 Open Access
Open Access
ARTICLE
Rice E3 Ligase-Like Protein OsPIAL1 Positively Regulated the Drought Stress Response but Negatively Regulated the Salt Stress Response
Department of Bioscience and Bioinformatics, Myongji University, Yongin, 17058, Korea
* Corresponding Author: Sang Ik Song. Email:
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(7), 2017-2034. https://doi.org/10.32604/phyton.2023.027469
Received 31 October 2022; Accepted 06 March 2023; Issue published 29 May 2023
Abstract
Small ubiquitin-like modifier (SUMO) E3 ligases that facilitate the conjugation of SUMO proteins to target substrates contain an SP-RING domain which is like the RING domain found in ubiquitin E3 ligases. In this study, we isolated and characterized the Oryza sativa protein inhibitor of activated STAT like1 (OsPIAL1) containing SP-RING domains, as the rice homolog of Arabidopsis PIALs. OsPIAL1 interacts with OsSUMO proteins but does not interact with rice SUMO-conjugating enzymes (OsSCEs). An analysis of transgenic rice plant shows that OsPIAL1 is involved in SUMO conjugation to SCEs but not in SUMO conjugation to substrates. In addition, this OsPIAL1 activity requires drought stress conditions. Expression profiles show that the OsPIAL1 gene is induced by only drought stress in the leaves, whereas it is repressed by ABA and abiotic stresses in the roots. Salt stress leads to the fastest decrease in OsPIAL1 transcripts in the roots. Furthermore, the stress experiments indicate that the transgenic rice plants overexpressing OsPIAL1 exhibit a drought stress-tolerant phenotype but a salt stress hypersensitive phenotype. Our results and those from Arabidopsis pial mutants suggest that PIALs act as a positive regulator in the drought stress response but as a negative regulator in the salt stress response.Keywords
Nomenclature
| ABA | Abscisic acid |
| PIAL1 | Protein inhibitor of activated STAT like1 |
| qRT-PCR | Quantitative real-time PCR |
| SP-RING | (SIZ-PIAS)-RING |
| SUMO | Small ubiquitin-like modifier |
| SCE | SUMO-conjugating enzyme |
Protein posttranslational modifications by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins, such as small ubiquitin-like modifiers (SUMO), are involved in many biological processes through changes in biological activities, subcellular localization, and target protein stability [1–4]. SUMO conjugation (sumoylation) plays important roles in abiotic stresses, ABA signaling, flowering time, cell growth and development, and nitrogen assimilation in plants [5–13]. The sumoylation carries out through a three-step enzymatic process, including a SUMO-activating enzyme (E1, SAE1/SAE2), SUMO-conjugating enzyme (E2, SCE), and SUMO ligase (E3), as in ubiquitination [3,14]. This process is well characterized in yeast and animal cells.
In plants, the sumoylation enzymes have been studied in mostly Arabidopsis [15]. Results in the monocot plant rice were recently published [16–18]. SUMO links to the E1/SAE with thioester linkage. Next, the sumo is moved to the E2/SCE. The SUMO is conjugated to substrate with an isopeptide linkage by E2/SCE and E3 [14]. Most of SUMO substrate proteins contain sumoylation sites, Ψ-K-X-(D/E) (Ψ, a hydrophobic amino acid; X, any amino acid) [19]. It has been reported that the E2/SCE interacts with sumoylation sites and they play an important role in the stabilization of interactions [20]. The E2/SCE shows specific substrate recognition capabilities without E3/SUMO ligase [21]. Despite this ability, E3/SUMO ligases are key factors in SUMO substrate specificity [22].
Human protein inhibitor of activated STAT (PIAS) proteins and their yeast homologs (SIZ) were identified as SUMO E3 ligases. In plants, functionally identified SUMO E3 ligases are SIZ1, SIZ2 and HPY2/MMS21 [10,11,23]. These SUMO E3 ligases contain an SP(SIZ-PIAS)-RING (also called a zf-MIZ) domain, which is like the RING domain found in ubiquitin E3 ligases. Thus, the function of the SP-RING domain is proposed to recruit SUMO E2 into a complex with a substrate to facilitate conjugation [22]. The rice genome encodes four SP-RING domain-containing proteins, two SIZ1 types (OsSIZ1 and OsSIZ2), one HPY2/MMS21 type (Os05g48880), and one PIAS-like type (Os06g06870) [15]. OsSIZ1 and OsSIZ2 have been functionally characterized and are involved in stress responses and spikelet fertility [16,17]. The HPY2/MMS21 and PIAS-like genes have not yet been studied. Recently, Arabidopsis PIAL1 and PIAL2 proteins have been shown to contain an SP-RING domain, and they showed SUMO ligase activity forming the SUMO chain in vitro [24]. In this study, we characterized the rice protein inhibitor of activated STAT like1 (OsPIAL1). We showed that OsPIAL1 interacts with OsSUMO proteins. The expression patterns of OsPIAL1 gene were also analyzed. To know its function, the transgenic rice plants overexpressing OsPIAL1 gene were made and analyzed. Our results suggested that OsPIAL1 is involved in the drought stress response in rice.
2.1 The Identification of the Rice PIAL1 Gene
The rice gene encoding the OsPIAL1 protein was identified using BLAST searches of the Oryza sativa DNA sequences in the National Center for Biotechnology Information database with the amino acid sequences of rice OsSIZ1 and OsSIZ2 as a query. Clone Os06g0164000 was selected from among the matches. The cDNA clone was obtained by RT-PCR using the primer pairs PIAL1-F and PIAL1-R. Supplementary Table S1 listed the primers used in this study.
2.2 Two-Hybrid Assays and Transactivation in Yeast
The transactivation assay was carried out using the vector pAD-GAL4-2.1 (Stratagene) and yeast strain YRG2. The full-length OsPIAL1 and OsSIZ2 were inserted into the yeast GAL4 DNA-binding domain in frame. Transformants were plated onto minimal medium (–Leu/–His/+10 mM or 25 mM 3-Amino-1,2,4-triazole, 3AT) for the transactivation assay. The GAL4-based two-hybrid system (Stratagene) was used in the yeast two-hybrid assay. To construct the bait and prey vector, the coding region OsSCEs or OsSUMOs gene was inserted into the pBD-GAL4 Cam vector and OsPIAL1 gene was inserted into pAD-GAL4-2.1 vector, respectively. To confirm the interaction, minimal medium (–Leu/–Trp/–His/+3 mM 3AT) was used in selection. Supplementary Table S1 listed the primers used in the construction.
2.3 Quantitative Real-Time PCR Analysis
TRI Reagent® (Molecular Research Center, Cincinnati, OH, USA) was used to isolate total RNA according to the manufacturer’s descriptions. RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Burlington, Canada) was used to first strand cDNA synthesis with 5 μg of total RNA and oligo (dT)18 primers according to the manufacturer’s descriptions. The 2X Real-time PCR Pre-Mix with Evagreen (SolGent, Daejeon, Korea) and an Mx3000p Real-time PCR machine (Stratagene, La Jolla, CA, USA) were used to perform quantitative real-time PCR (qRT-PCR). Triplicate was used in each sample. Three repetitions were carried out in all experiments. The expression data normalization was performed using OsUbi1 gene (LOC4341860). Supplementary Table S1 listed the primers used in the qRT-PCR analyses.
2.4 Plant Materials and Growth Conditions
Oryza sativa subsp. japonica cv. Nakdong background were used in transgenic and wild-type (WT) rice plants. The husked and sterilized seeds were germinated in a growth chamber, and then the germinated seedlings were transplanted into soil pot as previously described [25]. The seedlings were grown in the greenhouse (25°C–28°C with 16 h of light and 8 h of darkness). The rice organ samples were obtained as previously described [26]. The leaf, internode, and root samples were prepared from 3-week-old plants, and the pre- and post-heading panicles from field-grown mature rice plants.
The overexpression vector construction was carried out as previously described [26]. The bar gene and the rice cytochrome c promoter were used as a selection marker gene and to drive constitutive overexpression of transgene, respectively. The OsPIAL1 gene was obtained using PCR. Supplementary Table S1 listed the primers used in the construction. The final constructed plasmid was confirmed by DNA sequencing. The triparental mating transformation of Agrobacterium tumefaciens LBA4404 and Agrobacterium-mediated rice transformation was performed as described previously [26].
2.6 Abiotic Stress, Sugar, and Hormone Treatments
The abiotic stress, sugar, and hormone treatment conditions and methods followed the previously publish [26]. Three days water adapted 3-week-old WT seedlings were air-dried or transferred to solution containing 100 mM glucose, 100 mM sucrose, 100 μM gibberellin, 100 μM ABA, or 250 mM NaCl. The Sampling time is indicated. To evaluate the drought and high-salinity tolerance, each transgenic line and WT seedling was grown in 3 cm × 3 cm × 5 cm pot for 4 weeks. To test for drought tolerance, after not supplying water for 2 days, they were observed while supplying water again for 10 days in a greenhouse [26]. To test for high-salinity tolerance, after 4 days of 250 mM NaCl solution treatment, they were observed while supplying fresh water again for 34 days in a greenhouse [26].
2.7 Immunoblotting and Protein Detection
For the analysis of SUMO conjugates, total protein was extracted from the roots of 14-day-old seedlings grown under drought stress conditions. Plant tissues (0.2 g) were extracted in grinding buffer [100 mm Na-MOPS, pH 7.5, 10 mm NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), pH 8.0, 10% sucrose, 5% b-mercaptoethanol, 4% sodium dodecyl sulfate (SDS), 2 mM PMSF, and a proteinase inhibitor cocktail (Roche)] using a mortar and pestle at room temperature. The Bio-Rad Protein Assay Kit (Bio-Rad) was used to measure the protein concentration. The 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the proteins. Separated proteins were transferred onto a polyvinylidene fluoride membrane (Millipore, Burlington, MA, USA). The anti-SUMO1 antibody (Abcam, Ltd., Cambridge, UK) and ECL Western blot detection system (Amersham Biosciences, Buckinghamshire, UK) were used in probing and detection, respectively.
Each experiment was carried out in triplicate. The displayed data are the means ± SD. Intergroup comparisons between the two tested groups were carried out using a paired t-test with Microsoft Office Excel ver. 2007 (Microsoft Corporation, Richmond, WA, USA). A p-value of <0.05 was considered statistically significant.
3.1 Rice Encodes One PIAS-Like Protein
To identify rice SUMO E3 ligase genes, we performed BLAST searches against the National Center for Biotechnology Information database using OsSIZ1 (Os05g0125000) and OsSIZ2 (Os03g0719100) as queries. Using TBLASTN, the rice Os06g0164000 gene was identified to have the highest sequence identity to OsSIZ1 and OsSIZ2 around the SP-RING domain (also called a zf-MIZ domain), which is required for SUMO E3 ligase activity (Fig. 1A and Supplementary Fig. S1). PIAS and PIAS-like proteins share an SP-RING domain in the central region [27]. They can enhance sumoylation [28,29]. However, Os06g0164000 does not contain other domains that are conserved among SIZ/PIAS-type SUMO E3 ligases, such as a plant homeodomain-finger (PHD) domain involved in nuclear retention and an SAP (scaffold attachment factor A/B/acinus/PIAS) domain involved in DNA binding [30,31] (Fig. 1B).
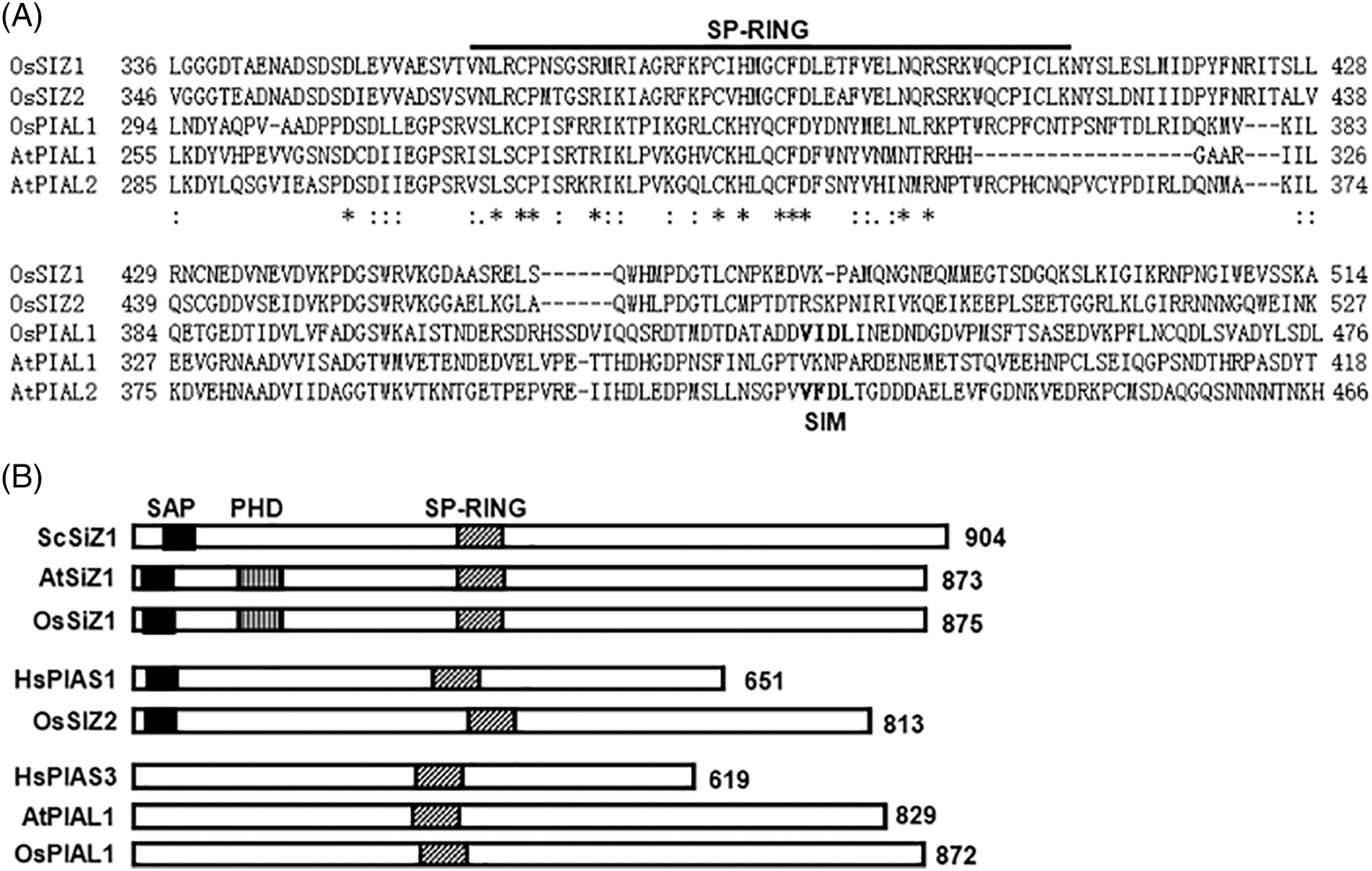
Figure 1: The structure of the OsPIAL1 gene. (A) Sequence alignment of the OsSIZs and PIALs. The SP-RING domain was identified in Pfam, and it is underlined in above the sequence. Putative SUMO-interacting motifs (SIM) are indicated. Residue numbers are shown. The conserved amino acids were indicated asterisks. The dashes denote gaps. At, Arabidopsis thaliana; Hs, Homo sapiens; Os, Oryza sativa; Sc, Saccharomyces cerevisiae. (B) The gene structure of OsPIAL1. The SP-RING, plant homeodomain-finger (PHD) and scaffold attachment factor A/B/acinus/PIAS (SAP) domains are indicated. The total amino acid residues of each protein are indicated
Recently, Arabidopsis PIAL1 and PIAL2 genes have been reported [15,24]. The Os06g0164000 (OsPIAL1) shows high amino acid sequence identity with Arabidopsis PIAL1 and PIAL2 (Supplementary Fig. S2). To infer the functions, the conserved amino acid motif sequences were scanned in OsPIAL1. It contains an SP-RING domain (318–367), two SUMO interaction motifs (SIM) (amino acids 158–161 and 435–438), and three putative sumoylation sites (amino acids 24, 174, and 214).
3.2 OsPIAL1 Has SUMO Binding Activity but No Transactivation Activity
The transactivation activity of OsPIAL1 was analyzed. The OsPIAL1 showed no transactivation activity, while OsSIZ2 showed transactivation activity in yeast (Fig. 2A). Arabidopsis PIAL1 and PIAL2 exhibit SUMO ligase activity in SUMO chain formation. The SUMO-modified E2/SCE1 is required for their optimal activity [24]. The interaction between OsPIAL1 and SUMO was investigated in yeast (Fig. 2B). OsPIAL1 interacted with OsSUMO3. It also weakly interacted with OsSUMO1. Next, we examined the interaction between OsPIAL1 and rice SCEs. They showed no interaction. The results suggested that OsPIAL1 interacts with only OsSUMOs.
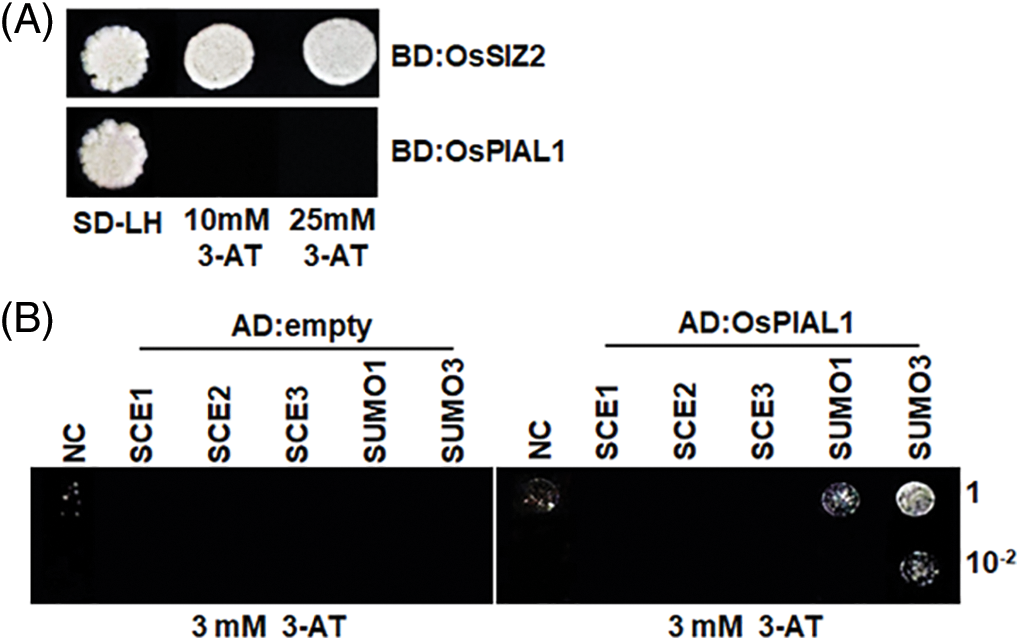
Figure 2: Transactivation activity and interaction assays for OsPIAL1. (A) Transactivation activity assay for OsPIAL1 and OsSIZ2. The transactivation activity of BD: OsSIZ2 and BD: OsPIAL1 fusion proteins were analyzed in the yeast strain YRG2. (B) Yeast two-hybrid assays for OsPIAL1 with OsSCEs or OsSUMOs
3.3 Response of OsPIAL1 Expression Levels under Abiotic Stress
To determine whether the OsPIAL1 gene shows tissue-specific expression, qRT-PCR analysis was conducted in various rice tissues (Fig. 3A). Transcripts of OsPIAL were detected in all tested tissues. The OsPIAL1 gene was expressed primarily in the panicles and had relatively low expression levels in the leaves. To evaluate the relationship between the expression of the OsPIAL1 gene and abiotic stress in rice, the transcript levels of this gene in compliance with ABA, drought, high salinity, and H2O2 treatments were checked (Fig. 3B). Interestingly, OsPIAL1 transcripts were increased by only drought. In response to drought, the OsPIAL1 transcripts showed a 10-fold increase within 2 h. There was no significant difference in OsPIAL1 transcripts in response to exogenous GA and glucose treatments (Fig. 3C). These results suggest that only drought positively regulates the expression of the OsPIAL1 gene in the leaves. In contrast, OsPIAL1 transcripts were decreased in the roots under ABA and abiotic stresses (Fig. 3D). Interestingly, the 250 mM NaCl treatments induced the fastest decrease in OsPIAL1 transcripts in the roots.
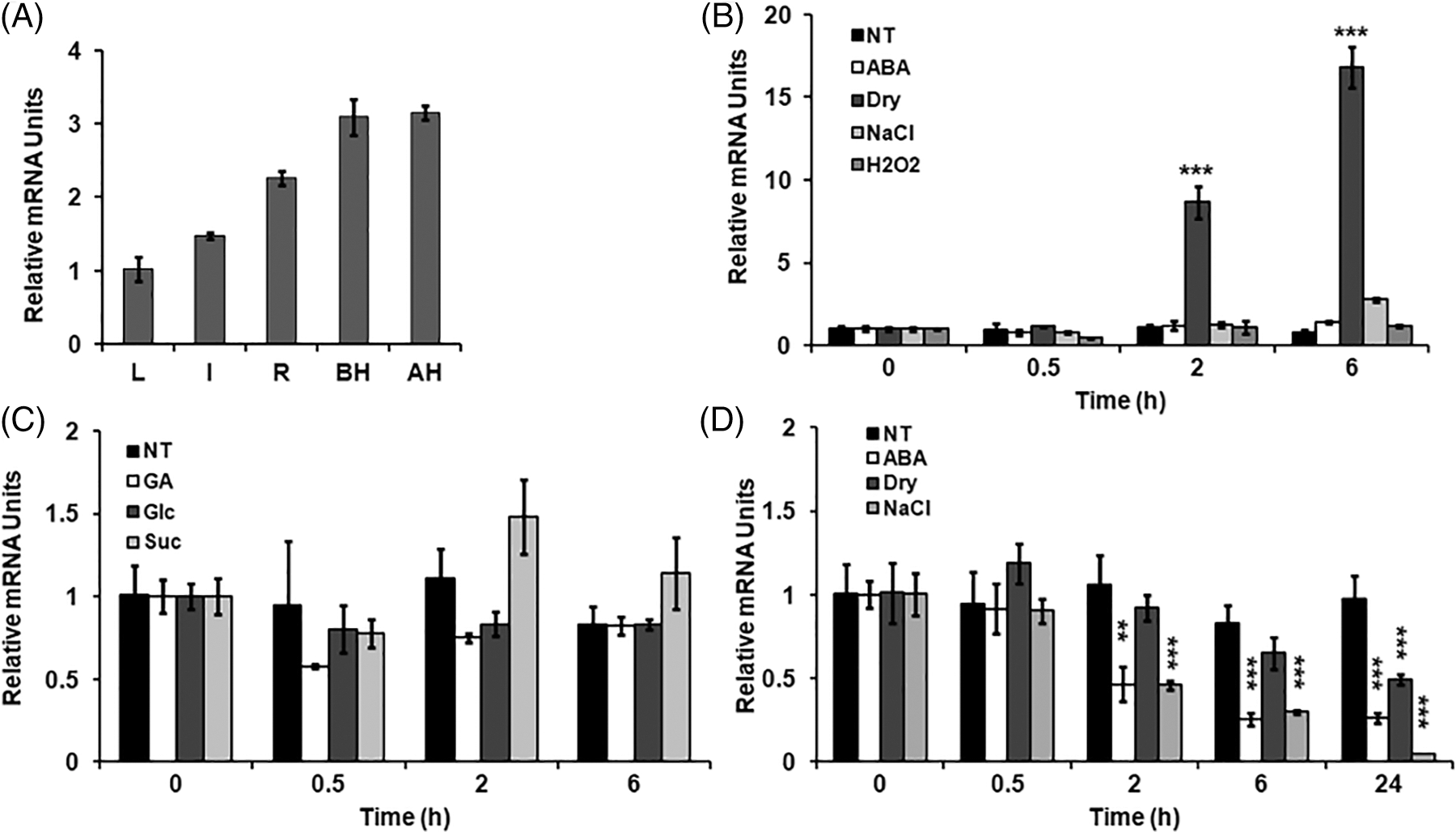
Figure 3: Expression analysis of the OsPIAL1 gene. (A) Expression analysis of the OsPIAL1 gene was determined using qRT-PCR in several organs. leaves (L), roots (R), and internode (I) were prepared from 14-day-old plants. The pre-pollinated spikelets (BH) and post-pollinated spikelets (AH) were collected from mature plants. (B) Transcript analysis of the OsPIAL1 gene in ABA, drought (dry), high salinity (NaCl), or H2O2 treatments in the leaves. (C) The relative transcript levels of the OsPIAL1 gene were determined in response to glucose (Glc), sucrose (Suc), or gibberellin (GA) in the leaves. (D) The relative transcript levels of the OsPIAL1 gene were determined in response to ABA, drought, or high-salinity treatments in the roots. The transcript levels of rice OsUbi1 gene were used to normalize. The data show the means ± SE from biological repeat two experiments. Statistically significant differences are indicated by asterisks, as calculated using Student’s t-test. **, p < 0.005; ***, p < 0.001
3.4 The Overexpression of the OsPIAL1 Gene Improved Drought Stress Tolerance
To know the functions of OsPIAL1 in rice, transgenic rice plants overexpressing OsPIAL1 (OsPIAL1-OX) were constructed (Fig. 4A). The rice cytochrome c promoter was used for constitutive overexpression of the OsPIAL1 gene [25]. The overexpression of the OsPIAL1 gene in OsPIAL1-OX lines was checked (Fig. 4B). The OsPIAL1-OX lines exhibited higher expression levels of OsPIAL1 gene than WT plants. Lines 2, 3 and 9 showed higher expression of the OsPIAL1 gene. The homozygous T4 seeds of these three lines were used in this study.
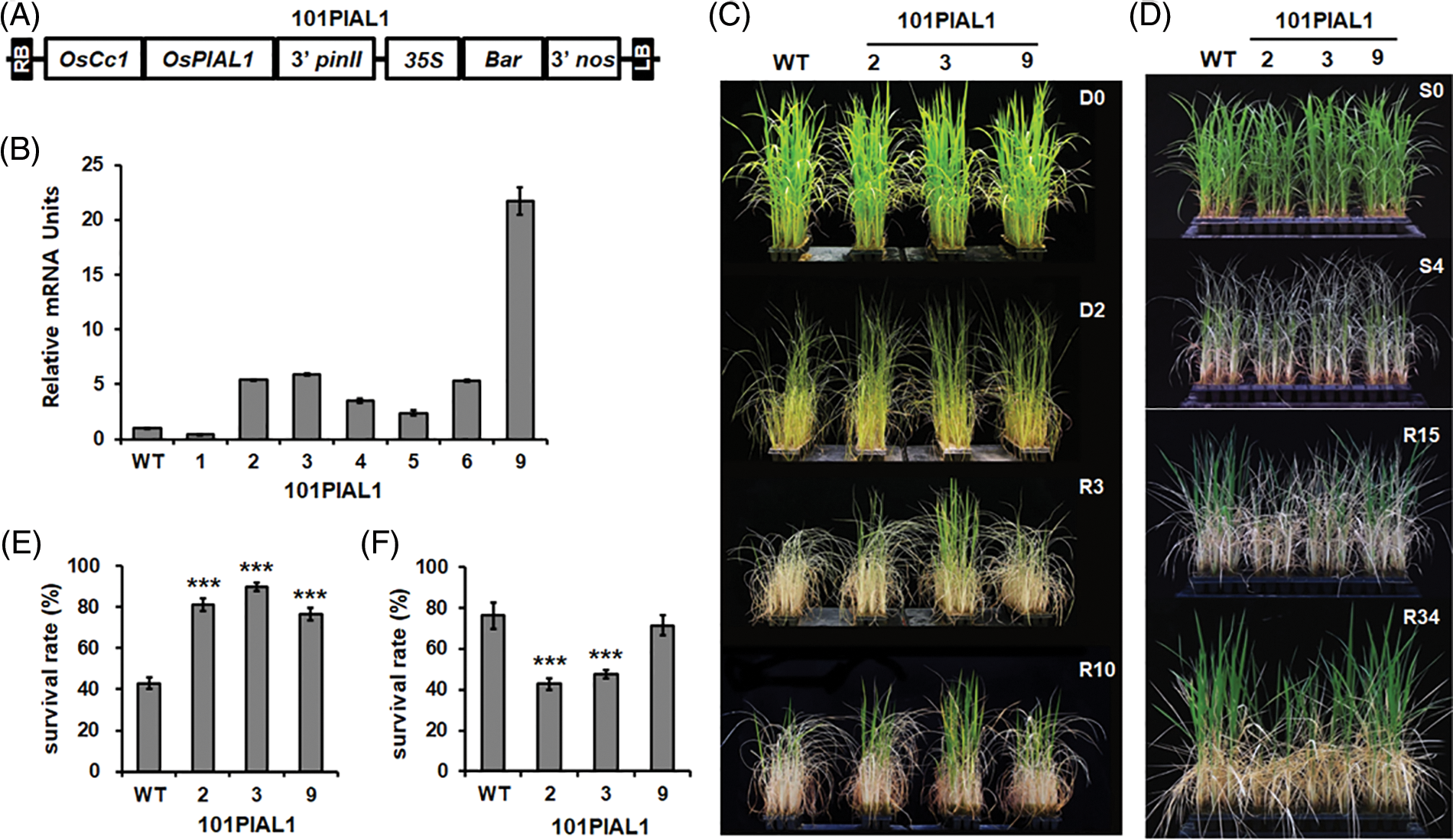
Figure 4: Drought and high salinity tolerance analysis of transgenic plants overexpressing the OsPIAL1 gene. (A) The diagram of overexpression plasmid. (B) The expression levels of the OsPIAL1 gene in the OsPIAL1-OX lines. (C) Drought stress tolerance of OsPIAL1-OX lines. D0 and D2 indicate 0 and 2 days after water draining, respectively. R3 and R10 indicate 3 and 10 days after rewatering, respectively. (D) High-salinity stress assay of OsPIAL1-OX transgenic rice plants. Pictures were taken at 0 and 4 days after transfer to a 250 mM NaCl solution (S0 and S4) and at 15 and 34 days after fresh watering (R15 and R34). (E) The survival rates after drought treatment. (F) The survival rates after high salinity treatment. Statistically significant differences are indicated by asterisks, as calculated using Student’s t-test. ***, p < 0.001.
It has been reported that SUMO conjugation is involved in plant responses to environmental stresses, and the OsPIAL1 gene was induced by only drought stress (Fig. 3B). To examine the drought response of the OsPIAL1-OX plants, the transgenic and WT seedlings were drought-treated for two days and then supplied with water again (Fig. 4C, R3 and R10). During rewatering, the OsPIAL1-OX transgenic lines exhibited faster recovery and more promoted growth than WT plants. The OsPIAL1-OX lines showed 76% to 90% of the survival rates, but WT plants showed only 43% (Fig. 4E).
Our results indicated that high salinity suppressed OsPIAL1 gene expression in the roots (Fig. 3D). To test the high salinity response of the OsPIAL1-OX lines, transgenic and WT seedlings were treated with 250 mM NaCl for four days, and then transferred into fresh water (Fig. 4D, R15 and R34). Most of the OsPIAL1-OX plants were less able to recover than WT plants. The WT seedlings showed 76% of the survival rates, but the OsPIAL1-OX lines 2 and 3 seedlings showed only 43% and 47%, respectively (Fig. 4F). OsPIAL-OX line 9 seedlings showed a further 6% decrease in survival rates compared to the WT seedlings. These results suggested that the overexpression of OsPIAL1 gene confers drought tolerance, but results in a hypersensitive phenotype under high salinity.
3.5 SUMO Conjugates Accumulation in OsPIAL1-OX Lines
To check the alteration of SUMO conjugate accumulation in OsPIAL1-overexpressing transgenic lines, immunoblot assay was performed with root total protein extract. The anti-AtSUMO1 antibody was used in the immunoblot analysis. It has been reported that rice SUMO can be detected specifically by the anti-AtSUMO1 antibody [17]. The formation of SUMO conjugates was not different in the transgenic lines and the WT controls (Fig. 5, 0 h). This result suggests that OsPIAL1 does not stimulate the formation of SUMO conjugates in rice. However, drought treatments significantly increased the approximately 40- and 55-kDa bands within 1 h in the OsPIAL1-OX lines but not in the WT (Fig. 5, 1 h).
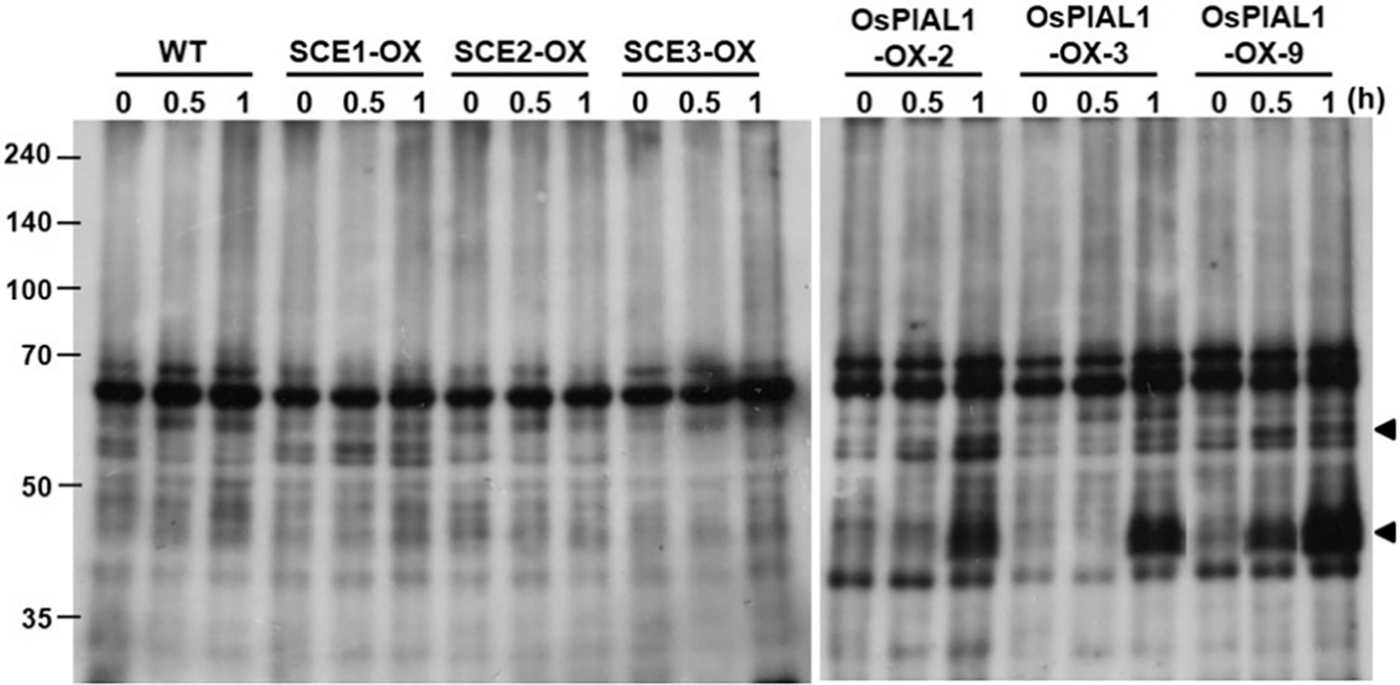
Figure 5: Sumoylation profiles in the OsPIAL1-OX transgenic rice plants under drought stress conditions. Crude extracts from the transgenic rice plants overexpressing OsSCE1 (SCE1), OsSCE2 (SCE2), OsSCE3 (SCE3), or OsPIAL1 (PIAL1-2, 3, 9) and WT plants were subjected to immunoblot analyses with anti-AtSUMO1. The arrowheads indicate the putative SUMO-SCE conjugates
To determine whether the increase in the 40- and 55-kDa bands was an OsPIAL1 gene-specific response, transgenic rice plants overexpressing OsSCEs were also analyzed. There was no increase in the 40- and 55-kDa bands in the transgenic rice plants overexpressing OsSCE genes (Fig. 5). Arabidopsis PIAL2 catalyzes increased SUMO conjugation to SCEs, which has a molecular weight of approximately 40 kDa [24]. The molecular weights of OsSCEs and OsSUMOs are similar to those of Arabidopsis SCE1 and AtSUMOs, respectively. These results suggested that the 40-kDa band may be a SUMO-SCE conjugate. Our results suggested that OsPIAL1 is involved in SUMO conjugation to SCEs but not in SUMO conjugation to substrates.
Substrate specificity in the SUMO pathway has remained unclear. Unlike in the ubiquitin pathway, which uses many kinds of E2 and E3 enzymes to regulate substrate selection, only a few E2 and E3 enzymes have been known [15,21]. SIZ1, SIZ2, methyl methanesulfonate-sensitivity protein 21 (Mms21) and molecular zipper protein 3 (Zip3) in yeast and PIAS1, PIAS3, PIASxα, PIASxβ and PIASy in humans have been identified as SUMO E3 ligases [21]. In plants, known SUMO E3 ligases include SIZ1, SIZ2, and MMS21/HPY2 [2,4,15]. These ligases contain an SP-RING domain. In this work, we characterized a putative rice SUMO E3 ligase, OsPIAL1, which contains an SP-RING domain. Recently, Arabidopsis PIAL1 and PIAL2 have been characterized, and it was suggested that PIALs may specialize in extending single SUMO residues on substrates into chains [24].
PIAS proteins contain four structural motifs: an N-terminal SAP motif, a PINIT (Pro-Ile-Asn-Ile-Thr) motif, an SP-RING zinc finger domain, and a SUMO-interacting motif [6,29,32]. The SP-RING domain is required for the SUMO E3 ligase activity of several PIAS proteins [33–35] and likely interacts with E2 Ubc9 in a manner similar to that observed in other ubiquitin E2:E3 complexes [22]. OsPIAL1 also contains an SP-RING domain similar to the Siz/PIAS E3 ligase family (Fig. 1B), but it does not interact with OsSCEs in yeast (Fig. 2B). In addition, OsSIZ2 showed transactivation activity in yeast, but OsPIAL did not (Fig. 2A). These results suggested that the biological function of OsPIAL1 differs from that of OsSIZ2 in rice.
The Siz/PIAS E3 proteins also contain SIMs in their C-terminal domains. SIMs are the mediators of noncovalent interactions between SUMO and SUMO-binding proteins [15]. SIM has also been thought to be involved in SUMO selection or the selection of downstream effectors of the target [21,36]. In Siz1, this motif is not required for E3 activity [22]. OsPIAL1 contains two putative SIMs (amino acids158–161 and 435–438). We tested the interaction between OsPIAL1 and OsSUMO1/4 (Fig. 2B). OsPIAL1 interacted with OsSUMO3. OsPIAL1 also weakly interacted with OsSUMO1.
Posttranslational protein modifications by the SUMO protein family are involved in diverse cellular processes, including development, hormonal responses, and biotic and abiotic stress signaling [4,15,37]. The sumoylation has been known to play important roles in drought response and ABA signaling in Arabidopsis [5,8,38–40]. In the monocot plant rice, the studies of SUMO conjugation were published recently. Rice SIZ1 and SIZ2 are able to function in the Arabidopsis SUMO conjugation pathway, and they are related to stress responses and stress adaptation [17]. The overexpression of OsSIZ1 in creeping bentgrass enhances abiotic stress tolerance by modifying the expression of stress-related genes [41]. OsSIZ1 is involved in the phosphate and nitrogen responses [42]. However, the biological function of the putative rice SUMO E3 ligase OsPIAL1 has not yet been studied.
To verify the function of OsPIAL1 in rice plants, we constructed transgenic rice plants overexpressing the OsPIAL1 gene. Interestingly, the OsPIAL1 transcripts were increased by only drought stress in the leaves. Additionally, the OsPIAL1 transcripts were decreased by abiotic stresses in the roots (Fig. 2). We evaluated the responses of the OsPIAL1-OX transgenic rice plants to drought and high-salinity stresses. The OsPIAL1-OX lines exhibited better and faster recovery than the WT (Fig. 4C). The survival rate of OsPIAL1-OX transgenic lines was significantly higher than that of WT plants. In contrast, most of the OsPIAL1-OX plants were less able to recover from the presence of 250 mM NaCl compared to WT plants (Fig. 4D). The survival rates of OsPIAL1-OX transgenic lines were lower than the WT. Our results are consistent with those from Arabidopsis pial mutants. Arabidopsis mutant pial2 and pial1 pial2 plants exhibit better growth than the WT plants in the presence of 150 mM NaCl. In contrast, these plants are less able to maintain growth under osmotic stress (300 mM mannitol) [24]. Our results and those from Arabidopsis pial mutants suggest that PIAL1s act as a positive regulator in the drought-stress response but a negative regulator in high-salinity stress responses. It remains to be studied the drought stress specific mechanism of OsPIAL1. Drought stress strongly induced the OsPIAL1 expression only in leaves (Fig. 3B). In contrast, high-salinity stress strongly repressed the OsPIAL1 expression only in roots (Fig. 3D). Basis on the regulation patterns of the OsPIAL1 expression, phenotype of OsPIAL1-OX and Arabidopsis pial mutants [24], it could be assumed that OsPIAL1 works positively for drought stress response in the leaves, but negatively for high-salinity stress response in roots. The simplest putative explanation for this is that the OsPIAL1-OX transgenic plants reduce water loss rate and/or increase water holding capacity in leaves by produce some compatible solutes or proteins, which. confer osmotic stress tolerance. However, they do not involve in the sodium ion toxicity, or do negative effect on high concentration ion toxicity inhibition mechanisms.
The function of Arabidopsis PIAL1 and PIAL2 is suggested to control and enhance SUMO chain-forming activity because PIAL-mediated enhancement of SUMO conjugation to a number of previously identified substrates was unsuccessful in vitro [24]. We tested the SUMO conjugate patterns of OsPIAL1-OX transgenic and WT plants under normal and drought stress conditions (Fig. 5) because OsPIAL1 transcripts were induced by only drought (Fig. 3B). OsPIAL1-OX transgenic rice plants show a similar SUMO conjugate pattern to that of WT plants under normal conditions (Fig. 5 at 0 h). This result coincides with those from Arabidopsis pial1 pial2 double mutants. Arabidopsis pial1 pial2 double mutants show similar SUMO conjugate levels than WT plants under both heat stress and normal conditions [24]. Under drought stress conditions, there was a significant increase in the approximately 40- and 55-kDa bands within 1 h in the OsPIAL1-OX transgenic rice plants (Fig. 5). Except for these two bands, there was no difference in the SUMO conjugate pattern between WT and OsPIAL1-OX transgenic rice plants. This increase in the 40- and 55-kDa bands is an OsPIAL1 overexpression-specific phenotype because it is not detected in transgenic rice plants overexpressing OsSCE genes.
Recently, it was suggested that Arabidopsis PIAL activity enhances SUMO ligation to SCE1 and/or extends single SUMO residues on substrates into chains. Arabidopsis PIAL2 catalyzes increased SUMO conjugation to SCE1, which has a molecular weight of approximately 40 kDa [24]. The molecular weights of OsSUMO (OsSUMO3, 12.43 kDa) and OsSCE (OsSCE1, 18.05 kDa) are similar to those of AtSUMO (AtSUMO1, 10.98 kDa) and AtSCE1 (17.99 kDa), respectively. These results suggested that 40-kDa bands, which were increased in OsPIAL1-OX transgenic rice plants, may be OsSUMO-OsSCE conjugates.
OsPIAL1 preferentially interacts with OsSUMO3, which contains the consensus SUMO modification site ψKxD/E in the N-terminal region. However, OsSUMO1 and OsSUMO2 do not contain this sequence. Human SUMO2 and SUMO3 have ψKxD/E, and this sequence promotes poly-SUMO chain formation in SUMO2 and SUMO3 but not in SUMO1 [43]. Maize SUMO1a also built poly-SUMO chains in E. coli, and most of the accessible Lys residues were detected as SUMO attachment sites [44]. In addition, AtSUMO1 efficiently built poly-SUMO chains and was conjugated to AtSCE1 [45]. Our results suggest that the approximately 55-kDa band that was increased may be poly-OsSUMO3-OsSCE conjugates (Fig. 5) and that OsPIAL1 activity enhances poly-SUMO conjugation to OsSCEs.
The overexpression of OsPIAL1 does not enhance SUMO conjugation under normal conditions. However, drought stress induced an increase in the putative OsSUMO-OsSCE conjugates in the OsPIAL1-OX transgenic rice plants. Therefore, drought stress conditions may induce posttranslational modifications of OsPIAL1, which are required for the activation of OsPIAL1. OsPIAL1 contains three putative sumoylation sites (amino acids 24, 174, and 214). The sumoylation of OsPIAL1 could be one of the posttranslational modifications contributing to its activation.
Our results suggest that OsPIAL1 catalyzes increased SUMO conjugation to SCE1 and that drought stress induced this activity of OsPIAL1. Furthermore, our results and those from Arabidopsis pial mutants suggest that PIALs act as a positive regulator in the drought-stress response but a negative regulator in high-salinity stress responses. Recently, it was reported that PIAL1 and PIAl2 act as components of the MORPHEUS’ MOLECULE1 (MOM1)-containing complex to mediate transcriptional silencing [46]. The mutation in the RING domain of PIAL2 abolished the SUMO ligase activity, and the mutation in the SIM domain partially reduced the SUMO ligase activity. Interestingly, the SUMO ligase activity of PIAL2 is not required for the function of PIAL2 in transcriptional silencing [46]. It is possible that the drought tolerant and high-salinity sensitive responses caused by OsPIAL1 overexpression can result from the modification of OsSUMO-OsSCE conjugation and/or transcriptional silencing.
Acknowledgement: I thank Dr. Jong Soo Joo for helping the rice transformation.
Funding Statement: This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant No. 2017R1D1A1B03030725).
Author Contributions: Sang Ik Song designed and performed the experiments. Sang Ik Song drafted and revised the manuscript.
Conflicts of Interest: The author declares that they have no conflicts of interest to report regarding the present study.
References
1. Dohmen, R. J. (2004). SUMO protein modification. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1695(1–3), 113–131. https://doi.org/10.1016/j.bbamcr.2004.09.021 [Google Scholar] [PubMed] [CrossRef]
2. Novatchkova, M., Budhiraja, R., Coupland, G., Eisenhaber, F., Bachmair, A. (2004). SUMO conjugation in plants. Planta, 220(1), 1–8. https://doi.org/10.1007/s00425-004-1370-y [Google Scholar] [PubMed] [CrossRef]
3. Kerscher, O., Felberbaum, R., Hochstrasser, M. (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annual Review of Cell and Developmental Biology, 22(1), 159–180. https://doi.org/10.1146/annurev.cellbio.22.010605.093503 [Google Scholar] [PubMed] [CrossRef]
4. Park, H. J., Kim, W. Y., Park, H. C., Lee, S. Y., Bohnert, H. J. et al. (2011). SUMO and SUMOylation in plants. Molecules and Cells, 32(4), 305–316. https://doi.org/10.1007/s10059-011-0122-7 [Google Scholar] [PubMed] [CrossRef]
5. Kurepa, J., Walker, J. M., Smalle, J., Gosink, M. M., Davis, S. J. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: Accumulation of SUMO1 and -2 conjugates is increased by stress. Journal of Biological Chemistry, 278(9), 6862–6872. https://doi.org/10.1074/jbc.M209694200 [Google Scholar] [PubMed] [CrossRef]
6. Miura, K., Jin, J. B., Hasegawa, P. M. (2007). Sumoylation, a post-translational regulatory process in plants. Current Opinion in Plant Biology, 10(5), 495–502. https://doi.org/10.1016/j.pbi.2007.07.002 [Google Scholar] [PubMed] [CrossRef]
7. Conti, L., Price, G., O’Donnell, E., Schwessinger, B., Dominy, P. et al. (2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. The Plant Cell, 20(10), 2894–2908. https://doi.org/10.1105/tpc.108.058669 [Google Scholar] [PubMed] [CrossRef]
8. Lois, L. M., Lima, C. D., Chua, N. H. (2003). Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. The Plant Cell, 15(6), 1347–1359. https://doi.org/10.1105/tpc.009902 [Google Scholar] [PubMed] [CrossRef]
9. Miura, K., Lee, J., Jin, J. B., Yoo, C. Y., Miura, T. et al. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proceedings of the National Academy of Sciences, 106(13), 5418–5423. https://doi.org/10.1073/pnas.0811088106 [Google Scholar] [PubMed] [CrossRef]
10. Huang, L., Yang, S., Zhang, S., Liu, M., Lai, J. (2009). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. The Plant Journal, 60(4), 666–678. https://doi.org/10.1111/j.1365-313X.2009.03992.x [Google Scholar] [PubMed] [CrossRef]
11. Ishida, T., Fujiwara, S., Miura, K., Stacey, N., Yoshimura, M. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. The Plant Cell, 21(8), 2284–2297. https://doi.org/10.1105/tpc.109.068072 [Google Scholar] [PubMed] [CrossRef]
12. Murtas, G., Reeves, P. H., Fu, Y. F., Bancroft, I., Dean, C. et al. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. The Plant Cell, 15(10), 2308–2319. https://doi.org/10.1105/tpc.015487 [Google Scholar] [PubMed] [CrossRef]
13. Park, B. S., Song, J. T., Seo, H. S. (2011). Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nature Communications, 2(1), 1–10. https://doi.org/10.1038/ncomms1408 [Google Scholar] [PubMed] [CrossRef]
14. Johnson, E. S. (2004). Protein modification by SUMO. Annual review of Biochemistry, 73(1), 355–382. https://doi.org/10.1146/annurev.biochem.73.011303.074118 [Google Scholar] [PubMed] [CrossRef]
15. Novatchkova, M., Tomanov, K., Hofmann, K., Stuible, H. P., Bachmair, A. (2012). Update on sumoylation: Defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytologist, 195(1), 23–31. https://doi.org/10.1111/j.1469-8137.2012.04135.x [Google Scholar] [PubMed] [CrossRef]
16. Thangasamy, S., Guo, C. L., Chuang, M. H., Lai, M. H., Chen, J. et al. (2011). Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence. New Phytologist, 189(3), 869–882. https://doi.org/10.1111/j.1469-8137.2010.03538.x [Google Scholar] [PubMed] [CrossRef]
17. Park, H. C., Kim, H. U. N., Koo, S. C., Park, H. J., Cheong, M. S. (2010). Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant, Cell & Environment, 33(11), 1923–1934. https://doi.org/10.1111/j.1365-3040.2010.02195.x [Google Scholar] [PubMed] [CrossRef]
18. Srivastava, A. K., Zhang, C., Yates, G., Bailey, M., Brown, A. et al. (2016). SUMO is a critical regulator of salt stress responses in rice. Plant Physiology, 170(4), 2378–2391. https://doi.org/10.1104/pp.15.01530 [Google Scholar] [PubMed] [CrossRef]
19. Rodriguez, M. S., Dargemont, C., Hay, R. T. (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. Journal of Biological Chemistry, 276(16), 12654–12659. https://doi.org/10.1074/jbc.M009476200 [Google Scholar] [PubMed] [CrossRef]
20. Sampson, D. A., Wang, M., Matunis, M. J. (2001). The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. Journal of Biological Chemistry, 276(24), 21664–21669. https://doi.org/10.1074/jbc.M100006200 [Google Scholar] [PubMed] [CrossRef]
21. Gareau, J. R., Lima, C. D. (2010). The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nature Reviews Molecular Cell Biology, 11(12), 861–871. https://doi.org/10.1038/nrm3011 [Google Scholar] [PubMed] [CrossRef]
22. Yunus, A. A., Lima, C. D. (2009). Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Molecular Cell, 35(5), 669–682. https://doi.org/10.1016/j.molcel.2009.07.013 [Google Scholar] [PubMed] [CrossRef]
23. Miura, K., Rus, A., Sharkhuu, A., Yokoi, S., Karthikeyan, A. S. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proceedings of the National Academy of Sciences, 102(21), 7760–7765. https://doi.org/10.1073/pnas.0500778102 [Google Scholar] [PubMed] [CrossRef]
24. Tomanov, K., Zeschmann, A., Hermkes, R., Eifler, K., Ziba, I. (2014). Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. The Plant Cell, 26(11), 4547–4560. https://doi.org/10.1105/tpc.114.131300 [Google Scholar] [PubMed] [CrossRef]
25. Joo, J., Choi, H. J., Lee, Y. H., Kim, Y. K., Song, S. I. (2013). A transcriptional repressor of the ERF family confers drought tolerance to rice and regulates genes preferentially located on chromosome 11. Planta, 238(1), 155–170. https://doi.org/10.1007/s00425-013-1880-6 [Google Scholar] [PubMed] [CrossRef]
26. Joo, J., Lee, Y. H., Kim, Y. K., Nahm, B. H., Song, S. I. (2013). Abiotic stress responsive rice ASR1 and ASR3 exhibit different tissue-dependent sugar and hormone-sensitivities. Molecules and Cells, 35(5), 421–435. https://doi.org/10.1007/s10059-013-0036-7 [Google Scholar] [PubMed] [CrossRef]
27. Wu, L., Wu, H., Sangiorgi, F., Wu, N., Bell, J. R. et al. (1997). Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mechanisms of Development, 65(1–2), 3–17. https://doi.org/10.1016/S0925-4773(97)00032-4 [Google Scholar] [PubMed] [CrossRef]
28. Sharma, M., Li, X., Wang, Y., Zarnegar, M., Huang, C. Y. (2003). hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. The EMBO Journal, 22(22), 6101–6114. https://doi.org/10.1093/emboj/cdg585 [Google Scholar] [PubMed] [CrossRef]
29. Palvimo, J. J. (2007). PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochemical Society Transactions, 35(6), 1405–1408. https://doi.org/10.1042/BST0351405 [Google Scholar] [PubMed] [CrossRef]
30. Garcia-Dominguez, M., March-Diaz, R., Reyes, J. C. (2008). The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. Journal of Biological Chemistry, 283(31), 21469–21477. https://doi.org/10.1074/jbc.M708176200 [Google Scholar] [PubMed] [CrossRef]
31. Suzuki, R., Shindo, H., Tase, A., Kikuchi, Y., Shimizu, M. et al. (2009). Solution structures and DNA binding properties of the N-terminal SAP domains of SUMO E3 ligases from Saccharomyces cerevisiae and Oryza sativa. Proteins: Structure, Function, and Bioinformatics, 75(2), 336–347. https://doi.org/10.1002/prot.22243 [Google Scholar] [PubMed] [CrossRef]
32. Sharrocks, A. D. (2006). PIAS proteins and transcriptional regulation—more than just SUMO E3 ligases? Genes & Development, 20(7), 754–758. https://doi.org/10.1101/gad.1421006 [Google Scholar] [PubMed] [CrossRef]
33. Takahashi, Y., Kikuchi, Y. (2005). Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. Journal of Biological Chemistry, 280(43), 35822–35828. https://doi.org/10.1074/jbc.M506794200 [Google Scholar] [PubMed] [CrossRef]
34. Reindle, A., Belichenko, I., Bylebyl, G. R., Chen, X. L., Gandhi, N. et al. (2006). Multiple domains in Siz SUMO ligases contribute to substrate selectivity. Journal of Cell Science, 119(22), 4749–4757. https://doi.org/10.1242/jcs.03243 [Google Scholar] [PubMed] [CrossRef]
35. Cheong, M. S., Park, H. C., Hong, M. J., Lee, J., Choi, W. (2009). Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiology, 151(4), 1930–1942. [Google Scholar] [PubMed]
36. Geiss-Friedlander, R., Melchior, F. (2007). Concepts in sumoylation: A decade on. Nature Reviews Molecular Cell Biology, 8(12), 947–956. https://doi.org/10.1038/nrm2293 [Google Scholar] [PubMed] [CrossRef]
37. Miura, K., Hasegawa, P. M. (2010). Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends in Cell Biology, 20(4), 223–232. https://doi.org/10.1016/j.tcb.2010.01.007 [Google Scholar] [PubMed] [CrossRef]
38. Yoo, C. Y., Miura, K., Jin, J. B., Lee, J., Park, H. C. (2006). SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiology, 142(4), 1548–1558. https://doi.org/10.1104/pp.106.088831 [Google Scholar] [PubMed] [CrossRef]
39. Catala, R., Ouyang, J., Abreu, I. A., Hu, Y., Seo, H. et al. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. The Plant Cell, 19(9), 2952–2966. https://doi.org/10.1105/tpc.106.049981 [Google Scholar] [PubMed] [CrossRef]
40. Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. The Plant Cell, 19(4), 1403–1414. https://doi.org/10.1105/tpc.106.048397 [Google Scholar] [PubMed] [CrossRef]
41. Li, Z., Hu, Q., Zhou, M., Vandenbrink, J., Li, D. (2013). Heterologous expression of OsSIZ 1, a rice SUMO E 3 ligase, enhances broad abiotic stress tolerance in transgenic creeping bentgrass. Plant Biotechnology Journal, 11(4), 432–445. https://doi.org/10.1111/pbi.12030 [Google Scholar] [PubMed] [CrossRef]
42. Wang, H., Sun, R., Cao, Y., Pei, W., Sun, Y. (2015). OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice. Plant and Cell Physiology, 56(12), 2381–2395. https://doi.org/10.1093/pcp/pcv162 [Google Scholar] [PubMed] [CrossRef]
43. Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H. et al. (2001). Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. Journal of Biological Chemistry, 276(38), 35368–35374. https://doi.org/10.1074/jbc.M104214200 [Google Scholar] [PubMed] [CrossRef]
44. Augustine, R. C., York, S. L., Rytz, T. C., Vierstra, R. D. (2016). Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress. Plant Physiology, 171(3), 2191–2210. https://doi.org/10.1104/pp.16.00353 [Google Scholar] [PubMed] [CrossRef]
45. Castaño-Miquel, L., Seguí, J., Lois, L. M. (2011). Distinctive properties of Arabidopsis SUMO paralogues support the in vivo predominant role of AtSUMO1/2 isoforms. Biochemical Journal, 436(3), 581–590. https://doi.org/10.1042/BJ20101446 [Google Scholar] [PubMed] [CrossRef]
46. Han, Y. F., Zhao, Q. Y., Dang, L. L., Luo, Y. X., Chen, S. S. (2016). The SUMO E3 ligase-like proteins PIAL1 and PIAL2 interact with MOM1 and form a novel complex required for transcriptional silencing. The Plant Cell, 28(5), 1215–1229. https://doi.org/10.1105/tpc.15.00997 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials


Figure S1: Amino acid sequence alignment of the OsSIZ1, OsSIZ2, and PIAL1(Os06g0164000) in rice. MIZ/SP-RING domain was identified in Pfam and is indicated below the sequence by underline. Residue numbers are shown for each polypeptide. Asterisks indicate conserved amino acids. Dashes denote gaps


Figure S2: Amino acid sequence alignment of PIALs. MIZ/SP-RING domain was identified in Pfam and is indicated below the sequence by underline. Residue numbers are shown for each polypeptide. Asterisks and dots indicate conserved and similar amino acids, respectively. Dashes denote gaps
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools