and red fluorescence signals excited by P6-GFP and CBL1, respectively. These observations indicated that P6 proteins are presented in both cytomembrane and nuclear membrane (Fig. 2A). To further study the pathogenicity of p6 proteins, expression vectors PVX and PVX-P6 were transformed into Agrobacterium GV3101, which were then inoculated into N. benthamiana, respectively. After inoculation with PVX-P6 for 14 days, the upper leaves of N. benthamiana showed no significant changes, while the upper leaves of N. benthamiana inoculated with PVX only showed slight mosaic symptoms (Fig. 2B). RT-PCR analysis of viral gene expression (primers in Table S1) showed that PVX was detected in all N. benthamiana inoculated with PVX and PVX-P6, while P6 was only detected in N. benthamiana inoculated with PVX-P6 (Fig. 2C). These results suggested that heterologous expression of P6 in N. benthamiana could not enhanced the pathogenicity of PVX.
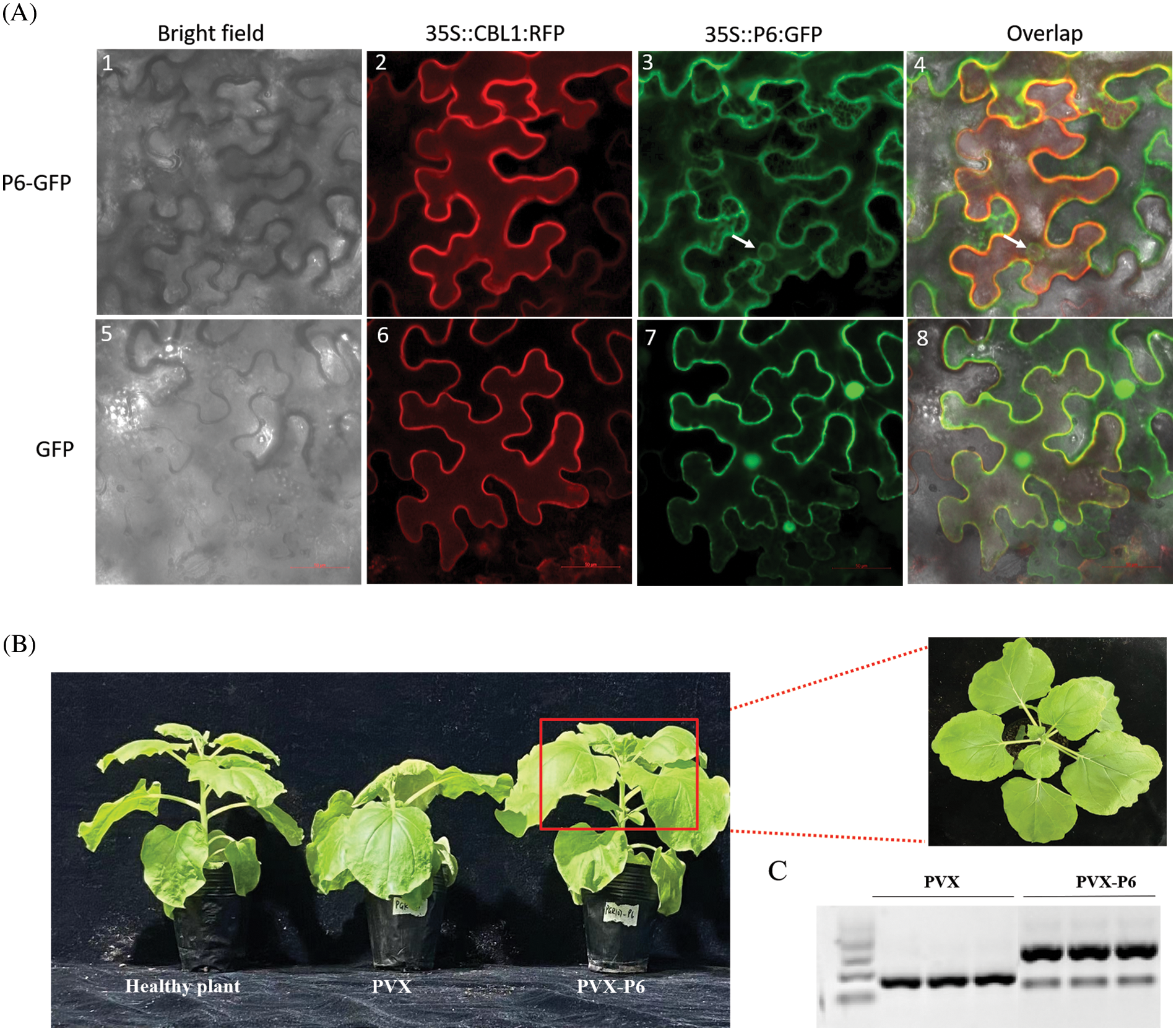
Figure 2: Subcellular localization and pathogenicity characteristics of P6 proteins in plant cells. (A) Subcellular localization of P6 proteins in plant cells. White arrows indicate the nuclear membrane. Bars, 50 μm; (B) Symptom analysis of infiltration of N. benthamiana with the PVX and PVX-P6, respectively. (C) RT-PCR analysis of viral gene expression
PBNSPaV belongs to the family Closteroviridae and possesses the largest and most complex genomes (up to 20 kb) of all positive-sense single-stranded RNA plant-infecting viruses. PBNSPaV is distributed worldwide, affecting a broad range of Prunus species [9,11]. However, prevention and treatment of PBNSPaV infection are not well explored due to the lack of theoretical and practical studies. Therefore, analyzing the biological functions of viral proteins would contribute to understanding PBNSPaV infection mechanisms, the interactions between the virus and its plant hosts, and the prevention of viral diseases. In this study, we cloned the P6 gene of PBNSPaV from a sweet cherry isolate in Shandong province and found that it is 174 bp long and encodes a protein of approximately 6.3 kD with a transmembrane domain. All P6 proteins have comparatively high similarity in amino acid sequences, and P6 genes of all PBNSPaV isolates are highly conservative in phylogenetic development (Fig. 1A). In addition to PBNSPaV, other members of the family Closteroviridae also encode small molecular weight proteins (P3.5, P6, and P8) [13–17]. However, these proteins are different from the P6 protein in this study in coding position and homology with P6 in this study. In contrast to PBNSPaV-P6, the subcellular localization of other small proteins encoded by viruses in the family Closteroviridae might be different. For example, the P6 protein encoded by Cucurbit chlorotic yellows virus (CCYV) is distributed in the cytoplasm and nucleus [13], and P5 protein encoded by Lettuce infectious yellows virus (LIYV) is an endoplasmic reticulum (ER) localized integral membrane protein capable of inducing ER stress [17].
Furthermore, several proteins of the same family could also induce necrosis in host plants and play important roles in viral infection, spread, and pathogenicity. PVX vector-mediated overexpression of P22, a viral gene silencing suppressor that plays an essential role during virus infection, causes plant necrosis [18]. P5 and P9 proteins encoded by LIYV are produced in LIYV-infected plants and are essential for efficient viral infectivity [17]. However, our study indicates that p6 may not be a potential pathogenic factor associated with the causing of symptoms. These findings suggest that these small proteins are similar in certain aspects, but they may function differently during virus infection, and the specific function and mode of action of PBNSPaV-P6 protein remain to be further studied.
Acknowledgement: PVX vector (pGR107) was a gift from Professor Xiaofei Cheng in Northeast Agricultural University. The authors would like to thank the funding from the National Natural Science Foundation of China and Shandong Province Natural Sciences Foundation of China and TopEdit (www.topeditsci.com) for linguistic assistance during the preparation of this manuscript.
Funding Statement: This study was funded by the National Natural Science Foundation of China (32102143), Shandong Province Natural Sciences Foundation of China (ZR2019PC011 and ZR2020QC122), Scientific Research Foundation for Ph.D. Programs of Zaozhuang University (2018BS040 and 2018BS042), and Science and Technology Program of Zaozhuang (2019NS03).
Author Contributions: DW and XY designed the experiment, performed analyses, and drafted the manuscript; DW and XY contributed to data acquisition and analysis; YL and JM contributed to subcellular localization and pathogenicity of Plum bark necrosis stem-pitting associated virus protein P6; HL contributed to collection of samples; DW and QL substantively revised the paper. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Cui, H. G., Hong, N., Xu, W. X., Zhou, J. F., Wang, G. P. (2011). First report of Plum bark necrosis stem pitting-associated virus in stone fruit trees in China. Plant Disease, 95(11), 1483. [Google Scholar] [PubMed]
2. Villamor, D. E., Mekuria, T. A., Pillai, S. S., Eastwell, K. C. (2016). High-throughput sequencing identifies novel viruses in nectarine: Insights to the etiology of stem-pitting disease. Phytopathology, 106(5), 519–527. [Google Scholar] [PubMed]
3. Wang, J., Zhai, Y., Liu, W., Zhu, D., Pappu, H. R. et al. (2016). Complete genomic characterization of Plum bark necrosis stem pitting-associated virus infecting sweet cherry in China. Genome Announcements, 4(3), e00413–e00416. [Google Scholar] [PubMed]
4. Yang, Y., Sun, Y., Li, Q., Wu, Y., Wang, D. (2020). First report of Plum bark necrosis stem pitting-associated virus infecting grapevine in China. Virology Journal, 17(1), 181. [Google Scholar] [PubMed]
5. Boscia, D., Myrta, A., Uyemoto, J. K. (2010). Plum bark necrosis stem pitting associated virus. In: Virus and virus-like diseases of pome and stone fruits, pp. 177–183. St. Paul, MN, USA: APS Press. [Google Scholar]
6. Amenduni, T., Hobeika, C., Minafra, A., Savino, V. (2003). Detection of Plum bark necrosis stem pitting-associated virus (PBNSPaV) from different stone fruit species and optimisation of diagnostic tools. Acta Horticulturae, XIX International Symposium on Virus and Virus-Like Diseases of Temperate Fruit Crops-Fruit Tree Diseases, vol. 657, pp. 93–97. [Google Scholar]
7. Meng, X., Rao, Y., Tao, T., Dong, S., Jia, A. L. et al. (2021). A review of plant breeders’ rights application and granting for fruit trees in China from 2000 to 2019. Scientia Horticulturae, 276, 109749. [Google Scholar]
8. Fan, Y., Cheng, Z., Zhang, Q., Xiong, Y., Li, B. et al. (2022). Meizi-consuming culture that fostered the sustainable use of plum resources in Dali of China: An Ethnobotanical Study. Biology, 11(6), 832. [Google Scholar] [PubMed]
9. Guo, M., Zhong, J., Wang, C., Chen, Y., Yang, Z. et al. (2022). First report of Plum bark necrosis stem pitting associated virus affecting sweet cherry in Yunnan, China. Journal of Plant Pathology, 104, 829–830. [Google Scholar]
10. Zhang, Y., Zhou, J., Zhan, B., Li, S., Zhang, Z. (2021). First report of Peach leaf pitting-associated virus (PLPaVplum bark necrosis stem pitting-associated virus (PBNSPaVand Mume virus A (MuVA) from Mei (Prunus mume) in China. Plant Disease, 105(8), 2259. [Google Scholar]
11. Maliogka, V. I., Minafra, A., Saldarelli, P., Ruiz-García, A. B., Glasa, M. et al. (2018). Recent advances on detection and characterization of fruit tree vruses using high-throughput sequencing technologies. Viruses, 10(8), 436. [Google Scholar] [PubMed]
12. Liu, J., Liu, Y., Fang, Y., Zhang, L., Yu, K. et al. (2021). Evaluation of potato virus X resistance in potato cultivars and identification of an innate immunity-independent resistance phenotype. Phytopathology Research, 3(1), 1–12. [Google Scholar]
13. Tu, L., Gan, S., Wu, S., Ren, C., Cheng, Z. et al. (2021). Characterization of the subcellular localization and pathogenicity of P6 encoded by Cucurbit chlorotic yellows virus. Acta Horticulturae Sinica, 48(8), 1531–1540. [Google Scholar]
14. Tahzima, R., Foucart, Y., Peusens, G., Beliën, T., Massart, S. et al. (2019). High-throughput sequencing assists studies in genomic variability and epidemiology of Little cherry virus 1 and 2 infecting Prunus spp. in Belgium. Viruses, 11(7), 592. [Google Scholar] [PubMed]
15. Ruiz-García, A. B., Candresse, T., Canales, C., Morán, F., Machado de Oliveira, C. et al. (2020). Molecular characterization of the complete coding sequence of Olive leaf yellowing-associated virus. Plants, 9(10), 1272. [Google Scholar]
16. Ruiz, L., Simón, A., García, C., Velasco, L., Janssen, D. (2018). First natural crossover recombination between two distinct species of the family Closteroviridae leads to the emergence of a new disease. PLoS One, 13(9), e0198228. [Google Scholar] [PubMed]
17. Qiao, W., Helpio, E. L., Falk, B. W. (2018). Two crinivirus-conserved small proteins, P5 and P9, are indispensable for efficient Lettuce infectious yellows virus infectivity in plants. Viruses, 10(9), 459. [Google Scholar] [PubMed]
18. Landeo-Ríos, Y., Navas-Castillo, J., Moriones, E., Cañizares, M. C. (2017). The heterologous expression of the p22 RNA silencing suppressor of the Crinivirus tomato chlorosis virus from Tobacco rattle virus and Potato virus X enhances disease severity but does not complement suppressor-defective mutant viruses. Viruses, 9(12), 358. [Google Scholar]
Supplementary Materials
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools